- Zhejiang Provincial Key Laboratory of Germplasm Innovation and Utilization for Garden Plants, School of Landscape and Architecture, Zhejiang A&F University, Hangzhou, Zhejiang, China
Flower appearance stands as a key characteristic of flowering plants and is closely linked to their ornamental value. Phytohormone Gibberellin (GA), essential for plant growth and development are widely reported for expansion in flower. DELLA proteins are known to negatively regulate GA signaling and influences plant growth and development through the regulation of cell expansion. However, the specific biological function of DELLA proteins in the woody plant Osmanthus fragrans remains unclear. In this study, O. fragrans ‘Sijigui’ was utilized as the experimental material, and OfRGA was isolated using the PCR method. OfRGA is expressed in various tissues and is localized in the nucleus. A negative association was observed between OfRGA expression and petal size across four different Osmanthus fragrans cultivars. Transformation experiments in tobacco revealed that transgenic plants overexpressing OfRGA exhibited increased plant height, greater node spacing, shorter leaf length, and wider leaves during the vegetative phase. Notably, the flower organs of transgenic tobacco plants displayed noticeable alterations, including reduced petal size, shorter corolla tubes, pedicels, male and female stamens, and lighter petal color. Furthermore, a decrease in the length and area of petal and corolla tube cells was observed as well. DEGs were found in RNA-seq studies of OfRGA transgenic plants. Subsequent investigation revealed a considerable quantity of down-regulated genes were associated with cell wall synthesis genes and expansion genes, such as CesA1, XEH, and EXPB1, as well as genes related to anthocyanin biosynthesis. Overall, our findings suggest that OfRGA undermines tobacco petal size by influencing cell expansion. The present study offers a fundamental comprehension of the role of DELLA protein in the organ development in Osmanthus fragrans.
1 Introduction
Floral organ development has profound significance in the plant life cycle, which is directly related to a plant’s ability to reproduce and the ecological adaptability of plants (Chahtane et al., 2023; Mā, 2005). This process is both finely regulated by internal physiological mechanisms and also greatly influenced by the external environment (Coen and Meyerowitz, 1991; Yue et al., 2023). The phytohormone gibberellin regulates the development of floral organs, according to rencent studies (Li et al., 2017, 2021).
Tetracyclic diterpenoids known as gibberellins are crucial for various critical processes in plant development, including fruit ripening, leaf expansion, floral organ development, stem elongation, seed germination, and flowering induction (Fleet and Sun, 2005). Previous research has identified that in mutants of Arabidopsis thaliana with deficiencies in GA synthesis, such as ga1-1, ga1-3, and ga1-6, the petals and stamens become damaged, and anthers become stunted in the absence of mature pollen, which causes male sterility in severe cases. However, exogenous GA can return the floral developmental phenotypes of ga1-3 mutants to their normal state (Gotô and Pharis, 1999; Koornneef and van der Veen, 1980; Wilson et al., 1992). Likewise, in tomato, GA-deficient mutant ga-2 has impaired flower development due to the lack of corolla and stamen elongation and the style deformation or elongation, leading to plant sterility. But the completion of bud development and fruit set was achieved by applying GA1 to developing inflorescences (Joan and Jan, 1988). The above results show that GA can control the flower development of plants. DELLA proteins are numbers of the plant-specific GRAS family and identified as negative regulators of GA. They have an inhibitory influence on plant development and growth. DELLA proteins are able to bind to GID1 to form a GA-GID1-DELLA trimer, which is labeled by the SCF polymers and subsequently eliminated by the ubiquitin 26S proteasome. Arabidopsis thaliana contains five DELLA proteins, identified as gibberellin insensitive (GAI), repressors of GA1-3 (RGA), RGA-Like1 (RGL1), RGL2, and RGL3 (Tyler et al., 2004). Both overlapping and specific functions are present in these five proteins. Among them, the growth of stamens, petals, and anthers in Arabidopsis is co-regulated by RGA, RGL1, and RGL2 (Cheng et al., 2004). Further studies revealed that GA degrades DELLA primarily through the GA-GID1-DELLA pathway, thereby relieving the inhibitory effect on plant growth and development. In Arabidopsis thaliana, the inhibition of DELLA proteins RGL1, RGL2, and RGA by GA leads to the expression up-regulation of the floral homologous genes AP3, PI, and AG, which promote the normal development of floral organs (Cheng et al., 2004; Yu et al., 2004). CsGAIP, a homologue of DELLA protein in cucumber, inhibits the production of stamens in male cucumber flowers by downregulating the transcription of class B MADS-box genes PI and AP3 (Zhang et al., 2014). Furthermore, the GA-DELLA-OsMS188 module controls rice anther development and integrates gibberellin hormone signaling into the anther genetic program (Jin et al., 2021).
Early studies have also found that biologically active gibberellins (GA) regulate plant floral organ development by modulating cell proliferation and expansion. In Gerbera hybrida, GA promotes petal elongation by enhancing cell expansion (Zhang et al., 2012). The phloem and xylem tissues in the peduncle of the “Wonhwang” oriental pear also expanded rapidly under the stimulation of exogenous GA (Park and Park, 2017). DELLA proteins, as key factors in the GA pathway, also play a role in regulating plant cell proliferation and expansion. DELLAs inhibited leaf growth through a dual mechanism, first by changing the rate of cell division during the proliferative phase of leaf development and then by effecting the rate of cell expansion while in the expansion phase (Achard et al., 2009). For example, cell proliferation in the meristematic zone might be inhibited by MpDELLA, which could restrict the growth of the liverwort Marchantia polymorpha (Hernández-García et al., 2021). In roses, RhGAI1, a target of RhEIN3-3, being silenced in rose petals has promoted cell expansion, which has resulted in enlarged petal size (Luo et al., 2013). In addition, DELLA proteins are not only solely responsible for regulating flower organ development but also have a certain effect on flower color. Research has found that TAP-GAId17 overexpression led to a significant rise in anthocyanins in transgenic Arabidopsis (Zhang et al., 2017). MdRGL2a can promote anthocyanin accumulation under the protection of the protein kinase MdCIPK20 in apple (An et al., 2023). The findings suggest that the DELLAs serve as positive regulators in the anthocyanin accumulation. This was investigated further and revealed that RGA interacts with JAZ and MYBL2 in the MYB/bHLH/WD40 complex to release the bHLH/MYB subunit, which in turn forms an active MBW complex and hence anthocyanin biosynthesis (Xie et al., 2016). Currently, there are few studies on the regulatory mechanisms of DELLA proteins on the development of floral organs and flower color, which need to be further explored.
In ‘Sijigui’, we identified a gene encoding a DELLA protein (OfRGA), cloned the OfRGA gene by RT-PCR, and examined the patterns of OfRGA expression in various tissues and in various ‘Sijigui’ cultivars. We further heterologously transformed OfRGA into tobacco and investigated the gene function of OfRGA. The cell size of transgenic petals was observed by temporary sectioning. To gain a more profound understanding of the regulation mechanism of OfRGA on the development of floral organs, transcriptome sequencing was carried out. These investigations advance our knowledge of the molecular pathways through which OfRGA controls the development of floral organs.
2 Materials and methods
2.1 Plant materials and growing conditions
Four different cultivars of Osmanthus fragrans (‘Tianxiang Taige’, ‘Rixiang Gui’, ‘Sijigui’, and ‘Tiannu Sanhua’) plants were produced in a natural environment and provided through the resource garden at Zhejiang A&F University (Wang et al., 2022). The roots, stems, leaves, flower buds, leaf buds, petioles, stem segments, and other tissues of ‘Sijigui’ were gathered and kept at -80°C for subsequent experiments. The Zhejiang Provincial Key Laboratory of Garden Plant Germplasm Innovation and Utilization preserves tobacco of the wild type. Plants of the transgenic varieties and the wild type were cultivated at 23°C in a greenhouse with a 12 hours of light followed by 12 hours of darkness.
2.2 Gene cloning and bioinformatics analysis
There is a DELLA protein in Osmanthus fragrans, which is called OfRGA. The ORF sequence of OfRGA was obtained from transcriptome data. Gene-specific primers (listed in Supplementary Table S1) were used to amplify OfRGA. First-strand cDNA synthesized from O. fragrans ‘Sijigui’ was used as the PCR template. The amplification process consisted of an initial denaturation at 95°C for 3 minutes, followed by 32 cycles of denaturation at 95°C for 15 seconds, annealing at 60°C for 15 seconds, and extension at 72°C for 2 minutes. A final extension step at 72°C for 5 minutes completed the process.
The PCR product that had been gel-purified was cloned into the pMD18-T vector (TaKaRa, Dalian, China) and validated by forwarding it to the company for sequencing (Zhejiang Youkang Biotechnology Co. Ltd., Hangzhou, China).
Gene sequence homology was further analyzed using BLAST in the NCBI GenBank database (https://blast.ncbi.nlm.nih.gov). The phylogenetic tree was made with the MEGA 7.0 software. With the DNAMAN program, multiple sequence alignments and analyses were carried out (https://www.lynnon.com/dnaman.html) (Kumar et al., 2018).
2.3 Gene expression analysis by quantitative real-time PCR
The roots, flower buds, stems, leaf buds, leaves, petioles, and shoot apex of O. fragrans ‘Sijigui’ were collected as previously described. Total RNA was individually isolated from each tissue through the RNAprep Pure Plant Kit (Vazyme, Nanjing, China). Next, HiScript® Reverse Transcriptase was utilized to generate the cDNA of the first strand (Vazyme, Nanjing, China) in accordance with the instructions provided by the manufacturer. Supplementary Table S1 lists the specific primers that were created. RT-qPCR was carried out with a 10 uL PCR mixture that included 5 μL of SYBR Premix Ex Taq, 2 μL of cDNA, 0.4 μL each of PCR forward primer and reverse primer, and 2.2 uL of double-distilled (ddH2O). Then the internal reference gene and the target gene were added independently to a 96-well plate with three copies of each sample. The relative gene expression was computed using the 2-Ct method (Livak and Schmittgen, 2001).
2.4 Subcellular localization
The ORF of OfRGA lacking the stop codon was ligated into the pORER4-35S-GFP vector. And using the freeze-thaw method, the control, nuclear marker, and OfRGA-GFP expression plasmids were transferred into the Agrobacterium rhizogenes strain GV3101, where Agrobacterium expressing the plasmid and the control plasmid were mixed with Agrobacterium with the nuclear marker (35S::D53-RFP), respectively, and co-injected into the back of the tobacco leaves. The GFP fluorescence of tobacco leaves was detected after infiltration for 2 days to identify the protein’s position in the cells using a confocal microscope (Olympus Corporation, Tokyo, Japan) (Liu et al., 2023).
2.5 Tobacco transformation
Similarly, the ORF of OfRGA had been inserted into the vector pORER4 behind the 35S promoter, and the resulting construct 35S:OfRGA was transferred into the Agrobacterium tumefaciens strain GV3101 and then transformed into tobacco by the leaf disk method (Sparkes et al., 2006). Leaves were cultured in MS medium for 3 days. They were then moved to MS Medium, which included the following: 250 mg/L of carbenicillin disodium (Carb) + 2.25 mg/L of 6-benzyladenine (6-BA) + 0.3 mg/L of 1-naphthylacetic acid (NAA) + 100 mg/L of kana (a pH of 5.8). After being separated from the callus, the regenerated shoots were placed on the rooting media (MS + 100 mg/L Kana + 250 mg/L Carb) (a pH of 5.8). After rooting, the seedlings were moved to soil pots for cultivation. In order to observe phenotypic traits and compile data, transgenic plants that have been identified and WT control plants were grown in identical environments.
2.6 Microscopy examination and cell measurements
To observe the upper epidermal cells of the petals, the petals were collected and preserved for 24 hours at 4°C using a formaldehyde-acetic acid-alcohol (FAA) fixative (5% acetic acid, 5% formaldehyde, and 70% alcohol), followed by an alcohol rinse. Petal cells were then photographed using a confocal microscope (Olympus Corporation, Tokyo, Japan). The upper portion of the tobacco corolla tube was excised and transversely sectioned into several thin slices using a clean double-sided blade. These slices were then observed under the previously described confocal microscope. The area of the AbsE cells was measured using the ImageJ software (Rueden et al., 2017).
2.7 RNA sequencing and differential expression gene analysis
Using cDNA created from RNA extracted from tobacco flower bracts, differentially expressed genes (DEGs) between WT and transgenic lines of OfGRA were found using an Illumina sequencing platform (Beijing Bemac Biotechnology Co., Ltd., Beijing, China). Next, the Osmanthus fragrans reference geneome was mapped to the clean reads, and DEGs were examined for GO and KEGG enrichment. On the NCBI sequence read archive (SRA), the clean reads were submitted. (http://www.ncbi.nlm.nih.gov/sra/), accession number: PRJNA1122233. (https://www.ncbi.nlm.nih.gov/sra/PRJNA1122233).
2.8 Statistical analysis
Statistical significance was assessed using *p < 0.05, **p < 0.01, and ***p < 0.001 after data were processed with SPSS statistics and Excel software. There were three biological duplicates for each experiment.
3 Results
3.1 Sequence alignment and phylogenetic analysis
The ORF of the OfRGA gene is 1776 bp in length and encodes 592 amino acids. Multiple sequence comparisons using DNAMAN software showed that all DELLA protein sequences have DELLA and TVHYNP motifs at the n-terminus and GRAS structural domains at the C-terminus, suggesting high consistency among DELLA proteins (Figure 1A). To further explore the evolutionary connection between OfRGA and other DELLA protein members, we compared 16 protein sequences from eight representative plant species and constructed a phylogenetic tree. According to the phylogenetic tree, OfRGA and the evolutionary history of OeDELLA1 in Olea europaea are tightly linked (Figure 1B).
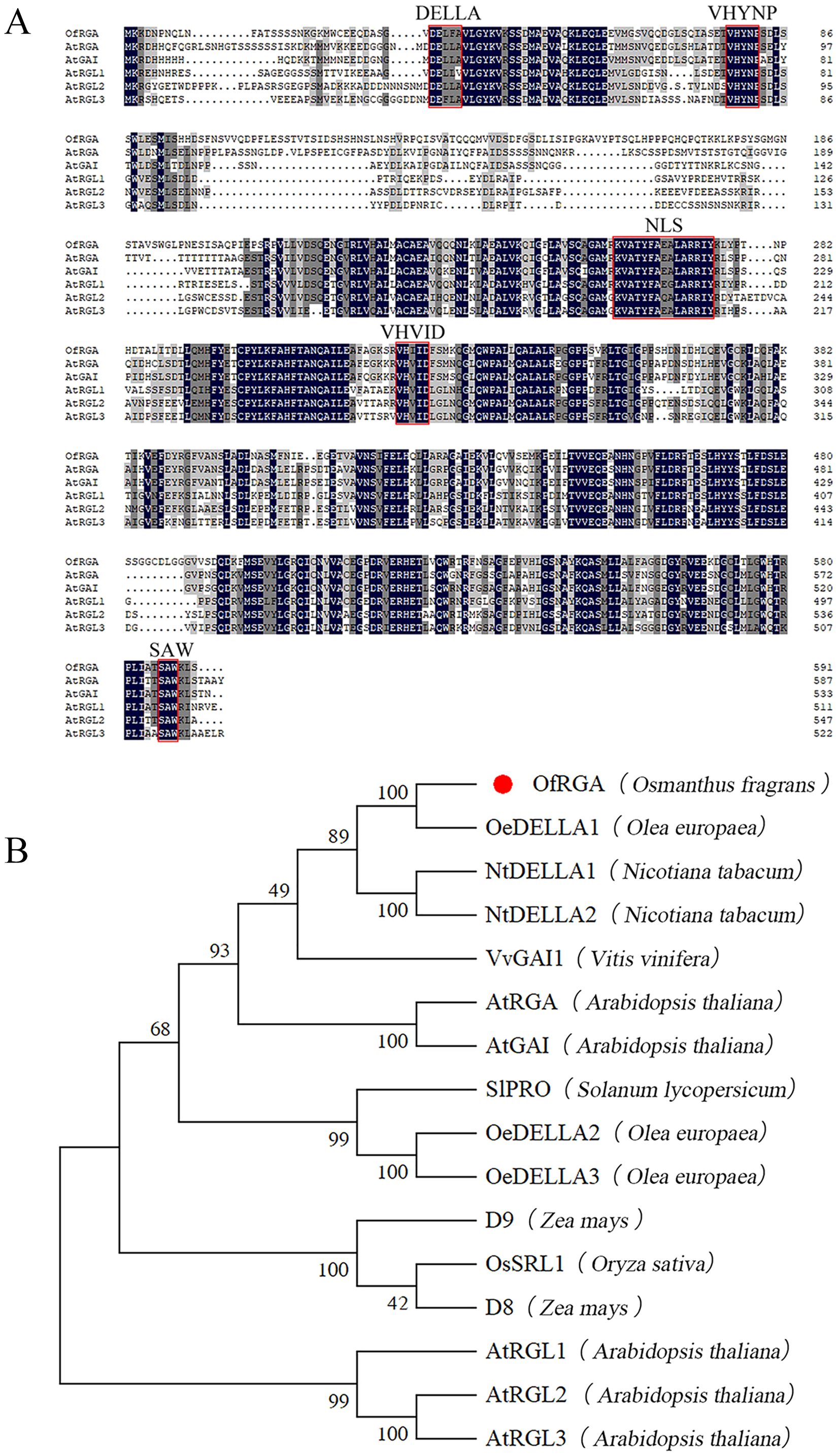
Figure 1. Sequence analysis of OfRGA. (A) Sequence alignment comparing AtDELLAs to OfRGA. DNAMAN was used for the alignment of amino acid sequences. Red lines surround the conserved domains of the DELLA protein. (B) The evolutionary connection between homologous genes and OfRGA. The symbol for OfRGA is a red circle. The following are the protein sequences: OfRGA (Osmanthus fragrans), OeDELLA1 (Olea europaea XP_022855476.1), OeDELLA2 (Olea europaea XP_022844159.1), OeDELLA3 (Olea europaea XP_022884592.1), NtDELLA1 (Nicotiana tabacum XP_016452673.1), NtDELLA2 (Nicotiana tabacum XP_016441385.1), VvGAI1 (Vitis vinifera NP_001384785.1), AtRGA (Arabidopsis thaliala NP_178266.1), AtGAI (Arabidopsis thaliala NP_172945.1), AtRGL1 (Arabidopsis thaliala NP_176809.1), AtRGL2 (Arabidopsis thaliala NP_186995.1), AtRGL3 (Arabidopsis thaliala NP_197251.1), OsSRL1 (Oryza sativa NP_001405437.1), ZmD8 (Zea mays NP_001354393.1), ZmD9 (Zea mays NP_001296780.1), SlPRO (Solaum lycopersicum NP_001234365.1).
3.2 Expression and subcellular localization of OfRGA
In order to further elucidate the role of OfRGA, qRT-PCR is used to detect the expression of OfRGA in the stems, roots, flower buds, petioles, leaves, leaf buds, and shoot apex of O. fragrans ‘Sijigui’ (Figure 2A). The results indicate that OfRGA has the maximum degree of expression in the shoot apex. According to subcellular localization experiments, the control GFP signal was dispersed throughout the cell, whereas OfRGA-GFP showed strong fluorescence signals in the nucleus. The finding demonstrated that the nucleus was where the OfRGA fusion protein was located (Figure 2B).
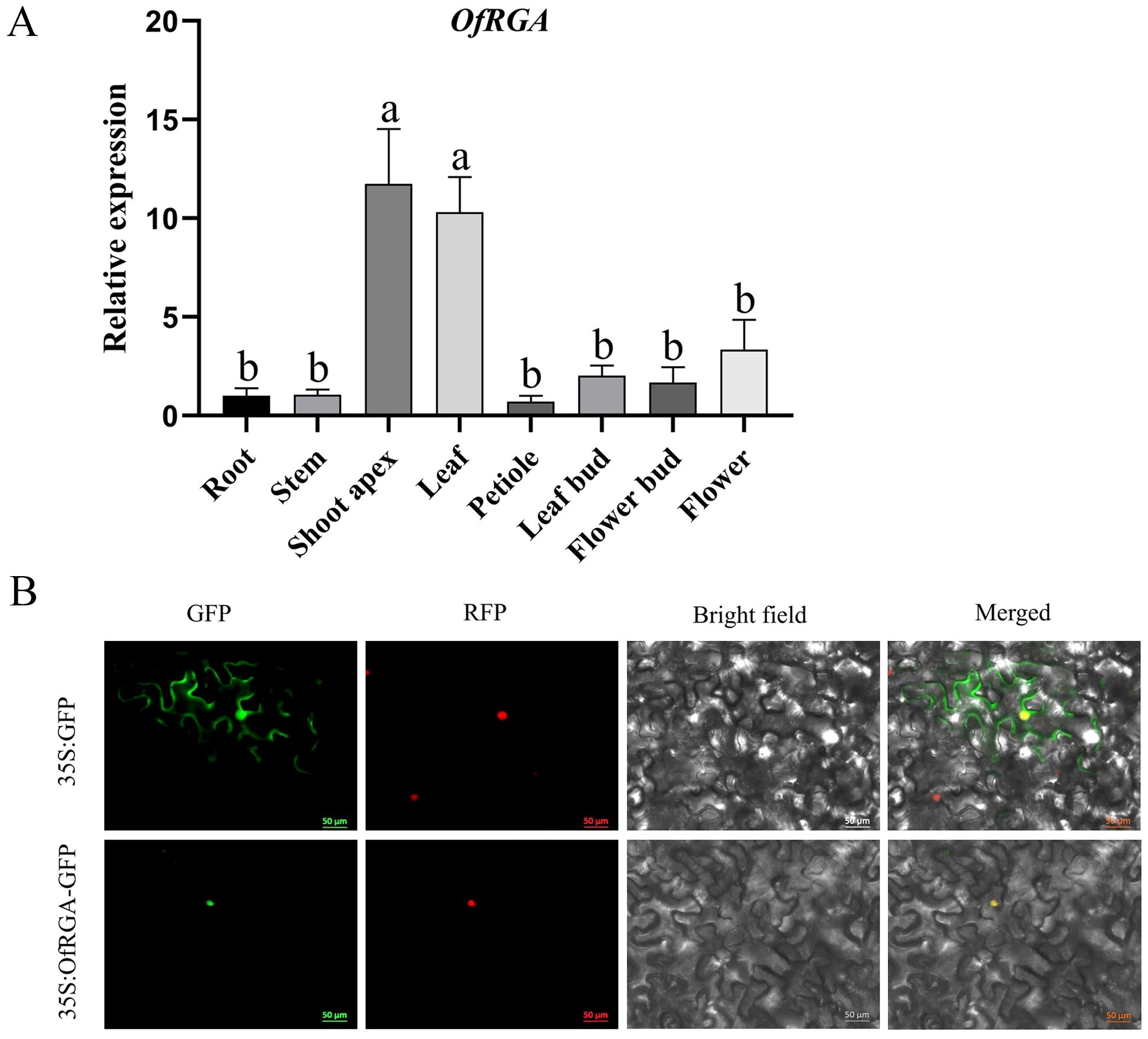
Figure 2. Expression and Subcellular Localization of OfRGA. (A) Profile of expression for OfRGA in Osmanthus fragrans. Quantitative real-time PCR examination of OfRGA in the leaf bud, petiole, stem, shoot apex, flower bud, root, and leaf of Osmanthus fragrans. Data are the means ± SD, n = 3, P < 0.05. (B) Location of OfRGA subcellularly in tobacco leaves. The nuclear marker signal is red, whereas the green fluorescent protein is green. Scale bars = 50 μm. a,b means α=0.05.
3.3 Analysis of OfRGA expression in the petals of different varieties of ‘Sijigui’
Under the same growing environment, we observed a significant difference in petal size among four different cultivars of ‘Sijigui’. In order to understand this difference, four varieties, ‘Tianxiang Taige’, ‘Rixiang Gui’, ‘Putong Sijigui’, and ‘Tiannu Sanhua’, were selected for observation and measurement (Figures 3A, C). The results showed that the ‘Tianxing Taige’ variety had the largest petals, followed by ‘Rixiang Gui’, while ‘Putong Sijigui’ and ‘Tiannu Sanhu’ had approximately the same petal size. In order to investigate whether petal size is associated with the OfRGA gene, we quantitatively analyzed the petals of four different varieties of ‘Sijigui’ (Figure 3B). The analysis showed varying levels of OfRGA gene expression among the petals of the four different varieties, with a negative correlation between OfRGA expression and petal size. These findings suggest that OfRGA may play a role in limiting cell proliferation, thereby contributing to reduced petal size.
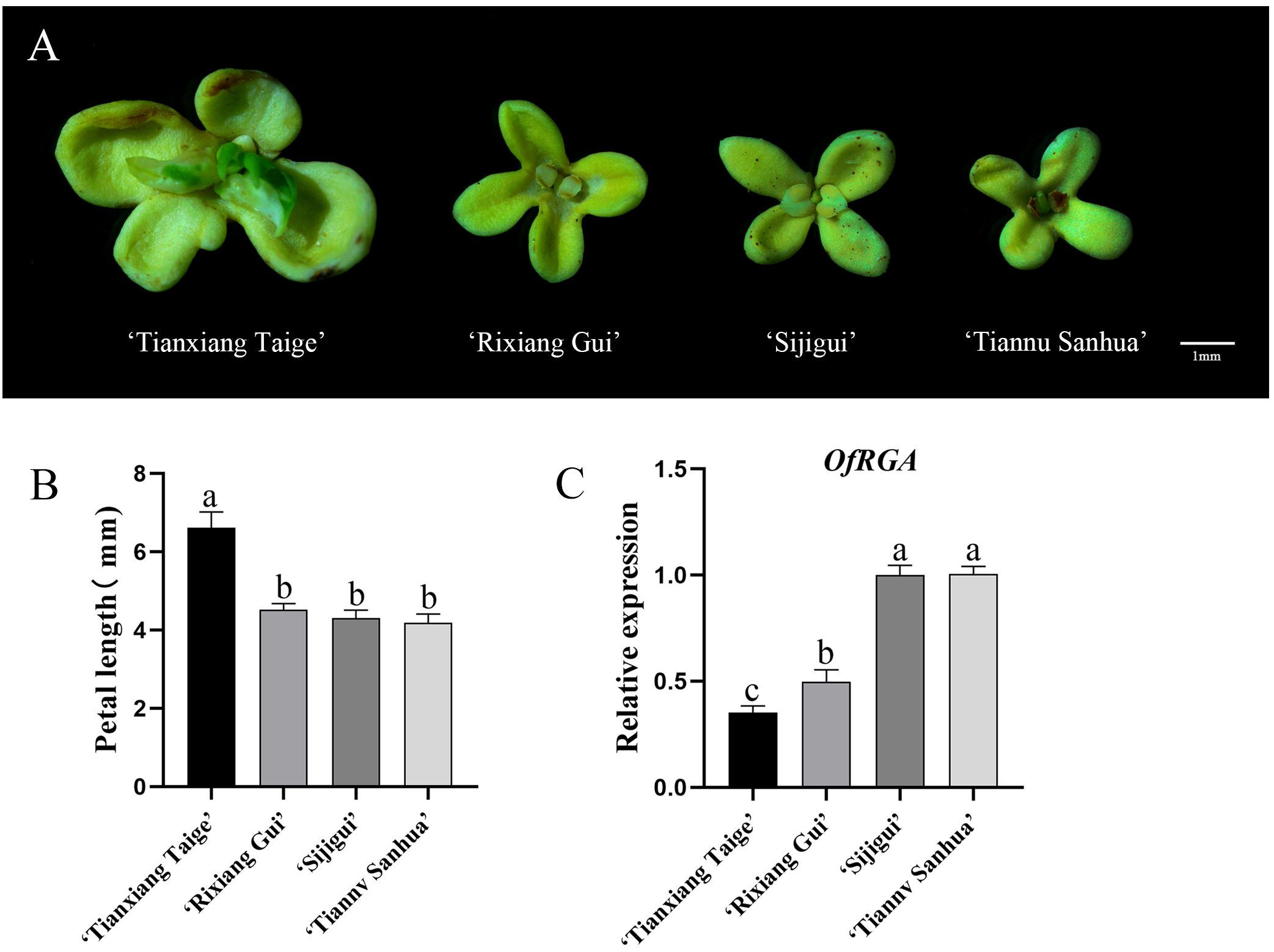
Figure 3. Analysis of OfRGA expression in the petals of various ‘Sijigui’ cultivars. (A) Flowers of different cultivars of ‘Sijigui’ (scale bar = 1 mm). (B) Measurement of petal length in different cultivars of ‘Sijgui’. (C) Gene expression of OfRGA in petals of different varieties. Data are the means ± SD, n = 3, P < 0.05. a,b,c means α=0.05.
3.4 OfRGA overexpression alters tobacco development
Agrobacterium-mediated transformation was used to create tobacco transgenic overexpression lines in order to further explore the gene function of OfRGA. From each construct, over ten distinct transgenic tobacco lines (35S::OfRGA) were produced, the vast majority of which had significantly altered phenotypes. For phenotyping purposes, we chose two transgenic lines that exhibit elevated levels of expression. The transgenic lines of OfRGA were all found to exhibit phenotypes of increased plant height, increased internode spacing, shorter leaf length, wider leaf width, and varying degrees of curling of the leaf margins when comparing the transgenic lines to the wild type (Figures 4A–G). These findings indicate that the overexpression of OfRGA promotes plant development and growth.
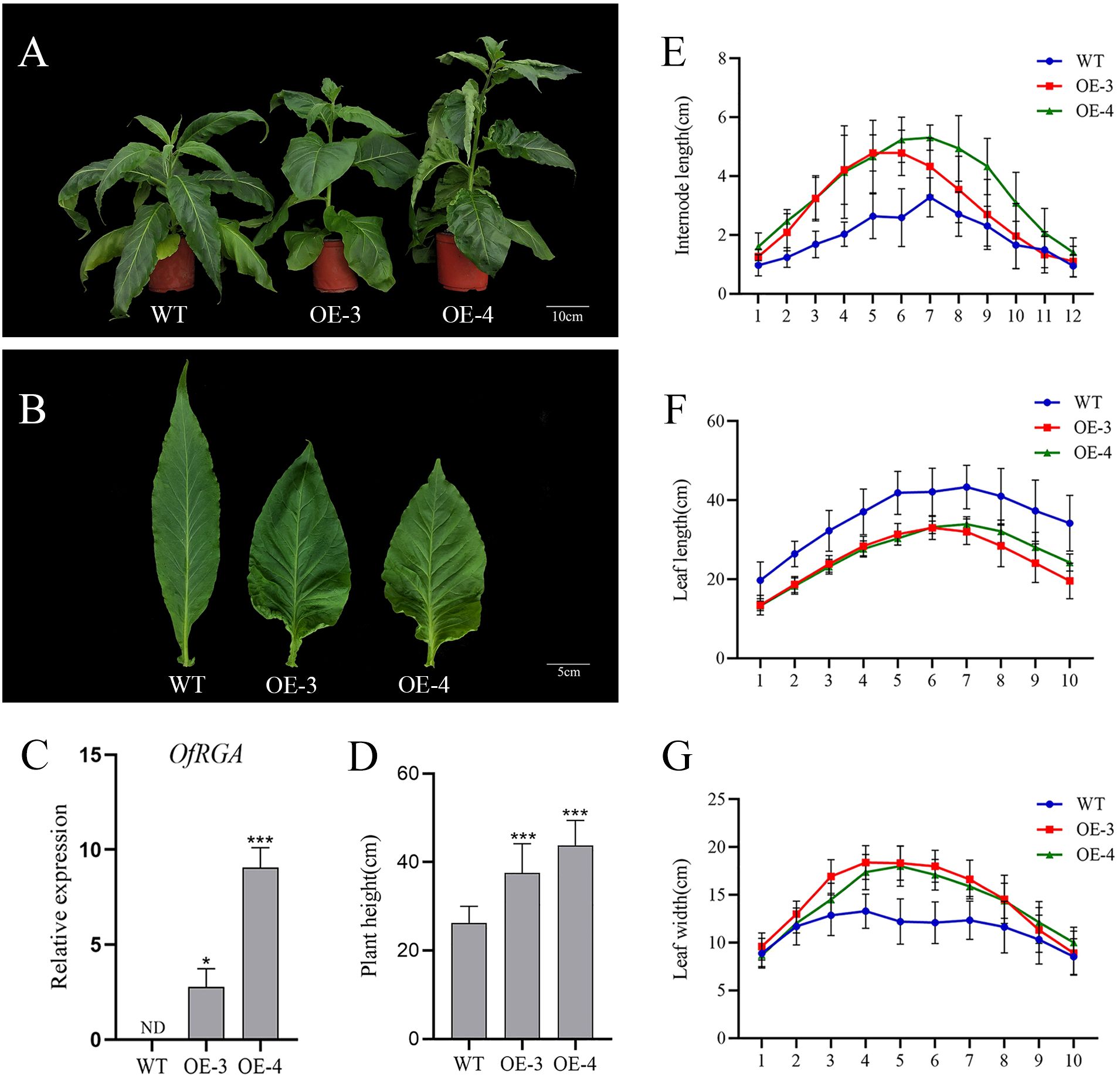
Figure 4. (G) Morphological phenotypes of tobacco overexpressing OfRGA. (A) Phenotypes of 73-day-old wild-type and transgenic plants. Scale bar = 10 cm. (B) Abnormal development of plant leaves. Scale bar = 5 cm. (C) Expression analysis of the OfRGA gene in transgenic lines. (D–F) Measure the plant height, internode length, leaf length, and leaf width of the wild type and transgenic lines. I1-I12, internode1-internode12; L1-L10, leaf1-leaf10. Error bars: ± SD. n = 10, *p < 0.05, ***P <0.001.
3.5 Development of floral organs is impacted by OfRGA overexpression
After blooming, the phenotypes were further observed in the transgenic tobacco lines, and the outcomes demonstrated that the petals of the OfRGA transgenic line were significantly smaller and lighter in color than those of the wild type, further verification of the hypothesis that OfRGA inhibits cell expansion and reduces petal size (Figures 5A, B). In addition, to determine whether different floral organs’ growth and development were impacted, we also measured the width and length of the wild-type and transgenic corolla tubes, as well as the lengths of the stamens, pistils, and flower stalks (Figures 5C–F). We found that compared to the wild type, the petal diameter of OE-3 and OE-4 was reduced by 0.92 cm and 0.74 cm, the corolla tube diameter was reduced by 0.86 mm and 0.76 mm, the length of the corolla tube was reduced by 0.68 cm and 0.64 cm, the length of the pedicel was reduced by 2.7 mm and 3.17 mm, the length of the pistil was reduced by 0.91 cm and 0.82 cm, and the length of the stamen was reduced by 0.88 cm and 0.80 cm. The results indicate that the corolla tube of the transgenic strain became shorter and narrower, with reduced lengths of stamens, pistils, and pedicels compared to the wild type (Figures 5G–L). These findings confirm that the OE-OfRGA transgenic line exhibited significant reductions in the size and dimensions of these floral structures. In summary, the overexpression of OfRGA led to the shortening of floral organs.
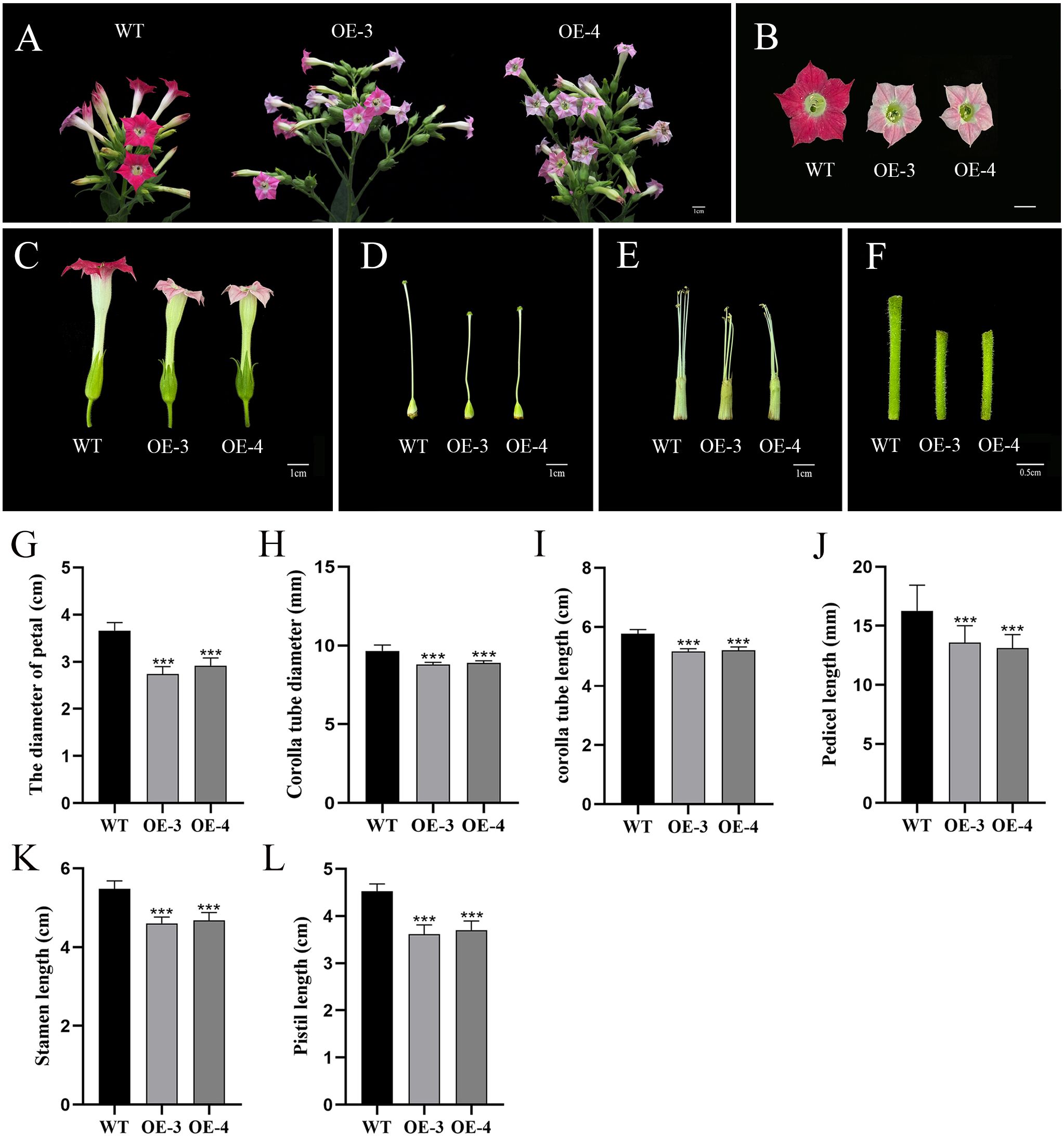
Figure 5. Effects of floral organ development modulated by overexpression of OfRGA in tobacco. (A–F) Phenotypic comparison of inflorescence, petals, corolla tube, pistil, stamen, and pedicel between wild-type and transgenic plants. (G–L) Comparison of petal diameter, corolla tube diameter and length, pedicel length, stamen length, and pistil length between plants that overexpress OfRGA and WT. Data were the means ± SD, n = 20, ***P <0.001.
To look into why the corolla tube is shorter and the petals are smaller, we conducted measurements on the cell area of both wild-type (WT) and transgenic petals, as well as corolla tubes (Figures 6A, E–J). We found that petal cell area decreased by 33.04% and 20.60%, as well as cell length decreased by 19.45% and 10.13% in OE-3 and OE-4, compared with the wild type (Figures 6B–D). It suggests that OfRGA may inhibit the growth of petals and corolla tubes by inhibiting cell expansion. Suggesting that OfRGA may inhibit petal and corolla tube growth by suppressing cell expansion.
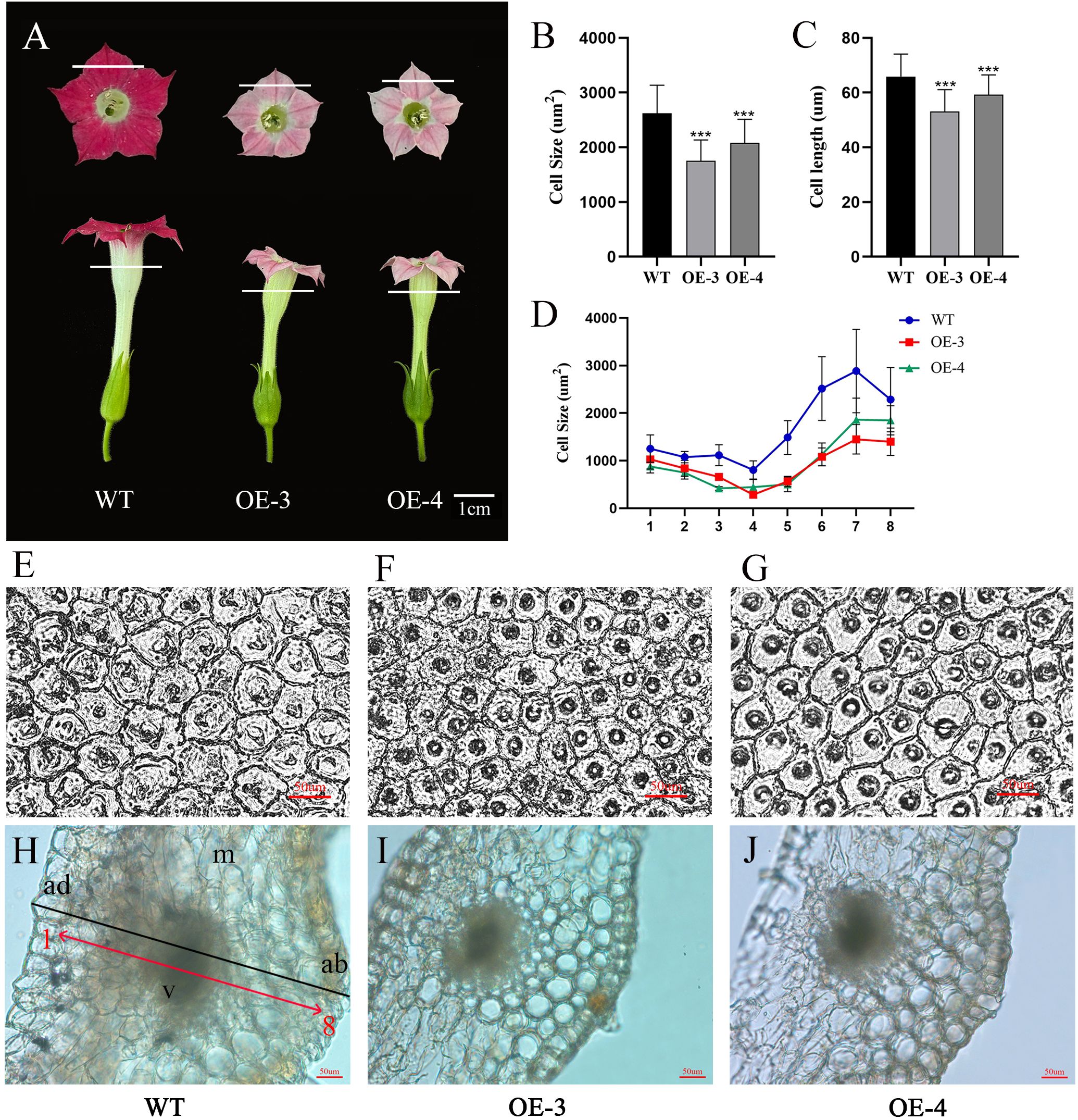
Figure 6. Overexpression of OfRGA can be attributed to a reduction in cell area and length, leading to a smaller petal. (A) Phenotypic petal and corolla tube. (B, C) Comparison of petal cell size and length in WT and overexpressing OfRGA plants. (D) Comparison of corolla tube cell size between WT and OfRGA overexpressing plants. (E–G) Using an optical microscope, the center portions of the petals of both WT and transgenic plants’ abaxial epidermal cells were studied. A 50 μm scale bar is used. (H–J) Cross-sections of the upper part of the corolla tube from WT and OfRGA overexpressing plants. ad, adaxial epidermal cell; ab, abaxial epidermis cell; m, mesophyll; v, vascular bundle. Bar = 50 µm. ***P <0.001.
3.6 RNA-seq revealed that OfRGA is targeted at multiple downstream genes engaged in the development of floral organs
To gain additional insight on the phenomenon through which OfRGA may inhibit the growth of petals and corolla tubes, as well as the causes of the lightening of flower color and the related genes through which OfRGA acts, differentially expressed genes (DEGs) were found using RNA-seq analysis. Transcriptomes from three biological replicates of WT and OfRGA overexpression transgenic plants were obtained for further bioinformatics analysis. Nine samples yielded 54 G of clean data, which was then used for additional bioinformatic analysis. 96.73% of the readings were successfully mapped to the Nicotiana genome. The DEG screen was conducted based on the criteria of a log2 fold change ≥ 1 and a Q value ≤ 0.05. 2,676 genes showed up-regulation and 2,074 genes showed down-regulation in WT compared to the OE-3 group, according to the analysis. Similarly, 4,331 genes showed up-regulation and 3,521 genes showed down-regulation in WT compared to the OE-4 group (Figures 7A, D). Taken together, genes that were up-regulated outnumbered those that were down-regulated. The GO analysis of WT versus OE-3 and OE-4 revealed that the up-regulated genes showed a high enrichment in molecular functions such as ‘DNA-binding transcription factor activity’ and ‘cytoskeletal motor activity’. In terms of cellular components, these genes were significantly enriched in the ‘nucleus’ and ‘cell wall’. The most significant enrichment was observed in the biological processes of ‘cell cycle’ and ‘multicellular organism development’ (Figure 7B). The molecular functions of the down-regulated genes were primarily abundant in ‘transporter activity’, the cellular components were primarily abundant in ‘plasma membrane’ and ‘extracellular region’, and the biological processes were mainly enriched in ‘transport’ and ‘cellular homeostasis’ (Figure 7E). According to KEGG enrichment analysis, the majority of the up-regulated genes were enriched in ‘Chromosome and associated proteins’, ‘Cytoskeleton proteins’, ‘Transcription factors’, and ‘Protein families: signaling and cellular processes’ (Figure 7C). while the down-regulated genes were gratetly enriched in these pathways, such as ‘Starch and Sucrose Metabolism’, ‘Carbohydrate Metabolism’, ‘Transporters’, and ‘Protein Families: Signaling and Cellular Processes’ (Figure 7F). These results indicate that genes and DNA-binding transcription factors involved in regulating the cell wall and cell cycle could be crucial in the organ development and cell expansion of transgenic tobacco flowers.
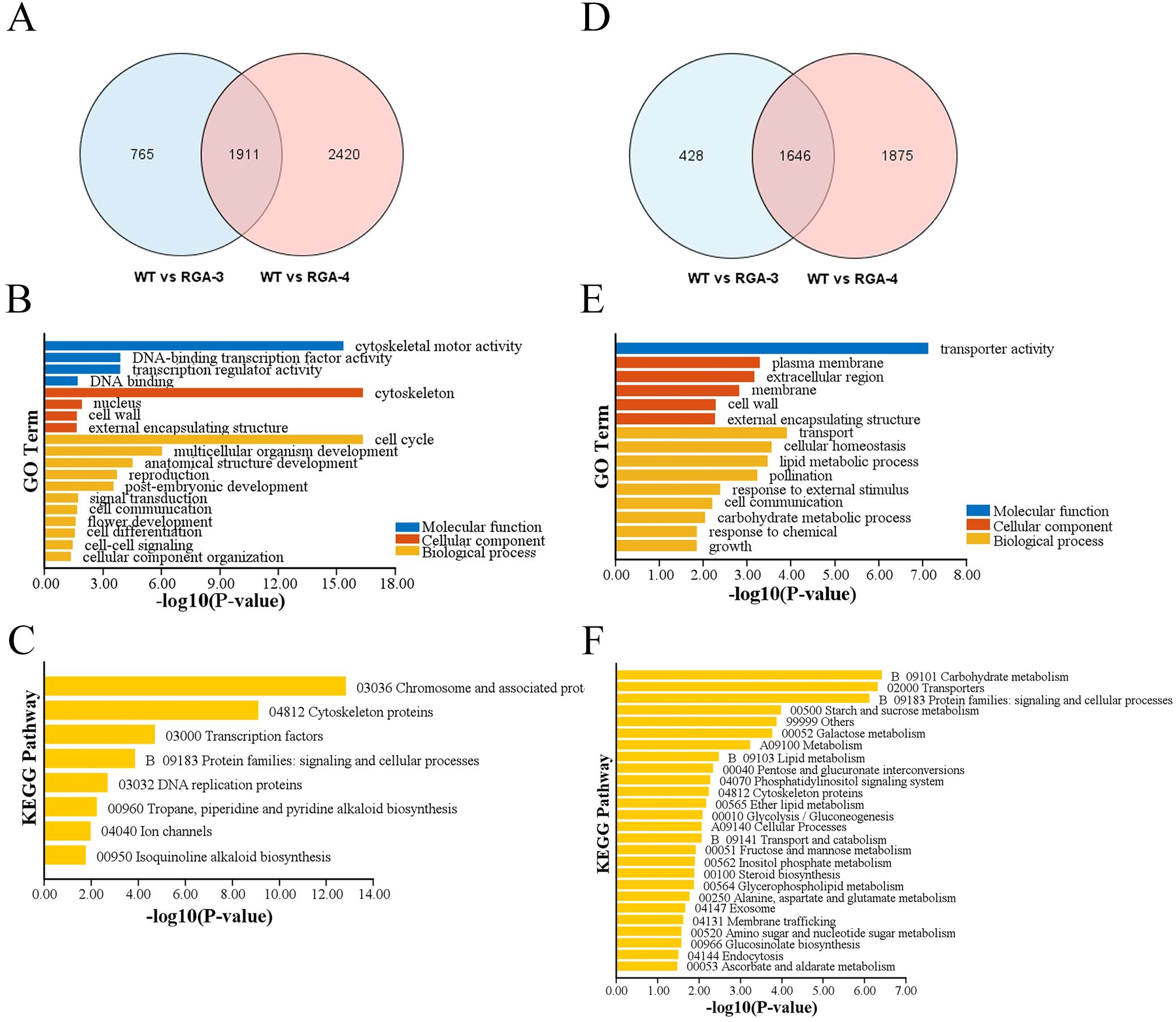
Figure 7. DEGs of transgenic tobacco with the OfRGA gene compared with WT. (A) Venn diagram showing how many up-regulated DEGs are shared and unique across the OfRGA-3 and OfRGA-4 groups. (B, C) GO and KEGG analysis of shared up-regulated DEGs between OfRGA-3 and OfRGA-4. (D) Venn diagram showing how many down-regulated DEGs are shared and unique across the OfRGA-3 and OfRGA-4 groups. (E, F) GO and KEGG analysis of shared down-regulated DEGs between OfRGA-3 and OfRGA-4.
A total of 13 genes associated with the development of floral organs were identified from differentially expressed genes (DEGs), including the MADS-box gene family, AP2/ERF gene family, xyloglucan endotransglucosylase/hydrolase (XTH), Cellulose Synthase A1 (CESA1), Expansin B1 (EXPB1), NAC56, and Plasma Membrane Intrinsic Protein2-7 (PIP2-7). Among these, the MADS-box gene family and AP2/ERF gene family-related genes were significantly upregulated, while XTH, CesA1, EXPB1, NAC56, PIP2-7, and other genes participating in cell wall synthesis and cell expansion were significantly down-regulated (Figure 8A). These findings indicate that genes related to cell expansion are essential to the regulation of floral organ development. Furthermore, several DEGs associated with anthocyanin synthesis and regulation were identified. Genes involved in anthocyanin synthesis, such as Chalcone Isomerase (CHI), 4-Coumarate: Coenzyme A Ligase 3 (4CL3), MYB transcription factors MYB12, and MYB57 were significantly down-regulated, whereas genes related to anthocyanin regulation, like MYB transcription factors MYB26, MYB5, MYB32, MYB3, and bHLH transcription factor bHLH9, were markedly up-regulated (Figure 8B). These results suggest that the color fading of transgenic plants may be due to the declined expression of anthocyanin biosynthesis-related genes and the upregulation of transcription factors such as MYB and bHLH. The exact results still need to be further verified through more experiments.
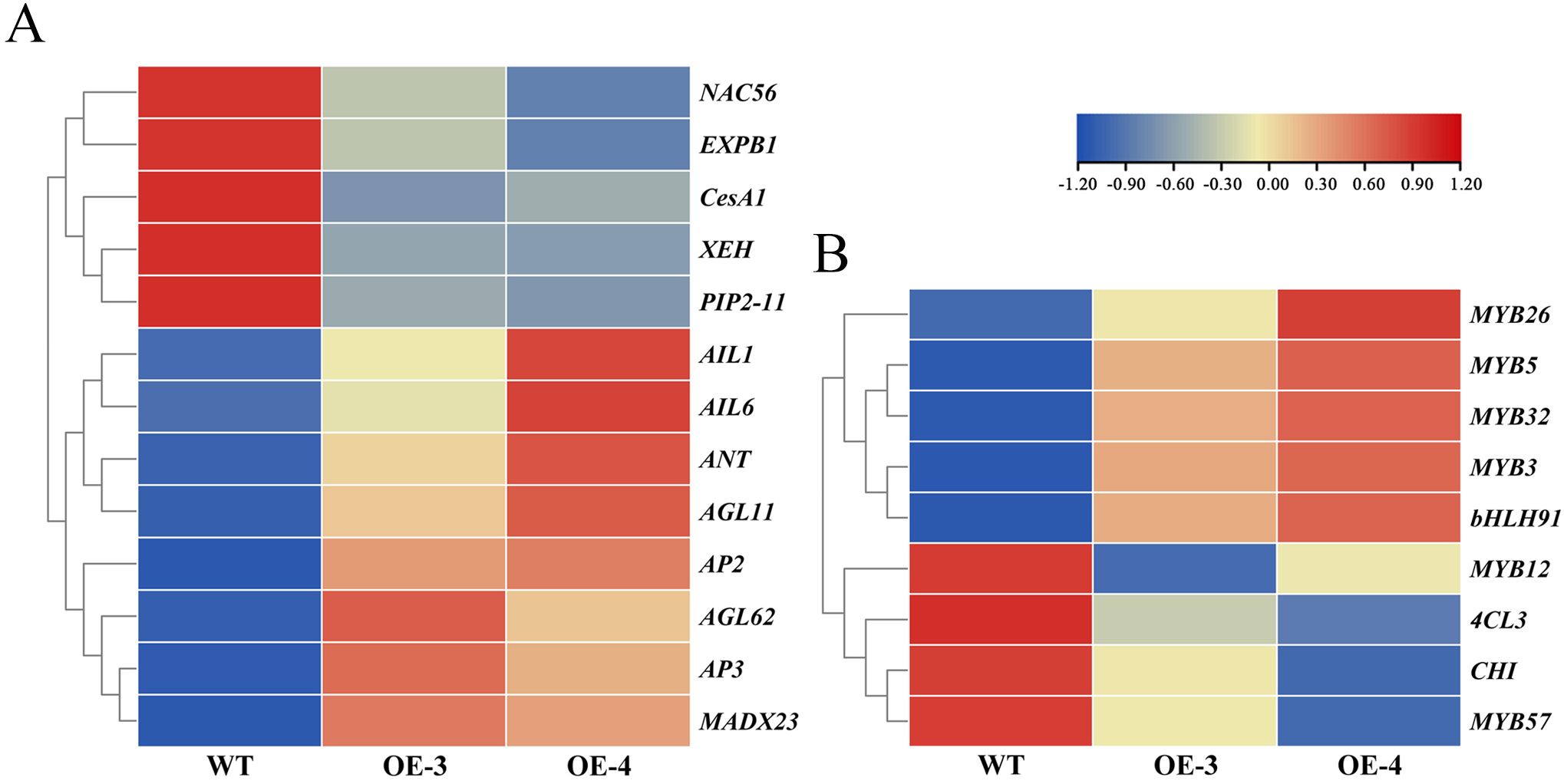
Figure 8. Expression heatmap of the DEGs. (A) Heatmap of floral organ development. (B) Heatmap of anthocyanin synthesis-related genes.
4 Discussion
One of the most attractive features of Osmanthus fragrans is its floral organs. The regulation of floral organ development is significantly influenced by phytohormones. DELLA protein, as a central element of the phytohormone regulatory network, is engaged in the communication and control of several hormones (Navarro et al., 2008). Here, we analyzed the DELLA protein OfRGA transcript levels in the petals of four different ‘Sijigui’ varieties, indicating that the DELLA protein OfRGA may play a role in limiting cell proliferation, thereby contributing to reduced petal size. In a previous study, DELLA proteins were found to take part in nearly all plant growth and development processes (Thomas et al., 2018). AtRGL2 plays a role in regulating floral development in Arabidopsis thaliana. The expression of a dominant GA-resistant version of RGL2, resulted in a phenotype characterized by reduced stamen and petal size, as well as inhibited anther dehiscence (Gómez et al., 2019). In poplar, overexpressing PmRGL2 from Japanese apricots delayed the beginning of bud dormancy and produced dwarfed plants (Lv et al., 2018). The tomato DELLA protein SlPRO participates in fruit development, flower development, and floral induction in tomatoes (Martí et al., 2007; Silva et al., 2018). However, information regarding the precise role of DELLA proteins in “Sijigui” is lacking.
With tobacco that has been heterologously transformed, the function of the OfRGA gene has been further investigated. The findings demonstrated that the OfRGA transgene displayed shorter and wider leaves, as well as increased plant height and node spacing when compared to the wild type (Figure 4). Early research has demonstrated that transgenic tobacco produces a dwarf phenotype when AtGAI and DoDELLA are overexpressed (Hynes et al., 2003; Zhou et al., 2022). And the same dwarfing phenotype was also observed in Liriodendron hybrids (Liu et al., 2020). The OfRGA transgenic plant height phenotypes differed from those reported in the existing species, but the protein sequences were found to be not significantly different by amino acid sequence comparison with those of the model plant Arabidopsis thaliana (Figure 1A). Therefore, it is speculated that it may be because of the different regulation of downstream structural genes by OfRGA during the growth and developmental stages of tobacco. This suggests a potential different regulation of downstream structural genes by OfRGA during the growth and developmental stages of tobacco. Furthermore, OfRGA was found to be involved in flower development in tobacco; the OE-OfRGA transgenic lines exhibited a significant reduction in the size and dimensions of these floral structures (Figure 5). This is similar with previous research findings on DELLA in other species. The DELLA homologue CsGAIP in cucumber suppresses stamen development by repressing homozygous heterozygous genes in class B flowers (Zhang et al., 2014). Arabidopsis RGA, RGL1, and RGL2 all function to inhibit stamen, petal, and anther development in Arabidopsis (Cheng et al., 2004). Coordinated cell division and expansion during flower formation determine the size of floral organs (Mizukami and Fischer, 2000). Therefore, to further investigate the reasons for the smaller floral organs in overexpression of OfRGA transgenic tobacco, we performed freehand sections of petals as well as corolla tubes. The results showed a decrease in petal area as well as length in the transverse section of the upper portion of the corolla tube compared to WT (Figure 6). The above results show that the smaller size of transgenic tobacco floral organs may be due to a reduction in cell area and length. Further exploration of the function of the OfRGA gene requires in vivo verification through the acquisition of transgenic Osmanthus plants.
The entire set and quantity of transcripts present in a cell at a specific physiological state or developmental stage is known as the transcriptome, which is significant for understanding the level of gene expression, and exposing the molecular mechanism of biological gene expression regulation research (Qi et al., 2011). Using transcriptome technology, the primary mechanism of calcium-induced anthocyanin production in grapevine pericarp was discovered (Yu et al., 2020). Ninebark transcriptome analysis postulated that BgSEP2b is a center gene, interacting with multiple transcription factors and carpel-related genes, indicating that it might be an important gene for the development of ninebark carpels (Zhang et al., 2022). To further investigate the specific molecular mechanism by which OfRGA overexpression in tobacco leads to smaller floral organs, we examined transgenic tobacco using RNA-seq sequencing. And the transcriptome data demonstrated that genes associated with the expression of cell wall synthesis and proliferation, such as CesA1, XEH, and EXPB, were notably down-regulated in the transgenic strain when contrasted with the wild type (Figure 8). The size of plant organ is regulated by cell proliferation and expansion (Tsukaya, 2006). In the model plant Arabidopsis thaliana, AtEXPA15 and AtERF regulate petal and fruit size by promoting cell division and thereby regulating petal and fruit size (Bernal-Gallardo et al., 2024; Lee et al., 2023). Furthermore, OfXTH genes might take part in the opening process of Osmanthus flowers by regulating cellular petal expansion (Yang et al., 2022). Earlier studies revealed that DELLA proteins control cell expansion and proliferation, which contribute to plant growth and development. RhGAI1 overexpression inhibits petal cell expansion in Arabidopsis thaliana, and RhGAI1 is engaged in the regulation of rose petal size by inhibiting the expression of RhCesA2 (Luo et al., 2013). BraRGL1 is a DELLA protein in Brassica rapa that represses the transcriptional activation of BraXTH by BraSOC1 to inhibit the expansion of stalk (Wang et al., 2023). Together, these results imply that OfRGA may inhibit cell expansion by lowering the level of gene expression such as CesA1, XEH, and EXPB, resulting in smaller petals and corolla tubes of transgenic tobacco. However, it is still unclear whether the differences in petal size among four different varieties of ‘Sijigui’ are caused by OfRGA regulating the EXPB, XEH, and CesA1 genes, and further experiments are needed to verify this.
Previous studies have found that the DELLA protein in Arabidopsis promotes anthocyanin biosynthesis by isolating the JAZ and MYBL2 genes in the MBW complex or by interacting to enhance the transcriptional activity of PAP1/MYB75 (Xie et al., 2016; Zhang et al., 2017). Nevertheless, in this paper, the transgenic petals were significantly lighter in flower color. And further transcriptome analysis demonstrated that following OfRGA overexpression, the expression of the anthocyanin synthesis gene CHI was markedly down-regulated, whereas the anthocyanin-regulated transcription factors MYB and bHLH were significantly up-regulated (Figure 8). The CHI gene is an important structural gene in the anthocyanin synthesis process (Sun et al., 2019). Given the results above, it is speculated that introducing OfRGA through heterologous transformation could impact the expression of endogenous anthocyanin synthesis and regulatory genes in tobacco. Furthermore, studies have reported that Osmanthus fragrans contains anthocyanins (Shi et al., 2021). However, whether OfRGA mediates the synthesis pathway of anthocyanins in Osmanthus fragrans requires further research.
5 Conclusion
This research quantitatively examined the expression levels of OfRGA in the petals of four different ‘Sijigui’ varieties and observed the phenotypic changes in OfRGA transgenic tobacco, revealing the potential role of OfRGA in controlling the size of plant floral organs. Further results of slicing experiments and analysis of transgenic plants’ transcriptome data revealed that OfRGA might influence the size of plant petals by regulating genes related to cell expansion, such as CesA1, XEH, and EXPB1. These findings suggest that OfRGA might be crucial for the development of the organs in the Osmanthus fragrans flower and establish the framework for additional revelation of the molecular mechanism of OfRGA in Osmanthus fragrans flower organ development.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA1122233.
Author contributions
QW: Data curation, Formal analysis, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing, Software. ML: Data curation, Software, Validation, Writing – review & editing. GJ: Data curation, Formal analysis, Validation, Writing – review & editing. JS: Formal analysis, Investigation, Writing – review & editing. TC: Methodology, Software, Writing – review & editing. LY: Investigation, Writing – review & editing. IK: Formal analysis, Writing – review & editing. JD: Methodology, Writing – review & editing. SZ: Software, Writing – review & editing. YW: Supervision, Writing – review & editing. ZX: Resources, Supervision, Writing – review & editing. QF: Formal analysis, Methodology, Resources, Supervision, Writing – original draft, Writing – review & editing. HZ: Data curation, Formal analysis, Funding acquisition, Methodology, Resources, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The study was funded by the National Natural Science Foundation of China (no. 32072615 and 32201617) and the Major Scientific and Technological Project of Zhejiang Province (2021C02071).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1502347/full#supplementary-material
References
Achard, P., Gusti, A., Cheminant, S., Alioua, M., Dhondt, S., Coppens, F., et al. (2009). Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr. Biol. 19, 1188–1193. doi: 10.1016/j.cub.2009.05.059
An, J., Zhang, X., Li, H., Wang, D., You, C., Han, Y. (2023). The E3 ubiquitin ligases SINA1 and SINA2 integrate with the protein kinase CIPK20 to regulate the stability of RGL2a, a positive regulator of anthocyanin biosynthesis. New Phytol. 239, 1332–1352. doi: 10.1111/nph.18997
Bernal-Gallardo, J. J., González-Aguilera, K. L., Folter, S. (2024). EXPANSIN15 is involved in flower and fruit development in Arabidopsis. Plant Reprod. 37, 259–270. doi: 10.1007/s00497-023-00493-4
Chahtane, H., Lai, X., Tichtinsky, G., Rieu, P., Arnoux-Courseaux, M., Cancé, C., et al. (2023). Flower development in arabidopsis. Methods Mol. Biol. 2686, 3–38. doi: 10.1007/978-1-0716-3299-4_1
Cheng, H., Qin, L., Lee, S., Fu, X., Richards, D. E., Cao, D., et al. (2004). Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131, 1055–1064. doi: 10.1242/dev.00992
Coen, E., Meyerowitz, E. M. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353, 31–37. doi: 10.1038/353031a0
Fleet, C. M., Sun, T. (2005). A DELLA cate balance: the role of gibberellin in plant morphogenesis. Curr. Opin. Plant Biol. 8, 77–85. doi: 10.1016/j.pbi.2004.11.015
Gómez, M. D., Fuster-Almunia, C., Ocaña-Cuesta, J., Alonso, J. M., Pérez-Amador, M. A. (2019). RGL2 controls flower development, ovule number and fertility in Arabidopsis. Plant Sci. 281, 82–92. doi: 10.1016/j.plantsci.2019.01.014
Gotô, N., Pharis, R. P. (1999). Role of gibberellins in the development of floral organs of the gibberellin-deficient mutant, ga1-1, of Arabidopsis thaliana. Can. J. Bot. 77, 944–954. doi: 10.1139/b99-090
Hernández-García, J., Sun, R., Serrano-Mislata, A., Inoue, K., Vargas-Chávez, C., Esteve-Bruna, D., et al. (2021). Coordination between growth and stress responses by DELLA in the liverwort Marchantia polymorpha. Curr. Biol. 31, 3678–3686. doi: 10.1016/j.cub.2021.06.010
Hynes, L. W., Peng, J., Richards, D. E., Harberd, N. P. (2003). Transgenic expression of the Arabidopsis DELLA proteins GAI and gai confers altered gibberellin response in tobacco. Transgenic Res. 12, 707–714. doi: 10.1023/b:trag.0000005145.68017.6e
Jin, Y., Song, X., Chang, H., Zhao, Y., Cao, C., Qiu, C., et al. (2021). The GA-DELLA-OsMS188 module controls male reproductive development in rice. New Phytol. 233, 2629–2642. doi: 10.1111/nph.17939
Joan, E. N., Jan, A. D. Z. (1988). Flower development in normal tomato and a gibberellin-deficient (ga-2) mutant. Am. J. Bot. 75, 45–45. doi: 10.2307/2443904
Koornneef, M., van der Veen, J. H. (1980). Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) heynh. Theor. Appl. Genet. 58, 257–263. doi: 10.1007/bf00265176
Kumar, S., Stecher, G., Li, M., Knyazm, C., Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lee, P., Zhan, Y., Wang, J., Cheng, Y., Hsu, W., Hus, H., et al. (2023). The AtERF19 gene regulates meristem activity and flower organ size in plants. Plant J. 118, 1232–1232. doi: 10.1111/tpj.16196
Li, D., Guo, Z., Liu, C., Li, J., Xu, W., Chen, Y. (2017). Quantification of near-attomole gibberellins in floral organs dissected from a single Arabidopsis thaliana flower. Plant J. 91, 547–557. doi: 10.1111/tpj.13580
Li, Q., Li, J., Zhang, L., Pan, C., Yang, N., Sun, K., et al. (2021). Gibberellins are required for dimorphic flower development in Viola philippica. Plant Sci. 303, 110749–110749. doi: 10.1016/j.plantsci.2020.110749
Liu, X., Wang, Q., Jiang, G., Wan, Q., Dong, B., Lu, M., et al. (2023). Temperature-responsive module of OfAP1 and OfLFY regulates floral transition and floral organ identity in Osmanthus fragrans. Plant Physiol. Biochem. 203, 108076–108076. doi: 10.1016/j.plaphy.2023.108076
Liu, Y., Wang, P., Yan, S., Liu, X., Lu, L., Chen, X., et al. (2020). Molecular cloning and functional characterization of the DELLA gene family in Liriodendron hybrids. Forests 11, 1363–1363. doi: 10.3390/f11121363
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.3779/j.issn.1009-3419.2011.12.07
Luo, J., Ma, N., Pei, H., Chen, J., Li, J., Gao, J. (2013). A DELLA gene, RhGAI1, is a direct target of EIN3 and mediates ethylene-regulated rose petal cell expansion via repressing the expression of RhCesA2. J. Exp. Bot. 64, 5075–5084. doi: 10.1093/jxb/ert296
Lv, L., Huo, X., Wen, L., Gao, Z., Khalil-ur-Rehman, M. (2018). Isolation and role of PmRGL2 in GA-mediated floral bud dormancy release in Japanese apricot (Prunus mume Siebold et Zucc.). Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00027
Mā, H. (2005). Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 56, 393–434. doi: 10.1146/annurev.arplant.55.031903.141717
Martí, C., Orzáez, D., Ellul, P., Moreno, V., Carbonell, J., Granell, A. (2007). Silencing of DELLA induces facultative parthenocarpy in tomato fruits. Plant J. 52, 865–876. doi: 10.1111/j.1365-313x.2007.03282.x
Mizukami, Y., Fischer, R. L. (2000). Plant organ size control: Aintegumenta regulates growth and cell numbers during, organogenesis. Curr. Opin. Plant Biol. 3, 174–175. doi: 10.1016/s1369-5266(00)80047-3
Navarro, L., Bari, R., Achard, P., Lisón, P., Nemri, A., Harberd, N. P., et al. (2008). DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr. Biol. 18, 650–655. doi: 10.1016/j.cub.2008.03.060
Park, Y., Park, H. (2017). Microstructural changes in the fruit and pedicel of ‘Wonhwang’ oriental pear induced by exogenous gibberellins. Scientia Hortic. 222, 1–6. doi: 10.1016/j.scienta.2017.05.009
Qi, Y., Liu, Y., Rong, W. (2011). RNA-Seq and its applications: a new technology for transcriptomics. Yichuan 33, 1191–1202. doi: 10.3724/sp.j.1005.2011.01191
Rueden, C., Schindelin, J., Hiner, M., DeZonia, B. E., Walter, A. E., Arena, E. T., et al. (2017). ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinf. 18, 529. doi: 10.1186/s12859-017-1934-z
Shi, Y., Xia, H., Cheng, X., Zhang, L. (2021). Genome-wide miRNA analysis and integrated network for flavonoid biosynthesis in Osmanthus fragrans. BMC Genomics 22, 141. doi: 10.1186/s12864-021-07439-y
Silva, G. F. F., Silva, E. M., Correa, P., Vicente, M. H., Jiang, N., Notini, M. M., et al. (2018). Tomato floral induction and flower development are orchestrated by the interplay between gibberellin and two unrelated microRNA-controlled modules. New Phytol. 221, 1328–1344. doi: 10.1111/nph.15492
Sparkes, I. A., Runions, J., Kearns, A., Hawes, C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1, 2019–2025. doi: 10.1038/nprot.2006.286
Sun, W., Shen, H., Xu, H., Tang, X., Tang, M., Ju, Z., et al. (2019). Chalcone Isomerase a key enzyme for Anthocyanin Biosynthesis in Ophiorrhiza japonica. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00865
Thomas, S. G., Blázquez, M. Á., Alabadı́, D. (2018). Della proteins: master regulators of gibberellin-responsive growth and development. Annu. Plant Rev. Online 49, 189–227. doi: 10.1002/9781119312994.apr0536
Tsukaya, H. (2006). Mechanism of leaf-shape determination. Annu. Rev. Plant Biol. 57, 477–496. doi: 10.1146/annurev.arplant.57.032905.105320
Tyler, L., Thomas, S. G., Hu, J., Dill, A., Alonso, J. M., Ecker, J. R., et al. (2004). DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 135, 1008–1019. doi: 10.1104/pp.104.039578
Wang, Q., Gao, G., Chen, X., Liu, X., Dong, B., Wang, Y., et al. (2022). Genetic studies on continuous flowering in woody plant Osmanthus fragrans. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1049479
Wang, Y., Song, S., Hao, Y., Chen, C., Ou, X., Li, T., et al. (2023). Role of BraRGL1 in regulation of Brassica rapa bolting and flowering. Hortic Res. 10, uhad119. doi: 10.1093/hr/uhad119
Wilson, R. N., Heckman, J. W., Somerville, C. (1992). Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol. 100, 403–408. doi: 10.1104/pp.100.1.403
Xie, Y., Tan, H., Ma, Z., Huang, J. (2016). DELLA proteins promote anthocyanin biosynthesis via sequestering MYBL2 and JAZ suppressors of the MYB/bHLH/WD40 complex in Arabidopsis thaliana. Mol. Plant 9, 711–721. doi: 10.1016/j.molp.2016.01.014
Yang, Y., Miao, Y., Zhong, S., Fang, Q., Wang, Y., Dong, B., et al. (2022). Genome-wide identification and expression analysis of XTH gene family during flower-opening stages in Osmanthus fragrans. Plants 11, 1015–1015. doi: 10.3390/plants11081015
Yu, H., Ito, T., Zhao, Y., Peng, J., Kumar, P. P., Meyerowitz, E. M. (2004). Floral homeotic genes are targets of gibberellin signaling in flower development. Proc. Natl. Acad. Sci. United States America 101, 7827–7832. doi: 10.1073/pnas.0402377101
Yu, J., Zhu, M., Wang, M., Xu, Y., Chen, W., Yang, G. (2020). Transcriptome analysis of calcium-induced accumulation of anthocyanins in grape skin. Scientia Hortic. 260, 108871. doi: 10.1016/j.scienta.2019.108871
Yue, Y., Zhu, W., Shen, H., Wang, H., Du, J., Wang, L., et al. (2023). DNA-Binding one finger transcription factor PhDof28 regulates petal size in Petunia. Int. J. Mol. Sci. 24, 11999. doi: 10.3390/ijms241511999
Zhang, L., Li, L., Wu, J., Peng, J., Zhang, L., Wang, X. (2012). Cell expansion and microtubule behavior in ray floret petals of Gerbera hybrida: Responses to light and gibberellic acid. Photochem Photobiol Sci. 11, 279–288. doi: 10.1039/c1pp05218
Zhang, Y., Liu, Z., Liu, J., Lin, S., Wang, J., Lin, W., et al. (2017). GA-DELLA pathway is involved in regulation of nitrogen deficiency-induced anthocyanin accumulation. Plant Cell Rep. 36, 557–569. doi: 10.1007/s00299-017-2102-7
Zhang, J., Zhang, Y., Liu, B., Yang, S., An, J., Chen, C., et al. (2014). A cucumber DELLA homolog CsGAIP may inhibit staminate development through transcriptional repression of B class floral homeotic genes. PloS One 9, e91804. doi: 10.1371/journal.pone.0091804
Zhang, W., Zhou, Q., Lin, J., Ma, X., Dong, F., Yan, H., et al. (2022). Transcriptome analyses shed light on floral organ morphogenesis and bract color formation in Bougainvillea. BMC Plant Biol. 22, 97. doi: 10.1186/s12870-022-03478-z
Keywords: Osmanthus fragrans, OfRGA, floral organ development, floral size, cell expansion
Citation: Wan Q, Lu M, Jiang G, Shao J, Chen T, Yang L, Khan IA, Deng J, Zhong S, Wang Y, Xiao Z, Fang Q and Zhao H (2024) The characterization of OfRGA in regulation of flower size through tuning cell expansion genes. Front. Plant Sci. 15:1502347. doi: 10.3389/fpls.2024.1502347
Received: 26 September 2024; Accepted: 09 December 2024;
Published: 20 December 2024.
Edited by:
Changlin Liu, Chinese Academy of Agricultural Sciences (CAAS), ChinaReviewed by:
Langlang Ma, Maize Research Institute of Sichuan Agricultural University, ChinaYuanzheng Yue, Nanjing Forestry University, China
Copyright © 2024 Wan, Lu, Jiang, Shao, Chen, Yang, Khan, Deng, Zhong, Wang, Xiao, Fang and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiu Fang, ZmFuZ3FpdUB6YWZ1LmVkdS5jbg==; Hongbo Zhao, emhhb2hiQHphZnUuZWR1LmNu
†These authors share first authorship
 Qianqian Wan†
Qianqian Wan† Irshad Ahmad Khan
Irshad Ahmad Khan Yiguang Wang
Yiguang Wang Zheng Xiao
Zheng Xiao Hongbo Zhao
Hongbo Zhao