
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 26 September 2024
Sec. Functional and Applied Plant Genomics
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1467232
 Jing-Jing Zou1,2,3†
Jing-Jing Zou1,2,3† Jun Zhang4†
Jun Zhang4† Xiaoqian Wang1,2,5†
Xiaoqian Wang1,2,5† Hui Xia1,2
Hui Xia1,2 Xiangling Zeng1,2,3
Xiangling Zeng1,2,3 Xuan Cai1,2,3
Xuan Cai1,2,3 Jie Yang1,2,3
Jie Yang1,2,3 Jin Zeng1,2
Jin Zeng1,2 Zeqing Li1,2,4*
Zeqing Li1,2,4* Guifu Zhang1,2*
Guifu Zhang1,2* Hongguo Chen1,2,3*
Hongguo Chen1,2,3*Osmanthus fragrans is an ethylene-sensitive flower, and flower senescence was mediated by ethylene-responsive transcription factors (OfERFs). A total of 227 OfERFs were identified from O. fragrans, which were classified into five subfamilies: AP2 (35), DREB (57), ERF (125), RAV (6), and Soloist (4). Gene composition and structural analysis indicate that members of different subfamilies have different gene structures and conserved domains. Their gene promoter contains various functional responsive elements, including auxin, jasmonic acid, and other responsive elements. Among them, 124 OfAP2/ERF genes have expressed at any stage of flowering, and 10 of them may play roles in flowering or senescence. By comparative transcriptome analysis, OfAP2/ERFs affected by ethephon (ETH) and 5′-azacytidine (Aza) treatment were divided into three categories, which have various target gene sets. Importantly, these target gene sets participate in similar or different biological processes and metabolic pathways, suggesting that ethylene and DNA hypomethylation have crosstalk and a unique mechanism in regulating the flower senescence of O. fragrans. Co-expression analysis revealed that several key OfAP2/ERFs played a central role in organic acid metabolism and biosynthesis of branched-chain amino acids (BcAAs), among which OfERF017 was selected for further functional analysis. Overexpression of OfERF017 leads to significant enrichment of genes in organic acid metabolism pathways, which leads to a decrease in organic acid levels and promoting the flower senescence of O. fragrans. Together, these results give insights into the characteristics and functional analysis of OfAP2/ERF genes in O. fragrans.
Sweet osmanthus (Osmanthus fragrans) is an ornamental plant in the Osmanthus family. As one of the top 10 famous traditional flowers in China cultivated for more than 2,500 years, it has been widely used in streets or gardens (Shi et al., 2024; Yue et al., 2024). Because of its unique aroma, sweet osmanthus is also an important economic fragrant flower in China and widely used in food additives or skincare products (Pan et al., 2021; Wu et al., 2022; Zhou et al., 2024). However, the short flowering period with an optimal harvesting time of 2–3 days has greatly limited both its ornamental and economic value (Zou et al., 2014). Therefore, understanding the flower senescence mechanism of sweet osmanthus is important for the harvest utilization and improvement of industrial products.
Flower senescence in ornamental plants is often associated with endogenous ethylene climacteric induced by pollination (Luangsuwalai et al., 2011; Sun et al., 2021), while, economically, harvest cultivars of sweet osmanthus without fruits are also sensitive to ethylene and non-pollination-induced endogenous ethylene climacteric is an important regulator for flower senescence (Zhou et al., 2008; Zou et al., 2014). The visible flower senescence characteristics coincide with the peak of the endogenous ethylene production. Exogenous ethylene treatment has not only significantly accelerated the abscission and wilting of cut flower petals, but also led to the central vacuole rupture, cell deformation, DNA degradation, and membrane lipid peroxidation in O. fragrans (Zou et al., 2014, 2017). Therefore, ethylene is an important regulatory factor in the flower senescence of O. fragrans. It was also found that DNA methylation decreased during the flower opening and senescence of O. fragrans, and the number of ethylene-responsive transcription factors (ERFs) associated with hypomethylated differentially methylated regions (Hypo-DMRs) was significantly enriched, suggesting that DNA hypomethylation participates in the flower senescence mediated by OfERFs through the ethylene response pathways (Zou et al., 2023). However, the specific regulatory networks of OfAP2/ERFs on flower senescence are still unclear.
APETALA2/ethylene-responsive factor (AP2/ERF) are plant-specific transcription factors and play important roles in plant growth, development, maturation, and senescence, as well as biotic and abiotic stress processes (Feng et al., 2020). AP2/ERF family members contain an AP2/ERF domain composed of 60–70 amino acids. According to the number and type of structural domains, the AP2/ERF family can be divided into five subfamilies: AP2, ERF, DREB, Soloist, and RAV. Among them, members of the AP2 subfamily contain two AP2/ERF domains, which have important functions in the regulation of developmental processes (Zhang et al., 2021a). Members of the ERF, DREB, and Soloist subfamilies usually only have one AP2/ERF domain. The members of the RAV subfamily contain not only one AP2/ERF domain, but also one B3 domain, which are important in the signal transduction of plant hormones (Alonso et al., 2003; Li et al., 2022a). Based on the conserved amino acid and binding sequences, the ERF subfamily with the 14th and 19th amino acids alanine (A) and aspartic acid (D), respectively, specifically bind to the cis-acting element GCC box with a conserved sequence of AGCCGCC and participate in ethylene response and abiotic stress (Li et al., 2022b). The DREB subfamily with the 14th and 19th amino acids valine (V) and glutamate (E), respectively, specifically bind to dehydration-responsive elements (DRE)/C-repeat (CRT) elements and are involved in response to drought and cold stress (Sakuma et al., 2002; Yang et al., 2021). Thus, the systematic identification of OfERFs in O. fragrans will help in the further exploration of their response patterns to flower senescence.
Based on the genome of O. fragrans “Liuye Jingui”, the OfAP2/ERFs family was identified. Combined with the transcriptomic data for flowers in different developmental stages and cut flowers treated with 5′-azacytidine (Aza) and ethephon (ETH), we analyzed the ethylene response members in the OfAP2/ERF family, as well as their potential downstream regulatory networks. Then, the function of OfERF017 in organic acid decomposition metabolism was characterized. These results not only provide a foundation for the further exploration of the biological functions of genes in the OfAP2/ERF family, but also provide clues for unraveling the ethylene regulatory mechanism in the flower senescence of sweet osmanthus.
Plant tissues (root, stem, leaf, and flower) of O. fragrans “Liuye Jingui” was collected from the Huazhong Agricultural University campus (Wuhan, China) (30°29′N, 114°21′W). Flowers were collected at six flowering stages (S1–S6: bud stage, initial flowering stage, early full flowering stage, full flowering stage, late full flowering stage, and abscission stage) as described in previous studies (Chen et al., 2021; Zou et al., 2023). Samples were weighed and conserved in an ice box or frozen in liquid nitrogen immediately for testing or stored at −80°C until further analysis.
Detached branches with floral buds at the S1 stage (bud stage) were treated with 10 mM Aza and 50 mg L−1 ETH dissolved in distilled water. As a control, cut flowers were placed in individual dishes containing distilled water. After 24 h, all samples were transferred to distilled water and kept at 25°C under a 12 h photoperiod with cool-white fluorescent lighting at an intensity of 10 mol m−2 s−1, in accordance with a previous study (Zou et al., 2014).
To identify the OfAP2/ERF gene family members in the sweet osmanthus genome, the protein sequences of AtAP2/ERF in Arabidopsis were downloaded from the PlantTFDB (Plant Transcription Factor Database; https://planttfdb.gao-lab.org/). Potential OfAP2/ERFs were isolated from the genome of O. fragrans “Liuye Jingui” using SwissProt and ITAK annotations. Then, a Hidden Markov Model (HMM) of PF00847 (AP2 domain) and PF02362 (B3 domain) was performed from the Pfam database (http://pfam.sanger.ac.uk/) to obtain candidate members of the OfAP2/ERFs family (Finn et al., 2011; Xu et al., 2015; Chen et al., 2021).
For phylogenetic analysis, MEGA11 (Kumar et al., 2018) was used to investigate the phylogenetic interactions of OfAP2/ERFs between O. fragrans and Arabidopsis. The Arabidopsis and O. fragrans OfAP2/ERF protein sequences were aligned by the MUSCLE program with default parameters. The multiple sequence alignment generated was analyzed and then used to build phylogenetic trees with the neighbor-joining method (with 1,000 bootstrap replicates), and evolutionary distances were computed using Poisson correction with pairwise deletion.
Circos v0.63 (http://circos.ca/) was employed to investigate their tandem duplication and synteny relationships as previously described (Li et al., 2013; Fan et al., 2017). Tandem duplication OfAP2/ERF genes was identified according to their physical locations on individual chromosomes in the sweet osmanthus genome. Adjacent homologous OfAP2/ERF genes on the same sweet osmanthus chromosome with no more than one intervening gene were considered tandem duplicates. The synteny blocks among different sweet osmanthus chromosomes were identified using the mcscan software (Wang et al., 2012).
For the gene structural analysis, gene models were downloaded from the sweet osmanthus genome (Chen et al., 2021). They were then submitted into the Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/) for structural analyses (Hu et al., 2015). Additionally, the OfAP2/ERF protein sequences were employed to investigate conserved motifs with the MEME Suite platform (http://meme-suite.org/), and 30 motifs were found within each gene family. Analysis of Gene Ontology (GO) terms was performed with the online database (http://www.geneontology.org).
A sequence 2,000 bp upstream of the transcription start site of each OfAP2/ERF gene was selected to predict CREs, using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html) (Magali et al., 2002; Xia et al., 2024).
The genome and transcriptome sequencing data in different tissues (root, stem, and leaf) and nature flowering stages (S1–S6) of O. fragrans ‘Liuye Jingui’ are the same as those used in a previous study (NCBI accession number PRJNA679852) (Chen et al., 2021).
Cut flowers treated with 50 mg L−1 ETH, 10 mM Aza, and distilled water in different days were collected for RNA preparation. RNA sequencing libraries were sequenced and analyzed by IGENEBOOK Biotechnology Co. Ltd as previously described (Chen et al., 2021). The transcript levels were calculated with the fragments per kilobase of exon per million fragments mapped (FPKM) method. The transcriptome sequencing data using different treatments—50 mg L−1 ETH, 10 mM Aza, and distilled water—have been deposited into the NCBI BioProject under the accession number PRJNA798559.
The STRING database (v12.0) was used to construct a protein–protein interaction (PPI) network (Szklarczyk et al., 2023). Based on the whole genome sequence and transcriptome data, 124 OfAP2/ERF members of O. fragrans were used to predict the PPI network. The minimum required interaction score was set to medium confidence (0.4). Cytoscape (v3.10.0) was used for image processing (Shannon et al., 2003).
TBtools (v2.052) was used to extract promoter sequences from the genome of O. fragrans ‘Liuye Jingui’ (Chen et al., 2023). Using the FIMO program (v5.5.5), genes containing GCCGCC (GCC box) and G/A (R) CCGAC (DRE/CRT core) were scanned from the whole genome promoter, with a match p-value of 0.001.
Flowers at the initial flowering stage (S2) of O. fragrans ‘Sijigui’ were collected for the transgenic experiment. Full-length OfERF017 cloned into the pCAMBIA2300s vector and the empty pCAMBIA2300s vector was used as the negative control. The transgenic conditions are the same as those in a previous study (Xi et al., 2021). Three days after the inoculation, the flowers were collected for qRT-PCR and transcriptome sequencing analysis.
qRT-PCR was conducted utilizing Applied Biosystems 7500 sequence detection system (ABI7500; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The experimental conditions and methods for qRT-PCR are the same as those used in previous studies, and the expression level of Actin served as the reference (Chen et al., 2021; Xia et al., 2024). The primers for qRT-PCR and full-length OfERF017 were designed using Prime Premier 5 (Supplementary Table S1).
Endogenous ethylene production was measured as previously described (Zou et al., 2023). Cut flowers were treated with Aza, ETH, and ddH2O, and flowers of transgenic O. fragrans were analyzed for ethylene production using a gas chromatograph (Agilent Technologies 7890B). The concentration was expressed as nanograms of ethylene per kilogram of plant fresh weight per second (ng kg−1 s−1).
Widely targeted metabolites were quantified as previously described (Chen et al., 2013). The widely targeted metabolites of flowers during different stages (S1–S6) and transgenic O. fragrans were quantified by MetWare (Wuhan, China) using the ultraperformance liquid chromatography–tandem mass spectrometry (UPLC-MS/MS) platform following standard procedures.
Three replicates were performed per treatment in each experiment. All the data are presented as means ± standard errors. Significant differences were obtained using SPSS with Duncan’s test at p < 0.05.
In this study, a total of 227 OfAP2/ERFs genes were identified in the O. fragrans ‘Liuye Jingui’ genome according to the gene annotation and AP2 domain (PF00847) information. The protein sequences of these OfAP2/ERFs genes were extracted and compared with 172 AtAP2/ERF proteins in Arabidopsis. Then, the physical and chemical characterization of OfAP2/ERFs with deduced amino acid sequence was analyzed using ProtParam. The gene length of OfAP2/ERF ranged from 100 to 640 aa, the protein molecular weight (MW) ranged from 11.74 to 71.28 kDa, and the isoelectric point (pI) ranged from 4.24 to 9.8 (Supplementary Table S2). Among the 227 OfAP2/ERF members, 212 genes were successfully anchored to 23 chromosomes. From the distribution of OfERF genes, some of them displayed close locations from each other (Figure 1), which imply that the tandem duplication events might have occurred in this gene family.
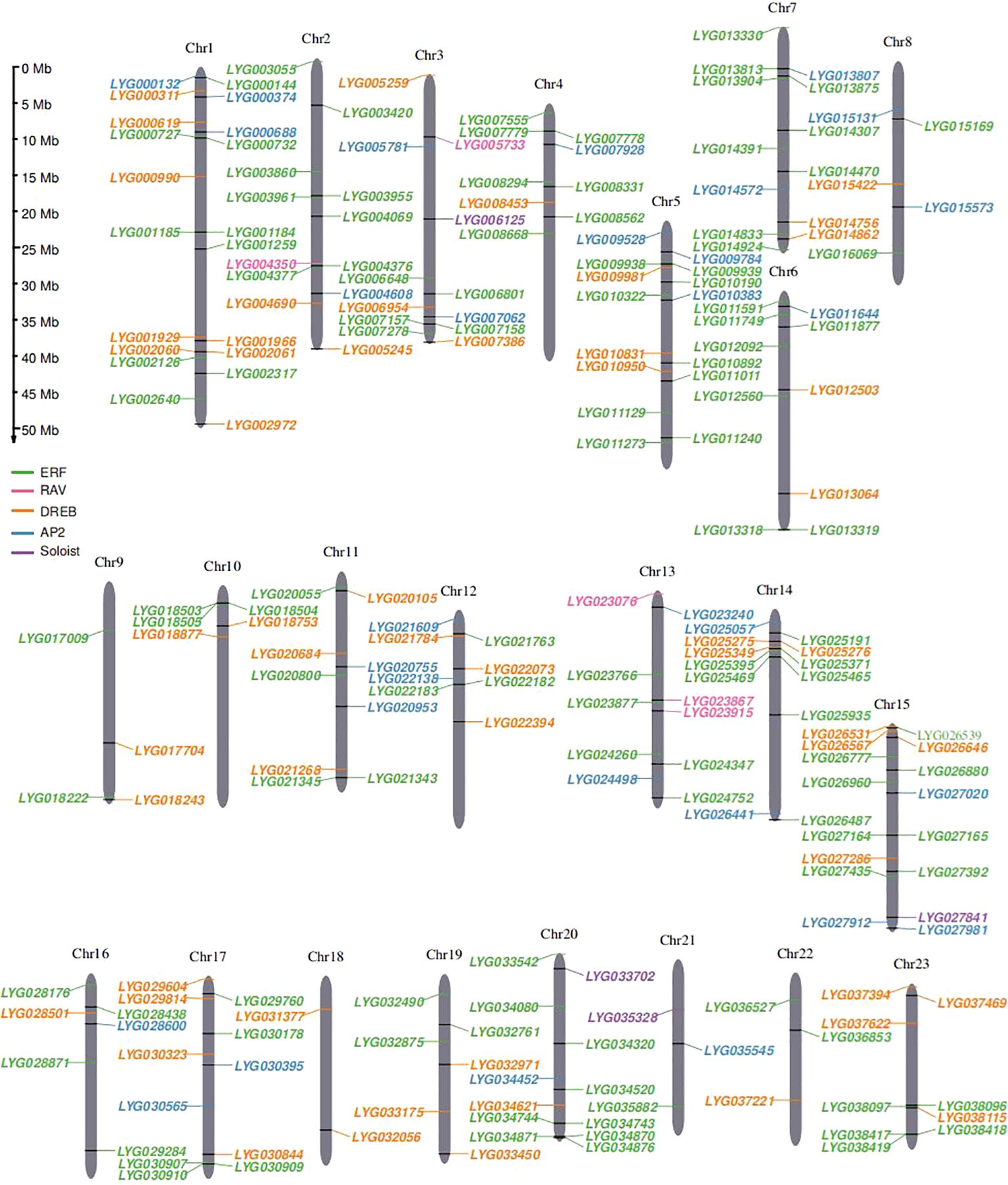
Figure 1. Chromosomal distribution of 227 OfAP2/ERFs genes on 23 O. fragrans chromosomes. The size of each chromosome and its corresponding OfAP2/ERFs gene distribution. Chr1–Chr23 are abbreviations for chromosome numbers 1–23, and black rectangles refer to gene pairs of tandem duplications.
In order to investigate the phylogenetic relationship between O. fragrans and Arabidopsis, multiple alignment analysis of 227 OfAP2/ERF proteins from O. fragrans and 172 AtAP2/ERF proteins from Arabidopsis protein was performed through conserved amino acid sequences obtained from the PlantTFDB (v5.0) (Figure 2A). The phylogenetic tree analysis has shown that each OfAP2/ERFs in O. fragrans can be found as a homologous AtAP2/ERF in Arabidopsis (Figure 2A). Based on the number of AP2 domains and sequence similarity, the 227 OfAP2/ERFs in O. fragrans are divided into five categories: the AP2 subfamily (35 members) that contains two AP2/ERF domains, the DREB subfamily (57 members), the ERF subfamily (125 members), the RAV subfamily (6 members) that contains one AP2/ERF domain and one B3 domain, and Soloist (4 members) (Figure 2B).
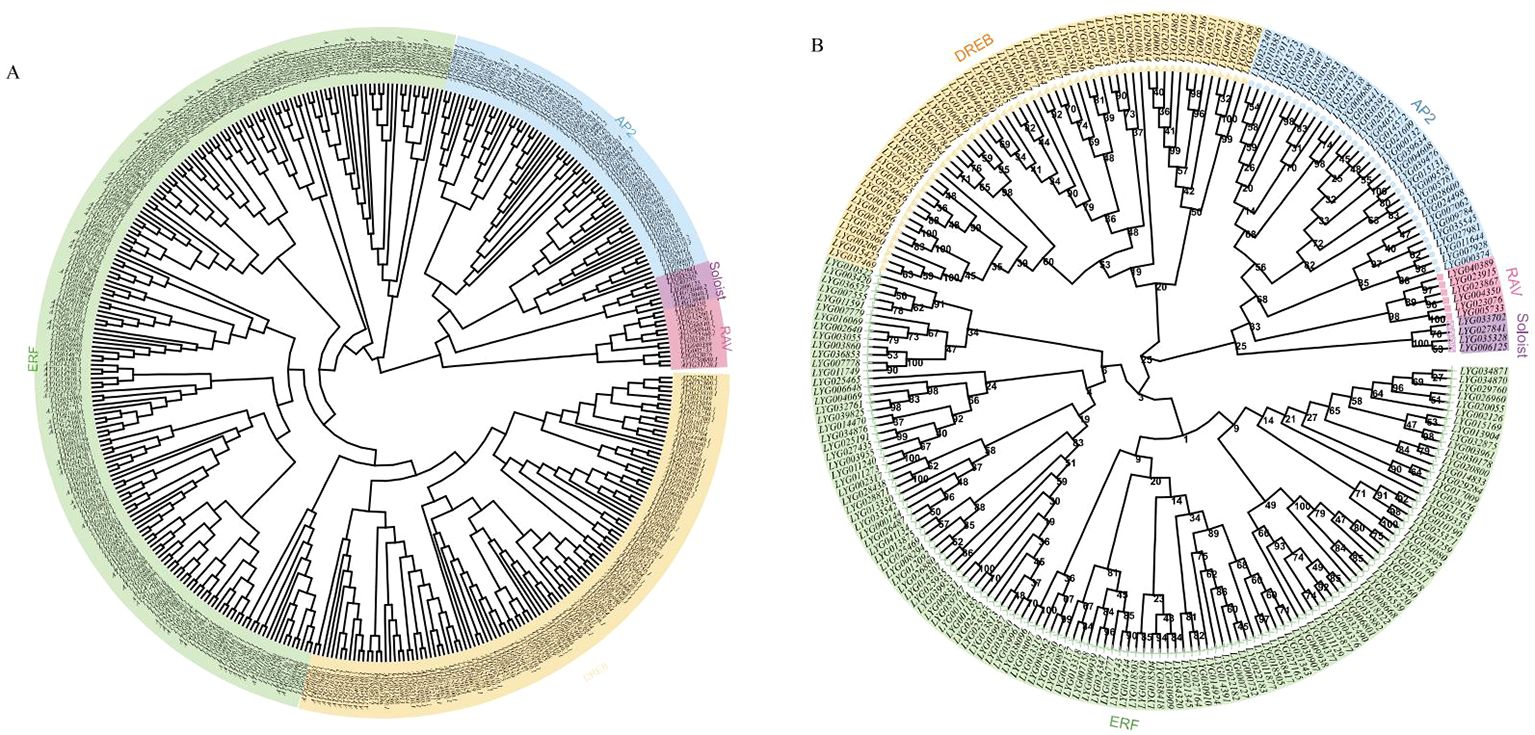
Figure 2. Phylogenetic analysis of OfAP2/ERFs in O. fragrans and Arabidopsis. (A) Phylogenetic analysis of OfAP2/ERFs in O. fragrans and Arabidopsis. (B) Phylogenetic analysis of the subfamily of OfAP2/ERFs genes.
Gene duplication events play key roles in the evolution of a gene family. On the genome chromosomes of O. fragrans, there are a total of 160 pairs of genes exhibiting collinearity (Supplementary Figure S1A). These genes belong to the subfamilies AP2, DREB, ERF, RAV, and Soloist, with respective counts of 19, 43, 92, 1, and 5. Additionally, in interspecific collinearity analysis, there are 100 OfAP2/ERFs, forming 158 pairs of collinear gene pairs with genes in Arabidopsis thaliana (Supplementary Figure S1B). Among these, there are 61 ERF subfamily genes, 31 DREB subfamily genes, and 4 AP2 subfamily genes (LYG015131, LYG035545, LYG000374, and LYG022138), as well as 2 genes each in RAV (LYG023867 and LYG004350) and Soloist (LYG027841 and LYG033702). This indicates that the AP2/ERFs of O. fragrans have undergone gene expansion.
The average number of introns in all OfAP2/ERF genes is 1.5, ranging from 0 to 9, while the number of coding sequences (CDS) ranges from 1 to 10 (Supplementary Figure S2). Among them, LYG020953 belonging to the AP2 subfamily has the highest number of CDS, with 10. In different subfamilies, AP2 and Soloist genes have relatively more introns and CDS, averaging 5.3 and 6.3, and 5.8 and 5.8, respectively (Supplementary Figures S2B, C). In contrast, other subfamily genes mostly contain 0–2 introns and 1–2 CDS, with exceptions such as LYG005733 in RAV containing 5 introns (Supplementary Figure S2A), and LYG010322, LYG010892, and LYG013319 in ERF containing 3–5 introns and CDS (Supplementary Figure S2E). Many genes in the DREB subfamily do not even contain introns (Supplementary Figure S2D). The genes with less intron could have a rapid transcription and response to environmental change, which suggested a high-efficiency response of these OfAP2/ERFs to external threats.
Conserved motif analyses could imply functional differences among the family members. Among the 30 different motifs identified, motifs 1, 2, and 4 are present in all subfamilies. All genes contain one to nine different motifs, with the maximum observed in genes LYG021609 and LYG022138, and the minimum in LYG013330, which belongs to the ERF subfamily (Supplementary Figure S2). Specifically, the AP2 subfamily contains 10 different motifs, namely, motifs 1, 2, 3, 4, 6, 9, 12, 13, 25, and 26, with the first three being shared among all AP2 genes (Supplementary Figure S2C). The number of motifs in ERF subfamily genes ranges from 1 to 6, with a total of 17 different motifs (Supplementary Figure S2E). Motifs 1, 2, 3, and 4 are present in most genes, with motif 1 being shared among all genes except LYG013330. In the DREB subfamily, there are 12 different motifs, with motifs 1 and 2 being shared by all genes (Supplementary Figure S2D). In the RAV and Soloist subfamilies, motifs 3, 4, 11, and 14 are conserved in eight different motifs in RAV, while motif 21 is a conserved motif specific to the Soloist subfamily among its four motifs (Supplementary Figures S2A, B). Most of the genes in the same subfamily showed similar motif components.
To reveal the underlying regulatory mechanism of OfAP2/ERFs, cis-elements of OfAP2/ERFs were analyzed. The elements are mainly categorized into four major classes: growth and development, light, hormones (auxin, gibberellin, salicylic, abscisic, and MeJA), and stress response (drought, temperature, anaerobic, etc.), with 355, 1,841, 2,901, and 768 elements, respectively (Supplementary Figure S3). There are differences in both the total number and types of elements contained among different subfamilies. The ERF and DREB subfamilies have a higher proportion of elements in all four major categories, while RAV and Soloists have fewer. Specifically, the ERF subfamily genes have the highest proportion of elements in the light response category, reaching 55.57%, followed by DREB (34.75%). The number of elements in different genes ranges from 11 (LYG025191) to 44 (LYG007779). In terms of light response elements, genes in the ERF subfamily, such as LYG026539 (27), LYG032056 (26), and LYG029604 (25), have relatively more elements, while LYG020755 has the fewest, with only 4 elements. In hormone response elements, the gene LYG029284, also belonging to the ERF subfamily, has the most elements (24), while the gene LYG024347 has only 1 element. Additionally, all these elements comprise 62 different types, with the light response category having the highest number of types, with 34, of which the DREB subfamily has the most types (31), followed by ERF. These findings indicated that OfAP2/ERFs might play roles in a variety of pathways.
Gene expression patterns in various tissues can give clues to predict gene function. Using RNA-seq, we analyzed the expression profile of OfAP2/ERFs in different tissues (root, stem, and leaf) and flowering stages (S1 to S6) of O. fragrans. Using FPKM > 1 as effective expression, it was found that the highest number of 124 OfAP2/ERFs is expressed in the flower [at least at any stage (S1 to S6)], followed by the root (109), stem (95), and leaf (79) (Figure 3A). These have indicated that OfAP2/ERFs may play important roles in flowers. In order to further investigate the key role that OfAP2/ERFs may play in flowers, we constructed a PPI network between 124 OfAP2/ERF members that are expressed in flowers. After filtering, 71 OfAP2/ERFs have been left in the network, among which LYG027841, LYG033702, LYG035328, LYG006125, LYG007062, LYG004350, LYG002640, LYG003055, LYG007779, and LYG034743 are at the central position, indicating their key roles in flowering (Figure 3B).
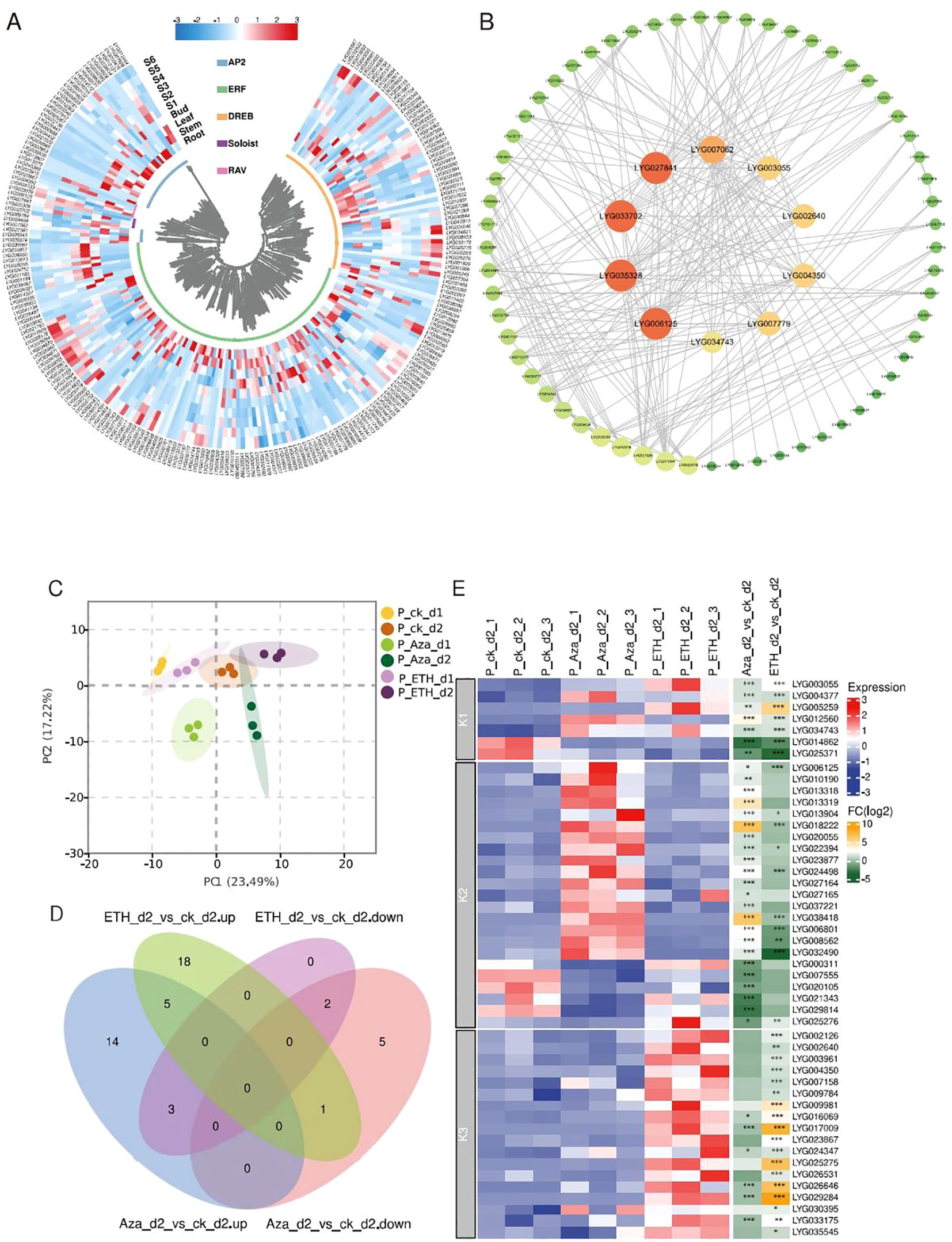
Figure 3. Expression profile and protein–protein interaction of OfAP2/ERFs. (A) Heatmap showing the transcript level of OfAP2/ERFs in the root, stem, leaf, and flowering stages (S1 to S6). (B) Protein–protein interaction. The orange and big circles represent more interaction pairs, and green and small circles represent fewer interaction pairs. (C) The principal component analysis (PCA) of the OfAP2/ERFs expression level under ETH and Aza treatment. (D) Venn diagram of the up- and downregulated genes in ETH_d2 or Aza_d2 relative to ck_d2, respectively. (E) Heatmap showing the transcript level of OfAP2/ERFs under ETH and Aza treatment and the fold change in ETH_d2_vs_ ck_d2 and Aza_d2_vs_ ck_d2 comparison.
To investigate the similarities and differences of the response pattern of OfAP2/ERFs to ethylene in sweet osmanthus, we performed RNA-seq in cut flowers treated with 10 mM Aza and 50 mg L−1 ETH, both of which have caused earlier flower senescence and an increase of endogenous ethylene production (Supplementary Figures S4A, B). On day 1 and day 2, the endogenous ethylene production has increased to 39.20 and 31.33 ng−1 kg−1 s−1 under ETH treatment; the corresponding values are 6.29 and 5.29 ng−1 kg−1 s−1 under control (Supplementary Figure S4B). Correspondingly, Aza treatment increased the endogenous ethylene release to 16.98 ng−1 kg−1 s−1 on day 1 and to 14.49 ng−1 kg−1 s−1 on day 2 (Supplementary Figure S4B).
By the principal component analysis (PCA) of all genes, the samples have been only separated from each other treated by different days, but have not been separated from each other among different treatments (Supplementary Figure S5A). On the other hand, consistent with the increase in endogenous ethylene production, PCA shows that significant changes have occurred in ethylene biosynthesis genes and OfAP2/ERFs among the different treated samples (Supplementary Figure S5B; Figure 3C). A total of 15 and 24 OfAP2/ERFs have upregulated on day 1 and day 2 under ETH treatment, while 28 and 22 OfAP2/ERFs have upregulated on day 1 and day 2 under Aza treatment. The differentially expressed OfAP2/ERFs members on day 2 were used to perform the Venn analysis (Figure 3D). The results showed that the expression pattern of seven OfAP2/ERF members was consistent under ETH and Aza treatment, which were defined as Aza-mediated ethylene-dependent response factors (K1-ERFs). Four OfAP2/ERFs showed opposite changes under ETH and Aza treatments, while 19 OfAP2/ERFs were only affected by Aza treatments. These 23 genes are defined as Aza-dependent response factors (K2-ERFs). Additionally, 18 OfAP2/ERFs were only affected by ETH treatment and were defined as ethylene-dependent response factors (K3-ERFs). The heatmap clearly displays the expression patterns and variation relationships of these three types of OfAP2/ERFs (Figure 3E).
As the ETH and Aza treatments have led to a different expression of OfAP2/ERFs, the downstream target genes of three types of OfAP2/ERFs were analyzed. The ERF protein binds to target genes by recognizing the GCC box or DRE/CRT cis-elements in the promoter. We scanned 6,626 promoter genes containing the GCC box and 23,561 promoter genes containing DRE/CRT from the genome of O. fragrans ‘Liuye Jingui’, respectively. The Venn plot shows that 4,388 gene promoters contain both the GCC box and DRE/CRT cis-elements (Figure 4A), which were used for further analysis. A total of 829 downstream genes of K1-ERFs, 2,161 downstream genes of K2-ERFs, and 1,090 downstream genes of K3-ERFs were obtained after felting with a threshold |cor| > 0.8 and p < 0.05 (Figure 4B). The downstream target genes of the three types of ERFs have overlapped in a considerable proportion (693/2,316), indicating that Aza and ethylene have similar pathways in regulating flower senescence in O. fragrans. Compared to the K1-ERFs and K3-ERFs groups, the downstream target genes in the K2-ERFs group have the most unique genes, suggesting a special regulatory pathway for DNA hypomethylation to regulate downstream gene function through OfAP2/ERFs.
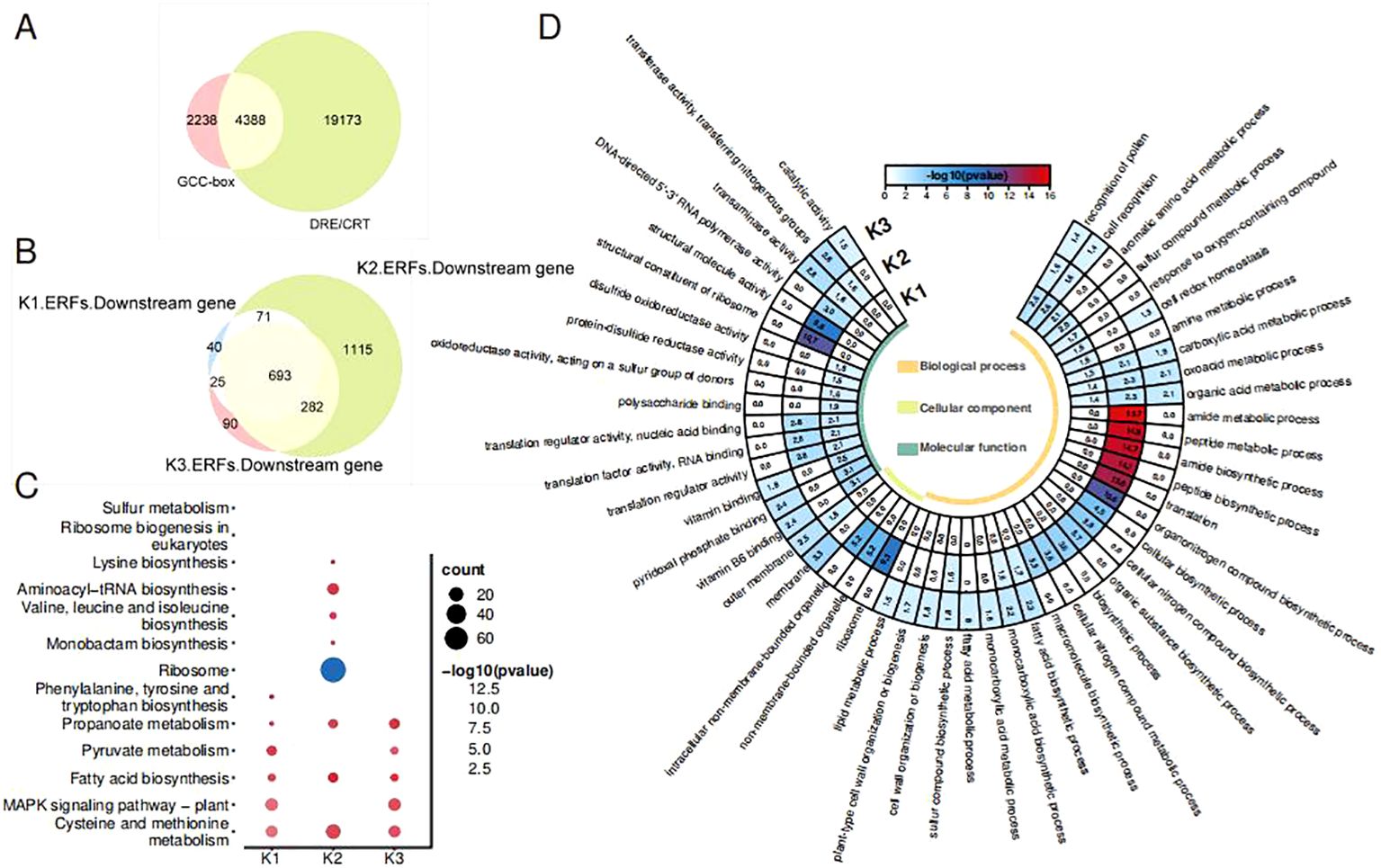
Figure 4. OfAP2/ERF downstream gene analysis. (A) Venn diagram of the promoter includes the GCC box or DRE/CRT genes. (B) Venn diagram of the three cluster ERF downstream genes. (C) GO analysis of the three types of ERF downstream genes. (D) KEGG analysis of the three types of ERF downstream genes.
To further investigate the pathways regulated by OfAP2/ERFs under ETH and Aza treatment, we conducted GO and KEGG enrichment analysis on downstream target genes of three types of OfERFs. GO enrichment has shown that downstream genes were involved in pollen recognition, cell recognition, the carboxylic acid metabolic process, the oxoacid metabolic process, and the organic acid metabolic process (Figure 4C). However, downstream genes of the K2-ERFs group are involved in more unique biological processes, such as translation, amide and peptide biosynthetic and metabolic processes, and organic and cellular nitrogen compound biosynthetic processes.
According to the KEGG enrichment, downstream genes of all three types of OfERFs are involved in cysteine and methionine metabolism, propanoate metabolism, and fatty acid biosynthesis (Figure 4D). Additionally, downstream genes of the K1-ERFs and K3-ERFs groups are involved in the MAPK signaling pathway plant, which is known to be associated with ethylene signaling transduction (Xu and Zhang, 2014; Chen et al., 2017). However, the downstream genes of the K2-ERFs group are independently involved in aminoacyl-tRNA biosynthesis and valine, leucine, and isoleucine biosynthesis. A recent study has shown that CHH hypermethylation induces petal senescence in carnations by reducing the accumulation of branched-chain amino acids (BcAAs, including valine, leucine, and isoleucine) (Feng et al., 2024). These results indicate that the Aza treatment-mediated DNA demethylation affects the decomposition metabolism of carboxylic acids, oxygen-containing acids, and organic acids, as well as the biosynthesis of fatty acids, through the ethylene signaling pathway. On the other hand, independent ethylene signaling pathways affect nitrogen metabolism, amide tRNA biosynthesis, and BcAA biosynthesis, mediating the flowering and senescence of sweet osmanthus.
Because of the GO enrichment analysis of downstream genes in K1-ERF, K2-ERF, and K3-ERF types, which simultaneously point to organic acid metabolism pathways (Figure 4C), we then constructed a transcriptional regulatory network for organic acid metabolism (Figure 5A). It regulates a total of 46 organic acid metabolism genes, of which 15 are simultaneously regulated by all three types of OfERFs, suggesting their similar functions. K1-ERFs regulate 18 organic acid metabolism genes, of which 16 are also regulated by K3-ERFs. Additionally, K2-ERFs independently regulate 19 organic acid metabolism genes. These results indicate that the flower senescence induced by ETH or Aza may both be achieved through the organic acid metabolism pathway mediated by OfERFs.
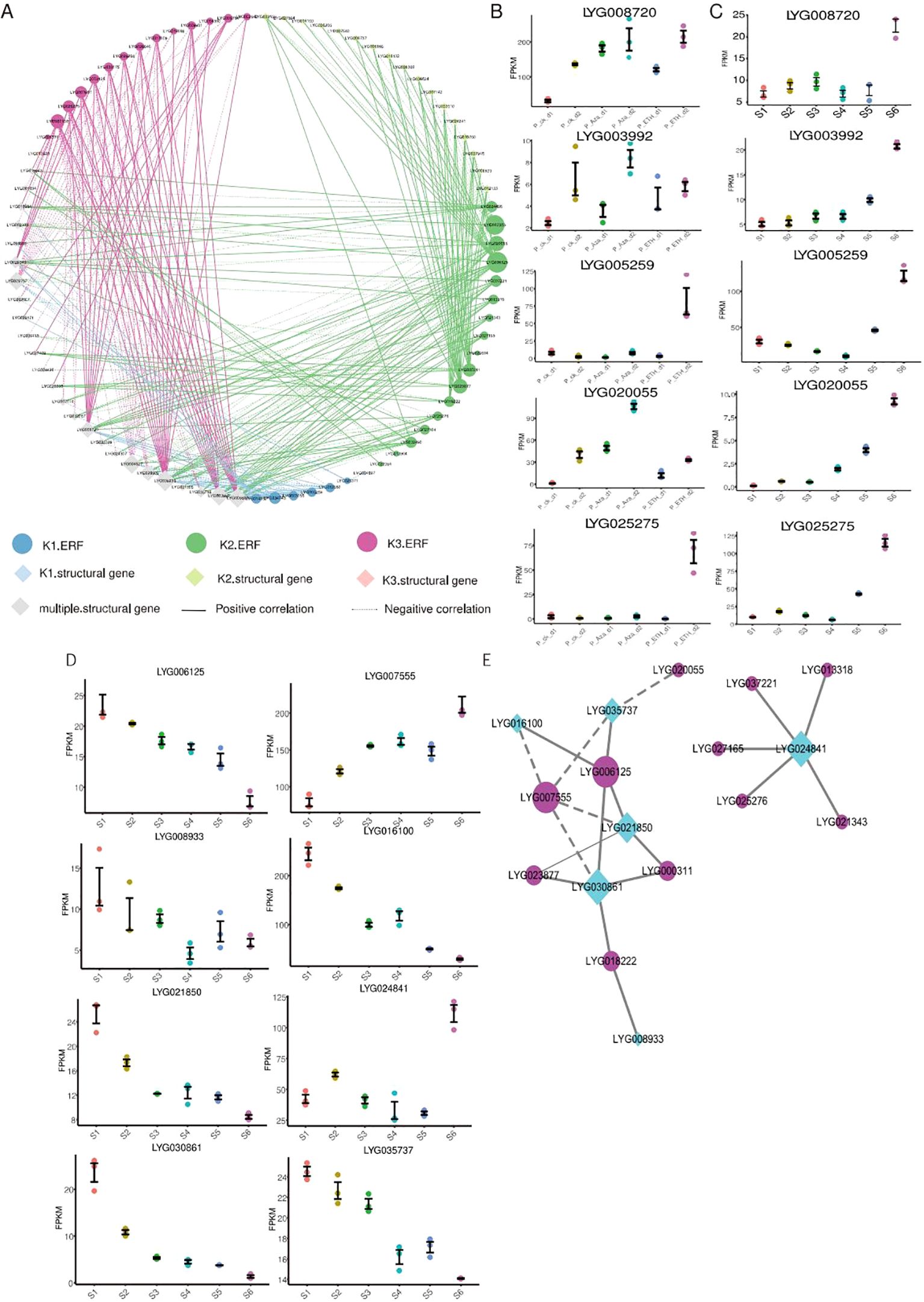
Figure 5. Analysis of genes involved in organic acid metabolism and branched-chain amino acid biosynthesis. (A) Transcriptional regulatory network of the organic acid metabolism process. The ‘multiple structural gene’ represent these genes are downstream cluster of ERFs.. (B) Expression of key genes involved in organic acid metabolism at six flowering stages. (C) Expression of key genes involved in organic acid metabolism under ETH and Aza treatment. (D) Expression of key genes involved in branched-chain amino acid biosynthesis at six flowering stages. (E) Transcriptional regulatory network of branched-chain amino acid biosynthesis.
In the whole transcriptional regulatory network, LYG039238, LYG008720, LYG006602, LYG028609, LYG003992, LYG009757, and LYG004527 are the key genes of organic acid metabolism (Figure 5A), and three of them upregulated during the flower senescence of sweet osmanthus. Among them, LYG008720 and LYG003992 are also induced by Aza and ETH treatments (Figures 5B, C). LYG034743, LYG014862, LYG005259, LYG006125, LYG020055, LYG000311, LYG007555, LYG023877, LYG003961, LYG007158, and LYG025275 are the key OfERFs in this network (Figure 5A), and most of them have upregulated during flower senescence. Among them, LYG005259, LYG020055, and LYG025275 belong to the K1-ERFs, K2-ERFs, and K3-ERFs groups, respectively. LYG005259 was induced by both Aza and ETH treatments, while LYG020055 was induced by Aza treatment, and LYG025275 was induced by ETH treatment (Figures 5B, C).
During the flower senescence of carnations, the accumulation of BcAAs decreases. Exogenous supplementation of BcAAs significantly delays the flower senescence of carnations (Feng et al., 2024), indicating that the decrease in BcAAs may be a hallmark of flower senescence. In our study, K2-ERFs regulated a total of six genes involved in BcAA biosynthesis. Interestingly, five of them downregulate within flower senescence (Figure 5D), which may lead to a decrease in the content of BcAAs during flower senescence. To understand the ERFs that regulate the BcAA biosynthesis, we constructed a transcriptional regulatory network from downstream genes of K2-ERFs (Figure 5E). The whole network can be divided into two sub-networks, one of which is composed of LYG024841 and five ERFs, and the other sub-network consists of six ERFs and five BcAA synthase genes. LYG030861 and LYG021850 are the key BcAA synthase genes, while LYG006125 and LYG007555 are the key ERF genes. LYG030861 is a homologous DHAD protein of Arabidopsis, which is almost not expressed during the abscission stage (S6 stage) of sweet osmanthus. LYG007555 encodes an ERF109 protein, which is upregulated during flower senescence (Figure 5D). In Arabidopsis, the mutation erf109 causes a decrease in ROS and delayed plant senescence (Lin et al., 2020). Silencing BrERF109 in Chinese cabbage promotes flavonoid biosynthesis and delays leaf senescence (Yue et al., 2023). These results suggest that LYG007555 may be a negative regulatory factor for flower senescence, delaying flower senescence by inhibiting the expression of LYG030861 and reducing the content of BcAAs.
It was found that OfERF017 (LYG025275) participates in the organic acid metabolism network, and its expression increases with flower senescence (Figures 5A, C), consistent with the increase of ethylene production (Zou et al., 2023) and the decrease of total organic acid in S1 to S6 (Supplementary Table S3). Thus, OfERF017 was chosen as an example to investigate its function in organic acid decomposition metabolism and flower senescence.
The results of qPCR showed that OfERF017 was mainly expressed in S5 and S6 and reached its peak in S6 (Figure 6A). Overexpression of OfERF017 in transgenic flowers of O. fragrans (OfERF017-OE) (Figure 6B) led to a significant increase of endogenous ethylene production (Figure 6C) and an earlier senescence phenotype (Figure 6D). After 1 day of inoculation, OfERF017-OE exhibited partial flower browning, with only a small number of flowers browning in CK and P2300. By the third day, all flowers of OfERF017-OE showed browning, while some flowers of WT and P2300 remained yellow without decay (Figure 6D). These results indicate that OfERF017 promotes flower senescence in transgenic O. fragrans.
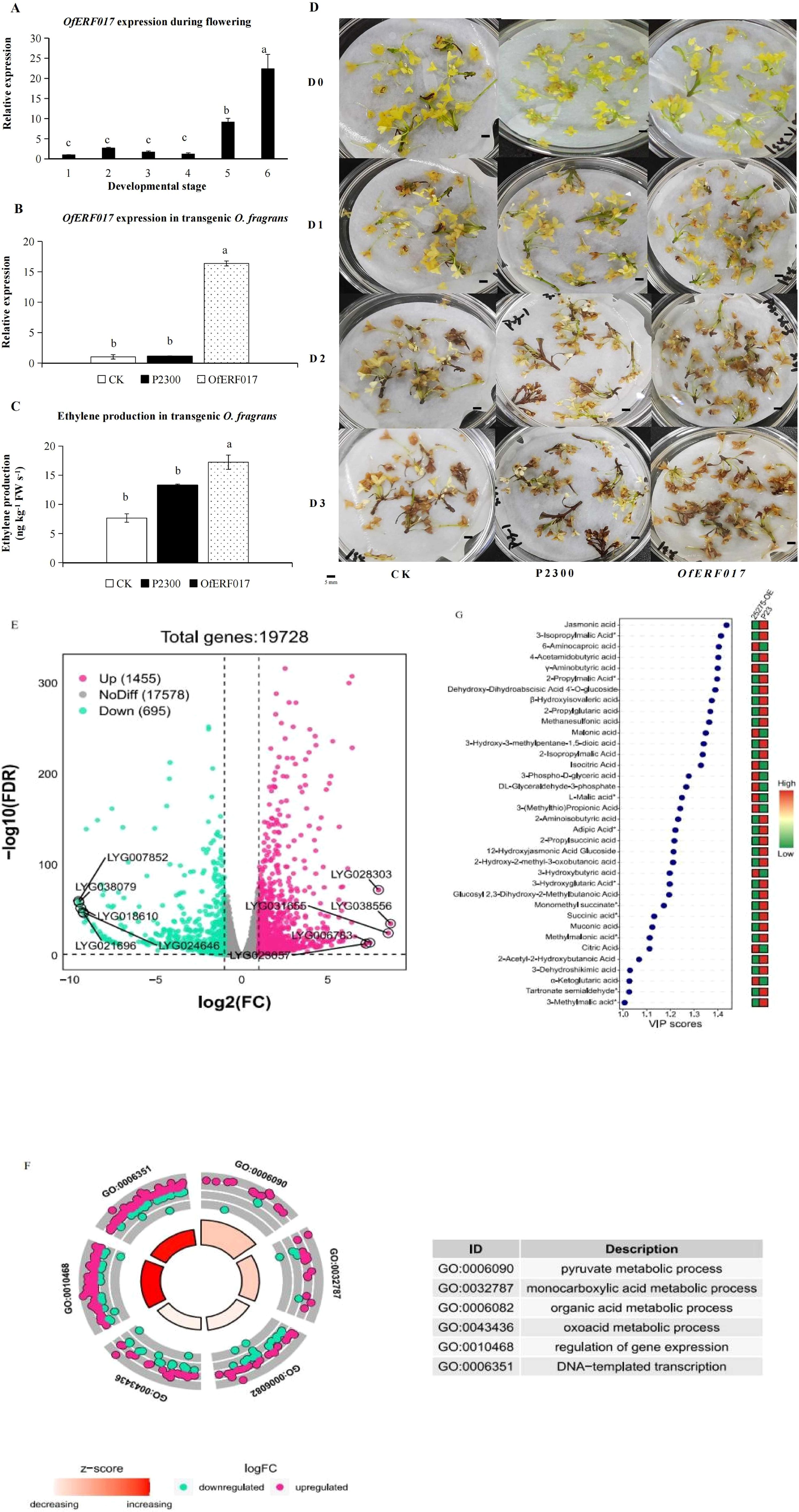
Figure 6. Functional characterization of OfERF017 in transgenic O. fragrans. (A) The expression analysis of OfERF017 during flowering. (B) The expression analysis of OfERF017 in transgenic O. fragrans. (C) Ethylene production in OfERF017-overexpressing transgenic O. fragrans. (D) The phenotype of OfERF017-overexpressing transgenic flowers in O. fragrans. (E) Volcano map of differentially expressed genes (DEGs) in OfERF017-overexpressing transgenic flowers. (F) Gene Ontology enrichment results of DEGs in OfERF017-overexpressing transgenic flowers. (G) Heatmap of differential organic acid content in OfERF017-overexpressing transgenic flowers.
We then performed transcriptome analysis in OfERF017-OE and P2300 flowers. Based on |log2fold change| > 1 and FDR < 0.05, 2,150 differentially expressed genes (DEGs) were identified from OfERF017-OE, including 1,455 upregulated DEGs and 695 downregulated DEGs (Figure 6E; Supplementary Table S4). GO enrichment showed that these DEGs are involved in various biological processes (Supplementary Table S5). It is worth noting that the terms “DNA template transcription” and “regulation of gene expression” are significantly enriched (p-values of 9.89 × 10−3 and 4.69 × 10−3, respectively), representing the function of OfERF017 as a transcription factor (Figure 6F). In addition, the “pyruvate metabolism process” (p-value of 1.62 × 10−4), “monocarboxylate metabolism process” (p-value of 1.05 × 10−2), “oxygenic acid metabolism process” (p-value of 3.88 × 10−2), and “organic acid metabolism process” (p-value of 2.79 × 10−2) were significantly enriched. It is interesting that the number of upregulated DEGs during these processes is greater than that of downregulated DEGs (Figure 6F). LC-MS/MS was used to detect organic acids in OfERF017-OE and P2300. A total of 65 organic acids were detected, with which the total content was downregulated from 2.00E+08 to 1.71E+08 in OfERF017-OE (Figure 6G). In summary, our research findings indicate that OfERF017 responds to ethylene by inducing high expression of genes involved in organic acid metabolism pathways, leading to a decrease in organic acid levels and promoting the flower senescence of O. fragrans.
Ethylene is an important regulatory factor in the flower senescence of O. fragrans (Zou et al., 2014), and DNA hypomethylation participates in the flower senescence mediated by OfERFs through the ethylene response pathways (Zou et al., 2023). In this study, a total of 227 OfAP2/ERFs genes were identified in the O. fragrans “Liuye Jingui” genome. Chromosome 1 contained the greatest numbers of OfERFs genes (20 OfERFs, 9.43%), while chromosome 18 contained the lowest numbers (only 2 OfERFs, 0.94%). There are only four OfERFs on chromosome 9 (1.89%) and three OfERFs on chromosomes 21 and 22 (1.42%). OfERF genes on other chromosomes are generally between 5 and 16 (84.9%) (Figure 1). It can be seen from this that the number of OfERFs distributed on each chromosome is not positively correlated with chromosome length. OfAP2/ERFs in O. fragrans are higher than those in most plant species, such as A. thaliana (172) (Sakuma et al., 2002), Populus trichocarpa (200) (Zhuang et al., 2008), Hypericum perforatum (101) (Zhang et al., 2021b), Salix arbutifolia (173) (Rao et al., 2015), Camellia sinensis (178) (Liu et al., 2023), Rose chinensis (137) (Li et al., 2020), and Cucurbita moschia (212) (Li et al., 2022a), and most of them have played an important role in abiotic stress. Especially in the ERF subfamily, there are 125 OfERFs, far exceeding 88 SaERFs, 88 CsERFs, and 92 CmoERFs. Additionally, collinearity analysis showed that the majority of OfERF gene pairs are segmental duplicates, with five pairs being tandem duplicates. Segmental duplicates generate homologous genes, and tandem duplicates produce gene clusters (Zhang and Li, 2018), which leads to an expansion in the number of OfAP2/ERFs genes.
To investigate the similarities and differences of the response pattern of OfAP2/ERFs to ethylene in sweet osmanthus, RNA-seq of cut flowers treated with Aza and ETH was performed, as both of them caused earlier flower senescence and an increase in endogenous ethylene production. According to the different expression patterns of OfAP2/ERFs in treatments, OfAP2/ERFs were divided into K1 to K3 groups, and the downstream target genes of the three types of OfAP2/ERFs were analyzed. GO enrichment analysis has shown that downstream genes were involved in pollen recognition, cell recognition, the carboxylic acid metabolic process, the oxoacid metabolic process, and the organic acid metabolic process (Figure 4C). Organic acids widely exist in plants and degrade during the development and storage of fruits and flowers (Zhang et al., 2014). It has been reported that nitrogen nutrition is closely related to ethylene signaling and plant senescence, and low nitrogen conditions can induce early leaf senescence in Arabidopsis (Cheng et al., 2023; Fan et al., 2023; Ma et al., 2023).
Citric acid, humic acid, and salicylic acid in organic acids are often used as preservatives for fresh cut flowers (Fan et al., 2015; Aziz et al., 2020; Rahbar et al., 2023), which cleared the ROS accumulated by senescent damage or by retaining cellular water. We constructed a transcriptional regulatory network for organic acid metabolism (Figure 5A). Among them, LYG008720 is a homologous protein of pyruvate kinase (PK) in Arabidopsis, which plays an important role in cellular senescence metabolic reprogramming (Dou et al., 2023; Wu et al., 2023). PKs are closely related to growth, development, and stress resistance in plants (Wulfert et al., 2020). LYG003992 is a homologous protein of sphingosine-1-phosphate lyase in Arabidopsis, involved in regulating the levels of long-chain bases and their phosphorylated derivatives (LCB-P), and may play a role in senescence of Arabidopsis (Chen et al., 2009). At the same time, the protein also plays a role in water deficiency stress (Nakagawa et al., 2012), which is in concordance with water loss during the flower senescence of O. fragrans. LYG005259, LYG020055, and LYG025275 are the key OfERFs. LYG005259 and LYG025275 are homologous proteins of ERF017 in Arabidopsis that have been associated with nitrogen metabolism (Gaudinier et al., 2018) and fruit peel degreening (Han et al., 2018). LYG020055 is a homologous protein of AtERF114 in Arabidopsis that is involved in nitrogen metabolism (Gaudinier et al., 2018) and disease resistance (Li et al., 2022b). Then, LYG025275 (OfERF017) was chosen as an example to investigate its function in organic acid decomposition metabolism and flower senescence via transgenic flowers of O. fragrans. The results showed that overexpression of OfERF017 induced the high expression of genes involved in organic acid metabolism pathways, leading to a decrease in organic acid levels and promoting the flower senescence of O. fragrans, which confirms the previous analysis that OfAP2/ERFs may regulate the flower senescence of O. fragrans through organic acid metabolism.
This is the first comprehensive analysis of the AP2/ERF superfamily in O. fragrans ‘Liuye Jingui’. The phylogenetic relationship, gene structures, protein interaction network, and expression patterns in various tissues and flowering stages were analyzed. By comparative transcriptome analysis, the downstream genes of OfAP2/ERFs affected by ETH and Aza treatment were analyzed, and these target gene sets participate in similar or different biological processes and metabolic pathways, suggesting that ethylene and DNA hypomethylation have crosstalk and a unique mechanism in regulating the flower senescence of O. fragrans. Co-expression analysis revealed that several key OfAP2/ERFs played a central role in organic acid metabolism and biosynthesis of BcAAs, among which OfERF017 was selected for further functional analysis. Overexpression of OfERF017 increased the endogenous ethylene production, leading to an earlier senescence phenotype and a decrease in organic acid levels and promoting the flower senescence of O. fragrans. Overall, these findings will lay a valuable foundation to elucidate the functions of OfAP2/ERFs in O. fragrans.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
J-JZ: Conceptualization, Project administration, Writing – original draft. JuZ: Formal analysis, Writing – original draft. XW: Investigation, Writing – review & editing. HX: Visualization, Writing – review & editing. XZ: Writing – review & editing, Investigation. XC: Methodology, Writing – review & editing. JY: Formal analysis, Writing – review & editing. JiZ: Software, Writing – review & editing. ZL: Resources, Writing – review & editing. GZ: Project administration, Writing – review & editing. HC: Funding acquisition, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by research grants provided by the National Natural Science Foundation of China (32271951, 32101581, and 32372754), the Hubei Provincial Central Leading Local Special Project (2022BGE263), the Hubei Province Natural Science Foundation (2023AFB1063 and 2024AFB1057), the Innovation Team Project from Hubei University of Science and Technology (2022T02), and PhD grant from Hubei University of Science and Technology (BK202419 and BK202002).
Authors JuZ and ZL were employed by the company IGENEBOOK Biotechnology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1467232/full#supplementary-material
Supplementary Figure 1 | Synteny analysis of OfAP2/ERFs in O. fragrans and Arabidopsis. (A) Collinearity analysis of 227 OfAP2/ERFs in O. fragrans. The gray lines represent collinearity among all gene pairs in the O. fragrans genome, while the red lines indicate collinearity relationships between genes of the OfAP2/ERFs family. (B) Collinearity analysis between 172 OfAP2/ERFs of O. fragrans and Arabidopsis genes. The gray lines denote all collinear gene pairs between the O. fragrans and Arabidopsis genomes, whereas the purple lines represent the OfAP2/ERFs genes of O. fragrans. Genes labeled with different colors represent distinct subfamilies of OfAP2/ERFs.
Supplementary Figure 2 | Gene sequence and motif analysis of OfAP2/ERF in sweet osmanthus. (A) RAV subfamily; (B) Soloist subfamily; (C) AP2 subfamily; (D) DREB subfamily; and (E) ERF subfamily. To reveal the underlying regulatory mechanism of OfAP2/ERFs, cis-elements of OfAP2/ERFs.
Supplementary Figure 3 | Analysis of cis-elements in the promoters of OfAp2/ERF genes. (A) AP2 subfamily; (B) DREB subfamily; (C) RAV subfamily; (D) Soloist subfamily; (E) ERF subfamily. For panels A to E, the heatmap on the left displays the number of four major categories of cis-elements on different genes, where deeper shades of red indicate a greater quantity represented, while greener shades indicate fewer. The middle plot illustrates the positional distribution of different cis-elements on each gene. The stacked bar chart on the right represents the quantity of different types of elements on each gene.
Supplementary Figure 4 | An earlier appearance of senescence and increase of endogenous ethylene production is triggered by Aza and ETH treatment. (A) Morphology of cut flowers treated with 10 mM 5′-azacytidine (Aza), distilled water (CK), and 50 mg L−1 ethephon (ETH) showing that the senescence process was accelerated in response to Aza and ETH treatment. (B) Aza and ETH treatment significantly increased the release of endogenous ethylene production of cut flowers (D1 and D2). Error bars indicate the SE. Asterisks indicate the significance at p < 0.05. Three biological replicates under Aza or distilled water treatment were used. ns, nonsignificant.
Supplementary Figure 5 | The principal component analysis (PCA) of all genes and ethylene biosynthesis genes. (A) The PCA of samples based on all genes only separate in different vase life but not separate from each treatment. (B) The PCA of samples based on ethylene biosynthesis genes separate in both different vase life and treatment.
Alonso, J. M., Stepanova, A. N., Solano, R., Wisman, E., Ferrari, S., Ausubel, F. M., et al. (2003). Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 100, 2992–2997. doi: 10.1073/pnas.0438070100
Aziz, S., Younis, A., Jaskani, M. J., Ahmad, R. (2020). Effect of PGRs on antioxidant activity and phytochemical in delay senescence of lily cut flowers. Agronomy. 10, 1704. doi: 10.3390/agronomy10111704
Chen, J., Zhang, Q., Wang, Q., Feng, M., Li, Y., Meng, Y., et al. (2017). RhMKK9, a rose MAP KINASE KINASE gene, is involved in rehydration-triggered ethylene production in rose gynoecia. BMC Plant Biol. 17, 51. doi: 10.1186/s12870-017-0999-1
Chen, M., Cahoon, E. B., Saucedo-García, M., Plasencia, J., Gavilanes-Ruíz, M. (2009). Plant Sphingolipids: Structure, Synthesis and Function. In: Wada, H., Murata, N. (eds) Lipids in Photosynthesis. Advances in Photosynthesis and Respiration, vol 30. Dordrecht: Springer. doi: 10.1007/978-90-481-2863-1_5
Chen, W., Gong, L., Guo, Z., Wang, W., Zhang, H., Liu, X., et al. (2013). A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Mol. Plant 6, 1769–1780. doi: 10.1093/mp/sst080
Chen, C., Wu, Y., Li, J., Wang, X., Zeng, Z., Xu, J., et al. (2023). TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 16, 1733–1742. doi: 10.1016/j.molp.2023.09.010
Chen, H. G., Zeng, X. L., Yang, J., Cai, X., Shi, Y. M., Zheng, R. R., et al. (2021). Whole-genome resequencing of Osmanthus fragrans provides insights into flower color evolution. Hortic. Res. 8, 98. doi: 10.1038/s41438-021-00531-0
Cheng, S. L. H., Xu, H., Ng, J. H. T., Chua, N. H. (2023). Systemic movement of long non-coding RNA ELENA1 attenuates leaf senescence under nitrogen deficiency. Nat. Plants 9, 1598–1606. doi: 10.1038/s41477-023-01521-x
Dou, X., Fu, Q., Long, Q., Liu, S., Zou, Y., Fu, D., et al. (2023). PDK4-dependent hypercatabolism and lactate production of senescent cells promotes cancer Malignancy. Nat. Metab. 5, 1887–1910. doi: 10.1038/s42255-023-00912-w
Fan, H. M., Li, T., Sun, X., Sun, X. Z., Zheng, C. S. (2015). Effects of humic acid derived from sediments on the postharvest vase life extension in cut chrysanthemum flowers. Postharvest Biol. Tec. 101, 82–87. doi: 10.1016/j.postharvbio.2014.09.019
Fan, H., Quan, S., Ye, Q., YI, K., Crawford, N. M., Wang, Y. (2023). A molecular framework underlying low-nitrogen-induced early leaf senescence in Arabidopsis thaliana. Mol. Plant 16, 756–774. doi: 10.1016/j.molp.2023.03.006
Fan, S., Zhang, D., Gao, C., Zhao, M., Wu, H., Li, Y. (2017). Identification, classification, and expression analysis of GRAS gene family in Malus domestica. Front. Physiol. 8. doi: 10.3389/fphys.2017.00253
Feng, K., Hou, X., Xing, G., Liu, J., Duan, A., Xu, Z., et al. (2020). Advances in AP2/ERF super-family transcription factors in plant. Crit. Rev. Biotechnol. 40, 1–27. doi: 10.1080/07388551.2020.1768509
Feng, S., Jiang, X., Huang, Z., Li, F., Wang, R., Yuan, X., et al. (2024). DNA mEthylation remodeled amino acids biosynthesis regulates flower senescence in carnation (Dianthus caryophyllus). New Phytologist. 241, 1605–1620. doi: 10.1111/nph.19499
Finn, R. D., Clements, J., Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. doi: 10.1093/nar/gkr367
Gaudinier, A., Rodriguez-Medina, J., Zhang, L., Olson, A., Liseron-Monfils, C., Bågman, A. M., et al. (2018). Transcriptional regulation of nitrogen-associated metabolism and growth. Nature. 563, 259–264. doi: 10.1038/s41586-018-0656-3
Han, Z., Hu, Y., Lv, Y., Rose, J. K. C., Sun, Y., Shen, F., et al. (2018). Natural variation underlies differences in ETHYLENE RESPONSE FACTOR17 activity in fruit peel degreening. Plant Physiol. 176, 2292–2304. doi: 10.1104/pp.17.01320
Hu, B., Jin, J., Guo, A.-Y., Zhang, H., Luo, J., Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evolution. 5, 1547–1549. doi: 10.1093/molbev/msy096
Li, J., Hou, H., Li, X., Xiang, J., Yin, X., Gao, H., et al. (2013). Genome-wide identification and analysis of the SBP-box family genes in apple (Malus× domestica Borkh.). Plant Physiol. Biochem. 70, 100–114. doi: 10.1016/j.plaphy.2013.05.021
Li, D., Liu, X., Shu, S., Zhang, H., ZHang, S., Song, Y., et al. (2020). Global analysis of the AP2/ERF gene family in rose (Rosa chinensis) genome unveils the role of RcERF099 in Botrytis resistance. BMC Plant Biol. 20, 533. doi: 10.1186/s12870-020-02740-6
Li, Q., Zhang, L., Chen, P., Wu, C., Zhang, H., Yuan, J., et al. (2022a). Genome-wide identification of APETALA2/ETHYLENE RESPONSIVE FACTOR transcription factors in cucurbita moschata and their involvement in ethylene response. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.847754
Li, Z., Zhang, Y., Ren, J., Jia, F., Zeng, H., Li, G., et al. (2022b). Ethylene-responsive factor ERF114 mediates fungal pathogen effector PevD1-induced disease resistance in Arabidopsis thaliana. Mol. Plant Pathology. 23, 819–831. doi: 10.1111/mpp.13208
Lin, W., Zhang, H., Huang, D., Schenke, D., Cai, D., Wu, B., et al. (2020). Dual-localized WHIRLY1 affects salicylic acid biosynthesis via coordination of isochorismate synthase1, phenylalanine ammonia lyase1, and s-adenosyl-l-mEthionine-dependent mEthyltransferase1. Plant Physiol. 184 (4), 1884–1899. doi: 10.1104/pp.20.00964
Liu, Y., Chen, S., Chen, J., Wang, J., Wei, M., Tian, X., et al. (2023). Comprehensive analysis and expression profiles of the AP2/ERF gene family during spring bud break in tea plant (Camellia sinensis). BMC Plant Biol. 23, 206. doi: 10.1186/s12870-023-04221-y
Luangsuwalai, K., Ketsa, S., Van Doorn, W. G. (2011). Ethylene-regulated hastening of perianth senescence after pollination in Dendrobium flowers is not due to an increase in perianth Ethylene production. Postharvest Biol. Tec. 62, 338–341. doi: 10.1016/j.postharvbio.2011.07.011
Ma, B., Ma, T., Xian, W., Hu, B., Chu, C. (2023). Interplay between Ethylene and nitrogen nutrition: How Ethylene orchestrates nitrogen responses in plants. J. Integr. Plant Biol. 2, 399–407. doi: 10.1111/jipb.13355
Magali, L., Patrice, D., Gert, T., Kathleen, M., Yves, M., Yves, V. D. P., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Nakagawa, N., Kato, M., Takahashi, Y., Shimazaki, K. I., Tamura, K., Tokuji, Y. (2012). Degradation of long-chain base 1-phosphate (LCBP) in Arabidopsis: functional characterization of LCBP phosphatase involved in the dehydration stress response. J. Plant Res. 125, 439–449. doi: 10.1007/s10265-011-0451-9
Pan, C., Zhao, L., Zhao, D. (2021). Microwave-assisted green extraction of antioxidant components from Osmanthus fragrans (Lour) flower using natural deep eutectic solvents. J. Appl. Res. Med. Aroma. Plants. 20, 100285. doi: 10.1016/j.jarmap.2020.100285
Rahbar, F. G., Vaziri, A., Asil, M. H., Sasani, S. T., Olfati, J. (2023). Effects of humic acid on antioxidant defense system and senescence-related gene expression in leaves of longiflorum × asiatic of lilies (LA lilium hybrid). J. Soil Sci. Plant Nutr. 23, 3500–3507. doi: 10.1007/s42729-023-01266-8
Rao, G., Sui, J., Zeng, Y., He, C., Zheng, J. (2015). Genome-wide analysis of the AP2/ERF gene family in Salix arbutifolia. FEBS Open Bio. 5, 132–137. doi: 10.1016/j.fob.2015.02.002
Sakuma, Y., Liu, Q., Dubouzet, J. G., Abe, H., Shinozaki, K., Yamaguchi-Shinozaki, K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophy. Res. Commu. 290, 998–1009. doi: 10.1006/bbrc.2001.6299
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shi, T., Zhou, L., Ye, Y., Yang, X., WANg, L., Yue, Y. (2024). Characterization of YABBY transcription factors in Osmanthus fragrans and functional analysis of OfYABBY12 in floral scent formation and leaf morphology. BMC Plant Biol. 24, 589. doi: 10.1186/s12870-024-05047-y
Sun, X., Qin, M., Yu, Q., Huang, Z., Xiao, Y., Li, Y., et al. (2021). Molecular understanding of postharvest flower opening and senescence. Mol. Hortic. 1, 7–7. doi: 10.1186/s43897-021-00015-8
Szklarczyk, D., Rebecca, K., Mikaela, K., Katerina, N., Farrokh, M., Hachilif, R., et al. (2023). The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51 (D1), D638–D646. doi: 10.1093/nar/gkac1000
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Wu, L., Liu, J., Huang, W., Wang, Y., Chen, Q., Lu, B. (2022). Exploration of Osmanthus fragrans Lour.’s composition, nutraceutical functions and applications. Food Chem. 377, 131853. doi: 10.1016/j.foodchem.2021.131853
Wu, Y., Tang, L., Huang, H., Yu, Q., Hu, B., Wang, G., et al. (2023). Phosphoglycerate dehydrogenase activates PKM2 to phosphorylate histone H3T11 and attenuate cellular senescence. Nat. Commun. 14, 1323. doi: 10.1038/s41467-023-37094-8
Wulfert, S., Schilasky, S., Krueger, S. (2020). Transcriptional and biochemical characterization of cytosolic pyruvate kinases in arabidopsis thaliana. Plants. 9, 353. doi: 10.3390/plants9030353
Xi, W., He, Y., Zhu, L., Hu, S., Xiong, S., Zhnag, Y., et al. (2021). CPTA treatment reveals potential transcription factors associated with carotenoid metabolism in flowers of. Osmanthus fragrans. Hortic. Plant J. 7, 1–9. doi: 10.1016/j.hpj.2021.03.002
Xia, H., Zhang, Y. T., Chen, X., Zeng, X. L., Cai, X., Li, Z. Q., et al. (2024). Genome-Wide Identification of Osmanthus fragrans Histone Modification Genes and Analysis of Their Expression during the Flowering Process and under Azacytidine and Ethylene Treatments. Plants. 13, 777. doi: 10.3390/plants13060777
Xu, J., Xu, H., Liu, Y., Wang, X., Xu, Q., Deng, X. (2015). Genome-wide identification of sweet orange (Citrus sinensis) histone modification gene families and their expression analysis during the fruit development and fruit-blue mold infection process. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00607
Xu, J., Zhang, S. (2014). Regulation of ethylene biosynthesis and signaling by protein kinases and phosphatases. Mol. Plant 7 (6), 939–942. doi: 10.1093/mp/ssu059
Yang, H., Sun, Y., Wang, H., Zhao, T., Xu, X., Jiang, J., et al. (2021). Genome-wide identification and functional analysis of the ERF2 gene family in response to disease resistance against Stemphylium lycopersici in tomato. BMC Plant Biol. 21, 72. doi: 10.1186/s12870-021-02848-3
Yue, Y., Wang, H., Wang, T., Zheng, D., Thomas, H. R., Yang, X., et al. (2024). Integrated physiological, hormonal, and transcriptomic analyses reveal a novel E3 ubiquitin ligase-mediated cold acclimation mechanism for the acquisition of cold tolerance in sweet osmanthus. Ind. Crops Products. 220, 119171–119171. doi: 10.1016/j.indcrop.2024.119171
Yue, L., Kang, Y., Zhong, M., Kang, D., Zhao, P., Chai, X., et al. (2023). Melatonin delays postharvest senescence through suppressing the inhibition of BrERF2/BrERF109 on flavonoid biosynthesis in flowering chinese cabbage. Int. J. Mol. Sci. 24 (3), 2933. doi: 10.3390/ijms24032933
Zhang, Y., Hu, C. X., Tan, Q. L., Zheng, C. S., Gui, H. P., Zeng, W. N., et al. (2014). Plant nutrition status yield and quality of satsuma mandarin (Citrus unshiu, Marc.) under soil application of Fe-EDDHA and combination with zinc and manganese in calcareous soil. Sci. Hortic. 174, 46–53. doi: 10.1016/j.scienta.2014.05.005
Zhang, Z., Li, X. (2018). Genome-wide identification of AP2/ERF superfamily genes and their expression during fruit ripening of Chinese jujube. Sci. Rep. 8, 15612. doi: 10.1038/s41598-018-33744-w
Zhang, H., Pan, X., Liu, S., Lin, W., Li, Y., Zhang, X. (2021a). Genome-wide analysis of AP2/ERF transcription factors in pineapple reveals functional divergence during flowering induction mediated by ethylene and floral organ development. Genomics. 113, 474–489. doi: 10.1016/j.ygeno.2020.10.040
Zhang, Q., Zhou, W., Li, B., Li, L., Fu, M., Zhou, L., et al. (2021b). Genome-wide analysis and the expression pattern of the ERF gene family in hypericum perforatum. Plants 10, 133. doi: 10.3390/plants10010133
Zhou, L., Tian, Q., Ding, W., Hu, H., Shi, T., Ning, G., et al. (2024). The OfMYB1R114-OfSDIR1-like-OfCCD4 module regulates β-ionone synthesis in Osmanthus fragrans. Ind. Crops Products 217, 118879. doi: 10.1016/j.indcrop.2024.118879
Zhou, Y., Wang, C. Y., Cheng, Z. W. (2008). Effects of exogenous Ethylene and Ethylene inhibitor on longevity and petal senescence of sweet osmanthus. Acta Hortic. 768, 487–493. doi: 10.17660/ActaHortic.2008.768.65
Zhuang, J., Cai, B., Peng, R. H., Zhu, B., Jin, X. F., Xue, Y., et al. (2008). Genome-wide analysis of the AP2/ERF gene family in Populus trichocarpa. Biochem. Biophys. Res. Commun. 4, 371(3):468–74. doi: 10.1016/j.bbrc.2008.04.087
Zou, J. J., Cai, X., Wang, C. Y. (2017). The spatial and temporal distribution of programmed cell death (PCD) during petal senescence of Osmanthus fragrans. Acta Hortic. 1185, 315–324. doi: 10.17660/ActaHortic.2017.1185.39
Zou, J. J., Cai, X., Yang, J., Zeng, X., Liu, D. X., Huang, S., et al. (2023). DNA hypomEthylation mediates flower opening and senescence in sweet osmanthus through auxin and Ethylene responsive pathways. Postharvest Biol. Tec. 198, 112250. doi: 10.1016/j.postharvbio.2023.112250
Keywords: sweet osmanthus (Osmanthus fragrans), ethylene-responsive transcription factor (ERF), organic acid decomposition metabolism, flower senescence, gene expression patterns
Citation: Zou J-J, Zhang J, Wang X, Xia H, Zeng X, Cai X, Yang J, Zeng J, Li Z, Zhang G and Chen H (2024) Comprehensive transcriptome analysis of AP2/ERFs in Osmanthus fragrans reveals the role of OfERF017-mediated organic acid metabolism pathway in flower senescence. Front. Plant Sci. 15:1467232. doi: 10.3389/fpls.2024.1467232
Received: 19 July 2024; Accepted: 26 August 2024;
Published: 26 September 2024.
Edited by:
Peng Wang, Jiangsu Province and Chinese Academy of Sciences, ChinaReviewed by:
Yuanzheng Yue, Nanjing Forestry University, ChinaCopyright © 2024 Zou, Zhang, Wang, Xia, Zeng, Cai, Yang, Zeng, Li, Zhang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zeqing Li, bGl6ZXFpbmdAaGJ1c3QuZWR1LmNu; Guifu Zhang, WmhhbmdfR3VpZnUxOTY4QDE2My5jb20=; Hongguo Chen, Q2hlbl9Ib25nZ3VvMTk2OUAxNjMuY29t
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.