
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 09 January 2025
Sec. Plant Pathogen Interactions
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1519039
This article is part of the Research Topic Innovative Molecular Strategies for Enhancing Plant Defense Against Biotic Stresses View all 9 articles
 Win Tun1
Win Tun1 Kieu Thi Xuan Vo1
Kieu Thi Xuan Vo1 Behnam Derakhshani1
Behnam Derakhshani1 Jinmi Yoon2
Jinmi Yoon2 Lae-Hyeon Cho3
Lae-Hyeon Cho3 Kay Tha Ye Soe Win1
Kay Tha Ye Soe Win1 Sang-Won Lee1
Sang-Won Lee1 Ki-Hong Jung1
Ki-Hong Jung1 Jong-Seong Jeon1*
Jong-Seong Jeon1* Gynheung An1*
Gynheung An1*Plants are susceptible to infection by various pathogens with high epidemic potential. Xanthomonas oryzae pv. oryzae (Xoo) causes bacterial blight in rice, one of the most significant diseases in both temperate and tropical regions. In this study, we report the identification and characterization of OsWRKY26, a sucrose-inducible transcription factor, that plays a role in the plant defense responses following Xoo infection. We found that mutant plants with defective OsWRKY26 showed enhanced defense response specifically to Xoo, indicating that this transcription factor acts as a negative defense regulator. In contrast, mutant plants did not exhibit higher resistance compared to wild-type (WT) plants when infected with the rice blast fungal pathogen Magnaporthe oryzae. Transcriptomic analysis of mutant and WT plants revealed that several pathogen resistance genes were upregulated in mutants. Of these, we selected OsXa39 for further analysis. Transient expression experiments in rice protoplasts showed that OsWRKY26 repressed the expression of a Luciferase reporter gene driven by the OsXa39 promoter. Chromatin immunoprecipitation analysis revealed that OsWRKY26 binds directly to the promoter region of OsXa39. These findings suggest that OsWRKY26 negatively regulates the defense response during Xoo infection by repressing OsXa39 as well as other pathogen-related genes such as OsXa47, OsBBR1, OsRSR1, OsPR1a, OsPR1-11, OsPR2, and OsPR4c.
Rice is one of the most important staple food crops throughout the world, but its production is significantly threatened by various pathogens, including bacterial blight Xanthomonas oryzae pv. oryzae (Xoo) (Mondal et al., 2021). In general, plants defend themselves against pathogens via two primary immune mechanisms: pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI) (Vo et al., 2023). PTI acts as an early line of defense by recognizing common pathogen molecules through pattern-recognition receptors (PRRs) located on the membrane, which include receptor-like protein (RLPs) and receptor-like kinases (RLKs). RLKs have a single transmembrane domain to recognize pathogen molecules and intracellular kinase domain for signal transduction. Upon recognition, PRRs activate downstream pathways, including the MAPK cascade, to trigger immune responses such as ROS generation, cell death, and increased cytoplasmic calcium ion concentration (Yu et al., 2024). In contrast, ETI provides a stronger and more specific response by detecting pathogen effectors through intracellular resistance (R) proteins (Yuan et al., 2021; Vo et al., 2023). ETI plays a critical role in Xoo resistance, as specific R genes recognize Xoo effectors and trigger corresponding defense responses (Zhao et al., 2009; Lee et al., 2011; Fiyaz et al., 2022). Among the R genes, the nucleotide-binding and leucine-rich repeat (NLR) genes, along with variable amino-terminal domains, are particularly important as the family consists of the largest group in rice. These proteins mediate the recognition of transcription-activator-like effectors (TALEs) produced by Xanthomonas. TALE effectors facilitate pathogenicity because they induce the expression of susceptibility (S) genes, which are involved in normal plant physiological processes. Moreover, specific manipulations have been found to alter R gene-mediated resistance to Xanthomonas (Hammond-Kosack and Kanyuka, 2007; Lee et al., 2011; Nowack et al., 2022; Xu et al., 2022).
The accumulation of R gene transcripts can also confer resistance to specific pathogen strains, while mutations in R genes lead to susceptibility. For example, the R gene OsXa39, a member of the NLR gene family, is known to confer resistance to 21 Xoo strains. Previous studied using the rice line H471, which carries OsXa39, has revealed that it triggers a robust hypersensitive response involving programmed cell death upon infection with Xoo strains (Shi et al., 2022; Zhang et al., 2015a, 2015b; Zhang et al., 2015c). These findings highlight its effectiveness in providing durable resistance to a varierty of Xoo strains. Knockout mutants of another NLR gene, Xa47, are more susceptible to Xoo, whereas the overexpression of Xa47 in the susceptible rice variety JG30 has been found to increase Xoo resistance (Lu et al., 2022). Similarly, the overexpression of the NLR gene OsBBR1, was demonstrated to confer moderate resistance to the Xoo strains PXO86 and PXO341 via upregulation of pathogenesis-related (PR) gene expression (Wang et al., 2013). Comparable resistance phenomena have also been found for the NLR gene Xa1 and several of its alleles (e.g., Xa1-2 and Xa14), as well as for Xa31 (Zhang et al., 2020). Although many NLR genes may be involved in resistance to Xoo, at present the ways in which these genes are controlled by upstream regulatory elements remain poorly understood.
WRKY transcription factors (TFs) are known to mediate several signaling pathways and regulatory networks involved in defense responses (Xie et al., 2021; Javed and Gao, 2023). For example, in rice a RNAi-mediated knockdown of OsWRKY6 was found to reduce resistance to Xoo infection, and was accompanied by decreased expression of the defense-related genes OsPR1a and OsPR1b (Im et al., 2022). In addition, the OsWRKY6 protein activates other WRKY genes such as OsWRKY45 and OsWRKY47 (Im et al., 2022). Recent work has also shown that CRISPR/Cas9 mutations in OsWRKY7 result in longer lesions in plants infected with Magnaporthe oryzae and Xoo than is found in wild-type (WT) plants. In contrast, overexpression of this gene is associated with increased expression of several pathogenesis-related (PR) genes, including OsPR1a, OsPR1b, and OsPR10a, which leads to pathogen resistance (Tun et al., 2023; Zheng et al., 2023). In another study, OsWRKY10 was found to enhance resistance to M. oryzae via direct activation of diterpenoid biosynthesis genes (Wang et al., 2023). Overexpression of OsWRKY11 confers resistance to Xoo strains by directly binding to the promoter region of CHITINASE2, while silencing of the WRKY gene results in reduced resistance (Lee et al., 2018). Other studies focusing on ectopic expression of OsWRKY22 found that it enhanced resistance to M. oryzae, whereas knockout mutant lines showed higher pathogen susceptibility (Abbruscato et al., 2012). Furthermore, constitutive activation of OsWRKY30 enhances defense responses to Xoo via mediation of the salicylic acid (SA) signaling pathway (Han et al., 2013). Overexpression of OsWRKY31 resulted in enhanced resistance to M. oryzae, accompanied by increased expression of defense-related genes PBZ1 and OsSci2 (Zhang et al., 2008). Moreover, RNA silencing of OsWRKY67 was found to reduce the levels of SA as well as the expression levels of SA- and PR-associated genes, including OsPR1a and OsPR10a. This resulted in higher susceptibility to both M. oryzae and Xoo, whereas overexpressing the WRKY gene enhanced resistance to both pathogens (Liu et al., 2018).
Some WRKY genes function as negative regulators of plant defense responses to bacterial and fungal pathogens. For example, one study showed that constitutive expression of OsWRKY28 impaired resistance to M. oryzae (Chujo et al., 2013), while another showed that overexpression of OsWRKY72 led to reduced levels of endogenous jasmonic acid (JA) as well as lowered expression of JA- and PR-related genes, thereby resulting in increased susceptibility to Xoo infection (Hou et al., 2019). Similarly, overexpression of OsWRKY76 has been found to increase susceptibility to M. oryzae by reducing the expression levels of PRs and phytoalexin biosynthesis genes via the suppression of W-box elements (Yokotani et al., 2013).
Interestingly, some WRKY TFs seem to have multiple functions. During Xoo invasion, OsWRKY53 functions as a negative regulator by suppressing OsMYB63, which strengthens the sclerenchyma cell walls of vascular tissues via the activation of cellulose synthases, including OsCesA4, OsCesA7, and OsCesA9 (Xie et al., 2021). In contrast, OsWRKY53 functions as a transcriptional activator by inducing the expression of PR genes, thus improving resistance to M. oryzae (Chujo et al., 2007, 2014). Moreover, overexpression of OsWRKY62 is known to suppress the expression of several defense-related genes, which results in enhanced susceptibility to infection by both Xoo and M. oryzae (Peng et al., 2008; Xu et al., 2022). However, when OsWRKY62 forms a heterodimer with OsWRKY45, the dimer functions as a positive regulatory element in inducing diterpenoid phytoalexin biosynthesis genes to enhance defense against pathogens (Fukushima et al., 2016).
In general, WRKY proteins interact with numerous other proteins, such as kinases, receptors, and other transcription factors, to form transcriptional regulatory networks (Chen et al., 2019). These proteins can either stimulate or inhibit the expression of various downstream genes through TGAC core sequences, including the W-box motif, W-box like elements 1 (WLE1), and ASF1MOTIF (Maleck et al., 2001; Hwang et al., 2008). However, some WRKY proteins can also bind to other core sequences within the promoter regions of downstream genes (Van Verk et al., 2008; Machens et al., 2014).
OsWRKY26, also referred to as OsWRKY59 and DLN19 (https://rapdb.dna.affrc.go.jp; Singh et al., 2019) belongs to the group II WRKY family and is upregulated by M. oryzae infection (Li et al., 2021). Interestingly, both compatible and incompatible strains of Xoo induce the expression of OsWRKY26 (Choi et al., 2017), which suggests that it plays a role in pathogen defense. However, to date the molecular mechanisms responsible for its involvement in plant defense responses remain unclear. In this study, we show that OsWRKY26 functions as a negative regulator of defense against Xoo by directly binding to the promoter of OsXa39, a known broad-spectrum NLR gene, and suppressing its expression.
For all experiments, we used Oryza sativa var. japonica, cultivar “Dongjin” to generate transgenic lines and act as a WT control. Seeds were germinated on the ½ Murashige and Skoog (MS) medium, supplemented with 3% sucrose and 0.3% agar, and seedlings were maintained in the medium until 10 days after germination (DAG). The plants were then transferred to growth rooms with controlled conditions (14-h-light at 28°C/10-h-dark at 22°C, with relative humidity maintained at 50% –70%), greenhouses, or the paddy field, depending on the experimental requirements.
We generated oswrky26 mutants by clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated nuclease 9 (Cas9)-mediated gene editing. To do so, we designed two single guide RNA (sgRNA) targets using two web-based tools: http://crispr.dbcls.jp and http://crispor.org (Naito et al., 2015; Concordet and Haeussler, 2018). The structures of the RNA scaffolds of the target sequences were verified using the bioinformatic tool (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi). The specificity of target sequences was confirmed using CRISPR RGEN Tools (http://www.rgenome.net/cas-offinder/).
For transformation, synthesized and annealed sgRNA sequences were first ligated into the BsaI-digested pRGEB32 vector (Xie et al., 2015). The resulting plasmids were then transformed into Agrobacterium tumefaciens strain LBA4404 using the freeze-thaw method (An, 1987). These were subsequently transformed into rice embryonic calli according to a previously published method (Lee et al., 1999). Transgenic T0 plants were then selected on ½ MS agar medium including 50 mg L−1 hygromycin. After selection, genomic DNA was extracted from transformed plants, and the target sequence was PCR amplified using primers flanking the target site. Finally, the resulting PCR products were subcloned into T-easy vectors, and five colonies from each line were sequenced to validate sequence identities.
For the overexpression of OsWRKY26 in transgenic plants, the full-length cDNA of OsWRKY26 was amplified by PCR using primers (Supplementary Table 1) with leaf cDNA as a template and inserted into the binary vector pGA3438 (Kim et al., 2009) using the 5x In-fusion HD enzyme (STO345, Takara). The resulting construct was then introduced into A. tumefaciens LBA4404 and transformed into rice. As a control, transgenic plants expressing the Myc tag alone were also generated. To confirm the expression of the target gene, total protein was extracted from the leaf blades of six transgenic T1 lines at 50 DAG, and exogenous OsWRKY26 was detected by western blotting using a Myc antibody (9B11 with HRP conjugate; Cell Signaling Technology). Additionally, OsWRKY26 transcript levels were analyzed by qRT-PCR, with OsUbi5 as the reference gene.
The OsWRKY26 promoter:β-glucuronidase (GUS) vector (pWRKY26:GUS) was created by insertion of a fragment 3,000 bp upstream of the start codon into a pGA3519 vector (Yoon et al., 2014) through the KpnI and HpaI restriction enzymes sites. pWRKY26:GUS transgenic plants were then stained using GUS solution (50 mM sodium phosphate pH 7.0, 1 mM potassium ferricyanide, 1 mM potassium ferrocyanide, 0.1% triton X-100, 10 mM EDTA, pH 8.0, 0.1% X-Glu dissolved in 1% DMSO and 5% methanol). Plants showing a strong GUS signal were selected for further analysis.
RNA was isolated, and transcript levels were quantified following the method as previously reported (Tun et al., 2023). Transcript levels of specific genes were examined at different time points; these levels were normalized to those of rice Ubiquitin 5 (OsUbi5, LOC_Os01g22490) (Jain et al., 2006), which was used as an internal control. At least three biological replicates were performed for each time point. Relative expression levels were calculated using the ΔΔCt method; primer specificity was verified by observing a single sharp peak in a melting curve analysis (Schmittgen and Livak, 2008; Cho et al., 2016). The primers used in this study are listed in Supplementary Table 1.
M. oryzae inoculation was conducted as a previously described method (Tun et al., 2023). Briefly, M. oryzae isolate RO1-1 was first grown on V8 juice agar plates (80 mL L−l V8 Juice [Campbell’s Soup Company, Camden, NJ], 15 g L−1 agar, pH 6.8) for 2 weeks under continuous fluorescent light. Conidia were collected, examined, and quantified under a microscope, then suspended in water to reach a final concentration of 5 x 106 mL−1. Leaves from 50 DAG plants were used for spot inoculation (Kanzaki et al., 2002). Particularly, 3 μl of conidia suspension was applied to 2 mm press-injured spots on the leaves. After inoculation, the infected plants were placed in a closed box to maintain 100% relative humidity at 25°C in the dark for 24 h before being transferred to a moist incubator. Infected leaves were harvested at 9 days post inoculation (DPI) for lesion length quantification and photographed.
The Xoo strain PXO99 was used for bacterial infection analyses. Xoo inoculation on the leaves were carried out using a previously described leaf clipping method (Ke et al., 2017). For the calculation of colony-forming units (CFUs), leaf fragments (6 cm in length) from below the Xoo lesion area were collected at 12 DPI. CFUs were calculated according to the previous method (Ke et al., 2017). Lesion length was measured at 14 DPI to assess the degree of disease symptoms.
The second leaves from WT and OsWRKY26 mutant plants at 50 DAG were used for transcriptome analyses. Total RNA was extracted in triplicates from each line and subjected to RNA sequencing. Sequencing data quality was assessed using FastQC v0.11.9 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). To obtain high-quality clean reads, low-quality bases from both 5′ and 3′ ends of the paired-end reads were trimmed using Trimmomatic v0.39 (Bolger et al., 2014). The rice reference genome was downloaded from the Rice Genome Annotation Project database (http://rice.plantbiology.msu.edu/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/pseudomolecules/version_7.0/all.dir/) (Kawahara et al., 2013). Index construction for the reference genome and alignment of the cleaned reads to the reference genome were both performed using Hisat2 v2.2.1 using the default parameters (Kim et al., 2015). Aligned SAM files were then converted to BAM format and sorted using SAMtools v1.10 (Danecek et al., 2021). Raw read counts were generated from the alignment files with featureCounts v2.0.0, which is part of the Subread package v1.6.2 (Liao et al., 2014). Post-normalizations of read counts were calculated as Fragments Per Kilobase of transcript per Million mapped reads (FPKM) to facilitate the estimation of gene expression levels of each sample. Differentially expressed genes were then identified using the DEseq2 package v1.38.3 (Love et al., 2014), using an adjusted p-value threshold of 0.05 and an absolute log2 fold change ≥1. Functional annotations of genes were obtained by performing a BLASTP search against the NCBI Nr protein database with an E-value cutoff of 1.0e-8. Using the MSU7 functional annotation files, description, Gene Ontology (GO) terms, and Pfam IDs were assigned to each gene. GO enrichment analysis of all upregulated genes in the biological process category was carried out using the BiNGO tool, employing a hypergeometric test and Benjamini & Hochberg False Discovery Rate correction to obtain an adjusted p-value cutoff of < 0.05 (Maere et al., 2005).
The cDNA sequence of the OsWRKY26 coding region was inserted into the BamHI and KpnI-digested pGA3452 vector containing the ZmUbi1 promoter, a green fluorescence protein (GFP) coding region, and the nos terminator (Kim et al., 2009). The resulting fusion construct was then co-transformed with a vector expressing a nuclear localization signal (NLS)-red fluorescence protein (RFP) conjugate under the control of the ZmUbi1 promoter (NLS-RFP) into protoplasts isolated from rice Oc cells (Baba et al., 1986). Fluorescence signals were observed using a confocal microscope (K1-Fluo, Nanoscope, Korea) 14 h after transformation. GFP and RFP were excited at 488 nm and 561 nm, respectively, and their emission signals were collected using 525–550 nm and 561 nm long-pass filters, respectively.
The full-length coding sequence of OsWRKY26 was inserted between the ZmUbi1 promoter and the HA tag through the HpaI and KpnI restriction enzymes into pGA3698 (Kim et al., 2009) using In-fusion HD enzymes. The resulting vector was used as an effector construct. For luciferase reporter assays, different lengths of the OsXa39 promoter sequence were subcloned into a pGL4.23 vector that consisted of a minimal promoter and the Luciferase coding sequence (E8411, Promega). To normalize luciferase activity, the ZmUbi : GUS vector (Cho et al., 2008) was used as an internal control.
Chromatin immunoprecipitation (ChIP) experiments were performed as a previously described protocol (Haring et al., 2007). Briefly, fresh leaves from OsWRKY26-OX-9 and Myc tag alone transgenic plants were harvested at 10 DAG. Proteins and chromatin complexes were cross-linked with 1% formaldehyde in solution A (10 mM Tris pH 8.0, 400 mM sucrose, 0.1 mM PMSF, and 5 mM β-mercaptoethanol). After 10 min of vacuum infiltration, 125 mM glycine was added to quench the reaction. Following nuclei isolation, extracted DNA was sonicated to fragments of approximately 200-1,000 bp in length. Immunoprecipitation was performed using an anti-Myc antibody (9B11, cell signaling) and agarose beads conjugated with protein G and A (EMD Millipore). Washing and reverse cross-linking steps were then carried out according to the protocol of Haring et al. (2007). The enriched chromatin was analyzed by PCR using the primer sets listed in Supplementary Table 1.
We previously reported that the expression of OsWRKY26 in rice Oc cells was induced by supplementation with sucrose within 2 h, reached its highest level after 4 h, and then rapidly declined (Tun et al., 2023). To further characterize the gene, its expression pattern was studied during plant development. The transcript level of OsWRKY26 gradually increased during the vegetative growth stage and reached the maximum level at 50 DAG (Figure 1A). The gene remained highly expressed until 70 DAG, after which transcript levels dropped rapidly at the heading stage (80-90 DAG).
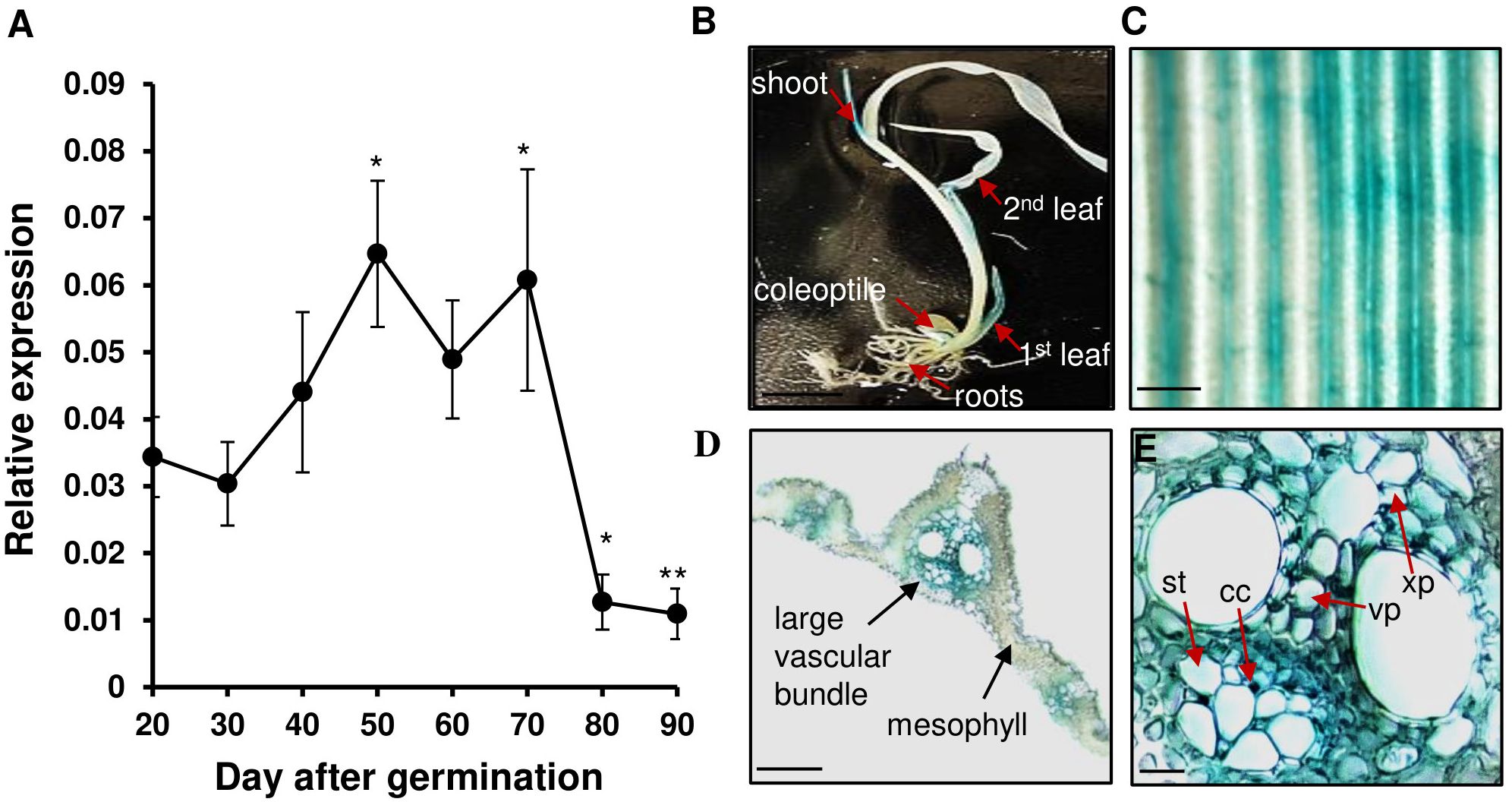
Figure 1. Expression level and GUS signal of OsWRKY26. (A) The transcript levels of OsWRKY26 were measured by qRT-PCR. The second leaf from the top was collected at ZT 4 between 20 and 90 DAG. Error bars represent the standard deviation of four replicates. The transcript levels of OsWRKY26 were normalized to OsUbi5. Two-tailed Student’s t-test was used for the statistical analyses, and statistical significance is indicated by * (p < 0.05) ** (p < 0.01). 20 DAG was used as a control to compare with other time points. (B) GUS signal of pOsWRKY26:GUS seedlings at 10 DAG. Scale bar = 1 cm. (C) GUS activity in a leaf blade at 55 DAG. Scale bar = 1 mm. (D, E) Cross sections of the leaf blade at 55 DAG. Scale bar = 1 mm (in Figure (D)) or 0.2 mm (in Figure (E)). cc, companion cells; st, sieve tube; xp, xylem parenchyma; vp, vascular parenchyma.
To investigate the tissue-specific expression pattern of OsWRKY26, a 3,000 bp sequence upstream of the start codon of OsWRKY26 was cloned to a GUS reporter vector, and the resulting pOsWRKY26:GUS vector was transformed into rice plants. GUS staining of tissues from transgenic plants at various developmental stages showed that this signal was detected mainly in leaf vascular bundles (Figures 1B–E). In addition, weak GUS staining was found in spikelets (Supplementary Figure 1A). However, the staining was not detected in roots (Supplementary Figures 1B, C).
To investigate the functional role of OsWRKY26, null mutants of the gene were generated by the CRISPR/Cas9 methods. Among several transgenic plants, two mutant lines were selected for further study (Figure 2A). In the first mutant line, oswrky26-1 (wr26-1), 4 nucleotides were deleted along with a 12-nucleotide substitution at the sgRNA binding site. The second mutant line, oswrky26-2 (wr26-2), carried biallelic mutations: a five base pair deletion in the target site on one chromosome and a one base pair deletion on another chromosome. Both mutant lines were found to grow and show normal head morphology (Figure 2B). However, plant height and grain yield were slightly reduced in mutant plants compared to the WT control (Supplementary Figure 2 and Supplementary Table 2).
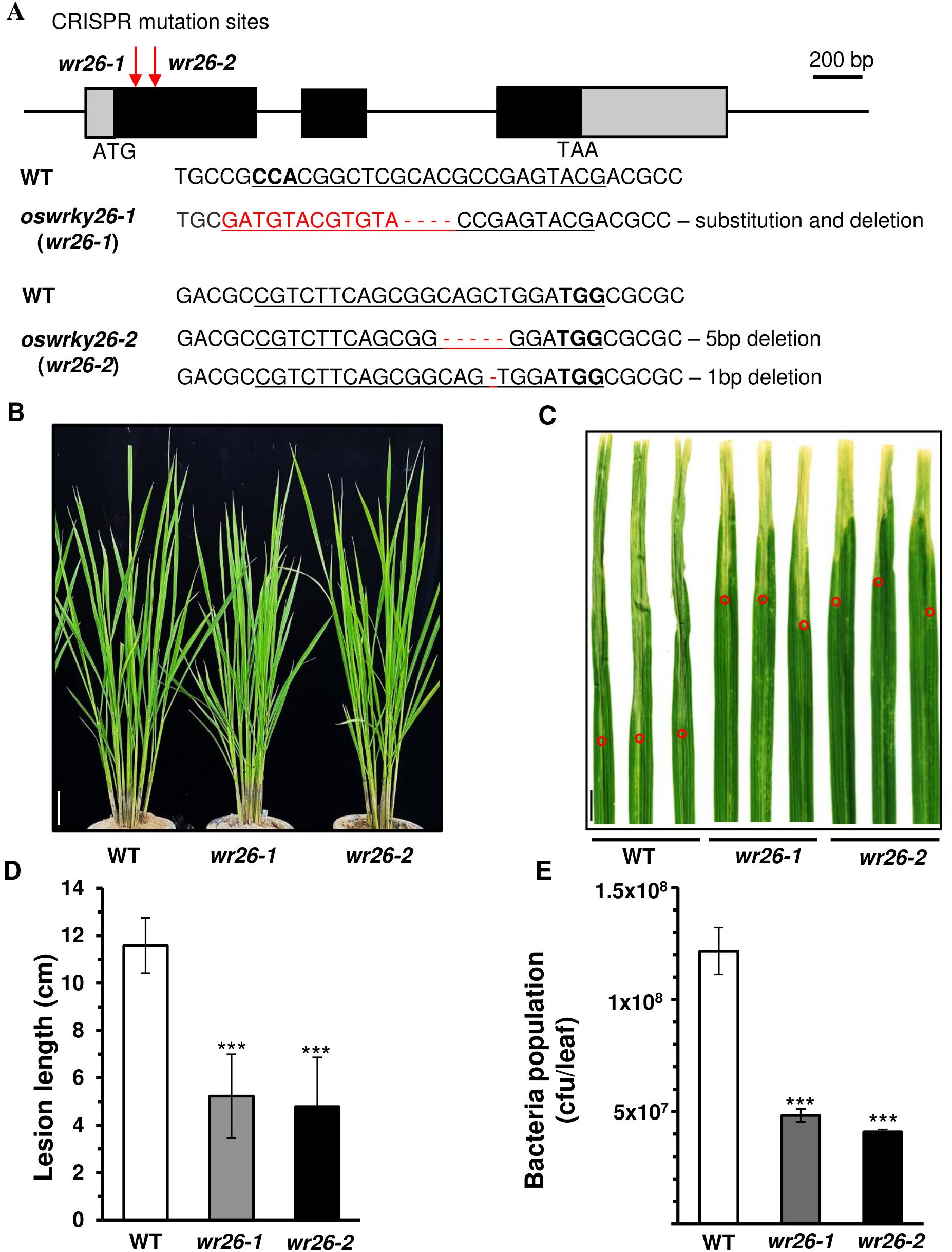
Figure 2. Phenotypes of oswrky26 mutants created by CRISPR/Cas9 gene editing. (A) Schematic diagram of OsWRKY26 and sequence comparisons of the mutation sites. Gray boxes indicate 5’-UTR and 3’-UTR; black boxes represent exons; lines between boxes represent introns. Red arrows indicate CRISPR target sites. Scale bar = 200 bp. All PAM sites (bold) and target sequences are underlined. Deleted and exchanged sequences in the mutants are highlighted in red. (B) Phenotypes of WT, wr26-1, and wr26-2 plants in the T2 generation from paddy fields at 50 DAG. Scale bar = 10 cm. (C) Lesions developed by 14 d after infection with Xoo strain PXO99. Leaves of WT and mutant plants at 50 DAG were used for Xoo inoculation. Scale bar = 1 cm. Red open circles indicate the end of lesion area in each leaf. (D) Lesion lengths at 14 d after infection. Error bars represent standard deviation; n = 20. (E) Xoo population at 12 d after infection. Three infected leaves were used to generate bacterial population counts. Error bars represent the standard deviation of three replicates. Two-tailed Student’s t-test was used for all statistical analyses, and statistical significance is indicated by *** (p < 0.001).
It has been previously reported that OsWRKY26 expression can be induced by Xoo and M. oryzae infection (Choi et al., 2017; Li et al., 2021). We therefore examined whether these mutant lines showed altered responses to Xoo infection. Bacterial inoculation of leaves during the juvenile stage (30 DAG) resulted in slightly reduced lesion lengths in mutant lines compared to WT (Supplementary Figures 3A, B), with no significant differences observed in heading stage plants (90 DAG) (Supplementary Figures 3C, D). However, significant levels of resistance were found in mutant plants at the adult vegetative stage (50 DAG), as evidenced by shorter lesion lengths (Figures 2C, D). The bacterial populations were also reduced in the lesions of wr26 lines compared to the WT (Figure 2E). Taken together, these results suggest that enhanced resistance to Xoo in the mutants was observed when OsWRKY26 expression was highest. Notably, no changes in resistance were observed at the heading stage, when OsWRKY26 expression was low, suggesting that its impact on resistance likely depends on expression levels.
To determine whether OsWRKY26 also responded to M. oryzae infection, we inoculated fungal spores on the second leaf blades of WT, wr26-1, and wr26-2 plants at 50 DAG. Lesion measurements nine days after infection showed similar lesion lengths between WT and mutant plants, suggesting that OsWRKY26 does not play a significant role in defense against M. oryzae (Supplementary Figures 4A, B).
OsWRKY26 belongs to the WRKY family of TFs, which are known to regulate responses to biotic and abiotic stress, senescence, and various developmental processes (Li et al., 2021; Wang et al., 2023; Javed and Gao, 2023). To confirm the subcellular localization of OsWRKY26, GFP signals from OsWRKY26-GFP and RFP from a nucleus localization signal (NLS-RFP) marker were tracked in protoplasts derived from Oc cells. As a control, the NLS was connected to RFP under the ZmUbi1 promoter (NLS-RFP). Both constructs were co-introduced into rice protoplasts and transiently expressed for 14 h. Visualization of the expressed proteins under a fluorescence microscope showed that the OsWRKY26-GFP protein co-localized with NLS-RFP within the nucleus, indicating that OsWRKY26 is a nuclear-localized protein (Supplementary Figure 5).
To investigate the potential targets of OsWRKY26, we performed transcriptome analyses of wr26-1 and WT leaves at the adult vegetative stage. Analysis of three biological replicates revealed 1,298 differentially expressed transcripts between mutant and WT plants. Among them, 777 transcripts were upregulated and 521 were downregulated in the mutant plants (Figure 3A, Supplementary Table 3). GO analysis revealed that stress- and stimulus-related transcripts were among the most abundantly induced categories in the mutants (Figure 3B, Supplementary Table 4). Based on their annotated functions, we focused on five groups: Group 1 - NLR genes, Group 2 - PR genes, Group 3 - WRKY genes, Group 4 - hormone-related genes, and Group 5 - sugar allocation genes (Figure 3C).
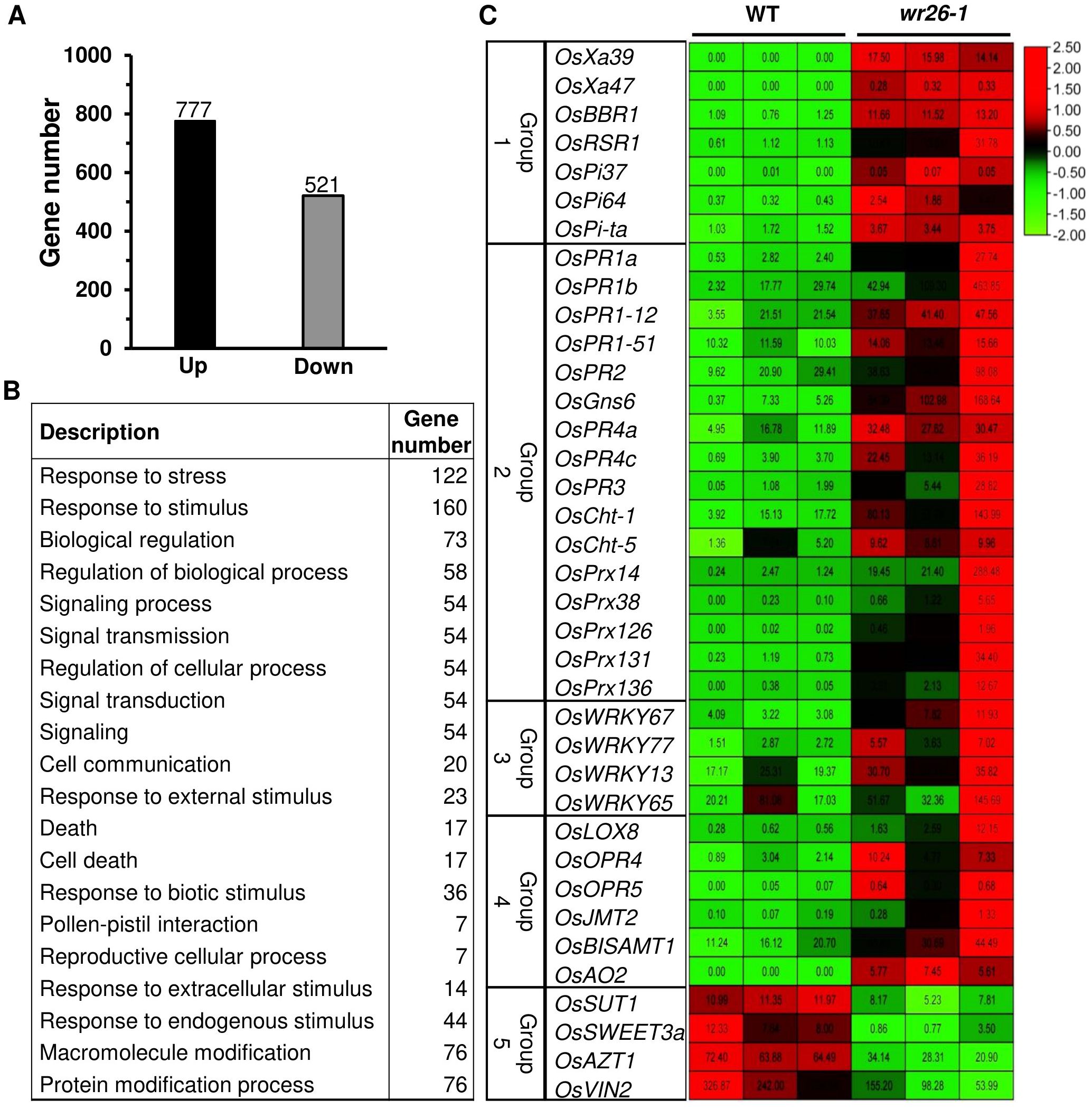
Figure 3. Transcriptomic analyses of three replicates of WT and wr26 plants. (A) Number of up- and downregulated genes in mutant plants. (B) Gene Ontology analysis of the biological processes associated with upregulated genes in mutant plants. GO terms are arranged by corrected p <0.05. (C) Heatmap of disease-related and sugar allocation genes. Numbers within boxes show FPKM values. Red indicates upregulated genes and green indicates downregulated genes.
Since wr26 mutant plants showed enhanced resistance to bacterial pathogen infection, we chose eight genes from Group 1 and Group 2 that were related to disease resistance for further analysis. The expression levels of several NLR genes, OsXa39, OsXa47, OsBBR1, and OsRSR1, and PR genes, OsPR1a, OsPR1b, OsPR2, and OsPR4c, were significantly higher in wr26 mutant leaves compared to the WT (Figures 4A–H). In contrast, RNA-seq data revealed that transcript levels of genes involved in sugar allocation were significantly lower in mutant plants compared to the WT (Figures 4I–L). These results indicate that OsWRKY26 plays a significant role in regulating defense-related genes, thereby enhancing resistance to Xoo infection, while also affecting sugar metabolism pathways.
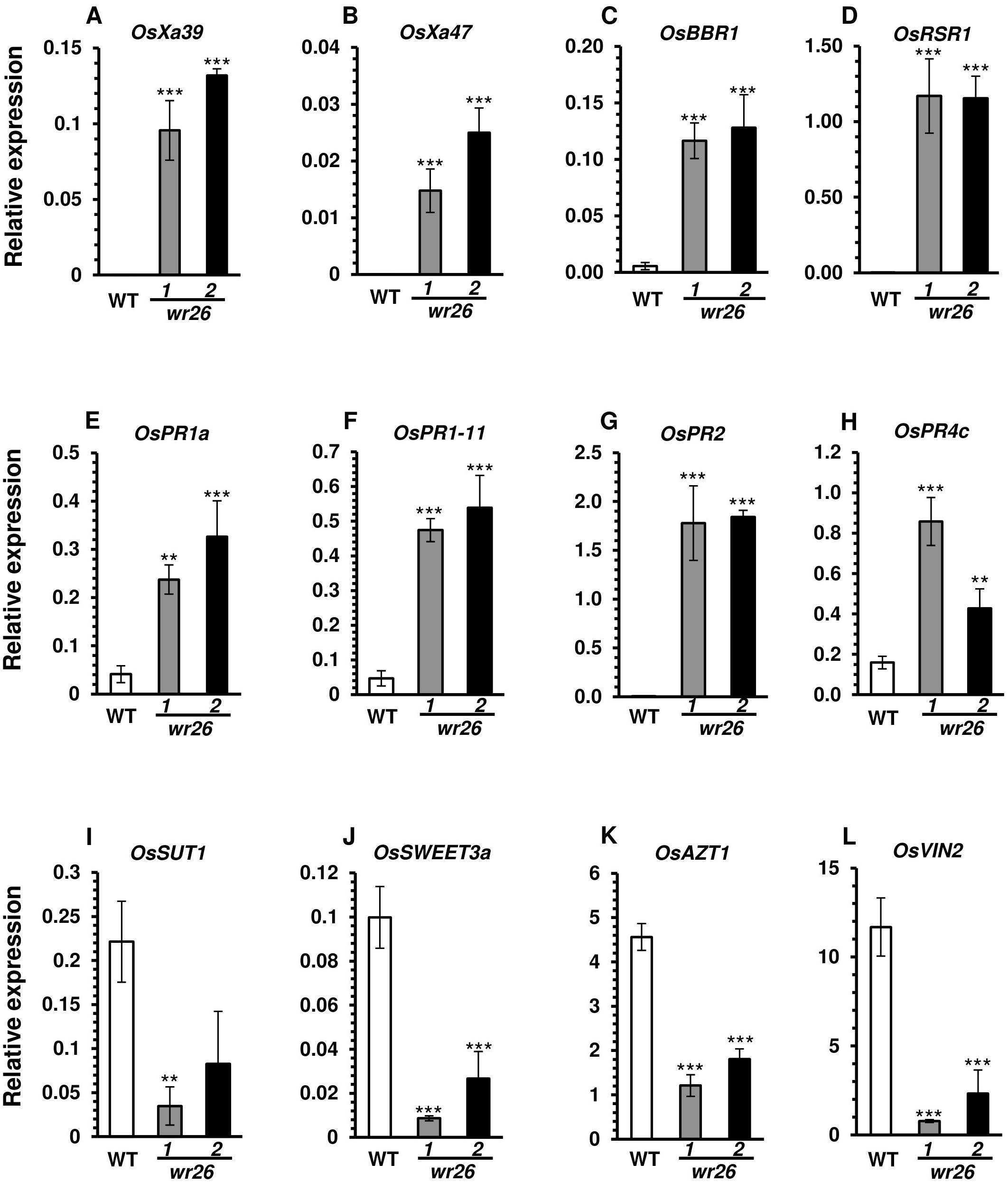
Figure 4. Validation of transcriptomic analyses. (A–D) NLR domain-containing genes. (E–H) Pathogen-related genes. (I–L) Sugar allocation genes. Leaf blades of WT, wr26-1, and wr26-2 plants were collected at 50 DAG. RNA concentrations were normalized to those of OsUbi5. Errors bars represent standard deviation. n = 4. Statistical significance is indicated by ** (p <0.01) and *** (p < 0.001).
As NLR genes are preferentially respond to Xoo strains (Lu et al., 2022), we focused on the Group I from Figure 1C. Because OsXa39 confers broad-spectrum resistance to various Xoo strains compared with other NLR genes (Shi et al., 2022; Zhang et al., 2015a, 2015b; Zhang et al., 2015c), we investigated whether this gene is a direct target of OsWRKY26 by employing a transient expression assay. The OsXa39 promoter region (−2,572 to −63 bp upstream of the start ATG codon) was fused to the Luciferase gene (Figure 5A). As a reference, the GUS reporter was driven by the ZmUbi1 promoter. Both constructs were co-transformed into rice protoplasts along with an OsWRKY26-HA fusion protein-expressing plasmid (Figure 5A). As a control, a plasmid expressing only the HA tag was also introduced. After 15 h of incubation, the luciferase and GUS activities were measured. The results showed that luciferase activity driven by the OsXa39 promoter was significantly reduced when co-expressed with OsWRKY26 compared to the HA alone (Figure 5B). As a further control, we introduced OsWRKY7, a gene involved in resistance to both M. oryzae and Xoo, and found that this did not affect OsXa39 expression (Figure 5B). These results indicated that OsXa39 promoter activity was specifically repressed by OsWRKY26.
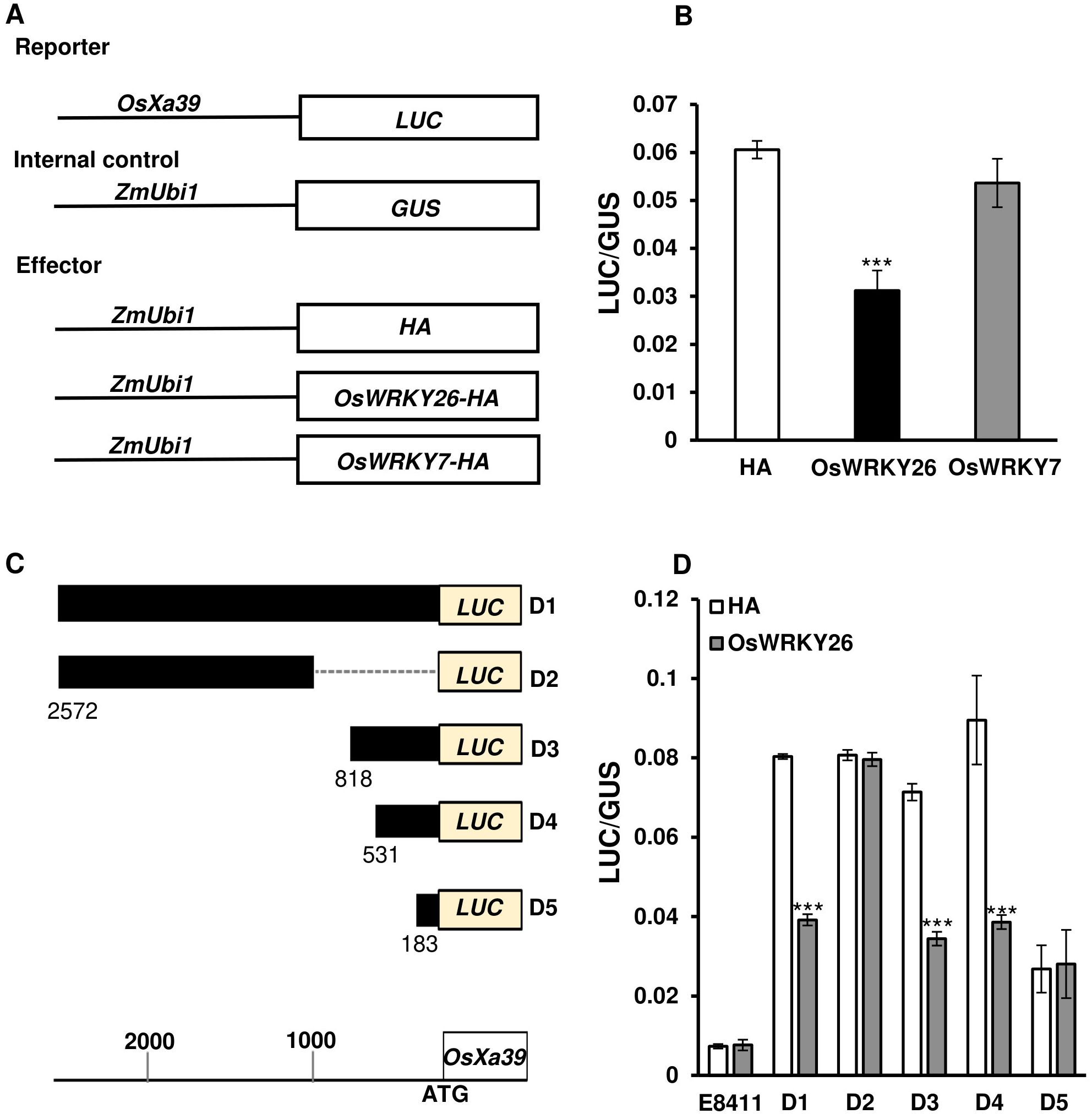
Figure 5. Transient expression analyses of the OsXa39 promoter. (A) Schematic diagrams of OsXa39 reporter and effector constructs. (B) Luciferase activity driven by the OsXa39 promoter, normalized to GUS activity driven by the ZmUbi1 promoter. Effector constructs, HA, OsWRKY26, or OsWRKY7 were connected to the ZmUbi1 promoter then co-introduced to Oc cell protoplasts alongside reporter constructs. Error bars represent standard deviation; n = 3. (C) Schematic diagrams of reporter constructs showing different regions of the OsXa39 promoter and Luciferase (LUC) gene. D1, 2,572 to 63 bp; D2, 2,572 to 1,022 bp; D3, 818 to 63 bp; D4, 531 to 63 bp; D5, 183 to 63 bp upstream of the ATG start codon. Scale bar = 200 bp. (D) Luciferase assay used to identify the repressive binding region of OsWRKY26 to the OsXa39 promoter. ZmUbi : GUS was used as an internal control. LUC/GUS represents the ratio of luciferase activity driven by the OsXa39 promoter to the GUS activity of the control. Error bars represent standard deviation; n = 4. E8411 (pGL4.23) is the original reporter vector. Experiments were repeated three times. Statistical significance is indicated by *** (p < 0.001).
To locate the region responsible for the repression, two constructs were generated: one containing an upstream region (−2,572 to −1,022 bp) and the other containing a downstream region (−818 to −63 bp) (Figure 5C). These regions were connected to the Luciferase gene and introduced into rice protoplasts along with OsWRKY26-HA or with only the HA tag. Measurement of luciferase activity showed that expression of Luciferase driven by the upstream region was not inhibited by OsWRKY26 (Figure 5D). However, the marker expression linked to the downstream region was significantly suppressed by OsWRKY26 (Figure 5D). To further narrow down the repressor region, two additional fragments (−531 to −63 bp and −183 to −63 bp) were tested (Figure 5C). Transient assay results revealed that the longer fragment retained the repressor activity, whereas the shorter fragment did not (Figure 5D). Therefore, the OsWRKY26 repressor region is likely located between −531 and −183 bp.
OsWRKY26 may bind to and interfere with an unspecified positive regulatory transcription factor that induces expression of OsXa39. To find potential TFs interacting with OsWRKY26, we conducted a yeast two-hybrid screen, which identified 19 interacting proteins, including E3 ligases, enzymes, hormone-related genes, and other proteins of unknown function. However, no TFs known to be involved in Xoo resistance were identified (Supplementary Table 5). These results suggest that OsWRKY26 may directly bind to the promoter region of OsXa39 and repress its expression.
To confirm whether OsWRKY26 directly binds to the OsXa39 promoter, we performed ChIP-PCR analysis. Overexpression lines of OsWRKY26 were generated using a construct containing the OsWRKY26 coding region between ZmUbi1 promoter and a Myc tag (Supplementary Figure 6A). Leaf samples from six transgenic plants were harvested to measure OsWRKY26-Myc protein levels by western blot analysis (Supplementary Figure 6B). Three transgenic plants expressed the fusion protein at high levels, and the transcript levels of lines #9 and #10 were further confirmed (Supplementary Figure 6C). These transgenic plants did not show altered responses to Xoo (Supplementary Figures 6D, E).
Transgenic plants expressing OsWRKY26 at high levels (OsWRKY26-OX-9) and those expressing only the Myc tag were used for ChIP-PCR. Chromatins from transgenic nuclei were sonicated and immunoprecipitated with Myc antibody. Precipitated DNA was amplified with fifteen overlapping primer sets spanning 96-397 bp of the promoter region of OsXa39 (Figure 6A). In these tests, OsUbi5 was used as a negative control to test for non-specific binding of the antibody. The analysis showed that three regions, P12 (−680 to −531 bp), P13 (−554 to −324 bp), and P14 (−348 to −191 bp), were enriched with OsWRKY26, indicating that OsWRKY26 binds directly to the OsXa39 promoter region between −680 and −191 bp upstream of the start codon (Figure 6B). We also noted that this region overlapped with the OsWRKY26 repressor region (−531 and −183 bp) identified by the transient expression assay using the Luciferase marker. These results highlight the repressive activity of OsWRKY26 on the OsXa39 promoter.
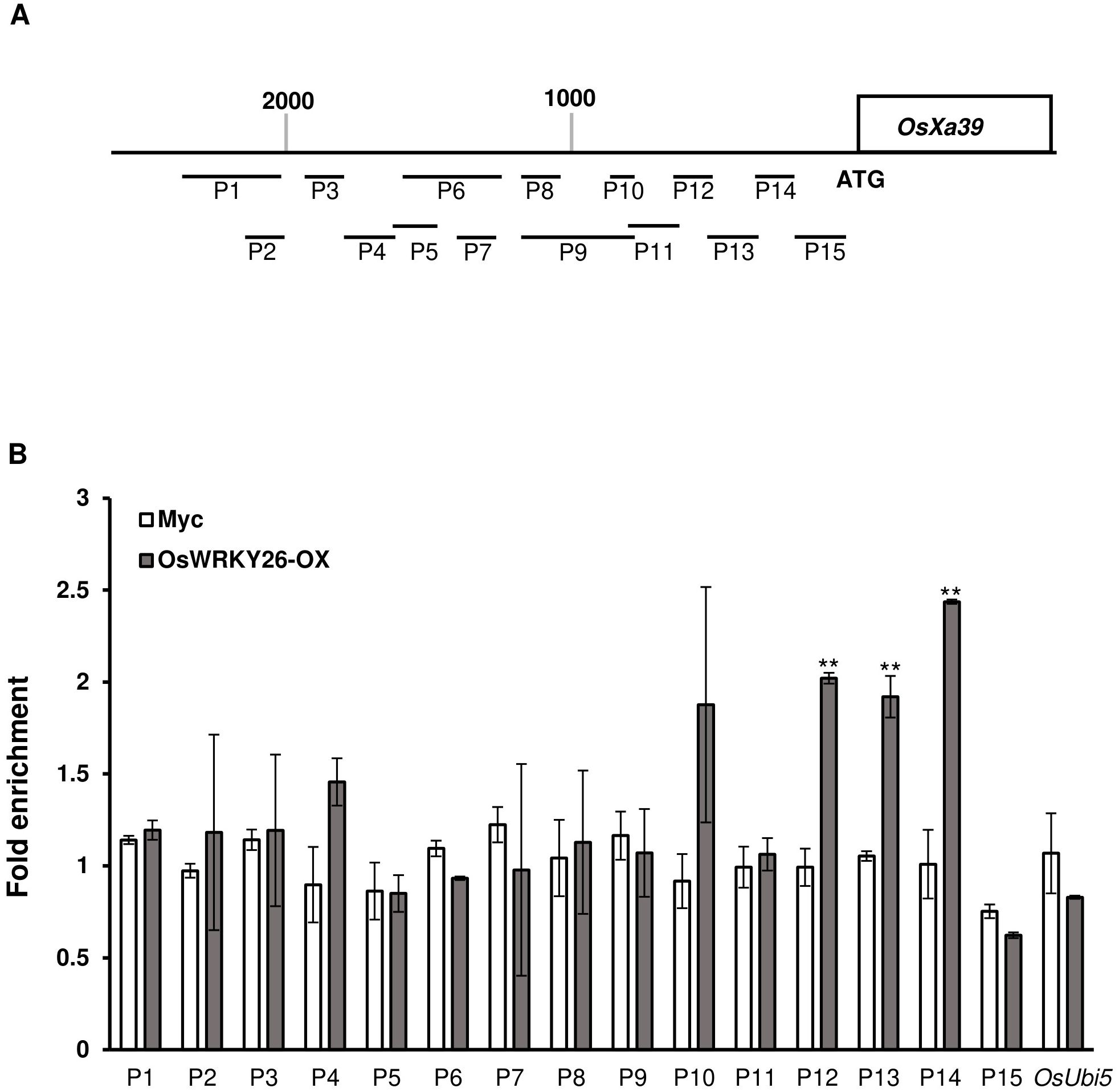
Figure 6. Chromatin immunoprecipitation assay of the promoter region of OsXa39. (A) Schematic diagram of the OsXa39 promoter and amplicon locations. Fifteen amplicons and OsUbi5 were used for this analysis: P1, 2,414 to 2,017 bp; P2, 2,163 to 2,017 bp; P3,1,906 to 1,782 bp; P4,1,805 to 1,611 bp; P5, 1,631 to 1,481 bp; P6, 1,631 to 1,236 bp; P7, 1,372 to 1,236 bp; P8, 1,164 to 1,022 bp, P9, 1,164 to 818 bp; P10, 914 to 818 bp; P11, 840 to 658 bp; P12, 680 to 531 bp; P13, 554 to 324 bp; P14 348 to 191 bp; and P15 201 to 1 bp upstream of the ATG start codon. Scale bar = 200 bp. (B) Fold enrichment of OsWRKY26 binding to fifteen regions of the OsXa39 promoter. Chromatins from OsWRKY26-OX plants at 10 DAG was immunoprecipitated with Myc antibody, and the enriched DNA was quantified by qRT-PCR. Error bars represent standard deviation; n = 2. Experiments were repeated three times. Statistical significance is indicated by ** (p < 0.01).
WRKY TFs are pivotal regulators of plant defense mechanisms, modulating responses against various biotic stresses. Their functional diversity enables plants to mount appropriate responses to different pathogens with distinct infection strategies. WRKY proteins can act as both positive and negative regulators of defense, depending on the pathogen and context. For example, OsWRKY7 and OsWRKY67 are known to induce PR gene expression, which enhances defense against Xoo and M. oryzae (Liu et al., 2018; Tun et al., 2023; Vo et al., 2018; Zhao et al., 2024; Zheng et al., 2023). On the other hand, several WRKY proteins act as negative regulators of defense. For example, OsWRKY62 represses defense pathways, leading to increased vulnerability to Xoo (Peng et al., 2008).
We found that OsWRKY26 is preferentially expressed in vascular bundles, and its expression pattern during plant development correlates with key growth stages, peaking at 50 DAG. Importantly, OsWRKY26 expression is rapidly induced by sucrose supplementation and remains high during vegetative growth, suggesting a functional link between sugar signaling and defense response. This finding aligns with the function of OsWRKY26 in regulating defense against vascular-targeting pathogens like Xoo.
Several WRKY genes exhibit multifunctionality, playing roles in defense against different biotic stressors. For example, OsWRKY45 is a gene with dual functions: its ectopic expression enhances resistance to both Xoo and M. oryzae, while loss-of-function mutations confer resistance against the necrotrophic pathogen Rhizoctonia solani and the piercing-sucking insect Nilaparvata lugens (Shimono et al., 2012; Huangfu et al., 2016). Interestingly, some defensive responses involve tradeoffs; for example, overexpression of OsWRKY70 enhances resistance to the chewing herbivore Chilo suppressalis as well as increased susceptibility to the brown planthopper N. lugens (Li et al., 2015). This study revealed that OsWRKY70 promotes resistance to C. suppressalis by enhancing JA biosynthesis and suppressing GA signaling, while its GA-dependent regulation increases susceptibility to N. lugens, highlighting the tradeoff between defense and growth. These examples highlight the flexibility of WRKY genes like OsWRKY45 and OsWRKY70 in modulating different defense pathways. Similarly, OsWRKY53 has been found to affect both bacterial and fungal resistance. For example, mutations in OsWRKY53 lead to thickened sclerenchyma cell walls and increased resistance to Xoo, while its overexpression enhances resistance to M. oryzae through activation of PR genes (Chujo et al., 2007, 2014; Xie et al., 2021). This suggests that OsWRKY53 can regulate both structural defenses and defense signaling pathways. Previously, we mentioned that OsWRKY26 were strongly induced by sugars. Mutation of OsWRKY26 increased resistance to Xoo infection and up-regulated the expression level of several defense related genes, on the other hands, reduced some sugar related genes, suggesting that OsWRKY26 might control the trade-offs mechanisms between defense mechanisms and sugar allocation during the plant development.
Our findings indicate that while OsWRKY26 is induced by both Xoo and M. oryzae (Choi et al., 2017; Li et al., 2021; Tun et al., 2023), it plays a distinct role specifically in defense against Xoo. This pathogen-specific regulation may reflect differences in infection strategies employed by these two pathogens. Xoo, as an obligate biotroph, primarily targets vascular tissues, while M. oryzae transitions from a biotrophic phase to necrotrophy during infection. The lack of enhanced resistance to M. oryzae in wr26 mutants suggests that OsWRKY26 is more closely linked to vascular-based defense, which are crucial for combating Xoo.
Transcriptome analysis of wr26 mutants revealed that the loss of OsWRKY26 function leads to the upregulation of various genes linked to resistance, including NLRs and PR genes. For example, among NLR genes, OsXa39 is a gene that is known to confer resistance to 21 Xoo strains via hypersensitive response mechanisms (Zhang et al., 2015c). Moreover, OsXa47 is involved in Xoo resistance (Lu et al., 2022), with overexpression of this gene causing the induction of OsNPR1, OsPR1a, and OsPR10a. In addition, knockout mutations of OsXa47 has been found to lead to the downregulation of PR genes. Overexpression of OsBBR1 has been previously found to moderately increase resistance to the Xoo strains PXO86 and PXO341 (Wang et al., 2013), and OsRSR1 was found to contribute to sheath blight resistance by ROS modulation and antioxidation activity in rice (Wang et al., 2021).
Several PR genes were also significantly induced in wr26 mutants. These include OsPR1a, which is induced by infection by Xoo as well as by the fungal strains, M. oryzae and Rhizoctonia solani. RNA silencing of OsPR1a has been found to result in longer lesion lengths (Yan et al., 2022), whereas its overexpression shortens lesion length (Liu et al., 2021). Overexpression of another gene, OsPR1-11 (also known as OsPR1b), has been found to strengthen resistance to Xoo (Luan and Zhou, 2015). More generally, the OsPR2 protein, which encodes a β- 1,3-glucanase, is known to be involved in pathogen defense response, where it degrades fungal cell wall components (Kaur et al., 2017), and OsPR4c, part of the PR4 family, is induced by M. oryzae infection and has several antifungal properties (Wang et al., 2011). This finding is also consistent with previous reports, which found that WRKY TFs regulate these types of defense genes (Javed and Gao, 2023). One notable finding is the upregulation of OsXa39, a broad-spectrum resistance gene that confers resistance to multiple Xoo strains (Zhang et al., 2015c). The strong upregulation of OsXa39 in wr26 mutants suggests that OsWRKY26 functions as a repressor of this gene, and can fine-tune its expression under normal conditions to prevent excessive defense activation. This regulation is important because excessive or untimely defense activation can lead to unnecessary energy expenditures, which affect overall plant growth and fitness (Huot et al., 2014). Further studies on other NLR and PR genes regulated by OsWRKY26 will provide deeper insights into its role in disease resistance.
It was recently reported that the TGAC(N)TGAC sequence is bound by WRKY transcription factors with higher efficiency compared to the TGAC(N)ACGT sequence. While the ACGT motif is activated under stress conditions, the TGAC motif is generally downregulated. This suggests that the TGAC motif may serve as a binding site for negative regulators or repressors (Dhatterwal et al., 2019). Molecular analyses, including ChIP-PCR and promoter deletion assays, identified a repressor-binding region located between −531 and −183 bp upstream of the OsXa39 start codon. This region contains three TGAC motifs, which have been associated with repressor binding under non-biotic stress conditions (Dhatterwal et al., 2019). Our findings suggest that OsWRKY26 binds directly to these motifs to suppress OsXa39 expression. This repressive regulation likely plays a critical role in balancing host defense and growth, thereby enabling the plant to conserve energy during non-infectious conditions.
Interestingly, our data also suggest that OsWRKY26 regulates sugar allocation in vascular tissues. Several genes involved in sucrose transport and allocation, such as SUT1, VIN2, SWEET3a, and TMT2, were downregulated in wr26 mutants. Xoo is known to hijack plant sugar transport pathways by activating SWEET genes through TAL effectors. TAL proteins contain key domains: a type III secretion signal for host cell entry, a central repeat region binding to effector binding elements in SWEET gene promoters, a nuclear localization signal, and an activation domain. As sucrose cannot passively diffuse through the plasma membrane, SWEET proteins mediate its translocation to the apoplasm of phloem cells, providing nutrients in the cell wall space where Xoo propagates (Mondal et al., 2021; Gupta et al., 2021; Verdier et al., 2012). The reduced expression of SWEET3a and other sugar transporters in wr26 mutants may result in a reduction in the resources available for Xoo growth, thereby contributing to the enhanced resistance observed in these plants. Overall, our data therefore suggest that OsWRKY26 plays a dual role in regulating both sugar transport and defense responses.
The slight reduction in plant height and grain yield observed in wr26 mutants suggests that OsWRKY26 also plays a role in growth regulation, likely by balancing defense and growth pathways. As many defense-related genes were upregulated in the mutants, it is possible that the activation of these pathways diverts energy and resources away from growth, resulting in the observed phenotype. In general, tradeoffs between defense activation and growth are common in plants, with enhanced defense often coming at the expense of growth and reproductive success (Yang et al., 2023). Alternatively, the lack of OsWRKY26 reduced sucrose allocation to sink tissues, causing the growth reduction and yield loss. Further study is needed to investigate the role of OsWRKY26 in plant growth and development.
In conclusion, OsWRKY26 functions as a negative regulator of defense responses against Xoo and plays a crucial role in balancing defense and growth through its regulation of key resistance genes such as OsXa39 and sugar transport pathways. By fine-tuning immune responses, OsWRKY26 enables plants to respond effectively to bacterial pathogens while minimizing negative impacts on growth and energy resources. The dual role of OsWRKY26 in modulating both defense and sugar allocation provides new insights into the complex regulatory networks that govern plant immunity and growth. Further research is required to explore how OsWRKY26 coordinates these processes by interacting with other signaling pathways. A deeper understanding of these mechanisms could support the development of rice cultivars with enhanced Xoo resistance that maintain overall plant fitness.
The data presented in the study are deposited in the NCBI repository, accession number PRJNA1201887.
WT: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. KV: Data curation, Investigation, Methodology, Supervision, Visualization, Writing – review & editing. BD: Formal analysis, Investigation, Methodology, Software, Writing – review & editing. JY: Conceptualization, Data curation, Formal analysis, Investigation, Resources, Supervision, Validation, Visualization, Writing – review & editing. L-HC: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Supervision, Validation, Visualization, Writing – review & editing. KW: Data curation, Methodology, Writing – review & editing. S-WL: Conceptualization, Data curation, Investigation, Supervision, Writing – review & editing. K-HJ: Conceptualization, Investigation, Methodology, Software, Supervision, Writing – review & editing. J-SJ: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Visualization, Writing – original draft, Writing – review & editing. GA: Conceptualization, Data curation, Funding acquisition, Investigation, Project administration, Resources, Supervision, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the National Research Foundation of Korea (RS-2020-NR048757 to GA) and the Rural Development Administration, Korea (RS-2024-00322378 to J-SJ).
We are thankful to Kyungsook An for maintenance of experimental plants.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1519039/full#supplementary-material
CFU, Colony-forming units; CRISPR, Clustered regularly interspaced short palindromic repeats; DAG, Days after germination; DEG, Differentially expressed genes; DPI, Days post inoculation; ETI, Effector-triggered immunity; GFP, Green fluorescence protein; GO, Gene Ontology; JA, Jasmonic acid; MS, Murashige and Skoog; NLR, Nucleotide-binding and leucine-rich repeat; NLS, Nuclear localization signal; PR, Pathogenesis-related; PRRs, Pattern-recognition receptors; RLPs, Receptor-like proteins; RLKs, Receptor-like kinases; RFP, Red fluorescence protein; SA, Salicylic acid; TALE, Transcription-activator-like effectors; TF, Transcription factors; WT, Wild-type.
Abbruscato, P., Nepusz, T., Mizzl, L., Del Corvo, M., Morandini, P., Fumasoni, I., et al. (2012). OsWRKY22, a monocot WRKY gene, plays a role in the resistance response to blast. Mol. Plant Pathol. 13, 828–841. doi: 10.1111/j.1364-3703.2012.00795.x
An, G. (1987). Binary Ti vectors for plant transformation and promoter analysis. Methods Enzymol. 153, 292–305. doi: 10.1016/0076-6879(87)53060-9
Baba, A., Hasezawa, S., Syōno, K. (1986). Cultivation of rice protoplasts and their transformation mediated by Agrobacterium Spheroplasts. Plant Cell Physiol. 27, 463–471. doi: 10.1093/oxfordjournals.pcp.a077122
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Chen, X., Li, C., Wang, H., Guo, Z. (2019). WRKY transcription factors: evolution, binding, and action. Phytopathol. Res. 1, 13. doi: 10.1186/s42483-019-0022-x
Cho, J.-I., Ryoo, N., Eom, J.-S., Lee, D.-W., Kim, H.-B., Jeong, S.-W., et al. (2008). Role of rice hexokinases OsHXK5 and OsHXK6 as glucose sensors. Plant Physiol. 149, 745–759. doi: 10.1104/pp.108.131227
Cho, L.-H., Yoon, J., Pasriga, R., An, G. (2016). Homodimerization of Ehd1 is required to induce flowering in rice. Plant Physiol. 170, 2159–2171. doi: 10.1104/pp.15.01723
Choi, N.-Y., Lee, E., Lee, S., Choi, C., Park, S., Ahn, I., et al. (2017). Genome-wide expression profiling of OsWRKY superfamily genes during infection with Xanthomonas oryzae pv. oryzae using real-time PCR. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01628
Chujo, T., Miyamoto, K., Ogawa, S., Masuda, Y., Shimizu, T., Kishi-Kaboshi, M., et al. (2014). Overexpression of phosphomimic mutated OsWRKY53 leads to enhanced blast resistance in rice. PLoS One 9, e98737. doi: 10.1371/journal.pone.0098737
Chujo, T., Miyamoto, K., Shimogawa, T., Shimizu, T., Otake, Y., Yokotani, N., et al. (2013). OsWRKY28, a PAMP-responsive transrepressor, negatively regulates innate immune responses in rice against rice blast fungus. Plant Mol. Biol. 82, 23–37. doi: 10.1007/s11103-013-0032-5
Chujo, T., Takai, R., Akimoto-Tomiyama, C., Ando, S., Minami, E., Nagamura, Y., et al. (2007). Involvement of the elicitor-induced gene OsWRKY53 in the expression of defense-related genes in rice. Biochim. Biophys. Acta BBA - Gene Struct. Expr. 1769, 497–505. doi: 10.1016/j.bbaexp.2007.04.006
Concordet, J.-P., Haeussler, M. (2018). CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 46, 242–245. doi: 10.1093/nar/gky354
Danecek, P., Bonfield, J. K., Liddle, J., Marshall, J., Ohan, V., Pollard, M. O., et al. (2021). Twelve years of SAMtools and BCFtools. GigaScience. 10, giab008. doi: 10.1093/gigascience/giab008
Dhatterwal, P., Basu, S., Mehrotra, S., Mehrotra, R. (2019). Genome wide analysis of W-box element in Arabidopsis thaliana reveals TGAC motif with genes down regulated by heat and salinity. Sci. Rep. 9, 1681. doi: 10.1038/s41598-019-38757-7
Fiyaz, R. ,. A., Shivani, D., Chaithanya, K., Mounika, K., Chiranjeevi, M. C., Laha, G., et al. (2022). Genetic improvement of rice for bacterial blight resistance: Present Status and Future Prospects. Rice Sci. 29, 118–132. doi: 10.1016/j.rsci.2021.08.002
Fukushima, S., Mori, M., Sugano, S., Takatsuji, H. (2016). Transcription factor WRKY62 plays a role in pathogen defense and hypoxia-responsive gene expression in rice. Plant Cell Physiol. 57, 2541–2551. doi: 10.1093/pcp/pcw185
Gupta, P. K., Balyan, H. S., Gautam, T. (2021). SWEET genes and TAL effectors for disease resistance in plants: Present status and future prospects. Mol. Plant Pathol. 22, 1014–1026. doi: 10.1111/mpp.13075
Hammond-Kosack, K., Kanyuka, K. (2007). “Resistance genes (R Genes) in plants,” in Encyclo of Life Sci. Hoboken, NJ, USA: John Wiley & Sons, Ltd. doi: 10.1002/9780470015902.a0020119
Han, M., Ryu, H.-S., Kim, C.-Y., Park, D.-S., Ahn, Y.-K., Jeon, J.-S. (2013). OsWRKY30 is a transcription activator that enhances rice resistance to the Xanthomonas oryzae pathovar oryzae. J. Plant Biol. 56, 258–265. doi: 10.1007/s12374-013-0160-0
Haring, M., Offermann, S., Danker, T., Horst, I., Peterhansel, C., Stam, M. (2007). Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods 3, 11. doi: 10.1186/1746-4811-3-11
Hou, Y., Wang, Y., Tang, L., Tong, X., Wang, L., Liu, L., et al. (2019). SAPK10-mediated phosphorylation on WRKY72 releases its suppression on jasmonic acid biosynthesis and bacterial blight resistance. Science. 16, 499–510. doi: 10.1016/j.isci.2019.06.009
Huangfu, J., Li, J., Li, R., Ye, M., Kuai, P., Zhang, T. (2016). The transcription factor OsWRKY45 negatively modulates the resistance of rice to the brown planthopper Nilaparvata lugens. Int. J. Mol. Sci. 17 (6), 697. doi: 10.3390/ijms17060697
Huot, B., Yao, J., Montgomery, B. L., He, S. Y. (2014). Growth-defense trade-offs in plants: a balancing act to optimize fitness. Mol. Plant 7, 1267–1287. doi: 10.1093/mp/ssu049
Hwang, S.-H., Lee, I. A., Yie, S. W., Hwang, D.-J. (2008). Identification of an OsPR10a promoter region responsive to salicylic acid. Planta. 227, 1141–1150. doi: 10.1007/s00425-007-0687-8
Im, J. H., Choi, C., Park, S. R., Hwang, D.-J. (2022). The OsWRKY6 transcriptional cascade functions in basal defense and Xa1-mediated defense of rice against. Xanthomonas oryzae pv. oryzae. Planta. 255, 47. doi: 10.1007/s00425-022-03830-5
Jain, M., Nijhawan, A., Tyagi, A. K., Khurana, J. P. (2006). Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 345, 646–651. doi: 10.1016/j.bbrc.2006.04.140
Javed, T., Gao, S.-J. (2023). WRKY transcription factors in plant defense. Trends Genet. 39, 787–801. doi: 10.1016/j.tig.2023.07.001
Kanzaki, H., Nirasawa, S., Saitoh, H., Ito, M., Nishihara, M., Terauchi, R., et al. (2002). Overexpression of the wasabi defensin gene confers enhanced resistance to blast fungus (Magnaporthe grisea) in transgenic rice. Theor. Appl. Genet. 105, 809–814. doi: 10.1007/s00122-001-0817-9
Kaur, A., Pati, P. K., Pati, A. M., Nagpal, A. K. (2017). In-silico analysis of cis-acting regulatory elements of pathogenesis-related proteins of Arabidopsis thaliana and Oryza sativa. PLoS One 12, e0184523. doi: 10.1371/journal.pone.0184523
Kawahara, Y., Bastide, M., Hamilton, J. P., Kanamori, H., McCombie, W. R., Ouyang, S., et al. (2013). Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice. 6, 4. doi: 10.1186/1939-8433-6-4
Ke, Y., Hui, S., Yuan, M. (2017). Xanthomonas oryzae pv. oryzae inoculation and growth rate on rice by leaf clipping method. Bio-Protoc. 7, e2568. doi: 10.21769/BioProtoc.2568
Kim, D., Langmead, B., Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kim, S.-R., Lee, D.-Y., Yang, J.-I., Moon, S., An, G. (2009). Cloning vectors for rice. J. Plant Biol. 52, 73. doi: 10.1007/s12374-008-9008-4
Lee, H., Cha, J., Choi, C., Choi, N., Ji, H.-S., Park, S. R., et al. (2018). Rice WRKY11 plays a role in pathogen defense and drought tolerance. Rice. 11, 5. doi: 10.1186/s12284-018-0199-0
Lee, S., Jeon, J.-S., Jung, K.-H., An, G. (1999). Binary vectors for efficient transformation of rice. J. Plant Biol. 42, 310–316. doi: 10.1007/BF03030346
Lee, S. W., Han, M., Park, C. J., Seo, Y. S., Jeon, J. S. (2011). The molecular mechanisms of rice resistance to the bacterial blight pathogen, Xanthomonas oryzae pathovar oryzae. Adv. Botani. Res. 60, 51–87. doi: 10.1016/B978-0-12-385851-1.00002-0
Li, Y., Liao, S., Mei, P., Pan, Y., Zhang, Y., Zheng, X., et al. (2021). OsWRKY93 dually functions between leaf senescence and in response to biotic stress in rice. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.643011
Li, R., Zhang, J., Li, J., Zhou, G., Wang, Q., Bian, W., et al. (2015). Prioritizing plant defence over growth through WRKY regulation facilitates infestation by non-target herbivores. eLife. 4, e04805. doi: 10.7554/eLife.04805
Liao, Y., Smyth, G. K., Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30, 923–930. doi: 10.1093/bioinformatics/btt656
Liu, Q., Li, X., Yan, S., Yu, T., Yang, J., Dong, J., et al. (2018). OsWRKY67 positively regulates blast and bacteria blight resistance by direct activation of PR genes in rice. BMC Plant Biol. 18, 257–257. doi: 10.1186/s12870-018-1479-y
Liu, Y., Yan, G., Zhang, T., Lan, J., Guo, Y., Li, L., et al. (2021). Overexpression of OsPR1A enhanced Xa21-mediated resistance to rice bacterial blight. Sci. Agric. Sin. 54, 4933–4942. doi: 10.3864/j.issn.0578-1752.2021.23.001
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Lu, Y., Zhong, Q., Xiao, S., Wang, B., Ke, X., Zhang, Y., et al. (2022). A new NLR disease resistance gene Xa47 confers durable and broad-spectrum resistance to bacterial blight in rice. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1037901
Luan, Z., Zhou, D. W. (2015). Screening of rice (Oryza sativa L.) OsPR1b-interacting factors and their roles in resisting bacterial blight. Genet. Mol. Res. 14, 1868–1874. doi: 10.4238/2015.March.13.15
Machens, F., Becker, M., Umrath, F., Hehl, R. (2014). Identification of a novel type of WRKY transcription factor binding site in elicitor-responsive cis-sequences from Arabidopsis thaliana. Plant Mol. Biol. 84, 371–385. doi: 10.1007/s11103-013-0136-y
Maere, S., Heymans, K., Kuiper, M. (2005). BiNGO: a cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 21, 3448–3449. doi: 10.1093/bioinformatics/bti551
Maleck, K., Levine, A., Eulgem, T., Morgan, A., Schmid, J., Lawton, K., et al. (2001). The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 26, 403–410. doi: 10.1038/82521
Mondal, K. K., Rashmi, E. R., Ghoshal, T. (2021). Rice SWEET proteins: the key targets by the bacterial blight pathogen, Xanthomonas oryzae pv. oryzae to ensure nutrition in the rice apoplast during infection. Indian Phytopathol. 74, 323–331. doi: 10.1007/s42360-021-00369-5
Naito, Y., Hino, K., Bono, H., Ui-Tei, K. (2015). CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics. 31, 1120–1123. doi: 10.1093/bioinformatics/btu743
Nowack, M. K., Holmes, D. R., Lahaye, T. (2022). TALE-induced cell death executors: an origin outside immunity? Trends Plant Sci. 27, 536–548. doi: 10.1016/j.tplants.2021.11.003
Peng, Y., Bartley, L. E., Chen, X., Dardick, C., Chern, M., Ruan, R., et al. (2008). OsWRKY62 is a negative regulator of basal and Xa21-mediated defense against Xanthomonas oryzae pv. oryzae in rice. Mol. Plant 1, 446–458. doi: 10.1093/mp/ssn024
Schmittgen, T. D., Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Shi, Y., Zeng, W., Xie, J., Zhang, F., Zhuo, D., Gao, L., et al. (2022). Marker-assisted introgression of Xa39 improve rice resistance to five Xanthomonas oryzae pv. oryzae strains. J. Plant Pathol. 104, 801–806. doi: 10.1007/s42161-022-01101-6
Shimono, M., Kiga, H., Akagi, A., Hayashi, N., Goto, S., Sawada, M., et al. (2012). Rice WRKY45 plays important roles in fungal and bacterial disease resistance. Mol. Plant Pathol. 13, 83–94. doi: 10.1111/j.1364-3703.2011.00732.x
Singh, P., Mathew, I. E., Verma, A., Tyagi, A. K., Agarwal, P. (2019). Analysis of rice proteins with DLN repressor motif. Int. J. Mol. Sci. 20 (7), 1600. doi: 10.3390/ijms20071600
Tun, W., Yoon, J., Vo, K. T. X., Cho, L.-H., Hoang, T. V., Peng, X., et al. (2023). Sucrose preferentially promotes expression of OsWRKY7 and OsPR10a to enhance defense response to blast fungus in rice. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1117023
Van Verk, M. C., Pappaioannou, D., Neeleman, L., Bol, J. F., Linthorst, H. J. M. (2008). A novel WRKY transcription factor is required for induction of PR1a gene expression by salicylic acid and bacterial elicitors. Plant Physiol. 146, 1983–1995. doi: 10.1104/pp.107.112789
Verdier, V., Triplett, L. R., Hummel, A. W., Corral, R., Cernadas, R. A., Schmidt, C. L., et al. (2012). Transcription activator-like (TAL) effectors targeting OsSWEET genes enhance virulence on diverse rice (Oryza sativa) varieties when expressed individually in a TAL effector-deficient strain of Xanthomonas oryzae. New Phytol. 196, 1197–1207. doi: 10.1111/j.1469-8137.2012.04367.x
Vo, K., Kim, C.-Y., Hoang, T., Lee, S.-K., Shirsekar, G., Seo, Y.-S., et al. (2018). OsWRKY67 plays a positive role in basal and XA21-mediated resistance in rice. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.02220
Vo, K. T. X., Yi, Q., Jeon, J.-S. (2023). Engineering effector-triggered immunity in rice: obstacles and perspectives. Plant Cell Environ. 46, 1143–1156. doi: 10.1111/pce.14477
Wang, H., Chen, W., Xu, Z., Chen, M., Yu, D. (2023). Functions of WRKYs in plant growth and development. Trends Plant Sci. 28, 630–645. doi: 10.1016/j.tplants.2022.12.012
Wang, X., Chen, J., Yang, Y., Zhou, J., Qiu, Y., Yu, C., et al. (2013). Characterization of a novel NBS-LRR gene involved in bacterial blight resistance in rice. Plant Mol. Biol. Rep. 31, 649–656. doi: 10.1007/s11105-012-0537-0
Wang, A., Shu, X., Jing, X., Jiao, C., Chen, L., Zhang, J., et al. (2021). Identification of rice (Oryza sativa L.) genes involved in sheath blight resistance via a genome-wide association study. Plant Biotechnol. J. 19, 1553–1566. doi: 10.1111/pbi.13569
Wang, N., Xiao, B., Xiong, L. (2011). Identification of a cluster of PR4-like genes involved in stress responses in rice. J. Plant Physiol. 168, 2212–2224. doi: 10.1016/j.jplph.2011.07.013
Xie, W., Ke, Y., Cao, J., Wang, S., Yuan, M. (2021). Knock out of transcription factor WRKY53 thickens sclerenchyma cell walls, confers bacterial blight resistance. Plant Physiol. 187, 1746–1761. doi: 10.1093/plphys/kiab400
Xie, K., Minkenberg, B., Yang, Y. (2015). Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. U. S. A. 112 (11), 3570–5. doi: 10.1073/pnas.1420294112
Xu, X., Wang, H., Liu, J., Han, S., Lin, M., Guo, Z., et al. (2022). OsWRKY62 and OsWRKY76 interact with importin α1s for negative regulation of defensive responses in rice nucleus. Rice 15, 12. doi: 10.1186/s12284-022-00558-4
Yan, G., Liu, Y., Lan, J., Zhang, T., Wang, T., Li, L., et al. (2022). The rapid induction of OsPR1a protein is crucial in Xa21-mediated rice bacterial blight resistance. J. Plant Pathol. 104, 969–978. doi: 10.1007/s42161-022-01105-2
Yang, S., Fu, Y., Zhang, Y., Peng Yuan, D., Li, S., Kumar, V., et al. (2023). Rhizoctonia solani transcriptional activator interacts with rice WRKY53 and grassy tiller 1 to activate SWEET transporters for nutrition. J. Adv. Res. 50, 1–12. doi: 10.1016/j.jare.2022.10.001
Yokotani, N., Sato, Y., Tanabe, S., Chujo, T., Shimizu, T., Okada, K., et al. (2013). WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J. Exp. Bot. 64, 5085–5097. doi: 10.1093/jxb/ert298
Yoon, J., Cho, L.-H., Kim, S. L., Choi, H., Koh, H.-J., An, G. (2014). The BEL1-type homeobox gene 5 induces seed shattering by enhancing abscission-zone development and inhibiting lignin biosynthesis. Plant J. 79, 717–728. doi: 10.1111/tpj.12581
Yu, X.-Q., Niu, H.-Q., Liu, C., Wang, H.-L., Yin, W., Xia, X. (2024). PTI-ETI synergistic signal mechanisms in plant immunity. Plant Biotechnol. J. 22, 2113–2128. doi: 10.1111/pbi.14332
Yuan, M., Jiang, Z., Bi, G., Nomura, K., Liu, M., Wang, Y., et al. (2021). Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature. 592, 105–109. doi: 10.1038/s41586-021-03316-6
Zhang, F., Huang, L.-Y., Zhang, F., Ali, J., Cruz, C. V., Zhuo, D.-L., et al. (2015b). Comparative transcriptome profiling of a rice line carrying Xa39 and its parents triggered by Xanthomonas oryzae pv. oryzae provides novel insights into the broad-spectrum hypersensitive response. BMC Genomics 16, 111. doi: 10.1186/s12864-015-1329-3
Zhang, F., Huang, L., Zhang, F., Hu, D., Wu, W., Wang, W., et al. (2015a). Interacting transcriptomes revealing molecular mechanisms underlying Xa39 mediated broad spectrum resistance of rice to bacterial blight. Plant Genome. 8. doi: 10.3835/plantgenome2014.12.0094
Zhang, J., Peng, Y., Guo, Z. (2008). Constitutive expression of pathogen-inducible OsWRKY31 enhances disease resistance and affects root growth and auxin response in transgenic rice plants. Cell Res. 18, 508–521. doi: 10.1038/cr.2007.104
Zhang, B., Zhang, H., Li, F., Ouyang, Y., Yuan, M., Li, X. (2020). Multiple alleles encoding atypical NLRs with unique central tandem repeats in rice confer resistance to Xanthomonas oryzae pv. oryzae. Plant Commun. 1, 100088. doi: 10.1016/j.xplc.2020.100088
Zhang, F., Zhuo, D.-L., Zhang, F., Huang, L.-Y., Wang, W.-S., Xu, J.-L., et al. (2015c). Xa39, a novel dominant gene conferring broad-spectrum resistance to Xanthomonas oryzae pv. oryzae in rice. Plant Pathol. 64, 568–575. doi: 10.1111/ppa.12283
Zhao, J., Fu, J., Li, X., Xu, C., Wang, S. (2009). Dissection of the factors affecting development-controlled and race-specific disease resistance conferred by leucine-rich repeat receptor kinase-type R genes in rice. Theor. Appl. Genet. 119, 231–239. doi: 10.1007/s00122-009-1032-3
Zhao, Y., Zhu, X., Shi, C.-M., Xu, G., Zuo, S., Shi, Y., et al. (2024). OsEIL2 balances rice immune responses against hemi-biotrophic and necrotrophic pathogens via the salicylic acid and jasmonic acid synergism. New Phytol. 243, 362–380. doi: 10.1111/nph.19809
Keywords: disease, OsWRKY26, OsXa39, rice, Xanthomonas oryzae
Citation: Tun W, Vo KTX, Derakhshani B, Yoon J, Cho L-H, Win KTYS, Lee S-W, Jung K-H, Jeon J-S and An G (2025) OsWRKY26 negatively regulates bacterial blight resistance by suppressing OsXa39 expression. Front. Plant Sci. 15:1519039. doi: 10.3389/fpls.2024.1519039
Received: 29 October 2024; Accepted: 05 December 2024;
Published: 09 January 2025.
Edited by:
Antonio Figueira, University of São Paulo, BrazilReviewed by:
Anil Kumar, Iowa State University, United StatesCopyright © 2025 Tun, Vo, Derakhshani, Yoon, Cho, Win, Lee, Jung, Jeon and An. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gynheung An, Z2VuZWFuQGtodS5hYy5rcg==; Jong-Seong Jeon, amplb25Aa2h1LmFjLmty
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.