- 1Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen, China
- 2Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen, China
- 3Department of Botany, University of British Columbia, Vancouver, BC, Canada
- 4Zhejiang Provincial Key Laboratory of Biotechnology on Specialty Economic Plants, College of Life Sciences, Zhejiang Normal University, Jinhua, Zhejiang, China
- 5Key Laboratory of Bio-resource and Eco-environment of Ministry of Education, College of Life Sciences, Sichuan University, Chengdu, China
Systemic acquired resistance (SAR) is activated by local infection and confers enhanced resistance against subsequent pathogen invasion. Salicylic acid (SA) and N-hydroxypipecolic acid (NHP) are two key signaling molecules in SAR and their levels accumulate during SAR activation. Two members of plant-specific Calmodulin-Binding Protein 60 (CBP60) transcription factor family, CBP60g and SARD1, regulate the expression of biosynthetic genes of SA and NHP. CBP60g and SARD1 function as master regulators of plant immunity and their expression levels are tightly controlled. Although there are numerous reports on regulation of their expression, the specific mechanisms by which SARD1 and CBP60g respond to pathogen infection are not yet fully understood. This study identifies and characterizes the role of the LAZARUS 1 (LAZ1) and its homolog LAZ1H1 in plant immunity. A forward genetic screen was conducted in the sard1-1 mutant background to identify mutants with enhanced SAR-deficient phenotypes (sard mutants), leading to the discovery of sard6-1, which maps to the LAZ1 gene. LAZ1 and its homolog LAZ1H1 were found to be positive regulators of SAR through regulating the expression of CBP60g and SARD1 as well as biosynthetic genes of SA and NHP. Furthermore, Overexpression of LAZ1, LAZ1H1 and its homologs from Nicotiana benthamiana and potato enhanced resistance in N. benthamiana against Phytophthora pathogens. These findings indicate that LAZ1 and LAZ1H1 are evolutionarily conserved proteins that play critical roles in plant immunity.
Introduction
Plant immunity relies on two major classes of immune receptors, located on the cell surface or intracellularly, which recognize a wide range of pathogens, including viruses, bacteria, fungi, oomycetes, insects, and nematodes, and activation of the plant’s immune system for self-defense (Jones et al., 2024; Man et al., 2022). The cell surface immune receptors, referred to as Pattern-Recognition Receptors (PRRs) detect pathogen-associated molecular patterns (PAMPs), activating pattern-triggered immunity (PTI), which restricts pathogen invasion (Jones and Dangl, 2006; Zhou and Zhang, 2020). Pathogens secrete effectors into host cells to suppress PTI and disrupt normal physiological processes, facilitating invasion (Jones and Dangl, 2006; Zhou and Zhang, 2020). Intracellular immune receptors, mainly a group of proteins with nucleotide-binding sites and leucine-rich repeat domains (NLRs), recognize effectors secreted by pathogens, activating effector-triggered immunity (ETI) (Jones and Dangl, 2006; Man et al., 2022; Zhou and Zhang, 2020). As a result, both PTI and ETI responses lead to the accumulation of defense signaling molecules, such as salicylic acid (SA) and N-hydroxypipecolic acid (NHP), and trigger secondary immune responses in distant tissues, known as systemic acquired resistance (SAR), which confer enhanced resistance against subsequent pathogen invasion (Chen et al., 2018; Fu and Dong, 2013; Hartmann and Zeier, 2019; Sun and Zhang, 2021).
SA and NHP are two plant defense signaling molecules involved in PTI, ETI and SAR (Hartmann and Zeier, 2019; Peng et al., 2021). Upon pathogen invasion, SA and NHP levels escalate in both local and systemic plant tissues (Hartmann and Zeier, 2019). Application of exogenous SA or NHP on plants enhances their disease resistance (Chen et al., 2018; Hartmann et al., 2018; Peng et al., 2021). In Arabidopsis, the perception of SA predominantly depends on the Non-expressor of PR genes 1 (NPR1) and its homologs, NPR1-LIKE proteins 3 and 4 (NPR3/4), leading to upregulating the expression of genes associated with immune responses (Ding et al., 2018; Fu et al., 2012; Wu et al., 2012). Although perception of SA by NPR1 and NPR3/NPR4 is required for NHP-induced resistance in Arabidopsis, NPR proteins fail to bind to NHP (Liu et al., 2020), implying that SA and NHP signaling might occur via distinct pathways.
Biosynthesis processes of both SA and NHP are well illustrated. SA biosynthesis in plants is mediated by the isochorismate synthase (ICS) and phenylalanine (Phe) ammonia-lyase (PAL) pathways (Peng et al., 2021). In Arabidopsis thaliana, the ICS pathway contributes predominantly to SA levels. The ICS pathway include ICS1, the MATE transporter EDS5 and the aminotransferase PBS3 (Rekhter et al., 2019; Torrens-Spence et al., 2019). ICS1 is the rate limiting enzyme of the ICS pathway and its expression level is tightly regulated by various transcription factors (Huang et al., 2020; Wildermuth et al., 2001). The NHP biosynthetic process involves three enzymatic steps performed by the aminotransferase ALD1, the reductase SARD4 and the monooxygenase FMO1, catalyzing the conversion of lysine into NHP (Chen S. et al., 2021; Ding et al., 2016; Navarova et al., 2012). Expression of these NHP biosynthetic genes is also dynamically controlled during plant defense (Huang et al., 2020).
Two members of plant-specific Calmodulin-Binding Protein 60 (CBP60) transcription factor family, CBP60g and SARD1, regulate expression of biosynthesis genes of both SA and NHP upon pathogen infection (Sun et al., 2015; Wang et al., 2009; Zhang et al., 2010). Despite their common ancestry within the same protein family, CBP60g and SARD1 operate through separate pathways. The loss of either SARD1 or CBP60g results in a significant reduction in the levels of ICS1 and SA, while in the sard1-1 cbp60g-1 double mutant, the induction of ICS1 expression and the biosynthesis of SA are both blocked, suggesting that SARD1 and CBP60g regulate ICS1 expression through two parallel pathways (Wang et al., 2011; Zhang et al., 2010). Expression of SARD1 and CBP60g is also tightly regulated by various transcription factors, including positive regulators such as TGA1/4, NPR1, CBP60b, WRKY54/79 and GBPL3, as well as negative ones, including CAMTA1/2/3, NPR3/4 and HDA6 (Chen et al., 2021; Ding et al., 2018; Huang et al., 2021; Kim et al., 2022; Li et al., 2021; Sun et al., 2018, 2020; Wu et al., 2021). CBP60g is also regulated post-translationally. CALMODULIN (CAM) TOUCH3 and its homologs CAM1/4/6 cooperate with calcium-dependent protein kinases (CPK4/5/6/11) to phosphorylate and activate CBP60g (Sun et al., 2022). Although significant advances as mentioned above have been made, the specific mechanisms by which SARD1 and CBP60g respond to pathogen infection are not yet fully understood.
In pursuit of a deeper comprehension of CBP60g’s role in modulating plant immune responses, we conducted a forward genetic screen in sard1-1 mutant background to look for mutants with enhanced SAR-deficient (sard) phenotype using the SAR assay developed by our group (Zhang et al., 2010). After two rounds of SAR screen, about 80 mutants show inheritable enhanced sard phenotype. The candidate mutants are further narrowed down to about 40 through direct sequencing known SAR genes, including CBP60g, ICS1, EDS5, PBS3, ALD1 and FMO1 etc. In this study, we characterized and identified one of sard1-1 enhancer mutants, namely sard1-1 sard6-1, using bulked-segregant analysis sequencing (BSA-Seq) and genetic complementation, confirming that SARD6 encodes LAZARUS1 (LAZ1, AT4G38360). LAZ1 encodes a protein with a domain of unknown function (DUF300) and has been previously shown to modulate brassinosteroid and programmed cell death signaling pathways (Liu et al., 2018; Malinovsky et al., 2010). Here, we show that LAZ1 and its homolog LAZ1 HOMOLOG1 (LAZ1H1, AT1G77220) are positive regulators of plant immunity and SAR. In addition, LAZ1 and LAZ1H1 are conserved proteins and overexpression of their homologs from Nicotiana benthamiana (Nb) and Solanum tuberosum in Nb leaf showed enhanced resistance against Phytophthora pthogens. These results suggest that LAZ1 and LAZ1H1 are evolutionarily conserved and play a positive role in immunity.
Materials and methods
Plant material and growth environment
Arabidopsis plants were grown in soil at 23°C/21 °C day/night under 16/8-h light/dark cycles in a growth chamber with 40% relative humidity (RH) (Bi et al., 2010). The N. benthamiana plants were sowed and grown in a controlled environment room (CER) at 22 °C and 45–65% humidity with a 16/8-h light/dark cycles (Lin et al., 2023). Four-week-old plants were used for assay. The potato plants were grown in an artificial climate chamber at 25 ± 2 °C and 58–67% relative humidity under a 16/8-h light/dark photoperiod (Yang et al., 2023).
Mapping‐by‐sequencing
Mapping‐by‐sequencing involves combining next-generation sequencing with classical genetic mapping to identify candidate mutations associated with a phenotype was carried out as previously described (Sun et al., 2020). The mutant phenotype of the selected F2 lines were confirmed by examining the self‐fertilized F3 progeny. Leaves were collected from the F3 progeny of 30 F2 lines with confirmed mutant-like phenotype. Genomic DNA was extracted from the mixed tissue and sent for WGS. WGS reads were aligned with the TAIR10 reference genome. SNPs were identified and the ratios of SNPs were plotted and used for linkage analysis. Genes containing nonsynonymous mutations in the linkage region were selected as candidate genes for knockout analysis.
Mutant generation
The laz1 mutations were generated by targeting AT4G38360 in sard1-1 and Col using the egg cell- specific promoter-controlled CRISPR/Cas9 system (Wang et al., 2015). The laz1h1 mutations were generated by targeting AT1G77220 in sard1-1 and Col, respectively, using the same CRISPR/Cas9 system. The sard1-1 sard6-2 F2 mutant was obtained by crossing sard1-1 with SALK_023954C (laz1-7). The sard1-1mutant was reported (Zhang et al., 2010). Refer to Supplementary Table S1 for a comprehensive list of all primers utilized in this process.
Quantitative PCR
Total RNA was extracted from various tissues using TRIzol reagent (Invitrogen). Complementary DNAs (cDNAs) were synthesized using a ReverTra Ace kit (Toyobo) and served as templates for quantitative reverse transcription polymerase chain reaction (qRT-PCR), which was conducted with a SYBR Premix ExTaq kit (Takara) on a Bio-Rad iQ2 system. The procedure was as follows: initial polymerase activation for 30 s at 95°C followed by 40 cycles of 95°C for 5 s and 60°C for 20 s (Chen Y. et al., 2021). Each sample underwent three biological replicates and three technical replicates. The expression levels of the target genes were normalized to those of the actin gene. Primers used for qPCR can be found in Supplementary Table S1.
Pathogen infection assays
The obligate pathogen Hyaloperonospora arabidopsidis (Hpa) Noco2 spore suspension in water was weekly propagated on Col seedlings at 18 °C and 60-80% humidity with a 12/12-h light/dark cycles (Zhang et al., 2010; Bi et al., 2010). The Phytophthora infestans (P. infestans) strain 1306 were cultured on Rye A agar medium at 18°C in the dark. The Phytophthora capsici (P. capsici) strain BYA5 was cultured on Rye A agar medium at 24°C in the dark (Abrahamian et al., 2016; Wang et al., 2019). The bacterial pathogen Pseudomonas syringae pv maculicola (Psm) ES4326 was cultured on King’s B medium Agar plate supplemented with 50 μg/mL streptomycin at 28°C incubator (Zhang et al., 2010).
For infection with Psm ES4326 (diluted in 10 mM MgCl2 to OD600 as indicated below) or 10 mM MgCl2, leaves of 3-week-old plants were infiltrated with the bacteria at a dose of OD600 = 0.0025-0.005 for SAR and OD600 = 0.001 for gene expression. For SAR assay (Zhang et al., 2010), the Hpa Noco2 infection assay was carried out on 3-week-old soil-grown seedlings two days after infection with Psm ES4326, by spraying plants with Hpa Noco2 spore suspension at a concentration of 5 x 104 spores/mL. Inoculated plants were covered with a clean dome and grown at 18 °C under 12/12-h light/dark cycles in a growth chamber and growth of Hpa Noco2 was quantified seven days later. For genes expression, infected leaves were collected at two days after inoculation, two or three infected leaves of different plants were collected as one sample, and three samples were used for each genotype (Bi et al., 2010; Lan et al., 2023).
For inoculation assay with P. infestans strain 1306 (Abrahamian et al., 2016) and P. capsici strain BYA5 (Wang et al., 2019) on Nb leaves, Agrobacterium tumefaciens cultures were resuspended in infiltration buffer (10 mM MgCl2, 10 mM MES [pH 5.6], and 150 μM acetosyringone) at a final concentration of OD600 = 0.6 and infiltrated into leaves for transient expression of interested genes in planta. The leaves were detached 48h after agroinfiltration, then inoculated with P. capsici BYA5 mycelium (r = 2.5 mm) or 10 μL of the P. infestans 1306 zoospore suspension (200 zoospores/μL). The lesion areas (cm2) of P. capsici-inoculated leaves were measured under UV light at 48 h after inoculation. The P. infestans-inoculated leaves were incubated in a growth chamber at 18°C, and lesion areas were scored 3-4 days after infection.
Determination of SA concentrations
The total SA was extracted following a modified method previously described for extraction of phenolic compounds (Zhang et al., 2012). About 100 mg of leaf tissue from 3- or 4-week-old plants 2 days after inoculation with Psm ES4326 (OD600 = 0.001) was collected, in four biological replicates from independent plants for each genotype. The rosette leaves were ground in liquid nitrogen. Around 100mg powders were added into 1ml 80% MeOH in a 2ml eppendorf tube. Then the eppendorf tube was agitated for 2hr at 4°C, and then centrifuged at 13,000g at 4°C for 10 min. The supernatant was transferred into a new eppendorf tube, and the sediment was re-extracted with 500μl 100% MeOH. Both extracts were combined and blow-dryed by nitrogen gas, then was resolved by 500μl sodium acetate (0.1M, pH 5.5). The resuspension was added with 10 μl β-glucosidase (1Uμl-1) and hydrolyzed at 37°C for 2hr in the water bath. After the hydrolysate was heated in boiling water for 5 min and centrifuged at 13,000g at 4°C for 10 min, the supernatant was used for analyzing total SA by HPLC as mentioned previously (Zhang et al., 2012). SA was detected at 296-nm excitation and 410-nm emission by using fluorescence detector. According to the standard curve, the concentration of SA is calculated by the HPLC peak area.
Results
Identification of sard1-1 sard6-1 mutants
As shown in Figures 1A, B, wild-type (Col) plants were susceptible to the virulent isolate of Hyaloperonospora arabidopsidis (Hpa) Noco2. After treatment with the bacterial pathogen Pseudomonas syringae pv maculicola (Psm) ES4326, Col plants became resistant against Hpa Noco2, suggesting an robust SAR response induced by Psm infection. The sard1-1 plants showed a mild SAR-compromised phenotype, while the sard1-1 sard6-1 double mutant exhibited an exacerbated SAR-deficient phenotype. We examined Psm-induced expression of the defense marker genes PR1 and PR2 in those lines and found that induction of both genes in sard1-1 sard6-1 double mutant by Psm treatment was significantly reduced compared to that in sard1-1 mutant (Figures 1C, D). We also detected a further compromised induction of critical genes involved in SA biosynthesis CBP60g and ICS1 in the sard1-1 sard6-1 mutant compared to sard1-1 mutant (Figures 1E, F). Next, we quantified SA levels in Col, sard1-1 and the sard1-1 sard6-1 mutant plants. Following treatment with Psm, the total SA levels in the sard1-1 sard6-1 mutant was significantly reduced compared to those in the Col or sard1-1 plants (Figure 1G). These findings indicate that the systemic resistance in the sard1-1 sard6-1 mutant may be impeded due to the impact on the induction of CBP60g and SA biosynthesis.
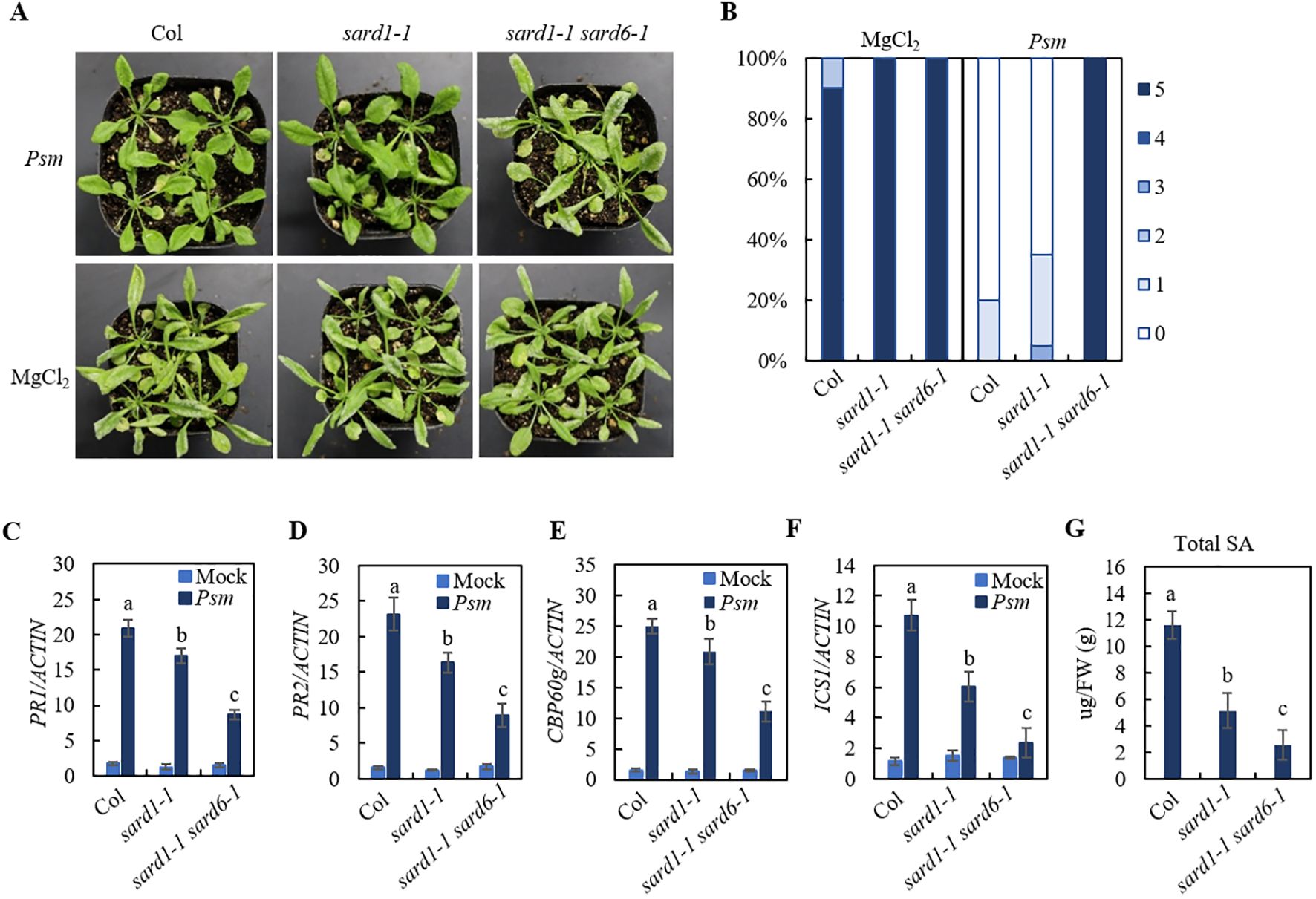
Figure 1. Identification of sard1-1 sard6-1 mutant lines of Arabidopsis. (A) Growth of Hpa Noco2 on the distal leaves of wild-type Col, sard1-1 and sard1-1 sard6-1 plants in a SAR assay. Two primary leaves of 3-week-old plants were infiltrated with Psm ES4326 (OD600 = 0.0025) or 10 mM MgCl2 (mock) 2 d before the plants were sprayed with Hpa Noco2 spore suspension (50,000 spores/mL in water). (B) SAR phenotypic statistics of wild-type Col, sard1-1 and sard1-1 sard6-1 plants. Disease ratings are as follows: 0, no conidiophores on plants; 1, one leaf is infected with no more than five conidiophores; 2, one leaf is infected with more than five conidiophores; 3, two leaves are infected but with no more than five conidiophores on each infected leaf; 4, two leaves are infected with more than five conidiophores on each infected leaf; 5, more than two leaves are infected with more than five conidiophores. The experiment was repeated three times with independently grown plants, yielding similar results. (C–F) Expression of PR1, PR2 CBP60g and ICS1. Total RNA was extracted from the leaves of 3-week-old plants 2 d after infiltration with Psm ES4326 (OD600 = 0.001) or 10 mM MgCl2 (mock). Data were normalized relative to the expression of the AtActin gene. Error bars means ± SD of 3 biological replicates. Significant differences indicated by different letters were calculated using the Duncan’s new multiple range test. (G) total SA levels in leaves of Col, sard1-1 and sard1-1 sard6-1 2 days after inoculation with Psm ES4326 (OD600 = 0.001). Bars represent means ± SD (n = 3). Statistically significant differences among the samples are labelled with different letters (one-way ANOVA with Tukey’s multiple comparisons test, P< 0.05).
SARD6 encodes LAZARUS1
To identify sard6, we performed combining next-generation sequencing with classical genetic mapping to identify candidate mutations associated with a phenotype on sard1-1 sard6-1 mutant. The sard1-1 sard6-1 mutant was backcrossed with the sard1-1 line and resulting F1 plants exhibit sard1-like sard phenotype (Figure 2A; Supplementary Figure S1A), suggesting sard6-1 is a recessive mutant. In the F2 generation, lines with sard1-1 sard6-1-like and sard1-like sard phenotype were kept and validated in the F3 progeny, respectively. Pooled genomic DNA from each segregant population (30 confirmed lines) was subjected to whole-genome next-generation sequencing (WGS). Analysis of the single nucleotide polymorphism (SNP) frequency distribution across the genome unveiled a linked genetic region on chromosome 4 (Supplementary Figure S1B). Within this chromosomal segment, three genes, AT4G30790, AT4G30990 and AT4G38360, using linkage analysis, we detected G to A transitions that resulted in missense mutations (Figure 2B). To ascertain the gene correlating with the sard1-1 sard6-1 phenotype, we generated deletion mutant for each gene in sard1-1 background using CRISPR/Cas9 technology (Figure 2C). Upon subsequent SAR analysis of the homozygous lines for the three candidate genes, only sard1-1 AT4G38360-cr double mutant phenocopied sard1-1 sard6-1, indicating that AT4G38360, alias LAZARUS1 (LAZ1) (Malinovsky et al., 2010), is the gene of interest (Figure 2D).
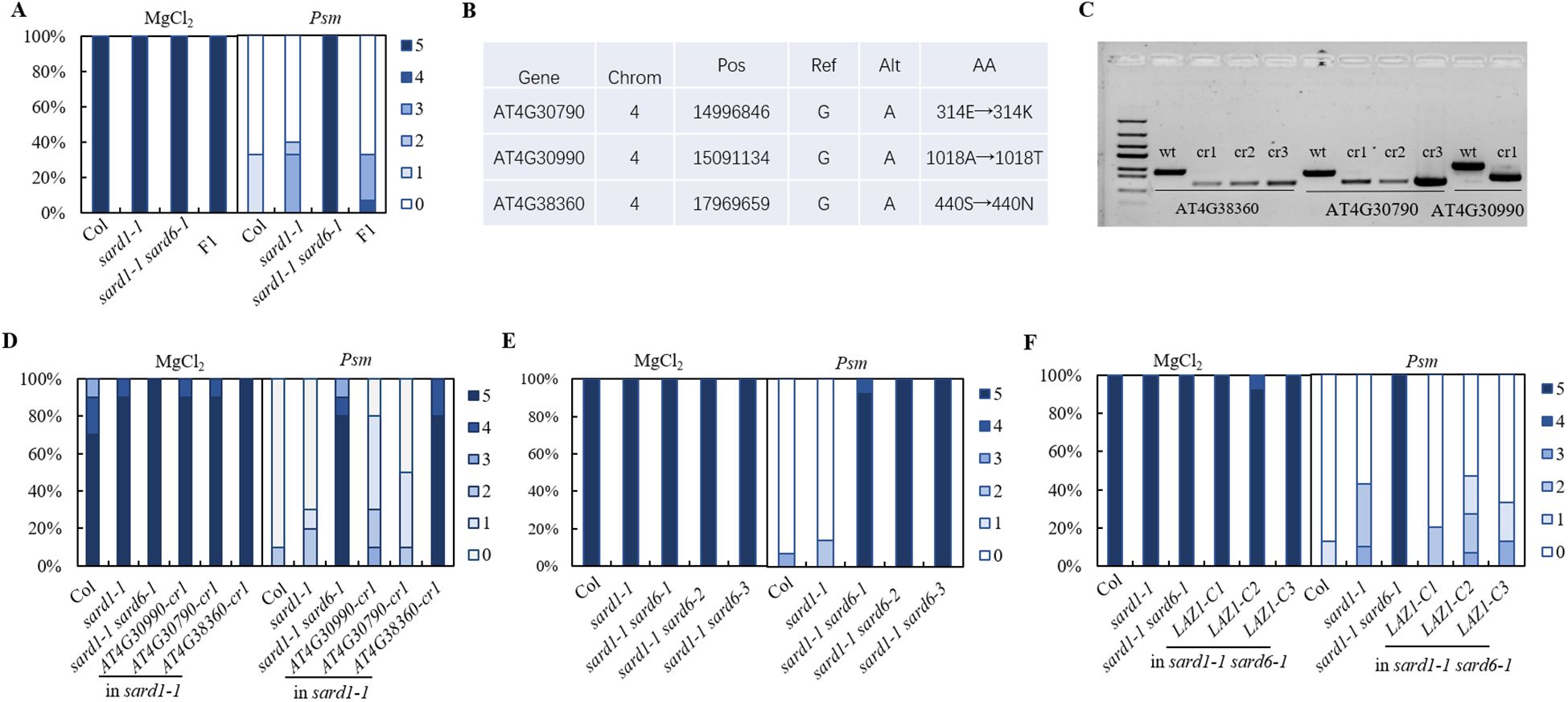
Figure 2. Positional cloning and gene verification of sard6. (A) SAR phenotypic statistics of wild-type Col, sard1-1, sard1-1 sard6-1 double mutants and the F1 progeny of sard1-1 crossed with sard1-1 sard6-1 plants. (B) Compilation of candidate genes with annotations including Chromosome (Chrom), Position (Pos), Reference allele (Ref), Alternate allele (Alt), and Amino acid (AA) changes. (C) Electropherograms depicting the wild-type and knockout genotypes for the candidate genes. The wild-type (WT) lanes are aligned alongside those representing the respective deletion mutants generated using CRISPR/Cas9 system. cr1 lines are in sard1-1 background and used in Figure 1D. (D) SAR phenotypic statistics comparing the wild-type Col, sard1-1, sard1-1 sard6-1 double mutants, and deletion lines in sard1-1 background: AT4G30990-cr1 (deletion mutation in AT4G30990), AT4G30790-cr1 (deletion in AT4G30790), and AT4G38360-cr1 (deletion in AT4G38360). (E) SAR phenotypic statistics of wild-type Col, sard1-1, sard1-1 sard6-1 and sard1-1 sard6-2, sard1-1 sard6-3 plants. sard6-2 and sard6-3 are T-DNA allele and deletion allele for AT4G38360, respectively; see details in Supplementary Figures S2A, B. (F) SAR phenotypic statistics of wild-type Col, sard1-1, sard1-1 sard6-1 and sard1-1 laz1-C plants. sard1-1 laz1-C: complementation lines of AT4G38360 in sard1-1 sard6-1.
To further ascertain the association of the mutation in AT4G38360 with the sard1-1 sard6-1 phenotype, we identified the sard1-1 sard6-2 double mutant from F2 progeny of a cross between sard1-1 and sard6-2, a T-DNA mutant SALK_023954C targeting the At4G38360 locus and silencing the gene (Supplementary Figures S2A, S2C), and confirmed DNA fragment deletion in sard1-1 AT4G38360-cr double mutant (reassigned as sard1-1 sard6-3) using Sanger sequencing (Supplementary Figure S2B). Subsequent SAR verification revealed that both sard1-1 sard6-2 and the sard1-1 sard6-3 lines exhibited sard1-1 sard6-1-like sard phenotype (Figure 2E). Additionally, we performed genetic complementation by agrobacteria mediated transformation of a 4.3 kb fragment containing LAZ1 coding region into the sard1-1 sard6-1 mutant. Three independent lines with LAZ1 expression were chosen and tested for SAR phenotype (Supplementary Figure S2D). As shown in Figure 2F, these three lines exhibited sard1-like sard phenotype, indicating that expressing LAZ1 revert the enhanced sard phenotype of sard1-1 sard6-1. These findings support that the SARD6 locus corresponds to AT4G38360/LAZ1, encoding a protein with a DUF300 domain. This protein is implicated in vacuolar transport and appears to modulate brassinosteroid signaling pathways (Malinovsky et al., 2010). For simplicity and consistency, sard6-1, sard6-2 and sard6-3 is reassigned to laz1-6, laz1-7 and laz1-8, respectively (Supplementary Figure S2A).
LAZ1 HOMOLOGO1 positively regulates SAR
In Arabidopsis, LAZ1 has a close homolog, LAZ1 Homolog 1 (LAZ1H1, AT1G77220). Quantitative PCR analysis of the expression of LAZ1 and LAZ1H1 showed that both genes were induced after Psm ES4326 treatment (Supplementary Figures S3A, B), indicating that, like LAZ1, LAZ1H1 may also play a role in plant immunity. To check whether LAZ1H1 contributes to SAR, we employed CRISPR/Cas9 technology to generate knockout mutants for LAZ1H1 in both the Col and sard1-1 backgrounds. Utilizing PCR amplification and sanger sequencing, we identified three homozygous deletion lines in the sard1-1 background and three deletion lines in Col, designated as sard1-1 laz1h1-1, sard1-1 laz1h1-2, sard1-1 laz1h1-3, laz1h1-4, laz1h1-5 and laz1h1-6, respectively (Supplementary Figure S3C). Upon verification of SAR response in the sard1-1 laz1h1 lines, we observed that they exhibited the same phenotype as the sard1-1 laz1-8 double knockout, while laz1h1 mutants showed minimal sard phenotype (Supplementary Figure S3D), suggesting that LAZ1H1 also positively regulates SAR.
LAZ1 and LAZ1H1 play overlapping roles in plant immunity
To investigate the roles of LAZ1 and LAZ1H1 in SAR, we employed CRISPR/Cas9 technology to generate knockout mutants for laz1 in Col background. Utilizing PCR amplification and sanger sequencing, we identified two homozygous deletion lines in Col, designated as laz1-9, laz1-10 respectively (Supplementary Figure S2B), and generated laz1-7 laz1h1-5 double mutant from F3 progeny of a cross between laz1-7 and laz1h1-5. Psm-induced SAR in Col, laz1-7, laz1-9, laz1-10, laz1h1-5, laz1-7 laz1h1-5 plants were performed. Compared to Col, the mutants laz1-7, laz1-9 and laz1-10, laz1h1-5 exhibited a weak sard phenotype, laz1-7 laz1h1-5 exhibited a stronger sard phenotype (Figure 3A).
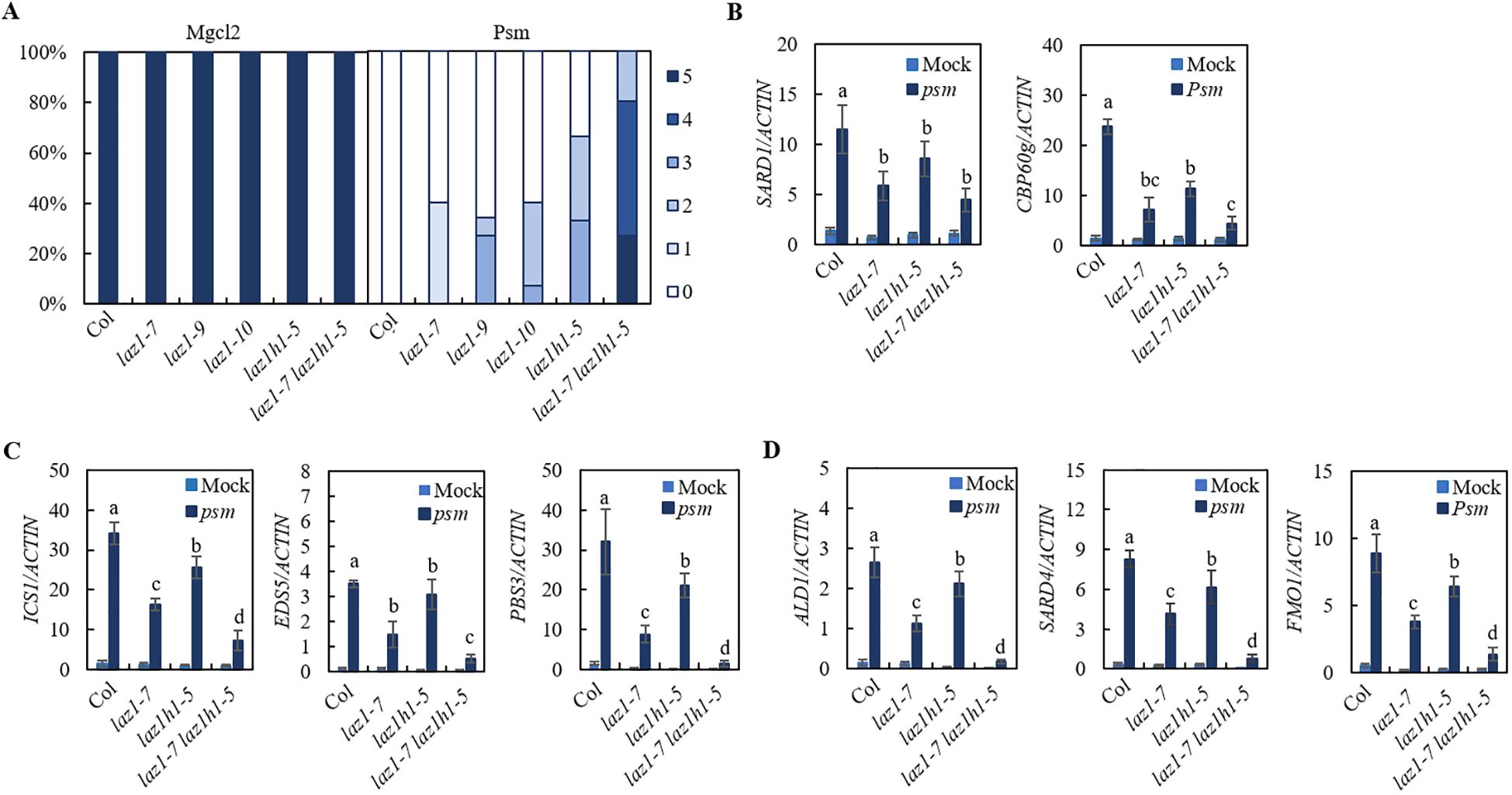
Figure 3. SAR phenotype in laz1 and laz1h1 mutants. (A) SAR phenotypic statistics in Col, laz1-7, laz1-9, laz1-10, laz1h1-5 and laz1-7 laz1h1-5 plants. (B–D) Expression of SARD1, CBP60g, ICS1, EDS5, PBS3, ALD1, SARD4 and FMO1. Total RNA was extracted from the leaves of 3-week-old plants 2d after infiltration with Psm ES4326 (OD600 = 0.001) or 10 mM MgCl2 (mock). Data were normalized relative to the expression of the AtActin gene. Error bars means ± SD of 3 biological replicates. Significant differences indicated by different letters were calculated using the Duncan’s new multiple range test.
We examined Psm-induced expression of SARD1 and CBP60g as well as biosynthetic genes of SA and NHP in Col, laz1-7, laz1h1-5 and laz1-7 laz1h1-5 lines and found that induction of these genes in three mutants were significantly reduced compared to that in Col, and the expression levels of ICS1, EDS5, PBS3, ALD1, SARD4 and FMO1 were lower in laz1-7 laz1h1-5 (Figures 3B–D). The results showed that laz1 and laz1h1 play overlapping roles in plant immunity.
LAZ1 is not required for NHP-induced immunity
Given that NHP acts as the mobile signal for SAR (Chen et al., 2018; Hartmann et al., 2018) and that the LAZ1/LAZ1H1 are putative channel proteins, we investigated whether LAZ1/LAZ1H1 is necessary for NHP-induced immune responses. Col, along with the laz1-7, laz1-9, laz1-10, laz1h1-4, laz1h1-5, laz1h1-6 and laz1-7 laz1h1-5 mutants were utilized as experimental materials for verification. Initially, we infiltrated the primary leaves with 1 mM or 0.3 mM NHP and subsequently spray-inoculated the entire plants with a spore suspension of Hpa Noco2, separately. As depicted in Supplementary Figure S4, minimal pathogen growth was observed on Col pretreated with NHP, indicating that NHP confers robust resistance against Hpa Noco2. Similar outcomes were observed in the NHP-pretreated seven mutant lines. These results suggest that laz1 and laz1h1 are not involved in the regulation of NHP-induced immunity.
Overexpression of LAZ1 and LAZ1H1 enhances the resistance of N. benthamiana to Phytophthora
The results mentioned above suggest that LAZ1 and LAZ1H1 positively regulate immunity against the obligate oomycete Hpa Noco2. To determine if LAZ1 and LAZ1H1 could augment resistance against different Phytophthora pathogens, we individually inserted the genomic sequences of LAZ1 and LAZ1H1, including 35S promoters, coding regions, and terminators, into the pCAMBIA1300 vector. These constructs were transformed into agrobacterium and used for agrobacterium-mediated transient overexpression of the respective proteins in Nicotiana benthamiana (Nb) leaves. After 48 hours post-infiltration of agrobacterium strain carrying 35S-LAZ1, 35S-LAZ1H1 or empty vector, the Nb leaves were inoculated with spores of P.infestans strain 1306 or mycelium of P.capsici strain BYA5. The resulting lesion areas were evaluated 3-4 days following infection. In Figure 4A, Nb leaf areas overexpressing LAZ1 exhibited significantly reduced lesion sizes compared to those with an empty-vector (EV) control after inoculation with P.infestans strain 1306, with statistically significant differences in lesion sizes observed (Figure 4B). Overexpression of LAZ1H1 yielded analogous results (Figures 4C, D).
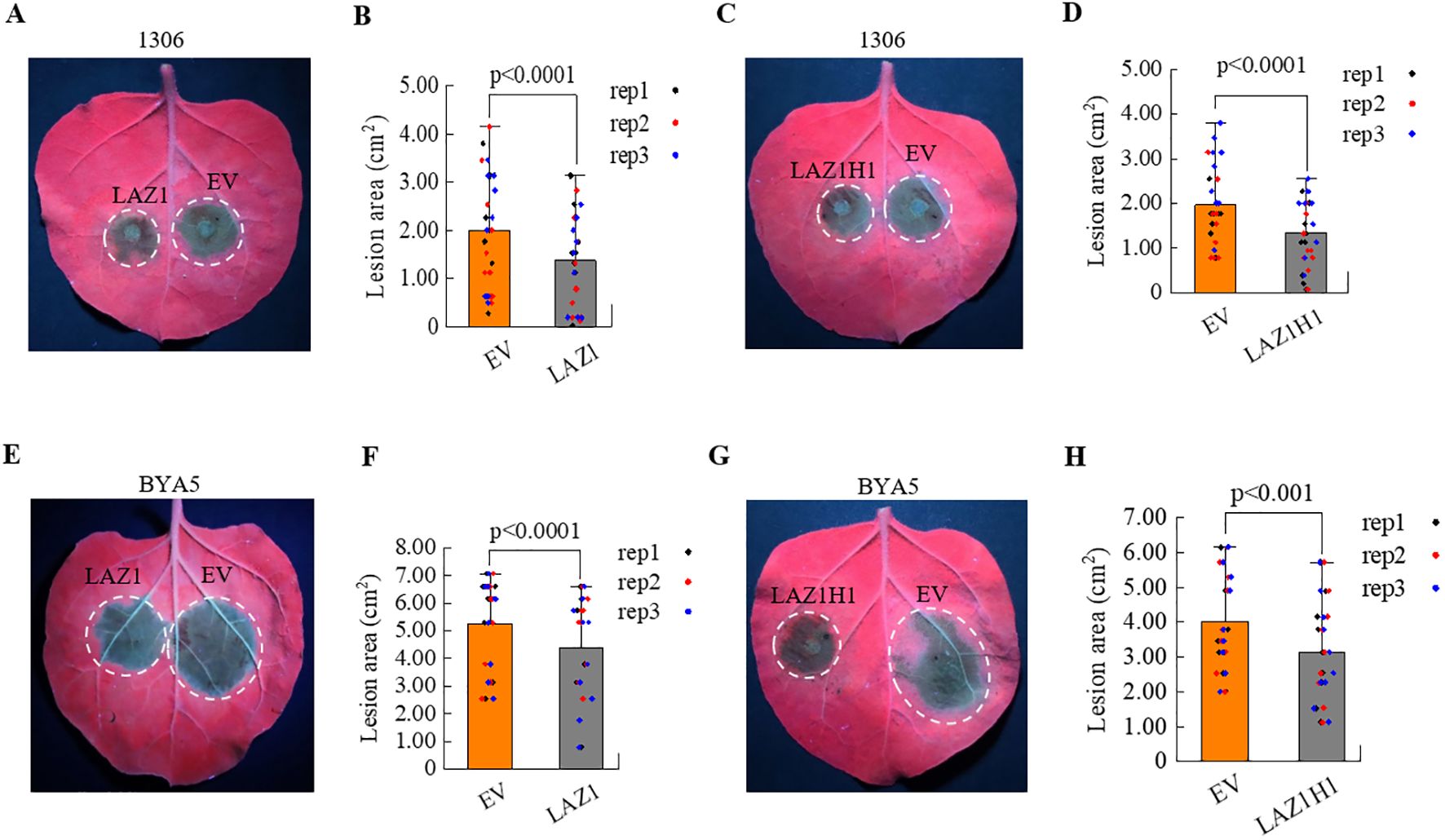
Figure 4. Impact of LAZ1 and LAZ1H1 on N. benthamiana. (A) Infection assays on N. benthamiana (Nb) leaves transient expression of LAZ1 or EV with P. infestans strain 1306. Two days after infiltration with Agrobacterium carrying 35S-LAZ1 or empty vector (EV), leaf areas were inoculated with P. infestans 1306 zoospore suspension. Detached leaves were incubated in a growth chamber at 18°C, and lesion areas were scored under UV light 3–4 days after infection. (B) Statistical analyses on lesion sizes of Nb leaves in Figure 4A (n=10-15 from three replicates). (C) Infection assays on Nb leaves transient expression of LAZ1H1 or EV with P. infestans strain 1306, following the same procedure as in Figure 4A. (D) Statistical analyses on lesion sizes of Nb leaves in Figure 4C (n=10-15 from three replicates). (E–H) Infection assays on Nb leaves transient expression of LAZ1, LAZ1H1 or EV with P. capsici strain BYA5. Two days after infiltration with Agrobacterium carrying 35S-LAZ1 (E), 35S-LAZ1H1 (G) or empty vector (EV), leaf areas were inoculated with P. capsici BYA5 mycelium. Detached leaves were incubated in a growth chamber at 25°C, and lesion areas were measured under UV light 36 h after inoculation. Statistical analyses on lesion sizes were shown in (F) (n=10-15 from three replicates) and (H) (n=10-15 from three replicates), respectively. In (B, D, F, H) data were normally distributed and were shown as means ± SD. Outliers were identifed and removed using Grubbs test. Statistical significance was determined by Student’s t test.
Similarly, overexpression of LAZ1 or LAZ1H1 lead to reduced lesion sizes compared to the EV control, following infection with P.capsici strain BYA5 (Figures 4E–H). These findings indicate that overexpression of LAZ1 or LAZ1H1 in Nb leaves can enhance resistance to P.infestans and P.capsici.
Overexpression of homologous genes of LAZ1 enhances the resistance of N. benthamiana to P.infestans
Homologs of LAZ1 were found in various plants. To understand whether homologs of the LAZ1 gene in N.benthamiana and Solanum tuberosum (potato) can enhance resistance to Phytophthora, we identified sequences with high homology to LAZ1 from the N.benthamiana genome database (https://nbenthamiana.jp/nbrowser/anno) (Kurotani et al., 2023) and the genome database of the diploid potato inbreeding line A157 (Zhang et al., 2021). The protein sequences of Nbe.v1.s00130g02480 (NB00130g02480) and Nbe.v1.s00150g07560 (NB00150g07560) in N.benthamiana and the protein sequences of A157_07G018790 and A157_12G022720 in A157 showed the highest homology to LAZ1 protein (Figure 5A). Phylogenetic analysis grouped these four genes into a single cluster, suggesting a close evolutionary relationship, and LAZ1 is closely related to these four proteins (Supplementary Figure S5A). The coding sequences (CDS) of these four homologs gene were amplified and individually inserted into the binary pCAMBIA1300 vector under the 35S promoter. Transient expression in Nb leaves followed by inoculation with P.infestans strain 1306 revealed that the lesion areas at sites of overexpression of these four genes were significantly reduced compared to the empty-vector (EV) control (Figure 5B), with statistically significant differences observed (Figure 5C). Validation through western blot analysis confirmed the presence of protein products encoded by these genes in Nb leaves (Supplementary Figure S5B). These findings indicate that overexpression of these LAZ1 homologs significantly enhances the resistance of N. benthamiana to P.infestans.
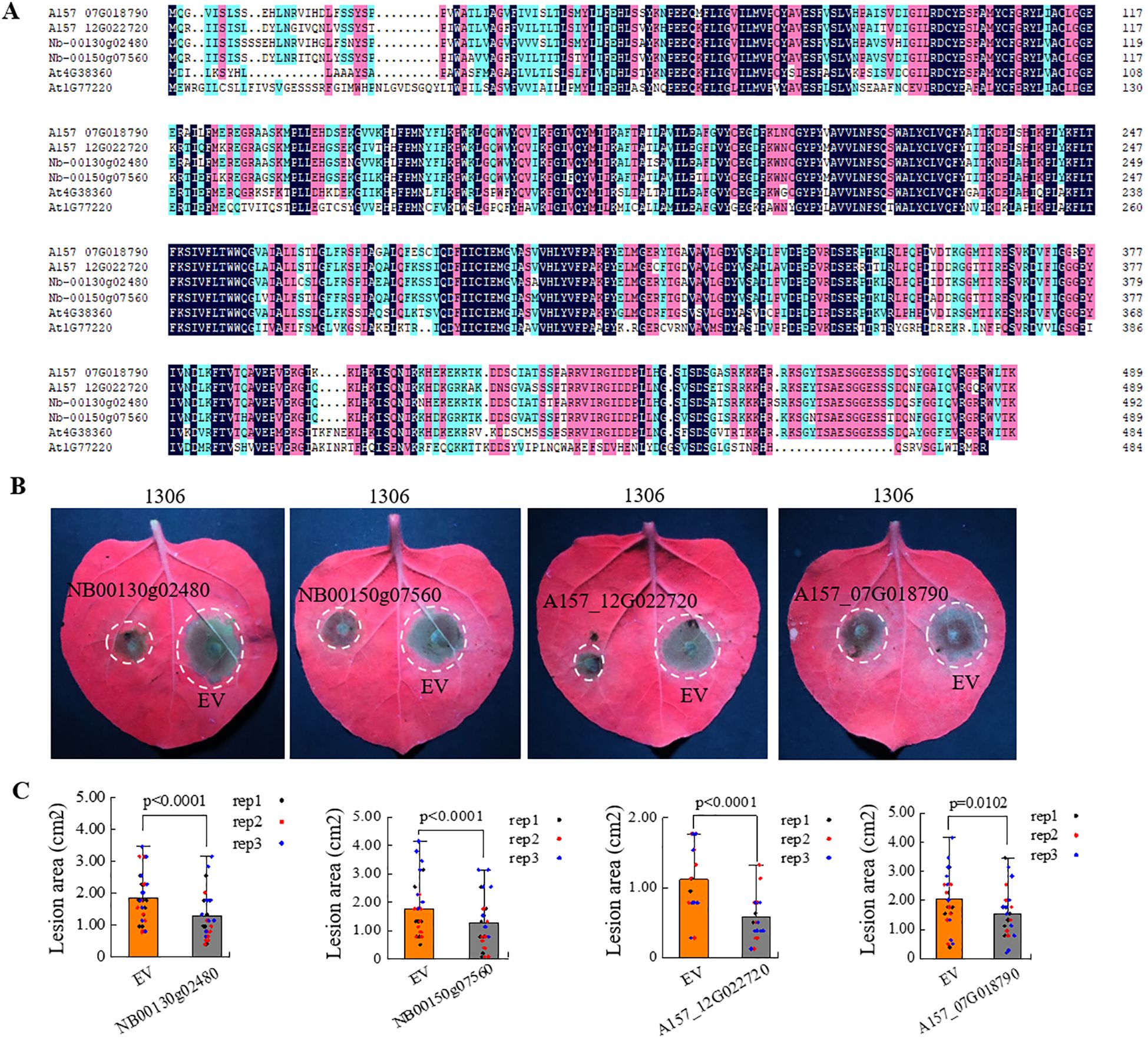
Figure 5. Influence of LAZ1 homologous genes from N. benthamiana and potato on N. benthamiana immunity. (A) Protein sequence alignment with the highest homology of LAZ1 in N. benthamiana and potato. (B) Infection assays were performed on Nb leaves overexpressing the LAZ1 homologous genes from N. benthamiana (NB00130g02480 and NB00150g07560) and potato (A157_12G022720 and A157_07G018790), challenged with P. infestans strain 1306. (C), Data processing and statistical analysis of lesion sizes in (B). Data were normally distributed and were shown as means ± SD (n=10-15 from three replicates). Outliers were identified and removed using Grubbs test. Statistical significance was determined by Student’s t test.
Discussion
LAZ1/LAZ1H1 are positive regulators of SAR
LAZ1 and LAZ1H1 belong to the evolutionarily conserved DUF300 family of transmembrane proteins in eukaryotes. LAZ1 serves as a regulatory factor for certain Hypersensitive Response (HR) cell deaths conditioned by the TIR-NB-LRR protein RPS4 and by the CC-NB-LRR protein RPM1 (Malinovsky et al., 2010). LAZ1 and LAZ1H1 have been shown to play a pivotal role in maintaining vacuole membrane integrity, which is crucial for proper Brassinosteroid (BR) signaling (Liu et al., 2018). Despite these insights, the roles of LAZ1 and LAZ1H1 in the systemic acquired resistance (SAR) pathway remain unexplored. In this report, we show that LAZ1 and LAZ1H1 are positive regulators of SAR.
Employing a forward genetic strategy, we isolated the sard1-1 sard6-1 mutant as one of sard enhancers of sard1-1. This sard1-1 sard6-1 mutant exhibited severely impaired systemic resistance (Figures 1A, B). Through mapping-by-sequence and gene complementation, we identified that SARD6 encodes LAZ1 (Figure 2). We also found that loss-of-function of LAZ1H1 leads to enhanced SAR deficiency in sard1-1 background (Supplementary Figure S3). Furthermore, we showed that the laz1 laz1h1 double mutant exhibited stronger sard phenotype compared to laz1 and laz1h1 single mutants (Figure 3A), indicating a functional redundancy between LAZ1 and LAZ1H1 in regulation of SAR.
In this study, we have used Psm ES4326, a virulent pathogen which does not trigger HR, to induce SAR and found that Laz1 and laz1h1 mutants are compromised in SAR, suggesting that LAZ1/LAZ1H1 play an HR-independent role in SAR.
LAZ1/LAZ1H1 regulate SAR by affecting expression of biosynthesis genes of SA and NHP
SA and NHP are two key signaling molecules in SAR and expression levels of their biosynthetic genes are tightly controlled during plant immunity (Hartmann and Zeier, 2019; Peng et al., 2021). SARD1 and CBP60g are master transcriptional regulators in plant defense and positively regulate biosynthetic genes of SA and NHP (Sun et al., 2020). Psm-induced expression of CBP60g and ICS1 in sard1-1 sard6-1 was further diminished compared to that in sard1-1 (Figures 1E, F) and total SA levels was lower in sard1-1 sard6-1 than that in sard1-1 (Figure 1G), indicating that the involvement of LAZ1 in SAR may be attributed to its regulatory effects on the expression of CBP60g and ICS1, as well as the accumulation of SA.
laz1 and laz1h1 single mutants showed minor sard phenotype while laz1 laz1h1 double mutant exhibited stronger sard phenotype (Figure 3A). Accordingly, Psm-induced expression of biosynthesis genes of SA and NHP in laz1 and laz1h1 single mutants was reduced compared to that in Col, and further diminished in laz1 laz1h1 double mutant (Figures 3C, D), suggesting that LAZ1 and LAZ1H1 have an overlapping function in SAR and that they positively regulate SAR by modulating the expression of biosynthetic genes of SA and NHP. Since Psm-induced expression of SARD1 and CBP60g was diminished in laz1 laz1h1 double mutant (Figure 3B), it is possible that LAZ1 and LAZ1H1 modulate the expression of biosynthetic genes of SA and NHP through regulating expression of SARD1 and CBP60g. In addition, we showed that NHP-induced resistance against Hpa Noco2 was not significantly affected in laz1, laz1h1 or laz1 laz1h1 double mutant (Supplementary Figure S4). These results suggest that LAZ1 and LAZ1H1 regulate SAR mainly through affecting the expression of biosynthesis genes of SA and NHP. However, the underlying mechanism necessitates further investigation.
Recently, LAZ1 homologs in maize, ZmLAZ1-4 and ZmLAZ1-8, were predicted to bind metal ions including Zn2+, Mg2+, or Ca2+ and ZmLAZ1-4 protein was shown to act as a Zinc transporter that modulate Zinc homeostasis on plasma and vacuolar membrane (Liu et al., 2022), it will be interesting to test whether LAZ1 and LAZ1H1 combine Ca2+ and regulate the expression of defense genes through modulating calcium homeostasis during plant immunity.
LAZ1 and LAZ1H1 are evolutionarily conserved
LAZ1 and LAZ1H1 are conserved proteins with their homologs found in various plants. We showed overexpression of LAZ1 and LAZ1H1 as well as their close homologs from N.benthamiana and potato in Nb leaves leads to enhanced resistance to Phytophthora species (Figures 4, 5), suggesting that LAZ1 and LAZ1H1 are evolutionarily conserved in positive regulation of plant defense, thereby emphasizing the significance and utility of these genes in investigating plant-pathogen interactions. Presently, in potato, the cultivation of disease-resistant varieties is mainly to isolate disease resistance genes from wild species and introduce them into cultivated varieties through genetic transformation. The homologs of LAZ1 in potato were found to similarly bolster the plants’ resistance to Phytophthora (Figures 5B, C). This study provides an important genetic resource for potato disease resistance breeding.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
YC: Funding acquisition, Investigation, Writing – original draft, Data curation, Formal analysis, Visualization, Writing – review & editing. YHa: Data curation, Validation, Resources, Writing – review & editing. WH: Data curation, Methodology, Writing – review & editing. YaZ: Data curation, Methodology, Writing – review & editing. XC: Resources, Validation, Writing – review & editing. DL: Methodology, Writing – review & editing. YHo: Methodology, Writing – review & editing. HG: Methodology, Writing – review & editing. KZ: Supervision, Writing – review & editing. YuZ: Supervision, Writing – review & editing, Conceptualization. TS: Conceptualization, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Guangdong Basic and Applied Basic Research Foundation (2023A1515110409 to YC) and by Science, Technology and Innovation Commission of Shenzhen Municipality, China (to TS).
Acknowledgments
We sincerely thank Jinmei Ding and Dr. Qing Kong (National Institute of Biological Sciences, Beijing) for assistance with the sard1-1 suppressor screen, Dr. Qijun Chen (China Agricultural University) for sharing the pHEE401E constructs, Dr. Suomeng Dong (Nanjing Agricultural University) for Phytophthora infestans strain 1306 and Dr. Xili Liu (Northwest Agriculture & Forestry University) for Phytophthora capsici strain BYA5.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1490466/full#supplementary-material
References
Abrahamian, M., Ah-Fong., A. M. V., Davis., C., Andreeva., K., Judelson., H. S. (2016). Gene expression and silencing studies in phytophthora infestans reveal infection-specific nutrient transporters and a role for the nitrate reductase pathway in plant pathogenesis. PloS Pathog. 12, e1006097. doi: 10.1371/journal.ppat.1006097
Bi, D., Cheng, Y. T., Li, X., Zhang, Y. (2010). Activation of plant immune re sponses by a gain-of-function mutation in an atypical receptor-like kinase. Plant Physiol. 153, 1771–1779. doi: 10.1104/pp.110.158501
Chen, S., Ding, Y., Tian, H., Wang, S., Zhang, Y. (2021). WRKY54 and WRKY70 positively regulate SARD1 and CBP60g expression in plant immunity. Plant Signal Behav. 16, 1932142. doi: 10.1080/15592324.2021.1932142
Chen, Y. C., Holmes, E. C., Rajniak, J., Kim, J. G., Tang, S., Fischer, C. R., et al. (2018). N-Hydroxy-pipecolic acid is a mobile metabolite that induces systemic disease resistance in Ara bidopsis. Proc. Natl. Acad. Sci. U.S.A. 115, E4920–E4929. doi: 10.1073/pnas.1805291115
Chen, Y., Zhong, D., Yang, X., Zhao, Y., Dai, L., Zeng, D., et al. (2021). ZmFdC2 encoding a ferredoxin protein with C-terminus extension is indispensable for maize growth. Front. Plant Sci. 12, 646359. doi: 10.3389/fpls.2021.646359
Ding, P., Rekhter, D., Ding, Y., Feussner, K., Busta, L., Haroth, S., et al. (2016). Characterization of a pipecolic acid biosynthesis pathway required for systemic acquired resistance. Plant Cell 28, 2603–2615. doi: 10.1105/tpc.16.00486
Ding, Y., Sun, T., Ao, K., Peng, Y., Zhang, Y., Li, X., et al. (2018). Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in tran scriptional regulation of plant immunity. Cell 6, 1454–1467. doi: 10.1016/j.cell.2018.03.044
Fu, Z. Q., Dong, X. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. doi: 10.1146/annurev-arplant-042811-105606
Fu, Z. Q., Yan, S., Saleh, A., Wang, W., Ruble, J., Oka, N., et al. (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486, 228–236. doi: 10.1038/nature11162
Hartmann, M., Zeier, J. (2019). N-hydroxypipecolic acid and salicylic acid: a metabolic duo for systemic acquired resistance. Curr. Opin. Plant Biol. 50, 44–57. doi: 10.1016/j.pbi.2019.02.006
Hartmann, M., Zeier, T., Bernsdorff, F., Reichel-Deland, V., Kim, D., Hohmann, M., et al. (2018). Flavin monooxygenase-generated N-hydroxypipecolic acid is a critical element of plant systemic immunity. Cell 173, 456–469. doi: 10.1016/j.cell.2018.02.049
Huang, W., Wang, Y., Li, X., Zhang, Y. (2020). Biosynthesis and regulation of salicylic acid and N-hydroxypipecolic acid in plant. Mol. Plant 13, 31–41. doi: 10.1016/j.molp.2019.12.008
Huang, W., Wu, Z., Tian, H., Li, X., Zhang, Y. (2021). Arabidopsis CALMODULIN-BINDING PROTEIN 60b plays dual roles in plant immunity. Plant Commun. 2, 100213. doi: 10.1016/j.xplc.2021.100213
Jones, J. D. G., Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Jones, J. D. G., Staskawicz, B. J., Dangl, J. L. (2024). The plant immune system: From discovery to deployment. Cell 187, 2095–2116. doi: 10.1016/j.cell.2024.03.045
Kim, J. H., Castroverde, C. D. M., Huang, S., Li, C., Hilleary, R., Seroka, A., et al. (2022). Increasing the resilience of plant immunity to a warming climate. Nature 607, 339–344. doi: 10.1038/s41586-022-04902-y
Kurotani, K.I., Hirakawa, H., Shirasawa, K., Tanizawa, Y., Nakamura, Y., Isobe, S., et al. (2023). Genome sequence and analysis of nicotiana benthamiana, the model plant for interactions between organisms. Plant Cell Physiol. 64, 248–257. doi: 10.1093/pcp/pcac168
Lan, J., Chen, Y., Pico, J., Ao, K., Xia, S., Wang, C., et al. (2023). Epigenetic regulation of N-hydroxypipecolic acid biosynthesis by the AIPP3-PHD2-CPL2 complex. J. Integr. Plant Biol. 65, 2660–2671. doi: 10.1111/jipb.v65.12
Li, L. S., Ying, J., Li, E., Ma, T., Li, M., Gong, L. M., et al. (2021). Arabidopsis CBP60b is a central transcriptional activator of immunity. Plant Physiol. 186, 1645–1659. doi: 10.1093/plphys/kiab164
Lin, X., Jia, Y., Heal, R., Prokchorchik, M., Sindalovskaya, M., Olave-Achury, A., et al. (2023). Solanum americanum genome-assisted discovery of immune receptors that detect potato late blight pathogen effectors. Nat. Genet. 55, 1579–1588. doi: 10.1038/s41588-023-01486-9
Liu, Y., Sun, T., Sun, Y. (2020). Diverse roles of the salicylic acid receptors NPR1 and NPR3/NPR4 in plant immunity. Plant Cell 32, 4002–4016. doi: 10.1105/tpc.20.00499
Liu, Q., Vain, T., Viotti, C., Doyle, S. M., Hofius, D. (2018). Vacuole integrity maintained by DUF300 proteins is required for brassinosteroid signaling regulation. Mol. Plant 11, 553–567. doi: 10.1016/j.molp.2017.12.015
Liu, B., Yu, H, Yang, Q., Ding, L., Sun, F., Qu, J., et al. (2022). Zinc transporter zmLAZ1-4 modulates zinc homeostasis on plasma and vacuolar membrane in maize. Front. Plant Sci. 13, 1–12. doi: 10.3389/fpls.2022.881055
Malinovsky, F. G., Peter, B., Katrine, F. B., Vig, M. K. L., Stephan, T., Martina, B., et al. (2010). Lazarus1, a DUF300 protein, contributes to programmed cell death associated with arabidopsis acd11 and the hypersensitive response. PloS One 5, e12586. doi: 10.1371/journal.pone.0012586
Man, N. B. P., Pingtao, D., Jones, J. D. G. (2022). Thirty years of resistance: Zig-zag through the plant immune system. Plant Cell 34, 1447–1478. doi: 10.1093/plcell/koac041
Navarova, H., Bernsdorff, F., Doring, A. C., Zeier, J. (2012). Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell 24, 5123–5141. doi: 10.1105/tpc.112.103564
Peng, Y., Yang, J., Li, X., Zhang, Y. (2021). Salicylic acid: biosynthesis and signaling. Annu. Rev. Plant Biol. 72, 761–791. doi: 10.1146/annurev-arplant-081320-092855
Rekhter, D., Lüdke, D., Ding, Y., Feussner, K., Zienkiewicz, K., Lipka, V., et al. (2019). Isochorismate-derived biosynthesis of the plant stress hormone salicylic acid. Science 365, 498–502. doi: 10.1126/science.aaw1720
Sun, T., Busta, L., Zhang, Q., Ding, P., Jetter, R., Zhang, Y. (2018). TGACG-BINDING FACTOR 1 (TGA1) and TGA4 regulate salicylic acid and pipecolic acid biosynthesis by modulating the expression of SYSTEMIC ACQUIRED RESISTANCE DEFICIENT 1 (SARD1) and CALMODULIN-BINDING PROTEIN 60g (CBP60g). New Phytol. 217, 344–354. doi: 10.1111/nph.2018.217.issue-1
Sun, T., Huang, J., Xu, Y., Verma, V., Jing, B., Sun, Y., et al. (2020). Redundant CAMTA transcription factors negatively regulate the biosynthesis of salicylic acid and N -hydroxypipecolic acid by modulating the expression of SARD1 and CBP60g. Mol. Plant 13, 144–156. doi: 10.1016/j.molp.2019.10.016
Sun, L., Qin, J., Wu, X., Zhang, J., Zhang, J. (2022). TOUCH 3 and CALMODULIN 1/4/6 cooperate with calcium-dependent protein kinases to trigger calcium-dependent activation of CAM-BINDING PROTEIN 60-LIKE G and regulate fungal resistance in plants. Plant Cell 34, 4088–4104. doi: 10.1093/plcell/koac209
Sun, T., Zhang, Y. (2021). Short-and long-distance signaling in plant defense. Plant J. 105, 505–517. doi: 10.1111/tpj.v105.2
Sun, T., Zhang, Y., Li, Y., Zhang, Q., Ding, Y., Zhang, Y. (2015). ChIP-seq reveals broad roles of SARD1 and CBP60g in regulating plant immunity. Nat. Commun. 6, 10159. doi: 10.1038/ncomms10159
Torrens-Spence, M. P., Bobokalonova, A., Carballo, V., Glinkerman, C. M., Pluskal, T., Shen, A., et al. (2019). PBS3 and EPS1 complete salicylic acid biosynthesis from isochorismate in arabidopsis. Mol. Plant 12, 1577–1586. doi: 10.1016/j.molp.2019.11.005
Wang, L., Tsuda, K., Sato, M., Cohen, J. D., Katagiri, F., Glazebrook, J. (2009). Arabidopsis CaM Binding Protein CBP60g Contributes to MAMP-Induced SA Accumulation and Is Involved in Disease Resistance against Pseudomonas syringae. PloS Pathog. 5, e1000301. doi: 10.1371/journal.ppat.1000301
Wang, L., Tsuda, K., Truman, W., Sato, M., Nguyen, L. V., Katagiri, F., et al. (2011). CBP60g and SARD1 play partially redundant critical roles in salicylic acid signaling. Plant J. 67, 1029–1041. doi: 10.1111/j.1365-313X.2011.04655.x
Wang, Z. P., Xing, H. L., Dong, L., Zhang, H. Y., Han, C. Y., Wang, X. C., et al. (2015). Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 16, 144. doi: 10.1186/s13059-015-0715-0
Wang, W. Z., Xue, Z. L., Miao, Q. J., Cai, M., Zhang, C., Li, J. T., et al. (2019). PcMuORP1, an oxathiapiprolin-resistance gene, functions as a novel selection marker for phytophthora transformation and CRISPR/cas9 mediated genome editing. Front. Microbiol. 10, 2402. doi: 10.3389/fmicb.2019.02402
Wildermuth, M. C., Dewdney, J., Wu, G., Ausubel, F. M. (2001). Isochoris mate synthase is required to synthesize salicylic acid for plant defence. Nature 414, 562–565. doi: 10.1038/35107108
Wu, Z., He, L., Jin, Y., Chen, J., Shi, H., Wang, Y., et al. (2021). HISTONE DEACETYLASE 6 suppresses salicylic acid biosynthesis to repress autoimmunity1. Plant Physiol. 187, 2592–2607. doi: 10.1093/plphys/kiab408
Wu, Y., Zhang, D., Chu, J. Y., Boyle, P., Wang, Y., Brindle, I. D., et al. (2012). The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 1, 639–647. doi: 10.1016/j.celrep.2012.05.008
Yang, W., Liu., C., Fu., Q., Jia., X., Deng., L., Feng., C., et al. (2023). Knockout of SlbZIP68 reduces late blight resistance in tomato. Plant Sci. 336, 111861. doi: 10.1016/j.plantsci.2023.111861
Zhang, K., Bhuiya, M. W., Pazo, J. R., Miao, Y., Kim, H., Ralph, J., et al. (2012). An engineered monolignol 4-o-methyltransferase depresses lignin biosynthesis and confers novel metabolic capability in Arabidopsis. Plant Cell 24, 3135–3152. doi: 10.1105/tpc.112.101287
Zhang, Y., Xu, S., Ding, P., Wang, D., Cheng, Y., Jing, H., et al. (2010). Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc. Natl. Acad. Sci. U.S.A. 107, 18220–18225. doi: 10.1073/pnas.1005225107
Zhang, C., Yang, Z., Tang, D., Zhu, Y., Huang, S. (2021). Genome design of hybrid potato. Cell 184, 3873–3883. doi: 10.1016/j.cell.2021.06.006
Keywords: salicylic acid, N-hydroxypipecolic acid, CBP60g, SARD1, LAZ1
Citation: Chen Y, Han Y, Huang W, Zhang Y, Chen X, Li D, Hong Y, Gao H, Zhang K, Zhang Y and Sun T (2024) LAZARUS 1 functions as a positive regulator of plant immunity and systemic acquired resistance. Front. Plant Sci. 15:1490466. doi: 10.3389/fpls.2024.1490466
Received: 03 September 2024; Accepted: 30 October 2024;
Published: 20 November 2024.
Edited by:
Yasser Nehela, University of Florida, United StatesReviewed by:
Aswin Nair, Umeå University, SwedenToshik Iarley Da Silva, Federal University of the Recôncavo of Bahia, Brazil
Qi Zheng, Shandong University, China
Copyright © 2024 Chen, Han, Huang, Zhang, Chen, Li, Hong, Gao, Zhang, Zhang and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tongjun Sun, c3VudG9uZ2p1bkBjYWFzLmNu
 Yue Chen1,2
Yue Chen1,2 Yanjun Zhang
Yanjun Zhang Kewei Zhang
Kewei Zhang Tongjun Sun
Tongjun Sun