
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci. , 15 January 2025
Sec. Functional and Applied Plant Genomics
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1495069
This article is part of the Research Topic Genomics-Driven Advances in Crop Productivity and Stress Resilience View all 7 articles
 Daiana A. Zhernova1*
Daiana A. Zhernova1* Elena N. Pushkova1
Elena N. Pushkova1 Tatiana A. Rozhmina2
Tatiana A. Rozhmina2 Elena V. Borkhert1
Elena V. Borkhert1 Alexander A. Arkhipov1,3
Alexander A. Arkhipov1,3 Elizaveta A. Sigova1,4
Elizaveta A. Sigova1,4 Ekaterina M. Dvorianinova1,4
Ekaterina M. Dvorianinova1,4 Alexey A. Dmitriev1,4
Alexey A. Dmitriev1,4 Nataliya V. Melnikova1*
Nataliya V. Melnikova1*Flax (Linum usitatissimum L.) is known as a dual-purpose crop, producing both fiber and oil, which have a wide range of uses. Successful flax breeding requires knowledge on the genetic determinants of flax traits. The former identification of molecular markers for valuable traits used labor-intensive and sometimes poorly reproducible approaches. However, they allowed an assessment of the genetic diversity of flax and its relatives, the construction of linkage maps, and the identification of some markers for important characteristics. The sequencing of flax whole genome triggered the development of genome-wide association studies (GWAS) and quantitative trait locus (QTL) mapping. QTLs and quantitative trait nucleotides (QTNs) were identified for valuable seed- and fiber-related features and for resistance to biotic and abiotic stressors. Cost-effective and accurate analysis of large number of genotypes for multiple markers simultaneously using microarrays or targeted deep sequencing became available, as well as HRM, TaqMan, KASP, and other fluorescence-based high-throughput methods for detecting DNA polymorphisms. However, most DNA markers identified in flax are ambiguously linked to trait expression and are not universally applicable. A major challenge remains the lack of knowledge on functional polymorphisms. To date, only a few are known, mainly mutations in the FAD3 genes responsible for reduced linolenic acid content in linseed oil. For the further development of marker-assisted and genomic selection of flax, it is necessary to analyze exhaustively phenotyped sample sets, to identify DNA polymorphisms that determine valuable traits, and to develop efficient DNA test systems.
Flax (Linum usitatissimum L.) is one of the world’s ancient crops (family Linaceae DC. ex Perleb), known since ancient Egypt and Mesopotamia (Saha and Hazra, 2004), although it was domesticated only 8,000-10,000 years ago. Flax is a dicotyledonous self-pollinated annual herbaceous plant (diploid chromosome set 2n = 2x = 30, genome size ~ 450 Mb (Green et al., 2008; Soto-Cerda et al., 2013; Dvorianinova et al., 2022; You et al., 2023). Flax is known as a dual-purpose crop, producing both fiber and oil. L. usitatissimum is one of the three most important textile crops in the world and one of the five most important oilseed crops (Deng et al., 2011; Ottai et al., 2011). Flax seed is a rich source of ω-3 (linolenic) and ω-6 (linoleic) fatty acids, easily digestible dietary fiber, and lignans, which have beneficial effects on the human body. In particular, they prevent and reduce the risk of cardiovascular diseases, various types of cancer, diabetes, atherosclerosis, arthritis, osteoporosis, autoimmune diseases, reduce cholesterol levels, stimulate blood flow, have a beneficial effect on the nervous system, and have antioxidant, anti-inflammatory, and antimicrobial effects (Kezimana et al., 2018; Campos et al., 2019; Parikh et al., 2019; Saini et al., 2021; Al-Madhagy et al., 2023). In addition, flax seeds are widely used in pharmaceuticals, animal feed, and for the production of environmentally friendly paints, varnishes, lubricants, linoleum, biofuels, etc (Singh et al., 2011; Corino et al., 2014; Goyal et al., 2014; Campos et al., 2019). Another valuable product derived from flax is fiber, which is used in the textile industry and for the production of paper and a variety of composite materials (Jhala and Hall, 2010; Fombuena et al., 2019; Asyraf et al., 2022).
The aim of our review is to describe the history of development of flax DNA markers and to propose the promising directions of their future in flax breeding for creation of varieties with a complex of valuable traits.
The beginning of the new millennium was characterized by a great interest in the use of molecular markers for plant studies. The first studies on flax used mainly dominant (Random Amplified Polymorphic DNA – RAPD, Amplified Fragment Length Polymorphism – AFLP, Inter Simple Sequence Repeat – ISSR, Inter-Retrotransposon Amplified Polymorphism – IRAP, Retrotransposon Microsatellite Amplified Polymorphism – REMAP, Sequence-Specific Amplified Polymorphism – SSAP, and Inter-Primer Binding Site – iPBS) and codominant (Restriction Fragment Length Polymorphism – RFLP and microsatellites or Simple Sequence Repeats – SSRs) markers.
RFLP markers were used in studies of flax varieties and hybrids to identify molecular markers for valuable traits and to characterize genetic diversity (Schneeberger and Cullis, 1991; Lawrence et al., 1993; Oh et al., 2000).
RAPD method was applied in studies of flax varieties, hybrids, landraces, and related to L. usitatissimum species for genotyping, characterization of genetic diversity, determination of phylogenetic relationships, and identification of molecular markers for valuable traits (Cullis et al., 1999; Stegniî et al., 2000; Bo et al., 2002; Fu et al., 2002a, 2002b, 2003a, 2003b; Muravenko et al., 2003; Fu, 2005; Diederichsen and Fu, 2006; Bo et al., 2008; Muravenko et al., 2009; Singh et al., 2009; Bibi et al., 2015; Nagabhushanam et al., 2021).
AFLP approach also enabled genotyping flax varieties, analyzing their genetic diversity, assessing genetic relationships between genotypes, and identifying markers for valuable flax traits using mapping populations (Spielmeyer et al., 1998; Everaert et al., 2001; Van Treuren et al., 2001; Wakjira et al., 2005; Adugna et al., 2006; Chandrawati et al., 2014).
SSR markers were widely used in flax studies to genotype varieties, evaluate their genetic diversity, assess their phylogenetic relationships, create their genetic maps, and identify markers for valuable traits using mapping populations and accession collections (Ellegren, 2004; Rashid and Duguid, 2005; Roose‐Amsaleg et al., 2006; Cloutier et al., 2009; Fu and Peterson, 2010; Deng et al., 2011; Rachinskaya et al., 2011; Soto-Cerda et al., 2011; Cloutier et al., 2012; Soto-Cerda et al., 2012; Asgarinia et al., 2013; Soto-Cerda et al., 2013, 2014; Chandrawati et al., 2017b; Choudhary et al., 2017a, 2017; Sudarshan et al., 2017; Wu et al., 2017; Chandrawati et al., 2017a; Saha et al., 2019; Soto-Cerda et al., 2019; Singh et al., 2021; Saroha et al., 2022b; Chen and Liu, 2024).
ISSR markers were applied to characterize the genetic diversity and relationships among flax varieties and L. usitatissimum wild ancestor Linum bienne Mill. and to identify markers for valuable traits (Wiesnerova and Wiesner, 2004; Rajwade et al., 2010; Uysal et al., 2010; El Sayed et al., 2018; Kocak et al., 2023).
Markers based on the analysis of sequences derived from retrotransposons were also used in flax research. SSAP, IRAP, REMAP, and iPBS markers allowed evaluation of genetic diversity and genotyping of flax varieties, characterization of relationships between species of the genus Linum, and assessment of activity of LTR retrotransposons in flax plants exposed to stressors (Smýkal et al., 2011; Melnikova et al., 2014; Habibollahi et al., 2015; Abbasi Holasou et al., 2016; Lancíková and Žiarovská, 2020; Žiarovská et al., 2022).
The combination of different types of dominant and codominant markers was utilized in flax studies for linkage map development, genotyping, and evaluation of genetic diversity and relationships among varieties (Oh et al., 2000; Kumari et al., 2018; Mhiret and Heslop-Harrison, 2018; Osman et al., 2021).
The molecular markers described above have significant weaknesses: insufficient number in the genome, inapplicability for complex polygenic traits, poor reproducibility, and labor-intensive experiments. The period of active use of dominant and codominant markers was replaced by the era of Single Nucleotide Polymorphisms (SNPs), which became possible due to advances in DNA sequencing technologies. The phylogenetic relationships of species in the genus Linum were studied using sequencing of non-coding regions of chloroplast DNA, and some hypotheses on the origin of cultivated flax were confirmed (Fu and Allaby, 2010). The combined use of EST (expressed sequence tag)-SSR and SNP markers on a doubled haploid population allowed the development of a linkage map for flax and the localization of markers for seed traits on chromosomes (Cloutier et al., 2011). However, a new level of flax research was opened by the transition from sequencing single loci to sequencing whole genomes or large sets of genomic regions. The sequencing of about 44 thousand BAC clones of the variety CDC Bethune enabled the construction of a genome-wide physical map of flax and the characterization of its genome (Ragupathy et al., 2011). The further development of flax molecular markers was largely driven by next-generation sequencing (NGS) technologies. Sequencing of reduced representation libraries for eight flax varieties identified about 55 thousand SNPs that could be useful for flax research and breeding (Kumar et al., 2012).
Quantitative Trait Locus (QTL) mapping and Genome-Wide Association Study (GWAS) allow the identification of genomic regions responsible for valuable traits. QTL mapping uses biparental populations, while GWAS is based on the analysis of unrelated individuals (Gupta et al., 2019). These approaches can greatly advance the identification of DNA markers for valuable traits and accelerate marker-assisted and genomic selection (Adlak et al., 2023; Veerendrakumar et al., 2024). In 2012, the whole genome of flax was sequenced and assembled (Wang et al., 2012), facilitating GWAS and QTL mapping in this crop. We describe the relevant studies below and summarized the data on the identified QTLs in Supplementary Table 1. Supplementary Table 1 lists QTLs associated with seed-related traits, fiber-related traits, resistance to biotic and abiotic stressors, and some other valuable flax traits. QTL coordinates on the reference genome and references to the works in which the QTLs were identified are also listed.
In many studies, authors identified hundreds or thousands of QTLs associated with different flax traits. In Supplementary Table 1, we included only the most significant QTLs, if they were highlighted by the authors of the studies.
Seed-related traits, which are particularly important for linseed, were in the focus of a significant number of works. QTLs for seed oil content and composition, seed protein content, seed yield and weight, and days to maturity were identified using the recombinant inbred line (RIL) population (Kumar et al., 2015). QTLs for seed oil traits, seed yield, and days to maturity were revealed using genome sequencing of biparental mapping populations (You et al., 2018a). Yield-related QTLs (thousand seed weight, capsule number, number of branches, fatty acid content) were also determined using Specific-Locus Amplified Fragment Sequencing (SLAF-seq) of the flax core collection (Xie et al., 2017, 2018, 2019). In addition, QTLs for seed size and weight were revealed by GWAS of the flax core collection (Guo et al., 2019), and Quantitative Trait Nucleotides (QTNs) for seed weight were identified using multi-locus GWAS (Saroha et al., 2023). QTLs associated with fatty acid composition of oil and its content in flax seeds were identified by Genotyping-By-Sequencing (GBS) of the RIL population (Zhao et al., 2020). SNPs associated with seed weight, seed oil content, days to flowering, and plant branching were revealed by GBS of a set of linseed varieties (Singh et al., 2019). Furthermore, QTLs associated with flowering time (Soto-Cerda et al., 2021) and mucilage and hull content (Soto-Cerda et al., 2018) and QTNs associated with flowering and maturity time (Saroha et al., 2022a) were identified by GWAS analysis of the flax core collections. In addition, SNPs associated with male sterility (Zhao et al., 2023) and QTLs for petal color (Guo et al., 2024) were revealed in flax using GWAS analysis.
The search for QTLs for fiber-related traits is another major direction in flax research, which is particularly important for fiber flax. QTLs for plant height, technical length, and fiber content were identified using SLAF-seq of the flax core collection (Xie et al., 2017, 2018). QTLs for plant height, technical length, straw weight, fiber content, and fiber yield were revealed using genome sequencing of biparental mapping populations (Wu et al., 2018; You et al., 2018a). QTLs for plant height and technical length were also identified using GBS of the RIL population (Zhang et al., 2018). SNPs associated with flax plant type (fiber flax or linseed) were revealed by the analysis of whole-genome sequencing data for the flax core collection (Povkhova et al., 2021). QTNs for fiber traits and plant height were revealed using GWAS (Kanapin et al., 2022; Saroha et al., 2022a).
In addition to traits related to seed and fiber, the resistance of flax plants to biotic and especially abiotic stressors is of interest. QTNs for drought resistance were identified by GWAS (Soto-Cerda et al., 2020; Sertse et al., 2021). QTLs for salt tolerance were also revealed (Li et al., 2022). QTNs for resistance to Fusarium wilt (Fusarium oxysporum f. sp. lini) were identified using GWAS (Kanapin et al., 2021) and RIL population analysis (Cloutier et al., 2024). QTLs for resistance to pasmo (Septoria linicola) were also revealed using GWAS (He et al., 2019, 2023). In addition, QTLs and QTNs for resistance to powdery mildew (Oidium lini) were identified by GWAS (Speck et al., 2022; You et al., 2022).
An important study to systematize the available data on flax QTLs and to determine their location on chromosomes of the variety CDC Bethune was published in 2020 (You and Cloutier, 2020). QTLs associated with flax resistance to biotic and abiotic stressors were summarized in the review of 2022 (Yadav et al., 2022). Fiber-related QTLs were also compiled in the study of 2024 (Gudi et al., 2024). Identification and systematization of flax QTLs are necessary to understand the genetic basis of flax traits and to effectively develop improved varieties using marker-assisted and genomic selection and genome editing. Currently, hundreds of flax genomes were sequenced, and sequencing data are available in databases. However, the lack of available and detailed descriptions of phenotypes of the sequenced samples is now becoming a more pressing problem. Such data would allow comprehensive analyses to identify the genetic determinants of valuable flax traits and to assess their diversity. In addition, almost all the studies described above used the genome assembly of the variety СDС Bethune (You et al., 2018b) as a reference. However, this genome was assembled without the use of long-read sequencing data and is less complete and accurate than flax genomes obtained later using third-generation sequencing platforms from Oxford Nanopore Technologies or Pacific Biosciences (Sa et al., 2021; Dvorianinova et al., 2022). Therefore, reanalysis of the genome sequencing data for flax samples using a more complete and accurate genome as a reference may improve the results of the QTL search. In addition, the construction of the flax pan-genome could also lead to the efficient localization of QTLs and QTNs. Furthermore, the identification of genes that play a role in the determination of valuable flax traits could be based on transcriptome data. Genes with high expression levels in specific tissues and developmental stages are likely to be involved in the processes occurring there and deserve attention for further research (Galinousky et al., 2020; Dvorianinova et al., 2023; Gorshkova et al., 2023).
Many QTLs were identified for valuable flax traits. However, QTLs for the same traits revealed in different studies often do not overlap, suggesting that the studied genotypes, the experimental design, and the data analysis have a significant influence on the obtained results. This hinders the widespread use of QTLs in flax breeding. However, some functional DNA markers, which are not only associated but specifically define valuable flax traits, were identified.
It was shown that nonsense and missense mutations in the FAD3A and FAD3B genes decrease linolenic acid and increase linoleic acid in linseed oil (Vrinten et al., 2005; Banik et al., 2011; Thambugala et al., 2013; Rajwade et al., 2016; Porokhovinova et al., 2019; Dmitriev et al., 2020). These markers are useful and are already used in linseed breeding to develop food varieties whose oil is more resistant to oxidation compared to traditional linseed varieties (Povkhova et al., 2022).
The researchers also revealed functional SNPs that determine the color of flax seeds. Mutations in the second exon of the FLAVONOID 3′5′ HYDROXYLASE (F3′5′H) gene negatively affect the synthesis of proanthocyanidins, resulting in a yellow seed coat (Sudarshan et al., 2017). Some of the SNPs in the glutathione S-transferase gene, which lead to four amino acid substitutions, are also likely to result in a yellow color of flax seed coats (Young et al., 2022). These markers are useful for flax breeding because yellow seeds may be more attractive for the use in food products (Abtahi and Mirlohi, 2024).
In addition, S-lectin receptor-like kinase (SRLK) (Lus10025891) was proposed as a candidate for a major flax resistance gene to Fusarium wilt. The SNP and indel, which resulted in amino acid substitutions in SRLK, distinguished resistant flax varieties from susceptible ones and probably determined the resistance (Cloutier et al., 2024). These markers could be used in the development of flax varieties with resistance to the harmful flax pathogen – Fusarium oxysporum f. sp. lini. Moreover, ethyl methanesulfonate induced a mutation in LuALS1 that conferred resistance to sulfonylurea herbicides in flax plants (Liu et al., 2023).
The use of functional DNA polymorphisms in plant breeding requires the utilization of simple and reliable assays. Cleaved Amplified Polymorphic Sequence (CAPS) markers were efficiently used in marker-assisted selection of crops (Shavrukov, 2014, 2016), including flax (Povkhova et al., 2022). However, less labor-intensive and more rapid methods are preferred for the analysis of large sample sets, and approaches based on fluorescence detection are promising. High-Resolution Melting (HRM) allows the identification of DNA polymorphisms based on the comparison of amplicon melting curves and is suitable for high-throughput genotyping of plants (Simko, 2016), including flax (Povkhova et al., 2022). HRM requires a DNA-intercalating dye, as opposed to fluorophore-labeled SNP-specific probes in the TaqMan method (Broccanello et al., 2018; Ayalew et al., 2019). TaqMan assays also allow for rapid and accurate genotyping of large sample sets (Woodward, 2014). Another method for fluorescence detection of DNA polymorphisms is Kompetitive Allele-Specific PCR (KASP) (He et al., 2014), which uses universal Fluorescence Resonance Energy Transfer (FRET) cassettes. FRET cassettes greatly simplify and/or cheapen genotyping, so KASP was used effectively for many crops (Dipta et al., 2024). The KASP markers were involved in QTL mapping and identification of the gene likely responsible for flax resistance to Fusarium wilt (Cloutier et al., 2024). Thus, KASP and similar approaches utilizing FRET cassettes are promising for the identification of SNPs and indels that define valuable flax traits, including in marker-assisted selection.
Microarrays were used extensively in recent years in large-scale studies where samples were needed to be analyzed for a large number of markers simultaneously. The technology allows DNA to be tested for the presence of target sequences by hybridizing to probes attached to a solid platform. This method is convenient for mass use when the same traits are analyzed in many samples, but it is disadvantageous for single use (Yang and Wei, 2015). Studies on flax using microarrays are rare (Roach and Deyholos, 2007; 2008; Fenart et al., 2010).
Targeted amplicon sequencing allows the complete sequence of a gene or locus of interest to be obtained and SNPs/indels to be analyzed (Senapathy et al., 2010; Cronn et al., 2012; Akhmetshina et al., 2020). This approach requires target enrichment, for instance by amplification or hybridization. Targeted sequencing expands horizons in areas such as plant breeding, genetics, evolutionary and phylogenetic studies (Mertes et al., 2011; Cronn et al., 2012; Andermann et al., 2020).
The above methods can be applied in flax breeding. A scheme for marker-assisted selection in flax, in which different approaches can be used to identify nucleotides at sites of interest, is shown in Figure 1. It involves collecting material from individual plants, isolating DNA, testing samples for the presence of target DNA polymorphisms, and selecting promising plants for breeding based on the obtained results. The use of these technologies will increase the efficiency of creating improved flax varieties. At the same time, marker-assisted selection in flax requires the identification of a larger number of functional polymorphisms that determine the desired traits. This can be facilitated by extensive phenotyping of those flax samples for which genome sequencing data are available. As a result, rapid and accurate determination of flax plant traits based on effective DNA test systems will take flax breeding to the next level.
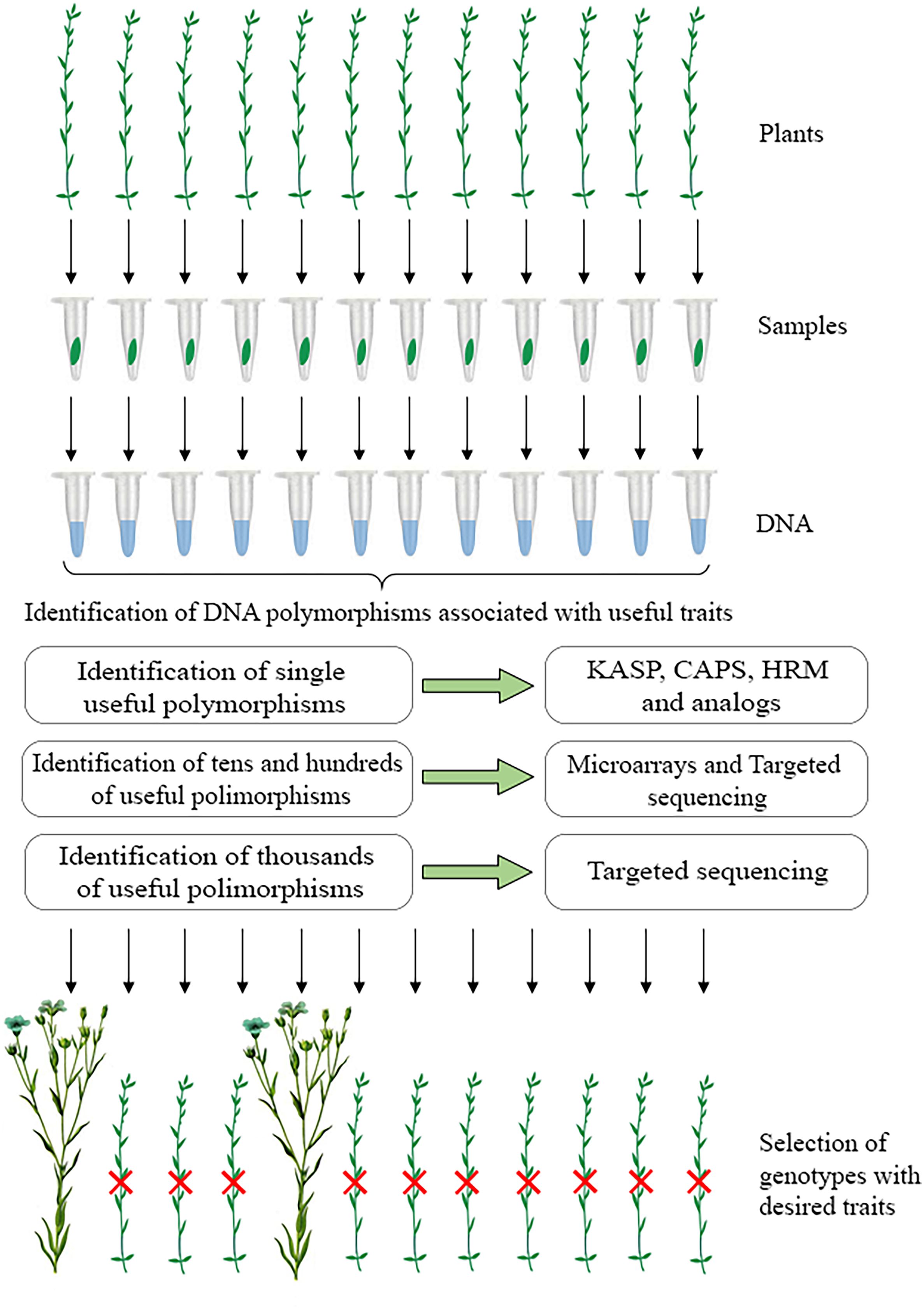
Figure 1. A scheme for marker-assisted selection in flax: collection of material from individual plants, isolation of DNA, testing samples for the presence of target DNA polymorphisms using various approaches, and selection of promising plants for breeding based on the obtained results.
DZ: Writing – original draft, Writing – review & editing. EP: Writing – original draft, Writing – review & editing. TR: Writing – original draft, Writing – review & editing. EB: Writing – original draft, Writing – review & editing. AA: Writing – original draft, Writing – review & editing. ES: Writing – original draft, Writing – review & editing. ED: Writing – original draft, Writing – review & editing. AD: Writing – original draft, Writing – review & editing. NM: Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Part of this work (review of DNA markers of valuable fiber flax traits) was financially supported by the Russian Science Foundation, grant number 24-64-00033, https://rscf.ru/project/24-64-00033/. Part of this work (review of DNA markers of valuable linseed traits) was financially supported by the Ministry of Science and Higher Education of the Russian Federation, grant number 075-15-2021-1064.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1495069/full#supplementary-material
Abbasi Holasou, H., Abdollahi Mandoulakani, B., Jafari, M., Bernousi, I. (2016). Use of IRAP and REMAP markers to interpret the population structure of Linum usitatissimum from Iran. Biologia 71, 305–315. doi: 10.1515/biolog-2016-0042
Abtahi, M., Mirlohi, A. (2024). Quality assessment of flax advanced breeding lines varying in seed coat color and their potential use in the food and industrial applications. BMC Plant Biol. 24, 60. doi: 10.1186/s12870-024-04733-1
Adlak, T., Tiwari, S., Rathore, M. S., Tripathi, N., Tiwari, P. N., Tripathi, M. (2023). Biotechnological approaches for genetic improvement of crops. book: Cutting Edge Res. Biol. 7, 63–84. doi: 10.9734/bpi/cerb/v7/5376C
Adugna, W., Labuschagne, M., Viljoen, C. (2006). The use of morphological and AFLP markers in diversity analysis of linseed. Biodiversity Conserv. 15, 3193–3205. doi: 10.1007/s10531-005-6970-8
Akhmetshina, A. O., Strygina, K. V., Khlestkina, E. K., Porokhovinova, E. A., Brutch, N. B. (2020). High-throughput sequencing techniques to flax genetics and breeding. Ecol. Genet. 18, 103–124. doi: 10.17816/ecogen16126
Al-Madhagy, S., Ashmawy, N. S., Mamdouh, A., Eldahshan, O. A., Farag, M. A. (2023). A comprehensive review of the health benefits of flaxseed oil in relation to its chemical composition and comparison with other omega-3-rich oils. Eur. J. Med. Res. 28, 240. doi: 10.1186/s40001-023-01203-6
Andermann, T., Torres Jiménez, M. F., Matos-Maraví, P., Batista, R., Blanco-Pastor, J. L., Gustafsson, A. L. S., et al. (2020). A guide to carrying out a phylogenomic target sequence capture project. Front. Genet. 10, 1407. doi: 10.3389/fgene.2019.01407
Asgarinia, P., Cloutier, S., Duguid, S., Rashid, K., Mirlohi, A., Banik, M., et al. (2013). Mapping quantitative trait loci for powdery mildew resistance in flax (Linum usitatissimum L.). Crop Sci. 53, 2462–2472. doi: 10.2135/cropsci2013.05.0298
Asyraf, M. R. M., Ishak, M. R., Norrrahim, M. N. F., Amir, A. L., Nurazzi, N. M., Ilyas, R. A., et al. (2022). Potential of flax fiber reinforced biopolymer composites for cross-arm application in transmission tower: a review. Fibers Polymers 23, 853–877. doi: 10.1007/s12221-022-4383-x
Ayalew, H., Tsang, P. W., Chu, C., Wang, J., Liu, S., Chen, C., et al. (2019). Comparison of TaqMan, KASP and rhAmp SNP genotyping platforms in hexaploid wheat. PloS One 14, e0217222. doi: 10.1371/journal.pone.0217222
Banik, M., Duguid, S., Cloutier, S. (2011). Transcript profiling and gene characterization of three fatty acid desaturase genes in high, moderate, and low linolenic acid genotypes of flax (Linum usitatissimum L.) and their role in linolenic acid accumulation. Genome 54, 471–483. doi: 10.1139/g11-013
Bibi, T., Mustafa, H. S. B., Hasan, E., Rauf, S., Mahmood, T., Ali, Q. (2015). Analysis of genetic diversity in linseed using molecular markers. Life Sci. J. 12, 28–37.
Bo, T., Ma, J., Chen, J., Miao, T., Zhai, W. (2008). Identification of specific molecular markers linked to the rust resistance gene M4 in flax. Australas. Plant Pathol. 37, 417–420. doi: 10.1071/AP08032
Bo, T. Y., Ye, H. Z., Wang, S. Q., Yang, J. C., Li, X. B., Zhai, W. X. (2002). Specific molecular markers of the rust resistance gene M4 in flax. Yi Chuan Xue Bao 29, 922–927.
Broccanello, C., Chiodi, C., Funk, A., McGrath, J. M., Panella, L., Stevanato, P. (2018). Comparison of three PCR-based assays for SNP genotyping in plants. Plant Methods 14, 28. doi: 10.1186/s13007-018-0295-6
Campos, J. R., Severino, P., Ferreira, C. S., Zielinska, A., Santini, A., Souto, S. B., et al. (2019). Linseed essential oil–source of lipids as active ingredients for pharmaceuticals and nutraceuticals. Curr. Medicinal Chem. 26, 4537–4558. doi: 10.2174/0929867325666181031105603
Chandrawati, D., Singh, N., Kumar, R., Kumar, S., Ranade, S., Kumar Yadav, H. (2017a). Agro-morphological traits and microsatellite marker's based genetic diversity in Indian genotypes of linseed (Linum usitatissimum L.). J. Agricul. Sci. Technol. 19, 707–718.
Chandrawati, D., Singh, N., Kumar, R., Kumar, S., Singh, P., Yadav, V., et al. (2017b). Genetic diversity, population structure and association analysis in linseed (Linum usitatissimum L.). Physiol. Mol. Biol. Plants 23, 207–219. doi: 10.1007/s12298-016-0408-5
Chandrawati, Maurya, R., Singh, P., Ranade, S., Yadav, H. K. (2014). Diversity analysis in Indian genotypes of linseed (Linum usitatissimum L.) using AFLP markers. Gene 549, 171–178. doi: 10.1016/j.gene.2014.07.067
Chen, C., Liu, Y. (2024). Genetic diversity and distinctness of flax (Linum usitatissimum L.) based on morphological and simple sequence repeat (SSR) markers. Genet. Resour. Crop Evol. 71, 4763–4777. doi: 10.1007/s10722-024-01933-4
Choudhary, S. B., Sharma, H. K., Kumar, A. A., Chowdhury, I., Maruthi, R., Kak, A. (2017a). Genetic diversity spectrum and marker trait association for agronomic traits in global accessions of Linum usitatissimum L. Ind. Crops Products 108, 604–615. doi: 10.1016/j.indcrop.2017.07.003
Choudhary, S. B., Sharma, H. K., Kumar, A. A., Maruthi, R. T., Mitra, J., Chowdhury, I., et al. (2017b). SSR and morphological trait based population structure analysis of 130 diverse flax (Linum usitatissimum L.) accessions. Comptes Rendus Biol. 340, 65–75. doi: 10.1016/j.crvi.2016.12.002
Cloutier, S., Edwards, T., Zheng, C., Booker, H. M., Islam, T., Nabetani, K., et al. (2024). Fine-mapping of a major locus for Fusarium wilt resistance in flax (Linum usitatissimum L.). Theor. Appl. Genet. 137, 27. doi: 10.1007/s00122-023-04528-2
Cloutier, S., Niu, Z., Datla, R., Duguid, S. (2009). Development and analysis of EST-SSRs for flax (Linum usitatissimum L.). Theor. Appl. Genet. 119, 53–63. doi: 10.1007/s00122-009-1016-3
Cloutier, S., Ragupathy, R., Miranda, E., Radovanovic, N., Reimer, E., Walichnowski, A., et al. (2012). Integrated consensus genetic and physical maps of flax (Linum usitatissimum L.). Theor. Appl. Genet. 125, 1783–1795. doi: 10.1007/s00122-012-1953-0
Cloutier, S., Ragupathy, R., Niu, Z., Duguid, S. (2011). SSR-based linkage map of flax (Linum usitatissimum L.) and mapping of QTLs underlying fatty acid composition traits. Mol. Breed. 28, 437–451. doi: 10.1007/s11032-010-9494-1
Corino, C., Rossi, R., Cannata, S., Ratti, S. (2014). Effect of dietary linseed on the nutritional value and quality of pork and pork products: Systematic review and meta-analysis. Meat Sci. 98, 679–688. doi: 10.1016/j.meatsci.2014.06.041
Cronn, R., Knaus, B. J., Liston, A., Maughan, P. J., Parks, M., Syring, J. V., et al. (2012). Targeted enrichment strategies for next-generation plant biology. Am. J. Bot. 99, 291–311. doi: 10.3732/ajb.1100356
Cullis, C. A., Swami, S., Song, Y. (1999). RAPD polymorphisms detected among the flax genotrophs. Plant Mol. Biol. 41, 795–800. doi: 10.1023/a:1006385606163
Deng, X., Long, S., He, D., Li, X., Wang, Y., Hao, D., et al. (2011). Isolation and characterization of polymorphic microsatellite markers from flax (Linum usitatissimum L.). Afr. J. Biotechnol. 10, 734–739.
Diederichsen, A., Fu, Y.-B. (2006). Phenotypic and molecular (RAPD) differentiation of four infraspecific groups of cultivated flax (Linum usitatissimum L. subsp. usitatissimum). Genet. Resour. Crop Evol. 53, 77–90. doi: 10.1007/s10722-004-0579-8
Dipta, B., Sood, S., Mangal, V., Bhardwaj, V., Thakur, A. K., Kumar, V., et al. (2024). KASP: a high-throughput genotyping system and its applications in major crop plants for biotic and abiotic stress tolerance. Mol. Biol. Rep. 51, 508. doi: 10.1007/s11033-024-09455-z
Dmitriev, A. A., Kezimana, P., Rozhmina, T. A., Zhuchenko, A. A., Povkhova, L. V., Pushkova, E. N., et al. (2020). Genetic diversity of SAD and FAD genes responsible for the fatty acid composition in flax cultivars and lines. BMC Plant Biol. 20, 301. doi: 10.1186/s12870-020-02499-w
Dvorianinova, E. M., Bolsheva, N. L., Pushkova, E. N., Rozhmina, T. A., Zhuchenko, A. A., Novakovskiy, R. O., et al. (2022). Isolating Linum usitatissimum L. nuclear DNA enabled assembling high-quality genome. Int. J. Mol. Sci. 23, 13244. doi: 10.3390/ijms232113244
Dvorianinova, E. M., Zinovieva, O. L., Pushkova, E. N., Zhernova, D. A., Rozhmina, T. A., Povkhova, L. V., et al. (2023). Key FAD2, FAD3, and SAD genes involved in the fatty acid synthesis in flax identified based on genomic and transcriptomic data. Int. J. Mol. Sci. 24, 14885. doi: 10.3390/ijms241914885
Ellegren, H. (2004). Microsatellites: simple sequences with complex evolution. Nat. Rev. Genet. 5, 435–445. doi: 10.1038/nrg1348
El Sayed, A., Ezzat, S. M., Mostafa, S., Zedan, S., Abdel-Sattar, E., El Tanbouly, N. (2018). Inter simple sequence repeat analysis of genetic diversity and relationship in four Egyptian flaxseed genotypes. Pharmacognosy Research. 10, 166–172. doi: 10.4103/pr.pr_126_17
Everaert, I., Riek, J. D., Loose, M. D., Waes, J. V., Bockstaele, E. V. (2001). Most similar variety grouping for distinctness evaluation of flax and linseed (Linum usitatissimum L.) varieties by means of AFLP and morphological data. Plant Varieties and Seeds, 14, 69–87.
Fenart, S., Ndong, Y.-P. A., Duarte, J., Rivière, N., Wilmer, J., van Wuytswinkel, O., et al. (2010). Development and validation of a flax (Linum usitatissimum L.) gene expression oligo microarray. BMC Genomics 11, 592. doi: 10.1186/1471-2164-11-592
Fombuena, V., Petrucci, R., Dominici, F., Jordá-Vilaplana, A., Montanes, N., Torre, L. (2019). Maleinized linseed oil as epoxy resin hardener for composites with high bio content obtained from linen byproducts. Polymers 11, 301. doi: 10.3390/polym11020301
Fu, Y.-B. (2005). Geographic patterns of RAPD variation in cultivated flax. Crop Sci. 45, 1084–1091. doi: 10.2135/cropsci2004.0345
Fu, Y.-B., Allaby, R. G. (2010). Phylogenetic network of Linum species as revealed by non-coding chloroplast DNA sequences. Genet. Resour. Crop Evol. 57, 667–677. doi: 10.1007/s10722-009-9502-7
Fu, Y.-B., Diederichsen, A., Richards, K., Peterson, G. (2002a). Genetic diversity within a range of cultivars and landraces of flax (Linum usitatissimum L.) as revealed by RAPDs. Genet. Resour. Crop Evol. 49, 167–174. doi: 10.1023/A:1014716031095
Fu, Y.-B., Guerin, S., Peterson, G. W., Carlson, J. E., Richards, K. (2003a). Assessment of bulking strategies for RAPD analyses of flax germplasm. Genet. Resour. Crop Evol. 50, 743–746. doi: 10.1023/A:1025015401382
Fu, Y.-B., Peterson, G. W. (2010). Characterization of expressed sequence tag-derived simple sequence repeat markers for 17 Linum species. Botany 88, 537–543. doi: 10.1139/B10-019
Fu, Y.-B., Peterson, G., Diederichsen, A., Richards, K. (2002b). RAPD analysis of genetic relationships of seven flax species in the genus Linum L. Genet. Resour. Crop Evol. 49, 253–259. doi: 10.1023/A:1015571700673
Fu, Y. B., Rowland, G. G., Duguid, S. D., Richards, K. W. (2003b). RAPD analysis of 54 North American flax cultivars. Crop Sci. 43, 1510–1515. doi: 10.2135/cropsci2003.1510
Galinousky, D., Mokshina, N., Padvitski, T., Ageeva, M., Bogdan, V., Kilchevsky, A., et al. (2020). The toolbox for fiber flax breeding: A pipeline from gene expression to fiber quality. Front. Genet. 11, 589881. doi: 10.3389/fgene.2020.589881
Gorshkova, T., Mokshina, N., Mitsuda, N., Gorshkov, O. (2023). “Key Stages of Flax Bast Fiber Development Through the Prism of Transcriptomics,” in The Flax Genome. Eds. You, F. M., Fofana, B. (
Goyal, A., Sharma, V., Upadhyay, N., Gill, S., Sihag, M. (2014). Flax and flaxseed oil: an ancient medicine & modern functional food. J. Food Sci. Technol. 51, 1633–1653. doi: 10.1007/s13197-013-1247-9
Green, A. G., Chen, Y., Singh, S. P., Dribnenki, J.C. P. (2008). “Flax”, in Compendium of Transgenic Crop Plants: Transgenic Oilseed Crops. Eds. Kole, C., Hall, T. C. (Oxford, UK: Blackwell Publishing), 199–226.
Gudi, S., M, P., Alagappan, P., Raigar, O. P., Halladakeri, P., Gowda, R. S. R., et al. (2024). Fashion meets science: how advanced breeding approaches could revolutionize the textile industry. Crit. Rev. Biotechnol. 44, 1653–1679. doi: 10.1080/07388551.2024.2314309
Guo, D., Jiang, H., Xie, L. (2024). An R2R3-MYB transcriptional factor luMYB314 associated with the loss of petal pigmentation in flax (Linum usitatissimum L.). Genes (Basel) 15, 511. doi: 10.3390/genes15040511
Guo, D., Jiang, H., Yan, W., Yang, L., Ye, J., Wang, Y., et al. (2019). Resequencing 200 flax cultivated accessions identifies candidate genes related to seed size and weight and reveals signatures of artificial selection. Front. Plant Sci. 11, 1682. doi: 10.3389/fpls.2019.01682
Gupta, P. K., Kulwal, P. L., Jaiswal, V. (2019). Association mapping in plants in the post-GWAS genomics era. Adv. Genet. 104, 75–154. doi: 10.1016/bs.adgen.2018.12.001
Habibollahi, H., Noormohammadi, Z., Sheidai, M., Farahani, F. (2015). Genetic structure of cultivated flax (Linum usitatissimum L.) based on retrotransposon-based markers. Genetika 47, 1111–1122. doi: 10.2298/GENSR1503111H
He, C., Holme, J., Anthony, J. (2014). SNP genotyping: the KASP assay. Methods Mol. Biol. 1145, 75–86. doi: 10.1007/978-1-4939-0446-4_7
He, L., Sui, Y., Che, Y., Wang, H., Rashid, K. Y., Cloutier, S., et al. (2023). Genome-wide association studies using multi-models and multi-SNP datasets provide new insights into pasmo resistance in flax. Front. Plant Sci. 14, 1229457. doi: 10.3389/fpls.2023.1229457
He, L., Xiao, J., Rashid, K. Y., Yao, Z., Li, P., Jia, G., et al. (2019). Genome-wide association studies for pasmo resistance in flax (Linum usitatissimum L.). Front. Plant Sci. 9, 1982. doi: 10.3389/fpls.2018.01982
Jhala, A. J., Hall, L. M. (2010). Flax (Linum usitatissimum L.): current uses and future applications. Aust. J. Basic Appl. Sci. 4, 4304–4312.
Kanapin, A., Bankin, M., Rozhmina, T., Samsonova, A., Samsonova, M. (2021). Genomic regions associated with Fusarium wilt resistance in flax. Int. J. Mol. Sci. 22, 12383. doi: 10.3390/ijms222212383
Kanapin, A., Rozhmina, T., Bankin, M., Surkova, S., Duk, M., Osyagina, E., et al. (2022). Genetic determinants of fiber-associated traits in flax identified by omics data integration. Int. J. Mol. Sci. 23, 14536. doi: 10.3390/ijms232314536
Kezimana, P., Dmitriev, A. A., Kudryavtseva, A. V., Romanova, E. V., Melnikova, N. V. (2018). Secoisolariciresinol diglucoside of flaxseed and its metabolites: biosynthesis and potential for nutraceuticals. Front. Genet. 9, 641. doi: 10.3389/fgene.2018.00641
Kocak, M. Z., Kaysim, M. G., Aydın, A., Erdinc, C., Kulak, M. (2023). Genetic diversity of flax genotypes (Linum usitatissimum L.) by using agro-morphological properties and molecular markers. Genet. Resour. Crop Evol. 70, 2279–2306. doi: 10.1007/s10722-023-01608-6
Kumar, S., You, F. M., Cloutier, S. (2012). Genome wide SNP discovery in flax through next generation sequencing of reduced representation libraries. BMC Genomics 13, 684. doi: 10.1186/1471-2164-13-684
Kumar, S., You, F. M., Duguid, S., Booker, H., Rowland, G., Cloutier, S. (2015). QTL for fatty acid composition and yield in linseed (Linum usitatissimum L.). Theor. Appl. Genet. 128, 965–984. doi: 10.1007/s00122-015-2483-3
Kumari, A., Paul, S., Sharma, V. (2018). Genetic diversity analysis using RAPD and ISSR markers revealed discrete genetic makeup in relation to fibre and oil content in Linum usitatissimum L. genotypes. Nucleus 61, 45–53. doi: 10.1007/s13237-017-0206-7
Lancíková, V., Žiarovská, J. (2020). Inter-retrotransposon amplified polymorphism markers revealed long terminal repeat retrotransposon insertion polymorphism in flax cultivated on the experimental fields around Chernobyl. J. Environ. Sci. Health Part A 55, 957–963. doi: 10.1080/10934529.2020.1760016
Lawrence, G., Finnegan, J., Ellis, J. (1993). Instability of the L6 gene for rust resistance in flax is correlated with the presence of a linked Ac element. Plant J. 4, 659–669. doi: 10.1046/j.1365-313x.1993.04040659.x
Li, X., Guo, D., Xue, M., Li, G., Yan, Q., Jiang, H., et al. (2022). Genome-Wide Association Study of Salt Tolerance at the Seed Germination Stage in Flax (Linum usitatissimum L.). Genes (Basel) 13, 486. doi: 10.3390/genes13030486
Liu, C., Zhang, T., Yang, X., Wang, L., Long, Y., Hasi, A., et al. (2023). A luALS mutation with high sulfonylurea herbicide resistance in Linum usitatissimum L. Int. J. Mol. Sci. 24, 2820. doi: 10.3390/ijms24032820
Melnikova, N. V., Kudryavtseva, A. V., Zelenin, A. V., Lakunina, V. A., Yurkevich, O. Y., Speranskaya, A. S., et al. (2014). Retrotransposon-based molecular markers for analysis of genetic diversity within the genus Linum. BioMed. Res. Int. 2014, 231589. doi: 10.1155/2014/231589
Mertes, F., ElSharawy, A., Sauer, S., van Helvoort, J. M., van der Zaag, P., Franke, A., et al. (2011). Targeted enrichment of genomic DNA regions for next-generation sequencing. Briefings Funct. Genomics 10, 374–386. doi: 10.1093/bfgp/elr033
Mhiret, W. N., Heslop-Harrison, J. S. (2018). Biodiversity in Ethiopian linseed (Linum usitatissimum L.): molecular characterization of landraces and some wild species. Genet. Resour. Crop Evol. 65, 1603–1614. doi: 10.1007/s10722-018-0636-3
Muravenko, O. V., Lemesh, V. A., Samatadze, T. E., Amosova, A. V., Grushetskaya, Z. E., Popov, K. V., et al. (2003). Genome comparisons with chromosomal and molecular markers for three closely related flax species and their hybrids. Russian J. Genet. 39, 414–421. doi: 10.1023/A:1023309831454
Muravenko, O. V., Yurkevich, O. Y., Bolsheva, N. L., Samatadze, T. E., Nosova, I. V., Zelenina, D. A., et al. (2009). Comparison of genomes of eight species of sections Linum and Adenolinum from the genus Linum based on chromosome banding, molecular markers and RAPD analysis. Genetica 135, 245–255. doi: 10.1007/s10709-008-9273-7
Nagabhushanam, B., Mir, M. I., Nagaraju, M., Sujatha, E., Devi, B. R., Kumar, B. K. (2021). Genetic diversity analysis of Linseed (Linum usitatissimum L.) accessions using RAPD Markers. Emirates J. Food Agric. (EJFA) 33, 589–599. doi: 10.9755/ejfa.2021.v33.i7.2736
Oh, T., Gorman, M., Cullis, C. (2000). RFLP and RAPD mapping in flax (Linum usitatissimum). Theor. Appl. Genet. 101, 590–593. doi: 10.1007/s001220051520
Osman, M. H., Badawi, M., Zayed, E., Abd El-Maksoud, R. M., Radwan, K. (2021). Genetic diversity of selected flax genotypes (Linum usitatissimum L.) based on scot, issr and ramp markers. Egyptian J. Genet. And Cytology 50, 95–117.
Ottai, M., Al-Kordy, M. A., Afiah, S. (2011). Evaluation, correlation and path coefficient analysis among seed yield and its attributes of oil flax (Linum usitatissimum L.) genotypes. Australian Journal of Basic and Applied Sciences 5, 252–258.
Parikh, M., Maddaford, T. G., Austria, J. A., Aliani, M., Netticadan, T., Pierce, G. N. (2019). Dietary flaxseed as a strategy for improving human health. Nutrients 11, 1171. doi: 10.3390/nu11051171
Porokhovinova, E. A., Shelenga, T. V., Matveeva, T. V., Pavlov, A. V., Grigorieva, E. A., Brutch, N. B. (2019). Polymorphism of genes controlling low level of linolenic acid in lines from VIR flax genetic collection. Ecol. Genet. 17, 5–19. doi: 10.17816/ecogen1725-19
Povkhova, L. V., Melnikova, N. V., Rozhmina, T. A., Novakovskiy, R. O., Pushkova, E. N., Dvorianinova, E. M., et al. (2021). Genes associated with the flax plant type (oil or fiber) identified based on genome and transcriptome sequencing data. Plants 10, 2616. doi: 10.3390/plants10122616
Povkhova, L. V., Pushkova, E. N., Rozhmina, T. A., Zhuchenko, A. A., Frykin, R. I., Novakovskiy, R. O., et al. (2022). Development and complex application of methods for the identification of mutations in the FAD3A and FAD3B genes resulting in the reduced content of linolenic acid in flax oil. Plants (Basel) 12, 95. doi: 10.3390/plants12010095
Rachinskaya, O. A., Lemesh, V. A., Muravenko, O. V., Yurkevich, O. Y., Guzenko, E. V., Bol’sheva, N. L., et al. (2011). Genetic polymorphism of flax Linum usitatissimum based on the use of molecular cytogenetic markers. Russian J. Genet. 47, 56–65. doi: 10.1134/S1022795411010108
Ragupathy, R., Rathinavelu, R., Cloutier, S. (2011). Physical mapping and BAC-end sequence analysis provide initial insights into the flax (Linum usitatissimum L.) genome. BMC Genomics 12, 217. doi: 10.1186/1471-2164-12-217
Rajwade, A. V., Arora, R. S., Kadoo, N. Y., Harsulkar, A. M., Ghorpade, P. B., Gupta, V. S. (2010). Relatedness of Indian flax genotypes (Linum usitatissimum L.): an inter-simple sequence repeat (ISSR) primer assay. Mol. Biotechnol. 45, 161–170. doi: 10.1007/s12033-010-9256-7
Rajwade, A. V., Joshi, R. S., Kadoo, N. Y., Gupta, V. S. (2016). Sequence characterization and in silico structure prediction of fatty acid desaturases in linseed varieties with differential fatty acid composition. J. Sci. Food Agric. 96, 4896–4906. doi: 10.1002/jsfa.7775
Rashid, K., Duguid, S. (2005). Inheritance of resistance to powdery mildew in flax. Can. J. Plant Pathol. 27, 404–409. doi: 10.1080/07060660509507239
Roach, M. J., Deyholos, M. K. (2007). Microarray analysis of flax (Linum usitatissimum L.) stems identifies transcripts enriched in fibre-bearing phloem tissues. Mol. Genet. Genomics 278, 149–165. doi: 10.1007/s00438-007-0241-1
Roach, M. J., Deyholos, M. K. (2008). Microarray analysis of developing flax hypocotyls identifies novel transcripts correlated with specific stages of phloem fibre differentiation. Ann. Bot. 102, 317–330. doi: 10.1093/aob/mcn110
Roose-Amsaleg, C., Cariou-Pham, E., Vautrin, D., Tavernier, R., Solignac, M. (2006). Polymorphic microsatellite loci in Linum usitatissimum. Mol. Ecol. Notes 6, 796–799. doi: 10.1111/j.1471-8286.2006.01348.x
Sa, R., Yi, L., Siqin, B., An, M., Bao, H., Song, X., et al. (2021). Chromosome-level genome assembly and annotation of the fiber flax (Linum usitatissimum) genome. Front. Genet. 12, 735690. doi: 10.3389/fgene.2021.735690
Saha, D., Hazra, C (2004). Agrihistorical genesis of flax (Linum usitatissimum LINN.). J. Agril. 9, 1–8.
Saha, D., Rana, R. S., Das, S., Datta, S., Mitra, J., Cloutier, S. J., et al. (2019). Genome-wide regulatory gene-derived SSRs reveal genetic differentiation and population structure in fiber flax genotypes. J. Appl. Genet. 60, 13–25. doi: 10.1007/s13353-018-0476-z
Saini, R. K., Prasad, P., Sreedhar, R. V., Akhilender Naidu, K., Shang, X., Keum, Y.-S. (2021). Omega–3 polyunsaturated fatty acids (PUFAs): Emerging plant and microbial sources, oxidative stability, bioavailability, and health benefits—A review. Antioxidants 10, 1627. doi: 10.3390/antiox10101627
Saroha, A., Gomashe, S. S., Kaur, V., Pal, D., Ujjainwal, S., Aravind, J., et al. (2023). Genetic dissection of thousand-seed weight in linseed (Linum usitatissimum L.) using multi-locus genome-wide association study. Front. Plant Sci. 14, 1166728. doi: 10.3389/fpls.2023.1166728
Saroha, A., Pal, D., Gomashe, S. S., Akash, Kaur, V., Ujjainwal, S., et al. (2022a). Identification of QTNs associated with flowering time, maturity, and plant height traits in Linum usitatissimum L. using genome-wide association study. Front. Genet. 13, 811924. doi: 10.3389/fgene.2022.811924
Saroha, A., Pal, D., Kaur, V., Kumar, S., Bartwal, A., Aravind, J., et al. (2022b). Agro-morphological variability and genetic diversity in linseed (Linum usitatissimum L.) germplasm accessions with emphasis on flowering and maturity time. Genet. Resour. Crop Evol. 69, 315–333. doi: 10.1007/s10722-021-01231-3
Schneeberger, R. G., Cullis, C. A. (1991). Specific DNA alterations associated with the environmental induction of heritable changes in flax. Genetics 128, 619–630. doi: 10.1093/genetics/128.3.619
Senapathy, P., Bhasi, A., Mattox, J., Dhandapany, P. S., Sadayappan, S. (2010). Targeted genome-wide enrichment of functional regions. PloS One 5, e11138. doi: 10.1371/journal.pone.0011138
Sertse, D., You, F. M., Ravichandran, S., Soto-Cerda, B. J., Duguid, S., Cloutier, S. (2021). Loci harboring genes with important role in drought and related abiotic stress responses in flax revealed by multiple GWAS models. Theor. Appl. Genet. 134, 191–212. doi: 10.1007/s00122-020-03691-0
Shavrukov, Y. (2014). Cleaved Amplified Polymorphic Sequences (CAPS) Markers in Plant Biology (New York, NY, USA: Nova Science Publishers).
Shavrukov, Y. (2016). CAPS markers in plant biology. Russian J. Genetics: Appl. Res. 6, 279–287. doi: 10.1134/S2079059716030114
Simko, I. (2016). High-resolution DNA melting analysis in plant research. Trends Plant Sci. 21, 528–537. doi: 10.1016/j.tplants.2016.01.004
Singh, N., Agarwal, N., Yadav, H. K. (2019). Genome-wide SNP-based diversity analysis and association mapping in linseed (Linum usitatissimum L.). Euphytica 215, 1–11. doi: 10.1007/s10681-019-2462-x
Singh, P., Akram, M., Srivastava, R. (2009). Genetic diversity in linseed (Linum usitatissimum) cultivars based on RAPD-analysis. Indian J. Agric. Sci. 79, 1046–1049.
Singh, N., Kumar, R., Kumar, S., Singh, P. K., Yadav, H. K. (2021). Mapping QTLs for Alternaria blight in Linseed (Linum usitatissimum L.). 3 Biotech. 11, 91. doi: 10.1007/s13205-020-02638-y
Singh, K., Mridula, D., Rehal, J., Barnwal, P. (2011). Flaxseed: a potential source of food, feed and fiber. Crit. Rev. Food Sci. Nutr. 51, 210–222. doi: 10.1080/10408390903537241
Smýkal, P., Bačová-Kerteszová, N., Kalendar, R., Corander, J., Schulman, A. H., Pavelek, M. (2011). Genetic diversity of cultivated flax (Linum usitatissimum L.) germplasm assessed by retrotransposon-based markers. Theor. Appl. Genet. 122, 1385–1397. doi: 10.1007/s00122-011-1539-2
Soto-Cerda, B. J., Aravena, G., Cloutier, S. (2021). Genetic dissection of flowering time in flax (Linum usitatissimum L.) through single- and multi-locus genome-wide association studies. Mol. Genet. Genomics 296, 877–891. doi: 10.1007/s00438-021-01785-y
Soto-Cerda, B. J., Cloutier, S., Gajardo, H. A., Aravena, G., Quian, R. (2019). Identifying drought-resilient flax genotypes and related-candidate genes based on stress indices, root traits and selective sweep. Euphytica 215, 1–16. doi: 10.1007/s10681-019-2362-0
Soto-Cerda, B. J., Cloutier, S., Gajardo, H. A., Aravena, G., Quian, R., You, F. M. (2020). Drought response of flax accessions and identification of quantitative trait nucleotides (QTNs) governing agronomic and root traits by genome-wide association analysis. Mol. Breed. 40, 1–24. doi: 10.1007/s11032-019-1096-y
Soto-Cerda, B. J., Cloutier, S., Quian, R., Gajardo, H. A., Olivos, M., You, F. M. (2018). Genome-wide association analysis of mucilage and hull content in flax (Linum usitatissimum L.) seeds. Int. J. Mol. Sci. 19, 2870. doi: 10.3390/ijms19102870
Soto-Cerda, B. J., Diederichsen, A., Duguid, S., Booker, H., Rowland, G., Cloutier, S. (2014). The potential of pale flax as a source of useful genetic variation for cultivated flax revealed through molecular diversity and association analyses. Mol. Breed. 34, 2091–2107. doi: 10.1007/s11032-014-0165-5
Soto-Cerda, B. J., Diederichsen, A., Ragupathy, R., Cloutier, S. (2013). Genetic characterization of a core collection of flax (Linum usitatissimum L.) suitable for association mapping studies and evidence of divergent selection between fiber and linseed types. BMC Plant Biol. 13, 78. doi: 10.1186/1471-2229-13-78
Soto-Cerda, B. J., Maureira-Butler, I., Muñoz, G., Rupayan, A., Cloutier, S. (2012). SSR-based population structure, molecular diversity and linkage disequilibrium analysis of a collection of flax (Linum usitatissimum L.) varying for mucilage seed-coat content. Mol. Breed. 30, 875–888. doi: 10.1007/s11032-011-9670-y
Soto-Cerda, B. J., Saavedra, H. U., Navarro, C. N., Ortega, P. M. (2011). Characterization of novel genic SSR markers in Linum usitatissimum (L.) and their transferability across eleven Linum species. Electronic J. Biotechnol. 14, 4–4. doi: 10.2225/vol14-issue2-fulltext-6
Speck, A., Trouve, J. P., Enjalbert, J., Geffroy, V., Joets, J., Moreau, L. (2022). Genetic architecture of powdery mildew resistance revealed by a genome-wide association study of a worldwide collection of flax (Linum usitatissimum L.). Front. Plant Sci. 13, 871633. doi: 10.3389/fpls.2022.871633
Spielmeyer, W., Green, A., Bittisnich, D., Mendham, N., Lagudah, E. (1998). Identification of quantitative trait loci contributing to Fusarium wilt resistance on an AFLP linkage map of flax (Linum usitatissimum). Theor. Appl. Genet. 97, 633–641. doi: 10.1007/s001220050939
Stegniî, V., IuV, C., Salina, E. (2000). RAPD analysis of the flax (Linum usitatissimum L.) varieties and hybrids of various productivity. Genetika 36, 1370–1373.
Sudarshan, G. P., Kulkarni, M., Akhov, L., Ashe, P., Shaterian, H., Cloutier, S., et al. (2017). QTL mapping and molecular characterization of the classical D locus controlling seed and flower color in Linum usitatissimum (flax). Sci. Rep. 7, 15751. doi: 10.1038/s41598-017-11565-7
Thambugala, D., Duguid, S., Loewen, E., Rowland, G., Booker, H., You, F. M., et al. (2013). Genetic variation of six desaturase genes in flax and their impact on fatty acid composition. Theor. Appl. Genet. 126, 2627–2641. doi: 10.1007/s00122-013-2161-2
Uysal, H., Fu, Y.-B., Kurt, O., Peterson, G. W., Diederichsen, A., Kusters, P. (2010). Genetic diversity of cultivated flax (Linum usitatissimum L.) and its wild progenitor pale flax (Linum bienne Mill.) as revealed by ISSR markers. Genet. Resour. Crop Evol. 57, 1109–1119. doi: 10.1007/s10722-010-9551-y
Van Treuren, R., Van Soest, L., Van Hintum, T. J. (2001). Marker-assisted rationalisation of genetic resource collections: a case study in flax using AFLPs. Theor. Appl. Genet. 103, 144–152. doi: 10.1007/s001220100537
Veerendrakumar, H., Barmukh, R., Shah, P., Bomireddy, D., Jamedar, H. V. R., Roorkiwal, M., et al. (2024). “Genomic Selection in Crop Improvement,” in Frontier Technologies for Crop Improvement Eds. Pandey, M. K., Bentley, A., Desmae, H., Roorkiwal, M., Varshney, R. K. (Singapore: Springer, Singapore), 187–222.
Vrinten, P., Hu, Z., Munchinsky, M.-A., Rowland, G., Qiu, X. (2005). Two FAD3 desaturase genes control the level of linolenic acid in flax seed. Plant Physiol. 139, 79–87. doi: 10.1104/pp.105.064451
Wakjira, A., Viljoen, C., Labuschagne, M. (2005). Analysis of genetic diversity in linseed using AFLP markers. SINET: Ethiopian J. Sci. 28, 41–50. doi: 10.4314/sinet.v28i1.18230
Wang, Z., Hobson, N., Galindo, L., Zhu, S., Shi, D., McDill, J., et al. (2012). The genome of flax (Linum usitatissimum) assembled de novo from short shotgun sequence reads. Plant J. 72, 461–473. doi: 10.1111/j.1365-313X.2012.05093.x
Wiesnerova, D., Wiesner, I. (2004). ISSR-based clustering of cultivated flax germplasm is statistically correlated to thousand seed mass. Mol. Biotechnol. 26, 207–214. doi: 10.1385/MB:26:3:207
Woodward, J. (2014). Bi-allelic SNP genotyping using the TaqMan® assay. Crop Breeding: Methods Protoc. Eds. Fleury, D., Whitford, R. (New York, NY, USA: Springer), 67–74. doi: 10.1007/978-1-4939-0446-4_6
Wu, J., Zhao, Q., Wu, G., Zhang, S., Jiang, T. (2017). Development of novel SSR markers for flax (Linum usitatissimum L.) using reduced-representation genome sequencing. Front. Plant Sci. 7, 2018. doi: 10.3389/fpls.2016.02018
Wu, J., Zhao, Q., Zhang, L., Li, S., Ma, Y., Pan, L., et al. (2018). QTL mapping of fiber-related traits based on a high-density genetic map in flax (Linum usitatissimum L.). Front. Plant Sci. 9, 885. doi: 10.3389/fpls.2018.00885
Xie, D., Dai, Z., Yang, Z., Sun, J., Zhao, D., Yang, X., et al. (2017). Genome-wide association study identifying candidate genes influencing important agronomic traits of flax (Linum usitatissimum L.) using SLAF-seq. Front. Plant Sci. 8, 2232. doi: 10.3389/fpls.2017.02232
Xie, D., Dai, Z., Yang, Z., Tang, Q., Deng, C., Xu, Y., et al. (2019). Combined genome-wide association analysis and transcriptome sequencing to identify candidate genes for flax seed fatty acid metabolism. Plant Sci. 286, 98–107. doi: 10.1016/j.plantsci.2019.06.004
Xie, D., Dai, Z., Yang, Z., Tang, Q., Sun, J., Yang, X., et al. (2018). Genomic variations and association study of agronomic traits in flax. BMC Genomics 19, 512. doi: 10.1186/s12864-018-4899-z
Yadav, B., Kaur, V., Narayan, O. P., Yadav, S. K., Kumar, A., Wankhede, D. P. (2022). Integrated omics approaches for flax improvement under abiotic and biotic stress: Current status and future prospects. Front. Plant Sci. 13, 931275. doi: 10.3389/fpls.2022.931275
Yang, C., Wei, H. (2015). Designing microarray and RNA-Seq experiments for greater systems biology discovery in modern plant genomics. Mol. Plant 8, 196–206. doi: 10.1016/j.molp.2014.11.012
You, F. M., Cloutier, S. (2020). Mapping quantitative trait loci onto chromosome-scale pseudomolecules in flax. Methods Protoc. 3, 28. doi: 10.3390/mps3020028
You, F. M., Moumen, I., Khan, N., Cloutier, S. (2023). “Reference genome sequence of flax,” in The Flax Genome. Eds You, F. M., Fofana, B. (Cham, Switzerland: Springer), 1–17.
You, F. M., Rashid, K. Y., Zheng, C., Khan, N., Li, P., Xiao, J., et al. (2022). Insights into the Genetic Architecture and Genomic Prediction of Powdery Mildew Resistance in Flax (Linum usitatissimum L.). Int. J. Mol. Sci. 23, 4960. doi: 10.3390/ijms23094960
You, F. M., Xiao, J., Li, P., Yao, Z., Jia, G., He, L., et al. (2018a). Genome-wide association study and selection signatures detect genomic regions associated with seed yield and oil quality in flax. Int. J. Mol. Sci. 19, 2303. doi: 10.3390/ijms19082303
You, F. M., Xiao, J., Li, P., Yao, Z., Jia, G., He, L., et al. (2018b). Chromosome-scale pseudomolecules refined by optical, physical and genetic maps in flax. Plant J. 95, 371–384. doi: 10.1111/tpj.2018.95.issue-2
Young, L., Akhov, L., Kulkarni, M., You, F., Booker, H. (2022). Fine-mapping of a putative glutathione S-transferase (GST) gene responsible for yellow seed colour in flax (Linum usitatissimum). BMC Res. Notes 15, 72. doi: 10.1186/s13104-022-05964-x
Zhang, J., Long, Y., Wang, L., Dang, Z., Zhang, T., Song, X., et al. (2018). Consensus genetic linkage map construction and QTL mapping for plant height-related traits in linseed flax (Linum usitatissimum L.). BMC Plant Biol. 18, 160. doi: 10.1186/s12870-018-1366-6
Zhao, X., Yi, L., Zuo, Y., Gao, F., Cheng, Y., Zhang, H., et al. (2023). High-quality genome assembly and genome-wide association study of male sterility provide resources for flax improvement. Plants (Basel) 12, 2773. doi: 10.3390/plants12152773
Zhao, L., Zhao, W., Wang, L., Wang, B., Dang, Z., Li, W. (2020). Construction of A High-Density SNP Genetic Map by Genotyping-By-Sequencing and Location of Seed Quality traits in Flax (Linum usitatisimum L.). Res. Square. doi: 10.21203/rs.3.rs-70683/v1
Keywords: flax, Linum usitatissimum L., DNA polymorphisms, DNA markers, GWAS, QTL, QTN, marker-assisted selection
Citation: Zhernova DA, Pushkova EN, Rozhmina TA, Borkhert EV, Arkhipov AA, Sigova EA, Dvorianinova EM, Dmitriev AA and Melnikova NV (2025) History and prospects of flax genetic markers. Front. Plant Sci. 15:1495069. doi: 10.3389/fpls.2024.1495069
Received: 12 September 2024; Accepted: 10 December 2024;
Published: 15 January 2025.
Edited by:
Parimalan Rangan, Indian Council of Agricultural Research (ICAR), IndiaReviewed by:
Dhammaprakash Pandhari Wankhede, Indian Council of Agricultural Research (ICAR), IndiaCopyright © 2025 Zhernova, Pushkova, Rozhmina, Borkhert, Arkhipov, Sigova, Dvorianinova, Dmitriev and Melnikova. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daiana A. Zhernova, emhlcm5vdmEuZEB5YS5ydQ==; Nataliya V. Melnikova, bW52LTQ1MjkyNjRAeWFuZGV4LnJ1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.