- School of Life Sciences, Qufu Normal University, Qufu, Shandong, China
Objective: This study aimed to screen the differentially expressed genes (DEGs) of Gossypium hirsutum under UV-B stress and identify the significant pathways based on gene enrichment analysis results.
Methods: In this study, the allotetraploid crop G. hirsutum was used to examine changes in various physiological indexes under UV-B stress, and screened out all DEGs under UV-B stress (16 kJ m-2 d-1) based on six leaf transcriptomes. The main enrichment pathways of DEGs were analyzed according to gene annotation. Finally, we predicted the regulatory genes of phenylpropanoid pathway under UV-B stress by co-expression network analysis, and selected GhMYB4 for verification.
Results: Gene annotation analysis revealed that DEGs were predominantly enriched in pathways related to photosynthesis and secondary metabolism. Further analysis revealed that UV-B stress impaired photosynthesis mainly by damaging photosystem II (PSII) and inhibiting electron transport, whereas G. hirsutum responded to UV-B stress by synthesizing secondary metabolites such as anthocyanins and lignin. We selected the regulatory genes GhMYB4 for verification. It was found to be an anthocyanin negative regulator in response to UV-B stress.
Conclusions: UV-B stress impaired photosynthesis mainly by damaging photosystem II (PSII) and inhibiting electron transport, whereas G. hirsutum responded to UV-B stress by synthesizing secondary metabolites such as anthocyanins and lignin.
1 Introduction
Ongoing changes in global climate and the stratospheric ozone layer have resulted in changes in temperature, solar ultraviolet radiation, and other factors (Bornman et al., 2020). Based on the wavelength range, UV (ultraviolet) light is divided into three types: UV-A radiations (from 315 to 400 nm), UV-B (from 280 to 315 nm), and UV-C (from 100 to 280 nm) (Iasnogorodskii, 2005). The stratospheric ozone layer can absorb the entire UV-C radiation, most UV-B radiation, and a small portion of UV-A radiation. Although only a small portion of UV-B radiation reaches the Earth’s surface, it can significantly affects plants owing to its high energy (Vanhaelewyn et al., 2020).
Low-level UV-B radiation promotes plant photomorphogenesis and adaptation. However, high-levels of UV-B radiation affect the growth of terrestrial plants, causing DNA damage, reactive oxygen species (ROS) accumulation, and photosynthetic impairment. High-level UV-B radiation, known as UV-B stress, can damage plants and lead to abnormal growth and development. Plant growth, physiological characteristics, and secondary metabolism undergo significant changes under UV-B stress. Excessive UV-B radiation can lead to plant cell death, and its physiological effects include leaf wilting, yellowing, and bleaching (Jenkins, 2009). During photosynthesis, UV-B radiation directly damages photosystem II (PSII) by degrading the PSII proteins D1 and D2 and damaging the photosynthetic machinery (Zhou et al., 2024). In addition to photosystem damage, UV-B can promote chlorophyll degradation and reduce chlorophyll content, resulting in low photosynthetic capacity of plants (Rahimzadeh Karvansara and Razavi, 2019). UV-B exposure inhibits primary root elongation by reducing cell proliferation in the meristematic zone of Arabidopsis (Sheridan et al., 2022). Plant height, tiller number, and root length decreases in both indica and japonica rice plants under UV-B stress (Mathur et al., 2024). Seed germination and seedling development in Anatolian black pines decrease with increasing exposure time to UV radiation (Ozel et al., 2021). UV-B radiation can cause DNA and protein damage at the molecular level. UV-B stress induces DNA damage by producing cyclobutene pyrimidine dimers (CPDs) and pyrimidine (6-4) and pyrimidinone photoproducts (6-4 PPs), that affect DNA replication and transcription, thereby inhibiting plant development and metabolism (Chen et al., 2022). In addition to plant morphology, UV-B radiation also has a significant effect on plant metabolism. Phenylpropanoids, including flavonoids and polyphenols, accumulate in the epidermal cells to protect plants from potentially harmful UV-B radiation. UV-B radiation promotes flavonoid and phenol production (Liu et al., 2020; Lee et al., 2021; Li et al., 2023a). Phenylpropanoid, lignin, flavonoid production, phenolic, flavonoid, antioxidant, and anthocyanin concentrations significantly increase after UV-B irradiation (Lee et al., 2021; Gong et al., 2024). The phenylpropanoid biosynthesis pathway is the general pathway for producing compounds such as lignin, flavonoids, and anthocyanins. The phenylpropanoid pathway has two metabolic branches, flavonoid and lignin biosynthesis, that play a role in the stress response (Li et al., 2023a). UV-B stress promoted lignin accumulation, which can alleviate the damage caused by UV-B stress to Rhododendron chrysanthum (Gong et al., 2024). Morphological dissection revealed that the thickness of quinoa epidermal cell wall increased under UV-B stress due to the accumulation of lignin (Hilal et al., 2004). Lignin deposition in epidermal tissues of quinoa is a resistance mechanism against UV-B radiation. The carboxymethyl cellulose-lignin composite film from oil palm empty fruit bunch prepared by utilizing lignin resistance has excellent ultraviolet resistance (Haqiqi et al., 2021). After UV-B treatment, the anthocyanin content in the leaves of Lycium ruthenicum increased significantly, and the expression of 24 structural genes in anthocyanin synthesis pathway was up-regulated (Chen et al., 2024). Exposure to UV-B increased anthocyanin and flavone levels in the cotyledons of Fagopyrum esculentum, and inhibited hypocotyl elongation (Dębski et al., 2016). Higher resistance to UV-B radiation in F. esculentum may be attributed to its higher anthocyanin content. Plants commonly resist abiotic stress, and the effects of abiotic stress on plants are complex and involve physiological, biochemical, and molecular pathways. Therefore, a comprehensive understanding of these pathways and their interactions is essential for understanding plant stress resistance.
AtMYB4 is a key regulator of phenylpropanoid pathway gene expression, and is the first example of a MYB protein that functions as a transcriptional repressor (Hemm et al., 2001). AtMYB4 is one of the first proteins to transcriptionally regulate general phenylpropanoid pathway genes at the physiological level as a transcriptional repressor. It has shown that Arabidopsis MYB (AtMYB4) regulates the accumulation of the UV-protective compound sinapoylmalate by inhibiting the transcription of the gene encoding the phenylpropyl enzyme cinnamate 4-hydroxylase (C4H) (Hemm et al., 2001). The conclusion was verified in Arabidopsis AtMYB4 mutant. The AtMYB4 mutant line is more tolerant of UV-B irradiation than wild type (Jin, 2000). The increased accumulation of sinapate ester in the mutants was associated with enhanced expression of the gene encoding C4H, thereby increasing the resistance of the mutant (Jin, 2000). The defect in the mutants blocked the conversion of ferulate to L-hydroxyferulate in the general phenylpropanoid pathway (Chapple et al., 1992). As a result, the lignin of the mutant lacks the sinapic acid-derived components. SlMYB7 inhibited anthocyanin accumulation and reduced expression of anthocyanin synthase genes in Solanum lycopersicum fruits (Zhang et al., 2023). Sequence similarity comparison revealed that AtMYB4 and SlMYB7 are homologous genes of G. hirsutum GhMYB4 in Arabidopsis thaliana and Solanum lycopersicum. The transcription factors (TFs) VcMYBPA1, MYBPA2.1, MYB114, MYBA2, MYBF, and MYB102 from blueberry (Vaccinium corymbosum) are activators, whereas MYB20, VcMYB14, MYB44, and VcMYB4a are inhibitors of the flavonoid biosynthetic pathway (Song et al., 2022). BrMYB4 from turnip (Brassica rapa), as a negative transcriptional regulator of BrC4H, mediates UV-B-dependent phenylpropanoid biosynthesis (Zhang et al., 2014). The anthocyanin biosynthesis process is regulated by the MYB-bHLH-WD40 regulatory complexes at the transcription level (Schaart et al., 2012), and the R2R3-MYB subfamily is considered to be the main MYB regulator involved in this pathway. In our previous study, we explored and characterized the potential MYB regulatory genes in the anthocyanin biosynthesis pathway in G. hirsutum by co-expression regulatory network analysis (Zhu and Bao, 2021). It was found that MYBs are involved in the regulation of structural genes and multiple reaction processes of anthocyanin biosynthesis pathway.
With changes in stratospheric ozone and climate over the past few years, crops, including G. hirsutum, face an increasingly pronounced threat from UV-B radiation. G. hirsutum, produced by hybridizing two diploid species with AA and DD genomes, is an important fiber crop (Mansoor and Paterson, 2012). Plant leaves, the primary photosynthetic and transpiration organs, determine radiation interception and play a crucial role in the exchange of heat, water, and gas between plants and the external environment (Taleisnik et al., 2009). This process converts light energy and inorganic nutrients into chemical energy and organic compounds, that serve as cellular components supporting overall plant growth. Due to the large surface area, leaves are particularly vulnerable to biotic and abiotic stresses (Taleisnik et al., 2009). For instance, salinity reduces leaf expansion by affecting cell division and elongation (Taleisnik et al., 2009). Histological studies on Vitis vinifera leaves revealed increased leaf thickness and disordered thylakoid structures under heat stress (Salem-Fnayou et al., 2010). Additionally, water stress decreased stomatal density, size, and aperture in V. vinifera leaves (Ju et al., 2018). In this study, we used G. hirsutum as the experimental material to elucidate gene expression changes and related metabolic pathways under UV-B stress. These analyses provide more insight into the molecular mechanisms underlying the adaptation of polyploid plants to environmental changes. We identified differentially expressed genes (DEGs) and analyzed the associated metabolic pathways in G. hirsutum under UV-B stress through transcriptome analysis. The results offer insights for future research on plant responses to UV-B stress and the selection of tolerant crop varieties.
2 Materials and methods
2.1 Plant materials and growth conditions
From sowing to three-leaf stage, G. hirsutum cv. Acala Maxxa plants were cultivated in greenhouse with a long-day (LD, 16 h light/8 h dark) photoperiod at temperature of 23 ± 1°C in Qufu Normal University, China. Seeds were sown in plastic pots (10×10 cm) containing a mixture of nutrient soil and vermiculite in a ratio of 3:1, with two seeds per pot.
UV-B does simulated 30% depletion of stratospheric ozone (16 kJ m-2 d-1) (Kakani, 2003). To examine the UV-B stress response of the cotton seedlings, each of six plants were placed under normal condition (CK) or 6 h of UV-B radiation (UV-B, 16 kJ m-2 d-1), respectively. The UV-B lamp (Beijing Zhongyi Boteng Technology Co., LTD) was used to generate UV-B radiation. After treatment, the leaves from control and treated plants were collected for all experiments. All the treatments were conducted in triplicates.
The seeds of Arabidopsis thaliana ecotype Columbia (Col-0) were surface sterilized in 10% (v/v) sodium hypochlorite for 20 mins, washed three times with sterilized water, and then grown on medium (pH=5.8) after one day vernalization in darkness at 4°C, and then transferred to long-day (16 h light–8 h dark) growth conditions at 22°C.
2.2 Extraction and quantification of chlorophylls and anthocyanins
Chlorophylls were extracted with 80% (v/v) acetone (Porra et al., 1989). Two leaf disks of 1cm diameter were soaked in 2ml dimethyl sulfoxide (DMSO) at 65 °C in the dark for one hour until all leaves turned white. After cooling to room temperature, 8mL 80% (v/v) acetone was added to extract for 30 mins, and then both extracts (CK and UV-B groups) were analyzed by UV-visible scanning spectrophotometry (UV-5500, METASH, Shanghai, China) at 646.6 and 663.6 nm.
Anthocyanins were extracted with a methanol–HCl method (Kuhnen et al., 2011). The 2 g Maxxa leaves or 0.1g Arabidopsis leaves were soaked in 5 ml or 2ml 1% (v/v) methanol–HCL and extracted overnight at 24°C in darkness. Both extracts (CK and UV-B groups) were analyzed by UV–visible scanning spectrophotometry (UV-5500, METASH, Shanghai, China) at 530, 620, and 650 nm. All measurements were done in triplicate.
2.3 RNA extraction and transcriptome sequencing
Total RNAs extracted from leves of control and UV-B treatment by RNAprep Pure Plant Kits (Cat. No 4992237, TIANGEN, Beijing, China) following the manufacturer’s instructions, respectively. Purity and integrity of RNA were assessed using the RNANano 6000Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). Then, mRNA was purified from total RNA by using poly-T oligo-attached magnetic beads. The cDNA was synthesized through reverse transcription using random hexamer primer. Then the PCR product was purified by AMPure XP beads (BeckmanCoulter, Beverly, USA), and the library was finally obtained. After the library was qualified, six cDNA were sequenced by the Illumina NovaSeq 6000. Library construction and transcriptome sequencing were achieved by Novogene Bioinformatics Technology Co. Ltd (Novogene Co., Ltd., Beijing).
2.4 Gene annotation and DEG analysis
Clean data (clean reads) were obtained by removing reads containing adapter, reads containing Nbase and low quality reads from raw data using fastp (v. 0.19.7) software (Chen et al., 2018). All data were uploaded to NCBI (https://www.ncbi.nlm.nih.gov/) under the accession number PRJNA893188. Paired-end clean reads were aligned to the cotton reference genome (UTX_v2.1) using Hisat2 (v2.0.5) (Mortazavi et al., 2008). The featureCounts v1.5.0-p3 (Liao et al., 2014) was used to count the reads numbers mapped to each gene. And then FPKM (Fragments Per Kilobase of transcript per Million reads mapped) that represented the gene expression value of RNA-seq in subsequent analysis was calculated based on the length of the gene and reads count mapped to this gene.
Differential expression analysis of UV-B and CK groups (three biological replicates per condition) was performed using the DESeq2 R package (1.20.0) (Love et al., 2014). The padj ≤ 0.05 and |log2(foldchange)| ≥ 1 were set as the threshold for significantly differential expression genes.
2.5 Functional annotation and classification
The protein sequence file (Ghirsutum_527_v2.1.protein.fa) was downloaded from the cottongen website (https://www.cottongen.org/), and then submitted to the Mercator 4 website (https://www.plabipd.de/mercator_main.html) to obtain the MapMan mapping file. The log2FoldChange data file of DEGs obtained by comparative transcriptome analysis. The two files were submitted to the MapMan software (Thimm et al., 2004) for gene function classification and comprehensive pathway analysis of DEGs.
Kyoto Encyclopedia of genes and genomes (KEGG) enrichment analysis of DEGs was implemented by the clusterProfiler R package (3.8.1) (Yu et al., 2012). KEGG enrichment analyses used padj ≤ 0.05 as the threshold for significant enrichment. Subsequently, the top 20 most significant pathways of KEGG enrichment analysis were analyzed, respectively.
The structural genes, as target genes, submitted to the ccNET database (http://structuralbiology.cau.edu.cn/gossypium/compare.php) to search for their potential regulatory genes (You et al., 2017). Then, we downloaded all PCC and MR values of co-expression data of the structural genes. Based on the PCC and MR values, we constructed several co-expression regulatory networks to trace back to the regulatory genes in the pathway. Several co-expression networks were integrally generated based on threshold, and were visualized using Cytoscape software (v.3.8.0) (Shannon et al., 2003).
2.6 Real-time quantitative polymerase chain reaction analysis
Total RNA was extracted from leaves and the cDNA was synthesized using the HiScript II 1st Strand cDNA Synthesis Kit (+gDNA wiper) (Cat. No R212-01, Vazyme, Nanjing, Jiangsu Province, China), primers designed with Oligo are shown in Supplementary Table S1. The qRT-PCR experiment was carried out using an CFX Connect Real-time System (BIO-RAD, California, USA) and SYBR Green Premix Pro Taq HS qPCR Kit (Cat. No AG11701, Accurate Biology, Hangzhou, Zhejiang Province, China). The reaction system was as follows: 95°C for 30s, followed by 40 cycles of 95°C for 5 s, and 60°C for 30 s. Each sample was repeated three times, and gene GhPU1 (Gohir.A01G131900) was used as internal control (Xiao et al., 2007). Each primer pair was validated the specifcity by melt curve analysis, and relative expression of these genes were calculated according to the 2–ΔΔCT method (Schmittgen and Livak, 2008). T-test was applied for significant differences among treatments at P < 0.05 level. The results were shown as the means ± standard error of three replicates.
2.7 Functional verification
The anthocyanin biosynthesis process is considered to be regulated by the MYB-bHLH-WD40 regulatory complexes. And our previous study explored the potential MYB genes in anthocyanin biosynthesis in G. hirsutum. Therefore, we selected a MYB gene (Gohir.A01G153200, GhMYB4) for functional verification. The GhMYB4 coding sequences were retrieved from CottonFGD (https://cottonfgd.org/). Genes (Supplementary Table S2) were amplified from the Maxxa cDNA library and the PCR products were cloned into pCAMBIA1300-GFP vector according to the instructions of the ClonExpress II One Step Cloning Kit (Cat. No C112-01/02, Vazyme, Nanjing, Jiangsu Province, China). The vector was subsequently transformed into Arabidopsis Col-0 plants via the Agrobacterium-mediated foral-dip method. The relative expression levels of GhMYB4 and GhC4H genes under UV-B stress and overexpression lines were measured via qRT-PCR (Supplementary Table S2). Each sample was repeated three times, and gene UBQ10 (AT4G05320) was used as internal control.
3 Results
3.1 UV-B treatment triggers physiological changes in G. hirsutum
Chlorophyll and anthocyanin contents in the leaves of G. hirsutum were measured immediately after 6 h of UV-B treatment, and detailed physiological responses were observed. We found that Ca, Cb, and CT contents significantly decreased following UV-B treatment, conversely anthocyanin content significantly increased (Figures 1A, B). Anthocyanins, among the most common stress-resistant compounds in plants, are likely synthesized by G. hirsutum to resist UV-B stress. After two days of UV-B treatment, the leaves exhibited visible signs of drying and damage compared to that of the control plants (Figure 1C). This indicates that UV-B stress (16 kJ m-2 d-1, lasting 6 h) caused severe damage to the cotton plants (Figure 1D).

Figure 1. Stress treatment, and physiological index analysis of G. hirsutum. (A), The chlorophyll content (Ca, Cb, CT) in leaves. (B), The anthocyanins content in leaves. (C, D), Leaves and plant phenotypes under natural condition for 6 h (CK) or UV-B (16 kJ m-2 d-1) (UV-B) for 6 h. Ca, Chlorophyll a; Cb, Chlorophyll b; CT, Total chlorophyll. **P < 0.01.
3.2 Transcriptome analyses and DEGs identification of G. hirsutum under UV-B stress
To investigate the potential pathways and genes of cotton response to UV-B stress, RNA-seq analysis was conducted on leaves under normal growth conditions and UV-B stress (three-leaf stage, UV-B, 16 kJ m-2 d-1). Six high-quality transcriptomes were obtained (Supplementary Table S3). The raw reads of the libraries have been deposited in the NCBI Sequence Read Archive (SRA) database (accession number: PRJNA893188). The FPKM value was calculated for each unigene, and padj ≤ 0.05 and |log2(foldchange)| ≥ 1 were set as the threshold for DEGs. A total of 24,257 DEGs were detected in the CK/UV-B comparison, comprising 12,645 upregulated and 11,612 downregulated.
To ensure the reliability of transcriptome sequencing data, we evaluated the expression patterns of 18 randomly selected genes associated with UV-B stress response using qRT-PCR. These genes showed different functions and were involved in a variety of pathways, including four genes (4CL1, 4CLL9, F6’H1, and PER29) involved in the phenylpropanoid biosynthesis pathway, one gene (CSLG3) involved in cellulose synthesis and 13 TFs (Figure 2). The expression trends of these genes under UV-B stress were highly consistent with the RNA-seq results, confirming the reliability of RNA sequencing.
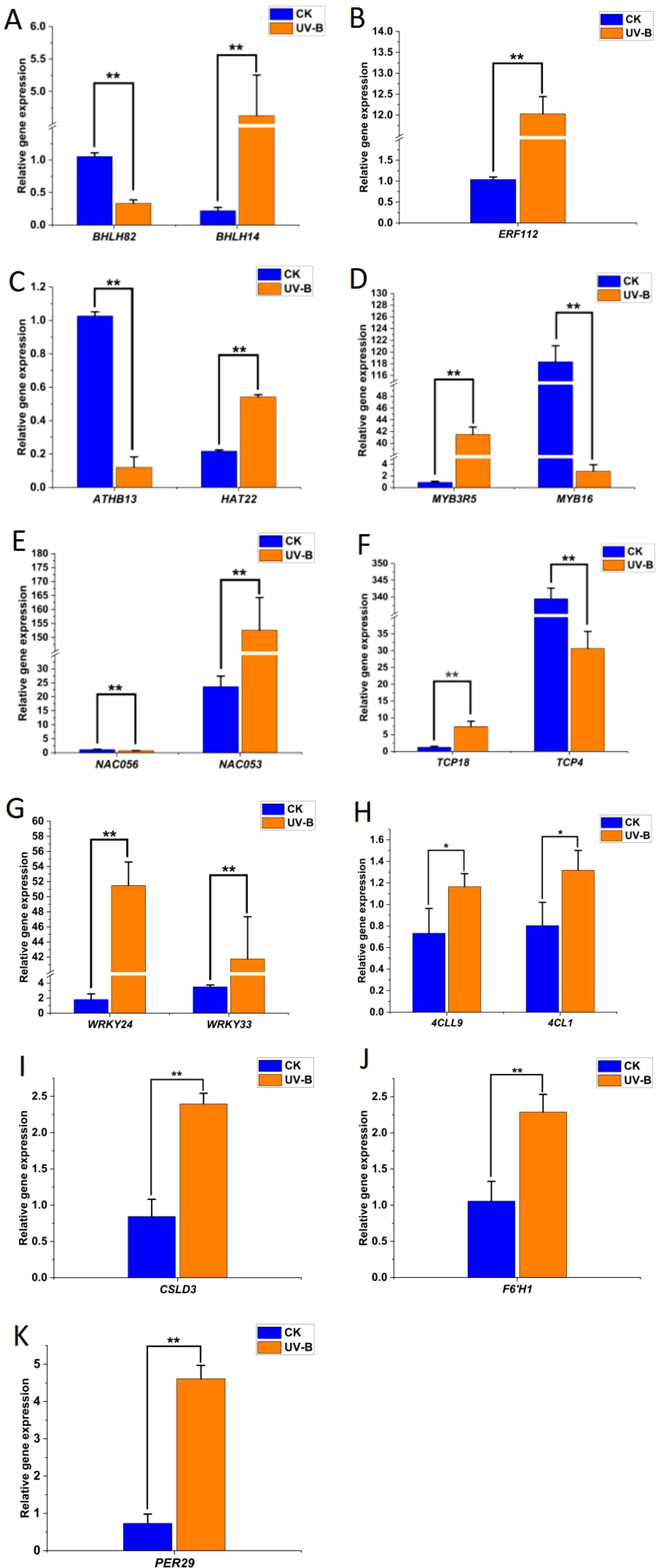
Figure 2. qRT-PCR analysis of expression changes of representative genes in the phenylpropanoid pathway under UV-B stress. (A–K), The relative gene expression of bHLH, ERF, HD-Zip, MYB, NAC, TCP, WRKY, 4CL, CSLD, F6’H, and PER. Statistical significance was calculated using T-test; *P < 0.05, **P < 0.01.
3.3 MapMan analysis of DEGs based on signaling and metabolic pathways
MapMan ontology analysis was employed to construct a genome-wide map of G. hirsutum gene expression by identifying pathways associated with defense-related signaling and metabolic responses. We conducted MapMan analysis of total 24,257 DEGs in the CK vs. UV-B comparison group. Analysis results revealed that genes involved in abiotic stress, pathogen recognition, phytohormones, cell wall metabolism, redox state, metabolic processes, and secondary metabolism (Figure 3). In addition, the DEGs also encoded for transcription factors (TFs) and large enzyme families.
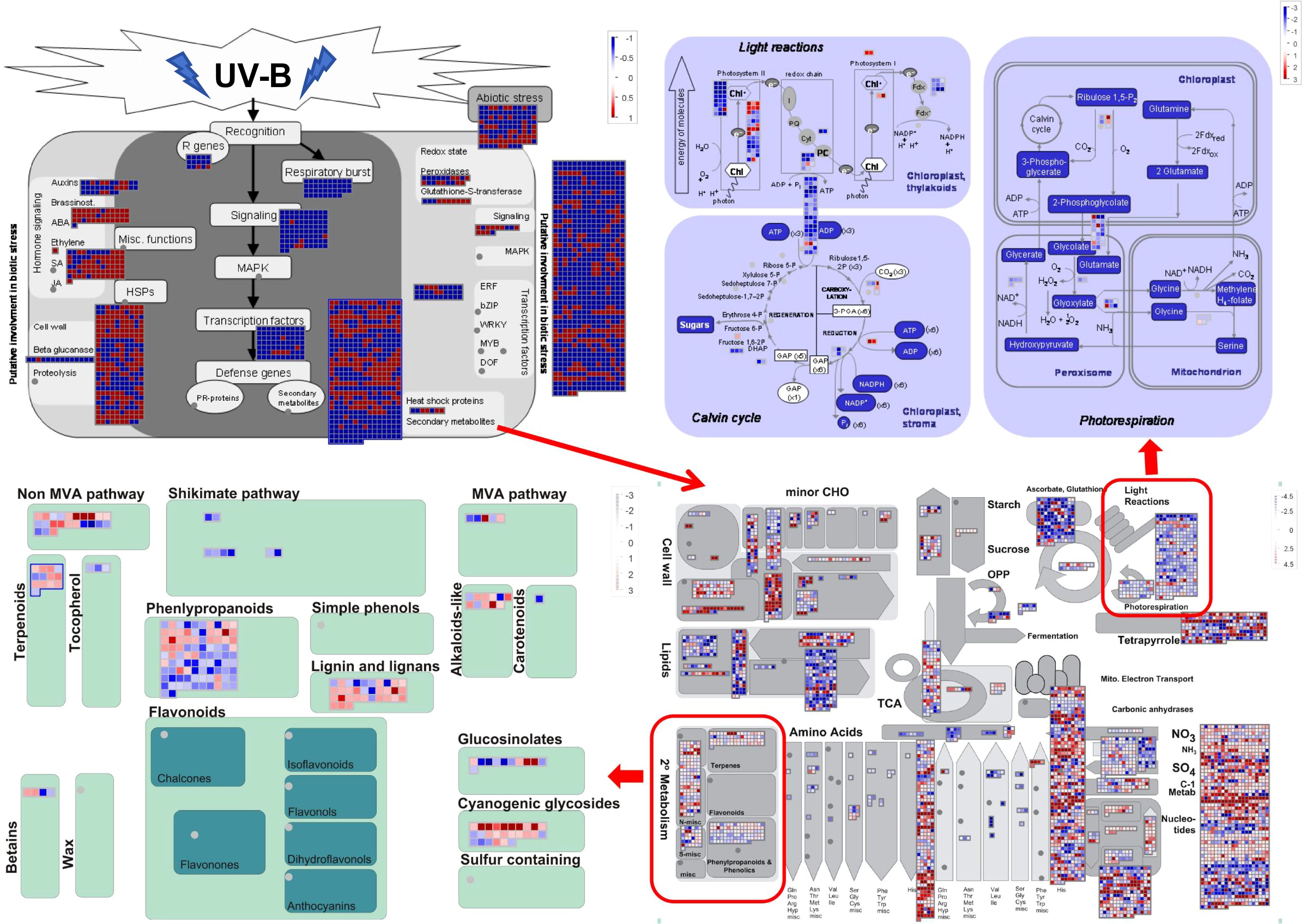
Figure 3. MapMan diagram of G. hirsutum genes involved in the response to UV-B stress. Overview of DEGs expression patterns in UV-B stress leaves relative to untreated controls. Dots show the DEGs encoding proteins related to a certain step in the defense response. Red and blue dots indicate upregulation and downregulation, respectively.
Under UV-B stress, the expression of various genes exhibits distinct regulatory patterns. Genes related to phytohormones (e.g. brassinosteroids, ethylene, and SA), cell wall, abiotic stress, and secondary metabolites (e.g. phenylpropanoid, lignin and lignans, cyanogenic glycosides, terpenoids, and alkaloids) were upregulated under UV-B stress. In contrast, genes associated with other phytohormones (e.g. auxins), respiratory burst, and signaling pathways were downregulated (Figure 3). Similarly, genes encoding TFs, such as the Ethylene Response Factor (ERF), were also downregulated. Analysis of biotic stress pathways revealed that DEGs were mainly enriched in cell wall-related pathways, secondary metabolism, and TFs. Further analysis of the secondary metabolic pathways indicated that the DEGs were primarily enriched in the phenylpropanoid- and lignin-related pathways. The phenylpropanoid pathway serves as an upstream pathway involved in lignin and anthocyanin biosynthesis.
Specifically, coumaroyl-CoA in the phenylpropanoid pathway contributes to flavonoid biosynthesis, while p-coumaryl alcohol, coniferyl-alcohol, 5-hydroxy-coniferyl alcohol, and sinapyl alcohol are involved in synthesizing various types of lignin (H-lignin, G-lignin, and S-lignin) (Figure 4). Similarly, a detailed analysis of the photosynthesis-related DEGs revealed that most of these genes were downregulated (Figure 3). Enrichment analysis indicated that UV-B stress mainly affected PSII and ATP synthesis during light reactions. These light reactions occur in the thylakoid membrane, where photosynthetic pigments absorb light energy for hydrolysis. The downregulated genes inhibit the light reaction process, thereby inhibiting the production of photosynthetic pigments. Therefore, the chlorophyll content decreased significantly after UV-B stress. MapMan analysis demonstrated that UV-B stress influenced photosynthesis in G. hirsutum, mainly affecting PSII and ATP production. It also alleviates the adverse effects of UV-B radiation by generating flavonoids and lignins (Figure 3). Here, We describe the same physiological processes from different perspectives and provide a deeper understanding of the molecular mechanisms underlying UV-B tolerance in G. hirsutum.
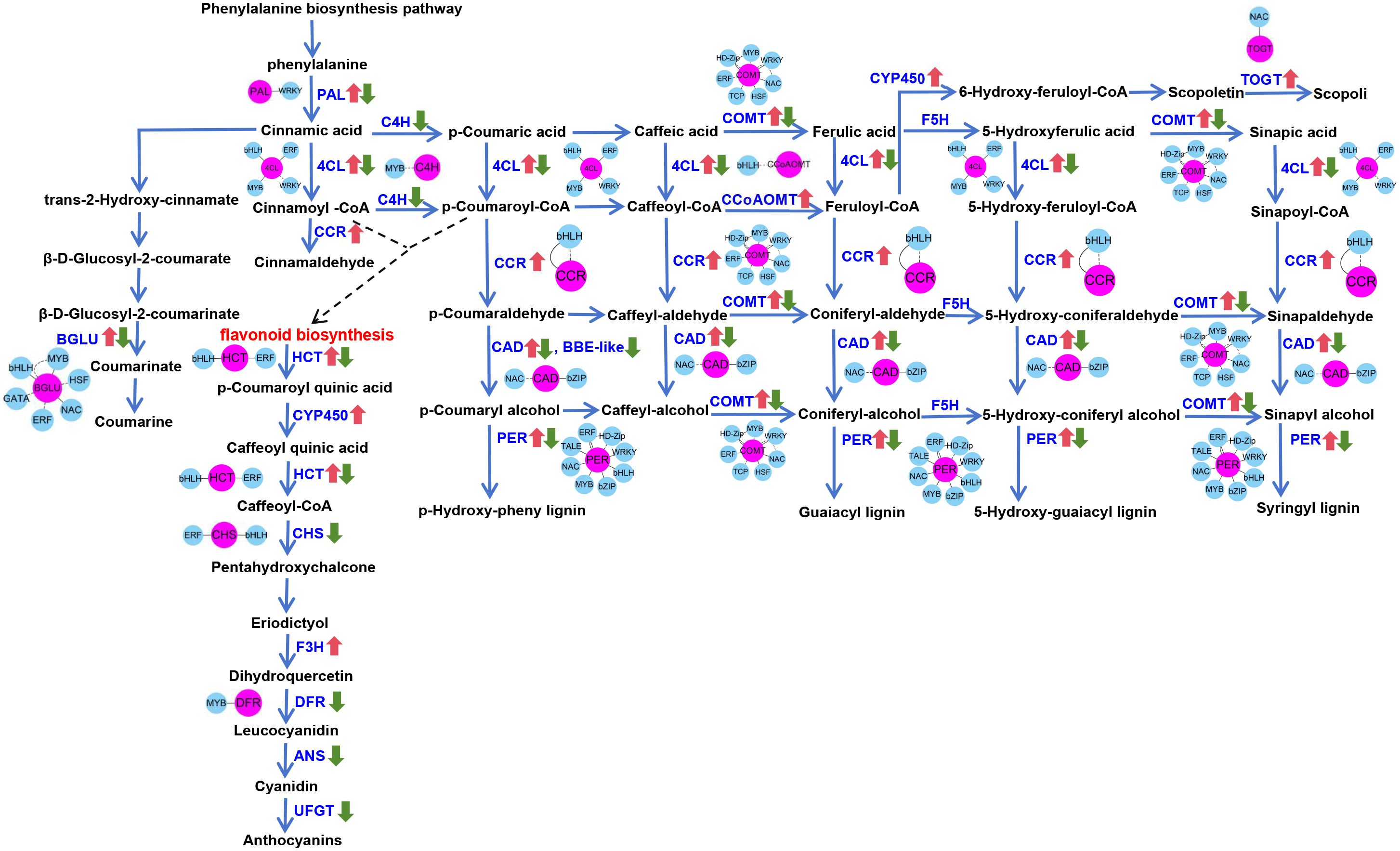
Figure 4. Pathway diagram of structural and regulatory genes related to the phenylpropanoid pathway. The blue words indicate the enzymes encoded by the structural genes. The red arrow indicates the upregulated genes, and the green arrow indicates the downregulated genes. Red dots represent structural genes, and blue dots represent regulatory genes.
3.4 KEGG analysis of DEGs in G. hirsutum under UV-B stress
To further determine the function of DEGs, we performed KEGG enrichment analysis of DEGs. Due to KEGG functional annotations could not be found for some DEGs, these genes were removed from the KEGG enrichment analysis. Under UV-B stress, 4,689 DEGs were eventually annotated with 30 KEGG pathways. The top 20 KEGG terms were screened out based on P-value.
Based on the number of genes, a larger number of genes were found to be enriched in terms of MAPK signaling pathway-plant (ghi04016), starch and sucrose metabolism (ghi00500), amino sugar and nucleotide sugar metabolism (ghi00520), and phenylpropanoid biosynthesis (ghi00940), among others (Supplementary Figure S1). This indicates that UV-B stress affects both primary and secondary metabolism in G. hirsutum. The phenylpropanoid biosynthesis pathway is an upstream process in the synthesis of lignins, flavonoids, and other substances that play important roles in plant responses to abiotic stress. Consistent with the KEGG results, MapMan analysis showed that DEGs were enriched in the phenylpropanoid biosynthesis pathway of the secondary metabolic pathway. Therefore, we analyzed the phenylpropanoid pathway in detail. The DEGs related to phenylpropanoid biosynthesis were mapped using the KEGG database (Figure 4).
Comparative transcriptome analysis revealed that the phenylpropanoid biosynthesis pathway was enriched with a higher number of unigenes (131 DEGs), 69 genes upregulated and 62 genes downregulated (Supplementary Table S4). All the enzymes are encoded by different gene families, each containing more than one gene member. Gene annotation identified 81 DEGs encoding key enzymes, including 4-coumarate–CoA ligase (4CL), berberine bridge enzyme-like (BBE-like), cinnamate 4-monooxygenase (C4H), cinnamyl alcohol dehydrogenase (CAD), caffeoyl-CoA O-methyltransferase (CCoAOMT), cinnamoyl-CoA reductase (CCR), caffeic acid 3-O-methyltransferase (COMT), phenylalanine ammonia lyase (PAL), and peroxidase (PER), all of which are ultimately involved in the synthesis of various types of lignin (H-lignin, G-lignin, S-lignin) and flavonoid bioanabolic pathways (Figure 4). Twenty-seven DEGs were identified that encoded leucoanthocyanidin dioxygenase (ANS), chalcone synthase (CHS), Cytochrome P450 (CYP450), dihydrofavonol-4-reductase (DFR), flavanone 3-hydroxylase (F3H), shikimate O-hydroxycinnamoyltransferase (HCT), and UDP-glucose flavonoid 3-O-glucosyltransferase (UFGT), which are ultimately involved in the anthocyanin biosynthetic pathway (Figure 4). A total of 23 DEGs (Beta-glucosidase BGLU, scopoletin glucosyltransferase TOGT) were involved in the synthesis of the down-stream products of phenylpropanoid metabolism, including coumarin, cinnamaldehyde, and scopolin (Figure 4). The expression patterns of these DEGs varied, and different members of the same gene family had opposite expression patterns under UV-B stress. Among these DEGs, the genes encoding CCoAOMT, CCR, CYP450, F3H, and TOGT were upregulated; the genes encoding ANS, BBE-like, C4H, CHS, DFR, and UFGT were downregulated. But the members encoding 4CL, BGLU, CAD, COMT, HCT, PAL, and PER were both upregulated and downregulated. Changes in the expression levels of these DEGs were associated with different physiological processes. Some of these genes are involved in lignin and flavonoid bioanabolic pathways, which may lead to changes in the content of total lignin, lignin monomers, and anthocyanins. Anthocyanins were the most important flavonoids detected. We determined the anthocyanin content in G. hirsutum leaves and found that it increased significantly under UV-B stress. These results suggest that G. hirsutum responds to UV-B stress by regulating lignin and anthocyanin synthesis, leading to UV-B resistance.
To understand the regulatory networks of the phenylpropanoid metabolism pathway under UV-B stress, we employed co-expression network analysis to identify potential regulatory genes of different structural genes. Structural genes were used as targets to identify potential regulatory genes in this pathway. Finally, 40 structural genes (three 4CL, five BGLU, two C4H, two CAD, one CCoAOMT, two CCR, three CHS, five COMT, three DFR, two HCT, one PAL, ten PER, and one TOGT) were ultimately found to be associated with 60 regulatory genes (13 bHLH, two bZIP, ten ERF, one GATA, six HD-Zip, two HSF, ten MYB, seven NAC, one TALE, two TCP, and six WRKY) (Supplementary Table S5). Among the regulatory genes, members of the MYB gene family were particularly prominent, with GhMYB4 (Gohir.A01G153200) emerging as a key regulator that we previously predicted to regulate anthocyanin biosynthesis. Therefore, we selected a MYB family gene (GhMYB4, Gohir.A01G153200) for functional verification by Arabidopsis transformation.
3.5 Overexpression of GhMYB4 reduced the UV-B tolerance in Arabidopsis
Co-expression network analysis revealed that GhMYB4 was associated with GhC4H (Gohir.A13G235200). qRT-PCR analysis revealed that the relative expression levels of GhMYB4 and GhC4H significantly decreased under UV-B stress (Figure 5A). To further examine the role of GhMYB4 in response to UV-B stress, we produced Arabidopsis lines overexpression (OE) GhMYB4 and analyzed their physiology. We assessed the transgenic efficiency in homozygous transgenic Arabidopsis lines from the T3 generation (OE-6) (Figure 5B). The growth of GhMYB4 overexpression lines (GhMYB4-OE4 and GhMYB4-OE6) and Col-0 plants was compared following UV-B treatment. There was almost no difference in the growth of Col-0, OE4, and OE6 plants under normal growth conditions (Figure 5C). However, the growth of OE4 and OE6 plants was significantly slower than that of Col-0 plants after UV-B treatment (Figure 5C). The anthocyanin content of OE4, OE6, and Col-0 plants after UV-B treatment was also compared (Figure 5D). The anthocyanin content of the GhMYB4-OE lines decreased significantly under both normal growth conditions and UV-B stress (Figure 5E). The relative expression levels of genes AtC4H, AtANS, AtCHS, and GhMYB4 related to phenylpropanoid biosynthesis pathway were quantified. The relative gene expression of AtC4H, AtANS, and AtCHS was all downregulated in overexpression lines (OE4 and OE6) compared to CK (Figure 5F). These findings suggested that GhMYB4 reduces UV-B stress tolerance in Arabidopsis by negatively regulating anthocyanin synthesis.
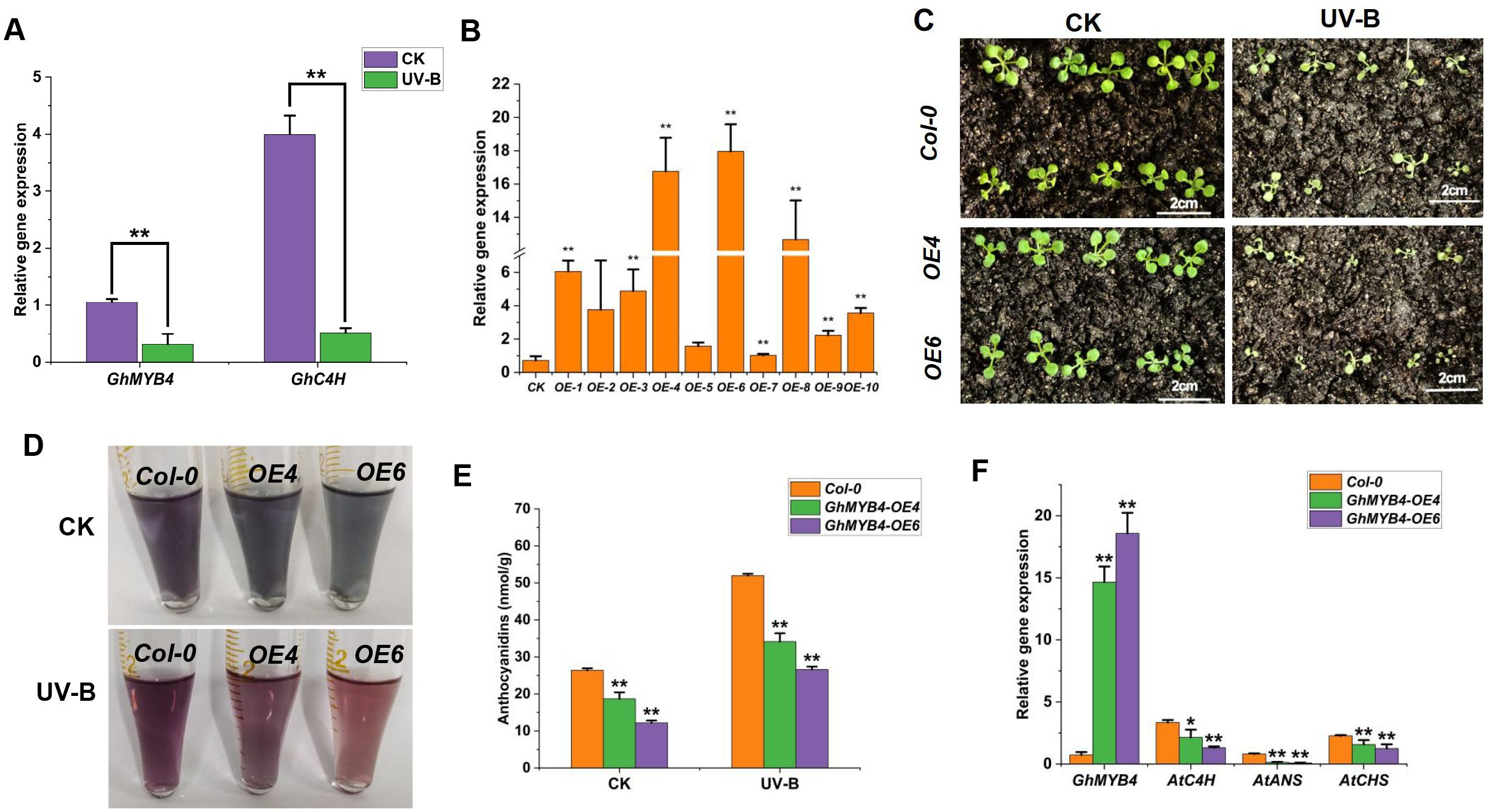
Figure 5. The phenotype and anthocyanins content of Arabidopsis transgenic overexpression lines under UV-B stress. (A), The relative gene expression of GhMYB4 and its associated gene GhC4H under UV-B stress. (B), qRT-qPCR detection of relative expression of GhMYB4 in WT and GhMYB4-OE transgenic lines. (C), The growth of Col-0, OE-4, and OE6 under normal growth conditions (CK) and UV-B stress. (D, E), Detection of anthocyanin relative content in Col-0, OE-4, and OE-6 under UV-B stress. (F), The relative gene expression of AtC4H, AtANS, and AtCHS in overexpression lines (OE4 and OE6) compared to CK. Statistical significance was calculated using T-test; *P < 0.05, **P < 0.01.
4 Discussion
Under UV-B stress, the leaves of G. hirsutum exhibited obvious phenotypic damage and physiological responses. Plants have evolved various strategies to adapt to abiotic stress. These mechanisms involve complex physiological, biochemical, and molecular responses affecting plant growth. Therefore, a comprehensive understanding of these pathways and their interactions is crucial. Through comparative transcriptome analysis, we identified DEGs in G. hirstium under UV-B stress and conducted enrichment analyses and functional studies of these genes.
4.1 UV-B stress weakened the photosynthesis of G. hirsutum
It was found that UV-B stress damaged the photosynthetic system, significantly decreasing the chlorophyll content of G. hirsutum (Figure 6). This phenomenon has been observed in various plants, where enhanced UV-B exposure negatively affects chlorophyll content (Rahimzadeh Karvansara and Razavi, 2019; Tripathi et al., 2021). Chloroplasts are crucial organelles involved in plant photosynthesis and environmental sensing. Photoreactions occur on the thylakoid membrane, where light-excited electrons are transferred from the reaction center to chlorophyll (Rochaix, 2011). Light-driven charge separation occurs in PSII and PSI, where electrons flow along the electron transport chain and enter the thylakoid lumen via proton pumps (Rochaix, 2011). Subsequently, a proton motive force is generated, driving ATP synthesis. UV-B stress weakens photosynthesis by damaging the PSII electron donors, inhibiting electron transfer, and reducing the efficiency of light energy utilization in photosynthetic organs (Zhou et al., 2024). Enrichment analysis revealed that UV-B stress altered the expression of genes related to photosystem and light responses. This may weaken photosynthesis by damaging PSII, inhibiting electron transfer, and reducing the efficiency of light energy utilization (Figure 3). The Calvin cycle plays a vital role in photosynthesis. A previous study showed that the dihydroxyacetone phosphate content in the Calvin cycle significantly increased after UV-B stress (Zhou et al., 2024). Dihydroxyacetone phosphate reacts with glyceraldehyde 3-phosphate to form D-fructose 1, 6-diphosphate. UV-B radiation also induces amino acid accumulation in Rhododendron chrysanthum (Zhou et al., 2024). The findings of this study are consistent with those of previous studies, showing that enhanced UV-B radiation negatively influences photosynthetic mechanisms and chlorophyll content.
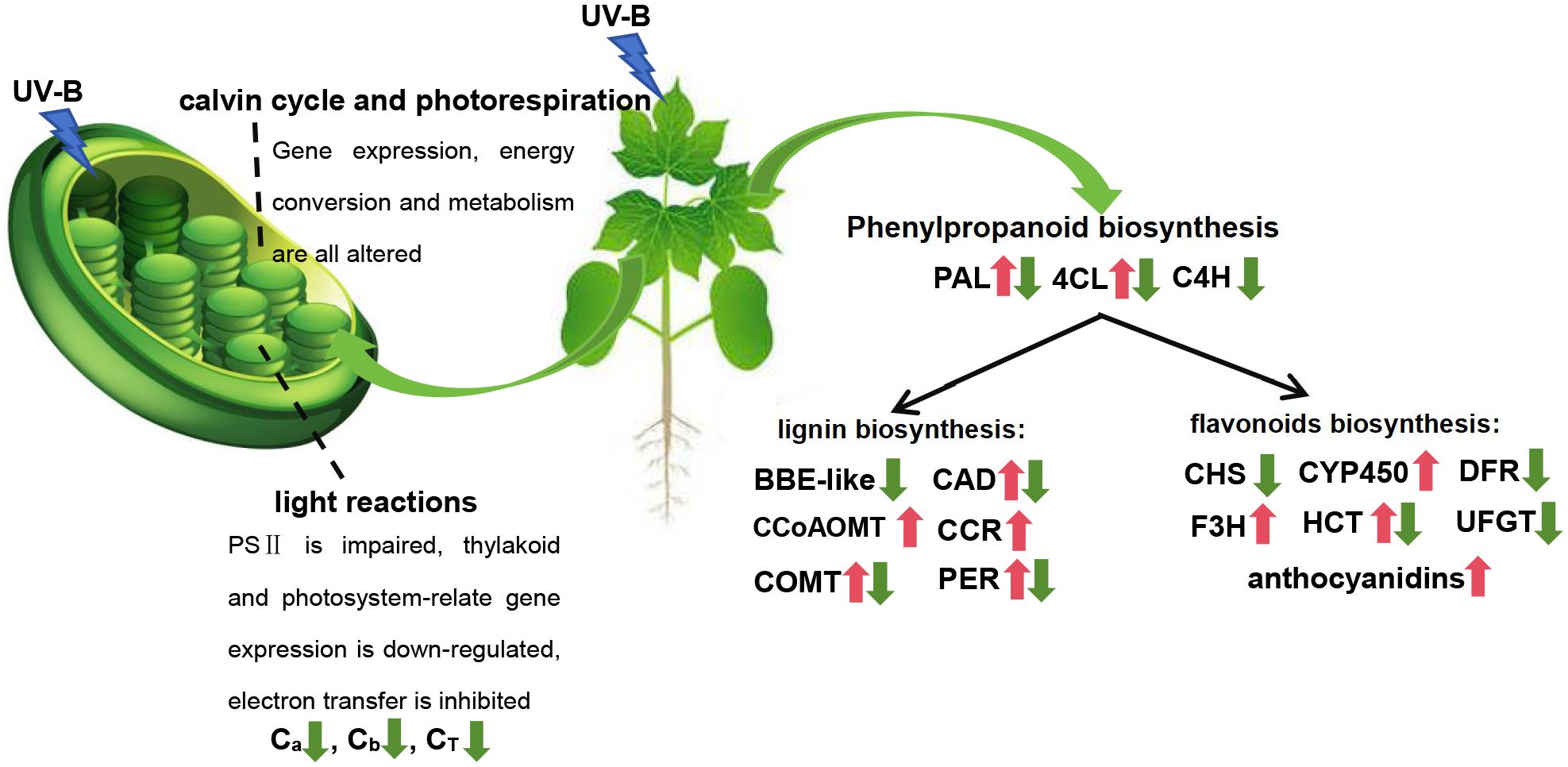
Figure 6. Response of G. hirsutum leaves to UV-B stress. The red arrow indicates upregulation and the green arrow indicates downregulation.
4.2 UV-B stress affected two common metabolic branches of the phenylpropanoid pathway: flavonoids and lignin biosynthesis pathway
The levels of stress-resistant compounds, including phenylpropanoids, flavonoids, phenols, carotenoids, and anthocyanins, significantly increased under UV-B stress (Lee et al., 2021; Tripathi et al., 2021). We also found that the anthocyanin content significantly increased after UV-B treatment compared to that in the control. This finding was consistent with the that of the previous studies. To further investigate the formation and regulatory mechanisms of these stress-resistant compounds, a model was constructed using MapMan and KEGG analysis to visualize the expression of DEGs involved in defense-related signaling and metabolic pathways in G. hirsutum under UV-B stress. MapMan analysis revealed significant enrichment of DEGs associated with secondary metabolism. Further analysis of secondary metabolism-related pathways revealed that the DEGs were primarily enriched in phenylpropanoids, lignin and lignans (Figure 6). The phenylpropanoid pathway is also involved in plant stress resistance. It plays a critical role in regulating drought resistance during foxtail millet germination (Yu et al., 2020). The accumulated metabolites and DEGs associated with phenylpropanoid biosynthesis were significantly altered in tomato roots under salt stress (Jia et al., 2022). Quantification of cell morphology revealed that the cultures supplemented with phenylpropanoid-related precursors in vitro suffered less damage than the control group under stress (Commisso et al., 2016). Similarly in this study, DEGs associated with phenylpropanoid metabolism were enriched using MapMan secondary metabolic pathway analysis. Additionally, the phenylpropanoid biosynthesis pathway was significantly enriched in the KEGG analysis, suggesting that phenylpropanoid biosynthesis may play a typical role in the resistance to different plant stresses. The phenylpropanoid biosynthesis pathway is a precursor pathway for synthesizing various substances that produce many secondary metabolites, such as coumarin, lignin, methyleugenol, flavonoids, and anthocyanins. These secondary metabolites are also stress-resistant.
The flavonoid and lignin biosynthesis pathways, crucial in stress response, are two common metabolic branches of the phenylpropanoid pathway (Li et al., 2023a). Flavonoid and lignin biosynthesis share the same precursors and early steps (Figure 4). The common precursors are phenylalanine, cinnamic acid, cinnamoyl-CoA, p-coumaric acid, and p-coumaroyl-CoA (Figure 4). Enzymes such as 4CL, C4H, and PAL, transform and generate substances during this process. Lignin is a complex biopolymer that typically surrounds the polysaccharide components of plant cell walls (Zhong et al., 2010). As a mechanical tissue, it provides structural support and protection. Lignin biosynthesis is an important step in the phenylpropanoid metabolic pathway and plays an essential role in responding to various abiotic stresses. Various abiotic stressors, such as salt stress, drought, UV-B radiation, and low temperatures, induce changes in lignin content of several plants (Zhao et al., 2021; Li et al., 2023a, 2023b; Yang et al., 2023). In Medicago sativa, the phenylpropanoid pathway regulates lignin biosynthesis in response to osmotic stress (Yang et al., 2023). Similarly, the lignin and flavonoid biosynthesis pathways contribute to the response of Morus alba to UV-B stress (Li et al., 2023a). Changes in lignin content are attributed to variations in the levels of phenylpropanoid lignin precursors and the activities of related enzymes (Moura et al., 2010). Osmotic stress enhances lignin accumulation by increasing the guaiacyl and syringyl content, associated with alterations in the activity of enzymes involved in the metabolism of eight intermediate metabolites (Yang et al., 2023). PoCCoAOMT promoted lignin synthesis and ROS scavenging to resist drought stress in P. ostia (Zhao et al., 2021). PAL, a key enzyme in plant resistance to abiotic stress, mediates the drought tolerance in Fritillaria unibracteata by inducing the biosynthesis and accumulation of salicylic acid and lignin (Qin et al., 2022). Eight structural genes (4CL, CAD, CCR, COMT, F5H, CYP73A, CCoAOMT, and C3′H) were identified as significant regulators of lignin biosynthesis (Li et al., 2023b). In this study, we observed changes in the expression of various structural genes (4CL, BBE-like, C4H, CAD, CCoAOMT, CCR, COMT, PAL, and PER) involved in lignin synthesis, indicating that lignin biosynthesis undergoes systematic changes under stressful conditions (Figure 6). Genes encoding enzymes associated with secondary metabolic pathways were significantly altered, resulting in changes in compound levels.
The flavonoid biosynthetic pathway is an important branch of the phenylpropanoid metabolic pathway. Plants accumulate flavonoids in response to biological stressors (Li et al., 2023a). Flavonoids, a diverse class of phenylpropanoids, are believed to resist the penetration of UV-B radiation into sensitive leaf tissues and are involved in protecting plants from damage caused by stratospheric ozone layer depletion. Anthocyanins, a group of flavonoid pigments, are crucial for pollination by absorbing light and protecting plants from UV radiation-induced damage and cold stress (Mattioli et al., 2020). Environmental stimuli and phytohormones promote anthocyanin biosynthesis. These pigments serve as stress protectants, induced by biotic and abiotic stress conditions, and help regulate stress-induced damage. Anthocyanin content increases under high light stress to improve the tolerance in Brassica napus (Luo et al., 2021). Additionally, anthocyanin biosynthesis is enhanced by altering the expression levels of synthase-related genes, thereby improving cold and freezing stress tolerance in Brassica rapa (Ahmed et al., 2014). Similarly, the induction of anthocyanin accumulation can improve the tolerance of various plants to drought, salinity, heavy metals, nutrient limitations, and diseases (Naing and Kim, 2021; Xiang et al., 2021; Shi et al., 2023). Anthocyanin accumulation is often associated with the expression of enzymatic modification genes. Similar gene sequences from different species were compared, and their respective functions revealed a high degree of homology (Ahmed et al., 2014). In this study, we found that the expression of enzymatic modification genes (CHS, CYP450, DFR, F3H, HCT, and UFGT) related to anthocyanin biosynthesis was upregulated or downregulated under UV-B stress (Figure 6). These findings suggest that the anthocyanin biosynthetic pathways of different species respond similarly to various stress conditions.
4.3 MYB regulators play an important role in anthocyanin and flavonoid biosynthesis
Phenylpropanoids, the largest class of natural products including flavonoids, anthocyanins, lignins and tannins, perform multiple functions including biotic and abiotic stress responses (Pratyusha and Sarada, 2022). The plant-specific transcription factor MYB regulates the expression of structural genes involved in anthocyanin and flavonoid biosynthesis. Recent studies have revealed that MYBs can positively or negatively regulate anthocyanin biosynthesis. MYB4 acts as a transcriptional repressor of key flavonoid biosynthesis genes and reduces flavonoid accumulation following UV-B exposure (Zhang et al., 2014; Banerjee et al., 2024). Additionally, it has been found that MYB20, MYB14, MYB44 and MYB4a inhibit the flavonoid biosynthesis pathway in blueberry under UV-B radiation (Song et al., 2022). LcMYBx competes with LcMYB1 for LcbHLHs, preventing the activation of LcDFR by LcMYB1-LcbHLHs complex, and negatively regulating anthocyanin biosynthesis (Zhao et al., 2022). BrMYBL2.1-G actively inhibits pigment accumulation by inhibiting the transcription of anthocyanin biosynthesis genes (Kim et al., 2022). In our previous study, 31 potential R2R3-MYB regulatory genes involved in the anthocyanin biosynthesis pathway of G. hirsutum were predicted using co-expression network analysis (Zhu and Bao, 2021). Our results are consistent with those of previous studies showing that GhMYB4 directly regulates the expression of C4H in the phenylpropanoid and flavonoid pathways, ultimately affecting anthocyanin synthesis. However, the functions of other predicted regulatory genes require further verification.
In summary, the MapMan and KEGG enrichment analyses demonstrated that UV-B stress affected the morphology, physiology, and secondary metabolism of G. hirsutum. Further analysis revealed that G. hirsutum primarily responded to UV-B stress by affecting the phenylpropanoid metabolic pathway. This pathway played a key role in UV-B stress resistance by producing necessary precursors of lignin and flavonoids biosynthesis, ultimately leading to the synthesis of lignin and anthocyanidins, respectively. Our findings further revealed the complex physiological and metabolic changes, and intricate regulatory networks, involved in G. hirsutum’s response to UV-B stress. GhMYB4 was selected for Arabidopsis transformation, and was found to respond to UV-B stress by negatively regulating anthocyanin synthesis. Further functional characterization of other candidate genes involved in the defense of G. hirsutum against UV-B stress is warranted. This study provides a valuable foundation for further analyses of the molecular mechanisms underlying UV-B stress resistance in G. hirsutum and offers insights for breeding stress-resistant G. hirsutum cultivars.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
XS: Conceptualization, Data curation, Formal analysis, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. YZ: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Writing – original draft. YB: Conceptualization, Data curation, Methodology, Validation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. We thank the National Natural Science Foundation of China (32070246) for providing the funds necessary for conducting this research.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1529912/full#supplementary-material
Supplementary Figure 1 | KEGG analysis (top 20 terms) of DEGs in G. hirsutum under UV-B stress.
References
Ahmed, N. U., Park, J.-I., Jung, H.-J., Hur, Y., Nou, I.-S. (2014). Anthocyanin biosynthesis for cold and freezing stress tolerance and desirable color in Brassica rapa. Funct. Integr. Genomics 15, 383–394. doi: 10.1007/s10142-014-0427-7
Banerjee, S., Agarwal, P., Choudhury, S. R., Roy, S. (2024). MYB4, a member of R2R3-subfamily of MYB transcription factor functions as a repressor of key genes involved in flavonoid biosynthesis and repair of UV-B induced DNA double strand breaks in Arabidopsis. Plant Physiol. Biochem. 211, 108698. doi: 10.1016/j.plaphy.2024.108698
Bornman, J. F., Barnes, P. W., Robson, T. M., Robinson, S. A., Jansen, M. A. K., Ballaré, C. L., et al. (2020). Linkages between stratospheric ozone, UV radiation and climate change and their implications for terrestrial ecosystems. Photochem. Photobiol. Sci. 18, 681–716. doi: 10.1039/c8pp90061b
Chapple, C. C., Vogt, T., Ellis, B. E., Somerville, C. R. (1992). An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell. 4, 1413–1424. doi: 10.1105/tpc.4.11.1413
Chen, S., Xu, Y., Zhao, W., Shi, G., Wang, S., He, T. (2024). UV-B irradiation promotes anthocyanin biosynthesis in the leaves of Lycium ruthenicum Murray. PeerJ. 12, e18199. doi: 10.7717/peerj.18199
Chen, S., Zhou, Y., Chen, Y., Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Chen, Z., Dong, Y., Huang, X. (2022). Plant responses to UV-B radiation: signaling, acclimation and stress tolerance. Stress Biol. 2, 51. doi: 10.1007/s44154-022-00076-9
Commisso, M., Toffali, K., Strazzer, P., Stocchero, M., Ceoldo, S., Baldan, B., et al. (2016). Impact of phenylpropanoid compounds on heat stress tolerance in carrot cell cultures. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01439
Dębski, H., Szwed, M., Wiczkowski, W., Szawara-Nowak, D., Bączek, N., Horbowicz, M. (2016). UV-B radiation increases anthocyanin levels in cotyledons and inhibits the growth of common buckwheat seedlings. Acta Biol. Hung. 67, 403–411. doi: 10.1556/018.67.2016.4.6
Gong, F., Yu, W., Cao, K., Xu, H., Zhou, X. (2024). RcTRP5 Transcription Factor Mediates the Molecular Mechanism of Lignin Biosynthesis Regulation in R. chrysanthum against UV-B Stress. Int. J. Mol. Sci. 25, 9205. doi: 10.3390/ijms25179205
Haqiqi, M. T., Bankeeree, W., Lotrakul, P., Pattananuwat, P., Punnapayak, H., Ramadhan, R., et al. (2021). Antioxidant and UV-blocking properties of a carboxymethyl cellulose–lignin composite film produced from oil palm empty fruit bunch. ACS Omega. 6, 9653–9666. doi: 10.1021/acsomega.1c00249
Hemm, M. R., Herrmann, K. M., Chapple, C. (2001). AtMYB4: a transcription factor general in the battle against UV. Trends Plant Sci. 6, 135–136. doi: 10.1016/s1360-1385(01)01915-x
Hilal, M., Parrado, M. F., Rosa, M., Gallardo, M., Orce, L., Massa, E. M., et al. (2004). Epidermal lignin deposition in quinoa cotyledons in response to UV-B radiation¶. Photochem. Photobiol. 79, 205–210. doi: 10.1562/0031-8655(2004)079<0205:Eldiqc>2.0.Co;2
Iasnogorodskii, V. G. (2005). Ultraviolet radiation. Vopr Kurortol Fizioter Lech Fiz Kult. 1, 30–31.
Jenkins, G. I. (2009). Signal transduction in responses to UV-B radiation. Annu. Rev. Plant Biol. 60, 407–431. doi: 10.1146/annurev.arplant.59.032607.092953
Jia, C., Guo, B., Wang, B., Li, X., Yang, T., Li, N., et al. (2022). Integrated metabolomic and transcriptomic analysis reveals the role of phenylpropanoid biosynthesis pathway in tomato roots during salt stress. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1023696
Jin, H. (2000). Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19, 6150–6161. doi: 10.1093/emboj/19.22.6150
Ju, Y.-l., Yue, X.-F., Zhao, X.-F., Zhao, H., Fang, Y.-L. (2018). Physiological, micro-morphological and metabolomic analysis of grapevine (Vitis vinifera L.) leaf of plants under water stress. Plant Physiol. Biochem. 130, 501–510. doi: 10.1016/j.plaphy.2018.07.036
Kakani, V. G. (2003). Effects of ultraviolet-B radiation on cotton (Gossypium hirsutum L.) morphology and anatomy. Ann. Bot. 91, 817–826. doi: 10.1093/aob/mcg086
Kim, J., Kim, D.-H., Lee, J.-Y., Lim, S.-H. (2022). The R3-type MYB transcription factor BrMYBL2.1 negatively regulates anthocyanin biosynthesis in Chinese cabbage (Brassica rapa L.) by repressing MYB–bHLH–WD40 complex activity. Int. J. Mol. Sci. 23, 3382. doi: 10.3390/ijms23063382
Kuhnen, S., Menel Lemos, P. M., Campestrini, L. H., Ogliari, J. B., Dias, P. F., Maraschin, M. (2011). Carotenoid and anthocyanin contents of grains of Brazilian maize landraces. J. Sci. Food Agric. 91, 1548–1553. doi: 10.1002/jsfa.4346
Lee, J.-H., Shibata, S., Goto, E. (2021). Time-course of changes in photosynthesis and secondary metabolites in canola (Brassica napus) under different UV-B irradiation levels in a plant factory with artificial light. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.786555
Li, Y., Liu, S., Zhang, D., Liu, A., Zhu, W., Zhang, J., et al. (2023a). Integrative Omic Analysis Reveals the Dynamic Change in Phenylpropanoid Metabolism in Morus alba under Different Stress. Plants (Basel). 12, 3265. doi: 10.3390/plants12183265
Li, Y., Zhang, T., Kang, Y., Wang, P., Yu, W., Wang, J., et al. (2023b). Integrated metabolome, transcriptome analysis, and multi-flux full-length sequencing offer novel insights into the function of lignin biosynthesis as a Sesuvium portulacastrum response to salt stress. Int. J. Biol. Macromol. 237, 124222. doi: 10.1016/j.ijbiomac.2023.124222
Liao, Y., Smyth, G. K., Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30, 923–930. doi: 10.1093/bioinformatics/btt656
Liu, Y., Liu, J., Abozeid, A., Wu, K.-X., Guo, X.-R., Mu, L.-Q., et al. (2020). UV-B radiation largely promoted the transformation of primary metabolites to phenols in astragalus mongholicus seedlings. Biomolecules. 10, 504. doi: 10.3390/biom10040504
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Luo, Y., Teng, S., Yin, H., Zhang, S., Tuo, X., Tran, L.-S. P. (2021). Transcriptome analysis reveals roles of anthocyanin- and jasmonic acid-biosynthetic pathways in rapeseed in response to high light stress. IInt J. Mol. Sci. 22, 13027. doi: 10.3390/ijms222313027
Mansoor, S., Paterson, A. H. (2012). Genomes for jeans: cotton genomics for engineering superior fiber. Trends Biotechnol. 30, 521–527. doi: 10.1016/j.tibtech.2012.06.003
Mathur, S., Bheemanahalli, R., Jumaa, S. H., Kakar, N., Reddy, V. R., Gao, W., et al. (2024). Impact of ultraviolet-B radiation on early-season morpho-physiological traits of indica and japonica rice genotypes. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1369397
Mattioli, R., Francioso, A., Mosca, L., Silva, P. (2020). Anthocyanins: A comprehensive review of their chemical properties and health effects on cardiovascular and neurodegenerative diseases. Molecules. 25, 3809. doi: 10.3390/molecules25173809
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Moura, J. C. M. S., Bonine, C. A. V., De Oliveira Fernandes Viana, J., Dornelas, M. C., Mazzafera, P. (2010). Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol. 52, 360–376. doi: 10.1111/j.1744-7909.2010.00892.x
Naing, A. H., Kim, C. K. (2021). Abiotic stress-induced anthocyanins in plants: Their role in tolerance to abiotic stresses. Physiol. Plant 172, 1711–1723. doi: 10.1111/ppl.13373
Ozel, H. B., Abo Aisha, A. E. S., Cetin, M., Sevik, H., Zeren Cetin, I. (2021). The effects of increased exposure time to UV-B radiation on germination and seedling development of Anatolian black pine seeds. Environ. Monit Assess. 193, 388. doi: 10.1007/s10661-021-09178-9
Porra, R. J., Thompson, W. A., Kriedemann, P. E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochimica et Biophysica Acta. 975, 384–394. doi: 10.1016/S0005-2728(89)80347-0
Pratyusha, D. S., Sarada, D. V. L. (2022). MYB transcription factors—master regulators of phenylpropanoid biosynthesis and diverse developmental and stress responses. Plant Cell Rep. 41, 2245–2260. doi: 10.1007/s00299-022-02927-1
Qin, Y., Li, Q., An, Q., Li, D., Huang, S., Zhao, Y., et al. (2022). A phenylalanine ammonia lyase from Fritillaria unibracteata promotes drought tolerance by regulating lignin biosynthesis and SA signaling pathway. Int. J. Biol. Macromol. 213, 574–588. doi: 10.1016/j.ijbiomac.2022.05.161
Rahimzadeh Karvansara, P., Razavi, S. M. (2019). Physiological and biochemical responses of sugar beet (Beta vulgaris L) to ultraviolet-B radiation. PeerJ. 7, e6790. doi: 10.7717/peerj.6790
Rochaix, J.-D. (2011). Regulation of photosynthetic electron transport. Biochim. Biophys. Acta 1807, 375–383. doi: 10.1016/j.bbabio.2010.11.010
Salem-Fnayou, A. B., Bouamama, B., Ghorbel, A., Mliki, A. (2010). Investigations on the leaf anatomy and ultrastructure of grapevine (Vitis vinifera) under heat stress. Microsc Res. Tech. 74, 756–762. doi: 10.1002/jemt.20955
Schaart, J. G., Dubos, C., Romero de la Fuente, I., van Houwelingen, A. M. M. L., de Vos, R. C. H., Jonker, H. H., et al. (2012). Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria × ananassa) fruits. New Phytol. 197, 454–467. doi: 10.1111/nph.12017
Schmittgen, T. D., Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Sheridan, M. L., Simonelli, L., Giustozzi, M., Casati, P. (2022). Ultraviolet-B radiation represses primary root elongation by inhibiting cell proliferation in the meristematic zone of arabidopsis seedlings. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.829336
Shi, L., Li, X., Fu, Y., Li, C. (2023). Environmental stimuli and phytohormones in anthocyanin biosynthesis: A comprehensive review. Int. J. Mol. Sci. 24, 16415. doi: 10.3390/ijms242216415
Song, Y., Ma, B., Guo, Q., Zhou, L., Lv, C., Liu, X., et al. (2022). UV-B induces the expression of flavonoid biosynthetic pathways in blueberry (Vaccinium corymbosum) calli. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1079087
Taleisnik, E., Rodríguez, A. A., Bustos, D., Erdei, L., Ortega, L., Senn, M. E. (2009). Leaf expansion in grasses under salt stress. J. Plant Physiol. 166, 1123–1140. doi: 10.1016/j.jplph.2009.03.015
Thimm, O., Bläsing, O., Gibon, Y., Nagel, A., Meyer, S., Krüger, P., et al. (2004). mapman: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939. doi: 10.1111/j.1365-313X.2004.02016.x
Tripathi, D., Meena, R. P., Pandey-Rai, S. (2021). Short term UV-B radiation mediated modulation of physiological traits and withanolides production in Withania coagulans (L.) Dunal under in-vitro condition. Physiol. Mol. Biol. Plants. 27, 1823–1835. doi: 10.1007/s12298-021-01046-7
Vanhaelewyn, L., van der Straeten, D., De Coninck, B., Vandenbussche, F. (2020). Ultraviolet radiation from a plant perspective: the plant-microorganism context. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.597642
Xiang, M., Ding, W., Wu, C., Wang, W., Ye, S., Cai, C., et al. (2021). Production of purple Ma bamboo (Dendrocalamus latiflorus Munro) with enhanced drought and cold stress tolerance by engineering anthocyanin biosynthesis. Planta. 254, 50. doi: 10.1007/s00425-021-03696-z
Xiao, Y.-H., Zhang, Z.-S., Yin, M.-H., Luo, M., Li, X.-B., Hou, L., et al. (2007). Cotton flavonoid structural genes related to the pigmentation in brown fibers. Biochem. Biophys. Res. Commun. 358, 73–78. doi: 10.1016/j.bbrc.2007.04.084
Yang, J., Song, J., Feng, Y., Cao, Y., Fu, B., Zhang, Z., et al. (2023). Osmotic stress-induced lignin synthesis is regulated at multiple levels in alfalfa (Medicago sativa L.). Int. J. Biol. Macromol. 246, 125501. doi: 10.1016/j.ijbiomac.2023.125501
You, Q., Xu, W., Zhang, K., Zhang, L., Yi, X., Yao, D., et al. (2017). ccNET: Database of co-expression networks with functional modules for diploid and polyploidGossypium. Nucleic Acids Res. 45, D1090–D1099. doi: 10.1093/nar/gkw910
Yu, A., Zhao, J., Wang, Z., Cheng, K., Zhang, P., Tian, G., et al. (2020). Transcriptome and metabolite analysis reveal the drought tolerance of foxtail millet significantly correlated with phenylpropanoids-related pathways during germination process under PEG stress. BMC Plant Biol. 20, 274. doi: 10.1186/s12870-020-02483-4
Yu, G., Wang, L.-G., Han, Y., He, Q.-Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 16, 284–287. doi: 10.1089/omi.2011.0118
Zhang, L., Duan, Z., Ma, S., Sun, S., Sun, M., Xiao, Y., et al. (2023). SlMYB7, an atMYB4-like R2R3-MYB transcription factor, inhibits anthocyanin accumulation in solanum lycopersicum fruits. J. Agric. Food Chem. 71, 18758–18768. doi: 10.1021/acs.jafc.3c05185
Zhang, L., Wang, Y., Sun, M., Wang, J., Kawabata, S., Li, Y. (2014). BrMYB4, a Suppressor of Genes for Phenylpropanoid and Anthocyanin Biosynthesis, is Down-Regulated by UV-B but not by Pigment-Inducing Sunlight in Turnip cv. Tsuda. Plant Cell Physiol. 55, 2092–2101. doi: 10.1093/pcp/pcu137
Zhao, D., Luan, Y., Shi, W., Zhang, X., Meng, J., Tao, J. (2021). A Paeonia ostii caffeoyl-CoA O-methyltransferase confers drought stress tolerance by promoting lignin synthesis and ROS scavenging. Plant Sci. 303, 110765. doi: 10.1016/j.plantsci.2020.110765
Zhao, J., Chen, L., Ma, A., Wang, D., Lu, H., Chen, L., et al. (2022). R3-MYB transcription factor LcMYBx from Litchi chinensis negatively regulates anthocyanin biosynthesis by ectopic expression in tobacco. Gene. 812, 146105. doi: 10.1016/j.gene.2021.146105
Zhong, R., Lee, C., Ye, Z.-H. (2010). Evolutionary conservation of the transcriptional network regulating secondary cell wall biosynthesis. Trends Plant Sci. 15, 625–632. doi: 10.1016/j.tplants.2010.08.007
Zhou, X., Yu, W., Gong, F., Xu, H., Lyu, J., Zhou, X. (2024). Golden 2-like transcription factors regulate photosynthesis under UV-B stress by regulating the calvin cycle. Plants (Basel). 13, 1856. doi: 10.3390/plants13131856
Keywords: photosynthesis, UV-B stress, phenylpropanoid metabolism, Gossypium hirsutum, co-expression network, anthocyanin biosynthesis
Citation: Song X, Zhu Y and Bao Y (2025) Identification and characteristics of differentially expressed genes under UV-B stress in Gossypium hirsutum. Front. Plant Sci. 15:1529912. doi: 10.3389/fpls.2024.1529912
Received: 18 November 2024; Accepted: 24 December 2024;
Published: 15 January 2025.
Edited by:
Guangqiang Zhang, Heze University, ChinaReviewed by:
Guang-Long Wang, Huaiyin Institute of Technology, ChinaQingming Li, Chinese Academy of Agricultural Sciences, China
Pichang Gong, Chinese Academy of Sciences (CAS), China
Copyright © 2025 Song, Zhu and Bao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Bao, YmFveWluZ0BxZm51LmVkdS5jbg==
 Xiaolin Song
Xiaolin Song Yingjie Zhu
Yingjie Zhu Ying Bao
Ying Bao