
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 02 April 2025
Sec. Plant Breeding
Volume 16 - 2025 | https://doi.org/10.3389/fpls.2025.1532216
Introduction: Lodging has become an important factor limiting oats production.
Methods: To understand the relationship between oat lodging and stem growth, we selected three oat cultivars with different lodging resistance traits and conducted a detailed analysis of their stem physicochemical properties, and transcriptome sequencing on them at different growth stages were performed.
Results: Important plant characteristics like: length of the second stem internode, stem wall thickness, breaking force, mechanical strength, soluble sugar, starch, lignin, and silicon content were closely related to oat lodging performance. With the growth of the second stem internode at the base of oats, the number of coexpressed differentially expressed genes (DEGs) increased. And DEGs were specifically enriched in starch and sucrose metabolism, phenylpropanoidbiosynthesis, MAPK signaling pathway-plant and carbon metabolism. There were many TF family types among the different comparison groups, and p450, Myb_DNA-binding, WRKY, and AP2 families accounted for the most. Additionally, there was a specific high expression of genes related to the synthesis of cellulose(CesA9, CesA7, and CesA4) and lignin (CCR1, 4CL8, and 4CL3) in lodgingresistant cultivar and middle lodging-resistant cultivar. WGCNA analysis identified genes closely related to lodging resistance, namely MBF1c, SKP1, and CAND1, which were specifically up-regulated on the 35th day of growth in the second stem internode of the highly resistant ‘LENA’. These genes can all serve as positive regulatory factors for oat lodging.
Discussion: Ultimately, our work analyzed the transcriptional network regulatory relationships, laying the foundation for elucidating the physiological and genetic mechanisms of oat lodging resistance, and providing excellent genetic resources for oat and other crop breeding.
Oat (Avena sativa) is an annual Avena crop from the Gramineae family that is known for its excellent characteristics, such as drought resistance, barren tolerance, saline-alkali tolerance, and rich nutritional value (Singh et al., 2013; Nan, 2021). It has become one of the most important forage crops in the world. In particular, its straw is used as a high-quality forage grass for developing animal husbandry and raising livestock (Ahmad et al., 2014). In recent years, with the increasing development of grassland supporting construction in pastoral areas, the planting area of oat forage base has been expanding, and it has become the dominant forage species for farmers and herdsmen in Qinghai-Tibet Plateau. It is also a leading crop of forage production base and “disaster resistance and livestock protection” high-quality forage in high-altitude pastoral areas, which plays an important role in the stable development of grassland animal husbandry on the Qinghai-Tibet Plateau. However, due to the frequent rainy and windy weather after heading, oats lodge in a large area, affecting the mechanized harvest of forage fields and reducing the yield and quality of forage grass, which has become the main factor affecting the high yield and quality of oats. It is also one of the key problems to be solved in the genetic improvement of oats. Crop lodging is the most common natural phenomenon in the late growth and development of cereal crops, which mainly refers to the phenomenon where stems go from a natural upright state to a permanent dislocation that is caused by the interaction of many factors and is limiting factor in crop production (Shah et al., 2017). Cloninger et al. (1970) classified crops into two categories based on the location of lodging: stem lodging and root lodging. Oat lodging is mainly stem lodging, which is consistent with the research results on lodging of wheat (Triticum aestivum). Lodging mainly occurs in the second internode of the base, and the internal causes of lodging are stem dysplasia and reduced mechanical strength of the stem in the internode (Berry and Spink, 2012).
Additionally, plant morphology and microscopic characteristics played an important role in lodging resistance, especially the biological characteristics of root, stem and ear (Silveira et al., 2022). Indeed, the mechanical properties of stem, carbohydrate content and anabolism of structural substances are also factors that determine the lodging resistance of oat (Nan, 2021). In particular, cellulose and lignin are the main components of stem cell wall that played an important role in strengthening it, while also increasing stem compressive strength and bending properties, which can provide mechanical support for the whole culm. Previous research has shown that changes in cellulose and lignin content are closely related to the lodging resistance of oats (Baucher et al., 1998). For instance, Ookawa and Ishihara (1993) found that the cellulose and lignin contents from the stems of lodging-resistant rice (Oryza sativa) were higher than those of lodging-susceptible cultivar. Nan (2021) also found that a major contributor for the decrease in stem bending resistance and mechanical strength of oats was low lignin content in the basal internode I and II. It has also been shown that the occurrence of plant lodging is closely related to the growth characteristics of the stem internodes and their physicochemical components.
In recent years, with the continuous improvement of biological technologies, researchers have tried to analyze the regulatory genes related to lodging resistance of oat at the molecular level. At present, many genes coding for enzymes involved in cellulose and lignin biosynthesis pathways related to plant lodging have been cloned and basic functional studies have been carried out. For example, cellulose is synthesized by the cellulose synthase (CesA) complex embedded in the plasma membrane that is connected to form microfibers through hydrogen bonds and van der Waals forces along the microtubule direction and comprises the main polysaccharide in the cell wall. Zhao et al. (2012) found that the activity of cellulose synthase (CesA) is closely related to plant cellulose content. Specifically, CesA is a supergene family that participates in many stages of plant cell wall development. For Arabidopsis thaliana, AtCesA1,-A3 and-A6 play a role in the synthesis of the primary cell wall, while AtCesA4,-A7 and-A8 are directly related to the formation of the secondary cell wall. Yan et al. (2007) found that cellulose content decreases in rice brittle culm. Here, they used a Brittle Culm (BC) mutant (BC1,-3,-5,-7,-10) and barley (Hordeum vulgare), and found the brittle culm mutant showed abnormal CesA gene expression with a missense mutation. The missense mutation of the secondary cell wall cellulose synthase monomers OsCESA9 and OsCESA4 in rice directly led to the hindrance and decrease of the cellulose synthesis in the secondary cell wall, and the stem was easy to break. Taken together, these genes control the content and structure of stem secondary cell wall components, and can make the secondary wall of thick-walled cells thin or loose in structure, reduce the bending stress of the stem, and make the stem brittle and easy to breakage. Lignin also enhances the toughness and mechanical supporting ability of cell walls, and its content is closely related to the mechanical strength and lodging resistance of the stem. Phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxylase (C4H), 4-coumarate:CoA lig-ase (4CL), cinnamoyl-CoA reductase (CCR1/2), and caffeic acid O-methyl-transferase (COMT) are the main genes coding for enzymes in the lignin biosynthesis pathway. Its expression is closely related to the synthesis and accumulation of lignin, which can effectively regulate the lodging resistance of crops (Liu et al., 2019). Yan et al. (2021) identified the dwarf gene (Dwarf) that regulates root and stem growth in oats and developed corresponding molecular markers that can effectively reduce the risk of oat lodging.
In addition, plant lodging resistance is also regulated by a complex transcription factor network (Wei and Lan, 2022). For instance, Ohashi-Ito and Fukuda (2010) found in Arabidopsis that the NAC family vascular bundle related NAC-domain 6 and 7 (VND6 and-7) induce vascular bundle cells to transfer to xylem tissue. The NST1, -2 and NST3/SND1 subfamily have also been shown to be key regulators of secondary cell wall formation in xylem tissue. Zhou et al. (2009) found that the overexpression of AtMYB58 and AtMYB63 can specifically activate lignin biosynthesis genes, accompanied by ectopic deposition of lignin in normal unamplified cells. However, others have found that the loss-of-function mutations in AtMYB75 can up-regulated the expression levels of lignin biosynthesis and cellulose-related genes (Bhargava et al., 2010). Wang et al. (2010) also found that mutations in WRKY family increased the biosynthesis of xylan, cellulose, and lignin, which are needed for secondary wall thickening, and result in a 50% increase in stem biomass. To sum up, these genes and TFs affect the lodging resistance of plant stems by regulating many physicochemical properties.
At present, it has been recognized that the morphological characteristics, physicochemical properties, and gene expression regulation of stem internodes have important effects on crop stem mechanical strength and lodging resistance, but their specific relationship and influence mechanism are still unclear. In alpine pastoral areas, culm lodging mainly occurs during oat planting, a transcriptional study on lodging resistance of oat has not been reported, therefore, it is particularly necessary to study the mechanism of oat resistance to lodging. Our study selected oat cultivars with different lodging resistance to investigate physiological indicators, such as stem morphology and physicochemical properties, as well as comparative transcriptome data, to accurately locate genes related to oat stem strength and lodging resistance. Our goal was to understand the molecular basis of functional performance for lodging-resistant cultivars and their related main control genes are expressed. Here, we show a network of regulatory relationships at the transcriptional level that will provide basic data to control the genetic maintenance of excellent traits at the gene expression level for lodging-resistant species and accelerate the development of animal husbandry in alpine pastoral areas.
Based on previous research (Zhang et al., 2020), three oat cultivars, ‘LENA’ (Avena sativa L. cv. LENA, high lodging-resistant cultivar, LN), ‘Qingyin No.1’ (middle lodging-resistant cultivar, QH) and ‘Qingyin No.2’ (lodging-susceptible cultivar, QY), were sown in Lusha Town, Huangzhong County, Qinghai Province, Qinghai Tibet Plateau, China in April 2023 and 2024, respectively. The geographical location was 101°37’ E, 36°28’ N, with an elevation of 2620 m, average annual precipitation 481 mm, and is characterized as a flat terrain with good fertility. The selection of materials is based on field experiments from the past ten years.
A single factor random block design was used for experiments. Each material was sown in a plot with a length of 5 m, a width of 3 m, and an area of 5 m × 3 m = 15 m2. Strip planting was used with a sowing rate of 225 kg·hm2, a depth of 3-4 cm, and at a rate of 3.75 million stems per hectare.
During the flowering stage, 10 plants were randomly selected from each plot of the three oat cultivars. Their plant, center of gravity, and ear height were recorded with a tape measure. Here, ear position coefficient = ear height/plant height. The fresh weight of each plant was weighed immediately after root removal. The second stem internode length and internode below the panicle length were measured with a ruler.
The incidence of lodging was determined and the actual area of lodging measured at flowering and before the harvest stage. The actual lodging rate is calculated using the following formula:
Randomly select 10 plants from each plot of three oat cultivars. The length from the base of the second stem internode to the top of the ear and the weight from the base of the second stem internode to the top of the ear were measured with a tape measure and balance. The stem diameter and wall thickness of the second stem internode were measured with a vernier caliper. The compression strength, puncture strength, breaking force, and mechanical strength of the second stem internode were measured by plant stem strength tester (YYD-1) (Wang et al., 2019). The bending moment is calculated using the following formula:
Bending moment = the fresh weight from the base of the internode to the top of the ear × the length from the base of the internode to the top of the ear, in g·cm-1 (Lei et al., 2013).
The collected samples were processed by removing the ears, leaves and roots. The stems were blanched at 105°C for 30 min and then dried at 65°C for 48 h, and then crushed. The contents of lignin, soluble sugar and starch, as well as the contents of calcium, potassium and silicon were determined using the Solabio kit (Beijing, China). For each index, 0.05~0.1 g was weighed, with three biological replicates. The specific steps were followed as per the instructions.
Samples were collected on the 21st, 28th, and 35th day after the growth of the second stem internode at the base of oats, and three biological repeats were set at each time point. Extracted sample RNA and Illumina’s NEB Next® Ultra™ RNA Library Prep Kit for Illumina® kit were used to build a cDNA library for testing. After passing library inspection, different libraries were sequenced by the Illumina NovaSeq 6000 after pooling, according to the effective concentration and the target off-machine data demand.
The original image data obtained from the high-throughput sequencing was transformed into raw reads using CASAVA base recognition. Clean reads were filtered and spliced. End reads were aligned using the reference genome (https://plants.ensembl.org/index.html) from HISAT2 v2.0.5 (Mortazavi et al., 2008). After, the samples were analyzed by principal component analysis (PCA).
We used DESeq2 (Love et al., 2014) to analyze differential expression between sample groups. DEGs were obtained based on screening criteria of a difference multiple |log2Fold Change| ≥ 1 and a false discovery rate (FDR)< 0.05. GO enrichment analysis and KEGG enrichment analysis of differentially expressed genes were realized using clusterProfiler (3.8.1) software.
Plant TF prediction was carried out using iTAK (Zheng et al., 2016) software, which integrates two databases, PlnTFDB and PlantTFDB. Following analysis, it identifies and classifies data according to methods from Pérez-Rodríguez et al. (2010).
In order to explore the expression trend of DEGs in different growth stages, the scale () function of R was used to standardize all DEGs with union FPKM. The processed DEGs were then analyzed by Kmeans cluster analysis.
Our co-expression network was constructed using the WGCNA package of R (Langfelder and Horvath, 2008), and samples are clustered and analyzed using the methods from Qin et al. (2020). The soft threshold β parameter is calculated by using the pickSoftThreshold function in WGCNA to meet the prerequisite of scale-free network distribution. Select the threshold parameter β when R2 reaches 0.8, calculate the dissimilarity coefficient between all genes to create the adjacency matrix, use the dissimilarity degree dissimilarity between genes to cluster the genes in the network, and establish a hierarchical clustering tree, and then use the dynamic cutting method to cut the tree into different modules (the minimum number of genes is 30), and merge the modules whose correlation coefficient are greater than 0.75, those are, the dissimilarity coefficient are less than 0.25 (Wu et al., 2022). Protein-protein interaction (PPI) network was constructed using STRING database. Cytoscape (version 3.8.2) (Shannon et al., 2003) is used to visualize the PPI network, using the plug-in cytoHubba to determine the central gene by Maximal Clique Centrality (MCC) calculation.
Analysis of variance (ANOVA) was performed on all data, and Windows version of SPSS 24.0 (SPSS Inc., New York, NY, USA) was used, and the mean separation was performed with Fisher’s protected least significant difference (LSD) test at 0.05 significance level. SPSS 24.0 was used for Pearson correlation analysis.
As shown in Table 1, ‘Qingyin No.1’ showed the highest plant height, center of gravity height, and ear height in 2023 and 2024, and was significantly higher than ‘Qingyin No.2’ and ‘LENA’ (P< 0.05). The mechanical strength of ‘LENA’ was the highest, and was significantly higher than that of ‘Qingyin No.1’ and ‘Qingyin No.2’ (P< 0.05). The second stem internode length, center of gravity height coefficient, and ear height of ‘Qingyin No.2’ was the highest, while ‘LENA’ was the lowest, and was significantly lower than the other two cultivars (P< 0.05). In 2023, ‘LENA’ had the highest single plant fresh weight, second stem internode stem diameter, stem wall thickness, compression strength, puncture strength, breaking force, and bending moment was significantly higher than the other two cultivars (P< 0.05). However, single plant fresh weight was the lowest in ‘LENA’, while ‘Qingyin No.2’ had the lowest value for the other indicators. In 2024, ‘Qingyin No.1’ had the highest single plant fresh weight, second stem internode stem diameter, stem wall thickness, compression strength, puncture strength, breaking force, and bending moment. However, it did not have the lowest second stem internode stem diameter and stem wall thickness. Still, there were no significant differences compared to ‘LENA’, and all other indicators were significantly higher than the other cultivars (P< 0.05). Here, ‘Qingyin No.2’ had the lowest for these indicators.
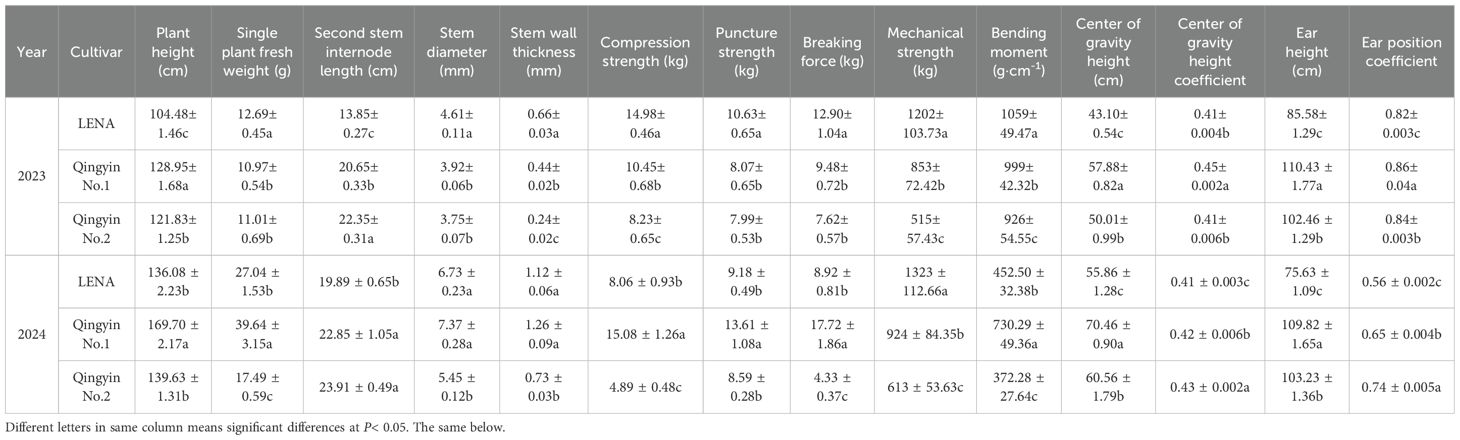
Table 1. Comparative analysis of field traits and stem mechanical characteristics in the second stem internode from three different lodging-resistant oat cultivars in 2023 and 2024.
We determined that there are differences in the soluble sugar, starch, lignin, potassium, calcium, and silicon content among the different oat cultivars during the flowering period. The soluble sugar, potassium, calcium, and silicon contents from ‘Qingyin No.2’ were the highest, except for the silicon content, which had no significant difference with ‘LENA’. All other indicators were significantly higher than the other two cultivars (P< 0.05). ‘LENA’ had the second highest content of these indicators, while ‘Qingyin No.1’ had the lowest content (Figures 1A, D–F). The starch content and lignin content in ‘LENA’ were the highest, followed by ‘Qingyin No.1’. There was no significant difference in starch content between ‘LENA’ and ‘Qingyin No.1’ (P > 0.05), while ‘Qingyin No.2’ had the lowest content, and was significantly lower than the other two cultivars (P< 0.05) (Figures 1B, C).
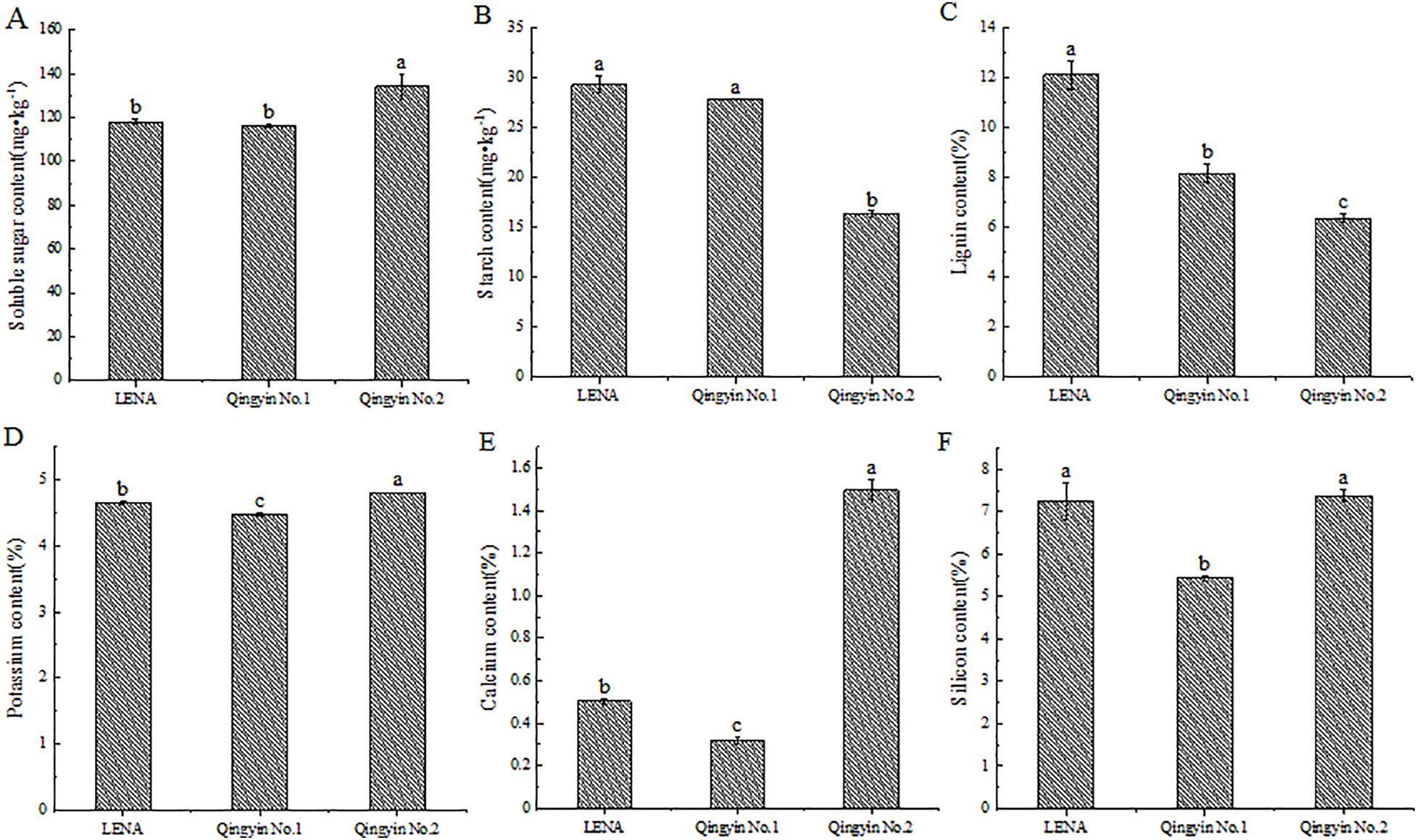
Figure 1. Changes in soluble sugar (A), starch (B), lignin (C), potassium (D), calcium (E) and silicon (F) contents in stems from different oat cultivars. Different lowercase letters in the figure represent significant difference at the 0.05 level.
The differences between the lodging index in the second stem internode during the flowering period and the actual lodging rate were extremely significant (P< 0.01), and were significantly positively correlated. The higher the value of both, the weaker the lodging resistance. The lodging index and actual lodging rate of the second stem internode were highly significant positively correlated with the second stem internode length (P< 0.01). However, it was highly significant negatively correlated with stem wall thickness, compression strength, puncture strength, breaking force, and mechanical strength, as well as soluble sugar, starch, lignin, and silicon content from the second stem internode (P< 0.01), and significantly negatively correlated with potassium content (P< 0.05). We also found that the lodging index of the second stem internode were significantly positively correlated with plant height, ear height, and ear position coefficient (P< 0.05), but were significantly negatively correlated with calcium content (P< 0.01). In addition, the actual lodging rate were highly significant negatively correlated with stem wall thickness in the second stem internode stem (P< 0.01) and significantly negatively correlated with calcium content (P< 0.05) (Table 2).
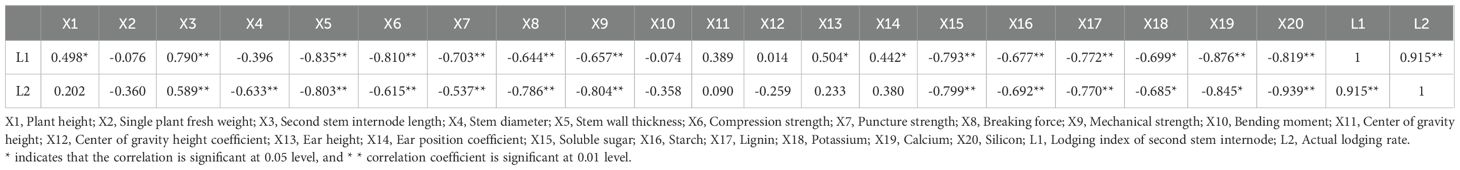
Table 2. Correlation analysis between field traits, stem mechanical characteristics, physiological indicators, lodging index, and lodging rate in oats.
Each filtered clean read had a total of 293.22 Gb, while clean bases were above 9 Gb. The total number of bases G and C as a percentage of the total number of bases (GC content) was 53.87% and 55.75%, respectively, while the reads compared to the reference genome ranged from 94.88% to 97.18% (Supplementary Table S1). PCA analysis revealed that the different treatments for the three oat cultivars were clustered together, and the three biological replicates of each cultivar were clustered together (Supplementary Figure S1). Together, this indicates the consistency of the biological replicates, as well as the quality of transcriptome sequencing data, which can be used for subsequent biological analysis.
During the growth of oat stems, the expression of many genes were significantly induced or inhibited. LN_35 vs QY_35, QH_35 vs QY_35 and QH_28 vs LN_28 had the most DEGs. This was followed by LN_28 vs QY_28, QH_28 vs QY_28 and QH_35 vs LN_35, respectively (Figure 2B, C). The number of DEGs for LN_21 vs QY_21, QH_21 vs QY_21, and QH_21 vs LN_21 was the least (Figure 2A). Our results showed that there were more genes involved in oat stem growth, and especially during the later stage of oat stem growth.
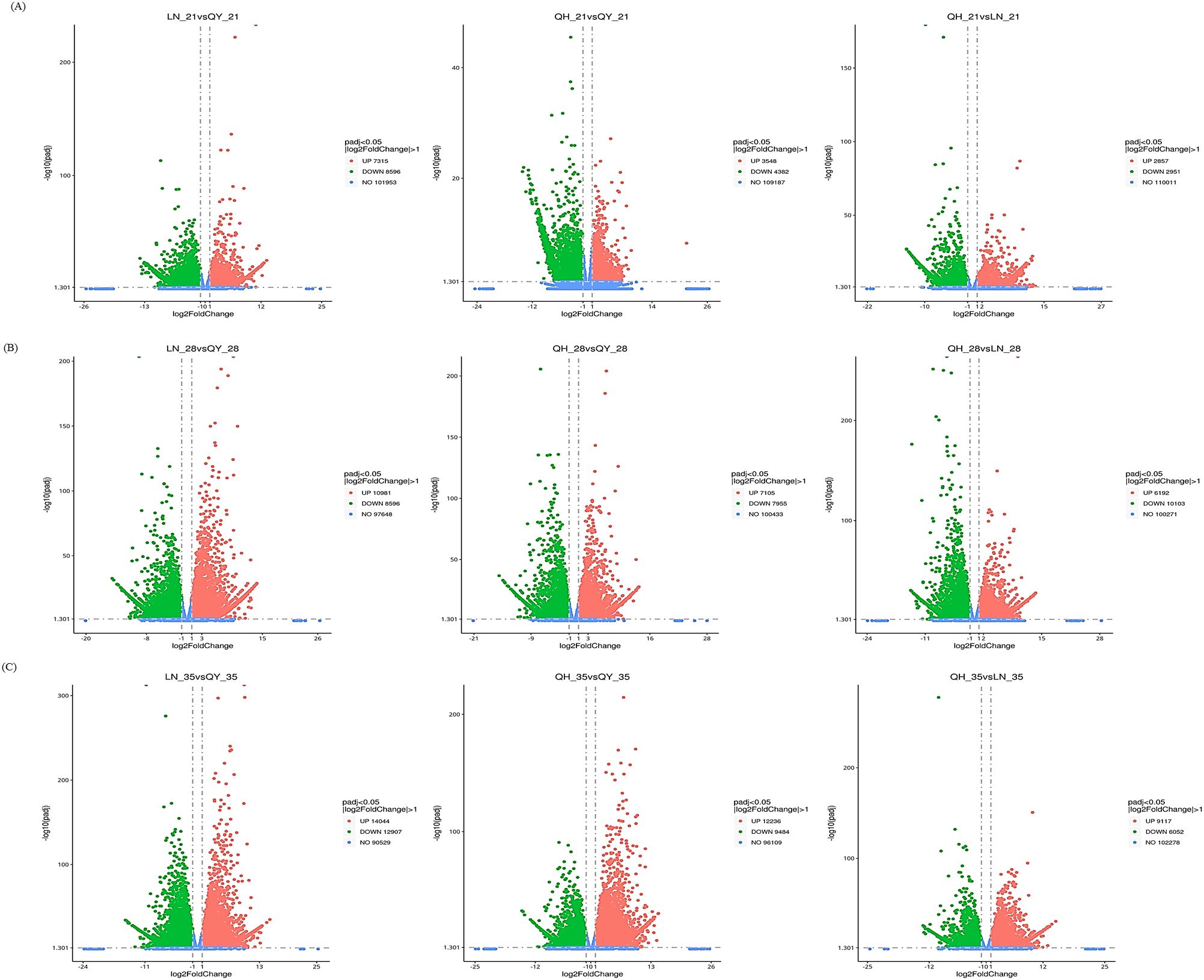
Figure 2. Volcanic diagram analysis of DEGs on the 21st (A), 28th (B), and 35th day (C) of growth in the second stem internode at the base for different oat cultivars. Red indicates up-regulated DEGs, green indicates down-regulated DEGs, and blue indicates unexpressed DEGs.
The 21st day of growth at the internode of the second stem at the base, 297 DEGs were co-expressed among the three oat cultivars (Figure 3A). 1437 DEGs were co-expressed on the 28th day (Figure 3B), and 2899 DEGs were co-expressed on the 35th day (Figure 3C). It can be seen that with the growth of the second stem internode at the base of oats, with an increase in co-expressed DEGs, the more complex the expression regulation pathway tends to be.
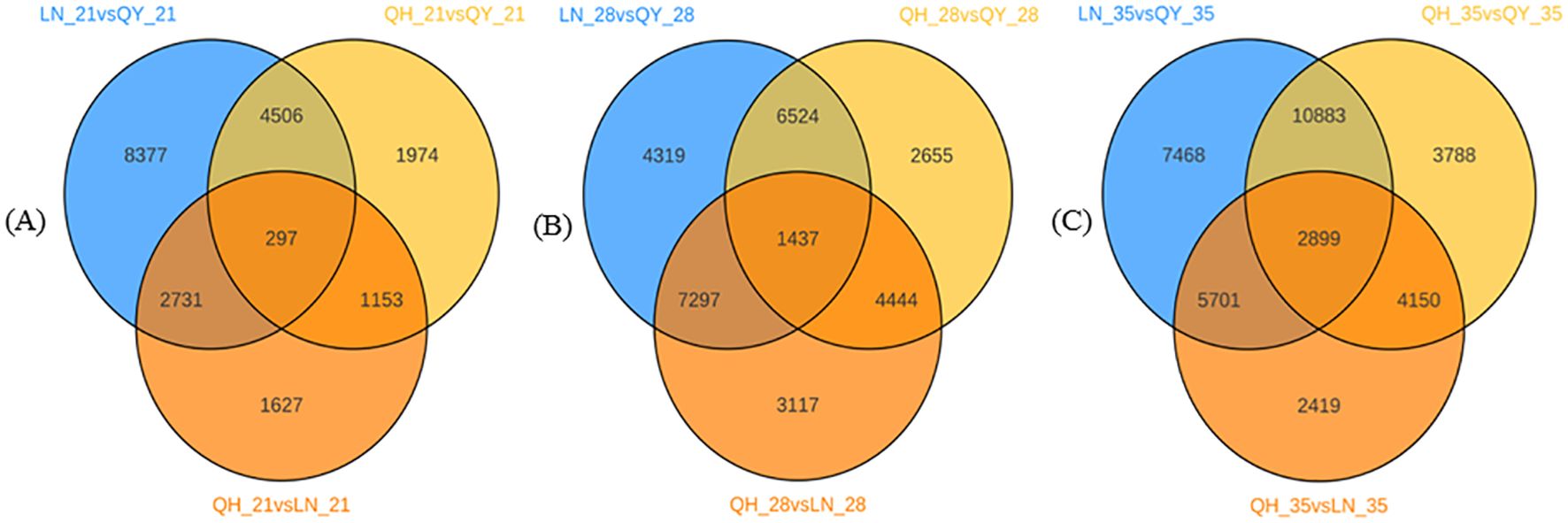
Figure 3. Venn diagram analysis of DEGs on the 21st (A), 28th (B), and 35th day (C) of growth in the basal second stem internode for different oat cultivars.
Gene ontology (GO) function enrichment analysis showed that the 21st and 28th days of growth at the internode of the second stem at the base of oat, the DEGs from LN_21 vs QY_21 and LN_28 vs QY_28 were significantly enriched in response to drug, drug transport, drug transmembrane transport, and other pathways (Figure 4A, B). Meanwhile, on the 35th day, the DEGs from LN_35 vs QY_35, QH_35 vs QY_35 and QH_35 vs LN_35 were significantly enriched in response to biotic stimulus and defense response. Among them, the DEGs from LN_35 vs QY_35 were specifically enriched in anion transport, organic anion transport, organic acid transport and other pathways (Figure 4C).
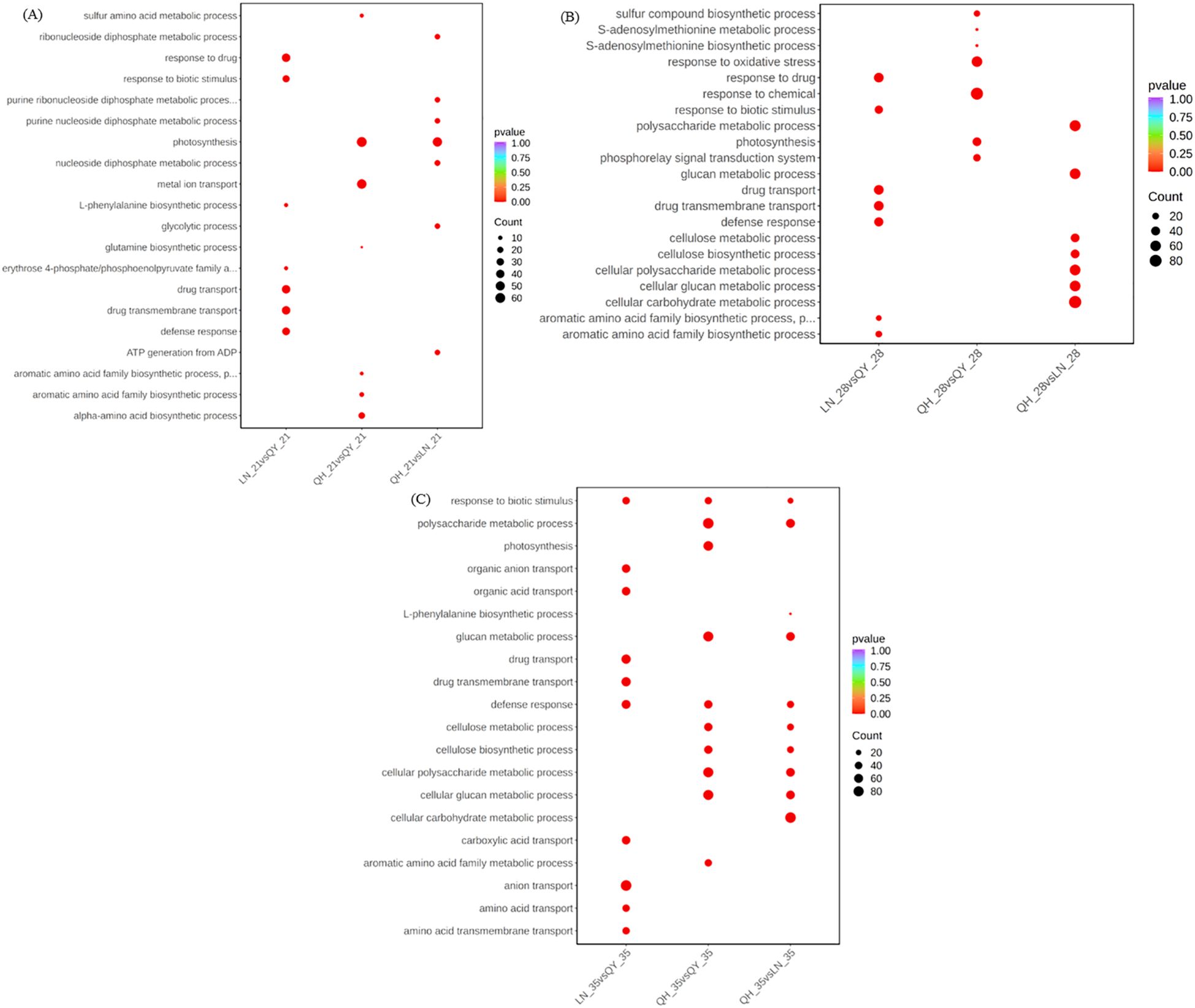
Figure 4. GO enrichment and scatter plots for DEGs on the 21st (A), 28th (B) and 35th day (C) of growth in the basal second stem internode from different oat cultivars. The smaller the q-value, the higher the enrichment degree. Same as below.
In order to further explore the metabolic pathways involved, kyoto encyclopedia of genes and genomes (KEGG) enrichment analysis showed that the DEGs from LN_21 vs QY_21 were significantly enriched in plant hormone signal transduction, MAPK signaling pathway-plant, glutathione metabolism and other pathways on the 21st day after the formation (Figure 5A). On the 28th day, the DEGs from LN_28 vs QY_28, QH_28 vs QY_28 and QH_28 vs LN_28 showed abundant pathways in starch and sucrose metabolism, phenylpropanoid biosynthesis, cyanoamino acid metabolism, among them. Meanwhile, the DEGs from LN_28 vs QY_28 were specifically enriched in plant-pathogen interaction, glutathione metabolism, as well as glycine, serine, and threonine metabolism (Figure 5B). On the 35th day, the DEGs from LN_35 vs QY_35, QH_35 vs QY_35 and QH_35 vs LN_35 were significantly enriched in fructose and mannose metabolism and cyanoamino acid metabolism. Among them, most DEGs were specifically enriched in starch and sucrose metabolism, phenylpropanoid biosynthesis, MAPK signaling pathway-plant and carbon metabolism (Figures 5A–C). And a large amount of DEGs are enriched in these pathways, indicating that these KEGG pathways are closely related to the lodging resistance of oat stems.
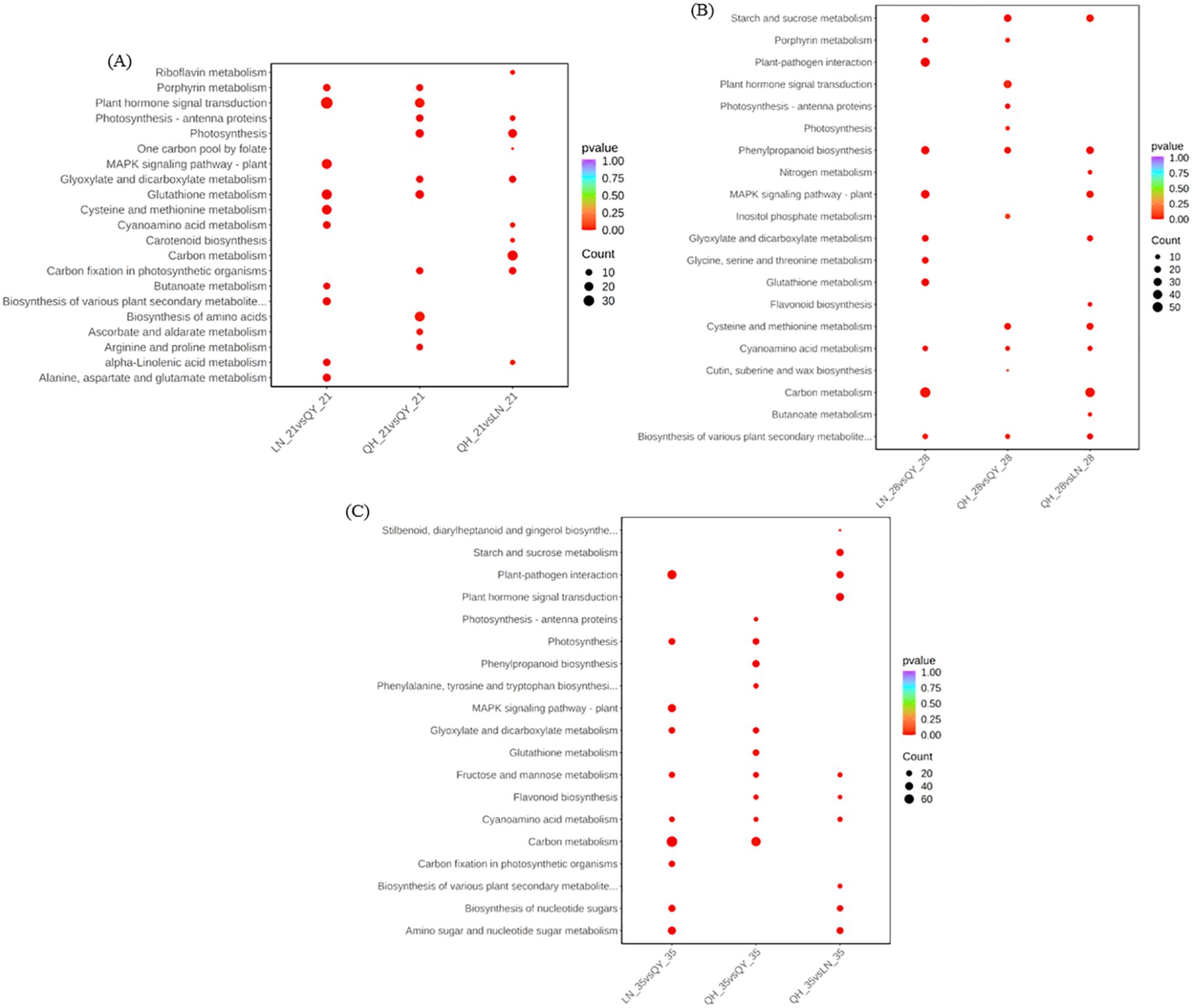
Figure 5. KEGG enrichment scatter plots for DEGs on the 21st (A), 28th (B), and 35th day (C) of growth in the second stem internode at the base of different oat cultivars.
Our RNA-seq analysis indicate that many TFs are involved in regulating the growth of oat stems. On the 21st day, the TF families with the highest proportion of differential groups were p450, Myb_DNA-binding, AP2, F-box, and WRKY (Figure 6A). On the 28th and 35th days, the TF families with the highest proportion were p450, Myb_DNA-binding, F-box, RRM1, AP2, and WRKY (Figures 6B, C). The up-regulated TFs were mainly enriched in the Myb_DNA-binding family, while the down-regulated TFs were mainly found in the p450, WRKY, F-box, RRM1, and AP2 families. There were many TF family types among the different comparison groups, and p450, Myb_DNA-binding, WRKY, and AP2 families accounted for the most.
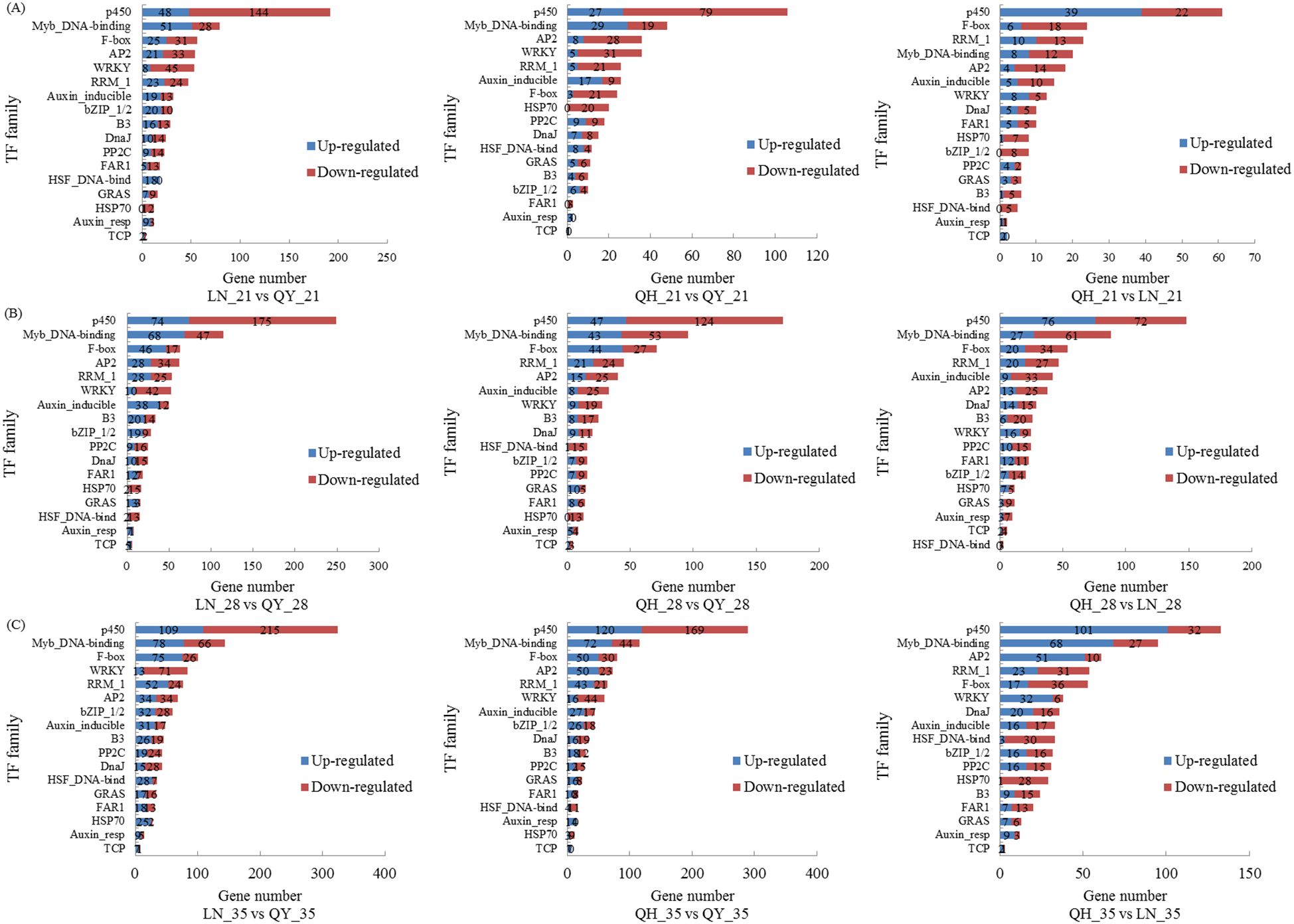
Figure 6. Taxonomic statistics for transcription factors on the 21st (A), 28th (B) and 35th day (C) of growth in the basal second stem internode of different oat cultivars. Figure shows the 17 TF families with the largest number of DEGs.
Based on the number of DEGs, the value of log2FoldChange (Log2FC), and the level of expression (FPKM), the 35th day comparison group was selected for further analysis of TFs. The results showed that the expression of MYB86, ODORANT1, MYBHv33, MYB308, and MYB39 in the Myb_DNA-binding family; CYP89A9, CYP75A5, CYP98A1, CYP710A1 and CYP714B2 in the p450 family; ERF055, DREB3 and ERF034 in the AP2 family were significantly up-regulated in lodging-resistant cultivar (‘LENA’ and ‘Qingyin No.1’). And the log2FC of multiple TFs was more than 3 times that of ‘Qingyin No.2’; the expression of WRKY71, WRKY46, WRKY57 and WRKY6 of the WRKY family were significantly down-regulated compared to the ‘Qingyin No.2’, while WRKY22 showed up-regulated; The ERF1A of the AP2 family were down-regulated in LN_35 vs QY_35 and QH_35 vs QY_35, and up-regulated in QH_35 vs LN_35, the DREB1H were down-regulated in LN_35 vs QY_35 and up-regulated in QH_35 vs QY_35 and QH_35 vs LN_35 (Table 3).
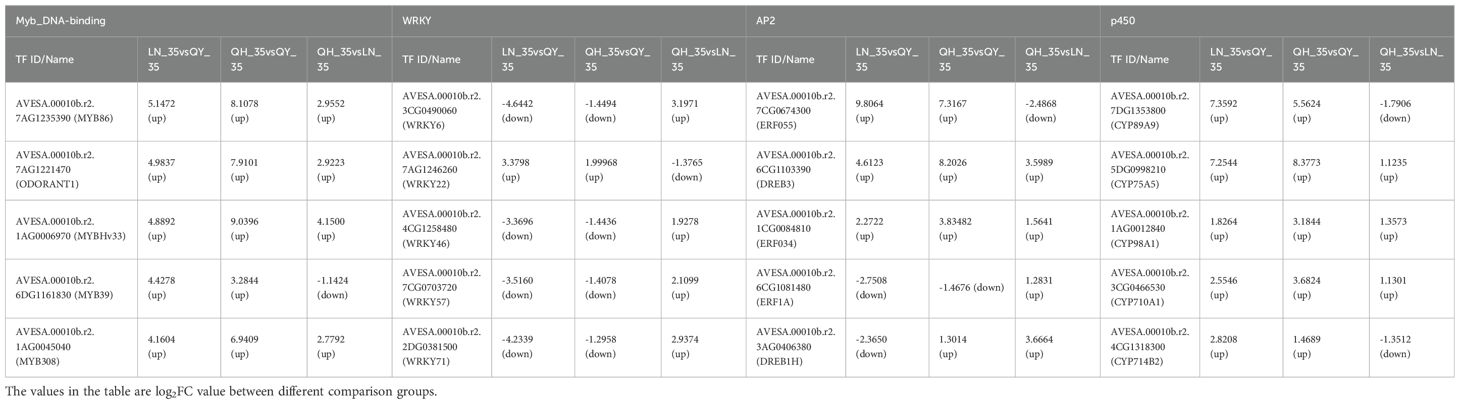
Table 3. Transcriptional factor screening on the 35th day of internode growth of the second stem of different oat cultivars.
Through DEG cluster analysis, we found that there were significant differences in the expression of each gene in different materials and/or different growth stages (Figure 7A). In order to further study the gene expression profiles, the DEGs from the different growth stages were clustered into 8 Profiles by STEM software. Our results showed that Profile 1 and 6 were significantly expressed in ‘LENA’ (P< 0.05). Profile1 gene expression continuously decreased on the 21st and 28th day of growth, but then remained stable on the 35th day. Profile 6 showed that gene expression continuously increased on the 21st and 28th day of the growth, but then remained stable on the 35th day (Figure 7B).
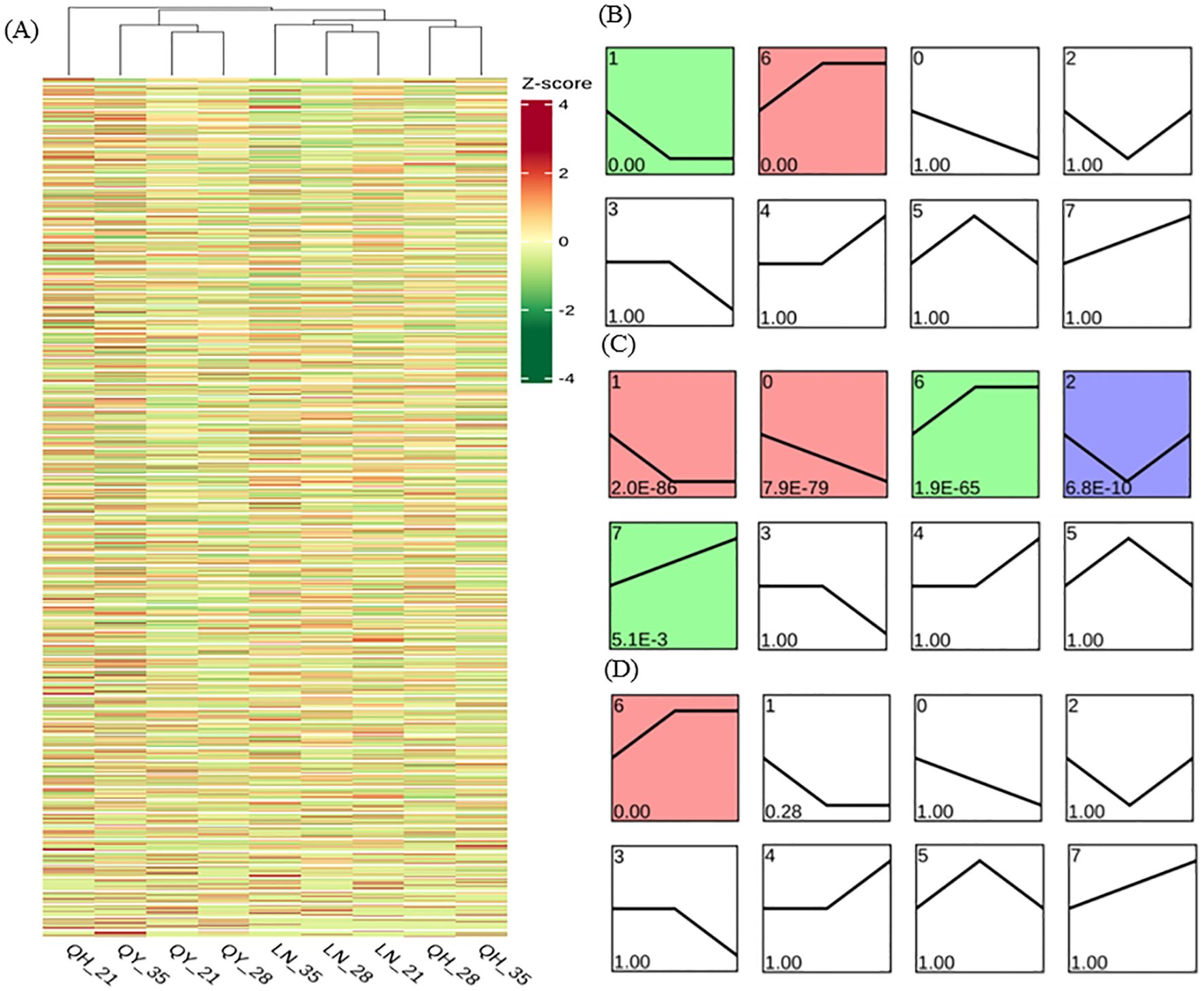
Figure 7. Gene expression trend analysis based on FPKM using log2 (fold change) hierarchical clustering analysis for DEG expression (A), LENA (B), Qingyin No.1 (C), and Qingyin No.2 (D) at different time points during the growth in the second stem internode at the base of oats. The numbers in the upper left corner of the box represent different gene expression trend patterns. Numbers below the box are the p-values.
In addition to Profile 1 and 6, Profile 0, 2 and 7 were also significantly expressed in ‘Qingyin No.1’ (P< 0.05). Profile 0 indicated that gene expression decreased during the different growth stages; Profile 2 pattern indicates that gene expression levels show a trend of first decreasing and then increasing at different growth stages of the second stem internode at the base of oats; Profile 7 gene expression showed an upward trend during the growth stages (Figure 7C). One major difference we noticed was that only Profile 6 were significantly expressed in ‘Qingyin No.2’ (P< 0.05), while Profile 1 were not enriched (Figure 7D). Our comprehensive analysis also showed that more than 50% of the DEGs were enriched into these five gene expression trend patterns.
Based on the results of venn diagram analysis, we found that the three oat cultivars had the most co-expressed genes on the 35th day of the growth of the second internode at the base. Therefore, we used the DEGs in this period to screen samples with the condition |log2Fold Change| ≥ 2 and FDR< 0.05, according to the expression level of DEGs and the results of KEGG function enrichment analysis. Our genetic screen found genes related to oat lodging resistance that involved photosynthesis (FDX6, PSBR and PETE), plant-pathogen interaction (HSP82), carbon metabolism (GCSH2, SGT, SDH4, RPE, GAPB, PGDH2, PGDH2.1, AACT2, HPR, PSAT2, GLO1, PGLP1B, ACOX3 and MTHFR), cyanoamino acid metabolism (ASRGL1 and BGLU16), amino sugar and nucleotide sugar metabolism (MPG1), phenylpropanoid biosynthesis (CCR1 and 4CL3) and glyoxylate and dicarboxylate metabolism (4CL8), as well as lignin synthase(CesA9, CesA7 and CesA4) (Figure 8).
Figure 8 also shows that there are some differences in the expression levels of the above genes among different cultivars of oats. The expression of most genes were up-regulated in the high lodging-resistant cultivar ‘LENA’ and the middle lodging-resistant cultivar ‘Qingyin No.1’. Among them, three key genes coding for enzymes (CCR1, 4CL8 and 4CL3) were found to affect the content and composition of lignin in varying degrees, and played an important role in regulating crops lodging resistance. In addition, CesA9, CesA7 and CesA4 were specifically up-regulated in the middle lodging-resistant cultivar ‘Qingyin No.1’. These three genes are genes encoding enzymes related to cellulose synthesis, which played an important role in the formation of plant cell wall and can improve the mechanical support and resistance to environmental stress of plants. Meanwhile, AACT2 and ACOX3 were specifically up-regulated in the high lodging-resistant cultivar ‘LENA’, which suggested that these genes might play a role in improving lodging resistance in oats. Therefore, CCR1, 4CL8 and 4CL3, as well as CesA9, CesA7, CesA4, AACT2 and ACOX3, which are specific and highly expressed in high lodging-resistant and medium lodging-resistant cultivars, were selected as candidate genes related to lodging resistance for further experiments.
Our WGCNA analysis found that when the fitting curve exceeds 0.8, the soft threshold β = 8 can be determined (Supplementary Figure S2A). This integrated expression matrix was used to screen 34937 genes, and the dynamic tree cutting method was employed to select the optimal soft threshold β = 8 to construct gene co-expression network for oat lodging resistance. The network were constructed by filtered probes, and a total of 26 modules were identified. In these modules, the turquoise module had the largest number of DEGs (7573), while the dark-orange module had the least (39). Agrey module is a set of genes that are not assigned to other modules (Supplementary Figure S2B).
Through clustering and heat map analysis between the modules, we found that 26 modules are clustered in three branches, and that the relationships between the various branches are relatively close (Figure 9A). In the gene co-expression network of oat lodging resistance, the correlation coefficients between module characteristic genes and samples were tested to explore the relationship between the identified modules and oat lodging resistance traits. Among the 26 modules, only 4 modules were significantly correlated with lodging resistance in oats. Both the dark-red module and blue module were positively correlated with the 21st day of the growth in ‘Qingyin No.1’ (r2 = 0.83, p = 8e-08; r2 = 0.67, p = 1e-04). In addition, there was a significant positive correlation between the midnight-blue module and the 35th day of the growth and ‘Qingyin No.1’ (r2 = 0.61, p = 7e-04). Likewise, there was a significant positive correlation between the black module and the 35th day of the growth in the ‘LENA’ cultivar (r2 = 0.60, p = 0.001) (Figure 9B). By comprehensively considering the correlation between the modules and the correlation between module characteristic genes and samples, the blue, black, midnight-blue and dark-red module were selected as the key modules related to lodging resistance in oats that can be further mined and analyzed.
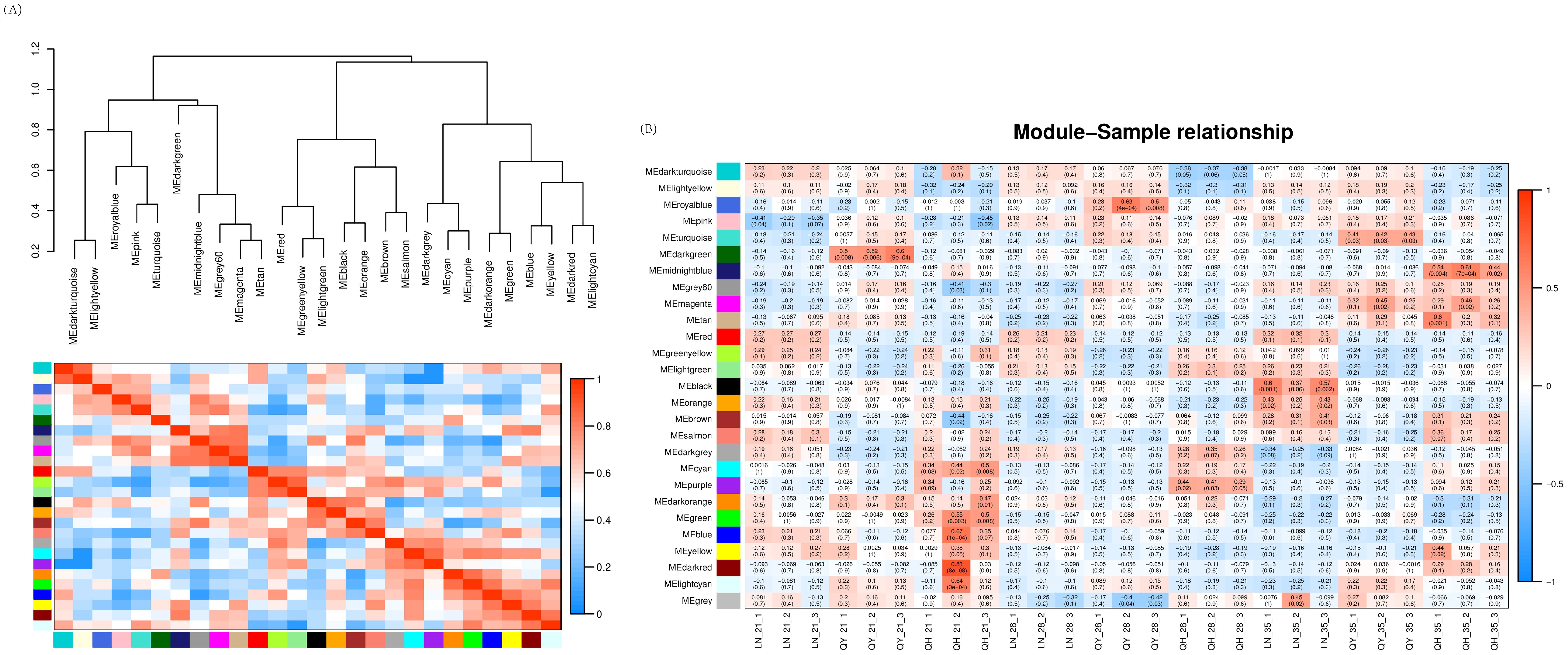
Figure 9. Clustering and heatmap analysis between modules (A), with the clustering tree between modules above and the heatmap and modules below. Correlation heatmap (B) between modules and the second stem internode of three oat cultivars at different growth stages. The numbers in the figure represent the correlation between modules and samples, while the numbers in parentheses indicate the p-value of the correlation. The more red the heatmap, the more positively correlated it is. The more blue the heatmap, the more negatively correlated it is.
From our module data, the GO terms significantly enriched in genes in the blue module are GO: 0044281 (small molecular metabolic process) and GO: 0072521 (purine containing compound metabolic process) (Figure 10A). The GO terms with significantly enriched genes in the black module are GO: 0051716 (cellular response to stimulus) and GO: 0050896 (response to stimulus) (Figure 10B). The GO terms enriched in genes in the midnightblue module are GO: 0006412 (translation) and GO: 0043043 (peptide biosynthetic process) (Figure 10C). Finally, the GO terms enriched in the darkred module are GO: 0050662 (coenzyme binding) and GO: 0048037 (cofactor binding), etc (Figure 10D).
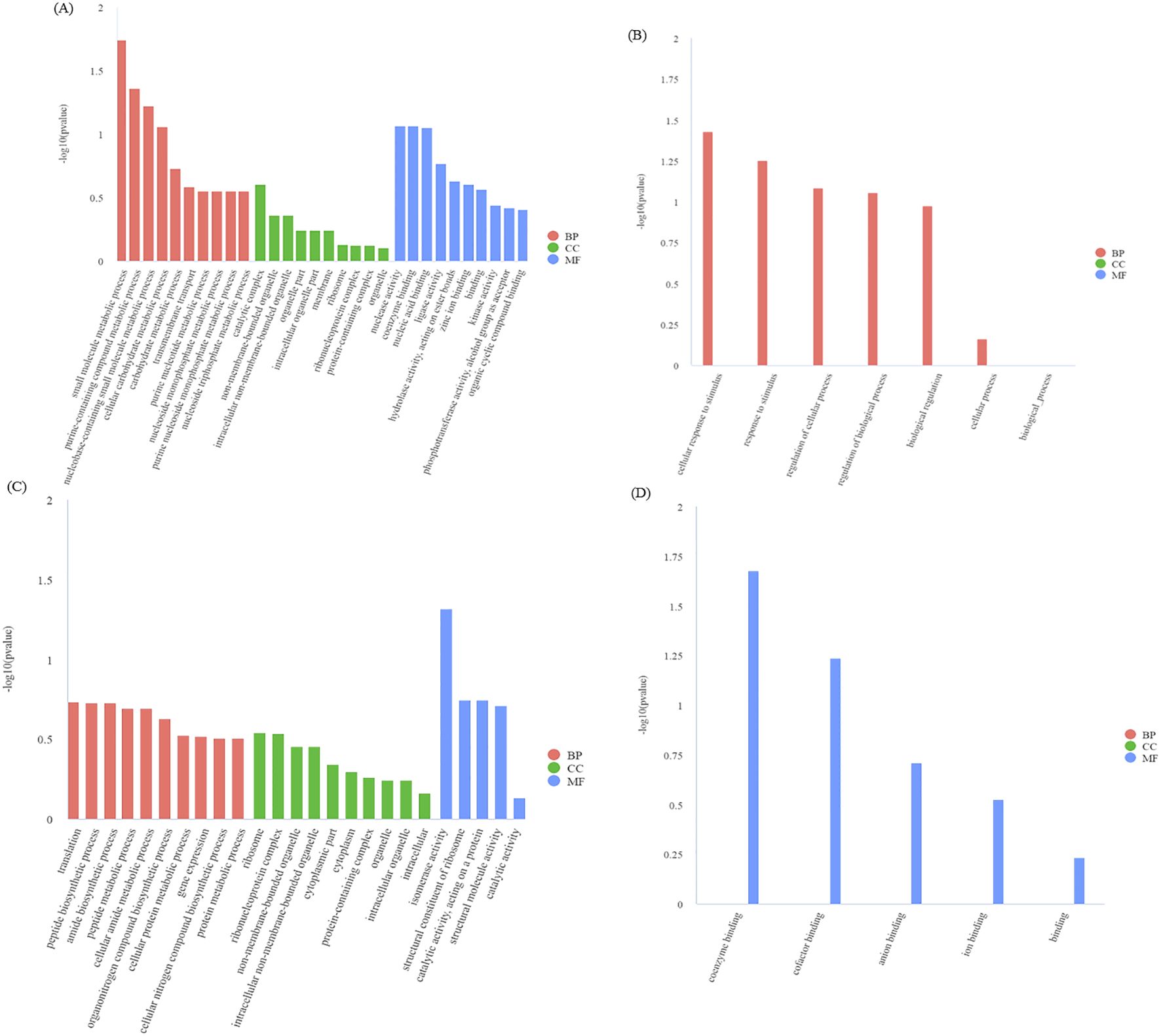
Figure 10. GO enrichment analysis of DEGs in the blue module (A), black module (B), midnight-blue module (C), and dark-red module (D). BP, biological process; CC, cellular component; MF, molecular function. The smaller the q-value, the higher the degree of enrichment. Same as below.
Our KEGG pathway analysis showed that 112, 49, 32 and 11 enriched KEGG pathways were identified by from the DEGs in the four modules. DEGs in the blue module were significantly enriched in sulfur metabolism and purine metabolism (Figure 11A), while DEGs in the black module were significantly enriched in protein processing in endoplasmic reticulum and spliceosome (Figure 11B). DEGs in the midnight-blue module were mainly enriched in plant-pathogen interaction and linoleic acid metabolism (Figure 11C), while DEGs in the dark-red module were significantly enriched in proteasome and lysine biosynthesis (Figure 11D).
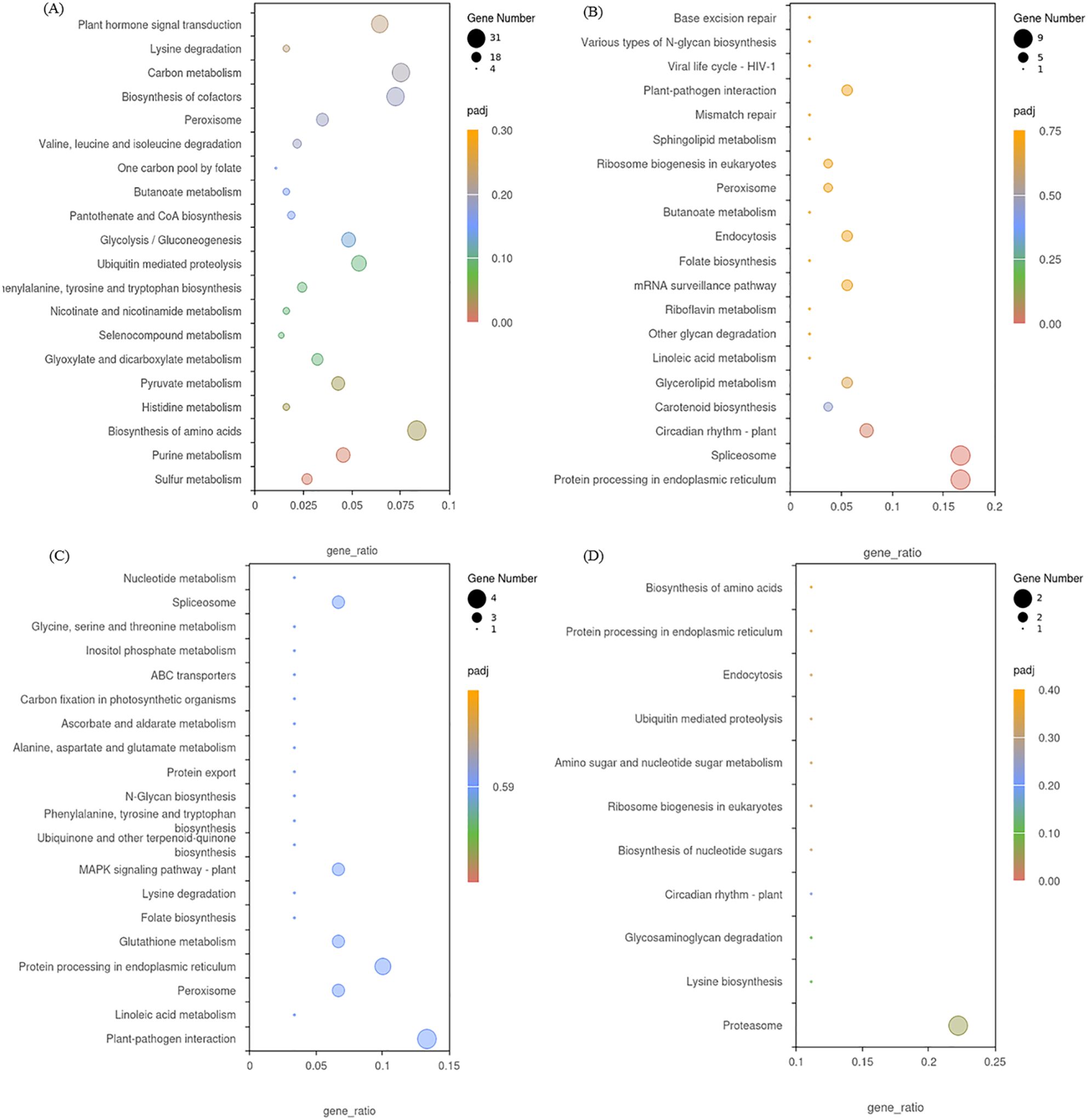
Figure 11. KEGG enrichment analysis of DEGs in the blue module (A), black module (B), midnight-blue module (C), and dark-red module (D).
In total, 20 genes with high connectivity in blue and black modules were identified as central genes (Figures 12A, B), while midnight-blue and dark-red modules were not built into the PPI network. Genes such as 30s ribosomal protein S13 (AVESA.00010b.r2.5DG0967260, RPS13), citrate synthase 4 (AVESA.00010b.r2.6AG1012620, CS4) and DNA-directed RNA polymerases II, IV and V subunit 9A (AVESA.00010b.r2.6AG1056760, RPB9) in blue module may be related to plant lodging resistance. In addition, the expression of the RPS13 gene is the highest on the 35th day of basal second stem internode growth in the high lodging-resistant cultivar ‘LENA’, while the expression of CS4 gene was the highest on the 35th for the middle lodging-resistant cultivar ‘Qingyin No.1’. Furthermore, the expression of RPB9 gene was the highest on the 21st day of the growth for the high lodging-resistant cultivar ‘LENA’ (Figure 12A).

Figure 12. The co-expression network of genes in the blue (A) and black module (B). The expression pattern of Hub genes were analyzed. The central circle represents the Hub genes and the genes with high connectivity in the corresponding network are shown in the larger circle.
Genes in the black module, such as multiprotein-bridging factor 1c (AVESA.00010b.r2.4CG1283660, MBF1c), SKP1-like protein 1B (AVESA.00010b.r2.5DG0952470, SKP1), and cullin-associated NEDD8-dissociated protein 1 (AVESA.00010b.r2.6CG1111830, CAND1) were highly expressed on the 35th day after the growth in the high lodging-resistant cultivar ‘LENA’ (Figure 12B), which were also previously speculated to have a certain regulatory effect on the lodging resistance of oats.
Plant lodging decreases quality and mechanical harvest efficiency, as well as increases grain dry rate by as much as 20%, which has a serious impact on production efficiency, and has become a key problem that restricts grain production (Wen et al., 2019). Oat is one of the main cultivars in the Qinghai-Tibet Plateau, the climatic conditions and soil types in this area are suitable for the growth of oats, but during the growth and development process of oats in high-altitude regions, lodging is an inevitable phenomenon, this has had a great impact on the development of animal husbandry in high-altitude pastoral areas. Plant phenotypic morphological characteristics are one of the most important traits that determine crop lodging resistance, and it is known as the most intuitive and easily identifiable feature to determine lodging. The stem is the main supporting organ for plant growth and development, and its internode length, stem thickness, and stem wall thickness are closely related to the lodging condition of the plant. It has been shown that plant lodging is mainly related to plant height and center of gravity height (Ahmad et al., 2023), and the cultivars with large height of stem center of gravity, unbalanced allocation of each internode, long basal second internode and large proportion of ear height, but has thin stems, thin wall, low density and plumpness had poor stem quality, low folding resistance and large lodging index. Lodging is prone to occur in the field, which is similar to the results of this study, which found that the phenotypic morphological characteristics of oat stem are closely related to its lodging resistance, and there is a significant positive correlation between the lodging index of the second stem internode and the actual lodging rate at flowering stage, the higher the value of both, the weaker the ability to resist lodging. The lodging index of the second stem internode showed a significant positive correlation with plant height, the length of the second stem internode, the height of ear height and the coefficient of ear position, and a significant negative correlation with the stem wall thickness of the second stem internode. This shows that the larger the plant height and the length of the second stem internode, the more prone to lodging, and increasing the stem wall thickness of the second stem internode will reduce the risk of oat lodging.
In addition, the stem internode composition of the stem was most closely related to the lodging resistance of the first and second internode of the main stem and the subpanicle internode. The structural characteristics, coordination degree and allocation ratio of the basal internodes jointly determined the lodging resistance of oat plants (Nan, 2021). In particular, Liu (2005) found that cultivars with strong lodging resistance had short basal internodes, thick culm walls, longer flag leaves, smaller leaf area, higher mechanical strength, and greater elasticity in their stems. This study found that there was a significant positive correlation between the actual lodging rate of oat and the length of the second stem internode, and a significant negative correlation between the second stem internode lodging index and compression strength, puncture strength, breaking force and mechanical strength. The second stem internode length of ‘Qingyin No.2’ was higher than that of ‘LENA’ and ‘Qingyin No.1’. The compression strength, puncture strength, breaking force and mechanical strength of ‘LENA’ and ‘Qingyin No.1’ were higher than those of ‘Qingyin No.2’, which were similar to that of Liu (2005), indicated that the oat cultivars with strong lodging resistance had shorter basal internodes and higher mechanical strength and compression strength. Nan (2021) found that the oat cultivars with short internodes, thick culm walls, more lignin in the cell wall, high hardness and plumpness in the cell wall, as well as a small pulp cavity volume created a strong resistance to lodging with high stem quality. Liang et al. (2019) also found that the stem diameter, stem diameter coefficient, and internode length of the second stem internode at the base of oat stem were significantly higher than those of the first stem internode. However, the stem wall thickness and mechanical properties of the first stem internode were significantly higher than those of the second stem internode, and stem base traits were the main factors affecting oat lodging. Based on previous analysis, this study found that oat lodging mainly occurs at the second stem internode at the base, the actual lodging rate of the second stem internode were significantly positively correlated with the length of the second stem internode, and significantly negatively correlated with the stem thickness, stem wall thickness, compression strength, puncture strength, breaking force, and mechanical strength of the second stem internode, this is similar to the results of Nan (2021) and Liang et al. (2019). Oats with strong lodging resistance have lower plant height and height of center of gravity, base stem thick and stem wall thick, shorter internodes, and better mechanical properties, these traits collectively affect the lodging resistance of oats. Based on these results, subsequent lodging studies in oat can focus on the second stem internode at the base to enhance the strength and toughness of the stem internode, and improve the lodging resistance. Plant height should also be investigated in lodging research, since the lodging resistance of oats can be increased by reducing plant height.
The physicochemical properties of plant stems are closely related to their lodging resistance. If some or a certain component in the stems is out of proportion or physiological disorders occur, it will increase the risk of lodging. Generally, the most abundant substances in the stem include trace mineral elements, lignin, cellulose, polysaccharides, and other non-structural carbohydrates, which mainly exist in the thin-walled cells of the stem. These substances coordinate and maintain the “source-sink” at different stages of plant growth and development, which affect the process of grain filling. Fan et al. (2017) found that the content of mineral elements, cellulose, and lignin in rice stem tissue cells maintains stable stem mechanical properties and support strength. They also found that increasing cellulose and lignin content improves stem mechanical strength and surface cell wall hardness to support the cell wall structure, which is more conducive to enhancing lodging resistance. This study found that the physiological indicators of the stems of three different lodging-resistant oat cultivars were extremely significant/significantly negative correlated with the lodging index and actual lodging rate of the second stem internode during flowering. Analysis of their lignin content revealed that the lodging index and actual lodging rate of the second stem internode during flowering were significantly negatively correlated with lignin content. The lignin content in the stems of lodging-resistant cultivars was higher than that of lodging susceptible cultivar, which is similar to the research results of Fan et al. (2017), indicating that higher lignin content can enhance the mechanical strength and epidermal cell wall hardness of oat stems, thereby reducing the risk of lodging.
Nan (2021) also found that the amount of potassium, calcium, silicon, and magnesium in the basal internodes of the resistant cultivars were significantly higher than those of the lodging susceptible cultivar. Low nitrogen and high potassium, calcium, silicon, and magnesium content are the key to enhance the lodging resistance in oat stems, which is similar to the results of this study. This study found that the lodging index of the second stem internode during oat flowering were extremely significant negative correlated with stem calcium and silicon content, and significantly negatively correlated with potassium content, the actual lodging rate during the flowering period were extremely significant negative correlated with stem silicon content, and significantly negatively correlated with potassium and calcium content. This indicating that the higher the content of potassium, calcium and silicon in oat stem, the stronger the lodging resistance, and the higher the content of potassium, calcium and silicon will enhance the mechanical strength and toughness of oat stem, make the stem harder, thus enhance the lodging resistance.
In addition, non-structural carbohydrates are the main enrichment substances in crop stems, especially polysaccharides such as starch and soluble sugars, which can promote the formation of cellulose and hemicellulose, thereby helping to maintain the mechanical strength of the stem. Zhang et al. (2010) found in their study on rice that the soluble sugar content in the stem and sheath in lodging-resistant cultivars was higher during the late flowering stage. However, there was not a correlation between soluble sugar content in the stem and lodging resistance during the milky stage. Ishimaru et al. (2008) also showed that the rice stem was easily bent by the wind, and that the cultivars with higher starch content in the stem were more beneficial to return to the normal growth state. Zhang et al. (2020) found that starch content were significantly positively correlated with cellulose content and hemicellulose content, which suggests that higher starch content in oat stems could promote the synthesis of cellulose and hemicellulose to improve their lodging resistance, which is similar to the results of this study. In this paper, based on the analysis of soluble sugar and starch content in oat stem, it was found that the lodging index and actual lodging rate of the second stem internode at flowering stage were extremely significant negative correlated with stem soluble sugar and starch content, and the soluble sugar and starch content in plant stem could affect the bending strength of oat stalk. This shows that increasing the content of soluble sugar and starch in oat stem can effectively enhance the lodging resistance of the species.
As an important regulatory protein, TFs specifically bind to cis-acting elements in the promoter region of eukaryotic genes to regulate the expression of target genes. They also played a central role in regulatory networks and signaling pathways during plant development, as well as respond to abiotic stress (Cui et al., 2016). Li et al. (2024) found that NAC and MYB family can regulate the development of plant secondary walls, especially the synthesis and accumulation of lignin, playing an important regulatory role in plant secondary growth and lodging resistance. Xiao et al. (2018) found that NAC can enhance CesA gene expression by promoting MYB transcription to enhance cellulose synthesis, and improve the mechanical strength and toughness of maize (Zea mays) stalks. This study found that TFs of the MYB family account for a relatively large proportion in the growth of the second stem internode at the base of different oat cultivars, which is consistent with previous research results. In addition, Wang et al. (2024) found that MYB110 can function to regulate plant height and lodging resistance, since MYB110 is the target gene of OsPHR2, which is the core regulator in the rice phosphorus signal transduction network. Here, they found lodging resistance was positively regulated by OsPHR2, which suppresses rice plant height. Although they saw plant height increased and stem lignin content decreased, their myb110 mutants had stronger lodging resistance since their stems were stronger than wild-type plants. In this study, we found that TFs such as MYB86, ODORANT1, MYBHv33, MYB308, and MYB39 were significantly up-regulated in lodging-resistant cultivar. It is speculated that these TFs may also affect plant height and stem strength by regulating cell division and differentiation, thereby regulating the synthesis of cellulose and lignin and affecting the stiffness and toughness of oat stems, directly affecting the lodging resistance of oats. Besides, Lv et al. (2024) showed that OsTCP19 of the TCP TF family can regulate the synthesis of cellulose and lignin in rice, thereby affecting the stiffness and toughness of stems. Our study also found significant differences in a small number of TCP TFs among different lodging-resistant oat cultivars, similar to previous research results.
Cytochrome P450 (CYP450) catalyzes the 3-hydroxylation of phenylpropane compounds to modulate the precursors of secondary metabolites like lignin and anthocyanins, which may affect the lignin content in plant stems and thus stem strength (Morant et al., 2007). Kim et al. (2005) found that most cyp85a mutants in plants exhibit a super compact phenotype with shortened internodes that lead to dwarfism and a reduced risk of lodging. Our study also discovered a family of cytochrome P450 TFs, including CYP89A9, CYP75A5, CYP98A1, CYP710A1, and CYP714B2, which were up-regulated during the growth of the second stem internode of the high lodging-resistant oat ‘LENA’. And the plant height of ‘LENA’ was significantly lower than the other two oat varieties, suggesting that these TFs may improve its lodging resistance by regulating the content of enriched substances in the ‘LENA’ stem and shortening internode length.
WRKY family can affect secondary cell wall thickening by binding to the promoters of other TFs that activate secondary wall synthesis (Wang et al., 2010). Lan et al. (2020) also showed that WRKY36 inhibits gibberellins (GAs) signaling by stabilizing the expression of the GAs inhibitor SLR1 to decrease rice grain size and plant height and reduce the occurrence of lodging. Similarly, multiple WRKY transcription factors were found to be differentially expressed during the growth process of the second stem internode in different lodging-resistant oat cultivars. Among them, the expression of WRKY6, WRKY46, WRKY57, and WRKY71 were down-regulated in lodging-resistant cultivars, while WRKY22 was up-regulated. It is speculated that these TFs can reduce the risk of oats lodging by regulating the thickness of the secondary cell wall of oat stems or reducing their plant height. AP2/ERF TF family played an important role in plant growth and development and stress response. Du (2021) and Qi et al. (2011) found that OsRPH1 and OsRPH2 of the AP2/ERF family can regulate the expression levels of genes related to GA synthesis metabolism, which reduce endogenous GA activity in rice and negatively regulate plant height to improve the lodging resistance and yield of rice. In this study, it was found that the TFs of the AP2 family also account for a large proportion in comparison groups of oats. Among them, the expression of ERF055, DREB3, and ERF034 were up-regulated in lodging-resistant oats. It is speculated that these TFs may have similar roles in regulating oat lodging resistance.
In recent years, high-throughput sequencing technology (RNA-seq) has rapidly developed, which can effectively understand the regulatory rules of gene expression, and identify key genes related to plant function regulation (Qiong et al., 2019). Liu et al. (2022) studied the lodging resistance traits of maize by integrating transcriptome and metabolome analysis, KEGG enrichment analysis found that DEGs were mainly enriched in phenylpropanoid and secondary metabolites biosynthesis pathways, and several functional and regulatory genes related to lodging resistance traits were screened, these genes mainly participate in cell wall assembly, lignin biosynthetic process, and hormone metabolic process. In present study, transcriptome was conducted on the different lodging-resistant oat cultivars and found that the three oat cultivars co-expressed the most genes on the 35th day of growth between the second stem internodes at the base, and these DEGs were significantly enriched in pathways such as starch and sucrose metabolism, phenylpropanoid biosynthesis, MAPK signaling pathway-plant and carbon metabolism.Genes related to cellulose synthesis and lignin synthesis were found in these pathways, which is similar to previous research results. Among them, the cellulose synthesis genes CesA4, CesA7, and CesA9 are specifically up-regulated in the middle lodging-resistant cultivar ‘Qingyin No.1’. Studies have shown that the expression of CesA4, CesA7, and CesA9 genes in constitutes the synthase complex necessary for the synthesis of secondary cell wall cellulose (Maleki et al., 2016). Li et al. (2018) found in their study on rice that mutations at the CESA4 P-CR site can affect cell wall characteristics, particularly cellulose structure, thereby enhancing biomass digestion and lodging resistance; Kumar et al. (2017) and Deng et al. (2019) also showed that a deletion mutant in cesa7 decreased cellulose content in the stem and created defects in secondary cell wall formation in the xylem, which led to the collapse of xylem constituent elements. Li et al. (2017) also found that mutant OsCesA9 reduced plant height and showed thinner stems with significantly increased tiller numbers compared to the wild type. It was speculated that high lodging-resistant in this mutant may be attributed to its plant height, single tiller fresh weight, and low cellulose crystallinity. These results indicate that different genes in the CesA family related to cellulose synthesis contribute to cellulose synthesis in primary and secondary cell walls during plant development, thereby enhancing plant lodging resistance. Therefore, we speculate that these three genes are closely related to the lodging resistance characteristics of oat stems.
In addition, lignin enhances the mechanical strength of the stem by participating in cell wall biosynthesis to improve the lodging resistance in crops. Hu et al. (2015) showed that the lignin biosynthesis related gene 4CL in the stem of Fagopyrum esculentum reached its highest expression during the full flowering stage, and that the expression level in lodging-resistant cultivar were significantly higher than that in lodging-susceptible cultivar, this is consistent with the research findings of this paper. In this paper, 4CL8 and 4CL3 genes related to lignin synthesis were screened, these two genes were specifically up-regulated in high lodging-resistant cultivar ‘LENA’ and middle lodging-resistant cultivar ‘Qingyin No.1’. The expression level in lodging-resistant cultivars were significantly higher than that in lodging-susceptible cultivar, which indicated that lignin synthesis related genes played an important role in the regulation of crops lodging resistance. CCR is the starting point in the special branching pathway for the synthesis of lignin in the phenylpropane metabolic pathway. Here, it acts as a key enzyme to control the content and quality of lignin, and changing CCR gene expression levels will affect the total lignin content in plants (Meester et al., 2020). In this paper, another gene related to lignin synthesis, CCR1, was also screened, which was specifically up-regulated in lodging- resistant cultivars, and the expression level in lodging-resistant cultivar were significantly higher than that in lodging-susceptible cultivar, Zhou et al. (2010) found that the growth rate and lignin content of Medicago truncatula decreased significantly after inhibiting the expression of CCR1; Hu et al. (2017) found that the lodging resistance of buckwheat increases with the relative expression levels of genes such as PAL, 4CL, C4H, CAD, and CCR, this indicates that up-regulated of lignin synthesis related genes in plants promotes an increase in lignin content, thereby enhancing the lodging resistance of plants. In addition, in the high lodging-resistant cultivar ‘LENA’, AACT2 and ACOX3 were specifically up-regulated, suggesting that these genes may play a certain role in improving the lodging resistance of oats.
WGCNA analysis based on large sample transcriptome data has become the most widely used co-expression network analysis method, not only for constructing gene networks, but also for detecting hub genes in gene modules and identifying them (Langfelder and Horvath, 2008). It will indicate the similarity of expression patterns between genes and whether they are in the same expression module, which is suitable for complex multi sample transcriptome data. It can quickly study the overall expression pattern of genes and provide feedback on the interaction patterns between genes in the sample (Zhang and Horvath, 2005). At present, the WGCNA analysis method has been widely used in the screening of stress resistance related genes in various plants. Chen et al. (2022) conducted WGCNA analysis on transcriptome data sequenced from cotton (Gossypium hirsutum) under salt and drought stress, and identified that GhWRKY46 gene can improve drought and salt tolerance of cotton. Wang et al. (2023) used WGCNA to construct a gene co-expression network and identified 20 hormone related candidate genes associated with salt stress in tomato (Solanum lycopersicum). In this study, the hub genes closely related to the lodging resistance traits of oats, such as MBF1c, SKP1 and CAND1, were screened by WGCNA analysis, and it was found that they were specifically expressed on the 35th day of internode growth of the second stem at the base of the lodging resistance cultivar ‘LENA’. Among them, MBF1c is a transcription co-activator that has been shown to affect the stem stiffness and toughness of plants by regulating the synthesis of plant cell wall components (Liu et al., 2023), which is consistent with the KEGG enrichment pathway results; In addition, the protein encoded by SKP1 can bind to Cullin protein and F-box protein, forming the core part of SCF complex, enhanced expression of this gene can improve plant resistance (Farrás et al., 2001); CAND1 is a protein associated with NEDD8 modification, and the SlCAND1-RNAi line were significantly dwarfed compared with wild-type plants during the vegetative growth stage of tomato, which effectively reduced the risk of lodging (Cheng et al., 2020). Based on phenotype data, we found that the lodging-resistant cultivar ‘LENA’ has lower plant height and stronger lodging resistance, and the high expression of this gene in ‘LENA’ is further confirmed the efficiency and accuracy of the WGCNA analysis method. This study comprehensively utilized transcriptome analysis and WGCNA analysis to obtain multiple genes related to oat lodging resistance, providing important genetic resources for the breeding of lodging-resistant oat cultivars. At present, molecular marker assisted breeding and CRISPR-Cas9 gene editing technology have brought revolutionary changes to biological breeding, they can accurately locate and modify target genes, thereby quickly cultivating crop cultivars with excellent traits. Applying these genes to these cutting-edge technologies is expected to open up new avenues for cultivating new cultivars of lodging-resistant oats and promote innovative development in oat breeding.
Our study conducted transcriptome analysis on the field traits and stem physiological indicators of three oat cultivars with different lodging resistance abilities during the key period of growth for the second stem internode at the base. We found that the lodging resistance of oats was closely related to the changes in stem growth characteristics and physicochemical component content. On the 35th day of growth, there are more co-expressed genes, which indicates that the expression of regulatory pathways may become more complex. The growth process of the second stem internode at the base of this species is mainly related to the KEGG pathway, such as starch and sucrose metabolism, phenylpropanoid biosynthesis, MAPK signaling pathway-plant and carbon metabolism. TFs from families like p450, Myb_DNA-binding, WRKY, and AP2, along with cellulose synthesis related genes CesA9, CesA7, and CesA4, lignin synthesis related genes CCR1, 4CL8, and 4CL3, as well as genes AACT2, ACOX3, MBF1c, SKP1, and CAND1 that are specifically up-regulated in the high lodging-resistant cultivar ‘LENA’, jointly regulate the growth process of the second stem internode at the base of oats. It is speculated that these TFs and genes have a certain regulatory effect on the lodging resistance of oats. Further research on the functions of these genes will help reveal the molecular mechanisms of oat lodging resistance and provide excellent genetic resources for lodging resistance breeding in oats and other crops.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA1186259.
JY: Data curation, Writing – original draft. RZ: Data curation, Funding acquisition, Methodology, Writing – review & editing. XM: Data curation, Funding acquisition, Methodology, Writing – review & editing. ZJ: Methodology, Resources, Writing – review & editing. XL: Supervision, Writing – review & editing. YL: Validation, Visualization, Writing – review & editing. HL: Funding acquisition, Writing – review & editing.
The author(s) declare that financial support was received for the research and/or publication of this article. This study was financially supported by National Science Foundation for Young Scientists of China (32301491); National Natural Science Foundation of China (32360344); Qinghai Province Basic Research Program Project (2023-ZJ-750).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1532216/full#supplementary-material
Ahmad, I., Batyrbek, M., Ikram, K., Ahmad, S., Kamran, M., Khan, R. S., et al. (2023). Nitrogen management improves lodging resistance and production in maize (Zea mays L.) at a high plant density. J. Integr. Agric. 22, 417–433. doi: 10.1016/J.JIA.2022.08.074
Ahmad, M., Dar, Z. A., Habib, M. (2014). A review on oat (Avena sativa L.) as a dual-purpose crop. Sci. Res. Essays. 9, 52–59. doi: 10.5897/SRE2014.5820
Baucher, M., Monties, B., Montagu, M. V., Boerjan, W. (1998). Biosynthesis and genetic engineering of lignin. Crit. Rev. Plant Sci. 17, 125–197. doi: 10.1016/s0735-2689(98)00360-8
Berry, P. M., Spink, J. (2012). Predicting yield losses caused by lodging in wheat. Field Crops Res. 137, 19–26. doi: 10.1016/j.fcr.2012.07.019
Bhargava, A., Mansfield, S. D., Hall, H. C., Douglas, C. J., Ellis, B. E. (2010). MYB75 functions in regulation of secondary cell wall formation in the Arabidopsis inflorescence stem. Plant Physiol. 154, 1428–1438. doi: 10.1104/pp.110.162735
Chen, P., Wei, F., Jian, H., Hu, T., Wang, B., Lv, X., et al. (2022). A comprehensive gene co-expression network analysis reveals a role of GhWRKY46 in responding to drought and salt stresses. Int. J. Mol. Sci. 23, 12181. doi: 10.3390/ijms232012181
Cheng, W., Yin, S., Tu, Y., Mei, H., Wang, Y., Yang, Y. (2020). SlCAND1, encoding cullin-associated Nedd8-dissociated protein 1, regulates plant height, flowering time, seed germination, and root architecture in tomato. Plant Mol. Biol. 102, 537–551. doi: 10.1007/s11103-020-00963-7
Cloninger, F. D., Zuber, M. S., Calvert, O. H., Loesch, J. P.J. (1970). Methods of evaluating stalk quality in corn. Phytopathology 60, 295–300. doi: 10.1094/Phyto-60-295
Cui, L., Feng, K., Wang, M., Wang, M., Deng, P., Song, W., et al. (2016). Genome-wide identification, phylogeny and expression analysis of AP2/ERF transcription factors family in Brachypodium distachyon. BMC Genomics 17, 1–19. doi: 10.1186/s12864-016-2968-8
Deng, Q., Kong, Z., Wu, X., Ma, S., Yuan, Y., Jia, H., et al. (2019). Cloning of a COBL gene determining brittleness in diploid wheat using a MapRseq approach. Plant Sci. 285, 141–150. doi: 10.1016/j.plantsci.2019.05.011
Du, L. (2021). Study on the function of rice plant height-related AP2 familytranscription factor OsRPH2. (Changchun: Jilin University). doi: 10.27162/d.cnki.gjlin.2021.004869
Fan, Y. Y., Yang, G. T., Chen, J., Jiang, F., Chen, Y. J., Hu, Y. G. (2017). The physical, chemical characters and lodging resistance of rice stem with silicon potassium collocation application. Acta Bot. Boreali-Occident. Sin. 37, 751–757. doi: 10.7606/j.issn.1000-4025.2017.04.0751
Farrás, R., Ferrando, A., Jásik, J., Kleinow, T., Ökrész, L., Tiburcio, A., et al. (2001). SKP1–SnRK protein kinase interactions mediate proteasomal binding of a plant SCF ubiquitin ligase. EMBO. J. 20, 2742–2756. doi: 10.1093/emboj/20.11.2742
Hu, D., Liu, X. B., She, H. Z., Gao, Z., Ruan, R. W., Wu, D. Q., et al. (2017). The lignin synthesis related genes and lodging resistance of Fagopyrum esculentum. Biol. Plant 61, 138–146. doi: 10.1007/s10535-016-0685-4
Hu, D., Liu, X. B., Wang, C., Yang, H., Li, H. X., Ruan, R. W., et al. (2015). Expression analysis of key enzyme genes in lignin synthesis of culm among different lodging resistances of common buckwheat (Fagopyrum esculentum Moench). Sci. Agric. Sin. 48, 1864–1872. doi: 10.3864/j.issn.0578-1752.2015.09.20
Ishimaru, K., Togawa, E., Ookawa, T., Kashiwagi, T., Madoka, Y., Hirotsu, N. (2008). New target for rice lodging resistance and its effect in a typhoon. Planta 227, 601–609. doi: 10.1007/S00425-007-0642-8
Kim, T. W., Hwang, J. Y., Kim, Y. S., Joo, S. H., Chang, S. C., Lee, J. S., et al. (2005). Arabidopsis CYP85A2, a cytochrome P450, mediates the Baeyer-Villiger oxidation of castasterone to brassinolide in brassinosteroid biosynthesis. Plant Cell 17, 2397–2412. doi: 10.1105/tpc.105.033738
Kumar, M., Atanassov, I., Turner, S. (2017). Functional analysis of cellulose synthase (CESA) protein class specificity. Plant Physiol. 173, 970–983. doi: 10.1104/pp.16.01642
Lan, J., Lin, Q., Zhou, C., Ren, Y., Liu, X., Miao, R., et al. (2020). Small grain and semi-dwarf 3, a WRKY transcription factor, negatively regulates plant height and grain size by stabilizing SLR1 expression in rice. Plant Mol. Biol. 104, 429–450. doi: 10.1007/s11103-020-01049-0
Langfelder, P., Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 9, 1–13. doi: 10.1186/1471-2105-9-559
Lei, X. L., Liu, L., Gou, W., Ma, R. C., Ren, W. J. (2013). Effects of planting methods on culm lodging resistance of indica hybrid rice (Oryza sativa L.). Acta Agron. Sin. 39, 1814–1825. doi: 10.3724/SP.J.1006.2013.01814
Li, X., Li, N., Wen, D., Yu, J., Hong, J., Wu, M., et al. (2024). Identification of Stress Responsive NAC Genes in Casuarina equisetifolia L. and Its Expression Analysis under Abiotic Stresses. Agronomy 14, 535. doi: 10.3390/AGRONOMY14030535
Li, F., Liu, S., Xu, H., Xu, Q. (2018). A novel FC17/CESA4 mutation causes increased biomass saccharification and lodging resistance by remodeling cell wall in rice. Biotechnol. Biofuels 11, 1–13. doi: 10.1186/s13068-018-1298-2
Li, F., Xie, G., Huang, J., Zhang, R., Li, Y., Zhang, M., et al. (2017). OsCESA9 conserved-site mutation leads to largely enhanced plant lodging resistance and biomass enzymatic saccharification by reducing cellulose DP and crystallinity in rice. Plant Biotechnol. 15, 1093–1104. doi: 10.1111/pbi.12700
Liang, G. L., Liu, W. H., Qin, Y., Wei, X. X., Liu, K. Q., Yang, J. (2019). Study of correlation between biomass composition, stem traits and lodging in different oats resources. Acta Agrestia Sin. 27, 1339–1346. doi: 10.11733/j.issn.1007-0435.2019.05.028
Liu, J. (2005). The reserach on lodging extra-intro inducing factors and stem dynamics character in spring wheat. (Haerbin: Northeast Agricultural University). doi: 10.7666/d.y785564
Liu, X., Chen, H., Li, S., Lecourieux, D., Duan, W., Fan, P., et al. (2023). Natural variations of HSFA2 enhance thermotolerance in grapevine. Hortic. Res. 10, uhac250. doi: 10.1093/hr/uhac250
Liu, L., Liu, S., Lu, H., Tian, Z., Zhao, H., Wei, D., et al. (2022). Integration of transcriptome and metabolome analyses reveals key lodging-resistance-related genes and metabolic pathways in maize. Front. Genet. 13. doi: 10.3389/fgene.2022.1001195
Liu, X. T., Wang, J., Hou, X. F., Li, Q., Jia, D. H., Gu, Y. G. (2019). Progress of functions of lignin and relevant genes in plant lodging resistance. Mol. Plant Breed. 17, 655–662. doi: 10.13271/j.mpb.017.000655
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21. doi: 10.1186/s13059-014-0550-8
Lv, S., Lin, Z., Shen, J., Luo, L., Xu, Q., Li, L., et al. (2024). OsTCP19 coordinates inhibition of lignin biosynthesis and promotion of cellulose biosynthesis to modify lodging resistance in rice. J. Exp. Bot. 75, 123–136. doi: 10.1093/jxb/erad367
Maleki, S. S., Mohammadi, K., Ji, K. S. (2016). Characterization of cellulose synthesis in plant cells. Sci. World J. 2016, 8641373. doi: 10.1155/2016/8641373
Meester, B. D., Barbara, M. C., Vries, L. D., Pollier, J., Goeminne, G., Doorsselaere, J. V. (2020). Tailoring poplar lignin without yield penalty by combining a null and haploinsufficient cinnamoyl-coa reductase2 allele. Nat. Commun. 11, 5020. doi: 10.1038/s41467-020-18822-w
Morant, M., Schoch, G. A., Ullmann, P., Ertunç, T., Little, D., Olsen, C. E., et al. (2007). Catalytic activity, duplication and evolution of the CYP98 cytochrome P450 family in wheat. Plant Mol. Biol. 63, 1–19. doi: 10.1007/s11103-006-9028-8
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Nan, M. (2021). Study expression of genes in stem lignin synthesis and intrinsic physiological mechanism of lodging resistancein oat (Avena sativa L.) (Lanzhou: Gansu Agricultural University). doi: 10.27025/d.cnki.ggsnu.2021.000011
Ohashi-Ito, K., Fukuda, O. H. (2010). Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 22, 3461–3473. doi: 10.1105/tpc.110.075036
Ookawa, T., Ishihara, K. (1993). Varietal difference of the cell wall components affecting the bending stress of the culm in relation to the lodging resistance in paddy rice. Jpn. J. Crop Sci. 62, 378–384. doi: 10.1626/jcs.62.378
Pérez-Rodríguez, P., Riano-Pachon, D. M., Corrêa, L. G. G., Rensing, S. A., Kersten, B., Mueller-Roeber, B. (2010). PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Res. 38, D822–D827. doi: 10.1093/nar/gkp805
Qi, W., Sun, F., Wang, Q., Chen, M., Huang, Y., Feng, Y. Q., et al. (2011). Rice ethylene-response AP2/ERF factor OsEATB restricts internode elongation by down-regulating a gibberellin biosynthetic gene. Plant Physiol. 157, 216–228. doi: 10.1104/pp.111.179945
Qin, T. Y., Sun, C., Bi, Z. Z. (2020). Identification of drought-related co-expression modules and hub genes in potato roots based on WGCNA. Acta Agron. Sin. 46, 1033–1051. doi: 10.3724/sp.j.1006.2020.94130
Qiong, L. I., Yali, H. E., Mingyue, T. U., Junhui, Y. A. N., Liangliang, Y. U., Weiwei, Q. I., et al. (2019). Transcriptome sequencing of two kentucky bluegrass (Poa pratensis L.) genotypes in response to heat stress. Not. Bot. Horti Agrobot. Cluj-Napoca. 47, 328–338. doi: 10.15835/nbha47111365
Shah, A. N., Tanveer, M., Rehman, A. U., Anjum, S. A., Iqbal, J., Ahmad, R. (2017). Lodging stress in cereal-effects and management: an overview. Environ. Sci. pollut. Res. 24, 5222–5237. doi: 10.1007/s11356-016-8237-1
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Silveira, D. C., Basso, S. M. S., Ebone, L. A., Caverzan, A., MaChado, J. M., Schaeffer, A. H., et al. (2022). Morphological traits of stem to indirect selection of resistance to lodging in Avena sativa L. J. Crop Sci. Biotechnol. 25, 39–50. doi: 10.1007/S12892-021-00111-7
Singh, R., De, S., Belkheir, A. (2013). Avena sativa (Oat), a potential neutraceutical and therapeutic agent: an overview. Crit. Rev. Food Sci. Nutr. 53, 126–144. doi: 10.1080/10408398.2010.526725
Wang, H., Avci, U., Nakashima, J., Hahn, M. G., Chen, F., Dixon, R. A. (2010). Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants. Proc. Natl. Acad. Sci. 107, 22338–22343. doi: 10.1073/pnas.1016436107
Wang, T., Jin, Y., Deng, L., Li, F., Wang, Z., Zhu, Y., et al. (2024). The transcription factor MYB110 regulates plant height, lodging resistance, and grain yield in rice. Plant Cell 36, 298–323. doi: 10.1093/plcell/koad268
Wang, B., Wang, J., Yang, T., Wang, J., Dai, Q., Zhang, F., et al. (2023). The transcriptional regulatory network of hormones and genes under salt stress in tomato plants (Solanum lycopersicum L.). Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1115593
Wang, X., Zhang, R., Shi, Z., Zhang, Y., Sun, X., Ji, Y., et al. (2019). Multi-omics analysis of the development and fracture resistance for maize internode. Sci. Rep. 9, 1–11. doi: 10.1038/s41598-019-44690-6
Wei, X. X., Lan, H. Y. (2022). Advances in the regulation of plant MYB transcription factors in secondary metabolism and stress response. Biotechnol. Bull. 38, 12–23. doi: 10.13560/j.cnki.biotech.bull.1985.2021-1350
Wen, W., Gu, S., Xiao, B., Wang, C., Wang, J., Ma, L., et al. (2019). In situ evaluation of stalk lodging resistance for different maize (Zea mays L.) cultivars using a mobile wind machine. Plant Methods 15, 1–16. doi: 10.1186/s13007-019-0481-1
Wu, Q., Pan, Y. B., Su, Y., Zou, W., Xu, F., Sun, T., et al. (2022). WGCNA identifies a comprehensive and dynamic gene co-expression network that associates with smut resistance in sugarcane. Int. J. Mol. Sci. 23, 10770. doi: 10.3390/ijms231810770
Xiao, W., Yang, Y., Yu, J. (2018). ZmNST3 and ZmNST4 are master switches for secondary wall deposition in maize (Zea mays L.). Plant Sci. 266, 83–94. doi: 10.1016/j.plantsci.2017.03.012
Yan, C., Yan, S., Zeng, X., Zhang, Z., Gu, M. (2007). Fine mapping and isolation of Bc7 (t), allelic to OsCesA4. J. Genet. Genomics 34, 1019–1027. doi: 10.1016/S1673-8527(07)60115-5
Yan, H., Yu, K., Xu, Y., Zhou, P., Zhao, J., Li, Y., et al. (2021). Position validation of the dwarfing gene Dw6 in oat (Avena sativa L.) and its correlated effects on agronomic traits. Front. Plant Sci. 12. doi: 10.3389/FPLS.2021.668847
Zhang, B., Horvath, S. (2005). A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 4, 17. doi: 10.2202/1544-6115.1128
Zhang, R., Jia, Z., Ma, X., Ma, H., Zhao, Y. (2020). Characterising the morphological characters and carbohydrate metabolism of oat culms and their association with lodging resistance. Plant Biol. 22, 267–276. doi: 10.1111/plb.13058
Zhang, F. Z., Jin, Z. X., Ma, G. H., Wan, Y. Z., Liu, H. Y., Xu, M. L. (2010). Correlation analysis between lodging resistance and morphological characters of physical and chemical components in rice’s culms. Crops 4, 15–19. doi: 10.16035/j.issn.1001-7283.2010.04.012
Zhao, D., Han, C., Tao, J., Wang, J., Hao, Z., Geng, Q., et al. (2012). Effects of inflorescence stem structure and cell wall components on the mechanical strength of inflorescence stem in herbaceous peony. Int. J. Mol. Sci. 13, 4993–5009. doi: 10.3390/ijms13044993
Zheng, Y., Jiao, C., Sun, H., Rosli, H. G., Pombo, M. A., Zhang, P., et al. (2016). iTAK: a program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol. Plant 9, 1667–1670. doi: 10.1016/j.molp.2016.09.014
Zhou, R., Jackson, L., Shadle, G., Nakashima, J., Temple, S., Chen, F., et al. (2010). Distinct cinnamoyl CoA reductases involved in parallel routes to lignin in Medicago truncatula. Proc. Natl. Acad. Sci. 107, 17803–17808. doi: 10.1073/pnas.1012900107
Keywords: lodging-resistant, transcriptome sequencing, differentially expressed genes, weighted gene co-expression network analysis (WGCNA), Avena sativa
Citation: Yu J, Zhang R, Ma X, Jia Z, Li X, Li Y and Liu H (2025) Physiological mechanism of lodging resistance of oat stalk and analysis of transcriptome differences. Front. Plant Sci. 16:1532216. doi: 10.3389/fpls.2025.1532216
Received: 21 November 2024; Accepted: 14 March 2025;
Published: 02 April 2025.
Edited by:
Fatemeh Maghuly, University of Natural Resources and Life Sciences Vienna, AustriaReviewed by:
Iman Samiei Mosleh, University of Natural Resources and Life Sciences Vienna, AustriaCopyright © 2025 Yu, Zhang, Ma, Jia, Li, Li and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoxia Li, bGl4aWFveGlhQGNhZi5hYy5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.