- 1Key Laboratory of Germplasm Innovation and Genetic Improvement of Grain and Oil Crops (Co-construction by Ministry and Province), Ministry of Agriculture and Rural Affairs, Crop Breeding and Cultivation Research Institute, Shanghai Academy of Agricultural Sciences, Shanghai, China
- 2Shanghai Agricultural Products Preservation Processing Engineering Technology Research Center, Shanghai, China
- 3Zhongken Seed Industry Co., Ltd., Shanghai, China
- 4Shanghai Agricultural Science and Technology Service Center, Shanghai, China
Introduction: Huruan1212 (HR1212) is well-regarded for its superior eating and cooking quality in the lower reaches of the Yangtze River in China. Still, its high susceptibility to rice panicle blast and lack of fragrance have limited its further spread and utilization. Pigm and Pi-ta are two dominant genes known for their stable broad-spectrum resistance against rice blast fungus Magnaporthe oryzae, while badh2 is the crucial gene that regulates rice aroma.
Methods: In this study, we utilized a molecular marker-assisted selection backcrossing strategy to introduce Pigm, Pi-ta, and badh2 into introgressed lines employing re-sequencing for precise genetic background selection.
Results: Finally, we selected three introgressed lines, including two that carry Pigm with the highest background recovery rates, showing eating and cooking qualities similar to those of HR1212, and one line that pyramids Pigm, Pi-ta, and badh2, which features a strong aroma. They all displayed significantly enhanced resistance to panicle blast and improved yield compared to HR1212.
Discussion: In conclusion, this study expanded the germplasm resources of japonica, providing a material foundation for enhancing breeding programs aimed at developing rice blast-resistant and high-quality fragrant japonica varieties. Additionally, the study demonstrated that integrating molecular markers and re-sequencing can inform breeders’ decision-making more precisely and efficiently.
1 Introduction
Rice is the staple food for over half of the world’s population and is one of the major food crops in China (Wing et al., 2018). According to 2022 statistics, its cultivation area accounts for a quarter of the annual planting area of grain crops in China (http://www.stats.gov.cn/). The development of new rice varieties is a key driver of rice production. To date, breeding high-yield, fine-quality, and disease-resistant varieties have remained rice breeders’ primary objective (Ye et al., 2022).
Rice blast disease caused by Magnaporthe oryzae (M. oryzae) is one of the most devastating worldwide. It is estimated that rice blast accounts for an annual loss of about 10%-30% in global rice yield, posing a significant threat to stable rice production (Hwang et al., 1987; Talbot, 2003). Rice blast manifests with different symptoms according to the affected organ, including leaf blast and panicle blast, the latter being particularly detrimental to yield (Gao et al., 2023). Recent studies have elucidated the genetic basis of rice blast resistance, and a total of 31 genes have been cloned, including two notable dominant genes, Pigm and Pi-ta, which confer durable, broad-spectrum resistance to panicle blast (Li et al., 2019, 2020; Song et al., 2024; Xiao et al., 2020). Pigm, located on chromosome 6, is either tightly linked or allelic to resistance gene Pi2 and Pi9 (Deng et al., 2006). Notably, Pigm has been shown to exhibit long-lasting resistance to M. oryzae without yield penalty (Deng et al., 2017; Wu et al., 2017; Zhai et al., 2022). A functional marker, M143104, was developed for Pigm and applied to breeding programs (Li, 2022; Wang et al., 2023).
Pi-ta is a single-copy gene near the centromere of chromosome 12, and the SNP at 2752 nt is the key to distinguishing the resistant Pi-ta allele from the susceptible pi-ta allele (Bryan et al., 2000). Based on this, several dominant markers were developed for genotyping Pi-ta (Huang et al., 2023; Jia et al., 2002, 2004; Wang et al., 2007), among which molecular markers YL155/YL87 and YL183/YL87 have been widely used in blast resistant germplasm screening (Khan et al., 2018; Liang et al., 2017; Ma et al., 2023; Terensan et al., 2021). Primers YL155 and YL183 were located in the intron region of the gene with the difference in their last four nucleotides, with YL183 specific for the susceptible allele, while YL155 specific for the resistant allele. However, the application of Pi-ta and Pigm in rice varieties remains limited. A study revealed that only 33%-55% of landraces or bred varieties in China carry the resistant genotypes of Pi-ta (Xu et al., 2020), and Pigm was not present in japonica varieties released in Jiangsu Province until 2020 (Wang et al., 2020), which highlighted the necessity of introducing the resistant allele(s) into more commercial rice varieties through breeding programs.
Aroma (fragrance) is a crucial component of end-use quality in rice and is largely controlled by allelic variation at the badh2 gene (Bradbury et al., 2005). A mutation in the BADH2 gene, which encodes betaine aldehyde dehydrogenase, leads to a loss of enzyme activity, resulting in the accumulation of 2-acetyl-1-pyrroline (2-AP). 2-AP is a potent flavor compound responsible for the distinctive aromas of both basmati and jasmine rice (Buttery et al., 1982). To date, 15 functional haplotypes have been identified within the coding region of BADH2, each with distinct geographical distribution patterns. The badh2-E2.1 haplotype is predominantly found in japonica rice varieties in Jiangsu Province, with the 2-AP concentration in these varieties reaching approximately 660 ng/g (Pan et al., 2021). A specific molecular marker, InDel-E2, was developed to identify the badh2-E2.1 haplotype, based on a 7-bp deletion in exon 2 of badh2 (Wang et al., 2008).
Huruan 1212 (HR1212) is a japonica variety known for its exceptional taste quality, developed by the Crop Research Institute of the Shanghai Academy of Agricultural Sciences in 2017. It was awarded the Gold Medal in the first session of the Taste Quality Evaluation of High-Quality Japonica Varieties in China in 2018. Its outstanding eating and cooking quality (ECQ) has contributed to its popularity in Shanghai.
However, severe panicle blast symptoms have been observed in HR1212 in the breeding practices of recent years, which resulted in substantial yield losses. We analyzed the whole-genome variation of HR1212 and found that nine genes related to rice blast resistance (Pid2, Pid3, Pi5-1, Pi-ta, RGA4, RGA5, Ptr, LHCB5, OsCERK1) were all in susceptible genotypes, highlighting the necessity of improving blast resistance in HR1212 by introducing favorable alleles. Another drawback of HR1212 is its lack of desirable aroma, which may disadvantage it in the market among consumers seeking high-quality fragrant rice.
In this study, we aimed to enhance panicle blast resistance and aroma in HR1212 by developing introgressed lines through a molecular marker-assisted backcrossing strategy. Molecular markers were used for foreground selection, and re-sequencing was employed for background selection. The favorable alleles of Pigm, Pi-ta, and badh2 were introgressed into HR1212, and superior introgressed lines were selected based on agronomic performance. As a result, three introgressed lines were successfully developed, exhibiting significantly improved panicle blast resistance, better yield performance, and high eating quality.
2 Materials and methods
2.1 Plant materials and field trials
HR1212 was the recurrent parent, lacking the favorable genotypes of Pigm, Pi-ta, and badh2. The donor parents included the japonica varieties Wuxianggeng 19 (WXG19) (carrying Pi-ta and badh2), Nangeng 46 (NG46) (carrying badh2), Songzaoxiang 1 (SZX1) (carrying Pi-ta and badh2), and a near-isogenic line (NIL) of Nangeng 9108 (hereafter referred to as the NIL) (carrying Pigm). NG46 has been widely used as a core parent in the lower Yangtze River region due to its excellent eating quality and high yield. SZX1 is a japonica variety known for its fragrant and glutinous texture. The NIL exhibits moderate resistance to rice blast disease. All parent lines were sourced from our genetic resources center.
Hybridization and backcrossing experiments were conducted from 2019 to 2022 in Fengxian District, Shanghai (30°54’N, 121°24’E), and Lingshui County, Hainan Province (18°30’N, 110°1’E). After backcrossing and marker-assisted foreground selection, selected lines were planted in Hainan and Shanghai in 2022 and 2023. The field trials followed local management practices, with row and column spacings set at 25 cm and 12 cm, respectively.
2.2 Molecular marker assays
Genomic DNA was extracted from fresh leaves of individual plants using the cetyltrimethylammonium bromide method. The genotypes of Pigm and badh2 were identified using the markers M143104 (Wang et al., 2023) and InDel-E2 (Wang et al., 2008), respectively. The genotypes of Pi-ta were determined using two dominant molecular markers YL155/YL87 for Pi-ta and YL183/YL87 for pi-ta (Jia et al., 2002, 2004). The detailed information on these markers was listed in Supplementary Table 1. All of them were synthesized by Tsingke Biotech. Co. Ltd, Shanghai, China. The PCR assays were conducted in a total volume of 10 μL containing 1 μL of genomic DNA template (about 50 ng), 5 μL of 2×EasyTaq PCR SuperMix (TransGen Biotech Co. Ltd, Beijing, China), 0.5 μL of each primer (1 μmol/L), and 3 μL of ddH2O. PCR amplification was performed with the following profile: 94°C for 5 min, 35 cycles of 94°C for 45 s, primer annealing at different temperatures for 45 s (Supplementary Table 1), and 72°C for 90 s, and 72°C for 10 min. The PCR products of markers M143104, YL155/YL87, and YL183/YL87 were visualized by electrophoresis on 1%-2% agarose gels in 1×TAE buffer at 130 V for 15 to 30 min. The PCR products of InDel-E2 were separated in 8% non-denatured polyacrylamide gel (PAGE) in 1.0×TBE buffer followed by silver staining.
2.3 Whole-genome sequencing and variant identification
Sequencing libraries were constructed using 200 ng of DNA per sample. Paired-end 150 bp single-index sequencing was carried out using reagents from the MGISEQ-2000RS high-throughput sequencing instrument. FastQC (v.0.10.1) (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used to assess the quality of the short sequencing reads, and Trimmomatic (v.0.36) (Bolger et al., 2014) was employed to trim the reads. The Burrows-Wheeler Aligner software (BWA, v.0.7.8) (Li and Durbin, 2009) was used to align high-quality paired-end clean reads to the Nipponbare reference genome (MSU V7.0). The alignment results were processed using SAMtools (v.0.1.19) (Li et al., 2009). Variant detection was performed using GATK software (McKenna et al., 2010). Firstly, the default parameters were used for filtering. Then remove low-quality variants that meet the following criteria: QD < 20.0, QUAL < Mean QUAL, ReadPosRankSum < -8.0, FS > 10.0. Then variants are required to meet the following conditions: minor allele frequency (MAF) ≥ 4%, missing data rate < 20%.
2.4 Identification of genomic fragments in pedigree
The sliding window method was used to calculate the SNP similarity of genomic fragments between individuals and parents. The window size was set to 1,210 SNPs (with an average physical distance of 300 kb), and the step size was 121 SNPs. A sequence with more than 90% identical SNPs within a window was considered to originate from the parent. The distribution of putative genomic fragments was visualized using the circlize package in R (Gu et al., 2014).
2.5 Phylogenetic tree and genetic distance
The phylogenetic tree was constructed using FastTree software with the ‘JC+CAT’ model (Price et al., 2010). The identical-by-state (IBS) genetic distance between each pair of individuals was calculated using PLINK (v.1.9) (Purcell et al., 2007), and the results were visualized as a heat map using pheatmap package in R.
2.6 Evaluation of panicle blast resistance at the booting stage
At the early booting stage, the plants were injected with a mixed spore suspension of M. oryzae isolates, consisting of five representative strains (2023-35, 2023-197, 2023-207, 2023-261, and 2023-296) in equal proportions, with 30-40 spores per field of view under a 10 × 10 magnification microscope. The spore suspension was provided by the Institute of Plant Protection, Jiangsu Academy of Agricultural Sciences. Each plot was inoculated with 10 panicles, applying 1 mL of the suspension per panicle. The experiment was conducted at the Zhuanghang Experimental Station in Fengxian District, Shanghai. At maturity, the number of diseased grains and total grains per tagged panicle were recorded. The average percentage of symptomatic grains (PSG) was used to evaluate the severity of the panicle blast.
2.7 Identification of agronomic traits and yield-related traits
The phenotypic evaluation was conducted in Shanghai in 2023. Each line was planted in ten rows, with seven plants per row. The heading date was recorded when approximately half of the plants in each plot had headed. Plant height, number of grains per panicle, and seed-setting rate were measured according to the standard evaluation systems for rice (Yu and Wei, 2010) using five representative plants from the central plot. Yield per plant was calculated as the average yield of ten consecutive plants from the central plot.
2.8 Measurements of physicochemical properties related to ECQ
After harvest, the grain samples were stored for three months in a sample drying room. Approximately 100 g of rice grain samples were shelled using a seed hulling machine (JLG-II, Zhonggu Mechanical Instrument Co., Ltd., Zhengzhou, China) and then milled with a machine (JNM-III, China Grain Storage Chengdu Storage Research Institute Co., Ltd., Chengdu, China). The milled rice samples were kept in an environmentally controlled room at approximately 28°C (humidity <10%) for 7 days. Subsequently, part of the milled rice was ground into powder and screened through a 100-mesh sieve for further analysis.
Gel consistency and amylose content were determined according to departmental standard NY/T 83-2017 and national standard GB/T 15683-2008, respectively. The pasting properties of starch were assessed using a rapid viscosity analyzer (RVA4500, PerkinElmer Inc., United States) following regulations of the American Association of Cereal Chemists (Cereals & Grains Association, 2000) and the manufacturer’s instructions.
The concentration of 2-acetyl-1-pyrroline (2-AP) was measured using a GCMS-QP2020 NX gas chromatograph-mass spectrometer (Shimadzu, Japan), in accordance with departmental standard NY/T 4350-2023. The percentage of chalky grains (CGP) was determined using approximately 200 randomly selected whole-milled grains analyzed by the QM3 Grain Rice Analyzer (VIBE Imagine Analytics, Israel). CGP was calculated as follows: CGP (%) = (Number of grains with at least 20% whitish area/Total number of grains) × 100%.
2.9 Statistical analysis
The Chi-square test was performed using SAS 9.4 software. Analysis of variance was conducted with R software, and the least significant difference method was employed for multiple comparisons.
3 Results
3.1 Development of introgressed lines for Pigm, Pi-ta, and badh2
Three populations were developed with HR1212 as the recurrent parent (Supplementary Figure 1). Population 1 (P1) was developed through continuous backcrossing of HR1212 with a stable line derived from the cross between NG46 and WXG19. The segregating BC1F1 population was screened using markers of Pi-ta and badh2. Since HR1212 lacks the favorable alleles of Pigm, Pi-ta, and badh2, these desirable genotypes were initially introduced as heterozygotes during the backcrossing process. Individual plants carrying heterozygotes of Pi-ta or badh2 and having agronomic performance similar to HR1212 were chosen for further backcrossing. Population 2 (P2) was derived from the backcross of HR1212 to SZX1 with a similar selection process (Supplementary Figure 2, Supplementary Table 2). Population 3 (P3) was formed by backcrossing HR1212 with NIL, and the offspring individuals were screened using the molecular marker for Pigm (Supplementary Figure 2). Individual plants carrying heterozygotes of Pigm and having agronomic performance similar to HR1212 were chosen for further backcrossing. As selection based on agronomic performance began in the early generations, genotype segregation did not conform to the expected theoretical ratios (Supplementary Table 3).
The BC5F1 of P3 continued to backcross with HR1212 to produce BC6F1. Simultaneously, BC5F1 of P3 was crossed with BC5F1 of P1 and BC5F1 of P2. The progeny from these crosses were tested using molecular markers (Supplementary Table 4). Based on agronomic performance, 41 individual plants (C01-C41) were selected for further analysis (Supplementary Table 5).
3.2 Comparison of the genetic background between the introgressed individuals and HR1212
To investigate the recovery of the genomic background, we conducted whole-genome sequencing with an average depth of 13.86× for 41 introgressed individuals (C01-C41) and deep resequencing for the recurrent parent HR1212 (Supplementary Table 6). SNP similarities between the 41 introgressed individuals and HR1212, based on 1,130,432 SNPs distributed across 12 chromosomes, showed a normal distribution (Figure 1A; Supplementary Figure 3). Among the three crosses, BC6F1 of P3 (P3-BC6F1) exhibited the highest genetic background similarity with HR1212 (Figure 1B). Three individuals, including two (C05 and C12) from P3-BC6F1 and one (C34) from P1-BC5F1×P3-BC5F1, showed the highest degree of genetic background recovery (Figure 1B). Phylogenetic tree and genetic distance analyses also showed that these three individuals were genetically closest to HR1212 (Supplementary Figure 4).
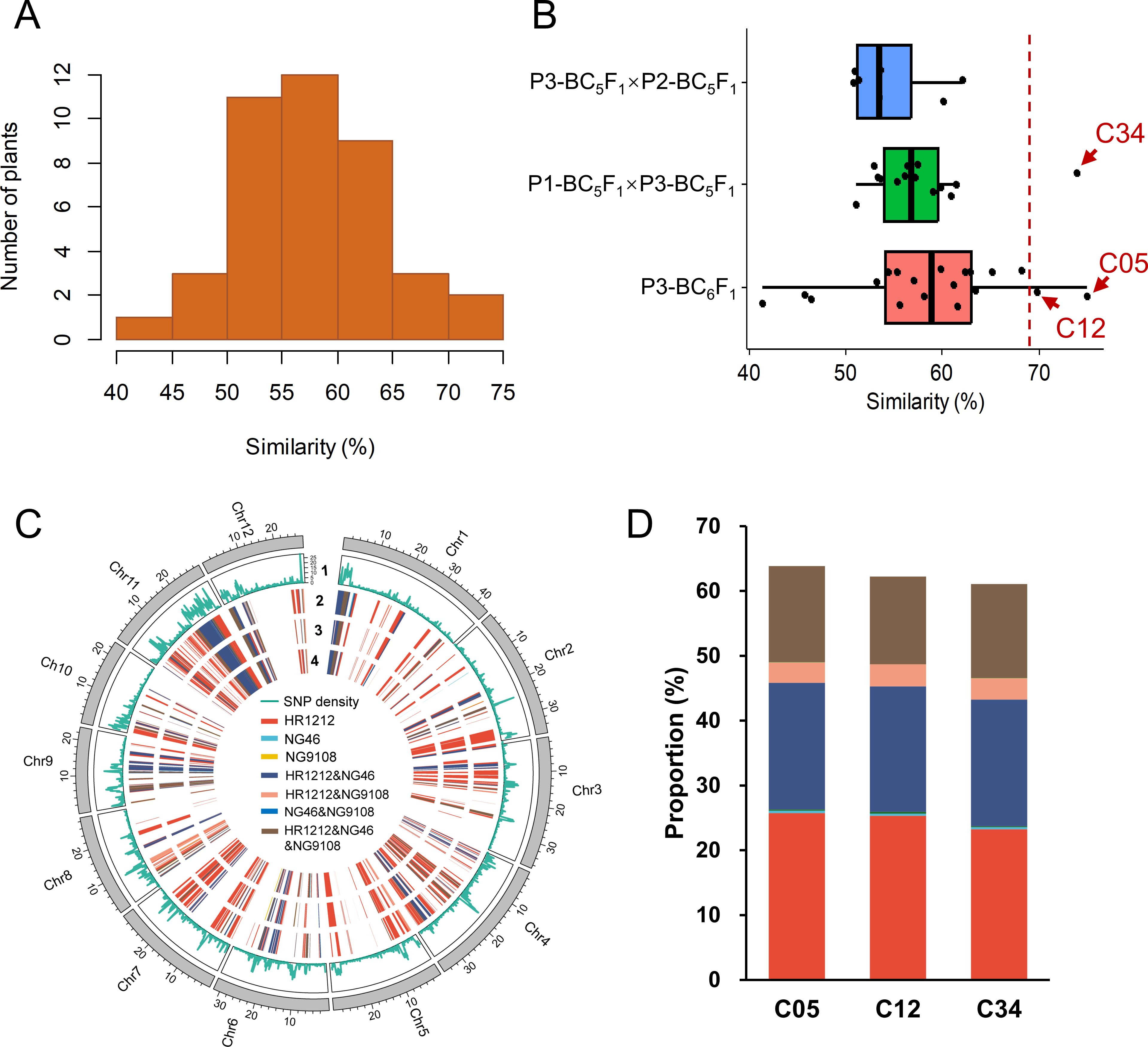
Figure 1. Analysis of genetic similarity between introgressed individuals and recurrent parent HR1212. (A) The distribution of SNP similarity between 41 individuals and recurrent parent. (B) Origin of individuals with various SNP similarities. The vertical red dotted line is located at 69% similarity. Three individuals with the highest similarity were C05 (74.91%), C34 (73.89%), and C12 (69.82%). (C) Distribution of genetic introgressed segments from parents on 12 chromosomes of three individuals. 1 indicates SNP density. From 2 to 4, the tracks represent individuals C34, C12, and C05, respectively. (D) The genomic contribution of parents to the three individuals. The same color scheme was used as (C).
To elucidate the genomic composition of these three individuals, we analyzed the whole-genome variation of their parents—NG46, NG9108, and HR1212. Despite their different pedigrees, the genomic compositions of the three individuals were quite similar (Figure 1C). About 61.8% of the genomic segments of the three individuals were derived from HR1212, of which HR1212-specific fragments accounted for about 24.8%, followed by shared genetic components from HR1212 and NG46, constituting about 19.5%. The common genetic framework from all three parents represented around 14.3% (Figure 1D).
3.3 Enhanced panicle blast resistance of introgressed lines
Two strategies were employed to select the introgressed lines: one focused on genetic background recovery following foreground selection, while the other prioritized the number of favorable alleles being pyramided. In December 2022, five targeted individuals were selected and planted into separate lines in Hainan (Figure 2). These included three individuals (C05, C12, and C34) with the closest genetic backgrounds to HR1212, as well as two individuals (C40 and C41) that pyramided heterozygotes of Pigm, Pi-ta, and badh2 (Figure 1B). For the segregating population, further screening was conducted through genotyping and agronomic performance assessment in April 2023. Consequently, ten individuals were selected and planted into ten final lines in June in Shanghai. These comprised seven monogenic lines (L01-L07) with Pigm and three lines (L08-L10) that pyramided Pigm, Pi-ta, and badh2 (Figure 2; Supplementary Figure 5).
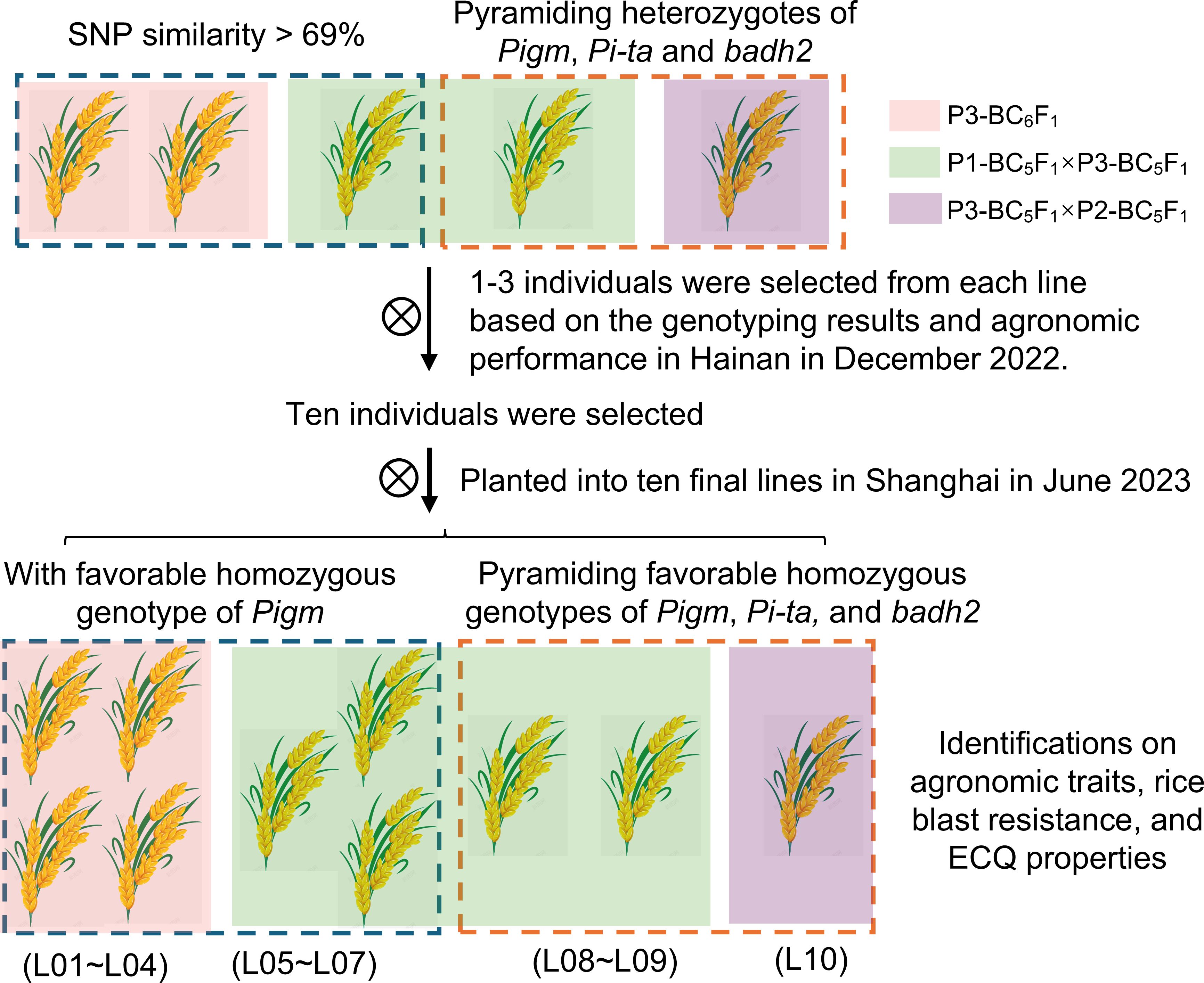
Figure 2. Flowchart of introgressed lines selection using two strategies. Three background colors of individuals or lines mean that they came from different hybrid combinations. The dashed boxes of different colors indicate the two strategies used for selecting the introgressed lines.
To evaluate the panicle blast resistance of the final introgressed lines, ten introgressed lines and five parent lines were inoculated with a mixture of M. oryzae isolates at the booting stage. The results indicated that all introgressed lines (L01-L10) displayed significantly improved panicle blast resistance compared to HR1212 (Figure 3). Notably, L03 and L04 exhibited significantly higher resistance than the donor parent NIL. Six lines (L02, L05, L06, L08, L09, L10) showed no significant differences in panicle blast resistance compared to NIL (Figure 3).
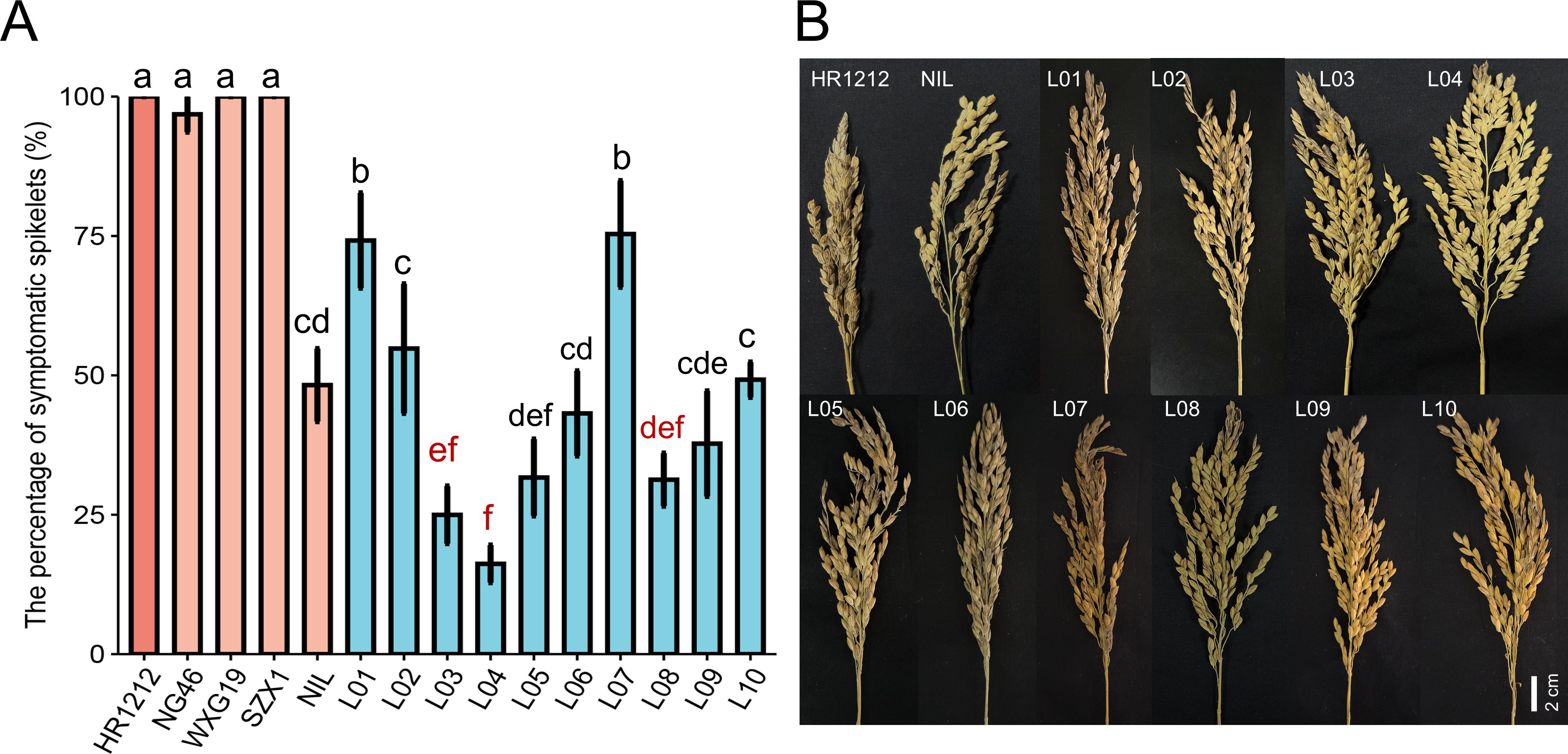
Figure 3. Panicle blast severity of the introgressed lines and the parents. (A) The incidence of introgressed lines (L01-L10) and five parents in the panicle blast evaluation. The red letters correspond to the three most resistant introgressed lines for panicle blast. Error bars, s.e. (n=10). Multiple comparison was conducted using the least significant difference (LSD) test. Different letters above the bars indicate significant differences among them (P<0.05). (B) Panicle blast symptom illustrations.
3.4 Comparison of agronomic traits between introgressed lines and HR1212
In Shanghai, heading dates for the ten introgressed lines ranged from 101 to 105 days. We focused on three lines (L03, L04, and L08) known for their strong resistance to panicle blast. Compared to the recurrent parent HR1212, both L04 and L08 exhibited a significant increase in plant height (Supplementary Figure 6). For grain yield-related traits, all three lines showed a significant increase in the number of secondary branches compared to HR1212, with L04 and L08 also demonstrating a notable increase in the number of grains per panicle. Additionally, the grain length-to-width ratio was higher in L03 and L08. The average yield per plant of the three introgressed lines exceeded that of HR1212 by more than 12% (Supplementary Figure 7).
3.5 Comparison of ECQ between introgressed lines and HR1212
To evaluate the ECQ of the introgressed lines, several key physicochemical characteristics were assessed. Compared to HR1212, L03 exhibited significantly lower amylose content, gel consistency, and chalky grain percentage, while L04 and L08 showed significantly reduced protein content (Figure 4). The RVA profile indicated that the parameters for L03 and L04 were closer to HR1212 than those for L08 (Supplementary Figure 8). The CGP for L03 was 13.86%, slightly lower than HR1212’s 15.5%, while L04 and L08 had significantly higher CGP values (Figure 4).

Figure 4. Quality traits evaluation of the introgressed lines and the parents. The red letters correspond to the three most resistant introgressed lines for rice blast. Error bars, s.e. (n=2). Multiple comparison was conducted using the least significant difference (LSD) test. Different letters above the bars indicate significant differences among them (P<0.05).
Additionally, the 2-acetyl-1-pyrroline (2-AP) content in the milled rice revealed that the pyramiding line L08 had a 2-AP concentration of 711.83 ng/g, surpassing that of its fragrance gene donor, WXG19 (558.24 ng/g) (Supplementary Figure 9).
In conclusion, among the three introgressed lines exhibiting the highest resistance to panicle blast, the ECQ of L03 and L04 was more similar to that of HR1212 based on their physicochemical properties, with L03 outperforming HR1212 due to its lower CGP. L08 had lower protein content and similar amylose content compared to HR1212. Although its starch viscosity was inferior to that of L03 and L04, it possessed a strong fragrance (Figure 4; Supplementary Figure 9).
4 Discussion
High yield, good quality, and disease resistance are primary objectives in rice breeding. Recently, excellent eating quality has become a key determinant in the breeding of japonica varieties in China, driven by rising living standards. However, japonica varieties known for their superior eating quality have historically been susceptible to rice blast disease. Previous studies have identified genetic constraints, particularly the reallocation of genetic resources during hybridization and genetic linkage—especially those on chromosome 6 (a deleterious linkage between Pigm/Piz-t and Wxmp)—that impede the simultaneous achievement of high eating quality and rice blast resistance (Xiao et al., 2020, 2021). For example, we analyzed the whole-genome data and found that HR1212, a major japonica variety in Shanghai, has favorable alleles Wxmp and ALKb for eating quality, but also carries susceptible alleles of Pigm, Pid2, and Pid3 for rice blast on chromosome 6. This highlights the necessity to overcome genetic linkage drag and introduce blast-resistant alleles.
During the backcrossing process, no negative drag was observed in this study, suggesting that unfavorable linkages, such as the Pigm-Wxmp linkage, might be broken through continuous positive selection, thereby providing a promising approach for refining elite varieties with minor drawbacks (Nihad et al., 2024; Tian et al., 2021). This strategy has been successfully applied to rice and other crops (Hu et al., 2023; Lv et al., 2014; Xiao et al., 2019). We observed that while all selected lines possessed the same superior alleles for Wx and ALK as HR1212, those selected based on genetic background recovery exhibited better eating quality compared to those selected solely for the number of pyramided alleles. This suggests that, in addition to the major genes Wx and ALK, HR1212’s genome might contain other genes influencing eating quality. Additionally, the RVA profile provides a more accurate assessment of eating quality than amylose content and protein content.
Previous studies have shown that the resistance conferred by pyramiding lines is not merely the sum of the resistance effects of individual genes (Fukuoka et al., 2015; Tabien et al., 2000). This is consistent with our findings, which indicate no significant difference in panicle blast resistance between the pyramiding lines of Pigm and Pi-ta and the Pigm monogenic lines. Several improved lines exhibited enhanced resistance to panicle blast compared to the donor NIL, possibly resulting from differences in genetic background between the improved lines and NIL (Feng et al., 2022). Some research suggests that lines with pyramided resistance alleles may experience reduced yield or other negative effects (Xiao et al., 2019). However, our study found no significant disadvantage in yield or eating quality for the pyramiding lines, likely due to the effective recovery of the genetic background achieved through backcrossing.
The reduced cost of sequencing has facilitated more efficient detection of genetic backgrounds using next-generation sequencing (NGS). Previous studies typically employed hundreds of SSR markers evenly distributed across the 12 chromosomes for background selection (Liu et al., 2016; Sabar et al., 2019; Wang et al., 2017; Xiao et al., 2019). In contrast, we used NGS to identify genome-wide variants and compared genetic backgrounds with greater accuracy and efficiency. This approach allowed us to precisely uncover the genomic contributions of both donor and recurrent parent genotypes. Additionally, our analysis revealed that the genomic components shared by HR1212 and NG46 accounted for approximately half of the genetic introgression, which aligns with the pedigree relationship of HR1212.
In this study, multiple backcrosses were conducted to ensure that the introgressed lines were as close as possible to the recurrent parent HR1212. However, a limitation is that the introgressed lines were only self-crossed twice. To develop exceptionally stable lines and further breed new varieties, additional self-crossing will be necessary.
In the genetic improvement, some major varieties may be replaced due to some shortcomings that fail to cater to contemporary breeders’ requirements. The present study offers valuable insight into enhancing the varieties of this sort. During backcrossing, those unfavorable traits were addressed using molecular markers, and a high recovery rate of the genetic background was ensured through NGS. This work demonstrates the optimization of breeding strategies, ultimately contributing to the improvement of a variety with high eating and cooking qualities. Furthermore, it points towards a promising future direction for breeding endeavors that are more precise, efficient, and sustainable.
Data availability statement
Raw genome re-sequencing reads are available under the National Center for Biotechnology Information sequence read archive with the BioProject ID: PRJNA1116583. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
JY: Data curation, Investigation, Software, Visualization, Writing – original draft, Writing – review & editing. KW: Data curation, Writing – review & editing, Formal analysis, Project administration. YW: Investigation, Writing – review & editing. ZZ: Resources, Writing – review & editing. YY: Methodology, Writing – review & editing. HY: Investigation, Methodology, Writing – review & editing. LZ: Methodology, Validation, Writing – review & editing. ZH: Formal analysis, Investigation, Writing – review & editing. ZS: Writing – review & editing. DS: Methodology, Writing – review & editing. JB: Investigation, Supervision, Writing – review & editing. LC: Conceptualization, Supervision, Writing – review & editing. SW: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Agriculture Research System of Shanghai, China (No. 202403), and the Outstanding Team Project from Shanghai Agricultural Academy of Science (No. 2022A006).
Acknowledgments
We are grateful to Prof. Yong Zhou (Yangzhou University) and Dr. Yaolong Yang (China National Rice Research Institute) for their critical reading and kindly providing help with the writing improvement of this manuscript. Thank Prof. Zhenying Shi (Shanghai Academy of Agricultural Sciences) for her contributions to the revision of this article. We thank TopEdit LLC for the linguistic editing and proofreading during the preparation of this manuscript.
Conflict of interest
Authors YW was employed by Zhongken Seed Industry Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1507827/full#supplementary-material
References
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bradbury, L. M. T., Fitzgerald, T. L., Henry, R. J., Jin, Q., Waters, D. L. (2005). The gene for fragrance in rice. Plant Biotechnol. J. 3, 363–370. doi: 10.1111/j.1467-7652.2005.00131.x
Bryan, G. T., Wu, K.-S., Farrall, L., Jia, Y., Hershey, H. P., McAdams, S. A., et al. (2000). A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell 12, 2033–2045. doi: 10.1105/tpc.12.11.2033
Buttery, R. G., Ling, L. C., Juliano, B. O. (1982). 2-Acetyl-1-pyrroline: an important aroma component of cooked rice. Chem. Industry (London) 12, 958–959.
Cereals & Grains Association. (2000). Approved Methods of the American Association of Cereal Chemists (10th Ed.). St. Paul, MN, U.S.A. doi: 10.1094/AACCIntMethod-61-02.01
Deng, Y. W., Zhai, K. R., Xie, Z., Yang, D. Y., Zhu, X. D., Liu, J. Z., et al. (2017). Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 355, 962–965. doi: 10.1126/science.aai8898
Deng, Y. W., Zhu, X. D., Shen, Y., He, Z. H. (2006). Genetic characterization and fine mapping of the blast resistance locus Pigm(t) tightly linked to Pi2 and Pi9 in a broad-spectrum resistant Chinese variety. Theor. Appl. Genet. 113, 705–713. doi: 10.1007/s00122-006-0338-7
Feng, Z. M., Li, M. Y., Xu, Z. W., Gao, P., Wu, Y. Y., Wu, K. T., et al. (2022). Development of rice variety with durable and broad-spectrum resistance to blast disease through marker-assisted introduction of Pigm Gene. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.937767
Fukuoka, S., Saka, N., Mizukami, Y., Koga, H., Yamanouchi, U., Yoshioka, Y., et al. (2015). Gene pyramiding enhances durable blast disease resistance in rice. Sci. Rep. 5, 7773. doi: 10.1038/srep07773
Gao, P., Li, M. Y., Wang, X. Q., Xu, Z. W., Wu, K. T., Sun, Q. Y., et al. (2023). Identification of elite R-gene combinations against blast disease in Geng rice varieties. Int. J. Mol. Sci. 24, 3984. doi: 10.3390/ijms24043984
Gu, Z., Gu, L., Eils, R., Schlesner, M., Brors, B. (2014). Circlize implements and enhances circular visualization in R. Bioinformatics 30, 2811–2812. doi: 10.1093/bioinformatics/btu393
Hu, W. J., Fu, L. P., Gao, D. R., Li, D. S., Liao, S., Lu, C. B. (2023). Marker-assisted selection to pyramid Fusarium head blight resistance loci Fhb1 and Fhb2 in the high-quality soft wheat cultivar Yangmai 15. J. Integr. Agr. 22, 360–370. doi: 10.1016/j.jia.2022.08.057
Huang, H., Lu, Y., Zhu, Y., Sun, X., Liu, J. (2023). Co-evolutionary characteristics and development of the dominant markers of the resistance genes Pi-ta and Ptr. Mol. Plant Breed. 21, 7399–7407. doi: 10.13271/j.mpb.021.007399
Hwang, B. K., Koh, Y. J., Chung, H. S. (1987). Effects of adult-plant resistance on blast severity and yield of rice. Plant Dis. 71, 1035–1038. doi: 10.1094/PD-71-1035
Jia, Y. L., Wang, Z. H., Fjellstrom, R. G., Moldenhauer, K. A. K., Azam, M. A., Correll, J., et al. (2004). Rice Pi-ta gene confers resistance to the major pathotypes of the rice blast fungus in the United States. Phytopathology 94, 296–301. doi: 10.1094/PHYTO.2004.94.3.296
Jia, Y. L., Wang, Z. H., Singh, P. (2002). Development of dominant rice blast Pi-ta resistance gene markers. Crop Sci. 42, 2145–2149. doi: 10.2135/cropsci2002.2145
Khan, G. H., Shikari, A. B., Vaishnavi, R., Najeeb, S., Padder, B. A., Bhat, Z. A., et al. (2018). Marker-assisted introgression of three dominant blast resistance genes into an aromatic rice cultivar Mushk Budji. Sci. Rep. 8, 4091. doi: 10.1038/s41598-018-22246-4
Li, J. (2022). Molecular marker-assisted selection of rice recovery lines resistant to rice blast and brown planthopper. Jiangxi Agriculture University, Nanchang (Jiangxi.
Li, W. T., Chern, M. S., Yin, J. J., Wang, J., Chen, X. W. (2019). Recent advances in broad-spectrum resistance to the rice blast disease. Curr. Opin. Plant Biol. 50, 114–120. doi: 10.1016/j.pbi.2019.03.015
Li, W., Deng, Y. W., Ning, Y. S., He, Z. H., Wang, G. L. (2020). Exploiting broad-spectrum disease resistance in crops: from molecular dissection to breeding. Annu. Rev. Plant Biol. 71, 575–603. doi: 10.1146/annurev-arplant-010720-022215
Li, H., Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Liang, Y., Yan, B. Y., Peng, Y. L., Ji, Z. J., Zeng, Y. X., Wu, H. L., et al. (2017). Molecular screening of blast resistance genes in rice germplasms resistant to Magnaporthe oryzae. Rice Sci. 24, 41–47. doi: 10.1016/j.rsci.2016.07.004
Liu, Y. L., Chen, L. M., Liu, Y. Q., Dai, H. M., He, J., Kang, H. Y., et al. (2016). Marker assisted pyramiding of two brown planthopper resistance genes, Bph3 and Bph27 (t), into elite rice cultivars. Rice 9, 27. doi: 10.1186/s12284-016-0096-3
Lv, C., Song, Y. X., Gao, L. F., Yao, Q., Zhou, R. H., Xu, R. G., et al. (2014). Integration of QTL detection and marker assisted selection for improving resistance to Fusarium head blight and important agronomic traits in wheat. Crop J. 2, 70–78. doi: 10.1016/j.cj.2013.10.004
Ma, Z. B., Wang, L. L., Zhang, L. K., Gu, S., Wang, H., Sui, G. M., et al. (2023). A molecular identification and resistance evaluation of the blast resistance genes in japonica rice in Northern China. Agron. Basel 13, 2662. doi: 10.3390/agronomy13102662
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110
Nihad, S. A. I., Hasan, M. A., Anik, T. R., Rashid, M. M., Khan, M. A. I., Islam, M. R., et al. (2024). Pyramiding of blast and bacterial blight resistance genes in premium quality rice variety, BRRI dhan63 through marker-assisted breeding approach. Euphytica 220, 13. doi: 10.1007/s10681-023-03255-5
Pan, Y., Huang, D., Wang, C., Li, H., Zhou, D., Wang, Z., et al. (2021). Research advances of haplotype variation at Badh2 gene and 2-acetyl-1-pyrroline biosynthetic pathway in aromatic rice. Guangdong Agric. Sci. 48, 9–16. doi: 10.16768/j.issn.1004-874X.2021.07.002
Price, M. N., Dehal, P. S., Arkin, A. P. (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PloS One 5, e9490. doi: 10.1371/journal.pone.0009490
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A. R., Bender, D., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. doi: 10.1086/519795
Sabar, M., Akhter, M., Bibi, T., Riaz, A., Haider, Z., Khan, A. R., et al. (2019). Basmati rice lines development carrying multiple bacterial blight resistance genes pyramided using the marker-assisted backcross breeding approach. Mol. Breed. 39, 155. doi: 10.1007/s11032-019-1047-7
Song, F., Zhou, R., Muhammad, A. H., Gan, Q., Lin, C., Ten, B., et al. (2024). Analysis of rice blast resistance dominant genes and their combinations in japonica rice. Physiol. Plantarum 176, e14156. doi: 10.1111/ppl.14156
Tabien, R. E., Li, Z., Paterson, A. H., Marchetti, M. A., Stansel, J. W., Pinson, S. R. M., et al. (2000). Mapping of four major rice blast resistance genes from ‘Lemont’ and ‘Teqing’ and evaluation of their combinatorial effect for field resistance. Theor. Appl. Genet. 101, 1215–1225. doi: 10.1007/s001220051600
Talbot, N. J. (2003). On the trail of a cereal killer: Exploring the biology of Magnaporthe grisea. Annu. Rev. Microbiol. 57, 177–202. doi: 10.1146/annurev.micro.57.030502.090957
Terensan, S., Fernando, H. N. S., Silva, J. N., Perera, S., Kottearachchi, N. S., Weerasena, O. (2021). In silico molecular and morphological analysis of rice blast resistant gene Pi-ta in Sri Lankan rice germplasm. J. Genet. Eng. Biotechnol. 19, 163. doi: 10.1186/s43141-021-00239-7
Tian, D. G., Chen, Z. Q., Lin, Y., Chen, Z. J., Luo, J. M., Ji, P. S., et al. (2021). Two novel gene-specific markers at the Pik locus facilitate the application of rice blast resistant alleles in breeding. J. Integr. Agr. 20, 1554–1562. doi: 10.1016/S2095-3119(20)63272-5
Wang, X., Du, H., Chen, X., Li, M., Wang, J., Xu, Z., et al. (2020). Analysis of blast resistance genes and neck blast resistance of japonica rice varieties/lines recently developed in Jiangsu province. Chin. J. Rice Sci. 34, 413–424. doi: 10.16819/j.1001-7216.2020.0208
Wang, Z., Jia, Y., Rutger, J. N., Xia, Y. (2007). Rapid survey for presence of a blast resistance gene Pi-ta in rice cultivars using the dominant DNA markers derived from portions of the Pi-ta gene. Plant Breed. 126, 36–42. doi: 10.1111/j.1439-0523.2007.01304.x
Wang, Y., Jiang, W. H., Liu, H. M., Zeng, Y., Du, B., Zhu, L. L., et al. (2017). Marker assisted pyramiding of Bph6 and Bph9 into elite restorer line 93-11 and development of functional marker for Bph9. Rice 10, 51. doi: 10.1186/s12284-017-0194-x
Wang, Y., Sun, Q., Du, H., Xu, Z., Wu, K., Yin, L., et al. (2023). Improvement of the resistance of Nanjing 9108 to blast and sheath blight by pyramiding resistance gene Pigm and quantitative trait genes qSB-9TQ and qSB-11HJX. Chin. J. Rice Sci. 37, 125–132. doi: 10.16819/j.1001-7216.2023.220413
Wang, J., Yang, J., Chen, Z. D., Zhong, W. G. (2008). Development and application of fragrance gene markers in rice. Mol. Plant Breed. 6, 1209–1212. doi: 10.3969/j.issn.1672-416X.2008.06.030
Wing, R. A., Purugganan, M. D., Zhang, Q. (2018). The rice genome revolution: from an ancient grain to Green Super Rice. Nat. Rev. Genet. 19, 505–517. doi: 10.1038/s41576-018-0024-z
Wu, Y. Y., Chen, Y., Pan, C. H., Xiao, N., Yu, L., Li, Y. H., et al. (2017). Development and evaluation of near-isogenic lines with different blast resistance alleles at the Piz Locus in japonica rice from the lower region of the Yangtze River, China. Plant Dis. 101, 1283–1291. doi: 10.1094/PDIS-12-16-1855-RE
Xiao, N., Pan, C. H., Li, Y. H., Wu, Y. Y., Cai, Y., Lu, Y., et al. (2021). Genomic insight into balancing high yield, good quality, and blast resistance of japonica rice. Genome Biol. 22, 283–304. doi: 10.1186/s13059-021-02488-8
Xiao, N., Wu, Y. Y., Li, A. H. (2020). Strategy for use of rice blast resistance genes in rice molecular breeding. Rice Sci. 27, 263–277. doi: 10.1016/j.rsci.2020.05.003
Xiao, W. M., Yang, Q. Y., Huang, M., Guo, T., Liu, Y. Z., Wang, J. F., et al. (2019). Improvement of rice blast resistance by developing monogenic lines, two-gene pyramids and three-gene pyramid through MAS. Rice 12, 78. doi: 10.1186/s12284-019-0336-4
Xu, Y., Wang, F. Q., Chen, Z. H., Wang, J., Li, W. Q., Fan, F. J., et al. (2020). Intron-targeted gene insertion in rice using CRISPR/Cas9: A case study of the Pi-ta gene. Crop J. 8, 424–431. doi: 10.1016/j.cj.2019.03.006
Ye, J. H., Zhang, M. C., Yuan, X. P., Hu, D. X., Zhang, Y. Y., Xu, S. L., et al. (2022). Genomic insight into genetic changes and shaping of major inbred rice cultivars in China. New Phytol. 236, 2311–2326. doi: 10.1111/nph.18500
Yu, H. Y., Wei, X. H. (2010). Principle and method of testing new rice varieties (Hangzhou: Zhejiang Science and Technology Press).
Keywords: pyramiding, panicle blast resistance, fragrance, eating and cooking quality, re-sequencing, rice
Citation: Ye J, Wang K, Wang Y, Zhao Z, Yan Y, Yang H, Zhang L, Hu Z, Shi Z, Sun D, Bai J, Cao L and Wu S (2025) Improving panicle blast resistance and fragrance in a high-quality japonica rice variety through breeding. Front. Plant Sci. 15:1507827. doi: 10.3389/fpls.2024.1507827
Received: 08 October 2024; Accepted: 06 December 2024;
Published: 13 January 2025.
Edited by:
Elena Corredoira, Spanish National Research Council (CSIC), SpainReviewed by:
Sathish Kumar Ponniah, University of Arkansas at Pine Bluff, United StatesBiaolin Hu, Jiangxi Academy of Agricultural Sciences, China
Copyright © 2025 Ye, Wang, Wang, Zhao, Yan, Yang, Zhang, Hu, Shi, Sun, Bai, Cao and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shujun Wu, d3VzaHVqdW5Ac2Fhcy5zaC5jbg==
†These authors have contributed equally to this work
 Junhua Ye
Junhua Ye Kai Wang
Kai Wang Yi Wang3
Yi Wang3 Lixia Zhang
Lixia Zhang Zejun Hu
Zejun Hu Shujun Wu
Shujun Wu