- 1Environment Friendly Crop Germplasm Innovation and Genetic Improvement Key Laboratory of Sichuan Province, Key Laboratory of Wheat Biology and Genetic Improvement in Southwestern China, Crop Research Institute, Sichuan Academy of Agricultural Sciences, Chengdu, Sichuan, China
- 2Department of Plant Pathology, Washington State University, Pullman, WA, United States
- 3US Department of Agriculture, Agricultural Research Service, Wheat Health, Genetics, and Quality Research Unit, Pullman, WA, United States
- 4Triticeae Research Institute, Sichuan Agricultural University, Chengdu, Sichuan, China
- 5State Key Laboratory of Crop Gene Exploitation and Utilization in Southwest China, Sichuan Agricultural University, Chengdu, Sichuan, China
Stripe rust of wheat is a serious disease caused by Puccinia striiformis f. sp. tritici (Pst). Growing resistant cultivars is the most preferred approach to control the disease. To identify wheat genotypes with quantitative trait loci (QTL) for durable resistance to stripe rust, 465 winter wheat entries that were presumed to have high-temperature adult-plant (HTAP) resistance were used in this study. In the greenhouse seedling tests with seven Pst races, 16 entries were resistant to all the tested races. The 465 entries were also phenotyped for stripe rust responses at the adult-plant stage under natural infection of Pst in multiple field locations from 2018 to 2021 in the Washington state, and 345 entries were found to have stable resistance. The contrast of the susceptibility in the greenhouse seedling tests and the resistance in the field adult-plant stage for most of the entries indicated predominantly HTAP resistance in this panel. The durability of the resistance was demonstrated by a subset of 175 entries that were tested in multiple locations from 2007 to 2021. The 465 entries were genotyped through genotyping by multiplexed sequencing of single-nucleotide polymorphism (SNP) markers. Combining the stripe rust response and SNP marker data, a genome-wide association study (GWAS) was conducted, resulting in 143 marker–trait associations, from which 28 QTL that were detected at least with two races or in two field environments were identified, including seven for all-stage resistance and 21 for HTAP resistance. These QTL each explained 6.0% to 40.0% of the phenotypic variation. Compared with previously reported Yr genes and QTL based on their genomic positions, five QTL including two for HTAP resistance were identified as new. A total of 10 user-friendly Kompetitive allele specific PCR (KASP) markers were developed for eight of the HTAP resistance loci. In addition, molecular markers were used to detect 13 previously reported HTAP resistance genes/QTL, including two also identified in the GWAS analyses, and their frequencies ranged from 0.86% to 88.17% in the panel. The durable resistant genotypes, the genes/QTL identified, and the KASP markers developed in this study should be useful to develop wheat cultivars with long-lasting resistance to stripe rust.
1 Introduction
Stripe rust, also called yellow rust (Yr), caused by Puccinia striiformis f. sp. tritici (Pst), is an important disease of wheat worldwide (Stubbs, 1985; Chen, 2005, 2020). In the United States, wheat stripe rust caused damages mostly in the states west of the Rocky Mountains before 2000 and has become a serious problem throughout the inland US since 2000 (Line, 2002; Chen, 2005, 2007; Chen and Kang, 2017). The disease can cause huge damage in terms of yield losses and/or cost of fungicide application—for example, yield losses of 10%–30% were reported in Washington and/or Idaho, Montana, and Oregon in 1959–1961, 1974, 1976, and 1980. The worst stripe rust epidemic in the western United States was in 1981, which severely damaged over 70% of the wheat acreages in these Pacific Northwest states and led to the first widespread use of chemicals to reduce yield losses but still caused more than one million tons of yield loss, including 10% yield loss in the Washington state alone (Line, 2002; Chen, 2020). Although the yield losses in these years were very high, they counted for only 0.84% (1959) to 2.17% (1976) of the national wheat production, as the disease occurred mainly in the western states. Whenever the Great Plains and the eastern states had severe stripe rust, the national yield losses were much higher. The national potential yield losses reached 3.66% (including 25% in California and 10% in Kansas and Nebraska) in 2003, 4.16% (including 10% in Kansas and Texas) in 2010, 8.7% (including 25% in Oklahoma and 15% in Kansas, South Dakota, and Minnesota) in 2015, and 5.61% (including 18% in Oklahoma and 9% in Kansas) in 2016 or 2,421,121, 2,608,720, 4,509,149, and 3,526,926 metric tons, respectively (Chen, 2020). Although the actual yield losses were much reduced from these estimates, the reductions of the yield losses by fungicide applications cost millions of dollars in each year.
To control stripe rust, the best approach is to develop and plant resistant cultivars, especially cultivars combining effective all-stage resistance (ASR) and high-temperature adult-plant (HTAP) resistance to achieve high-level and durable resistance (Chen, 2013; Liu et al., 2018, 2020). ASR is also known as seedling resistance as it can be detected at the seedling stage but expressed throughout all growth stages. It typically shows high-level resistance when effective and is not affected much by temperatures and rust pressure. As ASR is usually controlled by single genes and qualitatively inherited, it can be easily incorporated into new cultivars. However, ASR is mostly race-specific and vulnerable to new virulent races of the pathogen. In contrast, HTAP resistance starts expressing or increases the level of resistance when the weather becomes warm, and the plants passes the seedling stage. This type of resistance is usually controlled by quantitative trait locus or loci (QTL) and less easy for use in breeding programs than ASR. HTAP resistance is usually partial, and the resistance level can be affected by temperature, growth stage, and rust pressure, and as such, HTAP resistance may not be adequate for the complete protection of the crop (Chen, 2013; Liu et al., 2019a, b; Li et al., 2023, 2024). When cultivars do not have adequate resistance, fungicide application is needed to control a disease (Chen, 2014; Liu et al., 2019a). Therefore, the best strategy is to develop wheat cultivars with pyramided genes for high-level HTAP resistance or combined both ASR and HTAP resistance to increase the level and durability of resistance (Chen, 2013, 2020; Liu et al., 2018, 2019).
Up to now, 87 permanently named Yr genes and hundreds of temporarily named genes or QTL have been reported for stripe rust resistance in wheat (Wang and Chen, 2017; Feng et al., 2023; Zhu et al., 2023; Sharma et al., 2024). Among the 87 named Yr genes, 60 were ASR genes and 27 were adult-plant resistance (APR) or HTAP resistance genes, and 13 were cloned (Wang and Chen, 2017; Long et al., 2024; Sharma et al., 2024; Zhou et al., 2024). Breeders prefer genes already present in high-yielding and adapted wheat cultivars. Unfortunately, with the emergence of new Pst races, many resistance genes widely deployed in wheat cultivars have been overcome. Among the 60 ASR genes, only a few, such as Yr5, Yr15, Yr64, and Yr65, are still effective against the Pst populations in the United States and other countries (Cheng et al., 2014; Qie et al., 2019; Wan and Chen, 2014; Wan et al., 2016; Chen et al., 2021; Wang et al., 2022). Many mapped genes or QTL for APR or HTAP resistance have small and variable effects. Often because their effects are too small or too variable, minor genes or QTL are too difficult to be used in breeding programs. Genes or QTL with strong effects for durable type of resistance like HTAP resistance are more useful in breeding programs. Therefore, the identification of new genes for effective ASR and high-level HTAP resistance is essential to develop wheat cultivars with adequate and durable resistance.
Genome-wide association study (GWAS) has been widely used to identify marker–trait associations (MTAs) for various traits in major crops, including wheat resistance to stripe rust. Several studies on stripe rust resistance in wheat germplasm using the GWAS approach have been reported (Maccaferri et al., 2015; Bulli et al., 2016; Liu et al., 2017a, b, c; Liu et al., 2019c, 2020; Mu et al., 2020; Yao et al., 2020; Khan et al., 2024; Marone et al., 2024). These studies used various panels of germplasms including spring and winter hexaploid wheat (Triticum aestivum), durum wheat (T. turgidum ssp. durum), emmer wheat (T. turgidum ssp. dicoccum), and other tetraploid wheats (T. turgidum ssp. ssp. turanicum, spp. turgidum, ssp. polonicum, ssp. carthlicum, ssp. dicoccum, and ssp. dicoccoides). Many QTL for response to stripe rust have been identified, but only a few were well characterized as for durable resistance. From a winter wheat panel of world wheat germplasm collection, Maccaferri et al. (2015) detected a locus for APR on the short arm of chromosome 6B (QYr.ucw-6B) in several wheat germplasm accessions. This locus was validated through molecular mapping using several bi-parental populations and permanently named as Yr78 (Dong et al., 2017). Yao et al. (2021) detected five novel stripe rust resistance loci using a panel of Chinese wheat landraces. Using a spring wheat panel and a winter wheat panel consisting of cultivars, breeding lines, and genetic stocks from various wheat production regions in the United States, Liu et al. (2020) and Mu et al. (2020) identified 37 and 51 loci for resistance to stripe rust, respectively, including 10 novel loci in each of the studies, and determined different genes for different types of resistance used in different regions. These studies demonstrated that GWAS is a powerful approach to detect genes in many germplasm accessions and to identify new genes for stripe rust resistance.
In the present study, we identified stripe rust resistance loci in a panel of 465 winter wheat entries using the GWAS approach. The nursery of these entries was previously tested for many years to characterize stripe rust resistance, and the data indicated that most of the entries have HTAP resistance. Our objectives of this study were (1) to evaluate the stripe rust resistance of the 465 entries by testing them at the seedling stage with various Pst races and at the adult-plant stage in the fields under natural infections to identify entries with durable resistance under multiple environments, (2) to conduct genome-wide association mapping using genome-wide SNP markers to identify loci associated with stripe rust responses, (3) to identify new resistance loci by comparing the loci identified in the present study with previously reported loci, and (4) to convert SNP markers for some of the HTAP resistance loci into Kompetitive allele-specific PCR (KASP) markers to be used in wheat breeding programs. The wheat genotypes identified to have durable HTAP resistance, the identified resistance QTL, and the developed KASP markers should be useful in breeding programs to develop new wheat cultivars with adequate and durable resistance to stripe rust.
2 Materials and methods
2.1 Plant materials
A panel of winter wheat (T. aestivum) entries was assembled by selecting genotypes (genetic stocks, landraces, cultivars, and breeding lines) presumably with HTAP resistance from our initial screening of wheat germplasm for stripe rust resistance in the greenhouse and fields and started being tested in the fields as a winter high-temperature adult-plant resistant wheat nursery (WHAN) since 2007 (https://striperust.wsu.edu). The majority of the entries were previously tested in the greenhouse at both the seedling stage under the low-temperature profile (diurnal temperature gradually changing from 4°C at 2:00 p.m. to 20°C at 2:00 p.m.) and the adult-plant stage under the high-temperature profile (diurnal temperature gradually changing from 10°C at 2:00 p.m. to 30°C at 2:00 p.m.) (Chen, 2013). The number of entries in the panel was increased over the years and fixed to 465 for testing with recently predominant races of Pst at the seedling stage in the greenhouse and at the adult-plant stage in the fields from 2018 to 2021 to facilitate the GWAS analysis. Spring wheat ‘Avocet S’ (AvS), which is highly susceptible throughout the growth stages, and winter wheat ‘Nugaines’, which is highly susceptible in the seedling stage, were used for the reproduction of urediniospores of selected Pst isolates in the greenhouse. Winter wheat ‘PS 279’, which is highly susceptible throughout the growth stages and does not have any known genes for resistance to stripe rust, was used as a susceptible check in the greenhouse tests and the field tests and as a spreader surrounding the fields to create a uniform and high-level stripe rust pressure. The 18 Yr single-gene lines, which are used to differentiate Pst races (Wan and Chen, 2014; Wan et al., 2016; Wang et al., 2022), were used to confirm the races of the isolates used in the greenhouse tests. Information on the 465 entries is provided in Supplementary Table S1.
2.2 Pst races
Seven Pst races—PSTv-4, PSTv-14, PSTv-18, PSTv-37, PSTv-40, PSTv-51, and PSTv-198—which were predominant in the United States over the past decade except PSTv-51 that has the broadest virulence spectrum (Wan and Chen, 2014; Wan et al., 2016; Wang et al., 2022), were selected to phenotype the 465 entries for stripe rust responses at the seedling stage in the greenhouse. PSTv-37 is the most widely distributed and most frequent race throughout the United States. PSTv-4, PSTv-14, and PSTv-40 are popular races in the US Pacific Northwest. PSTv-198 is also an epidemic race with significant frequencies in some years. PSTv-18 is the oldest race but has occurred in the Pacific Northwest with a significant frequency almost every year. This race is avirulent to all Yr single-gene differentials but virulent to some wheat cultivars/lines like Nugaines and PS 279. The virulence/avirulence formulae of the seven races on the 18 Yr single-gene differentials and the isolates used to represent these races are given in Supplementary Table S2. Compared to the Yr gene symbols used in Wan and Chen (2014), YrTye was replaced by Yr76 and YrTr1 was replaced by Yr85 as these genes were permanently named by Xiang et al. (2016) and Feng et al. (2023), respectively.
2.3 Seedling tests in the greenhouse
The 465 wheat entries were evaluated at the seedling stage for stripe rust responses under controlled greenhouse conditions. The seven races (PSTv-4, PSTv-14, PSTv-18, PSTv-37, PSTv-40, PSTv-51, and PSTv-198) were used separately in the tests. The urediniospores of each race were increased using Nugaines or selected Yr single-gene differentials (Supplementary Table S2) and confirmed for race identity by testing the reproduced urediniospores on the set of 18 Yr single-gene differentials (Wan and Chen, 2014; Wan et al., 2016). Five to seven seeds of each entry and susceptible check AvS were planted in each well of plastic trays filled with a soil mixture and grown under the previously described conditions (Chen et al., 2002). After having been grown for 10 days to the two-leaf stage, the seedlings were uniformly dust-inoculated with a mixture of urediniospores and talc at a 1:20 ratio. After incubating in a dew chamber at 10°C for 24 h in the darkness, the seedlings were transferred to a growth chamber set at a diurnal cycle changing from 4°C at 2:00 a.m. to 20°C at 2:00 p.m. and 8-h dark/16-h light (Chen and Line, 1992). Infection type (IT) was recorded 18–20 days after inoculation based on the 0–9 scale (Line and Qayoum, 1992) when Pst was fully sporulating on the susceptible check.
2.4 Adult-plant tests in the fields
The 465 entries were evaluated from 2018 to 2021 for stripe rust response in the fields at Mount Vernon (48°25′12″ N, 122°19′34″ W) in the northwestern Washington and Palouse Conservation Field Station (PCFS, 46°43′59″ N, 117°10′00″ W) and Spillman Farm (SP, 46°43′47.1972″ N, 117°10′54.2568″ W) near Pullman in the southeastern Washington, referred to as year–location environments 18-21MV, 18-20PCFS, and 19-20SP, respectively. As the nursery was assembled and tested over the years, 175 of the 465 entries were also tested for stripe rust responses in the three locations from 2007 to 2017, referred to as environments 07-17MV, 07-17PCFS, and 08-17SP, respectively. In each environment, each entry of the nursery was planted in a single row of 50 cm long and 30 cm between rows in late October or early November. PS 279 was planted after each 20 rows and around the plots as a susceptible check and stripe rust spreader. The field tests were conducted under rainfed conditions without irrigation. All field tests were under the natural infection of Pst, except the Pullman locations in 2014, 2019, and 2021, where the nursery was inoculated before the boot stage with talc mixture of urediniospores collected from the same field in the previous years and stored in liquid nitrogen. The infection type and disease severity (DS) were recorded when the susceptible check PS 279 was fully infected with DS of 80%–100%. DS was assessed visually as the percentage of infected leaf area (0%–100%). To improve the normality of the DS data, relative DS (rDS) was calculated using the formula: rDS (%) = DS/DSPS279 × 100, where DSPS279 is the average DS value of PS 279 in the same environment. IT was recorded based on the 0–9 scales as described in Line and Qayoum (1992).
2.5 Phenotypic data analyses
For all tests, including the 465 entries tested in the recent nine environments and with the seven Pst races and the 175 entries tested in 32 environments, violin plots were drawn to show the distribution of the IT and rDS data using the ggplots package in R V3.6.2 (Wickham, 2016). The best linear unbiased estimate (BLUE) was calculated for the IT and rDS data separately assuming fixed effects for genotypes using IciMapping V4.0 (Meng et al., 2015). The minimum, maximum, and mean values, standard deviations, and coefficient of variation of stripe rust responses were calculated using EXCEL (Microsoft, Redmond, WA, USA). Pearson’s correlation coefficients (r) of pairwise environments were calculated and graphed using the corrplot package in R V3.6.2. Broad-sense heritability (H2) was estimated using SAS V8.0 (SAS Institute Inc., Cary, NC, USA) and the formula: H2 = σ2G/[σ2G + (σ2E + σ2E×G + σ2e)/n], where σ2G is the variance of genotypes, σ2E is the variance of environments, σ2G×E is the variance of the interaction between genotype and environment, σ2e is the variance of residuals, and n is the number of environments. Genotype, environment, and the genotype×environment interactions were treated as random factors.
2.6 Genotyping
A leaf sample was collected from one single seedling plant for each entry grown in the greenhouse, and genomic DNA was extracted from the sample using the cetyltrimethyl ammonium bromide method (Stewart and Via, 1993). The DNA concentration was determined using a BioTek Synergy 2 Microplate reader (BioTek, Winooski, VT, USA) and diluted to the concentration of 20 ng/μL. The 465 entries were genotyped using genotyping by multiplex sequencing (GMS) on an Ion Proton system (Life Technologies Inc., Carlsbad, CA, USA) based on the protocol developed by Ruff et al. (2020). In addition, one sequence-tagged site (STS) marker, 10 simple nucleotide repeat (SSR) markers, and nine KASP markers, which are closely linked to or diagnostic of 13 previously reported Yr genes or QTL for HTAP resistance, were used to genotype the 465 winter wheat entries. The 13 genes/QTL were Yr16 (Agenbag et al., 2012), Yr17 (Liu et al., 2018), Yr18 (Lagudah et al., 2009), Yr30 (Spielmeyer et al., 2003), Yr36 (Uauy et al., 2005), Yr46 (Forrest et al., 2014), Yr52 (Ren et al., 2012), Yr59 (Zhou et al., 2014b), Yr62 (Lu et al., 2014), Yr78 (Dong et al., 2017), Qyr.wgp-1B.1 (Naruoka et al., 2015), and QYrsk.wgp-3BS and QYrsk.wgp-4BL (Liu et al., 2019b). The markers for race-specific ASR gene Yr17 were included as the gene is linked to the HTAP resistance locus YrM1225 (Liu et al., 2018; Li et al., 2023). These markers, their representing Yr genes or QTL, and the primer sequences are provided in Supplementary Table S3.
Polymerase chain reaction (PCR) amplifications and KASP assays were conducted as described in Liu et al. (2019a, 2020). The PCR products of SSR and STS were detected using an ABI3730 DNA fragment analyzer (Applied Biosystems, Grand Island, NY, USA), and those of the KASP markers were detected using a Light-Cycler 480 real-time PCR system (Roche Applied Science, Indianapolis, IN, USA). The SSR alleles were scored using software GeneMarker v2.2.0 (Soft Genetics, State College, PA, USA).
2.7 Genetic diversity and population structure
After quality control, the polymorphism markers with missing rate ≤50% and minor allele frequency (MAF) ≥0.05 were used for genetic diversity and population structure analyses. Genetic diversity was estimated using Power-Marker V3.25 (Liu and Muse, 2005). Population structure was analyzed using software STRUCTHRE V2.3.4 with the Bayesian clustering algorithm (Pritchard et al., 2000) and using the parameter settings described in Yao et al. (2020). The optimum number (K) of subpopulations was determined using the software STRUCTURE HARVESTER (http://taylor0.biology.ucla.edu/structureHarvester/) (Evanno et al., 2005).
2.8 Linkage disequilibrium
To determine the interval defining a QTL and compare it with the previously reported Yr genes and QTL on the integrated map, the linkage disequilibrium (LD) decay distance was estimated as squared allele frequency correlation coefficient (r2) between intra-chromosomal marker pairs using the software Tassel V3.0 (Bulli et al., 2016). All SNP markers with known chromosomal positions were used to calculate the LD decay distance. To estimate the LD distance, a locally weighted polynomial regression (LOESS)-based curve was fitted on the scatter plot using the intra-chromosomal pairwise r2 values against the genetic distance. The genetic distance at which the LD decay curve intersects with the critical value r2 = 0.1 was used as the threshold to determine the confidence interval of significant QTL (Yao et al., 2020).
2.9 Genome-wide association study
Two association analyses were conducted using the Genome Association and Prediction Integrated Tool (GAPIT) R package version 2.0 (Tang et al., 2016). The first GWAS analysis was performed using the adult-plant stage IT and rDS data of the 465 entries in 10 field environments (including the BLUE values) and the seedling IT data of the greenhouse tests with the seven races. The second GWAS analysis was conducted using the IT and rDS data of the 175 entries in 32 field environments from 2007 to 2017 and the BLUE values across these environments. The genotype data used for the two GWAS analysis were filtrated according to the number of entries with the criteria of missing rate <50% and MAF >0.05. To reduce the false positive associations from the type I error, a mixed linear model (MLM) (Yu et al., 2006) with both kinship (K) and population structure (Q) matrices as covariates were used for the GWAS analyses of both the 465 and 175 entries. Loci that were significant in at least two of the field environments or of the race tests or located within the confidence intervals of LD decay (r2 ≤ 0.1) were considered as one QTL. Manhattan plots were drawn using the CMplot package in R V3.6.2 (https://github.com/YinLiLin/CMplot).
2.10 Comparison of QTL identified in the present study with previously reported Yr genes or QTL
The QTL detected in the present study were compared with previously reported Yr genes and QTL in the integrated map (Maccaferri et al., 2015; Wang et al., 2014). The positions of the polymorphic SNPs identified through GMS in the present study were 100% in agreement with their positions in the integrated map generated with the 90K SNP chip data. Therefore, we were able to compare the positions of our QTL with the previously reported Yr genes and QTL. With the LD decay distance (r2 ≤ 0.10) as the interval distance, we considered our QTL the same as a previously reported gene/QTL if the marker(s) associated with our QTL were mapped to the same interval of the previous one.
2.11 Developing new KASP markers for HTAP resistance QTL identified in the present study
After HTAP resistance QTL were mapped through GWAS, KASP markers were developed for the QTL with relatively large effects following the methods described in Liu et al. (2018). Their SNP markers were converted to KASP markers using the primer sequences of CerealsDB (http://www.cerealsdb.uk.net/cerealgenomics/CerealsDB/KASP_primers_for_iSelect.php). The primers were synthesized by Sigma-Aldrich (St. Louis, MO, USA). The KASP markers were tested on the 465 entries for validation following the method described above.
2.12 Determining the frequencies of resistance gene/QTL
The frequencies of the resistance genes or QTL in the panel of 465 entries were determined using their markers. To reduce false positives, entries that had the resistance allele but were as susceptible as the susceptible check were not considered to have the gene/QTL (Liu et al., 2020; Mu et al., 2020).
3 Results
3.1 Stripe rust resistance and broad-sense heritability
The 465 winter wheat entries were tested at the seedling stage with seven Pst races (PSTv-4, PSTv-14, PSTv-18, PSTv-37, PSTv-40, PSTv-51, and PSTv-198) and at the adult-plant stage in nine field environments (18MV, 18PFS, 19MV, 19PCFS, 19SP, 20MV, 20PFS, 20SP, and 21MV). Most of the entries were susceptible in the greenhouse seedling tests but showed resistance at the adult-plant stage in the field tests (Figure 1). Except for the seedling test with the least virulent race, PSTv-18, the majority of entries (306–375 entries; 65.81%–80.65%) were highly susceptible (IT 8-9) in the seedling tests with different races (Figure 1A, Table 1; Supplementary Table S1). Of the 465 entries, 16 (3.44%) were resistant to all seven races, indicating that they have genes or gene combinations effective against the diverse Pst races (Table 1; Supplementary Table S1). In contrast, most of the 465 entries were resistant at the adult-plant stage in the fields, as 345 entries (74.19%) had BLUE_IT ≤5 and 428 (92.04%) had BLUE_rDS ≤50%. The contrast responses indicated that most of the entries have various levels of HTAP resistance. Combining the BLUE_IT and BLUE_rDS results, 345 entries (74.19%), including the 16 entries with ASR effective against all races tested in the seedling stage, showed stably moderate to high levels of resistance across the nine field environments (Figures 1B, C; Table 1). These entries with effective ASR and/or high level of HTAP resistance could be used in breeding programs.
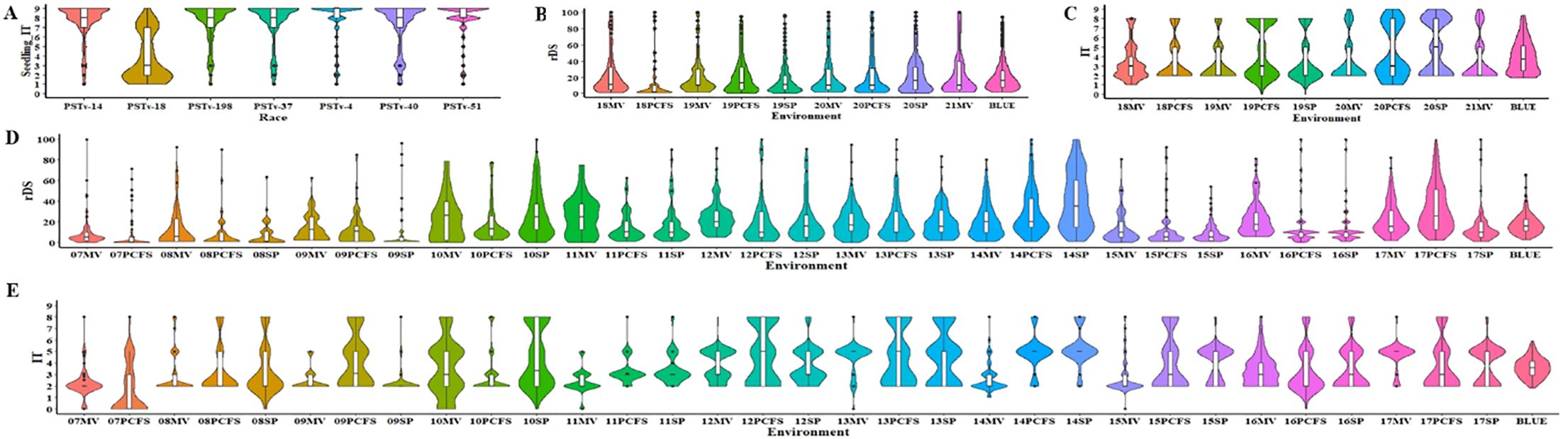
Figure 1. Violin plots showing the stripe rust response distributions in the winter wheat panel. (A) Infection types (IT) of 465 entries of the whole panel tested with seven races of Puccinia striiformis f. sp. tritici in the seedling stage at the low-diurnal-temperature cycle (4°C –20°C) in the greenhouse. (B) Relative disease severity (rDS) values and (C) IT values of the 465 entries in the whole panel at the adult-plant stage tested in different environments (year and location) from 2018 (18) to 2021 (21) at Mount Vernon (MV) in western Washington and Palouse Conservation Farm Station (PCFS) and Spillman Farm (SP) near Pullman in eastern Washington. (D) rDS values and (E) IT values of 175 entries of a sub-panel at the adult-plant stage in various environments from 2007 (07) to 2017 (17). BLUE, best linear unbiased estimator across all field environments.
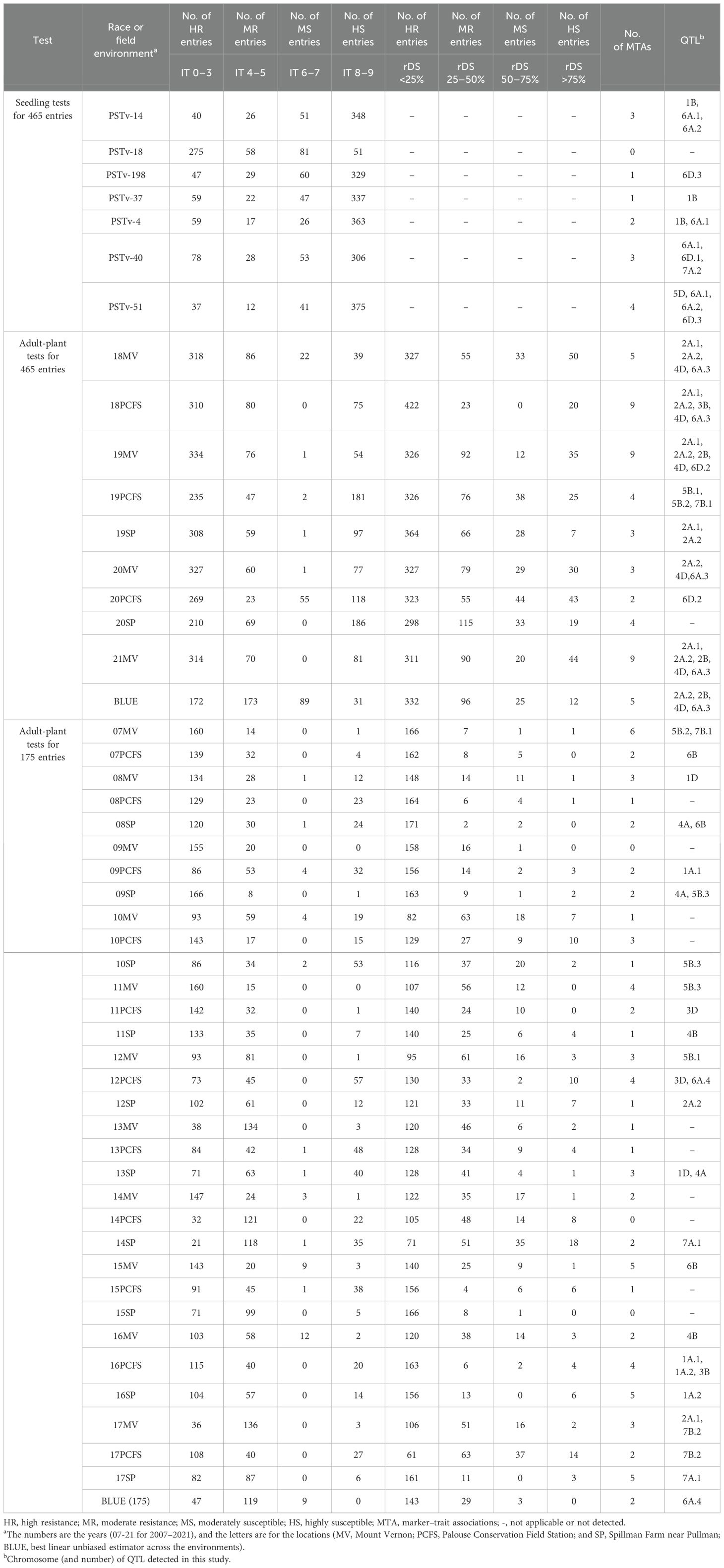
Table 1. Distribution of stripe rust responses and QTL detected from the greenhouse seedling tests and in adult-plant stage from the field environments for the 465 or 175 wheat entries.
For the 175 entries that were also tested from 2007 to 2017, 165 (94.29%) showed moderate to high levels of resistance (BLUE_rDS ≤50%, BLUE_IT ≤5) across the 32 environments (Figures 1D, E, Table 1; Supplementary Table S4). The rDS values under these environments were consistently low, except for some variations in resistance levels in the environments of 10MV, 10SP, 11MV, 14SP, and 17PCFS. These results further demonstrated the durability of the HTAP resistance in most of the entries.
Positive correlations in stripe rust response were observed across all tests (Figure 2; Supplementary Table S5). In the field adult-plant tests, the correlation coefficients ranged from 0.40 to 0.80 based on the rDS data and ranged from 0.28 to 0.78 based on the IT data across the environments. The correlation coefficients between the rDS and IT data across the environments ranged from 0.28 to 0.91 and usually relatively high within the same environments and relatively low between different environments. In the greenhouse seedling tests, the correlation coefficients were from 0.32 to 0.79 across the different race tests. These correlations were at moderate to high levels. However, the correlation coefficients between the greenhouse seedling tests and the field environments were low, ranging from 0.09 to 0.42 between the IT data in the seedling tests and rDS data in the field tests and from 0.02 to 0.43 between the IT data in the seedling tests and IT data in the field tests. These results indicated that the resistance values observed at the seedling stage under the controlled greenhouse conditions (at low temperatures) and at the adult-plant stage under the field conditions (mostly at high temperatures) are mostly controlled by different genes for different types of resistance. Broad-sense heritability (H2) was calculated as 0.89 and 0.92 for the IT and rDS data for the field tests, respectively (Table 2), indicating that the resistance, mostly HTAP resistance, detected at the adult-plant stage across the different environments was consistent and highly inheritable.
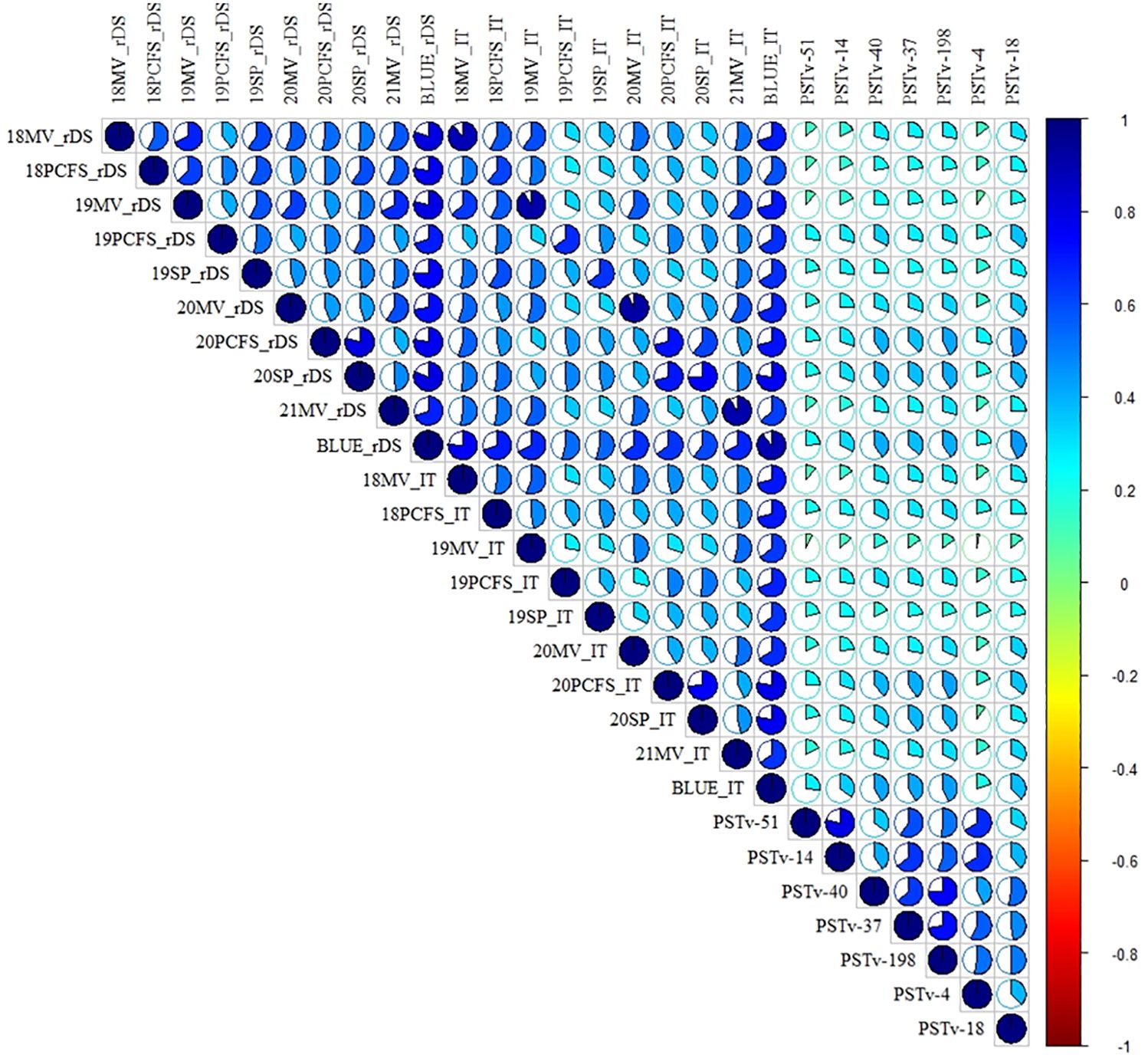
Figure 2. Heatmap of the correlation coefficients of stripe rust responses at the adult-plant and seedling stages of the 465 entries in the whole winter wheat panel. Positive to negative correlations are displayed in blue to red colors. The color intensity and the scale of the ellipse chart are proportional to the correlation coefficients. rDS, relative disease severity; IT, infection type; 18-21, 2018–2021; BLUE, best linear unbiased estimator across all field environments; MV, Mount Vernon; PCFS, Palouse Conservation Field Station; SP, Spillman Farm.
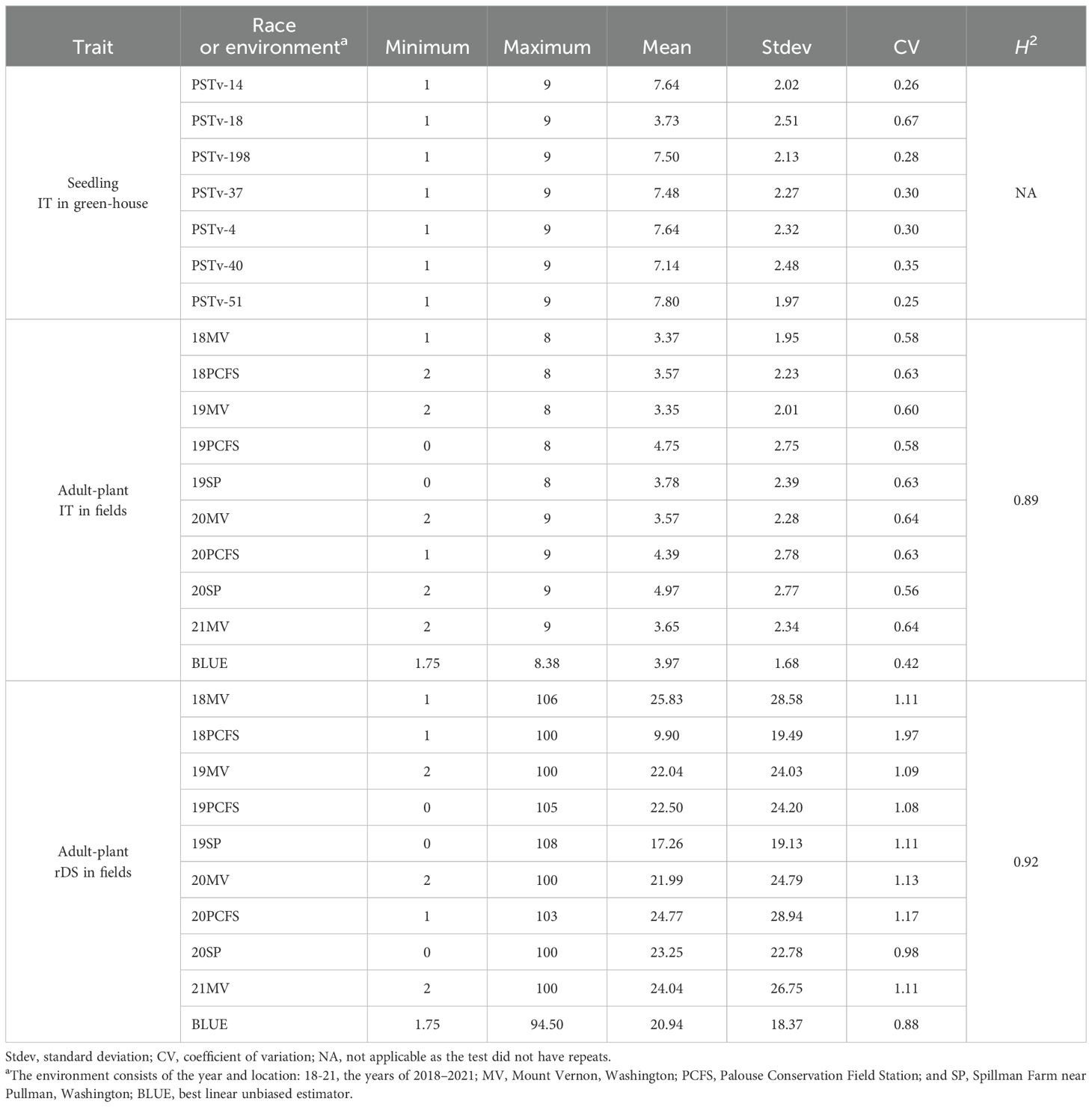
Table 2. Summary of stripe rust infection type (IT) and relative disease severity (rDS, %) data of the winter wheat panel (465 entries) in the tests with seven races of Puccinia striiformis f. sp. tritici at the seedling stage under the controlled greenhouse conditions and at the adult-plant stages under natural infection of the pathogen in various year-location environments and broad-sense heritability (H2) in the field tests.
3.2 Population structure, genetic diversity, and LD
After filtering the SNPs with missing data >50% and MAF <0.05, 1,278 polymorphic SNPs from GMS were obtained and used in the analysis of population structures (Supplementary Table S6). Based on the results from the STRUCTURE HARVESTER analysis (Figure 3A) and STRUCTURE V2.3.4 analysis (Figure 3B), the 465 entries were classified into two sub-populations (sub-1 and sub-2). Sub-1 consisted of 165 entries, while Sub-2 had 300, and these results were consistent with the neighbor-joining phylogenetic tree analysis (Figure 3C).
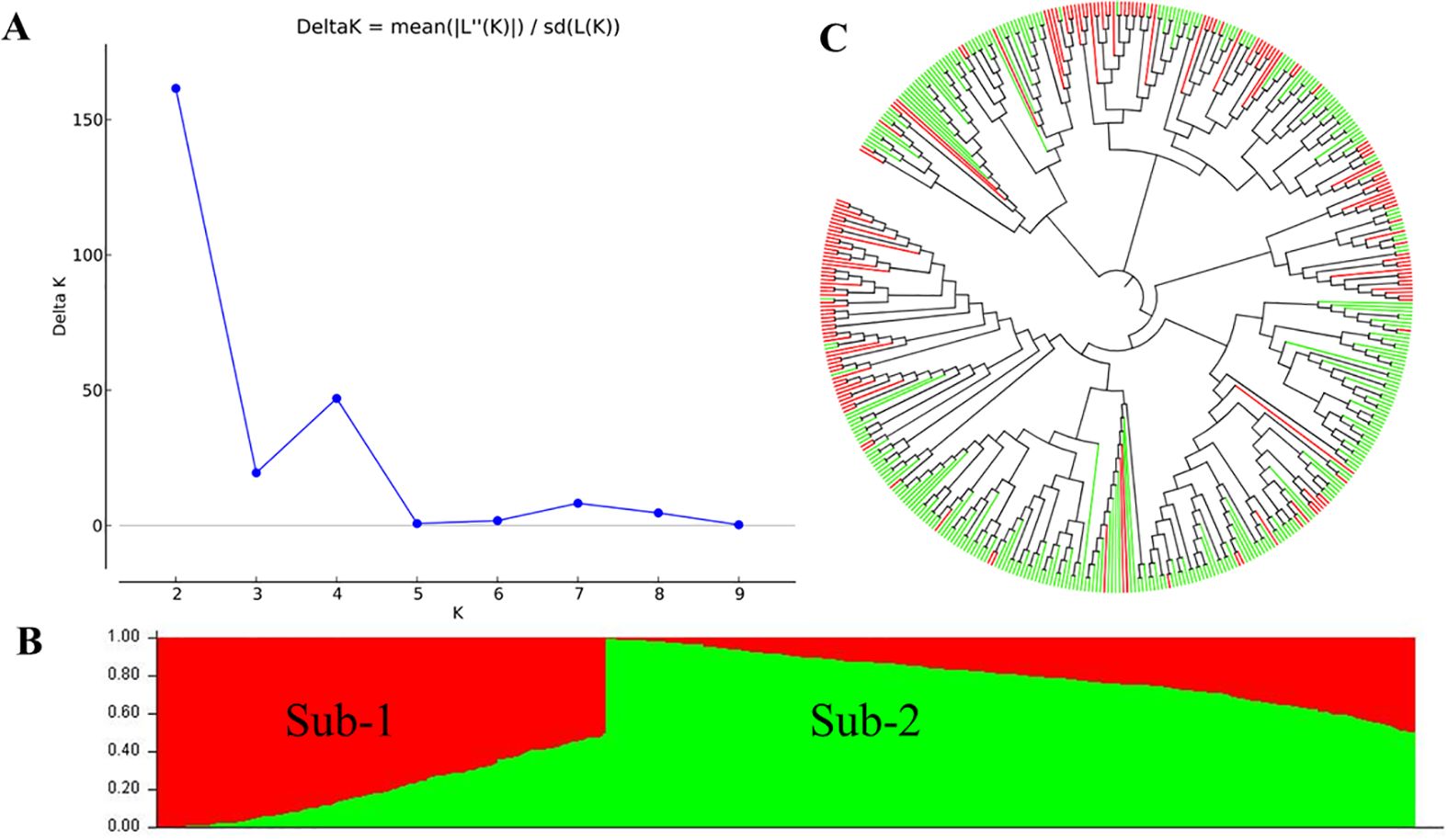
Figure 3. Population structure of the whole winter wheat panel consisting of 465 entries. (A) Estimated ΔK of the structure inferred by STRUCTURE HARVESTER. (B) Two subpopulations inferred by structural analysis using the software STRUCTURE V2.3.4. The red and green parts represent sub-1 and sub-2, respectively. (C) Neighbor-joining phylogenetic tree. The red and green branches represent the sub-1 and sub-2 accessions corresponding to the structural analysis.
Among the 1,278 polymorphic SNPs, 1,088 with known genomic positions were used to estimate the genetic diversity and LD. The panel had a high gene diversity value of 0.32 and polymorphism information content (PIC) value of 0.26 for the three genomes (Table 3). Among the chromosomes, the gene diversity and PIC values were quite similar, ranging from 0.26 on 7D to 0.36 on 5B for gene diversity and from 0.21 on 7D to 0.29 on 5B for PIC. The relatively even coverages of the genomes and chromosomes indicate that the polymorphic SNPs are suitable for GWAS. Regarding the two subpopulations, the gene diversity and PIC values of sub-2 were higher than those of sub-1 across the genomes and individual chromosomes (Figure 4). The difference in genetic diversity may be due to the different numbers of entries and geographic origins. The sub-1 entries were mainly from the US Pacific Northwest, while the entries in sub-2 were from numerous countries including other regions of the United States.
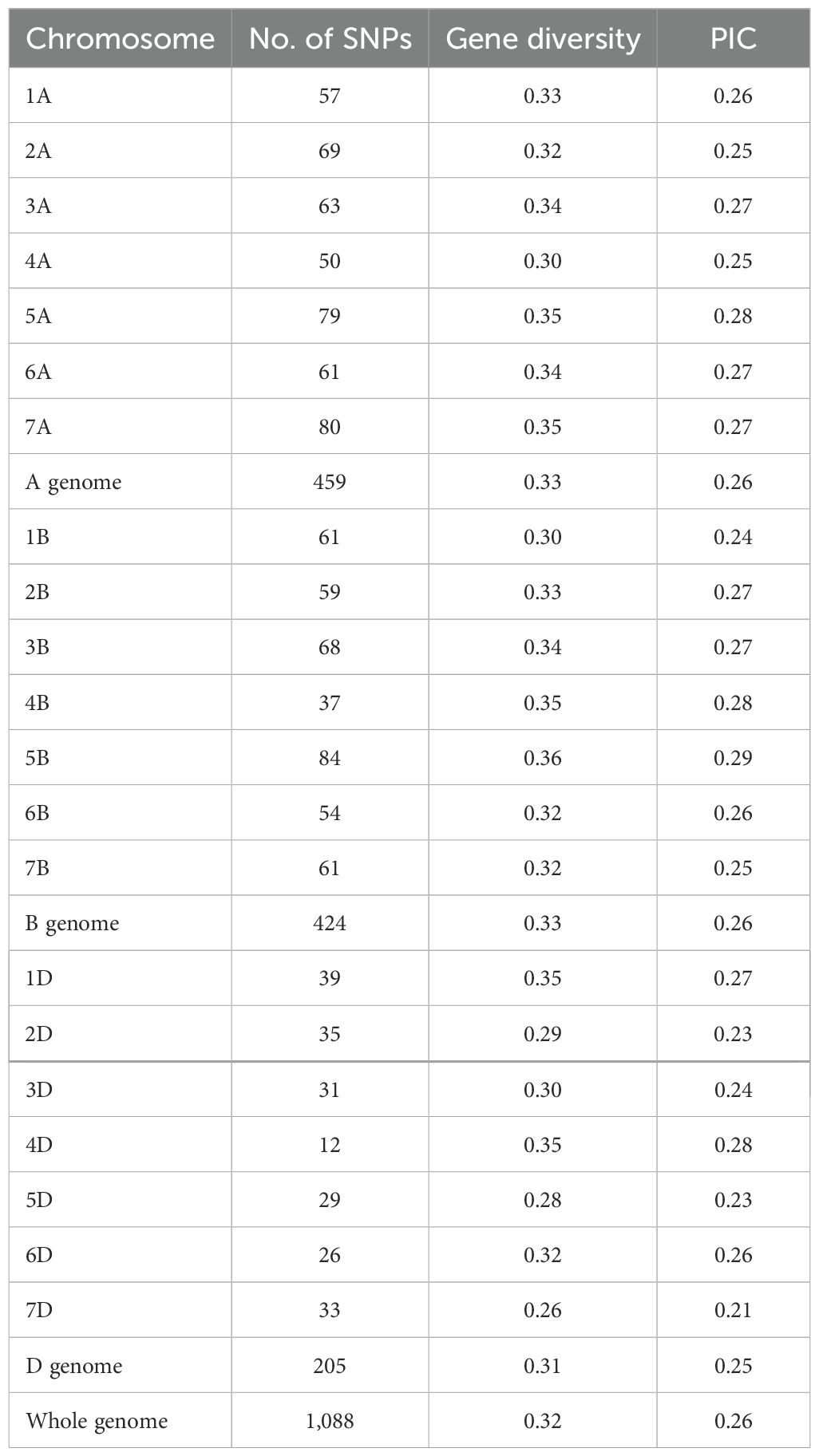
Table 3. Number of polymorphic SNP markers across the chromosomes and genomes identified through genotyping by multiplex sequencing, gene diversity, and polymorphism information content (PIC) of the winter wheat panel.
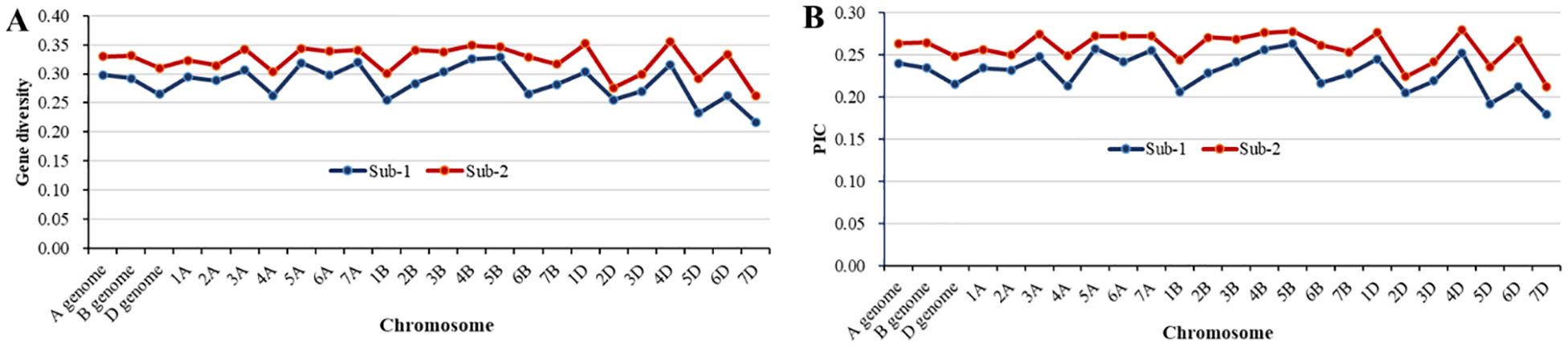
Figure 4. Comparison of the genetic diversity between sub-1 and sub-2. (A) Gene diversity and (B) polymorphism information content (PIC).
The LD analysis was conducted based on the pairwise squared allele frequency correlations (r2) for all intra-chromosomal SNPs. In total, 31,549 pairwise comparisons for the 1,088 SNP markers were used for the LD analysis. A scatter plot and a decay curve of LD r2 against the genetic distance are presented in Figure 5. With the increasing distance between intra-chromosomal markers, the correlation r2 dropped rapidly to 0.10 and then decreased slowly. Thus, 0.10 was used as the critical value for the significance of r2, and the distance of 6.44 cM in the curve that intercepted the critical r2 was determined as the mean LD decay. Therefore, SNPs significantly associated with stripe rust response in the same chromosome within 6.44 cM were considered in the same QTL.
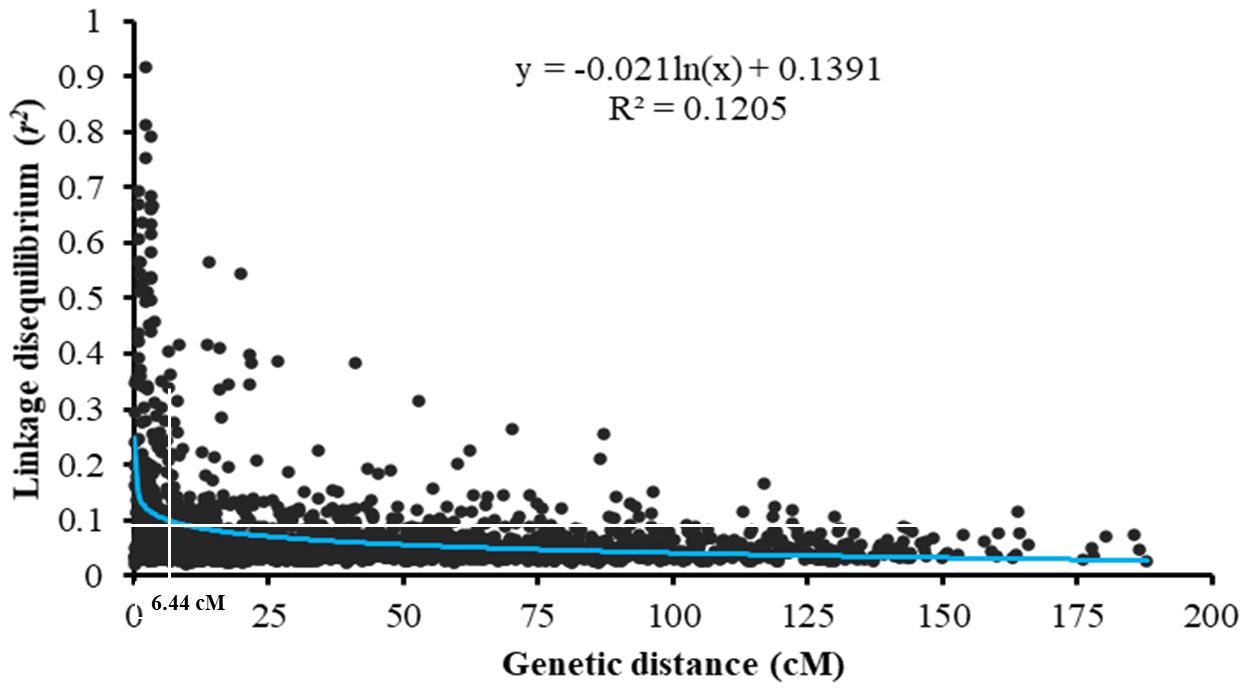
Figure 5. Genome-wide linkage disequilibrium (LD) decays over the genetic distance of the whole winter wheat panel consisting of 465 entries. The blue curve represents the model fitting LD decays. The red line represents the standard critical r2 = 0.10 used to establish QTL confidence intervals.
3.3 Significant QTL identified through GWAS analyses
Two sets of GWAS were conducted. The first set of analysis was done using 1,278 polymorphic SNPs (MAF ≥ 0.05) and the stripe rust IT and rDS data at the adult-plant stage of 465 wheat entries tested in nine field environments from 2018 to 2021 plus the BLUE data across environments and the seedling IT data tested with seven Pst races in the greenhouse. The second set of analysis was done using 1,200 polymorphism SNPs (MAF ≥ 0.05) and the IT and rDS data of 175 entries from 32 environments from 2007 to 2017 plus the BLUE data across the environments. Using the MLM (Q+K) with the significant threshold P ≤ 0.001 [-log10(P) ≥ 3.0], a total of 143 significant MTAs were identified from the two sets of GWAS analyses, and the number of MTA detected in each race test or field environment is given in Table 1. The distributions of the MTAs across the 21 chromosomes associated to the IT data from the seedling tests with the seven Pst races in the greenhouse and the rDS and IT data from the field environments are illustrated in Figures 6A–C, respectively. When the MTAs were detected in two or more race tests/field environments or located within the LD decay distance (6.44 cM), they were treated as a single QTL. Thus, 28 QTL were obtained, and they were on chromosomes 1A (2), 1B, 1D, 2A (2), 2B, 3B, 3D, 4A, 4B, 4D, 5B (3), 5D, 6A (4), 6B, 6D (3), 7A (2), and 7B (2) (Table 4). These QTL were named as QYrWW.wgp-1A.1, QYrWW.wgp-1A.2, QYrWW.wgp-1B, QYrWW.wgp-1D, QYrWW.wgp-2A.1, QYrWW.wgp-2A.2, QYrWW.wgp-2B, QYrWW.wgp-3B, QYrWW.wgp-3D, QYrWW.wgp-4A, QYrWW.wgp-4B, QYrWW.wgp-4D, QYrWW.wgp-5B.1, QYrWW.wgp-5B.2, QYrWW.wgp-5B.3, QYrWW.wgp-5D, QYrWW.wgp-6A.1, QYrWW.wgp-6A.2, QYrWW.wgp-6A.3, QYrWW.wgp-6A.4, QYrWW.wgp-6B, QYrWW.wgp-6D.1, QYrWW.wgp-6D.2, QYrWW.wgp-6D.3, QYrWW.wgp-7A.1, QYrWW.wgp-7A.2, QYrWW.wgp-7B.1, and QYrWW.wgp-7B.2, respectively. Each of the QTL explained 6%–40% of the phenotypic variation (R2).
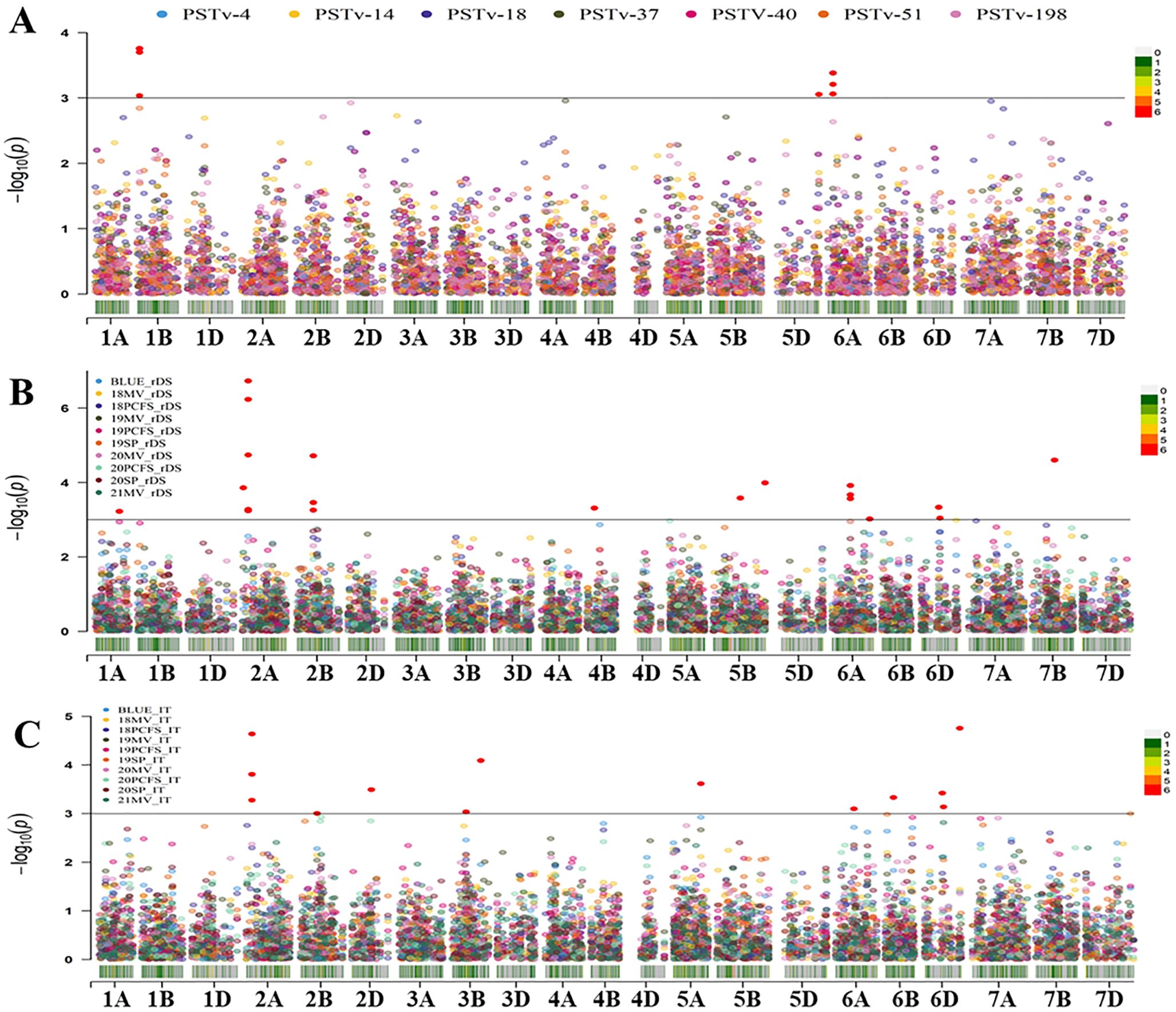
Figure 6. Distribution of the marker–trait associations (MTAs) for stripe rust response of the whole winter wheat panel consisting of 465 entries on the 21 chromosomes based on the infection type data of the seedling tests with seven Puccinia striiformis f. sp. tritici races (A), relative disease severity (rDS) data (B), and infection type (IT) data (C) at the adult-plant stage in various environments (year and location). BLUE, best linear unbiased estimator across all field environments; 18-21, the years 2018–2021; MV, Mount Vernon; PCFS, Palouse Conservation Field Station; SP, Spillman Farm.
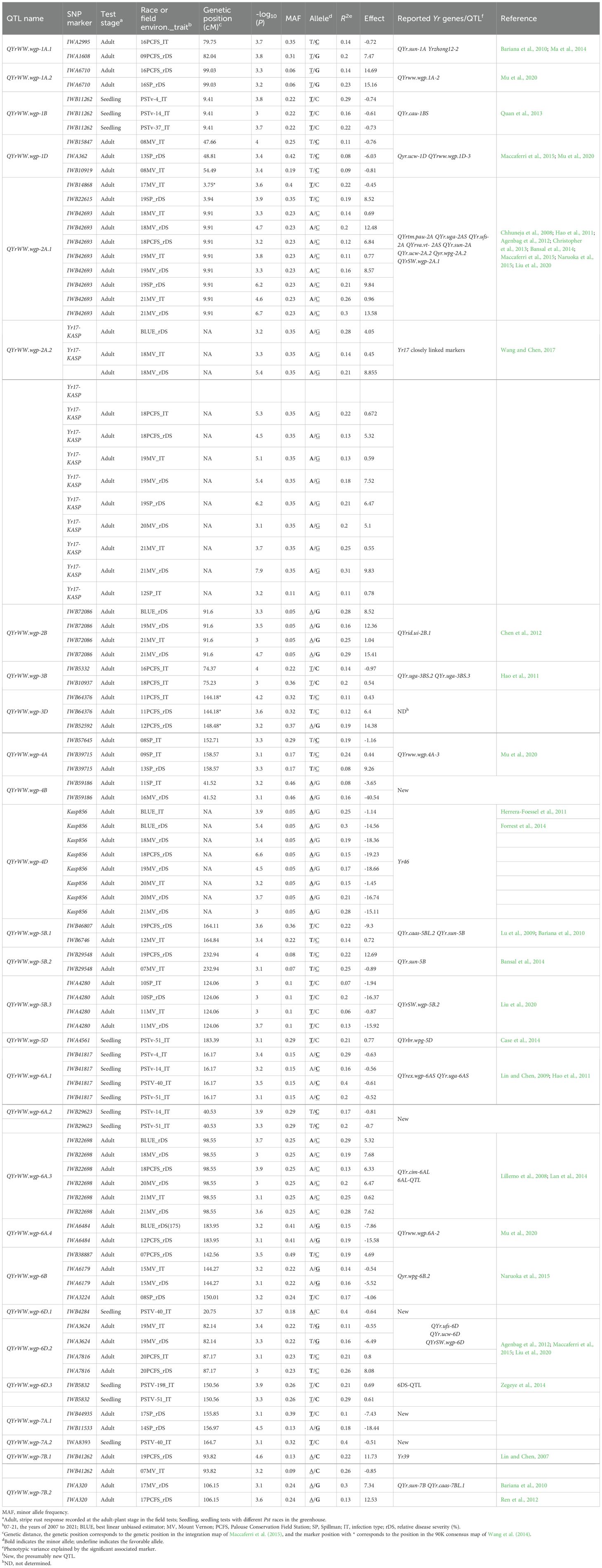
Table 4. QTL significantly associated to stripe rust resistance at the seedling stage tested with races of Puccinia striiformis f. sp. tritici (Pst) and at the adult-plant stage under the natural infection of the pathogen in various field environments (years and locations) in the winter wheat panel.
Among the 28 QTL, seven were detected at the seedling stage and considered for race-specific ASR and 21 QTL were detected only at the adult-plant stage in the fields and considered as HTAP resistance QTL. Among the seven ASR QTL, QYrWW.wgp-1B was detected with races PSTv-4, PSTv-14, and PSTv-37, QYrWW.wgp-5D with PSTv-51, QYrWW.wgp-6A.2 with PSTv-14 and PSTv-51, QYrWW.wgp-6A.3 with PSTv-4, PSTv-14, PSTV-40, and PSTv-51, QYrWW.wgp-6D.1 with PSTv-40, QYrWW.wgp-6D.3 with PSTv-198 and PSTv-51, and QYrWW.wgp-7A.2 with PSTv-40. Among the 21 HTAP resistance QTL, 12 (QYrWW.wgp-1A.1, QYrWW.wgp-1A.2, QYrWW.wgp-1D, QYrWW.wgp-3D, QYrWW.wgp-4A, QYrWW.wgp-4B, QYrWW.wgp-5B.3, QYrWW.wgp-6A.4, QYrWW.wgp-6B, QYrWW.wgp-7A.1, QYrWW.wgp-7B.1, and QYrWW.wgp-7B.2) were detected with the 175 entries in the 2007–2017 environments, four (QYrWW.wgp-2B, QYrWW.wgp-4D, QYrWW.wgp-6A.3, and QYrWW.wgp-6D.2) with the 465 entries in the 2018-2021 environments, and five (QYrWW.wgp-2A.1, QYrWW.wgp-2A.2, QYrWW.wgp-3B, QYrWW.wgp-5B.1, and QYrWW.wgp-5B.2) with both 175 and 465 entries in the 2007–2021 environments. Two of the QTL were detected to be significantly associated with previously reported Yr17 (QYrWW.wgp-2A.2) and Yr46 (QYrWW.wgp-4D) with R2 range of 11%–31% and 11%–26%, respectively (Table 4).
3.4 Comparison of significant QTL with known Pst resistance genes
The 28 QTL were compared with the previously reported Yr genes/QTL using the integration map (Wang et al., 2014; Maccaferri et al., 2015). Based on the chromosomal/genome locations, resistance type, and wheat variety origin, 23 QTL were overlapped (6.44 cM) with the previously reported Yr genes/QTL. Five QTL (QYrWW.wgp-4B, QYrWW.wgp-6A.2, QYrWW.wgp-6D.1, QYrWW.wgp-7A.1, and QYrWW.wgp-7A.2) were located far away from the previously reported Yr genes/QTL on chromosomes 4B, 6A, 6D, and 7A, and therefore they were considered new in the present study (Table 4).
3.5 KASP markers developed for eight QTL for HTAP resistance
A total of 10 KASP markers were developed for eight of the QTL for HTAP resistance, and their associations with stripe rust responses identical to their SNPs were confirmed through testing the 465 entries. The primer sequences are shown in Table 5. These polymorphic KASP markers should be useful in marker-assisted selections to develop stripe rust-resistant cultivars with the resistance QTL.
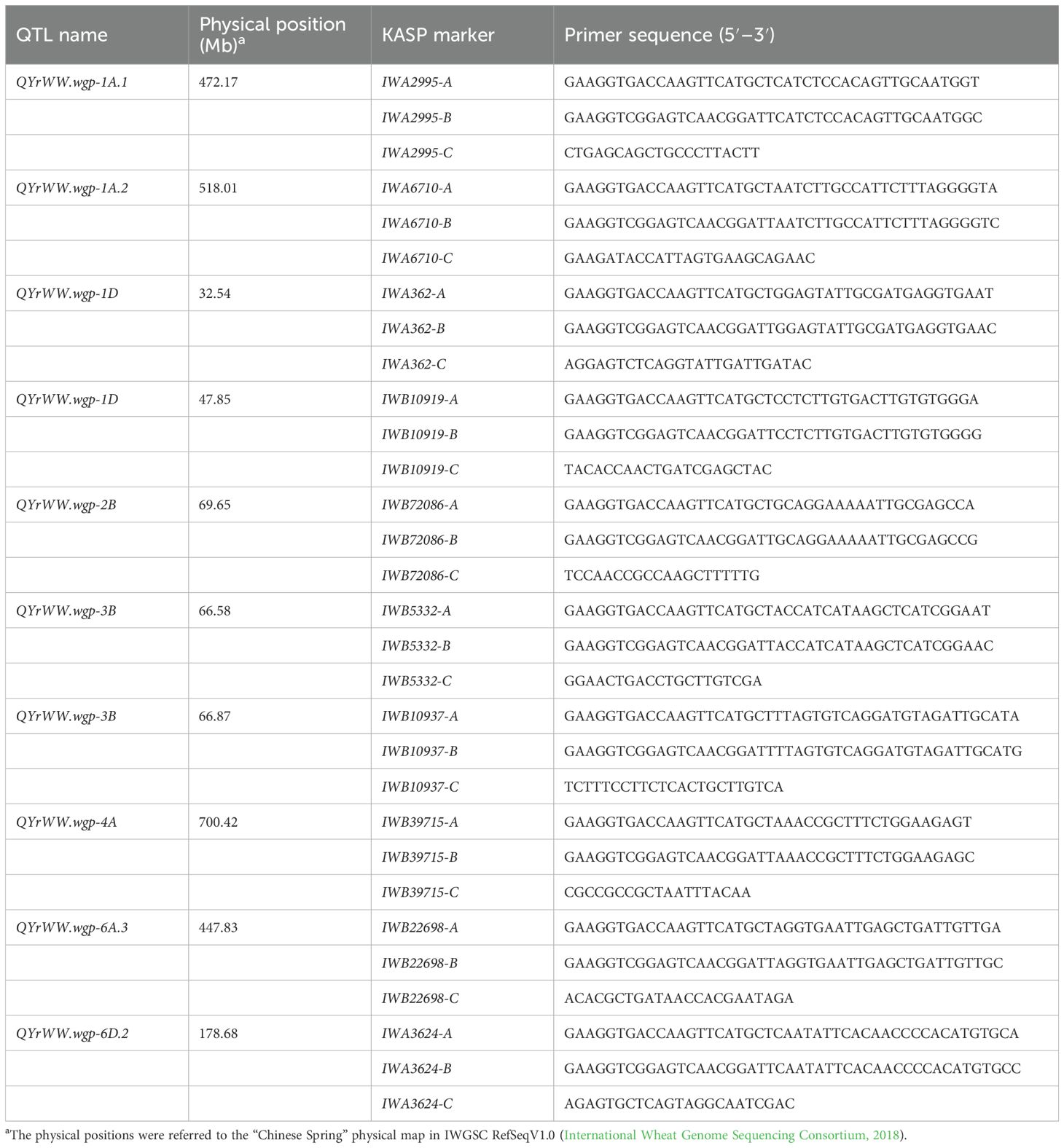
Table 5. Primer sequences of KASP markers developed from SNP markers significantly associated with eight QTL for high-temperature adult-plant resistance to stripe rust.
3.6 Frequencies of the Yr genes/QTL detected by markers
The presence (+) and absence (-) of the 13 Yr genes/QTL identified using previously reported markers and the 28 QTL identified through GWAS in the 456 wheat entries are provided in Supplementary Table S7. Entries considered having each of the 41 resistance genes/QTL are listed in Supplementary Table S8. The frequencies of the genes/QTL varied from 0.86% (for Yr36) to 95.91% (for QYrWW.wgp-1A.2). A total of 16 genes/QTL (Yr36, Yr62, QYrWW.wgp-6D.3, Yr30, Yr46, QYrWW.wgp-4D, Yr16, QYrWW.wgp-3B, QYrWW.wgp-6A.1, QYrWW.wgp-1B, Yr59, Yr78, QYrWW.wgp-6A.2, Yr17, QYrWW.wgp-2A.2, and QYrWW.wgp-6B) had low frequencies (<10%). There were 17 genes/QTL (QYrsk.wgp-3BS, QYrWW.wgp-1D, QYrWW.wgp-5B.3, QYrWW.wgp-7A.2, QYrWW.wgp-1A.1, QYrsk.wgp-4BL, QYrWW.wgp-6D.1, QYrWW.wgp-6D.2, Qyr.wgp-1B.1, QYrWW.wgp-2A.1, QYrWW.wgp-3D, QYrWW.wgp-4A, QYrWW.wgp-7B.2, QYrWW.wgp-5B.1, QYrWW.wgp-7A.1, QYrWW.wgp-5D, and Yr52) that had moderate frequencies (13.98%–61.08%). Eight genes/QTL (QYrWW.wgp-6A.3, QYrWW.wgp-6A.4, Yr18, QYrWW.wgp-4B, QYrWW.wgp-2B, QYrWW.wgp-7B.1, QYrWW.wgp-1A.2, and QYrWW.wgp-5B.2) had relatively high frequencies (74.84%–95.91%).
3.7 Pyramiding effect of resistance genes/QTL
The number of resistance genes/QTL in each entry based on the marker haplotypes was used to assess the pyramiding effect of resistant genes/QTL on stripe rust response. For the resistance observed in the field tests, the BLUE value of IT and rDS values across nine environments were regressed against the numbers of resistant genes/QTL. Since most of the 465 entries have HTAP resistance, with 74.19% of the BLUE_IT lower than 5% and 92.04% of the BLUE_rDS lower than 50%, the distributions of the entries were mainly concentrated in the resistant side, while very few entries were scattered in the susceptible side (Figure 7). The number of resistance genes/QTL present in an entry ranged from 0 to 31, and more than 90% of the entries have 11–27 of the favorable alleles (Figure 7; Supplementary Table S7). With the number of resistance genes/QTL increasing, the stripe rust phenotype (BLUE_rDS and BLUE_IT) was gradually decreasing, indicating that more genes/QTL improved the resistance to stripe rust.
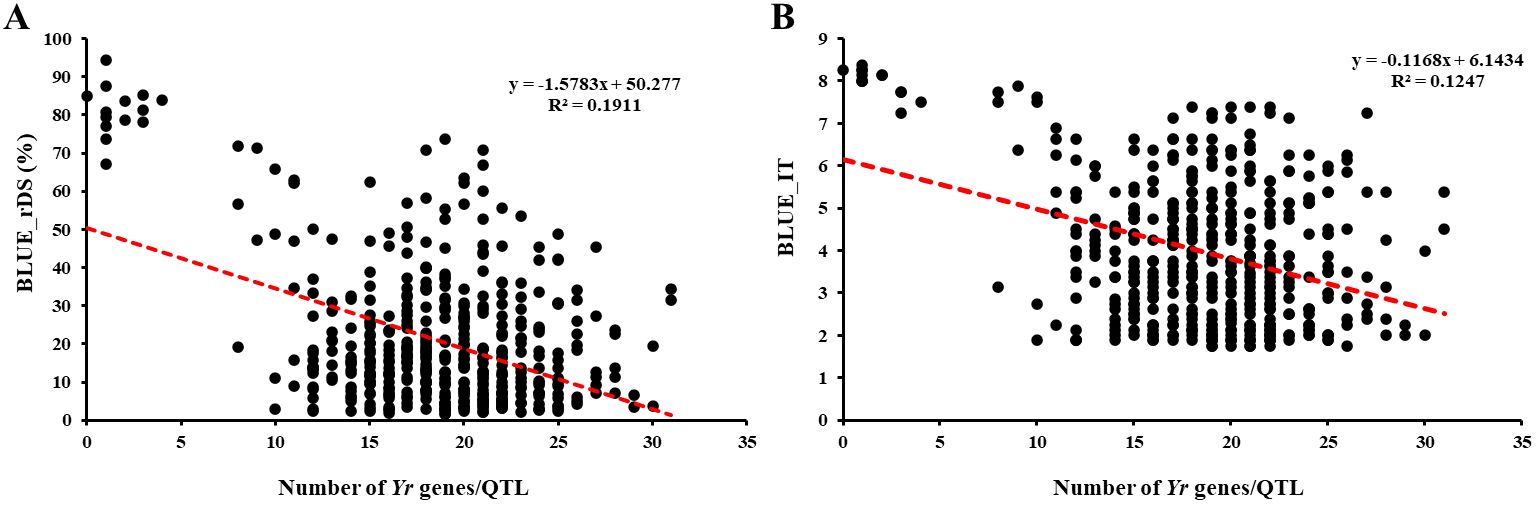
Figure 7. Effects of the number of Yr genes/QTL on stripe rust responses (A) for relative disease severity (rDS) and (B) for infection type (IT) in the 465 winter wheat entries. BLUE, best linear unbiased estimator.
4 Discussion
Stripe rust is a serious disease that threatens wheat production in the world (Stubbs, 1985; Chen, 2005, 2020; Wellings, 2011). In the United States, the disease has caused frequent epidemics in the regions west of the Rocky Mountains and has become a major disease in the regions east of the mountain range (Chen, 2007; Chen et al., 2010). This is mainly due to the virulence changes within the pathogen population and the new isolates with significant adaptation to the warm climates (Chen et al., 2002; Chen, 2005, 2020; Milus et al., 2009). To mitigate the problem, it is essential to continuously identify and utilize new resources of stripe rust resistance, especially durable types of resistance. In the present study, a panel of 465 winter wheat entries presumably with HTAP resistance was used to identify stable and potentially new QTL for stripe rust resistance through GWAS and determine the presence of previously reported Yr genes or QTL for HTAP resistance through marker-assisted detection.
The 465 entries were tested in different field environments for many years, especially the first part of 175 entries that have been tested for stripe rust response since 2007. A large number of entries with stable resistance that could be used in breeding programs were identified. The 465 entries were also tested at the seedling stage with seven contemporary Pst races in the present study. Since these entries were selected with HTAP resistance based on previous greenhouse seedling and field adult-plant tests, most of them should be susceptible at the seedling stage but resistant at the adult-plant stage. The results of the present study prove the significant levels of HTAP in most of the entries. The consistent resistant phenotypes of most entries across different environments indicate their durable resistance.
The 465 entries were genotyped using the GMS platform (Ruff et al., 2020). Previously, Liu et al. (2020) and Mu et al. (2020) used the platform in their studies of US spring and winter wheat panels through GWAS and identified 37 and 51 genes/QTL, respectively. In the present study, we used the same approach, but much more years of field data and identified 28 QTL in the winter wheat HTAP resistance panel. Among the 28 QTL, 21 conferred HTAP resistance and seven for ASR. After comparing with the previously reported Yr genes/QTL, five QTL (QYrWW.wgp-4B, QYrWW.wgp-6A.2, QYrWW.wgp-6D.1, QYrWW.wgp-7A.1, and QYrWW.wgp-7A.2) are newly identified loci for stripe rust resistance. The relationships of the QTL detected in this study and the previously reported Yr genes/QTL are discussed below.
Four QTL were identified in homoeologous group 1, namely, QYrWW.wgp-1A.1, QYrWW.wgp-1A.2, QYrWW.wgp-1B, and QYrWW.wgp-1D. Except for QYrWW.wgp-1B, an ASR locus, the other three QTL were all HTAP loci detected in the field environments, and each of them explained 8% to 29% of phenotypic variation. On the group 1 chromosomes, many genes/OTL for stripe rust resistance have been reported. Ma et al. (2014) reported a race-specific resistance gene on chromosome 1A, provisionally named Yrzhong12-2. Its most closely linked marker, Xcfd2129, was 1.31 and 3.60 cM away from the association markers IWA2995 and IWA1608 of QYrWW.wgp-1A.1 identified in the present study, respectively. As the distance between the two loci was within the LD decay distance 6.44 cM, they are likely located in the same chromosomal region. However, QYrWW.wgp-1A.1 confers HTAP resistance, while Yrzhong12.2 is a race-specific resistance gene. The relationship of QYrWW.wgp-1A.1 and Yrzhong12-2 needs to be determined by further studies. Bariana et al. (2010) reported an APR locus in the Australian cultivar Janz, QYr.sun-1A, which is adjacent to IWA2995 and IWA1608. Therefore, QYr.sun-1A and QYrWW.wgp-1A.1 should be the same QTL. QYrWW.wgp-1A.2 was significantly associated with IWA6710 in two environments (16PFS_rDS and 16SP_rDS), and it was also mapped to chromosome 1A. In a previous study, Mu et al. (2020) identified an APR QTL, QYrww.wgp.1A-2, also significantly associated with IWA6710 in a winter wheat panel. The same marker indicates that QYrWW.wgp-1A.2 and QYrww.wgp.1A-2 are the same QTL. On chromosome 1B, QYrWW.wgp-1B was an ASR locus identified with races PSTv-4, PSTv-14, and PSTv-37 and significantly associated with IWB11262 with the negative effect of seedling IT (-0.74, -0.61, and -0.73, respectively). Quan et al. (2013) reported that QYr.cau-1BS affected the latent period of Pst infection at the seedling stage and mapped it to the interval gwm374-gwm264-barc194 on chromosome 1B. On the integrated map, gwm374 is only 0.64 cM away from IWB11262 associated to QYrWW.wgp-1B. Thus, QYrWW.wgp-1B and QYr.cau-1BS are most likely the same locus. On chromosome 1D, QYrWW.wgp-1D was significantly associated with IWA362, IWB15847, and IWB10919 at the position of 47.66–55.49 cM on chromosome 1D. Maccaferri et al. (2015) reported a locus on chromosome 1D that was significantly associated with IWA980, and Mu et al. (2020) reported QYrww.wgp.1D-3 to be significantly associated with IWA3446. On the integrated map, IWA980 and IWA3446 were located at 49.26 and 45.09 cM on chromosome 1D, similar to the position of QYrWW.wgp-1D. Therefore, these three QTL are likely the same.
Three QTL for HTAP resistance were detected in homoeologous group 2, namely, QYrWW.wgp-2A.1, QYrWW.wgp-2A.2, and QYrWW.wgp-2B. QYrWW.wgp-2A.1 was significantly associated with IWB14868, IWB22615, and IWB42693 on the short arm of chromosome 2A in nine field environments. In fact, there was a hot spot on chromosome 2AS, on which many genes/QTL for stripe rust resistance were reported, including QYrtm.pau-2A (Chhuneja et al., 2008), QYr.uga-2AS (Hao et al., 2011), QYr.ufs-2A (Agenbag et al., 2012), QYrva.vt-2AS (Christopher et al., 2013), QYr.sun-2A (Bansal et al., 2014), QYr.ucw‐2A.2 (Maccaferri et al., 2015), Qyr.wpg-2A.2 (Naruoka et al., 2015), and QYrSW.wgp-2A.1 (Liu et al., 2020). These QTL were reported in an interval of 5.70–13.40 cM on the integrated map, which is within the confidence interval of QYrWW.wgp-2A.1. Among them, QYrtm.pau-2A was derived from einkorn wheat (T. monococcum) line pau14087, QYrSW.wgp-2A.1 was an ASR locus (Liu et al., 2020), while the other loci were all APR loci. Though they all fall in a hot spot of stripe rust resistance on chromosome 2A, they still have differences. Therefore, further studies are needed to determine the relationship among these loci. QYrWW.wgp-2A.2 was detected at the adult-plant stage by the closely linked KASP molecular marker Yr17-Kasp of Yr17. Yr17 is an ASR gene, but the KASP marker was only detected at the adult-plant stage in the field experiments. This is consistent with the results of Liu et al. (2020). In fact, Yr17 has a closely linked HTAP resistance gene, YrM1225, on chromosome 2AS (Li et al., 2023). Therefore, QYrWW.wgp-2A.2 detected by the Yr17-Kasp marker should be YrM1225. On chromosome 2B, QYrWW.wgp-2B conferred HTAP resistance and was identified with IWB72086 in the 19MV and 21MV and BLUE environments. The ASR gene Yr31 (Jighly et al., 2015), ASR locus QYrlu.cau-2BS1 (Guo et al., 2008), and the APR locus QYrid.ui-2B.1 (Chen et al., 2012) were close to QYrWW.wgp-2B based on their marker locations on the integrated map. Based on the type of resistance, QYrWW.wgp-2B is likely the same as QYrid.ui-2B.1, which was identified for HTAP resistance from the Idaho winter wheat breeding line IDO444 (Chen et al., 2012).
QYrWW.wgp-3B and QYrWW.wgp-3D were identified in homoeologous group 3. QYrWW.wgp-3B was detected at 74.37–75.23 cM on chromosome 3B of the integrated map with IWB5332 and IWB10937 in the 16PCFS and 18PCFS environments. On chromosome 3B, QYr.uga-3BS.2 and QYr.uga-3BS.3 were reported as two minor loci from AGS 2000 in a recombinant inbred population with a 2.4-cM interval (Hao et al., 2011), which could be considered the same locus based on the confidence interval (6.44 cM) of the present study. Thus, these two QTL are likely the same as QYrWW.wgp-3B. Another resistance gene, Yrwh2, was located at 72.95–75.78 cM of the integrated map, also within the confidence interval of QYrWW.wgp-3B, but the resistance type was different, indicating a different locus (Zhou et al., 2014a). Since the genetic position of QYrWW.wgp-3D was not found on the integrated map, its relationship with previously reported Yr genes/QTL could not be determined.
Three HTAP resistance QTL (QYrWW.wgp-4A, QYrWW.wgp-4B, and QYrWW.wgp-4D) were identified in homoeologous group 4. QYrWW.wgp-4A was detected with IWB57645 (152.71 cM) and IWB39715 (158.57 cM) on the integrated map, which is overlapped with QYrww.wgp.4A-3 detected with IWA559 (154.26 cM) in another winter wheat panel (Mu et al., 2020). Therefore, QYrWW.wgp-4A and QYrww.wgp.4A-3 should be the same. QYrWW.wgp-4B was detected at 41.52 cM on chromosome 4B and was found to be a new resistance locus based on its unique position from the previously reported Yr genes/QTL reported on chromosome 4B. QYrWW.wgp-4D was identified with KASP marker KASP856 in multiple environments (BLUE_IT, BLUE_rDS, 18MV_rDS, 18PCFS_rDS, 19MV_rDS, 20MV_IT, 20MV_rDS, and 21MV_rDS) on chromosome 4D. This QTL had a relatively large PVE value (15%–30%) and a negative effect value (IT corresponds to -1.14 to -1.45; rDS corresponds from -14.56 to -19.23). KASP856 was developed for the pleiotropic APR gene Yr46/Lr67 (for resistance to stripe rust and leaf rust) by Forrest et al. (2014). Therefore, QYrWW.wgp-4D should be Yr46/Lr67. The frequency of this resistance gene in the HTAP resistance panel was only 2.15% (Supplementary Table 8). Although the marker was also used in previous studies, Liu et al. (2020) and Mu et al. (2020) did not detect the gene in their panels consisting of 616 spring wheat and 857 winter wheat entries, respectively, mostly from the United States. The present study indicates that this gene has been started to be used in breeding programs.
Four HTAP QTL (QYrWW.wgp-5B.1, QYrWW.wgp-5B.2, QYrWW.wgp-5B.3, and QYrWW.wgp-5D) were identified in homoeologous group 5. QYrWW.wgp-5B.1 was detected with IWB46807 (164.11 cM) and IWB6746 (164.84 cM) on the integrated map, which overlapped with QYr.sun-5B detected with wPt-3030 (158.56 cM) from Janz (Bariana et al., 2010) and QYr.caas-5BL.2 detected with gwm604 (166.70 cM) (Lu et al., 2009), indicating them as the same QTL. QYrWW.wgp-5B.2 was significantly associated with IWB29548 (232.94 cM) and was approximately 68 cM apart from QYrWW.wgp-5B.1. Bansal et al. (2014) reported that QYr.sun-5B was closely linked to wPt-0837 and located at 226.95 cM of the integrated map, which was overlapped with the position of QYrWW.wgp-5B.2. Bansal et al. (2014) found that QYr.sun-5B was far away from QYr.sun-5B and QYr.caas-5BL.2 reported by Bariana et al. (2010) and Lu et al. (2009), which was consistent with the results of the present study. QYrWW.wgp-5B.3 was an HTAP resistance QTL detected in the SP and MV field environments and overlapped with an ASR QTL QYrSW.wgp-5B.2 detected in a spring wheat panel (Liu et al., 2020). Due to the different resistance types, these QTL should be different, but their genetic relationship needs further studies. QYrWW.wgp-5D was detected with IWA4561 (183.39 cM) for seedling reaction to PSTv-51 in the present study, and QYrbr.wpg-5D was detected between Xbarc144 and Xwmc765 (179.39–183.55 cM) for seedling reaction to PST-114 (Case et al., 2014). Based on their similar chromosomal locations, these two QTL should be the same.
Four ASR QTL (QYrWW.wgp-6A.1, QYrWW.wgp-6A.2, QYrWW.wgp-6D.1, and QYrWW.wgp-6D.3) and four HTAP resistance QTL (QYrWW.wgp-6A.3, QYrWW.wgp-6A.4, QYrWW.wgp-6B, and QYrWW.wgp-6D.2) were detected in homoeologous group 6. Lin and Chen (2009) reported a major QTL (QYrex.wgp-6AS) linked with Xgwm334 and Xwgp56. Hao et al. (2011) reported a minor QTL (QYr.uga-6AS) linked with DArT markers wPt-671561 and wPt-7840 and adjacent to SSR markers Xgwm459 and Xgwm334. In the present study, the significant association marker IWB41817 (16.17 cM) of QYrWW.wgp-6A.1 was 3.78, 1.83, and 1.04 cM away from wPt-7840, Xgwm459, and Xgwm334 on the integrated map, respectively, indicating that they are in the same position on chromosome 6A. However, QYrWW.wgp-6A.1 is an ASR locus. Further studies are needed to determine the relationship between these QTL. QYrWW.wgp-6A.2 was identified with races PSTv-14 and PSTv-51 and significantly associated with IWB29623. This locus should be new since no other genes were reported near the region. The significant association marker IWB22698 (98.55 cM) of QYrWW.wgp-6A.3 was 4.21 and 6.13 cM away from the flanker markers gwm356 and barc3 of QYr.cim-6AL (Lan et al., 2014) and 6AL-QTL (Lillemo et al., 2008), respectively. Based on the close positions, these three QTL are likely the same. QYrWW.wgp-6A.4 was significantly associated with IWA6484 (183.95 cM) and was 0.72 cM apart from the significant association marker IWA214 for QYrww.wgp.6A-2 identified in the previous US winter wheat germplasm panel (Mu et al., 2020). QYrWW.wgp-6B was significantly associated with three markers (IWB38887, IWA6179, and IWA3224) at 142.56–150.01 cM on chromosome 6B. Qyr.wpg-6B.2 was significantly associated with IWA3222 (150.00 cM) on the long arm of chromosome 6B in a US Pacific Northwest winter wheat panel (Naruoka et al., 2015), which is close to the location of QYrWW.wgp-6B, indicating that these two QTL are the same. QYrWW.wgp-6D.1 was an ASR locus identified with PSTv-40 and significantly associated with IWB4284 (20.75 cM) at the end of the 6D short arm. QYrWW.wgp-6D.1 should be a new locus since no other genes were reported near the region. QYrWW.wgp-6D.2 was detected at the 82.14–87.17 cM position of chromosome 6D, which is adjacent to QYr.ufs-6D (Agenbag et al., 2012), QYr.ucw-6D (Maccaferri et al., 2015), and QYrSW.wgp-6D (Liu et al., 2020), and therefore these QTL should be the same. QYrWW.wgp-6D.3 was an ASR locus identified with races PSTv-51 and PSTv-198 and significantly associated with IWB5832 at 150.56 cM of the integrated map. Zegeye et al. (2014) reported a ASR QTL (wsnp_Ex_c62371_62036044) located at 155.56 cM on the integrated map of chromosome 6DS, which was located within the LD decay distance (6.44 cM). Therefore, these loci are likely the same.
Four APR QTL (QYrWW.wgp-7A.1, QYrWW.wgp-7A.2, QYrWW.wgp-7B.1, and QYrWW.wgp-7B.2) were detected in homoeologous group 7. Zegeye et al. (2014) reported an ASR QTL located at 145.14 cM on the integrated map of chromosome 7A, which was more than 10 cM away from the HTAP resistance QTL QYrWW.wgp-7A.1 (155.85–156.97 cM) and ASR gene QYrWW.wgp-7A.2 (164.7 cM) identified in the present study. Therefore, QYrWW.wgp-7A.1 and QYrWW.wgp-7A.2 should be new QTL for stripe rust resistance. QYrWW.wgp-7B.1 (IWB41262) and QYrWW.wgp-7B.2 (IWA320) were 12.33 cM apart on the chromosome 7B integrated map and were named as two different QTL. The significant association marker IWB41262 of QYrWW.wgp-7B.1 was 0.67 cM away from one of the flanking markers (gwm131) of Yr39 (Lin and Chen, 2007), indicating that QYrWW.wgp-7B.1 is likely Yr39. The significant association marker IWA320 of QYrWW.wgp-7B.2 was 2.92 cM away from flanker marker wPt-8106 of QYr.caas-7BL.1 (Ren et al., 2012) and 2.65 cM away from the peak marker wPt-3723 of QYr.sun-7B (Bariana et al., 2010), indicating that these three QTL are likely the same. Interestingly, Ren et al. (2012) reported that Yr39 and QYr.caas-7BL.1 should be the same or closely linked loci. Through comparative analysis in the present study, we found that these loci were in a hot spot region of chromosome 7BL and were relatively close to each other, so further studies were needed to determine their relationships.
As the present study was aimed to identify genes/QTL associated to HTAP resistance to stripe rust and GWAS might miss resistance alleles either present at very high or very low frequencies, we tested the 465 entries with molecular markers for 13 Yr genes/QTL. Among the 13 Yr genes/QTL identified using previously reported markers, only Yr17-indicated HTAP locus and Yr46 were detected, as discussed above, while the remaining 11 HTAP loci were not detected through GWAS. The remaining 11 genes/QTL were found at either very high or very low frequencies. Yr18 was found in 410 (88.17%) entries. The high frequency was expected as Yr18 has been found to be widespread in wheat landraces and cultivars throughout the world (Lagudah et al., 2009; Krattinger et al., 2011), but with frequencies that were much higher than those found in the US spring and winter wheat cultivars and breeding lines (Liu et al., 2020; Mu et al., 2020). The frequency differences might be due to the different entries of the panels. It was a little surprising that Yr52 was found in 284 (61.08%) entries. Yr52 conferring a high level of HTAP resistance to stripe rust was first identified in wheat germplasm accession PI 183527 (Ren et al., 2012), which was originally from India and deposited in the USDA-ARS National Small Grains Collection (NSGC) in 1949 (http://www.ars-grin.gov/cgi-bin/npgs/acc/search.pl?accid=PI?183527). The high frequency was not too different from the frequency detected among 74 US wheat germplasms (Ren et al., 2012). These results indicate that the gene has been widely used in developing contemporary cultivars and breeding lines as well as is widely presented in landraces (Supplementary Table S8). Three QTL, Qyr.wgp-1B.1, QYrsk.wgp-4BL, and QYrsk.wgp-3BS, which were previously identified in the US Pacific Northwest wheat cultivars (Naruoka et al., 2015; Liu et al., 2019c), were identified in 27.74%, 23.01%, and 13.98%, respectively. The results indicate that these QTL have been used in breeding programs not only in the Pacific Northwest but also in other regions of the United States. Six HTAP resistance genes, Yr59 (6.45%), Yr78 (6.45%), Yr16 (2.37%), Yr30 (2.15%), Yr62 (1.94%), and Yr36 (0.86%), had frequencies below 10% in the HTAP-resistant winter wheat panel (Supplementary Table S8), indicating that these genes have not been widely used in US wheat breeding programs, and they should be used in future breeding efforts to diversify HTAP resistance genes. The 13 genes/QTL as well as those identified from the GWAS should be valuable to develop wheat cultivars with durable resistance to stripe rust. As found in the present study and previous studies (Liu et al., 2020; Mu et al., 2020), pyramiding several HTAP resistance can enhance the level of the durable type of resistance. In the present study, we developed 10 KASP markers for eight QTL. Further efforts should be taken to develop KASP for other QTL to be used in marker-assisted selection to incorporate the useful QTL into new wheat cultivars in various regions.
5 Conclusions
In the present study, 465 HTAP-resistant winter wheat entries were tested to identify stripe rust resistance sources and genes. After testing with the seven predominant or most virulent Pst races at the seedling stage under low temperatures, 16 entries resistant to all the tested races were identified, making them excellent resources for use in developing new cultivars with stripe rust resistance. The 465 entries were also tested in nine field environments at the adult-plant stage under natural infection conditions. As a result, 172 entries were found to have stably high resistance, including the 16 entries identified in the seedling tests. Combining the stripe rust response and SNP genotype data, 28 QTL were identified in the HTAP-resistant winter wheat panel, including seven for ASR and 21 for HTAP resistance. After comparing with the previously reported Yr genes/QTL, five QTL, QYrWW.wgp-4B, QYrWW.wgp-6A.2, QYrWW.wgp-6D.1, QYrWW.wgp-7A.1, and QYrWW.wgp-7A.2, were identified as new loci for stripe rust resistance. A total of 10 polymorphic KASP markers were developed for eight randomly selected HTAP resistance QTL. Further studies are needed to develop KASP markers for other QTL identified in the present study without KASP markers, especially for the five new QTL. In addition to the QTL identified through GWAS, 13 HTAP resistance genes/QTL were detected using their molecular markers reported in previous studies. Except for two genes, these genes/QTL were not detected in the GWAS analyses. The frequencies of all genes/QTL identified in the present study were determined in the HTAP-resistant winter wheat panel. This study provides winter wheat entries with stable ASR and HTAP resistance, information on QTL, their effects, and frequencies, and user-friendly KASP markers to breeding programs to develop new resistant cultivars to improve the control of stripe rust.
Authors’ note
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. USDA is an equal opportunity provider and employer.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
FY: Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. MW: Data curation, Investigation, Methodology, Writing – review & editing. DS: Data curation, Methodology, Software, Writing – review & editing. EY: Formal analysis, Methodology, Supervision, Writing – review & editing. GC: Methodology, Supervision, Writing – review & editing. XC: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported in part by US Department of Agriculture, Agricultural Research Service In-house Project to XC (Project No. 2090-22000-020-00D); Washington Grain Commission fund to XC (Project No. 13C-3061-3144); the Funding for the Introduction and Training of High-Level Talents of Sichuan Academy of Agricultural Sciences to FY (NKYRCZX2024001); and the Sichuan Province Natural Science Foundation to FY (24NSFSC6494).
Acknowledgments
We would like to thank David Wood, Kent Evans, Jason Sprott, and Yumei Liu for preparing seeds and fields, planting, and harvesting of the nursery over the years as well as Travis Ruff, Marcus Hooker, Karol Marlowe, and Yan Liu for performing genotyping by multiplex sequencing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1514926/full#supplementary-material
Supplementary Table 4 | Information on the 175 US winter wheat entries and their stripe rust data in various locations from 2007 to 2017 (note: four entries showing seedling stage resistance to all the seven races of Puccinia striiformis f. sp. tritici are highlighted in green).
Supplementary Table 7 | Presence (+) and absence (-) of 13 high-temperature adult-plant (HTAP) resistance Yr genes/QTL and 28 QTL for all-state or HTAP resistance detected through GWAS in the HTAP panel of 465 winter wheat entries.
References
Agenbag, G. M., Pretorius, Z. A., Boyd, L. A., Bender, C. M., Prins, R. (2012). Identification of adult plant resistance to stripe rust in the wheat cultivar Cappelle-Desprez. Theor. Appl. Genet. 125, 109–120. doi: 10.1007/s00122-012-1819-5
Bansal, U. K., Kazi, A. G., Singh, B., Hare, R. A., Bariana, H. S. (2014). Mapping of durable stripe rust resistance in a durum wheat cultivar Wollaroi. Mol. Breed. 33, 51–59. doi: 10.1007/s11032-013-9933-x
Bariana, H. S., Bansal, U. K., Schmidt, A., Lehmensiek, A., Kaur, J., Miah, H., et al. (2010). Molecular mapping of adult plant stripe rust resistance in wheat and identification of pyramided QTL genotypes. Euphytica 176, 251–260. doi: 10.1007/s10681-010-0240-x
Bulli, P., Zhang, J., Chao, S., Chen, X. M., Pumphrey, M. (2016). Genetic architecture of resistance to stripe rust in a global winter wheat germplasm collection. G3. 6, 2237–2253. doi: 10.1534/g3.116.028407
Case, A. J., Naruoka, Y., Chen, X. M., Garland-Campbell, K. A., Zemetra, R. S., Carter, A. H. (2014). Mapping stripe rust resistance in a Brundage × Coda winter wheat recombinant inbred line population. PloS One 9, e91753. doi: 10.1371/journal.pone.0091758
Chen, X. M. (2005). Epidemiology and control of stripe rust [Puccinia striiformis f. sp. tritici] on wheat. Can. J. Plant Pathol. 27, 314–337. doi: 10.1080/07060660509507230
Chen, X. M. (2007). Challenges and solutions for stripe rust control in the United States. Aus. J. Agric. Res. 58, 648–655. doi: 10.1071/AR07045
Chen, X. M. (2013). Review article: high-temperature adult-plant resistance, key for sustainable control of stripe rust. Am. J. Plant Sci. 4, 608–627. doi: 10.4236/ajps.2013.43080
Chen, X. M. (2014). Integration of cultivar resistance and fungicide application for control of wheat stripe rust. Can. J. Plant Pathol. 36, 311–326. doi: 10.1080/07060661.2014.924560
Chen, X. M. (2020). Pathogens which threaten food security: Puccinia striiformis, the wheat stripe rust pathogen. Food Secur. 12, 239–251. doi: 10.1007/s12571-020-01016-z
Chen, J. L., Chu, C., Souza, E. J., Guttieri, M. J., Chen, X. M., Xu, S., et al. (2012). Genome-wide identification of QTL conferring high-temperature adult-plant (HTAP) resistance to stripe rust (Puccinia striiformis f. sp. tritici) in wheat. Mol. Breed. 29, 791–800. doi: 10.1007/s11032-011-9590-x
Chen, X. M., Kang, Z. S. (2017). Stripe Rust (Dordrecht: Springer), 719 pp. doi: 10.1007/978-94-024-1111-9
Chen, X. M., Line, R. F. (1992). Inheritance of stripe rust resistance in wheat cultivars used to differentiate races of Puccinia striiformis in North America. Phytopathology 82, 633–637. doi: 10.1094/Phyto-82-633
Chen, X. M., Moore, M., Milus, E. A., Long, D. L., Line, R. F., Marshall, D., et al. (2002). Wheat stripe rust epidemics and races of Puccinia striiformis f. sp. tritici in the United States in 2000. Plant Dis. 86, 39–46. doi: 10.1094/PDIS.2002.86.1.39
Chen, X. M., Penman, L., Wan, A. M., Cheng, P. (2010). Virulence races of Puccinia striiformis f. sp. tritici in 2006 and 2007 and development of wheat stripe rust and distributions, dynamics, and evolutionary relationships of races from 2000 to 2007 in the United States. Can. J. Plant Pathol. 32, 315–333. doi: 10.1080/07060661.2010.499271
Chen, X. M., Wang, M. N., Wan, A. M., Bai, Q., Li, M. J., López, P. F., et al. (2021). Virulence characterization of Puccinia striiformis f. sp. tritici collections from six countries in 2013 to 2020. Can. J. Plant Pathol. 43, S308–S322. doi: 10.1080/07060661.2021.1958259
Cheng, P., Xu, L. S., Wang, M. N., See, D. R., Chen, X. M. (2014). Molecular mapping of genes Yr64 and Yr65 for stripe rust resistance in hexaploid derivatives of durum wheat accessions PI 331260 and PI 480016. Theor. Appl. Genet. 127, 2267–2277. doi: 10.1007/s00122-014-2378-8
Chhuneja, P., Kaur, S., Garg, T., Ghai, M., Kaur, S., Prashar, M., et al. (2008). Mapping of adult plant stripe rust resistance genes in diploid A genome wheat species and their transfer to bread wheat. Theor. Appl. Genet. 116, 313–324. doi: 10.1007/s00122-007-0668-0
Christopher, M. D., Liu, S., Hall, M. D., Marshall, D. S., Fountain, M. O., Johnson, J. W., et al. (2013). Identification and mapping of adult-plant stripe rust resistance in soft red winter wheat cultivar “USG 3555. Plant Breed. 132, 53–60. doi: 10.1111/pbr.12015
Dong, Z. Z., Hegarty, J. M., Zhang, J. L., Zhang, W. J., Chao, S., Chen, X. M., et al. (2017). Validation and characterization of a QTL for adult plant resistance to stripe rust on wheat chromosome arm 6BS (Yr78). Theor. Appl. Genet. 130, 2127–2137. doi: 10.1007/s00122-017-2946-9
Evanno, G., Regnaut, S., Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Feng, J. Y., Yao, F. J., Wang, M. N., See, D. R., Chen, X. M. (2023). Molecular mapping of Yr85 and comparison with other genes for resistance to stripe rust on wheat chromosome 1B. Plant Dis. 107, 3585–3591. doi: 10.1094/PDIS-11-22-2600-RE
Forrest, K., Pujol, V., Bulli, P., Pumphrey, M., Wellings, C., Herrera-Foessel, S., et al. (2014). Development of a SNP marker assay for the Lr67 gene of wheat using a genotyping by sequencing approach. Mol. Breed. 34, 2109–2118. doi: 10.1007/s11032-014-0166-4
Guo, Q., Zhang, Z. J., Xu, Y. B., Li, G. H., Feng, J., Zhou, Y. (2008). Quantitative trait loci for high-temperature, adult-plant and slow-rusting resistance to Puccinia striiformis f. sp. tritici in wheat cultivars. Phytopathology 98, 803–809. doi: 10.1094/PHYTO-98-7-0803
Hao, Y. F., Chen, Z. B., Wang, Y. Y., Bland, D., Buck, J., Brown-Guedira, G., et al. (2011). Characterization of a major QTL for adult plant resistance to stripe rust in US soft red winter wheat. Theor. Appl. Genet. 123, 1401–1411. doi: 10.1007/s00122-011-1675-8
Herrera-Foessel, S. A., Lagudah, E. S., Huerta-Espino, J., Hayden, M. J., Bariana, H. S., Singh, D., et al. (2011). New slow-rusting leaf rust and stripe rust resistance genes Lr67 and Yr46 in wheat are pleiotropic or closely linked. Theor. Appl. Genet. 122, 239–249. doi: 10.1007/s11032-014-0166-4
International Wheat Genome Sequencing Consortium. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, 6403. doi: 10.1126/science.aar7191
Jighly, A., Oyiga, B. C., Makdis, F., Nazari, K., Youssef, O., Tadesse, W., et al. (2015). Genome-wide DArT and SNP scan for QTL associated with resistance to stripe rust (Puccinia striiformis f. sp. tritici) in elite ICARDA wheat (Triticum aestivum L.) germplasm. Theor. Appl. Genet. 128, 1277–1295. doi: 10.1007/s00122-015-2504-2
Khan, H., Krishnappa, G., Kumar, S., Devate, N. B., Rathan, N. D., Kumar, S., et al. (2024). Genome-wide association study identifies novel loci and candidate genes for rust resistance in wheat (Triticum aestivum L.). BMC Plant Biol. 24, 411. doi: 10.1186/s12870-024-05124-2
Krattinger, S. G., Lagudah, E. S., Wicker, T., Risk, J. M., Ashton, A. R., Selter, L. L., et al. (2011). Lr34 multi-pathogen resistance ABC transporter: molecular analysis of homoeologous and orthologous genes in hexaploid wheat and other grass species. Plant J. 65, 392–403. doi: 10.1111/j.1365-313X.2010.04430.x
Lagudah, E. S., Krattinger, S. G., Herrera-Foessel., S., Singh, R. P., Huerta-Espino, J., Spielmeyer, Q., et al. (2009). Gene-specific markers for the wheat gene Lr34/Yr18/Pm38 which confers resistance to multiple fungal disease. Theor. Appl. Genet. 119, 889–898. doi: 10.1007/s00122-009-1097-z
Lan, C. X., Rosewarne, G. M., Singh, R. P., Herrera-Foessel, S. A., Huerta-Espino, J., Basnet, B. R., et al. (2014). QTL characterization of resistance to leaf rust and stripe rust in the spring wheat line Francolin 1. Mol. Breed. 34, 789–803. doi: 10.1007/s11032-014-0075-6
Li, Y. X., Liu, L., Wang, M. N., Ruff, T., See, D. R., Hu, X. P., et al. (2023). Characterization and molecular mapping of a gene conferring high-temperature adult-plant resistance to wheat stripe rust originally from Aegilops ventricosa. Plant Dis. 107, 431–442. doi: 10.1094/PDIS-06-22-1419-RE
Li, Y. X., Wang, M. N., Hu, X. P., Chen, X. M. (2024). Identification of a locus for high-temperature adult-plant resistance to stripe rust in the wheat Yr8 near-isogenic line through mutagenesis and molecular mapping. Plant Dis. 108, 1261–1269. doi: 10.1094/PDIS-10-23-2037-RE
Lillemo, M., Asalf, B., Singh, R. P., Huerta-Espino, J., Chen, X. M., He, Z. H., et al. (2008). The adult plant rust resistance loci Lr34/Yr18 and Lr46/Yr29 are important determinants of partial resistance to powdery mildew in bread wheat line Saar. Theor. Appl. Genet. 116, 1155–1166. doi: 10.1007/s00122-008-0743-1
Lin, F., Chen, X. M. (2007). Genetics and molecular mapping of genes for race-specific all-stage resistance and non-race-specific high-temperature adult-plant resistance to stripe rust in spring wheat cultivar Alpowa. Theor. Appl. Genet. 114, 1277–1287. doi: 10.1007/s00122-007-0518-0
Lin, F., Chen, X. M. (2009). Quantitative trait loci for non-race-specific, high-temperature adult-plant resistance to stripe rust in wheat cultivar Express. Theor. Appl. Genet. 118, 631–642. doi: 10.1007/s00122-008-0894-0
Line, R. F. (2002). Stripe rust of wheat and barley in North America: a retrospective historical review. Annu. Rev. Phytopathol. 40, 75–118. doi: 10.1146/annurev.phyto.40.020102.111645
Line, R. F., Qayoum, A. (1992). Virulence, aggressiveness, evolution and distribution of races of Puccinia striiformis (the cause of stripe rust of wheat) in North America 1968-87. US Dep. Agric. Tech. Bull., 44 pp. No. 1788. Available at: https://catalog.hathitrust.org/Record/009786514.
Liu, W. Z., Kolmer, J., Rynearson, S., Chen, X. M., Gao, L., Anderson, J., et al. (2019c). Identifying loci conferring resistance to leaf and stripe rusts in a spring wheat population (Triticum aestivum L.) via genome-wide association mapping. Phytopathology 109, 1932–1940. doi: 10.1094/PHYTO-04-19-0143-R
Liu, W. Z., Macaferri, M., Bulli, P., Rynearson, S., Tuberosa, R., Chen, X. M., et al. (2017a). Genome-wide association mapping of seedling and field resistance to Puccinia striiformis f. sp. tritici in elite global durum wheat. Theor. Appl. Genet. 130, 649–667. doi: 10.1007/s00122-016-2841-9
Liu, W. Z., Macaferri, M., Rynearson, S., Letta, T., Zegeye, H., Tuberosa, R., et al. (2017c). Novel sources of stripe rust resistance identified by genome-wide association mapping in Ethiopian durum wheat (Triticum turgidum ssp. durum). Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00774
Liu, W. Z., Maccaferri, M., Chen, X. M., Pumphrey, M., Laghetti, G., Pignone, D., et al. (2017b). Genome-wide association mapping reveals a rich genetic architecture of stripe rust resistance loci in emmer wheat (Triticum turgidum ssp. dicoccum). Theor. Appl. Genet. 130, 2249–2270. doi: 10.1007/s00122-017-2957-6
Liu, K., Muse, S. V. (2005). PowerMaker: An integrated analysis environment for genetic maker analysis. Bioinformatics 21, 2128–2129. doi: 10.1093/bioinformatics/bti282
Liu, L., Wang, M. N., Feng, J. Y., See, D. R., Chao, S. M., Chen, X. M. (2018). Combination of all-stage and high-temperature adult-plant resistance QTL confers high level, durable resistance to stripe rust in winter wheat cultivar Madsen. Theor. Appl. Genet. 131, 1835–1849. doi: 10.1007/s00122-018-3116-4
Liu, L., Wang, M. N., Feng, J. Y., See, D. R., Chen, X. M. (2019a). Whole-genome mapping of stripe rust resistance quantitative trait loci and race-specificity related to resistance reduction in winter wheat cultivar Eltan. Phytopathology 109, 1226–1235. doi: 10.1094/PHYTO-10-18-0385-R
Liu, L., Wang, M. N., Zhang, Z. W., See, D. R., Chen, X. M. (2020). Identification of stripe rust resistance loci in U.S. spring wheat cultivars and breeding lines using genome-wide association mapping and Yr gene markers. Plant Dis. 104, 218–2192. doi: 10.1094/PDIS-11-19-2402-RE
Liu, L., Yuan, C. Y., Wang, M. N., See, D. R., Zemetra, R. S., Chen, X. M. (2019b). QTL analysis of durable stripe rust resistance in the North American winter wheat cultivar Skiles. Theor. Appl. Genet. 132, 1677–1691. doi: 10.1007/s00122-019-03307-2
Long, L., Li, J., Huang, L. Y., Jin, H. L., Guan, F. N., Zhang, H. P., et al. (2024). Fine mapping and characterization of stripe rust resistance gene YrAYH in near-isogenic lines derived from a cross involving wheat landrace Anyuehong. Crop J. 12, 826–835. doi: 10.1016/j.cj.2024.03.009
Lu, Y. M., Lan, C. X., Liang, S. S., Zhou, X. L., Liu, D., Zhou, G., et al. (2009). QTL mapping for adult-plant resistance to stripe rust in Italian common wheat cultivars Libellula and Strampelli. Theor. Appl. Genet. 119, 1349–1359. doi: 10.1007/s00122-009-1139-6
Lu, Y., Wang, M. N., Chen, X. M., See, D. R., Chao, S. M., Jing, J. X. (2014). Mapping of Yr62 and a small-effect QTL for high-temperature adult-plant resistance to stripe rust in spring wheat PI 192252. Theor. Appl. Genet. 127, 1449–1459. doi: 10.1007/s00122-014-2312-0
Ma, D. F., Peng, F., Fang, Z. W., Fang, Z. W., Chao, K. X., Jing, J. X., et al. (2014). Genetic and molecular mapping of stripe rust resistance genes in wheat cultivar Zhongliang 12. J. Phytopathol. 163, 98–104. doi: 10.1111/jph.12284
Maccaferri, M., Zhang, J., Bulli, P., Abate, Z., Chao, S., Cantu, D., et al. (2015). A genome-wide association study of resistance to stripe rust (Puccinia striiformis f. sp. tritici) in a worldwide collection of hexaploid spring wheat (Triticum aestivum L.). G3. 5, 449–465. doi: 10.1534/g3.114.014563
Marone, D., Laidò, G., Saccomanno, A., Petruzzino, G., Azevedo, C. V. G., De Vita, P., et al. (2024). Genome wide association study of common resistance to rust species in tetraploid wheat. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1290643
Meng, L., Li, H. H., Zhang, L. Y., Wang, J. K. (2015). QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 3, 269–283. doi: 10.1016/j.cj.2015.01.001
Milus, E. A., Kristensen, K., Hovmøller, M. S. (2009). Evidence for increased aggressiveness in a recent widespread strain of Puccinia striiformis f. sp. tritici causing stripe rust of wheat. Phytopathology 99, 89–94. doi: 10.1094/PHYTO-99-1-0089
Mu, J. M., Liu, L., Liu, Y., Wang, M. N., See, D. R., Han, D. J., et al. (2020). Genome-wide association study and gene specific markers identified 51 genes or QTL for resistance to stripe rust in U.S. winter wheat cultivars and breeding lines. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00998
Naruoka, Y., Garland-Campbell, K. A., Carter, A. H. (2015). Genome-wide association mapping for stripe rust (Puccinia striiformis f. sp. tritici) in US Pacific Northwest winter wheat (Triticum aestivum L.). Theor. Appl. Genet. 128, 1083–1101. doi: 10.1007/s00122-015-2492-2
Pritchard, J. K., Stephens, M., Rosenberg, N. A., Donnelly, P. (2000). Association mapping in structured populations. Am. J. Hum. Genet. 67, 170–181. doi: 10.1086/302959
Qie, Y. M., Liu, Y., Wang, M. N., Li, X., See, D. R., An, D. G., et al. (2019). Development, validation, and re-selection of wheat lines with pyramided genes Yr64 and Yr15 linked on the short arm of chromosome 1B for resistance to stripe rust. Plant Dis. 103, 51–58. doi: 10.1094/PDIS-03-18-0470-RE
Quan, W., Hou, G. L., Chen, J., Du, Z. Y., Lin, F., Guo, Y., et al. (2013). Mapping of QTL lengthening the latent period of Puccinia striiformis in winter wheat at the tillering growth stage. Eur. J. Plant Pathol. 136, 715–727. doi: 10.1007/s10658-013-0201-z
Ren, R. S., Wang, M. N., Chen, X. M., Zhang, Z. J. (2012). Characterization and molecular mapping of Yr52 for high-temperature adult-plant resistance to stripe rust in spring wheat germplasm PI 183527. Theor. Appl. Genet. 125, 847–857. doi: 10.1007/s00122-012-1877-8
Ruff, T. M., Marston, E. J., Eagle, J. D., Sthapit, S. R., Hooker, M. A., Skinner, D. Z., et al. (2020). Genotyping by multiplexed sequencing (GMS): A customizable platform for genomic selection. PloS One 15, e0229207. doi: 10.1371/journal.pone.0229207
Sharma, D., Avni, R., Gutierrez-Gonzalez, J., Kumar, R., Sela, H., Prusty, M. R., et al. (2024). A single NLR gene confers resistance to leaf and stripe rust in wheat. Nat. Commun. 15, 9925. doi: 10.1038/s41467-024-54068-6
Spielmeyer, W., Sharp, P. J., Lagudah, E. S. (2003). Identification and validation of markers linked to broad-spectrum stem rust resistance gene Sr2 in wheat (Triticum aestivum L.). Crop Sci. 43, 333–336. doi: 10.2135/cropsci2003.3330
Stewart, C. N., Jr., Via, L. E. (1993). A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. BioTechniques 14, 748–750. Available at https://www.scribd.com/document/80909788/A-Rapid-CTAB-DNA-Isolation.
Stubbs, R. W. (1985). Stripe rust. Cereal Rusts Vol. II: Diseases, Distribution, Epidemiology, and Control. Eds. Roelfs, A. P., Bushnell, W. R. (Orlando: Academic Press), 61–101. doi: 10.1016/b978-0-12-148402-650011-0
Tang, Y., Liu, X. L., Wang, J. B., Li, M., Wang, Q. S., Tian, F., et al. (2016). GAPIT Version 2: An enhanced integrated tool for genomic association and prediction. Plant Genome 9, 2. doi: 10.3835/plantgenome2015.11.0120
Uauy, C., Brevis, J. C., Chen, X. M., Khan, I., Jackson, L., Chicaiza, O., et al. (2005). High-temperature adult-plant stripe rust resistance gene Yr36 from Triticum turgidum ssp. dicoccoides is closely linked to the grain protein content locus Gpc-B1. Theor. Appl. Genet. 11, 97–105. doi: 10.1007/s00122-005-0109-x
Wan, A. M., Chen, X. M. (2014). Virulence characterization of Puccinia striiformis f. sp. tritici using a new set of Yr single-gene line differentials in the United States in 2010. Plant Dis. 98, 1534–1542. doi: 10.1094/PDIS-01-14-0071-RE
Wan, A. M., Chen, X. M., Yuen, J. (2016). Races of Puccinia striiformis f. sp. tritici in the United States in 2011 and 2012 and comparison with races in 2010. Plant Dis. 100, 966–975. doi: 10.1094/PDIS-10-15-1122-RE
Wang, M. N., Chen, X. M. (2017). “Stripe rust resistance,” in Stripe Rust. Eds. Chen, X. M., Kang, Z. S. (Springer, Dordrecht), 353–558. doi: 10.1007/978-94-024-1111-9
Wang, M. N., Wan, A. M., Chen, X. M. (2022). Race characterization of Puccinia striiformis f. sp. tritici in the United States from 2013 to 2017. Plant Dis. 106, 1462–1473. doi: 10.1094/PDIS-11-21-2499-RE
Wang, S. C., Wong, D., Forrest, K., Allen, A., Chao, S., Huang, B. E., et al. (2014). Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnol. J. 12, 787–796. doi: 10.1111/pbi.12183
Wellings, C. R. (2011). Global status of stripe rust: a review of historical and current threats. Euphytica 179, 129–141. doi: 10.1007/s10681-011-0360-y
Wickham, H. (2016). Ggplot2 elegant graphics for data analysis, seceond edition (Dordrecht: Springer). 260. doi: 10.1007/978-3-319-24277-4
Xiang, C., Feng, J. Y., Wang, M. N., Chen, X. M., See, D. R., Wan, A. M., et al. (2016). Molecular mapping of Yr76 for resistance to stripe rust in winter club wheat cultivar Tyee. Phytopathology 106, 1186–1193. doi: 10.1094/PHYTO-01-16-0045-FI
Yao, F. J., Guan, F. N., Duan, L. Y., Long, L., Tang, H., Jiang, Y. F., et al. (2021). Genome-wide association analysis of stable stripe rust resistance loci in a Chinese wheat landrace panel using the 660K SNP array. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.783830
Yao, F. J., Long, L., Wang, Y. Q., Duan, L. Y., Zhao, X. Y., Jiang, Y. F., et al. (2020). Population structure and genetic basis of the stripe rust resistance of 140 Chinese wheat landraces revealed by a genome-wide association study. Plant Sci. 301, 110688. doi: 10.1016/j.plantsci.2020.110688
Yu, J. M., Pressoir, G., Briggs, W. H., Bi, I. V., Yamasaki, M., Doebley, J. F., et al. (2006). A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38, 203–208. doi: 10.1038/ng1702
Zegeye, H., Rasheed, A., Makdis, F., Badebo, A., Ogbonnaya, F. C. (2014). Genome-wide association mapping for seedling and adult plant resistance to stripe rust in synthetic hexaploid wheat. PloS One 9, e105593. doi: 10.1371/journal.pone.0105593
Zhou, X. L., Han, D. J., Gou, H. L., Wang, Q. L., Zeng, Q. D., Yuan, F. P., et al. (2014a). Molecular mapping of a stripe rust resistance gene in wheat cultivar Wuhan 2. Euphytica 196, 251–259. doi: 10.1007/s10681-013-1028-6
Zhou, X. L., Jia, G. Y., Luo, Y. Q., Li, X., Cai, L., Chen, X. M., et al. (2024). Fine mapping of QYrsv.swust-1BL for resistance to stripe rust in durum wheat Svevo. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1395223
Zhou, X. L., Wang, M. N., Chen, X. M., Lu, Y., Kang, Z. S., Jing, J. X. (2014b). Identification of Yr59 conferring high-temperature adult-plant resistance to stripe rust in wheat germplasm PI 178759. Theor. Appl. Genet. 127, 935–945. doi: 10.1007/s00122-014-2269-z
Keywords: wheat, stripe rust, durable resistance, genome-wide association study, marker-assisted detection
Citation: Yao F, Wang M, See DR, Yang E, Chen G and Chen X (2025) Identification of 39 stripe rust resistance loci in a panel of 465 winter wheat entries presumed to have high-temperature adult-plant resistance through genome-wide association mapping and marker-assisted detection. Front. Plant Sci. 15:1514926. doi: 10.3389/fpls.2024.1514926
Received: 21 October 2024; Accepted: 27 November 2024;
Published: 07 January 2025.
Edited by:
Guangqiang Zhang, Heze University, ChinaReviewed by:
Jindong Liu, Chinese Academy of Agricultural Sciences, ChinaJingzhong Xie, Chinese Academy of Sciences (CAS), China
Yinghui Li, University of Haifa, Israel
You-Liang Zheng, Sichuan Agricultural University, China
Copyright © 2025 Yao, Wang, See, Yang, Chen and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xianming Chen, eGlhbm1pbmcuY2hlbkB1c2RhLmdvdg==
 Fangjie Yao
Fangjie Yao Meinan Wang
Meinan Wang Deven R. See2,3
Deven R. See2,3 Guoyue Chen
Guoyue Chen Xianming Chen
Xianming Chen