
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Plant Sci. , 22 January 2025
Sec. Plant Breeding
Volume 16 - 2025 | https://doi.org/10.3389/fpls.2025.1489244
Wide hybridization is an important plant breeding strategy that can be used to expand the available genetic variation in present-day crops towards breeding for enhanced agronomic performance. The primary challenge in wide hybridization is the presence of reproductive barriers and genetic incompatibilities that limit the transfer of desirable wild or distant alleles in the genetic background of cultivated plant species. Here we provide perspectives on the possible role of hybrid sterility and gametocidal genes on the observed preferential introgression in exotic germplasm of rice. We argue that while these aberrant introgression and segregation behavior of wild or distant chromosomes presents significant barriers in exploiting ancestral germplasm in breeding, the same mechanisms can also be exploited to enhance the transfer of wild alleles in a cultivated genetic background. Understanding the genetic basis of preferential introgression and segregation in wide hybrids will have serious implications in our ability to capture ancestral genetic variation that can add significant agronomic value to staple crops like rice.
The end of the last ice age 10,000-12,000 years ago began humankind’s continuing pursuit to tame the wild. As vegetation flourished and humans shifted to gathering grains from large-seeded grasses, they observed that some plants of the same wild grass species have better traits than others and favored growing them every season. This cycle of artificial selection for traits that were deemed beneficial initiated the protracted process of plant domestication.
In time, domesticated plants lost many traits that were essential for their survival in the wild like seed shattering, dormancy, photoperiodicity and indeterminate habit (Meyer and Purugganan, 2013). With it, reproductive barriers and genetic incompatibilities evolved that would eventually distinguish the domesticated plant as a distinct species from its wild progenitor. Despite species boundaries however, gene flow between the domesticates and wild relatives remains possible, to varying extents (Khush and Brar, 1992).
Plant breeders have capitalized on the semi-permeable nature of species boundaries to access the genetic variation left behind in the distant relatives of crops during domestication. Using wide hybridization, or the artificial crossing between cultivated plants and their wild or distant relatives, existing gene pools of crops have been enriched and agronomic performances improved. Indeed, many success stories in plant breeding have much to attribute to novel genetic recombinations produced from wide hybridizations. In rice, the introgression in mega-varieties of the broad-spectrum bacterial blight resistance Xa21 allele from the wild O. longistaminata (Khush et al., 1990) and the flooding tolerance Sub1A allele from the landrance FR13A (Septiningsih et al., 2009) are just two noteworthy examples of how exotic alleles can add significant value to crops. Despite these gains, there remains a tremendous amount of genetic potential in wild or distant crop relatives that is yet to be exploited for breeding purposes.
The incorporation of wild alleles in the genetic background of plant domesticates is not always straightforward. Introgressions are usually stochastic and at the same time differential, with some loci having higher propensity for introgression than others. Generally, loci with alleles conferring advantageous traits tend to introgress more frequently, whereas loci associated with reproductive isolation will introgress less (Harrison and Larson, 2014; Suarez-Gonzalez et al., 2018). Oftentimes, this manifests in the preferential retention and transmission of a specific loci from one generation to another. The nature of such phenomena has been established as a function of genomic regions as opposed to the whole genome, where chromosome segments that are consistently transmitted from one generation to another possibly carry genes that ensure its own stable transmission (Harrison and Larson, 2014). In the context of plant breeding, differential introgression creates challenges when alleles that are advantageous to the wild relative but not necessarily to the cultivated species, provide fitness advantage in the wide hybrids. Desirable trait introgression becomes more complicated when wild loci conferring deleterious traits in a cultivated background are tightly linked to traits of agronomic interests. More so when wild introgressions are regulated by genetic factors localized in the wild genome itself.
In rice (Oryza sativa L.), preferential retention and transmission of chromosome segments have been reported in progenies of interspecific crosses between cultivars and distant relatives. In this paper, we provide perspectives on possible mechanisms underlying the aberrant chromosome introgression in exotic rice germplasm derived from O. nivara and O. latifolia (Furuta et al., 2016; Angeles-Shim et al., 2014). We drew parallelisms between the genetic drivers of preferential chromosome transfer that have been previously reported and the observed behavior of chromosome introgressions in our rice germplasm.
Twenty-seven diploid lines that segregated from monosomic alien addition lines (MAAL) of the allopolyploid O. latifolia in the genetic background of the elite breeding line IR31917 were genotyped to have overlapping fragments of O. latifolia chromosome 6 but not a single introgression from chromosome 7 (Figure 1A) (Angeles-Shim et al., 2014). Mapping of the location and extent of O. latifolia introgression in the MAAL-derived introgression lines (MDILs) using a combination of SNPs, SSRs, STS and indel markers identified 32 wild introgressions distributed across the genome except in chromosome 7. The chromosome 6 introgression mapped between 9.42 and 22.72 Mb, with each MDIL carrying an overlapping segment that spans 9.42 Mb. Evaluation of the 27 diploid lines for a suite of agronomic traits showed no penalty on panicle fertility of the wild introgressions.
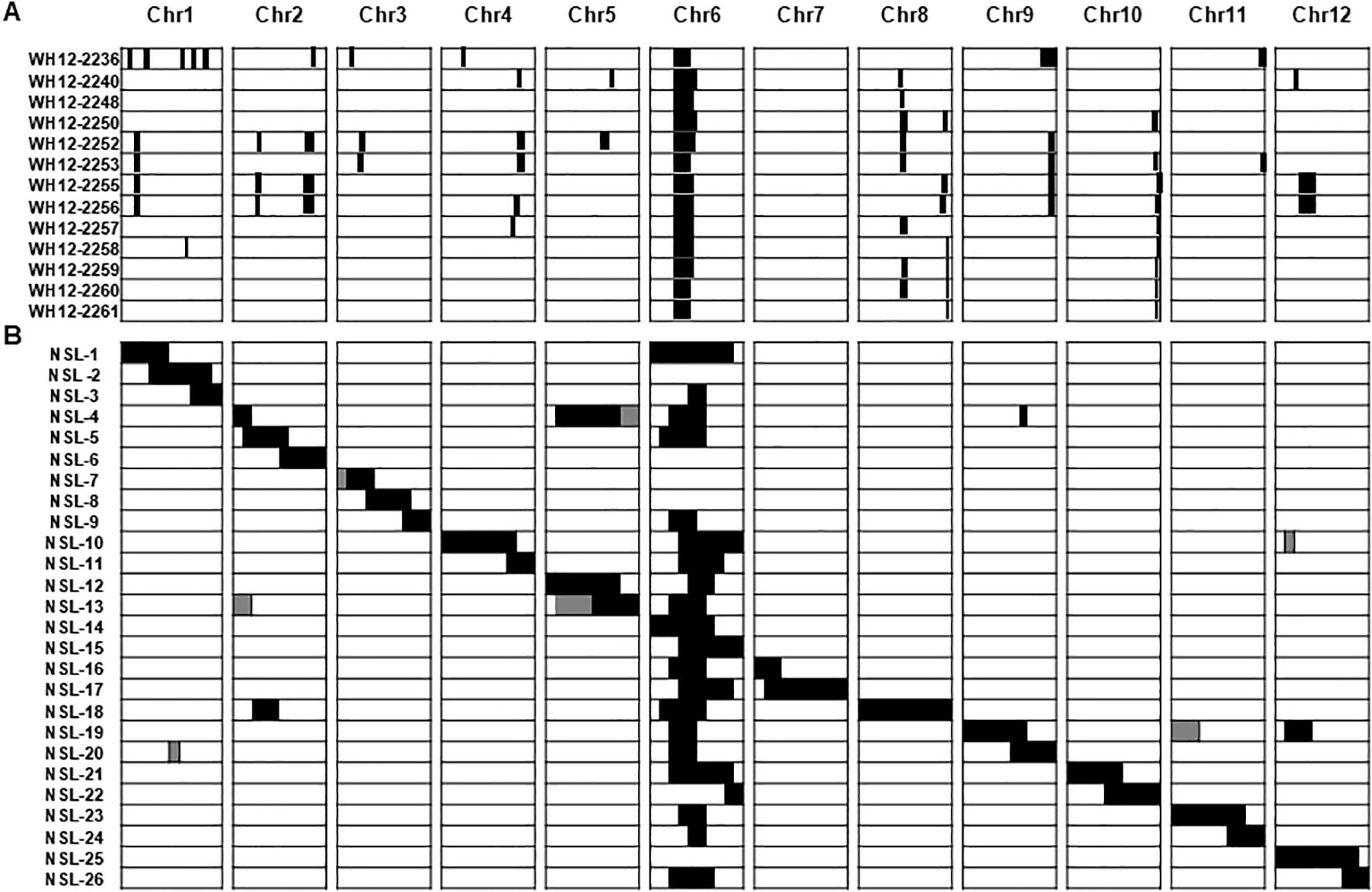
Figure 1. Patterns of preferential chromosome transfer in exotic rice germplasm. (A) Representative MDILs of O. latifolia in the background of O. sativa elite breeding line IR31917 showing stable transmission of O. latifolia chromosome 6 segments. Figure was adapted from Angeles-Shim et al., 2014. (B) Chromosome segment substitution lines of O. nivara in the background of O. sativa cv. Nipponbare (NSL) showing stable transmission of O. nivara chromosome 6 segments in twenty-one lines. Figure was adapted from Furuta et al., 2016. Black areas in (A, B) indicate wild introgression. chr, Chromosome.
A similar pattern of preferential transmission of chromosome 6 segment was observed in O. nivara CSSLs in the genetic background of the elite japonica cultivar Koshihikari (NSLs) (Figure 1B) (Furuta et al., 2016). Wild introgressions were identified using SNP markers that are evenly distributed across the genome at an average of 2.4 Mb intervals. The full O. nivara genome was represented in 26 NSLs, each carrying a contiguous or overlapping chromosome fragment from the wild donor. Of the 26 NSLs, 21 have substituted segments of O. nivara in chromosome 6. The substituted chromosome 6 segments span the full chromosome, with overlaps between 13.7 to 20.2 Mb.
Hybrid sterility is a post-zygotic reproductive barrier that has been widely reported in crosses between the Asian (O. sativa) and African cultivated rice (O. glaberrima), as well as between the cultivated rice and its wild relatives (Li et al., 2020). Locus controlling hybrid sterility (S) functions as a gamete eliminator, aborting gametes carrying the opposite allele in heterozygotes and promoting the frequent transmission of advantageous allele at this locus (Koide et al., 2008). In rice, more than 50 S loci controlling hybrid sterility have been identified from various Oryza species and have been mapped across the different chromosomes in the rice genome (Li et al., 2020). Several studies have investigated hybrid sterility between O. sativa and O. glaberrima. Notable S loci such as S19, S20, S37(t), S38(t) and S39(t) show a transmission advantage for the O. glaberrima allele (Xu et al., 2014). Even intra-specific hybrid sterility loci between subgroups of the Asian cultivated rice typically exhibit a transmission advantage of specific alleles. S67 is a recently reported hybrid sterility locus between two subgroups of temperate japonica rice and basmati, demonstrating preferential transmission of temperate japonica allele (Lv et al., 2024).
Chromosome 6 harbors the hybrid sterility locus S1 and S6 (Yang et al., 2016; Koide et al., 2008). S1 maps in the distal end of the short arm of chromosome 6, whereas S6 is located near the centromeric region at 18.7 Mb. The S1 locus was first discovered in O. glaberrima (Sano et al., 1979), although a similar hybrid sterility locus that maps between 1.76 and 2.88 Mb in chromosome 6 have also been identified in O. nivara, O. rufipogon and O. barthii. Conversely, S6 was originally discovered in crosses between O. rufipogon and O. sativa subsp. japonica cv. T65 (Koide et al., 2008). Like other S locus identified in Oryza species, S1 and S6 act as gamete eliminator, selectively favoring gametes with the wild S allele over those carrying the O. sativa S allele in heterozygotes. The control of hybrid sterility by the S1 locus is observed in both male and female gametes, a pattern also seen with the S6 locus (Zhou et al., 2010). The selective pressure imposed by S6 in both male and female gametes reinforce its preferential retention and transmission in hybrid generations (Yang et al., 2016; Koide et al., 2008).
In the case of both MDILs and NSLs, the introgressed fragments from chromosome 6 of O. latifolia and O. nivara, respectively, overlap with the mapped position of the S6 locus. This indicates the possibility of a novel hybrid sterility locus in both wild relatives that is allelic to the reported S6 from O. rufipogon (S6rufipogon). Like S6rufipogon, it is possible that S6latifolia and S6nivara alleles function as gamete eliminator, promoting their preferential transmission and retention in the O. latifolia MDILs and O. nivara NSLs. Whether the hybrid sterility locus occurs in either one or both gametes is yet to be determined.
Incidentally, similar patterns of preferential transfer of chromosome 6 segment carrying the S6 locus have not been observed in other CSSLs developed using O. barthii, O. longistaminata, O. rufipogon and O. glaberrima (Bessho-Uehara et al., 2017; Ramos et al., 2016; Furuta et al., 2014; Angeles-Shim et al., 2010). To date, the S6 locus has only been identified in O. rufipogon and putatively in the O. nivara NSLs and O. latifolia MDILs. This contrasts with the reported microco-linearity of other hybrid sterility locus like the S1 in at least 4 Oryza species with AA genome (Yang et al., 2016). To determine the conservation of the S6 locus in Oryza species with AA genome, syntenic analysis was carried out for O. sativa cv. Nipponbare, O. rufipogon, O. barthii, O. glaberrima and O. nivara chromosome 6 using data from the rice genome hub (https://rice-genome-hub.southgreen.fr/synvisio) (Bandi and Gutwin, 2020).
Syntenic analysis revealed variations in the structural arrangement of chromosome 6 across the four Oryza species (Figure 2). O. rufipogon maintained complete synteny with O. sativa cv. Nipponbare, while both O. barthii and O. glaberrima exhibited large-scale inversion events in the centromeric region for chromosome 6. O. rufipogon also shared synteny with O. barthii and O. glaberrima across chromosome 6 except for the centromeric region. The lack of synteny between O. rufipogon, O. barthii and O. glaberrima suggests that the S6 locus may be absent or transcriptionally inactive in the latter two, which would explain the normal chromosome transmission in previously reported O. barthii and O. glaberrima CSSLs (Bessho-Uehara et al., 2017; Angeles-Shim et al., 2010). Incidentally, the lack of systematic transmission of chromosome 6 segment in the O. rufipogon CSSLs developed by Furuta et al. (2014) contradicts with the original discovery of the S6 locus in another accession of O. rufipogon (Koide et al., 2008). This may be due to genetic background effects similar to what has been reported for the S24 sterility locus where male semi-sterility was only observed in S24 heterozygote in a japonica background but not in an indica background (Kubo et al., 2011).
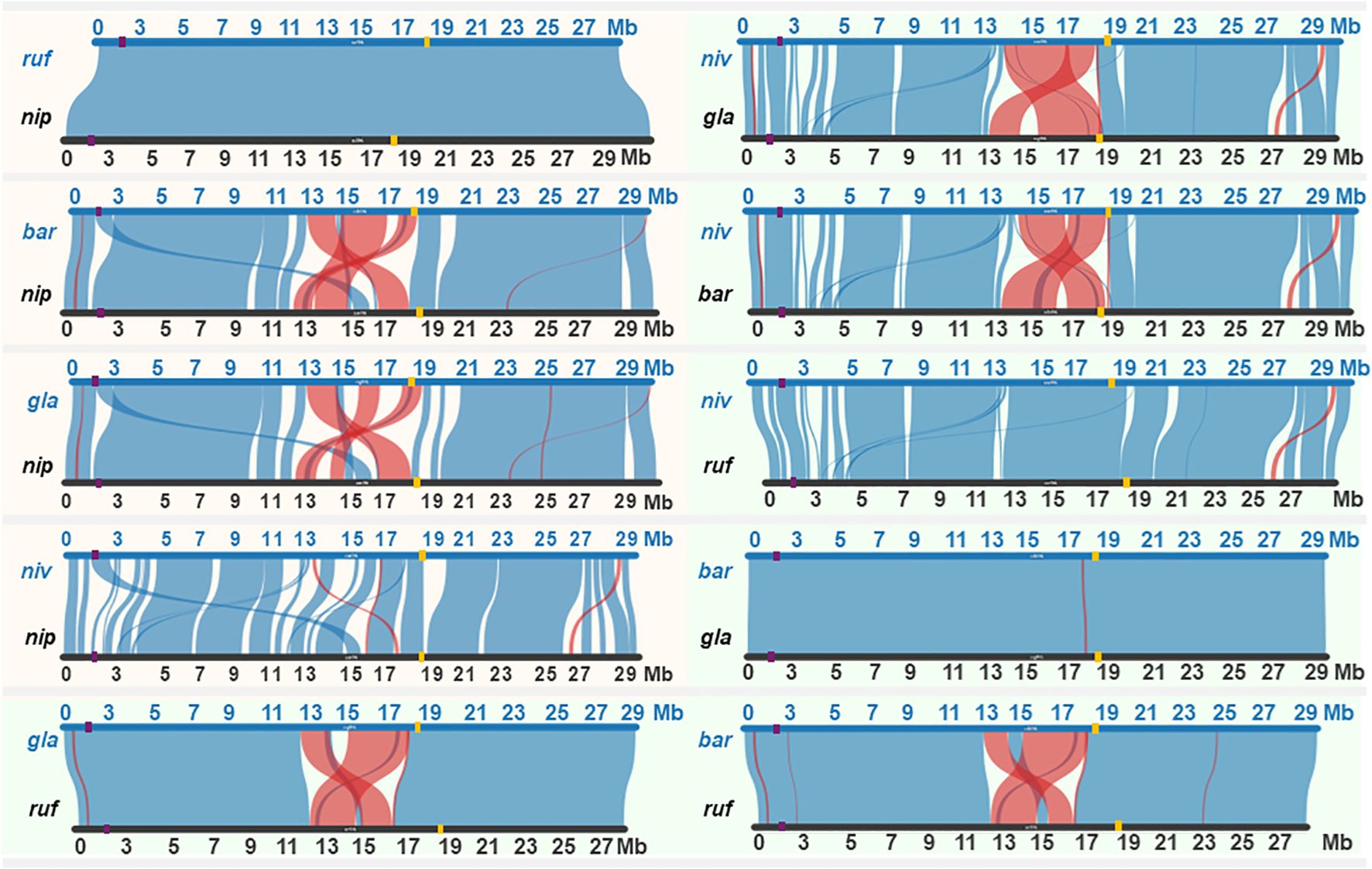
Figure 2. Synteny across chromosome 6 of wild rice species with AA genome. Orange shaded region highlights synteny between the cultivated Nipponbare (nip) and its wild relatives, while the green shaded region illustrates synteny among the wild species: O. barthii (bar), O. rufipogon (ruf), O. glaberrima (gla), and O. nivara (niv). Purple and yellow bars on chromosome represent the position of S1 and S6 locus, respectively.
O. nivara displayed minor syntenic disruptions at the centromeric region compared to cv. Nipponbare and showed inversion events at the centromeric region between 13 and 17 Mb relative to O. barthii and O. glaberrima but not O. rufipogon. Notably, O. nivara and O. rufipogon showed synteny at the S6 locus, supporting the proposed presence of a functional S6 locus in O. nivara that could have facilitated the stable inheritance of the wild chromosome 6 segment in the NSLs. Unlike O. rufipogon however, O. nivara exhibits unique structural variations compared to O. sativa that can potentially harbor genes or regulatory elements that enhance its PCT potential when combined with the functional S6 locus. We hypothesize that the interplay between the S6 locus and these distinct genomic features in chromosome 6 of O. nivara drives its heightened propensity for PCT. In the presence of linkage, the S6 locus will be inherited together with other proximal loci. With generation advance, distortion in transmission ratio exerted by S6 would extend beyond the locus itself, favoring the transmission of overlapping chromosome 6 segments in advanced progenies. In the development of the NSLs, it appears that the selective advantage imparted by the S6 locus in fact led to the accumulation of overlapping segments on chromosome 6 despite repeated backcrossing. The consistent retention of these overlapping segments, driven by the transmission bias of the S6 locus could effectively integrate O. nivara alleles across chromosome 6 in the Koshihikari background.
In silico analysis of the annotated genes within the 13-18 Mb of the introgressed chromosome 6 segment of O. nivara identified genes involved in fatty acid metabolism and cell death regulation, both of which have been reported to play crucial roles in pollen viability and gametophyte competitiveness under varying environmental conditions (Zhang et al., 2021; Wang et al., 2024). For example, fatty acid metabolism is necessary to maintain cell membrane integrity, which can support pollen resilience and function. The regulation of cell death within this region might contribute to selective elimination processes under competitive fertilization, promoting the preferential transfer of certain chromosome segments (Koide et al., 2008).
The MDILs showed normal spikelet fertility whereas two out of the 26 NSLs (nos. 16 and 17) expressed approximately 50% reduction in the number of grains per panicle. Both NSLs have overlapping, substituted segments in chromosome 6 and 7 of O. nivara. However, phenotype-genotype association using the full NSL set putatively associates the spikelet fertility reduction in lines 16 and 17 with the 3.9–7.9 Mb introgressed segment from chromosome 7 of O. nivara.
The observed fertility reduction in NSLs 16 and 17, marked by a 50% decrease in grain number per panicle, is likely linked to a gametocidal-like gene (Gc)-like. Gametocidal gene systems, as seen in hybrids of Triticum aestivum and Aegilops sharonensis, typically involve two elements: a “breaker,” which induces DNA breaks in gametes lacking specific chromosomal segments, and an “inhibitor,” which mitigates the breaker’s effects to preserve gamete viability (Knight et al., 2015). However, in the case of NSLs 16 and 17, there appears to be no evidence of an inhibitor-breaker like element, suggesting that the observed fertility reduction may arise from a gene(s) locus/loci acting independently.
Gene ontology (GO) analysis of the chromosome 7 region in NSL 16 and 17 revealed several pathways potentially contributing to reduced fertility. Key genes associated with lipid metabolism, cell wall biosynthesis, and DNA damage responses were identified, indicating the presence of mechanism that are associated with gamete stability and viability. For instance, the gene ONIVA07G13230.1, which encodes a fatty acyl-CoA reductase essential for lipid biosynthesis, likely plays a critical role in maintaining membrane stability and pollen viability (Zhang et al., 2021; Wang et al., 2024; Yang et al., 2017). Disruption of this pathway could lead to unstable pollen and selective gamete loss. Similarly, genes involved in cell wall biosynthesis, such as those encoding mannan synthase and cellulose synthase, are crucial for maintaining the structural integrity of pollen cell walls. Weakened cell walls in gametes lacking the protective chromosome segment may render them more susceptible to environmental and metabolic stress, further reducing fertility.
In addition to structural and metabolic disruptions, genes linked to DNA degradation and hypersensitive responses appear to exacerbate gamete instability. GO terms associated with exonuclease activity and defense responses suggest that DNA damage pathways may be hyperactivated in gametes without the chromosome 7 segment, leading to selective cell death. Furthermore, genes encoding AAI domain-containing proteins and AP2/ERF transcription factors within the introgressed region may influence pollen development and stress responses, indirectly affecting fertility (Long et al., 2023; Zhang et al., 2024). These proteins regulate lipid transport and developmental processes critical for gamete viability, and their altered function in NSLs 16 and 17 could explain the observed fertility reduction.
The observed 50% reduction in spikelet fertility is not mutually exclusive to the breaker-inducer complex as mentioned above. Our findings align with discoveries in other studies where reduced pollen viability significantly impacts spikelet fertility. This is well-illustrated in the reproductive behaviors associated with the S6 locus in rice hybrids (Koide et al., 2008). Histological analyses of female gametogenesis in S6rufipigom/S6sativa heterozygotes demonstrated that nearly half of the embryo sacs exhibited structural abnormalities. This aligns with the 50% fertility reduction observed in affected hybrids. While male gamete dysfunction is not visually apparent at the pollen grain stage, genetic evidence strongly suggests post-pollination competition or incompatibility, further exacerbating fertility losses. The absence of viable S6sativa gametes in progeny confirms the decisive role of selective gamete abortion mediated by the S6rufipigom locus. However, in our findings we can see a reduction of spikelet fertility under a homozygous state. Nonetheless, we can assume that the introgressed region of chromosome 7 (NSL 16 and NSL 17) might harbor a functional gene that directly leads to the 50% reduction in spikelet fertility in these NSLs.
The absence of an inhibitor element in this system suggests a simpler mechanism than the classic Gc model, where the breaker-like locus alone drives selective gamete loss. This fertility distortion aligns with the preferential transmission of the chromosome 7 segment and may provide a selective advantage in specific contexts. The cumulative evidence points to a multifaceted mechanism of fertility reduction involving disruptions in cell wall integrity, lipid metabolism, and DNA stability. Further research is necessary to identify the specific genetic contributors within the substituted chromosome 7 segment and to elucidate how these genes interact to influence fertility in hybrid genetic backgrounds. Understanding these mechanisms could have significant implications for introgression breeding and the regulation of hybrid fertility.
Exotic germplasm offers significant potential for plant improvement, but its use in breeding is often constrained by the biased transmission of distant genomic regions. From an evolutionary perspective, this biased transmission where one genotype is favored over another at the expense of overall fitness may seem paradoxical. This concept mirrors the early misconceptions about non-coding DNA, which was once considered “junk” but is now recognized for its crucial role in regulating gene expression. Similarly, this PCT associated with sterility loci can be exploited as a mechanism to access novel genetic variations from wild or distant crop relatives, rather than being viewed as a barrier for beneficial wild introgressions. By manipulating hybrid sterility genes, we can efficiently introduce these novel alleles into breeding programs without compromising their stability. The sterility locus ensures the retention of viable gametes, although its heterozygous state often leads to reduced gamete viability.
However, if the sterility locus is tightly linked to genes that negatively impact agronomic traits, we may need to develop strategies to mitigate these effects. While no such complications have been observed in studies involving sterility loci to date, it remains essential to understand the genetic mechanisms controlling these loci. Such knowledge will be crucial for integrating distant relatives into breeding programs, ultimately helping to develop more robust rice cultivars with broader genetic diversity.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
AS: Conceptualization, Formal analysis, Visualization, Writing – original draft, Writing – review & editing. CS: Conceptualization, Visualization, Writing – original draft, Writing – review & editing. RA-S: Conceptualization, Supervision, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Angeles-Shim, R., Angeles, E., Ashikari, M., Takashi, T. (2010). Development and evaluation of Oryza glaberrima Steud. chromosome segment substitution lines (CSSLs) in the background of O. sativa L. cv. Koshihikari. Breed. Sci. 60, 613–619. doi: 10.1270/jsbbs.63.468
Angeles-Shim, R. B., Vinarao, R. B., Marathi, B., Jena, K. K. (2014). Molecular analysis of Oryza latifolia Desv.(CCDD genome)-derived introgression lines and identification of value-added traits for rice (O. sativa L.) improvement. J. Hered. 105, 676–689. doi: 10.1093/jhered/esu032
Bandi, V., Gutwin, C. (2020). Interactive exploration of genomic conservation. Graphics Interface 2020. Available at: https://openreview.net/forum?id=7-C5VJWbnI (Accessed June 10, 2024).
Bessho-Uehara, K., Furuta, T., Masuda, K., Yamada, S., Angeles-Shim, R. B., Ashikari, M., et al. (2017). Construction of rice chromosome segment substitution lines harboring Oryza barthii genome and evaluation of yield-related traits. Breed. Sci. 67, 408–415. doi: 10.1270/jsbbs.17022
Furuta, T., Uehara, K., Angeles-Shim, R. B., Shim, J., Ashikari, M., Takashi, T. (2014). Development and evaluation of chromosome segment substitution lines (CSSLs) carrying chromosome segments derived from Oryza rufipogon in the genetic background of Oryza sativa L. Breed. Sci. 63, 468–475. doi: 10.1270/jsbbs.63.468
Furuta, T., Uehara, K., Angeles-Shim, R. B., Shim, J., Nagai, K., Ashikari, M., et al. (2016). Development of chromosome segment substitution lines harboring Oryza nivara genomic segments in Koshihikari and evaluation of yield-related traits. Breed. Sci. 66, 845–850. doi: 10.1270/jsbbs.16131
Harrison, R. G., Larson, E. L. (2014). Hybridization, introgression, and the nature of species boundaries. J. Heredity. 105, 795–809. doi: 10.1093/jhered/esu033
Khush, G., Bacalangco, E., Ogawa, T. (1990). A new gene for resistance to bacterial blight from O. longistaminata. Rice Genet. News Lett. 7, 121–122.
Khush, G., Brar, D. (1992). Overcoming the barriers in hybridization. Distant hybridization of crop plants. Monographs on TAG. Springer 16, 47–61. doi: 10.1007/978-3-642-84306-8_4
Knight, E., Binnie, A., Draeger, T., Moscou, M., Rey, M.-D., Sucher, J., et al. (2015). Mapping the ‘breaker’element of the gametocidal locus proximal to a block of sub-telomeric heterochromatin on the long arm of chromosome 4S sh of Aegilops sharonensis. Theor. Appl. Genet. 128, 1049–1059. doi: 10.1007/s00122-015-2489-x
Koide, Y., Onishi, K., Nishimoto, D., Baruah, A. R., Kanazawa, A., Sano, Y. (2008). Sex-independent transmission ratio distortion system responsible for reproductive barriers between Asian and African rice species. New Phytol. 179, 888–900. doi: 10.1111/j.1469-8137.2008.02490.x
Kubo, T., Yoshimura, A., Kurata, N. (2011). Hybrid male sterility in rice is due to epistatic interactions with a pollen killer locus. Genetics 189, 1083–1092. doi: 10.1534/genetics.111.132035
Li, J., Zhou, J., Zhang, Y., Yang, Y., Pu, Q., Tao, D. (2020). New insights into the nature of internspecific hybrid sterility in rice. Front. Plant Sci. 11, 555572. doi: 10.3389/fpls.2020.555572
Long, Q., Qiu, S., Man, J., Ren, D., Xu, N., Luo, R. (2023). OsAAI1 increases rice yield and drought tolerance dependent on ABA-mediated regulatory and ROS scavenging pathway. Rice 16, 35. doi: 10.1186/s12284-023-00650-3
Lv, Y., Li, J., Yang, Y., Pu, Q., Zhou, J., Deng, X., et al. (2024). Identification of a novel hybrid sterility locus S67 between temperate japonica subgroup and basmati subgroup in Oryza sativa L. Sci. Rep. 14, 28619. doi: 10.1038/s41598-024-80011-2
Meyer, R. S., Purugganan, M. D. (2013). Evolution of crop species: genetics of domestication and diversification. Nat. Rev. Genet. 14, 840–852. doi: 10.1038/nrg3605
Ramos, J. M., Furuta, T., Uehara, K., Niwa, C., Angeles-Shim, R. B., Shim, J., et al. (2016). Development and evaluation of chromosome segment substitution lines (CSSLs) of Oryza longsitaminata A. Chev. & Röhr in the background of the elite japonica cultivar, Taichung 65. Euphytica 210, 151–163. doi: 10.1007/s10681-016-1685-3
Sano, Y., Chu, Y. E., Oka, H. I. (1979). Genetic studies of speciation in cultivated rice, 1. Genic analysis for the F1 sterility between O. sativa L. and O. glaberrima Steud. Japan. J. Genet. 54, 121–332. doi: 10.1266/jjg.54.121
Septiningsih, E. M., Pamplona, A. M., Sanchez, D. L., Neeraja, C. N., Vergara, G. V., Heuer, S., et al. (2009). Development of submergence-tolerant rice cultivars: the Sub1 locus and beyond. Ann. Bot. 103, 151–160. doi: 10.1093/aob/mcn206
Suarez-Gonzalez, A., Lexer, C., Cronk, Q. C. (2018). Adaptive introgression: a plant perspective. Biol. Lett. 14, 20170688. doi: 10.1098/rsbl.2017.0688
Wang, Y. C., Liu, X. L., Zhang, Z., Zhou, L., Zhang, Y. F., Zhu, B. S., et al. (2024). The residual activity of fatty acyl-coA reductase underlies thermo-sensitive genic male sterility in rice. Plant. Cell Environment. 48 (2), 1273–1285. doi: 10.1111/pce.15230
Xu, P., Zhou, J., Li, J., Hu, F., Deng, X., Feng, S., et al. (2014). Mapping three new interspecific hybrid sterile loci between Oryza sativa and O. glaberrima. Breed. Sci. 63, pp.476–pp.482. doi: 10.1270/jsbbs.63.476
Yang, X., Liang, W., Chen, M., Zhang, D., Zhao, X., Shi, J. (2017). Rice fatty acyl-CoA synthetase OsACOS12 is required for tapetum programmed cell death and male fertility. Planta 246, 105–122. doi: 10.1007/s00425-017-2691-y
Yang, Y., Zhou, J., Li, J., Xu, P., Zhang, Y., Tao, D. (2016). Mapping QTLs for hybrid sterility in three AA genome wild species of Oryza. Breed. Sci. 66, 367–371. doi: 10.1270/jsbbs.15048
Zhang, S., Wu, S., Niu, C., Liu, D., Yan, T., Tian, Y., et al. (2021). ZmMs25 encoding a plastid-localized fatty acyl reductase is critical for anther and pollen development in maize. J. Exp. Bot. 72, 4298–4318. doi: 10.1093/jxb/erab142
Zhang, X., Peng, W., Chen, H., Xing, H. (2024). BnAP2-12 overexpression delays ramie flowering: evidence from AP2/ERF gene expression. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1367837
Keywords: wide hybridization, preferential chromosome transfer, chromosome introgression, gametocidal genes, distant relatives
Citation: Shrestha A, Stephens CJ and Angeles-Shim RB (2025) Taming wild genomes: perspectives on mechanisms governing differential introgression in exotic rice germplasm and their applications in breeding. Front. Plant Sci. 16:1489244. doi: 10.3389/fpls.2025.1489244
Received: 31 August 2024; Accepted: 02 January 2025;
Published: 22 January 2025.
Edited by:
Dayun Tao, Yunnan Academy of Agricultural Sciences, ChinaReviewed by:
Jiawu Zhou, Yunnan Academy of Agricultural Sciences, ChinaCopyright © 2025 Shrestha, Stephens and Angeles-Shim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rosalyn B. Angeles-Shim, cm9zYWx5bi5zaGltQHR0dS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.