- 1Department of Genetics and Plant Breeding, Centre for Plant Breeding and Genetics, Tamil Nadu Agricultural University, Coimbatore, Tamil Nadu, India
- 2Department of Plant Breeding, Swedish University of Agricultural Sciences, Lomma, Sweden
- 3Department of Plant Biotechnology, Centre for Plant Molecular Biology and Biotechnology, Tamil Nadu Agricultural University, Coimbatore, Tamil Nadu, India
- 4Centre for Plant Breeding and Genetics, Tamil Nadu Agricultural University, Coimbatore, Tamil Nadu, India
Global protein consumption is increasing exponentially, which requires efficient identification of potential, healthy, and simple protein sources to fulfil the demands. The existing sources of animal proteins are high in fat and low in fiber composition, which might cause serious health risks when consumed regularly. Moreover, protein production from animal sources can negatively affect the environment, as it often requires more energy and natural resources and contributes to greenhouse gas emissions. Thus, finding alternative plant-based protein sources becomes indispensable. Rapeseed is an important oilseed crop and the world’s third leading oil source. Rapeseed byproducts, such as seed cakes or meals, are considered the best alternative protein source after soybean owing to their promising protein profile (30%–60% crude protein) to supplement dietary requirements. After oil extraction, these rapeseed byproducts can be utilized as food for human consumption and animal feed. However, anti-nutritional factors (ANFs) like glucosinolates, phytic acid, tannins, and sinapines make them unsuitable for direct consumption. Techniques like microbial fermentation, advanced breeding, and genome editing can improve protein quality, reduce ANFs in rapeseed byproducts, and facilitate their usage in the food and feed industry. This review summarizes these approaches and offers the best bio-nutrition breakthroughs to develop nutrient-rich rapeseed byproducts as plant-based protein sources.
1 Introduction
United Nations predicts that the global population will exceed 9.5 billion by 2050. Agri-food Innovation Council (2019) estimates that global protein consumption will double within the next 40 years, increasing from 473 Metric tons (Mt) to 944 Mt by 2054 (Agri-Food Innovation Council, 2019; available at https://nrc.canada.ca/sites/default/files/2019-10/Plant_protein_industry_market_analysis_summary.pdf). This substantial increase in protein demand necessitates an annual increment of 14% in protein production. Hence, identifying novel alternative protein sources is crucial to supplement the growing protein needs. In addition to animal proteins, potential alternative plant protein sources must be exploited to meet the population’s growing needs. Traditional animal protein sources such as eggs, poultry, fish, and dairy provide complete protein diets containing all the essential amino acids. However, its high fat and low fiber composition poses significant health risks such as obesity, gastro-esophageal reflux disease (GERD), cardiovascular diseases, type 2 diabetes, sleep disorders, and non-alcoholic fatty liver disease (Neuenschwander et al., 2023). Moreover, the production of animal-based proteins is environmentally unsustainable, consuming high levels of energy and natural resources and increasing the emission of greenhouse gases. These factors have increased the demand for vegan-friendly and non-animal protein sources in food, beverages, and snacks. Also, consumers are interested in healthy dietary choices with high protein and low fat content, which has driven the market for plant-based proteins to gain momentum in recent years.
Rapeseed (Brassica napus L.) emerges as a promising alternative plant-based protein source, ranking second after soybean with promising nutrient profiles (Bell, 1993; Feng and Zuo, 2007; Nega and Woldes, 2018). It is the third leading source of vegetable oil globally, followed by soybean and palm oil; rapeseed meal (RSM) is the second most produced protein after soybean meal. Rapeseed has versatile uses, including oil for human consumption, protein-rich seed cakes or meals for animal feed, biodiesel extraction, and various industrial applications. The oil of commercial rapeseed varieties comprises 6%–14% α-linolenic acid, 50%–66% oleic acid, and less than 7% saturated fatty acids and is considered a healthy choice for human consumption (Ghazani and Marangoni, 2013). The byproducts of rapeseed oil extraction, such as cake and meal, are potential protein sources for human and animal consumption, with crude protein content ranging from 30%–60%. Compared to cereals, RSM contains higher concentrations of essential amino acids such as tryptophan, threonine, lysine, and sulfur-containing methionine and cysteine, making it the best alternative source for demanding protein needs (Le Thanh et al., 2019). However, anti-nutritional factors (ANFs) in the seed cake or meal limit its implementation in human diets and animal feeds.
2 Nutrient profile of rapeseed byproduct
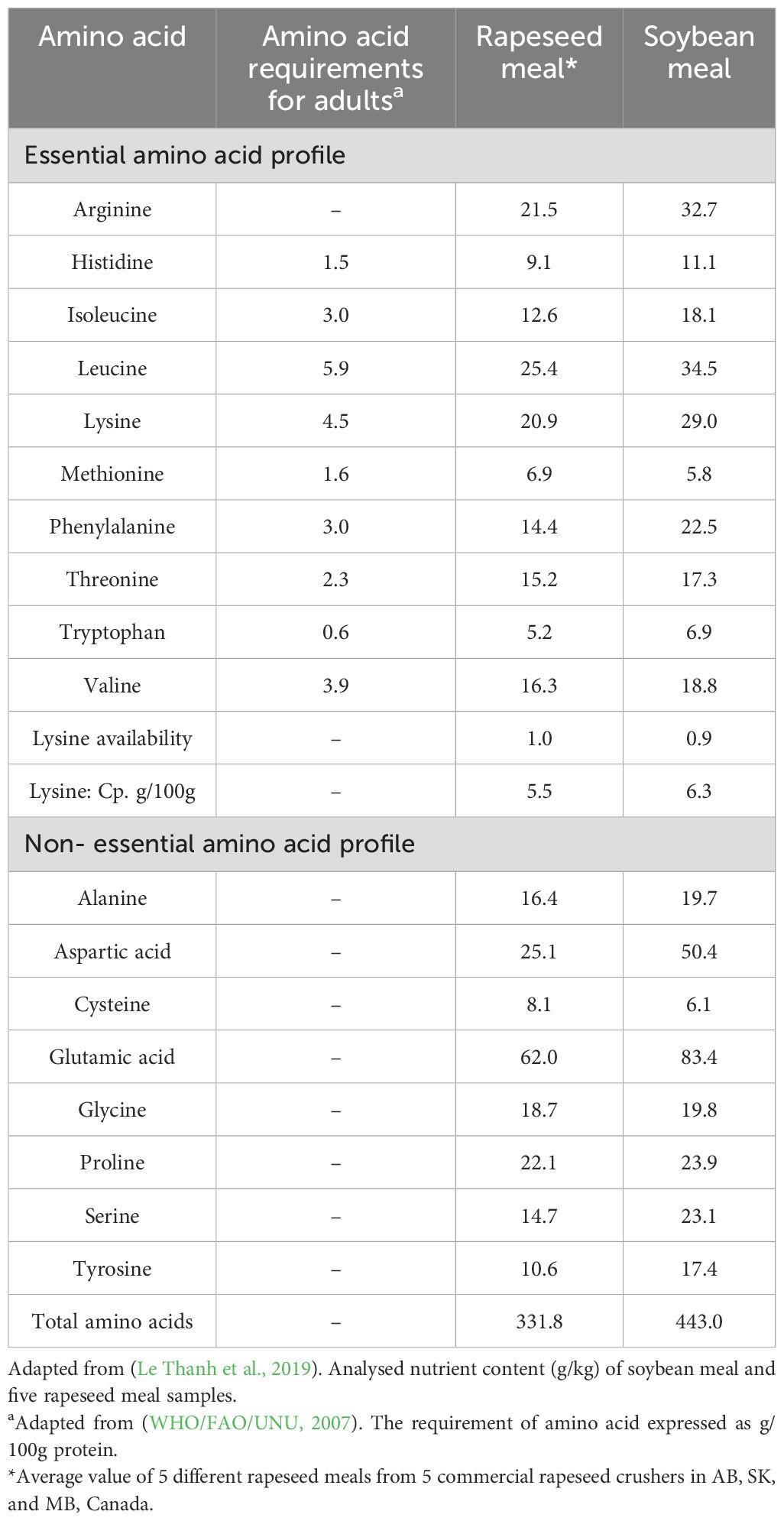
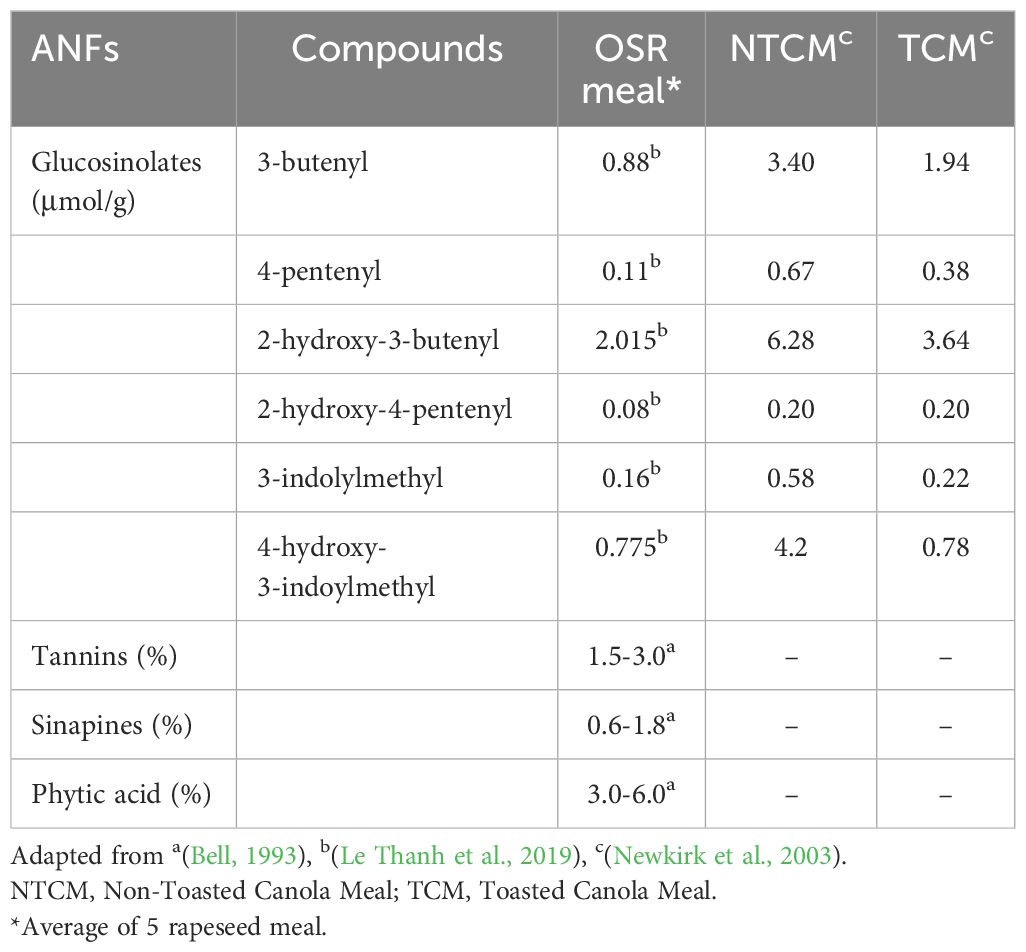
The nutrient profile of rapeseed byproducts is comparable to soybean meal. RSM’s crude protein content indicates its immense potential as a substitute for the plant-based protein diet for food and feed (Table 1).
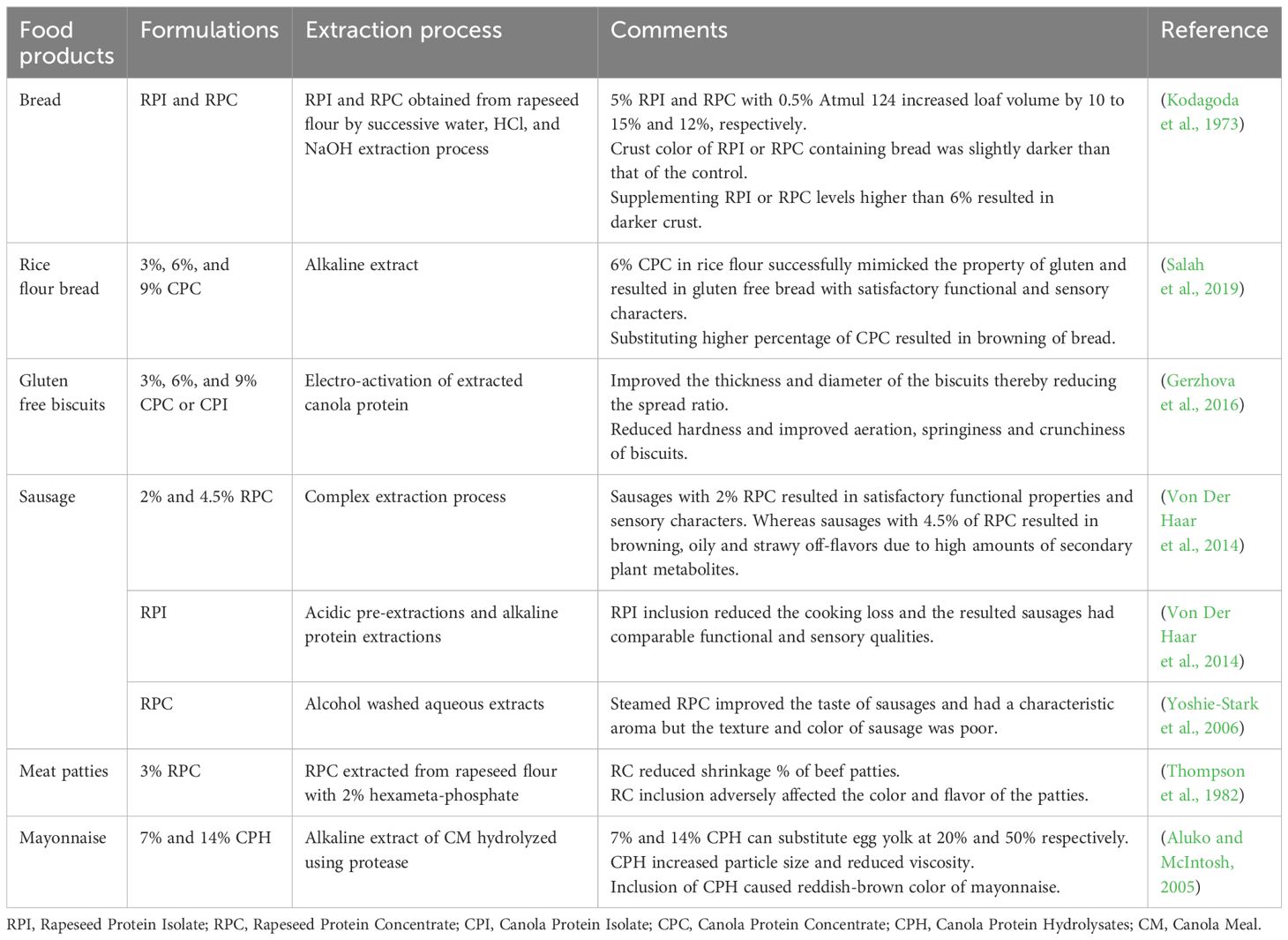
The postprandial biological value of rapeseed protein isolate (RPI) is 84%, which is higher than milk (82%) and soy protein (80%) (Bos et al., 2007). The balanced amino acid profile (Table 2) contributes to its biological value. Legumes such as green gram, peas and lentils are the primary plant-based protein sources for the human diet. Legumes are richer in lysine than cereals. However, they are lower in lysine and sulfur-rich amino acids than animal proteins (Yang et al., 2023). Nevertheless, unlike soybean meal, RSMs provide sulfur-rich amino acids (Table 2). However, high temperatures in the protein extraction methods might invoke the Maillard reaction and influence the level of available lysine content in rapeseed cakes or meals (Newkirk et al., 2003; Almeida et al., 2014; Salazar-Villanea et al., 2016).
The digestibility of the protein determines the extent of protein utilization in human food. Rapeseed proteins showed poor in vitro digestibility (83%) compared to casein (97%) (Savoie et al., 1988). The real ileal digestibility (RID) of RPI is the lowest (84%) when compared to milk (95%) and soybean (91.5%) (Bos et al., 2007). The high fiber content in the rapeseed cake or meal is a concern for their use as feed in animal husbandry as it is four times higher than the soybean meal (Table 1) (Bell, 1993; Feng and Zuo, 2007), affecting meal digestibility. Additionally, the gross energy of RSMs and metabolizable energy (ME) is lower than that of soybean meal (Blair and Reichert, 1984; Bell, 1993). ME of RSM in swine and poultry is reported to be 9.33 MJ/Kg and 7.41 MJ/Kg, respectively (Feng and Zuo, 2007). However, the ME of soybean meal was reported to be ~15-18 MJ/Kg (Lopez et al., 2020). The higher lignin content reduces the feed digestibility in RSMs, which is a prominent limitation of its application.
3 Rapeseed byproducts as a novel protein source in food and feed
Seed storage proteins (SSPs) are unique proteins found in the seeds of rapeseed and accumulated during seed maturation. Almost 90% of rapeseed proteins are storage proteins, made up of 11S cruciferin of the cupin superfamily and 2S napin of the prolamin superfamily (Wanasundara, 2011). About 32%-53% of the proteins in the rapeseed seeds are made up of cruciferin, a globulin-type protein (Malabat et al., 2003). Cruciferin is characterized by its large hexameric structure, with each subunit connected by a disulfide-bridged α and β subunits (Wanasundara et al., 2017). Napins are albumin proteins and account for 25%-45% of the total proteins in the rapeseed seeds (Malabat et al., 2003). The primary structure of a napin protein comprises 111 to 180 amino acids, while the secondary structure comprises four helices. Two subunits, small 4.5 kDa and large 10 kDa, are linked by two disulfide bonds (Wanasundara, 2011).
The properties of these proteins have immense potential for the food industry. The solubility of proteins is a functional requirement for a food-based protein system (Wanasundara et al., 2017). The solubility of napin is more than 90% in the pH range of 2-10, whereas cruciferin exhibits lower solubility in this pH range in relation to napin (Wanasundara et al., 2017). Cruciferin has better emulsifying properties than napin, whereas napin has better foaming capability than cruciferin. The emulsifying ability facilitates the use of rapeseed proteins in the production of mayonnaise and salad dressings. The poor gelling ability of the proteins limits their use in the food industry by reducing the firmness of the products (Thompson et al., 1982). However, enzymatic modification of canola protein isolate (CPI) with transglutaminase can improve the gelling ability (Pinterits and Arntfield, 2008). Food-grade canola protein products are commercially available under the names Puratein® (precipitated micelle protein of near neutral pH protein extracted with salt, >90% protein, cruciferin protein mainly), Supertein™ (protein remained soluble after micelle formation; Burcon Nutrascience, >90% protein, napin mainly). The Protein Digestibility Corrected Amino Acid Score (PDCASS) values of Supertein™ and Puratein® (Burcon Nutrascience protein products) were 0.61 (61%) and 0.64 (64%), respectively (Wanasundara et al., 2017). Utilizing rapeseed protein products in foods could result in a balanced protein diet (Table 3).
Despite their potential in the food industry, the ANFs significantly impact the sensory characteristics of rapeseed proteins, including flavor and color. Moreover, the extraction procedures highly affect the protein’s functional and sensory characteristics, which limits its usage in food formulations (Von Der Haar et al., 2014). Additionally, the presence of allergens such as napins and 2S albumins, which can cause IgE-mediated allergic responses (Puumalainen et al., 2006; Moreno and Clemente, 2008), limits their usage in food products (Monsalve et al., 2001; Murtagh et al., 2003; Puumalainen et al., 2006). Because of their stable disulfide bridges, napins and 2S albumins can resist proteolysis, high temperatures, low pH, and stay intact (Murtagh et al., 2003; Jyothi et al., 2007). Due to their resistance to degradation, they can remain in the gut for prolonged periods and may stimulate an immunological response, leading to allergic reactions (Pantoja-Uceda et al., 2004; Puumalainen et al., 2006). This resistance demands techniques to modify the protein structures to improve digestibility, which might decrease its allergic reactions and increase its potential use in the food industry. However, few rapeseed protein isolates (RPIs) have been considered safe for consumption by the Food and Drug Administration (FDA) (So and Duncan, 2021), and further research is needed to enhance their applications.
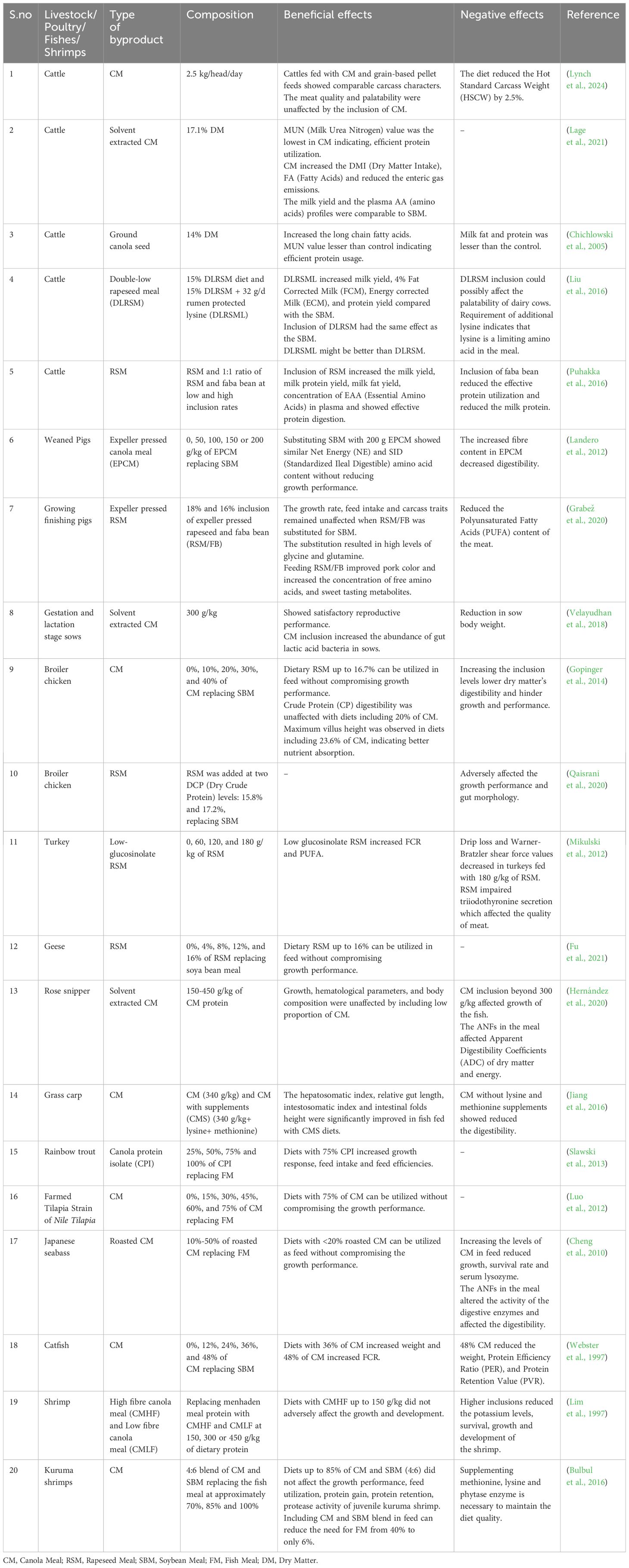
Rapeseed byproducts offer a cheap source of protein supplements for animal diets (Table 4). However, the presence of ANFs produces off-flavors and odors, which reduce the palatability of these byproducts and adversely affect the animal’s growth and development.
4 ANFs in rapeseed byproducts
ANFs are secondary metabolites and play an important role in plant defense against biotic and abiotic stresses. Any external environmental stress triggers an increase in secondary metabolite levels as a defense mechanism to cope with stress. However, they are the major problem in implementing rapeseed byproducts for food and feed consumption (Table 5). The ANFs at higher concentrations bind with the nutrients and reduce their bioavailability (Makkar, 2003) (Figure 1), causing adverse effects on the animal’s growth and development (Jansman, 1993; Mawson et al., 1994; Makkar, 2003). The major ANFs in rapeseed byproducts include glucosinolates, phytic acid, tannins, and sinapines.
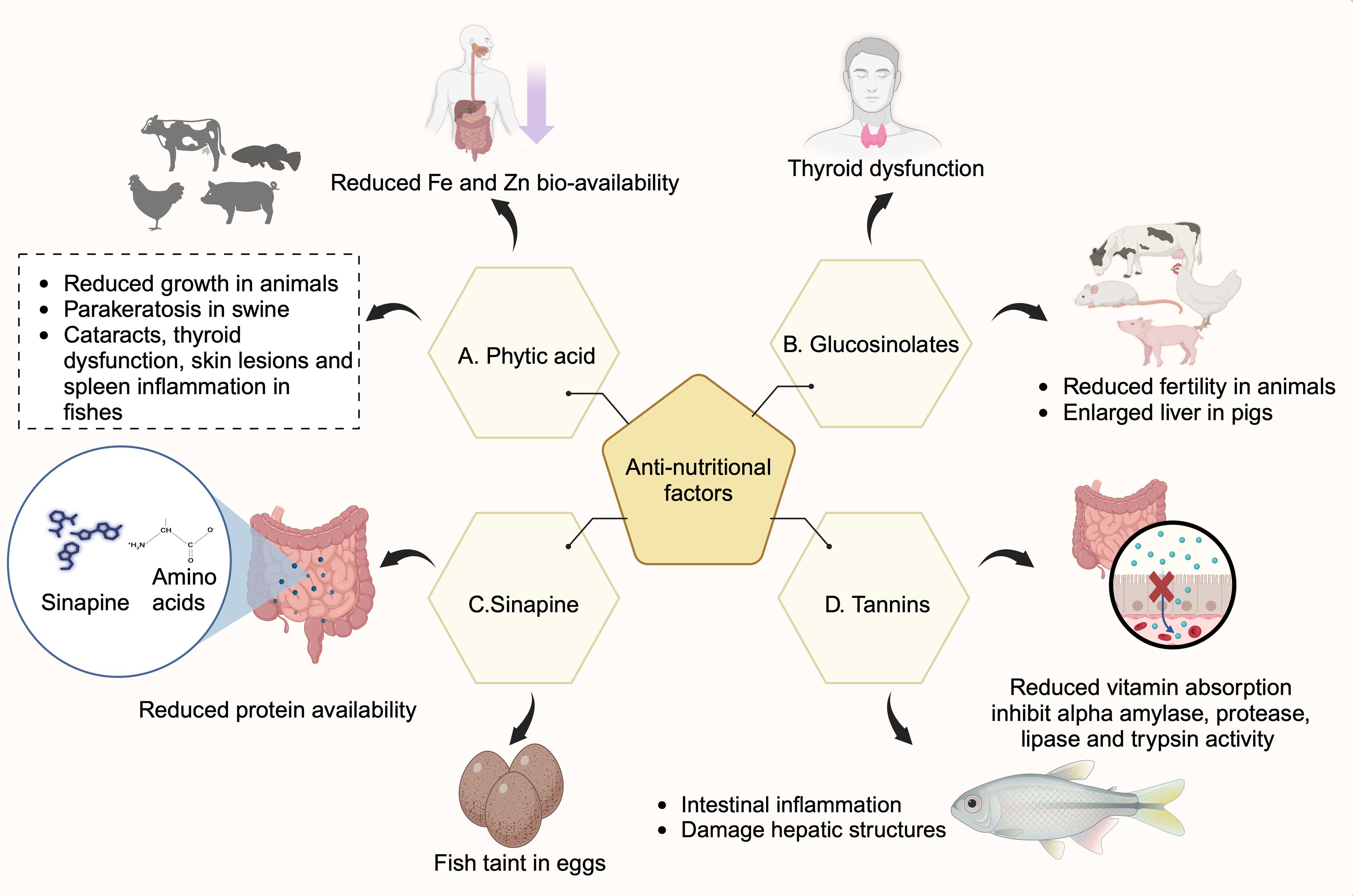
Figure 1. Effects of anti-nutritional factors (ANFs) in humans and animals. Rapeseed meal contains ANFs such as phytic acid, glucosinolates, tannins, and sinapines. (A) Phytic acid binds to metal ions such as iron and zinc and reduces their bioavailability. They cause thyroid dysfunction, cataracts, skin lesions, spleen inflammation in fish, and parakeratosis in swine. (B) Sinapines bind to amino acids and hinder their gut absorption, thereby reducing bioavailability. They also cause fish taints in eggs. (C) Glucosinolates cause thyroid dysfunction in humans, reduce fertility in animals and cause enlarged liver in pigs. (D) Tannins inhibit alpha-amylase, protease, lipase, and trypsin activities and affect protein digestion. Tannins cause intestinal inflammation, damage hepatic structures in fish, and impair vitamin absorption in rats (Created with BioRender.com).
4.1 Glucosinolates
Glucosinolates (GSLs) are sulfur and nitrogen-containing compounds derived from glucose and amino acids as precursors, consisting of thioglucose and a sulfonated oxime attached to the chain-elongated amino acid (Blazevic et al., 2020). GSLs are classified as aliphatic, aromatic, or indolic based on the amino acid moiety type of side chain group (Halkier and Du, 1997). These are commonly found in Brassica family members, including rapeseed, cabbage, and mustard. The enzyme myrosinase hydrolyzes GSLs, yielding isothiocyanates and epi-thionitriles (Wittstock et al., 2003).
GSLs are responsible for the pungency of these plants, which reduces their consumer preferences and palatability as fodder. GSLs can alter thyroid hormone levels in humans and animals (Tripathi and Mishra, 2007; Felker et al., 2016). However, hydrolyzed GSL products are beneficial to humans. For example, sulforaphane and indole-3-carbinol exhibit anti-cancerous properties (Keck and Finley, 2004), while glucoraphasatin and glucoerucin are known to have anti-oxidant and anti-carcinogenic properties (Singh et al., 2020; Cedrowski et al., 2021; Sundaram et al., 2022). Also, glucoberteroin prevents non-alcoholic fatty liver disease (Kim et al., 2021). Conversely, the hydrolyzed products are highly toxic to animals, and RSM consumption reduces their fertility. These effects result from thyroid dysfunction in the mother, reduced iodine transfer to the fetus, transfer of goitrogenic compounds to the fetus, or a combined action of all these factors (Mawson et al., 1994). Furthermore, goitrogenic effects of GSLs, including reduced feed intake and enlarged liver, have been reported in pigs (Velayudhan et al., 2017; Lee and Woyengo, 2018; Lee et al., 2020).
4.2 Phytic acid (phytate)
Phytic acid (myo-inositol hexakisphosphate) was first identified in 1855 (Oatway et al., 2007). In mature seeds, phosphorous is stored mainly in the form of phytic acid (phytate) (Erdman, 1979), which acts as the primary phosphorus reserve in cereals and legumes and plays a vital role in germination and growth (Feizollahi et al., 2021). During germination, the phytase enzyme degrades phytates to release phosphorous, thus supplementing the growth and development (Pramitha et al., 2021). Phytate levels increase at the ripening stage and are usually located in the aleurone layer and pericarp of cereal grains, as well as in the endosperm of maize (O’Dell et al., 1972; Bohn et al., 2007). In oilseeds, phytate is distributed throughout the seed and inside the protein body membrane (Erdman, 1979).
Humans and monogastric animals lack the phytase enzyme, which is necessary to digest phytates. About 70% of phosphorous ingested by monogastric animals is excreted (Jorquera et al., 2008). Insoluble phytate–metal ion complexes are formed at neutral and basic pH, with the concentration of metals exceeding that of phytate (Maenz, 2001). Moreover, phytic acid is a potent chelating agent that binds to monovalent and divalent cations, reducing their bioavailability (Erdman, 1979; Schlemmer et al., 2009; Moran and Bedford, 2024). This chelation substantially limits the bioavailability of essential minerals such as iron and zinc in humans, leading to nutrient deficiencies (Kim et al., 1993; Reddy et al., 1996; Kim et al., 2007; Hunt and Beiseigel, 2009). For example, a diet high in phytic acid might increase the risk of anemia and decrease zinc absorption, especially in women. The elimination of phytic acid increased zinc bioaccessibility by 18.19% in low-protein soymilk and increased the bioaccessibility of calcium and iron by 31.20% and 30.03%, respectively, in high-protein soymilk (Lv et al., 2024).
Phytates inhibit the action of α-amylase and affect starch digestion (Knuckles and Betschart, 2006). Phytins in the aleurone layer of grains, when exposed to low gastric pH dissociate as phytic acid and free Ca2+ ions. The free Ca2+ ions bind with pepsin and compromise proteolysis (Moran and Bedford, 2024). By reducing the activity of digestive enzymes, phytate makes proteins resistant to proteolytic digestion, affecting amino acid absorption (Knuckles et al., 1985; Liu et al., 2009). In animal nutrition, dietary phytic acid reduces growth and development in broilers and pigs (Woyengo and Nyachoti, 2013). Broilers, when fed with casein and 1g of phytic acid, increased the excretion of Ca (187%), Mg (39%), Mn (87%), and Na (174%) (Cowieson et al., 2006). Also, phytic acid reduced the ileal amino acid digestibility in broiler chickens (Ravindran et al., 2006). In pigs, the addition of phytic acid to their diet has been associated with symptoms of parakeratosis and growth depression (Oberleas et al., 1962). Similarly, supplementing 2% phytic acid in feed reduced the body weight gain and feed intake and the growth of pigs up to 37% (Woyengo et al., 2012; Woyengo and Nyachoti, 2013). In Chinook salmon, diets that included phytic acid have been observed to affect thyroid, kidney and alimentary tract morphology. Also, cataracts and hypertrophy of pyloric caeca were increased (Kaiser et al., 2022). Similarly, grass carp fed with phytate lowered antimicrobial activity, caused head, kidney, and spleen inflammation, and increased skin lesions (Zhong et al., 2020). Thus, the presence of phytic acid in feed negatively impacts the growth and development of animals. However, adding phytase to the diet can hydrolyze the phytic acid and improve the bioavailability of nutrients.
4.3 Tannins
Rapeseed hulls mainly contain condensable tannins (Shahidi and Naczk, 1992). They can be categorized as hydrolyzable and condensed tannins. Tannins are known for their anti-viral, anti-bacterial, anti-oxidant, anti-parasitic, and anti-diarrheic properties (Tong et al., 2021). Their natural occurrence helps to control bloating in ruminants (Mangan, 1988). However, tannins are known to impart a bitter taste in feed and reduce its palatability. Also, tannins inhibit the activity of digestive enzymes such as trypsin, protease, lipase, and α-amylase (Horigome et al., 1988; Maitra and Ray, 2003; Xiao et al., 2015), resulting in reduced protein digestibility. The inclusion of 1.25% tannins in aquafeeds resulted in impaired digestion and protein metabolism (Yao et al., 2019). Similarly, including 0.6% tannic acid in the feed diets of Xiangdong black goats, significantly reduced the crude protein digestibility (Wang et al., 2022). Also, including 0.75% tannins in feed induced oxidative stress and injured hepatic antioxidant ability, resulting in intestinal inflammation in grass carp (Yao et al., 2022). Also, diets that include tannins reduce feed utilization and immunity and damage the hepatic structures of carp (Liu et al., 2021b). Additionally, a diet supplemented with tannins significantly decreases milk urea nitrogen (MUN) content in lactating dairy cows (Zhang et al., 2019). Moreover, the presence of tannins in animal diets is associated with reduced feed conversion efficiency and retard growth (Jansman, 1993). Also, tannins have a negative impact on vitamin and mineral metabolism. For example, feeding rats with 3.2% tannic acid reduced vitamin A and cobalamin absorption (Jansman, 1993). These findings highlight the challenges of incorporating tannins into animal feed without compromising nutritional quality.
4.4 Sinapines
Sinapine, a choline ester of sinapic acid, is a major phenolic component in RSMs, which ranges from 1%-2.5% under different Canadian conditions (Mueller et al., 1978). About 80% of total phenols are constituted by sinapines (Krygier et al., 1982). Sinapine results in a bitter taste, off-flavors, dark color, and astringency to the RSM, which reduces its palatability (Josefsson and Uppstrom, 1976). Sinapines are not subjected to hydrolytic cleavages in plants and are catabolized only during germination (Sosulski, 1979). However, alkali hydrolysis extraction yields free phenolic acid and sinapic acid, which is known for its bitter taste (Durkee and Thivierge, 1975), and sinapine is one of the major phenolic components in rapeseed hulls (Durkee and Thivierge, 1975).
Previous studies have confirmed the presence of fishy taint in brown eggs when the chickens are fed RSM (Goh et al., 1979). In the gut, sinapine is hydrolyzed by microbes, releasing sinapic acid and trimethylamine. Sinapic acid reduces protein digestibility in RSMs (Qiao and Classen, 2003; Qiao et al., 2008). Trimethylamine is oxidized by trimethylamine oxidase in the liver and kidney and then excreted (Butler et al., 1982). However, birds with low concentrations of trimethylamine oxidase tend to develop a fishy taint in eggs. The reduction of enzyme concentration is related to ANFs such as tannins, glucosinolates, and progoitrin. The breakdown products of tannins and progoitrins, such as goitrogen and soluble tannins, reduce the activity of trimethylamine oxidase, resulting in a foul smell in the eggs. Moreover, liver damage caused by breakdown products of tannins and glucosinolates hinders the possibility of using RSM for poultry.
Thus, ANFs pose severe physiological health impairments in both humans and animals, requiring strict intake limitations. Reducing ANFs is essential to improving RSM quality and establishing its implications in the food industry. This review focuses on different strategies to achieve these goals, such as microbial fermentation, breeding, and biotechnological interventions.
5 Strategies to reduce ANFs in rapeseed byproducts
The quality of rapeseed byproducts can be enhanced by the following two strategies (Figure 2):
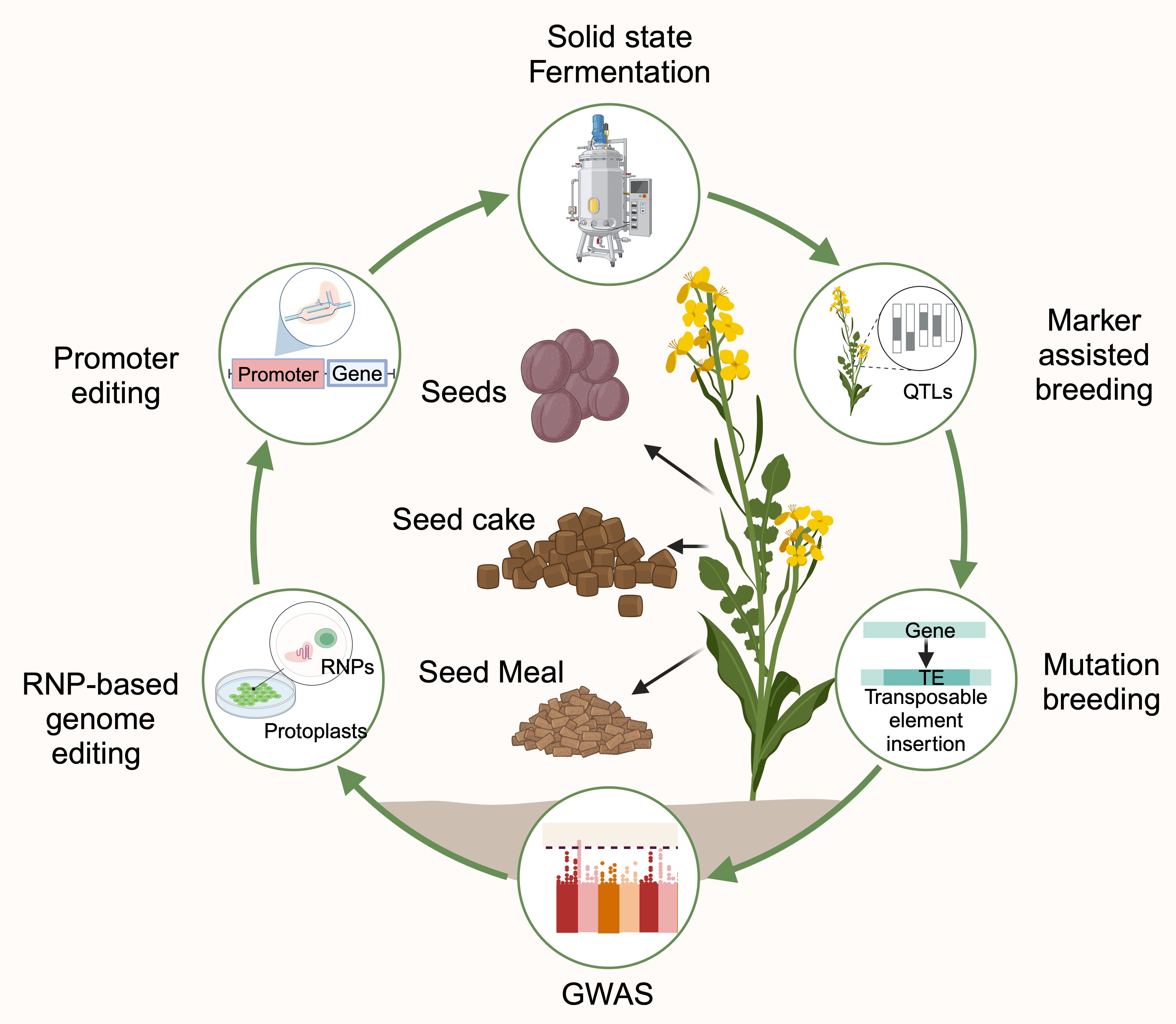
Figure 2. Schematic representation of rapeseed byproducts inclusion in human food and animal feed (Created with BioRender.com).
1. The first strategy focuses on reducing the ANFs in rapeseed byproducts through microbial fermentation to enhance the bioavailability of nutrients and palatability.
2. The second strategy focuses on improving protein content by manipulating protein profiles and reducing ANFs through advanced breeding and biotechnological approaches.
However, improving the protein profile of rapeseed is challenging due to its heritability, association with several nutritional characteristics, and sensitivity to environmental conditions (Wu et al., 2005; Poisson et al., 2019). It is also important to achieve balanced ANF profiles without compromising plant defense mechanisms (Barbehenn and Peter Constabel, 2011; Burow and Halkier, 2017). Reducing the ANFs should be a primary breeding objective to include RSM in the existing food system. Improving the protein profile without reducing ANFs will hinder its implementation in the food chain by affecting its sensory characteristics and acceptance.
5.1 Microbial fermentation to improve RSM quality
Various chemical, thermal, mechanical and biological treatments have been investigated to optimize the reduction of ANFs and enhance the nutritional quality of RSM for use as food and feed material. These treatments aim to balance efficiency and feasibility on a large scale without compromising benefits. Physical treatments include thermal, radiation, hulling and others. Thermal processing effectively reduces ANFs but negatively impacts protein content and digestibility (Jensen et al., 1995; Hansen et al., 2017; Tayyab et al., 2017). Irradiating rapeseed cake (RC) using gamma radiation is reported to reduce the ANFs (Anwar et al., 2015; Xiong et al., 2024). However, biohazards and the cost involved with the process limit its utilization. Hulling reduces the seeds’ fiber content but has no significant effects on all other ANFs (Kracht et al., 2004). Similarly, chemical treatments, including acid or alkali supplemented with alcoholic solvents, reduce the ANFs in the meal. However, chemical treatments adversely affect the protein structure and digestibility.
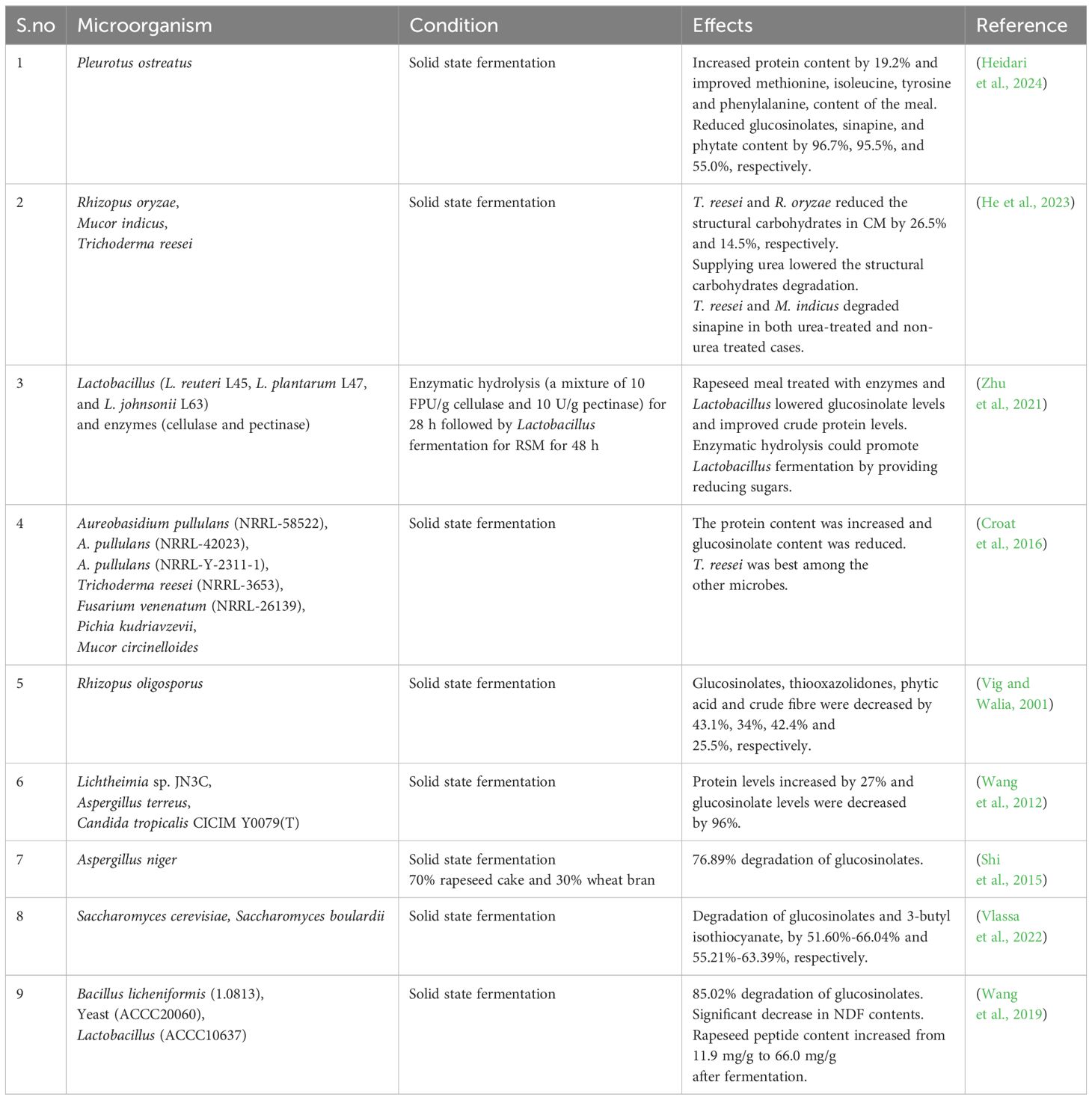
Microbial fermentation involving bacteria, fungi and yeast is a well-established and effective natural method for reducing ANFs while improving the nutritional quality of rapeseed (Vig and Walia, 2001; Wang et al., 2019; Boroojeni et al., 2022; Zaworska-Zakrzewska et al., 2023) (Table 6). Solid state fermentation (SSF), which occurs on the surface of water-insoluble materials with or without soluble nutrients and without free-flowing liquid, offers several advantages over other methods. These include low energy consumption, mild reaction conditions (Reddy et al., 2008) and reduced water content in the final product (Xie et al., 2015). The lower moisture content in SSF compared to submerged liquid fermentation facilitates a more accessible commercialization (Chen, 2013).
SSF is more commonly applied to RSM for ANFs reduction than liquid-phase fermentation. It prevents strong drying after fermentation, which often occurs in high-pressure and heat sterilization, leading to rapeseed protein denaturation and reduced conversion efficiency (He et al., 2014). Fermentation processes enhance protein content, vitamin and mineral levels, and digestibility while improving sensory profiles and nutrient utilization (He et al., 2012; Shi et al., 2016). Furthermore, fermentation increases proteolytic enzyme activity, promoting the hydrolysis of ANFs into low molecular weight peptides and detoxifying the RSM (He et al., 2012). These low molecular weight peptides derived from rapeseed exhibit improved protein functional properties, antioxidant activity, and other beneficial biological activities (Pinterits and Arntfield, 2008).
Several studies have explored the reduction of glucosinolates, phytic acid, and other ANFs through SSF (Wang et al., 2012). Due to their broad metabolic activities, fungal fermentations are particularly promising for reducing ANF components and biofortifying substrates with positive health effects (Wang et al., 2012; Shi et al., 2015). The filamentous fungus Aspergillus, recognized as a safe microorganism (GRAS- Generally Recognized as Safe), produces various degradation enzymes, including protease, phytase and hydrolases, which effectively degrade glucosinolates, phytic acid and other ANF components (El-Batal and Karem, 2001; Shi et al., 2015). Similarly, Rhizopus oligosporus, a GRAS organism commonly used as a “tempeh” starter, degrades glucosinolates and other phenolic compounds (Bau et al., 1994; Lücke et al., 2019). R. oligosporus has been shown to reduce glucosinolate levels by 47%, lignin and other polyphenolics by 30%, phytic acid by 42.4%, and total fiber content by 25.5% while simultaneously increasing the nitrogen and protein content of RSM (Lomascolo et al., 2012). Members of the Aspergillus genus are also widely recognized for their ability to degrade phytic acid in rapeseed during SSF (Al-Asheh and Duvnjak, 1995; Ebune et al., 1995a; Ebune et al., 1995b). In addition, white rot fungi, such as Pleurotus spp., commonly known as oyster mushrooms, are effective at degrading secondary metabolites. They have been reported to reduce sinapic acid (sinapines) by up to 99%.
Bacterial fermentation is extensively applied to degrade ANFs and enhance sensory properties in food products. Among the fermentative bacteria, Lactobacillus species are considered safe (GRAS) organisms and the most popular and traditional fermentation bacteria used as probiotics. These bacteria produce lactic acid, which lowers the pH of the substrate, thereby enhancing flavor and nutritional profile while inhibiting the growth of pathogens. Lactobacillus salivarious fermentation has been shown to reduce glucosinolate and fiber content in rapeseed (Aljuobori et al., 2014). Similarly, L. plantarum degrades the glucosinolates by up to 80% and reduces phytic acid in RSMs by secretion of extracellular enzymes. This process reduces the ANFs and enhances the soluble protein content and bioavailability, thereby improving the overall nutritional quality of the RSM (Chen et al., 2024).
Similarly, yeast fermentation effectively reduces the content of ANFs and improves nutrient quality. Various Saccharomyces species, such as Saccharomyces cariocanus, Saccharomyces cerevisiae, Saccharomyces boulardii and Wickerhamomyces anomalus, have been extensively explored in yeast fermentation to reduce ANFs and enhance the nutritional properties of rapeseed (Vlassa et al., 2022). Fermentation with S. cerevisiae or S. boulardii has been shown to minimize glucosinolate content by 51%-66%. Additionally, yeast fermentation decreases the carbohydrates, phenolic acids, organic acids, sinapic acid, tannins and phytic acid contents while increasing the protein content of RSM (Bau et al., 1994; Vlassa et al., 2022).
Co-inoculation with specific bacterial species such as Lactobacillus and Bacillus (including Bacillus cereus, Bacillus subtilis, and Bacillus licheniformis) has proven to be an efficient method for reducing glucosinolate content and enhancing protein content (Wang et al., 2019). Similarly, the combination of bacteria, fungi, and yeast strains, resulting in a diverse pool of enzymes, has shown superior ANFs degradation capabilities compared to single-strain fermentations (Xu et al., 2012; Yusuf et al., 2021). This mixed-strain approach has significantly improved ANFs degradation efficiency, particularly for glucosinolates, achieving up to 83% reduction (Yusuf et al., 2021).
Despite its benefits, fermentation has several challenges in the food industry. The process is prone to inconsistent product quality due to variations in microbial activity and prolonged fermentation (Zentek and Boroojeni, 2020). This leads to potential contamination and the growth of unwanted pathogens, which introduce health risks and affect product quality (Capozzi et al., 2017). Scaling up fermentation to industrial levels introduces complexities in maintaining consistent microbial activity, impacting the uniformity of the final product. This scaling requires specialized equipment, costly infrastructure, increased operational costs, and stringent environmental controls (Tamang et al., 2020).
Fermented foods must comply with food safety regulations and require continuous monitoring and testing to ensure safety and quality. The sensory changes fermentation induces can sometimes produce undesirable characteristics, affecting consumer acceptance (Marco et al., 2017). Therefore, addressing these challenges is crucial for successful fermentation processes.
Hence, microbial fermentation is a practical, robust, and versatile method for reducing ANFs in rapeseed. By strategically employing specific microorganisms in monoculture or combination with their fermentation techniques, it is possible to enhance rapeseed’s nutritional profile, sensory properties, and usability. This approach effectively addresses the challenges posed by ANFs. It adds value to rapeseed as a sustainable and nutritious resource, making it a valuable ingredient in both food and feed industries.
5.2 Classical breeding
B. napus, commonly known as rapeseed (2n = 38, AACC), is an allotetraploid crop from natural interspecific hybridization between B. rapa (2n = 20, AA) and B. oleracea (2n = 18, CC). Like other crops, the primary breeding objective in rapeseed is to increase oil yield. However, the high erucic acid (~40%) and glucosinolate (80 µmol/g) content hindered the use of rapeseed oil and meal for consumption (Daun, 1986). Intense breeding efforts successfully reduced these contents, and the first low erucic aid and low glucosinolate variety, Tower, was released by Canada and was named a double zero rapeseed variety. Breeding strategies for yield improvement, (a)biotic stress resistance, and nutritional improvement were primarily prioritized. Recently, the focus has shifted towards meal quality by modifying their protein profiles, which have been given importance for their use in the food industry (Wanasundara et al., 2017).
Selecting yellow seed coats can effectively reduce the fiber and tannin contents while improving the protein content of rapeseed. Yellow-seeded genotypes have thinner seed coats, lower hull content, and large embryos with higher oil and protein content in the seeds (Bell and Shires, 1982). Pro-anthocyanidins and tannins determine the seed coat color; darker seed coat colors are associated with high tannin content, which reduces the meal digestibility (Bell and Shires, 1982). Moreover, brown and black-seeded varieties have higher fiber content than yellow-seeded varieties, reducing meal digestibility (Shirzadegan and Röbbelen, 1985). The lack of natural yellow-seeded types in B. napus lead to development of varieties by interspecific crosses involving B. rapa, B. juncea, B. carinata and B. campestris spp. alboglabra, B.oleraceae (Rashid et al., 1994; Meng et al., 1998; Zhang et al., 2004; Rahman, 2008). The progenies from these interspecific crosses were subjected to further breeding techniques like pedigree and backcrossing to develop the yellow-seeded rapeseed lines, which are subsequently developed into varieties.
5.2.1 Inheritance of seed coat color and seed storage proteins of Brassica
Understanding the genetic control behind the seed color is essential for breeding rapeseed genotypes with increased protein and reduced fiber contents. Two independent dominant genes, Br1 and Br3, are responsible for seed coat color. Dominance at the Br1 locus results in brown seeds, while the dominant Br3 locus with a homozygous recessive Br1 locus results in yellow-brown seeds. Both loci in a homozygous recessive condition produce yellow seeds (Stringam, 1980).
In B. napus, three genes are responsible for the seed coat color expression (Rahman et al., 2010). The seed coat colors are black to dark brown, light brown, dark yellow, light yellow and yellow. The genes BSNap1 and BSNap2 control black/brown seed coat color, while the gene DKYSNap3 influences dark/light yellow seed coat color (Rahman et al., 2010). Homozygous recessive condition across all three genes results in yellow seeds. Studies on the seed coat color inheritance in F2, BC1 (Back Cross 1) and the DH (Double Haploid) population revealed the epistatic interactions between the dominant gene for yellow seed color and two independently segregating dominant genes for black color seeds (Liu et al., 2008).
Quantitative trait loci (QTLs) mapping identified two QTLs responsible for seed coat color, which accounted for 46% and 30.9% variation (Liu et al., 2006). Further discoveries of QTLs for seed coat color, protein, and fiber contents concluded that many genes control these traits and are highly influenced by the environment. For example, temperatures can affect the yellow and black-seeded rapeseed varieties. High temperatures might disrupt pigment synthesis, resulting in yellow-seeded genotypes (Deynze et al., 1993).
The discovery of QTLs significantly affects marker-assisted selections (MAS) and genomic selections. Identification of molecular markers associated with QTL of desirable traits facilitates MAS. This enables early selection, pyramiding favorable QTLs, and reduces the duration of the breeding cycle to get favorable phenotypes with improved performance. However, the QTL analysis for seed quality traits in rapeseed requires trials in multiple environments to minimize the environmental influence and determine stable QTLs across environments. Only upon validation of such major and consensus QTLs, they can be recruited in MAS programs.
As mentioned above, SSPs have immense potential in the food processing industry. However, breeding for favorable genotypes requires understanding the trait’s genetic basis. SSP content is a quantitative trait that involves complex gene regulation and biosynthesis. The SSPs, cruciferin and napin are coded by multiple genes, particularly 9-12 genes involved in the cruciferin biosynthesis (Wanasundara, 2011), and 10 genes are responsible for the napin biosynthesis (Josefsson et al., 1987). SSP content within the seed also depends on post-translational processes and the transfer of assembled SSP into storage vacuoles (So and Duncan, 2021). Several QTLs are associated with SSP content. QTLs Nap-1, Nap-2, Nap-3, Cru-1, Cru-2, Nap/Cru-1, Nap/Cru-2, and Nap/Cru-3 were identified in the double haploid populations of winter oilseed rape (Schatzki et al., 2014). Moreover, transcription factors such as Abscisic acid insensitive 3 (ABI3), Fusca 3 (FUS3), Leafy Cotyledon 1 (LEC1) and Leafy Cotyledon 2 (LEC2) are involved in the SSP regulation (Vicient et al., 2000; Kroj et al., 2003; So and Duncan, 2021). Hence, understanding the genetic control and regulatory mechanisms behind SSPs is crucial for modern breeding strategies to enhance SSP content for improved food processing applications.
5.2.2 Mutation breeding
Classical mutation breeding techniques have often focused on increasing rapeseed oil yield and quality (Röbbelen, 1990). Nonetheless, mutation breeding also has immense potential for reducing ANFs (Harloff et al., 2012) and increasing the protein fractions in RSM. Double haploid mutant lines produced from seeds or microspores are a promising option for producing successful mutations. The National Research Council of Canada (NRC) utilized this approach and recently reported a mutant line with a 7% increase in protein content (National Research Council Canada, 2023; available at: https://nrc.canada.ca/en/stories/boosting-canola-crop-value-mutation-breeding).
Similarly, transposon element (TE) insertions provide a biological means of inducing plant mutations. Transposons are mobile elements in plants that were once considered junk DNA and are now a valuable tool for breeders to create favorable plant variations. TE insertions in the coding regions will lead to loss-of-function mutations. In contrast, insertion in intron regions can result in new isoforms through exonization, alternative splicing, truncation, or a combination of these mechanisms (Dubin et al., 2018). Moreover, TE insertions can potentially disrupt enhancers or regulatory promoter elements outside the transcribing regions, thereby altering the transcriptional level (Dubin et al., 2018). TE activation can be achieved through toxins that inhibit DNA methylation, and stress-mediated TE activation has also been reported in different crops (Ito, 2022), for example, in vitro regeneration stress results in TE element activation in rice (Hirochika et al., 1996) and tobacco (Hirochika, 1993). However, TE-mediated mutagenesis requires robust methods to induce TE activation and transgenerational inheritance of TE insertions for further breeding implications (Kirov, 2023).
TE insertions in rapeseed have improved silique length and seed weight and reduced pod shattering. CACTA-like TE (3.9 kb) inserted in the upstream of BnaA9.CYP78A9 increased the expression of the loci, resulting in elongated siliques (Shi et al., 2019). Similarly, long-term repeat retrotransposon insertion (4803 bp) in the promotor of BnSHP1.A9 repressed its expression and conferred resistance to pod shattering (Liu et al., 2019). However, such transposons-mediated mutagenic studies have yet to be conducted to improve seed meal quality traits.
Mutation breeding could be essential as it avoids legislative pressure and regulations. Despite its potential, mutation breeding faces challenges due to a low frequency of mutation occurrence, unspecific mutations causing undesirable mutations, and safety hazards associated with the use of dangerous physical and chemical mutagenic agents.
5.3 Molecular breeding strategies
Modern breeding techniques offer numerous advantages, such as improved efficiency in breeding, precision to improve the target traits and reduced breeding cycles. The novel tools of biotechnology and genomics have transformed the approaches for breeding complex characteristics that are quantitative, pleiotropic and sensitive to environmental factors, which were challenging for the traditional breeding approaches to address. The introduction of molecular markers made the first significant leap in breeding strategies. PCR-based DNA markers have become integral in almost all areas, like pre-breeding to broaden the genetic base, aid in effective selections, introgression of desired characters, determining the heterotic potentials, and improving the quantitative traits.
Biparental mapping populations, which include F2 populations, recombinant inbred lines (RILs), double haploid lines (DH), backcross populations (BC), and near-isogenic lines (NILs), are developed from crossing two parents. The greater the contrast traits expressed by the parents, the more variations can be retrieved in the mapping populations. These populations are generally used for QTL detections, and the size of the population also determines the reliability of the identified QTLs. Several mapping populations have been created to detect QTLs for agronomic and seed traits in rapeseed (Chen et al., 2007, Chen et al., 2010; Fattahi et al., 2018; Yusuf and Möllers, 2024).
However, the time and cost of generating and maintaining biparental populations far exceed those required for association mapping (AM). Biparental mapping populations consider only the allelic variations between two parents. This narrows the allelic variations and in turn, limits the QTL detections for trait improvement (Singh et al., 2015). This limitation highlights the need for alternative methods like AM, which can incorporate a broader genetic base and provide more comprehensive insights into trait variation and improvement.
5.3.1 Genome-wide association studies
Genomics in breeding enables the exploration of various advanced tools to obtain accurate population genotypic data. Genome sequencing technologies provide high throughput genome sequencing data with high accuracy and reliability. The recent advances in the discovery of single-nucleotide polymorphism (SNP) markers through genome-wide association studies (GWAS) improved the efficiency of correlating the phenotypic and genotypic data and selection efficiency for precision breeding. Brassica napus 60K Illumina Infinium™ SNP array allows multiplex SNP genotyping, enabling simultaneous surveys of thousands of SNPs and increasing the efficiency of the genotypic data (Wang et al., 2014; Mason et al., 2017; Zhou et al., 2021).
AM approach facilitates the identification of markers closely linked to the genes of interest. Unlike linkage mapping, which relies on recombination events between the gene and the marker in a bi-parental cross, AM utilizes all the recombination events that would have happened in the past in the population (Singh and Singh, 2015). The post-GWAS era offers new approaches for processing GWAS data to extract meaningful information by identifying and functionally characterizing candidate genes using bioinformatics and reverse genetics approaches. Targeting-induced local lesions in genomes (TILLING) approaches functionally characterize candidate genes identified via GWAS in mutagenized populations. The Eco-TILLING approach is used in germplasms or natural populations (Gupta et al., 2019). GWAS studies have been successfully utilized for several traits in rapeseed, including disease resistance, flowering-related traits, salt tolerance, aluminum tolerance and silique length (Helal et al., 2021; Roy et al., 2021; Wang et al., 2021; Zhang et al., 2022; Zhou et al., 2022; Starosta et al., 2024; Zhang et al., 2024). Similarly, GWAS has been successfully applied to rapeseed for various seed quality traits like fiber traits, ANFs, oil, and amino acid content (Wang et al., 2015; Wei et al., 2019; Bhinder et al., 2022; Tan et al., 2022; Zhao et al., 2022). However, studies focusing on seed storage proteins seem to be lacking (So and Duncan, 2021). Even though GWAS has identified many QTLs for essential traits in rapeseed, this technique faces challenges such as poor reproducibility and false positives, indicating the need for further improvement in this crop.
5.3.2 Marker-assisted selection
Traditional breeding focuses on phenotypic selection for genetic improvement. However, recent developments in genomics led to the identification of several QTLs useful for marker-assisted selection (MAS). MAS facilitates an indirect selection based on markers linked to the target traits. These markers are highly reliable, unaffected by the environment, and non-specific to plant growth stages, promoting early selection and reducing the breeding cycle. Combining MAS with speed breeding techniques can reduce the breeding cycles, thus promoting the development of new varieties in short periods. However, focusing solely on genetic composition might mislead the selection of agronomically poor phenotypes. The phenotypic selection combined with MAS is always preferred to achieve all the agronomic and quality traits.
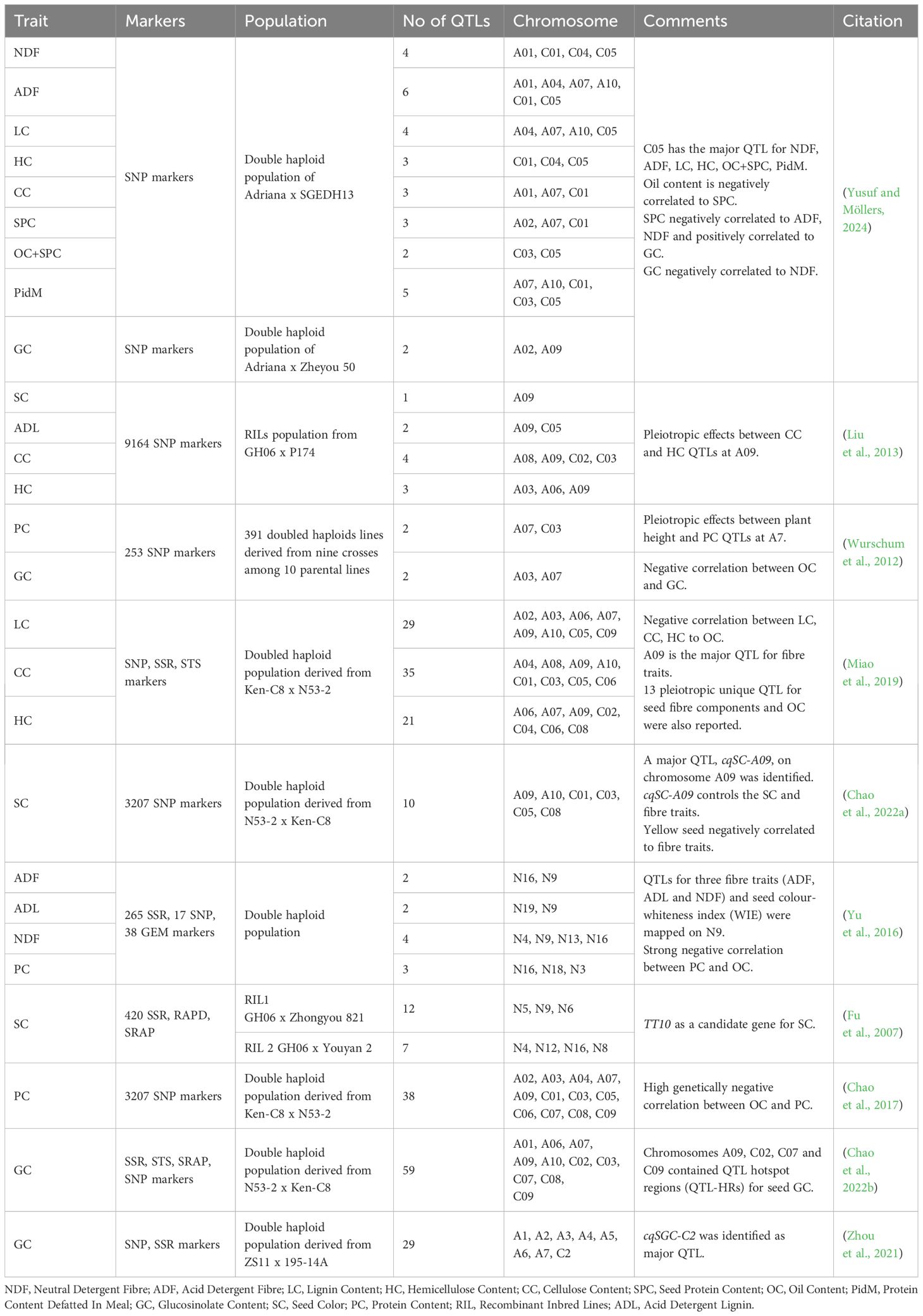
The trait of yellow seeds was confirmed to be quantitative after intense research by various scientists (Liu et al., 2006; Fu et al., 2007; Chao et al., 2022a). The QTL discovery for fiber content and its correlation with the yellow seeds paved the way for genetic manipulation (Badani et al., 2006). The fiber content of the seeds compromises acid detergent fiber (ADF), neutral detergent fiber (NDF), and acid detergent lignin (ADL). Hemicellulose, cellulose, and lignin predominate in NDF, while the ADF comprises cellulose and lignin. ADL represents the nondigestible lignins in the seeds (Behnke et al., 2018). Altering these fractions could be beneficial for animal husbandry.
Different DNA-based molecular marker systems have been used for MAS to improve these quality traits. The yellow-seeded trait associated with 8 RAPD markers co-segregated with a single major gene (Pigment 1) responsible for the yellow seed coat color trait (Somers et al., 2001). Other markers, including restricted fragment length polymorphism (RFLP), amplified fragment polymorphism (AFLP), cleaved amplified polymorphic sequences (CAPS), sequence related amplified polymorphism (SRAP), sequence characterized amplified region (SCAR), and simple sequence repeat (SSR) markers (Deynze et al., 1995; Liu et al., 2006; Rahman et al., 2010; Zhang et al., 2010b; Huang et al., 2016) were also identified for breeding yellow seeds and low fiber content. Pleiotropy arises when a single gene or genetic locus affects multiple phenotypic traits. Pleiotropic effects between the target traits are advantageous as they facilitate breeding. Selecting one QTL might give the desired phenotype for the other targeted trait.
The QTL analysis by various scientists (Table 7) clearly states that QTL at A09 is a major QTL for seed color and fiber traits (Liu et al., 2013; Miao et al., 2019; Chao et al., 2022a). The QTL at C05 is a major QTL for fiber and protein traits (Yusuf and Möllers, 2024). Focusing on these loci could provide desirable phenotypes. An apparent pleiotropic effect exists between seed fiber traits, plant height, and protein content (Wurschum et al., 2012; Liu et al., 2013; Miao et al., 2019). Additionally, a negative correlation exists between oil content and fiber characteristics, indicating that reducing the fiber content could improve the oil content. However, enhancing protein profiles will decrease the oil content due to their negative correlation (Yu et al., 2016; Chao et al., 2017, Yusuf and Möllers, 2024).
5.4 Genome editing as an advanced biotechnological intervention
Genome editing has revolutionized breeding strategies by focusing on molecular manipulation to achieve desirable plant ideotypes. This technology offers promising solutions for important breeding objectives such as climate-resilient crop breeding, nutritional improvement, and biotic and abiotic stress tolerance within a short time compared to conventional breeding techniques (Dhugga, 2022). The precision and specificity of the editing tool are essential for successful genetic manipulation. The discovery of CRISPR (clustered regularly interspaced short palindromic repeats) Cas9 (CRISPR-associated protein 9) has revolutionized the field of agriculture with its high specificity and precision. CRISPR/Cas9 technology originated from type II CRISPR/Cas systems, an adaptive immunity mechanism in bacteria, providing defense against viruses and plasmids (Doudna and Charpentier, 2014). CRISPR/Cas9 uses a guide RNA sequence targeting specific DNA sequences and endonuclease Cas9 cleaving the targeted site’s DNA. The protospacer adjacent motif (PAM) sequence adjacent to the single guide RNA (sgRNA) is critical for DNA binding by Cas9. Without the PAM sequence, sgRNA, even if fully complementary to the target sequences, is not recognized by the Cas9 protein (Doudna and Charpentier, 2014).
5.4.1 Ribonucleoprotein-based genome editing
The delivery of the CRISPR/Cas9 system into the host requires a vector. Agrobacterium-mediated delivery of sgRNA and Cas9 protein is widely deployed in genome editing. Agrobacterium tumefaciens, a gram-negative soil bacterium, is responsible for causing gall growth in plants. It possesses a Ti plasmid, which makes it a perfect biological tool for vector constructs. The T-DNA region of a Ti plasmid is approximately 15–30 kb in size (Rahman et al., 2023). The DNA insert size is also limited in this type of gene transfer, thus limiting the possibility of the multiplexing techniques. The transfection process is mediated by inserting the gene of interest by replacing the T-DNA region and co-cultivating the bacteria with the explant. Successful Agrobacterium-mediated genome editing was done in rapeseed, targeting seed coat color and tannins (Cheng et al., 2023; Li et al., 2024).
However, transformation efficiency highly depends on the transformation protocols, the nature of the explant, co-cultivation periods, and the markers used in the transformation (Rahman et al., 2023). The random integration of T-DNA from bacterial plasmids into the host raises regulatory concerns, affecting the marketability of the products due to reduced consumer acceptance (Figure 3). T-DNA components such as Cas9, gRNA, and selection markers can be effectively removed by selfing or backcrossing with wild-type to obtain edited progeny that is free of foreign DNA.
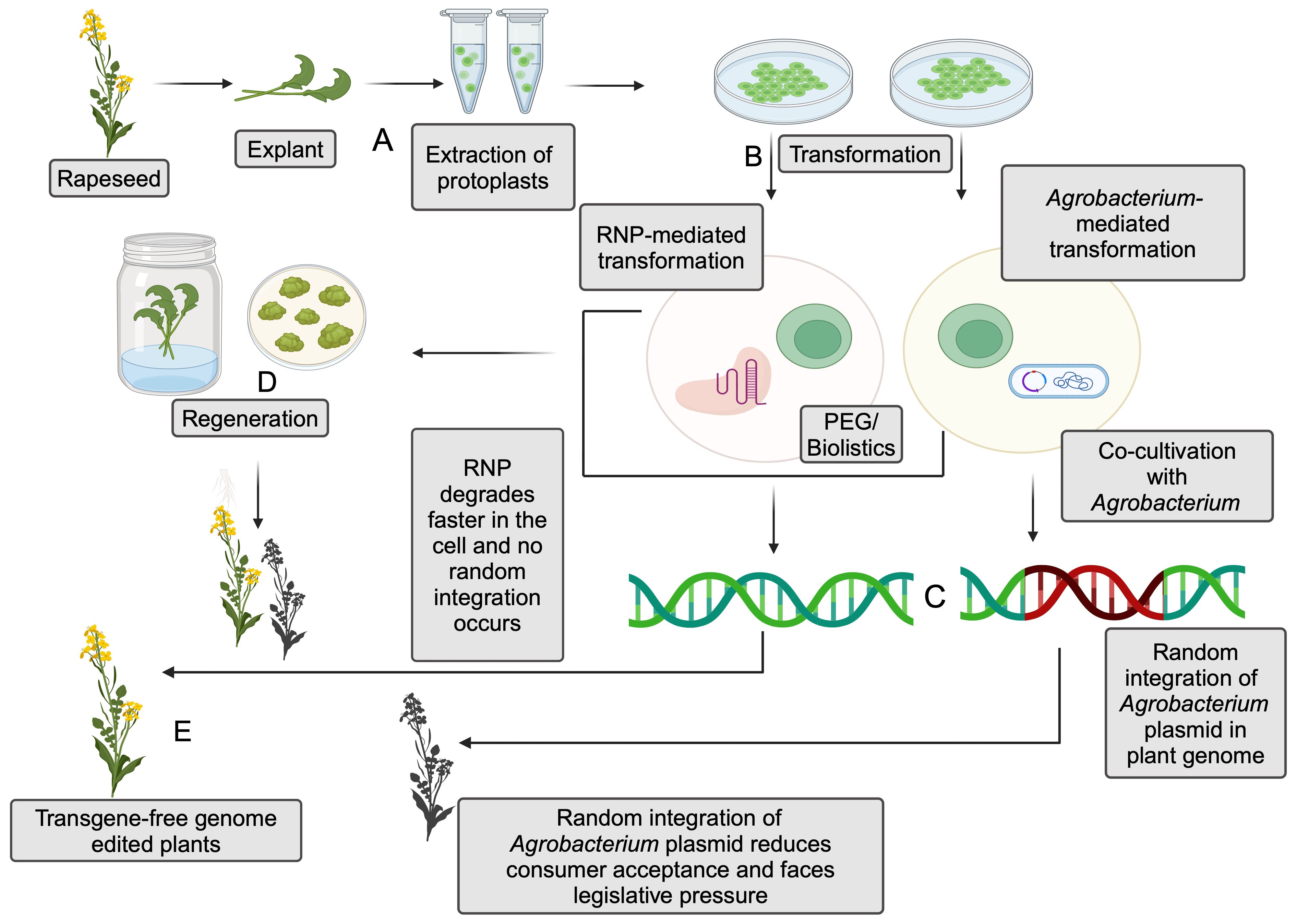
Figure 3. Comparison between ribonucleoprotein (RNP)-mediated and Agrobacterium-mediated genome editing in the protoplasts. (A) Extraction of protoplasts from explant. (B) Transformation of protoplasts with RNPs or Agrobacterium- mediated transformation. Polyethylene Glycol (PEG) or biolistics are utilized to achieve transformation via RNP, and co-cultivation is generally utilized for Agrobacterium-mediated transformation. (C) Random integration of plasmid DNA in Agrobacterium-mediated transformation and absence of such integration in RNP-mediated transformation. (D) Regeneration of the plants after transformation. (E) Transgene-free genome-editing plants are obtained from the regenerated plants (Created with BioRender.com).
Another promising alternative for eliminating the foreign DNA integration with the host genome is RNPs, which ensures transgene-free genome editing (Figure 3). This technique eliminates the need for a vector to deliver the sgRNA sequence and the protein while still being able to manipulate the DNA. Unlike Agrobacterium-mediated CRISPR/Cas9 editing, RNP-mediated editing contains Cas9 enzyme coupled with guide-RNA. The first step in RNP-mediated editing is isolating the protoplasts, followed by the transfection process, usually using polyethylene glycol (PEG) electrophoresis or biolistics for RNP delivery. The protoplasts lack cell walls, thereby facilitating the entry of RNPs into the host cells. The use of RNP complexes can reduce off-target effects and the risk of random DNA integration compared to plasmid-based delivery methods. This is due to the transient nature of RNPs in cells, which limits the time for unintended editing (Woo et al., 2015). However, carefully selecting guide RNAs can further minimize off-target effects in both Agrobacterium-mediated and RNP-based gene editing (Young et al., 2019).
Successful RNP-mediated genome editing using the biolistic approach has been shown in various crops (Svitashev et al., 2016; Liang et al., 2017; Banakar et al., 2020). Plasmid and RNP-based gene editing using protoplasts has been successfully demonstrated in rapeseed (Li et al., 2021; Sidorov et al., 2022). However, genomic instability (change in ploidy level due to endoreduplication and aneuploidy) associated with protoplast regeneration has been reported in various crops (Newell et al., 1984; Olin-Fatih, 1996; Sheng et al., 2011; Fossi et al., 2019).
5.4.2 Promoter editing
As mentioned before, ANFs play a vital role in plant defense. The complete elimination of ANFs by knocking out the target genes through genome editing may be detrimental to the survival of the plants in some cases. So, reducing their levels instead of eliminating them could keep the defense system intact. Promoter editing (Figure 4) could be used to target ANF profiles by altering the gene expression. The promoter regulates the gene expression and is located upstream of the region to be transcribed (Porto et al., 2014). It is a region where RNA polymerase binds to initiate transcription.
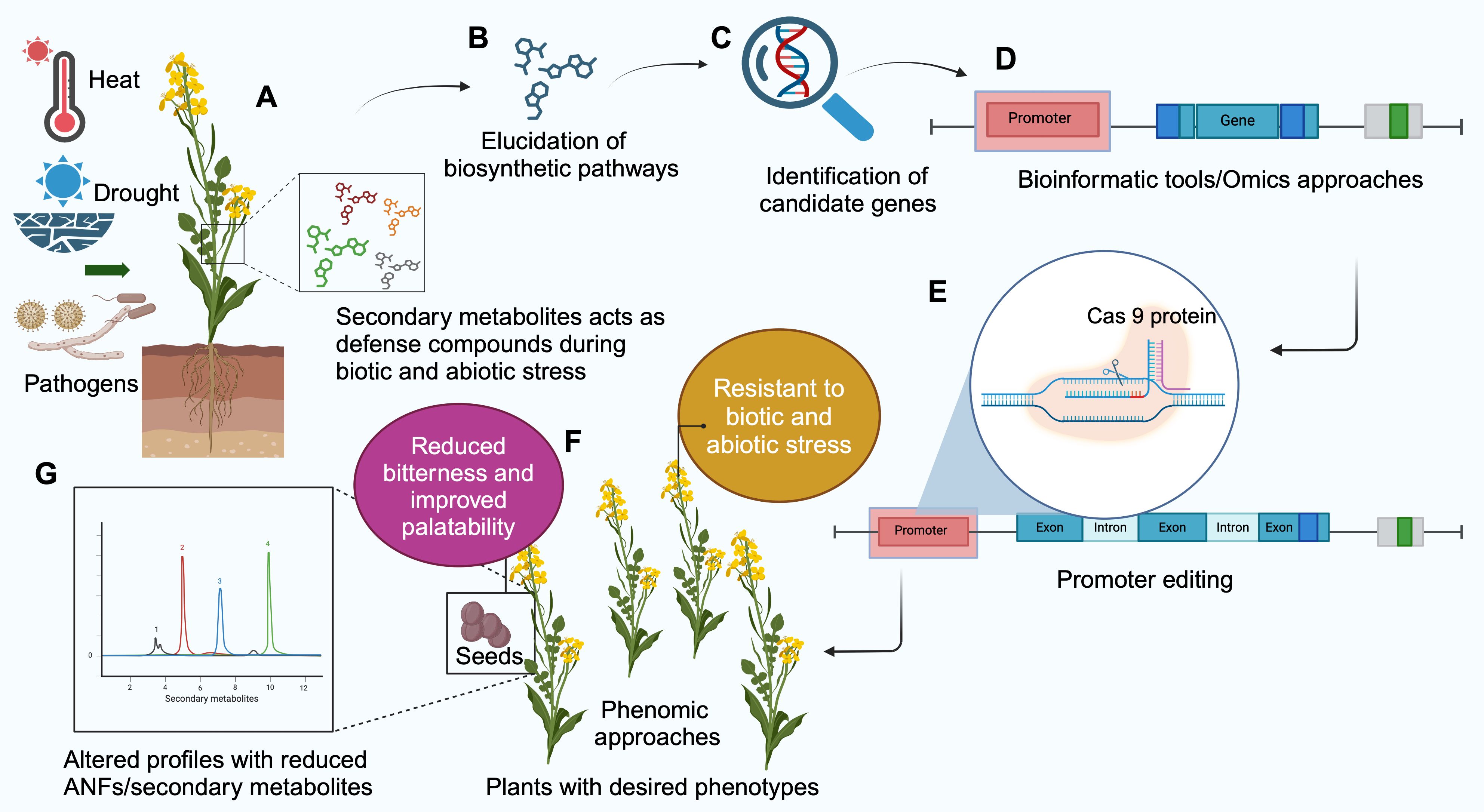
Figure 4. Schematic representation of promoter editing to alter the secondary metabolite/antinutritional factor (ANF) profile in plants. (A) Secondary metabolites protect plants during biotic and abiotic stresses. Knocking out the biosynthesis breaks the plant defense against such stress. Promoter editing begins with (B) Study of biosynthesis pathway of the metabolite, (C) Discovering the genes involved in biosynthesis. (D) Identifying the gene sequences of the promoter using bioinformatics tools. (E) Promoter region is edited using CRISPR/Cas9 technique. (F) The edited plants are subjected to phenomics approaches to identify desired phenotypes with reduced secondary metabolites/ANF without compromising the defense abilities of the plants. (G) Metabolomics approaches such as High Performance Liquid Chromatography (HPLC) and Liquid Chromatography-Mass Spectrometry (LCMS) can be used for assessing the profile of metabolites in plants (Created with BioRender.com).
Promoter editing begins with assessing the secondary metabolites/ANF compounds that are present in the crop. This is followed by studying the metabolic pathways, enzymes, and compounds involved. Bioinformatics and omics approaches are essential for identifying the genes involved in these pathways and for pre-determining the candidate genes and their promoter regions that must be targeted to alter the ANF profiles. Once the editing is successful, plant phenotyping is essential for the morphological and agronomical assessment of the crop. Screening the phenotypes for disease resistance is also vital to determine that the defense system is not compromised.
Successful promotor editing has been done in various crops (Cui et al., 2020; Huang et al., 2020; Zeng et al., 2020; Liu et al., 2021a; Song et al., 2022b). However, Agrobacterium-mediated editing has been followed in promoter editing, which raises biosafety concerns. In this regard, RNP-based promoter editing would be an alternative approach to producing transgene-free mutants to overcome legislative issues.
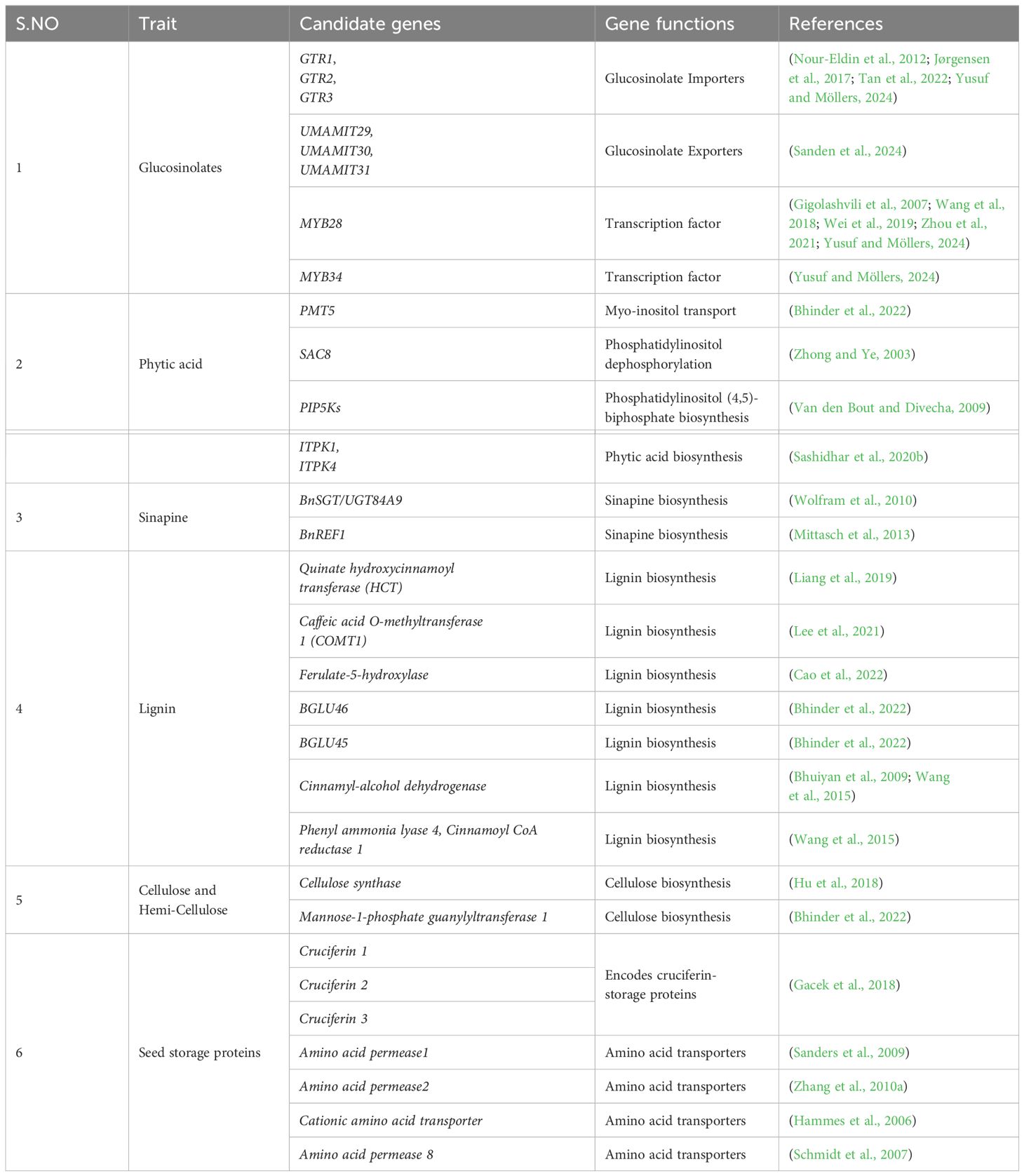
The candidate genes that could be considered for genome editing to alter the ANF profiles are discussed below (Table 8).
5.4.3 Glucosinolates
MYB28 is a transcription factor responsible for activating genes involved in aliphatic GSL biosynthesis. Downregulating MYB28 might reduce the aliphatic GSL biosynthesis. RNAi-mediated knockdown of the MYB28 decreased aliphatic GSL profiles, but the indolic GSLs remained unchanged (Gigolashvili et al., 2007). MYB28, MYB76, MYB29 activate genes in the aliphatic GSL biosynthetic pathways, such as MAM3, CYP79F1, and CYP83A1, without affecting genes involved in the indolic GSL pathway. Conversely, MYB34 and MYB51 activate genes for the indolic GSL biosynthesis (Gigolashvili et al., 2007). The downregulation of these transcription factors might affect genes such as CYP79B2 and CYP83B1 and prevent their conversion from tryptophan to GSLs. However, this might negatively affect the plant’s auxin homeostasis due to the interconnected nature of these pathways.
Moreover, the role of GSLs in defense must be addressed. Knocking down all transcription factors might harm the plants due to their interconnection with phytohormones, defense, growth and development. Thus, manipulating GSL profiles in specific tissues could be a better approach to maintaining the plant defense intact.
Biosynthesis and accumulation of GSLs show a distinct spatiotemporal pattern (Jorgensen et al., 2015) facilitated by glucosinolate transporters (GTR) transporting GSLs to various tissues. GTR proteins belong to the family of nitrate transporter 1/peptide transporter (NRT1/PTR). GTR1, GTR2, and GTR3 were recently identified, and elaborate information on GSLs mobilization and accumulation in sulfur-rich cells (S-cells) for storage in A. thaliana was described by (Schuster et al., 2006; Nour-Eldin et al., 2012). The manipulation of the GSL profiles via their transporters could be a better option. The gtr1gtr2 double knockout mutant failed to transport GSLs to seeds, resulting in a tenfold accumulation of GSLs in source tissues such as leaves and silique walls. The absence of aliphatic and indole GSLs in the gtr1gtr2 double knockout mutant seeds indicates their role in phloem loading and long-distance GSL transport. Thus, disrupting these transporters effectively reduces GSLs in the seeds without compromising plant defense.
This was successfully demonstrated in Brassica juncea (Mann et al., 2023). Knocking out GTR1 and GTR2 reduced the seed GSL content in B. juncea from 146.09 μmoles/g DW to 6.21 μmoles/g DW without compromising the defense (Mann et al., 2023). Similarly, the knockout in Camelina sativa reduced the seed GSL content up to 85%-88% (Hölzl et al., 2023). A clear picture of the GSL transporter is given in Figure 5 (Xu et al., 2023; Sanden et al., 2024). They have reported that the GTRs function as importers, and UMAMIT (Usually Multiple Amino Acids Move in And Out Transporter) functions as an exporters; together, they function as GSL transporters.
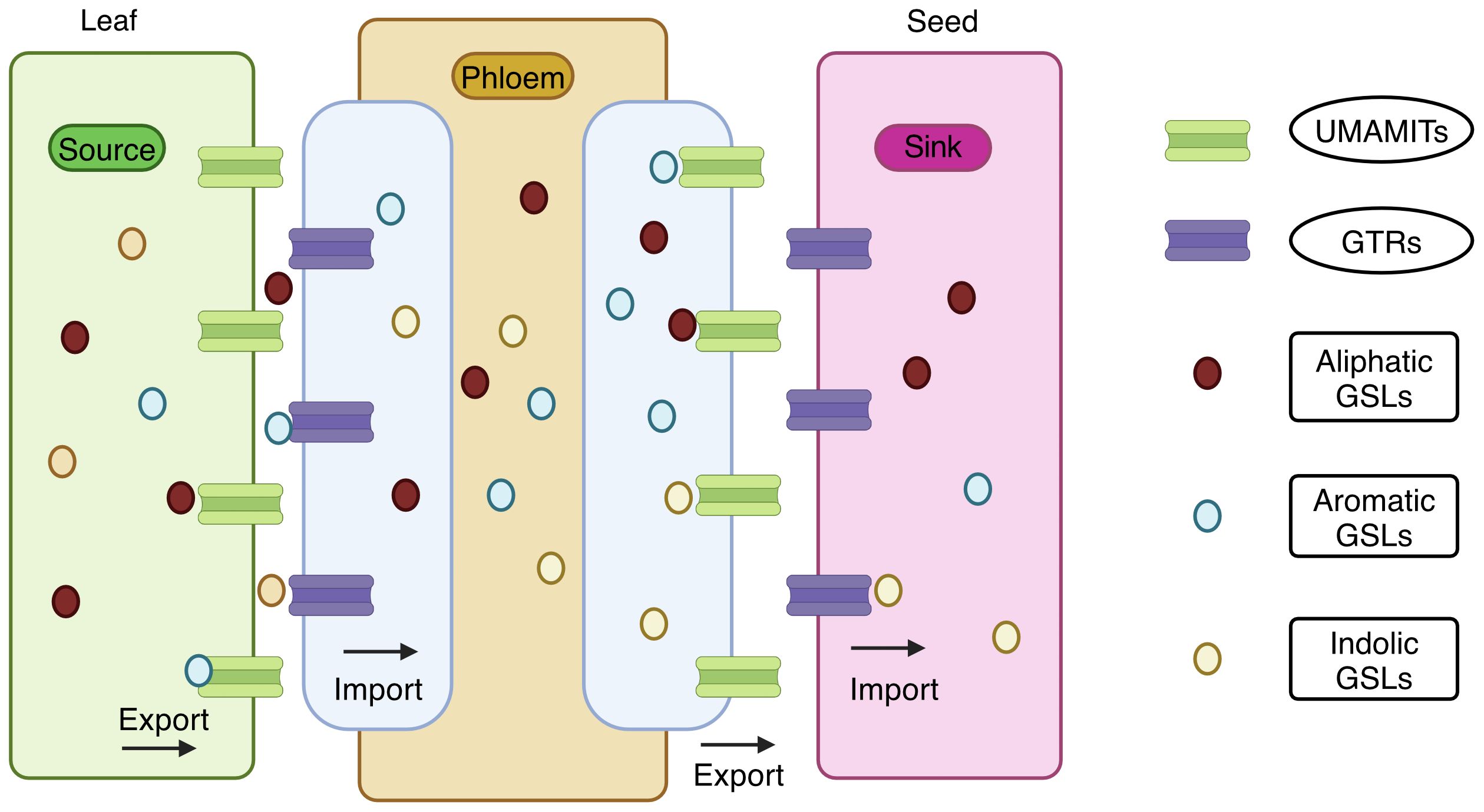
Figure 5. Transport of glucosinolates from source tissues to sink tissue. Usually Multiple Amino Acids Move in And Out Transporter (UMAMIT) acts as an exporter and mobilizes the glucosinolates from the source tissues into the apoplast. Glucosinolate transporters (GTR) acts as an importer and imports the glucosinolates into the phloem channel. Again, UMAMIT exports the glucosinolates into the apoplast at the unloading zone, and GTR imports them into the sink tissues. Both UMAMIT and GTR are involved in the transport of aliphatic, aromatic, and indolic glucosinolates in the plant (Created with BioRender.com).
UMAMIT are localized in the plasma membrane and export GSLs into the apoplast from the source tissue. In contrast, GTRs in phloem companion cells import GSLs into the phloem, facilitating the transport to sink tissues. At the phloem unloading zone, GSLs are exported by UMAMIT to the apoplast, and the GTRs import the GSLs into the seed. Both transporters transport all three types of GSLs: aliphatic, aromatic and indolic. Targeting UMAMIT genes impairs GSL export and prevents deposition in the sink tissues. UMAMIT29, UMAMIT30, and UMAMIT3 are exporters found in the Arabidopsis thaliana (Xu et al., 2023). The umamit29 umamit30 umamit31 triple mutants exhibited low seed GSLs. In the seeds, mutant alleles of UMAMIT29 showed an 80% reduction in total GSL levels. Thus, UMAMIT29 could be the optimal target gene for reducing the GSL export.
5.4.4 Phytic acid
Reducing phytic acid content in rapeseed is essential for improving the nutrient’s bioavailability. Phytic acid is synthesized through lipid-dependent and lipid-independent myo-inositol pathways. PMT5 and PIP5Ks play a vital role in the lipid-dependent biosynthesis pathway, whereas ITPK is pivotal in the lipid-independent biosynthesis pathway. SAC8 acts as a phosphatase and regulates phosphoinositide (Zhong and Ye, 2003). PMT5 (Polyol/monosaccharide transporter) is involved in transporting myo-inositol, glycerol, and several hexoses and pentoses, including ribose (Klepek et al., 2005). Thus, regulating the transporter could reduce the phytic acid content in the seeds. However, a major limitation lies in the multiple roles of the transporter. The knockdown mutants might result in poor seed development since PMT5 transports other compounds like glycerol and ribose. Thus, knockdown of the biosynthesis genes could be a better solution.
Phosphatidylinositol 4-phosphate 5-kinases (PIP5Ks) are a group of kinases that catalyze the production of phosphatidyl inositol (4,5)-bisphosphate (Van den Bout and Divecha, 2009). The ITPK gene family in B. napus holds four paralogues, namely BnITPK1, BnITPK2, BnITPK3, and BnITPK4. Ethyl methanesulphonate (EMS)-induced mutant lines targeting six genes in the phytic acid biosynthesis pathway were done by (Sashidhar et al., 2020a). However, due to the polyploid nature of rapeseed, the desired phenotype carrying all the mutant alleles requires laborious crossing. To overcome these challenges, BnITPK1 and BnITPK4 were knocked out by using CRISPR, resulting in a 35% phytate reduction (Sashidhar et al., 2020b). Similar phytate reduction has also been reported in soybean (Song et al., 2022a) and wheat (Ibrahim et al., 2022) using the CRISPR/Cas9 system.
5.4.5 Sinapines, lignins, cellulose and hemicellulose
Sinapine and lignin biosynthesis are interconnected and produced through the phenylpropanoid pathway (Boerjan et al., 2003; Milkowski and Strack, 2010). This interconnection provides a strategic advantage to alter both profiles by editing specific genes involved in biosynthesis. The enzymes involved in sinapine biosynthesis are UDP glucose sinapate glucosyltransferase (SGT), sinapoyl glucose choline sinapoyl transferase (SCT), sinapoyl glucose L-malate sinapoyl transferase (SMT) and sinapoyl choline (sinapine) esterase (SCE) (Milkowski and Strack, 2010). RNAi-mediated silencing was done in B. napus for the UGT84A9 gene, which disrupted the amounts and nature of the phenylpropanoid end products (Wolfram et al., 2010). Similarly, RNAi-mediated BnREF1 suppression resulted in reduced levels of sinapate esters, conjugated monolignols, dilignols, and trilignols and altered the minor conjugates of ferulate, caffeate, and 5hydroxyferulate (Mittasch et al., 2013).
The polymerization of monolignols like p-coumaryl alcohol, coniferyl alcohol, and sinapyl alcohol is essential for lignin biosynthesis (Xie et al., 2018). The monolignols are synthesized in the cytoplasm and transported to the cell wall, where they are polymerized into lignin, creating the p-hydroxyphenyl (H), guaiacyl (G) and syringyl (S) units (Boerjan et al., 2003). Targeting genes involved in this biosynthetic pathway, such as quinate hydroxycinnamoyl transferase (HCT), caffeic acid O-methyltransferase1 (COMT1), ferulate-5-hydroxylase, BGLU46, BGLU45, cinnamyl alcohol dehydrogenase, phenyl ammonia lyase 4 and cinnamoyl-CoA reductase 1 could reduce the lignin contents. The mutant obtained through CRISPR/Cas9 editing for caffeic acid O-methyltransferase 1 reduced lignin content by 14% and 34% increase in higher fermentable glucose recovery rate than the wild-type (Lee et al., 2021).
Additionally, cellulose synthase and mannose-1-phosphate guanylyltransferase 1 are involved in cellulose and hemicellulose biosynthesis, respectively (Lukowitz et al., 2001; Hu et al., 2018; Bhinder et al., 2022). This type of genetic modification improves the use of rapeseed in biofuel production by altering its fiber fraction, which comprises lignin, cellulose and hemicellulose. These compounds are involved in plant defense and maintain cell wall integrity to provide mechanical support to the plants. In this regard, promoter editing could be used to alter their levels rather than knocking down the genes completely, to maintain the essential structural integrity and defense while optimizing biosynthetic pathways.
5.4.6 Seed storage proteins
Recent efforts have been focused on altering SSPs to improve the meal quality of rapeseed. Unlike napin, a cysteine-rich SSP in rapeseed, the SSP of Brazil nut is rich in methionine. A chimeric gene with a methionine-rich 2S SSP coding sequence with soybean lectin promoter was used to improve the methionine content in rapeseed (Guerche et al., 1990). A similar attempt was made using a phaseolin promoter, resulting in an increase of 33% in the methionine content (Altenbach et al., 1992).
Improving the transport of the amino acids to the seed (sink) is another possible strategy. In arabidopsis, CAT6, AAP1, AAP2 and AAP8 have been reported to transport the amino acids from the source to the sink. CAT6 preferentially is known to transport neutral and cationic amino acids (Hammes et al., 2006). The mutant aap1 reduced total nitrogen (N) but showed a substantial increase in free amino acids in the seed coat and endosperm. This indicates that AAP1 is involved in the import of amino acids into the embryo (Sanders et al., 2009). AAP2 is involved in the phloem loading of amino acids; the aap2 mutant showed decreased amino acids in phloem sap and increased N content, leading to more branches (Zhang et al., 2010a). Meanwhile, AAP8 is crucial in transporting amino acids into the endosperm to nourish the embryo during early embryogenesis, with a high affinity towards acidic amino acids (Schmidt et al., 2007).
Altering a particular transporter could increase specific amino acid content. For instance, upregulating the expression of AtCAT6 could improve the transport of cationic amino acids like lysine and neutral amino acids like methionine and cysteine. Thus, increasing the expression of transporters in seeds could enhance the levels of essential amino acids such as lysine and methionine, which are often limited in plant-based diets (Yang et al., 2023).
Silencing cruciferin genes in rapeseed increased SSP cysteine, lysine, and methionine levels (Kohno-Murase et al., 1995). Conversely, silencing the napins in rapeseed resulted in higher levels of cruciferins and lower amounts of cysteine and lysine (Kohno-Murase et al., 1994). The inhibition of one type of SSP results in the accumulation of free amino acids, which can be directly allocated to synthesizing another SSP (Kohno-Murase et al., 1995). This suggests that protein profiles can be altered by targeting the specific genes involved in the biosynthesis of SSPs. However, these alterations did not affect the seed’s overall protein or oil content (Rolletschek et al., 2020). Thus, targeting SSP biosynthesis genes could modify the proportion of cruciferin and napins and improve their implications in food processing industries.
6 Conclusion
The increasing need for alternative protein sources due to rising population growth demands the exploration of new and sustainable options. Plant-based diets are gaining popularity, highlighting the need for alternative plant-derived proteins. Rapeseed byproducts hold promising potential as an alternative protein candidate for human food and animal feed. Their balanced amino acid profiles and functional properties make them suitable for various applications. However, the presence of ANFs poses challenges to their utilization. Strategies to reduce ANFs, including genetic, biotechnological and microbial methodologies, are essential to enhance rapeseed byproducts’ nutritional quality and acceptability. Therefore, further research and development are needed to optimize these strategies and fully utilize the potential of rapeseed as a sustainable protein source.
Author contributions
AM: Visualization, Writing – review & editing, Methodology, Writing – original draft, Investigation. SMu: Conceptualization, Methodology, Visualization, Writing – review & editing. EW: Writing – review & editing, Methodology. EI: Writing – review & editing, Supervision. SMa: Writing – review & editing. RS: Writing – review & editing. MN: Writing – review & editing. L-HZ: Writing – review & editing, Funding acquisition, Resources. RR: Writing – review & editing, Supervision. SK: Writing – review & editing, Methodology, Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Visualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Financial support for this research is highly acknowledged and was provided by Formas—A Swedish Research Council for Sustainable Development (grant numbers 2018-01301 and 2022-01483); SLU-Grogrund- Centre for Breeding of Food Crops; the Martha and Dagny Larssons Foundation; the Erik and Philip -Sörensens Foundation; The Royal Physiographic Society of Lund; The Crafoord Foundation; Einar and Inga Nilssons Foundation; and Magnus Bergvalls Foundation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Al-Asheh, S., Duvnjak, Z. (1995). Phytase production and decrease of phytic acid content in canola meal by Aspergillus carbonarius in solid-state fermentation. World J. Microbiol. Biotechnol. 11, 228–231. doi: 10.1007/BF00704655
Aljuobori, A., Abdullah, N., Zulkifli, I., Soleimani, A., Liang, J., Oskoueian, E. (2014). Lactobacillus salivarius fermentation reduced glucosinolate and fibre in canola meal. J. Food Sci. Vol. 3, 95–102. doi: 10.5539/jfr.v3n5p95
Almeida, F., Htoo, J., Thomson, J., Stein, H. (2014). Effects of heat treatment on the apparent and standardized ileal digestibility of amino acids in canola meal fed to growing pigs. Anim. Feed Sci. Technol. 187, 44–52. doi: 10.1016/j.anifeedsci.2013.09.009
Altenbach, S. B., Kuo, C.-C., Staraci, L. C., Pearson, K. W., Wainwright, C., Georgescu, A., et al. (1992). Accumulation of a Brazil nut albumin in seeds of transgenic canola results in enhanced levels of seed protein methionine. Plant Mol. Biol. 18, 235–245. doi: 10.1007/BF00034952
Aluko, R., McIntosh, T. (2005). Limited enzymatic proteolysis increases the level of incorporation of canola proteins into mayonnaise. Innov. Food Sci. Emerg. Technol. 6, 195–202. doi: 10.1016/j.ifset.2004.11.003
Anwar, M. M., Ali, S. E., Nasr, E. H. (2015). Improving the nutritional value of canola seed by gamma irradiation. J. Radiat. Res. Appl. Sci. 8, 328–333. doi: 10.1016/j.jrras.2015.05.007
Badani, A. G., Snowdon, R. J., Wittkop, B., Lipsa, F. D., Baetzel, R., Horn, R., et al. (2006). Colocalization of a partially dominant gene for yellow seed colour with a major QTL influencing acid detergent fibre (ADF) content in different crosses of oilseed rape (Brassica napus). Genome 49, 1499–1509. doi: 10.1139/g06-091
Banakar, R., Schubert, M., Collingwood, M., Vakulskas, C., Eggenberger, A. L., Wang, K. (2020). Comparison of CRISPR-Cas9/Cas12a ribonucleoprotein complexes for genome editing efficiency in the rice phytoene desaturase (OsPDS) gene. Rice 13, 1–7. doi: 10.1186/s12284-019-0365-z
Barbehenn, R. V., Peter Constabel, C. (2011). Tannins in plant-herbivore interactions. Phytochemistry 72, 1551–1565. doi: 10.1016/j.phytochem.2011.01.040
Bau, H. M., Villaume, C., Lin, C. F., Evrard, J., Quemener, B., Nicolas, J. P., et al. (1994). Effect of a solid-state fermentation using Rhizopus oligosporus sp. T-3 on elimination of antinutritional substances and modification of biochemical constituents of defatted rapeseed meal. J. Sci. Food Agric. 65, 315–322. doi: 10.1002/jsfa.2740650309
Behnke, N., Suprianto, E., Mollers, C. (2018). A major QTL on chromosome C05 significantly reduces acid detergent lignin (ADL) content and increases seed oil and protein content in oilseed rape (Brassica napus L.). Theor. Appl. Genet. 131, 2477–2492. doi: 10.1007/s00122-018-3167-6
Bell, J. (1993). Factors affecting the nutritional value of canola meal: a review. Can. J. Anim. Sci. 73, 689–697. doi: 10.4141/cjas93-075
Bell, J. M., Shires, A. (1982). Composition and digestibility by pigs of hull fractions from rapeseed cultivars with yellow or brown seed coats. Can. J. Anim. Sci. 62, 557–565. doi: 10.4141/cjas82-065
Bhinder, G., Sharma, S., Kaur, H., Akhatar, J., Mittal, M., Sandhu, S. (2022). Genomic regions associated with seed meal quality traits in Brassica napus germplasm. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.882766
Bhuiyan, N. H., Selvaraj, G., Wei, Y., King, J. (2009). Gene expression profiling and silencing reveal that monolignol biosynthesis plays a critical role in penetration defence in wheat against powdery mildew invasion. J. Exp. Bot. 60, 509–521. doi: 10.1093/jxb/ern290
Blair, R., Reichert, R. D. (1984). Carbohydrate and phenolic constituents in a comprehensive range of rapeseed and canola fractions: nutritional significance for animals. J. Sci. Food Agric. 35, 29–35. doi: 10.1002/jsfa.2740350106
Blazevic, I., Montaut, S., Burcul, F., Olsen, C. E., Burow, M., Rollin, P., et al. (2020). Glucosinolate structural diversity, identification, chemical synthesis and metabolism in plants. Phytochemistry 169, 112100. doi: 10.1016/j.phytochem.2019.112100
Boerjan, W., Ralph, J., Baucher, M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54, 519–546. doi: 10.1146/annurev.arplant.54.031902.134938
Bohn, L., Josefsen, L., Meyer, A. S., Rasmussen, S. K. (2007). Quantitative analysis of phytate globoids isolated from wheat bran and characterization of their sequential dephosphorylation by wheat phytase. J. Agric. Food Chem. 55, 7547–7552. doi: 10.1021/jf071191t
Boroojeni, F. G., Männer, K., Boros, D., Wiśniewska, M., Kühnel, S., Beckmann, K., et al. (2022). Spontaneous and enzymatic fermentation of rapeseed cake for broiler nutrition. Anim. Feed Sci. Technol. 284, 115135. doi: 10.1016/j.anifeedsci.2021.115135
Bos, C., Airinei, G., Mariotti, F., Benamouzig, R., Bérot, S., Evrard, J., et al. (2007). The poor digestibility of rapeseed protein is balanced by its very high metabolic utilization in humans. J. Nutr. 137, 594–600. doi: 10.1093/jn/137.3.594
Bulbul, M., Kader, M. A., Asaduzzaman, M., Ambak, M. A., Chowdhury, A. J. K., Hossain, M. S., et al. (2016). Can canola meal and soybean meal be used as major dietary protein sources for kuruma shrimp, Marsupenaeus japonicus? Aquaculture 452, 194–199. doi: 10.1016/j.aquaculture.2015.10.036
Burow, M., Halkier, B. A. (2017). How does a plant orchestrate defense in time and space? Using glucosinolates in Arabidopsis as case study. Curr. Opin. Plant Biol. 38, 142–147. doi: 10.1016/j.pbi.2017.04.009
Butler, E. J., Pearson, A. W., Fenwick, G. R. (1982). Problems which limit the use of rapeseed meal as a protein source in poultry diets. J. Sci. Food Agric. 33, 866–875. doi: 10.1002/jsfa.2740330909
Cao, Y., Yan, X., Ran, S., Ralph, J., Smith, R. A., Chen, X., et al. (2022). Knockout of the lignin pathway gene BnF5H decreases the S/G lignin compositional ratio and improves Sclerotinia sclerotiorum resistance in Brassica napus. Plant Cell Environ. 45, 248–261. doi: 10.1111/pce.14208
Capozzi, V., Fragasso, M., Romaniello, R., Berbegal, C., Russo, P., Spano, G. (2017). Spontaneous food fermentations and potential risks for human health. Fermentation 3, 49. doi: 10.3390/fermentation3040049
Cedrowski, J., Dąbrowa, K., Przybylski, P., Krogul-Sobczak, A., Litwinienko, G. (2021). Antioxidant activity of two edible isothiocyanates: Sulforaphane and erucin is due to their thermal decomposition to sulfenic acids and methylsulfinyl radicals. Food Chem. 353, 129213. doi: 10.1016/j.foodchem.2021.129213
Chao, H., Guo, L., Zhao, W., Li, H., Li, M. (2022a). A major yellow-seed QTL on chromosome A09 significantly increases the oil content and reduces the fiber content of seed in Brassica napus. Theor. Appl. Genet. 135, 1293–1305. doi: 10.1007/s00122-022-04031-0
Chao, H., Li, H., Yan, S., Zhao, W., Chen, K., Wang, H., et al. (2022b). Further insight into decreases in seed glucosinolate content based on QTL mapping and RNA-seq in Brassica napus L. Theor. Appl. Genet. 135, 2969–2991. doi: 10.1007/s00122-022-04161-5
Chao, H., Wang, H., Wang, X., Guo, L., Gu, J., Zhao, W., et al. (2017). Genetic dissection of seed oil and protein content and identification of networks associated with oil content in Brassica napus. Sci. Rep. 7, 46295. doi: 10.1038/srep46295
Chen, G., Geng, J., Rahman, M., Liu, X., Tu, J., Fu, T., et al. (2010). Identification of QTL for oil content, seed yield, and flowering time in oilseed rape (Brassica napus). Euphytica 175, 161–174. doi: 10.1007/s10681-010-0144-9
Chen, H. (2013). Modern solid state fermentation:Theory and Practice. (Springer Dordrecht, Netherlands).
Chen, W., Zhang, Y., Liu, X., Chen, B., Tu, J., Tingdong, F. (2007). Detection of QTL for six yield-related traits in oilseed rape (Brassica napus) using DH and immortalized F(2) populations. Theor. Appl. Genet. 115, 849–858. doi: 10.1007/s00122-007-0613-2
Chen, Y., Xie, C., Bilal, M., Wang, P., Yang, R. (2024). Screening of glucosinolates degrading lactic acid bacteria and their utilization in rapeseed meal fermentation. Grain Oil Sci. Technol. 7, 168–176. doi: 10.1016/j.gaost.2024.05.005
Cheng, H., Cai, S., Hao, M., Cai, Y., Wen, Y., Huang, W., et al. (2023). Targeted mutagenesis of BnTTG1 homologues generated yellow-seeded rapeseed with increased oil content and seed germination under abiotic stress. Plant Physiol. Biochem. 108302. doi: 10.1016/j.plaphy.2023.108302
Cheng, Z., Ai, Q., Mai, K., Xu, W., Ma, H., Li, Y., et al. (2010). Effects of dietary canola meal on growth performance, digestion and metabolism of Japanese seabass, Lateolabrax japonicus. Aquaculture 305, 102–108. doi: 10.1016/j.aquaculture.2010.03.031
Chichlowski, M., Schroeder, J., Park, C., Keller, W., Schimek, D. (2005). Altering the fatty acids in milk fat by including canola seed in dairy cattle diets. J. Dairy Sci. 88, 3084–3094. doi: 10.3168/jds.S0022-0302(05)72990-8
Cowieson, A., Acamovic, T., Bedford, M. (2006). Phytic acid and phytase: implications for protein utilization by poultry. Poult. Sci. 85, 878–885. doi: 10.1093/ps/85.5.878
Croat, J. R., Berhow, M., Karki, B., Muthukumarappan, K., Gibbons, W. R. (2016). Conversion of canola meal into a high-protein feed additive via solid-state fungal incubation process. J. Am. Oil Chem. Soc 93, 499–507. doi: 10.1007/s11746-016-2796-7
Cui, Y., Hu, X., Liang, G., Feng, A., Wang, F., Ruan, S., et al. (2020). Production of novel beneficial alleles of a rice yield-related QTL by CRISPR/Cas9. Plant Biotechnol. J. 18, 1987–1989. doi: 10.1111/pbi.13370
Daun, J. (1986). Glucosinolate levels in western Canadian rapeseed and canola. J. Am. Oil Chem. Soc 63, 639–643. doi: 10.1007/BF02638228
Deynze, A. V., Beversdorf, W. D., Pauls, K. P. (1993). Temperature effects on seed color in black-and yellow-seeded rapeseed. Can. J. Plant Sci. 73, 383–387. doi: 10.4141/cjps93-057
Deynze, A. V., Landry, B. S., Pauls, K. P. (1995). The identification of restriction fragment length polymorphisms linked to seed colour genes in Brassica napus. Genome 38, 534–542. doi: 10.1139/g95-069
Dhugga, K. S. (2022). Gene editing to accelerate crop breeding. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.889995
Doudna, J. A., Charpentier, E. (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096. doi: 10.1126/science.1258096
Dubin, M. J., Mittelsten Scheid, O., Becker, C. (2018). Transposons: a blessing curse. Curr. Opin. Plant Biol. 42, 23–29. doi: 10.1016/j.pbi.2018.01.003
Durkee, A. B., Thivierge, P. A. (1975). Bound phenolic acids in Brassica and Sinapis oilseeds. J. Food Sci. 40, 820–822. doi: 10.1111/j.1365-2621.1975.tb00565.x
Ebune, A., Al-Asheh, S., Duvnjak, Z. (1995a). Effects of phosphate, surfactants and glucose on phytase production and hydrolysis of phytic acid in canola meal by Aspergillus ficuum during solid-state fermentation. Bioresour. Technol. 54, 241–247. doi: 10.1016/0960-8524(95)00133-6
Ebune, A., Al-Asheh, S., Duvnjak, Z. (1995b). Production of phytase during solid state fermentation using Aspergillus ficuum NRRL 3135 in canola meal. Bioresour. Technol. 53, 7–12. doi: 10.1016/0960-8524(95)00041-C
El-Batal, A., Karem, H. A. (2001). Phytase production and phytic acid reduction in rapeseed meal by Aspergillus Niger during solid state fermentation. Food Res. Int. 34, 715–720. doi: 10.1016/S0963-9969(01)00093-X
Erdman, J. (1979). Oilseed phytates: nutritional implications. J. Am. Oil Chem. Soc 56, 736–741. doi: 10.1007/BF02663052
Fattahi, F., Fakheri, B. A., Solouki, M., Möllers, C., Rezaizad, A. (2018). Mapping QTL controlling agronomic traits in a doubled haploid population of winter oilseed rape (Brassica napus L.). J. Genet. 97, 1389–1406. doi: 10.1007/s12041-018-1044-3
Feizollahi, E., Mirmahdi, R. S., Zoghi, A., Zijlstra, R. T., Roopesh, M. S., Vasanthan, T. (2021). Review of the beneficial and anti-nutritional qualities of phytic acid, and procedures for removing it from food products. Food Res. Int. 143, 110284. doi: 10.1016/j.foodres.2021.110284
Felker, P., Bunch, R., Leung, A. M. (2016). Concentrations of thiocyanate and goitrin in human plasma, their precursor concentrations in brassica vegetables, and associated potential risk for hypothyroidism. Nutr. Rev. 74, 248–258. doi: 10.1093/nutrit/nuv110
Feng, D., Zuo, J. (2007). “Nutritional and anti-nutritional composition of rapeseed meal and its utilization as a feed ingredient for animal,” in Feed and industrial raw material: feed. (USA: Science Press USA Inc.), 265–270.
Fossi, M., Amundson, K., Kuppu, S., Britt, A., Comai, L. (2019). Regeneration of Solanum tuberosum plants from protoplasts induces widespread genome instability. Plant Physiol. 180, 78–86. doi: 10.1104/pp.18.00906
Fu, F. Y., Liu, L. Z., Chai, Y. R., Chen, L., Yang, T., Jin, M. Y., et al. (2007). Localization of QTLs for seed color using recombinant inbred lines of Brassica napus in different environments. Genome 50, 840–854. doi: 10.1139/g07-068
Fu, Z., Su, G., Yang, H., Sun, Q., Zhong, T., Wang, Z. (2021). Effects of dietary rapeseed meal on growth performance, carcass traits, serum parameters, and intestinal development of geese. Animals 11. doi: 10.3390/ani11061488
Gacek, K., Bartkowiak-Broda, I., Batley, J. (2018). Genetic and molecular regulation of Seed Storage Proteins (SSPs) to improve protein nutritional value of oilseed rape (Brassica napus L.) seeds. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00890
Gerzhova, A., Mondor, M., Benali, M., Aider, M. (2016). Incorporation of canola proteins extracted by electroactivated solutions in gluten-free biscuit formulation of rice–buckwheat flour blend: assessment of quality characteristics and textural properties of the product. Int. J. Food Sci. Technol. 51, 814–827. doi: 10.1111/ijfs.13034
Ghazani, S. M., Marangoni, A. G. (2013). Minor components in canola oil and effects of refining on these constituents: A review. J. Am. Oil Chem. Soc 90, 923–932. doi: 10.1007/s11746-013-2254-8
Gigolashvili, T., Yatusevich, R., Berger, B., Muller, C., Flugge, U. I. (2007). The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 51, 247–261. doi: 10.1111/j.1365-313X.2007.03133.x
Goh, Y., Clandinin, D., Robblee, A., Darlington, K. (1979). The effect of level of sinapine in a laying ration on the incidence of fishy odor in eggs from brown-shelled egg layers. Can. J. Anim. Sci. 59, 313–316. doi: 10.4141/cjas79-037
Gopinger, E., Xavier, E., Elias, M., Catalan, A., Castro, M., Nunes, A., et al. (2014). The effect of different dietary levels of canola meal on growth performance, nutrient digestibility, and gut morphology of broiler chickens. Poult. Sci. 93, 1130–1136. doi: 10.3382/ps.2013-03426
Grabež, V., Egelandsdal, B., Kjos, N. P., Håkenåsen, I. M., Mydland, L. T., Vik, J. O., et al. (2020). Replacing soybean meal with rapeseed meal and faba beans in a growing-finishing pig diet: Effect on growth performance, meat quality and metabolite changes. Meat. Sci. 166, 108134. doi: 10.1016/j.meatsci.2020.108134
Guerche, P., De Almeida, E. R., Schwarztein, M. A., Gander, E., Krebbers, E., Pelletier, G. (1990). Expression of the 2S albumin from Bertholletia excelsa in Brassica napus. Mol. Gen. Genet. 221, 306–314. doi: 10.1007/BF00259393
Gupta, P. K., Kulwal, P. L., Jaiswal, V. (2019). Association mapping in plants in the post-GWAS genomics era. Adv. Genet. 104, 75–154. doi: 10.1016/bs.adgen.2018.12.001
Halkier, B. A., Du, L. (1997). The biosynthesis of glucosinolates. Trends Plant Sci. 2, 425–431. doi: 10.1016/S1360-1385(97)90026-1
Hammes, U. Z., Nielsen, E., Honaas, L. A., Taylor, C. G., Schachtman, D. P. (2006). AtCAT6, a sink-tissue-localized transporter for essential amino acids in Arabidopsis. Plant J. 48, 414–426. doi: 10.1111/j.1365-313X.2006.02880.x
Hansen, J.Ø., Skrede, A., Mydland, L. T., Øverland, M. (2017). Fractionation of rapeseed meal by milling, sieving and air classification—Effect on crude protein, amino acids and fiber content and digestibility. Anim. Feed Sci. Technol. 230, 143–153. doi: 10.1016/j.anifeedsci.2017.05.007
Harloff, H. J., Lemcke, S., Mittasch, J., Frolov, A., Wu, J. G., Dreyer, F., et al. (2012). A mutation screening platform for rapeseed (Brassica napus L.) and the detection of sinapine biosynthesis mutants. Theor. Appl. Genet. 124, 957–969. doi: 10.1007/s00122-011-1760-z
He, R., He, H.-Y., Chao, D., Ju, X., Aluko, R. (2014). Effects of high pressure and heat treatments on physicochemical and gelation properties of rapeseed protein isolate. Food Bioproc. Tech. 7, 1344–1353. doi: 10.1007/s11947-013-1139-z
He, R., Ju, X., Yuan, J., Wang, L., Girgih, A. T., Aluko, R. E. (2012). Antioxidant activities of rapeseed peptides produced by solid state fermentation. Food Res. Int. 49, 432–438. doi: 10.1016/j.foodres.2012.08.023
He, S., Sun, X., Wei, W., Hu, B. (2023). Upcycling canola meal as compared to soybean meal by fungal bioconversion for potential monogastric animal nutrition. Waste Biomass Valori. 14, 795–804. doi: 10.1007/s12649-022-01889-5
Heidari, F., Øverland, M., Hansen, J.Ø., Mydland, L. T., Urriola, P. E., Chen, C., et al. (2024). Enhancing the nutritional value of canola meal through solid culture with Pleurotus ostreatus. Anim. Feed Sci. Technol. 309, 115893. doi: 10.1016/j.anifeedsci.2024.115893
Helal, M., Gill, R. A., Tang, M., Yang, L., Hu, M., Yang, L., et al. (2021). SNP-and haplotype-based GWAS of flowering-related traits in Brassica napus. Plants 10, 2475. doi: 10.3390/plants10112475
Hernández, C., Olmeda-Guerrero, L., Chávez-Sánchez, M., Ibarra-Castro, L., Gaxiola-Cortez, G., Martínez-Cárdenas, L. (2020). Nutritional evaluation of canola meal as fish meal replacement for juvenile spotted rose snapper (Lutjanus guttatus): Effects on growth performance, hematological parameters, body composition, and nutrient digestibility. Anim. Feed Sci. Technol. 269, 114683. doi: 10.1016/j.anifeedsci.2020.114683
Hirochika, H. (1993). Activation of tobacco retrotransposons during tissue culture. EMBO.J. 12, 2521–2528. doi: 10.1002/j.1460-2075.1993.tb05907.x
Hirochika, H., Sugimoto, K., Otsuki, Y., Tsugawa, H., Kanda, M. (1996). Retrotransposons of rice involved in mutations induced by tissue culture. Proc. Natl. Acad. Sci. U.S.A 93, 7783–7788. doi: 10.1073/pnas.93.15.7783
Hölzl, G., Rezaeva, B. R., Kumlehn, J., Dörmann, P. (2023). Ablation of glucosinolate accumulation in the oil crop Camelina sativa by targeted mutagenesis of genes encoding the transporters GTR1 and GTR2 and regulators of biosynthesis MYB28 and MYB29. Plant Biotechnol. J. 21, 189–201. doi: 10.1111/pbi.13936
Horigome, T., Kumar, R., Okamoto, K. (1988). Effects of condensed tannins prepared from leaves of fodder plants on digestive enzymes in vitro and in the intestine of rats. Br. J. Nutr. 60, 275–285. doi: 10.1079/bjn19880099
Hu, H., Zhang, R., Tao, Z., Li, X., Li, Y., Huang, J., et al. (2018). Cellulose synthase mutants distinctively affect cell growth and cell wall integrity for plant biomass production in Arabidopsis. Plant Cell Physiol. 59, 1144–1157. doi: 10.1093/pcp/pcy050
Huang, X.-Q., Huang, T., Hou, G.-Z., Li, L., Hou, Y., Lu, Y.-H. (2016). Identification of QTLs for seed quality traits in rapeseed (Brassica napus L.) using recombinant inbred lines (RILs). Euphytica 210, 1–16. doi: 10.1007/s10681-016-1675-5
Huang, L., Li, Q., Zhang, C., Chu, R., Gu, Z., Tan, H., et al. (2020). Creating novel Wx alleles with fine-tuned amylose levels and improved grain quality in rice by promoter editing using CRISPR/Cas9 system. Plant Biotechnol. J. 18, 2164–2166. doi: 10.1111/pbi.13391
Hunt, J. R., Beiseigel, J. M. (2009). Dietary calcium does not exacerbate phytate inhibition of zinc absorption by women from conventional diets. Am. J. Clin. Nutr. 89, 839–843. doi: 10.3945/ajcn.2008.27175
Ibrahim, S., Saleem, B., Rehman, N., Zafar, S. A., Naeem, M. K., Khan, M. R. (2022). CRISPR/Cas9 mediated disruption of Inositol Pentakisphosphate 2-Kinase 1 (TaIPK1) reduces phytic acid and improves iron and zinc accumulation in wheat grains. J. Adv. Res. 37, 33–41. doi: 10.1016/j.jare.2021.07.006
Ito, H. (2022). Environmental stress and transposons in plants. Genes Genet. Syst. 97, 169–175. doi: 10.1266/ggs.22-00045
Jansman, A. J. (1993). Tannins in feedstuffs for simple-stomached animals. Nutr. Res. Rev. 6, 209–236. doi: 10.1079/NRR19930013
Jensen, S. K., Liu, Y.-G., Eggum, B. (1995). The effect of heat treatment on glucosinolates and nutritional value of rapeseed meal in rats. Anim. Feed Sci. Technol. 53, 17–28. doi: 10.1016/0377-8401(94)00740-Z
Jia, W., Rodriguez-Alonso, E., Bianeis, M., Keppler, J. K., van der Goot, A. J. (2021). Assessing functional properties of rapeseed protein concentrate versus isolate for food applications. Innov. Food Sci. Emerg. Technol. 68, 102636. doi: 10.1016/j.ifset.2021.102636
Jiang, J., Shi, D., Zhou, X. Q., Feng, L., Liu, Y., Jiang, W. D., et al. (2016). Effects of lysine and methionine supplementation on growth, body composition and digestive function of grass carp (Ctenopharyngodon idella) fed plant protein diets using high-level canola meal. Aquacult. Nutr. 22, 1126–1133. doi: 10.1111/anu.12339
Jorgensen, M. E., Nour-Eldin, H. H., Halkier, B. A. (2015). Transport of defense compounds from source to sink: lessons learned from glucosinolates. Trends Plant Sci. 20, 508–514. doi: 10.1016/j.tplants.2015.04.006
Jørgensen, M. E., Xu, D., Crocoll, C., Ernst, H. A., Ramírez, D., Motawia, M. S., et al. (2017). Origin and evolution of transporter substrate specificity within the NPF family. eLife 6, e19466. doi: 10.7554/eLife.19466
Jorquera, M., Martinez, O., Maruyama, F., Marschner, P., de la Luz Mora, M. (2008). Current and future biotechnological applications of bacterial phytases and phytase-producing bacteria. Microbes Environ. 23, 182–191. doi: 10.1264/jsme2.23.182
Josefsson, L. G., Lenman, M., Ericson, M. L., Rask, L. (1987). Structure of a gene encoding the 1.7 S storage protein, napin, from Brassica napus. J. Biol. Chem. 262, 12196–12201. doi: 10.1016/s0021-9258(18)45336-7
Josefsson, E., Uppstrom, B. (1976). Influence of sinapine and p-hydroxybenzylglucosinolate on the nutritional value of rapeseed and white mustard meals. J. Sci. Food Agric. 27, 438–442. doi: 10.1002/jsfa.2740270509
Jyothi, T., Singh, S. A., Appu Rao, A. (2007). Conformation of napin (Brassica juncea) in salts and monohydric alcohols: Contribution of electrostatic and hydrophobic interactions. J. Agric. Food Chem. 55, 4229–4236. doi: 10.1021/jf0700935
Kaiser, F., Harbach, H., Schulz, C. (2022). Rapeseed proteins as fishmeal alternatives: A review. Rev. Aquacult. 14, 1887–1911. doi: 10.1111/raq.12678
Keck, A.-S., Finley, J. W. (2004). Cruciferous vegetables: cancer protective mechanisms of glucosinolate hydrolysis products and selenium. Integr. Cancer Ther. 3, 5–12. doi: 10.1177/1534735403261831
Kim, Y., Carpenter, C. E., Mahoney, A. W. (1993). Gastric acid production, iron status and dietary phytate alter enhancement by meat of iron absorption in rats. J. Nutr. 123, 940–946. doi: 10.1093/jn/123.5.940
Kim, J., Paik, H. Y., Joung, H., Woodhouse, L. R., Li, S., King, J. C. (2007). Effect of dietary phytate on zinc homeostasis in young and elderly Korean women. J. Am. Coll. Nutr. 26, 1–9. doi: 10.1080/07315724.2007.10719579
Kim, Y. J., Park, S. Y., Lee, J.-H. (2021). Berteroin ameliorates lipid accumulation through AMPK-mediated regulation of hepatic lipid metabolism and inhibition of adipocyte differentiation. Life Sci. 282, 119668. doi: 10.1016/j.lfs.2021.119668
Kirov, I. (2023). Toward transgene-free transposon-mediated biological mutagenesis for plant breeding. Int. J. Mol. Sci. 24:17054. doi: 10.3390/ijms242317054
Klepek, Y. S., Geiger, D., Stadler, R., Klebl, F., Landouar-Arsivaud, L., Lemoine, R., et al. (2005). Arabidopsis polyol transporter 5, a new member of the monosaccharide transporter-like superfamily, mediates H+-Symport of numerous substrates, including myo-inositol, glycerol, and ribose. Plant Cell 17, 204–218. doi: 10.1105/tpc.104.026641
Knuckles, B. E., Betschart, A. A. (2006). Effect of phytate and other myo-inositol phosphate esters on α-amylase digestion of starch. J. Food Sci. 52, 719–721. doi: 10.1111/j.1365-2621.1987.tb06710.x
Knuckles, B., Kuzmicky, D., Betschart, A. (1985). Effect of phytate and partially hydrolyzed phytate on in vitro protein digestibility. J. Food Sci. 50, 1080–1082. doi: 10.1111/j.1365-2621.1985.tb13016.x
Kodagoda, L., Nakai, S., Powrie, W. (1973). Some functional properties of rapeseed protein isolates and concentrates. Can. Inst. Food Technol. J. 6, 266–269. doi: 10.1016/S0315-5463(73)74038-4
Kohno-Murase, J., Murase, M., Ichikawa, H., Imamura, J. (1994). Effects of an antisense napin gene on seed storage compounds in transgenic Brassica napus seeds. Plant Mol. Biol. 26, 1115–1124. doi: 10.1007/BF00040693
Kohno-Murase, J., Murase, M., Ichikawa, H., Imamura, J. (1995). Improvement in the quality of seed storage protein by transformation of Brassica napus with an antisense gene for cruciferin. Theor. Appl. Genet. 91, 627–631. doi: 10.1007/BF00223289
Kracht, W., Dänicke, S., Kluge, H., Keller, K., Matzke, W., Hennig, U., et al. (2004). Effect of dehulling of rapeseed on feed value and nutrient digestibility of rape products in pigs. Arch. Anim. Nutr. 58, 389–404. doi: 10.1080/00039420400005018
Kroj, T., Savino, G., Valon, C., Giraudat, J., Parcy, F. (2003). Regulation of storage protein gene expression in Arabidopsis. Development 130, 6065–6073. doi: 10.1242/dev.00814
Krygier, K., Sosulski, F., Hogge, L. (1982). Free, esterified, and insoluble-bound phenolic acids. 1. Extraction and purification procedure. J. Agric. Food Chem. 30, 330–334. doi: 10.1021/jf00110a028
Lage, C., Räisänen, S., Stefenoni, H., Melgar, A., Chen, X., Oh, J., et al. (2021). Lactational performance, enteric gas emissions, and plasma amino acid profile of dairy cows fed diets with soybean or canola meals included on an equal protein basis. J. Dairy Sci. 104, 3052–3066. doi: 10.3168/jds.2020-18851
Landero, J., Beltranena, E., Cervantes, M., Araiza, A., Zijlstra, R. (2012). The effect of feeding expeller-pressed canola meal on growth performance and diet nutrient digestibility in weaned pigs. Anim. Feed Sci. Technol. 171, 240–245. doi: 10.1016/j.anifeedsci.2011.11.004
Lee, J. W., Wang, S., Huang, Y., Seefeldt, T., Donkor, A., Logue, B. A., et al. (2020). Toxicity of canola-derived glucosinolates in pigs fed resistant starch-based diets. J. Anim. Sci. 98, skaa111. doi: 10.1093/jas/skaa111
Lee, J. H., Won, H. J., Hoang Nguyen Tran, P., Lee, S.m., Kim, H. Y., Jung, J. H. (2021). Improving lignocellulosic biofuel production by CRISPR/Cas9-mediated lignin modification in barley. Glob. Change Biol. Bioenergy 13, 742–752. doi: 10.1111/gcbb.12808
Lee, J. W., Woyengo, T. A. (2018). Growth performance, organ weights, and blood parameters of nursery pigs fed diets containing increasing levels of cold-pressed canola cake. J. Anim. Sci. 96, 4704–4712. doi: 10.1093/jas/sky317
Le Thanh, B. V., Beltranena, E., Zhou, X., Wang, L. F., Zijlstra, R. T. (2019). Amino acid and energy digestibility of Brassica napus canola meal from different crushing plants fed to ileal-cannulated grower pigs. Anim. Feed Sci. Technol. 252, 83–91. doi: 10.1016/j.anifeedsci.2019.04.008
Li, X., Sandgrind, S., Moss, O., Guan, R., Ivarson, E., Wang, E. S., et al. (2021). Efficient protoplast regeneration protocol and Crispr/Cas9-mediated editing of Glucosinolate Transporter (GTR) genes in rapeseed (Brassica napus L.). Front. Plant Sci. 12. doi: 10.3389/fpls.2021.680859
Li, H., Yu, K., Zhang, Z., Yu, Y., Wan, J., He, H., et al. (2024). Targeted mutagenesis of flavonoid biosynthesis pathway genes reveals functional divergence in seed coat colour, oil content and fatty acid composition in Brassica napus L. Plant Biotechnol. J. 22, 445–459. doi: 10.1111/pbi.14197
Liang, Z., Chen, K., Li, T., Zhang, Y., Wang, Y., Zhao, Q., et al. (2017). Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat. Commun. 8, 14261. doi: 10.1038/ncomms14261
Liang, Y., Eudes, A., Yogiswara, S., Jing, B., Benites, V. T., Yamanaka, R., et al. (2019). A screening method to identify efficient sgRNAs in Arabidopsis, used in conjunction with cell-specific lignin reduction. Biotechnol. Biofuels 12, 130. doi: 10.1186/s13068-019-1467-y
Lim, C., Beames, R., Eales, J., Prendergast, A., McLeese, J., Shearer, K., et al. (1997). Nutritive values of low and high fibre canola meals for shrimp (Penaeus vannamei). Aquacult. Nutr. 3, 269–279. doi: 10.1046/j.1365-2095.1997.00048.x
Liu, L., Gallagher, J., Arevalo, E. D., Chen, R., Skopelitis, T., Wu, Q., et al. (2021a). Enhancing grain-yield-related traits by CRISPR–Cas9 promoter editing of maize CLE genes. Nat. Plants 7, 287–294. doi: 10.1038/s41477-021-00858-5
Liu, L., Liang, X. F., Fang, J., Yuan, X., Li, B., Li, J. (2021b). Tannase alleviates the adverse physiological and toxicological impacts of supplementing tannin in the diet of grass carp (Ctenopharyngodon idellus). Aquacult. Nutr. 27, 1612–1625. doi: 10.1111/anu.13301
Liu, G., Ma, Z., Shan, A., Wang, L., Bi, Z. (2016). Effects of dietary rumen-protected lysine on milk yield and composition in lactating cows fed diets containing double-low rapeseed meal. Int. J. Dairy Technol. 69, 380–385. doi: 10.1111/1471-0307.12270
Liu, L. Z., Meng, J. L., Lin, N., Chen, L., Tang, Z. L., Zhang, X. K., et al. (2006). QTL mapping of seed coat color for yellow seeded Brassica napus. Acta Genet. Sin. 33, 181–187. doi: 10.1016/S0379-4172(06)60037-1
Liu, L., Qu, C., Wittkop, B., Yi, B., Xiao, Y., He, Y., et al. (2013). A high-density SNP map for accurate mapping of seed fibre QTL in Brassica napus L. PloS One 8, e83052. doi: 10.1371/journal.pone.0083052
Liu, N., Ru, Y. J., Li, F. D., Wang, J.-P., Lei, X.-Q. (2009). Effect of dietary phytate and phytase on proteolytic digestion and growth regulation of broilers. Arch. Anim. Nutr. 63, 292–303. doi: 10.1080/17450390903020422
Liu, X. P., Tu, J. X., Chen, B. Y., Fu, T. D. (2008). Identification and inheritance of a partially dominant gene for yellow seed colour in Brassica napus. Plant Breed. 124, 9–12. doi: 10.1111/j.1439-0523.2004.01051.x
Liu, J., Zhou, R., Wang, W., Wang, H., Qiu, Y., Rosy, R., et al. (2019). An Epigenetic LTR-retrotransposon insertion in the upstream region of BnSHP1. A9 controls quantitative pod shattering resistance in Brassica napus. bioRxiv 858407. doi: 10.1101/858407
Lomascolo, A., Uzan-Boukhris, E., Sigoillot, J.-C., Fine, F. (2012). Rapeseed and sunflower meal: a review on biotechnology status and challenges. Appl. Microbiol. Biotechnol. 95, 1105–1114. doi: 10.1007/s00253-012-4250-6
Lopez, D. A., Lagos, L. V., Stein, H. H. (2020). Digestible and metabolizable energy in soybean meal sourced from different countries and fed to pigs. Anim. Feed Sci. Technol. 268, 114600. doi: 10.1016/j.anifeedsci.2020.114600
Lücke, F.-K., Fritz, V., Tannhäuser, K., Arya, A. (2019). Controlled fermentation of rapeseed presscake by Rhizopus, and its effect on some components with relevance to human nutrition. Food Res. Int. 120, 726–732. doi: 10.1016/j.foodres.2018.11.031
Lukowitz, W., Nickle, T. C., Meinke, D. W., Last, R. L., Conklin, P. L., Somerville, C. R. (2001). Arabidopsis cyt1 mutants are deficient in a mannose-1-phosphate guanylyltransferase and point to a requirement of N-linked glycosylation for cellulose biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 98, 2262–2267. doi: 10.1073/pnas.051625798
Luo, Z., Liu, C. X., Wen, H. (2012). Effect of dietary fish meal replacement by canola meal on growth performance and hepatic intermediary metabolism of genetically improved farmed tilapia strain of Nile tilapia, Oreochromis niloticus, reared in freshwater. J. World. Aquac. Soc 43, 670–678. doi: 10.1111/j.1749-7345.2012.00601.x
Lv, W., Chen, W., Tan, S., Ba, G., Sun, C., Feng, F., et al. (2024). Effects of removing phytic acid on the bioaccessibility of Ca/Fe/Zn and protein digestion in soymilk. J. Sci. Food Agric. 104, 5262–5273. doi: 10.1002/jsfa.13367
Lynch, E. E., Clayton, E. H., Holman, B. W., Hopkins, D. L., Polkinghorne, R. J., Campbell, M. A. (2024). Canola meal as a supplement for grass-fed beef cattle: Effects on growth rates, carcase and meat quality, and consumer sensory evaluations. Meat. Sci. 207, 109363. doi: 10.1016/j.meatsci.2023.109363
Maenz, D. (2001). Enzymatic characteristics of phytases as they relate to their use in animal feeds (Wallingford,UK: CABI).
Maitra, S., Ray, A. (2003). Inhibition of digestive enzymes in rohu, Labeo rohita (Hamilton), fingerlings by tannin: an in vitro study. Aquac. Res. 34, 93–95. doi: 10.1046/j.1365-2109.2003.00792.x
Makkar, H. P. S. (2003). Effects and fate of tannins in ruminant animals, adaptation to tannins, and strategies to overcome detrimental effects of feeding tannin-rich feeds. Small Rumin. Res. 49, 241–256. doi: 10.1016/s0921-4488(03)00142-1
Malabat, C., Atterby, H., Chaudhry, Q., Renard, M., Guéguen, J. (2003). “Genetic variability of rapeseed protein composition,” in Proceedings of the 11th International Rapeseed Congress, Royal Veterinary and Agricultural University. (Copenhagen, Denmark: Global Council for Innovation in Rapeseed and Canola) 1, 205–208.
Mangan, J. L. (1988). Nutritional effects of tannins in animal feeds. Nutr. Res. Rev. 1, 209–231. doi: 10.1079/NRR19880015
Mann, A., Kumari, J., Kumar, R., Kumar, P., Pradhan, A. K., Pental, D., et al. (2023). Targeted editing of multiple homologues of GTR1 and GTR2 genes provides the ideal low-seed, high-leaf glucosinolate oilseed mustard with uncompromised defence and yield. Plant Biotechnol. J. 21, 2182–2195. doi: 10.1111/pbi.14121
Marco, M. L., Heeney, D., Binda, S., Cifelli, C. J., Cotter, P. D., Foligné, B., et al. (2017). Health benefits of fermented foods: Microbiota and beyond. Curr. Opin. Biotechnol. 44, 94–102. doi: 10.1016/j.copbio.2016.11.010
Mason, A. S., Higgins, E. E., Snowdon, R. J., Batley, J., Stein, A., Werner, C., et al. (2017). A user guide to the Brassica 60K Illumina Infinium SNP genotyping array. Theor. Appl. Genet. 130, 621–633. doi: 10.1007/s00122-016-2849-1
Mawson, R., Heaney, R. K., Zdunczyk, Z., Kozlowska, H. (1994). Rapeseed meal-glucosinolates and their antinutritional effects. Part 5. Animal reproduction. Food/Nahrung 38, 588–598. doi: 10.1002/food.19940380607
Meng, J., Shi, S., Gan, L., Li, Z., Qu, X. (1998). The production of yellow-seeded Brassica napus (AACC) through crossing interspecific hybrids of B. campestris (AA) and B. carinata (BBCC) with B.napus. Euphytica 103, 329–333. doi: 10.1023/A:1018646223643
Miao, L., Chao, H., Chen, L., Wang, H., Zhao, W., Li, B., et al. (2019). Stable and novel QTL identification and new insights into the genetic networks affecting seed fiber traits in Brassica napus. Theor. Appl. Genet. 132, 1761–1775. doi: 10.1007/s00122-019-03313-4
Mikulski, D., Jankowski, J., Zdunczyk, Z., Juskiewicz, J., Slominski, B. (2012). The effect of different dietary levels of rapeseed meal on growth performance, carcass traits, and meat quality in Turkeys. Poult. Sci. 91, 215–223. doi: 10.3382/ps.2011-01587
Milkowski, C., Strack, D. (2010). Sinapate esters in brassicaceous plants: biochemistry, molecular biology, evolution and metabolic engineering. Planta 232, 19–35. doi: 10.1007/s00425-010-1168-z
Mittasch, J., Bottcher, C., Frolov, A., Strack, D., Milkowski, C. (2013). Reprogramming the phenylpropanoid metabolism in seeds of oilseed rape by suppressing the orthologs of reduced epidermal fluorescence1. Plant Physiol. 161, 1656–1669. doi: 10.1104/pp.113.215491
Monsalve, R., Villalba, M., Rodríguez, R. (2001). Allergy to mustard seeds: the importance of 2S albumins as food allergens. Internet Symposium Food Allergens 3, 57–69. Available online at: http://www.food-allergens.de/password/PDF-downloads/complete-articles/3-2-monsalve-et-al.pdf
Moran, E. T., Bedford, M. R. (2024). Basis for the diversity and extent in loss of digestible nutrients created by dietary phytin: Emphasis on fowl and swine. Anim. Nutr. 16, 422–428. doi: 10.1016/j.aninu.2023.11.010
Moreno, F. J., Clemente, A. (2008). 2S albumin storage proteins: what makes them food allergens? Open Biochem. J. 2, 16–28. doi: 10.2174/1874091X00802010016
Mueller, M., Ryl, E. B., Fenton, T., Clandinin, D. (1978). Cultivar and growing location differences on the sinapine content of rapeseed. Can. J. Anim. Sci. 58, 579–583. doi: 10.4141/cjas78-076
Murtagh, G., Archer, D., Dumoulin, M., Ridout, S., Matthews, S., Arshad, S., et al. (2003). In vitro stability and immunoreactivity of the native and recombinant plant food 2S albumins Ber e 1 and SFA-8. Clin. Exp. Allergy 33, 1147–1152. doi: 10.1046/j.1365-2222.2003.01736.x
Nega, T., Woldes, Y. (2018). Review on nutritional limitations and opportunities of using rapeseed meal and other rapeseed by - products in animal feeding. J. Nutr. Health Food Eng. 8, 43–48. doi: 10.15406/jnhfe.2018.08.00254
Neuenschwander, M., Stadelmaier, J., Eble, J., Grummich, K., Szczerba, E., Kiesswetter, E., et al. (2023). Substitution of animal-based with plant-based foods on cardiometabolic health and all-cause mortality: a systematic review and meta-analysis of prospective studies. BMC Med. 21, 404. doi: 10.1186/s12916-023-03093-1
Newell, C., Rhoads, M., Bidney, D. (1984). Cytogenetic analysis of plants regenerated from tissue explants and mesophyll protoplasts of winter rape, Brassica napus L. Can. J. Genet. Cytol. 26, 752–761. doi: 10.1139/g84-119
Newkirk, R., Classen, H., Scott, T., Edney, M. (2003). The digestibility and content of amino acids in toasted and non-toasted canola meals. Can. J. Anim. Sci. 83, 131–139. doi: 10.4141/A02-028
Nour-Eldin, H. H., Andersen, T. G., Burow, M., Madsen, S. R., Jorgensen, M. E., Olsen, C. E., et al. (2012). NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds. Nature 488, 531–534. doi: 10.1038/nature11285
O’Dell, B. L., De Boland, A. R., Koirtyohann, S. R. (1972). Distribution of phytate and nutritionally important elements among the morphological components of cereal grains. J. Agric. Food Chem. 20, 718–723. doi: 10.1021/jf60181a021
Oatway, L., Vasanthan, T., Helm, J. H. (2007). Phytic acid. Food Rev. Int. 17, 419–431. doi: 10.1081/fri-100108531
Oberleas, D., Muhrer, M., O’dell, B. (1962). Effects of phytic acid on zinc availability and parakeratosis in swine. J. Anim. Sci. 21, 57–61. doi: 10.2527/jas1962.21157x
Olin-Fatih, M. (1996). The morphology, cytology, and C-banded karyotypes of Brassica campestris, B. oleracea, and B. napus plants regenerated from protoplasts. Theor. Appl. Genet. 93, 414–420. doi: 10.1007/BF00223184
Pantoja-Uceda, D., Palomares, O., Bruix, M., Villalba, M., Rodríguez, R., Rico, M., et al. (2004). Solution structure and stability against digestion of rproBnIb, a recombinant 2S albumin from rapeseed: relationship to its allergenic properties. Biochemistry 43, 16036–16045. doi: 10.1021/bi048069x
Pinterits, A., Arntfield, S. D. (2008). Improvement of canola protein gelation properties through enzymatic modification with transglutaminase. LWT-Food Sci. Technol. 41, 128–138. doi: 10.1016/j.lwt.2007.01.011
Poisson, E., Trouverie, J., Brunel-Muguet, S., Akmouche, Y., Pontet, C., Pinochet, X., et al. (2019). Seed yield components and seed quality of oilseed rape are impacted by sulfur fertilization and its interactions with nitrogen fertilization. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00458
Porto, M. S., Pinheiro, M. P. N., Batista, V. G. L., dos Santos, R. C., de Albuquerque Melo Filho, P., de Lima, L. M. (2014). Plant promoters: an approach of structure and function. Mol. Biotechnol. 56, 38–49. doi: 10.1007/s12033-013-9713-1
Pramitha, J. L., Rana, S., Aggarwal, P. R., Ravikesavan, R., Joel, A. J., Muthamilarasan, M. (2021). Diverse role of phytic acid in plants and approaches to develop low-phytate grains to enhance bioavailability of micronutrients. Adv. Genet. 107, 89–120. doi: 10.1016/bs.adgen.2020.11.003
Puhakka, L., Jaakkola, S., Simpura, I., Kokkonen, T., Vanhatalo, A. (2016). Effects of replacing rapeseed meal with fava bean at 2 concentrate crude protein levels on feed intake, nutrient digestion, and milk production in cows fed grass silage–based diets. J. Dairy Sci. 99, 7993–8006. doi: 10.3168/jds.2016-10925
Puumalainen, T. J., Poikonen, S., Kotovuori, A., Vaali, K., Kalkkinen, N., Reunala, T., et al. (2006). Napins, 2S albumins, are major allergens in oilseed rape and turnip rape. J. Allergy Clin. Immunol. 117, 426–432. doi: 10.1016/j.jaci.2005.10.004
Qaisrani, S., Van Krimpen, M., Verstegen, M., Hendriks, W., Kwakkel, R. (2020). Effects of three major protein sources on performance, gut morphology and fermentation characteristics in broilers. Br. Poult. Sci. 61, 43–50. doi: 10.1080/00071668.2019.1671958
Qiao, H., Classen, H. L. (2003). Nutritional and physiological effects of rapeseed meal sinapine in broiler chickens and its metabolism in the digestive tract. J. Sci. Food Agric. 83, 1430–1438. doi: 10.1002/jsfa.1559
Qiao, H., Dahiya, J., Classen, H. (2008). Nutritional and physiological effects of dietary sinapic acid (4-hydroxy-3, 5-dimethoxy-cinnamic acid) in broiler chickens and its metabolism in the digestive tract. Poult. Sci. 87, 719–726. doi: 10.3382/ps.2007-00357
Rahman, M. H. (2008). Production of yellow-seeded Brassica napus through interspecific crosses. Plant Breed. 120, 463–472. doi: 10.1046/j.1439-0523.2001.00640.x
Rahman, S. U., Khan, M. O., Ullah, R., Ahmad, F., Raza, G. (2023). Agrobacterium-mediated transformation for the development of transgenic crops; present and future prospects. Mol. Biotechnol. 66, 1836–1852. doi: 10.1007/s12033-023-00826-8
Rahman, M., Li, G., Schroeder, D., McVetty, P. B. E. (2010). Inheritance of seed coat color genes in Brassica napus (L.) and tagging the genes using SRAP, SCAR and SNP molecular markers. Mol. Breed. 26, 439–453. doi: 10.1007/s11032-009-9384-6
Rashid, A., Rakow, G., Downey, R. K. (1994). Development of yellow seeded Brassica napus through interspecific crosses. Plant Breed. 112, 127–134. doi: 10.1111/j.1439-0523.1994.tb00660.x
Ravindran, V., Morel, P., Partridge, G., Hruby, M., Sands, J. (2006). Influence of an Escherichia coli-derived phytase on nutrient utilization in broiler starters fed diets containing varying concentrations of phytic acid. Poult. Sci. 85, 82–89. doi: 10.1093/ps/85.1.82
Reddy, M. B., Hurrell, R. F., Juillerat, M. A., Cook, J. D. (1996). The influence of different protein sources on phytate inhibition of nonheme-iron absorption in humans. Am. J. Clin. Nutr. 63, 203–207. doi: 10.1093/ajcn/63.2.203
Reddy, L., Wee, Y.-J., Yun, J.-S., Ryu, H.-W. (2008). Optimization of alkaline protease production by batch culture of Bacillus sp. RKY3 through Plackett–Burman and response surface methodological approaches. Bioresour. Technol. 99, 2242–2249. doi: 10.1016/j.biortech.2007.05.006
Röbbelen, G. (1990) Mutation breeding for quality improvment: a case of study for oilseeds crops. Mutation Breeding Review No. 6. FAO/IAEA. Vienna: Division of Nuclear Techniques in Food and Agriculture. Available at: https://inis.iaea.org/collection/NCLCollectionStore/_Public/24/053/24053434.pdf?r=1
Rolletschek, H., Schwender, J., Konig, C., Chapman, K. D., Romsdahl, T., Lorenz, C., et al. (2020). Cellular plasticity in response to suppression of storage proteins in the Brassica napus embryo. Plant Cell 32, 2383–2401. doi: 10.1105/tpc.19.00879
Roy, J., Shaikh, T., del Río Mendoza, L., Hosain, S., Chapara, V., Rahman, M. (2021). Genome-wide association mapping and genomic prediction for adult stage Sclerotinia stem rot resistance in Brassica napus (L) under field environments. Sci. Rep. 11, 21773. doi: 10.1038/s41598-021-01272-9
Salah, K., Olkhovatov, E. A., Aïder, M. (2019). Effect of canola proteins on rice flour bread and mathematical modelling of the baking process. J. Food Sci. Technol. 56, 3744–3753. doi: 10.1007/s13197-019-03842-2
Salazar-Villanea, S., Bruininx, E. M., Gruppen, H., Hendriks, W. H., Carré, P., Quinsac, A., et al. (2016). Physical and chemical changes of rapeseed meal proteins during toasting and their effects on in vitro digestibility. J. Anim. Sci. Biotechnol. 7, 1–11. doi: 10.1186/s40104-016-0120-x
Sanden, N. C. H., Kanstrup, C., Crocoll, C., Schulz, A., Nour-Eldin, H. H., Halkier, B. A., et al (2024). An UMAMIT-GTR transporter cascade controls glucosinolate seed loading in Arabidopsis. Nat. Plants. 10, 172–179. doi: 10.1038/s41477-023-01598-4
Sanders, A., Collier, R., Trethewy, A., Gould, G., Sieker, R., Tegeder, M. (2009). AAP1 regulates import of amino acids into developing Arabidopsis embryos. Plant J. 59, 540–552. doi: 10.1111/j.1365-313X.2009.03890.x
Sashidhar, N., Harloff, H. J., Jung, C. (2020a). Identification of phytic acid mutants in oilseed rape (Brassica napus) by large-scale screening of mutant populations through amplicon sequencing. New Phytol. 225, 2022–2034. doi: 10.1111/nph.16281
Sashidhar, N., Harloff, H. J., Potgieter, L., Jung, C. (2020b). Gene editing of three BnITPK genes in tetraploid oilseed rape leads to significant reduction of phytic acid in seeds. Plant Biotechnol. J. 18, 2241–2250. doi: 10.1111/pbi.13380
Savoie, L., Galibois, I., Parent, G., Charbonneau, R. (1988). Sequential release of amino acids and peptides during in vitro digestion of casein and rapeseed proteins. Nutr. Res. 8, 1319–1326. doi: 10.1016/S0271-5317(05)80094-6
Schatzki, J., Ecke, W., Becker, H. C., Mollers, C. (2014). Mapping of QTL for the seed storage proteins cruciferin and napin in a winter oilseed rape doubled haploid population and their inheritance in relation to other seed traits. Theor. Appl. Genet. 127, 1213–1222. doi: 10.1007/s00122-014-2292-0
Schlemmer, U., Frolich, W., Prieto, R. M., Grases, F. (2009). Phytate in foods and significance for humans: food sources, intake, processing, bioavailability, protective role and analysis. Mol. Nutr. Food Res. 53 Suppl 2, S330–S375. doi: 10.1002/mnfr.200900099
Schmidt, R., Stransky, H., Koch, W. (2007). The amino acid permease AAP8 is important for early seed development in Arabidopsis thaliana. Planta 226, 805–813. doi: 10.1007/s00425-007-0527-x
Schuster, J., Knill, T., Reichelt, M., Gershenzon, J., Binder, S. (2006). Branched-chain aminotransferase 4 is part of the chain elongation pathway in the biosynthesis of methionine-derived glucosinolates in Arabidopsis. Plant Cell 18, 2664–2679. doi: 10.1105/tpc.105.039339
Shahidi, F., Naczk, M. (1992). An overview of the phenolics of canola and rapeseed: chemical, sensory and nutritional significance. J. Am. Oil Chem. Soc 69, 917–924. doi: 10.1007/BF02636344
Sheng, X., Zhao, Z., Yu, H., Wang, J., Xiaohui, Z., Gu, H. (2011). Protoplast isolation and plant regeneration of different doubled haploid lines of cauliflower (Brassica oleracea var. botrytis). Plant Cell Tiss. Org. Cult. 107, 513–520. doi: 10.1007/s11240-011-0002-z
Shi, C., He, J., Wang, J., Yu, J., Yu, B., Mao, X., et al. (2016). Effects of Aspergillus Niger fermented rapeseed meal on nutrient digestibility, growth performance and serum parameters in growing pigs. Anim. Sci. J. 87, 557–563. doi: 10.1111/asj.12457
Shi, C., He, J., Yu, J., Yu, B., Huang, Z., Mao, X., et al. (2015). Solid state fermentation of rapeseed cake with Aspergillus Niger for degrading glucosinolates and upgrading nutritional value. J. Anim. Sci. Biotechnol. 6, 1–7. doi: 10.1186/s40104-015-0015-2
Shi, L., Song, J., Guo, C., Wang, B., Guan, Z., Yang, P., et al. (2019). A CACTA-like transposable element in the upstream region of BnaA9.CYP78A9 acts as an enhancer to increase silique length and seed weight in rapeseed. Plant J. 98, 524–539. doi: 10.1111/tpj.14236
Shirzadegan, M., Röbbelen, G. (1985). Influence of seed color and hull proportion on quality properties of seeds in Brassica napus L. Fette Seifen Anstrichmittel 87, 235–237. doi: 10.1002/lipi.19850870605
Sidorov, V., Wang, D., Nagy, E. D., Armstrong, C., Beach, S., Zhang, Y., et al. (2022). Heritable DNA-free genome editing of canola (Brassica napus L.) using PEG-mediated transfection of isolated protoplasts. In Vitro Cell. Dev. Biol. Plant 58, 447–456. doi: 10.1007/s11627-021-10236-7
Singh, D., Arora, R., Bhatia, A., Singh, H., Singh, B., Arora, S. (2020). Molecular targets in cancer prevention by 4-(methylthio) butyl isothiocyanate-A comprehensive review. Life Sci. 241, 117061. doi: 10.1016/j.lfs.2019.117061
Singh, B. D., Singh, A. K. (2015). “Association mapping,” in Marker-assisted plant breeding: Principles and practices (Springer, New Delhi), 217–255.
Singh, B., Singh, A., Singh, B., Singh, A. (2015). “Mapping of quantitative trait loci,” in Marker-assisted plant breeding: principles and practices. (Springer, New Delhi), 125–150.
Slawski, H., Nagel, F., Wysujack, K., Balke, D., Franz, P., Schulz, C. (2013). Total fish meal replacement with canola protein isolate in diets fed to rainbow trout (Oncorhynchus mykiss W.). Aquacult. Nutr. 19, 535–542. doi: 10.1111/anu.12005
So, K. K. Y., Duncan, R. W. (2021). Breeding canola (Brassica napus L.) for protein in feed and food. Plants (Basel) 10, 2220. doi: 10.3390/plants10102220
Somers, D. J., Rakow, G., Prabhu, V. K., Friesen, K. R. D. (2001). Identification of a major gene and RAPD markers for yellow seed coat colour in Brassica napus. Genome 44, 1077–1082. doi: 10.1139/gen-44-6-1077
Song, X., Meng, X., Guo, H., Cheng, Q., Jing, Y., Chen, M., et al. (2022b). Targeting a gene regulatory element enhances rice grain yield by decoupling panicle number and size. Nat. Biotechnol. 40, 1403–1411. doi: 10.1038/s41587-022-01281-7
Song, J. H., Shin, G., Kim, H. J., Lee, S. B., Moon, J. Y., Jeong, J. C., et al. (2022a). Mutation of GmIPK1 gene using CRISPR/Cas9 reduced phytic acid content in soybean seeds. Int. J. Mol. Sci. 23, 10583. doi: 10.3390/ijms231810583
Sosulski, F. (1979). Organoleptic and nutritional effects of phenolic compounds on oilseed protein products: a review. J. Am. Oil Chem. Soc 56, 711–715. doi: 10.1007/BF02663047
Starosta, E., Jamruszka, T., Szwarc, J., Bocianowski, J., Jędryczka, M., Grynia, M., et al. (2024). DArTseq-based, high-throughput identification of novel molecular markers for the detection of blackleg (Leptosphaeria Spp.) resistance in rapeseed. Int. J. Mol. Sci. 25, 8415. doi: 10.3390/ijms25158415
Stringam, G. (1980). Inheritance of seed color in turnip rape. Can. J. Plant Sci. 60, 331–335. doi: 10.4141/cjps80-054
Sundaram, M. K., Preetha, R., Haque, S., Akhter, N., Khan, S., Ahmad, S., et al. (2022). Dietary isothiocyanates inhibit cancer progression by modulation of epigenome. Semin. Cancer Biol. 83, 353–376. doi: 10.1016/j.semcancer.2020.12.021
Svitashev, S., Schwartz, C., Lenderts, B., Young, J. K., Mark Cigan, A. (2016). Genome editing in maize directed by CRISPR–Cas9 ribonucleoprotein complexes. Nat. Commun. 7, 1–7. doi: 10.1038/ncomms13274
Tamang, J. P., Cotter, P. D., Endo, A., Han, N. S., Kort, R., Liu, S. Q., et al. (2020). Fermented foods in a global age: East meets West. Compr. Rev. Food Sci. Food Saf. 19, 184–217. doi: 10.1111/1541-4337.12520
Tan, Z., Xie, Z., Dai, L., Zhang, Y., Zhao, H., Tang, S., et al. (2022). Genome-and transcriptome-wide association studies reveal the genetic basis and the breeding history of seed glucosinolate content in Brassica napus. Plant Biotechnol. J. 20, 211–225. doi: 10.1111/pbi.13707
Tayyab, K., Afzal, M., Iqbal, M., Arshad, A., Aslam, S., Batool, M. (2017). Enhanced digestibility of phytase treated canola meal based diet for Labeo rohita fingerlings. J. Anim. Plant Sci. 27, 1386–1393. Available online at: https://www.thejaps.org.pk/docs/v-27-04/43.pdf.
Thompson, L., Liu, R., Jones, J. (1982). Functional properties and food applications of rapeseed protein concentrate. J. Food Sci. 47, 1175–1180. doi: 10.1111/j.1365-2621.1982.tb07643.x
Tong, Z., He, W., Fan, X., Guo, A. (2021). Biological function of plant tannin and its application in animal health. Front. Vet. Sci. 8. doi: 10.3389/fvets.2021.803657
Tripathi, M., Mishra, A. (2007). Glucosinolates in animal nutrition: A review. Anim. Feed Sci. Technol. 132, 1–27. doi: 10.1016/j.anifeedsci.2006.03.003
Van den Bout, I., Divecha, N. (2009). PIP5K-driven PtdIns(4,5)P2 synthesis: regulation and cellular functions. J. Cell Sci. 122, 3837–3850. doi: 10.1242/jcs.056127
Velayudhan, D., Hossain, M., Regassa, A., Nyachoti, C. (2018). Effect of canola meal inclusion as a major protein source in gestation and lactation sow diets with or without enzymes on reproductive performance, milk composition, fecal bacterial profile and nutrient digestibility. Anim. Feed Sci. Technol. 241, 141–150. doi: 10.1016/j.anifeedsci.2018.05.001
Velayudhan, D., Schuh, K., Woyengo, T., Sands, J., Nyachoti, C. (2017). Effect of expeller extracted canola meal on growth performance, organ weights, and blood parameters of growing pigs. J. Anim. Sci. 95, 302–307. doi: 10.2527/jas.2016.1046
Vicient, C. M., Bies-Etheve, N., Delseny, M. (2000). Changes in gene expression in the leafy cotyledon1 (lec1) and fusca3 (fus3) mutants of Arabidopsis thaliana L. J. Exp. Bot. 51, 995–1003. doi: 10.1093/jexbot/51.347.995
Vig, A. P., Walia, A. (2001). Beneficial effects of Rhizopus oligosporus fermentation on reduction of glucosinolates, fibre and phytic acid in rapeseed (Brassica napus) meal. Bioresour. Technol. 78, 309–312. doi: 10.1016/S0960-8524(01)00030-X
Vlassa, M., Filip, M., ăranu, I., Marin, D., Untea, A. E., Ropotă, M., et al. (2022). The yeast fermentation effect on content of bioactive, nutritional and anti-nutritional factors in rapeseed meal. Foods 11, 2972. doi: 10.3390/foods11192972
Von Der Haar, D., Müller, K., Bader-Mittermaier, S., Eisner, P. (2014). Rapeseed proteins–Production methods and possible application ranges. OCL 21, D104. doi: 10.1051/ocl/2013038
Wanasundara, J. P. (2011). Proteins of Brassicaceae oilseeds and their potential as a plant protein source. Crit. Rev. Food Sci. Nutr. 51, 635–677. doi: 10.1080/10408391003749942
Wanasundara, J. P. D., Tan, S., Alashi, A. M., Pudel, F., Blanchard, C. (2017). “Proteins from canola/rapeseed,” in Sustainable protein sources (Academic press, Cambridge, USA), 285–304.
Wang, J., Fan, Y., Mao, L., Qu, C., Lu, K., Li, J., et al. (2021). Genome-wide association study and transcriptome analysis dissect the genetic control of silique length in Brassica napus L. Biotechnol. Biofuels 14, 1–14. doi: 10.1186/s13068-021-02064-z
Wang, J., Jian, H., Wei, L., Qu, C., Xu, X., Lu, K., et al. (2015). Genome-wide analysis of seed acid detergent lignin (ADL) and hull content in rapeseed (Brassica napus L.). PloS One 10, e0145045. doi: 10.1371/journal.pone.0145045
Wang, X., Jin, Q., Wang, T., Huang, J., Xia, Y., Yao, L., et al. (2012). Screening of glucosinolate-degrading strains and its application in improving the quality of rapeseed meal. Ann. Microbiol. 62, 1013–1020. doi: 10.1007/s13213-011-0341-3
Wang, N., Li, F., Chen, B., Xu, K., Yan, G., Qiao, J., et al. (2014). Genome-wide investigation of genetic changes during modern breeding of Brassica napus. Theor. Appl. Genet. 127, 1817–1829. doi: 10.1007/s00122-014-2343-6
Wang, Y., Liu, J., Wei, F., Liu, X., Yi, C., Zhang, Y. (2019). Improvement of the nutritional value, sensory properties and bioavailability of rapeseed meal fermented with mixed microorganisms. LWT-Food Sci. Technol. 112, 108238. doi: 10.1016/j.lwt.2019.06.005
Wang, B., Wu, Z., Li, Z., Zhang, Q., Hu, J., Xiao, Y., et al. (2018). Dissection of the genetic architecture of three seed-quality traits and consequences for breeding in Brassica napus. Plant Biotechnol. J. 16, 1336–1348. doi: 10.1111/pbi.12873
Wang, Z., Yin, L., Liu, L., Lan, X., He, J., Wan, F., et al. (2022). Tannic acid reduced apparent protein digestibility and induced oxidative stress and inflammatory response without altering growth performance and ruminal microbiota diversity of Xiangdong black goats. Front. Vet. Sci. 9. doi: 10.3389/fvets.2022.1004841
Webster, C. D., Tiu, L. G., Tidwell, J. H., Grizzle, J. M. (1997). Growth and body composition of channel catfish (Ictalurus punctatus) fed diets containing various percentages of canola meal. Aquaculture 150, 103–112. doi: 10.1016/S0044-8486(96)01471-8
Wei, D., Cui, Y., Mei, J., Qian, L., Lu, K., Wang, Z. M., et al. (2019). Genome-wide identification of loci affecting seed glucosinolate contents in Brassica napus L. J. Integr. Plant Biol. 61, 611–623. doi: 10.1111/jipb.12717
WHO/FAO/UNU (2007). “Protein and amino acid requirements in human nutrition,” in World Health Organization technical report series (Geneva, Switzerland: World Health Organization), vol. 935. , 1–265.
Wittstock, U., Kliebenstein, D. J., Lambrix, V., Reichelt, M., Gershenzon, J. (2003). “Glucosinolate hydrolysis and its impact on generalist and specialist insect herbivores,” in Recent advances in phytochemistry. Ed. Romeo, J. T. (Elsevier, Amsterdam), 101–125.
Wolfram, K., Schmidt, J., Wray, V., Milkowski, C., Schliemann, W., Strack, D. (2010). Profiling of phenylpropanoids in transgenic low-sinapine oilseed rape (Brassica napus). Phytochemistry 71, 1076–1084. doi: 10.1016/j.phytochem.2010.04.007
Woo, J. W., Kim, J., Kwon, S. I., Corvalán, C., Cho, S. W., Kim, H., et al. (2015). DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 33, 1162–1164. doi: 10.1038/nbt.3389
Woyengo, T. A., Nyachoti, C. M. (2013). Review: Anti-nutritional effects of phytic acid in diets for pigs and poultry – current knowledge and directions for future research. Can. J. Anim. Sci. 93, 9–21. doi: 10.4141/cjas2012-017
Woyengo, T., Weihrauch, D., Nyachoti, C. (2012). Effect of dietary phytic acid on performance and nutrient uptake in the small intestine of piglets. J. Anim. Sci. 90, 543–549. doi: 10.2527/jas.2011-4001
Wu, J. G., Shi, C. H., Zhang, H. Z. (2005). Genetic analysis of embryo, cytoplasmic, and maternal effects and their environment interactions for protein content in Brassica napus L. Aust. J. Agric. Res. 56, 69–73. doi: 10.1071/ar04089
Wurschum, T., Liu, W., Maurer, H. P., Abel, S., Reif, J. C. (2012). Dissecting the genetic architecture of agronomic traits in multiple segregating populations in rapeseed (Brassica napus L.). Theor. Appl. Genet. 124, 153–161. doi: 10.1007/s00122-011-1694-5
Xiao, H., Liu, B., Mo, H., Liang, G. (2015). Comparative evaluation of tannic acid inhibiting α-glucosidase and trypsin. Food Res. Int. 76, 605–610. doi: 10.1016/j.foodres.2015.07.029
Xie, H., Wang, Y., Zhang, J., Chen, J., Wu, D., Wang, L. (2015). Study of the fermentation conditions and the antiproliferative activity of rapeseed peptides by bacterial and enzymatic cooperation. Int. J. Food Sci. Technol. 50, 619–625. doi: 10.1111/ijfs.12682
Xie, M., Zhang, J., Tschaplinski, T. J., Tuskan, G. A., Chen, J. G., Muchero, W. (2018). Regulation of lignin biosynthesis and its role in growth-defense tradeoffs. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01427
Xiong, C., Zou, X., Phan, C.-W., Huang, W., Zhu, Y. (2024). Enhancing the potential of rapeseed cake as protein-source food by gamma irradiation. Biosci. Rep. 44, BSR20231807. doi: 10.1042/BSR20231807
Xu, D., Sanden, N. C. H., Hansen, L. L., Belew, Z. M., Madsen, S. R., Meyer, L., et al. (2023). Export of defensive glucosinolates is key for their accumulation in seeds. Nature 617, 132–138. doi: 10.1038/s41586-023-05969-x
Xu, F., Zeng, X., Ding, X. (2012). Effects of replacing soybean meal with fermented rapeseed meal on performance, serum biochemical variables and intestinal morphology of broilers. Asian Australas. J. Anim. Sci. 25, 1734–1741. doi: 10.5713/ajas.2012.12249
Yang, Y., Zheng, Y., Ma, W., Zhang, Y., Sun, C., Fang, Y. (2023). Meat and plant-based meat analogs: Nutritional profile and in vitro digestion comparison. Food Hydrocoll. 143, 108886. doi: 10.1016/j.foodhyd.2023.108886
Yao, J., Chen, P., Apraku, A., Zhang, G., Huang, Z., Hua, X. (2019). Hydrolysable tannin supplementation alters digestibility and utilization of dietary protein, lipid, and carbohydrate in grass carp (Ctenopharyngodon idellus). Front. Nutr. 6. doi: 10.3389/fnut.2019.00183
Yao, J., Hang, Y., Hua, X., Li, N., Li, X. (2022). Hepatopancreas-intestinal health in grass carp (Ctenopharyngodon idella) fed with hydrolyzable tannin or rapeseed meal. Aquacult. Nutr. 2022, 6746201. doi: 10.1155/2022/6746201
Yoshie-Stark, Y., Wada, Y., Schott, M., Wäsche, A. (2006). Functional and bioactive properties of rapeseed protein concentrates and sensory analysis of food application with rapeseed protein concentrates. LWT-Food Sci. Technol. 39, 503–512. doi: 10.1016/j.lwt.2005.03.006
Young, J., Zastrow-Hayes, G., Deschamps, S., Svitashev, S., Zaremba, M., Acharya, A., et al. (2019). CRISPR-Cas9 editing in maize: systematic evaluation of off-target activity and its relevance in crop improvement. Sci. Rep. 9, 6729. doi: 10.1038/s41598-019-43141-6
Yu, B., Boyle, K., Zhang, W., Robinson, S. J., Higgins, E., Ehman, L., et al. (2016). Multi-trait and multi-environment QTL analysis reveals the impact of seed colour on seed composition traits in Brassica napus. Mol. Breed. 36, 111. doi: 10.1007/s11032-016-0521-8
Yusuf, A. O., Möllers, C. (2024). Inheritance of cellulose, hemicellulose and lignin content in relation to seed oil and protein content in oilseed rape. Euphytica 220, 5. doi: 10.1007/s10681-023-03264-4
Yusuf, H. A., Piao, M., Ma, T., Huo, R., Tu, Y. (2021). Enhancing the quality of total mixed ration containing cottonseed or rapeseed meal by optimization of fermentation conditions. Fermentation 7, 234. doi: 10.3390/fermentation7040234
Zaworska-Zakrzewska, A., Kasprowicz-Potocka, M., Kierończyk, B., Józefiak, D. (2023). The effect of solid-state fermentation on the nutritive value of rapeseed cakes and performance of broiler chickens. Fermentation 9, 435. doi: 10.3390/fermentation9050435
Zeng, D., Liu, T., Ma, X., Wang, B., Zheng, Z., Zhang, Y., et al. (2020). Quantitative regulation of waxy expression by CRISPR/Cas9-based promoter and 5’UTR-intron editing improves grain quality in rice. Plant Biotechnol. J. 18, 2385–2387. doi: 10.1111/pbi.13427
Zentek, J., Boroojeni, F. G. (2020). (Bio) Technological processing of poultry and pig feed: Impact on the composition, digestibility, anti-nutritional factors and hygiene. Anim. Feed Sci. Technol. 268, 114576. doi: 10.1016/j.anifeedsci.2020.114576
Zhang, Y., Li, X., Chen, W., Yi, B., Wen, J., Shen, J., et al. (2010b). Identification of two major QTL for yellow seed color in two crosses of resynthesized Brassica napus line No. 2127-17. Mol. Breed. 28, 335–342. doi: 10.1007/s11032-010-9486-1
Zhang, Y., Li, P., Zhang, J., Li, Y., Xu, A., Huang, Z. (2022). Genome-wide association studies of salt tolerance at the seed germination stage and yield-related traits in Brassica napus L. Int. J. Mol. Sci. 23, 15892. doi: 10.3390/ijms232415892
Zhang, L., Tan, Q., Lee, R., Trethewy, A., Lee, Y. H., Tegeder, M. (2010a). Altered xylem-phloem transfer of amino acids affects metabolism and leads to increased seed yield and oil content in Arabidopsis. Plant Cell 22, 3603–3620. doi: 10.1105/tpc.110.073833
Zhang, G. Q., Tang, G. X., Song, W. J., Zhou, W. J. (2004). Resynthesizing Brassica napus from interspecific hybridization between Brassica rapa and B. oleracea through ovary culture. Euphytica 140, 181–187. doi: 10.1007/s10681-004-3034-1
Zhang, J., Xu, X., Cao, Z., Wang, Y., Yang, H., Azarfar, A., et al. (2019). Effect of different tannin sources on nutrient intake, digestibility, performance, nitrogen utilization, and blood parameters in dairy cows. Anim. (Basel) 9, 507. doi: 10.3390/ani9080507
Zhang, L., Yang, B., Li, X., Chen, S., Zhang, C., Xiang, S., et al. (2024). Integrating GWAS, RNA-Seq and functional analysis revealed that BnaA02. SE mediates silique elongation by affecting cell proliferation and expansion in Brassica napus. Plant Biotechnol. J. 22, 2907–2920. doi: 10.1111/pbi.14413
Zhao, C., Xie, M., Liang, L., Yang, L., Han, H., Qin, X., et al. (2022). Genome-wide association analysis combined with quantitative trait loci mapping and dynamic transcriptome unveil the genetic control of seed oil content in Brassica napus L. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.929197
Zhong, J.-R., Wu, P., Feng, L., Jiang, W.-D., Liu, Y., Kuang, S.-Y., et al. (2020). Dietary phytic acid weakened the antimicrobial activity and aggravated the inflammatory status of head kidney, spleen and skin in on-growing grass carp (Ctenopharyngodon idella). Fish Shellfish Immunol. 103, 256–265. doi: 10.1016/j.fsi.2020.05.037
Zhong, R., Ye, Z.-H. (2003). The SAC domain-containing protein gene family in Arabidopsis. Plant Physiol. 132, 544–555. doi: 10.1104/pp.103.021444
Zhou, X., Dai, L., Wang, P., Liu, Y., Xie, Z., Zhang, H., et al. (2021). Mining favorable alleles for five agronomic traits from the elite rapeseed cultivar zhongshuang 11 by QTL mapping and integration. Crop J. 9, 1449–1459. doi: 10.1016/j.cj.2020.12.008
Zhou, H., Xiao, X., Asjad, A., Han, D., Zheng, W., Xiao, G., et al. (2022). Integration of GWAS and transcriptome analyses to identify SNPs and candidate genes for aluminum tolerance in rapeseed (Brassica napus L.). BMC Plant Biol. 22, 130. doi: 10.1186/s12870-022-03508-w
Zhu, X., Wang, L., Zhang, Z., Ding, L., Hang, S. (2021). Combination of fiber-degrading enzymatic hydrolysis and Lactobacilli fermentation enhances utilization of fiber and protein in rapeseed meal as revealed in simulated pig digestion and fermentation in vitro. Anim. Feed Sci. Technol. 278, 115001. doi: 10.1016/j.anifeedsci.2021.115001
Keywords: rapeseed byproducts, nutritional enhancement, anti-nutritional factors, microbial fermentation, seed storage proteins, breeding strategies, genome editing
Citation: Manikandan A, Muthusamy S, Wang ES, Ivarson E, Manickam S, Sivakami R, Narayanan MB, Zhu L-H, Rajasekaran R and Kanagarajan S (2024) Breeding and biotechnology approaches to enhance the nutritional quality of rapeseed byproducts for sustainable alternative protein sources- a critical review. Front. Plant Sci. 15:1468675. doi: 10.3389/fpls.2024.1468675
Received: 22 July 2024; Accepted: 30 September 2024;
Published: 11 November 2024.
Edited by:
Kanwarpal S. Dhugga, HiBioS Inc., United StatesCopyright © 2024 Manikandan, Muthusamy, Wang, Ivarson, Manickam, Sivakami, Narayanan, Zhu, Rajasekaran and Kanagarajan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ravikesavan Rajasekaran, cmF2aWtlc2F2YW5AdG5hdS5hYy5pbg==; Selvaraju Kanagarajan, c2VsdmFyYWp1LmthbmFnYXJhamFuQHNsdS5zZQ==
 Anandhavalli Manikandan1,2
Anandhavalli Manikandan1,2 Saraladevi Muthusamy
Saraladevi Muthusamy Eu Sheng Wang
Eu Sheng Wang Emelie Ivarson
Emelie Ivarson Sudha Manickam
Sudha Manickam Manikanda Boopathi Narayanan
Manikanda Boopathi Narayanan Li-Hua Zhu
Li-Hua Zhu Selvaraju Kanagarajan
Selvaraju Kanagarajan