
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 20 January 2025
Sec. Plant Pathogen Interactions
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1513430
This article is part of the Research TopicFruit Trees Under Stress: Physiological, Biochemical, and Molecular MechanismsView all 5 articles
Citrus bacterial canker (CBC) disease, caused by Xanthomonas citri subsp. citri (Xcc), is one of the major diseases that seriously endanger citrus production. Citrus regulates the balance of endogenous plant hormones to resist CBC through multiple synthetic pathways, including the demethylation pathways of methyl salicylate (MeSA), methyl jasmonate (MeJA) and methyl indole-3-acetic acid (MeIAA). Here, four methylesterase (MES) genes, MES1.1, MES17.3, MES10.2, and MES1.5 were screened in the transcriptomes of CBC-resistant and CBC-susceptible varieties after Xcc inoculation. Among these MES genes, the expression levels of MES10.2, MES1.1, and MES1.5 were up-regulated in CBC-resistant varieties, while MES17.3 was down-regulated in both CBC-resistant and susceptible varieties. Subcellular localization analysis showed that the four MES-encoding proteins were localized in the cytoplasm. Overexpression of CmMES1.1 and CmMES1.5 from calamondin (Citrofortunella microcarpa) significantly enhanced CBC resistance and increased the salicylic acid (SA) content in calamondin. Conversely, overexpression of CmMES10.2 and CmMES17.3 significantly reduced CBC resistance and increased the contents of jasmonic acid (JA) and indole-3-acetic acid (IAA), respectively. We concluded that the resistant varieties confer CBC-resistance by regulating the expression of CmMES1.1 and CmMES1.5 to increase SA content, and regulating CmMES10.2 and CmMES17.3 to inhibit the synthesis of JA and IAA, respectively. Their ability to regulate the endogenous SA, JA and IAA content through the demethylation pathway was an attractive breeding target for conferring CBC resistance.
Citrus bacterial canker (CBC) is a bacterial disease caused by Xanthomonas citri subsp. citri (Xcc). The pathogen mainly invades citrus plants through wounds or stomas and presents with pustule or cork-like necrotic lesions on young tissues, including leaves, fruit, and stems. In severe cases, symptoms such as leaf drop, branch dieback, and early fruit drop may occur. The appearance and quality of susceptible fruits deteriorate, and yields decrease, causing serious economic losses (Gochez et al., 2020; Das, 2003; Liu et al., 2024). The application of copper bactericides is the main measure for controlling CBC, but it seriously pollutes the environment and affects the quality of citrus (Behlau et al., 2017; Martinez et al., 2016). At present, Xcc can infect most citrus cultivars, such as lime (Citrus aurantifolia), sweet orange (C. sinensis), and grapefruit (C. paradisi). However, ‘Meiwa’ kumquat (Fortunella crassifolia), ‘Marumi’ kumquat (F. japonica), ‘Nagami’ kumquat (F. margarita) and calamondin (Citrofortunella microcarpa) are less susceptible to the disease (Ference et al., 2020; Long et al., 2019; Duan et al., 2022). Intensive studies on the mechanism differences of citrus varieties in response to CBC will provide a theoretical basis and contribute to the improvement of CBC resistance breeding.
Plant hormones are closely related to the pathogenesis of CBC in citrus. Salicylic acid (SA) played a positive role in resistance to CBC, exogenous treatments with SA increase the resistance of CBC in susceptible citrus cultivars (Wang and Liu, 2012). It is noteworthy that overexpression of the Arabidopsis NPR1 increased the CBC resistance of susceptible citrus cultivars (Zhang et al., 2020), and most recent studies showed the NPR1-like genes, as receptors for SA, could be stimulated by Xcc infection, implying their responsiveness to CBC challenges (Ali et al., 2024). In addition, SA could regulate plant resistance through antagonism and synergism with other plant hormones (Robert-Seilaniantz et al., 2011). For example, JA could antagonize SA-mediated immune responses, promoting plant susceptibility to pathogens (Yang et al., 2017). Similarly, MeJA treatment resulted in CBC susceptibility, and SA treatment significantly enhanced the CBC resistance in Wanjincheng orange (C. sinensis). However, the accumulated JA inhibited effective SA-mediated defense and promoted CBC symptom formation (Long et al., 2019). Furthermore, auxin promoted CBC susceptibility in citrus, while the inhibitor of gibberellin synthesis, chlorocholine chloride (CCC), antagonized auxin signaling and inhibited CBC symptom formation (Cernadas and Benedetti, 2009).
The demethylation pathway of plant hormones played an important role in plant immune response, the demethylation of the methyl esters of IAA, SA, and JA was catalyzed by methylesteras (MES), which belonged to the α/β hydrolase superfamily (Nardini and Dijkstra, 1999). SABP2 (salicylic acid binding protein 2), a tobacco methyl salicylate (MeSA) esterase, was essential for the development of systemic acquired resistance (SAR) (Kumar and Klessig, 2003; Forouhar et al., 2005; Gong et al., 2023). Previous studies showed that the common beans PvMES1, soybean GmSABP2-1, potato StMES1 and Lycium chinense LcSABP also had salicylate methyl esterase activity, which could convert methyl salicylate to SA and participate in stress response (Xue et al., 2021; Lin et al., 2024; Manosalva et al., 2010; Li et al., 2019). In Arabidopsis thaliana genome, twenty MESs homologous to SABP2 were identified by protein homology analysis and named AtMES1-AtMES20 (Yang et al., 2008). AtMES1/7/9 were induced during pathogen infection, and overexpression of AtMES1/7/9 in SABP2-silenced tobacco could restore SAR deficiency while silencing them could lead to MeSA accumulation (Vlot et al., 2008; Gao et al., 2021). Similarly, MES family genes from Brassica oleracea var. Capitata actively responded to Plasmodiophora brassicae infection (Manoharan et al., 2016). FvMES2 from strawberry (Fragaria vesca) were involved in MeSA demethylation and responded significantly to Botrytis cinerea stress through the SA signaling pathway (Jia et al., 2024). Furthermore, some MES proteins showed specificity and preference to the specific substrate, while some MES proteins shared multiple methylesterase activity. The substrate specificity test of AtMES proteins showed that five, six and eight AtMES proteins had salicylate methyl esterase activity, jasmonate methyl esterase activity and methyl IAA esterase activity, respectively (Yang et al., 2008; Vlot et al., 2008). AtMES17 had only IAA methylesterase activity, while AtMES1 had higher SA methylesterase activity and lower JA and IAA methylesterase activity (Yang et al., 2008; Vlot et al., 2008). VvMES5 from grape (Vitis Vinifera) showed similar jasmonate methyl esterase activity as its homologue AtMES10 (Zhao et al., 2016). PpMES1 from peach (Prunus persica L. Batsch) only had methyl jasmonate esterase activity, while PpMES2 had methyl jasmonate and salicylate methyl esterase activity (Cao et al., 2019). CsMES1 from sweet orange (C. sinensis), a homologue of tobacco SABP2, could convert MeSA to SA, and its inhibitor promoted CBC symptom formation, suggesting that CsMES1 might play a positive role in CBC resistance (Lima Silva et al., 2019). However, the activity of most MES family genes and their function in CBC remain unclear.
To understand the function of MES family genes responded to CBC in citrus, four MES family genes were obtained by comparing the transcriptomes of CBC- resistant and CBC-susceptible varieties. The cytoplasmic localization of the four candidate genes from calamondin was determined through the transient transformation of Nicotiana benthamiana leaves. The results of Xcc inoculation and plant hormones determination of transiently overexpressing calamondin and ‘Taoyecheng’ sweet orange leaves showed that overexpression of CmMES1.1 and CmMES1.5 enhanced CBC resistance and increased SA content, while overexpression of CmMES17.3 and CmMES10.2 promoted citrus canker disease development and increased auxin and jasmonic acid content, respectively. These MES genes might be important genetic resources for screening and breeding CBC-resistant varieties.
The calamondin and ‘Taoyecheng’ sweet orange were grown in a greenhouse at 25 ± 1°C in Wuhan, China. The leaves of calamondin and ‘Taoyecheng’ sweet orange were utilized in Xcc inoculation experiments and transient overexpression analyses. The Xcc strain was routinely cultured at 28°C in an Lysogeny Broth (LB) solid culture medium.
The transcriptome data of CBC-resistant ‘Meiwa’ kumquat (F. crassifolia) and CBC-susceptible ‘Mexican’ lime (C. aurantifolia) responding to Xcc infection at 24 hpi, 48 hpi and 72 hpi were obtained through CitrusKB database (http://bioinfo.deinfo.uepg.br/citrus/) (Supplementary Table S1).
In addition, the young leaves of calamondin were selected for Xcc in vivo inoculation. Xcc was cultured overnight at 28 °C. The activated bacterial solution was diluted to OD=0.6 (108 cfu/ml) with sterile water and further diluted to 104 cfu/ml for infiltration inoculation. Sterile water inoculation was used as a blank control. Subsequently, leaf samples were collected at different time points 1 dpi, 3 dpi and 5 dpi for transcriptome analysis and differentially expressed genes (DEGs) analysis. (Supplementary Table S2).
The whole MES proteins of ‘Hong Kong’ kumquat (F. hindsii) were downloaded from the CPBD database (http://citrus.hzau.edu.cn/index.php, accessed on 13 May 2024), and 20 MES proteins of A. thaliana were obtained from the TAIR database (https://www.arabidopsis.org/, accessed on 13 May 2024). Four candidate MES proteins from calamondin were cloned and sequenced; the primers were listed in Supplementary Table S3. Then, the phylogenetic relationships of the MES proteins were constructed using the neighbor-joining method by MEGA 11.0 software with the following parameters: Poisson model, pairwise deletion, and 1000 bootstrap replications. The phylogenetic trees were visualized using the iTOL web package. All the gene accession numbers and protein sequences are listed in Supplementary Tables S4.
The structure of MES proteins from F. hindsii was analyzed using the MEME Suite 5.5.2 online program (Multiple Em for Motif Elicitation 5.5.2, http://alternate.meme-suite.org/tools/meme, accessed on) and TBtools (https://github.com/CJChen/TBtools, accessed on). The 2000-bp promoter sequences of the upstream regions of MES proteins were obtained. Cis-acting elements were predicted via PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on), and the responsive regulatory elements were analyzed via TBtools.
Due to CBC mainly infects young leaves, stems and fruits of citrus, young and mature leaves, stem and fruit were used for tissue-specific analysis of four methylesterase family genes.
Young leaves of calamondin were selected for plant hormone response analysis. 500 µM SA, 100 mg/L IAA, 5 mg/L GA3, 200µM mg/L methyl Jasmonate, and 100 mM abscisic acid (ABA) were used to spray the calamondin leaves. Hormone-treated leaf samples were collected 1, 2, and 3 days after spraying, and untreated leaf samples were used as a control.
The total RNA of calamondin leaves was extracted using the RNA prep Pure Plant Kit (Tiangen, Beijing, China), and the cDNA was obtained by the Monad MonScriptTMRTIII Super Mix with dsDNase (Two-Step) (Monad, Suzhou, China). The Monad MonAmpTM SYBR® Green qPCR Mix (Vazyme, Nanjing, China) and Applied Biosystems PCR-7500 (ABI, USA) were used for amplification and the qRT-PCR analysis. The primers used for the qRT-PCR are shown in Supplementary Table S3. Three biological replicates were conducted.
The CDS sequences of CmMES1.1 (441 bp), CmMES17.3 (885 bp), CmMES10.2 (642 bp) and CmMES1.5 (804 bp) from calamondin were inserted into the PK7203-RFP vector to generate a fusion protein of the target gene with RFP. First, PK7203-RFP vector was digested by Sall-HF enzyme, and then the coding sequence of each MES gene was cloned into the vector by homologous recombination cloning technology. The overexpression vector includes an enhanced green fluorescent protein (EGFP) activated by minimal CaMV 35S promoter as a screening marker, and an MES protein fusion mRFP promoted by minimal CaMV 35S promoter for subcellular localization. Please refer to Supplementary Figure S1 for detailed map. The specific steps were as follows: first, the single colony of each Agrobacterium strain EHA105 was cultured on LB medium at 28 °C overnight; then resuspended with infiltration buffer (10 mM 2-(N-morpholino) ethanesulfonic acid, pH 5.85; 10 mM MgCl2; 30 mg/L acetosyringone) to OD=0.6; finally the infiltration buffer was injected into the leaves of Nicotiana benthamiana, which was then cultured in the dark for 12 hours and 16-h/8-h light/dark photocycle for 2 days. The fluorescence was observed through a laser scanning confocal microscope (TCS-SP8 SR, Leica, Wetzlar, Germany). The primers used for the construction of the overexpression vector are shown in Supplementary Table S3.
The above four EHA105 strains for subcellular localization analysis were used in transient transformation assay, with the leaves of calamondin and ‘Taoyecheng’ sweet orange (C.sinensis) as explants. The optimized transient transformation method was used to obtain overexpressed calamondin leaves. The specific steps were as follows: first, the single colony of each Agrobacterium strain EHA105 was grown at 28 °C overnight; then resuspended with infiltration buffer (10 mM 2-(N-morpholino) ethanesulfonic acid, pH 5.85; 10 mM MgCl2; 30 mg/L acetosyringone) to OD=1.0-1.5; finally injected the infiltration buffer into the young calamondin leaves and ‘Taoyecheng’ sweet orange with a needleless syringe, and injected repeatedly for three times at one-hour intervals and cultured in the dark for 2 days. Finally, the instantaneous transformation of Agrobacterium was determined by green fluorescence observation (Supplementary Figures S2).
To determine the CBC resistance ability of the four key MES genes, leaves were picked from the transiently transformed calamondin and ‘Taoyecheng’ sweet orange leaves for in vitro Xcc inoculation. 0.1 mL Xcc (108 cfu/ml) was infiltrated into the calamondin leaves, and 5 µL Xcc (108 cfu/ml) was dripped onto each puncture site made with a pin (0.5 mm in diameter) on’Taoyecheng’sweet orange leaves. The leaves were then cultured in an incubator at 28°C, with 80% relative humidity and a 16-h/8-h light/dark photocycle. CBC symptom and resistance degree were evaluated at 5 and 15 days post-inoculation (dpi) of calamondin and ‘Taoyecheng’ sweet orange leaves, respectively. Lesion area was analyzed by ImageJ software, and all experiments were repeated at least three times.
Approximately 100 mg of frozen calamondin leaves were ground and extracted with 1 ml ice-cold 50% aqueous acetonitrile (vol/vol). Subsequently, the samples were sonicated for 3 min and extracted for 30 min at 4°C. The supernatant was obtained after centrifugation (10 min, 12,000 rpm, 4°C) and purified using C18 reversed-phase. After this solid phase extraction, the samples were blown dry by nitrogen and dissolved in 200 μl of 30% acetonitrile (vol/vol). Ultra-efficient liquid chromatography (Vanquish, UPLC, Thermo, USA) and high-resolution mass spectrometry (Q Exactive, Thermo, USA) were used to determine the plant hormone levels. The analytical conditions were as follows: column: Waters HSS T3 (50×2.1 mm, 1.8 μm); mobile phase: phase A is ultra-pure water (containing 0.1% acetic acid) and Phase B is acetonitrile (containing 0.1% acetic acid). The data were collected using the Q Exactive high-resolution mass spectrometry system from Thermo Fisher Scientific and processed using TraceFinder Software. Triplicates were conducted for determine each plant hormone.
All data analyses were performed using GraphPad Prism 6.0 (GraphPad, USA), the results were presented as means ± standard deviation (SD), and comparisons were made using ANOVAs with Duncan’s multiple range test.
The transcriptome data of CBC-resistant ‘Meiwa’ kumquat (F. crassifolia) and calamondin (Citrofortunella microcarpa), and CBC-susceptible ‘Mexican’ lime (C. aurantifolia) in response to Xcc infection were re-analyzed with Tbtools. Eight differentially expressed MES genes were screened from CBC-resistant calamondin, including MES1.1, MES1.4, MES17.3, MES17.1, MES10.2, MES1.5, MES1.3 and MES11.5. Among them, MES1.1, MES17.3, MES10.2 and MES1.5 were shared by CBC-resistant ‘Meiwa’ kumquat and calamondin and CBC-susceptible ‘Mexican’ lime. Compared with the control, the expression of MES1.1, MES10.2 and MES1.5 were down-regulated at 24 hpi after Xcc inoculation in CBC-susceptible ‘Mexican’ lime, but up-regulated at 24 hpi hpi, 48 hpi and 72 hpi after Xcc inoculation in CBC-resistant ‘Meiwa’ kumquat and up-regulated at 1 dpi and/or 3 dpi and/or 5dpi after Xcc inoculation in CBC-resistant calamondin. The expression of MES17.3 was down-regulated in all three citrus species. The expression level of MES17.3 was decreased by 7.5 and 3.1 times at 48 hpi and 72 hpi after Xcc inoculation in CBC-susceptible ‘Mexican’ lime, respectively (Figure 1A). However, it was decreased by 16.8, 54.2 and 26.9 times at 24 hpi, 48 hpi and 72 hpi in CBC-resistant ‘Meiwa’ kumquat and 1.8, 1.5 and 8.5 times at 1 dpi, 3 dpi and 5 dpi in CBC-resistant calamondin (Figures 1B, C). These results indicated that MES family genes played an important role in CBC resistance, among which MES1.1, MES17.3, MES10.2 and MES1.5 might be key regulatory genes.
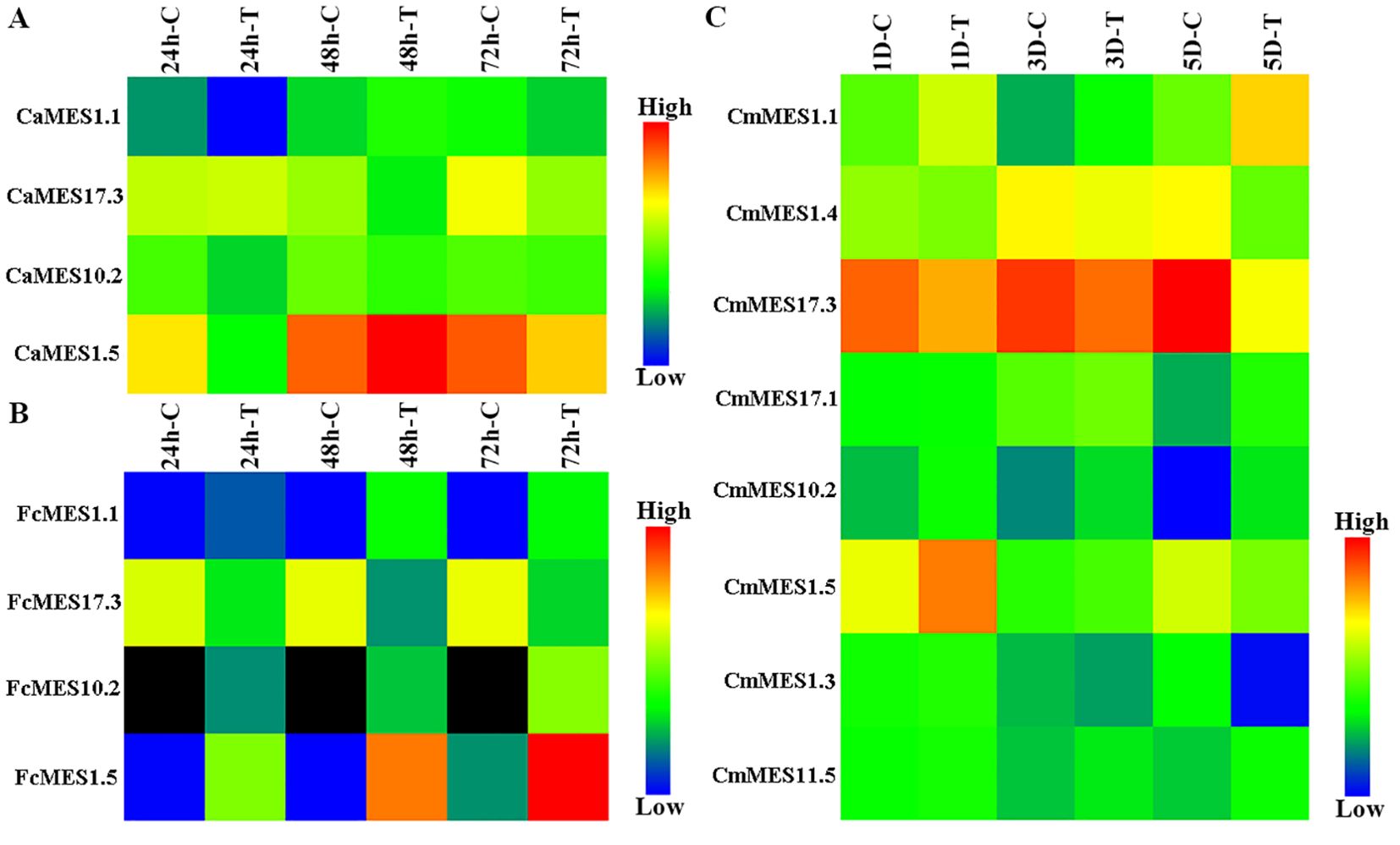
Figure 1. Expression heatmap of MES family genes after Xcc infection. (A) ‘Mexican’ lime (C. aurantifolia). (B) ‘Meiwa’ kumquat (F. crassifolia). (C) Calamondin (Citrofortunella microcarpa). hpi, hour post-inoculation; dpi, day post-inoculation; C, control; T, treatment. Black indicates no detection.
The expression patterns of four MES genes (CmMES1.1, CmMES17.3, CmMES10.2, and CmMES1.5) that responded to Xcc were further analyzed by qRT-PCR in calamondin. Young leaves of calamondin were infiltrated with low (104 cfu/ml) and high (108 cfu/ml) concentrations of Xcc suspension and analyzed at four stages (1 dpi, 3 dpi, 5 dpi, and 7 dpi) after Xcc inoculation. Compared with sterile water inoculation used as a blank control, the leaves of calamondin inoculated with a low concentration of Xcc showed no obvious symptoms within 7 dpi, while the leaves inoculated with a high concentration of Xcc had obvious pustule symptoms after 5 dpi and appeared hypersensitive necrotic after 7 dpi (Figure 2A). When inoculated with a high concentration of Xcc, the expression levels of CmMES1.1, CmMES10.2 and CmMES1.5 were all significantly down-regulated at an early stage (1 dpi and 3 dpi) and up-regulated at a later stage (5 dpi and/or 7 dpi) (Figure 2B). However, the expression level of CmMES17.3 was significantly down-regulated at the whole stage, particularly at the later stage (Figure 2B). The expression levels of the four MES genes were only slightly regulated by low concentration Xcc infection. For example, all the four MES genes were slightly up or down regulated at the early stage (1 dpi and 3 dpi). Although CmMES1.1 and CmMES10.2 were significantly up-regulated at 5 dpi, CmMES17.3 and CmMES1.5 were significantly down regulated at 5 dpi and 7 dpi, their up-regulation and down-regulation levels were significantly lower than those with high concentration Xcc infection. These results indicated that the function of these MES genes might be strongly activated at the later stage upon Xcc infection associated with the development of the symptoms.
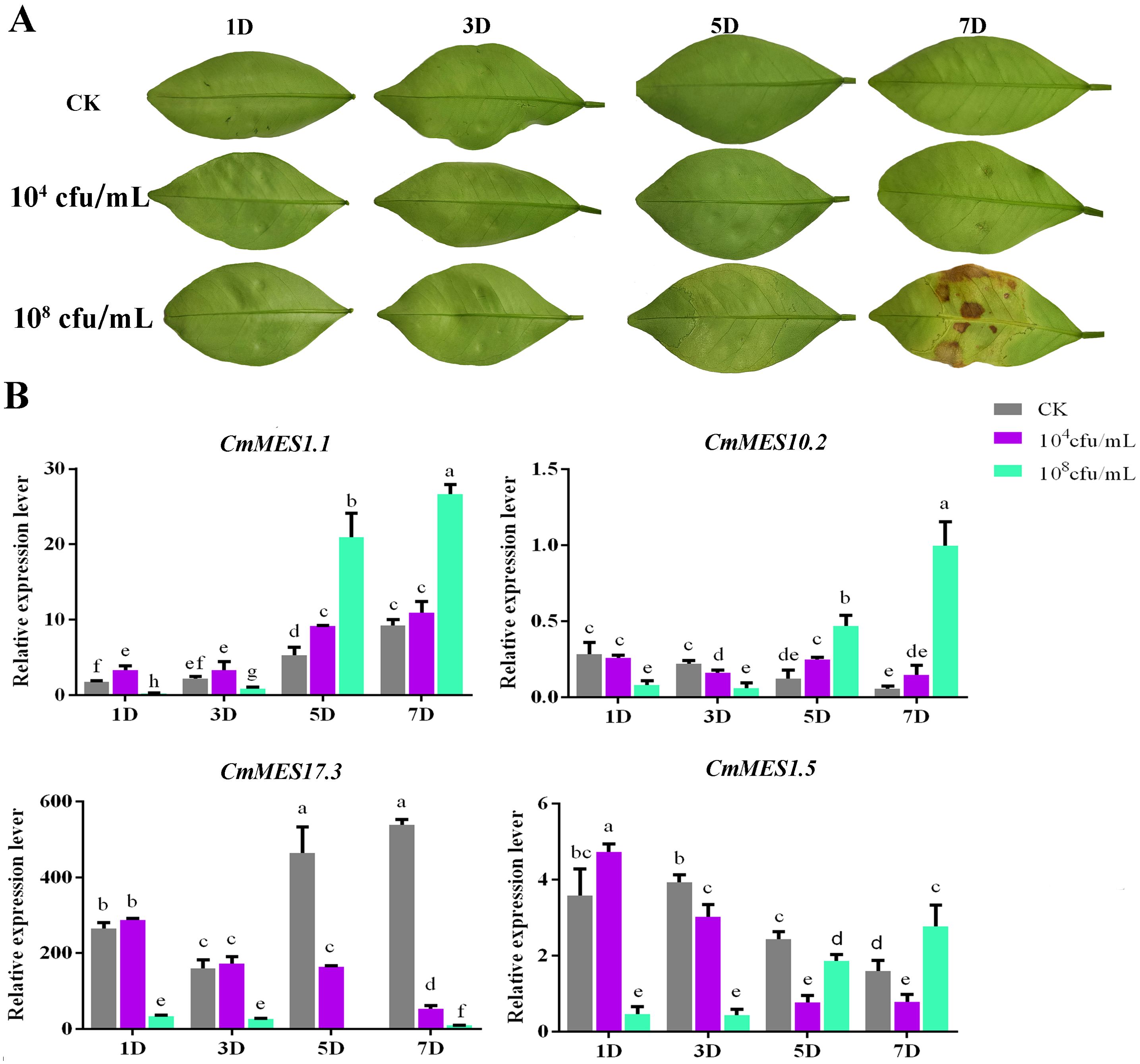
Figure 2. The response of four key MES genes upon Xcc infection. (A) Phenotypic observation of young leaves of calamondin inoculated with low (104 cfu/ml) and high (108 cfu/ml) concentrations of Xcc. (B) The expression patterns of four key MES genes inoculated with low and high concentrations of Xcc (p < 0.05, ANOVAs with Tukey’s multiple range test).
To explore the phylogenetic relationships of kumquat MES proteins, we constructed a phylogenetic tree based on the amino acid sequences of 12 kumquat MES proteins and 20 A.thaliana MES proteins. In the kumquat (F. hindsii) genome, Sjg142100, Sjg239980, and Sjg214930 contained two spliceosomes, Sjg072760 had three spliceosomes, and the rest had only one spliceosome. Based on comparison analysis with A.thaliana MES proteins, the kumquat MES family genes were mainly divided into six groups, namely group AtMES1 (FhMES1.1-FhMES1.5), group AtMES10 (FhMES10.1 and FhMES10.2), group AtMES11 (FhMES11.1-FhMES11.5), group AtMES14 (FhMES14), group AtMES17 (FhMES17.1-FhMES17.3) and group Sjg190120 (Figure 3). In addition, four candidate CBC resistance-related MES genes from calamondin were cloned and sequenced, and they belonged to FhMES1.1, FhMES17.3, FhMES10.2 and FhMES1.5 (Supplementary Figure S3). Gene structure analysis showed that the kumquat MES family genes contained 2 - 6 exons. Promoter element analysis showed that the promoters of kumquat MES family genes contained a large number of plant hormone-responsive elements (SA, auxin, methyl jasmonate, ABA (abscisic acid), and gibberellin), stress (low temperature and drought) and defense-related response elements (Figure 3).
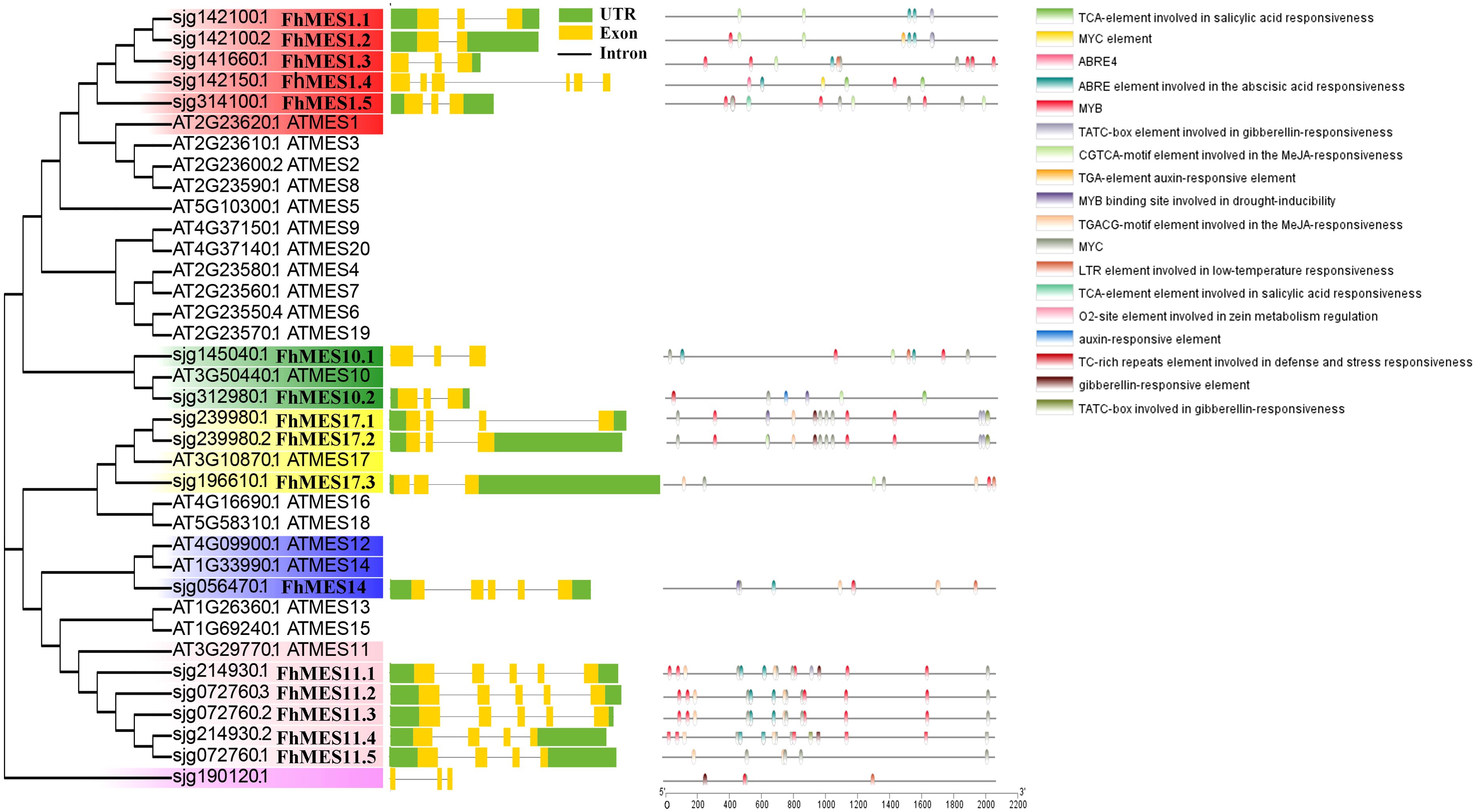
Figure 3. The phylogenetic relationship, gene structure and promoter element analysis of the MES family proteins in kumquat. Conserved MES proteins from kumquat and A. thaliana were aligned using the ClustalW function of MEGA11. The phylogenetic tree (with 1000 replicates) was constructed by NJ method and bootstrapping analysis. Different colors represent different types of MES proteins in kumquat. Gene structure of MES genes in kumquat. The yellow bar indicates the coding sequence (CDS), the line indicates the intron, and the green bar indicates the untranslated region (UTRs). The cis-acting element within the 2000-bp upstream sequence of the kumquat MES gene. This study used the database PlantCARE to predict the motif, different colors represent different promoter elements.
Therefore, the MES genes might be involved in response of plant hormones including SA, IAA, JA, ABA or GA.
The stems, leaves, and fruits of calamondin at different growth stages were used to analyze the tissue-specific expression patterns of CmMES1.1, CmMES17.3, CmMES10.2, and CmMES1.5. The results indicated that CmMES1.1 was mainly expressed in fruits. However, the expression level was higher in the young tissues of leaves and stems than in mature tissues. CmMES17.3 was mainly expressed in stems, especially in mature stems. CmMES10.2 and CmMES1.5 were highly expressed in young tissues, particularly in young fruits (Figure 4A). Furthermore, the four MES genes were regulated by different plant hormones. CmMES1.1 and CmMES1.5 were down-regulated by SA treatment and up-regulated by JA treatment, meanwhile, CmMES1.5 was also significantly up-regulated by ABA treatment. CmMES17.3 was mainly up-regulated by JA treatment and CmMES10.2 was mainly up-regulated by ABA treatment (Figure 4B). These results revealed that the four MES genes might prone to express in young tissues and be involved in different plant hormone pathways.
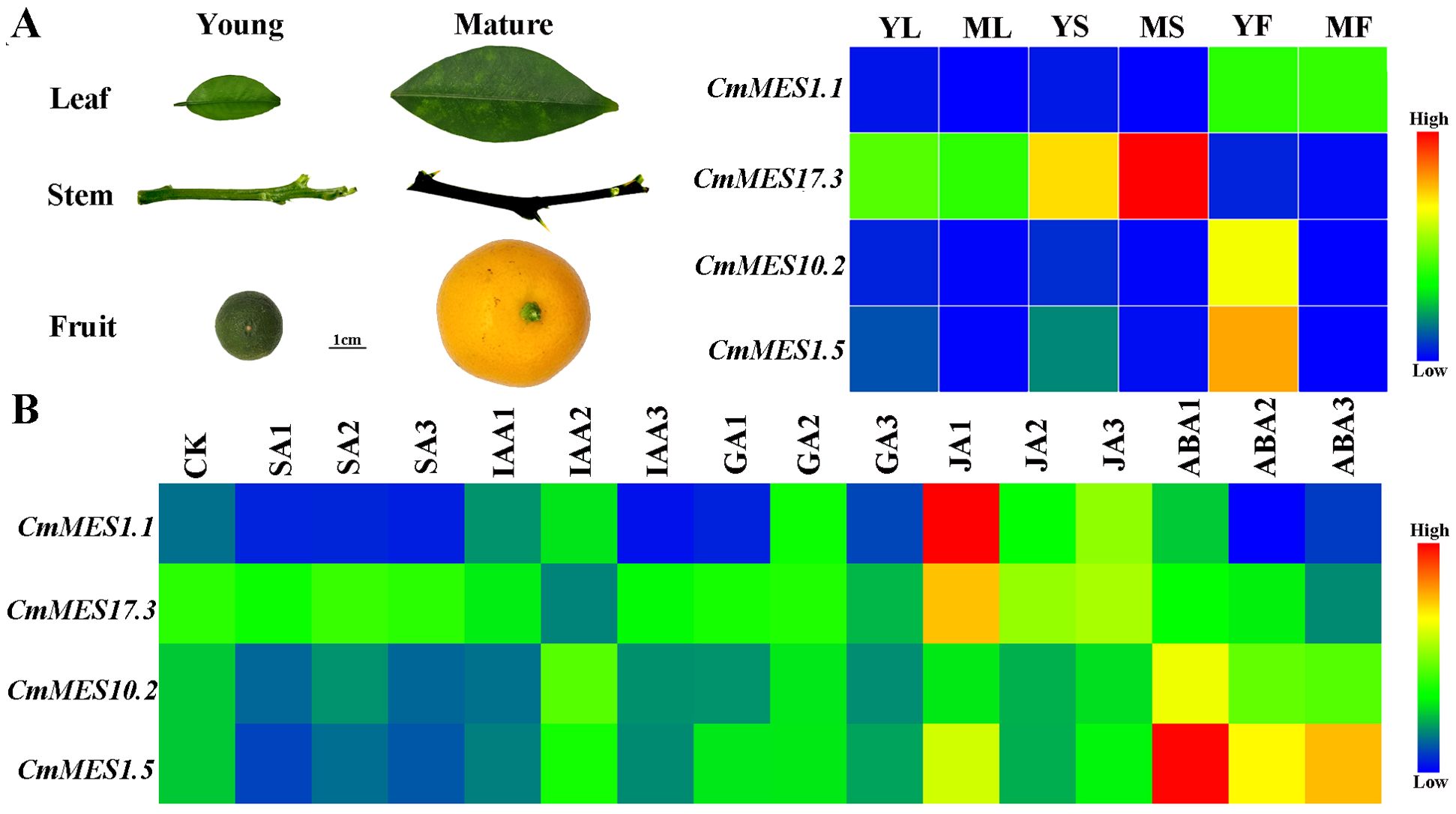
Figure 4. The expression patterns of four calamondin MES genes in different tissues and in response to different hormone treatments. (A) The tissue-specific expression patterns of four MES genes. YL, Young leaf; ML, Mature leaf; YS, Young stem; MS, Mature stem; YF, Young fruit; MF, Mature fruit. (B) The expression patterns of four MES genes at 1, 2, and 3 days after different plant hormone treatments. CK, samples untreated for 0 days; SA, 500µM SA treatment; IAA, 100 mg/L indole-3-acetic acid treatment; GA, 5 mg/L GA3 treatment; JA, 200µM methyl Jasmonate treatment; ABA, 100 mM abscisic acid treatment.
To examine the subcellular localization of the four MES proteins, the positive control (RFP), CmMES1.5-RFP, CmMES1.5-RFP, CmMES10.2-RFP, and CmMES17.3-RFP were introduced into N. benthamiana leaves by Agrobacterium-mediated transient transformation. Three days later, fluorescence of all four MES genes fused to RFP could be observed in the cytoplasm (Figure 5). The result indicated that the four MES proteins might function in the cytoplasm.
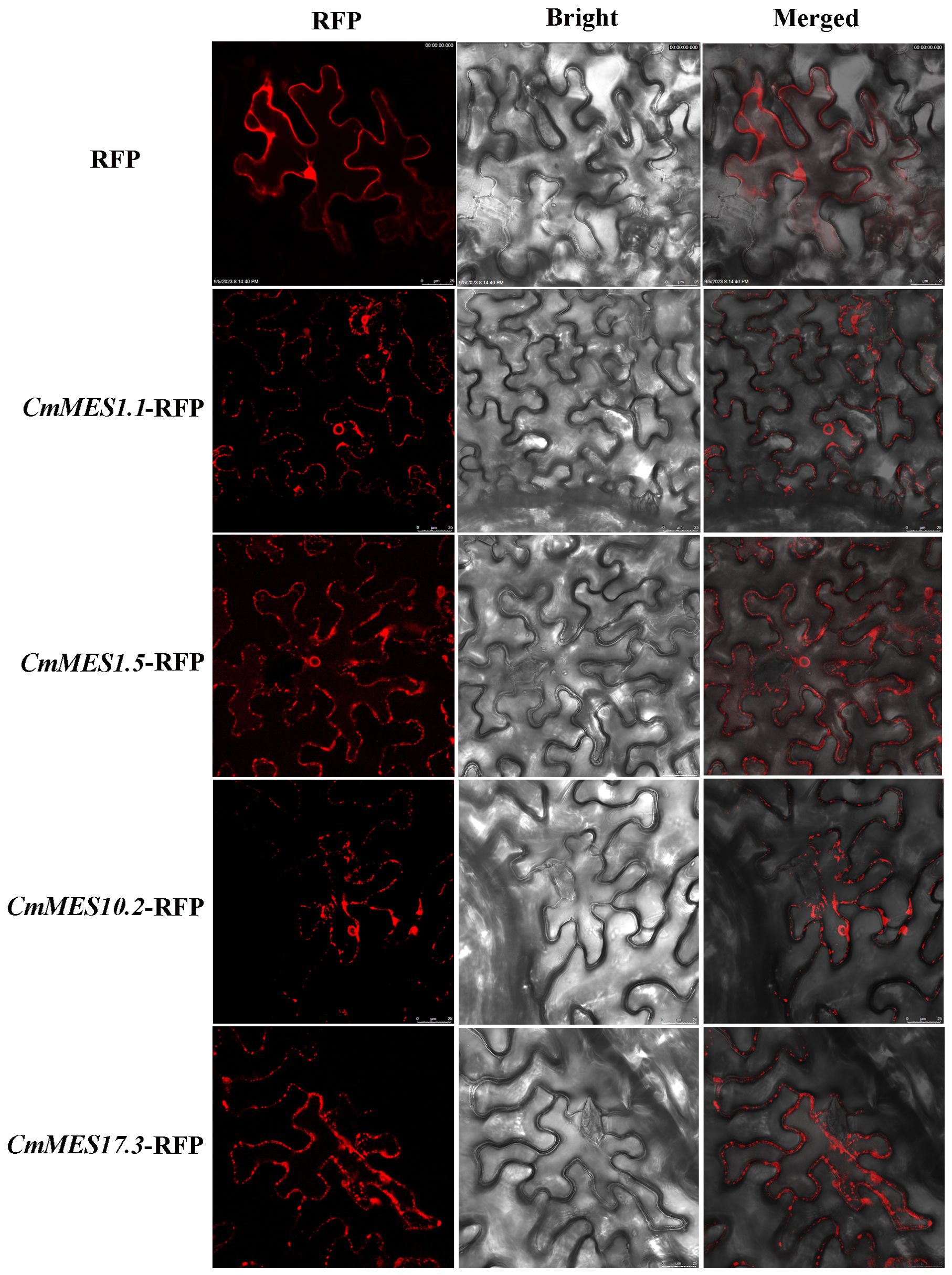
Figure 5. Subcellular localization of four calamondin MES genes in N. benthamiana leaves. Fields of view are shown as fluorescence field, bright field, and merged images, scale bar = 25 μm.
Transient overexpression in leaves of calamondin and ‘Taoyecheng’ sweet orange by infiltration were employed to verify the functions of the four MES genes. Green fluorescence observation showed that four key MES genes were successfully overexpressed in the leaves (Figures 6A, B). The quantitative analysis results showed that the expression levels of the four methylesterase genes were significantly increased in the transiently overexpressed citrus leaves compared with the control (Supplementary Figures S4). To identify the function of four key MES genes associated with CBC-resistance, transiently overexpressed leaves were in vitro inoculated with high concentrations (108 cfu/ml) of Xcc suspension. The results of phenotypic observation and disease area statistics showed that overexpression of CmMES17.3 and CmMES10.2 increased the disease area, while overexpression of CmMES1.1 and CmMES1.5 decreased the disease area compared with the controls (Figures 6B, C). These results indicated that CmMES17.3 and CmMES10.2 might be negative regulatory factors for CBC resistance, while CmMES1.1 and CmMES1.5 might be positive regulatory factors.
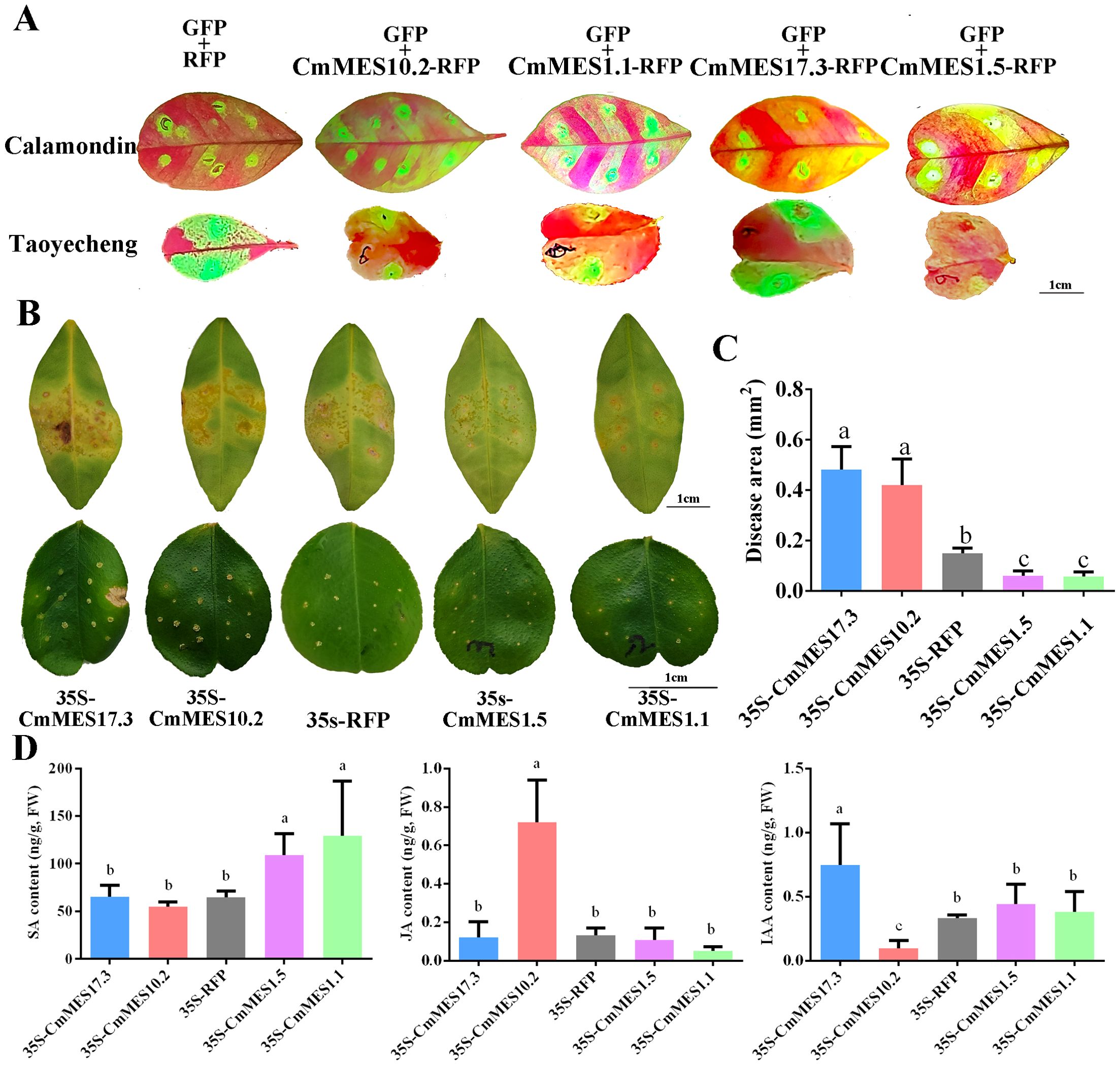
Figure 6. Functional identification of four citrus MES genes associated with CBC resistance. (A) Identification of transiently transgenic citrus leaves by green fluorescence observation. The overexpression vector contains two separate promoter regions. RFP is fused to the target gene for subcellular localization, and GFP is driven alone for fluorescence verification. (B) Citrus canker symptoms on transiently transgenic citrus leaves after in vitro infiltration inoculation with Xcc. The leaves above are calamondin leaves. 0.1mL 108 cfu/ml of Xcc was injected into the leaves, and the water-soaked protrusions on the leaf surface were observed to evaluate CBC resistance. The leaves below are ‘Taoyecheng’ sweet orange. The CBC resistance was determined by dripping with 5 µL Xcc (108 cfu/ml) to each puncture site made with a pin (0.5 mm in diameter), and the CBC resistance was evaluated by counting the lesion area. Scale bar = 1.0 cm. (C) Disease area on leaves of transiently transgenic citrus after Xcc inoculation (p < 0.05, ANOVAs with Tukey’s multiple range test). (D) Determination of SA, JA and IAA contents in calamondin leaves overexpressing four key MES genes. Different letters indicate significant differences (p < 0.05, ANOVAs with Tukey’s multiple range test). Data are the mean ± SD (n=3).
The transiently overexpressed calamondin leaves were further employed to calculate the contents of SA, JA and IAA. The results showed that overexpression of CmMES1.1 and CmMES1.5 significantly increased the content of SA, and CmMES17.3 increased the content of IAA, however, CmMES10.2 increased the JA level (Figure 6D).
CBC, as a major disease, seriously threatens the development of the citrus industry (Martins et al., 2020). Citrus endogenous hormones are important substances in response to CBC, especially SA, JA, and auxin. MES family genes participate in the demethylation pathway of MeSA, MeJA, and MeIAA, and produce active plant endogenous hormones to regulate plant resistance (Yang et al., 2008). In this study, we identified 13 FcMES genes which were divided into six groups in CBC-resistant kumquat. Among them, the MES genes in five groups were homologous MES genes of Arabidopsis, namely AtMES1, AtMES10, AtMES11, AtMES12/14, and AtMES17. In the meantime, AtMES1, AtMES10, AtMES11, AtMES14, and AtMES17 also have homologous genes in grapes (Zhao et al., 2016). Furthermore, we demonstrated that CmMES1.1 and CmMES1.5 (homolog of AtMES1), CmMES10.2 (homolog of AtMES10) and CmMES17.3 (homolog of AtMES17) have a similar cytoplasmic localization as NtSABP2 (homolog of AtMES1) in tobacco and AtMES7 in Arabidopsis (Soares et al., 2022; Gao et al., 2021). These results indicated that the MES genes might have conserved functions during plant evolution.
Previous researches revealed that MES family genes played an important role in response to pathogen infection. For example, GmSABP2-1 encodes methyl salicylate esterase and functions in soybean defense against soybean cyst nematode, and Citrus sinensis CmMES1 play a positive role in the defense against CBC (Lin et al., 2024; Lima Silva et al., 2019). In this study, MES1.1, MES1.5 and MES10.2 were substantially upregulated at a later stage after Xcc infection in CBC-resistant varieties, but downregulated at a later stage after Xcc infection in CBC-susceptible varieties. MES17.3 was down-regulated after Xcc infection, but the down-regulated level in CBC-resistant varieties was higher than that in CBC-susceptible varieties. These results implied that the expression of these MES genes might be associated with CBC resistance. In addition, the expression of the four MES genes was strongly activated or inhibited by high-concentration Xcc infection, but slightly activated or inhibited by low-concentration Xcc infection. Besides, the expression was strongly activated or inhibited at later stage (5D and 7D) upon Xcc infection. According to the symptomatic reactions, the leaves inoculated with high concentration Xcc showed severe water soaking at 5D, followed by hypersensitive necrosis at 7D. However, only slight water soaking was shown at the 7D after low concentration Xcc infection. In addition, MES family genes contain a large number of hormone response elements, such as ABRE which can be bound by abscisic acid responsive element (ABRE)-binding factor (ABF) (Han et al., 2024). Although these hormone response elements have been shown to respond to certain plant hormone treatments, the upstream regulatory mechanisms of many response elements are still unclear. Studies have shown that the content of ABA, SA, and JA will increase after Xcc infection, we hypothesized that the related transcription factors, such as ABFs would change with increasing plant hormone contents, which activate or inhibit the expression of MES family genes (Long et al., 2019). These results revealed that the expression of these MES genes might be associated with the development of CBC symptoms. Furthermore, these MES genes tended to be more highly expressed in young citrus tissues (including stems, leaves, and fruits) which were the major Xcc infection sites. Taken together, these MES genes might play crucial roles in CBC.
The MES family genes had catalytic activity for MeSA, MeJA, and MeIAA, and were usually important regulators of plant endogenous hormones in response to pathogen infection in plants (Vlot et al., 2008; Yang et al., 2008). In A. thaliana, AtMES1 mainly had the MeSA methylesterase activity, while the methylesterase activity of MeIAA and MeJA of AtMES1 was only 2% and 8%, respectively (Vlot et al., 2008). In sweet orange, the inhibitor of CsMES1 decreased the SA content and inhibited CBC resistance (Lima Silva et al., 2019). In this study, CmMES1.1 and CmMES1.5 were significantly up-regulated in CBC-resistant varieties but down-regulated in CBC-susceptible varieties at later stage after Xcc infection. Overexpression of CmMES1.1 and CmMES1.5 enhanced CBC resistance and significantly increased the SA content, but did not significantly increase IAA and JA content. Our results might explain why CBC-resistant varieties accumulated more SA than CBC-susceptible varieties after Xcc infection (Long et al., 2019). Therefore, CmMES1.1 and CmMES1.5 might be important genetic loci for CBC-resistant varieties to resist CBC. Furthermore, IAA could negatively regulate disease resistance by antagonizing the SA signaling pathway via JA (Xu et al., 2024). The previous study revealed that in the initial stage of citrus canker, exogenous NAA, an auxin analog, could significantly promote lesions formation (Cernadas and Benedetti, 2009). In Arabidopsis, AtMES17 had IAA methylesterase activity and could enhance the production of active IAA (Yang et al., 2008). In this study, the expression of CmMES17.3 were substantially inhibited by Xcc infection. Overexpression of CmMES17.3 promoted CBC susceptibility and increased the content of IAA. These results indicated that citrus might resist Xcc infection by inhibiting the expression of CmMES17.3 to reduce the production of MeIAA methylesterase, hindering the conversion of inactive MeIAA into active IAA. In addition, JA could antagonize the function of SA, and negatively regulate plant resistance (Jiao et al., 2022; Huang et al., 2023). For example, JA could antagonize the function of SA to regulate Arabidopsis immunity and promote Pseudomonas syringae infection (Gupta et al., 2020). In this study, overexpression of CmMES10.2 promoted CBC susceptibility and increased the content of JA. At present, only one S gene LATERAL ORGAN BOUNDARIES 1 (LOB1) had been identified to be induced by Xcc, which enhanced CBC resistance after CRISPR gene editing and antisense oligonucleotide silencing (Hu et al., 2014; Jia et al., 2016; Peng et al., 2017; Su et al., 2023; de Lima et al., 2024). Therefore, CmMES17.3 and CmMES10.2 might provide alternative genes for gene editing to breed CBC-resistant varieties.
Thus, a model was proposed to explain how MESs confer CBC resistance by manipulating plant endogenous hormone balance in citrus. Xcc infection might lead to increased expression of CmMES1.1, and CmMES1.5, which promoted SA production, and decreased expression of CmMES17.3, which inhibited IAA production (Figure 7).
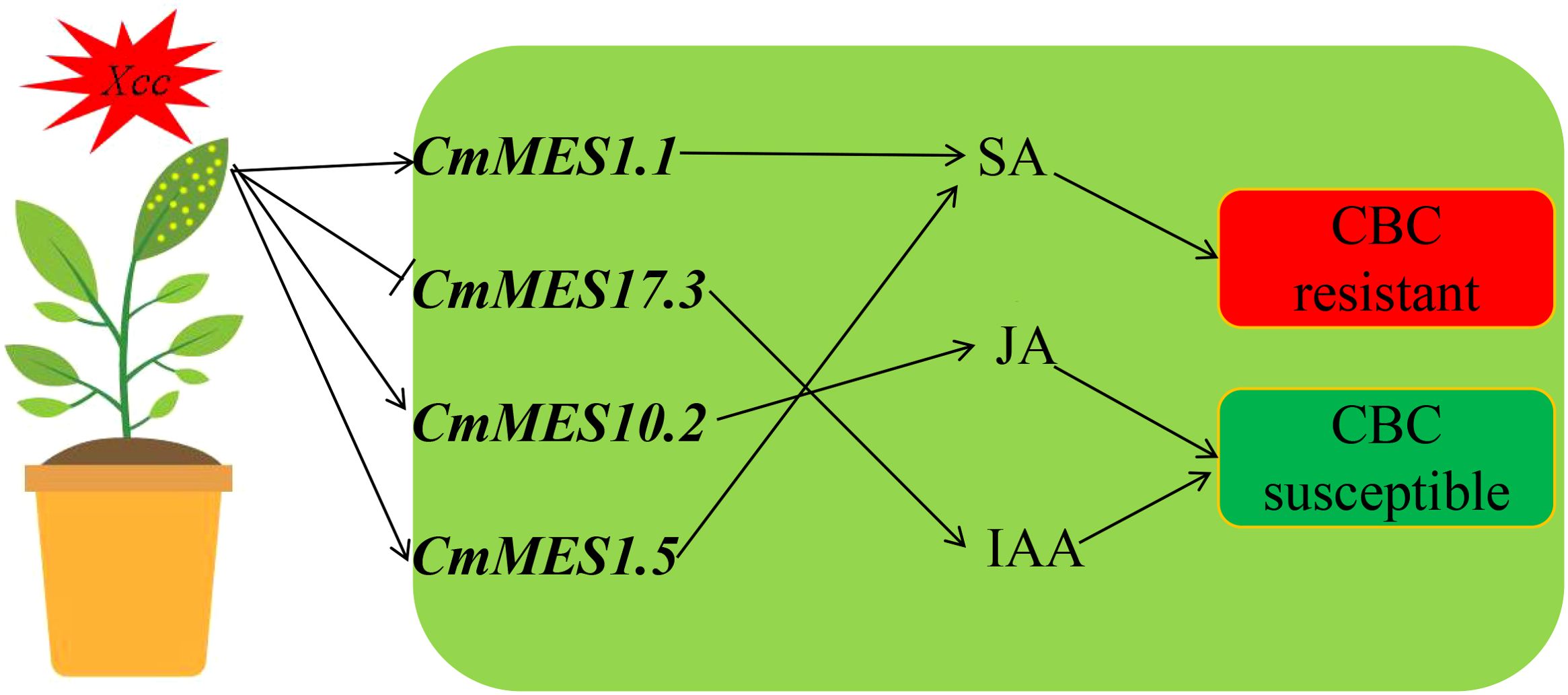
Figure 7. Model for CmMES1.1, CmMES17.3, CmMES10.2, and CmMES1.5 regulating CBC resistance during infection with Xcc. After Xcc infection, the transcript levels of CmMES1.1 and CmMES1.5 in CBC-resistant varieties were significantly increased, increasing SA content, while the transcript level of CmMES17.3 was significantly decreased, reducing IAA content, thereby enhancing canker resistance. The transcript level of CmMES10.2 was also induced to increase, increasing the JA content, increasing the JA content and enhancing canker sensitivity.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Y-XX: Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft. CX: Investigation, Methodology, Resources, Writing – review & editing. ZT: Methodology, Resources, Validation, Writing – review & editing. X-JH: Investigation, Methodology, Resources, Writing – original draft. Z-QW: Investigation, Methodology, Writing – original draft. H-YZ: Data curation, Investigation, Writing – original draft. W-MQ: Conceptualization, Data curation, Funding acquisition, Investigation, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Natural Science Foundation of Hubei Province, China (2024AFB879), the Leading Talent Cultivation Program of Hubei Academy of Agricultural Sciences (LJ20230504), Natural Science Youth Foundation of Hubei Academy of Agricultural Sciences (2024NKYJJ24), Hubei Innovation Center of Agricultural Science and Technology (2024-620-000-001-023).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1513430/full#supplementary-material
Ali, M., Shafiq, M., Haider, M. Z., Sami, A., Alam, P., Albalawi, T., et al. (2024). Genome-wide analysis of NPR1-like genes in citrus species and expression analysis in response to citrus canker (Xanthomonas axonopodis pv. citri). Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1333286
Behlau, F., Scandelai, L. H. M., da Silva Junior, G. J., Lanza, F. E. (2017). Soluble and insoluble copper formulations and metallic copper rate for control of citrus canker on sweet orange trees. Crop Prot. 94, 185–191. doi: 10.1016/j.cropro.2017.01.003
Cao, X., Duan, W., Wei, C., Chen, K., Grierson, D., Zhang, B. (2019). Genome-wide identification and functional analysis of carboxylesterase and MES gene families in peach (Prunus persica L. Batsch). Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01511
Cernadas, R. A., Benedetti, C. E. (2009). Role of auxin and gibberellin in citrus canker development and in the transcriptional control of cell-wall remodeling genes modulated by xanthomonas axonopodis pv. citri. Plant Sci. 177, 190–195. doi: 10.1016/j.plantsci.2009.05.006
Das, A. K. (2003). Citrus canker-a review. J. Appl. Hortic. 5, 52–60. doi: 10.37855/jah.2003.v05i01.15
de Lima, L. F. F., Carvalho, I. G. B., de Souza-Neto, R. R., Dos Santos, L. D. S., Nascimento, C. A., Takita, M. A., et al. (2024). Antisense oligonucleotide as a new technology application for CsLOB1 gene silencing aiming citrus canker resistance. Phytopathology 114, 1802–1809. doi: 10.1094/PHYTO-02-24-0058-KC
Duan, S., Long, Y., Cheng, S., Li, J., Ouyang, Z., Wang, N. (2022). Rapid Evaluation of the Resistance of Citrus Germplasms Against Xanthomonas citri subsp. citri. Phytopathology 112, 765–774. doi: 10.1094/PHYTO-04-21-0175-R
Ference, C. M., Manthey, J. A., Narciso, J. A., Jones, J. B., Baldwin, E. A. (2020). Detection of phenylpropanoids in citrus leaves produced in response to Xanthomonas citri subsp. citri. Phytopathology 110, 287–296. doi: 10.1094/PHYTO-06-19-0219-R
Forouhar, F., Yang, Y., Kumar, D., Chen, Y., Fridman, E., Park, S. W., et al. (2005). Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in plant innate immunity. Proc. Natl. Acad. Sci. U.S.A. 102, 1773–1778. doi: 10.1073/pnas.0409227102
Gao, W., Liu, Y., Huang, J., Chen, Y., Chen, C., Lu, L., et al. (2021). MES7 modulates seed germination via regulating salicylic acid content in Arabidopsis. Plants-Basel 10, 903. doi: 10.3390/plants10050903
Gochez, A. M., Behlau, F., Singh, R., Ong, K., Jones, J. B. (2020). Panorama of citrus canker in the United States. Trop. Plant Pathol. 45, 192–199. doi: 10.1007/s40858-020-00355-8
Gong, Q., Wang, Y., He, L., Huang, F., Zhang, D., Wang, Y., et al. (2023). Molecular basis of methyl-salicylate-mediated plant airborne defence. Nature 622, 139–148. doi: 10.1038/s41586-023-06533-3
Gupta, A., Bhardwaj, M., Tran, L. P. (2020). Jasmonic acid at the crossroads of plant immunity and pseudomonas syringae Virulence. Int. J. Mol. Sci. 21, 7482. doi: 10.3390/ijms21207482
Han, Y., Luo, F., Liang, A., Xu, D., Zhang, H., Liu, T., et al. (2024). Aquaporin CmPIP2;3 links H2O2 signal and antioxidation to modulate trehalose-induced cold tolerance in melon seedlings. Plant Physiol. 197, kiae477. doi: 10.1093/plphys/kiae477
Hu, Y., Zhang, J., Jia, H., Sosso, D., Li, T., Frommer, W. B., et al. (2014). Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc. Natl. Acad. Sci. U.S.A. 111, E521–E529. doi: 10.1073/pnas.1313271111
Huang, P. C., Tate, M., Berg-Falloure, K. M., Christensen, S. A., Zhang, J., Schirawski, J., et al. (2023). A non-JA producing oxophytodienoate reductase functions in salicylic acid-mediated antagonism with jasmonic acid during pathogen attack. Mol. Plant Pathol. 24, 725–741. doi: 10.1111/mpp.13299
Jia, H., Orbovic, V., Jones, J. B., Wang, N. (2016). Modification of the PthA4 effector binding elements in Type I CsLOB1 promoter using Cas9/sgRNA to produce transgenic Duncan grapefruit alleviating XccΔpthA4:dCsLOB1.3 infection. Plant Biotechnol. J. 14, 1291–1301. doi: 10.1111/pbi.12495
Jia, R., Xing, K., Tian, L., Dong, X., Yu, L., Shen, X., et al. (2024). Analysis of Methylesterase Gene Family in Fragaria vesca Unveils Novel Insights into the Role of FvMES2 in Methyl Salicylate-Mediated Resistance against Strawberry Gray Mold. J. Agric. Food Chem. 72, 11392–11404. doi: 10.1021/acs.jafc.4c01447
Jiao, L., Bian, L., Luo, Z., Li, Z., Xiu, C., Fu, N., et al. (2022). Enhanced volatile emissions and anti-herbivore functions mediated by the synergism between jasmonic acid and salicylic acid pathways in tea plants. Hortic. Res. 9, uhac144. doi: 10.1093/hr/uhac144
Kumar, D., Klessig, D. F. (2003). High-affinity salicylic acid-binding protein 2 is required for plant innate immunity and has salicylic acid-stimulated lipase activity. Proc. Natl.Acad. Sci. U.S.A. 100, 16101–16106. doi: 10.1073/pnas.0307162100
Li, Q., Wang, G., Guan, C., Yang, D., Wang, Y., Zhang, Y., et al. (2019). Overexpression of LcSABP, an orthologous gene for salicylic acid binding protein 2, enhances drought stress tolerance in transgenic tobacco. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00200
Lima Silva, C. C., Shimo, H. M., de Felício, R., Mercaldi, G. F., Rocco, S. A., Benedetti, C. E. (2019). Structure-function relationship of a citrus salicylate MES and role of salicylic acid in citrus canker resistance. Sci. Rep. 9, 3901. doi: 10.1038/s41598-019-40552-3
Lin, J., Wang, W., Mazarei, M., Zhao, N., Chen, X., Pantalone, V. R., et al. (2024). GmSABP2-1 encodes methyl salicylate esterase and functions in soybean defense against soybean cyst nematode. Plant Cell Rep. 43, 138. doi: 10.1007/s00299-024-03224-9
Liu, L., Liu, X., Liu, L., Zhu, T., Ye, R., Chen, H., et al. (2024). Clarification of the infection pattern of Xanthomonas citri subsp. citri on citrus fruit by artificial inoculation. Plant Methods 20, 65. doi: 10.1186/s13007-024-01190-7
Long, Q., Xie, Y., He, Y., Li, Q., Zou, X., Chen, S. (2019). Abscisic acid promotes jasmonic acid accumulation and plays a key role in citrus canker development. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01634
Manoharan, R. K., Shanmugam, A., Hwang, I., Park, J. I., Nou, I. S. (2016). Expression of salicylic acid-related genes in Brassica oleracea var. capitata during Plasmodiophora brassicae infection. Genome 59, 379–391. doi: 10.1139/gen-2016-0018
Manosalva, P. M., Park, S. W., Forouhar, F., Tong, L., Fry, W. E., Klessig, D. F. (2010). Methyl esterase 1 (StMES1) is required for systemic acquired resistance in potato. Mol. Plant Microbe Interact. 23, 1151–1163. doi: 10.1094/MPMI-23-9-1151
Martinez, J. G., Paran, G. P., Rizon, R., Meester, N. D., Moens, T. (2016). Copper effects on soil nematodes and their possible impact on leaf litter decomposition: a microcosm approach. Eur. J. Soil Biol. 73, 1–7. doi: 10.1016/j.ejsobi.2015.12.004
Martins, P. M. M., Andrade, M. D. O., Benedetti, C. E., Souza, A. A. D. (2020). Xanthomonas citri subsp. citri: host interaction and control strategies. Trop. Plant Pathol. 45, 213–236. doi: 10.1007/s40858-020-00376-3
Nardini, M., Dijkstra, B. W. (1999). Alpha/beta hydrolase fold enzymes: the family keeps growing. Curr. Opin. Struct. Biol. 9, 732–737. doi: 10.1016/s0959-440x(99)00037-8
Peng, A., Chen, S., Lei, T., Xu, L., He, Y., Wu, L., et al. (2017). Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 15, 1509–1519. doi: 10.1111/pbi.12733
Robert-Seilaniantz, A., Grant, M., Jones, J. D. (2011). Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 49, 317–343. doi: 10.1146/annurev-phyto-073009-114447
Soares, J. M., Weber, K. C., Qiu, W., Mahmoud, L. M., Grosser, J. W., Dutt, M. (2022). Overexpression of the salicylic acid binding protein 2 (SABP2) from tobacco enhances tolerance against Huanglongbing in transgenic citrus. Plant Cell Rep. 41, 2305–2320. doi: 10.1007/s00299-022-02922-6
Su, H., Wang, Y., Xu, J., Omar, A. A., Grosser, J. W., Calovic, M., et al. (2023). Generation of the transgene-free canker-resistant Citrus sinensis using Cas12a/crRNA ribonucleoprotein in the T0 generation. Nat. Commun. 14, 3957. doi: 10.1038/s41467-023-39714-9
Vlot, A. C., Liu, P. P., Cameron, R. K., Park, S. W., Yang, Y., Kumar, D., et al. (2008). Identification of likely orthologs of tobacco salicylic acid-binding protein 2 and their role in systemic acquired resistance in Arabidopsis thaliana. Plant J. 56, 445–456. doi: 10.1111/j.1365-313X.2008.03618.x
Wang, Y., Liu, J. H. (2012). Exogenous treatment with salicylic acid attenuates occurrence of citrus canker in susceptible navel orange (Citrus sinensis Osbeck). J. Plant Physiol. 169, 1143–1149. doi: 10.1016/j.jplph.2012.03.018
Xu, T., Zheng, X., Yang, Y., Yang, S., Yi, X., Yu, C., et al. (2024). Indole-3 acetic acid negatively regulates rose black spot disease resistance through antagonizing the salicylic acid signaling pathway via jasmonic acid. Planta 259, 129. doi: 10.1007/s00425-024-04406-1
Xue, R., Feng, M., Chen, J., Ge, W., Blair, M. W. (2021). A methyl esterase 1 (PvMES1) promotes the salicylic acid pathway and enhances Fusarium wilt resistance in common beans. TAG. Theor. Appl. Genet. Theor. Appl. Genet. 134, 2379–2398. doi: 10.1007/s00122-021-03830-1
Yang, L., Teixeira, P. J., Biswas, S., Finkel, O. M., He, Y., Salas-Gonzalez, I., et al. (2017). Pseudomonas syringae type III effector hopBB1 promotes host transcriptional repressor degradation to regulate phytohormone responses and virulence. Cell Host Microbe 21, 156–168. doi: 10.1016/j.chom.2017.01.003
Yang, Y., Xu, R., Ma, C. J., Vlot, A. C., Klessig, D. F., Pichersky, E. (2008). Inactive methyl indole-3-acetic acid ester can be hydrolyzed and activated by several esterases belonging to the AtMES esterase family of Arabidopsis. Plant Physiol. 147, 1034–1045. doi: 10.1104/pp.108.118224
Zhang, X., Francis, M. I., Dawson, W. O., Graham, J. H., Orbović, V., Triplett, E. W., et al. (2020). Over-expression of the arabidopsis npr1 gene in citrus increases resistance to citrus canker. Eur. J. Plant Pathol. 128, 91–100. doi: 10.1007/s10658-010-9633-x
Keywords: calamondin, citrus bacterial canker (CBC), methylesterase (MES) genes, salicylic acid (SA), jasmonic acid (JA), indole-3-acetic acid (IAA)
Citation: Xiao Y-X, Xiao C, Tong Z, He X-J, Wang Z-Q, Zhang H-Y and Qiu W-M (2025) Four MES genes from calamondin (Citrofortunella microcarpa) regulated citrus bacterial canker resistance through the plant hormone pathway. Front. Plant Sci. 15:1513430. doi: 10.3389/fpls.2024.1513430
Received: 15 November 2024; Accepted: 30 December 2024;
Published: 20 January 2025.
Edited by:
Michael V. Kolomiets, Texas A and M University, United StatesReviewed by:
Manikandan Ramasamy, Texas A&M AgriLife Research & Extension Center, United StatesCopyright © 2025 Xiao, Xiao, Tong, He, Wang, Zhang and Qiu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen-Ming Qiu, cWl1d21AaGJhYXMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.