- Maize Research Institute, Sichuan Agricultural University, Chengdu, China
A lot of endogenous genes, as well as genes from related species, are transformed back into crops for overexpression to improve their corresponding traits. However, almost all of these transgenic events remain at the testing stage. Most of the singular transgenic events of crops approved for commercial release are developed by the transformation and heterologous expression of exogenous genes from distant species. To detect the differences in expression, protein accumulation, and enzyme activity between transformed exogenous and endogenous genes, the coding sequences (CDSs) of the alcohol dehydrogenase (ADH) genes were cloned from dicotyledonous Arabidopsis, monocotyledonous maize, and prokaryotic Escherichia coli, constructed into expression vector pBI121-cMycNY, and used to transform wild-type Arabidopsis, respectively. Three homozygous T3 lines with a single integration site were screened for each of the three transformed genes by antibiotic screening, polymerase chain reaction (PCR) identification, and genomic DNA resequencing. Real-time quantitative PCR (RT-qPCR) analysis showed that the relative expression levels of the transformed exogenous ZmADH and EcADH genes were ten or tens of times higher than that of the transformed endogenous AtADH gene. After confirming the encoded proteins of these transformed genes by Western blotting, enzyme-linked immunosorbent assay (ELISA) showed that the accumulation levels of the proteins encoded by the transformed genes ZmADH and EcADH were significantly higher than that encoded by the transformed endogenous gene AtADH. Enzyme reaction assay showed that the ADH activities of the T3 lines transformed by the exogenous genes ZmADH and EcADH were also significantly higher than that transformed by the endogenous gene AtADH as well as the wild-type control. These results indicated that exogenous genes were more conducive to transgenic improvement of crops.
1 Introduction
Transgenic technology accelerates crop improvement, not only enabling it to acquire biotic and abiotic resistances but also effectively meeting various requirements such as higher yield and nutritional value (Raymond Park et al., 2011; Kamthan et al., 2016). As of 2023, a total of 440 singular transgenic events have been approved for commercial release, and the planting area of genetically modified crops has risen sharply to 20,626 ha, accounting for 80.4%, 73.7%, 32%, 23.8%, and 11.4% of global planting areas of cotton, soybean, maize, rapeseed, and sugar beet, respectively (International Service for the Acquisition of Agri-biotech Applications, 2023). The huge benefits not only come from increased production but also from reduced inputs of pesticides, labor, and machinery (Karalis et al., 2020; Kobayashi et al., 2023).
Of the 472 singular transgenic events approved for commercial release, 332 events were developed by the heterologous expression of transformed exogenous genes, 29 events were transformed by synthetic sequences transcribing antisense or double-stranded RNAs for the interference of disease viruses, and only three events were transformed by the mutated copies of endogenous genes for suppressed expression of undesirable genes (Yu et al., 2022; International Service for the Acquisition of Agri-biotech Applications, 2024). By a strategy of overexpression of endogenous genes, numerous transgenic events have also been developed and reported to improve target agronomic traits, such as the transformation of endogenous genes SOD (superoxide dismutase), VP1 (vacuolar proton-pumping pyrophosphatase), BADH (betaine aldehyde dehydrogenase), LEA (late embryogenesis-abundant proteins), DREB (dehydration-responsive element binding), NF-Y, and WRKY, in an attempt to improve drought tolerance (Kasuga et al., 1999; Nelson et al., 2007; Xiao et al., 2007; Century et al., 2008; Schilling et al., 2017; Gao et al., 2018). However, all of these efforts remained at the testing stage. Of the five singular transgenic events approved for commercial release with the significant improvement of drought tolerance (International Service for the Acquisition of Agri-biotech Applications, 2024), one was transformed by the exogenous cold shock protein gene CspB from genetically distant Bacillus subtilis and three by the exogenous choline dehydrogenase genes BetA from genetically distant Escherichia coli and Rhizobium meliloti, respectively, although the endogenous genes of cold shock proteins are also found in many eukaryotic species (Castiglioni et al., 2008; Tollefson, 2011). Only one was transformed by the exogenous transcription factor gene Hahb-4 from a sexually incompatible sunflower (Helianthus annuus) (Ribichich et al., 2020).
The most successful achievements of transgenic improvement are none other than herbicide and insect resistance of cotton, maize, soybean, canola (Brassica napus), and some other crops (Padgette et al., 1995; Bates et al., 2005; Raymond Park et al., 2011; Klumper and Qaim, 2014; Kamthan et al., 2016). The 291 and 241 singular transgenic events approved for commercial release were transformed by exogenous genes from genetically distant species (Thompson et al., 1987; Stalker et al., 1988; Ghareyazie et al., 1997; Castle et al., 2004; Bates et al., 2005; Showalter et al., 2009; Cui et al., 2011; Guo et al., 2015; Karthik et al., 2020; International Service for the Acquisition of Agri-biotech Applications, 2024). The same strategy was successfully applied to the transgenic manipulation of “Golden” rice and potato for biofortification of vitamin A (Ye et al., 2000; Paine et al., 2005; Chitchumroonchokchai et al., 2017; Napier et al., 2019; Yang et al., 2021) and the transgenic improvement of canola for oil quality (Napier et al., 2019; Kinney et al., 2022; International Service for the Acquisition of Agri-biotech Applications, 2024).
In plants, alcohol dehydrogenase (ADH) catalyzes the reversible reduction of aldehydes that is key to the establishment of the fermentative metabolism and to the constant supply of nicotinamide adenine dinucleotide (NADH) (Chung and Ferl, 1999). In response of plants to oxygen shortage under abiotic or biotic stresses such as waterlogging, submergence, saline-alkaline, low temperature, and pathogen infection, the fermentative metabolism allows ATP production when mitochondrial respiration is hampered (Millar and Dennis, 1996; Shi et al., 2017; Song et al., 2019; Su et al., 2020; Ventura et al., 2020; Zhou et al., 2020; Shen et al., 2021). In the Arabidopsis and maize genome, the ADH gene exists as a single copy and double copies, respectively (Chang and Meyerowitz, 1986; Peters and Frenkel, 2004). On the basis of constitutive expression, their expression is also induced by various stress conditions such as oxidation, dehydration, and low temperature through signal transduction pathways such as abscisic acid (Dolferus et al., 1994; Peters and Frenkel, 2004). In the genome of Escherichia coli, the ADH gene exists as a single copy (Blattner et al., 1997). The activity of ADH is easy to be detected by using commercial kits.
In this study, the CDSs of the ADH genes were cloned from Arabidopsis, maize, and Escherichia coli and transformed into wild-type Arabidopsis, which has only a single copy of the ADH gene in its genome. Homozygous T3 lines with a single integration site of the T-DNA were screened and used to detect the differences in relative expression levels, protein accumulation, and enzyme activity, in order to provide reference for strategy choice of transgenic improvement of crops.
2 Materials and methods
2.1 CDS cloning and transformation
The CDSs of the ADH genes were amplified from the cDNA samples of wild-type Arabidopsis (Col-0) and maize inbred line B73, as well as a genomic DNA sample of E. coli with homologous recombination primers (Supplementary Table S1), separated by 1% agarose gel electrophoresis, purified using the Universal DNA Purification Kit (Tiangen, Beijing, China), and constructed into dicotyledonous expression vector pBI121-cMyc-NY (Supplementary Figure S1) using homologous recombination kit ClonExpress II (Vazyme, Nanjing, China), respectively. The recombined vectors were transformed into competent cells, Trans-T1 of E. coli (TransGen, Beijing, China), incubated in LB liquid medium, and screened on LB plates containing 50 mg/l kanamycin. The vector plasmids were purified using the TIANprep Mini Plasmid Kit (Tiangen, Beijing, China) and then used to transform Agrobacterium tumefaciens competent cells Gv3101-pSoup-19 (Angyubio, Shanghai, China). After screening on LB plates containing 50 mg/l kanamycin and rifampicin and identification by bacterial PCR and sequencing at Kangyou Biotech (Hangzhou, China), the positive monoclonals were incubated in LB liquid medium, recovered, and used to transform wild-type Arabidopsis (Col-0) by floral dipping. Homozygous T3 lines were screened on MS plates containing 50 mg/l kanamycin, identified by PCR amplification using the same primers as shown in Supplementary Table S1, and calculated for transformation rate.
2.2 Identification of T-DNA integration sites
The genomic DNA of each homozygous T3 line was extracted using CTAB buffer. After detection for concentration and purity in an NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific Ltd., Paisley, United Kingdom) and identification for integrity by 1% agarose gel electrophoresis, the samples were resequenced (10 × deep) on a DNBSEQ-T7 platform at Geneplus (Shenzhen, China). After preliminary evaluation of the raw data for quality, clean reads were filtered by the FASTP software (https://github.com/OpenGene/fastp) and aligned against the sequences of the Arabidopsis genome TAIR10 (http://www.arabidopsis.org) and the expression vector pBI121-cMyc-NY to find out the integration sites of T-DNA. As described by Zhang et al. (2011), specific primers (Supplementary Table S2) were designed using online software Primer-BLAST (https://blast.ncbi.nlm.nih.gov) and used to amplify the flanking sequences of the left (from 1000 bp upstream of the integration sites to 1000 bp of the T-DNA) and right (from 1000 bp of the T-DNA to 1000 bp downstream of the integration sites) borders of the T-DNA integration sites from the DNA samples extracted as above from each of the homozygous T3 lines. The amplified products were separated by 1% agarose gel electrophoresis, recovered, and sequenced at Kangyou Biotech (Hangzhou, China). The sequenced results were aligned against the sequences of the Arabidopsis genome TAIR10 (http://www.arabidopsis.org) and the expression vector pBI121-cMyc-NY again for confirmation of the number and integration sites of T-DNA.
2.3 Expression detection of transformed genes
Three biological replates of seedlings of the homozygous T3 lines and their type wild control were incubated in a phytotron for 30 days. Consistent plants were sampled from each replicate. RNA was extracted from one-third of each sample using RNAiso Plus (TaKaRa, Dalian, China) and reverse transcribed into cDNA using ABScript Neo RT Master Mix for qPCR with gDNA Remover (ABclonal, Wuhan, China). Specific primers (Supplementary Table S3) were designed using online software Primer-Blast (https://blast.ncbi.nlm.nih.gov) and used for real-time quantitative PCR (RT-qPCR) amplification of the transformed genes AtADH, ZmADH, and EcADH, as well as the reference gene AtUBQ5 using the Universal SYBR Green Fast qPCR Mix (Abclonal, Wuhan, China) in a CFX96 MangerTM Real-Time System (Bio-Rad, CA, USA) with four technical replicates. The relative expression levels of the three transformed genes were calculated, normalized, and analyzed for statistical significance using the 2−ΔΔCT method of the CFX96 Manger™ software v 2.0.
2.4 Western blotting
Total protein was extracted from another one-third of each of the T3 seedling samples using the Plant Total Protein Extraction Kit (Coolaber, Beijing, China), detected for concentration using the BCA Protein Assay Kit (Coolaber, Beijing, China), and diluted to gradient standard samples with the extraction buffer. The OD562 of the gradient standard samples were detected in a microplate reader Varioskan LUX (Thermo Fisher Scientific Ltd., Paisley, United Kingdom) and used to establish the standard curve. The concentration of total protein in the samples was calculated and diluted to 5 μg/μl with the extraction buffer. Half of each of the diluted protein samples were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using the SDS-PAGE Gel Preparation Kit (Biosharp, Hefei, China) and transferred onto a polyvinylidene fluoride membrane (Millipore, Billerica, USA). After blocking and rinsing, the membrane was incubated in Myc-Tag pAb antibody (Abclonal, Wuhan, China) solution at 4°C for 3 h, incubated in anti-rabbit IgG (H+L) antibody (Jackson, MS, USA) solution at room temperature for 30 min, rinsed with rinsing buffer for 3 × 10 min, colored using Ultra-sensitive ECL Chemiluminescent Substrate Kit (Powerful Biology, Wuhan, China), and imaged in Gel Doc™ EZ System (Bio-Rad, Hercules, CA, USA).
2.5 Enzyme-linked immunosorbent assay
The ADH standard sample of the ADH ELISA Kit (MLBio, Shanghai, China) was diluted to gradient standard samples and added into the wells of a 96-well plate with three technical replicates. The other half of each diluted protein sample was added into the other wells of the same plate with three replicates. One hundred microliter of HRP antibody was added into each of these wells. The plate was sealed with film, incubated at 37°C for 1 h, and dried by removing the solution in the wells. The wells were washed 5 times by being filled with the diluted washing buffer, incubated at room temperature for 1 min, and dried by removing the solution, added with 50 μl of 0.01 H2O2 and 0.1% tetramethylbenzidine, shaken well, incubated at 37°C in the dark for 15 min, added with 50 μl termination solution, shaken well, and read for OD450 in a microplate reader Varioskan LUX (Thermo Fisher Scientific Ltd., Paisley, United Kingdom). A standard curve was established based on the concentration of the standard samples and their OD450 and used to calculate the concentrations of the enzyme proteins. Due to the different molecular weights of the AtADH, EcADH, and ZmADH proteins, the concentrations were divided by their molecular weight to convert to the accumulation of each enzyme in ng/ml/kDa.
2.6 Enzyme reaction assay
The other one-third of each sample was ground in liquid nitrogen, transferred into an Enppendoff tube, weighed (W), added with 1 ml of the extraction solution of the ADH reaction assay kit CAS:9031-72-5 (Solarbio, Beijing, China), incubated on ice for 20 min, and centrifuged at 4°C and 16000 r/min for 20 min. The supernatant was used to be detected for ADH activity (U/g) using the same assay kit in an ultraviolet spectrophotometer UV8000 (Shimadzu, Japan). In the reversible reaction between ethanol and acetaldehyde, ADH catalyzes the reduction of acetaldehyde by NADH to produce ethanol and nicotinamide adenine dinucleotide (NAD+). NADH has an absorption peak at 340 nm, while NAD+ does not. According to the manual of the assay kit, the ADH activity was calculated as
Where, ΔA340 was the difference of absorbance at 340 nm before and after reaction, ϵ (molar extinction coefficient of NADH) = 6.22 × 103 l/mol/cm, d (optical diameter of colorimetric dish) = 1 cm, VReaction total (total volume of reaction) = 1 ml, VSample total (the added volume of the ADH extraction solution) = 1 ml, T (reaction time) = 1 min, VSample (the added volume of the supernatant) = 100 μl, and W was the weight (g) of the ground sample.
3 Results
3.1 Homozygous T3 lines and T-DNA integration sites
The results of the PCR amplification showed that the CDSs of the AtADH, ZmADH, and EcADH genes were successfully cloned (Supplementary Figure S2), conducted into dicotyledonous expression vector pBI121-cMyc-NY, and transformed into Agrobacterium tumefaciens (Supplementary Figure S3). After transformation, 1.2%, 1.4%, and 1.3% T0 plants were screened on the resistant plates (Supplementary Figure S4A). Each of these T1 lines is segregated in a 3:1 ratio (Supplementary Figure S4B). Untill T3 generation, five, three, and six homozygous lines transformed by the AtADH, ZmADH, and EcADH genes, respectively, were screened on the resistant plates (Supplementary Figure S4C) and identified by the PCR amplification (Figure 1).
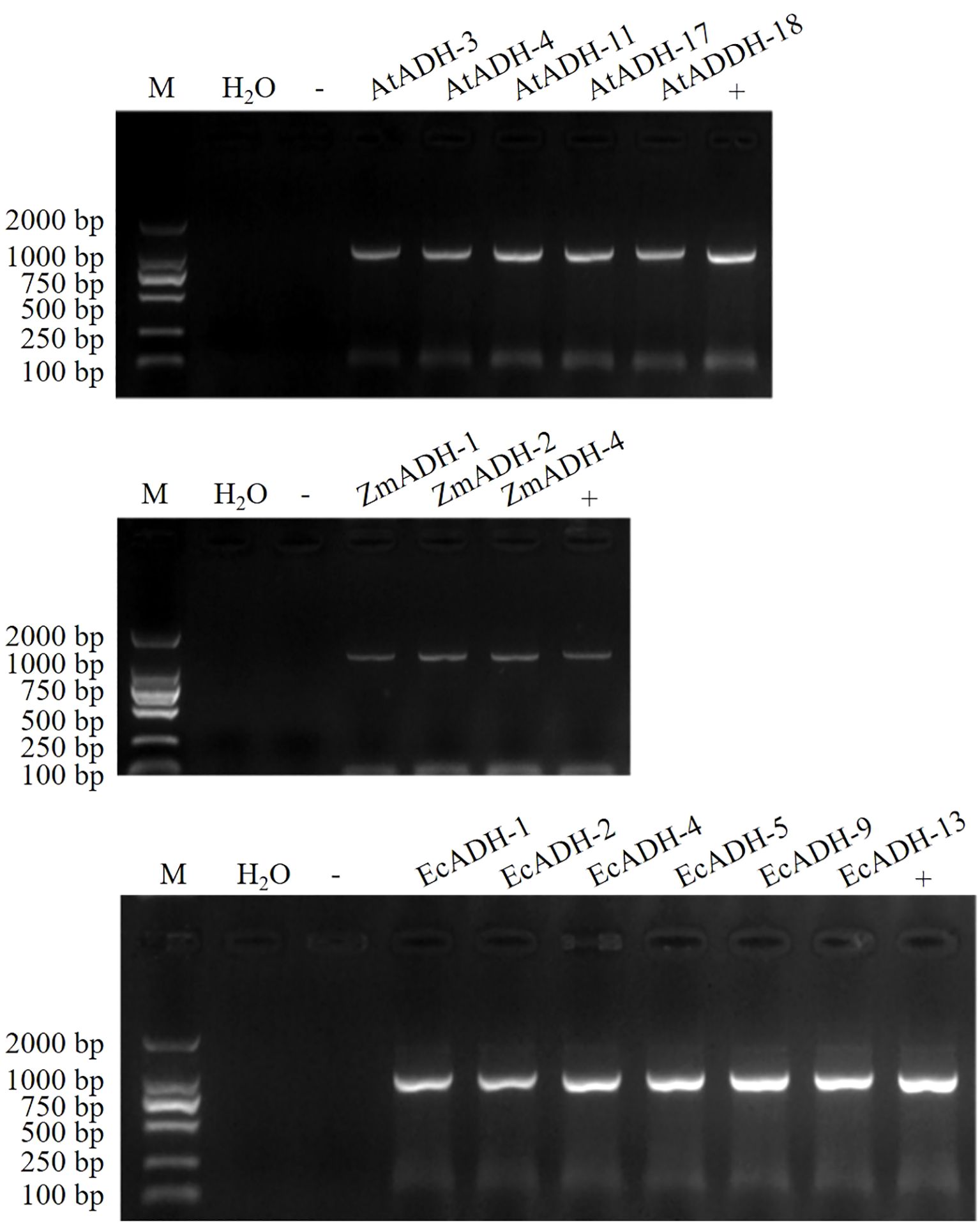
Figure 1. PCR identification of homozygous T3 lines. M: DNA marker DL2000, -: Negative control (Wild type), +: Positive control (pBI121-cMyc-NY), the other lanes: amplified CDSs of genes AtADH, ZmADH, and EcADH, respectively, from the T3 lines.
By resequencing the genomic DNA of these 14 T3 lines and alignment against the sequences of the Arabidopsis genome and T-DNA, a single T-DNA integrated site was found on chromosomes 5, 5, 1, 3, 4, 1, 3, 5, and 5 of lines AtADH-11, AtADH-17, AtADH-18, ZmADH-1, ZmADH-2, ZmADH-4, EcADH-4, EcADH-5, and EcADH-9, respectively. Each of the integrated sequences contained a complete expression structure of 4404, 4024, 2778, 4500, 13131, 4279, 13124, 11887, and 13114 bp, respectively (Supplementary Table S4). By using the primers (Supplementary Table S2), the sequences flanking the left or right borders, or both, of these integration sites were successfully amplified, sequenced, and aligned against the T-DNA sequences of the expression vector and the Arabidopsis genome with high confidence (E value ≤ 2.90E-40 and similarity ≥ 93.60%) (Supplementary Table S5). Single integrated sites of the T-DNA containing each of the transformed genes AtADH, ZmADH, and EcADH were confirmed on chromosome 5 of line AtADH-11, chromosome 5 of line AtADH-17, chromosome 1 of line AtADH-18, chromosome 3 of line ZmADH-1, chromosome 4 of line ZmADH-2, chromosome 1 of line ZmADH-4, chromosome 3 of line EcADH-4, chromosome 5 of line EcADH-5, and chromosome 5 of line EcADH-9, respectively (Figure 2). These nine homozygous T3 lines were used for the following detections.
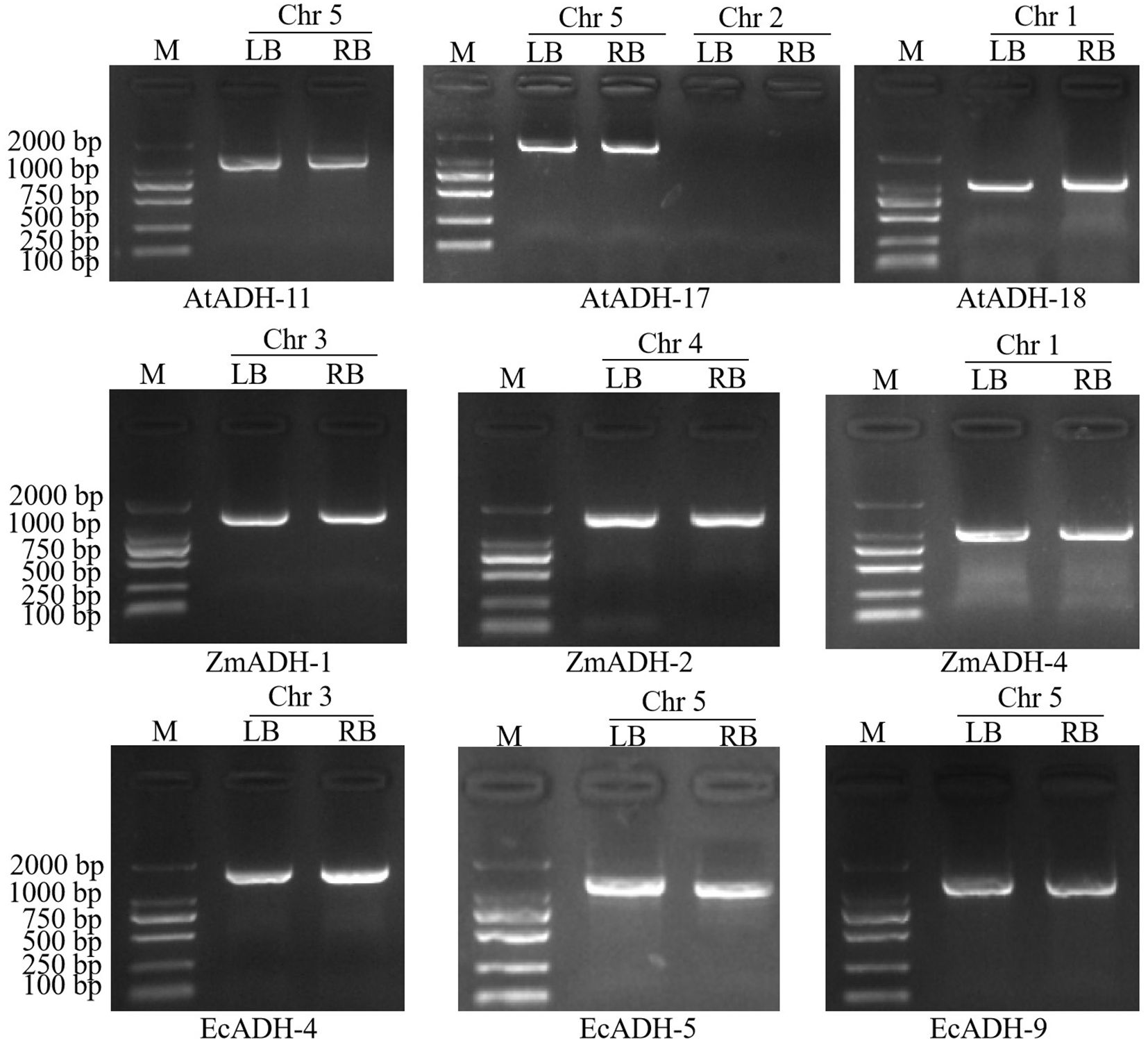
Figure 2. Agarose electrophoresis of amplified products of T-DNA flanking sequences in T3 lines. M: DNA marker DL2000, Chr: chromosome, LB: left border, RB right border.
3.2 Relative expression levels of transformed genes
The result of RT-qPCR showed that the transformed endogenous gene AtADH was overexpressed under the drive of the CaMV35S promoter in all three T3 lines (AtADH-11, AtADH-17, and AtADH-18) transformed by the endogenous gene AtADH, and the relative expression levels were more than 15 times higher than the wild control with only the endogenous AtADH gene (Figure 3A). In the three T3 lines ZmADH-1, ZmADH-2, and ZmADH-4, and the three T3 lines EcADH-4, EcADH-5, and EcADH-9, the transformed exogenous genes ZmADH and EcADH were overexpressed under the drive of the CaMV35S promoter, and their relative expression levels were several or tens of times higher than those in the three T3 lines transformed by the endogenous gene AtADH (Figure 3B). Taken together, the averages of the relative expression levels of the three T3 lines transformed by the ZmADH gene and the three T3 lines transformed by the EcADH gene were both tens of times higher than the average of the relative expression levels of the three T3 lines transformed by the endogenous AtADH gene (Figure 3C). All the above differences were greatly significant (Figure 3).
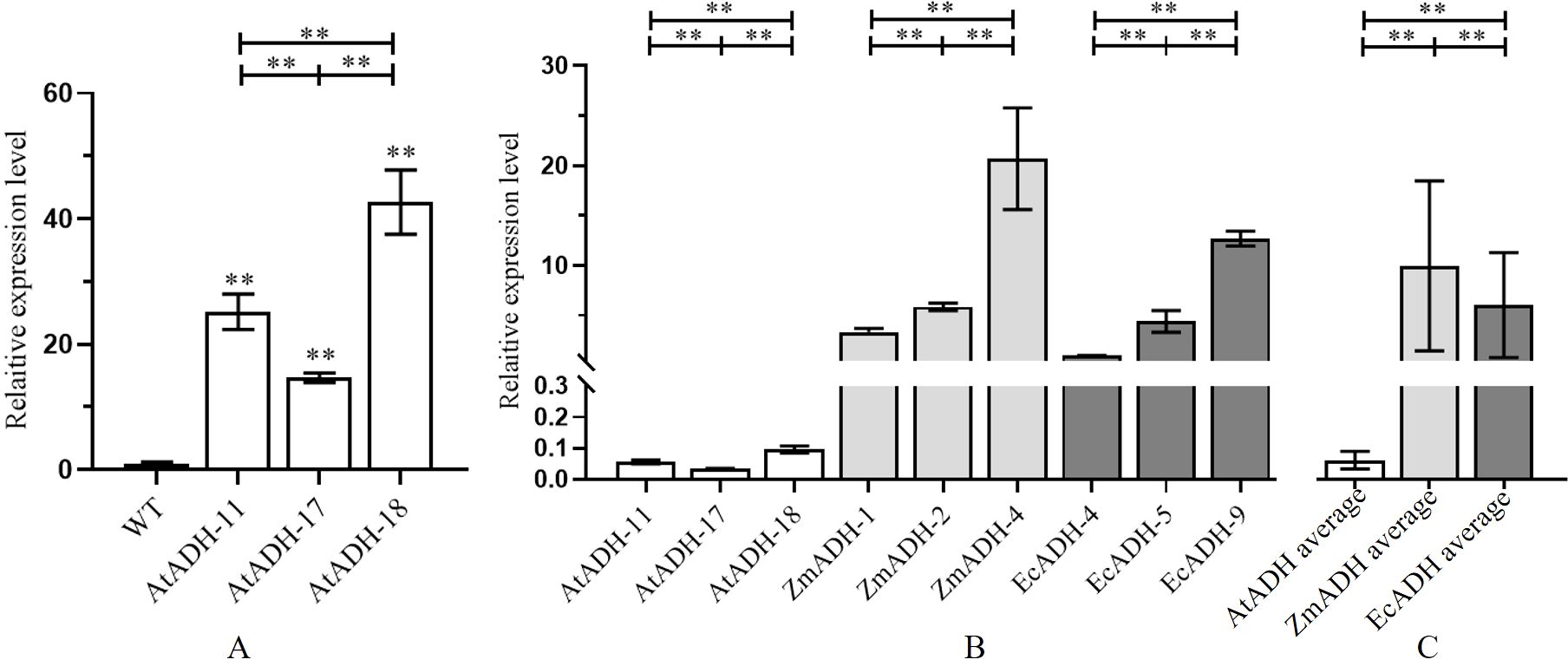
Figure 3. Relative expression levels of transformed genes AtADH, ZmADH, and EcADH in homozygous T3 lines. (A) The expression level of AtADH in overexpressing AtADH lines. (B) Comparison of relative expression level of AtADH, ZmADH, and EcADH in each line. (C) Average relative expression level of AtADH, ZmADH, and EcADH in transgenic lines. The dark, white, dark gray, and light gray columns represent the wild-type control, and the T3 lines transformed by AtADH, ZmADH, and EcADH, respectively. The double asterisk (**) at the top of the figure indicates significant differences at p < 0.01 among the three T3 lines transformed by the same gene and among the average of the three lines transformed by the same gene. The double asterisk (**) at the top of each column indicates significant differences at p < 0.01 between each of the T3 lines and the wild-type control.
3.3 Accumulation of proteins encoded by transformed genes
By Western blotting, the ADH proteins encoded by each of the AtADH, ZmADH, and EcADH genes were detected in the corresponding T3 lines with a single integration site of the T-DNA (Figure 4). The result of ELISA showed that the accumulation of the ADH protein in the wild-type control was 0.1621 ng/ml/kDa. In the three T3 lines (AtADH-11, AtADH-17, and AtADH-18) transformed by the endogenous AtADH gene, the accumulation was 0.2237, 0.2462, and 0.2416 ng/ml/kDa, respectively, with no significant difference among them but greatly significantly higher than that in the wild-type control (Figure 5A). The accumulation of the ADH protein in the three T3 lines (ZmADH-1, ZmADH-2, and ZmADH-4) transformed by the exogenous ZmADH gene was 0.2180, 0.2489, and 0.4984 ng/ml/kDa, respectively. The difference between lines ZmADH-1 and ZmADH-2 was not significant, while the difference between these two lines and line ZmADH-4 was significant with a p-value less than 0.01. However, all three lines were greatly significantly or significantly higher than that of the wild-type control (Figure 5A). The accumulation of the ADH protein in the three T3 lines (EcADH-4, EcADH-5, and EcADH-9) transformed by the exogenous EcADH genes was 0.5201, 0.3481, and 0.2660 ng/ml/kDa, respectively, with significant or greatly significant differences among them, but all were significantly higher than that of the wild-type control (Figure 5A). However, the accumulation of the ADH proteins in each of the six T3 lines transformed by the exogenous ZmADH and EcADH genes, respectively, as well as the average accumulations of the ADH protein in the three T3 lines transformed by the exogenous ZmADH gene and average accumulation of the ADH protein in the three T3 lines transformed by the exogenous EcADH genes, were both significantly or greatly higher than the average accumulation of the three T3 lines transformed by the endogenous AtADH gene (Figure 5B).
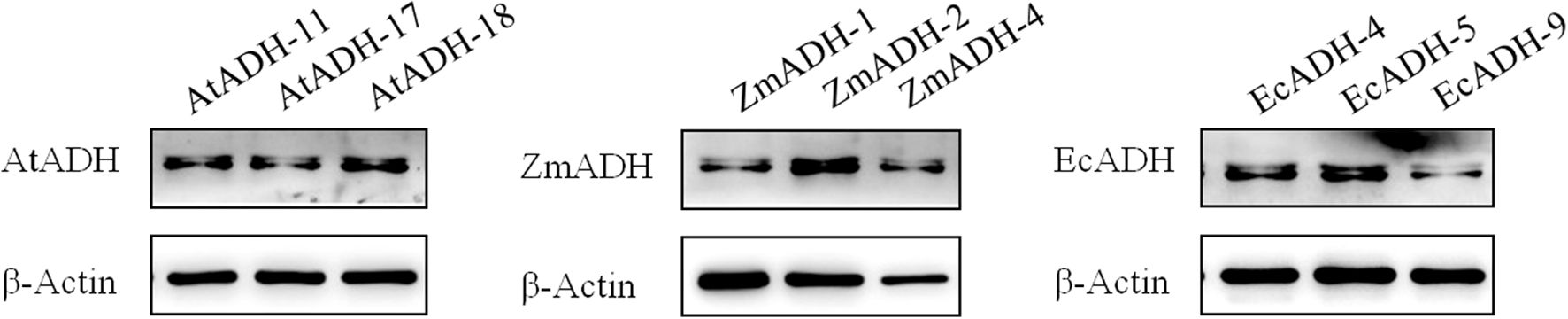
Figure 4. Western blot of AtADH, ZmADH, and EcADH proteins in each of the three T3 lines transformed by the AtADH, ZmADH, and EcADH genes, respectively.
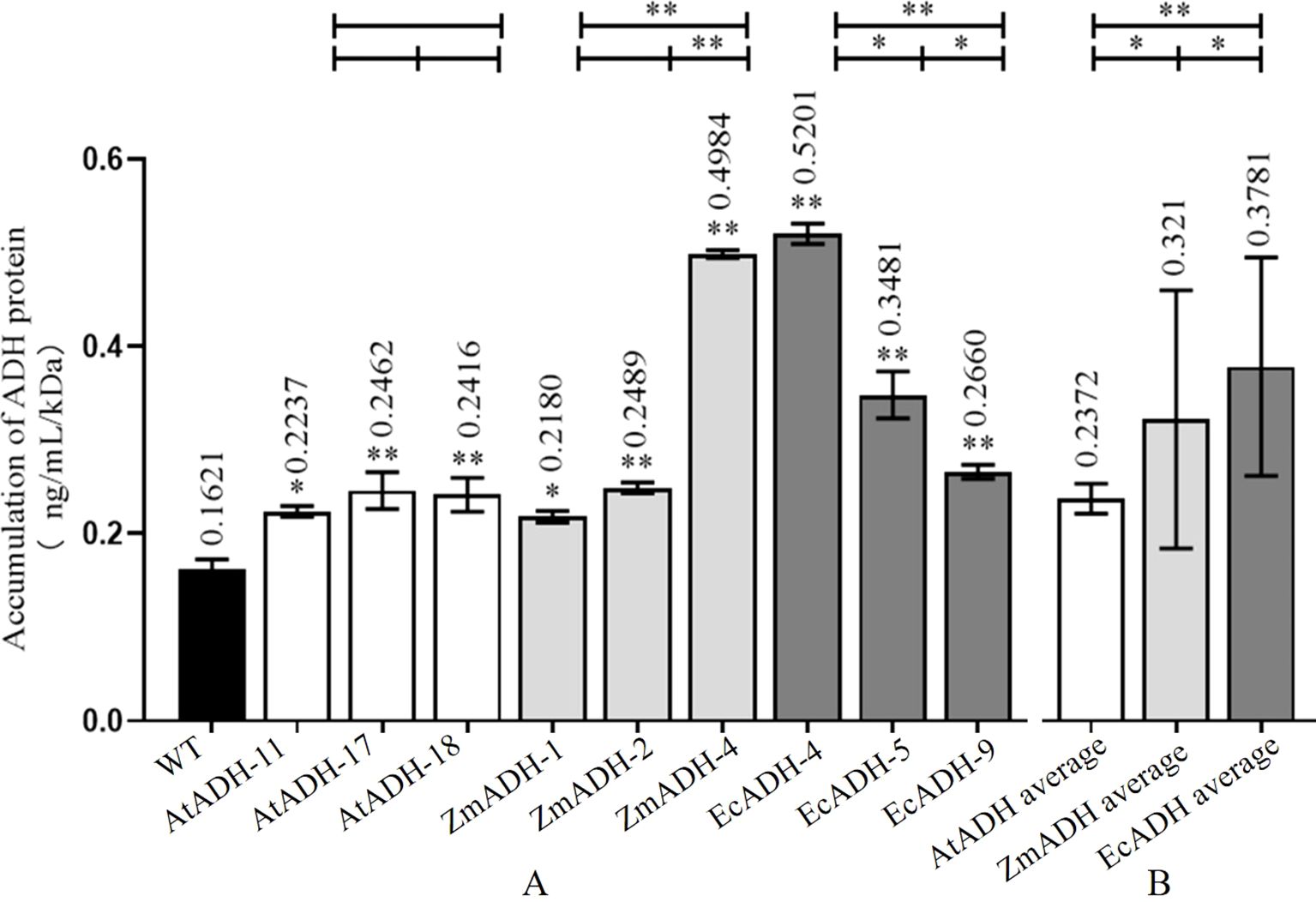
Figure 5. Accumulation of ADH protein in homozygous T3 lines. (A) ADH amount in each transgenic line. (B) Average of ADH amount of different transgenic lines of each gene. The asterisk (*) and double asterisk (**) at the top of each column indicate significant differences between the T3 lines and the wild-type (WT) at p < 0.05 and p < 0.01, respectively. The dark, white, dark gray, and light gray columns represent the wild-type control, and the T3 lines transformed by AtADH, ZmADH, and EcADH, respectively. The asterisk (*) and double asterisk (**) at the top of the figure indicate significant differences among the three T3 lines transformed by each of the three genes at p < 0.05 and p < 0.01, respectively.
3.4 ADH enzyme activity
In the wild-type control, the activity of the ADH enzyme was as low as 0.0306 U/g. In the three T3 lines (AtADH-11, AtADH-17, and AtADH-18) transformed by the endogenous AtADH gene, the activities were 0.0784, 0.0436, and 0.1502 U/g, respectively, with only line AtADH-18 significantly higher than that of the wild-type control. However, the activities of the ADH enzyme in the three T3 lines (ZmADH-1, ZmADH-2, and ZmADH-4) transformed with the exogenous ZmADH gene were 1.2860, 4.3368, and 5.3720 U/g, respectively, with significant differences among them, but all were significantly higher than that of the wild-type control. The activities of the ADH enzyme in the three T3 lines (EcADH-4, EcADH-5, and EcADH-9) transformed by the exogenous EcADH gene were 0.0750, 0.4458, and 1.1046 U/g, respectively, with greatly significant or significant differences among them. The latter two lines (EcADH-5 and EcADH-9) were significantly higher than the wild-type control. However, the average activity of the ADH enzyme in the three T3 lines transformed by the exogenous ZmADH gene and the three T3 lines transformed by the exogenous EcADH gene were significantly higher than the average activity of the ADH enzyme in the three T3 lines transformed by the endogenous AtADH gene (Figure 6).
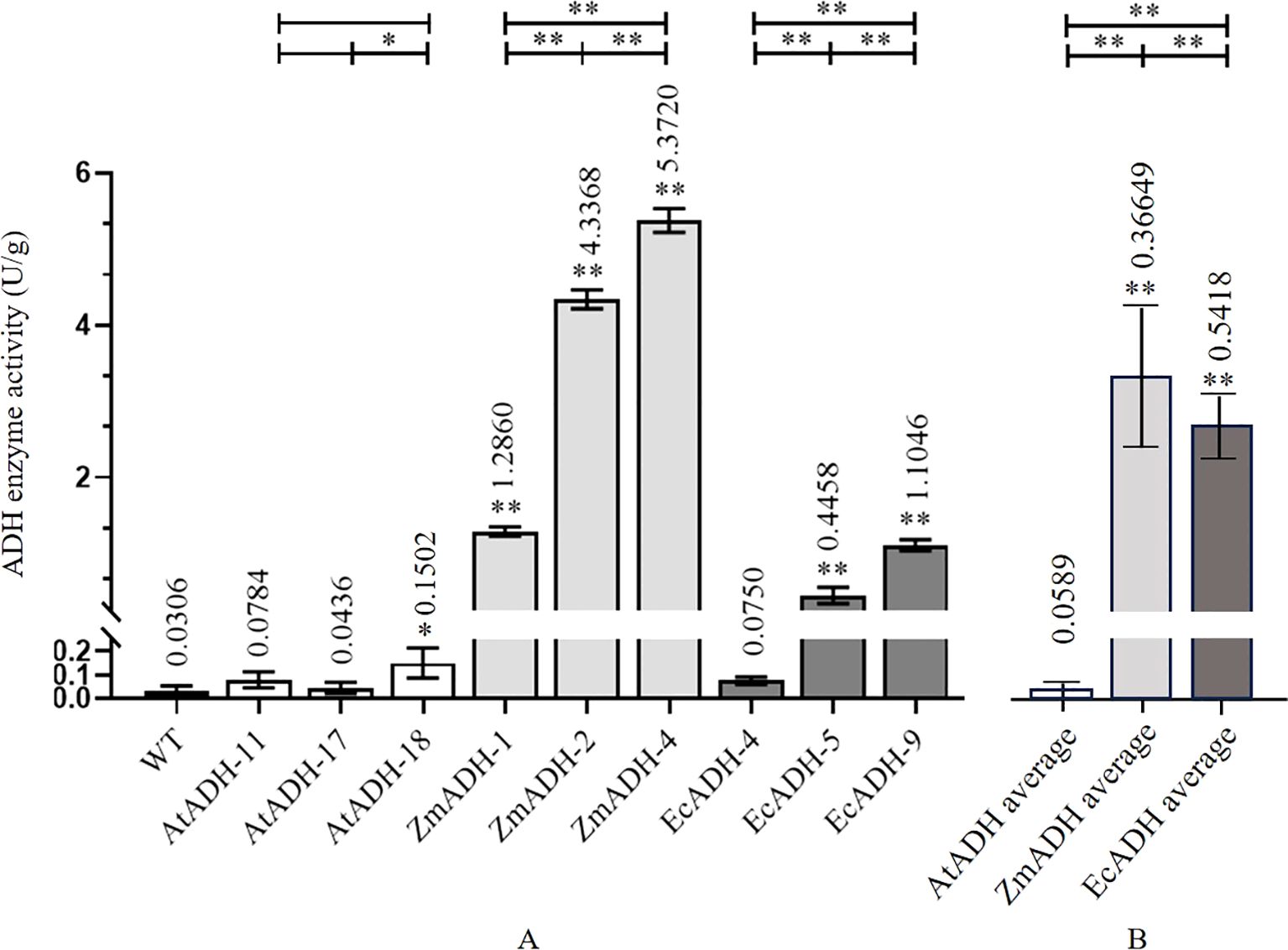
Figure 6. ADH enzyme activity in homozygous T3 lines. (A) ADH activity in each transgenic line. (B) Average of ADH activity of different transgenic lines of each gene. The dark, white, dark gray, and light gray columns represent the wild-type control, and the T3 lines transformed by AtADH, ZmADH, and EcADH, respectively. The asterisk (*) and double asterisk (**) at the top of each column indicate significant differences between the T3 lines and the wild-type (WT) at p < 0.05 and p < 0.01, respectively. The asterisk (*) and double asterisk (**) at the top of the figure indicate significant differences among the three T3 lines transformed by each of the three genes at p < 0.05 and p < 0.01, respectively.
4 Discussion
4.1 Less regulation of endogenous mechanisms to the expression of exogenous genes
In the three T3 lines transformed by the endogenous AtADH gene, the relative expression levels of the transformed genes detected by RT-qPCR inevitably included the expression of the endogenous AtADH gene of the recipient wild-type Arabidopsis. However, the transformed exogenous AtADH gene, driven by the constitutive strong promoter CaMV35S, significantly increased their expression levels together with the expression of the endogenous AtADH gene. The relative expression levels were more than 15 times higher than those in the wild-type control (Figure 3A). In the six T3 lines transformed by the exogenous ZmADH and EcADH genes, the expression levels of the exogenous genes were also significantly increased by ten to tens of times compared to the three T3 lines transformed by the endogenous AtADH genes (Figure 3B). This result indicated that the expression of the transformed exogenous genes was less regulated by endogenous mechanisms and more mRNA could be accumulated, although they were under the drive of the same constitutive strong promoter as the transformed endogenous genes. The same results of database and literature research were also reviewed by Khush (2001); Showalter et al. (2009); Chen et al. (2019), and Yu et al. (2022). The significant differences in the relative expression levels of the transformed genes among the different T3 lines transformed with the same gene could be due to the different integration sites of the transformed genes in the Arabidopsis genome and their impact on the expression of the transformed genes (Figure 2). As we discussed in a previous review (Yu et al., 2022), the transformed exogenous genes overexpress under the control of constitutive promoters and confer recipient crops with novel traits that are usually absent or function in different pathways in the recipient crops themselves. In addition to the influence of the genetic background, growth stage, and environment, the heterologous expression of the transformed exogenous genes in recipient crops is usually less regulated at the transcriptional level (Khush, 2001; Showalter et al., 2009; Chen et al., 2019).
4.2 More accumulation of proteins encoded by exogenous genes
By Western blotting, the ADH proteins encoded by each of the transformed AtADH, ZmADH, and EcADH genes were detected in each of the three corresponding T3 lines (Figure 4). The result of ELISA showed that the accumulation of the ADH protein in the three T3 lines transformed by the endogenous AtADH gene was significantly higher than that in the wild-type control (Figure 5A). The most important was that the accumulation of the ADH proteins in each of the six T3 lines transformed by the exogenous ZmADH and EcADH genes, respectively, as well as the average accumulations of the ADH protein in each of the three T3 lines transformed by the exogenous ZmADH and EcADH genes, respectively, were both significantly higher than the average accumulation of the three T3 lines transformed by the endogenous AtADH gene (Figure 5B). This result indicated that the accumulation of the proteins encoded by exogenous genes was higher than that encoded by endogenous genes. The proteins encoded by the transformed exogenous genes accumulate and function independently of the endogenous metabolic pathways and confer recipient crops with novel or enhanced traits (Adamczyk and Sumerford, 2001; Palma et al., 2014; Melo et al., 2016). In the 13 commercial cotton varieties, the accumulation of the Cry protein was also found to nonlinearly correlate with the heterologous transcription levels of the transformed exogenous Cry1Ac gene (Adamczyk and Sumerford, 2001). This indicates that there are few endogenous factors that affect the accumulation of exogenous proteins, which can function independently of the endogenous metabolic pathways of recipient crops (Palma et al., 2014; Melo et al., 2016). However, endogenous proteins encoded by the overexpressed endogenous genes are usually regulated in complex networks with functional redundance or replaceable pathways and are difficult to confer the desirable phenotypes significantly (Winkel, 2004; Fiehn and Weckwerth, 2010; Wan et al., 2017).
4.3 Higher enzyme activity of proteins encoded by exogenous genes
The result of the enzyme reaction assay showed that the activities of the ADH enzyme in the three T3 lines transformed by the exogenous ZmADH gene and two T3 lines transformed by the exogenous EcADH gene were greatly significantly higher than that of the wild-type control (Figure 6A). Moreover, the average ADH enzyme activity in the three T3 lines transformed by the exogenous ZmADH gene and in the three T3 lines transformed by the exogenous EcADH gene were both significantly higher than the average ADH enzyme activity in the three T3 lines transformed by the endogenous AtADH gene (Figure 6B). This result indicated that the enzyme activity of the proteins encoded by exogenous genes was higher than that of the proteins encoded by endogenous genes.
4.4 Limitaions to be improved
One of the T3 lines transformed by the exogenous EcADH gene did not show a significant increase in enzyme activity (Figure 6A), although its relative expression level of and accumulation of the EcADH protein encoded by the transformed exogenous EcADH gene were significantly increased (Figures 3B, 5A). This exception should be investigated further. In addition, more homozygous transgenic lines should be transformed and screened for elimination of potential biases probably caused by variable T-DNA integration sites or operational deviation on specific methodologies like RT-qPCR, ELISA, and enzyme activity assay.
5 Conclusion
The relative expression levels of the transformed exogenous ZmADH and EcADH genes were ten or tens of times higher than that of the transformed endogenous AtADH gene. The accumulation levels of the proteins encoded by the transformed ZmADH and EcADH were significantly higher than that encoded by the transformed endogenous AtADH. The ADH activities of the T3 lines transformed by the exogenous ZmADH and EcADH were also significantly higher than that transformed by the endogenous AtAHD as well as the wild-type control. According to these results, along with database and literature searches, it can be concluded that exogenous genes are more conducive to transgenic improvement of crop varieties. It is suggested that more attention should be paid to the transformation of exogenous genes from distant species for transgenic improvement of crops, especially for abiotic and biotic tolerance as well as mechanism pathways.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
HY: Investigation, Visualization, Writing – original draft, Writing – review & editing, Funding acquisition. HL: Data curation, Formal analysis, Writing – original draft. FL: Data curation, Writing – original draft. QY: Methodology, Resources, Writing – original draft. HD: Formal analysis, Validation, Visualization, Software, Writing – review & editing. WL: Conceptualization, Supervision, Writing – original draft. FF: Project administration, Conceptualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (32001552) and the Department of Science and Technology of Sichuan Province (2022YFH0067).
Acknowledgments
We are grateful to the Key Laboratory of Biology and Genetic Improvement of Maize in Southwest Region, Ministry of Agriculture, for its technical support.
Conflict of interest
The authors declare that the study was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1476754/full#supplementary-material
References
Adamczyk, J. J., Jr., Sumerford, D. V. (2001). Potential factors impacting season-long expression of Cry1Ac in 13 commercial varieties of Bollgard cotton. J. Insect Sci. 1, 13. doi: 10.1673/031.001.1301
Bates, S. L., Zhao, J. Z., Roush, R. T., Shelton, A. M. (2005). Insect resistance management in GM crops: past, present and future. Nat. Biotechnol. 23, 57–62. doi: 10.1038/nbt1056
Blattner, F. R., Plunkett, G., 3rd, Bloch, C. A., Perna, N. T., Burland, V., Riley, M., et al. (1997). The complete genome sequence of Escherichia coli K-12. Science 277, 1453–1462. doi: 10.1126/science.277.5331.1453
Castiglioni, P., Warner, D., Bensen, R. R., Anstrom, D. D., Harrison, J., Stoecker, M., et al. (2008). Bacterial RNA chaperones confer abiotic stress tolerance in plants and improved grain yield in maize under water-limited conditions. Plant Physiol. 147, 446–455. doi: 10.1104/pp.108.118828
Castle, L. A., Siehl, D. L., Gorton, R., Patten, P. A., Chen, Y. H., Bertain, S., et al. (2004). Discovery and directed evolution of a glyphosate tolerance gene. Science 304, 1151–1114. doi: 10.1126/science.1096770
Century, K., Reuber, T. L., Ratcliffe, O. J. (2008). Regulating the regulators: the future prospects for transcription-factor based agricultural biotechnology products. Plant Physiol. 147, 20–29. doi: 10.1104/pp.108.117887
Chang, C., Meyerowitz, E. M. (1986). Molecular cloning and DNA sequence of the Arabidopsis thaliana alcohol dehydrogenase gene. Proc. Natl. Acad. Sci. U.S.A. 83, 1408–1412. doi: 10.1073/pnas.83.5.1408
Chen, Y., Li, Y., Zhou, M., Cai, Z., Tambel, L. I. M., et al. (2019). Nitrogen deficit decreases seed Cry1Ac endotoxin expression in Bt transgenic cotton. Plant Physiol. Biochem. 141, 114–121. doi: 10.1016/j.plaphy.2019.05.017
Chitchumroonchokchai, C., Diretto, G., Parisi, B., Giuliano, G., Failla, M. L. (2017). Potential of golden potatoes to improve vitamin A and vitamin E status in developing countries. PloS One 12, e0187102. doi: 10.1371/journal.pone.0187102
Chung, H. J., Ferl, R. J. (1999). Arabidopsis alcohol dehydrogenase expression in both shoots and roots is conditioned by root growth environment. Plant Physiol. 121, 429–436. doi: 10.1104/pp.121.2.429
Cui, J., Luo, J., van der Werf, W., Ma, Y., Xia, J. (2011). Effect of pyramiding Bt and CpTI genes on resistance of cotton to Helicoverpa armigera (Lepidoptera: Noctuidae) under laboratory and field conditions. J. Econ. Entomol. 104, 673–684. doi: 10.1603/ec09228
Dolferus, R., Jacobs, M., Peacock, W. J., Dennis, E. S. (1994). Differential interactions of promoter elements in stress responses of the Arabidopsis ADH gene. Plant Physiol. 105, 1075–1087. doi: 10.1104/pp.105.4.1075
Fiehn, O., Weckwerth, W. (2010). Deciphering metabolic networks. FEBS J. 270, 579–588. doi: 10.1046/j.1432-1033.2003.03427.x
Gao, H., Wang, Y., Xu, P., Zhang, Z. (2018). Overexpression of a WRKY transcription factor TaWRKY2 enhances drought stress tolerance in transgenic wheat. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00997
Ghareyazie, B., Alinia, F., Menguito, C. A., Rubia, L. G., de Palma, J. M., Liwanag, E. A. (1997). Enhanced resistance to two stem borers in an aromatic rice containing a synthetic cryIA(b) gene. Mol. Breed. 3, 401–414. doi: 10.1023/A:1009695324100
Guo, B., Guo, Y., Hong, H., Jin, L., Zhang, L., Chang, R. Z., et al. (2015). Co-expression of G2-EPSPS and glyphosate acetyltransferase GAT genes conferring high tolerance to glyphosate in soybean. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00847
International Service for the Acquisition of Agri-biotech Applications (2023). ISAAA in 2023: accomplishment report. Available online at: https://www.isaaa.org/resources/publications/annualreport/2023/default.asp (Accessed January 15, 2024).
International Service for the Acquisition of Agri-biotech Applications (2024). GM approval database. Available online at: https://www.isaaa.org/gmapprovaldatabase/default.asp (Accessed July 15, 2024).
Kamthan, A., Chaudhuri, A., Kamthan, M., Datta, A. (2016). Genetically modified (GM) crops: milestones and new advances in crop improvement. Theor. Appl. Genet. 129, 1639–1655. doi: 10.1007/s00122-016-2747-6
Karalis, D. T., Karalis, T., Karalis, S., Kleisiari, A. S. (2020). Genetically modified products, perspectives and challenges. Cureus 12, e7306. doi: 10.7759/cureus.7306
Karthik, K., Nandiganti, M., Thangaraj, A., Singh, S., Mishra, P., Rathinam, M., et al. (2020). Transgenic cotton (Gossypium hirsutum L.) to combat weed vagaries: utility of an apical meristem-targeted in planta transformation strategy to introgress a modified CP4-EPSPS gene for glyphosate tolerance. Front. Plant Sci. 11, 768. doi: 10.3389/fpls.2020.00768
Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, Y., Shinozaki, K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17, 287–291. doi: 10.1038/7036
Khush, G. S. (2001). Green revolution: the way forward. Nat. Rev. Genet. 2, 815–822. doi: 10.1038/35093585
Kinney, A. J., Cahoon, E. B., Hitz, W. D. (2022). Manipulating desaturase activities in transgenic crop plants. Biochem. Soc Trans. 30, 1099–1103. doi: 10.1042/bst0301099
Klumper, W., Qaim, M. (2014). Ameta-analysis of the impacts of genetically modified crops. PloS One 9, e111629. doi: 10.1371/journal.pone.0111629
Kobayashi, K., Wang, X., Wang, W. (2023). Genetically modified rice is associated with hunger, health, and climate resilience. Foods 12, 2776. doi: 10.3390/foods12142776
Melo, A. L., Soccol, V. T., Soccol, C. R. (2016). Bacillus thuringiensis: mechanism of action, resistance, and new applications: a review. Crit. Rev. Biotechnol. 36, 317–326. doi: 10.3109/07388551.2014.960793
Millar, A. A., Dennis, E. S. (1996). The alcohol dehydrogenase genes of cotton. Plant Mol. Biol. 31, 897–904. doi: 10.1007/BF00019476
Napier, J. A., Haslam, R. P., Tsalavouta, M., Sayanova, O. (2019). The challenges of delivering genetically modified crops with nutritional enhancement traits. Nat. Plants 5, 563–567. doi: 10.1038/s41477-019-0430-z
Nelson, D. D., Repetti, P. P., Adams, T. T., Creelman, R. R., Wu, J., Warner, D. D., et al. (2007). Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc. Natl. Acad. Sci. U.S.A. 104, 16450–16455. doi: 10.1073/pnas.0707193104
Padgette, S. R., Kolacz, K. H., Delannay, X., Re, D. B., Lavallee, B. G., Tinius, C. N., et al. (1995). Development, identification, and characterization of a glyphosate-tolerant soybean line. Crop Sci. 35, 1451–1461. doi: 10.2135/cropsci1995.0011183X003500050032x
Paine, J. A., Shipton, C. A., Chaggar, S., Howells, R. M., Kennedy, M. J., Vernon, G., et al. (2005). Improving the nutritional value of Golden Rice through increased pro-vitamin A content. Nat. Biotechnol. 23, 482–487. doi: 10.1038/nbt1082
Palma, L., Muñoz, D., Berry, C., Murillo, J., Caballero, P. (2014). Bacillus thuringiensis toxins: an overview of their biocidal activity. Toxins (Basel) 6, 3296–3325. doi: 10.3390/toxins6123296
Peters, J. S., Frenkel, C. (2004). Relationship between alcohol dehydrogenase activity and low-temperature in two maize genotypes, Silverado F1 and Adh1-Adh2- doubly null. Plant Physiol. Biochem. 42, 841–846. doi: 10.1016/j.plaphy.2004.10.004
Raymond Park, J., McFarlane, I., Hartley Phipps, R., Ceddia, G. (2011). The role of transgenic crops in sustainable development. Plant Biotechnol. J. 9, 2–21. doi: 10.1111/j.1467-7652.2010.00565.x
Ribichich, K. F., Chiozza, M., Avalos-Britez, S., Cabello, J. V., Arce, A. L., Watson, G., et al. (2020). Successful field performance in warm and dry environments of soybean expressing the sunflower transcription factor HaAb4. J. Exp. Bot. 71, 3142–3156. doi: 10.1093/jxb/eraa064
Schilling, R. K., Tester, M., Marschner, P., Plett, D. C., Roy, S. J. (2017). AVP1: one protein, many roles. Trends Plant Sci. 22, 154–162. doi: 10.1016/j.tplants.2016.11.012
Shen, C., Yuan, J., Ou, X., Ren, X., Li, X. (2021). Genome-wide identification of alcohol dehydrogenase (ADH) gene family under waterlogging stress in wheat (Triticum aestivum). Peer J. 9, e11861. doi: 10.7717/peerj.11861
Shi, H., Liu, W., Yao, Y., Wei, Y., Chan, Z. (2017). Alcohol dehydrogenase 1 (ADH1) confers both abiotic and biotic stress resistance in Arabidopsis. Plant Sci. 262, 24–31. doi: 10.1016/j.plantsci.2017.05.013
Showalter, A. M., Heuberger, S., Tabashnik, B. E., Carriere, Y., Coates, B. (2009). A primer for using transgenic insecticidal cotton in developing countries. J. Insect Sci. 9, 22. doi: 10.1673/031.009.2201
Song, Y., Liu, L., Ma, X. (2019). CbADH1 improves plant cold tolerance. Plant Signal. Behav. 14, 1612680. doi: 10.1080/15592324.2019.1612680
Stalker, D. M., Mcbride, K. E., Malyj, L. D. (1988). Herbicide resistance in transgenic plants expressing a bacterial detoxification gene. Science 242, 419–423. doi: 10.1126/science.242.4877.419
Su, W., Ren, Y., Wang, D., Su, Y., Feng, J., Zhang, C., et al. (2020). The alcohol dehydrogenase gene family in sugarcane and its involvement in cold stress regulation. BMC Genomics 21, 521. doi: 10.1186/s12864-020-06929-9
Thompson, C. J., Movva, N. R., Tizard, R., Crameri, R., Davies, J. E., Lauwereys, M., et al. (1987). Characterization of the herbicide-resistance gene bar from Streptomyces hygroscopicus. EMBO J. 6, 2519–2523. doi: 10.1002/j.1460-2075
Ventura, I., Brunello, L., Iacopino, S., Valeri, M. C., Novi, G., Dornbusch, T., et al. (2020). Arabidopsis phenotyping reveals the importance of alcohol dehydrogenase and pyruvate decarboxylase for aerobic plant growth. Sci. Rep. 10, 16669. doi: 10.1038/s41598-020-73704-x
Wan, H., Cui, Y., Ding, Y., Mei, J., Dong, H., Zhang, W., et al. (2017). Time-series analyses of transcriptomes and proteomes reveal molecular networks underlying oil accumulation in canola. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.02007
Winkel, B. S. (2004). Metabolic channeling in plants. Annu. Rev. Plant Biol. 55, 85–107. doi: 10.1146/annurev.arplant.55.031903
Xiao, B., Huang, Y., Tang, N., Xiong, L. (2007). Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor. Appl. Genet. 115, 35–46. doi: 10.1007/s00122-007-0538-9
Yang, F., Debatosh, D., Song, T., Zhang, J. H. (2021). Retraction note: light harvesting-like protein 3 interacts with phytoene synthase and is necessary for carotenoid and chlorophyll biosynthesis in rice. Rice 14, 41. doi: 10.1186/s12284021-00484-x
Ye, X., Al-Babili, S., Kloti, A., Zhang, J., Lucca, P., Beyer, P., et al. (2000). Engineering the provitamin A (beta-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 287, 303–305. doi: 10.1126/science.287.5451.303
Yu, H., Yang, Q., Fu, F., Li, W. (2022). Three strategies of transgenic manipulation for crop improvement. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.948518
Zhang, Z. Y., Yang, L., Zhou, S. F., Wang, H. G., Li, W. C., Fu, F. L. (2011). Improvement of resistance to maize dwarf mosaic virus mediated by transgenic RNA interference. J. Biotechnol. 153, 181–187. doi: 10.1016/j.jbiotec.2011.03.019
Keywords: endogenous gene, enzyme activity, exogenous gene, protein accumulation, transgenics
Citation: Yu H, Lv H, Lu F, Yang Q, Duan H, Li W and Fu F (2025) Expression evaluation of exogenous and endogenous alcohol dehydrogenase genes in transgenic Arabidopsis. Front. Plant Sci. 15:1476754. doi: 10.3389/fpls.2024.1476754
Received: 06 August 2024; Accepted: 27 December 2024;
Published: 22 January 2025.
Edited by:
Avinash Mishra, Central Salt & Marine Chemicals Research Institute (CSIR), IndiaReviewed by:
Joorie Bhattacharya, International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), IndiaMd. Mohiuddin Kabir, East West University, Bangladesh
Copyright © 2025 Yu, Lv, Lu, Yang, Duan, Li and Fu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wanchen Li, YXVtZHltc0BzaWNhdS5lZHUuY24=; Fengling Fu, ZmZsQHNpY2F1LmVkdS5jbg==
†These authors have contributed equally to this work
 Haoqiang Yu
Haoqiang Yu Hong Lv†
Hong Lv† Fengzhong Lu
Fengzhong Lu Wanchen Li
Wanchen Li