- 1State Key Laboratory of Rice Biology and Breeding, National Center of Rice Improvement, China National Rice Research Institute, Hangzhou, China
- 2National Nanfan Research Institute (Sanya), Chinese Academy of Agricultural Sciences, Sanya, China
- 3Jiangxi Academy of Agricultural Sciences, Nanchang, China
- 4State Key Laboratory of Subtropical Silviculture, Zhejiang A&F University, Hangzhou, Zhejiang, China
The formation of the anther wall and the development of pollen processes, central to rice fertility and yield, are highly dependent on the synthesis and accumulation of lipid polymers. Although several regulatory factors related to lipid biosynthesis during pollen wall development have been identified, the molecular mechanisms controlling these processes remain poorly understood. In this study, a male-sterile rice mutant, lap3, was identified, characterized by normal vegetative growth but complete male sterility due to delayed programmed cell death (PCD) in tapetal cells and defects in anther cuticle and pollen exine formation. Map-based cloning revealed that OsLAP3 is a new allele of the strictosidine synthase-like gene, OsSTRL2. Functional analysis, including complementation and CRISPR/Cas9-based gene editing, confirmed that the 2-nucleotide deletion in the OsLAP3 is responsible for the male sterility phenotype. OsLAP3 is homologous to the maize ZmMS45, the core recessive nuclear sterile gene of maize Seed Production Technology (SPT), and localizes to the endoplasmic reticulum and plays a conserved role in anther development and pollenformation. Gene expression analysis revealed a significant downregulation of key genes involved in anther development and sporopollenin biosynthesis in lap3 anthers. Furthermore, lipid profiling demonstrated a marked reduction in both wax and cutin content. These findings establish OsLAP3 as a critical regulator of fatty acid synthesis and highlight its role in anther cuticle formation and pollen exine development. The findings of this study provide valuable insights into the molecular regulation of lipid biosynthesis during rice male reproductive development and offer potential applications for hybrid rice breeding.
1 Introduction
Male sterility in rice is a critical aspect of reproductive development and forms the foundation for hybrid rice breeding, which significantly enhances crop yield (Yuan, 2014). Multiple factors contribute to male sterility, involving key processes such as microspore mother cell development, meiosis, tapetum development and degradation, nutrient synthesis and transport within the anther wall, pollen wall formation, starch accumulation, and anther dehiscence (Fu et al., 2014; Shi et al., 2011; Mu et al., 2009; Ranjan et al., 2017). Among these processes, the programmed cell death (PCD) and synthesis and transport of nutrients in the anther wall have been the most extensively studied (Zhang and Yang, 2014). These events are crucial for the proper formation and maturation of pollen cells during the mid-to-late stages of anther development and directly impact male reproductive success in rice.
Anther development in rice is governed by a complex network of transcription factors (TFs) that regulate PCD in the tapetum, a necessary process for pollen development and fertility. Several TFs, including Tapetum Degeneration Retardation (TDR) (Li et al., 2006), Eternal Tapetum 1 (EAT1) (Niu et al., 2013), and TDR Interacting Protein 2 (TIP2) (Fu et al., 2014), play crucial roles in regulating tapetal PCD and pollen wall formation (Abbas et al., 2021). TDR, a basic helix-loop-helix TF, is responsible for the regulation of genes involved in tapetal cell degradation, such as OsCP1, and mutations in TDR result in premature degradation of the tapetum and pollen sterility (Ji et al., 2013). EAT1 and TIP2 work synergistically with TDR to coordinate PCD timing and support pollen exine development, which is essential for male fertility in rice (Ko et al., 2014). TDR directly regulates genes involved in pollen wall development, including OsC6 and ADF, by binding to their promoters (Li et al., 2015; Tariq et al., 2023). EAT1 works cooperatively with TDR to modulate the expression of AP25 and AP37, regulators of tapetal PCD (Niu et al., 2013). TIP2 (bHLH142), which acts upstream of TDR and EAT1, coordinates tapetal degradation and pollen formation, and tip2 mutants exhibiting delayed meiosis, defective tapetal PCD, and male Sterility (Fu et al., 2014; Ko et al., 2014). Additionally, the SLR1-MYB188 module integrates gibberellin signaling with PCD regulation, coordinating the expression of genes such as CYP703A3, DPW, ABCG15, and PKS1, all critical for pollen wall development (Jin et al., 2022). This regulatory interaction ensures that PCD occurs at the appropriate stage, allowing for proper deposition of sporopollenin and the formation of the pollen exine. Other transcription factors such as UDT1, OsMS1 and GAMYB are also involved in these regulatory pathways, with mutations leading to disruptions in tapetal development, resulting in pollen sterility (Jung et al., 2005; Liu et al., 2010; Yang et al., 2019).
In rice, nutrient synthesis and transport in the anther wall are vital for male fertility (Shi et al., 2011; Zhang et al., 2010). The most thoroughly researched directions collectively indicate that the formation of viable pollen depends on the synthesis and transport of lipid-based biomolecules, such as sporopollenin, Ubisch bodies, cuticular waxes, and cutin monomers. These compounds are essential for the proper formation of the pollen exine, which protects the pollen and ensures viability (Quilichini et al., 2015; Qin et al., 2013; Niu et al., 2013). Cytochrome P450 enzymes, including CYP704B2 and CYP703A3, hydroxylate fatty acids essential for anther cuticle and pollen exine development (Li et al., 2010; Yang et al., 2014, 2018). Mutations in these enzymes disrupt pollen wall structure, leading to sterility. Similarly, DPW and DPW2 regulate lipid accumulation and anther wax formation (Shi et al., 2011; Xu et al., 2017). Various ATP-binding cassette (ABC) transporters, such as ABCG15, ABCG26, and OsABCG3, are involved in the transport of lipid precursors for pollen exine and cuticle formation (Chang et al., 2018; Niu et al., 2013; Qin et al., 2013; Wu et al., 2014). Mutations in these transporters impair lipid transport and disrupt anther development, resulting in sterility (Zhang et al., 2010). Recent studies have further revealed that ROS homeostasis are jointly involved in the development of rice anther and panicles, namely, DPS1 and SUBSrP1 genes are considered to be crucial regulators of anther cuticle formation and ROS-mediated cell death in rice. DPS1, a cystathionine β-synthase (CBS) domain protein, maintains ROS homeostasis and cuticle integrity, with its loss-of-function mutants exhibiting increased ROS, early tapetal PCD, defective anther cuticles, and sterility (Zafar et al., 2020). Similarly, SUBSrP1, a subtilisin-like serine protease, is essential for anther cuticle biosynthesis in apical spikelets. Disruption of SUBSrP1 leads to excessive ROS, cell death, impaired cuticle formation, and panicle degeneration (Ali et al., 2022). These studies collectively expand and demonstrate the importance of ROS homeostasis in male reproductive development and fruiting in rice.
Lipid metabolism genes such as OsGPAT3 and RSM2 are also integral to anther tapetal PCD regulation. OsGPAT3 influences lipid metabolism within the anther, while RSM2, a GDSL lipase gene, impacts lipid metabolism in the anther wall (Men et al., 2017; Sun et al., 2018; Zhao et al., 2020). Other key genes, such as WDA1, OsACOS12, and OsNP1, are involved in sporopollenin biosynthesis and deposition, essential for pollen exine and anther cuticle formation. Mutations in these genes reduce the number of Ubisch bodies and oil bodies in the anther, leading to sterility (Chang et al., 2016a; Jung et al., 2006; Li et al., 2016; Liu et al., 2017). Additionally, OsPKS1 and OsPKS2, homologs of Arabidopsis PKSA/LAP6 and PKSB/LAP5, are critical for sporopollenin precursor biosynthesis. Mutations in OsPKS2 result in sterility due to failed exine formation (Zhu et al., 2017; Zou et al., 2018).
In Arabidopsis thaliana, strictosidine synthase-like (STR-like) genes also play significant roles in plant metabolism, defense, and secondary metabolite pathways (Sohani et al., 2009). Glu-309, a key catalytic residue in strictosidine synthase, is crucial for catalysis, functioning as a proton donor. Some STR-like genes, including AtSSL7 and AtSSL14, have diverged from their ancestral enzymatic functions and may serve regulatory roles in reproductive development (Kibble et al., 2009). Similarly, in rice, OsSTRL2, a homolog of the Arabidopsis STR-like genes, contributes to reproductive processes, particularly tapetal cell function and pollen development (Zou et al., 2017).
In this study, we identified a rice strictosidine synthase gene, OsLAP3, which is a new allele of the OsSTRL2 gene and plays a critical role in anther cuticle formation and pollen exine patterning. Mutation of OsLAP3 results in delayed tapetum degradation and defective pollen exine. Gene expression analyses revealed that OsLAP3 is involved in lipid biosynthesis and transport, which are necessary for proper anther and pollen development. Although the OsSTRL2 gene has been implicated in pollen development, its specific contributions to key biological processes such as lipid metabolism and tapetum degradation remain underexplored. In our study, we reported the OsSTRL2/OsLAP3 gene through map-based cloning, and these findings not only provide a more comprehensive understanding of the molecular mechanisms of male sterility in rice but also offer a new potential gene resource for SPT system and practical application during in rice hybrid seed production.
2 Materials and methods
2.1 Plant materials and growth conditions
The lap3 mutant was identified from a population of indica rice cv. Zhonghui8015 (Zh8015), generated through 60Coγ-ray radiation. For genetic analysis and mapping, the lap3 mutant (indica species) was as the pollen acceptor in crosses with wild type (WT) Zh8015 and 02428 (japonica species). Heterozygous F1 plants were self-pollinated to produce BC1F2 and an F2 population, which were further utilized for segregation analysis. All rice plants were grown under natural conditions in paddy fields located at the China National Rice Research Institute, Hangzhou, Zhejiang Province and Lingshui, Hainan Province.
2.2 Phenotypic and cytological analysis
The phenotypic characteristics of the lap3 mutant, including plant height, tiller number, spikelet morphology, and floral organs, were documented using a Nikon Coolpix E995 digital camera (Nikon, Tokyo, Japan) and a Carl Zeiss SteREO Lumar V12 stereo fluorescence stereomicroscope (Markku Saari, Jena, Germany). Pollen fertility was assessed by 1.0% I2/KI staining under a Leica DM2500 microscope. For histological analysis, anthers at various developmental stages were collected and processed for semi-thin sectioning, scanning electron microscopy (SEM), and transmission electron microscopy (TEM) (Sun et al., 2018, 2023).
2.3 Molecular cloning of OsLAP3 and gene complementation analysis
For map-based cloning of OsLAP3, a mapping population of 1,196 male-sterile plants was selected from the F2 segregation population obtained by crossing lap3 with 02428 (Supplementary Table S2). The modified cetyl trimethyl ammonium bromide (CTAB) method was used for DNA extraction (Allen et al., 2006). InDel markers were developed based on polymorphisms between the genome sequences of japonica Nipponpare and indica 9311 (http://www.gramene.org and http://blast.ncbi.nlm.nih.gov).
To verify the identity of the OsLAP3 gene, a 5,745 bp genomic fragment was amplified, including a 2,643 bp promoter, a 2,238 bp coding region, and an 864 bp downstream region. The fragment was cloned into the binary vector pCAMBIA1300 (CAMBIA; hygromycin resistance) using the in-fusion HD cloning kit (Takara Bio USA Inc., Mountain View, CA, USA) and introduced into the lap3 mutant through Agrobacterium-mediated transformation for complementation analysis. A CRISPR/Cas9 vector (pC1300-Ubi::Cas9-SK-5G), carrying rice Ubi promoter and a single-guide RNA (sgRNA), was constructed for targeting OsLAP3 to develop its knock-out lines. A target sequence (5-GCTCGTGTCGGTGAAGAACA-3) with protospacer adjacent motif (PAM) sequence CCA was identified in the second exon of OsLAP3. Two oligos were designed using an online CRISPR-GE tool (http://skl.scau.edu.cn/). The possible off-target effects were prevented by BLAST search using target sequence in Gramene (www.gramene.org) database. Positive transgenic lines were identified by PCR, sequenced for confirmation, and analyzed for phenotypic changes. Primers used for vector construction and sequencing are listed in Supplementary Table S3.
2.4 RNA extraction and qPCR analysis
Total RNA was extracted from various rice tissues, including roots, stems, leaves, and anthers at different developmental stages, using the TRIzol reagent (Invitrogen, USA). cDNA synthesis was performed using a PrimeScript RT reagent kit (TaKaRa, Japan). Quantitative real-time PCR (qRT-PCR) was conducted using SYBR Premix Ex Taq II (TaKaRa, Japan) on a Roche LightCycler 480 system. Gene expression was normalized to OsACTIN1 (Os03g0234350), and three biological replicates were included. Specific primers for each target gene are listed in Supplementary Table S3.
2.5 GUS histochemical staining
The promoter region of OsLAP3 (2.2 kb upstream) was cloned into the pCAMBIA1305 vector to drive the expression of the GUS reporter gene. The pOsLAP3:GUS construct was introduced into Zh8015 via Agrobacterium tumefaciens-mediated transformation. GUS activity was detected in spikelets at various developmental stages by staining with a GUS staining kit. Stained anthers were decolorized using 70% ethanol and subjected to transverse sectioning for microscopic analysis. Primers for pOsLAP3:GUS construction are listed in Supplementary Table S3.
2.6 Phylogenetic analysis
The conserved domain of OsLAP3 protein was identified using the NCBI CD-search tool (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Homologous protein sequences were retrieved from the NCBI database, and multiple sequence alignments were conducted using ClustalW. A phylogenetic tree was constructed using the neighbor-joining method with MEGA10 software (bootstrap = 1000) to assess the evolutionary relationship between OsLAP3 and its homologs in other species.
2.7 Subcellular localization
To investigate subcellular localization, the full-length coding sequence of OsLAP3, excluding the stop codon, was cloned into the pCAMBIA1305 vector to generate an OsLAP3-GFP fusion protein. Rice mesophyll protoplasts were isolated from 2-week-old Zh8015 plants, and plasmids with either the recombinant or empty vector were transformed into the protoplasts. After incubation for 36 to 48 hours in the dark, GFP signals were examined using a Zeiss LSM700 confocal laser scanning microscope (Zeiss, Oberkochen, Germany). Co-localization analysis was performed using the ER marker PDI-mCherry. Primers for subcellular localization are listed in Supplementary Table S3.
2.8 TUNEL assay
To detect DNA fragmentation in the anthers in WT and lap3 mutant plants, the TUNEL (terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling) assay was performed according to a previously described method (Chen et al., 2018). Selected paraffin-embedded anther sections at different developmental stages were dewaxed in xylene and rehydrated in an ethanol series. The assay was conducted using an in vitro DeadEnd™ Fluorometric TUNEL System, using fluorescein (Promega, Wisconsin, USA), following the manufacturer’s instructions. The signals were observed and imaged using a fluorescence confocal scanner microscope (ZEISS LSM 700, Oberkochen, Germany) under consistent settings.
2.9 Analysis of wax and cutin components
Anthers from WT and lap3 plants at the mature pollen stage, prior to heading, were collected for was cutin component analysis, with five biological replicates per sample. Extraction was performed as previously described in the Zhang Dabing Laboratory at Shanghai Jiaotong University (Li et al., 2010). The components were analyzed using gas chromatography–mass spectrometry (GC-MS), and the wax and cutin content between WT and mutant anthers was compared based on mass spectrometry results.
3 Results and analysis
3.1 The lap3 mutant exhibits normal vegetative growth but complete male sterility
The lap3 mutant displayed normal vegetative growth patterns, including comparable plant height, tiller number, and spikelet count to the WT Zh8015 (Figure 1A; Supplementary Table S1). Similarly, except for the morphology of the anthers, the number of flower organs in the mutant and the morphology of other flower organs, such as the lemma, palea and ovary, are basically the same as those in the wild type (Figure 1B). However, key differences were observed closer to the heading stage. In WT plants, the anthers were plump, golden, and shorter, while the stigmas were small, surrounded by filaments (Figure 1B). By contrast, the lap3 mutant displayed significantly elongated filaments, larger stigmas, and smaller, shriveled, and white anthers (Figure 1C). Furthermore, I2/KI staining revealed that over 99% of WT pollen grains were fertile, as indicated by dark staining. In contrast, lap3 mutants produced 100% abortive pollen, with no detectable staining (Figures 1D, E). These observations indicate that lap3 mutants, while maintaining normal vegetative growth, exhibit complete male sterility due to defective anther and pollen development.
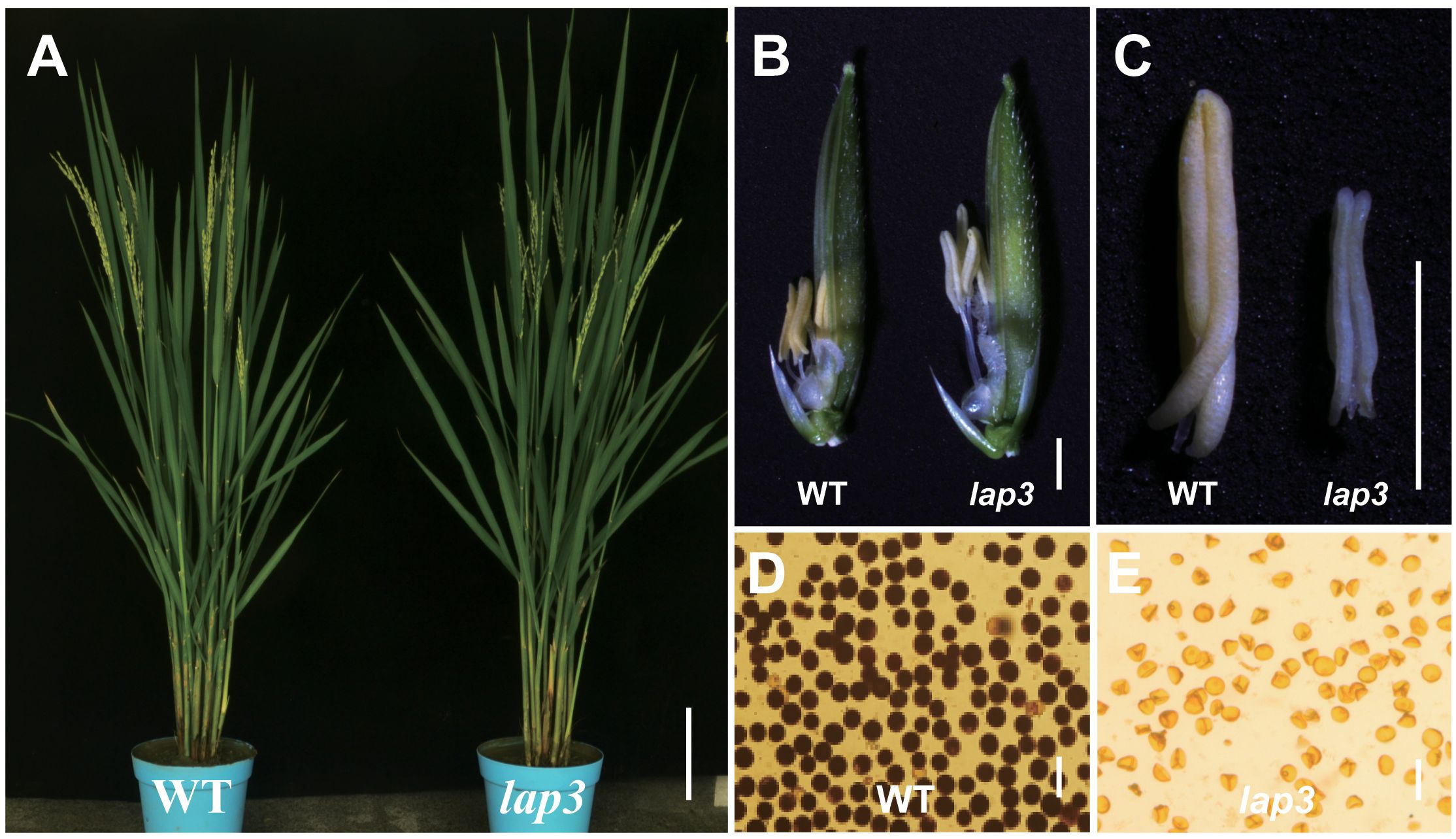
Figure 1. Phenotypes of the lap3 mutant. (A) Wild type (WT) and the lap3 mutant plants at anthesis stage. (B) Mature spikelets of WT Zh8015 and lap3 mutant at anthesis; lemmas were removed for clarity. (C) Mature anther of WT and lap3. (D, E) Pollen fertility of WT and lap3 by I2/KI staining. Bar = 15 cm (A), 1.5 mm (B), 500 µm (C), and 10 µm (D, E).
3.2 Abnormal tapetum and microspore development in lap3
To further investigate the defects in the lap3 anthers, semi-thin sections were analyzed at various developmental stages. At stage 7 (S7), no discernible differences were observed between WT (Zh8015) and lap3 anthers; both contained well-formed microspore mother cells (Figures 2A, B; Supplementary Figure S1). However, by stage 8 (S8), the first signs of abnormal development appeared. While WT underwent normal meiosis to form tetrads (Figures 2C, E), lap3 mutants showed irregular tetrad formation and failed to narrow and condense their tapetal cells (Figures 2D, F). At S9, WT tapetal cells underwent clear programmed cell death (PCD), leading to fragmentation and disintegration of the cell layer, whereas the lap3 tapetal cells remained structurally similar to those observed at S8b (Figures 2E, G, H).
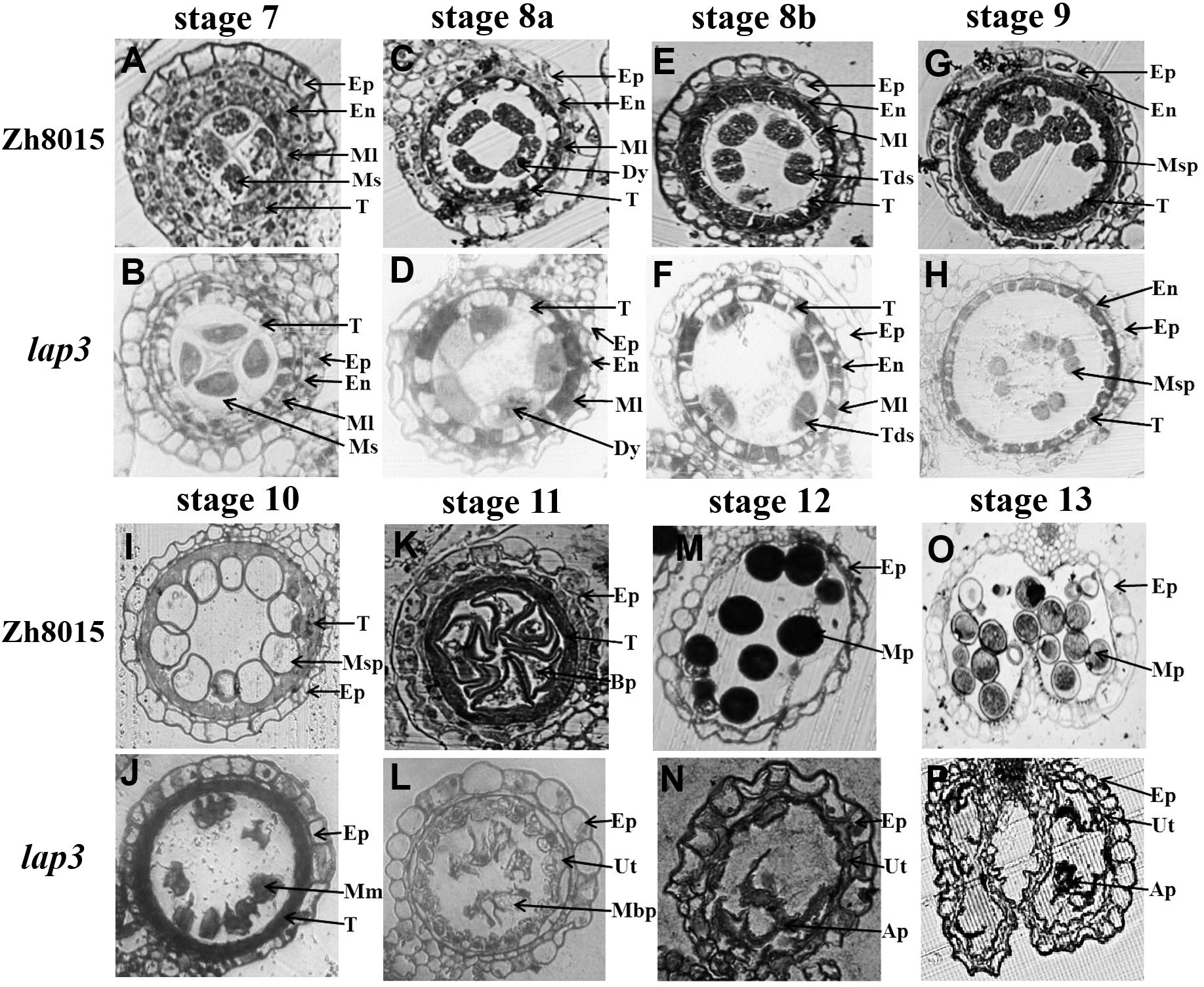
Figure 2. Transverse section analysis of anther development in WT and lap3. (A, C, E, G, H, I, K, M, O) and (B, D, F, H, J, L, N, P) are locules from anther sections of WT and lap3 from stage 7 to stage 13, respectively. Ep, epidermis; En, endothecium; Ml, middle layer; T, tapetum; Ms, microsporocyte; Dy, dyad cell; Msp, microspore; BP, bicellular pollen; MP, mature pollen; Tds, tetrads; Mm, malformed microspore; Ap, abortive pollen; Ut, undegraded tapetum. Bars = 50 μm.
By S10, the differences were more pronounced. WT microspores became vacuolated, spherical, and were closely associated with the tapetal cells which had degraded (Figure 2I). In lap3, tapetal cells remained circular and microspores appeared irregularly shaped and scattered throughout the anther chamber (Figure 2J). By S11, the tapetum layer became more irregular and microspores had gradually degraded and underwent asymmetric mitosis, forming one generative cell and one vegetative cell in WT (Figure 2K). In contrast, the lap3 tapetum had just begun to rapidly degrade, and the deformation and shrinkage of the microspores became more apparent (Figure 2L). By S12, WT microspores had formed bicellular pollen and contained two sperm nuclei and a vegetative nucleus (Figure 2M), the anther epidermis cracked normally, preparing for pollination (Figure 2O), but lap3 anthers contained nearly hollow, collapsed pollen grains, with only remnants of microspores in the locule (Figures 2N, P).
3.3 Defective Ubisch body synthesis and abnormal pollen exine formation in lap3
Scanning electron microscopy (SEM) was used to analyze the anther and pollen surface lap3 mutant anthers were noticeably shorter (Figures 3A, B) and contained densely packed anther cuticle on anther epidermis cells than WT (Figures 3C–F). The inner walls of WT anthers showed a smooth, evenly distributed Ubisch bodies (Figures 3G, I). In contrast, the inner walls of lap3 anthers were covered with underdeveloped, fluffy Ubisch bodies, indicating impaired sporopollenin deposition (Figures 3H, J). WT pollen grains were full and spherical, with smooth outer surfaces (Figures 3K, M, O), whereas lap3 appeared shriveled and deformed, with an incomplete exine layer and abnormal sporopollenin deposition (Figures 3L, N, P).
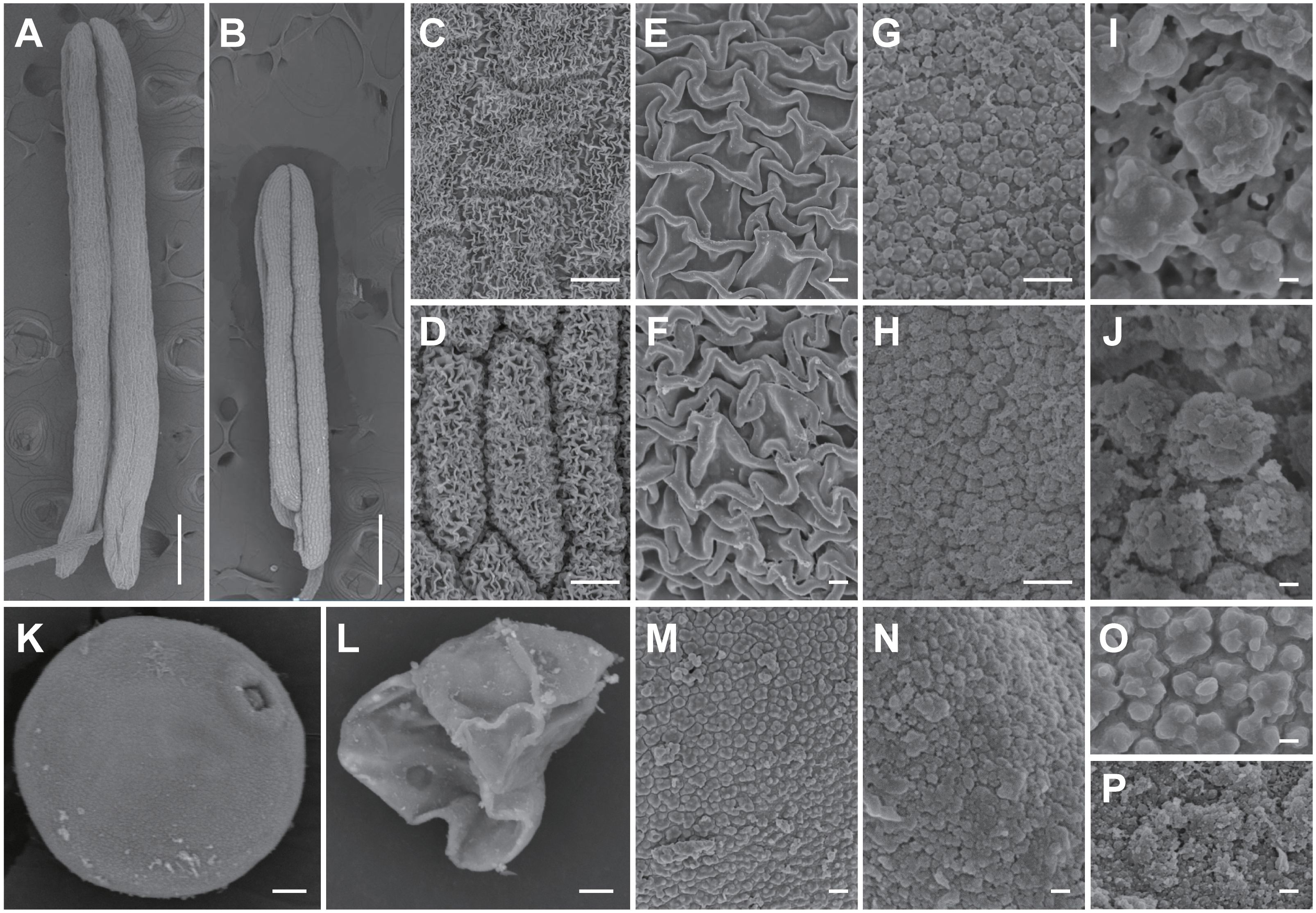
Figure 3. Scanning electron microscopy observation of anther and pollen grains in the WT and lap3. (A, B) Single anther of WT (A) and lap3 (B) at stage 13. (C-F) Observation of anther cuticle on anther epidermis cells in WT (C, E) and lap3 (D, F) at stage 13. (G-J) Observation on inner side of anther cells and Ubisch bodies in WT (G, I) and lap3 (H, J) at stage 13. (K, L) Pollen grains of WT (K) and lap3 (L) at stage 13. (M-P) Pollen exine and sporopollenin of WT (M, O) and lap3 (N, P) at stage 13. Bars = 1 mm (A, B); 20 µm (C-H); 10 µm (I-N); 200 nm (O, P).
3.4 Delayed tapetum degradation in lap3
To pinpoint the timing and location of defects in anther development in the lap3 mutant, semi-thin sectioning of anthers was performed at different developmental stages in both the WT and lap3, followed by TEM. At S7, the structural composition of the mutant anthers was similar to the WT, though the mutant’s tapetal cells were tightly packed with smaller vacuoles, while WT tapetal cells exhibited vacuoles occupying two-thirds of the cell. Nuclei appeared normal in both (Figures 4A–D). By S8b, WT anther epidermal cells had elongated shapes and orderly arrangement, whereas lap3 mutant displayed disordered cuboidal epidermal cells. Although vacuoles in the mutant tapetal cells slightly increased in size, they remained significantly smaller than those in the WT (Figures 4E, F). At S10, the WT tapetum was thin and organized, supporting microspore vacuolization. In contrast, lap3 tapetal cells exhibited large vacuoles and remained thicker, with irregularly shaped microspores distributed randomly in the locule (Figures 4G, H).
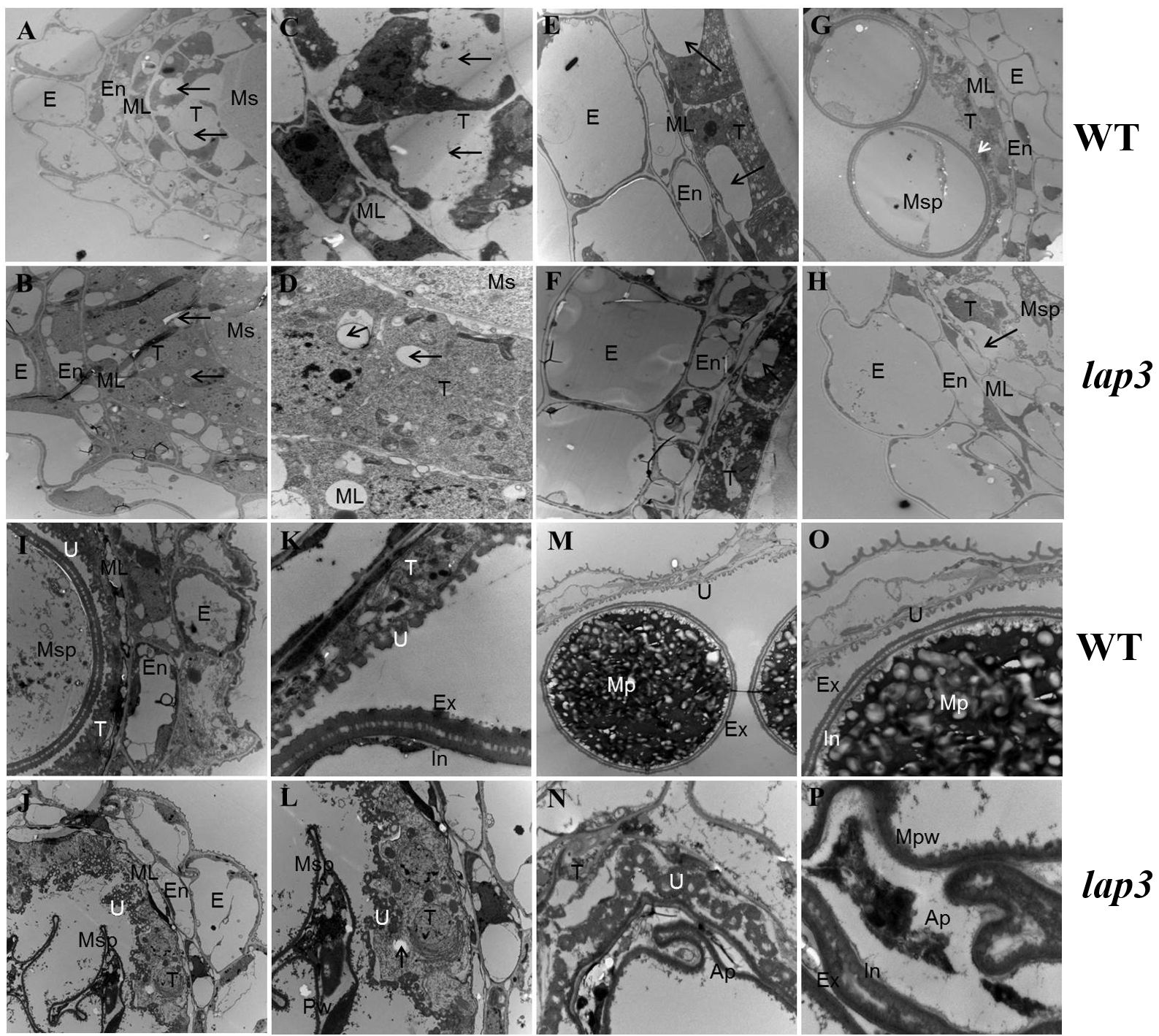
Figure 4. Transmission electron microscopy of anthers and pollen grains in WT and lap3. (A, C, E, G, H, I, K, M O) are from WT, and (B, D, F, H, J, L, N, P) are from lap3. (A, B) Anther wall layers and microsporocytes of WT (A) and lap3 (B) at S7. (C, D) Higher magnification of WT (C) and lap3 (D) tapetum at S7, showing nuclei and vacuoles. (E, F) Microsporocytes and tapetal cells with vacuole size differences (arrows) at S8b. (G, H) Anther wall structure of WT and lap3 at S10. (I, J) Anther and tapetum structures at S11. (K, L) Higher magnification of microspores and tapetal cells at S11. (M, N) Tapetum cells, Ubisch body, and pollen exine at S12. (O, P) Higher magnification of M and N, respectively. Ap, abortive pollen; E, epidermis; En, endothecium; Ex, exine; In, intine; ML, middle layer; Mpw, malformed pollen wall; Ms, microsporocyte; Msp, microspore; T, tapetum; U, Ubisch body. Bars = 5 µm (A, B, E-J, M, N); 0.5 µm (C, D, K, L, O, P).
By S11, most WT tapetum had degraded, leaving regular Ubisch bodies in close contact with microspores, aiding nutrient transfer. The pollen wall separated into two layers following the first mitosis (Figures 4I, K). In lap3, the tapetum remained thick and unevenly distributed, with irregular Ubisch bodies. Microspores were irregular and shrunken, with no clear distinction between intine and exine layers of the pollen wall (Figures 4J, L). At the final stage of pollen maturation, WT pollen was filled with starch granules, while lap3 pollen became hollow, with thin walls and visible tapetal cells (Figures 4M–P). These findings suggest delayed tapetum degradation in lap3 led to nutrient deficiencies, disrupting pollen development, and causing sterility.
To confirm delayed degradation, we performed TUNEL assays to detect DNA fragmentation, an indicator of PCD. At S7, no TUNEL signals were detected both in WT and lap3 mutant (Figures 5A, B). In WT, strong TUNEL signals were detected in the tapetum at S8a and S8b, peaking at S9 and diminishing by S10, indicating near complete tapetum degradation, almost completely extinguished at S11 and S12 (Figures 5C, E, G, I, K). In lap3, no TUNEL signals were detected at S8a (Figure 5D) and S8b (Figure 5F), TUNEL signals appeared at S9 and persisted until S12, confirming delayed PCD and tapetal cell degradation (Figures 5H, J, L). This delay likely disrupted sporopollenin precursor release, contributing to abnormal pollen wall formation and male sterility in the mutant.
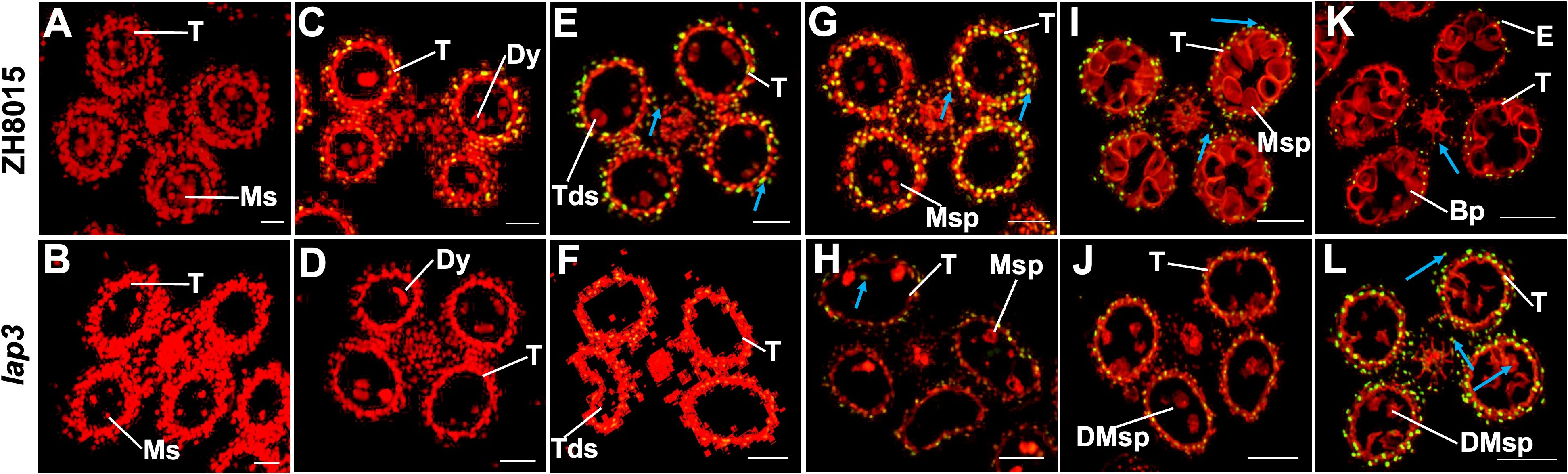
Figure 5. Detection of DNA fragmentation in WT(ZH8015) and lap3 anthers by TUNEL assay. (A, B) Anthers at stage 7. (C, D) Anthers at stage 8a. (E, F) Anthers at stage 8b. (G, H) Anthers at stage 9. (I, J) Anthers at stage 10. (K, L) Anthers at stage 12. Red signal indicates propidium iodide (PI) staining, and yellow and green fluorescence indicate TUNEL-positive signals. TUNEL-positive signals in tapetum cells are marked with white arrows, while those in outer cell layers (epidermis, endothecium, middle layer, and vascular bundle cells) are marked with blue arrows. BP, bicellular pollen; DMsp, degraded microspore; Dy, dyad cell; E, epidermis; Ms, microsporocyte; Msp, microspore; T, tapetum; Tds, tetrads. Bars = 50 µm.
3.5 Genetic analysis and positional cloning of OsLAP3
Genetic analysis of F1, BC1F2, and F2 populations from a cross between the lap3 and the WT showed that the male sterility phenotype in lap3 is controlled by a recessive nuclear gene. Segregation analysis of BC1F2 and F2 populations revealed a 3:1 ratio of fertile to sterile plants, consistent with a recessive nuclear site of the mutation (Supplementary Table S2).
For positional cloning of the OsLAP3 gene, 1,196 sterile plants from the F2 population were used for map-based cloning. Initial mapping with InDel markers localized lap3 to a region on the short arm of chromosome 3, between markers RM1338 and RM14723 (Figure 6A). Fine mapping further narrowed the region to a 20 kb interval between markers JS6 and JS7 (Figures 6B, C). Sequence analysis identified a two-nucleotide deletion in the Os03g0263600 gene (Figures 6D, E), which resulted in a frameshift and premature termination of the protein (Supplementary Figure S3). This confirmed Os03g0263600 as the candidate gene for OsLAP3. The gene has a full length of 2,083 bp, consisting of 4 exons and 3 introns (Supplementary Figure S3A). The coding region is 1,284 bp in length and encodes a 427 amino acid protein containing: a six-bladed beta-propeller TolB-like domain and a strictosidine synthase domain (Supplementary Figure S3B). The deletion in lap3 caused a substitution of position 390, changing serine (Ser) to arginine (Arg), resulting in premature termination of translation (Supplementary Figure S3C).
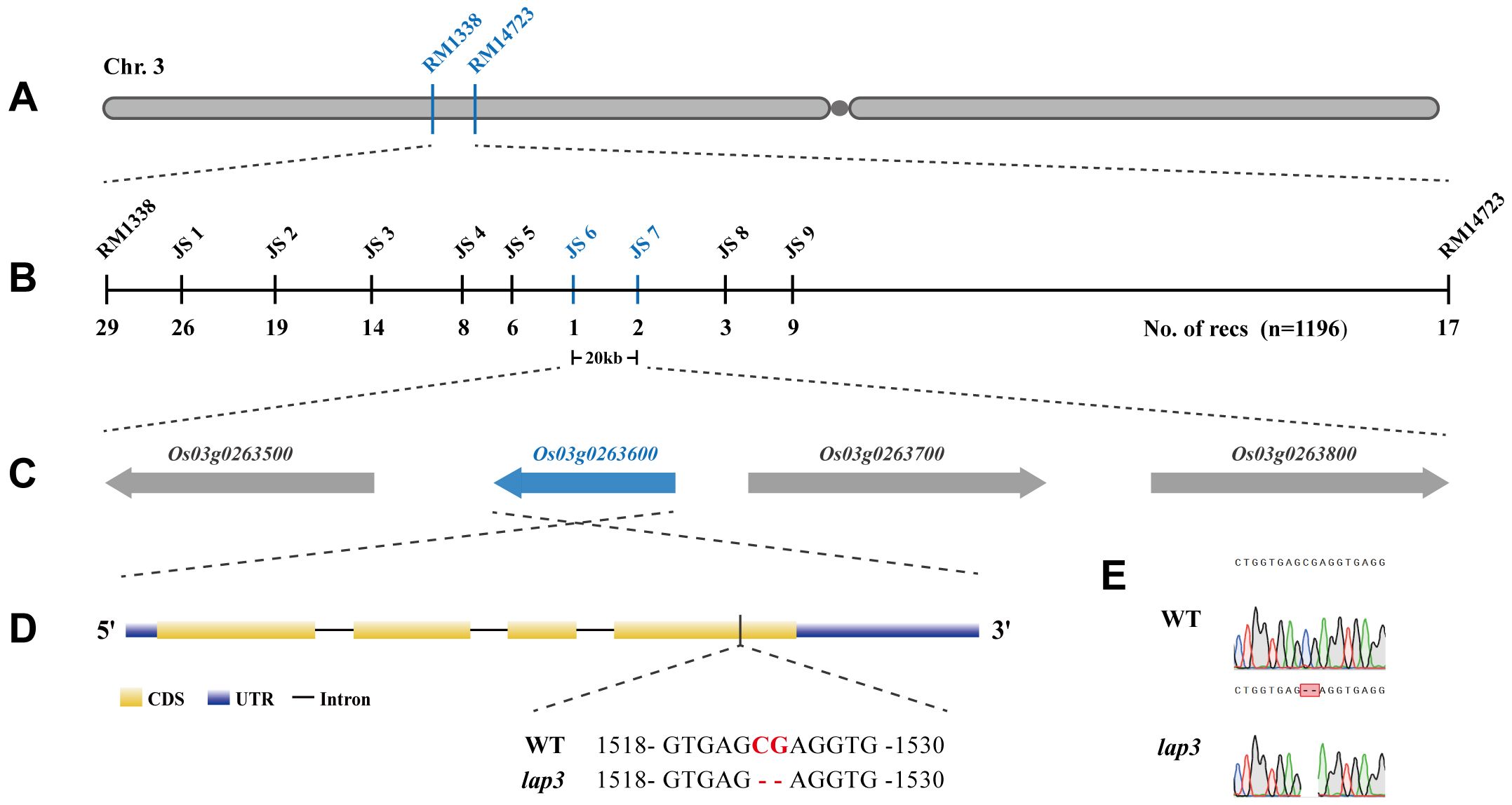
Figure 6. Fine mapping and molecular cloning of the OsLAP3 gene. (A) Genetic linkage map of the lap3 locus between RM1338 and RM14723 on the short arm of chromosome 3. (B) Fine mapping of lap3 to a 20 kb genomic region between JS6 and JS7. (C) Four ORFs in the target region; the red one was the target gene. (D, E) Sequence analysis of OsLAP3. Red capitals in Panel D and red frame in E represent the differential nucleotide base between WT and lap3 mutant.
3.6 Functional verification of OsLAP3
To confirm that OsLAP3 is responsible for the male sterility in lap3, a 5.75 kb genomic fragment containing the OsLAP3 coding region was introduced into lap3 mutants via Agrobacterium-mediated transformation (Figure 7A). Hygromycin resistance screening identified positive transgenic plants, which were subsequently grown in the field. Sampling and observation at the heading and flowering stages revealed that the complemented transgenic lines exhibited restored fertility, with yellow viable pollen, and similar seed set rates to WT plants (Figures 7B–E). SEM further confirmed the restoration of the WT pollen exine structure in the transgenic lines, verifying OsLAP3 function (Figures 7D, E).
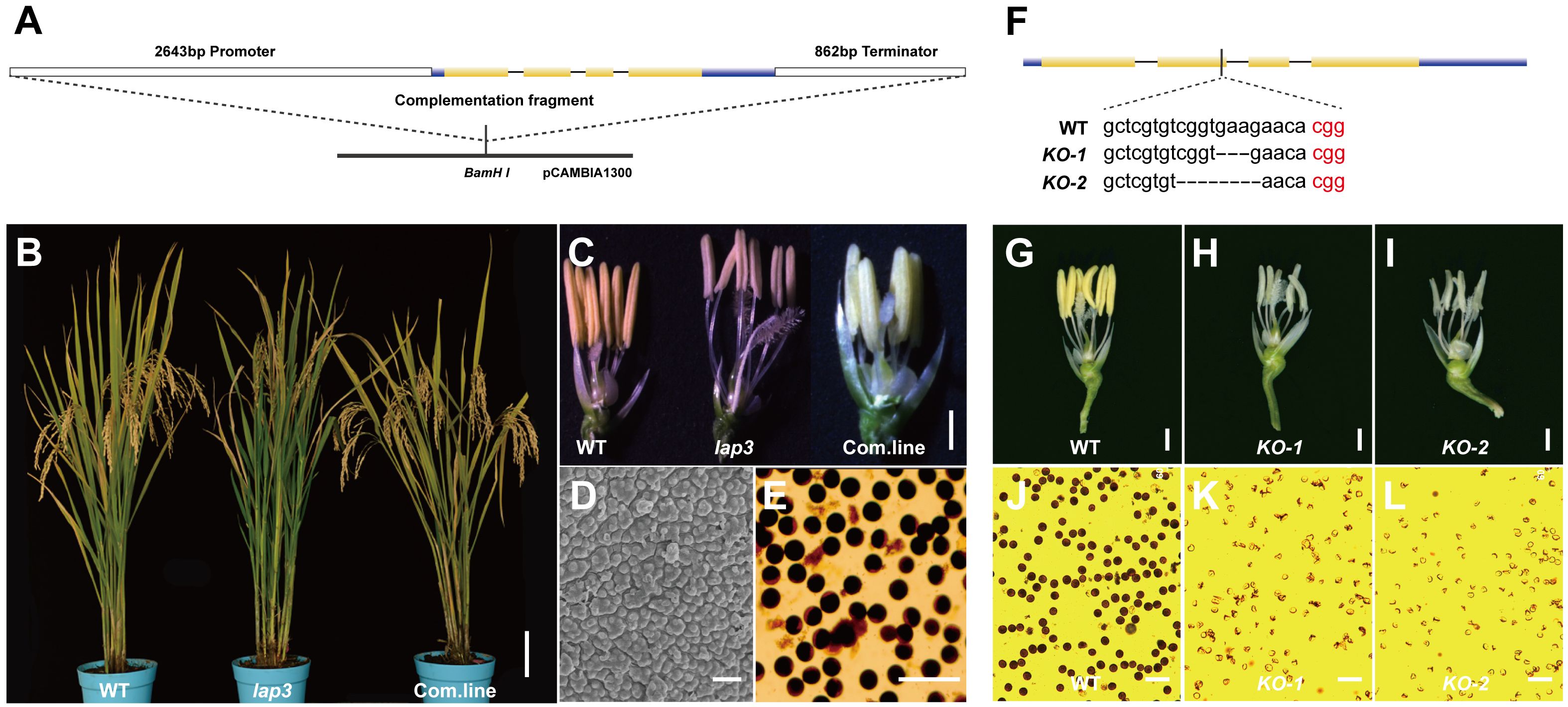
Figure 7. Function complementation of OsLAP3. (A) DNA fragment used for complementation. (B) Seed setting rate of spikelet in WT, lap3, and complementation plants. (C) Flowers and anther at stage 13 of WT, lap3, and complementation plant. (D) Pollen exine surface in complementation plants by SEM. (E) OsLAP3-complemented mutants stained with 1% I2-KI solution showing mature pollen grains. (F) OsLAP3 knockout target and mutation type of KO plants. (G-I) Flowers and anthers of WT and lap3-Cri plants at stage 13. (J-L) WT and lap3-Cri plants stained with 1% I2-KI solution showing mature pollen grains. Bars = 1 mm (C); 200 nm (D, E).
Additionally, the CRISPR-Cas9 knockout experiment of OsLAP3 generated lap3-Cri plants with 3 bp and 5 bp deletions (Figure 7F). Both KO-1 and KO-2 lines exhibited milk-yellow anthers (Figures 7H, I) and sterile pollen (Figures 7K, L) compared to wild-type yellow anthers (Figure 7G) and dark I2/KI stained pollen (Figure 7J). These results further demonstrated that OsLAP3, encoding strictosidine synthase domain protein, is responsible for the male sterile observed in the lap3 mutant.
3.7 Phylogenetic analysis of OsLAP3
Phylogenetic analysis of OsLAP3 and its homologs in other plant species revealed that OsLAP3 belongs to the strictosidine synthase gene family, situated within a distinct evolutionary branch of the Poaceae family among monocots. OsLAP3 is most closely related to the strictosidine synthase gene in Brachypodium distachyon, indicating a high degree of homology (Figures 8A, B). With the poaceae family, the only reported strictosidine synthase gene is ZmMS45, which primarily regulates the development of the anther tapetum and pollen exine (Zhang et al., 2014). This function is similar to the phenotype observed in the lap3 mutation. Additionally, its homologous gene AtLAP3 has been reported to have similar functions (Dobritsa et al., 2009), suggesting that OsLAP3 plays a conserved role in regulating male reproductive development across different plant species.
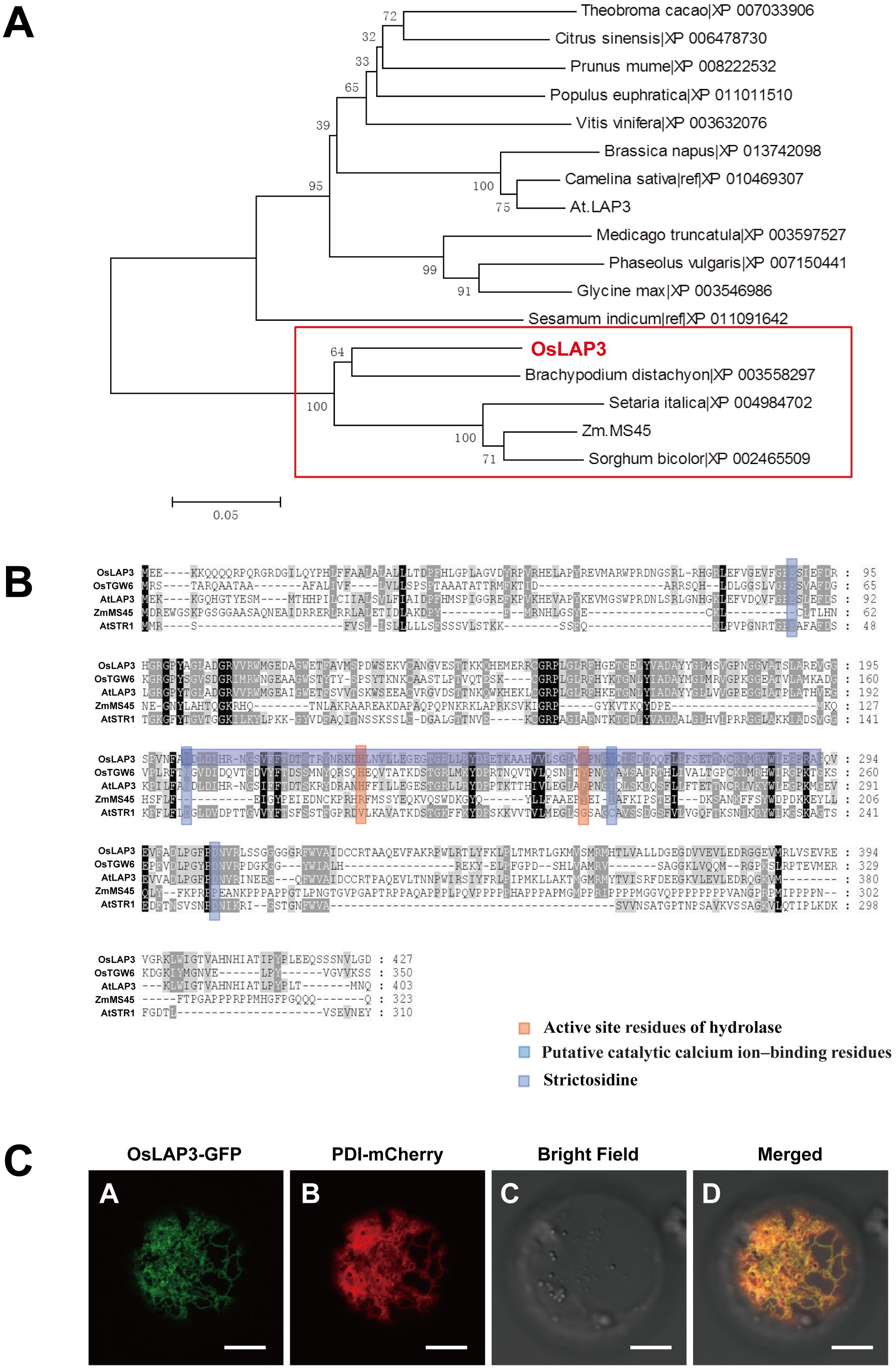
Figure 8. Functional analysis of OsLAP3 protein. (A) Homology analysis and protein sequence alignment of OsLAP3. (B) Evolutionary analysis of OsLAP3 and homologous proteins. (C) Subcellular localization of OsLAP3-GFP fusion protein and ER marker PDI-mCherry in rice protoplasts.
To determine the subcellular localization of OsLAP3, a GFP-tagged OsLAP3 construct was transiently expressed in rice protoplasts. Confocal laser scanning microscopy revealed that OsLAP3 predominantly localizes to the endoplasmic reticulum (Figure 8C; Supplementary Figure S4).
3.8 Expression analysis of OsLAP3
Given that the lap3 mutant specifically affects rice anther development and pollen fertility, with minimal impact on vegetative growth and floral organ development, quantitative real-time PCR (qRT-PCR), promoter-GUS fusion protein transgenic analysis, and in situ hybridization were employed to examine OsLAP3 expression during anther development. Initially, qRT-PCR analyzed OsLAP3 expression in various tissues developmental stages of the anther. OsLAP3 expression level was almost undetectable in low in roots, stems, flag leaves, and leaf sheaths (Figure 9A). However, expression was observed during meiosis and mitosis in anther development, with peak expression during mitosis. Further analysis of OsLAP3 expression in the anthers at different development stages in WT and mutants revealed that during the early meiosis phase (from S5 to S7), lap3 expression in the mutant was slightly higher than in the WT. After microsporocyte formation and the onset of mitosis, OsLAP3 expression in the WT significantly increased, while in the mutant, it markedly declined. This trend continued until stage S12 (Figure 9B), consistent with previous observations from semi-thin sections and electron microscopy.
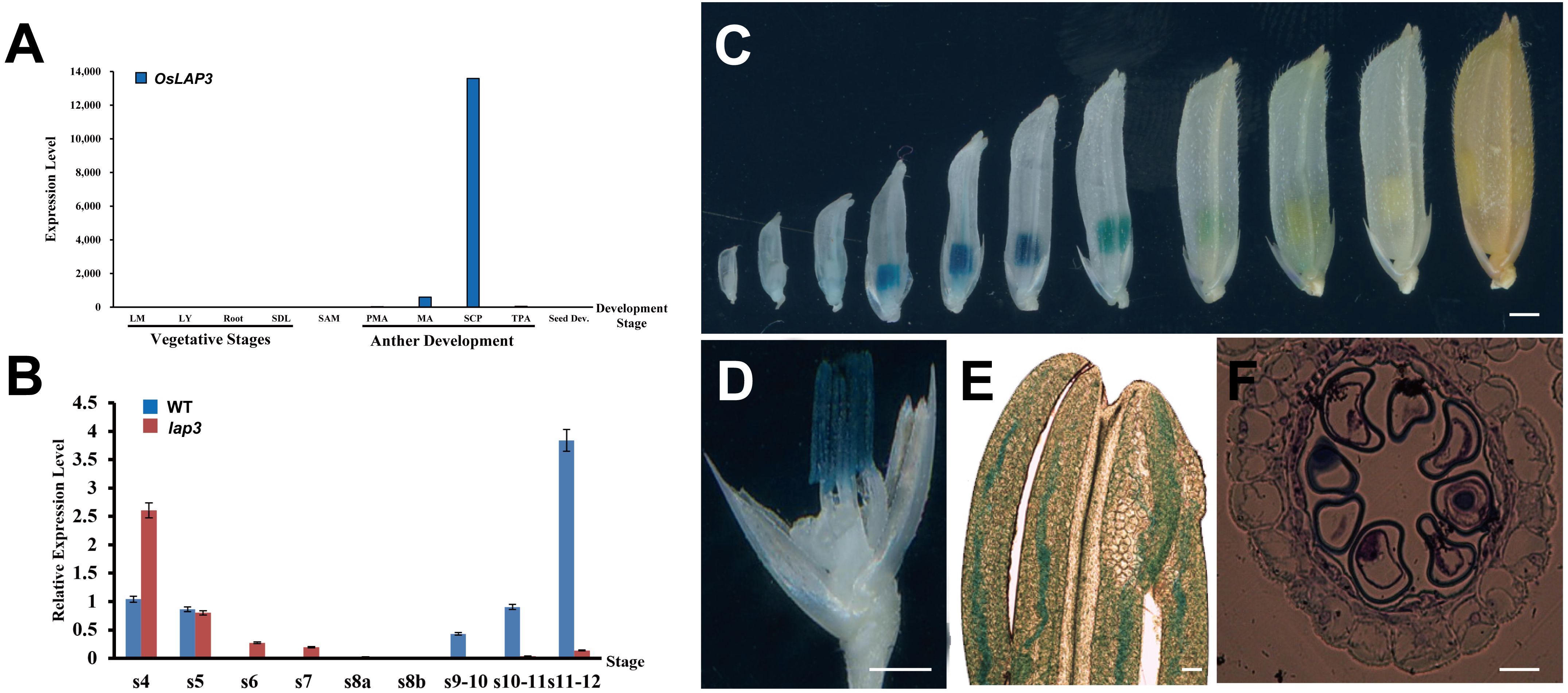
Figure 9. Expression analysis of OsLAP3. (A) Expression of OsLAP3 in different tissues. (B) Expression level in WT and lap3 mutant anthers at various development stages. (C) GUS expression patterns (blue staining) in the heterozygous spikelets of OsLAP3-promoter-GUS transgenic lines at various stages. (D) GUS staining of an 8 mm spikelet (palea and lemma removed, marked by a red arrow). (E) Phase-contrast microscope analysis of anther from (D, F) Transverse section of anther from (D) Bars = 0.5 mm (C-E); 20 µm (F).
GUS staining analysis of transgenic plants carrying an OsLAP3-promoter-GUS construct demonstrated that OsLAP3 was barely expressed in florets and anthers measuring 1–5 mm. However, strong expression was observed exclusively in the anthers of 6–9 mm florets, peaking at 7–8 mm florets (Figure 9C). Semi-thin section analysis indicated that this stage corresponded to the late S10 of anther development (Figures 9D–F). Staining was predominantly concentrated in the anther wall, with weaker staining in the microspores. As floret length exceeded 9 mm, OsLAP3 expression began to decrease, and in nearly mature florets, expression became undetectable. These findings suggest that OsLAP3 plays a crucial role in regulating anther wall development and pollen maturation during the mitotic phase.
3.9 lap3 mutant anthers show reduced wax and cutin content
The synthesis and accumulation of lipid polymers are essential for anther development and pollen function in rice. Cuticular wax and cutin, which are major components of the anther epidermis, contribute to maintaining structural integrity. Disruptions in these pathways often lead to male sterility due to defects in anther epidermis and pollen wall formation. To investigate the impact of the lap3 mutation on lipid metabolism in rice anthers, wax and cutin compositions were analyzed in both WT and lap3 anther (Supplementary Figure S2). The results showed notable changes in wax and cutin profiles in the lap3 mutant, highlighting the broad effects of this mutation on lipid biosynthesis. Total wax content in lap3 anthers was significantly reduced by 42.65%, with a level of 0.5391 µg/mm² compared to 0.9398 µg/mm² in WT anthers (Figure 10A). Among wax components, C18:0 FA (stearic acid) showed a significant reduction, while C24:0 FA (lignoceric acid) displayed an increase, potentially indicating a compensation mechanism in long-chain fatty acid metabolism. Additionally, plant sterols such as β-sitosterol and stigmasterol were significantly reduced in lap3, potentially affecting membrane stability (Figure 10B). Cutin content also showed a 22.95% reduction in lap3, from 0.3328µg/mm² in WT to 0.2564 µg/mm² in the mutant. Key cutin monomers, such as C18:1 ω-HFA and 9, 10, 16 Tri-OH C16FA, were significantly reduced in lap3 (Figure 10C), suggesting that the mutation affects specific steps in cutin biosynthesis.
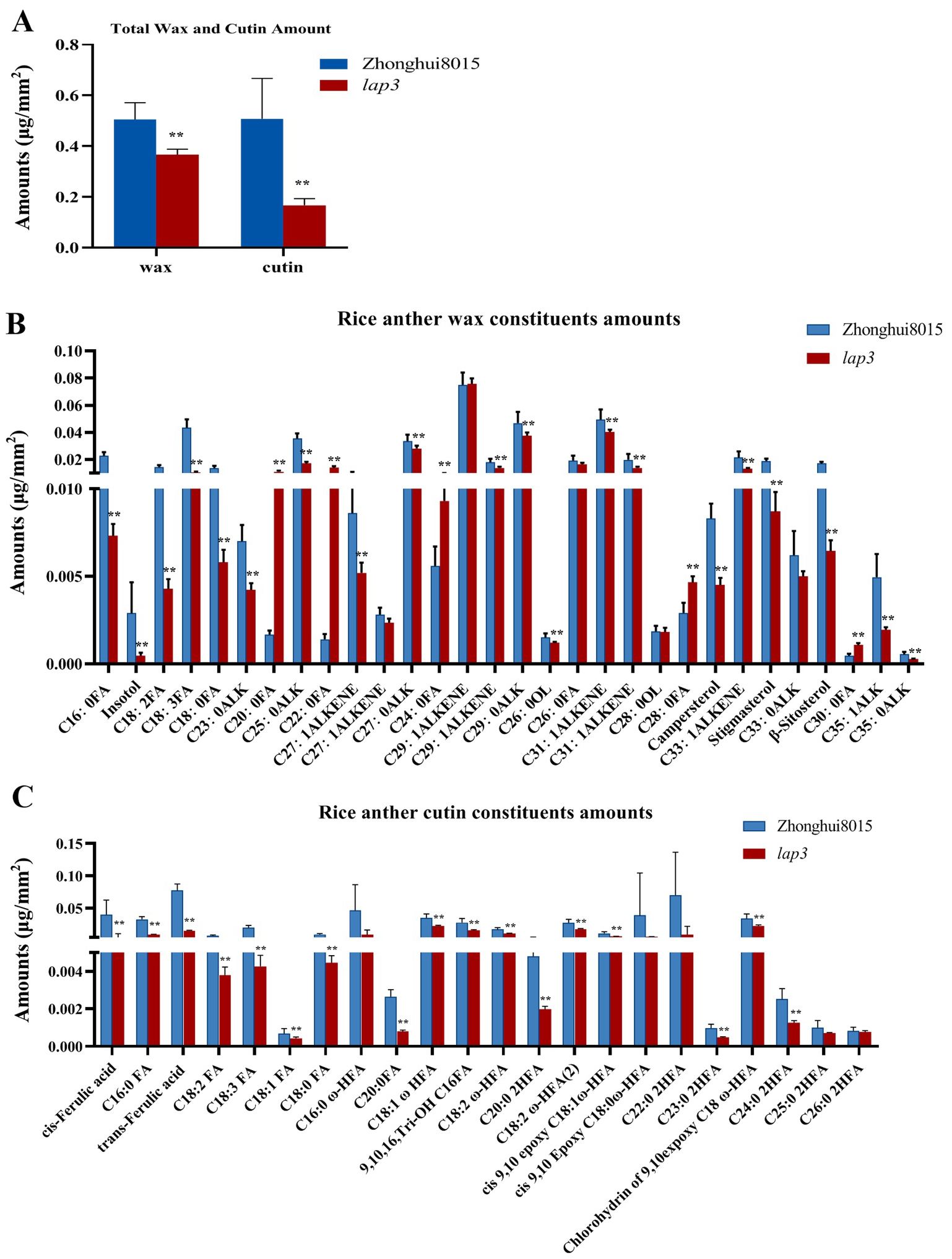
Figure 10. OsLAP3 mutation reduces wax and cutin content in anthers. (A) Total wax and cutin contents in WT and lap3 anthers. (B) Wax component contents. (C) Cutin component contents. Data are mean ± SD of five biological replicates (**p < 0.01). C16:0 FA, hexadecanoic acid; C18:0 FA, octadecanoic acid; C18:1 FA, oleic acid; C18:2 FA, linoleic acid; C18:3 FA, linolenic acid; C20:0 FA, eicosanoic acid; C22:0 2HFA, 2-hydroxy-docosanoic acid; C24:0 FA, tetracosanoic acid; C26:0 FA, hexacosanoic acid; C28:0 FA, octacosanoic acid; C30:0 FA, triacontanoic acid,; ALK, alkane; C24:0 2HFA, 2-hydroxy-tetracosanoic acid; C25:0 2HFA, 2-hydroxy-pentacosanoic acid; C18:0 ω-HFA, cis-9,10-epoxy 18-hydroxy-stearic acid; C18:2 ω-HFA, 18-hydroxy-linoleic acid; C18:1 ω-HFA, cis-9,10-epoxy-18-hydroxy-oleic acid; C26:0 2HFA, 2-hydroxy-hexacosanoic acid; C16:0 ω-HFA, 16-hydroxyhexadecanoic acid.
These data indicate that the lap3 mutation disrupts multiple lipid biosynthesis pathways, affecting both wax and cutin accumulation. The altered lipid profiles in lap3 suggest compromised anther epidermis integrity, which may contribute to its reproductive deficiencies. These findings highlight the importance of OsLAP3 in maintaining the balance of these components for normal anther and pollen development.
3.10 Expression analysis of OsLAP3
Gene expression patterns related to anther and pollen development were compared between both WT and lap3 mutant plants at different stages of anther development (S8b to S12) using qRT-PCR (Figure 11). The expression levels of 12 key genes involved in these processes, including GAMYB, OsCP1, ABCG15, WDA1, DPW, ABCG26, OsMADS3, UGP1, EAT1, CYP704B2, CYP703A3, and OsC6, were analyzed. In WT anthers, sharp increases in gene expression, peaking at S9 and S10. For example, GAMYB, OsCP1, and WDA1 exhibited the highest expression at S9, while OsC6 peaked at S10. This peak expression coincides with critical stages of tapetum development and pollen wall formation, indicating the involvement of these genes in these processes. In contrast, the lap3 mutant exhibited significantly lower expression of these genes throughout all stages, suggesting a disruption in transcriptional regulation.
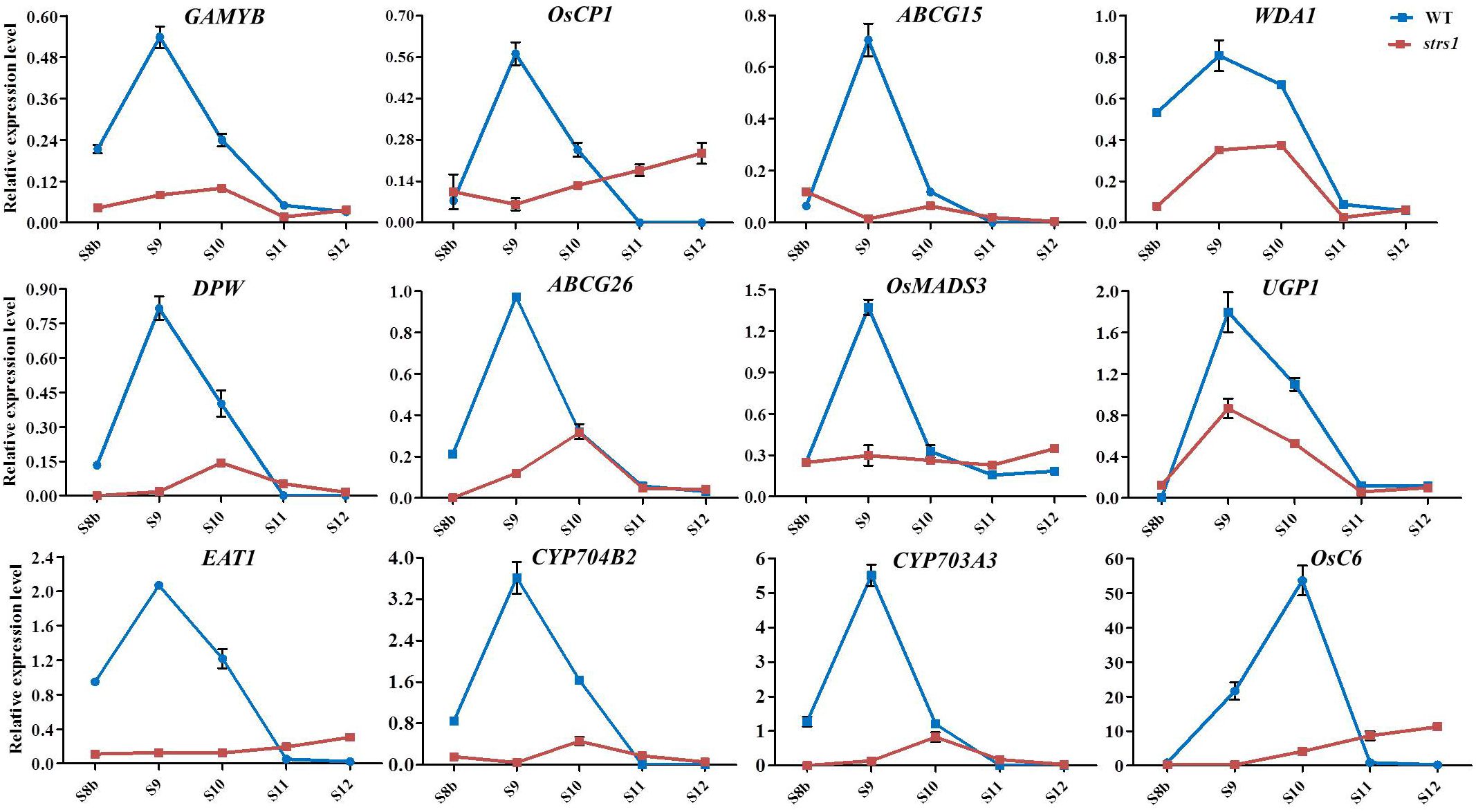
Figure 11. Expression profile of the genes involved in anther development in WT and lap3. OsActin1 was used as an internal control. Error bars show the SD (n = 3). S8b. stage 8b; S9. stage 9; S10. stage 10; S11. stage 11; S12. stage 12; WT. wild type.
Notably, genes involved in lipid biosynthesis for pollen exine formation, such as CYP704B2 and CYP703A3, and those in tapetum PCD, such as EAT1 and OsMADS3, showed much lower expression in lap3 compared to WT plants. Some genes, such as OsC6, exhibited slight increases in expression during later stages (S11 and S12) in lap3, possibly indicating a compensatory response to the developmental abnormalities. However, this late upregulation was insufficient to rescue the mutant phenotype, highlighting the importance of timely gene expression for proper anther and pollen development.
These expression patterns suggest that OsLAP3 plays a crucial role in regulating tapetum degradation and the biosynthesis and transport of lipids during function of strictosidine synthase genes in male reproductive development across plant species.
4 Discussion
In this study, OsLAP3 was characterized and identified as a gene essential for anther cuticle formation and pollen exine patterning in rice. The lap3 mutant exhibited normal vegetative growth but displayed complete male sterility, characterized by shrunken, sterile anthers and defective pollen. Through genetic, cytological, and molecular analyses, it was demonstrated that OsLAP3 plays a critical role in regulating lipid biosynthesis, tapetum degradation, and sporopollenin deposition during pollen development. These findings underscore the importance of OsLAP3 in maintaining tapetum integrity and enabling proper pollen maturation.
4.1 Functional comparative analysis
Distinct roles of OsLAP3 and OsSTRL2 in anther development were highlighted in this research, both of which belong to the strictosidine synthase-like family. OsLAP3 was found to significantly impact lipid metabolism and the timely degradation of tapetal cells, processes crucial for anther development and pollen viability. Mutations in OsLAP3 were shown to disrupt fatty acid biosynthesis pathways and delay PCD in tapetal cells, as confirmed by lipid profiling and TUNEL assays. This disruption led to aberrant lipid changes that impaired pollen exine formation. It was further established that key regulatory genes like CYP704B2, CYP703A3, and OsC6, which are integral to lipid biosynthesis and sporopollenin formation, are directly influenced by OsLAP3, establishing its critical role in pollen fertility. In contrast, OsSTRL2 was associated with broader anther dysplasia, without a specific link to lipid pathways, reflecting its atypical functional profile (Yang et al., 2018; Zou et al., 2017).
4.2 OsLAP3 is essential for lipid biosynthesis and pollen exine formation
OsLAP3 was found to be involved not only in fatty acid synthesis and transport but also in the PCD of tapetal cells. Future research will focus on identifying the upstream regulatory factors of OsLAP3. As an enzyme gene, OsLAP3 likely acts downstream, while its upstream regulators are thought to be transcription factors.
OsLAP3 was shown to coordinate multiple signaling pathways to regulate rice male reproductive development by modulating a series of genes. In the lap3 mutant, qPCR results and phenotypic analysis revealed that the expression of several lipid metabolism-related genes, such as CYP704B2, CYP703A3, and OsC6, was significantly downregulated. CYP704B2 and CYP703A3, which catalyze the hydroxylation of C16/C18 fatty acids and the modify lauric acid, respectively, play critical roles in sporopollenin biosynthesis (Li et al., 2010; Yang et al., 2014). Mutations in OsC6, a lipid transport protein, leads to abnormal Ubisch bodies and defective pollen wall structures, indicating that OsLAP3 regulates these genes to ensure proper anther development. The regulatory role of OsLAP3 was shown to be selective within lipid biosynthesis pathways. While genes associated with sporopollenin and essential wax and cutin components exhibited significant changes in expression and accumulation, other lipid components, such as C24:0 fatty acid, remained stable or even increased, likely due to compensatory adjustments in long-chain fatty acid metabolism. This selective regulation ensures that OsLAP3 primarily affects key components crucial for anther epidermis and pollen wall integrity, while non-essential components are maintained through redundant metabolic pathways or compensatory mechanisms, preserving basic metabolic functions.
Tapetum PCD is closely related to pollen wall formation, with evidence suggesting that OsLAP3 coordinates these processes by regulating PCD-related genes such as OsCP1 and OsC6. TUNEL assays indicated a significant delay in PCD in the lap3 mutant, suggesting a role for OsLAP3 in regulating the PCD process and, thus, impacting anther development. OsLAP3 may interact with PCD-related genes, such as TDR (Tapetum Degeneration Retardation), which controls the expression of genes including OsCP1 and OsC6, ensuring proper tapetal degradation and pollen wall formation (Li et al., 2015). Additionally, SLR1 (Slender Rice 1) was found to interact with MYB188 to regulate several downstream genes involved in sporopollenin biosynthesis, including CYP703A3, DPW, and ABCG15, which are critical to pollen wall formation (Jin et al., 2022). We hypothesize that OsLAP3 is regulated by multiple transcription factors, including bHLHs and the SLR1-MYB188 module, which will be the focus of future research. Through these regulatory mechanisms, OsLAP3 ensures timely tapetal PCD and the synthesis and transport of lipids during anther development, providing structural and nutritional support for normal pollen wall formation.
4.3 Functional conservation of strictosidine synthase domain proteins in reproductive development
Strictosidine synthase domain-containing proteins are known for their roles in secondary metabolite biosynthesis in plants, particularly in alkaloid production (Stöckigt et al., 2008). However, their function in reproductive development has been less explored. In this study, OsLAP3 was shown to share homology with strictosidine synthase proteins from various plant species, including Brachypodium distachyon and Zea mays, both involved in reproductive development. MS45 in Zea mays, for example, has been implicated in regulating tapetal development and pollen wall formation (Wang et al., 2022). Additionally, two strictosidine synthase genes, AtLAP3 and OsSTRL2, have been reported to be involved in male reproductive development (Zou et al., 2017). In our study, OsLAP3 is involved in lipid biosynthesis and pollen exine formation, showing the conserved function of strictosidine synthase-like genes in plant reproductive development. The conserved function of OsLAP3 in rice, similar to these genes, suggests its role as a core regulator in male sterility systems.
Comparing the roles of OsLAP3 and OsSTRL2 (Zou et al., 2017) in pollen wall formation and tapetal cell regulation revealed similarities and differences that enhance the understanding of their functions in rice reproductive biology. Both genes are key regulators of anther development and play essential roles in pollen exine formation. High conservation was observed with AtLAP3 and ZmMS45, both involved in sporopollenin biosynthesis, a crucial component of the pollen wall. OsLAP3 loss-of-function resulted in defective pollen development and male sterility, similar to OsSTRL2. However, OsLAP3 demonstrated a more refined regulatory mechanism compared to OsSTRL2, particularly in its coordination of tapetum PCD. This network is believed to operate more effectively in OsLAP3 due to its ability to interface with multiple pathways, enhancing its regulatory accuracy in pollen exine and cuticle formation. The expression patterns of OsLAP3, predominantly in anthers and particularly in tapetal cells and microspores during pollen development, further supported its significant role in integrating multiple developmental cues.
4.4 OsLAP3 acts as a potential target for GMS line breeding and rice hybrid seed production
The development of male sterility systems has been critical for hybrid seed production in rice, particularly in overcoming the limitations of conventional CMS (cytoplasmic male sterility) and P/TGMS (photo/thermo-sensitive genic male sterility) systems (Li et al., 2007). Despite the widespread use of CMS, low utilization efficiency has been reported due to the dependence on fertility restorer genes, while P/TGMS lines have been shown to be vulnerable to environmental fluctuations, leading to inconsistent seed purity. These limitations have prompted the exploration of more efficient sterility systems. To address these challenges, innovative technologies such as Seed Production Technology (SPT) have been developed to improve seed production efficiency and stability (Chang et al., 2016b). OsLAP3, given its functional similarity to ZmMS45, has emerged as a promising candidate for next-generation rice male sterility systems. ZmMS45 has been successfully used in maize hybrid seed production as a core component of the smart sterility system (Wu et al., 2016). Similarly, OsLAP3 was identified as playing a pivotal role in regulating tapetum PCD and pollen wall formation, making it an ideal target for developing hybrid rice production systems that leverage its regulatory functions. Collaborations with Boleyn Genetics have enabled the integration of OsLAP3 into SPT technology, aimed at developing stable, non-GMO male sterile lines for hybrid seed production. The potential of OsLAP3 in improving rice productivity through GMS line breeding and its application in SPT systems is significant, with future developments expected to further enhance hybrid rice breeding technologies.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
D-BC: Data curation, Formal analysis, Funding acquisition, Investigation, Writing – original draft. RZ: Data curation, Formal analysis, Investigation, Visualization, Writing – review & editing. H-MW: Resources, Writing – review & editing. P-PZ: Investigation, Methodology, Writing – review & editing. Z-FY: Investigation, Methodology, Writing – review & editing. D-DX: Data curation, Investigation, Writing – review & editing. Y-XZ: Funding acquisition, Resources, Writing – review & editing. X-DZ: Supervision, Writing – review & editing. L-YC: Funding acquisition, Supervision, Writing – review & editing. S-HC: Funding acquisition, Supervision, Writing – review & editing. L-PS: Funding acquisition, Supervision, Writing – review & editing, Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Software, Validation, Visualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by grants from the National Natural Science Foundation of China (32272191), PhD Scientific Research and Innovation Foundationof Sanya Yazhou Bay Science and Technology City (SHPHDSRF-2023-11-011), the National Key Research and Development Program of China (2022YFD1200800 and 2023YFD1201200), the China Agriculture Research System (CARS-01-11), and the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Science (CAAS-ASTIP-2013-CNRRI).
Acknowledgments
We would like to thank Prof. Dabing Zhang (Deleased), Prof. Wanqi Liang and Qian Luo (Shanghai Jiao Tong University, Shanghai, China) for their help in the determination of fatty acid content.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1508828/full#supplementary-material
References
Abbas, A., Yu, P., Sun, L. P., Yang, Z. F., Chen, D. B., Cheng, S. H., et al. (2021). Exploiting genic male sterility in rice: from molecular dissection to breeding applications. Front. Plant Sci. 12, 27. doi: 10.3389/fpls.2021.629314
Ali, A., Wu, T., Zhang, H., Xu, P., Zafar, S. A., Liao, Y., et al. (2022). A putative SUBTILISIN-LIKE SERINE PROTEASE 1 (SUBSrP1) regulates anther cuticle biosynthesis and panicle development in rice. J. advanced Res. 42, 273–287. doi: 10.1016/j.jare.2022.01.003
Allen, G. C., Flores-Vergara, M. A., Krasynanski, S., Kumar, S., Thompson, W. F. (2006). A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 1, 2320–2325. doi: 10.1038/nprot.2006.384
Chang, Z. Y., Chen, Z. F., Wang, N., Xie, G., Lu, J. W., Yan, W., et al. (2016a). Construction of a male sterility system for hybrid rice breeding and seed production using a nuclear male sterility gene. Proc. Natl. Acad. Sci. United States America 113, 14145–14150. doi: 10.1073/pnas.1613792113
Chang, Z. Y., Chen, Z. F., Yan, W., Xie, G., Lu, J. W., Wang, N., et al. (2016b). An ABC transporter, OsABCG26, is required for anther cuticle and pollen exine formation and pollen-pistil interactions in rice. Plant Sci. 253, 21–30. doi: 10.1016/j.plantsci.2016.09.006
Chang, Z. Y., Jin, M. N., Yan, W., Chen, H., Qiu, S. J., Fu, S., et al. (2018). The ATP-binding cassette (ABC) transporter OsABCG3 is essential for pollen development in rice. Rice 11. doi: 10.1186/s12284-018-0248-8
Chen, Z. S., Liu, X. F., Wang, D. H., Chen, R., Zhang, X. L., Xu, Z. H., et al. (2018). Transcription factor osTGA10 is a target of the MADS protein osMADS8 and is required for tapetum development. Plant Physiol. 176, 819–835. doi: 10.1104/pp.17.01419
Dobritsa, A. A., Nishikawa, S.-I., Preuss, D., Urbanczyk-Wochniak, E., Sumner, L. W., Hammond, A., et al. (2009). LAP3, a novel plant protein required for pollen development, is essential for proper exine formation. Sexual Plant Reprod. 22, 167–177. doi: 10.1007/s00497-009-0101-8
Fu, Z. Z., Yu, J., Cheng, X. W., Zong, X., Xu, J., Chen, M. J., et al. (2014). The rice basic helix-Loop-Helix transcription factor TDR INTERACTING PROTEIN2 is a central switch in early anther development. Plant Cell 26, 1512–1524. doi: 10.1105/tpc.114.123745
Ji, C., Li, H., Chen, L., Xie, M., Wang, F., Chen, Y., et al. (2013). A novel rice bHLH transcription factor, DTD, acts coordinately with TDR in controlling tapetum function and pollen development. Mol. Plant 6, 1715–1718. doi: 10.1093/mp/sst046
Jin, Y., Song, X., Chang, H., Zhao, Y., Cao, C., Qiu, X., et al. (2022). The GA–DELLA–OsMS188 module controls male reproductive development in rice. New Phytol. 233, 2629–2642. doi: 10.1111/nph.v233.6
Jung, K.-H., Han, M.-J., Lee, Y.-S., Kim, Y.-W., Hwang, I., Kim, M.-J., et al. (2005). Rice Undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell 17, 2705–2722. doi: 10.1105/tpc.105.034090
Jung, K.-H., Han, M.-J., Lee, D.-Y., Lee, Y.-S., Schreiber, L., Franke, R., et al. (2006). Wax-deficient anther1 is involved in cuticle and wax production in rice anther walls and is required for pollen development. Plant Cell 18, 3015–3032. doi: 10.1105/tpc.106.042044
Kibble, N. A., Sohani, M. M., Shirley, N., Byrt, C., Roessner, U., Bacic, A., et al. (2009). Phylogenetic analysis and functional characterisation of strictosidine synthase-like genes in Arabidopsis thaliana. Funct. Plant Biol. 36, 1098–1109. doi: 10.1071/FP09104
Ko, S.-S., Li, M.-J., Sun-Ben Ku, M., Ho, Y.-C., Lin, Y.-J., Chuang, M.-H., et al. (2014). The bHLH142 transcription factor coordinates with TDR1 to modulate the expression of EAT1 and regulate pollen development in rice. Plant Cell 26, 2486–2504. doi: 10.1105/tpc.114.126292
Li, Y., Li, D., Guo, Z., Shi, Q., Xiong, S., Zhang, C., et al. (2016). OsACOS12, an orthologue of Arabidopsis acyl-CoA synthetase5, plays an important role in pollen exine formation and anther development in rice. BMC Plant Biol. 16, 1–12. doi: 10.1186/s12870-016-0943-9
Li, L., Li, Y., Song, S., Deng, H., Li, N., Fu, X., et al. (2015). An anther development F-box (ADF) protein regulated by tapetum degeneration retardation (TDR) controls rice anther development. Planta 241, 157–166. doi: 10.1007/s00425-014-2160-9
Li, H., Pinot, F., Sauveplane, V., Werck-Reichhart, D., Diehl, P., Schreiber, L., et al. (2010). Cytochrome P450 family member CYP704B2 catalyzes the ω-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell 22, 173–190. doi: 10.1105/tpc.109.070326
Li, S., Yang, D., Zhu, Y. (2007). Characterization and use of male sterility in hybrid rice breeding. J. Integr. Plant Biol. 49, 791–804. doi: 10.1111/j.1744-7909.2007.00513.x
Li, N., Zhang, D.-S., Liu, H.-S., Yin, C.-S., Li, X.-x., Lian, W.-Q., et al. (2006). The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18, 2999–3014. doi: 10.1105/tpc.106.044107
Liu, Z., Bao, W., Liang, W., Yin, J., Zhang, D. (2010). Identification of gamyb4 and analysis of the regulatory role of GAMYB in rice anther development. J. Integr. Plant Biol. 52, 670–678. doi: 10.1111/j.1744-7909.2010.00959.x
Liu, Z., Lin, S., Shi, J., Yu, J., Zhu, L., Yang, X., et al. (2017). Rice No Pollen 1 (NP1) is required for anther cuticle formation and pollen exine patterning. Plant J. 91, 263–277. doi: 10.1111/tpj.2017.91.issue-2
Men, X., Shi, J. X., Liang, W. Q., Zhang, Q. F., Lian, G. B., Quan, S., et al. (2017). Glycerol-3-Phosphate Acyltransferase 3 (OsGPAT3) is required for anther development and male fertility in rice. J. Exp. Bot. 68, 512–525. doi: 10.1093/jxb/erw445
Mu, H., Ke, J., Liu, W., Zhuang, C., Yip, W. (2009). UDP-glucose pyrophosphorylase2 (OsUgp2), a pollen-preferential gene in rice, plays a critical role in starch accumulation during pollen maturation. Chin. Sci. Bull. 54, 234–243. doi: 10.1007/s11434-008-0568-y
Niu, N., Liang, W., Yang, X., Jin, W., Wilson, Z. A., Hu, J., et al. (2013). EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat. Commun. 4, 1445. doi: 10.1038/ncomms2396
Niu, B.-X., He, F.-R., He, M., Ren, D., Chen, L.-T., Liu,, Y.-G. (2013). The ATP-binding cassette transporter OsABCG15 is required for anther development and pollen fertility in rice. Journal of Integrative Plant Biology. 55 (8), 710–720. doi: 10.1111/jipb.12053
Qin, P., Tu, B., Wang, Y., Deng, L., Quilichini, T. D., Li, T., et al. (2013). ABCG15 encodes an ABC transporter protein, and is essential for post-meiotic anther and pollen exine development in rice. Plant Cell Physiol. 54, 138–154. doi: 10.1093/pcp/pcs162
Quilichini, T. D., Grienenberger, E., Douglas, C. J. (2015). The biosynthesis, composition and assembly of the outer pollen wall: A tough case to crack. Phytochemistry 113, 170–182. doi: 10.1016/j.phytochem.2014.05.002
Ranjan, R., Khurana, R., Malik, N., Badoni, S., Parida, S. K., Kapoor, S., et al. (2017). bHLH142 regulates various metabolic pathway-related genes to affect pollen development and anther dehiscence in rice. Sci. Rep. 7, 43397. doi: 10.1038/srep43397
Shi, J., Tan, H., Yu, X.-H., Liu, Y., Liang, W., Ranathunge, K., et al. (2011). Defective pollen wall is required for anther and microspore development in rice and encodes a fatty acyl carrier protein reductase. Plant Cell 23, 2225–2246. doi: 10.1105/tpc.111.087528
Sohani, M., Schenk, P., Schultz, C., Schmidt, O. (2009). Phylogenetic and transcriptional analysis of a strictosidine synthase-like gene family in Arabidopsis thaliana reveals involvement in plant defence responses. Plant Biol. 11, 105–117. doi: 10.1111/j.1438-8677.2008.00139.x
Stöckigt, J., Barleben, L., Panjikar, S., Loris, E. A. (2008). 3D-Structure and function of strictosidine synthase-the key enzyme of monoterpenoid indole alkaloid biosynthesis. Plant Physiol. Biochem. 46, 340–355. doi: 10.1016/j.plaphy.2007.12.011
Sun, Z. H., Liu, K. K., Chen, C., Chen, D. B., Peng, Z. Q., Zhou, R., et al. (2023). OsLDDT1, encoding a transmembrane structural DUF726 family protein, is essential for tapetum degradation and pollen formation in rice. Plant Sci. 329, 111596. doi: 10.1016/j.plantsci.2023.111596
Sun, L., Xiang, X., Yang, Z., Yu, P., Wen, X., Wang, H., et al. (2018). OsGPAT3 plays a critical role in anther wall programmed cell death and pollen development in rice. Int. J. Mol. Sci. 19. doi: 10.3390/ijms19124017
Tariq, N., Yaseen, M., Xu, D., Rehman, H., Bibi, M., Uzair, M. (2023). Rice anther tapetum: a vital reproductive cell layer for sporopollenin biosynthesis and pollen exine patterning. Plant Biol. 25, 233–245. doi: 10.1111/plb.13485
Wang, Y., Bao, J., Wei, X., Wu, S., Fang, C., Li, Z., et al. (2022). Genetic structure and molecular mechanisms underlying the formation of tassel, Anther, and pollen in the male inflorescence of maize (Zea Mays L.). Cells 11, 1753. doi: 10.3390/cells11111753
Wu, Y. Z., Fox, T. W., Trimnell, M. R., Wang, L. J., Xu, R. J., Cigan, A. M., et al. (2016). Development of a novel recessive genetic male sterility system for hybrid seed production in maize and other cross-pollinating crops. Plant Biotechnol. J. 14, 1046–1054. doi: 10.1111/pbi.2016.14.issue-3
Wu, L., Guan, Y., Wu, Z., Yang, K., Lv, J., Converse, R., et al. (2014). OsABCG15 encodes a membrane protein that plays an important role in anther cuticle and pollen exine formation in rice. Plant Cell Rep. 33, 1881–1899. doi: 10.1007/s00299-014-1666-8
Xu, Y., Liu, S., Liu, Y., Ling, S., Chen, C., Yao, J. (2017). HOTHEAD-like HTH1 is involved in anther cutin biosynthesis and is required for pollen fertility in rice. Plant Cell Physiol. 58, 1238–1248. doi: 10.1093/pcp/pcx063
Yang, Z., Liu, L., Sun, L., Yu, P., Zhang, P., Abbas, A., et al. (2019). OsMS1 functions as a transcriptional activator to regulate programmed tapetum development and pollen exine formation in rice. Plant Mol. Biol. 99, 175–191. doi: 10.1007/s11103-018-0811-0
Yang, X. J., Wu, D., Shi, J. X., He, Y., Pinot, F., Grausem, B., et al. (2014). Rice CYP703A3, a cytochrome P450 hydroxylase, is essential for development of anther cuticle and pollen exine. J. Integr. Plant Biol. 56, 979–994. doi: 10.1111/jipb.v56.10
Yang, Z., Zhang, Y., Sun, L., Zhang, P., Liu, L., Yu, P., et al. (2018). Identification of cyp703a3-3 and analysis of regulatory role of CYP703A3 in rice anther cuticle and pollen exine development. Gene 649, 63–73. doi: 10.1016/j.gene.2018.01.058
Yuan, L. P. (2014). Development of hybrid rice to ensure food security. Rice Sci. 21, 1–2. doi: 10.1016/S1672-6308(13)60167-5
Zafar, S. A., Patil, S. B., Uzair, M., Fang, J., Zhao, J., Guo, T., et al. (2020). DEGENERATED PANICLE AND PARTIAL STERILITY 1 (DPS1) encodes a cystathionine β-synthase domain containing protein required for anther cuticle and panicle development in rice. New Phytol. 225, 356–375. doi: 10.1111/nph.v225.1
Zhang, H., Egger, R. L., Kelliher, T., Morrow, D., Fernandes, J., Nan, G. L., et al. (2014). Transcriptomes and proteomes define gene expression progression in pre-meiotic maize anthers. G3: Genes Genomes Genet. 4, 993–1010. doi: 10.1534/g3.113.009738
Zhang, D., Liang, W., Yin, C., Zong, J., Gu, F., Zhang, D. (2010). OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiol. 154, 149–162. doi: 10.1104/pp.110.158865
Zhang, D. B., Yang, L. (2014). Specification of tapetum and microsporocyte cells within the anther. Curr. Opin. Plant Biol. 17, 49–55. doi: 10.1016/j.pbi.2013.11.001
Zhao, J., Long, T., Wang, Y., Tong, X., Tang, J., Li, J., et al. (2020). RMS2 encoding a GDSL lipase mediates lipid homeostasis in anthers to determine rice male fertility. Plant Physiol. 182, 2047–2064. doi: 10.1104/pp.19.01487
Zhu, X., Yu, J., Shi, J., Tohge, T., Fernie, A. R., Meir, S., et al. (2017). The polyketide synthase OsPKS2 is essential for pollen exine and Ubisch body patterning in rice. J. Integr. Plant Biol. 59, 612–628. doi: 10.1111/jipb.12574
Zou, T., Li, S. C., Liu, M. X., Wang, T., Xiao, Q., Chen, D., et al. (2017). An atypical strictosidine synthase, OsSTRL2, plays key roles in anther development and pollen wall formation in rice. Sci. Rep. 7, 6863. doi: 10.1038/s41598-017-07064-4
Keywords: rice, male sterility, OsLAP3, pollen exine, anther development
Citation: Chen D-b, Zhou R, Wang H-m, Zhang P-p, Yang Z-f, Xuan D-d, Zhang Y-x, Zhan X-d, Cao L-y, Cheng S-h and Sun L-p (2025) OsLAP3/OsSTRL2, encoding a rice strictosidine synthase, is required for anther cuticle formation and pollen exine patterning in rice. Front. Plant Sci. 15:1508828. doi: 10.3389/fpls.2024.1508828
Received: 09 October 2024; Accepted: 17 December 2024;
Published: 20 January 2025.
Edited by:
Akshaya Kumar Biswal, The University of Georgia, United StatesReviewed by:
Kenichi Nonomura, National Institute of Genetics, JapanAsif Ali, Sichuan Agricultural University, China
Copyright © 2025 Chen, Zhou, Wang, Zhang, Yang, Xuan, Zhang, Zhan, Cao, Cheng and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li-yong Cao, Y2FvbGl5b25nQGNhYXMuY24=; Shi-hua Cheng, Y2hlbmdzaGlodWFAY2Fhcy5jbg==; Lian-ping Sun, c3VubGlhbnBpbmdAY2Fhcy5jbg==
†These authors have contributed equally to this work
 Dai-bo Chen1†
Dai-bo Chen1† Pei-pei Zhang
Pei-pei Zhang Zheng-fu Yang
Zheng-fu Yang Ying-xin Zhang
Ying-xin Zhang Xiao-deng Zhan
Xiao-deng Zhan Lian-ping Sun
Lian-ping Sun