- 1Indian Council of Agricultural Research (ICAR)- National Institute for Plant Biotechnology (NIPB), New Delhi, India
- 2Department of Biotechnology, Guru Jambheshwar University of Science and Technology (GJUS&T), Hisar, Haryana, India
- 3Department of Botany, Institute of Science, Banaras Hindu University (BHU), Varanasi, Uttar Pradesh, India
- 4Veer Chandra Singh Garhwali Uttarakhand University of Horticulture & Forestry, Bharsar, Uttarakhand, India
- 5Crop Improvement Section, ICAR-Indian Institute of Oilseeds Research, Hyderabad, Telangana, India
- 6Division of Genetics, ICAR-Indian Agricultural Research Institute (IARI), New Delhi, India
- 7Applied Genomics Section, Bhabha Atomic Research Centre (BARC), Mumbai, India
- 8Division of Germplasm Evaluation, ICAR-National Bureau of Plant Genetic Resources (NBPGR), New Delhi, India
In India, amphidiploid Brassica juncea (AABB, 2n=36) is a significant oilseed crop, but its small gene pool limits its ability to develop traits of higher breeding and economic value. Through interspecific hybridization from various lines of the progenitor species, resynthesized B. juncea (RBJ) can provide breeders with additional resources for creating genetically diverse stress-tolerant and high-yielding cultivars. Three B. rapa accessions and eight B. nigra accessions were crossed in this study to develop 33 synthetic B. juncea lines. A total of 28 crosses were attempted, including the three-way crosses, but only the cross combinations with B. rapa cytoplasm led to successful embryonic development. Molecular diversity analysis of these lines in S2 generation revealed significant genetic diversity with higher levels of heterozygosity and allelic richness, along with significant variations for the yield-related traits. These results suggest that the synthesized lines could effectively enrich the genetic base of B. juncea and generate variability for agronomically important traits in a shorter time duration. The characterized variability in the synthetic lines needs to be utilized in hybridization, with already evolved genotypes, in early generations before it is lost due to chromosomal rearrangements, recombination and natural selection.
1 Introduction
Amphidiploid Brassica juncea holds a pivotal position as a vital oilseed crop in India and adjacent Asian countries, making a substantial contribution to the agricultural landscape of these countries. Specifically in India, 23.5% of the cultivated area is dedicated to oilseed crops and is responsible for approximately 24.2% of the total oilseed production within the country (Jat et al., 2019). The allotetraploid species, B. juncea has evolved from the hybridization of two different diploid progenitor species- B. rapa (AA, 2n = 20) and B. nigra (BB, 2n = 16), encompassing genetic diversity from both the progenitors and, potentially creating a genetic base for the developing cultivars (Jat et al., 2019). However, complex polyploidy, selective domestication, and modern breeding techniques restricted the genetic variability in Indian mustard (B. juncea) (Li et al., 2013; Zhang et al., 2022). It is also anticipated that during the process of evolution, the genetic diversity of both the parental species is not fully utilized, resulting in a narrow genetic base in the natural gene pool, ultimately limiting the potential for breeders to develop new promising cultivars with desired traits (Gepts and Papa, 2003; Hu et al., 2021; Singh et al., 2021).
One of the valuable approaches for developing a new gene pool is resynthesizing allotetraploid species that gives access to a new genetic diversity that may or may not exist in the original parental species, conferring desirable traits such as yield contributing traits, quality parameters, and tolerance to different biotic and abiotic stresses. This will allow the breeders to utilize the diverse parental diploid progenitor species to develop a diverse set of genetic stocks of B. juncea which will enrich its primary gene pool. This technique typically involves crossing the ancestral parental species and then inducing polyploidization in the resulting hybrid (Mohd Saad et al., 2021; Quezada-Martinez et al., 2021; Hu et al., 2021), which can be done through various means, such as chemical treatment or by crossing the hybrid with a closely related tetraploid species followed by the screening for desirable traits (Li et al., 2018; Petereit et al., 2022). This technique of upsurging genetic diversity provides the breeders with more opportunities to create cultivars for sustainable production of B. juncea and other allotetraploid species, benefiting the agriculture industry and food security (Parmar et al., 2017; Mohd Saad et al., 2021).
Previously, synthetic lines have been successfully resynthesized in various crop species to improve genetic diversity and enhance desirable traits (Eduardo et al., 2020). For instance, in wheat (Triticum aestivum), the resynthesis of hexaploid wheat was accomplished by crossing tetraploid durum wheat (T. turgidum) with diploid Aegilops tauschii, resulting in novel genotypes with improved yield potential and resistance to biotic and abiotic stresses (Ogbonnaya et al., 2013). All three amphidiploid species were resynthesized and utilized to generate new genetic variability and recover desirable traits in Brassica crops. The resynthesized B. napus and B. carinata lines have been used as a source of genetic diversity in breeding programs to improve the yield and quality of Brassica crops and to enhance their resistance to biotic and abiotic stresses (Ozminkowski and Jourdan, 1994; Zhang et al., 2004; Rahman et al., 2015).
The previous studies by Bansal et al. (2009); Katiyar et al. (1998); Yadav et al. (2009), and Sheng et al. (2012) have also utilized diploid progenitor species of B. juncea to generate variation in morpho-physiological traits. We want to highlight that most of the earlier efforts on re-synthesis of B. juncea have involved B. rapa variety yellow sarson due to high recovery/efficiency (Hinata and Konno, 1979; Bhat and Sarla, 2004). Owing to the poor efficiency and self-incompatible nature of B. rapa var. toria, it was rarely successful in developing synthetic B. juncea (Srinivasachar, 1964; Prakash, 1973). In this study, we have followed a new approach along with the earlier method, wherein B. rapa var. yellow sarson was crossed with B. rapa var. toria and the F1s derived from these crosses (including reciprocal) were mated with B genome donor (B. nigra). This will allow the unique genetic variability available in B. rapa var. toria to tap into resynthesizing B. juncea. This study also addresses the challenges associated with breeding synthetic B. juncea lines, i.e., tissue culture being the most time-consuming and tedious aspect of resynthesis. Here, crosses are attempted using B. rapa var. yellow sarson NRCPB rapa 8 (IC0623820), a novel B. rapa germplasm that bypasses the need to rescue the embryos and hence, tissue culture interventions are not needed (Rao et al., 2024). Furthermore, we noticed that in most of the earlier reports, one or few accessions of B. nigra were involved for this purpose (Prakash, 1973; Bhat and Sarla, 2004; Sheng et al., 2012), and therefore, we utilized the varied B. nigra accessions to generate the synthetic lines. Thus, a highly diverse set of materials and a novel approach were deployed for developing synthetic B. juncea lines. We propose that the observed diversity in the early generation provides valuable insights for mustard breeding programs, especially when aiming to exploit the maximum genetic potential inherited from the parental species. Genetic variability lying dormant in the synthetic lines needs to be characterized and utilized for hybridization with the already evolved B. juncea genotypes before it is lost due to the bottleneck effect of populations, chromosomal rearrangements, recombination, natural selection, etc. Therefore, we characterized the developed synthetic lines in the S2 generation to capture the maximum diversity inherited from the parental species. The main aim of this study was to (i) create, report and harness novel genetic variability for yield contributing traits at early generation, which was lost during domestication/evolution of B. juncea, and (ii) explore a novel approach for involving B. rapa var. toria (B. rapa var. yellow sarson/B. rapa var. toria/B. nigra) in developing synthetic B. juncea.
2 Materials and methods
2.1 Plant materials
Three accessions of B. rapa (var. yellow sarson and toria) and eight B. nigra were used for inter-specific crosses to develop resynthesized B. juncea (RBJ) (scheme in Figure 1), as listed in Table 1. A total of 28 cross combinations (Supplementary Table S1) were attempted using different accessions of B. rapa and B. nigra as both male and female parents. To infuse larger genetic variability in resynthesized lines, a three-way cross approach was used in which B. rapa var. yellow sarson and B. rapa var. toria were hybridized to generate F1 seed for further crossing with B. nigra populations. B. juncea cultivars, namely, Pusa Jaikisan (PJK), Pusa Vijay (P. Vijay), Pusa Mustard 28 (PM 28) and Varuna were used as controls to compare with the synthetic lines developed.
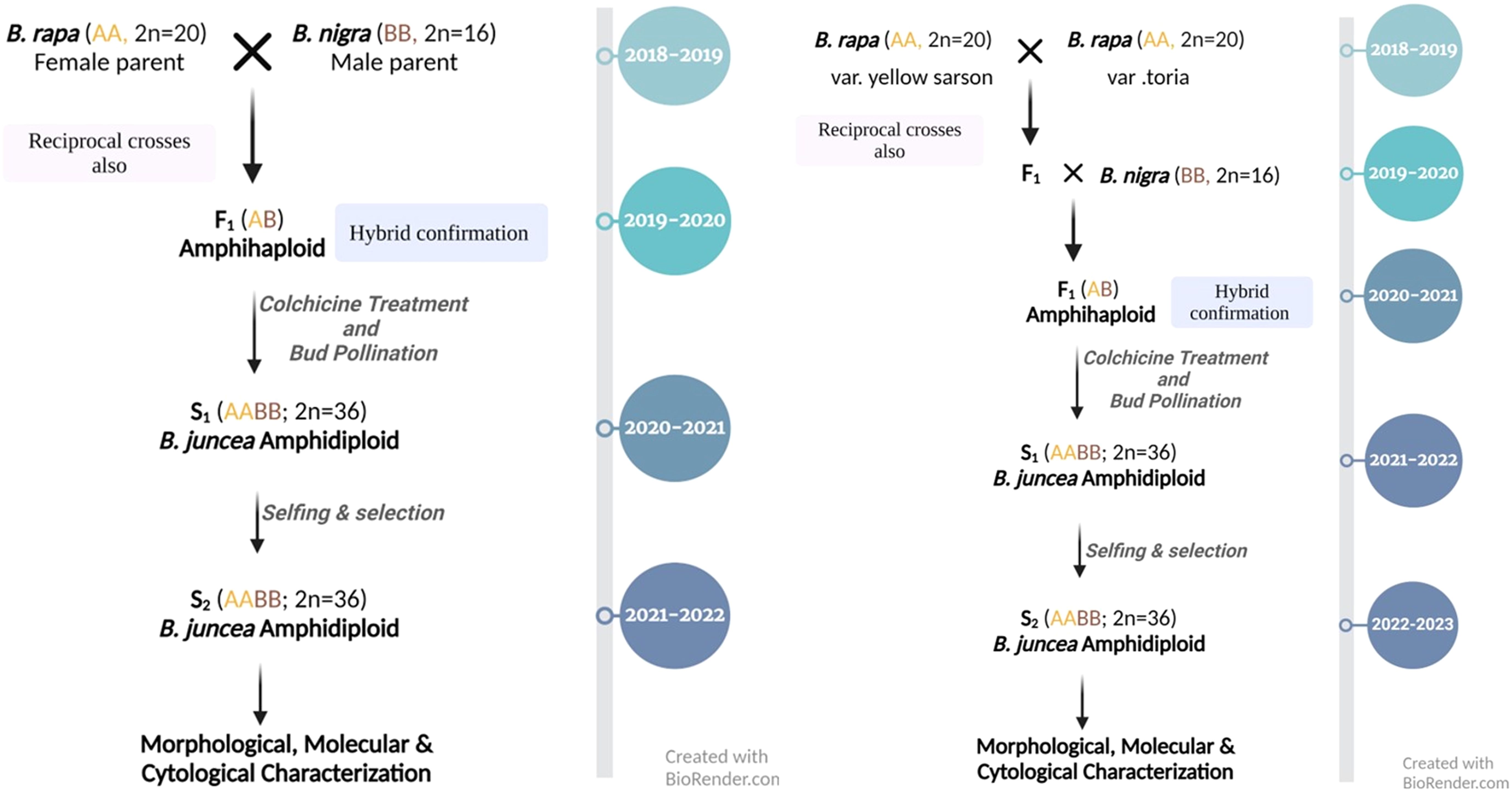
Figure 1. Year-wise scheme opted for synthesis of B. juncea by hybridization of B. rapa with B. nigra (left) and involving B. rapa var. yellow sarson, B. rapa var. toria and B. nigra in three-way crosses (right).
2.2 Emasculation and pollination
The mature, unopened buds were chosen for emasculation to avoid any self or foreign pollen contamination. All the anthers, sepals, and petals were removed collectively from the selected flower buds using forceps, adjoining younger buds were snipped off, and the inflorescence was bagged. The male parent’s inflorescence was bagged to avoid any pollen contamination (Seyis et al., 2005). The following day, collected pollens from the male parent were used to pollinate the female parent’s emasculated buds.
2.3 Chromosome doubling by colchicine treatment
To develop amphidiploids from amphihaploid inter-specific hybrids, non-absorbent cotton balls soaked in 0.2% colchicine were applied to the hybrid plants’ axillary buds and apical meristem on alternate days for 5-7 days (Rajcan et al., 2011; Manzoor et al., 2019).
2.4 Pollen viability test
The viability of pollens in the parental and interspecific hybrid plants was examined under a light microscope (Carl Zeiss Axiolab 5, Germany) using the protocol by Katche et al. (2021). The mature flower buds were collected in the ice container, and with the aid of forceps and a needle, the anther was removed and crushed in 1% acetocarmine. Three buds from each plant were inspected to determine the pollen viability, which was analyzed and expressed in terms of percentage. Round, plump, and stained pollens were regarded as viable, whereas shriveled or unstained pollens were sterile.
2.5 Cytogenetic analysis
The procedure of Snowdon et al. (1997) was followed to count the total number of diploid (2n) chromosomes in mitotic cells from root tissue by applying DAPI (4,6-diamidino-2-phenylindole) as fluorescent dye under the microscope. The root tips were harvested early in the morning in 0.002M 8-hydroxyquinoline solution and then fixed in Carnoy’s solution (3:1 ethanol: acetic acid solution), followed by transfer into 70% ethanol. For slide preparation, enzyme solution was added to root tips on a slide and incubated at 37°C for 45 minutes, followed by the addition of 10µL 45% acetic acid after removing the enzyme solution (Rao et al., 2024). The root tip was solubilized with a needle to release the cells, and then a coverslip was placed on top. The slide was fixed in liquid N2, and the coverslip was removed with the help of a scalpel blade and then allowed to air dry. After staining with 10µL DAPI, the chromosomes were seen under the fluorescent microscope (Carl Zeiss Axiolab 5, Germany).
2.6 Evaluation of morphological traits
Phenotypic data was collected from three selected plants in S2 generation of each line across the three replication plots for plant height (PH), main shoot length (MSL), silique length (SL), number of siliques on main shoot (SMS), number of primary branches (PB), seeds per silique (SS), oil content (OC), yield per plant (YP), and thousand seed weight (TSW). Oil content was estimated using the Near-Infrared Spectroscopy (NIRS) by Newport NMR analyzer (Model-4000) (Shruti et al., 2023). The morphological diversity was assessed for thirty-three synthetic B. juncea lines, four B. juncea cultivars, and diploid parental species, i.e., B. rapa (3 accessions) and B. nigra (8 accessions).
2.7 Confirmation of hybridity and molecular diversity analysis
A set of 94 SSR markers (Supplementary Table S3) pertaining to AA (B. rapa), BB (B. nigra) and AABB (B. juncea) genome were selected for the hybridity confirmation (Supplementary Table 3) in F1 generation along with genetic diversity analysis in S2 generation (Sudan et al., 2016). These polymorphic markers are available in the Brassicaceae database (BRAD) (http://Brassicadb.cn) (Chen et al., 2022) and are also reported by Lowe et al. (2004); Kim et al. (2009); Dhaka et al. (2017).
The total genomic DNA was isolated following DNA extraction protocol from fresh leaves (Doyle and Doyle, 1990). In a reaction volume of 20 µl, the PCR mixture contained 1 µl of template DNA (25 ng/µl), 1 µl of each forward and reverse primer (100 pmol/µl), 1µl of 10 mM dNTPs, 1.5 µl of 25mM MgCl2, 4 µl of 10x PCR buffer, and 0.5µl of 0.5U Taq polymerase and 10 µl of nuclease-free water. The PCR cycle was designed with an initial denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 54°C for 1 min and 15 sec, and extension at 72°C for 1 min 30 sec, before a final extension at 72°C for 10 min. The PCR products were processed in 1x TAE buffer and separated on a 2.5% agarose gel along with the 50 bp DNA ladder as a benchmark on both sides of the gel.
2.8 Data analysis
The statistical analysis was done using the metan package in R program v4.2.0 (Olivoto and Lúcio, 2020). Due to the presence of multi-collinear factors, the data was subjected to principal component analysis (PCA) based clustering. While conducting a cluster analysis with pair group distance and Euclidean similarity metrics, the factors corresponding to significant PCs were chosen. Using the DARwin software v6.0.021, the neighbor-joining tree was created (Perrier and Jacquemoud-Collet, 2006). The population structure was assessed using the Bayesian clustering model-based software STRUCTURE v2.3.4 (Pritchard et al., 2000; Falush et al., 2003). Five iterations were performed for each cluster, K = 2 to 8, with the length of the burn-in period and Markov Chain Monte Carlo (MCMC) replications set to 50,000 each. The most probable K value was determined using a web-based software StructureSelector (Li and Liu, 2018), which uses combined measures and estimators to select the best K-value (Evanno et al., 2005; Raj et al., 2014; Puechmaille, 2016) and integrates CLUMPAK program for graphical representation (Kopelman et al., 2015).
3 Results
3.1 B. rapa as the female parent yielded successful cross combinations with intermediate parental phenotypes in the progenies
Twenty-eight crosses attempted in this study yielded thirty-three lines of RBJ in twelve cross combinations, as given in Table 2. The crosses were successful for combinations having B. rapa as the female parent, and hence, no seeds were obtained for reciprocal crosses (B. nigra ×B. rapa). When compared with parents and control (Figures 2A–C), morphological variations for leaf architecture and size were observed in amphihaploid F1 plants (Figures 2D–F) and S1 generation (Figures 2G, H). In Figures 3A–K, a clear difference was observed for leaf tenderness in amphihaploid plants (Figures 3D–G) and robust and firm leaves in S1 plants (Figures 3H–K), which resembles the natural B. juncea cultivars (Figure 3C). Furthermore, the morphology of mature plants in S2 generation (Supplementary Figure S1) clearly indicates that the resynthesized plants differ morphologically from both parental lines. The plants in the S2 generation exhibited significant variability in terms of plant height, number of primary branches, seeds per silique, etc., and were found to be more similar to the B. juncea cultivar. This indicates that the process of resynthesis might have led to some novel genetic combinations.
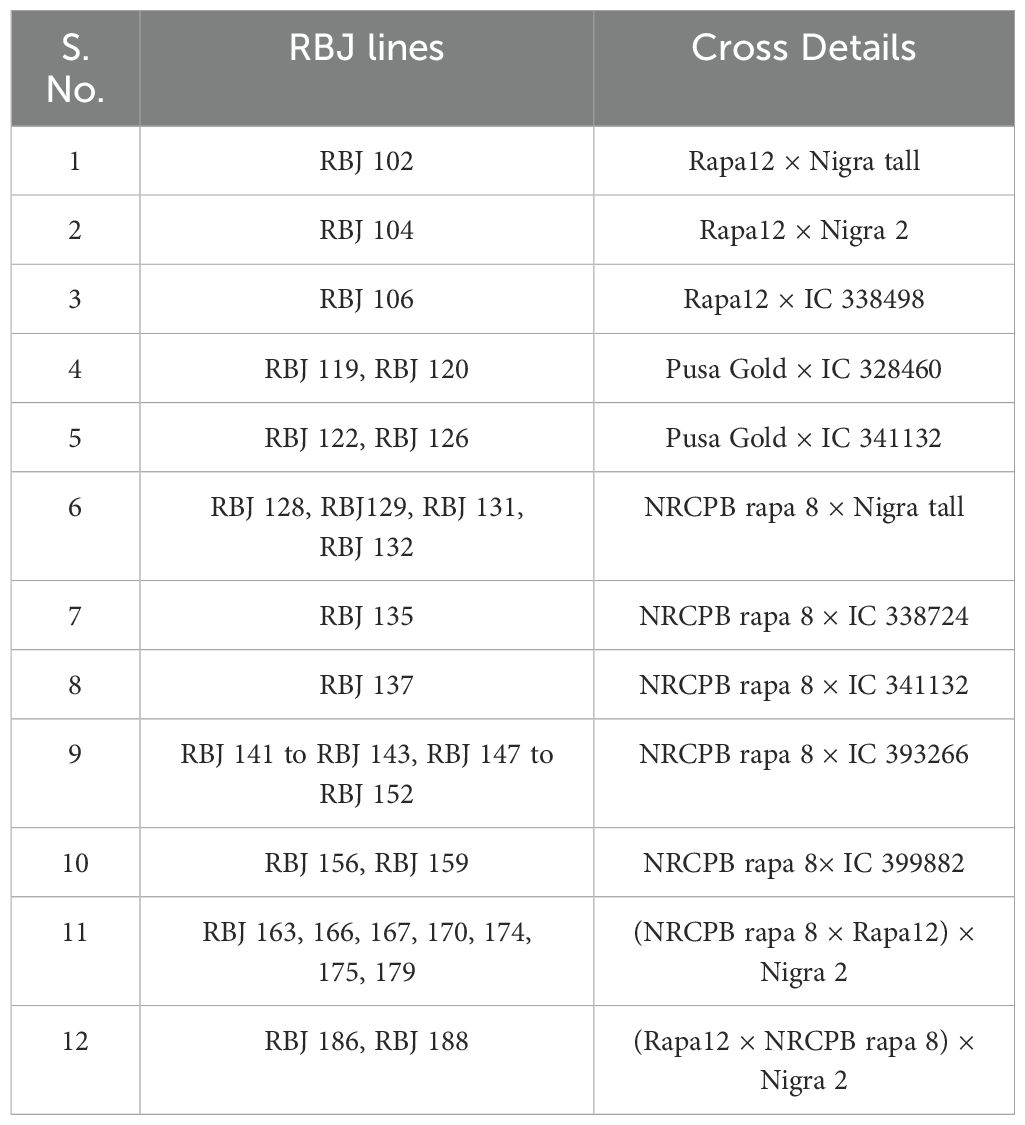
Table 2. List of resynthesized B. juncea (RBJ) lines developed using different accessions of parental diploid species.
Figure 2. Morphology of plants under controlled conditions (A) NRCPB rapa 8 (female parent), (B) B. nigra Dwarf (male parent), (C) Pusa Jaikisan (cultivar), (D–F), F1 and (G, H) S1 generations of synthesized B. juncea lines (RBJ 106, RBJ 135).

Figure 3. Leaf morphology of (A) NRCPB rapa 8 (female parent), (B) B. nigra Dwarf (male parent), (C) Pusa Jaikisan (cultivar), (D–G) F1 and (H–K) S1 generations of synthesized B. juncea lines (RBJ104, RBJ 119, RBJ 126, RBJ 147).
3.2 F1 true hybrids were fertile after chromosome doubling
The validation of hybridity in the developed amphihaploid was conducted utilizing a comprehensive set of simple sequence repeat (SSR) primers specific to the genomes involved. A total of 14 SSR primers targeting the A-genome, 18 targeting the B-genome, and 29 targeting the AB-genome were employed. The parental polymorphism was done, and the polymorphic primers were used for the hybridity confirmation. The analysis of amplification patterns obtained from these primers confirmed the hybrid nature of the developed amphihaploid. The presence of characteristic bands corresponding to the A, B, and AB genomes further substantiated the successful hybridization process (Supplementary Figure S2). The list of SSR primers used and hybridity assessed for each resynthesized line is given in Supplementary Tables S2, S3. Figures 4A–C show pollen viability of both diploid parents and the control. Pollen sterility in F1 hybrids was evident (nearing 100%) at the amphihaploid stage (Figures 4D–F), and fertility was reinstated in the S1 generation (Figures 4G–I) due to chromosome doubling with over 50% pollen stainability. Upon reaching the S2 generation, the synthesized lines displayed a discernible range of pollen fertility, spanning from 53% to nearly 100%, thereby culminating in an average of approximately 85% (Figures 4J–M).
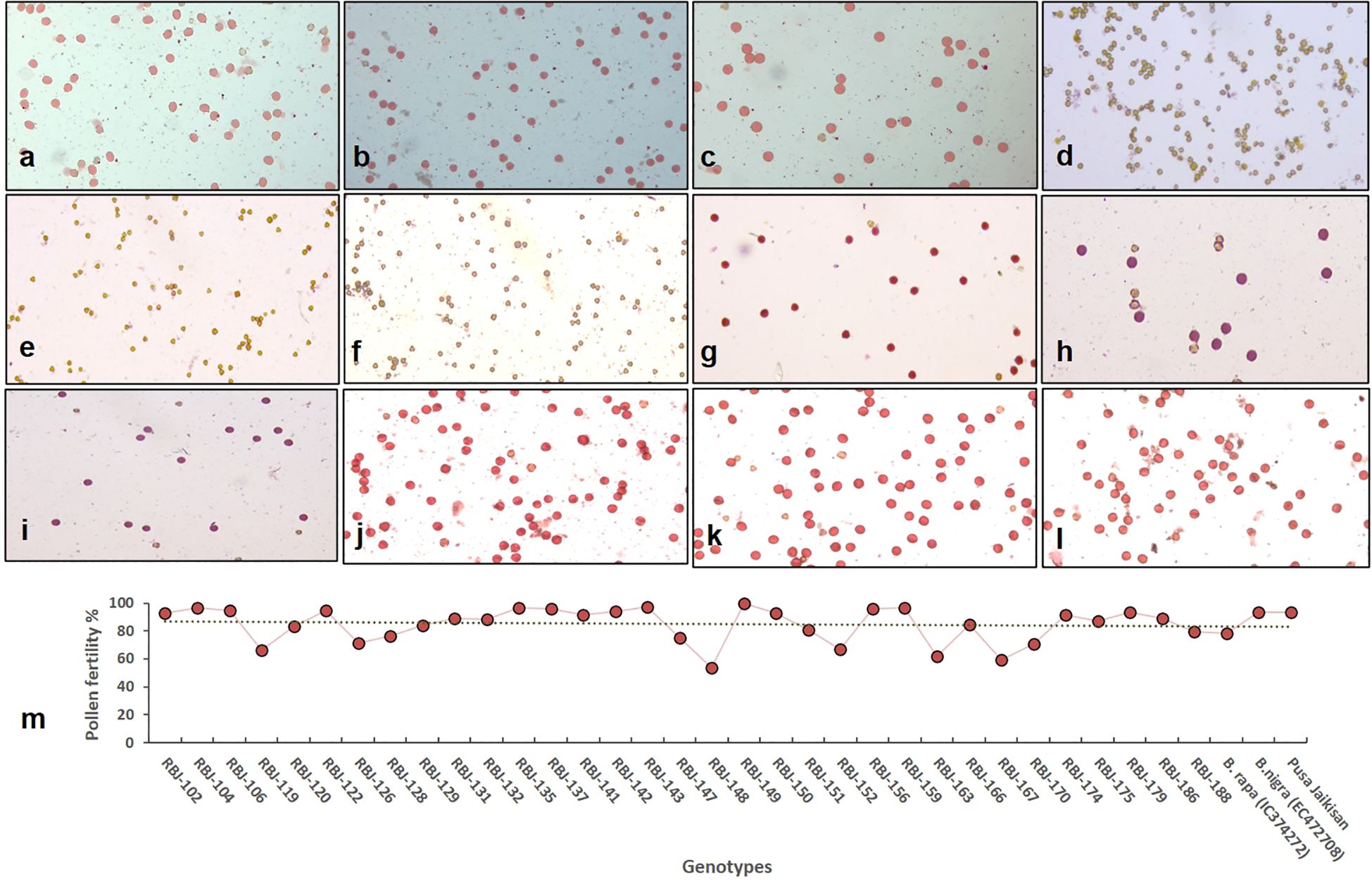
Figure 4. Pollen viability in (A) NRCPB rapa 8 (female parent), (B) EC472708 (B. nigra; male parent) and (C) Pusa Jaikisan (cultivar); (D–F) sterility in F1 generation and (G–I) fertility in S1 and (J–L) S2 generation of RBJ 159, RBJ 174, RBJ 179, respectively. (M) Graph representing the fertility of synthetic B. juncea lines in S2 generation along with parental diploid species viz., B. rapa (IC0623820) and B. nigra (EC472708), and B. juncea (PJK) cultivar.
The confirmed 33 true hybrids were assayed for chromosome number via mitotic configurations at the F1 stage, revealing 18 chromosomes under the microscope (Figure 5D), and the mitotic analysis of diploid parents, B. rapa and B. nigra, exhibited 20 and 16 chromosomes, respectively (Figures 5A, B). Thirty-six chromosomes were clearly visible in synthetic B. juncea lines (S2 generation), which was similar to the B. juncea cultivar Pusa Jaikisan (Figures 5C, E, F). The conducted cytogenetic studies established the successful development of synthetic B. juncea lines.
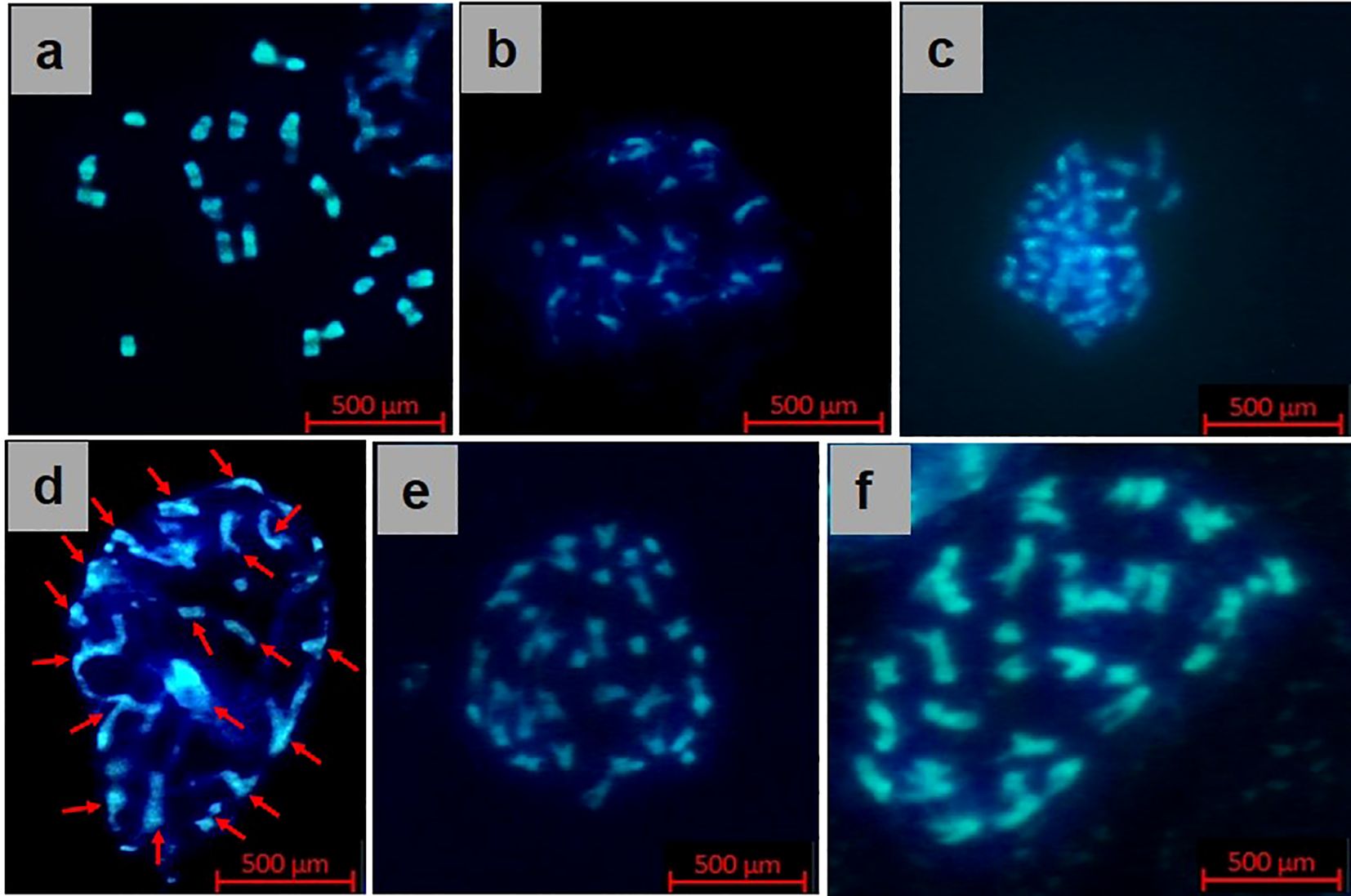
Figure 5. Cytogenetics (mitosis) of (A) B. rapa (female parent), (B) B. nigra (male parent), (C) Pusa Jaikisan (cultivar), (D) RBJ 122 (F1 generation), (E) RBJ 122 (S2 generation), and (F) RBJ 132 (S2 generation) of synthesized B. juncea lines.
3.3 Significant correlation observed between different morphological traits
The genetic variation among the resynthesized genotypes with diploid parents and Indian mustard cultivars was evaluated through Analysis of Variance (ANOVA) for various phenotypic traits (Table 3). The results highlight significant contributions from various factors in the study. The genotypes, traits and their interactions displayed significant variations of 2.16%, 89.61% and 7.90% respectively at P > 0.0001.
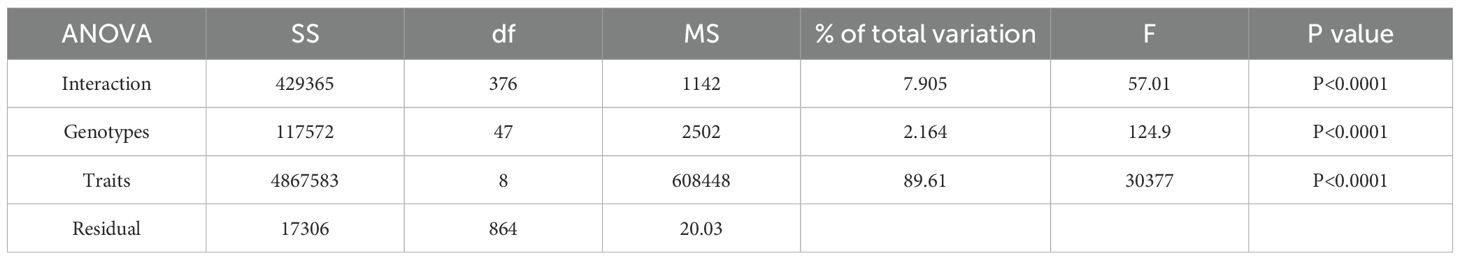
Table 3. Analysis of variance showing mean square values and level of significance for agro-morphological traits of RBJ lines.
The correlation between different agro-morphological traits was analyzed using Pearson’s correlation coefficient, and the results are presented in Figure 6A. Several significant correlations were observed among the traits. Firstly, YP exhibited positive correlations with PH (r = 0.52, p < 0.001), MSL (r = 0.51, p < 0.001), and TSW (r = 0.57, p < 0.001), indicating potential associations between yield-related parameters. TSW also demonstrated strong positive correlations with PH (r = 0.46, p < 0.01), MSL (r = 0.74, p < 0.001), SL (r = 0.72, p < 0.001), SS (r =0.38, p < 0.01) and oil content (r = 0.58, p < 0.001) suggesting their potential influence on seed weight. This suggests that taller plants with longer main shoots and siliqua length tend to have higher yields. Additionally, oil content showed a positive association with SL (r = 0.60, p < 0.001) and SS (r = 0.56, p < 0.001), indicating their potential contribution to oil accumulation. This indicates that longer main shoots and siliqua length are associated with higher oil content. PH and MSL are positively correlated with r = 0.53 at p < 0.001. The weak negative correlations observed between oil content and PH (r = -0.04, p = ns) and SMS (r = -0.18, p = ns) are found to be non-significant in our study. This may imply that either there is no correlation between these two variables or this may be a random association. The positive correlation between oil content and YP (r = 0.30, p < 0.05) suggests that higher seed yield may be associated with increased oil accumulation. Oil content and YP may not be directly associated with each other but may be dependent on a complex agronomic trait, seed size or TSW which is positively correlated with both YP (r= 0.57, p< 0.001) and oil content (r= 0.58, p< 0.001). Oil content is an economically important trait and, lines with a higher oil content can thus be selected by opting for lines with higher TSW or YP. These findings provide insights into the interdependence of these traits and can guide future breeding and selection strategies to enhance specific desirable traits in Indian mustard cultivars.
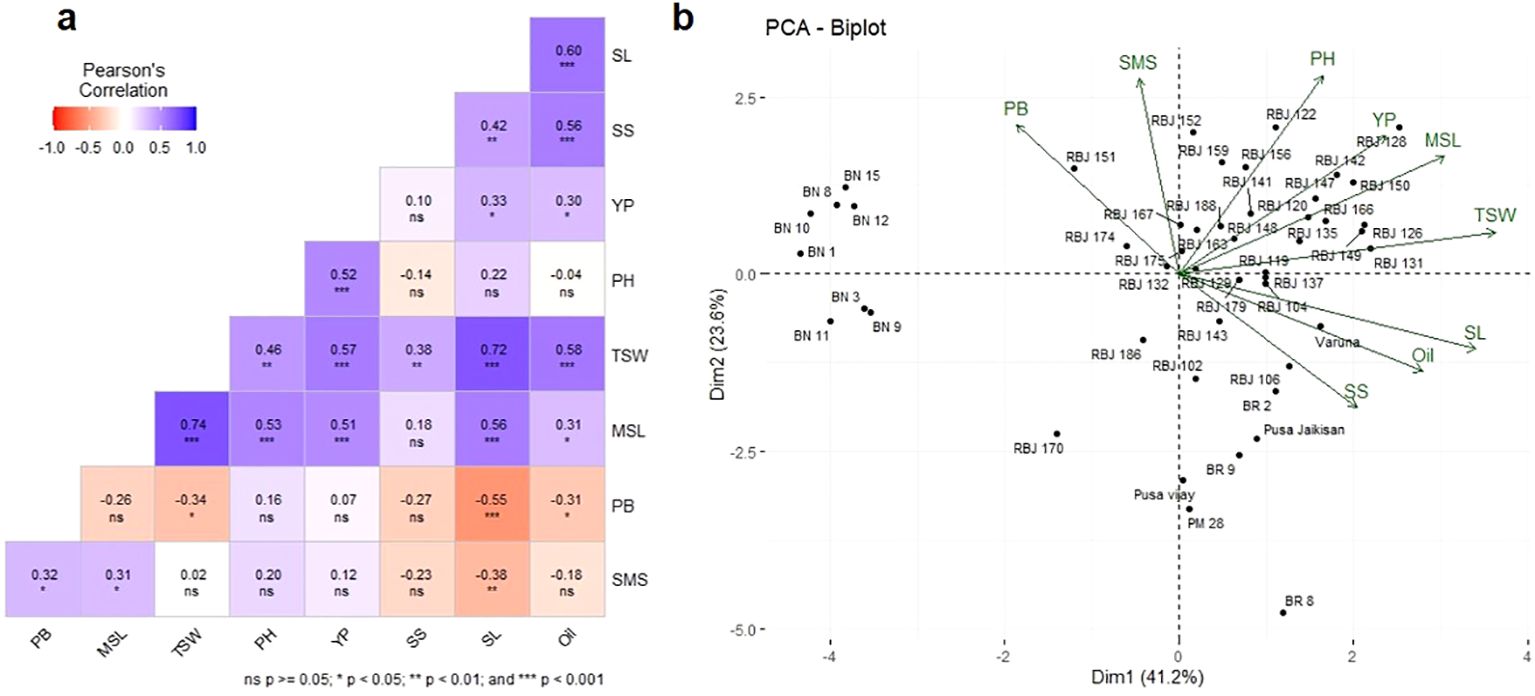
Figure 6. (A) Phenotypic correlation coefficients for evaluated morphological traits and oil content of RBJ lines. PH, Plant Height; MSL, Main Shoot Length; SL, Silique Length; SMS, Number of Siliques on Main Shoot; PB, Number of Primary Branches; SS, Seeds Per Silique; OC, Oil Content; and YP, Yield Per Plant; TSW, Thousand Seed Weight, (B) The biplot illustrating Principal Coordinate analysis of different morphological traits.
Principal component analysis (PCA) (Figure 6B; Supplementary Table S4) was computed to show each trait’s contribution to the overall morphological variations. Categorized under various clusters, along with their associated morphological traits, the PCs represent axes of variation that capture the morphological diversity within the genotypes. The importance of each PC is measured by its standard deviation, percentage of variance, and cumulative proportion of variance. These statistics indicate how much each PC contributes to the overall morphological diversity captured by the entire set of PCs. The cumulative proportion of variance demonstrates that the first four PCs contribute more than 80% to the total variance, with PC1 alone accounting for about 41.2% of the total variance. It highlights the significance of specific PCs in explaining variations in morphological traits and underscores the potential implications for crop enhancement and breeding strategies.
3.4 Molecular and morphological diversity reflected uniqueness in the resynthesized lines of B. juncea
The cluster analysis (Figure 7A) based on SSR markers showed that the RBJ lines were effectively distributed across all three discrete clusters. Within cluster IIb, a subset of resynthesized lines (specifically, RBJ 175, RBJ 179, RBJ 186, and RBJ 188) demonstrated close affinity with genotypes of parent B. rapa. This observation underscores a genomic-level congruence among these resynthesized lines and the B. rapa genotypes. Notably, the RBJ lines underwent subsequent sub-clustering, revealing the emergence of genomic-level diversity as a result of the resynthesis process. Moreover, the composition of cluster IIa included B. juncea cultivars exhibiting a more proximate phylogenetic alignment with B. nigra accessions. This proximity is likely attributed to historical processes of natural or artificial selection over extended timeframes.
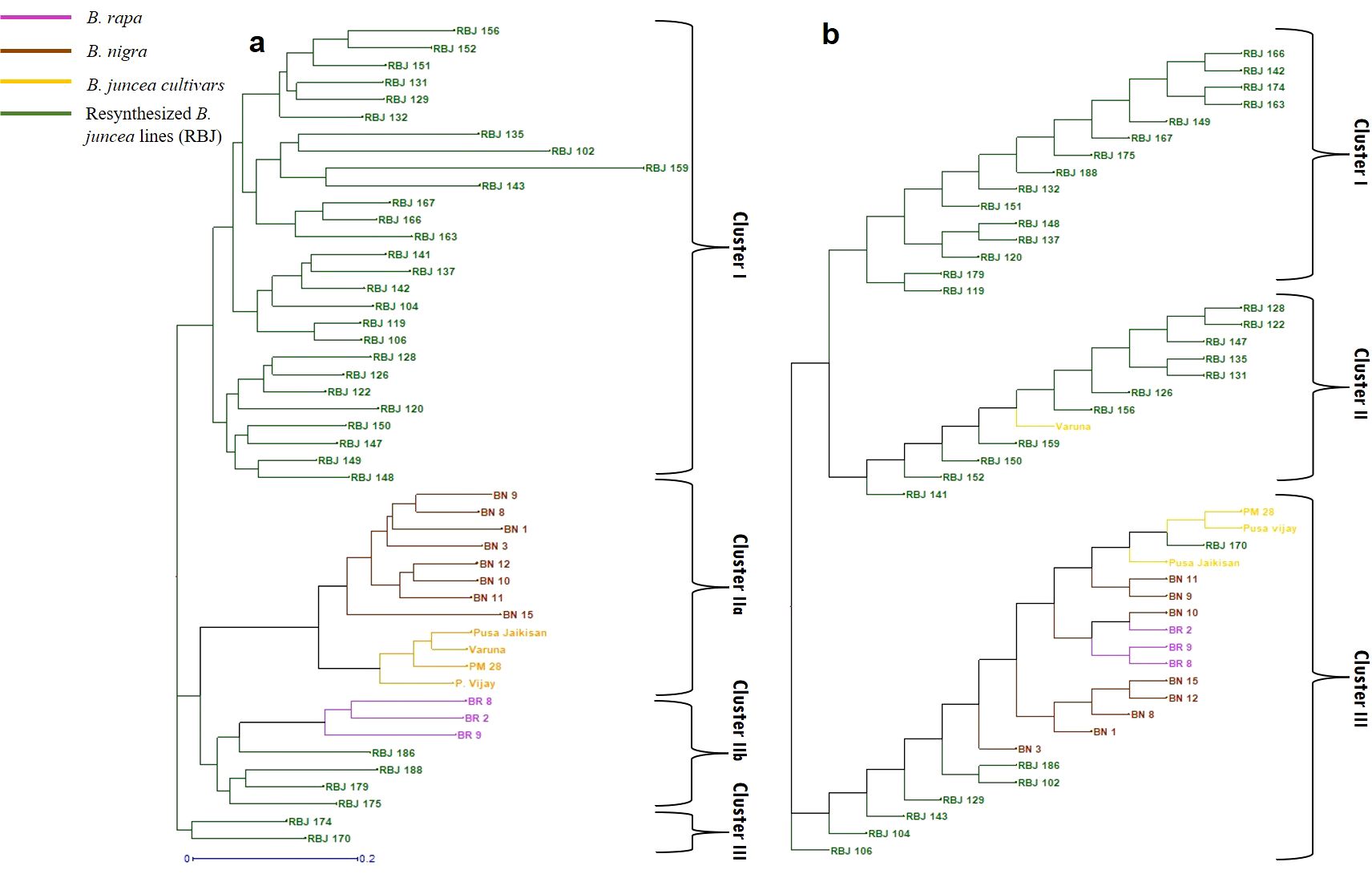
Figure 7. Diversity analysis based on (A) polymorphism generated by SSR markers and (B) morphological parameters. Abbreviations used: BR 2- Rapa 12, BR 8- NRCPB rapa 8, BR 9- Pusa gold, BN 1- Nigra tall, BN 3- EC426390, BN 8- IC 338498, BN 9- IC 338724, BN 10- IC 341132, BN 11- IC 393266, BN 12- IC 399882, BN 15- IC 328460.
The outcomes of the morphological cluster analysis also unveiled the presence of three prominent clusters (Figure 7B). The clusters I and II encompassed a substantial proportion of the synthetic B. juncea lines, along with the cultivar Varuna. The third cluster comprised B. nigra and B. rapa genotypes. In this cluster, a notable proximity was observed between the morphology of RBJ 170 and the cultivars Pusa Jaikisan, Pusa Vijay, and PM 28. Additionally, this cluster highlighted the close morphological proximity of RBJ 102 and RBJ 186 to the Nigra 2 genotype.
3.5 Population structure validates the results of genetic diversity analysis
The population structure of the Brassica genotypes was analyzed using STRUCTURE software. The optimal K value was determined by plotting the values of ΔK against the number of clusters (Figure 8A; Supplementary Table S5) that show the highest ΔK value at K=3. This indicates that the genotypes used in the study are divided into three subpopulations (pop 1, pop 2 and pop 3). The estimated Ln probability of data was -18391.7 (Figure 8B) with mean ln likelihood, variance of ln likelihood, and mean alpha value at -18118.9, 545.6, and 0.0381, respectively. Table 4 summarizes the overall proportion of membership (inferred clusters), mean fixation index (Fst), divergence among the two subpopulations, and number of genotypes in each population. The genotypes were assigned to either of the populations based on the Q values from both clusters. The inferred ancestry of all individuals can be seen in Figure 8C; Supplementary Table S5.
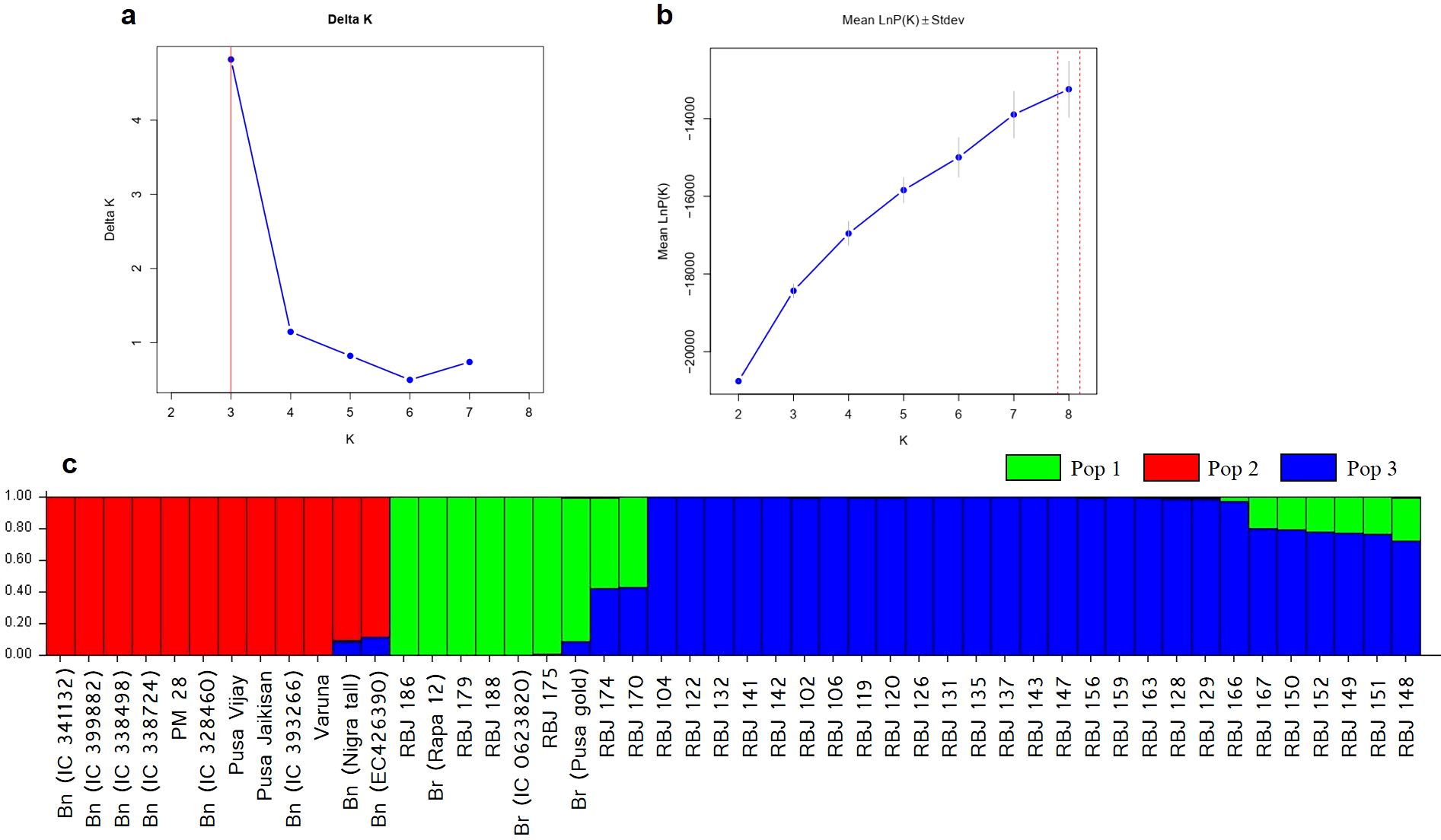
Figure 8. Population structure analysis (A) Delta K (ΔK) plot for varying K populations, (B) LnP D plot, and (C) Estimated population structure of Brassica genotypes used in the study at K=2. Br and Bn denote different accessions of B. rapa and B. nigra, i.e., parental lines.
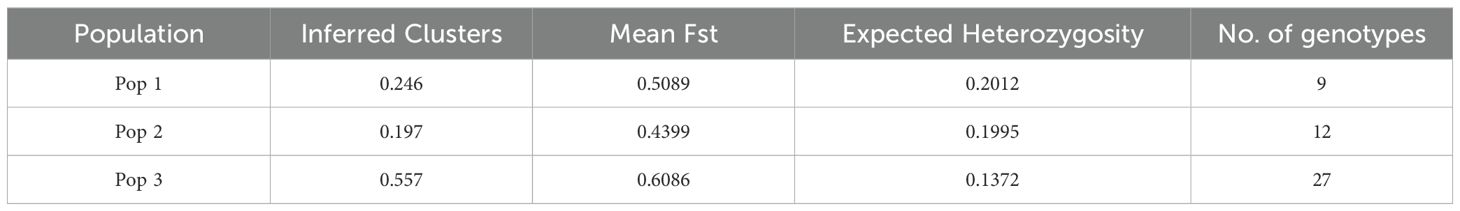
Table 4. The STRUCTURE results of Brassica accessions for Fst, expected heterozygosity and no. of genotypes in each population (major).
3.6 High-yielding lines were observed in resynthesized lines
A clear overview of variations observed in the developed RBJ lines is presented in Figure 9. The synthetic lines are seen to perform better than the four B. juncea cultivars in terms of plant height, main shoot length, no. of siliqua on main shoot, no. of seeds per siliqua, thousand seed weight and yield per plant. RBJ 163 and RBJ 142 recorded the highest plant height of 290 cm and 284 cm, while PM 28 was the shortest at 159.7 cm. RBJ 122 has the longest main shoot length (87.7 cm) with a higher number of siliquae on the main shoot, i.e., 81.7. RBJ 106 (18.7) and RBJ 135 (18) have more seeds per siliqua followed by RBJ 149, RBJ 102 and Varuna having the same mean of 17.7. RBJ 128 observed the highest thousand seed weight (4.8) as well as yield per plant (75.1 g), while RBJ 170 has the lowest values for both traits (1.40 and 7.3 g, respectively). Yield and TSW for B. juncea cultivars are as follows: PJK (20.7 g, 3.3 g), P.vijay (11.5 g, 2.8 g), PM 28 (10.3 g, 2.5 g) and Varuna (67.3 g, 2.9 g).
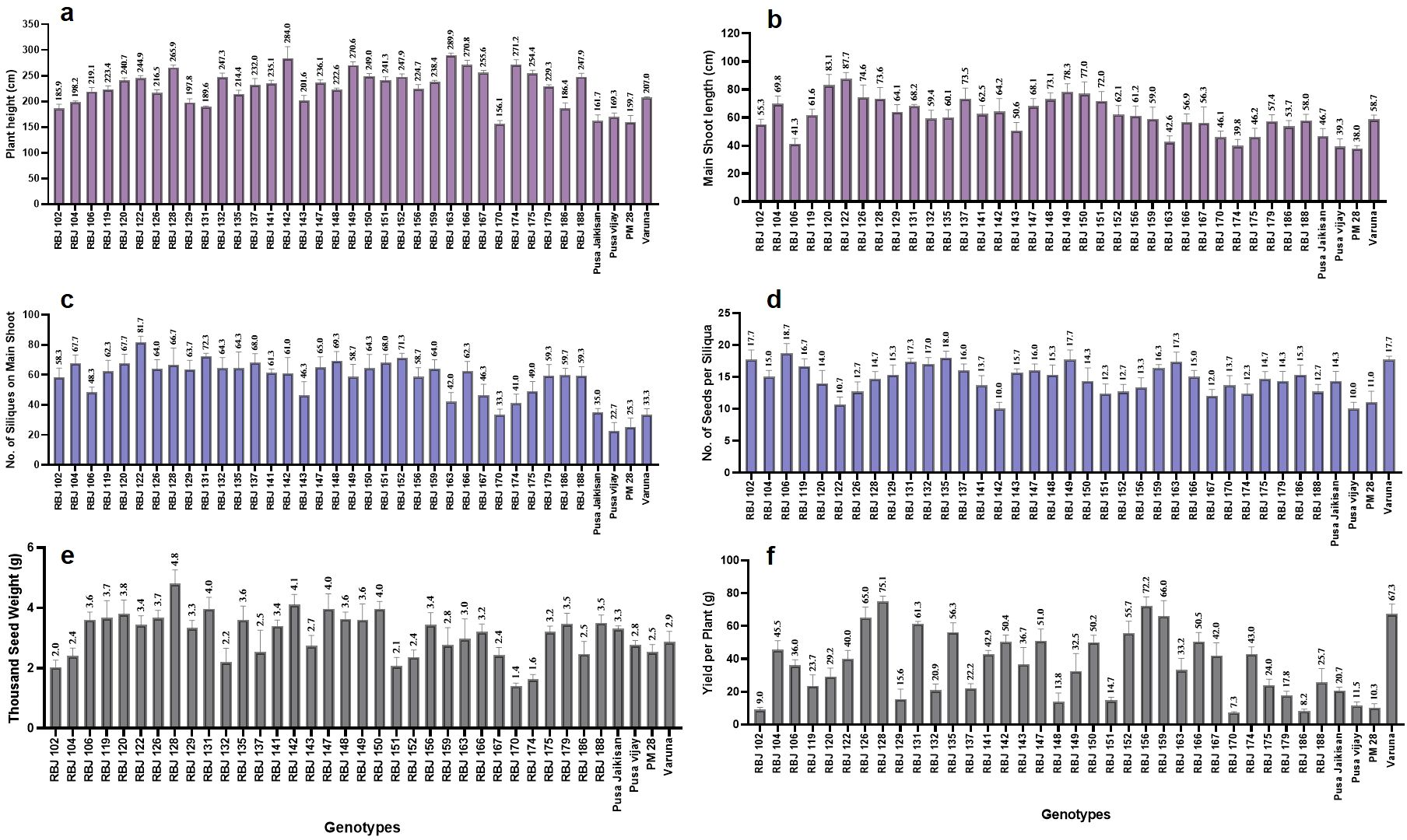
Figure 9. The performance of synthetic B. juncea lines (RBJ) in comparison to B. juncea cultivars for morpho-physiological traits viz., (A) plant height, (B) main shoot length, (C) no. of siliques on main shoot, (D) no. of seeds per siliqua, (E) thousand seed weight and (F) yield per plant under field conditions. Error bars: +/- 2 SE.
4 Discussion
Due to their limited genetic diversity and narrow gene pool, oilseed Brassica, especially B. juncea, are extremely susceptible and non-resilient to environmental influences. Resynthesizing B. juncea from the existing diploid progenitor species can enhance genetic diversity, even if initial yields are lower (Gupta et al., 2015; Seyis et al., 2003; Srivastava et al., 2004). Our objective encompassed the reconstitution of B. juncea with the intent of enhancing genetic diversity, thereby facilitating the comprehensive evaluation of these variants across a spectrum of morphogenetic parameters. Also, the early generation characterization will help breeders to utilize the traits of interest in a time-efficient manner. As per the available data, this is the first successful attempt where the self-incompatible B. rapa var. toria was involved in the synthesis of B. juncea by deploying B. rapa var. yellow sarson as a bridge. It has been reported that the seed set is very low when toria is used as a female parent (Srinivasachar, 1964; Prakash, 1973). The resynthesized lines were validated through the meticulous study of mitotic events and the confirmation of hybridity using molecular markers within the F1 generation. Earlier researchers also reported such attempts for artificial synthesis of B. juncea (Bansal et al., 2009; Bhat and Sarla, 2004; Srivastava et al., 2004; Yadav et al., 2009; Hasan and Rahman, 2018) and B. napus (Zhang et al., 2004; Hilgert-Delgado et al., 2015; Chatterjee et al., 2016) using their progenitor species. Interestingly, crosses attempted using B. nigra as a female parent were unsuccessful and did not bear any seeds. Prior investigations conducted by Bhat and Sarla (2004) concluded that if B. nigra is used as a female parent in the synthesis of B. juncea, it is necessary to use tissue culture interventions to overcome post-fertilization barriers.
We observed an intermediate phenotype of the newly synthesized B. juncea lines. Consistent with our observations, prior work has demonstrated the development of interspecific hybrids between B. juncea and autotetraploid B. fruticulosa, resulting in intermediate phenotypes (Song et al., 2018). They reported that the size and shape of the leaves of F1 hybrids were closer to B. juncea, the female parent. Recently, fertile allohexaploid Brassica hybrids were developed from the crosses between B. oleracea and B. juncea (Mwathi et al., 2020). Similar to their parents, the flowers in the hybrid plants were yellow; however, the leaf morphology was intermediate between the two parents. There are allotetraploid species like B. juncea (Olsson, 1960; Prakash et al., 1984; Bansal et al., 2009; 2012), B. carinata (Prakash et al., 1984), and B. napus (Seyis et al., 2003) in which the increased heterozygosity attained after intergenomic crossing-over show intermediate effect on phenotypes in both resynthesized and natural types. Cytological analysis of F1 hybrids (n=18) confirmed hybridity and parental homeology, aligned with the results reported by Kumar et al. (2018). In contrast, our study validated amphihaploid hybridity via SSR primers for A, B, and AB genomes. Mitotic analysis in S1 confirmed 36 chromosomes, and cytogenetics affirmed synthetic B. juncea (2n=36), similar to Pusa Jaikisan.
Genomic studies on Brassica crops show that the current Brassica diploids originally came from ancient polyploids. These polyploids underwent a natural diploidization process to become functional diploids. This diploidization process involves genetic exchanges, genome restructuring, the development of new functions, modular organization, and gene silencing within a shared nucleus. This concept was first highlighted by Warwick and Black (1991), further discussed by Lagercrantz (1998), and explored by Lysak et al. (2005). Additionally, the technique of derived amphiploidy, as introduced by Banga and Kaur (2009), is rooted in the natural notion of cyclic polyploidy in which a genome that has undergone diploidization may recurrently participate in multiple rounds of genome merging, duplication, and diploidization. Wang et al. (2021) also discussed about this concept in his review and how genome downsizing occurs and is selectively favored. The concept of whole genome duplication and post-polyploidy genome divergence is discussed by many researchers (Leitch and Bennett, 2004; Wendel, 2015; Zenil-Ferguson et al., 2016; Pellicer et al., 2018). This mechanism contributes to the intricate evolutionary processes observed in Brassica species. This can be observed in the SSR-based clustering analysis, where the resynthesized genotypes were grouped into a separate cluster, whereby we found that RBJ lines were more closely related to B. rapa. One might argue this to be the case when using B. rapa as a female parent. However, it is to be emphasized that the cytoplasm does not play a significant role in genetic clustering, and the resynthesized types with B. nigra cytoplasm may also show closer proximity with B. rapa in terms of genetic relatedness (Bansal et al., 2009). The resynthesized lines having one or both common diploid parents may not always fall in the same group. However, most of the resynthesized lines generated through three-way crosses involving a hybrid of B. rapa var. toria, B. rapa var. yellow sarson, and B. nigra (RBJ 170, RBJ 174, RBJ 175, RBJ 179, RBJ 186, RBJ 188) were found to be in the same cluster and were distantly placed from other resynthesized lines, thus indicating the contribution of B. rapa var. toria in generating this genetic diversity. The significance of a three-way cross in creating a gene pool with high genetic variance is well demonstrated in this study. The PCA and clustering analysis for agronomic traits also revealed the successful synthesis of B. juncea lines, which were highly diverse but similar to natural cultivars of B. juncea. Out of 33 RBJ lines, 11 lines were morphologically more similar to the B. juncea cultivar Varuna. Surprisingly, RBJ 170 was most closely related to PJK, Pusa Vijay and PM 28 in terms of agronomic traits.
Pandey et al. (2021) reported that the differing alleles among clusters can help in detecting the principal differences and thus can lead to the use of these genotypes for breeding successfully. According to Meirmans (2015), there is a chance of uncertainty in inferring K and hence, correlating these results with PCA and the phylogenetic tree obtained by DARwin is extremely helpful. The population genetics results determined by STRUCTURE at K=3 (Figure 8C) and DARwin (Figure 7A) perfectly align forming three clusters. Here also, based on the Q values, resynthesized lines obtained by three-way crosses are closer to the B. rapa accessions, as in Figure 7A. The studies conducted by Giri et al. (2017); Zhao et al. (2018); Zhou et al. (2018), and Luo et al. (2019) also reported similar observations.
The prior studies done by Scannell et al. (2007) suggests that the age/generation of the polyploids also play an important role in genome size, DNA loss and recombination frequencies as reviewed by Wang et al. (2021). Due to higher recombination frequency in early-generation polyploids (compared to stable and advanced polyploids) (Yant et al., 2013; Lloyd and Bomblies, 2016), these polyploids have a higher chromosomal and genetic diversity. Indeed, it has been hypothesized that a positive feedback loop exists, whereby homeologous recombination in young allopolyploids causes depletion in DNA mismatch repair proteins, which enhances aberrant recombination and DNA loss, leading to even more homeologous recombination in future generations (Comai, 2000). Also, it has been seen that DNA loss is more in early-stage polyploid as reported in Phlox drummondii, in which there was a decrease in genome size by one-fourth (Raina et al., 1994). This might offer a selective advantage. Selection from this diversity could favor variants with smaller GS (Wang et al., 2021).
5 Conclusion
We report the successful synthesis of allotetraploid B. juncea lines using the two diploid progenitor species. For the development of the synthetic amphidiploid B. juncea lines, B. rapa and B. nigra were crossed to make synthetic amphihaploids (AB, chromosome number 18), followed by the chromosome doubling and further selfing in the subsequent generations. Due to continuous selfing and selection in the subsequent generations, the chromosomal rearrangement and inter-genomic interactions for the stability and homozygosity will prevail leading to draining of the genetic variability in the synthetic B. juncea lines over the generations. Thus, characterizing and utilizing these new B. juncea lines is a promising strategy for harnessing maximum parental genomic diversity to improve Indian mustard. Involving early-stage synthetic lines in the breeding program can save time as the generations used for the advancement and stability of these lines can be utilized in the varietal development. Assessing the potential of these lines/traits for integration into B. juncea enhancement initiatives, particularly targeting seed and oil yield, shall open new crop breeding opportunities.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
PG: Writing – original draft, Conceptualization, Methodology, Writing – review & editing. ST: Conceptualization, Investigation, Writing – original draft, Writing – review & editing. AK: Data curation, Writing – review & editing. AA: Data curation, Formal analysis, Writing – review & editing. SK: Investigation, Methodology, Writing – review & editing. MS: Investigation, Methodology, Writing – review & editing. RK: Methodology, Writing – review & editing. SS: Data curation, Investigation, Methodology, Writing – review & editing. JS: Methodology, Writing – review & editing. RY: Software, Validation, Writing – review & editing. NG: Resources, Writing – review & editing. NS: Resources, Supervision, Validation, Writing – review & editing. RB: Funding acquisition, Resources, Supervision, Writing – review & editing. VC: Formal analysis, Supervision, Writing – review & editing. MR: Conceptualization, Funding acquisition, Investigation, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. We are thankful to DST-SERB (ECRA project file no. ECR/2017/002858) and ICAR-NP FGGM project for funding this research. The corresponding author acknowledges the support from the DST-SERB SIRE fellowship to MR (SIR/2022/ 000361) for a better understanding of the subject during the foreign experience at the University of Bonn, Germany.
Acknowledgments
The authors are thankful to Dr. Rakesh Bhardwaj, Principal Scientist, ICAR-NBPGR, Delhi-110012, for providing NMR spectroscopy facility for oil content analysis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1493618/full#supplementary-material
Supplementary Figure 1 | Morphology of plants in the field. NRCPB rapa 8 (P1); Nigra Dwarf (P2); Pusa Jaikisan (cultivar) (PJK) and S2 generation of resynthesized B. juncea lines (RBJ 104, RBJ 106, RBJ 131, RBJ 137, RBJ 141, RBJ 143, RBJ 170 and RBJ 188).
Supplementary Figure 2 | Representative image of hybridity confirmation of resynthesized amphihaploid hybrids (33) at F1 generation using SSR primer UGM 632 on 2.5% agarose gel and 100bp ladder (L).
References
Banga, S. S., Kaur, N. (2009). An alternate procedure for resynthesis of Brassica juncea. Proc. 16th Aust. Res. assembly Brassicas Ballarat 106, 1–4.
Bansal, P., Banga, S., Banga, S. S. (2012). Heterosis as investigated in terms of polyploidy and genetic diversity using designed Brassica juncea amphiploid and its progenitor diploid species. PLoS One 7, e29607. doi: 10.1371/journal.pone.0029607
Bansal, P., Kaur, P., Banga, S. K., Surinder, Banga, S. (2009). Augmenting Genetic Diversity in Brassica juncea through its Resynthesis using Purposely Selected Diploid Progenitors. Int. J. Plant Breed. 3, 41–45.
Bhat, S., Sarla, N. (2004). Identification and overcoming barriers between Brassica rapa L. em. Metzg. and B. nigra (L.) Koch crosses for the resynthesis of B. juncea (L.) Czern. Genet. Resour Crop Evol. 51, 455–469. doi: 10.1023/B:GRES.0000024154.19867.cd
Chatterjee, D., Banga, S., Gupta, M., Bharti, S., Salisbury, P. A., Banga, S. S. (2016). Resynthesis of Brassica napus through hybridization between B. juncea and B. carinata. Theor. Appl. Genet. 129, 977–990. doi: 10.1007/s00122-016-2677-3
Chen, H., Wang, T., He, X., Cai, X., Lin, R., Liang, J., et al. (2022). BRAD V3.0: an upgraded Brassicaceae database. Nucleic Acids Res. 50, D1432–D1441. doi: 10.1093/nar/gkab1057
Comai, L. (2000). “Genetic and epigenetic interactions in allopolyploid plants,” in Plant Gene Silencing (Springer Netherlands, Dordrecht), 267–279. doi: 10.1007/978-94-011-4183-3_19
Dhaka, N., Rout, K., Yadava, S. K., Sodhi, Y. S., Gupta, V., Pental, D., et al. (2017). Genetic dissection of seed weight by QTL analysis and detection of allelic variation in Indian and east European gene pool lines of Brassica juncea. Theor. Appl. Genet. 130, 293–307. doi: 10.1007/s00122-016-2811-2
Eduardo, I., Alegre, S., Alexiou, K. G., Arús, P. (2020). Resynthesis: marker-based partial reconstruction of elite genotypes in clonally-reproducing plant species. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.01205
Evanno, G., Regnaut, S., Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE : a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Falush, D., Stephens, M., Pritchard, J. K. (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587. doi: 10.1093/genetics/164.4.1567
Gepts, P., Papa, R. (2003). Possible effects of (trans)gene flow from crops on the genetic diversity from landraces and wild relatives. Environ. Biosafety Res. 2, 89–103. doi: 10.1051/ebr:2003009
Giri, L., Jugran, A. K., Bahukhandi, A., Dhyani, P., Bhatt, I. D., Rawal, R. S., et al. (2017). Population genetic structure and marker trait associations using morphological, phytochemical and molecular parameters in Habenaria edgeworthii—a threatened medicinal orchid of west Himalaya, India. Appl. Biochem. Biotechnol. 181, 267–282. doi: 10.1007/s12010-016-2211-8
Gupta, M., Gupta, S., Kumar, H., Kumar, N., Banga, S. S. (2015). Population structure and breeding value of a new type of Brassica juncea created by combining A and B genomes from related allotetraploids. Theor. Appl. Genet. 128, 221–234. doi: 10.1007/s00122-014-2423-7
Hasan, M. J., Rahman, H. (2018). Resynthesis of Brassica juncea for resistance to Plasmodiophora brassicae pathotype 3. Breed Sci. 68, 385–391. doi: 10.1270/jsbbs.18010
Hilgert-Delgado, A., Klíma, M., Viehmannová, I., Urban, M. O., Fernández-Cusimamani, E., Vyvadilová, M. (2015). Efficient resynthesis of oilseed rape (Brassica napus L.) from crosses of winter types B. rapa × B. oleracea via simple ovule culture and early hybrid verification. Plant Cell Tissue Organ Cult 120, 191–201. doi: 10.1007/s11240-014-0593-2
Hinata, K., Konno, N. (1979). Studies on a male sterile strain having the Brassica campestris nucleus and the Diplotaxis muralis cytoplasm. I On the breeding procedure and some characteristics of the male sterile strain. Japanese J. Breed. 29, 305. doi: 10.1270/jsbbs1951.29.305
Hu, D., Jing, J., Snowdon, R. J., Mason, A. S., Shen, J., Meng, J., et al. (2021). Exploring the gene pool of Brassica napus by genomics-based approaches. Plant Biotechnol. J. 19, 1693–1712. doi: 10.1111/pbi.13636
Jat, R. S., Singh, V. V., Sharma, P., Rai, P. K. (2019). Oilseed brassica in India: Demand, supply, policy perspective and future potential. Oilseeds fats Crops Lipids 26, 8. doi: 10.1051/ocl/2019005
Katche, E., Gaebelein, R., Idris, Z., Vasquez-Teuber, P., Lo, Y., Nugent, D., et al. (2021). Stable, fertile lines produced by hybridization between allotetraploids Brassica juncea (AABB) and Brassica carinata (BBCC) have merged the A and C genomes. New Phytol. 230, 1242–1257. doi: 10.1111/nph.17225
Katiyar, R. K., Chamola, R., Chopra, V. L. (1998). Tetralocular mustard, Brassica juncea : New promising variability through interspecific hybridization. Plant Breed. 117, 398–399. doi: 10.1111/j.1439-0523.1998.tb01962.x
Kim, H., Choi, S. R., Bae, J., Hong, C. P., Lee, S. Y., Hossain, M. J., et al. (2009). Sequenced BAC anchored reference genetic map that reconciles the ten individual chromosomes of Brassica rapa. BMC Genomics 10, 432. doi: 10.1186/1471-2164-10-432
Kopelman, N. M., Mayzel, J., Jakobsson, M., Rosenberg, N. A., Mayrose, I. (2015). Clumpak : a program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour 15, 1179–1191. doi: 10.1111/1755-0998.12387
Kumar, A., Meena, H. S., Ram, B., Priyamedha, Sharma, A., Yadav, S., et al. (2018). Some Cytomorphological Evidence for Synthesis of Interspecific Hybrids between Brassica juncea and Autotetraploid B. fruticulosa. Cytologia (Tokyo) 83, 421–426. doi: 10.1508/cytologia.83.421
Lagercrantz, U. (1998). Comparative Mapping Between Arabidopsis thaliana and Brassica nigra Indicates That Brassica Genomes Have Evolved Through Extensive Genome Replication Accompanied by Chromosome Fusions and Frequent Rearrangements. Genetics 150, 1217–1228. doi: 10.1093/genetics/150.3.1217
Leitch, I. J., Bennett, M. D. (2004). Genome downsizing in polyploid plants. Biol. J. Linn. Soc. 82, 651–663. doi: 10.1111/j.1095-8312.2004.00349.x
Li, Y., Liu, J. (2018). StructureSelector : A web-based software to select and visualize the optimal number of clusters using multiple methods. Mol. Ecol. Resour 18, 176–177. doi: 10.1111/1755-0998.12719
Li, A., Liu, D., Yang, W., Kishii, M., Mao, L. (2018). Synthetic hexaploid wheat: yesterday, today, and tomorrow. Engineering 4, 552–558. doi: 10.1016/j.eng.2018.07.001
Li, Q., Mei, J., Zhang, Y., Li, J., Ge, X., Li, Z., et al. (2013). A large-scale introgression of genomic components of Brassica rapa into B. napus by the bridge of hexaploid derived from hybridization between B. napus and B. oleracea. Theor. Appl. Genet. 126, 2073–2080. doi: 10.1007/s00122-013-2119-4
Lloyd, A., Bomblies, K. (2016). Meiosis in autopolyploid and allopolyploid Arabidopsis. Curr. Opin. Plant Biol. 30, 116–122. doi: 10.1016/j.pbi.2016.02.004
Lowe, A. J., Moule, C., Trick, M., Edwards, K. J. (2004). Efficient large-scale development of microsatellites for marker and mapping applications in Brassica crop species. Theor. Appl. Genet. 108, 1103–1112. doi: 10.1007/s00122-003-1522-7
Luo, Z., Brock, J., Dyer, J. M., Kutchan, T., Schachtman, D., Augustin, M., et al. (2019). Genetic diversity and population structure of a Camelina sativa spring panel. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00184
Lysak, M. A., Koch, M. A., Pecinka, A., Schubert, I. (2005). Chromosome triplication found across the tribe Brassiceae. Genome Res. 15, 516–525. doi: 10.1101/gr.3531105
Manzoor, A., Ahmad, T., Bashir, M., Hafiz, I., Silvestri, C. (2019). Studies on colchicine induced chromosome doubling for enhancement of quality traits in ornamental plants. Plants 8, 194. doi: 10.3390/plants8070194
Meirmans, P. G. (2015). Seven common mistakes in population genetics and how to avoid them. Mol. Ecol. 24, 3223–3231. doi: 10.1111/mec.13243
Mohd Saad, N. S., Severn-Ellis, A. A., Pradhan, A., Edwards, D., Batley, J. (2021). Genomics armed with diversity leads the way in brassica improvement in a changing global environment. Front. Genet. 12. doi: 10.3389/fgene.2021.600789
Mwathi, M. W., Gupta, M., Quezada-Martinez, D., Pradhan, A., Batley, J., Mason, A. S. (2020). Fertile allohexaploid Brassica hybrids obtained from crosses between B. oleracea and B. juncea via ovule rescue and colchicine treatment of cuttings. Plant Cell Tissue Organ Cult 140, 301–313. doi: 10.1007/s11240-019-01728-x
Ogbonnaya, F. C., Abdalla, O., Mujeeb-Kazi, A., Kazi, A. G., Xu, S. S., Gosman, N., et al. (2013). “Synthetic hexaploids: harnessing species of the primary gene pool for wheat improvement,” in Plant Breeding Reviews (Wiley), 35–122. doi: 10.1002/9781118497869.ch2
Olivoto, T., Lúcio, A. D. (2020). metan: An R package for multi-environment trial analysis. Methods Ecol. Evol. 11, 783–789. doi: 10.1111/2041-210X.13384
Olsson, G. (1960). Species crosses within the genus Brassica. I. Artificial Brassica juncea Coss. Hereditas 46, 171–223. doi: 10.1111/j.1601-5223.1960.tb03082.x
Ozminkowski, R. H., Jourdan, P. (1994). Comparing the resynthesis of Brassica napus L. by interspecific somatic and sexual hybridization. I. Producing and identifying hybrids. J. Am. Soc. Hortic. Sci. 119, 808–815. doi: 10.21273/JASHS.119.4.808
Pandey, J., Scheuring, D. C., Koym, J. W., Coombs, J., Novy, R. G., Thompson, A. L., et al. (2021). Genetic diversity and population structure of advanced clones selected over forty years by a potato breeding program in the USA. Sci. Rep. 11, 8344. doi: 10.1038/s41598-021-87284-x
Parmar, N., Singh, K. H., Sharma, D., Singh, L., Kumar, P., Nanjundan, J., et al. (2017). Genetic engineering strategies for biotic and abiotic stress tolerance and quality enhancement in horticultural crops: a comprehensive review. 3 Biotech. 7, 239. doi: 10.1007/s13205-017-0870-y
Pellicer, J., Hidalgo, O., Dodsworth, S., Leitch, I. (2018). Genome size diversity and its impact on the evolution of land plants. Genes (Basel) 9, 88. doi: 10.3390/genes9020088
Perrier, X., Jacquemoud-Collet, J. (2006). DARwin software: Dissimilarity analysis and representation for windows.
Petereit, J., Bayer, P. E., Thomas, W. J. W., Tay Fernandez, C. G., Amas, J., Zhang, Y., et al. (2022). Pangenomics and crop genome adaptation in a changing climate. Plants 11, 1949. doi: 10.3390/plants11151949
Prakash, S. (1973). Artificial synthesis of Brassica juncea Coss. Genetica 44, 249–263. doi: 10.1007/BF00119110
Prakash, S., Gupta, S., Raut, R. N., Kalra, A. (1984). Synthetic Brassica carinata–a preliminary report. Cruciferae Newsletter. 35.
Pritchard, J. K., Stephens, M., Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. doi: 10.1093/genetics/155.2.945
Puechmaille, S. J. (2016). The program STRUCTURE does not reliably recover the correct population structure when sampling is uneven: subsampling and new estimators alleviate the problem. Mol. Ecol. Resour 16, 608–627. doi: 10.1111/1755-0998.12512
Quezada-Martinez, D., Addo Nyarko, C. P., Schiessl, S. V., Mason, A. S. (2021). Using wild relatives and related species to build climate resilience in Brassica crops. Theor. Appl. Genet. 134, 1711–1728. doi: 10.1007/s00122-021-03793-3
Rahman, H., Bennett, R. A., Séguin-Swartz, G. (2015). Broadening genetic diversity in Brassica napus canola: Development of canola-quality spring B. napus from B. napus×B. oleracea var. alboglabra interspecific crosses. Can. J. Plant Sci. 95, 29–41. doi: 10.4141/cjps-2014-017
Raina, S. N., Parida, A., Koul, K. K., Salimath, S. S., Bisht, M. S., Raja, V., et al. (1994). Associated chromosomal DNA changes in polyploids. Genome 37, 560–564. doi: 10.1139/g94-080
Raj, A., Stephens, M., Pritchard, J. K. (2014). fastSTRUCTURE: Variational inference of population structure in large SNP data sets. Genetics 197, 573–589. doi: 10.1534/genetics.114.164350
Rajcan, I., Boersma, J. G., Shaw, E. J. (2011). “Plant genetic techniques,” in Comprehensive Biotechnology (Academic press, Burlington: Elsevier), 133–147. doi: 10.1016/B978-0-08-088504-9.00252-X
Rao, M., Kashyap, A., Garg, P., Sharma, S. S., Gupta, N. C., Chamola, R., et al. (2024). An indigenous germplasm of Brassica rapa var. yellow NRCPB rapa 8 enhanced resynthesis of Brassica juncea without in vitro intervention. Genet. Resour Crop Evol. doi: 10.1007/s10722-024-02052-w
Scannell, D. R., Frank, A. C., Conant, G. C., Byrne, K. P., Woolfit, M., Wolfe, K. H. (2007). Independent sorting-out of thousands of duplicated gene pairs in two yeast species descended from a whole-genome duplication. Proc. Natl. Acad. Sci. 104, 8397–8402. doi: 10.1073/pnas.0608218104
Seyis, F., Friedt, W., Luhs, W. (2005). Development of Resynthesized Rapeseed (Brassica napus L.) Forms with Low Erucic Acid Content Through in ovulum Culture. Asian J. Plant Sci. 4, 6–10. doi: 10.3923/ajps.2005.6.10
Seyis, F., Snowdon, R. J., Luhs, W., Friedt, W. (2003). Molecular characterization of novel resynthesized rapeseed (Brassica napus) lines and analysis of their genetic diversity in comparison with spring rapeseed cultivars*. Plant Breed. 122, 473–478. doi: 10.1111/j.1439-0523.2003.00859.x
Sheng, X., Wen, G., Guo, Y., Yan, H., Zhao, H., Liu, F. (2012). A semi-fertile interspecific hybrid of Brassica rapa and B. nigra and the cytogenetic analysis of its progeny. Genet. Resour Crop Evol. 59, 73–81. doi: 10.1007/s10722-011-9669-6
Shruti, Shukla, A., Rahman, S. S., Suneja, P., Yadav, R., Hussain, Z., et al. (2023). Developing an NIRS prediction model for oil, protein, amino acids and fatty acids in amaranth and buckwheat. Agriculture 13, 469. doi: 10.3390/agriculture13020469
Singh, K. P., Kumari, P., Rai, P. K. (2021). Current status of the disease-resistant gene(s)/QTLs, and strategies for improvement in Brassica juncea. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.617405
Snowdon, R. J., Köhler, W., Köhler, A. (1997). Chromosomal localization and characterization of rDNA loci in the Brassica A and C genomes. Genome 40, 582–587. doi: 10.1139/g97-076
Song, X., Ma, X., Li, C., Hu, J., Yang, Q., Wang, T., et al. (2018). Comprehensive analyses of the BES1 gene family in Brassica napus and examination of their evolutionary pattern in representative species. BMC Genomics 19, 346. doi: 10.1186/s12864-018-4744-4
Srinivasachar, D. (1964). Possibilities of transferring high branching habit of Brassica nigra Koch, to Brassica júncea Coss. through amphidiploidy. Curr. Sci. 33, 533–534. doi: 10.5555/19651604924
Srivastava, A., Mukhopadhyay, A., Arumugam, N., Gupta, V., Verma, J. K., Pental, D., et al. (2004). Resynthesis of Brassica juncea through interspecific crosses between B. rapa and B. nigra. Plant Breed. 123, 204–206. doi: 10.1046/j.1439-0523.2003.00933.x
Sudan, J., Khajuria, P., Gupta, S. K., Singh, R. (2016). Analysis of molecular diversity in Indian and Exotic genotypes of Brassica juncea using SSR markers. Indian J. Genet. Plant Breed. 76, 361. doi: 10.5958/0975-6906.2016.00054.7
Wang, X., Morton, J. A., Pellicer, J., Leitch, I. J., Leitch, A. R. (2021). Genome downsizing after polyploidy: mechanisms, rates and selection pressures. Plant J. 107, 1003–1015. doi: 10.1111/tpj.15363
Warwick, S. I., Black, L. D. (1991). Molecular systematics of Brassica and allied genera (Subtribe Brassicinae, Brassiceae) —chloroplast genome and cytodeme congruence. Theor. Appl. Genet. 82, 81–92. doi: 10.1007/BF00231281
Wendel, J. F. (2015). The wondrous cycles of polyploidy in plants. Am. J. Bot. 102, 1753–1756. doi: 10.3732/ajb.1500320
Yadav, P., Bhat, S. R., Prakash, S., Mishra, L. C., Chopra, V. L. (2009). Resynthesized Brassica juncea lines with novel organellar genome constitution obtained through protoplast fusion. J. Genet. 88, 109–112. doi: 10.1007/s12041-009-0016-z
Yant, L., Hollister, J. D., Wright, K. M., Arnold, B. J., Higgins, J. D., Franklin, F. C. H., et al. (2013). Meiotic adaptation to genome duplication in Arabidopsis arenosa. Curr. Biol. 23, 2151–2156. doi: 10.1016/j.cub.2013.08.059
Zenil-Ferguson, R., Ponciano, J. M., Burleigh, J. G. (2016). Evaluating the role of genome downsizing and size thresholds from genome size distributions in angiosperms. Am. J. Bot. 103, 1175–1186. doi: 10.3732/ajb.1500408
Zhang, L., Li, X., Chang, L., Wang, T., Liang, J., Lin, R., et al. (2022). Expanding the genetic variation of Brassica juncea by introgression of the Brassica rapa genome. Hortic. Res. 9, 1–9. doi: 10.1093/hr/uhab054
Zhang, G. Q., Tang, G. X., Song, W. J., Zhou, W. J. (2004). Resynthesizing Brassica napus from interspecific hybridization between Brassica rapa and B. oleracea through ovary culture. Euphytica 140, 181–187. doi: 10.1007/s10681-004-3034-1
Zhao, P., Zhou, H.-J., Potter, D., Hu, Y.-H., Feng, X.-J., Dang, M., et al. (2018). Population genetics, phylogenomics and hybrid speciation of Juglans in China determined from whole chloroplast genomes, transcriptomes, and genotyping-by-sequencing (GBS). Mol. Phylogenet Evol. 126, 250–265. doi: 10.1016/j.ympev.2018.04.014
Keywords: Brassica juncea, inter-specific hybridization, resynthesized lines, genetic diversity, SSR markers, pollen fertility, allelic richness
Citation: Garg P, Tripathi S, Kashyap A, Anil Kumar A, Kumari S, Singh M, Kushwaha R, Sharma SS, Sharma J, Yadav R, Gupta NC, Singh N, Bhattacharya R, Chhokar V and Rao M (2025) Insights into early generation synthetic amphidiploid Brassica juncea: a strategy to harness maximum parental genomic diversity for improving Indian mustard. Front. Plant Sci. 16:1493618. doi: 10.3389/fpls.2025.1493618
Received: 09 September 2024; Accepted: 06 January 2025;
Published: 13 February 2025.
Edited by:
Sergio J. Ochatt, INRA UMR1347 Agroécologie, FranceReviewed by:
Anupam Singh, Shree Guru Gobind Singh Tricentenary University, IndiaAjay Kumar Thakur, Central Potato Research Institute (ICAR), India
Copyright © 2025 Garg, Tripathi, Kashyap, Anil Kumar, Kumari, Singh, Kushwaha, Sharma, Sharma, Yadav, Gupta, Singh, Bhattacharya, Chhokar and Rao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mahesh Rao, bXJhb2ljYXJAZ21haWwuY29t; Vinod Chhokar, dmlub2RjaGhva2FyQHlhaG9vLmNvbQ==; Ramcharan Bhattacharya, cmNiaGF0dGFjaGFyeWExQGdtYWlsLmNvbQ==
†These authors have contributed equally to this work
 Pooja Garg
Pooja Garg Shikha Tripathi
Shikha Tripathi Anamika Kashyap
Anamika Kashyap A. Anil Kumar
A. Anil Kumar Sujata Kumari
Sujata Kumari Mandeep Singh
Mandeep Singh Ranjeet Kushwaha
Ranjeet Kushwaha Shiv Shankar Sharma
Shiv Shankar Sharma Jyoti Sharma
Jyoti Sharma Rashmi Yadav
Rashmi Yadav N. C. Gupta
N. C. Gupta Naveen Singh
Naveen Singh Ramcharan Bhattacharya
Ramcharan Bhattacharya Vinod Chhokar
Vinod Chhokar Mahesh Rao
Mahesh Rao