- 1Key Laboratory of Modern Agricultural Cultivation and Crop Germplasm Improvement of Heilongjiang Province, Agronomy College of Heilongjiang Bayi Agricultural University, Daqing, Heilongjiang, China
- 2Key Laboratory of Low Carbon Green Agriculture in Northeast Plain, Ministry of Agriculture and Rural Affairs, Daqing, Heilongjiang, China
Among the major abiotic stresses, salt and drought have considerably affected agricultural development globally by interfering with gene expression profiles and cell metabolism. Transcription factors play crucial roles in activating or inhibiting the expression of stress-related genes in response to abiotic stress in plants. In this study, the Zea mays L. SQUAMOSA promoter-binding protein gene (ZmSBP17) was identified, and the molecular regulatory mechanism of osmotic stress tolerance was analyzed. Phylogenetic analysis confirmed that ZmSBP17 is part of the SBP gene family and is closely related to OsSBP17. The ZmSBP17-GFP fusion protein exhibited green fluorescence in the nucleus, as determined via tobacco epidermal transient transformation system. Acting as a transcriptional activator, the overexpression of ZmSBP17 in Arabidopsis significantly enhanced the expression of genes encoding superoxide dismutases (CSD1/2, MSD1), catalases (CAT1/2), ascorbate peroxidase 1 (APX1), and myeloblastosis transcription factors (AtMYB53/65), which increased the activity of reactive oxygen species (ROS)-scavenging enzymes and reduced ROS levels. Additionally, the expression of abiotic stress-related genes, such as AtDREB2A and AtNHX1, was significantly upregulated by ZmSBP17. Furthermore, ZmSBP17 specifically bound to cis-acting elements containing GTAC core sequences in the promoters of stress-related genes, suggesting that ZmSBP17 regulates the transcription of certain genes by recognizing these sequences. These results indicate that the overexpression of ZmSBP17 in Arabidopsis thaliana significantly increased tolerance to osmotic stress during the germination and seedling stages, which may enhance our understanding of the biological functions of SBPs in maize under abiotic stresses.
1 Introduction
Salinization of soil increases ion toxicity, making it difficult for roots to absorb water and nutrients, leading to physiological drought and nutritional deficiencies (Zhao et al., 2020). Saline–alkali soil, an important soil type, is widely distributed in over 100 countries. The Songnen Plain in Northeast China is a key grain production area and commodity grain base, and it is the primary region in China with saline–alkali soil. Additionally, drought, particularly spring drought, is a major abiotic stress prevalent in this region. Various abiotic stresses, including salt, drought, and cold, can impose osmotic stress on crops (Vinocur and Altman, 2005). Maize (Zea mays L.), a critical global food crop, is used for human consumption, animal feed, biofuel production, and other industrial purposes (Serna-Saldivar and Carrillo, 2018). Therefore, to ensure national food security and sustainable agricultural development, it is crucial to focus on the molecular basis of maize’s response to osmotic stress.
Transcription factors (TFs) are crucial in regulating the abiotic stress signal transduction pathway because they can activate or inhibit the expression of stress-related genes by interacting with their cis-elements (Agarwal et al., 2017). The SQUAMOSA promoter-binding protein (SBP) family is one of the largest plant-specific TF gene families. Members of this family contain a highly conserved DNA-binding domain known as the SBP-box (Klein et al., 1996), which consists of approximately 76 amino acid residues forming two zinc finger motifs (C3H and C2HC) and a nuclear localization signal (Yamasaki et al., 2013). SBP TFs have been identified in many plants, including Arabidopsis thaliana (Cardon et al., 1999), Brassica napus (Cheng et al., 2016), Petunia (Zhou et al., 2018), and wheat (Li et al., 2020). These TFs are known to be involved in regulating growth, development, and responses to abiotic stress. For example, in A. thaliana, AtSPL3/8 influences floral development, with its expression induced by prolonged sunlight starting to increase before flowering (Unte et al., 2003; Cardon et al., 1997). AtSPL10/11/12 redundantly controls the proper development of lateral organs in relation to shoot maturation during the reproductive phase (Shikata et al., 2009). In rice, OsSPL10 negatively regulates salt tolerance but positively affects trichome formation (Lan et al., 2019), whereas OsSPL3/12 positively regulates crown root formation (Shao et al., 2019). Overexpression of VpSBP16 from grapes in A. thaliana significantly increases root length and seed germination rate under salt and drought stress (Hou et al., 2018). VpSBP5 contributes to resistance against Erysiphe necator by inducing salicylic acid and methyl jasmonate signaling pathways (Hou et al., 2013). Mao et al. (2016) provided a comprehensive overview of SBP TFs in maize, noting that the expression of certain SBP TFs increased significantly under abiotic stress. However, the biological functions of SBPs in maize under abiotic stresses remain unclear.
In this study, a novel SBP transcription factor gene, ZmSBP17, was isolated from the inbred line Hei maize. Overexpression of ZmSBP17 enhanced the plant’s tolerance to osmotic stress by increasing the expression of abiotic stress-related genes. The core GTAC sequence was found to be crucial for regulating gene expression mediated by ZmSBP17. This study enhances our understanding of the roles of maize SBP TFs and the mechanisms underlying plant tolerance to osmotic stress.
2 Materials and methods
2.1 Plant and osmotic stress treatments
Pre-germination treatments of maize seeds were performed and growth conditions applied as described in our previous study (Sun et al., 2018). Germinated seeds were sown in 20 × 22 cm plastic pots containing 2.5 kg vermiculite. All pots were watered with a half-strength Hoagland nutrient solution once daily. At the 3-leaf seedling stage of maize, 24 pots were selected and randomly divided into two equal sets. One set was used as a control by watering with a half-strength Hoagland nutrient solution, and the other set was treated with 200 mmol/L NaCl or 10% polyethylene glycol (PEG6000) in half-strength Hoagland nutrient solution. Leaves were harvested at 0, 2, 6, and 12 h post-stress treatment for RNA extraction to detect the expression pattern of ZmSBP17 under abiotic stress. Three biological samples (three pots for one time in one set) were collected from each tissue.
A. thaliana Columbia ecotype was used in this study. Growth conditions in soil and stress treatment methods were applied as described by He et al., 2019, 2024. Four-week-old gene-overexpressing Arabidopsis lines (OE) and wild-type (WT) plants were treated with water (control) or 200 mmol/L NaCl and 10% PEG6000 for 36 h. Then, leaves were collected to analyze the expression levels of abiotic stress-related genes and determine the activities of key reactive oxygen species (ROS)-scavenging enzymes.
2.2 Real-time quantitative RT-PCR analysis
Total RNA was extracted from the samples using the RNAiso Reagent Kit (TaKaRa, Dalian, China). The FastQuant RT Kit (TIANGEN, China) was used to synthesize the first-strand cDNA, following the procedure outlined in our previous studies (Sun et al., 2012; 2018). ZmActin1 (NM_001155179) was used as the endogenous control to compare the expression level of ZmSBP17. ACT8 (AT1G49240) was utilized as the internal control to analyze the expression levels of abiotic stress-related genes in Arabidopsis. All primers used for qRT-PCR assay are listed in Supplementary File 1. A standard SYBR® Premix Ex Taq™ kit (TaKaRa, Dalian, China), Bio-Rad CFX96 Real-Time System (Bio-Rad, USA, Hercules, CA, USA), and CFX Manager System software version 2.0 were utilized per the manufacturer’s instructions. At the end of PCR, Ct values for each sample were obtained to analyze the transcript levels of each gene using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
2.3 Gene isolation and sequence analysis of ZmSBP17
Based on the sequence of ZmSBP17 available in the NCBI (http://www.ncbi.nlm.nih.gov/Blast.cgi) and maize genome (www.plantgdb.org/ZmGDB/cgi-bin/blastGDB.pl) databases, gene-specific primers for RT-PCR were designed using the NCBI primer designer. The primer sequences are listed in Supplementary File 1. Following the procedure outlined in our previous study (Sun et al., 2012), the full-length coding sequence (CDS) of Hei ZmSBP17 was isolated from the first-strand cDNA, cloned into the pMD18-T cloning vector (TaKaRa, Dalian, China), and sequenced (BGI, China). NCBI’s Conserved Domain database (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) was used to analyze the sequences and identify the conserved regions. Phylogenetic trees of ZmSBP17 and sequences from homologous plants were then constructed using DNAMAN software.
2.4 Subcellular localization, trans-activation, and DNA-binding assays of ZmSBP17
The full-length CDS of ZmSBP17, lacking its termination codon, was inserted into the pCAMBIA2300-GFP vector using a combination of homologous recombination and restriction digestion (SacI and XbaI). This construct was then used to transform the Escherichia coli strain DH5α. Expression of ZmSBP17 was driven by a CaMV 35S promoter. The primer sequences used for amplification are listed in Supplementary File 1. The validated recombinant pCAMBIA2300-ZmSBP17-GFP and pCAMBIA2300-GFP (control) vector were transformed into EHA105 Agrobacterium competent cells for injecting into 5-6 leaf old N. benthamiana leaves epidermal cells, respectively, as previously reported (Chao et al., 2023). The transformed tobacco plants were incubated at room temperature in the dark for 12-16 h, and then imaged by laser scanning confocal microscopy (Leica SP8) after cultured for 2-3 days at normal condition.
To detect the transcriptional activation of ZmSBP17, the full-length and different truncated versions, which were amplified (primers listed in Supplementary File 1) from the pMD18-T-ZmSBP17 plasmid into the pGBKT7 vector. These constructs, along with the pGBKT7 control vector, were used to transform the yeast strain Y2HGold according to the manufacturer’s protocol (Clontech, USA). The transformants were then cultured on SD/-Trp and SD/-Trp/-His media for 3–5 days at 30°C.
For DNA-binding assays, the full-length ZmSBP17 CDS was amplified and cloned into the pGAD7 vector. The sequences of the SBP17-F4 and R4 primers, which contain NdeI and EcoRI sites, are listed in Supplementary File 1. The 5X GTAC cis-elements and the mutated GTACm version were inserted into the pHIS2 vector (Yu et al., 2017) and used to co-transform the Y2HGold cells. The transformants were streaked onto SD/-Trp and SD/-Trp/-His/-Ade media and incubated for 3–5 days at 30°C.
2.5 Generation of transgenic Arabidopsis plants overexpressing ZmSBP17
The pCAMBIA2300-ZmSBP17-GFP vector was introduced into the Agrobacterium tumefaciens strain EHA105, which was then used to transform A. thaliana via the floral dip method (Clough and Bent, 1998). Transgenic seeds (T0) were selected on MS inorganic salts (Murashige and Skoog, 1962) medium supplemented with 50 mg·L−1 kanamycin. At the 7–8 leaf seedling stage, the lowest leaves were used to identify PCR-positive plants using detection primers (Supplementary File 1). Homozygous lines were selected on MS medium containing 50 mg·L−1 kanamycin.
2.6 Osmotic treatment of transgenic Arabidopsis plants and phenotype analysis
For osmotic stress seed germination assays, WT and T3 homozygous OE seeds were sterilized and vernalized at 4°C for 3 days. They were then sown on half-strength MS medium supplemented with either 100 mM NaCl or 8% PEG. Germination rates were assessed after 7 days. All germination assays were performed in triplicate. To evaluate the sensitivity of adult plants to osmotic stress, 4-week-old WT and OE plants were irrigated with either 200 mM NaCl or 10% PEG at 3-day intervals for 7 days and subsequently photographed.
2.7 Histochemical analysis under osmotic treatment
For histochemical analysis, 4-week-old WT and OE plants were treated with 200 mM NaCl or 10% PEG for 36 h. The entire plants were then used for staining. DAB and NBT staining were performed as described by Zhang et al. (2011) with slight modifications.
2.8 Measurements of enzyme activity in transgenic Arabidopsis
For detecting the activity of ROS-scavenging enzymes, 4-week-old WT and OE plants were irrigated with 200 mM NaCl or 10% PEG for 36 h, and then rosette leaves were collected. The activities of peroxidase (POD), superoxide dismutase (SOD), and catalase (CAT) were measured as described by Wang et al. (2019). Ascorbate peroxidase (APX) was extracted as described by Erinle et al. (2018) with slight modifications.
3 Results
3.1 Analysis of ZmSBP17
Following the expression analysis of SBPs under salt stress by qRT-PCR in our previous study, a putative salt stress-related SBP from the Hei maize inbred line was isolated and named ZmSBP17 (GenBank accession no. NM_001308478) according to blastp results (Supplementary File 2). The open reading frame of ZmSBP17 is 1137 bp long, encoding a 379 amino acid polypeptide. Amino acid sequence alignment and phylogenetic analysis of ZmSBP17 homologs are shown in Figure 1. The average identity homology of ZmSBP17 with those in rice and Arabidopsis was 12.9%. Multiple sequence alignment by DNAMAN revealed it to have C2HC and C3H domains at the N terminus (Supplementary File 3). Phylogenetic analysis showed that ZmSBP17 was relatively close to OsSBP17, but ZmSBP17 was less identical to homologs other than OsSBP17, which suggested that ZmSBP17 markedly differs from other members. Analysis of the upstream 2000 bp promoter region of ZmSBP17 revealed the presence of multiple elements related to abiotic stress, including abscisic acid response element (ABRE), antioxidant response element (ARE), long terminal repeat (LTR), and MYB-binding sites (MBS) (Supplementary File 4).
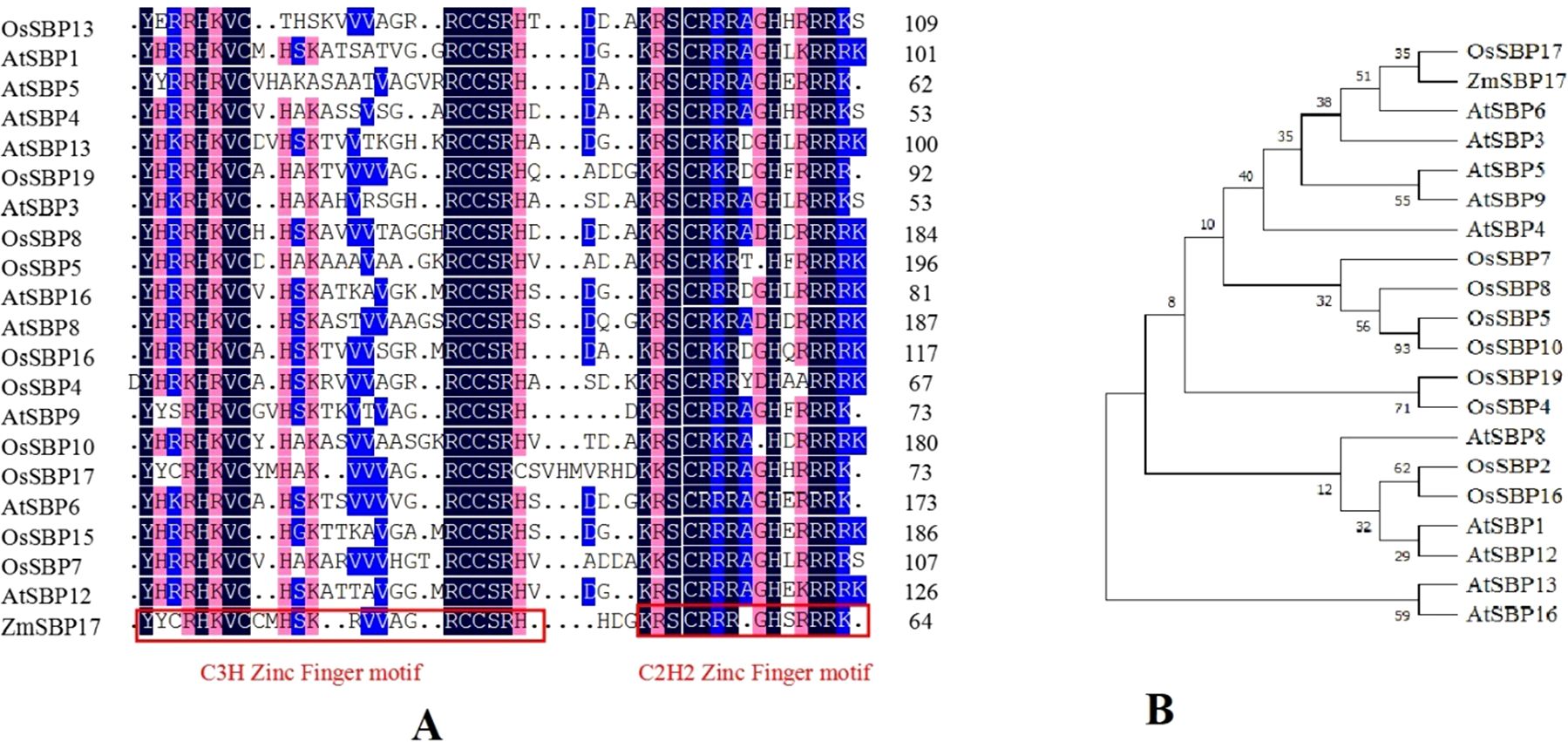
Figure 1. Multiple alignments and phylogenetic relationships of ZmSBP17 with orthologs in rice and Arabidopsis. (A) Multiple alignments of ZmSBP17 with homologous proteins in rice and Arabidopsis. Different background colors display similar acid sequences. Red boxes indicate conserved C3H and C2H2 zinc finger motifs, respectively. (B) Phylogenetic relationships of ZmSBP17 with orthologs in rice and Arabidopsis.
3.2 ZmSBP17 expression and protein localization
The expression pattern of ZmSBP17 under abiotic stress was investigated by subjecting the three-leaf stage maize seedlings to the indicated treatment. ZmSBP17 expression increased significantly and reached a peak at 12 and 2 h under salt and PEG stresses, respectively, indicating that it may participate in the osmotic stress responses in maize (Figure 2A; Supplementary File 5).
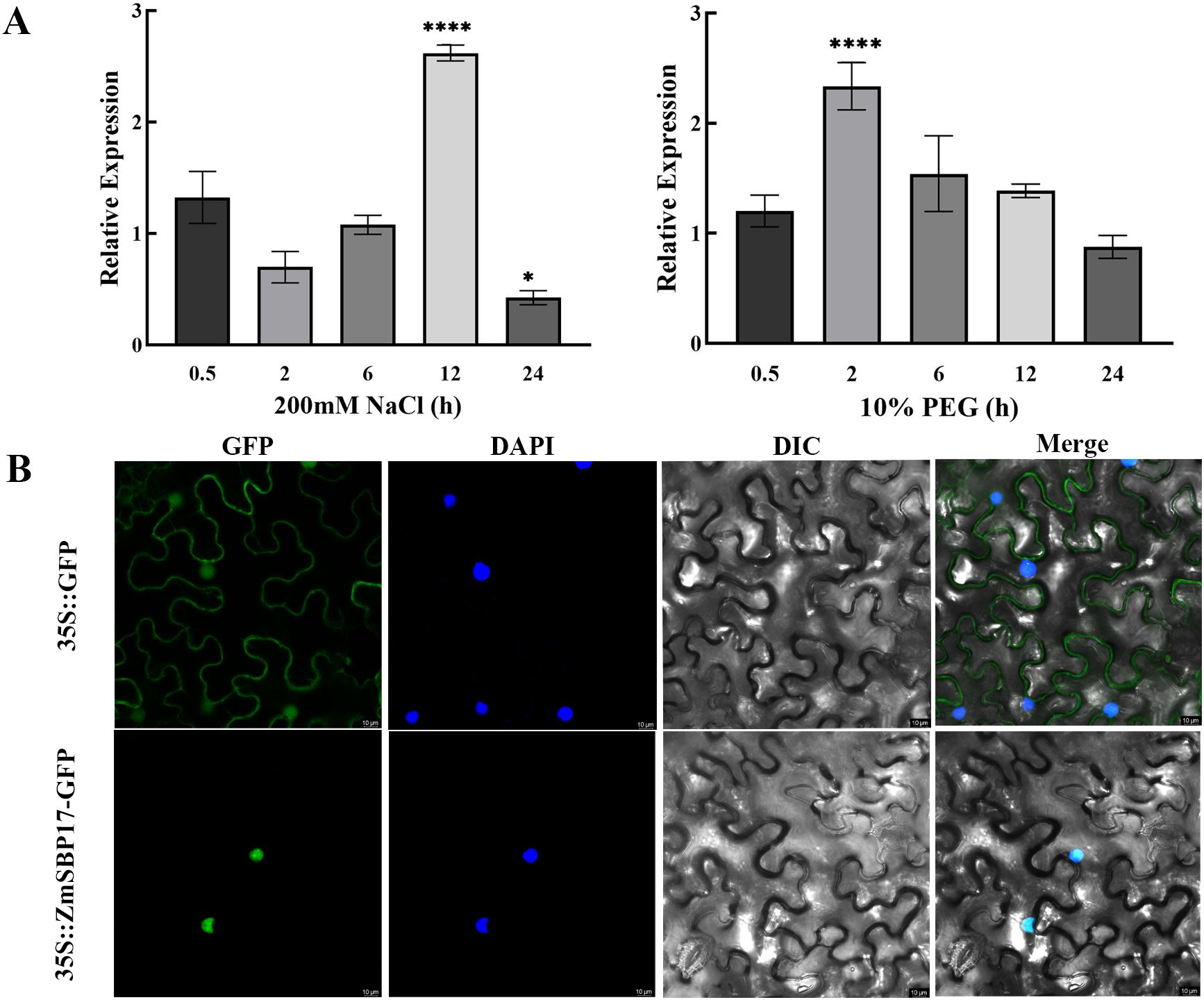
Figure 2. ZmSBP17 expression and subcellular localization. (A) Expression patterns of ZmSBP17 under different treatments. Expression pattern of ZmSBP17 in leaves. Relative expression levels were normalized to 1 in Hei plants without stress (0 h). Values are presented as mean (± SD) of three biological replicates. * and **** significantly higher values in treatment samples than in control plants (0 h) at p ≤ 0.05 and p ≤ 0.0001 as determined using Student’s t-test, respectively. (B) Subcellular localization of ZmSBP17 in the tobacco epidermal cells. GFP, green fluorescence protein images; DAPI, 4’, 6-diamidino-2-phenylindole (DAPI) stained images; DIC, brightfield images; Merged, merged brightfield and GFP images. Scale bar = 10 μm.
The subcellular localization of ZmSBP17 constructed transient expression vector pCAMBIA2300-ZmSBP17-GFP (Supplementary File 2) and pCAMBIA2300- GFP vectors were transformed into the tobacco epidermal cells. The green fluorescence signals of expressing GFP gene was monitored by a confocal laser scanning microscope. As shown in Figure 2B, the pCAMBIA2300-ZmSBP17-GFP fusion protein was specifically detected in the nucleus compare to the control distributed throughout the cell.
3.3 Transcriptional activity assay of ZmSBP17
The full-length ZmSBP17 was fused to the GAL4 (GAL4-BD) binding domain of pGBKT7 to confirm if ZmSBP17 has transcription activation ability. The constructed pGBKT7-ZmSBP17 vector is shown in Supplementary File 3. A comparison of the growth phenotype of the pGBKT7-ZmSBP17 and pGBKT7 recombinant yeast revealed that they can survive on SD/-Trp selective medium with the same colony size, indicating the successful transfer of each vector into the Y2H cells. However, only those containing the pGBKT7-ZmSBP17 vector grew normally on SD/-Trp-His selective medium, suggesting that ZmSBP17 was able to activate the HIS reporter gene for survival. Thus, ZmSBP17 acts as a transcriptional activator in yeast (Figure 3).
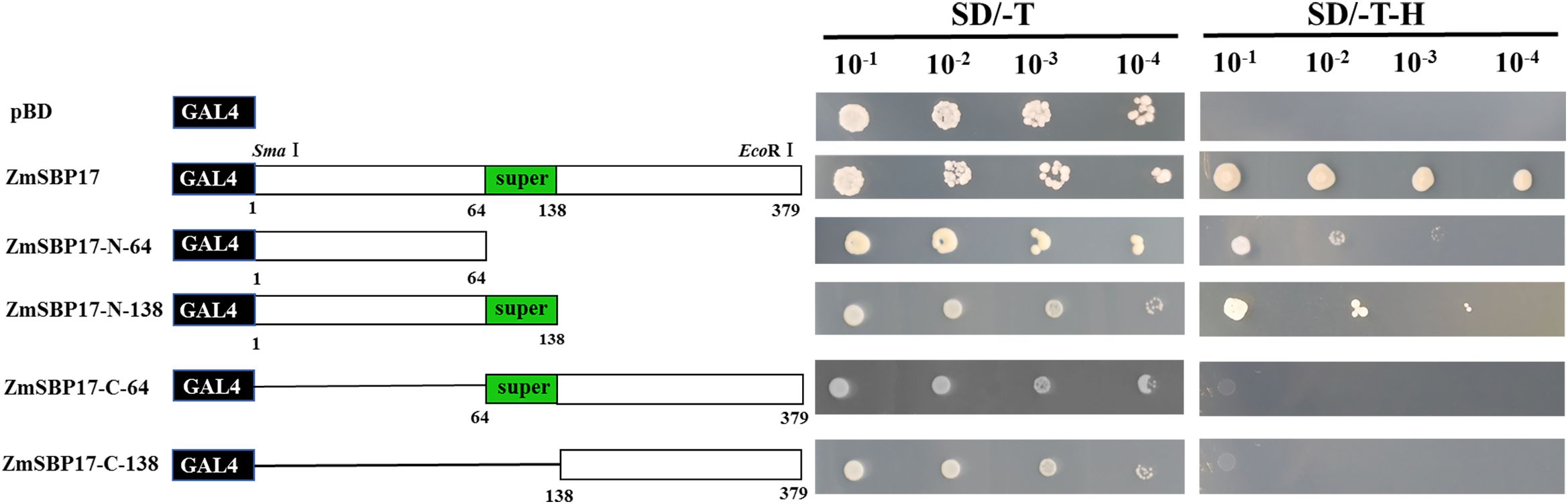
Figure 3. Analysis of the transcriptional activation capacity of the full-length or partially deleted ZmSBP17 in yeast. Numbers indicate amino acid positions. Transformants carrying the empty pGBKT7 plasmid were used as a negative control. Transformants were examined on SD/-Trp (growth control) and SD/-Trp/-His media.
Protein sequence analysis revealed that the SBP superfamily domains are present in the ZmSBP17 protein (from 64 to 138 amino acid residues). To determine which domain in ZmSBP17 was the transcriptional activator, four deletion fragments were inserted into pGBKT7 and used to transform Y2H cells. All transformants grew well on the SD/-Trp medium. Contrary to those carrying the N-64 and N-138 fragments, the yeast cells carrying the C-64 fragment of ZmSBP17 could not grow on the selection medium. Meanwhile, yeast cells carrying the N-138 SBP domain fragment (amino acids 64–138) grew better than those carrying the N-64 fragment on the selection medium. These results indicate that the SBP domain enhanced the transcription activation ability of ZmSBP17, and the 1–64 amino acid fragment was crucial for this ability.
3.4 ZmSBP17 positively regulates osmotic responses in plants
pCAMBIA2300-ZmSBP17 was used to transform the WT A. thaliana Columbia-0 plants to characterize whether ZmSBP17 is associated with the osmotic stress response. The transgenic plants were screened on kanamycin and identified by PCR (Supplementary Figure 4). The T3 plants were used for further analysis. No significant differences in seed germination rate were observed between transgenic and WT plants on MS medium. However, in the presence of 100 mM NaCl and 8% PEG, an obvious reduction in germination rate was detected in both transgenic and WT seeds but was significantly higher in the transgenic than WT seeds (Figures 4A, B; Supplementary File 6).
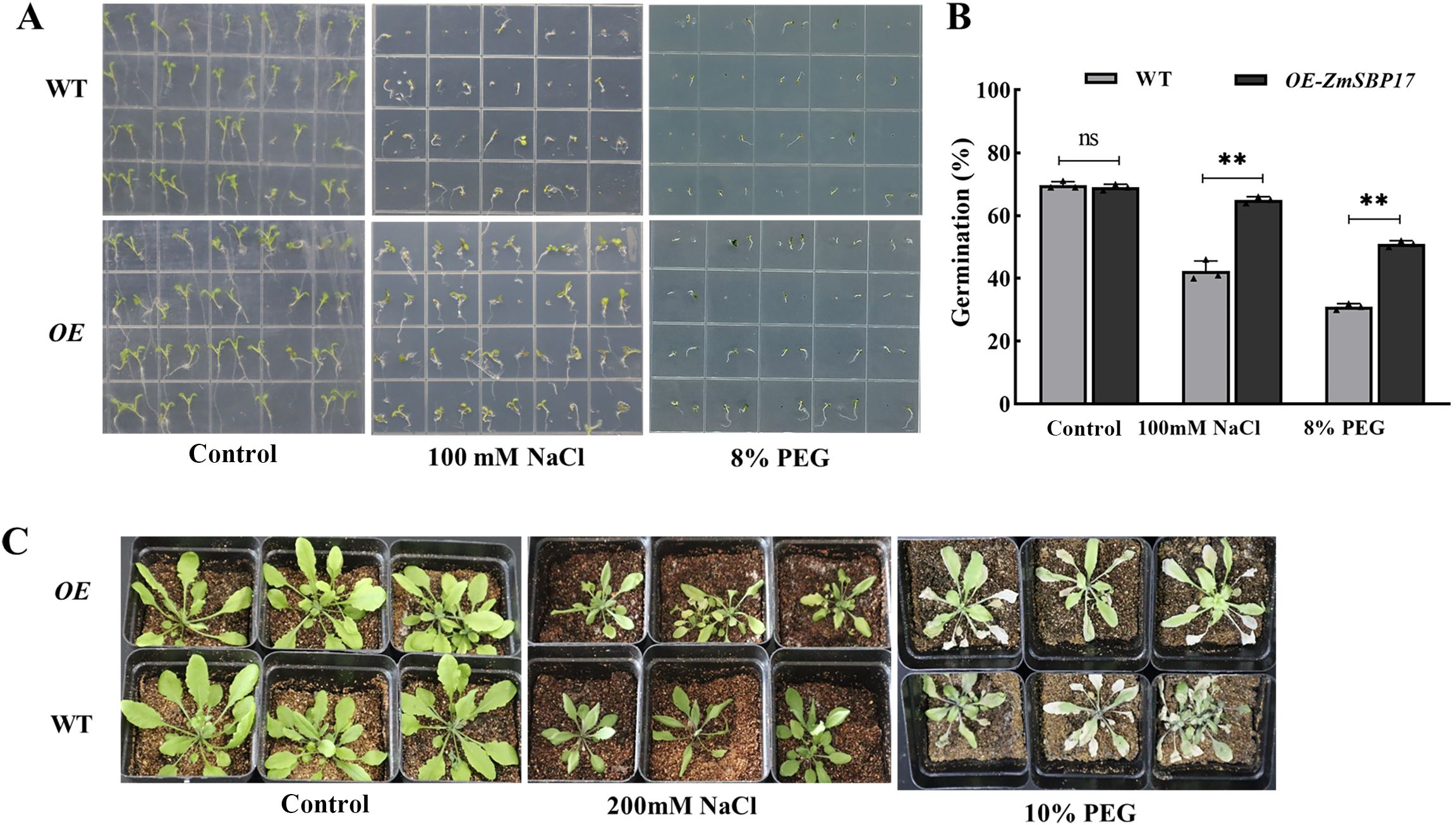
Figure 4. ZmSBP17 positively regulates osmotic stress response. (A) Germination assay. Seeds of the OE and WT plants were cultivated for 7 days on a half-strength MS medium supplemented with 100 mM NaCl and 5% PEG. (B) Statistical analysis of the germination rates; 72 seeds were used for each experiment. **P < 0.01 as determined by Student’s t-test. ns, no significance. (C) Phenotypic assay of soil-grown plants; 4-week-old OE and WT plants were treated with 200 mM NaCl and 10% PEG for 7 days.
For further functional analysis of ZmSBP17, the adult stage ZmSBP17-OE lines were examined in soil. Under normal conditions, no significant phenotypic variations between the OE lines and WT were observed. However, post-treatment with 200 mM NaCl and 10% PEG for 36 h, the stems and leaves of the WT showed obvious wilting, with some leaves turning yellow and drying. However, the wilting of transgenic A. thaliana was significantly less severe than that of the WT under PEG stress. The stems and leaves of the transgenic plants remained relatively upright, and leaf yellowing was also less pronounced compared with the WT. However, under salt stress, there was no apparent difference between the WT and transgenic plants (Figure 4C). These results indicate that ZmSBP17 may improve the tolerance of Arabidopsis seeds at various stages to PEG.
3.5 ZmSBP17 positively regulates ROS-scavenging systems under osmotic stress
The levels of H2O2 and O2¯ that are key ROS species were detected in ZmSBP17 OE line and WT plants using DAB and NBT staining, respectively to investigate whether ZmSBP17 is associated with the regulation of ROS scavenging under osmotic stress. After treatment with 200 mM NaCl and 10% PEG, the staining in the leaf tissues of the WT and OE lines increased significantly. However, the DAB staining was deeper in the WT than in the OE lines, and there was no significant difference in NBT intensity between the WT and OE lines under PEG stress. Under salt stress, the DAB and NBT staining was deeper in the WT than in the OE line (Figures 5A, B). Moreover, the activities of key ROS-scavenging enzymes POD, SOD, CAT, and APX were assessed in the OE and WT plants at 0 and 36 h after osmotic treatment. As shown in Figure 5C, the activities of all four increased significantly in the OE line compared to WT plants after stress treatment (Supplementary File 6). These results indicate that ZmSBP17 might play a role in modulating the ROS-scavenging system by increasing the activities of protective enzymes under osmotic stress.
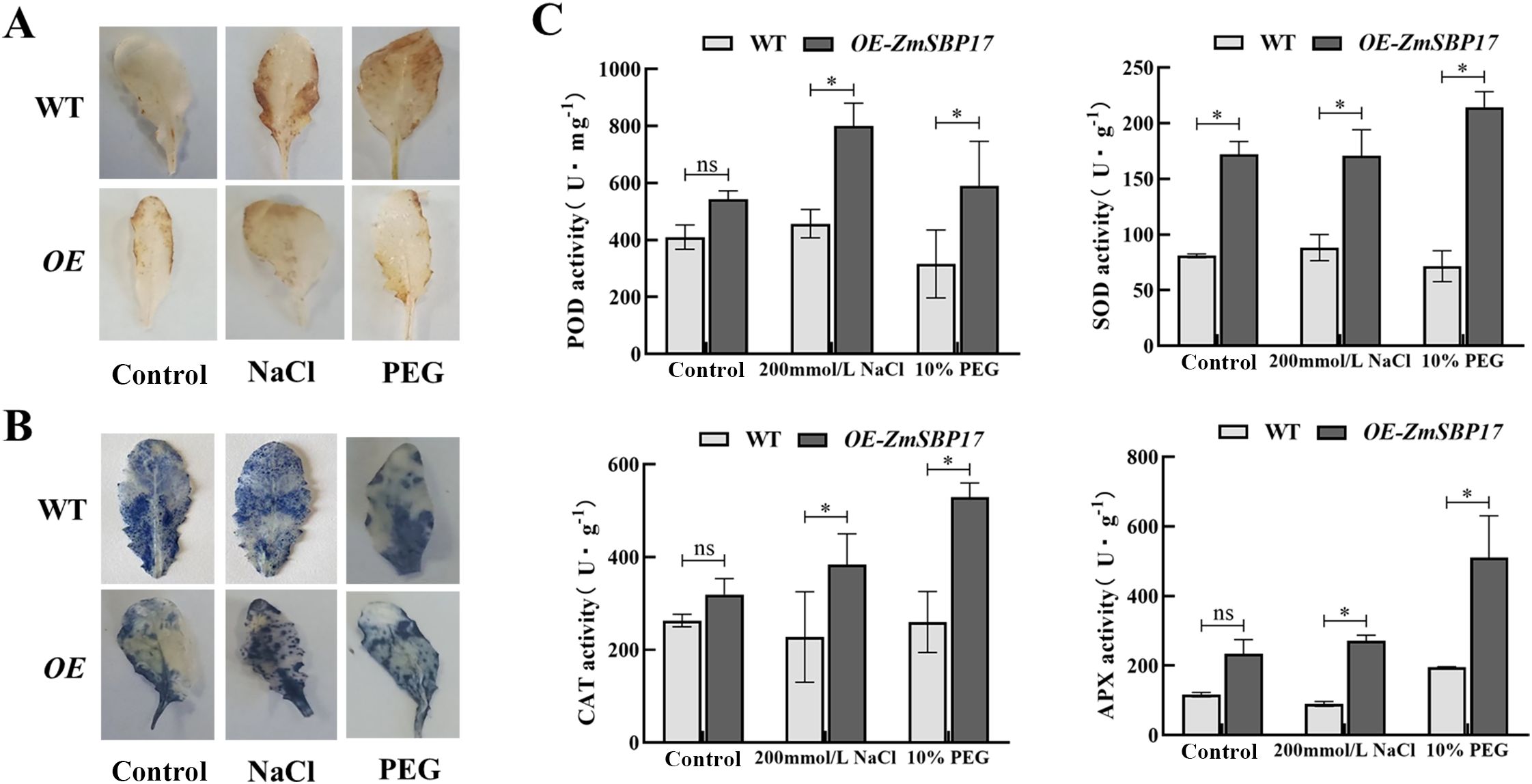
Figure 5. Detection of ROS accumulation and scavenging capability. (A, B) Analysis of H2O2 and O2- accumulation using DAB and NBT staining, respectively. (C) Measurement of POD, SOD, CAT, and APX activities. Values are presented as the mean (± SD) of three biological replicates. Upper or lower whiskers delineate maximum and minimum values, respectively. *P < 0.05 as determined by Student’s t-test. ns, no significance.
3.6 ZmSBP17 positively regulates osmotic stress-related gene expression in transgenic plants
The expression levels of the abiotic stress-responsive genes—CSD1, AT1G08830.1; CSD2, AT2G28190.1; MSD1, AT3G10920.1; CAT1, AT1G20630.1; CAT2, AT4G35090.1; APX1, AT1G07890.3; DREB2A, AT5G05410; NHX2, AAT3G05030; MYB65, AT3G1140; and MYB53, AT5G65230—were analyzed in WT and transgenic plants by real-time qRT-PCR to better understand the mechanistic basis of the improved osmotic stress tolerance exhibited by the ZmSBP17-transgenic plants. As shown in Figure 6, the expression levels of 10 genes increased significantly in transgenic plants compared to WT under salt or PEG stresses. For example, the expression levels of the genes encoding the ROS-scavenging enzymes, SODs (CSD1/2 and MSD1) and CATs (CAT1/2), were significantly elevated in the ZmSBP17-OE lines than that in the WT under PEG stress, while the expression levels of CAT1 and APX1 were significantly higher under salt stress. Moreover, the expression level of the drought-related gene (DREB2A) was significantly more than in WT under PEG stress. The expression levels of the Na+ or K+ transport-related gene (NHX1) were not only significantly lower in the WT than in transgenic plants under normal conditions but also significantly lower in the WT than in the transgenic plants after salt stress to maintain Na+ and K+ balance. Additionally, the expression level of the MYB TF family genes (MYB53/65) in the OE and WT plants was assessed. The expression level of MYB65 was significantly higher in the transgenic plants than in WT after PEG stress, while that of MYB53 was significantly enhanced in the transgenic plants than that in the WT after salt and PEG stress.
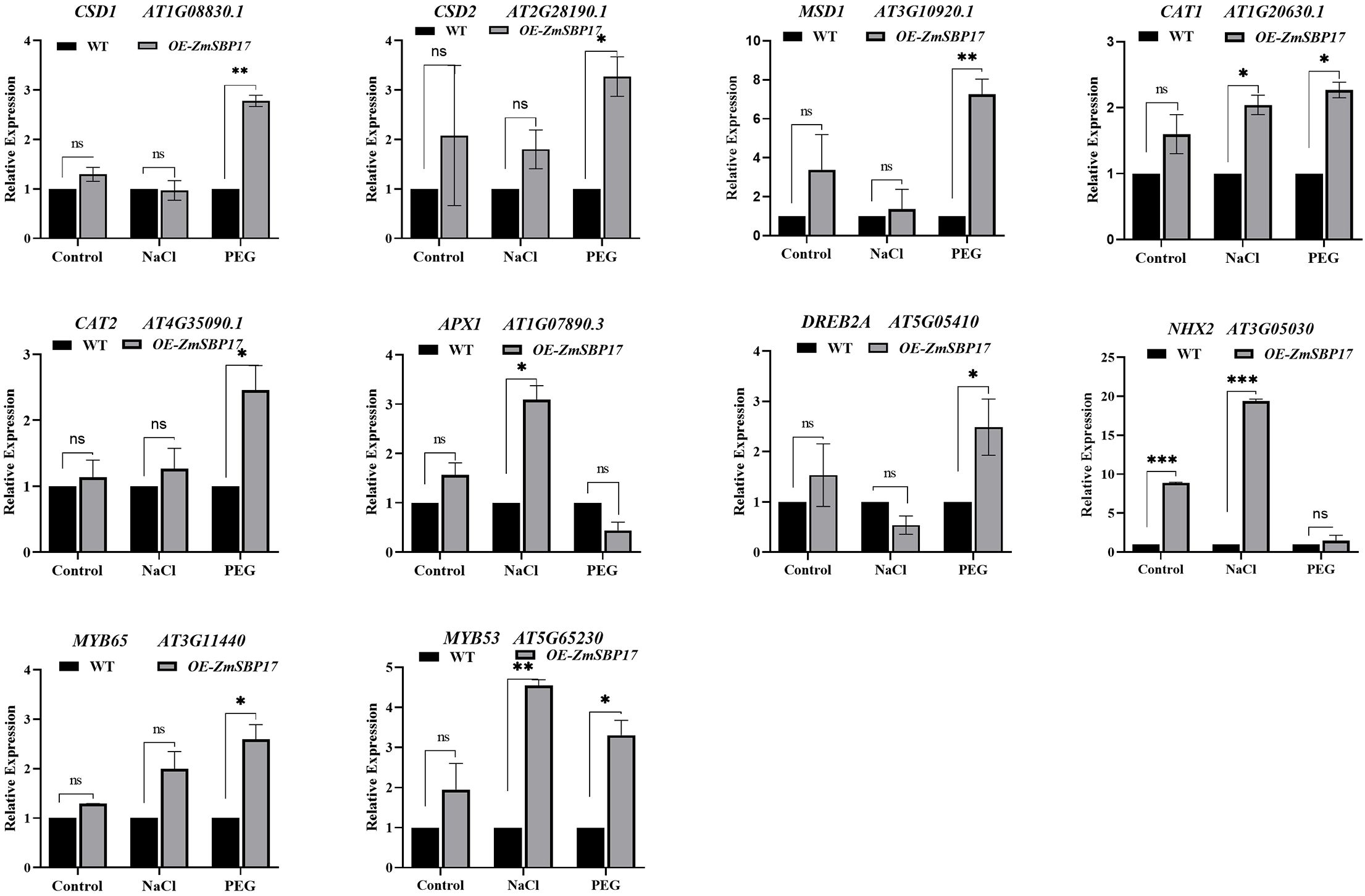
Figure 6. Relative expression levels of abiotic stress-responsive genes in OE and WT plants under salt and PEG stress. Expression of each gene in the WT was set to 1. Values are presented as the mean (± SD) of three biological replicates. *, ** and *** indicate significantly higher values in the treatment samples than in the WT at p ≤ 0.05, p ≤ 0.01 and p ≤ 0.001, respectively, determined using the Student’s t-test. ns, no significance.
These results indicate that the overexpression of ZmSBP17 may aid in improving osmotic stress tolerance in plants by elevating the expression of ROS-scavenging enzymes encoding, drought-related, Na+ or K+ transport-related, and MYB TF family genes.
3.7 ZmSBP17 binds to the GTAC ([G/T] [A/G/T]) core sequence
SBP TFs could bind to the GTAC-box ([C][C]GTAC[A/G]) cis-elements. Therefore, ZmSBP17 was cloned into pGADT7 while the GTAC core sequence and mutated elements were repeated five times and cloned separately into pHIS2. Growth assays revealed that the yeast co-transformed with GTAC and ZmSBP17 can grow better on the selection medium, while those containing the GTACm sequence displayed the same growth as the negative control pHis2 (Figure 7B). These results indicate that the core GTAC-box sequence is crucial for the ZmSBP17 protein to recognize its target genes.
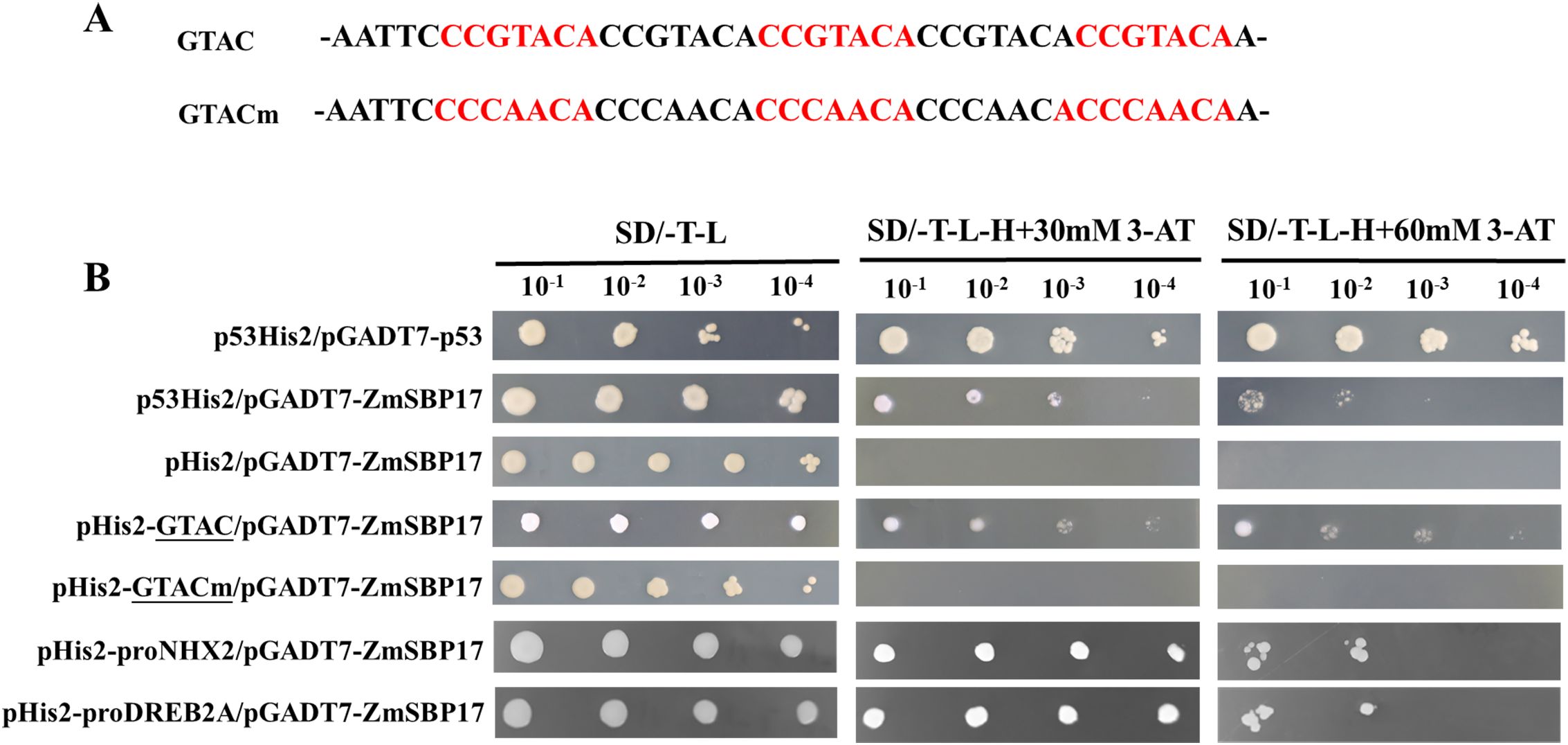
Figure 7. ZmSBP17 binds to GTAC core sequences and the promoters of target genes. (A) Sequences of the WT and mutated GTAC cis-elements; (B) Yeast one-hybrid assays displaying the binding ability of ZmSBP17 to the GTAC cis-element in the promoters of the target genes; yeast co-transformants with the effector vector and each reporter were analyzed by culturing serial dilutions (10−1, 10−2, 10−3, and 10−4) on a selection medium containing 3-AT. p53His2/pGADT7-p53: positive controls, pHis2/pGADT7-ZmSBP17: negative controls.
The promoters (from −1500 to −1) of 10 putative target genes that were significantly upregulated by ZmSBP17 were analyzed to investigate the distribution of the core GTAC-box sequences. The promoters of all putative target genes contained more than four GTAC boxes. The schematic diagrams of the promoters of these genes are shown in Supplementary File 7. The GTAC-box may be essential for ZmSBP17 to regulate its target genes.
The direct binding of ZmSBP17 to the promoters of the putative target genes containing GTAC-box core sites was verified. The promoter fragments of NHX2 and DREB2A containing the core GTAC-box sequences were isolated through genomic PCR and inserted into pHIS2 in yeast to identify the binding affinity to ZmSBP17 on the selection medium. The results show that all co-transformants exhibited binding affinity on the selection medium (Figure 7B), indicating that ZmSBP17 can directly interact with the NHX1 and DREB2A promoter fragments containing the core GTAC-box sequences.
4 Discussion
TFs can act as activators or repressors to regulate the expression of various downstream genes, thereby enhancing stress tolerance in plants (He et al., 2019; Sun et al., 2022). The SBP-box gene family, a group of plant-specific TFs, plays a significant role in the growth and development of many plants (Shao et al., 2019; Tong et al., 2020; Li et al., 2021). However, the activation mechanism of SBP-box genes related to abiotic stress in maize remains unexplored. In this study, an SBP TF, ZmSBP17, was isolated from maize based on the salt stress transcriptome database (Supplementary File 2). The SBP domains of ZmSBP17 were found to be highly similar, with a conserved sequence at specific positions, a nuclear localization signal, and two zinc finger domains (Klein et al., 1996; Yamasaki et al., 2006). This study confirmed that ZmSBP17 contains two zinc finger domains, similar to those in other plants (Figure 1). Additionally, it was demonstrated that ZmSBP17 possesses transcriptional activation ability, with the N-terminal region exhibiting stronger activity than the C-terminal region (Figure 3).
Moreover, the expression of ZmSBP17 was significantly induced by salt and PEG stress during the seedling stage of maize (Figure 2A). This induction may be attributed to the presence of cis-elements such as ABRE, ARE, LTR, and MBS in the ZmSBP17 promoter, which are associated with abiotic stress responses. In Arabidopsis, MBS cis-elements in the gene promoter are involved in responses to drought, salt, and low-temperature stress (Yao et al., 2022; Li et al., 2023). Similarly, the ABRE element in the Arabidopsis RD29A promoter plays a crucial role in abiotic stress responses by driving and regulating the expression of stress-related TFs AREB1 and AREB2 (Bihmidine et al., 2013). Therefore, this study focuses on elucidating the molecular mechanism of ZmSBP17 in response to drought and salt stress.
Numerous studies have shown that abiotic stress can lead to excessive accumulation of ROS in plant cells, resulting in cellular oxidative stress. This condition can limit growth, development, and metabolism, ultimately leading to cell death (Singh et al., 2018). However, plants have evolved complex defense mechanisms to counteract stress-induced damage, including antioxidant enzyme systems such as SOD, POD, CAT, and APX (Gelaw and Sanan-Mishra, 2024; Kiratli et al., 2024). For instance, overexpression of SbNAC2 from Sorghum in Arabidopsis and rice enhances antioxidant enzyme activities and induces stress-responsive genes, thereby improving tolerance to abiotic stress (Jin et al., 2021). Similarly, overexpression of NtWRKY65 in tobacco plants increases salinity tolerance by elevating antioxidant enzyme activities compared with WT plants (Zhang et al., 2024). In this study, under salt and PEG stress, overexpression of ZmSBP17 in Arabidopsis significantly improved the germination rate compared with the WT (Figure 4). At the seedling stage, the activities of SOD, POD, CAT, and APX in transgenic Arabidopsis plants were all significantly higher than those observed in WT plants (Figure 5).
TFs regulate gene expression by binding to cis-acting elements in the promoters of abiotic stress-related genes. For instance, SPL1, SPL3, SPL7, and SPL8 in Arabidopsis can bind to the AP1 and FUL promoters, which contain the GTAC core sequence (Cardon et al., 1997; Birkenbihl et al., 2005; Kropat et al., 2005; Shikata et al., 2009). Our study also revealed that ZmSBP17 specifically binds to the GTAC core sequence (Figure 7), suggesting that SBPs from different species can recognize similar core sequences to regulate biological functions. Additionally, we discovered that SBP-specific GTAC core sequences are widely present in the promoters of stress-related genes, including FSD3, CBF3, DREB1A/19, NHX1/2/6, and MYB53/65/99 (Supplementary File 7). Notably, DREB2A, NHX2, and MYB53/65 were significantly upregulated in ZmSBP17-OE plants under osmotic stress (Figure 6). DREB2A regulates abiotic stress responses by activating downstream genes (Agarwal et al., 2010; Sato et al., 2014; Chen et al., 2016). NHX2, a vacuolar Na+/H+ antiporter, enhances tolerance to drought and salt stresses by reducing Na+ content in leaves, showing improved root health under salt stress, and retaining more water during drought (Wang et al., 2016). High expression of NHX1 in Arabidopsis can enhance salt tolerance in saline–alkaline environments (Pérez-Martín et al., 2022; Eduardo et al., 2007). Additionally, several MYB family members, such as AtMYB41, AtMYB44, AtMYB88, and AtMYB102, positively contribute to Arabidopsis’s response to abiotic stress (Denekamp and Smeekens, 2003; Cominelli et al., 2008; Jung et al., 2008; Xie et al., 2010). Transgenic Arabidopsis and potato plants overexpressing a mutated version of MYB33/65/101 with altered miR159 recognition sites displayed hypersensitivity to ABA and relatively high tolerance to drought (Xue et al., 2017). Collectively, these results indicate that the overexpression of ZmSBP17 in Arabidopsis may enhance osmotic stress tolerance by upregulating stress-responsive genes, supporting ZmSBP17 as a positive regulator. However, further investigations are needed to identify the cis-elements and downstream genes targeted by ZmSBP17 in maize.
5 Conclusion
In conclusion, we demonstrated that the expression of ZmSBP17 in the inbred line Hei maize was induced by salt and PEG stress. ZmSBP17 was confirmed to be a transcriptional activator and was able to bind to the GTAC core sequences to downregulate target genes. Overexpression of ZmSBP17 in Arabidopsis significantly increased tolerance to osmotic stress during the germination and seedling stages. This enhanced tolerance to abiotic stress was likely caused by a decrease in ROS levels through improved ROS clearance ability. Another possible factor may be the altered expression of a series of stress-related genes in ZmSBP17-OE plants (Figure 8). These strategies may collectively provide a more robust defense against osmotic stress.
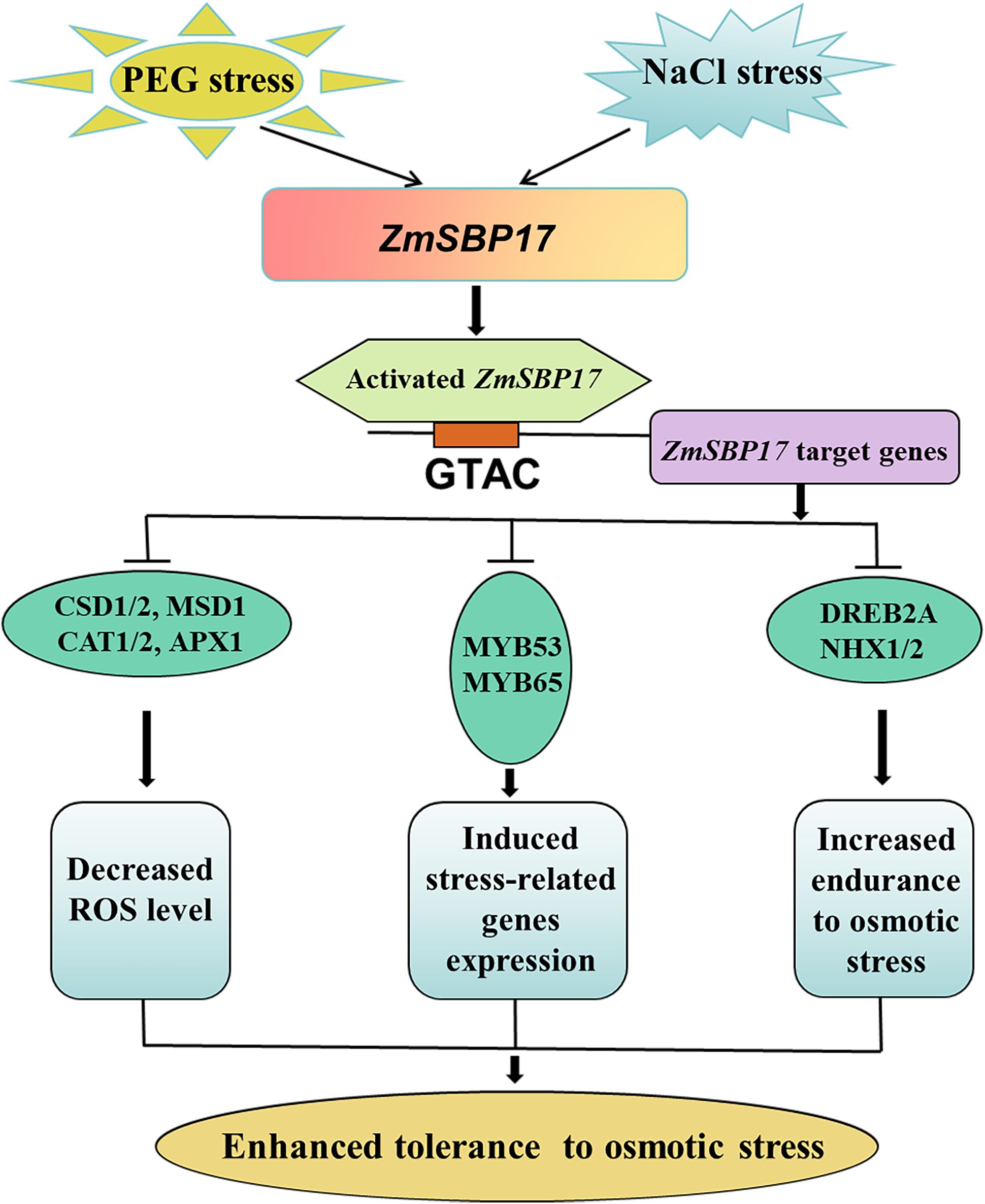
Figure 8. Working model of ZmSBP17 under osmotic stress. Salt or PEG stress induces the expression of ZmSBP17. Activated ZmSBP17 enhances stress-associated genes expression by binding to GTAC core sequences, which results in increasing ROS scavenging capability, induced stress-related genes expression, and increased endurance to osmotic stress. These physiological changes finally enhanced the tolerance of ZmSBP17 transgenic plants to osmotic stress.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
LS: Data curation, Funding acquisition, Writing – original draft, Writing – review & editing. LW: Data curation, Formal analysis, Investigation, Visualization, Writing – original draft. JN: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft. WY: Data curation, Formal analysis, Methodology, Writing – original draft. ZL: Formal analysis, Methodology, Writing – original draft. LL: Methodology, Visualization, Writing – original draft. SG: Project administration, Supervision, Writing – original draft, Funding acquisition.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Heilongjiang Provincial “Double First Class” New Round of Discipline Collaborative Innovation Achievement Construction Project of China (LJGXCG2022-107).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1483486/full#supplementary-material.
Supplementary File 1 | Primers used for qRT-PCR.
Supplementary File 2 | Expression levels of SBPs in maize under salt stress and sequence analysis of ZmSBP17.
Supplementary File 3 | Identification of the recombinant plasmid pGBKT7-ZmSBP17.
Supplementary File 4 | Cis-elements in the ZmSBP17 promoter related to abiotic stress.
Supplementary File 5 | Multiple sequence alignment of the SBP family.
Supplementary File 6 | qRT-PCR analysis of ZmSBP17 in maize plants under salt and PEG stress.
Supplementary File 7 | Overexpression of ZmSBP17 in Arabidopsis thaliana.
Supplementary File 8 | Germination rates of OE and WT seeds under salt or PEG stress and activities of ROS-scavenging enzymes.
Supplementary File 9 | Distribution of the GTAC-box core sequences in the promoters of genes regulated by ZmSBP17.
References
Agarwal, P., Agarwal, P. K., Joshi, A. J., Sopory, S. K., Reddy, M. K. (2010). Overexpression of PgDREB2A transcription factor enhances abiotic stress tolerance and activates downstream stress-responsive genes. Mol. Biol. Rep. 37, 1125–1135. doi: 10.1007/s11033-009-9885-8
Agarwal, P. K., Gupta, K., Lopato, S., Agarwal, P. (2017). Dehydration responsive element binding transcription factors and their applications for the engineering of stress tolerance. Journal of Experimental Botany 68, 2135–2148. doi: 10.1093/jxb/erx118
Bihmidine, S., Lin, J., Stone, J. M., Awada, T., Specht, J. E., Clemente, T. E. (2013). Activity of the Arabidopsis RD29A and RD29B promoter elements in soybean under water stress. Planta 237, 55–64. doi: 10.1007/s00425-012-1740-9
Birkenbihl, R. P., Jach, G., Saedler, H., Huijser, P. (2005). Functional dissection of the plant-specific SBP-domain: overlap of the DNA binding and nuclear localization domains. J. Mol. Biol. 352, 585–596. doi: 10.1016/j.jmb.2005.07.013
Cardon, G., Höhmann, S., Klein, J., Nettesheim, K., Saedler, H., Huijser, P. (1999). Molecular characterisation of the Arabidopsis SBP-box genes. Gene 237, 91–104. doi: 10.1016/s0378-1119(99)00308-x
Cardon, G. H., Höhmann, S., Nettesheim, K., Saedler, H., Huijser, P. (1997). Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: a novel gene involved in the floral transition. Plant J. 12, 367–377. doi: 10.1046/j.1365-313X.1997.12020367.x
Chao, M., Dong, J., Hu, G. H., Li, Y. Y., Huang, L., Zhang, J. B., et al. (2023). Phylogeny, gene structures, and expression patterns of the auxin response factor (GhARF2) in upland cotton (Gossypium hirsutum L.). Mol. Biol. Rep. 50, 1089–1099. doi: 10.1007/s11033-022-07999-6
Chen, H., Liu, L., Wang, L., Wang, S., Cheng, X. (2016). VrDREB2A, a DREB-binding transcription factor from Vigna radiata, increased drought and high-salt tolerance in transgenic Arabidopsis thaliana. J. Plant Res. 129, 263–273. doi: 10.1007/s10265-015-0773-0
Cheng, H., Hao, M., Wang, W., Mei, D., Tong, C., Wang, H., et al. (2016). Genomic identification, characterization and differential expression analysis of SBP-box gene family in Brassica napus. BMC Plant Biol. 16, 196. doi: 10.1186/s12870-016-0852-y
Clough, S., Bent, A. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Cominelli, E., Sala, T., Calvi, D., Gusmaroli, G., Tonelli, C. (2008). Over expression of the Arabidopsis AtMYB41 gene alter cell expansion and leaf surface permeability. Plant J. 53, 53–64. doi: 10.1111/j.1365-313X.2007.03310.x
Denekamp, M., Smeekens, S. C. (2003). Integration of wounding and osmotic stress signals determine the expression of the AtMYB102 transcription factor gene. Plant Physiol. 132, 1415–1423. doi: 10.1104/pp.102.019273
Eduardo, B., Yehoshua, S., Sottosanto, J. B. (2007). Impact of AtNHX1, a vacuolar Na+ /H+ antiporter, upon gene expression during short- and long-term salt stress in Arabidopsis thaliana. BMC Plant Biol. 7, 1–15. doi: 10.1186/1471-2229-7-18
Erinle, K. O., Jiang, Z., Ma, B., Ur-Rehman, K., Shahla, A., Zhang, Y. (2018). Physiological and molecular responses of pearl millet seedling to atrazine stress. Int. J. Phytoremediation 20, 1–10. doi: 10.1080/15226514.2017.1393385
Gelaw, T. A., Sanan-Mishra, N. (2024). Molecular priming with H2O2 and proline triggers antioxidant enzyme signals in maize seedlings during drought stress. BBA Gen. Subj. 1868. doi: 10.1016/j.bbagen.2024.130633
He, L., Bian, J., Xu, J. Y., Yang, K. J. (2019). Novel maize NAC transcriptional repressor zmNAC071 confers enhanced sensitivity to ABA and osmotic stress by downregulating stress-responsive genes in transgenic arabidopsis. J. Agric. Food Chem. 67, 8905–8918. doi: 10.1021/acs.jafc.9b02331
He, L., Wu, Z., Wang, X., Zhao, C., Cheng, D., Du, C., et al. (2024). A novel maize F-bZIP member, Zm bZIP76, functions as a positive regulator in ABA-mediated abiotic stress tolerance by6 binding to ACGT-containing elements. Plant Sci. 341. doi: 10.1016/j.plantsci.2023.111952
Hou, H., Jia, H., Yan, Q., Wang, X. (2018). Overexpression of a SBP-Box Gene (VpSBP16) from Chinese wild vitis species in Arabidopsis improves salinity and drought stress tolerance. Int. J. Mol. Sci. 19, 1–17. doi: 10.3390/ijms19040940
Hou, H., Yan, Q., Wang, X., Xu, H. (2013). A SBP-box gene vpSBP5 from chineseWild vitis species responds to erysiphe necator and defense signaling molecules. Plant Molecules Biol. Rep. 31, 1261–1270. doi: 10.1007/s11105-013-0591-2
Jin, X., Long, Y., Xiong, S., Yang, Z., Chen, W., Hawar, A., et al. (2021). SbNAC2 enhances abiotic stress tolerance by upregulating ROS scavenging activities and inducing stress-response genes in sorghum. Environ. Exp. Bot. 192, 1–13. doi: 10.1016/j.envexpbot.2021.104664
Jung, C., Seo, J. S., Han, S. W., Koo, Y. J., Kim, C. H., Song, S. I., et al. (2008). Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 146, 623–635. doi: 10.1104/pp.107.110981
Kiratli, H., Seymen, M., Kiymaci, G. (2024). Determination of tolerance to flooding stress in melon cultivars by agronomic and physio-biochemical responses. Genet. Resour. Crop Evol. 71, 1643–1657. doi: 10.1007/s10722-023-01728-z
Klein, J., Saedler, H., Huijser, P. (1996). A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Molecular&General Genet. MGG, 250 (1), 7–16. doi: 10.1007/BF02191820
Kropat, J., Tottey, S., Birkenbihl, R., Depège, N., Huijser, P., Merchant, S. (2005). A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc. Natl. Acad. Sci. United States America 102, 18730–18735. doi: 10.1073/pnas.0507693102
Lan, T., Zheng, Y., Su, Z., Yu, S., Song, H., Zheng, X., et al. (2019). OsSPL10, a SBP-box gene, plays a dual role in salt tolerance and trichome formation in rice (oryza sativa L.). G3: Genes|Genomes|Genetics 9, 4107–4114. doi: 10.1534/g3.119.400700
Li, J., Fan, R., Wu, B. D., Ji, X. Z., Hao, C. Y. (2021). Genome-wide identification and functional exploration of SBP-box gene family in black pepper (Piper nigrum L.). Genes 12. doi: 10.3390/genes12111740
Li, Y., Song, Q., Zhang, Y., Li, Z., Guo, J., Chen, X., et al. (2020). Genome-wide identification, characterization, and expression patterns analysis of the SBP-box gene family in wheat (Triticum aestivum L.). Sci. Rep. 10, 17250. doi: 10.1038/s41598-020-74417-x
Li, H., Yang, J., Ma, R., An, X., Pan, F., Zhang, S., et al. (2023). Genome-wide identification and expression analysis of MYB gene family in Cajanus cajan and CcMYB107 improves plant drought tolerance. Physiologia plantarum, 175. doi: 10.1111/ppl.13954
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 25, 402–408. doi: 10.1006/meth.1262
Mao, H. D., Yu, L. J., Li, Z. J., Yan, Y., Han, R., Liu, H., et al. (2016). Genome-wide analysis of the SPL family transcription factors and their responses to abiotic stresses in maize. Plant Gene 6, 1–12. doi: 10.1016/j.plgene.2016.03.003
Murashige, T., Skoog, F. (1962). A revised medium for rapid growth and bio-assays with tobacco tissue cultures. Physiol. Plant 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Pérez-Martín, L., Busoms, S., Almira, M. J., Azagury, N., Terés, J., Tolrà, R., et al. (2022). Evolution of salt tolerance in Arabidopsis thaliana on siliceous soils does not confer tolerance to saline calcareous soils. Plant Soil 476, 455–475. doi: 10.1007/s11104-022-05439-9
Sato, H., Mizoi, J., Tanaka, H., Maruyama, K., Qin, F., Osakabe, Y., et al. (2014). Arabidopsis DPB3-1, a DREB2A interactor, specifically enhances heat stress-induced gene expression by forming a heat stress-specific transcriptional complex with NF-Y subunits. Plant Cell 26, 4954–4973. doi: 10.1105/tpc.114.132928
Serna-Saldivar, S. O., Carrillo, E. P. (2018). Food uses of whole corn and dry-milled fractions. In Corn: Chemistry and technology, 3rd Edition, Elsevier pp. 435–467. doi: 10.1016/B978-0-12-811971-6.00016-4
Shao, Y., Zhou, H. Z., Wu, Y., Zhang, H., Lin, J., Jiang, X., et al. (2019). OsSPL3, an SBP-domain protein, regulates crown root development in rice. Plant Cell 31, 1257–1275. doi: 10.1105/tpc.19.00038
Shikata, M., Koyama, T., Mitsuda, N., Ohme-Takagi, M. (2009). Arabidopsis SBP-box genes SPL10, SPL11 and SPL2 control morphological change in association with shoot maturation in the reproductive phase. Plant Cell Physiol. 50, 2133–2145. doi: 10.1093/pcp/pcp148
Singh, S., Kumar, V., Chauhan, A., Datta, S., Wani, A. B., Singh, N., et al. (2018). Toxicity, degradation and analysis of the herbicide atrazine. Environ. Chem. Lett. 16, 211–237. doi: 10.1007/s10311-017-0665-8
Sun, L. F., Miao, X. F., Cui, J., Deng, J., Wang, X., Wang, Y. F., et al. (2018). Genome-wide high-resolution mapping of DNA methylation identifies epigenetic variation across different salt stress in maize (Zea mays L.). Euphytica 214, 1–15. doi: 10.1007/s10681-017-2076-0
Sun, M., Shen, Y., Chen, Y., et al. (2022). Osa-miR1320 targets the ERF transcription factor OsERF096 to regulate cold tolerance via JA-mediated signaling. Plant Physiol. 189, 2500–2516. doi: 10.1093/plphys/kiac208
Sun, L. F., Wu., Y., Su, S. Z., Liu, H. K., Yang, G., Li, S. P., et al. (2012). Differential gene expression of maize (Zea mays L.) inbred line H99 during somatic embryogenesis. Plant Cell Tissue Organ. Cult. 109, 271–286. doi: 10.1007/s11240-011-0093-6
Tong, T., Fang, Y., Zhang, X. X. D. (2020). Genome-wide identification, phylogenetic and expression analysis of SBP-box gene family in barley (Hordeum vulgare L.). Plant Growth Regul. 90, 137–149. doi: 10.1007/s10725-019-00559-2
Unte, U. S., Sorensen, A.-M., Pesaresi, P., Gandikota, M., Leister, D., Saedler, H., et al. (2003). SPL8, an SBP-box gene that affects pollen sac development in arabidopsis. Plant Cell 15, 1009–1019. doi: 10.1105/tpc.010678
Vinocur, B., Altman, A. (2005). Recent advances in engineering plant tolerance to abiotic stress: Achievement and limitations. Current Opinion in Biotechnology 16, 123–132. doi: 10.1016/j.copbio.2005.02.001
Wang, B., Zhai, H., He, S., Zhang, H., Ren, Z., Zhang, D., et al (2016). A vacuolar Na+/H+ antiporter gene, IbNHX2, enhances salt and drought tolerance in transgenic sweetpotato. Scientia Horticulturae 201, 153–166. doi: 10.1016/j.scienta.2016.01.027
Wang, X., Liu, H., Yu, F., Hu, B., Jia, Y., Sha, H., et al (2019). Differential activity of the antioxidant defence system and alterations in the accumulation of osmolyte and reactive oxygen species under drought stress and recovery in rice (Oryza sativa L.) tillering. Scientific Reports 9, 8543. doi: 10.1038/s41598-019-44958-x
Xie, Z., Li, D., Wang, L., Sack, F. D., Grotewold, E. (2010). Role of the stomatal development regulators FLP/MYB88 in abiotic stress responses. Plant J. 64, 731–739. doi: 10.1111/j.1365-313X.2010.04364.x
Xue, T., Liu, Z., Dai, X., Xiang, F. (2017). Primary root growth in Arabidopsis thaliana is inhibited by the miR159 mediated repression of MYB33, MYB65 and MYB101. Plant Sci. 262, 182–189. doi: 10.1016/j.plantsci.2017.06.008
Yamasaki, K., Kigawa, T., Inoue, M., Yamasaki, T., Yabuki, T., Aoki, M., et al. (2006). An Arabidopsis SBP-domain fragment with a disrupted C-terminal zinc-binding site retains its tertiary structure. FEBS Lett. 580, 2109–2116. doi: 10.1016/j.febslet.2006.03.014
Yamasaki, K., Kigawa, T., Seki, M., Shinozaki, K., Yokoyama, S. (2013). DNA-binding domains of plant-specific transcription factors: structure, function, and evolution. Trends Plant Sci. 18, 267–276. doi: 10.1016/j.tplants.2012.09.001
Yao, C., Li, W., Liang, X., Ren, C., Liu, W., Yang, G., et al. (2022). Molecular cloning and characterization of MbMYB108, a Malus baccata MYB transcription factor gene, with functions in tolerance to cold and drought stress in transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 23, 4846. doi: 10.3390/ijms23094846
Yu, Y., Duan, X., Ding, X., Chen, C., Zhu, D., Yin, K., et al. (2017). A novel AP2/ERF family transcription factor from Glycine soja, GsERF71, is a DNA binding protein that positively regulates alkaline stress tolerance in Arabidopsis. Plant Mol. Biol. 94, 509–530. doi: 10.1007/s11103-017-0623-7
Zhang, X., Wang, L., Meng, H., Wen, H., Fan, Y., Zhao, J. (2011). Maize ABP9 enhances tolerance to multiple stresses in transgenic Arabidopsis by modulating ABA signaling and cellular levels of reactive oxygen species. Plant Mol. Biol. 75, 365–378. doi: 10.1007/s11103-011-9732-x
Zhang, X., Zhang, Y., Li, M., Jia, H., Wei, F., Xia, Z., et al. (2024). Overexpression of the WRKY transcription factor gene NtWRKY65 enhances salt tolerance in tobacco (Nicotiana tabacum). BMC Plant Biol. 24, 326. doi: 10.1186/s12870-024-04966-0
Zhao, C., Zhang, H., Song, C., Zhu, J. K., Shabala, S. (2020). Mechanisms of Plant Responses and Adaptation to Soil Salinity. Innovation (Camb) 24, 100017. doi: 10.1016/j.xinn.2020.100017
Keywords: maize (Zea mays L.), osmotic stress, regulatory mechanism, SBP transcription factor, Arabidopsis thaliana
Citation: Sun L, Wang L, Niu J, Yang W, Li Z, Liu L and Gao S (2024) The maize gene ZmSBP17 encoding an SBP transcription factor confers osmotic resistance in transgenic Arabidopsis. Front. Plant Sci. 15:1483486. doi: 10.3389/fpls.2024.1483486
Received: 20 August 2024; Accepted: 07 October 2024;
Published: 07 November 2024.
Edited by:
Yong Jia, Murdoch University, AustraliaReviewed by:
Fulgencio Alatorre-Cobos, Centro de Investigación Científica de Yucatán, MexicoYong Zhou, Jiangxi Agricultural University, China
Copyright © 2024 Sun, Wang, Niu, Yang, Li, Liu and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuren Gao, Z2Fvc2hyMTA3QDEyNi5jb20=
 Lifang Sun
Lifang Sun Lijiao Wang
Lijiao Wang Jinping Niu
Jinping Niu Wei Yang1
Wei Yang1 Libin Liu
Libin Liu