- 1Tobacco Research Institute, Chinese Academy of Agricultural Sciences, Qingdao, China
- 2College of Agronomy, Qingdao Agricultural University, Qingdao, China
WRKY transcription factors (TFs) are one of the largest transcription factor families in plants and play important roles in plant processes, most notably in responding to diverse biotic and abiotic stresses. This article reviews the recent research progresses on WRKY TFs in regulating plant immunity, which includes both positive and negative regulation. WRKY TFs were shown to regulate plant defense against pathogens including fungi, bacteria, oomycetes, and viruses by modulating downstream pathogen resistance genes or interacting with other regulators. Plant signaling pathways or components involved in the regulatory network of WRKY-mediated plant immunity mainly involve the action of phytohormones, MAPKs (Mitogen-activated protein kinases), and other transcription factors. The interaction of WRKY TFs with these factors during pathogen resistance was discussed in this article, which may contribute to understanding the mechanisms of WRKY transcription factors in plant immunity.
Introduction
WRKY transcription factors (TFs) compose one of the largest transcription factor families in plants, and they play important roles in plant growth, development, and stress tolerance (Rushton et al., 2010). The first WRKY gene was isolated from sweet potato and named as SPF1 (sweet potato factor 1) in 1994 (Ishiguro and Nakamura, 1994). Subsequently, a vast amount of WRKY TFs (Supplementary Table S1) have been identified in various plant genera as well as non-plant species, such as fungi, diplomonads, and amoebae (Zhang and Wang, 2005; Rinerson et al., 2015). In the non-plant species Dictyostelium discoideum and Giardia lamblia, only one WRKY protein was identified, whereas many WRKY proteins were identified in flowering-plants, such as Brassica napus and Zea mays containing 153 and 180 WRKY proteins, respectively. Researchers proposed that the evolution of WRKY TFs includes early lateral gene transfer to non-plant organisms (Rinerson et al., 2015).
The most typical feature of WRKY TFs is the conserved N-terminal amino acid sequence WRKYGQK (Eulgem et al., 2000). In addition, they possess a typical zinc-finger structure at the C-terminus, with the sequence of CX4-5CX22-23HXH or CX7CX23HXC (Eulgem et al., 2000). In a few of WRKY proteins, the amino acid sequence WRKYGQK is replaced by WRRY, WSKY, WKRY, WVKY, or WKKY (Eulgem et al., 2000). In higher plants, the WRKY family has been divided into groups I, IIa, IIb, IIc, IId, IIe, and III based on their phylogenetic relationship and was proven to regulate the expression of downstream genes by binding to the W-box (TTGACC/T) in their promoters (Rushton et al., 2010). Thus, all genes, including WRKY genes containing W-box elements in the promoter, are assumed as the regulatory targets of WRKY TFs.
Over the past few decades, extensive and in-depth research has been conducted on WRKY TFs, which revealed the key roles of WRKY TFs in regulating plant growth, development, and stress response (Wang et al., 2022a; Khoso et al., 2022). Several previous articles have summarized the roles of WRKY TFs in regulating plant tolerance to biotic stresses (Eulgem and Somssich, 2007; Pandey and Somssich, 2009; Amorim et al., 2016; Finatto et al., 2018). In recent years, many novel works on the function of WRKY TFs in regulating plant resistance to biotic stresses have been reported. The current article summarizes recent progresses on the functions of WRKY TFs in regulating plant defense to pathogens including fungi, bacteria, oomycetes, and viruses, and aims to provide a comprehensive and systematic understanding of the regulatory network of WRKY TFs.
Role of WRKY TFs in plant resistance to fungal pathogens
Pathogens causing powdery mildew disease
Powdery mildew is a fungal disease caused by several different genera of fungi and affects many plant species, such as grape, wheat, and barley, to cause huge losses to agricultural production. In grape, VqWRKY56, VqWRKY31, VqWRKY6, and VqWRKY52 were shown to regulate the resistance to powdery mildew pathogen Erysiphe necator (Wang et al., 2017a; Yin et al., 2022; Zhang et al., 2022; Wang et al., 2023a). Overexpression of VqWRKY56 increased resistance to powdery mildew by increasing the accumulation of proanthocyanidins (PAs), reactive oxygen species (ROS), and salicylic acid (SA). VqWRKY56 could interact with transcript factor VqbZIPC22 and activate the expression of PA biosynthetic genes VvCHS3 (chalcone synthase 3), VvLAR1 (leucoanthocyanidin reductase 1), and VvANR (anthocyanidin reductase) to increase the accumulation of PAs (Wang et al., 2023a). VqWRKY56 binds to the promoters of VvCHS3, VvLAR1, and VvANR, while VqbZIPC22 binds to the promoter of VvANR during this process (Wang et al., 2023a). Co-expression of VqWRKY56 and VqbZIPC22 significantly increased the transcription levels of VvCHS3, VvLAR1, and VvANR in grapes to enhance the resistance to powdery mildew pathogen (Wang et al., 2023a). These findings revealed a complex and elaborate mechanism by which WRKY TFs regulate grape resistance against E. necator (Figure 1).
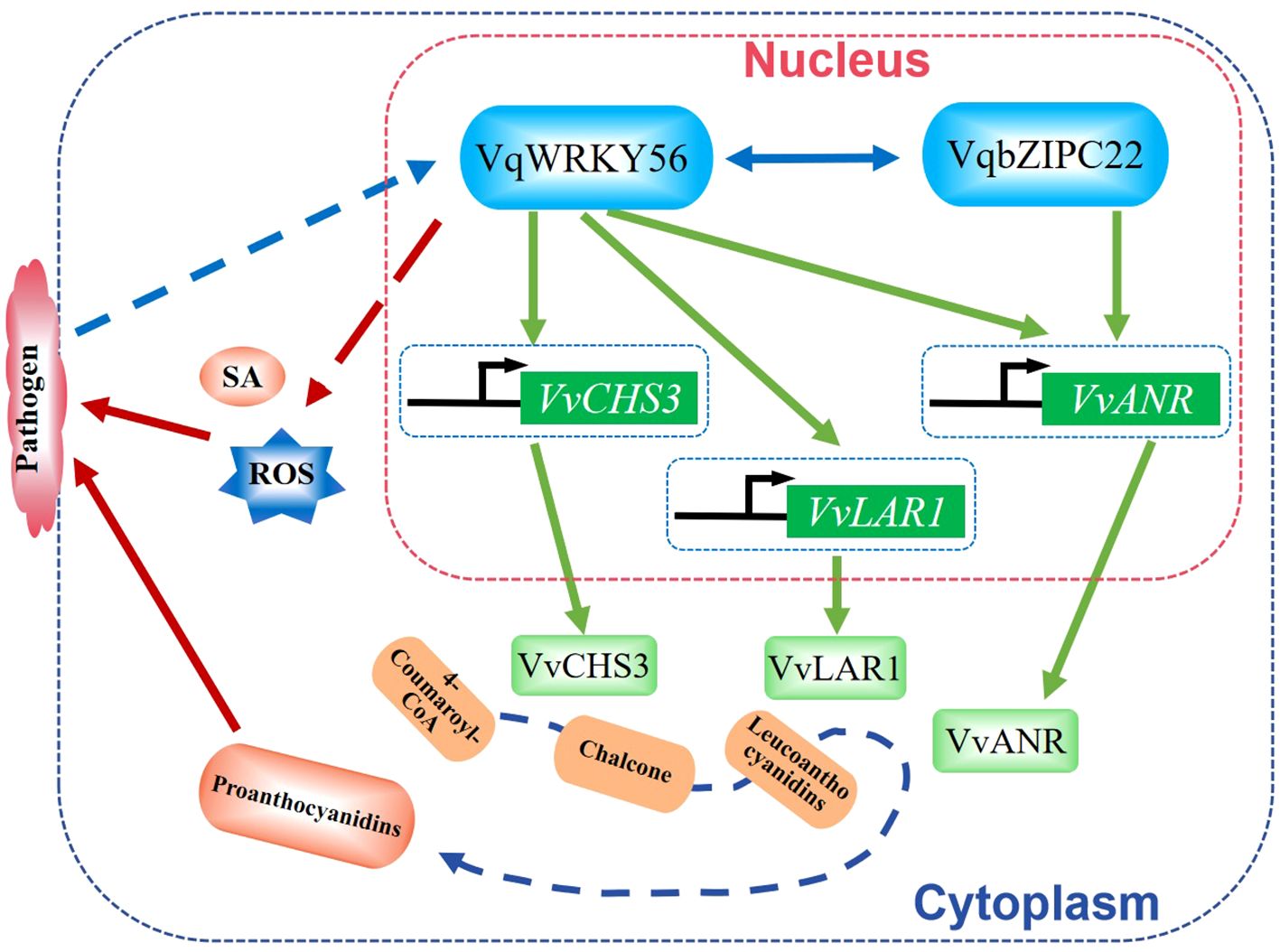
Figure 1. Schematic diagram of VqWRKY56 in regulating grape resistance against powdery mildew pathogen Erysiphe necator. VqWRKY56 interacts with VqbZIPC22 to activate the expression of PA (proanthocyanidin) biosynthetic genes VvCHS3, VvLAR1, and VvANR to promote the accumulation of PAs. The overexpression of VqWRKY56 can increase the accumulation of PAs, ROS, and SA, which provide resistance to powdery mildew pathogen E. necator (Wang et al., 2023a).
WRKY TFs may regulate plant resistance to powdery mildew pathogen via various other manners. For instance, VqWRKY31 activates the SA signaling and promotes the biosynthesis of secondary metabolites, such as stilbenes, flavonoids, and proanthocyanidins, to enhance mildew pathogen resistance (Yin et al., 2022). In vitro assays demonstrated that VqWRKY31 directly binds to the promoters of STS9 and STS48, two structural genes associated with stilbene synthesis, and activates their expression to regulate the resistance to mildew pathogen (Yin et al., 2022). VqWRKY6 interacts with VqbZIP1 to form a transcription factor complex and to activate the jasmonic acid (JA) signaling pathway, which enhances the accumulation of ROS and the expression of pathogenesis-related (PR) genes PR3 and PR4 to ultimately achieving the resistance against powdery mildew pathogens (Zhang et al., 2022). When ectopically expressed in Arabidopsis thaliana, VqWRKY52 positively regulates the resistance to powdery mildew pathogen through the SA signaling, but the underlying molecular mechanisms is yet to be revealed (Wang et al., 2017a). Similarly, ShWRKY81 positively regulates tomato resistance to powdery mildew pathogens by regulating the expression of SA-dependent genes and PR genes as well as the accumulation of hydrogen peroxide (H2O2) (Wang et al., 2023b). On the contrary, barley HvWRKY2 and wheat TaWRKY76-D negatively regulate host resistance to powdery mildew pathogens. HvWRKY2 targets a putative chitin receptor gene HvCEBiP and TaWRKY76-D interacts with a CC-NBS-LRR receptor protein Pm2b to promoting pathogen infection (Yu et al., 2023; Jin et al., 2022). These findings showed that WRKY TFs could affect plant resistance to pathogens causing powdery mildew diseases by altering phytohormone signaling, hydrogen peroxide production, or the function of receptor protein.
Penicillium digitatum
Penicillium digitatum is a fungus causing green mold disease, one of the most challenging and destructive diseases of citrus worldwide. This disease accounts for approximately 60~90% of postharvest diseases in citrus. So far, four WRKY TFs have been isolated from citrus, including CsWRKY23, CsWRKY25, CsWRKY65, and CsWRKY70, and all these WRKY TFs have been reported to positively regulate resistance to P. digitatum. Specifically, CsWRKY23 promotes the accumulation of SA and ROS and enhances the thickness of cell walls to improve pathogen resistance (Wang et al., 2023c). It could directly target 48 genes, most of which are associated with plant-pathogen interaction, phytohormone signaling, secondary metabolite biosynthesis and other metabolisms (Wang et al., 2023c). Similar functions were observed for CsWRKY25 and CsWRKY65 in regulating pathogen resistance, during which CsWRKY25 activates the expression of target genes RbohB (respiratory burst oxidase homolog protein), RbohD, and PR10 and CsWRKY65 activates the expression of CsRbohB, CsRbohD, CsCDPK33 (calcium dependent protein kinase), and CsPR10 (Wang et al., 2021a, b). CsWRKY70 activates the expression of a gene encoding salicylate carboxymethyltransferase (SAMT) involved in the SA biosynthesis and enhances host resistance to P. digitam (Deng et al., 2020). These reports indicated a complicated network of WRKY TFs in regulating citrus resistance to P. digitatum.
Fusarium solani
Fusarium solani is a soil-borne pathogen known to cause apple replant disease (ARD) in all major apple-growing regions worldwide. This pathogen hinders tree growth and reduces the yield and quality of apple (Xiang et al., 2022). Researches show that the WRKY TFs MdWRKY75 and MdWRKY74 positively regulate apple resistance to F. solani (Xiang et al., 2022; Liu et al., 2023). MdWRKY75 promotes the accumulation of lignin to enhance apple resistance to F. solani via the MdWRKY75-MdERF114-MdMYB8-MdPRX63 complex, while MdWRKY74 alone binds to the promoter of MdECHT (endochitinase gene) and enhances chitin accumulation to improve pathogen resistance (Xiang et al., 2022; Liu et al., 2023). Consistently, PnWRKY9 in Panax notoginseng (Burk) F.H. Chen, a rare and valuable Chinese herb, regulates resistance to the root rot caused by F. solani, which severely affects the yield and quality of this herb (Zheng et al., 2022). During this process, PnWRKY9 regulates JA signaling and binds to the promoter of PnDEFL1, a JA-responsive and F. solani resistance-associated gene, to obtain pathogen resistance (Zheng et al., 2022). On the contrary, sugarcane ScWRKY3, which is upregulated by ABA but downregulated by SA and MeJA, negatively regulates plant resistance against F. solani infection (Wang et al., 2018).
Fusarium oxysporum
Fusarium oxysporum is also a soil-borne pathogenic fungus with a wide range of hosts worldwide. It causes wilt diseases in more than 100 plants species, such as cotton, melons, bananas, legumes, and flowers etc (Wang et al., 2022b, 2022). In cotton, the group IIc WRKY TFs induce the GhMKK2-GhNTF6 pathway of mitogen-activated protein kinase (MAPK) cascade and increase the resistance to F. oxysporum by upregulating GhMYC2-mediated expression of flavonoid biosynthetic genes to promote the accumulation of flavonoids (Wang et al., 2022b). Similarly, PnWRKY22 of P. notoginseng upregulates SA levels to increase the resistance to F. oxysporum (Ning et al., 2021). In lily, LrWRKY2 and LrWRKY3 positively regulate the resistance to root rot, the most destructive disease caused by F. oxysporum (Li et al., 2021; Wang et al., 2022c). LrWRKY2 binds to the W-box in the promoter of LrCHI2 (a chitinase-encoding gene) and activates its expression to accentuate defense responses (Li et al., 2021). On the other hand, LrWRKY3 targets the antimicrobial peptide-encoding gene LrDef1 to increase pathogen resistance (Wang et al., 2022c). Overexpression of LrWRKY3 in tobacco increased the transcription of JA biosynthesis-related genes and the SA-mediated responses to modulate pathogen resistance (Wang et al., 2022c). In tomato, F. oxysporum causes the extremely destructive soil-borne disease Fusarium crown and root rot (FCRR). Transcriptome analysis showed that F. oxysporum infection induces the MAPK signaling to stimulate tomato WRKY genes and initiate the resistance reaction to FCRR (Sun et al., 2022a).
Alternaria alternata
Alternaria alternata is a fungus causing leaf spot and blight in many plants. Various WRKY TFs are known to regulate plant resistance to A. alternata. In Populus nigra, PsnWRKY70 enhances the resistance to A. alternata by activating the MAP kinase cascade genes, calcium channel protein-encoding genes, and calcium-dependent protein kinase genes (Wang et al., 2022d). It also directly binds to the promoters of homologous WRKY genes (such as WRKY6, WRKY18, WRKY22, and WRKY22-1) and those of LRR domain protein genes (such as LRR8, LRR-RLK, ADR1-like 2, and NB-ARC) to promote their expression and to enhance pathogen resistance (Wang et al., 2022d). On the contrary, apple MdWRKY120 negatively regulates the resistance to A. alternata in an unknown regulatory mechanism (Liu et al., 2022). Tobacco NaWRKY3 is a fine-tuned master regulator of the defense network against A. alternata by enhancing the expression of target genes involved in phytoalexin biosynthesis and the immune signaling conducted by phytohormones (JA and ET), ROS, and the A. alternata resistance long noncoding RNA L2 and berberine bridge-like gene NaBBL28 (Xu et al., 2023).
Colletotrichum gloeosporioides
The fungus Colletotrichum gloeosporioides causes walnut anthracnose, a catastrophic disease adversely affecting the walnut production in China (Zhou et al., 2022). JrWRKY21 in walnut positively regulates the resistance to C. gloeosporioides by interacting with PTI5L, the transcriptional activator of PR gene, to form a WRKY21-PTI5L protein complex that activates the expression of JrPR5L (Zhou et al., 2022).
Puccinia horiana Henn
The fungus Puccinia horiana Henn causes chrysanthemum white rust (CWR), which impairs the production and ornamental value of chrysanthemum. Recent study showed that CmWRKY15-1 positively regulates chrysanthemum defense response to P. horiana (Gao et al., 2022). CmWRKY15-1 interacts with CmNPR1 (non-expressor of pathogenesis-related gene 1) to activate the expression of downstream PR genes and enhance resistance through the SA pathway (Gao et al., 2022).
Botrytis cinerea
Botrytis cinerea is a fungus with a wide range of hosts and can infect seedlings, fruits, and storage products of plants, causing heavy losses in agricultural production (Fu et al., 2022). Several studies in Arabidopsis have reported the regulation of B. cinerea resistance by WRKY TFs, such as AtWRKY33, AtWRKY2/AtWRKY34 (close homologs of AtWRKY33), AtWRKY3, AtWRKY4, AtWRKY18, AtWRKY40, AtWRKY60, and AtWRKY70. A few articles have comprehensively discussed the regulatory roles of these WRKY TFs (Amorim et al., 2016; Chen et al., 2019).
Studies in Lillium. regale revealed that LrWRKY39, LrWRKY4, and LrWRKY12 positively regulate the immune response to B. cinerea, while LrWRKY41a was identified to play a negative role (Cui et al., 2018a; Fu et al., 2022). The distinct functions of these WRKY members during Botrytis resistance may be related to the transcriptional changes in SA- and JA-responsive genes. Consistently, tomato SlWRKY46 and SpWRKY6 play negative roles during B. cinerea infection by inhibiting the activities of antioxidant and defense-related enzymes that influence the SA- and JA-signaling, ROS homeostasis, and the biosynthesis of SA, JA, and ABA (Liu et al., 2016; Shu et al., 2021). In contrast, FvWRKY50 in woodland strawberry positively regulates the resistance to B. cinerea by adjusting the JA signaling and the expression of defense-related genes (Ma et al., 2021). In rubber (Hevea brasiliensis), HbWRKY40 positively regulates host resistance to B. cinerea (Yang et al., 2020). Notably, overexpression of HbWRKY40 significantly induced the reactive oxygen burst in Nicotiana benthamiana and increased the resistance against B. cinerea in Arabidopsis via regulating multiple downstream genes involved in pathogen resistance (Yang et al., 2020). Conversely, overexpression of PtrWRKY73 from Populus trichocarpa in Arabidopsis increased the sensitivity to B. cinerea, accompanied by reduced expression of the defense-related gene PAL4 and upregulation of the SA-mediated defense-associated genes PR1, PR2, and PAD4 (Duan et al., 2015). Accordingly, GbWRKY1 of cotton negatively regulates the resistance to B. cinerea and V. dahliae by activating the expression of JA signaling suppressor gene GhJAZ1 to attenuate the JA responses (Li et al., 2014).
Rhizopus stolonifer
Peach PpWRKY45 and PpWRKY70 have been demonstrated to regulate the resistance against Rhizopus stolonifer, which causes severe fruit rot disease (Ji et al., 2021a, b; Ji et al., 2022; Li et al., 2022a). PpWRKY46 and PpWRKY53 contribute to methyl jasmonate (MeJA)-primed defense by regulating the energy metabolism-related genes PpSDH and PpCOX15, while PpWRKY22 physically interacts with PpHOS1/PpTGA1 (PpHOS1: a RING E3 ubiquitin ligase; TGA1: transcript factor) to regulate several SA-responsive PR genes and to positively modulate pathogen resistance (Ji et al., 2022; Li et al., 2022a). Accordingly, PpWRKY45 and PpWRKY70 participate in the MeJA-primed defense response against R. stolonifera, which occurs via the activation of JA biosynthetic genes, PR genes, and those of phenylpropanoid metabolism (Ji et al., 2021a, b).
Zymoseptoria tritici
Septoria tritici blotch (STB) caused by the fungus Zymoseptoria tritici is currently the main threat to wheat production in the temperate zone. Recent studies have revealed the role of WRKY TFs in regulating wheat responses to this fungus. For instance, TaWRKY10, a novel modulator of JA responses, negatively regulates the expression of JA receptor gene TaCOI1 to affect JA response and resistance to Z. tritici (Campanaro et al., 2021).
Botryosphaeria dothidea
In apple, Botryosphaeria dothidea causes various diseases, which adversely affect yield. Several transcription factors regulating host responses against this pathogen have been identified. Particularly, MdWRKY15 plays a positive role in regulating the resistance to B. dothidea by activating the expression of MdICS1, encoding an isochorismate synthase that is essential in SA accumulation, to increase the SA accumulation and the expression of various disease response-related genes (Zhao et al., 2020).
Verticillium dahliae
The wilt caused by Verticillium dahliae is a destructive disease, resulting in severe yield and quality losses in cotton worldwide. GhWRKY41, GhWRKY1-like and GhWRKY70 are associated with wilt resistance in cotton (Xiong et al., 2019; Hu et al., 2021; Xiao et al., 2023). Among these WRKY TFs, GhWRKY41 physically interacts with itself and directly activates its transcription (Xiao et al., 2023). The GhWRKY41 homodimer further activates the expression of GhC4H (cinnamate 4-hydroxylase) and Gh4CL(cinnamate 4-hydroxylase) to influence the phenylpropanoid metabolism and to modulate the accumulation of lignin and flavonoid (Xiao et al., 2023). GhWRKY41 also forms a positive feedback regulatory loop and improves cotton defense response against V. dahliae by regulating the phenylpropanoid metabolism (Xiao et al., 2023). GhWRKY1-like positively regulates cotton resistance to V. dahliae via directly manipulating the lignin biosynthesis. GhWRKY1-like can interact with the lignin biosynthesis related genes GhPAL6 and GhCOMT1 and activates the expression of GhPAL6 and GhCOMT1 to enhance the biosynthesis of total lignin (Hu et al., 2021). On the other hand, GhWRKY70 negatively regulates cotton resistance to V. dahliae through its effect on the biosynthesis and associated signaling of phytohormones ET, JA, and SA (Xiong et al., 2019).
Colletotrichum musae
Colletotrichum musae is a major pathogen known to infect plant species of the Musa genus. Studies in banana has shown that WRKY TFs can take part in the plant defense regulation conducted by NACs (NAM/ATAF/CUC), a plant-specific transcription factor family that have received much attention. In banana, the NAC transcription factor MaNAC5 cooperates with MaWRKY1 and MaWRKY2 to activate the transcriptional activities of MaPR1-1, MaPR2, MaPR10c, and MaCHIL1 (chitinase-like 1) to regulate SA- and MeJA-induced resistance to C. musae (Shan et al., 2016).
Sclerotinia sclerotiorum
Sclerotinia sclerotiorum is a necrotrophic fungal pathogen causing the stem rot of Brassica napus, a major oil crop worldwide. B. napus BnWRKY33 positively regulates the resistance to S. sclerotiorum by enhancing the expression of genes involved in camalexin biosynthesis and those regulated by SA and JA (Liu et al., 2018a). In contrast, BnWRKY15 increases B. napus susceptibility to S. sclerotiorum by interacting with BnWRKY33 and inhibiting its transcriptional activation (Liu et al., 2018a).
Puccinia striiformis f. sp. tritici
The stripe rust caused by Puccinia striiformis f. sp. tritici (Pst) is a destructive disease of wheat. Studies have reported diverse roles of wheat TaWRKY49 and TaWRKY62 in the defense response against stripe rust (Wang et al., 2017b). TaWRKY49 negatively regulates wheat resistance to Pst by increasing the expression of SA- and JA-responsive genes TaPR1.1 and TaAOS and ROS-associated genes TaCAT and TaPOD and decreasing the expression of ethylene-responsive gene TaPIE1 (Wang et al., 2017b). In contrast, TaWRKY62 positively regulates wheat resistance to Pst by differentially controlling SA, JA, ET, and ROS mediated signaling (Wang et al., 2017b). TaWRKY19 could bind to a W-box element in the promoter of TaNOX10, which encodes an NADPH oxidase and is required for ROS generation, and repress ROS generation, resulting the decreased host resistance to Pst (Wang et al., 2022e).
In barley, the WRKY TFs HvWRKY6 and HvWRKY70 positively regulate the resistance to Pst and Blumeria graminis f. sp. tritici (Bgt) infection. Notably, overexpression of HvWRKY6 in barley improved resistance to leaf rust (Puccinia triticina, Pt), Fusarium crown rot (FCR, Fusarium pseudograminearum, Fpg), and sharp eyespot (Rhizoctonia cerealis, Rc) by activating the SA signaling and suppressing the ABA and JA signaling (Li et al., 2020; 2022b). The findings of earlier studies indicated that HvWRKY6 could offer a broad-spectrum resistance against upto five fungal pathogens.
Magnaporthe oryzae
The blast fungus Magnaporthe oryzae is one of the most destructive pathogens of rice. Studies have shown that many WRKY factors play regulatory roles in rice resistance against M. oryzae, including OsWRKY13, OsWRKY22, OsWRKY24, OsWRKY28, OsWRKY30, OsWRKY31, OsWRKY42, OsWRKY45-1, OsWRKY45-2, OsWRKY47, OsWRKY53, OsWRKY62, OsWRKY76, and OsWRKY89. These WRKY TFs, except for OsWRKY28, OsWRKY42, and OsWRKY76, positively regulate rice resistance to blast disease (Wang et al., 2007; Qiu et al., 2007; Peng et al., 2008; Chujo et al., 2013; Cheng et al., 2015; Yokotani et al., 2018; Xu et al., 2022).
The OsWRKY45-2-OsWRKY13-OsWRKY42 transcriptional regulatory cascade play important roles in rice resistance to M. oryzae. OsWRKY42 functions downstream of OsWRKY13 and OsWRKY45-2 to negatively regulate rice resistance to M. oryzae by directly or indirectly suppressing the JA biosynthetic genes and JA-dependent defense signaling, and can be directly suppressed by OsWRKY13 (Cheng et al., 2015). Consistently, the overexpression of OsWRKY28 enhanced rice susceptibility to M. oryzae (Chujo et al., 2013).
In contrast, OsWRKY89 contributes positively to the resistance against blast by regulating the wax content/deposition on leaf surface (Wang et al., 2007). The rice knockdown lines of OsWRKY89 showed reduced wax content and increased susceptibility to M. grisea (Wang et al., 2007). OsWRKY53 also positively regulates blast resistance, and its function is activated by OsMPK3/OsMPK6-mediated phosphorylation (Chujo et al., 2014). The regulation of rice resistance against M. oryzae by OsWRKY30 is related to the activation and expression of JA biosynthetic genes LOX and AOS2 and PR genes PR3 and PR10 (Peng et al., 2012). OsWRKY31 acts downstream of OsMKK10-2 in the OsMKK10-2–OsMPK3–OsWRKY31 cascade as a key regulator of rice defense to M. oryzae. OsMKK10-2 enhances the resistance against M. oryzae through increasing the JA and SA accumulation and decreasing the indole-3-acetic acid level (Wang et al., 2023d). Modification of OsWRKY31 by phosphorylation and ubiquitination is involved in the OsMKK10-2-mediated defense pathway (Wang et al., 2023d).
The WRKY TFs reported to regulate plant resistance against fungal pathogens are shown in Table 1.
Role of WRKY TFs in plant resistance to bacterial pathogens
Pseudomonas syringae
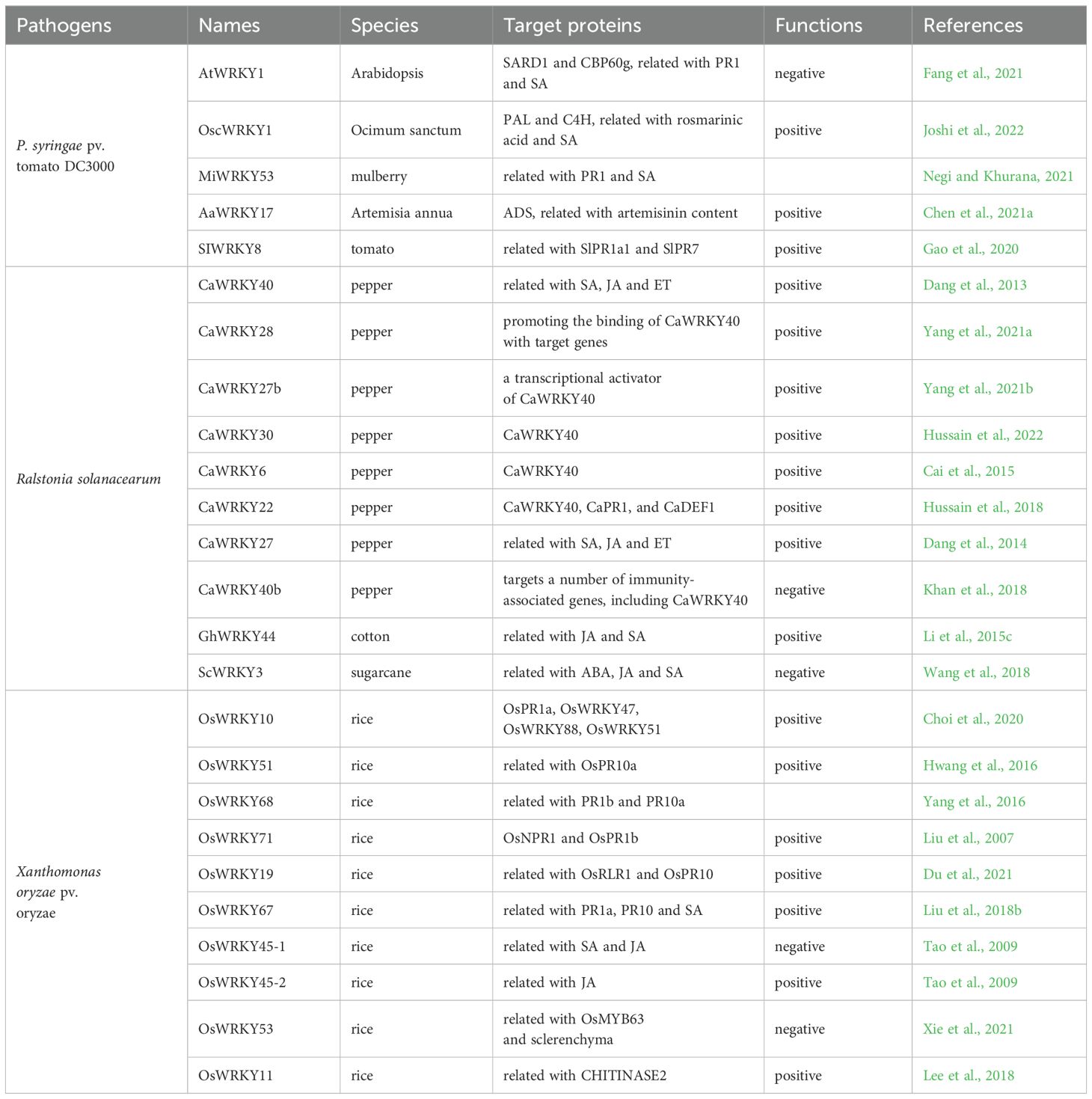
Pseudomonas syringae is a common gram-negative plant bacteria widely present in nature. Diseases caused by P. syringae rank the first tier among the plant bacterial diseases and cause huge economic losses to global agricultural production (Lai et al., 2008; Hu et al., 2012; Arrano-Salinas et al., 2018). Many WRKY TFs regulating P. syringae resistance have been isolated from Arabidopsis, such as AtWRKY11, AtWRKY17, AtWRKY18, AtWRKY40, AtWRKY60, AtWRKY25, AtWRKY3, AtWRKY4, AtWRKY38, AtWRKY62, AtWRKY46, AtWRKY70, AtWRKY53, and AtWRKY7/11/17 etc (Journot-Catalino et al., 2006; Xu et al., 2006; Zheng et al., 2007; Lai et al., 2008; Kim et al., 2008; Hu et al., 2012; Arrano-Salinas et al., 2018). The functions of these transcription factors have been discussed in previously published articles (Eulgem and Somssich, 2007; Pandey and Somssich, 2009; Amorim et al., 2016; Finatto et al., 2018). Recent studies found that AtWRKY1 negatively regulates Arabidopsis response to P. syringae pv. tomato DC3000 (Pto DC3000) by activating the SA signaling pathway and inhibiting the expression of PR1, PR2, and PR5 genes (Fang et al., 2021).
In Ocimum sanctum, the expression of OscWRKY1 gene was induced by JA, SA, and wound, and OscWRKY1 interacts with the W-box cis-element in the promoters of PAL and C4H to positively regulate phenylpropanoid metabolism, which leads to a change in the content of rosmarinic acid and an ultimate increase in resistance against Pto DC3000 (Joshi et al., 2022). Accordingly, mulberry MiWRKY53 regulates the resistance to Pto DC3000 through activating SA-mediated responses (Negi and Khurana, 2021). On the other hand, AaWRKY17 of Artemisia annua directly binds to the W-box motifs in the promoter of artemisinin biosynthetic gene ADS (amorpha-4,11-diene synthase) and promotes its expression to increases the artemisinin content, which enhances the resistance to Pto DC3000 (Chen et al., 2021a). Tomato SlWRKY8 increases the expression of pathogen-related genes SlPR1a1 and SlPR7 to improve the resistance to Pto DC3000 (Gao et al., 2020). Grape VqWRKY52, known as a positive regulator of powdery mildew resistance, enhances the resistance to Pto DC3000 but increases the susceptibility to Botrytis cinerea (Wang et al., 2017a). Populus PtrWRKY73 that negatively regulate the resistance to Botrytis cinerea could increase populus resistance to Pto DC3000 (Duan et al., 2015). These phenomena indicate that WRKY TFs regulate a broad-spectrum pathogen resistance in plants.
Ralstonia solanacearum
Bacterial wilt is a devastating soil-borne plant disease caused by Ralstonia solanacearum. So far, eight WRKY TFs related to bacterial wilt resistance have been isolated from chili peppers, including CaWRKY40, CaWRKY28, CaWRKY27b, CaWRKY30, CaWRKY6, CaWRKY27, CaWRKY22, and CaWRKY40b. Among them, CaWRKY40 is the first identified WRKY TF. Overexpression of CaWRKY40 enhanced the resistance to R.solanacearum in tobacco, while silencing of CaWRKY40 enhanced the susceptibility to R. solanacearum in pepper (Dang et al., 2013). CaWRKY40 is regulated by SA, JA and ET signaling and coordinates the responses mediated by these phytohormones to R. solanacearum (Dang et al., 2013). The other seven WRKY TFs, except CaWRKY40b, positively regulate pepper resistance to R. solanacearum. CaWRKY28 functions not by direct modulating the W-box containing immunity-related genes but by promoting the binding of CaWRKY40 to the immunity-related target genes, including CaPR1, CaNPR1, CaDEF1, and CaABR1, to activate their expression (Yang et al., 2021a). Similarly, CaWRKY27b is phosphorylated by CaCDPK29 and acts as a transcriptional activator of CaWRKY40 to initiate the response to R. solanacearum (Yang et al., 2021b). CaWRKY30 and CaWRKY6 also function in the R. solanacearum resistance by targeting CaWRKY40 (Cai et al., 2015; Hussain et al., 2022). Meanwhile, CaWRKY22 is regulated by SA, JA, and ET and directly binds to the promoters of CaPR1, CaDEF1 (a defense-related gene), and CaWRKY40 to controls host pathogen resistance (Hussain et al., 2018). Consistently, CaWRKY27 positively regulates tobacco resistance to R. solanacearum and its function is associated with the SA, JA, and ET mediated signaling (Dang et al., 2014). On the contrary, CaWRKY40b is a homolog of AtWRKY40 and negatively regulates pepper immunity against R. solanacearum (Figure 2) via the transcriptional modulation of a subset of immunity-associated genes as well as CaWRKY40 (Khan et al., 2018).
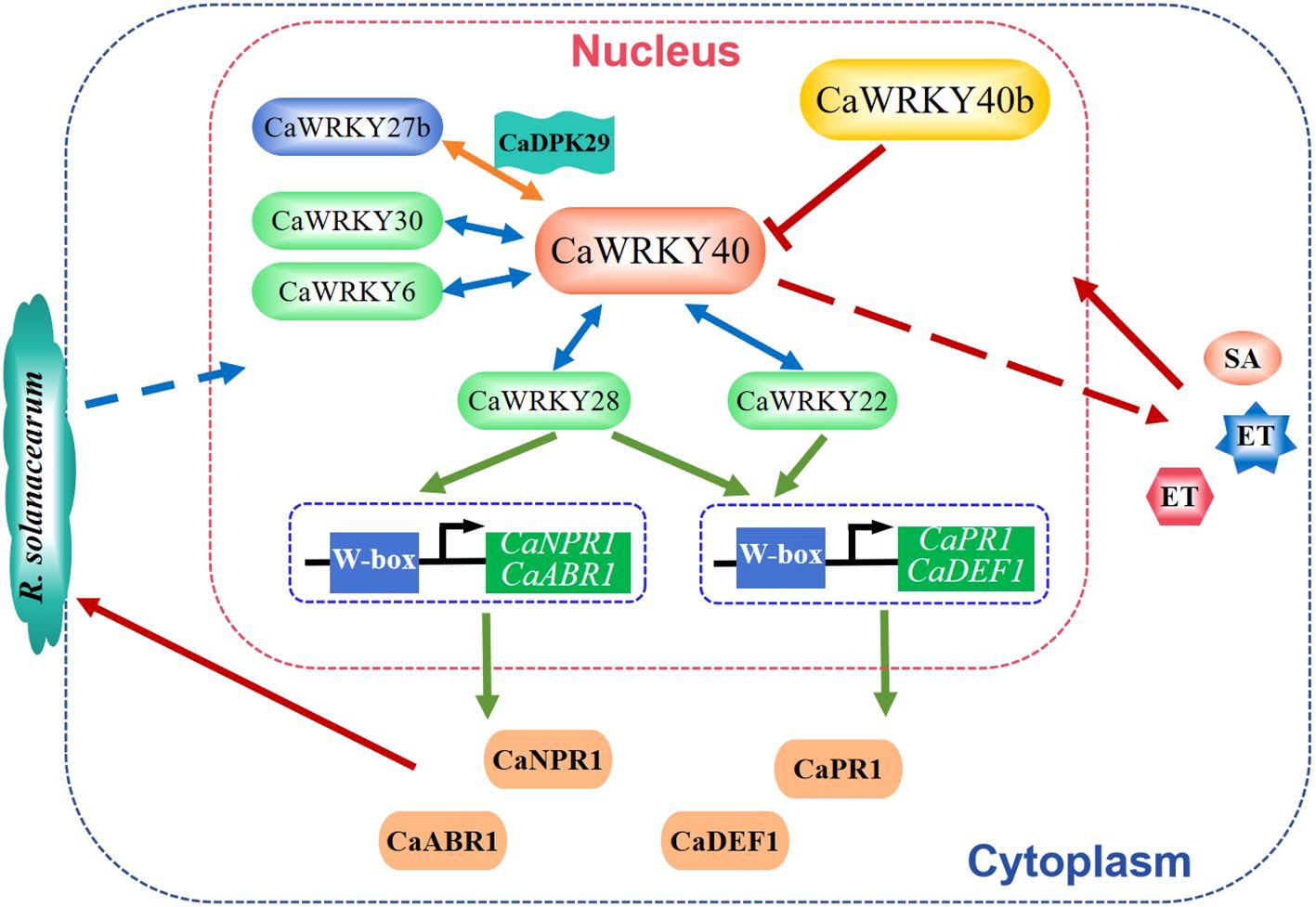
Figure 2. WRKY factors regulate pepper resistance against Ralstonia solanacearum. CaWRKY40 could interact with CaWRKY22, CaWRKY28, CaWRKY27b, CaWRKY30, and CaWRKY6 and affect the SA, JA and ET signaling to positively regulate pepper resistance to R. solanacearum. Yet, the function of CaWRKY40 is suppressed by CaWRKY40b (Dang et al., 2013; Khan et al., 2018; Yang et al., 2021a, 2021; Hussain et al., 2022).
In cotton, GhWRKY44 plays a positive role in regulating resistance to R. solanacearum infection. Overexpression of GhWRKY44 in cotton induced the expression of several defense-related genes, including PR-1, PR-2, PR-5, and NPR1 for SA signaling and PR-4 for JA signaling, to increase R. solanacearum resistance (Li et al., 2015c). ScWRKY3, a negative regulator of F. solani infection, also plays a negative regulation during the challenges by bacterial pathogen R. solanacearum (Wang et al., 2018). In 2022, 174 WRKY genes were identified from the genome of cultivated peanuts. Among the peanut WRKY TFs, 16 members were found to be upregulated by R. solanacearum infection, indicating an importance of these transcription factors in regulating peanut resistance (Yan et al., 2022b).
Xanthomonas oryzae pv. oryzae
Bacterial blight caused by Xanthomonas oryzae pv. oryzae (Xoo) is a major disease resulting in significant losses to rice production. Currently, multiple WRKY TFs related to bacterial blight have been isolated from rice, including OsWRKY10, OsWRKY11, OsWRKY13, OsWRKY19, OsWRKY45-1, OsWRKY45-2, OsWRKY47, OsWRKY51, OsWRKY53, OsWRKY62, OsWRKY67, OsWRKY68, OsWRKY71, and OsWRKY88. Two transcriptional regulatory cascades have been established in OsWRKY10-mediated blight disease resistance. In the first transcriptional regulatory cascade, OsWRKY47 acts downstream of OsWRKY10, while OsWRKY51 acts upstream of OsWRKY10 (Choi et al., 2020). In the second transcriptional regulatory cascade, OsWRKY47 acts downstream of OsWRKY10, and OsWRKY88 acts upstream of OsWRKY10. Specifically, OsWRKY10 binds to the promoter of OsPR1a to activate its expression and positively regulates the resistance to X. oryzae (Choi et al., 2020) as illustrated in Figure 3.
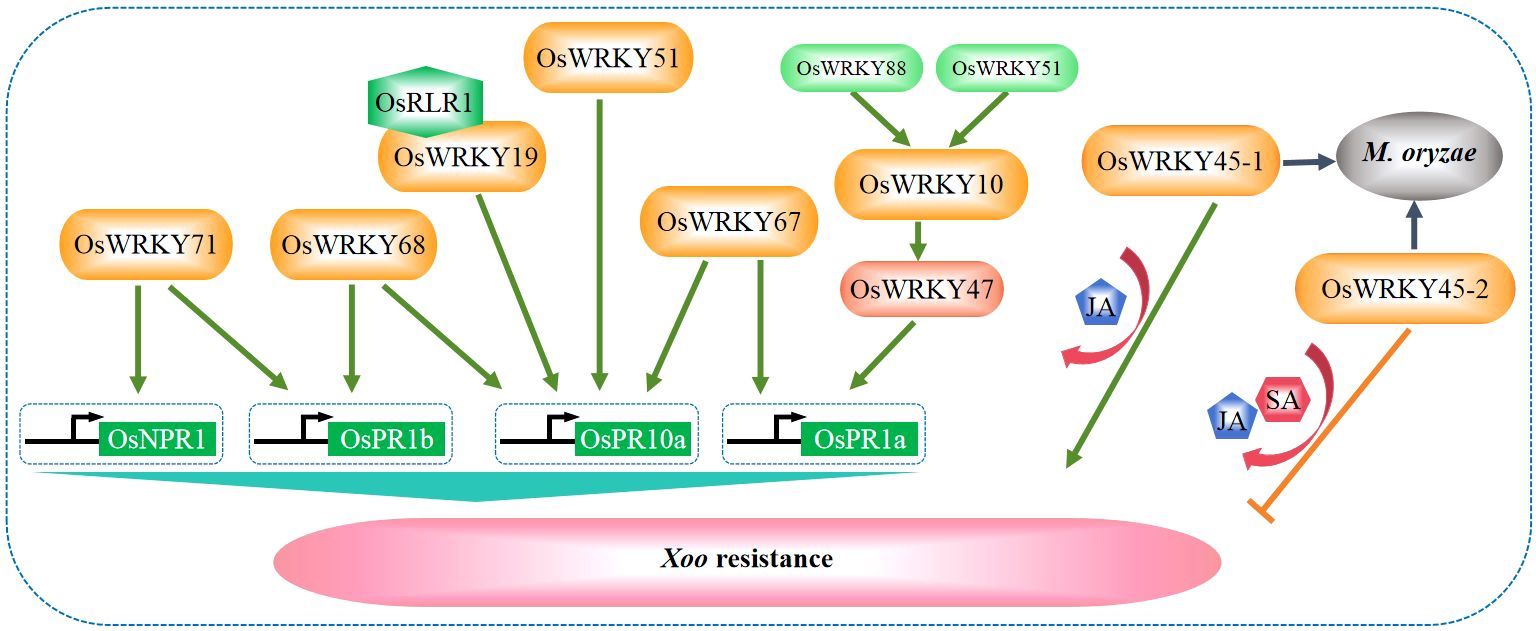
Figure 3. WRKY factors regulate rice resistance against Xanthomonas oryzae pv. Oryzae (Xoo). Rice WRKY TFs regulate the resistance to Xoo by directly modulating the expression of defense-related genes, mediating the phytohormone signaling of JA and SA, or interacting with other regulators including OsRLR1 (Tao et al., 2009; Yang et al., 2016; Liu et al., 2018b; Choi et al., 2020; Du et al., 2021).
During the interaction between rice and Xoo, OsWRKY68 is induced and binds to the W-boxes in the promoters of OsPR1b and OsPR10a to activate their expression (Yang et al., 2016). In contrast, OsWRKY71 functions as a transcriptional regulator upstream of OsNPR1 and OsPR1b and positively regulates the defense response against Xoo (Liu et al., 2007; Figure 3). OsWRKY51 also serves as a positive regulator to directly bind to the promoter of OsPR10a (Hwang et al., 2016). Recently, it was evidenced that the OsRLR1 (RPM1-like resistance gene 1)-OsWRKY19-OsPR10 complex plays a positive role in regulating Xoo resistance (Du et al., 2021). In addition, OsWRKY67 positively regulates rice responses to leaf blast, panicle blast, and bacterial blight. It activates the expression of OsPR1a and OsPR10 by directly binding to their promoters and induces the expression of various other defense-related genes (Figure 3), including those involved in the SA-dependent pathway (Liu et al., 2018b).
Two allelic genes OsWRKY45-1 and OsWRKY45-2, whose encoding proteins differ for 10 amino acids, play opposite roles in regulating rice resistance against Xoo. OsWRKY45-1 negatively regulates rice resistance to Xoo, while OsWRKY45-2 positively regulates the resistance (Tao et al., 2009). The OsWRKY45-1-mediated Xoo resistance is accompanied by increased accumulation of SA and JA and enhanced expression of a subset of defense-responsive genes. On the other hand, the OsWRKY45-2-mediated regulation of Xoo resistance is accompanied by increased accumulation of JA (but not SA) and enhanced expression of another subset of defense-responsive genes (Tao et al., 2009; Figure 3). Interestingly, both OsWRKY45-1 and OsWRKY45-2 are positive regulators in rice resistance against M. grisea.
Rice OsMYB63 promotes the expression of secondary cell wall-related cellulose synthase genes and the accumulation of cellulose, which results in thickened sclerenchyma cell walls, to provide resistance against pathogen infection (Xie et al., 2021). However, OsWRKY53 acts upstream of OsMYB63 to suppress its expression thus to negatively regulate the resistance to Xoo by weakening the sclerenchyma cell walls of vascular bundle (Xie et al., 2021). OsWRKY11 is also known to initiate defense responses to Xoo and it directly binds to the promoter of chitinase 2-encoding defense gene to activate its expression (Lee et al., 2018).
The WRKY TFs identified to regulate plant resistance to bacterial pathogens are listed in Table 2.
Role of WRKY TFs in plant resistance to oomycete pathogens
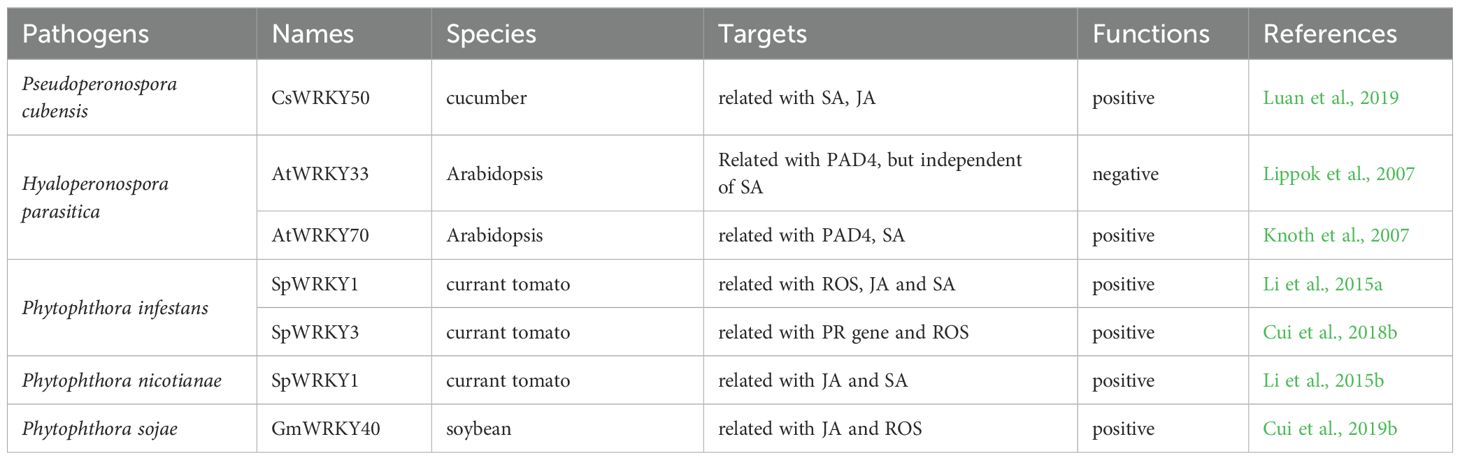
Pseudoperonospora cubensis, Hyaloperonospora parasitica and Phytophthora spp. are the major oomycete pathogens affecting plants. Various WRKY TFs have been identified to regulate plant responses to these oomycetes. In cucumber, CsWRKY50 positively regulates the resistance to H. parasitica via multiple signaling pathways to activate the expression of SA- and JA-responsive genes as well as the SA biosynthetic genes (Luan et al., 2019). In Arabidopsis, AtWRKY33 negatively regulates the resistance to H. parasitica and its gene expression is modulated by PAD4, a key component upstream of the SA signaling (Lippok et al., 2007). In addition, AtWRKY70 performs a positive role in the RPP4 (recognition of H. parasitica)-mediated resistance and basal defense against H. parasitica under the regulation by PAD4 and SA signaling (Knoth et al., 2007). In Solanum pimpinellifolium, SpWRKY1 and SpWRKY3 positively regulate the response to P. infestans. SpWRKY1 enhances the expression of ROS scavenging-related genes, SA/JA-responsive genes, and SA/JA biosynthetic genes (Li et al., 2015a; Cui et al., 2019a), while SpWRKY3 induces the expression of PR genes and reduces ROS accumulation to protect cell membrane from injury and to enhance the resistance (Cui et al., 2018b). SpWRKY1 also positively regulates the resistance to P. nicotianae via the SA and JA signaling (Li et al., 2015b). In soybean, GmWRKY40 functions as a positive regulator of plant response to P. sojae by modulating the JA signaling and hydrogen peroxide accumulation (Cui et al., 2019b)
The WRKY TFs identified to regulate plant resistance to oomycete pathogens are listed in Table 3.
Role of WRKY TFs in plant resistance to viruses and other pathogens
Studies have demonstrated the role of WRKY TFs in regulating plant resistance to viruses. CaWRKY-a of Capsicum annuum, NbWRKY1 of tobacco, and group III WRKY TFs of tomato regulate the resistance against tobacco mosaic virus, mulberry mosaic dwarf virus, and tomato yellow leaf curly virus (TYLCV), respectively (Park et al., 2006; Sun et al., 2022b; Huang et al., 2016), however, the detailed mechanisms underlying the function of these factors remain unclear.
Soybean is one of the plant species with the largest WRKY gene family, and at least three WRKY factors, including GmWRKY136, 53, and 86, have been identified to positively regulate the resistance to soybean cyst nematode (SCN; Heterodera glycines), which is the most devastating pathogen to soybean (Yang et al., 2017). Accordingly, wheat TaWRKY74 positively regulates the resistance to ‘Candidatus Phytoplasma tritici’ (‘Ca. P. tritici’), but it is destabilized by the pathogen effector SWP12, which weakens plant resistance and promotes virus colonization (Bai et al., 2022). Tomato SlWRKY45 could interact with most JA-ZIM domain family JAZ proteins, key repressors of the JA signaling, to attenuate JA biosynthesis and to repress the defense against root-knot nematodes (Huang et al., 2022).
Mechanisms underlying the regulation of plant immunity by WRKY factors
Numerous WRKY TFs have been identified to regulate plant resistance against pathogens. These WRKY factors can regulate plant pathogen resistance via multiple pathways, mainly involving the action of phytohormones, MAPK (mitogen-activated protein kinases) signaling cascades, and other signaling pathways.
In plants, phytohormones such as SA, JA, ABA, and ET are essential regulators of immunity. The WRKY factors specifically regulate the biosynthesis and signaling pathways of these phytohormones to modulate the expression of downstream disease resistance-related genes (Figure 4). For example, CsWRKY70 of citrus and MdWRKY15 of apple activate the expression of SAMT and MdICS1- encoding genes, respectively, to regulate plant pathogen resistance (Deng et al., 2020; Zhao et al., 2020). Additionally, OsWRKY30, OsWRKY42, PpWRKY45, PpWRKY70, and LrWRKY3 are involved in the biosynthesis of SA and JA, which in turn changes the plant resistance levels.
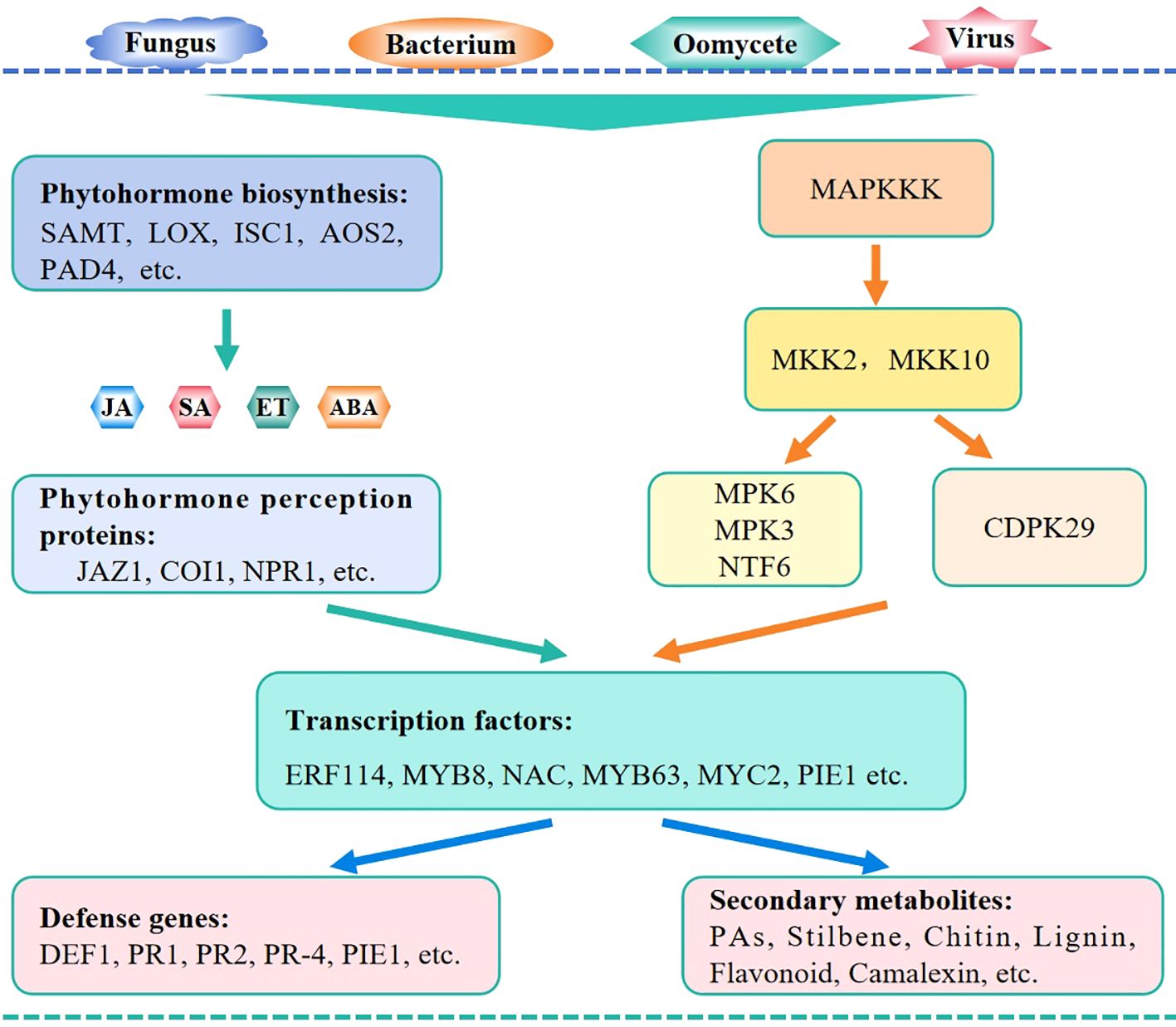
Figure 4. Mechanism module of WRKY TFs in regulating plant immunity against pathogens. WRKY TFs regulate plant pathogen resistance mainly via the action of phytohormone signaling, MAPK cascades, and other signaling pathways. During these processes, WRKY TFs may modulate the expression of phytohormone biosynthetic genes, affect the components in phytohormone signaling and MAPK cascades, or interact with other regulators including WRKY TFs.
NPR1 is a master regulator of the SA signaling pathway, while COI1 and JAZ are pivotal components of the JA signaling pathway. All these components are targets of WRKY TFs. Specifically, CmWRKY15-1, GhWRKY44, and OsWRKY71 function as transcriptional regulators upstream of NPR1 to regulate the plant defense response against pathogens. Moreover, TaWRKY10 and GbWRKY1 regulate the expression of genes encoding the JA-signaling components TaCOI1 and GhJAZ1, respectively, to regulate plant pathogen resistance. Among the WRKY TFs discussed in this article, almost 99% of them are associated with the JA and SA signaling, which is accompanied with changes in ROS accumulation and PR gene expression. In addition, other phytohormones, such as ET and ABA, are involved in the WRKY TFs-assisted immune network in regulating plant resistance against pathogens (Dang et al., 2014; Hussain et al., 2018; Wang et al., 2018).
Furthermore, WRKY TFs participate in the MAPK signaling pathways. MAPK signaling cascades link upstream receptors to downstream transcription factors via multiple phosphorylation events to function in the defensive responses in plants (Jagodzik et al., 2018). Group IIc WRKY TFs in cotton and PsnWRKY70 in Populus nigra both activate the MAP kinase cascade genes to enhance host resistance against pathogens. In tomato infected by F. oxysporum, the genes encoding MAPK and WRKY regulators are majorly enriched in the metabolic pathways related to disease resistance (Sun et al., 2022a). OsWRKY53 is activated by the OsMPK3/OsMPK6-mediated phosphorylation in the regulation of blast resistance (Chujo et al., 2014). OsWRKY31 is a key component in the MAPK signaling pathway involved in rice resistance to M. oryzae and is modulated by phosphorylation and ubiquitination via RING-finger E3 ubiquitin ligases (Wang et al., 2023d). CaWRKY27b is phosphorylated by CaCDPK29 to act as a transcriptional activator of CaWRKY40 and to initiate the response to R. solanacearum (Yang et al., 2021b).
Additionally, WRKY TFs may interact with other transcription factors to regulate plant pathogen resistance. The transcription factor complexes, such as MdWRKY75-MdERF114-MdMYB8-MdPRX63, MaNAC5-MaWRKY1-MaWRKY2, BnWRKY15-BnWRKY33, and CaWRKY40-CaWRKYs play important roles in regulating host plant resistance.
Conclusions and future perspectives
Plants employ multiple approaches to defend pathogen attack during their development processes, among which the WRKY TFs play crucial roles. Since the discovery of the first WRKY TF, lots of studies on the identification and function dissection of WRKY TFs have been reported (Supplementary Table S1). Recent studies showed that WRKY TFs coordinate a sophisticated network composed of phytohormone signaling, MAPK pathway, protein interaction, regulation of functional genes, and other immune responses in reprogramming plant responses against pathogens. WRKY TFs may exhibit either positive or negative role in regulating plant resistance to pathogens. Even though the majority of the identified WRKY TFs display positive functions, some of them show negative roles during plant responding to pathogen attack. For instance, 63 of the WRKY TFs reviewed in this article are positive regulators, whereas 21 of them are negative regulators. Furthermore, some WRKY TFs could mediate plant immunity to a broad spectrum of pathogens, such as HvWRKY6 regulating barley resistance to Pst, Bgt, Pt, Fpg, and Rc (Li et al., 2020; 2022b). Therefore, the function of WRKY TFs in regulating plant pathogen resistance is diverse and complex.
Plant genomes contain massive WRKY TFs to be investigated. The developing high-throughput transcriptomics, proteomics, metabolomics, gene editing and biosynthetic technologies will provide more efficient approaches to the identification and functional analysis of WRKY TFs. The function of WRKY TFs should be thoroughly revealed by future in-depth studies, and the mechanism underlying their regulation of pathogen resistance will be comprehensively understood. Thus, future studies may provide good choices for WRKY TFs of negative roles in regulating plant pathogen resistance to be targeted for gene editing or RNAi and those of positive roles in this process to be targeted for overexpression to improve plant disease resistance.
Author contributions
WW: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – original draft, Writing – review & editing. HC: Writing – review & editing. JW: Writing – review & editing. HZ: Funding acquisition, Project administration, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Key Laboratory of Tobacco Pest Monitoring Controlling & Integrated Management (KLTPMJMT2022-06); China National Tobacco Corporation (110202103013); Yunnan Tobacco Company (2022530000241011).
Conflict of interest
The authors declare that this study received funding from China National Tobacco Corporation and Yunnan Tobacco Company. These funders were not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1517595/full#supplementary-material
References
Amorim, L. L. B., dos Santos, R. D., Neto, J. P. B., Guida-Santos, M., Crovella, S., Benko-Iseppon, A. M. (2016). Transcription factors involved in plant resistance to pathogens. Curr. Protein Pept. Sci. 18, 335–351. doi: 10.2174/1389203717666160619185308
Arrano-Salinas, P., Dominguez-Figueroa, J., Herrera-Vasquez, A., Zavala, D., Medina, J., Vicente-Carbajosa, J., et al. (2018). WRKY7,-11and-17 transcription factors are modulators of the bZIP28 branch of the unfolded protein response during PAMP-triggered immunity in Arabidopsis thaliana. Plant Science. 277, 242–250. doi: 10.1016/j.plantsci.2018.09.019
Bai, B. X., Zhang, G. D., Li, Y., Wang, Y. B., Sujata, S., Zhang, X. D., et al. (2022). The 'Candidatus Phytoplasma tritici' effector SWP12 degrades the transcription factor TaWRKY74 to suppress wheat resistance. Plant J. 112, 1473–1488. doi: 10.1111/tpj.v112.6
Cai, H. Y., Yang, S. G., Yan, Y., Xiao, Z. L., Cheng, J. B., Wu, J., et al. (2015). CaWRKY6 transcriptionally activates CaWRKY40, regulates Ralstonia solanacearum resistance, and confers high-temperature and high-humidity tolerance in pepper. J. Exp. Botany. 66, 3163–3174. doi: 10.1093/jxb/erv125
Campanaro, A., Srivastava, A. K., Zhang, C. J., Lee, J., Millyard, L., Gatehouse, A. M. R., et al. (2021). TaWRKY10 transcription factor is a novel jasmonic acid signalling regulator involved in immunity against Septoria tritici blotch disease in wheat. Plant Pathol. 70, 1397–1408. doi: 10.1111/ppa.13388
Chen, T. T., Li, Y. P., Xie, L. H., Hao, X. L., Liu, H., Qin, W., et al. (2021a). AaWRKY17, a positive regulator of artemisinin biosynthesis, is involved in resistance to Pseudomonas syringae in Artemisia annua. Horticulture Res. 8, 1. doi: 10.1038/s41438-021-00652-6
Chen, X. J., Li, C., Wang, H., Guo, Z. J. (2019). WRKY transcription factors: evolution, binding, and action. Phytopathol. Res. 1, 13. doi: 10.1186/s42483-019-0022-x
Cheng, H. T., Liu, H. B., Deng, Y., Xiao, J. H., Li, X. H., Wang, S. P. (2015). The WRKY45-2 WRKY13 WRKY42 transcriptional regulatory cascade is required for rice resistance to fungal pathogen. Plant Physiol. 167, 1087–1099. doi: 10.1104/pp.114.256016
Choi, N., Im, J. H., Lee, E., Lee, J., Choi, C., Park, S. R., et al. (2020). WRKY10 transcriptional regulatory cascades in rice are involved in basal defense and Xa1-mediated resistance. J. Exp. Botany. 71, 3735–3748. doi: 10.1093/jxb/eraa135
Chujo, T., Miyamoto, K., Ogawa, S., Masuda, Y., Shimizu, T., Kishi-Kaboshi, M., et al. (2014). Overexpression of phosphomimic mutated OsWRKY53 leads to enhanced blast resistance in rice. PloS One 9, e98737. doi: 10.1371/journal.pone.0098737
Chujo, T., Miyamoto, K., Shimogawa, T., Shimizu, T., Otake, Y., Yokotani, N., et al. (2013). OsWRKY28, a PAMP-responsive transrepressor, negatively regulates innate immune responses in rice against rice blast fungus. Plant Mol. Biol. 82, 23–37. doi: 10.1007/s11103-013-0032-5
Cui, J., Meng, J., Yang, G. L., Liu, W. W., Zhou, X. X., Ma, N., et al. (2019a). LncRNA33732-respiratory burst oxidase module associated with WRKY1 in tomato- Phytophthora infestans interactions. Plant J. 97, 933–946. doi: 10.1111/tpj.2019.97.issue-5
Cui, J., Xu, P. S., Meng, J., Li, J. B., Jiang, N., Luan, Y. S. (2018b). Transcriptome signatures of tomato leaf induced by Phytophthora infestans and functional identification of transcription factor SpWRKY3. Theor. Appl. Genet. 131, 787–800. doi: 10.1007/s00122-017-3035-9
Cui, X. X., Yan, Q., Gan, S. P., Xue, D., Wang, H. T., Xing, H., et al. (2019b). GmWRKY40, a member of the WRKY transcription factor genes identified from Glycine max L., enhanced the resistance to Phytophthora sojae. BMC Plant Biol. 19, 1. doi: 10.1186/s12870-019-2132-0
Cui, Q., Yan, X., Gao, X., Zhang, D. M., He, H. B., Jia, G. X. (2018a). Analysis of WRKY transcription factors and characterization of two Botrytis cinerea-responsive LrWRKY genes from Lilium regale. Plant Physiol. Biochem. 127, 525–536. doi: 10.1016/j.plaphy.2018.04.027
Dang, F. F., Wang, Y. N., Eulgem, T., Lai, Y., Liu, Z. Q., Wang, X., et al. (2013). CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environment. 36, 757–774. doi: 10.1111/pce.2013.36.issue-4
Dang, F. F., Wang, Y. N., She, J. J., Lei, Y. F., Liu, Z. Q., Eulgem, T., et al. (2014). Overexpression of CaWRKY27, a subgroup IIe WRKY transcription factor of Capsicum annuum, positively regulates tobacco resistance to Ralstonia solanacearum infection. 2014 Physiologia Plantarum 150, 397–411. doi: 10.1111/ppl.2014.150.issue-3
Deng, B., Wang, W. J., Ruan, C. Q., Deng, L. L., Yao, S. X., Zeng, K. F. (2020). Involvement of CsWRKY70 in salicylic acid-induced citrus fruit resistance against Penicillium digitatum. Horticulture Res. 7, 157. doi: 10.1038/s41438-020-00377-y
Du, D., Zhang, C. W., Xing, Y. D., Lu, X., Cai, L. J., Yun, H., et al. (2021). The CC-NB-LRR OsRLR1 mediates rice disease resistance through interaction with OsWRKY19. Plant Biotechnol. J. 19, 1052–1064. doi: 10.1111/pbi.13530
Duan, Y. J., Jiang, Y. Z., Ye, S. L., Karim, A., Ling, Z. Y., He, Y. Q., et al. (2015). PtrWRKY73, a salicylic acid-inducible poplar WRKY transcription factor, is involved in disease resistance in Arabidopsis thaliana. Plant Cell Rep. 34, 831–841. doi: 10.1007/s00299-015-1745-5
Eulgem, T., Rushton, P. J., Robatzek, S., Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/S1360-1385(00)01600-9
Eulgem, T., Somssich, I. E. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10, 366–371. doi: 10.1016/j.pbi.2007.04.020
Fang, X., Meng, X. N., Cao, S. Q., Tang, X. F., Fan, T. T. (2021). AtWRKY1 negatively regulates the response of Arabidopsis thaliana to Pst. DC3000. Plant Physiol. Biochem. 166, 799–806. doi: 10.1016/j.plaphy.2021.06.044
Finatto, T., Viana, V. E., Woyann, L. G., Busanello, C., Maia, L. C. D., Oliveira, A. C. D. (2018). Can WRKY transcription factors help plants to overcome environmental challenges? Genet. Mol. Biol. 41, 533544. doi: 10.1590/1678-4685-GMB-2017-0232
Fu, Y. Y., Li, J., Wu, H., Jiang, S. J., Zhu, Y. Y., Liu, C. Y., et al. (2022). Analyses of Botrytis cinerea-responsive LrWRKY genes from Lilium regale reveal distinct roles of two LrWRKY transcription factors in mediating responses to B. cinerea. Plant Cell Rep. 41, 995–1012. doi: 10.1007/s00299-022-02833-6
Gao, G., Jin, R. B., Liu, D., Zhang, X., Sun, X. M., Zhu, P. F., et al. (2022). CmWRKY15-1 promotes resistance to Chrysanthemum white rust by regulating CmNPR1 expression. Fronties Plant science. 13, 865607. doi: 10.3389/fpls.2022.865607
Gao, Y. F., Liu, J. K., Yang, F. M., Zhang, G. Y., Wang, D., Zhang, L., et al. (2020). The WRKY transcription factor WRKY8 promotes resistance to pathogen infection and mediates drought and salt stress tolerance in Solanum lycopersicum. Pysiologia plantarum 168, 98–117. doi: 10.1111/ppl.v168.1
Hu, Y. R., Dong, Q. Y., Yu, D. Q. (2012). Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae. Plant Sci. 185, 288–297. doi: 10.1016/j.plantsci.2011.12.003
Hu, Q., Xiao, S. H., Wang, X. R., Ao, C. W., Zhang, X. L., Zhu, L. F. (2021). GhWRKY1-like enhances cotton resistance to Verticillium dahliae via an increase in defense-induced lignification and S monolignol content. Plant Sci. 305, 110833. doi: 10.1016/j.plantsci.2021.110833
Huang, Y., Li, M. Y., Wu, P., Xu, Z. S., Que, F., Wang, F., et al. (2016). Members of WRKY Group III transcription factors are important in TYLCV defense signaling pathway in tomato (Solanum lycopersicum). BMC Genomics 17, 2–18. doi: 10.1186/s12864-016-3123-2
Huang, H., Zhao, W. C., Qiao, H., Li, C. H., Sun, L. L., Yang, R., et al. (2022). SlWRKY45 interacts with jasmonate-ZIM domain proteins to negatively regulate defense against the root-knot nematode Meloidogyne incognita in tomato. Horticulture Res. 9, uhac197. doi: 10.1093/hr/uhac197
Hussain, A., Khan, M. I., Albaqami, M., Mahpara, S., Noorka, I. R., Ahmed, M. A. A., et al. (2022). CaWRKY30 positively regulates pepper immunity by targeting CaWRKY40 against Ralstonia solanacearum inoculation through modulating defense-related genes. Int. J. Mol. science. 22, 21. doi: 10.3390/ijms222112091
Hussain, A., Li, X., Weng, Y. H., Liu, Z. Q., Ashraf, M., Noman, A., et al. (2018). CaWRKY22 acts as a positive regulator in pepper response to Ralstonia Solanacearum by constituting networks with CaWRKY6, CaWRKY27, CaWRKY40, and CaWRKY58. Int. J. Mol. science. 19, 5. doi: 10.3390/ijms19051426
Hwang, S. H., Kwon, S. I., Jang, J. Y., Fang, I. L., Lee, H., Choi, C., et al. (2016). OsWRKY51, a rice transcription factor, functions as a positive regulator in defense response against Xanthomonas oryzae pv. Oryzae. Plant Cell Rep. 35, 1975–1985. doi: 10.1007/s00299-016-2012-0
Ishiguro, S., Nakamura, K. (1994). Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 50 upstream regions of genes coding for sporamin and beta-amylase from sweet potato. Mol. Gen. Genet. 244, 563–571. doi: 10.1007/BF00282746
Jagodzik, P., Tajdel-Zielinska., M., Ciesla., A., Marczak., M., Ludwikow., A. (2018). Mitogen-activated protein kinase cascades in plant hormone signaling. Fronties Plant Sci. 9, 1–26. doi: 10.3389/fpls.2018.01387
Ji, N. N., Li, Y. F., Wang, J., Zuo, X. X., Li, M. L., Jin, P., et al. (2022). Interaction of PpWRKY46 and PpWRKY53 regulates energy metabolism in MeJA primed disease resistance of peach fruit. Plant Physiol. Biochem. 171, 157–168. doi: 10.1016/j.plaphy.2021.12.035
Ji, N. N., Wang, J., Li, Y. F., Li, M. L., Jin, P., Zheng, Y. H. (2021b). Involvement of PpWRKY70 in the methyl jasmonate primed disease resistance against Rhizopus stolonifer of peaches via activating phenylpropanoid pathway. Postharvest Biol. Technol. 174, 11390. doi: 10.1016/j.postharvbio.2021.111466
Ji, N. N., Wang, J., Zuo, X. X., Li, Y. F., Li, M. L., Wang, K. T., et al. (2021a). PpWRKY45 is involved in methyl jasmonate primed disease resistance by enhancing the expression of jasmonate acid biosynthetic and pathogenesis-related genes of peach fruit. Postharvest Biol. Technol. 172, 11390. doi: 10.1016/j.postharvbio.2020.111390
Jin, Y. L., Liu, H., Gu, T. T., Xing, L. X., Han, G. H., Ma, P. T., et al. (2022). PM2b, a CC-NBS-LRR protein, interacts with TaWRKY76-D to regulate powdery mildew resistance in common wheat. Fronties Plant Sci. 13, 973065. doi: 10.3389/fpls.2022.973065
Joshi, A., Jeena, G. S., Kumar, R. S., Pandey, A., Shukla, R. K. (2022). Ocimum sanctum, OscWRKY1, regulates phenylpropanoid pathway genes and promotes resistance to pathogen infection in Arabidopsis. Plant Mol. Biol. 110, 235–251. doi: 10.1007/s11103-022-01297-2
Journot-Catalino, N., Somssich, I. E., Roby, D., Kroj, T. (2006). The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 18, 3289–3302. doi: 10.1105/tpc.106.044149
Khan, M. I., Zhang, Y. W., Liu, Z. Q., Hu, J., Liu, C. L., Yang, S. L., et al. (2018). CaWRKY40b in pepper acts as a negative regulator in response to Ralstonia solanacearum by directly modulating defense genes including CaWRKY40. Int. J. Mol. Sci. 19, 5. doi: 10.3390/ijms19051403
Khoso, M. A., Hussain, A., Ritonga, F. N., Ali, Q., Channa, M. M., Alshegaihi, R. M., et al. (2022). WRKY transcription factors (TFs): Molecular switches to regulate drought, temperature, and salinity stress in plants. Front. Plant Sci. 13, 1039329. doi: 10.3389/fpls.2022.1039329
Kim, K. C., Lai, Z. B., Fan, B. F., Chen, Z. X. (2008). Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell. 20, 2357–2371. doi: 10.1105/tpc.107.055566
Knoth, C., Ringler, J., Dangl, J. L., Eulgem, T. (2007). Arabidopsis WRKY70 is required for full RPP4-mediated disease resistance and basal defense against Hyaloperonospora parasitica. Mol. Plant-Microbe Interact. 20, 120–128. doi: 10.1094/MPMI-20-2-0120
Lai, Z. B., Vinod, K. M., Zheng, Z. Y., Fan, B. F., Chen, Z. X. (2008). Roles of Arabidopsis WRKY3 and WRKY4 transcription factors in plant responses to pathogens. BMC Plant Biol. 8, 68. doi: 10.1186/1471-2229-8-68
Lee, H., Cha, J., Choi, C., Choi, N., Ji, H. S., Park, S. R., et al. (2018). Rice WRKY11 plays a role in pathogen defense and drought tolerance. Rice 11, 5. doi: 10.1186/s12284-018-0199-0
Li, S., Hai, J., Wang, Z., Deng, J., Liang, T. T., Su, L. L., et al. (2021). Lilium regale Wilson WRKY2 regulates chitinase gene expression during the response to the root rot pathogen Fusarium oxysporum. Fronties Plant Sci. 12, 741463. doi: 10.3389/fpls.2021.741463
Li, C., He, X., Luo, X. Y., Xu, L., Liu, L. L., Min, L., et al. (2014). Cotton WRKY1 mediates the plant defense-to-development transition during infection of cotton by Verticillium dahliae by activating JASMONATE ZIM-DOMAIN1 expression. Plant Physiol. 166, 2179–2194. doi: 10.1104/pp.114.246694
Li, C. H., Lei, C. Y., Huang, Y. X., Zheng, Y. H., Wang, K. T. (2022a). PpWRKY22 physically interacts with PpHOS1 / PpTGA1 and positively regulates several SA-responsive PR genes to modulate disease resistance in BABA-primed peach fruit. Scientia Hortic. 290, 11047. doi: 10.1016/j.scienta.2021.110479
Li, J. B., Luan, Y. S., Liu, Z. (2015a). SpWRKY1 mediates resistance to Phytophthora infestans and tolerance to salt and drought stress by modulating reactive oxygen species homeostasis and expression of defense-related genes in tomato. Plant Cell Tissure Organ Culture 123, 67–81. doi: 10.1007/s11240-015-0815-2
Li, J. B., Luan, Y. S., Liu, Z. (2015b). Overexpression of SpWRKY1 promotes resistance to Phytophthora nicotianae and tolerance to salt and drought stress in transgenic tobacco. Physiologia Plantarum 155, 248–266. doi: 10.1111/ppl.2015.155.issue-3
Li, J., Wang, J., Wang, N. X., Guo, X., Gao, Z. (2015c). GhWRKY44, a WRKY transcription factor of cotton, mediates defense responses to pathogen infection in transgenic Nicotiana benthamiana. Plant Cell Tissure Organ Culture. 121, 127–140. doi: 10.1007/s11240-014-0688-9
Li, H. P., Wu, J. J., Shang, X. F., Geng, M. M., Gao, J., Zhao, S. Q., et al. (2020). WRKY transcription factors shared by BTH-Induced resistance and NPR1-Mediated acquired resistance improve broad-spectrum disease resistance in wheat. Mol. Plant-Microbe Interact. 33, 433–443. doi: 10.1094/MPMI-09-19-0257-R
Li, M. Y., Zhao, S. Q., Yang, J. Y., Ren, Y., Su, J., Zhao, J. J., et al. (2022b). Exogenous expression of barley HvWRKY6 in wheat improves broad-spectrum resistance to leaf rust, Fusarium crown rot, and sharp eyespot. Int. J. Of Biol. Macromolecules 218, 1002–1012. doi: 10.1016/j.ijbiomac.2022.07.138
Lippok, B., Birkenbihl, R. P., Rivory, G., Brümmer, J., Schmelzer, E., Logemann, E., et al. (2007). Expression of AtWRKY33 encoding a pathogen- or PAMP-responsive WRKY transcription factor is regulated by a composite DNA motif containing W box elements. Mol. Plant-Microbe Interactions. 20, 420–429. doi: 10.1094/MPMI-20-4-0420
Liu, X. Q., Bai, X. Q., Wang, X. J., Chu, C. C. (2007). OsWRKY71, a rice transcription factor, is involved in rice defense response. J. Plant Physiol. 164, 969–979. doi: 10.1016/j.jplph.2006.07.006
Liu, L. F., Li, X. M., Guo, W., Shi, J. J., Chen, W. J., Lei, Y. Y., et al. (2022). MdWRKY120 enhance apple susceptibility to Alternaria alternata. Plant-Basel 11, 3389. doi: 10.3390/plants11233389
Liu, F., Li, X. X., Wang, M. R., Wen, J., Yi, B., Shen, J. X., et al. (2018a). Interactions of WRKY15 and WRKY33 transcription factors and their roles in the resistance of oilseed rape to Sclerotinia infection. Plant Biotechnol. J. 16, 911–925. doi: 10.1111/pbi.2018.16.issue-4
Liu, Q., Li, X., Yan, S. J., Yu, T., Yang, J. Y., Dong, J. F., et al. (2018b). OsWRKY67 positively regulates blast and bacteria blight resistance by direct activation of PR genes in rice. BMC Plant Biol. 18, 2–13. doi: 10.1186/s12870-018-1479-y
Liu, Y. S., Liu, Q. W., Li, X. W., Zhang, Z. J., Ai, S. K., Liu, C., et al. (2023). MdERF114 enhances the resistance of apple roots to Fusarium solani by regulating the transcription of MdPRX63. Plant Physiol. 192, 2015–2029. doi: 10.1093/plphys/kiad057
Liu, Z., Luan, Y. S., Li, J. B. (2016). Molecular cloning and expression analysis of SpWRKY6 gene from Solanum pimpinellifolium. Biol. Plantarum 60, 226–234. doi: 10.1007/s10535-016-0582-x
Luan, Q. Q., Chen, C. H., Liu, M. Y., Li, Q., Wang, L. N., Ren, Z. (2019). CsWRKY50 mediates defense responses to Pseudoperonospora cubensis infection in Cucumis sativus. Plant Sci. 279, 59–69. doi: 10.1016/j.plantsci.2018.11.002
Ma, C. J., Xiong, J. S., Liang, M. R., Liu, X. Y., Lai, X. D., Bai, Y. B., et al. (2021). Strawberry WRKY transcription factor WRKY50 is required for resistance to necrotrophic fungal pathogen Botrytis cinerea. Agronomy-basel 11, 2377. doi: 10.3390/agronomy11122377
Negi, N., Khurana, P. (2021). A salicylic acid inducible mulberry WRKY transcription factor, MiWRKY53 is involved in plant defence response. Plant Cell Rep. 40, 2151–2171. doi: 10.1007/s00299-021-02710-8
Ning, K., Li, M. Z., Wei, G. F., Zhou, Y. X., Zhang, G. Z., Huai, H., et al. (2021). Genomic and transcriptomic analysis provide insights into root rot resistance in Panax notoginseng. Fronties Plant Sci. 12, 775019. doi: 10.3389/fpls.2021.775019
Pandey, S. P., Somssich, I. E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiol. 150, 1648–1655. doi: 10.1104/pp.109.138990
Park, C. J., Shin, Y. C., Lee, B. J., Kim, K. J., Kim, J. K., Paek, K. H. (2006). A hot pepper gene encoding WRKY transcription factor is induced during hypersensitive response to tobacco mosaic virus and Xanthomonas campestris. Planta 223, 168–179. doi: 10.1007/s00425-005-0067-1
Peng, Y., Bartley, L. E., Chen, X. W., Dardick, C., Chern, M. S., Ruan, R., et al. (2008). OsWRKY62 is a negative regulator of basal and Xa21-mediated defense against Xanthomonas oryzae pv. oryzae rice. Mol. Plant 1, 446–458. doi: 10.1093/mp/ssn024
Rinerson, C. I., Rabara, R. C., Tripathi, P., Shen, Q. J., Rushton, P. J. (2015). The evolution of WRKY transcription factors. BMC Plant Biol. 15, 66. doi: 10.1186/s12870-015-0456-y
Rushton, P. J., Somssich, I. E., Ringler, P., Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Science. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Shan, W., Chen, J. Y., Kuang, J. F., Lu, W. J. (2016). Banana fruit NAC transcription factor MaNAC5 cooperates with MaWRKYs to enhance the expression of pathogenesis-related genes against Colletotrichum musae. Mol. Plant Pathol. 17, 330–338. doi: 10.1111/mpp.2016.17.issue-3
Shu, P., Zhang, S. J., Li, Y. J., Wang, X. Y., Yao, L., Sheng, J. P., et al. (2021). Over-expression of SlWRKY46 in tomato plants increases susceptibility to Botrytis cinerea by modulating ROS homeostasis and SA and JA signaling pathways. Plant Physiol. Biochem. 166, 1–9. doi: 10.1016/j.plaphy.2021.05.021
Sun, S. S., Ren, Y. X., Wang, D. X., Farooq, T., He, Z. F., Zhang, C., et al. (2022b). A group I WRKY transcription factor regulates mulberry mosaic dwarf-associated virus-triggered cell death in Nicotiana benthamiana. Mol. Plant Pathol. 23, 237–253. doi: 10.1111/mpp.13156
Sun, Y. Q., Yang, H. H., Li, J. F. (2022a). Transcriptome analysis reveals the response mechanism of Frl-mediated resistance to Fusarium oxysporum f. sp. radicis-lycopersici (FORL) infection in tomato. Int. J. Mol. Sci. 23, 7078. doi: 10.3390/ijms23137078
Tao, Z., Liu, H. B., Qiu, D. Y., Zhou, Y., Li, X. H., Xu, C. G., et al. (2009). A pair of allelic WRKY genes play opposite roles in rice-bacteria interactions. Plant Physiol. 151, 936–948. doi: 10.1104/pp.109.145623
Wang, W., Bai, X. D., Chen, K., Gu, C. R., Yu, Q. B., Jiang, J., et al. (2022d). Roles of PsnWRKY70 in regulatory network response to infection with Alternaria alternata Fr. Keissl in populus. Int. J. Mol. Sci. 23, 7537. doi: 10.3390/ijms23147537
Wang, H. P., Chen, W. Q., Xu, Z. Y., Chen, M. F., Yu, D. Q. (2022a). Functions of WRKYs in plant growth and development. Trends Plant Sci. 28, 630–645. doi: 10.1016/j.tplants.2022.12.012
Wang, Z., Deng, J., Liang, T. T., Su, L. L., Zheng, L. L., Chen, H. J., et al. (2022c). Lilium regale Wilson WRKY3 modulates an antimicrobial peptide gene, LrDef1, during response to Fusarium oxysporum. BMC Plant Biol. 22, 257. doi: 10.1186/s12870-022-03649-y
Wang, N., Fan, X., He, M. Y., Hu, Z. Y., Tang, C. L., Zhang, S., et al. (2022e). Transcriptional repression of TaNOX10 by TaWRKY19 compromises ROS generation and enhances wheat susceptibility to stripe rust. THE Plant Cell 34, 1784–1803. doi: 10.1093/plcell/koac001
Wang, H., Gong, W. F., Wang, Y., Ma, Q. (2023b). Contribution of a WRKY transcription factor, ShWRKY81, to powdery mildew resistance in wild tomato. Int. J. Mol. science. 24, 2583. doi: 10.3390/ijms24032583
Wang, X. H., Guo, R. R., Tu, M. X., Wang, D. J., Guo, C. L., Wan, R., et al. (2017a). Ectopic expression of the wild grape WRKY transcription factor VqWRKY52 in Arabidopsis thaliana enhances resistance to the biotrophic pathogen powdery mildew but not to the necrotrophic pathogen Botrytis cinerea. Fronties Plant Sci. 8, 97. doi: 10.3389/fpls.2017.00097
Wang, L. J., Guo, D. Z., Zhao, G. D., Wang, J. Y., Zhang, S. X., Wang, C., et al. (2022b). Group IIc WRKY transcription factors regulate cotton resistance to Fusarium oxysporum by promoting GhMKK2-mediated flavonoid biosynthesis. New Phytologyist 236, 249–265. doi: 10.1111/nph.v236.1
Wang, S., Han, S. Y., Zhou, X. G., Zhao, C. J., Guo, L. A., Zhang, J. Q., et al. (2023d). Phosphorylation and ubiquitination of OsWRKY31 are integral to OsMKK10-2-mediated defense responses in rice. THE Plant Cell 35, 2391–2412. doi: 10.1093/plcell/koad064
Wang, H. H., Hao, J. J., Chen, X. J., Hao, Z. N., Wang, X., Lou, Y. G., et al. (2007). Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol. Biol. 65, 799–815. doi: 10.1007/s11103-007-9244-x
Wang, W. J., Li, T., Chen, Q., Deng, B., Deng, L. L., Zeng, K. F. (2021b). Transcription factor CsWRKY65 participates in the establishment of disease resistance of citrus fruits to Penicillium digitatum. J. Agric. Food Chem. 69, 5671–5682. doi: 10.1021/acs.jafc.1c01411
Wang, W. J., Li, T., Chen, Q., Yao, S. X., Deng, L. L., Zeng, K. F. (2021a). CsWRKY25 improves resistance of citrus fruit to Penicillium digitatum via modulating reactive oxygen species production. Front. Plant Sci. 12, 818198. doi: 10.3389/fpls.2021.818198
Wang, W. J., Li, T., Chen, Q., Yao, S. X., Zeng, K. F. (2023c). Transcriptional regulatory mechanism of a variant transcription factor CsWRKY23 in citrus fruit resistance to Penicillium digitatum. Food Chem. 413, 135573. doi: 10.1016/j.foodchem.2023.135573
Wang, L., Liu, F., Zhang, X., Wang, W. J., Sun, T. T., Chen, Y. F., et al. (2018). Expression characteristics and functional analysis of the ScWRKY3 gene from sugarcane. Int. J. Mol. Science. 19, 4059. doi: 10.3390/ijms19124059
Wang, J. J., Tao, F., Tian, W., Guo, Z. F., Chen, X. M., Xu, X. M., et al. (2017b). The wheat WRKY transcription factors TaWRKY49 and TaWRKY62 confer differential high-temperature seedling-plant resistance to Puccinia striiformis f. sp tritici. PloS One 12, 7. doi: 10.1371/journal.pone.0181963
Wang, Y., Wang, X. H., Fang, J. H., Yin, W. C., Yan, X. X., Tu, M. X., et al. (2023a). VqWRKY56 interacts with VqbZIPC22 in grapevine to promote proanthocyanidin biosynthesis and increase resistance to powdery mildew. New Phytol. 237, 1856–1875. doi: 10.1111/nph.v237.5
Xiang, L., Wang, M., Huang, J. X., Jiang, W. T., Yan, Z. B., Chen, X. S., et al. (2022). MdWRKY74 is involved in resistance response to apple replant disease. Plant Growth Regul. 96, 145–156. doi: 10.1007/s10725-021-00766-w
Xiao, S. H., Ming, Y. Q., Hu, Q., Ye, Z. X., Si, H., Liu, S. M., et al. (2023). GhWRKY41 forms a positive feedback regulation loop and increases cotton defence response against Verticillium dahliae by regulating phenylpropanoid metabolism. Plant Biotechnol. J. 21, 961–978. doi: 10.1111/pbi.14008
Xie, W. Y., Ke, Y. G., Cao, J. B., Wang, S. P., Yuan, M. (2021). Knock out of transcription factor WRKY53 thickens sclerenchyma cell walls, confers bacterial blight resistance. Plant Physiol. 187, 1746–1761. doi: 10.1093/plphys/kiab400
Xiong, X. P., Sun, S. C., Li, Y. J., Zhang, X. Y., Sun, J., Xue, F. (2019). The cotton WRKY transcription factor GhWRKY70 negatively regulates the defense response against Verticillium dahliae. Crop J. 7, 393–402. doi: 10.1016/j.cj.2018.10.005
Xu, X. P., Chen, C. H., Fan, B. F., Chen, Z. X. (2006). Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18, 1310–1326. doi: 10.1105/tpc.105.037523
Xu, X. H., Wang, H., Liu, J. Q., Han, S. Y., Lin, M. M., Guo, Z. J., et al. (2022). OsWRKY62 and OsWRKY76 interact with importin α1s for negative regulation of defensive responses in rice nucleus. Rice (NY) 1, 12. doi: 10.1186/s12284-022-00558-4
Xu, Z., Zhang, S. T., Wu, J. S. (2023). NaWRKY3 is a master transcriptional regulator of the defense network against brown spot disease in wild tobacco. J. Exp. Bot. 74, 4169–4188. doi: 10.1093/jxb/erad142
Yan, L., Jin, H. T., Raza, A., Huang, Y., Gu, D. P., Zou, X. Y. (2022b). WRKY genes provide novel insights into their role against Ralstonia solanacearum infection in cultivated peanut (Arachis hypogaea L.). Fronties Plant Sci. 13, 1–16. doi: 10.3389/fpls.2022.986673
Yang, S., Cai, W. W., Shen, L., Cao, J. S., Liu, C. L., Hu, J., et al. (2021b). A CaCDPK29-CaWRKY27b module promotes CaWRKY40-mediated thermo tolerance and immunity to Ralstonia solanacearum in pepper. New Phytol. 233, 1843–1863. doi: 10.1111/nph.17891
Yang, J., Wang, Q. N., Luo, H. L., He, C. Z., An, B. (2020). HbWRKY40 plays an important role in the regulation of pathogen resistance in Hevea brasiliensis. Plant Cell Rep. 39, 1095–1107. doi: 10.1007/s00299-020-02551-x
Yang, S., Zhang, Y. W., Cai, W. W., Liu, C. L., Hu, J., Shen, L., et al. (2021a). CaWRKY28 Cys249 is required for interaction with CaWRKY40 in the regulation of pepper immunity to Ralstonia solanacearum. Mol. Plant-Microbe Interact. 34, 733–745. doi: 10.1094/MPMI-12-20-0361-R
Yang, Y., Zhou, Y., Chi, Y. J., Fan, B. F., Che, Z. X. (2017). Characterization of Soybean WRKY gene family and identification of soybean WRKY genes that promote resistance to soybean cyst nematode. Sci. Rep. 7, 17804. doi: 10.1038/s41598-017-18235-8
Yang, S., Zhou, L., Miao, L. Y., Shi, J. N., Sun, C. Q., Fan, W., et al. (2016). The expression and binding properties of the rice WRKY68 protein in the Xa21-mediated resistance response to Xanthomonas oryzae pv. Oryzae. J. Integr. Agric. 15, 2451–2460. doi: 10.1016/S2095-3119(15)61265-5
Yin, W. C., Wang, X. H., Liu, H., Wang, Y., van Nocker, S., Tu, M. X., et al. (2022). Overexpression of VqWRKY31 enhances powdery mildew resistance in grapevine by promoting salicylic acid signaling and specific metabolite synthesis. Hortic. Resarch 9, 1–17. doi: 10.1093/hr/uhab064
Yokotani, N., Shikata, M., Ichikawa, H., Mitsuda, N., Ohme-Takagi, M., Minami, E., et al. (2018). OsWRKY24, a blast-disease responsive transcription factor, positively regulates rice disease resistance. J. Gen. Plant Pathol. 84, 85–91. doi: 10.1007/s10327-018-0768-5
Yu, D. H., Fan, R. C., Zhang, L., Xue, P. Y., Liao, L. B., Hu, M. Z., et al. (2023). HvWRKY2 acts as an immunity suppressor and targets HvCEBiP to regulate powdery mildew resistance in barley. Crop J. 11, 99–107. doi: 10.1016/j.cj.2022.05.010
Zhang, J., Jiang, C. Y., Wang, Y. J. (2022). Functional analysis of the interaction between transcription factors VqWRKY6 and VqBZIP1 in regulating the resistance to powdery mildew in Chinese wild Vitis quinquangularis. Scientia Agricultura Sin. 55, 4626–4639. doi: 10.1007/s00425-010-1258-y
Zhang, Y. J., Wang, L. J. (2005). The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evolutionary Biology BMC Evolutionary Biol. 5, 1. doi: 10.1186/1471-2148-5-1
Zhao, X. Y., Qi, C. H., Jiang, H., Zhong, M. S., You, C. X., Li, Y. Y., et al. (2020). MdWRKY15 improves resistance of apple to Botryosphaeria dothidea via the salicylic acid-mediated pathway by directly binding the MdICS1 promoter. J. Integr. Plant Biol. 62 4, 527–543. doi: 10.1111/jipb.12825
Zheng, Z. Y., Mosher, S. L., Fan, B. F., Klessig, D. F., Chen, Z. X. (2007). Functional analysis of Arabidopsis WRKY25 transcription factor in plant defense against Pseudomonas syringae. BMC Plant Biol. 7, 1–13. doi: 10.1186/1471-2229-7-2
Zheng, L. L., Qiu, B. L., Su, L. L., Wang, H. L., Cui, X. M., Ge, F., et al. (2022). Panax notoginseng WRKY transcription factor 9 is a positive regulator in responding to root rot pathogen Fusarium solani. Fronties Plant Sci. 13, 1–17. doi: 10.3389/fpls.2022.930644
Keywords: WRKY TFs, biotic stress, plant resistance, defense signaling, plant-pathogen interactions
Citation: Wang W, Cao H, Wang J and Zhang H (2025) Recent advances in functional assays of WRKY transcription factors in plant immunity against pathogens. Front. Plant Sci. 15:1517595. doi: 10.3389/fpls.2024.1517595
Received: 26 October 2024; Accepted: 26 December 2024;
Published: 23 January 2025.
Edited by:
Antonio Figueira, University of São Paulo, BrazilReviewed by:
Ramu Vemanna, Regional Centre for Biotechnology (RCB), IndiaSrayan Ghosh, Durham University, United Kingdom
Copyright © 2025 Wang, Cao, Wang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenjing Wang, d2FuZ3dlbmppbmdAY2Fhcy5jbg==
 Wenjing Wang
Wenjing Wang Haihui Cao
Haihui Cao Jiahao Wang1
Jiahao Wang1 Hongbo Zhang
Hongbo Zhang