
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 21 November 2024
Sec. Plant Development and EvoDevo
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1467596
Paeonia have diverse flower colors, rich flower types, varying bloom periods, and delightful fragrances, rendering them highly valuable for both ornamental and economic purposes in horticulture. Investigating the developmental mechanisms of morphology, flower color, flowering and floral fragrance in Paeonia holds significant value for enhancing their ornamental traits and conducting germplasm improvement. This review provides an overview of research progress on Paeonia flower morphology (including flower bud differentiation, classification, omics applications in shape studies, and functional genes regulating flower morphology), flower colors (omics applications in color research and functional genes regulating flower colors), bloom periods (flower bud dormancy, flowering time), and fragrances (preparation, analysis, components, and molecular biology research of flower fragrances) within the Paeonia. Additionally, it offers a comprehensive analysis of current research challenges and future directions.
Paeonia L. is the only genus in Paeoniaceae and it holds great ornamental and economic value in gardening. The flower is one of the key characteristics that contribute to the ornamental and economic value of Paeonia. Factors that influence the aesthetic and economic value of the flower include its morphology, color, flowering, and floral fragrance. Paeonia exhibit a diverse range of flower morphologies, with a total of 13 different forms (Fan et al., 2020, 2023a). This variety of flower morphologies provides people with a diverse visual experience, enhancing its appeal as an ornamental plant. The flower color of Paeonia plays an important role in determining its ornamental value. Herbaceous peony flowers are known to have nine major color series, including white, yellow, pink, red, purple, ink, green, orange, and multicolor (Figure 1). The flowering period of Paeonia can be divided into five categories: extremely early flowering, early flowering, mid-flowering, late flowering, and extremely late flowering. Paeonia vary greatly in their flowering time and duration. By strategically combining different cultivars in garden design, can create a Paeonia garden with an extended blooming season. Additionally, the fragrant flowers not only provide a delightful sensory experience but also have economic value for use in floral arrangements, sachets, and other decorative purposes.
Differences in flower morphology of Paeonia begin with flower bud differentiation. Methods commonly used to study the differences between flower types include transcriptomics and proteomics. These studies have identified transcription factors and functional genes that may be involved in regulating flower development, providing important insights into the molecular mechanisms underlying flower development in Paeonia (Fan et al., 2020). In the study of flower color, molecular biology approaches have been widely used to explore the regulation of anthocyanins, flavonoids, flavonols, and other flavonoid substances in Paeonia flowers, identifying the functions of related genes (Guo et al., 2019; Zhang et al., 2018a). In research on flowering time, genes regulating flowering, including bud dormancy, bud differentiation, and flower senescence, have been identified (Wu et al., 2018). Studies on floral fragrance have focused on identifying the aromatic components of Paeonia and uncovering genes that may be involved in regulating scent synthesis (Zhao et al., 2023). Although numerous scholars have conducted research on flower morphology, color, flowering time, and floral fragrance in Paeonia, the relationships between these studies are not yet clear. The interactions among identified genes remain unclear, and the research results on the floral organs of Paeonia are not yet systematic.
This review systematically summarizes important research related to floral traits in Paeonia, including flower morphology (flower bud differentiation, flower type classification, application of genomics in flower type research, and functional genes regulating flower morphology), flower color (application of genomics in flower color research, and functional genes regulating flower color), flowering time (flower bud dormancy, and flowering time), and floral fragrance (preparation, analysis, and components of floral fragrance, and molecular biology research on floral fragrance). The compilation in this review can serve as a reference for researchers in the field of Paeonia to conduct studies related to floral trait improvement.
In China, the classification of flower types within the Paeonia exhibits some differences compared to other countries. According to Qin and Li (1990), the flower types of Paeonia were divided into three levels according to the series, class, and type. According to the differences between the wild species of tree peony, cultivars were divided into peony series, spotted peony series, yellow peony series and purple peony series. According to the differences between wild species or populations of herbaceous peony, cultivars were divided into Chinese peony series and European peony series.
Based on the differences in flower structure, tree peonies and herbaceous peonies were further classified into two major categories within each group: single-flower type and double-flower type. Within the single-flower type, they were further divided into the Thousand-layer Subclass and the Lou-zi Subclass based on the origin of the petals, totaling nine flower types. These included Single form, Lotus form, Chrysanthemum form, Rose form, which belong to the Thousand-layer Subclass, and Golden-slamen form, Anemone form, Golden-circle form, Crown form, Globular form, which belong to the Lou-zi Subclass. The double-flower types were divided into the Thousand-layer Tai-ge Subclass and the Lou-zi Tai-ge Subclass, totaling four flower types, including Primary Proliferation form, Color-petalled Proliferation form, Stratified Proliferation form, and Globular Proliferation form. Primary Proliferation form belonged to the Thousand-layer Tai-ge Subclass, while Color-petalled Proliferation form, Stratified Proliferation form, and Globular Proliferation form belonged to the Lou-zi Tai-ge Subclass (Qin and Li, 1990).
Different flower types exhibit distinct morphological differences. Single-petal type: 2-3 rounds of petals, normal development of stamens and pistils (Figure 2A). Lotus type: 4-5 rounds of petals, normal development of stamens and pistils (Figure 2B). Chrysanthemum type: Over 6 rounds of petals, reduced number of stamens, some stamens petaloid (Figure 2C). Rose type: Extreme proliferation of petals, complete loss of stamens or several small stamens in the flower center (Figure 2D). Golden stamen type: Outer petals expanded, 2-3 rounds of petals, stamens dome-shaped (Figure 2E). Tulip type: Outer petals 2-3 rounds, stamens completely petaloid, pistils normal (Figure 2F). Golden ring type: Wide outer petals, stamens transformed into elongated inner petals, a circle of normal stamens remaining between inner and outer petals (Figure 2G). Crown type: Wide outer petals, stamens almost completely petaloid, rounded and prominent (Figure 2H). Hydrangea type: Stamens completely petaloid, forming petals, resembling a hydrangea (Figure 2I).
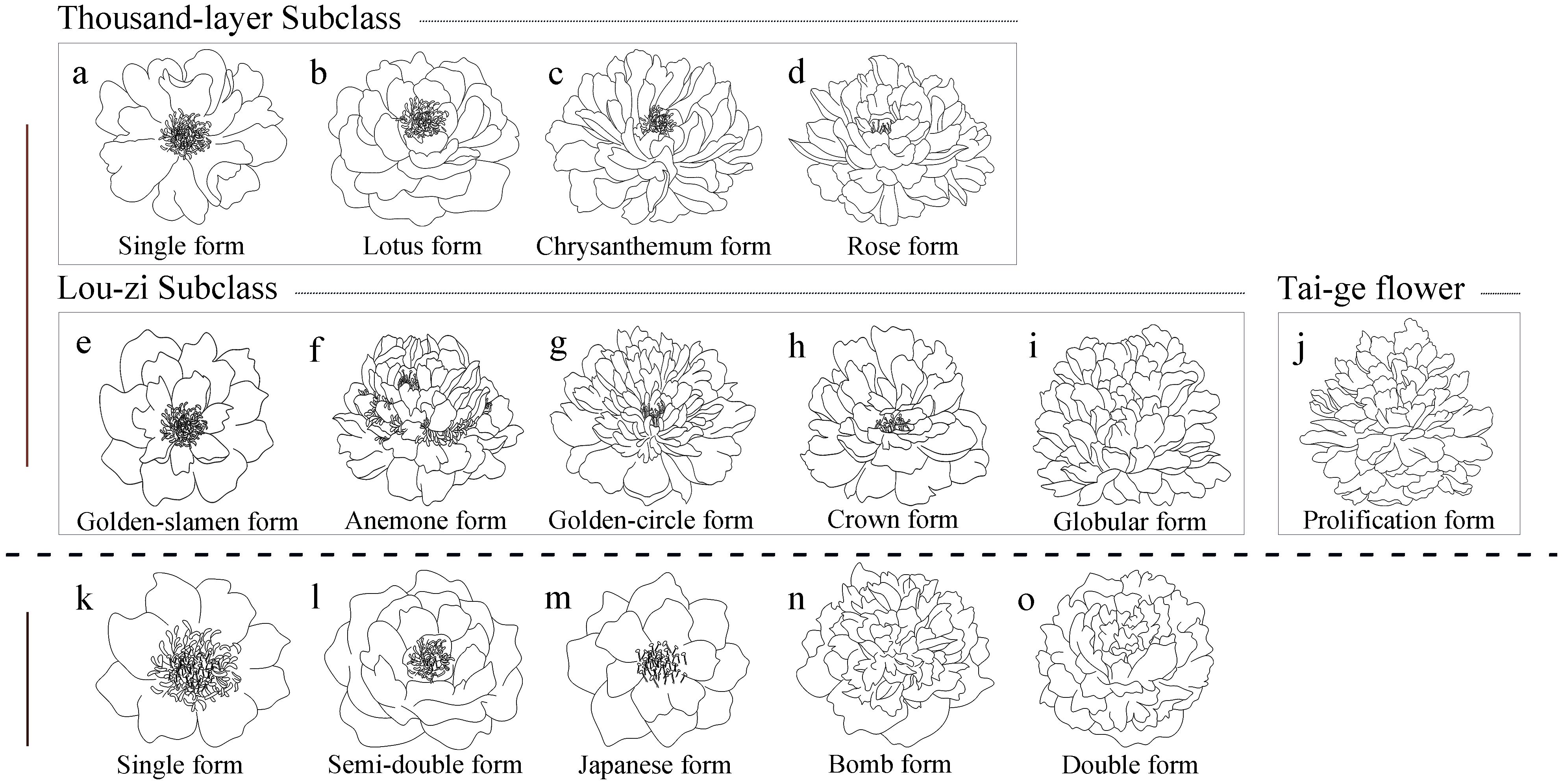
Figure 2. Flower type classification of Paeonia in China and America. (A-J) represent the flower type classification of Paeonia in China; (K-O) represent the flower type classification of Paeonia in America; a-d are the Thousand-layer Subclass; (E-I) are the Lou-zi Subclass, (J) is the Tai-ge flower.
Tai-ge flowers consist of two flowers: an upper flower and a lower flower (Figure 2J). In the initial Tai-ge type, both upper and lower flowers develop normal stamens and pistils, with little difference in petal morphology between inner and outer petals. In the variegated petal Tai-ge type, a few stamens are petalized, some stamens and pistils develop normally in the upper flower, while the pistils of the lower flower are almost completely petalized, with some differences in petal shapes between inner and outer petals. In the layered Tai-ge type, stamens in the upper flower are almost completely petalized or few remain, while pistils develop normally; in the lower flower, both stamens and pistils are almost completely petalized, with noticeable differences in petal shapes between inner and outer petals. In the spherical Tai-ge type, stamens and pistils in both upper and lower flowers are almost completely petalized, with differences in petal shapes between inner and outer petals. The classification of flower types is based on their growth and development stages; once the flowers are fully open, it becomes more difficult to accurately identify the type of Tai-ge flower.
The American Peony Society classifies the flower forms of the Paeonia into five types based on floral structure, including Single form (Figure 2K), Semi-double form (Figure 2L), Japanese form (Figure 2M), Bomb form (Figure 2N), and Double form (Figure 2O).
Flower bud differentiation refers to the process in which the growth status of a plant changes from vegetative bud to reproductive bud. The apical meristem of the bud undergoes a transition from vegetative growth state to reproductive growth state under suitable conditions. When the plant receives a signal to initiate flowering, the apical meristem of the bud transforms into an inflorescence meristem, and then the floral organs begin to develop (Coen and Meyerowitz, 1991). During the nutritional growth process of Paeonia, the apex of the stem (or bud) is pointed in the vegetative growth stage. Upon entering the reproductive growth stage, the apex of the stem (or bud) becomes relatively blunt. The protrusion formed at the edge of the apical meristem is the bract primordium. After the formation of the bract primordium, protuberances gradually appear on its inner side, which are the sepals primordium. As the floral bud continues to develop, the primordia of petals gradually appear on the inner side of the sepals primordium, and the layers of petal primordia correspond to the number of petals. The stamen primordium begins to appear after the formation of petal primordia. Before the stamen primordium is fully formed, the carpel primordium also begins to develop. The process can be divided into six stages: floral bud differentiation initiation stage, bract primordium differentiation stage, sepal primordium differentiation stage, petal primordium differentiation stage, stamen primordium differentiation stage, and carpel primordium differentiation stage (Table 1) (Zhang et al., 2019). Herbaceous peony and tree peony have the same floral bud differentiation process, but the tree peony floral bud differentiation period is earlier and the process takes less time compared to herbaceous peony (He et al., 2014). In terms of research methods, the main approaches used to observe floral bud differentiation include paraffin sectioning, cryosectioning, and stereomicroscopy (Zhang et al., 2019). Some studies suggest that paraffin sectioning provides clearer images than cryosectioning, while stereomicroscopy is the simplest and quickest among the three methods (Zhang et al., 2019). Research on floral bud differentiation not only allows for an understanding of the process in Paeonia from a morphological perspective but also summarizes the regularities in floral pattern formation. It provides reference for sampling in further molecular biology studies (Fan et al., 2021). Additionally, studying floral bud differentiation helps in understanding the floral types of Paeonia, providing guidance for selecting parents in hybrid breeding programs (He et al., 2014).
With the advancement of molecular biology techniques, research on flower organ development in Paeonia is increasing (Fan et al., 2020). The genome of Paeonia suffruticosa ‘Luo Shen Xiao Chun’ has been sequenced and published. Genome assembly and de novo transcriptome assembly identified a total of 77 MADS-box genes, including 44 type I and 33 type II genes. Among the type II genes, 18 were identified as ABCDE genes, comprising 5 A class genes, 5 B class genes, 2 C/D class genes, and 6 E class genes. Among the 5 B class genes, apart from the PISTILLATA, B-PI (PsuMADS6) gene, the other four B class genes showed high expression levels in flowers. The APETALA3 (AP3) gene (PsuMADS32 or TRINITYpsu14) and B-PI (TRINITYpsu13) exhibited similar expression levels in flowers and flower buds, suggesting that their encoded proteins form heterodimers during petal and stamen development. The TOMATO MADS-BOX 6, B-TM6 played a crucial role in stamen petaloidy and floral morphogenesis, indicating its partial functionality in C-class genes. Studies have shown that C-class gene expression levels are low in floral organs of P. suffruticosa ‘Luo shen xiao chun’, suggesting a potential association with observed stamen petaloidy in this peony cultivar. When C-class gene functions were limited, TM6 expression ensures proper carpel development. The research speculated that AP3/PI and TM6 genes together determined tree peony petal and stamen formation, as well as their functional interconversion when C-class gene function was restricted (Lv et al., 2020). Additionally, the genome of P. ostii has been sequenced, with a genome size exceeding 10 Gb. Studies indicated that ectopic expression of A-class gene APETALA1 (AP1) and reduced expression of C-class gene AGAMOUS (AG) might have contributed to petaloid stamen formation (Yuan et al., 2022).
Transcriptome, proteome, microRNA, and degradome studies have been extensively applied in the research of floral organ development (Table 2). In tree peonies, omics studies have identified several genes belonging to the MADS-box gene family, including PsAP1, APETALA2 (PsAP2), PsAP3, PsPI, PsAG, SEPALLATA3 (PsSEP3), and AGAMOUS-LIKE (AGLs), categorized as A, B, C, and E class genes within the ABCDE model of floral organ development (Wang et al., 2019). For instance, a transcriptome sequencing study of petals from P. suffruticosa ‘Luo yang hong’ at bud and full blooming stages identified 71 candidate genes across 4 groups potentially linked to tree peony petal formation or development. Specifically, Group 1 (MADS-box genes, SQUAMOSA promoter-binding-protein (SBP) genes, teosinte branched1/cycloidea/proliferating cell factors (TCP) genes) and Group 2 (trihelix genes, basic region/leucine zipper motif (bZIP) genes, B3 genes, cysteine-rich polycomb-like protein (CPP) genes) directly influence floral organ formation, while Group 3 (light signal transduction-related genes) and Group 4 (plant hormone-related genes) were involved in light signal transduction-mediated and plant hormone-mediated processes during floral organ development (Li et al., 2017). Furthermore, members of transcription factor families such as basic helix-loop-helix (bHLH), GRAS, homeodomain-leucine zipper (HD-ZIP), leucine-rich repeat (LRR), v-myb avian myeloblastosis viral oncogene homolog (MYB), NAC, Ring Finger Protein 1 (RING1), Trihelix, WD, and WRKY also played crucial roles in peony floral development (Liu et al., 2019; Guo et al., 2022).
In herbaceous peonies, floral organ development genes identified through genomic studies were predominantly members of the MADS-box gene family, including PlAP3, PlDEFA, PlPI2, PlAG-1, PlSEP3, PlSEP1-1, and PlSEP1-2. Additionally, nine other transcription factor families were also participate in herbaceous peony floral organ development, namely TCP, bHLH, ICE, LBD, NAC, BLH, PDF, GBF, and IIIA (Wu et al., 2018; Fan et al., 2021). Genomic research has identified candidate genes involved in Paeonia floral development, laying a foundation for further studies on gene function.
In studies related to functional genes involved in floral organ formation in tree peony, genes expressed in sepals included PsAP1, PsAGL6, and PsMADS5. Genes expressed in petals included PsAP1/PsAP1fd, PsAP2/PsAP2fd, PsAP3/PsAP3fd, PsPI/PsPIfd, PsMADS1, PsMADS5, PSMADS9, and PsAGL6. Genes expressed in stamens included PsAG/PsAGfd, PsAP3/PsAP3fd, PsMADS1, PsPI/PsPIfd, PsMADS5, PsAGL6, and PsTM6. Genes expressed in carpels/ovules included PsAP1, PsMADS1, PsAG/PsAGfd, PsMADS5, and PsAGL6. Additionally, E-class genes PsSEP1/PsSEP1fd, PsSEP3, and PsSEP4 were expressed in four floral organs. These genes collectively participate in the process of organ formation in peony flowers (Ren, 2011; Shu et al., 2012; Tang et al., 2018a; Zhang et al., 2015a; Wang et al., 2019; Zhou et al., 2022). Research indicated that in tree peonies, PsAP1 and PsAP2 were primarily expressed in sepals and bracts, with weak expression in petals and carpels, and almost no expression in stamens. PsAP3 and PsPI exhibit strong expression in petals and stamens, with lower expression in sepals and carpels. PsAG showed high expression in stamens and carpels. PsSEP1, PsSEP3, and PsSEP4 were expressed in all four floral organs. This suggested that PsAP1 and PsAP2 were involved in sepal development, PsAP3 and PsPI in petal and stamen development, and AG in stamen and carpel development. PsSEP1, PsSEP3, and PsSEP4 participated in the development of all four floral organs (Wang et al., 2019). In P. ostii ‘Feng Dan’, the A-class gene PsAP1fd was mainly expressed in sepals and petals, while PsAP2fd was predominantly expressed in tree peony petals, indicating their likely involvement in sepal development. The B-class genes PsAP3fd and PsPIfd exhibited strong expression in stamens and petals, respectively, with PsPIfd showing the highest expression in petals, suggesting their roles in petal and stamen development. The C-class gene PsAGfd showed the highest expression in carpels. The E-class gene PsSEP1fd was predominantly expressed in carpels (Zhou et al., 2022). The expression patterns of these genes in tree peonies demonstrate that the regulation of floral organ development follows a predictable pattern according to the ABCE model (Figure 3).
In herbaceous peonies, the expression levels of A-class gene PlAP1 was highest in sepals, lower in petals, and very low in stamens and carpels. The expression of PlAP2 gene was highest in carpels, lower in sepals, and very low in petals and stamens. Among B-class genes, PlAP3-2 and PlPI showed highest expression in stamens, lower in petals, and very low in sepals and carpels. PlAP3-1 exhibited descending expression levels across floral organs, highest in petals, followed by stamens, carpels, and sepals, with its expression in floral organs 100 times lower than that of PlAP3-2. PlSEP3 was expressed in all floral organs, with levels highest in sepals, followed by carpels, petals, and stamens (Ge et al., 2014). Additionally, research indicated that AP3-PI obligate heterodimerization played a crucial role in the determination of organ identity in corolla and stamens (Gong et al., 2017).
With deeper research, certain genes related to floral organ development in Paeonia have been isolated, while studies on functional genes governing tree peony flower development are more advanced compared to those on herbaceous peony. Although the functions of some genes have been verified to participate in the process of peony floral organ development (Table 3), their specific roles across different floral organs and their interactions with other genes remain unclear.
In Paeonia, the different flower colors are closely associated with the types of pigments present in petal cells, cellular pH values, epidermal cell shapes, and intracellular physiological environments. The main determinants of flower color are the types and concentrations of pigments. Paeonia flowers primarily contain anthocyanins, flavonoids, flavonols, and other flavonoid substances, among which anthocyanins play a key role in determining flower color (Wang et al., 2023). With the advancement of molecular biology technologies, applications of omics such as transcriptomics, proteomics, microRNA analysis, Simplified Genome-Specific Locus Fragment Sequencing (SLAF-seq), DNA methylation profiling, and others have provided effective research methods for elucidating the biosynthesis pathways and regulatory genes involved in flavonoid and anthocyanin glycoside production related to Paeonia flower colors. These approaches also facilitate the discovery of new genetic resources and the clarification of genetic evolution patterns.
In studies of flower color in Paeonia, transcriptomics was the most widely utilized approach, while proteomics, microRNA, DNA methylation, and SLAF-seq were less commonly applied. Research indicated that multiple genes were involved in tree peony flower color formation, including flavanone 3-hydroxylase gene (F3H), dihydroflavonol 4-reductase (DFR), anthocyanidin synthase (ANS), anthocyanidin 3-O-glucosyltransferase (3GT), THC 2′-glucosyltransferase (THC2’GT), SQUAMOSA promoter-binding protein-like (SPL2), flavone synthase, (FNS II), chalcone synthase (CHS), chalcone isomerase (CHI), F2H, flavanone-3-hydroxylase (F3′H) and PsDFR, among others. These genes regulated the synthesis of flavonoids and anthocyanins, thereby influencing flower color presentation (Shi et al., 2015; Luo et al., 2022; Zou et al., 2023; Zhang et al., 2018a). In addition, some transcription factors MYB, bHLH and WD40 were also involved in the regulation of floral color (Gu et al., 2019. PoMYB2 and PoSPL1 were reported to negatively regulate the accumulation of anthocyanins by modulating the activity of the MYB-bHLH-WDR complex (Gao et al., 2016). Among different-colored peonies, numerous genes showed differential expression, potentially directly or indirectly participated in the formation and regulation of floral color. UDP-glucosyltransferases (UGTs) such as PhUGT78A22 was reported to play important roles in floral color formation, participating in glycosylation reactions of flavonoids and anthocyanins (Li et al., 2022a). Methylation modifications played crucial regulatory roles in the formation of spots on tree peony petals, potentially influencing the expression of structural genes (Zhu et al., 2023) (Table 4). Key genes and regulatory mechanisms influencing the formation of flower color in herbaceous peonies primarily focus on the biosynthetic pathways of flavonoids and anthocyanins, as well as the regulatory roles of transcription factors. Genes such as phenylalanine ammonialyase gene (PlPAL), flavonol synthase gene (PlFLS), PlDFR, PlANS, Pl3GT, and anthocyanidin 5-O-glucosyltransferase gene (Pl5GT) were involved in the biosynthetic pathways of flavonoids and anthocyanins, playing crucial roles in the formation of yellow flower color (Zhao et al., 2014). Transcription factors PsMYB111, PsMYB4 and PsSPL9 regulated the expression of structural genes, influencing the synthesis and accumulation of pigments in herbaceous peony flowers. Research had found that by modulating the expression levels of specific genes, it was possible to induce or inhibit the accumulation of particular pigments, thereby affecting the presentation of herbaceous peony flower colors (Luo et al., 2021a). Transcriptome sequencing and association analysis have revealed SNPs closely associated with herbaceous peony flower color, providing crucial information for genetic improvement of herbaceous peony coloration (Liu et al., 2022). miR156e-3p might participate in the regulation of yellow flower color formation by modulating specific genes such as SPL1 (Zhao et al., 2017).
Research indicated that glutathione S-transferases transporter of anthocyanin, PsGSTF3, interacted with PsDFR, jointly promoting petal coloring in P. suffruticosa ‘Zhao Fen’ (Han et al., 2022). PsDFR and PsANS played crucial roles in the synthesis of flower pigments in P. suffruticosa, leading to the transformation of flower color from white to red (Figure 4A) (Zhao et al., 2015). PsMYB58 interacted with PsbHLH1 and PsbHLH3 in P. suffruticosa ‘Er qiao’. When expressed heterologously in tobacco, PsMYB58 promoted the expression of anthocyanin biosynthetic genes and upregulates the expression of the anthocyanin regulatory bHLH, NtAN1b, indicating that PsMYB58 functioned as a regulator of anthocyanin biosynthesis in tree peony. PsMYB58 played a significant role in the formation of tree peony flower color (Zhang et al., 2021a). PsMYB44 negatively regulated anthocyanin biosynthesis by directly binding to the PsDFR promoter and inhibiting petal spot formation in P. suffruticosa ‘High noon’, thus elucidating the molecular regulatory network mediated by anthocyanins in plant spot formation (Luan et al., 2023). In the petals of P. suffruticosa, PsMYBPA2 and PsbHLH1-3 activated the expression of PsF3H, providing precursor substrates for anthocyanin biosynthesis and blotch formation. Meanwhile, PsMYB21 activated the expression of both PsF3H and PsFLS, promoting flavonol biosynthesis. Its significantly high expression in the non-blotch region inhibited blotch formation by competing for substrates involved in anthocyanin biosynthesis. Conversely, its lower expression in the blotch region facilitated blotch development. Additionally, PsMYB308 interferes with the expression of PsDFR and PsMYBPA2 by competitively binding to PsbHLH1-3 with PsMYBPA2, thereby preventing anthocyanin biosynthesis and blotch formation in P. suffruticosa petals (Figure 4B) (Luan et al., 2024). Exogenous glucose improved the coloration of cut flowers of P. suffruticosa ‘Tai Yang’ by affecting the accumulation of flavonoids and anthocyanins through signal transduction pathways. Studies indicated that ectopic expression of PsMYB2 in tobacco led to strong pigmentation in petals and stamens, while in Arabidopsis, ectopic expression enhances the ability to accumulate anthocyanins induced by glucose in seedlings (Zhang et al., 2022b). anthocyanin O-methyltransferase (PsAOMT) and PtAOMT, two homologous genes, exhibited similar substrate preferences for anthocyanins in vitro. Transgenic tobacco expressing PsAOMT showed significant differences in flower color compared to wild-type tobacco. PsAOMT played a crucial role in flower color formation in P. suffruticosa ‘Gunpohden’ (purple-flowered) (Du et al., 2015).
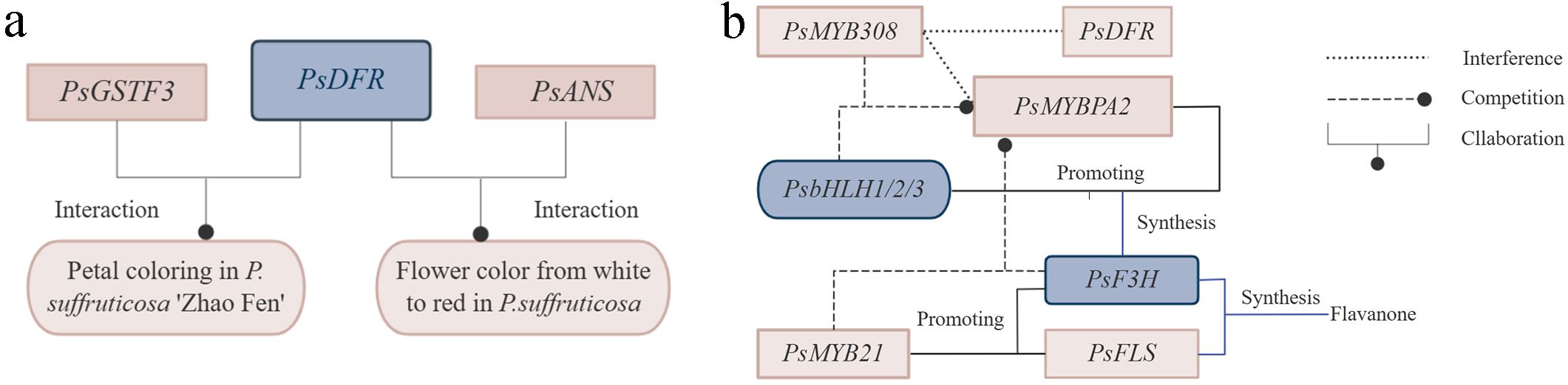
Figure 4. Genetic regulation of petal coloring, flower color and petal spot formation in tree peony. (A) Genetic regulation of petal coloring and flower color formation. (B) Genetic regulation of petal spot formation.
In P. lactiflora ‘Hong Yan Zheng Hui’ (purplish-red), ‘Huang Jin Lun’ (yellow), and ‘Yu Lou Hong Xing’ (white), research divided the flower development process into flower-bud stage, initiating bloom, bloom stage, and withering stage. Phenylalanine ammonia-lyase (PlPAL) showed high expression levels in the flower-bud stage and withering stage. PlCHS and PlCHI maintain high expression levels throughout all developmental stages. PlF3H, PlF3’H, PlDFR, PlANS, and UDP-glucose: flavonoid 5-o-glucosyltransferase (PlUF5GT) gradually decreased as the flowers develop. PlUF5GT exhibited a gradual increase during flower development. Among them, PlCHI promoted the accumulation of chalcones, playing a crucial role in the formation of yellow flowers. The significant expression of upstream genes and low expression of DFR led to the synthesis of abundant anthoxanthins and a small amount of colorless anthocyanins. Due to high expressions of PlDFR, PlANS, and PlUF3GT, numerous colored anthocyanins were converted from anthoxanthins (Zhao et al., 2012a). The reduced expression of PlF3′H, PlDFR, and PlANS following daminozide treatment might have decreased anthocyanin accumulation, ultimately resulting in reduced red color intensity in P. lactiflora ‘Fen Zhu Pan’ (Tang et al., 2018b). Research showed that ATP-citrate lyase (ACL) PlACLB2 promoted anthocyanin accumulation by increasing the abundance of its precursor substrate acetyl-CoA, thereby regulating the formation of red petals in P. lactiflora (Luan et al., 2022). Melatonin played a crucial role in the formation of herbaceous peony flower color. The tryptophan decarboxylase gene (TDC) gene was the rate-limiting gene in melatonin biosynthesis. Herbaceous peony petals were rich in melatonin, with the highest content found in white cultivars, followed by black, red, and pink cultivars. Melatonin levels peak during flowering, and the expression pattern of the TDC correlated positively with melatonin production (Zhao et al., 2018a). These studies can provided important theoretical foundations for understanding the molecular mechanisms regulating Paeonia flower coloration, thereby offering valuable insights for flower color improvement and breeding. Future research could further explore the differences in flower color formation among different cultivars, uncover additional key genes and transcription factors, and elucidate their regulatory networks.
Dormancy is the primary obstacle to flowering in Paeonia, and studying the dormancy mechanisms of tree peonies and herbaceous peonies is of great significance for understanding their flowering mechanisms. In tree peonies, Huang et al. (2008a) screened for genes that were highly expressed in dormancy-released buds, ultimately identifying 31 unique genes, of which 25 sequences were identified as potentially involved in the dormancy release process. DNA methylation was identified as an important epigenetic regulatory factor influencing gene expression. Chilling accumulation promoted bud dormancy release and sprouting through DNA methylation modification of specific genes (Zhang et al., 2021b). Studies indicated that the SUPPRESSOR OF OVEREXPRESSION OF CO1 (PsSOC1) exhibits high expression in early flowering leaves of tree peonies and showed a high hybridization signal in tender leaves of dormant buds. PsSOC1 accelerates bud dormancy release and flowering, and its overexpression in Arabidopsis resulted in early flowering (Zhang et al., 2015b). Long-term cold exposure (0-4°C) activated the Pentose Phosphate Pathway (PPP), promoting dormancy release in P. suffruticosa ‘Lu he hong’ (Zhang et al., 2018b). The mitochondrial phosphate transporter gene of P. suffruticosa ‘Lu He Hong’, mitochondrial phosphate transporters (PsMPT), when ectopically expressed in Arabidopsis, accelerated growth and advanced flowering time. PsMPT played a crucial role in the dormancy release process of peony buds (Huang et al., 2008b). The transcription factor MYB-like protein, PsMYB1, was expressed in all tissues of P. suffruticosa ‘Lu He Hong’ and responds to 4°C cold temperatures. PsMYB1 was implicated not only in dormancy release and eco-dormancy but also in tissue development (Zhang et al., 2015c). In P. suffruticosa ‘Lu He Hong’, PsmiR172b negatively regulated bud dormancy release, whereas TARGET OF EAT (PsTOE3) promoted bud dormancy release. Genes associated with bud dormancy release, including EARLY BUD-BREAK 1 (PsEBB1), EARLY BUD-BREAK 3 (PsEBB3), D-type cyclins (PsCYCD) and β-1,3-glucanase (PsBG6), were upregulated. Ectopic expression of PsmiR172b-PsTOE3 in Arabidopsis conservatively regulated flowering (Zhang et al., 2023).
In herbaceous peony, the molecular mechanisms underlying dormancy release in herbaceous peony buds were studied later compared to tree peony. Zhang et al. (2015d) pioneered the construction of transcriptome libraries using RNA-seq technology for different stages of underground bud dormancy in the low-chill P. lactiflora ‘Hang Bai Shao’, providing a comprehensive insight into the transcriptome profiles during dormancy release. A set of target genes related to bud dormancy release were identified, involving genes associated with environmental response, energy and substance metabolism, and cell growth and development. Subsequently, analysis focused on 66 candidate genes related to light and temperature during herbaceous peony bud dormancy release, with expression pattern analysis of 12 genes across different dormancy stages, suggesting the involvement of SOC1 and WRKY33 in the low-temperature dormancy release process of P. lactiflora ‘Hang Bai Shao’ buds (Zhang et al., 2017). Subsequently, researchers utilized P. lactiflora ‘Da Fu Gui’ to conduct transcriptome sequencing of underground buds at three different stages: physiological dormancy, ecological dormancy, and sprouting. They identified a set of differentially expressed genes related to dormancy and focused particularly on annotating and analyzing genes associated with hormones (Guo et al., 2017).
Treatment of P. suffruticosa ‘Qiu Fa No. 1’ with gibberellin (GA) plus nutrient can advance its autumn reflowering time by 38 days. During this process, GA induced peony reflowering, while the fertilizer provided sustained nutrients for the flowering process. At the transcriptional level, SUPPRESSOR OF CONSTANS OF OVEREXPRESSION1 (PsSOC1) and LEAFY (PsLFY) played critical integrating roles in gibberellin-induced bud differentiation and nutrient supply from fertilization, respectively. GA20 OXIDASE (PsGA20ox) was primarily involved in the GA signaling pathway, while GA INSENSITIVE (PsGAI) potentially modulated flower formation post-fertilization. Sugar signaling pathways also participated in this process, emphasizing the requirement for adequate nutrient supply during early gibberellin-induced autumn reflowering of tree peonies (Xue et al., 2022). SHORT VEGETATIVE PHASE (PsSVP) negatively regulated flowering and positively regulated leaf growth in P. suffruticosa ‘Luoyang Hong’. Gibberellic ACID 3 (GA3) improved floral bud development by suppressing PsSVP expression. High expression of the PsSVP gene may contributed to floral bud abortion in tree peonies (Wang et al., 2014).
The leaves of P. ostii ‘Fengdan’ were sprayed with brassinolide at concentrations of 25 μg/L, 50 μg/L, 100 μg/L, and 200 μg/L. Significant flowering delay was observed with brassinolide at 50 μg/L. Transcriptome and miRNA sequencing of P. ostii ‘Fengdan’ leaves treated with different brassinolide concentrations identified six miRNAs and their target genes potentially involved in regulating the flowering of P. ostii. These include four novel miRNAs targeting SPA1, miR156b targeting SPL, and miR172 targeting AP2 (Zhang et al., 2022c). Additionally, research indicated that in P. ostii ‘Fengdan’, transgenic plants with pCAMBIA2300-PoVIN3 exhibit significantly earlier flowering compared to the wild type, suggesting a role for the PoVIN3 gene in promoting flowering (Li et al., 2022b). Overexpression of the PoFLC gene in Arabidopsis resulted in significantly delayed bolting and flowering compared to the wild type, indicating an important role for the PoFLC gene in delaying flowering processes (Fan et al., 2023b). Overexpression of the gene CONSTANS (PlCO) from P. × lemoinei ‘High Noon’ in Arabidopsis resulted in earlier flowering, indicating its promotive role in tree peony flowering. PlCO protein localized to the nucleus and exhibited transcriptional activity, suggesting that PlCO may function as a transcription factor (Chang et al., 2022). Transcriptome sequencing was conducted on P. lemoinei ‘High Noon’ and non-reblooming P. suffruticosa ‘Luo Yang Hong’, identifying 59,275 and 63,962 unigenes, respectively. Integrated analysis and RT-PCR validation identified several genes potentially playing crucial roles in tree peony reblooming, including FLOWERING LOCUS T (PsFT), VERNALIZATION INSENSITIVE 3 (PsVIN3), PsCO and GIBBERELLIN 20-OXIDASE (PsGA20OX) (Zhou et al., 2013).
Dormancy breaking treatment of P. lactiflora ‘Taebaek’ in November (0°C for 6 weeks) advanced flowering to early spring. However, similar treatments in August, September, or October resulted in flower bud abortion or deformities. Research indicated that a 2-week pre-chilling at 10°C followed by dormancy breaking treatment was the optimal method for promoting autumn or winter flowering of herbaceous peonies without bud abortion (Park et al., 2015).
Research indicated that the duration of cut flower bloom period was closely related not only to harvest time and storage methods but also to the addition of exogenous substances post-harvest, which could enhance cut flower quality and prolong the bloom period. During the cut flower opening process of P. suffruticosa ‘Luoyang Hong’, studies have found that ethylene inhibits the expression of Ethylene resistant (Ps-ETR1-1) and Ethylene insensitive (Ps-EIN3-1) genes. Additionally, Ps-ETR1-1 expression was significantly inhibited by 1-MCP, with a slight induction observed in Ps-EIN3-1 gene expression. This suggests that the proteins encoded by Ps-ETR1-1 and Ps-EIN3-1 played important roles in ethylene sensitivity and response processes in tree peony cut flowers. These findings were crucial for extending the cut flower bloom period (Zhou et al., 2010). Furthermore, research indicated that PsEIL2, PsEIL3, and PsERF1 might also have played roles in regulating ethylene response in P. suffruticosa ‘Luoyang Hong’ (Wu et al., 2017).
Treatment of fresh-cut flowers of P. lactiflora ‘Hongyan Zhenghui’ with nano-silver increased the soluble protein content inside the flowers. This enhancement promoted the activities of antioxidant enzymes such as superoxide dismutase (SOD), catalase (CAT), and ascorbic acid peroxidase (APX), thereby reducing the accumulation of reactive oxygen species such as malondialdehyde (MDA), superoxide radicals (O2·−), and hydrogen peroxide (H2O2). Furthermore, nano-silver treatment exerted inhibitory effects on the growth and reproduction of microorganisms at the stem base of cut flowers, thereby delaying stem blockage. Additionally, AQUAPORIN genes (AQPs) induced by nano-silver treatment played a crucial role in maintaining water balance in cut flowers (Zhao et al., 2018b). Moreover, research has shown that both sucrose transporter genes and invertase genes were involved in maintaining vase quality in cut flowers, specifically in P. lactiflora ‘Yang Fei Chu Yu’ (Xue et al., 2018).
Silencing PlZFP in tobacco petals prolonged flower opening time, while overexpressing PlZFP shortened flower lifespan. This process involved downregulation of genes related to plant hormone biosynthesis, including PlACO1, PlACO3, PlACS1, PlNCED2 and PlAAO, along with increased transcript levels of PlGA3ox1. Conversely, overexpression of PlZFP in tobacco flowers exhibited opposite effects on these genes. Additionally, PlZFP directly bound to the PlNCED2 promoter. Silencing PlNCED2 delayed senescence in petal discs. Reduction of PlZFP decreased ethylene and ABA levels but increased GA levels in herbaceous peony petal discs. Furthermore, exogenous ABA, ethephon, and paclobutrazol (PAC), a GA inhibitor, accelerated senescence in PlZFP-silenced discs. Collectively, the PlZFP-PlNCED2 regulatory checkpoint likely enhanced ABA production and modulates the interplay of ABA with GA and ethylene during flower senescence in cut P. lactiflora ‘Hangshao’ (Ji et al., 2023).
Paeonia are known for their rich floral fragrances, with various aromatic profiles identified in tree peony flowers, including grassy scent, fruity scent, woody scent, rose scent, lily of the valley scent, phenolic scent, and unidentified scent (Li et al., 2012, 2021). In herbaceous peony fragrance research, six types of fragrances have been identified, namely woody scent, fruity scent, lily scent, rose scent, orange blossom scent, and mixed scent (Song et al., 2018; Zhao et al., 2023).
The composition of floral fragrance was complex and diverse, primarily including alcohols, aldehydes, terpenes, esters, and volatile phenols, which often exhibit significant diversity in plants (Li et al., 2012; Luo et al., 2020). Currently, common methods for aroma collection and component analysis in Paeonia included Headspace Solid-Phase Microextraction (HS-SPME) and Dynamic Headspace Sampling (DHS). Widely used analytical methods included artificial olfactory systems, GC-MS, and Automated Thermal Desorber-Gas Chromatography-Mass Spectrometry (ATD-GC/MS) (Table 5) (Zhao et al., 2012b; Li et al., 2023). The headspace technique was favored in fragrance preparation due to its automation, ability to minimize sample matrix interference, and simplicity in sampling, thus it was widely applied in volatile compound preparation. GC-MS offered high sensitivity and simultaneous qualitative and quantitative analysis capabilities, contributing to its widespread use in fragrance component analysis.
The fragrance composition of Paeonia was complex and diverse (Table 5). In tree peonies, the main components responsible for the grassy scent in petals were ocimene, for fruity scent it was 2-ethyl hexanol, for musky scent it was cis-ocimene/longifolene, for rose scent it was D-citronellol, for lily scent it was linalool, for phenolic scent it was 1,3,5-trimethoxybenzene, and for unknown scent it was Pentadecane (Li et al., 2012, 2021). In herbaceous peonies, the primary components responsible for woody scent were β-caryophyllene, for fruity scent it was phenylethyl alcohol, for lily scent it was linalool, for rose scent it was (R)-citronellol, and for orange blossom scent it was nerol (Song et al., 2018; Zhao et al., 2023). Additionally, terpenes were the most abundant compounds in peony petal volatiles (Li et al., 2012), and major fragrance components in peonies also include geraniol and 2-phenylethyl alcohol (Zhao et al., 2023). In conclusion, the fragrance components of Paeonia are complex and diverse, and further research is needed to understand their biosynthetic pathways and gene regulation mechanisms.
Paeonia flower fragrance is a quantitatively controlled trait influenced by multiple genes, with a complex genetic regulatory mechanism. Studying the biosynthetic pathways of floral fragrance compounds is crucial for understanding the molecular mechanisms underlying fragrance formation in peonies. In tree peonies, linalool was identified as one of the dominant volatile compounds in the fragrance of members of the subsection Delavayanae and subsect. Delavayanae hybrids. Five terpene synthases (TPSs) (PdTPS1, PdTPS2, PdTPS3, PdTPS4, and PdTPS5) were identified through global transcriptome sequencing analysis of 11 wild tree peony genotypes and 60 cultivars, potentially involved in the biosynthetic pathway of linalool. Among these, PdTPS1, PdTPS2, and PdTPS4 showed significant positive correlations with linalool emissions across tree peony cultivars. Biochemical data analyses confirmed that PdTPS1 and PdTPS4 were key genes determining linalool synthesis (Li et al., 2023). Furthermore, expression levels of enzymes crucial for terpene skeleton biosynthesis, including acetoacetyl-CoA thiolase (AACT), 3-Hydroxy-3-methylglutaryl-coenzyme A reductase (HMGR), phosphomevalonate kinase (PMK), 1-Deoxyxylulose-5-phosphate synthase (DXS), 1-deoxy-D-xylulose-5-phosphate reductoisomerase (DXR), 4-Hydroxy-3-methylbut-2-enyl diphosphate synthase (HDS), 4-hydroxy-3-methylbut-2-enyl diphosphate reductase (HDR), and geranylgeranyl diphosphate synthase (GGPS), were upregulated in the tree peony cultivar ‘Huangguan’ (strong fragrance) compared to the cultivar ‘Fengdan’ (faint fragrance). Additionally, transcription abundance of one linalool synthase gene (LIS) and one myrcene synthase gene (MYS) in ‘Huangguan’ petals was higher compared to ‘Fengdan’. Therefore, high expression of genes involved in terpene skeleton biosynthesis and monoterpene metabolism pathways was associated enhanced fragrance in tree peonies (Li et al., 2021).
In herbaceous peonies, analysis of aroma components and aroma thresholds across 17 cultivars identified linalool, geraniol, citronellol, and phenylethyl alcohol (2-PE) as primary aromatic substances. Subsequent qRT-PCR investigations into genes potentially regulating aroma synthesis in peony petals revealed significant candidates, including 1-DEOXY-D-XYLULOSE 5-PHOSPHATE SYNTHASE (PlDXS2), 1-deoxy-D-xylulose-5-phosphate reductoisomerase (PlDXR1), 2-C-METHYL-D-ERYTHRITOL-2,4-CYCLODIPHOSPHATE (PlMDS1), 1-HYDROXY-2-METHYL-2-(E)-BUTENYL-4-DIPHOSPHATE REDUCTASE (PlHDR1), GERANYL PYROPHOSPHATE SYNTHASE (PlGPPS3 and PlGPPS4), as well as LINALOOL SYNTHASE (LIS) and GERANIOL SYNTHASE (GES) genes. These findings underscored the critical role of differential gene expression in the monoterpene and 2-PE synthesis pathways in shaping the aroma profile of herbaceous peonies (Zhao et al., 2023).
Furthermore, improvement of floral fragrance traits in Paeonia requires support from genomic marker resources. Molecular marker studies on floral fragrance traits in Paeonia have been shown to promote the breeding of fragrant cultivars. Research based on tree peony genomic data has developed microsatellite markers associated with major floral fragrance components. A total of 8443 EST-SSRs were identified, with genetic diversity and population structure analyses conducted on 31 polymorphic EST-SSR markers, yielding 176 alleles. Polymorphic information content (PIC) ranged from 0.31 to 0.87. Single marker-trait association analysis using a mixed linear model (MLM) identified 29 significantly associated loci, involving 18 SSR loci associated with 5 floral fragrance components, with phenotypic variance explained ranging from 4.57% to 23.46% (Luo et al., 2021b).
While numerous scholars have identified genes potentially involved in Paeonia floral development, how these genes function remains unclear, and the expression patterns of these genes and their interactions have not been thoroughly investigated. The mechanisms and formation processes of floral development in Paeonia are not well understood. Future research should focus on elucidating the interactions between the genes that regulate floral development in peonies.
The formation of flower color in Paeonia is a complex process involving interactions among multiple genes and regulatory networks. Despite the identification of key genes and regulatory factors, the genetic regulatory network underlying flower color formation remains incompletely understood. Further research is needed to investigate the interactions among different genes and their regulatory mechanisms. Current studies have primarily focused on a few Paeonia cultivars, necessitating deeper exploration into the differences in flower color formation among different cultivars and the corresponding genetic variations. Future research should delve into the interactions among different genes to construct a comprehensive regulatory network for peony flower color formation. This will uncover more critical regulatory factors and pathways. Additionally, studying the impact mechanisms of environmental factors such as light and temperature on Paeonia flower color formation will provide more possibilities and approaches for regulating Paeonia flower color.
In the study of flowering in Paeonia, there is a lack of comprehensive understanding regarding the release of flower bud dormancy and flowering, and a need for deeper exploration into the functions of key genes and regulatory factors. The molecular mechanisms underlying physiological and ecological dormancy are also poorly understood. Furthermore, research on the influence of environmental factors such as temperature and light on the flowering of Paeonia is relatively scarce, and their regulatory mechanisms remain incompletely elucidated. Despite some understanding of cut flower quality, ethylene responses, and their regulatory mechanisms in Paeonia, there are still deficiencies. Future research efforts should focus on elucidating the molecular mechanisms governing the release of flower bud dormancy, exploring novel regulatory factors and gene families, and further clarifying the regulatory networks controlling the flowering in Paeonia. Additionally, there should be enhanced studies on the differences in flowering among different cultivars, and a deeper investigation into the effects of environmental factors on flowering to enhance understanding and optimize cultivation techniques in Paeonia. In terms of fresh cut flower preservation, there is a need for in-depth research into the cut flower characteristics of different peony species and their ethylene signaling pathways, to broaden our understanding of herbaceous peony cut flowers. Furthermore, research should investigate the mechanisms by which exogenous substances affect the quality of herbaceous peony cut flowers during storage, aiming to extend the storage period and prolong the bloom period.
Research on the complexity of fragrance components and the related biosynthetic pathways in the study of floral fragrance in Paeonia remains unclear. Additionally, further exploration is needed into the genetic regulatory networks governing floral fragrance traits, and the development and utilization of genomic marker resources for fragrance-related genes in Paeonia have not been fully realized. Future research can delve deeply into identifying fragrance-related genes in Paeonia, exploring details of fragrance biosynthetic pathways, enhancing genetic resource studies among cultivars, and employing advanced molecular biology and gene editing technologies for precise breeding to improve fragrance quality and yield in Paeonia.
Research has found that in terms of flower shape and petal quantity, tree peonies are more complex and abundant, while herbaceous peonies are relatively simpler. In terms of color diversity and vibrancy, tree peonies are generally more prominent. Tree peonies bloom earlier, while herbaceous peonies bloom slightly later. There are also differences in fragrance strength and type, with tree peonies typically having a more intense scent. Peonies exhibit a variety of floral scents and rich colors, as well as differences in blooming periods and fragrances. Comparing floral traits qualitatively or quantitatively among different species or varieties can be challenging. Future research could delve deeper into the mechanisms behind the differences in flower shape, color, blooming period, and fragrance among various species or varieties.
In summary, many scholars have identified key genes regulating flower shape, color, blooming, and fragrance in the Peony genus. However, the specific functions of these genes and their interactions require further investigation. Additionally, the key pathways and gene regulatory networks controlling flower development are not yet clear, and future research should focus on these areas. With the maturity of CRISPR/Cas9 technology, its application in plant research is becoming increasingly widespread, including flower development and plant growth and development (Fan et al., 2020). In the future, using CRISPR/Cas9 technology to artificially modify the floral organ traits of Paeonia is of great significance for screening and cultivating ornamental plants with high economic value, as well as for expanding the cultivation range and introducing new cultivars.
YF: Writing – original draft, Writing – review & editing. XJ: Writing – review & editing. MW: Formal analysis, Writing – review & editing. HDL: Writing – review & editing. WT: Formal analysis, Writing – review & editing. YX: Writing – review & editing. KW: Writing – review & editing. HL: Writing – review & editing. YW: Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by High-level Talent Scientific Research Startup Project of North China University of Water Resources and Electric Power (No. 202110007), Graduate Education Reform Project of Henan Province (2023SJGLX113Y) and Key Project of Henan Education Science Planning (2023JKZD10) and China State Construction Engineering Corporation Seventh Engineering Division 2023 Research Project (CSCEC7b-2023-Z-11).
Authors XJ, HDL, YX were employed by Construction Decoration Co., LTD of China Construction No.7 Engineering Bureau.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Barzilay, A., Zemah, H., Kamenetsky, R., Ran, I. (2002). Annual life cycle and floral development of ‘Sarah Bernhardt’ peony in Israel. HortScience 37, 300–303. doi: 10.21273/HORTSCI.37.2.300
Chang, Y., Zhang, W., Ma, Y., Xia, M., Fan, K., Jiang, Z., et al. (2022). Transcriptome analysis of floral bud development and function analysis of a novel CO gene in Paeonia × lemoinei ‘High Noon’. Sci. Rep. 12, 17281. doi: 10.1038/s41598-022-22195-z
Coen, E. S., Meyerowitz, E. M. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353, 31–37. doi: 10.1038/353031a0
Dong, X. X., Bie, P. T., Yuan, T. (2020). Changes of morphology and carbohydrate content in leaves of three tree peony during flower bud differentiation. J. Northeast For. Univ. 48, 34–39. doi: 10.13759/j.cnki.dlxb.2020.07.007
Du, H., Wu, J., Ji, K. X., Zeng, Q. Y., Bhuiya, M. W., Wang, L. S. (2015). Methylation mediated by an anthocyanin, O-methyltransferase, is involved in purple flower coloration in Paeonia. J. Exp. Bot. 66, 6563–6577. doi: 10.1093/jxb/erv365
Fan, M. Y., Chen, L. F., Wang, E. R., Xue, X., Guo, Q., Guo, D. L., et al. (2023b). Identification and characterization of flowering time regulatory gene FLC of Paeonia ostii ‘Fengdan’. Sci. Hortic. 310, 111748. doi: 10.1016/j.scienta.2022.111748
Fan, Y., Wang, Q., Dong, Z., Yin, Y., Teixeira da Silva, J. A., Yu, X. (2020). Advances in molecular biology of Paeonia L. Planta 251, 23. doi: 10.1007/s00425-019-03299-9
Fan, Y. M., Zheng, Y. Y., Chen, L., Xu, L. W., Teixeira da Silva, J. A., Wu, B. W., et al. (2023a). Transcriptomic and proteomic analyses reveal changes in the metabolic pathways of Paeonia lactiflora petaloid stamens. Sci. Hortic. 312, 111859. doi: 10.1016/j.scienta.2023.111859
Fan, Y. M., Zheng, Y. Y., Teixeira da Silva, J. A., Yu, X. N. (2021). Comparative transcriptomics and WGCNA reveal candidate genes involved in petaloid stamens in Paeonia lactiflora. J. Hortic. Sci. Biotechnol. 96, 588–603. doi: 10.1080/14620316.2021.1892535
Gao, L., Yang, H., Liu, H., Yang, J., Hu, Y. (2016). Extensive transcriptome changes underlying the flower color intensity variation in Paeonia ostii. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.01205
Ge, J., Zhao, D., Han, C., Wang, J., Hao, Z., Tao, J. (2014). Cloning and expression of floral organ development-related genes in herbaceous peony (Paeonia lactiflora Pall.). Mol. Biol. Rep. 41, 6493–6503. doi: 10.1007/s11033-014-3532-8
Gong, P., Ao, X., Liu, G., Cheng, F., He, C. (2017). Duplication and whorl-specific down-regulation of the obligate AP3-PI heterodimer genes explain the origin of Paeonia lactiflora plants with spontaneous corolla mutation. Plant Cell Physiol. 58, 411–425. doi: 10.1093/pcp/pcw204
Gu, Z., Zhu, J., Hao, Q., Yuan, Y. W., Duan, Y. W., Men, S., et al. (2019). A novel R2R3-MYB transcription factor contributes to petal blotch formation by regulating organ-specific expression of PsCHS in tree peony (Paeonia suffruticosa). Plant Cell Physiol. 60, 599–611. doi: 10.1093/pcp/pcy232
Guo, L., Li, Y., Zhang, C., Wang, Z., Carlson, J. E., Yin, W., et al. (2022). Integrated analysis of miRNAome transcriptome and degradome reveals miRNA-target modules governing floral florescence development and senescence across early- and late-flowering genotypes in tree peony. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1082415
Guo, L., Wang, Y., Teixeira da Silva, J. A., Fan, Y., Yu, X. (2019). Transcriptome and chemical analysis reveal putative genes involved in flower color change in Paeonia ‘Coral Sunset’. Plant Physiol. Bioch. 138, 130–139. doi: 10.1016/j.plaphy.2019.02.025
Guo, X., Hu, X., Ma, Y., Guo, J., Zang, D. (2017). Identification of differentially expressed genes during bud dormancy release in Paeonia lactiflora ‘Dafugui’. Acta Hortic. 1171, 163–174. doi: 10.17660/ActaHortic.2017.1171.23
Han, L., Zhou, L., Zou, H., Yuan, M., Wang, Y. (2022). PsGSTF3, an anthocyanin-related glutathione s-transferase gene, is essential for petal coloration in tree peony. Int. J. Mol. Sci. 23, 1423. doi: 10.3390/ijms23031423
He, D., Gao, X., Lv, B., Wang, X., He, S. (2014). Morphology research of flower bud differentiation of tree peony and herbaceous peony. J. Agric. Sci. 43, 117–120 + 124. doi: 10.15933/j.cnki.1004-3268.2014.12.026
Huang, X., Xue, T., Dai, S., Gai, S., Zheng, C., Zheng, G. (2008a). Genes associated with the release of dormant buds in tree peonies (Paeonia suffruticosa). Acta Physiol. Plant 30, 797–806. doi: 10.1007/s11738-008-0184-0
Huang, X., Zhu, W., Dai, S., Gai, S., Zheng, G., Zheng, C. (2008b). The involvement of mitochondrial phosphate transporter in accelerating bud dormancy release during chilling treatment of tree peony (Paeonia suffruticosa). Planta 228, 545–552. doi: 10.1007/s00425-008-0757-6
Ji, X. T., Yuan, Y. P., Bai, Z. Z., Wang, M. L., Niu, L. X., Sun, D. Y. (2023). PlZFP mediates the combinatorial interactions of abscisic acid with gibberellin and ethylene during flower senescence in cut herbaceous peony. Postharvest Biol. Technol. 195, 112130. doi: 10.1016/j.postharvbio.2022.112130
Li, S. S., Chen, L. G., Xu, Y. J., Wang, L. J., Wang, L. S. (2012). Identification of floral fragrances in tree peony cultivars by gas chromatography-mass spectrometry. Sci. Hortic. 142, 158–165. doi: 10.1016/j.scienta.2012.05.015
Li, Y., Kong, F., Liu, Z., Peng, L., Shu, Q. (2022a). PhUGT78A22, a novel glycosyltransferase in Paeonia ‘He Xie’, can catalyze the transfer of glucose to glucosylated anthocyanins during petal blotch formation. BMC Plant Biol. 22, 405. doi: 10.1186/s12870-022-03777-5
Li, R., Li, Z., Leng, P., Hu, Z., Wu, J., Dou, D. (2021). Transcriptome sequencing reveals terpene biosynthesis pathway genes accounting for volatile terpene of tree peony. Planta 254, 67. doi: 10.1007/s00425-021-03715-z
Li, Y., Lu, J., Chang, Y., Tang, W., Yang, Q. (2017). Comparative analysis of tree peony petal development by transcriptome sequencing. Acta Physiol. Plant 39, 216. doi: 10.1007/s11738-017-2520-8
Li, Y., Wang, C., Guo, Q., Song, C., Wang, X., Guo, L., et al. (2022b). Characteristics of PoVIN3, a key gene of vernalization pathway, affects flowering time. Int. J. Mol. Sci. 23, 14003. doi: 10.3390/ijms232214003
Li, S., Zhang, L., Sun, M., Lv, M., Yang, Y., Xu, W., et al. (2023). Biogenesis of flavor-related linalool is diverged and genetically conserved in tree peony (Paeonia × suffruticosa). Hortic. Res. 10, uhac253. doi: 10.1093/hr/uhac253
Liu, N., Cheng, F., Zhong, Y., Guo, X. (2019). Comparative transcriptome and coexpression network analysis of carpel quantitative variation in Paeonia rockii. BMC Genomics 20, 683. doi: 10.1186/s12864-019-6036-z
Liu, G., Li, Y., Sun, X., Guo, X., Jiang, N., Fang, Y., et al. (2022). Association study of SNP locus for color related traits in herbaceous peony (Paeonia lactiflora Pall.) using SLAF-seq. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1032449
Luan, Y., Chen, Z., Tang, Y., Sun, J., Meng, J., Tao, J., et al. (2023). Tree peony PsMYB44 negatively regulates petal blotch distribution by inhibiting dihydroflavonol-4-reductase gene expression. Ann. Bot. 131, 323–334. doi: 10.1093/aob/mcac155
Luan, Y., Chen, Z., Wang, X., Zhang, H., Tao, J., Zhao, D. (2022). Herbaceous peony PlACLB2 positively regulates red petal formation by promoting anthocyanin accumulation. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.992529
Luan, Y., Tao, J., Zhao, D. (2024). Synergistic actions of three MYB transcription factors underpin blotch formation in tree peony. Plant Physiol. 196, 1869–1886. doi: 10.1093/plphys/kiae420
Luo, X., Luo, S., Fu, Y., Kong, C., Wang, K., Sun, D., et al. (2022). Genome-wide identification and comparative profiling of micrornas reveal flavonoid biosynthesis in two contrasting flower color cultivars of tree peony. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.797799
Luo, X., Sun, D., Wang, S., Luo, S., Fu, Y., Niu, L., et al. (2021a). Integrating full-length transcriptomics and metabolomics reveals the regulatory mechanisms underlying yellow pigmentation in tree peony (Paeonia suffruticosa Andr.) flowers. Hortic. Res. 8, 235. doi: 10.1038/s41438-021-00666-0
Luo, L., Yang, Y., Zhao, H., Leng, P., Hu, Z., Wu, J., et al. (2021b). Development of EST-SSR markers and association analysis of floral scent in tree peony. Sci. Hortic. 289, 110409. doi: 10.1016/j.scienta.2021.110409
Luo, X. N., Yuan, M., Li, B. J., Li, C. Y., Zhang, Y. L., Shi, Q. Q. (2020). Variation of floral volatiles and fragrance reveals the phylogenetic relationship among nine wild tree peony species. Flavour Fragrance J. 35, 227–241. doi: 10.1002/ffj.3558
Lv, S., Cheng, S., Wang, Z., Li, S., Jin, X., Lan, L., et al. (2020). Draft genome of the famous ornamental plant Paeonia suffruticosa. Ecol. Evol. 10, 4518–4530. doi: 10.1002/ece3.5965
Park, J. H., Rhie, Y. H., Lee, S. Y., Kim, K. S. (2015). Pre-chilling promotes flowering in Paeonia lactiflora ‘Taebaek’ without flower bud abortion. Hortic. Environ. Biotechnol. 56, 1–8. doi: 10.1007/s13580-015-0050-0
Qin, K. J., Li, J. Y. (1990). Study on floral classification of tree peony and herbaceous peony cultivars. J. Beijing For. Univ. 1, 18–26. (in Chinese).
Ren, L. (2011). Cloning and expression of floral organ development related genes in tree peony. Chinese Academy of Forestry, Beijing.
Shi, Q., Zhou, L., Wang, Y., Li, K., Zheng, B., Miao, K. (2015). Transcriptomic analysis of Paeonia delavayi wild population flowers to identify differentially expressed genes involved in purple-red and yellow petal pigmentation. PloS One 10, e0135038. doi: 10.1371/journal.pone.0135038
Shu, Q., Wang, L., Wu, J., Du, H., Liu, Z., Ren, H., et al. (2012). Analysis of the formation of flower shapes in wild species and cultivars of tree peony using the MADS-box subfamily gene. Gene 493, 113–123. doi: 10.1016/j.gene.2011.11.008
Song, C. W., Wang, Q., Teixeira da Silva, J. A., Yu, X. N. (2018). Identification of floral fragrances and analysis of fragrance patterns in herbaceous peony cultivars. J. Am. Soc Hortic. Sci. 143, 248–258. doi: 10.21273/JASHS04420-18
Tang, W. L., Zhao, P. F., Li, Y. H., He, D., Lu, J. X. (2018a). Cloning and expression analysis of transcription factor gene PsAGL6 in tree peony (Paeonia suffruticosa L.). Northwest Plant J. 38, 439–444. doi: 10.7606/j.issn.1000-4025.2018.03.0439
Tang, Y., Zhao, D., Tao, J. (2018b). Daminozide reduces red color intensity in herbaceous peony (Paeonia lactiflora Pall.) flowers by inhibiting the expression of flavonoid biosynthetic genes. 3 Biotech. 8, 102. doi: 10.1007/s13205-018-1127-0
Wang, S., Gao, J., Xue, J., Xue, Y., Li, D., Guan, Y., et al. (2019). De novo sequencing of tree peony (Paeonia suffruticosa) transcriptome to identify critical genes involved in flowering and floral organ development. BMC Genomics 20, 572. doi: 10.1186/s12864-019-5857-0
Wang, S., Xue, J., Ahmadi, N., Holloway, P., Zhu, F., Ren, X., et al. (2014). Molecular characterization and expression patterns of PsSVP genes reveal distinct roles in flower bud abortion and flowering in tree peony (Paeonia suffruticosa). Can. J. Plant Sci. 94, 1181–1193. doi: 10.4141/cjps2013-360
Wang, Q., Zhu, J., Li, B., Li, S., Yang, Y., Wang, Q., et al. (2023). Functional identification of anthocyanin glucosyltransferase genes: a Ps3GT catalyzes pelargonidin to pelargonidin 3-O-glucoside painting the vivid red flower color of Paeonia. Planta 257, 65. doi: 10.1007/s00425-023-04095-2
Wu, Y. Q., Tang, Y. H., Jiang, Y., Zhao, D. Q., Shang, J. L., Tao, J. (2018). Combination of transcriptome sequencing and iTRAQ proteome reveals the molecular mechanisms determining petal shape in herbaceous peony (Paeonia lactiflora Pall.). Biosci. Rep. 38, BSR20181485. doi: 10.1042/BSR20181485
Wu, F., Zhang, C., Wang, X. Q., Guo, J., Dong, L. (2017). Ethylene-influenced development of tree peony cut flowers and characterization of genes involved in ethylene biosynthesis and perception. Postharvest Biol. Technol. 125, 150–160. doi: 10.1016/j.postharvbio.2016.11.014
Xue, J. Q., Tang, Y., Wang, S. L., Yang, R. W., Xue, Y. Q., Wu, C. H., et al. (2018). Assessment of vase quality and transcriptional regulation of sucrose transporter and invertase genes in cut peony (Paeonia lactiflora ‘Yang Fei Chu Yu’) treated by exogenous sucrose. Postharvest Biol. Technol. 143, 92–101. doi: 10.1016/j.postharvbio.2018.04.014
Xue, Y., Xue, J., Ren, X., Li, C., Sun, K., Cui, L., et al. (2022). Nutrient supply is essential for shifting tree peony reflowering ahead in autumn and sugar signaling is involved. Int. J. Mol. Sci. 23, 7703. doi: 10.3390/ijms23147703
Yuan, J., Jiang, S., Jian, J., Liu, M., Yue, Z., Xu, J., et al. (2022). Genomic basis of the giga-chromosomes and giga-genome of tree peony Paeonia ostii. Nat. Commun. 13, 7328. doi: 10.1038/s41467-022-35063-1
Zhang, Y., Gao, L., Wang, Y., Niu, D., Yuan, Y., Liu, C., et al. (2023). Dual functions of PsmiR172b-PsTOE3 module in dormancy release and flowering in tree peony (Paeonia suffruticosa). Hortic. Res. 10, uhad033. doi: 10.1093/hr/uhad033
Zhang, L., Li, Y. E., Guan, S. M., Liu, C. Y., Gai, S. P., Zhang, Y. X. (2015a). Expression analysis and functional identification of PsAP1 in tree peony. Acta Agric. Boreali−Sinica 30, 84–89. (in Chinese).
Zhang, J., Li, D., Shi, X., Zhang, D., Qiu, S., Wei, J., et al. (2017). Mining and expression analysis of candidate genes involved in regulating the chilling requirement fulfillment of Paeonia lactiflora ‘Hang Baishao’. BMC Plant Biol. 17, 262. doi: 10.1186/s12870-017-1205-1
Zhang, Y., Li, Y., Zhang, Y., Guan, S., Liu, C., Zheng, G., et al. (2015b). Isolation and characterization of a SOC1-like gene from tree peony (Paeonia suffruticosa). Plant Mol. Biol. Rep. 33, 855–866. doi: 10.1007/s11105-014-0800-7
Zhang, L., Song, C., Guo, D., Guo, L., Hou, X., Wang, H. (2022c). Identification of differentially expressed miRNAs and their target genes in response to brassinolide treatment on flowering of tree peony (Paeonia ostii). Plant Signaling Behav. 17, 2056364. doi: 10.1080/15592324.2022.2056364
Zhang, R., Tang, H., He, L. X. (2022a). Comparison of flower bud differentiation and endogenous hormone contents between single and double petals of Paeonia rockii from same plant. Acta Bot. Boreal. Occident. Sin. 42, 1152–1160. doi: 10.7606/j.issn.1000-4025.2022.07.1152
Zhang, J., Wu, Y., Li, D., Wang, G., Li, X., Xia, Y. (2015d). Transcriptomic analysis of the underground renewal buds during dormancy transition and release in ‘Hangbaishao’ peony (Paeonia lactiflora). PloS One 10, e0119118. doi: 10.1371/journal.pone.0119118
Zhang, Y., Xu, S., Ma, H., Duan, X., Gao, S., Zhou, X., et al. (2021a). The R2R3-MYB gene PsMYB58 positively regulates anthocyanin biosynthesis in tree peony flowers. Plant Physiol. Biochem. 164, 279–288. doi: 10.1016/j.plaphy.2021.04.034
Zhang, L., Yan, L., Zhang, C., Kong, X., Zheng, Y., Dong, L. (2022b). Glucose supply induces PsMYB2-mediated anthocyanin accumulation in Paeonia suffruticosa ‘Tai Yang’ cut flower. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.874526
Zhang, Y., Yu, D., Liu, C. Y., Gai, S. P. (2018b). Dynamic of carbohydrate metabolism and the related genes highlights PPP pathway activation during chilling induced bud dormancy release in tree peony (Paeonia suffruticosa). Sci. Hortic. 242, 36–43. doi: 10.1016/j.scienta.2018.07.022
Zhang, Y., Zhang, L., Gai, S., Liu, C., Lu, S. (2015c). Cloning and expression analysis of the R2R3-PsMYB1 gene associated with bud dormancy during chilling treatment in the tree peony (Paeonia suffruticosa). Plant Growth Regul. 75, 667–676. doi: 10.1007/s10725-014-9968-y
Zhang, Y., Zhang, T., Si, F., Wang, X., Liu, C., Yuan, Y., et al. (2021b). Changes of DNA methylation patterns reveal epigenetic modification of dormancy release-related genes is induced by chilling in tree peony. DNA Cell Biol. 40, 606–617. doi: 10.1089/dna.2020.6142
Zhang, X., Zhao, L., Xu, Z., Yu, X. (2018a). Transcriptome sequencing of Paeonia suffruticosa ‘Shima Nishiki’ to identify differentially expressed genes mediating double-color formation. Plant Physiol. Bioch. 123, 114–124. doi: 10.1016/j.plaphy.2017.12.009
Zhang, J., Zhu, W., Teixeira da Silva, J. A., Fan, Y., Yu, X. (2019). Comprehensive application of different methods of observation provides new insight into flower bud differentiation of double-flowered Paeonia lactiflora ‘Dafugui’. HortScience 54, 28–37. doi: 10.21273/HORTSCI13229-18
Zhao, D., Cheng, M., Tang, W., Liu, D., Zhou, S., Meng, J., et al. (2018b). Nano-silver modifies the vase life of cut herbaceous peony (Paeonia lactiflora Pall.) flowers. Protoplasma 255, 1001–1013. doi: 10.1007/s00709-018-1209-1
Zhao, Q., Gu, L., Li, Y., Zhi, H., Luo, J., Zhang, Y. (2023). Volatile composition and classification of Paeonia lactiflora flower aroma types and identification of the fragrance-related genes. Int. J. Mol. Sci. 24, 9410. doi: 10.3390/ijms24119410
Zhao, J., Hu, Z. H., Leng, P. S., Zhang, H. X., Cheng, F. Y. (2012b). Fragrance composition in six tree peony cultivars. Korean J. Hortic. Sci. Technol. 30, 617–625. doi: 10.7235/hort.2012.12055
Zhao, D., Jiang, Y., Ning, C., Meng, J., Lin, S., Ding, W., et al. (2014). Transcriptome sequencing of a chimaera reveals coordinated expression of anthocyanin biosynthetic genes mediating yellow formation in herbaceous peony (Paeonia lactiflora Pall.). BMC Genomics 15, 689. doi: 10.1186/1471-2164-15-689
Zhao, D., Tang, W., Hao, Z., Tao, J. (2015). Identification of flavonoids and expression of flavonoid biosynthetic genes in two coloured tree peony flowers. Biochem. Biophys. Res. Commun. 459, 450–456. doi: 10.1016/j.bbrc.2015.02.126
Zhao, D., Tao, J., Han, C., Ge, J. (2012a). Flower color diversity revealed by differential expression of flavonoid biosynthetic genes and flavonoid accumulation in herbaceous peony (Paeonia lactiflora Pall.). Mol. Biol. Rep. 39, 11263–11275. doi: 10.1007/s11033-012-2036-7
Zhao, D., Wang, R., Liu, D., Wu, Y., Sun, J., Tao, J. (2018a). Melatonin and expression of tryptophan decarboxylase gene (TDC) in herbaceous peony (Paeonia lactiflora Pall.) flowers. Molecules 23, 1164. doi: 10.3390/molecules23051164
Zhao, D., Wei, M., Shi, M., Hao, Z., Tao, J. (2017). Identification and comparative profiling of miRNAs in herbaceous peony (Paeonia lactiflora Pall.) with red/yellow bicoloured flowers. Sci. Rep. 7, 44926. doi: 10.1038/srep44926
Zhao, Q., Zhang, M., Gu, L., Yang, Z., Li, Y., Luo, J., et al. (2024). Transcriptome and volatile compounds analyses of floral development provide insight into floral scent formation in Paeonia lactiflora ‘Wu Hua Long Yu’. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1303156
Zhou, H., Cheng, F. Y., Wang, R., Zhong, Y., He, C. (2013). Transcriptome comparison reveals key candidate genes responsible for the unusual reblooming trait in tree peonies. PloS One 8, e79996. doi: 10.1371/journal.pone.0079996
Zhou, L., Dong, L., Jia, P. Y., Wang, W. R., Wang, L. Y. (2010). Expression of ethylene receptor and transcription factor genes, and ethylene response during flower opening in tree peony (Paeonia suffruticosa). Plant Growth Regul. 62, 171–179. doi: 10.1007/s10725-010-9503-8
Zhou, S., Ma, C., Gao, S. C., Hou, D. Y., Song, P., Guo, Z. Z., et al. (2022). Cloning and expression of floral organ identity genes in Paeonia ostii ‘Fengdan’. Pak. J. Bot. 54, 2115–2123. doi: 10.30848/PJB2022-6(27
Zhu, J., Wang, Y., Wang, Q., Li, B., Wang, X., Zhou, X., et al. (2023). The combination of DNA methylation and positive regulation of anthocyanin biosynthesis by MYB and bHLH transcription factors contributes to the petal blotch formation in Xibei tree peony. Hortic. Res. 10, uhad100. doi: 10.1093/hr/uhad100
Keywords: Paeonia, floral organ, flower development, regulatory mechanism, research progress
Citation: Fan Y, Jin X, Wang M, Liu H, Tian W, Xue Y, Wang K, Li H and Wu Y (2024) Flower morphology, flower color, flowering and floral fragrance in Paeonia L.. Front. Plant Sci. 15:1467596. doi: 10.3389/fpls.2024.1467596
Received: 20 July 2024; Accepted: 30 October 2024;
Published: 21 November 2024.
Edited by:
Zhongxiong Lai, Fujian Agriculture and Forestry University, ChinaReviewed by:
Xiyang Zhao, Jilin Agricultural University, ChinaCopyright © 2024 Fan, Jin, Wang, Liu, Tian, Xue, Wang, Li and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongming Fan, ZmFueW9uZ21pbmdAbmN3dS5lZHUuY24=; Hu Li, bGlodTgxQDE2My5jb20=; Yan Wu, d3V5YW4xMTExMTFAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.