
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 20 January 2025
Sec. Plant Pathogen Interactions
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1515060
This article is part of the Research TopicUnraveling Pathogen-Plant-Microbiome Interactions in Horticultural Crops Through Omics ApproachesView all 16 articles
Potato (Solanum tuberosum L.), as the world’s fourth largest food crop, plays a crucial role in ensuring food security through its disease resistance. The RPP13 gene family is known to play a pivotal role in plant disease resistance responses; however, its specific functions in potato remain unclear. In this study, we conducted the first comprehensive identification and analysis of 28 RPP13 gene family members in potato, examining their gene structures, chromosomal locations, expression patterns, and functional characteristics. Gene structure analysis revealed that most members contain the typical CC-NBS-LRR domains, with exon numbers ranging from 1 to 6. Phylogenetic analysis grouped these genes into four evolutionary clades, indicating a high level of conservation. Cis-regulatory element analysis identified that the promoter region of StRPP13-26 is enriched with pathogen-responsive elements such as the WUN-motif and MYC, suggesting its potential role in disease defense. Expression pattern analysis showed that StRPP13-8, StRPP13-10, and StRPP13-23 are highly expressed in various tissues, indicating their involvement in basic physiological functions, whereas StRPP13-6 and StRPP13-25 are mainly induced under specific pathogen infection conditions. Transcriptome and qRT-PCR analyses further revealed functional divergence of the RPP13 gene family in response to potato scab disease. Notably, StRPP13-11 was significantly downregulated in both resistant and susceptible cultivars, suggesting its crucial role in the early stages of pathogen recognition. Subcellular localization experiments showed that the StRPP13-11 protein is localized in the chloroplast. Combined with transcriptome-based functional enrichment analysis, this finding implies that StRPP13-11 may participate in disease defense by regulating photosynthesis-related genes and the dynamic balance of reactive oxygen species within the chloroplast. This study provides new insights into the potential functions of the RPP13 gene family in potato disease resistance mechanisms, offering valuable genetic resources and theoretical support for future disease-resistant breeding programs.
Potato (Solanum tuberosum L.) is the fourth most important food crop globally, playing a critical role in ensuring food security due to its high yield, broad adaptability, and rich nutritional content (Ugent, 1970; Zaheer and Akhtar, 2016). As a staple food and significant economic resource for farmers, the potato’s high production potential and nutritional value make it a key crop for addressing food shortages, particularly in the face of rapid population growth and climate change (Devaux et al., 2014, Devaux et al., 2021; Dongyu, 2022).
However, the sustainable expansion of potato cultivation is increasingly threatened by various soil- and air-borne diseases, which significantly compromise yield and tuber quality (Visser et al., 2009; Zhang et al., 2017). To date, approximately 40 soil-borne diseases have been identified as major threats to potato production (Arora and Khurana, 2004). Among these, soil-borne diseases such as common scab (Streptomyces scabies), late blight (Phytophthora infestans), and black scurf (Rhizoctonia solani) are particularly devastating, causing global economic losses estimated in billions of dollars annually (Hill and Lazarovits, 2005; Tscharntke et al., 2012; Fiers et al., 2012; Arshaghi et al., 2023). Enhancing potato resistance to these diseases is therefore a pressing priority for ensuring agricultural sustainability and food security.
Resistance (R) genes are critical components of plant immunity, representing one of the largest and most diverse gene families in plants. These genes play pivotal roles in recognizing pathogen-derived effectors and triggering immune responses (Jin et al., 2024; Qi et al., 2024a). Among them, the RPP13 gene family stands out for its ability to mediate pathogen-specific defense responses. In Arabidopsis thaliana, the RPP13-Nd gene was found to function independently of classical immune pathways like NDR1 and EDS1, highlighting its unique regulatory mechanism (Bittner-Eddy and Beynon, 2001). Furthermore, the high amino acid diversity of RPP13 genes was reported to enhance their adaptability to evolving pathogens (Rose et al., 2004). In wheat, the TaRPP13-3 gene, located on chromosome 7D and encoding a CC-NBS-LRR protein, was shown to confer resistance to powdery mildew, illustrating the potential of this gene family in crop protection (Liu et al., 2020). Zhang et al. (2022) further demonstrated that the truncated TaRPP13L1-3D gene positively regulated wheat resistance to powdery mildew via the RanGAP-WPP complex-mediated nucleocytoplasmic transport pathway. Recent studies also suggested that RPP13-like proteins were involved in abiotic stress responses, such as heat stress tolerance mediated by adenylyl cyclase activity in maize (Yang et al., 2021). These findings underscored the broad-spectrum functionality of RPP13 genes, making them attractive targets for improving disease resistance in crops.
Despite these advances, the functional roles of the RPP13 gene family in potatoes remain poorly understood. The lack of knowledge about their expression patterns, regulatory mechanisms, and specific contributions to disease resistance limits the ability to exploit these genes for breeding purposes. Addressing this gap is essential for leveraging RPP13 genes in developing disease-resistant potato varieties (Ortega and Lopez-Vizcon, 2012; Paluchowska et al., 2022).
In this study, we conducted the first comprehensive identification and characterization of the RPP13 gene family in potatoes. Specifically, we aimed to identify and annotate RPP13 gene family members in the potato genome, investigate their gene structures, chromosomal distributions, and expression patterns, and validate the functional roles of key RPP13 genes through subcellular localization experiments and transcriptomic analysis. These insights would pave the way for future molecular breeding programs, facilitating the development of potato varieties with enhanced disease resistance.
Using BLASTP analysis of the potato genome, 28 members of the RPP13 gene family were identified and designated as StRPP13-1 through StRPP13-28. These genes were distributed across 12 chromosomes (Figure 1), with chromosomes 1, 2, 4, and 8 each harboring four RPP13 genes (e.g., StRPP13-1 to StRPP13-4 on chromosome 1). The remaining genes were distributed across other chromosomes, such as chromosomes 9 and 12, which also contained multiple RPP13 genes (e.g., StRPP13-18 to StRPP13-20 and StRPP13-25 to StRPP13-28). This distribution pattern suggested that the RPP13 gene family exhibited a widespread and evolutionarily conserved organization within the potato genome.
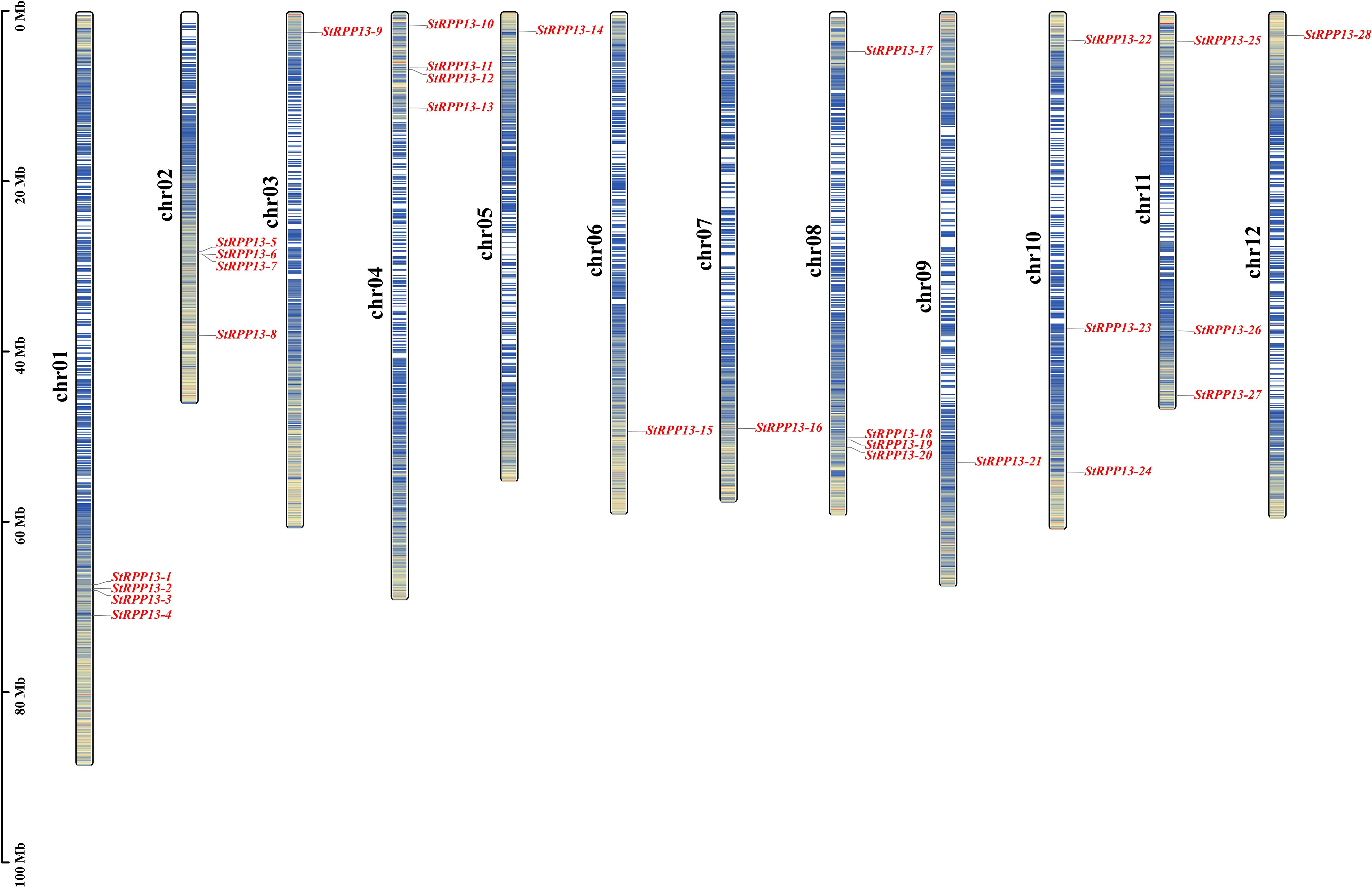
Figure 1. Chromosomal localization of the potato RPP13 genes. The chromosome numbers were indicated on the left, with the scale on the left representing genetic distance in megabases (Mb). A genetic interval of 100 kb was used to precisely map the distribution of the RPP13 genes. The color gradient from red to blue reflected gene density across the chromosomes, with darker regions indicating higher gene density and lighter regions representing lower gene density. Unmarked regions on the chromosomes denote areas for which no gene distribution data were available.
The physicochemical properties of the RPP13 proteins revealed amino acid lengths ranging from 781 to 1714, with molecular weights varying between 89,918.09 Da and 195,237.26 Da. The theoretical isoelectric points (pI) ranged from 5.42 to 9.00, indicating a broad spectrum of acid-base properties. Most RPP13 proteins had instability indices below 50, suggesting that they were relatively stable within the cellular environment. Moreover, the GRAVY (Grand Average of Hydropathicity) values for all RPP13 family proteins were negative, reflecting their hydrophilic nature. Subcellular localization predictions indicated that all RPP13 proteins were localized in the cytoplasm (Supplementary Table S1).
A phylogenetic analysis of the RPP13 gene family members from potato and Arabidopsis thaliana classified the 28 RPP13 genes into four evolutionary clades (Figure 2). Group I included genes such as StRPP13-1, StRPP13-4, and StRPP13-15. Group II contained StRPP13-18, StRPP13-9, along with two Arabidopsis homologs (AT3G50950.1 and AT3G14470.1), indicating strong evolutionary relationships. The members of Group III and Group IV demonstrated divergent evolutionary trajectories, with Group III genes such as StRPP13-3 and StRPP13-5 exhibiting unique conservation patterns. This classification revealed the evolutionary diversity of the RPP13 gene family in potato, providing a foundation for future functional studies.
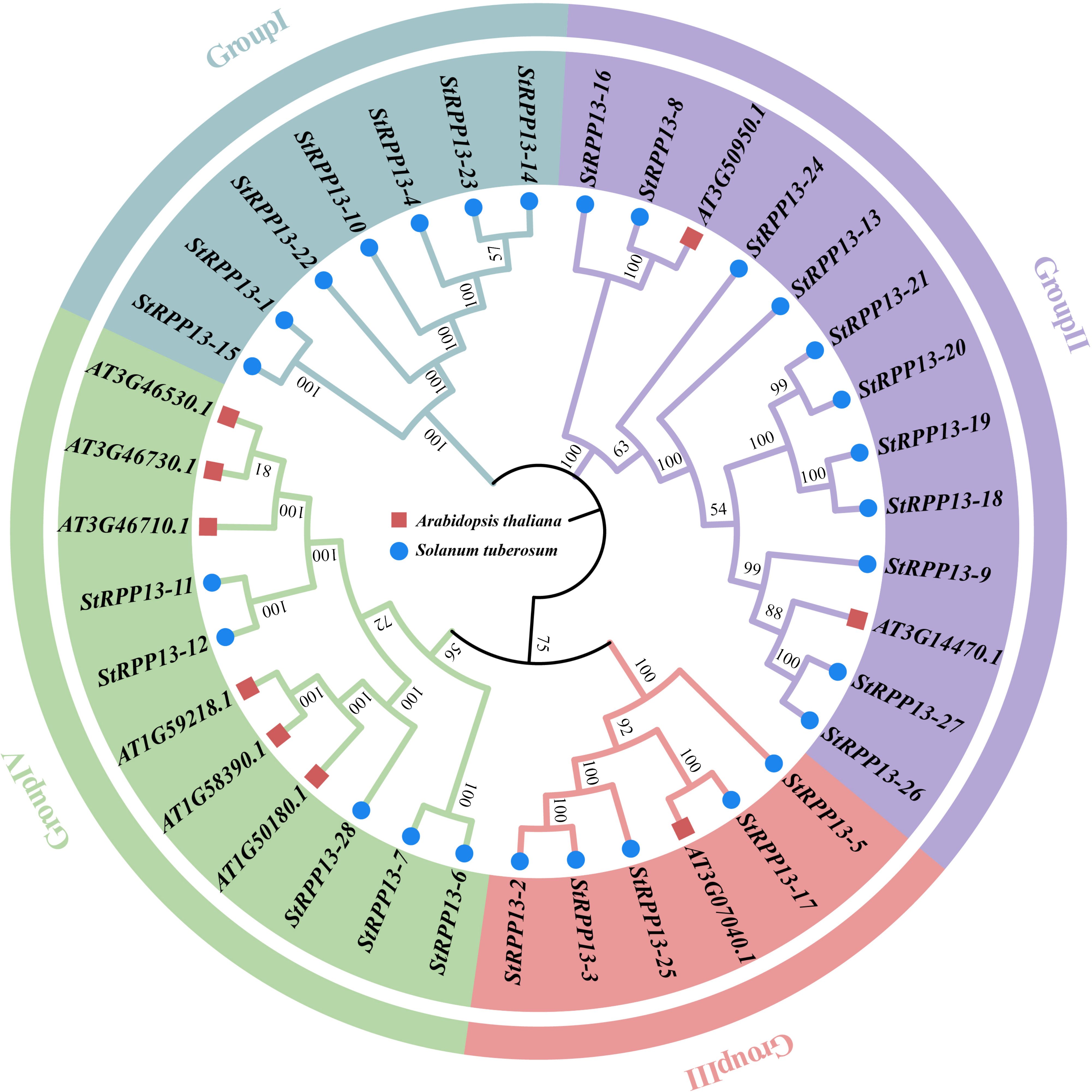
Figure 2. Phylogenetic analysis of potato and Arabidopsis thaliana RPP13 gene family members. The phylogenetic tree divides the 28 potato RPP13 genes and their Arabidopsis homologs into four groups. Potato RPP13 genes were indicated by blue circles, while Arabidopsis thaliana homologs were marked with red squares.
Gene structure analysis revealed that the number of exons in the RPP13 genes ranged from 1 to 6. Most genes, such as StRPP13-1 and StRPP13-5, contained 2 to 3 exons, while StRPP13-26 exhibited a more complex gene structure with 6 exons, suggesting that this gene might possess intricate transcriptional and functional regulation mechanisms. Additionally, StRPP13-9 was the longest gene in the family; despite having only 3 exons, it featured significantly extended introns, indicating that it might have unique regulatory characteristics (Figure 3A).

Figure 3. Gene structure, conserved motifs, and functional domains of potato RPP13 genes. (A) The gene structures of RPP13 family members, where green boxes represent untranslated regions (UTRs) at the 5’ and 3’ ends, yellow boxes represent coding sequences (CDS), and black lines represent introns. The numbers (0, 1, 2) indicate the intron phases. (B) Conserved motif patterns identified in the RPP13 gene family. Different motifs are indicated by colored boxes, with each motif numbered and aligned according to their position within the gene. (C) Functional domain distribution of RPP13 genes, where different colors correspond to distinct functional domains. The relative lengths of gene structures, motifs, and domains can be estimated using the scale bar at the bottom.
In the conserved motif analysis, 20 distinct conserved motifs were identified in the RPP13 proteins (Figure 3B). Motifs 1 through 4 were widely distributed across all RPP13 proteins, underscoring their evolutionary conservation. Motifs 16, 19, and 20 were predominantly present in Group III, suggesting their possible involvement in specific disease resistance functions. Functional domain analysis further confirmed the functional conservation of the RPP13 family proteins. All proteins were found to contain disease resistance-related domains such as NB-ARC, RX-CC, and LRR, with StRPP13-25 and StRPP13-26 in Group III also containing additional domains, including PLN00113 and the COG3903 superfamily domains, indicating that these proteins might play specialized roles in pathogen defense (Figure 3C).
Collinearity analysis revealed the conservation and evolutionary relationships of the RPP13 gene family both within and across species. Intraspecific collinearity analysis showed significant collinearity relationships of the RPP13 genes on chromosomes 1, 2, 3, 8, 9, and 12. Particularly notable were the collinearity observed between chromosomes 2 and 3, and between chromosomes 8 and 9, suggesting the occurrence of tandem duplications or segmental duplications, which might have contributed to the expansion of this gene family. However, in the central chromosomal regions, collinearity between StRPP13-19 and StRPP13-20 was the only observed segment, indicating that these genes might have been subjected to specific selective pressures during evolution (Figure 4A).
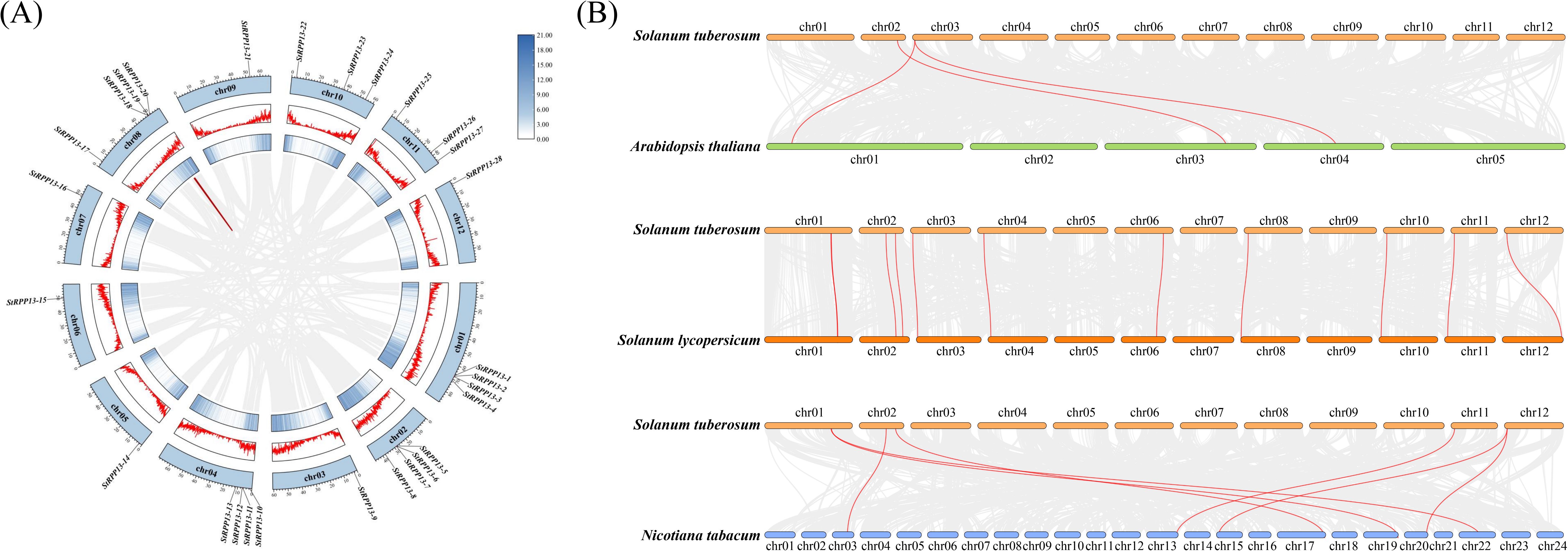
Figure 4. Collinearity analysis of the potato RPP13 genes within and across species. (A) The intraspecific collinearity of the RPP13 gene family within the potato genome. Collinear blocks across the entire genome were depicted with a grey background, while red curves connect pairs of duplicated RPP13 genes, highlighting regions of segmental duplications. (B) Interspecific collinearity analysis between potato and other species (Arabidopsis thaliana, Solanum lycopersicum, and Nicotiana tabacum). Gray lines in the background represent collinear gene pairs between species, while red lines specifically connect collinear RPP13 genes.
Interspecific collinearity analysis revealed a degree of collinearity between the potato RPP13 gene family and those in other Solanaceae species, such as tomato and tobacco, particularly on chromosomes 1, 2, and 12. This suggested that these gene families might have originated from a common ancestor and undergone conserved evolution within Solanaceae species. Specifically, synteny analysis with tomato suggested that these genes were highly aligned with chloroplast-related disease defense pathways, further supporting their conserved functional characteristics. In contrast, collinearity with Arabidopsis thaliana was limited, with only a few collinear segments observed on chromosomes 2 and 3. The collinearity with tobacco was more complex, indicating that multiple chromosomal rearrangement events might have occurred during evolution (Figure 4B).
Cis-acting element analysis of the promoter regions (2000 bp upstream) of 28 potato RPP13 genes revealed the presence of multiple elements associated with environmental stress responses, plant growth and development, hormone responses, and light responses (Figure 5). Among these, stress-responsive elements such as MYC, STRE, and ARE were particularly prominent, with the highest abundance observed in StRPP13-5, StRPP13-20, and StRPP13-26. Additionally, StRPP13-26 contained the largest number of WUN-motif, suggesting its involvement in wound response and pathogen defense mechanisms.
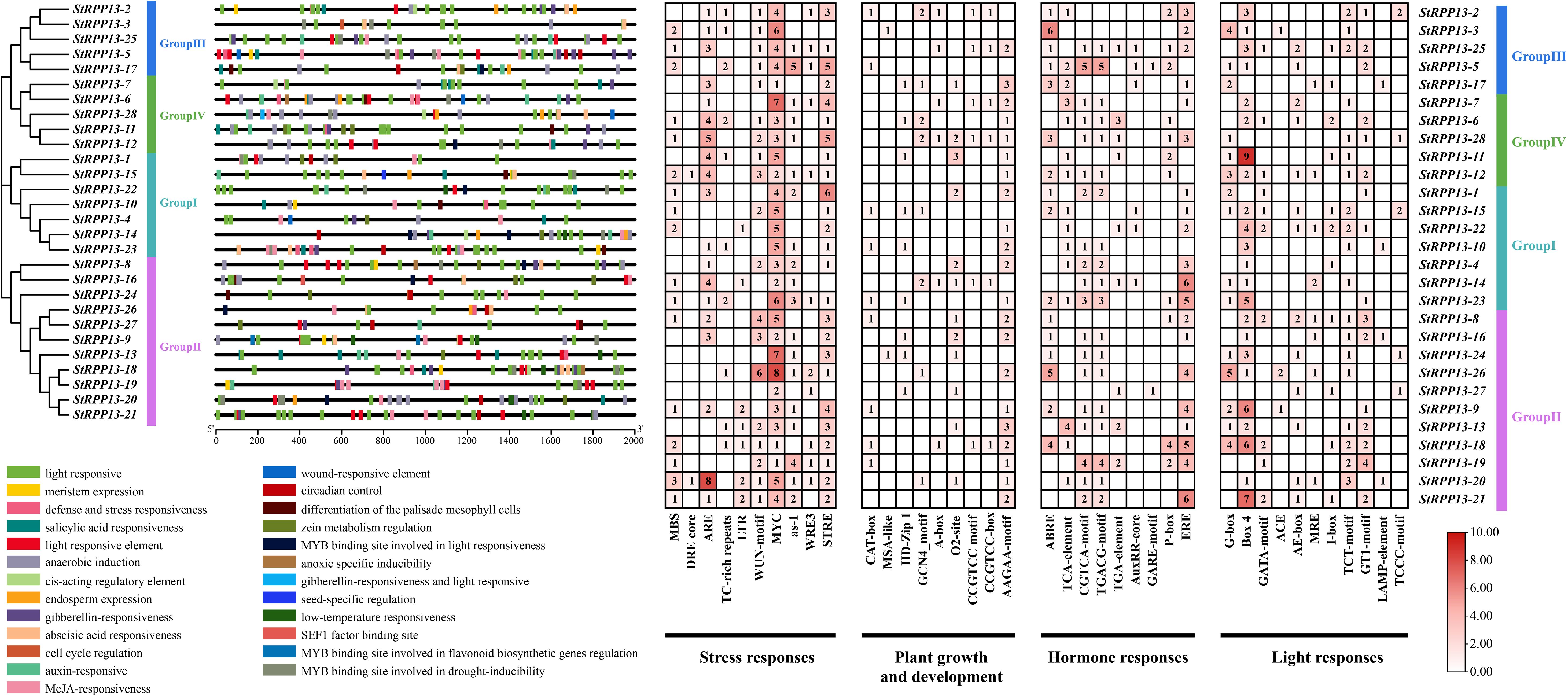
Figure 5. Cis-acting element analysis of RPP13 gene promoters in potato. The diagram on the left illustrated the distribution of various cis-acting elements across the 2000 bp upstream regions of the 28 RPP13 genes. Each colored box represented different functional elements, including light-responsive (green), meristem expression (light brown), defense and stress responsiveness (red), and hormone-responsive elements (orange), among others. On the right, heatmaps showed the abundance of stress response, plant growth and development, hormone response, and light response elements. Numbers within the boxes indicated the number of occurrences of each element in the respective RPP13 gene promoter. The color gradient from light pink to dark red represents the frequency of each element, with darker colors indicating higher abundance.
Plant growth and development-related elements, such as AAGAA-motif, O2-site, and GCN4-motif, were widely distributed among the genes, particularly in StRPP13-22 and StRPP13-15 in Group I and Group IV, indicating that these genes might play crucial roles in cell division and tissue differentiation.
Hormone response elements, such as ERE and ABRE, were highly enriched in StRPP13-14, StRPP13-21, and StRPP13-3, suggesting their involvement in hormone-regulated processes. Light response elements, including Box 4, G-box, and GT1-motif, were significantly increased in StRPP13-18, StRPP13-20, and StRPP13-21, indicating that these genes might function in light-regulated growth processes.
The cis-acting elements identified in the promoters of the RPP13 gene family highlighted their critical roles in potato’s disease resistance, environmental stress response, hormone regulation, and light-responsive processes. Furthermore, the analysis suggested that these genes not only participated in disease defense mechanisms but might also influence the plant’s light response by regulating photosynthesis-related pathways and chloroplast metabolism, providing a more comprehensive view of their function in both defense and broader physiological processes.
Using the gene expression tools from the EnsemblPlants database, the expression patterns of 28 RPP13 genes were analyzed across 14 different tissues, revealing significant variation in expression levels across tissues (Supplementary Figure 1). StRPP13-8, StRPP13-10, and StRPP13-23 exhibited high expression levels in all tissues, suggesting their involvement in basic physiological processes across various potato tissues. In contrast, StRPP13-6, StRPP13-7, and StRPP13-25 were nearly undetectable in most tissues, while StRPP13-18, StRPP13-19, StRPP13-26, and StRPP13-27 displayed low expression across all tissues, indicating their potential roles under specific stress conditions.
Further analysis revealed that StRPP13-2 and StRPP13-24 showed significant expression in the root, tuber sprout, and whole in vitro plant, implying that these genes might be crucial in root development and tuber formation. StRPP13-4 was exclusively expressed in the young tuber, while StRPP13-15 and StRPP13-28 were highly expressed only in the petiole. These findings suggested that the RPP13 gene family displayed clear tissue specificity during various developmental stages and in different tissues of the potato, highlighting their potential roles in regulating diverse biological functions.
Based on previous transcriptomic data, the expression changes of RPP13 genes in resistant cultivar Chunshu 10 and susceptible cultivar Chunshu 11 were analyzed after inoculation with the pathogen Streptomyces scabiei at 0 and 10 days post-inoculation (Figure 6A). Results showed significant differences in the expression of certain RPP13 genes between the resistant and susceptible cultivars. For example, the expression of StRPP13-22 decreased significantly in the resistant cultivar 10 days post-inoculation, while no noticeable change was observed in the susceptible cultivar.
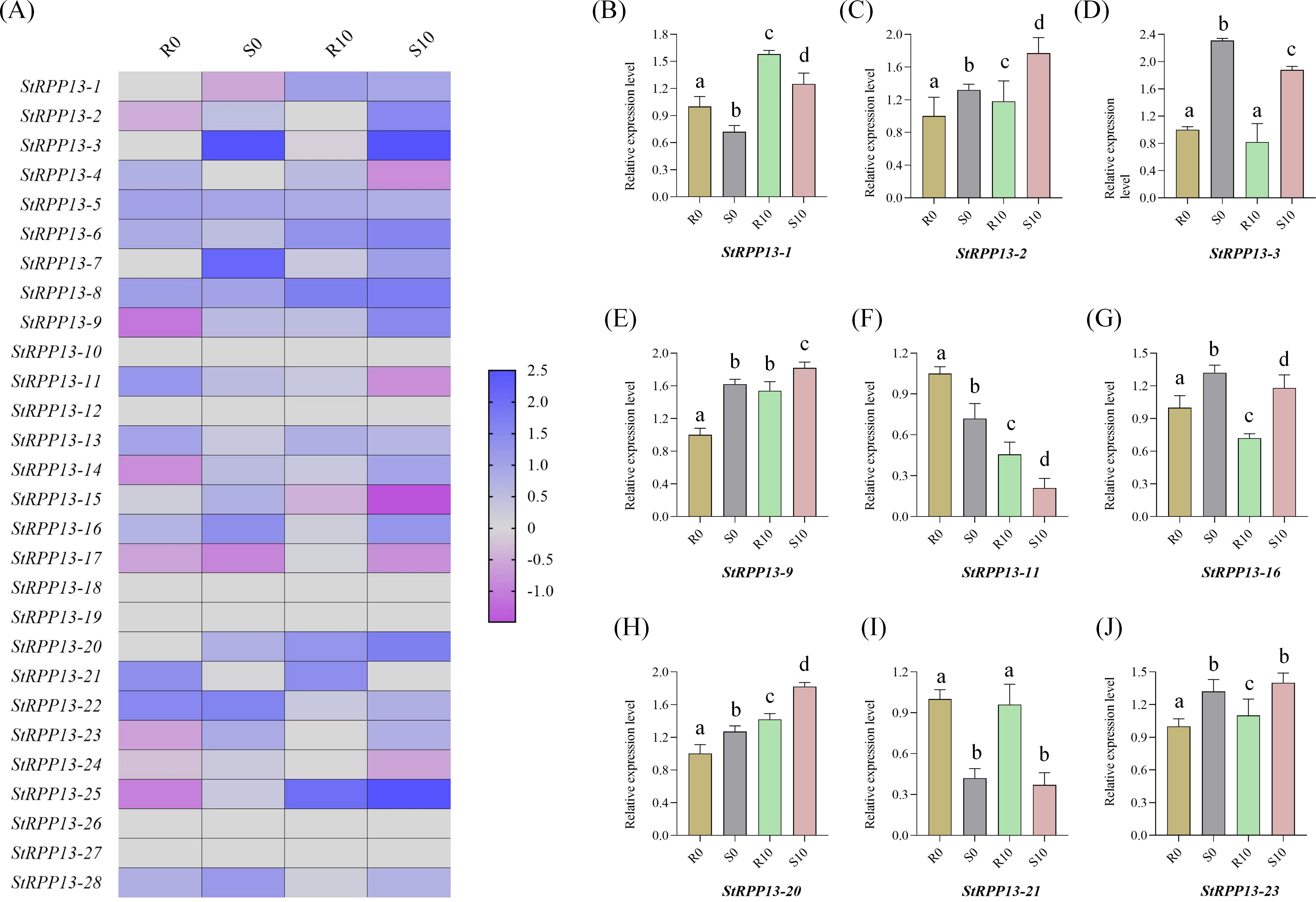
Figure 6. Expression pattern and qRT-PCR validation of the potato RPP13 gene family in resistant (Chunshu 10) and susceptible (Chunshu 11) cultivars after Streptomyces scabies infection. (A) Heatmap showing the expression levels of 28 RPP13 genes at 0 days (R0, S0) and 10 days (R10, S10) post-inoculation in resistant (R) and susceptible (S) cultivars. Expression values were represented as log2-transformed relative fold changes, with blue indicating higher expression and pink indicating lower expression. (B–J) qRT-PCR validation of selected RPP13 genes in resistant (R0, R10) and susceptible (S0, S10) cultivars at 0 and 10 days post-inoculation. Data were presented as the mean ± standard error of three biological replicates. The expression levels were normalized against a housekeeping gene, and the relative fold change in expression was shown for each condition. Different letters indicated statistically significant differences among groups (p < 0.05, Duncan’s multiple range test).
On the other hand, StRPP13-16 exhibited a marked downregulation in the susceptible cultivar post-infection, suggesting its involvement in the pathogen response of the susceptible plant. Notably, StRPP13-11 showed significant downregulation in both resistant and susceptible cultivars following pathogen infection, indicating that this gene might play a critical role in the early response to pathogen attack and could be a key regulator of resistance mechanisms. These results suggested that different members of the RPP13 gene family exhibited diverse response patterns during the potato’s defense against scab disease.
To further validate the expression patterns observed in the transcriptomic data, nine genes were randomly selected from the 28 RPP13 genes for qRT-PCR analysis. The results showed that the expression changes of these genes in resistant and susceptible cultivars after Streptomyces scabiei infection were highly consistent with the transcriptomic data (Figures 6B–J). For instance, StRPP13-2 and StRPP13-3 exhibited significant expression differences between resistant and susceptible cultivars, particularly StRPP13-11 and StRPP13-21, which showed marked downregulation 10 days post-inoculation. These findings further confirmed the differential expression patterns observed in the transcriptomic data, emphasizing their potential roles in the defense against disease.
To further investigate the specific subcellular localization of the key disease resistance gene StRPP13-11, a transient expression system using Arabidopsis protoplasts was employed. The Arabidopsis protoplast system, known for its efficient transient expression, allowed for the rapid evaluation of gene subcellular localization. The subcellular localization of the StRPP13-11 protein was determined by observing the GFP signal from the fusion protein under a laser confocal microscope.
The results showed that the GFP control (35S-GFP) was evenly distributed in the cytoplasm, whereas the GFP signal of the StRPP13-11-GFP fusion protein was mainly concentrated in the chloroplast region (Figure 7), with a high overlap with the red autofluorescence of chlorophyll. This localization result indicated that the StRPP13-11 protein is localized in the chloroplast. Chloroplasts are essential organelles in plant cells, responsible for photosynthesis and serving as hubs for numerous metabolic and signaling pathways. The chloroplast localization of StRPP13-11 suggested that it might exert direct influence on pathogen infection by regulating photosynthesis or chloroplast function as part of its defense mechanisms. This finding provided an important clue for further investigating its specific functions within the chloroplast.
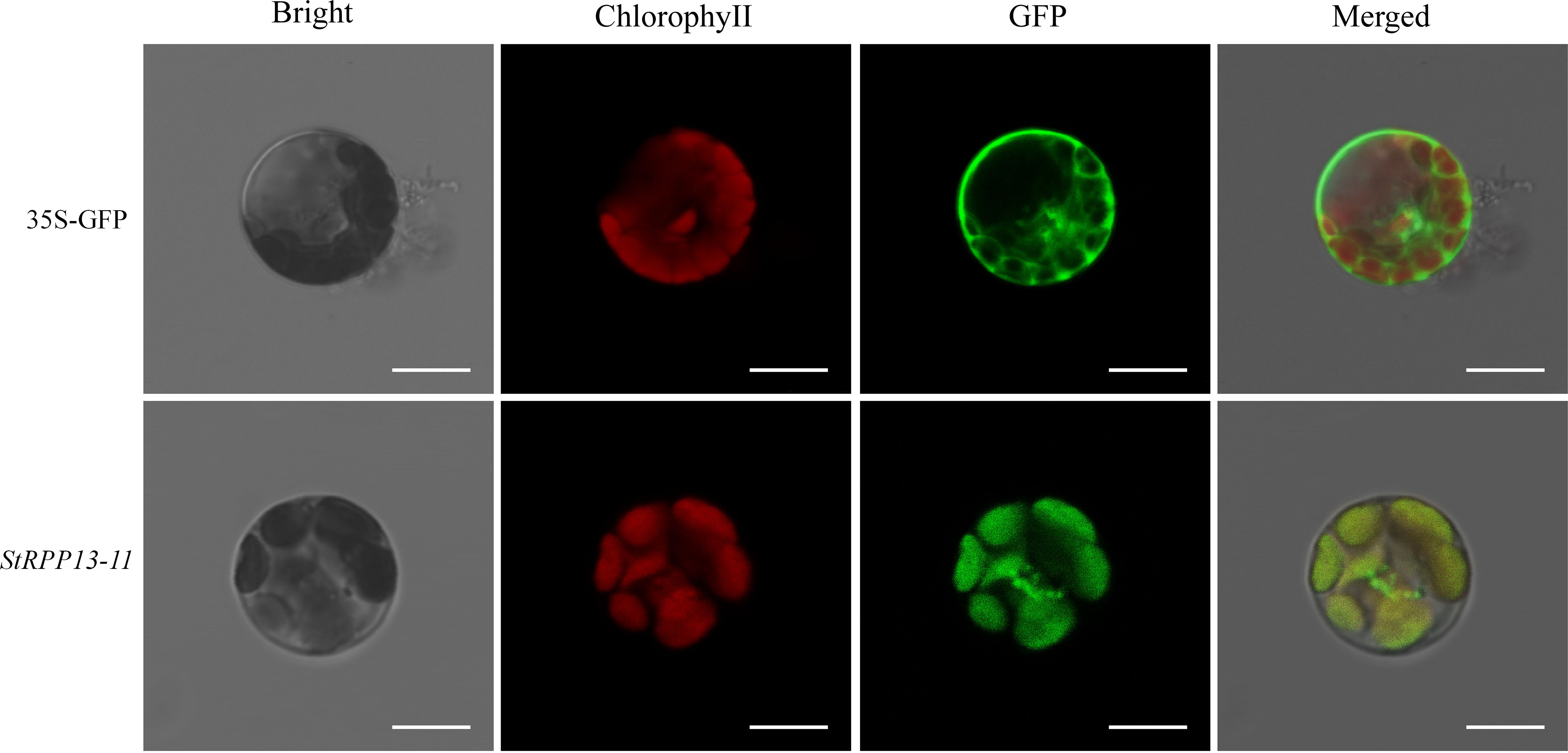
Figure 7. Subcellular localization of StRPP13-11 protein in Arabidopsis protoplasts using GFP fusion proteins. The control GFP (35S-GFP) was uniformly distributed throughout the cytoplasm. StRPP13-11-GFP fusion proteins showed a distinct GFP signal localized to the chloroplasts, co-localizing with the red autofluorescence of chlorophyll. The images in the “Bright” column represent bright field microscopy, the “Chlorophyll” column showed chlorophyll autofluorescence (red), the “GFP” column shows the GFP signal (green), and the “Merged” column presents an overlay of the GFP and chlorophyll signals. Scale bars = 10 µm.
The RPP13 gene family, as a typical class of CC-NBS-LRR resistance genes, plays a crucial role in plant disease resistance (van der Biezen and Jones, 1998). Its functional diversity and conservation are essential for understanding the mechanisms of plant immunity. In this study, we systematically identified 28 members of the RPP13 gene family in potatoes and revealed their evolutionary characteristics through phylogenetic analysis and chromosomal localization. The results indicated that segmental and tandem duplication events were the main driving forces behind the expansion of this gene family, consistent with findings in other species (Cannon et al., 2004; Cheng et al., 2018). Compared to diploid species such as Arabidopsis and tomato, the RPP13 gene family in potatoes exhibited greater diversity, which might be related to its complex tetraploid genome structure and specific environmental selection pressures (Meyers et al., 2003).
Phylogenetic analysis showed that most potato RPP13 genes were conserved within the Solanaceae family, particularly displaying collinearity with homologous genes in tomato and tobacco (Huang et al., 2005). This suggested that the RPP13 gene family might have originated from a common ancestor in these species and retained basic functional characteristics throughout evolution (Marla, 2017). However, some genes, such as StRPP13-14 and StRPP13-22, lacked clear homologs in other species, indicating that these genes might have undergone species-specific expansion in potatoes and acquired novel functional differentiation in disease resistance (Bakker et al., 2006). Further analysis of gene structure and conserved motifs revealed that most members of the RPP13 family possessed the typical CC-NBS-LRR domains, which is consistent with their roles in immune responses (Song et al., 2003). Despite experiencing gene expansion and diversification, the RPP13 gene family has maintained its core functions (Kuang et al., 2005). Nevertheless, certain gene domains have undergone variations, possibly associated with specific functional differentiation and environmental adaptation (van der Vossen et al., 2005). This combination of conservation and diversity provides a multi-layered molecular basis for potatoes to cope with various pathogenic stresses.
This study also provides a preliminary analysis of the functional divergence and disease resistance mechanisms of the RPP13 gene family in potatoes across different tissues and under the infection of the scab pathogen Streptomyces scabies. Cis-acting element analysis revealed that the promoter regions of genes such as StRPP13-5, StRPP13-20, and StRPP13-26 were enriched with pathogen-responsive elements, including MYC, STRE, and ARE, with StRPP13-26 showing the highest abundance of the WUN-motif (Rushton et al., 2010). This suggested that StRPP13-26 might play a crucial role in wound response and pathogen defense, a finding consistent with the pathogen recognition mechanisms of RPP13 genes in Arabidopsis and other species but highlighting a potentially unique pathogen-responsive function in potatoes (Meyers et al., 2003).
Gene expression pattern analysis indicated that StRPP13-8, StRPP13-10, and StRPP13-23 were highly expressed across all tissues, suggesting their involvement in fundamental physiological processes. In contrast, StRPP13-6, StRPP13-7, and StRPP13-25 showed minimal expression in most tissues, implying that these genes might be induced specifically under certain pathogen infection conditions (Sekhwal et al., 2015; Tang et al., 2017). This result differed from the expression characteristics of RPP13 genes in diploid species such as Arabidopsis and rice. In Arabidopsis, RPP13 genes were uniformly expressed across various tissues even under non-stress conditions, indicating their widespread role in immune surveillance (Mohr et al., 2010). In rice, the homologous RPP13 genes were predominantly expressed in leaves and roots, particularly showing strong responses under pathogen infection (Nandety et al., 2013). In contrast, some RPP13 genes in potatoes were activated only under specific pathogen infection conditions, reflecting a unique spatiotemporal regulatory pattern that might be linked to the tetraploid genome structure and the complex pathogen adaptation mechanisms of potatoes.
Transcriptome and qRT-PCR analyses further revealed the functional divergence of the RPP13 gene family in potato resistance to scab disease. For instance, StRPP13-22 showed significant downregulation in resistant cultivars after inoculation, while no substantial changes were observed in susceptible cultivars, suggesting its involvement in negative regulation of disease resistance. Meanwhile, StRPP13-11 exhibited pronounced downregulation in both resistant and susceptible cultivars, indicating its potential role in the early stages of pathogen recognition. The presence of a high density of W-box cis-acting elements in its promoter suggested that StRPP13-11 might interact with WRKY transcription factors to regulate downstream defense genes (Deslandes et al., 2003; Tsuda and Katagiri, 2010; Kuang et al., 2004). These findings suggested that the functional specificity of the potato RPP13 gene family might stem from its unique evolutionary background and regulatory mechanisms.
Additionally, StRPP13-11 was found to be localized in the chloroplast through subcellular localization experiments, suggesting that the chloroplast might play a crucial role in the disease resistance function of this gene family. Chloroplasts are not only the primary sites for photosynthesis and the production and scavenging of reactive oxygen species (ROS), but they also play a vital role in plant defense against pathogen infections (Song et al., 2021). Transcriptome analysis further supported this hypothesis, as photosynthesis-related pathways (photosynthesis pathway and chloroplast thylakoid membrane pathway) were significantly enriched in both GO and KEGG enrichment analyses, which aligned with the chloroplast localization of StRPP13-11. This suggested that the RPP13 proteins localized in the chloroplast might regulate plant defense responses by modulating the expression of photosynthesis-related genes and maintaining the dynamic balance of ROS in the chloroplast, thereby exerting their disease resistance function (De Torres Zabala et al., 2015; Li et al., 2024).
Recent research by Qi et al. (2024b) showed that the chloroplast elongation factors StTuA/B enhanced potato resistance to late blight and significantly increased crop yield by promoting chloroplast protein synthesis and photosynthesis efficiency. This finding highlighted the central role of chloroplasts in balancing growth and immunity and emphasized the importance of chloroplast pathways in potato disease defense, which was consistent with the chloroplast localization of StRPP13-11 observed in our study. It suggested that the RPP13 proteins in the chloroplast might interact with WRKY transcription factors to regulate the expression of downstream defense genes, thus playing a role in pathogen recognition and resistance response.
The comprehensive identification and functional characterization of the RPP13 gene family in potatoes revealed their critical roles in disease resistance, particularly against scab disease caused by Streptomyces scabies. Among the 28 identified genes, StRPP13-11 emerged as a key candidate, localized in the chloroplast and potentially involved in regulating photosynthesis and ROS homeostasis. This study not only established a robust theoretical foundation for breeding potato varieties with enhanced disease resistance but also highlighted the potential of RPP13 genes in breeding programs aimed at developing disease-resistant potato varieties. Future research should focus on validating the roles of key genes, such as StRPP13-11, and exploring their practical applications in crop improvement strategies.
This study presented the first comprehensive identification and characterization of the RPP13 gene family in potatoes, revealing 28 members with conserved CC-NBS-LRR domains and grouping them into four evolutionary clades through phylogenetic analysis. Functional characterization highlighted their roles in potato disease resistance, particularly against scab disease. Notably, StRPP13-11 was identified as a key gene, localized in the chloroplast and potentially involved in regulating photosynthesis and ROS homeostasis, underscoring its importance in early pathogen recognition. These findings offered a robust theoretical foundation and valuable genetic resources for future molecular breeding programs to enhance potato resistance to scab disease and other pathogens.
The potato genome data and its gff annotation file (DM 1-3 516 R44) were downloaded from the Spud DB online database (http://spuddb.uga.edu/). The amino acid sequences of Arabidopsis thaliana RPP13 genes were obtained from the TAIR database (https://www.arabidopsis.org/) and used as references for BLASTP analysis. The specific locus names of the Arabidopsis RPP13 genes used in this study are AT1G59218.1, AT1G58390.1, AT3G46530.1, AT3G46710.1, AT3G46730.1, AT1G50180.1, AT3G07040.1, AT3G50950.1, and AT3G14470.1. A local BLASTP analysis was conducted using the Diamond software (E-value set to 10^-3). The presence of characteristic domains of the RPP13 proteins, such as RX-CC, NB-ARC, and LRR, was confirmed using the Conserved Domain Database (CDD) of the National Center for Biotechnology Information (NCBI).
The physicochemical properties of the potato RPP13 proteins, including amino acid sequence length, theoretical isoelectric point (pI), molecular weight (Mw), instability index, aliphatic index, and grand average of hydropathicity (GRAVY) index, were calculated using the ProtParam tool (https://web.expasy.org/protparam/). Additionally, the subcellular localization of the RPP13 proteins was predicted using the CELLO online tool (http://cello.life.nctu.edu.tw/). Finally, the chromosomal locations of the RPP13 genes were extracted from the genome annotation file and visualized using the Gene Location Visualize from GTF/GFF module of the Tbtools software (v2.097).
The amino acid sequences of RPP13 proteins from potato and Arabidopsis thaliana were aligned using the Clustal algorithm. A phylogenetic tree was constructed using the Neighbor-Joining (NJ) method in MEGA11 software with 1000 bootstrap replicates to assess branch confidence. The constructed phylogenetic tree was visualized and enhanced using the iTOL tool (https://itol.embl.de/). Bootstrap values are displayed at the corresponding nodes to indicate the confidence level of each branch.
The conserved motifs of the RPP13 genes were analyzed using the MEME suite (http://meme-suite.org/) with the maximum number of motifs set to 20, the minimum width to 6, and the maximum width to 50. Gene structure analysis was performed by extracting the coding sequences (CDS) and untranslated regions (UTR) of the potato RPP13 genes, and the results were visualized using TBtools v2.101.
The BLASTP program was employed to identify homologous RPP13 genes in potato (E-value threshold < e-5). The collinearity relationships among potato RPP13 genes were analyzed using the MCScanX program, and the collinear blocks and duplicated gene pairs were visualized using TBtools.
Additionally, genome sequences and annotation files of Arabidopsis thaliana, tobacco, and tomato were downloaded from the Phytozome v13 (https://phytozome-next.jgi.doe.gov/), NCBI (https://www.ncbi.nlm.nih.gov/), and Sol Genomics Network (https://solgenomics.net/organism/solanum_lycopersicum/genome) websites, respectively. Collinearity between potato and Arabidopsis thaliana, tobacco, and tomato was analyzed using MCScanX, and the results were visualized with TBtools.
The 2000 bp sequences upstream of the start codon of each potato RPP13 gene were extracted and considered as the promoter regions. The promoter sequences were extracted using TBtools and analyzed using the PlantCARE database (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to predict cis-regulatory elements. The cis-regulatory elements associated with stress response, plant growth and development, hormone response, and light responsiveness were summarized and visualized using the Basic Biosequence View and HeatMap modules of TBtools.
The expression patterns of 28 RPP13 genes in 14 different tissues, including stolon, young tuber, flower, leaf, shoot apex, petiole, stem, mature tuber, root, tuber peel, tuber cortex, tuber pith, tuber sprout, and whole in vitro plant, were analyzed using the gene expression tool from the EnsemblPlants database (https://plants.ensembl.org/Solanum_tuberosum/Info/Index), and the results were visualized to show the expression differences across tissues.
Based on previous transcriptome sequencing data from our team, the expression changes of RPP13 genes were analyzed in the resistant cultivar Chunshu 10 and the susceptible cultivar Chunshu 11 after inoculation with scab pathogen Streptomyces scabies at 0 days and 10 days post-inoculation (De Torres Zabala et al., 2015). The two cultivars were grown in pots, and a spore suspension of 2×10^7 spores/mL was applied during the sprouting phase (Wanner, 2007). Samples were collected at 0 days (pre-inoculation) and 10 days post-inoculation, rapidly frozen in liquid nitrogen, and stored at -80°C for further analysis.
To further validate the expression patterns of RPP13 genes under scab infection, nine genes from the 28 RPP13 family members were randomly selected for qRT-PCR analysis. Primer design was performed using Primer 3 (https://primer3.ut.ee/), and all primer sequences used in this study are provided in Supplementary Table S2. Total RNA was extracted from the samples using the TaKaRa MiniBEST Plant RNA Extraction Kit, and cDNA was synthesized using PrimeScript™ RT Master Mix (TaKaRa) (Ye et al., 2018). qRT-PCR was performed on the Applied Biosystems QuantStudio 6 system, with StActin97 used as the reference gene. Each gene’s expression level was determined using three biological replicates, and relative expression levels were calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001).
The subcellular localization of the StRPP13-11 gene was analyzed using the Arabidopsis protoplast transient expression system. The StRPP13-11 gene was cloned into the 16318-hGFP vector, and seamless cloning was used to transfer the gene into Escherichia coli DH5α for cloning and plasmid extraction. The extracted plasmid was then introduced into Arabidopsis protoplasts via PEG-mediated transformation, followed by washing and purification with W5 solution. The transformed protoplasts were incubated overnight (Kobayashi et al., 2006).
After transformation, the subcellular localization of the StRPP13-11 protein was observed using a laser confocal microscope at excitation wavelengths of 405 nm and 488 nm to detect GFP fluorescence (Zhu et al., 2023).
Statistical analyses were performed using Microsoft Excel 2013 and SPSS 19.0. Gene expression data from qRT-PCR experiments and transcriptomic analyses were included in the analysis. Group comparisons were carried out using Duncan’s multiple range tests to evaluate differences among experimental groups. To ensure the validity of ANOVA results, assumptions of normality and homogeneity of variances were verified using the Shapiro-Wilk test and Levene’s test, respectively. Only data that met these assumptions were subjected. For repeated experiments, the mean values were calculated from three biological replicates, and data variability was expressed as the standard deviation (SD). Significant differences among groups were indicated with different letters. A p-value of less than 0.05 was considered statistically significant. Data visualization, including graphical representations of gene expression levels, was conducted using GraphPad Prism 9.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found here: https://www.ncbi.nlm.nih.gov/, accession number PRJNA1057135.
BY: Conceptualization, Formal analysis, Methodology, Validation, Writing – original draft. CL: Conceptualization, Formal analysis, Methodology, Validation, Writing – review & editing. QW: Data curation, Investigation, Methodology, Validation, Writing – review & editing. QY: Formal analysis, Software, Validation, Writing – review & editing. XG: Formal analysis, Software, Validation, Writing – review & editing. YZ: Formal analysis, Software, Validation, Writing – review & editing. ZW: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Jilin Provincial Agricultural Science and Technology Innovation Project (CXGC2023RCB008, CXGC2024ZD009) and the Jilin Provincial Science and Technology Department Key R&D Project (20220202005NC).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1515060/full#supplementary-material
Arora, R. K., Khurana, S. M. P. (2004). “Major fungal and bacterial diseases of potato and their management,” in Fruit and vegetable diseases. Ed. Mukerji, K. G. (Springer Netherlands, Dordrecht), 189–231.
Arshaghi, A., Ashourian, M., Ghabeli, L. (2023). Potato diseases detection and classification using deep learning methods. Multimed. Tools Appl. 82, 5725–5742. doi: 10.1007/s11042-022-13390-1
Bakker, E. G., Toomajian, C., Kreitman, M., Bergelson, J. (2006). A genome-wide survey of R gene polymorphisms in Arabidopsis. Plant Cell 18, 1803–1818. doi: 10.1105/tpc.106.042614
Bittner-Eddy, P. D., Beynon, J. L. (2001). The Arabidopsis downy mildew resistance gene, RPP13-Nd, functions independently of NDR1 and EDS1 and does not require the accumulation of salicylic acid. Mol. Plant-Microbe Interact. 14, 416–421. doi: 10.1094/MPMI.2001.14.3.416
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4, 10. doi: 10.1186/1471-2229-4-10
Cheng, J., Fan, H., Li, L., Hu, B., Liu, H., Liu, Z. (2018). Genome-wide identification and expression analyses of rpp13-like genes in barley. Biochip J. 12, 102–113. doi: 10.1007/s13206-017-2203-y
Deslandes, L., Olivier, J., Peeters, N., Feng, D. X., Khounlotham, M., Boucher, C., et al. (2003). Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. U.S.A. 100, 8024–8029. doi: 10.1073/pnas.1230660100
De Torres Zabala, M., Littlejohn, G., Jayaraman, S., Studholme, D., Bailey, T., Lawson, T., et al. (2015). Chloroplasts play a central role in plant defence and are targeted by pathogen effectors. Nat. Plants 1, 15074. doi: 10.1038/nplants.2015.74
Devaux, A., Goffart, J. P., Kromann, P., Andrade-Piedra, J., Polar, V., Hareau, G. (2021). The potato of the future: opportunities and challenges in sustainable agri-food systems. Potato Res. 64, 681–720. doi: 10.1007/s11540-021-09501-4
Devaux, A., Kromann, P., Ortiz, O. (2014). Potatoes for sustainable global food security. Potato Res. 57, 185–199. doi: 10.1007/s11540-014-9265-1
Dongyu, Q. (2022). Role and potential of potato in global food security. 11th ed (Dublin: World Potato Congress).
Fiers, M., Edel-Hermann, V., Chatot, C., Le Hingrat, Y., Alabouvette, C., Steinberg, C. (2012). Potato soil-borne diseases. A review. Agron. Sustain. Dev. 32, 93–132. doi: 10.1007/S13593-011-0035-Z
Hill, J., Lazarovits, G. (2005). A mail survey of growers to estimate potato common scab prevalence and economic loss in Canada. Can. J. Plant Pathol. 27, 46–52. doi: 10.1080/07060660509507192
Huang, S., van der Vossen, E. A., Kuang, H., Vleeshouwers, V. G., Zhang, N., Borm, T. J., et al. (2005). Comparative genomics enabled the isolation of the R3a late blight resistance gene in potato. Plant J. 42, 251–261. doi: 10.1111/j.1365-313X.2005.02365.x
Jin, X., Wang, Z., Ai, Q., Li, X., Yang, J., Zhang, N., et al. (2024). DNA-binding with one finger (Dof) transcription factor gene family study reveals differential stress-responsive transcription factors in contrasting drought tolerance potato species. Int. J. Mol. Sci. 25 (6), 3488. doi: 10.3390/ijms25063488
Kobayashi, M., Kawakita, K., Maeshima, M., Doke, N., Yoshioka, H. (2006). Subcellular localization of Strboh proteins and NADPH-dependent O2–generating activity in potato tuber tissues. J. Exp. Bot. 57, 1373–1379. doi: 10.1093/jxb/erj113
Kuang, H., Wei, F., Marano, M. R., Wirtz, U., Wang, X., Liu, J., et al. (2005). The R1 resistance gene cluster contains three groups of independently evolving, type I R1 homologues and shows substantial structural variation among haplotypes of Solanum demissum. Plant J. 44, 37–51. doi: 10.1111/j.1365-313X.2005.02506.x
Kuang, H., Woo, S. S., Meyers, B. C., Nevo, E., Michelmore, R. W. (2004). Multiple genetic processes result in heterogeneous rates of evolution within the major cluster disease resistance genes in lettuce. Plant Cell 16, 2870–2894. doi: 10.1105/tpc.104.025502
Li, C., Yuan, B., Zhang, C., Yao, Q., He, H., Wang, Q., et al. (2024). Revealing key genes and pathways in potato scab disease resistance through transcriptome analysis. Agronomy 14, 291. doi: 10.3390/agronomy14020291
Liu, X., Zhang, C., Zhang, L., Huang, J., Dang, C., Xie, C., et al. (2020). TaRPP13-3, a CC-NBS-LRR-like gene located on chr 7D, promotes disease resistance to wheat powdery mildew in Brock. J. Phytopathol. 168, 688–699. doi: 10.1111/jph.12949
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Marla, S. S. (2017). “Structural analysis of resistance (R) genes in potato (Solanum species) genome,” in The potato genome. Eds. Kumar Chakrabarti, S., Xie, C., Kumar Tiwari, J. (Springer, Cham), 269–281.
Meyers, B. C., Kozik, A., Griego, A., Kuang, H., Michelmore, R. W. (2003). Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15, 809–834. doi: 10.1105/tpc.009308
Mohr, T. J., Mammarella, N. D., Hoff, T., Woffenden, B. J., Jelesko, J. G., McDowell, J. M. (2010). ). The Arabidopsis downy mildew resistance gene RPP8 is induced by pathogens and salicylic acid and is regulated by W box cis elements. Mol. Plant-Microbe Interact. 23, 1303–1315. doi: 10.1094/MPMI-01-10-0022
Nandety, R. S., Caplan, J. L., Cavanaugh, K., Perroud, B., Wroblewski, T., Michelmore, R. W., et al. (2013). The role of TIR-NBS and TIR-X proteins in plant basal defense responses. Plant Physiol. 162, 1459–1472. doi: 10.1104/pp.113.219162
Ortega, F., Lopez-Vizcon, C. (2012). Application of molecular marker-assisted selection (MAS) for disease resistance in a practical potato breeding programme. Potato Res. 55, 1–13. doi: 10.1007/s11540-011-9202-5
Paluchowska, P., Śliwka, J., Yin, Z. (2022). Late blight resistance genes in potato breeding. Planta 255, 127. doi: 10.1007/s00425-022-03910-6
Qi, T., He, F., Zhang, X., Wang, J., Zhang, Z., Jiang, H., et al. (2024b). Genome-wide identification and expression profiling of potato (Solanum tuberosum L.) universal stress proteins reveal essential roles in mechanical damage and deoxynivalenol stress. Int. J. Mol. Sci. 25 (2), 1341. doi: 10.3390/ijms25021341
Qi, Y., Wu, J., Yang, Z., Li, H., Liu, L., Wang, H., et al. (2024a). Chloroplast elongation factors break the growth–immunity trade-off by simultaneously promoting yield and defence. Nat. Plants 10, 1576–1591. doi: 10.1038/s41477-024-01793-x
Rose, L. E., Bittner-Eddy, P. D., Langley, C. H., Holub, E. B., Michelmore, R. W., Beynon, J. L. (2004). The maintenance of extreme amino acid diversity at the disease resistance gene, RPP13, in Arabidopsis thaliana. Genetics 166, 1517–1527. doi: 10.1534/genetics.166.3.1517
Rushton, P. J., Somssich, I. E., Ringler, P., Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Sekhwal, M. K., Li, P., Lam, I., Wang, X., Cloutier, S., You, F. M. (2015). Disease resistance gene analogs (RGAs) in plants. Int. J. Mol. Sci. 16, 19248–19290. doi: 10.3390/ijms160819248
Song, J., Bradeen, J. M., Naess, S. K., Raasch, J. A., Wielgus, S. M., Haberlach, G. T., et al. (2003). Gene RB cloned from Solanum bulbocastanum confers broad spectrum resistance to potato late blight. Proc. Natl. Acad. Sci. U.S.A. 100, 9128–9133. doi: 10.1073/pnas.1533501100
Song, Y., Feng, L., Alyafei, M. A. M., Jaleel, A., Ren, M. (2021). Function of chloroplasts in plant stress responses. Int. J. Mol. Sci. 22, 13464. doi: 10.3390/ijms222413464
Tang, D., Wang, G., Zhou, J. M. (2017). Receptor kinases in plant-pathogen interactions: more than pattern recognition. Plant Cell 29, 618–637. doi: 10.1105/tpc.16.00891
Tscharntke, T., Clough, Y., Wanger, T. C., Jackson, L., Motzke, I., Perfecto, I., et al. (2012). Global food security, biodiversity conservation and the future of agricultural intensification. Biol. Conserv. 151, 53–59. doi: 10.1016/j.biocon.2012.01.068
Tsuda, K., Katagiri, F. (2010). Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 13, 459–465. doi: 10.1016/j.pbi.2010.04.006
Ugent, D. (1970). The Potato: What is the botanical origin of this important crop plant, and how did it first become domesticated? Science 170, 1161–1166. doi: 10.1126/science.170.3963.1161
van der Biezen, E. A., Jones, J. D. (1998). The NB-ARC domain: a novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr. Biol. 8, R226–R228. doi: 10.1016/S0960-9822(98)70145-9
van der Vossen, E. A., Gros, J., Sikkema, A., Muskens, M., Wouters, D., Wolters, P., et al. (2005). The Rpi-blb2 gene from Solanum bulbocastanum is an Mi-1 gene homolog conferring broad-spectrum late blight resistance in potato. Plant J. 44, 208–222. doi: 10.1111/j.1365-313X.2005.02527.x
Visser, R. G., Bachem, C. W., De Boer, J. M., Bryan, G. J., Chakrabati, S. K., Feingold, S., et al. (2009). Sequencing the potato genome: outline and first results to come from the elucidation of the sequence of the world’s third most important food crop. Am. J. Potato Res. 86, 417–429. doi: 10.1007/s12230-009-9097-8
Wanner, L. A. (2007). A new strain of Streptomyces causing common scab in potato. Plant Dis. 91, 352–359. doi: 10.1094/PDIS-91-4-0352
Yang, H., Zhao, Y., Chen, N., Liu, Y., Yang, S., Du, H., et al. (2021). A new adenylyl cyclase, putative disease-resistance RPP13-like protein 3, participates in abscisic acid-mediated resistance to heat stress in maize. J. Exp. Bot. 72, 283–301. doi: 10.1093/jxb/eraa431
Ye, M., Peng, Z., Tang, D., Yang, Z., Li, D., Xu, Y., et al. (2018). Generation of self-compatible diploid potato by knockout of S-RNase. Nat. Plants 4, 651–654. doi: 10.1038/s41477-018-0218-6
Zaheer, K., Akhtar, M. H. (2016). Potato production, usage, and nutrition—a review. Crit. Rev. Food Sci. Nutr. 56, 711–721. doi: 10.1080/10408398.2012.724479
Zhang, H., Fen, X., Yu, W., Hu, H. H., Dai, X. F. (2017). Progress of potato staple food research and industry development in China. J. Integr. Agric. 16, 2924–2932. doi: 10.1016/S2095-3119(17)61736-2
Zhang, X., Wang, G., Qu, X., Wang, M., Guo, H., Zhang, L., et al. (2022). A truncated CC-NB-ARC gene TaRPP13L1-3D positively regulates powdery mildew resistance in wheat via the RanGAP-WPP complex-mediated nucleocytoplasmic shuttle. Planta 255, 60. doi: 10.1007/s00425-022-03843-0
Keywords: potato, RPP13 gene family, disease resistance, gene expression, subcellular localization
Citation: Yuan B, Li C, Wang Q, Yao Q, Guo X, Zhang Y and Wang Z (2025) Identification and functional characterization of the RPP13 gene family in potato (Solanum tuberosum L.) for disease resistance. Front. Plant Sci. 15:1515060. doi: 10.3389/fpls.2024.1515060
Received: 18 November 2024; Accepted: 27 December 2024;
Published: 20 January 2025.
Edited by:
Abhay K. Pandey, North Bengal Regional R & D Center, IndiaReviewed by:
Qibin Wu, Chinese Academy of Tropical Agricultural Sciences, ChinaCopyright © 2025 Yuan, Li, Wang, Yao, Guo, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhongwei Wang, anpzd3p3QGNqYWFzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.