
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 16 October 2024
Sec. Plant Breeding
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1478854
Gibberellin 20-oxidases (GA20oxs) are multifunctional enzymes involved in regulating gibberellin (GA) biosynthesis and controlling plant growth. We identified and characterized the GA20ox1 gene in a plant height mutant of Sophora davidii, referred to as SdGA20ox1. This gene was expressed in root, stem, and leaf tissues of the adult S. davidii plant height mutant, with the highest expression observed in the stem. The expression of SdGA20ox1 was regulated by various exogenous hormones. Overexpression of SdGA20ox1 in Arabidopsis resulted in significant elongation of hypocotyl and root length in seedlings, earlier flowering, smaller leaves, reduced leaf chlorophyll content, lighter leaf color, a significant increase in adult plant height, and other phenotypes. Additionally, transgenic plants exhibited a substantial increase in biologically active endogenous GAs (GA1, GA3, and GA4) content, indicating that overexpression of SdGA20ox1 accelerates plant growth and development. Using a yeast two-hybrid (Y2H) screen, we identified two SdGA20ox1-interacting proteins: the ethylene receptor EIN4 (11430582) and the rbcS (11416005) protein. These interactions suggest a potential regulatory mechanism for S. davidii growth. Our findings provide new insights into the role of SdGA20ox1 and its interacting proteins in regulating the growth and development of S. davidii.
Plant height is a crucial component of plant structure, significantly impacting crop yield and quality (Wang et al., 2017). Many genes associated with plant height are closely linked to phytohormone synthesis, transportation, and signal transduction (Wang and Li, 2008; Ku et al., 2015). For example, the semidwarf and high-yield varieties of the “Green Revolution” resulted from changes in gibberellin (GA) biosynthesis and signaling pathways (Hedden, 2003). Gibberellin is a plant hormone that affects plant growth and development, playing a role in seed germination, stem elongation, leaf growth, and flowering (Yamaguchi, 2008). Gibberellin 20-oxidases (GA20oxs) are critical regulatory enzymes in the GA synthesis pathway, acting as soluble dioxygenases that regulate the synthesis of biologically active GA4 and GA1 in plants (Phillips et al., 1995). Many GA20oxs genes have been isolated and characterized in Arabidopsis (Arabidopsis thaliana), tobacco (Nicotiana tabacum), and other plants. For instance, overexpression of AtGA20ox1 in Arabidopsis thaliana led to earlier flowering and increased plant height (Coles et al., 1999). Similarly, overexpression of CrGA20ox1 promoted increased height in Camellia reticulata compared to wild-type plants (Wang et al., 2018), and overexpression of StGA20ox1 in Solanum tuberosum resulted in increased plant height and a requirement for longer daylight to produce tubers (Carrera et al., 2000). In contrast, overexpression of PdGA20ox1 in the woody plant Pinus densiflora significantly enhanced stem growth and increased stem weight (Jeon et al., 2016).
Sophora davidii (Franch.) Skeels (2n = 18) is a typical deciduous shrub or small tree found in the mountains of Guizhou Province, Southwest China (Zhao et al., 2022a). In recent decades, stone desertification in Southwest China has severely threatened the local ecology (Ding et al., 2020). S. davidii, with its deep and well-developed root system, has been widely used for vegetation restoration to repair damaged ecosystems (Zhao et al., 2022c, Zhao et al., 2024). The stems and leaves of S. davidii are tender, tasty, and rich in protein, vitamins, minerals, amino acids, alkaloids, and flavonoids, making it a high-quality local fodder with medicinal value (Zhao et al., 2022b). However, adult S. davidii plants can reach up to 2 meters in height, which hinders the normal feeding of shorter livestock such as goats. Conventional breeding methods to develop new dwarf cultivars are extremely difficult and time-consuming. However, by analyzing the functions of genes regulating the height of S. davidii plants, genome editing technologies can be used to accelerate the breeding of new semi-dwarf or dwarf phenotypes of S. davidii, promoting the development of local ecological livestock farming.
To date, no studies have investigated the mechanism of S. davidii seedling height growth. Our research team previously conducted preliminary studies on S. davidii plant height mutants obtained through mutagenesis and speculated that the GA20ox1 gene plays an important role in plant height regulation (Xie et al., 2023; Zhao et al., 2022b). In this study, we isolated SdGA20ox1 from S. davidii plant height mutants, analyzed its encoded protein sequence, and introduced it into Arabidopsis to validate its function. We constructed a yeast nuclear library and screened for proteins interacting with SdGA20ox1 using yeast two-hybrid (Y2H) and bimolecular fluorescence complementation (BiFC) techniques. To our knowledge, this is the first report on the isolation of the GA20-oxidase gene from S. davidii. The research results reveal the function of the SdGA20ox1 gene, enriching the knowledge of GA20ox1 and providing material support for the genetic engineering of plant height mutant varieties.
Wild-type (WT) Sophora davidii and radiation-induced plant height mutants grown in Guiyang, Guizhou, China, were used in this study, similar to previous studies (Zhao et al., 2022b). These plants were grown under natural light conditions with regular irrigation. Arabidopsis thaliana ecotype Columbia (Col-0) was used as the WT and grown alongside the second generation (T2) of pure transgenic Arabidopsis in a growth chamber set at 22 ± 2°C with 70% humidity and 1200 Lux light intensity, following a 16-hour daytime and 8-hour darkness cycle. For sampling, S. davidii and Arabidopsis samples were immediately immersed in liquid nitrogen and stored at -80°C.
Three-month-old S. davidii plant height mutants with similar growth status were selected for various treatments. Hormone treatments included 10 μM Gibberellin A3 (GA3, CAS: 77-06-5), 5 μM brassinolide (BR, CAS: 72962-43-7), 10 μM indole-3-acetic acid (IAA, CAS: 87-51-4), 10 μM uniconazole (CAS: 83657-22-1), and 10 μM Ethephon (substitute for ethylene, ETH, CAS: 16672-87-0) (Xie et al., 2023). All hormone solutions were first dissolved in a small amount of ethanol and then diluted with distilled water to the appropriate concentration. The hormones were sprayed on the plants until the solution started to run off the leaves. Stems from the various treatments were sampled at the indicated time points (0, 3, 6, 9, 12, 24, 36, 48 hours) for qRT-PCR analysis.
Total RNA was extracted from plant tissues as described previously (Zhao et al., 2022a). Reverse transcription was performed using the HiFiScript cDNA synthesis kit (Cwbiotech), and gene-specific primers were synthesized using the PerlPrimer program. qRT-PCR was performed on an Applied Biosystems CFX ConnectTM real-time PCR detection system using UltraSYBR® Mixture (Low ROX). The qRT-PCR thermal cycling parameters were: 95°C for 10 minutes followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute, in 20 μL volumes. Expression data were analyzed using the 2-ΔΔCt method. Each reaction was performed with three independent biological replicates and three technical replicates per sample. All primers used for qRT-PCR are listed in Supplementary Table S1.
Total RNA was extracted from S. davidii as described in Zhao et al. (2022b), and the coding region of SdGA20ox1 was amplified according to the manufacturer’s instructions based on the transcriptomic data previously obtained in our laboratory. The PCR products were subsequently cloned into the pGEM-T vector (Promega Corporation, Madison, WI, USA) and sequenced. Specific primer information for gene cloning is listed in Supplementary Table S1.. The 40 μL RT-PCR solution contained 5.0 μL of 2× Taq PCR Master Mix (Tian-gen, China), 1 μL forward and 1 μL reverse primers, 3.0 μL cDNA, and 15.0 μL deionized water. RT-PCR was performed with 30 cycles of 95°C for 30 seconds, 58°C for 30 seconds, and 72°C for 30 seconds, followed by 72°C for 10 minutes. The PCR product was then recovered and ligated into the pEASY-Blunt cloning vector (TransGen, Beijing, China), and sequenced by Sangon Biotech (Shanghai, China).
Amino acid sequences were obtained from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov), with the gene name SdGA20ox1 and GenBank accession number ON229923. MEGA 11 software (https://www.megasoftware.net/megamac.php) and the online tool Evolview (https://www.evolgenius.info) were used to create phylogenetic trees (Subramanian et al., 2019). A neighbor-joining phylogenetic tree was generated using MEGA 11 with the Poisson model, gamma-distributed rates, and 1,000 bootstrap replicates.
The BamHI (5’) and PstI (3’) restriction sites were added to the ends of the SdGA20ox1 CDS fragment, respectively, and the PCR products were cloned into the pHB vector and sequenced. The overexpression vector plasmid was transformed into Agrobacterium GV3101, and positive transgenic plants were identified with 50 μg/mL of kanamycin. The Agrobacterium strain carrying the pHB::SdGA20ox1 vector was transformed into Arabidopsis using the flower dip method (Clough and Bent, 1998). PCR primer sequences for transgenic plants are shown in Supplementary Table S1.. The transgenic Arabidopsis was screened for resistance using 50 μmol/L glufosinate until stable genetically pure T2 generations (OE2, OE5, and OE7) were obtained for subsequent experiments.
Wild-type and transgenic plants were grown under identical conditions, and seeds were collected simultaneously. Seed germination tests were conducted as described by Yan et al. (2017). Thirty seeds were sown on half-strength MS solid medium and incubated in the dark at 4°C for 4 days before being transferred to a growth chamber at 22°C. Seeds that completed germination were counted every 24 hours. The experiment was repeated three times. For seedling hypocotyl and root length determination, seeds were sown on half-strength MS solid medium. Plates were refrigerated at 4°C in the dark for 4 days, then moved to 22°C with a 16-hour light/8-hour dark cycle. On day 7, the hypocotyl lengths of approximately 10 seedlings were measured. On day 14, the root lengths of about 10 seedlings were measured. Arabidopsis seedlings grown on half-strength medium for 7 days were transplanted to pots for 45 days, and morphological characteristics (leaf length, plant height) were measured during the growing season.
One gram of leaf tissue from 45-day-old Arabidopsis and 3-month-old S. davidii plants was collected and ground to a powder in liquid nitrogen. The powdered leaves were placed in 10 ml of extraction solution (95% ethanol: 80% acetone = 1:1 by volume), mixed, and stored overnight at 4°C in the dark. Total chlorophyll (Chl) was measured according to the method described by Yang et al. (2020), and each measurement was repeated three times.
Samples of wild-type and transgenic Arabidopsis were collected and weighed for further analysis. Young stems (from stem tip to node) were collected for GA content determination. The assay was performed as described previously by Chen et al. (2012), with three independent biological replicates and three technical replicates measured for each sample.
To study the subcellular localization of SdGA20ox1, specific primers with the enzymatic cut site BsaI/Eco31I were designed. The pBWA(V)HS-SdGA20ox1-Glosgfp vector was constructed, transformed into Escherichia coli (DH5α), and then transformed into tobacco leaves. The fluorescence signal was observed using confocal laser scanning microscopy (CLSM, Leica TCS SP2) with excitation and emission light at 511 nm and 525 nm, respectively. For the bimolecular fluorescence complementation (BiFC) assay, full-length SdGA20ox1, 11430582, and 11416005 genes were amplified using primers (Supplementary Table S1), cloned into the PpSm35s vector (Engler et al., 2008), and transiently transformed into the lower epidermis of tobacco leaves. After 48 hours of transfection, interactions were observed using CLSM with excitation at 511 nm and emission at 525 nm.
To prevent self-activation of SdGA20ox1, a self-activation assay was performed. The decoy plasmid transformed recipient strain AH109 was cotransformed with pGBKT7-SdGA20ox1 and pGBKT7 using Saccharomyces cerevisiae. The transformed AH109 strain was plated on SD/-Trp plates deficient in tryptophan. Six randomly selected spots were verified by PCR with vector primers of pGBKT7. Three cloning sites were randomly selected and applied to culture medium deficient in tryptophan, histidine, and adenine, supplemented with X-α-Gal blue-displaying substrate (5-Bromo-4-chloro-3-indolyl-alpha-D-galactopyranoside), namely SD/-Trp, SD/-Trp/-His, SD/-Trp/-His/-Ade, and SD/-Trp/-His/-Ade+X-α-gal plates. Plates were incubated at 30°C for 3-5 days. pGBKT7 was used as the negative control, and the diameter and color of the colonies were observed and recorded.
To identify candidate interacting proteins of SdGA20ox1, a cDNA library of S. davidii mutants was constructed, and transformed cells were plated on SD/-Trp plates and cultured. Cells were then transferred to YPDA and cultured for 4–5 hours. Twenty microliters of culture were diluted and plated on SD/-TLH plates, incubated at 30°C for 3-7 days. Single colonies were picked and transferred to SD/-TLH plates for 3-5 days. Positive yeast clones were obtained by screening on SD/-TLH plates. Positive clones were amplified from yeast cells and sequenced. The positive clones were diluted and plated on SD/-TL, SD/-TLH, SD/-TLHA, and SD/-TLHA/+X-α-gal, respectively, and incubated at 30°C for 3 days. pGBKT7-p53 and pGADT7-largeT were co-transformed as positive controls, and pGBKT7-laminC and pGADT7-largeT were cotransformed as negative controls.
All experiments were performed at least three times. Error bars in each graph indicate the mean values ± SE of three replicates. Statistical significance between control and treatment groups was determined using Tukey’s test and Student’s t-test in SPSS Statistics software version 19 (IBM, USA). Different lowercase letters indicate significant differences (P < 0.05), with ** indicating P < 0.01 and * indicating P < 0.05.
Based on the S. davidii transcriptome data (SRA315988) previously obtained by our group, we isolated the full-length cDNA of SdGA20ox1 (GenBank accession number ON229923) from S. davidii plant height mutants using reverse transcriptase-polymerase chain reaction (RT-PCR). The full-length cDNA contained a coding sequence (CDS) of 1143 bp (Supplementary Figure S1). The primary structure of the SdGA20ox1 protein was deduced to contain 380 amino acids, with a predicted molecular mass of 43.16 kDa and an isoelectric point (pI) of 5.96. According to the National Center for Biotechnology Information (NCBI), the SdGA20ox1 protein contains a conserved 2OG-FeII_Oxy structural domain. Using the NCBI Conserved Domain database, we identified that the protein includes a 2OG-FeII_Oxy structural domain spanning amino acids 225–324, classifying SdGA20ox1 as part of the 2OG-FeII_Oxy protein superfamily (Figure 1A).
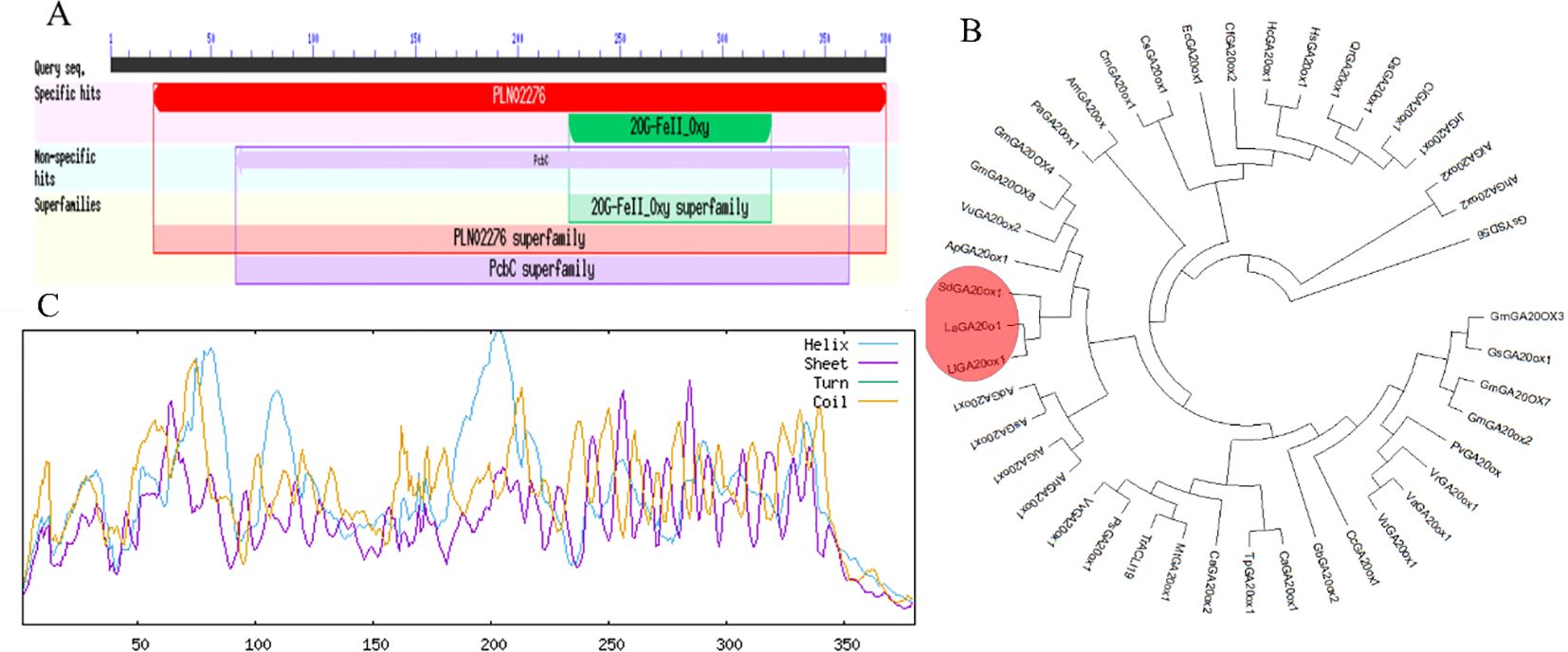
Figure 1. Structural features and phylogenetic analysis of the SdGA20ox1 protein. (A) Structure of the SdGA20ox1 protein. (B) Phylogenetic tree analysis of the SdGA20ox1 protein based on the amino acid sequence. The red boxes represent other GA20ox1 proteins with the highest identity to the SdGA20ox1 protein. The GenBank accession numbers are as follows: LlGA20ox1: Lupinus luteus (MG181995.1), LaGA20o1:Lupinus angustifolius (XM_019586625.1), GmGA20OX8:Glycine max (XM_003555313.5), GmGA20ox2:G. max (EU884445.1), GmGA20OX3:G. max (NM_001371636.1), GmGA20OX4:G. max (XR_416536.4), GmGA20OX7:G. max (NR_163976.2), GsGA20ox1:Glycine soja (XM_028324044.1), GsYSD56:G. soja (CP154586.1), VuGA20ox2:Vigna unguiculata (XM_028067520.1), VuGA20ox1:V. umbellate (XM_047326684.1), MtGA20ox1:Medicago truncatula (XM_003592315.4), PvGA20ox:Phaseolus vulgaris (U70530.1), VaGA20ox1:Vigna angularis (XM_017583418.2), VrGA20ox1:Vigna radiata var. radiata (XM_014649777.2), CaGA20ox2:Cicer arietinum (XM_004496578.3), CaGA20ox1:C. arietinum (XM_004513601.3), CcGA20ox1:Cajanus cajan (XM_020351662.2), AhGA20ox1:Arachis hypogaea (XM_025823797.2), AhGA20ox2:A. hypogaea (XM_025800912.2), AiGA20ox1:Arachis ipaensis (XM_016321927.2), AiGA20ox2:A. ipaensis (XM_016348678.2), AsGA20ox1:Arachis stenosperma (XM_057879779.1), ApGA20ox1:Abrus precatorius (XM_027511274.1), PaGA20ox1:Prosopis alba (XM_028900508.1), AdGA20ox1:Arachis duranensis (XM_016086148.3), PsGA20ox1:Pisum sativum (XM_051035601.1), VvGA20ox1:Vicia villosa (XM_058883955.1), AmGA20ox:Acacia mangium (EU252106.1), GbGA20ox2:Gastrolobium bilobum (XM_061494988.1), TpGA20ox1:Trifolium pratense (XM_045962335.1), QrGA20ox1:Quercus robur (XM_050397600.1), QsGA20ox1:Quercus suber (XM_024053726.2), CiGA20ox1:Carya illinoinensis (XM_043114810.1), JrGA20ox1:Juglans regia (XM_018972764.2), CfGA20ox2:Cornus florida (XM_059811028.1), CmGA20ox1:Cucumis melo (XM_008469033.3), CsGA20ox1:Cucumis sativus (XM_004143598.3), HcGA20ox1:Hibiscus cannabinus (KY399834.1), EcGA20ox1:Erigeron canadensis (XM_043758862.1), HsGA20ox1:Hibiscus syriacus (XM_039177100.1), TrACLI19:Trifolium repens (CP125836.1). (C) secondary structures comparison of the SdGA20ox1 protein. In the figure, Helix: Alpha helix (Hh), Sheet: Extended strand (Ee), Coil: Random coil (Cc), Turn: Beta turn (Tt).
To further characterize the SdGA20ox1 gene within the GA20oxs family, we performed a phylogenetic analysis of the amino acid sequences of GA20oxs from various plant species. The phylogenetic tree revealed that the amino acid sequences of GA20oxs from S. davidii, Lupinus luteus, and Lupinus angustifolius clustered together, indicating a higher homology of SdGA20ox1 with LaGA20ox1 and LlGA20ox1 proteins (Figure 1B). The SdGA20ox1 protein showed the highest identity with LaGA20ox1 from Lupinus angustifolius and LlGA20ox1 from Lupinus luteus (Figure 1C). Additionally, The SdGA20ox1 protein contains three types of helical structures, with alpha helices accounting for 33.68%, irregular coils accounting for 52.11%, and extended chains accounting for 14.21%, which indicated that its main components are α-helices and irregular coils (Figure 1C). Based on these findings, we named this gene SdGA20ox1.
The quantitative real-time fluorescence PCR (qRT-PCR) analysis results showed significant differences in SdGA20ox1 expression across various parts of S. davidii, influenced by exogenous phytohormones (Figure 2). The gene was significantly more expressed in leaves and stems than in roots (P < 0.05), with the highest expression in stems (Figure 2A). The expression of SdGA20ox1 decreased significantly after exogenous spraying of the gibberellin synthesis inhibitor uniconazole; however, it began to increase again after 12 hours, reaching a level not significantly different from 0 hours (Figure 2B). Following exogenous ETH treatment, the expression of SdGA20ox1 showed a trend of decreasing and then increasing, and it was significantly higher than at 0 hours after 24 hours of treatment (Figure 2C). After exogenous BR treatment, SdGA20ox1 expression peaked at 12 hours, then decreased, but remained not significantly different from pre-treatment levels (Figure 2D). After exogenous spraying of GA3, SdGA20ox1 expression continued to decline, eventually decreasing significantly at 48 hours (Figure 2E). Conversely, following exogenous spraying of IAA, SdGA20ox1 expression first increased and then decreased, peaking at 24 hours, which was significantly higher than at other treatment times (Figure 2F).
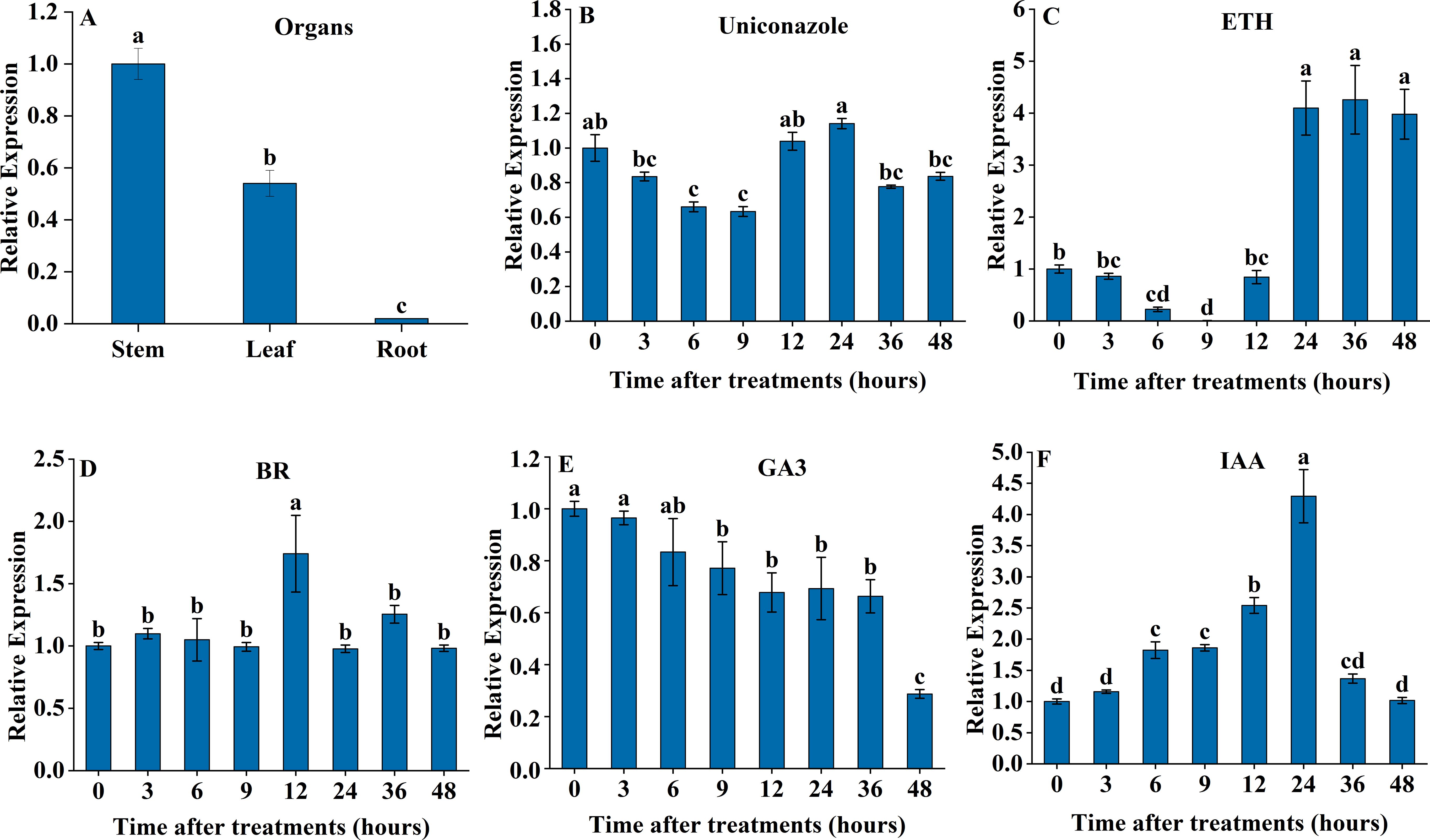
Figure 2. qRT-PCR was used to determine the expression pattern of the SdGA20ox1 gene in adult S. davidii mutants. (A) Expression of SdGA20ox1 in stem, leaf, and root. (B) Relative expression under 10 μM uniconazole treatment conditions. (C) Relative expression under 10 μM ETH treatment conditions. (D) Relative expression under 5 μM BR treatment conditions. (E) Relative expression under 10 μM GA3 treatment conditions. (F) Relative expression after 10 μM IAA treatment conditions. For each treatment, the expression level at 0 h was set as 1.0, and each value represents three replicate experiments. Values are shown as mean ± standard error (SE), and different lowercase letters indicate significant differences (P < 0.05).
The open reading frame (ORF) of SdGA20ox1 was fused to the N-terminal of the GFP protein in the CaMV35S promoter-driven pBWA(V)HS-SdGA20ox1-Glosgfp vector to construct the 35S::SdGA20ox1 vector, which was transiently transformed into tobacco leaves. Microscopic observation revealed that SdGA20ox1-GFP was distributed in the nucleus and cytoplasm, with weaker expression in the cell membrane and more concentration in the nucleus compared to the distribution of the GFP control (Figure 3).
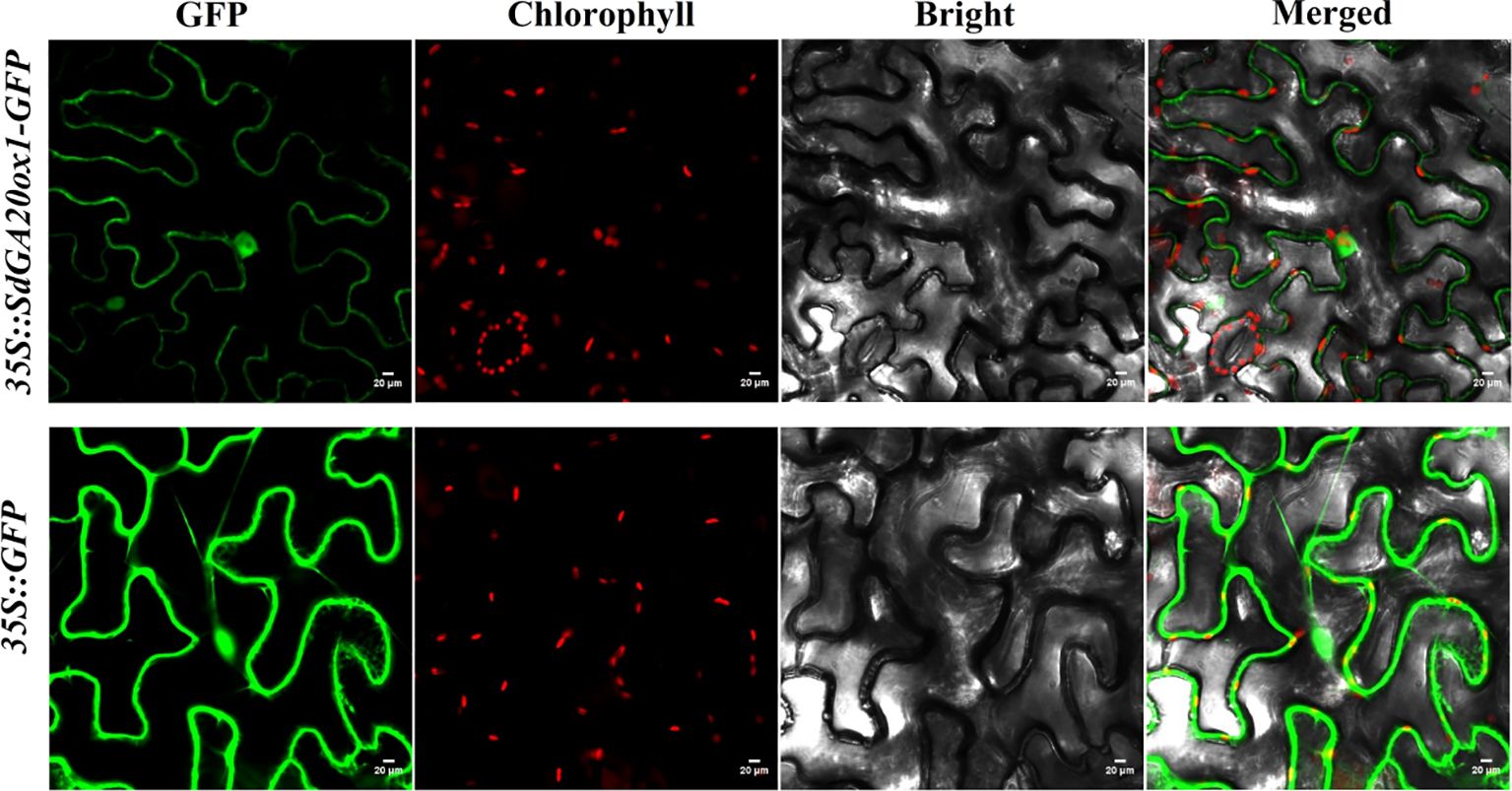
Figure 3. Subcellular localization of SdGA20ox1. GFP, green fluorescent protein; chlorophyll, chloroplast channel; bright, bright field; merged, GFP + chlorophyll + bright are shown. Scale bars = 20 μm.
PCR amplification of target fragments in Arabidopsis resistant plants showed consistency with the positive control band, but the target fragment was not detected in wild-type Arabidopsis (Supplementary Figure S2), indicating the successful acquisition of transgenic Arabidopsis. The T2 generation homozygous OE2, OE5, and OE7 lines were further screened for subsequent experiments. During seed germination, the germination rate of seeds from transgenic lines overexpressing (OE) SdGA20ox1 was significantly higher than that of the wild type (WT), with the germination time also earlier (Figures 4A, B). On the second day, the transgenic Arabidopsis seeds germinated in large quantities, with the germination rate of OE7 close to 80%, while the WT lines had a germination rate of less than 20%, significantly lower than that of the transgenic lines (P < 0.05).
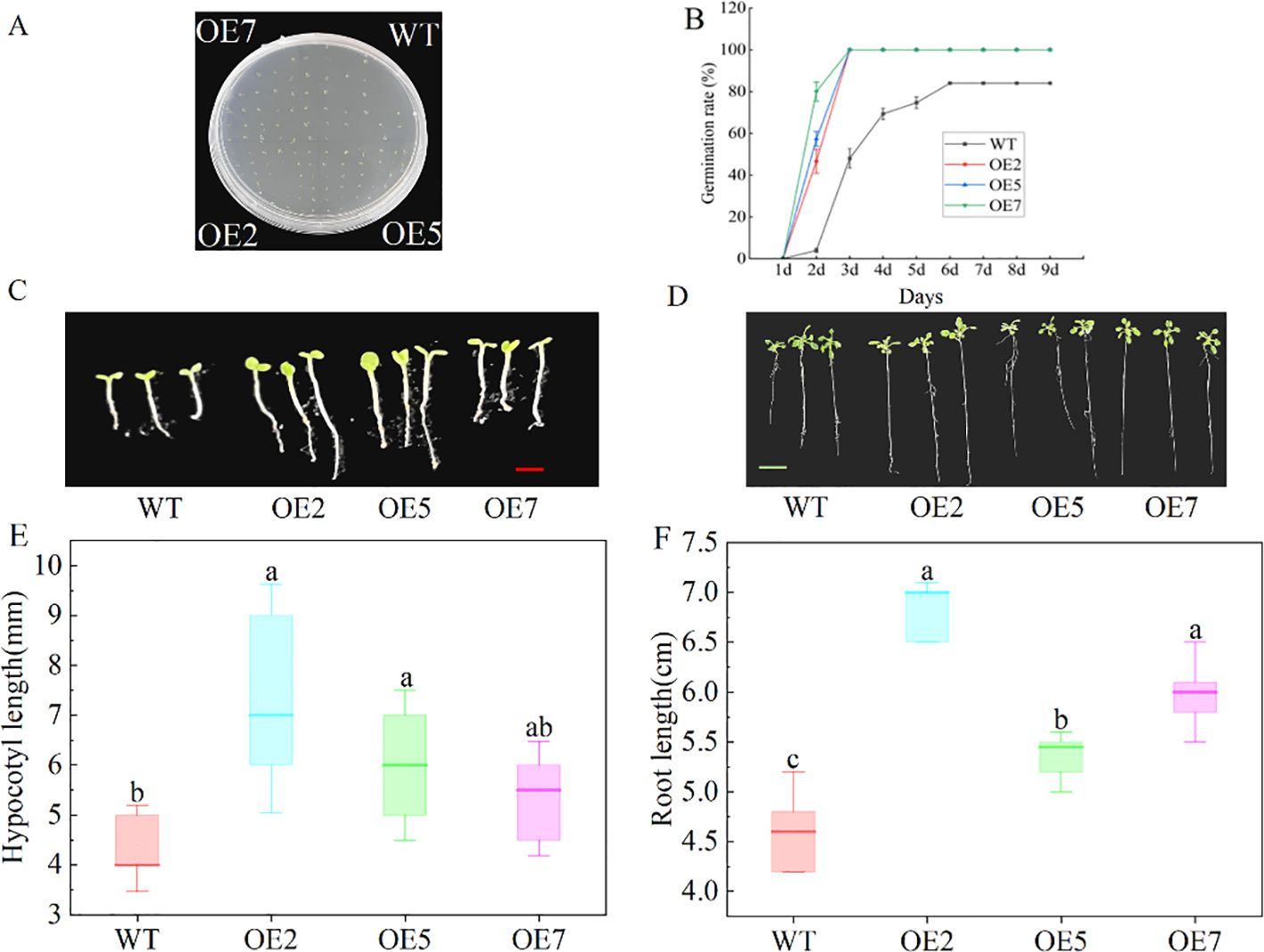
Figure 4. Effect of overexpression of SdGA20ox1 on seed germination and seedling growth of Arabidopsis. (A, B) Arabidopsis germination assay on half-strength MS medium (WT as control), using a total of 30 seeds per treatment and counting germination numbers from day 1 for 9 days. (C, E) Hypocotyl length of Arabidopsis seedlings after 7 days of growth on half-strength MS medium. (D, F) Root length of Arabidopsis seedlings after 14 days of growth on half-strength MS medium. WT: Col-0 wild-type Arabidopsis control; OE2, OE5, and OE7, separate overexpression lines. Each value represents three replicate experiments. Values are shown as mean ± standard error (SE), and different lowercase letters indicate significant differences (P < 0.05). The red bar in C: 0.5 mm, the green bar in D: 1.25cm.
Compared to the WT, transgenic Arabidopsis seeds sown on half-strength MS agar medium exhibited significantly faster vegetative growth (Figures 4C, D). After 7 days of growth on half-strength MS medium, the hypocotyl length of transgenic Arabidopsis seedlings was significantly greater than that of the WT (Figures 4C, E), and after 14 days, the root length of transgenic Arabidopsis was significantly greater than that of the WT (Figure 4F), indicating that overexpression of SdGA20ox1 resulted in increased hypocotyl and root lengths in Arabidopsis seedlings.
After transplanting plants grown in half-strength medium for 14 days into nutrient soil, we observed the time of first bud appearance in both WT and transgenic Arabidopsis, along with their plant heights at this time. The experiment showed that transgenic Arabidopsis flowered earlier, with the first bud appearing about 5 days earlier than in the WT (Figure 5A). Additionally, when the first bud appeared, the plant height of the transgenic Arabidopsis was significantly higher than that of the WT (P < 0.05) (Figure 5B). We also measured the leaf size and leaf chlorophyll content of 3-month-old S. davidii plant height mutants and Arabidopsis after 45 days of growth. The results showed that the leaf length and plant height of the S. davidii plant height mutant were significantly higher than those of the WT S. davidii (Figures 5F, G, 6A, B), and the leaf length of transgenic Arabidopsis was also significantly higher than that of the WT (Figures 5C, D). However, the chlorophyll content of the leaves in both the S. davidii plant height mutant and transgenic Arabidopsis was not significantly different from the WT (P > 0.05), although it was lower (Figures 5E, H). After 45 days of growth in nutrient soil, the plant height of transgenic Arabidopsis was significantly higher (P < 0.05) than that of WT Arabidopsis across the three overexpression lines (OE2, OE5, and OE7) (Figures 6C, D). The plant heights of the OE2, OE5, and OE7 lines increased by 37.0%, 34.8%, and 27.7%, respectively, compared to WT Arabidopsis.
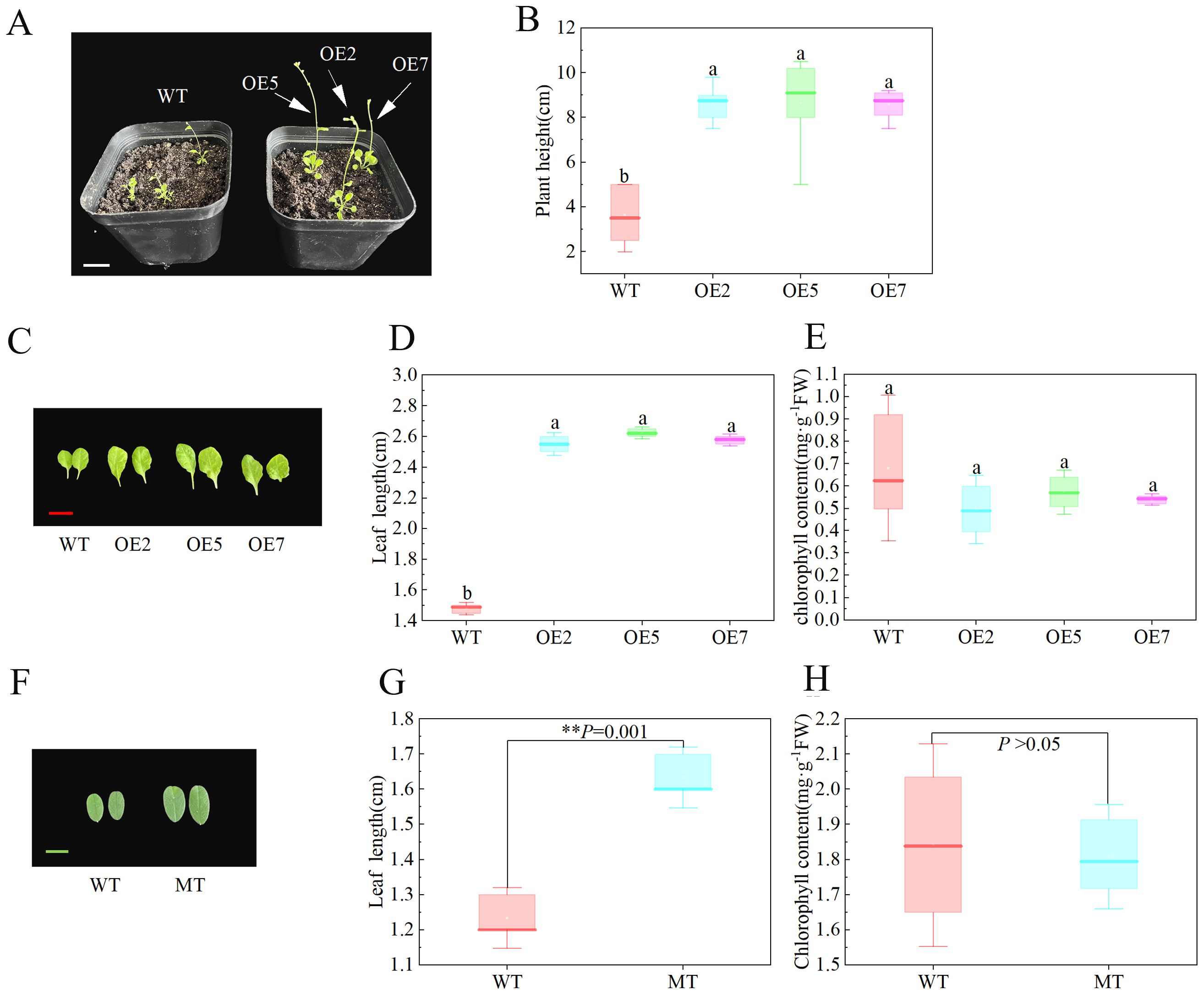
Figure 5. Flowering, leaf, and plant height observations of SdGA20ox1 overexpression in Arabidopsis. (A, B) Flowering stage and plant height observation of Arabidopsis plants at the appearance of the first bud, WT: Col-0 wild-type Arabidopsis control; OE2, OE5, and OE7, separate overexpression lines. (C–E): Leaf length and leaf chlorophyll content measurement of Arabidopsis plants after 45 days of growth, WT: Col-0 wild-type Arabidopsis control; OE2, OE5, and OE7, separate overexpression lines. (F–H): Leaf length and chlorophyll content measurements of 3-month-old S. davidii plants, WT: wild-type S. davidii, MT: S. davidii plant height mutant. Each value represents three replicate experiments. Values are shown as mean ± standard error (SE), different lowercase letters indicate significant differences (P < 0.05), and for (G, H), statistical significance between WT and MT was evaluated by t-tests: **P < 0.01. The white bar in A: 2cm, the red bar in C: 2cm, the green bar in E: 1cm.
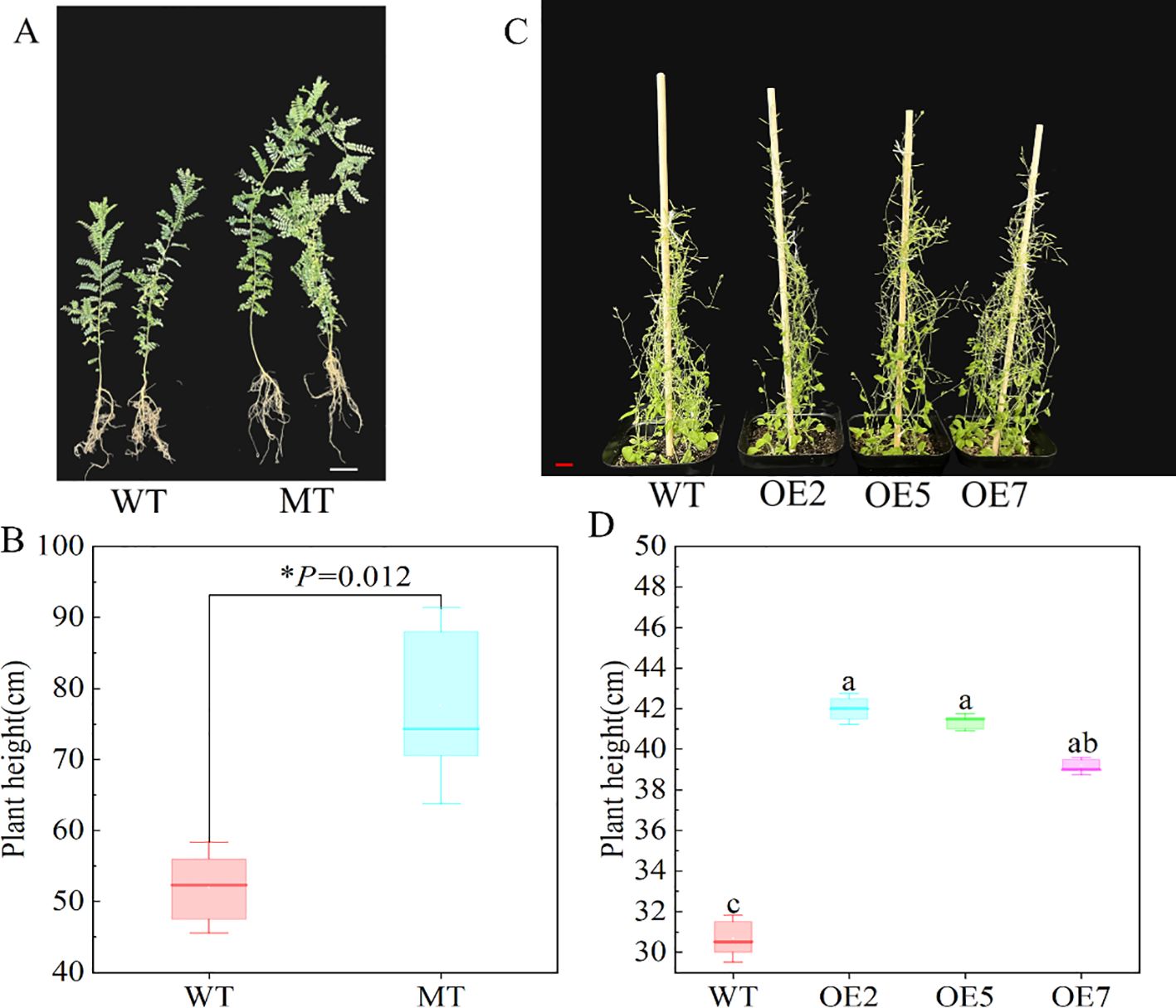
Figure 6. Observation of plant height in Arabidopsis and S. davidii. (A, B) 3-month-old S. davidii plant height, WT: wild type, MT: S. davidii plant height mutant. (C, D) Arabidopsis plant height after 45 days of growth, WT: Col-0 wild-type Arabidopsis control; OE2, OE5, and OE7, separate overexpression lines. Each value represents three replicate experiments, and values are shown as mean ± standard error (SE), with different lowercase letters indicating significant differences (P < 0.05), and for B, statistical significance between WT and MT was evaluated by t-tests: *P < 0.05. The white bar in A: 1cm, the red bar in C: 2cm.
We measured the biologically active GA1, GA3, and GA4 contents in young transgenic Arabidopsis (OE2, OE5, and OE7) stems grown for 40 days (Figure 7). The results showed that GA1 and GA3 contents in transgenic plants were significantly higher than in WT (P < 0.05), approximately 1.8-fold, 2.6-fold, and 1.6-fold for GA1 and 1.2-fold, 1.5-fold, and 1.2-fold for GA3, respectively. While GA4 content in transgenic plants was higher than in WT, only OE5 showed a significant difference (P < 0.05), being about 1.4-fold higher than WT.
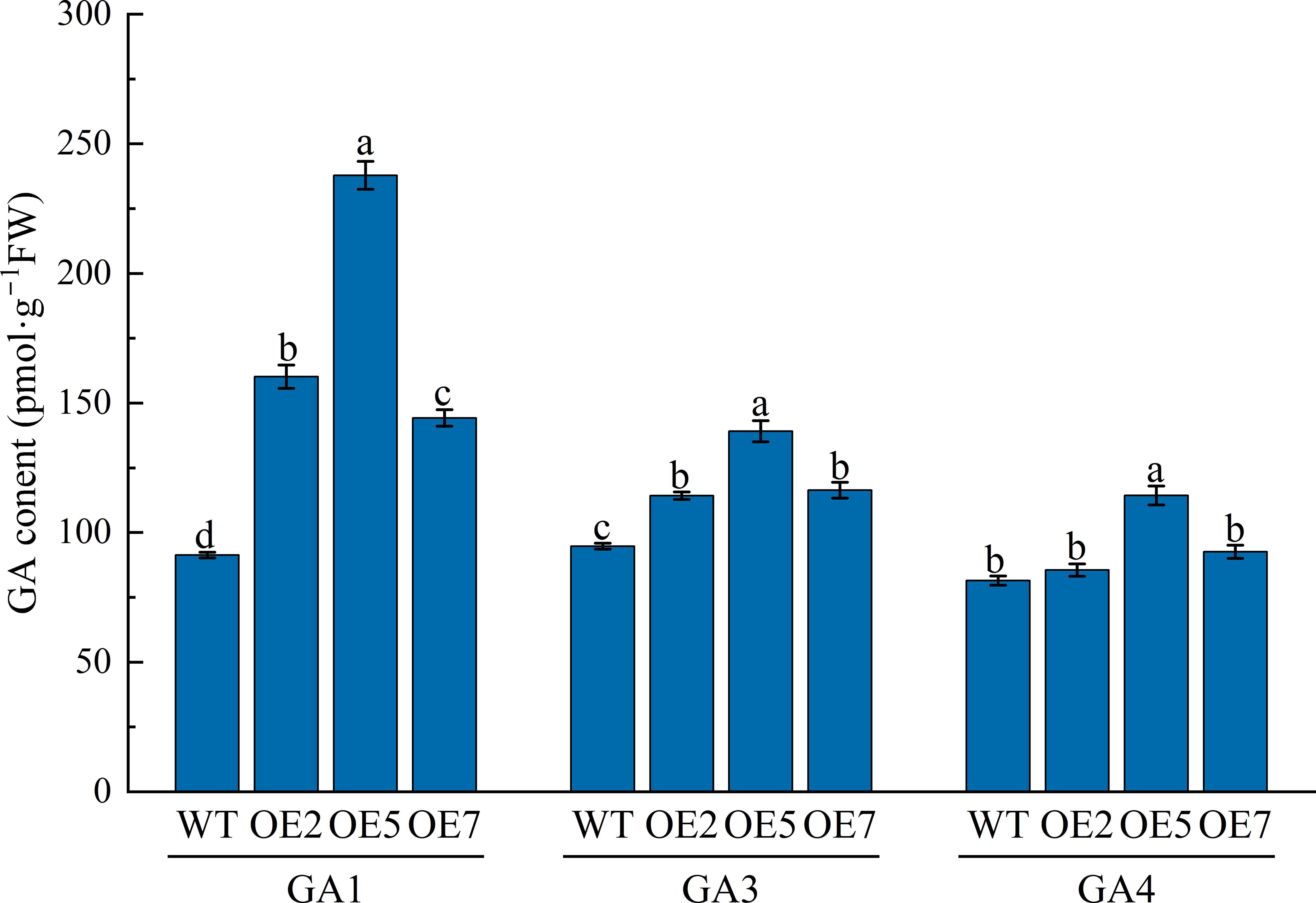
Figure 7. Analysis of endogenous active GA content in WT and transgenic Arabidopsis at 40 days of growth. WT: Col-0 wild-type Arabidopsis control; OE2, OE5, and OE7, separate overexpression lines. Each value represents three replicate experiments, and values are shown as mean ± standard error (SE), with different lowercase letters indicating significant differences (P < 0.05).
We constructed a cDNA library with a capacity of 5.84 × 107 CFU/ml and a total number of clones of 1.168 × 10⁸ CFU (Supplementary Figure S3). To analyze the band distribution structure of the constructed library, 24 randomly selected clones were subjected to colony PCR using T7-F/AD-R primers (Supplementary Table S1.). PCR electrophoresis results showed that the average size of the library fragments was 1000 bp (Supplementary Figure S4).
We used Y2H to screen for interacting proteins. Autoactivation analysis showed that pGBKT7-SdGA20ox1 had no autoactivation (Figure 8A). The AH109 yeast strain containing the pGBKT7-SdGA20ox1 decoy plasmid was used as a receptor to prepare the sensory state, into which the library plasmid pGADT7-S. davidii cDNA was subsequently transferred and plated on SD-TLH plates. Screening on SD-TLH plates yielded 192 positive yeast clones (Supplementary Figures S5, S6). We then amplified and DNA sequenced the 192 positive clones from yeast cells. BLAST analysis of the sequences in the GenBank database resulted in 28 reciprocal protein gene sequences (Supplementary Figure S7; Supplementary Table S2).
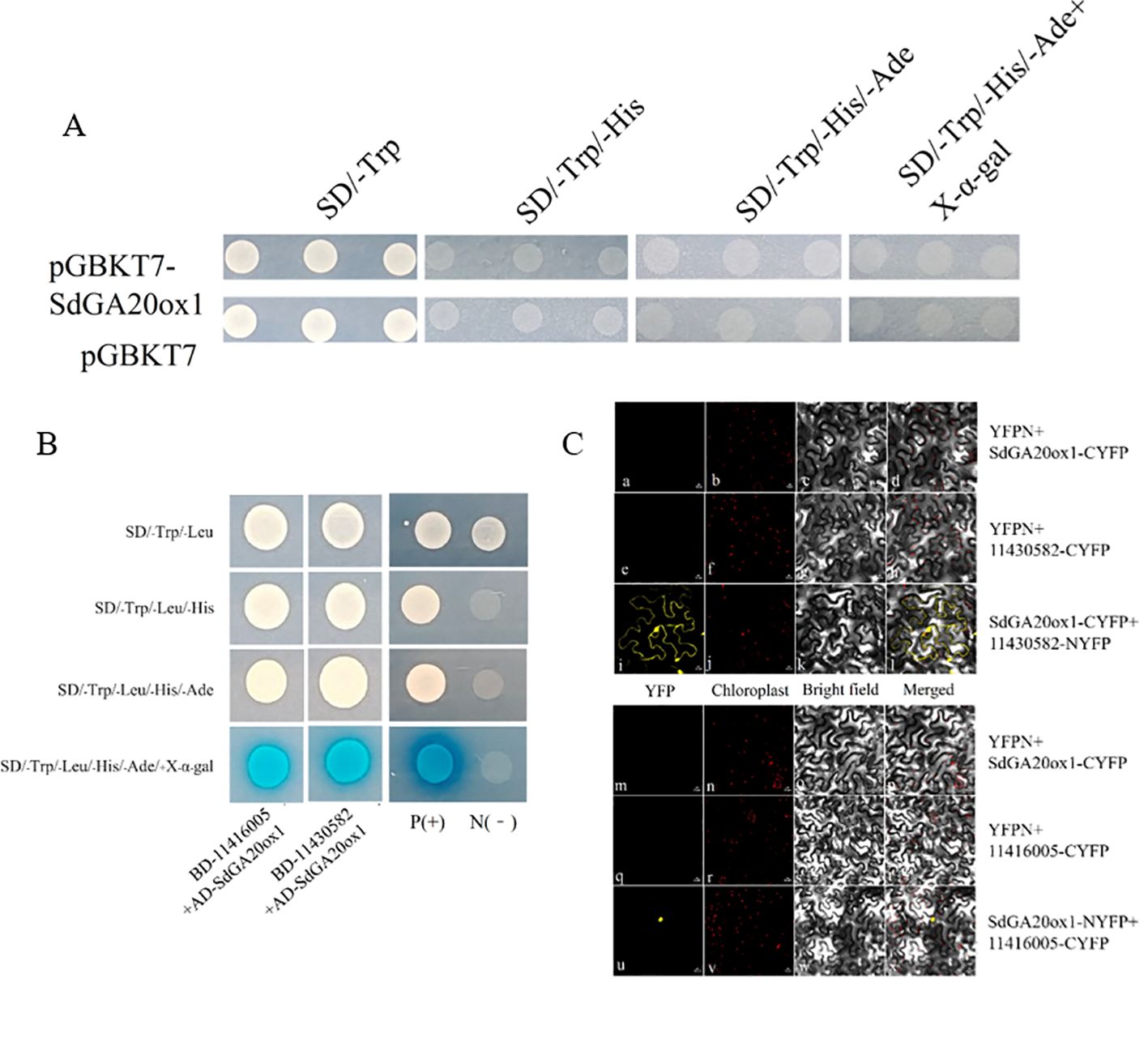
Figure 8. Validation of interaction protein of SdGA20ox1. (A) Analysis of the autoactivation and toxicity of pGBKT7-SdGA20ox1. pGBKT7 is the negative control. SD medium without tryptophan (SD/-Trp), without tryptophan and histidine (SD/-Trp/-His), without tryptophan, histidine, and adenine (SD/-Trp/-His/-Ade), and without tryptophan, histidine, adenine, and containing X-α-gal (SD/-Trp/-His/-Ade/+X-α-gal). (B) Co-transformed yeast cells were dropped onto SD medium without tryptophan and leucine (SD/-Trp/-Leu), without tryptophan, leucine, and histidine (SD/-Trp/-Leu/-His), without tryptophan, leucine, histidine, and adenine (SD/-Trp/-Leu/-His/-Ade), and without tryptophan, leucine, histidine, adenine, and containing 200 mg/mL X-α-gal (SD/-Trp/-Leu/-His/-Ade/+X-α-gal). pGADT7-largeT+pGBKT7-p53 was used as a positive control (P+) and pGADT7-largeT+pGBKT7-laminC was used as a negative control (N-). (C) For the assessment of PpSm35s-SdGA20ox1 and PpSm35s-11430582, BiFC analysis of the interaction between PpSm35s-11416005. (a, e, i, m, q, u) YFP fluorescence images; (b, f, j, n, r, v) chloroplast images; (c, g, k, o, s, w) bright-field images; and (d, h, l, p, t, x) fluorescence images merged with their respective bright-field images.
To further validate the possible interaction of candidate proteins with SdGA20ox1, we selected two genes from the 28 candidate genes with predicted functions related to plant growth and development: rbcS (11416005) and EIN4 (11430582). We constructed pGADT7-11416005 and pGADT7-11430582 vectors for point-to-point yeast transformation with pGBKT7-SdGA20ox1. The results showed that the positive control could grow on SD/-Trp/-Leu, SD/-Trp/-Leu/-His, SD/-Trp/-Leu/-His/-Ade, and SD/-Trp/-Leu/-His/-Ade/+X-α-gal plates, and displayed a blue color on SD/-Trp/-Leu/-His/-Ade/+X-α-gal plates. The negative control could grow only on SD/-Trp/-Leu plates. In the experimental group, the growth of pGBKT7-SdGA20ox1 + pGADT7-11416005 and pGBKT7-SdGA20ox1 + pGADT7-11430582 was consistent with the positive control, indicating that both 11416005 and 11430582 physically interact with SdGA20ox1 in yeast (Figure 8B).
11416005 is involved in plant photosynthesis, while 11430582 may function in the early stages of the ethylene signaling pathway. We used the BiFC assay to confirm the interaction between SdGA20ox1 and 11416005 and 11430582. A distinct YFP yellow fluorescent signal was observed in tobacco leaf pulp protoplast cells after cotransformation with N-terminal YFPN fusion 11430582 and C-terminal YFPC fusion SdGA20ox1, as well as after cotransformation with N-terminal YFPN fusion SdGA20ox1 and C-terminal YFPC fusion 11416005. However, no YFP signal was detected in the negative controls (YFPN and YFPC-SdGA20ox1, YFPN and 11430582-YFPC, YFPN and YFPC-SdGA20ox1, and YFPN and 11416005-YFPC) (Figure 8C). These results suggest that the SdGA20ox1 protein can physically interact with 11416005 and 11430582 in plants, and the three may affect endogenous gibberellin production through these interactions.
Sophora davidii is a common diploid cross-pollination plant in the subtropical karst region of southwest China, characterized by a complex genetic background. The absence of complete genome sequencing has made gene mining and isolation more challenging compared to other crops (Sun et al., 2022). As an excellent plant for soil and water conservation, with high nutritional and medicinal values, optimizing the genetic architecture of S. davidii is crucial for promoting local ecological animal husbandry and improving stone desertification soil. In this study, through a preliminary transcriptomic analysis of an S. davidii mutant, we cloned a gibberellin 20-oxidase homologue gene, SdGA20ox1, and predicted that the secondary structure of this protein is predominantly α-helix. By comparing the SdGA20ox1 protein with GA20ox proteins from legumes such as Lupinus luteus, Lupinus angustifolius, and Cicer arietinum, we found that they share the same conserved structural domain (Griggs et al., 1991), indicating the evolutionary conservation of GA20ox proteins in legumes. The subcellular localization of SdGA20ox1 to the nucleus and cytoplasm is similar to that of CoGA20ox1 (Wang et al., 2023). These results provide a reference for a more comprehensive study of gibberellins and the growth and development of S. davidii.
qRT-PCR results showed that SdGA20ox1 was expressed in the roots, stems, and leaves of S. davidii, with the highest expression in the stem, indicating that SdGA20ox1 expression in plant stems is a major factor determining the height of S. davidii plants. This finding aligns with the study by Wang et al. (2018), which reported significantly higher expression of CrGA20ox1 in Camellia reticulata stems compared to leaves. So, Gibberellin regulation may involve feedback from other plant hormones or growth regulators during plant development (Zhao et al., 2022b; Luo et al., 2015). In this study, the expression of SdGA20ox1 initially decreased upon exogenous application of the plant growth retardant uniconazole to S. davidii, then began to increase again. This may be due to uniconazole inhibiting the activity of upstream genes of GA20ox1, reducing the reaction substrate of the GA20ox1 gene, and consequently decreasing its expression initially. A previous study also reported a decrease in GA20ox expression after 24 hours of uniconazole treatment in Landoltia punctata. However, no effect on HaGA20ox1 gene expression was observed when sunflower was treated with gibberellin inhibitors (Carzoli et al., 2009). Conversely, SdGA20ox1 expression continued to decrease and was significantly lower than the control 6 hours after exogenous GA3 application. This is consistent with studies showing a continuous decrease in GA20ox gene expression after gibberellin treatment in sugarcane, eventually lower than the untreated control (Wu et al., 2009). These results suggest that exogenous gibberellins significantly inhibit GA20ox gene expression in different plants.
Exogenous ETH application significantly increased SdGA20ox1 expression in S. davidii from 24 to 48 hours, implying that ETH may positively regulate SdGA20ox1 expression. There are fewer studies on the relationship between ETH and GA20oxs, but ETH is known to play an important role in plant growth and development as a unique plant hormone (Dugardeyn and van der Straeten, 2008). Our study showed that SdGA20ox1 expression increased and then decreased after exogenous application of BR and IAA, with significant differences in the periods of highest expression. Exogenous BR application was able to upregulate GA20ox gene expression, as demonstrated by exogenous BR spraying in Arabidopsis mutants, which upregulated AtGA20ox1 expression (Ozga and Reinecke, 2003). Previous research found that AtGA20ox1 gene expression increased after IAA treatment of Arabidopsis seedlings under light conditions (Goda et al., 2004), and another study showed that low IAA concentration (30 mg/L) treatments transiently increased PsGA20ox1 gene expression levels in pea (Pisum sativum) pericarp (Ngo et al., 2002).
In this study, Arabidopsis seeds overexpressing SdGA20ox1 showed higher and earlier germination rates compared to wild-type (WT) seeds. Similar results were found by Gao and Ayele (2014), who demonstrated that GA20ox plays a crucial role in promoting wheat (Triticum aestivum) seed germination. This suggests that SdGA20ox1 overexpression increases endogenous gibberellin content in seeds, enhancing germination speed and rate. Our study also found that SdGA20ox1 overexpression significantly promoted hypocotyl and root elongation in Arabidopsis seedlings, increased plant height and leaf length, and led to earlier flowering in transgenic plants. These findings are consistent with previous research indicating that overexpression of GA20oxs results in hypocotyl and root elongation, increased plant height, and early flowering in transgenic Arabidopsis (Coles et al., 1999; Eriksson et al., 2000; Wang et al., 2023; Wu et al., 2023).
Additionally, the S. davidii mutant and transgenic Arabidopsis overexpressing SdGA20ox1 exhibited lighter leaf colors and lower chlorophyll content, though the difference in chlorophyll content was not significant. In contrast, overexpression of CoGA20ox1 and EguGA20ox1 significantly decreased chlorophyll content in Arabidopsis (Wang et al., 2023; Wu et al., 2023). These findings suggest that the regulation of leaf color and chlorophyll content by GA differs among plant species. In our study, the active GA content (GA1, GA3, and GA4) was significantly higher in transgenic plants than in WT, with GA1 showing the largest increase. This suggests that SdGA20ox1 may promote growth by increasing endogenous active GA content, particularly GA1, thereby altering plant phenotypes. Similarly, GA1 plays a crucial role in controlling branch elongation in peach (Prunus persica) and aspen (Populus tremula) (Zhang et al., 2022; Israelsson et al., 2005), while GA4 is the key regulatory factor in controlling shoot elongation in Camellia reticulata (Wang et al., 2018) and Oryza sativa (Qin et al., 2013). These research results demonstrate that the main biological activity GA varies at different stages of tissue development in some plant species (Hu et al., 2017).
The bioactive GA1 and GA4 are synthesized from GA12 through two parallel pathways (early-13-hydroxylation and non-13-hydroxylation) by two kinds of 2-oxoglutarate dependent dioxygenases in the cytoplasm, GA20 oxidase (GA20ox) and GA3 oxidase (GA3ox). The promotion of shoot and internode elongation is a common phenomenon in GA-overproducing or GA-treated plants. Overexpression of the SdGA20-oxidase gene increases levels of intermediates for bioactive GAs, promoting cell elongation and/or cell division, thereby increasing shoot heights. This effect has been observed in GA20ox transgenic plants of various species, such as Zea mays (Liu et al., 2024), Camellia reticulata (Wang et al., 2018), hybrid poplar (Populus tremula × P. alba) (Fladung, 2022), Camellia oleifera (Wang et al., 2023), and rice (Oryza sativa) (Qin et al., 2013). Conversely, semi-dwarf or dwarf phenotypes have been observed in transgenic plants overexpressing the GA20 oxidase gene from Torenia fournieri (Otani et al., 2014) and Cucurbita maxima (Radi et al., 2006), similar to GA20 oxidase-deficient rice mutants. This variation in phenotypes suggests different functions of GA20 oxidase genes in regulating plant height in different species, possibly due to feedback regulation of GA20 oxidase gene expression by bioactive GA.
The construction of an efficient yeast two-hybrid (Y2H) library supports protein interaction research and potential gene screening, providing indispensable conditions for an in-depth understanding of protein function and revealing mechanisms of action (Wang et al., 2018). Our homogenized full-length cDNA library of S. davidii, with a titer of 5.84 × 107 cfu/ml and an average insertion fragment length of 1000 bp, meets the requirements of high-quality cDNA libraries (Clarke and Carbon, 1976) and is reliable for subsequent yeast hybridization screening. Our analysis indicates that SdGA20ox1 protein can physically interact with rbcS (11416005) and EIN4 (11430582) proteins in yeast and tobacco cells.
The RbcS gene encodes a small subunit of the key enzyme RuBisCo (Ribulose-1,5-bisphosphate carboxylase/oxygenase), involved in photosynthetic carbon assimilation (Paritosh et al., 2013). The expression level of the RbcS gene correlates with Rubisco efficiency, ultimately affecting photosynthetic efficiency (Zhuang et al., 2020). This has been widely confirmed in pea (Pisum sativum) (Kuhlemeier et al, 1987), Zea mays (Silverthorne and Tobin, 1990), poplar (Populus deltoides × P. euramericana cv ‘Nanlin895’) (Wang et al., 2013), cotton (Gossypium hirsutum) (Paritosh et al., 2013), and tomato (Solanum lycopersicum) (Zhuang et al., 2020). A significant increase in RbcS gene expression levels was observed in tomatoes, which may help improve the photosynthetic efficiency and nutritional growth of tomatoes. At the same time, GA20 gene expression levels were similar to that of RbcS gene, significantly increased by three times, promoting tomato growth, and both jointly regulate tomato growth (Zhuang et al., 2020). In this study, both yeast two-hybrid and BiFC assays showed that SdGA20ox1 protein can interact with RbcS protein, potentially leading to increased photosynthesis and gibberellin synthesis, promoting better nutritional growth of S. davidii, similar to findings in tomatoes (Zhuang et al., 2020).
Ethylene plays crucial roles in various processes of plant growth, development, and stress responses (Gallie, 2015). Ethylene is perceived through its binding to receptors located in the endoplasmic reticulum (Shakeel et al., 2013). Once ethylene binds to the receptors, various responses are observed in seed germination, stem cell division, cell elongation and differentiation, root hair growth, fruit ripening, senescence, abscission, and responses to abiotic stresses, confirmed in Arabidopsis, tomato (Solanum lycopersicum), maize (Zea mays), and other plants (Hua and Meyerowitz, 1998; Tieman et al., 2000; Gallie and Young, 2004). EIN/EILs play an important role in the ethylene signaling pathway and can also cross regulate plant traits with other plant hormone signals. For example, in rice, EIN3 binds to the promoters of SK1 and SK2 to promote the expression of both, activating the gibberellin response required for stem elongation and regulating stem elongation in rice (Ayano et al., 2014). EIN4 is also one of the ethylene receptors. In this study, results indicate that SdGA20ox1 protein may interact with EIN4 protein, affecting ethylene response and regulating plant stem growth. However, further experimental verification is needed to confirm the specific pathway of this interaction.
Overexpression of SdGA20ox1 in Arabidopsis resulted in reduced chlorophyll accumulation in plant leaves, earlier flowering, elongation of seedling hypocotyl and root lengths, and increased plant height. These findings suggest that SdGA20ox1 could be a potential candidate gene for selecting new dwarf varieties of S. davidii using gene editing technology. By constructing a nuclear yeast library, we predicted and screened two proteins that interact with SdGA20ox1. Further yeast two-hybrid (Y2H) and bimolecular fluorescence complementation (BiFC) assays demonstrated that SdGA20ox1 interacts with the ethylene receptor EIN4 and the RBCS proteins, indicating its involvement in the regulation of plant growth and development.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
LZ: Writing – original draft, Data curation, Methodology. WX: Writing – original draft, Data curation. LH: Writing – original draft, Investigation. SL: Writing – original draft. WP: Writing – review & editing, Funding acquisition.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was funded through projects of Science and Technology Project of Guizhou Province (QKHPTRC(2021)5636, QKHPTRC-GCC(2022)022-1), Guizhou Provincial Department of Education Natural Science Research Project (QJJ(2022)076), and the National Natural Science Foundation of China (32060391, 32260340). So we thankfully acknowledge their support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1478854/full#supplementary-material
Ayano, M., Kani, T., Kojima, M., Sakakibara, H., Kitaoka, T., Kuroha, T., et al. (2014). Gibberellin biosynthesis and signal trans-duction is essential for internode elongation in deepwaterrice. Plant Cell Environ. 37, 2313–2324. doi: 10.1111/pce.12377
Carrera, E., Bou, J., Garcia-Martinez, J. L., Prat, S. (2000). Changes in ga 20-oxidase gene expression strongly affect stem length, tuber induction and tuber yield of potato plants. Plant Journal: For Cell Mol. Biol. 22, 247–256. doi: 10.1046/j.1365-313x.2000.00736.x
Carzoli, F. G., Michelotti, V., Fambrini, M., Salvini, M., Pugliesi, C. (2009). Molecular cloning and organ-specific expression of two gibberellin 20-oxidase genes of helianthus annuus. Plant Mol. Biol. Rep. 27, 144–152. doi: 10.1007/s11105-008-0066-z
Chen, M. L., Fu, X. M., Liu, J. Q., Ye, T. T., Hou, S. Y., Huang, Y. Q., et al. (2012). Highly sensitive and quantitative profiling of acidic phytohormones using derivatization approach coupled with nano-lc-esi-q-tof-ms analysis. J. Chromatogr. B 905, 67–74. doi: 10.1016/j.jchromb.2012.08.005
Clarke, L., Carbon, J. (1976). A colony bank containing synthetic coi ei hybrid plasmids representative of the entire e. Coli genome. Cell 9, 91–99. doi: 10.1016/0092-8674(76)90055-6
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of arabidopsis thaliana. Plant J.: For Cell Mol. Biol. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Coles, J. P., Phillips, A. L., Croker, S. J., Garcia-Lepe, R., Lewis, M. J., Hedden, P., et al. (1999). Modification of gibberellin production and plant development in arabidopsis by sense and antisense expression of gibberellin 20-oxidase genes. Plant J.: For Cell Mol. Biol. 17, 547–556. doi: 10.1046/j.1365-313X.1999.00410.x
Ding, L. L., Shang, Y. S., Zhang, W., Zhang, Y., Li, S. G., Wei, X., et al. (2020). Disentangling the effects of driving forces on soil bacterial and fungal communities under shrub encroachment on the guizhou plateau of China. Sci. Total Environ. 709. doi: 10.1016/j.scitotenv.2019.136207
Dugardeyn, J., van der Straeten, D. (2008). Ethylene: fine-tuning plant growth and development by stimulation and inhibition of elongation. Plant Sci. 175, 59–70. doi: 10.1016/j.plantsci.2008.02.003
Engler, C., Kandzia, R., Marillonnet, S. (2008). A one pot, one step, precision cloning method with high throughput capability. PloS One 3 (11), e3647. doi: 10.1371/journal.pone.0003647
Eriksson, M. E., Israelsson, M., Olsson, O., Moritz, T. (2000). Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fiber length. Nat. Biotechnol. 18, 784–788. doi: 10.1038/77355
Fladung, M. (2022). Xylem-specific overexpression of the gibberellin acid 20 oxidase gene (ga20-oxidase) from pine in hybrid poplar (populus tremula l. X p. Alba l.) Revealed reliable increase in growth and biomass production just in a single-copy-line. Gesunde Pflanzen 74, 239–248. doi: 10.1007/s10343-022-00653-y
Gallie, D. R. (2015). Appearance and elaboration of the ethylene receptor family during land plant evolution. Plant Mol. Biol. 87, 521–539. doi: 10.1007/s11103-015-0296-z
Gallie, D. R., Young, T. E. (2004). The ethylene biosynthetic and perception machinery is differentially expressed during endosperm and embryo development in maize. Mol. Genet. Genomics: Mgg 271, 267–281. doi: 10.1007/s00438-004-0977-9
Gao, F., Ayele, B. T. (2014). Functional genomics of seed dormancy in wheat: advances and prospects. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00458
Goda, H., Sawa, S., Asami, T., Fujioka, S., Shimada, Y., Yoshida, S., et al. (2004). Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in arabidopsis. Plant Physiol. 134, 1555–1573. doi: 10.1104/pp.103.034736
Griggs, D. L., Hedden, P., Lazarus, C. M. (1991). Partial purification of two gibberellin 2β-hydroxylases from cotyledons of phaseolus vulgaris. Phytochemistry 30, 2507–2512. doi: 10.1016/0031-9422(91)85090-M
Hedden, P. (2003). The genes of the green revolution. Trends Genet. 19, 5–9. doi: 10.1016/S0168-9525(02)00009-4
Hu, Y. X., Tao, Y. B., Xu, Z. F. (2017). Overexpression of jatropha gibberellin 2-oxidase 6 (jcga2ox6) induces dwarfism and smaller leaves, flowers and fruits in arabidopsis and jatropha. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.02103
Hua, J., Meyerowitz, E. M. (1998). Ethylene responses are negatively regulated by a receptor gene family in arabidopsis thaliana. Cell 94, 261–271. doi: 10.1016/S0092-8674(00)81425-7
Israelsson, M., Sundberg, B., Moritz, T. (2005). Tissue-specific localization of gibberellins and expression of gibberellin-biosynthetic and signaling genes in wood-forming tissues in aspen. Plant Journal: For Cell Mol. Biol. 44, 494–504. doi: 10.1111/j.1365-313X.2005.02547.x
Jeon, H. W., Cho, J. S., Park, E. J., Han, K. H., Choi, Y. I., Ko, J. H., et al. (2016). Developing xylem-preferential expression of pdga20ox1, a gibberellin 20-oxidase 1 from pinus densiflora, improves woody biomass production in a hybrid poplar. Plant Biotechnol. J. 14, 1161–1170. doi: 10.1111/pbi.12484
Ku, L., Zhang, L., Tian, Z., Guo, S., Su, H., Ren, Z. Z., et al. (2015). Dissection of the genetic architecture underlying the plant density response by mapping plant height-related traits in maize (zea mays l.). Mol. Genet. Genomics 290, 1223–1233. doi: 10.1007/s00438-014-0987-1
Kuhlemeier, C., Green, P. J., Chua, N. H. (1987). Regulation of gene expression in higher plants. Annu. Rev. Plant Biol. 38, 221–257. doi: 10.1146/annurev.pp.38.060187.001253
Liu, Y., Chen, Z. Q., Zhang, C., Guo, J., Liu, Q., Yin, Y., et al. (2024). Gene editing of zmga20ox3 improves plant architecture and drought tolerance in maize. Plant Cell Rep. 43. doi: 10.1007/s00299-023-03090-x
Luo, Y. H., Dong, X. W., Yu, T. Y., Shi, X., Li, Z. Y., Yang, W. C., et al. (2015). A single nucleotide deletion in gibberellin20-oxidase1 causes alpine dwarfism in arabidopsis. Plant Physiol. 168, 930. doi: 10.1104/pp.15.00005
Ngo, P., Ozga, J. A., Reinecke, D. M. (2002). Specificity of auxin regulation of gibberellin 20-oxidase gene expression in pea pericarp. Plant Mol. Biol. 49, 439–448. doi: 10.1023/A:1015522404586
Otani, M., Meguro, S., Gondaira, H., Hayashi, M., Saito, M., Han, D. S., et al. (2014). Overexpression of the gibberellin 20-oxidase or gibberellin 3-oxidase gene from torenia fournieri affecting plant morphology in transgenic. tricyrtis Acta Hortic. sp 1025, 23–30. doi: 10.17660/ActaHortic.2014.1025.2
Ozga, J. A., Reinecke, D. M. (2003). Hormonal interactions in fruit development. J. Plant Growth Regul. 22, 73–81. doi: 10.1007/s00344-003-0024-9
Paritosh, K., Pental, D., Burma, P. K. (2013). Structural and transcriptional characterization of rbcs genes of cotton (gossypium hirsutum). Plant Mol. Biol. Rep. 31, 1176–1183. doi: 10.1007/s11105-013-0576-1
Phillips, A. L., Ward, D. A., Uknes, S., Appleford, N. E., Lange, T., Huttly, A. K., et al. (1995). Isolation and expression of three gibberellin 20-oxidase cdna clones from arabidopsis. Plant Physiol. 108, 1049–1057. doi: 10.1104/pp.108.3.1049
Qin, X., Liu, J. H., Zhao, W. S., Chen, X. J., Guo, Z. J., Peng, Y. L., et al. (2013). Gibberellin 20-oxidase gene osga20ox3 regulates plant stature and disease development in rice. Mol. Plant Microbe Interact. 26, 227–239. doi: 10.1094/MPMI-05-12-0138-R
Radi, A., Lange, T., Niki, T., Koshioka, M., Lange, M. J. P. (2006). Ectopic expression of pumpkin gibberellin oxidases alters gibberellin biosynthesis and development of transgenic arabidopsis plants. Plant Physiol. 140, 528–536. doi: 10.1104/pp.105.073668
Shakeel, S. N., Wang, X. M., Binder, B. M., Schaller, G. E. (2013). Mechanisms of signal transduction by ethylene: overlapping and non-overlapping signalling roles in a receptor family. Aob Plants 5, plt010. doi: 10.1093/aobpla/plt010
Silverthorne, J., Tobin, E. M. (1990). Post-transcriptional regulation of organ-specific expression of individual rbcs mrnas in lemna gibba. Plant Cell 2, 1181–1190. doi: 10.2307/3869338
Subramanian, B., Gao, S. H., Lercher, M. J., Hu, S. N., Chen, W. H. (2019). Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic. Acids Res. 47, W270–W275. doi: 10.1093/nar/gkz357
Sun, H. G., Han, X. W., Ma, D. L., Li, Y., Kang, T., Sun, R., et al. (2022). The complete chloroplast genome of sophora davidii (fabaceae) and its phylogenetic implications. Mitochondrial DNA Part B-Resour. 7, 503–504. doi: 10.1080/23802359.2021.1999187
Tieman, D. M., Taylor, M. G., Ciardi, J. A., Klee, H. J. (2000). The tomato ethylene receptors nr and leetr4 are negative regulators of ethylene response and exhibit functional compensation within a multigene family. Proc. Natl. Acad. Sci. U. S. A. 97, 5663–5668. doi: 10.1073/pnas.090550597
Wang, Y. H., Li, J. Y. (2008). Molecular basis of plant architecture. Annu. Rev. Plant Biol. 59, 253–279. doi: 10.1146/annurev.arplant.59.032607.092902
Wang, Y., Li, J. A., Guo, P. R., Liu, Q., Ren, S. S., Juan, L. M., et al. (2023). Ectopic expression of camellia oleifera abel. gibberellin 20-oxidase gene increased plant height and promoted secondary cell walls deposition in arabidopsis. Planta 258 (3), 65. doi: 10.1007/s00425-023-04222-z
Wang, L. K., Li, L. S., Xu, L. N., Zhou, J., Zhuang, H. H., Gong, X. D., et al. (2013). Isolation and functional analysis of the poplar rbcs gene promoter. Plant Mol. Biol. Rep. 31, 120–127. doi: 10.1007/s11105-012-0482-y
Wang, J. Y., Wu, B., Li, J. Y., Yin, H. F., Fan, Z. Q., Li, X. L., et al. (2018). Overexpression and silent expression of crga20ox1 from camellia reticulata ‘hentiangao’ and its effect on morphological alterations in transgenic tobacco plants. Plant Breed. 137, 903–911. doi: 10.1111/pbr.12653
Wang, Y. J., Zhao, J., Lu, W. J., Deng, D. X. (2017). Gibberellin in plant height control: old player, new story. Plant Cell Rep. 36, 391–398. doi: 10.1007/s00299-017-2104-5
Wu, J. M., Li, Y. R., Wang, A. Q., Yang, L. T., Yang, L. (2009). Cloning and sequence analysis of sugarcane ga20-oxidase gene. Chin. J. Trop. Crops 30, 817–821.
Wu, W. F., Zhu, L. H., Wang, P., Liao, Y. W., Duan, L. J., Lin, K., et al. (2023). Transcriptome-based construction of the gibberellin metabolism and signaling pathways in eucalyptus grandis x e. Urophylla, and functional characterization of ga20ox and ga2ox in regulating plant development and abiotic stress adaptations. Int. J. Mol. Sci. 24 (8), 7051. doi: 10.3390/ijms24087051
Xie, W. H., Sun, X. F., Zhao, L. L., Huang, L. J., Wang, P. C. (2023). Exogenous hormone application regulates dwarf mutant plant height in sophora davidii (franch.) Skeels by changing endogenous hormone levels. Chil. J. Agric. Res. 83, 195–202. doi: 10.4067/S0718-58392023000200195
Yamaguchi, S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251. doi: 10.1146/annurev.arplant.59.032607.092804
Yan, J. D., Liao, X. Y., He, R. Q., Zhong, M., Feng, P. P., Li, X. M., et al. (2017). Ectopic expression of ga 2-oxidase 6 from rapeseed (brassica napus l.) Causes dwarfism, late flowering and enhanced chlorophyll accumulation in arabidopsis thaliana. Plant Physiol. Biochem. 111, 10–19. doi: 10.1016/j.plaphy.2016.11.008
Yang, P., Li, Y. X., He, C. S., Yan, J. D., Zhang, W., Li, X., et al. (2020). Phenotype and tmt-based quantitative proteomics analysis of brassica napus reveals new insight into chlorophyll synthesis and chloroplast structure. J. Proteomics 214. doi: 10.1016/j.jprot.2019.103621
Zhang, M. M., Ma, Y. T., Zheng, X. B., Tan, B., Ye, X., Wang, W., et al. (2022). The distribution of bioactive gibberellins along peach annual shoots is closely associated with ppga20ox and ppga2ox expression profiles. BMC Genomics 23 (1), 1–13. doi: 10.1186/s12864-022-08943-5
Zhao, X., Huang, L. J., Sun, X. F., Zhao, L. L., Wang, P. C. (2022a). Transcriptomic and metabolomic analyses reveal key metabolites, pathways and candidate genes in sophora davidii (franch.) Skeels seedlings under drought stress. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.785702
Zhao, X., Huang, L. J., Zhao, L. L., Wang, P. C., Sun, X. F. (2022c). Transcriptome analysis of sophora davidii leaves in response to low-phosphorus stress. J. Plant Growth Regul. 41, 1241–1253. doi: 10.1007/s00344-021-10382-7
Zhao, X., Sun, X. F., Zhao, L. L., Huang, L. J., Wang, P. C. (2022b). Morphological, transcriptomic and metabolomic analyses of sophora davidii mutants for plant height. BMC Plant Biol. 22, 1–17. doi: 10.1186/s12870-022-03503-1
Zhao, L. L., Wang, L. T., Chen, K. K., Sun, H., Wang, P. C. (2024). Effects of arbuscular mycorrhizal fungi on the growth and physiological performance of sophora davidii seedling under low-phosphorus stress. J. Plant Growth Regul. 43, 2383–2395. doi: 10.1007/s00344-024-11273-3
Keywords: Sophora davidii, gibberellin 20-oxidases, gibberellins, plant height, interacting protein
Citation: Zhao L, Xie W, Huang L, Long S and Wang P (2024) Characterization of the gibberellic oxidase gene SdGA20ox1 in Sophora davidii (Franch.) skeels and interaction protein screening. Front. Plant Sci. 15:1478854. doi: 10.3389/fpls.2024.1478854
Received: 11 August 2024; Accepted: 30 September 2024;
Published: 16 October 2024.
Edited by:
Linkai Huang, Sichuan Agricultural University, ChinaReviewed by:
Yuefei Xu, Northwest A&F University, ChinaCopyright © 2024 Zhao, Xie, Huang, Long and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Puchang Wang, d2FuZ3B1Y2hhbmdAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.