
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 27 March 2025
Sec. Plant Bioinformatics
Volume 16 - 2025 | https://doi.org/10.3389/fpls.2025.1531642
 Meini Song1,2
Meini Song1,2 Pengcheng Li1,2
Pengcheng Li1,2 Lirong Yao1,2
Lirong Yao1,2 Chengdao Li3
Chengdao Li3 Erjing Si1,2
Erjing Si1,2 Baochun Li1,4
Baochun Li1,4 Yaxiong Meng1,2
Yaxiong Meng1,2 Xiaole Ma1,2
Xiaole Ma1,2 Ke Yang1,2
Ke Yang1,2 Hong Zhang1,2
Hong Zhang1,2 Xunwu Shang2
Xunwu Shang2 Huajun Wang1,2*
Huajun Wang1,2* Juncheng Wang1,2*
Juncheng Wang1,2*Introduction: Inorganic phosphorus (Pi) is an indispensable nutrient for plant growth, with phosphate transporter proteins (PHTs) having key roles in Pi uptake, transport, and signal transduction in plants. However, a systematic and comprehensive genomic analysis of the wheat PHT family (covering PHT1-5 and PHO1) is lacking.
Methods: In view of this, we successfully identified 180 Triticum aestivum PHT (TaPHT) members in 6 PHT families using bioinformatics, and performed in-depth phylogenetic analyses between these protein sequences and PHT family proteins from Arabidopsis thaliana and an important rice crop.
Results: We observed that the TaPHT family could be subdivided into 6 phylogenetic clusters, specifically including 46 TaPHT1, 3 TaPHT2, 65 TaPHT3, 22 TaPHT4, 14 TaPHT5, and 30 TaPHO1 members. We also comprehensively profiled the phylogenetic relationships, structural features, conserved motifs, chromosomal localization, cis-acting elements and subcellular localization of these members. These features showed a high degree of conservation within each subfamily. In particular, in the 2000 bp sequence upstream of the TaPHT genes, we identified multiple cis-acting elements closely related to Pi responses, such as P1BS (PHR1 binding site), MBS (MYB binding site), and a W-box (WRKY binding site), which suggested that TaPHT genes were possibly involved in Pi signaling pathways. We screened 24 TaPHT genes by qRT-PCR (real-time quantitative PCR) and investigated their expression in roots and shoots of two wheat cultivars (Pi efficient material SW2 and Pi inefficient material SW14) under low Pi stress conditions. All genes showed up-regulated expression patterns associated with Pi nutritional status, with relative gene expression generally higher in the SW2 cultivar when compared to SW14. Particularly noteworthy was that TaPHT1;36 in the SW2 cultivar showed high and relative stable expression in wheat roots. Combining our bioinformatics and relative gene expression analyses, we preliminarily screened TaPHT1;36 as a candidate gene for low Pi tolerance and further confirmed its subcellular localization.
Discussion: Our work not only identified important TaPHT family roles in coping with low Pi stress, but it also provides a functional research basis and candidate gene resource for solving Pi deficiency-related problems.
Food security represents a significant global challenge, one that is fundamental to human survival and development (Cordell et al., 2009). As the global population continues to grow, the inelastic demand for food is so too increasing. Inorganic phosphorus (Pi) is a major nutrient required for crop growth and development. It is vital for living organisms, serving as a nucleic acid and cell membrane components and participating in several key biological processes (Muchhal et al., 1996; Westheimer, 1987). However, the majority of phosphates are rapidly immobilized following phosphate fertilizer application to soils, making them unavailable for absorption and use by crops (Davies et al., 2002). Consequently, specialized transporter proteins are required to facilitate Pi transport from the soil to plant roots for uptake (Wang et al., 2019b).
Phosphate transporter proteins (PHTs) are responsible for Pi uptake and transport in plants. They are typically divided into 6 different subclasses based on sequence similarity, structure, and function, and are referred to as PHT1-PHT5 and PHO1 (Ahmad et al., 2021; Rui et al., 2023). Since the earliest identification of the high-affinity PHT gene, PHO84, in yeast, there has been a significant increase in PHT gene numbers identified in Arabidopsis thaliana (Muchhal et al., 1996), Oryza sativa (Paszkowski et al., 2002), Malus domestica (Sun et al., 2017), Populus simonii (Zhang et al., 2016), Medicago truncatula (Harrison et al., 2002), Solanum tuberosum (Liu et al., 2017), Lupinus albus (Aslam et al., 2022), Spirodela polyrhiza (Zhao et al., 2021b), Brassica napus (Yang et al., 2020), Capsicum annuum (Ahmad et al., 2021) and Saccharum (Yang et al., 2020), amongst others.
PHT1 is one of the most studied subclasses in the PHT family, and is notably characterized by 12 transmembrane structural domains (Ullrich-Eberius et al., 1981). PHT2 is localized in the inner chloroplast membrane and is mainly expressed in green tissues and roots, with its main function being phosphate transport within the leaf (Daram et al., 1999; Harrison et al., 2002). As the first identified mitochondrial Pi transporter gene, PHT3 is highly conserved among the family and is essential for maintaining Pi homeostasis in plants under adverse conditions (Rausch and Bucher, 2002; Takabatake et al., 1999). PHT4 is primarily localized to the plasma membrane, periplasm, and Golgi apparatus, and plays a key role in Pi transport (Guo et al., 2008; Wan et al., 2020). PHT5, in addition to its role in these organelles, is involved in several biological processes. It is located in vesicle membranes and has important roles regulating Pi homeostasis in plants, which regulates Pi storage and acclimatization in vesicles (Liu et al., 2016). PHO1 is expressed in specific patterns in plants, predominantly in stellate cells in roots and the lower portion of hypocotyls. At these locations, PHO1 facilitates effective Pi transport from root epidermal cells to the xylem and back to the roots via the phloem, thereby ensuring effective Pi recycling and use in plants (Poirier and Bucher, 2002; Ticconi et al., 2001).
Wheat (Triticum aestivum L.) is a major global food crop and essential for food production and security (Godfray et al., 2010). To enhance wheat yields, substantial Pi fertilizers are used to facilitate maturation and augment agronomic quality (Shukla et al., 2016). Pi is readily immobilized and has low mobility in soil, which impedes efficient Pi absorption in wheat. This causes Pi deficiency which reduces wheat yields and quality. Phosphate transporters play a central role in coping with low phosphorus stress and improving phosphorus use efficiency in wheat, and in-depth studies of their functions provide an important theoretical basis for solving the problems of low phosphorus stress in agricultural production. By regulating the expression and function of phosphate transporters, we can not only improve the yield and quality of wheat in low-phosphorus soils, but also reduce the amount of phosphorus fertilizer applied, reduce the negative impact on the environment, and achieve the sustainable development of agriculture. Therefore, it is vital to identify and functionally analyze genes related to low Pi stress tolerance in wheat, to enhance effective Pi use, and achieve high and stable wheat yields. In this study, key genes related to efficient Pi use were explored in cultivated wheat to provide genetic resources and technical support for cultivating new low Pi-tolerant wheat varieties.
Arabidopsis and rice PHT family protein sequences were retrieved from TAIR and RiceData genome databases, respectively. Wheat whole genome data, protein sequences, and annotation files were downloaded from the Ensemble Plant database. BLAST comparisons were conducted using TBTools software to retrieve homologous PHT genes from the whole wheat genome based on Arabidopsis PHT protein sequences and identify wheat PHT family members. HMMER files for the conserved structural domain in PHT genes were downloaded from the PFAM database. Query sequences, PHT1 (PF00083), PHT2 (PF01384), PHT3 (PF00153), PHT4 (PF07690), PHT5 (PF07690 and PF03105) and PHO1 (PF03124 and PF03105), were downloaded from PFAM and used to search for PHT family members in the Wheat Protein Data Bank (E value ≤ 1×10-10). To guarantee data precision, the sequences underwent further verification in NCBI-CDD (National Center for Biotechnology Information-Conserved Domains Database) and Inter Pro databases, which confirmed defining structural domain characteristics in each subfamily. Following this, sequences were classified using established criteria. Based on this approach, genes with identical sequences and those lacking PHT structural domains were excluded. Arabidopsis and rice PHT protein sequences were obtained from the NCBI (National Center for Biotechnology Information) database and aligned with wheat sequences using multiple sequence alignments (MEGA 10 software). Alignments were then used to construct a phylogenetic tree using the Neighbor-Joining (NJ) method with 1000 bootstrap repeats to represent evolutionary relationships in the PHT family. The resulting “Newick” tree file was then exported and displayed on the Chiplot website (Supplementary Table S1 for URLs).
TaPHT physical and chemical properties were predicted using ProtParam, while hydrophilicity was assessed using ProtScale. Phosphorylation sites were identified using Server, while signal peptide and transmembrane regions were predicted using SignalP-5.0 and TMHMM, respectively. Additionally, secondary and tertiary protein structures and structural domains were predicted using SOPMA, Swiss-Model, and SMART.
SMART was used to sequentially predict secondary and tertiary protein structures and also structural domains, while subcellular localization was determined using WoLF PSORT. Gene density profiles were calculated using TBtools software, and chromosomal gene positions were visualized using wheat GFF3 files. TaPHT conserved motifs were analyzed using the MEME website. Genome sequences and GFF3 annotation files for Arabidopsis and barley (Hordeum vulgare. L) were downloaded from the Ensembl Plant website, and covariance between TaPHT genes was analyzed using One Step MCScanX. Co-linearity within TaPHT gene species was analyzed and beautified using the Advanced Circos function. A 2000 bp sequence upstream of the TaPHT start codon was identified as a promoter (TBtools) and submitted to the Plant-CARE website to predict cis-acting elements (type and number) in the promoter sequence (Supplementary Table S1 for all URLs).
The low phosphorus tolerant spring wheat material SW2 and phosphorus sensitive spring wheat material SW14 were provided by the State Key Laboratory of Aridland Crop Science/Gansu Key Lab of Crop Improvement and Germplasm Enhancement, and were derived from the results of previous research in this laboratory (Li et al., 2023). Seed surfaces were disinfected in 70%–75% alcohol for 30-60 s, quickly removed, and seeds rinsed 3–5 times in distilled water for 30 s each time. Then, seeds were transferred to Petri dishes for 1 h, after which 20 seeds were sown in each dish with the ventral groove facing downward. After 4 d, seedlings showing good growth and uniform development were selected and transplanted into a 900 mL hydroponic incubator, further incubated for 2 d, and then transferred to an artificial climate chamber with 60% relative humidity. White fluorescent lamps provided light conditions (irradiance = 300 µmol/m²/s) and Pi treatments were conducted over a day-night cycle: 16 h of light (25°C) and 8 h of darkness (18°C). The improved Hoagland nutrient solution was used in the wheat hydroponics test and was replaced every 3 days. The nutrient solution contains a large number of elements: 4 mM Ca(NO3)2·4H2O, 5 mM KNO3, 1 mM NH4NO3, 2 mM MgSO4·7H2O; and trace elements: 200 μM Na2-EDTA, 200 μM FeSO4·7H2O, 10 μM H3BO3, 13μM MnSO4·H2O, 0.01 μM CuSO4·5H2O, 3 μM ZnSO4·7H2O, 0.02 μM (NH4)6Mo7O24·4H2O. Normal phosphorus (NP) treatment: 500 mM KH2PO4; and low phosphorus (LP) treatments of 6.25 mM KH2PO4 and 247 mM K2SO4. Samples were collected at different time points after transplantation (0, 0.25, 0.5, 1, 3, 7, 14, and 21 d) and excess water of wheat roots were removed by gently pressing with filter paper. Then, roots and shoots samples were separately placed into 5 ml freezing tubes and quickly frozen in liquid nitrogen. Some samples were used for RNA extraction and the remainder were stored at -80°C.
To explore the positive correlation between TaPHT expression and phosphorus uptake under low and normal phosphorus conditions, total P concentration in plant samples was measured with 3 biological replicates for each treatment. The fresh roots and shoots were dried in an oven at 105 °C. In order to facilitate the determination of total P concentration by molybdate blue colorimetry, the dried samples were crushed according to the method designed by Li (Li et al., 2023) and digested with concentrated H2SO4 and H2O2.To investigate TaPHT genes expression in response to phosphate starvation, relative gene expression levels in SW2 and SW14 cultivars were compared using qRT-PCR under normal Pi (NP) and low Pi (LP) conditions. Total RNA was extracted using a Total RNA Extraction Kit for Polysaccharide-Polyphenol Plants (TIANGEN, Beijing, DP441). RNA quantity and concentration levels were measured using the UVITEC Multi-color Fluorescence/Chemiluminescence Gel Imaging Analysis System (UVITEC, UK) and the Pultton P100/P100+ Ultra-Micro Spectrophotometer. To synthesize cDNA, a qPCR RT Master Mix with gDNA Remover (TOYOBO, Japan) was used following manufacturer’s instructions. All cDNA primers are shown (Supplementary Table S2), with TaActin an internal reference gene. SYBR-Green Prime Script RT-PCR kits (Takara Biotechnology, Japan) were used on the QuantStudio™ 5 System Real-Time System for qRT-PCR analysis. The reaction system is shown (Supplementary Table S3). Comparative Cycle (Ct) threshold (ΔΔCt) analysis was performed in 0.2 mL 96-well plates. Thermal cycling parameters were as follows; Hold stage with initial denaturation at 95°C for 30 s, then PCR Stage with denaturation at 95°C for 5 s, annealing at 60°C for 30 s, and cycling 40 times. Melt Curve Stage with denaturation at 95°C for 15 s, annealing at 60°C for 60 s, and separation at 95°C for 15 s. Three biological replicates were performed per treatment, and gene expression levels were calculated as average signal intensity values from replicates using the Ct method. Data were represented as the mean ± standard deviation from replicates, with error bars representing the standard error. Differences in TaPHT expression after low-Pi treatment were assessed using least significant difference (LSD) tests (P < 0.05) using the GenStat 12th Edition.
Based on the TaPHT1;36 gene sequence in the wheat reference genome, specific primers were designed to amplify the full-length coding region (CDS) sequence using the cDNA of the phosphorus-efficient SW2 root system of wheat as a template. The vector pC1300-GFP was selected to construct the 35S::TaPHT1;36-GFP fusion protein expression plasmid, and the PEG-mediated method was used to transform rice protoplasts, and the subcellular localization signals were observed and photographed using a laser confocal microscope (Leica STELLARIS 8).
To investigate the evolutionary relationships among TaPHT gene families, we performed multiple sequence alignments using protein sequences from 32 Arabidopsis, 33 rice, and 180 wheat. Subsequently, a phylogenetic tree was constructed using the NJ method. As shown (Figure 1), Arabidopsis, rice, and wheat PHT gene families were grouped into 6 branches, with 46 TaPHT1 genes clustered in group F and 3 TaPHT2, 14 TaPHT5, and 30 TaPHO1 genes clustered in groups A and B. TaPHT2 was in a separate branch, and TaPHT5 and TaPHO1 were clustered in groups A and B due to the common PFAM number (PF03105); 22 TaPHT4 genes were clustered in group C; and 65 TaPHT3 genes were clustered in group D. TaPHT family genes were named according to Arabidopsis and rice, which were categorized for each subfamily and their location on the wheat chromosome (Supplementary Table S5).
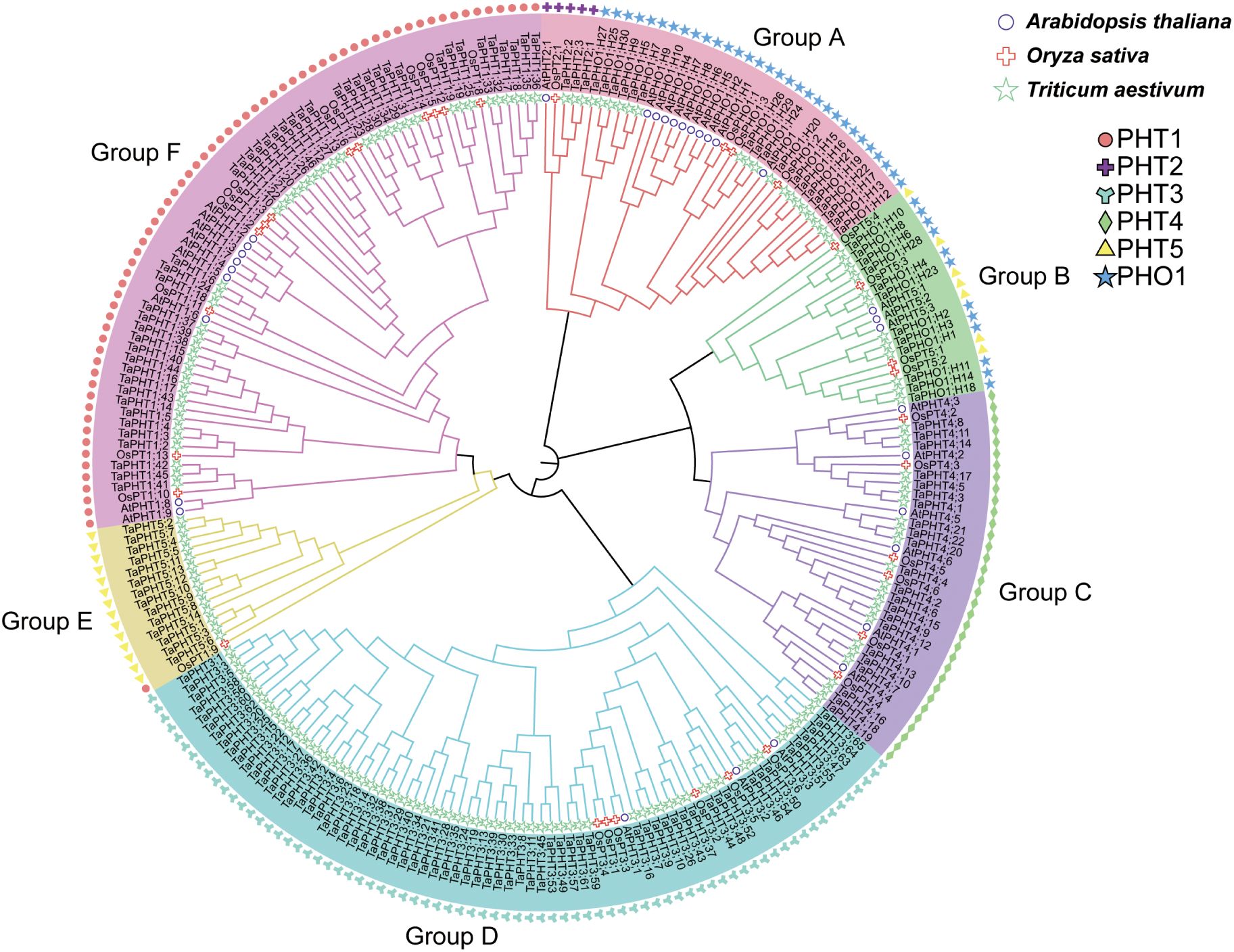
Figure 1. Phylogenetic analysis of PHT proteins in Arabidopsis thaliana, Oryza sativa, and Triticum aestivum.
Physicochemical TaPHT protein properties were analyzed using ProtParam (Supplementary Table S4). The following physiochemical ranges were identified; the encoded amino acid (aa) range was 276 (TaPHT3;37) to 855 (TaPHO1;H29); the molecular weight range was 29.9 (TaPHT3;37) to 97.1 (TaPHO1;H29) kDa; the theoretical pI range was 5.41 (TaPHT4;21) to 10.45 (TaPHT4;14); the instability index range was 21.52 (TaPHT3;4) to 52.97 (TaPHT4;22), the aliphatic index range was 76.75 (TaPHT3;40) to 118.65 (TaPHT5;5); and the GRAVY (Grand average of hydropathicity) range was -0.253 (TaPHT3;38) to 0.729 (TaPHT5;14). Subcellular localization predictions showed that 99/180 of TaPHT were localized to cell membranes.
In genes, exon–intron structures are important evolutionary features that provide clues to gene family functions. To investigate TaPHT structural diversity in wheat, we evaluated conserved exon–intron organization. Using TBtools, 180 TaPHT coding sequences (CDS) were compared with corresponding genomic sequences to generate a TaPHT structure visualization map (Figure 2). All TaPHT genes had introns and exons. Genes with similar exon–intron structures and closer phylogenetic relationships showed more similar conserved structural domain organization. Exon and intron numbers were clearly partitioned according to subfamilies, with data showing different exon and intron numbers according to subfamily. The TaPHT1 subfamily had 1-5 exons and 0-4 introns; the TaPHT2 subfamily had 3 exons and 2 introns; the TaPHT3 subfamily had 1–11 exons and 0-5 introns; the TaPHT4 subfamily had 1-11 exons and 0-5 introns; and the TaPHT5 subfamily had 1-11 exons and 0–10 introns. These findings suggested a high degree of variation among TaPHT genes, with comparable intron/exon distribution patterns within the same subfamily.
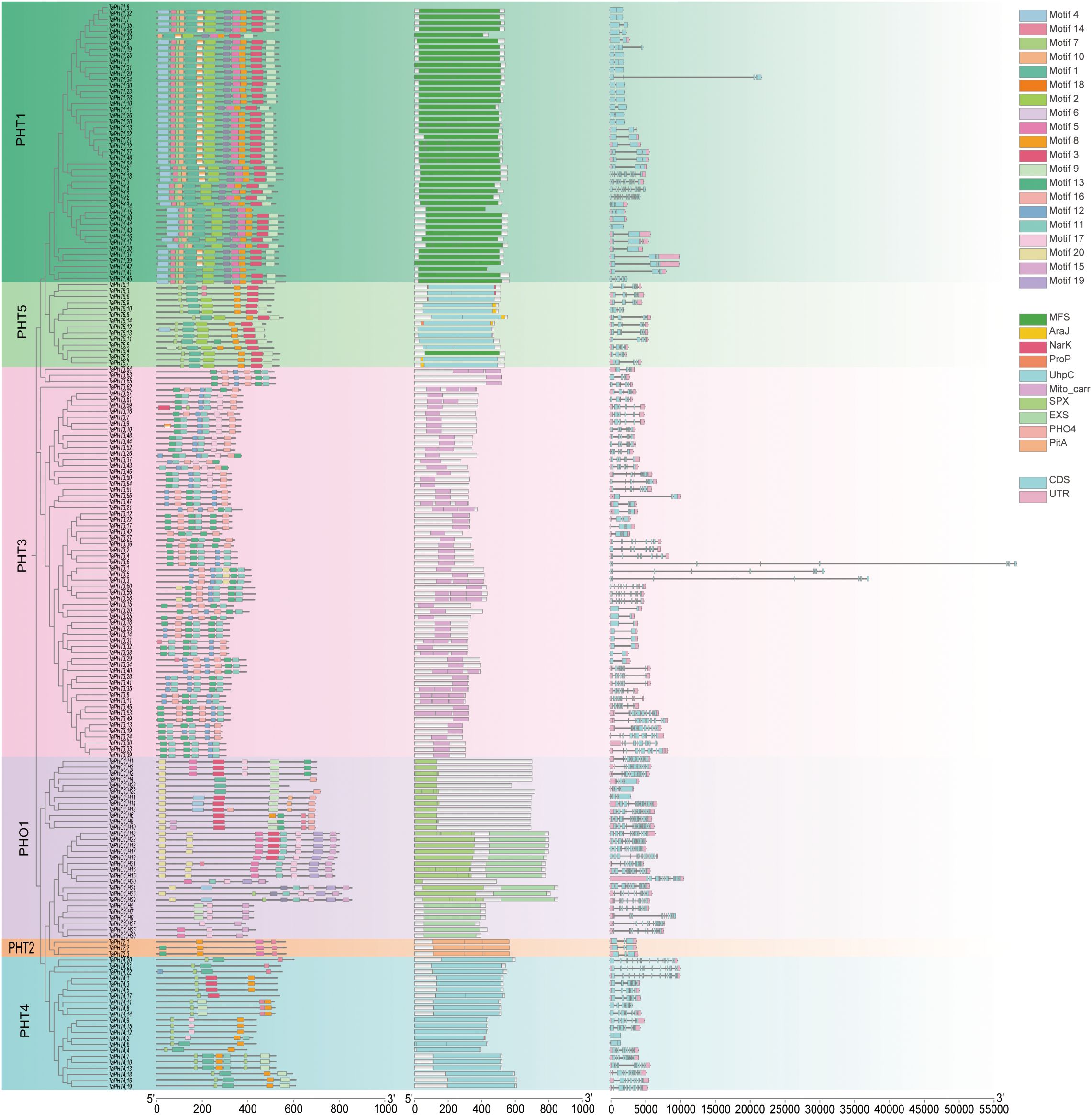
Figure 2. TaPHT structure analyses encompassing a phylogenetic analysis for each subfamily, the identification of conserved domains, and exon and intron distribution/counts for each TaPHT family member. Analyses also included an examination of conserved motifs in TaPHT. The scale (bottom of the graph) represents protein and nucleotide sequence lengths for each TaPHT member.
To structurally characterize TaPHT-conserved motif distribution, 20 conserved motifs were identified using protein sequence comparisons (MEME software) and labeled as 1-20, with MEME-8 having the highest E-value (Table 1; Supplementary File S1). Most TaPHT1 subfamily members exhibited motifs 1-0, 14, and 18; the TaPHT2 subfamily exhibited motifs 5, 10, 11, and 14; the TaPHT3 subfamily included motifs 3, 11, 12, 16, 17, and 20; the TaPHT4 subfamily included motifs 3, 7, 8, 9, 11, 14, and 17; the TaPHT5 subfamily included motifs 1, 2, 3, 7, 8, 9, 13, and 14; and the TaPHO1 subfamily included motifs 4, 9, 14, 15, 16, 17, 19, and 20.
Cis-acting elements are crucial for gene transcription and expression as they regulate plant adaptation to different environments via different functions (Marand et al., 2023). To further identify TaPHT characteristics, 17 species and 7709 cis-acting elements in the PHT family 2000 bp promoter region were identified using PlantCARE, and element distribution related to growth and development, hormone regulation, and low-Pi regulation were analyzed. TaPHT promoter analyses showed that the majority of genes possessed multiple cis-acting elements which were related to stress, hormone, and light responses (Supplementary Table S5; Figure 3). Most family genes contained PIBS, W-box, and MBS elements, suggesting that these genes were transcriptionally induced by PHR1, WRKY, and MYB transcription factors under low Pi conditions to generate more Pi. These data suggested that TaPHT expression potentially regulated wheat growth and development, cytological processes, and adverse stress responses.
Based on the physical locations in the GFF3 file, the localization results showed that the 180 TaPHT genes on wheat chromosomes were distributed on 21 chromosomes of wheat (Figure 4). In the 3 homologous chromosome groups A, B and D of wheat, TaPHT genes were distributed on 21 chromosomes of wheat. The 180 TaPHT genes were not evenly distributed on the wheat chromosome, but were more densely distributed on the lower end of the chromosome. Except for 2 genes that were not located on chromosomes, the most genes were located on chromosomes 4A and 21; the number of genes located on chromosomes 1A, 1B, 1D and 7D is minimal. Among them, four genes were located on chromosomes 1A, 1B, 1D and 7D.
To investigate evolutionary relationships among wheat, Arabidopsis, and barley PHT genes, intra- and inter-species collinearity analyses were performed using their genomes. We observed that the 180 TaPHT genes formed 159 collinearity gene pairs, with a denser distribution on chromosomes 4A–6D (Figure 5; Supplementary Table S6).
Wheat shared 25 covariates with Arabidopsis, mainly on Arabidopsis chromosomes 1–4 and wheat chromosomes 1A, 1B, 1D, 2B, 3A, 3B, 3D, 4A, 4B, 4D, 5A, 5D, 6A, 6B, and 6D, while it shared 142 covariates with barley, which were distributed on wheat chromosomes 1A–7D and barley chromosomes 1H–7H (Figure 6). These observations indicated that TaPHT genes exhibited obvious covariance with Arabidopsis and barley, with significant correlations. Thus, we hypothesized that these genes may have evolved from a same ancestor genome.
In order to study the correlation between TaPHTs expression and phosphorus absorption under low and normal phosphorus conditions, we observed that SW2 and SW14 low phosphorus stress significantly decreased in wheat roots after 14 days of low phosphorus stress (Figure 7). In wheat shoots, there were significant differences on days 3-14, and the root P concentration of SW2 reached its peak on days 14 of low P stress (P < 0.05). The results showed that SW2 was more adaptable to low P stress than SW14 during 14 days of P stress. To understand TaPHT transcriptional level responses to low Pi stress, 180 TaPHT genes were searched against the Wheat Expression Database (Wheat Omics 1.0), combining tissue expression patterns in wheat roots, leaves, stems, spikes, and seeds, and gene expression under low Pi treatments (Figures 8, 9). Among genes, TaPHT3;2, TaPHT3;4, TaPHT3;6, TaPHT3;30, TaPHT3;33, TaPHT3;39, TaPHT3;57, TaPHT3;59, TaPHT3;61, TaPHT5;8, TaPHT5;9, and TaPHT5;10 were expressed in all tested tissues. Under low Pi stress, TaPHT1 and TaPHT3 subfamilies were up-regulated in roots and down-regulated in shoots, whereas TaPHT2 and TaPHT4 subfamilies were up-regulated in shoots and down-regulated in roots, while most of the TaPHT5 subfamily was up-regulated in shoots, and most of the TaPHO1 subfamily was up-regulated in roots. Among the six subfamilies, the highest root expression level was observed for the TaPHT1 subfamily, while the highest expression level in shoots was observed for the TaPHT2 subfamily.
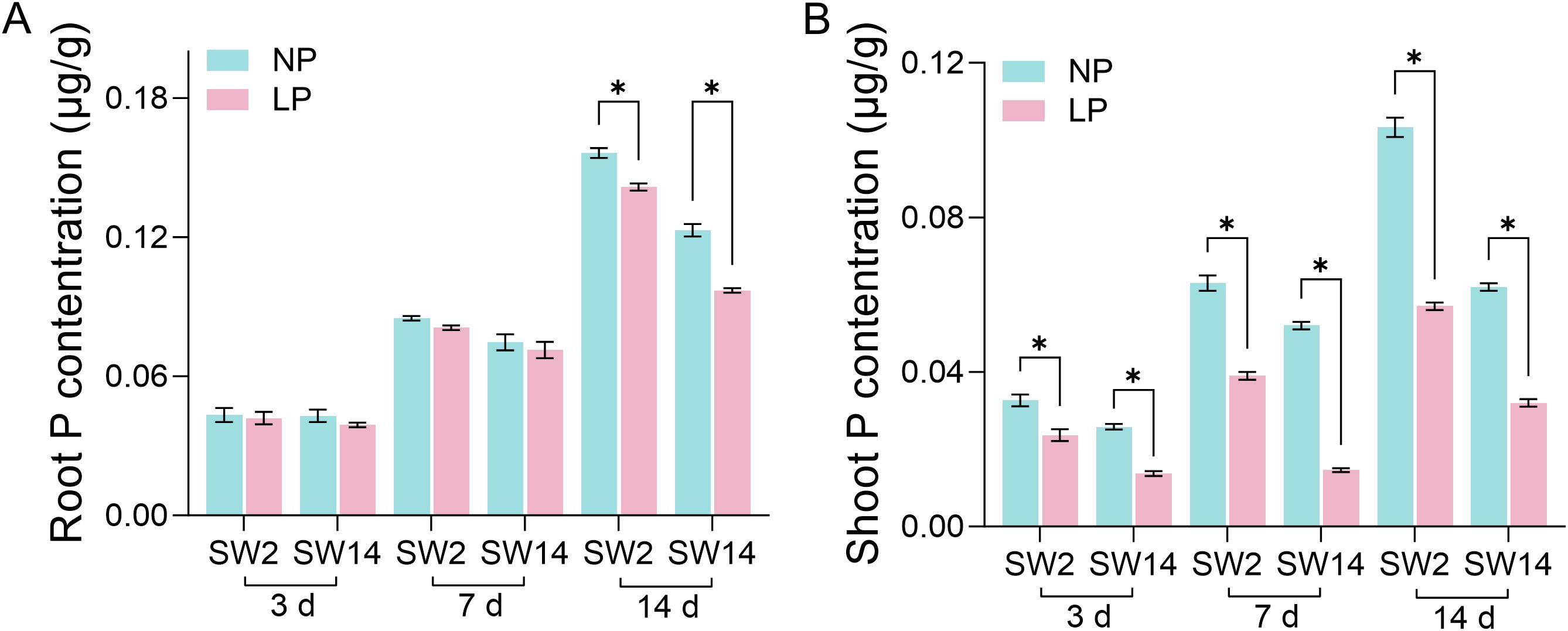
Figure 7. P concentration in root (A) and shoot (B) (μg/g). Student’s method was used to compare significant differences between LP and NP. *(P < 0.05) represents the significance of the difference between two treatments of the same variety at the same time point.
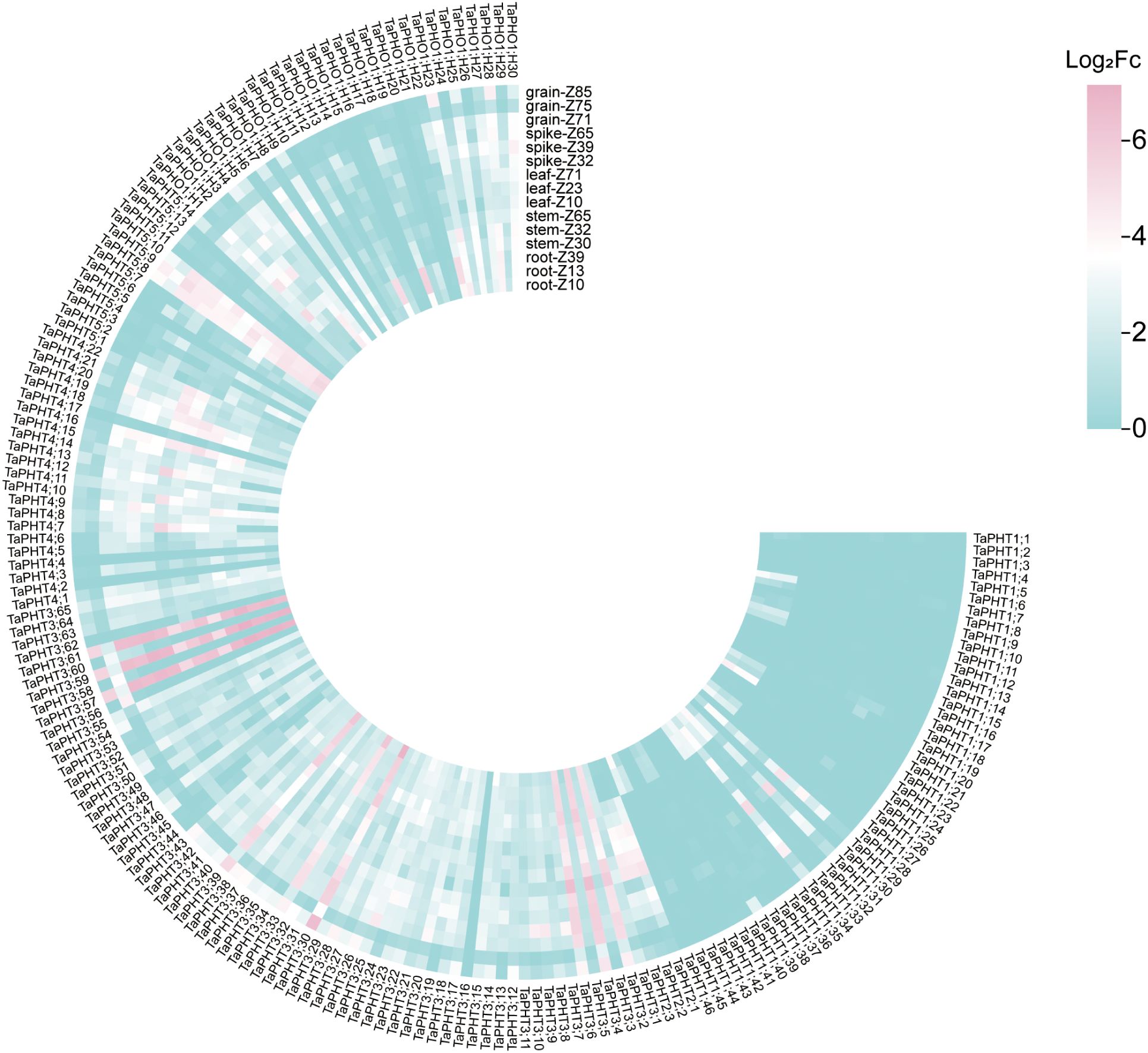
Figure 8. TaPHT expression patterns in different tissues. Grain_z71: seed 2d after anthesis; Grain_ z75: seed 14d after anthesis; Grain_z85: Seeds 20 days after flowering; Spike_z32: internode spike; Spike_z39: spike at visible flag leaf stage; Spike_z65: flowering spike; Leaf_z10: leaves of seedling stage; Leaf_z23: leaves in the three-tillering period; Leaf_z71: leaves 2d after flowering; Stem_z30: stem at heading stage; Stem_z32: stem interstitial; Stem_z65: flowering stem; Root_z10: root at seedling stage; Root_z13: root of trilobate stage; Root_z39: root of flag leaf visible period.
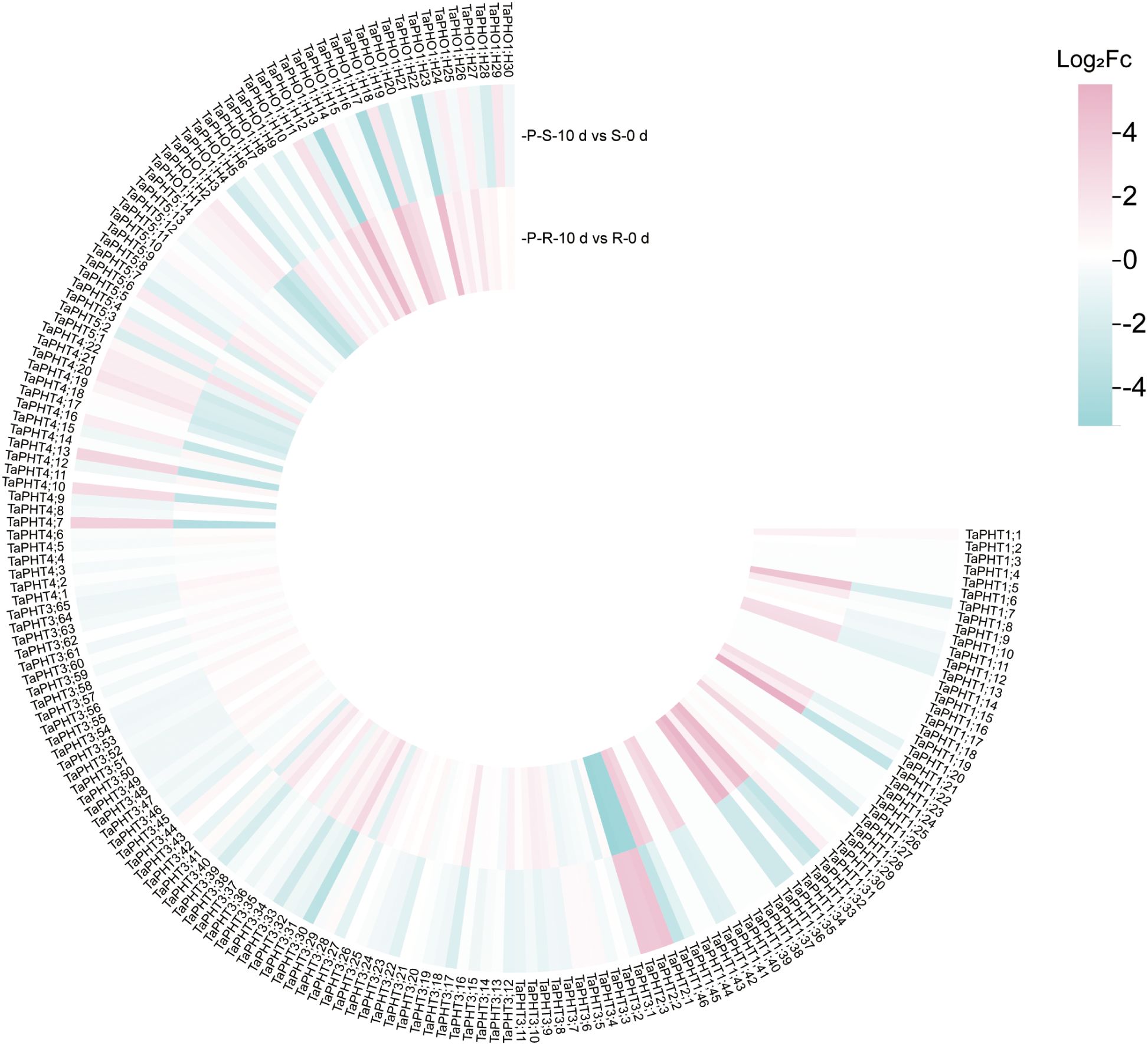
Figure 9. TaPHT expression patterns under low phosphorus (Pi) stress. -P-S-10 d: shoots with 10 d of low Pi stress; S-0 d: shoots with 0 d of low Pi stress; P-R-10 d: roots with 10 d of low Pi stress; R-0 d: roots with 0 d of low Pi stress.
We next selected 3–5 genes in each TaPHT subfamily based on predicted expression results, and examined the expression of 30 TaPHT genes in wheat roots and shoots, under low Pi stress, by qRT-PCR. Six genes were not expressed or abnormally expressed, with the remaining 24 analyzed in roots and shoots.
In the TaPHT1 subfamily, 0-3 d was the initial response period. Wheat root PHT1 family members rapidly responded to low Pi stress with increased relative expression, which kept rising as the stress continued, then slowly decreased from 14 - 21 d. TaPHT1;36 in roots had the highest relative expression of 114.727 at 7 d and then stabilized, suggesting its role in Pi uptake under long - term low Pi stress. The LSD value (Supplementary Figure S2) indicated that PHT1 family members in wheat roots responded rapidly to low Pi stress, with increased relative expression. The distance between the two points in Figure 9 is greater than the distance of the LSD line segment, indicating that there is a significant difference based on LSD (P < 0.05). The specific LSD values are shown in Supplementary Table S7. TaPHT2 subfamily levels in roots were slightly up-regulated in the 0–21 d period of low Pi stress, then gradually adapted to the low Pi environment after peaking at 7 d. In the TaPHT3 subfamily, except for TaPHT3;36 whose relative expression peaked at 2.207 at 7 d, the relative expression of all other genes increased slowly, suggesting that physiological stress caused by low Pi could be alleviated by slightly increased TaPHT3 expression. Under low Pi conditions, wheat optimizes Pi uptake, transport, and partitioning by adjusting the expression of TaPHT4 family members to adapt to the low Pi environment. Among the 5 TaPHT4 subfamily genes, TaPHT4;10 and TaPHT4;13 were highly expressed in wheat roots, suggesting that up-regulated TaPHT4 helped plants increase Pi uptake in low Pi environments. TaPHT5 was slightly up - regulated in roots, with TaPHT5;9 peaking at 3.841 after 7 d and others stabilizing. In the TaPHO1 subfamily, TaPHO1;H11 had higher relative expression in SW14 than SW2, likely due to cultivar differences. With low Pi stress, TaPHO1 expression in roots gradually stabilized and reduced Pi - nutrition dependence. Except for TaPHO1;H11, the SW2 cultivar had higher relative expression in the PHT gene family in wheat roots than SW14. These results suggested that up-regulated TaPHT genes were possibly involved in phosphate uptake when soil phosphate levels were limited, thus alleviating physiological stresses caused by low Pi.
In wheat shoots, relative TaPHT1;36 expression was close to 3, while relative TaPHT1 subfamily expression, except TaPHT1;36, was in the 0.2-1.2 range over the 0-21 d period (Supplementary Figure S3). This, together with relative expression in wheat roots, indicated that TaPHT1 subfamily genes were expressed in wheat roots. TaPHT2 subfamily expression in early low-Pi treatment in shoots had no significant change, but with longer treatment, it was up - regulated, with TaPHT2;1 peaking at 64.985 at 7 d and then stabilizing to adapt to low Pi stress and improve Pi transport to leaves. For the TaPHT3 subfamily, mainly involved in Pi transport and distribution, except for TaPHT3;36, other TaPHT3 genes had low relative expression in shoots. TaPHT4 subfamily expression didn’t change much at first in shoots, with TaPHT4;20 peaking at 7 d and then decreasing. TaPHT5 subfamily expression decreased, and TaPHT5;7 was higher in SW14 than SW2 after 3 d, likely due to factors like Pi transport and leaf physiology. In the TaPHO1 subfamily, TaPHO1;H11 had much higher relative expression. Shoots quickly sensed low Pi and TaPHO1 subfamily expression increased with treatment time, peaking at 7 d, then declined at late stages as wheat adapted and possibly initiated other metabolic pathways. Wheat shoots are not the main direct exposure sites to low P stress, so their gene expression responses are relatively delayed, but overall up-regulated TaPHT family expression in wheat shoots was higher in the Pi-efficient material SW2.
In conclusion, qRT-PCR analysis of 24 low phosphorus responsive TaPHT genes showed that in the low phosphorus tolerant material SW2, the relative expression of the TaPHT1;36 gene in wheat roots reached a peak of 114.727 at 7 d and then reached a more stable state with high and more stable relative expression. Thus, this further confirmed the subcellular location of TaPHT1;36.
To validate the results of online prediction of subcellular localization of gene expression proteins and to investigate the subcellular localization of TaPHT1;36 protein, the TaPHT1;36-GFP fusion protein expression vector was constructed and the empty vector pC1300S-GFP was used as a control. According to the results of rice protoplasts, GFP green fluorescence signals were detected throughout the cells of GFP empty control protoplasts with no obvious organelle localization specificity, and the GFP green fluorescence in the ligated target protein TaPHT1;36 was mainly distributed in the plasma membrane (Figure 10). The results indicate that TaPHT1;36 protein is localized to the plasma membrane, which is consistent with previous studies and online predictions.
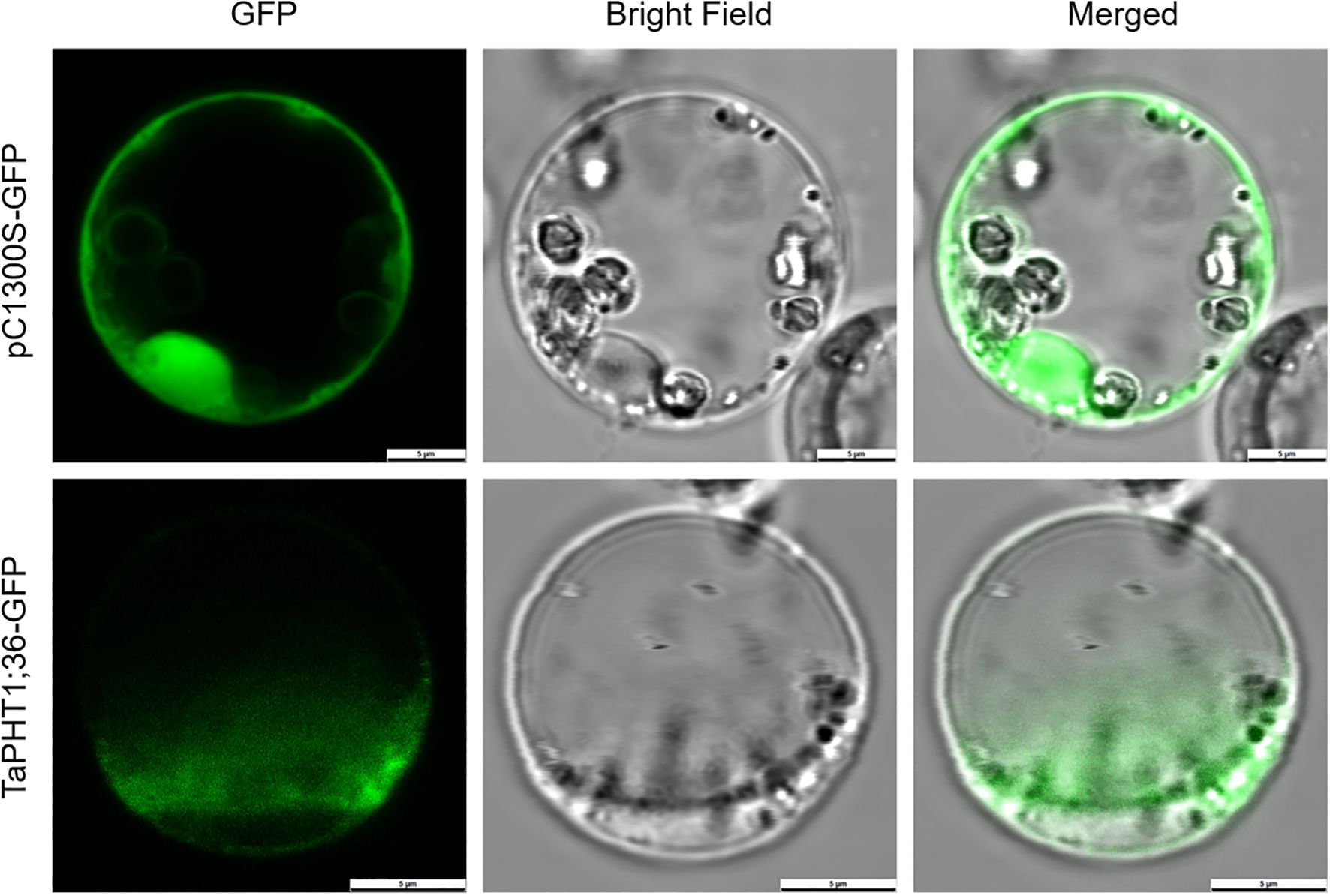
Figure 10. TaPHT1;36 subcellular localization after transient expression in rice protoplasts. Scale 5 μm.
Phosphate ore is a non-renewable resource, and with the growing demand for phosphate fertilizers in global agriculture, phosphate resources are increasingly at risk of depletion (Chowdhury et al., 2017). Pi is a vital macronutrient required for plant growth and development, and is absorbed by roots via PHT proteins, which have key roles supporting overall plant growth and development (Zhang et al., 2016). Reduced Pi availability, increased Pi fertilizer costs, and decreased plant Pi efficiency have highlighted a need to investigate wheat PHT protein families. Cereals are important Pi reservoirs for plant growth and development (Godfray et al., 2010). Wheat is a globally important food crop, occupies a prominent position in global cereal production, and uses a large amount of Pi, which is stored in seeds as PA (Phosphatidic acid) (Wang et al., 2019a). Increasing the expression efficiency of TaPHT under low phosphorus stress is a key factor in promoting their yield and quality. Currently, the sequencing of the full-length wheat genome is providing important genetic resources for further functional genomics research and genetic improvements in crops. Considering the close evolutionary relationship between wheat and its homologs in Arabidopsis and rice, we hypothesized that TaPHT genes could exhibit similar functions. To do this, we successfully identified TaPHT family members and comprehensively analyzed their phylogenetic relationships and expression patterns across various tissues and growth stages in wheat and in different cultivars during various growth stages under low Pi stress. This study not only lays the foundation for investigating the molecular mechanisms of inorganic phosphate transport in response to abiotic stress, but also has important implications for improving wheat yield under low phosphorus stress and addressing global challenges to sustainable agriculture.
In dicotyledonous plants, the number of PHT families varies, e.g., 23 PHT genes occur in sugarcane and 73 occur in S. polyrhiza (Mudge et al., 2002; Zhao et al., 2021b). Previous research involving the genome-wide identification and expression analysis of the PHT1 subfamily in wheat identified 36 TaPHT genes. It was noted that heterozygous hexaploid wheat expressed significantly more PHT1 genes when compared to diploid cereals like barley and rice, and not all PHT1 genes in wheat were isolated (Teng et al., 2017). A previous genome-wide PHT-based study identified only 23 wheat PHT (mainly TaPHT1–TaPHT4) and focused on tissue-specific transcriptional profiling and associations with phosphate partitioning in cereals (Shukla et al., 2016). Thus, detailed studies of the wheat PHT family based on de novo genome sequencing have not been reported. However, recent advances in wheat genome sequencing have enabled extensive bioinformatics analyses of the TaPHT family. In our study, we extended these findings based on 180 TaPHT genes identified in the latest wheat genome, including 46 PHT1, three PHT2, 65 PHT3, 22 PHT4, 14 PHT5, and 30 PHO1 genes. Proteins with high homology in a class/subfamily may have similar activities, so to characterize putative PHT sequences, we predicted conserved PHT protein structural domains using the NCBI -CDD (Liu et al., 2021). Consistent with previous studies, conserved structural domains were found in all proteins which were confirmed as typical PHT proteins (Aslam et al., 2022).
Notably, in contrast to the 46 PHT1 genes identified in wheat, 9 PHT1 genes were previously identified in Arabidopsis, 8 of which were expressed in roots and had key roles in Pi harvesting in low and high Pi environments. Also, 13 genes were identified in rice, where they had redundant roles in root-to-shoot Pi transport (Mudge et al., 2002; Ruili et al., 2020; Wan et al., 2020). Single genes for PHT2 were identified in many plants, including tomato (Rui et al., 2023), potato (Liu et al., 2017), and C. annuum (Ahmad et al., 2021). In contrast, 3 PHT genes (TaPHT2;1, TaPHT2;2, and TaPHT2;3) located on different chromosomes (6A, 6B, and 6D) and encoding 564–567 aa with predicted molecular weights of 58.2-58.8 kDa were detected in wheat, similar to a previous report showing that AtPHT2.1 cDNA encoded 587 aa with a predicted molecular weight of 61 kDa (Daram et al., 1999). So far, at least three AtPHT3 genes have been identified in Arabidopsis, 6 PHT3 genes in rice, and here, we identified 65 PHT3 family genes in wheat, evenly distributed on 21 chromosomes, wheat 1A-6D (Wang et al., 2017, 2020). Additionally, 6 PHT4 family genes were characterized in Arabidopsis, 6 in rice, and 22 in wheat (Ruili et al., 2020; Wang et al., 2017). It is noteworthy that PHT5 family genes have not been identified in most plants, including potato (Liu et al., 2017), sugarcane (Murugan et al., 2022), and poplar (Zhang et al., 2016). Previous studies have reported three PHT5 genes in Arabidopsis and four in rice, and in this study, we identified 14 in wheat (Wang et al., 2017). Additionally, 10 genes homologous to AtPHO1 occur in the Arabidopsis genome and only three PHO1 genes in rice, while 30 PHO1 genes were identified in wheat in this study (Secco et al., 2010; Wang et al., 2004).
Cis-acting elements in promoter regions are important for the regulation of stress-related gene expression and to help plants resist abiotic and biotic stresses (Li et al., 2021). Cis-regulatory elements also have key roles in gene transcription and subsequent expression, while their diverse functional regulatory mechanisms significantly enhance plant adaptation to different environmental conditions (Li et al., 2019; Wang et al., 2021). Previous studies have detailed cis-elements in PHO family members in wheat; however, cis-elements in all PHT family members in wheat remain to be identified (Shukla et al., 2016). TaPHT genes are transcriptionally induced at low Pi levels, probably because they contain P1BS (PHR1 binding site), W-box (WRKY binding site), and MBS (MYB binding site) in their promoter regions, which are involved in PHT gene regulation. These transcription factors are highly distributed in the upstream 2000 bp regions of TaPHT genes (Nash et al., 1990; Teng et al., 2017; Zhao et al., 2021a). In Arabidopsis, functional PHT expression is initiated downstream by binding to cis-acting elements in P1BS (GNATATNC), where phosphate starvation-regulated response factors have important roles in Pi starvation signaling (Zhang et al., 2024). The P1BS cis-acting element is present in the barley PHT1 promoter, with multiple copies in other genes (Schünmann et al., 2004). The TATA-box is a key component sequence in eukaryotic promoters that determines the onset of gene transcription, with increased TATA-box expression observed in Arabidopsis promoters in response to low Pi stress (Hammond et al., 2003). ABRE is a well-known cis-acting element that facilitates abscisic acid-dependent signaling, which has important effects on plant growth, development, and adversity responses (Baker et al., 1994) LTR is a DNA sequence in the plant genome involved in regulated low temperature responses (White et al., 1994). Additionally, TaPHT contain various responsive elements, including those related to light, antioxidants, gibberellin, drought stress, salicylic acid, and general stress responses. Therefore, these cis-acting elements are required to regulate plant gene expression and manage processes such as plant growth, development, and stress responses by reacting to different biotic and abiotic signals. Thus, cis-acting elements in TaPHT genes, along with their potential roles regulating gene expression, suggest that TaPHT genes have key roles in stress responses in wheat.
Among the 180 TaPHT genes screened against the public transcriptome database, 24 showed significantly higher expression in roots or aerial parts under low Pi stress conditions, suggesting important roles in Pi transport and regulation. Pi transport protein genes are induced in plants in response to Pi depletion, processes also reported in Arabidopsis, rice, B. napus, poplar, maize, barley, and other plants (Mudge et al., 2002; Rae et al., 2003; Shukla et al., 2016; Teng et al., 2017; Wan et al., 2020; Wang et al., 2014).
PHT family genes have different expression patterns in different tissues. Our qRT-PCR analysis of TaPHT showed that TaPHT1 was mainly expressed in roots, with previous studies showing that TaPHT1.1/9, TaPHT1.2, and TaPHT1.10 were mainly expressed in roots, and that low-Pi treatments in hydroponic cultures significantly induced TaPHT1 expression (Teng et al., 2017). Additionally, most PtPHT genes (from poplar) were highly expressed in P. simonii roots, whereas PtPHT1.5 and PtPHO9 were highly expressed only in Populus tremula roots and suckers (Zhang et al., 2016). TaPHT2 was predominantly expressed in shoots, consistent with previous studies, but most reported PHT2;1 genes were predominantly expressed in green tissues (Rausch et al., 2004; Versaw and Harrison, 2002), and all AtPHT genes showed detectable expression levels in roots. MdPHT3;3 (apple) showed high expression in roots, stems, and young fruits. Similarly, in the PHT3 subfamily of Populus, PtPHT3.1a is low expressed in root, freshly expanded and mature leaves, and high expressed in expanding young leaves. PtPHT3.1b is highly expressed in root and expanding young leaves, but low in freshly expanded and mature leaves. The expression of PtPHT3.2a was low in root, freshly expanded and mature leaves. There was no significant change in the expression of expanding young leaves. PtPHT3.2b is highly expressed in root and expanding young leaves. The expression was low in freshly expanded and mature leaves. PtPHT3.3a is highly expressed in root, freshly expanded and mature leaves. The expression was low in expanding young leaves. PtPHT3.3b is highly expressed in root, expanding young and mature leaves. There was no obvious change of expression in freshly expanded leaves (Zhang et al., 2016). In our study, TaPHT3;36 was more highly expressed in wheat shoots and less in roots, which was possibly due to species differences (Sun et al., 2017). In the TaPHT4 subfamily, relative TaPHT4;10 and TaPHT4;13 expression was high in wheat roots, while TaPHT4;20 was high in wheat shoots. Previous studies reported that AtPHT4;1 and AtPHT4;4 had the highest expression levels in leaves (Guo et al., 2008), and OsPT4;1, OsPT4;4, and OsPT4;5 were highly expressed in leaves, but OsPT4;2 was highly expressed in roots and stems at nodal growth stages, suggesting that PHT4 expression is varied across different species and tissues (Ruili et al., 2020). TaPHT5 and TaPHO1 were expressed in both shoots and roots, with levels up-regulated for a short period from 0–3 d, and then followed by relatively high expression levels at 7 d or 14 d in sustained Pi stress responses Similar to the low standing Physcomitrella patens, PpPHO1;1 and PpPHO1;7 expression was progressively increased over a 14 d Pi deficiency treatment period (Wang et al., 2007). In our study, relative TaPHT expression was up-regulated in both aboveground and roots by low Pi stress in SW2 and SW14 cultivars, consistent with observations in hairy camelina, maize, and three other grass crops (Xu et al., 2018). These results indicated that TaPHT family genes maintained normal plant growth and Pi homeostasis under low Pi stress.
The PHT1 family is a major Pi transporter family in plants and facilitates Pi transport from the soil to the plant. Under low Pi stress, plants may respond to Pi deficiency by up-regulating PHT1 family members to increase Pi uptake and transport capacity. Relative TaPHT1;36 expression in wheat roots was high and stable among the TaPHT family in the low Pi tolerant SW2 cultivar (by qRT-PCR), further confirming its subcellular location. Importantly, subcellular location prediction databases often differ and thus require validation based on experimental data (Gillani and Pollastri, 2024). In this study, Arabidopsis was transiently transformed with TaPHT1;36-GFP and empty control pC1300S-GFP vectors, examined under laser confocal microscopy, which showed that fluorescence signals from the empty vector were localized to the nucleus, cell membrane, and cytoplasm, while TaPHT1;36-GFP signals were localized to the plasma membrane. These observations suggested TaPHT1;36 involvement in Pi transport, consistent with subcellular localization data from Gene Sequences Online, which is of great importance to verify its response to low Pi stress. The BnPHT1-GFP fusion protein was constructed in B. napus, a kale-type oilseed rape, with the 35S:BnPht1;4-GFP fusion construct introduced into onion endoepidermal cells using the PDS-1000 helium biobody device. Fluorescence signals were localized to the cell periphery using confocal laser microscopy, showing that the BnPHT1 protein was located to the plasma membrane, consistent with TaPHT1;36 (Ren et al., 2014).
We comprehensively analyzed the TaPHT family in terms of gene structure, conserved motifs, cis-acting elements, and expression patterns. We identified 180 TaPHT genes, which were classified into 6 groups (TaPHT1–TaPHT5 and TaPHO1) based on phylogenetic relationships. Cis-acting element analysis of TaPHT promoters revealed multiple Pi-responsive elements. qRT-PCR analysis of TaPHT genes showed that relative expression in the low-Pi-tolerant wheat SW2 cultivar was higher than in the Pi-sensitive SW14 cultivar under low-Pi stress. We identified TaPHT1;36 which had the highest relative expression levels in wheat roots, which will be further analyzed at physiological and molecular levels in future work. Our whole genome TaPHT analysis has provided a platform for screening and verifying genes during low Pi stress and uptake, which may improve Pi use efficiency for greener agricultural production.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
MS: Conceptualization, Data curation, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. PL: Conceptualization, Investigation, Visualization, Writing – review & editing. LY: Writing – review & editing, Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources. CL: Writing – review & editing, Conceptualization, Formal Analysis, Investigation. ES: Writing – review & editing, Conceptualization, Formal Analysis, Funding acquisition, Methodology, Project administration, Resources. BL: Writing – review & editing, Data curation, Methodology, Supervision, Validation. YM: Writing – review & editing, Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources. XM: Writing – review & editing, Conceptualization, Data curation, Formal Analysis, Validation. KY: Writing – review & editing, Investigation, Methodology, Software. HZ: Methodology, Project administration, Visualization, Writing – review & editing. XS: Writing – review & editing, Conceptualization, Resources, Supervision. HW: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Writing – review & editing. JW: Writing – review & editing, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision.
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the State Key Laboratory of Aridland Crop Science, Gansu Agricultural University (GSCS-2021-05); 2023 Gansu Provincial Key Talent Project; Gansu Province Agricultural Science and Technology Support Project (KJZC-2023-2); Industrial Support Project of Colleges and Universities in Gansu Province (2021CYZC-12); Fuxi Talent Project of Gansu Agricultural University (Ganfx-03Y06; GAUfx-04Y011); Gansu Provincial Science and Technology Program (24CXNA038); Gansu Provincial Science and Technology Programme Project Natural Science Foundation Key Project (24JRRA637); Gansu Provincial Science; Gansu Provincial Seed Industry Key Project in 2025 and Technology Programme Project Joint Research Fund (24JRRA840).
We thank the professionals of BioMed Proofreading LLC for English corrections and copyediting this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1531642/full#supplementary-material
Ahmad, I., Rawoof, A., Islam, K., Momo, J., Ramchiary, N. (2021). Identification and expression analysis of phosphate transporter genes and metabolites in response to phosphate stress in Capsicum annuum. Environ. Exp. Bot. 190, 104597. doi: 10.1016/j.envexpbot.2021.104597
Aslam, M. M., Waseem, M., Weifeng, X., Qamar, M. (2022). Identification and expression analysis of phosphate transporter (PHT) gene family in Lupinus albus cluster root under phosphorus stress. Int. J. Biol. Macromolecules 205, 772–781. doi: 10.1016/j.ijbiomac.2022.03.085
Baker, S. S., Wilhelm, K. S., Thomashow, M. F. (1994). The 5’-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 24, 701–713. doi: 10.1007/bf00029852
Chowdhury, R. B., Moore, G. A., Weatherley, A. J., Arora, M. (2017). Key sustainability challenges for the global phosphorus resource, their implications for global food security, and options for mitigation. J. Cleaner Production 140, 945–963. doi: 10.1016/j.jclepro.2016.07.012
Cordell, D., Drangert, J.-O., White, S. (2009). The story of phosphorus: Global food security and food for thought. Global Environ. Change 19, 292–305. doi: 10.1016/j.gloenvcha.2008.10.009
Daram, P., Brunner, S., Rausch, C., Steiner, C., Amrhein, N., Bucher, M. (1999). Pht2;1 encodes a low-affinity phosphate transporter from Arabidopsis. Plant Cell 11, 2153–2166. doi: 10.1105/tpc.11.11.2153
Davies, T., Ying, J., Xu, Q., Li, Z., Li, J., Gordon-Weeks, R. (2002). Expression analysis of putative high-affinity phosphate transporters in Chinese winter wheats. Plant Cell Environ. 25, 1325–1339. doi: 10.1046/j.1365-3040.2002.00913.x
Gillani, M., Pollastri, G. (2024). Protein subcellular localization prediction tools. Comput. Struct. Biotechnol. J. 23, 1796–1807. doi: 10.1016/j.csbj.2024.04.032
Godfray, H. C. J., Beddington, J. R., Crute, I. R., Haddad, L., Lawrence, D., Muir, J. F., et al. (2010). Food security: the challenge of feeding 9 billion people. Science 327, 812–818. doi: 10.1126/science.1185383
Guo, B., Jin, Y., Wussler, C., Blancaflor, E. B., Motes, C. M., Versaw, W. K. (2008). Functional analysis of the Arabidopsis PHT4 family of intracellular phosphate transporters. New Phytol. 177, 889–898. doi: 10.1111/j.1469-8137.2007.02331.x
Hammond, J. P., Bennett, M. J., Bowen, H. C., Broadley, M. R., Eastwood, D. C., May, S. T., et al. (2003). Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol. 132, 578–596. doi: 10.1104/pp.103.020941
Harrison, M. J., Dewbre, G. R., Liu, J. (2002). A phosphate transporter from medicago truncatula involved in the acquisition of phosphate released by Arbuscular mycorrhizal fungi. Plant Cell 14, 2413–2429. doi: 10.1105/tpc.004861
Li, J., Han, G., Sun, C., Sui, N. (2019). Research advances of MYB transcription factors in plant stress resistance and breeding. Plant Signaling Behav. 14, 1613131. doi: 10.1080/15592324.2019.1613131
Li, J., Lin, K., Zhang, S., Wu, J., Fang, Y., Wang, Y. (2021). Genome-wide analysis of myeloblastosis-related genes in Brassica napus L. and positive modulation of osmotic tolerance by bnMRD107. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.678202
Li, P., Ma, X., Wang, J., Yao, L., Li, B., Meng, Y., et al. (2023). Integrated analysis of metabolome and transcriptome reveals insights for low phosphorus tolerance in wheat seedling. Int. J. Mol. Sci. 24, 14840. doi: 10.3390/ijms241914840
Liu, B., Zhao, S., Wu, X., Wang, X., Nan, Y., Wang, D., et al. (2017). Identification and characterization of phosphate transporter genes in potato. J. Biotechnol. 264, 17–28. doi: 10.1016/j.jbiotec.2017.10.012
Liu, H., Yang, Y., Zhang, L. (2021). Zinc finger-homeodomain transcriptional factors (ZF-HDs) in wheat (Triticum aestivum L.): identification, evolution, expression analysis and response to abiotic stresses. Plants 10, 593. doi: 10.3390/plants10030593
Liu, T.-Y., Huang, T.-K., Yang, S.-Y., Hong, Y.-T., Huang, S.-M., Wang, F.-N., et al. (2016). Identification of plant vacuolar transporters mediating phosphate storage. Nat. Commun. 7, 11095. doi: 10.1038/ncomms11095
Marand, A. P., Eveland, A. L., Kaufmann, K., Springer, N. M. (2023). cis-regulatory elements in plant development, adaptation, and evolution. Annu. Rev. Plant Biol. 74, 111–137. doi: 10.1146/annurev-arplant-070122-030236
Muchhal, U. S., Pardo, J. M., Raghothama, K. G. (1996). Phosphate transporters from the higher plant Arabidopsis thaliana. Proc. Natl. Acad. Sci. 93, 10519–10523. doi: 10.1073/pnas.93.19.10519
Mudge, S. R., Rae, A. L., Diatloff, E., Smith, F. W. (2002). Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J. 31, 341–353. doi: 10.1046/j.1365-313X.2002.01356.x
Murugan, N., Palanisamy, V., Channappa, M., Ramanathan, V., Ramaswamy, M., Govindakurup, H., et al. (2022). Genome-wide in silico identification, structural analysis, promoter analysis, and expression profiling of PHT gene family in sugarcane root under salinity stress. Sustainability 14, 15893. doi: 10.3390/su142315893
Nash, J., Luehrsen, K. R., Walbot, V. (1990). Bronze-2 gene of maize: reconstruction of a wild-type allele and analysis of transcription and splicing. Plant Cell 2, 1039–1049. doi: 10.1105/tpc.2.11.1039
Paszkowski, U., Kroken, S., Roux, C., Briggs, S. P. (2002). Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. 99, 13324–13329. doi: 10.1073/pnas.202474599
Poirier, Y., Bucher, M. (2002). “Phosphate transport and homeostasis in Arabidopsis,” in The Arabidopsis Book 2002. (Rockville, Maryland, USA: American Society of Plant Biologists (ASPB)).
Rae, A. L., Cybinski, D. H., Jarmey, J. M., Smith, F. W. (2003). Characterization of two phosphate transporters from barley; evidence for diverse function and kinetic properties among members of the Pht1 family. Plant Mol. Biol. 53, 27–36. doi: 10.1023/B:PLAN.0000009259.75314.15
Rausch, C., Bucher, M. (2002). Molecular mechanisms of phosphate transport in plants. Planta 216, 23–37. doi: 10.1007/s00425-002-0921-3
Rausch, C., Zimmermann, P., Amrhein, N., Bucher, M. (2004). Expression analysis suggests novel roles for the plastidic phosphate transporter Pht2;1 in auto- and heterotrophic tissues in potato and Arabidopsis. Plant J. 39, 13–28. doi: 10.1111/j.1365-313X.2004.02106.x
Ren, F., Zhao, C.-Z., Liu, C.-S., Huang, K.-L., Guo, Q.-Q., Chang, L.-L., et al. (2014). A Brassica napus PHT1 phosphate transporter, BnPht1;4, promotes phosphate uptake and affects roots architecture of transgenic Arabidopsis. Plant Mol. Biol. 86, 595–607. doi: 10.1007/s11103-014-0249-y
Rui, W., Ma, J., Wei, N., Zhu, X., Li, Z. (2023). Genome-wide analysis of the PHT gene family and its response to mycorrhizal symbiosis in tomatoes under phosphate starvation conditions. Int. J. Mol. Sci. 24, 10246. doi: 10.3390/ijms241210246
Ruili, L., Jiaoling, W., Lei, X., Meihao, S., Keke, Y., Hongyu, Z. (2020). Functional analysis of phosphate transporter osPHT4 family members in rice. Rice Sci. 27, 493–503. doi: 10.1016/j.rsci.2020.09.006
Schünmann, P. H. D., Richardson, A. E., Smith, F. W., Delhaize, E. (2004). Characterization of promoter expression patterns derived from the Pht1 phosphate transporter genes of barley (Hordeum vulgare L.). J. Exp. Bot. 55, 855–865. doi: 10.1093/jxb/erh103
Secco, D., Baumann, A., Poirier, Y. (2010). Characterization of the rice PHO1 gene family reveals a key role for osPHO1;2 in phosphate homeostasis and the evolution of a distinct clade in dicotyledons. Plant Physiol. 152, 1693–1704. doi: 10.1104/pp.109.149872
Shukla, V., Kaur, M., Aggarwal, S., Bhati, K. K., Kaur, J., Mantri, S., et al. (2016). Tissue specific transcript profiling of wheat phosphate transporter genes and its association with phosphate allocation in grains. Sci. Rep. 6, 39293. doi: 10.1038/srep39293
Sun, T., Li, M., Shao, Y., Yu, L., Ma, F. (2017). Comprehensive genomic identification and expression analysis of the phosphate transporter (PHT) gene family in apple. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00426
Takabatake, R., Hata, S., Taniguchi, M., Kouchi, H., Sugiyama, T., Izui, K. (1999). Isolation and characterization of cDNAs encoding mitochondrial phosphate transporters in soybean, maize, rice, and Arabidopsis. Plant Mol. Biol. 40, 479–486. doi: 10.1023/A:1006285009435
Teng, W., Zhao, Y.-Y., Zhao, X.-Q., He, X., Ma, W.-Y., Deng, Y., et al. (2017). Genome-wide identification, characterization, and expression analysis of PHT1 phosphate transporters in wheat. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00543
Ticconi, C. A., Delatorre, C. A., Abel, S. (2001). Attenuation of phosphate starvation responses by phosphite in Arabidopsis. Plant Physiol. 127, 963–972. doi: 10.1104/pp.010396
Ullrich-Eberius, C. I., Novacky, A., Fischer, E., Lüttge, U. (1981). Relationship between energy-dependent phosphate uptake and the electrical membrane potential in lemna gibba G1 1 2. Plant Physiol. 67, 797–801. doi: 10.1104/pp.67.4.797
Versaw, W. K., Harrison, M. J. (2002). A chloroplast phosphate transporter, PHT2;1, influences allocation of phosphate within the plant and phosphate-starvation responses. Plant Cell 14, 1751–1766. doi: 10.1105/tpc.002220
Wan, Y., Wang, Z., Xia, J., Shen, S., Guan, M., Zhu, M., et al. (2020). Genome-wide analysis of phosphorus transporter genes in brassica and their roles in heavy metal stress tolerance. Int. J. Mol. Sci. 21, 2209. doi: 10.3390/ijms21062209
Wang, D., Lv, S., Jiang, P., Li, Y. (2017). Roles, regulation, and agricultural application of plant phosphate transporters. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00817
Wang, X., Niu, Y., Zheng, Y. (2021). Multiple functions of MYB transcription factors in abiotic stress responses. Int. J. Mol. Sci. 22, 6125. doi: 10.3390/ijms22116125
Wang, J., Qin, Q., Pan, J., Sun, L., Sun, Y., Xue, Y., et al. (2019a). Transcriptome analysis in roots and leaves of wheat seedlings in response to low-phosphorus stress. Sci. Rep. 9, 19802. doi: 10.1038/s41598-019-56451-6
Wang, Y., Ribot, C., Rezzonico, E., Poirier, Y. (2004). Structure and expression profile of the Arabidopsis PHO1 gene family indicates a broad role in inorganic phosphate homeostasis. Plant Physiol. 135, 400–411. doi: 10.1104/pp.103.037945
Wang, Y., Secco, D., Poirier, Y. (2007). Characterization of the PHO1 gene family and the responses to phosphate deficiency of physcomitrella patens. Plant Physiol. 146, 323–324. doi: 10.1104/pp.107.108548
Wang, X., Wang, Y., Piñeros, M., Wang, Z., Wang, W., Li, C., et al. (2014). Phosphate transporters;9 and;10 are involved in phosphate uptake in rice. Plant Cell Environ. 37, 1159–1170. doi: 10.1111/pce.12224
Wang, L., Xiao, L., Yang, H., Chen, G., Zeng, H., Zhao, H., et al. (2020). Genome-wide identification, expression profiling, and evolution of phosphate transporter gene family in green algae. Front. Genet. 11. doi: 10.3389/fgene.2020.590947
Wang, J., Yang, Y., Liao, L., Xu, J., Liang, X., Liu, W. (2019b). Genome-wide identification and functional characterization of the phosphate transporter gene family in sorghum. Biomolecules 9, 670. doi: 10.3390/biom9110670
Westheimer, F. H. (1987). Why nature chose phosphates. Science 235, 1173–1178. doi: 10.1126/science.2434996
White, A. J., Alison Dunn, M., Brown, K., Hughes, M. A. (1994). Comparative analysis of genomic sequence and expression of a lipid transfer protein gene family in winter barley. J. Exp. Bot. 45, 1885–1892. doi: 10.1093/jxb/45.12.1885
Xu, Y., Liu, F., Han, G., Cheng, B. (2018). Genome-wide identification and comparative analysis of phosphate starvation-responsive transcription factors in maize and three other gramineous plants. Plant Cell Rep. 37, 711–726. doi: 10.1007/s00299-018-2262-0
Yang, J., Zhou, J., Zhou, H.-J., Wang, M.-M., Liu, M.-M., Ke, Y.-Z., et al. (2020). Global survey and expressions of the phosphate transporter gene families in Brassica napus and their roles in phosphorus response. Int. J. Mol. Sci. 21, 1752. doi: 10.3390/ijms21051752
Zhang, C., Meng, S., Li, M., Zhao, Z. (2016). Genomic identification and expression analysis of the phosphate transporter gene family in poplar. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01398
Zhang, J.-F., Wang, Y.-Y., He, L., Yan, J.-Y., Liu, Y.-Y., Ruan, Z.-Y., et al. (2024). PHR1 involved in the regulation of low phosphate-induced leaf senescence by modulating phosphorus homeostasis in Arabidopsis. Plant Cell Environ. 47, 799–816. doi: 10.1111/pce.14790
Zhao, X., Li, G., Sun, Z., Chen, Y., Guo, W., Li, Y., et al. (2021b). Identification, structure analysis, and transcript profiling of phosphate transporters under Pi deficiency in duckweeds. Int. J. Biol. Macromolecules 188, 595–608. doi: 10.1016/j.ijbiomac.2021.08.037
Keywords: wheat (Triticum aestivum L.), phosphate transporter protein, PHT gene family, gene expression, low phosphorus stress
Citation: Song M, Li P, Yao L, Li C, Si E, Li B, Meng Y, Ma X, Yang K, Zhang H, Shang X, Wang H and Wang J (2025) Characterizing the wheat (Triticum aestivum L.) phosphate transporter gene family and analyzing expression patterns in response to low phosphorus stress during the seedling stage. Front. Plant Sci. 16:1531642. doi: 10.3389/fpls.2025.1531642
Received: 20 November 2024; Accepted: 26 February 2025;
Published: 27 March 2025.
Edited by:
Yinghui Li, University of Haifa, IsraelReviewed by:
Karishma Seem, Indian Agricultural Research Institute (ICAR), IndiaCopyright © 2025 Song, Li, Yao, Li, Si, Li, Meng, Ma, Yang, Zhang, Shang, Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Juncheng Wang, d2FuZ2pjQGdzYXUuZWR1LmNu; Huajun Wang, d2FuZ2hqQGdzYXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.