- 1School of Traditional Chinese Medicine, Guangdong Pharmaceutical University, Guangzhou, China
- 2Yunnan General Administration of Forestry Seeds and Seedlings, Kunming, China
- 3College of Resources Environment and Chemistry, Chuxiong Normal University, Chuxiong, China
Background: Asparagus L. is a large genus widely distributed across the continents of the Old World. Among its members, approximately 14 species found in China are recognized as popular herbal medicines. However, accurate authentication of these medicinal species and their phylogenetic relationships with related taxa remains unresolved.
Methods: To identify simple sequence repeats (SSRs) and divergence hotspot regions appropriate for future authentication studies, as well as to infer the phylogenetic relationships among Asparagus species, we employed a plastid genome (plastome) dataset consisting of 25 Asparagus species (21 newly sequenced and four retrieved from GenBank), encompassing 12 Chinese medicinal species, for comparative and phylogenetic analyses.
Results: All Asparagus plastomes displayed a typical quadripartite structure with sizes ranging from 155,948 bp to 157,128 bp and harbored 114 unique genes (80 protein-coding genes, 30 tRNA genes, and four rRNA genes). IRscope and Mauve analyses indicated minimal structural variation among Asparagus plastomes. We detected between 79 to 95 SSRs across the plastomes; most were located in the large single-copy (LSC) region and primarily consisted of mono-nucleotide repeat sequences (especially A and T repeats). The genus displayed mono-, di-, tri-, tetra-, penta-, and hexa-nucleotide repeats, but with variations in types and numbers among different species. Additionally, we identified 12 special SSR motifs and seven divergent hotspot regions that may serve as potential molecular markers for future identification efforts. Phylogenetic analyses yielded a robust phylogeny for Asparagus taxa, which were split into Clades I, II, and III. Notably, medicinal Asparagus species were mainly found in Clade III. Although the phylogenetic relationships of most Asparagus species aligned with previous study findings, the phylogenetic positions of A. munitus, A. subscandens, A. gobicus, and A. dauricus were newly determined.
Conclusions: The plastomes of Asparagus are largely conserved in terms of genome structure, size, gene content, and arrangement. Nevertheless, SSRs analyses revealed significant interspecific polymorphism within Asparagus. In addition, special SSR motifs and divergent hotspot regions identified from Asparagus plastomes provided reference for subsequent identification investigations. The plastome-based phylogeny provided preliminary insights into the relationships among the Chinese group of medicinal species and related taxa within Asparagus. Overall, this study offers a wealth of informative genetic resources pertinent to Asparagus, thereby enhancing our understanding of its evolution and laying a foundation for species identification, assessment of genetic population diversity, as well as the exploration and conservation of germplasm resources.
Introduction
Asparagus L. (Asparagaceae), a genus comprising herbaceous and sub-shrubby members, as well as scramblers and climbers, encompasses 170 to 300 species that are widely distributed across the continents of the Old World (with only northern Australia in the Antipodes) (Stevens, 2024). Many of these species are known for their edible, ornamental, or medicinal applications (Amel et al., 2020). Currently, 31 Asparagus species (15 endemic, two introduced) have been documented in China (Chen and Tamanian, 2000), with approximately 14 species recognized for their significant medicinal value as herbal medicines (Editorial Committee of Chinese Materia Medica of the National Administration of Traditional Chinese Medicine, 1999; Yunnan Food and Drug Administration, 2005; Sichuan Food and Drug Administration, 2010). Tiandong, a well-known traditional Chinese medicine listed in the Chinese Pharmacopoeia, is derived from the dried root tubers of A. cochinchinensis (Lour.) Merr., which is effective in treating a wide range of ailments, including asthma, cough, constipation, thrombosis, and inflammatory diseases (Chinese Pharmacopoeia Commission, 2020; Wang et al., 2022a). Other medicinal Asparagus species, including A. filicinus Buch.-Ham. ex D.Don, A. meioclados H.Lév., A. gobicus Ivanova ex Grubov, A. subscandens F.T.Wang & S.C.Chen, A. brachyphyllus Turcz., A. lycopodineus (Baker) F.T.Wang & Tang, A. officinalis L., A. setaceus (Kunth) Jessop, A. racemosus Willd., A. myriacanthus F.T.Wang & S.C.Chen, A. kansuensis F.T. Wang & Tang, A. munitus F.T.Wang & S.C.Chen, and A. taliensis F.T.Wang & Tang ex S.C.Chen, are utilized as herbal medicines in traditional remedies; some of these exhibit effects similar to those of A. cochinchinensis or possess unique therapeutic efficacy (Editorial Committee of Chinese Materia Medica of the National Administration of Traditional Chinese Medicine, 1999; Yunnan Food and Drug Administration, 2005; Sichuan Food and Drug Administration, 2010; Lv and Chen, 2019; Parihar and Sharma, 2021; Li and Huang, 2021).
Nevertheless, the accurate authentication of A. cochinchinensis and other congeneric species continues to pose significant challenges. The abundant interspecific and even intraspecific morphological variations complicate species identification (Afroz et al., 2022). Moreover, the overlapping phenotypic traits of species within the genus to varying degrees often lead to confusion or admixture among congeneric species in practical applications. In certain regions of China, Asparagus species—particularly those with similar swollen roots, such as A. filicinus, A. meioclados, A. munitus, A. subscandens, A. taliensis, A. brachyphyllus, and A. lycopodineus—are frequently misidentified as A. cochinchinensis (Editorial Committee of Chinese Materia Medica of the National Administration of Traditional Chinese Medicine, 1999; Yunnan Food and Drug Administration, 2005; Sichuan Food and Drug Administration, 2010; Chen et al., 2000; Lv and Chen, 2019), undermining the quality control of Tiandong and posing potential risks to drug safety. Although several studies have aimed to differentiate medicinal species from other congeneric taxa, these efforts have not comprehensively addressed the issue due to inadequate molecular data (trnH-psbA, trnL-F) or insufficient sampling of medicinal species (Huang et al., 2010; Ou et al., 2010, Ou et al., 2013).
Furthermore, the evolutionary relationships between these medicinal species and related taxa of Asparagus distributed in China remain inadequately defined. Previous studies on this genus were generally taxonomic and did not specifically take into account the explicit phylogeny (Fukuda et al., 2005; Kubota et al., 2012; Norup et al., 2015). Meanwhile, the sparse or inadequate sampling has resulted in a poor understanding of interspecific relationships among Asparagus species (Sheng, 2022; Wong et al., 2022; Tian et al., 2023). Recently, Bentz et al. (2024) reconstructed the phylogeny of Asparagus based on a majority of plastid protein-coding genes. Despite including many Chinese Asparagus species, the taxonomic sampling of medicinal species still requires improvement. Additionally, excluding ycf1, ycf2, accD, and infA genes from phylogenetic analysis, as well as the lack of non-coding regions of plastid DNA commonly used in closely related species phylogenetic studies (Borsch et al., 2009), may result in some discrepancies in interspecific relationships. Therefore, to accurately identify species and further investigate the phylogenetic relationships between Chinese Asparagus medicinal species and related taxa, a broader sampling of medicinal species along with additional genomic data would be beneficial.
Despite being relatively conservative (Ravi et al., 2008; Jansen and Ruhlman, 2012), plastomes with abundant genetic variants have been widely recognized as a powerful tool for phylogenetic reconstruction across various taxonomic levels (Gitzendanner et al., 2018; Givnish et al., 2018; Wu et al., 2021; Ji et al., 2023; Ma et al., 2014; Guo et al., 2022). Meanwhile, the characteristics and structural patterns of the plastome obtained through comparative analysis also provide supplementary information for phylogenetic inference (Liu et al., 2018; Lyko and Wicke, 2021). Additionally, divergence hotspots and simple sequence repeats (SSRs) derived from plastome data may serve as candidate DNA barcodes or efficient molecular markers for species identification (Downie and Jansen, 2015; Zhou et al., 2023).
In this study, a dataset comprising 25 complete Asparagus plastomes (21 newly sequenced and four obtained from GenBank) was utilized for comparative and phylogenetic analyses, with 12 out of 14 Asparagus medicinal species included. Our primary objectives are to (1) investigate the plastome structure and sequence divergence of Asparagus plants; (2) identify SSRs and divergence hotspots of Asparagus; and (3) preliminarily elucidate the phylogenetic relationships among the Chinese group of medicinal species and related taxa within Asparagus.
Materials and methods
Plant sampling, DNA extraction and sequencing
Fresh leaves from 21 Asparagus species were collected and dried with silica gel. The identifications of the samples were conducted by Professor Heng Li. Voucher specimens were deposited in the Herbarium of Kunming Institute of Botany, CAS (KUN), Chinese Academy of Sciences, Yunnan, China, with collection details provided in Supplementary Table 1. Plastid genomic DNA was extracted from approximately 5 mg of silica gel-dried leaf tissues using the CTAB method (Doyle and Doyle, 1987). Total DNA was fragmented into 400 bp for pair-end library preparation utilizing a TruSeq DNA Sample Prep Kit (Illumina, Inc., San Diego, CA, USA) according to the manufacturer’s instructions. All genomic data were sequenced on the Illumina NovaSeq platform at Personalbio (Shanghai, China).
Plastome assembling and annotation
After filtering the raw data using fastP v0.15.0 (Chen et al., 2018) with parameters -n 10 and -q 15, high-quality reads were obtained. Subsequently, these high-quality reads were employed to assemble plastome through NOVOPlasty v2.6.2 (Dierckxsens et al., 2017) with the default settings, employing the rbcL sequence from A. officinalis (JQ273904) as a seed reference. The plastomes were annotated using the CPGAVAS2 (Shi et al., 2019) and further checked manually using Geneious v10.2.3 (Kearse et al., 2012). Additionally, the circular maps of the plastomes were generated using OrganellarGenomeDRAW (OGDRAW) (Greiner et al., 2019). GenBank accession numbers for the 21 Asparagus plastomes were documented in Supplementary Table 1. Together with six available plastomes obtained from NCBI (Supplementary Table 2), a total of 25 plastomes from Asparagus and two outgroup species were included in follow-up analyses.
Simple sequence repeat analysis
Simple sequence repeats (SSRs) were identified in each Asparagus plastome by using MISA (http://pgrc.ipk-gatersleben.de/misa/), with the thresholds for repeat units set at ten, five, four, three, three, and three, for mono-, di-, tri-, tetra-, penta- and hexa-nucleotide repeats, respectively.
Genomic comparison and divergence hotspots analysis
To investigate the structural variation of plastomes in Asparagus, we compared the expansion and contraction of inverted repeat (IR) regions across these plastomes using IRscope (Amiryousefi et al., 2018). After the removal of one copy of the IR region, the general structural characteristics of Asparagus plastome were further analyzed with Mauve v2.3.1 (Darling et al., 2004). Furthermore, the sequence divergence of Asparagus plastomes was assessed using the mVISTA (Frazer et al., 2004) in the Shuffle-LAGAN alignment program, with the plastome of A. cochinchinensis set as the reference. The nucleotide variability values (π) for the alignment of Asparagus plastomes were calculated using DnaSP v.5.0 (Librado and Rozas, 2009), with the window length and step size set to 600 bp and 200 bp, respectively. Coding and non-coding regions with π ≥ 0.010 were extracted using Geneious v10.2.3 (Kearse et al., 2012) and their number of variable sites and parsimony-informative (parsim-info) sites were evaluated by MEGA X (Kumar et al., 2018).
Phylogenetic analyses
To infer the phylogenetic relationships among Asparagus species, phylogenomic analyses were conducted to reconstruct phylogenetic trees based on 27 plastomes (Supplementary Table 1; Supplementary Table 2). Cordyline indivisa (G. Forst.) Endl. and Eustrephus latifolius R.Br. were utilized as outgroup species with the findings of Ji et al. (2023). All plastomes were aligned using MAFFT v7.450 (Katoh and Standley, 2013) with the default settings, and then manually adjusted in Geneious v10.2.3 (Amiryousefi et al., 2018). The characteristics of plastome alignments were analyzed using MEGA X (Kumar et al., 2018). Maximum likelihood (ML) analysis was conducted using the RAxML-HPC2 tool on XSEDE within the CIPRES Science Gateway platform (http://www.phylo.org/portal2), under a GTR + GAMMA model along with 1000 rapid bootstrap replicates. Bayesian Inference (BI) was executed by utilizing MrBayes v3.2.7a (Ronquist et al., 2012) with the most suitable substitution model (GTR + I + G) chosen through Modeltest v3.7 (Posada and Crandall, 1998). The independent run of Markov Chain Monte Carlo (MCMC) was conducted for 1,000,000 generations, with sampling performed every 100 generations and the first 25% of generations disregarded as burn-in for each matrix. Stationarity was attained when the average standard deviation of split frequencies fell below 0.01. The software FigTree v1.4.2 (Rambaut, 2014) was employed to visualize and modify the phylogenetic trees.
Results
Characteristics of the Asparagus plastomes
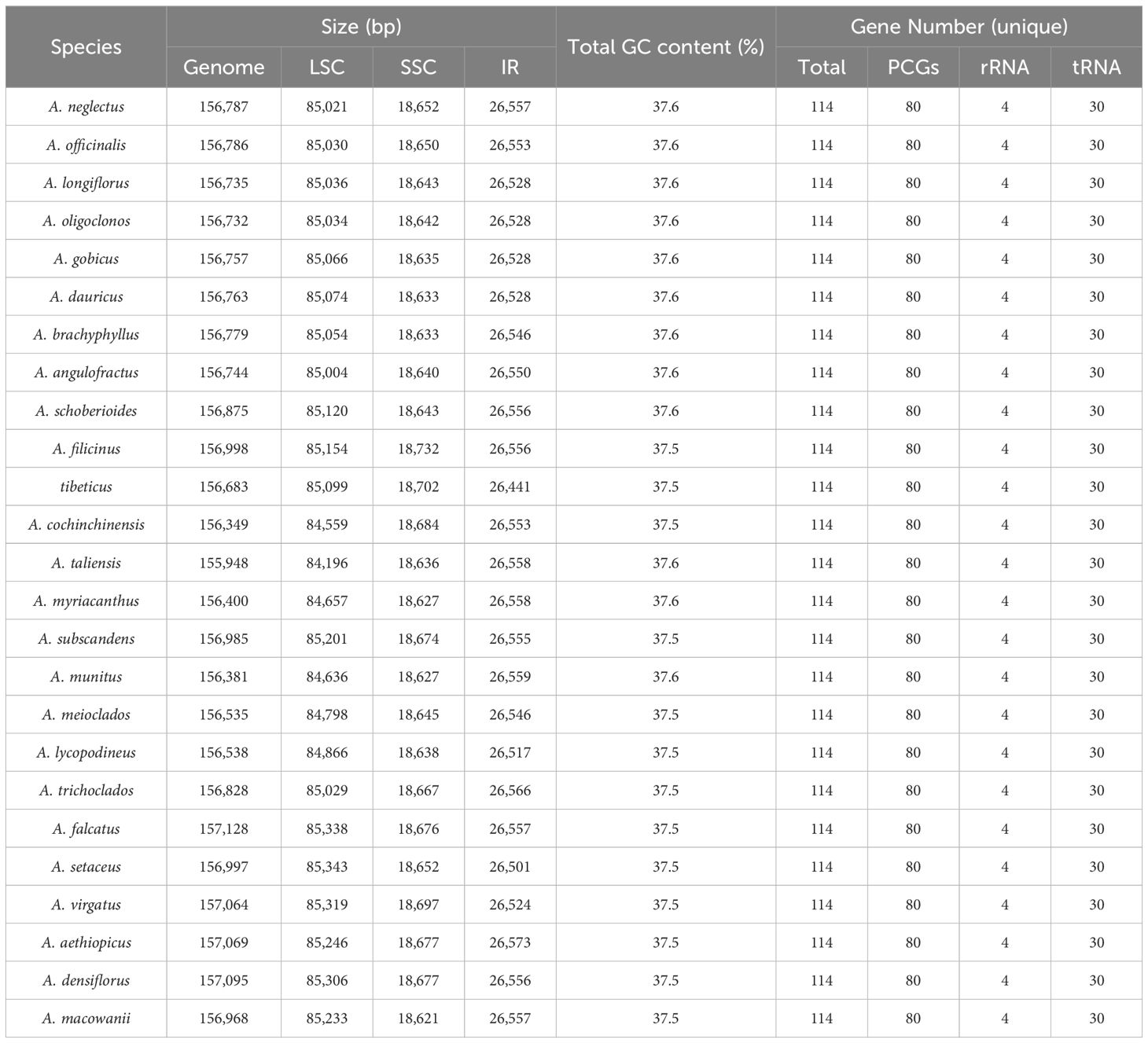
The summary of the low-coverage genome sequencing and assembly of plastomes is presented in Supplementary Table 3. The plastomes of Asparagus species exhibited sizes ranging from 155,948 bp (A. taliensis) to 157,128 bp (A. falcatus L.), which displayed a typical quadripartite structure, comprising two inverted repeat regions (IRs, 26,441–26,573 bp), a large single-copy (LSC) region (84,196–85,343 bp), and a small single-copy (SSC) region (18,621–18,732 bp) (Table 1; Figure 1). The total GC content of the plastomes varied between 37.5% to 37.6% (Table 1). Each plastome harbored 114 unique genes, including 80 protein-coding genes, 30 tRNA genes, and four rRNA genes (Table 1; Supplementary Table 4). Among them, the ycf15 gene was annotated as a pseudogene.
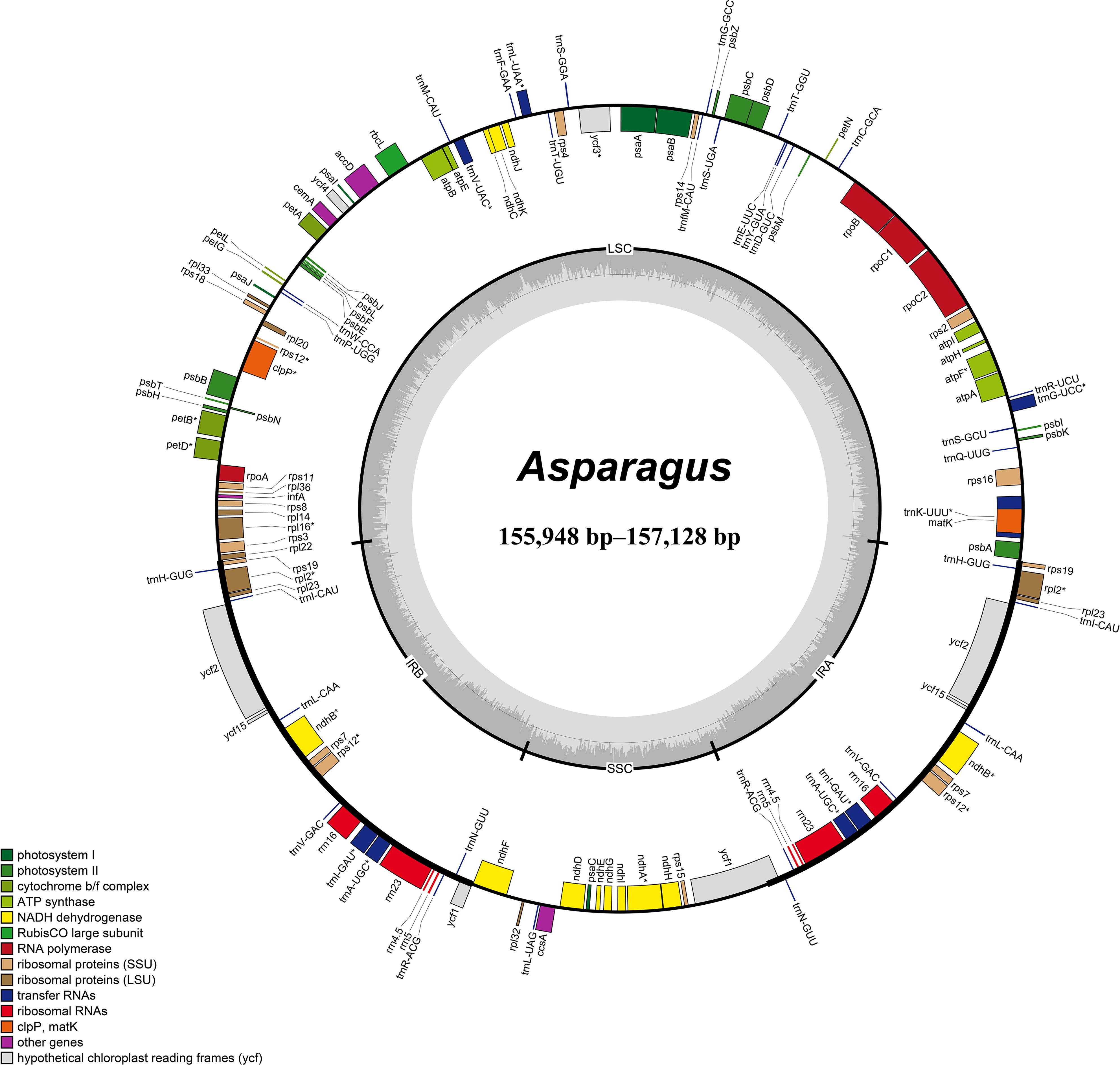
Figure 1. Circular maps of Asparagus plastomes. The inside and outside of the circle are genes transcribed clockwise and counterclockwise, respectively. The genes belonging to different functional groups are color-coded. The dashed area in the inner circle shows GC content.
Characterization of SSRs
A total of 2,142 SSRs were identified across the 25 Asparagus plastomes (Figure 2; Supplementary Table 5), with the number of SSRs per species ranging from 79 (A. munitus, A. angulofractus Iljin) to 95 (A. trichoclados (F.T.Wang & Tang) F.T.Wang & S.C.Chen), as illustrated in Figure 2A. All the Asparagus plastomes contained mono-, di-, tri-, and tetra-nucleotide repeats; however, penta-, and hexa-nucleotide repeats varied among different Asparagus species (Figure 2B). Specifically, penta-, and hexa-nucleotide repeats were absent in A. virgatus Baker, A. macowanii Baker, and A. angulofractus, while both of them were present in A. filicinus. In addition, hexa-nucleotide repeats could be observed in A. setaceus, A. densiflorus (Kunth) Jessop, A. aethiopicus L., A. falcatus, and A. schoberioides Kunth, and the penta-nucleotide repeats were only found in the remaining species. Among these SSRs, mono-nucleotide repeat type was the most dominant composition (48–63), followed by di-nucleotide repeats (12–15), tetra-nucleotide repeats (10–14), and tri-nucleotide repeats (4–8). Meanwhile, the A and T repeats were the majority of SSRs in each plastome (Supplementary Table 5). Concerning the distribution of SSRs, the majority were located in the LSC region across all Asparagus plastomes (Figure 2C).
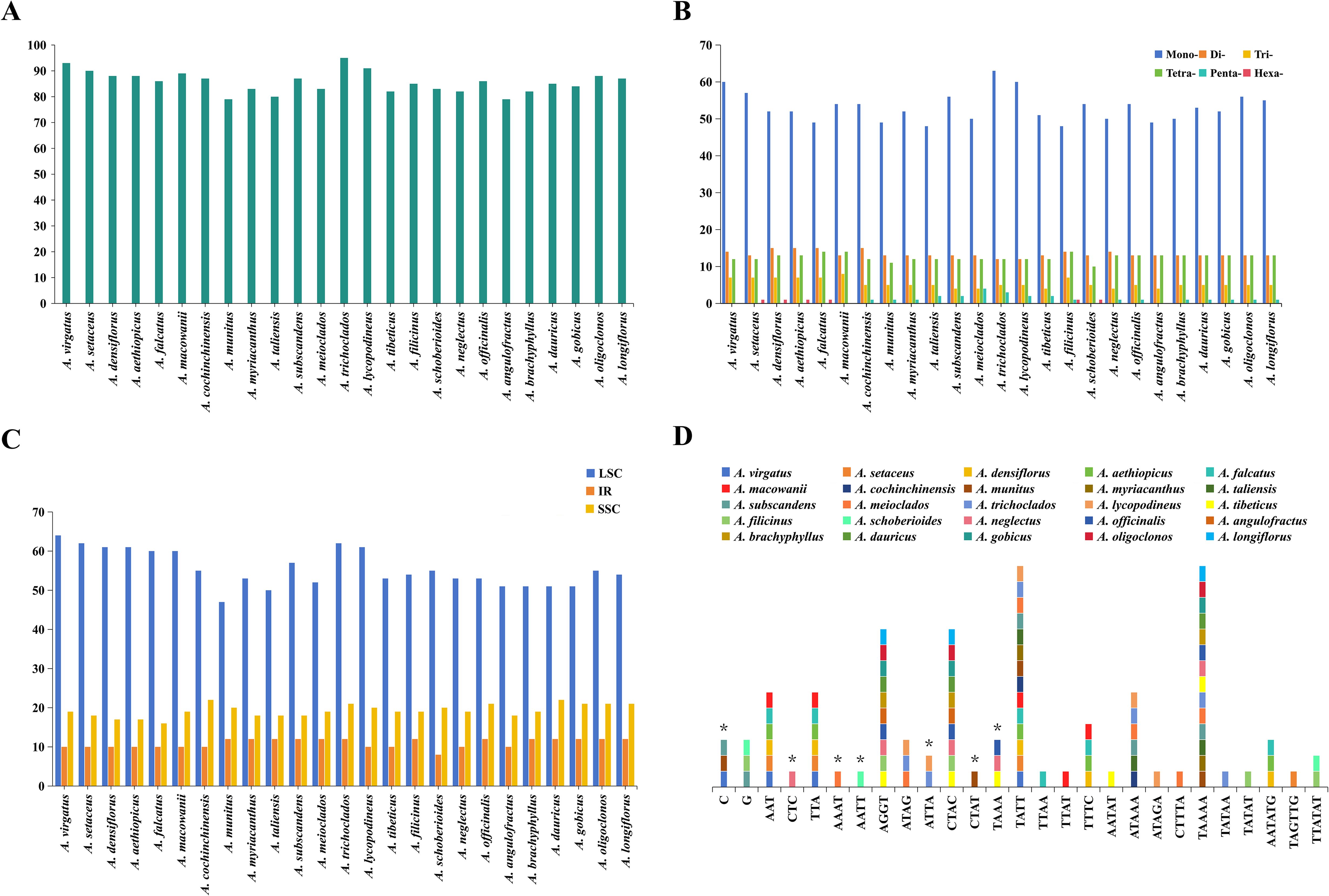
Figure 2. Analyses of simple sequence repeats (SSRs) in 25 Asparagus plastomes: (A) numbers of SSRs detected in each plastome; (B) numbers of different repeat types; (C) presence of SSRs in LSC, SSC, and IR; (D) the unique presence or absence of SSR motifs in one or several Asparagus species. The asterisk indicates the absence of SSR motifs.
To further investigate the differences between SSRs, 27 of 41 kinds of SSR motifs with unique presence or absence in one or several plastomes were ultimately recognized after excluding the shared repeat sequences (Figure 2D; Supplementary Table 5). Among these, the tetra-nucleotide repeats constituted the main component, followed by penta-nucleotide repeats. Notably, C repeat sequences were only absent in A. virgatus, A. munitus, and A. subscandens, while the G repeat sequences were exclusively found in A. subscandens, A. filicinus, and A. schoberioides. In addition, CTC, AAAT, AATT, and CTAT repeat sequences were absent in A. neglectus Kar. & Kir., A. meioclados, A. schoberioides, and A. munitus, respectively; TTAA, TTAT, AATAT, ATAGA, CTTTA, TATAA, TATAT, TAGTTG repeat sequences were detected solely in A. falcatus, A. macowanii, A. tibeticus F.T.Wang & S.C.Chen, A. lycopodineus, A. meioclados, A. trichoclados, A. filicinus, and A. setaceus, respectively. Apart from that, AAT/TTA repeat sequences were identified in A.setaceus, A. virgatus, A. macowanii, A. falcatus, A. densiflorus, and A. aethiopicus. CTAC/AGGT repeats were detected in A. filicinus, A. tibeticus, A. officinalis, A. neglectus, A. angulofractus, A. brachyphyllus, A. gobicus, A. dauricus Link, A. longiflorus Franch., and A. oligoclonos Maxim. The presence or absence of the remaining SSR motifs exhibited variability among Asparagus species.
Plastome comparison and hotspot identification
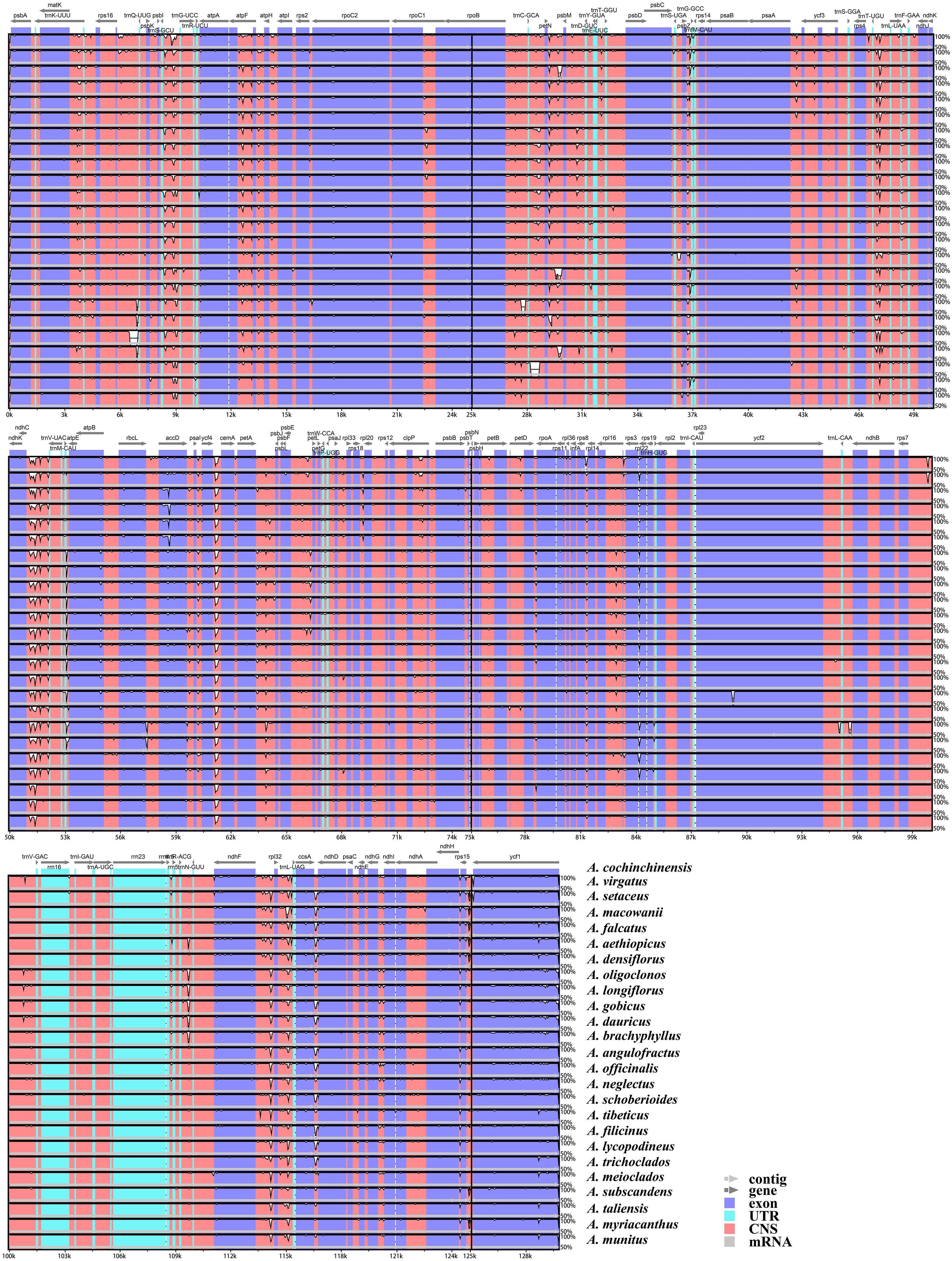
The expansion and contraction of the IRs were compared among the Asparagus species and the outgroups. Overall, no significant variation of IR length was detected within Asparagus plastomes. The boundaries of the LSC/IRb, IRb/SSC, SSC/IRa, and IRa/LSC are relatively conserved (Figure 3). Specifically, the LSC/IRb junctions were located between rpl22 and rps19; the boundaries of IRb/SSC were situated downstream from the ndhF but fell into ycf1 (the partially duplicated gene); the SSC/IRa borders were crossed by the ycf1; the junctions of IRa/LSC were positioned between rps19 and psbA. Furthermore, conservation in gene arrangement among Asparagus plastomes was confirmed with no evidence of gene rearrangement detected (Figure 4).
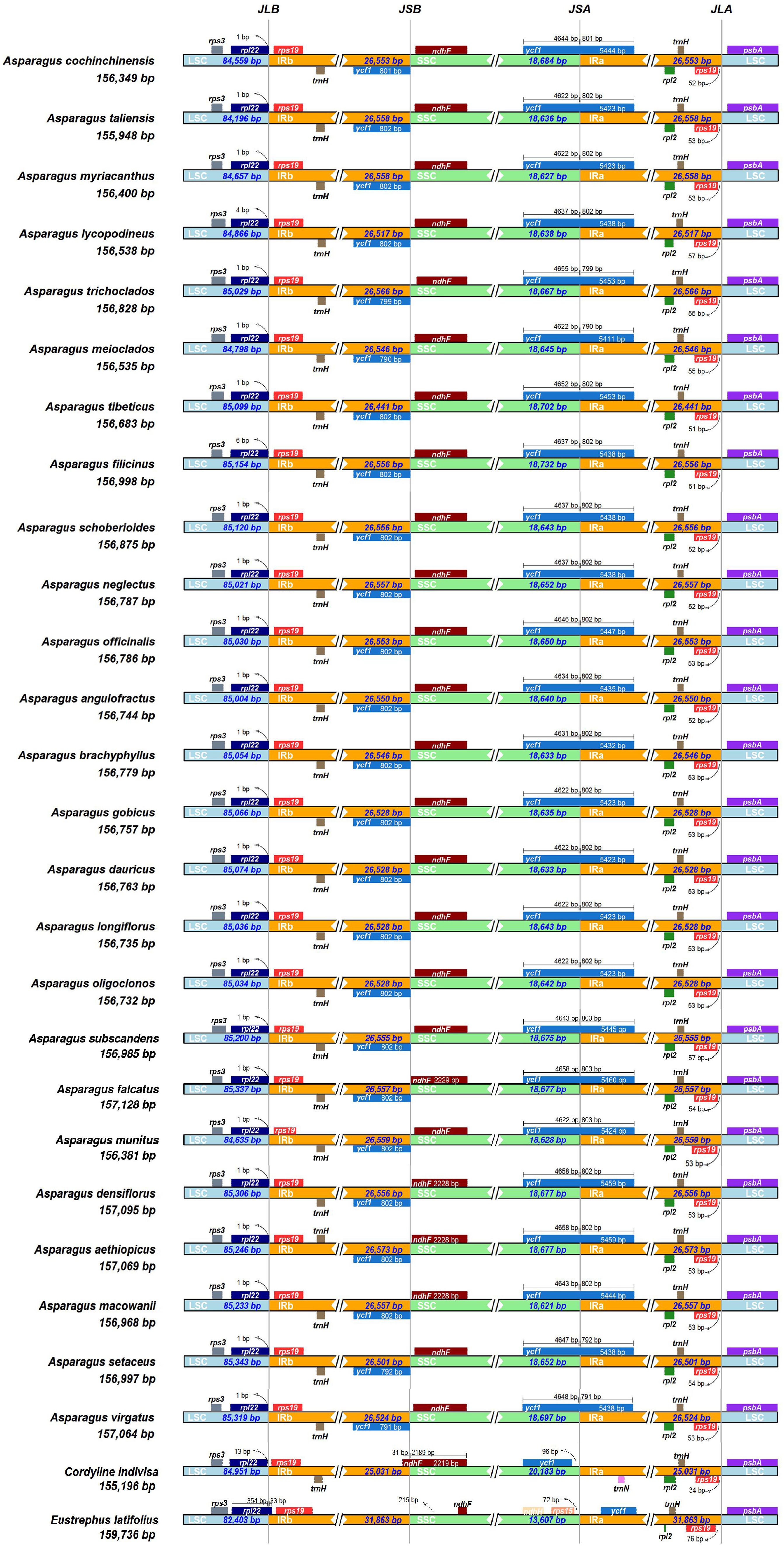
Figure 3. Comparison of the borders of the LSC, SSC, and IR regions among 25 Asparagus plastomes. JLB, junction line between LSC and IRb; JSB, junction line between IRb and SSC; JSA, junction line between SSC and IRa; JLA, junction line between IRa and LSC. These features are not to scale.

Figure 4. Mauve alignment of plastomes from 25 Asparagus species and outgroups. Rectangular blocks of the same color indicate collinear regions of sequences and the histograms within each block indicate the degree of sequence similarity.
The results of the mVISTA analysis indicated that the alignment of Asparagus plastomes exhibited high similarity in the sequence identity plots (Figure 5). Nonetheless, certain sequence variations among different Asparagus plastomes were still evident. Overall, the divergence levels in noncoding regions were much higher compared to those in coding regions. Among these, three coding regions (rbcL, accD, and ycf1) and 12 non-coding regions (trnS-trnG, rpoB-trnC, trnC-petN, petN-psbM, trnT-trnL, ndhC-trnV, petA-psbJ, psbE-petL, rpl16 intron, ndhF-rpl32, rpl32-trnL and ccsA-ndhD) displayed relatively significant difference. In the sliding-window analysis, the LSC and SSC regions exhibited greater nucleotide diversity with more peaks compared to the two IR regions (Figure 6A). Ultimately, seven regions—accD, rbcL, ycf1, rpl32-trnL, ccsA-ndhD, trnS-trnG, and ndhC-trnV—were identified as divergence hotspots with nucleotide variability values (π) ≥ 0.010. Among these, ycf1 contained the most variable sites, followed by ndhC-trnV, whereas ccsA-ndhD had the fewest. Additionally, ycf1 displayed the most abundance of parsim-info sites, followed by accD; however, ccsA-ndhD exhibited the smallest number of parsimony-informative (parsim-info) sites (Figure 6B; Supplementary Table 6).
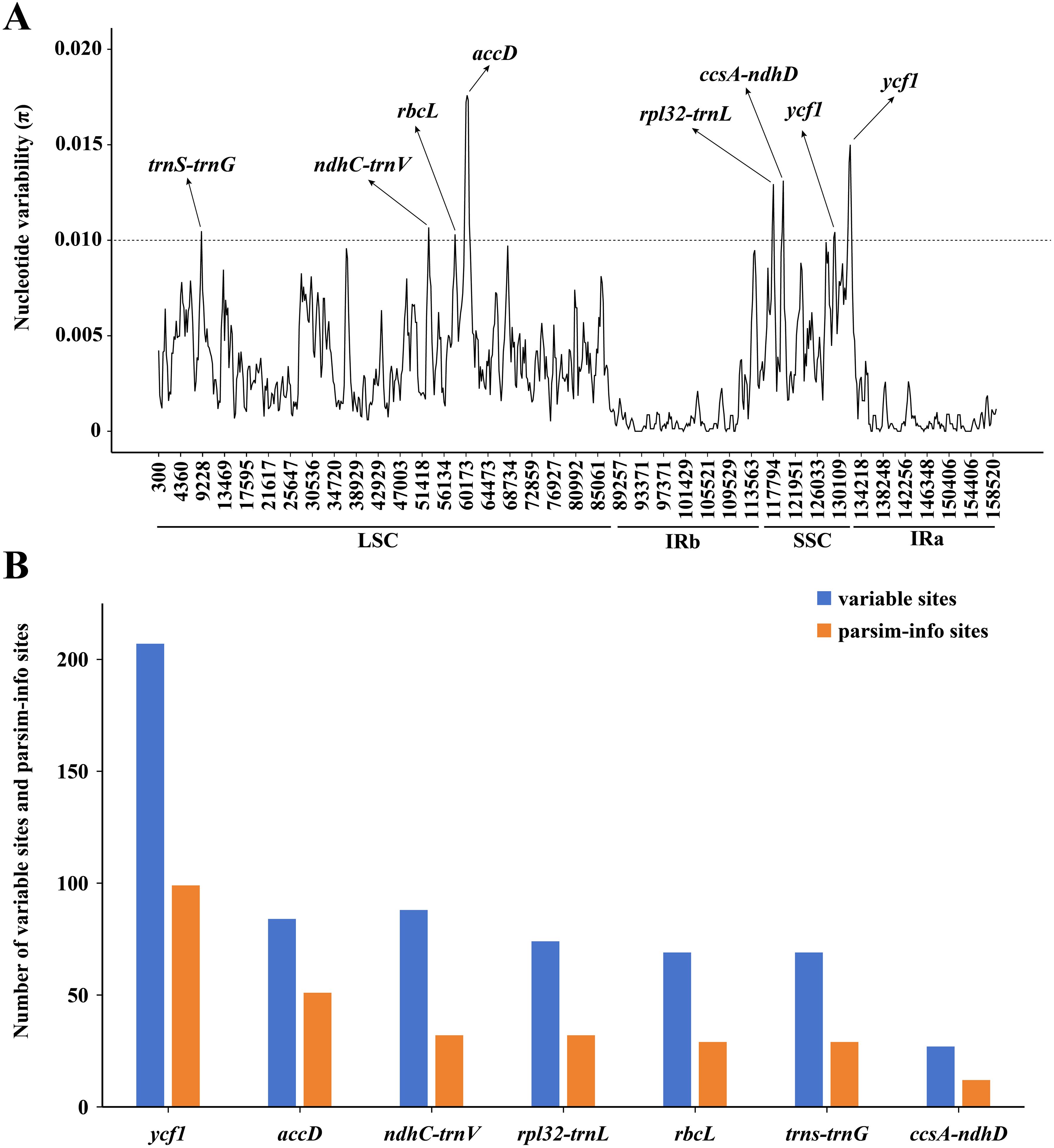
Figure 6. (A) Sliding window analysis of the 25 Asparagus plastomes (window length: 600 bp, step size: 200 bp) to show high variable regions (HVRs). X-axis: position of the midpoint of a window; Y-axis: nucleotide variability values (π) of each window. The dashed line indicates that the π value is equal to 0.010; (B) Number of variable sites and parsim-info sites in the alignment of divergence hotspots.
Phylogenetic analyses
The final alignment was 176,103 bp in length, in which we identified 18,001 variable sites (10.22%) with 4,139 (2.35%) being parsimoniously informative. Both ML and BI analyses yielded identical tree topologies with strong support (Bootstrap values (BS) = 100 and posterior probability (PP) = 1.00) (Figure 7). All Asparagus species were strongly supported as monophyletic (BS = 100; PP = 1.00) and resolved into three clades: (I) A.setaceus was clustered with A. virgatus (BS = 100; PP = 1.00) at the base of the phylogenetic tree; (II) A. macowanii and A. falcatus were successively sister to the clade comprising A. densiflorus and A. aethiopicus (BS = 100; PP = 1.00); (III) the remaining species formed a distinct clade (BS = 100; PP = 1.00), which was further divided into subclade 1, 2, and 3. Within subclade 1, A. cochinchinensis and A. myriacanthus were sequentially sister to the A. munitus + A. taliensis clade; while A. subscandens and A. meioclados were successively sister to the A. lycopodineus + A. trichoclados clade. Additionally, A. filicinus and A. tibeticus formed the subclade 2 (BS = 74; PP = 1.00) and were sister to the subclade 3, which included A. schoberioides, A. officinalis, A. neglectus, A. angulofractus, A. brachyphyllus, A. gobicus, A. dauricus, A. longiflorus, and A. oligoclonos (BS = 100; PP = 1.00). In subclade 3, A. schoberioides was the first to diverge from the remainders, followed by the A. officinalis + A. neglectus clade. Subsequently, A. angulofractus and A. schoberioide were successively sister to a clade in which the A. gobicus + A. dauricus clade was clustered with the A. longiflorus + A. oligoclonos clade.
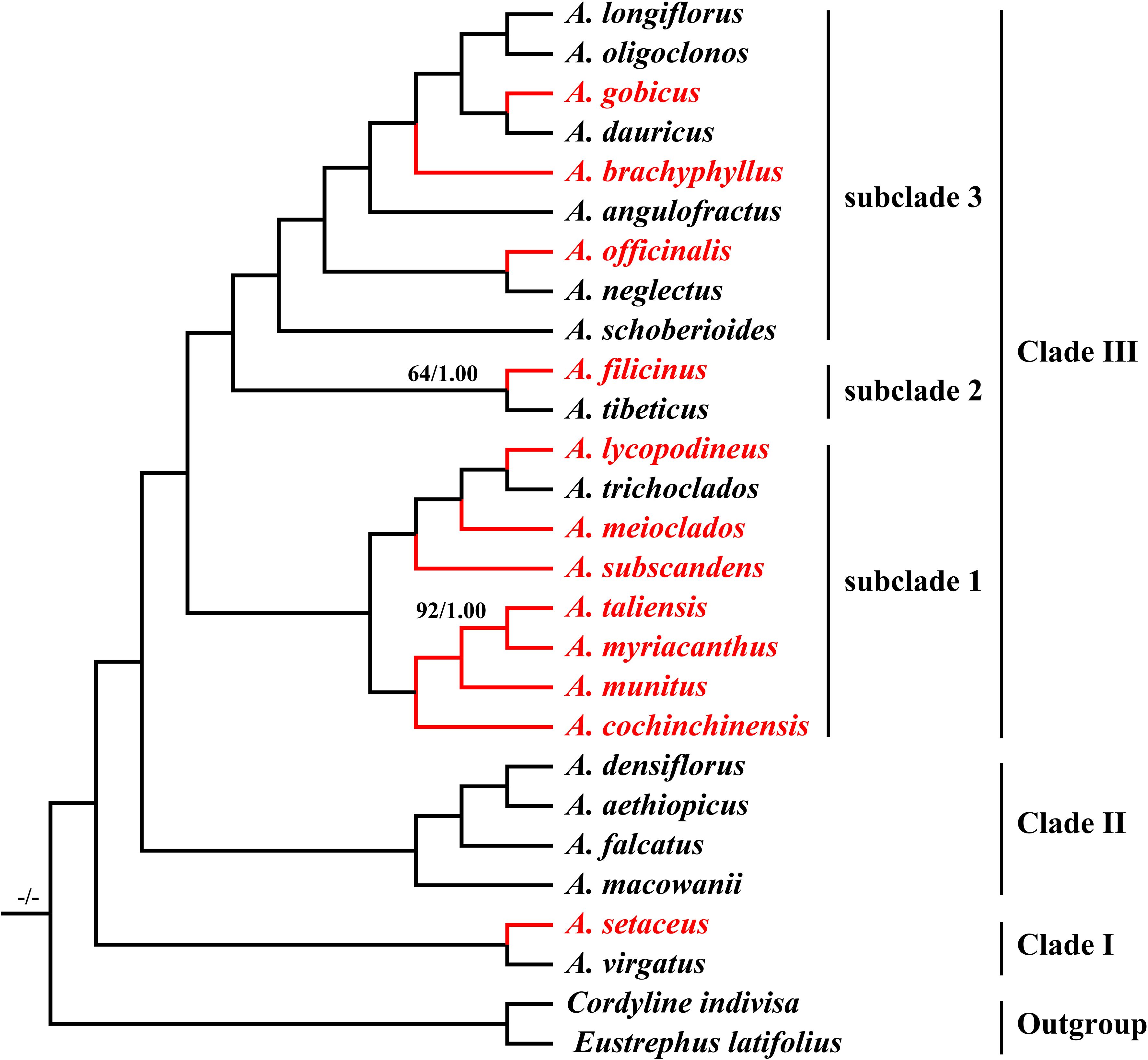
Figure 7. Phylogeny of the 25 Asparagus species inferred from maximum likelihood (ML) and Bayesian (BI) analyses based on the complete plastomes. The values above the nodes represent the ML bootstrap values (BS) and the posterior probabilities values (PP) and are shown only on branches with < 99% support. The highlighted red color represents the medicinal Asparagus species.
Discussion
Chloroplast genome structure
In this study, we conducted the sequencing, assembly, and annotation of 21 plastomes from Asparagus species and compared them with other four Asparagus plastomes. All Asparagus plastomes exhibited a typical quadripartite molecular structure consistent with those in Asparagaceae (Lu et al., 2022; Sheng, 2022; Van et al., 2022; Wang et al., 2022b; Wong et al., 2022; Huang et al., 2023; Tian et al., 2023; Wang et al., 2023). The pattern for the boundaries of the LSC/IRb, IRb/SSC, SSC/IRa, and IRa/LSC regions was also shared by all Asparagus plastomes, which was previously reported within Asparagaceae plastomes (Lu et al., 2022; Wang et al., 2022b; Huang et al., 2023). Furthermore, genome size, GC content, gene content, and order among Asparagus plastomes were highly similar. These phenomena indicated that the plastomes of Asparagus were highly conservative. Notably, the pseudogenization of the ycf15 gene, frequently documented across a wide range of lineages (Dong et al., 2013; Shi et al., 2013; Jiao et al., 2022), was found in all Asparagus plastomes, which may be the synapomorphy of the genus.
Simple sequence repeat and hotspot identification
In recent years, plastome-derived SSRs have been deemed a valuable resource for species authentication, clarifying evolutionary relationships, and assessing genetic diversity within plant populations (Provan et al., 2001; Ahmad et al., 2018; Zhou et al., 2023; Tu et al., 2024). Our study identified 79 to 95 SSRs in each Asparagus plastome, and types of SSRs, as well as most detected SSR motifs were also reported by previous studies (Sheng, 2022; Wong et al., 2022; Tian et al., 2023). On the whole, SSRs exhibited high levels of interspecific polymorphism across Asparagus plastomes. Firstly, the quantity of SSRs of the same type varied across different Asparagus species, especially for mono-nucleotide repeats. Secondly, there are differences in the composition of SSRs among Asparagus plastomes. Pentanucleotide repeats were found only in Clade III species, but not in all, while hexa-nucleotide repeats were relatively dispersed in Clade I to Clade III species. Moreover, even if the types of SSRs are the same, the specific SSR motifs in Asparagus plastomes were diverse (Supplementary Table 5). Through further comparison, a tatal of 12 special SSR motifs were found to be unique in specific species, which may serve as effective markers for species identification (Figure 2D).
Despite 15 regions showing significant sequence differences through mVISTA analysis, only seven regions (accD, rbcL, ycf1, trnS-trnG, ndhC-trnV, rpl32-trnL, ccsA-ndhD) corresponded to the results of the sliding-window analysis. Notably, the other regions exhibiting relatively high divergence yet low π values may be attributed to the presence of gaps within the compared matrix. Hence, we selected the above seven regions as candidate barcodes for Asparagus, with five (accD, ycf1, trnS-trnG, rpl32-trnL, and ndhC-trnV) reported by previous studies (Wong et al., 2022; Tian et al., 2023) and two (rbcL and ccsA-ndhD) firstly determined in this study. Noteworthy, despite exceeding 2,000 bp in length, the ycf1 sequence has been proposed as the most promising plastid DNA barcode of land plants (Dong et al., 2015). The successful design and implementation of taxon-specific PCR primers spanning the large locus ycf1 across different taxa (Neubig and Abbott, 2010; Parks et al., 2011; Dong et al., 2015) underscore its potential application as a marker for species identification. Nevertheless, the reliability of these sequences as DNA barcodes for species identification remains to be confirmed (Chao et al., 2023; Chac and Thinh, 2023) and requires more intra- and inter-species Asparagus sampling in future studies (Zhang et al., 2010; Gao et al., 2012).
Phylogeny inference
Although ITS and several plastome fragments have been widely utilized for the phylogenetic reconstruction of Asparagus, studies employing these data often produced phylogenetic trees with weak support or low resolution (Fukuda et al., 2005; Ou et al., 2010; Kubota et al., 2012; Ou et al., 2013). In this study, we performed phylogenetic analyses based on plastome data. As expected, the robust phylogenetic framework for Asparagus taxa was yielded, with high support values for nearly all nodes. All 25 Asparagus species were divided into three clades and the phylogenetic positions of most Asparagus species were largely congruent with findings from earlier researches (Kubota et al., 2012; Norup et al., 2015; Bentz et al., 2024). Apparently, medicinal Asparagus species were mainly distributed in Clade III, especially the subclade 1 (Figure 7). Notably, the close relationships between A. cochinchinensis, A. munitus, A. taliensis, and A. myriacanthus were identified, which was different from those of Wong et al. (2022) and Tian et al. (2023) but basically consistent with that of Bentz et al. (2024). Their affinities were further supported by similar morphological evidence—typically 3-angled cladodes, spinescent leaf spurs, and main stems with sharp spines. We also revealed the affinity between A. subscandens and the previously reported group including A. meioclados, A. lycopodineus, and A. trichoclados (Bentz et al., 2024). Similar leaf spur characteristics—either not spinescent or indistinctly spinescent—may justify their phylogenetic location found here. Additionally, it is worth noting that the swollen roots of many species in subclade 1, such as A. munitus, A. subscandens, A. lycopodineus, A. meioclados and A. taliensis, on the whole, have some similar medicinal value with that of A. cochinchinensis (Editorial Committee of Chinese Materia Medica of the National Administration of Traditional Chinese Medicine, 1999; Yunnan Food and Drug Administration, 2005; Sichuan Food and Drug Administration, 2010; Lv and Chen, 2019). We also identified a strong affinity between A. gobicus and A. dauricus (BS = 100; PP = 1.00). However, this finding was inconsistent with that of Bentz et al. (2024), who determined A. gobicus was closely related to A. kiusianus, a species endemic to Japan, and found A. dauricus to be sister to the A. breslerianus + A. longiflorus clade. These discrepancies may arise from differences in taxonomic sampling and molecular variations, highlighting the need for further investigation into their phylogenetic relationships. Overall, based on the robust phylogenetic framework, the relationships among the Chinese group of medicinal species and allied taxa within Asparagus were preliminarily elucidated.
Conclusion
In this study, we conducted more comprehensive analyses of the characteristics and structural variations of Asparagus plastomes than previously. These results indicate that the Asparagus plastomes are relatively conserved in terms of the structure, genome size, and gene content. Nevertheless, the quantities and types of SSRs exhibit high levels of interspecific polymorphism within Asparagus. The special SSR motifs and selected divergent hotspot regions from Asparagus plastomes may serve as potential molecular markers for future research on interspecific diversity and the identification of Asparagus. Our robust plastome-based phylogeny has provided preliminary insights into phylogenetic relationships among the Chinese group of medicinal species and related taxa within Asparagus. In conclusion, this study offers a wealth of informative genetic resources pertinent to Asparagus, significantly enhancing our comprehension of the evolution of this genus and laying a foundation for its identification, assessment of genetic population diversity, exploration and conservation of germplasm resources.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
PX: Data curation, Formal analysis, Investigation, Methodology, Resources, Visualization, Writing – original draft. TW: Data curation, Formal analysis, Investigation, Methodology, Resources, Visualization, Writing – original draft. JT: Data curation, Formal analysis, Investigation, Resources, Writing – review & editing. LF: Data curation, Resources, Investigation, Writing – review & editing. CL: Conceptualization, Project administration, Supervision, Writing – review & editing. HY: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Guangzhou Science and Technology Plan project (2024B03J1280).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1508898/full#supplementary-material
References
Afroz, S., Rahman, M. O., Hassan, M. A. (2022). Systematic studies of the genus asparagus tourn. Ex linn. (Liliaceae) in Bangladesh. Bangl J. Plant Taxon. 29, 97–107. doi: 10.3329/bjpt.v29i1.60451
Ahmad, A., Wang, J. D., Pan, Y. B., Sharif, R., Gao, S. J. (2018). Development and use of simple sequence repeats (SSRs) markers for sugarcane breeding and genetic studies. Agronomy. 8, 260. doi: 10.3390/agronomy8110260
Amel, H., Sara, J. C., Rocio, R. A., Ana, J. A., Rafael, G. B. (2020). “Chapter 8 - asparagus,” in Nutritional composition and antioxidant properties of fruits and vegetables. Ed. Jaiswal, A. K. (London, United Kingdom: Academic Press).
Amiryousefi, A., Hyvönen, J., Poczai, P. (2018). IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 34, 3030–3031. doi: 10.1093/bioinformatics/bty220
Bentz, P. C., Liu, Z., Yang, J. B., Zhang, L., Burrows, S., Burrows, J., et al. (2024). Young evolutionary origins of dioecy in the genus Asparagus. Am. J. Bot. 111, e16276. doi: 10.1002/ajb2.v111.2
Borsch, T., Quandt, D., Koch, M. (2009). Molecular evolution and phylogenetic utility of non-coding DNA: applications from species to deep level questions. Plant Syst. Evol. 282, 107–108. doi: 10.1007/s00606-008-0115-y
Chac, L. D., Thinh, B. B. (2023). Species identification through DNA barcoding and its applications: A review. Biol. Bull. 50, 1143–1156. doi: 10.1134/S106235902360229X
Chao, Z., Hong, X., Xie, X., Huang, R., Tian, E. (2023). Phylogenomic analysis of Bupleurum in Western Sichuan, China, including an overlooked new species. Front. Plant Sci. 14, 1294670. doi: 10.3389/fpls.2023.1294670
Chen, S., Zhou, Y., Chen, Y., Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Chen, X. Q., Tamanian, K. G. (2000). “Asparagu,” in Flora of China, vol 24 (Flagellariaceae through Marantaceae). Ed. Wu Raven, Z. Y. P. H. (Beijing: Science and MO Botanical Garden, USA: St. Louis, MO).
Chinese Pharmacopoeia Commission (2020). Pharmacopoeia of the people’s republic of China, vol 1 (Beijing: China Medical Science and Technology Press).
Darling, A. C., Mau, B., Blattner, F. R., Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
Dierckxsens, N., Mardulyn, P., Smits, G. (2017). NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45, e18. doi: 10.1093/nar/gkw955
Dong, W., Xu, C., Cheng, T., Zhou, S. (2013). Complete chloroplast genome of Sedum sarmentosum and chloroplast genome evolution in Saxifragales. PloS One 8, e77965. doi: 10.1371/journal.pone.0077965
Dong, W., Xu, C., Li, C., Sun, J., Zuo, Y., Shi, S., et al. (2015). ycf1, the most promising plastid DNA barcode of land plants. Sci. Rep. 5, 8348. doi: 10.1038/srep08348
Downie, S. R., Jansen, R. K. (2015). A comparative analysis of whole plastid genomes from the Apiales: Expansion and contraction of the inverted repeat, mitochondrial to plastid transfer of DNA, and identifcation of highly divergent noncoding regions. Syst. Bot. 40, 336–351. doi: 10.1600/036364415X686620
Doyle, J. J., Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15.
Editorial Committee of Chinese Materia Medica of the National Administration of Traditional Chinese Medicine (1999). Chinese matea medica (Zhonghua bencao), vol. 8 (Shanghai: Shanghai Science & Technology Press).
Frazer, K. A., Pachter, L., Poliakov, A., Rubin, E. M., Dubchak, I. (2004). VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32, W273–W279. doi: 10.1093/nar/gkh458
Fukuda, F., Ashizawa, H., Suzuki, R., Ochiai, T., Nakamura, T., Kanno, A., et al. (2005). Molecular phylogeny of the genus Asparagus (Asparagaceae) inferred from plastid petB intron and petD-rpoA intergenic spacer sequences. Plant Species Biol. 20, 121–132. doi: 10.1111/j.1442-1984.2005.00131.x
Gao, L. M., Jie, L. J., Cai, J., Yang, J. B., Zhang, T., Li, D. Z. (2012). A synopsis of technical notes on the standards for plant DNA barcoding. Plant Diversity 34, 592–606. doi: 10.3724/SP.J.1143.2012.12138
Gitzendanner, M. A., Soltis, P. S., Wong, G. K., Ruhfel, B. R., Soltis, D. E. (2018). Plastid phylogenomic analysis of green plants: a billion years of evolutionary history. Am. J. Bot. 105, 291–301. doi: 10.1002/ajb2.2018.105.issue-3
Givnish, T. J., Zuluaga, A., Spalink, D., Soto, Gomez, M., Lam, V. K. Y., Saarela, J. M., et al. (2018). Monocot plastid phylogenomics, timeline, net rates of species diversification, the power of multi-gene analyses, and a functional model for the origin of monocots. Am. J. Bot. 105, 1888–1910. doi: 10.1002/ajb2.2018.105.issue-11
Greiner, S., Lehwark, P., Bock, R. (2019). OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47, W59–W64. doi: 10.1093/nar/gkz238
Guo, X., Shi, N., Xie, P., Zhang, G., Liu, H., Ji, Y. (2022). Plastome sequencing for accurate and effective authentication of Polygonatum kingianum (Asparagaceae). Ind. Crops Prod. 184, 115056. doi: 10.1016/j.indcrop.2022.115056
Huang, J., Lu, Z., Lin, C., Xu, W., Liu, Y. (2023). Comprehensive comparative analyses of aspidistra chloroplast genomes: insights into interspecific plastid diversity and phylogeny. Genes. 14, 1894. doi: 10.3390/genes14101894
Huang, Y., Wang, Y. R., Tang, Y., Liu, J. M., Li, B., Ou, L. J. (2010). TrnL-F Sequence analysis of 4 species of Asparagus. Huaihua Xueyuan Xuebao. 29, 63–65.
Jansen, R. K., Ruhlman, T. A. (2012). “Plastid genomes of seed plants,” in Genomics of chloroplasts and mitochondria, vol. Vol 35 . Eds. Bock, R., Knoop, V. (Springer, Dordrecht. Netherlands).
Ji, Y., Landis, J. B., Yang, J., Wang, S., Zhou, N., Luo, Y., et al. (2023). Phylogeny and evolution of Asparagaceae subfamily Nolinoideae: new insights from plastid phylogenomics. Am. J. Bot. 131, 301–312. doi: 10.1093/aob/mcac144
Jiao, Y., Feng, G., Huang, L., Nie, G., Li, Z., Peng, Y., et al. (2022). Complete chloroplast genomes of 14 subspecies of D. glomerata: Phylogenetic and comparative genomic analyses. Genes. 13, 1621. doi: 10.3390/genes13091621
Katoh, K., Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kubota, S., Konno, I., Kanno, A. (2012). Molecular phylogeny of the genus Asparagus (Asparagaceae) explains interspecific crossability between the garden asparagus (A. officinalis) and other Asparagus species. Theor. Appl. Genet. 124, 345–354. doi: 10.1007/s00122-011-1709-2
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547. doi: 10.1093/molbev/msy096
Li, H. R., Huang, Y. (2021). A brief introduction of Yi medicines for treatment of “Sise” and their types of “Sise” treated. Chin. J. Ethn Med. 27, 47–52. doi: 10.3969/j.issn.1006-6810.2021.10.021
Librado, P., Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Liu, H., He, J., Ding, C., Lyu, R., Pei, L., Cheng, J., et al. (2018). Comparative analysis of complete chloroplast genomes of Anemoclema, Anemone, Pulsatilla, and Hepatica revealing structural variations among genera in Tribe Anemoneae (Ranunculaceae). Front. Pl Sci. 9, 1097. doi: 10.3389/fpls.2018.01097
Lu, Q. X., Chang, X., Gao, J., Wu, X., Wu, J., Qi, Z. C., et al. (2022). Evolutionary comparison of the complete chloroplast genomes in convallaria species and phylogenetic study of asparagaceae. Genes. 13, 1724. doi: 10.3390/genes13101724
Lv, X. Y., Chen, A. M. (2019). Research status and prospects on cultivation and processing technique of Asparagus cochinchinensis in Neijiang Area. Mod Agric. Sci. Technol. 14, 77–79. doi: 10.3969/j.issn.1007-5739.2019.14.047
Lyko, P., Wicke, S. (2021). Genomic reconfiguration in parasitic plants involves considerable gene losses alongside global genome size inflation and gene births. Plant Physiol. 186, 1412–1423. doi: 10.1093/plphys/kiab192
Ma, P. F., Zhang, Y. X., Zeng, C. X., Guo, Z. H., Li, D. Z. (2014). Chloroplast phylogenomic analyses resolve deep-level relationships of an intractable bamboo tribe Arundinarieae (Poaceae). Syst. Biol. 63, 933–950. doi: 10.1093/sysbio/syu054
Neubig, K. M., Abbott, J. R. (2010). Primer development for the plastid region ycf1 in Annonaceae and other magnoliids. Am. J. Bot. 97, e52–e55. doi: 10.3732/ajb.1000128
Norup, M. F., Petersen, G., Burrows, S., Bouchenak-Khelladi, Y., Leebens-Mack, J., Pires, J. C., et al. (2015). Evolution of Asparagus L. (Asparagaceae): Out-of-South-Africa and multiple origins of sexual dimorphism. Mol. Phylogenet Evol. 92, 25–44. doi: 10.1016/j.ympev.2015.06.002
Ou, L. J., Sun, H. J., Wei, G., Wang, Z. X. (2013). Comparison of trnH-psbA sequences of some species in Asparagus. Shizhen Guoyi Guoyao. 24, 2678–2679. doi: 10.3969/j.issn.1008-0805.2013.11.043
Ou, L. J., Ye, W., Zheng, G. P., Jiang, X. H., Shen, C. W., Xu, D., et al. (2010). Comparison of rDNA internal transcribed spacer sequences in Asparagus. Zhongyaocai. 33, 1542–1549.
Parihar, S., Sharma, D. (2021). A brief overview on Asparagus racemous. Int. J. Res. Anal. Rev. 8, 96–108.
Parks, M., Liston, A., Cronn, R. (2011). Newly developed primers for complete ycf1 amplification in Pinus (Pinaceae) chloroplasts with possible family-wide utility. Am. J. Bot. 98, e185–e188. doi: 10.3732/ajb.1100088
Posada, D., Crandall, K. A. (1998). Modeltest: testing the model of DNA substitution. Bioinformatics. 14, 817–818. doi: 10.1093/bioinformatics/14.9.817
Provan, J., Powell, W., Hollingsworth, P. M. (2001). Chloroplast microsatellites: New tools for studies in plant ecology and evolution. Trends Ecol. Evol. 16, 142–147. doi: 10.1016/S0169-5347(00)02097-8
Rambaut, A. (2014). FigTree v1. 4.2, a graphical viewer of phylogenetic trees. Available online at: http://tree.bio.ed.ac.uk/software/figtree/ (Accessed January 09 2023).
Ravi, V., Khurana, J. P., Tyagi, A. K., Khurana, P. (2008). An update on chloroplast genomes. Plant Syst. Evol. 271, 101–122. doi: 10.1007/s00606-007-0608-0
Ronquist, F., Teslenko, M., Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Sheng, W. (2022). The entire chloroplast genome sequence of Asparagus cochinchinensis and genetic comparison to Asparagus species. Open Life Sci. 17, 893–906. doi: 10.1515/biol-2022-0098
Shi, L., Chen, H., Jiang, M., Wang, L., Wu, X., Huang, L., et al. (2019). CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47, W65–W73. doi: 10.1093/nar/gkz345
Shi, C., Liu, Y., Huang, H., Xia, E. H., Zhang, H. B., Gao, L. Z. (2013). Contradiction between plastid gene transcription and function due to complex posttranscriptional splicing: an exemplary study of ycf15 function and evolution in angiosperms. PloS One 3), e59620. doi: 10.1371/journal.pone.0059620
Sichuan Food and Drug Administration (2010). Sichuan chinese materia medica standards. 2010 edition (Chengdu: Sichuan Science & Technology Press).
Stevens, P. F. (2024). Angiosperm phylogeny website. Available online at: http://www.mobot.org/MOBOT/research/APweb/ (Accessed 9 May 2024).
Tian, Y., Liu, X., Xu, Y., Yu, B., Wang, L., Qu, X. (2023). Comparative and phylogenetic analysis of Asparagus meioclados Levl. and Asparagus munitus Wang et SC Chen plastomes and utility of plastomes mutational hotspots. Sci. Rep. 13, 15622. doi: 10.1038/s41598-023-42945-x
Tu, X. D., Lin, W. J., Fu, H. H., Lin, Y. Z., Shen, J., Chen, S., et al. (2024). Comparative analysis of six complete plastomes of tripterospermum spp. Int. J. Mol. Sci. 25, 2534. doi: 10.3390/ijms25052534
Van, K., Larridon, I., Shah, T., Bauters, K., Asselman, P., Goetghebeur, P., et al. (2022). Plastid phylogenomics of the Sansevieria Clade of Dracaena (Asparagaceae) resolves a recent radiation. Mol. Phylogenet Evol. 169, 107404. doi: 10.1016/j.ympev.2022.107404
Wang, J., Qian, J., Jiang, Y., Chen, X., Zheng, B., Chen, S., et al. (2022b). Comparative analysis of chloroplast genome and new insights into phylogenetic relationships of polygonatum and tribe polygonateae. Front. Plant Sci. 13,882189. doi: 10.3389/fpls.2022.882189
Wang, M., Wang, S., Hu, W., Wang, Z., Yang, B., Kuang, H. (2022a). Asparagus cochinchinensis: A review of its botany, traditional uses, phytochemistry, pharmacology, and applications. Front. Pharmacol. 13, 1068858. doi: 10.3389/fphar.2022.1068858
Wang, S., Zhou, N., Shi, N., Zhang, G., Liu, H., Guo, X., et al. (2023). Testing and using complete plastomes for authentication of medicinal Polygonatum species (Asparagaceae). Ind. Crops Prod. 197, 116557. doi: 10.1016/j.indcrop.2023.116557
Wong, K. H., Kong, B. L. H., Siu, T. Y., Wu, H. Y., But, G. W. C., Shaw, P. C., et al. (2022). Complete chloroplast genomes of Asparagus aethiopicus L., A. densiflorus (Kunth) Jessop ‘Myers’, and A. cochinchinensis (Lour.) Merr.: Comparative and phylogenetic analysis with congenerics. PloS One 17, e0266376. doi: 10.1371/journal.pone.0266376
Wu, L., Nie, L., Wang, Q., Xu, Z., Wang, Y., He, C., et al. (2021). Comparative and phylogenetic analyses of the chloroplast genomes of species of Paeoniaceae. Sci. Rep. 11, 14643. doi: 10.1038/s41598-021-94137-0
Yunnan Food and Drug Administration (2005). Yunnan chinese materia medica standards. 2005 edition Vol. Vol. 3 (Kunming: Dai medicine). Yunnan Science & Technology Press).
Zhang, A. B., He, L. J., Crozier, R. H., Muster, C., Zhu, C. D. (2010). Estimating sample sizes for DNA barcoding. Mol. Phylogenet Evol. 54, 1035–1039. doi: 10.1016/j.ympev.2009.09.014
Keywords: Asparagus, plastid genome, simple sequence repeats, divergent hotspots, phylogenetic relationships, species identification
Citation: Xie P, Wang T, Tan J, Fan L, Liu C and Yan H (2025) Plastid genome comparison and phylogenetic analyses of the Chinese group of medicinal species and related taxa within Asparagus genus. Front. Plant Sci. 16:1508898. doi: 10.3389/fpls.2025.1508898
Received: 10 October 2024; Accepted: 02 January 2025;
Published: 27 January 2025.
Edited by:
Gabriele Casazza, University of Genoa, ItalyReviewed by:
Yunheng Ji, Chinese Academy of Sciences (CAS), ChinaZhi Chao, Southern Medical University, China
Wentao Sheng, Nanchang Normal University, China
Copyright © 2025 Xie, Wang, Tan, Fan, Liu and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hanjing Yan, eWFuaGFuamluZzEyMTFAMTYzLmNvbQ==; Changkun Liu, bGl1Y2hhbmdrdW5AY3h0Yy5lZHUuY24=
†These authors have contributed equally to this work
 Pingxuan Xie
Pingxuan Xie Tinglu Wang
Tinglu Wang Jiawei Tan1
Jiawei Tan1 Changkun Liu
Changkun Liu Hanjing Yan
Hanjing Yan