- 1Guangdong Engineering Technology Research Centre of Modern Eco-agriculture and Circular Agriculture, Department of Ecology, College of Natural Resources and Environment, South China Agricultural University, Guangzhou, China
- 2Key Laboratory of Arable Land Conservation (Middle and Lower Reaches of Yangtze River), Ministry of Agriculture, Micro-elements Research Center, College of Resource and Environment, Huazhong Agricultural University, Wuhan, China
- 3Department of Biology, Faculty of Science, University of Tabuk, Tabuk, Saudi Arabia
- 4Biodiversity Genomics Unit, Faculty of Science, University of Tabuk, Tabuk, Saudi Arabia
- 5College of Agriculture, South China Agricultural University, Guangzhou, China
- 6The Germplasm Bank of Wild Species, Yunnan Key Laboratory for Fungal Diversity and Green Development, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, Yunnan, China
- 7Guangdong Provincial Key Laboratory of Postharvest Science of Fruits and Vegetables/Engineering Research Center for Postharvest Technology of Horticultural Crops in South China, Ministry of Education, College of Horticulture, South China Agricultural University, Guangzhou, Guangdong, China
- 8Department of Botany and Microbiology, Faculty of Science, Assiut Universityt, Assiu, Egypt
Molybdenum (Mo) deficiency is a global problem in acidic soils, limiting plant growth, development, and nutrient availability. To address this, we carried out a field study with two treatments, i.e., Mo applied (+Mo) and without Mo (−Mo) treatment to explore the effects of Mo application on crop growth and development, microbial diversity, and metabolite variations in maize and soybean cropping systems. Our results indicated that the nutrient availability (N, P, K) was higher under Mo supply leading to improved biological yield and nutrient uptake efficiency in both crops. Microbial community analysis revealed that Proteobacteria and Acidobacteria were the dominant phyla in Mo treated (+Mo) soils for both maize and soybean. Both these phyla accounted together 39.43% and 57.74% in −Mo and +Mo, respectively, in soybean rhizosphere soil, while they accounted for 44.51% and 46.64% in maize rhizosphere soil. This indicates more variations among the treatments in soybean soil compared to maize soil. At a lower taxonomic level, the diverse responses of the genera indicated the specific bacterial community adaptations to fertilization. Candidatus Koribacter and Kaistobacter were commonly significantly higher in both crops under Mo-applied conditions in both cropping systems. These taxa, sharing similar functions, could serve as potential markers for nutrient availability and soil fertility. Metabolite profiling revealed 8 and 10 significantly differential metabolites in maize and soybean, respectively, under +Mo treatment, highlighting the critical role of Mo in metabolite variation. Overall, these findings emphasize the importance of Mo in shaping soil microbial diversity by altering metabolite composition, which in turn may enhance the nutrient availability, nutrient uptake, and plant performance.
1 Introduction
Molybdenum is essentially required by plants and rhizosphere microbiota for sustainable crop production. Mo deficiency is common in the acidic soils of China. Mo-deficient plants exhibit stunted growth and poor nutrient interaction is rhizosphere soil (Nie et al., 2015). Mo is an essential component of several enzymes, including nitrate reductase (NR), aldehyde oxidase (AO), and xanthine dehydrogenase (XDH), which play critical roles in the proper physiological functioning of various crops (Huang et al., 2022). Under Mo-deficient conditions, plants experience a variety of phenotypic changes that hinder their growth. These effects are primarily associated with the impaired activity of molybdoenzymes. Notably, these include key enzymes involved in N metabolism, such as NR and nitrogenase. Nitrogenase, in particular, plays a critical role in N fixation within the bacteroids of legume nodules. Other molybdoenzymes found in plants include XDH, which is essential for purine degradation and ureide synthesis in legumes; AO, which participates in the biosynthesis of abscisic acid; and sulfite oxidase, which converts sulfite to sulfate, a vital step in the metabolism of sulfur-containing amino acids (Mendel and Hänsch, 2002; Sauer and Frebort, 2003; Mendel and Kruse, 2012). The availability of Mo for plant uptake is significantly affected by soil pH, the presence of adsorbing oxides, such as iron oxides, water drainage conditions, and organic compounds in soil colloids. In alkaline soils with high pH, Mo becomes more soluble and is predominantly available to plants in its anionic form, MoO4−. In contrast, in acidic soils with a pH below 5.5, Mo availability decreases due to increased adsorption of the anion onto soil oxides (Brinza et al., 2008). Moreover, organic matter in the soil can interact with molybdenum to form complexes, thereby affecting its availability to plants. Soils with high organic matter content can improve Mo retention and enable its slow, sustained release providing a consistent supply for plant uptake. This controlled release can profoundly impact microbial activity and play a crucial role in supporting long-term nutrient cycling within the soil ecosystem. It indicates that Mo has a critical role in proper plant functioning and related biological processes happening in the rhizosphere soil (Rana et al., 2020a). Therefore, understanding the impact of long-term Mo supply on soil microbiota, rhizosphere metabolic profiling, and nutrient acquisition by different crops is highly essential for agriculture sustainability.
The nutrient status in soil can regulate the microbial diversity (Kang et al., 2024). Microbial populations exhibit significant dynamism and self-organization in response to the specific nutrients. As a result, both biotic factors, such as ecological interactions, and abiotic factors, like nutrient availability, shape microbial community composition. Studies on their spatial distribution have shown that distinct microbial arrangements emerge under varying levels of nutrient limitations (Mitri et al., 2016). In nutrient-limited conditions, cooperative behaviors, such as the secretion of metabolites, can maintain diversity (Gandhi et al., 2019; Kang et al., 2024). These shifts in microbial communities reflect the assembly processes that link environmental factors to community structure and function. The most recent development of high-throughput sequencing and culture-independent molecular technologies, using 16S rRNA that encodes the small ribosomal unit of RNA, has provided major insights into bacterial functional population diversity under different fertilizer management practices (Dinsdale et al., 2008; Spor et al., 2011). Microbial diversity analysis not only addresses variations in the function and composition of the bacterial community (Abia et al., 2018) but also describes the association among the external drivers and community components (Langenheder et al., 2010). Molecular tactics may answer the questions about the long-term fertilizer management (Mo-supply) effects on the alteration of soil bacterial diversity and abundance.
Root exudates play significant roles in nutrient acquisition and interaction with rhizosphere microorganisms (Selvakumar et al., 2012). Therefore, it is necessary to analyze root metabolites for an understanding of the interaction between soil microorganisms and roots. Previous studies have designated that plants shape and drive the surrounding microbiome through exudate secretion that specifically represses or stimulates distinctive microbial members of soil. The secretions of root exudates in the rhizosphere depend on the availability of nutrients and the physiological stage of plants (Hartmann et al., 2009; Badri et al., 2013; Chaparro et al., 2013). High nutrient availability positively influences the release of root exudates, whereas low nutrient availability restricts the allocation of plant resources to root exudation and alters the microbiology of the rhizosphere (Liu et al., 2011). For example, during N deficiency, less amino acid is secreted by roots of maize plants. This fluctuation in root exudates could impact the microbial community structure and nutrient uptake by plants (Carvalhais et al., 2013). Recently, “omics” approaches (e.g., LM-MS) are being widely used to describe the different mechanistic alterations happening in the rhizosphere soil for better understating of rhizosphere processes (Urano et al., 2010; Booth et al., 2011). The metabolomic approach can unravel the complex underlying mechanisms in the rhizosphere by profiling root exudates, which play a crucial role in various biochemical processes across biological systems (Patti et al., 2012).
Mo can exist in multiple oxidation states ranging from zero to VI, with VI being the most common in soils. Similar to other metals essential for plant growth, Mo plays a vital role in facilitating redox reactions through specific plant enzymes. However, Mo itself is not directly biologically active. Instead, it is primarily incorporated into an organic pterin complex known as the Mo co-factor. This co-factor is associated with Mo-dependent enzymes, or molybdoenzymes, found across the biological systems of plants, animals, and prokaryotes (Jean et al., 2013). Numerous studies have shown that Mo application enhances nutrient availability in the soil, which can contribute to improved plant performance. These outcomes indicated a synergistic interaction between the application of Mo and other essential elements (Nie et al., 2015). This study highlights the previously underexplored role of Mo in shaping rhizosphere soil biological processes, including variations in metabolites and shifts in bacterial community dynamics, under long-term Mo application. By examining the synergistic effects of Mo on nutrient acquisition, rhizosphere metabolites, and microbial composition in maize and soybean cropping system, the research offers fresh insights into Mo’s contribution to improving soil and crop health aligning seamlessly with the objective of unraveling the interaction between soil chemical and biological indicators.
2 Materials and methods
2.1 Experimental materials and treatment
A long-term experiment was set up in 2008 at the experimental area of Huazhong Agricultural University, Wuhan, China. Maize and soybean were cultivated in April in an average plot size of 18 m2 (9 m × 2 m) and harvested in August and September, respectively. The experiment consisted of two treatments for each crop, i.e., −Mo (Control) and +Mo, and each treatment consisted of three replicated plots. In maize and soybean, both treatments received the NPK@120:80:80 kg/ha as urea, superphosphate, and potassium chloride respectively. Phosphorus (P) and potassium (K) fertilizers were completely applied at the time of sowing, while N fertilizer was applied in two splits, i.e., before sowing and after sowing of crops. The +Mo treatment received the Mo fertilizer as ammonium molybdate (0.41 kg/ha) from June 2009 to October 2013. The available Mo content of soil showed a concentration of 1.80 mg/kg, which crossed the threshold normal demand of plant growth (1 mg/kg in soil) (He et al., 2005). Therefore, we closed the Mo fertilizer supply until the collection of soil samples. The test soil was yellow-brown (Alfisol) from Hubei Province (Xinzhou), China. Basic soil chemical characteristics were as follows: pH 5.64; organic matter, 15.5 g/kg; available N, 80.5 mg/kg; available P, 8.11 mg/kg; available K, 120.6 mg/kg; and available Mo, 0.112 mg/kg.
2.2 Sample collection
Plant and soil samples were collected during the maturity stages of crops. Rhizosphere soil samples from maize and soybean fields were collected by gently uprooting four to six plants from each plot, and the collected rhizosphere soils were combined to make a composite sample. The composite sample represents the single sample that was obtained after mixing of representative samples. The composite soil samples were then divided into two sets. One set of composite samples was transferred to ice boxes, where large organic debris was removed. The samples were then thoroughly homogenized, wrapped in aluminum foil, and stored in liquid nitrogen. These samples were transported to the laboratory in liquid nitrogen and stored at −80°C for molecular analysis. The other set of composite samples was air dried at room temperature and manually ground to pass through a 2-mm sieve for chemical analysis of the soil. The harvested plant samples were washed with distilled water and then oven dried (80°C) to achieve constant weight. The dried plant samples were then ground through a stainless-steel grinder for plant chemical analysis.
2.3 Soil and plant chemical analysis
The chemical properties of the soil were analyzed by following the methods of Bao (2002). The soil pH was measured in a 1:2.5 water suspension with a pH meter (Mettler Toledo, China) as described by Rana et al. (2018, 2020b). Soil organic matter (SOM) was determined through the dichromate digestion method. Available N was determined by the alkali hydrolysis–diffusion method, and P was determined using the Mo antimony colorimetric method by a spectrophotometer (Lu, 1999); available K was measured using the flame photometric method. The polarographic catalytic wave analysis technique was used to analyze the available Mo content in the soil (Wen et al., 2018). The total N concentrations were determined by following the method of Barbano and Clark (1990). Plant K concentration was determined using a flame photometer (Model 410, USA). P concentration in the digested tissues was determined colorimetrically followed by the Mo blue method (Lu, 1999). The Mo contents in plant samples were determined by polarographic catalytic wave analysis using a JP-2 oscilloscope polarograph according to Ismael et al. (2018); Imran et al. (2019a, 2019b), and Rana et al. (2020c, 2020d).
2.4 Bacterial community analysis
Through the PowerMax DNA isolation kit (MoBio Laboratories, Carlsbad, CA, USA), total genomic bacterial DNA was extracted from the soil sample according to the instructions of the manufacturer and stored at −20°C. The quantity and quality of extracted DNAs were measured using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and agarose gel electrophoresis, respectively.
The V4 region of bacterial 16S rRNA genes was subjected to PCR amplification through forward primer 515F (GTGCCAGCMGCCGCGGTAA) and reverse primer 806R (GGACTACHVGGGTWTCTAAT) (Caporaso et al., 2011; Ding et al., 2024). The PCR components consisted of 25 μl of PCR Master Mix, 3 μl of each forward and reverse primer (10 μM), 10 μl of DNA template, and 6 μl of ddH2O. Thermal cycling comprised a 30-s denaturation at 98°C, followed by 25 cycles consisting of denaturation at 98°C for 15 s, annealing at 58°C for 15 s, and extension at 72°C for 15 s, with a final 1-min extension at 72°C. Agencourt AMPure XP Beads (Beckman Coulter, Indianapolis, IN) were used to purify the PCR amplicons and quantified by the Kit (PicoGreen dsDNA Assay) (Invitrogen, Carlsbad, CA, USA). Amplicons were pooled in equal amounts after the individual quantification step, and then, using the Illumina HiSeq4000 platform, paired-end 2 × 150-bp sequencing was performed at GUHE Info technology Co., Ltd (Hangzhou, China). To process the data, the Quantitative Insights Into Microbial Ecology (QIIME, v1.9.0) pipeline was employed, as described by Caporaso et al. (2010). Briefly, raw sequencing reads that exactly matched the barcode were assigned to each sample and identified as valid sequences. Low-quality sequences were screened by the criteria explained by Gill et al. (2006) and Chen and Jiang (2014): sequences <150-bp length, sequences with an average Phred score of <20, sequences containing ambiguous bases, and sequences containing single nucleotide repeats of >8 bp. Paired-end reads were pulled together using FLASH (Magoč and Salzberg, 2011). OTU picking was performed using Vsearch v1.11.1, which included dereplication, cluster, and detection of chimeras (Rognes et al., 2016). Through default parameters, a representative sequence was selected from each OTU. VSEARCH search was used to perform OTU classification on representative sequences against the Greengenes database. An OTU table was also generated to record the abundance of each OTU in each sample and the classification of these OTUs. All those samples that contained less than 0.001% of the total sequence of OTUs were discarded. To minimize differences in sequencing depth between samples, an average, rounded OTU table was generated by averaging 100 uniformly resampled OTU subsets at 90% of the minimum sequencing depth for further analysis.
2.5 Metabolomics analysis
For analysis, 200-mg samples were used, and then 800 μl of methanol and 10 μl of internal standard (2.9 mg/mL, DL-o-chlorophenylalanine) were added. Thereafter, the samples were subjected to vortexing (30 s) and centrifugation (12,000 rpm) at 4°C for 15 min. The supernatant was collected and concentrated by adding 200 μl of methanol and moved to a vial for analysis. The instrument analysis platform was LC-MS (Thermo, Ultimate 3000LC, Orbitrap Elite), while the chromatographic column was C18 [Hypergod C18 (100 × 4.6 mm 3 µm)]. Chromatographic separation conditions were as follows: column temperature: 40°C, flow rate: 0.3 ml/min, mobile phase A: water + 0.1% formic acid, mobile phase B: acetonitrile + 0.1% formic acid, injection volume: 4 ml, automatic injector temperature: 4°C, ESI: heater temperature 300°C, sheath gas flow rate: 45 arb, auxiliary gas flow rate: 15 arb, sweep gas flow rate: 1 arb, spray voltage: 3.0 kV, capillary temperature: 350°C, and S-Lens RF level: 30%. The gradient of the mobile phase is shown in Supplementary Table S1.
2.6 Statistical analysis
The least significant difference (LSD) test at p < 0.05 was used for mean variances of the data. Statistix 8.1 software (Analytical Software, Tallahassee, FL, USA) was used for statistical analyses of data following analysis of variance. The analyses of sequence data were carried out through QIIME. The OTU table in QIIME was used to calculate the OTU level alpha diversity index. Generated abundance curves of OTU gradients were used to compare the richness and uniformity of OTUs between samples. UniFrac distance metrics were used for beta diversity analysis to study structural changes in microbial communities across samples (Lozupone and Knight, 2005; Lozupone et al., 2007) and visualized by principal coordinate analysis (PCoA) (Ramette, 2007). PCA was also performed based on the compositional profiles at the genus level (Ramette, 2007). The importance of microbiota structure differentiation between groups was evaluated through permutational multivariate analysis of variance (PERMANOVA) (McArdle and Anderson, 2001) using R package (vegan). GraPhlAn (Asnicar et al., 2015) and MEGAN were used to visualize the abundance and taxonomic composition (Huson et al., 2011). To see the unique and shared OTUs between the treatments, a Venn diagram was created using the R package that was constructed based on the presence of OTUs regardless of their relative abundance (Zaura et al., 2009). Based on high-quality sequences, microbial functions were predicted using phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt) (Langille et al., 2013). The output files were further analyzed using the statistical analysis of metagenomic profiles (STAMP) software package (Parks et al., 2014). Beta diversity and Meta-Storms distance-based functions were created using Parallel-META 3 (Jing et al., 2017). For the metabolite variations, the data were analyzed using feature extraction and pre-processed using SIEVE software, then normalized through Excel 2010 and edited into a two-dimensional data matrix, together with retention time (RT), compound molecular weight (compMW), observed values (sample), and peak intensity. The data were subjected to MVA (multivariate analysis) using the SIMCA-P software (Umetrics AB, Umea, Sweden). OPLS-DA model’s VIP (important variable in projection) value (threshold value >1) and t-test p value (p < 0.05) were used to find differentially expressed metabolites.
3 Results
3.1 Effect of Mo on soil physiochemical characteristics
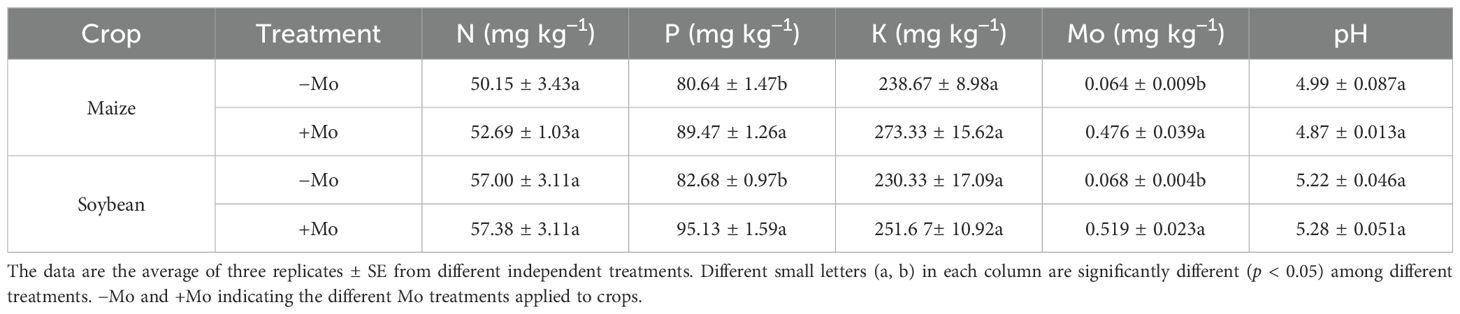
The chemical properties of soil changed substantially after long-term Mo applications (Table 1). Soil pH showed decreasing and increasing trends for both crops. In particular, there was no significant effect on soil pH. The pH increase was minor under the +Mo treatment in the soybean rhizosphere soil, while in the maize crop, it indicated a reduction difference of 0.12 U under the +Mo treatment. The changes in soil pH were slightly greater in maize rhizosphere compared to those in soybean rhizosphere (Table 1). The concentrations of N, P, K, and Mo were significantly (p < 0.05) higher in the soil that received treatment with Mo than those in the soil that received treatment without Mo. In addition, P and K concentrations in soybean soils were 15.05% and 9.26% higher in the +Mo treatment compared to those in the −Mo treatment. Conversely, N concentration decreased to 0.66%, whereas in maize rhizosphere soil, we found that Mo supply increased the N, P, and K concentrations up to 5.06%, 10.94%, and 14.52%, respectively.
3.2 Effect of Mo on concentration and accumulation of nutrients in different parts of the plants
Maize showed a higher N uptake in all parameters except for the leaves with the +Mo treatment, with significant differences observed in the stem (0.69%), bractea (0.72%), and grain (2.18%) (Figure 1A). A similar trend was observed in soybean, with significant N concentration differences in roots and seeds (Figure 1B), where N acquisition was higher in soybean than in maize. K concentration decreased in all maize parameters except for the leaves and spikestalk, while it increased in soybean leaves (1.62%) and grains (1.98%) under Mo treatment (Figures 1E, F). Mo supply significantly increased P concentration in all plant parts of both crops, except for the maize bractea and spikestalk (Figure 1C). Mo concentration also significantly increased in all physical parameters for both crops (Figures 1G, H). Nutrient concentrations were generally higher in soybean than in maize. Biomass was significantly higher in maize with Mo application, except for the roots and spikestalk (Figures 2A, B). In soybean, Mo supply significantly increased biomass in most plant parts resulting in a higher grain yield compared to those with −Mo treatment.
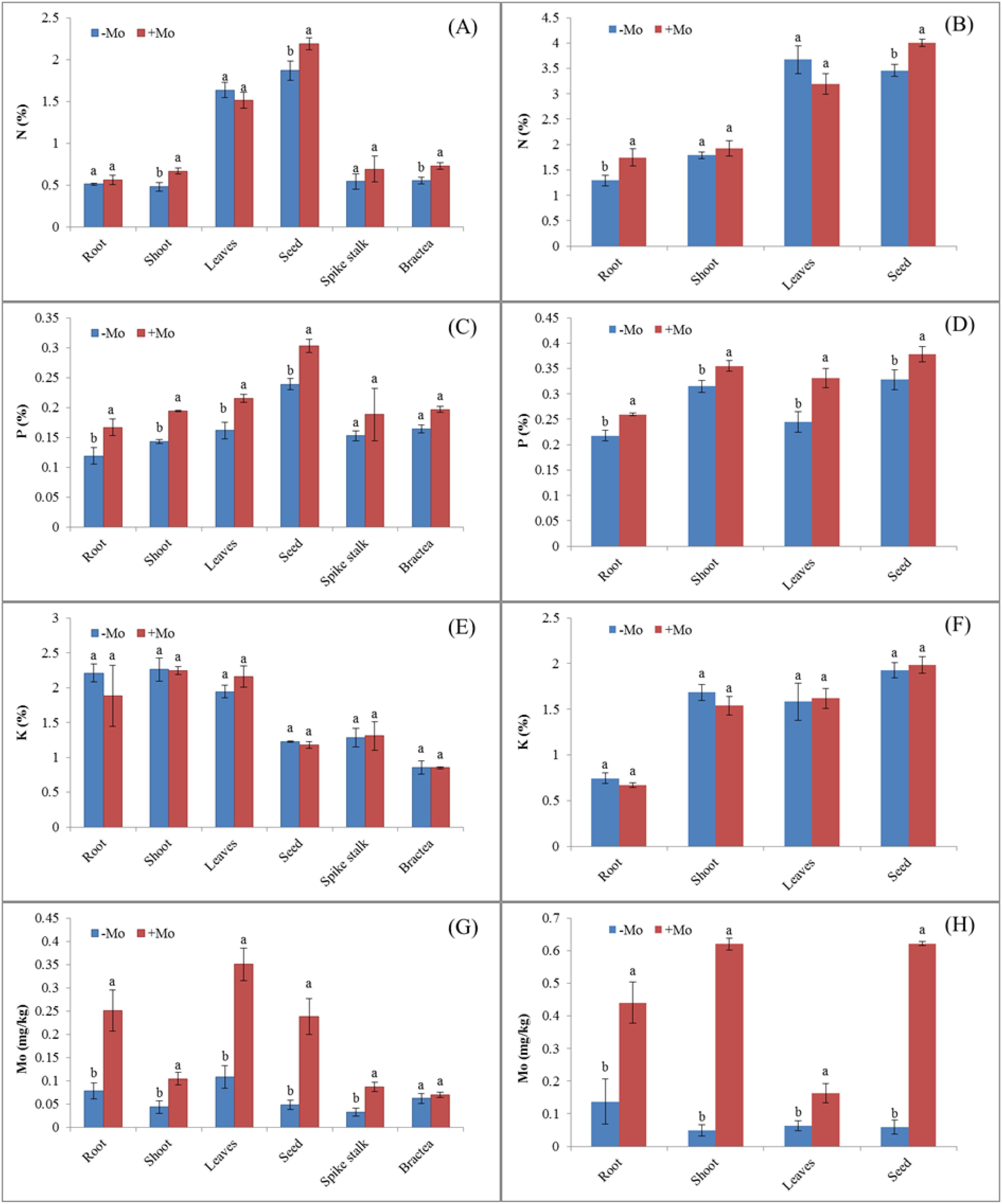
Figure 1. Nutrient acquisition in different parts of maize and soybean crops under Mo application. Treatments: −Mo (without Mo application) and +Mo (with Mo application). Vertical bars represent the standard error of three replicates. Different lowercase letters (a, b) indicate significant differences according to the LSD test (p < 0.05). (A, C, E, G), illustrate N, P, K, and Mo acquisition in maize, respectively, while (B, D, F, H), indicate these nutrient acquisitions in soybean crop.
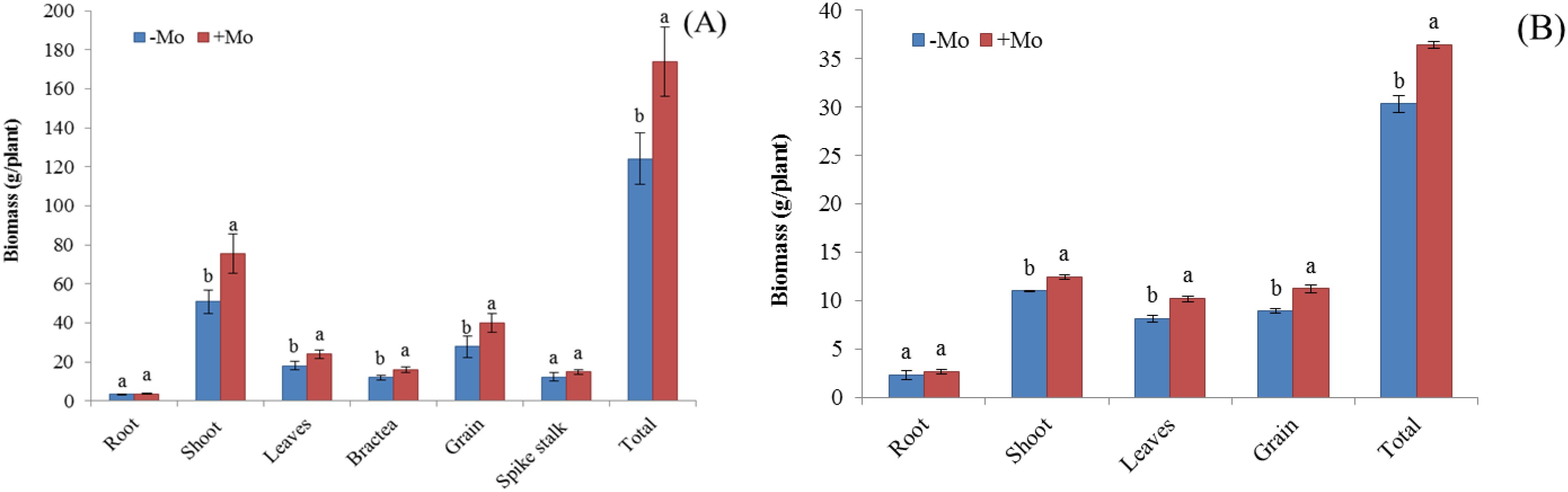
Figure 2. Effect of Mo application on biomass yield components of maize and soybean under +Mo (with Mo application) and −Mo (without Mo application) treatments. Vertical bars represent the standard error of three replicates. Different lowercase letters (a, b) indicate significant differences according to the LSD test (p < 0.05). (A) shows the treatment effects on maize growth, while (B) illustrates the effects on soybean growth.
Nitrogen accumulation in maize crop was significantly higher in stem, seed, and bractea in the +Mo treatment than those in the −Mo treatment. On the other hand, in soybean, a significant difference was observed only in the seeds, with higher nitrogen (N) accumulation in the +Mo treatment. In the other plant parts, N accumulation did not show significant differences between the two treatments (Supplementary Figure S1). In maize crops, the stems, leaves, seeds, and bracteas exhibited significantly higher P accumulation in Mo-applied treatment. In soybean, P accumulation was significantly high in all parameters except for the root under the +Mo treatment. No statistically significant difference was observed in K accumulation in the soybean under the Mo fertilizer. Mo accumulation was significantly higher in both crops in all the parameters except for the bractea and seed in maize and soybean, respectively (Supplementary Figure S1). Results also indicated that the concentration of nutrients in different parts were higher in the soybean crop than in the maize crop, while the nutrient accumulation results showed an opposite trend. The results indicated that Mo supply played an important role in enhancing nutrient acquisition and improved biomass compared to other treatments.
3.3 Response of species richness and bacterial community diversity under Mo supply
The average total valid OTUs were 2,328.66 in maize and 2,350.66 in soybean, with no significant difference observed between crops (ANOVA, p < 0.05). The highest OTU numbers were recorded under −Mo treatment in both crops. Alpha diversity analysis using the Shannon index showed no significant effect of fertilization, though values were higher with +Mo in maize and lower in soybean. Similar trends were observed with the Simpson index (Table 2). The Chao 1 estimator also indicated no significant differences between treatments but highlighted higher alpha diversity in soybean rhizosphere soil with Mo application. Rarefaction analysis was carried out with each sample to evaluate whether more sampling would add more OTUs. The rarefaction curve represented that in both crops, −Mo showed a steeper curve, while +Mo indicated a less steep slope (Figure 3). Although thousands of tags were observed in both crops, none of the curves appeared to be gentle or reach a plateau indicating high species diversity in the samples. 16S rRNA gene analysis indicated that crop and fertilization can influence the bacterial community taxonomic composition. A Venn diagram shows the degree of interaction of bacterial OTUs among the −Mo and +Mo treatment (Supplementary Figures S2A, B). The results clearly demonstrated that 1,080 OUTs were common in maize soil, while 1,086 OUT were similar in soybean crop under both treatments. This indicates that there was much similarity between the crops in the sharing of OTUs among the −Mo and +Mo treatments. In comparison between maize and soybean, it can be seen from the results that soybean exhibited a steeper slope compared to maize (Figures 3A, B).
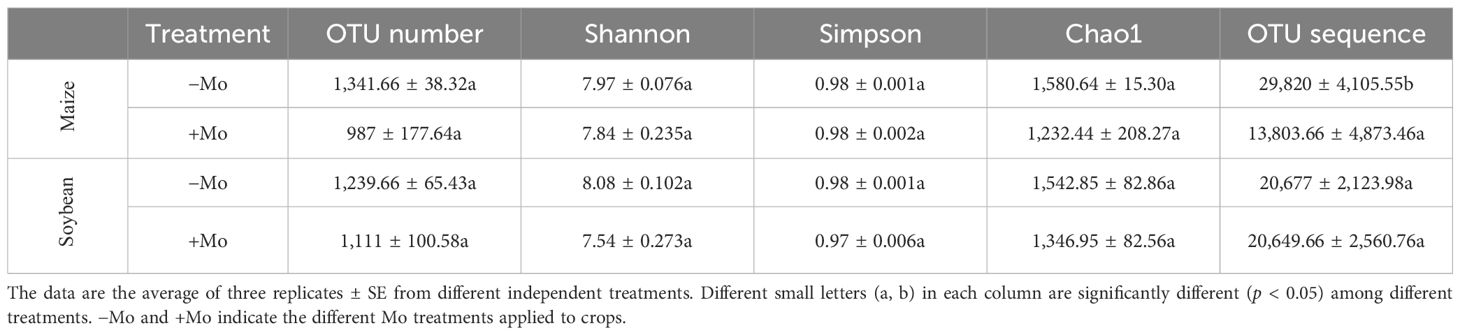
Table 2. Effect of the Mo supply on alpha diversity of bacterial community in miaze and soybean soil.
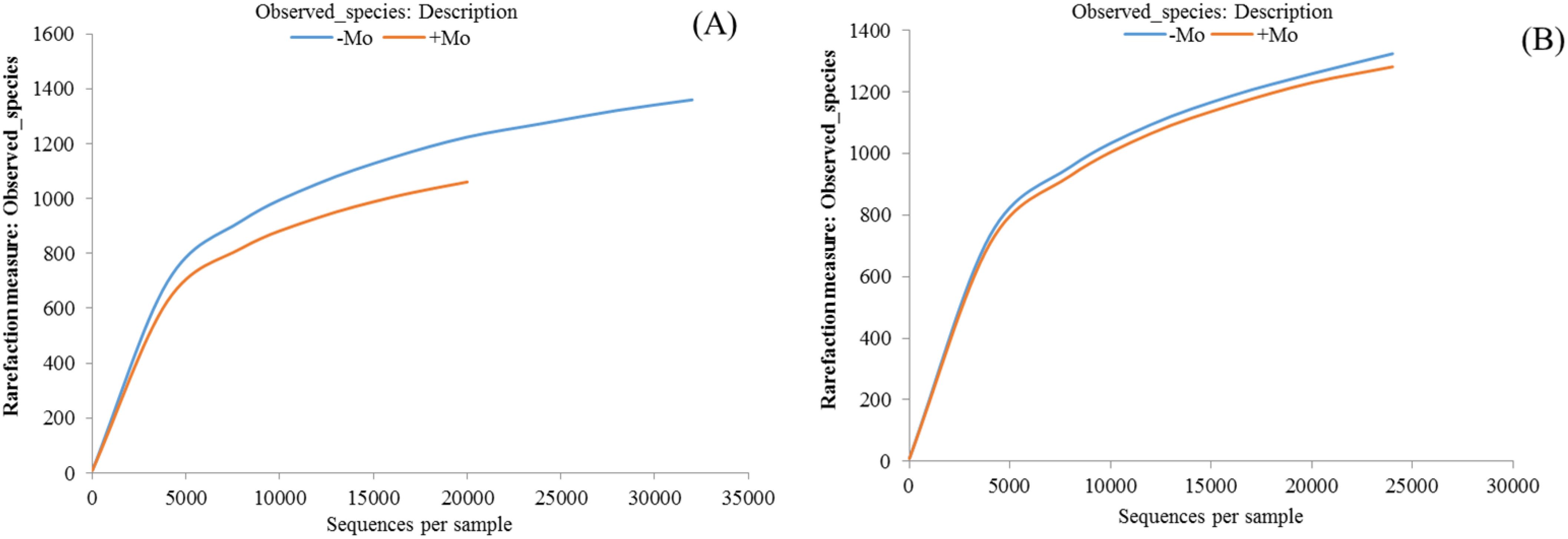
Figure 3. Rarefaction curves showing the observed operational taxonomic units (OTUs) in maize and soybean under +Mo (with Mo application) and −Mo (without Mo application) treatments. (A, B) indicate the maize and soybean crops, respectively. The abscissa in the refraction curve represents the number of sequences, while the ordinates indicate the number of OTUs observed. The abscissa position at the end point of the sample curve is the number of sequences of the sample.
3.4 Effect of Mo on bacterial community structure
Fertilizer treatments −Mo and +Mo had various effects on the composition of bacteria at the phylum level. The distribution of each phylum was different in each treatment for both crops. The distribution of the top 10 phyla is shown in Figures 4A, B. In soybean, Acidobacteria was the most abundant (22.30%–35.17%), while in maize, it ranged from 21.30% to 22.57%. In maize, the dominant phylum was Proteobacteria, which showed higher values compared to other phyla. In maize and soybean, +Mo showed higher percentages of Proteobacteria and Acidobacteria compared to the −Mo treatment. These two phyla were found to occupy the subsequent top phyla in all soil samples accounting for 39.43% and 57.74% in soybean with −Mo and +Mo treatments, respectively. In maize, these phyla accounted for 44.51% and 46.64% for both treatments. The Mo supply increased the percentage of Acidobacteria, Actinobacteria, Proteobacteria, and the Chloroflexi in soybean (Figure 4B). Moreover, in soybean, there was a significant difference among the treatments in the abundance of Acidobacteria, Actinobacteria, and Proteobacteria. In contrast, Chloroflexi was more prominent in maize with the Mo application. In maize, Actinobacteria and Gemmatimonadetes showed significant differences between the two treatments.
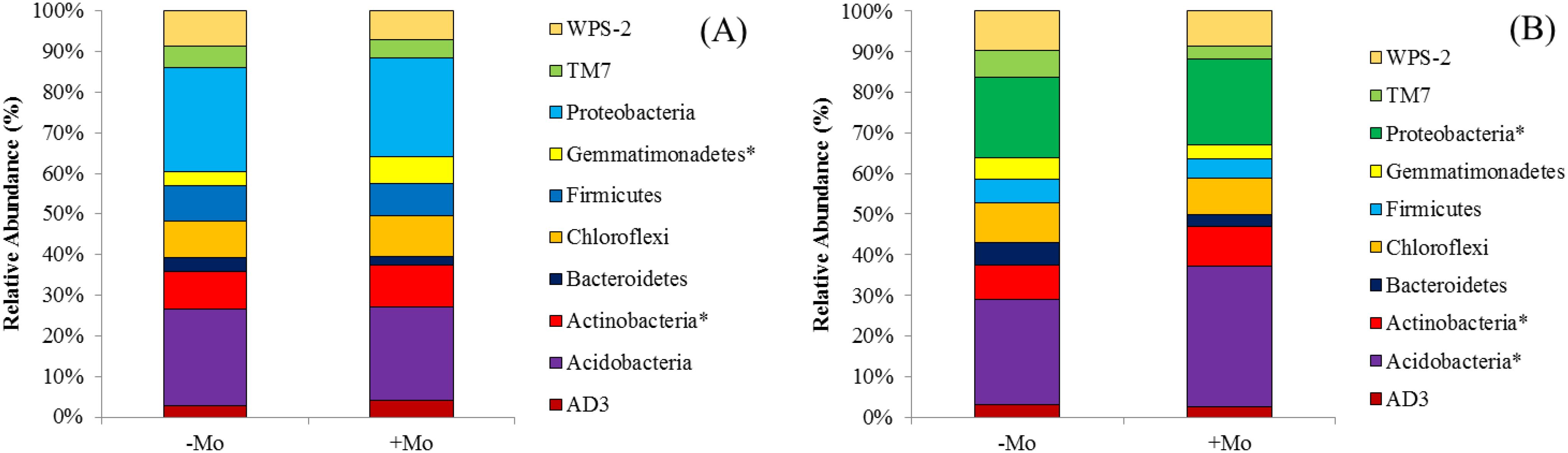
Figure 4. Proportional distribution of bacterial phyla in maize and soybean under +Mo (with Mo application) and −Mo (without Mo application) treatments. (A, B) indicate the maize and soybean crops, respectively. The star (*) represents significant differences using the LSD test (p < 0.05).
At the class level, there was a significant difference between the treatment for Acidobacteriia, Ktedonobacteria, iii-8, Actinobacteria, Thermomicrobia, Gemmatimonadetes, Deinococci, DA052, and α,β-proteobacteria in soybean crop. In soybean, four groups of Acidobacteriia were found in more abundance with the following order: Acidobacteriia > DA052 > Acidobacteria-6 > Solibacteres. Acidobacteriia, DA052, and Solibacteres was mostly enriched with the +Mo treatment, while the Acidobacreia-6 was higher in the −Mo treatment of soybean. In miaze, there was a significant difference in Acidobacteriia, Actinobacteria, Cytophagia, α, β, and δ-proteobacteria, TK-10, and 4C0d-2 between the −Mo and +Mo treatments. In maize, the increasing abundance order of Acidobacteria > DA052 > ABS-6 > Solibacteres showed a dominant percentage in +Mo treatment. The abundance of the Proteobacteria group, including α, β, γ, and δ-proteobacteria, was maximum in both crops (Supplementary Figures S3A, B). These classes of Proteobacteria phyla were higher with the Mo-applied treatments in maize and soybean. Acidobacteriia (20.73%), DA052 (10.67%), and α-proteobacteria (11.10%) in soybean and Acidobacteriia (12.97%), α-proteobacteria (12.90%), Ktedonobacteria (7.73%), and Bacilli (7.60%) in maize were prominently high with the Mo application (Supplementary Figures S3A, B).
At the genus level, this study showed that the bacterial community composition along with common 11 genera distribution varied between the samples of both crops (Figures 5A, B). A significant alteration was observed in the abundance of Candidatus koribacter, Rhodoplanes, and Kaistobacter in the soybean treatment, with these taxa being most abundant in the +Mo treatment. Besides this, Candidatus koribacter, Kaistobacter, Lactococcus, Rhodoplanes, and Burkholderia were significantly higher under +Mo in maize crop. Bacillus and Candidatus were the most abundant genera commonly found in all samples (Figures 5A, B). The phylogenetic dendrogram of specific bacteria showed significant differences between treatments indicating that Mo supply has an important role in bacterial community composition (Figures 6A, B). A plot of the most significant coordinates revealed a clear separation among the treatments (Figure 7). There was more variation among the treatments, but when we compared the maize and soybean crops, there was not much higher variation in beta diversity. The community structure of individual samples was compared through principal component and principal coordinate analyses using the weighted Unifrac metric. The PCoA scheme showed that the bacterial community structure in the −Mo treatment was different from that in the +Mo treatment. The first principle (PC1) component could highly decentralize the treatments that were with or without the application of Mo suggesting that the long-term application of Mo altered the bacterial community of rhizosphere soil. Fertilization management showed numerous impacts on bacterial composition at the phylum, genus, and class levels. It is clear form these results that Mo has a significant role in shaping the microbial community structure and diversity not even among the applied treatment but also between the maize and soybean plant soil.
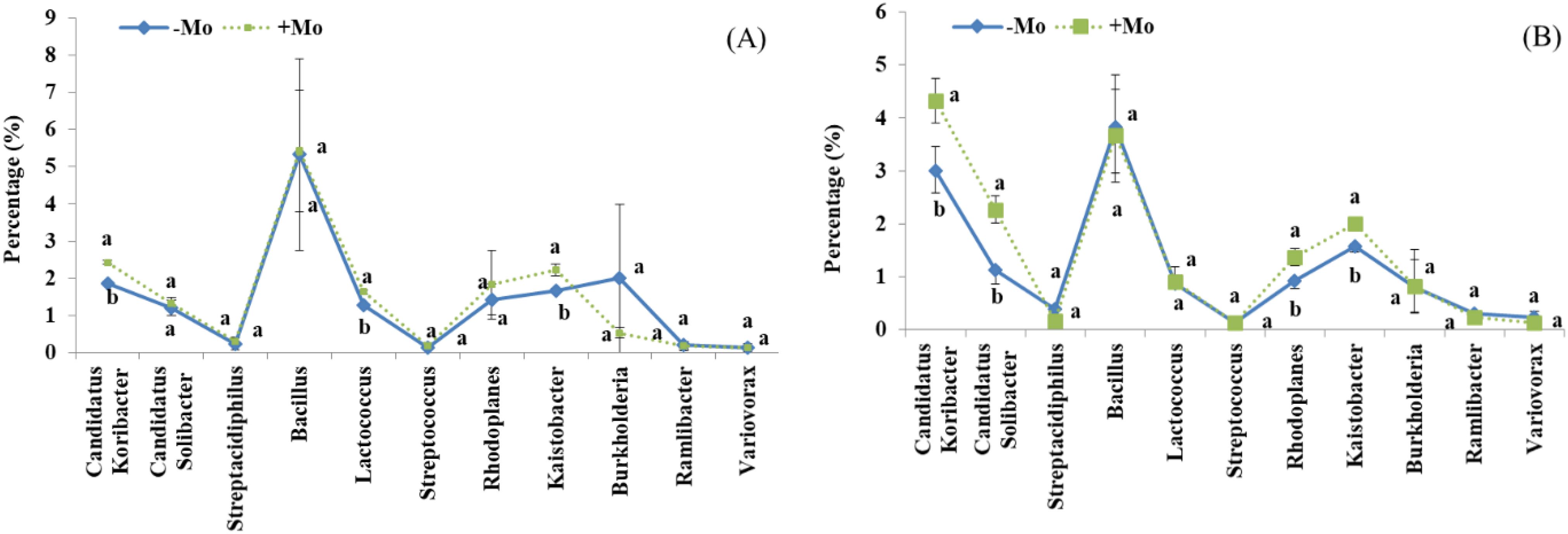
Figure 5. Proportional distribution (%) of the most abundant genera across all samples under +Mo (with Mo application) and −Mo (without Mo application) treatments. Vertical bars represent the standard error of three replicates. Different lowercase letters (a, b) indicate significant differences according to the LSD test (p < 0.05). (A, B) indicate the maize and soybean crops, respectively.
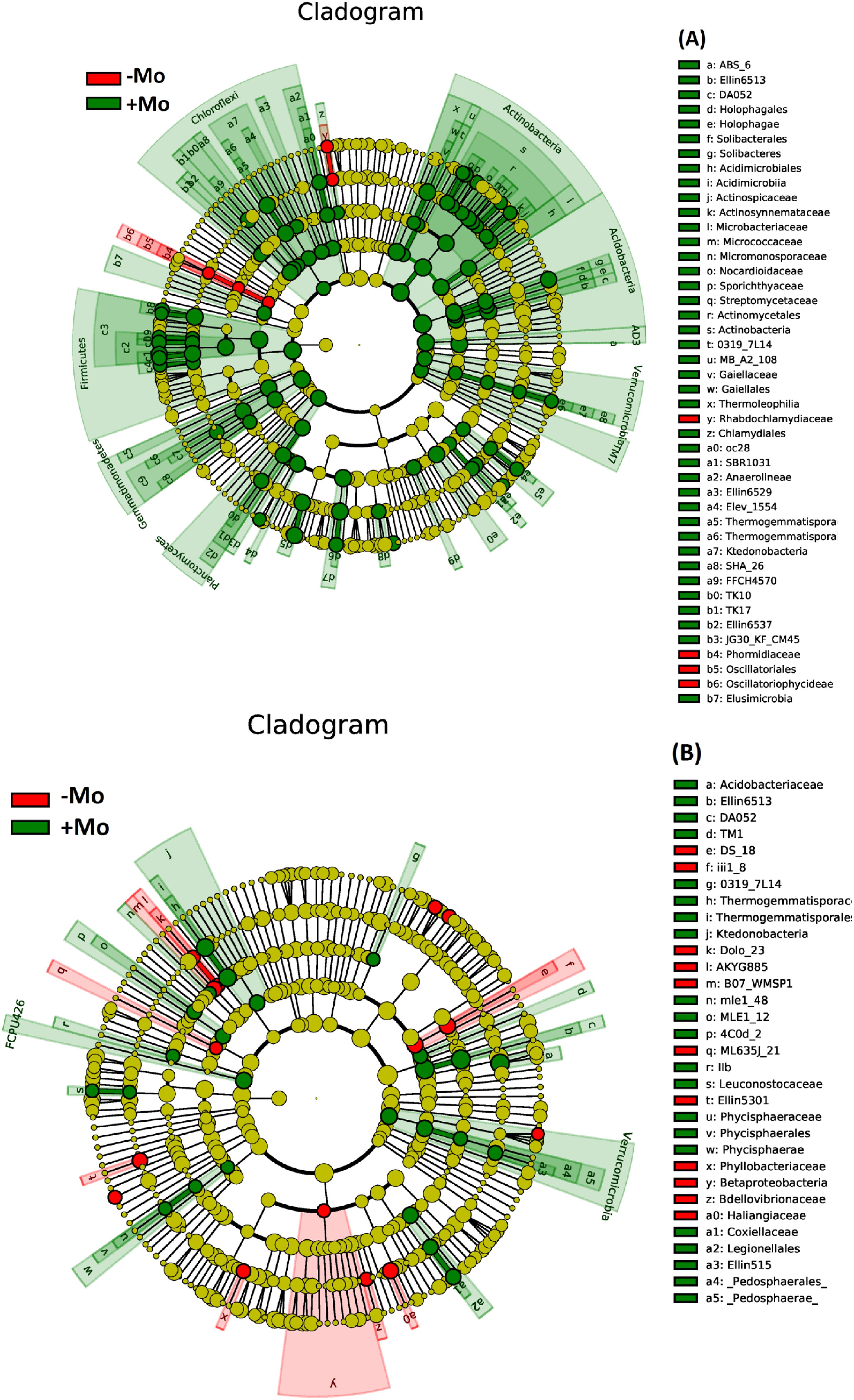
Figure 6. Phylogenetic dendrogram of specific bacteria in maize (A) and soybean (B) under +Mo (with Mo application) and −Mo (without Mo application) treatments. Different colors represent distinct bacterial groups, while the colored nodes in the dendrogram highlight bacterial groups that play key roles in the corresponding clusters. The species names represented by the letters are detailed in the legend on the right.

Figure 7. Principal component analysis (PCA) and principal coordinate analysis (PCoA) of beta diversity in soil samples of −Mo and +Mo treatments. (A, C, E), show the variations in maize, while (B, D, F), indicate these changes in soybean crop.
3.5 Effect of Mo on metabolite variations
To assess the impact of Mo application on different metabolites and identify those contributing to significant differences, PLS-DA and supervised OPLS-DA analyses were conducted. The key parameters for judging model quality were R2Y (which indicates the interpretation rate of the model) and the Q2 value (which is the prediction rate of the model). The score chart is shown in Figure 8 and Supplementary Figure S4. The treatments distinctively reacted to a majority of the deviations identified in the treatments across the two axis, and it also can be observed clearly that changes were also prominent under different treatments and crops. Fertilization and crop types resulted in a dramatic shift of metabolites in the rhizosphere. Overall, it could be concluded that fertilizer and crop management have a strong impact on the metabolite variations and their secretions. Different substances mainly included amino acids, organic acids, fatty acids, and so on. The significantly different metabolites in maize and soyabean among the two treatments are shown in Table 3. N-acetyl-p-benzoquinonimine, 5-hydroxyindol-2-carboxylic acid, and 4-carboxyphenylglycine were significantly higher in the maize crop under Mo applied treatment compared to those under −Mo treatment (Table 3). In soybean, L-serine, phenylacetic acid, PA(20:4(5Z,8Z,11Z,14Z)/22:1(11Z)), oxoglaucine, and Cer(t18:0/24:0(2–OH)) were significantly prominent in the treatment that received the Mo fertilizer compared to those in the −Mo treatment. When we compared the −Mo treatment between the maize and soybean crops, a total of 53 metabolites were found in number that significantly differed in both crops with the same treatment. Seven metabolites were upregulated in the maize crop, while all others were downregulated compared to those in the soybean crop (Supplementary Table S2). In the comparison of Mo application in both crops, 70 significantly different metabolites were identified in maize and soybean (Supplementary Table S3). Overall, it is concluded that fertilizer management and crops have a strong effect on the metabolites.
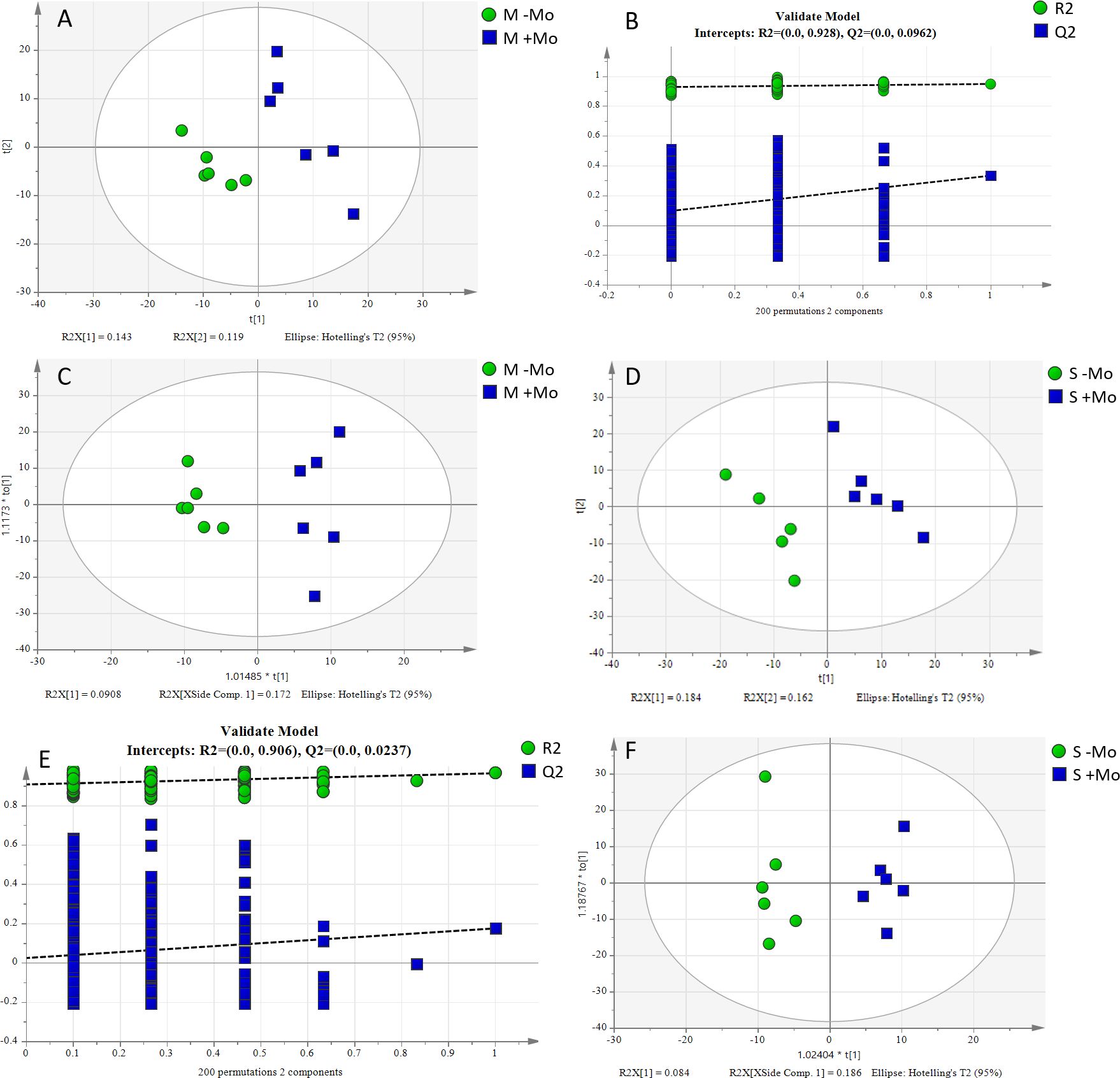
Figure 8. PLS-DA and OPLS-DA score plots and sorting validation diagrams for the −Mo (without Mo application) and +Mo (with Mo application) treatments. (A, B) indicate the PLS-DA score chart, while (C) represents the OPLS-DA score chart for the maize crop. Similarly, (D, E) indicate the PLS-DA score chart, while (F) represents the OPLS-DA score chart for the soybean crop.
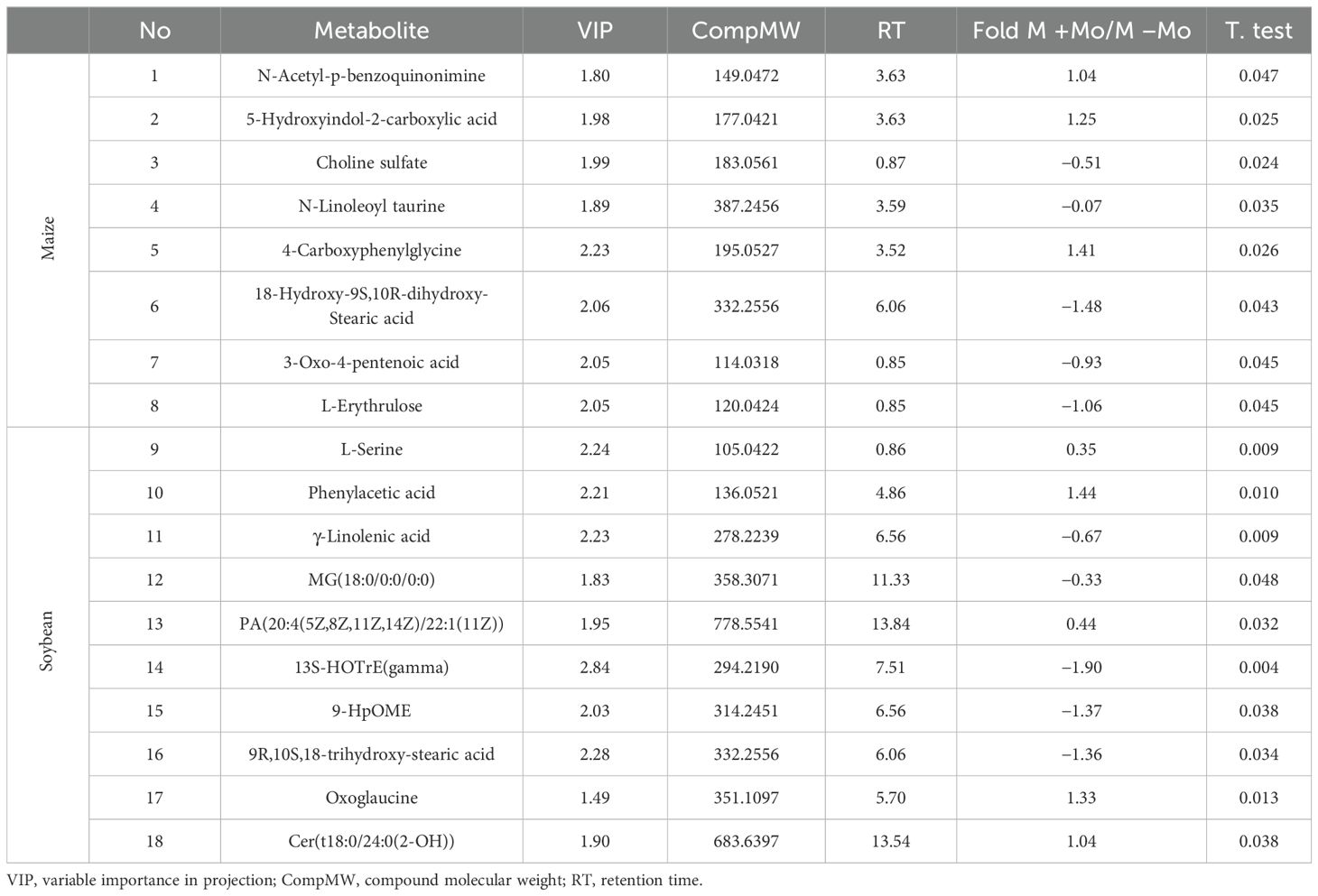
Table 3. Effect of Mo application on significantly altered metabolites in the rhizosphere soil of maize and soybean under −Mo and +Mo treatments.
4 Discussion
This study provides an effective investigation of the Mo influence on crop nutrient acquisition, metabolite variations, and alteration of rhizosphere microbial diversity in maize and soybean. Mo application significantly increased biological yield as well as the concentration and uptake of N, P, and Mo while having no significant effect on K uptake. Similar results were also reported in previous studies (Rawat et al., 2015; Solanki, 2015). Our results revealed that the low mineral nutrient (N, P, K, and Mo) uptake efficiency can be enhanced by Mo application. Notably, Mo application can alleviate the reduced uptake and availability of P in plants and soil by modifying soil chemical properties (Table 1). Mo nutrition significantly affects Mo and other nutrient concentrations in soil and plants (Kovács et al., 2015). This may be attributed to the fact that Mo is an essential component of many enzymes and microbes that play a key role in the mobility of mineral nutrients. In fact, our results indicated that Mo concentration in different parts of the plant was significantly correlated with soil Mo availability leading to higher biomass and gain yield. Mo supply significantly increased Mo concentrations in the shoots, leaves, bracteas, spikestalks, and roots in both crops, while the roots showed steadily higher Mo concentrations compared to the shoots. Kádár (1995) and Kovács et al. (2015) also obtained similar results. The steadily higher concentration of Mo in biological parameters indicates that this could be due to the higher N-assimilation, as it is important for the activity and stability of the NR. The interactive effect of Mo on K was significant in soil, which resulted in increased rhizosphere K availability. These results are similar to the findings of Markam et al. (2017).
Phosphorus efficiency increased in soil and plants through the application of Mo. This increase in P facilitated the increased N accumulation and acquisition in the shoots and other parts of the plant. Similar results were also verified in the bean and rice plants by (Høgh‐Jensen et al., 2002). A positive interaction exists between Mo and P due to the formation of anionic complexes and ligand exchange mechanism, which could account for the higher availability of P in rhizosphere soil (Barrow et al., 2005). The increased nutrient availability in the rhizosphere soil could be due to the organic acid secreted by the roots. Overall, it could be concluded that applied Mo increased P contents in maize and soybean crop by enhancing the mineral nutrient availability in rhizosphere soil. This is consistent with other researchers who have found that Mo increases the effectiveness of P in the hydroponic cultivation of rapeseed (Liu et al., 2010).
Synergistic interaction on biological yield among Mo and P fertilizers resulted in the highest biological yield. There was a significant difference in biomass among treatments. In maize, Mo application significantly increased biomass in most of the parameters, except for the roots and spikestalks, where no significant difference was observed. In soybean, Mo significantly increased biomass in all parts except for the roots. Grain yield was higher in the +Mo treatment for both crops compared to that in the −Mo treatment. One possible reason Mo supply enhanced seed yield is the increased Mo and P concentrations in the two crops. This improvement likely boosted compound concentrations, Mo- and P-related enzyme activities (NR, acid phosphatase, glutamine synthetase), photosynthesis, and additional nutrient metabolism ultimately contributing to higher yields (Liu et al., 2009; Yu, 2010; Khan et al., 2014; Manohar, 2014). Mo might also enhance the nitrogenase activity, thereby increasing the supply of N to plants through biological fixation N, thus improving growth and increasing the yield (Biswas et al., 2009). An increase in P availability may be attributed to cell division activity, consequently leading to higher plant dry weight (Tesfaye et al., 2007). These variations may have contributed to higher yield.
Molybdenum played a crucial role in enhancing microbial diversity and nutrient availability. This can be attributed to its function as a co-factor for molybdoenzymes, which are indispensable for key metabolic pathways in both microbes and plants (Mendel and Hänsch, 2002; Sauer and Frebort, 2003; Mendel and Kruse, 2012). Mo improved P availability by reducing its adsorption in acidic soils. This could be due to stimulation of phosphate-solubilizing microorganisms and increasing acid phosphatase activity, thereby releasing P in a form accessible to plants (Brinza et al., 2008; Nie et al., 2015). Furthermore, Mo plays an essential role in enhancing the activity of nitrogen-fixing bacteria and molybdoenzymes, such as nitrogenase and nitrate reductase, both of which are critical for nitrogen cycling and microbial metabolism (Kaiser et al., 2005). These multiple actions of Mo might explain the observed enhancements in nutrient availability, accumulation, and microbial diversity in this study. The Mo-induced secretion of organic acids and metabolites created a favorable environment for microbial growth further boosting nutrient availability and improving soil health. Collectively, these effects of Mo on microbial processes, enzymatic activities, and soil properties promote efficient nutrient cycling and enhance nutrient acquisition by plants.
We performed microbial diversity analysis to determine the composition and diversity of bacterial community responses. This method has been used to evaluate the fertilization effect on plant and rhizosphere soil (Zhao et al., 2014). As shown by the number of OTUs, our long-term fertilization experiments did not significantly affect the richness and diversity of the bacteria. Previous studies have shown that the effects of fertilization on soil bacterial diversity are inconsistent (Coolon et al., 2013). Overall, significant variations in bacterial community composition were observed at the phylum, genus, and class levels. Although thousands of tags were identified per sample, no rarefaction curve reached a plateau indicating that no reasonable bacterial community was sequenced (Sengupta and Dick, 2015; Daquiado et al., 2016). Several studies (Youssef and Elshahed, 2008; Morales et al., 2009) have revealed that the analyzed sequences number per sample affects the number of OTU predictions. Generally, the fewer sample sequences from the Mo treatment led toward a smaller curve progression and a fewer number of predicted OTUs. The variances in soil bacterial diversity might largely be explained by differences in effective nutrient concentrations in soil. Earlier studies have presented that nutrient availability (Eo and Park, 2016) and crop types are significant regulators of soil microbial community activity and composition (Suleiman et al., 2013).
The distinctive configuration of the bacterial community in −Mo and +Mo treatment in both crops inferred that fertilization deficiency in one of the constituents may alter the bacterial community composition potentially leading to differences in nutrient availability, uptake, and crop use efficiency. Soil microorganisms could facilitate nutrient mineralization. These aspects of microbiota could account for increased mineral nutrition and biomass yield in our experiment due to Mo supply. Proteobacteria and Acidobacteria were the dominant phyla in all the treatments in both maize and soybean especially in Mo-treated treatments. These results are in line with the findings of previous studies (Ahn et al., 2012), as they found a high abundance of these phyla with other nutrients. The higher abundance of Proteobacteria could be attributable to the fact that most of the bacterial groups that are functionally diverse belong to the leading phyla of Proteobacteria and Gram-negative bacteria (Gupta, 2000). Specifically, in the treatments receiving Mo fertilizer, the four Proteobacteria groups (α-proteobacteria, β-proteobacteria, γ-proteobacteria, and δ-proteobacteria) were the most abundant. Remarkably Actinobacteria are thought to actively participate in organic matter degradation (Ahn et al., 2012), and these were prominently high in treatments receiving Mo. Thus, the higher organic matter may play a role in the increased mineral nutrient availability. Metabolites produced by certain phyla may help in maintaining other bacterial populations enhancing nutrient acquisition and crop growth (Sun et al., 2014). Mo supply can improve soil chemical properties supporting beneficial microbial activity in the rhizosphere. Acidobacteria, known for thriving in resource-poor environments (Fierer et al., 2007), showed a slight increase with Mo application in our study. This phylum, difficult to cultivate and poorly understood (Ward et al., 2009), may respond variably to environmental changes (Fierer et al., 2007). Both Acidobacteria and Actinobacteria, along with Proteobacteria, increased significantly with Mo application, while they were lower in the −Mo treatments of soybean rhizosphere soil. In case of maize, Actinobacteria and Gemmatimonadetes were significantly increased with the Mo application than with the −Mo treatment. These phyla in soil environments have been consistently identified as the key divisions of bacteria (Janssen, 2006). The relative abundance response pattern of Firmicutes observed in this study was non-linear; such a response has not been previously observed. In +Mo treatment, the Firmicutes relative abundance was lower in soybean, while it increased in maize with the same treatment. It could be concluded that the crop type have an effect on relative abundance. Schreiter et al. (2014) described that this phylum in the rhizosphere is more abundant than in non-rhizosphere soil suggesting the constructive influence of plants. Interestingly, the percentage of TM7 was increased only with −Mo treatment, while it was lower with Mo supply in our study. This proposes that different groups of bacteria might play a parallel role in this treatment regarding the lower availability of nutrients in rhizosphere soil through negative effects. The candidate division TM7 and family Sphingomonadaceae (Phylum Proteobacteria) are known to be involved in toluene and benzene degradation (Xie et al., 2011). In terrestrial ecosystems, fluctuating nutrient supply and lower pH are determinants of microbial communities (Zhang et al., 2013). Bacteria belonging to Chloroflexi are the main degradation agents of polysaccharides (Schreiter et al., 2014). Fierer et al. (2012) described that after adding N, the proportion of bacteria belonging to these phyla decreased. However, in our results, it is shown that it increases with the application of Mo.
At a lower taxonomic level, the diverse responses of the genus reveal the specific adaptation of bacterial communities to fertilization. Candidatus Koribacter, Rhodoplan, and Kaistobacter were significantly higher in both crops under Mo-applied conditions. The higher abundance of these groups may have contributed to the distinctive biological activity of the communities compared to that in the −Mo treatment. Results suggested that the abundance and relative proportion of these similar taxa having the same function could be used as markers for nutrient availability and soil fertility (Soman et al., 2017). This shift in microbial community structure may have contributed to increased mineral nutrient availability in the rhizosphere soil, as well as higher grain yield and biomass in maize and soybean crops under the +Mo treatment. It has been reported that Actinobacteria, Alphaproteobacteria, and Betaproteobacteria follow copiotrophic lifestyles (Acosta-Martínez et al., 2010; Singh et al., 2010). The copiotrophic environment could be considered in the Mo applied system, which has resulted in higher nutrient cycling due to the activity of phosphatase and phosphodiesterase. The richness of these microbial groups can improve essential nutrient cycling, which may result in increased crop productivity and soil fertility (Lesaulnier et al., 2008; Daquiado et al., 2016). Our findings suggest that Mo application plays a pivotal role in shaping microbial community composition and abundance ultimately contributing to enhanced nutrient use efficiency and improved crop growth and development. Specifically, the shift from a −Mo to a +Mo system appears to alter bacterial community abundance, which is likely due to changes in nutrient availability in the rhizosphere of maize and soybean crops. These results also answer the questions about the mechanisms by which Mo application influences specific bacterial phyla, genera, and classes. One possible explanation is that Mo alters key soil chemical properties, such as O.M content, pH, and available P, creating a favorable environment for certain microbial groups. Another potential mechanism is the suppression of competing bacterial populations through biological activities, such as the production of inhibitory substances or competition for limited resources. Further studies are needed to disentangle the relative contributions of abiotic factors and biotic interactions in driving these changes.
It has been observed that Mo supply may influence the composition and quantity of root exudates impacting the nutritional status of plants and availability of mineral nutrients in the rhizosphere soil. This, in turn, led to alterations in the bacterial community composition in maize and soybean crops. Mo application enhanced the secretion of different metabolites in rhizosphere soil, which impacted the microbiome and promoted the transformation of insoluble nutrients into soluble form. Similar results are also reported in a previous study (Qin et al., 2023). It could be possible that variation in the nutrients resulting from Mo application may have altered the metabolites in the rhizosphere soil. Therefore, the alteration in metabolites may have led to changes in the bacterial community composition. The researcher has reported that Mo supply could enhance the availability of P in the soil through increased secretion of malic acid and succinic acid in different crops. In plants, the lack of L-serine-derived molecules can have serious consequences. For example, a deficiency in phosphatidylserine, a relatively small plant cell lipid, leads to altered microspore development. The mutant deficient in serine palmitoyl transferase, which condenses L-serine and palmitoyl-CoA, participates in the first step of sphingolipid biosynthesis showing embryonic and male gametophyte lethality. Due to a decrease in sphingolipid content, the homeostasis of plant mineral ions changes steadily, and the plant cannot survive (Chao et al., 2011). L-serine is also critical for the transfer regulation of the methyl group by giving tetrahydrofolate metabolism along with C1 units. Serine is also involved in phospholipid formation required for cell production. Phenylacetic acid has been found to be an active auxin (a type of plant hormone) (Wightman and Lighty, 1982). Amino acids secreted by the roots were high in the Mo treatment. 4-Carboxyphenylglycine, PA(20:4(5Z,8Z,11Z,14Z)/22:1(11Z)), oxoglaucine, and Cer(t18:0/24:0(2-OH)) were upregulated significantly in the Mo-applied treatment in maize and soybean. These metabolites mainly consisted of organic acids, amino acids, and lipids. Generally, plants compositionally yield a varied array of diverse low-molecular natural products (>100,000) identified as secondary metabolites (Bais et al., 2006). Early studies showed that gluconic acid, glucuronic acid, and aldehyde sugar acids can be complexed with Mo (Sawyer, 1964; Ramos et al., 1997). Organic acids chelate Mo in soil to reduce the leaching loss of Mo in soil (Wichard et al., 2009). Many of these metabolites in +Mo treatment have a carboxyl group structure suggesting that some of the compounds might be involved in the transport of molybdate. This mechanism could contribute to the higher biomass and grain yield observed in maize and soybean crops. These secondary metabolites and their functions in the rhizosphere of maize and soybean are not well understood. Organic substances released from the roots can also accelerate O.M degradation and stimulate the rhizosphere microorganisms to dissolve insoluble minerals. Root exudates can stimulate the transformation of O.M leading to improved availability of P (Chen et al., 2014). The stimulation of microbial activity in the rhizosphere could activate the nutrient cycling or can speed up the rhizosphere priming effect.
The findings of this research have significant implications for sustainable agriculture, particularly for maize and soybean cultivation in acidic soils. Molybdenum application emerges as a promising, eco-friendly strategy to enhance crop productivity by improving nutrient availability and uptake, especially P, a nutrient often limited in such soils. Mo plays an important role in N metabolism, particularly through its involvement in key enzymes (NR and nitrogenase), which can enhance N use efficiency. This is evident from the increased N availability and uptake observed in our study benefiting both leguminous (Soybean) and non-leguminous crops (Maize). Additionally, Mo application promoted a healthier rhizosphere by improving the microbial diversity and activity aligning with sustainable farming objectives by improving soil fertility, nutrient availability, and their uptake. Incorporating Mo into nutrient management strategies will offer a practical approach to achieve higher yields, preserving soil health, and contributing to global food security through environmentally sustainable practices.
This research provides valuable insights into how Mo influences crop yields, soil microbial diversity, and metabolite variations; however, it is important to recognize its limitations. First, the study was conducted in a specific region with acidic soil, which may restrict the broader applicability of the findings to other soil types or climatic conditions. Expanding future research to include a variety of soil types and environmental settings would enhance the generalizability of the results. Additionally, the 16S rRNA method was employed to analyze microbial communities. While this method is effective for taxonomic profiling, it does not capture the functional complexities of microbial processes affected by Mo. Employing advanced techniques, such as metagenomics or meta-transcriptomics, in future studies could provide deeper insights into the functional dynamics within microbial communities. Furthermore, although metabolite profiling revealed significant changes in response to Mo, the specific roles of these metabolites in plant–microbe interactions and nutrient uptake pathways remain unexplored. Further biochemical and molecular investigations are required to elucidate these mechanisms. Despite these limitations, the findings emphasize the critical role of Mo in improving soil nutrient availability and crop performance. They also highlight the need for continued research to address these gaps and expand our understanding of Mo’s contributions to agricultural ecosystems.
5 Conclusion
This study demonstrates the critical role of Mo in improving microbial diversity, nutrient availability, and nutrient uptake in the rhizosphere soils of maize and soybean. Mo application significantly increased biological yield and enhanced the concentrations and uptake of N, P, and Mo, while K uptake remained unaffected. The findings revealed that Mo amendments altered microbial community structures, particularly increasing the abundance of dominant phyla such as Proteobacteria and Acidobacteria. These changes were associated with enhanced metabolite secretion in the rhizosphere promoting microbial activity and nutrient availability. This led to improved nutrient acquisition and a substantial increase in grain yield. These results underscore the importance of Mo in optimizing soil–plant–microbe interactions and highlight its potential for improving crop performance in acidic soils. Future research should explore the effects of Mo on soil geochemical health, microbial functionality, and its application across diverse soil types and cropping systems.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
MR: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Visualization, Writing – original draft, Writing – review & editing. DA: Software, Validation, Writing – review & editing, Conceptualization. R-LW: Software, Validation, Writing – review & editing. MI: Data curation, Writing – review & editing, Formal analysis, Investigation, Methodology. YA: Data curation, Software, Writing – review & editing, Conceptualization. FR: Data curation, Investigation, Writing – review & editing, Software. MA: Conceptualization, Data curation, Writing – review & editing. HG: Data curation, Software, Writing – review & editing, Validation. AA: Conceptualization, Data curation, Investigation, Software, Writing – review & editing. C-XH: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Validation, Visualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Key Research and Development Program of China (2016YFD0200108), the National Natural Science Foundation of China (Program No. 41771329), and the 948Project from the Ministry of Agriculture of China (2016-X41).
Acknowledgments
We extend our appreciation to the Project funding (13445-Tabuk-2023-UT-R-3-1-SE) Research, Development, and Innovation Authority (RDIA)—Kingdom of Saudi Arabia.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1519540/full#supplementary-material
References
Abia, A. L. K., Alisoltani, A., Keshri, J., Ubomba-Jaswa, E. (2018). Metagenomic analysis of the bacterial communities and their functional profiles in water and sediments of the Apies River, South Africa, as a function of land use. Sci. Total Environ. 616, 326–334. doi: 10.1016/j.scitotenv.2017.10.322
Acosta-Martínez, V., Dowd, S., Sun, Y., Wester, D., Allen, V. (2010). Pyrosequencing analysis for characterization of soil bacterial populations as affected by an integrated livestock-cotton production system. Appl. Soil Ecol. 45, 13–25. doi: 10.1016/j.apsoil.2010.01.005
Ahn, J.-H., Song, J., Kim, B.-Y., Kim, M.-S., Joa, J.-H., Weon, H.-Y. (2012). Characterization of the bacterial and archaeal communities in rice field soils subjected to long-term fertilization practices. J. Microbiol. 50, 754–765. doi: 10.1007/s12275-012-2409-6
Asnicar, F., Weingart, G., Tickle, T. L., Huttenhower, C., Segata, N. (2015). Compact graphical representation of phylogenetic data and metadata with GraPhlAn. PeerJ 3, e1029. doi: 10.7717/peerj.1029
Badri, D. V., Zolla, G., Bakker, M. G., Manter, D. K., Vivanco, J. M. (2013). Potential impact of soil microbiomes on the leaf metabolome and on herbivore feeding behavior. New Phytol. 198, 264–273. doi: 10.1111/nph.2013.198.issue-1
Bais, H. P., Weir, T. L., Perry, L. G., Gilroy, S., Vivanco, J. M. (2006). The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 57, 233–266. doi: 10.1146/annurev.arplant.57.032905.105159
Bao, S. D. (2002). Soil and agricultural chemistry analysis, 3rd edn. (Beijing: China Agriculture Press), 43–149.
Barbano, D., Clark, J. (1990). Kjeldahl method for determination of total nitrogen content of milk: collaborative study. Journal-Association Off. Analytical Chem. 73, 849–859. doi: 10.1093/jaoac/73.6.849
Barrow, N. J., Cartes, P., Mora, M. (2005). Modifications to the Freundlich equation to describe anion sorption over a large range and to describe competition between pairs of ions. Eur. J. Soil Sci. 56, 601–606. doi: 10.1111/j.1365-2389.2005.00700.x
Biswas, P., Bhowmick, M., Bhattacharya, A. (2009). Effect of molybdenum and seed inoculation on nodulation, growth and yield in urdbean [Vigna mungo (L.) Hepper. J. Crop Weed 5, 141–144.
Booth, S. C., Workentine, M. L., Weljie, A. M., Turner, R. J. (2011). Metabolomics and its application to studying metal toxicity. Metallomics 3, 1142–1152. doi: 10.1039/c1mt00070e
Brinza, L., Benning, L. G., Statham, P. J. (2008). Adsorption studies of Mo and V onto ferrihydrite. Mineralogical Magazine 72, 385–388. doi: 10.1180/minmag.2008.072.1.385
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335. doi: 10.1038/nmeth.f.303
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Carvalhais, L. C., Dennis, P. G., Fan, B., Fedoseyenko, D., Kierul, K., Becker, A., et al. (2013). Linking plant nutritional status to plant-microbe interactions. PloS One 8, e68555. doi: 10.1371/journal.pone.0068555
Chao, D.-Y., Gable, K., Chen, M., Baxter, I., Dietrich, C. R., Cahoon, E. B., et al. (2011). Sphingolipids in the root play an important role in regulating the leaf ionome in Arabidopsis thaliana. Plant Cell 23, 1061–1081. doi: 10.1105/tpc.110.079095
Chaparro, J. M., Badri, D. V., Bakker, M. G., Sugiyama, A., Manter, D. K., Vivanco, J. M. (2013). Root exudation of phytochemicals in Arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PloS One 8, e55731. doi: 10.1371/annotation/51142aed-2d94-4195-8a8a-9cb24b3c733b
Chen, H., Jiang, W. (2014). Application of high-throughput sequencing in understanding human oral microbiome related with health and disease. Front. Microbiol. 5, 508. doi: 10.3389/fmicb.2014.00508
Chen, L., Luo, S., Li, X., Wan, Y., Chen, J., Liu, C. (2014). Interaction of Cd-hyperaccumulator Solanum nigrum L. and functional endophyte Pseudomonas sp. Lk9 on soil heavy metals uptake. Soil Biol. Biochem. 68, 300–308. doi: 10.1016/j.soilbio.2013.10.021
Coolon, J. D., Jones, K. L., Todd, T. C., Blair, J. M., Herman, M. A. (2013). Long-term nitrogen amendment alters the diversity and assemblage of soil bacterial communities in tallgrass prairie. PloS One 8, e67884. doi: 10.1371/journal.pone.0067884
Daquiado, A. R., Kuppusamy, S., Kim, S. Y., Kim, J. H., Yoon, Y.-E., Kim, P. J., et al. (2016). Pyrosequencing analysis of bacterial community diversity in long-term fertilized paddy field soil. Appl. Soil Ecol. 108, 84–91. doi: 10.1016/j.apsoil.2016.08.006
Ding, J.-J., Zhou, G.-J., Chen, X.-J., Xu, W., Gao, X.-M., Zhang, Y.-Z., et al. (2024). Analysis of microbial diversity and community structure of rhizosphere soil of three astragalus species grown in special high-cold environment of Northwestern Yunnan, China. Microorganisms 12, 539. doi: 10.3390/microorganisms12030539
Dinsdale, E. A., Edwards, R. A., Hall, D., Angly, F., Breitbart, M., Brulc, J. M., et al. (2008). Functional metagenomic profiling of nine biomes. Nature 452, 629. doi: 10.1038/nature06810
Eo, J., Park, K.-C. (2016). Long-term effects of imbalanced fertilization on the composition and diversity of soil bacterial community. Agriculture Ecosyst. Environ. 231, 176–182. doi: 10.1016/j.agee.2016.06.039
Fierer, N., Breitbart, M., Nulton, J., Salamon, P., Lozupone, C., Jones, R., et al. (2007). Metagenomic and small-subunit rRNA analyses reveal the genetic diversity of bacteria, archaea, fungi, and viruses in soil. Appl. Environ. Microbiol. 73, 7059–7066. doi: 10.1128/AEM.00358-07
Fierer, N., Lauber, C. L., Ramirez, K. S., Zaneveld, J., Bradford, M. A., Knight, R. (2012). Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 6, 1007. doi: 10.1038/ismej.2011.159
Gandhi, S. R., Korolev, K. S., Gore, J. (2019). Cooperation mitigates diversity loss in a spatially expanding microbial population. Proc. Natl. Acad. Sci. 116, 23582–23587. doi: 10.1073/pnas.1910075116
Gill, S. R., Pop, M., DeBoy, R. T., Eckburg, P. B., Turnbaugh, P. J., Samuel, B. S., et al. (2006). Metagenomic analysis of the human distal gut microbiome. science 312, 1355–1359. doi: 10.1126/science.1124234
Gupta, R. S. (2000). The phylogeny of proteobacteria: relationships to other eubacterial phyla and eukaryotes. FEMS Microbiol. Rev. 24, 367–402. doi: 10.1111/j.1574-6976.2000.tb00547.x
Hartmann, A., Schmid, M., Van Tuinen, D., Berg, G. (2009). Plant-driven selection of microbes. Plant Soil 321, 235–257. doi: 10.1007/s11104-008-9814-y
He, Z. L., Yang, X. E., Stoffella, P. J. (2005). Trace elements in agroecosystems and impacts on the environment. J. Trace elements Med. Biol. 19, 125–140. doi: 10.1016/j.jtemb.2005.02.010
Høgh-Jensen, H., Schjoerring, J. K., Soussana, J. F. (2002). The influence of phosphorus deficiency on growth and nitrogen fixation of white clover plants. Ann. Bot. 90, 745–753. doi: 10.1093/aob/mcf260
Huang, X.-Y., Hu, D.-W., Zhao, F.-J. (2022). Molybdenum: Mo re than an essential element. J. Exp. Bot. 73, 1766–1774. doi: 10.1093/jxb/erab534
Huson, D. H., Mitra, S., Ruscheweyh, H.-J., Weber, N., Schuster, S. C. (2011). Integrative analysis of environmental sequences using MEGAN4. Genome Res. 21, 1552–1560. doi: 10.1101/gr.120618.111
Imran, M., Hu, C., Hussain, S., Rana, M. S., Riaz, M., Afzal, J., et al. (2019a). Molybdenum-induced effects on photosynthetic efficacy of winter wheat (Triticum aestivum L.) under different nitrogen sources are associated with nitrogen assimilation. Plant Physiol. Biochem. 141, 154–163. doi: 10.1016/j.plaphy.2019.05.024
Imran, M., Sun, X., Hussain, S., Ali, U., Rana, M. S., Rasul, F., et al. (2019b). Molybdenum-induced effects on nitrogen metabolism enzymes and elemental profile of winter wheat (Triticum aestivum L.) under different nitrogen sources. Int. J. Mol. Sci. 20, 3009. doi: 10.3390/ijms20123009
Ismael, M., Elyamine, A., Zhao, Y., Moussa, M., Rana, M., Afzal, J., et al. (2018). Can selenium and molybdenum restrain cadmium toxicity to pollen grains in Brassica napus? Int. J. Mol. Sci. 19, 2163. doi: 10.3390/ijms19082163
Janssen, P. H. (2006). Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72, 1719–1728. doi: 10.1128/AEM.72.3.1719-1728.2006
Jean, M.-E., Phalyvong, K., Forest-Drolet, J., Bellenger, J.-P. (2013). Molybdenum and phosphorus limitation of asymbiotic nitrogen fixation in forests of Eastern Canada: influence of vegetative cover and seasonal variability. Soil Biol. Biochem. 67, 140–146. doi: 10.1016/j.soilbio.2013.08.018
Jing, G., Sun, Z., Wang, H., Gong, Y., Huang, S., Ning, K., et al. (2017). Parallel-META 3: Comprehensive taxonomical and functional analysis platform for efficient comparison of microbial communities. Sci. Rep. 7, 40371. doi: 10.1038/srep40371
Kádár, I. (1995). The Contamination of the Soil-Plant-Animal-Human Food Chain with Chemical Elements in Hungary (Budapest, Hungary: KTM; MTA TAKI).
Kaiser, B. N., Gridley, K. L., Ngaire Brady, J., Phillips, T., Tyerman, S. D. (2005). The role of molybdenum in agricultural plant production. Ann. Bot. 96, 745–754. doi: 10.1093/aob/mci226
Kang, H., Xue, Y., Cui, Y., Moorhead, D. L., Lambers, H., Wang, D. (2024). Nutrient limitation mediates soil microbial community structure and stability in forest restoration. Sci. Total Environ. 935, 173266. doi: 10.1016/j.scitotenv.2024.173266
Khan, M. S., Zaidi, A., Ahmad, E. (2014). “Mechanism of phosphate solubilization and physiological functions of phosphate-solubilizing microorganisms,” in Phosphate solubilizing microorganisms. eds. Khan, M., Zaidi, A., Musarrat, J. (Cham: Springer), 31–62. doi: 10.1007/978-3-319-08216-5_2
Kovács, B., Puskás-Preszner, A., Huzsvai, L., Lévai, L., Bódi, É. (2015). Effect of molybdenum treatment on molybdenum concentration and nitrate reduction in maize seedlings. Plant Physiol. Biochem. 96, 38–44. doi: 10.1016/j.plaphy.2015.07.013
Langenheder, S., Bulling, M. T., Solan, M., Prosser, J. I. (2010). Bacterial biodiversity-ecosystem functioning relations are modified by environmental complexity. PloS One 5, e10834. doi: 10.1371/journal.pone.0010834
Langille, M. G., Zaneveld, J., Caporaso, J. G., McDonald, D., Knights, D., Reyes, J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814. doi: 10.1038/nbt.2676
Lesaulnier, C., Papamichail, D., McCorkle, S., Ollivier, B., Skiena, S., Taghavi, S., et al. (2008). Elevated atmospheric CO2 affects soil microbial diversity associated with trembling aspen. Environ. Microbiol. 10, 926–941. doi: 10.1111/j.1462-2920.2007.01512.x
Liu, H., Hu, C., Hu, X., Nie, Z., Sun, X., Tan, Q., et al. (2010). Interaction of molybdenum and phosphorus supply on uptake and translocation of phosphorus and molybdenum by Brassica napus. J. Plant Nutr. 33, 1751–1760. doi: 10.1080/01904167.2010.503778
Liu, H., Hu, C., Sun, X., Tan, Q., Nie, Z., Su, J., et al. (2009). Interactive effects of molybdenum and phosphorus fertilizers on grain yield and quality of Brassica napus. J. Food Agric. Environ. 7, 266–269.
Liu, J., Wang, G., Jin, J., Liu, J., Liu, X. (2011). Effects of different concentrations of phosphorus on microbial communities in soybean rhizosphere grown in two types of soils. Ann. Microbiol. 61, 525–534. doi: 10.1007/s13213-010-0168-3
Lozupone, C. A., Hamady, M., Kelley, S. T., Knight, R. (2007). Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 73, 1576–1585. doi: 10.1128/AEM.01996-06
Lozupone, C., Knight, R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005
Lu, R. (1999). Analytical methods of soil agrochemistry. China Agric. Sci. Technol. Publishing House Beijing China, 18–99.
Magoč, T., Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Manohar, M. S. (2014). Effect of Sulphur and Molybdenum on Growth and Productivity of Fenugreek (Trigonella foenum–graecum L.). Diss., Krishikosh, India.
Markam, A., Swaroop, N., Thomas, T., Dhruw, S. S. (2017). Effects of Different Levels of NPK and molybdenum on soil physico-chemical properties of black gram (Vigna mungo L.) var. Shekhar-2. Int. J. Curr. Microbiol. App. Sci. 6, 1082–1088. doi: 10.20546/ijcmas.2017.608.134
McArdle, B. H., Anderson, M. J. (2001). Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology 82, 290–297. doi: 10.1890/0012-9658(2001)082[0290:FMMTCD]2.0.CO;2
Mendel, R. R., Hänsch, R. (2002). Molybdoenzymes and molybdenum cofactor in plants. J. Exp. Bot. 53, 1689–1698. doi: 10.1093/jxb/erf038
Mendel, R. R., Kruse, T. (2012). Cell biology of molybdenum in plants and humans. Biochim. Biophys. Acta (BBA)-Molecular Cell Res. 1823, 1568–1579. doi: 10.1016/j.bbamcr.2012.02.007
Mitri, S., Clarke, E., Foster, K. R. (2016). Resource limitation drives spatial organization in microbial groups. ISME J. 10, 1471–1482. doi: 10.1038/ismej.2015.208
Morales, S. E., Cosart, T. F., Johnson, J. V., Holben, W. E. (2009). Extensive phylogenetic analysis of a soil bacterial community illustrates extreme taxon evenness and the effects of amplicon length, degree of coverage, and DNA fractionation on classification and ecological parameters. Appl. Environ. Microbiol. 75, 668–675. doi: 10.1128/AEM.01757-08
Nie, Z., Li, S., Hu, C., Sun, X., Tan, Q., Liu, H. (2015). Effects of molybdenum and phosphorus fertilizers on cold resistance in winter wheat. J. Plant Nutr. 38, 808–820. doi: 10.1080/01904167.2014.939289
Parks, D. H., Tyson, G. W., Hugenholtz, P., Beiko, R. G. (2014). STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124. doi: 10.1093/bioinformatics/btu494
Patti, G. J., Yanes, O., Siuzdak, G. (2012). Innovation: Metabolomics: the apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 13, 263. doi: 10.1038/nrm3314
Qin, X., Hao, S., Hu, C., Yu, M., Shabala, S., Tan, Q., et al. (2023). Revealing the mechanistic basis of regulation of phosphorus uptake in soybean (Glycine max) roots by molybdenum: an integrated omics approach. J. Agric. Food Chem. 71, 13729–13744. doi: 10.1021/acs.jafc.3c04637
Ramette, A. (2007). Multivariate analyses in microbial ecology. FEMS Microbiol. Ecol. 62, 142–160. doi: 10.1111/j.1574-6941.2007.00375.x
Ramos, M. L., Caldeira, M. M., Gil, V. M. (1997). NMR spectroscopy study of the complexation of D-gluconic acid with tungsten (VI) and molybdenum (VI). Carbohydr. Res. 304, 97–109. doi: 10.1016/S0008-6215(97)00245-0
Rana, M. S., Hu, C. X., Shaaban, M., Imran, M., Afzal, J., Moussa, M. G., et al. (2020a). Soil phosphorus transformation characteristics in response to molybdenum supply in leguminous crops. J. Environ. Manage. 268, 110610. doi: 10.1016/j.jenvman.2020.110610
Rana, M. S., Murtaza, G., Hu, C.-X., Sun, X.-C., Imran, M., Afzal, J., et al. (2020b). Nitrate leaching losses reduction and optimization of N-use efficiency in Triticum aestivum L. and Oryza sativa L. crop rotation for enhancing crop productivity. Pakistan J. Agric. Sci. 57, 645–653. doi: 10.21162/PAKJAS/20.7264
Rana, M. S., Murtaza, G., Sun, X.-C., Imran, M., Afzal, J., Elyamine, A. M., et al. (2018). Nitrate leaching losses reduction, N-use efficiency optimization in Triticum aestivum L. and Oryza sativa L. rotation to improve production and soil reclamation rate with gypsum in salt-affected soil. Indian J. Agric. Res. 52. doi: 10.18805/IJARe.A-350
Rana, M. S., Sun, X., Imran, M., Ali, S., Shaaban, M., Moussa, M. G., et al. (2020c). Molybdenum-induced effects on leaf ultra-structure and rhizosphere phosphorus transformation in Triticum aestivum L. Plant Physiol. Biochem. 153, 20–29. doi: 10.1016/j.plaphy.2020.05.010
Rana, M. S., Sun, X., Imran, M., Khan, Z., Moussa, M. G., Abbas, M., et al. (2020d). Mo-inefficient wheat response toward molybdenum supply in terms of soil phosphorus availability. J. Soil Sci. Plant Nutr. 20, 1560–1573. doi: 10.1007/s42729-020-00298-8
Rawat, U., Rajput, R., Rawat, G., Garg, S. (2015). Effect of varieties and nutrient management on growth, yield and economics of clusterbean (Cyamopsis tetragonoloba L.). Res. Crops 16, 64–67. doi: 10.5958/2348-7542.2015.00009.1
Rognes, T., Flouri, T., Nichols, B., Quince, C., Mahé, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4, e2584. doi: 10.7717/peerj.2584
Sauer, P., Frebort, I. (2003). Molybdenum cofactor-containing oxidoreductase family in plants. Biol. plantarum 46, 481–490. doi: 10.1023/A:1024814007027
Schreiter, S., Ding, G.-C., Heuer, H., Neumann, G., Sandmann, M., Grosch, R., et al. (2014). Effect of the soil type on the microbiome in the rhizosphere of field-grown lettuce. Front. Microbiol. 5, 144. doi: 10.3389/fmicb.2014.00144
Selvakumar, G., Panneerselvam, P., Ganeshamurthy, A. N. (2012). “Bacterial mediated alleviation of abiotic stress in crops,” in Bacteria in agrobiology: Stress management, ed. D. K., Maheshwari (Berlin Heidelberg: Springer-Verlag), 205–224. doi: 10.1007/978-3-662-45795-5_10
Sengupta, A., Dick, W. A. (2015). Bacterial community diversity in soil under two tillage practices as determined by pyrosequencing. Microbial Ecol. 70, 853–859. doi: 10.1007/s00248-015-0609-4
Singh, B. K., Bardgett, R. D., Smith, P., Reay, D. S. (2010). Microorganisms and climate change: terrestrial feedbacks and mitigation options. Nat. Rev. Microbiol. 8, 779–790. doi: 10.1038/nrmicro2439
Solanki, R. B. (2015). Effect of Foliar Application of Iron on Clusterbean [Cyamopsis tetragonoloba (L.) Taub] Varieties (SKNAU) (Thesis). SKNAU, India.
Soman, C., Li, D., Wander, M. M., Kent, A. D. (2017). Long-term fertilizer and crop-rotation treatments differentially affect soil bacterial community structure. Plant Soil 413, 145–159. doi: 10.1007/s11104-016-3083-y
Spor, A., Koren, O., Ley, R. (2011). Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 9, 279. doi: 10.1038/nrmicro2540
Suleiman, A. K. A., Manoeli, L., Boldo, J. T., Pereira, M. G., Roesch, L. F. W. (2013). Shifts in soil bacterial community after eight years of land-use change. Systematic Appl. Microbiol. 36, 137–144. doi: 10.1016/j.syapm.2012.10.007
Sun, J., Zhang, Q., Zhou, J., Wei, Q. (2014). Pyrosequencing technology reveals the impact of different manure doses on the bacterial community in apple rhizosphere soil. Appl. Soil Ecol. 78, 28–36. doi: 10.1016/j.apsoil.2014.02.004
Tesfaye, M., Liu, J., Allan, D. L., Vance, C. P. (2007). Genomic and genetic control of phosphate stress in legumes. Plant Physiol. 144, 594–603. doi: 10.1104/pp.107.097386
Urano, K., Kurihara, Y., Seki, M., Shinozaki, K. (2010). [amp]]lsquo;Omics’ analyses of regulatory networks in plant abiotic stress responses. Curr. Opin. Plant Biol. 13, 132–138. doi: 10.1016/j.pbi.2009.12.006
Ward, N. L., Challacombe, J. F., Janssen, P. H., Henrissat, B., Coutinho, P. M., Wu, M., et al. (2009). Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl. Environ. Microbiol. 75, 2046–2056. doi: 10.1128/AEM.02294-08
Wen, X., Hu, C., Sun, X., Zhao, X., Tan, Q., Liu, P., et al. (2018). Characterization of vegetable nitrogen uptake and soil nitrogen transformation in response to continuous molybdenum application. J. Plant Nutr. Soil Sci. 181, 516–527. doi: 10.1002/jpln.201700556
Wichard, T., Mishra, B., Myneni, S. C., Bellenger, J.-P., Kraepiel, A. M. (2009). Storage and bioavailability of molybdenum in soils increased by organic matter complexation. Nat. Geosci. 2, 625. doi: 10.1038/ngeo589
Wightman, F., Lighty, D. L. (1982). Identification of phenylacetic acid as a natural auxin in the shoots of higher plants. Physiologia Plantarum 55, 17–24. doi: 10.1111/j.1399-3054.1982.tb00278.x
Xie, S., Sun, W., Luo, C., Cupples, A. M. (2011). Novel aerobic benzene degrading microorganisms identified in three soils by stable isotope probing. Biodegradation 22, 71–81. doi: 10.1007/s10532-010-9377-5
Youssef, N. H., Elshahed, M. S. (2008). Species richness in soil bacterial communities: a proposed approach to overcome sample size bias. J. microbiological Methods 75, 86–91. doi: 10.1016/j.mimet.2008.05.009
Yu, B. (2010). A minimum version of log-rank test for testing the existence of cancer cure using relative survival data. Biometrical J. 54, 45–60. doi: 10.1002/bimj.201100069
Zaura, E., Keijser, B. J., Huse, S. M., Crielaard, W. (2009). Defining the healthy" core microbiome" of oral microbial communities. BMC Microbiol. 9, 259. doi: 10.1186/1471-2180-9-259
Zhang, X., Liu, W., Schloter, M., Zhang, G., Chen, Q., Huang, J., et al. (2013). Response of the abundance of key soil microbial nitrogen-cycling genes to multi-factorial global changes. PloS One 8, e76500. doi: 10.1371/journal.pone.0076500
Keywords: maize, soybean, microbial diversity, metabolite variations, nutrient acquisition
Citation: Rana MS, Alshehri D, Wang R-L, Imran M, Abdellah YAY, Rahman FU, Alatawy M, Ghabban H, Abeed AHA and Hu C-x (2025) Effect of molybdenum supply on crop performance through rhizosphere soil microbial diversity and metabolite variation. Front. Plant Sci. 15:1519540. doi: 10.3389/fpls.2024.1519540
Received: 30 October 2024; Accepted: 30 December 2024;
Published: 28 January 2025.
Edited by:
Babar Iqbal, Jiangsu University, ChinaReviewed by:
Ahmad Ali, Florida Agricultural and Mechanical University, United StatesAdnan Mustafa, Brno University of Technology, Czechia
Shahbaz Khan, Huazhong Agricultural University, China
Dan Du, Southwest University, China
M. A. Iqbal, Louisiana Tech University, United States
Copyright © 2025 Rana, Alshehri, Wang, Imran, Abdellah, Rahman, Alatawy, Ghabban, Abeed and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cheng-xiao Hu, aHVjeEBtYWlsLmh6YXUuZWR1LmNu
 Muhammad Shoaib Rana
Muhammad Shoaib Rana Dikhnah Alshehri
Dikhnah Alshehri Rui-Long Wang
Rui-Long Wang Muhammad Imran
Muhammad Imran Yousif Abdelrahman Yousif Abdellah
Yousif Abdelrahman Yousif Abdellah Faiz Ur Rahman
Faiz Ur Rahman Marfat Alatawy3
Marfat Alatawy3 Hanaa Ghabban
Hanaa Ghabban Amany H. A. Abeed
Amany H. A. Abeed