- 1Department of Agriculture, Food and Environment, University of Pisa, Pisa, Italy
- 2Department of Biological and Environmental Sciences and Technologies, University of Salento, Lecce, Italy
- 3Department of Agriculture Technologies, Food, Environment and Forestry, University of Florence, Florence, Italy
- 4Regional Phytosanitary Service, Laboratory of Phytopathological Diagnostics and Molecular Biology, Pistoia, Italy
Flavescence dorée (FD) is a major grapevine disease in Europe, despite the quarantine status of its causal agent [FD phytoplasma (FDp)] and the mandatory monitoring and vector control practices. As alarming FD epidemic outbreaks continue to appear in Tuscany (Central Italy), a 4-year survey was carried out in the main wine-growing areas of the region, where FD presence was investigated in both primary and secondary FDp hosts and vectors, i.e., Vitis vinifera (VV), Clematis vitalba (CV), Alnus glutinosa (AG), Scaphoideus titanus (ST), and Dictyophara europaea (DE). This work i) confirmed FD diffusion in almost the whole of Tuscany and even with an increased occurrence rate (approximately 50% of the samples tested positive); ii) highlighted a complex FDp ecology also in the Tuscan vineyard agroecosystem, as FDp was reported not only in VV and ST but also in secondary vectors (DE) and hosts (AG and CV); iii) reported nine FDp strains (three of which were novel) belonging to all the three methionine aminopeptidase (map) clusters, i.e., map-FD1, map-FD2, and map-FD3, with the map-FD3/M51 genotype mostly reported in not only VV but also DE and CV; and iv) further confirmed a complex FDp ecology in the vineyard agroecosystem, also by phylogenetic analyses carried out at both Italian and European levels, which also showed some relations between the Tuscan FDp strains and those reported in the Balkan and the French scenarios. We believe that the outcomes reported here will be useful in preventing and controlling the spread of harmful FD.
1 Introduction
Flavescence dorée (FD) is a major disease of grapevine (Vitis vinifera) in Europe, causing both severe yield losses and lower grape quality, with harsh economic consequences in the main wine- and vine-producing countries (EFSA et al., 2020). It is associated with flavescence dorée phytoplasma (FDp), which is transmitted from vine to vine by infected Scaphoideus titanus (a monovoltine leafhopper accidentally introduced from North America in the 1950s) through its feeding activity on leaves (Chuche and Thiéry, 2014). FD symptoms develop gradually throughout the summer, reaching their peak appearance in late August–September. Affected grapevines exhibit a drooping appearance due to absent lignification, and leaves generally become crisp, brittle, and curled downward, with white and red grape varieties showing yellowing and reddening, respectively. FDp also leads to inflorescence and berry death (Boulent et al., 2020). FDp-infected plants die or become less productive over the years, also constituting a potential source of inoculum. These symptoms are identical to those of Bois noir (BN), another phytoplasma disease associated with ‘Candidatus phytoplasma solani’ (BNp); to distinguish FD from BN, both referred to as “grapevine yellows”, laboratory analyses are needed (Chuche and Thiéry, 2014). Despite the quarantine status of FDp (included in the A2 EPPO list; EPPO, 2024a) and the (costly) mandatory monitoring and vector control practices carried out with a high pesticide consumption (there is no phytoplasma control method and no cure for infected plants), FD is still spreading in several European countries (EFSA et al., 2020).
Recent studies have reported that the epidemiological cycle of FDp could be more complex than the vine-to-vine transmission by S. titanus, as i) FDp was also found in other plant hosts (mostly asymptomatic) located close to vineyards, such as Alnus glutinosa (Angelini et al., 2004), Clematis vitalba (Casati et al., 2017), Ailanthus altissima (Plavec et al., 2019), and Salix spp (Jarausch et al., 2023); ii) S. titanus can occasionally feed on other plant species that may act as FDp reservoirs, although the insect cannot complete its development on them (Trivellone et al., 2013), and these host plants may act as pathogen reservoirs (Chuche and Thiéry, 2014; Moussa et al., 2023); and iii) FDp can be transmitted to grapevines by other insects, such as Allygus spp., Dictyophara europaea, Orientus ishidae, and Phlogotettix cyclops (Ripamonti et al., 2020; Galetto et al., 2023). Overall, given the abovementioned alarming signs of FDp diffusion in many European viticultural areas and the ecological complexity of FDp, further investigations are needed to ascertain the causes and origins of new outbreaks and predict the route of disease spread, with a precise identification and typing of FDp strains playing a crucial role in this context (Arnaud et al., 2007).
Based on nucleotide sequence analysis of the gene 16S rRNA and the intergenic spacer between 16S and 23S rRNA, FDp has been assigned to the 16SrV taxonomic group (elm yellows group) and specifically to the subgroups 16SrV-C and 16SrV-D (Lee et al., 1998; Angelini et al., 2001; Davis and Dally, 2001; Lee et al., 2004; EPPO, 2024b). However, the variability of 16S rRNA is not sufficient to account for the abovementioned emerging ecological differences in FDp biological cycles (i.e., secondary vectors and hosts). In contrast, molecular and phylogenetic analyses of the epidemiologically informative genetic loci (i.e., methionine aminopeptidase, map; ribosomal proteins, rp; preprotein translocase membrane subunit, secY; subunit B of the exonuclease family protein, uvrB-degV; and adhesion-related proteins, vmp) allowed the identification of different FDp genetic clusters. Specifically, the map gene was shown to be the most auspicious marker for the genetic characterization of phytoplasma strains within the 16SrV group (Plavec and Music, 2023) and allowed the recognition of three FDp genetic clusters, namely, map-FD1 (16SrV-C), map-FD2 (16SrV-C and 16SrV-D), and map-FD3 (16SrV-C), each of which differs in genetic variability and more importantly is characterized by specific vectors, hosts, and geographical distribution (Arnaud et al., 2007; Malembic-Maher et al., 2020). To date, only map-FD1 and map-FD2 have been found in France and Switzerland (Arnaud et al., 2007; Casati et al., 2017; Malembic-Maher et al., 2020), only map-FD2 has been reported in Portugal (De Sousa et al., 2010), only map-FD2 and map-FD3 have been identified in Austria and Slovenia (Mehle et al., 2011; Reisenzein and Steffek, 2011), and map-FD3 has been exclusively found in Serbia (Malembic-Maher et al., 2020; Krnjajić et al., 2007; Holz et al., 2016; Krstić et al., 2018). All three genotype clusters have been reported in Croatia, Hungary, and Italy (Angelini et al., 2001; Martini et al., 2002; Rossi et al., 2019; Kriston et al., 2019; Plavec et al., 2019; Malembic-Maher et al., 2020).
Although FDp has been present for a long time in Italy (Belli et al., 2010)—beyond the fact that Italy is renowned for consistently ranking as the world’s largest wine producer and for producing some of the world’s most excellent wines (Ohana-Levi and Netzer, 2023)—studies that have focused on FDp diffusion and genetic typing of FDp strains have been mostly performed in regions of Northern Italy (Schubert, 2017; Angelini et al., 2001; Martini et al., 2002; Rossi et al., 2019), leaving out other major viticultural areas such as Tuscany (Central Italy), which is characterized by a warmer Mediterranean climate. The presence of FDp in Tuscany was first reported in a northern province more than two decades ago (Bertaccini et al., 2003) and confirmed more recently in other central and southern traditional viticultural areas by field surveys carried out from 2012 to 2015 (Rizzo et al., 2018) and in 2017 (Pierro et al., 2023), also reporting an increasing diffusion from 2%–11% to 17% of positive samples, respectively. However, only very few FDp strains from Tuscany have been characterized molecularly, mainly resulting in cluster map-FD1 strains (Arnaud et al., 2007; Malembic-Maher et al., 2011; Malembic-Maher et al., 2020). Actually, we carried out the first multi-locus sequence typing (MLST) of 15 FDp strains isolated in 2017, determined through the analysis of the 16S rRNA, rp, and secY genes, showing that i) the Tuscan FDp strains are within the subgroup 16SrV-C, with many strains resulting very close to others previously identified not only in Northern Italy but also in France, Slovenia, and Serbia; and ii) the collective genotype (16S rRNA/rp/secY) of the Tuscan FDp strains is unique and constitutes a new highly homogeneous lineage (Pierro et al., 2023). However, comprehensive of the population and molecular features of the Tuscan FDp strains by map gene typing (i.e., the most suitable approach for the genetic characterization of FDp, as stated above; Plavec and Music, 2023) has not been performed. Furthermore, a study of FDp ecology in the Tuscan vineyard agroecosystem (i.e., characterizing FDp strains collected also from secondary hosts and vectors) has never been carried out before the present study.
To address these knowledge gaps, and as alarming FD epidemic outbreaks continue to appear in Tuscany (as reported by the monitoring activity carried out by both the Department of Agriculture, Food and Environment of the University of Pisa, and the Regional Phytosanitary Service of the Region of Tuscany), also emerging in diverse habitats, this study aimed to i) elucidate the current FDp diffusion through a long-term field survey (2020–2023) carried out in the major viticultural areas of Tuscany, ii) investigate the FDp epidemiological cycle in the whole vineyard agroecosystem, iii) determine the genetic diversity among the collected FDp strains by map gene typing, and iv) elucidate the evolutionary relationships of Tuscan FDp strains with those reported in Italy, as well as in Europe. We anticipate that the outcomes reported here will be useful in preventing and controlling the spread of harmful FD.
2 Materials and methods
2.1 Field surveys and sampling of leaf and insect
Experimental activities were carried out from 2020 to 2023. In total, 12 vineyards located in the main wine-growing areas of Tuscany (Central Italy; two vineyards per province), i.e., Aulla and Licciana Nardi (Massa, MS), Capannori and Aquilea (Lucca, LU), Serravalle Pistoiese and Quarrata (Pistoia, PT), Gaiole in Chianti, Radda in Chianti (Siena, SI), Greve in Chianti, San Casciano in Val di Pesa (Florence, FI), and Bucine and Montevarchi (Arezzo, AR) were selected in collaboration with the Regional Phytosanitary Service of the Region of Tuscany (Figure 1). Leaf samples were collected at the end of August/September from V. vinifera cv. Sangiovese (VV), as well as from C. vitalba (CV) and A. glutinosa (AG) located at the edges of the examined vineyards. Concomitantly, S. titanus (ST) and D. europaea (DE) were collected using chromotropic traps placed in the vineyards at grapevine canopy height, as well as in the nearby wood margins. A total of 276 samples were collected, i.e., 218 VV, 24 ST, 20 DE, 10 CV, and 4 AG. The leaves were placed in coolers and quickly carried to the Plant Pathology Laboratory of the Department of Agriculture, Food, and Environment (DAFE) of the University of Pisa, where their midribs were dissected and stored at −20°C. The insects were recovered from traps, identified by experts of the Entomology Laboratory of DAFE, placed in 70% (v/v) ethanol, and stored at +4°C.
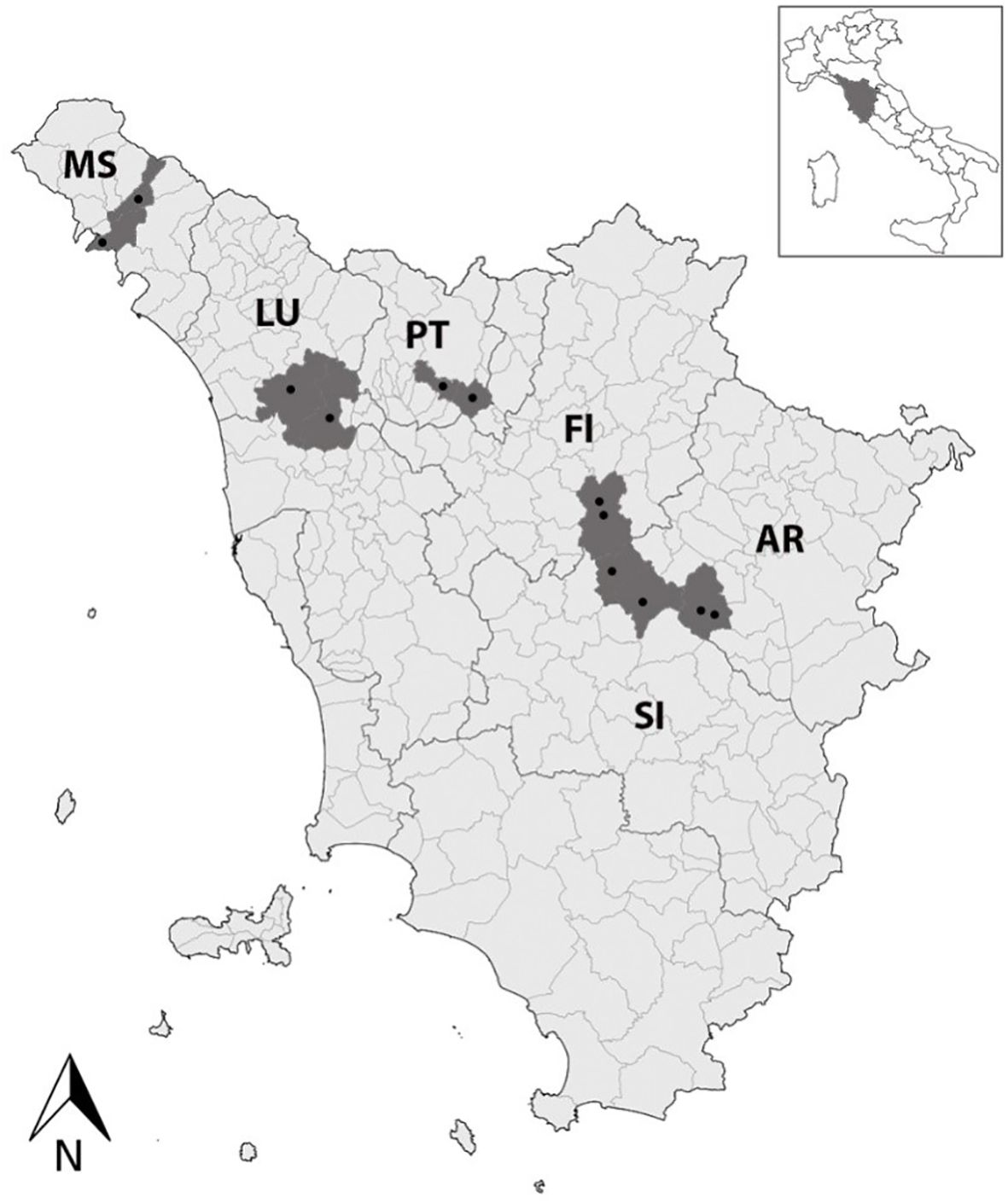
Figure 1. A map of Tuscany (Central Italy) showing the 12 investigated vineyards (black dots) located in the main wine-growing areas, i.e., Aulla and Licciana Nardi (Massa, MS), Capannori and Aquilea (Lucca, LU), Serravalle Pistoiese and Quarrata (Pistoia, PT), Gaiole in Chianti, Radda in Chianti (Siena, SI), Greve in Chianti, San Casciano in Val di Pesa (Florence, FI), and Bucine and Montevarchi (Arezzo, AR).
2.2 DNA isolation
A cetyltrimethylammonium bromide (CTAB) buffer procedure was used to extract DNA from leaves, according to Li et al. (2008), with minor modifications (Pedrelli et al., 2023a; Pedrelli et al., 2023c). Approximately 0.5 g of leaf midribs was powdered in liquid nitrogen, added to 5 mL of 2% (w/v) CTAB buffer, and incubated at +65°C for 15 min. DNA was extracted by one volume of chloroform:isoamyl alcohol (24:1, v/v) and precipitated with one volume of isopropanol. The pellets were then washed with 70% (v/v) ethanol, air-dried, and dissolved in 80 μL of RNase/DNase-free water.
In contrast, DNA from insects was extracted following Marzachi et al. (1998), with minor modifications. Every single insect was inserted into a 2.0-mL screw cap microcentrifuge tube containing six 2.8-mm ceramic (zirconium oxide) slingshot beads (Bertin Technologies SAS, Montigny-le-Bretonneux, France), and 400 μL of 2.5% (v/v) CTAB buffer was added. Then, tubes were centrifuged at 7,200 g for 10 s using Precellys Evolution Touch (Bertin Technologies SAS). The formed suspensions were incubated at +60°C for 30 min and then extracted with chloroform:isoamyl alcohol (24:1, v/v). DNA was precipitated by adding one volume of cold isopropanol and recovered by centrifugation at 14,000 g for 3 min, washed with 70% (v/v) ethanol, air-dried, and re-suspended in 80 μL of RNase/DNase-free water.
The DNA (extracted from both leaves and insects) was finally purified using the DNeasy PowerClean Pro Cleanup Kit (Qiagen, Venlo, The Netherlands) according to the manufacturer’s instructions, checked for their quality and quantity using a QIAExpert system (Qiagen), and stored at −80°C until molecular analysis.
2.3 16S rRNA phytoplasma subgroup V and XII detection and map gene amplification
Samples were tested for the presence of 16S rRNA phytoplasma subgroups V (16SrV) and XII (16SrXII) by real-time polymerase chain reaction (qPCR) using specific primer pairs (Angelini et al., 2007). The qPCR assays were carried out by a Rotor-Gene Q Thermocycler (Qiagen) using a 20-μL reaction volume containing a QuantiNova Probe (Qiagen). Moreover, phytoplasma identification was confirmed by end-point PCR for 16S rRNA using the universal P1/P7 primer pair (Deng and Hiruki, 1991), followed by the R16(V)F1/R1 and R16(XII)F1/R1 ones for the detection of 16SrV and 16SrXII, respectively (Lee, 1994). Differently, the protocol described by Arnaud et al. (2007) was adopted to amplify the map genetic loci. PCRs were conducted employing the FD9F5/MapR1 primer pair (direct PCR), followed by the FD9F6/MapR2 one (nested PCR). PCR and nested PCR were performed using a C1000 Touch® thermal cycler (Bio-Rad, Hercules, CA, USA) in reaction volumes of 25 μL and 50 μL, respectively, including DreamTaq PCR Master Mix and DreamTaq Green PCR Master Mix (Thermo Fisher Scientific, Waltham, MA, USA), respectively. PCR-amplified products were observed by electrophoresis through 1.2% (w/v) agarose gel and sequenced following the Sanger DNA method (Eurofins, Ebersberg, Germany). Positive, healthy, and no-template controls, were included in each molecular assay.
2.4 16S rRNA and map gene amplicon sequencing, characterization, and in silico analysis
Amplicons from 16SrV-positive samples and the corresponding map gene were sequenced following the Sanger DNA method by the Eurofins Genomics commercial service (Ebersberg, Germany). Group/subgroup assignment (16SrV-C/D; 16SrXII) of the 16S rRNA nucleotide sequences was accomplished using the online tool iPhyClassifier (Zhao et al., 2009). map gene amplicons were typed by restriction fragment length polymorphism (RFLP) analysis performed through the AluI and Eco72I restriction enzymes in order to characterize their map cluster (i.e., map-FD1, map-FD2, and map-FD3; Arnaud et al., 2007). Nucleotide and amino acid sequences were analyzed in silico using the BioEdit software (Hall, 1999), and nucleotide sequences were also compared in BLASTn (www.ncbi.nlm.ni.gov) and to the dataset provided by Malembic-Maher et al. (2020). Alignments were performed to observe the presence of non-synonymous (dN) and synonymous (dS) single-nucleotide and amino acid polymorphisms, and a sequence identity determination was carried out by the sequence identity matrix application of BioEdit. The role of natural selection in the FDp population using the Codon Z-test by the Nei–Gojobori method and the dN/dS ratio by the Felsenstein (1981) model was estimated using MEGA X (Pedrelli et al., 2023b). Phylogenetic analyses were carried out in MEGA X using the maximum likelihood (ML) and neighbor-joining methods (using the Jukes–Cantor and p-distance models, respectively) with 1,000 bootstrap replicates (Kumar et al., 2018). A BN isolate (AM990988) was used as an outgroup. The used map strain dataset is reported in Supplementary Table S1.
3 Results and discussion
3.1 Diffusion of FD was confirmed in almost the whole Tuscany and even with an increased occurrence rate
The present study first confirmed that, despite the regulation that requires FDp monitoring and vector control practices, along with the destruction of vineyards when they are infected with FDp, this threatening disease is still present in Tuscany (Central Italy; Bertaccini et al., 2003; Rizzo et al., 2018; Pierro et al., 2023), in the regions of Northern Italy (Rigamonti et al., 2023; Quaglino et al., 2024), and other European areas (e.g., Plavec et al., 2019; Krstić et al., 2022). The typical FD leaf symptoms, i.e., reddening and downward rolling, observed on all collected VV leaves were indeed confirmed by molecular assays, which reported the presence of FDp in a large number of samples (47%; Table 1), whereas only four VV samples tested positive for BNp (2%), in accordance with Pierro et al. (2018), and with no mixed infection. FDp-positive samples were collected in all the investigated Tuscan provinces (i.e., Arezzo, Florence, Lucca, Massa, Pistoia, and Siena; Figure 1). These outcomes confirm that the FD outbreak has extended to almost the whole of Tuscany (Pierro et al., 2023), but also suggest an alarming increase in the disease occurrence rate, given the rise in the percentage of FDp-positive samples from 2%–11% and 17% reported in previous surveys (Rizzo et al., 2018; Pierro et al., 2023) to almost 50% of the present study.
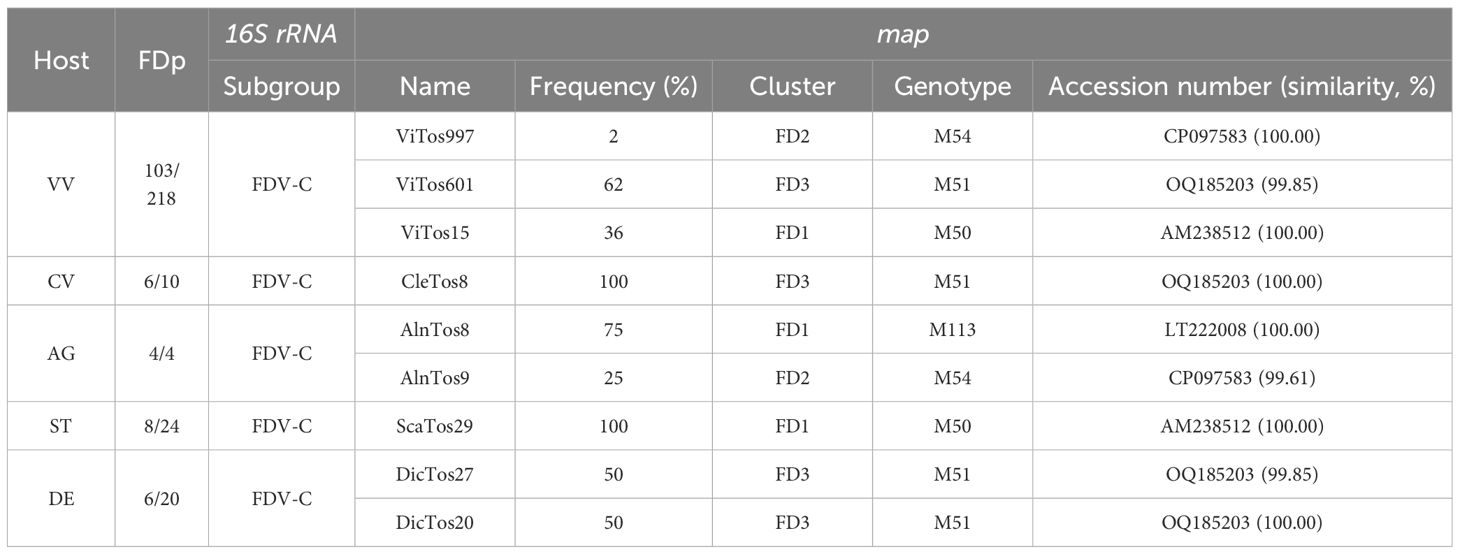
Table 1. Host (VV, Vitis vinifera; CV, Clematis vitalba; AG, Alnus glutinosa; ST, Scaphoideus titanus; DE, Dictyophara europaea) and samples tested positive for flavescence dorée phytoplasma (FDp)/samples investigated by PCR assay; subgroup obtained by iPhyClassifier of FDp strains identified by 16s rRNA typing; and name, frequency of occurrence, cluster, associated genotype, and accession number and related similarity obtained by BLASTn analysis (AM238512, isolate FD70; CP097583, isolate CH; LT222008, isolate AI-AL4; OQ185203, isolate 6023/2021) of FDp strains identified by map typing.
3.2 FDp was also reported in secondary vectors and hosts, highlighting a complex FDp ecology in Tuscany
A second major achievement of the present study was the first detection of FDp in multiple plants and insects collected inside and outside vineyards in Tuscany, although the presence in vineyard agroecosystems of different leafhopper and plant species infected with FDp was already reported in other regions of Northern Italy (Ripamonti et al., 2021; Rigamonti et al., 2023; Quaglino et al., 2024) and Europe (Rizzoli et al., 2021; Radonjić et al., 2023; Kogej Zwitter et al., 2023; Oggier et al., 2023). Although no macroscopic symptoms were reported on the collected CV and AG leaves, the molecular assays reported the presence of FDp in 60% of CV samples and all AG ones, and interestingly, FDp was also reported in 33% and 30% of ST and DE samples, respectively (Table 1). It is, therefore, reasonable to hypothesize that in Tuscany, the ecological cycle of FDp may be related exclusively not only to the grapevine-specific feeding of S. titanus, but also to that of other insect vectors and plant hosts (Casati et al., 2017). Previous studies have suggested that the vector competence of FDp transmission could be widespread and proposed the question of whether in a viticultural area the range/number of effective insect vector species could be determined by the availability of suitable host plant species in a system where FDp strains flow among many different hosts, inside and outside the vineyards (Bressan et al., 2006).
3.3 The map gene typing of nine FDp strains collected in Tuscany showed that they belong to all three map clusters, with the map-FD3/M51 genotype mostly reported not only in VV but also in DE and CV
The abovementioned diagnostic results were confirmed by end-point PCRs targeting the 16S rRNA gene, which also allowed us to obtain amplicons of the expected size. Specific primers for the map genetic loci enabled the generation of a comparable number of amplicons to those obtained from samples that previously tested positive for FDp. As expected, the iPhyClassifier, utilized using our 16S rRNA sequences as queries, revealed that the identified FDp strains belonged to the 16SrV-C subgroup (Table 1), not only those found in VV samples, which is in agreement with previous observations in Tuscany (Pierro et al., 2023), but also those reported in the other primary and secondary FDp insect vectors and plant hosts.
The alignment of the 127 map nucleotide sequences (657–763 bp) and their RFLP analysis allowed the identification of nine FDp strains, which (as expected) belonged to all the known three map clusters (Arnaud et al., 2007; Malembic-Maher et al., 2020; Plavec and Music, 2023): four to map-FD3, three to map-FD1, and two to map-FD2 (Table 1). Actually, the identification of two of our FDp strains belonging to the map-FD2 cluster disagrees with the association of all the 16S rRNA sequences to the 16SrV-C subgroup reported above (Arnaud et al., 2007; Zhao et al., 2009). However, the occurrence of these FDp strains changed depending on the type of samples collected in the Tuscan vineyard agroecosystem: genotypes collected in VV mostly belonged to the map-FD3 cluster (62% of FDp-positive samples), followed by map-FD1 (36%) and map-FD2 (2%), whereas CV was infected by only one genotype belonging to map-FD3 (100%), and the most frequent cluster in AG was map-FD1 (75%), followed by map-FD2 (25%). All FDp strains collected from ST and DE belonged to the map-FD1 and map-FD3 clusters, respectively.
BLASTn analysis of the nine map nucleotide sequences allowed the identification of three novel FDp strains each collected in VV, DE, and AG, and named ViTos601, DicTos27, and AlnTos9, respectively (Table 1). ViTos601 and DicTos27 showed a 99.85% similarity with isolate 6023/2021 (OQ185203) previously found in VV in the Czech Republic and were related to the M51 genotype; AlnTos9 was characterized by a 99.61% similarity with isolate CH (CP097583) previously found in an unidentified insect in Switzerland and was associated with the M54 genotype (Malembic-Maher et al., 2020). For the remaining six isolates, two were collected from VV (ViTos997 and ViTos15) and one each from CV, AG, ST, and DE (CleTos8, AlnTos8, ScaTos29, and DicTos20, respectively). All of them were identical (100.00% similarity) to previously identified isolates: ViTos997 to the abovementioned CH (CP097583, M54 genotype), ViTos15 and ScaTos29 to FD70 (AM238512, M50 genotype, previously reported in Catharanthus roseus in France), CleTos8 and DicTos20 to the abovementioned 6023/2021 (OQ185203, M51 genotype), and AlnTos8 to AI-AL4 (LT222008, M113 genotype, previously reported in AG in Tuscany); Malembic-Maher et al., 2020; Table 1). All the identified isolates were deposited in GenBank (www.ncbi.nlm.ni.gov; Supplementary Table S1).
Contrary to previous studies reporting the map-FD2/M54 FDp strain as the most prevalent genotype in Northern Italy (Rigamonti et al., 2023), here, map-FD3/M51 (ViTos601) was the most prevalent genotype reported in VV, followed by map-FD1/M50 (ViTos15), and only a few VV samples were infected with map-FD2/M54 (ViTos997). In fact, the map-FD1/M50 genotype (ViTos15) was also the only one reported in ST (ScaTos29), confirming the role of this insect as the main FDp vector in vineyards (Rizzoli et al., 2023). However, the fact that the novel map-FD3/M51 genotype, largely reported in VV (ViTos601) was also found in DE (DicTos27) hypothetically suggests that DE is also capable of (occasionally) transmitting FDp directly to VV (Radonjić et al., 2023). Furthermore, the association of both FDp strains reported in DE and (asymptomatic) CV (DicTos20 and CleTos8) with the same map-FD3/M51 genotype confirms the ability of FDp transmission between these secondary insect vectors and plant hosts, according to previous studies reporting the same genotype in other Italian and European regions (Angelini et al., 2004; Filippin et al., 2009; Linhartová et al., 2023; Quaglino et al., 2024). This prevalence of the map-FD3/M51 genotype in secondary FDp host plants and insect vectors also confirms a link between Tuscany and Eastern European nations like Croatia, Montenegro, and Serbia (Plavec et al., 2019; Krstić et al., 2022; Radonjić et al., 2023). Finally, AG was confirmed to be a potential (asymptomatic) reservoir of FDp, here, with the map-FD1/M113 and map-FD2/M54 related genotypes, confirming its capacity to host multiple FDp genotypes (Malembic-Maher et al., 2020). Numerous investigations have demonstrated the ability of insect vectors such as Allygus spp. and Oncopsis spp. to acquire FDp from AG and transfer it to other hosts, even if VV is not currently included (Maixner et al., 2000; Jarausch et al., 2021).
3.4 The phylogenetic analysis carried out at the Italian and European levels confirmed a complex FDp ecology, with some relations with the Balkan and the French scenarios
Identity levels between map nucleotide sequences ranged from 97.84% to 100.00%, with the maximum difference observed between ViTos997 and AlnTos8 and the minimum ones between ViTos601 and DicTos27, ViTos15, and ScaTos9, and CleTos8 and DicTos20 (Table 2; the 100.00% similarities are reported above). Overall, the genetic variability (π) was estimated to be 0.013 ± 0.003 (mean ± standard error) if using only the FDp strains reported in the present study and 0.010 ± 0.003 if including also the other FDp strains previously collected in Tuscany. These values are in agreement with those emerging from the analysis of FDp strains found in Veneto (n = 16, 0.017 ± 0.003) and Friuli Venezia Giulia (n = 6, 0.015 ± 0.003; other Italian regions were not taken into account because of the low number of FDp strains available). The amount of SNPs among the nine sequences of the FDp strains reported here was 54: 13 nt for ViTos997 with 3 dN, 11 nt for AlnTos9 with 4 dN, 10 nt each for ScaTos29 and ViTos15 with 3 dN each, 8 nt for AlnTos8 with 2 dN, and 1 nt each for CleTos8 and DicTos20 without dN. The overall dN/dS ratio was 0.08 (p = 0.001), taking into account only the FDp strains collected in the present study, 0.10 (p = 0.002), including also those previously reported in Tuscany, and 0.18 and 0.19, analyzing those from Veneto (p < 0.001) and Friuli Venezia Giulia (p = 0.001), respectively. These outcomes suggest a satisfactory adaptation of FDp in Tuscany, even more than in the other investigated Italian regions, a phenomenon that likely favors a high stability of the FDp genetic structure (Escriu, 2017; Pedrelli et al., 2024).
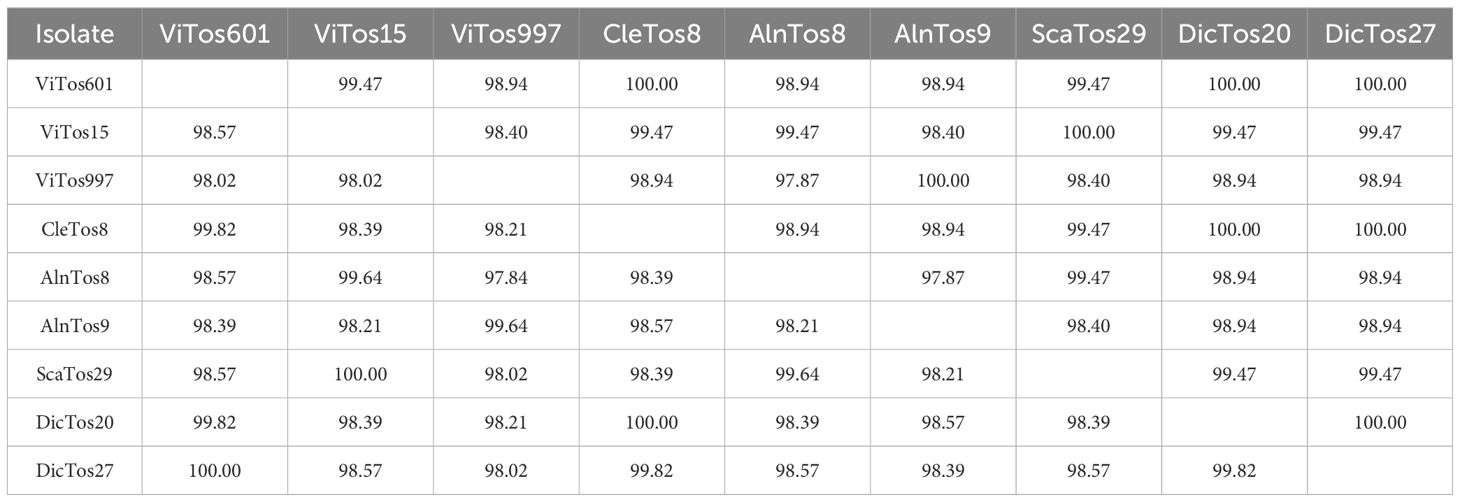
Table 2. Identity matrix between flavescence dorée phytoplasma (FDp) nucleotide (lower left) and deduced amino acid (upper right) sequences recovered in Tuscan vineyards located in the wine-growing areas.
The phylogenetic analysis conducted using the available Italian map nucleotide sequences (i.e., those reported here and previously submitted to NCBI; Supplementary Table S1) confirmed the occurrence of the three map clusters (Arnaud et al., 2007) with high bootstrap values (Figure 2; Supplementary Figure S1), with the nine FDp strains reported in the present study distributed among them. In the map-FD3 cluster, DicTos27 and ViTos601 (related to the M51 genotype) were close together and close to isolates previously reported in AG in the Northern Regions, i.e., Veneto (AI-365-07, LT221967, M72) and Piedmont (AI-040-08, LT221952, M57). Conversely, DicTos20 and CleTos8 (M51) were closely related to the isolate Vv-SI257 (FN811141, M51) found in VV in Tuscany, and interestingly, they also grouped with isolates CL-UD147 (FN811142) and CI-CL-UD147 (LT222014, M119) collected in CV in Friuli Venezia Giulia, i.e., the only isolates reported in CV at the Italian level. In fact, using all the available European map nucleotide sequences, DicTos27 and ViTos601, as along with DicTos20 and CleTos8, were also found to be related to other FDp strains collected in the Balkans (i.e., Montenegro and Serbia): the former was close to strains reported only in VV, and the latter was close to strains also found in CV (and A. altissima, an invasive alien species; Pisuttu et al., 2020, 2023; Figure 3; Supplementary Figure S2). These results confirm the relationship between DE and CV and highlight that CV is (so far) exclusively infected by map-FD3 genotypes also in Tuscany (Malembic-Maher et al., 2020; Radonjić et al., 2023).
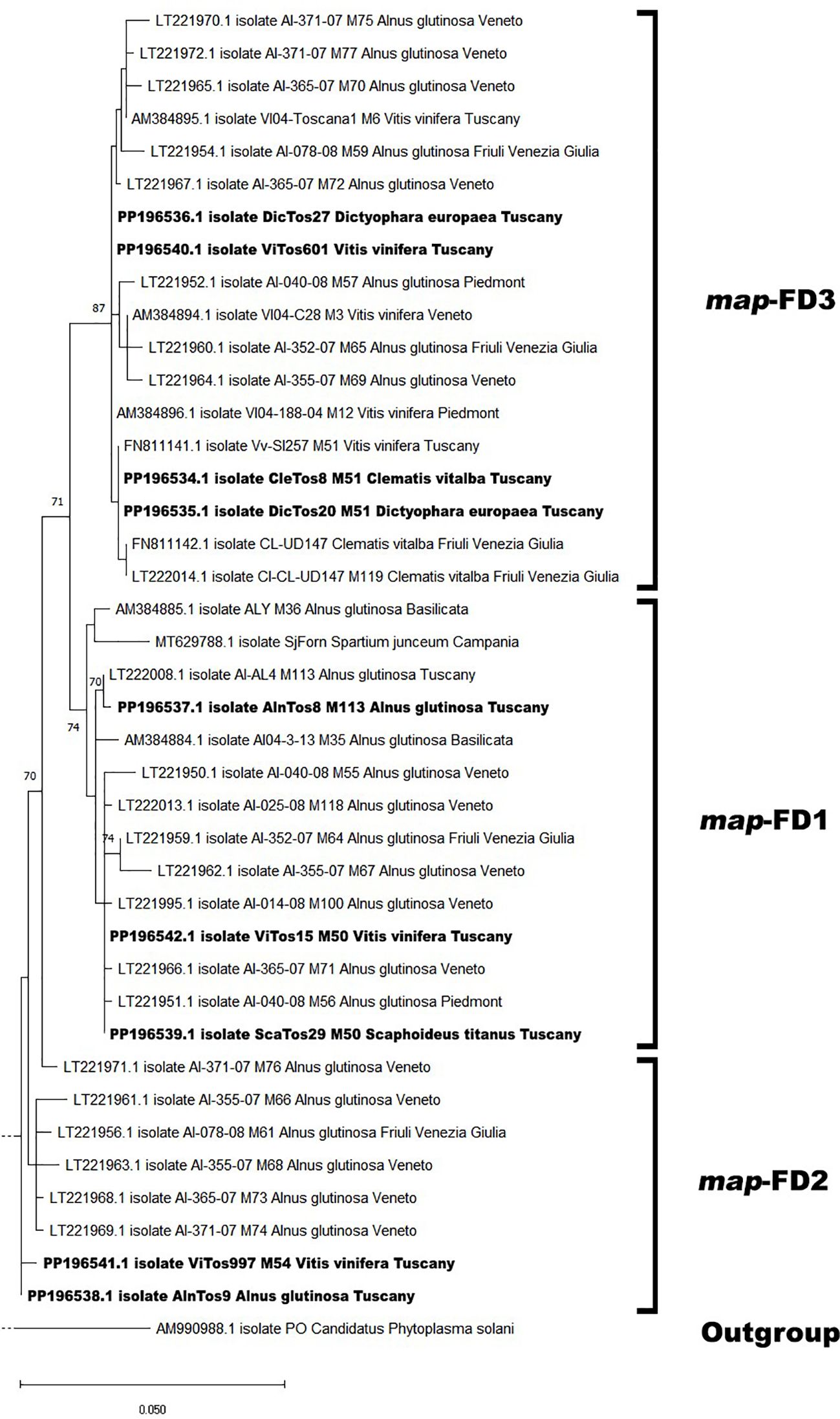
Figure 2. Phylogenetic tree of flavescence dorée phytoplasma (FDp) isolates from Italy reconstructed from partial map genes. The tree was generated by maximum likelihood (ML) method using the Jukes–Cantor model of evolution for nucleotides. The significance of each branch was evaluated by constructing 1,000 trees in bootstrap analysis. Bootstrap values >70 are shown. The scale represents a distance of 0.050 substitutions per site. The isolates sequenced in this study are in bold, and the subdivisions between clusters are reported on the right. The Bois noir phytoplasma (BNp) isolate (AM990988) was used as an outgroup. A more detailed version of the phylogenetic tree is reported in Supplementary Figure S1.
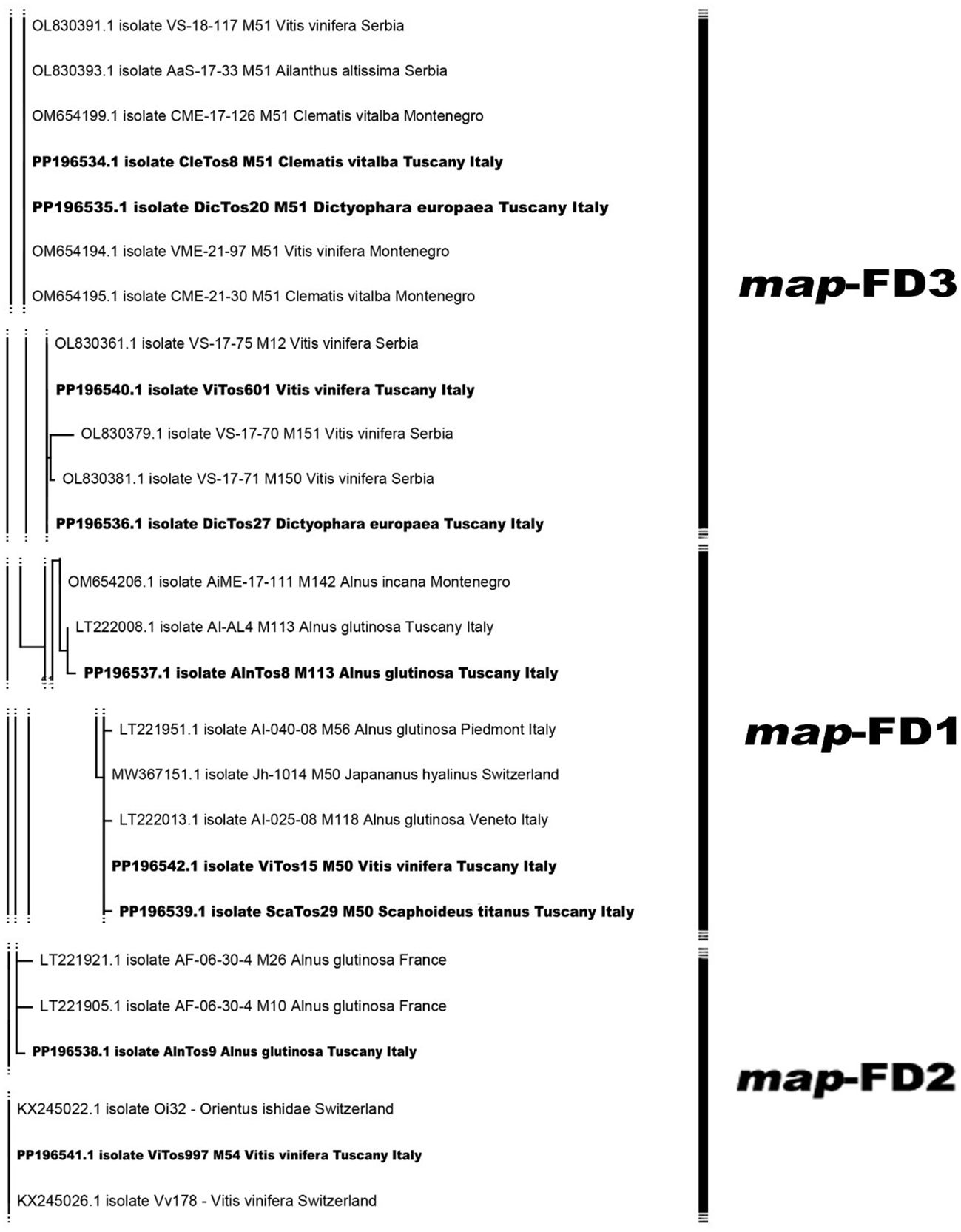
Figure 3. Phylogenetic tree of flavescence dorée phytoplasma (FDp) isolates from Europe reconstructed from partial map genes. The tree was generated by maximum likelihood (ML) method using the Jukes–Cantor model of evolution for nucleotides. The significance of each branch was evaluated by constructing 1,000 trees in bootstrap analysis. Bootstrap values >70 are shown. The scale represents a distance of 0.050 substitutions per site. The isolates sequenced in this study are in bold, and the subdivisions between clusters are reported on the right. The Bois noir phytoplasma (BNp) isolate (AM990988) was used as an outgroup. A more detailed version of the phylogenetic tree is reported in Supplementary Figure S2.
In contrast, the map-FD1 cluster was found to be mostly composed of isolates collected only in AG in Northern Regions and in the other secondary FDp host Spartium junceum in Campania (MT629788), and, indeed, included the FDp strain reported here, AlnTos8 (M113 genotype), which showed a high bootstrap value with isolate AI-AL4 (LT222008, M113) already reported in AG in Tuscany (Figure 2; Supplementary Figure S1). Accordingly, the phylogenetic analysis conducted at the European level also placed AlnTos8 close to a strain previously collected in Alnus incana, again in the Balkans (Montenegro; Figure 3). Interestingly, however, we also reported the occurrence in the map-FD1 cluster of FDp strains reported in VV and ST (ViTos15 and ScaTos29, M50 genotypes), which were also placed close to the isolates AI-40-08 (LT221951, M56), AI-365-07 (LT22966, M71), and AI-014-08 (LT221995, M100) found in AG in Piedmont and Veneto (Figure 2; Supplementary Figure S1). This first Italian submission of map-FD1/M50 genotypes collected in VV and ST confirms the complexity of the FDp ecology in the vineyard ecosystem also in Tuscany (Casati et al., 2017) and proposes a scenario close to the French one (Malembic-Maher et al., 2020; Rigamonti et al., 2023), as further confirmed by the close relation found between ViTos15 and ScaTos29 to FDp strains isolated from VV in France, highlighted by the European phylogenetic analysis (Figure 3; Supplementary Figure S2). Similarly, in the map-FD2 cluster characterized only by FDp strains collected in AG, we reported not only a novel FDp strain found in AG (AlnTos9, M54 related genotype; Figure 2; Supplementary Figure S1), which was also close to other FDp strains reported in AG in Europe (Figure 3; Supplementary Figure S2) but also an FDp strain collected in VV (ViTos997, M54; Figure 2). This interesting occurrence of a map-FD2/M54 genotype in VV was previously reported (Rigamonti et al., 2023) and was confirmed by the very close relation between ViTos997 and FDp strains previously collected in VV in Switzerland, as highlighted by the phylogenetic analysis conducted at the European level (Figure 3; Supplementary Figure S2). These outcomes further confirm the complexity of the FDp ecology, although the way in which the two hosts VV and AG interact in vineyard agroecosystems is still not clear, likely due to the lack of observations in secondary insect vectors.
Finally, the map-deduced amino acid sequences revealed an identity ranging from 97.87%, reported for AlnTos8 with ViTos997 and AlnTos9, to 100.00% for the others (Table 2), and since we obtained a longer portion of the map gene, a better evaluation of the amino acid variations was operable. Specifically, the alignment of the amino acid sequences revealed specific patterns for the isolates belonging to the different map clusters: F22, E26, M42, G114, F192, and N209 for map-FD3 (ViTos601, DicTos20, DicTos27, and CleTos8); L22, E26, M42, D114, V192, and T209 for map-FD2 (ViTos997 and AlnTos9); and F22, E26, I42, G114, V192, and T209 for map-FD1 (ViTos15 and ScaTos9), although AlnTos8, which also belongs to map-FD1, showed unique shorter F22, Q26, I42, and G114 patterns (Table 3). These outcomes confirm the potential power of elucidating the amino acid variations to better characterize FDp strains, an approach that has been little used for this phytoplasma (Quaglino et al., 2010), in contrast to that for other plant pathogens (e.g., Pedrelli et al., 2024).
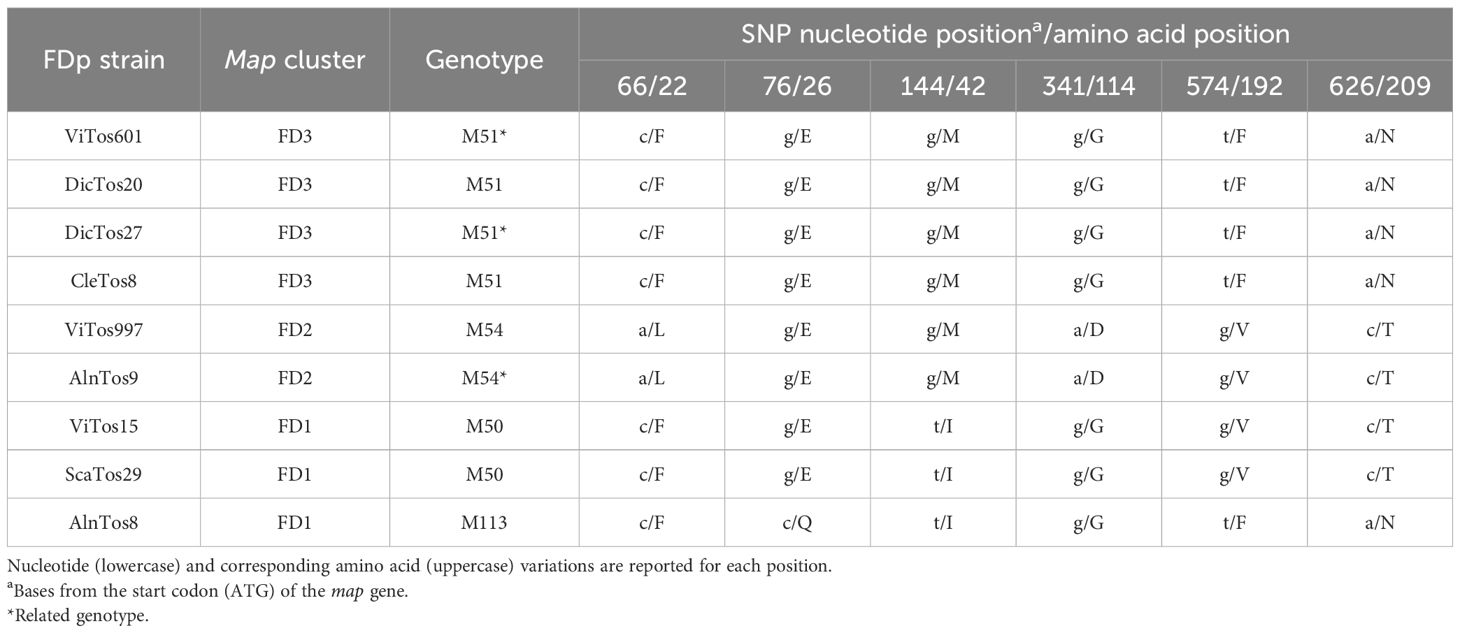
Table 3. Single-nucleotide polymorphisms (SNPs) and amino acid changes reported in the FDp strains identified by map typing (map cluster and associated genotype are reported).
4 Conclusions
The present study presents the first comprehensive determination of the population and molecular features of the Tuscan FDp strains by map gene typing, as well as of FDp ecology in the Tuscan vineyard agroecosystem. Based on a 4-year survey carried out in the main wine-growing areas of Tuscany, where FD presence was investigated in both primary and secondary FDp hosts and vectors, this work i) confirmed FD diffusion in almost the whole of Tuscany and even with an increased occurrence rate (approximately 50% of samples tested positive); ii) highlighted a complex FDp ecology also in the Tuscan vineyard agroecosystem, as FDp was reported not only in VV and ST, but also in secondary vectors (DE) and hosts (AG and CV); iii) reported nine FDp strains (three of which were novel) belonging to all the three map clusters, i.e., map-FD1, map-FD2, and map-FD3, with the map-FD3/M51 genotype mostly reported in not only VV but also DE and CV; and iv) further confirmed a complex FDp ecology also by phylogenetic analyses carried out at both Italian and European levels, which also showed some relations between the Tuscan FDp strains and those reported in the Balkan and French scenarios. We believe that the outcomes reported here will be useful to prevent and control the spread of the harmful FD in Tuscany, and in areas of the world where FD epidemic outbreaks continue to occur.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
APe: Data curation, Formal analysis, Investigation, Visualization, Writing – original draft. MC: Data curation, Formal analysis, Investigation, Visualization, Writing – original draft. APa: Data curation, Writing – review & editing. EP: Resources, Supervision, Visualization, Writing – review & editing. DR: Resources, Supervision, Writing – review & editing. CN: Project administration, Resources, Supervision, Writing – review & editing. LC: Conceptualization, Formal analysis, Methodology, Supervision, Visualization, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
The authors gratefully acknowledge Andrea Lucchi and the Entomology Lab of the University of Pisa for supplying insect samples, Piero Braccini for supporting field activities, and Roberto Pierro for helping with molecular analyses.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1489790/full#supplementary-material
Abbreviations
AG, Alnus glutinosa; BN, Bois noir; BNp, Candidatus phytoplasma solani; CV, Clematis vitalba; DE, Dictyophara europaea; FD, flavescence dorée; FDp, flavescence dorée phytoplasma; ST, Scaphoideus titanus; VV, V. vinifera cv. Sangiovese.
References
Angelini, E., Clair, D., Borgo, M., Bertaccini, A., Boudon-Padieu, E. (2001). Flavescence dorée in France and Italy–Occurrence of closely related phytoplasma isolates and their near relationships to Palatinate grapevine yellows and an alder yellows phytoplasma. Vitis 40, 79–86. doi: 10.5073/vitis.2001.40.79-86
Angelini, E., Luca Bianchi, G., Filippin, L., Morassutti, C., Borgo, M. (2007). A new TaqMan method for the identification of phytoplasmas associated with grapevine yellows by Real-Time PCR assay. J. Microbiol. Methods 68, 613–622. doi: 10.1016/j.mimet.2006.11.015
Angelini, E., Squizzato, F., Lucchetta, G., Borgo, M. (2004). Detection of a phytoplasma associated with grapevine flavescence dorée in Clematis vitalba. Eur. J. Plant Pathol. 110, 193–201. doi: 10.1023/B:EJPP.0000015361.95661.37
Arnaud, G., Malembic-Maher, S., Salar, P., Bonnet, P., Maixner, M., Marcone, C., et al. (2007). Multilocus sequence typing confirms the close genetic interrelatedness of three distinct flavescence dorée phytoplasma strain clusters and group 16SrV phytoplasmas infecting grapevine and alder in Europe. Appl. Environ. Microbiol. 73, 4001–4010. doi: 10.1128/AEM.02323-06
Belli, G., Bianco, P. A., Conti, M. (2010). Grapevine yellows in Italy: Past, present and future. J.l Plant Pathol. 92, 303–326.
Bertaccini, A., Botti, S., Tonola, A., Milano, C., Braccini, P., Sfalanga, A. (2003). Identificazione di fitoplasmi di flavescenza dorata in vigneti della Toscana. Informatore Agrar. 59, 65–67.
Boulent, J., St-Charles, P.-L., Foucher, S., Théau, J. (2020). Automatic detection of flavescence dorée symptoms across white grapevine varieties using deep learning. Front. Artif. Intell. 3. doi: 10.3389/frai.2020.564878
Bressan, A., Larrue, J., Boudon Padieu, E. (2006). Patterns of phytoplasma-infected and infective Scaphoideus titanus leafhoppers in vineyards with high incidence of flavescence dorée. Entomol. Exp. Appl. 119, 61–69. doi: 10.1111/j.1570-7458.2006.00391.x
Casati, P., Jermini, M., Quaglino, F., Corbani, G., Schaerer, S., Passera, A., et al. (2017). New insights on flavescence dorée phytoplasma ecology in the vineyard agro-ecosystem in southern Switzerland. Ann. Appl. Biol. 171, 37–51. doi: 10.1111/aab.12359
Chuche, J., Thiéry, D. (2014). Biology and ecology of the flavescence dorée vector Scaphoideus titanus: a review. Agron. Sustain. Dev. 34, 381–403. doi: 10.1007/s13593-014-0208-7
Davis, R. E., Dally, E. L. (2001). Revised subgroup classification of group 16SrV phytoplasmas and placement of flavescence dorée-associated phytoplasmas in two distinct subgroups. Plant Dis. 85, 790–797. doi: 10.1094/PDIS.2001.85.7.790
Deng, S., Hiruki, C. (1991). Genetic relatedness between two non culturable mycoplasma like organisms revealed by nucleic acid hybridization and polymerase chain reaction. Phytopathology 81, 1475. doi: 10.1094/Phyto-81-1475
De Sousa, E., Casati, P., Cardoso, F., Baltazar, C., Durante, G., Quaglino, F., et al. (2010). Flavescence dorée phytoplasma affecting grapevine (Vitis vinifera) newly reported in Portugal. Plant Pathol. 59, 398. doi: 10.1111/j.1365-3059.2009.02130.x
EFSA, Tramontini, S., Delbianco, A., Vos, S. (2020). Pest survey card on flavescence dorée phytoplasma and its vector Scaphoideus titanus. EFS3 17, 1–36. doi: 10.2903/sp.efsa.2020.EN-1909
EPPO (2024a). EPPO A2 List of Pests Recommended for Regulation as Quarantine Pests. Available online at: https://www.eppo.int/ACTIVITIES/plant_quarantine/A2_list (Accessed July 15, 2024).
EPPO (2024b). EPPO Global Database. Available online at: https://gd.eppo.int/taxon/PHYP64 (Accessed July 15, 2024).
Escriu, F. (2017). “Diversity of plant virus population: a valuable tool for epidemiological studies,” in Genetic diversity. Ed. Bitz, L. (Intechopen, London, United Kingdom), 150.
Felsenstein, J. (1981). Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376. doi: 10.1007/BF01734359
Filippin, L., Jović, J., Cvrković, T., Forte, V., Clair, D., Toševski, I., et al. (2009). Molecular characteristics of phytoplasmas associated with flavescence dorée in clematis and grapevine and preliminary results on the role of Dictyophara europaea as a vector. Plant Pathol. 58, 826–837. doi: 10.1111/j.1365-3059.2009.02092.x
Galetto, L., Pesando, M., Abbà, S., Bodino, N., Marzachì, C. (2023). Transcriptional profile of selected flavescence dorée phytoplasma genes during infection of insect vectors and host plants. J. Plant Pathol. 105, 393–407. doi: 10.1007/s42161-023-01310-7
Hall, T. A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Holz, S., Duduk, B., Büttner, C., Kube, M. (2016). Genetic variability of Alder yellows phytoplasma in Alnus glutinosa in its natural Spreewald habitat. For. Pathol. 46, 11–21. doi: 10.1111/efp.12206
Jarausch, B., Biancu, S., Kugler, S., Wetzel, T., Baumann, M., Winterhagen, P., et al. (2021). First report of flavescence dorée-related phytoplasma in a productive vineyard in Germany. Plant Dis. 105, 3285. doi: 10.1094/PDIS-02-21-0330-PDN
Jarausch, B., Markheiser, A., Jarausch, W., Biancu, S., Kugler, S., Runne, M., et al. (2023). Risk assessment for the spread of flavescence dorée-related phytoplasmas from alder to grapevine by alternative insect vectors in Germany. Microorganisms 11, 2766. doi: 10.3390/microorganisms11112766
Kogej Zwitter, Z., Seljak, G., Jakomin, T., Brodarič, J., Vučurović, A., Pedemay, S., et al. (2023). Epidemiology of flavescence dorée and hazelnut decline in Slovenia: geographical distribution and genetic diversity of the associated 16SrV phytoplasmas. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1217425
Kriston, É., Krizbai, L., Juhász, E., Melika, G. (2019). Status of grapevine “flavescence dorée” in Hungary. Phytopathogenic Mollicutes 9, 63–64. doi: 10.5958/2249-4677.2019.00032.X
Krnjajić, S., Mitrović, M., Cvrković, T., Jović, J., Petrović, A., Forte, V., et al. (2007). Occurrence and distribution of Scaphoideus titanus in multiple outbreaks of “flavescence dorée” in Serbia. Bull. Insectol. 60, 197–198.
Krstić, O., Cvrković, T., Marinković, S., Jakovljević, M., Mitrović, M., Toševski, I., et al. (2022). Genetic diversity of flavescence dorée phytoplasmas in vineyards of Serbia: From the widespread occurrence of autochthonous map-M51 to the emergence of endemic map-FD2 (vectotype II) and new map-FD3 (vectotype III) epidemic genotypes. Agronomy 12, 448. doi: 10.3390/agronomy12020448
Krstić, O., Cvrković, T., Mitrović, M., Radonjić, S., Hrnćić, S., Toševski, I., et al. (2018). Wolbachia infection in natural populations of Dictyophara europaea, an alternative vector of grapevine flavescence dorée phytoplasma: effects and interactions. Ann. Appl. Biol. 172, 47–64. doi: 10.1111/aab.12400
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lee, I.-M. (1994). Use of mycoplasma like organism (MLO) group-specific oligonucleotide primers for nested-PCR assays to detect mixed-MLO infections in a single host plant. Phytopathology 84, 559. doi: 10.1094/Phyto-84-559
Lee, I.-M., Gundersen-Rindal, D. E., Davis, R. E., Bartoszyk, I. M. (1998). Revised classification scheme of phytoplasmas based on RFLP analyses of 16rRNA and ribosomal protein gene sequences. Int. J. Syst. Evol. Microbiol. 48, 1153–1169. doi: 10.1099/00207713-48-4-1153
Lee, I. M., Martini, M., Marcone, C., Zhu, S. F. (2004). Classification of phytoplasma strains in the elm yellows group (16SrV) and proposal of 'Candidatus phytoplasma ulmi' for the phytoplasma associated with elm yellows. Int. J. Syst. Evol. Microbiol. 54, 337–347. doi: 10.1099/ijs.0.02697-0
Li, R., Mock, R., Huang, Q., Abad, J., Hartung, J., Kinard, G. (2008). A reliable and inexpensive method of nucleic acid extraction for the PCR-based detection of diverse plant pathogens. J. Virol. Methods 154, 48–55. doi: 10.1016/j.jviromet.2008.09.008
Linhartová, Š., Tománková, K., Orságová, H., Hejlová, M., Čermák, V., Beránek, J., et al. (2023). First report of grapevine flavescence dorée phytoplasma in the Czech Republic. Plant Dis. 107, 3273. doi: 10.1094/PDIS-02-23-0315-PDN
Maixner, M., Reinert, W., Darimont, H. (2000). Transmission of grapevine yellows by Oncopsis alni (Schrank) (Auchenorrhyncha: Macropsinae). Vitis 39, 83–84. doi: 10.5073/VITIS.2000.39.83-84
Malembic-Maher, S., Desqué, D., Khalil, D., Salar, P., Bergey, B., Danet, J.-L., et al. (2020). When a Palearctic bacterium meets a Nearctic insect vector: Genetic and ecological insights into the emergence of the grapevine flavescence dorée epidemics in Europe. PLoS Pathog. 16, 1–26. doi: 10.1371/journal.ppat.1007967
Malembic-Maher, S., Salar, P., Filippin, L., Carle, P., Angelini, E., Foissac, X. (2011). Genetic diversity of European phytoplasmas of the 16SrV taxonomic group and proposal of ‘Candidatus Phytoplasma rubi’. Int. J. Syst. Evol. Microbiol. 61, 2129–2134. doi: 10.1099/ijs.0.025411-0
Martini, M., Botti, S., Marcone, C., Marzachı, C., Casati, P., Bianco, P. A., et al. (2002). Genetic variability among flavescence dorée phytoplasmas from different origins in Italy and France. Mol. Cell. Probe. 16, 197–208. doi: 10.1006/mcpr.2002.0410
Marzachi, C., Veratti, F., Bosco, D. (1998). Direct PCR detection of phytoplasmas in experimentally infected insects. Ann. Appl. Biol. 133, 45–54. doi: 10.1111/j.1744-7348.1998.tb05801.x
Mehle, N., Rupar, M., Seljak, G., Ravnikar, M., Dermastia, M. (2011). Molecular diversity of ‘flavescence dorée’ phytoplasma strains in Slovenia. Bull. Insectol. 64, S29–S30.
Moussa, A., Guerrieri, E., Torcoli, S., Serina, F., Quaglino, F., Mori, N. (2023). Identification of phytoplasmas associated with grapevine bois noir and flavescence dorée in inter-row groundcover vegetation used for green manure in Franciacorta vineyards. J. Plant Pathol. 105, 1511–1519. doi: 10.1007/s42161-023-01474-2
Oggier, A., Conedera, M., Jermini, M., Debonneville, C., Schumpp, O., Rizzoli, A. (2023). Gone-wild grapevines in forests may act as a potential habitat for ‘Flavescence dorée’ phytoplasma vectors and inoculum. J. Appl. Entomol. 147, 777–789. doi: 10.1111/jen.13169
Ohana-Levi, N., Netzer, Y. (2023). Long-term trends of global wine market. Agriculture 13, 224. doi: 10.3390/agriculture13010224
Pedrelli, A., Panattoni, A., Cotrozzi, L. (2023a). First molecular characterization of plum pox virus strains in stone fruits of Tuscany (Central Italy). J. Plant Pathol. 105, 1045–1053. doi: 10.1007/s42161-023-01430-0
Pedrelli, A., Panattoni, A., Cotrozzi, L. (2024). First molecular analysis of wisteria vein mosaic virus in Italy: eight new variants reported in Wisteria sinensis. J. Plant Pathol. 106, 117–125. doi: 10.1007/s42161-023-01526-7
Pedrelli, A., Panattoni, A., Nali, C., Cotrozzi, L. (2023b). Occurrence of fig mosaic disease in Tuscany, Central Italy: Characterization of new fig mosaic virus isolates, and elucidation of physiochemical responses of infected common fig cv. Dottato. Sci. Hortic. 322, 112440. doi: 10.1016/j.scienta.2023.112440
Pedrelli, A., Ricci, G. P., Panattoni, A., Nali, C., Cotrozzi, L. (2023c). Physiological and biochemical responses induced by plum pox virus and plum bark necrosis steam pitting associated virus in Tuscany autochthonous plum cv. Coscia di Monaca. Plants 12, 3264. doi: 10.3390/plants12183264
Pierro, R., Bottner-Parker, K., Panattoni, A., Wei, W., Marcone, C., Rizzo, D., et al. (2023). Multilocus sequence typing of phytoplasmas associated with flavescence dorée disease in Tuscany vineyards identifies a highly homogeneous lineage in the subgroup 16SrV-C. Crop Prot. 163, 106–114. doi: 10.1016/j.cropro.2022.106114
Pierro, R., Passera, A., Panattoni, A., Casati, P., Luvisi, A., Rizzo, D., et al. (2018). Molecular typing of Bois noir phytoplasma strains in the Chianti Classico area (Tuscany, Central Italy) and their association with symptom severity in Vitis vinifera ‘Sangiovese.’. Phytopathology 108, 362–373. doi: 10.1094/PHYTO-06-17-0215-R
Pisuttu, C., Lo Piccolo, E., Paoli, L., Cotrozzi, L., Nali, C., Pellegrini, E., et al. (2023). Physiochemical responses of Ailanthus altissima under the challenge of Verticillium dahliae: elucidating the decline of one of the world’s worst invasive alien plant species. Biol. Invasions 25, 61–78. doi: 10.1007/s10530-022-02891-7
Pisuttu, C., Marchica, A., Bernardi, R., Calzone, A., Cotrozzi, L., Nali, C., et al. (2020). Verticillium wilt of Ailanthus altissima in Italy caused by V. dahliae: New outbreaks Tuscany. iForest 13, 238–245. doi: 10.3832/ifor3238-013
Plavec, J., Budinšćak, Ž., Križanac, I., Škorić, D., Foissac, X., Šeruga Musić, M. (2019). Multilocus sequence typing reveals the presence of three distinct flavescence dorée phytoplasma genetic clusters in Croatian vineyards. Plant Pathol. 68, 18–30. doi: 10.1111/ppa.12934
Plavec, J., Music, M. S. (2023). Differentiation of the flavescence dorée phytoplasma genetic clusters by multiplex real-time PCR assay targeting the map gene. J. Plant Pathol. 105, 409–416. doi: 10.1007/s42161-023-01331-2
Quaglino, F., Guerrieri, E., Torcoli, S., Barbieri, C., Serina, F., Bianco, P. A., et al. (2024). “Recent findings on flavescence dorée in Franciacorta (North Italy): prevalence of associated phytoplasma genotypes in symptomatic grapevines and in additional plant and insect hosts within and around vineyards,” in Proceedings of the European Workshop on Flavescence dorée. Ed. Bertaccini, A.(Verona, Italy), 15–16.
Quaglino, F., Casati, P., Bianco, P. A. (2010). Distinct rpsC single nucleotide polymorphism lineages of flavescence dorée subgroup 16SrV-d phytoplasma co-infect Vitis vinifera l. Folia Microbiol. 55, 251–257. doi: 10.1007/s12223-010-0037-2
Radonjić, S., Krstić, O., Cvrković, T., Hrnčić, S., Marinković, S., Mitrović, M., et al. (2023). The first report on the occurrence of flavescence dorée phytoplasma affecting grapevine in vineyards of Montenegro and an overview of epidemic genotypes in natural plant reservoirs. J. Plant Pathol. 105, 419–427. doi: 10.1007/s42161-023-01318-z
Reisenzein, H., Steffek, R. (2011). First outbreaks of grapevine ‘flavescence dorée’ in Austrian viticulture. Bull. Insectol. 64, S223–S224.
Rigamonti, I. E., Salvetti, M., Girgenti, P., Bianco, P. A., Quaglino, F. (2023). Investigation on flavescence dorée in North-western Italy identifies Map-M54 (16SrV-D/Map-FD2) as the only phytoplasma genotype in Vitis vinifera L. and reveals the presence of new putative reservoir Plants. Biology 12, 1216. doi: 10.3390/biology12091216
Ripamonti, M., Pegoraro, M., Rossi, M., Bodino, N., Beal, D., Panero, L., et al. (2020). Prevalence of flavescence dorée phytoplasma-infected Scaphoideus titanus in different vineyard agroecosystems of Northwestern Italy. Insects 11, 301. doi: 10.3390/insects11050301
Ripamonti, M., Pegoraro, M., Morabito, C., Gribaudo, I., Schubert, A., Bosco, D., et al. (2021). Susceptibility to flavescence dorée of different vitis vinifera genotypes from north-western italy. Plant Pathol. 70, 511–520. doi: 10.1111/ppa.13301
Rizzo, D., Materazzi, A., Stefani, L., Panattoni, A., Pierro, R., Marchi, G., et al. (2018). The monitoring program of grapevine phytoplasmas in Tuscany (Italy): results of a four year survey. Adv. Hortic. Sci. 32, 281–288. doi: 10.13128/AHS-22566
Rizzoli, A., Belgeri, E., Jermini, M., Conedera, M., Filippin, L., Angelini, E. (2021). Alnus glutinosa and Orientus ishidae (Matsumura 1902) share phytoplasma genotypes linked to the ‘flavescence dorée’ epidemics. J. Appl. Entomol. 145, 1015–1028. doi: 10.1111/jen.12933
Rizzoli, A., Oggier, A., Jermini, M., Battelli, R., Debonneville, C., Schumpp, O., et al. (2023). Adaptive management trials for the control of Scaphoideus titanus, main vector of “flavescence doree” phytoplasmas. Phytopathogenic Mollicutes. 13, 59–60. doi: 10.5958/2249-4677.2023.00030.0
Rossi, M., Pegoraro, M., Ripamonti, M., Abbà, S., Beal, D., Giraudo, A., et al. (2019). Genetic diversity of flavescence dorée phytoplasmas at the vineyard scale. Appl. Environ. Microbiol. 85, e03123–e03118. doi: 10.1128/AEM.03123-18
Schubert, A. (2017). Un approccio integrato alla lotta contro la flavescenza dorata della vite (INTEFLAVI) (Torino: Dipartimento di Scienze Agrarie, Forestali e Alimentari), 1–24.
Trivellone, V., Jermini, M., Linder, C., Cara, C., Delabays, N., Baumgartner, J. (2013). Role de la flore du vignoble sur la distribution de Scaphoideus titanus. Rev. suisse viticulture arboriculture horticulture. 45, 222–228.
Zhao, Y., Wei, W., Lee, I.-M., Shao, J., Suo, X., Davis, R. E. (2009). Construction of an interactive online phytoplasma classification tool, iPhyClassifier, and its application in analysis of the Peach X-disease phytoplasma group (16SrIII). Int. J. Syst. Evol. Microbiol. 59, 2582–2593. doi: 10.1099/ijs.0.010249-0
Keywords: Alnus glutinosa, Clematis vitalba, Dictiophara europaea, gene typing, grapevine yellows, Scaphoideus titanus, Vitis vinifera
Citation: Pedrelli A, Carli M, Panattoni A, Pellegrini E, Rizzo D, Nali C and Cotrozzi L (2024) Investigating a new alarming outbreak of flavescence dorée in Tuscany (Central Italy): molecular characterization and map gene typing elucidate the complex phytoplasma ecology in the vineyard agroecosystem. Front. Plant Sci. 15:1489790. doi: 10.3389/fpls.2024.1489790
Received: 01 September 2024; Accepted: 19 November 2024;
Published: 13 December 2024.
Edited by:
Andreia Figueiredo, University of Lisbon, PortugalReviewed by:
Duska Delic, University of Banjaluka, Bosnia and HerzegovinaFabio Quaglino, University of Milan, Italy
Copyright © 2024 Pedrelli, Carli, Panattoni, Pellegrini, Rizzo, Nali and Cotrozzi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alessandra Panattoni, YWxlc3NhbmRyYS5wYW5hdHRvbmlAdW5pcGkuaXQ=
†These authors share first authorship
 Athos Pedrelli
Athos Pedrelli Marco Carli1,3†
Marco Carli1,3† Alessandra Panattoni
Alessandra Panattoni Elisa Pellegrini
Elisa Pellegrini Domenico Rizzo
Domenico Rizzo Cristina Nali
Cristina Nali Lorenzo Cotrozzi
Lorenzo Cotrozzi