
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 06 March 2025
Sec. Plant Abiotic Stress
Volume 16 - 2025 | https://doi.org/10.3389/fpls.2025.1499244
This article is part of the Research TopicMechanisms of Stress Tolerance in Horticultural Crops: Physiological and Molecular InsightsView all 14 articles
 Ting Gao1†
Ting Gao1† Xiaojuan Zhou1†
Xiaojuan Zhou1† Mian Han1
Mian Han1 Yuexin Shen1
Yuexin Shen1 Yimeng Zhang1
Yimeng Zhang1 Qi Wu1
Qi Wu1 Haoyuan Dan1
Haoyuan Dan1 Tingyu Wang1
Tingyu Wang1 Hang Ye2
Hang Ye2 Li Liu1
Li Liu1 Min Chai1*
Min Chai1* Yiling Wang1*
Yiling Wang1*The TCP gene family plays pivotal roles in the development and abiotic stress responses of plants; however, no data has been provided for this gene family in Opisthopappus taihangensis. Based on O. taihangensis genome, 14 TCP genes were identified and divided into two classes (I and II). After tandem and segmental duplication/whole-genome duplication (WGD), more loss and less gain events of OtTCPs occurred, which might be related with the underwent purifying selection during the evolution. The conserved motifs and structures of OtTCP genes contained light response, growth and development, hormone response, and stress-related cis-acting elements. Different OtTCP genes, even duplicated gene pairs, could be expressed in different tissues, which implied that OtTCP genes had diverse function. Among OtTCPs, OtTCP4, 9 and 11 of CYC clade (Class II) presented a relative wide expression pattern with no or one intron. The three TCP genes could be regarded as important candidate factors for O. taihangensis in growth, development and stress response. These results provided some clues and references for the further in-depth exploration of O. taihangensis resistance mechanisms, as well as those of other unique eco-environment plants.
Transcription factors are one kind of proteins that play essential roles in the growth and development of plants by binding to specific gene promoters or enhancer regions (Katagiri and Chua, 1992). Based on the characteristics of their structural domains, transcription factors may be classified as WRKY (WRKYGQ), SPL (SQUAMOSA promoter-binding protein-like), NAC (NAM, ATAF, and CUC), AP2/ERF (Apetala2/Ethylene Responsive Factor), TCP (TEOSINTE BRANCHED1/CYCLOIDEA/PCF), and various other families (Lehti-Shiu et al., 2017). Among them, TCP transcription factors are specific to plants, cell growth, and cell proliferation (Zhan et al., 2023). Every TCP member has an atypical basic helix–loop–helix (bHLH) secondary structure that made up of two hydrophilic α-helices, a disordered loop, and about 60 amino acid residues (Cubas et al., 1999). This conserved domain is essential for DNA binding, protein interactions, and the regulation of downstream gene expression in the biological processes of plants (Martín-Trillo and Cubas, 2010; Manassero et al., 2013).
As ancient transcription factors that appeared ~650–800 million years ago (Navaud et al., 2007), the genes of TCP family are primarily categorized Class I and II, due to the deletion of four amino acids in the basic domain of Class II (Martín-Trillo and Cubas, 2010). Class I (also referred to as the TCP-P class) contains PCF genes (PCF1 and PCF2), can promote cell proliferation and growth. Class II is TCP-C genes, divided into CYC/TB1 and CIN branches. Different branches may have diverse function. The TB1 genes play an important role in inhibiting lateral branch growth and male flower formation (Doebley et al., 1997; Dixon et al., 2018), the CYC genes are generally involved in the expression of lateral regions of early floral organs and regulates floral symmetry (Luo et al., 1999; Hileman, 2014). Whereas the CIN genes are mainly related to leaf morphogenesis (Nath et al., 2003; Palatnik et al., 2003; Crawford et al., 2004; Walcher-Chevillet and Kramer, 2016). At the structure of Class II, some members contain an arginine-rich R domain with an unknown biological function aside from the TCP domain. Most CYC/TB1 members possess a conserved and functionally uncharacterized ECE motif (a sequence of glutamic acid-cysteine-glutamic acid) (Howarth and Donoghue, 2006; Martín-Trillo and Cubas, 2010). From the evolutionary viewpoint, the CYC/TB1 genes have not been discovered in lycophytes or other early-diverging land plants (Horn et al., 2015). Thus, CIN genes might have arisen earlier than CYC/TB1 genes (Palatnik et al., 2003; Koyama et al., 2007; Horn et al., 2015).
TCP genes typically form homodimers or heterodimers with each other to regulate the expressions of target genes (Li et al., 2017). All target genes of TCPs contain a highly conserved DNA motif (G(T/C) GGNCCCAC), specifically the core motif (TGGGCC, GCCCR, GG(A/T) CCC) (Kosugi and Ohashi, 2002; Li et al., 2005; Schommer et al., 2008; Aggarwal et al., 2010; Viola et al., 2012; Danisman et al., 2013; Parapunova et al., 2014). Further, TCPs engage with various other transcription factors, such as DELLAs, AS2, ABI4, MYBs, and bHLHs, that can promote flavonoid biosynthesis, trigger effector immunity, respond to abiotic stress, and mediate salicylic acid (SA), jasmonate (JA), auxin, cytokinin (CK), abscisic acid (ABA), and gibberellin (GA) responses (Pruneda-Paz et al., 2009; Li et al., 2012; Steiner et al., 2012; Li and Zachgo, 2013; Tao et al., 2013; Chen et al., 2014; Davière et al., 2014; Marín-De-La-Rosa et al., 2014; Mukhopadhyay and Tyagi, 2015). In Arabidopsis, TCP20 interacts with NIN-like proteins NLP6 and NLP7 to modulate signal transduction pathways, as well as to control root growth (Guan et al., 2017). AtTCP5, AtTCP13, and AtTCP17 positively regulate the responses of Arabidopsis under high-temperature stress (Han et al., 2019; Zhou et al., 2019). In maize, the natural variation in the ZmTCP42 promoter is significantly related to drought tolerance. The overexpression of ZmTCP42 can increase the sensitivity of transgenic Arabidopsis to abscisic acid (ABA) and increase its tolerance to drought stress (Ding et al., 2019). In moso bamboo, PeTCP10 enhances the salt stress tolerance (Xu et al., 2022). Nonetheless, these studies primarily concentrated on the functions and molecular mechanisms of TCPs in model plants (such as Arabidopsis thaliana) and agricultural species. Limited researches would be performed on wild and/or non-model plant species.
Opisthopappus taihangensis belongs to the family Asteraceae, it is endemic to the Taihang Mountains that span Henan and Shanxi Provinces and typically grows within steep cliff crevices, or on slopes up to ~1000 meters above sea level (Chai et al., 2018; Zhou et al., 2024). Being a cliff species, O. taihangensis exhibits good cold and drought resistance and has high ecological and ornamental value, with a large number of flowers and lengthy flowering period (Chen et al., 2022; Han et al., 2024; Zhang et al., 2024c). During drought stress, O. taihangensis presents decreased relative water and chlorophyll contents although having a high degree of proline accumulation (Gu et al., 2019). Under longer salt stress exposure times and at higher salt concentrations, O. taihangensis survives by engaging redox-regulated antioxidant enzyme mechanisms, including superoxide dismutase (SOD), peroxidase (POD), and catalase (CAT) (Zhou et al., 2024). More, the upregulated genes under salt stress are primarily participated in the processes related to amino acid metabolism, the regulation of transcription factors, ABA signaling pathway, osmolyte metabolism, and antioxidant enzyme activities (Gu et al., 2019; Yang et al., 2020). However, the roles of the transcription factors involved in these responses (e.g., TCPs) are unknown when O. taihangensis is under abiotic stress.
For this study, the characteristics of the TCP gene family, including their evolution and diversification in O. taihangensis were initially explored using bioinformatics and comparative analyses based on its whole genomics data. Subsequently, the expression levels of O. taihangensis TCP genes in distinct tissues under abiotic stress were investigated using RNA-seq data and qRT-PCR. Finally, the potential roles and regulatory pathways of TCP genes in response to abiotic stress for O. taihangensis were elucidated. The results provided important clues for the further investigation of the endurance mechanisms of O. taihangensis in cliff environments, which are foundational for the study of other unique cliff plant species.
With the O. taihangensis genome database obtained by our previous study (Ye et al., 2024; Zhou et al., 2024), various strategies were adopted to ensure the integrity (as much as possible) of the TCP gene family in O. taihangensis. Firstly, the protein sequences of TCP genes in A. thaliana were downloaded from TAIR website as queries, the O. taihangensis protein sequences were from its genome database. These protein sequences then were used to identify O. taihangensis TCP genes by BLAST program with an e value of 1 × 10-5, whereas the other parameters were set to default values (NumofThreads: 2, NumofHits: 500, NumofAligns: 250) in TBtools (Chen et al., 2020). After which, the TCP domain was retrieved based on the hidden Markov model (HMM) (PF03634) with Simple HMM Search. Finally, all TCP genes were analyzed by the NCBI Batch-CDD tool (Wang et al., 2023b) (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) combining the BLAST and HMM search results, and the genes containing the entire TCP domain were retained.
The identified TCPs were designated as OtTCP + numbers (Table 1). Subsequently, the ExPASy (ProtParam) (Duvaud et al., 2021) (http://www.expasy.org/tools/protparam.html) tool was used to evaluate the physicochemical characteristics of the OtTCP proteins, including the number of amino acids (aa), isoelectric point (pI), and molecular weights (MW). And then, the subcellular locations of the OtTCP proteins were predicted using WoLF PSORT (Horton et al., 2007) (https://wolfpsort.hgc.jp/).
Using the Clustal X in MEGA (Kumar et al., 2018) with defaulted parameters, multiple sequence alignment (MSA) was conducted by the protein sequences of TCPs in O. taihangensis, A. thaliana and Oryza sativa. The conserved regions of the obtained sequences were subsequently trimmed using trimAl in TBtools (Chen et al., 2020). Then, the rootless phylogenetic tree was constructed using IQ-TREE 2 software (Nguyen et al., 2014) with the maximum likelihood (ML) method and the bootstrap validation parameter 1000. All TCP proteins’ conserved domains and amino acid sequences were compared and examined using the GeneDoc program (Nicholas and Nicholas, 1997).
MEME tool in the MEME SUITE (https://meme-suite.org/meme/tools/meme) online website (Bailey et al., 2009) was employed to examine the motifs (number =10) of the TCP protein sequences of O. taihangensis. The relative genetic structural data was obtained from the O. taihangensis genome database (GFF file) based on our laboratory. The protein motifs and intron/exon organization were visualized using Gene Structure View (Advanced) in TBtools (Chen et al., 2020).
NOTUNG software (Chen et al., 2000; Stolzer et al., 2012) was used to perform the gene gain and loss events of TCP gene family in Asteraceae. The genomic data of other Asteraceae species was downloaded from NCBI (https://www.ncbi.nlm.nih.gov/).
A species tree was from the TIMETREE (http://www.timetree.org/) online website (Kumar et al., 2017), while the gene phylogenetic tree was developed utilizing IQ-tree software (Nguyen et al., 2014). The tree species and gene tree were imported into the NOTUNG software and analyzed by the Reconciliation Mode function of NOTUNG, in which A. thaliana and O. sativa were employed as an outgroup.
The chromosomal locations of OtTCP genes were visualized with TBtools software, using the GFF file of the O. taihangensis genome database.
To explore the potential evolutionary relationships of TCP genes, the collinearity analysis among A. thaliana, O. sativa and Asteraceae species (Helianthus annuus, Arctium lappa, Cynara cardunculus, Cichorium intybus, Centaurea solstitialis, Erigeron canadensis, Lactuca saligna, Lactuca virosa, Smallanthus sonchifolius, Mikania micrantha, and Tagetes erecta) were investigated using the Multiple Collinearity Scan Toolkit (MCScanX) in TBtools (Chen et al., 2020). The genomic data of A. thaliana (TAIR 10), O. sativa (IRGSP-1.0), H. annuus (HanXRQr2.0-SUNRISE), A. lappa (ASM2352574v1), C. cardunculus (CcrdV1.1), C. intybus (ASM2352571v1), C. solstitialis (ASM3016916v1), E. canadensis (C_canadensis_v1), L. saligna (Lactuca_saligna), L. virosa (Lvir_assembly_v4), S. sonchifolius (ASM2352597v1), M. micrantha (ASM936387v1), and T. erecta (ASM3086718v1) was downloaded from NCBI (https://www.ncbi.nlm.nih.gov/).
Gene repetition events (such as tandem replication and fragment replication) were performed using MCSCANX in TBtools (Wang et al., 2012; Chen et al., 2020). The TCP protein sequences of these species were aligned using Blastp program in TBtools (Chen et al., 2020), with an e value of 1 × 10−10, other parameters set to default values.
The Ka (nonsynonymous substitution per site) and Ks (synonymous substitution per site) (Zhang et al., 2006) between segmental and tandem duplicate gene pairs were calculated by the simple Ka/Ks Calculator in TBtools (Chen et al., 2020). The Ka/Ks value was further utilized to identify the selection mode of OtTCP genes.
The secondary and tertiary structures of OtTCP proteins were predicted and modelled using the SOPMA (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html) (Geourjon and Deléage, 1995), SWISS MODEL (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html) servers (Waterhouse et al., 2018), while the tertiary structures were examined by PyMOL (Rosignoli and Paiardini, 2022) (http://www.pymol.org/pymol).
The cis-acting elements were predicted using 2000 bp sequences upstream of OtTCP genes in PlantCARE (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/) online website (Lescot et al., 2002), the relative results were visualized with GSDS online website 2.0 (Hu et al., 2015).
The transcriptome data (PRJNA400848, PRJNA437359) of O. taihangensis under drought treatments were downloaded from NCBI (Gu et al., 2019; Yang et al., 2020). And the transcriptome sequencing data of O. taihangensis different tissues under salt stress were from our laboratory. Under 500 mM/L salt treatment, O. taihangensis individuals were treated for 0, 6, 24 and 48 h respectively. While under 24 h treatment, the sampled individuals were treated with 0 mM/L, 100 Mm/L, 300 Mm/L and 500 Mm/L salt respectively. Three replicates were set up for each treatment. After treatments, the sampled leaves from the same sites of each individual were frozen in liquid nitrogen for transcriptome sequencing (Han et al., 2024; Ye et al., 2024; Zhang et al., 2024c; Zhou et al., 2024). Based on the above, an expression heatmap of O. taihangensis under different treatments was generated using TBtools (Chen et al., 2020).
Finally, qRT-PCR was conducted to validate the expression patterns of randomly selected TCP genes of O. taihangensis. According to Peng et al (Peng et al., 2024), the internal reference genes were selected for evm. TU. Chr8.13443 (Han et al., 2024; Zhang et al., 2024c) and evm. TU. Chr8.39 (Zhou et al., 2024). Three technical replicates were performed for each selected gene. The PCR primers were designed using PRIMER 5.0 software (Supplementary Table S1), and the primer efficiency was evaluated from the amplification of three replicates based on Bello et al (Bello et al., 2017).
The qRT-PCR was performed with the UltraSYBR mixture (TaKaRa, Dalian, China) using an ABI7500 RT-PCR system. Reactions were done in 20 μl volume, the following qRT-PCR program was used: the template denaturation at 95°C for 3 min; followed by amplification for 40 cycles with a melting temperature of 95°C for 10s and an annealing temperature of 68°C for 15s. After 40 cycles, the melting curve analysis ranged from 60°C to 95°C, and the amplification efficiency was determined from the slope of the standard curve linear-log of target genes. All relative gene expression levels were calculated using 2−ΔΔCT (Penfield et al., 2001).
A total of 14 TCP genes with conserved domains were identified in O. taihangensis, which were designated OtTCP1 - OtTCP14 based on their locations on the chromosomes (Table 1).
As shown in Table 1, OtTCP proteins varied in their lengths, molecular weights, theoretical isoelectric points, and so on. Sequence analyses revealed that the 14 OtTCP proteins ranged from 241 (OtTCP13) to 426 amino acids (OtTCP5), with average lengths of 358 amino acids. The molecular weights ranged from 26398.93 to 45725.23 Da. For the theoretical pI, significant differences between the OtTCP proteins suggested that they might function under various acidic and basic conditions. The lowest (5.39) and highest (9.74) pI were OtTCP4 and OtTCP14, respectively. Thereinto, there were 7 OtTCP proteins (50%) with pI values of < 7.0, which indicated that they contained an abundance of acidic amino acids. All OtTCP proteins were unstable with a values of over 40 instability index (Guruprasad et al., 1990). More, almost all of the OtTCP proteins were hydrophilic that had a negative grand average of hydropathicity (GRAVY) values. Furthermore, all 14 OtTCP proteins were located within the nucleus.
A total of 70 complete protein sequences, including 33 AtTCPs (A. thaliana), 23 OsTCPs (O. sativa), and 14 OtTCPs, were used in the phylogenetic analysis. Based on a phylogenetic tree, all analyzed TCP genes were segregated into two main classes: Class I (PCF) and Class II (CIN and CYC/TB1) (Figure 1). Class I was the largest group, which contained five OtTCPs, ten OsTCPs, and fifteen AtTCPs. While class II included eight OtTCPs, three OsTCPs, and five AtTCPs. Interestingly, the CIN group in Class II contained only one OtTCP member (OtTCP6).
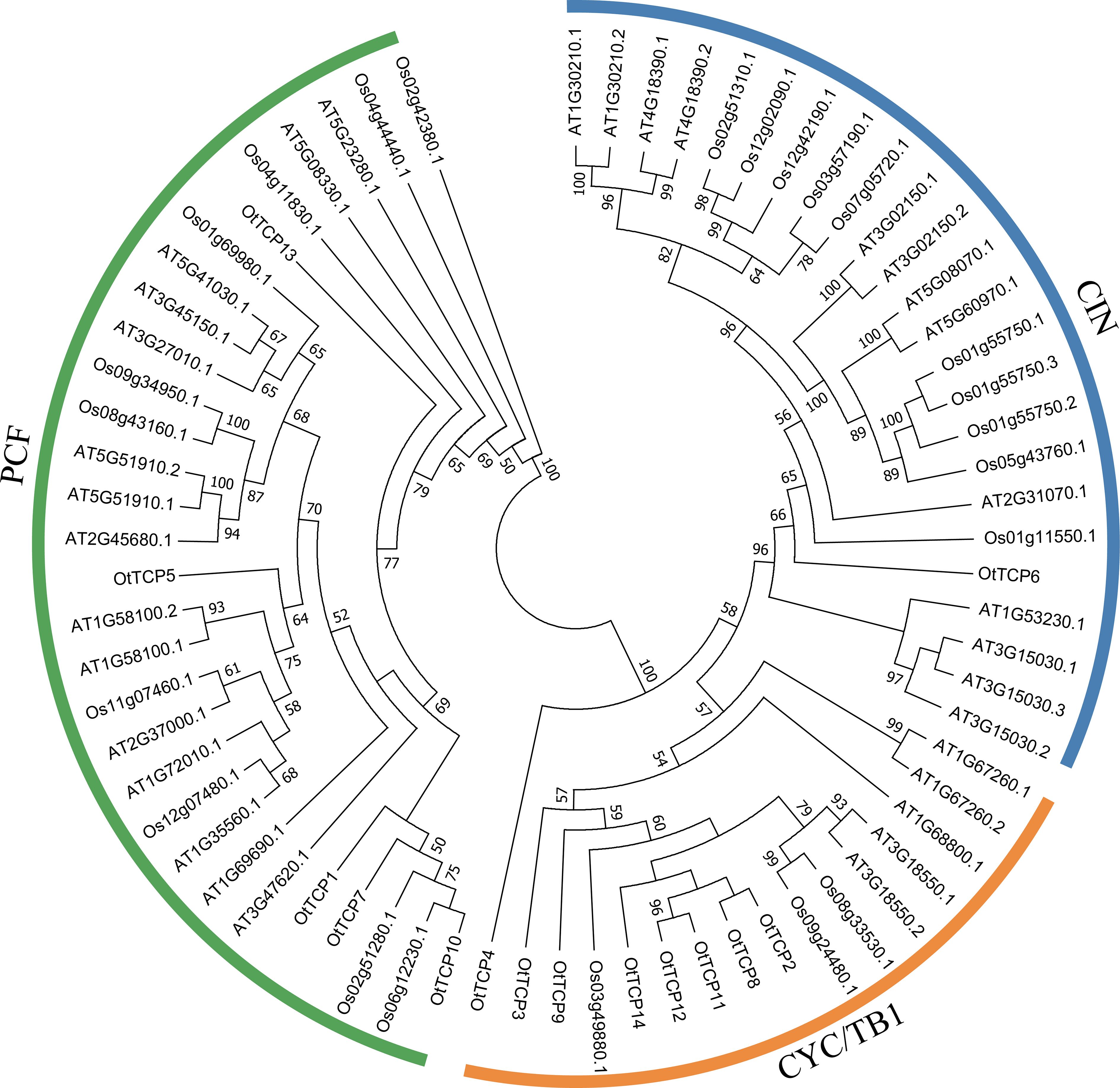
Figure 1. The constructed phylogenetic tree based on the TCP proteins of O. taihangensis (Ot), Arabidopsis thaliana (At), and Oryza sativa (Os). The TCP gene family was mainly divided into two clades: Class I (PCF) and Class II with possessing two subclades (CYC/TB1 and CIN).
Conserved domain sequence alignment analysis was conducted to gain further insights into the evolutionary relationships and structural characteristics of OtTCP genes. The results (Figure 2) revealed that all 14 OtTCPs possessed a conserved domain of 60 amino acid residues. This conserved domain included a primary region at the N-terminus and a HLH (helix–loop–helix) motif at the C-terminus, which was consistent with the TCPs’ structure observed in other plant species (Liu et al., 2022a; Jiang et al., 2023; Wu et al., 2023). Notably, the primary regions of Class I of OtTCPs contained four fewer amino acid residues than that of Class II (Figure 2).
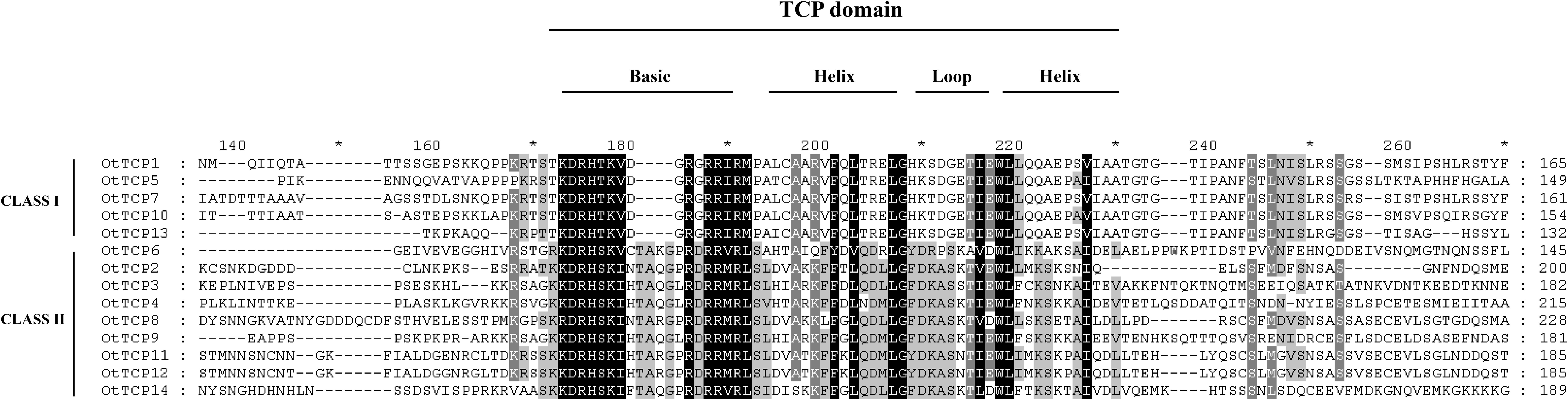
Figure 2. Multiple sequence alignment of OtTCP protein was divided into an alkaline region basic and Helix I–Loop–Helix II.
Regarding the OtTCPs exons and introns, six genes (42.8%) contained introns, while the remaining 8 TCP genes (57.2%) had none. The OtTCP genes of Class I possessed more introns than did Class II. Of the OtTCP genes, five possessed a single intron, whereas only one gene (OtTCP14) had two (Figure 3C).
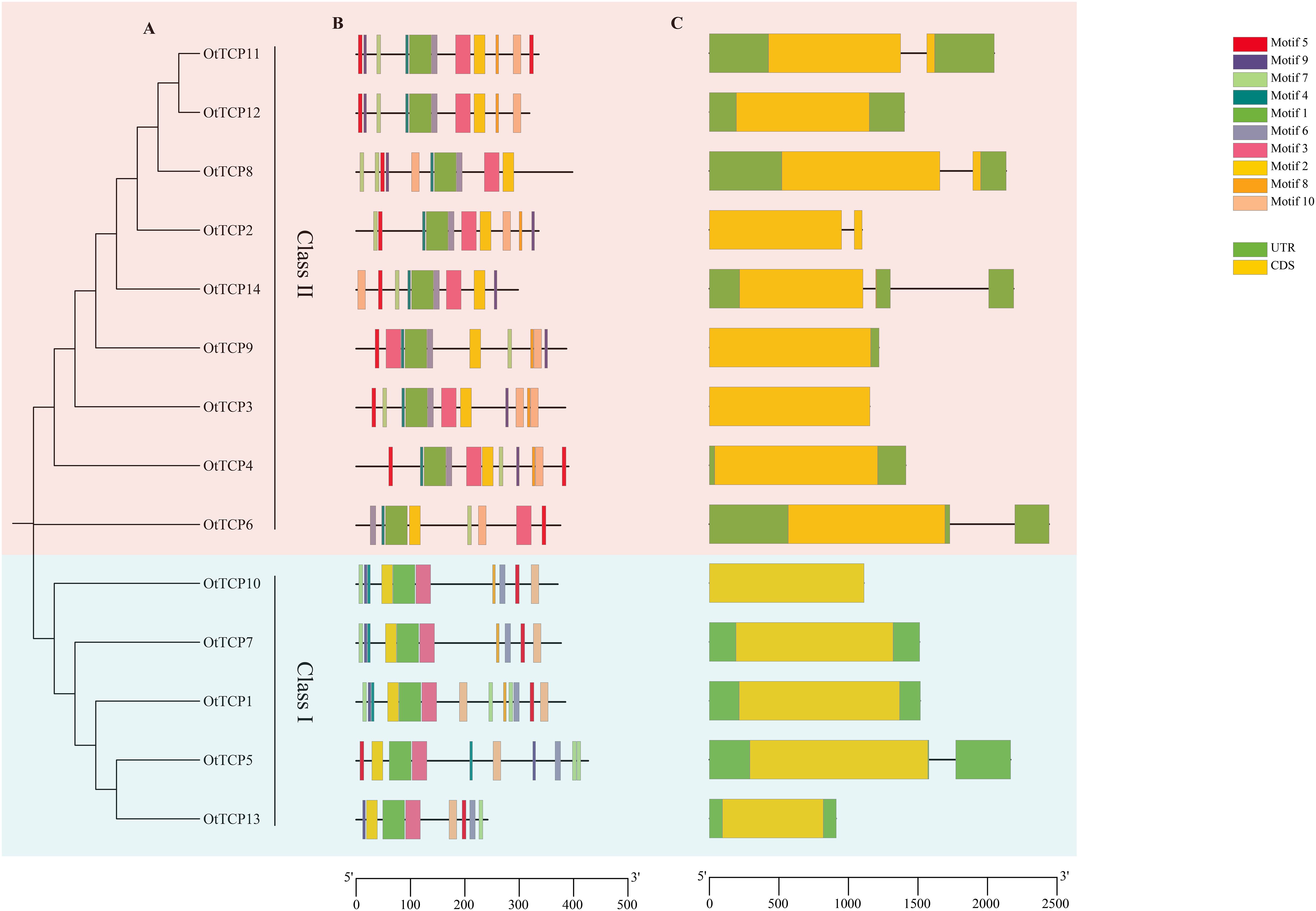
Figure 3. (A) Phylogenetic tree of the OtTCPs with two classes. (B) Conserved motifs of the OtTCP proteins. Different color represented different motif. (C) Exon-intron organization of OtTCP genes. Yellow boxes represented exons (CDS), green boxes represented UTR, and grey lines represented introns. The scale was the sizes of exon or intron.
Ten conserved motifs were identified and designated as motifs 1 - 10. In the CYC/TB1 group of Class II, most genes contained ten motifs except for OtTCP14. The CIN group of Class II did not include motifs 8 and 9, while in the PCF group of Class I, three OtTCPs contained all ten motifs. Further, OtTCP5 did not contain motif 8 and OtTCP13 did not include motifs 4 and 8. Overall, the genetic structures and conserved motifs of most OtTCPs within the same class were similar.
The locations of OtTCP genes on chromosomes were relatively dispersed (Figure 4). Chromosome 1 contained the most OtTCP genes (4 genes, ~28.6%), followed by chromosome 6 (3 genes, ~21%), while chromosomes 2, 3, and 5 had the least (1 gene, ~7%). Chromosomes 4 and 9 held the same number of OtTCP genes (2 each, ~14%), while chromosomes 7 and 8 had no OtTCP genes.
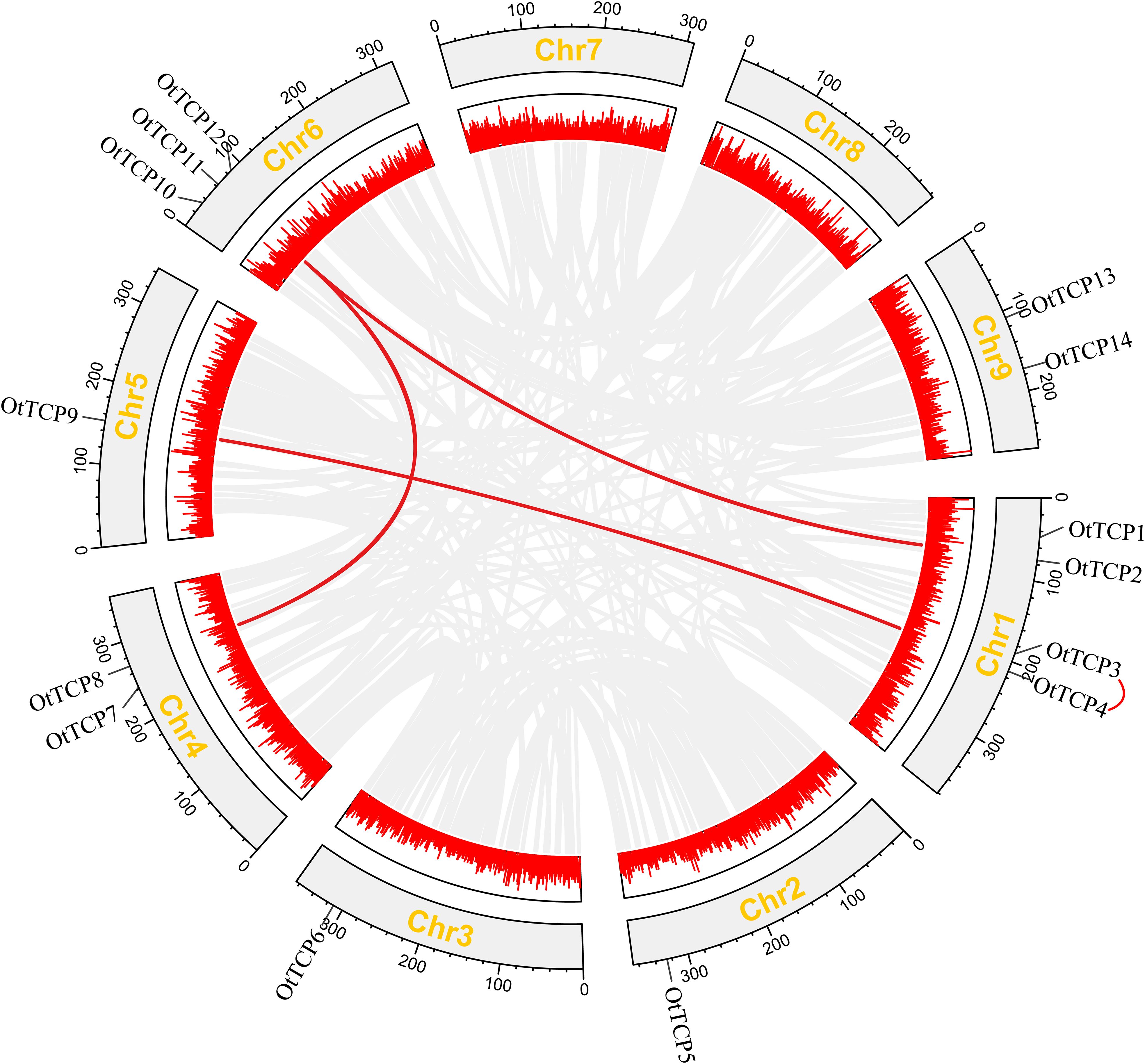
Figure 4. Replication events of OtTCP genes. Gray lines represented the duplicated genes, while the red lines represented the segmental duplicate TCP gene pairs. Additionally, the red lines connecting genes outside the chromosome represented the tandem duplicated pairs. Box with red line graph showed the gene densities. Gray rectangles represented the chromosomes, the corresponding names displayed externally for each chromosome.
The duplication events of OtTCP genes were analyzed, showing that only one tandem repeat gene pair (OtTCP3-OtTCP4) was found on chromosome 1. Three segmental duplication events (OtTCP8-OtTCP11, OtTCP2-OtTCP11 and OtTCP4-OtTCP9) were detected to be scattered across four chromosomes. These results suggested that tandem and segmental duplication events may play key roles in the OtTCP gene family.
The substitution Ka/Ks ratio was used to elucidate OtTCPs evolutionary processes and selection pressures, where a Ka/Ks value of 1 indicated neutral selection, < 1 denoted purification selection, and Ka/Ks > 1 signified positive selection. The Ka/Ks value for tandem duplication was 0.31778, while that for segmental duplication varied from 0.2681 to 0.3804 with a mean value 0.3292 (Supplementary Table S2). The Ka/Ks value for all duplication events was < 1, which implied that OtTCP genes evolved under the effects of purifying selection.
To further illustrate the potential evolutionary relationships of the OtTCP gene family, the comparative collinearity relationships were identified between O. taihangensis and the other 13 species (Figure 5).
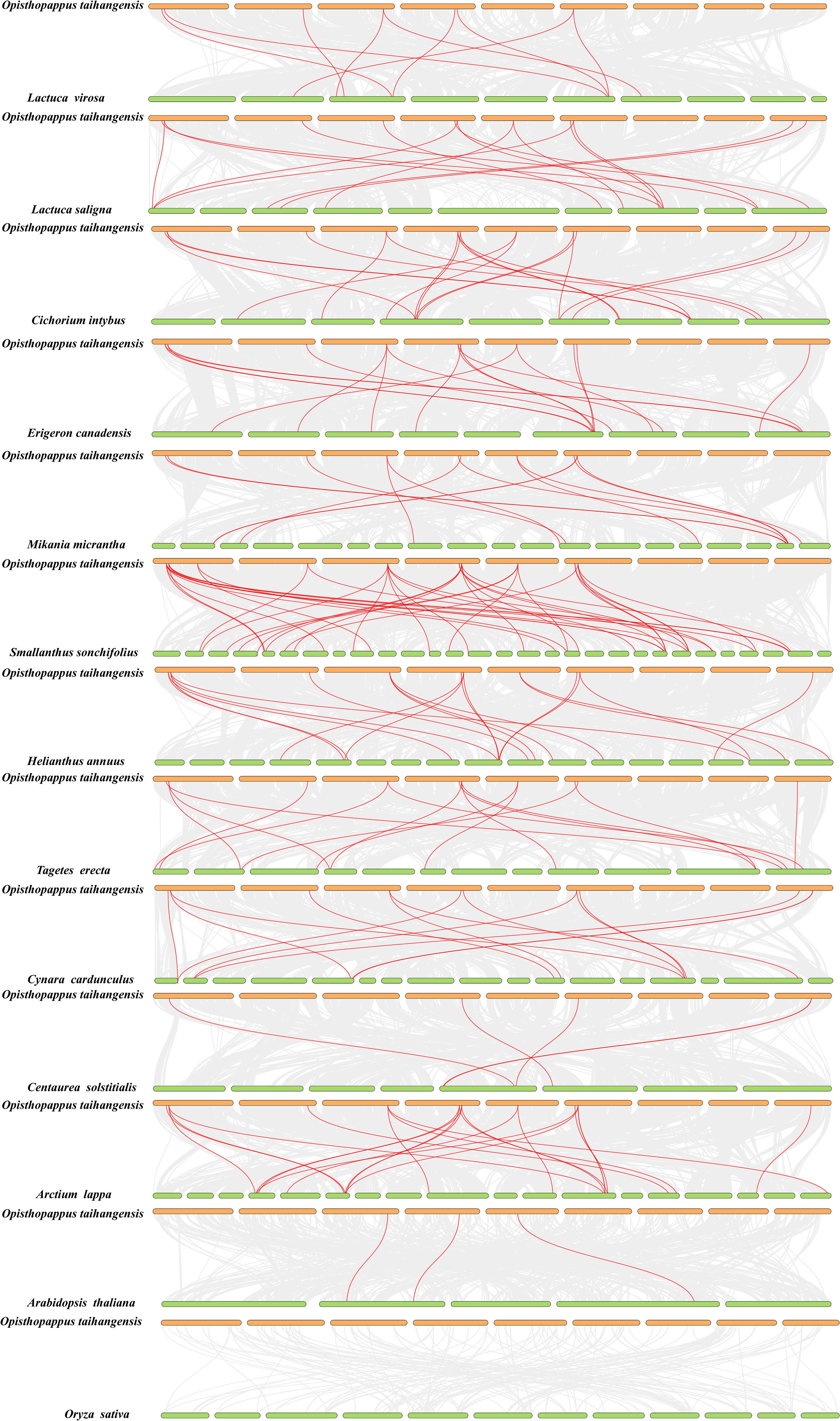
Figure 5. The collinearity analysis among A. thaliana, O. sativa and 11 Asteraceae species. The red line represents TCP collinear gene pairs in O. taihangensis and other genomes.
To further illustrate the potential evolutionary relationships of the OtTCP gene family, the comparative collinearity relationships were identified between O. taihangensis and the other 13 species (Figure 5). The collinear revealed that there were 3 collinear gene pairs between O. taihangensis and A. thaliana, no collinear genes were found in O. sativa. In Asteraceae, 9, 15, 18, 17, 13, 38, 22, 17, 14, 5, and 22 of collinear gene pairs were identified in O. taihangensis with L. virosa, L. saligna, C. intybus, E. canadensis, M. micrantha, S. sonchifolius, H. annuus, T. erecta, C. cardunculus, C. solstitialis, and A. lappa, respectively. The large number of collinear pairs between O. taihangensis and S. sonchifolius indicated a closely relationship among them. Notably, some OtTCP genes were found to have at least three collinear pairs (particularly between O. taihangensis and H. annuus), such as OtTCP2, OtTCP3, OtTCP6, and OtTCP8 (Supplementary Table S3).
A gene gain and loss analysis revealed that TCP genes underwent a dramatic dynamic change in Asteraceae (Figures 6; Supplementary Figure S1), as 103 gain and 243 loss events occurred (Figure 6). Differentiation from the outgroups, the common ancestor of Asteraceae TCPs underwent duplication (+31). Subsequently, loss events during Asteraceae evolution occurred that resulting in the TCP gene family continuously contracted. The most loss occurred in T. erecta (-22), followed H. annuus (-15) and E. canadensis (-12), while the lowest in L.virosa (-1), L. saligna (-2) and C. cardunculus (-3). For O. taihangensis, TCP genes underwent one duplication events and six loss events.
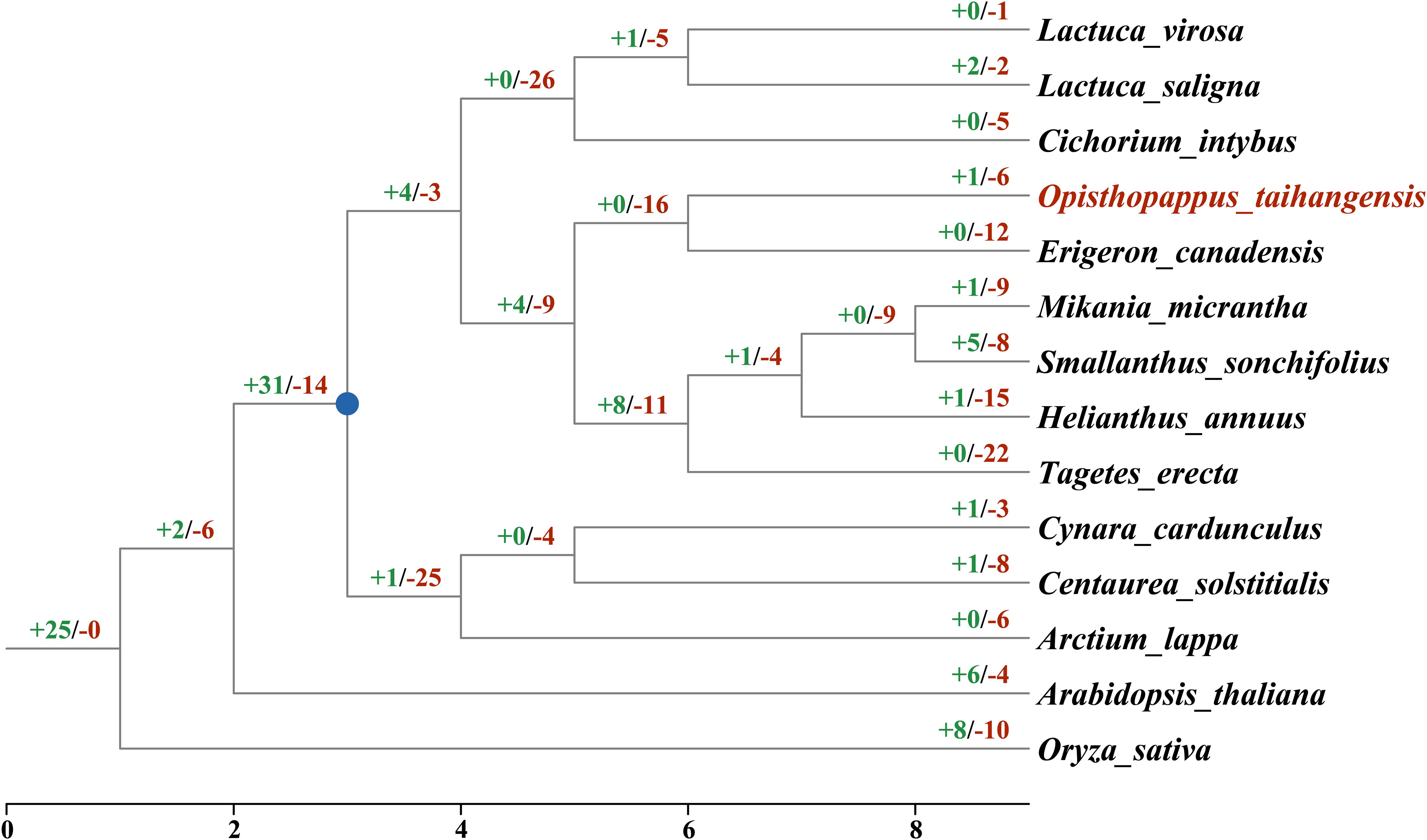
Figure 6. The gain and loss events of TCP genes in the Asteraceae. The numbers at the branch nodes represented the gained and lost genes. The blue nodes represented the possible common ancestor of the Asteraceae.
The results of investigations into the secondary structures of OtTCP proteins (Supplementary Table S4) indicated that they were primarily comprised of α-helices (12.27%–34.03%), extended strands (8.36%–16.60%), β-turns (1.19%–9.13%), and random coils (2.05%–73.33%). The tertiary structures contained α-helices, β-turns, and random coil structures (Supplementary Figure S2), which translated to distinct OtTCP protein conformations and implied their functional differentiation.
In total, 305 cis-acting elements attributed to 22 types were identified in OtTCP genes (Figure 7; Supplementary Table S5). These elements were segregated into four categories (light response, growth and development, hormone response, and stress-related cis-acting elements).
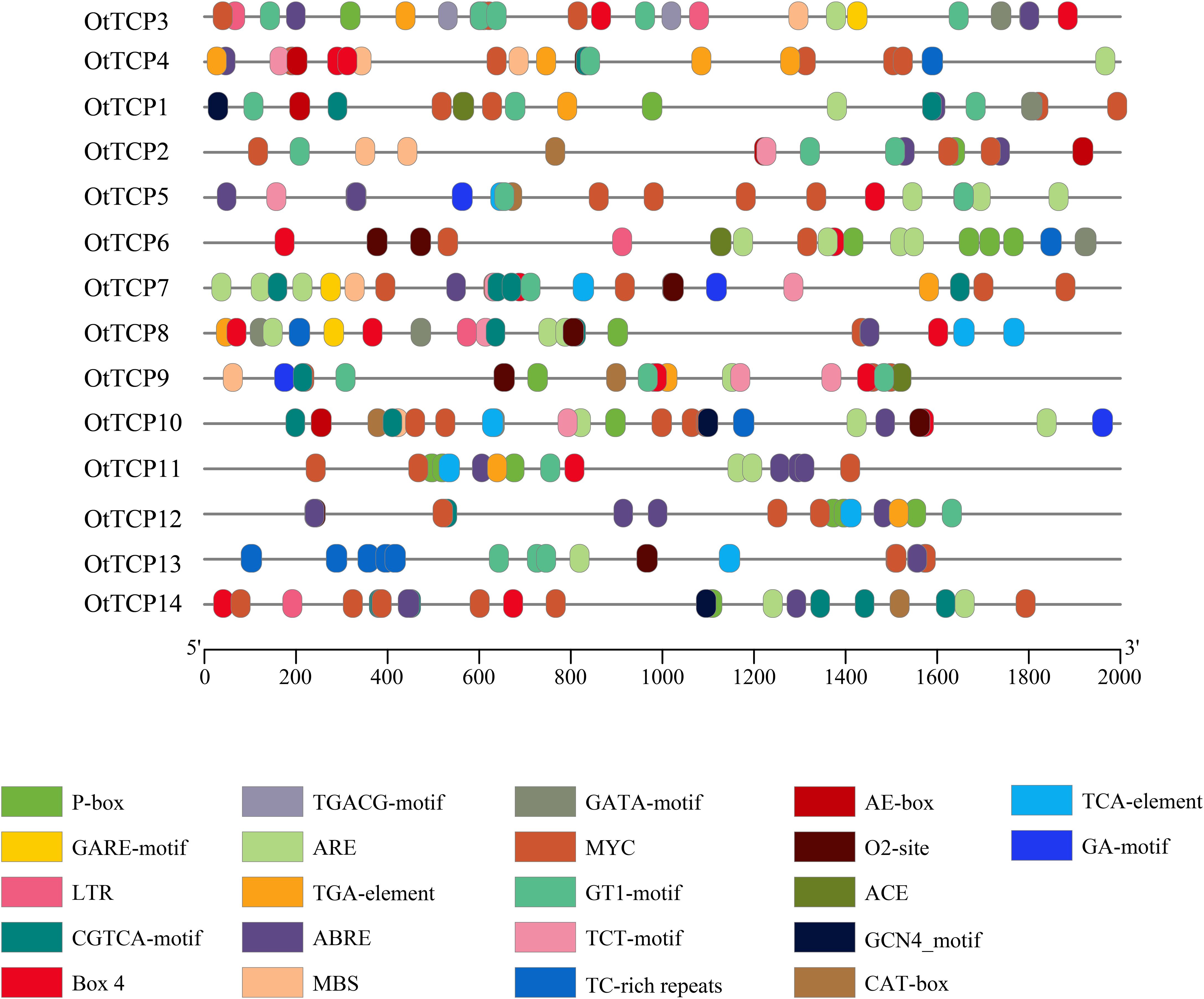
Figure 7. Cis-acting elements in the OtTCP promoter regions. Different colors represented various elements.
All OtTCP genes had light responses. However, the types and quantities of each OtTCP gene varied, which suggested that light signals may positively impact transcriptional regulation processes. Twelve OtTCP genes contained cis-acting elements related to growth and development, including the CGN4-motif, CAT-box, and O2-site. Hormone-responsive elements, such as ABRE (ABA-responsive element), TGA elements, CGTCA motifs, and TGACG motifs (elements involved in MeJA responsiveness), and Gibberellic acid-responsive elements (GAREs), were also screened. ABA-responsive elements (ABREs) were identified in 12 (86%) OtTCP genes (save for OtTCP6 and OtTCP9). Further, stress-related cis-regulatory elements including MBS (drought-induced response element), LTR (low-temperature response element), ARE (anaerobically induced response element), and TC-rich (defense and stress response element), were identified in the promoter regions of 14 OtTCP genes.
Using the download and our previous transcriptomic datasets (Ye et al., 2024), we analyzed the expressions of 14 OtTCP genes in different O. taihangensis tissues, including stems, leaves, roots, buds, and flowers (Gu et al., 2019; Yang et al., 2020) (Figure 8A; Supplementary Table S6). Tissue-specific expressions were predominantly observed for 8 OtTCP genes (OtTCP1, OtTCP7, OtTCP8, OtTCP9, OtTCP11, OtTCP12, OtTCP13, and OtTCP14) in stems; 4 OtTCP genes (OtTCP1, OtTCP3, OtTCP6, and OtTCP10) overrepresented in leaves; 3 OtTCP genes (OtTCP5, OtTCP7, and OtTCP14) highly expressed in roots; OtTCP2 mainly expressed in buds, and OtTCP4 primarily expressed in flowers.
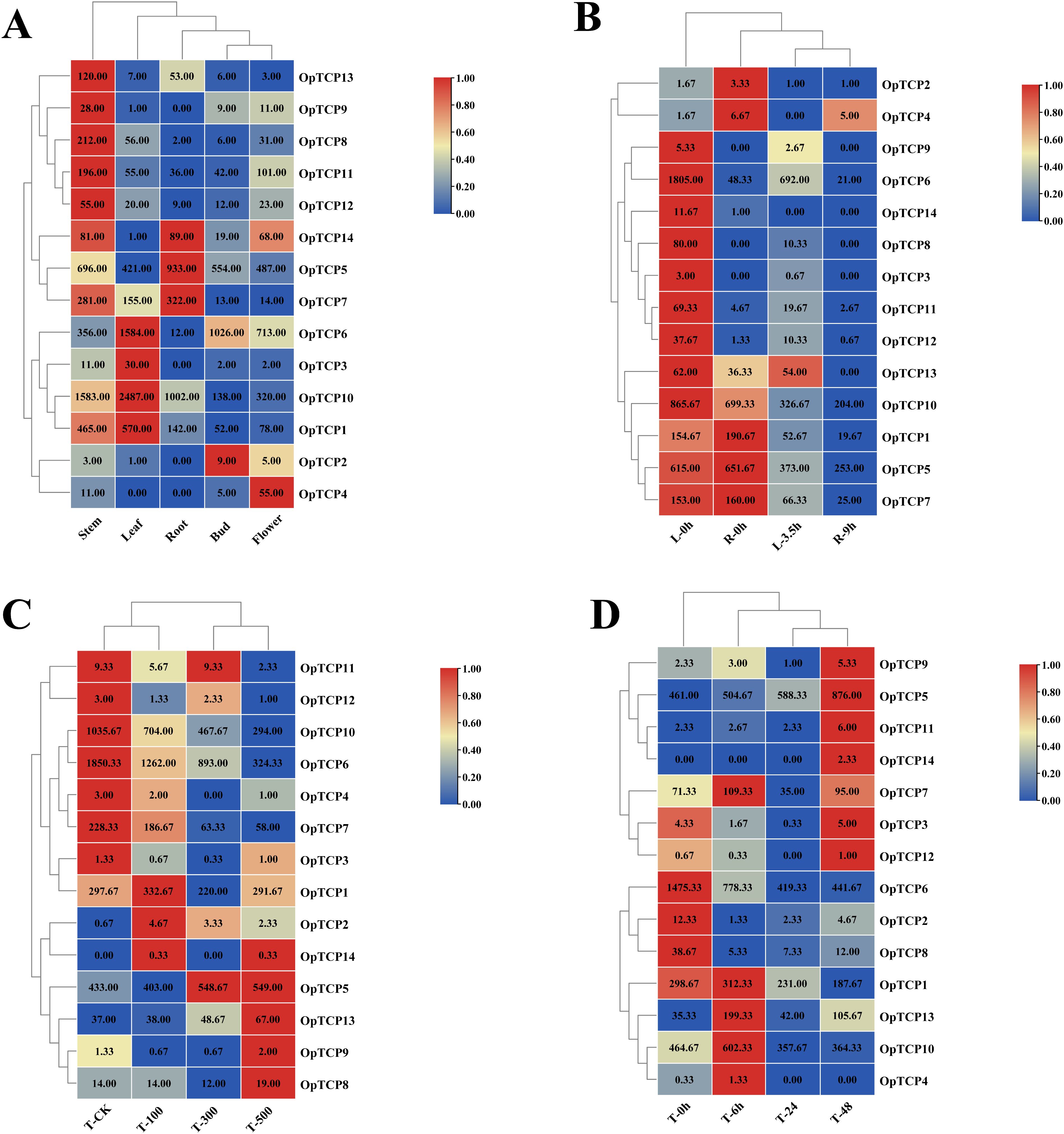
Figure 8. Expression patterns of OtTCPs. (A) Expression profiles of the OtTCPs in different tissues. (B) Leaves treated with 20% PEG6000 for 3.5 hours or untreated (0 hours); Roots treated with 20% PEG6000 for 9 hours or untreated (0 hours). (C) 100 Mm/L, 300 Mm/L, and 500 Mm/L mixed salt solution for 24h treatment. (D) 500 Mm/L mixed salt solution in leaves for 6h, 24h, and 48h treatment. The different colored boxes indicated the different log2 (FPKM) values, the red blocks indicated high relative expression levels and blue blocks indicated low relative expression levels.
To further explore the roles of these OtTCP genes under drought and salt stress, we compared the expression patterns across various treatments (Figure 8; Supplementary Tables S7, S8). The expression levels of OtTCP genes were diverse, with 12 genes being highly expressed in leaves, and seven being highly expressed in roots at 0h under a 20% PEG6000 treatment. All genes showed a downward trend when subjected to different levels of drought stress. Overall, the TCP genes showed high expression levels in leaves, but not in roots, which presented tissue-specific expression patterns.
The expressions of most OtTCP genes were altered under increasing salt concentrations (Figures 8C, D). Under the 100 mM and 300 mM treatments, the expressions of most genes were rapidly induced in the early stages (e.g., OtTCP1, OtTCP2, OtTCP3, OtTCP4, OtTCP5, OtTCP7, and OtTCP14). The expressions of OtTCP5, OtTCP8, OtTCP9, OtTCP13, and OtTCP14 peaked at 500 mM/L. Meanwhile, half of the OtTCP gene expressions increased over time gradients (Figure 8D).
The qRT-PCR showed the relative genes were significant induced or inhibited under various salt treatments, with their expression levels consistent with the previous results. These verified the precision of our analyses (Figure 9).
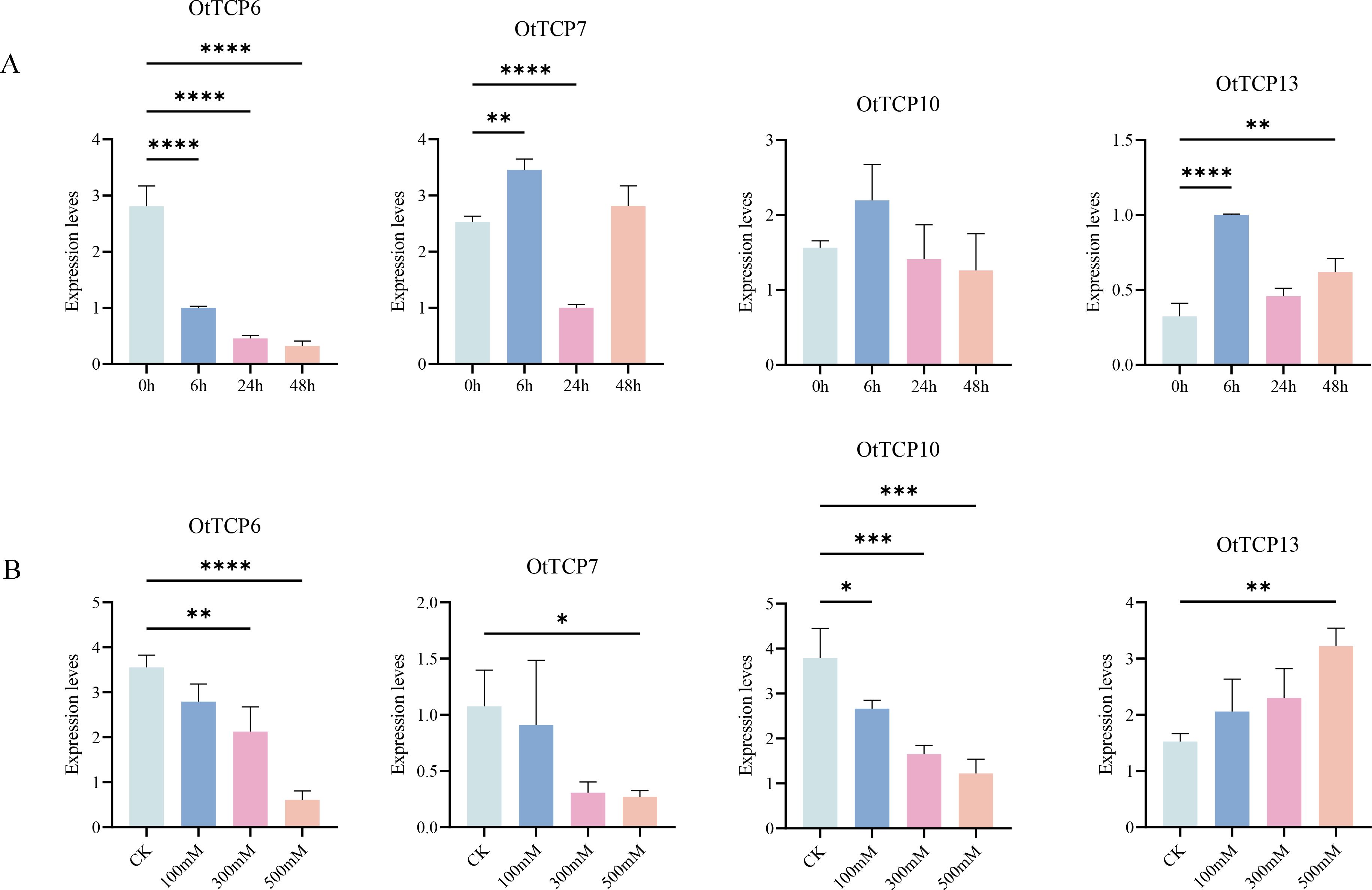
Figure 9. Expression patterns of OtTCP genes under qRT-PCR. (A) represented the treatment of 0h, 6h, 24h, and 48h of 500 Mm/L salt stress. (B) represented CK, 100 Mm/L, 300 Mm/L, and 500 Mm/L salt stress for 24h treatment. Vertical bars represented the mean ± SD of three biological replicates. Statistical significance was determined using one-way ANOVA (**** p < 0.0001 *** p < 0.001 ** p < 0.01* p < 0.05).
Multiple studies have established that TCP genes play a widespread role in diverse physiological and biological processes, encompassing plant growth and abiotic stress responses (Huo et al., 2019; Liu et al., 2022a; Panzade et al., 2024). In this study, we performed the whole genome and transcriptomic data to explore the TCP gene family in O. taihangensis.
A total of 14 identified TCP genes (OtTCP) in O. taihangensis were classified into two main clades (Class I and Class II) and three subfamilies (PCF, CIN, CYC/TB1). Within each subfamily, the TCP gene members were from O. taihangensis, A. thaliana, and O. sativa. This suggested that these genes originated from the common ancestors, which was consistent with preceding studies on other species (Jiang et al., 2023).
Gene structures and conserved motifs provide clues for the prediction of the evolution of genes and their corresponding proteins (Cao et al., 2019). The TCP genes in O. taihangensis lacked introns or had only single or two introns (Figure 3). This structural feature was also found in Camellia sinensis (Shang et al., 2022), Cymbidium goeringii (Liu et al., 2022b), and Dactylis glomerata (Wang et al., 2023a). With fewer introns, genes can rapidly generate more proteins and quickly respond to abiotic stresses (Ma et al., 2021). For OtTCP genes, the lack of introns might be a strategy for responding to abiotic stresses.
Previous studies revealed that the CIN clade was relatively ancient in the TCP gene family. In this study, the CIN clade contained only one gene (OtTCP6) that possessed one intron (Figure 3). OtTCP genes gradually lose introns during evolution, which might be due to inversion or homologous recombination with intron-containing genes (Wu et al., 2005). OtTCP1, OtTCP4, OtTCP7, OtTCP10, and OtTCP13 had no introns; thus, they could rapidly express under abiotic stress (Figures 7C, D). Smaller genes (such as OtTCP14) that contained more introns might be involved in biological processes such as mRNA output and alternative splicing, which could modify their functionalities to a certain extent (Roy and Gilbert, 2006).
Meanwhile, in terms of structure, ancient OtTCP6 contained no motifs 8 or 9. This suggested that certain members of Class I (PCF) and Class II (CYC/TB1) eventually experienced increases in motifs 8 and 9. These structural changes may either support original functionality or induce increased functional diversity.
In contrast to other plants, the number of TCP genes in O. taihangensis was lower than that in Chrysanthemum lavandulifolium (39) (Wu et al., 2023), Chrysanthemum nankingense (23) (Yu et al., 2022b), A. thaliana (33) (Yu et al., 2022a) and O. sativa (23) (Li et al., 2017).
Generally, there were 14 to 38 TCPs found in angiosperms (Liu et al., 2019). Moreover, the TCP gene number was increased with the evolution of species from early-diverging to later-diverging (Martín-Trillo and Cubas, 2010; Shang et al., 2022). For example, Amborella trichopoda (belonging to Amborella of Amborellaceae) was a species of basic angiosperm group and had 15 TCP genes. Some eudicots, such as Aquilegia coerulea (14), Citrus sinensis (15), Eucalyptus grandis (16), and Vitis vinifera (15), also had less TCP genes (Liu et al., 2019). In Asteraceae, Opisthopappus genus was regarded as a relative close ancestral group of Ajania (Zhao, 2007). O. taihangensis possessing 14 TCP genes may be related with its phylogenic position (Shen et al., 2021).
Through chromosome localization analysis, it was found that OtTCP genes were unevenly distributed across nine chromosomes in the O. taihangensis genome. The uneven distribution of genes in genomic chromosomes was closely related to extensive gene loss, which is pervasive in angiosperm (Sun et al., 2023).
As know, whole genome duplication (WGD) is one of the most important driving forces for genome evolution (Magadum et al., 2013). Large number of duplicated genes would be produced after WGD (Chen et al., 2023). Here, we identified OtTCP genes experienced by tandem and segmental duplication (Figure 4). In our other study (unpulished), O. taihangensis genome was detected undergone WGD event at 59 Mya. However, more loss (-6) and less gain (+1) events of OtTCP genes occurred during the evolution (Figures 4 and Supplementary Figure S1). Some studies showed that the frequency of gene loss is up to three times higher than the rate of gene gain (Koskiniemi et al., 2012; Puigbò et al., 2014; Nelson-Sathi et al., 2015). After WGD, some functionally important gene copies can be retained, whereas some functionally redundant gene copies would be lost or pseudogenized (Duan et al., 2014; Liang et al., 2016). For O. taihangensis TCP genes, the loss events should be post whole genome duplication, and only keep some important copies (Li et al., 2024). Based on Zhang et al. (2024a), CYC2 genes of TCP family were experienced the duplications that predated their gains during the evolution of florets and floral symmetry in Asteraceae. More, the loss of CYC2d were found in the formation of ligulate florets (Chen et al., 2018; Zhang et al., 2024a). These may support our results at a certain extent.
Gene loss can contribute to species’ adaptive evolution, particularly in response to environmental challenges (Albalat and Cañestro, 2016). Under selection, positive selective pressure facilitates gene expansion or functional differentiation, whereas purifying selective pressure often renders more conservative genes (Song and Nan, 2014). Indeed, purifying selection occurred during the evolution of OtTCP genes (with Ka/Ks values consistently < 1). This selective pressure may have ultimately led to the contraction or loss of OtTCPs (Wu et al., 2022). The gene loss in the OtTCP family might be an adaptive strategy for O. taihangensis on the cliff habitats.
On the other hand, the high collinearity (38 syntenic blocks) occurred between O. taihangensis (one member of Asterodae) and S. sonchifolius (one member of Helianthodae) in the studied Asteraceae species (Figure 5). It indicated that these genes located in corresponding syntenic blocks occurred before the divergence of O. taihangensis and S. sonchifolius. Asterodae and Helianthodae both were the members of Asteroideae and diverged about 57.71 Mya after ancient WGD event (Zhang et al., 2024a). High collinearity among the two species should be happened before 57.71 Mya.
As pivotal molecular switches, cis-regulatory elements participate in the transcriptional regulation of genes and control a variety of biological processes (Huang et al., 2021). OtTCPs were found that enriched with cis-regulatory elements associated with growth and development, hormone signaling, and stress responses (Figure 7).
All TCP genes of O. taihangensis possessed photonically responsive elements, indicating that OtTCP genes responded to light for the regulation of growth and development in O. taihangensis, which aligned with the results of C. goeringii (Liu et al., 2022b). A dozen of 14 OtTCP genes contained ABRE cis-regulatory elements associated with ABA responsiveness. ABRE-binding protein/ABRE-binding factor (AREB/ABF) can positively regulate the plant responses and enhance tolerance (Fujita et al., 2013; Yang et al., 2024), while the ABA signaling pathway is crucial for abiotic stress resistance. Plants challenged by water deficits, salinity, cold, or pathogen attacks induce the accumulation of ABA, which translates to gene expression via ABRE cis-acting elements to defend against these stresses (Dar et al., 2017). OsTCP19 gene from rice, which activated by salt, drought, and cold stresses, enhances ABA signal transduction by promoting the expression of ABA INSENSITIVE4, which interacts directly with relative encoded proteins (Tatematsu et al., 2008; Rueda-Romero et al., 2012). The TCP10 gene of Moso bamboo positively regulates early tolerance by regulating the ABA signaling pathway, which negatively regulates lateral root growth via the methyl jasmonate (Me-JA)-mediated signaling pathway (Xu et al., 2022). In A. thaliana, the TCP14 gene interacts with the DNA BINDING WITH ONE FINGER 6 transcription factor, inhibiting the activation of the ABA biosynthetic gene ABA DEFICIENT1 and other ABA-related stress genes, and then promoting the germination of Arabidopsis seeds (Tatematsu et al., 2008; Rueda-Romero et al., 2012).
Conversely, MBS (MYB binding site) is renowned for its key roles in stress signaling transduction and drought stress responses (Guo et al., 2023). TC-rich repeats are involved in defense and stress responses, while LTR elements engage low temperature stress responses. Six OtTCP genes (OtTCP2, OtTCP3, OtTCP4, OtTCP7, OtTCP9, and OtTCP10), five OtTCP genes (OtTCP4, OtTCP6, OtTCP8, OtTCP10, and OtTCP13), and four OtTCP genes (OtTCP3, OtTCP6, OtTCP8, and OtTCP14) contained MBS, TC-rich, and LTR cis-regulatory elements, respectively. This indicated that OtTCPs might utilize differential regulatory pathways to counter abiotic stresses.
It is widely recognized that the expression profiles of genes are intimately linked with their functionalities to a large extent. TCP genes in O. taihangensis exhibit significantly different expression patterns in different tissues and treatments (Figure 7). Generally, CYC members in TCP family regulate branching, such as TB1 in maize and BRC1 in Arabidopsis (Cubas et al., 1999; Aguilar-Martínez et al., 2007); and some involved in flower development (Balsemão-Pires et al., 2013). OtTCP2, 3, 4, 8, 9, 11, 12, 14 all were the genes of CYC clade. OtTCP2 up-expressed in bud, OtTCP3 in leaf, OtTCP4 in flower, OtTCP8, 9, 11, 12 in stem, and OtTCP14 in stem, root and flower (Figure 8A). The members in PCF clade of TCP family Class I also are involved in plant development (Liu et al., 2019). OtTCP1, 5, 7, 10 and 13 (PCF genes) mainly expressed in stem, root and leaf. Recently, PCF genes were demonstrated to participate in abiotic stresses (Liu et al., 2019). OtPCF genes high expressed under different salt treatments in this study (Figures 8C, D). OtTCP genes expression profiles indicated that their diverse functions, which may play important roles in the growth and development of O. taihangensis.
Additionally, some pairs of duplicate genes revealed similar or distinct expression patterns (Zhang et al., 2024b). For example, OtTCP8 and OtTCP11 exhibited a negative expression trend under drought treatments. However, under salt stress, OtTCP11 showed an upward trend, while OtTCP8 showed the converse. Duplicate genes responded to different stresses through functional diversity. Gene replication can drive the development of new biological functions, which was supported by the tertiary structures of OtTCP genes (Supplementary Figure S2).
Within different tissues and under salt stress and drought, OtTCP4, 9 and 11 presented a relative wide expression (such as OtTCP4 up-expressed in flower, R-9h, T-100 and T-6h, Figure 9). These three genes all were the member of CYC clade and had no or one intron, which contained ABRE and/or MYC/MYB cis-acting elements with the capacity to rapidly respond to stressors. Thus, OtTCP4, 9 and 11 could be considered as the candidates for the development, growth and responding to stresses of O. taihangensis, although further detailed research is necessary.
In O. taihangensis, 14 TCP genes were identified. Compared with other species, relative less TCP genes might be accsioated with its ancestral phylogenic position. The OtTCP gene family mainly underwent gene loss events after duplication, which could induce adaptive genetic changes. When challenged the stressors, those OtTCPs that lack introns can quickly respond primarily through different cis-regulatory elements. More, OtTCP genes exhibit different expression patterns in different tissues and treatments. Thereinto, OtTCP4, 9 and 11 could be recognized as important candidates for O. taihangensis with a wide expression model. These data may provide clues for the further exploration of the potential resistance mechanisms of O. taihangensis in the cliff environments of the Taihang Mountains.
The data presented in this study are available in the article and Supplementary Materials. The transcriptome data of O. taihangensis under drought treatment were downloaded from the NCBI with the accession number PRJNA400848 (leaf tissues, 20% PEG6000 treatment for 0 h and 3.5 h with three replicates), PRJNA437359 (root tissues, 20% PEG6000 treatment for 0 h and 9 h with three replicates). The specimens of O. taihangensis are stored in the Herbarium of Shanxi Normal University, with the storage number SNUP20230988.
TG: Writing – original draft, Data curation, Visualization, Software. XZ: Writing – original draft, Data curation, Visualization, Software. MH: Investigation, Validation, Writing – original draft. YS: Formal Analysis, Writing – original draft. YZ: Formal Analysis, Writing – original draft. QW: Data curation, Investigation, Writing – original draft. HD: Data curation, Investigation, Writing – original draft. TW: Investigation, Writing – original draft. HY: Investigation, Writing – original draft. LL: Writing – review & editing, Supervision. MC: Writing – review & editing, Supervision. YW: Funding acquisition, Writing – review & editing, Supervision.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (31970358).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1499244/full#supplementary-material
Aggarwal, P., Das Gupta, M., Joseph, A. P., Chatterjee, N., Srinivasan, N., Nath, U. (2010). Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis. Plant Cell 22, 1174–1189. doi: 10.1105/tpc.109.066647
Aguilar-Martínez, J. A., Poza-Carrión, C. S., Cubas, P. (2007). Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19, 458–472. doi: 10.1105/tpc.106.048934
Albalat, R., Cañestro, C. (2016). Evolution by gene loss. Nat. Rev. Genet. 17, 379–391. doi: 10.1038/nrg.2016.39
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Balsemão-Pires, E., Andrade, L. R., Sachetto-Martins, G. (2013). Functional study of TCP23 in Arabidopsis thaliana during plant development. Plant Physiol. Biochem. 67, 120–125. doi: 10.1016/j.plaphy.2013.03.009
Bello, M. A., Cubas, P., Álvarez, I., Sanjuanbenito, G., Fuertes-Aguilar, J. (2017). Evolution and expression patterns of CYC/TB1 genes in anacyclus: phylogenetic insights for floral symmetry genes in asteraceae. Front. Plant Sci. 8, 589. doi: 10.3389/fpls.2017.00589
Cao, S., Guo, M., Wang, C., Xu, W., Shi, T., Tong, G., et al. (2019). Genome-wide characterization of aspartic protease (AP) gene family in Populus trichocarpa and identification of the potential PtAPs involved in wood formation. BMC Plant Biol. 19, 276. doi: 10.1186/s12870-019-1865-0
Chai, M., Wang, S., He, J., Chen, W., Fan, Z., Li, J., et al. (2018). De novo assembly and transcriptome characterization of opisthopappus (Asteraceae) for population differentiation and adaption. Front. Genet. 9, 371. doi: 10.3389/fgene.2018.00371
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, H., Guo, M., Dong, S., Wu, X., Zhang, G., He, L., et al. (2023). A chromosome-scale genome assembly of Artemisia argyi reveals unbiased subgenome evolution and key contributions of gene duplication to volatile terpenoid diversity. Plant Commun. 4, 100516. doi: 10.1016/j.xplc.2023.100516
Chen, J., Shen, C. Z., Guo, Y. P., Rao, G. Y. (2018). Patterning the asteraceae capitulum: duplications and differential expression of the flower symmetry CYC2-like genes. Front. Plant Sci. 9, 551. doi: 10.3389/fpls.2018.00551
Chen, K., Durand, D., Farach-Colton, M. (2000). NOTUNG: a program for dating gene duplications and optimizing gene family trees. J. Comput. Biol. 7, 429–447. doi: 10.1089/106652700750050871
Chen, G. H., Sun, J. Y., Liu, M., Liu, J., Yang, W. C. (2014). SPOROCYTELESS is a novel embryophyte-specific transcription repressor that interacts with TPL and TCP proteins in Arabidopsis. J. Genet. Genomics 41, 617–625. doi: 10.1016/j.jgg.2014.08.009
Chen, N., Zhang, H., Zang, E., Liu, Z. X., Lan, Y. F., Hao, W. L., et al. (2022). Adaptation insights from comparative transcriptome analysis of two Opisthopappus species in the Taihang mountains. BMC Genomics 23, 466. doi: 10.1186/s12864-022-08703-5
Crawford, B. C., Nath, U., Carpenter, R., Coen, E. S. (2004). CINCINNATA controls both cell differentiation and growth in petal lobes and leaves of Antirrhinum. Plant Physiol. 135, 244–253. doi: 10.1104/pp.103.036368
Cubas, P., Lauter, N., Doebley, J., Coen, E. (1999). The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 18, 215–222. doi: 10.1046/j.1365-313X.1999.00444.x
Danisman, S., Van Dijk, A. D., Bimbo, A., van der Wal, F., Hennig, L., De Folter, S., et al. (2013). Analysis of functional redundancies within the Arabidopsis TCP transcription factor family. J. Exp. Bot. 64, 5673–5685. doi: 10.1093/jxb/ert337
Dar, N. A., Amin, I., Wani, W., Wani, S. A., Shikari, A. B., Wani, S. H., et al. (2017). Abscisic acid: A key regulator of abiotic stress tolerance in plants. Plant Gene 11, 106–111. doi: 10.1016/j.plgene.2017.07.003
Davière, J. M., Wild, M., Regnault, T., Baumberger, N., Eisler, H., Genschik, P., et al. (2014). Class I TCP-DELLA interactions in inflorescence shoot apex determine plant height. Curr. Biol. 24, 1923–1928. doi: 10.1016/j.cub.2014.07.012
Ding, S., Cai, Z., Du, H., Wang, H. (2019). Genome-wide analysis of TCP family genes in Zea mays L. Identified a role for ZmTCP42 in drought tolerance. Int. J. Mol. Sci. 20 (11), 2762. doi: 10.3390/ijms20112762
Dixon, L. E., Greenwood, J. R., Bencivenga, S., Zhang, P., Cockram, J., Mellers, G., et al. (2018). TEOSINTE BRANCHED1 regulates inflorescence architecture and development in bread wheat (Triticum aestivum). Plant Cell 30, 563–581. doi: 10.1105/tpc.17.00961
Doebley, J., Stec, A., Hubbard, L. (1997). The evolution of apical dominance in maize. Nature 386, 485–488. doi: 10.1038/386485a0
Duan, W., Song, X., Liu, T., Huang, Z., Ren, J., Hou, X., et al. (2014). Patterns of evolutionary conservation of ascorbic acid-related genes following whole-genome triplication in Brassica rapa. Genome Biol. Evol. 7, 299–313. doi: 10.1093/gbe/evu293
Duvaud, S., Gabella, C., Lisacek, F., Stockinger, H., Ioannidis, V., Durinx, C. (2021). Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 49, W216–w227. doi: 10.1093/nar/gkab225
Fujita, Y., Yoshida, T., Yamaguchi-Shinozaki, K. (2013). Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol. Plant 147, 15–27. doi: 10.1111/j.1399-3054.2012.01635.x
Geourjon, C., Deléage, G. (1995). SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics 11, 681–684. doi: 10.1093/bioinformatics/11.6.681
Gu, H., Yang, Y., Xing, M., Yue, C., Wei, F., Zhang, Y., et al. (2019). Physiological and transcriptome analyses of Opisthopappus taihangensis in response to drought stress. Cell Biosci. 9, 56. doi: 10.1186/s13578-019-0318-7
Guan, P., Ripoll, J. J., Wang, R., Vuong, L., Bailey-Steinitz, L. J., Ye, D., et al. (2017). Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proc. Natl. Acad. Sci. U.S.A. 114, 2419–2424. doi: 10.1073/pnas.1615676114
Guo, Z., He, L., Sun, X., Li, C., Su, J., Zhou, H., et al. (2023). Genome-wide analysis of the rhododendron AP2/ERF gene family: identification and expression profiles in response to cold, salt and drought stress. Plants (Basel) 12 (5), 994. doi: 10.3390/plants12050994
Guruprasad, K., Reddy, B. V., Pandit, M. W. (1990). Correlation between stability of a protein and its dipeptide composition: a novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng. 4, 155–161. doi: 10.1093/protein/4.2.155
Han, M., Niu, M., Gao, T., Shen, Y., Zhou, X., Zhang, Y., et al. (2024). Responsive alternative splicing events of opisthopappus species against salt stress. Int. J. Mol. Sci. 25, 1227. doi: 10.3390/ijms25021227
Han, X., Yu, H., Yuan, R., Yang, Y., An, F., Qin, G. (2019). Arabidopsis transcription factor TCP5 controls plant thermomorphogenesis by positively regulating PIF4 activity. iScience 15, 611–622. doi: 10.1016/j.isci.2019.04.005
Hileman, L. C. (2014). Bilateral flower symmetry–how, when and why? Curr. Opin. Plant Biol. 17, 146–152. doi: 10.1016/j.pbi.2013.12.002
Horn, S., Pabón-Mora, N., Theuß, V. S., Busch, A., Zachgo, S. (2015). Analysis of the CYC/TB1 class of TCP transcription factors in basal angiosperms and magnoliids. Plant J. 81, 559–571. doi: 10.1111/tpj.2015.81.issue-4
Horton, P., Park, K.-J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587. doi: 10.1093/nar/gkm259
Howarth, D. G., Donoghue, M. J. (2006). Phylogenetic analysis of the “ECE” (CYC/TB1) clade reveals duplications predating the core eudicots. Proc. Natl. Acad. Sci. U.S.A. 103, 9101–9106. doi: 10.1073/pnas.0602827103
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Huang, L., Li, Z., Fu, Q., Liang, C., Liu, Z., Liu, Q., et al. (2021). Genome-wide identification of CBL-CIPK gene family in honeysuckle (Lonicera japonica thunb.) and their regulated expression under salt stress. Front. Genet. 12, 751040. doi: 10.3389/fgene.2021.751040
Huo, Y., Xiong, W., Su, K., Li, Y., Yang, Y., Fu, C., et al. (2019). Genome-wide analysis of the TCP gene family in switchgrass (Panicum virgatum L.). Int. J. Genomics 2019, 8514928. doi: 10.1155/2019/8514928
Jiang, Y., Jiang, D., Xia, M., Gong, M., Li, H., Xing, H., et al. (2023). Genome-wide identification and expression analysis of the TCP gene family related to developmental and abiotic stress in ginger. Plants (Basel) 12 (19), 3389. doi: 10.3390/plants12193389
Katagiri, F., Chua, N. H. (1992). Plant transcription factors: present knowledge and future challenges. Trends Genet. 8, 22–27. doi: 10.1016/0168-9525(92)90020-5
Koskiniemi, S., Sun, S., Berg, O. G., Andersson, D. I. (2012). Selection-driven gene loss in bacteria. PloS Genet. 8, e1002787. doi: 10.1371/journal.pgen.1002787
Kosugi, S., Ohashi, Y. (2002). DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 30, 337–348. doi: 10.1046/j.1365-313X.2002.01294.x
Koyama, T., Furutani, M., Tasaka, M., Ohme-Takagi, M. (2007). TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19, 473–484. doi: 10.1105/tpc.106.044792
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kumar, S., Stecher, G., Suleski, M., Hedges, S. B. (2017). TimeTree: A resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 34, 1812–1819. doi: 10.1093/molbev/msx116
Lehti-Shiu, M. D., Panchy, N., Wang, P., Uygun, S., Shiu, S. H. (2017). Diversity, expansion, and evolutionary novelty of plant DNA-binding transcription factor families. Biochim. Biophys. Acta Gene Regul. Mech. 1860, 3–20. doi: 10.1016/j.bbagrm.2016.08.005
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van De Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, D., Li, H., Feng, H., Qi, P., Wu, Z. (2024). Unveiling kiwifruit TCP genes: evolution, functions, and expression insights. Plant Signaling Behav. 19, 1. doi: 10.1080/15592324.2024.2338985
Li, W., Li, D. D., Han, L. H., Tao, M., Hu, Q. Q., Wu, W. Y., et al. (2017). Genome-wide identification and characterization of TCP transcription factor genes in upland cotton (Gossypium hirsutum). Sci. Rep. 7, 10118. doi: 10.1038/s41598-017-10609-2
Li, Z., Li, B., Shen, W. H., Huang, H., Dong, A. (2012). TCP transcription factors interact with AS2 in the repression of class-I KNOX genes in Arabidopsis thaliana. Plant J. 71, 99–107. doi: 10.1111/j.1365-313X.2012.04973.x
Li, C., Potuschak, T., Colón-Carmona, A., Gutiérrez, R. A., Doerner, P. (2005). Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc. Natl. Acad. Sci. U.S.A. 102, 12978–12983. doi: 10.1073/pnas.0504039102
Li, S., Zachgo, S. (2013). TCP3 interacts with R2R3-MYB proteins, promotes flavonoid biosynthesis and negatively regulates the auxin response in Arabidopsis thaliana. Plant J. 76, 901–913. doi: 10.1111/tpj.2013.76.issue-6
Liang, Y., Xiong, Z., Zheng, J., Xu, D., Zhu, Z., Xiang, J., et al. (2016). Genome-wide identification, structural analysis and new insights into late embryogenesis abundant (LEA) gene family formation pattern in Brassica napus. Sci. Rep. 6, 24265. doi: 10.1038/srep24265
Liu, D. H., Luo, Y., Han, H., Liu, Y. Z., Alam, S. M., Zhao, H. X., et al. (2022a). Genome-wide analysis of citrus TCP transcription factors and their responses to abiotic stresses. BMC Plant Biol. 22, 325. doi: 10.1186/s12870-022-03709-3
Liu, M. M., Wang, M. M., Yang, J., Wen, J., Guo, P. C., Wu, Y. W., et al. (2019). Evolutionary and comparative expression analyses of TCP transcription factor gene family in land plants. Int. J. Mol. Sci. 20 (14), 3591. doi: 10.3390/ijms20143591
Liu, D. K., Zhang, C., Zhao, X., Ke, S., Li, Y., Zhang, D., et al. (2022b). Genome-wide analysis of the TCP gene family and their expression pattern in Cymbidium goeringii. Front. Plant Sci. 13, 1068969. doi: 10.3389/fpls.2022.1068969
Luo, D., Carpenter, R., Copsey, L., Vincent, C., Clark, J., Coen, E. (1999). Control of organ asymmetry in flowers of Antirrhinum. Cell 99, 367–376. doi: 10.1016/S0092-8674(00)81523-8
Ma, L., Zhu, T., Wang, H., Zhou, H., Shao, L., Ding, Q., et al. (2021). Genome-wide identification, phylogenetic analysis and expression profiling of the late embryogenesis-abundant (LEA) gene family in Brachypodium distachyon. Funct. Plant Biol. 48, 386–401. doi: 10.1071/FP20143
Magadum, S., Banerjee, U., Murugan, P., Gangapur, D., Ravikesavan, R. (2013). Gene duplication as a major force in evolution. J. Genet. 92, 155–161. doi: 10.1007/s12041-013-0212-8
Manassero, N. G., Viola, I. L., Welchen, E., Gonzalez, D. H. (2013). TCP transcription factors: architectures of plant form. Biomol Concepts 4, 111–127. doi: 10.1515/bmc-2012-0051
Marín-De-La-Rosa, N., Sotillo, B., Miskolczi, P., Gibbs, D. J., Vicente, J., Carbonero, P., et al. (2014). Large-scale identification of gibberellin-related transcription factors defines group VII ETHYLENE RESPONSE FACTORS as functional DELLA partners. Plant Physiol. 166, 1022–1032. doi: 10.1104/pp.114.244723
Martín-Trillo, M., Cubas, P. (2010). TCP genes: a family snapshot ten years later. Trends Plant Sci. 15, 31–39. doi: 10.1016/j.tplants.2009.11.003
Mukhopadhyay, P., Tyagi, A. K. (2015). OsTCP19 influences developmental and abiotic stress signaling by modulating ABI4-mediated pathways. Sci. Rep. 5, 9998. doi: 10.1038/srep09998
Nath, U., Crawford, B. C., Carpenter, R., Coen, E. (2003). Genetic control of surface curvature. Science 299, 1404–1407. doi: 10.1126/science.1079354
Navaud, O., Dabos, P., Carnus, E., Tremousaygue, D., Hervé, C. (2007). TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 65, 23–33. doi: 10.1007/s00239-006-0174-z
Nelson-Sathi, S., Sousa, F. L., Roettger, M., Lozada-Chávez, N., Thiergart, T., Janssen, A., et al. (2015). Origins of major archaeal clades correspond to gene acquisitions from bacteria. Nature 517, 77–80. doi: 10.1038/nature13805
Nguyen, L.-T., Schmidt, H. A., Von Haeseler, A., Minh, B. Q. (2014). IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nicholas, K., Nicholas, H. (1997). GeneDoc: a tool for editing and annotating multiple sequence alignments. Computer Science, Biology.
Palatnik, J. F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J. C., et al. (2003). Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. doi: 10.1038/nature01958
Panzade, K. P., Vishwakarma, H., Kharate, P. S., Azameti, M. K. (2024). Genome-wide analysis and expression profile of TCP gene family under drought and salinity stress condition in cowpea (Vigna unguiculata (L.) Walp.). 3 Biotech. 14, 138. doi: 10.1007/s13205-024-03976-x
Parapunova, V., Busscher, M., Busscher-Lange, J., Lammers, M., Karlova, R., Bovy, A. G., et al. (2014). Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biol. 14, 157. doi: 10.1186/1471-2229-14-157
Penfield, S., Meissner, R. C., Shoue, D. A., Carpita, N. C., Bevan, M. W. (2001). MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell 13, 2777–2791. doi: 10.1105/tpc.010265
Peng, S., Ali Sabir, I., Hu, X., Chen, J., Qin, Y. (2024). Advancements in reference gene selection for fruit trees: A comprehensive review. Int. J. Mol. Sci. 25 (2), 1142. doi: 10.3390/ijms25021142
Pruneda-Paz, J. L., Breton, G., Para, A., Kay, S. A. (2009). A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323, 1481–1485. doi: 10.1126/science.1167206
Puigbò, P., Lobkovsky, A. E., Kristensen, D. M., Wolf, Y. I., Koonin, E. V. (2014). Genomes in turmoil: quantification of genome dynamics in prokaryote supergenomes. BMC Biol. 12, 66. doi: 10.1186/s12915-014-0066-4
Rosignoli, S., Paiardini, A. (2022). Boosting the full potential of PyMOL with structural biology plugins. Biomolecules 12 (12), 1764. doi: 10.3390/biom12121764
Roy, S. W., Gilbert, W. (2006). The evolution of spliceosomal introns: patterns, puzzles and progress. Nat. Rev. Genet. 7, 211–221. doi: 10.1038/nrg1807
Rueda-Romero, P., Barrero-Sicilia, C., Gómez-Cadenas, A., Carbonero, P., Oñate-Sánchez, L. (2012). Arabidopsis thaliana DOF6 negatively affects germination in non-after-ripened seeds and interacts with TCP14. J. Exp. Bot. 63, 1937–1949. doi: 10.1093/jxb/err388
Schommer, C., Palatnik, J. F., Aggarwal, P., Chételat, A., Cubas, P., Farmer, E. E., et al. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PloS Biol. 6, e230. doi: 10.1371/journal.pbio.0060230
Shang, X., Han, Z., Zhang, D., Wang, Y., Qin, H., Zou, Z., et al. (2022). Genome-wide analysis of the TCP gene family and their expression pattern analysis in tea plant (Camellia sinensis). Front. Plant Sci. 13, 840350. doi: 10.3389/fpls.2022.840350
Shen, C. Z., Zhang, C. J., Chen, J., Guo, Y. P. (2021). Clarifying recent adaptive diversification of the chrysanthemum-group on the basis of an updated multilocus phylogeny of subtribe artemisiinae (Asteraceae: Anthemideae). Front. Plant Sci. 12, 648026. doi: 10.3389/fpls.2021.648026
Song, H., Nan, Z. (2014). Genome-wide identification and analysis of WRKY transcription factors in Medicago truncatula. Yi Chuan 36, 152–168. doi: 10.3724/SP.J.1005.2014.00152
Steiner, E., Efroni, I., Gopalraj, M., Saathoff, K., Tseng, T. S., Kieffer, M., et al. (2012). The Arabidopsis O-linked N-acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell 24, 96–108. doi: 10.1105/tpc.111.093518
Stolzer, M., Lai, H., Xu, M., Sathaye, D., Vernot, B., Durand, D. (2012). Inferring duplications, losses, transfers and incomplete lineage sorting with nonbinary species trees. Bioinformatics 28, i409–i415. doi: 10.1093/bioinformatics/bts386
Sun, B., Zhao, X., Gao, J., Li, J., Xin, Y., Zhao, Y., et al. (2023). Genome-wide identification and expression analysis of the GASA gene family in Chinese cabbage (Brassica rapa L. ssp. pekinensis). BMC Genomics 24 (1), 668. doi: 10.1186/s12864-023-09773-9
Tao, Q., Guo, D., Wei, B., Zhang, F., Pang, C., Jiang, H., et al. (2013). The TIE1 transcriptional repressor links TCP transcription factors with TOPLESS/TOPLESS-RELATED corepressors and modulates leaf development in Arabidopsis. Plant Cell 25, 421–437. doi: 10.1105/tpc.113.109223
Tatematsu, K., Nakabayashi, K., Kamiya, Y., Nambara, E. (2008). Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. Plant J. 53, 42–52. doi: 10.1111/j.1365-313X.2007.03308.x
Viola, I. L., Reinheimer, R., Ripoll, R., Manassero, N. G. U., Gonzalez, D. H. (2012). Determinants of the DNA binding specificity of class I and class II TCP transcription factors. J. Biol. Chem. 287, 347–356. doi: 10.1074/jbc.M111.256271
Walcher-Chevillet, C. L., Kramer, E. M. (2016). Breaking the mold: understanding the evolution and development of lateral organs in diverse plant models. Curr. Opin. Genet. Dev. 39, 79–84. doi: 10.1016/j.gde.2016.06.005
Wang, J., Chitsaz, F., Derbyshire, M. K., Gonzales, N. R., Gwadz, M., Lu, S., et al. (2023b). The conserved domain database in 2023. Nucleic Acids Res. 51, D384–d388. doi: 10.1093/nar/gkac1096
Wang, C., Feng, G., Xu, X., Huang, L., Nie, G., Li, D., et al. (2023a). Genome-wide identification, characterization, and expression of TCP genes family in orchardgrass. Genes (Basel) 14 (4), 925. doi: 10.3390/genes14040925
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi: 10.1093/nar/gky427
Wu, K. L., Guo, Z. J., Wang, H. H., Li, J. (2005). The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 12, 9–26. doi: 10.1093/dnares/12.1.9
Wu, X., Li, J., Wen, X., Zhang, Q., Dai, S. (2023). Genome-wide identification of the TCP gene family in Chrysanthemum lavandulifolium and its homologs expression patterns during flower development in different Chrysanthemum species. Front. Plant Sci. 14, 1276123. doi: 10.3389/fpls.2023.1276123
Wu, Y., Wu, S., Wang, X., Mao, T., Bao, M., Zhang, J., et al. (2022). Genome-wide identification and characterization of the bHLH gene family in an ornamental woody plant Prunus mume. Hortic. Plant J. 8, 531–544. doi: 10.1016/j.hpj.2022.01.004
Xu, Y., Wang, L., Liu, H., He, W., Jiang, N., Wu, M., et al. (2022). Identification of TCP family in moso bamboo (Phyllostachys edulis) and salt tolerance analysis of PheTCP9 in transgenic Arabidopsis. Planta 256, 5. doi: 10.1007/s00425-022-03917-z
Yang, Y., Guo, Y., Zhong, J., Zhang, T., Li, D., Ba, T., et al. (2020). Root physiological traits and transcriptome analyses reveal that root zone water retention confers drought tolerance to Opisthopappus taihangensis. Sci. Rep. 10, 2627. doi: 10.1038/s41598-020-59399-0
Yang, F., Sun, X., Wu, G., He, X., Liu, W., Wang, Y., et al. (2024). Genome-wide identification and expression profiling of the ABF transcription factor family in wheat (Triticum aestivum L.). Int. J. Mol. Sci. 25 (7), 3783. doi: 10.3390/ijms25073783
Ye, H., Liu, H., Han, M., Zhang, N., Feng, X., Gao, T., et al. (2024). The Genome-Wide Identification, Characterization, and Expression Profiles of the NADPH Oxidase (NOX) Gene Family under Drought and Salt Stress in Opisthopappus taihangensis (Asteraceae). Agronomy 14 (4), 653. doi: 10.3390/agronomy14040653
Yu, L., Chen, Q., Zheng, J., Xu, F., Ye, J., Zhang, W., et al. (2022a). Genome-wide identification and expression pattern analysis of the TCP transcription factor family in Ginkgo biloba. Plant Signal Behav. 17, 1994248. doi: 10.1080/15592324.2021.1994248
Yu, Z., Tian, C., Guan, Y., He, J., Wang, Z., Wang, L., et al. (2022b). Expression analysis of TCP transcription factor family in autopolyploids of chrysanthemum nankingense. Front. Plant Sci. 13, 860956. doi: 10.3389/fpls.2022.860956
Zhan, W., Cui, L., Guo, G., Zhang, Y. (2023). Genome-wide identification and functional analysis of the TCP gene family in rye (Secale cereale L.). Gene 854, 147104. doi: 10.1016/j.gene.2022.147104
Zhang, Z., Li, J., Zhao, X. Q., Wang, J., Wong, G. K., Yu, J. (2006). KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinf. 4, 259–263. doi: 10.1016/S1672-0229(07)60007-2
Zhang, P., Liu, D., Ma, J., Sun, C., Wang, Z., Zhu, Y., et al. (2024b). Genome-wide analysis and expression pattern of the ZoPP2C gene family in Zingiber officinale Roscoe. BMC Genomics 25, 83. doi: 10.1186/s12864-024-09966-w
Zhang, Y., Shen, Y., Han, M., Su, Y., Feng, X., Gao, T., et al. (2024c). Potential Response Patterns of Endogenous Hormones in Cliff Species Opisthopappus taihangensis and Opisthopappus longilobus under Salt Stress. Plants (Basel) 13 (4), 557. doi: 10.3390/plants13040557
Zhang, G., Yang, J., Zhang, C., Jiao, B., Panero, J. L., Cai, J., et al. (2024a). Nuclear phylogenomics of Asteraceae with increased sampling provides new insights into convergent morphological and molecular evolution. Plant Commun. 5, 100851. doi: 10.1016/j.xplc.2024.100851
Zhao, H. (2007). Phylogeny of tribe anthemideae (Asteraceae) from east asia and intergeneric cross between dendranthema×grandiflorum (Ramat.) Kitam. and Ajania pacifica (Nakai) K. Bremer & Humphries. Ph. D. Thesis. Nanjing Agricultural University, Nanjing, China.
Zhou, X., Gao, T., Zhang, Y., Han, M., Shen, Y., Su, Y., et al. (2024). Genome-wide identification, characterization and expression of C2H2 zinc finger gene family in Opisthopappus species under salt stress. BMC Genomics 25, 385. doi: 10.1186/s12864-024-10273-7
Keywords: Opisthopappus taihangensis, TCP gene family, abiotic stress, gene expression analysis, evolution
Citation: Gao T, Zhou X, Han M, Shen Y, Zhang Y, Wu Q, Dan H, Wang T, Ye H, Liu L, Chai M and Wang Y (2025) Identification and expression responses of TCP gene family in Opisthopappus taihangensis under abiotic stress. Front. Plant Sci. 16:1499244. doi: 10.3389/fpls.2025.1499244
Received: 20 September 2024; Accepted: 17 February 2025;
Published: 06 March 2025.
Edited by:
Rahul Kumar Tiwari, Indian Institute of Sugarcane Research (ICAR), IndiaReviewed by:
Joseph Gallagher, United States Department of Agriculture (USDA), United StatesCopyright © 2025 Gao, Zhou, Han, Shen, Zhang, Wu, Dan, Wang, Ye, Liu, Chai and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yiling Wang, eWx3YW5nYmpAaG90bWFpbC5jb20=; Min Chai, NzAzMzIyQHN4bnUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.