
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 06 February 2025
Sec. Aquatic Photosynthetic Organisms
Volume 16 - 2025 | https://doi.org/10.3389/fpls.2025.1497064
This article is part of the Research Topic Functional Response of Aquatic Plants to Environmental Stressors View all 12 articles
Introduction: The adaptation mechanisms of marine plants to the environments have garnered significant attention in recent years. Eelgrass (Zostera marina), a representative marine angiosperm, serves as an ideal model for investigating the mechanisms underlying salt tolerance.
Methods: This study integrated mRNA, sRNA, and degradome sequencing data to identify key genes associated with salt tolerance in eelgrass.
Results: The results indicate that a series of genes involved in biological processes such as “in response to water deprivation” and “biosynthesis of secondary metabolites” respond to salt stress. Analysis of cis-regulatory elements and expression similarities suggests that the ABA synthase 9-cis-epoxycarotenoid dioxygenase (NCED) may be regulated by ERF members, while phenylalanine ammonia-lyase (PAL) may be regulated by MYB members. At the post-transcriptional regulation level, miRNA156 and miRNA166 might be involved in the response by regulating potential target genes, such as members of the WRKY and HD-ZIP families. Additionally, eelgrass exhibits unique responses to salt, such as the up-regulation of genes involved in the “fucose biosynthetic process”. These findings enhance our understanding of how eelgrass adapts to the marine environment.
Discussion: As a marine monocotyledon, eelgrass is helpful to find conserved salt tolerance mechanisms by cross-species comparison. By examining the transcriptional responses of homologous genes in eelgrass, rice, and maize, we identified several groups of genes that are conserved in their response to salt stress. These conserved gene resources may provide targets for genetic engineering to improve the salt tolerance of crops.
Seagrass, a widely distributed coastal angiosperm, plays a crucial role in marine ecosystem functioning and global carbon sequestration (Ma X. et al., 2021). In order to adapt to the marine environment, seagrass has evolved different morphological (e.g. osmotic capacities), ultrastructural (e.g. absence of the real cuticle) and physiological characteristics (e.g. polyanionic, low-methylated pectins and sulfated galactans in cell walls) to tolerate salinity (Olsen et al., 2016; Wissler et al., 2011; Sandoval-Gil et al., 2023). Hence, salinity is considered as one of the major factors that condition the distribution, ecology and biology of seagrasses. Changes in salinity have the potential to impact on growth and biomass production of seagrass (Sandoval-Gil et al., 2023). In recent years, salinity levels in some seagrass beds are frequently influenced by extreme climate events such as continuous evaporation under high temperature or water exchange by rainfall (Koch et al., 2007; Tomasello et al., 2009), as well as human activities including the discharge from desalination factories (Cambridge et al., 2019), which are seriously threatened the maintenance of seagrass beds.
Eelgrass (Zostera marina), a member of the Angiosperms within the Monocotyledon group, is widely distributed in the coastal areas of the Northern Hemisphere (Zhang Y. et al., 2023). It is considered as one of the marine ecological model species for revealing adaptation to marine life. The activities of Na+/H+ exchangers in the plasma membrane of eelgrass have been involved in salinity tolerance (Rubio et al., 2011). Nitric oxide (NO) is also reported to play a crucial role in regulating the salinity tolerance of eelgrass by enhancing antioxidant defense mechanisms, as well as by modulating osmotic balance and energy metabolism (Wang X. et al., 2024). The analysis of the genome has revealed eelgrass exhibits some cell wall characteristics similar to those of macroalgae, which facilitate the retention of water and ions (Olsen et al., 2016). Genome-wide analysis in eelgrass revealed a reduced number of aquaporins and higher expression of plasma membrane intrinsic proteins (PIPs) compared to terrestrial plants (Shivaraj et al., 2017). These studies have revealed many genomic changes responsible for marine physiological adaptations, particularly regarding salinity tolerance.
Transcription factors (TFs) could activate or inhibit the transcription of target genes by binding specific DNA sequence (Yoon et al., 2020). Stress-responsive TFs such as the members of AP2/ERF and ARF, control the gene transcription in abscisic acid (ABA) and jasmonic acid (JA) signaling which are the key stress hormones in plant response to abiotic stress. Numerous studies have highlighted the regulatory functions of those two stress-responsive TFs in mediating terrestrial plants’ responses to salinity. For instance, ERF106MZ in rice or EREB57 in maize are reported to mediate ABA signaling under salinity stress (Chen et al., 2023; Zhu et al., 2023). The salt tolerance role of ARF5 in sweet potato has been functionally verified in transgenic Arabidopsis thaliana and is associated with ABA signaling (Kang et al., 2018). However, in many species, ARFs are down-regulated at high salinity, suggesting that members of this family may play a negative role (Ye et al., 2020; Wang C. et al., 2024). Besides, other TFs including MYB, NAC, bHLH and WRKY, are also predicted to be differentially expressed under salinity stress. For example, the AtMYB20 in A. thaliana is up-regulated by salt stress, and transgenic plants overexpressing AtMYB20 showed improved tolerance to salt stress (Cui et al., 2013). The heterologous overexpression of strawberry FaMYB63 in A. thaliana increased salt tolerance by reactive oxygen species (ROS) clearance (Wang S. et al., 2024). SlNAC4 in Suaeda liaotungensis was also verified to enhance tolerance to salt by regulating ABA metabolism through heterologous overexpressed (Liu et al., 2023). ThNAC13 in Tamarix hispida was proved to improve salt tolerance by binding to ThPP2 gene to enhance antioxidant enzyme activity (Liu R. et al., 2024). MYC2, a bHLH family member in A. thaliana and BpWRKY32 in Betula platyphylla were verified to increase salt tolerance (Verma et al., 2020; Liu Z. et al., 2024). In summary, these TFs primarily influence salt stress response and tolerance by modulating the expression of downstream genes, which participate in enhancing the antioxidant system, maintaining ion balance, regulating proline synthesis and ABA response. However, there is a notable scarcity of studies investigating these regulatory factors in seagrass species.
Plant microRNAs (miRNAs), typically consisting of 20-24 nucleotides, are also important post-transcriptional regulatory molecules (Begum, 2022). miRNAs could influence the expression of target genes. The function of stress-responsive miRNAs and their target under salt stress have been studied in model plants and some economic terrestrial crops. The expression of Tch-miR167 was negatively correlated to the expression of TcARF6, which indicated that TcARF6 increases salt tolerance in Tamarix chinensis by post-transcriptional regulation (Ye et al., 2020). The miR393 in A. thaliana regulated the salt-responsive pathway by scaffold protein RACK1A mediated ABA signaling pathways (Denver and Ullah, 2019). Overexpression of Osa-miR319a in transgenic creeping bentgrass Agrostis stolonifera inhibited the expression of AsPCF6, AsPCF8, AsTCP14, and AsNAC60, and modified the salt tolerance of transgenic plant (Zhou et al., 2013). The ABA receptor genes NtPYL2 and NtSAPK3 showed up-regulated expression in TaemiR408 tobacco lines, which indicated that TaemiR408 is associated with the miRNA modulation for ABA signaling to improve salt tolerance (Bai et al., 2018). miR172a and miR172b in cereal crops, miR398 in tomato and miR528-AO in rice were also proved to play a regulatory role in maintaining ROS homeostasis during salt stress (Cheng et al., 2021; He et al., 2021; Wang et al., 2021). In contrast, research on the role of miRNAs in aquatic plants under environmental stress conditions remains limited. Studies addressing the response of microRNAs to salt stress have been reported in species such as Dunaliella salina (Gao et al., 2019) and Spirulina platensis (Zhao et al., 2016). Some miRNAs in eelgrass have been identified by genomic analysis by Olsen et al. (2016) and Ma et al. (2021), which provide information for further research on the regulation mechanism of miRNA under abiotic stress.
The function of TFs and microRNAs in terrestrial plants have been identified as important regulatory elements that enable plants to withstand salt stress. Compared to terrestrial plants, the transcriptional and post-transcriptional response of those regulator in seagrass under stress remain elusive. Previous studies on eelgrass exhibited that high salinity not only inhibits seed germination but also effect the growth rate and survival of eelgrass (Zhang Y.-H. et al., 2023; Zhang Y. et al., 2023). In order to investigate the molecular mechanism and identify the potential regulatory factors, we performed a comprehensive analysis on the transcriptional response of eelgrass to salt stress using mRNA sequencing and small RNA sequencing data. Based on differential expression characteristics and similarity in expression trends, we identified various TFs, miRNAs, and functional target genes that may be associated with salt tolerance in eelgrass. The relationship between target genes and miRNAs was predicted and proved by degradome sequencing. This discovery provides more information about the molecular mechanisms underlying the adaptation of eelgrass to hypersalinity. Furthermore, we chose two terrestrial economic crops, rice and maize which also belong to Monocotyledon, to compare the homologous differential expression genes (DEGs) in response to salt stress. The comparative analysis not only sheds light on the conserved and specific mechanism in eelgrass adaption to hypersalinity but also provides gene resources for functional verification in terrestrial model species.
The eelgrass used in this study was sourced from a seedling factory in Qingdao, Shandong Province of China. Initially, the plants underwent a cleaning process and were then methodically arranged within a temporary tank. The base of the temporary storage tank comprised sand and stone that had undergone high-temperature sterilization, with the filtered seawater. The seawater was maintained with temperature of 17.5°C and salinity of 30, while the light intensity was approximately 120 µmol/(m²·s). Following three days of cultivation, the plants exhibiting robust growth were selected and transferred to experimental boxes No. 1 and No. 2, with seawater salinities of 30 and 50 (Xu et al., 2015), respectively. After 12 hours of treatment, the plants were removed from the boxes and promptly placed in liquid nitrogen for rapid freezing. A total of 6 plants were sampled in each treatment to build three biological replicates (with 2 plants being mixed to form a sample) for small RNA and mRNA sequencing. Different tissues were separated with sterile scissor. As the individual root samples did not provide enough material for sequencing, we mixed the stem and root together as one tissue (SR tissue). Besides, another 3 plants under high salt conditions were collected as a mixed sample for degradome sequencing.
The total RNA was extracted using the RNAprep Pure Plant Kit DP441 (Tiangen, Beijing, China) according to the kit instructions. Then, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in First Strand Synthesis Reaction Buffer. First strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase (RNase H-). Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of 3’ ends of DNA fragments, Adaptor with hairpin loop structure were ligated to prepare for hybridization. In order to select cDNA fragments of preferentially 370-420 bp in length, the library fragments were purified with AMPure XP system. Then PCR was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers and Index (X) Primer. At last, PCR products were purified. After the library construction is completed, preliminary quantification is carried out using Qubit, and the library is diluted to 1.5ng/ul. Subsequently, the inserted fragments of the library are detected. Once the inserted fragments meet expectations, the effective concentration of the library is accurately quantified using realtime PCR to ensure the quality of the library. The qualified libraries were pooled and sequenced on Illumina sequencing platforms, according to effective library concentration and data amount required.
The total RNA extracted was also used for small RNA sequencing (Zhang M. et al., 2023; Hu et al., 2021). We used NEB Next® Multiplex Small RNA Library Prep Set (NEB E7300L) for small RNA library construction. The 5 ‘end of small RNA has a complete phosphate group and the 3’ end of small RNA has a hydroxyl group. So special 3’ and 5’ adaptors were ligated to the 3’ and 5’ ends of small RNA based on the special structure of the 3’ and 5’ end of small RNA, respectively. Following this step, the first strand cDNA was synthesized subsequent to hybridizing with the reverse transcription primer. Subsequently, the double-stranded cDNA library was created via PCR enrichment. The libraries containing insertions ranging from 18 to 40 bp were then readied for sequencing after purification and size selection. Upon completion of the library construction process, the inserted fragments were quantified and the effective concentration (9~14.41 nmol/L) was assessed to ensure the quality of the libraries. The qualified libraries were pooled and sequenced on Illumina sequencing platforms, according to effective library concentration and data amount required. To gather more data for analysis, additional sequencing runs were conducted on four small RNA samples (L30-2, L30-3, L50-2, L50-3), which were collectively labeled as “batch2”.
The RNA-seq data was subjected to quality assessment using Fastqc and then preprocessed using trim-galore v0.6.10. Basic data information, such as sequence number and sequence length, was obtained using the seqkit v2.1.0. The preprocessed data was then aligned to the reference genome Zostera marina v3.1 from Phytozome13 using hisat2 v2.2.1. The results were sorted using samtools, and the quantification of gene loci was performed with featureCounts v2.0.6. Additionally, stringtie v2.2.2 was utilized to compute Fragments Per Kilobase of transcript per Million mapped reads (FPKM). Additionally, the sample R50-1 was excluded from further analysis due to its low mapping rate. Besides, we analyzed the PRJNA342750 data from NCBI related to the NaCl response.
The DESeq2 R package was used to analyze gene expression differences based on read count data, with the criteria for DEGs screening set as false discovery rate (FDR) ≤ 0.05 and fold change (FC) ≥ 2. The identified DEGs were classified according to their differential expression in different tissues, and the R package ComplexHeatmap was utilized to draw heat maps. The gene annotation was downloaded from Phytozome13, and complemented by eggNOG and KASS for GO and KEGG annotation. Mercator4 was utilized for mapman annotation of genes. Functional enrichment analysis was performed using the R package clusterProfiler (FDR ≤ 0.05).
To reveal the transcriptional regulatory network of eelgrass in response to high salt stress, we first conducted co-expression network analysis using the Weighted Gene Co-expression Network Analysis (WGCNA) package. Our dataset was combined with 58 additional samples obtained from the NCBI database (refer to Supplementary Data S2). In the initial stages of data preprocessing, we utilized Sratoolkit for data format conversion and Trim-galore for the identification and removal of adaptors and low-quality reads. Subsequently, the preprocessed data were aligned to a reference genome using hisat2. RSeQC was employed to ascertain the strand specificity of the mapped data. Following this, Stringtie was employed to compute the gene expression levels in fragments per kilobase of transcript per million mapped reads (FPKM), with genes exhibiting a Coefficient of Variation (CV) ≥ 1 retained for further analysis. Then, the gene expression values were log2-transformed (log2[FPKM+1]) for normalization. To construct an appropriate correlation weighting value (soft threshold) for co-expression network prediction, we utilized the pickSoftThreshold function in the R package WGCNA, with a criterion of SFT.R.sq ≥ 0.85 to ensure a scale-free network structure. Subsequently, we determined the soft threshold value of 8 for network construction. The one-step method (blockwiseModule function, parameters: networkType=“signed hybrid”; minModuleSize=30; mergeCutHeight=0.25) was then employed to build the network, and eigengene analysis was applied to illustrate the gene expression trends within each module. Finally, we used the R package Complex Heatmap to generate heat maps to visualize the distribution of DEGs in co-expression modules.
Small RNA data were aligned to the reference genome Zostera marina v3.1. The mapping files were merged to identify microRNAs using ShortStack v3.8.5 and miRDeep-P2 v1.1.4. Subsequently, bedtools v2.30.0 was employed to distinguish the same and unique MIRNA loci predicted by the two tools. The non-coding RNAs such as rRNA, tRNA, snRNA, and snoRNA were excluded from this analysis. For the identification of these non-coding RNAs, distinct prediction tools including barrnap, tRNAscan-SE, and Infernal-Rfam were utilized.
The pre-miRNA sequences were extracted based on the predicted MIRNA loci. Then, RNAfold (https://www.tbi.univie.ac.at/RNA/tutorial/) was used to examine the secondary structure of pre-miRNAs. Simultaneously, the predicted mature miRNA sequences were compared with those documented in miRbase (http://www.mirbase.org/) to determine miRNA families based on similarity. Meanwhile, we compared the MIRNA and gene location to classify the MIRNA into two types: the intragenic ones and the intergenic ones.
The read counts for miRNA and other sRNA loci were calculated using the ShortStack. According to the correlation analysis on the expression of small RNAs, we removed four samples in subsequent analysis, including L30.1, L50.1, SR30.3, and SR50.2. The R package EdgeR was used for counts per million (CPM) quantification. Differentially expressed miRNAs (DEmiRNAs) were identified using both EdgeR and DESeq2, with a threshold of p-value ≤ 0.05. The online tool psRNATarget (2017 release) was employed to predict miRNA target genes.
We utilized the SteadyPure Plant RNA Extraction Kit (Accurate Biotechnology, Changsha, Hunan, China) for mRNA extraction and the Evo M-MLV Reverse Transcription Kit (Accurate Biotechnology, Changsha, Hunan, China) for reverse transcription of RNA. For small RNA extraction, we utilized the SteadyPure Tissue and Cell Small RNA Extraction Kit (Accurate Biotechnology, Changsha, Hunan, China) followed by reverse transcription using the miRcute Enhanced miRNA cDNA First Strand Synthesis Kit (Tiangen, Beijing, China). The procedures were conducted according to the instructions. Quantitative real-time PCR (qRT-PCR) experiments were carried out using 7500 Fast Real-Time PCR equipment, with the following thermal cycling conditions: 95°C for 30s, followed by 40 cycles at 95°C for 5s and 60°C for 30s. In total, 9 DEGs and 4 DEmiRNAs were verified. The primers for these genes and miRNAs are listed in Supplementary Data S12. For normalization, 18S rRNA and U6 served as the reference gene, and relative expression changes were calculated using the 2−ΔΔCt method. The statistical analysis was performed using the function t.test in R software.
Total RNA was isolated and purified using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s procedure. The RNA amount and purity of each sample were quantified using NanoDrop ND-1000 (NanoDrop, Wilmington, DE, USA). The RNA integrity was assessed by Agilent 2100 with RIN number >7.0.
Poly(A) RNA is purified from plant total RNA (20μg) using poly-T oligo-attached magnetic beads using two rounds of purification. Because of the 3′ cleavage product of the mRNA contains a 5′-monophosphate, the 5’ adapters were ligated to the 5’ end of the 3′ cleavage product of the mRNA by the RNA ligase. The next step is reverse transcription to make the first strand of cDNA with a 3′-adapter random primer and size selection was performed with AMPureXP beads. Then the cDNA was amplified with PCR by the following conditions: initial denaturation at 95°C for 3 min; 15cycles of denaturation at 98°C for 15 s, annealing at 60°C for 15 s, and extension at 72°C for 30 s; and then final extension at 72°C for 5 min. The average insert size for the final cDNA library was 200-400 bp. At last, we performed the 50bp single-end sequencing on an Illumina Hiseq 2500 (LC Bio, China) following the vendor’s recommended protocol.
Degradome sequencing data were analyzed using ACGT101-DEG (LC Sciences, Houston, Texas, USA), which incorporates CleaveLand4. The analysis began with the preprocessing of sequences and followed by the creation of degradome density file. The miRNA sequences were aligned with the reference transcript sequence to identify potential clipping sites. Finally, the integration of sequencing data and comparison results allowed for the identification of targets and cutting sites which are supported by evidence of degradation fragments, and obtain the P-values.
The 1000 bp upstream sequence of DEGs and the 2000 bp upstream sequence of MIRNA loci were extracted, followed with cis-regulatory element prediction carried out using FIMO v4.11.2 (parameter: default) with the JASPAR2024_CORE_non-redundant database. The classification of cis-regulatory elements was based on the prediction results obtained from plantTFDB for the corresponding proteins documented in the JASPAR database. At the same time, we used the plantCARE online tool to make additional predictions.
The TFs of eelgrass were predicted using the Plant Transcription Factor Database (plantTFDB) available at http://planttfdb.cbi.pku.edu.cn/. The expression patterns of these TFs were compared with those predicted target genes. TFs with a Pearson correlation coefficient (PCC) of 0.8 or higher were selected as candidate regulators. Cytoscape v3.10.2 (https://cytoscape.org/) was employed for visualization of the transcriptional regulatory network.
Based on the results of functional enrichment analysis in eelgrass and references to genes involved in salt tolerance in terrestrial plants, we selected a set of genes for gene family analysis.
Aquaporins in eelgrass containing PF00230 domains were identified by hmmscan 3.4 (parameter:-E 10-5). Four classes of ion channel proteins, the NHX, KEA, and CHX family genes were gathered from the research of Olsen et al. (2016). Blast comparison was performed based on the protein sequences to find the corresponding gene IDs in Zostera marina v3.1. Additionally, genes possessing the PF01699 domain were classified as members of the Ca2+/H+exchanger antiporter (CAX) family using the hmmscan tool with a parameter setting of -E 10-5. Antioxidant enzyme genes, including superoxide dismutase (SOD), catalase (CAT), and glutathione peroxidase (GPX) collected based on the research of Olsen et al. (2016). Blast comparisons were carried out using the protein sequences to determine the corresponding gene IDs in Zostera marina v3.1. Moreover, a list of ascorbate peroxidase (APX) family genes from A. thaliana was retrieved from NCBI, and the homologous genes in eelgrass were identified using OrthoFinder. Moreover, OrthoFinder was utilized to identify homologous genes in eelgrass based on hormone-related genes in A. thaliana summarized by Altmann et al. (2020). Genes involved in the salt overly sensitive (SOS) pathway were identified by first compiling a list of A. thaliana genes associated with this pathway (Sandoval-Gil et al., 2023). Subsequently, using the A. thaliana gene IDs, the ortherFinder identification results were utilized to locate the corresponding genes in eelgrass.
Using protein sequences, we employed blastp v2.12.0 (parameters: -evalue 10-10, -max_target_seqs 5) to identify the top five homologous genes in A. thaliana, rice, and maize to construct phylogenetic tree. Besides, to compare the response to salinity across species, 100 rice sra data and 57 maize sra data (Supplementary Data S9) were downloaded from the NCBI. The reference genome was obtained from Ensembl Plants. Data preprocessing, sequence alignment, gene expression quantification, and identification of DEGs were performed following the same procedures as those for the samples of eelgrass. The GO annotation of genes in rice and maize was performed by eggNOG. To identify the set of genes that are widely up-regulated in rice and maize, we counted sample comparisons for up-regulated DEGs. Then, we selected the genes based on the top 10% percentile of comparison counts and performed functional enrichment analysis.
The raw data of RNA-seq includes 41.8-53.4 million reads, and the mapping rate of most samples is greater than 80%. We identified a total of 409 DEGs between the samples under salinity 50 and 30 treatment using DESeq2 (Supplementary Data S1; Supplementary Figure S1A), including 188 genes from leaf tissue and 236 genes from SR tissue (the mixed tissue of stem and root). Additionally, an analysis of the PRJNA342750 dataset revealed 334 DEGs in leaf tissues in response to NaCl. There was minimal overlap among the DEGs identified across the three groups (Supplementary Figure S1B), indicating distinct responses to salt stress in different tissues and under varying experimental conditions.
We performed functional enrichment analysis on the DEGs identified in our data, which revealed several enriched GO terms and KEGG pathways such as “response to water deprivation (GO:0009414)”, “MAPK signaling pathway-plant (map04016)”, “Biosynthesis of secondary metabolites (map01110)”, “Phenylpropanoid biosynthesis (map00940)” (Figure 1; Supplementary Figures S1C, D). DEGs in GO:0009414 were predominantly up-regulated in leaf, including members of the PP2C family (Zosma01g16060), ERF family members (Zosma05g33490 and Zosma03g37150), Late embryogenesis abundant protein (Zosma05g03710), and the UPF0057 family gene (Zosma03g14850). Conversely, DEGs in map04016 were primarily down-regulated in leaf, including family members of MAPKKK (Zosma03g00830, Zosma03g00810), ERF (Zosma06g29270), WRKY (Zosma06g26970), and CALM (Zosma06g17200). What’s more, DEGs in map01110 were mainly up-regulated in SR tissue, including Cinnamate 4-coumarate:CoA ligase (Zosma01g00600), trans-cinnamate 4-monooxygenase (Zosma02g22230), cinnamoyl-CoA reductase (Zosma03g12930), phenylalanine ammonia-lyase (Zosma03g22300), cinnamyl-alcohol dehydrogenase (Zosma06g25800), chalcone synthase (Zosma01g25890), and chalcone isomerase (Zosma06g21110). MapMan annotation and enrichment results indicated that salt stress may induce programmed cell death (Supplementary Figure S1C). Zosma03g26890 is a proline dehydrogenase and down-regulated under salt stress, indicating the proline accumulation might be enhanced in eelgrass. (Supplementary Figure S1D). Additionally, plant hormones such as ABA and gibberellin may play a role in the response to salt stress (Supplementary Figure S1D).
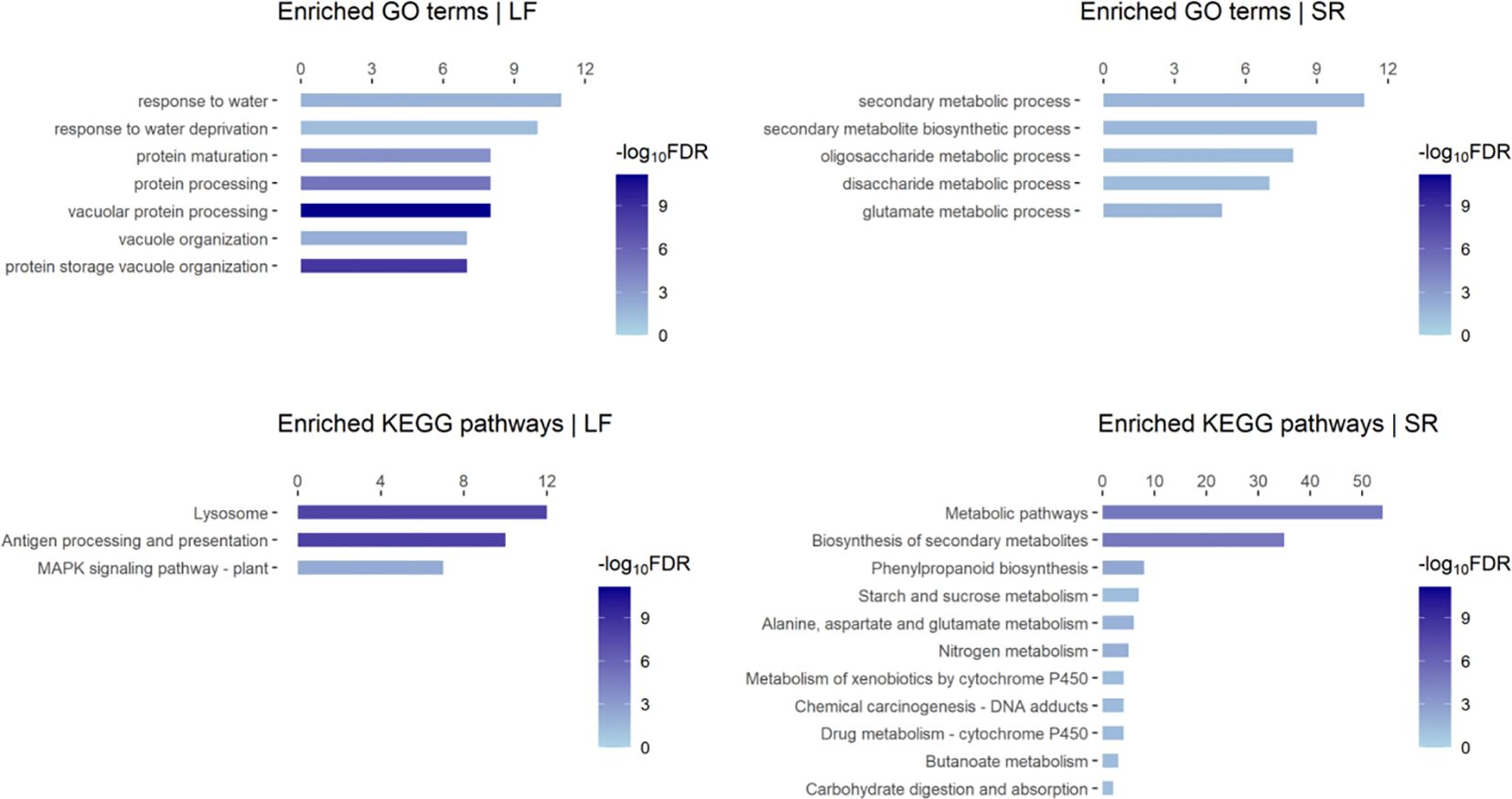
Figure 1. Enriched GO terms and KEGG pathways of responsive DEGs to high salt in eelgrass. “LF” refers to the leaf; “SR” refers to the mixed tissue of stem and root. The length of the bar refers to the count of DEGs in a Go-term or KEGG pathway.
By examining the overlapping DEGs, we identified four genes (Zosma02g20240, Zosma04g22590, Zosma05g04820, Zosma05g31200) that were up-regulated in all three comparisons (Supplementary Figure S1B). Zosma05g04820 is a member of the AP2/ERF family, and its homologous genes participate in abiotic stress such dehydration (Sakuma et al., 2002). Zosma04g22590 encodes NCED is a crucial enzyme in the biosynthesis of ABA (Qin and Zeevaart, 1999), of which homologous genes function in drought tolerance (Iuchi et al., 2001). Zosma02g20240 belongs to the CAX family of cation antiporters, of which the homologous gene is involved in calcium transport (Emery et al., 2012). Zosma05g31200 is a senescence-associated protein, of which homologous genes are involved in various abiotic stress including salt stress (Barajas-Lopez et al., 2021). The transcriptional response of these four DEGs was further verified through qRT-PCR assays (Figure 2).
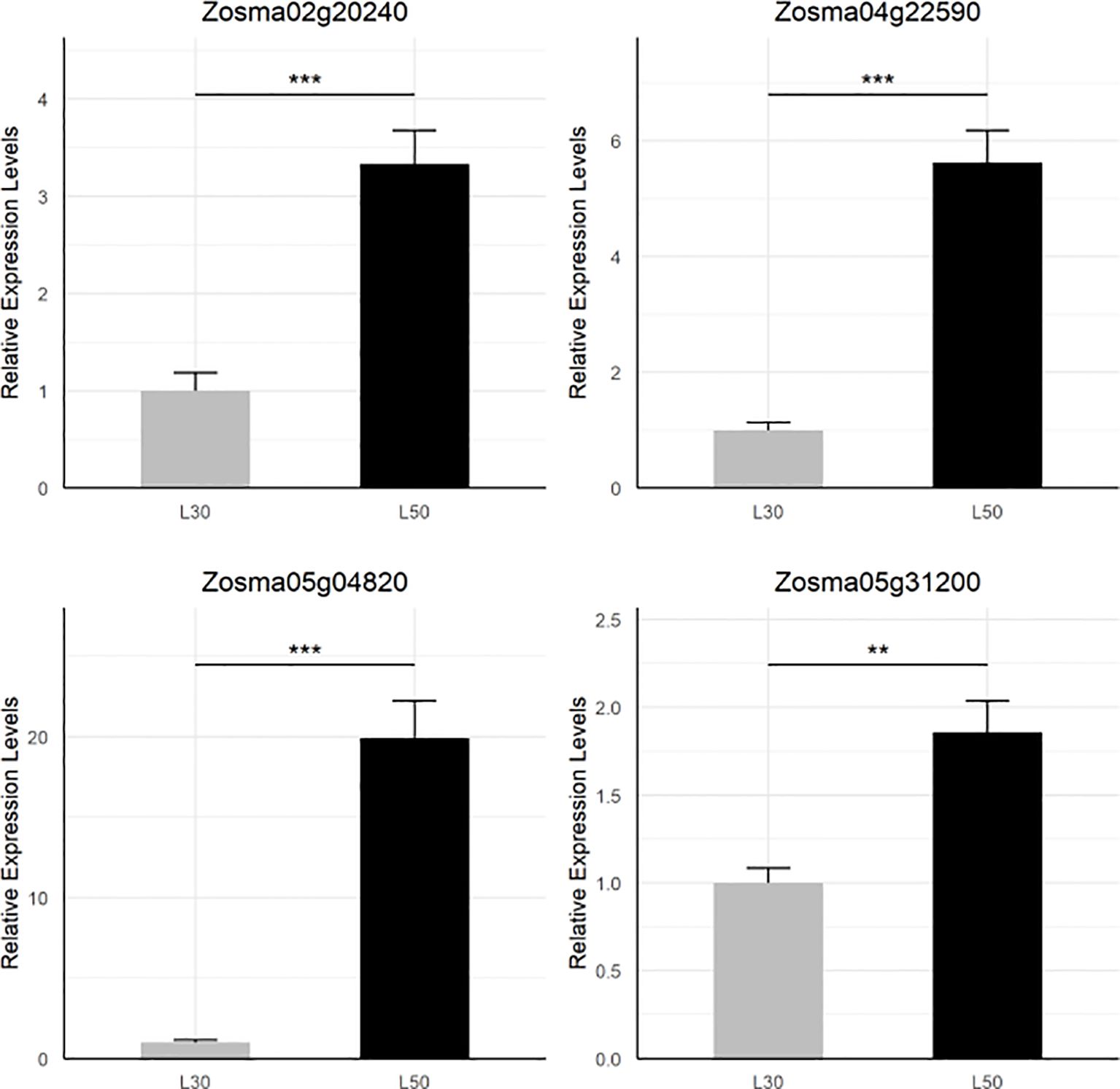
Figure 2. qRT-PCR validation of four DEGs that are up-regulated in all comparisons. The star marks on statistical charts represent the statistical significance: “*” stands for P < 0.05, “**” stands for P < 0.01, “***” stands for P < 0.001.
We observed that the tissue-specific responsive genes account for a large proportion of the DEGs identified by our data (Supplementary Figure S1B). Consistently, the results of functional enrichment analysis on these gene sets (Supplementary Figure S1E) were similar to those described above (Figure 1), which indicate enhanced and impaired biological processes in different tissues. Especially, we found that an enriched GO term “fucose biosynthetic process” was identified from up-regulated genes in leaf. In a previous study, fucose was found as the main component of extracted polysaccharides in Lessonia (Zou et al., 2019). This raises the question of whether fucose is involved in the adaptation of marine plants to high-salt environments. What’s more, we examined the specific DEGs in response to NaCl (identified by PRJNA342750 data): 118 genes were up-regulated, which were involved in biological processes such as “response to stimulus (GO: 0050896)”, “cobalamin biosynthetic process (GO: 0009236)” and “heme biosynthetic process (GO: 0006783)”; 167 genes were down-regulated, which were involved in the process such as “carbohydrate metabolic process (GO: 0005975)” and “mucilage metabolic process (GO: 0010191)”. These results help us to gain a deeper understanding of the response of eelgrass to salinity changes at the organ level.
Utilizing our data and transcriptome data from 58 samples of eelgrass obtained from NCBI (Supplementary Data S2), we constructed a gene co-expression network that revealed a total of 36 gene modules, each containing at least 30 genes (Supplementary Data S3; Supplementary Figures S2A, B). The analysis (Supplementary Figure S2C) indicated that the up-regulated genes under salt stress in leaf were predominantly distributed in the M3, M4, M6, and M7 modules, while the down-regulated genes in leaf were primarily found in the M1, M2, M4, and M6 modules. In SR tissue, most of the up-regulated genes were distributed in the M4 and M6 modules, while the down-regulated genes were mainly distributed in the M2, M4, M6, and M7 modules.
We focused on the salt-responsive DEGs in modules M4, M6, and M9 due to their expression patterns (Figure 3A; Supplementary Figure S2B): M4 exhibited a high expression primarily in leaf. M6 was notably expressed in leaf, stems, and roots. M9 showed predominant expression in stem and root. M4 harbored a number of metal transporters that were up-regulated (Supplementary Figure S2D), such as the mitochondrial iron transporter (Zosma03g13600) and several heavy metal-associated domain-containing proteins (Zosma04g04780, Zosma02g05210, Zosma02g16380). Conversely, many DEGs related to photosynthesis in M4 were significantly down-regulated (Supplementary Figure S2D), including the light-harvesting complex I chlorophyll a/b binding proteins (Zosma02g19650, Zosma03g33730, Zosma03g35370, Zosma05g20560) and the photosystem II reaction center PSB28 protein (Zosma03g24460). Furthermore, several nutrition-related DEGs in leaf were up-regulated (Supplementary Figure S2D), including nitrite reductase (Zosma02g18160, Zosma03g25160) in M4, nitrate transmembrane transporters (Zosma05g08330), proton/phosphate symporters (Zosma05g32160) and ferritin (Zosma02g21810) in M6. These findings suggest a shift in the nutritional status of eelgrass under salt stress. Additionally, M6 contains a few up-regulated DEGs associated with calcium-dependent signaling (Supplementary Figure S2D), such as calmodulin-related calcium sensor proteins (Zosma01g15140, Zosma01g36630, Zosma04g24750), indicating heightened calcium signaling activity in response to high salinity. A set of up-regulated DEGs in M9 were involved in secondary metabolism (Supplementary Figure S2D), such as phenylalanine ammonia-lyase (Zosma03g22300), cinnamoyl-CoA reductase (Zosma03g12930) and glutamate decarboxylase (Zosma02g19830), indicating that the plants might undergo secondary metabolic changes in stem and root.

Figure 3. Salt-responsive DEGs in WGCNA modules. (A) Up-regulated or down-regulated genes in response to high salinity assigned to M4, M6 and M9. (B) The transcriptional response of TFs to high salinity. The asterisk stands for a significant difference in the expression comparison.
TFs are potential regulators for genes in co-expression modules. The M4 module contains 12 salt-responsive TFs. Three of these TFs were up-regulated in leaf, including two bHLH family members (Zosma03g02210, Zosma06g18160) and one MYB family member (Zosma01g40890). Two of these TFs were up-regulated in SR tissue, including one ERF family member (Zosma01g29190) and one TALE family member (Zosma05g18780). The M6 module includes 14 salt-responsive TFs. Eight of these TFs were up-regulated in leaf, including four ERF family members (Zosma03g37150, Zosma05g04820, Zosma05g33490, Zosma02g05080), two bHLH family members (Zosma01g41170, Zosma05g10360), one C2H2 family member (Zosma01g25030), and one WRKY family member (Zosma01g05750). Two ERF family members (Zosma05g04820 and Zosma05g33490) were up-regulated in SR tissue. M9 module contains 2 salt-responsive TFs, which belong to MYB family members (Zosma06g17610 and Zosma06g17660) and show up-regulated expression in SR tissue. How these TFs regulate the response of eelgrass to salt stress is an issue that remains to be resolved. (Figure 3B; Supplementary Figure S2E; Supplementary Data S4). We analyzed the cis-regulatory elements located within the 1000 bp region upstream of DEGs. Our findings revealed that the motifs potentially binding TFs such as bHLH, ERF and MYB family members (Supplementary Figure S2F) are universally distributed in the promoter of DEGs. Moreover, predictions from the PlantCARE indicate that plant hormone signals, such as ABA and JA, may also contribute to the regulatory mechanisms in eelgrass (Supplementary Figure S2G).
In this study, a total of 172 MIRNA loci were identified through the analysis of small RNA sequencing data (Figure 4A). miRDeep-P2 exhibited a higher sensitivity compared to Shortstack, being able to identify 157 miRNAs independently. Additionally, ShortStack detected 6 miRNAs that were not identified by miRDeep-P2. Of these sites, 9 were found to be identified by both the Shortstack and miRDeep-P2 tools, which are considered the most reliable sites for MIRNA. The majority of these miRNAs were found to be located on chromosomes (Supplementary Figure S3A), with a few being situated on the scaffold. The length distribution of the identified mature sequences was predominantly 20-22 nt, aligning with the typical length characteristics of miRNAs in plants (Figure 4C).
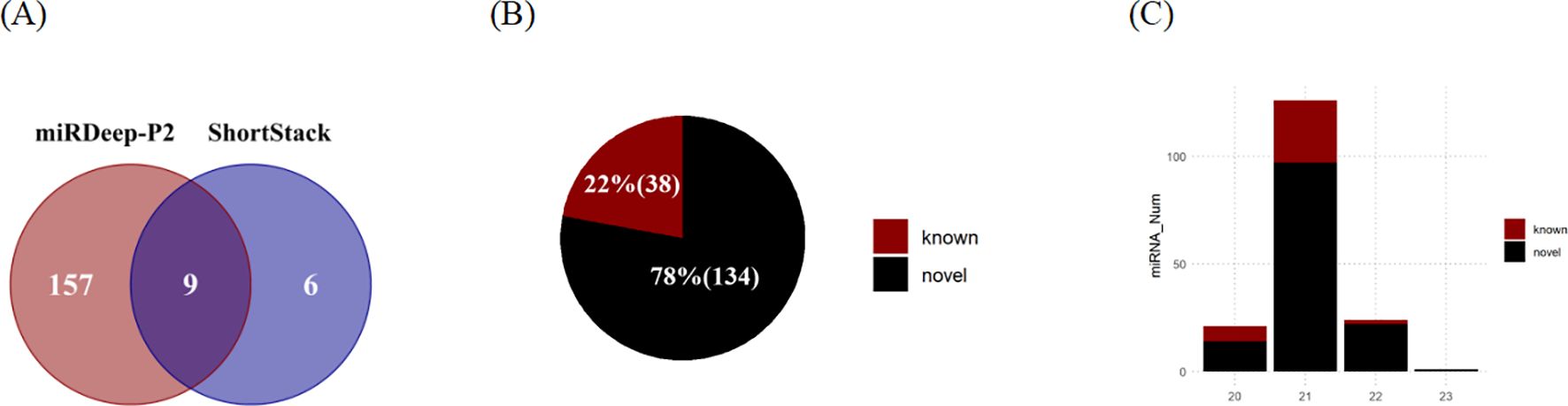
Figure 4. Identification of microRNAs in eelgrass. (A) The number of MIRNAs identified by ShortStack and miRDeep-P2. (B) The number of identified MIRNAs belonging to known and novel families. (C) The number of miRNAs (mature sequence) of different lengths.
Subsequently, a comparison was conducted between the mature sequences obtained from the predicted sites and those documented in miRbase (Figure 4B). This comparison led to the annotation of a total of 38 miRNAs belonging to known MIRNA families, while an additional 134 miRNAs were identified as de novo loci (Supplementary Data S5). Among the known families, MIR156 was identified to have the highest number of loci in eelgrass, specifically containing six members. Moreover, several other families, including MIR166, MIR172, MIR164, MIR396, MIR319, and MIR390, were found to have more than two members each.
Simultaneously, the relative positions of MIRNAs to neighboring genes were assessed. A total of 41 MIRNAs were categorized as intragenic, while 131 were categorized as intergenic (Supplementary Figure S3B). Among the intragenic MIRNAs, 31 were located within intron regions, 3 were found in exon regions, and 7 spanned both introns and exons (Supplementary Figure S3C). For 78.3% of the intergenic MIRNAs were located within 20,000 bp of an upstream gene (Supplementary Figure S3D).
We identified a total of 14 DEmiRNAs in response to salt stress using edgeR and DESeq2. There were 8 overlapped DEmiRNAs identified by both tools, including MIR166a, MIR166d, MIR172d and a series of de novo loci (Figure 5A; Supplementary Figure S3E; Supplementary Data S6). The transcriptional responses of miRNAs from these loci such as miR156, miR166 and NOVEL008 were further validated through qRT-PCR assays (Figure 5C; Supplementary Figure S3F). To uncover the potential mechanisms regulating miRNA expression, we analyzed the cis-regulatory elements within the 2000 bp upstream region of MIRNAs. The results showed that there were many potential TF binding sites upstream of the MIRNA. The binding sites for TFs such as bZIP, C2H2, and Dof are widely present (Supplementary Figures S4A, B). Additionally, predictions from plantCARE indicated that the upstream regions of DEmiRNAs were enriched in cis-regulatory elements associated with abiotic stress and hormone responses. It suggests that diverse signaling pathways, such as those related to osmotic stress (STRE) and JA (CGTCA-motif and TGACG-motif), play a role in regulating microRNA expression in eelgrass (Supplementary Figure S4C).
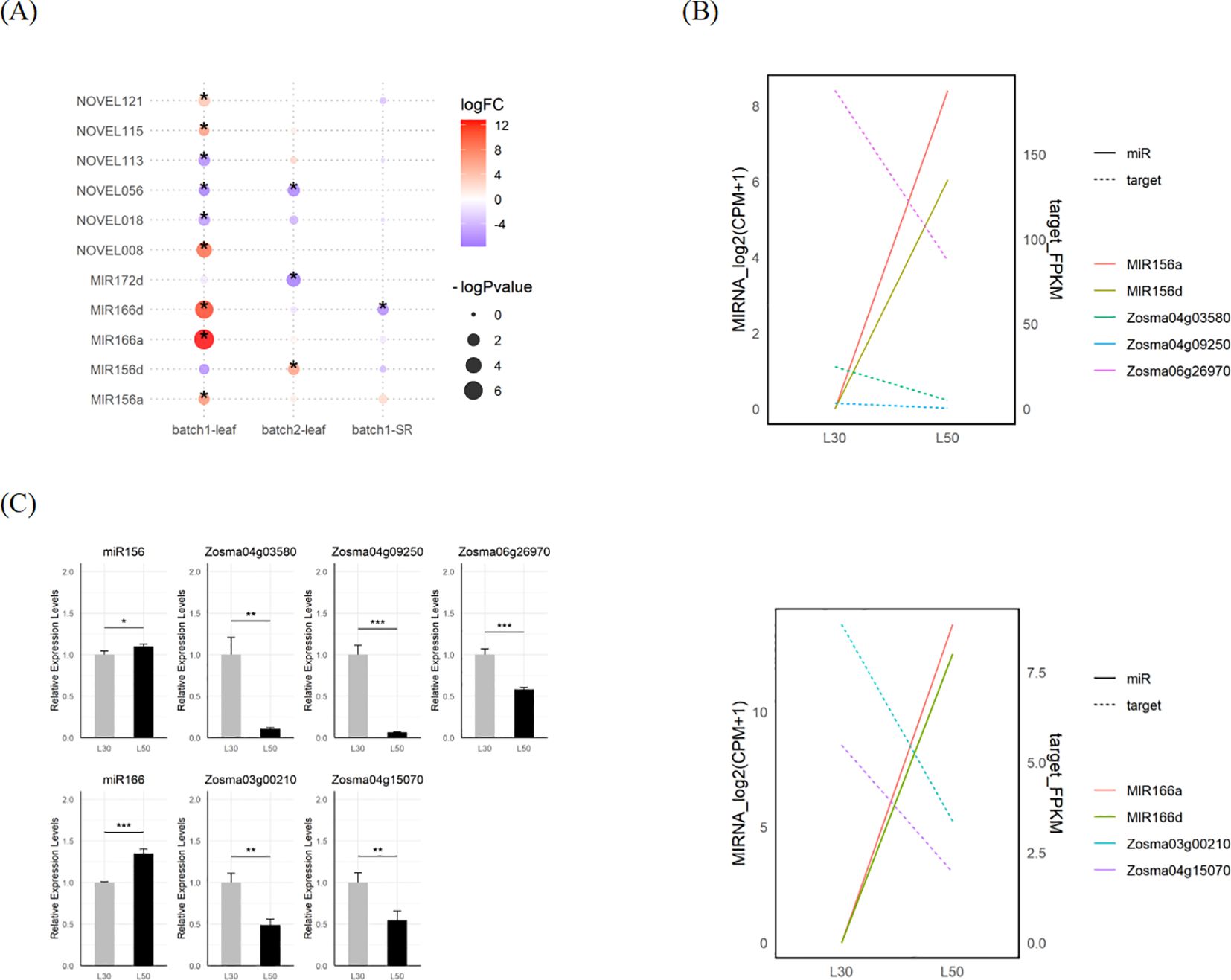
Figure 5. DEmicroRNAs and their potential targets in eelgrass. (A) The DEmiRNAs in response to salt stress in eelgrass identified by edgeR. (B) The expression of MIR156, MIR166 and their predicted targets in L30 and L50. (C) Validated expression of MIR156a/d and MIR166a/d mature sequences as well as their predicted targets using qRT-PCR. The gray bar stands for L30, while the black bar stands for L50. The star marks on statistical charts represent the statistical significance: “*” stands for P < 0.05, “**” stands for P < 0.01, “***” stands for P < 0.001.
We employed psRNATarget to predict microRNA target genes, of which 35 were DEGs in response to salt stress and potentially targeted by DEmiRNA (Supplementary Data S7). There are a number of target genes whose expression changes are negatively correlated with their corresponding miRNAs, such as miR156 and miR166. Among them, Zosma06g26970, is a member of WRKY family and identified as a target gene for miR156; Zosma03g00210, is a member of the HD-ZIP family and identified as a target gene for miR166 (Figure 5B). Both of them exhibit down-regulated expression at high salinity, of which homologous genes has been reported to be involved in salt stress (Jiang and Deyholos, 2009; Yadav et al., 2021). The responses of these miRNAs and their target genes were further validated through qRT-PCR assays (Figure 5C).
We also employed degradome sequencing to investigate the target sites of microRNAs (miRNAs), resulting in identification of 91 reliable miRNA-target pairs (Supplementary Data S8). 86.8% of these miRNA-target pairs were consistent with the predicted pairs identified by psRNATarget. Among the predicted targets, we found two genes were DEGs in response to salt stress. Zosma04g26100, a potential target of miRNA160 (Figure 6), exhibited up-regulation in SR tissue. The homolog of Zosma04g26100, athARF16 (AT4G30080) plays roles in root cap cell differentiation and is regulated by both miRNA160 and plant hormones (Wang et al., 2005; Bascom, 2023). The relatively insignificant response of miRNA160 might facilitate the increased expression of Zosma04g26100. Zosma05g29470, identified as a potential target for NOVEL056, displayed a predicted cutting site that is not located near the peak of degraded fragments (Supplementary Figure S5). The homologous gene athPAL1 (AT2G37040) of Zosma05g29470 is implicated in the biosynthesis of lignin and flavonoid (Olsen et al., 2008), suggesting that the down-regulation of Zosma05g29470 could adversely affect related metabolic functions.
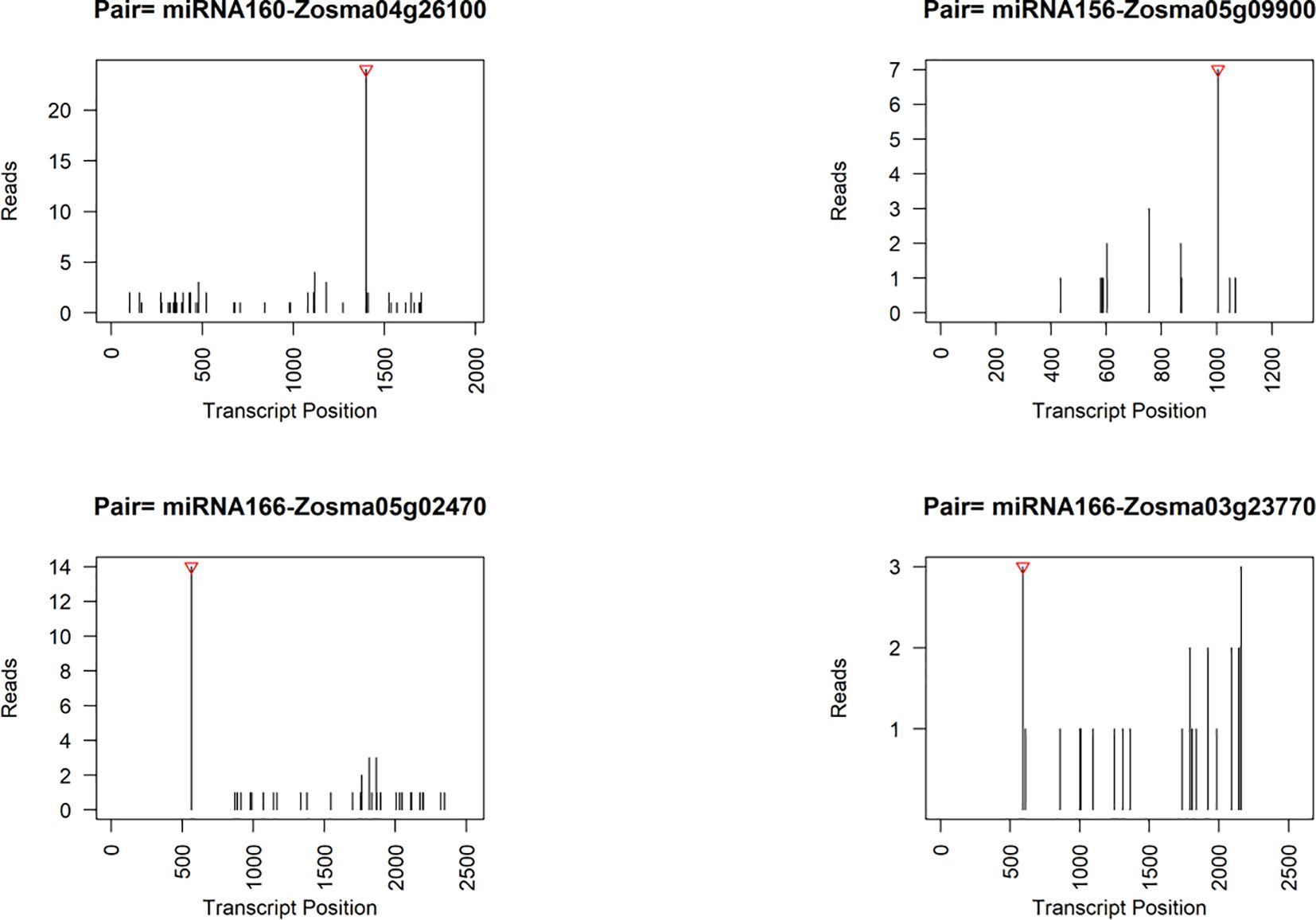
Figure 6. Representative target plots (t-plot) depicting categories of the cleaved mRNAs confirmed by degradome sequencing. The red triangle at the top represents the predicted cleavage location.
Although some genes did not exhibit obvious responses to salt stress, they were potentially regulated by microRNAs, as indicated by the degradome data and previous studies (Figure 6). For instance, the target gene of miRNA156, Zosma05g09900, is a member of the SPL family, of which homologous gene has been implicated in various processes such as plant morphogenesis (Ferreira e Silva et al., 2014), secondary metabolism (Gou et al., 2011), and signal transduction (Barrera-Rojas et al., 2023). Zosma05g02470 and Zosma03g23770, both belonging to the HD-ZIP family (Supplementary Figure S5), are predicted target genes of miR166. Their homologous genes in A. thaliana play crucial roles in regulating vascular development (Kim et al., 2005). Moreover, the degradome data provided evidence for additional miRNA-target pairs, including miR396-GRF (Zosma02g02440/Zosma01g12870), miR393-TIR (Zosma01g13700/Zosma02g25510), miR167-ARF (Zosma01g35720), miR319-TCP (Zosma03g04610), and miR164-NAC (Zosma05g19590) (Supplementary Figure S5), which have been reported to function in other plant species (Guo et al., 2005; Gutierrez et al., 2009; Liang et al., 2014; Koyama, 2017; Wang et al., 2018). The miRNA cleavage sites identified by the degradome are speculative and require future verification.
The enriched GO-term “response to water deprivation” suggests that eelgrass may experience osmotic stress similar to drought conditions in high salt environment. Most DEGs within this GO-term are significantly up-regulated in leaf, including Zosma01g16060, Zosma03g14850, Zosma03g23560, Zosma03g33910, Zosma03g37150, Zosma05g03710 and Zosma05g33490. Among these genes, Zosma01g16060 is a member of the PP2C family, of which homolog in A. thaliana, HAI2 (AT1G07430), whose mutant has been documented to enhance drought resistance by regulating proline and osmoregulatory solute accumulation (Bhaskara et al., 2012). The other two DEGs Zosma05g03710 and Zosma03g14850 in GO term of “response to water deprivation” also appear to be involved in salt stress. The homolog (AT1G01470) of Zosma05g03710 has been reported to exhibit significant protective effects under salt stress (Jia et al., 2014), while the homolog RCI2B (AT3G05890) of Zosma03g14850 is responsive to abiotic stresses such as low temperature and water scarcity, with its expression induced by ABA (Capel et al., 1997). Zosma05g33490 and Zosma03g37150 belong to the AP2/ERF family. The homologous gene DREB1A (AT4G25480) of Zosma05g33490 has been implicated in the response to abiotic stresses, including drought, across various plant species (Polizel et al., 2011; Ravikumar et al., 2014). Meanwhile, the homolog of Zosma03g37150 is known to participate in gene expression related to dehydration and cold stress (Sakuma et al., 2002). Besides, the collinear genes of Zosma01g16060, Zosma05g33490, and Zosma05g03710 in rice and maize also exhibit responses to salt stress, suggesting a conserved role among these “response to water deprivation” genes (Supplementary Data S10; Supplementary Figure S6). By comparing the enriched go-terms, we observed that the biological process “response to water deprivation” is conserved in three species (Supplementary Figure S6C). Within this go-term, some gene families, such as AP2, PP2C, and Annexin, are present in both eelgrass and rice, suggesting similarities in how marine and terrestrial plants respond to high salinity stress.
Aquaporins are the key regulators for regulating osmotic homeostasis in plants (Ren et al., 2021). Previous studies in other seagrass have suggested that aquaporins contribute to the adaptation in hypersaline environments (Sandoval-Gil et al., 2023), and one of the studies has shown that aquaporins are involved in the maintenance of osmotic balance (Serra et al., 2013). In this study, we identified a total of 25 aquaporins in eelgrass (Supplementary Data S11). The gene expression profile revealed that several aquaporin members, including Zosma01g17610, Zosma01g27210, Zosma02g03080, Zosma02g23130, and Zosma03g25250, exhibited high expression levels across multiple organs (Supplementary Figure S7A). The phylogenetic analysis (Supplementary Figure S7B) indicates that Zosma01g17610 belongs to the TIP branch, while Zosma01g27210, Zosma02g23130, and Zosma02g03080 belong to the SIP branch, and Zosma03g25250 is part of the NIP branch. Members of the TIP branch have been associated with the promotion of new lateral root primordia (Reinhardt et al., 2016), whereas SIP branch members are primarily localized in the endoplasmic reticulum, functioning as channels for water, small molecules, and ions (Ishikawa et al., 2005). Additionally, NIP branch members are implicated in the absorption of certain mineral ions and can influence hydrogen peroxide (H2O2) levels (Kamiya et al., 2009; Kamiya and Fujiwara, 2009; Wang et al., 2017). The widespread expression of these genes may facilitate the transport of water and small solutes across cell membranes in eelgrass. Simultaneously, we observed that three aquaporins (Zosma03g21300, Zosma01g24240, and Zosma03g27440) exhibited significantly down-regulated expression levels under high salt conditions. The phylogenetic tree reveals that Zosma01g24240 belongs to the PIP1 branch, Zosma03g21300 to the PIP2 branch, and Zosma03g27440 to the TIP branch. Members of the PIP1 branch are reported to be involved in water transport and immune responses (Postaire et al., 2010; Tian et al., 2016), while those in the PIP2 branch are associated with drought resistance (Chen et al., 2021, 2022) and the transport of H2O2 (Rodrigues et al., 2017; Wang et al., 2020). Zosma03g27440 and Zosma01g17610 share the same branch, suggesting potential functional redundancy. Furthermore, a number of their homologous genes in rice and maize, such as Zm00001eb096680, also exhibit reduced expression at high salinity (Supplementary Figure S7C). In terrestrial plants, drought or salt stress can suppress the expression of certain aquaporins such as PIPs, thereby reducing root hydraulic conductivity by limiting water loss (Yepes-Molina et al., 2020). The down-regulation of aquaporins in eelgrass might reflect a conserved character in plants responding to salt stress.
Plant adaptation to high-salinity environments depends in part on the maintenance of cell ion homeostasis, a process significantly facilitated by ion transporters (Malakar and Chattopadhyay, 2021). In this study, we identified several key ion transporter families in eelgrass, including five Na(+)/H(+) exchangers (NHX), five K(+) efflux antiporters (KEA), fifteen cation/H(+) exchangers (CHX), and eleven sodium/calcium exchanger proteins belonging to CAX family (Supplementary Data S11). The gene expression profile (Supplementary Figure S8A) indicates that most NHX and KEA genes are broadly expressed across multiple organs, with the exception of one NHX member (Zosma01g13710), which exhibits specific expression in floral organs. In contrast, the expression of CHX family exhibits organ specificity, particularly with higher expression levels in male flowers, suggesting a potential role in reproductive development. Approximately half of the CAX members are also widely expressed in various organs, including Zosma02g20240, which shows significant accumulation under high salt stress. While alterations in calcium levels are critical in the SOS pathway (Ali et al., 2023), there is currently no evidence linking CAX family members to this process. Previous research has indicated that certain CAX family members, such as athNCL (AT1G53210), contribute to plant salt tolerance (Wang et al., 2012). The genes of eelgrass in the same branch of athNCL include Zosma01g22450 and Zosma06g26160 (Supplementary Figure S8B), while Zosma02g20240 is positioned on a separate branch, indicating the need for further investigation into their roles in salt tolerance. Additionally, comparative analysis reveals that, unlike eelgrass, many ion channel protein genes in rice and maize respond to salt stress (Supplementary Figure S8C), highlighting the differences in ion regulation mechanisms between terrestrial and marine plants.
Osmotic stress and ionic stress induced by salt stress can lead to the production of ROS in plants (Hasanuzzaman et al., 2021). To mitigate the effects of ROS, plants can accumulate osmotic products and enhance the activity of antioxidant enzymes, thereby improving their tolerance to salt stress (Yang and Guo, 2018). In this study, several antioxidant enzymes were identified in eelgrass, including SOD, CAT, GPX, and APX (Supplementary Data S11). Gene expression profiles revealed that, while most antioxidant enzymes did not show a significant response to salt stress, they were highly expressed across multiple organs (Supplementary Figure S9A). Zhang et al. (2023) also discovered the upregulation of SOD, peroxidase and other antioxidant enzyme genes in germination of eelgrass, but with no significant difference. Olsen et al. (2016) reported many stress-resistance genes in eelgrass. The salinity and temperature fluctuation are usually gradual in coastal environments, we speculate that high constitutive expression of antioxidant enzyme genes in eelgrass might be the adaptation mechanism to reduce the ROS during the gradual change of abiotic stress. Notably, Zosma06g29150, a member of the GPX family, exhibited significant up-regulation in the leaf, indicating that this enzyme may function in reducing H2O2 by oxidizing glutathione (GSH) (Das and Roychoudhury, 2014), thereby enhancing salt tolerance. GPX was also reported as the most abundant DEGs and proteins in eelgrass under low salinity stimulation during germination (Zhang Y. et al., 2023), which is coincident with the result of our study. It indicated that GPX family is the main antioxidant enzyme for scavenging ROS in eelgrass. Furthermore, homologous genes of Zosma41g01010 in rice and maize, such as Os02g0664000 and Zm00001eb016270, also respond to salt stress (Supplementary Figures S9B, C).
Salt stress induces secondary metabolic changes in plants, especially the accumulation of phenolic compounds, including lignin (Dabravolski and Isayenkov, 2023) and flavonoids (Shomali et al., 2022). Lignin fortifies plant cell walls, thereby protecting membrane integrity under salt stress. Flavonoids play a role in reducing ROS accumulation to enhance plant resistance to adverse conditions (Li et al., 2018). The biosynthesis of many phenolic compounds occurs via the phenylpropanoid pathway (Salam et al., 2023). Transcriptional analysis in this study revealed that several genes associated with phenylpropanoid and flavonoid synthesis were responsive to high salt stress. This suggests that the synthesis of phenolic compounds in eelgrass is highly active under such conditions. Furthermore, homologous genes in rice and maize exhibited significant up-regulation (Supplementary Figure S10), supporting the conserved secondary metabolic responses to salt stress across plant species.
Plant hormones are important participants in the salt tolerance of plants (Yu et al., 2020). In eelgrass, we identified a series of hormone-related genes responding to salt stress, suggesting high salinity stimulates the activation of phytohormone signaling pathway.
We have mentioned in the result that the ABA synthesis gene, Zosma04g22590, a member of the NCED family, was consistently up-regulated across three comparisons. However, another NCED family member, Zosma01g20220, exhibited down-regulation. In rice and maize, most members of this family are up-regulated (Supplementary Figure S11), suggesting the conservation of ABA signaling in plant salt responses. This viewpoint is further supported by several potential downstream genes of the ABA signaling pathway, including dehydrin family protein (Zosma03g27730), protein kinases (Zosma03g25920), and PP2C family members (Zosma01g41250, Zosma02g21670). Their expressions were up-regulated at high salinity and homologous genes have been reported to be induced by ABA (Supplementary Figure S11). RAB18 is the homolog of Zosma03g27730 and reported to interact with PIP2B (Hernández-Sánchez et al., 2019), which suggests the regulation of aquaporin genes by ABA signaling. Zosma01g41250 and Zosma02g21670 are members of the PP2C family. Phylogenetic tree analysis shows that Zosma01g41250 is located on the branch with HAI1 and HAI2, while Zosma02g21670 is located on the branch with PP2C5 and AP2C1(Supplementary Figure S11). It is reported that HAI PP2C mutants had enhanced proline and osmoregulatory solute accumulation at low water potential (Bhaskara et al., 2012). Both PP2C5 and AP2C1 function as MAPK phosphatases, which are important to ABA signaling (Brock et al., 2010; Schweighofer et al., 2007). The up-regulation of PP2C members seems to be a conserved character under salt stress, as many homologous genes in rice and maize also show similar responses (Supplementary Figure S11). Terrestrial plants typically respond to osmotic stress caused by high salinity by regulating stomatal opening and closing via ABA (Hedrich and Shabala, 2018; Komatsu, 2020). Since eelgrass lacks stomata (Olsen et al., 2016), how ABA helps eelgrass cope with high-salinity environments needs further investigation.
In addition, JA, auxin, brassinosteroid (BR), cytokinin (CK) and gibberellin (GA) are also potential regulatory factors in the salt stress response of eelgrass. Research indicates that JA signaling in terrestrial plants is activated in response to salt stress, thereby enhancing salinity tolerance (Valenzuela et al., 2016; Zhao et al., 2014). Among the DEGs identified in eelgrass, Zosma01g30620, Zosma05g15520, and Zosma06g28020 are associated with JA signaling and down-regulated at high salinity. The homologous gene of Zosma01g30620, lipoxygenases (LOX3) is involved in JA synthesis (López-Cruz et al., 2014). The homolog of Zosma05g15520, jasmonic acid carboxyl methyltransferase (JMT) is a carboxyl methyltransferase that catalyzes the formation of methyl jasmonate from jasmonate (Seo et al., 2001). Zosma06g28020 is homologous to the jasmonic acid receptor COI1 (Song et al., 2021). Furthermore, we found that auxin-related genes Zosma03g17780 and Zosma05g12790 were down-regulated under high salt stress. The homolog of Zosma03g17780, PGP4 encodes an auxin effector transmembrane transporter, which is involved in auxin transport and inhibits root hair elongation (Cho et al., 2007). The homolog of Zosma05g12790, ARA-2 plays a role in the auxin signaling pathway and inhibits lateral root development (Koh et al., 2009). Exogenous application of BR can enhance the salt tolerance of terrestrial plants, indicating that BR may be involved in salt tolerance (Fu and Yang, 2023). Zosma03g35130 and Zosma03g30090 are identified as BR-related genes. Overexpression of the BKI1, a homolog of Zosma03g35130, leads to a salt-tolerant phenotype (Zhang et al., 2017). DOGT1, a homolog of Zosma03g30090, encodes a don-glucosyltransferase that regulates BR activity (Poppenberger et al., 2005). Moreover, Zosma06g28530 and Zosma01g36380, family members of two-component response regulator ARR-A, are associated with CK (Li et al., 2013; Ishida et al., 2008). These two genes are down-regulated at high salinity in eelgrass, most of which homologous genes in maize exhibited similar responses (Supplementary Figure S11). Among the down-regulated DEGs in SR tissue, we identified one gene related to gibberellin synthesis, Zosma06g07970 (ent-kaurene oxidase), suggesting that hypersalinity may regulate gibberellin levels in eelgrass.
In terrestrial plants, the actions of different phytohormones are interconnected. For instance, the key regulator jasmonate ZIM-domain proteins (JAZ) in JA could be regulated by the PYL6-MYC2 module of ABA (Aleman et al., 2016). CK receptor kinase significantly regulates ABA levels (Nishiyama et al., 2013). The interplay between ABA and GA signaling under abiotic stress conditions is significantly influenced by DELLA proteins (Aizaz et al., 2024). In the future, we aim to reveal the reasons for these transcriptional changes by studying the hormone content in eelgrass. Furthermore, exploring the potential interactions of phytohormones under salt stress in eelgrass will be valuable.
The SOS pathway is crucial for plant salt tolerance. Its core components include SOS1 (sodium-proton exchanger), SOS2 (CBL-interacting protein kinase), SOS3 (calcium-binding EF-hand family protein), and SCaBP8 (SOS3-like calcium-binding protein). Additional components associated with SOS pathway, such as HIS1-3, MPK4, PLATZ2, RSA1 at the transcriptional level, and UBC, as well as 14-3-3, BIN2, CIPK8, GI, PKS5, and PP2C at the post-transcriptional level, were also identified for plant salt tolerance (Ali et al., 2023). In our study, we utilized OrthoFinder (Supplementary Data S11) to identify SOS pathway genes in eelgrass. We found that none of these genes exhibited a significant response to salt stress, which was different from the findings in rice and maize (Supplementary Figure S12). Unlike terrestrial plants, which only have their roots in direct contact with soil salinity change, the whole plant of seagrass is exposed to the hypersalinity. The SOS pathway genes show high expression levels in various organs in eelgrass (Supplementary Figure S12A), which may provide a foundation for the protein-level responses of SOS pathway. Furthermore, several genes including Zosma01g22480 (SCaBP), Zosma05g05710 (14-3-3), and Zosma01g41780 (BIN2), were specifically expressed in floral organs, indicating their potential involvement in the reproductive development of eelgrass.
Salt stress can activate the MAPK signaling cascade, which involves protein kinases that phosphorylate various substrates (Taj et al., 2010). In this study, we observed that the expression of two MAPKKK genes, Zosma03g00830 and Zosma03g00810, was down-regulated under high salt conditions in leaf. Their A. thaliana homolog MAPKKK18 (AT1G05100) is induced by ABA and promotes leaf senescence (Matsuoka et al., 2015). This suggests that eelgrass may enhance its salinity tolerance by reducing the accumulation of these MAPKKK genes, thereby avoiding senescence. Additionally, the colinear gene Os05g0545300, corresponding to Zosma03g00810, also responded to salt stress, although it exhibited down-regulation in only one comparison (Supplementary Figure S13).
We identified a total of 172 MIRNA loci using small RNA-seq data. The mature sequences derived from these MIRNAs were predominantly 21 nucleotides in length. 83 (71.6%) of these microRNAs had uracil (U) as the first base at the 5’ end. Previous studies have demonstrated that the presence of uracil at the 5’ end can enhance the binding of microRNAs to the AGO1 protein (Mi et al., 2008). Based on the similarity of mature miRNAs, we identified 38 MIRNA belonging to known families, which is far fewer than the 93 known conserved miRNAs predicted genome-wide by Ma et al. (2021). The limited number of known miRNAs might be attributed to the restrictions in the plant tissues or RNA extraction method. The same kits for total RNA extraction were used for both mRNA and small RNA sequencing, as they have been employed in other researches (Zhang M. et al., 2023; Hu et al., 2021). In order to prove the accuracy of our study, we have experimentally validated the response of some DEmiRNAs using qRT-PCR. The results proved the existence of those miRNAs and the expression pattern of them was the same as the result from library construction.
Differential expression analysis of miRNAs revealed multiple miRNAs response to salt stress, including miR156, miR166, miR172, and a number of de novo miRNAs. miR156 has been reported to participate in salt tolerance across different plant species such as apple (Ma Y. et al., 2021), tobacco (Kang et al., 2019) and Alfalfa (Arshad et al., 2017). Similarly, miR166 has been shown to respond to salt stress in plants such as Hemerocallis fulva (Zhou et al., 2023), sugar beet (Zhang Z. et al., 2023), and potato (Kitazumi et al., 2015). It also plays a regulatory role in other abiotic stresses, including drought (Zhao et al., 2024) and low potassium conditions (Lei et al., 2023). In cereal crops, miR172 contributes to salt tolerance by maintaining ROS homeostasis (Cheng et al., 2021). Based on expression correlation analysis, we hypothesize that miR156 and miR166 are key regulators of the salt stress response in eelgrass. Specifically, miR156 was significantly up-regulated in leaf, while three of its target genes, WRKY family member Zosma06g26970, cellulose synthase Zosma04g03580, and heavy metal-related domain protein Zosma04g09250, were significantly down-regulated. Similarly, miR166 exhibited significant up-regulation in leaf, with two of its target genes, HD-ZIP family member Zosma03g00210 and S-locus lectin protein kinase family protein Zosma04g15070, also showing significant down-regulation. This negative correlation in expression patterns suggests the potential existence of miRNA-target modules involved in the salt stress response. Meanwhile, the degradome data predicted additional miRNA-target pairs (Figure 5; Supplementary Figure S5), which require further verification of their function in eelgrass.
Several microRNAs, including miR160 (Tang et al., 2022), miR164 (Lu et al., 2017), miR167 (Ye et al., 2020), miR172 (Cheng et al., 2021), miR319 (Zhou et al., 2013), miR390 (Chu et al., 2022), miR393 (Denver and Ullah, 2019), miR396 (Pegler et al., 2021), miR398 (He et al., 2021), miR399 (Pegler et al., 2020), and miR528 (Wang et al., 2021), have been reported to respond to salt stress or contribute to salt tolerance in various plant species. Although these miRNAs were identified in eelgrass, their expression levels did not exhibit significant differences under salt stress. However, some target genes of these miRNAs are responsive to salt stress (Supplementary Data S7), such as Zosma01g26740, a predictive target gene for miR167, and Zosma03g30130, a predictive target gene for miR319. Zosma01g26740 belongs to the ARF family and shows significantly reduced transcription levels under salt stress. ARF8 (AT5G37020), which is homologous to Zosma01g26740, has been reported to be regulated by miR167 and involved in root development (Gutierrez et al., 2009; Liao et al., 2023). Similarly, Zosma03g30130, a member of MYB family, exhibits down-regulated transcription levels under salt stress. MYB33 (AT5G06100), which is homologous to Zosma03g30130, is reported to be regulated by miR319 and specifically down-regulated in roots treated with ethylene, thereby influencing root growth (Zhang et al., 2016). The interaction between these prediction targets and miRNAs is worthy of further exploration in eelgrass.
The results revealed that 81% of salt-responsive TFs were distributed in eight modules: M1, M2, M3, M4, M5, M6, M7, and M9. A homologous analysis of these TFs indicated that several homologous genes from the bHLH, ERF, and MYB families in rice and maize exhibited similar expression patterns under high salt stress (Supplementary Figure S14). For instance, the bHLH family member Zosma03g02210, Os04g0300600, Os04g0301500, and Zm00001eb086370 are grouped in a branch. Similarly, ERF family members Zosma03g37150 and Os06g0166400 are found in the same branch, while ERF family members Zosma02g05080 and Os12g0168100 are collinear genes in one branch. Additionally, MYB family members Zosma06g17610, Zosma06g17660, Os03g0720800, Os11g0207600, and Zm00001eb168720 are grouped together in a branch. The consistent up-regulation of these genes suggests that they may function as conserved TFs involved in response to high salinity.
We focused on TFs that potentially regulate enriched biological processes and pathways, including GO:0009414 (response to water deprivation), map04016 (MAPK signaling pathway - plant), map00940 (phenylpropanoid biosynthesis), and map01110 (biosynthesis of secondary metabolites). To evaluate the expression correlation between DEGs, we calculated the Pearson correlation coefficient, considering values of ≥0.8 as indicative of similarity. The results revealed the potential regulatory roles of several TFs (Supplementary Figure S15). For instance, Zosma04g22590 (NCED) and Zosma02g25150 (arogenate/prephenate dehydratase) may be targeted by the ERF family member Zosma05g04820. Zosma03g22300 (phenylalanine ammonia-lyase) may be regulated by MYB family members Zosma06g17610 and Zosma06g17660. Furthermore, numerous potential binding sites for TFs are located in the upstream regions of microRNAs that respond to salt stress (Supplementary Figure S4). Further research is needed to determine whether these TFs can effectively regulate microRNA expression.
In this study, we performed a comprehensive analysis to reveal the response of eelgrass to high salt using transcriptome and degradome, and compared the result with other transcriptomes obtained from NCBI. In total, 14 DEmiRNAs and 691 DEGs including 53 TFs were identified. We further predicted the interactions between TFs, miRNAs, and their potential target genes. For instance, the up-regulated ERF members in the leaf may function by regulating the ABA synthase NCED, while MYB members might affect secondary metabolism by regulating PAL. Additionally, miRNA156-WRKY and miRNA166-HD-ZIP are supposed to be regulatory modules influencing the response on the post-transcriptional level.
Through the identification of homologous genes and expression profiling, we obtained a series of conserved responsive genes in eelgrass and two terrestrial plants rice and maize. The osmotic response related to the go-term “response to water deprivation” seems to be conserved in three species. Besides, we identified other conserved responses between rice and maize (Supplementary Figure S16), which suggest potential differences between terrestrial plants and marine plants in response to salt stress, such as the “gibberellin biosynthetic process”. Moreover, eelgrass exhibits specific responses to high salinity, as evidenced by the enriched go-terms such as fucose biosynthesis and vacuolar protein processing. These findings enhance our understanding of the molecular-level adaptability of seagrass to marine environments. In the future, we aim to validate the functions of these genes and their homologs in model organisms through experiments such as constructing overexpression plants. The conserved and specific responsive genes can serve as potential genetic resources for improving the salt tolerance of crops.
The RNA-seq, small RNA-seq and degradome data used in this study has been deposited into CNCB database (https://www.cncb.ac.cn/) with the accession number of CRA020957 (BioProject: PRJCA031998). The other datasets for detailed analysis were provided in supporting information files.
All methods were performed in accordance with the relevant Chinese national guidelines. All experimental studies on plants complied with relevant institutional, national, and international guidelines and legislation.
HZ: Writing – original draft, Writing – review & editing, Funding acquisition. CL: Writing – original draft, Writing – review & editing, Funding acquisition. XD: Writing – original draft. DY: Writing – review & editing. QG: Writing – original draft. PL: Writing – original draft.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was mainly supported by the Fund of Educational Department of Liaoning Province (LJKQZ20222354, LJ212410158026, 2024JBYBZ003).
We are very grateful to Professor Peidong Zhang’s research group for providing the resources of eelgrass plants for this work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1497064/full#supplementary-material
Aizaz, M., Lubna null Jan, R., Asaf, S., Bilal, S., Kim, K.-M., et al. (2024). Regulatory dynamics of plant hormones and transcription factors under salt stress. Biology 13, 673. doi: 10.3390/biology13090673
Aleman, F., Yazaki, J., Lee, M., Takahashi, Y., Kim, A. Y., Li, Z., et al. (2016). An ABA-increased interaction of the PYL6 ABA receptor with MYC2 Transcription Factor: A putative link of ABA and JA signaling. Sci. Rep. 6, 28941. doi: 10.1038/srep28941
Ali, A., Petrov, V., Yun, D.-J., Gechev, T. (2023). Revisiting plant salt tolerance: novel components of the SOS pathway. Trends Plant Sci. 28, 1060–1069. doi: 10.1016/j.tplants.2023.04.003
Altmann, M., Altmann, S., Rodriguez, P. A., Weller, B., Elorduy Vergara, L., Palme, J., et al. (2020). Extensive signal integration by the phytohormone protein network. Nature 583, 271–276. doi: 10.1038/s41586-020-2460-0
Arshad, M., Gruber, M. Y., Wall, K., Hannoufa, A. (2017). An Insight into microRNA156 Role in Salinity Stress Responses of Alfalfa. Front. Plant Sci. 8, 356. doi: 10.3389/fpls.2017.00356
Bai, Q., Wang, X., Chen, X., Shi, G., Liu, Z., Guo, C., et al. (2018). Wheat miRNA TaemiR408 Acts as an Essential Mediator in Plant Tolerance to Pi Deprivation and Salt Stress via Modulating Stress-Associated Physiological Processes. Front. Plant Sci. 9, 499. doi: 10.3389/fpls.2018.00499
Barajas-Lopez, J. D., Tiwari, A., Zarza, X., Shaw, M. W., Pascual, J. S., Punkkinen, M., et al. (2021). EARLY RESPONSE TO DEHYDRATION 7 remodels cell membrane lipid composition during cold stress in arabidopsis. Plant Cell Physiol. 62, 80–91. doi: 10.1093/pcp/pcaa139
Barrera-Rojas, C. H., Vicente, M. H., Pinheiro Brito, D. A., Silva, E. M., Lopez, A. M., Ferigolo, L. F., et al. (2023). Tomato miR156-targeted SlSBP15 represses shoot branching by modulating hormone dynamics and interacting with GOBLET and BRANCHED1b. J. Exp. Bot. 74, 5124–5139. doi: 10.1093/jxb/erad238
Bascom, C. (2023). Hormone synergy: Auxin and jasmonate boost abscisic acid signaling via ARF10 and ARF16. Plant Cell 35, 971–972. doi: 10.1093/plcell/koad012
Begum, Y. (2022). Regulatory role of microRNAs (miRNAs) in the recent development of abiotic stress tolerance of plants. Gene 821, 146283. doi: 10.1016/j.gene.2022.146283
Bhaskara, G. B., Nguyen, T. T., Verslues, P. E. (2012). Unique drought resistance functions of the highly ABA-induced clade A protein phosphatase 2Cs. Plant Physiol. 160, 379–395. doi: 10.1104/pp.112.202408
Brock, A. K., Willmann, R., Kolb, D., Grefen, L., Lajunen, H. M., Bethke, G., et al. (2010). The Arabidopsis mitogen-activated protein kinase phosphatase PP2C5 affects seed germination, stomatal aperture, and abscisic acid-inducible gene expression. Plant Physiol. 153, 1098–1111. doi: 10.1104/pp.110.156109
Cambridge, M. L., Zavala-Perez, A., Cawthray, G. R., Statton, J., Mondon, J., Kendrick, G. A. (2019). Effects of desalination brine and seawater with the same elevated salinity on growth, physiology and seedling development of the seagrass Posidonia australis. Mar. pollut. Bull. 140, 462–471. doi: 10.1016/j.marpolbul.2019.02.001
Capel, J., Jarillo, J. A., Salinas, J., Martínez-Zapater, J. M. (1997). Two homologous low-temperature-inducible genes from Arabidopsis encode highly hydrophobic proteins. Plant Physiol. 115, 569–576. doi: 10.1104/pp.115.2.569
Chen, Q., Liu, R., Wu, Y., Wei, S., Wang, Q., Zheng, Y., et al. (2021). ERAD-related E2 and E3 enzymes modulate the drought response by regulating the stability of PIP2 aquaporins. Plant Cell 33, 2883–2898. doi: 10.1093/plcell/koab141
Chen, S., Xu, K., Kong, D., Wu, L., Chen, Q., Ma, X., et al. (2022). Ubiquitin ligase OsRINGzf1 regulates drought resistance by controlling the turnover of OsPIP2;1. Plant Biotechnol. J. 20, 1743–1755. doi: 10.1111/pbi.13857
Chen, H. C., Huang, S.-C., Chen, Y.-F., Kuo, C.-W., Chen, Y.-H., Chang, M.-C. (2023). Overexpression of OsERF106MZ promotes parental root growth in rice seedlings by relieving the ABA-mediated inhibition of root growth under salinity stress conditions. BMC Plant Biol. 23, 144. doi: 10.1186/s12870-023-04136-8
Cheng, X., He, Q., Tang, S., Wang, H., Zhang, X., Lv, M., et al. (2021). The miR172/IDS1 signaling module confers salt tolerance through maintaining ROS homeostasis in cereal crops. New Phytol. 230, 1017–1033. doi: 10.1111/nph.17211
Cho, M., Lee, S. H., Cho, H.-T. (2007). P-glycoprotein4 displays auxin efflux transporter-like action in Arabidopsis root hair cells and tobacco cells. Plant Cell 19, 3930–3943. doi: 10.1105/tpc.107.054288
Chu, Y., Bai, W., Wang, P., Li, F., Zhan, J., Ge, X. (2022). The mir390-GhCEPR2 module confers salt tolerance in cotton and Arabidopsis. Ind. Crops Prod. 190, 115865. doi: 10.1016/j.indcrop.2022.115865
Cui, M. H., Yoo, K. S., Hyoung, S., Nguyen, H. T. K., Kim, Y. Y., Kim, H. J., et al. (2013). An Arabidopsis R2R3-MYB transcription factor, AtMYB20, negatively regulates type 2C serine/threonine protein phosphatases to enhance salt tolerance. FEBS Lett. 587, 1773–1778. doi: 10.1016/j.febslet.2013.04.028
Dabravolski, S. A., Isayenkov, S. V. (2023). The regulation of plant cell wall organisation under salt stress. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1118313
Das, K., Roychoudhury, A. (2014). Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2. doi: 10.3389/fenvs.2014.00053
Denver, J. B., Ullah, H. (2019). miR393s regulate salt stress response pathway in Arabidopsis thaliana through scaffold protein RACK1A mediated ABA signaling pathways. Plant Signaling Behav. 14, 1600394. doi: 10.1080/15592324.2019.1600394
Emery, L., Whelan, S., Hirschi, K. D., Pittman, J. K. (2012). Protein Phylogenetic Analysis of Ca(2+)/cation Antiporters and Insights into their Evolution in Plants. Front. Plant Sci. 3. doi: 10.3389/fpls.2012.00001
Ferreira e Silva, G. F., Silva, E. M., Azevedo da, M. S., Guivin, M. A. C., Ramiro, D. A., Figueiredo, C. R., et al. (2014). microRNA156-targeted SPL/SBP box transcription factors regulate tomato ovary and fruit development. Plant Journal: For Cell Mol. Biol. 78, 604–618. doi: 10.1111/tpj.12493
Fu, H., Yang, Y. (2023). How plants tolerate salt stress. Curr. Issues Mol. Biol. 45, 5914–5934. doi: 10.3390/cimb45070374
Gao, X., Cong, Y., Yue, J., Xing, Z., Wang, Y., Chai, X. (2019). Small RNA, transcriptome, and degradome sequencing to identify salinity stress responsive miRNAs and target genes in Dunaliella salina. J. Appl. Phycol. 31, 1175–1183. doi: 10.1007/s10811-018-1612-1
Gou, J.-Y., Felippes, F. F., Liu, C.-J., Weigel, D., Wang, J.-W. (2011). Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23, 1512–1522. doi: 10.1105/tpc.111.084525
Guo, H. S., Xie, Q., Fei, J.-F., Chua, N.-H. (2005). MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for arabidopsis lateral root development. Plant Cell 17, 1376–1386. doi: 10.1105/tpc.105.030841
Gutierrez, L., Bussell, J. D., Pacurar, D. I., Schwambach, J., Pacurar, M., Bellini, C. (2009). Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 21, 3119–3132. doi: 10.1105/tpc.108.064758
Hasanuzzaman, M., Raihan, M. R. H., Masud, A. A. C., Rahman, K., Nowroz, F., Rahman, M., et al. (2021). Regulation of reactive oxygen species and antioxidant defense in plants under salinity. Int. J. Mol. Sci. 22, 9326. doi: 10.3390/ijms22179326
He, Y., Zhou, J., Hu, Y., Fang, C., Yu, Y., Yang, J., et al. (2021). Overexpression of sly-miR398b increased salt sensitivity likely via regulating antioxidant system and photosynthesis in tomato. Environ. Exp. Bot. 181, 104273. doi: 10.1016/j.envexpbot.2020.104273
Hedrich, R., Shabala, S. (2018). Stomata in a saline world. Curr. Opin. Plant Biol. 46, 87–95. doi: 10.1016/j.pbi.2018.07.015
Hernández-Sánchez, I. E., Maruri-López, I., Molphe-Balch, E. P., Becerra-Flora, A., Jaimes-Miranda, F., Jiménez-Bremont, J. F. (2019). Evidence for in vivo interactions between dehydrins and the aquaporin AtPIP2B. Biochem. Biophys. Res. Commun. 510, 545–550. doi: 10.1016/j.bbrc.2019.01.095
Hu, Z., Nie, Z., Yan, C., Huang, H., Ma, X., Wang, Y., et al. (2021). Transcriptome and Degradome Profiling Reveals a Role of miR530 in the Circadian Regulation of Gene Expression in Kalanchoë marnieriana. Cells 10, 1526. doi: 10.3390/cells10061526
Ishida, K., Yamashino, T., Mizuno, T. (2008). Expression of the cytokinin-induced type-A response regulator gene ARR9 is regulated by the circadian clock in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 72, 3025–3029. doi: 10.1271/bbb.80402
Ishikawa, F., Suga, S., Uemura, T., Sato, M. H., Maeshima, M. (2005). Novel type aquaporin SIPs are mainly localized to the ER membrane and show cell-specific expression in Arabidopsis thaliana. FEBS Lett. 579, 5814–5820. doi: 10.1016/j.febslet.2005.09.076
Iuchi, S., Kobayashi, M., Taji, T., Naramoto, M., Seki, M., Kato, T., et al. (2001). Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant Journal: For Cell Mol. Biol. 27, 325–333. doi: 10.1046/j.1365-313x.2001.01096.x
Jia, F., Qi, S., Li, H., Liu, P., Li, P., Wu, C., et al. (2014). Overexpression of Late Embryogenesis Abundant 14 enhances Arabidopsis salt stress tolerance. Biochem. Biophys. Res. Commun. 454, 505–511. doi: 10.1016/j.bbrc.2014.10.136
Jiang, Y., Deyholos, M. K. (2009). Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 69, 91–105. doi: 10.1007/s11103-008-9408-3
Kamiya, T., Fujiwara, T. (2009). NIP1;1, an aquaporin homolog, determines the arsenite sensitivity of Arabidopsis thaliana. J. Biol. Chem. 284, 2114–2120. doi: 10.1074/jbc.M806881200
Kamiya, T., Fujiwara, T. (2009). Arabidopsis NIP1;1 transports antimonite and determines antimonite sensitivity. Plant Cell Physiol. 50, 1977–1981. doi: 10.1093/pcp/pcp130
Kang, C., He, S., Zhai, H., Li, R., Zhao, N., Liu, Q. (2018). A sweetpotato auxin response factor gene (IbARF5) is involved in carotenoid biosynthesis and salt and drought tolerance in transgenic arabidopsis. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01307
Kang, T., Yu, C.-Y., Liu, Y., Song, W.-M., Bao, Y., Guo, X.-T., et al. (2019). Subtly manipulated expression of zmmiR156 in tobacco improves drought and salt tolerance without changing the architecture of transgenic plants. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01664
Kim, J., Jung, J.-H., Reyes, J. L., Kim, Y.-S., Kim, S.-Y., Chung, K.-S., et al. (2005). microRNA-directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. Plant Journal: For Cell Mol. Biol. 42, 84–94. doi: 10.1111/j.1365-313X.2005.02354.x
Kitazumi, A., Kawahara, Y., Onda, T. S., De Koeyer, D., de los Reyes, B. G. (2015). Implications of miR166 and miR159 induction to the basal response mechanisms of an andigena potato (Solanum tuberosum subsp. andigena) to salinity stress, predicted from network models in Arabidopsis. Genome 58, 13–24. doi: 10.1139/gen-2015-0011
Koch, M. S., Schopmeyer, S. A., Holmer, M., Madden, C. J., Kyhn-Hansen, C. (2007). Thalassia testudinum response to the interactive stressors hypersalinity, sulfide and hypoxia. Aquat. Bot. 87, 104–110. doi: 10.1016/j.aquabot.2007.03.004
Koh, E. J., Kwon, Y.-R., Kim, K.-I., Hong, S.-W., Lee, H. (2009). Altered ARA2 (RABA1a) expression in Arabidopsis reveals the involvement of a Rab/YPT family member in auxin-mediated responses. Plant Mol. Biol. 70, 113–122. doi: 10.1007/s11103-009-9460-7
Komatsu, K. (2020). Decoding ABA and osmostress signalling in plants from an evolutionary point of view. Plant Cell Environ. 43, 2894–2911. doi: 10.1111/pce.13869
Koyama, T. (2017). Roles of miR319 and TCP transcription factors in leaf development. Plant Physiol. 175, 874–885. doi: 10.1104/pp.17.00732
Lei, X., Chen, M., Xu, K., Sun, R., Zhao, S., Wu, N. (2023). The miR166d/TaCPK7-D Signaling Module is a Critical Mediator of Wheat (Triticum aestivum L.) Tolerance to K+ Deficiency. Int. J. Mol. Sci. 24, 7926. doi: 10.3390/ijms24097926
Li, Y., Kurepa, J., Smalle, J. (2013). AXR1 promotes the Arabidopsis cytokinin response by facilitating ARR5 proteolysis. Plant Journal: For Cell Mol. Biol. 74, 13–24. doi: 10.1111/tpj.12098
Li, Q., Yu, H.-M., Meng, X.-F., Lin, J.-S., Li, Y.-J., Hou, B.-K. (2018). Ectopic expression of glycosyltransferase UGT76E11 increases flavonoid accumulation and enhances abiotic stress tolerance in Arabidopsis. Plant Biol. (Stuttgart Germany) 20, 10–19. doi: 10.1111/plb.12627
Liang, G., He, H., Li, Y., Wang, F., Yu, D. (2014). Molecular mechanism of microRNA396 mediating pistil development in Arabidopsis. Plant Physiol. 164, 249–258. doi: 10.1104/pp.113.225144
Liao, R., Wei, X., Zhao, Y., Xie, Z., Nath, U. K., Yang, S., et al. (2023). bra-miR167a Targets ARF8 and Negatively Regulates Arabidopsis thaliana Immunity against Plasmodiophora brassicae. Int. J. Mol. Sci. 24, 11850. doi: 10.3390/ijms241411850
Liu, J., Wang, H., Su, M., Li, Q., Xu, H., Song, J., et al. (2023). A Transcription Factor SlNAC4 Gene of Suaeda liaotungensis Enhances Salt and Drought Tolerance through Regulating ABA Synthesis. Plants (Basel) 12, 2951. doi: 10.3390/plants12162951
Liu, R., Meng, J.-H., Zuo, W.-T., Jin, W.-M., Wang, L.-Q., Sun, T.-T. (2024). The phloem protein 2 (PP2) is positively regulated by ThNAC13 that enhances salt tolerance of Tamarix. Environ. Exp. Bot. 224, 105784. doi: 10.1016/j.envexpbot.2024.105784
Liu, Z., Wang, P., Wang, Z., Wang, C., Wang, Y. (2024). Birch WRKY transcription factor, BpWRKY32, confers salt tolerance by mediating stomatal closing, proline accumulation, and reactive oxygen species scavenging. Plant Physiol. biochem.: PPB 210, 108599. doi: 10.1016/j.plaphy.2024.108599
López-Cruz, J., Finiti, I., Fernández-Crespo, E., Crespo-Salvador, O., García-Agustín, P., González-Bosch, C. (2014). Absence of endo-1,4-β-glucanase KOR1 alters the jasmonate-dependent defence response to Pseudomonas syringae in Arabidopsis. J. Plant Physiol. 171, 1524–1532. doi: 10.1016/j.jplph.2014.07.006
Lu, X., Dun, H., Lian, C., Zhang, X., Yin, W., Xia, X. (2017). The role of peu-miR164 and its target PeNAC genes in response to abiotic stress in Populus euphratica. Plant Physiol. biochem.: PPB 115, 418–438. doi: 10.1016/j.plaphy.2017.04.009
Ma, X., Olsen, J. L., Reusch, T. B.H., Procaccini, G., Kudrna, D., Williams, M., et al. (2021). Improved chromosome-level genome assembly and annotation of the seagrass, Zostera marina (eelgrass). F1000Research 10, 289. doi: 10.12688/f1000research.38156.1
Ma, Y., Xue, H., Zhang, F., Jiang, Q., Yang, S., Yue, P., et al. (2021). The miR156/SPL module regulates apple salt stress tolerance by activating MdWRKY100 expression. Plant Biotechnol. J. 19, 311–323. doi: 10.1111/pbi.13464
Malakar, Chattopadhyay, D. (2021). Adaptation of plants to salt stress: the role of the ion transporters. J. Plant Biochem. Biotechnol. 30, 668–683. doi: 10.1007/s13562-021-00741-6
Matsuoka, D., Yasufuku, T., Furuya, T., Nanmori, T. (2015). An abscisic acid inducible Arabidopsis MAPKKK, MAPKKK18 regulates leaf senescence via its kinase activity. Plant Mol. Biol. 87, 565–575. doi: 10.1007/s11103-015-0295-0
Mi, S., Cai, T., Hu, Y., Chen, Y., Hodges, E., Ni, F., et al. (2008). Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5’ terminal nucleotide. Cell 133, 116–127. doi: 10.1016/j.cell.2008.02.034
Nishiyama, R., Watanabe, Y., Leyva-Gonzalez, M. A., Ha, C. V., Fujita, Y., Tanaka, M., et al. (2013). Arabidopsis AHP2, AHP3, and AHP5 histidine phosphotransfer proteins function as redundant negative regulators of drought stress response. Proc. Natl. Acad. Sci. United States America 110, 4840–4845. doi: 10.1073/pnas.1302265110
Olsen, K. M., Lea, U. S., Slimestad, R., Verheul, M., Lillo, C. (2008). Differential expression of four Arabidopsis PAL genes; PAL1 and PAL2 have functional specialization in abiotic environmental-triggered flavonoid synthesis. J. Plant Physiol. 165, 1491–1499. doi: 10.1016/j.jplph.2007.11.005
Olsen, J. L., Rouzé, P., Verhelst, B., Lin, Y.-C., Bayer, T., Collen, J., et al. (2016). The genome of the seagrass Zostera marina reveals angiosperm adaptation to the sea. Nature 530, 331–335. doi: 10.1038/nature16548
Pegler, J. L., Oultram, J. M. J., Grof, C. P. L., Eamens, A. L. (2020). Molecular Manipulation of the miR399/PHO2 Expression Module Alters the Salt Stress Response of Arabidopsis thaliana. Plants (Basel Switzerland) 10, 73. doi: 10.3390/plants10010073
Pegler, J. L., Nguyen, D. Q., Oultram, J. M. J., Grof, C. P. L., Eamens, A. L. (2021). Molecular manipulation of the miR396/GRF expression module alters the salt stress response of arabidopsis thaliana. Agronomy 11, 1751. doi: 10.3390/agronomy11091751
Polizel, A. M., Medri, M. E., Nakashima, K., Yamanaka, N., Farias, J. R. B., De Oliveira, M. C. N., et al. (2011). Molecular, anatomical and physiological properties of a genetically modified soybean line transformed with rd29A:AtDREB1A for the improvement of drought tolerance. Genet. Mol. Res. 10, 3641–3656. doi: 10.4238/2011.October.21.4
Poppenberger, B., Fujioka, S., Soeno, K., George, G. L., Vaistij, F. E., Hiranuma, S., et al. (2005). The UGT73C5 of Arabidopsis thaliana glucosylates brassinosteroids. Proc. Natl. Acad. Sci. United States America 102, 15253–15258. doi: 10.1073/pnas.0504279102
Postaire, O., Tournaire-Roux, C., Grondin, A., Boursiac, Y., Morillon, R., Schäffner, A. R., et al. (2010). A PIP1 aquaporin contributes to hydrostatic pressure-induced water transport in both the root and rosette of Arabidopsis. Plant Physiol. 152, 1418–1430. doi: 10.1104/pp.109.145326
Qin, X., Zeevaart, J. A. (1999). The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proc. Natl. Acad. Sci. United States America 96, 15354–15361. doi: 10.1073/pnas.96.26.15354
Ravikumar, G., Manimaran, P., Voleti, S. R., Subrahmanyam, D., Sundaram, R. M., Bansal, K. C., et al. (2014). Stress-inducible expression of AtDREB1A transcription factor greatly improves drought stress tolerance in transgenic indica rice. Transgenic Res. 23, 421–439. doi: 10.1007/s11248-013-9776-6
Reinhardt, H., Hachez, C., Bienert, M. D., Beebo, A., Swarup, K., Voß, U., et al. (2016). Tonoplast aquaporins facilitate lateral root emergence. Plant Physiol. 170, 1640–1654. doi: 10.1104/pp.15.01635
Ren, J., Yang, X., Ma, C., Wang, Y., Zhao, J., Kang, L. (2021). Meta-analysis of the effect of the overexpression of aquaporin family genes on the drought stress response. Plant Biotechnol. Rep. 15, 139–150. doi: 10.1007/s11816-021-00666-5
Rodrigues, O., Reshetnyak, G., Grondin, A., Saijo, Y., Leonhardt, N., Maurel, C., et al. (2017). Aquaporins facilitate hydrogen peroxide entry into guard cells to mediate ABA- and pathogen-triggered stomatal closure. Proc. Natl. Acad. Sci. United States America 114, 9200–9205. doi: 10.1073/pnas.1704754114
Rubio, L., Belver, A., Venema, K., Jesús García-Sánchez, M., Fernández, J. A. (2011). Evidence for a sodium efflux mechanism in the leaf cells of the seagrass Zostera marina L. J. Exp. Mar. Biol. Ecol. 402, 56–64. doi: 10.1016/j.jembe.2011.03.016
Sakuma, Y., Liu, Q., Dubouzet, J. G., Abe, H., Shinozaki, K., Yamaguchi-Shinozaki, K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290, 998–1009. doi: 10.1006/bbrc.2001.6299
Salam, U., Ullah, S., Tang, Z.-H., Elateeq, A. A., Khan, Y., Khan, J., et al. (2023). Plant metabolomics: an overview of the role of primary and secondary metabolites against different environmental stress factors. Life (Basel Switzerland) 13, 706. doi: 10.3390/life13030706
Sandoval-Gil, J. M., Ruiz, J. M., Marín-Guirao, L. (2023). Advances in understanding multilevel responses of seagrasses to hypersalinity. Mar. Environ. Res. 183, 105809. doi: 10.1016/j.marenvres.2022.105809
Schweighofer, A., Kazanaviciute, V., Scheikl, E., Teige, M., Doczi, R., Hirt, H., et al. (2007). The PP2C-type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in Arabidopsis. Plant Cell 19, 2213–2224. doi: 10.1105/tpc.106.049585
Seo, H. S., Song, J. T., Cheong, J. J., Lee, Y. H., Lee, Y. W., Hwang, I., et al. (2001). Jasmonic acid carboxyl methyltransferase: a key enzyme for jasmonate-regulated plant responses. Proc. Natl. Acad. Sci. United States America 98, 4788–4793. doi: 10.1073/pnas.081557298
Serra, I. A., Nicastro, S., Mazzuca, S., Natali, L., Cavallini, A., Innocenti, A. M. (2013). Response to salt stress in seagrasses: PIP1;1 aquaporin antibody localization in Posidonia oceanica leaves. Aquat. Bot. 104, 213–219. doi: 10.1016/j.aquabot.2011.05.008
Shivaraj, S. M., Deshmukh, R., Bhat, J. A., Sonah, H., Bélanger, R. R. (2017). Understanding aquaporin transport system in eelgrass (Zostera marina L.), an aquatic plant species. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01334
Shomali, A., Das, S., Arif, N., Sarraf, M., Zahra, N., Yadav, V., et al. (2022). Diverse physiological roles of flavonoids in plant environmental stress responses and tolerance. Plants (Basel Switzerland) 11, 3158. doi: 10.3390/plants11223158
Song, S., Liu, B., Zhai, J., Zhang, Y., Wang, K., Qi, T. (2021). The intragenic suppressor mutation Leu59Phe compensates for the effect of detrimental mutations in the jasmonate receptor COI1. Plant Journal: For Cell Mol. Biol. 108, 690–704. doi: 10.1111/tpj.15464
Taj, G., Agarwal, P., Grant, M., Kumar, A. (2010). MAPK machinery in plants: recognition and response to different stresses through multiple signal transduction pathways. Plant Signaling Behav. 5, 1370–1378. doi: 10.4161/psb.5.11.13020
Tang, Y., Du, G., Xiang, J., Hu, C., Li, X., Wang, W., et al. (2022). Genome-wide identification of auxin response factor (ARF) gene family and the miR160-ARF18-mediated response to salt stress in peanut (Arachis hypogaea L.). Genomics 114, 171–184. doi: 10.1016/j.ygeno.2021.12.015
Tian, S., Wang, X., Li, P., Wang, H., Ji, H., Xie, J., et al. (2016). Plant aquaporin atPIP1;4 links apoplastic H2O2 induction to disease immunity pathways. Plant Physiol. 171, 1635–1650. doi: 10.1104/pp.15.01237
Tomasello, A., Di Maida, G., Calvo, S., Pirrotta, M., Borra, M., Procaccini, G. (2009). Seagrass meadows at the extreme of environmental tolerance: the case of Posidonia oceanica in a semi-enclosed coastal lagoon. Mar. Ecol. 30, 288–300. doi: 10.1111/j.1439-0485.2009.00285.x
Valenzuela, C. E., Acevedo-Acevedo, O., Miranda, G. S., Vergara-Barros, P., Holuigue, L., Figueroa, C. R., et al. (2016). Salt stress response triggers activation of the jasmonate signaling pathway leading to inhibition of cell elongation in Arabidopsis primary root. J. Exp. Bot. 67, 4209–4220. doi: 10.1093/jxb/erw202
Verma, D., Jalmi, S. K., Bhagat, P. K., Verma, N., Sinha, A. K. (2020). A bHLH transcription factor, MYC2, imparts salt intolerance by regulating proline biosynthesis in Arabidopsis. FEBS J. 287, 2560–2576. doi: 10.1111/febs.15157
Wang, J. W., Wang, L.-J., Mao, Y.-B., Cai, W.-J., Xue, H.-W., Chen, X.-Y. (2005). Control of root cap formation by MicroRNA-targeted auxin response factors in Arabidopsis. Plant Cell 17, 2204–2216. doi: 10.1105/tpc.105.033076
Wang, P., Li, Z., Wei, J., Zhao, Z., Sun, D., Cui, S. (2012). A Na+/Ca2+ exchanger-like protein (AtNCL) involved in salt stress in Arabidopsis. J. Biol. Chem. 287, 44062–44070. doi: 10.1074/jbc.M112.351643
Wang, Y., Li, R., Li, D., Jia, X., Zhou, D., Li, J., et al. (2017). NIP1;2 is a plasma membrane-localized transporter mediating aluminum uptake, translocation, and tolerance in Arabidopsis. Proc. Natl. Acad. Sci. United States America 114, 5047–5052. doi: 10.1073/pnas.1618557114
Wang, L., Liu, Z., Qiao, M., Xiang, F. (2018). miR393 inhibits in vitro shoot regeneration in Arabidopsis thaliana via repressing TIR1. Plant Sci.: Int. J. Exp. Plant Biol. 266, 1–8. doi: 10.1016/j.plantsci.2017.10.009
Wang, H., Schoebel, S., Schmitz, F., Dong, H., Hedfalk, K. (2020). Characterization of aquaporin-driven hydrogen peroxide transport. Biochim. Et Biophys. Acta Biomembr. 1862, 183065. doi: 10.1016/j.bbamem.2019.183065
Wang, M., Guo, W., Li, J., Pan, X., Pan, L., Zhao, J., et al. (2021). The miR528-AO module confers enhanced salt tolerance in rice by modulating the ascorbic acid and abscisic acid metabolism and ROS scavenging. J. Agric. Food Chem. 69, 8634–8648. doi: 10.1021/acs.jafc.1c01096
Wang, C., Li, X., Zhuang, Y., Sun, W., Cao, H., Xu, R., et al. (2024). A novel miR160a-GmARF16-GmMYC2 module determines soybean salt tolerance and adaptation. New Phytol. 241, 2176–2192. doi: 10.1111/nph.19503
Wang, S., Jiang, R., Feng, J., Zou, H., Han, X., Xie, X., et al. (2024). Overexpression of transcription factor FaMYB63 enhances salt tolerance by directly binding to the SOS1 promoter in Arabidopsis thaliana. Plant Mol. Biol. 114, 32. doi: 10.1007/s11103-024-01431-2
Wang, X., Wang, T., Yu, P., Li, Y., Lv, X. (2024). NO enhances the adaptability to high-salt environments by regulating osmotic balance, antioxidant defense, and ion homeostasis in eelgrass based on transcriptome and metabolome analysis. Front. Plant Sci. 15, 1343154. doi: 10.3389/fpls.2024.1343154
Wissler, L., Codoñer, F. M., Gu, J., Reusch, T. B., Olsen, J. L., Procaccini, G., et al. (2011). Back to the sea twice: identifying candidate plant genes for molecular evolution to marine life. BMC Evolution. Biol. 11, 8. doi: 10.1186/1471-2148-11-8
Xu, Q., Niu, S., Zhang, P., Zhang, X., Fang, C. (2015). Salinity suitability of Zostera marina seedlings. Chinese Journal of Ecology 34(11), 3146–3150. doi: 10.13292/j.1000-4890.20151023.013
Yadav, A., Kumar, S., Verma, R., Lata, C., Sanyal, I., Rai, S. P. (2021). microRNA 166: an evolutionarily conserved stress biomarker in land plants targeting HD-ZIP family. Physiol. Mol. Biol. Plants 27, 2471–2485. doi: 10.1007/s12298-021-01096-x
Yang, Y., Guo, Y. (2018). Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 217, 523–539. doi: 10.1111/nph.14920
Ye, Y., Wang, J., Wang, W., Xu, L.-A. (2020). ARF family identification in Tamarix chinensis reveals the salt responsive expression of TcARF6 targeted by miR167. PeerJ 8, e8829. doi: 10.7717/peerj.8829
Yepes-Molina, L., Bárzana, G., Carvajal, M. (2020). Controversial regulation of gene expression and protein transduction of aquaporins under drought and salinity stress. Plants (Basel Switzerland) 9, 1662. doi: 10.3390/plants9121662
Yoon, Y., Seo, D. H., Shin, H., Kim, H. J., Kim, C. M., Jang, G. (2020). The role of stress-responsive transcription factors in modulating abiotic stress tolerance in plants. Agronomy 10, 788. doi: 10.3390/agronomy10060788
Yu, Z., Duan, X., Luo, L., Dai, S., Ding, Z., Xia, G. (2020). How plant hormones mediate salt stress responses. Trends Plant Sci. 25, 1117–1130. doi: 10.1016/j.tplants.2020.06.008
Zhang, F., Wang, L., Lim, J. Y., Kim, T., Pyo, Y., Sung, S., et al. (2016). Phosphorylation of CBP20 links microRNA to root growth in the ethylene response. PloS Genet. 12, e1006437. doi: 10.1371/journal.pgen.1006437
Zhang, M., Zhao, J., Li, L., Gao, Y., Zhao, L., Patil, S. B., et al. (2017). The Arabidopsis U-box E3 ubiquitin ligase PUB30 negatively regulates salt tolerance by facilitating BRI1 kinase inhibitor 1 (BKI1) degradation. Plant Cell Environ. 40, 2831–2843. doi: 10.1111/pce.13064
Zhang, Y.-H., Li, J.-D., Yan, W.-J., Luo, F.-S., Wang, L., Zuo, L.-M., et al. (2023). The combined effect of seawater salinity and duration on the survival and growth of eelgrass Zostera marina. Aquatic Botany 187. doi: 10.1016/j.aquabot.2023.103652
Zhang, Y., Yue, S., Liu, M., Wang, X., Xu, S., Zhang, X., et al. (2023). Combined transcriptome and proteome analysis reveal the key physiological processes in seed germination stimulated by decreased salinity in the seagrass Zostera marina L. BMC Plant Biol. 23, 605. doi: 10.1186/s12870-023-04616-x
Zhang, Z., Fu, Z., Chen, W., Wang, L., Zhao, S., Yuanyuan, E., et al. (2023). Salt Stress Induces Complicated miRNA Responses of Sugar Beet (Beta vulgaris L.). Sugar Tech. 26, 1297–1305. doi: 10.1007/s12355-023-01341-5
Zhang, M., Zhang, X., Wang, R., Zang, R., Guo, L., Qi, T., et al. (2023). Heat-responsive microRNAs participate in regulating the pollen fertility stability of CMS-D2 restorer line under high-temperature stress. Biol Res 56, 58. doi: 10.1186/s40659-023-00465-y
Zhao, Y., Dong, W., Zhang, N., Ai, X., Wang, M., Huang, Z., et al. (2014). A wheat allene oxide cyclase gene enhances salinity tolerance via jasmonate signaling. Plant Physiol. 164, 1068–1076. doi: 10.1104/pp.113.227595
Zhao, X., Wang, X., Li, F., Dahlgren, R. A., Wang, H. (2016). Identification of microRNA-size sRNAs Related to Salt Tolerance in Spirulina platensis. Plant Mol. Biol. Rep. 34, 539–555. doi: 10.1007/s11105-015-0938-y
Zhao, C., Ma, J., Yan, C., Jiang, Y., Zhang, Y., Lu, Y., et al. (2024). Drought-triggered repression of miR166 promotes drought tolerance in soybean. Crop J. 12, 154–163. doi: 10.1016/j.cj.2023.12.005
Zhou, M., Li, D., Li, Z., Hu, Q., Yang, C., Zhu, L., et al. (2013). Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Physiol. 161, 1375–1391. doi: 10.1104/pp.112.208702
Zhou, B., Gao, X., Zhao, F. (2023). Integration of mRNA and miRNA Analysis Reveals the Post-Transcriptional Regulation of Salt Stress Response in Hemerocallis fulva. Int. J. Mol. Sci. 24, 7290. doi: 10.3390/ijms24087290
Zhu, J., Wei, X., Yin, C., Zhou, H., Yan, J., He, W., et al. (2023). ZmEREB57 regulates OPDA synthesis and enhances salt stress tolerance through two distinct signalling pathways in Zea mays. Plant Cell Environ. 46, 2867–2883. doi: 10.1111/pce.14644
Zou, P., Lu, X., Zhao, H., Yuan, Y., Meng, L., Zhang, C., et al. (2019). Polysaccharides derived from the brown algae lessonia nigrescens enhance salt stress tolerance to wheat seedlings by enhancing the antioxidant system and modulating intracellular ion concentration. Front. Plant Sci. 10, 48. doi: 10.3389/fpls.2019.00048
Keywords: Zostera marina, salt stress response, miRNA, transcription factor, comparative transcriptome, degradome sequencing, gene regulation
Citation: Zhao H, Dong X, Yang D, Ge Q, Lu P and Liu C (2025) New insights into the salt-responsive regulation in eelgrass at transcriptional and post-transcriptional levels. Front. Plant Sci. 16:1497064. doi: 10.3389/fpls.2025.1497064
Received: 16 September 2024; Accepted: 17 January 2025;
Published: 06 February 2025.
Edited by:
Yi Zhou, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Vesselin Baev, Plovdiv University “Paisii Hilendarski”, BulgariaCopyright © 2025 Zhao, Dong, Yang, Ge, Lu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chang Liu, bGl1Y2hhbmdfYmlvQGZveG1haWwuY29t
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.