
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 13 March 2025
Sec. Plant Bioinformatics
Volume 16 - 2025 | https://doi.org/10.3389/fpls.2025.1500429
Zinc finger protein (ZFP) represent a significant class of transcription factors in plants, involved in various functions, including tissue development, signal transduction, and responses to both biotic and abiotic stresses. ZFPs are categorized into 10 distinct subfamilies, among which the C3H gene family is recognized as a functionally significant group of transcription factors.To date, no studies have been reported regarding the C3H gene family in melon (Cucumis melo). In this study, 38 CmC3H genes were identified in the melon genome, and these genes are unevenly distributed across the 12 chromosomes. Phylogenetic analysis classified the C3H family members into four groups, with significant differences observed in sequence, protein motifs, and gene structure among CmC3H genes within the same group. The CmC3H family contains one pair of segmentally duplicated genes and shares 20, 7, 39, and 38 pairs of homologous C3H genes with Arabidopsis thaliana, rice (Oryza sativa), cucumber (Cucumis sativus), and watermelon (Citrullus lanatus), respectively. Promoter region analysis revealed a high abundance of cis-elements associated with growth and development, hormone regulation, and stress responses. Expression profiling revealed that CmC3H family members exhibit significant tissue-specific expression patterns. Quantitative PCR analysis indicated that six genes (CmC3H4, CmC3H7, CmC3H13, CmC3H24, CmC3H33, and CmC3H38) may play roles in melon’s drought stress resistance. Heavy metal lead stress appears to suppress the expression of CmC3H genes. The genes CmC3H24 and CmC3H33 may be involved in regulating melon’s resistance to Fusarium wilt infection. CmC3H11 and CmC3H21 can be considered as the key candidate genes for improving the melon’s ability to resist both biotic and abiotic stresses.This study provides preliminary insights into the expression profiles of CmC3H genes under drought stress, heavy metal lead stress, and Fusarium wilt infection, offering a theoretical foundation for the molecular mechanisms underlying melon improvement and stress resistance.
Zinc finger proteins (ZFPs) represent one of the largest and most specific families of transcription factors in plants. ZFPs are classified into 10 major types based on the number of cysteine and histidine residues and the spacing of amino acids between them: C2H2, C2HC, C2HC5, C2C2, C3H, C3HC4, C4, C4HC3, C6, and C8 (Kamaliyan and Clarke, 2024). The C3H gene family, a significant subgroup of this group, is characterized by the presence of one or more C3H-type zinc finger domains, each consisting of three cysteine and one histidine residue bound to a zinc ion. This characteristic C3H-type zinc finger motif is commonly found in eukaryotes and is named for its distinctive structure (Bogamuwa and Jang, 2014; Liu et al., 2022). Initially, the consensus sequence of the C3H motif was defined as C-X6-14-C-X4-5-C-X3-H (where X represents any amino acid), based on variations in the spacing of amino acids between cysteine and histidine in the motif. Subsequent studies have redefined the C3H motif as C-X4-17-C-X4-6-C-X3-H (Berg and Shi, 1996; Rameneni et al., 2018). C3H genes play crucial roles in both plants and animals by specifically binding to RNA or DNA, participating in various biological processes, including the regulation of gene expression, RNA processing, disease response, and adaptation to environmental stresses (Cheng et al., 2020; Velásquez et al., 2024). Research has demonstrated that members of the C3H gene family are essential for processes such as photoperiod regulation, hormone signaling, and disease resistance in plants, particularly in modulating responses to stresses like drought, salinity, and pathogen infection (Han et al., 2021). Advancements in genomics and functional genomics technologies have facilitated the identification of additional C3H gene family members, providing new insights into their roles in plant physiology and molecular mechanisms (Ai et al., 2022).
To date, the C3H gene family has been studied in several plants, including A. thaliana (Wang et al., 2008), rice (Wang et al., 2008), maize (Peng et al., 2012), alfalfa (Zhang et al., 2013), citrus (Liu et al., 2014), and poplar (Chai et al., 2012), with the functions of several C3H genes validated across species. In A. thaliana, the overexpression of AtC3H17, AtC3H29, and AtC3H47 has been shown to enhance salt tolerance (Seok et al., 2018; Sun et al., 2007), whereas the overexpression of AtC3H49 improves tolerance to drought as well as salt stress (Lee et al., 2012). Overexpression of GhZFP1 in transgenic tobacco significantly enhances salt tolerance by influencing Na+ homeostasis and K+ acquisition (Guo et al., 2009). The overexpression of BoC3H improves salt tolerance in transgenic cabbage (Jiang et al., 2017), while PeC3H74 transgenic Arabidopsis plants exhibit stronger drought tolerance compared to wild-type plants (Chen et al., 2020a). Regarding biotic stress in plants, the nuclear protein GHZFP1 in cotton interacts with GZIRD21A and GZIPR5, contributing to enhanced fungal disease resistance (Guo et al., 2009). In chili pepper, the C3H gene CaC3H1 modulates the antagonistic interaction between salicylic acid (SA) and jasmonic acid (JA)/ethylene (ET) signaling pathways, thus enhancing resistance to bacterial wilt caused by Ralstonia solanacearum (Lei, 2014). In A. thaliana, the C3H15 gene has been found to negatively regulate cell elongation by inhibiting brassinosteroid signaling (Chai et al., 2022). In rice, the C3H-type genes OsLIC and OsDOS are involved in the regulation of brassinosteroid signaling, affecting structural development and delaying leaf senescence, respectively (Wang et al., 2008; Kong et al., 2006). ZmC3H9 (maize) is implicated in the regulation of phenolic compound biosynthesis (Abnave et al., 2024). The GhTZF1 gene regulates leaf senescence in cotton (Zhou et al., 2014). PEI1 plays a crucial role during Arabidopsis embryogenesis and is an embryo-specific C3H zinc finger gene that primarily functions in the apical domain of the embryo (Li and Thomas, 1998). The C3H gene AtC3H14 (At1g66810) has been identified as a key regulator of secondary cell wall biosynthesis in A. thaliana (Ko et al., 2009).
Melon (C. melo L.) is a member of the Cucurbitaceae family and is valued for its juicy fruit, rich in carbohydrates and nutrients, making it economically important. However, melon cultivation is often hindered by various stress factors that reduce fruit yield and quality. In Henan Province, China, melon is widely cultivated, but its production is frequently threatened by Fusarium wilt, which can cause plant mortality. Moreover, Henan Luoyang, situated in the middle reaches of the Yellow River, experiences a semi-arid to semi-humid climate(Shen et al., 2023). Melon cultivation in this region is highly susceptible to drought stress. Industrial development has also led to heavy metal pollution in some soils, further compromising melon growth and development. Despite these challenges, no research has been conducted on the C3H gene family in melon. Melon, the second Cucurbitaceae crop to have its genome sequenced, has a chromosome number of 2n = 2x = 24 and an approximate genome size of 450 Mb (Garcia-Mas et al., 2012). The updated, high-quality melon genome sequence provides an opportunity for the identification and characterization of the CmC3H family. Thus, this study aims to identify members of the C3H gene family in melon and perform bioinformatic analyses, including assessments of protein physicochemical properties, phylogeny, conserved motifs, gene structure, gene duplication, cis-elements, and expression patterns. Additionally, the study investigates the expression of 10 selected CmC3H genes in the ‘Super Sweet White Sugar’ melon variety under drought stress, lead (Pb) heavy metal stress, and Fusarium wilt infection using quantitative PCR analysis. This research provides valuable insights into the structure and function of CmC3H genes.
The melon genome sequence DHL92 v4.0 was downloaded from the CuGenDB database (http://cucurbitgenomics.org/) to identify C3H genes (Garcia-Mas et al., 2012). Using the Hidden Markov Model (HMM) profile PF00642 (https://www.ebi.ac.uk/interpro/entry/pfam/#table), a genome-wide search was conducted on the melon genome protein data using the HMMER tool, retaining genes with an e-value of less than or equal to e−5 (Potter et al., 2018). Additionally, 50 Arabidopsis C3H protein sequences were downloaded from the UniProt database (https://www.uniprot.org/) (UniProt Consortium, 2021) and subjected to a BLAST search against the Arabidopsis C3H protein sequences with an e-value of less than or equal to e−5 (Ye et al., 2006). After removing redundant sequences, the remaining dataset could be used for further analysis. Ultimately, 38 C3H genes were identified in the melon genome, and their chromosomal distribution was mapped using the TBtools (V2.131) (Chen et al., 2020b). Following the same methodology, 35 and 38 C3H members were identified in cucumber and watermelon, respectively. Protein physicochemical properties were analyzed using ExPASy (http://web.expasy.org/protparam/) (Wilkins et al., 1999), and subcellular localization was predicted using WoLF PSORT II (https://www.genscript.com/wolf-psort.html?src=leftbar) (Horton et al., 2007). Finally, a phylogenetic tree of the melon C3H gene family was constructed following a previous study (Deng et al., 2023). This process involved using MEGA 11 software to perform a ClustalW multiple sequence alignment of the CmC3H and AtC3H sequences and constructing the phylogenetic tree with the Neighbor-Joining (NJ) method, with 1,000 bootstrap replications, using the Poisson correction model and the pairwise deletion option (Deng et al., 2023).
Conserved motifs were identified in the 38 CmC3H protein sequences using the MEME tool (http://meme-suite.org/tools/meme), specifying a maximum of 10 motifs with default parameters (Bailey et al., 2009). The positions and numbers of C3H domains within the CmC3H protein sequences were analyzed using the NCBI CDD online tool (https://www.ncbi.nlm.nih.gov/cdd/) (Marchler-Bauer et al., 2015). Exon and intron positions and numbers within the CmC3H genes were identified based on melon genome data. The phylogenetic tree, conserved motifs, domains, and gene structures of CmC3H family members were subsequently clustered and visualized through TBtools (V2.131).
Gene duplication, including segmental and tandem duplications, among the 38 CmC3H members was analyzed using the MCScanX tool (Wang et al., 2012). Constructing chromosome homology maps using SRplot online (https://www.bioinformatics.com.cn/?keywords=circos) Rcircos circle diagram tool (Tang et al., 2023a). Homologous C3H gene pairs between melon and Arabidopsis, rice, cucumber, and watermelon were identified. The Ka (nonsynonymous substitution rate), Ks (synonymous substitution rate), and Ka/Ks ratios of all duplicated genes were computed through the KaKs_Calculator tool(Version 3.0) (Zhang et al., 2006).
Promoter sequences (2,000 bp upstream) of the 38 CmC3H genes were extracted and annotated to identify cis-elements through the PlantCARE online tool (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002). The positional distribution of cis-elements related to abiotic and biotic stresses, phytohormone responsiveness, and plant growth and development was mapped according to previous studies (Zhang et al., 2024).
The expression patterns of the CmC3H family members were analyzed by retrieving FPKM (Fragments Per Kilobase of exon model per Million mapped fragments) values from the melon transcriptome database (https://melonet-db.dna.affrc.go.jp/ap/top) across eleven distinct tissues: dry seeds, root, middle stem, upper stem, leaves, tendril, petal, stigma, ovary at 4 Days After Flowering (DAF4), fruit flesh at 50 Days After Flowering (DAF50), and fruit epicarp at 50 Days After Flowering (DAF50) (Yu et al., 2023). The expression pattern of the CmC3H family was visualized using TBtools (V2.131). In this figure, the expression levels are represented by color intensity, with darker colors corresponding to higher expression levels.
The protein sequences of CmC3H family members were uploaded to the STRING database (https://string-db.org/) for node comparison, with interactions among CmC3H members predicted based on Arabidopsis protein interaction data (Szklarczyk et al., 2023). The binding motif profile for the Arabidopsis C3H12 transcription factor (MA1756.2) was obtained from the JASPAR Plantae database (https://jaspar.elixir.no/search?q=&collection=CORE&tax_group=plants) (Castro-Mondragon et al., 2022). Subsequently, the 2,000 bp promoter sequences of all genes in the melon genome were extracted and analyzed using the motif FIMO tool (https://meme-suite.org/meme/) to detect genes potentially binding with the C3H transcription factor. Finally, target gene domain prediction was performed using the PFAM database (Mistry et al., 2021), and KEGG (Kyoto Encyclopedia of Genes and Genomes) and GO (Gene Ontology) enrichment analyses were conducted on the target genes using OmicShare Tools (https://www.omicshare.com/tools).
The seeds of the melon variety ‘Super Sweet White Sugar Jar’ were soaked, germinated, and then sown. The plants were grown in a greenhouse under a 16-hour light/8-hour dark photoperiod, with temperatures maintained at 28°C during the day and 18°C at night, until they reached the three-leaf stage. Fusarium oxysporum was isolated from wilted melon plants, and a spore suspension with a concentration of 1×108 spores/mL was prepared. The plants were inoculated via root irrigation, applying 10 mL of spore suspension per plant. Root, middle stem, and first true leaf samples were collected at 0 hours (CK), 12 hours, 24 hours, and 48 hours post-inoculation.
Drought stress treatment was applied by placing the three-leaf stage melon plants in a 15% PEG6000 solution, and leaf samples were collected at 0 hours (CK), 12 hours, 24 hours, and 48 hours. For heavy metal lead stress treatment, Pb (NO3)2 was dissolved in distilled water and applied to the pots, bringing the Pb ion concentration in the soil to 2000 mg/kg. Leaf samples were collected at 0 hours (CK), 12 hours, 24 hours, and 48 hours. All experiments were conducted with three biological replicates, and the collected samples were immediately placed in liquid nitrogen and stored at -80°C for further analysis.
The expression levels of 10 CmC3H genes at four time points (0 hours, 12 hours, 24 hours, and 48 hours) under drought stress (leaves; three biological replicates), heavy metal lead stress (leaves; three biological replicates), and Fusarium wilt infection (roots, stems, and leaves; three biological replicates) were analyzed using quantitative real-time PCR (qRT-PCR).The qRT-PCR primers for the selected CmC3H genes were designed using Primer Premier 5 (Singh et al., 1998). Total RNA was extracted from all samples using the Spectrum™ Plant Total RNA Kit (Merck KGaA), with the quality and concentration assessed using a Nanodrop 2000. First-strand cDNA was synthesized using the SweScript All-in-One First Strand cDNA Synthesis kit (TRANS, G3337). The qPCR reactions were performed using a CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA) with 2×SYBR Green qPCR Master Mix (no ROX) (TRANS, G3320). The cycling conditions were set as follows: 95°C for 30 seconds of pre-denaturation, followed by 40 cycles of 95°C for 15 seconds and 60°C for 30 seconds. The LOC103490203 gene was used as the internal reference gene in melon, and the relative expression levels of the target genes were calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001). Differences among treatments were assessed by the least significance difference using one-way analysis in SPSS 19.0 (SPSS China, Beijing, China).
The subcellular localization of the target gene was predicted using the WOLF PSORT website. The coding sequence of CmC3H21 was cloned into the pBI121-GFP vector to construct the pBI121-CmC3H21-GFP recombinant plasmid. Subsequently, the pBI121-CmC3H21-GFP plasmid and mCherry (a nuclear marker) were introduced into Agrobacterium tumefaciens strain GV3101. The Agrobacterium culture was activated and grown until the OD600 reached 1.5, followed by centrifugation to remove the supernatant. The pellet was resuspended in an infiltration buffer (10 mM MES, 10 mM MgCl2, and 200 µM acetosyringone) and vortexed to adjust the OD600 to 1.0. The resulting suspension was infiltrated into the abaxial side of leaves from approximately five-week-old tobacco (Nicotiana benthamiana) seedlings. After infiltration, the plants were kept in the dark at 22°C for 48 hours. The infiltrated leaves were then excised and imaged using a confocal laser scanning microscope (TCS SP8, Leica) to visualize the subcellular localization of the fluorescently tagged proteins.
A total of 38 C3H genes were identified in the melon genome, and these are unevenly distributed across 12 chromosomes (Figure 1). Specifically, the genes are distributed as follows: 5, 6, 2, 5, 1, 3, 4, 5, 1, 1, 3, and 2 on chromosomes Chr01 to Chr12, respectively. These genes were renamed CmC3H1 through CmC3H38 based on their chromosomal locations. The physicochemical properties of the CmC3H family members were further analyzed (Supplementary Table S1) (“aa” denotes amino acids, and “Da” refers to Daltons). The analysis indicated that the number of amino acids in CmC3H members ranges from 202 aa (CmC3H1) to 1034 aa (CmC3H25), with molecular weights varying from 22128.07 Da (CmC3H1) to 114623.34 Da (CmC3H25). The Instability Index ranges from 28.7 (CmC3H16) to 82.26 (CmC3H29), while the Aliphatic Index varies from 36.53 (CmC3H31) to 78.96 (CmC3H16). All CmC3H members are hydrophilic proteins. The theoretical isoelectric points (pI) range from 4.92 to 9.65, with 17 members having a pI below 7 and 21 members having a pI above 7. Subcellular localization analysis revealed that three genes, CmC3H9, CmC3H20, and CmC3H31, lack a start codon, preventing the prediction of their subcellular localization. Among the remaining members, 31 are predicted to localize in the nucleus, 3 in the chloroplast, and 1 in the cytoplasm.
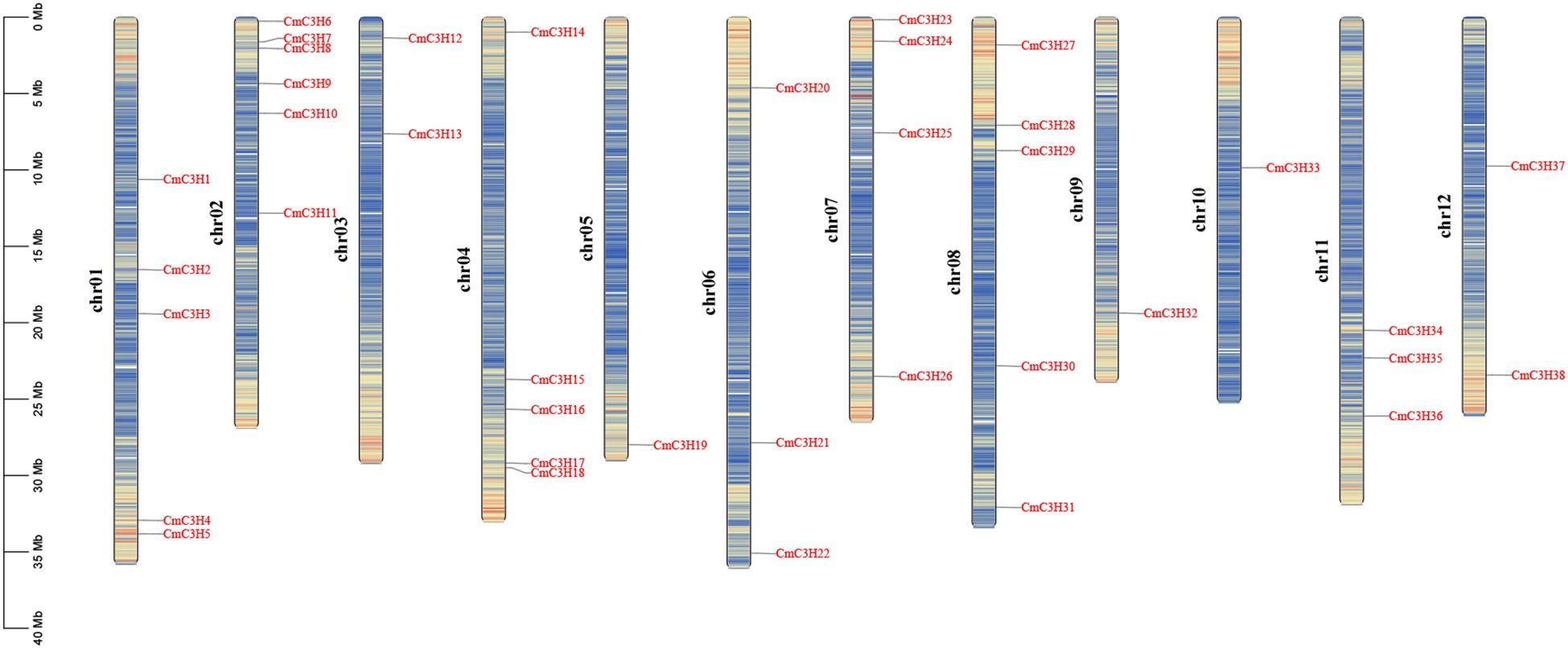
Figure 1. Chromosomal distribution of CmC3H family members. The left scale represents the chromosome length, with “chr” denoting the chromosome. Gene density on each chromosome was calculated over a genetic distance of 200 kb and is represented by a gradient color from blue (low gene density) to red (high gene density). White areas indicate regions lacking gene distribution information.
A Neighbor-Joining phylogenetic tree of C3H family members from A. thaliana, melon (C. melo), cucumber (C. sativus), and watermelon (C. lanatus) was constructed using MEGA 11. The resulting tree was categorized into four groups: Group I, Group II, Group III, and Group IV (Figure 2; Supplementary Table S2).Groups I, II, III, and IV contained 42, 33, 26, and 60 members, respectively. The CmC3H genes were distributed across these groups, with 10, 9, 5, and 14 genes in Groups I to IV, respectively, accounting for 26.3%, 23.7%, 13.2%, and 36.8% of the total. The C3H family consisted of 38 members, while cucumber and watermelon had 35 and 38 C3H genes, respectively, indicating similar gene numbers among the three species. Clustering analysis revealed that C3H genes in melon, cucumber, and watermelon were highly conserved, suggesting strong evolutionary conservation among these closely related species. Analysis of motifs and domains in the CmC3H family revealed that members within the same subgroup shared similar motif types and domain structures. These findings support the phylogenetic subgroup classification based on evolutionary analysis and further validate the rationale behind subgroup categorization in the evolutionary tree (Liu et al., 2024).
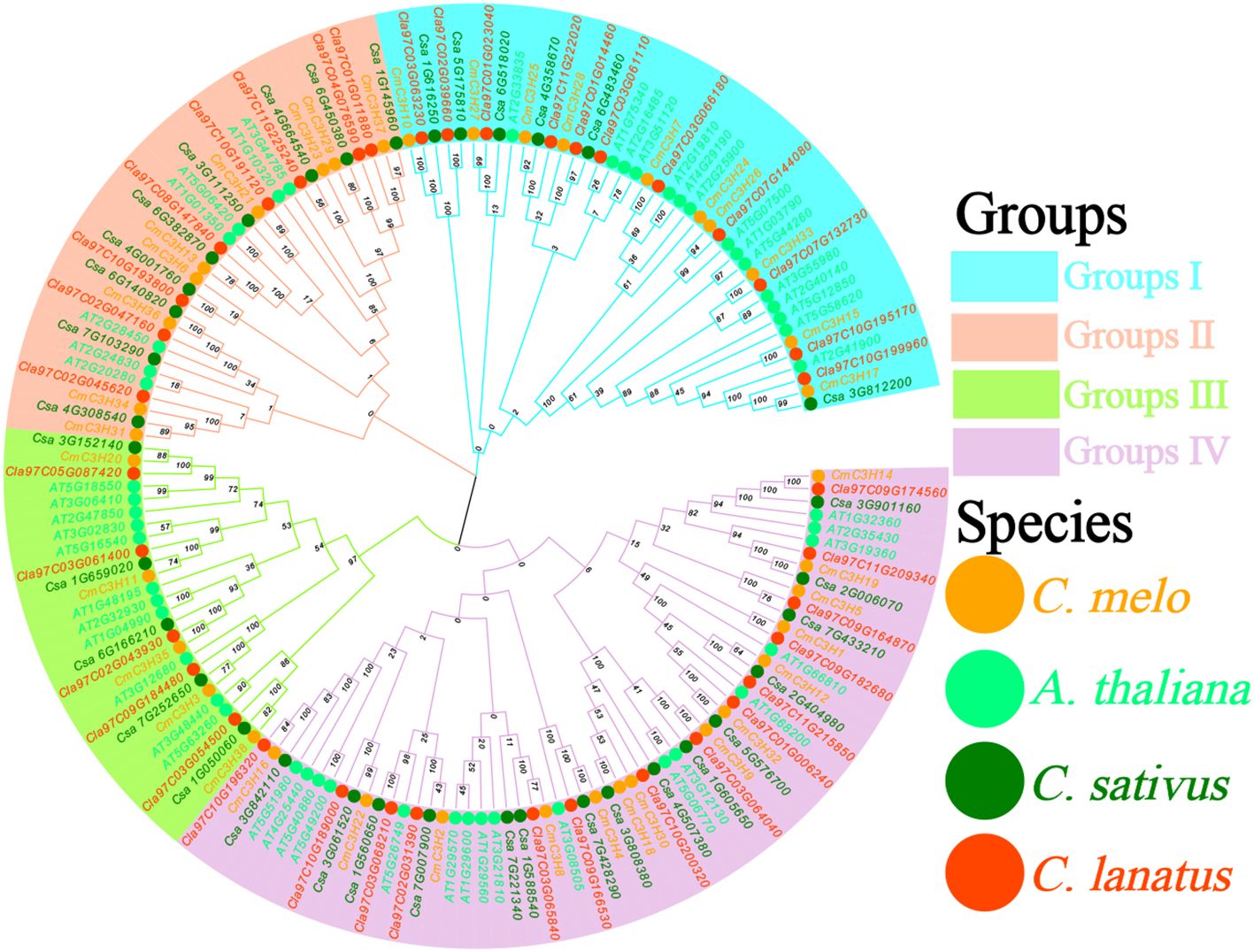
Figure 2. Neighbor-Joining phylogenetic tree of C3H family members from A. thaliana, C. melo, C. sativus, and C. lanatus. The four differently colored regions correspond to the four groups, and the solid circles of different colors represent the four species.
A total of 10 motifs were annotated in the protein sequences of CmC3H family members. A Neighbor-Joining phylogenetic tree was further constructed specifically for CmC3H family members (Figure 3A). The analysis indicated that, unlike the broader phylogenetic distribution shown in Figure 2, the CmC3H members within the same group were not strictly clustered within the same major branches. Additionally, significant differences in the types and numbers of conserved protein motifs were observed among CmC3H members across the four groups (Figure 3B). As illustrated in Figure 3B, the protein motifs in the 10 CmC3H members of Group I were primarily motifs 1, 2, 3, and 10. The 9 CmC3H members of Group II mainly contained motifs 1, 4, 6, and 7. The 5 CmC3H members in Group III predominantly featured motifs 5 and 6. The 14 CmC3H members in Group IV primarily exhibited motifs 1 and 4. Moreover, according to the protein domain annotation results, each CmC3H family member contained at least one C3H domain (Figure 3C). In Group III, four CmC3H members contained five C3H domains situated at different positions within their protein sequences, and one member contained six C3H domains. For the other members, most of the C3H domains were distributed across one to three locations.
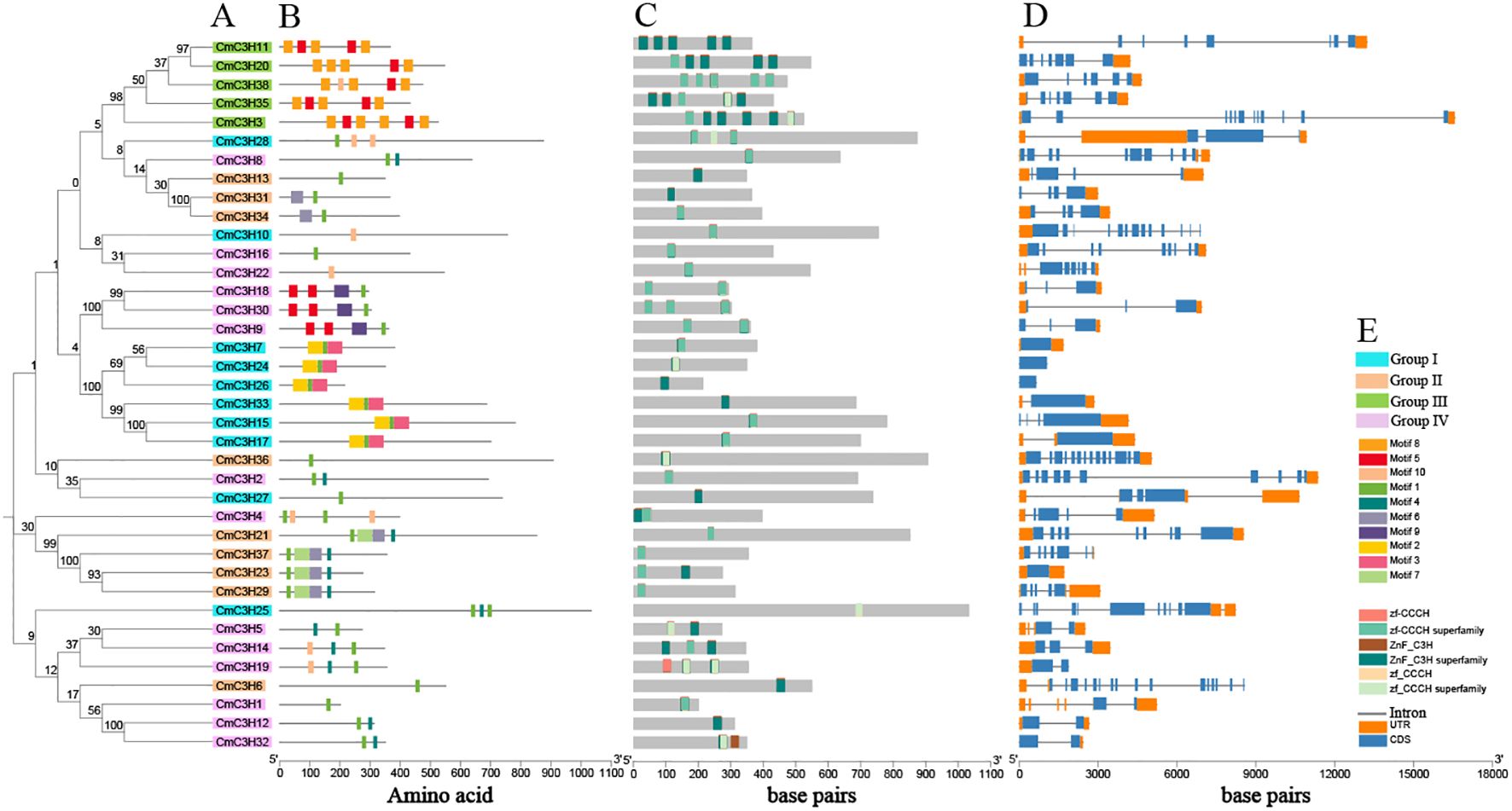
Figure 3. Analysis of the phylogenetic tree, motifs, domains, and gene structure of CmC3H family members. (A) Neighbor-Joining phylogenetic tree. (B) Conserved protein motifs. (C) Distribution of GATA domains. (D) Exon and intron positions. (E) Color-coded rectangles and their corresponding types.
The gene structure of CmC3H family members is highly diverse, with the number of exons ranging from 1 to 16 and the number of introns ranging from 0 to 15 (Figure 3D). Significant differences were observed in the number of exons and introns among CmC3H members across the four groups, and also within members of the same group. In Group I, eight members had fewer than five exons, and two members had more than ten exons. In Group II, eight members had fewer than ten exons, and one member had more than ten exons. In Group III, one member had six exons, three members had seven exons, and one member had twelve exons. In Group IV, all 14 members had fewer than ten exons. Overall, the diversity in protein motifs and gene structures suggests that CmC3H family members may have diverse functions in regulating melon growth and development.
A pair of segmental duplication genes was identified within the CmC3H family (CmC3H31/CmC3H34) (Figure 4A). Additionally, the C3H family members in melon were identified to have 20, 7, 39, and 38 homologous gene pairs with A. thaliana, rice, cucumber, and watermelon, respectively (Figure 4B; Supplementary Table S3). Furthermore, the Ka/Ks values for both segmental duplication gene pairs and homologous gene pairs were less than 1, except for 6 homologous gene pairs between melon and rice, for which Ka/Ks values were not calculable (Supplementary Table S3). These results indicate that C3H genes in dicotyledonous plants are likely to be highly conserved, whereas the conservation between monocots and dicots may be lower. Notably, the C3H homologous gene pairs between melon and cucumber, as well as between melon and watermelon, primarily exhibit a 1:1 distribution, indicating a high level of conservation of C3H family members across these three species during evolution.
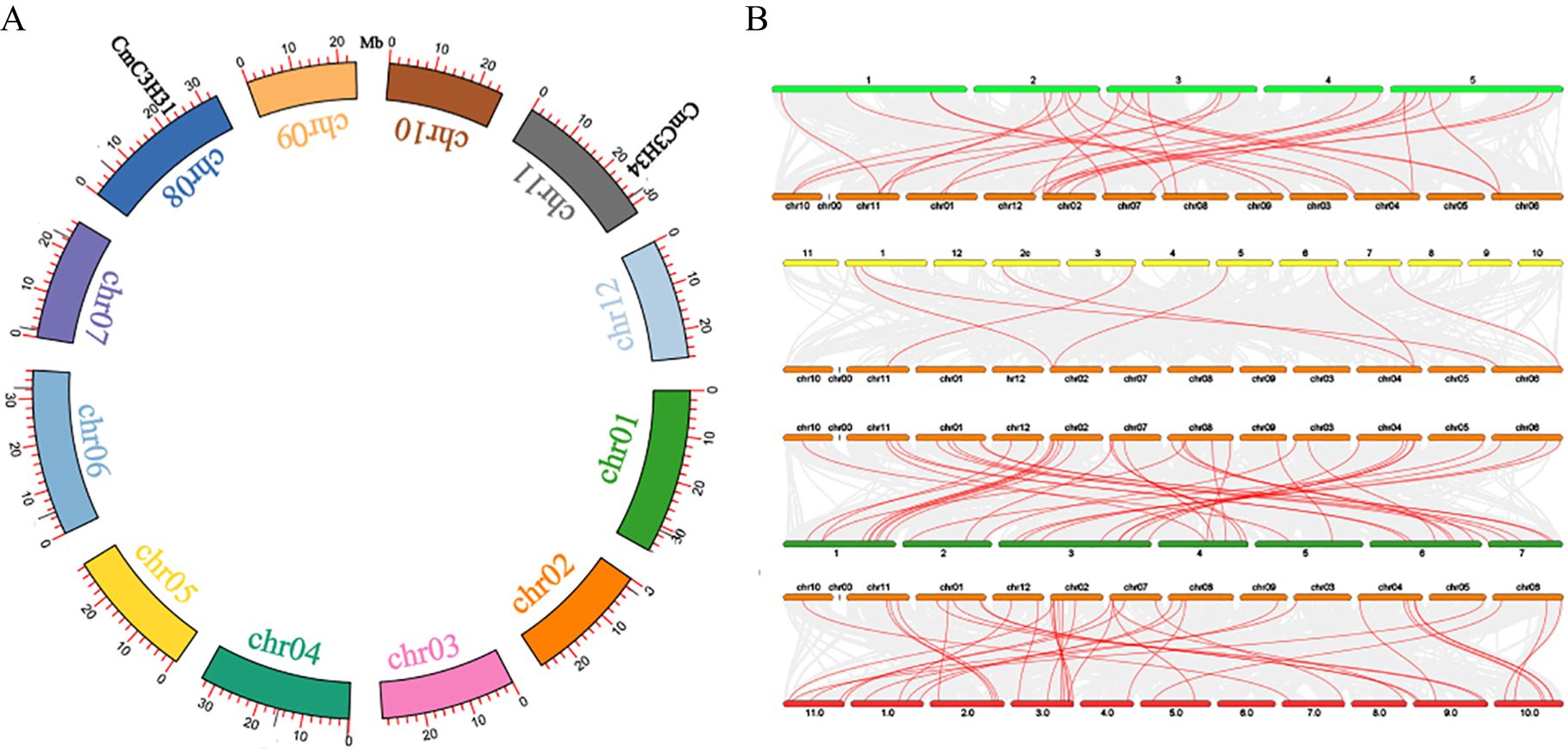
Figure 4. Synteny and homologous gene pair analysis of CmC3H family members. (A) Synteny circos plot of CmC3H family members within chromosomes. Red lines represent segmental duplication gene pairs. (B) Homologous gene pairs between melon and A. thaliana, rice, cucumber, and watermelon C3H family members. Red lines indicate homologous gene pairs.
Potential protein-protein interactions among CmC3H family members were predicted using the Arabidopsis database in the STRING tool (https://cn.string-db.org/) (Figures 5A, B; Supplementary Table S6). Potential interactions were identified among 29 CmC3H members, connected by 76 edges, forming a complex interaction network. For example, CmC3H26 potentially interacts with CmC3H27, CmC3H30, and other members. Conversely, some members, such as CmC3H28, show potential interaction with only a single member, CmC3H36. Notably, CmC3H13 was predicted to be a core gene, interacting with 13 other members. GO enrichment analysis revealed that CmC3H family members are involved in various protein functions, including nucleus (GO:0005634), ribonucleoprotein complex (GO:1990904), DNA binding (GO:0003677), metal ion binding (GO:0046872), nucleic acid binding (GO:0003676), and RNA processing (GO:0006396).
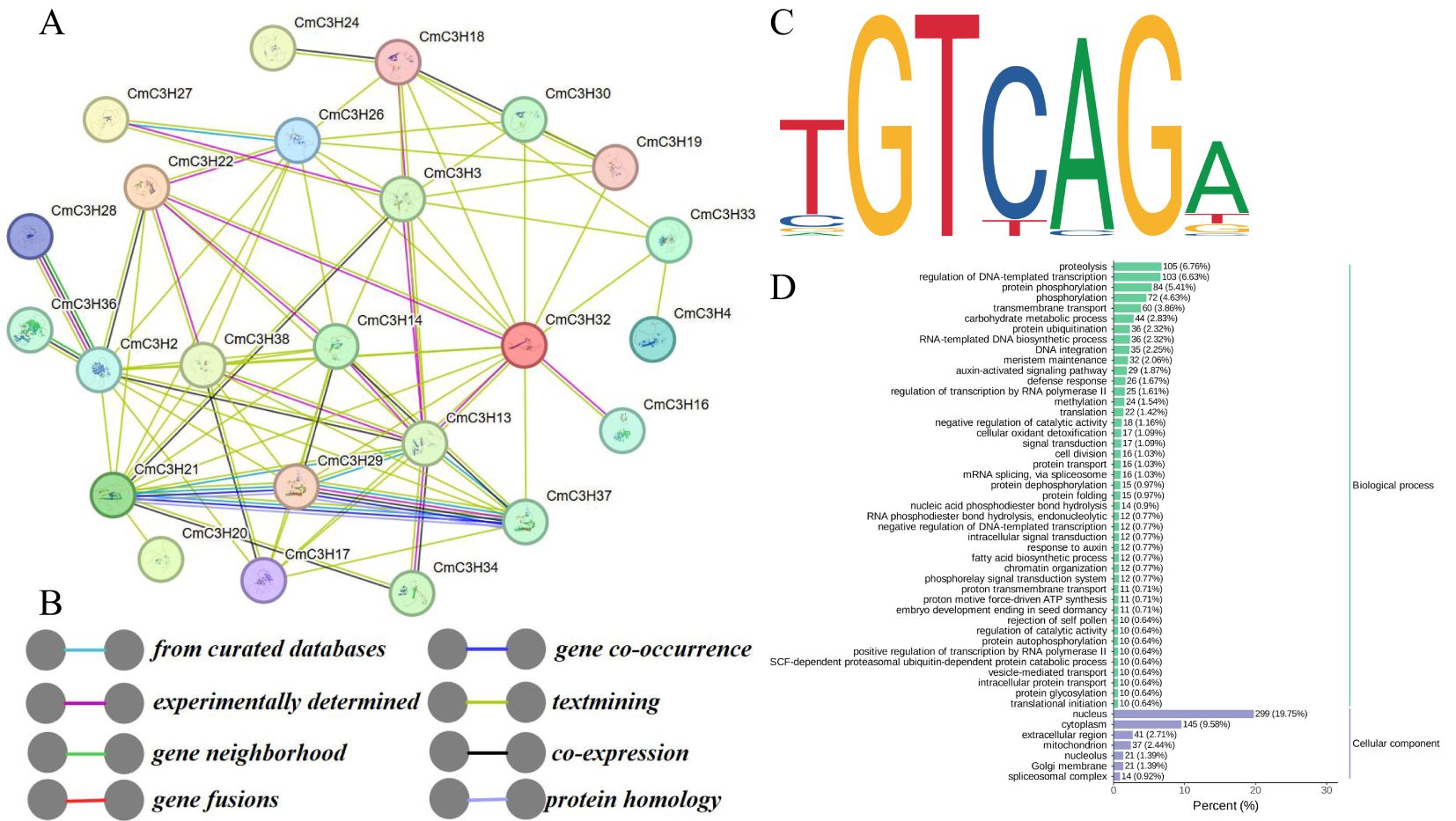
Figure 5. Protein interaction and target gene analysis of CmC3H family members. (A) Protein interaction network of CmC3H family members. (B) The meaning of different colored lines in the interaction network. (C) GRF9 (C3H) transcription factor binding profile. (D) GO enrichment results for target genes.
Using the binding motif profile of the Arabidopsis C3H12 transcription factor (MA1756.2), 3,243 target genes were identified in the melon genome, all of which matched the TGTCAGA sequence type (Figure 5C; Supplementary Table S7). GO enrichment analysis of these target genes (Figure 5D) indicated that, in the biological process category, most target genes were enriched in proteolysis (GO:0006508), regulation of DNA-templated transcription (GO:0006355), and protein phosphorylation. Five protein functional categories in the biological process category, including proteolysis (GO:0006508), regulation of DNA-templated transcription (GO:0006355), protein phosphorylation (GO:0006468), transmembrane transport (GO:0055085), and carbohydrate metabolic process (GO:0005975), were enriched with more than 40 target genes, based on the number of target genes enriched in Gene Ontology (GO) terms (Supplementary Table S8). In the cellular component category, the target genes were primarily enriched in the nucleus (GO:0005634), cytoplasm (GO:0005737), and membrane (GO:0016020). Overall, the potential interactions among CmC3H members suggest a coordinated regulatory network in melon, and the target gene analysis indicates that CmC3H genes may regulate a wide range of genes involved in various growth and developmental processes.
A total of 102 types of cis-elements were identified in the CmC3H family members. In addition to numerous light-responsive elements (such as G-box, MRE, and ATCT-motif), three major categories of cis-elements were classified: hormone regulation, stress response, and growth and development (Figure 6A; Supplementary Table S4). Hormone-regulatory cis-elements include ten types: TGA-element, TATC-box, TCA-element, ABRE, AuxRR-core, CGTCA-motif, TGACG-motif, GARE-motif, P-box, and TGA-box. Stress-responsive cis-elements include six types: TC-rich repeats, LTR, ARE, GC-motif, MBS, and WUN-motif. Growth and development-related cis-elements comprise nine types: MSA-like, circadian, RY-element, CAT-box, motif I, GCN4_motif, HD-Zip 1, AACA_motif, and CellCycle-1b.
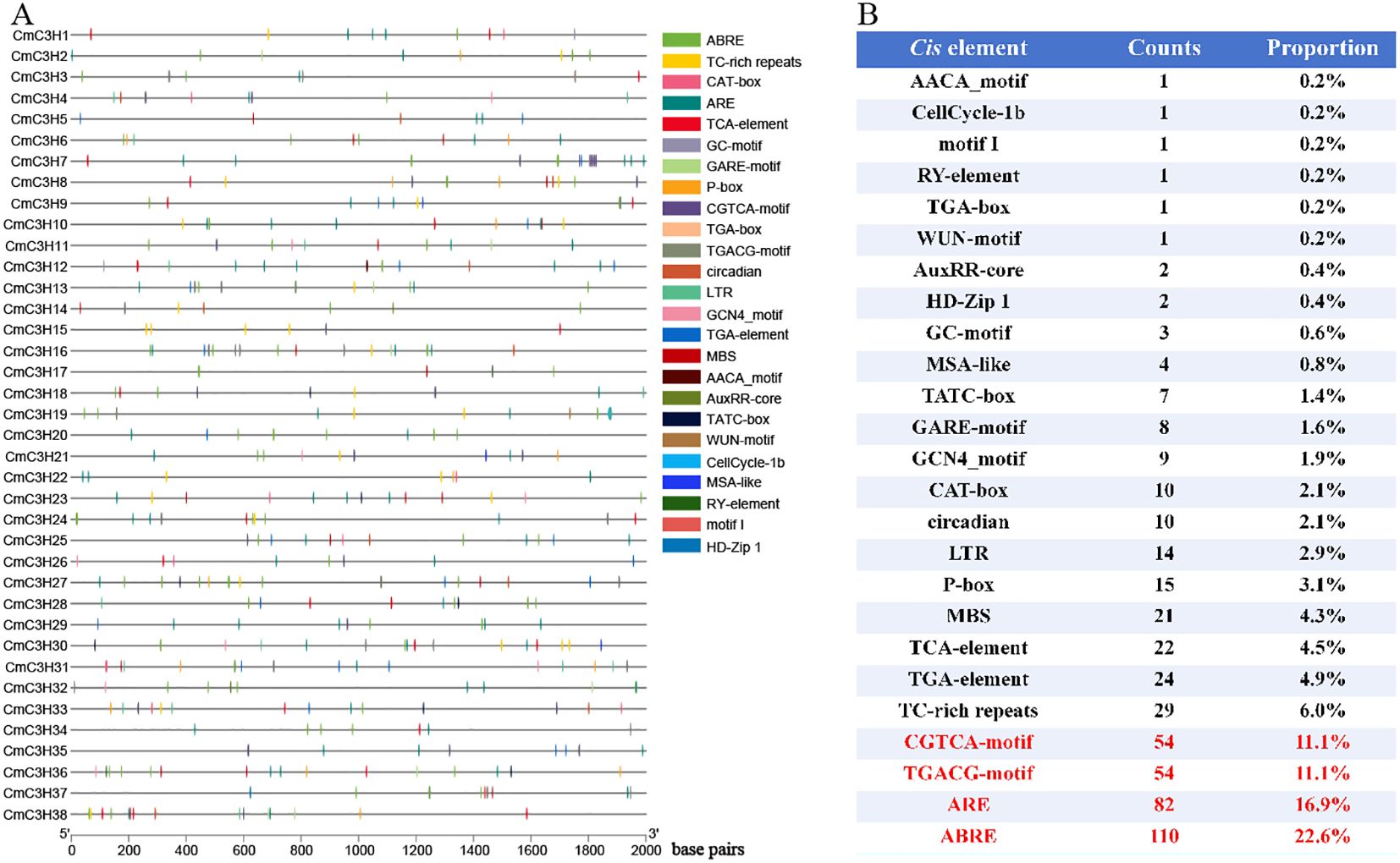
Figure 6. Cis-elements in CmC3H family members. (A) Distribution of hormone regulation, stress response, and growth and development-related cis-elements in CmC3H family members. (B) Quantitative distribution of hormone regulation, stress response, and growth and development-related cis-elements in CmC3H family members.
Furthermore, the total number of hormone regulation, stress response, and growth and development-related cis-elements within the CmC3H family members was quantified. The CGTCA-motif (cis-element involved in MeJA-responsiveness), TGACG-motif (cis-element involved in MeJA-responsiveness), ARE (cis-element essential for anaerobic induction), and ABRE (cis-element involved in abscisic acid responsiveness) were particularly abundant, with 54, 54, 82, and 110 occurrences, respectively (Figure 6B). Overall, the presence of these cis-elements suggests that CmC3H family members may play a significant role in various physiological processes in melon, contributing to the normal growth and development of tissues under diverse conditions.
The FPKM values for 36 CmC3H family members were obtained across eleven melon tissues: dry seeds, root, middle stem, upside stem, leaves, tendril,petal, stigma, ovary at 4 Days After Flowering (DAF4), fruit flesh at 50 DaysAfter Flowering (DAF50), and fruit epicarp at 50 Days After Flowering (DAF50). A heatmap was generated to visualize their expression patterns (Figure 7; Supplementary Table S5). The results indicate that the expression levels of 13 genes (CmC3H7, CmC3H9, CmC3H16, CmC3H18, CmC3H19, CmC3H22, CmC3H23, CmC3H25, CmC3H26, CmC3H30, CmC3H31, CmC3H34, and CmC3H37) show significant expression in dry seeds. The expression levels of seven genes (CmC3H8, CmC3H10, CmC3H11, CmC3H12, CmC3H17, CmC3H28, and CmC3H38) are significantly expressed in root. The expression levels of twelve genes (CmC3H3, CmC3H8, CmC3H11, CmC3H12, CmC3H13, CmC3H15, CmC3H17, CmC3H22, CmC3H28, CmC3H29, CmC3H32, and CmC3H38) are significantly expressed in middle stem. The expression levels of five genes (CmC3H1, CmC3H2, CmC3H9, CmC3H29, and CmC3H36) are significantly expressed in upside stem.Only CmC3H1 shows significant expression in leaves. The expression levels of three genes (CmC3H1, CmC3H4, and CmC3H32) are significantly expressed in tendril. The expression levels of three genes (CmC3H4, CmC3H14, and CmC3H27)are significantly expressed in petal. The expression levels of eight genes (CmC3H4, CmC3H13, CmC3H17, CmC3H21, CmC3H28, CmC3H32, CmC3H36, and CmC3H38) are significantly expressed in stigma. The expression levels of eight genes (CmC3H2, CmC3H5, CmC3H10, CmC3H14, CmC3H15, CmC3H22, CmC3H27, and CmC3H29) show significant expression in ovary at DAF4. The expression levels of five genes (CmC3H16, CmC3H24, CmC3H30, CmC3H33, and CmC3H35) are significantly expressed in fruit flesh at DAF50. The expression levels of five genes (CmC3H5, CmC3H13, CmC3H21, CmC3H24, and CmC3H34) are significantly expressed in fruit epicarp at DAF50. Notably, several genes, such as CmC3H4, CmC3H11, CmC3H17, and CmC3H29, are significantly expressed in multiple tissues. Overall, CmC3H family members exhibit distinct tissue-specific expression patterns in melon.
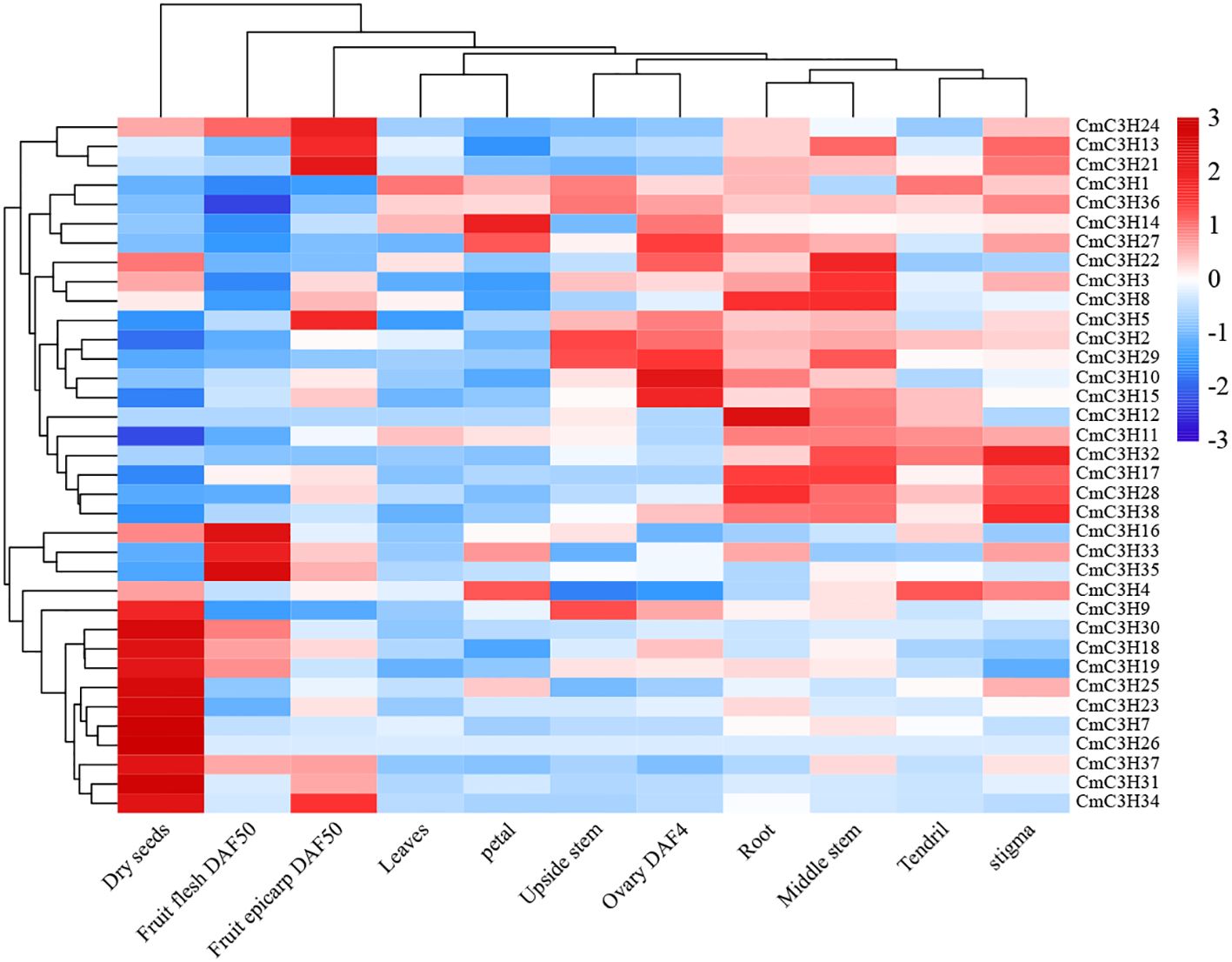
Figure 7. Heatmap of expression patterns of CmC3H family members in different tissues. Expression levels were normalized across rows, with color ranging from blue to red, representing low to high expression levels, respectively.
The expression levels of 10 CmC3H genes in melon leaves were measured over time under drought stress treatment (Figure 8A; Supplementary Table S9). The results showed that the expression levels of six genes (CmC3H4, CmC3H7, CmC3H13, CmC3H24, CmC3H33, and CmC3H38) exhibited significant changes at 48 hours, with expression levels ranging from 2 to 5 times those at 0 hours. Notably, the expression levels of CmC3H11 and CmC3H21 increased over time, particularly at 48 hours, where their expression levels were over 50 times higher than at 0 hours. In contrast, following lead stress treatment, the expression levels of the 10 CmC3H genes in melon leaves exhibited relatively minor changes over time, showing a general trend of decreasing expression, indicating that expression was suppressed (Figure 8B; Supplementary Table S9). Expression changes of the 10 CmC3H genes were further examined in the roots, stems, and leaves of melon under Fusarium wilt treatment (Figure 8C; Supplementary Table S9). The results showed that the expression levels of five genes (CmC3H4, CmC3H15, CmC3H33, CmC3H21, and CmC3H11) increased significantly in roots at 12 and 24 hours, particularly CmC3H4, whose expression increased by approximately 348 and 126 times at 12 and 24 hours, respectively, compared to 0 hours. In the stems, the expression levels of eight genes (CmC3H7, CmC3H35, CmC3H38, CmC3H33, CmC3H21, CmC3H11, CmC3H13, and CmC3H24) increased significantly at 24 hours, with CmC3H24 and CmC3H33 increasing approximately 669 and 153 times, respectively, compared to 0 hours. Additionally, CmC3H33 expression at 48 hours increased approximately 696 times compared to 0 hours. In the leaves, the expression levels of nine genes(CmC3H7, CmC3H35, CmC3H38, CmC3H15, CmC3H33, CmC3H21, CmC3H11, CmC3H13, and CmC3H24) significantly increased at 24 hours following Fusarium wilt treatment.
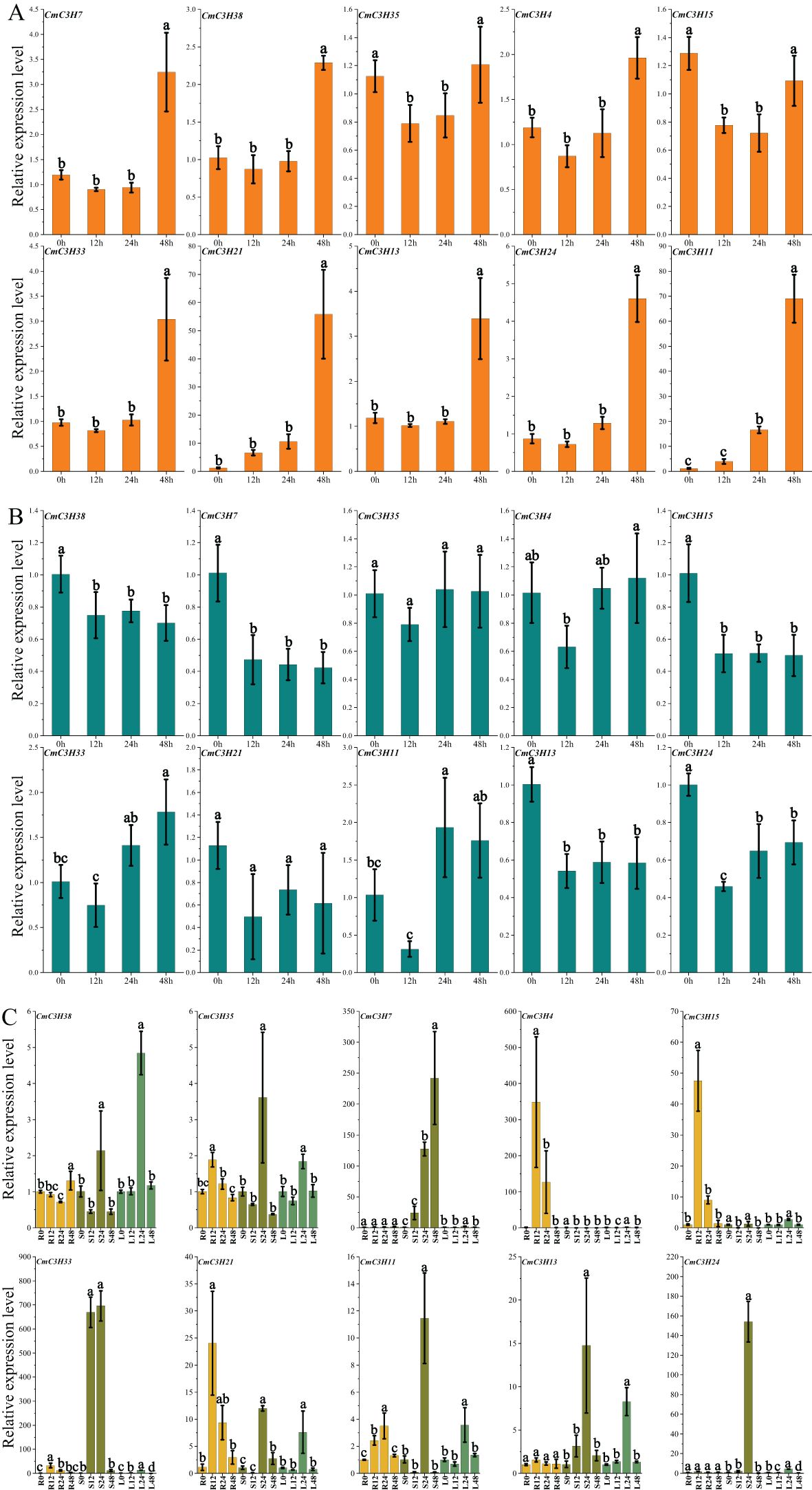
Figure 8. Quantitative Expression Levels of 10 CmC3H Genes at Four Time Points. (A) Expression levels under drought stress. (B) Expression levels under lead stress. (C) Expression levels in roots (R), stems (S), and leaves (L) following Fusarium wilt infection. Bars with lowercase letters above the columns indicate significant differences among the treatments at p < 0.05.
Based on the aforementioned results, we selected CmC3H21 for further subcellular localization experiments. According to bioinformatics predictions, the CmC3H21 protein is likely localized in the nucleus (Supplementary Table S1). To validate this prediction, we employed the Agrobacterium infiltration method to introduce the protein fusion into tobacco leaves to observe the subcellular localization of CmC3H21. The results showed that CmC3H21-GFP was co-expressed with mCherry in the nucleus(Figure 9), indicating that CmC3H21 is expressed in the nucleus.
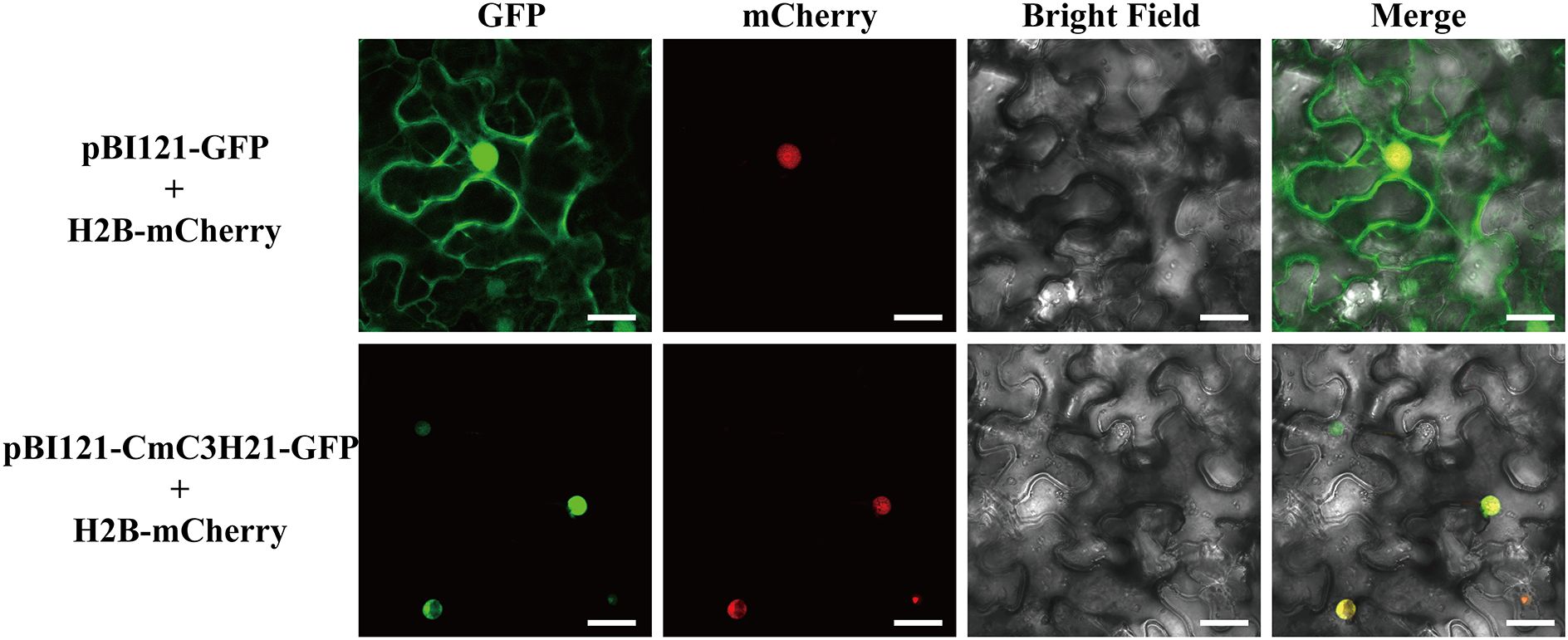
Figure 9. Subcellular localization of CmC3H21, with green (pBI121-CmC3H21-GFP) and red (nuclear marker H2B-mCherry) fluorescence signals (scale bar = 25 μm).
Zinc finger protein C3H genes are extensively involved in plant development, signaling, and responses to both biotic and abiotic stresses (Han et al., 2021). To date, numerous studies have investigated the C3H gene family in various plant species. However, research on the C3H gene family in melon remains underexplored. This study aims to address this gap by investigating the C3H gene family in melon. A total of 38 CmC3H genes were identified in melon, a number comparable to closely related species such as cucumber and watermelon, but significantly fewer than those in A. thaliana (68) (Wang et al., 2008), potato (50) (Deng et al., 2023), and Pyrus betulaefolia (17) (Liu et al., 2020). The CmC3H family members are unevenly distributed across the 12 chromosomes, with no significant correlation between chromosome length and the number of CmC3H genes. These findings are consistent with the distribution patterns of C3H genes observed in other species (Deng et al., 2023; Liu et al., 2020). Additionally, 17 CmC3H genes are located in regions characterized by low gene density on the chromosomes. No studies have examined the distribution of C3H family members across regions of varying gene density in other species. CmC3H members exhibit considerable variation in the number of amino acids, protein molecular weight, theoretical isoelectric point, instability index, and aliphatic index. Subcellular localization results predominantly indicate nuclear localization, consistent with findings for C3H family members in species like potato (Deng et al., 2023). The phylogenetic analysis did not reveal distinct classifications within the C3H gene family (Zhang et al., 2013; Liu et al., 2014). Consequently, the CmC3H members were classified into four groups (Group I, Group II, Group III, and Group IV) based on the phylogenetic tree of the C3H family in potato. High clustering conservation was observed among the C3H members from melon, cucumber, and watermelon within the same group. Additionally, significant sequence diversity exists among the CmC3H members within the same group. Gene function is closely linked to conserved motifs and gene structure within their protein sequences. Motif analysis of CmC3H family members revealed substantial diversity in gene sequences and significant differences in the number of exons and introns among the genes. These findings are consistent with previous reports on C3H family members in other species (Liu et al., 2020). In conclusion, the results suggest that CmC3H gene sequences have undergone significant differentiation during evolution, likely due to functional divergence driven by environmental adaptation.
Gene duplication is a major factor in the expansion and evolution of gene family members, contributing to the adaptation of species to environmental changes and the maintenance of normal life processes. 5, 15, 16, and 24 pairs of segmentally duplicated genes have been identified in the C3H family in four species: Solanum tuberosum (Deng et al., 2023), Zea mays (Peng et al., 2012), Panicum virgatum (Yuan et al., 2015), and Vitis vinifera (Wang et al., 2014). In the CmC3H family, only one pair of segmentally duplicated genes and no tandemly duplicated genes were identified, suggesting that gene duplication may not be the main factor driving the expansion of CmC3H family members. Melon shares 20, 7, 39, and 38 pairs of C3H homologous genes with A. thaliana, rice, cucumber, and watermelon, respectively, indicating a clear distinction between dicotyledonous and monocotyledonous plants. Furthermore, the C3H genes of closely related species exhibit greater conservation. The Ka/Ks ratio serves as an effective method for studying the evolutionary selection of duplicated genes. The Ka/Ks ratios for both segmentally duplicated and homologous gene pairs were found to be less than 1, indicating that these duplicated genes have mainly undergone purifying selection and maintain a high level of functional conservation (Song et al., 2024). Transcription factors often regulate various response processes through protein-protein interactions (Alves et al., 2014). It was predicted that 29 CmC3H members might have potential protein interactions, offering a reference for future exploration of CmC3H interactions. Transcription factors regulate gene expression by binding to specific cis-regulatory sequences within the promoters of target genes. Research on the target genes of C3H transcription factors in plants is limited. In this study, 3,243 target genes were identified in the melon genome based on the binding sites of the Arabidopsis GRF9 (C3H) transcription factor. These target genes are enriched in diverse GO functions and may play roles in regulating various response processes in melon, laying a foundation for further research into the interactions between CmC3H genes and other genes.
cis-elements within promoter regions regulate gene expression. The results of cis-element analysis indicate that CmC3H family members are involved in various functions, including melon growth and development, stress responses, and hormone regulation. Investigating gene expression during tissue development and under adverse environmental conditions is crucial for understanding molecular developmental mechanisms (Page and Minocha, 2005). In various species such as A. thaliana, rice, potato, and banana (Mazumdar et al., 2017), members of the C3H family exhibit tissue-specific expression patterns in tissues such as leaves, floral organs, and fruits. Similarly, CmC3H family members in melon exhibit significant tissue-specific expression across eleven tissues, including dry seeds, root, middle stem, upside stem, leaves, tendril, petal, stigma, ovary at DAF4, fruit flesh at DAF50, and fruit epicarp at DAF50. This suggests that these genes may play roles in the growth, development, and functional regulation of different melon tissues. C3H genes are widely involved in plant responses to both biotic and abiotic stresses, as shown by significant findings. In pepper, the C3H-type genes PEPTY4 and PEPTY5 are induced by heat stress (Tang et al., 2023b). The overexpression of the BoC3H gene can enhance salt tolerance in transgenic cabbage (Jiang et al., 2017). Transgenic expression of PeC3H74 in Arabidopsis significantly improves drought resistance (Chen et al., 2020a). PvC3H72 enhances cold tolerance in transgenic switchgrass by regulating the expression of genes in the ICE1-CBF-COR complex and ABA signaling pathways (Xie et al., 2019). GhZFP1 overexpression in tobacco significantly increases salt tolerance and disease resistance in transgenic plants (Guo et al., 2009). DgC3H1 overexpression improves cold tolerance in chrysanthemum (Bai et al., 2021). Similarly, six genes (CmC3H4, CmC3H7, CmC3H13, CmC3H24, CmC3H33, and CmC3H38) exhibited significant upregulation in melon leaves 48 hours after drought stress treatment, with CmC3H11 and CmC3H21 showing particularly marked increases. Notably, under lead stress, the expression levels of these ten genes declined, suggesting that CmC3H genes may not enhance melon’s ability to respond to ionic stress. Interactions among the transcription factors GHZFP1, GZIRD21A, and GZIPR5 can enhance fungal disease tolerance in cotton (Guo et al., 2009). Several CmC3H genes in melon roots, stems, and leaves showed significantly increased expression levels 24 hours after Fusarium wilt infection, with CmC3H24 and CmC3H33 displaying particularly high expression levels. Notably, CmC3H11 and CmC3H21 exhibited significant expression responses in roots, stems, and leaves under both drought and Fusarium wilt stress, indicating that these two genes may contribute to enhanced resistance to both biotic and abiotic stresses in melon. Quantitative PCR can be used to detect gene expression levels and predict their potential functions (Yang et al., 2022). Our results suggest that CmC3H24 and CmC3H33 are promising candidate genes for improving resistance to biotic stress, particularly Fusarium wilt, while CmC3H11 and CmC3H21 are key candidate genes for enhancing resistance to both biotic and abiotic stresses in melon. The expression levels of CmC3H genes were validated under both biotic and abiotic stress conditions using quantitative fluorescence analysis, which identified candidate genes that serve as a foundation for subsequent functional studies. Transgenic experiments and subcellular localization assays are crucial for the validation of gene functions. However, these experiments were not conducted in this study due to certain limitations. Future research will prioritize these experimental validations to thoroughly elucidate the function of the CmC3H genes.
In this study, 38 CmC3H genes were identified based on the melon DHL92 v4.0 genome, and various aspects of the CmC3H family members were analyzed, including protein physicochemical properties, phylogeny, gene structure, protein motifs, gene duplication, and expression patterns. Phylogenetic analysis categorized the CmC3H family members into four groups, with members within each group showing high conservation in gene structure and protein motifs. Gene duplication may not be significant in the expansion of CmC3H family members. Finally, quantitative fluorescence techniques were employed to examine the expression patterns of ten CmC3H genes in melon under drought stress, heavy metal lead stress, and Fusarium wilt infection at four different time points. This analysis revealed several key candidate genes that may play an important role in enhancing melon resistance to both biotic and abiotic stresses. The results of this study provide important insights for exploring the characteristics and functions of CmC3H family members.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
LZ: Conceptualization, Data curation, Software, Visualization, Writing – original draft, Writing – review & editing. HD: Formal analysis, Methodology, Writing – review & editing. YM: Formal analysis, Software, Writing – original draft. JL: Data curation, Supervision, Writing – review & editing. YC: Funding acquisition, Supervision, Visualization, Writing – review & editing. JH: Funding acquisition, Formal analysis, Methodology, Writing – review & editing.
The author(s) declare that financial support was received for the research and/or publication of this article. The work was funded by the Henan Science and Technology Research Program (242102110326)—Study on the Molecular Mechanism of Yellow Flower Formation in the Endangered Wild Species Paeonia ludlowii Based on Transcriptomics and Metabolomics; the Henan Science and Technology Research Program (232102110227)—Functional Analysis of the Peony TPS Gene and Its Application in Fragrance Breeding; the Science and Technology Key Project of Henan Province (Project No. 232102110046); and the Luoyang Core Technology Research and Development Public Welfare Special Project (Project No. 2302036A).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1500429/full#supplementary-material
Abnave, A., John, J., Grotewold, E., Doseff, A. I., Gray, J. (2024). Upper level and cross hierarchical regulation of predominantly expressed phenolic genes in maize. Curr. Plant Biol. 39, 100364. doi: 10.1016/j.cpb.2024.100364
Ai, Q., Pan, W., Zeng, Y., Li, Y., Cui, L. (2022). CCCH zinc finger genes in barley: genome-wide identification, evolution, expression, and haplotype analysis. BMC Plant Biol. 22, 117. doi: 10.1186/s12870-022-03533-5
Alves, M. S., Dadalto, S. P., Gonçalves, A. B., de Souza, G. B., Barros, V. A., Fietto, L. G. (2014). Transcription factor functional protein-protein interactions in plant defense responses. Proteomes 2, 85–106. doi: 10.3390/proteomes2010085
Bai, H., Lin, P., Li, X., Liao, X., Wan, L., Yang, X., et al. (2021). DgC3H1, a CCCH zinc finger protein gene, confers cold tolerance in transgenic chrysanthemum. Sci. Hortic. 281, 109901. doi: 10.1016/j.scienta.2021.109901
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Berg, J. M., Shi, Y. (1996). The galvanization of biology: a growing appreciation for the roles of zinc. Science 271, 1081–1085. doi: 10.1126/science.271.5259.1081
Bogamuwa, S. P., Jang, J. C. (2014). Tandem CCCH zinc finger proteins in plant growth, development and stress response. Plant Cell Physiol. 55, 1367–1375. doi: 10.1093/pcp/pcu083
Castro-Mondragon, J. A., Riudavets-Puig, R., Rauluseviciute, I., Lemma, R. B., Turchi, L., Blanc-Mathieu, R., et al. (2022). JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 50, D165–D173. doi: 10.1093/nar/gkab1113
Chai, G., Hu, R., Zhang, D., Qi, G., Zuo, R., Cao, Y. (2012). Comprehensive analysis of CCCH zinc finger family in poplar (Populus trichocarpa). BMC Genomics 13, 253. doi: 10.1186/1471-2164-13-253
Chai, G., Qi, G., Wang, D., Zhuang, Y., Xu, H., Bai, Z. (2022). The CCCH zinc finger protein C3H15 negatively regulates cell elongation by inhibiting brassinosteroid signaling. Plant Physiol. 189, 285–300. doi: 10.1093/plphys/kiac046
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y. (2020b). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, F., Liu, H. L., Wang, K., Gao, Y. M., Wu, M., Xiang, Y. (2020a). Identification of CCCH zinc finger proteins family in moso bamboo (Phyllostachys edulis), and PeC3H74 confers drought tolerance to transgenic plants. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.579255
Cheng, X., Cao, J., Gao, C., Gao, W., Yan, S., Yao, H., et al. (2020). Identification of the wheat C3H gene family and expression analysis of candidates associated with seed dormancy and germination. Plant Physiol. Biochem. 156, 524–537. doi: 10.1016/j.plaphy.2020.09.032
Deng, Z., Yang, Z., Liu, X., Dai, X., Zhang, J., Deng, K. (2023). Genome-wide identification and expression analysis of C3H zinc finger family in potato (Solanum tuberosum L.). Int. J. Mol. Sci. 24, 12888. doi: 10.3390/ijms241612888
Garcia-Mas, J., Benjak, A., Sanseverino, W., Bourgeois, M., Mir, G., González, V. M., et al. (2012). The genome of melon (Cucumis melo L.). Proc. Natl. Acad. Sci. U.S.A. 109, 11872–11877. doi: 10.1073/pnas.1205415109
Guo, Y. H., Yu, Y. P., Wang, D., Wu, C. A., Yang, G. D., Huang, J. G., et al. (2009). GhZFP1, a novel CCCH-type zinc finger protein from cotton, enhances salt stress tolerance and fungal disease resistance in transgenic tobacco by interacting with GZIRD21A and GZIPR5. New Phytol. 183, 62–75. doi: 10.1111/j.1469-8137.2009.02839.x
Han, G., Qiao, Z., Li, Y., Wang, C., Wang, B. (2021). The roles of CCCH zinc-finger proteins in plant abiotic stress tolerance. Int. J. Mol. Sci. 22, 8327. doi: 10.3390/ijms22168327
Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587. doi: 10.1093/nar/gkm259
Jiang, M., Jiang, J. J., Miao, L. X., He, C. (2017). Over-expression of a C3H-type zinc finger gene contributes to salt stress tolerance in transgenic broccoli plants. Plant Cell Tissue Organ Cult. 130, 239–254. doi: 10.1007/s11240-017-1225-9
Kamaliyan, Z., Clarke, T. L. (2024). Zinc finger proteins: guardians of genome stability. Front. Cell Dev. Biol. 12. doi: 10.3389/fcdb.2024.1448789
Ko, J. H., Kim, W. C., Han, K. H. (2009). Ectopic expression of MYB46 identifies transcriptional regulatory genes involved in secondary wall biosynthesis in Arabidopsis. Plant J. 60, 649–665. doi: 10.1111/j.1365-313X.2009.03950.x
Kong, Z., Li, M., Yang, W., Xu, W., Xue, Y. (2006). A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol. 141, 1376–1388. doi: 10.1104/pp.106.080823
Lee, S. J., Jung, H. J., Kang, H. (2012). Arabidopsis zinc finger proteins AtC3H49/AtTZF3 and AtC3H20/AtTZF2 are involved in ABA and JA responses. Plant Cell Physiol. 53, 673–686. doi: 10.1093/pcp/pcs025
Lei, Y. F. (2014). The role of Capsicum CaC3H1 in response to Ralstonia solanacearum infection and salt stress. Fuzhou, Fujian, China: Fujian Agriculture and Forestry University.
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, Z., Thomas, T. L. (1998). PEI1, an embryo-specific zinc finger protein gene required for heart-stage embryo formation in Arabidopsis. Plant Cell 10, 383–398. doi: 10.1105/tpc.10.3.383
Liu, S., Khan, M. R., Li, Y., Zhang, J., Hu, C. (2014). Comprehensive analysis of CCCH-type zinc finger gene family in citrus (Clementine mandarin) by genome-wide characterization. Mol. Genet. Genomics 289, 855–872. doi: 10.1007/s00438-014-0893-8
Liu, A., Lu, J., Song, H., Wang, X., Wang, M., Lei, Z., et al. (2024). Comparative genomics and transcriptomics analysis of the bHLH gene family indicate their roles in regulating flavonoid biosynthesis in Sophora flavescens. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1445488
Liu, H., Xiao, S., Sui, S., Huang, R., Wang, X., Wu, H., et al. (2022). A tandem CCCH type zinc finger protein gene CpC3H3 from Chimonanthus praecox promotes flowering and enhances drought tolerance in Arabidopsis. BMC Plant Biol. 22, 506. doi: 10.1186/s12870-022-03521-2
Liu, C., Xu, X., Kan, J., Cheng, Z. M., Chang, Y., Lin, J., et al. (2020). Genome-wide analysis of the C3H zinc finger family reveals its functions in salt stress responses of Pyrus betulaefolia. Peer J. 8, e9328. doi: 10.7717/peerj.9328
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Marchler-Bauer, A., Derbyshire, M. K., Gonzales, N. R., Lu, S., Chitsaz, F., Geer, L. Y., et al. (2015). CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43, D222–D226. doi: 10.1093/nar/gku1221
Mazumdar, P., Lau, S. E., Wee, W. Y., Singh, P., Harikrishna, J. A. (2017). Genome-wide analysis of the CCCH zinc-finger gene family in banana (Musa acuminata): An insight into motif and gene structure arrangement, evolution, and salt stress responses. Trop. Plant Biol. 10, 177–193. doi: 10.1007/s12042-017-9194-0
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
Page, A. F., Minocha, S. C. (2005). Analysis of gene expression in transgenic plants. Methods Mol. Biol. 286, 291–312. doi: 10.1385/1-59259-827-7:291
Peng, X., Zhao, Y., Cao, J., Zhang, W., Jiang, H., Li, X., et al. (2012). CCCH-type zinc finger family in maize: genome-wide identification, classification and expression profiling under abscisic acid and drought treatments. PloS One 7, e40120. doi: 10.1371/journal.pone.0040120
Potter, S. C., Luciani, A., Eddy, S. R., Park, Y., Lopez, R., Finn, R. D. (2018). HMMER web server: 2018 update. Nucleic Acids Res. 46, W200–W204. doi: 10.1093/nar/gky448
Rameneni, J. J., Dhandapani, V., Paul, P., Devaraj, S. P., Choi, S. R., Yi, S. Y., et al. (2018). Comprehensive analysis of CCCH zinc-finger-type transcription factors in the Brassica rapa genome. Hortic. Environ. Biotechnol. 59, 729–747. doi: 10.1007/s13580-018-0052-0
Seok, H. Y., Nguyen, L. V., Park, H. Y., Tarte, V. N., Ha, J., Lee, S. Y., et al. (2018). Arabidopsis non-TZF gene AtC3H17 functions as a positive regulator in salt stress response. Biochem. Biophys. Res. Commun. 498, 954–959. doi: 10.1016/j.bbrc.2018.03.130
Shen, W., Li, Y., Qin, Y. (2023). Research on the influencing factors and multi-scale regulatory pathway of ecosystem health: A case study in the Middle Reaches of the Yellow River, China. J. Clean. Prod. 406, 137038. doi: 10.1016/j.jclepro.2023.137038
Singh, V. K., Mangalam, A. K., Dwivedi, S., Naik, S. (1998). Primer premier: program for design of degenerate primers from a protein sequence. Biotechniques 24, 318–319. doi: 10.2144/98242pf02
Song, H., Ji, X., Wang, M., Li, J., Wang, X., Meng, L., et al. (2024). Genome-wide identification and expression analysis of the Dof gene family reveals their involvement in hormone response and abiotic stresses in sunflower (Helianthus annuus L.). Gene 910, 148336. doi: 10.1016/j.gene.2024.148336
Sun, J., Jiang, H., Xu, Y., Li, H., Wu, X., Xie, Q., et al. (2007). The CCCH-type zinc finger proteins AtSZF1 and AtSZF2 regulate salt stress responses in Arabidopsis. Plant Cell Physiol. 48 8, 1148–1158. doi: 10.1093/pcp/pcm088
Szklarczyk, D., Kirsch, R., Koutrouli, M., Nastou, K., Mehryary, F., Hachilif, R., et al. (2023). The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51, D638–D646. doi: 10.1093/nar/gkad937
Tang, D., Chen, M., Huang, X., Zhang, G., Zeng, L., Zhang, G., et al. (2023a). SRplot: A free online platform for data visualization and graphing. PloS One 18, e0294236. doi: 10.1371/journal.pone.0294236
Tang, W., Hao, Y., Ma, X., Shi, Y., Dang, Y., Dong, Z., et al. (2023b). Genome-wide analysis and identification of stress-responsive genes of the CCCH zinc finger family in Capsicum annuum L. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1189038
UniProt Consortium (2021). UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 49, D480–D489. doi: 10.1093/nar/gkaa1100
Velásquez, C. V., Moustafa, M. A. M., Rocha, S. C., Parveen, N. (2024). Borrelia burgdorferi colonizes the mammary glands of lactating C3H mice: does not cause congenital Lyme disease. Microbes Infect. 26, 105241. doi: 10.1016/j.micinf.2024.105241
Wang, D., Guo, Y., Wu, C., Yang, G., Li, Y., Zheng, C. (2008). Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice. BMC Genomics 9, 44. doi: 10.1186/1471-2164-9-44
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Wang, X. L., Zhong, Y., Cheng, Z. M. (2014). Evolution and expression analysis of the CCCH zinc finger gene family in Vitis vinifera. Plant Genome 7, plantgenome2014.05.0019. doi: 10.3835/plantgenome2014.05.0019
Wilkins, M. R., Gasteiger, E., Bairoch, A., Sanchez, J. C., Williams, K. L., Appel, R. D., et al. (1999). Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552. doi: 10.1385/1-59259-584-7:531
Xie, Z., Lin, W., Yu, G., Cheng, Q., Xu, B., Huang, B. (2019). Improved cold tolerance in switchgrass by a novel CCCH-type zinc finger transcription factor gene, PvC3H72, associated with ICE1-CBF-COR regulon and ABA-responsive genes. Biotechnol. Biofuels 12, 224. doi: 10.1186/s13068-019-1567-2
Yang, Y., Wang, X., Zheng, J., Men, Y., Zhang, Y., Liu, L., et al. (2022). Amino acid transporter (AAT) gene family in Tartary buckwheat (Fagopyrum tataricum L. Gaertn.): Characterization, expression analysis and functional prediction. Int. J. Biol. Macromol. 217, 330–344. doi: 10.1016/j.ijbiomac.2022.07.059
Ye, J., McGinnis, S., Madden, T. L. (2006). BLAST: improvements for better sequence analysis. Nucleic Acids Res. 34, W6–W9. doi: 10.1093/nar/gkl151
Yu, J., Wu, S., Sun, H., Wang, X., Tang, X., Guo, S., et al. (2023). CuGenDBv2: an updated database for cucurbit genomics. Nucleic Acids Res. 51, D1457–D1464. doi: 10.1093/nar/gkac1058
Yuan, S., Xu, B., Zhang, J., Xie, Z., Cheng, Q., Yang, Z., et al. (2015). Comprehensive analysis of CCCH-type zinc finger family genes facilitates functional gene discovery and reflects recent allopolyploidization event in tetraploid switchgrass. BMC Genomics 16, 129. doi: 10.1186/s12864-015-1330-4
Zhang, Z., Li, J., Zhao, X. Q., Wang, J., Wong, G. K., Yu, J. (2006). KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinf. 4, 259–263. doi: 10.1016/S1672-0229(07)60007-2
Zhang, D., Yu, Z., Zeng, B., Liu, X. (2024). Genome-wide analysis of the ABC gene family in almond and functional predictions during flower development, freezing stress, and salt stress. BMC Plant Biol. 24, 12. doi: 10.1186/s12870-023-04315-9
Zhang, C., Zhang, H., Zhao, Y., Jiang, H., Zhu, S., Cheng, B., et al. (2013). Genome-wide analysis of the CCCH zinc finger gene family in Medicago truncatula. Plant Cell Rep. 32, 1543–1555. doi: 10.1007/s00299-013-1460-0
Keywords: Cucumis melo, C3H gene family, drought stress, heavy metal lead stress, fusarium wilt infection
Citation: Zheng L, Dai H, Mu Y, Li J, Cheng Y and Han J (2025) Genome-wide identification and expression analysis of C3H gene family in melon. Front. Plant Sci. 16:1500429. doi: 10.3389/fpls.2025.1500429
Received: 23 September 2024; Accepted: 18 February 2025;
Published: 13 March 2025.
Edited by:
Ake Liu, Changzhi University, ChinaReviewed by:
Jun Tang, Jiangsu Academy of Agricultural Sciences, ChinaCopyright © 2025 Zheng, Dai, Mu, Li, Cheng and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianming Han, aGFuamlhbm1pbmdAbHludS5lZHUuY24=; Yanwei Cheng, Y2hlbmd5YW53ZWlAbHludS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.