
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 05 September 2024
Sec. Plant Abiotic Stress
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1467015
This article is part of the Research Topic Advanced Breeding for Abiotic Stress Tolerance in Crops, Volume II View all 18 articles
 Jie Zheng1
Jie Zheng1 Yunxiao Ma1
Yunxiao Ma1 Yu Liang1
Yu Liang1 Tianhan Zhang1
Tianhan Zhang1 Chang Chen1
Chang Chen1 Aduragbemi Amo2,3
Aduragbemi Amo2,3 Wenyu Wang4
Wenyu Wang4 Fangfang Ma1
Fangfang Ma1 Yuanhuai Han1
Yuanhuai Han1 Hongying Li1
Hongying Li1 Siyu Hou1*
Siyu Hou1* Yang Yang1*
Yang Yang1*Essential mineral elements such as zinc and iron play a crucial role in maintaining crop growth and development, as well as ensuring human health. Foxtail millet is an ancient food crop rich in mineral elements and constitutes an important dietary supplement for nutrient-deficient populations. The ZIP (ZRT, IRT-like protein) transporters are primarily responsible for the absorption, transportation and accumulation of Zn, Fe and other metal ions in plants. Here, we identified 14 ZIP transporters in foxtail millet (SiZIP) and systematically characterized their phylogenetic relationships, expression characteristics, sequence variations, and responses to various abiotic stresses. As a result, SiZIPs display rich spatiotemporal expression characteristics in foxtail millet. Multiple SiZIPs demonstrated significant responses to Fe, Cd, Na, and K metal ions, as well as drought and cold stresses. Based on homologous comparisons, expression characteristics and previous studies, the functions of SiZIPs were predicted as being classified into several categories: absorption/efflux, transport/distribution and accumulation of metal ions. Simultaneously, a schematic diagram of SiZIP was drawn. In general, SiZIPs have diverse functions and extensively involve in the transport of metal ions and osmotic regulation under abiotic stresses. This work provides a fundamental framework for the transport and accumulation of mineral elements and will facilitate the quality improvement of foxtail millet.
As essential mineral elements, iron (Fe) and zinc (Zn) exert a significant role in sustaining the growth, development, and physiological metabolism of plants. Zn serves as a cofactor for over 300 enzymes, encompassing zinc finger binding protein, ring finger protein, single serine protein kinase, etc., and acts as a binding factor for the normal functionalities of more than 2000 transcription factors, influencing biological metabolism, biomembrane stability, and gene expression (Coleman, 1998). Zn deficiency can have a substantial impact on plant growth and development, such as inducing oxidative damage (Cakmak, 2000); inhibiting auxin synthesis (Broadley et al., 2012); reducing RNA polymerase activity, decreasing the number of ribosomes, and lowering the rate and content of protein synthesis (Ho, 2004). Fe, existing in two ionic forms of Fe2+ and Fe3+, participates in the synthesis of chlorophyll precursor ALA (δ-Aminolevulinic Acid) (Pushnik et al., 1984) and the synthesis of crucial plant proteins like Fe-S proteins (Rouault, 2015), which are indispensable for maintaining the normal operation of various physiological processes in plants, including photosynthesis, respiration, and nitrogen fixation (Broadley et al., 2012). A moderate increase of Zn and Fe in crops has a positive influence on yield and quality, while their deficiency can result in stunted plants and a considerable reduction in the Zn and Fe content of grains (Sadeghzadeh, 2013; Pinson et al., 2015).
In addition to being significant mineral elements that directly impact plant growth and development, Zn and Fe are also indispensable trace elements for humans, and their accumulation within the body is closely associated with human health. More than half of the global population has a seriously insufficient intake of Fe, Zn, and selenium (Se) (White and Broadley, 2009; Trijatmiko et al., 2016). The ‘Hidden Hunger’ caused by the deficiency of essential vitamins and micronutrients has affected two billion people worldwide and emerges as a global challenge (Khush et al., 2012). Insufficient intake of essential micronutrients, aside from causing a weakened immune system, also leads to poor physical and mental development in children and even fatal diseases (Bourke et al., 2016). Moreover, with the advancement of industry and agriculture, such as mining, metal smelting and excessive application of pesticides and fertilizers, there is severe heavy metal pollution such as cadmium (Cd) in the soil, which can readily enter the human body through the food chain and directly threaten human health (Pan et al., 2016; Yang et al., 2021c). Due to the similar geochemical behavior of metal elements and the poor specificity of plant metal transporters, there exists a certain synergism/antagonism between trace metal elements and harmful heavy metal elements in the uptake, transport, and accumulation of crops (Bolan et al., 2003; Baxter, 2015; Wang et al., 2022).
The zinc-regulated transporters, iron-regulated transporter-like protein (ZIP) family acts as the primary functional proteins in plants responsible for absorbing Zn and Fe ions from the soil. It comprises two major categories of protein members: zinc-regulated transporter (ZRT) and iron-regulated transporter (IRT)-like proteins. Members of the ZIP family are widely distributed in various tissues of plants and play crucial roles in the absorption, transport, distribution and utilization of Zn and Fe ions (Eide, 2006). AtIRT1 was first discovered in Arabidopsis to specifically transport Fe and Zn ions in roots and was induced by Fe deficiency (Eide et al., 1996); Compared to the wild type, the irt1 mutant displayed a 70% reduction in Fe content in leaves and exhibited a chlorotic phenotype (Vert et al., 2002), and overexpression of AtIRT1 in the irt1 mutant could alleviate the chlorotic phenotype (Krämer et al., 2007); AtIRT2 also demonstrated a similar function to AtIRT1 in transporting Fe (Vert et al., 2001). AtZIP1 and AtZIP2 are crucial for plants to absorb manganese (Mn) and Zn through the root system and transport them from the roots to the leaves (Milner et al., 2013). Although both are reported as major Zn transporter genes, their subcellular localizations and tissue expression patterns are slightly different, indicating that these two genes have distinct functions. AtZIP1 is primarily expressed on the tonoplast of leaf veins and root cells, while AtZIP2 is highly expressed on the plasma membrane of root column cells. AtZIP1 and AtZIP2 are respectively involved in the reactivation of metal ions from vacuoles to the cytoplasm and the absorption of Mn and Zn by roots, respectively (Milner et al., 2013). Other studies have also confirmed that AtZIP genes are induced by Zn deficiency, and some AtZIP genes are directly involved in the accumulation of Zn in the edible parts of plants (Ramegowda et al., 2013; Gaitan-Solis et al., 2015). In rice (Oryza sativa), both OsIRT1 and OsIRT2 are highly expressed in roots for Fe transport and are significantly induced by Fe deficiency (Li et al., 2019). Overexpression of both OsIRT1 and OsIRT2 enhances the resistance of plants to Fe deficiency stress and concurrently increases the content of Fe and Zn in grains (Bughio et al., 2002; Ishimaru et al., 2007a). Overexpression of OsZIP1 increases the accumulation of Zn and Fe in roots, buds and seeds (Ishimaru et al., 2006), while overexpression of OsZIP4 and OsZIP5 significantly elevates the Zn content in roots of transgenic plants, but not in grains (Ishimaru et al., 2007b; Lee et al., 2010a). It has been verified that ZIP transporters are also involved in the transportation of bivalent metal cations such as Mn2+ and Cd2+ (Chen et al., 2008; Fan et al., 2021). For instance, the expression levels of ZIP2 and ZIP3 in Chinese cabbage (Brassica chinensis), as well as the expression level of SlZIP4 in leaves of tomato (Solanum lycopersicum), are closely related to Cd transport and concentration (Yu et al., 2017; Wu et al., 2019). Generally, different ZIP transporters influence plant growth and development as well as the accumulation of trace elements in grains by mediating the absorption, transportation and distribution of divalent metal ions such as Fe and Zn. Therefore, studying the transportation and accumulation of metal ions such as Zn, Fe and Cd mediated by different ZIP transporters is crucial for improving food quality and ensuring human health.
Food crops are the main dietary source of energy and essential trace elements for human intake. Due to the relatively low contents of Zn and Fe in maize and rice, the risk of deficiency in nutritional elements such as Fe and Zn among people who mainly consume corn and rice, especially those in poverty-stricken areas with simple dietary structures, has significantly increased (Wessells and Brown, 2012). Currently, it is widely believed that the solutions to address the ‘hidden hunger’ caused by the deficiency of trace elements such as Zn and Fe in humans include measures such as dietary diversification, functional nutritional supplements for food, or biofortification of staple foods (Yan et al., 2023). Foxtail millet, as one of the earliest domesticated cereal crops by humans, is widely cultivated in arid and semi-arid regions of Asia and Africa, including China, India, and Nigeria. The hulled product of foxtail millet, known as millet, is rich in various nutrients such as amino acids, vitamins, and minerals that are essential for the human body. The contents of Fe and Zn are much higher than those of staple crops such as rice and maize (Zea mays), and it can serve as a good dietary supplement for people with nutrient deficiencies (He et al., 2022). However, the mechanism of nutrient enrichment is still unclear. Although ZIP transporters have been investigated in Arabidopsis, rice, maize and other staple crops, scarce knowledge is available regarding the ZIP transporters in foxtail millet (Eide et al., 1996; Eide, 2006; Ishimaru et al., 2006, 2007; Li et al., 2013). Here, we systematically identified and characterized ZIP transporters in foxtail millet using the latest genomic data, and predicted their functions using a strategy integrating homology comparison, expression characteristic analysis and previous studies. The goals of this study are to: (1) accurately identify and characterize the ZIP gene family in foxtail millet; (2) elucidate the characteristics of family expansion and functional divergence; (3) explore SiZIP expression patterns in different tissues and under metal ion and abiotic stresses; (4) clarify the variation characteristics of SiZIPs at the sequence and expression levels, and explore their values in breeding and improvement of foxtail millet; (5) predict the potential functions of SiZIPs. This study will lay the foundation for further functional research on foxtail millet ZIP genes, and will also contribute to the elucidation of the molecular mechanisms underlying metal ion transport and accumulation in foxtail millet.
The genomic sequence, protein sequence and genomic annotation file of the foxtail millet material ‘xiaomi’ were obtained from the Multi-omics Database for Setaria italica (MDSi, http://foxtail-millet.biocloud.net/) (Yang et al., 2020). The genomic information of barley (Hordeum vulgare), sorghum (Sorghum bicolor), maize, rice, Arabidopsis, and potato (Solanum tuberosum) was downloaded from the Ensemble Plants database (http://plants.ensembl.org/index.html). The ZIP proteins in barley were obtained from published papers and were used together with ZIP proteins from rice and Arabidopsis as seed sequences for BLASTP alignment (BLAST+, version 2.12.0) of the protein databases of foxtail millet, sorghum, maize, and potato (Deng et al., 2022). Protein sequences with a similarity greater than 50% and an e-value less than 10-5 were considered as candidates for ZIP. The conserved domain (PF02535) of all candidate genes was screened using the HMMsearch tool (version 3.3.2) with a threshold of an e-value less than 10-10, and only those candidate genes containing the complete domain were retained. The putative candidate ZIPs were obtained by manually removing different transcripts of the same gene. Subsequently, all ZIPs were renamed according to their positions on the chromosome, such as SiZIP1, SiZIP2.
The basic information of these ZIP genes and the proteins they encode was extracted from the genomic annotation files. The information such as protein molecular weight and isoelectric point, transmembrane domains and subcellular localization was obtained respectively from the computer pI/MW tool in the ExPASy database (https://web.expasy.org/compute_pi/), the TMHMM Server 2.0 (http://www.cbs.dtu.dk/services/TMHMM/), and Cell-PLoc 2.0 (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/).
To investigate the expansion pattern of the ZIP gene family in foxtail millet, the Multiple Collinearity Scan toolkit (MCscanX) was employed to analyze the duplication relationship of ZIP genes in foxtail millet. Meanwhile, for the duplicated gene pairs, the ratio of non-synonymous mutations (Ka) to synonymous mutations (Ks), Ka/Ks, was calculated using KaKs_Calculator 3.0. Finally, the SiZIP genes and their duplication relationships on the foxtail millet chromosomes were visualized using the TBtools (version 2.119) (Zhang, 2022; Chen et al., 2023a).
To clarify the relationships among the members of the ZIP gene family in foxtail millet, the protein sequences of ZIP transporters from six species were selected to construct a phylogenetic tree together with SiZIPs, including the monocotyledonous gramineous crops rice, sorghum, maize, barley, and the dicotyledonous plants Arabidopsis and potato. Multiple sequence alignment was performed using MEGA 6.06, and the maximum likelihood method was used to construct the phylogenetic tree. The phylogenetic tree was visualized by Interactive Tree of Life (iTOL v6, https://itol.embl.de/).
To analyze the similarities and differences among different SiZIP transporters, the Motif-based sequence analysis Server (MEME version 5.5.1, http://meme-suite.org) was used to predict the conserved motifs of these SiZIP proteins. The motif width was set from 6-200aa, the number of predicted motifs was 10, and they could be displayed repeatedly. The gene structure information of these SiZIP genes was extracted from the genomic annotation file. MEGA 6.06 was used for multiple sequence alignment of all SiZIP sequences and a phylogenetic tree was constructed based on the maximum likelihood method. TBtools was used to visualize protein conserved domains, gene structure, and phylogenetic trees (Sievers et al., 2011). Meanwhile, Escript 3.0 was used to visualize the results of multiple sequence alignment of SiZIP proteins and analyze the relationship between conserved motifs and transmembrane (TM) regions.
To elucidate the sequence characteristics of the promoter regions of SiZIPs, we used TBtools to extract the sequences 2000 bp upstream of the start codon of these genes for cis-acting element prediction. All sequences were uploaded to the PlantCare website (version 1.0, http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Yang et al., 2023). After the prediction was completed, the cis-acting elements in the promoter regions were counted and classified to study the response patterns of SiZIPs.
The spatiotemporal expression profiles of SiZIPs in different tissues during the whole growth period of foxtail millet were obtained through the foxtail millet multi-omics website (MDSi, version 1.0, http://foxtail-millet.biocloud.net/home). These datasets included RNA-seq data of various tissues, including roots, stems, leaves, flowers, and grains of the foxtail millet model material ‘xiaomi’ from the seedling stage to the mature stage. The expression levels of all genes were standardized by TPM values.
To investigate the responses of SiZIPs to Cd, Fe, and K ions, the genomic sequencing material ‘xiaomi’ was selected for seedling experiment and subjected to stresses of Cd, Fe, and K. The detailed steps for seedling planting and Cd treatment at three different concentrations (5 µM, Cd1; 10 µM, Cd2, 30 µM, Cd3) were referred to our previous article (Yang et al., 2023). As for Fe and K stresses, five treatments were set by controlling the concentrations of Fe and K ions in Hoagland’s nutrient solution: normal Fe and K (CK), Fe deficiency (0 µM, LFe), high Fe (600 µM, HFe), low K (0.1 mM, LK) and high K (10 mM, HK). The seedlings and roots were rapidly frozen in liquid nitrogen and sent to PERSONAL GENE TECHNOLOGY Co., Ltd. (Nanjing, China) for transcriptome sequencing. cDNA library construction and transcriptome sequencing were performed on the BGISEQ500 sequencing platform and followed the company’s standard sequencing and analysis procedures (Yang et al., 2023). The sequencing raw data was deposited in the NCBI Sequence Read Archive (SRA) with the accession number PRJNA1146895. Meanwhile, the seedlings and roots after Cd treatments were also used for ionome determination of 9 metal ions including Cd, Fe, Zn, Mn, Cu, Mg, Ca, K, Na and Mo on the Thermo Fisher Scientific iCAP TQ ICPMS/MS platform.
Furthermore, to clarify the expression characteristics of SiZIPs under salt stress, cold stress and drought stress, we added gene expression datasets obtained from some public databases and unpublished transcriptomes, including the transcriptomes of foxtail millet seedlings after treatment with 150 mM NaCl for 0.5 h and 2 h (unpublished), the transcriptomes of foxtail millet seedlings after cold stress for 1 h, 3 h and 6 h (SRA accession: PRJNA343268), and the transcriptomes of foxtail millet seedlings during the day/night after long-term drought (unpublished). The gene expression levels under all these different treatments were standardized using TPM values (Transcripts Per Kilobase of exon model per Million mapped reads).
To explore the sequence variation and expression level variation of SiZIPs in natural populations and further evaluate the role of SiZIPs in the quality improvement of foxtail millet. We analyzed the previously completed genomic resequencing data of 360 foxtail millet genotypes and the transcriptome data of grains at the filling stage (Li et al., 2022), and obtained the sequence variation information and expression data of SiZIPs in 360 genotypes based on manual screening.
Considering the great progress of rice in the study of gene functions and the widespread convergent evolutionary characteristics of gramineous crops, a foxtail millet-rice homology comparison strategy was used to further analyze the functions of SiZIPs. The orthologs of SiZIPs in rice were detected by MCscanX and the phylogenetic tree. On this basis, we conducted an in-depth and detailed literature survey to search for the reported functions of OsZIPs. Additionally, the expression patterns of these genes across different tissues were obtained from the Rice Expression Database (https://rapdb.dna.affrc.go.jp/) (Chen et al., 2023b). By comparing the expression characteristics of ZIP genes in foxtail millet and rice, along with the reported functions of OsZIP genes, we inferred the potential functions of SiZIP genes. In addition, the PPI (protein-protein interaction) networks of SiZIPs were predicted through the STRING database (Search Tool for the Retrieval of Interacting Genes/Proteins, version 12.0, https://cn.string-db.org/).
Through local BLASTP and after eliminating sequences of different transcripts from the same gene as well as those containing incomplete conserved domains, 14, 14, 12, and 12 ZIP genes were identified in foxtail millet, sorghum, maize, potato, respectively. Compared with 13 in rice and 15 in Arabidopsis, the number of ZIPs did not exhibit significant disparities between monocotyledons and dicotyledons, and no conspicuous uneven expansion was discerned among different species.
Based on their locations on the chromosomes, the SiZIPs were designated as SiZIP1 to SiZIP14 and the detailed information was listed in Table 1. Overall, SiZIPs are relatively short with lengths ranging from 1062 bp for SiZIP3 to 2637 bp for SiZIP8. Except for SiZIP8 with 10 introns, the number of introns in other SiZIPs varies from 1 to 3. The 14 SiZIP proteins range in size from a minimum of 271 amino acids (aa) to a maximum of 474 aa and the corresponding molecular weights range from a minimum of 28678.23 Da to a maximum of 43731.21 Da. As typical transmembrane transporters, all SiZIPs contain varying numbers of TM regions with the number of TMs ranging from 4 to 9. The subcellular localization results based on Loctree 3 and Cell-PLoc indicated that all SiZIPs were localized on the membrane. In general, SiZIPs exhibited rich diversity in terms of gene structure, protein physicochemical properties, and TM regions.
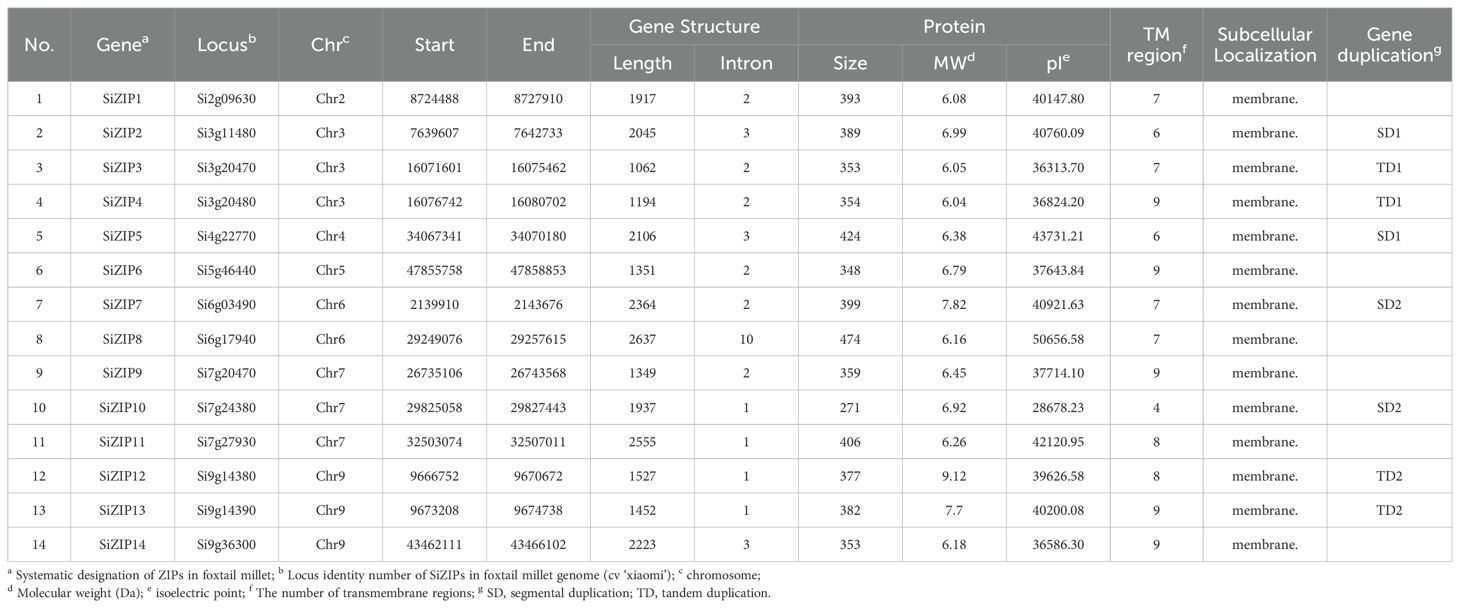
Table 1. The detail information and sequence characterization of 14 putative ZIP genes in foxtail millet.
All 14 SiZIPs were unevenly distributed across 7 chromosomes with chromosomes 3, 7, and 9 each having the highest number of 3 genes. Chromosome 6 contains 2 genes and the remaining genes are distributed on chromosomes 2, 4, and 5 (Supplementary Figure S1). Overall, SiZIPs were mainly located at both ends of the chromosomes, which in line with the distribution characteristics of foxtail millet genes on chromosomes. Gene duplication analysis showed that 8 SiZIPs participated in tandem duplication (TD) and segmental duplication (SD) events, forming two TD gene pairs (SiZIP3 & SiZIP4, TD1; SiZIP12 & SiZIP13, TD2) and two SD gene pairs (SiZIP2 & SiZIP5, SD1; SiZIP7 & SiZIP10, SD2). Although the Ka/Ks values of all duplicated gene pairs were less than 1, the TD gene pairs (0.32 and 0.43) had higher Ka/Ks values than the SD gene pairs (0.09 and 0.24). These results indicated that both TD and SD have contributed to the expansion of ZIP gene family in foxtail millet.
To clarify the phylogenetic relationships of the ZIP family in different species, we constructed a phylogenetic tree based on protein sequences for 99 ZIP proteins from seven species (dicots: Arabidopsis and potato; monocots: foxtail millet, sorghum, maize, rice, and barley). The results showed that all ZIP proteins were divided into four major clades, Class I-IV, and all clades contained ZIP transporters from dicots and monocots, indicating that the ZIP gene family was formed before the differentiation of dicots and monocots (Figure 1).
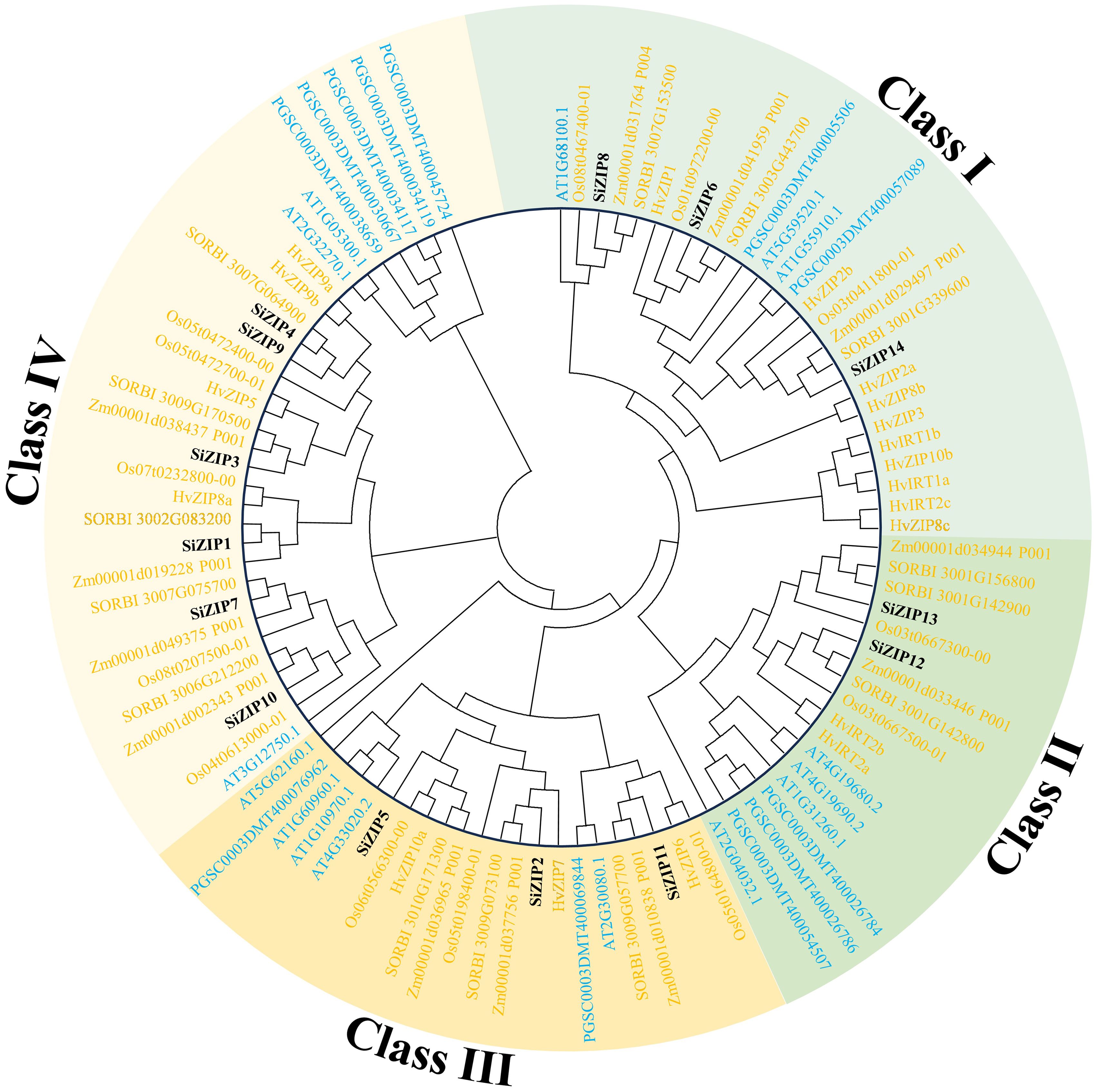
Figure 1. The phylogenetic tree of ZIP transporters in seven plants. The phylogenetic tree is constructed by ZIP proteins of foxtail millet, sorghum, maize, rice, barley, Arabidopsis and potato using MEGA 6 with maximum likelihood method. Different clades and ZIPs from monocots and dicots are distinguished by different colors.
Class I consisted of 27 ZIPs, including 3 SiZIPs of foxtail millet, 3 OsZIPs of rice, 3 ZmZIPs of maize, 3 SbZIPs of sorghum, 10 HvZIPs of barley, 3 AtZIPs and 2 StZIPs of potato, forming three orthologous clusters (OCs, OC1–3) (Table 2). Except for the significant expansion of HvZIPs in Class I, ZIPs of other species were relatively conserved in monocots and dicots. Class II contained 18 ZIPs from different species, including 2 SiZIPs, 2 OsZIPs, 2 ZmZIPs, 3 SbZIPs, 2 HvZIPs, 4 AtZIPs and 3 StZIPs. Although the number of ZIPs of different species in Class II did not show a significant difference, ZIPs of monocots and dicots in Class II were located in different branches. Furthermore, Class II included multiple tandem duplicate gene pairs of monocots and dicots, which confirmed that the ZIP genes in Class II were formed by independent duplication after the differentiation of monocots and dicots (OC 4-1 and OC 4-2). Class III comprised 22 ZIP proteins, including 3 SiZIPs, 3 OsZIPs, 3 ZmZIPs, 3 SbZIPs, 3 HvZIPs, 5 AtZIPs and 2 StZIPs, forming an OC (OC5) containing ZIP proteins from seven species and two OCs (OC6-1 and OC6-2) consisting of five monocotyledonous ZIP proteins. Moreover, the tandem duplication of AtZIPs in Class III also promotes the expansion of ZIP gene family in Arabidopsis. Class IV consisted of 32 ZIPs. Besides including one orthologous gene cluster OC 7 composed of ZIPs of monocots and dicots, it also contained 4 homologous gene clusters (OC 8-1 to OC 8-4) specific to monocots. The remaining ZIPs in Arabidopsis and potato were formed after the divergence of monocots and dicots. Overall, ZIP proteins, apart from those of Class I being highly conserved, displayed a certain level of unequal duplication and expansion in monocots and dicots in other classes, forming multiple ZIP clusters unique to monocots.
To compare the characteristics of different SiZIP genes, we analyzed their gene structures and protein conserved domains (Figure 2). Overall, SiZIP genes within the same class exhibit relative conservation in gene structure. For instance, SiZIP3, SiZIP4, SiZIP9 and SiZIP1 in Class IV contained two introns, while SiZIP2 and SiZIP5 in Class III had three introns. However, some SiZIP genes also displayed some variations, such as SiZIP6 and SiZIP14. Furthermore, the two genes of paralogous gene pairs also showed characteristics of the coexistence of similarity in conservation and diversity in gene structure. For example, the gene structures of SiZIP2 and SiZIP5 (SD1) were similar, while SiZIP7 and SiZIP10 (SD2) showed differences. In terms of the conserved domains, some motifs were conserved in all SiZIP transporters, such as Motif 5, Motif 3 and Motif 6, while some motifs were specific in some SiZIPs. For instance, Motif 9 only exists in SiZIP6 and SiZIP14. Regarding paralogous gene pairs, the conserved domains of SiZIP12 and SiZIP13 (TD2) were basically the same, while SiZIP10 lacked three motifs compared to SiZIP7. In general, SiZIPs maintained a certain level of conservation in both gene structure and protein conserved domains, while also generating novel variations, especially among paralogous gene pairs.
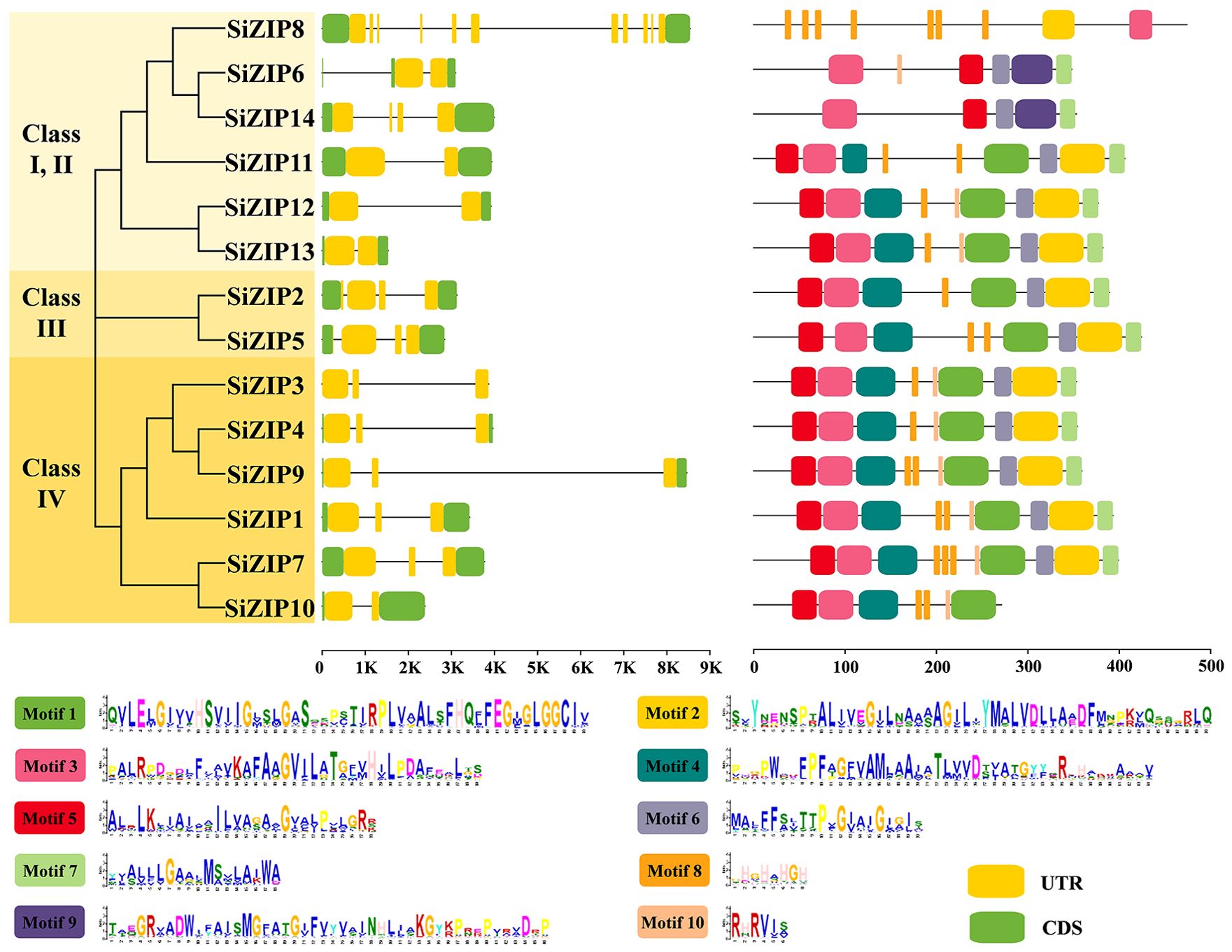
Figure 2. Gene structure and conserved motifs of SiZIPs in foxtail millet. Different classes are distinguished by different colors, and the gene structure and conserved motifs are derived from genome annotation file and MEME prediction, respectively.
The relationship between the TM regions and conserved motifs of these SiZIP proteins was further compared (Figure 3). The results showed that a large number of conserved motifs highly coincided with the TM regions, such as Motif 5 - TM 2, Motif 4 - TM 4, Motif 6 - TM 7 and Motif 7 - TM 9. Some other motifs contained TM regions and some intra- and extra-membrane structures. For example, Motif 3 contained TM 3 and the intra-membrane part, and Motif 1 contained TM regions TM 5 and TM 6 and the extra-membrane part in the middle. The absence of conserved motifs in SiZIP proteins was directly related to the number of their TM regions.
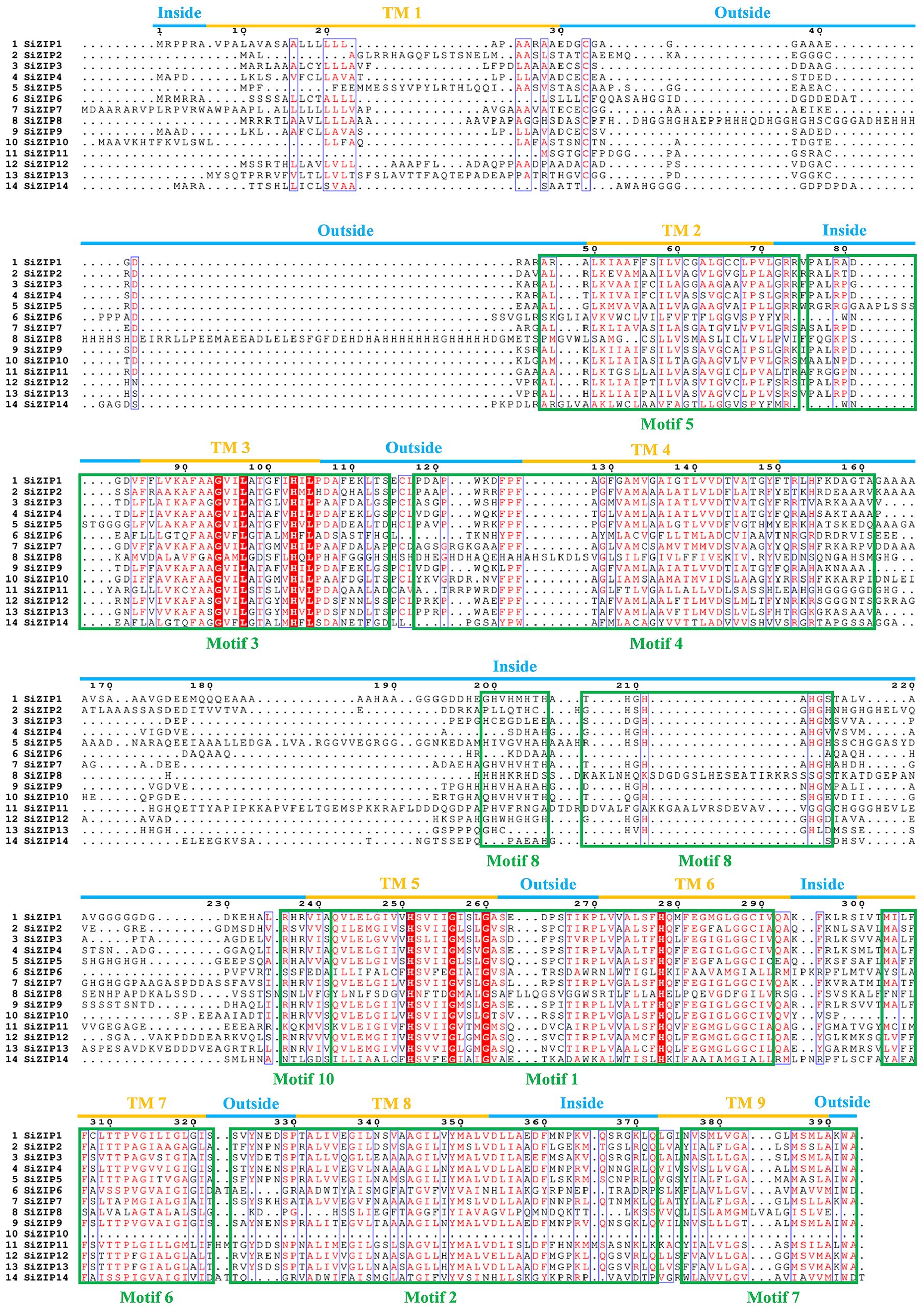
Figure 3. The multiple-sequence alignment of SiZIPs in foxtail millet. Clustal Omega and Escript 3.0 are used to construct the multiple sequence alignment and visualization respectively. The transmembrane (TM) regions and conserved motifs are labeled by blue/orange lines and green boxes, respectively.
We predicted and analyzed the cis-acting elements in the promoter regions of all SiZIP genes (Figure 4). The promoter regions of SiZIPs mainly contained five types of cis-acting elements: hormone response, light response, MYB transcription factor binding, stress response and tissue-specific expression. The hormone response elements included abscisic acid response, auxin response, gibberellin response, MeJA response and salicylic acid response elements. The abscisic acid response elements and MeJA response elements were the most abundant, followed by gibberellin response elements and salicylic acid response elements, indicating that these SiZIP genes are regulated by plant growth regulators to varying degrees. Multiple MYB transcription factor binding sites were predicted in the promoter regions of SiZIPs such as SiZIP13 and SiZIP9, suggesting that these genes may be regulated by MYB transcription factors. In addition, the stress response elements included various stresses such as anaerobic induction, drought, low temperature, and salt, confirming that these genes might be involved in the response to various osmotic stresses. Finally, The presence of tissue-specific expression elements, including endosperm-, root-, meristem-, and seed-specific expression elements, was likely a significant factor contributing to the diversity of tissue-specific expression in SiZIPs.
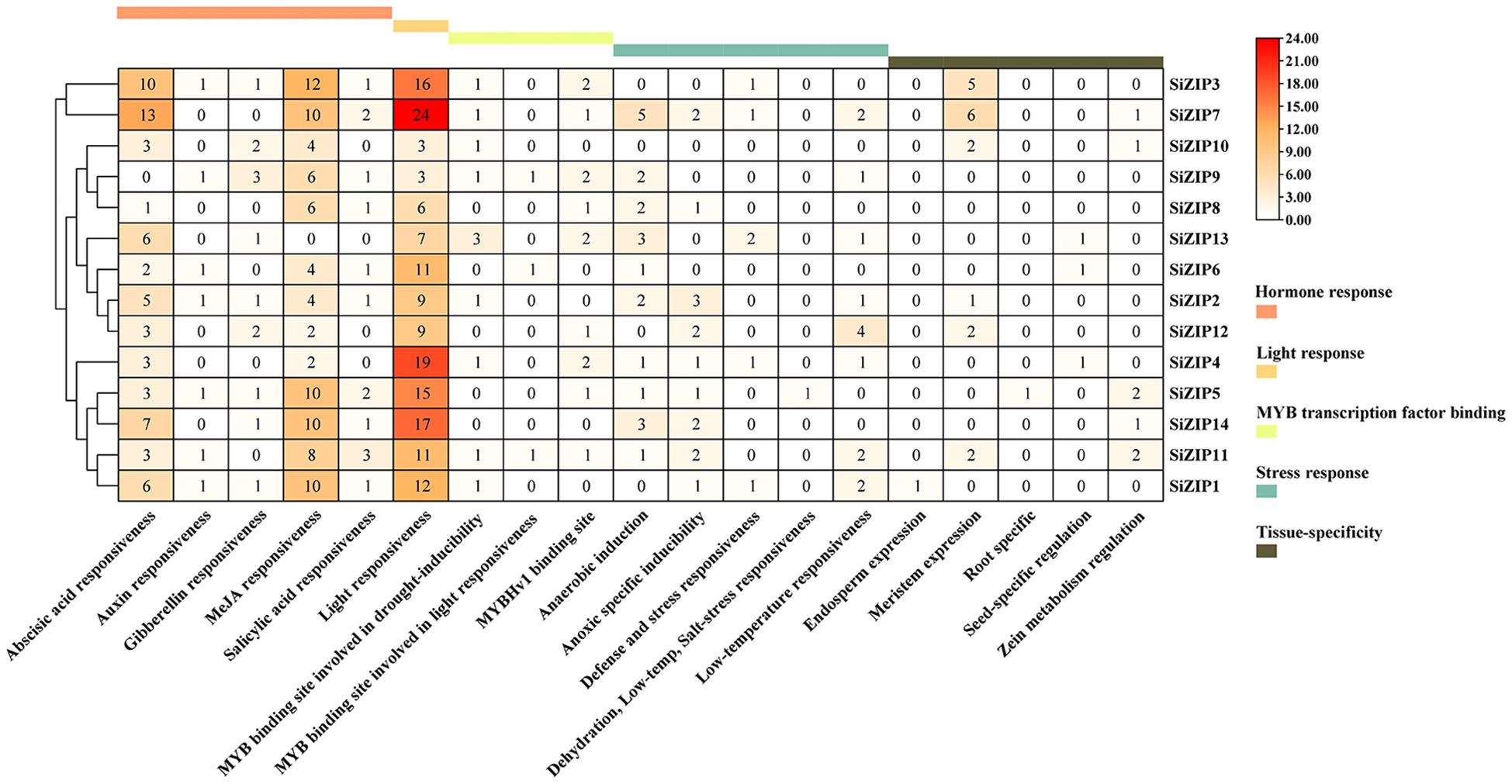
Figure 4. The quantity distribution of different cis-acting elements in the promoter regions of SiZIPs. All cis-acting are predicted online by PlantCare and manually classified. The numbers in the figure represent the quantity of cis-acting elements in the promoter region of the SiZIP genes.
SiZIPs exhibited diverse expression patterns throughout the whole growth period of foxtail millet, demonstrating clear temporal and spatial specificity (Figure 5; Supplementary Figure S2). Overall, the expression patterns of SiZIPs could be broadly classified into three categories. The first category was highly expressed during the germination and the subsequent seedling growth, with SiZIP9, SiZIP4 and SiZIP13 specifically expressed in germinating seeds and SiZIP6 expressed in both germinating seeds and seedlings. The second category was primarily expressed in panicles and leaves during the heading and flowering stages, such as SiZIP8 and SiZIP3. The third category was expressed in various tissues during the grain-filling stage, such as SiZIP14 and SiZIP7. Regarding tissue-specific expression, SiZIP10 was specifically expressed in stems, SiZIP5 and SiZIP1 were primarily expressed in leaves, SiZIP12 was expressed in panicles during multiple stages, and SiZIP2, SiZIP7 and SiZIP11 were highly expressed in roots. Furthermore, paralogous gene pairs also exhibited differences in expression patterns. For example, in SD1, SiZIP2 was expressed in multiple tissues including roots, stems and leaves, while its paralog SiZIP5 was primarily expressed in leaves. In TD1, SiZIP3 was primarily expressed in panicles, while its paralog SiZIP4 was specifically expressed in germinating seeds. The diverse temporal and spatial expression patterns exhibited by different members of the SiZIP gene family, along with the emergence of novel expression patterns in newly duplicated genes, contributed significantly to the functional diversification of SiZIPs.
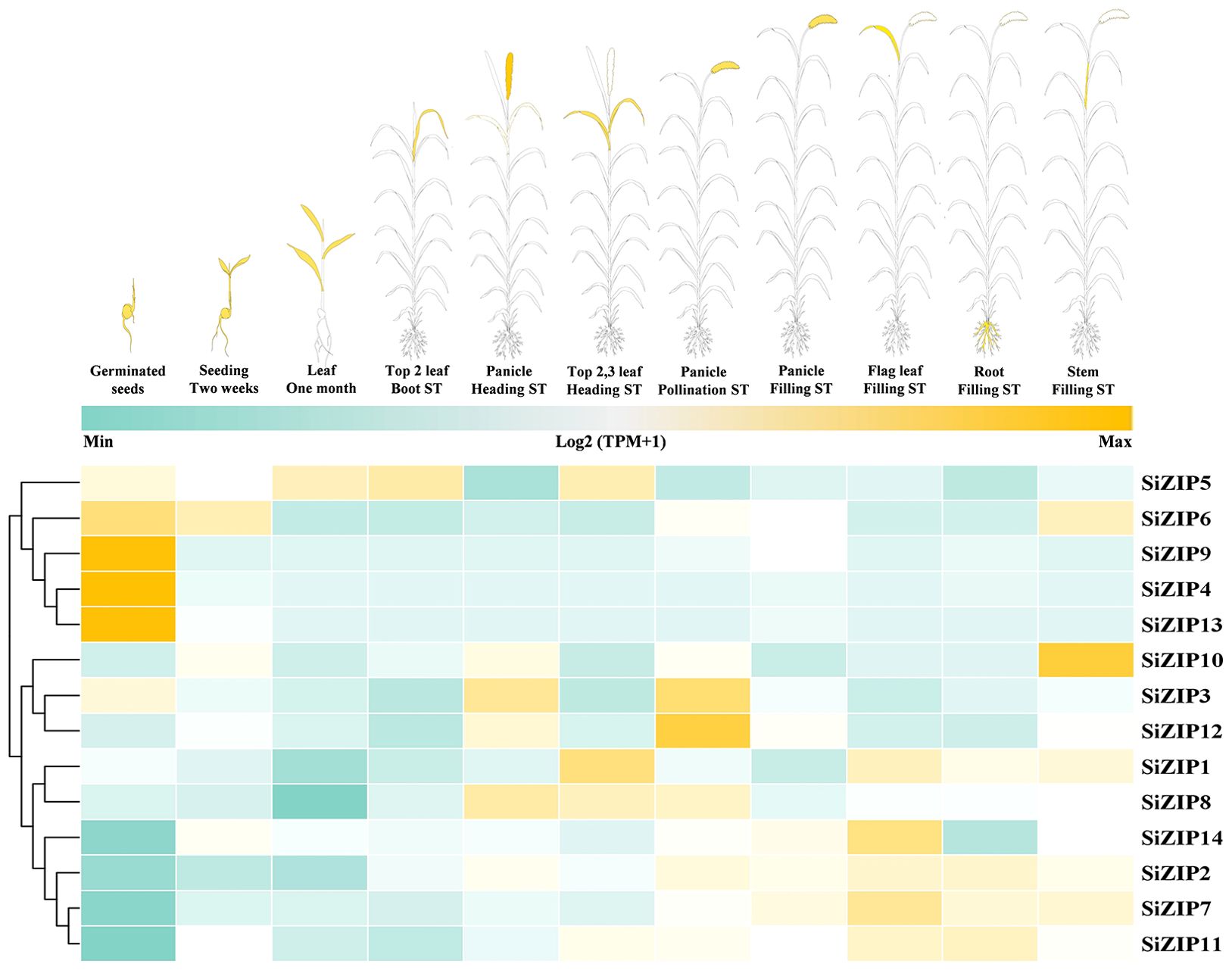
Figure 5. The spatiotemporal expression patterns of SiZIPs in multiple tissues during whole growth period in foxtail millet. The expression matrices (TPM values) are obtained from foxtail millet multi-omics database (MDSi). ST represents stage.
SiZIPs exhibited distinct expression patterns in different tissues in response to varying concentrations of Cd, Fe, and K stress (Figure 6). Except for SiZIP10, SiZIP8 and SiZIP11, which did not respond to Cd, Fe and K treatments, the expression of the remaining 11 SiZIPs was inhibited or induced by different metal ions. As important Zn and Fe transporters, 8 genes (SiZIP2, SiZIP5, SiZIP7, SiZIP3, SiZIP4, SiZIP13, SiZIP1 and SiZIP6) showed significantly upregulated expression under high Fe treatment. Interestingly, some SiZIPs were induced to be expressed by both high Fe and high K simultaneously. While SiZIP12 and SiZIP9 were significantly suppressed under different concentrations of Fe treatment but significantly upregulated under different K treatments. In addition, the expression patterns of duplicated genes under different metal treatments were also diverse. For example, SiZIP7 in SD2 was significantly induced under high Fe and high K treatments, while SiZIP10 did not respond to any metal ions. In TD1, SiZIP3 was expressed in both roots and leaves and was not significantly induced by K, while SiZIP4 was only expressed in roots and was significantly induced and upregulated by K. Overall, the SiZIP genes exhibited diverse responses and expression patterns under different ionic stresses.
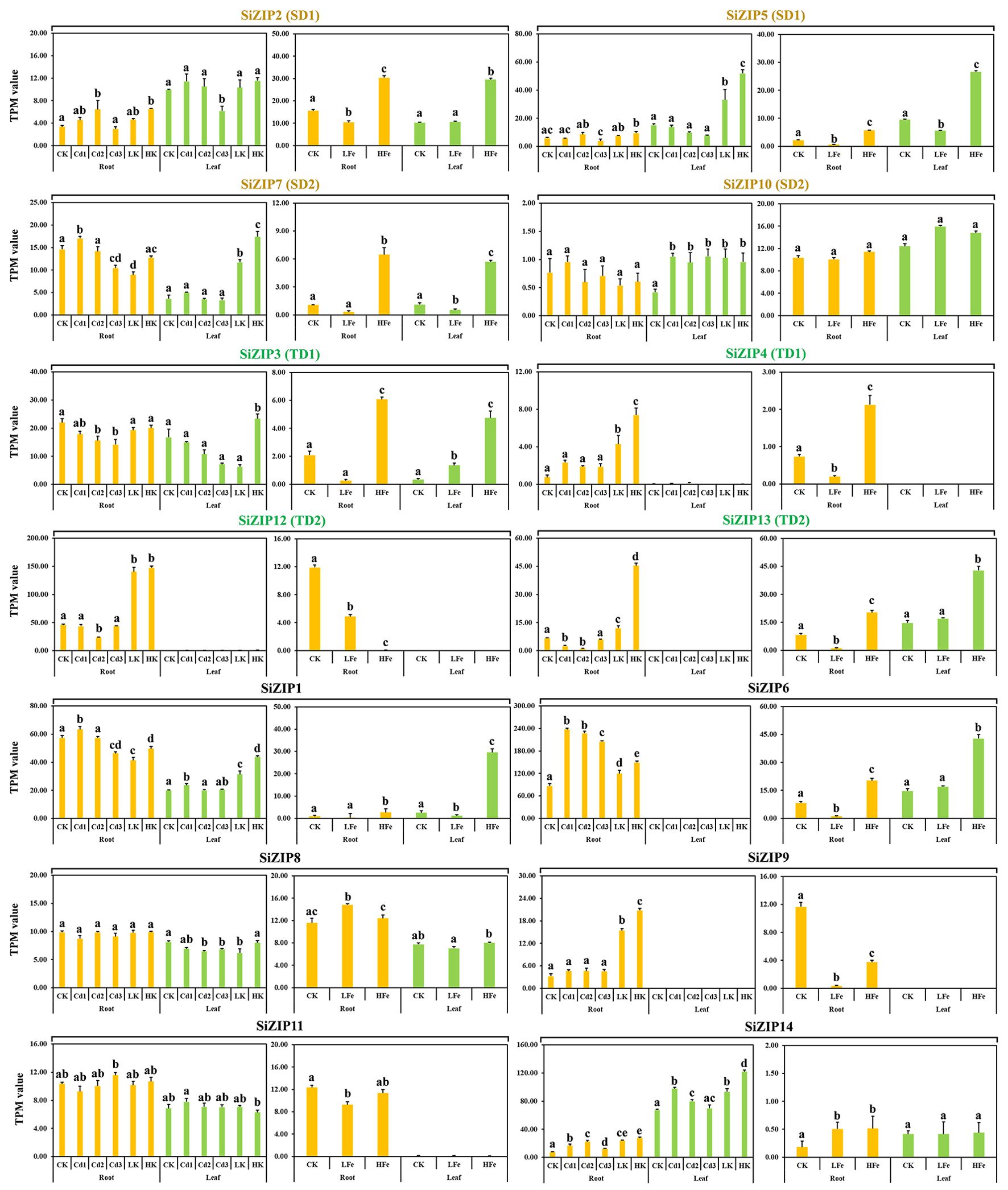
Figure 6. The expression patterns of 14 SiZIPs under Cd, Fe and K treatments. The Y-axis represents the TPM value based on RNA-seq. Bars represent the mean values of three replicates ± standard deviation (SD). Cd1, Cd2, Cd3, LK, HK, LFe and HFe represent 5 µM Cd treatment, 10 µM Cd treatment, 30 µM Cd treatment, 0.1 mM K treatment, 10 mM K treatment, 0 uM Fe treatment and 600 µM Fe treatment, respectively. Significant differences (P < 0.05) between groups are marked with different letters.
To clarify the correlation between SiZIP gene expression and metal ion transport, we measured the contents of 10 metal ions (Cd, K, Fe, Ca, Na, Mg, Mn, Cu, Zn and Mo) in the roots and leaves of foxtail millet seedlings after Cd stresses (Supplementary Figure S3A). Among these ions, besides Cd, Fe (318.9-657.6 mg/kg), Ca (345.0-1880.4 mg/kg), K (2249.8-14230.1 mg/kg) and Na (31.5-193.1 mg/kg) all showed certain differences between Cd treatments and CK. The correlation analysis revealed significant correlations between the expression levels of different SiZIPs in various tissues and the contents of these metal ions. Specifically, SiZIP4, SiZIP6, SiZIP7 and SiZIP9 showed significant positive and negative with Fe (at least 0.57) and K (0.81-0.90) content, respectively, which directly correlated with their induced expression under Fe and K stresses (Supplementary Figure S3B). SiZIP12, which was induced by K, exhibited a significant negative correlation (0.85) with K content. Although the dataset used for correlation analysis was not large, it suggested to a certain extent that SiZIPs were closely related to the transport and accumulation of different metal ions in foxtail millet, especially Fe.
In addition to analyzing the response of SiZIPs to metal ions, we have also analyzed their response to various abiotic stresses (Figure 7). Except for SiZIP6, all other SiZIPs were induced to up-regulate their expression under salt stress. The weak response of SiZIP6 to Na and K and its strong up-regulation under Fe and Cd stresses suggested that it might be involved in the transport of Fe and Cd in foxtail millet simultaneously. Multiple SiZIPs, including SiZIP3, SiZIP10, SiZIP7, SiZIP14 and SiZIP8, were induced to up-regulate their expression under cold stress, especially in the early stage (1 h). SiZIP13, SiZIP3, SiZIP11, SiZIP5 and SiZIP1 were induced to up-regulate their expression after drought stress. The results indicated that these SiZIPs might enhance the tolerance of seedlings to abiotic stresses by altering the transport and distribution of metal ions in foxtail millet and regulating the osmotic potential inside and outside the membrane under abiotic stresses. Furthermore, The PPI results showed that except for a certain degree of interaction between SiZIP13, SiZIP8, SiZIP11, and SiZIP12, the functions of other SiZIP proteins were relatively independent (Figure 8). However, interactions between SiZIPs and MTPs (Metal Tolerance Protein), potassium channel protein, EXO (Exocyst subunit Exo70 family protein), calumenin protein, other Zn transporters, and MYB transcription factors were discovered. For example, SiZIP8 interacted with ZTP29, MTP1 and calumenin-B, SiZIP13 interacted with MYB4 and EXO70B1. This suggested that SiZIPs might play a role in metal transport in conjunction with other metal transporters and exosome related proteins, and these transporters were also regulated by transcription factors.
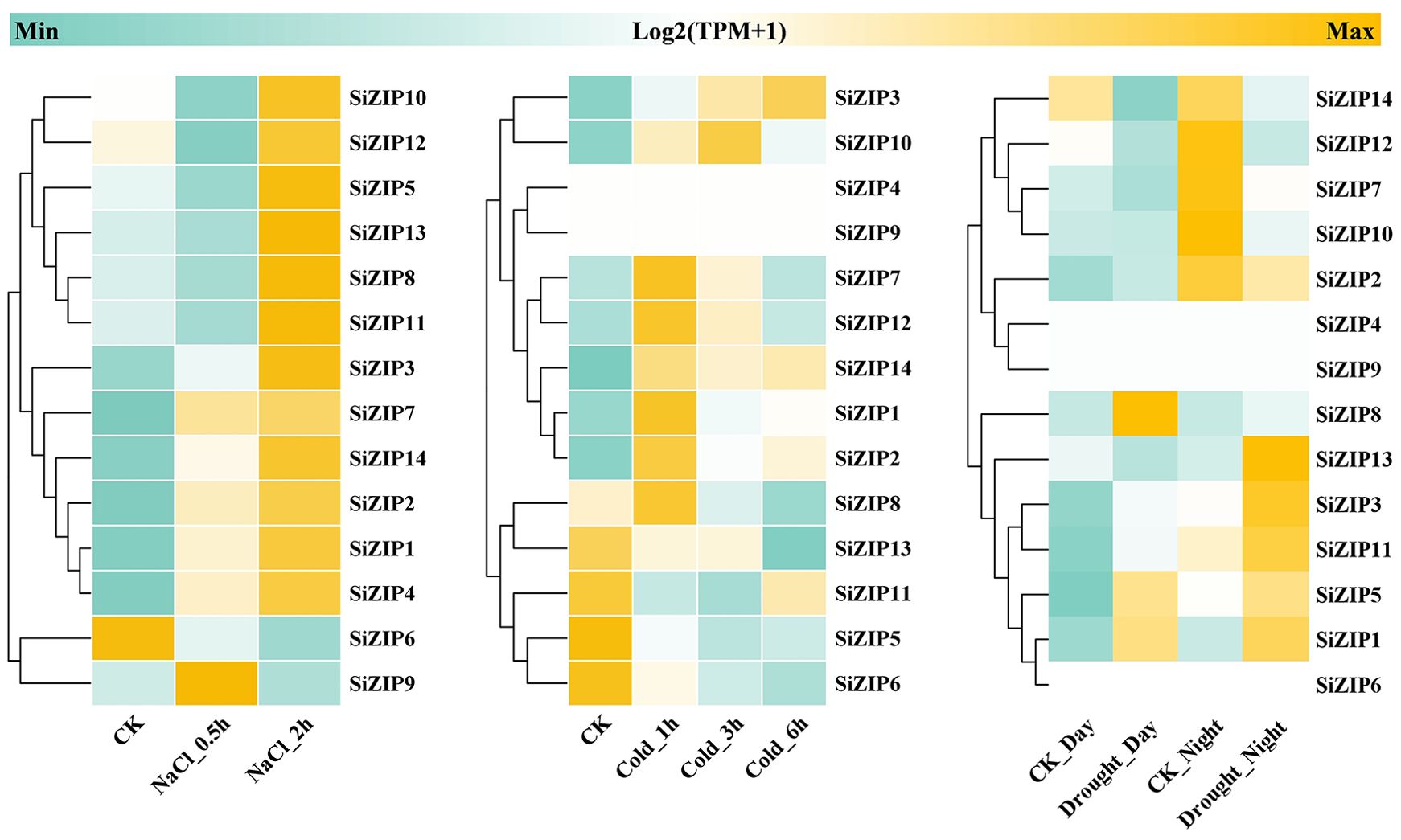
Figure 7. The heatmaps of SiZIP expression under salt stress, cold stress and drought stress. From left to right are heatmaps of expression levels of SiZIPs in foxtail millet seedlings under salt stress (150 mM NaCl), cold stress (4°C) and drought stress. All expression data are obtained from RNA-seq and the gene expression levels are standardized using TPM values (Transcripts Per Kilobase of exon model per Million mapped reads). The log2 (TPM+1) values are used to display the expression levels.
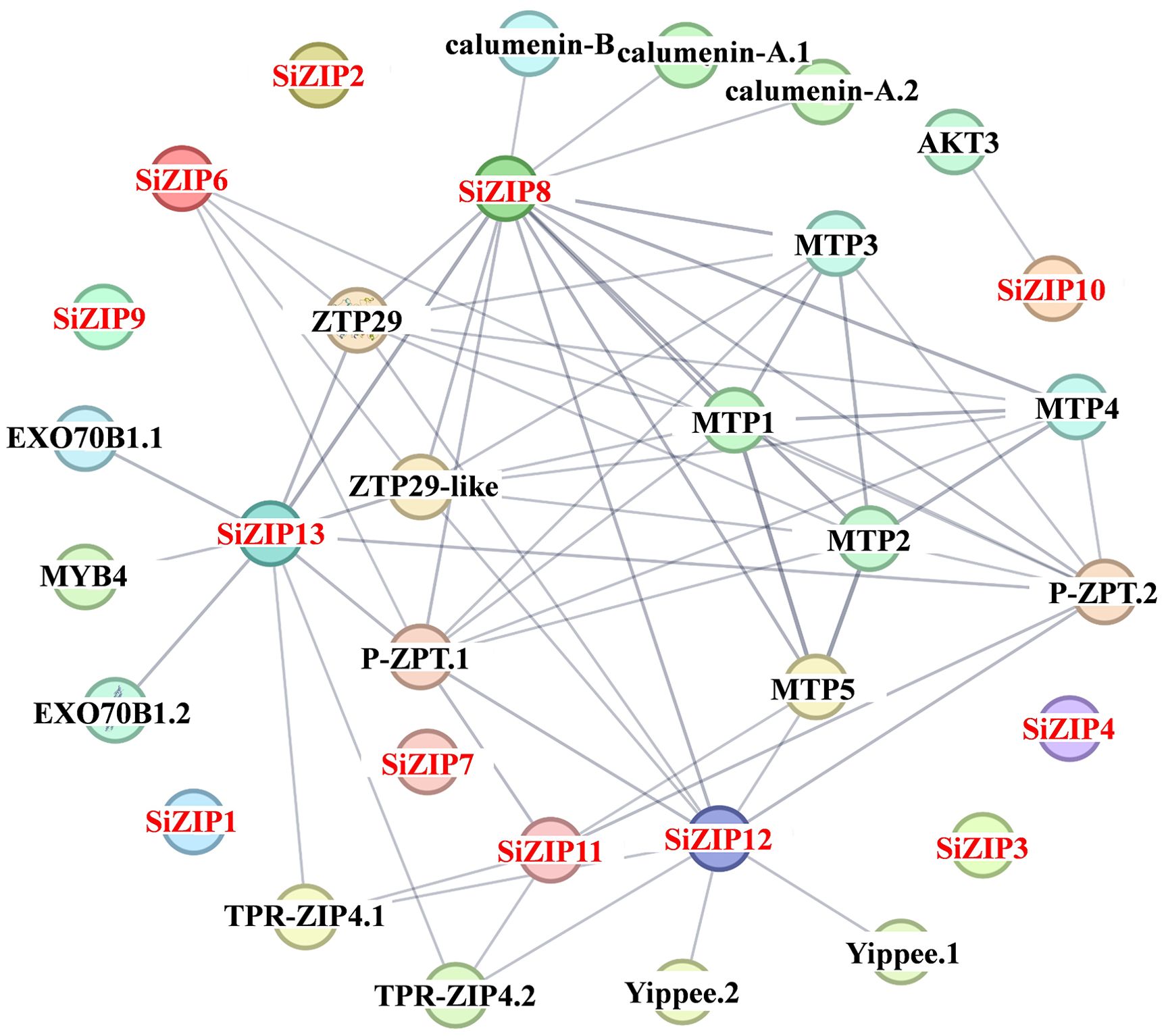
Figure 8. The protein-protein interaction networks of SiZIPs with other proteins in foxtail millet. The protein-protein interaction networks of SiZIPs were predicted by the STRING database (Search Tool for the Retrieval of Interacting Genes/Proteins, https://cn.string-db.org/). The thickness of the line represents the degree of interaction between different proteins. MTP: metal tolerance protein; AKT3: potassium channel AKT3; EXO70B1: exocyst subunit Exo70 family protein; ZTP29/ZTP29-like: zinc transporter ZTP/ZTP29-like protein; TPR-ZIP4: TPR repeat-containing protein ZIP4; P-ZRT: putative zinc transporter; Yippee: yippee family putative zinc-binding protein.
A total of 1718 genetic variation loci were identified among 360 foxtail millet genotypes for 14 SiZIP genes, including 1525 SNPs and 193 Indels (Table 3). Among them, 54 variations were non-synonymous SNPs, and 235 variations were located in the promoter regions. To explore the expression variations of SiZIPs in different genotypes, we investigated the expression levels of SiZIPs in grains during the filling stage across 360 genotypes (Supplementary Figure S4) (Li et al., 2022). The expression levels of SiZIP genes showed significant differences among 360 accessions. For example, SiZIP14 was expressed at a low level (TPM < 25) in most foxtail millet genotypes, but up to a high level in a few genotypes (TPM = 80). The results confirmed the extensive sequence and expression variations of SiZIPs in the natural foxtail millet population, suggesting their genetic selection potential in foxtail millet breeding.
Extensive studies have confirmed the existence of convergent selection among gramineae crops (Chen et al., 2022). Based on the analysis of orthologous relationships and expression characteristics between different species, several important yield-related genes have been discovered in foxtail millet and wheat (Yang et al., 2021a; Liu et al., 2022). Rice, as a model crop in the family of gramineae, has undergone intensive research on the functions of multiple OsZIPs, while SiZIPs have not been reported yet. Therefore, we preliminarily predicted the functions of SiZIPs through literature investigation and comparison of the expression patterns of orthologous ZIPs in foxtail millet and rice (Table 4). Out of the 14 SiZIP genes, 12 orthologs have been identified in rice, and 8 of them exhibited similar expression patterns in foxtail millet and rice, such as SiZIP10, SiZIP3 and SiZIP2. SiZIP10 was specifically highly expressed in the stems of foxtail millet, while its ortholog OsZIP3 in rice is reported to be responsible for Zn unloading and distribution in the enlarged vascular bundle xylem. SiZIP3 was specifically highly expressed in the panicles of foxtail millet at multiple stages, and its ortholog OsZIP5 was mainly expressed in the panicles and participates in Zn homeostasis. SiZIP2 was highly expressed in the roots, stems, and panicles of foxtail millet, and its ortholog OsZIP7 has been proven to be highly expressed in the same tissues in rice, responsible for the loading of Zn/Cd in the root xylem and their transfer from the stem node vascular bundles to the grains. The highly conserved expression patterns of ZIP genes in foxtail millet and rice suggested that the functions of these genes in gramineae, at least in foxtail millet and rice, were relatively conserved. In addition to sharing similar tissue expression patterns with their orthologs in rice, some SiZIP genes also expressed in other tissues. For example, SiZIP12, besides highly expressing in the roots for Zn/Fe absorption and transport similar to its ortholog OsIRT1, also exhibited high expression in the panicles, suggesting that it may have evolved new functions in foxtail millet while maintaining its essential roles. Moreover, apart from conserved and partially conserved genes, there were a few SiZIP genes that exhibited completely different expression patterns compared to their orthologs in rice. For instance, SiZIP5 was specifically highly expressed in the leaves of foxtail millet throughout all developmental stages, whereas its ortholog OsZIP12 was primarily expressed in the grains. It suggested that the functions of ZIP12 may differ in foxtail millet and rice.
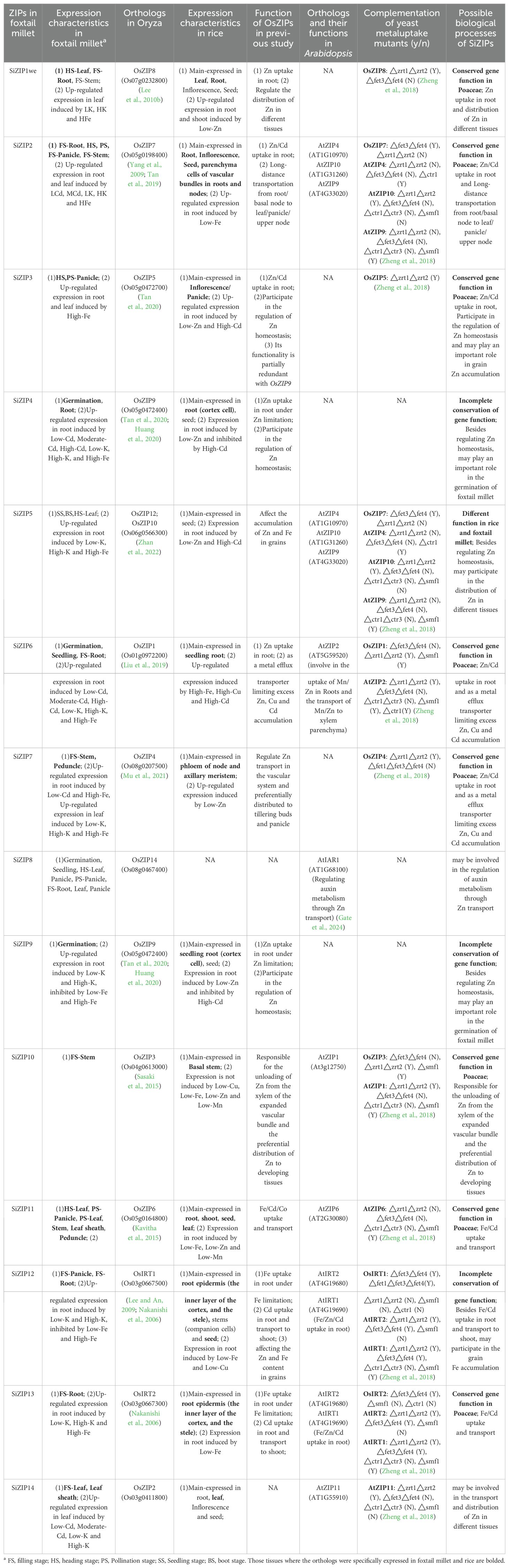
Table 4. Functional prediction of ZIPs in foxtail millet based on homology relationship, expression characteristics and previous studies.
Based on the homology relationships and expression patterns of ZIPs between foxtail millet, rice, and Arabidopsis, we predicted the functions of most SiZIP genes. Ultimately, we created a preliminary model diagram of the SiZIP genes in foxtail millet, integrating gene function predictions and expression characteristics in response to metal ions (Figure 9). According to the expression levels of the genes in different tissues and previous studies, the functions of the SiZIP genes were mainly classified into four categories: 1) absorption and efflux of metal ions such as Zn, Fe, and Cd in roots; 2) loading, unloading, and distribution of metal ions in stems; 3) intracellular homeostasis and utilization of metal ions in tissues and organs like leaves; 4) accumulation of metal ions in grains. The localization of conserved SiZIP genes in different tissues and cells was also depicted as precisely as possible, such as SiZIP2 (OsZIP7) likely localized in the root stele, and SiZIP7 (OsZIP4) potentially positioned in the stem or phloem of leaf axillary meristems. Additionally, the expression changes of ZIP genes under non-stress conditions of Fe, Cd, K, and Na were also characterized. In addition to having significant responses to different concentrations of Fe and Cd, the SiZIP genes also exhibited general responses to K and Na, which was likely closely related to the complex synergistic and antagonistic relationships between different metal ions and their transporters.
Trace mineral elements such as Fe and Zn are crucial for crop growth, development, quality formation, and human health (Pushnik et al., 1984; Broadley et al., 2012; Pinson et al., 2015; Wang et al., 2024), while excessive accumulation of heavy metals like Cd in crops can easily enter the human body through the food chain, leading to severe health issues (Pan et al., 2016; Yang et al., 2021c). Numerous studies have confirmed that essential trace elements like Zn and Fe, and harmful heavy metals like Cd, are simultaneously absorbed, transported, and accumulated by crops due to their similar geochemical behaviors (Zheng et al., 2018). The ZIP family, as an important metal ion transporter in crops, is directly responsible for the absorption, transport, and distribution of metal ions such as Zn, Fe, and Cd (Eide, 2006). Therefore, they can serve as potential targets to improve the absorption and utilization efficiency of Zn and Fe in crops, ensure food security, and improve nutritional quality. Although ZIP genes have been reported in Arabidopsis, rice, wheat and other crops, little is known about them in foxtail millet (Zheng et al., 2018; Li et al., 2021). In this study, we identified 14 ZIP genes in foxtail millet, which did not show significant differences in number compared to monocot grasses like sorghum (14), maize (12), rice (13), and dicotyledonous plants like Arabidopsis (15) and potato (12). Phylogenetic analysis revealed that ZIP genes from these species were mixedly distributed into four class, indicating that the formation of the ZIP family predates the divergence of monocots and dicots (Figure 1). Although the number of ZIP genes in different species was similar, except for the relatively conserved ZIP genes in Class I across monocots and dicots, ZIP genes in other class showed significant uneven expansion. Unique gene clusters formed in Class III and Class IV, such as OC8-1 to OC8-4, were specific to monocots, which may be an important reason for the significant difference in Zn and Fe absorption between monocots and dicots (Table 2) (Zheng et al., 2018). Through an in-depth assessment of phylogenetic relationships, we clarified the orthologous relationships of ZIP genes. ZIP genes in orthologous gene clusters across multiple species may retain the most basic functions from their ancestor before the divergence of monocots and dicots. On the other hand, paralogous genes formed after divergence have greatly enriched the functional diversity of the ZIP gene family, playing an important role in the environmental adaptability of different species.
The expansion and functional divergence of gene families often rely on early duplication events and the emergence of novel genes and functions that follow thereafter (Yang et al., 2021b). More than 50% (8/14) of the genes in the foxtail millet ZIP family were involved in gene duplication events, forming two pairs of TDs and two pairs of SDs (Supplementary Figure S1), which confirmed the contribution of duplication events to the expansion of the foxtail millet ZIP gene family. Although the Ka/Ks values of all duplicated gene pairs were less than 1, the TD gene pairs had relatively higher values, indicating that under the premise of conserving important functions, new genes generated by TD may be more crucial for the functional divergence of the SiZIP gene family, similar to reports in other species (Gao et al., 2022). In addition to differences in sequences, duplicated gene pairs also exhibited new variations in gene structure, protein conserved domains, and transmembrane regions. SiZIP5 and SiZIP1 (SD1), SiZIP7 and SiZIP10 (SD2) had deletions in conserved motifs (Figure 2), while SiZIP7 and SiZIP10 (SD2), SiZIP3 and SiZIP4 (TD1), and SiZIP12 and SiZIP13 (TD2) showed variations in the number of TM domains (Table 1). As important transmembrane transporters, the TM regions are crucial for protein function (Yang et al., 2022). Moreover, the duplicated gene pairs also showed significant differences in tissue expression patterns (Figure 5, Supplementary Figure S2). The SiZIP2 gene in SD1 was highly expressed in panicles and roots during multiple stages of foxtail millet growth, while SiZIP5 was mainly expressed in leaves. In TD1, SiZIP3 was specifically expressed in panicles, while SiZIP4 was highly expressed in germinating seeds. In TD2, SiZIP13 was specifically expressed in roots, while SiZIP12 was highly expressed in both roots and panicles. Additionally, we found that the responses of these genes to different metal ions have also changed. For example, the expression level of SiZIP7 in SD2 was significantly induced by high Fe, while SiZIP10 did not respond. In conclusion, while gene duplication drives the expansion of the SiZIP family, the changes in sequence, protein structure, and expression characteristics of the newly generated genes directly promote the functional divergence of the SiZIP family.
The spatiotemporal expression characteristics and responses to different stresses of members within the same gene family often exhibit significant differences, providing fundamental information for understanding the functions of important genes (Yang et al., 2021b). The expression patterns of SiZIP genes showed clear temporal and spatial specificity throughout the growth period of foxtail millet (Figure 5, Supplementary Figure S2). Specifically, SiZIP9, SiZIP4, and SiZIP6 were highly expressed in germinating seeds and seedlings, likely participating in seed germination and seedling growth by regulating the absorption and transport of Zn, Fe, and other ions. In contrast, SiZIP8 and SiZIP5 were primarily expressed in the panicles, stems, and leaves during the later stages of foxtail millet growth and development, potentially involved in the transport and redistribution of metal ions. Furthermore, some SiZIP genes exhibited tissue-specific expression patterns, such as SiZIP13, SiZIP14, SiZIP10, and SiZIP3, which were specifically expressed in roots, leaves, stems, and panicles, respectively, indicating more specialized functions. ZIP genes are not only responsible for the migration of Zn and Fe in plants but also for the absorption and transport of other metals (Zheng et al., 2018).
Most SiZIP genes responded to different metal ion stresses, including Fe, Cd, K, and Na (Figures 6, 7). Over 50% of SiZIP genes (8/14) were significantly upregulated under high Fe stress, similar to the response pattern of OsZIP in rice (Zheng et al., 2018). SiZIP6 and SiZIP4 were significantly upregulated under Cd stress, which was consistent with the response pattern of their orthologs OsZIP1 and OsZIP9 (Liu et al., 2019; Huang et al., 2020; Tan et al., 2020). Interestingly, SiZIP genes show widespread responses to K and Na, and some of them exhibited inconsistent response patterns with Fe, such as SiZIP12 and SiZIP9. After Cd treatment, significant changes occurred in the accumulation of Fe, Ca, K, and Na in the roots and aboveground tissues of foxtail millet seedlings. At the same time, there was a significant correlation between SiZIP gene expression and metal ion content, especially with Fe and K (Supplementary Figure S3). These results suggested that SiZIP genes, in addition to their previously reported role in transporting divalent metal ions, also had indirect synergistic or antagonistic relationships with the transport of monovalent metal ions such as Na and K. Additionally, some SiZIP genes, such as SiZIP1 and SiZIP3, were upregulated under cold and drought stress. These genes may repair oxidative damage and maintain membrane stability through the transport of metal ions, thus resisting plant damage caused by stress (Coleman, 1998; Cakmak, 2000). Additionally, a large number of cis-acting elements involved in stress response and tissue expression specificity were found in the promoter regions of multiple SiZIPs (Figure 4). These elements may be related to the expression levels of SiZIPs. For example, seed-specific expression elements were found in the upstream of SiZIP4 and SiZIP6, which is consistent with the specific high expression of these two genes in germinating seeds (Supplementary Figure S2). The upstream of SiZIP12 contains four low-temperature response elements, which may be related to its rapid up-regulated expression under low-temperature stress (Figure 7). The multiple stress response elements upstream of SiZIP13 may be closely related to its extensive response to various abiotic stresses. A large number of studies have confirmed that transcription factors such as MYB affect biotic and abiotic stress responses by regulating the expression of metal transport genes (Liu et al., 2024). Although we did not find cis-acting elements directly related to metal ion transport in the upstream of SiZIPs, the presence of multiple MYB transcription factor binding elements may be related to their metal transport. In general, we have found the possible associations between multiple cis-acting elements in the upstream of SiZIPs and their expression levels. A more in-depth and accurate regulatory mechanism needs to be further verified through experiments.
Analyzing genetic variations in important functional genes in germplasm resources is an important means to clarify their value for breeding improvement (Liu et al., 2022). We identified a total of 1718 SNPs and Indels distributed across SiZIP genes (including upstream and downstream regions) in 360 foxtail millet genotypes, including 235 mutations located in promoter regions and 54 non-synonymous SNPs (Table 3). Further association analysis between grain metal ion content and variation sites revealed 102 SNPs significantly associated with Fe, Zn, Cd, and other metal ion contents (unpublished data). Additionally, the expression levels of SiZIP genes showed significant differences in grains of different foxtail millet genotypes (Supplementary Figure S4). The above results confirmed the significant genetic selection potential of SiZIP genes in the improvement of foxtail millet quality.
During the domestication process of gramineae crops, there has been a large-scale convergent selection, with multiple important genes including OsKRN2, TaGS-D1, TaCKX2, TaTGW6, and TaCWI reported to regulate traits such as yield in maize, rice, and wheat through similar pathways (Zhang et al., 2011, 2014; Jiang et al., 2015; Hanif et al., 2016; Chen et al., 2022; He et al., 2024). Meanwhile, by comparing the orthologous relationships and expression characteristics of functional genes in rice, several important genes related to wheat and foxtail millet yield have been discovered (Yang et al., 2021a; Liu et al., 2022). These findings confirmed that through homologous comparison, expression analysis, and in-depth research on functional genes of leading model species, functional genes in less-studied secondary crops such as foxtail millet can be rapidly identified. Based on this, we combined the research on ZIP genes in rice and Arabidopsis, and predicted the function of ZIP genes in foxtail millet under the premise of fully considering the expression characteristics of orthologous genes (Table 4). Multiple SiZIPs and their orthologous genes exhibited highly similar expression patterns in foxtail millet and rice. SiZIP7 was specifically highly expressed in stems and peduncles of foxtail millet, and its orthologous gene OsZIP4 has been proven in rice to be highly expressed in the vascular bundle system of stem nodes and axillary meristems to regulate the preferential distribution of Zn in different organs (Mu et al., 2021), suggesting that SiZIP7 may have similar functions in foxtail millet. SiZIP13 was specifically expressed in foxtail millet roots, while its orthologous gene OsIRT2 was localized in the stele and epidermis of rice roots and was responsible for Fe/Cd uptake in roots (Nakanishi et al., 2006). SiZIP3 and its orthologous gene OsZIP5 were both specifically highly expressed in panicles, and OsZIP5 has been shown to be involved in the regulation of Zn homeostasis, indicating that the functions of SiZIP3/OsZIP5 may be relatively conserved (Tan et al., 2020). OsZIP1, as a well-documented important metal efflux transporter that was specifically expressed in the roots of seedlings, exhibited a similar expression pattern in its foxtail millet ortholog SiZIP6, suggesting that they may share similar functions. SiZIP1 was highly expressed in the leaves and roots of foxtail millet, and overexpression of its orthologous gene OsZIP8 with the same expression characteristics resulted in a decrease in Zn content in the shoots and an increase in Zn content in the roots of transgenic rice plants, suggesting that SiZIP1 may be involved in the distribution of Zn in the shoots similar to OsZIP8 (Lee et al., 2010b). We found that at least 8 SiZIP genes have similar expression patterns with their rice orthologous genes, and the functions of these genes may be highly conserved between foxtail millet and rice, similar to the previous reports of hundreds of genes being convergently selected in maize and rice (Chen et al., 2022). In addition to these conserved genes, a few genes showed inconsistent expression patterns, which may have some foxtail millet-specific functions. Overall, based on the expression patterns, orthologous relationships, and previous reports of these genes, we have predicted the functions of SiZIP genes as much as possible and classified them into four categories: root absorption/efflux, stem transport/distribution, utilization in tissues such as leaves, and grain development and accumulation (Figure 9). Some SiZIP genes were only involved in one of these processes, while others played roles in multiple processes. In general, although the functions of individual SiZIP genes were not studied in depth in this study, and there is a lack of relevant results on their response to Zn, a basic research framework for SiZIP genes in foxtail millet has been constructed through the combined strategy of systematic characterization, homologous comparison, and expression characteristic analysis. More in-depth functional studies will be the focus of our next work.
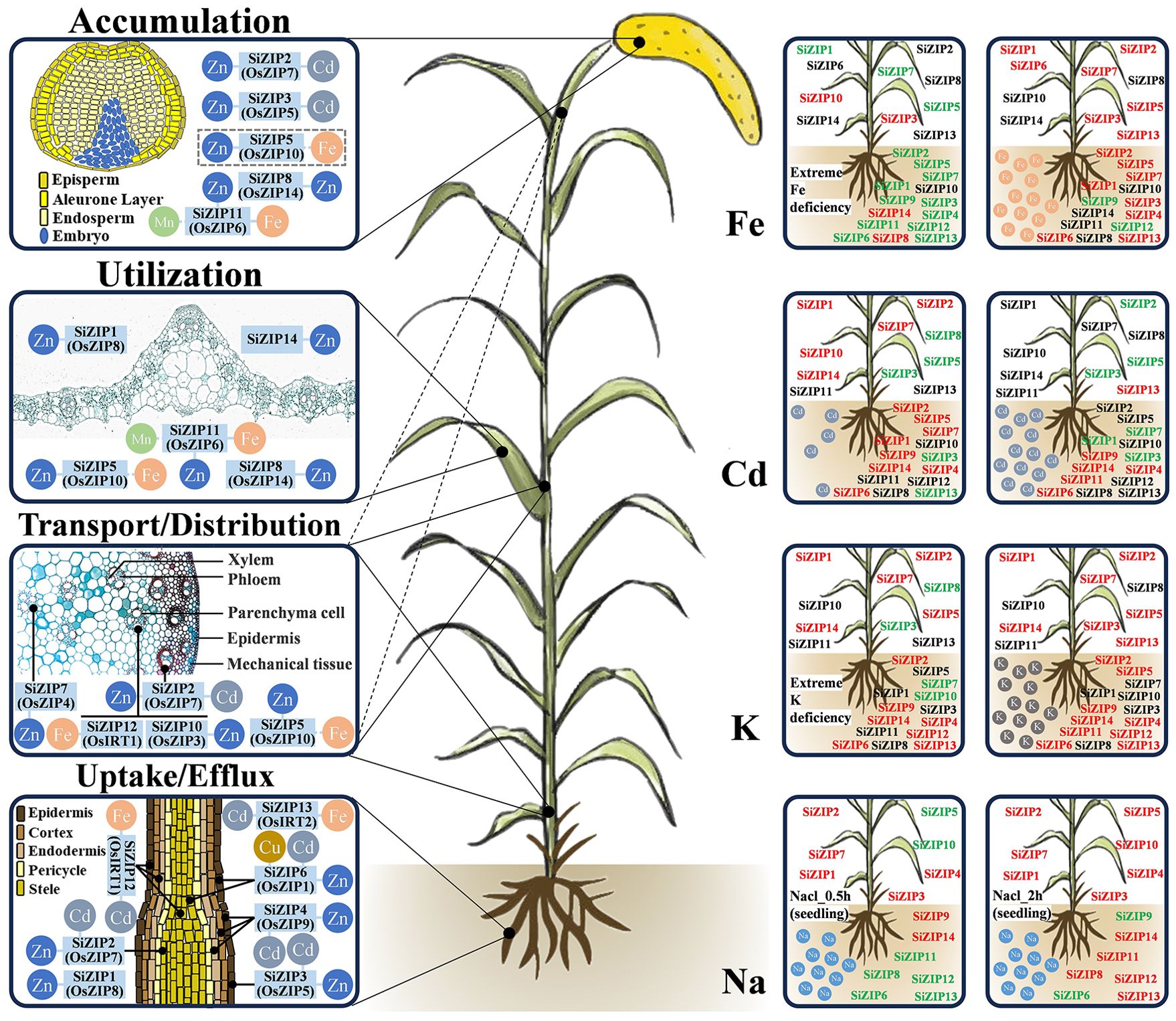
Figure 9. The schematic diagram of predicted functions and response to metal ions of SiZIPs in foxtail millet. Functional predictions of SiZIPs are derived from homology comparisons, expression profiling and previous studies. SiZIPs and their conserved orthologs in rice are labeled together. The left side shows the main functions involved by SiZIPs, while the right side shows the response of SiZIPs to different metal ions. Red represents up-regulation and green represents downregulation. Except for Na treatment, the expression levels of SiZIPs between aboveground and underground parts were distinguished.
In this study, 14 ZIP transporters were accurately identified and characterized in foxtail millet. Based on the phylogenetic relationships of ZIPs from six species, SiZIPs were divided into four evolutionary branches and their homologous relationships with ZIPs of staple crops were clarified. We demonstrated that gene duplication and subsequent multi-level variations jointly promote the expansion and functional differentiation of the ZIP family in foxtail millet. These SiZIP genes showed abundant spatiotemporal expression characteristics in different tissues of foxtail millet during the whole growth period, and exhibited positive responses to Fe, Cd, K, Na, drought and cold stresses. It’s indicated that they might be extensively involved in the transport of metal ions and osmotic regulation under abiotic stresses. Additionally, we analyzed the sequence variation and expression variation of SiZIP in different genotypes of foxtail millet, and evaluated its genetic potential in improving foxtail millet quality. Finally, we predicted the function of SiZIP gene in foxtail millet and preliminarily drew its functional pattern diagram by integrating homologous comparison, expression analysis and previous studies. This study will lay the foundation for further functional research on SiZIPs, and will also contribute to the elucidation of the molecular mechanisms underlying metal ion transport and accumulation in foxtail millet and the quality improvement.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author/s.
JZ: Methodology, Writing – original draft. YM: Software, Visualization, Writing – original draft. YL: Validation, Visualization, Writing – original draft. TZ: Software, Validation, Writing – original draft. CC: Investigation, Software, Writing – original draft. AA: Formal analysis, Writing – review & editing. WW: Project administration, Validation, Writing – review & editing. FM: Data curation, Writing – review & editing. YH: Resources, Supervision, Writing – review & editing. HL: Resources, Writing – review & editing. SH: Conceptualization, Writing – review & editing. YY: Conceptualization, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by Fundamental Research Program of Shanxi Province (202103021223157), National guides local science and technology development fund projects(YDZJSX2022B007, National Important Specialty Crop foxtail millet Breeding Joint Research Project (gzlg-01), and the Scientific and Technological Innovation Programs of Shanxi Agricultural University (2021BQ80).
Author WW were employed by Join Hope Seed Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1467015/full#supplementary-material
Baxter, I. (2015). Should we treat the ionome as a combination of individual elements, or should we be deriving novel combined traits? J. Exp. Bot. 66, 2127–2131. doi: 10.1093/jxb/erv040
Bolan, N. S., Adriano, D. C., Curtin, D. (2003). Soil acidification and liming interactions with nutrient and heavy metal transformation and bioavailability. Adv. Agron. 78, 215–272. doi: 10.1016/S0065-2113(02)78006-1
Bourke, C. D., Berkley, J. A., Prendergast, A. J. (2016). Immune dysfunction as a cause and consequence of malnutrition. Trends Immunol. 37, 386–398. doi: 10.1016/j.it.2016.04.003
Broadley, M., Brown, P., Cakmak, I., Rengel, Z., Zhao, F. (2012). Function of nutrients: micronutrients. Marschner’s Mineral Nutr. Higher Plants, 191-248. doi: 10.1016/B978-0-12-384905-2.00007-8
Bughio, N., Yamaguchi, H., Nishizawa, N. K., Nakanishi, H., Mori, S. (2002). Cloning an iron-regulated metal transporter from rice. J. Exp. Bot. 53, 1677–1682. doi: 10.1093/jxb/erf004
Cakmak, I. (2000). Tansley Review No. 111: Possible roles of zinc in protecting plant cells from damage by reactive oxygen species. New Phytol. 146, 185–205. doi: 10.1046/j.1469-8137.2000.00630.x
Chen, C., Wu, Y., Li, J., Wang, X., Zeng, Z., Xu, J., et al. (2023a). TBtools-II: A ‘one for all, all for one’ bioinformatics platform for biological big-data mining. Mol. Plant 16, 1733–1742. doi: 10.1016/j.molp.2023.09.010
Chen, L., Yang, Y., Zhao, Z., Lu, S., Lu, Q., Cui, C., et al. (2023b). Genome wide identification and comparative analyses of key genes involved in C4 photosynthesis in five main gramineous crops. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1134170
Chen, W., Chen, L., Zhang, X., Yang, N., Guo, J., Wang, M., et al. (2022). Convergent selection of a WD40 protein that enhances grain yield in maize and rice. Science 375, eabg7985. doi: 10.1126/science.abg7985
Chen, W. R., Feng, Y., Chao, Y. E., Chen, W. R. (2008). Genomic analysis and expression pattern of OsZIP1, OsZIP3, and OsZIP4 in two rice (Oryza sativa L.) genotypes with different zinc efficiency. Russian J. Plant Physiol. 55, 400–409. doi: 10.1134/S1021443708030175
Coleman, J. E. (1998). Zinc enzymes. Curr. Opin. Chem. Biol. 2, 222–234. doi: 10.1016/S1367-5931(98)80064-1
Deng, P., Tao, T., Ji, W., Zhang, G., Wu, L., Wu, D. (2022). Population-level transcriptomes reveal gene expression and splicing underlying cadmium accumulation in barley. Plant J. 112, 847–859. doi: 10.1111/tpj.15986
Eide, D. J. (2006). Zinc transporters and the cellular trafficking of zinc. BBA-Mol. Cell Res. 1763, 711–722. doi: 10.1016/j.bbamcr.2006.03.005
Eide, D., Broderius, M., Fett, J., Guerinot, M. L. (1996). A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl. Acad. Sci. U. S. A. 93, 5624–5628. doi: 10.1073/pnas.93.11.5624
Fan, W., Liu, C., Cao, B., Ma, S., Hu, J., Xiang, Z., et al. (2021). A meta-analysis of transcriptomic profiles reveals molecular pathways response to cadmium stress of Gramineae. Ecotoxicol. Environ. Saf. 209, 111816. doi: 10.1016/j.ecoenv.2020.111816
Gaitan-Solis, E., Taylor, N. J., Siritunga, D., Stevens, W., Schachtman, D. P. (2015). Overexpression of the transporters AtZIP1 and AtMTP1 in cassava changes zinc accumulation and partitioning. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00492
Gao, F., Li, J., Zhang, J., Li, N., Tang, C., Bakpa, E. P., et al. (2022). Genome-wide identification of the ZIP gene family in lettuce (Lactuca sativa L.) and expression analysis under different element stress. PloS One 17, e0274319. doi: 10.1371/journal.pone.0274319
Gate, T., Hill, L., Miller, A. J., Sanders, D. (2024). AtIAR1 is a Zn transporter that regulates auxin metabolism in Arabidopsis thaliana. J. Exp. Bot. 75, 1437–1450. doi: 10.1093/jxb/erad468
Hanif, M., Gao, F., Liu, J., Wen, W., Zhang, Y., Rasheed, A., et al. (2016). TaTGW6-A1, an ortholog of rice TGW6, is associated with grain weight and yield in bread wheat. Mol. Breed. 36, 1. doi: 10.1007/s11032-015-0425-z
He, L., Cheng, L., Wang, J., Liu, J., Cheng, J., Yang, Z., et al. (2022). Carotenoid cleavage dioxygenase 1 catalyzes lutein degradation to influence carotenoid accumulation and color development in Foxtail millet grains. J. Agr Food Chem. 70, 9283–9294. doi: 10.1021/acs.jafc.2c01951
He, Z., Zhang, P., Jia., H., Zhang, S., Nishawy, E., Sun, X., et al. (2024). Regulatory mechanisms and breeding strategies for crop drought resistance. New Crops 1, 100029. doi: 10.1016/j.ncrops.2024.100029
Ho, E. (2004). Zinc deficiency, DNA damage and cancer risk. J. Nutr. Biochem. 15, 572–578. doi: 10.1016/j.jnutbio.2004.07.005
Huang, S., Sasaki, A., Yamaji, N., Okada, H., Mitani-Ueno, N., Ma, J. (2020). The ZIP transporter family member OsZIP9 contributes to root zinc uptake in rice under zinc-limited conditions. Plant Physiol. 183, 1224–1234. doi: 10.1104/pp.20.00125
Ishimaru, Y., Kim, S., Tsukamoto, T., Oki, H., Kobayashi, T., Watanabe, S., et al. (2007a). Mutational reconstructed ferric chelate reductase confers enhanced tolerance in rice to iron deficiency in calcareous soil. Proc. Natl. Acad. Sci. U. S. A. 104, 7373–7378. doi: 10.1073/pnas.0610555104
Ishimaru, Y., Masuda, H., Suzuki, M., Bashir, K., Takahashi, M., Nakanishi, H., et al. (2007b). Overexpression of the OsZIP4 zinc transporter confers disarrangement of zinc distribution in rice plants. J. Exp. Bot. 58, 2909–2915. doi: 10.1093/jxb/erm147
Ishimaru, Y., Suzuki, M., Tsukamoto, T., Suzuki, K., Nakazono, M., Kobayashi, T., et al. (2006). Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+. Plant J. 45, 335–346. doi: 10.1111/j.1365-313X.2005.02624.x
Jiang, Y., Jiang, Q., Hao, C., Hou, J., Wang, L., Zhang, H., et al. (2015). A yield-associated gene TaCWI, in wheat: its function, selection and evolution in global breeding revealed by haplotype analysis. Theor. Appl. Genet. 128, 131–143. doi: 10.1007/s00122-014-2417-5
Kavitha, P. G., Kuruvilla, S., Mathew, M. K. (2015). Functional characterization of a transition metal ion transporter, OsZIP6 from rice (Oryza sativa L.). Plant Physiol. Bioch. 97, 165–174. doi: 10.1016/j.plaphy.2015.10.005
Khush, G. S., Lee, S., Cho, J. I., Jeon, J. S. (2012). Biofortification of crops for reducing malnutrition. Plant Biotechnol. Rep. 6, 195–202. doi: 10.1007/s11816-012-0216-5
Krämer, U., Talke, I. N., Hanikenne, M. (2007). Transition metal transport. FEBS Lett. 581, 2263–2272. doi: 10.1016/j.febslet.2007.04.010
Lee, S., An, G. (2009). Over-expression of OsIRT1 leads to increased iron and zinc accumulations in rice. Plant Cell Environ. 32, 408–416. doi: 10.1111/j.1365-3040.2009.01935.x
Lee, S., Jeong, H. J., Kim, S. A., Lee, J., Guerinot, M. L., An, G. (2010a). OsZIP5 is a plasma membrane zinc transporter in rice. Plant Mol. Biol. 73, 507–517. doi: 10.1007/s11103-010-9637-0
Lee, S., Kim, S. A., Lee, J., Guerinot, M. L., An, G. (2010b). Zinc deficiency-inducible OsZIP8 encodes a plasma membrane-localized zinc transporter in rice. Mol. Cells 29, 551–558. doi: 10.1007/s10059-010-0069-0
Li, Q., Chen, L., Yang, A. (2019). The molecular mechanisms underlying iron deficiency responses in rice. Int. J. Mol. Sci. 21, 43. doi: 10.3390/ijms21010043
Li, X., Gao, J., Song, J., Guo, K., Hou, S., Wang, X., et al. (2022). Multi-omics analyses of 398 foxtail millet accessions reveal genomic regions associated with domestication, metabolite traits and anti-inflammatory effects. Mol. Plant 15, 1367–1383. doi: 10.1016/j.molp.2022.07.003
Li, S., Liu, Z., Guo, L., Li, H., Nie, X., Chai, S., et al. (2021). Genome-wide identification of wheat ZIP gene family and functional characterization of the TaZIP13-B in plants. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.748146
Li, S., Zhou, X., Huang, Y., Zhu, L., Zhang, S., Zhao, Y., et al. (2013). Identification and characterization of the zinc-regulated transporters, iron-regulated transporter-like protein (ZIP) gene family in maize. BMC Plant Biol. 13, 114. doi: 10.1186/1471-2229-13-114
Liu, X., Feng, S., Zhang, B., Wang, M., Cao, H., Rono, J. K., et al. (2019). OsZIP1 functions as a metal efflux transporter limiting excess zinc, copper and cadmium accumulation in rice. BMC Plant Biol. 19, 1–16. doi: 10.1186/s12870-019-1899-3
Liu, F., Xi, M., Liu, T., Wu, X., Ju, L., Wang, D. (2024). The central role of transcription factors in bridging biotic and abiotic stress responses for plants’ resilience. New. Crops. 1, 100005. doi: 10.1016/j.ncrops.2023.11.003
Liu, X., Yang, Y., Hou, S., Men, Y., Han, Y. (2022). The integration of genome-wide association study and homology analysis to explore the genomic regions and candidate genes for panicle-related traits in foxtail millet. Int. J. Mol. Sci. 23, 14735. doi: 10.3390/ijms232314735
Milner, M. J., Seamon, J., Craft, E., Kochian, L. V. (2013). Transport properties of members of the ZIP family in plants and their role in Zn and Mn homeostasis. J. Exp. Bot. 64, 369–381. doi: 10.1093/jxb/ers315
Mu, S., Yamaji, N., Sasaki, A., Luo, L., Du, B., Che, J., et al. (2021). A transporter for delivering zinc to the developing tiller bud and panicle in rice. Plant J. 105, 786–799. doi: 10.1111/tpj.15073
Nakanishi, H., Ogawa, I., Ishimaru, Y., Mori, S., Nishizawa, N. K. (2006). Iron deficiency enhances cadmium uptake and translocation mediated by the Fe2+ transporters OsIRT1 and OsIRT2 in rice. Soil Sci. Plant Nutr. 52, 464–469. doi: 10.1111/j.1747-0765.2006.00055.x
Pan, L., Ma, J., Hu, Y., Su, B., Fang, G., Wang, Y., et al. (2016). Assessments of levels, potential ecological risk, and human health risk of heavy metals in the soils from a typical county in Shanxi Province, China. Environ. Sci. pollut. R. 23, 19330–19340. doi: 10.1007/s11356-016-7044-z
Pinson, S. R., Tarpley, L., Yan, W., Yeater, K., Lahner, B., Yakubov, E., et al. (2015). Worldwide genetic diversity for mineral element concentrations in rice grain. Crop Sci. 55, 294–311. doi: 10.2135/cropsci2013.10.0656
Pushnik, J. C., Miller, G. W., Manwaring, J. H. (1984). The role of iron in higher plant chlorophyll biosynthesis, maintenance and chloroplast biogenesis. J. Plant Nutr. 7, 733–758. doi: 10.1080/01904168409363238
Ramegowda, Y., Venkategowda, R., Jagadish, P., Govind, G., Hanumanthareddy, R. R., Makarla, U., et al. (2013). Expression of a rice Zn transporter, OsZIP1, increases Zn concentration in tobacco and finger millet transgenic plants. Plant Biotechnol. Rep. 7, 309–319. doi: 10.1007/s11816-012-0264-x
Rouault, T. A. (2015). Iron-sulfur proteins hiding in plain sight. Nat. Chem. Biol. 11, 442–445. doi: 10.1038/nchembio.1843
Sadeghzadeh, B. (2013). A review of zinc nutrition and plant breeding. J. Soil Sci. Plant Nutr. 13, 905–927. doi: 10.4067/S0718-95162013005000072
Sasaki, A., Yamaji, N., Mitani-Ueno, N., Kashino, M., Ma, J. (2015). A node-localized transporter OsZIP3 is responsible for the preferential distribution of Zn to developing tissues in rice. Plant J. 84, 374–384. doi: 10.1111/tpj.13005
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. doi: 10.1038/msb.2011.75
Tan, L., Qu, M., Zhu, Y., Peng, C., Wang, J., Gao, D., et al. (2020). ZINC TRANSPORTER5 and ZINC TRANSPORTER9 function synergistically in zinc/cadmium uptake. Plant Physiol. 183, 1235–1249. doi: 10.1104/pp.19.01569
Tan, L., Zhu, Y., Fan, T., Peng, C., Wang, J., Sun, L., et al. (2019). OsZIP7 functions in xylem loading in roots and inter-vascular transfer in nodes to deliver Zn/Cd to grain in rice. Biochem. Biophys. Res. Commun. 512, 112–118. doi: 10.1016/j.bbrc.2019.03.024
Trijatmiko, K. R., Dueñas, C., Tsakirpaloglou, N., Torrizo, L., Arines, F. M., Adeva, C., et al. (2016). Biofortified indica rice attains iron and zinc nutrition dietary targets in the field. Sci. Rep. 6, 19792. doi: 10.1038/srep19792
Vert, G., Briat, J. F., Curie, C. (2001). Arabidopsis IRT2 gene encodes a root-periphery iron transporter. Plant J. 26, 181–189. doi: 10.1046/j.1365-313x.2001.01018.x
Vert, G., Grotz, N., Dédaldéchamp, F., Gaymard, F., Guerinot, M. L., Briat, J. F., et al. (2002). IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14, 1223–1233. doi: 10.1105/tpc.001388
Wang, M., Ma, W., Chaney, R. L., Green, C. E., Chen, W. (2022). Effects of Mn2+ on Cd accumulation and ionome in rice and spinach. J. Environ. Qual. 51, 890–898. doi: 10.1002/jeq2.20358
Wang, L., Wang, Y., Yin, P., Jiang, C., Zhang, M. (2024). ZmHAK17 encodes a Na+-selective transporter that promotes maize seed germination under salt conditions. New Crops 1, 100024. doi: 10.1016/j.ncrops.2024.100024
Wessells, K. R., Brown, K. H. (2012). Estimating the global prevalence of zinc deficiency: results based on zinc availability in national food supplies and the prevalence of stunting. PloS One 7, 50568. doi: 10.1371/journal.pone.0050568
White, P. J., Broadley, M. R. (2009). Biofortification of crops with seven mineral elements often lacking in human diets-iron, zinc, copper, calcium, magnesium, selenium and iodine. New Phytol. 182, 49–84. doi: 10.1111/j.1469-8137.2008.02738.x
Wu, X., Zhu, Z., Chen, J., Huang, Y., Liu, Z., Zou, J., et al. (2019). Transcriptome analysis revealed pivotal transporters involved in the reduction of cadmium accumulation in pak choi (Brassica chinensis L.) by exogenous hydrogen-rich water. Chemosphere 216, 684–697. doi: 10.1016/j.chemosphere.2018.10.152
Yan, P., Du, Q., Chen, H., Guo, Z., Wang, Z., Tang, J., et al. (2023). Biofortification of iron content by regulating a NAC transcription factor in maize. Science 382, 1159–1165. doi: 10.1126/science.adf3256
Yang, Y., Amo, A., Wei, D., Chai, Y., Zheng, J., Qiao, P., et al. (2021a). Large−scale integration of meta−QTL and genome−wide association study discovers the genomic regions and candidate genes for yield and yield−related traits in bread wheat. Theor. Appl. Genet. 134, 3083–3109. doi: 10.1007/s00122-021-03881-4
Yang, Y., Chai, Y., Liu, J., Zheng, J., Zhao, Z., Amo, A., et al. (2021b). Amino acid transporter (AAT) gene family in foxtail millet (Setaria italica L.): widespread family expansion, functional differentiation, roles in quality formation and response to abiotic stresses. BMC Genomics 22, 1–23. doi: 10.1186/s12864-021-07779-9
Yang, X., Huang, J., Jiang, Y., Zhang, H. (2009). Cloning and functional identification of two members of the ZIP (Zrt, Irt-like protein) gene family in rice (Oryza sativa L.). Mol. Biol. Rep. 36, 281–287. doi: 10.1007/s11033-007-9177-0
Yang, Y., Wang, X., Zheng, J., Men, Y., Zhang, Y., Liu, L., et al. (2022). Amino acid transporter (AAT) gene family in Tartary buckwheat (Fagopyrum tataricum L. Gaertn.): Characterization, expression analysis and functional prediction. Int. J. Biol. Macromol. 217, 330–344. doi: 10.1016/j.ijbiomac.2022.07.059
Yang, Z., Yang, F., Liu, J., Wu, H., Yang, H., Shi, Y., et al. (2021c). Heavy metal transporters: Functional mechanisms, regulation, and application in phytoremediation. Sci. Total Environ. 809, 151099. doi: 10.1016/j.scitotenv.2021.151099
Yang, Z., Zhang, H., Li, X., Shen, H., Gao, J., Hou, S., et al. (2020). A mini-foxtail millet with an Arabidopsis-like life cycle as a C4 model system. Nat. Plants 6, 1167–1178. doi: 10.1038/s41477-020-0747-7
Yang, Y., Zheng, J., Liang, Y., Wang, X., Li, K., Chen, L., et al. (2023). Natural resistance-associated macrophage protein (Nramp) family in foxtail millet (Setaria italica): Characterization, expression analysis and relationship with metal content under Cd stress. Agronomy 13, 2000. doi: 10.3390/agronomy13082000
Yu, R., Li, D., Du, X., Xia, S., Liu, C., Shi, G. (2017). Comparative transcriptome analysis reveals key cadmium transport-related genes in roots of two pak choi (Brassica rapa L. ssp chinensis) cultivars. BMC Genomics 18, 14. doi: 10.1186/s12864-017-3973-3982
Zhan, J., Zou, W., Li, S., Tang, J., Lu, X., Meng, L., et al. (2022). OsNAC15 regulates tolerance to zinc deficiency and cadmium by binding to OsZIP7 and OsZIP10 in rice. Int. J. Mol. Sci. 23, 11771. doi: 10.3390/ijms231911771
Zhang, Z. (2022). KaKs_Calculator 3.0: Calculating selective pressure on coding and non-coding sequences. Genomics Proteomics Bioinf. 20, 536–540. doi: 10.1016/j.gpb.2021.12.002
Zhang, Y., Liu, J., Xia, X., He, Z. (2014). TaGS-D1, an ortholog of rice OsGS3, is associated with grain weight and grain length in common wheat. Mol. Breed. 34, 1097–1107. doi: 10.1007/s11032-014-0102-7
Zhang, J., Liu, W., Yang, X., Gao, A., Li, X., Wu, X., et al. (2011). Isolation and characterization of two putative cytokinin oxidase genes related to grain number per spike phenotype in wheat. Mol. Biol. Rep. 38, 2337–2347. doi: 10.1007/s11033-010-0367-9
Keywords: foxtail millet, ZIP transporter, homologous comparison, expression characteristics, functional prediction
Citation: Zheng J, Ma Y, Liang Y, Zhang T, Chen C, Amo A, Wang W, Ma F, Han Y, Li H, Hou S and Yang Y (2024) An integration of genome-wide survey, homologous comparison and gene expression analysis provides a basic framework for the ZRT, IRT-like protein (ZIP) in foxtail millet. Front. Plant Sci. 15:1467015. doi: 10.3389/fpls.2024.1467015
Received: 19 July 2024; Accepted: 20 August 2024;
Published: 05 September 2024.
Edited by:
Meng Jiang, Zhejiang University, ChinaReviewed by:
Hengyu Yan, Qingdao Agricultural University, ChinaCopyright © 2024 Zheng, Ma, Liang, Zhang, Chen, Amo, Wang, Ma, Han, Li, Hou and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Yang, eWFuZzExNkBzeGF1LmVkdS5jbg==; Siyu Hou, c2l5dWhvdUBzeGF1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.