
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 21 January 2025
Sec. Plant Bioinformatics
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1485757
Introduction: Licorice stands out as an exceptional medicinal resource with a long history of application, attributed to its substantial pharmacological potential. The basic helix-loop-helix (bHLH) transcription factors (TFs) gene family, being the second-largest in plants, is vital for plant development and adapting to environmental shifts. Despite this, the comprehensive characteristics of licorice bHLH gene family are not well-documented.
Results: In this study, a detailed and thorough genome-wide identification and expression analysis of Glycyrrhiza uralensis bHLH gene family was carried out, resulting in the identification of 139 licorice bHLH members. Our duplication analysis highlighted the significant contribution of segmental duplications to the expansion of G. uralensis bHLH genes, with GubHLH genes experiencing negative selection throughout evolution. It was discovered that GubHLH64 and GubHLH38 could be importantly linked to the licorice trichome initiation and anthocyanin biosynthesis and GubHLH64 was also involved in the abiotic stress response. Additionally, certain subfamily III (d+e) GubHLH members could be implicated in the licorice drought response. GubHLH108, GubHLH109, and GubHLH116 were suggested to form a tightly related cluster, initiating transcriptional responses via JA signaling pathway.
Discussion: In summary, our findings furnish a foundational understanding for future investigations of GubHLH gene functions and regulation mechanisms, shedding light on the potential applications of licorice in medicine and agriculture.
Glycyrrhiza uralensis Fisch. (G. uralensis), which belongs to the Leguminosae family, is a perennial herb naturally thriving in sandy or saline soil within arid and semiarid regions of North China, Northeast China, Northwest China, and several other countries in Central Asia and Europe (Ding et al., 2022). With its history spanning more than 2000 years, G. uralensis has been a key component in traditional Chinese medicine. Initially documented in “Shen Nong’s Materia Medica”, this herb has found applications in 29.87% of preparations listed in the Pharmacopoeia of the People’s Republic of China (2020 edition), showcasing its significance (Jiang et al., 2020; Ding et al., 2022). It has been demonstrated to possess capabilities in invigoration, detoxification, analgesia, relieving coughs and harmonizing the effects of other medicines in traditional medicine (Li T. et al., 2020; Yang et al., 2015). Modern pharmacological studies have found that various active components, mainly flavonoids and triterpenoids, contribute to diverse pharmacological benefits in licorice (Hosseinzadeh and Nassiri-Asl, 2015). Licorice compounds have also shown promise as inhibitors of COVID-19 (Kim et al., 2023). Beyond medicinal applications, licorice and its extracts have found use in industries like food, tobacco, and household chemicals (Kao et al., 2014).
Acting as the central switches of all cellular regulation networks, TFs can bind the proper DNA elements and recruit additional proteins to perform transcription transactions, in turn, modulating gene expression and numerous biological processes involving plant growth and environment adaptation (Strader et al., 2022). Among TFs, the bHLH family comprises the second-largest plant gene family following the MYB gene family, playing pivotal roles (Feller et al., 2011). The first bHLH motif, with the capacity of dimerization and DNA binding, was identified in murine E12 and E47 proteins (Murre et al., 1989). The plant bHLH protein was originally found to be engaged in the maize anthocyanin synthesis (Ludwig et al., 1989).
The bHLH TFs derive their name from the structural characteristics of their conserved bHLH domain, approximately 50-60 amino acid residues in length, which is composed of two linked subregions. The N-terminal basic region with the length of 15-20 amino acids serves as a DNA-binding domain being in charge of recognization and binding of TFs to appropriate DNA sequences (Atchley et al., 1999). And they have been shown to recognize the hexanucleotide cis-regulatory element called E-box (5′-CANNTG-3′) in target gene promoter regions through certain conserved residues within the basic region, with G-box (5’-CACGTG-3′) being the commonest form (Li et al., 2006). The other one is the C-terminal HLH region with variable length of 40-50 amino acids, which form two α-helices with a variable length intervening loop by hydrophobic residues, facilitating the homodimers or heterodimers formation as well as the regulation of target genes (Murre et al., 1989; Nair and Burley, 2000). Whereas sequences of bHLH TFs outside the conserved domain represent considerable diversity (Atchley et al., 1999). The bHLH TFs in metazoans were reported to be classified into 6 main groups (A-F) according to their DNA contact motifs, evolutionary relationships, the preferent DNA binding sequence and function properties (Simionato et al., 2007). In plants, the majority (more than 50%) of bHLHs evolved from group B, and a few of them (8%) own the group A characteristics, while group C, E, and F characteristics are absent in plant bHLHs. There are also some plant proteins (11%) absent in animals (Pires and Dolan, 2010). Owing to the large variation, the plants bHLHs cannot be classified clearly complying with the classification in animals. In current studies, plant bHLHs are usually classified based on the results of Pires and Dolan (2010) and Carretero-Paulet et al. (2010), in which bHLHs were clustered into 26 and 32 subfamilies, respectively.
Plant bHLH TFs have been shown to be responsible for heterogeneous growth and metabolic processes, including seed germination (Oh et al., 2006), flowering (Kumar et al., 2012), stomatal fate (Raissig et al., 2016) and the development of awn and grain (Luo et al., 2013). It has also been widely reported that bHLH TFs are associated with the trichome development. The bHLH TF CsGL3 in tea is involved in the trichome regulation through forming complex with MYB TF CsMYB1 and CsWD40 (Li et al., 2022). A bHLH TF in Artemisia annua, AaMYC2-type, was found to play an important part in regulating trichome development (Khan et al., 2024). Tomato SlMYC1 plays a vital role in type VI glandular trichomes formation (Xu et al., 2018). Meanwhile, previous studies have highlighted the engagement of bHLH TFs in secondary metabolites biosynthesis such as artemisinin (Yuan et al., 2023) and triterpene saponin (Mertens et al., 2016). In G. uralensis, the GubHLH3 was reported to play a role in soyasaponin biosynthesis, which is a kind of triterpenoids (Tamura et al., 2018). Furthermore, since the first plant bHLH protein was identified to regulate anthocyanin synthesis in maize, a growing comprehension of bHLH participating in anthocyanin production regulation has been reported. The apple MdbHLH3 was validated to bind to anthocyanin biosynthesis related genes promoters, leading to enhanced anthocyanin content and fruit coloration (Xie et al., 2012). The mulberry bHLH3 is an essential regulator in anthocyanin biosynthesis and flavonoid homeostasis, thus affecting the fruit coloration (Li H. et al., 2020). The bHLH TF VvMYC1 was found to take part in the transcriptional cascade regulating the anthocyanin accumulation in Vitis vinifera (Hichri et al., 2010). The bHLH proteins TT8 (Transparent Testa8, AtbHLH42), GL3 (Glabra3, AtbHLH1) and EGL3 (Enhancer of Glabra3, AtbHLH2) are critical for trichome formation and anthocyanin biosynthesis in Arabidopsis thaliana (Qi et al., 2011). And the AtGL3 homologue in Brassica napus, BnGL3-1 owns the conserved functions as AtGL3, promoting anthocyanin production and flowers trichome formation (Gao et al., 2018). The bHLH2 in Ipomoea purpurea modulates seed trichome formation, proanthocyanidin and phytomelanins for pigmentation in seeds, as well as anthocyanin production (Park et al., 2007). FhGL3L and FhTT8L, two IIIf subfamily bHLHs from Freesia hybrida, were confirmed to partake in regulating the trichome patterning and anthocyanin accumulation (Li Y. et al., 2016). Moreover, the bHLHs have been found to exert functions in the regulation of environmental and abiotic responses. Overexpression of AtbHLH68 can enhance adaptation to drought via ABA signaling in Arabidopsis (Le Hir et al., 2017). TabHLH49 in wheat can improve drought tolerance via regulating the dehydrin gene (Liu et al., 2020). TabHLH27-A1 was identified to balance drought tolerance and growth, enhancing wheat water use efficiency (Wang et al., 2024). Overexpressed ZmPTF1 in maize improve the drought resistance, along with the enhanced yield and abscisic acid (ABA) synthesis (Li et al., 2019). OsbHLH062 in rice controls the transcription of target ion transporters in response to JA treatment, thereby altering the salt tolerance (Wu et al., 2015). The CabHLH035 in Capsicum annuum can increase salt tolerance by lowering intracellular Na+/K+ ratio and enhancing proline biosynthesis (Zhang et al., 2022). AhbHLH121 can improve salt tolerance by improving the antioxidant enzyme activity in peanut (Zhao et al., 2024). FIT (bHLH29, FER-like Iron Deficiency Induced Transcription Factor), a bHLH member from IIIa subfamily, interacts with bHLH38, bHLH39, bHLH100 and bHLH101, playing a core role in Fe deficiency response and homeostasis (Wang et al., 2013).
In the wake of the accomplishment of whole-genome sequencing and assembly for various plant species, bHLH proteins in multitudinous plants have been characterized. There were 162 bHLH members in Arabidopsis (Bailey et al., 2003), 251 in Brassica rapa (Miao et al., 2020), 440 in Brassica napus, 268 in Brassica oleracea, 208 in maize (Zhang et al., 2018), 167 in rice (Li et al., 2006), 437 in cotton (Lu et al., 2018) and 319 in soybean identified (Hudson and Hudson, 2015). A comprehensive genome-wide identification of G. uralensis bHLH gene family has not been explored. This study systematically analyzed the G. uralensis bHLH TF gene family, identifying 139 GubHLH members. And their physicochemical properties, evolution relationship and expression patterns were further investigated, providing a meaningful understanding for the characteristics and functions of the G. uralensis bHLH gene family.
The whole-genome sequence of G. uralensis was assembled by Wuhan Benagen Technology Co., Ltd. (Wuhan, China). The bHLH domain (PF00010) hidden Markov model (HMM) was downloaded from the Pfam database (http://pfam.xfam.org/, accessed on 18 April 2023) and employed to search against the local G. uralensis protein database through HMMER v3.2 (http://hmmer.janelia.org/) software with the e-value of 10-5. Furthermore, the protein sequences of Arabidopsis bHLHs members were acquired from the Arabidopsis resource database (https://www.arabidopsis.org/, accessed on 18 April 2023) and used as the queries for a local BLASTP (blast v2.9.0) search against the G. uralensis protein database. Then the hits from two ways of search methods were combined and redundant genes were eliminated to create the GubHLH candidates. Then the NCBI Conserved Domain Database (CDD, https://www.ncbi.nlm.nih.gov/cdd/, accessed on 19 April 2023) and the SMART (http://smart.embl-heidelberg.de/, accessed on 19 April 2023) were utilized to verify the candidates with the bHLH domain. Subcellular localization predictions were performed using Plant-mPLoc program in Cell-PLoc 2.0 (Chou and Shen, 2008), and their physicochemical parameters were predicted using the ExPASy ProtParam tool (Gasteiger et al., 2003).
The protein sequences of G. uralensis bHLHs and Arabidopsis bHLHs were combined and a multiple sequence alignment was generated using ClustalX v1.83 software (Thompson et al., 1997). A phylogenetic tree on the basis of the alignment was constructed by the MEGA7.0 employing the Neighbor-Joining (NJ) method. The parameter bootstrap was set as 1000 and amino acid substitution model was Jones-Taylor-Thornton (JTT) model (Kumar et al., 2016). The phylogenetic tree was then edited and visualized by iTol v5 (https://itol.embl.de/) (Letunic and Bork, 2021). The conserved bHLH domains sequences of GubHLH proteins were aligned and edited through Jalview v2.11.2.0 software (Waterhouse et al., 2009). The GubHLH domains logos were generated using TBtools v1.09876 (Chen et al., 2020). Tertiary structures of GubHLH bHLH domain were predicted through homology modeling on the SWISS-MODEL website (https://www.swissmodel.expasy.org/) website and visualized by PyMOL v2.4.0 (Seeliger and de Groot, 2010).
Information regarding the intron-exon distribution of G. uralensis bHLH genes was extracted from the G. uralensis GFF annotation file. The conservative motifs in GubHLH proteins were identified utilizing the MEME online program v5.5.2 (Bailey et al., 2015). The motif number was 10 with the width from 6 to 50 residues. The analysis of conserved domains in G. uralensis bHLH proteins was conducted via the CDD. These results were visualized by TBtools v1.09876 (Chen et al., 2020). The GubHLH proteins secondary structures were analyzed through SOPMA server (Geourjon and Deléage, 1995).
The physical location information of GubHLH genes on G. uralensis chromosomes was retrieved from the GFF file and visualized by TBtools. Duplication events analysis of GubHLHs was performed based on their amino acid sequences and chromosome locations utilizing Multicollinearity Scanning Toolkit (MCscanX) software (Wang et al., 2012), with visualizations and editing accomplished using Advance Circos program within TBtools v1.09876. Arabidopsis genomic data were accessed from the TAIR10 website (http://www.arabidopsis.org/index.jsp). Rice, maize, M. truncatula and Glycine max genomic data were downloaded from the JGI website (https://phytozome-next.jgi.doe.gov/). Collinearity analysis of licorice with these five species was performed by the One Step MCScanx program and visualized through TBtools Dual Systeny Plotter program in TBtools v1.09876. The synonymous substitution rate (Ks), nonsynonymous substitution rate (Ka), and Ka/Ks ratio between homologous GubHLH gene pairs were calculated applying KaKs_Calculator v3.0 through the NG method (Zhang, 2022).
The 2000 bp upstream sequences of GubHLH coding sequences were retrieved using TBtools, relying on the GubHLHs DNA sequences, which were then submitted to the PlantCare database for the prediction of cis-elements in the GubHLHs promoters (accessed on 17 June 2023) (Lescot et al., 2002).
The protein sequences of GubHLHs were submitted to KOBAS3.0 (accessed on 20 June 2023) (Bu et al., 2021) and the KEGG database (https://www.kegg.jp/, accessed on 20 June 2023) to acquire the Gene Ontology (GO) terms and KEGG pathway annotations, respectively. Additionally, protein-protein interaction (PPI) analysis was conducted on the STRING website using the orthologs in Arabidopsis as references (http://string-db.org, accessed on 26 September 2023) (Szklarczyk et al., 2023).
The coding sequence of GubHLHs without termination codons were cloned into pCambia1300-GFP vector to construct the recombinant vectors, which was then transformed into Agrobacterium strain GV3101 and infiltrated into Nicotiana benthamiana leaves along with the nuclear marker (GhBES1-mCherry) strain. The empty pCambia1300-GFP vector was served as the control. The fluorescence information was obtained through laser confocal microscopy (Leica TCSSP8, Wetzlar, Germany).
The GuMYB75 and GuTTG1 were obtained as the orthologs of AtMYB75 and AtTTG1 through the BLASTP search against the G. uralensis genome database using AtMYB75 and AtTTG1 as queries, respectively. And the genes with the highest bitscores were chosen. The full-length CDSs of GubHLH64, GubHLH38 and GuMYB75 were individually incorporated into the pGADT7 vectors as bait construct. Meanwhile, GuMYB75 and GuTTG1 were cloned into pGBKT7 vectors as prey construct. The bait and prey vectors were then co-transformed into the Y2HGold yeast strain and selected on synthetic dropout (SD) medium lacking leucine (-L) and tryptophan (-T). The transformed yeast cells on the SD-T-L mediums were isolated and dotted on SD-T-L-H (histidine) and SD-T-L-H-A (adenine) medium. The primers were listed in Supplementary Table S1.
The seeds of licorice underwent dormancy breakage through a 50-minute treatment with 98% concentrated H2SO4, followed by three washes with sterilized distilled water. Subsequently, the treated seeds were germinated and cultured in plastic pots filled with nutrient soil and vermiculite mixed in the proportion of 2:1 (v:v) within an automatic climate chamber maintaining steady conditions of 16 h/28°C at the day and 8 h/25°C during night as well as the relative humidity of 50-55%. After 60 days of cultivation, the seedlings were transferred to Hoagland solution medium containing 10% PEG6000 and 150 mM NaCl for abiotic treatment. Licorice roots were collected at different time points after NaCl and drought treatment (0 h, 2 h, 6 h, and 12 h). Simultaneously, 60-day-old seedlings were subjected to treatments with 50 mM ABA, 100 µM auxin (IAA), 100 µM gibberellin (GA), and 100 µM methyl jasmonate (MeJA) in the Hoagland solution medium for varying durations (0 h, 2 h, 6 h, and 12 h). Three biological replicates were collected for each experimental condition, with each replicate comprising roots from 15 seedlings, which were then promptly frozen in liquid nitrogen and stored at -80°C.
The RNA-seq raw data of G. uralensis roots under drought treatment were retrieved from the National Center for Biotechnology Information (NCBI) database (BioProject number: PRJNA810509) (Yao et al., 2022) and utilized to assess the expression levels of G. uralensis bHLH genes. After trimming and quality control by fastp v0.19.5 (Chen et al., 2018), the clean data were mapped to the G. uralensis genome using the HISAT2 software v2.1.0 (Kim et al., 2019). And RSEM software v1.3.3 (Li and Dewey, 2011) was used to obtain the gene read counts and normalize the expression to transcripts per million (TPM) values. DESeq2 (v1.10.0) (Love et al., 2014) was used to perform the differential expression analysis with the criteria |log2 (Fold change)| > 1.0 and p values < 0.05. Total RNA from G. uralensis roots were extracted using the RNAprep Pure Plant kit (TIANGEN BIOTECH, Beijing, China) which were applied to synthesize the first strand cDNAs utilizing the EasyScript One-step gDNA Removal and cDNA Synthesis SuperMix (Vazyme, Nanjing, China) following the manufacturer’s instructions. Primer Premier 5.0 was applied to design the qRT-PCR primers which were listed in Supplementary Table S2. The qRT-PCR analysis were conducted on a Bio-RAD CFX96 Real-Time system (Hercules, CA, USA) using the 2×ChamQ SYBR qPCR Master Mix (Vazyme, Nanjing, China). The expression levels of GubHLHs were calculated using 2−ΔΔCt method with Guactin (NCBI accession number: EU190972.1) as an internal control.
Through searches against the G. uralensis whole-genome protein database based on the hidden Markov model called bHLH domain (PF00010) and the queries AtbHLH protein sequences as well as the subsequent bHLH domain integrality verification for the combined candidate GubHLH members, we obtained 139 bHLH gene family members in G. uralensis genome (Supplementary Table S3). They were then designated in the light of their positional order on G. uralensis chromosomes as GubHLH1-GubHLH139, and their sequences were list in Supplementary Table S4. Furthermore, the physicochemical properties of GubHLH proteins were analyzed. These GubHLH proteins varied from 73 (GubHLH69) to 777 (GubHLH10) amino acids in length, with molecular weights ranging from 8.29 kDa (GubHLH69) to 85.16 kDa (GubHLH61) and an average of 41.19 kDa. The theoretical isoelectric points (pI) of GubHLH proteins spanned from 4.55 (GubHLH49) to 10.29 (GubHLH87). The negative grand average of hydropathicity (GRAVY) values, distributing from -1.068 to -0.012, highlighted their hydrophilicity and solubility features which may correlate with their underlying function as TFs. The instability index predictions suggested that nearly all GubHLH proteins were likely unstable in vitro, with an instability index higher than 40, excluding GubHLH131, GubHLH127, and GubHLH59. And the aliphatic index values of GubHLH proteins were between 49.33 (GubHLH42) and 103.91 (GubHLH119), indicating a substantial variation in thermal stability. Moreover, the subcellular localization predictions showed that 138 out of the 139 GubHLH proteins were located in the nucleus, with GubHLH56 being found in both Golgi apparatus and nucleus (Supplementary Table S3).
A multiple alignment analysis amino acid sequences in the bHLH domain of GubHLH proteins was performed to explore their features (Supplementary Figure S1). The analysis revealed that G. uralensis bHLH family proteins contained the typical conserved bHLH domain, and four conserved regions were presented within these domains, including one basic region, the first helix, the loop and the second helix region (Figures 1A, B). Within these bHLH domains, 23 conserved amino acids with the consensus ratio exceeding 50% existed and 7 out of them (Glu-9, Arg-10, Arg-12, Arg-13, Leu-23, Pro-28, Leu-58) exhibited a consensus ratio surpassing 75% (Figures 1B, C). The basic region being crucial for DNA-binding activity to target genes featured six conserved residues (His-5, Ala-8, Glu-9, Arg-10, Arg-12, and Arg-13) in GubHLH proteins. The first helix region contained 5 conserved amino acid residues (Ile-16, Asn-17, Leu-23, Leu-26, and Pro-28). The loop region exhibited conservation at Lys-41 and Asp-43. Ten residues (Ala-45, Ser-46, Leu-48, Ala-51, Ile-52, Tyr-54, Lys-56, Leu-58, Gln-59, and Leu-65) were identified to be conserved in the second helix region. Notably, the residues Leu-23 and Pro-28 showed particular high conservation within the 139 G. uralensis bHLH proteins, emphasizing their notability in the formation of bHLH proteins dimers.
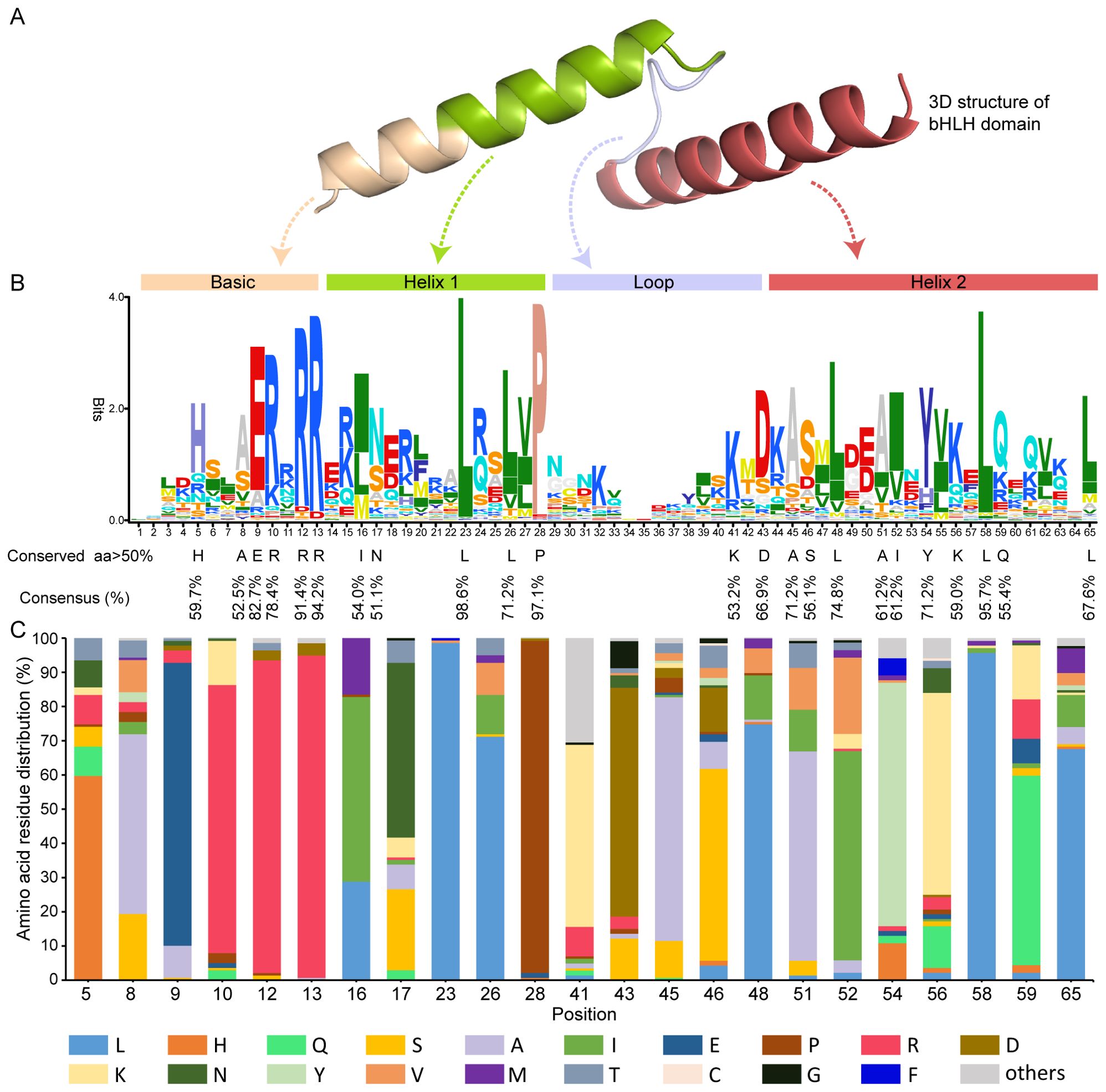
Figure 1. The bHLH domain sequences characterizations in the GubHLHs. (A) Three-dimensional structure of the GubHLH12 bHLH domain. (B) Sequence logo of conserved bHLH domain of GubHLHs. The amino acid height indicated the frequency of the sequence observed in GubHLH proteins at that site. The conservation sites with a consensus ratio exceeding 50% in the bHLH domain were highlighted by the black letters below. (C) The amino acids distribution at the 23 sites with a consensus ratio exceeding 50% in GubHLH bHLH domains. The numerals at the bottom represent the amino acid residues positions.
To classify and examine the phylogenetic relationships among GubHLH members, a Neighbor-Joining (NJ) phylogenetic tree was constructed using the protein sequences of 139 bHLH proteins from G. uralensis and 144 from A. thaliana. As depicted in Figure 2, all bHLH proteins were organized into 26 subfamilies based on the tree’s topological structure and a classification method recommended by previous studies (Heim et al., 2003; Pires and Dolan, 2010), which include Ia, Ib (1), Ib (2), II, III (a+c), IIIb, III (d+e), IIIf, IVa, IVb, IVc, IVd, Va, Vb, VI, VII (a+b), VIIIa, VIIIb, VIIIc, IX, X, XI, XII, XIII, XIV and XV. And GubHLHs were distributed across 25 subfamilies, and notably, they did not cluster together with AtbHLHs in subfamily X. Among them, subfamily IVa contained the highest number with 16 GubHLHs, followed by subfamilies VII (a+b) and XII, each with 11 GubHLHs. While the subfamilies Ib (1), IVb, VI, VIIIa, and XIV contained only one GubHLH each, making them the smallest subfamilies (Figure 2; Supplementary Figure S2). Within most subfamilies, comparable numbers of bHLH proteins were distributed in Arabidopsis and G. uralensis. There are also varied abundance of bHLH protein between two species within some subfamilies, which might be attributed to the gene expansion or loss during evolution. Subfamily IVa and Vb contained four and five AtbHLH proteins, and the numbers of GubHLH proteins increased to sixteen and ten, respectively, indicating the possible expansion of the GubHLH proteins in IVa and Vb subfamily. The absence of subfamily X in G. uralensis might indicate the loss or undifferentiation of GubHLH proteins throughout evolution. And these results suggest that the bHLH members in IVa and Vb might have species-special functions in G. uralensis and the function of bHLHs in subfamily X might be replaced by other bHLHs.
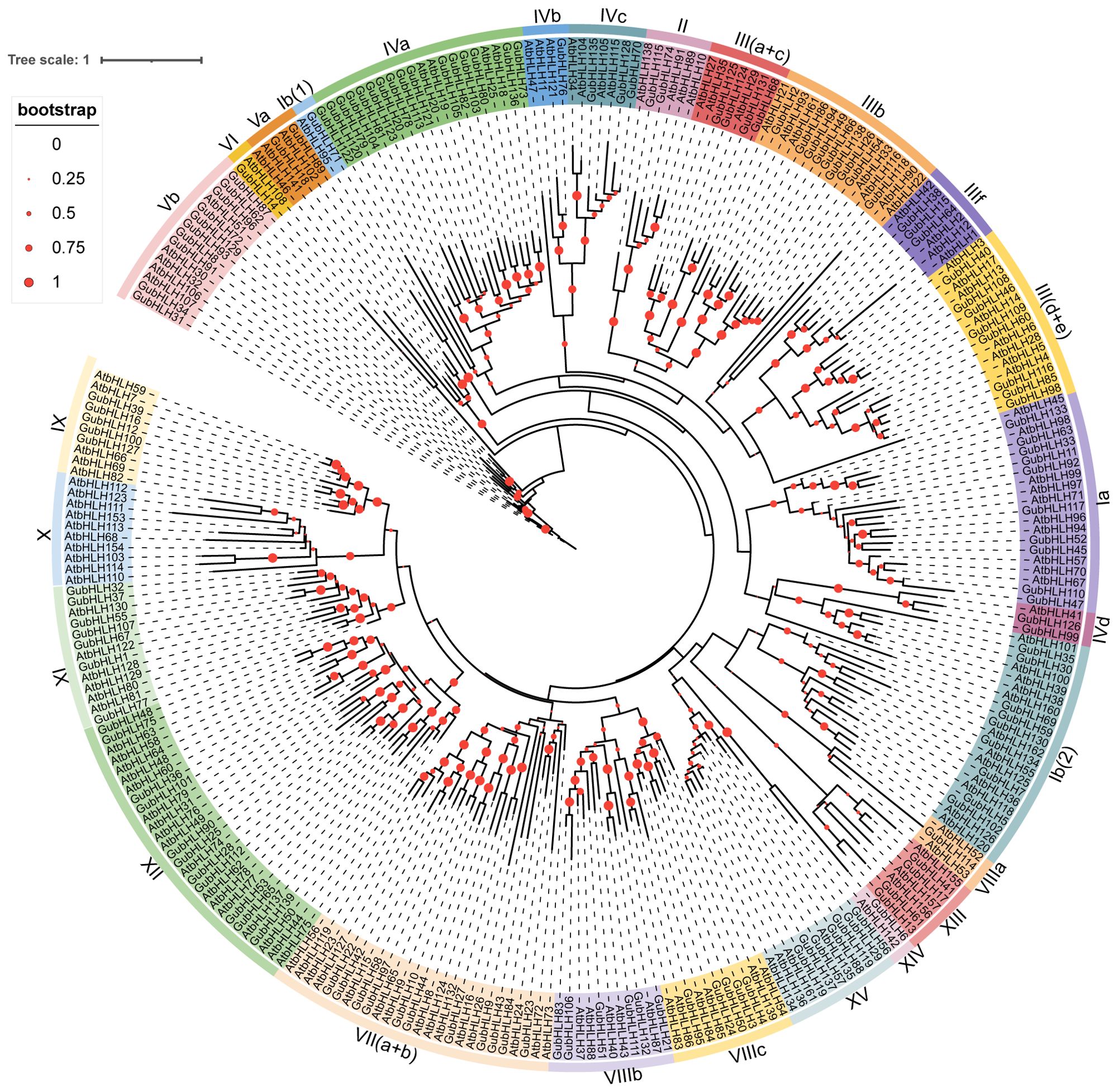
Figure 2. The phylogenetic tree of bHLH family members in G. uralensis and A. thaliana. Clustalx and MEGA 7 were used to generated the tree through NJ method with 1,000 bootstrap replicates. The bHLH proteins were clustered into different subgroups according to the priority classification rule of AtbHLHs, which was presented by differently colored branches.
A phylogenetic tree was constructed using the 139 GubHLH protein sequences (Figure 3A), in which the classification of GubHLHs aligned with the results in Figure 2. To broaden the understanding of structural features of GubHLH proteins, the composition of conserved motifs, distribution of domains and exons/introns of GubHLHs were analyzed (Figures 3B-D). MEME results revealed 10 conserved motifs within the GubHLHs. The motifs sequences were submitted to the Pfam and InterProScan databases for annotation. Motifs 1 and 2 were annotated to the bHLH domain, and motifs 7 and 9 were bHLH-MYC and R2R3-MYB transcription factor N-termini (Supplementary Table S5). All GubHLH proteins contained bHLH domain related motifs. Motifs 7 and 9 were distributed in subfamilies III (d+e) and IIIf. GubHLH2, GubHLH118 and all GubHLHs in subfamilies XIII and IVd contained motif 9. The distribution of these motifs was generally conserved within subfamilies, indicating their potential functional similarities.
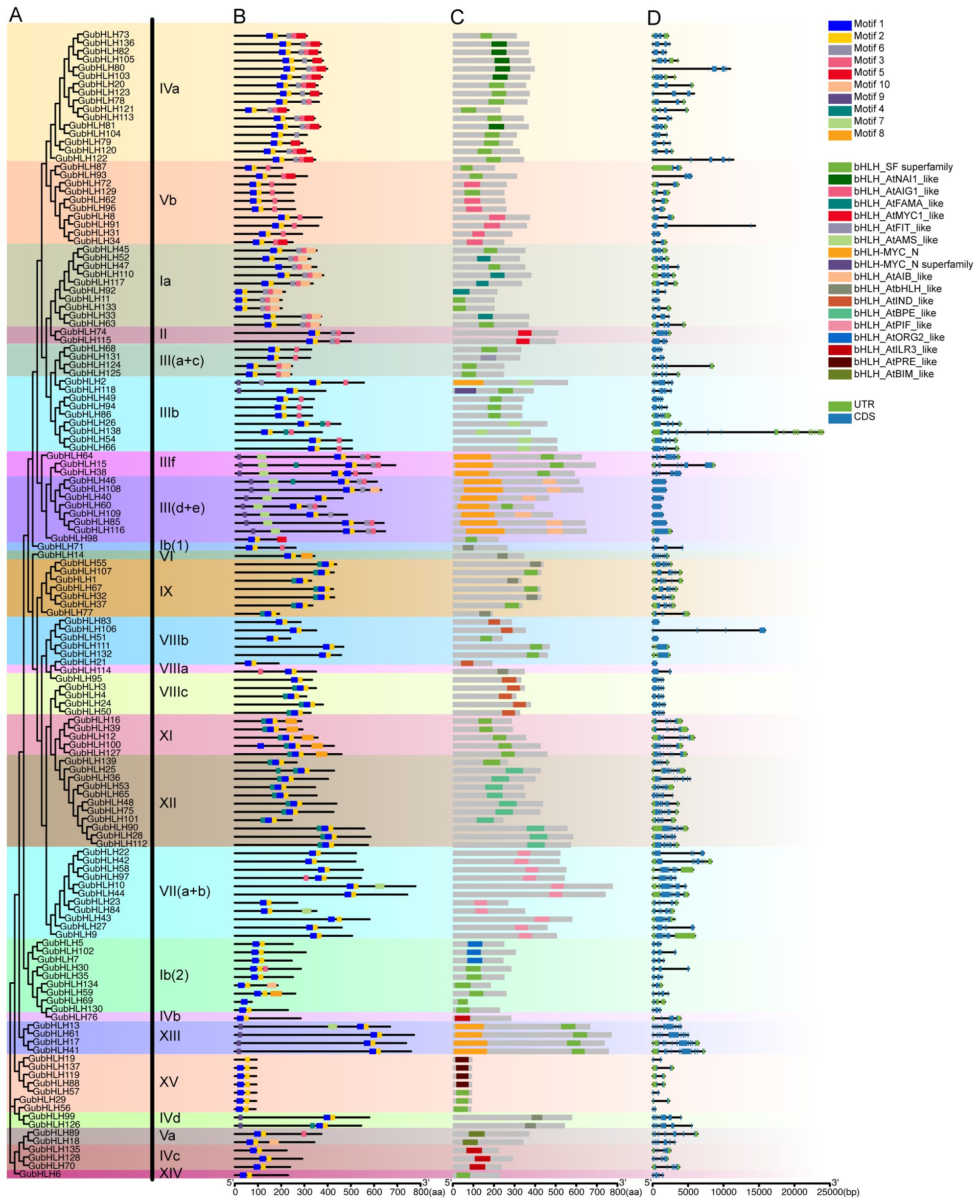
Figure 3. The phylogenetic tree, conserved motifs distribution, domains composition and gene structure of GubHLHs. (A) The neighbor-joining (NJ) phylogenetic tree of GubHLH proteins was generated using protein sequences with 1000 bootstrap replicates; (B) The distribution of conserved motifs in GubHLH proteins. (C) The distribution of bHLH domains in GubHLHs; (D) The gene structures of the GubHLHs, in which green rectangles indicated the untranslated regions (UTRs), blue rectangles indicated exons and black lines indicated the introns.
Meanwhile, the analysis of conserved domains using the CDD identified 18 types of bHLH domains in GubHLHs, highlighting their extensive diversity. Similar bHLH domain types were found within the same subfamily, corroborating the subfamily classification evidently in the phylogenetic tree (Figure 3C). GubHLH2, GubHLH118 and the GubHLH proteins in subfamilies III (d+e), IIIf and XIII were each characterized by the presence of a bHLH-MYC_N domain, strongly supporting the motif distribution in MEME results.
The exon-intron structures across GubHLH genes disclosed substantial variations. The subfamily XIII members had the highest exon count, featuring eleven exons in GubHLH13/17/41 and ten in GubHLH61. Conversely, most members of subfamily III (d+e) and VIIIb each contained only one exon. The majority members in subfamily III (d+e) had no introns. Except for GubHLH138 possessing 19 introns in subfamily IIIb, the subfamily XIII GubHLHs contained the greatest number of introns, ranging from 9 to 11. GubHLH138 had the most number of UTRs, with one in 3’ UTR and 14 in 5’ UTR. The similar exons/introns distribution within subfamilies supported the notion that closely related evolutionary relationships are linked to similar gene structures.
Furthermore, the secondary protein structures of GubHLHs widely influencing protein sequence, structure, activity, stability, and abundance, were predicted and the results were exhibited in Supplementary Table S6. GubHLH proteins mainly consisted of alpha helix (16.44-67.39%), random coil (30.11-73.82%), extended strand (0-20.27%) and beta turn (0-6.46%). The VII (a+b) subfamily members owned the largest proportion of random coil (54.06-73.45%) and the minimum proportion of alpha helix (16.44-31.88%). While XV subfamily GubHLHs had the largest proportion of alpha helix (64.04-67.39%) and the minimum proportion of random coil (30.11-35.96%).
The genomic distribution of G. uralensis bHLH genes was visualized on chromosomes, revealing an uneven distribution across 8 chromosomes (Supplementary Figure S3). Among them, chr2 owned the biggest amount of GubHLHs with 44 genes (31.65%). Chr1, chr6, chr5, chr7, chr3, chr4 and chr8 harbored 22, 22, 18, 17, 16, 16 and 3 GubHLHs, respectively.
Segmental and tandem duplications occupy important positions in gene family evolution. In the case of GubHLHs, 61 gene duplication events were identified, comprising 55 segmental and 6 tandem duplications. This suggested that segmental duplications were primarily responsible for the GubHLH gene family’ evolution (Figure 4; Supplementary Table S7). The calculation of the Ka/Ks ratio for these gene pairs yielded values less than 1, ranging from 0.045 to 0.566 (Supplementary Table S8). These findings implied the purifying selection throughout GubHLHs’ evolution, aiding in the preservation of their conserved structures.
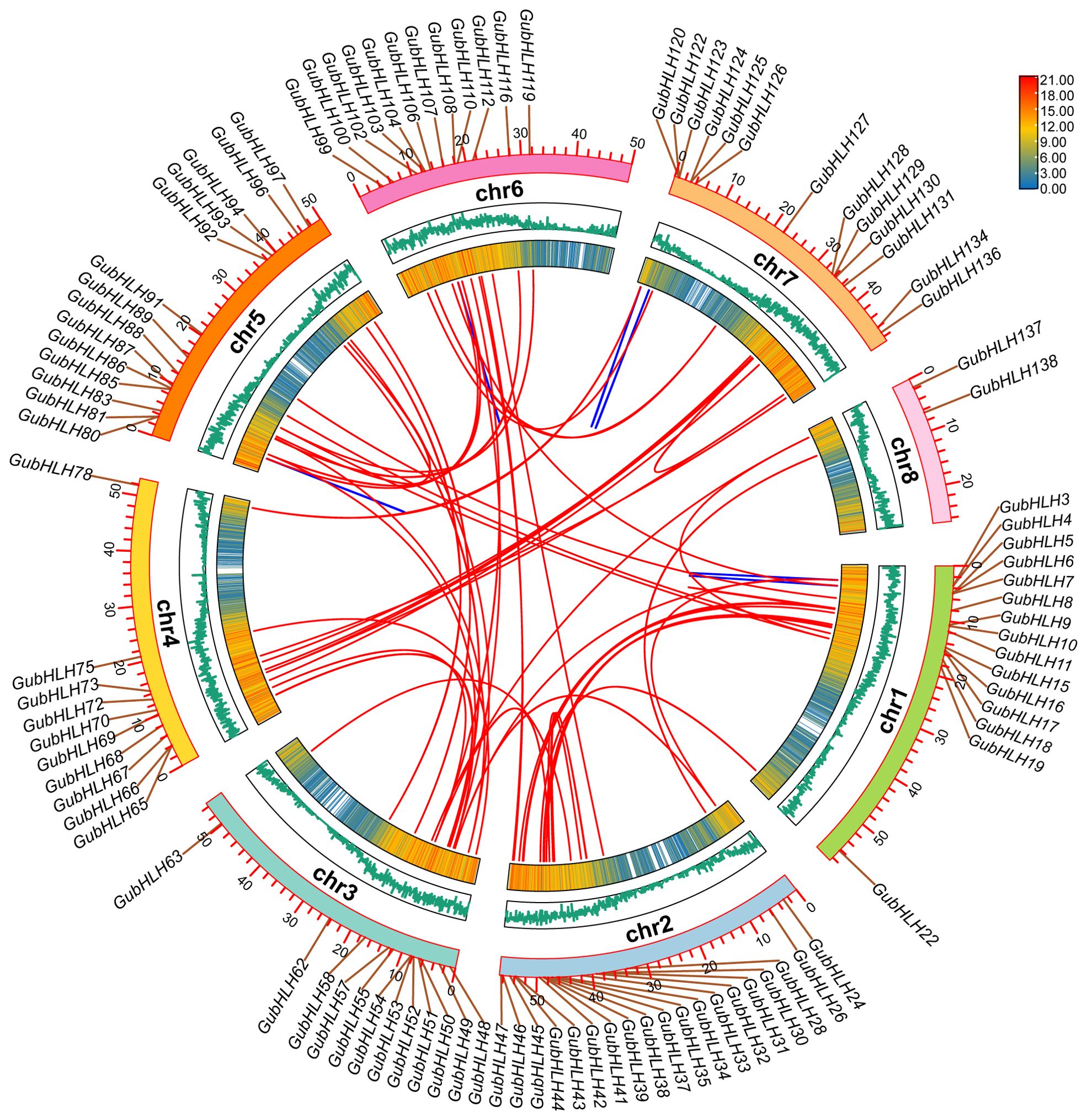
Figure 4. The collinearity of GubHLHs in G. uralensis genome. The blue lines represent the tandem duplication gene pairs and the red lines represent segmental duplication gene pairs relationship between GubHLHs.
For further exploration of the GubHLH genes duplication timing and the underlying phylogenetic mechanism, collinearity analysis between G. uralensis and 5 representative species, including 2 monocots (rice and maize) and 3 dicots (Arabidopsis, G. max and M. truncatula) was performed (Supplementary Figure S4). The results revealed that a total of 127 bHLH genes of G. uralensis were collinearly associated with those of G. max, followed by 120 GubHLH genes with M. truncatula, 91 with Arabidopsis, 44 with O. sativa, and 34 with Z. may (Supplementary Table S9). Compared to other plants, G. uralensis shared significantly larger number of collinear pairs with G. max and M. truncatula, which belong to Fabaceae family along with G. uralensis. Additionally, 24 GubHLHs exhibited collinear associations with all other five species, which might have existed before their ancestral divergence and play important roles in bHLH evolution (Supplementary Figure S5).
GO enrichment analysis was conducted to elucidate the potential functions of the GubHLHs, which were categorized into three main groups (Supplementary Figure S6A, Supplementary Table S10). In biological process, there were 109 GubHLHs enriched in regulation of biological process and biological regulation term, representing 78.42% of the GubHLHs. Metabolic process and cellular process were the second-largest terms, involving 107 genes each (76.98%). A total of 62 GubHLHs were enriched in single-organism process, 51 in response to stimulus, 48 in developmental process, 46 in multicellular organismal process, 27 in reproduction and reproductive process each, 18 in positive regulation of biological process, 17 in signaling, 12 in negative regulation of biological process and multi-organism process each, 7 in cellular component organization or biogenesis, 2 in growth and rhythmic process each. GubHLH106 was linked to localization. As for the molecular functions, almost all of the GubHLHs were enriched in binding except GubHLH14 and GubHLH22, followed by 133 GubHLHs in nucleic acid binding transcription factor activity and 2 GubHLHs in catalytic activity. In terms of cellular component, the majority GubHLHs were enriched in cell, organelle, and cell part, each with 138 members. And 138 out of 139 GubHLHs were enriched in nucleus, with the exception of GubHLH22. Seven GubHLHs were enriched in macromolecular complex and organelle part. GubHLH27 was enriched in cell junction and symplast.
Furthermore, KEGG enrichment analysis was performed (Supplementary Figure S6B). A total of 70 GubHLHs were enriched in the signal transduction pathway, 43 of which were engaged in MAPK signaling pathway and 64 of which were engaged in plant hormone signal transduction. Additionally, 42 GubHLHs were associated with environmental adaptation pathways, including 24 involved in plant-pathogen interaction and 18 involved in circadian rhythm (Supplementary Table S11).
In addition, a PPI network was built to explore the potential interactions among the GubHLH proteins (Figure 5A). A total of 137 GubHLH proteins had homologous proteins in Arabidopsis, in which 89 GubHLHs were predicted to have interactions. Several noteworthy interactions were predicted. For instance, GubHLH58/97 (homologous with PIF4, Phytochrome Interacting Factor4) were predicted to interact with GubHLH42 (homolog of PIF1/PIL5, PIF3-Like 5, PIF1/bHLH015), GubHLH10/44 (homolog of PIF3), and phytochromes (PHYA and PHYB). GubHLH26/54/66/138 (homologous with SCRM, SCREAM, also known as ICE1, Inducer of CBF Expression1) were predicted to collaborate with GubHLH133 (homolog of MUTE), GubHLH110 (homolog of FAMA), and GubHLH33/63 (homolog of SPCH, Speechless). The GubHLH106 (homolog of HEC1, HECATE1), GubHLH51/83 (homolog of HEC2), and GubHLH111 (homolog of HEC3) were predicted to interact with GubHLH23/84 (homolog of SPT, Spatula). GubHLH30 (homolog of AtbHLH100) was predicted to interact with GubHLH68/131 (homolog of FIT). GubHLH116 (homolog of MYC2), GubHLH108 (homolog of MYC3) and GubHLH109 (homolog of MYC4) were predicted to cluster into one functional module. The WD-repeat protein TTG1 (Transparent Testa Glabra1) was predicted to interact with TT8 (homolog of GubHLH38), GL3 (homolog of GubHLH64) and MYB75. TTG1, GL3, EGL3, and MYB75 have been reported to work in concert to modulate the accumulation of anthocyanin and initiation of trichome through JA signaling (Qi et al., 2011).
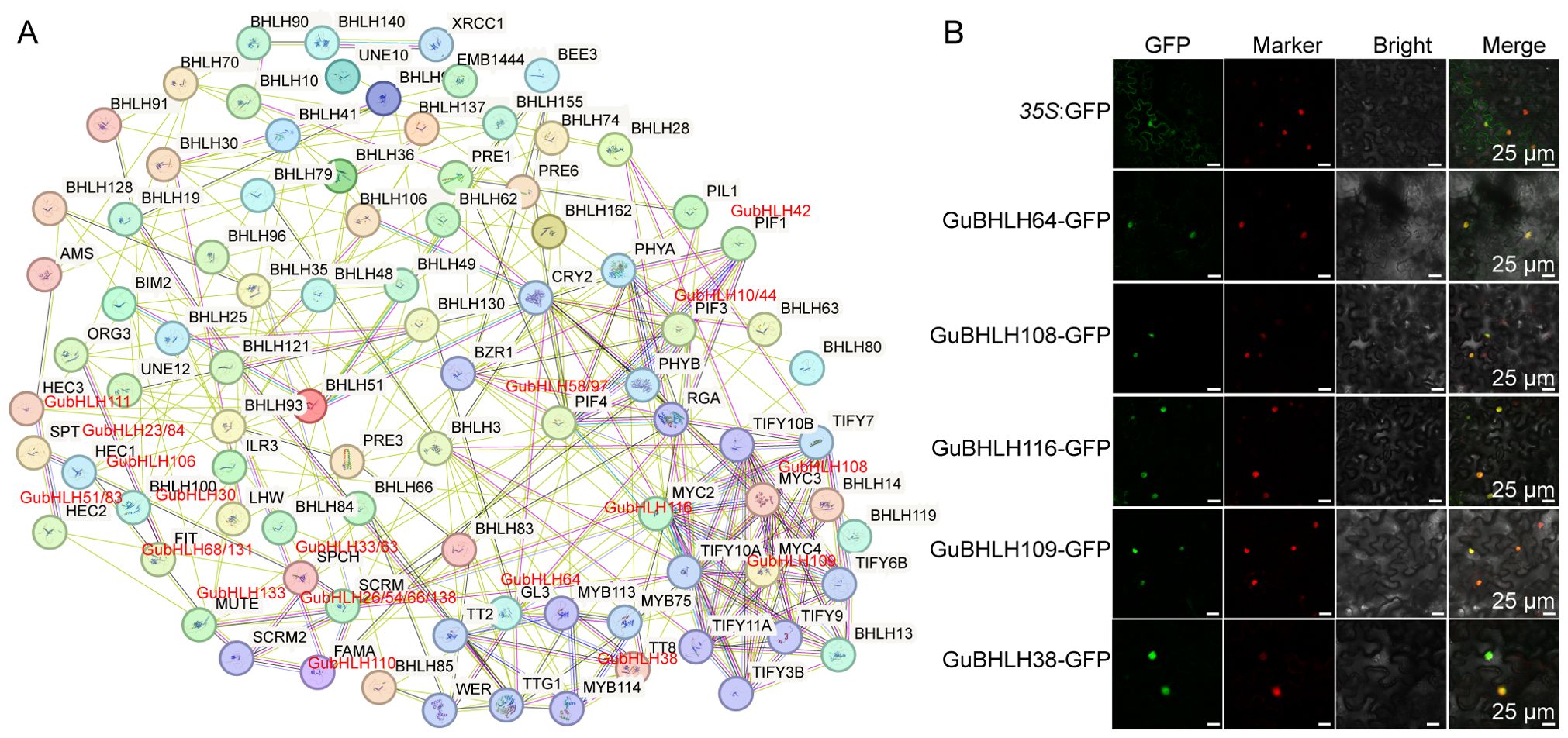
Figure 5. Protein-Protein interaction (PPI) network for GubHLHs and subcellular locations of GubHLH proteins. (A) PPI network for GubHLHs based on their orthologs in Arabidopsis. (B) Subcellular locations of GubHLH108, GubHLH109, GubHLH116, GubHLH64 and GubHLH38. The GhBES1-mCherry was used as the nuclear marker. The scale bars were 25 μm.
GubHLH64 interacted with GubHLH38, and GubHLH108, 109, 116 were clustered into one functional module according to the PPI network. In order to further explore their functions, subcellular localization assays of GubHLH38, 64, 108, 109, 116 were conducted using fluorescent reporter genes (GFP). The results showed that the fluorescence of 35S::GubHLH38-GFP, 35S::GubHLH64-GFP, 35S::GubHLH108-GFP, 35S::GubHLH109-GFP, and 35S::GubHLH116-GFP was present in the nucleus of N. benthamiana (Figure 5B), suggesting that GubHLH38, 64, 108, 109, 116 are specifically localized to the nucleus. In present PPI network, TTG1 interacted with GL3 (homolog of GubHLH64), TT8 (homolog of GubHLH38) and MYB75. The yeast two-hybrid assays were performed to elucidate the potential interactions between GuTTG1, GubHLH64, GubHLH38 and GuMYB75 in G. uralensis, which are homologs of TTG1, GL3, TT8 and MYB75, respectively. These assays revealed that GubHLH38, GubHLH64, and GuMYB75 directly interact with GuTTG1 (Figure 6A). Whereas GuMYB75 was found not to interact directly with GubHLH38 and GubHLH64 (Figure 6B).
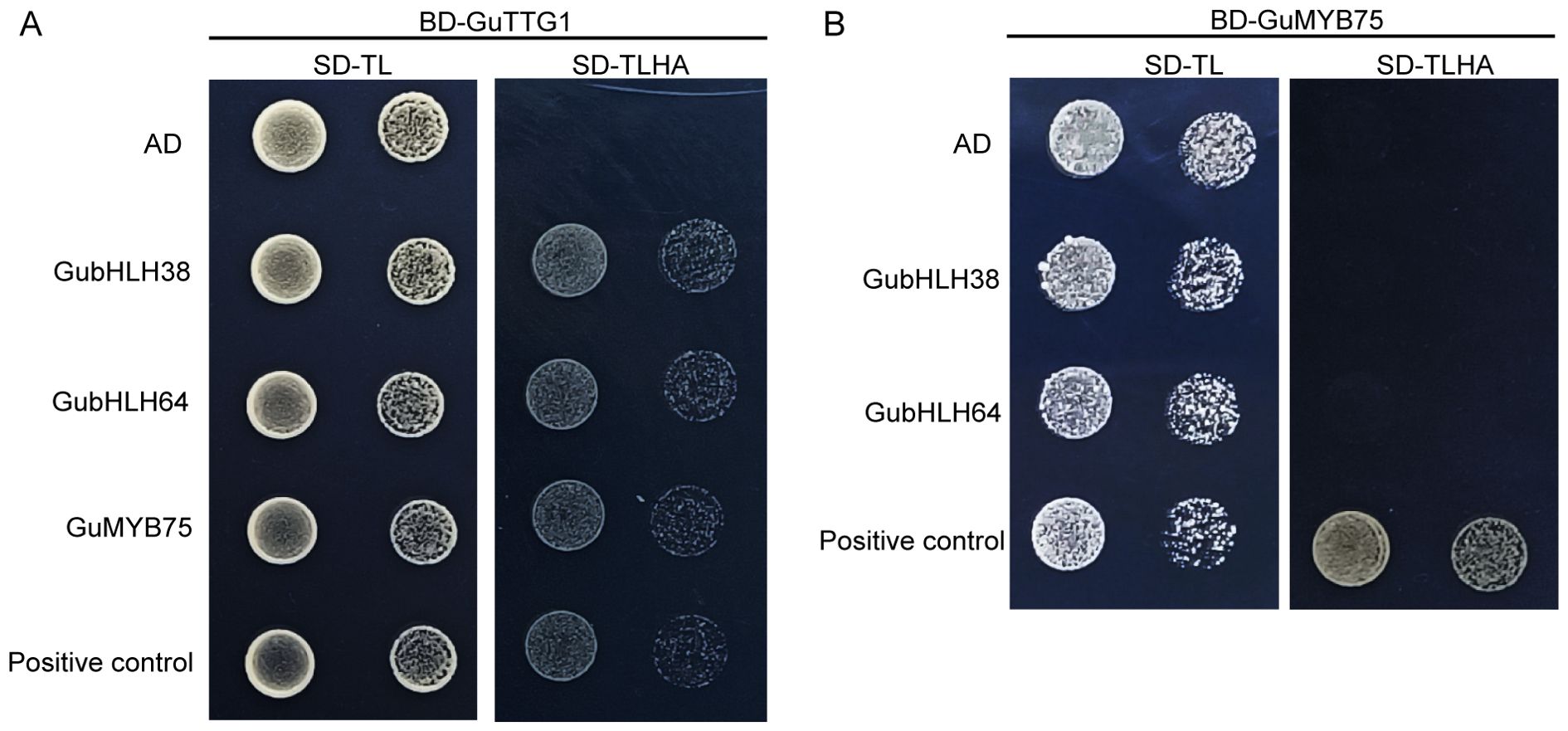
Figure 6. The Y2H assay of proteins in the anthocyanin biosynthesis and trichome formation associated network. (A) Interaction validation of GubHLH64, GubHLH38 and GuMYB75 with GuTTG1 via yeast two-hybrid assay. (B) Interaction validation of GubHLH38, GubHLH64 with GuMYB75 via yeast two-hybrid assay.
The expression profiles of GubHLHs under PEG treatment was analyzed according to the RNA-seq data (Figure 7A; Supplementary Table S12). GubHLH47, GubHLH78, GubHLH60, GubHLH126, GubHLH136, GubHLH80, GubHLH40, GubHLH46, GubHLH124, GubHLH103, GubHLH116, GubHLH64, GubHLH112, and GubHLH100 were obviously increased by PEG at 2 h. Conversely, GubHLH13, GubHLH32, GubHLH34, GubHLH123, and GubHLH74 were downregulated at 2 h. GubHLH47, GubHLH120, GubHLH9, GubHLH136, GubHLH73, GubHLH60, GubHLH56, GubHLH40, GubHLH100, GubHLH112, and GubHLH94 showed increased expression at 6 h. GubHLH9, GubHLH56, GubHLH73, GubHLH78, GubHLH64, GubHLH46, GubHLH49, GubHLH136, GubHLH94, GubHLH112, GubHLH80, and GubHLH5 were upregulated at 12 h. Furthermore, qRT-PCR was performed on 21 selected GubHLHs to validate their expressions under PEG treatment (Figure 7B). Most of the GubHLHs exhibited similar expression patterns in the RNA-seq and qRT-PCR analyses. GubHLH103, GubHLH116, GubHLH124, and GubHLH20 were induced by 3.75, 2.11, 6.21, and 3.37 folds at 2 h, respectively. GubHLH40 was induced significantly by 8.41 folds after 6 h treatment. GubHLH136 was up to 2.16 fold after 6 h and 12 h treatment, respectively. GubHLH5 showed inductions of 2.53, 1.34, and 2.56 folds at 2, 6, and 12 h. GubHLH64 exhibited increased expressions by 2.44, 5.63, and 14.08 folds at 2, 6, and 12 h. GubHLH60, GubHLH73, GubHLH108, GubHLH109, and GubHLH126 were also upregulated at three time points. GubHLH46 and GubHLH100 were markedly induced after treatment of 2 h and 6 h. GubHLH78 and GubHLH80 were significantly upregulated at 2 h and 12 h, respectively.
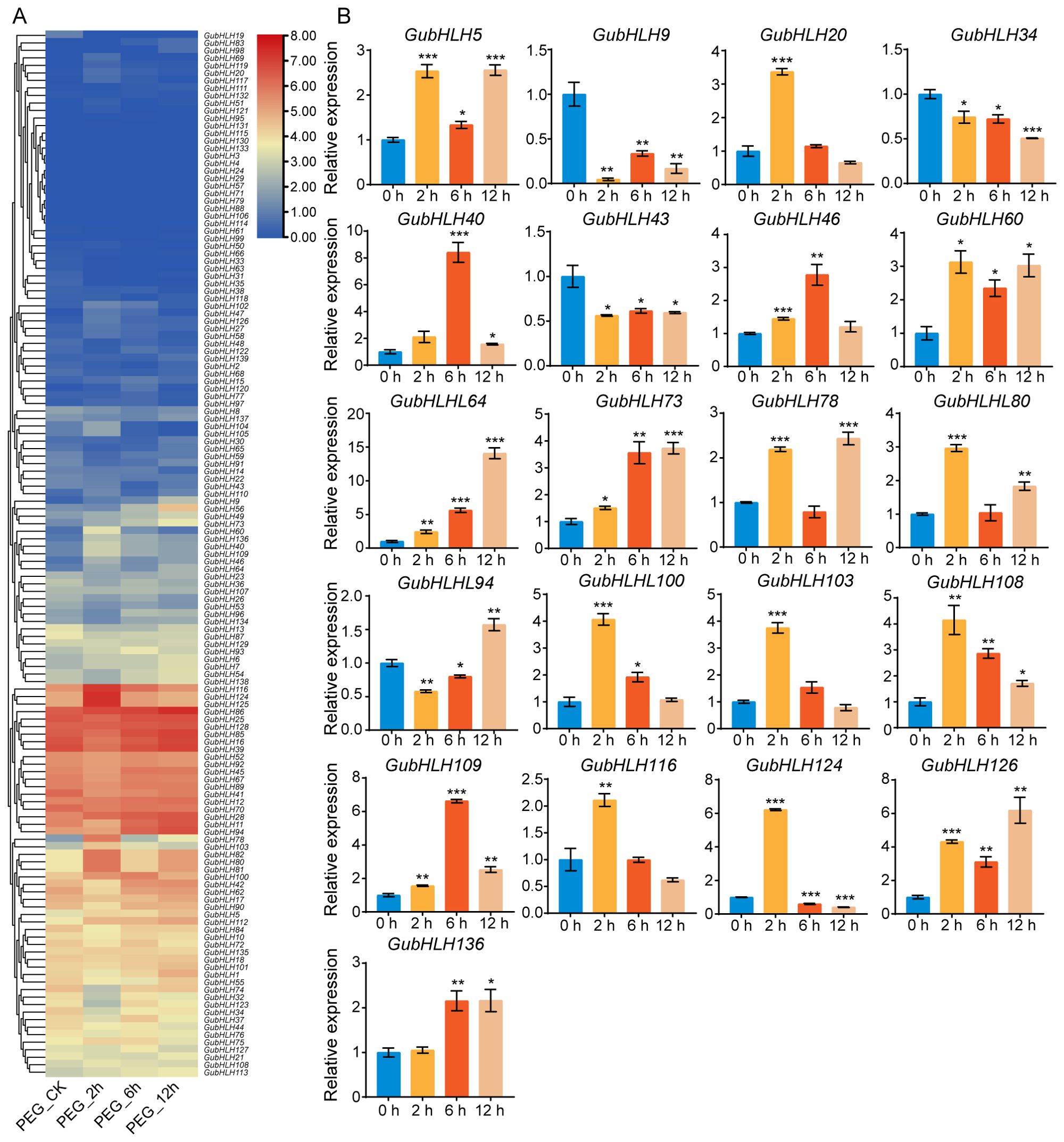
Figure 7. The expression profiles of GubHLHs under PEG treatments. (A) Heatmap of GubHLHs expression patterns under PEG treatment. (B) The selected GubHLHs expression levels in the licorice roots under PEG treatments via qRT-PCR. Student’s t-test was used to assess significant differences. Significance levels: *p < 0.05; **p < 0.01,***p < 0.001.
Moreover, the expression of these genes in licorice roots under NaCl treatment was also investigated via qRT-PCR (Figure 8). The results showed that GubHLH40, GubHLH43, GubHLH80, GubHLH94, GubHLH103, GubHLH109, GubHLH116, and GubHLH126 were reduced by NaCl. GubHLH5, GubHLH34, GubHLH78, and GubHLH100 showed increased expression trends after 2 h and 6 h of treatments, followed by a decrease at 12 h, with the highest expression occurring at 6 h, with 3.62-, 10.79-, 1.65-, and 3.69-fold increases, respectively. GubHLH9 was induced by 7.78 and 2.65 folds after 2 h and 6 h NaCl treatment, respectively. GubHLH20, GubHLH60, and GubHLH136 were induced at three time points. The expression of GubHLH124 was increased to 1.28 folds at 2 h. GubHLH64 exhibited the increase of 2.28 and 2.97 folds at 6 h and 12 h, respectively.
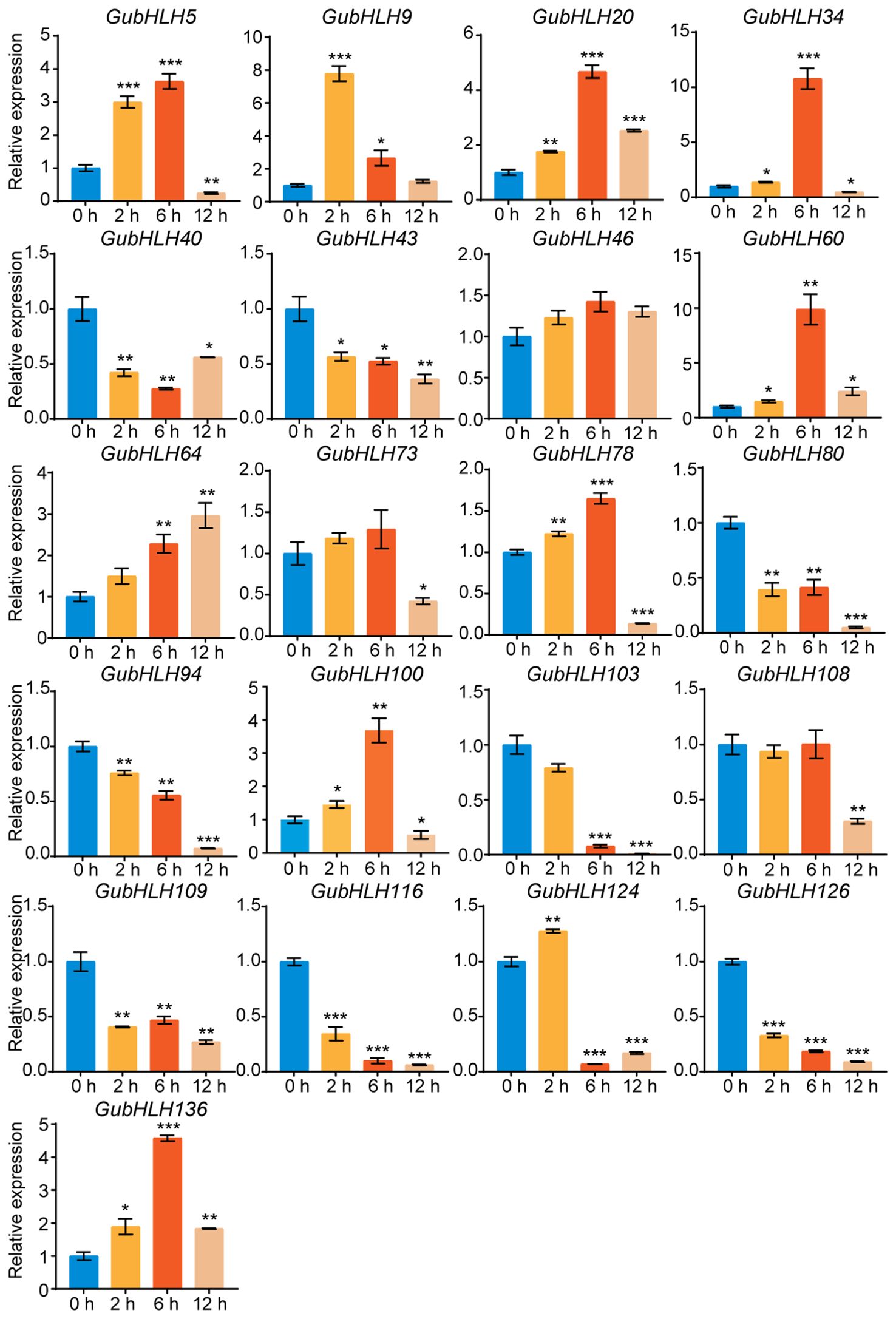
Figure 8. The selected GubHLHs expression levels in the licorice roots under NaCl treatments via qRT-PCR. Significant differences were evaluated by student’s t-test. Significance levels: *p < 0.05; **p < 0.01,***p < 0.001.
Phytohormones play substantial roles in plant growth, development and defense. We further investigated the response of these GubHLHs to four kinds of phytohormones, namely, IAA, GA, ABA, and MeJA, via qRT-PCR, and multifarious temporal-spatial expression profiles were acquired (Figure 9). Under IAA treatment, GubHLH5, GubHLH40, GubHLH73, GubHLH78, GubHLH80, and GubHLH103 reached the highest expression at 2 h, with increases of 17.55, 2.30, 3.44, 4.05, 5.10, and 2.12 folds, respectively. The expressions of GubHLH34, GubHLH43, GubHLH46, GubHLH109, GubHLH124, and GubHLH126 peaked at 6 h, showing increases of 6.21, 3.27, 7.45, 2.01, 3.63, and 17.48 folds, respectively. GubHLH20, GubHLH64 and GubHLH108 expressions were 6.73, 4.57 and 4.73 folds higher after being treated for 12 h, respectively. GubHLH9, GubHLH100, and GubHLH116 were reduced by IAA. Under GA treatment, GubHLH9, GubHLH20, GubHLH34, GubHLH100, and GubHLH116 exhibited downregulation. GubHLH43, GubHLH78, GubHLH80, and GubHLH124 showed increases at 2 and 6 h. GubHLH46, GubHLH64, GubHLH73, and GubHLH108 were upregulated at all three time points. GubHLH109 and GubHLH126 were induced at 6 h and 12 h. GubHLH5 was induced at 6 h, whereas GubHLH103 and GubHLH136 were induced at 2 h. Under ABA treatment, GubHLH9, GubHLH43, GubHLH78, GubHLH80, and GubHLH103 were induced by 1.88, 4.17, 1.68, 5.99, and 1.80 folds at 2 h, respectively. GubHLH20, GubHLH60, and GubHLH109 were up to 2.08, 3.33 and 2.31 folds at 12 h, respectively. GubHLH40, GubHLH46, GubHLH73, GubHLH100, GubHLH124, and GubHLH126 were upregulated at 2, 6, and 12 h. GubHLH5, GubHLH34, and GubHLH136 exhibited increases at 2 h and 6 h. GubHLH124 showed the highest expression after being treated for 2 h with a 63.91-fold rise. GubHLH94 and GubHLH116 expressions were reduced by ABA. Regarding the MeJA treatment, GubHLH9, GubHLH34, GubHLH43, GubHLH60, and GubHLH94 were significantly reduced by MeJA. The expression of GubHLH5 was increased by 4.79 folds after 2 h of treatment. GubHLH40 was induced by 1.72 folds after 12 h of treatment. The expression of GubHLH73, GubHLH78, GubHLH80, GubHLH124, GubHLH126, and GubHLH136 were upregulated at 2 h and 6 h. Among them, GubHLH78 and GubHLH80 showed the greatest increase at 2 h and reached 21.98 and 56.67 folds higher levels, respectively, than those in the control. GubHLH73, GubHLH124, GubHLH126, and GubHLH136 were induced by 2.56, 22.12, 6.52, and 2.81 folds at 6 h. GubHLH20, GubHLH46, GubHLH64, GubHLH103, GubHLH108, GubHLH109, and GubHLH116 were induced at all three time points. Among them, the expression levels of GubHLH103 and GubHLH116 peaked at 2 h, and were 36.44 and 5.02 folds higher than those in the control, respectively. The expression of GubHLH20 peaked at 6 h and was upregulated 33.04 folds. The expression of GubHLH46, GubHLH64, GubHLH108, and GubHLH109 peaked at 12 h, with increases of 13.20, 6.63, 32.70 and 10.78 folds, respectively. These analyses demonstrated the presumable function of these GubHLHs in G. uralensis phytohormone responses.
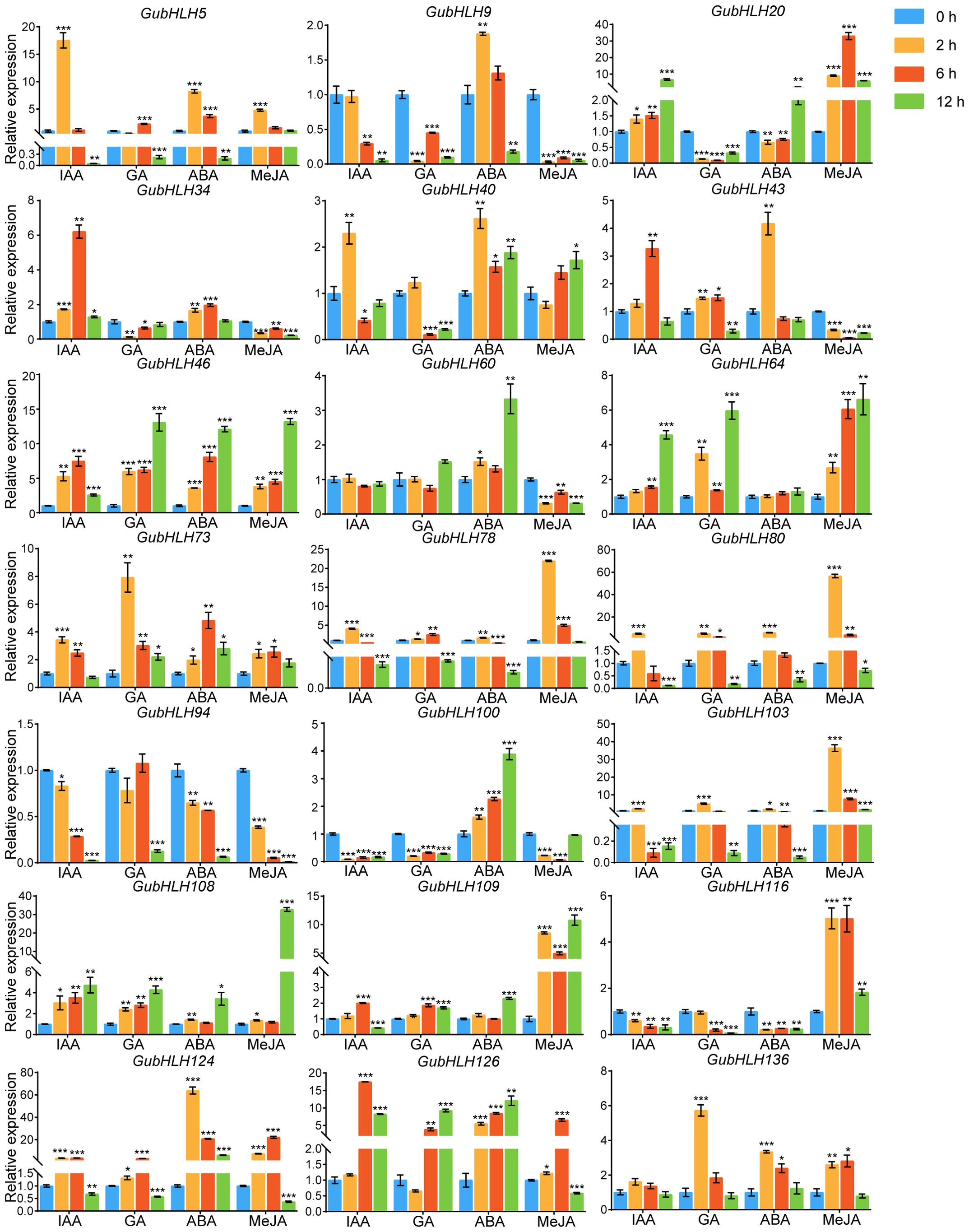
Figure 9. Expression profiles of GubHLHs in response to IAA, MeJA, ABA and GA treatments based on qRT-PCR analysis. Samples were collected at 0 h, 2 h, 6 h and 12 h after the four kinds of hormone treatments. Significant differences were estimated through student’s t-test. Significance levels: *p < 0.05; **p < 0.01,***p < 0.001.
Promoter cis-element analysis always contribute to the understanding of gene regulation mechanisms and expression profiles. The 2000 bp upstream promoter sequences of GubHLHs were extracted for cis-acting elements analysis. The results revealed the presence of 19089 cis-acting elements in their promoters (Supplementary Table S13). Among them, a variety of cis-acting elements associated with stress responses (2571, 13.47%), phytohormone responses (1325, 6.94%) and plant growth and development (251, 1.31%) were identified (Supplementary Figure S7). The abiotic/biotic stress related cis-acting elements were found in all GubHLH promoters. Among them, MYB elements were the most prevalent, with 624 instances, followed by MYC (548), STRE (383), ARE (256), as-1 (170), W box (104), WUN-motif (100), WRE3 (93), MBS (92), TC-rich repeats (63), LTR (60), MYB recognition site (42), GC-motif (26), and MBSI (10) (Supplementary Figures S7A, B). The MBS elements involved in drought-inducibility were identified in the promoters of 64 GubHLHs including GubHLH9, GubHLH20, GubHLH34, GubHLH40, GubHLH47, GubHLH73, GubHLH94, GubHLH112, GubHLH123 and GubHLH126, whose responses to drought might be related to these elements. And the phytohormone responsive related cis-elements were also found in these GubHLHs promoters. The IAA responsive elements (52 TGA-element, 19 AuxRR-core) were predicted in 54 GubHLHs. And the response of GubHLH5, GubHLH34, GubHLH40, GubHLH73, GubHLH78, GubHLH80, GubHLH109 and GubHLH116 to IAA might be related to the IAA responsive elements. Eighty-one GubHLHs contained gibberellin responsive elements (62 P-box, 34 GARE-motif, 29 TATC-box). The GA responses of GubHLH5, GubHLH40, GubHLH43, GubHLH46, GubHLH64, GubHLH73, GubHLH94 and GubHLH136 might be relevant to these elements. ABA responsive elements (365 ABRE, 66 ABRE3a, 66 ABRE4) were presented in promoters of 112 GubHLH genes. The expressions of GubHLH5, GubHLH9, GubHLH34, GubHLH40, GubHLH43, GubHLH46, GubHLH60, GubHLH73, GubHLH78, GubHLH80, GubHLH94, GubHLH100, GubHLH103, GubHLH109, GubHLH116, GubHLH124 and GubHLH136 were increased or decreased significantly under ABA treatment. Moreover, 170 TGACG-motif and 170 CGTCA-motif which are involved in MeJA response were distributed in the promoters of 88 GubHLHs. GubHLH5, GubHLH9, GubHLH34, GubHLH40, GubHLH43, GubHLH46, GubHLH60, GubHLH73, GubHLH80, GubHLH94, GubHLH100, GubHLH109, GubHLH124 and GubHLH136 were induced or reduced by MeJA. Additionally, the cis-acting elements involved in circadian regulation (31 circadian), seed-specific regulation (13 RY-element), meristem expression (68 CAT-box, 18 CCGTCC-box), zein metabolism regulation (81 O2-site), differentiation of palisade mesophyll cells (9 HD-Zip 1), and endosperm expression (31 GCN4_motif) were also identified.
The bHLH TFs represent one of the maximum TF families and are engaged in a wide array of growth, development, and environmental adaptation processes. The expeditious advancement of plant genome sequencing technology has facilitated the studies of plant bHLH gene families. Identifying various bHLH isoforms and investigating their expression patterns are fundamental to comprehend their functions. Nonetheless, limited research has been conducted on the medicinal plant licorice, which garners significant attention for its medicinal and industrial applications. In present study, a systematic and comprehensive investigation of G. uralensis bHLH family members was undertaken. A total of 139 genes encoding G. uralensis bHLH TFs were identified, with protein lengths varying from 73 to 777 amino acids and pI ranging from 4.55 to 10.29, suggesting potential structural and functional diversity. And 138 out of 139 GubHLH proteins were predicted to localize in the nucleus, emphasizing their primary roles in regulation and interaction within the nucleus. This localization pattern was generally in accord with the GO enrichment results, where the majority of GubHLHs were enriched in the nucleus at the cellular component level. Multiple alignment analysis revealed four conserved regions within the bHLH domain, aligning with the characteristics of bHLH TFs (Atchley et al., 1999; Nair and Burley, 2000). A total of 23 conserved amino acids were found with a consensus ratio greater than 50%, and seven out of these showed a consensus ratio greater than 75%. These findings are consistent with previous studies (Li et al., 2006; Zhang et al., 2018). The extremely conserved Leu-23 and Pro-28, respectively accounting for 98.6% and 97.1%, underscored their significance in dimerization formation (Atchley et al., 1999). And in the basic region, the conserved H5-E9-R13 region was present in GubHLH proteins, being in line with the characteristic of most plants (Heim et al., 2003; Qian et al., 2007; Pires and Dolan, 2010). A total of 403 G-box cis-acting elements were identified in the GubHLH gene promoters and GubHLH23 possessed the highest number of 19 of them. And 624 MYB cis-acting elements were detected that were existed in all GubHLH gene promoters. These results indicated that these GubHLH TFs may function through interacting with MYB TFs or among themselves, as also supported by the GO annotations of GubHLH TFs with binding activity.
The plant bHLH TF family comprises approximately 14-32 subfamilies (Heim et al., 2003; Toledo-Ortiz et al., 2003; Li et al., 2006; Carretero-Paulet et al., 2010; Pires and Dolan, 2010). Phylogenetic analysis classified GubHLHs into 25 subfamilies, with varying subfamily sizes ranging from 1 to 16 members. And it was shown that some gene pairs exhibited one-to-one orthologous relationships, such as AtbHLH71 and GubHLH117 in subfamily Ia, AtbHLH142 and GubHLH6 in subfamily XIV, AtbHLH95 and GubHLH71 in subfamily Ib (1), AtbHLH102 and GubHLH89 in subfamily Va, as well as AtbHLH108 and GubHLH14 in subfamily VI. In more cases, bHLH proteins from the same species were inclined to cluster together. Moreover, phylogenetic tree indicated that subfamily IVa contained six pairs of duplication GubHLHs, including three segmental duplication gene pairs (GubHLH73/136, GubHLH80/103, GubHLH78/120) and three tandem duplication gene pairs (GubHLH80/81, GubHLH103/104, GubHLH122/123). Subfamily Vb contained five pairs of segmental duplication GubHLHs (GubHLH87/93, GubHLH62/96, GubHLH72/129, GubHLH8/91, GubHLH31/34). These results indicated that species-specific duplication events are of significant importance in gene family evolution. And the specific genes might play significant roles in licorice. GubHLH20, GubHLH73, GubHLH78, GubHLH80 and GubHLH136 in subfamily IVa showed obvious up-regulated or down-regulated trends under PEG, NaCl and four phytohormones treatments, indicating that these GubHLHs play roles in the abiotic and phytohormones responses. Their regulatory mechanisms and functions should be further confirmed. MEME analysis revealed the presence of conserved motifs crucial for the structure and function specificity of bHLH domain in all GubHLH proteins (Feller et al., 2011). The varied exon/intron structures of the members within gene families are notable clues to apprehend their evolution and diversified functions, which are achieved via three main ways, insertion/deletion, exon/intron gain/loss, and exonization/pseudoexonization (Xu et al., 2012). A varying number of introns ranging from 0 to 19 was revealed in GubHLHs. which was similar to apple bHLH genes (Yang et al., 2017). It may be concluded that exon/intron gain/loss occurred during the evolution process of the G. uralensis bHLH family. With 15 UTRs, GubHLH138 may own the most complicated alternatively spliced form, and similar results were also obtained in passion fruit (Liang et al., 2023). These results indicated that closely related bHLH members are inclined to exhibit similar exon/intron structures, motif compositions and secondary protein structures. Gene duplication events are indispensable for the gene family expansion in the evolutionary process and the new members and functions generation (Flagel and Wendel, 2009). Segmental duplication played a prominent role, resulting in 55 pairs of segmentally duplicated genes. Forty-one pairs of these genes owned differential gene structures. For instance, GubHLH83 contained one exon, while its paralog, GubHLH106, had four exons, indicating the acquisition of three exons during evolutionary progression, a phenomenon similarly observed in Raphanus sativus and passion fruit (Wang et al., 2022; Liang et al., 2023). The purifying selection is highlighted by the Ka/Ks values exceeding 1 for all duplicated gene pairs, reflecting selective pressure throughout GubHLH genes evolution. This observation aligns with findings in other species, including Brassica oleracea, Brassica napus, and Brassica rapa (Miao et al., 2020), and rice (Li et al., 2006). Comparative analyses with other plant species demonstrated a higher number of collinear pairs between G. uralensis and G. max, and between G. uralensis and M. truncatula, indicative of their closer evolutionary connections.
At present, environmental stresses such as drought and salt stress have been the influencing factors of the plant progression and ecological environment (Liu et al., 2014). Expression pattern analysis is conducive to understanding gene function and characteristics. bHLH TFs were indicated to be engaged in regulating abiotic stress tolerance. AtbHLH112 transcript levels were shown to be positively related to PEG and NaCl tolerance, enhancing stress tolerance (Liu et al., 2015). AtbHLH122 was indicated to play a notable role in positively regulating drought, NaCl and osmotic resistance (Liu et al., 2014). The rd22BP1 (AtbHLH6) protein is significantly induced by dehydration, and ABA can activate the transcription of the dehydration-responsive rd22 gene (Abe et al., 1997). The AtbHLH17 (AtAIB) expression was markedly increased under drought stress and ABA treatment, and the coexpression of AtbHLH17 with AtWRKY28 can strengthen the transcription of downstream stress-responsive genes, in turn, enhance stress tolerance (Babitha et al., 2013). MfbHLH38 was shown to heighten drought and salinity resistance and to be involved in the ABA-dependent pathway (Qiu et al., 2020). Several licorice bHLHs were induced or reduced by PEG treatment, which might indicate their involvement in regulating the PEG response. GubHLH40, GubHLH46, GubHLH60, GubHLH108, GubHLH109, and GubHLH116, which clustered into subfamily III (d+e) with AtbHLH6 and AtbHLH17, showed increased trends after PEG treatment in both RNA-seq and qRT-PCR data. Moreover, GubHLH100, which belonged to subfamily XI along with AtbHLH122, was induced obviously by PEG treatment. These results might indicate the potential involvement of these genes in the modulation of drought tolerance and the improvement of adaptability to the environment. Additionally, ABA accumulation is necessary for the expression inductions of some gene under drought stress (Zhu, 2002). In our study, the expressions of GubHLH40, GubHLH46, GubHLH60, GubHLH108, and GubHLH109 tended to increase under ABA treatment, coinciding with the findings for AtbHLH17 and AtbHLH6. Whereas the GubHLH116 was reduced by ABA, which requires further in-depth exploration. In addition, significant induction of GubHLH64 expression was detected upon both PEG and NaCl treatments, which might show the involvement of GubHLH64 in licorice drought and salt stress.
The sophisticated interactions between GubHLH proteins and other proteins potentially enable their involvement in regulating multiple biological processes. Typically, three types of TFs, WD-repeat, bHLH, and MYB, can form complexes and modulate the anthocyanin accumulation and trichome development in a series of plants (Qi et al., 2011). The Arabidopsis WD40 repeat protein TTG1 occupies a central position in this complex, serving as a scaffold for the interactions between the other two TFs (Ramsay and Glover, 2005). The Medicago truncatula bHLH TF MtTT8 can form a complex with MtWD40-1 and MtLAP1 or MtPAR, regulating the synthesis of anthocyanins and proanthocyanidin through activating the downstream anthocyanidin reductase (MtANS) and anthocyanidin synthase (MtANR) (Li P. et al., 2016). In Prunus avium, PavWD40, PavMYB10.1, and PavbHLH interact with each other and regulate the cherry anthocyanin biosynthesis (Jin et al., 2016). JA, a significant plant hormone, mediates multifarious developmental processes in plants, functioning as a regulatory molecule (Li et al., 2021). JA has been previously reported to favor the anthocyanin accumulation (Shan et al., 2009) and trichome initiation (Traw and Bergelson, 2003). Arabidopsis GL3, TT8 and EGL3 have been shown to interact with MYB75 and TTG1, forming WD-repeat/bHLH/MYB complexes and modulating the accumulation of anthocyanin and initiation of trichome through JA signaling (Qi et al., 2011). In present PPI network, TT8, homologous with GubHLH38, and GL3 (homolog of GubHLH64), along with TTG1 and MYB75, were clustered into an interconnected functional module. Phylogenetic analysis showed that GubHLH64 and GubHLH38 are clustered into the same branch (subfamily IIIf) together with GL3, TT8 and EGL3, indicating that these bHLHs may exert similar functions. Subcellular localization assays confirmed that GubHLH64 and GubHLH38 are localized in nucleus. The expression of GubHLH64 was prominently induced by MeJA. Yeast two-hybrid assays verified that GubHLH38, GubHLH64, and GuMYB75 directly interact with GuTTG1. These results indicate that GubHLH64 and GubHLH38 might be closely relevant to the regulation of trichome initiation and anthocyanin biosynthesis in G. uralensis. And their precise regulatory mechanisms and functions should be further confirmed.
MYC2, MYC3 and MYC4 can form homo- and heterodimers among themselves and also interact with JAZ proteins, playing a prominent role in activating the JA regulated transcriptional response (Fernández-Calvo et al., 2011). MYC2/MYC3/MYC4 were also found to be able to originate a hierarchical network of a series of downstream TFs, establishing a core MYC2/MYC3/MYC4-dependent “regulon” (Van Moerkercke et al., 2019). In the PPI network, MYC2 (homolog of GubHLH116), MYC3 (homolog of GubHLH108), and MYC4 (homolog of GubHLH109) were clustered into one functional module, forming a closely interconnected cluster. Further investigation into the subcellular localization of GubHLH108, GubHLH109, and GubHLH116 confirmed their presence in the nucleus, aligning with their roles as TFs. The expression of GubHLH108, GubHLH109, and GubHLH116 were found to increase significantly after MeJA treatment. These results might imply the noteworthy functions of these three GubHLHs in a JA-regulated manner.
A total of 139 G. uralensis bHLH genes were identified and categorized into 25 subfamilies on the basis of the phylogenetic relationships with Arabidopsis bHLH genes. All GubHLH proteins exhibited the typical conserved bHLH domain, comprising four conserved regions with 23 amino acid residues. The analysis of motifs and domain composition, in conjunction with gene structure assessment, illustrated the relative conservation within specific subfamilies. Collinearity analysis emphasized the predominant contribution of segmental duplication to the expansion of GubHLH gene family, which experienced purifying selection throughout its evolution. The divergent exon/intron structures, observed in some of the duplication gene pairs might clarify their function diversification in evolution. The temporospatial expression profiles of GubHLHs following treatment with PEG, NaCl, and various phytohormones demonstrated their involvement in regulation of abiotic and phytohormones responses. GubHLH64 and GubHLH38 might be closely pertinent to the G. uralensis trichome initiation and anthocyanin biosynthesis, and GubHLH64 also related to the abiotic responses such as drought and salt. Several bHLH members in subfamily III (d+e) might participate in the G. uralensis drought response. GubHLH108, GubHLH109, and GubHLH116 might exploit noteworthy functions in a JA-dependent manner. The results gained in current study can favor the deeper insight into the characteristics, evolution, and expression patterns of GubHLH proteins, offering a foundation for future GubHLHs biological function explorations.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
GD: Formal analysis, Investigation, Writing – original draft, Writing – review & editing. YS: Investigation, Validation, Writing – review & editing. KX: Investigation, Validation, Writing – review & editing. HL: Conceptualization, Funding acquisition, Supervision, Writing – review & editing. GX: Conceptualization, Funding acquisition, Investigation, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by Science and Technology Project of Bingtuan (2023AB052), International Science and Technology Cooperation Project of Bingtuan (2020BC002), Science and Technology Project of Shihezi University (CXPY202006).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1485757/full#supplementary-material
Abe, H., Yamaguchi-Shinozaki, K., Urao, T., Iwasaki, T., Hosokawa, D., Shinozaki, K. (1997). Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9, 1859–1868. doi: 10.1105/tpc.9.10.1859
Atchley, W. R., Terhalle, W., Dress, A. (1999). Positional dependence, cliques, and predictive motifs in the bHLH protein domain. J. Mol. Evol. 48, 501–516. doi: 10.1007/pl00006494
Babitha, K. C., Ramu, S. V., Pruthvi, V., Mahesh, P., Nataraja, K. N., Udayakumar, M. (2013). Co-expression of AtbHLH17 and AtWRKY28 confers resistance to abiotic stress in Arabidopsis. Transgenic Res. 22, 327–341. doi: 10.1007/s11248-012-9645-8
Bailey, T. L., Johnson, J., Grant, C. E., Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Bailey, P. C., Martin, C., Toledo-Ortiz, G., Quail, P. H., Huq, E., Heim, M. A., et al. (2003). Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell 15, 2497–2502. doi: 10.1105/tpc.151140
Bu, D., Luo, H., Huo, P., Wang, Z., Zhang, S., He, Z., et al. (2021). KOBAS-i: intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 49, W317–W325. doi: 10.1093/nar/gkab447
Carretero-Paulet, L., Galstyan, A., Roig-Villanova, I., Martínez-García, J. F., Bilbao-Castro, J. R., Robertson, D. L. (2010). Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 153, 1398–1412. doi: 10.1104/pp.110.153593
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, S., Zhou, Y., Chen, Y., Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Chou, K. C., Shen, H. B. (2008). Cell-PLoc: a package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 3, 153–162. doi: 10.1038/nprot.2007.494
Ding, Y., Brand, E., Wang, W., Zhao, Z. (2022). Licorice: Resources, applications in ancient and modern times. J. Ethnopharmacol. 298, 115594. doi: 10.1016/j.jep.2022.115594
Feller, A., Machemer, K., Braun, E. L., Grotewold, E. (2011). Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 66, 94–116. doi: 10.1111/j.1365-313X.2010.04459.x
Fernández-Calvo, P., Chini, A., Fernández-Barbero, G., Chico, J. M., Gimenez-Ibanez, S., Geerinck, J., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23, 701–715. doi: 10.1105/tpc.110.080788
Flagel, L. E., Wendel, J. F. (2009). Gene duplication and evolutionary novelty in plants. New Phytol. 183, 557–564. doi: 10.1111/j.1469-8137.2009.02923.x
Gao, C., Guo, Y., Wang, J., Li, D., Liu, K., Qi, S., et al. (2018). Brassica napus GLABRA3-1 promotes anthocyanin biosynthesis and trichome formation in true leaves when expressed in Arabidopsis thaliana. Plant Biol. (Stuttg). 20, 3–9. doi: 10.1111/plb.12633
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., Bairoch, A. (2003). ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Geourjon, C., Deléage, G. (1995). SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci. 11, 681–684. doi: 10.1093/bioinformatics/11.6.681
Heim, M. A., Jakoby, M., Werber, M., Martin, C., Weisshaar, B., Bailey, P. C. (2003). The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20, 735–747. doi: 10.1093/molbev/msg088
Hichri, I., Heppel, S. C., Pillet, J., Léon, C., Czemmel, S., Delrot, S., et al. (2010). The basic helix-loop-helix transcription factor MYC1 is involved in the regulation of the flavonoid biosynthesis pathway in grapevine. Mol. Plant 3, 509–523. doi: 10.1093/mp/ssp118
Hosseinzadeh, H., Nassiri-Asl, M. (2015). Pharmacological effects of Glycyrrhiza spp. and its bioactive constituents: update and review. Phytother. Res. 29, 1868–1886. doi: 10.1002/ptr.5487
Hudson, K. A., Hudson, M. E. (2015). A classification of basic helix-loop-helix transcription factors of soybean. Int. J. Genomics 2015, 603182. doi: 10.1155/2015/603182
Jiang, M., Zhao, S., Yang, S., Lin, X., He, X., Wei, X., et al. (2020). An “essential herbal medicine”—licorice: A review of phytochemicals and its effects in combination preparations. J. Ethnopharmacol. 249, 112439. doi: 10.1016/j.jep.2019.112439
Jin, W., Wang, H., Li, M., Wang, J., Yang, Y., Zhang, X., et al. (2016). The R2R3 MYB transcription factor PavMYB10.1 involves in anthocyanin biosynthesis and determines fruit skin colour in sweet cherry (Prunus avium L.). Plant Biotechnol. J. 14, 2120–2133. doi: 10.1111/pbi.12568
Kao, T. C., Wu, C. H., Yen, G. C. (2014). Bioactivity and potential health benefits of licorice. J. Agric. Food Chem. 62, 542–553. doi: 10.1021/jf404939f
Khan, R. A., Kumar, A., Abbas, N. (2024). A bHLH transcription factor AaMYC2-type positively regulates glandular trichome density and artemisinin biosynthesis in Artemisia annua. Physiol. Plant 176, e14581. doi: 10.1111/ppl.14581
Kim, D., Paggi, J. M., Park, C., Bennett, C., Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Kim, J. H., Park, Y. I., Hur, M., Park, W. T., Moon, Y. H., Huh, Y. C., et al. (2023). Inhibition by components of Glycyrrhiza uralensis of 3CLpro and HCoV-OC43 proliferation. J. Enzyme Inhib. Med. Chem. 38, 2242704. doi: 10.1080/14756366.2023.2242704
Kumar, S. V., Lucyshyn, D., Jaeger, K. E., Alós, E., Alvey, E., Harberd, N. P., et al. (2012). Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484, 242–245. doi: 10.1038/nature10928
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Le Hir, R., Castelain, M., Chakraborti, D., Moritz, T., Dinant, S., Bellini, C. (2017). AtbHLH68 transcription factor contributes to the regulation of ABA homeostasis and drought stress tolerance in Arabidopsis thaliana. Physiol. Plant 160, 312–327. doi: 10.1111/ppl.12549
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., Bork, P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Li, P., Chen, B., Zhang, G., Chen, L., Dong, Q., Wen, J., et al. (2016). Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bHLH transcription factor MtTT8. New Phytol. 210, 905–921. doi: 10.1111/nph.13816
Li, B., Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 12, 323. doi: 10.1186/1471-2105/12/323
Li, X., Duan, X., Jiang, H., Sun, Y., Tang, Y., Yuan, Z., et al. (2006). Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 141, 1167–1184. doi: 10.1104/pp.106.080580
Li, P., Fu, J., Xu, Y., Shen, Y., Zhang, Y., Ye, Z., et al. (2022). CsMYB1 integrates the regulation of trichome development and catechins biosynthesis in tea plant domestication. New Phytol. 234, 902–917. doi: 10.1111/nph.18026
Li, T., Hua, S., Ma, J., Dong, L., Xu, F., Fu, X. (2020). Spectrum-effect relationships of flavonoids in Glycyrrhiza uralensis Fisch. J. Anal. Methods Chem. 2020, 8838290. doi: 10.1155/2020/8838290
Li, Z., Liu, C., Zhang, Y., Wang, B., Ran, Q., Zhang, J. (2019). The bHLH family member ZmPTF1 regulates drought tolerance in maize by promoting root development and abscisic acid synthesis. J. Exp. Bot. 70, 5471–5486. doi: 10.1093/jxb/erz307
Li, Y., Shan, X., Gao, R., Yang, S., Wang, S., Gao, X., et al. (2016). Two IIIf Clade-bHLHs from Freesia hybrida play divergent roles in flavonoid biosynthesis and trichome formation when ectopically expressed in Arabidopsis. Sci. Rep. 6, 30514. doi: 10.1038/srep30514
Li, H., Yang, Z., Zeng, Q., Wang, S., Luo, Y., Huang, Y., et al. (2020). Abnormal expression of bHLH3 disrupts a flavonoid homeostasis network, causing differences in pigment composition among mulberry fruits. Hortic. Res. 7, 83. doi: 10.1038/s41438-020-0302-8
Li, M., Yu, G., Cao, C., Liu, P. (2021). Metabolism, signaling, and transport of jasmonates. Plant Commun. 2, 100231. doi: 10.1016/j.xplc.2021.100231
Liang, J., Fang, Y., An, C., Yao, Y., Wang, X., Zhang, W., et al. (2023). Genome-wide identification and expression analysis of the bHLH gene family in passion fruit (Passiflora edulis) and its response to abiotic stress. Int. J. Biol. Macromol. 225, 389–403. doi: 10.1016/j.ijbiomac.2022.11.076
Liu, Y., Ji, X., Nie, X., Qu, M., Zheng, L., Tan, Z., et al. (2015). Arabidopsis AtbHLH112 regulates the expression of genes involved in abiotic stress tolerance by binding to their E-box and GCG-box motifs. New Phytol. 207, 692–709. doi: 10.1111/nph.13387
Liu, W., Tai, H., Li, S., Gao, W., Zhao, M., Xie, C., et al. (2014). bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. New Phytol. 201, 1192–1204. doi: 10.1111/nph.12607
Liu, H., Yang, Y., Liu, D., Wang, X., Zhang, L. (2020). Transcription factor TabHLH49 positively regulates dehydrin WZY2 gene expression and enhances drought stress tolerance in wheat. BMC Plant Biol. 20, 259. doi: 10.1186/s12870-020-02474-5
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Lu, R., Zhang, J., Liu, D., Wei, Y. L., Wang, Y., Li, X. B. (2018). Characterization of bHLH/HLH genes that are involved in brassinosteroid (BR) signaling in fiber development of cotton (Gossypium hirsutum). BMC Plant Biol. 18, 304. doi: 10.1186/s12870-018-1523-y
Ludwig, S. R., Habera, L. F., Dellaporta, S. L., Wessler, S. R. (1989). Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proc. Natl. Acad. Sci. U. S. A. 86, 7092–7096. doi: 10.1073/pnas.86.18.7092
Luo, J., Liu, H., Zhou, T., Gu, B., Huang, X., Shangguan, Y., et al. (2013). An-1 encodes a basic helix-loop-helix protein that regulates awn development, grain size, and grain number in rice. Plant Cell 25, 3360–3376. doi: 10.1105/tpc.113.113589
Mertens, J., Pollier, J., Vanden Bossche, R., Lopez-Vidriero, I., Franco-Zorrilla, J. M., Goossens, A. (2016). The bHLH transcription factors TSAR1 and TSAR2 regulate triterpene saponin biosynthesis in Medicago truncatula. Plant Physiol. 170, 194–210. doi: 10.1104/pp.15.01645
Miao, L., Gao, Y., Zhao, K., Kong, L., Yu, S., Li, R., et al. (2020). Comparative analysis of basic helix-loop-helix gene family among Brassica oleracea, Brassica rapa, and Brassica napus. BMC Genomics 21, 178. doi: 10.1186/s12864-020-6572-6
Murre, C., McCaw, P. S., Baltimore, D. (1989). A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56, 777–783. doi: 10.1016/0092-8674(89)90682-x
Nair, S. K., Burley, S. K. (2000). Recognizing DNA in the library. Nature 404, 717–718. doi: 10.1038/35008182
Oh, E., Yamaguchi, S., Kamiya, Y., Bae, G., Chung, W. I., Choi, G. (2006). Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 47, 124–139. doi: 10.1111/j.1365-313X.2006.02773.x
Park, K. I., Ishikawa, N., Morita, Y., Choi, J. D., Hoshino, A., Iida, S. (2007). A bHLH regulatory gene in the common morning glory, Ipomoea purpurea, controls anthocyanin biosynthesis in flowers, proanthocyanidin and phytomelanin pigmentation in seeds, and seed trichome formation. Plant J. 49, 641–654. doi: 10.1111/j.1365-313X.2006.02988.x
Pires, N., Dolan, L. (2010). Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 27, 862–874. doi: 10.1093/molbev/msp288
Qi, T., Song, S., Ren, Q., Wu, D., Huang, H., Chen, Y., et al. (2011). The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23, 1795–1814. doi: 10.1105/tpc.111.083261
Qian, W., Tan, G., Liu, H., He, S., Gao, Y., An, C. (2007). Identification of a bHLH-type G-box binding factor and its regulation activity with G-box and Box I elements of the PsCHS1 promoter. Plant Cell Rep. 26, 85–93. doi: 10.1007/s00299-006-0202-x
Qiu, J. R., Huang, Z., Xiang, X. Y., Xu, W. X., Wang, J. T., Chen, J., et al. (2020). MfbHLH38, a Myrothamnus flabellifolia bHLH transcription factor, confers tolerance to drought and salinity stresses in Arabidopsis. BMC Plant Biol. 20, 542. doi: 10.1186/s12870-020-02732-6
Raissig, M. T., Abrash, E., Bettadapur, A., Vogel, J. P., Bergmann, D. C. (2016). Grasses use an alternatively wired bHLH transcription factor network to establish stomatal identity. Proc. Natl. Acad. Sci. U. S. A. 113, 8326–8331. doi: 10.1073/pnas.1606728113
Ramsay, N. A., Glover, B. J. (2005). MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10, 63–70. doi: 10.1016/j.tplants.2004.12.011
Seeliger, D., de Groot, B. L. (2010). Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput.-Aided Mol. Des. 24, 417–422. doi: 10.1007/s10822-010-9352-6
Shan, X., Zhang, Y., Peng, W., Wang, Z., Xie, D. (2009). Molecular mechanism for jasmonate-induction of anthocyanin accumulation in Arabidopsis. J. Exp. Bot. 60, 3849–3860. doi: 10.1093/jxb/erp223
Simionato, E., Ledent, V., Richards, G., Thomas-Chollier, M., Kerner, P., Coornaert, D., et al. (2007). Origin and diversification of the basic helix-loop-helix gene family in metazoans: insights from comparative genomics. BMC Evol. Biol. 7, 33. doi: 10.1186/1471-2148-7-33
Strader, L., Weijers, D., Wagner, D. (2022). Plant transcription factors - being in the right place with the right company. Curr. Opin. Plant Biol. 65, 102136. doi: 10.1016/j.pbi.2021.102136
Szklarczyk, D., Kirsch, R., Koutrouli, M., Nastou, K., Mehryary, F., Hachilif, R., et al. (2023). The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51, D638–D646. doi: 10.1093/nar/gkac1000
Tamura, K., Yoshida, K., Hiraoka, Y., Sakaguchi, D., Chikugo, A., Mochida, K., et al. (2018). The basic helix-loop-helix transcription factor GubHLH3 positively regulates soyasaponin biosynthetic genes in Glycyrrhiza uralensis. Plant Cell Physiol. 59, 778–791. doi: 10.1093/pcp/pcy046
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Toledo-Ortiz, G., Huq, E., Quail, P. H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15, 1749–1770. doi: 10.1105/tpc.013839
Traw, M. B., Bergelson, J. (2003). Interactive effects of jasmonic acid, salicylic acid, and gibberellin on induction of trichomes in Arabidopsis. Plant Physiol. 133, 1367–1375. doi: 10.1104/pp.103.027086
Van Moerkercke, A., Duncan, O., Zander, M., Šimura, J., Broda, M., Vanden Bossche, R., et al. (2019). A MYC2/MYC3/MYC4-dependent transcription factor network regulates water spray-responsive gene expression and jasmonate levels. Proc. Natl. Acad. Sci. U. S. A. 116, 23345–23356. doi: 10.1073/pnas.1911758116
Wang, N., Cui, Y., Liu, Y., Fan, H., Du, J., Huang, Z., et al. (2013). Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana. Mol. Plant 6, 503–513. doi: 10.1093/mp/sss089
Wang, R., Li, Y., Gao, M., Han, M., Liu, H. (2022). Genome-wide identification and characterization of the bHLH gene family and analysis of their potential relevance to chlorophyll metabolism in Raphanus sativus L. BMC Genomics 23, 548. doi: 10.1186/s12864-022-08782-4
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Wang, D., Zhang, X., Cao, Y., Batool, A., Xu, Y., Qiao, Y., et al. (2024). TabHLH27 orchestrates root growth and drought tolerance to enhance water use efficiency in wheat. J. Integr. Plant Biol. 66, 1295–1312. doi: 10.1111/jipb.13670
Waterhouse, A. M., Procter, J. B., Martin, D. M., Clamp, M., Barton, G. J. (2009). Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. doi: 10.1093/bioinformatics/btp033
Wu, H., Ye, H., Yao, R., Zhang, T., Xiong, L. (2015). OsJAZ9 acts as a transcriptional regulator in jasmonate signaling and modulates salt stress tolerance in rice. Plant Sci. 232, 1–12. doi: 10.1016/j.plantsci.2014.12.010
Xie, X. B., Li, S., Zhang, R. F., Zhao, J., Chen, Y. C., Zhao, Q., et al. (2012). The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ. 35, 1884–1897. doi: 10.1111/j.1365-3040.2012.02523.x
Xu, G., Guo, C., Shan, H., Kong, H. (2012). Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. U. S. A. 109, 1187–1192. doi: 10.1073/pnas.1109047109
Xu, J., van Herwijnen, Z. O., Dräger, D. B., Sui, C., Haring, M. A., Schuurink, R. C. (2018). SlMYC1 regulates type VI glandular trichome formation and terpene biosynthesis in tomato glandular cells. Plant Cell 30, 2988–3005. doi: 10.1105/tpc.18.00571
Yang, J., Gao, M., Huang, L., Wang, Y., van Nocker, S., Wan, R., et al. (2017). Identification and expression analysis of the apple (Malus × domestica) basic helix-loop-helix transcription factor family. Sci. Rep. 7, 28. doi: 10.1038/s41598-017-00040-y
Yang, R., Wang, L.-q., Yuan, B.-c., Liu, Y. (2015). The pharmacological activities of licorice. Planta Med. 81, 1654–1669. doi: 10.1055/s-0035-1557893
Yao, H., Wang, F., Bi, Q., Liu, H., Liu, L., Xiao, G., et al. (2022). Combined analysis of pharmaceutical active ingredients and transcriptomes of Glycyrrhiza uralensis under PEG6000-induced drought stress revealed glycyrrhizic acid and flavonoids accumulation via JA-mediated signaling. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.920172
Yuan, M., Shu, G., Zhou, J., He, P., Xiang, L., Yang, C., et al. (2023). AabHLH113 integrates jasmonic acid and abscisic acid signaling to positively regulate artemisinin biosynthesis in Artemisia annua. New Phytol. 237, 885–899. doi: 10.1111/nph.18567
Zhang, Z. (2022). KaKs_Calculator 3.0: Calculating selective pressure on coding and non-coding sequences. Genom. Proteom. Bioinf. 20, 536–540. doi: 10.1016/j.gpb.2021.12.002
Zhang, H., Guo, J., Chen, X., Zhou, Y., Pei, Y., Chen, L., et al. (2022). Pepper bHLH transcription factor CabHLH035 contributes to salt tolerance by modulating ion homeostasis and proline biosynthesis. Hortic. Res. 9, uhac203. doi: 10.1093/hr/uhac203
Zhang, T., Lv, W., Zhang, H., Ma, L., Li, P., Ge, L., et al. (2018). Genome-wide analysis of the basic helix-loop-helix (bHLH) transcription factor family in maize. BMC Plant Biol. 18, 235. doi: 10.1186/s12870-018-1441-z
Zhao, X., Wang, Q., Yan, C., Sun, Q., Wang, J., Li, C., et al. (2024). The bHLH transcription factor AhbHLH121 improves salt tolerance in peanut. Int. J. Biol. Macromol. 256, 128492. doi: 10.1016/j.ijbiomac.2023.128492
Keywords: bHLH gene family, licorice, evolutionary analyses, expression patterns, stress responses, phytohormones
Citation: Ding G, Shi Y, Xie K, Li H and Xiao G (2025) Genome-wide identification and expression analysis of bHLH gene family revealed their potential roles in abiotic stress response, anthocyanin biosynthesis and trichome formation in Glycyrrhiza uralensis. Front. Plant Sci. 15:1485757. doi: 10.3389/fpls.2024.1485757
Received: 24 August 2024; Accepted: 27 December 2024;
Published: 21 January 2025.
Edited by:
George V. Popescu, Mississippi State University, United StatesCopyright © 2025 Ding, Shi, Xie, Li and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongbin Li, bGloYkBzaHp1LmVkdS5jbg==; Guanghui Xiao, Z3VhbmdodWl4QHNubnUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.