- Key Laboratory of Marine Genetics and Breeding (Ministry of Education), College of Marine Life Sciences, Ocean University of China, Qingdao, China
The basic leucine zipper (bZIP) family consists of conserved transcription factors which are widely present in eukaryotes and play important regulatory roles in plant growth, development, and stress responses. Neopyropia yezoensis is a red marine macroalga of significant economic importance; however, their bZIP family members and functions have not been systematically identified and analyzed. In the present study, the bZIP gene family in Ny. yezoensis was characterized by investigating gene structures, conserved motifs, phylogenetic relationships, chromosomal localizations, gene duplication events, cis-regulatory elements, and expression profiles. Twenty-three Ny. yezoensis bZIP (NyybZIP) genes were identified and sorted into 13 out of 30 groups, which were classified based on the bZIPs of Ny. yezoensis and 15 other red algae species. Phylogenetic analysis revealed that bZIP genes may have a complex evolutionary pattern in red algae. Cross-species collinearity analysis indicated that the bZIP genes in Ny. yezoensis, Neoporphyra haitanensis, and Porphyra umbilicalis are highly evolutionarily conserved. In addition, we identified four main categories of cis-elements, including development-related, light-responsive, phytohormone-responsive and stress-responsive promoter sequences in NyybZIP genes. Finally, RNA sequencing data and quantitative real-time PCR (qRT-PCR) showed that NyybZIP genes exhibited different expression patterns depending on the life stage. NyybZIP genes were also found to be involved in the nitrogen stress response. We thought that bZIP genes may be involved in Ny. yezoensis growth and development, and play a significant role in nitrogen deficiency response. Taken together, our findings provide new insights into the roles of the bZIP gene family and provide a basis for additional research into its evolutionary history and biological functions.
1 Introduction
A significant portion of eukaryotic genomes consists of transcription factor (TF) genes, which are predominantly categorized into a few distinct and often sizable gene families based on the type of DNA-binding domain they encode (Riechmann and Ratcliffe, 2000). In plants, TFs play crucial roles in numerous biological pathways (Wu et al., 2023). Among the eukaryotic TF gene families, the basic region-leucine zipper (bZIP) family is the most extensive and heterogeneous group [Hurst, 1995; Wang et al., 2011]. Named after its highly conserved domain of approximately 60–80 amino acids, the bZIP includes two functional regions: a basic region and a leucine zipper region (Jakoby et al., 2002). Following systematic identification and classification in Arabidopsis thaliana (Dröge-Laser et al., 2018), 78 bZIP family members were classified into 13 groups (designated A–M) based on the homology of the basic region, and additional conserved motifs were proposed (Dröge-Laser et al., 2018). To date, a comprehensive understanding of bZIP genes have been attained in several plant species, including A. thaliana (Dröge-Laser et al., 2018), Manihot esculenta (Hu et al., 2016), Cucumis sativus (Baloglu et al., 2014), Triticum aestivum (Agarwal et al., 2019), and Neoporphyra haitanensis (Wang et al., 2023).
The bZIP genes play an indispensable role in the regulation of growth, development, and abiotic stress responses (Landschulz et al., 1988). In A. thaliana, AtbZIP1 plays a role in the coordination of starvation responses triggered by darkness and NO3- signaling pathways (Para et al., 2014). AtbZIP29 is expressed in proliferating tissues (Van Leene et al., 2016), and AtbZIP59(PosF21) has been functionally implicated in auxin-mediated callus development and plant regeneration processes (Xu et al., 2018). In Capsicum annuum, CabZIP25 improves salt tolerance by preserving chlorophyll stability (Gai et al., 2020). In Nelumbo nucifera, NnbZIP36 facilitates anthocyanin accumulation (Zhou et al., 2023), while PpbZIP44 in Pyrus pyrifolia affects the sugar:acid ratio (Wang et al., 2023). In Aquilaria sinensis, both AsbZIP14 and AsbZIP41 respond to ethylene and agarwood inducers (Zhang et al., 2023). In Rosa, RcbZIP17 modulates B. cinerea resistance using both virus-induced gene silencing (VIGS) and overexpression (OE) approaches (Li et al., 2023). However, bZIP genes from red algae have not been a primary focus of investigation.
Red algae (Rhodophyta) represent a highly evolved and significant branch within eukaryotic photosynthetic organisms and constitute an independent lineage that numbers over 7000 species according to AlgaeBase (http://www.algaebase.org/), placing them among the most archaic of such groups (Lopez-Bautista, 2010). Red algae are categorized into seven distinct classes: Cyanidiophyceae, Bangiophyceae, Florideophyceae, Compsopogonophyceae, Porphyridiophyceae, Rhodellophyceae, and Stylonematophyceae, which collectively occupy diverse ecological niches ranging from hot springs and acidic sulfur vents, to freshwater systems, deep-sea abysses, and intertidal areas. Morphologically, red algae exhibit remarkable diversity, appearing as single-celled organisms, filamentous structures, leaf-like thalli, parenchymatous blades, and tubular forms composed of a single layer of cells (Yoon et al., 2010). Neopyropia yezoensis belongs to Bangiophyceae (Rhodophyta), which is the most genetically variable group of red algae and a key species in the commercial cultivation of edible marine macroalgae (Wang et al., 2016).
Ny. yezoensis possesses a complex life cycle characterized by alternating between two distinct multicellular phases: the haploid gametophytic stage and the diploid sporophytic stage. This lifecycle is crucial for the large-scale cultivation of Ny. yezoensis and its subsequent establishment and growth (Blouin et al., 2011). Ny. yezoensis inhabits the intertidal zone and exhibits remarkable stress tolerance, highlighting its adaptability to challenging environments (Burritt et al., 2002; Nishikawa et al., 2007; Pearson et al., 2010). For example, the growth, quality, and yield of Ny. yezoensis responds rapidly to the level of nitrogen in seawater (Kakinuma et al., 2008; Kakinuma et al., 2016; Li ChengZe et al., 2019). These attributes make Ny. yezoensis an ideal model organism for elucidating the evolutionary trajectory of interconnected metabolic and regulatory pathways governing growth, reproduction, and stress adaptation (Blouin et al., 2011). The high-quality chromosomal genome of Ny. yezoensis has enabled comprehensive characterization of its bZIP genes (Wang et al., 2020). Furthermore, the analysis of existing transcriptomic datasets has facilitated an in-depth investigation into the complex molecular functions performed by these bZIP genes (Sun et al., 2015; Sun et al., 2019; Tang et al., 2019).
In this study, we conducted a comprehensive analysis of the bZIP gene family in Ny. yezoensis by examining the chromosomal localization, duplication events, cis-regulatory elements, gene architecture, conserved motifs, evolutionary affiliations, expression patterns, protein interaction networks, and functional annotations using gene ontology. In addition, we utilized bioinformatic techniques to identify bZIP gene family members in 15 additional species of red algae and performed phylogenetic analyses, thereby establishing a foundation for the bZIP gene family classification within red algae. Furthermore, we examined the response of NyybZIP genes to nitrogen stress conditions and different life cycle phases, and validated their expression patterns using transcriptomic and qRT-PCRanalyses. Taken together, our results establish a framework for future functional investigations into NyybZIP genes.
2 Materials and methods
2.1 Genome-wide identification and characterization of the bZIP gene families in Ny. yezoensis and other red algae
Fifteen types of red algae and one representative species each from green algae, streptophyte algae, and bryophyte were selected for bZIP protein identification. The Cyanidioschyzon merolae, Chondrus crispus, Gracilariopsis chorda, Galdieria sulphuraria, Porphyridium purpureum, Ny. yezoensis, Porphyra umbilicalis, Chlamydomonas reinhardtii, Marchantia_polymorpha reference genome assemblies and protein sequences were obtained from GenBank using accessions GCA_000091205.1 (Nozaki et al., 2007), GCA_000350225.2 (Janouškovec et al., 2013), GCA_003194525.1 (Lee et al., 2018), GCA_000341285.1 (Schönknecht et al., 2013), GCA_008690995.1 (Lee et al., 2019), GCA_009829735.1 (Wang et al., 2020), GCA_002049455.2 (Brawley et al., 2017), GCA_000002595.3 (Merchant et al., 2007), and GCA_037833965.1 respectively. Additional red algal and Chara braunii protein sequences were downloaded from the EukProt database (https://doi.org/10.6084/m9.figshare.12417881.v3) (Richter et al., 2022). A. thaliana bZIP protein sequences were downloaded from the Pytozome database (Goodstein et al., 2012). No. haitanensis bZIP protein sequences were obtained from Wang et al (Wang et al., 2023). To obtain the bZIP protein sequences from these 15 types of red algae and three representative species of Viridiplantae. the Hidden Markov model (HMM) profile (Eddy, 2004) of the bZIP_1 (PF00170) and bZIP_2 (PF07716) domain was downloaded from the Pfam database (Browse - InterPro (ebi.ac.uk)), and potential proteins were identified using local HMMER software (E-value <1e-5). Next, a local BLASTP search (E-value <1e-5) was applied to identify possible bZIPs using A. thaliana and No. haitanensis as well as the PlnTFDB database (http://plntfdb.bio.uni-potsdam.de/v3.0/). Finally, InterProScan (http://www.ebi.ac.uk/interpro/result/InterProScan/), SMART (http://smart.embl-heidelberg.de/), and CDD (https://www.ncbi.nlm.nih.gov/cdd) databases (Marchler-Bauer et al., 2017) were used for additional screening and verification of the bZIP domain. Except for No. haitanensis, which used the name from the original manuscript (Wang et al., 2023), the naming of bZIP proteins in other red algae was based on molecular weight. The ProtParam tool (Gasteiger et al., 2003) was used to determine the molecular weight, instability index, and theoretical isoelectric point (pI) of these red algae bZIPs. The subcellular localization of Ny. yezoensis bZIPs was predicted using Cell-PLoc-2 (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/) (Chou and Shen, 2010), TBtools software (Chen et al., 2020) and the online website MEME (http://meme-suite.org/) (Bailey et al., 2006) were utilized to obtain the exon-intron structures, motifs, and visualize the chromosomal locations of the bZIP genes using Ny. yezoensis genome sequences and annotation files. The bZIP transcription factor family domains logos of Ny. yezoensis were generated using WebLogo (Crooks et al., 2004).
2.2 Phylogenetic and motif analyses of the bZIP gene families in Ny. yezoensis and other red algae
The conserved domains of bZIP protein sequences of red algae, A. thaliana, and other three representative species of Viridiplantae were imported into the MUSCLE program in the MEGA 11 software (Tamura et al., 2021). Then, the maximum likelihood (ML) phylogenetic tree was constructed using IQ-TREE (Minh et al., 2020) with the Ultrafast bootstrap (Hoang et al., 2018) set to 1000. ModelFinder (Kalyaanamoorthy et al., 2017) in IQ-TREE was then used to find the best-fit substitution model, and the tree figure was constructed using tvBOT (Xie et al., 2023). The same approach was used to construct a phylogenetic tree using bZIP protein sequences from 16 species of red algae and their conserved patterns in the bZIP gene family was determined using the online website MEME (http://meme-suite.org/) (Bailey et al., 2006).
2.3 Collinearity analysis of the bZIP gene families in Ny. yezoensis
Tandem and segmental duplication events of the red algal bZIP gene family and collinearity of the bZIP genes of Ny. yezoensis were analyzed using the One-Step MCScanX program in TBtools (Chen et al., 2020). To determine the selection pressure, the rates of nonsynonymous (Ka) and synonymous (Ks) substitutions (Zhang et al., 2006) were calculated using TBtools.
2.4 Cis-regulatory element analysis of the bZIP gene families in Ny. yezoensis
To predict the cis-acting elements present in Ny. yezoensis bZIP genes, TBtools software was used to extract the upstream 2000 bp promoter sequences of the Ny.yezoensis bZIP genes. Then, these sequences were submitted to PlantCARE (Lescot et al., 2002). Based on the annotated functions, the detected cis-acting regulatory elements were classified into different response types. A heat map was used to display the number of cis-acting regulatory elements detected.
2.5 Investigation of NyybZIP gene expression patterns at different life stages
To investigate the expression patterns of NyybZIP genes at different life stages, RNA-sequencing (RNA-Seq) data were obtained from a previous study (gametophytes and sporophytes; NCBI SRA: SRR10527930–SRR10527937). Gene expression levels were quantified as fragments per kilobase of transcript per million fragments mapped (FPKM), and expression heat maps were created using TBtools software based on log2 transformed FPKM values. Next, qRT-PCR was performed using established protocols from a previous study (Yu et al., 2020). We utilized the LightCycler® 480 Real-Time PCR System to confirm the identity and expression of the selected genes. Ubiquitin-conjugating enzyme (UBC) genes were used as a reference (Gao et al., 2018). The primer sequences are listed in Supplementary Table S1. The comparative 2^(-ΔΔCt) method was used to determine the relative gene expression levels.
2.6 Expression profile analysis of Ny. yezoensis bZIP genes under nitrogen stress
To investigate the expression patterns of NyybZIP genes under nitrogen stress, a lab-cultured pure RZ Ny. yezoensis line was used in this study. Leafy gametophytes (thalli) of the Ny. yezoensis were maintained in 2 L aerated bottles with the Provasoli’s enrichment solution (PES) medium under the following conditions: 10°C with 60 μmol photons·m−2 s−1 and a 12 h:12 h light:dark (L:D) photoperiod, until the thallus length reached 5-8 cm. Then we set up the nitrogen-deficiency experiment consisted of two treatment groups: zero-nitrogen (N0) and control (C). The NaNO3 content in the Provasoli’s enrichment solution (PES) medium was as follows: 0 μM NaNO3 (N0) and 500 μM NaNO3 (C). All other cultivation conditions remained consistent across the treatment groups, and three biological replicates were used for each treatment group. After an eight-day cultivation period, the samples were collected in liquid nitrogen then sent to the company for RNA-seq data. And used DESeq2 (Love et al., 2014) to perform RNAs differential expression analysis, we identify DEGs (Fold Change > = 2 and FDR < 0.05). In addition, total RNA was extracted using the RNeasy Plant Mini Kit (OMEGA) according to the manufacturer’s instructions. Next, approximately 1 μg of the obtained total RNA was utilized for the synthesis of first-strand cDNA using the HiScript® III RT SuperMix for qPCR (+gDNA wiper) Kit (Vazyme Biotech Co., Ltd., Nanjing, China). qRT–PCR was then performed using the primers listed in Supplementary Table S1.
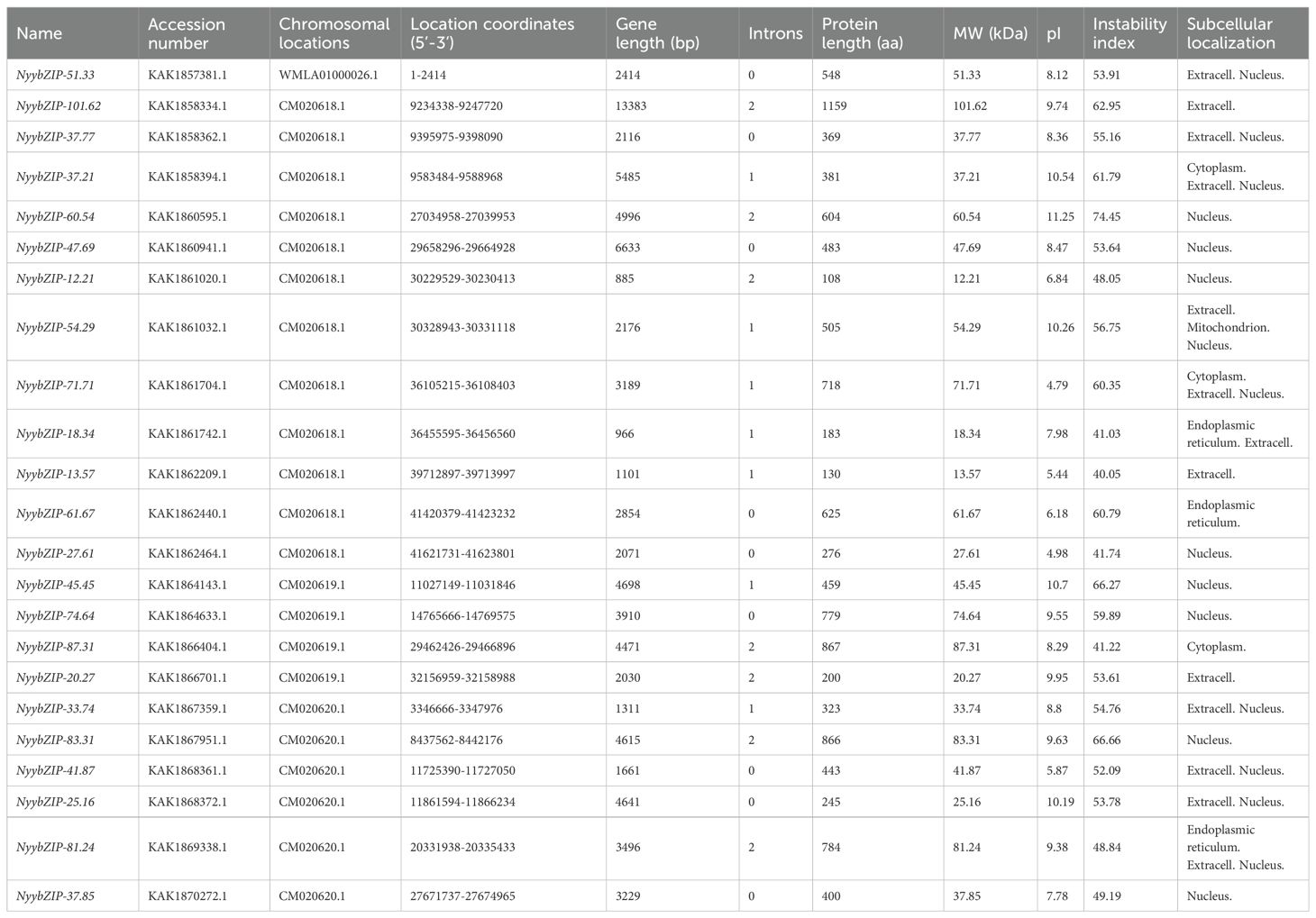
3 Results
3.1 Identification of bZIPs in Ny. yezoensis and other red algae
To identify the bZIP genes in Ny. yezoensis, HMM, BLASTP, and PlnTFDB databases (http://plntfdb.bio.uni-potsdam.de/v3.0/) were used to identify and authenticate bZIP protein sequences in Ny. yezoensis. Next, Pfam, SMART, and CDD were used to determine the integrity of the bZIP domain. The amino acid positions of the conserved structural domains of bZIP were visualized by multiple sequence alignment of the protein sequences in Ny yezoensis bZIP family members (Figure 1). Evidence indicated that the bZIP domain consisted of a basic DNA-binding region and an adjacent leucine zipper structure. As shown in Figure 1, the basic DNA-binding region had an invariable N-X7-R/N motif and the adjacent leucine zipper structure created an amphipathic helix with heptapeptide repeat of Leucine (L) or related hydrophobic amino acid. Methionine, alanine, valine, etc. can replace highly conserved leucine residues in some cases. Such results of Ny. yezoensis bZIPs were consistent with those of previous studies in other plants (Jakoby et al., 2002; Dröge-Laser et al., 2018; Zhao et al., 2021). In total, 23 non-redundant genes were determined to be Ny. yezoensis bZIP genes and were named according to their molecular weight (Table 1). The following ranges were observed: gene size: 885–13383 bps; the number of introns: 0–2; number of amino acids residues: 108– 1159; isoelectric point (pI): 4.79–11.25; molecular weight (MW): 12.21–101.62 kDa; and instability index: 40.05–74.45. To investigate the functions and diversification of bZIP protein sequences in Ny yezoensis, we used the MEME software to predict their conserved motifs. Among them, motif1 was identified as the bZIP domain, which is present in all 23 NyybZIPs, whereas the other motifs lack specific annotation information (Figure 2A). Of the 23 NyybZIP genes identified, nine were intron-less, seven possess one intron, and seven possess two introns (Table 1; Figure 2A). Based on their chromosomal locations, 22 NyybZIP genes exhibited uneven distribution across the three nori chromosomes, and only one NyybZIP gene was mapped to WMLA01000026.1 (Figure 2B). Specifically, CM020618.1, CM020619.1, and CM020620.1 contained twelve, four, and six NyybZIP genes, respectively. Protein information of the bZIP genes in the other 14 red algae species is detailed in Supplementary Table S2.
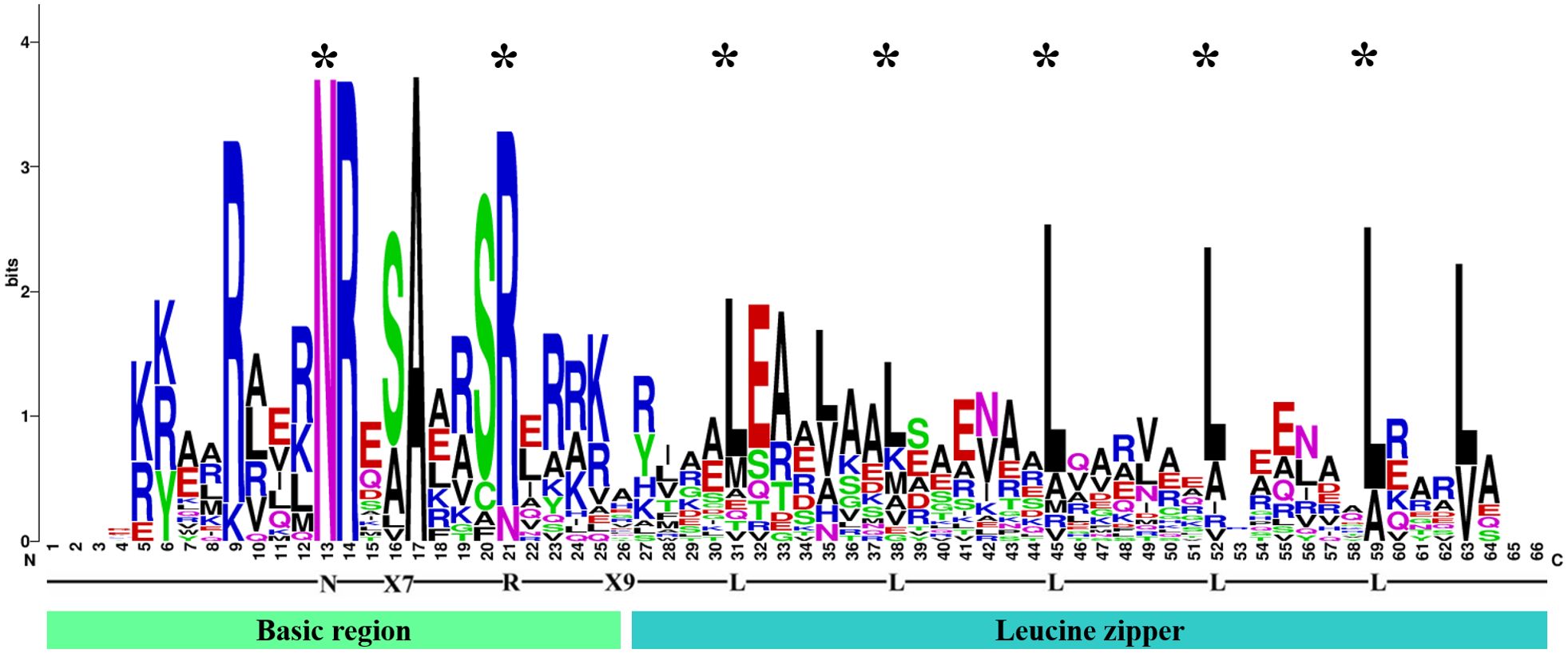
Figure 1. Visualization of multiple sequence alignment of the Ny yezoensis basic leucine zipper transcription factor family domains. The total height of the letter piles at each position indicates the conservation of the sequence at that position (measured in bits). The height of a single letter in the letter piles represents the relative frequency of the corresponding amino acid at that position. The symbol * means the conserved site.
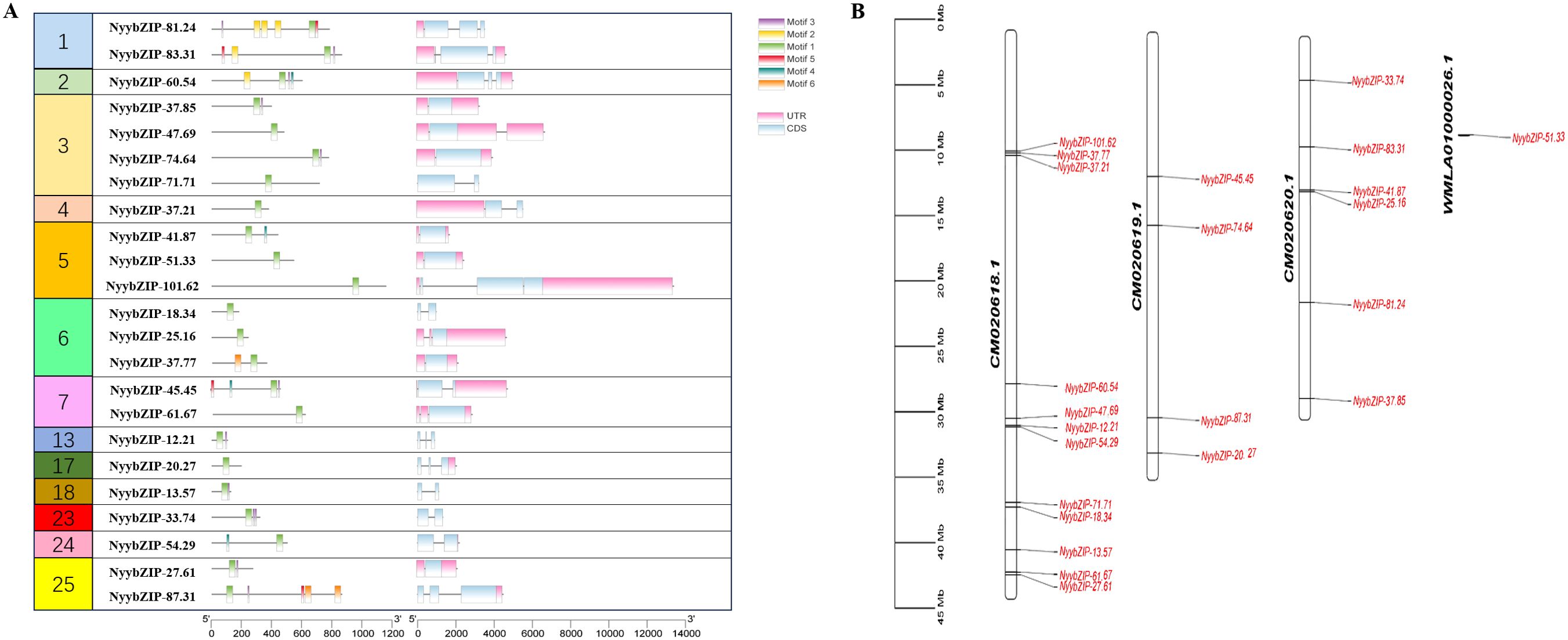
Figure 2. (A) Motifs and gene structures of the NyybZIP genes. (B) Chromosome distribution of the Ny. yezoensis bZIP genes. UTR, untranslated region; CDS, coding sequence.
3.2 Phylogenetic and motif analyses of Ny. yezoensis and other red algae bZIP gene families
To explore the evolutionary and classification relationships of the red algae bZIP families, we constructed a Maximum Likelihood (ML) phylogenetic tree using the bZIP domains of 16 types of red algae and 4 representative species of Viridiplantae bZIP protein sequences (Supplementary Figure S1). We found that bZIP protein sequences of bryophyte, green algae, and streptophyte algae could cluster with A. thaliana, and only a small portion of the red algae bZIP protein sequences could be classified into the A. thaliana bZIP groups; the majority of the red algae bZIP sequences formed distinct group, which we designated as Group R1 and R2. Among the NyybZIP protein sequences, only NyybZIP-18.34 and NyybZIP-20.27 were present in group M and classified with A. thaliana, whereas the other 21 sequences were distributed within Group R1 and R2. Therefore, we constructed a new phylogenetic tree using the complete sequences of bZIP proteins from 16 different species of red algae, which were classified based on the evolutionary relationships displayed in the constructed phylogenetic tree as well as the motif structures present (Figure 3). The bZIP proteins were divided into 30 groups (1–30), and phylogenetic analysis showed that NyybZIP-81.24 and NyybZIP-83.31 were in group 1 with motif 11; NyybZIP-60.54 was in group 2; NyybZIP-71.71, NyybZIP-47.69, and NyybZIP-74.64 were in group 3; NyybZIP-37.21 was in group 4; NyybZIP-41.87, NyybZIP-101.62, and NyybZIP-51.33 were in group 5; NyybZIP-18.34, NyybZIP-37.77, and NyybZIP-25.16 were in group 6; NyybZIP-45.45 and NyybZIP-61.67 were in group 7; NyybZIP-12.21 was in group 13; NyybZIP-20.27 was in group 17; NyybZIP-13.57 was in group 18; NyybZIP-33.74 was in group 23; NyybZIP-54.29 was in group 24; and NyybZIP-87.31 and NyybZIP-27.61 were in group 25.
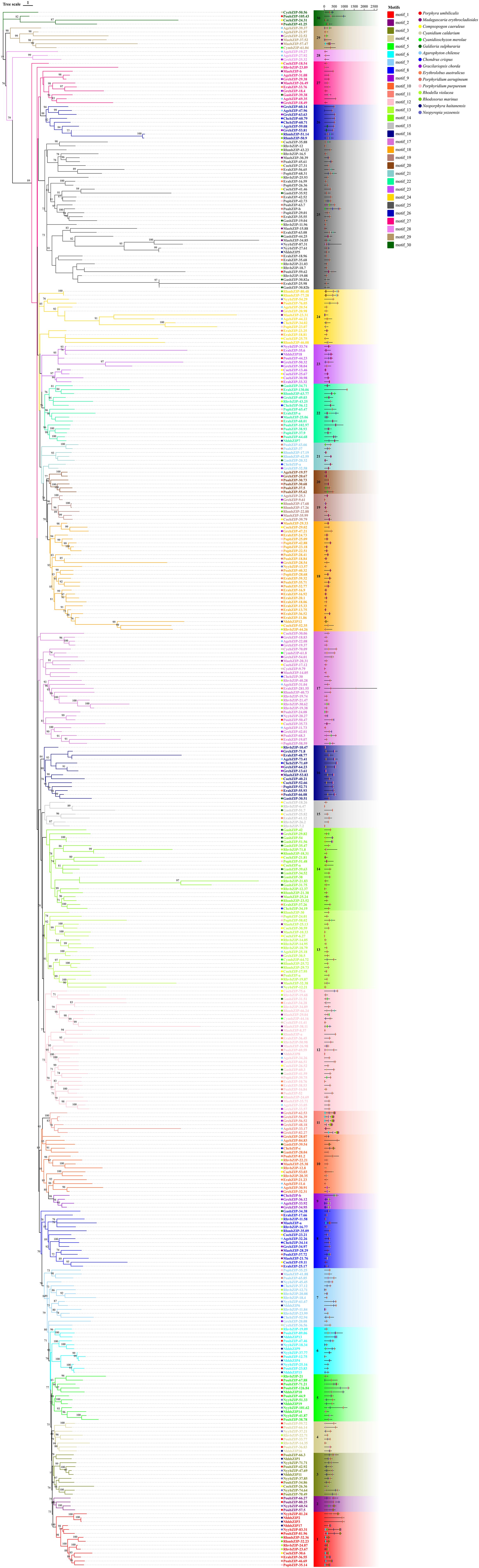
Figure 3. Phylogenetic relationship and conserved motifs of the bZIP proteins from 16 species of red algae. Different motifs are represented by different colored boxes. Conserved motifs were scanned with MEME.
In addition, phylogenetic analysis showed that motif 1 was the basic DNA-binding region of bZIP domain, motif 3 and motif 4 were the common Leucine zipper structure of bZIP domain found in red algae, and other motifs such as motifs 10, 15, and 16 were frequently observed in red algae bZIP protein sequences. Notably, due to their unique structural characteristics, motifs 11, 12, 13, 14, and 17 were employed as discriminative elements in the phylogenetic classification of red algae bZIP protein sequences (Figure 4). Many motifs exist in specific groups, which might be related to specific biological functions. Throughout the first evolutionary tree (Supplementary Figure S1), NyybZIPs did not cluster with the AtbZIP members, the similarity of bZIP proteins between Ny. yezoensis and A. thaliana showed low amino acid conservation which may be due to their relatively distant evolutionary relationship. In the second evolutionary tree (Figure 3), the bZIP genes of Ny. yezoensis showed good similarity with that of the other 15 red algae. And NyybZIPs clustered together with PoubZIPs and NhhbZIPs, which is consistent with the evolutionary relationships of red algae species.
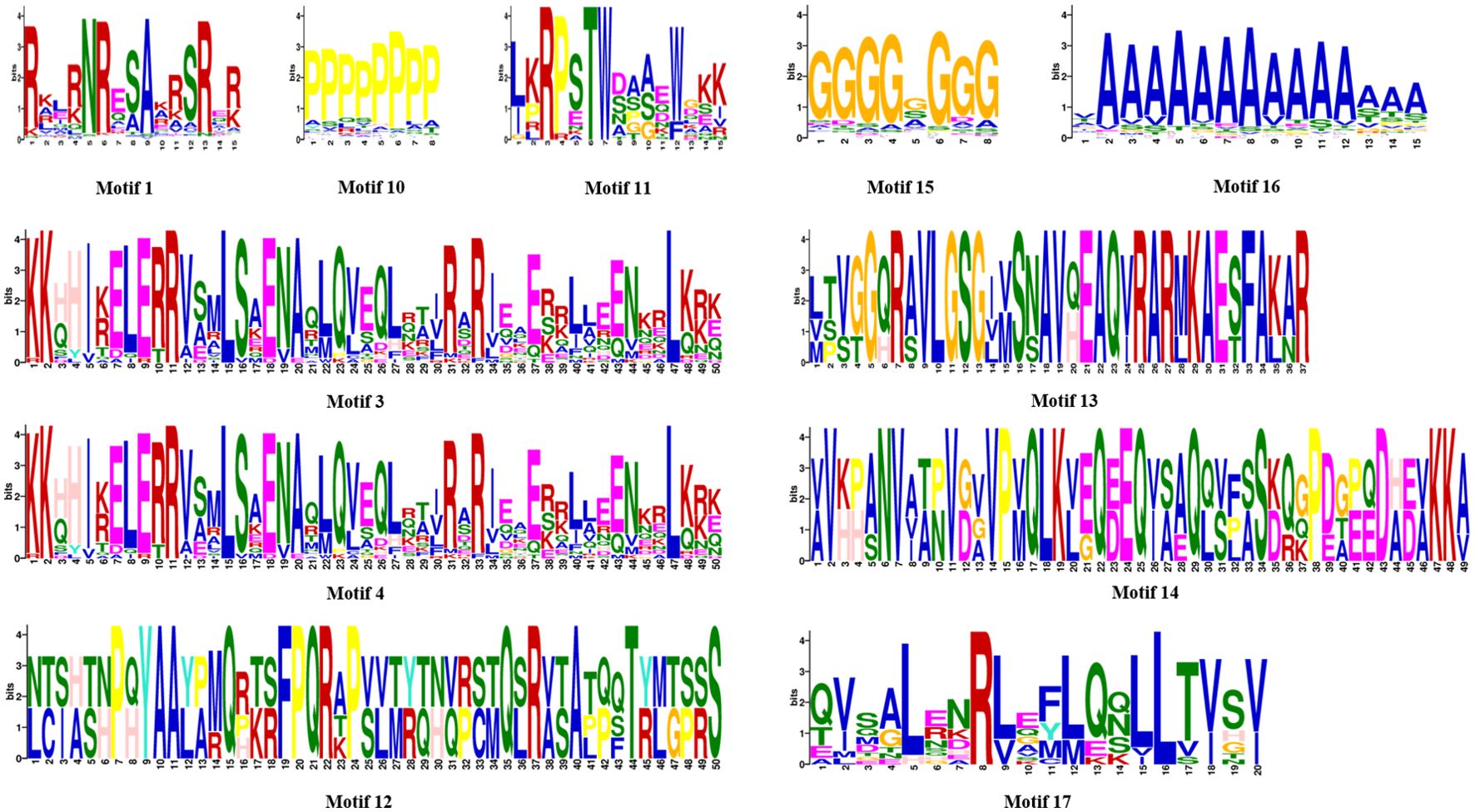
Figure 4. Discriminative motifs in the phylogenetic classification of Ny. yezoensis bZIP protein sequences.
3.3 Collinearity analysis of the Ny. yezoensis bZIP gene families
The One-Step MCScanX module within TBtools was used to identify and analyze tandem and segmental duplication events. And no tandem and segmental duplication events were detected in the bZIP gene family members of all these red algae species. Gene collinearity analysis was carried out between Ny. yezoensis and the other red algae with gene files and annotation files, the results showed that only the bZIP genes of No. haitanensis and P. umbilicalis have collinearity with the ones of Ny. yezoensis (Figures 5A, B). Twenty and eight NyybZIP genes were orthologous to the bZIP genes in No. haitanensis and P. umbilicalis, respectively. The ratio between the non-synonymous (Ka) and synonymous (Ks) has been widely used as an index for measuring the strength and direction of selection pressure. Ka/Ks > 1 means positive selection, Ka/Ks < 1 means purifying selection, and Ka/Ks = 1 means neutral selection. We found that all identified collinear gene pairs exhibited Ka/Ks ratio values well below one (Supplementary Table S3), confirming strong purifying selection during the evolution of the bZIP gene family in the Bangiaceae family. This observation aligns with our findings from the unrooted phylogenetic tree (Figure 3), suggesting that these genes may play important roles in the evolution of the red algae bZIP gene family.
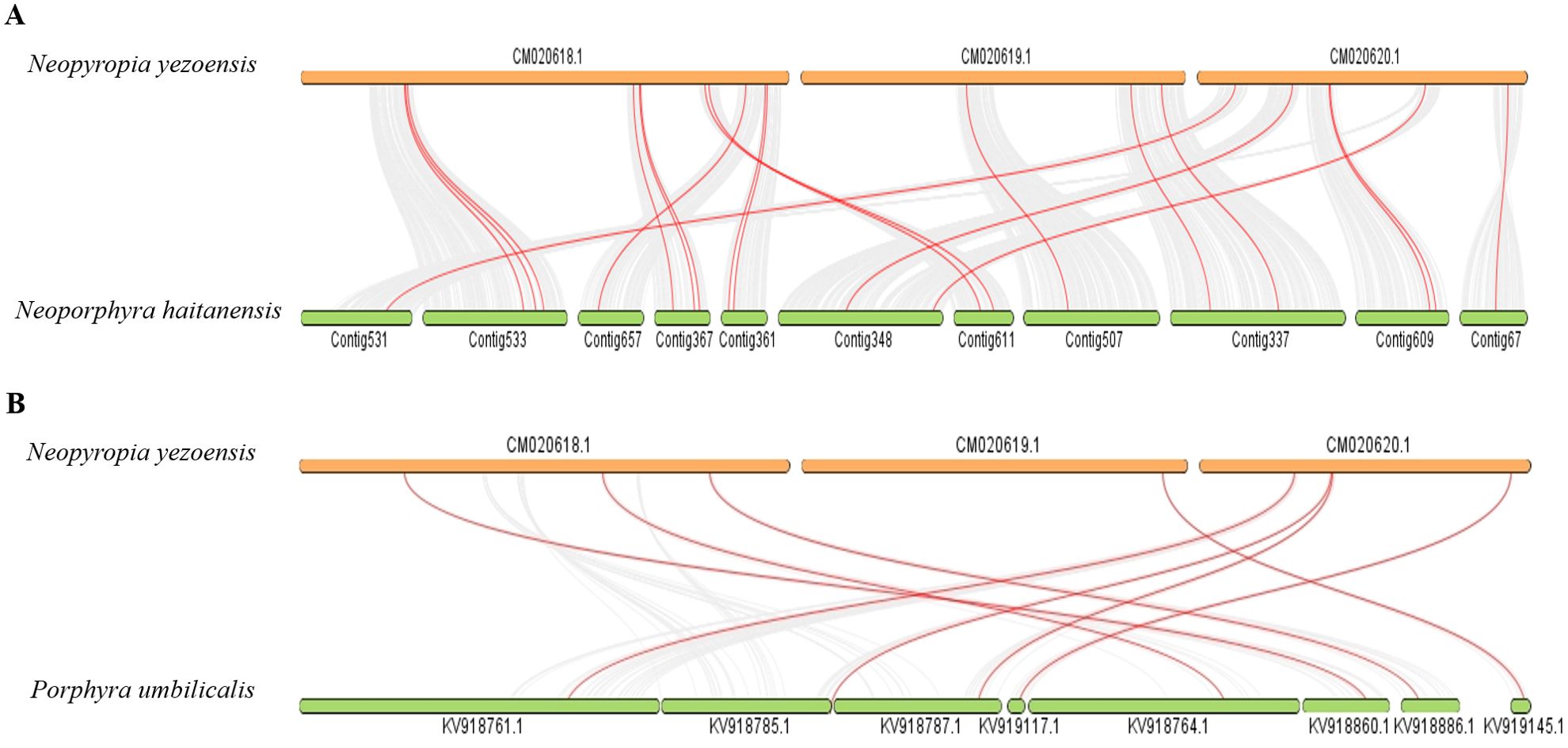
Figure 5. (A) Synteny analysis of the bZIP genes between Ny. yezoensis and No. haitanensis. (B) Synteny analysis of the bZIP genes between Ny. yezoensis and P. umbilicalis. The gray lines in the background indicate collinear blocks within the three red algal genomes. The red lines highlight syntenic bZIP gene pairs.
3.4 Cis-regulatory element analysis of the Ny. yezoensis bZIP gene families
Cis-regulatory elements play critical roles in the transcriptional regulation of gene expression and control various biological processes (Yamaguchi-Shinozaki and Shinozaki, 2005). To further explore the regulatory mechanisms of the bZIP genes in Ny. yezoensis growth, development, and stress responses, we analyzed cis-elements within the promoter sequences (2 kb) of 23 Ny. yezoensis bZIP genes by the PlantCARE database. Our analysis identified four main categories of cis-elements: development-related, light-responsive, phytohormone-responsive, and stress-responsive (Figure 6). Among these categories, phytohormone-responsive elements were the most abundant (41.77%), and included ABRE (involved in abscisic acid responsiveness), AuxRR-core and the TGA-element (involved in auxin responsiveness), CGTCA- and TGACG-motifs (involved in MeJA-responsiveness), and the GARE-motif and P-box (involved in gibberellin-responsiveness). Light-responsive elements were the second most abundant (36.38%), followed by the stress-responsive elements (17.18%). Stress-responsive elements included ARE (essential for anaerobic induction), the GC-motif (involved in anoxic-specific inducibility), LTR (involved in low-temperature responsiveness), MBS (involved in drought-inducibility), and TC-rich repeats (involved in defense and stress responsiveness). Development-related elements (4.66%) were the least abundant and included circadian (involved in circadian control), CAT-box (related to meristem expression), MSA-like (involved in cell cycle regulation), and O2-site (cis-acting regulatory element involved in zein metabolism regulation). Overall, analysis of cis-acting elements showed that NyybZIP genes have diverse functions in the transcriptional regulation of gene expression and control various biological processes.
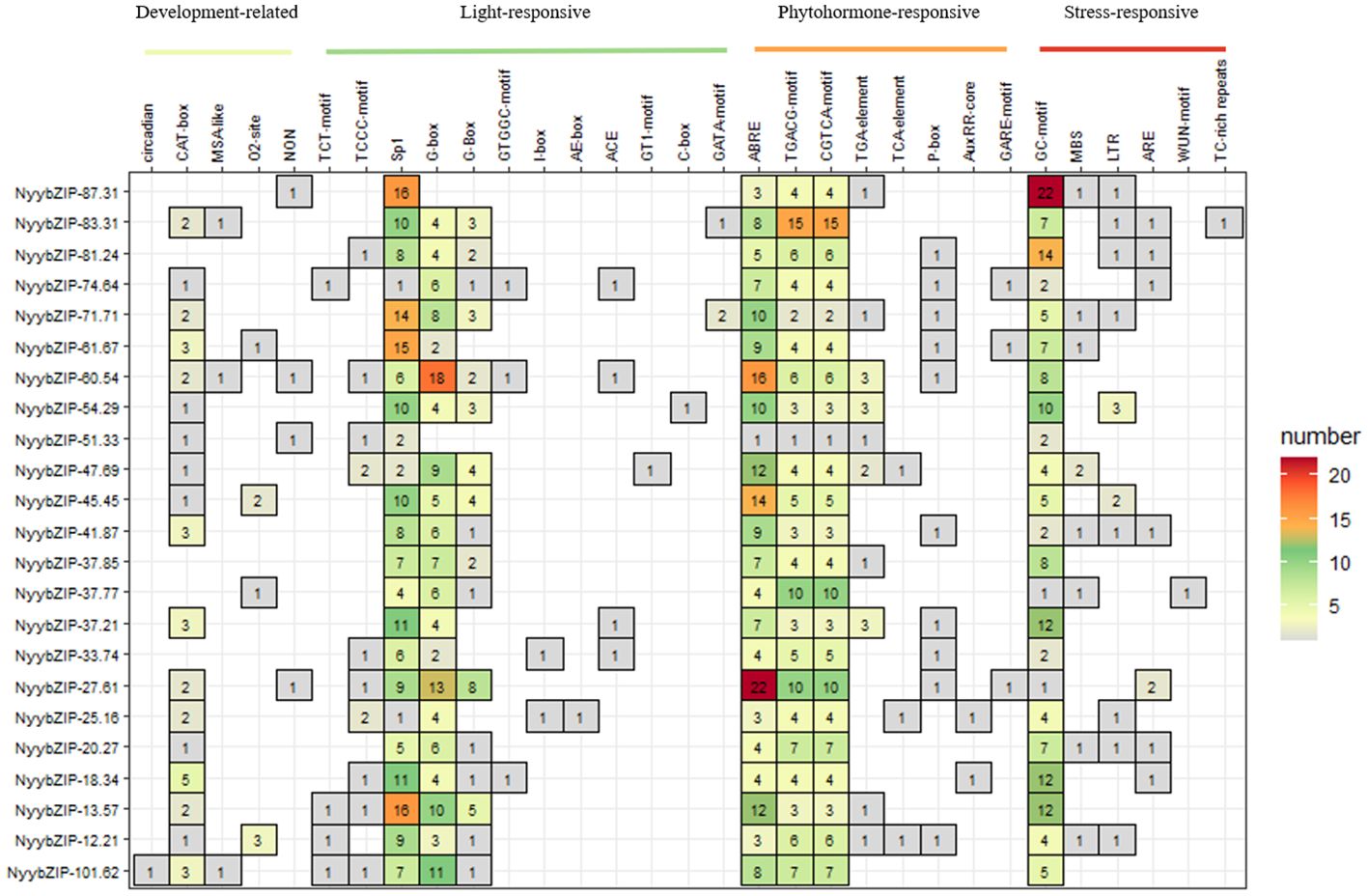
Figure 6. Analysis of 2 kb upstream cis-acting elements found in red algae bZIP genes. The different colors and numbers of the grid indicate the numbers of different promoter elements.
3.5 NyybZIP gene expression patterns at different life stages
Ny. yezoensis exhibits a sophisticated life cycle characterized by recurrent alternation between two distinct multicellular phases: gametophytic haploid blades (GAM) and sporophytic diploid filaments (SPO). Figure 7A illustrates the expression patterns of the 23 NyybZIP genes across these two life cycle stages. NyybZIP genes were expressed in both the sporophyte and gametophyte stages and demonstrated different expression levels. NyybZIP-41.87, NyybZIP-47.69, NyybZIP-37.21, NyybZIP-37.85, NyybZIP-33.74, NyybZIP-18.34, NyybZIP-60.54, and NyybZIP-61.67 exhibited higher expression levels in gametophytes than in sporophytes. In contrast, other NyybZIP genes exhibited lower expression levels in gametophytes than in sporophytes. These findings indicate distinct expression of bZIP genes in Ny. yezoensis across different stages of growth and development, suggesting specific regulatory roles where genes highly expressed in one stage may undergo significant changes in expression levels during another phase. These observations suggest that various bZIP genes play critical roles in the growth and development of Ny. yezoensis throughout its life cycle stages.
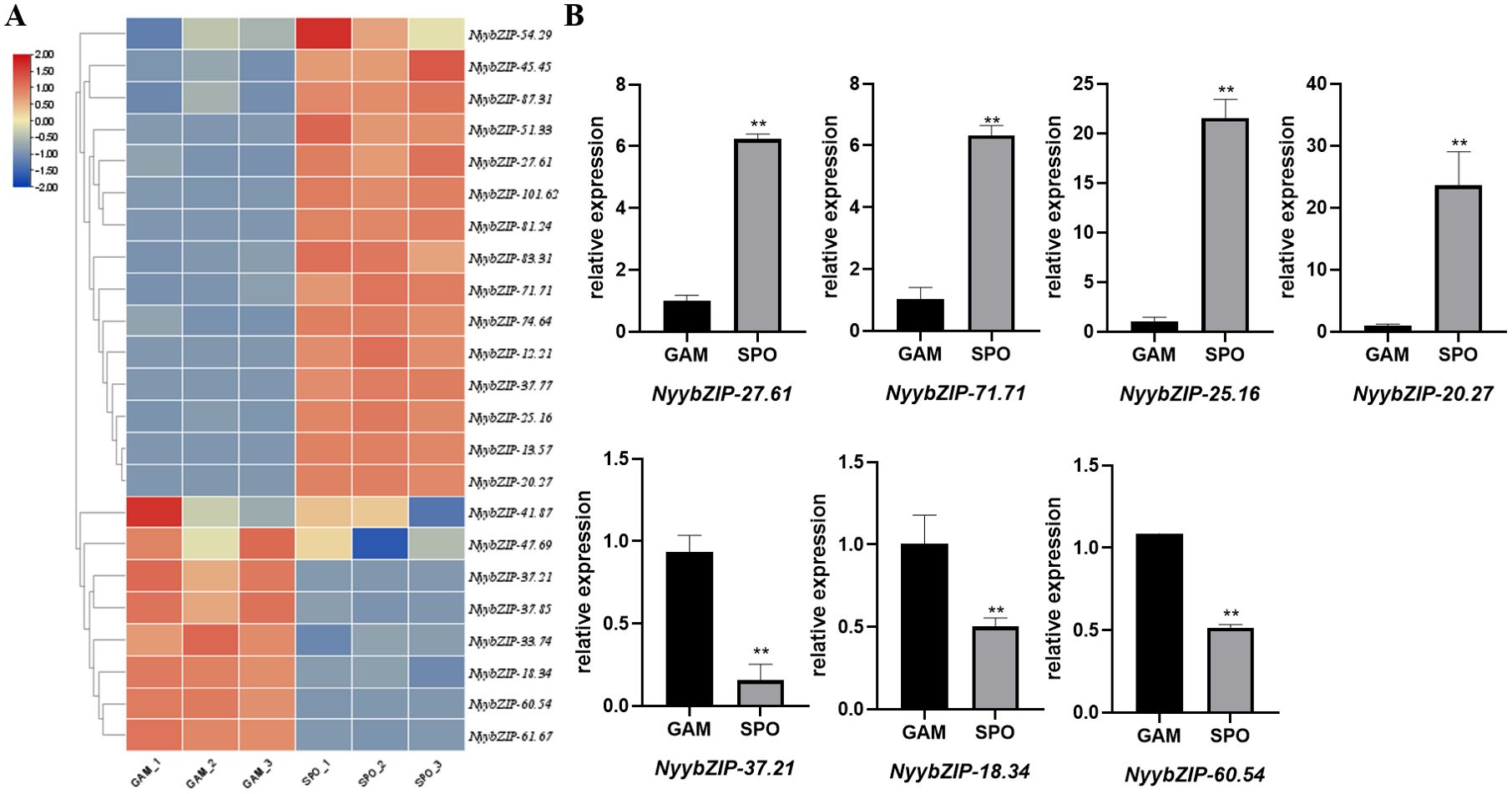
Figure 7. (A) NyybZIP gene expression patterns at different life stages. (B) Expression analysis of seven representative genes from the Ny. yezoensis bZIP family at different life stages. GAM, gametophytic haploid blades; SPO, sporophytic diploid filaments; **indicates a significant difference of p<0.01.
Next, we designed primers for seven NyybZIPs and performed quantitative real-time polymerase chain reaction (qRT–PCR) analysis (Figure 7B). Nyy-27.81, Nyy-71.71, Nyy-25.16, and Nyy-20.27 were highly expressed in SPO and exhibited lower levels in GAM. Conversely, Nyy-37.21, Nyy-18.34, and Nyy-60.54 exhibited low expression in SPO but were highly expressed in GAM. The qRT-PCR results validate the authenticity and reliability of the NyybZIP RNA-seq data in the two life cycle stages and provide a foundation for further investigation into the regulatory mechanisms underlying the differential expression patterns of bZIP genes in Ny. yezoensis at different life stages.
3.6 NyybZIP gene expression profile analysis under nitrogen stress
To explore the impact of nitrogen stress on Ny. yezoensis, we investigated the differential expression patterns of NyybZIP family members in response to abiotic stress. Based on the RNA-seq data from nitrogen stress treatments, we identified a subset of NyybZIP genes that exhibited differential expression levels. A heat map was constructed to visualize these findings (Figure 8A). Most of the NyybZIP genes exhibited higher expression levels under low nitrogen stress, with NyybZIP-54.29, NyybZIP-83.31, NyybZIP-101.62, NyybZIP-61.67, NyybZIP-41.87, NyybZIP-18.34, NyybZIP-33.74, NyybZIP-60.54, NyybZIP-71.71, and NyybZIP-20.27 demonstrating higher expression levels in the N0 group compared to those of the C group. However, NyybZIP-47.69 and NyybZIP-25.16 had lower expression levels in the N0 group compared to those in the C group. These results suggest that NyybZIP genes can exhibit responsive expression under nitrogen stress, which is consistent with observations in previous studies on A. thaliana (Sun et al., 2012; Alvarez et al., 2014).
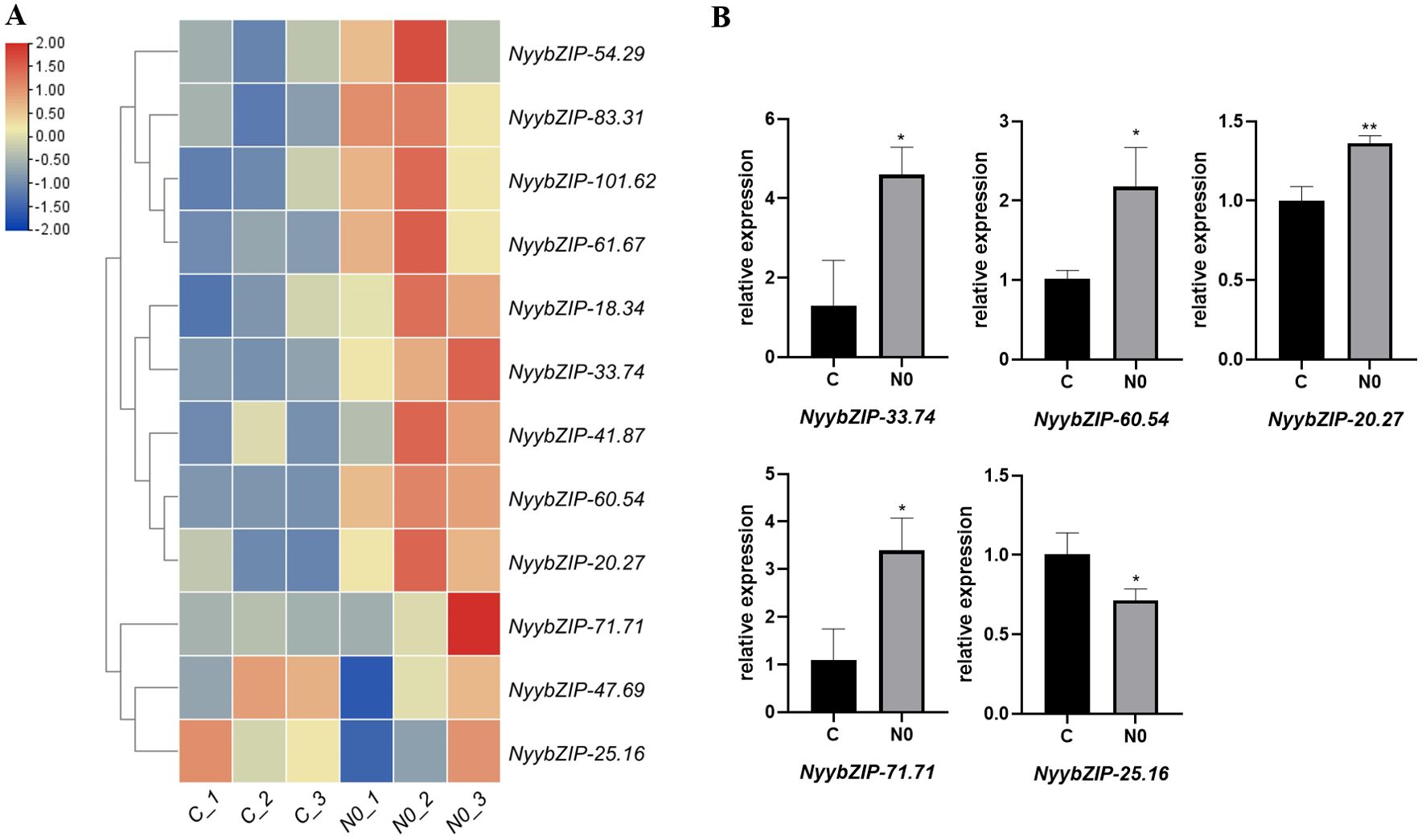
Figure 8. (A) The significant differential expression patterns of select NyybZIP genes under nitrogen stress. (B) Expression analysis of five representative genes from the Ny. yezoensis bZIP family under nitrogen stress. N0, zero nitrogen group; C, control group. ** and * indicate significant differences of p<0.01 and p<0.05, respectively.
Next, we designed primers for five representative NyybZIPs and conducted qRT-PCR analysis (Figure 8B). Our results revealed that Nyy-33.74, Nyy-60.54, Nyy-20.27, and Nyy-71.71 were highly expressed in the N0 group and exhibited lower expression levels in the C group. However, Nyy-25.16 exhibited low expression in the N0 group but was highly expressed in the C group. The qRT-PCR results validated the RNA-seq data, suggesting that under low nitrogen stress conditions, NyybZIP genes can respond to stress by altering expression levels. These results provide a foundation for further investigations into the potential roles of these genes under nitrogen stress and their associated functions.
4 Discussion
4.1 Identification and characterization of the Ny. yezoensis bZIP gene families
Ny. yezoensis, a key species in the commercial cultivation of edible marine macroalgae, has a sophisticated life cycle (Blouin et al., 2011). Like other plants, Ny. yezoensis is subjected to abiotic stresses, including nitrogen. The bZIP genes play an important role in the regulation of growth, development, and abiotic stress responses (Landschulz et al., 1988). The number of bZIP family members varies among different plant species, including 78 members in A. thaliana (Dröge-Laser et al., 2018), 19 in No. haitanensis (Wang et al., 2023), and 92 in rice (Corrêa et al., 2008). Despite extensive research on bZIP families in numerous plant species, relatively few studies have characterized bZIP families in red algae. From previous research in No. haitanensis (Wang et al., 2023), we found that the bZIP protein sequences of some red algae did not fit well into the classification established for A. thaliana (Dröge-Laser et al., 2018), a considerable fraction of them cannot be classified effectively. Moreover, the red algae species selected in the article are limited. So, in the present study, we selected the annotation results, gene sequence and protein sequence uploaded by Ny. yezoensis in NCBI as the data support, and gathered protein information from NCBI and EukProt (Brawley et al., 2017) from 14 representative species covering all seven classes of Rhodophyta. We identified 23 bZIP family members in Ny. yezoensis and found that the number of bZIP family members in these red algae ranged from 4 to 45. This count is notably lower when compared to A. thaliana and other plants, which is consistent with previous research (Bhattacharya et al., 2018). We observed no direct correlation between the size of red algal genomes and the number of bZIP family members.
In our analysis of bZIP family genes in Ny. yezoensis, we investigated the chromosome locations, number of introns, protein lengths, molecular weights, pI values, instability indices, and subcellular localizations. Our findings revealed that the number of introns ranged from 0 to 2, and the gene structure was relatively simple, which is consistent with observations in Malus domestica (Zhao et al., 2016). The presence of absence of introns has been linked to the upregulation of genes in response to diverse stressors (Jeffares et al., 2008), suggesting that the intron number plays a significant role in the diversification within multigene families (Wang et al., 2015). Genes with fewer introns are often associated with rapid activation in response to various stress conditions (Jeffares et al., 2008). The instability index estimates protein stability as a predictive measure, where values less than 40 generally indicate stable proteins (Gasteiger et al., 2005). Interestingly, our results showed that the instability indices of all Ny. yezoensis bZIPs were greater than 40, indicating that they were unstable proteins. This instability is regarded as a universal characteristic of stress-responsive proteins (Gao et al., 2022). However, the subcellular localization of proteins is closely related to their function, and proteins can only perform their proper functions at specific subcellular sites (Wang et al., 2014). Taken together, our results provide a reference for further investigation into the functional aspects of the bZIP protein family in Ny. yezoensis.
4.2 Phylogenetic classification of red algae bZIPs
We also investigated red algae bZIPs to classify and analyze the origin of their gene families. In T. aestivum, Glycine max and other plants, classification standards often use A. thaliana bZIP proteins based on the homology of their basic region, and additional conserved motifs have been proposed (Dröge-Laser et al., 2018). However, research on bZIP proteins in No. haitanensis (Wang et al., 2023) and our study found that most sequences of red algae bZIPs did not align with the classification system based on A. thaliana. This situation may be due to the early separation of the red algae with Viridiplantae. Thus, we classified bZIP protein sequences from 16 species of red algae into 30 groups using phylogenetic and motif analyses and provides a basis for the classification of Ny. yezoensis. However, owing to the lack of functional studies related to bZIP proteins in red algae, we can only conduct theoretical classification based on the currently available red algae protein sequence data. Nonetheless, this study can serve as a reference for future research on bZIP proteins in red algae and contribute to the investigation of their functions and roles.
4.3 Gene replication and collinearity analyses of the Ny. yezoensis bZIPs
Research indicates that genomic and gene duplication events play significant roles in plant evolution (Flagel and Wendel, 2009). Tandem and segmental duplications have also been frequently observed in the bZIP family genes within both M. domestica (Wang et al., 2021) and P. pyrifolia (Ma et al., 2021), and have contributed to the expansion and diversification of gene families. However, in Ny. yezoensis and other red algae, tandem duplications and segmental duplications were not detected, which is consistent with studies conducted on the red algae species No. haitanensis (Wang et al., 2023). Furthermore, we analyzed the collinearity between the bZIP genes of Ny. yezoensis and other red algae. The results proved that the bZIP genes of Ny. yezoensis had collinear gene pairs with No. haitanensis and P. umbilicalis. These three species had relatively close evolutionary relationships. And the calculated values of the Ka/Ks ratio for such gene pairs were less than 1, indicating that the bZIP gene family in these species likely underwent strong purifying selection pressure during evolution.
4.4 Expression and potential functions of the Ny. yezoensis bZIP gene families
Cis-regulatory elements play critical roles in the transcriptional regulation of gene expression and control various biological processes (Yamaguchi-Shinozaki and Shinozaki, 2005), making them essential tools for exploring the functions of bZIP proteins across numerous species (Hsieh et al., 2010; Wang et al., 2017). For example, under ABA and abiotic stress treatments, A. thaliana and Solanum lycopersicum plants exhibit overexpression of SlAREB, which regulates genes associated with stress responses, and overexpression of ABP9 in cotton results in increased sensitivity to exogenous ABA at seed germination, root growth, stomatal aperture, and stomatal density. All plant genes using the ABRE system require bZIP proteins for expression (Liao et al., 2008). In our study, ABRE was identified in all of the NyybZIP gene promoter sequences, suggesting that ABRE-mediated regulation may contribute to the involvement of NyybZIP genes in response to nitrogen stress. Previous articles in A. thaliana found that different bZIP genes affected the growth and development of different tissues (Murmu et al., 2010; Maier et al., 2011; Iglesias-Fernández et al., 2013). And in this study, we discovered that all NyybZIP genes displayed distinct expression levels at different life stages. So, we thought that these genes are involved in Ny. yezoensis growth and development.
Research has shown that nitrogen deficiency had a great impact on plant yield and quality. At the same time, nitrogen played an equally important role in Ny. yezoensis. When the cultivation area was short of nitrogen, severe thallus discoloration (“iroochi”) would occur, resulting in lower quality of Ny. yezoensis (Oyama et al., 2008). Based on A. thaliana research, bZIP protein sequences have been shown to play significant roles in nitrate stress, such as nitrate uptake, and nitrate responses (Sun et al., 2012). In A. thaliana roots, bZIP transcription factor mRNAs accumulated strongly after nitrate treatment, and regulated the expression of nitrate transporter genes NRT2.1 and NRT2.2 (Alvarez et al., 2014). Therefore, bZIP transcription factor played an important role in nitrogen absorption and transport Additional plant studies have demonstrated that multiple bZIP transcription factors are associated with nitrogen stress responses. For example, in G. max, the overexpression of GmbZIP44, GmbZIP62, and GmbZIP78 can notably increase nitrogen stress tolerance and they were negative regulators of ABA signaling and function in stress (Liao et al., 2008). In T. aestivum, lower the expression of TabZIP60 by RNA interference can increase NADH-dependent glutamate synthase (NADH-GOGAT) activity, N uptake and so on, while overexpression of TabZIP60 had the opposite effects (Yang et al., 2019). However, the role of bZIP sequences in Ny. yezoensis needs to be further explored. In the present study, under nitrogen deficiency conditions, there were differences in gene expression levels, which was consistent with previous studies in A. thaliana and other plants. So, we thought that NyybZIP genes may play a significant role in nitrogen deficiency response of Ny. yezoensis. However, additional studies are required to elucidate the roles of specific genes.
5 Conclusions
In the present study, we identified 23 bZIP genes in Ny. yezoensis and performed a comprehensive analysis of NyybZIP genes based on gene structures, conserved motifs, phylogenetic relationships and so on. Then, they were identified and sorted into 13 out of 30 groups, which were classified based on the bZIPs of Ny. yezoensis and 15 other red algae species. Phylogenetic analysis revealed that bZIP genes may have a complex evolutionary pattern in red algae. In addition, cross-species collinearity analysis indicated that the bZIP genes in Ny. yezoensis, No. haitanensis and P. umbilicalis are highly evolutionarily conserved. Finally, we identified four main categories of cis-elements, including development-related, light-responsive, phytohormone-responsive and stress-responsive promoter sequences in NyybZIP genes and also explored the expression profiles of NyybZIP genes at different life stages and under nitrogen stress using RNA-seq data and performed qRT–PCR analysis of these genes. This study aids in developing a more thorough understanding of NyybZIP genes and creates a foundation for future functional characterization efforts. In addition, our study provides new insights into the classification of bZIP gene families in red algae and contributes to our understanding of the adaptability of Ny. yezoensis to nitrogen stress and the significant differences in gametophytic and sporophytic stages.
Data availability statement
To investigate the expression patterns of NyybZIP genes under nitrogen stress, the transcriptome data has deposited in the SRA database of NCBl (accession number: SRR30899373– SRR30899378).
Author contributions
XZ: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Resources, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. TG: Writing – review & editing, Resources, Methodology, Conceptualization. KB: Writing – review & editing, Visualization, Investigation, Data curation. CM: Writing – review & editing, Software, Methodology, Investigation. XT: Writing – review & editing, Visualization, Software, Resources, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. YM: Writing – review & editing, Supervision, Resources, Methodology, Conceptualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (Grant No.42276148) and National Key R&D Program of China (Nos.2023YFD2400105).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1461922/full#supplementary-material
Supplementary Figure S1 | A Maximum Likelihood (ML) phylogenetic tree using the bZIP domains of 16 types of red algae and 4 representative species of Viridiplantae.
References
Agarwal, P., Baranwal, V. K., Khurana, P. (2019). Genome-wide analysis of bZIP transcription factors in wheat and functional characterization of a TabZIP under abiotic stress. Sci. Rep. 9, 4608. doi: 10.1038/s41598-019-40659-7
Alvarez, J. M., Riveras, E., Vidal, E. A., Gras, D. E., Contreras-López, O., Tamayo, K.P., et al. (2014). Systems approach identifies TGA 1 and TGA 4 transcription factors as important regulatory components of the nitrate response of A rabidopsis thaliana roots. Plant J. 80, 1–13. doi: 10.1111/tpj.2014.80.issue-1
Bailey, T. L., Williams, N., Misleh, C., Li, W. W. (2006). MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–W373. doi: 10.1093/nar/gkl198
Baloglu, M. C., Eldem, V., Hajyzadeh, M., Unver, T. (2014). Genome-wide analysis of the bZIP transcription factors in cucumber. PloS One 9, e96014. doi: 10.1371/journal.pone.0096014
Bhattacharya, D., Qiu, H., Lee, J. M., Lee, J. M., Yoon, H. S., Weber, A. P. M., et al. (2018). When less is more: red algae as models for studying gene loss and genome evolution in eukaryotes. Crit. Rev. Plant Sci. 37, 81–99. doi: 10.1080/07352689.2018.1482364
Blouin, N. A., Brodie, J. A., Grossman, A. C., Xu, P., Brawley, S. H. (2011). Porphyra: a marine crop shaped by stress. Trends Plant Sci. 16, 29–37. doi: 10.1016/j.tplants.2010.10.004
Brawley, S. H., Blouin, N. A., Ficko-Blean, E., Prochnik, S. E. (2017). Insights into the red algae and eukaryotic evolution from the genome of Porphyra umbilicalis (Bangiophyceae, Rhodophyta). Proc. Natl. Acad. Sci. 114, E6361–E6370. doi: 10.1073/pnas.1703088114
Burritt, D. J., Larkindale, J., Hurd, C. L. (2002). Antioxidant metabolism in the intertidal red seaweed Stictosiphonia arbuscula following desiccation. Planta 215, 829–838. doi: 10.1007/s00425-002-0805-6
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chou, K. C., Shen, H. B. (2010). Cell-PLoc 2.0: an improved package of web-servers for predicting subcellular localization of proteins in various organisms. Natural Sci. 2, 1090. doi: 10.4236/ns.2010.210136
Corrêa, L. G. G., Riaño-Pachón, D. M., Schrago, C. G., Santos, R. V., Mueller-Roeber, B., Vincentz, M. (2008). The role of bZIP transcription factors in green plant evolution: adaptive features emerging from four founder genes. PloS One 3, e2944. doi: 10.1371/journal.pone.0002944
Crooks, G. E., Hon, G., Chandonia, J. M., Brennr, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Dröge-Laser, W., Snoek, B. L., Snel, B., Weiste, C. (2018). The Arabidopsis bZIP transcription factor family—an update. Curr. Opin. Plant Biol. 45, 36–49. doi: 10.1016/j.pbi.2018.05.001
Eddy, S. R. (2004). What is a hidden Markov model? Nat. Biotechnol. 22, 1315–1316. doi: 10.1038/nbt1004-1315
Flagel, L. E., Wendel, J. F. (2009). Gene duplication and evolutionary novelty in plants. New Phytol. 183, 557–564. doi: 10.1111/j.1469-8137.2009.02923.x
Gai, W. X., Ma, X., Qiao, Y. M., Shi, B. H., Haq, S., Li, Q. H., et al. (2020). Characterization of the bZIP transcription factor family in pepper (Capsicum annuum L.): CabZIP25 positively modulates the salt tolerance. Front. Plant Sci. 11, 139. doi: 10.3389/fpls.2020.00139
Gao, D., Kong, F., Sun, P., Bi, G., Mao, Y. (2018). Transcriptome-wide identification of optimal reference genes for expression analysis of Pyropia yezoensis responses to abiotic stress. BMC Genomics 19, 1–14. doi: 10.1186/s12864-018-4643-8
Gao, T., Mo, Z., Tang, L., Yu, X., Du, G., Mao, Y. (2022). Heat shock protein 20 gene superfamilies in red algae: Evolutionary and functional diversities. Front. Plant Sci. 13, 817852. doi: 10.3389/fpls.2022.817852
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., Bairoch, A. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D., et al. (2005). Protein identification and analysis tools on the ExPASy server. In: Walker, J. M. (eds) The Proteomics Protocols Handbook. Springer Protocols Handbooks. Humana Press. doi: 10.1385/1-59259-890-0:571
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Hoang, D. T., Chernomor, O., Von Haeseler, A., Minh, B. Q., Vinh, L. S. (2018). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522. doi: 10.1093/molbev/msx281
Hsieh, T. H., Li, C. W., Su, R. C., Cheng, C. P., Sanjaya, Tsai, Y. C., et al. (2010). A tomato bZIP transcription factor, SlAREB, is involved in water deficit and salt stress response. Planta 231, 1459–1473. doi: 10.1007/s00425-010-1147-4
Hu, W., Yang, H., Yan, Y., Wei, Y., Tie, W., Ding, Z., et al. (2016). Genome-wide characterization and analysis of bZIP transcription factor gene family related to abiotic stress in cassava. Sci. Rep. 6, 22783. doi: 10.1038/srep22783
Iglesias-Fernández, R., Barrero-Sicilia, C., Carrillo-Barral, N., Oñate-Sánchez, L., Carbonero, P. (2013). Arabidopsis thaliana bZIP 44: a transcription factor affecting seed germination and expression of the mannanase-encoding gene AtMAN7. Plant J. 74, 767–780. doi: 10.1111/tpj.12162
Jakoby, M., Weisshaar, B., Dröge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., et al. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7, 106–111. doi: 10.1016/S1360-1385(01)02223-3
Janouškovec, J., Liu, S. L., Martone, P. T., Carré, W., Leblanc, C., Collén, J., et al. (2013). Evolution of red algal plastid genomes: ancient architectures, introns, horizontal gene transfer, and taxonomic utility of plastid markers. PloS One 8, e59001. doi: 10.1371/journal.pone.0059001
Jeffares, D. C., Penkett, C. J., Bähler, J. (2008). Rapidly regulated genes are intron poor. Trends Genet. 24, 375–378. doi: 10.1016/j.tig.2008.05.006
Kakinuma, M., Coury, D. A., Nakamoto, C., Sakaguchi, K., Amano, H. (2008). Molecular analysis of physiological responses to changes in nitrogen in a marine macroalga, Porphyra yezoensis (Rhodophyta). Cell Biol. Toxicol. 24, 629–639. doi: 10.1007/s10565-007-9053-7
Kakinuma, M., Suzuki, K., Iwata, S., Coury, D. A., Iwade, S., Mikami, K. (2016). Isolation and characterization of a new DUR3-like gene, PyDUR3.3, from the marine macroalga Pyropia yezoensis (Rhodophyta). Fisheries Sci. 82, 171–184. doi: 10.1007/s12562-015-0947-7
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., Haeseler, A., Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Landschulz, W. H., Johnson, P. F., McKnight, S. L. (1988). The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240, 1759–1764. doi: 10.1126/science.3289117
Lee, J. M., Kim, D., Bhattacharya, D., Yoon, H. S. (2019). Expansion of phycobilisome linker gene families in mesophilic red algae. Nat. Commun. 10, 4823. doi: 10.1038/s41467-019-12779-1
Lee, J. M., Yang, E. C., Graf, L., Yang, J. H., Qiu, H., Zelzion, U., et al. (2018). Analysis of the draft genome of the red seaweed Gracilariopsis chorda provides insights into genome size evolution in Rhodophyta. Mol. Biol. Evol. 35, 1869–1886. doi: 10.1093/molbev/msy081
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Peer, Y. V., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, D., Li, X., Liu, X., Zhang, Z. (2023). Comprehensive analysis of bZIP gene family and function of RcbZIP17 on Botrytis resistance in rose (Rosa chinensis). Gene 849, 146867. doi: 10.1016/j.gene.2022.146867
Liao, Y., Zou, H. F., Wei, W., Hao, Y. J., Tian, A. G., Huang, J., et al. (2008). Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta 228, 225–240. doi: 10.1007/s00425-008-0731-3
Li, C., Ariga, I., Mikami, K. (2019). Difference in nitrogen starvation-inducible expression patterns among phylogenetically diverse ammonium transporter genes in the red seaweed Pyropia yezoensis. Am J Plant Sci. 10 (8), 1325–1349. doi: 10.4236/ajps.2019.108096
Lopez-Bautista, J. M. (2010). Red Algal Genomics: A Synopsis. In: Seckbach, J., Chapman, D. (eds) Red Algae in the Genomic Age. Cellular Origin, Life in Extreme Habitats and Astrobiology, vol 13. Springer, Dordrecht, 227–240. doi: 10.1007/978-90-481-3795-4_12
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Ma, M., Chen, Q., Dong, H., Zhang, S., Huang, X. (2021). Genome-wide identification and expression analysis of the bZIP transcription factors, and functional analysis in response to drought and cold stresses in pear (Pyrus breschneideri). BMC Plant Biol. 21, 583. doi: 10.1186/s12870-021-03356-0
Maier, A., Stehling-Sun, S., Lohmann, J., Offenburger, S. (2011). The bZIP transcription factor PERIANTHIA: a multifunctional hub for meristem control. Front. Plant Sci. 2, 16827. doi: 10.3389/fpls.2011.00079
Marchler-Bauer, A., Bo, Y., Han, L., He, J., Lanczycki, C. J., Lu, S., et al. (2017). CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45, D200–D203. doi: 10.1093/nar/gkw1129
Merchant, S. S., Prochnik, S. E., Vallon, O., Harris, E. H., Karpowicz, S. J., Witman, G. B., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–250. doi: 10.1126/science.1143609
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Murmu, J., Bush, M. J., DeLong, C., Li, S., Xu, M., Khan, M., et al. (2010). Arabidopsis basic leucine-zipper transcription factors TGA9 and TGA10 interact with floral glutaredoxins ROXY1 and ROXY2 and are redundantly required for another development. Plant Physiol. 154, 1492–1504. doi: 10.1104/pp.110.159111
Nishikawa, T., Hori, Y., Tanida, K., Imai, I. (2007). Population dynamics of the harmful diatom Eucampia zodiacus Ehrenberg causing bleachings of Porphyra thalli in aquaculture in Harima-Nada, the Seto Inland Sea, Japan. Harmful algae 6, 763–773. doi: 10.1016/j.hal.2007.04.005
Nozaki, H., Takano, H., Misumi, O., Terasawa, K., Matsuzaki, M., Maruyama, S., et al. (2007). A 100%-complete sequence reveals unusually simple genomic features in the hot-spring red alga Cyanidioschyzon merolae. BMC Biol. 5, 1–8. doi: 10.1186/1741-7007-5-28
Oyama, K., Yoshimatsu, S., Honda, K., Abe, Y., Fujisawa, T. (2008). Bloom of a large diatom Chaetoceros densus in the coastal area of Kagawa Prefecture from Harima-Nada to Bisan-Seto, the Seto Inland Sea, in February 2005: environmental features during the bloom and influence on Nori Porphyra yezoensis cultures. Nippon Suisan Gakkaishi. 74, 660–670. doi: 10.2331/suisan.74.660
Para, A., Li, Y., Marshall-Colón, A., Coruzzi, G. M. (2014). Hit-and-run transcriptional control by bZIP1 mediates rapid nutrient signaling in Arabidopsis. Proc. Natl. Acad. Sci. 111, 10371–10376. doi: 10.1073/pnas.1404657111
Pearson, G. A., Hoarau, G., Lago-Leston, A., Coyer, J. A., Kube, M., Reinhardt, R., et al. (2010). An expressed sequence tag analysis of the intertidal brown seaweeds Fucus serratus (L.) and F. vesiculosus (L.) (Heterokontophyta, Phaeophyceae) in response to abiotic stressors. Mar. Biotechnol. 12, 195–213. doi: 10.1007/s10126-009-9208-z
Richter, D. J., Berney, C., Strassert, J. F. H., Burki, F., de Vargas, C. (2022). EukProt: a database of genome-scale predicted proteins across the diversity of eukaryotes. Peer Community J. 2. doi: 10.24072/pcjournal.173
Riechmann, J. L., Ratcliffe, O. J. (2000). A genomic perspective on plant transcription factors. Curr. Opin. Plant Biol. 3, 423–434. doi: 10.1016/S1369-5266(00)00107-2
Schönknecht, G., Chen, W. H., Ternes, C. M., Barbier, G. G., Shrestha, R. P., Stanke, M., et al. (2013). Gene transfer from bacteria and archaea facilitated evolution of an extremophilic eukaryote. Science 339, 1207–1210. doi: 10.1126/science.1231707
Sun, X., Li, Y., Cai, H., Bai, X., Ji, W., Ding, X., et al. (2012). The Arabidopsis AtbZIP1 transcription factor is a positive regulator of plant tolerance to salt, osmotic and drought stresses. J. Plant Res. 125, 429–438. doi: 10.1007/s10265-011-0448-4
Sun, P., Mao, Y., Li, G., Cao, M., Kong, F., Wang, L., et al. (2015). Comparative transcriptome profiling of Pyropia yezoensis (Ueda) MS Hwang & HG Choi in response to temperature stresses. BMC Genomics 16, 1–16. doi: 10.1186/s12864-015-1586-1
Sun, P., Tang, X., Bi, G., Xu, K., Kong, F., Mao, Y., et al. (2019). Gene expression profiles of Pyropia yezoensis in response to dehydration and rehydration stresses. Mar. Genomics 43, 43–49. doi: 10.1016/j.margen.2018.09.005
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tang, L., Qiu, L., Liu, C., Du, G., Mo, Z., Tang, X., et al. (2019). Transcriptomic insights into innate immunity responding to red rot disease in red alga Pyropia yezoensis. Int. J. Mol. Sci. 20, 5970. doi: 10.3390/ijms20235970
Van Leene, J., Blomme, J., Kulkarni, S. R., Cannoot, B., Winne, N. D., Eeckhout, D., et al. (2016). Functional characterization of the Arabidopsis transcription factor bZIP29 reveals its role in leaf and root development. J. Exp. Bot. 67, 5825–5840. doi: 10.1093/jxb/erw347
Wang, C., Lu, G., Hao, Y., Guo, H., Guo, Y., Zhao, J., et al. (2017). ABP9, a maize bZIP transcription factor, enhances tolerance to salt and drought in transgenic cotton. Planta 246, 453–469. doi: 10.1007/s00425-017-2704-x
Wang, L., Mo, Z., Yu, X., Mao, Y. (2023). Characterization of the basic leucine zipper transcription factor family of Neoporphyra haitanensis and its role in acclimation to dehydration stress. BMC Plant Biol. 23, 617. doi: 10.1186/s12870-023-04636-7
Wang, H., Wang, H., Shao, H., Tang, X. (2016). Recent advances in utilizing transcription factors to improve plant abiotic stress tolerance by transgenic technology. Front. Plant Sci. 7, 67. doi: 10.3389/fpls.2016.00067
Wang, H., Xu, K., Li, X., Blanco-Ulate, B., Yang, Q., Yao, G., et al. (2023). A pear S1-bZIP transcription factor PpbZIP44 modulates carbohydrate metabolism, amino acid, and flavonoid accumulation in fruits. Horticulture Res. 10, uhad140. doi: 10.1093/hr/uhad140
Wang, L., Yin, X., Cheng, C., Wang, H., Guo, R., Xu, X., et al. (2015). Evolutionary and expression analysis of a MADS-box gene superfamily involved in ovule development of seeded and seedless grapevines. Mol. Genet. Genomics 290, 825–846. doi: 10.1007/s00438-014-0961-y
Wang, D., Yu, X., Xu, K., Bi, G., Cao, M., Zelzion, E., et al. (2020). Pyropia yezoensis genome reveals diverse mechanisms of carbon acquisition in the intertidal environment. Nat. Commun. 11, 4028. doi: 10.1038/s41467-020-17689-1
Wang, S., Zhang, R., Zhang, Z., Zhao, T., Zhang, D., Sofkova, S., et al. (2021). Genome-wide analysis of the bZIP gene lineage in apple and functional analysis of MhABF in Malus halliana. Planta 254, 1–18. doi: 10.1007/s00425-021-03724-y
Wang, J., Zhou, J., Zhang, B., Vanitha, J., Ramachandran, S., Jiang, S. Y. (2011). Genome-wide expansion and expression divergence of the basic leucine zipper transcription factors in higher plants with an emphasis on sorghum F. J. Integr. Plant Biol. 53, 212–231. doi: 10.1111/j.1744-7909.2010.01017.x
Wang, Z., Zou, Q., Jiang, Y., Ju, Y., Zeng, X. (2014). Review of protein subcellular localization prediction. Curr. Bioinf. 9, 331–342. doi: 10.2174/1574893609666140212000304
Wu, Z., Fu, D., Gao, X., Zeng, Q., Chen, X., Wu, J., et al. (2023). Characterization and expression profiles of the B-box gene family during plant growth and under low-nitrogen stress in Saccharum. BMC Genomics 24, 79. doi: 10.1186/s12864-023-09185-9
Xie, J., Chen, Y., Cai, G., Cai, R., Hu, Z., Wang, H., et al. (2023). Tree visualization by one table (tvBOT): a web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res. 51 (W1), W587–W59. doi: 10.1093/nar/gkad359
Xu, C., Cao, H., Zhang, Q., Wang, H., Xin, W., Xu, E., et al. (2018). Control of auxin-induced callus formation by bZIP59–LBD complex in Arabidopsis regeneration. Nat. Plants 4, 108–115. doi: 10.1038/s41477-017-0095-4
Yamaguchi-Shinozaki, K., Shinozaki, K. (2005). Organization of cis-acting regulatory elements in osmotic-and cold-stress-responsive promoters. Trends Plant Sci. 10, 88–94. doi: 10.1016/j.tplants.2004.12.012
Yang, J., Wang, M., Li, W., He, X., Teng, W., Ma, W., et al. (2019). Reducing expression of a nitrate-responsive bZIP transcription factor increases grain yield and N use in wheat. Plant Biotechnol. J. 17, 1823–1833. doi: 10.1111/pbi.13103
Yoon, H. S., Zuccarello, G. C., Bhattacharya, D. (2010). “Evolutionary history and taxonomy of red algae,” in Red algae in the genomic age (Springer Netherlands, Dordrecht), 25–42.
Yu, X., Wang, L., Xu, K., Kong, F., Wang, D., Tang, X., et al. (2020). Fine mapping to identify the functional genetic locus for red coloration in Pyropia yezoensis thallus. Front. Plant Sci. 11, 867. doi: 10.3389/fpls.2020.00867
Zhang, H., Ding, X., Wang, H., Chen, H., Dong, W., Zhu, J., et al. (2023). Systematic evolution of bZIP transcription factors in Malvales and functional exploration of AsbZIP14 and AsbZIP41 in Aquilaria sinensis. Front. Plant Sci. 14, 1243323. doi: 10.3389/fpls.2023.1243323
Zhang, Z., Li, J., Zhao, X. Q., Wang, J., Wong, G. K. S., Yu, J. (2006). KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinf. 4, 259–263. doi: 10.1016/S1672-0229(07)60007-2
Zhao, K., Chen, S., Yao, W., Cheng, Z., Zhou, B., Jiang, T. (2021). Genome-wide analysis and expression profile of the bZIP gene family in poplar. BMC Plant Biol. 21, 1–16. doi: 10.1186/s12870-021-02879-w
Zhao, J., Guo, R., Guo, C., Hou, H., Wang, X., Gao, H. (2016). Evolutionary and expression analyses of the apple basic leucine zipper transcription factor family. Front. Plant Sci. 7, 376. doi: 10.3389/fpls.2016.00376
Keywords: Neopyropia yezoensis, basic leucine zipper family, phylogenetic analysis, expression profile, nitrogen stress
Citation: Zhu X, Gao T, Bian K, Meng C, Tang X and Mao Y (2024) Genome-wide analysis and expression profile of the bZIP gene family in Neopyropia yezoensis. Front. Plant Sci. 15:1461922. doi: 10.3389/fpls.2024.1461922
Received: 09 July 2024; Accepted: 02 October 2024;
Published: 21 October 2024.
Edited by:
Anna Torelli, University of Parma, ItalyReviewed by:
Amita Pandey, Shriram Institute for Industrial Research, IndiaElisabeth Fitzek, Bielefeld University, Germany
Copyright © 2024 Zhu, Gao, Bian, Meng, Tang and Mao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xianghai Tang, dHhpYW5naGFpQG91Yy5lZHUuY24=
 Xinyu Zhu
Xinyu Zhu Tian Gao
Tian Gao Ka Bian
Ka Bian Xianghai Tang
Xianghai Tang Yunxiang Mao
Yunxiang Mao