- 1College of Food and Biological Engineering, Chengdu University, Chengdu, China
- 2Chengdu Institute of Biology, Chinese Academy of Sciences, Chengdu, China
Introduction: Rhodiola crenulata (Hook.f. & Thomson) H.Ohba, a member of the Crassulaceae family, is a traditional Chinese medicine recognized as the original source of Rhodiolae Crenulatae Radix et Rhizoma in the 2020 edition of the China Pharmacopoeia. It has been widely used in both Asia and Europe to enhance stress resistance and reduce fatigue. However, the classification of Rhodiola species can lead to confusion, raising safety concerns in the herbal medicine market.
Methods: The cleaved amplified polymorphic sequence (CAPS) RT-PCR was used to identify the single nucleotide polymorphism (SNP) sites, based on which the loop-mediated isothermal amplification (LAMP) was employed to develop the method in Rh. crenulata identification. The specific loop backward primers, reaction temperature, reaction time, and color indicators were screened and optimized.
Results: Single nucleotide polymorphism (SNP) sites were identified between Rh. crenulata and two closely related species. Based on the identified SNP sites, the optimal real-time fluorescence LAMP system to identify Rh. crenulata was constructed with the most efficient specific loop backward primers, reaction temperature. The final detection system exhibited a sensitivity of up to 1,000 copies of the target DNA, maintaining a constant reaction temperature of 62°C within 35 minutes. To facilitate visualization, we incorporated two color indicators, hydroxy naphthol blue (HNB) and neutral red (N-red), into the reaction system.
Discussion: Collectively, we developed a simple, rapid, specific, sensitive, and visible method to distinguish Rh. crenulata from other two Rhodiola species and Rh. crenulata hybrids. This approach has significant potential for applications in pharmaceutical industry.
1 Introduction
Rhodiola plants, belonging to the Crassulaceae family, are important medicinal resources known for their dried roots and rhizomes. In China, 73 species of Rhodiola are recorded, primarily in high-altitude and cold regions. Among them, Rhodiola crenulata (Hook.f. & Thomson) H.Ohba is uniquely recognized as the original source of Rhodiolae Crenulatae Radix et Rhizoma in the Chinese Pharmacopoeia, 2005 to 2020 edition, while several Rhodiola species, such as Rhodiola kirilowii (Regel) Maxim., play significant roles in traditional medicine and are noted in Xizang monographs (Tao et al., 2019). Rhodiola species possess a variety of medicinal properties and have been traditionally used to enhance physical resistance to stress (Fan et al., 2016; Li et al., 2017; Tinsley et al., 2024) and to reduce fatigue (Jiao et al., 2023; Shikov et al., 2014; Yue et al., 2022) across both Asia and Europe, particularly in Xizang (Tao et al., 2019). Additionally, these plants are recognized as tonics or functional food (Recio et al., 2016). The significant demand for Rhodiola species has led to resource depletion due to overexploitation. Moreover, the presence of counterfeits and substitutes, including Rhodiola fastigiata (Hook.f. & Thomson) S.H.Fu, Rh. kirilowii, and other similarly named Rhodiola species, has caused confusion in the herbal medicine market (Tao et al., 2019).
The active ingredients and their concentrations vary among different Rhodiola species (Panossian et al., 2010). Mixing these medicinal materials can directly affect clinical safety and efficacy (Xin et al., 2015). However, distinguishing between Rhodiola species based on morphology and microscopic characteristics is challenging due to similarities in their roots and rhizomes. While liquid chromatography–mass spectrometry (LS-MS) analysis is effective for identifying medicinal plants, it can be time-consuming (Ganzera and Sturm, 2018; Kuwahara et al., 2019). Thus, there is an urgent need for a rapid and accurate method to differentiate Rh. crenulata from other Rhodiola species.
DNA-based molecular techniques have gained popularity for species identification due to their accuracy and efficiency (Ganie et al., 2015; Grazina et al., 2020). DNA barcoding methods, such as the nuclear ribosomal internal transcribed spacer 2 (ITS2), the large subunit of ribulose-bisphosphate carboxylase (rbcL), maturase K (matK), and psbA-trnH, have received considerable attention (Feng et al., 2015; Intharuksa et al., 2020; Mahadani and Ghosh, 2013; Mishra et al., 2016; Xu et al., 2022). Chen et al. (2010) first proposed ITS2 as a universal DNA barcode for identifying medicinal plants. Typically, distinguishing species within the same genus relies on detecting single nucleotide polymorphism (SNP) sites. In such cases, techniques like polymerase chain reaction (PCR) (Han et al., 2018), restriction fragment length polymorphism (RFLP) (Xu et al., 2022), cleaved amplified polymorphic sequence (CAPS) reverse transcription polymerase chain reaction (RT-PCR) (Hao et al., 2020), and derived CAPS (dCAPS) RT-PCR (Hao et al., 2023) are commonly used. However, these methods can be time-consuming and dependent on thermal cyclers. In contrast, loop-mediated isothermal amplification (LAMP) is a thermal cycler-free method known for its high sensitivity and rapid performance at a constant temperature. LAMP employs a DNA polymerase with strand-displacement activity and two pairs of primers for sequence amplification (Soroka et al., 2021). To enhance the reaction rate and visualize results, loop primer and colorimetric sensors, such as hydroxy naphthol blue (HNB) and neutral red (N-red), can be incorporated (Saejung et al., 2024; Yang et al., 2024). Recently, LAMP has gained traction for SNP detection and genotyping (Ding et al., 2019; Hyman et al., 2022; Sharafdarkolaee et al., 2019), demonstrating great potential for identifying Rhodiola species.
In this study, we identified several SNPs in the ITS2 region among various Rhodiola species. Based on one of these SNP sites and the LAMP technique, we developed a rapid, convenient, and efficient method for visually distinguishing Rh. crenulata from the common substitutes Rh. kirilowii and Rh. fastigiata, as well as Rh. crenulata hybrids, showcasing significant future application prospects.
2 Materials and methods
2.1 Plant materials
The plant materials for this study included Rh. crenulata [voucher specimen: Y.S. Chen et al. 13-0349 (PE)], Rh. kirilowii [voucher specimen: FLPH Xizang Expedition 12-0465 (PE)], Rh. fastigiata [voucher specimen: PE Xizang Expedition PE6468 (PE)], and a hybrid (Rh. crenulata × Rh. fastigiata). These species were provided by the Xizang Rhodiola Pharmaceutical Holding Company, Xizang, China, and were identified by professor Yuehua Wang of Chengdu University. Detailed collection information is provided in Supplementary Table S1. The 12 batches of Rhodiola decoction pieces were purchased from the Lotus Pond Chinese herbal medicine market in Chengdu, Sichuan province, China.
2.2 SNP analysis among Rh. crenulata, Rh. Kirilowii, and Rh. fastigiata
A total of 149 ITS2 sequences of Rhodiola species, including Rh. crenulata, Rh. kirilowii, and Rh. fastigiata, were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/), with accession numbers listed in Supplementary Table S2. The sequences were aligned and trimmed using MEGA 11, and nucleotide diversity (Pi) was calculated using DnaSP6, with a window width and step size set to 1. The Pi values were used to identify potential SNP sites and were visualized using GraphPad Prism 9. The concentrated region (from position 162 bp to 177 bp) containing the potential SNP sites was analyzed for base preference using the R package ggseqlogo with default parameters.
Genomic DNA was extracted from the leaves and decoction pieces of the Rhodiola species using the Plant Genomic DNA Kit (TIANGEN Biotech Co., Ltd., Beijing, China). The concentration of the extracted DNA was measured using a NanoDrop spectrophotometer (Thermo Fisher Scientific Corporation, USA). The ITS2 fragments were amplified with primers RhITS1-F and RhITS4-R (Supplementary Table S3) under standard PCR conditions and sequenced by Sangon Biotech Co., Ltd. (Shanghai, China). The sequencing results were aligned in DNAMAN 7.0 software to confirm the interspecific SNP sites between Rh. crenulata and the other two Rhodiola species.
2.3 CAPS analysis
The ITS2 fragments were amplified from the extracted DNA using primers RhITS1-F and RhITS4-R (Supplementary Table S3) in a reaction volume of 20 μL. The PCR products were then purified and digested with BglI (New England Biolabs, USA). Following a digestion period of 2 h, the products were separated by 2% agarose gel electrophoresis for band detection.
2.4 Plasmid construction and template preparation for LAMP reaction
The ITS2 sequences from Rh. crenulata and Rh. fastigiata were cloned into the pTOPO vector using the CV15-Zero Background pTOPO-TA Simple Cloning Kit (Aidlab Biotechnologies Co., Ltd., Beijing, China). The recombined plasmids were subsequently sequenced and verified by Sangon Biotech Co., Ltd. (Shanghai, China). Following purification with the HighPure Plasmid Mini Kit (Aidlab Biotechnologies Co., Ltd., Beijing, China) and concentration measurement using a NanoDrop spectrophotometer (Thermo Fisher Scientific Corporation, USA), the plasmids were diluted to different copy numbers (from 108 to 101) based on the following formula:
2.5 LAMP primer design
The LAMP primers, including F3, B3, F1C-F2 (FIP), B1C-B2 (BIP), and the backward-side loop primer (LB), were designed based on the SNP region in the ITS2 sequence using the online software PrimerExplorer V5 (http://primerexplorer.jp/lampv5e/index.html). The conventional LAMP primer set consisted of two inner primers (FIP and BIP) and two outer primers (F3 and B3). The SNP sites were flanked by the inner primers (B1 and B2) and located in the loop domain of the LAMP products. Specific LB primers (LBRc1/2/3 for Rh. crenulata and LBRh1/2/3 for the other two species) were designed based on the first SNP-concentrated region. LBRc1/2 and LBRh1/2 differed in the number of SNPs but were similar in length [11 nucleotides (nt)] and had SNPs located at the 5′ and 3′ ends, as well as in the middle of the primers. LBRc3 and LBRh3, each 17 nt long, contained four SNP sites distributed at the 5′ end and the middle of the primers. All primers are listed in Supplementary Table S3.
2.6 Real-time fluorescence LAMP reaction
The total volume of the real-time fluorescence LAMP reaction was 25 μL, which included the following components: 10 × Isothermal Amplification Buffer, 8 mM MgSO4, 1.4 mM dNTPs, 1.6 μM of each FIP and BIP primer, 0.2 μM of each F3 and B3 primer, 0.4 μM LB primer, 8 U Bst 2.0 WarmStart DNA polymerase (New England Biolabs, USA), 0.5 μL of 50 × LAMP fluorescence dye (New England Biolabs, USA), and 1 μL of plasmid template (104 copies). The amplification reaction was performed at a constant temperature for 90 min using the QuantStudio 3 Real-Time PCR System (Applied Biosystems, California, USA). All real-time fluorescence LAMP reactions were performed with three biological replicates, each consisting of three technical replicates.
2.7 The sensitivity and specificity analysis of the LAMP method
The sensitivity of the LAMP reaction was evaluated using a 10-fold serial dilution of the plasmid template, ranging from 108 to 101 copies. The specificity of LBRh3 was assessed by mixing plasmid templates with the same total copy number (106) but varying the ratios of RfITS2 to RcITS2 (100%, 10%, 1%, 0.1%, and 0%).
2.8 Visualization of LAMP detection
HNB and N-red were used as colorimetric indicators for visualized LAMP detection. In the reaction system, the 50 × LAMP fluorescence dye was replaced with either 0.12 mM HNB or 0.1 mM N-red, along with 1 μL of plasmid template (104 copies) or 0.5 μL of DNA (20 ng/μL) extracted from Rhodiola species. The remaining components were consistent with those in the real-time fluorescence LAMP reaction system described previously. The reaction was conducted at 62°C for 35 min using a VeritiPro 96-Well Thermal Cycler (Applied Biosystems, California, USA). The final visualized results were captured using a Sony α6000 camera (Sony, Japan) against a white background. All visualized LAMP reactions were performed in triplicate.
2.9 Statistical analysis
The real-time fluorescence signals of the LAMP reactions were collected using the QuantStudio 3 Real-Time PCR System and visualized with GraphPad Prism 9 or OriginPro 2022. The cycle quantification (Cq) value was defined as the detection time. All experiments were conducted in triplicate. Statistical analyses were performed using Duncan’s multiple range test via one-way ANOVA or two-tailed Student’s t-test in SPSS 25.
3 Results
3.1 SNP identification between Rh. crenulata and the other two Rhodiola species
To differentiate Rh. crenulata from its counterfeits and substitutes, we focused on the ITS2 gene to identify potential SNPs. We obtained a total of 149 sequences of three Rhodiola species from GenBank, including Rh. crenulata, Rh. fastigiata, and Rh. kirilowii (Supplementary Table S2). Nucleotide diversity (Pi) was used to access interspecific SNPs between Rh. crenulata and the other species. We identified multiple sites with higher interspecific Pi values (>0.37) and lower intraspecific Pi values (<0.19) (Figure 1A; Supplementary Table S4), which were considered candidate SNPs. These sites were concentrated in two fragments, each 50 nt long (Figure 1A).

Figure 1. SNP analysis and identification of Rh. crenulata and two other Rhodiola species. (A) Nucleotide diversity (Pi) of the ITS2 region in Rh. crenulata and two other Rhodiola species. A high Pi value indicated significant hypervariability in nucleotides, either intra- or interspecifically. SNP-concentrated regions are marked with blue double bracket. (B) Base preference analysis of the first SNP-concentrated region (162 to 177 bp). Potential SNP sites are highlighted with red stars. (C) Sequencing peak map of PCR products from four Rhodiola species. The SNP site is indicated by a red box. (D) Schematic diagram illustrating the BglI restriction site in the ITS2 region across Rhodiola species. (E) CAPS analysis of PCR products from the four Rhodiola species. Products from Rh. crenulata were cleaved into two fragments (232 and 462 bp), while those from Rh. crenulata hybrid (Rh. crenulata × Rh. fastigiata) were cleaved into three fragments (232, 462, and 694 bp). The PCR products from Rh. fastigiata and Rh. kirilowii remained uncleaved. Marker, DL2000 DNA ladder.
To further verify the differentiation among the Rhodiola species, we conducted a base preference analysis. This revealed five specific nucleotide preferences in Rh. crenulata within the first SNP-concentrated region (position 162 bp to 177 bp): C162, C166, C167, A172, and T177. In contrast, Rh. fastigiata and Rh. kirilowii were more likely to have T162, T166, T167, G172, and A177 at these positions (Figure 1B). A more dispersed nucleotide divergence was observed between Rh. crenulata and the other two Rhodiola species in the second SNP-concentrated region (Supplementary Figure S1A). Consequently, the five SNP sites in the first region were considered critical for distinguishing Rh. crenulata.
To determine whether these SNP sites were homozygous, we amplified ITS2 fragments from the DNA of each Rhodiola species and sequenced the PCR products. All sites in the first SNP-concentrated region exhibited single peaks, confirming their homozygosity in Rh. crenulata, Rh. fastigiata, and Rh. kirilowii (Figure 1C; Supplementary Figure S1B). However, double peaks were observed at the SNP positions in the Rh. crenulata hybrid (Figure 1C). Notably, a key restriction enzyme site (BglI) was formed by the specific bases C166 and C167 in Rh. crenulata (Figure 1D). CAPS analysis demonstrated that the PCR products amplified from Rh. crenulata were cleaved into two fragments (462 and 232 bp) by BglI, while those from Rh. fastigiata and Rh. kirilowii were not cleaved (Figure 1E). Moreover, we amplified the same 694-bp ITS2 fragment from the Rh. crenulata hybrid (Rh. crenulata × Rh. fastigiata) (Supplementary Figure S1C), which resulted in three fragments (232 bp, 462 bp, and 694 bp) following digestion with BglI (Figure 1E). These results indicated that positions 166 and 167 were key SNP sites for effectively distinguishing Rh. crenulata from the other two Rhodiola species and its hybrid.
3.2 Application design of the LAMP technique in Rh. crenulata identification
To rapidly distinguish Rh. crenulata from the other two Rhodiola species, we employed the LAMP technique, which eliminated the need for thermocycling equipment and simplified the procedures. The primers and strategy for differentiating Rh. crenulata are illustrated in Figure 2. Conventional LAMP primers (FIP, BIP, F3, and B3) were designed to amplify the ITS2 fragments in Rhodiola species, while specific LB primers targeting the first SNP-concentrated region were used to enhance the LAMP reaction. The design and binding characteristics of the LB primers significantly influence the amplification efficiency. The LBRc primers (LBRc1/2/3) were perfectly complementary to the SNP region of Rh. crenulata but exhibited mismatches at the 5′ and 3′ ends, as well as in the middle regions, preventing effective binding to the SNP regions of the other two Rhodiola species (Figure 2B). Consequently, Rh. crenulata was expected to be detected earlier than the other two species using the specific LBRc primer. Similarly, the specific LBRh primer facilitated the earlier detection of Rh. fastigiata and Rh. kirilowii. This allowed for the identification of optimal reaction times to distinguish different Rhodiola species within the LAMP system. To enhance detection visibility, colorimetric indicators such as HNB and N-red were incorporated into the LAMP reaction.
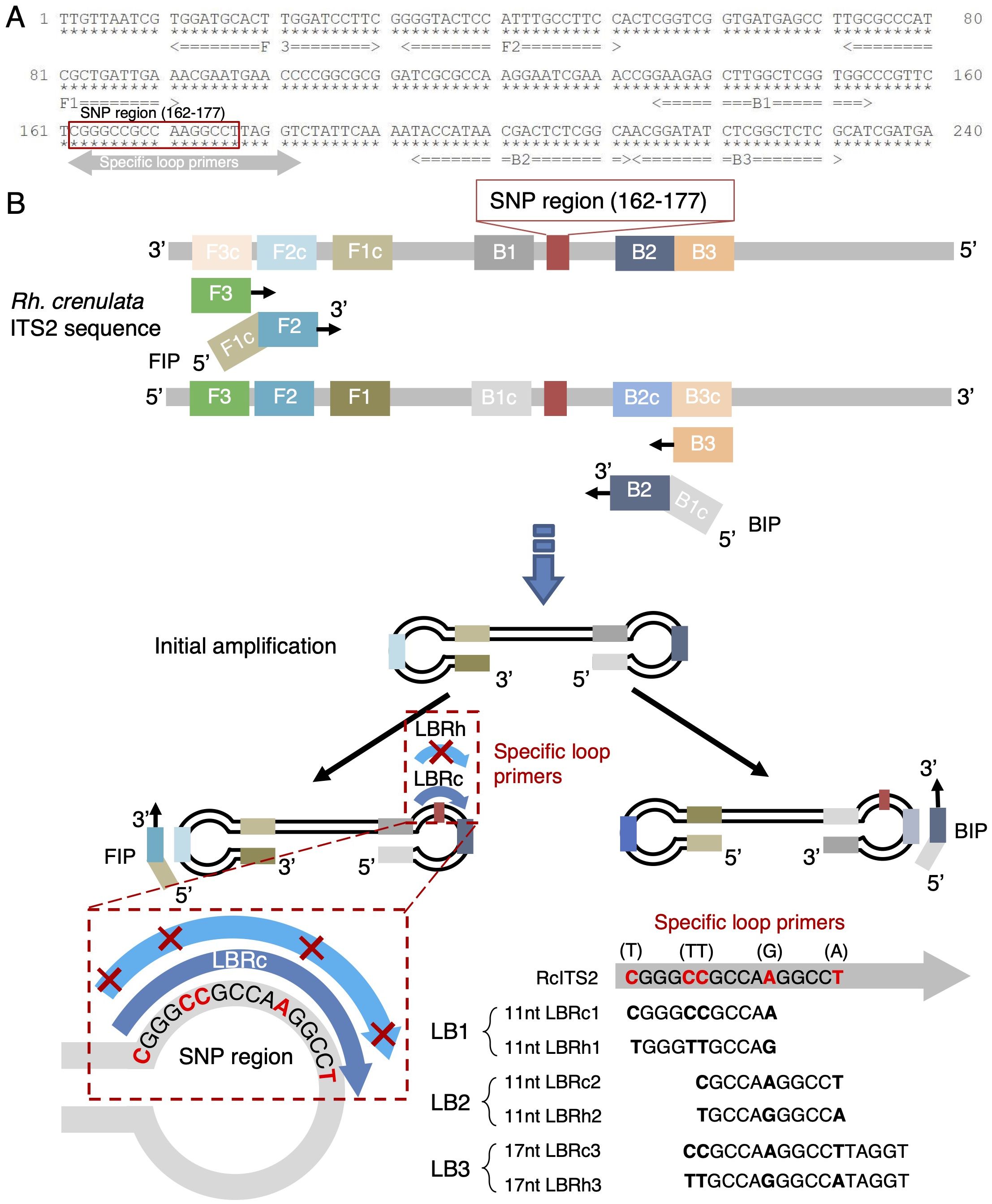
Figure 2. Schematic diagram of LAMP detection of Rhodiola species. (A) Location of LAMP primers within the ITS2 region of Rh. crenulata. The red box indicates the SNP region, while the double-headed gray arrow shows the positions of the specific loop primers. The stars under the nucleotides are for position clarification. (B) Overview of LAMP detection for Rhodiola species. The SNP region is highlighted in red, with bolded SNP sites shown in the specific loop primers. The symbol “✕” in LB-Rh represents non-complementary paring sites.
3.3 Optimization of the LAMP reactions
To optimize the LAMP reaction conditions for identifying Rh. crenulata, we constructed two plasmids containing the ITS2 sequences from Rh. crenulata and Rh. fastigiata, which served as reaction templates. We then conducted a series of real-time fluorescence LAMP reactions using specific primers at temperatures ranging from 56°C to 64°C in 2°C intervals to verify the most effective LB primer and reaction temperature for identifying Rhodiola species. The Cq value was used to indicate the detection time of the target species. Results revealed that the mean Cq value for the LBRc3 primer was consistently lower than that of LBRc1/2 across all reaction temperatures, particularly at 62°C, where its Cq value was significantly lower than at other temperatures (Figure 3A; Supplementary Table S5). Similarly, LBRh3 exhibited a lower mean Cq value at 62°C, with significant differences compared to other temperatures (56°C, 58°C, and 64°C), except at 60°C (Figure 3B; Supplementary Table S5). These findings indicated that the 17-nt LBRc3/Rh3 primers, which contained four SNP sites, demonstrated a high amplification rate at 62°C, identifying it as the optimal reaction temperature for the LAMP system.

Figure 3. Construction of the LAMP reaction system for Rhodiola species identification. (A) Detection time (Cq value) of RcITS2 using LB-Rc primers at different temperatures in a real-time fluorescence LAMP reaction. (B) Detection time (Cq value) of RfITS2 using LB-Rh primers at varying temperatures in a real-time fluorescence LAMP reaction. Statistical analyses were performed using Duncan’s multiple range test within a one-way ANOVA framework (*p < 0.05, **p < 0.01). Error bars indicate means ± standard deviation (SD) from three independent replicates. (C, D) Discrimination plot from the real-time fluorescence LAMP reaction using RcITS2 (C) or RfITS2 (D) plasmids as templates at 62°C. The plot was the representative of three independent experiments.
We validated the influence of the reaction template on amplification rates. When amplifying RcITS2, the amplification rates for the three LBRc primers were significantly higher than those for the LBRh primers, which contained mismatches, across the temperature range of 56°C to 64°C (Figures 3C; Supplementary Figure S2). Conversely, using a plasmid containing RfITS2 as the template resulted in lower amplification efficiency for the LBRc primers compared to the LBRh primers at all temperature gradients (Figures 3D; Supplementary Figure S2).
In particular, except for LBRc2, the maximum differences in Cq values across different templates were observed at 62°C (Figures 4; Supplementary Figures S3, S4). The time gap (ΔCq) exceeded 40 min when detecting RcITS2 and RfITS2 using LBRc1 or LBRc3 at this temperature (Figures 4A; Supplementary Figure S3A), which was particularly evident in 3D representations (Figures 4B; Supplementary Figures S4A, S4B). When LBRh3 was used, the time gap exceeded 55 min at 62°C (Figures 4C, D; Supplementary Figure S3B). Overall, both LBRc3 and LBRh3 effectively identified Rh. crenulata, Rh. fastigiata, and Rh. kirilowii in the LAMP system at 62°C.
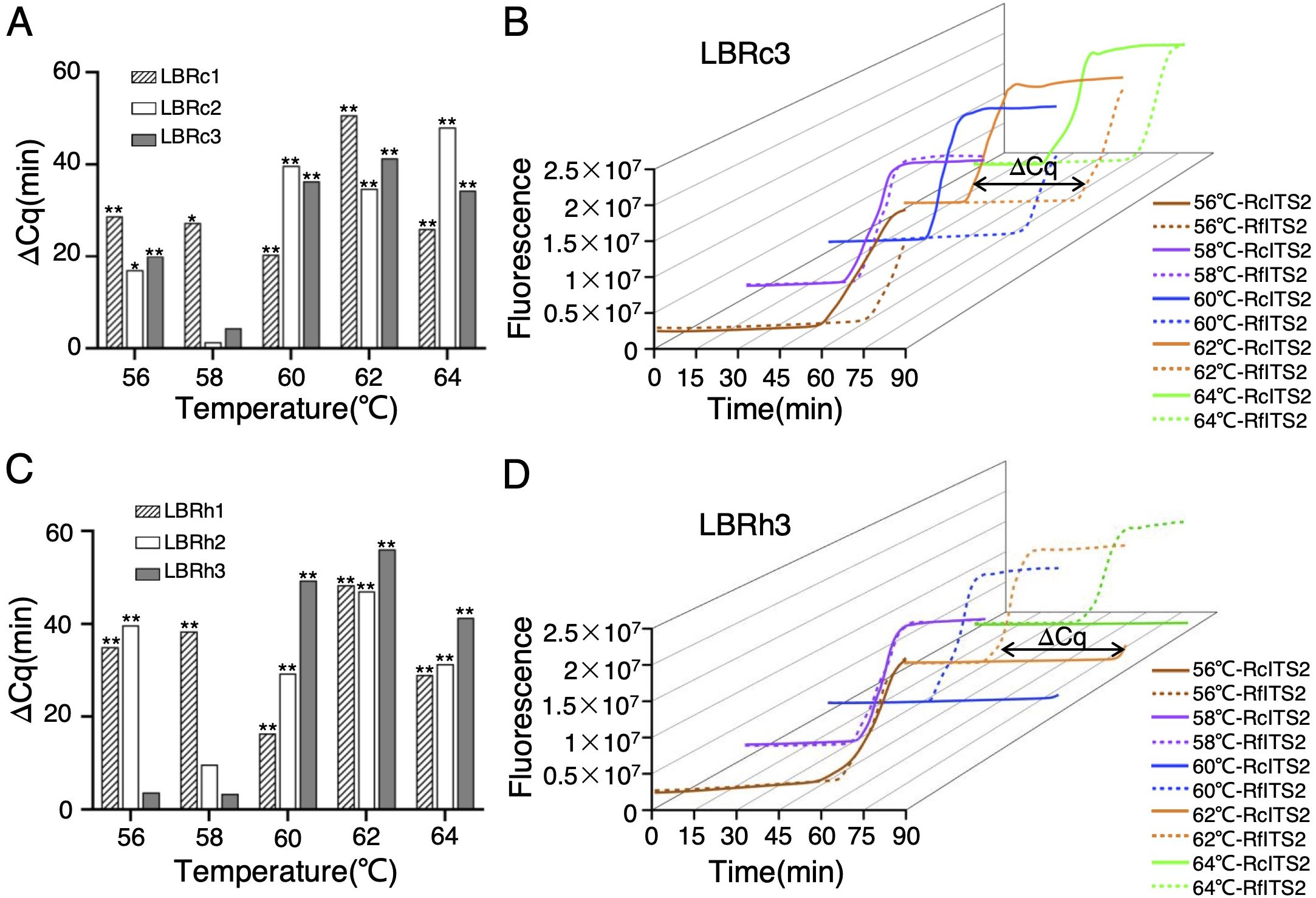
Figure 4. Time gaps between the detection of Rh. crenulata and Rh. fastigiata using specific LB primers. (A, C) Time gaps (ΔCq) between RcITS2 and RfITS2 detection using LBRc primers (A) and LBRh primers (C) at different temperatures. (B, D) Three-dimensional (3D) view of the time gap (ΔCq) between RcITS2 and RfITS2 detection using LBRc3 (B) and LBRh3 (D) at varying temperatures. All experiments were conducted in triplicate. Significance analysis was determined using a two-tailed Student’s t-test in SPSS 25 (*p < 0.05, **p < 0.01).
3.4 Sensitivity and specificity analysis of the LAMP system
Although the LAMP reaction with LBRc3 or LBRh3 at 62°C effectively distinguished between the RcITS2 and RfITS2 plasmid (at 104 copies), the presence of considerable impurities in the samples highlighted the need for a LAMP method with high sensitivity and specificity for practical detection.
In the medicinal materials of Rh. crenulata, it is common for adulterants to be mixed in. Consequently, we focused on evaluating the sensitivity of the LBRh3 primer to effectively distinguish these adulterants from Rh. crenulata. To assess this sensitivity, we utilized plasmid templates with copy number gradients ranging from 108 to 101. Real-time LAMP reaction results showed that the lowest copy number of the RcITS2 plasmid, efficiently amplified by LBRc3, was 103, while the lowest copy number of the RfITS2 plasmid, amplified by LBRh3, was 102 at 62°C (Figures 5A, B). The LAMP system demonstrated sufficient sensitivity to handle extracted genomic DNA samples with concentrations between 104 and 105 copies. Additionally, a linear correlation was established between the Cq value and plasmid concentration (Figures 5C, D), indicating that template concentration could be estimated based on the Cq value.
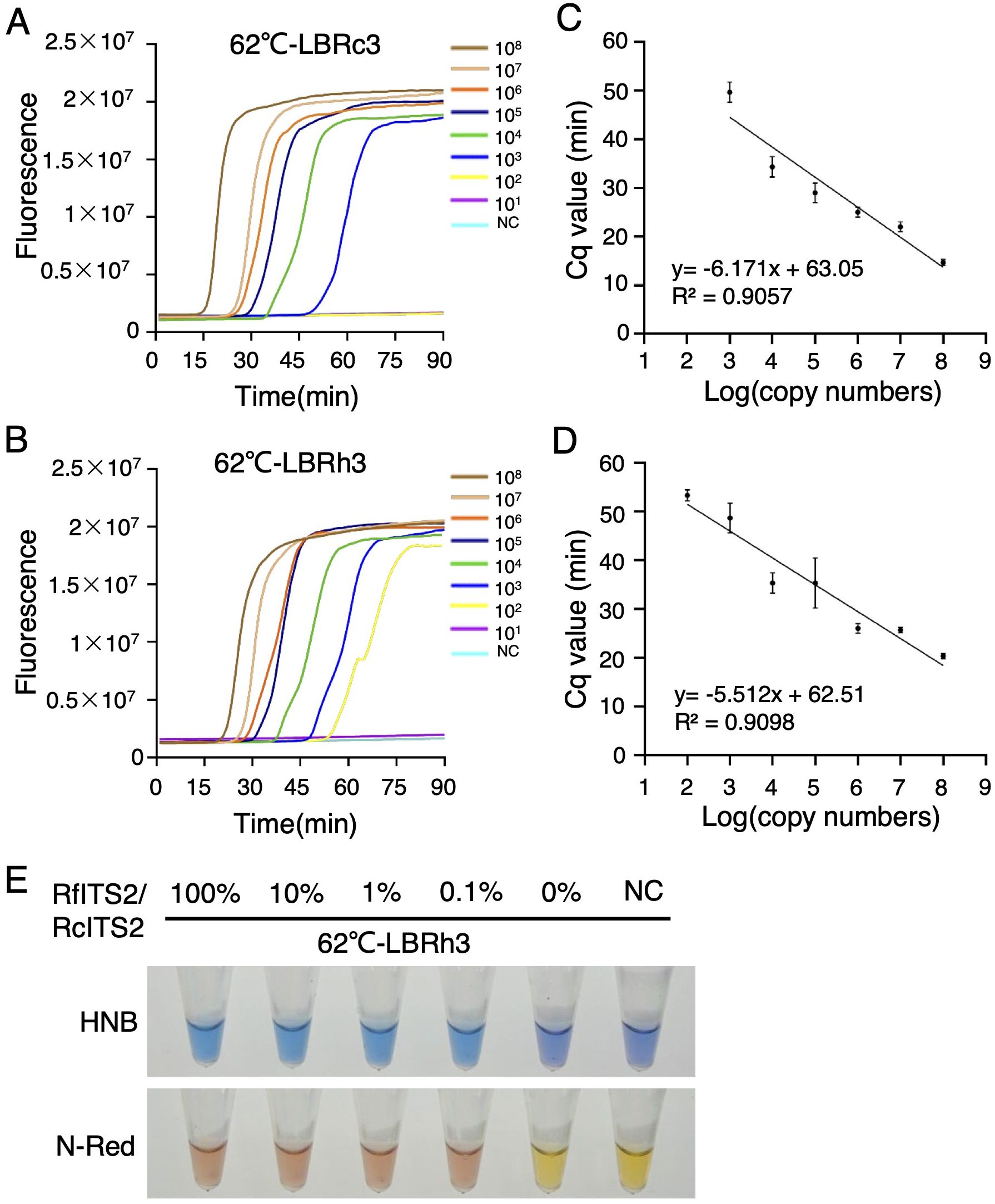
Figure 5. Sensitivity and specificity of the LAMP system. (A, B) Real-time fluorescence LAMP detection with different copy numbers of plasmid templates using LBRc3 (A) and LBRh3 (B) at 62°C. (C, D) Linear correlation between detection time (Cq value) and template copy number for LBRc3 (C) and LBRh3 (D). Error bars represent mean ± standard deviation (SD) from three independent biological replicates. (E) Specificity analysis of LBRh3 in detecting RfITS2 using colorimetric indicators in the LAMP system. All experiments were repeated three times.
In the herbal medicine market, Rh. crenulata is often adulterated with Rh. fastigiata and Rh. kirilowii, raising safety and quality concerns. Thus, the specificity and sensitivity of LBRh3 in identifying these Rhodiola species, particularly Rh. fastigiata and Rh. kirilowii, is crucial. To enhance the practicality and applicability of the LAMP detection system, we replaced fluorescence dyes with colorimetric indicators such as HNB and N-red. The color of the reaction system changed when the target gene was efficiently amplified. We generated a set of mixed plasmid templates with identical copy numbers (106) but varying RfITS2/RcITS2 ratios (100%, 10%, 1%, 0.1%, and 0%). Notably, a significant color change was observed by the naked eye in all experimental groups with RfITS2/RcITS2 ratios ranging from 100% to 0.1%, except in the group without the RfITS2 plasmid (Figure 5E).
3.5 Application of the LAMP system in Rhodiola species identification
We first validated the visualized detection of the LAMP reaction using plasmid templates. Based on the Cq values obtained for LBRc3/Rh3 in the amplification of RcITS2/RfITS2 at 62°C (Figures 3C, D, 4B, D), we set the reaction time for visualized detection to 35 min. Compared to the control group without plasmid templates, the color of the reaction system changed to light blue (with HNB) or pink (N-red) after 35 min for the following samples: RcITS2 plasmid + LBRc primer, RfITS2 plasmid + LBRh primer, RcITS2 plasmid + RfITS2 plasmid +LBRc primer, and RcITS2 plasmid + RfITS2 plasmid + LBRh primer (Figure 6A). These color changes were clearly observable to the naked eye and were consistent across both colorimetric indicators (Figure 6A).
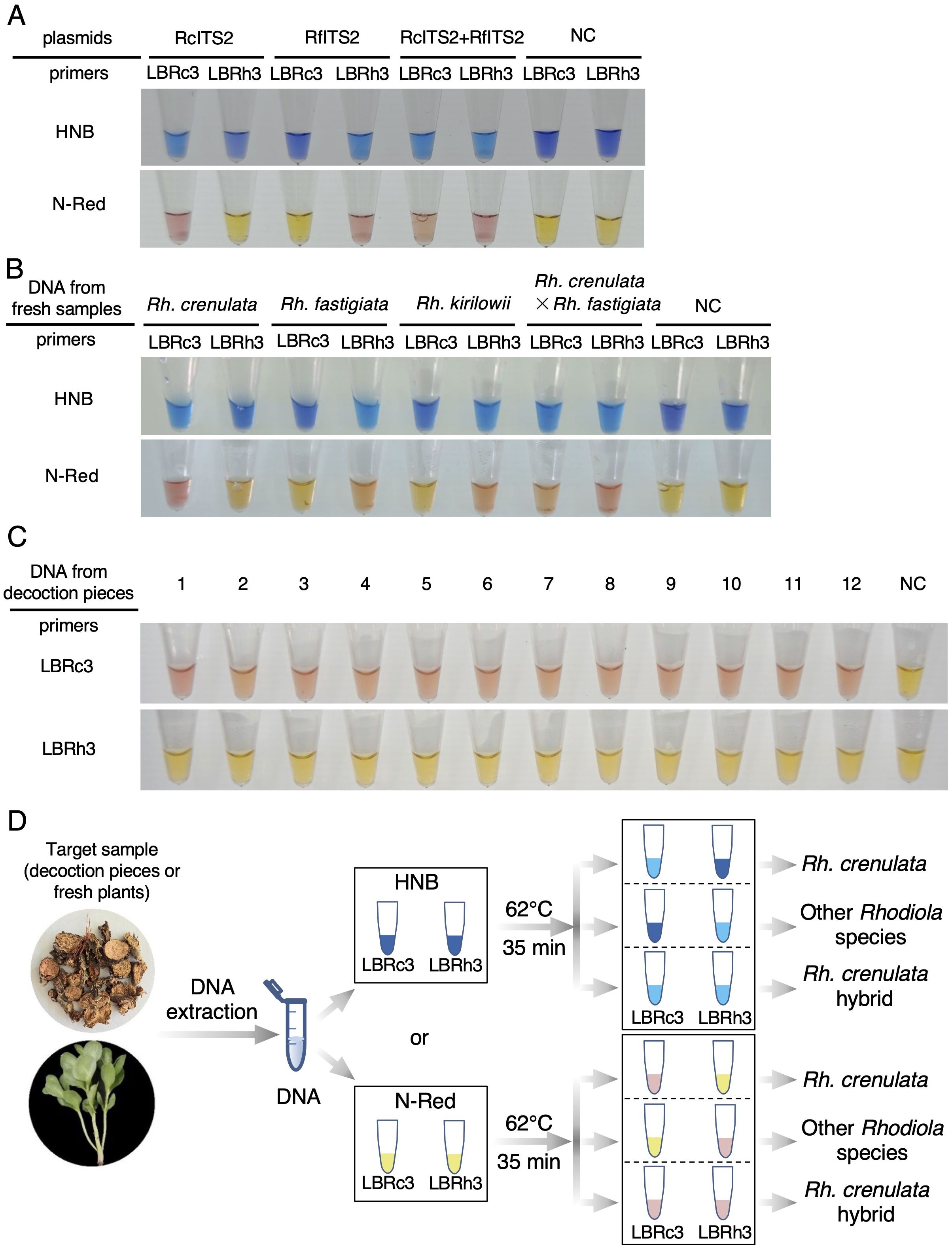
Figure 6. Visualized detection of Rh. crenulata using the LAMP system. (A) LAMP detection using HNB and N-red with plasmid templates. (B) Application of visual detection in DNA samples from four Rhodiola species. NC, negative control. Each experiment was repeated three times. (C) Application of the visual LAMP system with N-red in identifying Rhodiola decoction pieces. NC, negative control. Each experiment was repeated three times. (D) Flowchart illustrating the process of identifying Rh. crenulata using the visual LAMP system.
To test the LAMP detection system for identifying Rhodiola species, we used DNA extracted from leaves of four Rhodiola species as templates in the LAMP reaction. After 35 min at 62°C, a color change was observed in the reaction tubes containing DNA templates from Rh. crenulata or Rh. crenulata hybrids when LBRc3 was used as the primer (Figure 6B). Conversely, when LBRh3 served as the primer, the reaction tubes with DNA templates from Rh. fastigiata, Rh. kirilowii, or Rh. crenulata hybrids also exhibited a color change (Figure 6B).
To further verify the LAMP detection system for decoction piece identification, we performed reactions using DNA extracted from 12 batches of market samples with the LBRc3 and LBRh3 primers. After a 35-min reaction at 62°C, a color change was noted in the reaction tubes with LBRc3 primer, which turned light blue or pink. In contrast, those with the LBRh3 primer showed no color change, remaining blue or yellow (Figure 6C; Supplementary Figure S5). Based on these results, all the market samples were identified as Rh. crenulata.
In summary, the identification of target Rhodiola species requires simultaneous LAMP reactions using either the LBRc3 or LBRh3 primer, with two available calorimetric dyes (HNB and N-red). Three distinct color change scenarios can be observed in the reaction tubes. If the tubes with the LBRc3 primer show light blue (with HNB) or pink (N-red) while the LBRh3 tubes remain blue or yellow, the target species is identified as Rh. crenulata. If the LBRc3 tubes remain invariant in color (blue or yellow) while those with LBRh3 change to light blue or pink, the detected species is identified as another Rhodiola species, excluding Rh. crenulata. If both reaction tubes exhibit color changes, the target species is considered a hybrid of Rh. crenulata (Figure 6D).
Taken together, we have developed a rapid and convenient method using the LAMP system to distinguish Rh. crenulata within 35 min, with results that were visibly identifiable to the naked eye. This method has been validated with various materials including fresh samples of Rh. crenulata, Rh. fastigiata, Rh. kirilowii, and Rh. crenulata hybrids, and decoction pieces of Rh. crenulata.
4 Discussion
Rhodiola species have a long history as medicinal plants in Asia and Europe. Among them, Rh. crenulata and Rh. kirilowii are documented in various Chinese monographs, but only Rh. crenulata is included in the Chinese Pharmacopoeia 2020. Because of its significant clinical efficacy and high commercial value, Rh. crenulata is classified as a rare and endangered plant in China, primarily attributed to exploitation. Furthermore, the frequent mixing of different Rhodiola species in the herbal medicine market raises concerns regarding efficacy and safety. The pharmaceutical compositions and their contents vary among Rhodiola species, which can pose risks to food and medicine safety. Therefore, the rapid and accurate identification method developed in this study is crucial for enhancing market supervision within the pharmaceutical industry and ensuring the proper use of Rh. crenulata.
The ITS2 region is an effective DNA barcode for species identification, including for Rhodiola rosea L. and Rh. crenulata (Zhu et al., 2017). SNPs have emerged as efficient tools for identifying both intraspecific and interspecific variations in medicinal plants (Chen et al., 2024; Guo et al., 2017; Xu et al., 2021). A previous study identified 35 variable sites in 32 ITS2 sequences from 10 Rhodiola species (Xin et al., 2015). In this study, we identified two 50-nt regions within the ITS2 that contain multiple SNP sites among three Rhodiola species (Figure 1A), demonstrating sufficient interspecific divergence to serve as potential DNA markers. Reliable and stable SNP sites are essential for unambiguous differentiation between species (Mishra et al., 2016). Various technologies exist for SNP detection, including DNA sequencing, PCR, and PCR-derived methods. We employed two methods to validate the SNP sites between Rh. crenulata and the other two Rhodiola species. The first method, known as CAPS, involved traditional molecular approaches using the restriction endonuclease BglI (Figure 1E). However, this process is time-consuming, requiring multiple steps such as DNA extraction, PCR amplification, enzyme digestion, and gel electrophoresis, which are dependent on thermocyclers.
The second technique we utilized was the LAMP method, a highly sensitive and cost-effective technology that leverages primer mismatches and differentiated amplification rates. The hybridization reaction between the loop primer and the target sequence is significantly influenced by the characteristics of mismatched bases—especially in their number, type, and location. Generally, mismatches involving C–G pairs have a more pronounced impact on the reaction compared to A–T pairs. Regardless of the mismatch type, probe primers with a mismatch at the center demonstrate lower amplification efficiency than those with mismatches at the 5′ or 3′ ends (Ding et al., 2019; Duan et al., 2010). Specifically, at the probe center, T → A transitions exert a stronger effect on signal intensity compared to C → T transitions (Duan et al., 2010). Likewise, the LBRc3/Rh3 primers, which contained a C → T mismatch at the 5′ end and a T → A mismatch at the center, exhibited faster amplification rates than the LBRc1/Rh1 primers, which had two mismatches at the center (Figures 2B, 3A, B).
Since the initial identification of medicinal plants using DNA barcoding-targeted LAMP techniques in 2007 (Sasaki and Nagumo, 2007), this approach has been successfully applied to identify several species, including Hedyotis diffusa Willd (Li et al., 2013), Taraxacum formosanum Kitam (Lai et al., 2015), Crocus sativus L (Zhao et al., 2016), Dendrobium officinale Kimura & Migo (Yang et al., 2019), and Portulaca oleracea L (Xu et al., 2023). The high sensitivity and specificity of the LAMP technique are particularly important for detecting low-quality DNA samples, which may be extracted using simple methods or from mixed DNA sources. In our study, the species-specific primers LBRc3 and LBRh3 amplified as few as 103 copies of RcITS2 and 102 copies of RfITS2, respectively (Figures 5A, C), demonstrating performance comparable to previously reported methods (Ding et al., 2019) and high-resolution melting (HRM) analysis (Ren et al., 2017). This sensitivity helps to mitigate false negatives that may occur during practical applications. Furthermore, LBRh3 was able to successfully distinguish a 0.1% RfITS2 plasmid (approximately 103 copies) from a 99.9% RcITS2 plasmid (Figure 5E), indicating that the LAMP detection system developed in this study is effective for identifying mixed samples. To confirm the broad applicability of our LAMP system, we obtained ITS2 sequences of various Rhodiola species from GenBank, including Rhodiola yunnanensis (Franch.) S.H.Fu, Rhodiola tangutica (Maxim.) S.H.Fu, Rhodiola sacra (Prain ex Raym.-Hamet) S.H.Fu, Rh. rosea, and eight other Rhodiola species from GenBank. Base preference analysis illustrated that the ITS2 sequences from these species shared similar sequences in the SNP region (162–177 bp) with Rh. kirilowii and Rh. fastigiata, except for Rh. crenulata (Supplementary Figure S6). Therefore, our LAMP system can theoretically distinguish Rh. crenulata from the analyzed Rhodiola species.
In addition to sensitivity and specificity, the time required for identification methods is a significant consideration. LAMP is a rapid and convenient technique widely used in pathogen detection (Shen et al., 2020; Yang et al., 2018), disease diagnosis (Besuschio et al., 2017; Gawande et al., 2022), and herbal medicine identification (Xu et al., 2023; Yang et al., 2019). Most LAMP reactions typically require approximately 40 min. In this study, we successfully identified Rh. crenulata using the LAMP system in just 35 min, significantly shorter than the 2-h duration needed for traditional PCR. However, the complete identification process, including DNA extraction, took about 1.5 h. High-quality genomic DNA is essential for ensuring an efficient LAMP reaction. Common DNA extraction methods, such as the CTAB method and extraction kits, are often time-consuming. Thus, there is a pressing need to develop a rapid DNA extraction method.
Moreover, we improved the LAMP system by incorporating pre-added colorimetric indicators, which provided visible detection and significantly reduced costs. This visualized LAMP system offers a reliable and scalable technology for identifying medicinal plants compared to the CAPS method (Figure 1E), providing advantages in speed and convenience. In the future, combining the LAMP technique with DNA barcoding holds great promise for accurately identifying medicinal materials and resolving species confusion in the herbal medicine market.
As a rapid and sensitive molecular identification technology, LAMP amplification produces high concentrations of products, which increases the risk of false positives due to aerosol contamination. It is essential to design multiple primers based on SNP region analysis to minimize false positives. Additionally, traditional CAPS analysis can serve as a supplementary verification method by digesting products with specific restriction enzymes. However, the sensitivity of LAMP technology may limit its application, as low concentrations of adulterants in samples could remain undetected, leading to false negatives. To enhance the accuracy, it is advisable to combine LAMP with more precise identification methods.
5 Conclusion
In this study, we developed a rapid, convenient, sensitive, and visual method for identifying Rh. crenulata in just 35 min using SNP sites and the LAMP system, showcasing significant potential for application in the herbal medicine industry.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
LH: Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Validation, Writing – original draft. XS: Data curation, Formal analysis, Software, Visualization, Writing – review & editing. SW: Formal analysis, Writing – review & editing. CY: Formal analysis, Writing – review & editing. YC: Formal analysis, Writing – review & editing. SY: Formal analysis, Writing – review & editing. JC: Formal analysis, Writing – review & editing. KL: Formal analysis, Writing – review & editing. BL: Resources, Writing – review & editing. YS: Writing – review & editing. YZ: Conceptualization, Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Chengdu Economic Development Zone (Longquanyi District) technological innovation research and development project (2024LQYF0024) and the Science and Technology Department of Sichuan Province (2024NSFSC1330).
Acknowledgments
We are grateful to all researchers who unselfishly shared the sequences of ITS2 online. Great thanks to Xizang Rhodiola Pharmaceutical Holding Company for providing the plant materials and to Prof. Yuehua Wang for identifying these materials.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1492083/full#supplementary-material
References
Besuschio, S. A., Llano Murcia, M., Benatar, A. F., Monnerat, S., Cruz, I., Picado, A., et al. (2017). Analytical sensitivity and specificity of a loop-mediated isothermal amplification (LAMP) kit prototype for detection of Trypanosoma cruzi DNA in human blood samples. PloS Negl. Trop. Dis. 11, e0005779. doi: 10.1371/journal.pntd.0005779
Chen, P., Liu, J., Tang, Q., Zhou, T., Guo, L., Xu, Y., et al. (2024). Genetic identification of medicinal citrus cultivar ‘Local Juhong’ using molecular markers and genomics. Genes (Basel) 15, 719. doi: 10.3390/genes15060719
Chen, S., Yao, H., Han, J., Liu, C., Song, J., Shi, L., et al. (2010). Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PloS One 5, e8613. doi: 10.1371/journal.pone.0008613
Ding, S., Chen, R., Chen, G., Li, M., Wang, J., Zou, J., et al. (2019). One-step colorimetric genotyping of single nucleotide polymorphism using probe-enhanced loop-mediated isothermal amplification (PE-LAMP). Theranostics 9, 3723–3731. doi: 10.7150/thno.33980
Duan, F., Pauley, M. A., Spindel, E. R., Zhang, L., Norgren, R. B., Jr. (2010). Large scale analysis of positional effects of single-base mismatches on microarray gene expression data. BioData Min. 3, 2. doi: 10.1186/1756-0381-3-2
Fan, X. J., Wang, Y., Wang, L., Zhu, M. (2016). Salidroside induces apoptosis and autophagy in human colorectal cancer cells through inhibition of PI3K/Akt/mTOR pathway. Oncol. Rep. 36, 3559–3567. doi: 10.3892/or.2016.5138
Feng, S., Jiang, Y., Wang, S., Jiang, M., Chen, Z., Ying, Q., et al. (2015). Molecular identification of Dendrobium species (Orchidaceae) based on the DNA barcode ITS2 region and its application for phylogenetic study. Int. J. Mol. Sci. 16, 21975–21988. doi: 10.3390/ijms160921975
Ganie, S. H., Upadhyay, P., Das, S., Prasad Sharma, M. (2015). Authentication of medicinal plants by DNA markers. Plant Gene 4, 83–99. doi: 10.1016/j.plgene.2015.10.002
Ganzera, M., Sturm, S. (2018). Recent advances on HPLC/MS in medicinal plant analysis-an update covering 2011-2016. J. Pharm. Biomed. Anal. 147, 211–233. doi: 10.1016/j.jpba.2017.07.038
Gawande, S. P., Raghavendra, K. P., Monga, D., Nagrale, D. T., Prabhulinga, T., Hiremani, N., et al. (2022). Development of loop mediated isothermal amplification (LAMP): A new tool for rapid diagnosis of cotton leaf curl viral disease. J. Virol. Methods 306, 114541. doi: 10.1016/j.jviromet.2022.114541
Grazina, L., Amaral, J. S., Mafra, I. (2020). Botanical origin authentication of dietary supplements by DNA-based approaches. Compr. Rev. Food Sci. Food Saf. 19, 1080–1109. doi: 10.1111/1541-4337.12551
Guo, M., Ren, L., Pang, X. (2017). Inspecting the true identity of herbal materials from Cynanchum uing ITS2 barcode. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01945
Han, K., Wang, M., Zhang, L., Wang, C. (2018). Application of molecular methods in the identification of ingredients in Chinese herbal medicines. Molecules 23, 2728. doi: 10.3390/molecules23102728
Hao, L., Wang, S., Zhang, Y., Xu, C., Yu, Y., Xiang, L., et al. (2023). Long-distance transport of the pear HMGR1 mRNA via the phloem is associated with enhanced salt tolerance. Plant Sci. 332, 111705. doi: 10.1016/j.plantsci.2023.111705
Hao, L., Zhang, Y., Wang, S., Zhang, W., Wang, S., Xu, C., et al. (2020). A constitutive and drought-responsive mRNA undergoes long-distance transport in pear (Pyrus betulaefolia) phloem. Plant Sci. 293, 110419. doi: 10.1016/j.plantsci.2020.110419
Hyman, L. B., Christopher, C. R., Romero, P. A. (2022). Competitive SNP-LAMP probes for rapid and robust single-nucleotide polymorphism detection. Cell Rep. Methods 2, 100242. doi: 10.1016/j.crmeth.2022.100242
Intharuksa, A., Sasaki, Y., Ando, H., Charoensup, W., Suksathan, R., Kertsawang, K., et al. (2020). The combination of ITS2 and psbA-trnH region is powerful DNA barcode markers for authentication of medicinal Terminalia plants from Thailand. J. Nat. Med. 74, 282–293. doi: 10.1007/s11418-019-01365-w
Jiao, Y., Zhao, Z., Li, X., Li, L., Xiao, D., Wan, S., et al. (2023). Salidroside ameliorates memory impairment following long-term ethanol intake in rats by modulating the altered intestinal microbiota content and hippocampal gene expression. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1172936
Kuwahara, Y., Nakajima, D., Shinpo, S., Nakamura, M., Kawano, N., Kawahara, N., et al. (2019). Identification of potential genes involved in triterpenoid saponins biosynthesis in Gleditsia sinensis by transcriptome and metabolome analyses. J. Nat. Med. 73, 369–380. doi: 10.1007/s11418-018-1270-2
Lai, G. H., Chao, J., Lin, M. K., Chang, W. T., Peng, W. H., Sun, F. C., et al. (2015). Rapid and sensitive identification of the herbal tea ingredient Taraxacum formosanum using loop-mediated isothermal amplification. Int. J. Mol. Sci. 16, 1562–1575. doi: 10.3390/ijms16011562
Li, Y., Pham, V., Bui, M., Song, L., Wu, C., Walia, A., et al. (2017). Rhodiola rosea L.: an herb with anti-stress, anti-aging, and immunostimulating properties for cancer chemoprevention. Curr. Pharmacol. Rep. 3, 384–395. doi: 10.1007/s40495-017-0106-1
Li, M., Wong, Y. L., Jiang, L. L., Wong, K. L., Wong, Y. T., Lau, C. B. S., et al. (2013). Application of novel loop-mediated isothermal amplification (LAMP) for rapid authentication of the herbal tea ingredient Hedyotis diffusa Willd. Food Chem. 141, 2522–2525. doi: 10.1016/j.foodchem.2013.05.085
Mahadani, P., Ghosh, S. K. (2013). DNA Barcoding: A tool for species identification from herbal juices. DNA Barcodes 1, 35–38. doi: 10.2478/dna-2013-0002
Mishra, P., Kumar, A., Nagireddy, A., Mani, D. N., Shukla, A. K., Tiwari, R., et al. (2016). DNA barcoding: an efficient tool to overcome authentication challenges in the herbal market. Plant Biotechnol. J. 14, 8–21. doi: 10.1111/pbi.12419
Panossian, A., Wikman, G., Sarris, J. (2010). Rosenroot (Rhodiola rosea): traditional use, chemical composition, pharmacology and clinical efficacy. Phytomedicine 17, 481–493. doi: 10.1016/j.phymed.2010.02.002
Recio, M. C., Giner, R. M., Máñez, S. (2016). Immunmodulatory and antiproliferative properties of Rhodiola Species. Planta Med. 82, 952–960. doi: 10.1055/s-0042-107254
Ren, X., Fu, Y., Xu, C., Feng, Z., Li, M., Zhang, L., et al. (2017). High resolution melting (HRM) analysis as a new tool for rapid identification of Salmonella enterica serovar Gallinarum biovars Pullorum and Gallinarum. Poult. Sci. 96, 1088–1093. doi: 10.3382/ps/pew400
Saejung, W., Khumtong, K., Rapichai, W., Ratanabunyong, S., Rattanasrisomporn, A., Choowongkomon, K., et al. (2024). Detection of feline immunodeficiency virus by neutral red-based loop-mediated isothermal amplification assay. Vet. World 17, 72–81. doi: 10.14202/vetworld.2024.72-81
Sasaki, Y., Nagumo, S. (2007). Rapid identification of Curcuma longa and C. aromatica by LAMP. Biol. Pharm. Bull. 30, 2229–2230. doi: 10.1248/bpb.30.2229
Sharafdarkolaee, S. H., Gill, P., Motovali-Bashi, M., Sharafdarkolaee, F. H. (2019). Isothermal amplification methods for the SNP genotyping. Curr. Mol. Med. 19, 461–472. doi: 10.2174/1566524019666190527083947
Shen, H., Wen, J., Liao, X., Lin, Q., Zhang, J., Chen, K., et al. (2020). A sensitive, highly specific novel isothermal amplification method based on single-nucleotide polymorphism for the rapid detection of Salmonella pullorum. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.560791
Shikov, A. N., Pozharitskaya, O. N., Makarov, V. G., Wagner, H., Verpoorte, R., Heinrich, M. (2014). Medicinal plants of the Russian Pharmacopoeia; their history and applications. J. Ethnopharmacol. 154, 481–536. doi: 10.1016/j.jep.2014.04.007
Soroka, M., Wasowicz, B., Rymaszewska, A. (2021). Loop-mediated isothermal amplification (LAMP): The better sibling of PCR? Cells 10, 1931. doi: 10.3390/cells10081931
Tao, H., Wu, X., Cao, J., Peng, Y., Wang, A., Pei, J., et al. (2019). Rhodiola species: A comprehensive review of traditional use, phytochemistry, pharmacology, toxicity, and clinical study. Med. Res. Rev. 39, 1779–1850. doi: 10.1002/med.21564
Tinsley, G. M., Jagim, A. R., Potter, G. D. M., Garner, D., Galpin, A. J. (2024). Rhodiola rosea as an adaptogen to enhance exercise performance: a review of the literature. Br. J. Nutr. 131, 461–473. doi: 10.1017/S0007114523001988
Xin, T., Li, X., Yao, H., Lin, Y., Ma, X., Cheng, R., et al. (2015). Survey of commercial Rhodiola products revealed species diversity and potential safety issues. Sci. Rep. 5, 8337. doi: 10.1038/srep08337
Xu, M. R., Sun, F. C., Yang, B. C., Chen, H. J., Lin, C. H., Cheng, J. H., et al. (2023). Genetic authentication of the medicinal plant Portulaca oleracea using a quick, precise, and sensitive isothermal DNA amplification assay. Int. J. Mol. Sci. 24, 10730. doi: 10.3390/ijms241310730
Xu, Y., Tian, S., Li, R., Huang, X., Li, F., Ge, F., et al. (2021). Transcriptome characterization and identification of molecular markers (SNP, SSR, and indels) in the medicinal plant Sarcandra glabra spp. Biomed. Res. Int, 2021. doi: 10.1155/2021/9990910
Xu, M. R., Yang, B. C., Chang, H. C., Kuo, C. L., Lin, C. H., Chen, H. J., et al. (2022). Molecular authentication of the medicinal crop Portulaca oleracea and discrimination from its adulterants in herbal markets using PCR-restriction fragment length polymorphism (PCR-RFLP) analysis. Ind. Crops. Prod. 183, 114934. doi: 10.1016/j.indcrop.2022.114934
Yang, Q., Domesle, K. J., Ge, B. (2018). Loop-mediated isothermal amplification for Salmonella detection in food and feed: current applications and future directions. Foodborne Pathog. Dis. 15, 309–331. doi: 10.1089/fpd.2018.2445
Yang, L., Wu, W. R., Zhou, H., Lai, H. L., Fu, F. (2019). Rapid identification of Dendrobium officinale using loop-mediated isothermal amplification (LAMP) method. Chin. J. Nat. Med. 17, 337–345. doi: 10.1016/S1875-5364(19)30039-1
Yang, C., Zhen, Y., Hou, J., Mi, T. (2024). Development of a rapid detection method to prorocentrum lima by loop-mediated isothermal amplification with hydroxy naphthol blue. Mar. Biotechnol. (NY) 26, 475–487. doi: 10.1007/s10126-024-10310-2
Yue, H., Wang, L., Jiang, S., Banma, C., Jia, W., Tao, Y., et al. (2022). Hypoglycemic effects of Rhodiola crenulata (HK. f. et. Thoms) H. Ohba in vitro and in vivo and its ingredient identification by UPLC-triple-TOF/MS. Food Funct. 13, 1659–1667. doi: 10.1039/d1fo03436g
Zhao, M., Shi, Y., Wu, L., Guo, L., Liu, W., Xiong, C., et al. (2016). Rapid authentication of the precious herb saffron by loop-mediated isothermal amplification (LAMP) based on internal transcribed spacer 2 (ITS2) sequence. Sci. Rep. 6, 25370. doi: 10.1038/srep25370
Keywords: medicinal herb, original plant identification, molecular authentication method, 21 naked-eye detection, ITS sequences, using only visual inspection enhance the reaction rate
Citation: Hao L, Shi X, Wen S, Yang C, Chen Y, Yue S, Chen J, Luo K, Liu B, Sun Y and Zhang Y (2025) Single nucleotide polymorphism-based visual identification of Rhodiola crenulata using the loop-mediated isothermal amplification technique. Front. Plant Sci. 15:1492083. doi: 10.3389/fpls.2024.1492083
Received: 16 September 2024; Accepted: 23 December 2024;
Published: 16 January 2025.
Edited by:
Gulmira Khassanova, S. Seifullin Kazakh AgroTechnical Research University, KazakhstanReviewed by:
Zhi Chao, Southern Medical University, ChinaKaiser Iqbal Wani, Aligarh Muslim University, India
Copyright © 2025 Hao, Shi, Wen, Yang, Chen, Yue, Chen, Luo, Liu, Sun and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanxia Sun, c3VueWFueGlhMTk3NkBjZHUuZWR1LmNu; Yi Zhang, emhhbmd5aTFAY2liLmFjLmNu
 Li Hao1
Li Hao1 Xin Shi
Xin Shi Bingliang Liu
Bingliang Liu Yi Zhang
Yi Zhang