- 1College of Agriculture and Biology, Liaocheng University, Liaocheng, China
- 2Economic Crop Research Institute, Puyang Academy of Agriculture and Forestry Sciences, Puyang, China
The wall-associated kinase (WAK) gene family encodes functional cell wall-related proteins. These genes are widely presented in plants and serve as the receptors of plant cell membranes, which perceive the external environment changes and activate signaling pathways to participate in plant growth, development, defense, and stress response. However, the WAK gene family and the encoded proteins in soybean (Glycine max (L.) Merr) have not been systematically investigated. In this study, the soybean WAK genes (GmWAK) were identified based on genome-wide sequence information, the basic characteristics, chromosome location, gene replication, expression pattern, and responses to stress were comprehensively analyzed. A total of 74 GmWAK genes were identified and mapped to 19 different chromosomes in the soybean genome. Seventy-four GmWAK genes were divided into four groups, and GmWAK genes in the same group shared similar gene structures and conserved motifs. Thirty-seven duplicate pairs were identified in 74 GmWAK genes. Segmental duplication (SD) was critical in soybean WAK gene family expansion, and purification selection occurred during evolution. The promoter cis-element analysis displayed many hormone- and stress-related response elements in the promoter regions of GmWAK genes. GmWAK genes were diversely expressed in different organs and tissues, with most actively responding to cold, heat, salt, drought, and heavy metal stresses, suggesting that GmWAK genes could exhibit relevant roles in various bioprocesses.
1 Introduction
The cell wall is a thick, tough, and slightly elastic structure surrounding the cell membrane. Plant cell walls are composed of cellulose, hemicellulose, pectin, and small amounts of structural proteins, which are critical in maintaining cell morphology and resisting pathogen invasion (Kohorn et al., 2006; Underwood, 2012). Receptor-like kinases (RLKs) are transmembrane proteins located on the cell membrane that act as receptors for signaling molecules which play important regulatory roles in almost all life activities (Soltabayeva et al., 2022; Mehla et al., 2024; Ye et al., 2017). According to the different extracellular domains, RLKs can be divided into more than 40 subfamilies, including WAK-RLKs, Lec-RLKs, LRR-RLKs, etc (Afzal et al., 2008; Lehti-Shiu et al., 2009). Among them, wall-associated kinases (WAKs) comprise a specific RLK gene family associated with the cell wall pectin (Decreux and Messiaen, 2005; Kanneganti and Gupta, 2008). WAK proteins display typical structural domains, including the GUB_WAK_bind domain, epidermal growth factor (EGF) domain, transmembrane domain, and Pkinase domain (He et al., 1999). The GUB_WAK_bind and EGF domains are located outside the cell and assist cells in perceiving external signals (He et al., 1999). The Pkinase domain is located in the cell and is the starting point of the downstream signaling cascade in the cytoplasm (Tocquard et al., 2014; Wagner and Kohorn, 2001; Weremczuk et al., 2020). The WAK gene family is a subfamily of RLKs, which act as the information transmission link between the cell wall, cell membrane, and cytoplasm, being important in regulating plant growth, development, and response to environmental stress (Zuo et al., 2015; Sun et al., 2020; Dou et al., 2021).
Currently, the WAK gene families have been identified in a number of species. For instance, 27 WAK genes are present in Arabidopsis (Arabidopsis thaliana), 125 in rice (Oryza sativa), 29 in cotton (Gossypium hirsutum), 6 in wheat (Triticum aestivum), 68 in rose (Rosa chinensis), 44 in apple (Malus domestica), 41 in walnut (Juglans regia), and 29 in potato (Solanum tuberosum) (Verica and He, 2002; Zhang et al., 2005; Dou et al., 2021; Liu et al., 2006, 2021; Zuo et al., 2019; Li et al., 2022; Yu et al., 2022). WAK genes are important in plant growth and development. In Arabidopsis, AtGRP3 interacts with the extracellular domain of the receptor-like kinase AtWAK1 and inhibits root cell expansion. In addition, its gene knockout mutant shows increasing root length (Kohorn, 2001; Mangeon et al., 2016). The antisense expression of AtWAK4 inhibits cell elongation and altered root development (Lally et al., 2001). In rice, the wall-associated receptor-like kinase gene OsDEES1 regulates early embryonic sac development, and RNA interference silencing (RNAi) of OsDEES1 causes a high rate of female sterility (Wang et al., 2012). HvWAK1 is specifically expressed in barley roots, and a shorter root was observed in HvWAK1 Ds mutants than in wild-type specimens (Kaur et al., 2013; Tripathi et al., 2021). SlWAKL2 is homologous to AtWAKL14 in Arabidopsis and is specifically expressed in tomato vascular tissue. SlWAKL2 RNAi plants had small fruits, few seeds, and few vascular bundles (Ma et al., 2024).
WAK genes were also involved in the responses to biotic and abiotic stresses. The expression of OsWAK112 in rice was inhibited by salt stress, and decreased S-adenosyl-L-methionine synthetase (SAMS) content, ethylene content, and plant survival rate were observed in OsWAK112-overexpressing plants (Lin et al., 2021). OsWAK11 was upregulated by heavy metal stress in rice. OsWAK11 appears to be involved in Cu2+ detoxification, mediating Cu2+ accumulation in the cell wall, by regulating cell wall methylesterification and alleviating metal ion toxicity (Hu et al., 2014; Xia et al., 2018). TaWAK20 was a positive regulatory factor of cadmium (Cd) stress, and its overexpression improved the Cd tolerance of transgenic plants by increasing the activities of antioxidant enzymes and H2O2 content (Li, 2023). OsWAK2 participates in the signaling pathway of rice blast disease resistance. Transgenic plants overexpressing OsWAK2 exhibited enhanced disease resistance, whereas RNAi plants showed a loss of resistance (Li et al., 2004). ZmWAK02 encodes an RD-WAK protein in maize. Transgenic lines, mutants, and complementation lines confirmed that ZmWAK02 was the resistance gene for maize gray spot disease (Dai et al., 2024).
Soybean (Glycine max (L.) Merr) is an important oil crop and a primary source of plant-based proteins for human and animal feed, playing a crucial role in global food security. The soybean yield and quality are affected by abiotic stresses, such as salt, temperature, drought, and heavy metal stress (Nagajyoti et al., 2010; Hussain et al., 2023; Xu et al., 2023). The emergence rate of soybeans decreases by 5%–10% under cold stress (Staniak et al., 2021). The yield, harvest index, and seed quality of soybeans are reduced under heat and drought stress conditions (Carrera and Dardanelli, 2017; Djanaguiraman et al., 2013; Mourtzinis et al., 2015; Rasheed et al., 2022). Salt stress decreases the chlorophyll content and photosynthetic rate of soybeans and inhibits photosynthetic carbon metabolism (Weisany et al., 2011). Under aluminum (Al) stress, the Ca2+ homeostasis and signal transduction in soybeans are disrupted, reactive oxygen species (ROS) is increased, membrane peroxidation is accelerated, and root growth of soybeans is restrained (Chandra and Keshavkant, 2021; Chauhan et al., 2021; Ma, 2007). Under Cd stress, the photosynthetic system, cell membrane, and respiratory metabolism are damaged, inhibiting soybean growth and development (Liu et al., 2024; Shi et al., 2019).
The WAK gene family was identified in multiple species and was recognized as critical for plant growth, development, and stress tolerance (Verica and He, 2002; Zhang et al., 2005; Dou et al., 2021; Liu et al., 2006, 2021). However, the WAK gene family in soybeans has not yet been systematically investigated. In this study, the soybean WAK gene family was identified. Domain structure, duplication events, and phylogenetic relationships were analyzed. The tissue-specific expression of these genes was detected using RNA-Seq, and analysis of expression patterns under various abiotic stresses was performed by qRT-PCR. This study will provide potential candidate genes for further functional investigation at the molecular level and the molecular breeding of soybeans with stress tolerance.
2 Materials and methods
2.1 Identification of WAK genes in the soybean genome
The genome data of soybean (glyma.Wm82.a4.v1) was downloaded from the Ensembl Plants database (http://ensemblgenomes.org/). The AtWAK genes of Arabidopsis were downloaded from the TAIR database (https://www.arabidopsis.org/) (Verica and He, 2002). The AtWAK proteins were the query sequence, which blasted with the soybean genome in TBtools-II software. A total of 852 sequences were aligned at the threshold value 1e−5. Fifty-nine potential WAK genes were identified in the soybean genome based on the Swiss-Prot database.
The WAK domains, such as GUB_WAK_bind (PF13947), EGF (PF07645), and Pkinase (PF00069), were downloaded from the Pfam database (http://pfam.xfam.org/). The WAK sequences were used as the query in TBtools-II software, and an HMMER search was performed to identify WAK genes in the soybean genome (Chen et al., 2023). WAK genes were identified based on a threshold value (e−3). A total of 7, 100, and 2,231 genes were identified containing EGF, GUB_WAK_bind, or the Pkinase domain, respectively. There were 69 genes, including two types of domains: GUB_WAK_bind or EGF and Pkinase.
Ninety-four potential WAK genes were identified in the soybean genome through blast and HMMER searches. Further screening for the potential WAK genes was conducted in the Pfam (http://pfam.xfam.org/), SMART (http://smart.embl-heidelberg.de/), and NCBI CDD database (http://www.ncbi.nlm.nih.gov/cdd). Finally, 74 WAK genes were confirmed in the soybean genome, which contained at least two types of domains: GUB_WAK_bind or EGF and Pkinase domains.
The length and position information for 74 GmWAK genes were obtained from the gene annotations of the soybean genome and visualized using MapChart (Voorrips, 2002). The molecular weight (MW), isoelectric point (IP), instability index, and grand average of hydropathicity (GRAVY) were predicted with the Protein Parameter Calc in TBtools-II (Chen et al., 2023). The exon–intron structure was analyzed by GXF Stat in TBtools-II. The subcellular location of GmWAK genes was predicted using the online software DeepLoc-2.0 (https://services.healthtech.dtu.dk/services/DeepLoc-2.0/) (Almagro et al., 2017).
2.2 Gene phylogenetic, structure, and conserved motif analyses
The multiple sequences of GmWAKs and AtWAKs/WAKLs were aligned using the MUSCLE tool. The neighbor-joining method was used to build the phylogenetic tree using MEGA11 (Tamura et al., 2021). The parameters were set as follows: bootstrap method, Poisson model, 1,000 bootstrap replications, and partial deletion (Tamura et al., 2021). The phylogenetic tree was drawn on the website of EvolView v2 (He et al., 2016).
The Simple MEME Wrapper tool in TBtools-II was used to search for conserved motifs within GmWAK proteins (Chen et al., 2023). The parameter settings were as shown below: the maximum number of motifs was 10, the motif length was 6–50 amino acids, the max E-value was e−10, and the model was any number of occurrences per seq. The conserved domain analysis of GmWAKs was performed using the Batch SMART tool in TBtools-II (Chen et al., 2023). Visualizing the phylogenetic tree, gene structure, conserved motifs, and domains for GmWAKs was made within the Gene Structure View of TBtools-II (Chen et al., 2023).
2.3 Gene duplication and synteny analyses
Gene duplication (GD) was analyzed and visualized using TBtools-II (Chen et al., 2023). The simple Ka/Ks calculator was used to compute the non-synonymous substitution rate (Ka) and synonymous substitution rate (Ks) of duplicated gene pairs to evaluate GD events (Chen et al., 2023). The occurrence time of GD events was calculated using the following formula: T = (Ks/2λ) × 10−6, where λ = 6.161029 × 10−9 (Lynch and Conery, 2000). The syntenic relationships of different species were analyzed and visualized with One Step MCScanX and Multiple Synteny Plot in TBtools-II.
2.4 Cis-acting regulatory element analysis
The promoter sequence of the GmWAK genes was downloaded from the soybean genome, which covered the 2,000 bp upstream of the transcriptional start site. The cis-acting elements analysis was conducted using Plant CARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). The visualization of cis-acting elements was executed in TBtools-II and Adobe Photoshop 2023 (Chen et al., 2023).
2.5 Tissue-specific expression patterns of GmWAK genes
The RNA-Seq datasets of soybeans were obtained from the NCBI SRA database (http://www.ncbi.nlm.nih.gov/sra), including the gene expression information of different tissues at different developmental stages. The expression heat map of the GmWAK genes was drawn in TBtools-II.
2.6 Plant growth and stress treatment of soybean seedlings
The cultivated soybean variety Williams82 was employed to examine the gene profiles in response to stress treatments. Soybean seeds were sown in a double layer of absorbent paper and transferred to a half-strength Hoagland solution after 3 days. The conditions of the artificial illumination incubator were as follows: photoperiod 16 h light/8 h dark and temperature 25°C/18°C (day/night). After 10 days of cultivation, the seedlings were treated with multiple stress stimuli, such as cold (4°C), heat (40°C), salt (150 mM NaCl), drought (20% w/v PEG-6000), Al (100 μM of AlCl3), and Cd (100 μM of CdCl2). The root and leaves of seedlings were collected at 0, 3, 6, 12, and 24 h after treatment. Three replicates of three seedlings were collected for each treatment. All samples were rapidly frozen in liquid nitrogen and stored at −80°C for further analysis.
2.7 qRT-PCR analysis
The total RNA of the samples was extracted using a Total RNA Isolation Kit (Vazyme, RC411, China). The first strand of cDNA was synthesized using a reverse transcription kit (Vazyme, R312, China) according to the instructions. Quantitative real-time polymerase chain reaction (qRT-PCR) was performed on a LightCycler® 480 system (Roche, Basel, Switzerland). The qRT-PCR system consisted of 2 µl of cDNA, 0.8 µl of primer (10 µμ), 7.2 µl of ddH2O, and 10 µl of SYBR qPCR master mix (Cat. No. Q711, Vazyme, China). The qRT-PCR reaction program was 95°C for 30 s, 40 cycles at 95°C for 10 s and 60°C for 30 s, 95°C for 15 s, 60°C for 60 s, and 95°C for 15 s. Relative quantification of gene expression was calculated using the 2−ΔΔCT method, and GmGAPDH was used as a reference gene in qRT-PCR analysis (Livak and Schmittgen, 2001; Huang et al., 2015). The significance test of the gene expression was assessed by Tukey’s pairwise comparison test in SPSS software. The data were painted into graphs using GraphPad prism9.5 (https://www.graphpad.com/). The sequences of the gene-specific primers used in this study are listed in Supplementary Table S1.
3 Results
3.1 Identification, chromosomal localization, and physicochemical property analysis of the WAK gene family in soybeans
We identified 74 dependable GmWAK genes in the soybean genome using homology comparison and domain analysis. Sixty-one GmWAK genes contained GUB_WAK_bind or EGF and Pkinase conserved domains, and 13 genes contained GUB_WAK_bind, EGF, and Pkinase conserved domains. According to their positions on the chromosome, GmWAK genes were named GmWAK1–GmWAK74 (Figure 1; Supplementary Table S2). Chromosomal mapping showed that the members of the WAK gene family were unevenly distributed on the 19 chromosomes, ranging from 1 to 10, with none on chromosome 20. The maximum number of WAK genes was located on Gm07, Gm09, Gm13, and Gm14 chromosomes containing 8, 9, 10, and 9 WAK genes, respectively. The length of the GmWAK proteins was extremely varied. The shortest was GmWAK60, which encoded 476 amino acids, and the longest was GmWAK39, which encoded 987 amino acids. The molecular weight (MW) ranged from 53.60 kDa to 111.10 kDa. Among them, 48 GmWAK proteins belonged to acidic proteins (PI < 7.0), and 26 GmWAK proteins belonged to basic proteins (PI > 7.0). The instability index (InI) values ranged from 25.79 to 48.29. Thirty-two GmWAK proteins with InI values were greater than 40, indicating that they might be more unstable than the other 42 GmWAK proteins. The grand average of hydropathicity (GRAVY) index showed that 72 WAK proteins were hydrophilic (GRAVY < 0), and the rest were hydrophobic (GRAVY > 0). The subcellular location predicted that the GmWAK proteins were localized to the cell membrane, and almost all contained signal peptides and transmembrane domains.
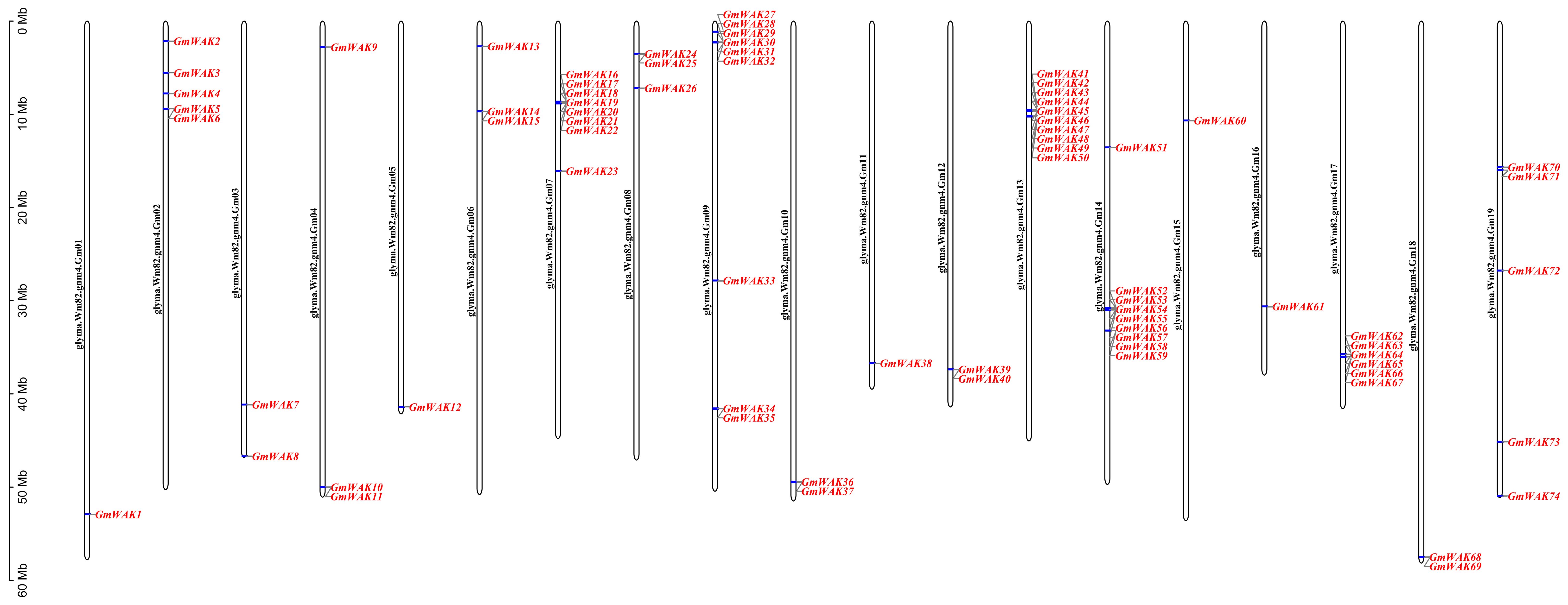
Figure 1. Chromosomal location of GmWAK genes in the soybean genome. Chromosome numbers are shown beside each chromosome. The scale on the left represents the chromosome length. The chromosome Gm 20 is not displayed, because no WAK gene was localized to that chromosome.
3.2 Phylogenetic, structural, and conserved motif characteristics of GmWAK genes
A phylogenetic tree was built using the neighbor-joining method with WAK proteins from soybean and Arabidopsis to explore the evolution of the WAK gene family. The WAK genes were divided into four groups: group I, group II, group III, and group IV (Figure 2). Group I clustered 13 GmWAK genes together with 7 AtWAK/WAKL genes. Group II included 7 GmWAK genes and 14 AtWAKL genes. Likewise, group IV contained 20 GmWAK genes and 4 AtWAKL genes. Group III was the largest one, containing 34 GmWAK genes without any member from Arabidopsis included within this group.
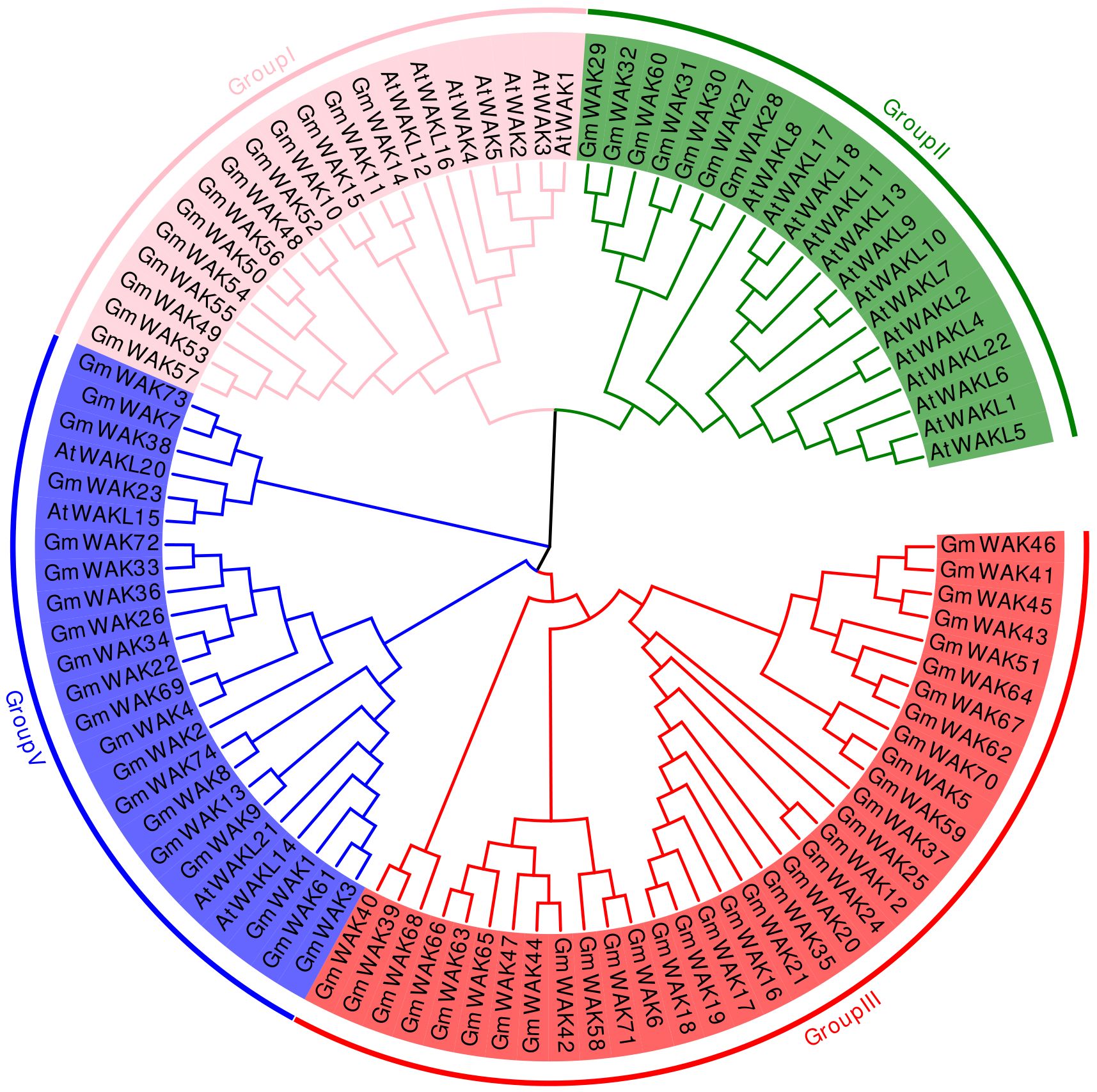
Figure 2. Phylogenetic tree analysis of the WAK gene family from Arabidopsis and soybean. Distinct color blocks represent different groups.
Based on gene structure analysis, one to eight exons were observed in the GmWAK genes, with a mean of 3.43 exons per gene (Figure 3; Supplementary Table S2). The proportion of GmWAK genes containing one, two, three, four, six, and eight exons was 4.05%, 12.16%, 39.18%, 39.18%, 1.35%, and 4.05%, respectively. The number of exons in groups I and II was three or four. In group III, we detected two to eight exons in GmWAK genes, and 41.17% of genes in group III were composed of three exons. A similar variation of exons was observed in group IV, and 65% of the GmWAK genes included four exons. We also found that the range of GmWAK gene lengths was from 2,068 to 20,003 bp, and the coding sequences (CDS) were 1,428 to 2,961 amino acid residues. There was an extensive length variation in GmWAK genes due to differences in the number and length of introns.
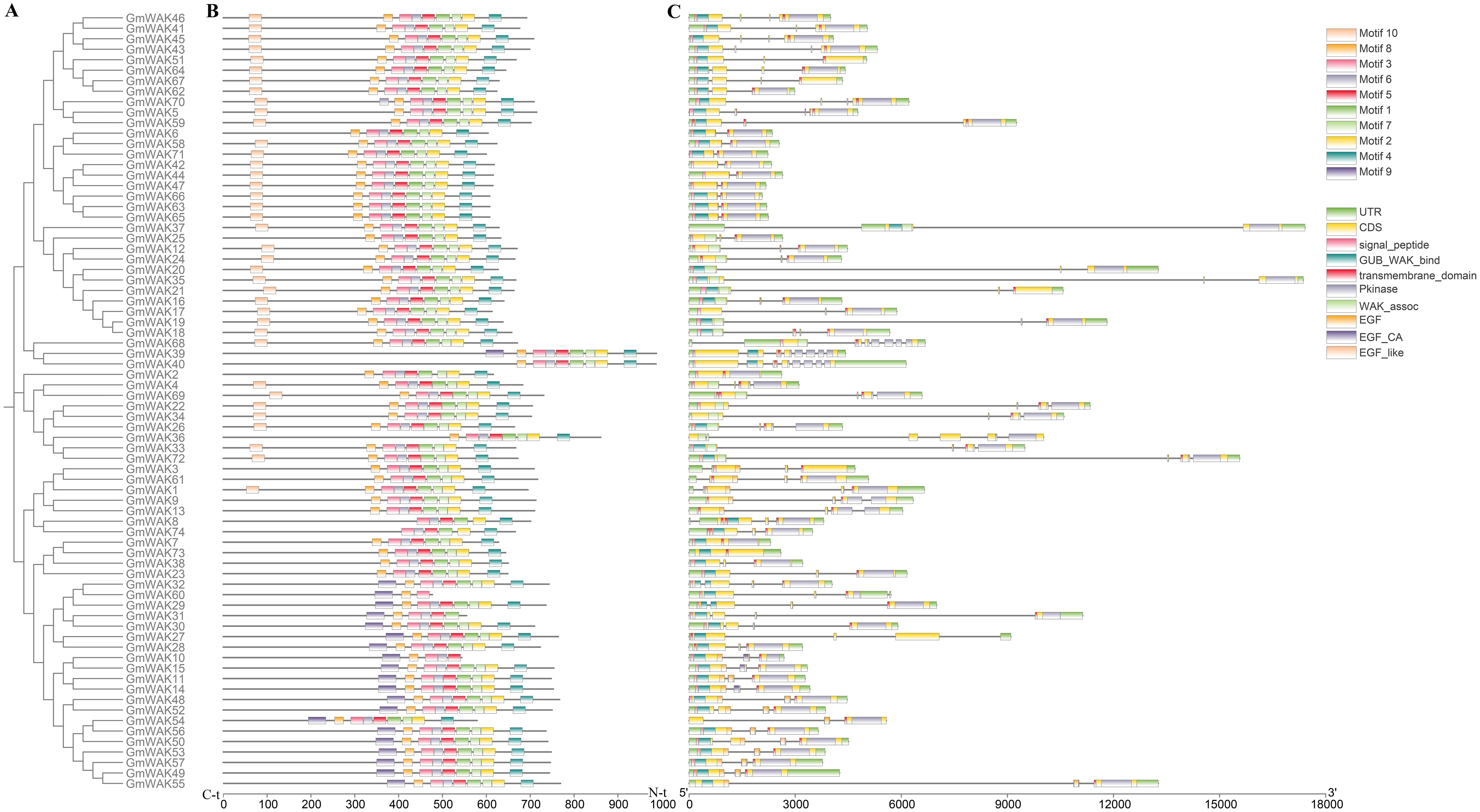
Figure 3. Gene structure analysis of GmWAK proteins. (A) The phylogenetic tree of the 74 GmWAK proteins. (B) The motif distribution of the 74 GmWAK proteins. (C) The intron–exon structure and conservative domain of the GmWAK protein sequences.
We analyzed the motif distribution of GmWAK proteins, and 10 motifs were predicted (Figure 3; Supplementary Tables S3, S4). Similar motif distributions were observed among GmWAK members. Approximately 93.2% (69) of GmWAK proteins shared eight conserved motifs (motifs 1–8), and 6.75% (5) of GmWAK proteins were observed to have two to six motifs from motifs 1 to 8. Furthermore, motif 9 was specific to group I and II members, and motif 10 was present in the GmWAK proteins from groups III and IV (Supplementary Table S3).
According to the conserved domain analysis, the GmWAK proteins contained a highly conserved domain structure. All GmWAK proteins had the Pkinase and transmembrane domains at the C-terminal end, and 77.02% of GmWAK proteins had signal peptides at the N-terminal end (Supplementary Table S2; Figure 3). The WAK proteins had the GUB_WAK_bind and EGF domains. All GmWAK proteins contained the GUB_WAK_bind and EGF domains in group I, except GmWAK54. In groups II and III, all GmWAK proteins contained only the GUB_WAK_bind domain without the EGF domain. In group IV, 70% and 20% of the GmWAK proteins included either the GUB_WAK_bind or the EGF-related domains, respectively. The remaining 10% of GmWAK proteins simultaneously contained the two domains. The GmWAK proteins in the same group displayed similar conserved domains and structures, indicating they might have comparable functions.
3.3 Duplication and synteny analysis of GmWAK genes
GD events, which are the main reason for the expansion of gene family members, are usually classified into segmental duplication (SD), tandem duplication (TD), and proximal duplication (PD). We conducted a GD analysis of the soybean WAK gene family. A total of 51 GmWAK genes participated in the expansion process of the GmWAK gene family, which included 21 pairs of SD, nine pairs of TD, and six pairs of PD (Table 1; Figure 4). The non-synonymous (Ka) and synonymous (Ks) substitution rates were calculated to investigate the selection pressures after GD. The Ka values ranged from 0.005 to 0.612, the Ks values ranged from 0.008 to 3.58, and the Ka/Ks values of 34 gene pairs ranged from 0.12 to 0.94, indicating that the GmWAKs evolved in purifying selection after GD (Ka/Ks < 1).
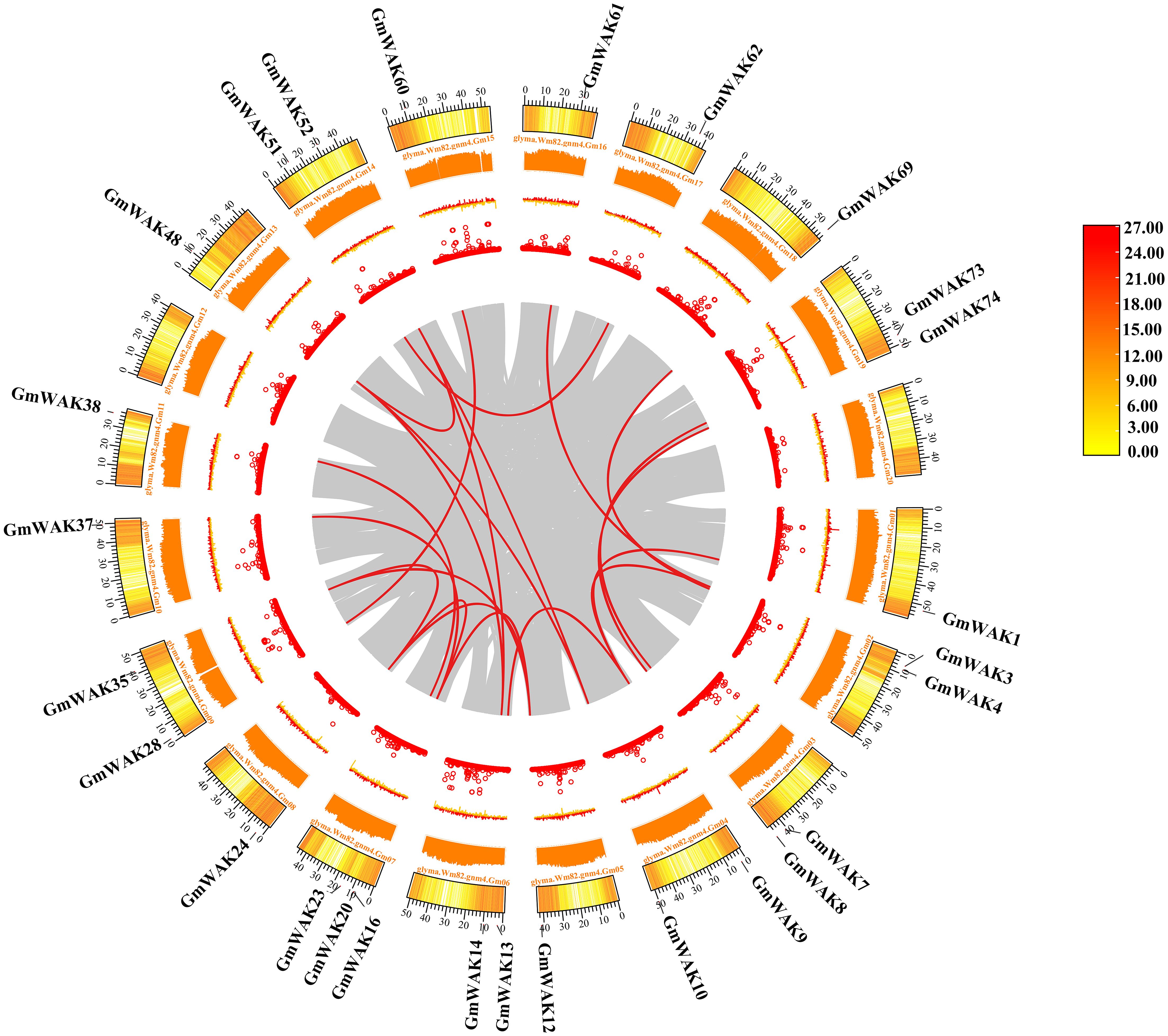
Figure 4. Synteny analysis of the GmWAK genes. The gray lines represent the collinear gene pairs in the whole soybean genome, and the red lines represent the collinear pair of GmWAK genes. The circles represent gene density, N ratio, GC skew, and GC ratio from inside to outside, respectively.
Additionally, Ks was used to estimate the divergence time in evolution. GD events occurred between 0.61 Mya and 290.33 Mya, averaging 91.23 Mya. We performed the synteny analysis using MCScanX in TBtools-II to further explore the evolutionary relationship of the WAK gene family in different species. The comparison of soybeans with Arabidopsis and rice at the genome level showed that 31,623 collinear pairs were observed between soybeans and Arabidopsis, and 10,953 collinear pairs were observed between soybeans and rice, among which the number of collinear pairs belonging to the WAK genes were eight and two, respectively (Figure 5; Supplementary Tables S5, S6).
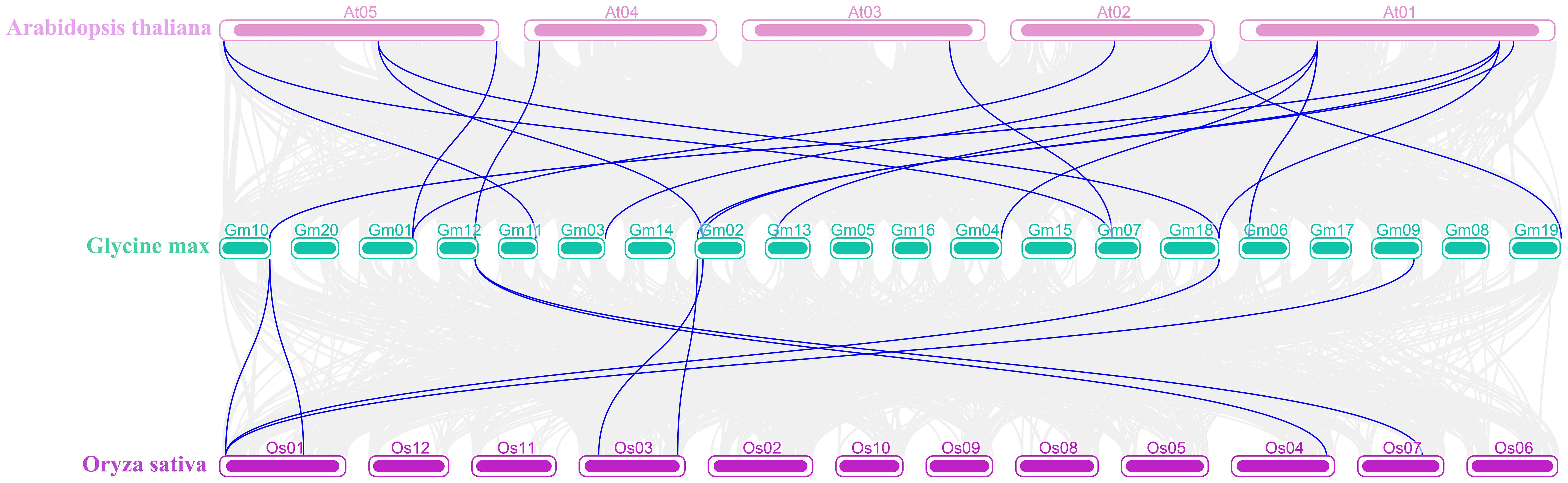
Figure 5. Synteny analysis of soybean with Arabidopsis and rice. The gray lines represent the collinear gene pairs of soybean with Arabidopsis and rice in the whole genome, and the blue lines represent the collinear pairs of the WAK gene family from those three species.
3.4 Cis-acting element analysis of the GmWAK genes
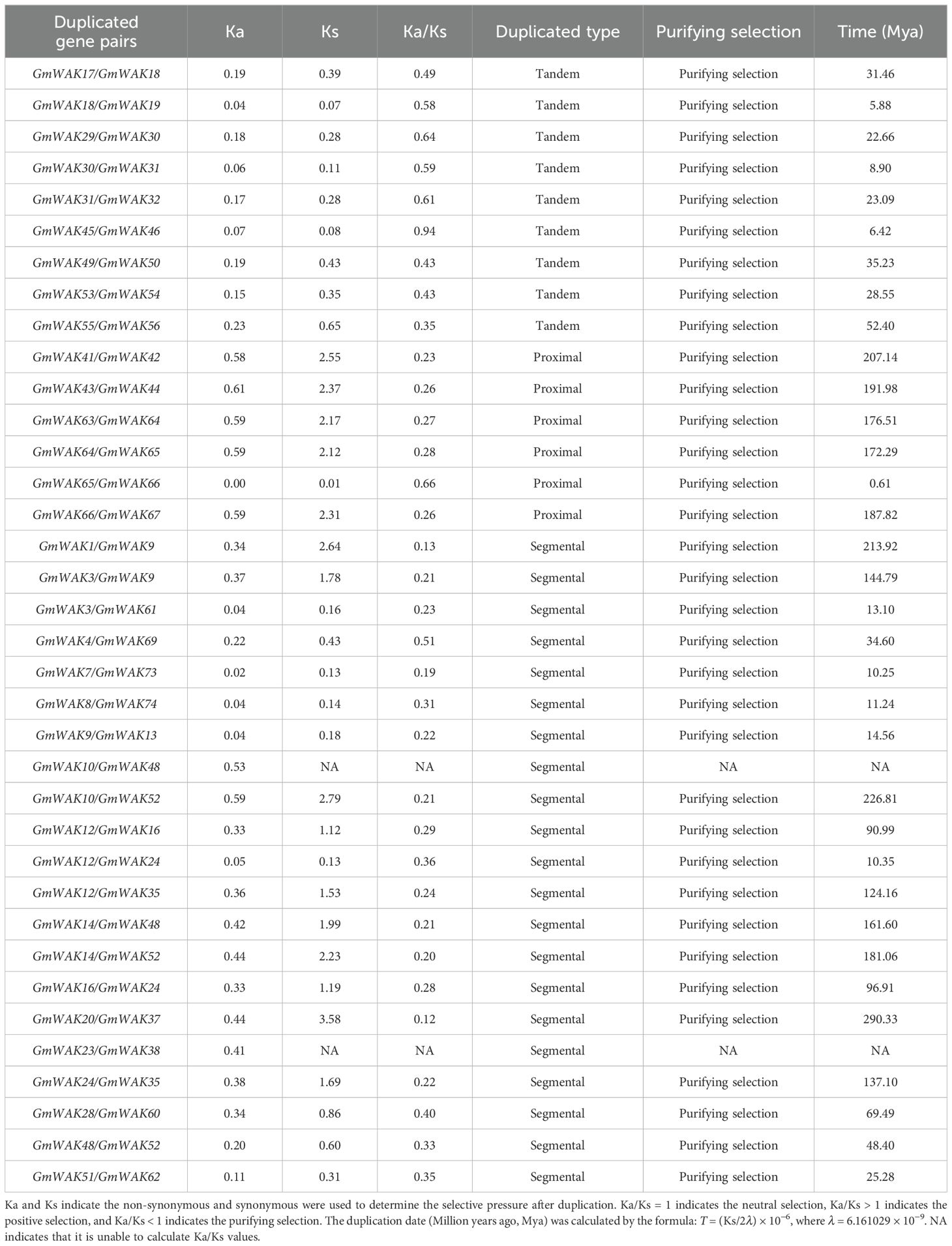
After cis-acting analysis of the GmWAK genes, 96 types of elements were identified in promoter regions, which were divided into six subgroups: core promoter elements, hormone-responsive elements, stress-responsive elements, light-responsive elements, metabolic- and growth-related elements, and unknown function elements (Figure 6; Supplementary Tables S7, S8). The core promoter elements (e.g., CAAT-box, TATA-box, AT~TATA-box) were identified in the promoter regions of all GmWAK genes. There were 19 types of hormone-responsive elements, which included MJA and JA (4), ABA (4), ETH (3), GA (3), IAA (3), and SA (2). Among the hormone-responsive elements, MJA- and JA-responsive elements occurred most frequently at 412 times and were distributed on the promoter regions of all GmWAK genes except GmWAK33. ETH-responsive elements were identified in promoter regions of GmWAK genes, where MYC- and ERE-responsive elements appeared 365 times. ABA and SA responsive elements were separately found in 81.08% and 71.62% of the GmWAK genes. Among them, the elements ABRE and W box were the most frequent. GA- and IAA-responsive elements were identified in 58.1% and 40.50% of the GmWAK genes. A total of 18 types of stress-responsive elements were found in the GmWAK genes. The drought-related elements MYB, Myb, and MBS were the most frequent, followed by the anaerobic-responsive elements (ARE), heat-responsive elements (STRE), and trauma-responsive elements (WUN-motif, WRE3). Approximately 29 light-responsive elements were near the GmWAK genes, among which Box 4 and G-box occurred in 98% of the GmWAK genes. Sixteen different elements were associated with the metabolism and growth of the plant. CAT-box, O2-site, and circadian were the most frequent elements related to meristem, gliadin metabolism, and circadian regulation. Furthermore, 11 unknown functional elements were predicted in the soybean WAK gene family.
3.5 Tissue expression profile analysis of the soybean WAK gene family
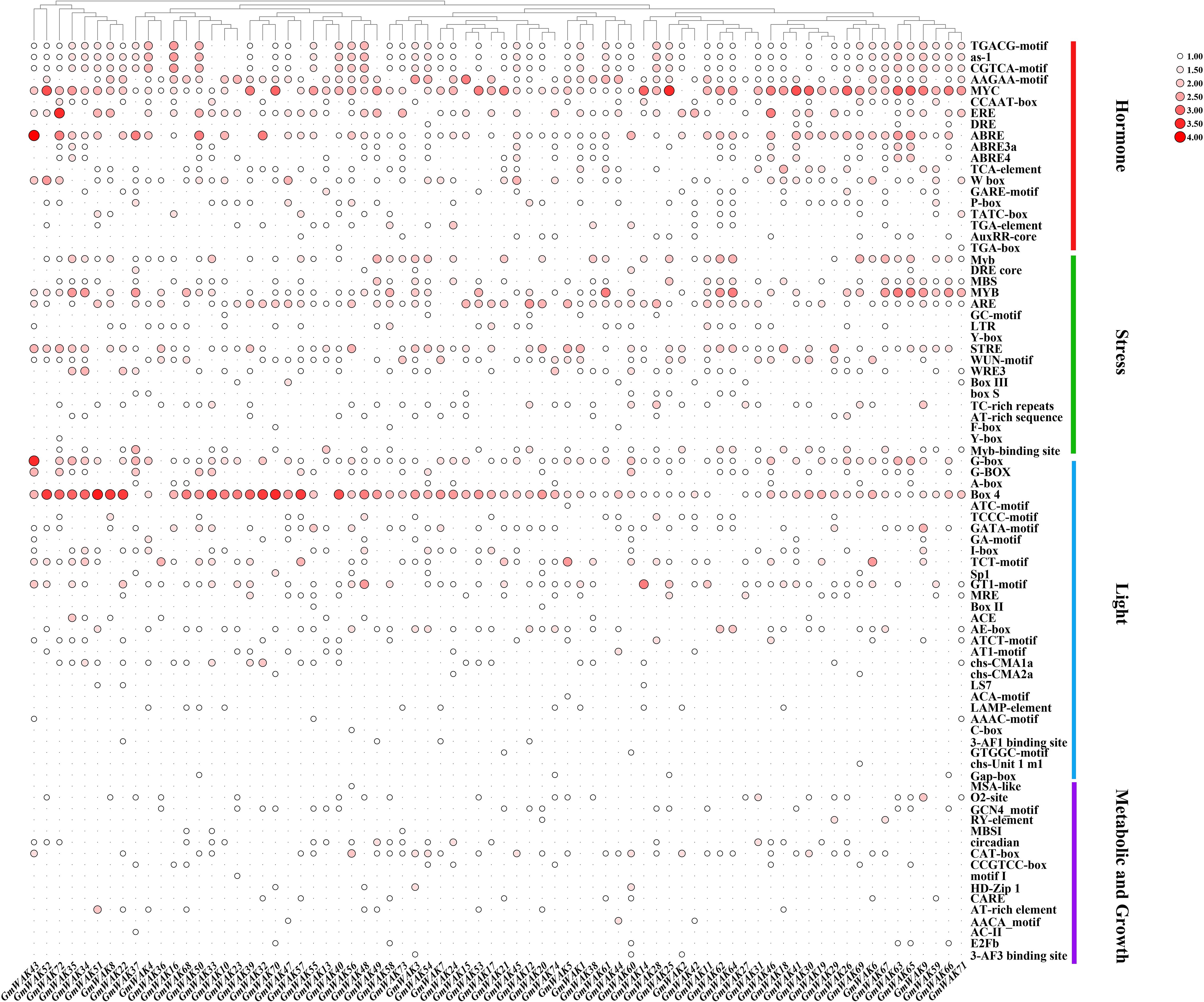
We obtained the RNA-Seq data of seven tissues from the NCBI database (https://www.ncbi.nlm.nih.gov/sra). We conducted a tissue expression analysis of 74 GmWAK genes (Figure 7; Supplementary Table S9) to explore the putative function of GmWAK genes in soybean development. Forty-seven GmWAK genes were expressed in soybean tissues, while 25 genes were almost not expressed by its fragments per kilobase of exon per million fragments mapped (FPKM) values. There were 1, 13, 18, 26, 29, and 38 GmWAK genes expressed in the seed, flower, pod, stem, leaf, and root, respectively (FPKM > 1). Among these, six GmWAK genes showed high expression (FPKM > 10), such as GmWAK15 in the root, leaf, and pod; GmWAK34, GmWAK38, and GmWAK53 in the root; and GmWAK40 and GmWAK45 in the stem or leaf.
3.6 Expression profile analysis of GmWAK genes under different abiotic stresses
We treated soybean seedlings with various abiotic stresses, including salt (150 mM of NaCl), drought (20% PEG6000), cold (4°C), heat (40°C), Al (100 µM of AlCl3), and Cd (100 µM of CdCl2) to further validate the response pattern of the GmWAK genes. Then, we detected the expression of 18 GmWAK genes using the qRT-PCR method.
3.6.1 Responses of GmWAK genes to cold stress
Seventeen differentially expressed GmWAK genes were identified under cold stress. The expression of these genes in cold conditions changed more than two-fold than that in the control group (Figure 8). A total of 10 GmWAK genes were significantly upregulated in soybean roots. Among them, GmWAK1/2/12/15/39 peaked after 3 h of cold treatment and were 2.3–5.3 times the control. GmWAK25/40/47/52/71 reached the maximum after 24 h and were 2.4–8.6 times the control. Five GmWAK genes were significantly downregulated in soybean roots. GmWAK38/41/42/48/50 decreased to 9.8%–38.2% of the control after 12 h of treatment. The expression level of GmWAK47 in soybean leaves was up to 2.6 times that of the control at 3 h after treatment. Fourteen significantly downregulated GmWAK genes were identified. GmWAK1 was 41.3% of the control after 3 h of treatment, and GmWAK25 was 22.2% of the control after 6 h. The remaining 12 GmWAK genes were reduced to 2.2%–52.8% of the control group after 24 h of treatment. Among the 17 significantly differentially expressed GmWAK genes, the relative expression levels of GmWAK2/15/25/40/45/46 were high, which were significantly increased by 4.1–8.6 times in the root and yet were significantly decreased by 71.5%–94.4% in the leaves.
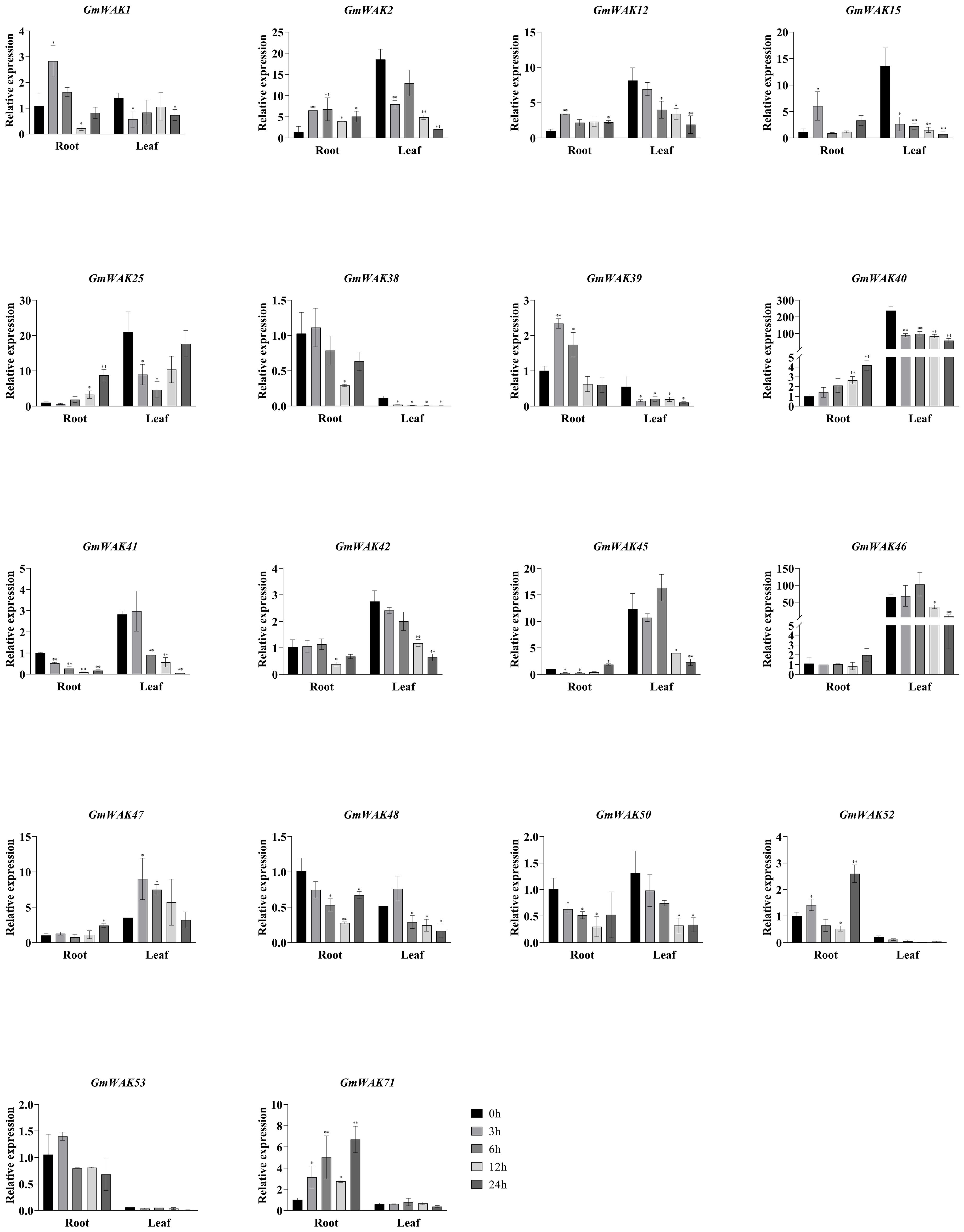
Figure 8. The expression levels of 18 GmWAK genes in soybean seedlings after cold (4°C) treatment. The abscissa indicates the time points of the leaf and root after the stress treatments. Data represent the average of three independent biological replicates ± SD. “*” and “**” indicate “P < 0.05” and “P < 0.01,” respectively, and mean significant difference occurred after treatment.
3.6.2 Responses of GmWAK genes to heat stress
Eighteen differentially expressed GmWAK genes were identified in soybeans under heat stress, which changed more than two-fold than that in the control group (Figure 9). Fifteen significantly upregulated GmWAK genes were detected in soybean roots under heat stress, GmWAK1/2/38/40/41/48 were up to 2.2–23.3 times the control at 3 h, GmWAK15/46/52/53/71 were up to 6.7–28.3 times the control at 6 h, and GmWAK25/39/42/45 were up to 2.4–7.9 times the control at 12 h. One GmWAK gene showed an expression pattern of first decreasing and then increasing. GmWAK50 significantly decreased by 79.2% after 3 h of treatment and increased by 82.0% after 24 h. Eleven differentially expressed GmWAK genes were identified under heat stress in soybean leaves. The expression level of GmWAK53 was sharply increased to 43,506 times that in the control after 3 h of treatment. GmWAK1/2/12/25/41/42/45/46/47/71 increased to 3.7–57.3 times the control group after 6 h of treatment. One GmWAK gene was significantly downregulated under heat stress. GmWAK40 was reduced to 6.2% of the control group after 6 h of treatment. Among the 18 significantly differentially expressed GmWAK genes, there were nine GmWAK genes with high expression levels, GmWAK1/42/45/46/47/52/53/71 were significantly increased by 6.4–43506 times in heat stress conditions, and GmWAK40 was upregulated by 14.5 times in the roots and yet was decreased by 93.8% in the leaves.
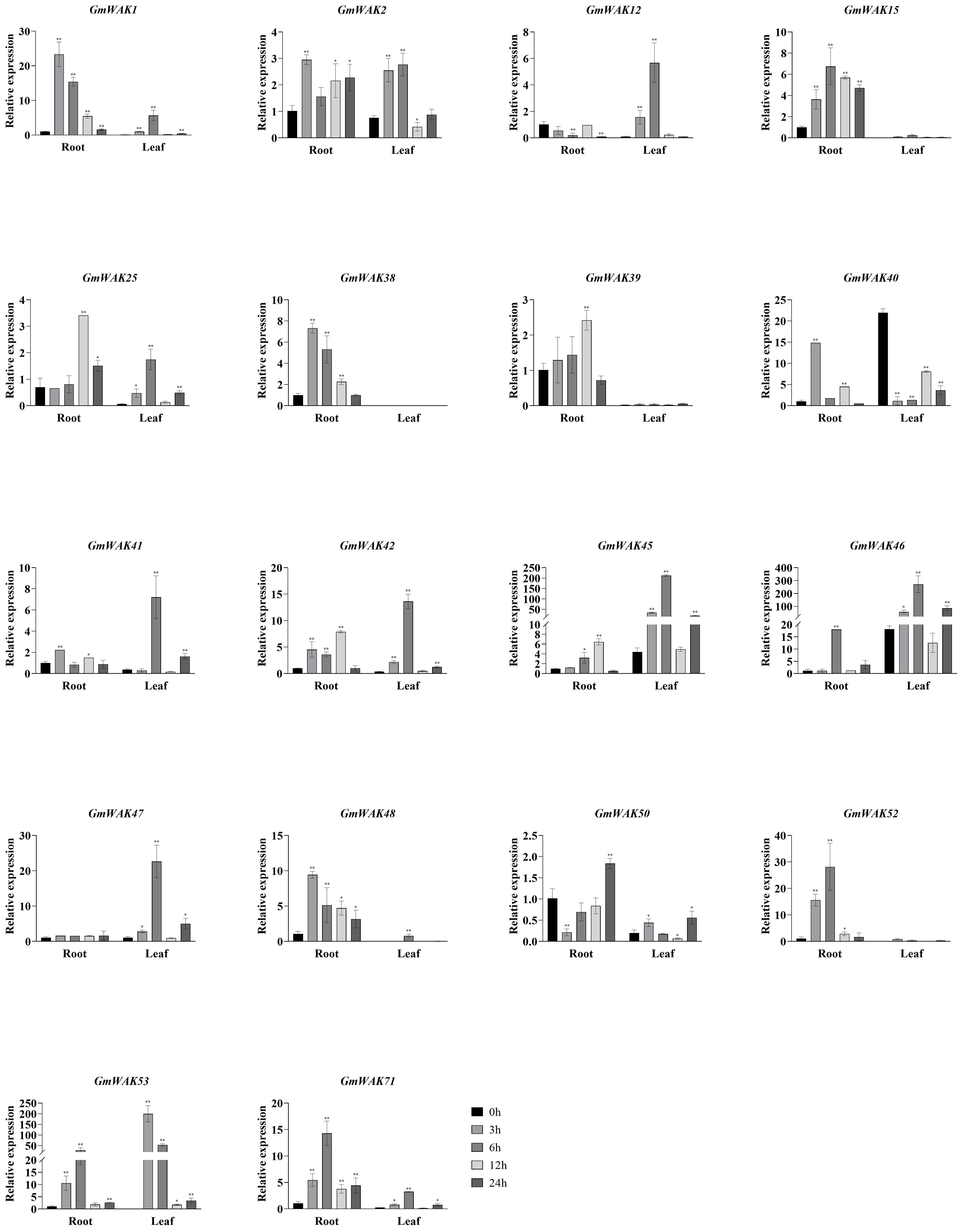
Figure 9. The expression levels of 18 GmWAK genes in soybean seedlings after heat (40°C) treatment. The abscissa indicates the time points of the leaf and root after the stress treatments. Data represent the average of three independent biological replicates ± SD. “*” and “**” indicate “P < 0.05” and “P < 0.01,” respectively, and mean significant difference occurred after treatment.
3.6.3 Responses of GmWAK genes to drought stress
Seventeen differentially expressed GmWAK genes were identified in soybeans under drought stress. These genes changed more than two-fold than those in the control group (Figure 10). There were 10 GmWAK genes significantly upregulated under drought stress in soybean roots. The expression levels of GmWAK1/2/38/39/40/52 reached their maximum at 3 h after treatment and were 2.1–10.6 times that of the control. GmWAK25/45/48 increased to 5.2–9.9 times the control at 6 h. GmWAK46 increased to 6.9 times the control at 24 h. Six GmWAK genes were significantly downregulated in soybean roots. The expression levels of GmWAK41/42 were reduced by 61.4% and 70% after 3 h of treatment, and GmWAK12/50/53/71 were reduced by 53.8%–99.7% at 12 h after treatment. Eleven GmWAK genes showed a significant increase under drought stress in soybean leaves. The expression of GmWAK1/2/12/25/41/42/45/46/47/48 increased to 1.6–21.9 times that of the control at 3 h after treatment. GmWAK50 was upregulated 8.1 times over the control at 6 h. Two GmWAK genes showed a significant decrease under drought stress, and the expression of GmWAK71 and GmWAK39 decreased by 83.7% and 99.7% at 6 h and 12 h, respectively. Five GmWAK genes had high expression levels among the 17 significantly differentially expressed genes. GmWAK25/45/46/47 were significantly increased by 2.0–21.9 times under heat stress, and GmWAK42 was downregulated by 70.0% in soybean roots and increased by 6.1 times in soybean leaves.
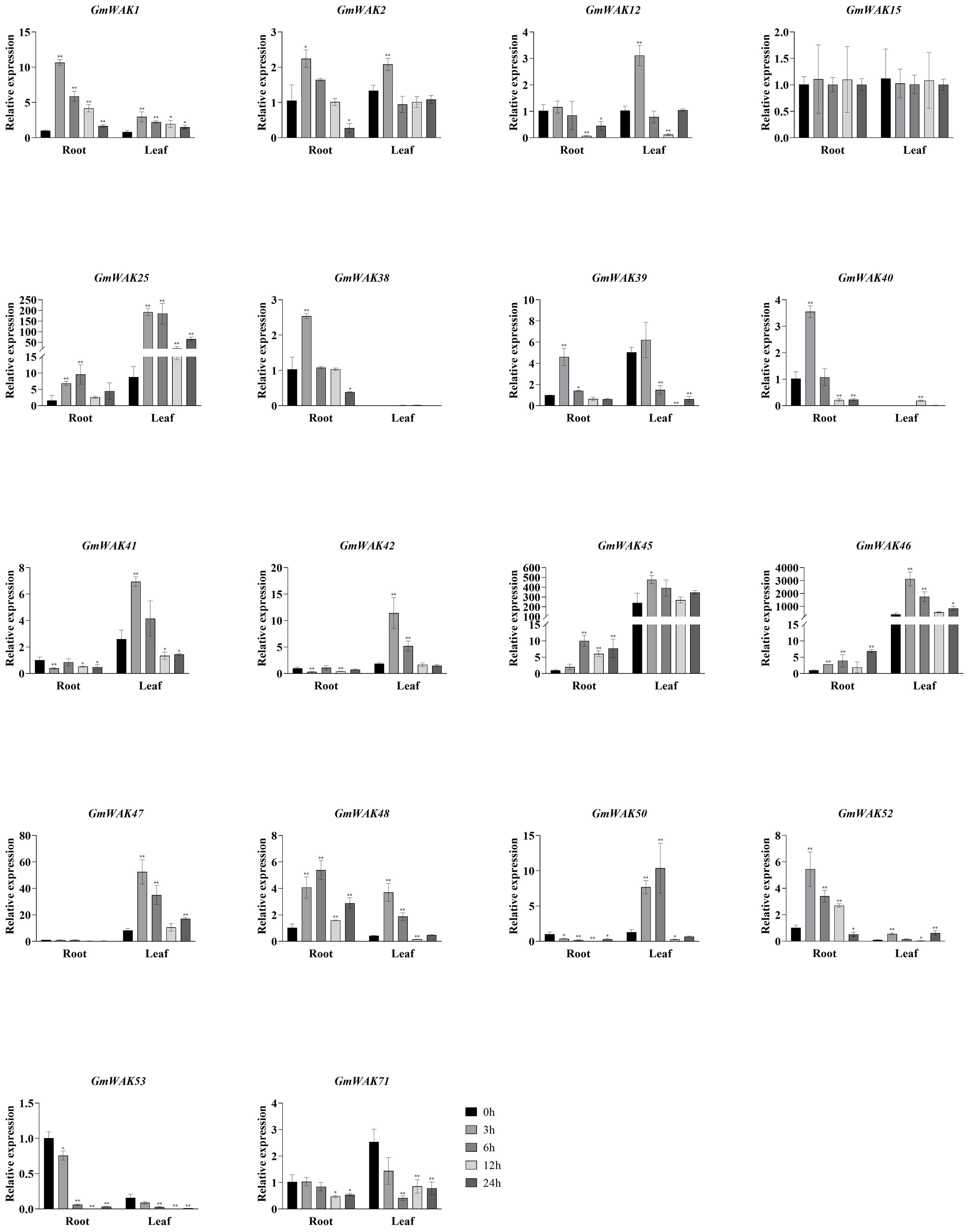
Figure 10. The expression levels of 18 GmWAK genes in soybean seedlings after drought treatment. The abscissa indicates the time points of the leaf and root after the stress treatments. Data represent the average of three independent biological replicates ± SD. “*” and “**” indicate “P < 0.05” and “P < 0.01,” respectively, and mean significant difference occurred after treatment.
3.6.4 Responses of GmWAK genes to salt stress
There were 18 differentially expressed GmWAK genes identified in soybeans, whose expression level was more than twice that of the control under salt stress (Figure 11). Eight GmWAK genes showed a significant increase in soybean roots under high salt treatment. The expression of GmWAK12 increased to 5.7 times the control at 3 h after treatment, GmWAK1/2/45/46 were up to 2.3–2.8 times the control at 6 h, GmWAK25 achieved 27.4 times of the control at 12 h, and GmWAK40/50 increased by 4.9 and 13.4 times at 24 h, respectively. Six GmWAK genes showed a significant decrease under high-salt treatment. GmWAK39/53 decreased by 80% and 87.2% at 12 h after treatment. GmWAK38/48/52/71 decreased by 35.6%–90.2% at 24 h. Nine GmWAK genes significantly increased under high salt treatment in soybean leaves. The expression of GmWAK40/42/45/46/47/48 increased by 1.7–3.9 times that of the control at 3 h after treatment. GmWAK15, GmWAK 25, and GmWAK 41 increased by 4.5, 10.7, and 3.3 times at 6 h, 12 h, and 24 h, respectively. Five GmWAK genes showed a significant decrease in soybean leaves. The expression of GmWAK1/2/12/50/71 was reduced to 4.6%–34.1% of the control group at 12 h after treatment. Among the 18 significantly differentially expressed genes, the relative expression of GmWAK25/40/45/46/47 was at high levels, significantly increasing by 2.4–27.4 times and 1.7–10.7 times in the roots and leaves, respectively.
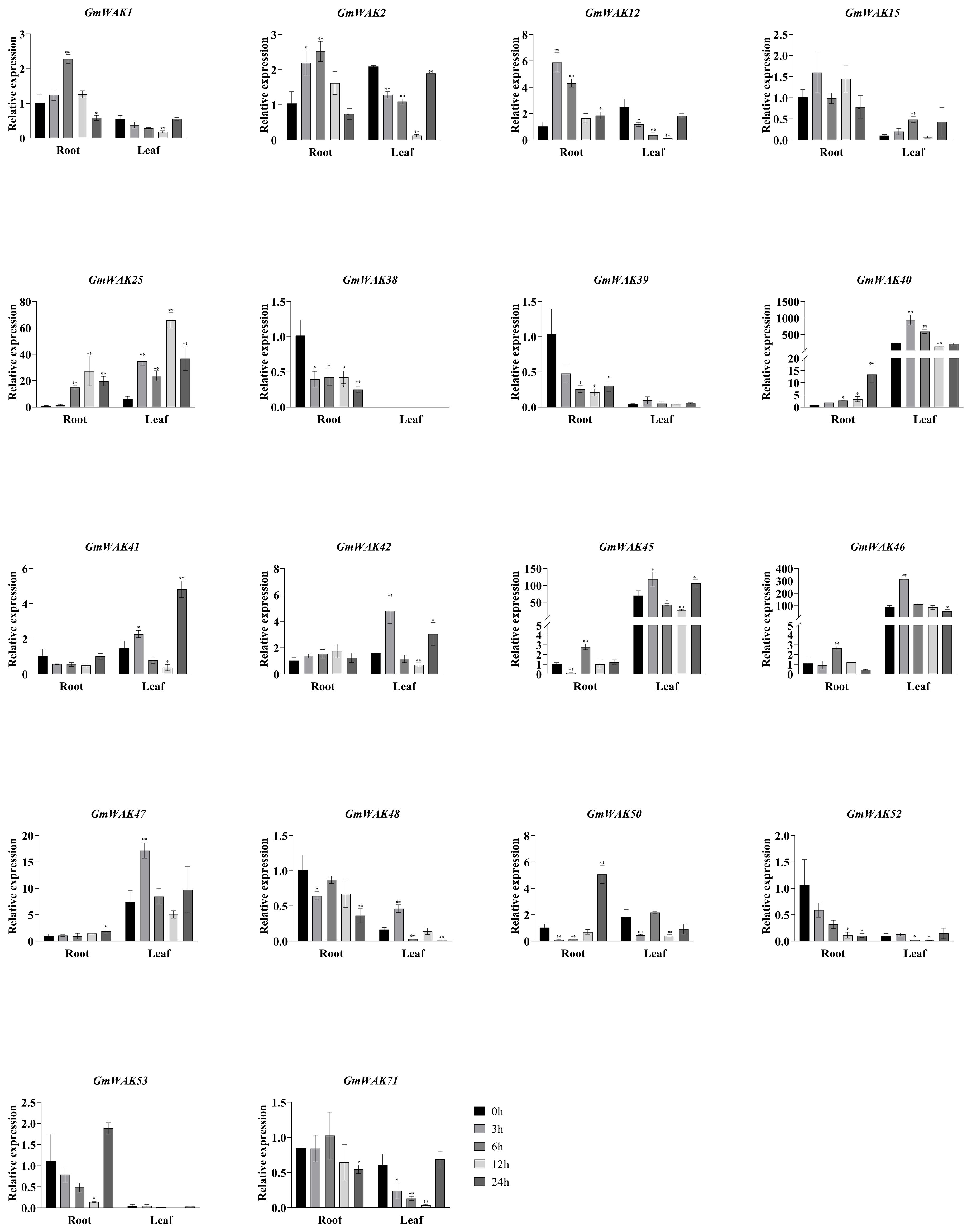
Figure 11. The expression levels of 18 GmWAK genes in soybean seedlings after salt treatment. The abscissa indicates the time points of the leaf and root after the stress treatments. Data represent the average of three independent biological replicates ± SD. “*” and “**” indicate “P < 0.05” and “P < 0.01,” respectively, and mean significant difference occurred after treatment.
3.6.5 Responses of GmWAK genes to Al stress
Eighteen differentially expressed GmWAK genes were observed in soybeans under Al stress. These genes changed more than two-fold than those in the control group (Figure 12). Fifteen GmWAK genes in soybean roots were significantly upregulated under Al stress. The expression of GmWAK46/50 increased 3.4 and 2.5 times, respectively, at 6 h after treatment. The remaining 13 GmWAK genes increased 1.5–13.2 times after 3 h of treatment. Three GmWAK genes were significantly downregulated in soybean roots. The expression of GmWAK12/42 was reduced by 87.4% and 91.9% at 12 h after treatment, and GmWAK40 was decreased by 73.7% at 24 h. Three GmWAK genes in soybean leaves were significantly upregulated by Al stress. The expression of GmWAK46/53 was increased to 1.5 and 2.8 times that of the control at 3 h after treatment. GmWAK52 was increased eight times at 24 h. Fifteen GmWAK genes were significantly downregulated in soybean leaves. GmWAK12/15/25/46/48 expression was reduced by 79.8%–99.8% at 6 h after treatment. GmWAK1/2/39/40/41/42/45/47/50/71 reduced by 89.0%–99.3% at 12 h. Among the 18 significantly differentially expressed genes, seven GmWAK genes had high expression levels in soybeans. The expression of GmWAK1/2/45/46/47 was significantly increased by 1.5–13.2 times in the roots but decreased by 81.7%–99.7% in the leaves. GmWAK12/40 decreased by 73.7%–99.8% in soybean roots and leaves.
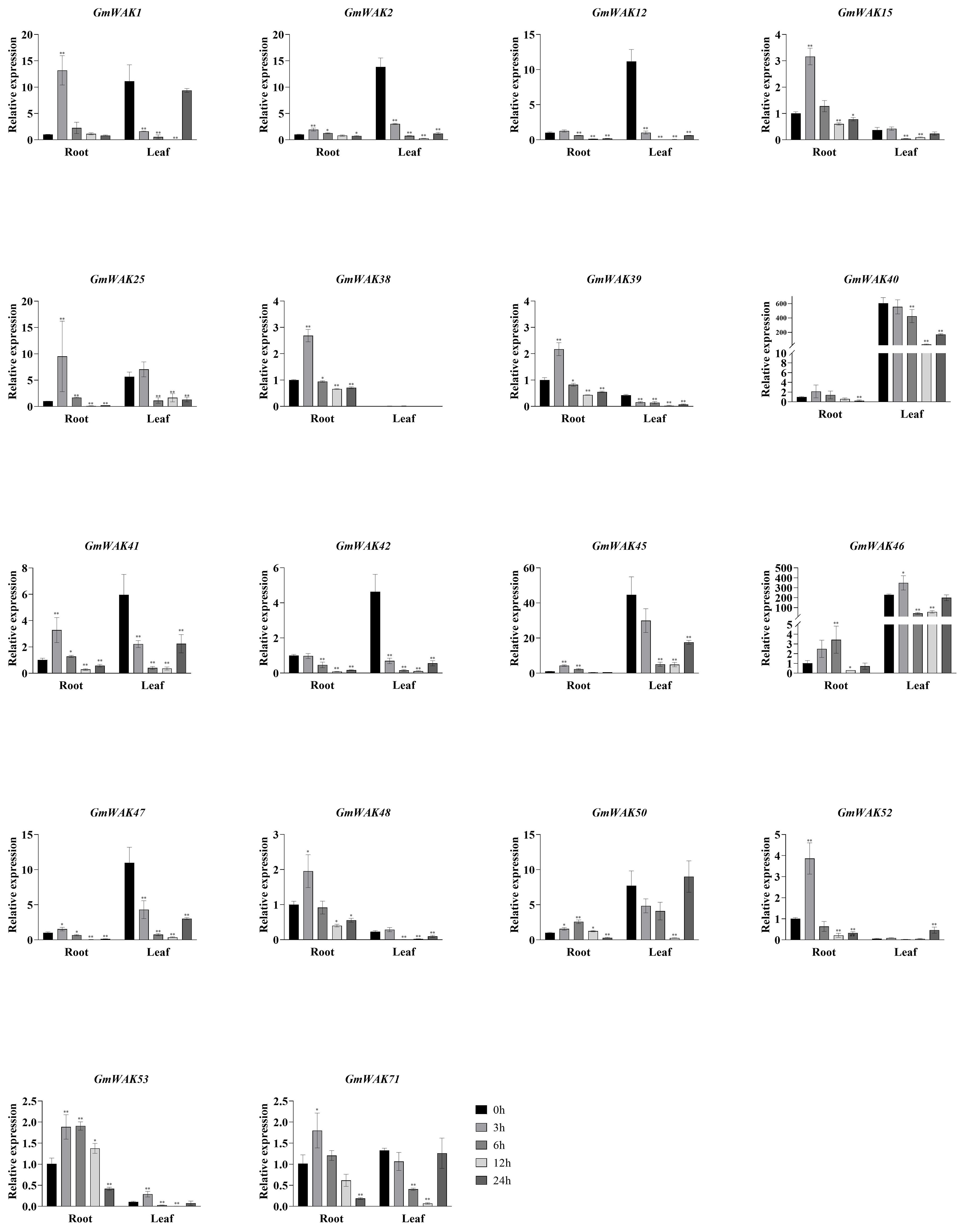
Figure 12. The expression levels of 18 GmWAK genes in soybean seedlings after Al treatment. The abscissa indicates the time points of the leaf and root after the stress treatments. Data represent the average of three independent biological replicates ± SD. “*” and “**” indicate “P < 0.05” and “P < 0.01,” respectively, and mean significant difference occurred after treatment.
3.6.6 Responses of GmWAK genes to Cd stress
Eighteen differentially expressed GmWAK genes were detected in soybeans under Cd stress. The changes in these genes in the treatment group were more than twice that of the control group (Figure 13). Six GmWAK genes were significantly upregulated under Cd stress in soybean roots. The expression of GmWAK50/53 was increased by 3.7 and 1.7 times at 6 h after treatment, and GmWAK1/48/52/71 increased by 3.7–8.1 times at 24 h. Eleven GmWAK genes were significantly downregulated in soybean roots. The expression of GmWAK2/12/25/38/41/42/45/46/47 reduced by 67.8%–94.8% at 12 h after treatment. GmWAK39/40 decreased by 83.1% and 89.5%, respectively, at 24 h. Nine GmWAK genes were significantly upregulated by Cd stress in soybean leaves. The expression of GmWAK1/15/40/41/42/45/46 increased by 1.9–9.5 times that of the control at 6 h after treatment. GmWAK2/12 increased to 5.4 and 3.4 times, respectively, after 24 h. Five GmWAK genes were significantly downregulated in soybean leaves. The expression of GmWAK48 was reduced by 96% 3 h after treatment. GmWAK25/47/50/71 were reduced by 65.6%–85.0% at 12 h. Among the 18 significantly differentially expressed genes, the relative expression of GmWAK40/45/46 was at high levels in soybeans, which were significantly decreased by 67.8%–89.5% in the roots and increased by 2.7–5.0 times in the leaves.
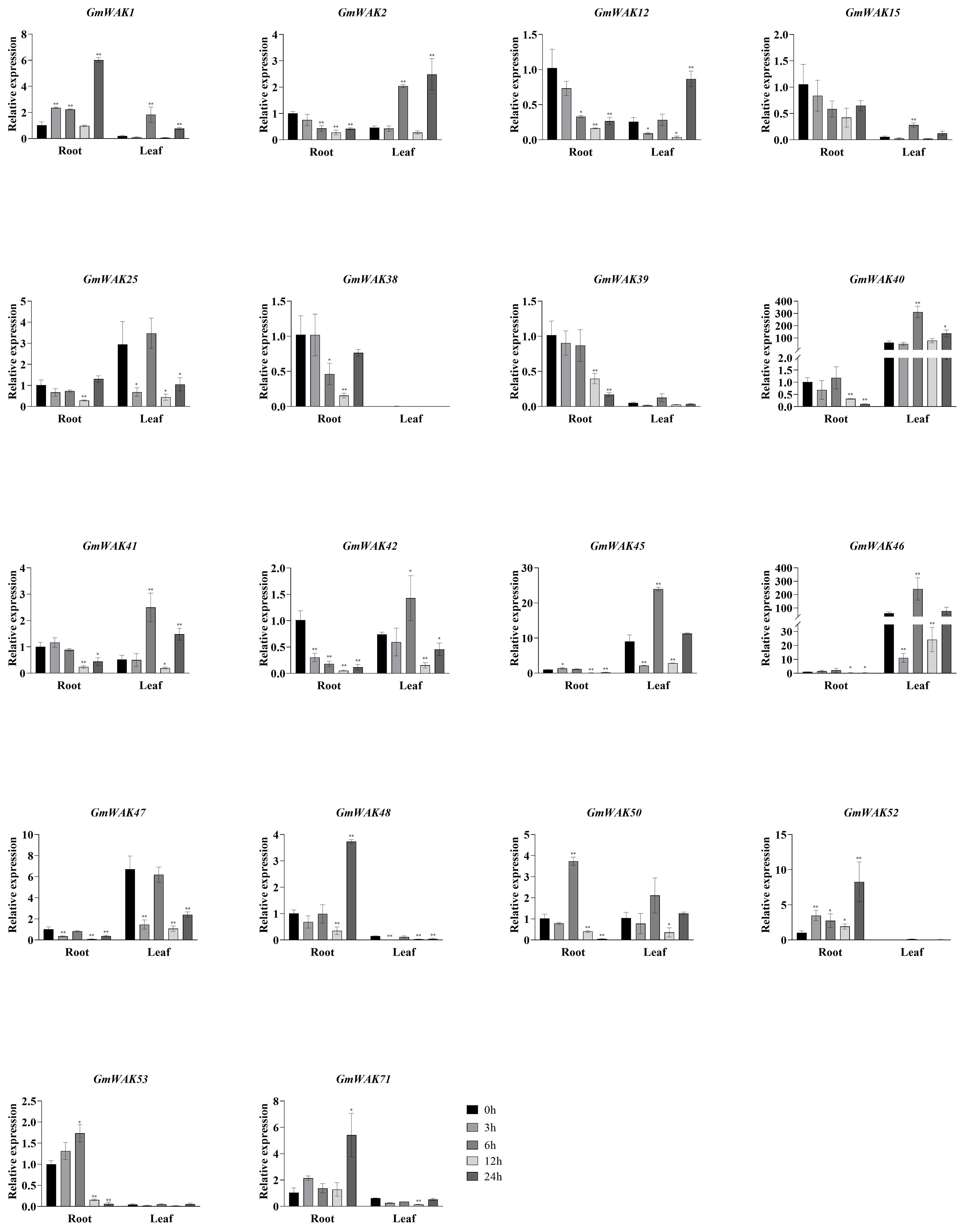
Figure 13. The expression levels of 18 GmWAK genes in soybean seedlings after Cd treatment. The abscissa indicates the time points of the leaf and root after the stress treatments. Data represent the average of three independent biological replicates ± SD. “*” and “**” indicate “P < 0.05” and “P < 0.01,” respectively, and mean significant difference occurred after treatment.
4 Discussion
WAK is a family of receptor-like kinases crucial in signal transduction between the cell wall and the cytoplasm (Nakhamchik et al., 2004). WAK genes participate in several physiological processes in plant growth and development, such as cell expansion and elongation, pathogen resistance, and metal tolerance (Lally et al., 2001; Zuo et al., 2015; Xia et al., 2018). WAK genes are highly conserved. The typical domains of WAK genes include the transmembrane, EGF, GUB_WAK _bind, and Pkinase domains (He et al., 1999; Zhang et al., 2005; Zuo et al., 2019; Tripathi et al., 2021). At present, 27, 125, 6, 91, 29, 29, 29, 44, 68, and 27 WAK/WAKL genes have been identified in Arabidopsis, rice, wheat, barley, cotton, tomato, potato, apple, rose, and walnut, respectively (Vaid et al., 2012; De et al., 2014; Liu et al., 2006; Tripathi et al., 2021; Dou et al., 2021; Sun et al., 2020; Yu et al., 2022; Zuo et al., 2019; Liu et al., 2021; Li et al., 2022). However, the WAK gene family of soybeans has not yet been systematically identified and characterized. In this study, 74 GmWAK genes were identified in the soybean genome (Figure 1). There were more WAK genes in soybean than in Arabidopsis (27 genes) but much less than in rice (125 genes) (Vaid et al., 2012; De et al., 2014). This indicated a wide variation of the WAK gene family in plants. The expansion of the WAK gene family in plants has contributed to genome-wide duplication (Vaid et al., 2012; De et al., 2014). The increasing amount of WAK genes in soybeans was due to SD (27 genes), TD (15 genes), and PD (9 genes) events in genome-wide duplication (Table 1). The WAK gene family expanded through TD and large-scale duplication in Arabidopsis, whereas this gene family expanded through TD in rice (Shiu et al., 2004). This suggests differences among plants in the WAK gene family expansion. GD analysis showed that the TD events of GmWAK genes occurred approximately 23.84 Mya (5.88–52.4 Mya), the PD events of GmWAK genes occurred approximately 156.05 Mya (0.61–207.14 Mya), and the SD events were approximately 102.10 Mya (10.35–290.33 Mya) (Table 1). This result suggests that the expansion of the soybean WAK gene family is PD first, followed by SD, and finally TD.
Neofunctionalized or subfunctionalized genes occur in GD events (Lynch and Conery, 2000; Roulin et al., 2013). Neofunctionalization generated new functional genes, and subfunctionalization divided the functions of the ancestral gene (Conant et al., 2014; Gout and Lynch, 2015). The subfunctionalized genes coexisted on the genome and worked together in the dose-balance model to ensure the expression of genes at normal levels (Lan and Pritchard, 2016). The neofunctionalized gene pairs experienced positive selection (Ka/Ks > 1), while the subfunctionalized gene pairs experienced purifying selection (Ka/Ks < 1) (Lynch and Conery, 2000; Roulin et al., 2013). The Ka/Ks values of 34 duplication gene pairs were lower than 1 in the soybean WAK gene family, suggesting that these genes underwent purifying selection and might be subfunctionalized (Table 1).
The phylogenetic analysis showed that 74 WAK genes in soybeans were divided into four subgroups (Figure 2). Similar divisions were observed in other plants, such as Arabidopsis, tomato, barley, and cotton (Verica and He, 2002; Sun et al., 2020; Tripathi et al., 2021; Dou et al., 2021). Approximately 78.4% of GmWAK genes obtained three to four exons, and 16.2% of GmWAK genes obtained one to two exons (Figure 3; Supplementary Table S2). In other plants, 79.3%–89.7% of the WAK genes had three to four exons, and 3.5%–13.8% of genes had one to two exons (Sun et al., 2020; Dou et al., 2021; Yu et al., 2022). The structural similarity of WAK genes among different plants suggests that the WAK gene family was evolutionarily conserved. Additionally, WAK genes in the same subgroup had similar intron–exon distribution, conserved motifs, and domains, implying that they might share similar functions.
GUB_WAK_bind, EGF, transmembrane domain, and Pkinase domains are typical domains of the WAK gene family (Verica and He, 2002; Tang et al., 2022). The GUB_WAK_bind domain is rich in cysteine residues and constitutes the extracellular domain of Ser/Thr protein kinase, which can bind to cross-linked pectin, oligo-galacturonides, injury-induced pectin fragments, or pathogens, regulating cell expansion and activating stress responses (Ridley et al., 2001; Kohorn and Kohorn, 2012; Kohorn, 2015, 2016). The EGF domain is located in extracellular space. It is tightly connected to the cell wall and interacts with different proteins in the cell wall (Hu and Lou, 2010). The transmembrane domain transfers signals to downstream molecules through in-vivo phosphorylation (Verica and He, 2002). The Pkinase domain is located inside the cell and is the key domain in which the WAK gene functions (Verica and He, 2002. Fourteen WAK genes in the soybean WAK gene family possessed four typical domains, indicating that these genes might be able to perceive and recognize external environmental signals. Fifty-six GmWAK genes contained three typical domains (e.g., GUB_WAK_bind/EGF, transmembrane_domain, and Pkinase domain), and four GmWAK genes contained two typical domains (e.g., GUB_WAK_bind and Pkinase domain) (Supplementary Table S2). These genes included the Pkinase domain but lacked transmembrane or EGF domains, which might synergistically participate in extracellular signal transduction with other WAK genes. Nineteen hormone-responsive elements, 18 stress-response elements, 29 light-response elements, and 16 metabolic- and growth-related elements were identified in the promoter regions of soybean WAK genes (Supplementary Tables S7, S8). Hormone- and stress-related elements have been recognized in most GmWAK genes, such as MJA, JA, SA, ABA, ETH, GA, IAA, drought, heat, anaerobism, and trauma. MJA, JA, and SA are important signal molecules involved in plant pathogen defense. Such molecules improve plant resistance by promoting cell wall synthesis and maintaining cell wall integrity (Takahashi et al., 2004). ABA, ETH, GA, and IAA participate in plant growth, development, and abiotic stress response (Zhao et al., 2016; Luo, 2024; Colebrook et al., 2014; Xu and Chen, 2023). Previous studies showed that the AtWAK gene was highly expressed in vigorous growth and differentiation tissues when the cell size and morphology of the Atwak mutant of Arabidopsis changed (Wagner and Kohorn, 2001; Kohorn et al., 2006). AtWAK1 showed a typical on-and-off pattern, with a first peak at 3 h and a complete disappearance after 9 h of Al exposure. Transgenic plants overexpressing ATWAK1 enhance Al tolerance (Sivaguru et al., 2003). ZmWAK enhanced the resistance of maize to head smut disease (Zuo et al., 2015). Therefore, we speculate that the WAK gene family might be involved in the physiological process of plant growth and development and biotic and abiotic stresses by responding to multiple hormone signals.
The tissue expression pattern of a gene is closely related to gene function. Forty-seven GmWAK genes were expressed in different soybean tissues, such as the roots, stems, leaves, flowers, pods, and seeds (FPKM > 1) (Figure 7; Supplementary Table S9). Among them, the expression levels of six GmWAK genes were high: GmWAK34/38/53 were highly expressed in the roots, GmWAK40/45 were highly expressed in the stems or leaves, and GmWAK15 was highly expressed in the roots, leaves, and pods. This result indicates that they might play an important role in the growth and development of soybean plants.
WAKs could perceive and transmit environmental signals to cells and play vital regulating roles in plant growth, development, and response to environmental stresses (Sun et al., 2020; Zhang et al., 2021). In this study, 12 GmWAK genes were expressed at high levels under abiotic conditions (Figures 8–13). Among them, GmWAK40/45/46/47 were simultaneously observed in various abiotic stresses. GmWAK40 was homologous with AT4G03230 in A. thaliana. AT4G03230 encoded a G-type lectin s-receptor-like serine/threonine-kinase, and its T-DNA insertion lines showed enhancing resistance to IAA, NPA, NaCl, and mannitol compared with the wild type (Ten et al., 2011). GmWAK40 was highly expressed in soybean leaves. The expression levels of GmWAK40 were significantly reduced by 76.1%, 94.9%, and 94.7% under cold, heat, and Al stresses, respectively. In contrast, they were significantly increased by 3.9 and 5.0 times after 3 h or 6 h, respectively, under salt or Cd stress. Many responsive elements, such as MJA, JA, ethylene, GA, cold, and heat, were identified in the promoter region of GmWAK40. MJA and JA are natural physiologically active substances and have been proven to regulate the adaptive responses of plants to various environmental stresses (Rahman et al., 2024). Multiple abiotic stresses were significantly induced by GmWAK40, suggesting that GmWAK40 might be related to the response to different abiotic stresses. GmWAK45/46 were a pair of duplicated genes that encoded the glycerol phosphodiesterases. Glycerol phosphodiesterase is a highly conserved enzyme in prokaryotes and eukaryotes, which participates in glycerol phospholipid metabolism by catalyzing glycerophosphodiester to generate glycerophosphate and alcohol (Patton-Vogt, 2007). Glycerol phospholipid metabolism participated in responding to various abiotic stresses in previous studies, such as the low nitrogen stress tolerance of sorghum, the cold stress of loquat, and the salt-alkali stress of Suaeda salsa (Wang et al., 2024; Xu et al., 2021; Sun, 2022). GmWAK45/46 were highly expressed in soybean leaves. The expression of GmWAK45/46 was significantly increased by 1.69–48.5 times under heat, salt, drought, and Cd stresses but was decreased by 72.5%–88.6% under cold and Al stresses. These results indicate that GmWAK45/46 might participate in response to abiotic stresses with glycerol phospholipid metabolism. GmWAK47 was homologous to AT1G66980 in Arabidopsis. AT1G66980 encoded an atypical receptor-like kinase, SNC4, which was a suppressor of NPR1-1 and was related to the Arabidopsis defense response. Its mutant snc4-1D displays a constitutive activation of defense responses (Zhang et al., 2014). The expression of GmWAK47 in soybeans showed significant changes under abiotic stresses. Decreases of 96.5% and 83.9% were observed under Al and Cd stresses, and an increase of 2.3–21.8 times was observed in the cold, heat, drought, and salt conditions. These results imply that GmWAK47 is possibly involved in the response of plants to abiotic stresses.
5 Conclusion
We conducted genome-wide identification of the WAK gene family in soybeans in this study. Seventy-four GmWAK genes were detected in the soybean genome and unevenly distributed on 19 chromosomes. The 74 GmWAK genes could be divided into four subgroups based on phylogenetic analysis. The genes within the same subgroup shared similar gene structures, conserved motifs, and domains. SD was critical in expanding the soybean WAK gene family, and purification selection was conducted in evolution. Many hormone- and stress-response-related elements were predicted in promoter regions of the GmWAK genes. Eighteen GmWAK genes displayed significant changes under abiotic stresses. Among them, GmWAK40/45/46/47 were simultaneously observed in various stresses, implying that they might be crucial for generating stress tolerance in soybeans. These genes could be potential candidates for investigating the molecular mechanisms of stress resistance in soybeans and as a genetic resource to cultivate new soybean varieties with enhanced stress resistance.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
XL: Conceptualization, Data curation, Investigation, Methodology, Writing – original draft, Writing – review & editing. SQ: Data curation, Investigation, Writing – review & editing. LM: Formal analysis, Methodology, Writing – review & editing. PS: Data curation, Supervision, Writing – review & editing. YWS: Data curation, Writing – review & editing. NL: Formal analysis, Writing – review & editing. DW: Investigation, Writing – review & editing. YF: Conceptualization, Writing – review & editing. YS: Funding acquisition, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Doctoral Research Start-up Foundation of Liaocheng University (318052325), the National Natural Science Foundation of China (32101788), and the Natural Foundation of Shandong Province (ZR2020MC034).
Acknowledgments
We thank Chengjie Chen for providing technical assistance in bioinformatics, and we appreciate the linguistic assistance provided by Charlesworth during the preparation of this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1511681/full#supplementary-material
References
Afzal, A. J., Wood, A. J., Lightfoot, D. A. (2008). Plant receptor-like serine threonine kinases: roles in signaling and plant defense. Mol. Plant Microbe Interact. 21, 507–517. doi: 10.1094/mpmi-21-5-0507
Almagro, A. J.J., Sønderby, C. K., Sønderby, S. K., Nielsen, H., Winther, O. (2017). DeepLoc: prediction of protein subcellular localization using deep learning. Bioinformatics. 33, 3387–3395. doi: 10.1093/bioinformatics/btx548
Carrera, C. S., Dardanelli, J. L. (2017). Water deficit modulates the relationship between temperature and unsaturated fatty acid profile in soybean seed oil. Crop Sci. 57, 3179–3189. doi: 10.2135/cropsci2017.04.0214
Chandra, J., Keshavkant, S. (2021). Mechanisms underlying the phytotoxicity and genotoxicity of aluminum and their alleviation strategies: A review. Chemosphere. 278, 130384. doi: 10.1016/j.chemosphere.2021.130384
Chauhan, D. K., Yadav, V., Vaculík, M., Gassmann, W., Pike, S., Arif, N., et al. (2021). Aluminum toxicity and aluminum stress-induced physiological tolerance responses in higher plants. Crit. Rev. Biotechnol. 41, 715–730. doi: 10.1080/07388551.2021.1874282
Chen, C., Wu, Y., Li, J., Wang, X., Zeng, Z., Xu, J., et al. (2023). TBtools-II: A "one for all, all for one" bioinformatics platform for biological big-data mining. Mol. Plant 16, 1733–1742. doi: 10.1016/j.molp.2023.09.010
Colebrook, E. H., Thomas, S. G., Phillips, A. L., Hedden, P. (2014). The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 217, 67–75. doi: 10.1242/jeb.089938
Conant, G. C., Bircher, J. A., Pires, J. C. (2014). Dosage, duplication and diploidization: clarifying the interplay of multiple models for duplicate gene evolution over time. Curr. Opin. Plant Biol. 19, 91–98. doi: 10.1016/j.pbi.2014.05.008
Dai, Z., Pi, Q., Liu, Y., Hu, L., Li, B., Zhang, B., et al. (2024). ZmWAK02 encoding an RD‐WAK protein confers maize resistance against gray leaf spot. New Phytologist 241. doi: 10.1111/nph.19465
De Oliveira, L. F. V., Christoff, A. P., Lima., J. C., Ross, B. C. F., Sachetto-Martins, G., Margis-Pinheiro, M., et al. (2014). The wall-associated kinase gene family in rice genomes. Plant Sci. 229, 181–192. doi: 10.1016/j.plantsci.2014.09.007
Decreux, A., Messiaen, J. (2005). Wall-associated kinase WAK1 interacts with cell wall pectins in a calcium-induced conformation. Plant Cell Physiol. 46, 268–278. doi: 10.1093/pcp/pci026
Djanaguiraman, M., Prasad, P. V. V., Boyle, D. L., Schapaugh, W. T. (2013). Soybean pollen anatomy, viability and pod set under high temperature stress. J. Agron. Crop Sci. 199, 171–177. doi: 10.1111/jac.12005
Dou, L., Li, Z., Shen, Q., Shi, H., Li, H., Wang, W., et al. (2021). Genome-wide characterization of the WAK gene family and expression analysis under plant hormone treatment in cotton. BMC Genomics 22, 85. doi: 10.1186/s12864-021-07378-8
Gout, J. F., Lynch, M. (2015). Maintenance and loss of duplicated genes by dosage subfunctionalization. Mol. Biol. Evol. 32, 2141–2148. doi: 10.1093/molbev/msv095
He, Z., Cheeseman, I., He, D., Kohorn, B. D. (1999). A cluster of five cell wall-associated receptor kinase genes, Wak1–5, are expressed in specific organs of Arabidopsis. Plant Mol. Biol. 39, 1189–1196. doi: 10.1023/a:1006197318246
He, Z., Zhang, H., Gao, S., Lercher, M. J., Chen, W. H., Hu, S. (2016). Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 44, W236–W241. doi: 10.1093/nar/gkw370
Hu, L. F., Lou, Y. G. (2010). Role of WAK gene family in signal transduction and plant resistance. Natl. Symposium Chem. Ecology., 84–85.
Hu, W., Lv, Y., Lei, W., Li, X., Chen, Y., Zheng, L., et al. (2014). Cloning and characterization of the Oryza sativa wall-associated kinase gene OsWAK11 and its transcriptional response to abiotic stresses. Plant Soil. 384, 335–346. doi: 10.1007/s11104-014-2204-8
Huang, S. C., Liu, A. R., Ye, M. R. (2015). Expression characterization of soybean GmDnaJ1 in response to heavy metal stresses. Guihaia. 35, 288–294. doi: 10.11931/guihaia.Gxzw201408020
Hussain, M. A., Li, S., Gao, H., Feng, C., Sun, P. Y., Sui, X. P., et al. (2023). Comparative analysis of physiological variations and genetic architecture for cold stress response in soybean germplasm. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1095335
Kanneganti, V., Gupta, A. K. (2008). Wall associated kinases from plants - an overview. Physiol. Mol. Biol. Plants. 14, 109–118. doi: 10.1007/s12298-008-0010-6
Kaur, R., Singh, K., Singh, J. (2013). A root-specific wall-associated kinase gene, HvWAK1, regulates root growth and is highly divergent in barley and other cereals. Funct. Integr. Genomics 13, 167–177. doi: 10.1007/s10142-013-0310-y
Kohorn, B. D. (2001). WAKs; cell wall associated kinases. Curr. Opin. Cell Biol. 13, 529–533. doi: 10.1016/S0955-0674(00)00247-7
Kohorn, B. D. (2015). The state of cell wall pectin monitored by wall associated kinases. A Model. Plant Signal Behav. 10, e1035854. doi: 10.1080/15592324.2015.1035854
Kohorn, B. D. (2016). Cell wall-associated kinase and pectin perception. Exp. Bot. 67, 489–494. doi: 10.1093/jxb/erv467
Kohorn, B. D., Kobayashi, M., Johansen, S., Riese, J., Huang, L., Koch, K. E., et al. (2006). An Arabidopsis cell wall-associated kinase required for invertase activity and cell growth. Plant Mol. Biol. 46, 307–316. doi: 10.1080/15592324.2015.1035854
Kohorn, B. D., Kohorn, S. L. (2012). The cell wall-associated kinases, WAKs, as pectin receptors. Front. Plant Sci. 3. doi: 10.3389/fpls.2012.00088
Lally, D., Ingmire, P., Tong, H. Y., He, Z. H. (2001). Antisense expression of a cell wall-associated protein kinase, WAK4, inhibits cell elongation and alters morphology. Plant Cell. 13, 1317–1331. doi: 10.1105/tpc.13.6.1317
Lan, X., Pritchard, J. K. (2016). Coregulation of tandem duplicate genes slows evolution of subfunctionalization in mammals. Science. 352, 1009–1013. doi: 10.1126/science.aad8411
Lehti-Shiu, M. D., Zou, C., Hanada, K., Shiu, S. H. (2009). Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes. Plant Physiol. 150, 12–26. doi: 10.1104/pp.108.134353
Li, H., Zhou, S. Y., Zhao, W. S., Fan, J., Peng, Y. L. (2004). Cloning and function analysis of a rice wall associated receptor like kinase gene involved in resistance against blast. Chin. Soc. Plant Pathology., 572–573.
Li, M., Ma, J., Liu, H., Ou, M., Ye, H., Zhao, P. (2022). Identification and characterization of wall-associated kinase (WAK) and wak-like (WAKL) gene family in Juglans regia and its wild related species Juglans mandshurica. Genes. 13, 134. doi: 10.3390/genes13010134
Li, M. Z. (2023). Molecular mechanism of TaWAK20 in Triticum aestivum in response to cadmium stress. Guizhou Normal University. doi: 10.27048/d.cnki.ggzsu.2023.001236
Lin, W., Wang, Y., Liu, X., Shang, J. X., Zhao, L. (2021). OsWAK112, A wall-associated kinase, negatively regulates salt stress responses by inhibiting ethylene production. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.751965
Liu, H. Y., Cen, K., Liu, Y. L., Lou, X. Y., Zhang, Y. T., Wu, R. X., et al. (2024). Photosynthetic repair of Glycine max (Linn.) Merr, by compound fungus agents and immobilization effect under cadmium stress. Acta Agronomica Sinica. 50, 2538–2549. doi: 10.3724/SP.J.1006.2024.44046
Liu, X., Wang, Z., Tian, Y., Zhang, S., Li, D., Dong, W., et al. (2021). Characterization of wall-associated kinase/wall-associated kinase-like (WAK/WAKL) family in rose (Rosa chinensis) reveals the role of RcWAK4 in Botrytis resistance. BMC Plant Biol. 21, 1–12. doi: 10.1186/s12870-021-03307-9
Liu, Y., Liu, D., Zhang, H., Gao, H., Zhang, A. (2006). Isolation and characterisation of six putative wheat cell wall-associated kinases. Funct. Plant Biol. 33, 811–821. doi: 10.1071/FP06041
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25, 402–408. doi: 10.1006/meth.2001.1262
Luo, X. J. (2024). Abscisic acid inhibits root growth in Arabidopsis through inducing ethylene biosynthesis (China Agricultural University). doi: CNKI:CDMD:1.1014.221235
Lynch, M., Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science. 290, 1151–1155. doi: 10.1126/science.290.5494.1151
Ma, J. F. (2007). Syndrome of aluminum toxicity and diversity of aluminum resistance in higher plants. Int. Rev. Cytol. 264, 225–252. doi: 10.1016/S0074-7696(07)64005-4
Ma, Y., Wang, Z., Humphries, J., Ratcliffe, J., Bacic, A., Johnson, K. L., et al. (2024). WALL-ASSOCIATED KINASE Like 14 regulates vascular tissue development in Arabidopsis and tomato. Plant Sci. 341, 112013. doi: 10.1016/j.plantsci.2024.112013
Mangeon, A., Pardal, R., Menezes-Salgueiro, A. D., Duarte, G. L., de, S. R., Cruz, F. P., et al. (2016). AtGRP3 is implicated in root size and aluminum response pathways in Arabidopsis. PloS One 11, e0150583. doi: 10.1371/journal.pone.0150583
Mehla, S., Singh, Y., Kumar, U., Balyan, P., Singh, K. P., Dhankher, O. P. (2024). Overexpression of rice lectin receptor-like kinase, OsLec-RLK, confers salinity stress tolerance and increases seed yield in pigeon pea (Cajanus cajan (L.) Millsp.). Plant Cell Rep. 43, 230. doi: 10.1007/s00299-024-03314-8
Mourtzinis, S., Specht, J. E., Lindsey, L. E., Wiebold, W. J., Ross, J., Nafziger, E. D., et al. (2015). Climate-induced reduction in US-wide soybean yields underpinned by region-and in-season-specific responses. Nat. Plants. 1, 14026. doi: 10.1038/nplants.2014.26
Nagajyoti, P. C., Lee, K. D., Sreekanth, T. (2010). Heavy metals, occurrence and toxicity for plants: a review. Environ. Chem. Lett. 8, 199–216. doi: 10.1007/s10311-010-0297-8
Nakhamchik, A., Zhao, Z., Provart, N. J., Shiu, S. H., Keatley, S. K., Cameron, R. K., et al. (2004). A comprehensive expression analysis of the Arabidopsis proline-rich extensin-like receptor kinase gene family using bioinformatic and experimental approaches. Plant Cell Physiol. 45, 1875–1881. doi: 10.1093/pcp/pch206
Patton-Vogt, J. (2007). Transport and metabolism of glycerophosphodiesters produced through phospholipid diacylation. Biochim. Biophys. Acta 1771, 337–342. doi: 10.1016/j.bbalip.2006.04.013
Rahman, M. M., Mostofa, M. G., Keya, S. S., Ghosh, P. K., Abdelrahman, M., Anik, T. R., et al. (2024). Jasmonic acid priming augments antioxidant defense and photosynthesis in soybean to alleviate combined heat and drought stress effects. Plant Physiol. Biochem. 206, 108193. doi: 10.1016/j.plaphy.2023.108193
Rasheed, A., Mahmood, A., Maqbool, R., Albaqami, M., Sher, A., Sattar, A., et al. (2022). Key insights to develop drought-resilient soybean: a review. J. King Saud Univ. Sci. 34, 102089. doi: 10.1016/j.jksus.2022.102089
Ridley, B. L., O’Neill, M. A., Mohnen, D. (2001). Pectins: structure, biosynthesis, and oligogalacturonide- related signaling. Phytochemistry. 57, 929–967. doi: 10.1016/s0031-9422(01)00113-3
Roulin, A., Auer, P. L., Libault, M., Schlueter, J., Farmer, A., May, G., et al. (2013). The fate of duplicated genes in a polyploid plant genome. Plant J. 73, 143–153. doi: 10.1111/tpj.12026
Shi, W., Zhang, Y., Chen, S., Polle, A., Rennenberg, H., Luo, Z. B. (2019). Physiological and molecular mechanisms of heavy metal accumulation in nonmycorrhizal versus mycorrhizal plants. Plant Cell & Environment. 42. doi: 10.1111/pce.13471
Shiu, S., Karlowski, W., Pan, R., Tzeng, Y., Mayer, K., Li, W. (2004). Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell. 16, 1220–1234. doi: 10.1105/tpc.020834
Sivaguru, M., Ezaki, B., He, Z. H., Tong, H. Y., Osawa, H., Baluška, F., et al. (2003). Aluminum-induced gene expression and protein location of a cell wall-associated receptor kinase in Arabidopsis. Plant Physiol. 132, 2256–2266. doi: 10.1104/pp.103.022129
Soltabayeva, A., Dauletova, N., Serik, S., Sandybek, M., Omondi, J. O., Kurmanbayeva, A., et al. (2022). Receptor-like kinases (LRR-RLKs) in response of plants to biotic and abiotic stresses. Plants (Basel Switzerland) 11, 2660. doi: 10.3390/plants11192660
Staniak, M., Stpień-Warda, A., Czopek, K., Kocira, A., Baca, E. (2021). Seeds quality and quantity of soybean [Glycine max (L.) Merr.] cultivars in response to cold stress. Agronomy. 11, 520. doi: 10.3390/AGRONOMY11030520
Sun, Y. H. (2022). Composition of root exudates from Suaeda Glauca and their effects on soil microorganisms and soil nutrients. Northeast Agricultural University. doi: 10.27010/d.cnki.gdbnu.2022.000349
Sun, Z. Y., Song, Y. P., Chen, D., Zang, Y. D., Zhang, Q. L., Yi, Y. T., et al. (2020). Genome-wide identification, classification, characterization, and expression analysis of the wall-associated kinase family during fruit development and under wound stress in tomato (Solanum lycopersicum L.). Genes. 11, 1186. doi: 10.3390/genes11101186
Takahashi, H., Kanayama, Y., Ming, S. Z., Kusano, T., Shah, J. (2004). Antagonistic interaction between the SA and JA signaling pathways in Arabidopsis modulate expression of defense genes and gene for gene resistance to cucumber mosaic virus. Plant Cell Physiol. 45, 803–809. doi: 10.1093/pcp/pch085
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tang, Y., Chen, H., Deng, T., Chang, Y., Sun, K., Ditta, A., et al. (2022). Genome-wide identification and analysis of the GUB_WAK_bind gene family in Gossypium hirsutum. Mol. Biol. Rep. 49, 6405–6413. doi: 10.1007/s11033-022-07449-3
Ten, H. C.A., Bochdanovits, Z., Jansweijer, V. M., Koning, F. G., Berke, L., Sanchez-Perez, G. F., et al. (2011). Probing the roles of LRR RLK genes in Arabidopsis thaliana roots using a custom T-DNA insertion set. Plant Mol. Biol. 76, 69–83. doi: 10.1007/s11103-011-9769-x
Tocquard, K., Lafon-Placette, C., Auguin, D., Muries, B., Bronner, G., Lopez, D., et al. (2014). In silico study of wall-associated kinase family reveals large-scale genomic expansion potentially connected with functional diversification in Populus. Tree Genet. Genomes. 10, 1135–1147. doi: 10.1007/s11295-014-0748-7
Tripathi, R., Aguirre, J., Singh, J. (2021). Genome-wide analysis of wall associated kinase (WAK) gene family in barley. Genomics. 113, 523–530. doi: 10.1016/j.ygeno.2020.09.045
Underwood, W. (2012). The plant cell wall: a dynamic barrier against pathogen invasion. Front. Plant Sci. 3. doi: 10.3389/fpls.2012.00085
Vaid, N., Pandey, P. K., Tuteja, N. (2012). Genome-wide analysis of lectin receptor-like kinase family from Arabidopsis and rice. Plant Mol. Biol. 80, 365–388. doi: 10.1007/s11103-012-9952-8
Verica, J. A., He, Z. H. (2002). The cell wall-associated kinase (WAK) and WAK-like kinase gene family. Plant Physiol. 129, 455–459. doi: 10.1104/pp.011028
Voorrips, R. E. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Wagner, T. A., Kohorn, B. D. (2001). Wall-associated kinases are expressed throughout plant development and are required for cell expansion. Plant Cell. 13, 303–318. doi: 10.1105/tpc.13.2.303
Wang, N., Huang, H., Ren, S. T., Li, J. J., Sun, Y., Sun, D. Y., et al. (2012). The rice wall-associated receptor-like kinase gene OsDEES1 plays a role in female gametophyte development. Plant Physiol. 160, 696–707. doi: 10.1104/pp.112.203943
Wang, R., Zhang, F. Y., Zhan, P. J., Chu, J. Q., Jin, M. S., Zhao, W. J., et al. (2024). Identification of candidate genes implicated in low-nitrogen-stress tolerance based on RNA-Seq in sorghum. Acta Agronomica Sinica. 50, 669–685. doi: 10.3724/SP.J.1006.2024.34055
Weisany, W., Sohrabi, Y., Heidari, G., Siosemardeh, A., Ghassemi-Golezani, K. (2011). Physiological responses of soybean (Glycine max L.) to zinc application under salinity stress. Aust. J. Crop Science. 5, 1441–1447. doi: 10.1016/j.fcr.2011.04.016
Weremczuk, A., Papierniak, A., Kozak, K., Willats, W. G. T., Antosiewicz, D. M. (2020). Contribution of NtZIP1-like, NtZIP11 and a WAK-pectin based mechanism to the formation of Zn-related lesions in tobacco leaves. Environ. Exp. Bot. 176, 104074. doi: 10.1016/j.envexpbot.2020.104074
Xia, Y., Yin, S., Zhang, K., Shi, X., Shen, Z. (2018). OsWAK11, a rice wall-associated kinase, regulates Cu detoxification by alteration the immobilization of Cu in cell walls. Environ. Exp. Botany. 150, 99–105. doi: 10.1016/j.envexpbot.2018.03.005
Xu, H. F., Chen, X. (2023). Current opinions on auxin research and its application in soybean breeding. Scientia Sin. Vitae. 54, 247–259. doi: 10.1360/ssv-2023-0069
Xu, H. X., Zhou, H. F., Li, X. Y., Jang, L. H., Chen, J. W. (2021). Comparative transcriptome analysis of different developmental stages of flowers and fruits in loquat under low temperature stress. Acta Hortic. Sinica. 048, 1680–1694. doi: 10.16420/j.issn.0513-353x.2021-0219
Xu, Y., Song, D., Qi, X., Asad, M., Wang, S., Tong, X., et al. (2023). Physiological responses and transcriptome analysis of soybean under gradual water deficit. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1269884
Ye, Y., Ding, Y., Jiang, Q., Wang, F., Sun, J., Zhu, C. (2017). The role of receptor-like protein kinases (RLKs) in abiotic stress response in plants. Plant Cell Rep. 36, 235–242. doi: 10.1007/s00299-016-2084-x
Yu, H., Zhang, W., Kang, Y., Fan, Y., Yang, X., Shi, M., et al. (2022). Genome wide identification and expression analysis of wall associated kinase (WAK) gene family in potato (Solanum tuberosum L.). Plant Biotechnol. Rep. 16, 317–331. doi: 10.1007/s11816-021-00739-5
Zhang, S., Chen, C., Li, L., Meng, L., Singh, J., Jiang, N., et al. (2005). Evolutionary expansion, gene structure, and expression of the rice wall-associated kinase gene family. Plant Physiol. 139, 1107–1124. doi: 10.1104/pp.105.069005
Zhang, Z., Liu, Y., Ding, P., Li, Y., Kong, Q., Zhang, Y. (2014). Splicing of receptor-like kinase-encoding SNC4 and CERK1 is regulated by two conserved splicing factors that are required for plant immunity. Mol. Plant 12, 1766–1775. doi: 10.1093/mp/ssu103
Zhang, Z. Q., Ma, W. Y., Ren, Z. Y., Wang, X. X., Zhao, J., Pei, X., et al. (2021). Characterization and expression analysis of wall-associated kinase (WAK) and WAK-like family in cotton. International. J. Biol. Macromolecules 187, 867–879. doi: 10.1016/j.ijbiomac.2021.07.163
Zhao, H., Chen, S. Y., Zhang, J. S. (2016). Ethylene signaling pathway in regulating plant response to abiotic stress. Biotechnol. Bulletin. 32, 1–10. doi: 10.13560/j.cnki.biotech.bull.1985.2016.10.001
Zuo, W., Chao, Q., Zhang, N., Ye, J., Tan, G., Li, B., et al. (2015). A maize wall-associated kinase confers quantitative resistance to head smut. Nat. Genet. 47, 151–157. doi: 10.1038/ng.3170
Zuo, C. W., Liu, Y., Guo, Z. G., Mao, J. C., Chu, M. Y., Chen, B. H. (2019). Genome-wide annotation and expression responses to biotic stresses of the wall-associated kinase-receptor-like kinase (WAK-RLK) gene family in Apple (Malus domestica). Eur. J. Plant Pathol. 153, 771–785. doi: 10.1007/s10658-018-1591-8
Keywords: abiotic stresses, expression analysis, genome-wide identification, soybean, wall-associated kinase
Citation: Li X, Qi S, Meng L, Su P, Sun Y, Li N, Wang D, Fan Y and Song Y (2025) Genome-wide identification of the wall-associated kinase gene family and their expression patterns under various abiotic stresses in soybean (Glycine max (L.) Merr). Front. Plant Sci. 15:1511681. doi: 10.3389/fpls.2024.1511681
Received: 15 October 2024; Accepted: 02 December 2024;
Published: 16 January 2025.
Edited by:
Omar Borsani, Universidad de la República, UruguayReviewed by:
Andrea Luciana Fleitas, Universidad de la República, UruguayVitor Amorim-Silva, University of Málaga–Spanish National Research Council (IHSM-UMA-CSIC), Spain
Copyright © 2025 Li, Qi, Meng, Su, Sun, Li, Wang, Fan and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yinglun Fan, ZmFueWluZ2x1bkBsY3UuZWR1LmNu; Yong Song, c29uZ3lvbmdAbGN1LmVkdS5jbg==
 Xiangnan Li
Xiangnan Li Sifei Qi
Sifei Qi Lingzhi Meng
Lingzhi Meng Peisen Su
Peisen Su Yongwang Sun
Yongwang Sun Nan Li
Nan Li Dan Wang2
Dan Wang2 Yong Song
Yong Song