- Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
Recently, due to the widespread use of the acetolactate synthase (ALS)-inhibiting herbicide penoxsulam in paddy fields in China, Echinochloa crus-galli (L.) P. Beauv. has become a problematic grass weed that is frequently not controlled, posing a threat to weed management and rice yield. There are many reports on target-site mutations of ALS inhibiting herbicides; however, the detailed penoxsulam resistance mechanism in E. crus-galli remains to be determined. Greenhouse and laboratory studies were conducted to characterize target-site resistance mechanisms in JL-R, AH-R, and HLJ-R suspected resistant populations of E. crus-galli survived the field-recommended dose of penoxsulam. The whole-plant dose–response testing of E. crus-galli to penoxsulam confirmed the evolution of moderate-level resistance in two populations, JL-R (9.88-fold) and HLJ-R (8.66-fold), and a high-level resistance in AH-R (59.71-fold) population. ALS gene sequencing identified specific mutations in resistant populations, including Pro-197-His in ALS1 for JL-R, Trp-574-Leu in ALS1 for AH-R, and Pro-197-Leu in ALS2 for HLJ-R. In vitro ALS activity assays demonstrated a significantly higher activity in AH-R compared to the susceptible population (YN-S). Molecular docking studies revealed that Trp-574-Leu mutation primarily reduced the enzyme’s ability to bind to the triazole-pyrimidine ring of penoxsulam due to decreased π–π stacking interactions, while Pro-197-His/Leu mutations impaired binding to the benzene ring by altering hydrogen bonds and hydrophobic interactions. Additionally, the Pro-197-His/Leu amino acid residue changes resulted in alterations in the shape of the active channel, impeding the efficient entry of penoxsulam into the binding site in the ALS protein. The three mutant ALS proteins expressed via the Bac-to-Bac baculovirus system exhibited notably lower activity inhibition rates than the non-mutant ALS proteins to penoxsulam, indicating all three ALS mutations reduce sensitivity to penoxsulam. This study elucidated the distinct impacts of the Pro-197-His/Leu and Trp-574-Leu mutations in E. crus-galli to penoxsulam resistance. Notably, the Trp-574-Leu mutation conferred stronger resistance to penoxsulam compared to the Pro-197-His/Leu mutations in E. crus-galli. The Pro-197-His/Leu mutations were first detected in E. crus-galli conferring penoxsulam resistance. These findings provide deeper insights into the molecular mechanisms underlying target-site resistance to penoxsulam in E. crus-galli.
1 Introduction
Weed infestation poses the most significant biotic threat to global food security, leading to yield reductions of up to 34% in agriculture (Oerke, 2006). With their pervasive presence in virtually every crop field, effective management and control of these weeds are imperative to ensure high crop yields. A variety of methods are available for weed control in agricultural fields, including manual and mechanical techniques, chemical control, crop rotation, and biological approaches (Adkins and Shabbir, 2014; Latif et al., 2015). Synthetic chemical herbicides serve as the primary tools employed worldwide (Brun et al., 2022). The application of herbicides with different modes of action has facilitated weed control, playing an important role in yields and qualities of crops (Kraehmer et al., 2014). Among these, acetolactate synthase (ALS, EC 4.1.3.18) inhibitors possess several distinctive characteristics in agricultural applications. These features encompass low application quantities, broad spectrum, expansive application windows, and low mammalian toxicity, coupled with high crop safety margins (Mazur and Falco, 1989; Singh et al., 2019).
Acetolactate synthase, also known as acetohydroxyacid synthase (AHAS, EC 2.2.1.6), plays a pivotal role as an enzyme in the biosynthesis of branched-chain amino acids, such as leucine, isoleucine, and valine in plants (Devine and Shukla, 2000; Duggleby et al., 2008). ALS can catalyze the conversion of two molecules of pyruvate into 2-acetolactate and the transformation of one molecule of 2-ketobutyrate and one molecule of pyruvate into 2-aceto-2-hydroxybutyrate (Duggleby et al., 2008; Zhou et al., 2007). The ALS enzyme consists of a catalytic subunit and a regulatory subunit, with necessary cofactors including thiamine diphosphate (ThDP or TPP), flavin adenine dinucleotide (FAD), and divalent metal ion (Mg2+) for the catalytic activity of the catalytic subunit (Chipman et al., 1998). ALS inhibitor herbicides are primarily categorized into five groups based on differences in the chemical structure of compounds: triazolopyrimidines (TPs), imidazolinones (IMIs), sulfonylureas (SUs), pyrimidinyl-thiobenzoates (PTBs), and sulfonyl-aminocarbonyl-triazolinones (SCTs). However, the excessive use of these chemicals has led to the evolution of herbicide-resistant weeds. Up to now, 174 weed species developed resistance to ALS inhibitors globally (Heap, 2024).
Herbicide resistance mechanisms are broadly categorized into target-site resistance (TSR) and nontarget-site resistance (NTSR) (Comont et al., 2020; Délye, 2013; Délye et al., 2013; Gaines et al., 2020). TSR involves the mutation of amino acids in the target enzyme through gene mutations or variations in gene copy numbers (Pan et al., 2022). A total of 24 amino acid sites are associated with resistance to ALS inhibitors in weeds, yeast, and bacteria (Yu and Powles, 2014). In relation to ALS inhibitors, various weed species have reported 31 types of amino acid mutations at nine conserved positions (Ala-122, Pro-197, Ala-205, Phe-206, Asp-376, Arg-377, Trp-574, Ser-653, and Gly-654), numbered according to the corresponding Arabidopsis thaliana (L.) Heynh. Sequence (Fang et al., 2022; Li et al., 2022; Powles and Yu, 2010; Tranel et al., 2024). Various mutations in the ALS gene have been demonstrated to confer distinct resistance patterns in weed populations (Yu and Powles, 2014). Mutations occurring at different amino acid positions can grant varying degrees of resistance within the same weed species (Cao et al., 2022; Fang et al., 2019b; Riar et al., 2013; Wang et al., 2021). Additionally, different mutations of the same amino acid can result in diverse cross-resistance patterns (Deng et al., 2014). It is noteworthy that the same mutation may lead to differing resistance in different weed species (Li et al., 2022). These resistance patterns are intricately tied to the structure of the ALS protein and the properties of the mutated amino acids. Various ALS gene mutations can accumulate within individual plants through cross-pollination, as evidenced by the simultaneous identification of the Pro-197-Thr and Trp-574-Leu mutations in individual Descurainia sophia L. plants (Deng et al., 2017). Accumulation of mutations within the same weed species may confer resistance to multiple herbicides, thereby adding complexity to the challenges of weed resistance management.
Echinochloa crus-galli (L.) P. Beauv., a pervasive weed in paddy rice (Oryza sativa L.) fields worldwide, has similar morphology and growth habits with rice and is difficult to be recognized and identified especially in its seedling stage. Being a C4 plant, it competes strongly in photosynthesis and may act as an intermediary host for certain pests and diseases, significantly affecting rice yield and quality (Chauhan and Johnson, 2011; Zhang et al., 2017). The ALS inhibitor penoxsulam is commonly used for post-emergence weed control, especially against E. crus-galli in rice field, in various types of culture, methods of planting, and cultivars of the crop in China. However, the overreliance on penoxsulam has led to the development of resistant weeds related to TSR and NTSR (Fang et al., 2019a, Fang et al., 2022, Fang et al., 2019b). To date, 12 different ALS gene mutations have been reported to endow resistance to ALS inhibitors in Echinochloa spp. (Table 1) for TSR mechanism (Amaro-Blanco et al., 2021; Délye et al., 2015; Fang et al., 2019a, Fang et al., 2022, Fang et al., 2019b; Feng et al., 2022; Liu et al., 2019; Panozzo et al., 2017; Tranel et al., 2024). Among these, mutations at positions Pro-197 and Trp-574 have commonly been documented in resistant weeds (Tranel et al., 2024). Clear evidence has shown that Trp-574-Arg/Leu mutations confer penoxsulam resistance in E. crus-galli, whereas mutations at position 197 have not been reported to confer penoxsulam resistance in E. crus-galli (Fang et al., 2019b; Feng et al., 2022).
The objective of this study is to fully investigate the TSR mechanism based on the suspected populations of E. crus-galli collected from rice fields in China, aimed to (1) demonstrate the resistance levels among different mutation populations to penoxsulam, (2) explore the underlying mechanisms of TSR, and (3) clarify the distinctions in resistance between Pro-197-His/Leu and Trp-574-Leu mutations.
2 Materials and methods
2.1 Plant materials and herbicide
In this research, four populations of E. crus-galli were investigated, each distributed in different geographical regions from China. The phenotypically resistant populations, JL-R (Tonghua city, Jilin province; 42.62°N, 126.07°E), AH-R (Hefei city, Anhui province; 31.25°N, 117.20°E), and HLJ-R (Hegang city, Heilongjiang province; 47.50°N, 130.86°E), were collected from rice fields where penoxsulam had proven ineffectiveness in weed control. In contrast, the susceptible population, YN-S (coordinates 30.91°N, 118.80°E), was obtained from a non-cultivated field close to rice paddies in Yunnan Province, where no herbicides had been previously used. For each of these four E. crus-galli populations, seeds were collected from a minimum of 40 individual plants and stored in a well-ventilated shelf. Penoxsulam (2-(2,2-difluoroethoxy)-N-(5,8-dimethoxy-[1,2,4]triazolo[1,5-c]pyrimidin-2-yl)-6-(trifluoromethyl)benzenesulfonamide, 25 g/L oil dispersion) was obtained from Dow AgroSciences.
2.2 Whole-plant dose–response bioassay
Seeds from each of the JL-R, AH-R, HLJ-R, and YN-S populations were randomly selected and germinated in Petri dishes with damp filter papers, then incubated in a growth chamber with a temperature of 30°C/25°C and a 12-h photoperiod. After the shoot length reached approximately 1 cm, seven seedlings were transplanted into four replicate pots filled with a commercial potting soil mixture for each herbicide rate. These pots were placed in a growth chamber set at 30°C/25°C with a 12-h day/night cycle. Penoxsulam was applied at the rates 0 g a.i. ha−1, 1.67 g a.i. ha−1, 5 g a.i. ha−1, 15 g a.i. ha−1, 45 g a.i. ha−1, 135 g a.i. ha−1, and 405 g a.i. ha−1 for the JL-R and HLJ-R populations; 0 g a.i. ha−1, 5 g a.i. ha−1, 15 g a.i. ha−1, 45 g a.i. ha−1, 135 g a.i. ha−1, 405 g a.i. ha−1, and 1,215 g a.i. ha−1 for the AH-R population; and 0 g a.i. ha−1, 0.19 g a.i. ha−1, 0.56 g a.i. ha−1, 1.67 g a.i. ha−1, 5 g a.i. ha−1, 15 g a.i. ha−1, and 45 g a.i. ha−1 for the YN-S population at three- to four-leaf stages of E. crus-galli using an experimental moving-boom sprayer (Model ASS-4, Beijing Research Center for Information Technology in Agriculture, China), equipped with a TeeJet XR8002VS flat-fan nozzle and a pressure of 0.275 MPa, delivering the volume of 450 L ha−1 liquid. E. crus-galli growing in the pots were all harvested, and their dry weight biomass (after 3 days of 75°C oven-dried) was measured 3 weeks after herbicide treatment. The whole-plant dose–response experiment was conducted twice.
2.3 ALS-cDNA and ALS-DNA sequencing
Seeds were cultured to three- to four-leaf stage as described in Section 2.2. Young plant leaf tissue, weighing 100 mg for each, was sampled from a minimum of 20 plants of each population. DNA was extracted using the DNAsecure Plant Kit (TianGen Biotech, Co. Ltd., Beijing, China), and total RNA was isolated using the RNA Easy Fast Plant Tissue Kit (TianGen Biotech, Co. Ltd., Beijing, China). First-strand cDNA was synthesized using the HiScript® III 1st Strand cDNA Synthesis Kit (Vazyme Biotech Co., Ltd., Nanjing, China).
E. crus-galli, an allohexaploid grass weed, harbors at least three ALS genes (Fang et al., 2022; Iwakami et al., 2015; Panozzo et al., 2021). Primers ALS-1 (forward, 5′-ATCCCCCATCCTCTCCTT-3′; reverse, 5′-GGTCCAGAGTTCACACCCTAG-3′) and primers ALS-2 (forward, 5′-CACCCTCCCCAAACCC-3′; reverse, 5′-CACGGAAACAACAGACTACAT-3′) were designed based on the ALS mRNA sequences of E. crus-galli LC006058.1 and LC006059.1 to amplify the complete ALS1 and ALS2 genes. Additionally, primers ALS-3 (forward, 5′-CCCCAATCCCCCATCCAT-3′; reverse, 5′-GCACCGCTCGCTGAATAC-3′) reported by Iwakami and coworkers were used to specific amplification of ALS3 gene (Iwakami et al., 2015). The fragments of ALS1-3 cover the known eight resistance-conferring mutation sites. For polymerase chain reaction (PCR), LA Taq® was used with GC Buffer (Takara Biomedical Technology Co., Ltd., Beijing, China) according to the manufacturer’s instructions. The purification and TA cloning of the PCR products were conducted using the methods outlined in a previous report (Sun et al., 2023). A minimum of eight white colonies were carefully selected and then sent to Tsingke Biotechnology Co., Ltd. (Beijing, China) for Sanger sequencing. The resulting sequences were aligned with the documented ALS genes of E. crus-galli (accession numbers, LC006058.1, LC006059.1, and LC006063.1) using NCBI-BLAST and analyzed using DNAMEN 9.0.1 (Lynnon Corporation, Quebec, Canada) and SeqMan Pro (v7.1.0, DNASTAR Lasergene) (Clewley, 1995).
2.4 In vitro assay of ALS activity
The extraction and evaluation of the ALS enzyme were conducted following the methods outlined by Yu with coworkers (Yu et al., 2004) and Han with coworkers (Han et al., 2012) with minor modifications. E. crus-galli seedlings (purified and ensured to carry the special mutation for all plants) were cultivated to the three- to four-leaf stage following the aforementioned method. Approximately 4 g of leaf tissue was sampled from each population, with a minimum of 30 seedlings harvested in each sample. The frozen leaf material was homogenized using a mortar and pestle in 8 mL of grinding buffer. Subsequently, an equal volume of 100% (NH4)2SO4 was added dropwise to the solution, followed by centrifugation. The resulting ALS pellet was redissolved in 3.5 mL resuspension buffer. ALS protein was desalted using a Sephadex G25 column with a 5-mL elution buffer.
For the enzyme assays, 100 μL of desalted enzyme extraction solution and 100 μL of penoxsulam at different concentrations were added to a 1.5-mL centrifugation tube. The dosage of penoxsulam was set at 0 μmol·L−1, 0.00001 μmol·L−1, 0.0001 μmol·L−1, 0.001 μmol·L−1, 0.01 μmol·L−1, 0.1 μmol·L−1, 1 μmol·L−1, 10 μmol·L−1, and 100 μmol·L−1 for YN-S population, while it was set at 0 μmol·L−1, 0.0001 μmol·L−1, 0.001 μmol·L−1, 0.01 μmol·L−1, 0.1 μmol·L−1, 1 μmol·L−1, 10 μmol·L−1, 100 μmol·L−1, and 1,000 μmol·L−1 for the JL-R, AH-R, and HLJ-R populations, respectively. The ALS enzyme concentration was determined using the Easy Protein Quantitative Kit (Bradford) (TransGen Biotech, Beijing, China), and the absorbance at 530 nm was measured using the FlexStation 3 full-wavelength scanning multifunctional enzyme marker (MD Electronics, USA) to determine ALS enzyme activity. The assay was conducted twice, with each treatment tested in three replicates.
2.5 ALS gene expression
The F1 generation of bagged selfed seeds from E. crus-galli mutated plants were all harvested for this experiment. After E. crus-galli seedlings reached three- to four-leaf stage, each tested population was divided into two subgroups. One subgroup was treated with 15 g a.i. ha−1 penoxsulam, while the other was treated with water. The leaf tissues were collected at 0 d, 1 d, 2 d, and 3 d after treatment with 10 plants sampled each day, respectively. The samples were immediately flash frozen in liquid nitrogen. Total RNA extraction and first-strand cDNA was synthesized as previously described. The ALS gene and β-actin reference gene were according to previous reports (Fang et al., 2022, Fang et al., 2019b). qRT-PCR was conducted following the manufacturer’s instructions for Taq Pro Universal SYBR qPCR Master Mix (Vazyme Biotech Co., Ltd., Nanjing, China), and the qRT-PCR program was executed using an ABI 7500 Fast Real-Time PCR System (Applied Biosystems, Waltham, MA, USA). The relative quantification of the ALS gene was calculated using the 2−ΔΔCt method, where ΔCT =ALS (Mean CT) − Actin (Mean CT), ΔΔCT = ΔCT (treatment) − ΔCT (control) (Bustin et al., 2009; Livak and Schmittgen, 2001). ALS expression level was normalized to the Ct values for the YN-S population at 0 d (water treatment). Each process was repeated three times. Independent sample T-test was performed using SPSS v25.0 (IBM, Armonk, NY, USA) software to determine significant differences in the expression levels.
2.6 Homology modeling and molecular docking
The ALS gene sequence alignment revealed three different mutations in the populations of E. crus-galli: JL-R (Pro-197-His, ALS1), AH-R (Trp-574-Leu, ALS1), and HLJ-R (Trp-197-Leu, ALS2). The amino acid sequences of these three mutated proteins, along with two non-mutated proteins (listed in Supplementary Table S1), were employed as query templates to the homology model using SWISS-MODEL (https://swissmodel.expasy.org/) (Waterhouse et al., 2018). The ALS protein (PDB ID 3e9y) from A. thaliana was used as the template protein. The homology modeling results were uploaded to SAVES (http://services.mbi.ucla.edu/SAVES/) to evaluate by PROCHECK. The chemical structure of penoxsulam was downloaded from the PubChem database (CID: 11784975) (https://pubchem.ncbi.nlm.nih.gov/). Molecule docking simulations were performed by Autodock Vina (Trott and Olson, 2010), according to previously reported method with a slight modification (Butt et al., 2020). The docking box was positioned at the active site, as reported in previous literature (Fang et al., 2022; Wang et al., 2009), where the protein bound with the penoxsulam ligand. The box size was set to 15 Å × 15 Å × 15 Å, with the active site coordinates as follows: center_x = 55.026, center_y = 50.923, and center_z = 46.274. The parameter settings were exhaustiveness = 400 and num_modes = 20, and other parameters were set to their default values.
2.7 Heterologous expression of ALS protein and activity assay
Three mutant and two non-mutant ALS protein-coding sequences (CDS) were codon optimized, synthesized, and confirmed through Sanger sequencing by Sangon (Sangon Biotech (Shanghai) Co., Ltd.). The Bac-to-Bac baculovirus expression system and the purification of the five target proteins were conducted following the methods described in previous literature (Fang et al., 2022). The expression vector for the target gene was constructed based on Fang’s method and transformed into DH5α cells for amplification. Plasmid extraction and PCR verification were performed to confirm correctness. The plasmid was then transferred into DH10Bac competent cells via a transposition reaction, following the instructions of the Bac-to-Bac® TOPO® Expression System (Thermo Fisher Technology Co. Ltd, Shanghai, China). SF9 cells were inoculated at a density of 1 × 106 cells mL−1 in 1-L culture flasks, totaling 200 mL of cells. To infect the cells, 0.8 mL of P4 viral stock was added to the 200-mL cell suspension. After 3 d, cells were collected and centrifuged at 4°C. The supernatant was then collected for ALS protein purification. The quantification of the expressed ALS proteins (diluted in elution buffer) was performed as mentioned above. The protocol used for the in vitro heterologous ALS activity assay closely resembled that for ALS isolated from E. crus-galli, with the exception that in the former case, the penoxsulam concentration was set at 1 μM. Each treatment was performed with three replicates.
2.8 Data analysis
The whole-plant dose–response bioassay data were subjected to a four-parameter log-logistic equation analysis using SigmaPlot 12.5 software (Systat Software, Inc., San Jose, CA, USA) to calculate the GR50 value (the dose causing a 50% reduction in above-ground dry weight). The regression equation used is as follows:
In this equation, y represents the ratio of dry weight at x herbicide dose to that of the control, x represents the herbicide treatment dose, C represents the lower limit of the dose–response, D represents the upper limit of the dose–response, and b denotes the slope of the curve.
Based on the GR50 values obtained for each population, the resistance index (RI) was calculated using the following formula:
The same analysis calculated the herbicide concentration required to inhibit 50% of ALS activity (I50) and its corresponding RI value. The resistance classification criteria were as follows: RI < 2, indicating a susceptibility; 2 ≤ RI < 5, indicating a low-level resistance; 5 ≤ RI < 10, indicating a moderate-level resistance; and RI > 10, indicating a high-level resistance.
3 Results
3.1 Whole-plant dose–response to penoxsulam
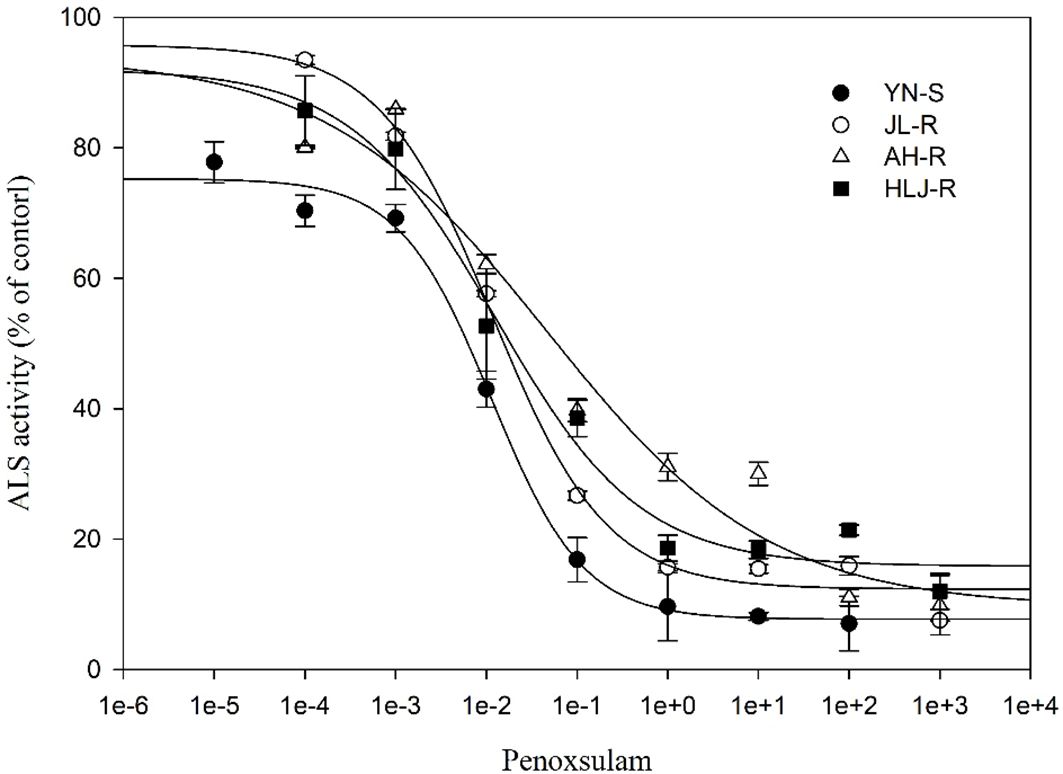
All seedlings of the S population, YN-S, were completely inhibited at 15 g a.i. ha−1 penoxsulam with a GR50 value of 2.76 g a.i. ha−1. The GR50 values of the JL-R, HLJ-R, and AH-R populations (R) were 27.26 g a.i. ha−1, 23.91 g a.i. ha−1, and 164.81 g a.i. ha−1, respectively, corresponding to R/S GR50 ratios (RI) of 9.88-fold, 8.66-fold, and 59.71-fold (Table 2; Figure 1) compared to the YN-S population, which revealed that the JL-R and HLJ-R populations had evolved moderate-level resistance and AH-R population showed high-level resistance to penoxsulam.
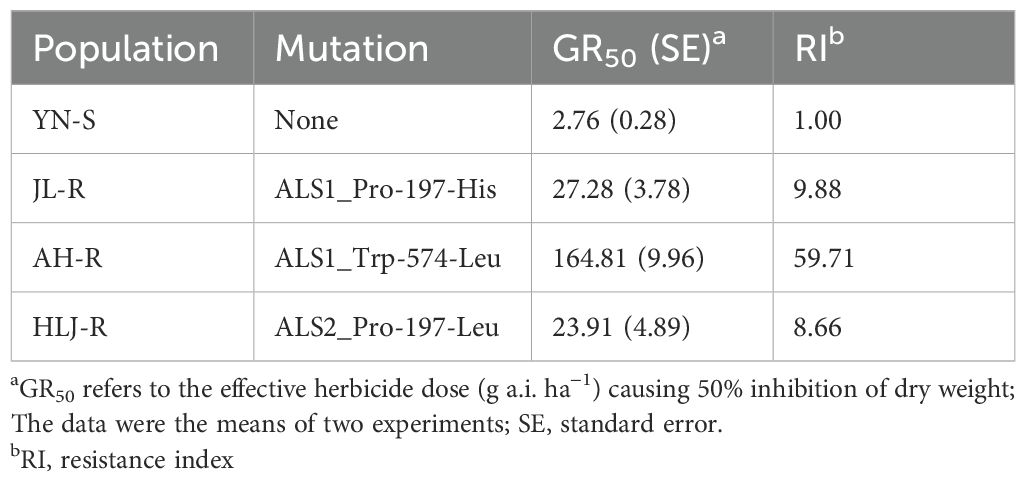
Table 2. The resistance levels of YN-S, JL-R, AH-R, and HLJ-R populations with distinct mutations of Echinochloa crus-galli to penoxsulam.
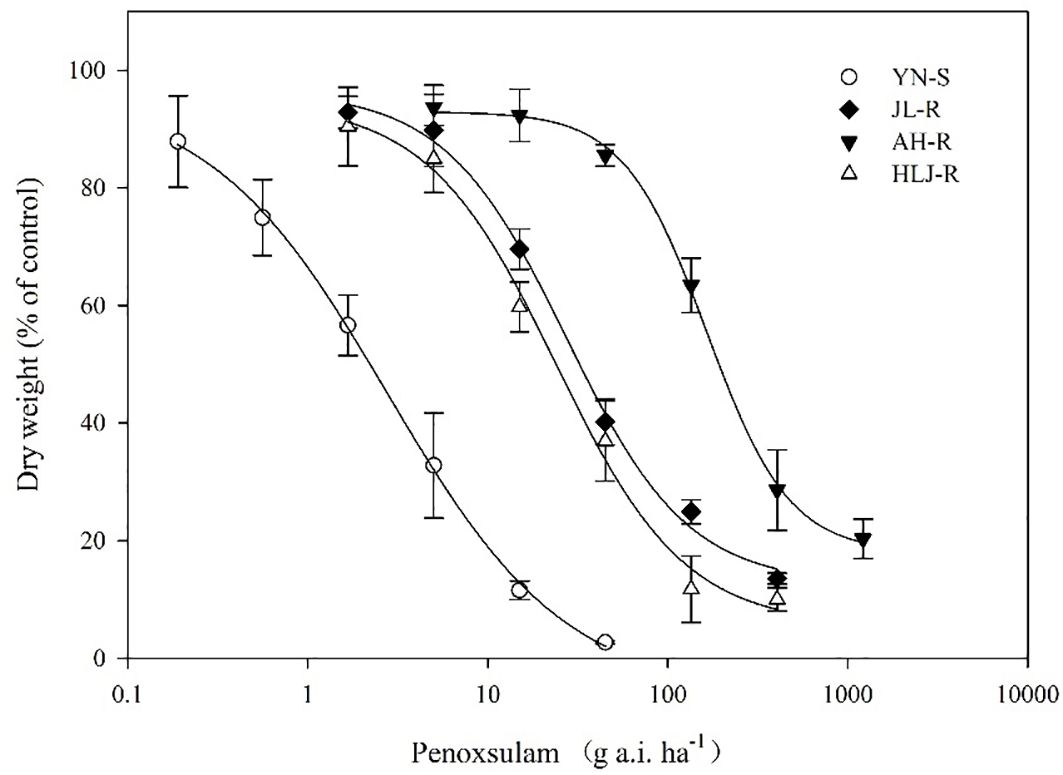
Figure 1. Dose–response curves for dry weights of YN-S, JL-R, AH-R, and HLJ-R populations of Echinochloa crus-galli to penoxsulam.
3.2 ALS gene sequencing and comparison
Sequencing confirmed that the ALS genes in E. crus-galli have no introns. Three ALS gene sequences were obtained, with lengths of 1,929 bp (ALS1), 1,932 bp (ALS2), and 1,935 bp (ALS3), encoding 643, 644, and 645 amino acids, respectively. Furthermore, sequence alignment analysis conducted using the COBALT tool on NCBI (https://www.ncbi.nlm.nih.gov/tools/cobalt/) identified five conserved amino acid mutations at specific positions within three distinct ALS copies (Figures 2, 3). Three nonsynonymous nucleotide mutations were identified in three resistant E. crus-galli populations. The Pro-197-His (CCC to CAC), Trp-574-Leu (TGG to TTG), and Pro-197-Leu (CCC to CTC) mutations caused by a nucleotide mutation were detected in ALS1 in JL-R population, in ALS1 in AH-R population, and in ALS2 in HLJ-R population, respectively (Figure 4). The mutation frequency in all three populations was 100%, and all colonies tested from each population showed consistent results. Additionally, Pro-197-His/Leu mutations were first identified in penoxsulam resistant E. crus-galli, with Pro-197-His mutation being reported for the first time in Echinochloa spp.
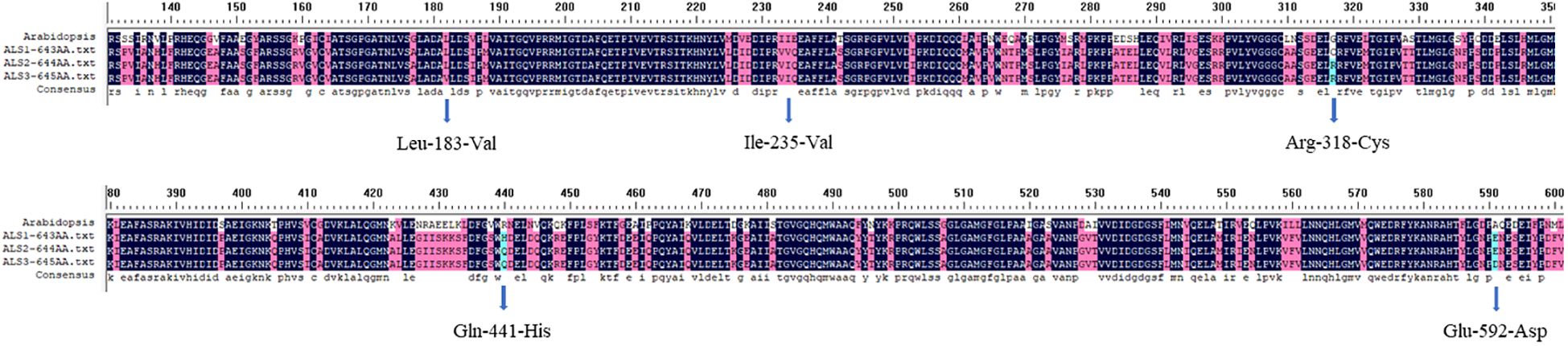
Figure 3. Alignment of the amino acid sequences of three E. crus-galli ALS copies with localization based on Arabidopsis thaliana ALS amino acid positions.
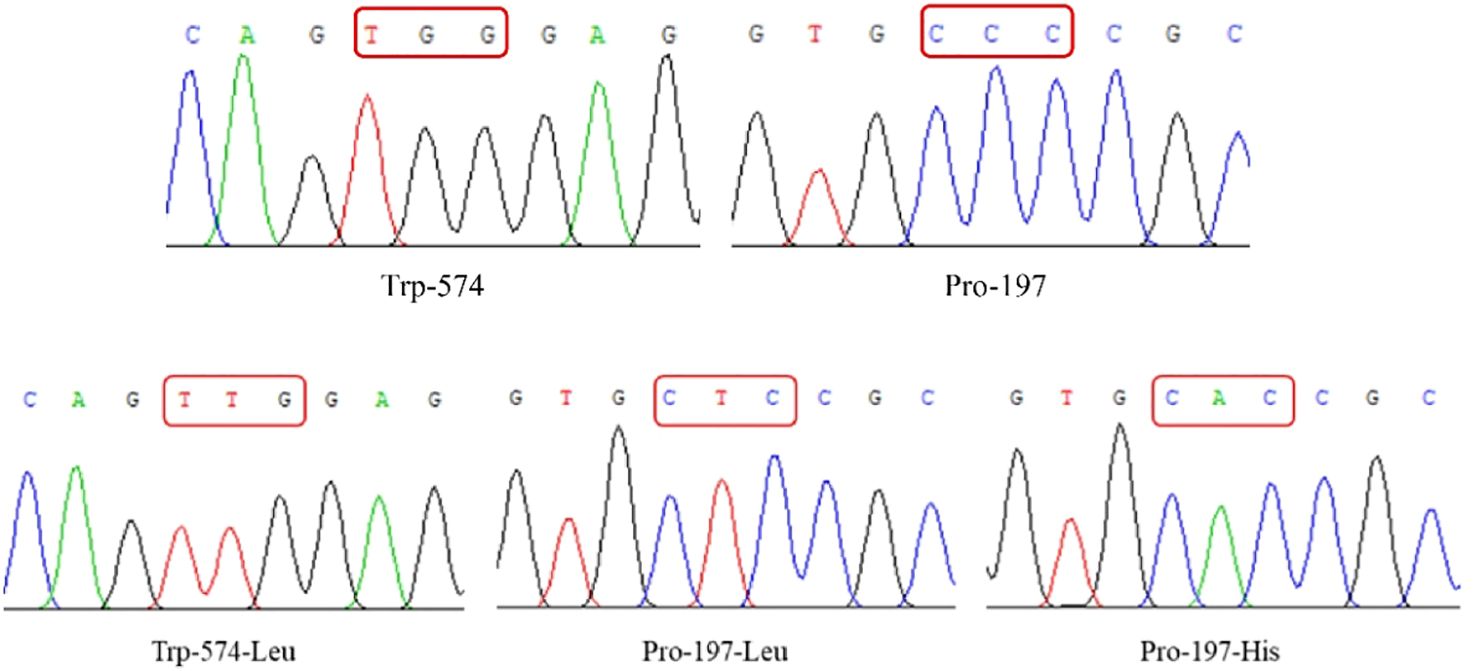
Figure 4. The spectrum of Trp-574-Leu and Pro-197-Leu/His mutations. The wild-type ALS gene is Leu at position 574 and Pro at position 197.
3.3 In vitro assay of ALS activity
The in vitro assay of E. crus-galli ALS activity showed no significant differences (p > 0.05) in total catalytic activity between the resistant populations JL-R (Pro-197-His), AH-R (Trp-574-Leu), and HLJ-R (Pro-197-Leu) compared to the susceptible YN-S population (Table 3). As shown in the dose–response curves (Figure 5), when the penoxsulam dose was 0.01 μmol·L−1, the ALS activity in the YN-S population was significantly inhibited, with an inhibition rate exceeding 50%, while the R populations showed less inhibition. Specifically, the I50 value for JL-R and HLJ-R population was 0.0118 μmol·L−1 and 0.0126 μmol·L−1, only 1.04- and 1.11-fold higher than that of the YN-S population, respectively. However, the I50 value for AH-R population was 0.0470 μmol·L−1, which was 4.12-fold higher than that of the YN-S, indicating a significant decrease in ALS sensitivity to penoxsulam in the AH-R population. Therefore, resistance in the HLJ-R and JL-R populations to penoxsulam may not be associated with a decrease in ALS sensitivity, whereas the reduced sensitivity of ALS in the AH-R population appears to be one of the key factors contributing to resistance development.
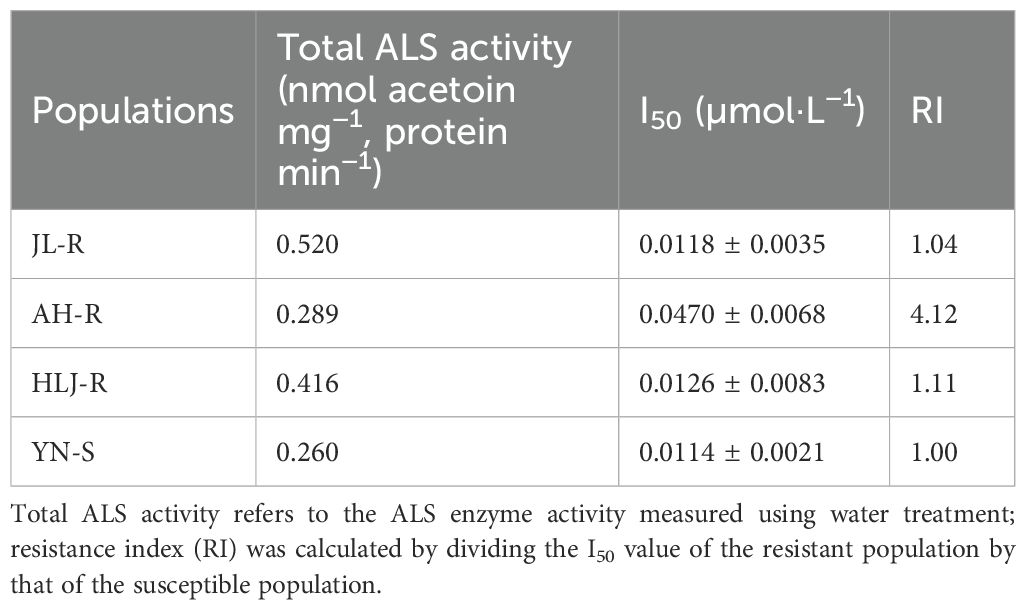
Table 3. The sensitivity of in vitro ALS extracted from different Echinochloa crus-galli populations to penoxsulam.
3.4 ALS gene expression
In this study, the transcriptional expression differences in ALS genes were compared among the resistant populations JL-R (Pro-197-His), AH-R (Trp-574-Leu), HLJ-R (Pro-197-Leu), and the susceptible population YN-S. The results of ALS gene expression levels, as shown in Figures 6A, B, indicated that there were no significant differences in ALS gene expression at 0 d, 1 d, 2 d, and 3 d after water treatment between the resistant populations JL-R, AH-R, HLJ-R, and the susceptible population YN-S (Figure 6A). Furthermore, there were no significant differences in ALS gene expression among these populations with the treatment of penoxsulam at 15 g a.i. ha−1 (Figure 6B). Therefore, the resistance to penoxsulam in JL-R, AH-R, and HLJ-R populations appears to be unrelated to the transcriptional level of ALS gene.

Figure 6. Relative expression levels of the ALS gene in different mutant populations in response to penoxsulam. (A) represents treatment with water only, while (B) represents treatment with penoxsulam.
3.5 Homology modeling and molecular docking
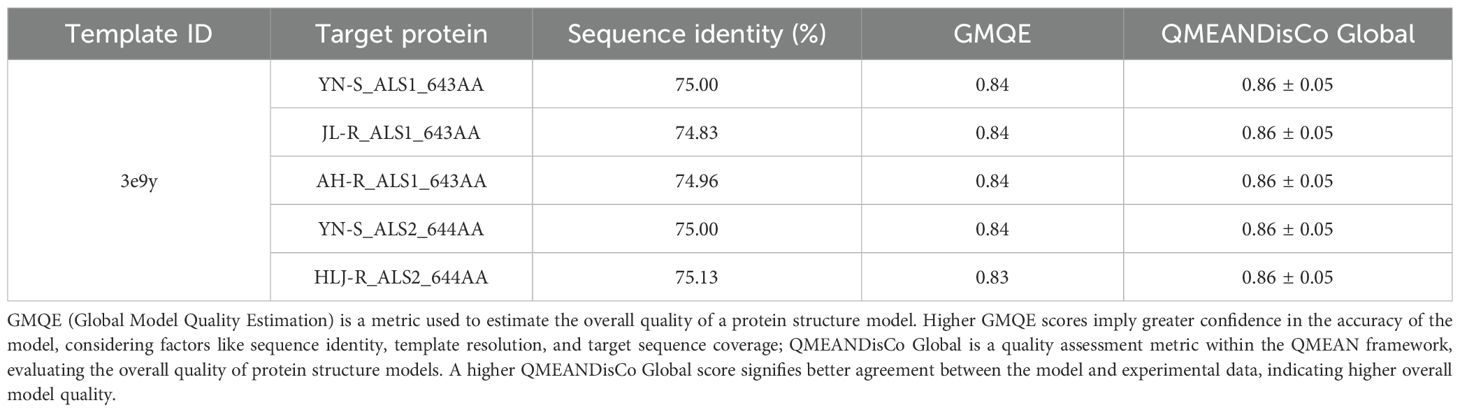
To gain a deeper understanding of the potential mechanism driving the differences in herbicide binding specificity to penoxsulam between the non-mutated and the three mutated ALS proteins in E. crus-galli, homology models of the ALS proteins were constructed, and comparative structural analyses were performed. The template protein 3e9y exhibited the highest sequence identity with the target ALS proteins from E. crus-galli, at approximately 75.00% (Table 4). The results of the Ramachandran plot indicated that more than 99% of the amino acid residues in the five different target ALS proteins were situated in the most favored and additional allowed regions (Table 5; Supplementary Figure S1). This validation confirmed that the protein model structures were of high quality and could be reliably used for subsequent molecular docking studies.
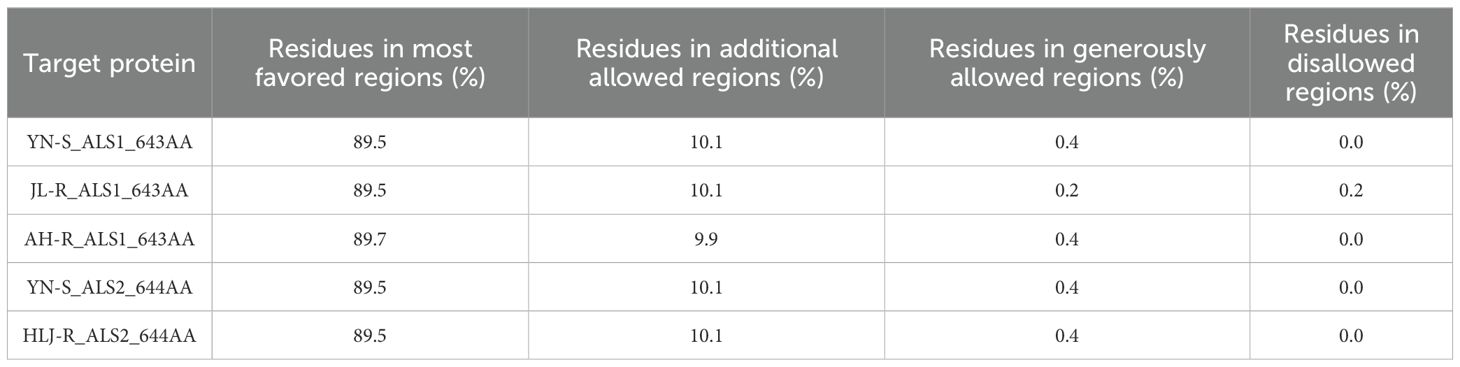
Table 5. The data of Ramachandran plot analysis of the five different target ALS proteins in E. crus-galli.
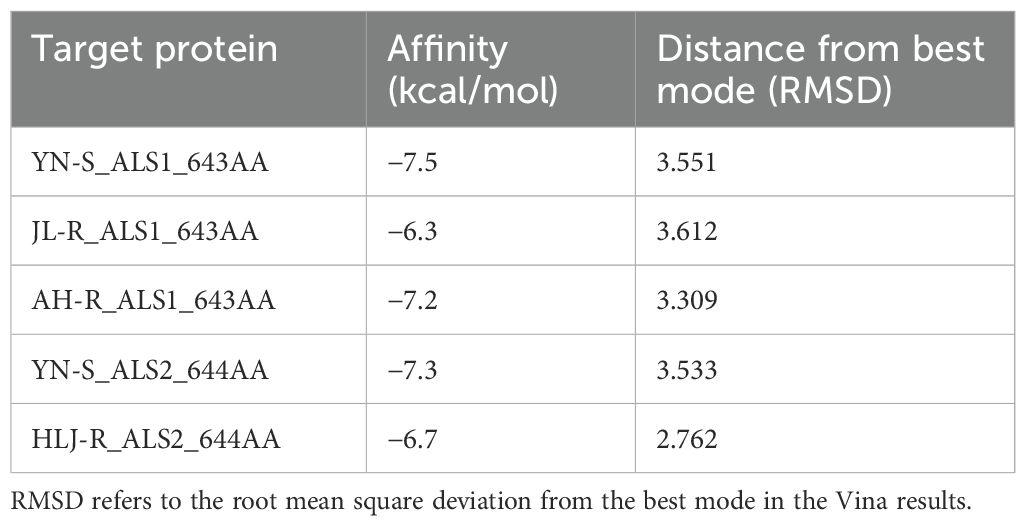
The homology modeling of ALS proteins included six ligands: two FAD molecules, two magnesium ions (Mg2+), and two 2-(cyclohexylamino)ethanesulfonic acid (CHE) molecules. Molecular docking simulations demonstrated consistent binding interactions of penoxsulam with two non-mutated ALS proteins, YN-S_ALS1_643AA and YN-S_ALS2_644AA with binding energies of −7.5 kcal mol−1 and −7.3 kcal mol−1, respectively (Table 6). The binding site of penoxsulam was situated close to the FAD-binding channel within the two subunits of E. crus-galli ALS. In this binding mode, the triazole-pyrimidine ring inserted into the interior of the channel, while the benzene ring structure was located on the outer surface of the channel (Figure 7). However, mutations occurred at H170 (equivalent to A. thaliana H197) in the E. crus-galli ALS1 and L171 (equivalent to A. thaliana L197) in the E. crus-galli ALS2, resulting in changes to the docking mode of penoxsulam with ALS1 and ALS2, with binding energies of −6.3 kcal mol−1 and −6.7 kcal mol−1, respectively (Table 6; Figure 8A). An increase in binding energy values indicates a reduced affinity of the ALS protein for penoxsulam. Specifically, the H170 mutation resulted in the formation of hydrogen bonds between the H170 residue and the benzene ring, disrupting pre-existing hydrophobic interactions. The penoxsulam benzene ring and sulfonyl group rotated, leading to changes in interactions with residues like F179, V169, and S626 (equivalent to A. thaliana F206, V196, and S653) (Figures 8A, 9A, B). Similarly, the ALS2_L171 mutation resulted in changes in the hydrophobic interactions involving the P171 residue in ALS2, leading to alterations in the binding conformation of penoxsulam with ALS2 and changes in interactions with surrounding amino acid residues (Figures 8B, 9D, E). Additionally, the mutation at L547 (equivalent to A. thaliana L574) in E. crus-galli ALS1 resulted in a binding energy of −7.2 kcal mol−1 with penoxsulam. The ALS1_L547 mutation disrupted the π–π stacking interaction between W547 and the pyrimidine ring, leading to altered interactions between penoxsulam and the surrounding residues. Notably, the pyrimidine ring of the penoxsulam molecule underwent a significant rotation (Figures 8A, 9C).
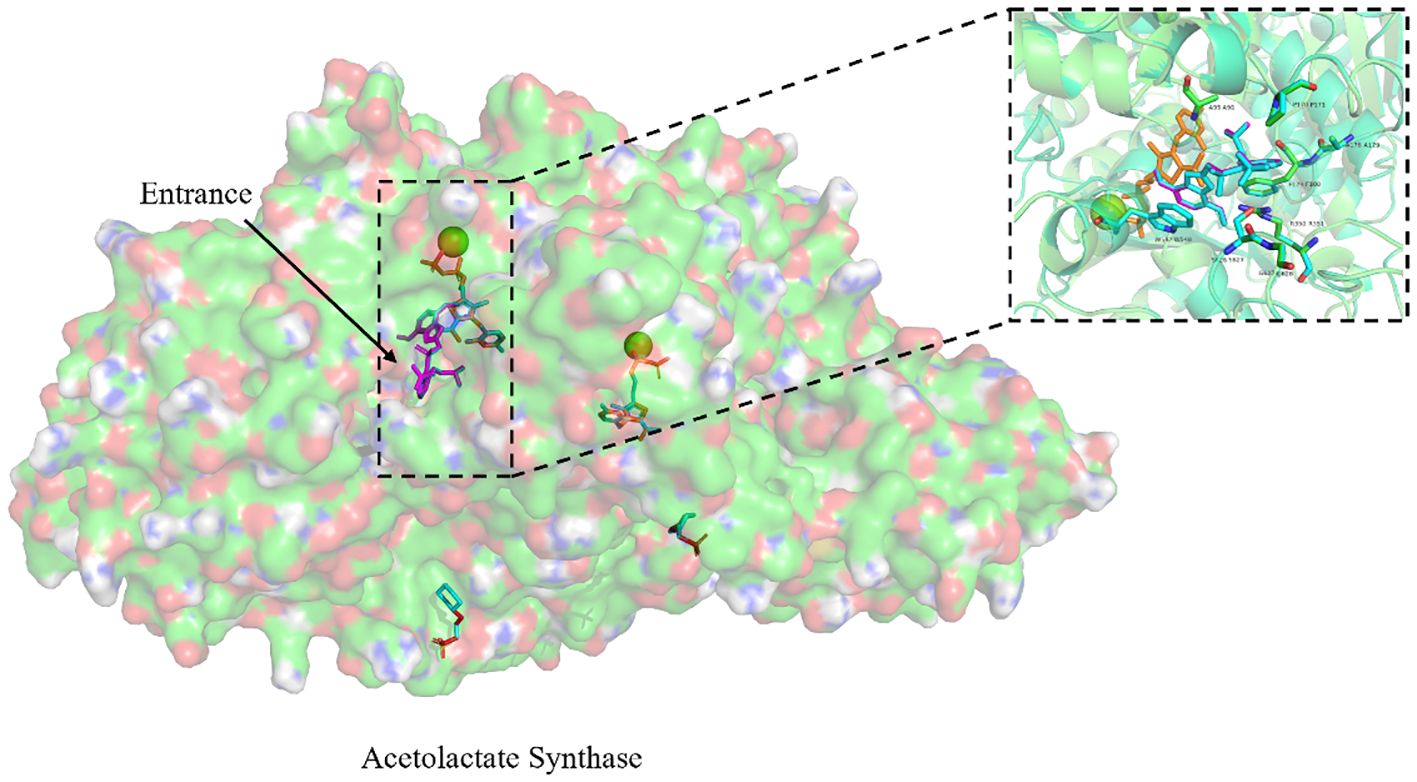
Figure 7. Exploring the binding conformation of penoxsulam with ALS proteins (YN-S_ALS1_643AA and YN-S_ALS2_644AA) in E. crus-galli through molecular docking. The green spheres represent Mg2+, the orange ligand represents FAD, and the binding positions of penoxsulam molecules in YN-S_ALS1_643AA and YN-S_ALS2_644AA proteins are consistent. Amino acids in the vicinity of the binding site are named based on the Arabidopsis thaliana ALS sequence for localization.
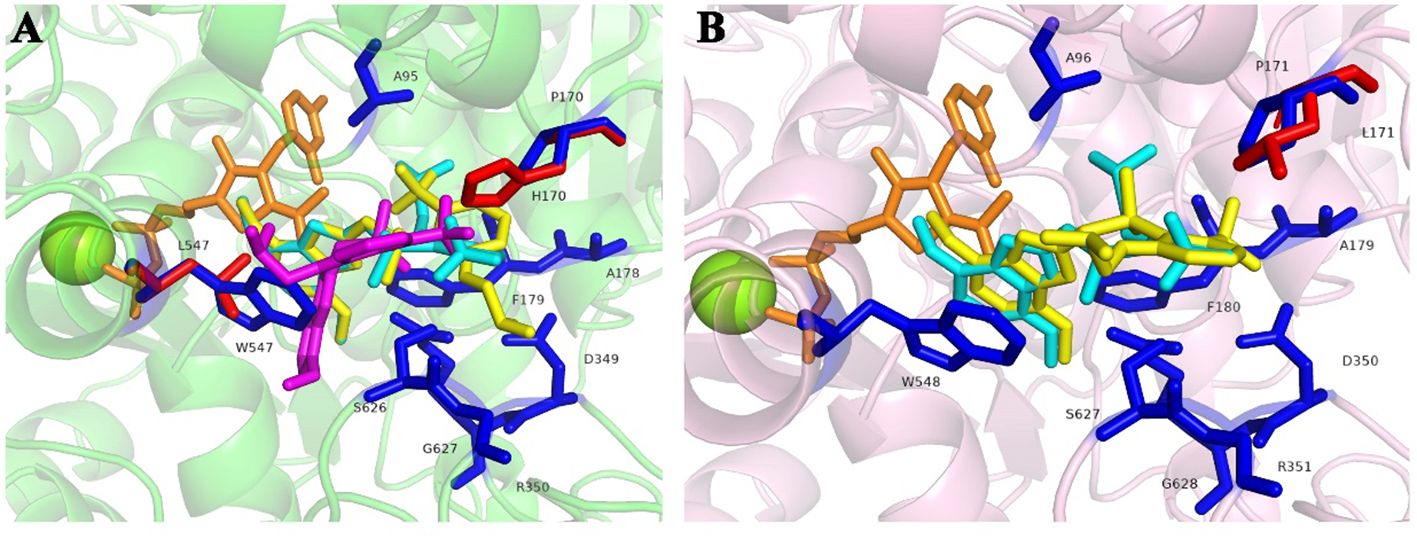
Figure 8. Molecular docking conformation of penoxsulam and ALS proteins in E. crus-galli: (A) The docking conformation of penoxsulam and three ALS1_643AA proteins: the residues of YN-S_ALS1_643AA are represented in blue, and the docking interaction with penoxsulam appeared in a peacock blue shade. The residues of JL-R_ALS1_643AA are the same as YN-S_ALS1_643AA, except for H170 highlighted in red, and the docking interaction with penoxsulam is shown in yellow. The residues of AH-R_ALS1_643AA are the same as YN-S_ALS1_643AA, except for L547 highlighted in red, and the docking interaction with penoxsulam is shown in purple. (B) The docking conformation of penoxsulam and two ALS2_644AA proteins: the residues of YN-S_ALS2_644AA are represented in blue, and the docking interaction with penoxsulam appears in a peacock blue shade. The residues of HLJ-R_ALS2_644AA are the same as YN-S_ALS2_644AA, except for H171 highlighted in red, and the docking interaction with penoxsulam is shown in yellow.
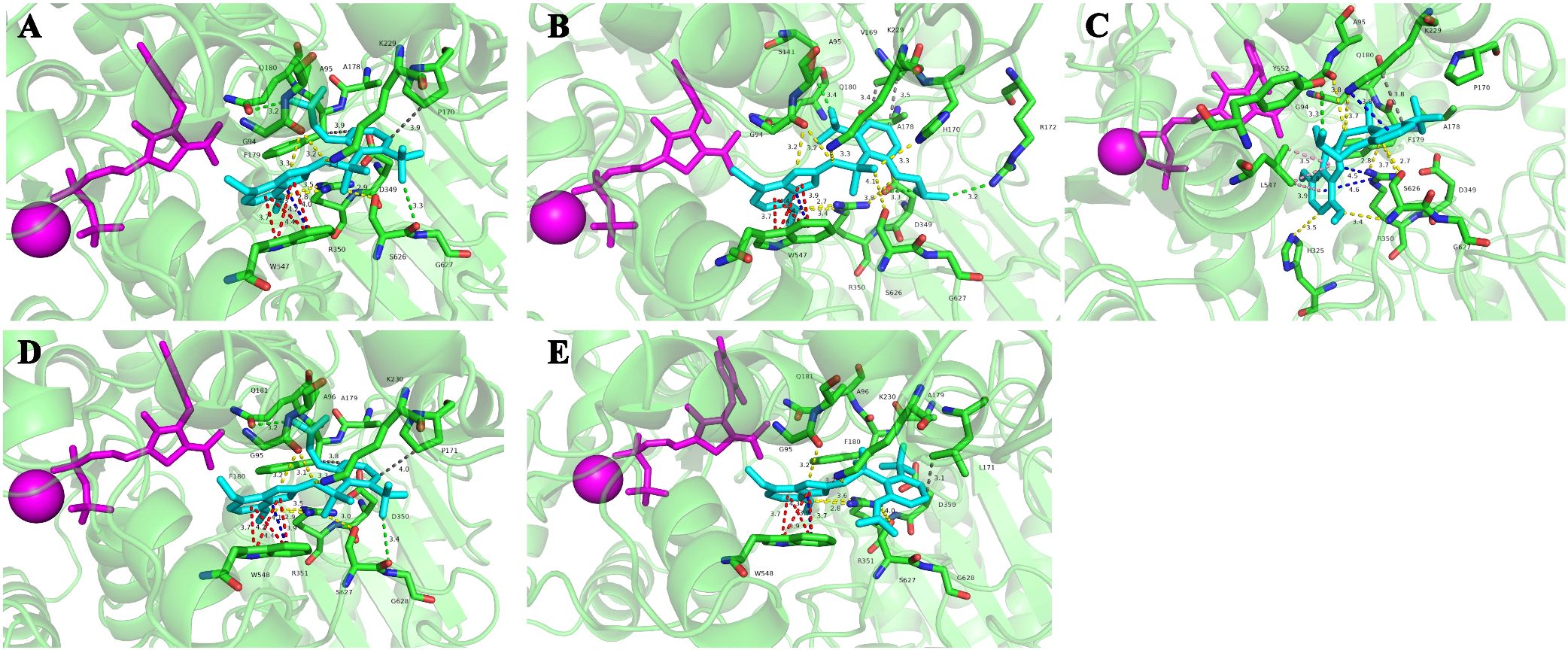
Figure 9. Molecular docking conformations of the mutated ALS proteins, Pro-197-His/Leu, and Trp-574-Leu, alongside the non-mutated E crus-galli ALS protein, to penoxsulam. The purple spherical ligand and small molecule ligand represent Mg2+ and FAD, respectively. Penoxsulam is depicted in peacock blue by stick model, and the amino acid residues interacting with penoxsulam are named using the single-letter amino acid code followed by the residue number, such as Pro170 written as P170. (A) The docking simulation conformation of penoxsulam with YN-S_ALS1_643AA. (B) The docking simulation conformation of penoxsulam with JL-R_ALS1_643AA. (C) The docking simulation conformation of penoxsulam with AH-R_ALS1_643AA. (D) The docking simulation conformation of penoxsulam with YN-S_ALS2_644AA. (E) The docking simulation conformation of penoxsulam with HLJ-R_ALS2_644AA. Gray line: hydrophobic interaction. Yellow line: hydrogen bond. Red line: π–π stacking interaction. Blue line: π–cation interaction. Green line: halogen bond. Pink line: π–sigma interaction.
3.6 Heterologous expression of ALS protein and activity to penoxsulam
The recombinant ALS proteins were expressed by the Bac-to-Bac baculovirus expression system with His-tags. The theoretical molecular weight of ALS1 and ALS2 is approximately 69 kDa. Western blot analysis revealed a distinct band approximately 72 kDa, confirming the successful expression of the ALS proteins. The results of purified ALS protein activity to penoxsulam are presented in Table 7. When treated with 1 μmol·L−1 penoxsulam, the various ALS proteins exhibited distinct responses. Specifically, WT_ALS1_643AA and WT_ALS2_644AA displayed significant inhibition, with ALS activity inhibition rates of 86.81% and 79.26%, respectively. In contrast, ALS proteins with different mutations showed considerably lower activity inhibition rates compared to non-mutated ALS proteins. Notably, the ALS protein with the Trp-574-Leu mutation exhibited the lowest level of inhibition, consistent with our previous in vitro ALS enzyme activity results.

Table 7. The activity assay of ALS protein expressed via baculovirus system in response to penoxsulam.
4 Discussion
A single mutation in the target enzyme is regarded as the most common cause of resistance evolution to ALS inhibiting herbicides. So far, there are 12 types of mutations documented in ALS inhibitors in Echinochloa spp. In the present study, three types of mutations (Pro-197-His, Trp-574-Leu, and Pro-197-Leu) were confirmed from three E. crus-galli populations; Pro-197-Leu and Trp-574-Leu mutations have been previously reported, and Pro-197-His mutation was first detected in E. crus-galli.
Mutations in the target enzyme can induce structural changes in its spatial conformation, leading to impaired or weakened binding between the target enzyme and herbicides (Duggleby et al., 2008; Yu and Powles, 2014). The point mutations in the target site of action are widely reported TSR mechanisms. For instance, the Asp-2078-Glu mutation in acetyl-coenzyme A carboxylase (ACCase) conferred resistance to ACCase herbicides in E. crus-galli (Fang et al., 2020), the Pro-197, Asp-376, and Trp-574 mutations in ALS conferred resistance to ALS inhibitors in D. sophia L (Deng et al., 2017; Wang et al., 2021; Yang et al., 2018). As widely recognized, E. crus-galli, a hexaploid plant, possesses at least three copies of the ALS gene in its genome. Following the sequencing of these three distinct ALS copies, three types of mutations were identified in the JL-R, AH-R, and HLJ-R populations (Figure 4). According to our knowledge, the Pro-197-His mutation in ALS1 was detected in E. crus-galli for the first time. This specific mutation has been demonstrated to confer resistance to ALS inhibitors in both D. sophia (Deng et al., 2015) and Galium aparine var. Tenerum Gren.et (Godr.) Rebb (Deng et al., 2019). This study found that E. crus-galli with the ALS-197-His/Leu mutation exhibited moderate-level resistance to penoxsulam. In contrast, Cyperus difformis L. with the ALS-197-His mutation and Amaranthus retroflexus L. with the ALS-197-Leu mutation displayed high levels of resistance to TPs inhibitors (Sibony et al., 2001; Tehranchian et al., 2015). These findings suggest that research on ALS mutations should be specific to the weed species involved. Therefore, we conducted an in-depth investigation of the newly identified mutations in E. crus-galli.
The overexpression of the target enzyme gene is considered one of the mechanisms in TSR. Panozzo et al. found that the expression levels of mutated ALS gene copies in E. crus-galli and E. oryzicola were significantly higher than that of non-mutated ALS gene copies (Panozzo et al., 2021). Additionally, ACCase gene overexpression was shown to confer resistance to ACCase inhibitors in Digitaria sanguinalis L (Laforest et al., 2017). However, there is no direct evidence of a correlation between the expression level and mutations in the ALS gene. In a population of Capsella bursa-pastoris L. Medik. with Pro-197-Ser and Pro-197-His mutations in the ALS gene copies, Wang et al. discovered that the ALS gene expression level did not significantly differ from that of the sensitive population (Wang et al., 2019). Our study conducted differential expression analysis on ALS genes and found no significant differences in expression between ALS genes in populations with different mutations (Pro-197-His, Trp-574-Leu, and Pro-197-Leu) and ALS genes in the sensitive population. This suggests that the resistance of the JL-R, AH-R, and HLJ-R populations to penoxsulam was not related to the overexpression of the ALS gene.
The determination of ALS enzyme activity through the colorimetric reaction of creatine and α-naphthol with 2-aceto-2-hydroxybutyrate is the most commonly used method in ALS in vitro enzyme assays (Fang et al., 2022; Yang et al., 2018). In vitro ALS enzyme activity analysis provides a rapid means of identifying weed resistance to ALS inhibitors. Cao et al. demonstrated that a reduced sensitivity of the ALS enzyme to imazethapyr was a crucial factor in conferring resistance to Chenopodium album L. against imazethapyr (Cao et al., 2022). In this study, the ALS in vitro enzyme activities of purified F1 generation plants from JL-R, AH-R, and HLJ-R were analyzed. The present research revealed that the I50 value of the AH-R (Trp-574-Leu) population was 4.12 times higher than that of the sensitive population, indicating that the decreased sensitivity of the ALS in the AH-R population was one of the reasons for its resistance to penoxsulam. The I50 values of the JL-R (Pro-197-His) and HLJ-R (Pro-197-Leu) populations exhibited no significant differences compared to the sensitive population. The populations of JL-R and HLJ-R exhibited moderate resistance to penoxsulam, with mutations occurring solely in one copy of the ALS gene. This observation may account for the lack of significant differences in their ALS in vitro enzyme activity.
Homology modeling and molecular docking techniques were extensively used in previous studies of protein–small molecule interactions (Joshi, 2016; Palma-Bautista et al., 2022; Zhao et al., 2022). The mutation of amino acids at specific positions in ALS has been verified to modify the binding forces between ALS and inhibitors, resulting in ALS resistance. For example, the double ALS gene mutation (Pro-197-Ser plus Trp-574-Leu) in C. bursa-pastoris induced alterations in H-bond, π–π, and π–sulfur interactions between ALS and herbicide, leading to high resistance to mesosulfuron-methyl (Lu et al., 2023). Similarly, the ALS mutation at position 206 (Phe-206-Leu) has been confirmed to confer penoxsulam resistance in E. crus-galli due to the disappearance of π–π interaction between the 206 site position and penoxsulam (Fang et al., 2022). ALS consists of catalytic and regulatory subunits, with the latter providing feedback inhibition (Duggleby et al., 2008). ALS inhibitors generally do not directly bind to the catalytic site; instead, they bind to the region located at the entrance of the ALS active site channel, where the Pro-197 and Trp-574 sites are situated (McCourt et al., 2006). The side chain of proline at the Pro-197 site is a saturated hydrocarbon, and this saturated hydrocarbon side chain imposes certain restrictions on the spatial structure of the protein. Liu and coworkers used molecular docking to discover that the Pro-197-Ser mutation in the ALS of E. phyllopogon confers resistance to various ALS inhibitors (Liu et al., 2019). This resistance is attributed to changes in the spatial structure of the substrate center due to the variation of the residue at the Pro-197 site, resulting in resistance to TPs, SUs, and SCTs, while remaining sensitive to PTBs and IMIs (Liu et al., 2019). In this study, we found that penoxsulam bound to E. crus-galli ALS by inserting the double heterocyclic ring of triazole and pyrimidine into the ALS channel. The π–π stacking formed by the aromatic rings of Phe-206 and Trp-574 with the double heterocyclic ring of penoxsulam was disrupted by the Trp-574-Leu mutation. The Trp-574-Leu mutation impacted contacts of surrounding residues with penoxsulam. This disruption reduced the binding strength between penoxsulam and ALS, consistent with previous research (Fang et al., 2022). Additionally, Trp-574 served as the key residue defining the shape of the substrate access tunnel, and its mutation to another residue was expected to significantly weaken the binding of IMIs (Duggleby et al., 2008). In the present study, although the binding energy between the ALS1 protein and penoxsulam was not significantly reduced following the Trp-574-Leu mutation, the smaller molecular weight of Leu compared to Trp enlarged the binding pocket at the channel entrance. As a result, the inhibitor could not effectively prevent the substrate from reaching the active site, leading to resistance to penoxsulam. Mutations at the Pro-197 site to Leu and His modify the spatial structure of this region due to changes in the amino acid side chains. This alteration may hinder the ability of herbicides to enter the binding channel. Although the affinities of the ALS protein with the Pro-197-His and Pro-197-Leu mutations for penoxsulam are lower than that of the Trp-574-Leu mutated ALS protein, the binding mode of penoxsulam remains largely unchanged. Consequently, it continues to effectively obstruct the substrate from accessing the active site, resulting in a lower level of resistance to penoxsulam compared to the ALS protein with the Trp-574-Leu mutation.
The previous study revealed that the impact of resistant alleles could diminish with the increase in the percentage of susceptible allele transcripts in polyploid weeds (Iwakami et al., 2012). In this current study, the expression of mutated ALS alleles might have experienced a dilution effect from sensitive alleles, potentially reducing the contribution to resistance. Therefore, the Bac-to-Bac baculovirus expression system was employed to individually express the mutant ALS protein and analyze its activity with penoxsulam. This system allows cells to grow in suspension, facilitating large-scale cultivation and demonstrating the capability for simultaneous expression of multiple genes with enhanced protein modification processing abilities (Fang et al., 2022; Van Oers et al., 2015). In this study, we successfully obtained wild-type (WT) and three mutant ALS proteins. Upon treatment with penoxsulam, the results revealed that, compared to the WT ALS proteins (WT_ALS1_643AA and WT_ALS2_644AA), the activity inhibition rates of the three mutant ALS proteins (197-His_ALS1_643AA, 574-Leu_ALS1_643AA, and 197-Leu_ALS2_644AA) were reduced at 1 μmol·L−1 penoxsulam. This finding was consistent with our differential analysis of ALS genes and molecular docking results, providing protein-level evidence that mutations in E. crus-galli ALS (Trp-574-Leu, Pro-197-Leu, and Pro-197-His) induce resistance to penoxsulam. Additionally, this also explained the impact of the dilution effect of sensitive allele genes in the in vitro detection of mutant plant ALS activity.
The ALS inhibitor penoxsulam, developed by Dow AgroSciences, is a trizolopyrimidine herbicide applied in paddy fields with the wide herbicidal spectrum. It has great effect not only on aquatic weed control but also on E. crus-galli, which has resistance to herbicides like quinclorac, propanil, and sulfonylurea. It was registered in China in 2007 and developed as a key product in weeds control in paddy field since then. However, the overreliance on the herbicide has led to the development of resistant weeds related to TSR and NTSR (Fang et al., 2019a, Fang et al., 2022, Fang et al., 2019b; Feng et al., 2022). In the past decade, E. crus-galli has been reported to evolve resistance to several sites of action of herbicides (Fang et al., 2019a, Fang et al., 2022, Fang et al., 2019b; Pan et al., 2022; Sun et al., 2023; Vázquez-García et al., 2021). Under the continuous selective pressure of herbicides, herbicide resistance evolution in weeds has become an inevitable consequence. Mutations in the target site of action in weeds frequently result not only in resistance to specific herbicides but also in cross-resistance to several herbicides (Han et al., 2012; Singh et al., 2019; Vital Silva et al., 2022). Through interspecies cross-pollination, mutation genes swiftly disseminate within the species. Therefore, there is an urgent need for exploring the mechanisms of weed resistance and the formulation of innovative, sustainable weed management methods to effectively delay and manage the evolution of herbicide resistance (Beckie et al., 2019; Pan et al., 2022).
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
PS: Data curation, Investigation, Methodology, Writing – original draft. LN: Investigation, Writing – original draft. PH: Investigation, Writing – original draft. HY: Methodology, Writing – original draft. JC: Methodology, Writing – original draft. HC: Supervision, Writing – original draft. XL: Conceptualization, Funding acquisition, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. We gratefully acknowledge the financial support provided by the National Natural Science Foundation of China (U20A2038) and Nanfan Special Project, CAAS (SWAQ03).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1488976/full#supplementary-material
References
Adkins, S., Shabbir, A. (2014). Biology, ecology and management of the invasive parthenium weed (Parthenium hysterophorus L.). Pest Manage Sci. 70, 1023–1029. doi: 10.1002/ps.3708
Amaro-Blanco, I., Romano, Y., Palmerin, J. A., Gordo, R., Palma-Bautista, C., De Prado, R., et al. (2021). Different mutations providing target site resistance to ALS-and ACCase-inhibiting herbicides in Echinochloa spp. from rice fields. Agriculture 11, 382. doi: 10.3390/agriculture11050382
Beckie, H. J., Ashworth, M. B., Flower, K. C. (2019). Herbicide resistance management: Recent developments and trends. Plants 8, 161. doi: 10.3390/plants8060161
Brun, T., Rabuske, J. E., Confortin, T. C., Luft, L., Todero, I., Fischer, M., et al. (2022). Weed control by metabolites produced from Diaporthe schini. Environ. Technol. 43, 139–148. doi: 10.1080/09593330.2020.1780477
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE Guidelines: M inimum I nformation for Publication of Q uantitative Real-Time PCR E xperiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Butt, S. S., Badshah, Y., Shabbir, M., Rafiq, M. (2020). Molecular docking using chimera and autodock vina software for nonbioinformaticians. JMIR Bioinform. Biotech. 1, e14232. doi: 10.2196/14232
Cao, Y., Zhou, X., Wei, S., Huang, H., Lan, Y., Li, W., et al. (2022). Multiple resistance to ALS-inhibiting and PPO-inhibiting herbicides in Chenopodium album L. from China. Pestic Biochem. Physiol. 186, 105155. doi: 10.1016/j.pestbp.2022.105155
Chauhan, B., Johnson, D. (2011). Ecological studies on Echinochloa crus-galli and the implications for weed management in direct-seeded rice. Crop Prot 30, 1385–1391. doi: 10.1016/j.cropro.2011.07.013
Chipman, D., Barak, Z., Schloss, J. V. (1998). Biosynthesis of 2-aceto-2-hydroxy acids: acetolactate synthases and acetohydroxyacid synthases. Biochim. Biophys. Acta 1385, 401–419. doi: 10.1016/S0167-4838(98)00083-1
Clewley, J. P. (1995). Macintosh sequence analysis software: DNAStar’s LaserGene. Mol. Biotechnol. 3, 221–224. doi: 10.1007/BF02789332
Comont, D., Lowe, C., Hull, R., Crook, L., Hicks, H. L., Onkokesung, N., et al. (2020). Evolution of generalist resistance to herbicide mixtures reveals a trade-off in resistance management. Nat. Commun. 11, 3086. doi: 10.1038/s41467-020-16896-0
Délye, C. (2013). Unravelling the genetic bases of non-target-site-based resistance (NTSR) to herbicides: a major challenge for weed science in the forthcoming decade. Pest Manage Sci. 69, 176–187. doi: 10.1002/ps.3318
Délye, C., Causse, R., Gautier, V., Poncet, C., Michel, S. (2015). Using next-generation sequencing to detect mutations endowing resistance to pesticides: application to acetolactate-synthase (ALS)-based resistance in barnyard grass, a polyploid grass weed. Pest Manage Sci. 71, 675–685. doi: 10.1002/ps.3818
Délye, C., Jasieniuk, M., Le Corre, V. (2013). Deciphering the evolution of herbicide resistance in weeds. Trends Genet. 29, 649–658. doi: 10.1016/j.tig.2013.06.001
Deng, W., Cao, Y., Yang, Q., Liu, M. J., Mei, Y., Zheng, M. Q. (2014). Different cross-resistance patterns to AHAS herbicides of two tribenuron-methyl resistant flixweed (Descurainia sophia L.) biotypes in China. Pestic Biochem. Physiol. 112, 26–32. doi: 10.1016/j.pestbp.2014.05.003
Deng, W., Di, Y., Cai, J., Chen, Y., Yuan, S. (2019). Target-site resistance mechanisms to tribenuron-methyl and cross-resistance patterns to ALS-inhibiting herbicides of catchweed bedstraw (Galium aparine) with different ALS mutations. Weed Sci. 67, 183–188. doi: 10.1017/wsc.2018.70
Deng, W., Liu, M. J., Yang, Q., Mei, Y., Li, X. F., Zheng, M. Q. (2015). Tribenuron-methyl resistance and mutation diversity of Pro197 in flixweed (Descurainia sophia L.) accessions from China. Pestic Biochem. Physiol. 117, 68–74. doi: 10.1016/j.pestbp.2014.10.012
Deng, W., Yang, Q., Zhang, Y., Jiao, H., Mei, Y., Li, X., et al. (2017). Cross-resistance patterns to acetolactate synthase (ALS)-inhibiting herbicides of flixweed (Descurainia sophia L.) conferred by different combinations of ALS isozymes with a Pro-197-Thr mutation or a novel Trp-574-Leu mutation. Pestic Biochem. Physiol. 136, 41–45. doi: 10.1016/j.pestbp.2016.08.006
Devine, M. D., Shukla, A. (2000). Altered target sites as a mechanism of herbicide resistance. Crop Prot 19, 881–889. doi: 10.1016/S0261-2194(00)00123-X
Duggleby, R. G., McCourt, J. A., Guddat, L. W. (2008). Structure and mechanism of inhibition of plant acetohydroxyacid synthase. Plant Physiol. Biochem. 46, 309–324. doi: 10.1016/j.plaphy.2007.12.004
Fang, J., He, Z., Liu, T., Li, J., Dong, L. (2020). A novel mutation Asp-2078-Glu in ACCase confers resistance to ACCase herbicides in barnyardgrass (Echinochloa crus-galli). Pestic Biochem. Physiol. 168, 104634. doi: 10.1016/j.pestbp.2020.104634
Fang, J., Liu, T., Zhang, Y., Li, J., Dong, L. (2019a). Target site–based penoxsulam resistance in barnyardgrass (Echinochloa crus-galli) from China. Weed Sci. 67, 281–287. doi: 10.1017/wsc.2019.5
Fang, J., Yang, D., Zhao, Z., Chen, J., Dong, L. (2022). A novel Phe-206-Leu mutation in acetolactate synthase confers resistance to penoxsulam in barnyardgrass (Echinochloa crus-galli (L.) P. Beauv). Pest Manage Sci. 78, 2560–2570. doi: 10.1002/ps.6887
Fang, J., Zhang, Y., Liu, T., Yan, B., Li, J., Dong, L. (2019b). Target-site and metabolic resistance mechanisms to penoxsulam in barnyardgrass (Echinochloa crus-galli (L.) P. Beauv). J. Agric. Food Chem. 67, 8085–8095. doi: 10.1021/acs.jafc.9b01641
Feng, T., Peng, Q., Wang, L., Xie, Y., Ouyang, K., Li, F., et al. (2022). Multiple resistance mechanisms to penoxsulam in Echinochloa crus-galli from China. Pesticide Biochem. Physiol. 187, 105211. doi: 10.1016/j.pestbp.2022.105211
Gaines, T. A., Duke, S. O., Morran, S., Rigon, C. A., Tranel, P. J., Küpper, A., et al. (2020). Mechanisms of evolved herbicide resistance. J. Biol. Chem. 295, 10307–10330. doi: 10.1074/jbc.REV120.013572
Han, H., Yu, Q., Purba, E., Li, M., Walsh, M., Friesen, S., et al. (2012). A novel amino acid substitution Ala-122-Tyr in ALS confers high-level and broad resistance across ALS-inhibiting herbicides. Pest Manage Sci. 68, 1164–1170. doi: 10.1002/ps.3278
Heap, I. M. (2024). The International Herbicide-Resistant Weed Database. Available online at: http://www.weedscience.org (accessed April 1, 2024).
Iwakami, S., Hashimoto, M., Matsushima, K.-I., Watanabe, H., Hamamura, K., Uchino, A. (2015). Multiple-herbicide resistance in Echinochloa crus-galli var. formosensis, an allohexaploid weed species, in dry-seeded rice. Pestic Biochem. Physiol. 119, 1–8. doi: 10.1016/j.pestbp.2015.02.007
Iwakami, S., Uchino, A., Watanabe, H., Yamasue, Y., Inamura, T. (2012). Isolation and expression of genes for acetolactate synthase and acetyl-CoA carboxylase in Echinochloa phyllopogon, a polyploid weed species. Pest Manage Sci. 68, 1098–1106. doi: 10.1002/ps.3287
Joshi, S. (2016). Elucidating modes of activation and herbicide resistance by sequence assembly and molecular modelling of the Acetolactate synthase complex in sugarcane. J. Theor. Biol. 407, 184–197. doi: 10.1016/j.jtbi.2016.07.025
Kraehmer, H., Laber, B., Rosinger, C., Schulz, A. (2014). Herbicides as weed control agents: state of the art: I. Weed control research and safener technology: the path to modern agriculture. Plant Physiol. 166, 1119–1131. doi: 10.1104/pp.114.241901
Laforest, M., Soufiane, B., Simard, M. J., Obeid, K., Page, E., Nurse, R. E. (2017). Acetyl-CoA carboxylase overexpression in herbicide-resistant large crabgrass (Digitaria sanguinalis). Pest Manage Sci. 73, 2227–2235. doi: 10.1002/ps.4675
Latif, A., Rao, A. Q., Khan, M. A. U., Shahid, N., Bajwa, K. S., Ashraf, M. A., et al. (2015). Herbicide-resistant cotton (Gossypium hirsutum) plants: an alternative way of manual weed removal. BMC Res. Notes 8, 1–8. doi: 10.1186/s13104-015-1397-0
Li, J., Li, Y., Fang, F., Xue, D., Li, R., Gao, X., et al. (2022). A novel naturally Phe206Tyr mutation confers tolerance to ALS-inhibiting herbicides in Alopecurus myosuroides. Pestic Biochem. Physiol. 186, 105156. doi: 10.1016/j.pestbp.2022.105156
Liu, J., Fang, J., He, Z., Li, J., Dong, L. (2019). Target site–based resistance to penoxsulam in late watergrass (Echinochloa phyllopogon) from China. Weed Sci. 67, 380–388. doi: 10.1017/wsc.2019.14
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, H., Liu, Y., Bu, D., Yang, F., Zhang, Z., Qiang, S. (2023). A double mutation in the ALS gene confers a high level of resistance to mesosulfuron-methyl in shepherd’s-purse. Plants 12, 2730. doi: 10.3390/plants12142730
Mazur, B. J., Falco, S. C. (1989). The development of herbicide resistant crops. Annu. Rev. Plant Biol. 40, 441–470. doi: 10.1146/annurev.pp.40.060189.002301
McCourt, J. A., Pang, S. S., King-Scott, J., Guddat, L. W., Duggleby, R. G. (2006). Herbicide-binding sites revealed in the structure of plant acetohydroxyacid synthase. Proc. Natl. Acad. Sci. U.S.A. 103, 569–573. doi: 10.1073/pnas.0508701103
Oerke, E.-C. (2006). Crop losses to pests. J. Agric. Sci. 144, 31–43. doi: 10.1017/S0021859605005708
Palma-Bautista, C., Portugal, J., Vázquez-García, J. G., Osuna, M. D., Torra, J., Lozano-Juste, J., et al. (2022). Tribenuron-methyl metabolism and the rare Pro197Phe double mutation together with 2, 4-D metabolism and reduced absorption can evolve in Papaver rhoeas with multiple and cross herbicide resistance to ALS inhibitors and auxin mimics. Pestic Biochem. Physiol. 188, 105226. doi: 10.1016/j.pestbp.2022.105226
Pan, L., Guo, Q., Wang, J., Shi, L., Yang, X., Zhou, Y., et al. (2022). CYP81A68 confers metabolic resistance to ALS and ACCase-inhibiting herbicides and its epigenetic regulation in Echinochloa crus-galli. J. Hazard Mater 428, 128225. doi: 10.1016/j.jhazmat.2022.128225
Panozzo, S., Mascanzoni, E., Scarabel, L., Milani, A., Dalazen, G., Merotto, A. J., et al. (2021). Target-site mutations and expression of ALS gene copies vary according to Echinochloa species. Genes 12, 1841. doi: 10.3390/genes12111841
Panozzo, S., Scarabel, L., Rosan, V., Sattin, M. (2017). A new Ala-122-Asn amino acid change confers decreased fitness to ALS-resistant Echinochloa crus-galli. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.02042
Powles, S. B., Yu, Q. (2010). Evolution in action: plants resistant to herbicides. Annu. Rev. Plant Biol. 61, 317–347. doi: 10.1146/annurev-arplant-042809-112119
Riar, D. S., Norsworthy, J. K., Srivastava, V., Nandula, V., Bond, J. A., Scott, R. C. (2013). Physiological and molecular basis of acetolactate synthase-inhibiting herbicide resistance in barnyardgrass (Echinochloa crus-galli). J. Agric. Food Chem. 61, 278–289. doi: 10.1021/jf304675j
Sibony, M., Michel, A., Haas, H., Rubin, B., Hurle, K. (2001). Sulfometuron-resistant Amaranthus retroflexus: cross-resistance and molecular basis for resistance to acetolactate synthase-inhibiting herbicides. Weed Res. 41, 509–522. doi: 10.1046/j.1365-3180.2001.00254.x
Singh, S., Singh, V., Salas-Perez, R. A., Bagavathiannan, M. V., Lawton-Rauh, A., Roma-Burgos, N. (2019). Target-site mutation accumulation among ALS inhibitor-resistant Palmer amaranth. Pest Manage Sci. 75, 1131–1139. doi: 10.1002/ps.5232
Sun, P., Niu, L., Lan, X., Yu, H., Cui, H., Chen, J., et al. (2023). Enhanced metabolic resistance mechanism endows resistance to metamifop in Echinochloa crus-galli (L.) P. Beauv. Pestic Biochem. Physiol., 105656. doi: 10.1016/j.pestbp.2023.105656
Tehranchian, P., Riar, D. S., Norsworthy, J. K., Nandula, V., McElroy, S., Chen, S., et al. (2015). ALS-resistant smallflower umbrella sedge (Cyperus difformis) in Arkansas rice: physiological and molecular basis of resistance. Weed Sci. 63, 561–568. doi: 10.1614/WS-D-14-00147.1
Tranel, P. J., Wright, T. R., Heap, I. M. (2024). Mutations in Herbicide-resistant Weeds to ALS Inhibitors. Available online at: http://www.weedscience.com (accessed April 1, 2024).
Trott, O., Olson, A. J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461. doi: 10.1002/jcc.21334
Van Oers, M. M., Pijlman, G. P., Vlak, J. M. (2015). Thirty years of baculovirus–insect cell protein expression: from dark horse to mainstream technology. J. Gen. Virol. 96, 6–23. doi: 10.1099/vir.0.067108-0
Vázquez-García, J. G., Rojano-Delgado, A. M., Alcántara-de la Cruz, R., Torra, J., Dellaferrera, I., Portugal, J., et al. (2021). Distribution of glyphosate-resistance in Echinochloa crus-galli across agriculture areas in the Iberian Peninsula. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.617040
Vital Silva, V., Mendes, R., Suzukawa, A., Adegas, F., Marcelino-Guimaraes, F., Oliveira, R., Jr. (2022). A target-site mutation confers cross-resistance to ALS-inhibiting herbicides in Erigeron sumatrensis from Brazil. Plants 11, 467. doi: 10.3390/plants11040467
Wang, J. G., Lee, P. K. M., Dong, Y. H., Pang, S. S., Duggleby, R. G., Li, Z. M., et al. (2009). Crystal structures of two novel sulfonylurea herbicides in complex with Arabidopsis thaliana acetohydroxyacid synthase. FEBS J. 276, 1282–1290. doi: 10.1111/j.1742-4658.2009.06863.x
Wang, H., Sun, P., Guo, W., Dong, X., Liu, W., Wang, J. (2021). Florasulam resistance status of flixweed (Descurainia sophia L.) and alternative herbicides for its chemical control in the North China plain. Pestic Biochem. Physiol. 172, 104748. doi: 10.1016/j.pestbp.2020.104748
Wang, H., Zhang, L., Li, W., Bai, S., Zhang, X., Wu, C., et al. (2019). Isolation and expression of acetolactate synthase genes that have a rare mutation in shepherd’s purse (Capsella bursa-pastoris (L.) Medik.). Pestic Biochem. Physiol. 155, 119–125. doi: 10.1016/j.pestbp.2019.01.013
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi: 10.1093/nar/gky427
Yang, Q., Deng, W., Wang, S., Liu, H., Li, X., Zheng, M. (2018). Effects of resistance mutations of Pro197, Asp376 and Trp574 on the characteristics of acetohydroxyacid synthase (AHAS) isozymes. Pest Manage Sci. 74, 1870–1879. doi: 10.1002/ps.4889
Yu, Q., Friesen, L. S., Zhang, X. Q., Powles, S. B. (2004). Tolerance to acetolactate synthase and acetyl-coenzyme A carboxylase inhibiting herbicides in Vulpia bromoides is conferred by two co-existing resistance mechanisms. Pestic Biochem. Physiol. 78, 21–30. doi: 10.1016/j.pestbp.2003.07.004
Yu, Q., Powles, S. B. (2014). Resistance to AHAS inhibitor herbicides: current understanding. Pest Manage Sci. 70, 1340–1350. doi: 10.1002/ps.3710
Zhang, Z., Gu, T., Zhao, B., Yang, X., Peng, Q., Li, Y., et al. (2017). Effects of common Echinochloa varieties on grain yield and grain quality of rice. Field Crop Res. 203, 163–172. doi: 10.1016/j.fcr.2016.12.003
Zhao, N., Yan, Y., Liu, W., Wang, J. (2022). Cytochrome P450 CYP709C56 metabolizing mesosulfuron-methyl confers herbicide resistance in Alopecurus aequalis. Cell Mol. Life Sci. 79, 205. doi: 10.1007/s00018-022-04171-y
Keywords: Echinochloa crus-galli, mutation, penoxsulam, molecular docking, resistance
Citation: Sun P, Niu L, He P, Yu H, Chen J, Cui H and Li X (2024) Trp-574-Leu and the novel Pro-197-His/Leu mutations contribute to penoxsulam resistance in Echinochloa crus-galli (L.) P. Beauv.. Front. Plant Sci. 15:1488976. doi: 10.3389/fpls.2024.1488976
Received: 31 August 2024; Accepted: 04 November 2024;
Published: 25 November 2024.
Edited by:
Muthusamy Ramakrishnan, Nanjing Forestry University, ChinaReviewed by:
Bhabesh Borphukan, Washington State University, United StatesDonald James, Kerala Forest Research Institute, India
Copyright © 2024 Sun, Niu, He, Yu, Chen, Cui and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangju Li, eGpsaUBpcHBjYWFzLmNu
 Penglei Sun
Penglei Sun Liangliang Niu
Liangliang Niu Haiyan Yu
Haiyan Yu Jingchao Chen
Jingchao Chen Hailan Cui
Hailan Cui