Abstract
Drought stress inhibits Salvia miltiorrhiza Bunge (S. miltiorrhiza) seedling growth and yield. Here, we studied the effects of drought stress on the different parts of S. miltiorrhiza seedlings through physiological, transcriptomic, and metabolomics analyses, and identified key genes and metabolites related to drought tolerance. Physiological analysis showed that drought stress increased the accumulation of hydrogen peroxide (H2O2), enhanced the activity of peroxidase (POD), decreased the activity of catalase (CAT) and the contents of chlorophyll b and total chlorophyll, reduced the degree of photosynthesis, enhanced oxidative damage in S. miltiorrhiza seedlings, and inhibited the growth of S. miltiorrhiza plants. Transcriptome analyses revealed 383 genes encoding transcription factors and 80 genes encoding plant hormones as hypothetical regulators of drought resistance in S. miltiorrhiza plants. Moreover, differentially expressed genes (DEGs) and differentially expressed metabolites (DEMs) are involved in a variety of biological processes, such as proline and glycine betaine metabolism, and biosynthesis of tanshinones and phenolic acids. Additionally, it has barely been reported that the AHL gene family may be involved in regulating the neocryptotanshinone biosynthesis. In conclusion, our results suggest that drought stress inhibits S. miltiorrhiza seedling growth by enhancing membrane lipid peroxidation, attenuating the antioxidant system, photosynthesis, and regulating proline and glycine betaine metabolism, transcription factors and plant hormones, and tanshinones and phenolic acid metabolism pathways. This study provides new insights into the complex mechanisms by which S. miltiorrhiza responds to drought stress.
Introduction
One of the most challenging natural dangers to measure is a drought environment, which affects soil moisture content, evapotranspiration, atmospheric evaporative demand, and vegetation conditions (Vicente-Serrano et al., 2022). Plants sense the water depletion signal mainly via roots and leaves, and controlling stomatal aperture and closure, which involves some key signaling molecules, including plant hormones, transcription factors (TFs), protein amino acids, reactive oxygen species (ROS), and calcium ion (Ca2+) (Liu H et al., 2022; Qiu J et al., 2020; Sharma et al., 2019).
Drought triggers a cascade of physiological and biochemical responses in plants. Photosynthesis is the first mechanism impacted by drought stress, and drought considerably disrupted temperature sensitivities, CO2 assimilation rates, gas exchange water-use efficiency, and photosynthetic pigments (Fang et al., 2022; Razi and Muneer, 2021). The antioxidant system is essential for shielding plants from drought stresses via scavenging the ROS overproduced. Peroxidase (POD), superoxide dismutase (SOD), CAT, glutathione S-transferase (GST), and aldehyde dehydrogenase (ALDHs) have been found to respond to drought stress (Li R et al., 2022; Han L. et al., 2020). Drought stress renders the maintenance of osmoregulation highly crucial, which depends on the production and buildup of osmolytes including soluble proteins, proline, and sugars (Ozturk et al., 2020; Furlan et al., 2020). TFs are also central regulators of transcriptional reprogramming, and drought stress affected the expression of TF genes and may be involved in metabolite biosynthesis (Thirumalaikumar et al., 2017; Wang et al., 2021; Jadhao et al., 2023). For example, the phenolic acids and anthocyanins were antagonistically regulated by SmbHLH60 and SmMYC2 in Salvia miltiorrhiza; SmSPL7 inhibits the production of phenolic acid and facilitates the development of anthocyanins (Liu S et al., 2022; Chen R et al., 2021; Yao et al., 2022; Liu et al., 2021). Moreover, plants can produce hormones to respond to drought stress, which may be closely related to secondary metabolite synthesis (Jafari and Shahsavar, 2021), such as methyl jasmonate (MeJA) stimulating the accumulation of salvianolic acids and tanshinones and triggering the activation of genes in S. miltiorrhiza (Pei et al., 2018). Finally, drought stress induces the reduction of yield and quality in plants (Gervais et al., 2021; Han G et al., 2020; Hu et al., 2019). At present, most research focuses on the effects of drought stress on a single system, while the mechanism of multi-system joint resistance to drought stress remains unclear, particularly in the interaction of TFs, plant hormones, and secondary metabolites under drought stress.
S. miltiorrhiza is one of the most frequently used herbal remedies with 1,000 years of clinical application in China (Huang et al., 2021). The majority of research on S. miltiorrhiza primarily focuses on clinical application, pharmacological activity, and active compounds (Wang et al., 2020; Lu T et al., 2022; MEIm et al., 2019). In northwest China, S. miltiorrhiza is extensively cultivated and suffered from the drought environment throughout the year (Chen J et al., 2021). Most importantly, physiological systems, ROS scavenging systems, proline metabolism, TFs, and plant hormones in plants during each development stage are specifically sensitive to drought stress environment (Dietz et al., 2021). The research of drought-stressed S. miltiorrhiza focuses on a single system or tissue, lacking comprehensive studies throughout the multi-system and multi-plant tissues (Duan et al., 2023). Therefore, exploring the mechanism of the S. miltiorrhiza seedlings’ response to drought stress is crucial for optimizing their growth and yield.
At present, the correlations between MYB, NAC, tanshinones, and phenolic acids in S. miltiorrhiza determined through a combination of transcriptomic–metabolomic analysis have been reported (Li S et al., 2022; Yin et al., 2020), but the mechanism of the comprehensive drought resistance is still unclear and incomplete (Zhou et al., 2024). Consequently, in this experiment, to explore the drought response of S. miltiorrhiza seedlings, a comprehensive analysis of physiologies, transcriptomics, and metabolomics data was performed. We presumed that (i) drought stress can reduce the degree of photosynthesis and proline and glycine betaine metabolism, enhance oxidative damage, and inhibit the growth in S. miltiorrhiza seedlings (Altaf et al., 2022); (ii) the response of TFs and plant hormones to drought stress is closely related (Bhagat et al., 2021); and (iii) there are key gene regulations in the biosynthesis of tanshinones and phenolic acids under drought stress (Zhou et al., 2024). This study provides insight to comprehensively elucidate the molecular mechanism underlying the drought resistance of S. miltiorrhiza seedlings. This could serve as a theoretical foundation for further research on the molecular mechanism, breeding, cultivation, and genetic regulation of drought stress resistance in S. miltiorrhiza.
Materials and methods
Plant materials and treatments
Plants of S. miltiorrhiza were grown in a greenhouse under the following conditions: 22°C, light intensity 100 µmol m−2 s−1, 16/8 h light/dark cycle, and 60% relative humidity. S. miltiorrhiza seedlings with essentially the same growth trend were chosen in our study for treatment. A weighing method (saturation moisture content control) was used to simulate water-deficit stress. The experiment was conducted using a randomized design with the following treatments: (i) control group (CK), watering to 75% field capacity; (ii) drought group (CL), water content maintained at 45%. There were 320 seedlings in total and 160 seedlings in each group. After 15 days of treatment, the aerial parts (Y) and underground parts (G) of S. miltiorrhiza in each treatment group were collected separately (15 days based on the previous research of the research group) (Chen J et al., 2021). Some samples were used to measure growth and physiological and biochemical characteristics, and the remaining samples were promptly refrigerated at −80°C for transcriptome and metabolome sequencing.
Measurement of physiological and biochemical characteristics
The contents of chlorophyll, H2O2, superoxide (O2−), and proline (Pro), and the activities of SOD, POD, and CAT were determined using reagent kits (Beijing Boxbio Science and Technology Co., Ltd., product numbers 1012311035, 10123122110, 10123112010, 1012311145, 10123112850, 10123112030, and 10123112510). The determination was carried out strictly according to the product instruction.
Transcriptome analysis
Aerial and underground parts were collected from the same position on the S. miltiorrhiza seedlings in each treatment group. The RNA extraction, detection, and cDNA library construction and sequencing were completed by Beijing Nuohezhiyuan Technology Co., Ltd. (Beijing, China). Transcriptome determination was conducted on three biological replicates. The specific methods were as described by Cheng et al. (2023). After the library passed the quality inspection, it was sequenced on an Illumina HiSeq™ 4000 sequencing platform (San Diego, CA, USA). Fragments per kilobase of transcript sequence per millions of base pairs sequenced (FPKM) were used to evaluate the expression level of genes or transcripts. Genes with a fold change > 1 and p < 0.05 found via DESeq were determined to be differentially expressed. GO functional enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed using the clusterProfiler 4.2.0 software. Significance threshold for GO functional and KEGG pathways enrichment was both set at padj < 0.05. Gene set enrichment analysis (GSEA) set the threshold values with p < 0.05 and FDR < 0.25 using the local version of the GSEA tool (http://www.broadinstitute.org/gsea/index.jsp), and GO and KEGG datasets were used for GSEA independently. As for the correlation analysis of TFs and plant hormones, the differentially expressed genes (DEGs) encoding TFs and plant hormones were imported into the Cytoscape v3.9.1 software.
Metabolomics analysis
The aerial and underground parts of the S. miltiorrhiza seedlings in each treatment group were shipped to Beijing Nuohezhiyuan Technology to conduct qualitative and quantitative analyses of the broad-target metabolome in 24 samples using the LC-QTRAP platform. The specific methods were as described by Cheng et al. (2023). A grinder (MM 400, Retsch, Haan, Germany) was used to crush all tissues to powder (30 Hz, 1.5 min). After being weighed, 100 mg of powder was dissolved in 1.2 mL of 70% methanol. The samples were put at 4°C and vortexed six times, once every 30 min. A microporous membrane with a pore size of 0.22 μm was used to filter the samples after centrifugation (12,000 rpm, 10 min) and the supernatant was aspirated. The samples were then placed in a sample vial for analysis using ultraperformance liquid chromatography–tandem mass spectrometry (UPLC-MS/MS). The secondary spectrum data were used to characterize the metabolites based on a self-constructed database. During the analysis, the repeated and isotopic signals were eliminated. Triple-quadrupole mass spectrometry (TQ-MS) in the multiple reaction monitoring (MRM) mode was used to quantitatively examine the metabolites. Hierarchical cluster analysis (HCA) was used to examine the pattern of metabolite accumulation across the various samples using R software (www.r-project.org). Differentially expressed metabolites (DEMs) were defined as metabolites with VIP > 1 and p < 0.05 and fold change ≥ 2 or FC ≤ 0.5, and were annotated, classified, and analyzed by the KEGG database.
DEG and DEM network analysis
In order to conduct a correlation analysis, the sample sizes must first be consistent. In order to eliminate the impact of magnitude, the data were standardized. The correlation analysis between DEGs and DEMs is based on the Pearson statistical method, calculating the correlation coefficient r2 and p-value. In the correlation heatmap analysis, the top 50 DEGs and DEMs were determined (sorted by p-value from smallest to largest), the top 5 metabolites were selected from the metabolomics enrichment results and the top 10 genes from the transcriptomics, and then the mixOmics package in R software was used to draw a correlation network diagram.
Statistical analyses
All statistical analyses were performed using GraphPad 9.0 Statistics. Three replications were presented in all treatments; the specific methods of one-way analysis of variance (ANOVA), Duncan’s multiple range tests, and statistical significance were as described by Ming et al. (2023).
Results
General description of transcriptome data and metabolome data
The transcriptomic and metabolomic changes in S. miltiorrhiza seedlings under different groups were compared. Detailed statements of transcriptome data are presented in Supplementary Table S1. The 8,588 and 6,327 DEGs were screened in G_CL vs. G_CK groups (3,657 upregulated and 4,931 downregulated), and Y_CL vs. Y_CK groups (2,699 upregulated and 3,628 downregulated), respectively (Supplementary Figure S1A). The heatmap visualized distinct hierarchical clustering of genes, suggesting that these genes have different expression patterns in response to the drought stress (Supplementary Figure S1B). A heatmap of DEMs was constructed in two-ion mode, which clearly reflects the different impacts of drought stress on the aerial and underground parts of S. miltiorrhiza seedlings (Supplementary Figures S1C, D). Moreover, we identified 80 common DEMs between the G_CL vs. G_CK and Y_CL vs. Y_CK groups in positive ion mode, and 34 in negative ion mode (Supplementary Figures S1E, F).
Plant growth parameters and photosynthesis
The change of morphological characteristics in stressed S. miltiorrhiza seedlings was performed (Figure 1A). Relative to the control group, drought stress inhibited the growth of S. miltiorrhiza seedlings by decreasing fiber root length, chlorophyll b, total chlorophyll, and fresh weight, but chlorophyll a did not show significant changes (Figure 1B). The four genes encoding LHC (photosystem I complex) and one gene encoding ATP synthase were downregulated in the Y-CL vs. Y-CK comparison (Figures 1C, D). Moreover, metabolomic analysis showed that the content of NADP+ decreased in the Y-CL vs. Y-CK comparison (Figure 1C).
Figure 1
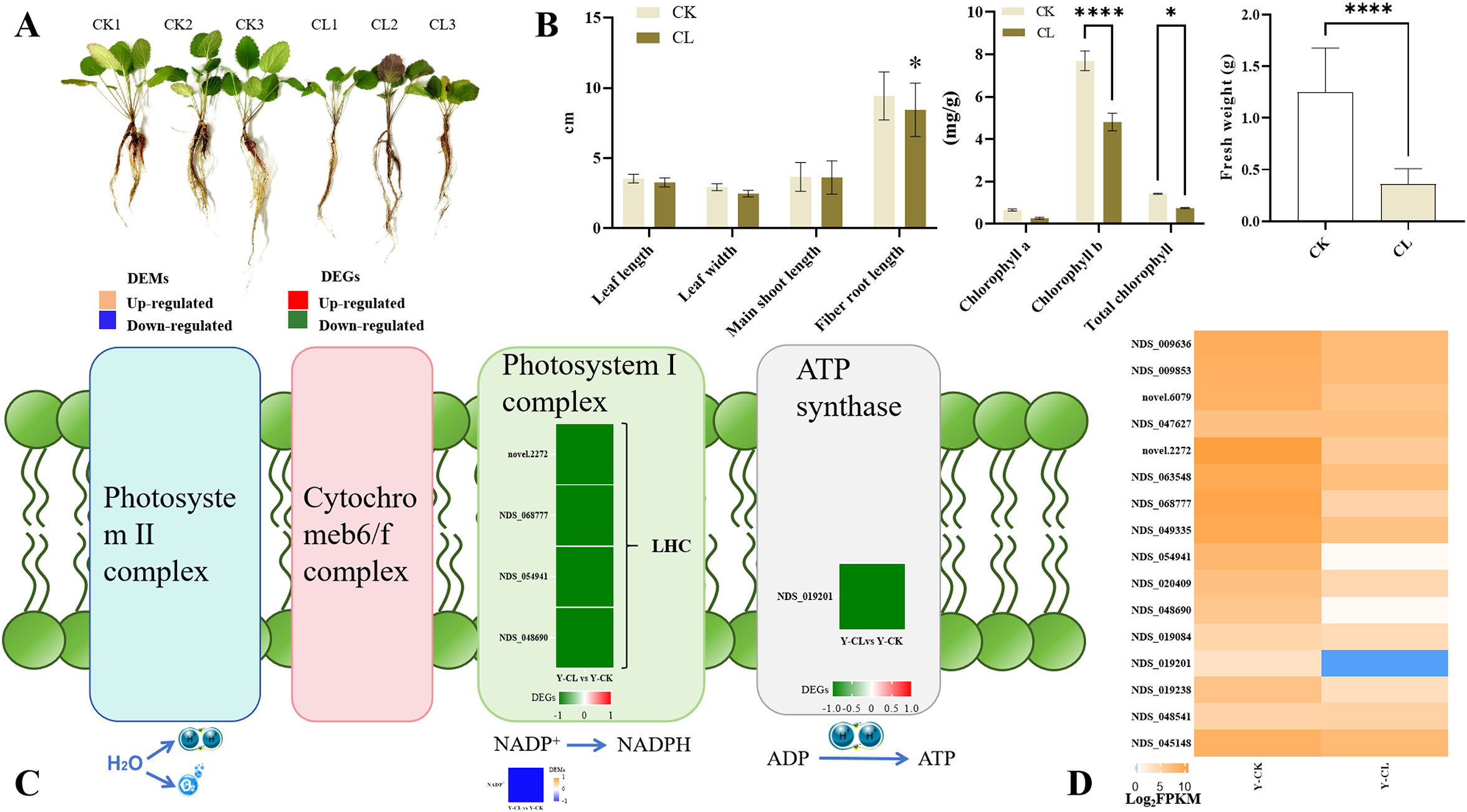
Effects of drought stress on growth and photosynthesis in S.miltiorrhiza seedling. (A) Growth situation in different conditions in S.miltiorrhiza seedlings. (B) Growth index and photosynthesis index in different conditions. (C) Expression patterns of the differentially expressed genes (DEGs) and differentially expressed metabolites (DEMs) related to photosynthesis. (D) Expression patterns of the genes related to photosynthesis. Values are the mean ± SE(n=3). Genes with p<0.05 and fold change≥1.5 were considered differentially expressed, metabolites with p<0.05, and variable importance in projection (VIP)>1, and fold change≥1 were considered differentially expressed. The “ * “ stands for the significant difference at p<0.05. The “ **** “ stands for the significant difference at p<0.001.
ROS scavenging process
The contents of O2− and H2O2 significantly increased in the G-CL vs. G-CK comparison, and the content of H2O2 also significantly increased in the Y-CL vs. Y-CK comparison (Figure 2A). The activity of CAT significantly decreased in the Y-CL vs. Y-CK comparison; however, the activity of POD significantly increased in the G-CL vs. G-CK comparison (Figure 2A). The genes of different groups encoding the ROS scavenging process were differentially expressed (Figures 2B, C). Two genes encoding SOD and five genes encoding POD were upregulated; inversely, eight genes encoding POD were downregulated in the G-CL vs. G-CK comparison and 1 gene encoding SOD was upregulated; in contrast, one gene encoding SOD and two genes encoding POD were downregulated in the Y-CL vs. Y-CK comparison (Figure 2B).
Figure 2
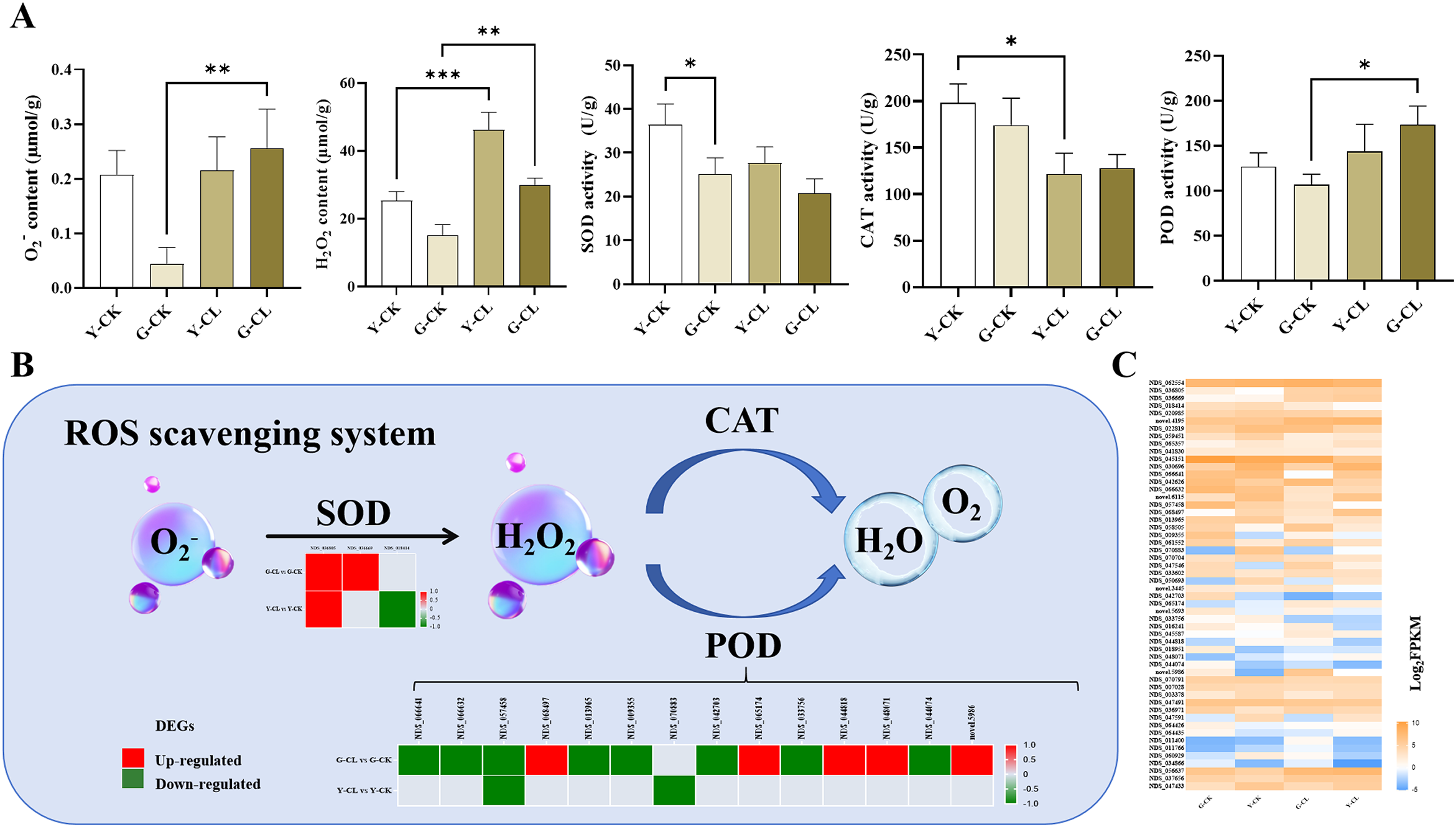
Effects of drought stress on ROS scavenging process in S.miltiorrhiza seedling. (A) the contents of ROS scavenging process markers and activities of the antioxidant enzymes. (B) Expression patterns of the DEGs related to antioxidant enzymes. (C) Expression patterns of the genes related to ROS scavenging process. Values are the mean ± SE(n=3). Genes with p<0.05 and fold change≥1.5 were considered differentially expressed. The “ * “ stands for the significant difference at p<0.05. The “ ** “ stands for the significant difference at p<0.01. The “ *** “ stands for the significant difference at p<0.005.
Proline and glycine betaine metabolism
Physiological data were combined with transcriptomic and metabolomic data to obtain a detailed pathway for proline and glycine betaine metabolism (Figure 3). The two genes encoding ProDH were downregulated in the G-CL vs. G-CK comparison, and one gene encoding ProDH was downregulated in the Y-CL vs. Y-CK comparison (Figure 3C). Moreover, the metabolomic analysis showed that the content of ornithine decreased in the G-CL vs. G-CK comparison, and the contents of glutamate (Glu) and serine decreased in the Y-CL vs. Y-CK comparison (Figure 3C). Inversely, the content of proline increased in both comparisons (Figure 3C).
Figure 3
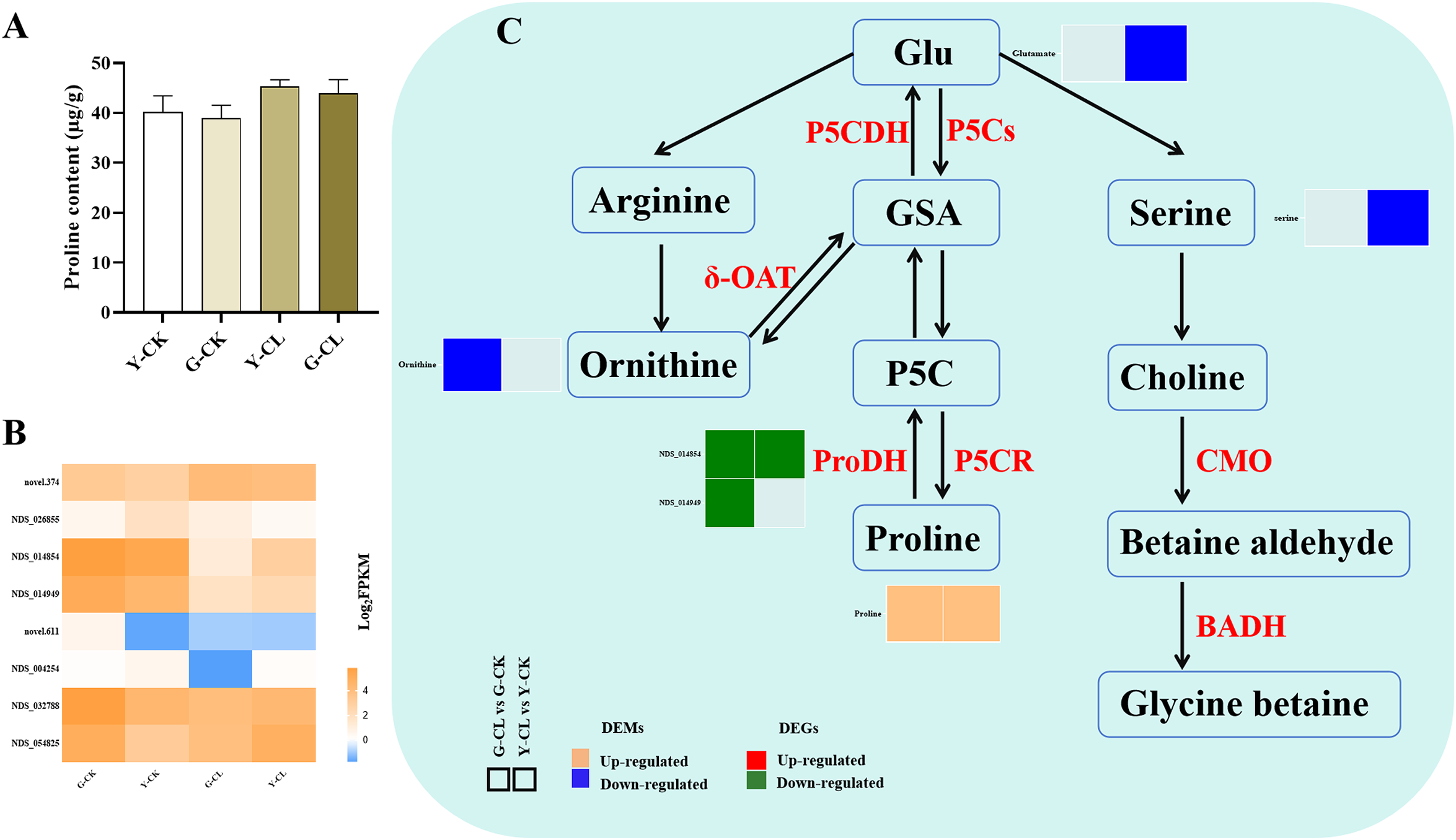
Effects of drought stress on proline and glycine betaine metabolism in S.miltiorrhiza seedling. (A) the content of proline. (B) Expression patterns of the genes related to proline and glycine betaine metabolism. (C) Expression patterns of the DEGs and DEMs related to proline and glycine betaine metabolism. Values are the mean ± SE (n=3). Genes with p<0.05 and fold change≥1.5 were considered differentially expressed, metabolites with p<0.05, and variable importance in projection (VIP)>1, and fold change≥1 were considered differentially expressed.
Transcription factors and plant hormones
TFs and plant hormones play an important regulatory role in plant responses to drought stress. In this study, a total of 383 TFs and 80 plant hormones were identified as hypothetical regulators of drought resistance in S. miltiorrhiza (Figure 4). TFs include 73 genes encoding basic helix-loop-helix (bHLHs), 25 genes encoding far-red impaired response 1 (FAR1s), 15 genes encoding TCP proteins (TCPs), 42 genes encoding basic region-leucine zipper (bZIPs), 43 genes encoding MYB domain proteins (MYBs), 53 genes encoding WRKY proteins (WRKYs), 85 genes encoding Cys3His zinc finger proteins (C3Hs), and 32 genes encoding NAC proteins (NACs) (Figure 4A). Plant hormones include 15 genes encoding abscisic acid (ABA), 9 genes encoding auxin, 2 genes encoding cytokinin (CTK), 1 gene encoding brassinolide (BR), and 53 genes encoding ethylene (Figure 4C).
Figure 4
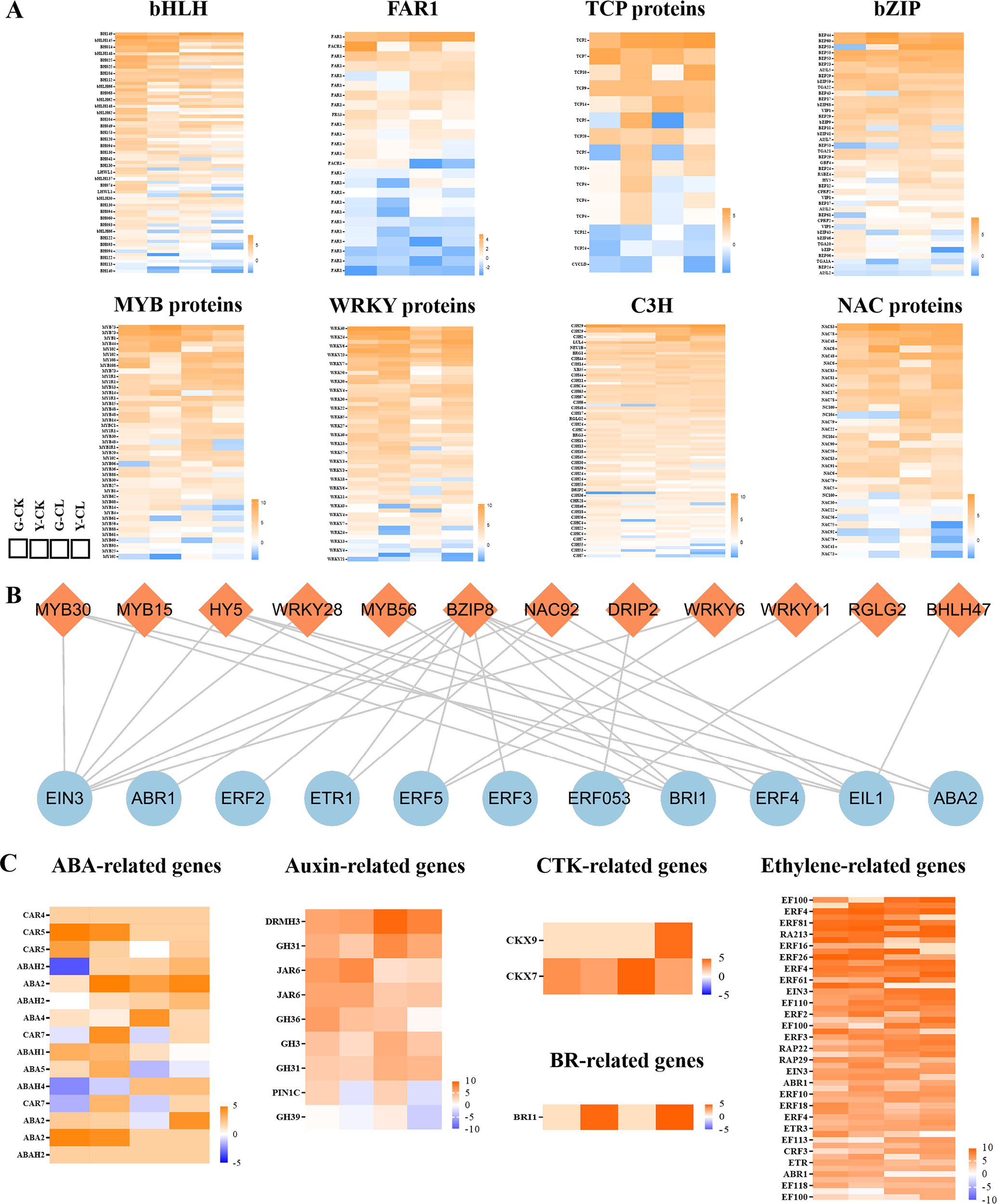
Effects of drought stress on transcription factors and plant hormones in S.miltiorrhiza seedling. (A) Expression patterns of the genes related to transcription factors. (B) the correlation of transcription factors and plant hormones. (C) Expression patterns of the genes related to plant hormones.
Moreover, the correlation of TFs and plant hormones was analyzed (Figure 4B). The genes encoding ethylene were correlated with genes encoding bZIP, bHLH, C3H, NAC, MYB, and WRKY. Genes encoding BR were correlated with genes encoding MYB and bZIP. In addition, genes encoding ABA were correlated with genes encoding bZIP.
Biosynthesis of tanshinones and phenolic acids
Figure 5 displays the tanshinones biosynthesis pathway. Transcriptomic analysis showed that the two genes encoding GGPPS and one gene encoding KSL were downregulated in the G-CL vs. G-CK comparison, respectively, and one gene encoding CYP76AK1 was upregulated in the Y-CL vs. Y-CK comparison.
Figure 5
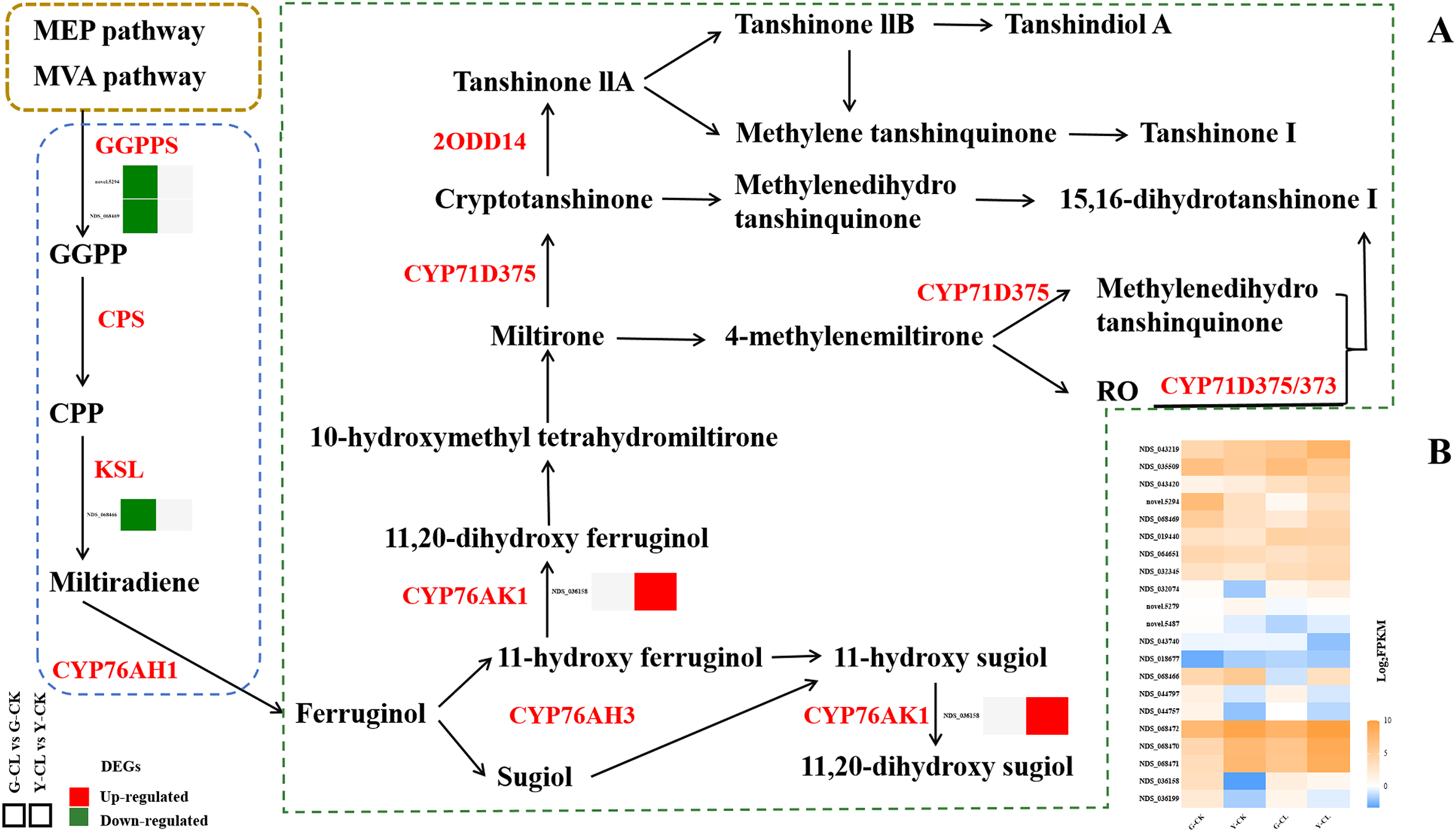
Effects of drought stress on tanshinones biosynthesis. (A) Expression patterns of the DEGs related to tanshinones biosynthesis. (B) Expression patterns of the genes related to tanshinones biosynthesis. Genes with p<0.05 and fold change≥1.5 were considered differentially expressed.
Figure 6 displays salvianolic acid B (Figures 6A, B) and phenolic acid (Figure 6C) pathways. Transcriptomic analysis showed that one gene encoding 4CL, one gene encoding CSE, two genes encoding CCR, and two genes encoding CAD were downregulated, but one gene encoding 4CL, two genes encoding HCT, one gene encoding CSE, two genes encoding CCR, two genes encoding RAS, and two genes encoding TAT were upregulated in the G-CL vs. G-CK comparison (Figure 6B). In the Y-CL vs. Y-CK comparison, one gene encoding HCT, one gene encoding CSE, and one gene encoding RAS were downregulated; inversely, one gene encoding 4CL and one gene encoding TAT were upregulated (Figure 6B). Moreover, the metabolomic analysis showed that the content of L-tyrosine, salvianolic acid D, danshensu, and caffeic acid increased, but the content of protocatechualdehyde decreased in the G-CL vs. G-CK comparison; the content of L-phenylalanine, L-tyrosine, and danshensu increased in the Y-CL vs. Y-CK comparison (Figures 6B, C).
Figure 6
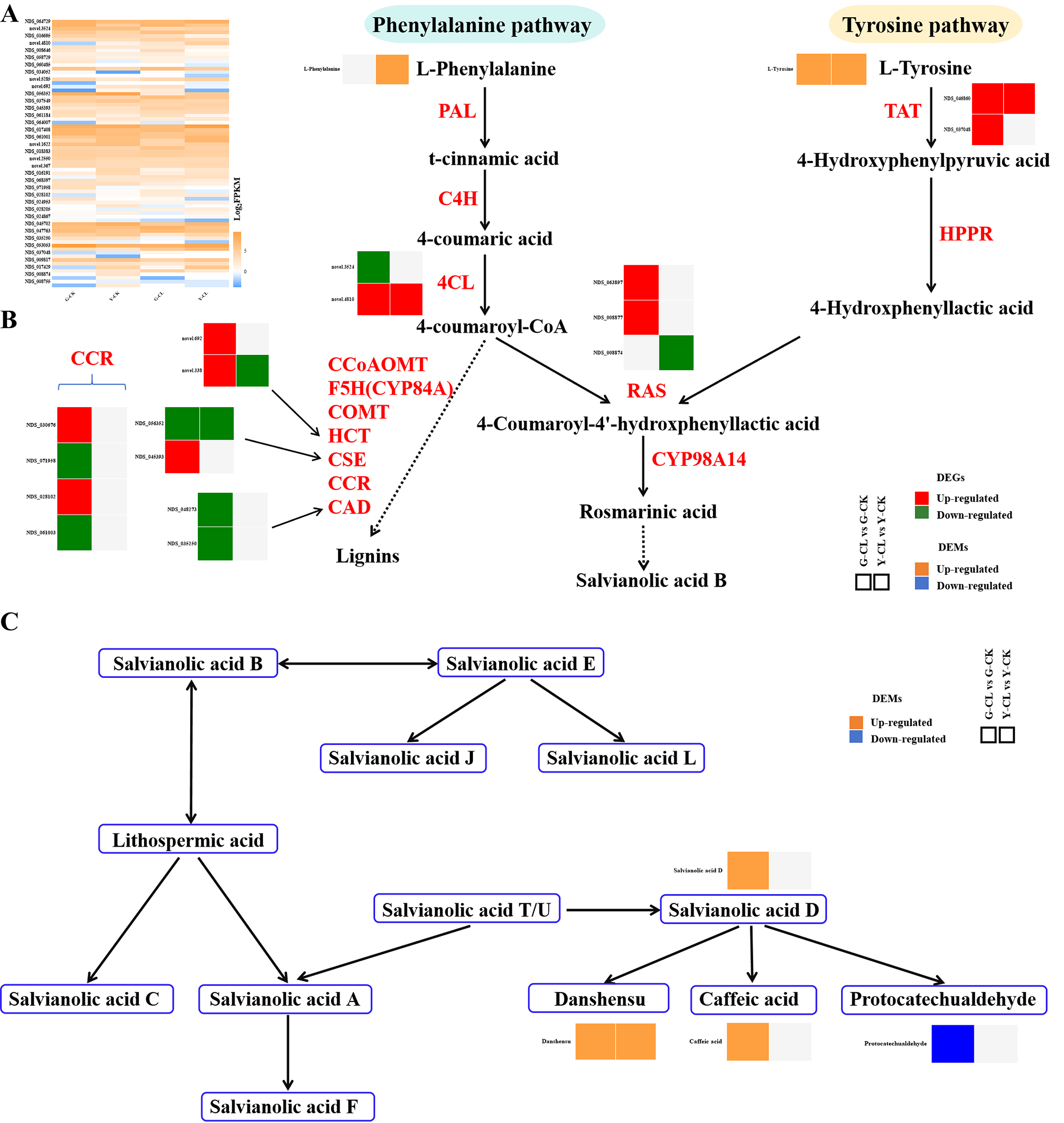
Effects of drought stress on phenolic acids biosynthesis. (A) Expression patterns of the genes related to phenolic acids biosynthesis. (B, C) Expression patterns of the DEGs and DEMs related to phenolic acids biosynthesis. Genes with p<0.05 and fold change≥1.5 were considered differentially expressed, metabolites with p<0.05, and variable importance in projection (VIP)>1, and fold change≥1 were considered differentially expressed.
The transcriptome and metabolome network analysis
In the G-CL vs. G-CK comparison (negative ion mode) (Figure 7A), danshensu was negatively correlated with 18 genes and positively correlated with 28 genes, with 1 gene encoding CYP450 (novel.1263) negatively correlated and 1 gene encoding PILS2 (NDS_047582) and 1 gene encoding MLO6 (NDS_068781) positively correlated; salvianolic acid D was negatively correlated with 18 genes and positively correlated with 30 genes. In the G-CL vs. G-CK comparison (positive ion mode) (Figure 7B), proline was negatively correlated with 18 genes and positively correlated with 29 genes, with 1 gene encoding CYP450 (novel.1263) negatively correlated and 1 gene encoding PILS2 (NDS_047582) and 1 gene encoding MLO6 (NDS_068781) positively correlated.
Figure 7
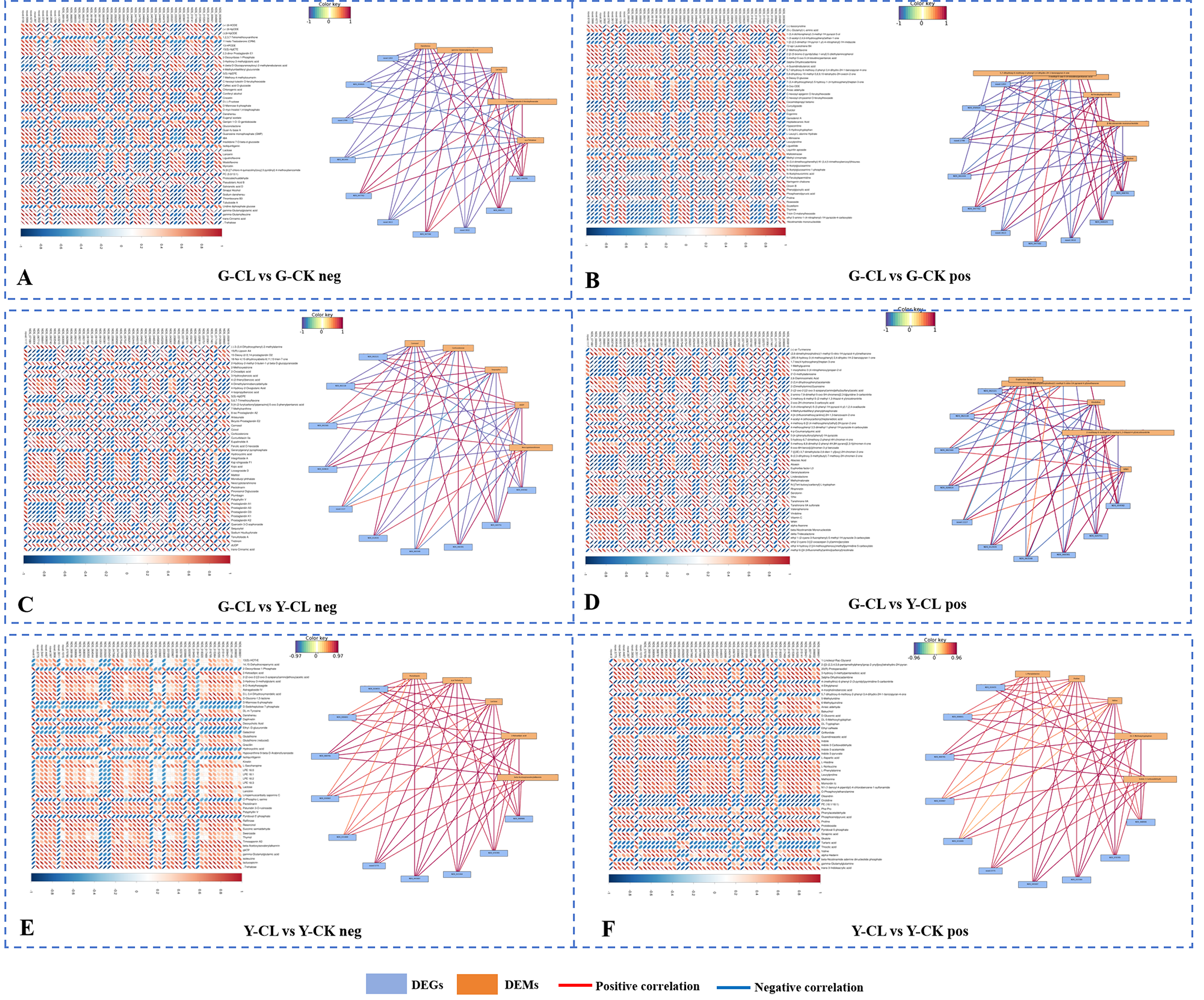
Association analysis of DEGs and DEMs. Heatmap of the expression of Top 50 DEGs (horizontal) and Top 50 DEMs (vertical) that differ in positive ion mode (positive) and negative ion mode (negative). (A, B) G-CL vs G-CK in negative and positive mode. (C, D) G-CL vs Y-CL in negative and positive mode. (E, F) Y-CL vs Y-CK in negative and positive mode. Network of the expression of Top 10 DEGs (blue) and Top 5 DEMs (yellow) that differ in positive ion mode (positive) and negative ion mode (negative). The positive correlation is red, and negative correlation is blue. Genes with p<0.05 and fold change≥1.5 were considered differentially expressed, metabolites with p<0.05, and variable importance in projection (VIP)>1, and fold change≥1 were considered differentially expressed. The p-values are ordered from smallest to largest.
In the G-CL vs. Y-CL comparison (negative ion mode) (Figure 7C), carnosol was negatively correlated with two genes encoding NLTP2 (NDS_062121; NDS_062118), one gene encoding Y1457 (NDS_020819), one gene encoding CB4A (NDS_063548), and one gene encoding GEDH1 (NDS_059582); inversely, carnosol was positively correlated with one gene encoding AAP3 (NDS_012939), one gene encoding AHL20 (NDS_060381), and one gene encoding PTR6Z (NDS_020751). Moreover, the correlation of neocryptotanshinone (NCTS) was the same as that of carnosol. In the G-CL vs. Y-CL comparison (positive ion mode) (Figure 7D), ABA was negatively correlated with 24 genes and positively correlated with 15 genes, and tanshinone IIA was negatively correlated with 24 genes and positively correlated with 21 genes.
In the Y-CL vs. Y-CK comparison (negative ion mode) (Figure 7E), danshensu was negatively correlated with 2 genes and positively correlated with 22 genes. α,α-trehalose was positively correlated with one gene encoding ADH1 (NDS_019455), one gene encoding EXLB1 (NDS_006601), one gene encoding MLO6 (NDS_068781), two genes encoding UGT2 (NDS_030967 and NDS_031009), one gene encoding amino acid permease 7 (novel.5775), one gene encoding TDT (NDS_045007), one gene encoding HSP7C (NDS_015364), one gene encoding EDL3 (NDS_070785), and one gene encoding SKU5 (NDS_068906). In the Y-CL vs. Y-CK comparison (positive ion mode) (Figure 7F), proline was negatively correlated with 5 genes and positively correlated with 15 genes. In particular, the correlation results of proline and valine were the same as that of α,α-trehalose.
GO and KEGG enrichment analysis
The GO terms “oxidoreductase activity” and “transcription regulator activity” were highly enriched among gene sets in the G_CL vs. G_CK group, and the GO terms “photosystem”, “photosynthetic membrane”, and “thylakoid part” were highly enriched among gene sets in Y_CL vs. Y_CK (Supplementary Figures S2A, B). The KEGG terms “Plant hormone signal transduction”, “MAPK signaling pathway - plant”, and “Peroxisome” were the significantly enriched different genes in the G_CL vs. G_CK group, and the KEGG terms “Plant hormone signal transduction”, “MAPK signaling pathway-plant”, “Photosynthesis”, and “Photosynthesis - antenna proteins” were the significantly enriched different genes in Y_CL vs. Y_CK (Supplementary Figures S2C, D).
Gene set enrichment analysis
GSEAs of the photosynthesis pathway (ATH00195), peroxisome pathway (ATH04146), MAPK signaling pathway (ATH04016), plant hormone signal transduction pathway (ATH04075), and biosynthesis of secondary metabolites pathway (ATH01110) were performed to ascertain the function changes in response to drought stress in S. miltiorrhiza seedlings (Figure 8). In the Y-CL vs. Y-CK comparison, the photosynthesis pathway was significantly enriched and downregulated (Figure 8A). However, peroxisome, MAPK signaling pathway, plant hormone signal transduction, and biosynthesis of secondary metabolites pathways were overall upregulated in two comparisons (Figures 8B-G).
Figure 8
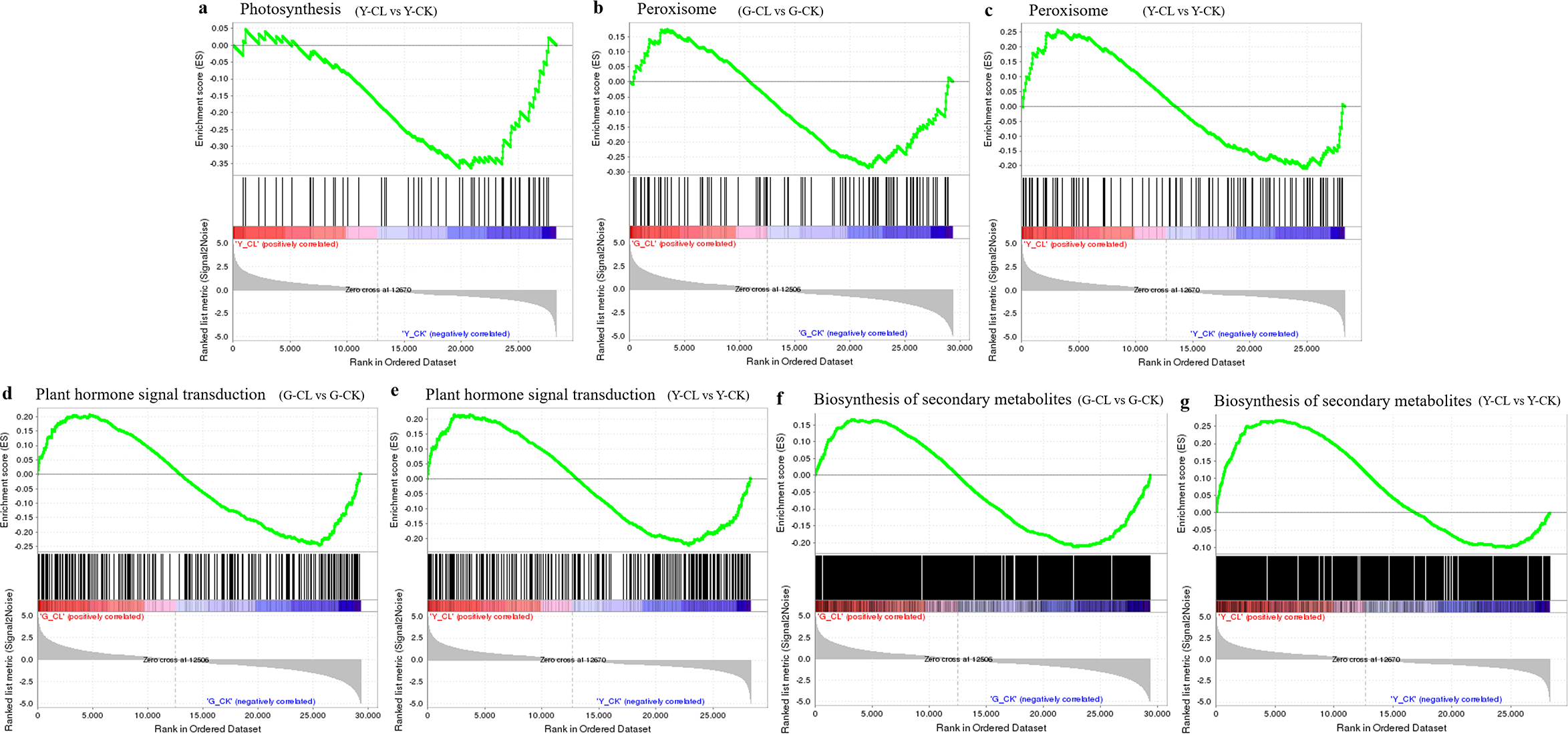
GSEA analysis of photosynthesis (A), peroxisome (B, C), plant hormone signal transduction (D, E), and biosynthesis of secondary metabolites pathway (F, G) in different treatments with p<0.05, FDR <0.25.
Discussion
Drought stress affects growth indexes and photosynthesis of S. miltiorrhiza seedlings
During plant cultivation, drought stress affects morphological and physiological changes in plants, which has a great impact on its growth and productivity (Lim et al., 2022). In this research, drought stress decreased fiber root length and fresh weight in S. miltiorrhiza seedlings (Figure 1B), so we predicted that root morphology would be firstly negatively affected under drought stress (Verbraeken et al., 2021; Gervais et al., 2021; Altaf et al., 2022). Moreover, drought stress decreased the contents of chlorophyll b, total chlorophyll, and NADP+, and downregulated the genes of encoding LHC and ATP synthase (Figures 1B, C), which were similar to the results of the GO, KEGG, and GSEA (Figure 8A; Supplementary Figures S2B, D). PSI and the ATP synthase are embedded where solar energy is collected and converted into NADPH and ATP (Jonwal et al., 2022). LHC proteins collect light energy and photoprotection and regulate chlorophyll synthesis in plants (Rochaix and Bassi, 2019). Therefore, in our research, drought stress inhibited the expression of genes encoding LHC and NADP+ supply, reduced light harvest, and thereby decreased the content of chlorophyll, which slowed down the growth of S. miltiorrhiza seedlings (Zhang et al., 2022).
Drought stress affects the ROS scavenging process of S. miltiorrhiza seedlings
Plants benefit from the ROS scavenging mechanism during drought stress, because it allows them to modify their metabolism and mount an appropriate acclimation response (Gao et al., 2022). Our KEGG and GSEA results showed that the peroxisome pathway was significantly enriched and upregulated (Figures 8B, C; Supplementary Figure S2C), which is consistent with the changes of SOD and POD (Figures 2A, B), and may be involved in ROS detoxification by accelerating the dismutation of O2− into H2O2 and scavenging the H2O2 under drought stress mainly by the increase in PODs rather than CATs (Ritonga et al., 2021; Balfagón et al., 2021; Hou et al., 2021). Interestingly, the results of transcriptomic analysis and enzyme activity on POD in the G-CL vs. G-CK comparison were not consistent (Figures 2A, B), indicating that these genes might not be key genes for the regulation of POD activity.
Drought stress affects the proline and glycine betaine metabolism of S. miltiorrhiza seedlings
Drought stress renders the maintenance of osmoregulation highly vital (Tan et al., 2022). Specifically, biosynthesis of proline and glycine betaine in plants occurs through Glu pathways, catalyzed by three pathways (Figure 3C). In our study, drought stress increased the content of proline and downregulated the expression of genes encoding ProDH (Figure 3C); the result might be due to drought stress mainly generating pyrroline-5-carboxylate (P5C) and then further reduced by P5CR to generate proline, rather than proline degradation pathway by ProDH (Ingrisano et al., 2023). Moreover, the content of ornithine and serine decreased due to drought stress (Figure 3C), and it was further proved that drought stress mainly provided sufficient substrate for proline by GSA, and the decrease in the ornithine and serine content might be due to one part being used to synthesize proline (Shafiq et al., 2021). Interestingly, we found in the G-CL vs. G-CK comparison (positive ion mode) (Figure 7B) that proline was positively correlated with one gene encoding PILS2 (NDS_047582). PILS can give a homeostatic feedback on the auxin signaling pathway, and proline and auxin signal transduction pathways are also crucial mechanisms for coping with drought stress, probably indicating that proline and auxin combined to respond to drought stress in S. miltiorrhiza seedlings (Feraru et al., 2022).
Drought stress affects the transcription factors and plant hormones of S. miltiorrhiza seedlings
Stress-responsive genes are expressed due to TFs, which are a crucial component of the adaptive stress process (Lu X et al., 2022; Ju et al., 2019; Joo et al., 2021; Han G et al., 2020; Xuan et al., 2022). In our study, bHLHs, WRKYs, and C3Hs are more sensitive to drought stress in S. miltiorrhiza seedlings (Figure 4A), because of which bHLHs, WRKYs, and C3Hs could regulate cellular activities and membrane-lipid stability, increase drought tolerance, and promote root growth and water retention (Qiu X. et al., 2020; Xiang et al., 2021). In addition, plant hormone homeostasis is critical to regulating plant growth and development under stress conditions, and multiple levels are regulated (Zhang et al., 2023; Wang and Qiao, 2020; Li et al., 2020; Waadt et al., 2022). In our study, drought stress regulated expression genes of ethylenes, ABAs, auxins, CTKs, and BRs, of which ABAs and ethylenes were encoded the most (Figure 4C), which was consistent with KEGG and GSEA results (Figures 8D, E; Supplementary Figure S2C). ABA and ethylene play key roles in mediating vessel size, cell wall thickness, water loss, and oxidative damage in plant responses to drought stress (Gambhir et al., 2022; Auler et al., 2020; Kong et al., 2023).
Interestingly, the correlation results of TFs and plant hormones showed that NAC92 was correlated with EIN3, ETR1, and EIL1 (Figure 4B), which were involved in regulating chlorophyll metabolism, the carotenoid biosynthetic process, and ethylene pathways (He et al., 2022; Luo et al., 2019). We found that EIN3 and EIL1 can bind to several gene members of MYB and bHLH (Figure 4B), which may mainly the regulate ethylene signaling pathway, thereby promoting root hair formation (Wen et al., 2023; Song et al., 2022). Moreover, we found that bZIP8 may be a key gene, because it involves most genes encoding plant hormones (Figure 4B), but little was known. Currently, it is known that BRI1 and ABAs are related to bZIP8 and regulate cell communication, membrane signaling, plant development, photomorphogenesis, flavonoid biosynthesis, nutrient acquisition, and response to drought stress (Lozano-Elena and Caño-Delgado, 2019; Bhagat et al., 2021; Xiao et al., 2022; Nie et al., 2022). According to these reports and our study results, the combination of key TFs and plant hormones might play a specific role in drought-stressed S. miltiorrhiza seedlings. These key genes are involved in regulating seedling growth, photosynthesis, the ROS scavenging system, proline metabolism, etc., and they are worthy of further research and development.
Drought stress affects the biosynthesis of tanshinones and phenolic acids of S. miltiorrhiza seedlings
Tanshinones (liposoluble) and phenolic acids (water-soluble) are a series of crucial compounds in S. miltiorrhiza (Shi et al., 2022). In our study, we only found that drought stress downregulated some genes encoding GGPPS and KSL in the tanshinones biosynthesis pathway (Figure 5); this result indicates that the effect of the tanshinones biosynthesis pathway in the present seedlings growth stage is relatively minimal, or the effect of drought stress on the tanshinones biosynthesis pathway is not significant (Loyola et al., 2011). Surprisingly, in the G-CL vs. Y-CL comparison (negative ion mode) (Figure 7C), we found that NCTS was correlated with genes encoding NLTP2, Y1457, CB4A, GEDH1, AAP3, AHL20, and PTR6Z, which related to the salicylic acid biosynthesis pathway and nutrient transport (Silverio-Gómez et al., 2021; Zhang et al., 2021). At present, the effect of drought stress on NCTS and AHL gene family has not been reported in S. miltiorrhiza seedlings (Ma et al., 2023). Studies have shown that NCTS is a diterpenoid with a similar structure to these tanshinones, and it has anti-inflammatory and blood-activating effects (Yang et al., 2022). AHL family members play a critical role in stress resistance regulation by DNA and protein interactions in plant biological processes (Zhao et al., 2020; Kumar et al., 2023). Therefore, the finding of NCTS and AHL gene family can provide more evidence and potential on the tanshinones pathway and growth and development in S. miltiorrhiza.
The phenolic acids biosynthesis pathway was significantly regulated under drought stress (Figure 6). Interestingly, in the present study, drought stress was significantly involved in lignin synthesis, indicating that lignin synthesis was sensitive to drought stress and lignin deposition in the plant secondary cell wall as a protection to drought attacks (Sharma et al., 2023). Moreover, genes encoding RAS upregulated in the G-CL vs. G-CK comparison (Figure 6B), which might accelerate the synthesis of salvianolic acid B in S. miltiorrhiza and upregulate the genes’ expression encoding TAT in the tyrosine pathway (Zhong et al., 2023; Wagay et al., 2023). In our study, the contents of salvianolic acid D, danshensu, and caffeic acid increased in the G-CL vs. G-CK comparison (Figure 6C), indicating that correlations were observed between drought stress and salvianolic acid accumulation, and this chemical is also the most responsive to drought stress. Above all, the secondary metabolite pathway was significantly enriched in two comparisons and was upregulated (Figures 8F, G). Interestingly, danshensu was negatively correlated with one gene encoding CYP450 (novel.1263) in the G-CL vs. G-CK comparison (negative ion mode) (Figure 7A), but CYPs played a continuous catalytic role in the tanshinones biosynthesis pathway (Mao et al., 2020); therefore, this gene could redirect both salvianolic acids and tanshinones biosynthesis possibly through bidirectional regulation. Danshensu was positively correlated with one gene encoding PILS2 (NDS_047582) and one gene encoding MLO6 (NDS_068781) (Figure 7A), which indicates that drought stress may produce the change of network system related to gene PILS, auxin, and danshensu (Bogaert et al., 2021; Li Q. et al., 2022). In addition, we also found that salvianolic acid D was negatively correlated with 18 genes and positively correlated with 30 genes (Figure 7A). However, at present, there are only a few reports on these genes and phenolic acids in plants; their response to drought stress in S. miltiorrhiza seedlings needs to be clarified in future research.
Conclusion
This study found that S. miltiorrhiza seedlings can quickly regulate gene expression, enzyme activity, and metabolism content, which is involved in photosynthesis, the ROS scavenging process, proline and glycine betaine metabolism, TFs, plant hormones, biosynthesis of tanshinones and phenolic acids, and adapting to drought stress conditions. We found that bZIP8 may be a key gene as it involves most genes encoding plant hormones, which regulate cell communication, membrane signaling, plant development, and response to drought stress. The relationship between NCTS and the AHL gene family plays pivotal roles in S. miltiorrhiza’s response to drought stress at the tanshinones pathway and various growth stages. These findings substantially enhance our understanding of the mechanisms underlying S. miltiorrhiza’s response to drought stress and provide new insights into the functions of relevant genes and metabolites in drought tolerance.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
YZ: Writing – original draft. HZ: Investigation, Writing – review & editing. YRZ: Formal analysis, Writing – review & editing. DW: Visualization, Writing – review & editing. XM: Data curation, Writing – review & editing. JC: Methodology, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Key Research and Development Program of Shaanxi (Program Nos. 2023-YBSF-471/491, 2023-YBSF-471/491, and 2024SF-YBXM-470), the project of Shaanxi Administration of Traditional Chinese Medicine (Program Nos. SZY-KJCYC-2023-052 and SZY-KJCYC-2023-026/052), the Natural Science Foundation of Shaanxi Province (2019JQ-995), and the Xi’an S&T Plan Project (22YXYJ0114).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1484688/full#supplementary-material
References
1
Altaf M. A. Shahid R. Ren M. X. Naz S. (2022). Melatonin improves drought stress tolerance of tomato by modulating plant growth, root architecture, photosynthesis, and antioxidant defense system. Antioxidants11, 309–324. doi: 10.3390/antiox11020309
2
Auler P. A. Nogueira A. M. Rossatto T. Lopez C. R. Milech C. (2020). Metabolism of abscisic acid in two contrasting rice genotypes submitted to recurrent water deficit. Physiol. Plantarum172, 304–316. doi: 10.1111/ppl.13126
3
Balfagón D. Terán F. de O. Tadeu D. R. Santa-Catarina C. Gómez-Cadenas A. (2021). Citrus rootstocks modify scion antioxidant system under drought and heat stress combination. Plant Cell Rep.41, 593–602. doi: 10.1007/s00299-021-02744-y
4
Bhagat P. K. Verma D. Sharma D. Sinha A. K. (2021). HY5 and ABI5 transcription factors physically interact to fine tune light and ABA signaling in Arabidopsis. Plant Mol. Biol.107, 117–127. doi: 10.1007/s11103-021-01187-z
5
Bogaert K. A. Blomme J. Beeckman T. De Clerck O. (2021). Auxin’s origin: do PILS hold the key? Trends Plant Sci.27, 227–236. doi: 10.1016/j.tplants.2021.09.008
6
Chen J. Li N. Wang X. Meng X. Cui X. Chen Z. et al . (2021). Late embryogenesis abundant (LEA) gene family in Salvia miltiorrhiza: identification, expression analysis, and response to drought stress. Plant Signal. Behav.16, 1–10. doi: 10.1080/15592324.2021.1891769
7
Chen R. Cao Y. Wang W. Li Y. Wang D. (2021). Transcription factor SmSPL7 promotes anthocyanin accumulation and negatively regulates phenolic acid biosynthesis in Salvia miltiorrhiza. Plant Sci.310. doi: 10.1016/j.plantsci.2021.110993
8
Cheng M. Zheng J. Cui K. Luo X. Yang T. Pan Z. et al . (2023). Transcriptomics integrated with metabolomics provides a new strategy for mining key genes in response to low temperature stress in Helictotrichon virescens. Int. J. Biol. Macromol.242, 125070. doi: 10.1016/j.ijbiomac.2023.125070
9
Dietz K. J. Zörb C. Geilfus C. M. (2021). Drought and crop yield. Plant Biol.23, 881–893. doi: 10.1111/plb.13304
10
Duan S. Zhang M. Zeng H. Song J. Zhang M. Gao S. et al . (2023). Integrated proteomics and phosphoproteomics profiling reveals the cardioprotective mechanism of bioactive compounds derived from Salvia miltiorrhiza Burge. Phytomedicine117, 154897. doi: 10.1016/j.phymed.2023.154897
11
Fang L. Yin X. van der Putten P. E. L. Martre P. Struik P. C. (2022). Drought exerts a greater influence than growth temperature on the temperature response of leaf day respiration in wheat (Triticum aestivum). Plant Cell Environ.45, 2062–2077. doi: 10.1111/pce.14324
12
Feraru E. Feraru M. I. Moulinier-Anzola J. Schwihla M. Ferreira Da Silva Santos J. (2022). PILS proteins provide a homeostatic feedback on auxin signaling output. Development. 149, dev200929. doi: 10.1242/dev.200929
13
Furlan A. L. Bianucci E. Giordano W. Castro S. Becker D. F. (2020). Proline metabolic dynamics and implications in drought tolerance of peanut plants. Plant Physiol. Bioch.151, 566–578. doi: 10.1016/j.plaphy
14
Gambhir P. Singh V. Parida A. Raghuvanshi U. Kumar R. (2022). Ethylene response factor ERF.D7 activates auxin response factor 2 paralogs to regulate tomato fruit ripening. Plant Physiol. 190, 2775–2796. doi: 10.1093/plphys/kiac441
15
Gao H. Cui J. Liu S. Wang S. Lian Y. (2022). Natural variations of ZmSRO1d modulate the trade-off between drought resistance and yield by affecting ZmRBOHC-mediated stomatal ROS production in maize. Mol. Plant. 15, 1558–1574. doi: 10.1016/j.molp.2022.08.009
16
Gervais T. Creelman A. Li X. Q. Bizimungu B. De Koeyer D. Dahal K. (2021). Potato response to drought stress: physiological and growth basis. Front. Plant Sci.12. doi: 10.3389/fpls.2021.698060
17
Han L. M. Hua W. P. Cao X. Y. Yan J. A. Chen C. (2020). Genome-wide identification and expression analysis of the superoxide dismutase (SOD) gene family in Salvia miltiorrhiza. Gene742, 144603. doi: 10.1016/j.gene.2020.144603
18
Han G. Lu C. Guo J. Qiao Z. Sui Na. (2020). C2H2 zinc finger proteins: master regulators of abiotic stress responses in plants. Front. Plant Sci.154, 1114. doi: 10.3389/fpls.2020.00115
19
He C. Liew L. C. Yin L. Lewsey M. G. (2022). The retrograde signaling regulator ANAC017 recruits the MKK9-MPK3/6, ethylene, and auxin signaling pathways to balance mitochondrial dysfunction with growth. Plant Cell34, 3460–3481. doi: 10.1093/plcell/koac177
20
Hou P. Wang F. Luo B. Li A. (2021). Antioxidant enzymatic activity and osmotic adjustment as components of the drought tolerance mechanism in. Plants-Basel10, 436–456. doi: 10.3390/plants10030436
21
Hu W. Liu Y. Loka D. A. Zahoor R. Wang S. (2019). Drought limits pollen tube growth rate by altering carbohydrate metabolism in cotton (Gossypium hirsutum) pistils. Plant Sci.286, 108–117. doi: 10.1016/j.plantsci.2019.06.003
22
Huang Y. Yu S. H. Zhen W. X. Cheng T. Wang D. Lin J. B. et al . (2021). Tanshinone I, a new EZH2 inhibitor restricts normal and Malignant hematopoiesis through upregulation of MMP9 and ABCG2. Theranostics11, 6891–6904. doi: 10.7150/thno.53170
23
Ingrisano R. Tosato E. Trost P. Gurrieri L. Sparla F. (2023). Proline, cysteine and branched-chain amino acids in abiotic stress response of land plants and microalgae. Plants-Basel12, 3410–3425. doi: 10.3390/plants12193410
24
Jadhao K. R. Kale S. S. Chavan N. S. Janjal P. H. (2023). Genome-wide analysis of the SPL transcription factor family and its response to water stress in sunflower (Helianthus annuus). Cell Stress Chaperon.28, 943–958. doi: 10.1007/s12192-023-01388-z
25
Jafari M. Shahsavar A. (2021). The effect of foliar application of melatonin on changes in secondary metabolite contents in two species under drought stress conditions. Front. Plant Sci.12. doi: 10.3389/fpls.2021.692735
26
Jonwal S. Verma N. Sinha A. K. (2022). Regulation of photosynthetic light reaction proteins via reversible phosphorylation. Plant Sci. 321, 111312. doi: 10.1016/j.plantsci.2022.111312
27
Joo H. Baek W. Lim C. W. Lee S. C. (2021). Post-translational modifications of bZIP transcription factors in abscisic acid signaling and drought responses. Curr. Genomics22, 4–15. doi: 10.2174/1389202921999201130112116
28
Ju Y. L. Yue X. F. Min Z. Wang X. H. Fang Y. L. (2019). VvNAC17, a novel stress-responsive grapevine (Vitis vinifera L.) NAC transcription factor, increases sensitivity to abscisic acid and enhances salinity, freezing, and drought tolerance in transgenic Arabidopsis. Plant Physiol. Biochem.146, 98–111. doi: 10.1016/j.plaphy.2019.11.002
29
Kong H. Hou M. Ma B. Xie Z. Wang J. (2023). Calcium-dependent protein kinase GhCDPK4 plays a role in drought and abscisic acid stress responses. Plant Sci.332, 111704. doi: 10.1016/j.plantsci.2023.111704
30
Kumar A. Singh S. Mishra A. (2023). Genome-wide identification and analyses of the AHL gene family in rice (Oryza sativa). 3 Biotech.13, 248. doi: 10.1007/s13205-023-03666-0
31
Li Qi Fang X. Zhao Y. Cao R. Dong J. (2022). The SmMYB36-SmERF6/SmERF115 module regulates the biosynthesis of tanshinones and phenolic acids in hairy roots. Hortic. Res.10, uhac238. doi: 10.1093/hr/uhac238
32
Li S. Lin D. Zhang Y. (2022). Genome-edited powdery mildew resistance in wheat without growth penalties. Nature602, 455–460. doi: 10.1038/s41586-022-04395-9
33
Li R. Su X. Zhou R. Zhang Y. Wang T. (2022). Molecular mechanism of mulberry response to drought stress revealed by complementary transcriptomic and iTRAQ analyses. BMC Plant Biol.22, 36–54. doi: 10.1186/s12870-021-03410-x
34
Li S. M. Zheng H. X. Zhang X. S. Sui N. (2020). Cytokinins as central regulators during plant growth and stress response. Plant Cell Rep.40, 271–282. doi: 10.1007/s00299-020-02612-1
35
Lim C. Kang K. Shim Y. Yoo S. C. Paek N. C. (2022). Inactivating transcription factor OsWRKY5 enhances drought tolerance through abscisic acid signaling pathways. Plant Physiol.188, 1900–1916. doi: 10.1093/plphys/kiab492
36
Liu F. Ali T. Liu Z. (2021). Comparative transcriptomic analysis reveals the effects of drought on the biosynthesis of methyleugenol in asarum sieboldii miq. Biomolecules11, 1233–1249. doi: 10.3390/biom11081233
37
Liu H. Song S. Zhang H. Li Y. Niu L. Zhang J. et al . (2022). Signaling transduction of ABA, ROS, and ca2+ in plant stomatal closure in response to drought. Int. J. Mol. Sci.23, 14824. doi: 10.3390/ijms232314824
38
Liu S. Wang Y. Shi M. Maoz I. Gao X. (2022). SmbHLH60 and SmMYC2 antagonistically regulate phenolic acids and anthocyanins biosynthesis in Salvia miltiorrhiza. J. Adv. Res.42, 205–219. doi: 10.1016/j.jare.2022.02.005
39
Loyola J. Verdugo I. González E. Casaretto J. A. Ruiz-Lara S. (2011). Plastidic isoprenoid biosynthesis in tomato: physiological and molecular analysis in genotypes resistant and sensitive to drought stress. Plant Biol.14, 149–156. doi: 10.1111/j.1438-8677.2011.00465.x
40
Lozano-Elena F. Caño-Delgado A. I. (2019). Emerging roles of vascular brassinosteroid receptors of the BRI1-like family. Curr. Opin. Plant Biol.51, 105–113. doi: 10.1016/j.pbi.2019.06.006
41
Lu T. C. Wu Y. H. Chen W. Y. Hung Y. C. (2022). Targeting oxidative stress and endothelial dysfunction using tanshinone IIA for the treatment of tissue inflammation and fibrosis. Oxid. Med. Cell Longev.2022, 2811789. doi: 10.1155/2022/2811789
42
Lu X. Zhang H. Hu J. Nie G. Khan I. (2022). Genome-wide identification and characterization of bHLH family genes from orchardgrass and the functional characterization of DgbHLH46 and DgbHLH128 in drought and salt tolerance. Funct. Integr. Genomics22, 1331–1344. doi: 10.1007/s10142-022-00890-4
43
Luo F. Cai J. H. Kong X. M. Zhou Q. Zhou X. (2019). Transcriptome profiling reveals the roles of pigment mechanisms in postharvest broccoli yellowing. Hortic. Res.6, 74–88. doi: 10.1038/s41438-019-0155-1
44
Ma L. Shao M. Cheng W. Jiang J. Chen X. (2023). Neocryptotanshinone ameliorates insufficient energy production in heart failure by targeting retinoid X receptor alpha. Biomed. Pharmacother.163, 114868. doi: 10.1016/j.biopha.2023.114868
45
Mao Y. Ma Y. Chen T. Ma X. Xu Y. (2020). Functional integration of two CYP450 genes involved in biosynthesis of tanshinones for improved diterpenoid production by synthetic biology. ACS Synth. Biol.9, 1763–1770. doi: 10.1021/acssynbio.0c00136
46
MEIm X. D. Cao Y. F. Che Y. Y. Li J. Shang Z. P. Zhao W. J. et al . (2019). Danshen: a phytochemical and pharmacological overview. Chin. J. Nat. Med.17, 59–80. doi: 10.1016/S1875-5364(19)30010-X
47
Ming F. Enhe Z. Fengxia Q. G. (2023). Silicon improves the growth of Glycyrrhiza uralensis fisch. under salt and/or drought stress by regulating respiration metabolism. Plant Growth Regul.101, 743–767. doi: 10.1007/s10725-023-01054-5
48
Nie K. Zhao H. Wang X. Niu Y. Zhou H. (2022). The MIEL1-ABI5/MYB30 regulatory module fine tunes abscisic acid signaling during seed germination. J. Integr. Plant Biol.64, 930–941. doi: 10.1111/jipb.13234
49
Ozturk M. Turkyilmaz Unal B. García-Caparrós P. Khursheed A. Gul A. (2020). Osmoregulation and its actions during the drought stress in plants. Physiol. Plant172, 1321–1335. doi: 10.1111/ppl.13297
50
Pei T. Ma P. Ding K. Liu S. Jia Y. (2018). SmJAZ8 acts as a core repressor regulating JA-induced biosynthesis of salvianolic acids and tanshinones in Salvia miltiorrhiza hairy roots. J. Exp. Bot.69, 1663–1678. doi: 10.1093/jxb/erx484
51
Qiu J. R. Huang Z. Xiang X. Y. Xu W. X. Wang J. T. (2020). MfbHLH38, a Myrothamnus flabellifolia bHLH transcription factor, confers tolerance to drought and salinity stresses in Arabidopsis. BMC Plant Biol.20, 542. doi: 10.1186/s12870-020-02732-6
52
Qiu X. M. Sun Y. Y. Ye X. Y. Li Z. G. (2020). Signaling role of glutamate in plants. Front. Plant Sci.10. doi: 10.3389/fpls.2019.01743
53
Razi K. Muneer S. (2021). Drought stress-induced physiological mechanisms, signaling pathways and molecular response of chloroplasts in common vegetable crops. Crit. Rev. Biotechnol.41, 1–40. doi: 10.1080/07388551.2021.1874280
54
Ritonga F. N. Ngatia J. N. Song R. X. Farooq U. Somadona S. Lestari A. T. (2021). Abiotic stresses induced physiological, biochemical, and molecular changes in Betula platyphylla: a review. Silva Fenn.55, 24. doi: 10.14214/sf.10516
55
Rochaix J. D. Bassi R. (2019). LHC-like proteins involved in stress responses and biogenesis/repair of the photosynthetic apparatus. Biochem. J.476, 581–593. doi: 10.1042/BCJ20180718
56
Shafiq S. Akram N. A. Ashraf M. García-Caparrós P. Ali O. M. (2021). Influence of glycine betaine (Natural and synthetic) on growth, metabolism and yield production of drought-stressed maize (Zea mays L.) plants. Plants-Basel10, 2540–2558. doi: 10.3390/plants10112540
57
Sharma A. Shahzad B. Rehman A. Bhardwaj R. Landi M. Zheng B. (2019). Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules24, 2452. doi: 10.3390/molecules24132452
58
Sharma N. K. Yadav S. Gupta S. K. Irulappan V. Francis A. (2023). MicroRNA397 regulates tolerance to drought and fungal infection by regulating lignin deposition in chickpea root. Plant Cell Environ.46, 3501–3517. doi: 10.1111/pce.14666
59
Shi M. Zhu R. Zhang Y. Zhang S. Liu T. (2022). A novel WRKY34-bZIP3 module regulates phenolic acid and tanshinone biosynthesis in Salvia miltiorrhiza. Metab. Eng.73, 182–191. doi: 10.1016/j.ymben.2022.08.002
60
Silverio-Gómez C. Vega-Arreguín J. Nic-Matos G. Narváez-Cab M. Sáenz-Carbonell L. (2021). Transcriptome analysis reveals key defense-related genes upon SA induction in Cocos nucifera L. Genes Genom.44, 197–210. doi: 10.1007/s13258-021-01068-1
61
Song S. Liu B. Song J. Pang S. Song T. (2022). A molecular framework for signaling crosstalk between jasmonate and ethylene in anthocyanin biosynthesis, trichome development, and defenses against insect herbivores in Arabidopsis. J. Integr. Plant Biol.64, 1770–1788. doi: 10.1111/jipb.13319
62
Tan M. Hassan M. J. Peng Y. Feng G. Huang L. (2022). Polyamines metabolism interacts with γ-aminobutyric acid, proline and nitrogen metabolisms to affect drought tolerance of creeping bentgrass. Int. J. Mol. Sci.23, 2779. doi: 10.3390/ijms23052779
63
Thirumalaikumar V. P. Devkar V. Mehterov N. (2017). NAC transcription factor JUNGBRUNNEN1 enhances drought tolerance in tomato. Plant Biotechnol. J.16, 354–366. doi: 10.1111/pbi.12776
64
Verbraeken L. Wuyts N. Mertens S. (2021). Drought affects the rate and duration of organ growth but not inter-organ growth coordination. Plant Physiol.186, 1336–1353. doi: 10.1093/plphys/kiab155
65
Vicente-Serrano S. M. Peña-Angulo D. Beguería S. Domínguez-Castro F. Tomás-Burguera M. Noguera I. et al . (2022). Global drought trends and future projections. Philos. Trans. A. Math. Phys. Eng. Sci.380, 20210285. doi: 10.1098/rsta.2023.0172
66
Waadt R. Seller C. A. Hsu P. K. Takahashi Y. (2022). Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 23, 680–694. doi: 10.1038/s41580-022-00479-6
67
Wagay N. A. Rafiq S. Khan A. Kaloo Z. A. Malik A. R. Pulate P. V. (2023). Impact of phenolics on drought stress and expression of phenylpropanoid pathway genes. Plant Phenolics Abiotic Stress Management., 265–285 doi: 10.1038/s41598-020-61140-w
68
Wang X. Niu Y. Zheng Y. (2021). Multiple functions of MYB transcription factors in abiotic stress responses. Int. J. Mol. Sci.22, 6125. doi: 10.3390/ijms22116125
69
Wang L. Qiao H. (2020). Chromatin regulation in plant hormone and plant stress responses. Curr. Opin. Plant Biol.57, 164–170. doi: 10.1016/j.pbi.2020.08.007
70
Wang X. Yang Y. Liu X. Gao X. (2020). Pharmacological properties of tanshinones, thenatural products from Salvia miltiorrhiza. Adv. Pharmacol.87, 43–70. doi: 10.1016/bs.apha.2019.10.001
71
Wen X. Chen Z. Yang Z. (2023). A comprehensive overview of cotton genomics, biotechnology and molecular biological studies. Sci. China Life Sci.66, 2214–2256. doi: 10.1007/s11427-022-2278-0
72
Xiang X.-Y. Chen J. Xu W.-X. Qiu J.-R. Song Li (2021). Dehydration-induced WRKY transcriptional factor mfWRKY70 of myrothamnus flabellifolia enhanced drought and salinity tolerance in arabidopsis. Biomolecules11, 327. doi: 10.3390/biom11020327
73
Xiao Y. Chu L. Zhang Y. Bian Y. Xiao J. (2022). HY5: A pivotal regulator of light-dependent development in higher plants. Front. Plant Sci.12. doi: 10.3389/fpls.2021.800989
74
Xuan H. Huang Y. Zhou L. Deng S. Wang C. (2022). Key soybean seedlings drought-responsive genes and pathways revealed by comparative transcriptome analyses of two cultivars. Int. J. Mol. Sci.23, 2893. doi: 10.3390/ijms23052893
75
Yang Ye Shao M. Yao J. Yang S. Cheng W. (2022). Neocryptotanshinone protects against myocardial ischemia-reperfusion injury by promoting autolysosome degradation of protein aggregates via the ERK1/2-Nrf2-LAMP2 pathway. Phytomedicine110, 154625. doi: 10.1016/j.phymed.2022.154625
76
Yao H. Wang F. Bi Q. Liu H. (2022). Combined Analysis of Pharmaceutical Active Ingredients and Transcriptomes of Glycyrrhiza uralensis Under PEG6000-Induced Drought Stress Revealed Glycyrrhizic Acid and Flavonoids Accumulation via JA-Mediated Signaling. Front. Plant Sci.13. doi: 10.3389/fpls.2022.920172
77
Yin X. Fan H. Chen Y. Li L.-Z. (2020). Integrative omic and transgenic analyses reveal the positive effect of ultraviolet-B irradiation on salvianolic acid biosynthesis through upregulation of SmNAC1. Plant J.104, 781–799. doi: 10.1111/tpj.14952
78
Zhang Y. Berman A. Shani E. (2023). Plant hormone transport and localization: signaling molecules on the move. Annu. Rev. Plant Biol.74, 453–479. doi: 10.1146/annurev-arplant-070722-015329
79
Zhang Q. Liu X. Xu D. Hong Z. Zhang N. (2022). Effects of drought and host on the growth of seedlings in pot culture. Int. J. Mol. Sci.23, 11241. doi: 10.3390/ijms231911241
80
Zhang H. Rutherford S. Qi S. (2021). Transcriptome profiling of Arabidopsis thaliana roots in response to allelopathic effects of Conyza canadensis. Ecotoxicology31, 53–63. doi: 10.1007/s10646-021-02489-0
81
Zhao L. Lü Y. Chen W. (2020). Genome-wide identification and analyses of the AHL gene family in cotton (Gossypium). BMC Genomics21, 69. doi: 10.1186/s12864-019-6406-6
82
Zhong M. Zhang L. Yu H. Liao J. Jiang Y. (2023). Identification and characterization of a novel tyrosine aminotransferase gene (SmTAT3-2) promotes the biosynthesis of phenolic acids in Salvia miltiorrhiza Bunge. Int. J. Biol. Macromol.254, 127858. doi: 10.1016/j.ijbiomac.2023.127858
83
Zhou Y. Bai Y.-H. Han F.-X. Chen X. (2024). Transcriptome sequencing and metabolome analysis reveal the molecular mechanism of Salvia miltiorrhiza in response to drought stress. BMC Plant Biol.24, 446. doi: 10.1186/s12870-024-05006-7
Summary
Keywords
Salvia miltiorrhiza Bunge, drought stress, transcriptomic analysis, metabolomic analysis, AHL gene family
Citation
Zhang Y, Zhang H, Zhang Y, Wang D, Meng X and Chen J (2025) Utilizing physiologies, transcriptomics, and metabolomics to unravel key genes and metabolites of Salvia miltiorrhiza Bge. seedlings in response to drought stress. Front. Plant Sci. 15:1484688. doi: 10.3389/fpls.2024.1484688
Received
22 August 2024
Accepted
16 December 2024
Published
14 January 2025
Volume
15 - 2024
Edited by
Jose Sebastian, Indian Institute of Science Education and Research Berhampur (IISER), India
Reviewed by
Hakim Manghwar, Lushan Botanical Garden (CAS), China
Michael Ackah, Jiangsu University, China
Updates
Copyright
© 2025 Zhang, Zhang, Zhang, Wang, Meng and Chen.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Juan Chen, 158501243@qq.com
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.