- 1National Key Laboratory for Tropical Crop Breeding, College of Breeding and Multiplication, Sanya Institute of Breeding and Multiplication, Hainan University, Sanya, China
- 2College of Tropical Agriculture and Forestry, Hainan University, Danzhou, China
Class III peroxidases are plant-specific glycoproteins and widely distributed among plant species, that play a crucial role in plant resistance to different stresses, such as salt, heat, cold and metal toxicity. The present study is the first comprehensive and systematic report to characterize the NcPOD gene family in water lily (Nymphaea colorata). In this study, 94 NcPOD genes in water lily were identified, each possessing a conserved POD domain, which are dispersed unevenly across the genome. Through comparative maximum-likelihood phylogenetic analysis, these genes were categorized into 10 groups, along with two other species, Arabidopsis thaliana and Nymphaea thermarum. Notably, the largest group, group-c, comprised 32 distinct types of NcPOD proteins. These genes exhibited uneven distribution on 11 of the 14 chromosomes of water lily. Exon-intron and motif analyses exhibited the structural and functional diversity among the sub-groups. The Examination of duplication patterns suggests that tandem duplication has contributed to the expansion of NcPOD genes. The analysis of promoter cis-acting elements indicated the presence of regulatory elements associated with various responses such as ABA, MeJA, light responsiveness, anaerobic conditions, and drought inducibility. Finally, the RT-qPCR based expression and enzymes activity of ten NcPOD genes depicted the dynamically differential response to NaCl, heat, cold, and heavy metals (CuSO4 and CdCl2) stresses. These findings provide valuable insights for future exploration of NcPOD functions in water lily growth and stress tolerance, laying a foundation for further comparative genomics and functional studies of this important class of antioxidant genes.
1 Introduction
Peroxidases, often referred to as PODs, represent a diverse group of enzymes responsible for catalyzing the oxidation of numerous substrates by utilizing hydrogen peroxide (H2O2) as an oxidizing mediator. These enzymes are found in all living organisms and can be categorized into different families based on their structural and catalytic characteristics (Pandey et al., 2017). Among them, one particular family, class III peroxidases (EC 1.11.1.7), is unique to the plant kingdom. Enormous varieties of POD isozymes encoded by a multigene family has been reported within higher plants (Passardi et al., 2004b; Kidwai et al., 2020). However, the abbreviation used in the papers differs as POD or PRX. Though, in this study, POD was used to distinguish it from another group of peroxidases.
Over the past few decades, extensive research has been conducted to unravel the functions of POD genes across various plant species. These genes have been confirmed to play a vital role in a wide range of physiological and developmental processes. Their functions include cell elongation, the creation of cross-links within cell wall components, the formation of lignin and suberin, the scavenging of reactive oxygen species (ROS), wound healing, phytoalexin synthesis, and defence responses against both abiotic and biotic stresses (Hiraga et al., 2001; Gabaldón et al., 2005; Passardi et al., 2005; Daudi et al., 2012; Qiang et al., 2023). Additionally, PODs have the capability to catalyze the phytohormone auxin and produce hydroxyl radicals and H2O2, which play intricate roles in defence signalling (Gaspar et al., 1985; Joo et al., 2001). Numerous functional studies have previously highlighted the role of PODs in enhancing crop plants’ tolerance to various environmental stresses (Obaid et al., 2024). For instance, when Arabidopsis peroxidase 64 (AtPrx64) was overexpressed in transgenic tobacco plants, they exhibited enhanced tolerance to aluminium stress (Wu et al., 2017). Similarly, the cold-inducible gene RCI3 (AtPrx3) encoded a POD in A. thaliana, providing increased tolerance to salt and drought stresses (Llorente et al., 2002). The germination rate was improved by overexpressing the peroxidase genes extracted from Catharanthus roseus (CrPrx & CrPrx1) against cold, salt and dehydration stresses (Kumar et al., 2012). In tomatoes, reduced susceptibility to bacterial fleck was achieved through the down-regulation of the Ep5C gene, encoding a POD (Coego et al., 2005). Knock-out lines of pepper lacking the extracellular gene CaPO2 displayed increased vulnerability to bacterial pathogens, while overexpression of CaPO2 conferred enhanced resistance (Choi et al., 2007). Furthermore, transgenic carrot plants overexpressing the rice cationic POD gene OsPrx114 exhibited heightened resistance to fungal diseases (Wally and Punja, 2010). Some maize POD genes are under the regulation of hormones and pathogen elicitors (Mika et al., 2010). Similarly, the overexpression of GsPRX9 in soybean composite seedlings led to improved tolerance to salt stress (Jin et al., 2019). These collective findings accentuate the positive contribution of class III plant peroxidases in responding to both biotic and abiotic stresses.
In various plant species, comprehensive whole-genome analyses have unveiled the members of the class III POD family, highlighting their essential roles in both defence responses and the regulation of plant growth and development. For example, tobacco (Nicotiana tabacum) comprised (210) (Cheng et al., 2022), Arabidopsis found 73 PODs (Tognolli et al., 2002; Duroux and Welinder, 2003; Valério et al., 2004), rice (Oryza sativa) contained 138 PODs (Passardi et al., 2004a), Populus trichocarpa had 93 PODs (Ren et al., 2014), Medicago sativa possessed 102 PODs (Behr et al., 2015), maize encompassed 119 PODs (Wang et al., 2015), and 94 PODs were identified in Pyrus bretschneideri (Cao et al., 2016).
Water lilies are aquatic plants of global horticultural and economic importance and among the earliest diverging angiosperms – Darwin’s abominable mystery –150 million years ago (Mya) (Zhang et al., 2020). The water lily (order Nymphaeales), comprises around 100 species of aquatic herbs, and plays a crucial role in evolutionary biology (Xiong et al., 2023; Khan et al., 2024). These plants are classified within the ornamental-rich genus Nymphaea (Borsch et al., 2007). Numerous Nymphaea species are globally recognized as important aquatic plants, known for their different colors, prolonged blooming periods, high stress tolerance, and adaptability (Chen et al., 2017). Their flowers are mostly used in tea making, as fresh cut flowers, and as well as in textile production (Khan et al., 2023). The water lily specie Nymphaea colorata, commonly known as blue pygmy or colorata, is a highly valued ornamental aquatic plant admired for its vibrant flower colours and attractive shapes. Its small size and nonviviparous nature make it ideal for small- to medium-sized water gardens. The plant feature leaves around 10 cm in diameter, which are green on the upper surface and have a bluish-violet color on the underside of the leaf. The flowers, are cup-shaped, and violet-blue, with a paler tone at the base of the petals and stamens (Zhang et al., 2020; Khan et al., 2024). They are also slightly fragrant, increasing their ornamental demand. These distinctive traits have contributed to the growing popularity of N. colorata in aquatic gardens worldwide. Furthermore, the species has been utilized in breeding programs across the globe. The first water lily (N. colorata) genome sequence was released in nature in 2020 (Zhang et al., 2020), though the POD gene family has not yet been identified in any species of water lily.
In the current study, we conducted a comprehensive bioinformatics analysis of the POD gene family of water lily and elucidated their roles in responding to various abiotic stressors, including NaCl, heat, cold, and heavy metals. We successfully identified a total of 94 genes within the water lily genome for the first time, subjecting them to thorough genome-wide analysis. This analysis encompassed the examination of physicochemical properties, phylogenetic relationships, motif composition, gene structure, promoter sequences, RT-qPCR, and enzymes accumulation in response to abiotic stresses. The conclusions of this study are expected to serve as a valuable resource for future research on water lily species and lay the base for the functional characterization of the POD gene family.
2 Materials and methods
2.1 Sequence data retrieval and identification of NcPOD genes
All NcPODs including genomic and coding sequences were obtained from the Phytozome database. The 73 POD genes of Arabidopsis (http://www.arabidopsis.org/index.jsp accessed on 15 March 2024) had their protein sequences obtained from the corresponding database (Lamesch et al., 2012). To identify POD genes in the water lily genome, the Arabidopsis POD sequences were used as queries. Additionally, an HMMER-based search was conducted to predict POD proteins in the water lily genome (Potter et al., 2018). Then, all the predicted POD sequences underwent further validation through BLAST searches using the Pfam (PF00141) (http://pfam.xfam.org/accessed on 19 March 2024), and conserved domain (CDD) databases (Finn et al., 2007; Marchler-Bauer et al., 2015). In last all, the POD genes obtained from HMMER and BLAST were scrutinized using SMART (http://smart.embl-heidelberg.de/ accessed on 20 March 2024) to confirm the presence of POD conserved domains (Letunic et al., 2012). Finally, any redundant proteins or proteins identified as ascorbate peroxidases were manually reviewed and excluded.
2.2 Examination of protein characteristics and in-silico subcellular localization
The ExPASy proteomics server (http://web.expasy.org/protparam/) was used to assess the physiochemical properties of NcPOD proteins. This analysis included parameters such as the number of amino acids, chromosome length, molecular weight (MW), isoelectric points (pI), and the grand average of hydropathicity (GRAVY) (Gasteiger et al., 2003). To predict the subcellular localization of POD proteins, the WoLF PSORT (https://wolfpsort.hgc.jp/ accessed on 25 March 2024) (Edgar, 2004) and the ProtComp 9.0 server (http://linux1.softberry.com/ accessed on 30 March 2024) was employed (Khan et al., 2023).
2.3 Analysis of conserved motifs and NcPOD domains
The MEME program with default settings (https://meme-suite.org/meme/db/motifs accessed on 5 April 2024) was used to identify conserved motifs within all protein sequences (Bailey et al., 2009). Subsequently, TBtools (V 1.068, https://github.com/CJ-Chen/TBtools/ accessed on 7 April 2024) was employed to visualize the phylogenetic tree, conserved motifs, and conserved domains of the NcPOD family genes (Chen et al., 2020).
2.4 Phylogenetic analysis of POD proteins
To create a phylogenetic tree, the POD protein sequences identified in N. colorata, A. thaliana, and N. thermarum were used. Alignment of the PODs was conducted employing the CLUSTALW method (Thompson et al., 1994). Subsequently, the phylogenetic tree for the POD family proteins was constructed using the maximum-likelihood model in MEGA-X (https://megasoftware.net/home accessed on 15 April 2024) (Kumar et al., 2016) with 1000 bootstrap replicates. Finally, the resulting phylogenetic tree was improved through the online tool iTOL (https://itol.embl.de/; accessed on 17 April 2024) (Letunic and Bork, 2021).
2.5 Mining cis-regulatory elements in NcPODs
The 2000 bp upstream regions was mined from the transcription start sites for all NcPODs, obtaining the data from the Phytozome database. These sequences were then subjected to a search for potential cis-acting regulatory elements through the PlantCARE tool (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/ accessed on 27 April 2024) (Lescot et al., 2002). The results of this analysis were manually processed and visualized using the ‘Basic Biosequence View’ feature in TBtools v1.108 software (Chen et al., 2020).
2.6 Gene structure, chromosomal location and synteny analysis
The CDS sequences of PODs were obtained from the genomic structure information (GFF) of the genome and then visually examined their intron and exon structures utilizing the Gene Structure Display Server (GSDS; http://gsds.cbi.pku.edu.cn/index.php accessed on 29 April 2024) (Guo et al., 2007). To generate a schematic diagram, TBtools were employed. The chromosomal locations of NcPOD family genes in the N. colorata genome were obtained and visualized using the GFF3 annotation file with TBtools v.2.069 software (Chen et al., 2020). Genome assemblies of both N. colorata and A. thaliana were utilized to calculate synteny and collinearity and create a dual synteny plot using the MCScanS program within TBTools (V 1.068, https://github.com/CJ-Chen/TBtools/ accessed on 1 May 2024) (v1.0692).
2.7 Plant materials and abiotic stresses
To investigate the response of N. colorata POD antioxidant genes to various abiotic stresses, including salinity (250 mM), heat (42°C), cold (08°C), and heavy metals (Cu; CuSO4 200 µM and Cd; 2.5 mM CdCl2), experiments were conducted (Khan et al., 2023). The water lily (N. colorata) plants were obtained from National Key Laboratory for Tropical Crop Breeding, College of breeding and multiplication, Sanya Institute of Breeding and Multiplication, Hainan University, Sanya, China. The mature N. colorata plants were grown in a water tub filled with tap water under an open atmosphere. Each treatment consisted of three separate biological replicates, with samples collected from at least five individual plants. Leaves from the plantlets were collected at 0, 2, 4, and 6 h for the heat, salt, cold, and heavy metal stress. After collection, all samples were immediately frozen in liquid nitrogen and stored at −80°C until total RNA extraction.
2.8 RNA extraction and real-time quantitative PCR expression analysis
RNA was extracted utilizing the RNAprep Pure Plant Kit (TIANGEN, Beijing, China), and the concentration of the samples was measured with a NanoDrop 2000 C spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Removal of genomic DNA was accomplished through DNase I treatment, followed by cDNA synthesis using the QuantiTect Reverse Transcription Kit (Qiagen, Shanghai, China). The RT-qPCR expression was performed on the Roche LightCycler 96 PCR system, following the recommended guidelines for the ChamQTM SYBR RT-qPCR Master Mix (Vazyme Biotech Co., Ltd., Sanya, China). Gene-specific primers (S1) for NcPOD and Nc-Actin were designed using the online NCBI Primer-BLASTProgram and their specificity was confirmed utilising the Oligo Calculator online tool (http://mcb.berkeley.edu/labs/krantz/tools/oligocalc.html accessed on 10 June 2024). Three biological replicates for each sample were employed for RT-qPCR. The 2 −ΔΔCT method was used to analyze gene expression (Jozefczuk and Adjaye, 2011).
2.9 Antioxidant POD enzyme activity of N. colorata
To evaluate the POD antioxidant enzyme activity, frozen samples (leaf, 6 h stressed) weighing 0.5 g were finely ground using liquid nitrogen and then homogenized with 4.5 ml of normal saline (0.154 M), following the protocol in the provided kit. The homogenate was subjected to centrifugation at 12,000 rpm and 4°C for 10 minutes. The resulting supernatant was transferred to a separate falcon tube for subsequent enzyme analysis. For the determination of POD activity, the instructions from kits (A084-3, Nanjing Jiancheng Bioengineering Institute in Nanjing, China) were exactly followed. The wavelength of POD activity was noted at 420 nm using a photometer (Lambda 25 UV/VIS Spectrophotometer).
2.10 Statistical analysis
Statistical analysis was accomplished using SPSS software (version 9.1, SPSS Institute Inc. Cary, NC, USA). Different stars and alphabets indicate a significant difference between the concentrations (p ≤ 0.05). For three replications of each treatment, the represented data are the means ± standard errors (SE). Fisher’s least significant difference (LSD) test was employed (p ≤ 0.05), to analyze the differences among the concentration.
3 Results
3.1 Characterization of NcPOD gene family in water lily (N. colorata)
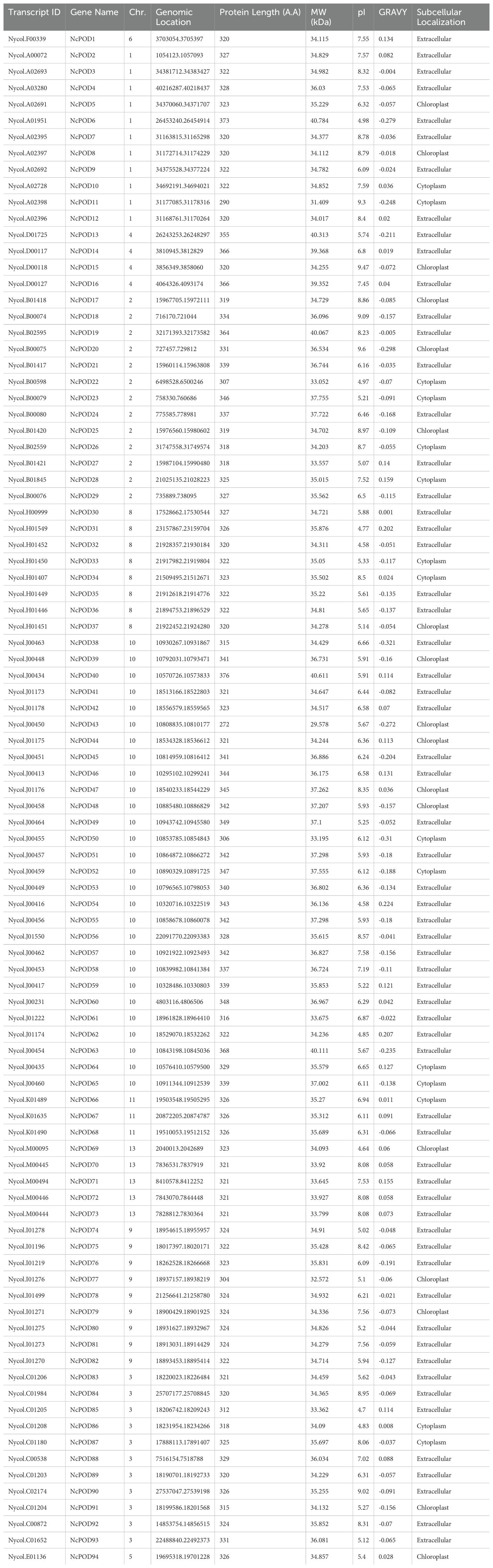
To identify NcPOD family members in water lily, BLASTP and HMMER based methods were utilized to search for NcPODs within the water lily genome database. The 73 Arabidopsis POD sequences were used as queries (Tognolli et al., 2002) and the candidate sequences were confirmed through conserved domain analysis based on CDD (Conserved Domain Database). The 94 POD genes (NcPOD) were identified within the water lily genome and the genes were renamed from NcPOD1 to NcPOD94 (Table 1). The overall number of POD family genes in water lily was lower than that in tobacco (N. tabacum) (210), S. spontaneum (113), sorghum (150), and rice (126), but slightly higher than in A. thaliana (73). Similarly, the analysis of various essential information regarding the NcPODs, including protein identifiers, chromosomal localization, and several physicochemical properties such as protein length (aa), molecular weight (kDa), isoelectric point (pI), grand average of hydropathicity (GRAVY), and in-silico subcellular localization were conducted. The protein length of NcPODs exhibited considerable variability, ranging from 272 (NcPOD 43) to 376 (NcPOD 40) amino acids, with predicted molecular weights spanning from 29.578 to 40.784 kDa. The theoretical pI values ranged from 4.7 to 9.47, respectively. Moreover, there was diversity among most of the genes regarding GRAVY, indicating predominantly hydrophilic properties, with only a few exhibiting hydrophobic characteristics, as evidenced by positive values. Furthermore, the in-silico subcellular locations of these NcPODs were found in cytoplasm, chloroplast and mainly in extracellular.
3.2 Analysis of conserved amino acid motifs
Molecular Evolutionary Genetics Analysis Version X (MEGA-X) was employed to make a rectangular phylogenetic tree of the 94 NcPOD proteins. The tree was constructed using the maximum likelihood method, with bootstrap value of 1000 replicates and it was organized into 10 distinct groups (Figure 1A). The structural characteristics of NcPODs were examined by identifying 10 conserved motifs through the MEME database, aligning them with the phylogenetic relationships (Figure 1B). The present analysis revealed that closely related NcPODs shared common motif compositions, suggesting potential functional similarities within the same NcPOD sub-group. Motifs 1–9 were highly prevalent in NcPODs, present in 90–98% of the proteins, while motif 10 exhibited variation across different groups, inferring at their potential significance in NcPOD proteins. The detailed information about their identified motifs, containing their names, sequences and widths is shown in S3. Additionally, an NCBI domain search and interproscan were conducted to perform a domain-based analysis of all POD proteins and used TBtools to create a comprehensive structural representation (Figure 1C).
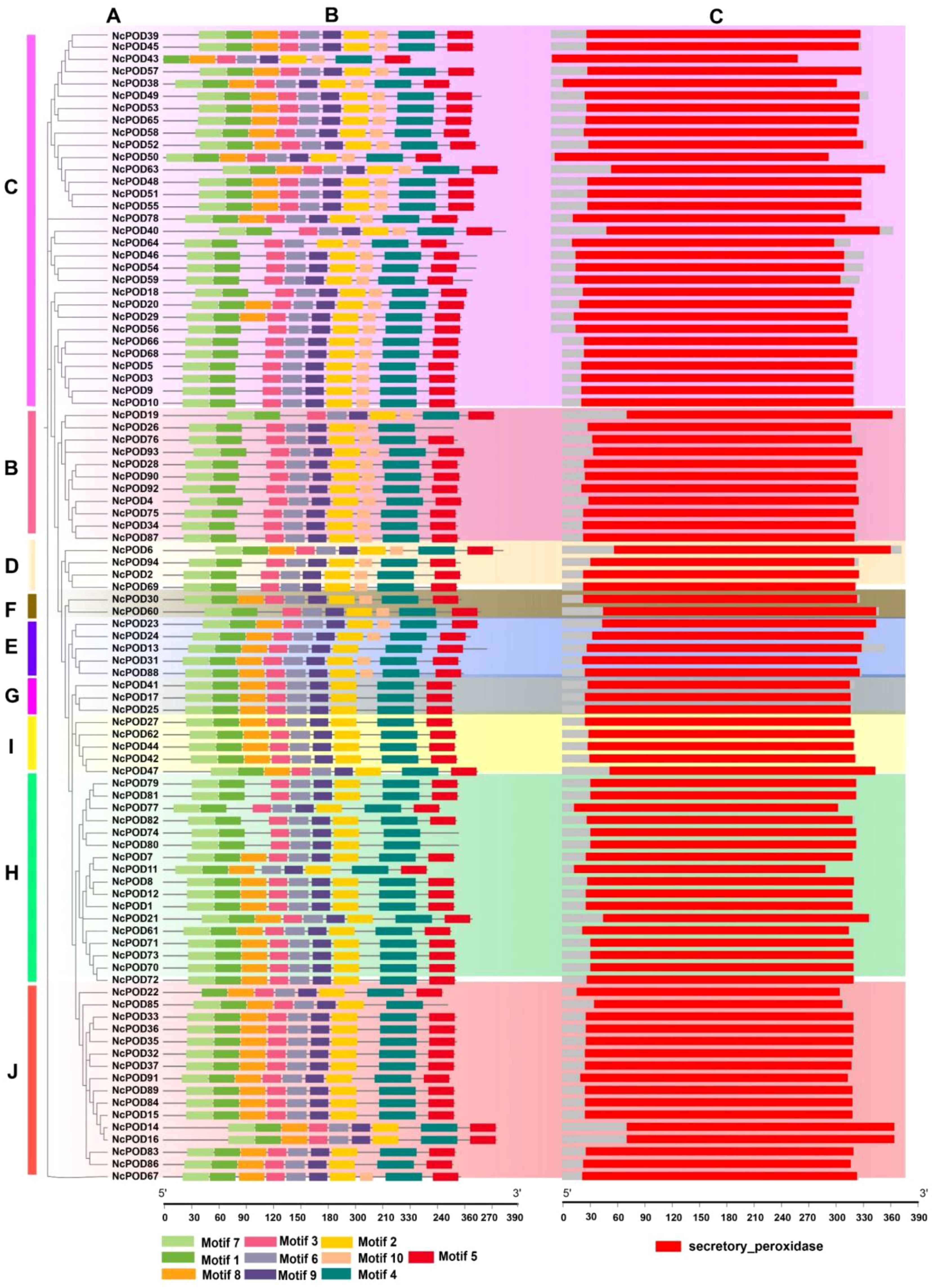
Figure 1. (A) Representation of the phylogenetic tree illustrating the relationships among NcPOD proteins (B) Motif composition of POD in the water lily, highlighted in various colors representing motifs 1–10, with non-conserved sequences indicated by grey lines. (C) Visualization of the conserved domains within NcPOD proteins. The presentation of motif composition and conserved domain structure was generated using TBtools software. The relative positioning is proportionally depicted on a kilobase scale at the bottom of the figure.
3.3 Phylogenetic and comparative analyses of PODs in water lily
In order to examine the evolutionary relations, 94 POD genes were used from Nymphaea colorata, 72 from Arabidopsis thaliana, and 85 from Nymphaea thermarum to create a maximum likelihood phylogenetic tree using MEGA-X. The POD family genes were categorized into ten distinct groups (A-J), as represented in Figure 2. Notably, group C was the largest, comprising 65 POD members, with 32 from N. colorata, 25 from N. thermarum, and 8 from A. thaliana. Groups B, H, and J also exhibited substantial number of genes, with 53, 28, and 34 genes, respectively. Conversely, group G was the smallest, encompassing just 5 members, including 3 of N. colorata and 2 of N. thermarum, while A. thaliana members were notably absent from this group. Interestingly, group A contained only members of A. thaliana and had no members of N. colorata and N. thermarum. Furthermore, a total of 56 orthologous pairs between N. colorata and N. thermarum were identified. However, we did not find any orthologous pairs between N. colorata and A. thaliana. This discrepancy in orthologous pairs is most likely due to the closer evolutionary relationship between the two water lily species, N. colorata and N. thermarum, as compared to the more distant relationship with A. thaliana.
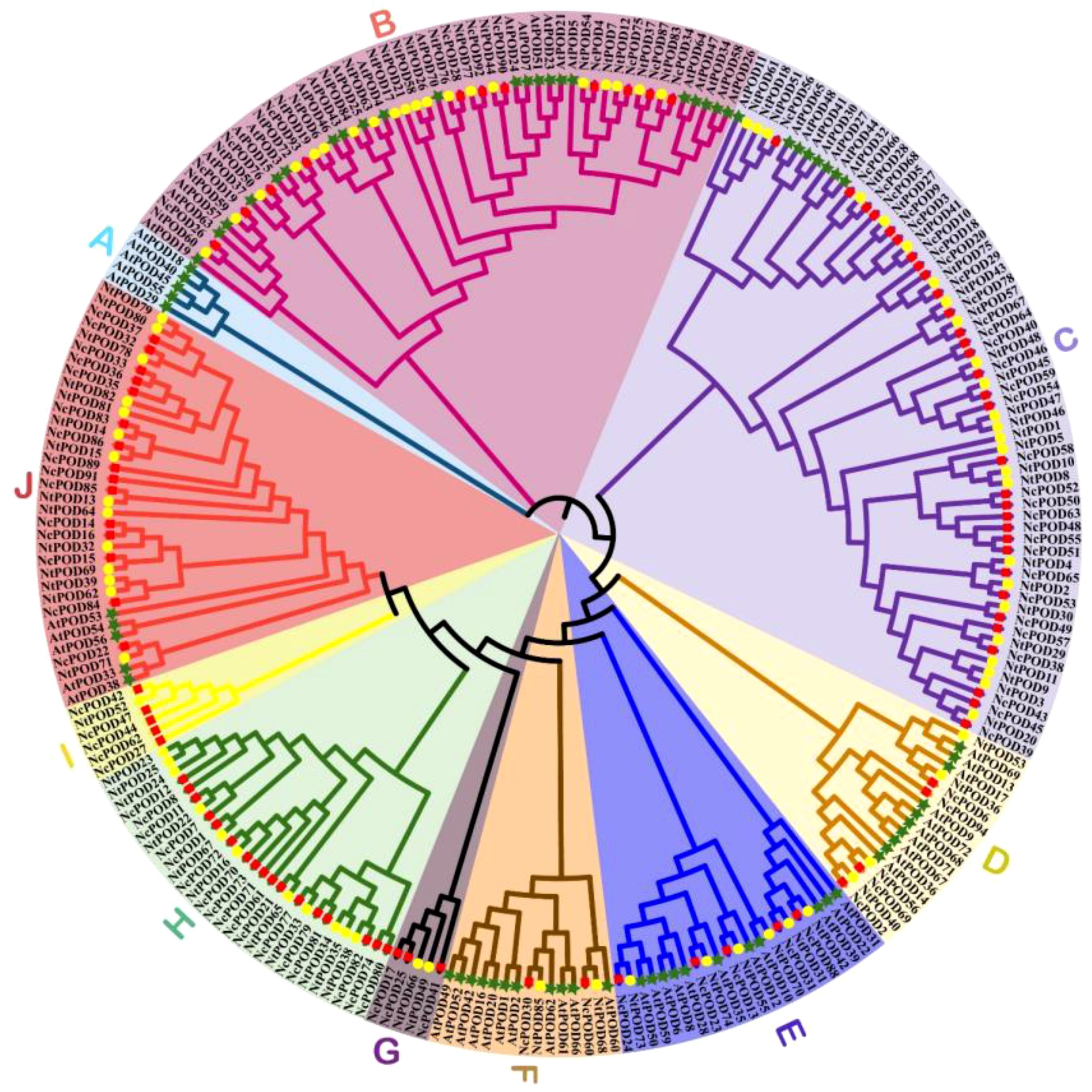
Figure 2. Phylogenetic relationships among Nymphaea colorata (red rectangle), Arabidopsis thaliana (green stars) and Nymphaea thermarum POD genes (yellow circles) based on the amino acid sequence alignment. The phylogenetic tree was generated using MEGAX with the Maximum Likelihood Method, and bootstrap support was assessed with 1000 replicates. Clusters were categorized into distinct groups (A–J) based on their evolutionary relationships.
3.4 Cis-acting elements in the promoter region of NcPODs
In current analysis, the cis-acting elements present in the upstream promoter region of the NcPOD genes were explored, which play a crucial role in regulating gene expression. The 2 kb region starting from the gene initiation site was focused on, and the PlantCARE database was used for cis-acting element analysis. The identified cis-acting elements were subsequently categorized into three distinct groups based on their putative functions: light-related, stress-related, and hormone-related elements. Notably, the largest group of regulatory elements was associated with abiotic stress, followed by elements involved in light regulation, including key regulatory motifs such as GT1-motif, G-Box, GATA-motif, and AE-Box. Additionally, hormone-related elements, encompassing CGTCA-motif, TGACG-motif, ABRE, and GARE-motif, were also prevalent (S4). This comprehensive analysis unveiled the diverse roles of POD gene members, highlighting their indirect contributions to various biological processes, including responses to biotic and abiotic stresses, as well as participation in hormone signalling pathways (Figure 3).
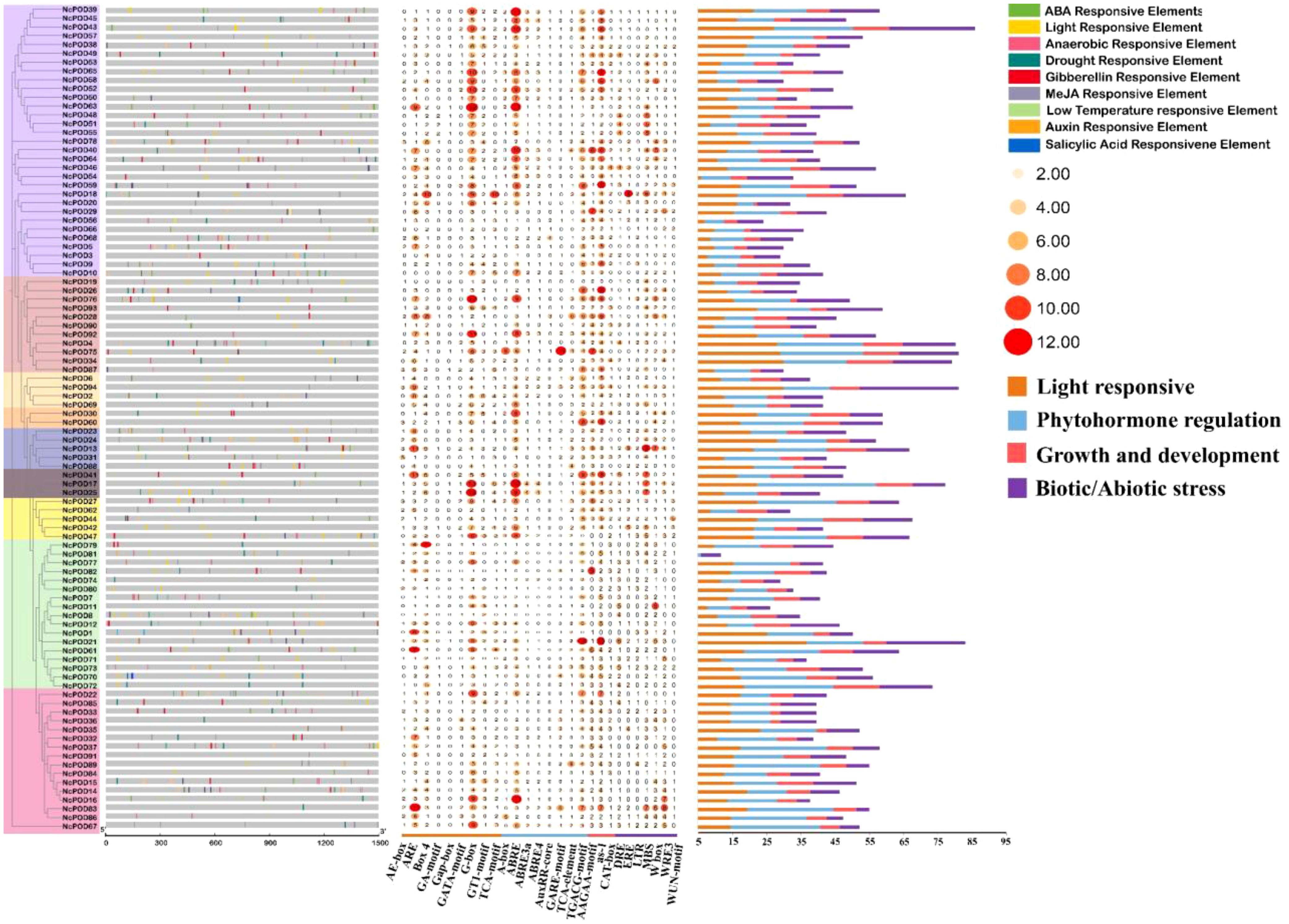
Figure 3. Examination of cis-regulatory elements within the 2000 bp region upstream of the transcription start site of NcPOD genes, with an illustration of their distribution along the promoter sequence. The cis-regulatory elements were classified based on their functional significance; including elements associated with light, stress, and hormone responsiveness. The motifs were identified using data from the PlantCARE database.
3.5 Gene structure analyses of NcPOD genes
This analysis provided valuable insights into the evolutionary development of structural diversity within the POD gene family. The exon-intron arrangement of the 94 NcPOD genes were examined to gain a better understanding of their structural characteristics, as illustrated in Figure 4. The present observations indicated that NcPOD genes exhibited conserved exon-intron distributions, with introns ranging from 1 to 3 and varying exon numbers between 2 and 6. While genes sharing similar exon/intron structures were grouped together, there were also occurrences of structural variation among these NcPOD genes. Gene structure analysis proposed that the NcPOD gene family generally maintained a conserved exon/intron organization.
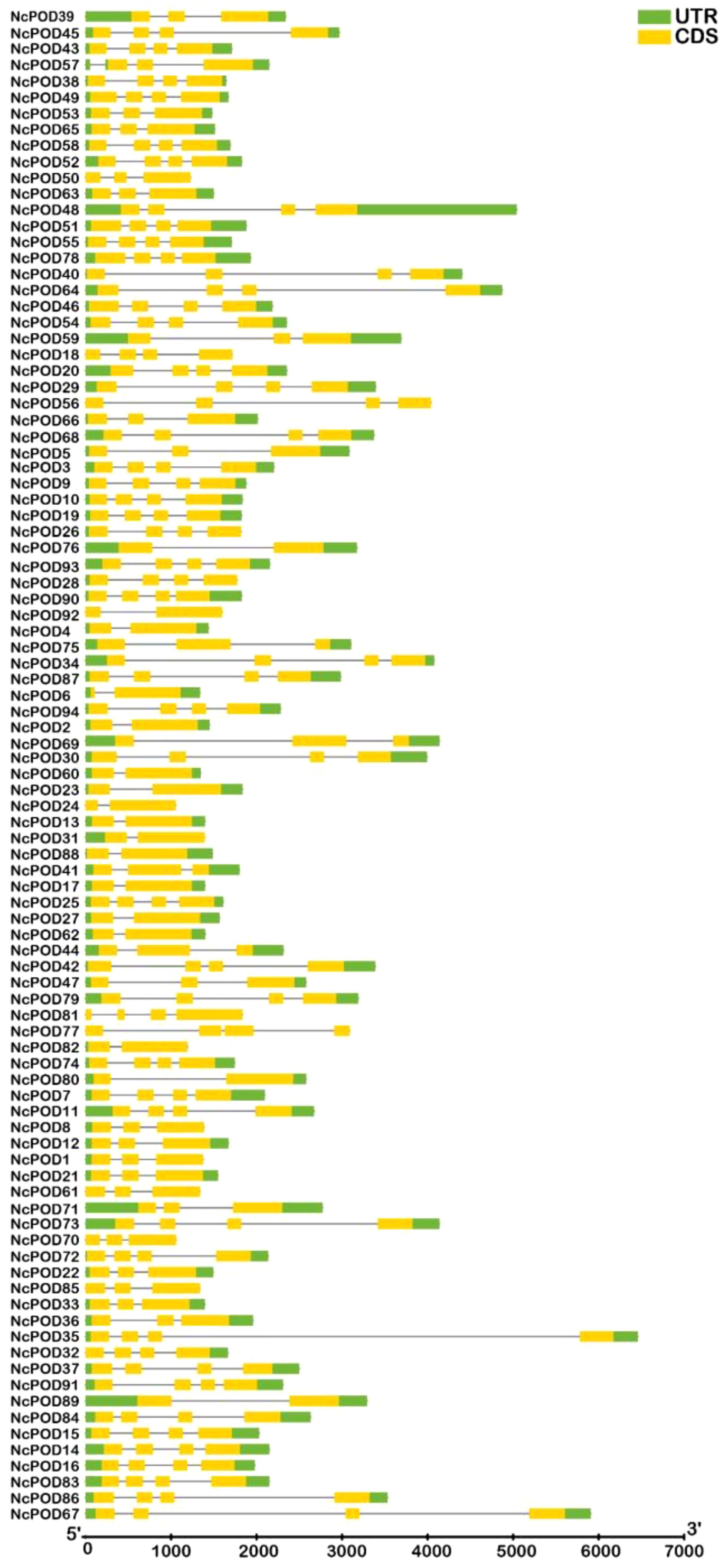
Figure 4. Exon/intron organization of NcPOD genes. Yellow boxes denote exons and black lines denote introns. The untranslated regions (UTRs) are showed by green boxes. The sizes of exons and introns can be assessed using the scale at the bottom.
3.6 Chromosomal locations and dual synteny analysis of NcPOD genes
To elucidate the genome organization and chromosome distribution of NcPOD genes in the water lily, a chromosomal map was generated using TBtools. In this map, a total of 94 NcPOD genes on the N. colorata chromosomes were identified (Figure 5). Notably, there was not found the distribution of any POD gene on chromosomes 7, 9 and 14. The distribution of NcPOD genes across the 11 chromosomes was uneven. Amongst them, the highest number of NcPOD genes (29) was located on chromosome 10, followed by chromosomes 2 (13), and chromosomes 1 and 3 had same number of genes (11). Conversely, only a few NcPOD genes were found on chromosomes 4 (4), 8 (8), 11 (3), 12 (9) and 13 (5), while the chromosome 5 and 6 contained only single gene. Additionally, some chromosomes showed dense clusters of NcPOD genes, particularly near the telomeric regions of chromosomes 2, 8, and 12. By mapping these patterns of uneven distribution and gene clustering, the chromosomal positioning was due to the evolutionary history of this species. The existence of clustered NcPOD genes suggests regions of the genome that may have experienced tandem duplication events, potentially contributing to the rapid diversification and functional expansion of this gene family.
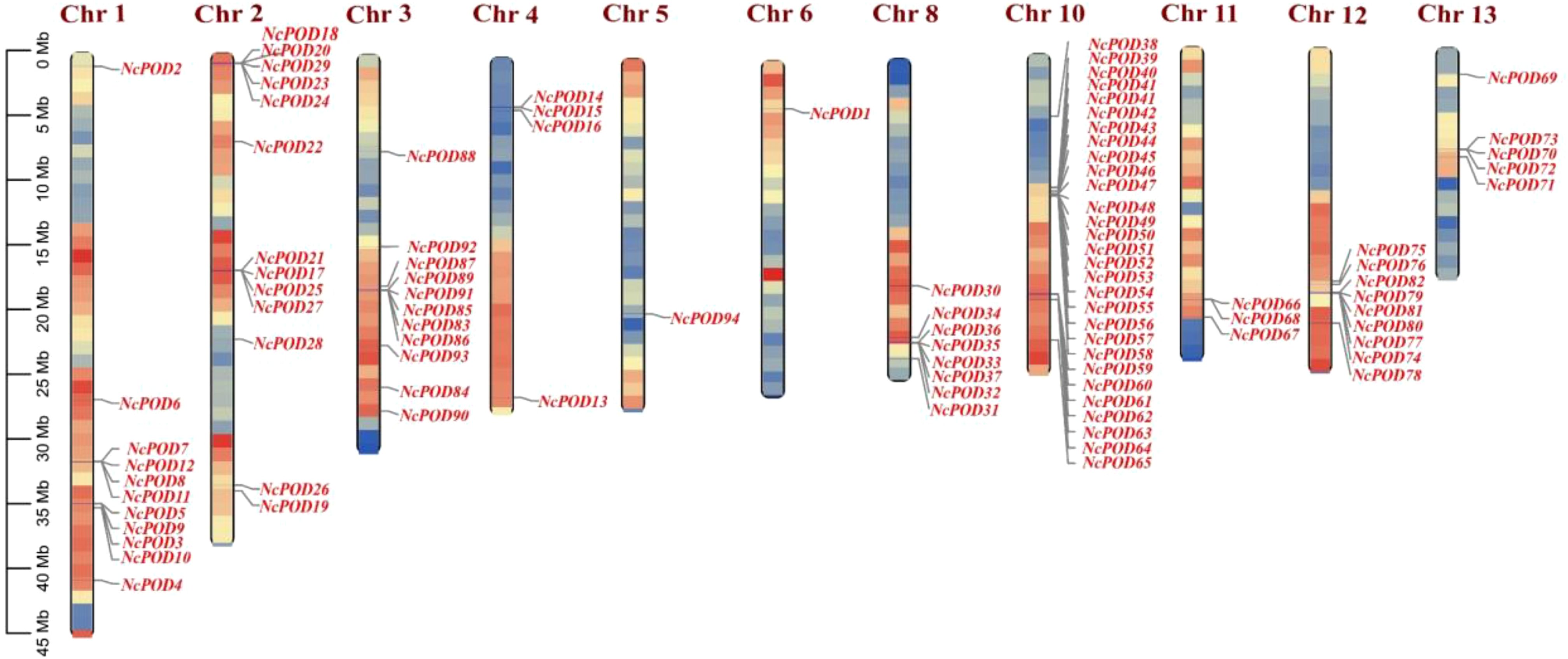
Figure 5. Genomic localization and distribution patterns of NcPOD genes in the N. colorata genome. The scale on the left presents the length of N. colorata chromosomes (Mb). The colors on the chromosomes denote gene density.
A dual synteny plot-based in-depth comparative analysis of PODs between N. colorata and A. thaliana genomes was conducted. In summary, the genes presented on Chr01, Chr02, Chr04, and Chr11, demonstrated syntenic relations with A. thaliana, respectively. In contrast, Chr03, Ch05, Ch06, Ch07, Ch08, Ch09, Ch10, Ch12, Ch13, and Ch14, did not show any syntenic relations with A. thaliana (Figure 6). The results indicated that water lilies and Arabidopsis do not have a close relationship.
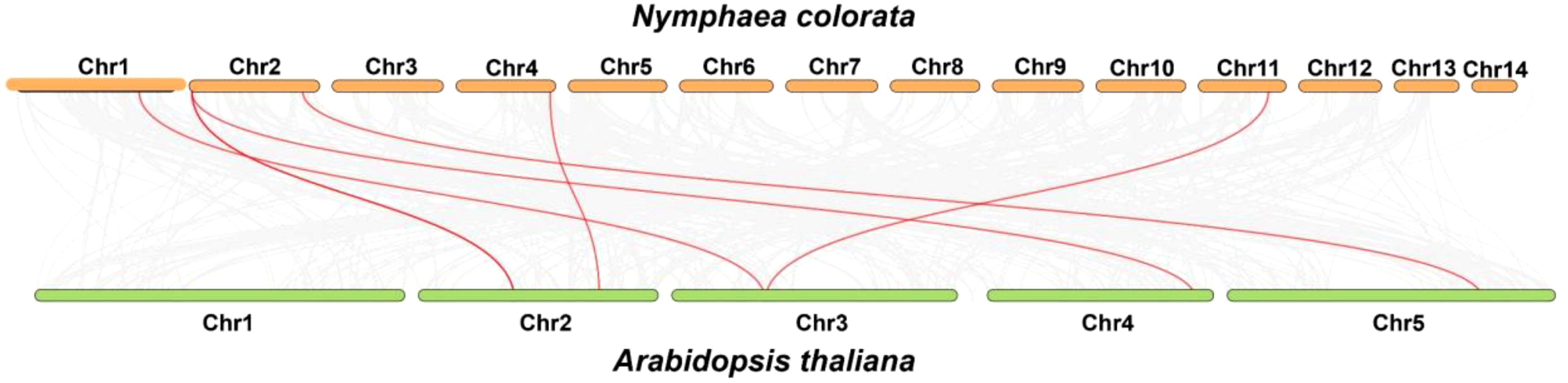
Figure 6. Genome-scale dual synteny plot between N. colorata and A. thaliana genomes, with syntenic POD orthologous genes shown in red connecting lines. Grey lines highlight the collinear blocks between the N. colorata and A. thaliana genomes.
3.7 Expression analysis of NcPOD genes under various abiotic stresses
Antioxidant enzymes play a vital role in scavenging reactive oxygen species and defending cells against oxidative stress from both biotic and abiotic stressors (Li et al., 2018). Peroxidase (POD) enzymes are a vital group of antioxidant enzymes that play a crucial role in protecting cells from oxidative damage caused by reactive free radicals (González-Gordo et al., 2023). In this context, the expression patterns and responses of POD antioxidant genes in N. colorata under various abiotic stress conditions were investigated using RT-qPCR analysis.
During salinity stress, the NcPODs showed variable expressions. Among them the NcPOD7, NcPOD12, NcPOD25, NcPOD39, NcPOD40 and NcPOD88 exhibited initially decreased expression and then reached their highest peak at 6 h. Similarly, a single NcPOD27 gene was found downregulated (Figure 7A).
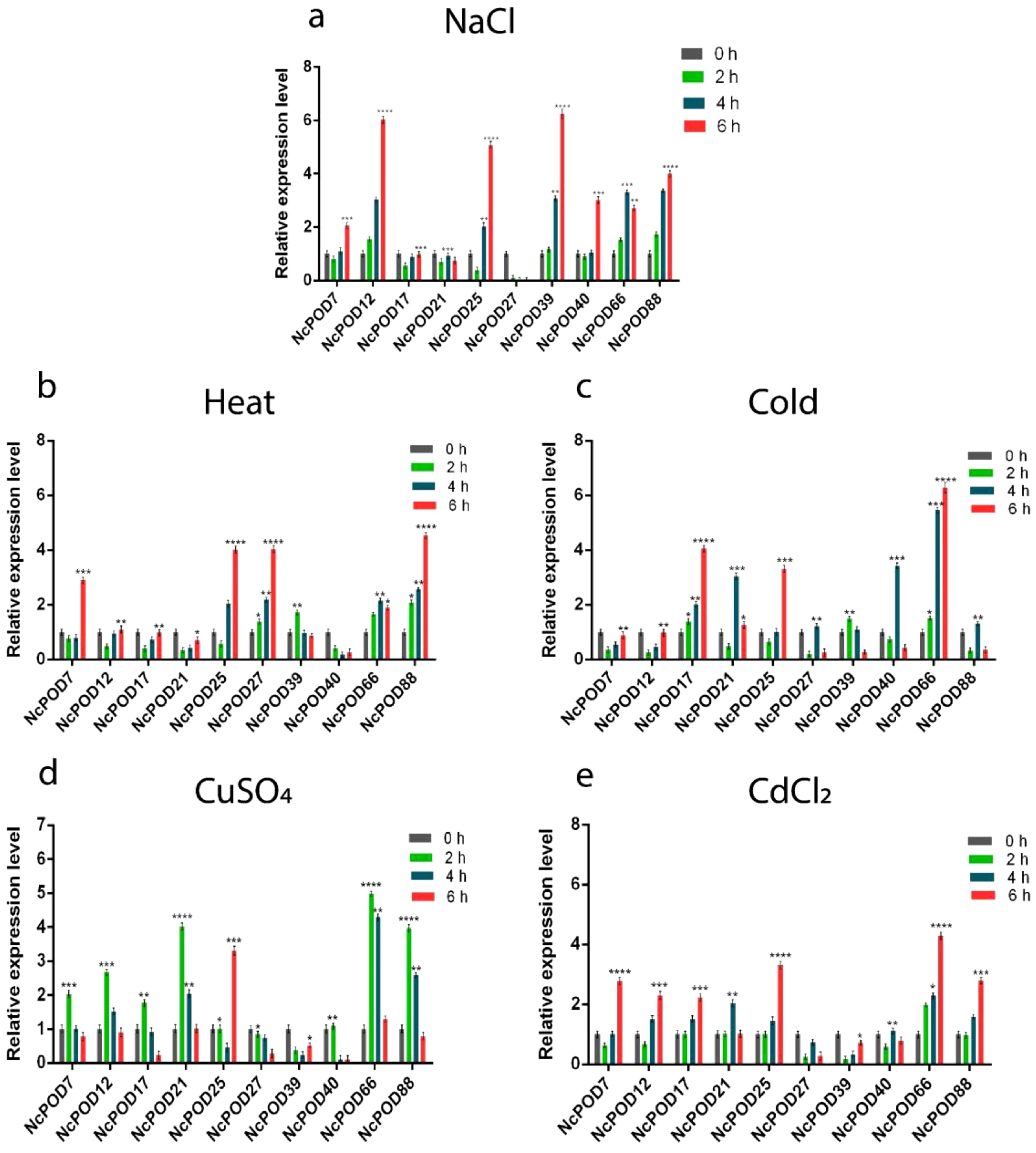
Figure 7. The expression pattern of POD genes in water lily leaves under various abiotic stresses. POD genes expression in N. colorata under (A) NaCl; (B) heat; (C) cold; (D) CuSO4; and (E) CdCl2 stress at different time intervals (0 (CK), 2, 4, and 6 h). Data presented as means, ± standard error, n = 3; statistically significant differences are exhibited by asterisks (p ≤ 0.05). The (*) symbols indicated statistically significant differences between treatments. Following the values of each star. (p < 0.05 => *, p < 0.01 => **, p < 0.001 =>***, p < 0.0001 =>****).
In response to heat stress, approximately 55% of the total genes exhibited elevated expression levels, while the remaining genes displayed either moderate or low expression. The NcPOD7, NcPOD25, and NcPOD88 showed a significant upregulation over time, with peak expression at 6 h. NcPOD39 exhibited higher expression at 2 h, which then decreased at 4 and 6 h. A single NcPOD66 gene was upregulated at 4 h (Figure 7B).
When the NcPOD genes were exposed to low temperature, fluctuations in expression were observed among the genes, with significant expression recorded at 6 h. Like the NcPOD17, NcPOD25, and NcPOD66 exhibited temporal upregulation expression. Additionally, NcPOD21, NcPOD40, and NcPOD88 were upregulated and exhibited a higher expression at 4 h (Figure 7C).
During heavy metals treatment, the POD genes exhibited dynamic expression. In response to CuSO4 treatment, the NcPOD genes showed differential expression level. Among the 10 genes studied, NcPOD7, NcPOD12, NcPOD17, NcPOD21, NcPOD66, and NcPOD88 exhibited high expression levels at 2 h, followed by a decrease in expression by 6 h. However, a single gene NcPOD25 revealed the highest expression at 6 h (Figure 7D).
Under CdCl2 treatment the NcPOD7, NcPOD12, NcPOD17, NcPOD25, NcPOD66, and NcPOD88 showed an upregulated temporal expression. However, the NcPOD21 and NcPOD40 exhibited a higher expression at 4 h (Figure 7E).
3.8 Enzymatic activity of NcPOD genes of N. colorata
In addition to the gene expression measurements, analyses of the 6 h stressed enzyme activity of NcPOD genes of N. colorata plant subjected to various abiotic stresses were conducted. The total enzyme content of POD was assessed. In particular, the comprehensive POD activity of N. colorata displayed a pattern of enhancement, demonstrating an accumulation of 200.8%, 242.16%, 278.4%, 74.4%, and 139.6% in the aforementioned stresses as compared to control (Figure 8). These results were found almost consistent with the RT-qPCR based expression analysis of POD genes suggesting that stress response was under genetic control in water lily specie.
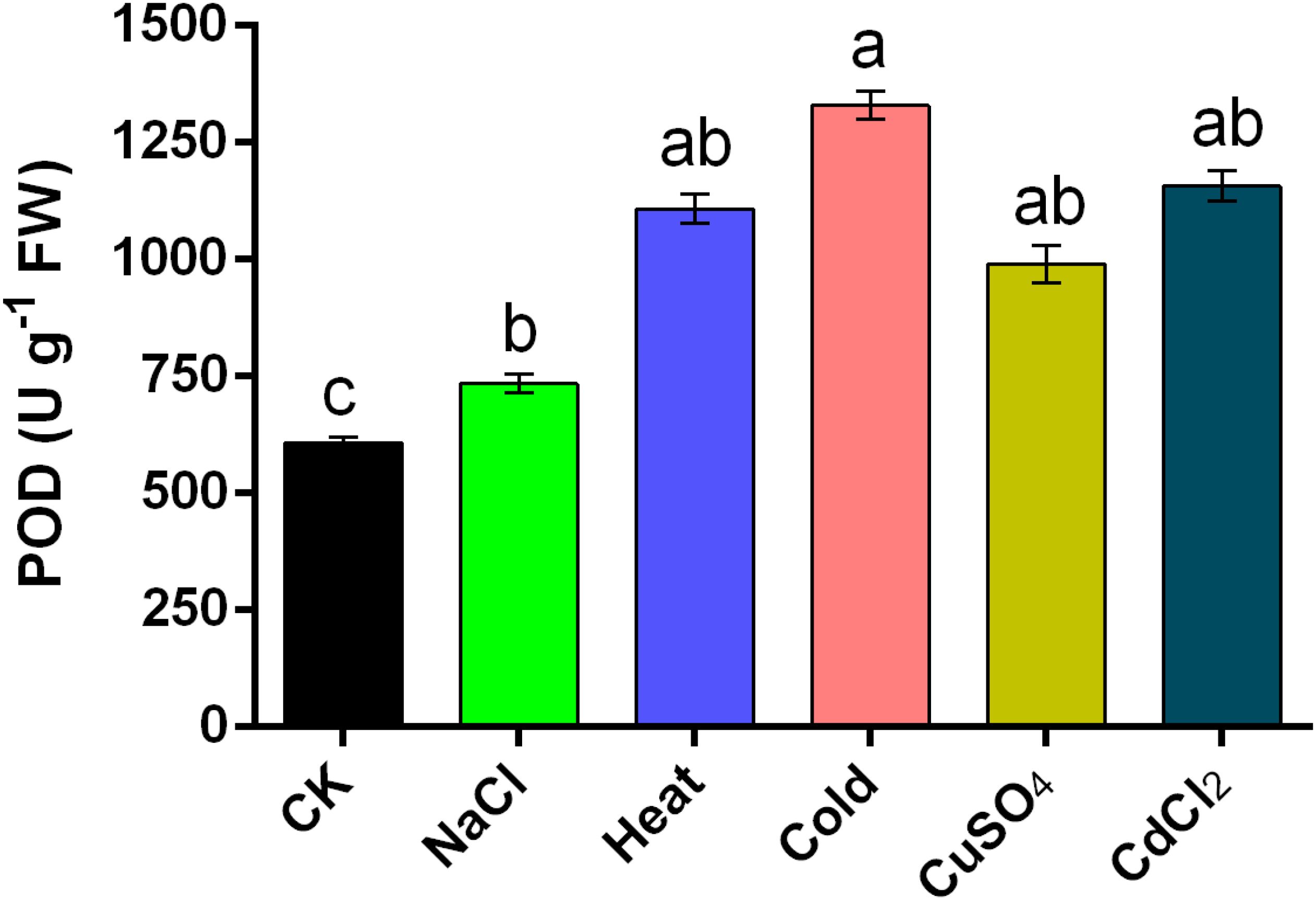
Figure 8. Concentration of POD enzymes under multiple abiotic stresses, such as salinity, heat, cold, CuSO4, and CdCl2 in Nymphaea colorata. The standard error of the means (n = 6) represented by the bars. Different lower-case letters represent significant differences among treatments at P ≤ 0.05, according to LSD test.
4 Discussion
Plants grown in natural environments constantly face various survival stress. Abiotic stress usually refers to adversity growth conditions like drought, salinity, extreme heat, cold, and ultraviolet (UV) which can affect plant growth (Han et al., 2023). Class III Peroxidases are known to be involved in various physiological processes within plants (Tognolli et al., 2002; Ren et al., 2014) and perform a crucial role in both biological and abiotic stress responses during plant development (Wang et al., 2015). Therefore, it is essential to systematically and comprehensively characterize the POD gene family at the genome scale in plant species to understand their contributions to plant growth and defense responses. Although POD gene families have been studied in A. thaliana (Tognolli et al., 2002), Populus trichocarpa (Ren et al., 2014), Zea mays (Wang et al., 2015), and Oryza sativa (Passardi et al., 2004a), there is a lack of information regarding the identification and function of the POD gene family in water lilies. Fortunately, with the completion of the full genome sequence of N. colorata, it has become possible to conduct bioinformatics analyses of the POD gene family at the genome level (Zhang et al., 2020). This study aimed to systematically characterize the POD family on a genome-wide scale in N. colorata and verify its role in abiotic stress responses. The analysis revealed the identification of 94 PODs in the N. colorata genome through a comprehensive genome-wide approach. This number surpasses those reported in Arabidopsis thaliana (73), cassava (91), and P. trichocarpa (93) (Lüthje et al., 2011), but is lower compared to tobacco N. tabacum (210) and O. sativa (138) (Passardi et al., 2004a). This suggests a significant expansion of the POD gene family in N. colorata, N. tabacum and O. sativa compared to other species. The study encompassed a thorough analysis of physicochemical properties, phylogenetic relationships, gene synteny, motif composition, gene structure organization, and cis-regulatory elements. Additionally, RT-qPCR analysis and enzyme concentrations in response to abiotic stresses (NaCl, heat, cold, and heavy metals) provided extensive information on gene functions and expression dynamics related to applied stress responses in water lily.
Comparative phylogenetic examination of N. colorata, N. thermarum, and A. thaliana POD family genes were divided into ten distinct groups (A-J). The analysis revealed a robust clustering relationship with closely related species, indicating a conserved evolution of this gene family. This suggests that this gene family has experienced relatively conserved evolution, almost consistent with findings on cassava (Wu et al., 2019).
In current analysis, ten highly conserved motifs present in the 94 NcPOD proteins were identified. Notably, there were slight variations in both the number and type of these conserved motifs among the proteins. Interestingly, the majority of NcPOD proteins contained all these conserved motifs, suggesting their potential involvement in the fundamental functions of POD proteins. The diversity in gene structure is known to play a significant role in the evolution of gene families (Muthamilarasan et al., 2014; Han et al., 2016). This study examined into the structure of NcPOD genes, revealing variations in the number of exons and introns. It is noteworthy that stress-related genes typically exhibit fewer introns (Jeffares et al., 2008). Consistent with this observation, NcPOD genes were found to have a maximum of three or fewer than three introns, confirming this conceptual framework. This is consistent with previous reports on POD genes in various plant species, such as maize (Wang et al., 2015), cassava (Wu et al., 2019), wheat (Yan et al., 2019), and Chinese pear (Cao et al., 2016), which also showed a lower number of introns. The similarities in gene structure and motif composition within each NcPOD subgroup support the phylogenetic classification proposed in this study.
As part of the characterization of the NcPOD genes, the presence of cis-regulatory elements in 2000 bp upstream regions from the transcription starting point of the 94 NcPOD genes was evaluated. The identified cis-acting elements were then classified into three different groups based on their potential functions i.e related to light, stress, and hormones. Particularly, the predominant group of regulatory elements was linked to abiotic stress, followed by elements associated with light regulation, which included significant regulatory motifs such as GT1-motif, G-Box, GATA-motif, and AE-Box. These cis-elements are likely contributors to the regulation of gene expression under various stress conditions. Similar findings have been reported in diverse plant species, such as Pepper (Capsicum annuum L.) (González-Gordo et al., 2023), sugarcane (Shang et al., 2023), cassava (Wu et al., 2019) and tobacco (Cheng et al., 2022).
Gene family expansion usually occurs through three main mechanisms: segmental, tandem and whole-genome duplication (Freeling, 2009; Cao and Shi, 2012). To explore the duplication modes of POD genes in the water lily, the chromosomal locations of the NcPOD genes were determined. Chromosomal mapping displayed that these genes are spread across 11 of the 14 chromosomes in N. colorata (Figure 5). Accumulated evidence has shown that duplication events play a crucial role in the expansion of genes within the POD family. This wide distribution is consistent with the chromosomal distribution of POD genes observed in other plants such as Arabidopsis, rice, Populus trichocarpa, maize and Pyrus bretschneideri (Tognolli et al., 2002; Passardi et al., 2004a; Ren et al., 2014; Wang et al., 2015; Cao et al., 2016).
Numerous researchers have underscored the involvement of plant peroxidases in a various of cellular processes throughout plant growth and development, and its consequences to abiotic and biotic stress have been reported over the years (Xue et al., 2008; Wang et al., 2015). Notably, NcPOD genes demonstrated significant variations in their expression profiles, highlighting their roles in various stress and defense responses in N. colorata. Meanwhile, individual POD genes typically exhibited sensitivity to specific external stresses, with few genes responding broadly to a variety of biotic and abiotic stresses. For instance, all selected NcPOD genes revealed responsiveness to numerous applied stresses, including heat, cold, salt, and heavy metals. Under salinity stress, among the genes of N. colorata a single gene (NcPOD88) exhibited the highest expression. These findings are consonant with earlier studies that highlighted differential expression patterns in grapevine (Vitis vinifera L.) under conditions of salinity stress (Xiao et al., 2020). The results unveiled diverse responses across all genes under heat stress, exhibiting varying levels of high, moderate or low expression in comparison with control. The genes i.e (NcPOD7, NcPOD12, NcPOD17, NcPOD25, NcPOD27, and NcPOD88) also exhibited significant expression. Comparable outcomes were observed in the context of heat stress treatment in potato (Solanum tuberosum L.), further affirming the consistency of these findings (Yang et al., 2020). Upon exposure to cold treatment, the NcPOD genes revealed upregulation expression pattern. Similar findings were observed in Cassava, under cold treatment (Wu et al., 2019). During heavy metal stress, the N. colorata exhibited variable expressions. Specifically, during CuSO4 treatment, a single gene (NcPOD25) exhibited a highest expression at 6 h. Under CdCl2 treatment as shown in the figure (Figure 7E), the PODs were significantly expressed and reached their expression at 6 h respectively. In this study, the expression of genes were in agreement with those detected in white clover (Trifolium repens) under Cd treatment (Zhang et al., 2015). The enzymatic activity of PODs in N. colorata was found to be almost consistent with the RT-qPCR-based expression analysis of antioxidant genes, indicating that the stress response in this water lily specie is under genetic control.
5 Conclusion
In this study, a genome-wide examination and comprehensive analysis of the POD gene family in water lily was conducted. In conclusion, a total of 94 POD genes were systematically identified in water lily (N. colorata) and categorized into 10 sub-groups, as supported by phylogenetic analysis. The structural diversity of NcPODs may reflect their functional diversity. The analysis of expression patterns of NcPOD genes and enzyme accumulation showed that these genes were expressed distinctly, and some might be linked to stress responses. Consequently, the present findings contribute significantly to the understanding of POD genes in water lilies, serving as a robust foundation for genetic enhancement in other aquatic plants. Future investigations will focus on gene engineering and comprehensive analysis, incorporating genomics, transcriptomics, proteomics, and metabolomics to further elucidate the functions of NcPODs.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author/s.
Ethics statement
The experiments did not involve endangered or protected species. The authors declare that all the permissions or licenses were acquired to collect the Water lily (Nymphaea colorata) and that all studies obey with relevant institutional, national, and international guidelines and legislation for plant ethics in the methods section.
Author contributions
WK: Conceptualization, Data curation, Investigation, Methodology, Writing – original draft, Writing – review & editing, Formal analysis, Resources, Software, Validation, Visualization. LK: Data curation, Formal analysis, Writing – review & editing. NM: Formal analysis, Writing – review & editing, Software. JZ: Data curation, Formal analysis, Software, Writing – review & editing. WW: Data curation, Formal analysis, Investigation, Resources, Validation, Writing – review & editing. FC: Data curation, Investigation, Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The National Natural Science Foundation of China (32172614), Hainan Province Science and Technology Special Fund (ZDYF2023XDNY050), Hainan Provincial Natural Science Foundation of China (324RC452), and the Project of National Key Laboratory supported this work for Tropical Crop Breeding (NO. NKLTCB202337).
Acknowledgments
We thank the editor and reviewers for their insightful comments and suggestions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1524657/full#supplementary-material
References
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Behr, M., Legay, S., Hausman, J.-F., Guerriero, G. (2015). Analysis of cell wall-related genes in organs of Medicago sativa L. under different abiotic stresses. Int. J. Mol. Sci. 16, 16104–16124. doi: 10.3390/ijms160716104
Borsch, T., Hilu, K. W., Wiersema, J. H., Löhne, C., Barthlott, W., Wilde, V. (2007). Phylogeny of Nymphaea (Nymphaeaceae): evidence from substitutions and microstructural changes in the chloroplast trnT-trnF region. Int. J. Plant Sci. 168, 639–671. doi: 10.1086/513476
Cao, Y., Han, Y., Meng, D., Li, D., Jin, Q., Lin, Y., et al. (2016). Structural, evolutionary, and functional analysis of the class III peroxidase gene family in Chinese pear (Pyrus bretschneideri). Front. Plant Sci. 7, 1874. doi: 10.3389/fpls.2016.01874
Cao, J., Shi, F. (2012). Evolution of the RALF gene family in plants: gene duplication and selection patterns. Evol. Bioinf. 8, EBO–S9652. doi: 10.4137/EBO.S9652
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, F., Liu, X., Yu, C., Chen, Y., Tang, H., Zhang, L. (2017). Water lilies as emerging models for Darwin’s abominable mystery. Horticult. Res. 4, 17051. doi: 10.1038/hortres.2017.51
Cheng, L., Ma, L., Meng, L., Shang, H., Cao, P., Jin, J. (2022). Genome-wide identification and analysis of the class III peroxidase gene family in tobacco (Nicotiana tabacum). Front. Genet. 13. doi: 10.3389/fgene.916867
Choi, H. W., Kim, Y. J., Lee, S. C., Hong, J. K., Hwang, B. K. (2007). Hydrogen peroxide generation by the pepper extracellular peroxidase CaPO2 activates local and systemic cell death and defense response to bacterial pathogens. Plant Physiol. 145, 890–904. doi: 10.1104/pp.107.103325
Coego, A., Ramirez, V., Ellul, P., Mayda, E., Vera, P. (2005). The H2O2-regulated Ep5C gene encodes a peroxidase required for bacterial speck susceptibility in tomato. Plant J. 42, 283–293. doi: 10.1111/j.1365-313X.2005.02372.x
Daudi, A., Cheng, Z., O’Brien, J. A., Mammarella, N., Khan, S., Ausubel, F. M., et al. (2012). The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern-triggered immunity. Plant Cell 24, 275–287. doi: 10.1105/tpc.111.093039
Duroux, L., Welinder, K. G. (2003). The peroxidase gene family in plants: a phylogenetic overview. J. Mol. Evol. 57, 397–407. doi: 10.1007/s00239-003-2489-3
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with improved accuracy and speed. Nucleic Acids Research. 32 (5), 728–729. doi: 10.1093/nar/gkh340
Finn, R. D., Tate, J., Mistry, J., Coggill, P. C., Sammut, S. J., Hotz, H.-R., et al. (2007). The Pfam protein families database. Nucleic Acids Res. 36, D281–D288. doi: 10.1093/nar/gkm960
Freeling, M. (2009). Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Biol. 60, 433–453. doi: 10.1146/annurev.arplant.043008.092122
Gabaldón, C., López-Serrano, M.a., Pedreño, M., Barceló, A. R. (2005). Cloning and molecular characterization of the basic peroxidase isoenzyme from Zinnia elegans, an enzyme involved in lignin biosynthesis. Plant Physiol. 139, 1138–1154.
Gaspar, T., Penel, C., Castillo, F. J., Greppin, H. (1985). A two-step control of basic and acidic peroxidases and its significance for growth and development. Physiol. plant. 64, 418–423. doi: 10.1111/j.1399-3054.1985.tb03362.x
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., Bairoch, A. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
González-Gordo, S., Muñoz-Vargas, M. A., Palma, J. M., Corpas, F. J. (2023). Class III Peroxidases (POD) in Pepper (Capsicum annuum L.): Genome-Wide Identification and Regulation during Nitric Oxide (NO)-Influenced Fruit Ripening. Antioxidants 12, 1013.
Guo, A.-Y., Zhu, Q.-H., Chen, X., Luo, J.-C. (2007). GSDS: a gene structure display server. Yi chuan= Hereditas 29, 1023–1026. doi: 10.1360/yc-007-1023
Han, Y., Ding, T., Su, B., Jiang, H. (2016). Genome-wide identification, characterization and expression analysis of the chalcone synthase family in maize. Int. J. Mol. Sci. 17, 161. doi: 10.3390/ijms17020161
Han, F., Wang, P., Chen, X., Zhao, H., Zhu, Q., Song, Y., et al. (2023). An ethylene-induced NAC transcription factor acts as a multiple abiotic stress responsor in conifer. Horticult. Res. 10, uhad130. doi: 10.1093/hr/uhad130
Hiraga, S., Sasaki, K., Ito, H., Ohashi, Y., Matsui, H. (2001). A large family of class III plant peroxidases. Plant Cell Physiol. 42, 462–468. doi: 10.1093/pcp/pce061
Jeffares, D. C., Penkett, C. J., Bähler, J. (2008). Rapidly regulated genes are intron poor. Trends Genet. 24, 375–378. doi: 10.1016/j.tig.2008.05.006
Jin, T., Sun, Y., Zhao, R., Shan, Z., Gai, J., Li, Y. (2019). Overexpression of peroxidase gene GsPRX9 confers salt tolerance in soybean. Int. J. Mol. Sci. 20, 3745. doi: 10.3390/ijms20153745
Joo, J. H., Bae, Y. S., Lee, J. S. (2001). Role of auxin-induced reactive oxygen species in root gravitropism. Plant Physiol. 126, 1055–1060. doi: 10.1104/pp.126.3.1055
Jozefczuk, J., Adjaye, J. (2011). “Quantitative real-time PCR-based analysis of gene expression,” in Methods in enzymology (Elsevier), 99–109.
Khan, W. U., Khan, L. U., Chen, D., Chen, F. (2023). Comparative analyses of superoxide dismutase (SOD) gene family and expression profiling under multiple abiotic stresses in water lilies. Horticulturae 9, 781. doi: 10.3390/horticulturae9070781
Khan, W. U., Khan, L. U., Khan, N. M., Wenquan, W., Chen, F. (2024). Evolutionary insights and expression dynamics of the glutathione peroxidase (GPX) gene family in water lily (Nymphaea colorata) in response to multiple abiotic stresses. Plant Stress. 14, 1–12 doi: 10.1016/j.stress.100699
Kidwai, M., Ahmad, I. Z., Chakrabarty, D. (2020). Class III peroxidase: An indispensable enzyme for biotic/abiotic stress tolerance and a potent candidate for crop improvement. Plant Cell Rep. 39, 1381–1393. doi: 10.1007/s00299-020-02588-y
Kumar, S., Jaggi, M., Sinha, A. K. (2012). Ectopic overexpression of vacuolar and apoplastic Catharanthus roseus peroxidases confers differential tolerance to salt and dehydration stress in transgenic tobacco. Protoplasma 249, 423–432. doi: 10.1007/s00709-011-0294-1
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lamesch, P., Berardini, T. Z., Li, D., Swarbreck, D., Wilks, C., Sasidharan, R., et al. (2012). The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40, D1202–D1210. doi: 10.1093/nar/gkr1090
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., Bork, P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Letunic, I., Doerks, T., Bork, P. (2012). SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40, D302–D305. doi: 10.1093/nar/gkr931
Li, X.-L., Zhang, J.-K., Li, M.-J., Zhou, B.-B., Zhang, Q., Wei, Q.-P. (2018). Genome-wide analysis of antioxidant enzyme gene families involved in drought and low-temperature responses in Apple (Malus domestica). J. Hortic. Sci. Biotechnol. 93, 337–346. doi: 10.1080/14620316.2017.1382314
Llorente, F., López-Cobollo, R. M., Catalá, R., Martínez-Zapater, J. M., Salinas, J. (2002). A novel cold-inducible gene from Arabidopsis, RCI3, encodes a peroxidase that constitutes a component for stress tolerance. Plant J. 32, 13–24. doi: 10.1046/j.1365-313X.2002.01398.x
Lüthje, S., Meisrimler, C.-N., Hopff, D., Möller, B. (2011). Phylogeny, topology, structure and functions of membrane-bound class III peroxidases in vascular plants. Phytochemistry 72, 1124–1135. doi: 10.1016/j.phytochem.2010.11.023
Marchler-Bauer, A., Derbyshire, M. K., Gonzales, N. R., Lu, S., Chitsaz, F., Geer, L. Y., et al. (2015). CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43, D222–D226. doi: 10.1093/nar/gku1221
Mika, A., Boenisch, M. J., Hopff, D., Lüthje, S. (2010). Membrane-bound guaiacol peroxidases from maize (Zea mays L.) roots are regulated by methyl jasmonate, salicylic acid, and pathogen elicitors. J. Exp. Bot. 61, 831–841. doi: 10.1093/jxb/erp353
Muthamilarasan, M., Khandelwal, R., Yadav, C. B., Bonthala, V. S., Khan, Y., Prasad, M. (2014). Identification and molecular characterization of MYB transcription factor superfamily in C4 model plant foxtail millet (Setaria italica L.). PloS One 9, e109920. doi: 10.1371/journal.pone.0109920
Obaid, U. S., Latif, U. K., Sana, B., Lingling, Z., Muhammad, I., Jiantao, P., et al. (2024). Genome-wide investigation of class III peroxidase genes in brassica napus reveals their responsiveness to abiotic stresses. Plants 13, 942. doi: 10.3390/plants13070942
Pandey, V. P., Awasthi, M., Singh, S., Tiwari, S., Dwivedi, U. N. (2017). A comprehensive review on function and application of plant peroxidases. Biochem. Anal. Biochem. 6, 308. doi: 10.4172/2161-1009.1000308
Passardi, F., Cosio, C., Penel, C., Dunand, C. (2005). Peroxidases have more functions than a Swiss army knife. Plant Cell Rep. 24, 255–265. doi: 10.1007/s00299-005-0972-6
Passardi, F., Longet, D., Penel, C., Dunand, C. (2004a). The class III peroxidase multigenic family in rice and its evolution in land plants. Phytochemistry 65, 1879–1893. doi: 10.1016/j.phytochem.2004.06.023
Passardi, F., Penel, C., Dunand, C. (2004b). Performing the paradoxical: how plant peroxidases modify the cell wall. Trends Plant Sci. 9, 534–540. doi: 10.1016/j.tplants.2004.09.002
Potter, S. C., Luciani, A., Eddy, S. R., Park, Y., Lopez, R., Finn, R. D. (2018). HMMER web server: 2018 update. Nucleic Acids Res. 46, W200–W204. doi: 10.1093/nar/gky448
Qiang, L., Xiujuan, Q., Miao, Z., Qiyuan, Y., Ruirui, J., Fan., J., et al. (2023). CsBZIP40 confers resistance against citrus bacterial canker by repressing CsWRKY43-CsPrx53/CsSOD13 cascade mediated ROS scavenging. Horticult. Res. 10, uhad138. doi: 10.1093/hr/uhad138
Ren, L.-L., Liu, Y.-J., Liu, H.-J., Qian, T.-T., Qi, L.-W., Wang, X.-R., et al. (2014). Subcellular relocalization and positive selection play key roles in the retention of duplicate genes of Populus class III peroxidase family. Plant Cell 26, 2404–2419. doi: 10.1105/tpc.114.124750
Shang, H., Fang, L., Qin, L., Jiang, H., Duan, Z., Zhang, H., et al. (2023). Genome-wide identification of the class III peroxidase gene family of sugarcane and its expression profiles under stresses. Front. Plant Sci. 14, 1101665. doi: 10.3389/fpls.2023.1101665
Thompson, J. D., Higgins, D. G., Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Tognolli, M., Penel, C., Greppin, H., Simon, P. (2002). Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene 288, 129–138. doi: 10.1016/S0378-1119(02)00465-1
Valério, L., De Meyer, M., Penel, C., Dunand, C. (2004). Expression analysis of the Arabidopsis peroxidase multigenic family. Phytochemistry 65, 1331–1342. doi: 10.1016/j.phytochem.2004.04.017
Wally, O., Punja, Z. K. (2010). Enhanced disease resistance in transgenic carrot (Daucus carota L.) plants over-expressing a rice cationic peroxidase. Planta 232, 1229–1239. doi: 10.1007/s00425-010-1252-4
Wang, Y., Wang, Q., Zhao, Y., Han, G., Zhu, S. (2015). Systematic analysis of maize class III peroxidase gene family reveals a conserved subfamily involved in abiotic stress response. Gene 566, 95–108. doi: 10.1016/j.gene.2015.04.041
Wu, C., Ding, X., Ding, Z., Tie, W., Yan, Y., Wang, Y., et al. (2019). The class III peroxidase (POD) gene family in cassava: identification, phylogeny, duplication, and expression. Int. J. Mol. Sci. 20, 2730. doi: 10.3390/ijms20112730
Wu, Y., Yang, Z., How, J., Xu, H., Chen, L., Li, K. (2017). Overexpression of a peroxidase gene (AtPrx64) of Arabidopsis thaliana in tobacco improves plant’s tolerance to aluminum stress. Plant Mol. Biol. 95, 157–168. doi: 10.1007/s11103-017-0644-2
Xiao, H., Wang, C., Khan, N., Chen, M., Fu, W., Guan, L., et al. (2020). Genome-wide identification of the class III POD gene family and their expression profiling in grapevine (Vitis vinifera L). BMC Genomics 21, 1–13. doi: 10.1186/s12864-020-06828-z
Xiong, X., Zhang, J., Yang, Y., Chen, Y., Su, Q., Zhao, Y., et al. (2023). Water lily research: Past, present, and future. Trop. Plants 2, 1–8. doi: 10.48130/TP-2023-0001
Xue, Y. J., Tao, L., Yang, Z. M. (2008). Aluminum-induced cell wall peroxidase activity and lignin synthesis are differentially regulated by jasmonate and nitric oxide. J. Agric. Food Chem. 56, 9676–9684. doi: 10.1021/jf802001v
Yan, J., Su, P., Li, W., Xiao, G., Zhao, Y., Ma, X., et al. (2019). Genome-wide and evolutionary analysis of the class III peroxidase gene family in wheat and Aegilops tauschii reveals that some members are involved in stress responses. BMC Genomics 20, 1–19. doi: 10.1186/s12864-019-6006-5
Yang, X., Yuan, J., Luo, W., Qin, M., Yang, J., Wu, W., et al. (2020). Genome-wide identification and expression analysis of the class III peroxidase gene family in potato (Solanum tuberosum L.). Front. Genet. 11, 593577. doi: 10.3389/fgene.2020.593577
Zhang, L., Chen, F., Zhang, X., Li, Z., Zhao, Y., Lohaus, R., et al. (2020). The water lily genome and the early evolution of flowering plants. Nature 577, 79–84. doi: 10.1038/s41586-019-1852-5
Keywords: plant peroxidases, water lily, expression pattern, phylogenetic analysis, stresses
Citation: Khan WU, Khan LU, Khan NM, Zhang J, Wenquan W and Chen F (2025) Comprehensive identification, characterization and expression analyses of the class III POD gene family in water lily (Nymphaea colorata). Front. Plant Sci. 15:1524657. doi: 10.3389/fpls.2024.1524657
Received: 08 November 2024; Accepted: 30 December 2024;
Published: 20 January 2025.
Edited by:
Umashankar Chandrasekaran, Kangwon National University, Republic of KoreaReviewed by:
Qibin Wu, Chinese Academy of Tropical Agricultural Sciences, ChinaSami Abou Fayssal, Sofia, Bulgaria
Muhammad Aamir Manzoor, Shanghai Jiao Tong University, China
Copyright © 2025 Khan, Khan, Khan, Zhang, Wenquan and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fei Chen, ZmVpY2hlbkBoYWluYW51LmVkdS5jbg==
 Wasi Ullah Khan
Wasi Ullah Khan Latif Ullah Khan
Latif Ullah Khan Noor Muhammad Khan
Noor Muhammad Khan Ji Zhang
Ji Zhang Wang Wenquan
Wang Wenquan Fei Chen
Fei Chen