
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 03 September 2024
Sec. Functional and Applied Plant Genomics
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1461322
This article is part of the Research TopicInsights in Functional and Applied Plant Genomics: 2023View all 9 articles
 Lian Duan1,2
Lian Duan1,2 Zhihui Hou1,2
Zhihui Hou1,2 Wuhua Zhang1,2
Wuhua Zhang1,2 Shuang Liang1,2
Shuang Liang1,2 Minge Huangfu1,2
Minge Huangfu1,2 Jinzhu Zhang1,2
Jinzhu Zhang1,2 Tao Yang1,2
Tao Yang1,2 Jie Dong1,2*
Jie Dong1,2* Daidi Che1,2*
Daidi Che1,2*WOXs are a class of plant-specific transcription factors that play key roles in plant growth and stress responses. However, the mechanism by which WOXs influence adventitious root development in Rosa hybrida remains unclear. In this study, RcWOX gene family in rose was identified and phylogenetically analyzed using bioinformatics analysis. A total of 381 RcWOX gene members were localized on seven chromosomes except of nine members. The main cis-acting elements involved in hormonal, light, developmental, and abiotic stress responses were identified in the promoters of RcWOX genes, suggesting their regulation by these signals. Nine RhWOX genes had significant different expression during rooting process of rose. RhWOX331, RhWOX308, RhWOX318 were positive with the formation of rose roots. RhWOX331 was positively involved in the formation of adventitious root primordia, which gene coding a transcription factor localized in the nucleus. The HOX conserved domain in the protein contributed to the self-activating activity of RhWOX331. We obtained genetically modified Arabidopsis to validate the function of RhWOX331. Overexpression of RhWOX331 gene alleviated the inhibition of root length of A. thaliana primary roots by high concentration of IBA and NPA, and significantly increased the number of lateral roots on the primary roots, as well as the height of A. thaliana plants. Additionally, RhWOX331 promoted adventitious root formation in A. thaliana and mitigated hormonal inhibition by exogenous 6-BA, NPA, and GA3. The RhWOX331 promoter contained cis-acting elements such as ABRE, Box 4 and CGTCA-motif et.al. GUS activity analysis showed that the gene acted at the cotyledon attachment site. Taken together, these studies identified a significant expansion of the RcWOX gene family, inferred roles of certain branch members in adventitious root formation, elucidated the function of RhWOX331 in adventitious root initiation, and laid the foundation for further research on the function of WOX gene family in roses.
In agriculture, forestry and horticulture, plant organ regeneration was often utilized in cuttings propagation practices to obtain a large number of plants that retained the parent’s good traits (De Klerk et al., 1999) For woody plants, The incidence of adventitious roots (ARs) during the propagation of cuttings determined the survival and efficiency of propagation of the species. ARs can be initiated from column sheath cells in the hypocotyl, thin-walled cells in the phloem or xylem, young secondary phloem cells, or cells of the inter bundle formation layer close to phloem cells (Bellini et al., 2014). The formation of adventitious roots was regulated by a combination of external environment, endogenous substances, and other factors, including light, water, spike age, and phytohormones (Bannoud and Bellini, 2021).
The WUSCHEL (WUS) homeobox transcription factor was a plant-specific transcription factor with a conserved “helix-loop-helix-turn-helix” motif comprising 60-66 amino acid residues (van der Graaff et al., 2009). During the phylogenetic process of higher plants, the WOX genes had evolved into three major classical clades: the modern/WUS clade, the ancient clade, and the intermediate clade. The modern evolutionary clade included WUS, WOX1~7, totaling 8 members, and the intermediate clade included 4 members, WOX8, WOX9, WOX11 and WOX12. The ancient clade members contained three genes, WOX10, WOX13 and WOX14 (Liu and Xu, 2018). Studies of the WOX gene family in Arabidopsis thaliana (Ohmori et al., 2013), Populus trichocarpa (Shuang et al., 2019), and Picea abies (Palovaara et al., 2010) had revealed that members of WOX gene family in each clade interacted with hormones to regulate plant growth and development processes. The WOX gene family played crucial regulatory roles during key stages of plant development such as embryo formation, stem cell maintenance, and organogenesis (Tanaka et al., 2015; Zhang et al., 2017), which were mediated by promoting cell division or inhibiting premature cell differentiation (Laux et al., 1996). These regulatory effects were likely achieved through interactions between WOX genes and hormones.
In modern clade, AtWUS regulated anther and ovule development (Reiser et al., 1995), and it also interacted with CLAVATA3 (CLV3) to maintain the balance between proliferation and differentiation of stem tip meristems (Laux et al., 1996). CsWUS overexpression increased the number of sepals, petals and carpels in Cucumis sativus (Che et al., 2020). AtWOX2 gene was expressed mainly in the apical cells of early embryonic development and regulated embryo formation (Liu and Xu, 2018). Overexpression of AtWOX4 gene promoted radial growth of primary roots (Zhang et al., 2019). Among the genes in the intermediate clade, AtWOX8 and AtWOX9 were co-expressed in the pituitary cells of the embryo, promoted embryo development, and also functioned to maintain cell proliferation in the apical and root tip meristematic tissues (Liu and Xu, 2018). Overexpressing PeWOX11a or PeWOX11b in poplar not only enhanced adventitious root formation on the plugs, but also induced ectopic rooting in the aboveground part of transgenic poplar (Li et al., 2018). OsWOX11 gene was expressed in the region of proliferative root tip cells and regulated the emergence and growth of crown roots in rice (Cheng et al., 2014). There were fewer members in the ancient clade, among which AtWOX13 functioned in early stages of root development and in organs with high proliferation, AtWOX14 gene expressed in A. thaliana primary roots, lateral root primordia, and floral organs, and inhibited cell differentiation (Deyhle et al., 2007). Expression of SkWOX13B in stone pine plants was closely related to root organogenesis (Ge et al., 2016).
As the premier among the world’s top four cut flowers, Rosa hybrida exhibits exceptionally high commercial value and possesses a unique cultural significance. In the genus Rosa, the ability to generate adventitious roots directly influences cutting survival and is a decisive factor in the garden application of Rosa species. WOX genes also regulated the growth and development of Rosa genus. The RcaWOX1 gene from Rosa canina was induced by auxin and expressed at the early stage of healing tissue formation, overexpressing this gene increased the number of lateral roots and induced the up-regulated expression of AtPIN1 and AtPIN7 in A. thaliana (Gao et al., 2014). Overexpression of RcWUS induced the transformation of parenchyma cells in the root cortex into meristematic tissue cells, leading to the ectopic occurrence of adventitious shoots at the root tip (Jiang et al., 2012). The rooting ability of rose was influenced by factors such as genotype, lignification level of the cuttings, hormones, and environmental conditions. However, the molecular mechanisms underlying rose rooting remained unclear. This study provided a comprehensive overview of the WOX gene family in roses, investigating the expression patterns and functions of RhWOX331 in adventitious rooting. It established a solid theoretical basis for further research on RhWOX genes involved in organogenesis in roses.
Genome and protein sequences of Rosa chinensis, Rosa rugosa and Rosa multiflora were obtained from the Rosaceae Genome Database (GDR) (Raymond et al., 2018; Jung et al., 2019). All WOXs in rose were identified using the Pfam protein family database (Mistry et al., 2021) by downloading the Hidden Markov Model (HMM) file for the WOX structural domain (PF00046) and setting a threshold of 1e-5. The core sequences of RcWOXs were verified using the SMART program and conserved domain database (CDD) (Wang et al., 2023). The protparam tool from the Expasy website (https://web.expasy.org/protparam/) was used to predict basic characteristics (amino acid length, amino acid composition, isoelectric point, etc.) of the obtained WOX family members (Walker, 2005). Each RcWOX gene family member was named according to their position on the chromosome using TBtools II (Chen et al., 2023). The sequences of WOXs in A. thaliana (Lamesch et al., 2012), Nicotiana tabacum, and Populus trichocarpa were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/). MEGA11 (Tamura et al., 2021) was used to perform multiple sequence comparisons under default parameters, and a phylogenetic tree was constructed using the Neighbor-Joining method (bootstrap: 1000).
Multiple sequence comparisons were performed using ClustalW in MEGA11 under default parameters to further analyze the characteristic structural domains of RcWOX proteins and manually adjust the amino acid sequences. GSDS (Hu et al., 2015) was used for exon-intron structure visualization of RcWOX genes. RcWOXs protein motifs were analyzed using MEME (Bailey et al., 2009) under the parameter maximum motif number of 20.
Localization of all RcWOX genes to rose chromosomes based on physical location information using TBtools II (Chen et al., 2023). Utilizing TBtools II for collinearity analysis of the WOX gene family in rose with the WOX gene families of A. thaliana and P. trichocarpa. Promoter cis-acting regulatory elements were analyzed in the 2 Kb region upstream of the rose WOXs using PlantCARE (Lescot et al., 2002), and WOX gene family was visualized by TBtools II.
R. ‘The Fairy’ and Nicotiana benthamiana were grown in the Northeast Agricultural University (Harbin City, Heilongjiang Province, China) under a 16 h light/8 h dark at 25°C cycle. A. thaliana was grown under 14 h light/10 h dark conditions with a temperature range of 22-23°C and relative humidity between 40-60%.
Total RNA of plants leaves and roots was isolated with the FastPure Universal Plant Total RNA Isolation Kit (Vazyme Biotech Co., Ltd., Nanjing, China), and transcribed into cDNA using the HiScript III 1st Strand cDNA Synthesis Kit (+gDNA wiper) (Vazyme Biotech Co., Ltd., Nanjing, China). HiScript II QRT SuperMix for qPCR (Vazyme Biotech Co., Ltd., Nanjing, China) was used for qPCR. The determination of gene expression levels refers to previous research descriptions (Dong et al., 2021). The 2−ΔΔCT quantification method (Schmittgen and Livak, 2008) was used to calculate the relative expression levels. RhActin (Fan et al., 2023) were selected as reference genes in Rosa hybrida. All experiments were conducted with three biological replicates, each containing three technical repeats. Define a total of 8 stages from US to CS7 based on the cutting time of cuttings. US: 0 d; CS1: 15 min; CS2: 1 d; CS3: 3 d; CS4: 5 d; CS5: 10 d; CS6: 15 d; CS7: 20 d. Primers used for RT-qPCR were listed in Supplementary Table S2.
The full-length RhWOX331 gene, lacking a stop codon, was inserted into KpnI and BamHI sites (Takara, Beijing, China) of the pGAMBIA1300-sGFP vector using the pEASY®-Basic Seamless Cloning and Assembly Kit (TransGen Biotech, Beijing, China). The constructed vector pGAMBIA1300-RhWOX331-sGFP was transformed into Agrobacterium tumefaciens GV3101 (WeiDi Biotechnology, Shanghai, China), and subcellular localization was performed according to the previous research (Li et al., 2023). The infection solution (200 μM acetosyringone (AS), 10 mM 2-morpholinoethanesulphonic acid (MES), and 10 mmol/l MgCl2) containing either pGAMBIA1300-RhWOX331-sGFP or pGAMBIA1300-sGFP were injected into the subepidermal cells of 4-week-old Nicotiana benthamiana leaves. After 2 days of dark incubation at 23 °C, the subcellular localization of RhWOX331 was visualized and photographed using a laser-scanning confocal microscope (FV3000, Olympus, Japan) at 488 nm.
The full-length RhWOX331 gene was inserted into NdeI and EcoRI (Takara, Beijing, China) sites of the pGBKT7 vector using the pEASY®-Basic Seamless Cloning and Assembly Kit (TransGen Biotech, Beijing, China). The pGADT7-T+pGBKT7-p53 (positive control), pGADT7-T+pGBKT7-lam (negative control), and pGBKT7-WOX331-1, pGBKT7-WOX331-2, pGBKT7-WOX331-3 plasmids were transformed into Y2HGold yeast competent cells (WeiDi Biotechnology, Shanghai, China). After 2 days of cultivation at 28°C, yeast colonies were selected and cultured in SD/-Trp/-Leu liquid yeast medium at 28°C and 200 rpm. Centrifuge yeast at 4000 rpm for 1 minute to collect the yeast cells. The Y2HGold yeast containing the recombinant plasmid was resuspended in sterile water until its OD600 reached 0.2. The suspended culture was diluted to 1X, 10X, and 100X concentrations. The positive control and negative control diluted yeast solution was placed on SD/-Trp/-Leu/-His/-Ade/X-α-gal solid medium, the diluted yeast solution transforming pGBKT7-WOX331-1, pGBKT7-WOX331-2, pGBKT7-WOX331-3 was placed on SD/-Trp/-His/X-α-gal solid medium and cultured at 28°C. After 36-48 h of incubation, the self-activating activity of RhWOX331 was assessed based on the blue coloration of the yeast.
pGAMBIA1300-RhWOX331-sGFP was transformed in A. thaliana with floral dip transformation method (Bent, 2006). Seeds of A. thaliana were collected and sown, until obtaining T3 generation plants. Transgenic A. thaliana were identified by PCR using WOX331F and WOX331R as primers (Supplementary Table S2). The seeds of transgenic A. thaliana were sterilized and sown in 1/2 MS medium (20 g/L sucrose + 8 g/L agar), and different hormones were added to the medium according to different treatments: CK: no hormone; IBA: 0.25 mg/L IBA; 6-BA: 0.5 mg/L 6-BA; GA3: 1 mg/L GA3; NPA: 10 μM NPA. The phenotypes of the primary roots of A. thaliana were determined after 14 days. At 14d, the main roots were removed and transferred to B5 medium (30 g/L sucrose + 8 g/L agar), and different hormones were added to the medium according to different treatments (hormone concentration as above), and the phenotypic changes of adventitious roots were observed.
The 2113 bp sequence upstream of the start codon of RhWOX331 was divided into three segments. pWOX331 replaced 35S in PBI121 and construct pWOX331-1::GUS, pWOX331-2::GUS and pWOX331-3::GUS vectors with BamHI and HindIII restriction site. Primers were listed in Supplementary Table S2. Transgenic A. thaliana overexpressing pWOX331-1::GUS, pWOX331-2::GUS and pWOX331-3::GUS were immersed in GUS staining solution (Coolaber, Beijing, China) and kept warm at 37°C for 1 h. Using 70% ethanol for decolorization 2~3 times, and the material was observed under the in vitro microscope (Olympus SZX2-ILLTQ). P1, P2, and P3 represent A. thaliana transformed with pWOX331-1:: GUS, pWOX331-2:: GUS, and pWOX331-3:: GUS, respectively. GUS activity was assessed using the previous method (Koo et al., 2007).
Statistical analyses were conducted with IBM SPSS v25.0 (SPSS Inc., Chicago, IL, USA). Least Significant Difference (LSD) test was performed in order to compare the statistical validity of data. Significance was set at p< 0.05. Three biological replicates were used for each assay. TBtoolsII software was used to create the conserved domains, motifs, gene structure. GraphPad Prism 8.0.0 (GraphPad Software San Diego, California USA) were used to plot graphs.
The 381 members of the rose WOX gene family were finally identified in the whole rose genome, and they were named RcWOX1-RcWOX381 based on their positions on the chromosome (Supplementary Table S1). The physicochemical properties of the 381 WOX genes revealed that the number of amino acids ranged from 81 to 400, and the theoretical isoelectric points ranged from 4.56 to 10.55, with 87.9% of them having isoelectric points lower than 7, indicating that they were mostly acidic proteins. The instability coefficients ranged from 36.55% to 86.47%, with 2.1% of the members having instability coefficients lower than 40%, and most of the WOX proteins were unstable proteins. The relative molecular mass of RcWOX335 was the largest at 44.66 KDa, and the relative molecular mass of RcWOX275 was the smallest at 9.87 KDa.
In order to explore the phylogenetic relationships of WOXs in rose and other model plants, a phylogenetic tree was constructed based on the sequences of 453 WOX proteins from rose (381), P. trichocarpa (26), N. tabacum (28) and A. thaliana (18) (Figure 1). The phylogenetic tree analysis showed that the 453 genes were clearly divided into eight clades: ancient clade, intermediate clade, modern/WUS clade, clade I, clade II, clade III, clade IV, clade V. Among these, the RcWOXs in the classical clades including ancient, intermediate, and modern/WUS clades were more closely related to N. tabacum, A. thaliana, and P. trichocarpa WOXs. On the contrary, WOX genes in rose belonging to clades I to V had no homologous genes with P. trichocarpa, N. tabacum and A. thaliana WOX genes. Ancient clade contained 2 genes in rose, 3 genes in A. thaliana, 6 genes in P. trichocarpa, 6 genes in N. tabacum. Intermediate clade contained 2 genes in rose, 7 genes in A. thaliana, 6 genes in P. trichocarpa, 6 genes in N. tabacum. Modern/WUS clade contained 13 genes in rose, 8 genes in A. thaliana, 14 genes in P. trichocarpa, 16 genes in N. tabacum. Clades I to V contained 364 members, all of which originated from rose. Clade V was the largest clade, containing 265 members. The results show that there are a large number of similar redundant genes in rose WOX gene family, and they are distantly homologous to the WOX family members of the ancient, intermediate, and modern/WUS clades.
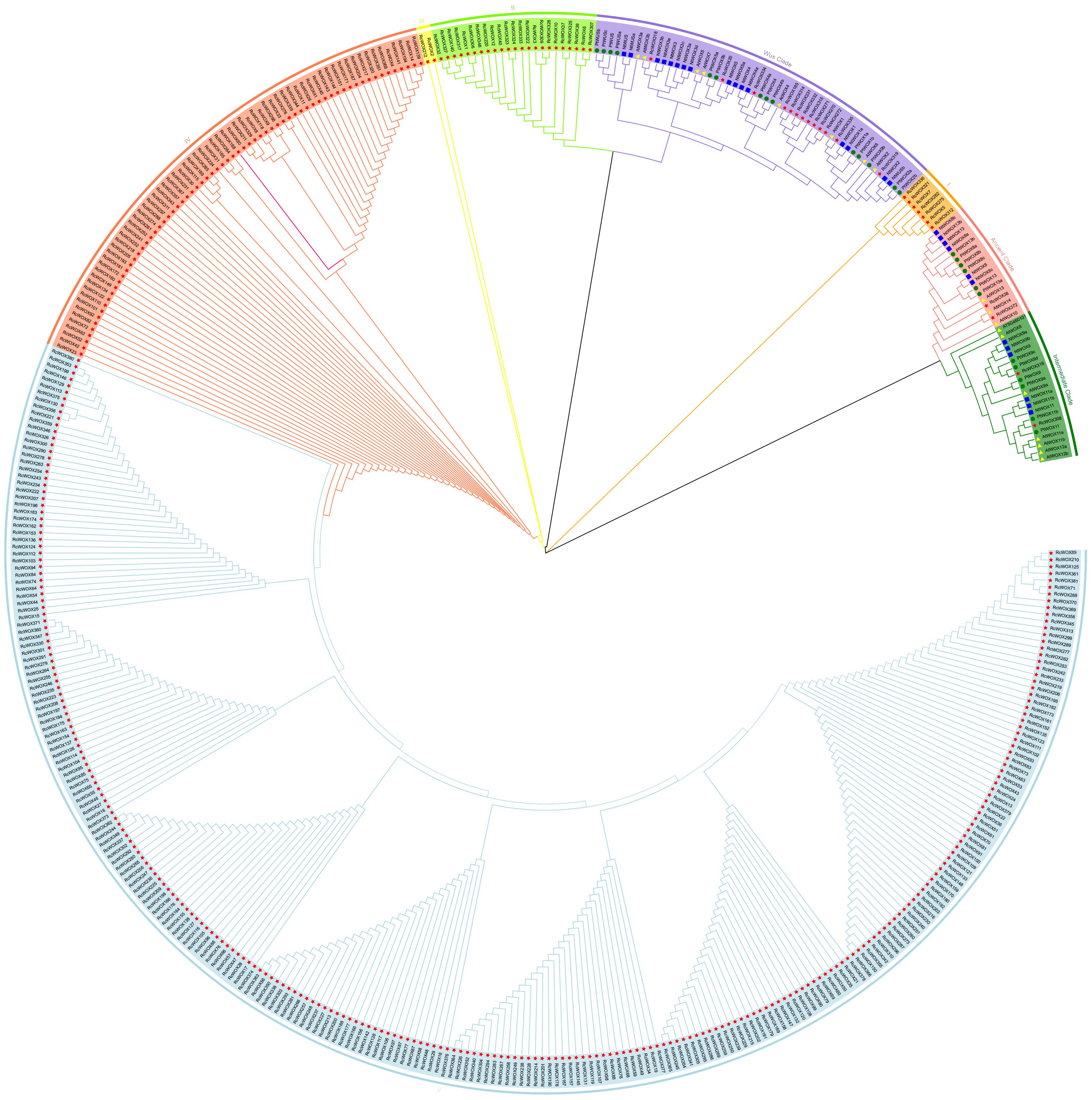
Figure 1. Phylogenetic analysis of the RcWOX gene family of R. chinensis and N. tabacum, A. thaliana, P. trichocarpa. Proteins from R. chinensis, N. tabacum, A. thaliana and P. trichocarpa were respectively denoted by the prefixes Rc, Nt, At, and Pt, respectively. They were divided into eight major phylogenetic clusters: ancient clade, intermediate clade, WUS clade, clade I, clade II, clade III;, clade IV, and clade V. Each clade was indicated by different colors. Bootstrap:1000.
Based on the chromosomal location information of RcWOXs in R. chinensis, the positions of 381 RcWOX members on the chromosomes were visualized and analyzed (Figure 2). RcWOXs were distributed on all seven chromosomes, with a total of 226 RcWOX genes on chromosome 2, 46 RcWOX genes on chromosome 3, 38 RcWOX genes on chromosome 7, 36 RcWOX genes on chromosome 1, 10 RcWOX genes on chromosome 5, 9 RcWOX genes on chromosome 6, 7 RcWOX genes on chromosome 4, and 9 RcWOX genes not localized on any chromosome. WOX genes were most densely distributed on chromosome 2.
Conserved domain analysis of 381 RcWOX family members revealed the presence of two conserved domains: Homeodomain superfamily and Homeobox (Supplementary Figure S1B). In order to study the structure of RcWOXs, a figure depicting the RcWOX structure was created (Supplementary Figure S1A), which showed that motifs 10 and 15 were present in all members of RcWOXs of classical clades. In contrast, the vast majority of the members in clades I to V contained motifs 1, 2 and 4. The gene structure figure also indicated that 86.8% of members in clades I to V and 29.4% of members in classical clades lacked UTRs (Supplementary Figure S1C). Analysis of the amino acid sequences of clade V revealed that most of the proteins contained the amino acid domains SIMEQRGBYHQBIBTLPLFPMHGEDILGNMKTTSEGGGGGYGG and G/DSHISLELSLNSYRDADMA, corresponding to motifs 2 and 4. For clade IV of WOXs in rose, most proteins contained the amino acid domains HQEIETLMHGEDI and YGQIEDKNVFFWFQNLKA, which were absent in classical clades. These findings suggest significant differences in amino acid sequences, conserved domains, and intron distribution between WOX members of classical clades and clades I to V, implying potential functional distinctions.
The cis-acting elements within the upstream 2000bp of the initiation codon of 381 WOX genes in rose were involved in hormone, environment, growth and development (Supplementary Figure S2). Hormone-related cis-acting elements were salicylic acid-induced (W-box), jasmonic acid signaling pathway (MYC), and gibberellin response element (P-box). Cis-acting elements involved in environment including light-responsive (G-box, Sp1, TGACG-motif, TCCC-motif) and trauma response (WUN-motif). MYB, GCN4-motif, Circadian clock belonged to growth and development-related cis-acting elements. It was observed that WOX genes within the same clade of the phylogenetic tree exhibited similar cis-acting elements. These findings suggest that WOX genes in rose may be regulated by a diverse array of phytohormones, biotic and abiotic stimuli, influencing plant growth and development.
In order to further investigate the interspecific evolutionary relationship of WOXs, intergenic collinearity analysis was performed between roses and model plants, such as A. thaliana (Figure 3A) and P. trichocarpa (Figure 3B). It was found that there were 12 homologous gene pairs between 381 WOX genes of rose and 15 AtWOX genes of A. thaliana, and 21 homologous gene pairs between 381 WOX gene family members of rose and 26 PtWOX genes of P. trichocarpa. These results suggest a high number of homologous gene pairs between rose WOX genes and both AtWOXs and PtWOXs.
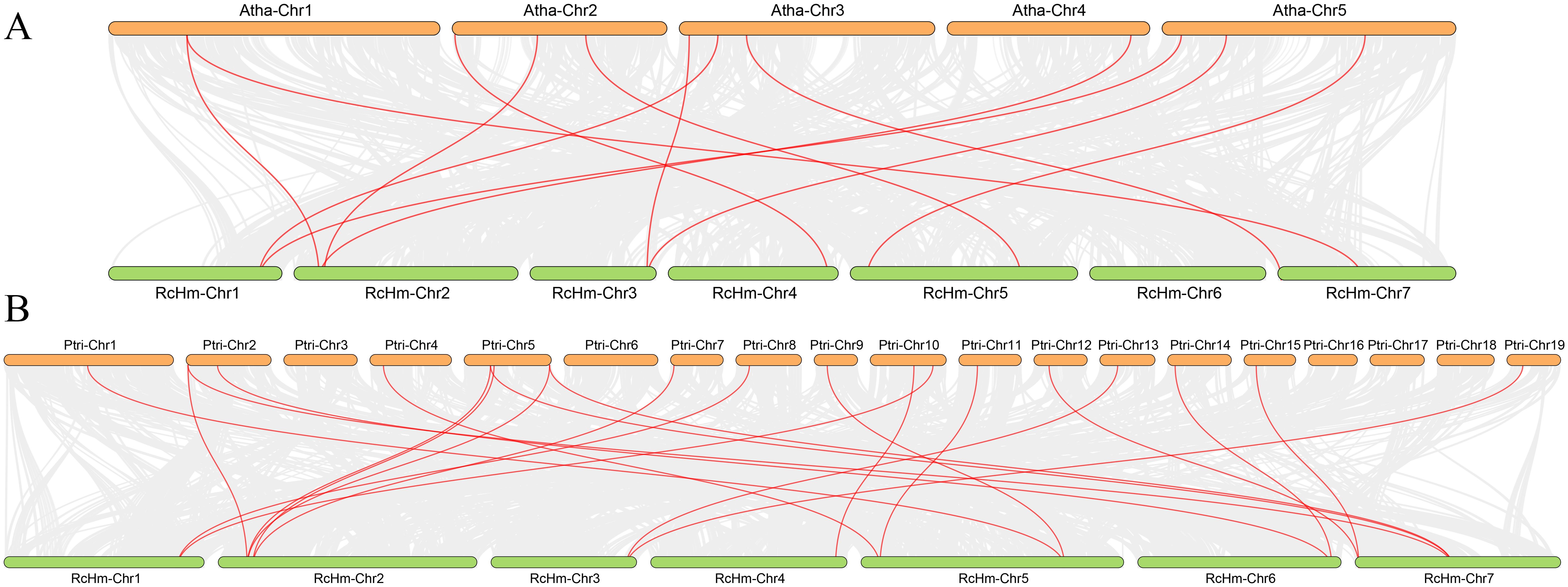
Figure 3. Collinearity analysis of the WOXs between R. chinensis both A. thaliana and P. trichocarpa. (A) Collinearity analysis of the WOXs between R. chinensis and A. thaliana. (B) Collinearity analysis of the WOXs between R. chinensis and P. trichocarpa.
The expression levels of WOX genes during adventitious root formation in roses indicated relatively higher expression levels in classical clades, with almost all members in clades I to IV showing no expression. Therefore, we selected RhWOX genes in ancient, intermediate, and modern/WUS clades. Combining the expression data of WOXs transcripts during adventitious rooting process of R. hybrida, nine RhWOXs were finally identified (Figure 4). Expression analysis of RhWOXs gene family members during rooting of single-node spikes of rose showed that RhWOX284 was down-regulated 15 minutes after cutting, and up-regulated during leaf production. RhWOX372, RhWOX316 and RhWOX271 were up-regulated in the mid-root stage of CS3~CS5, and down-regulated in the root elongation stage. RhWOX308 and RhWOX270 were initially down-regulated after pruning, and these genes were significantly up-regulated as the stem cells divided and root primordia formed. RhWOX318 gradually activated during root tip formation, exhibiting peak activity during root elongation. RhWOX185 showed significant up-regulation during CS2 stage. RhWOX331 remained low until root primordium formation (CS1~CS4), exhibited significant up-regulation during CS4~CS5, and then down-regulated during the period of root tip formation and root elongation, showing strong correlation with root primordium development. RT-qPCR data corroborated RNA sequencing results, with RhWOX331 showing significant positive correlation with root primordium differentiation, Thus, we speculate that RhWOX331 gene play a key role in the development of adventitious roots in R. hybrida.
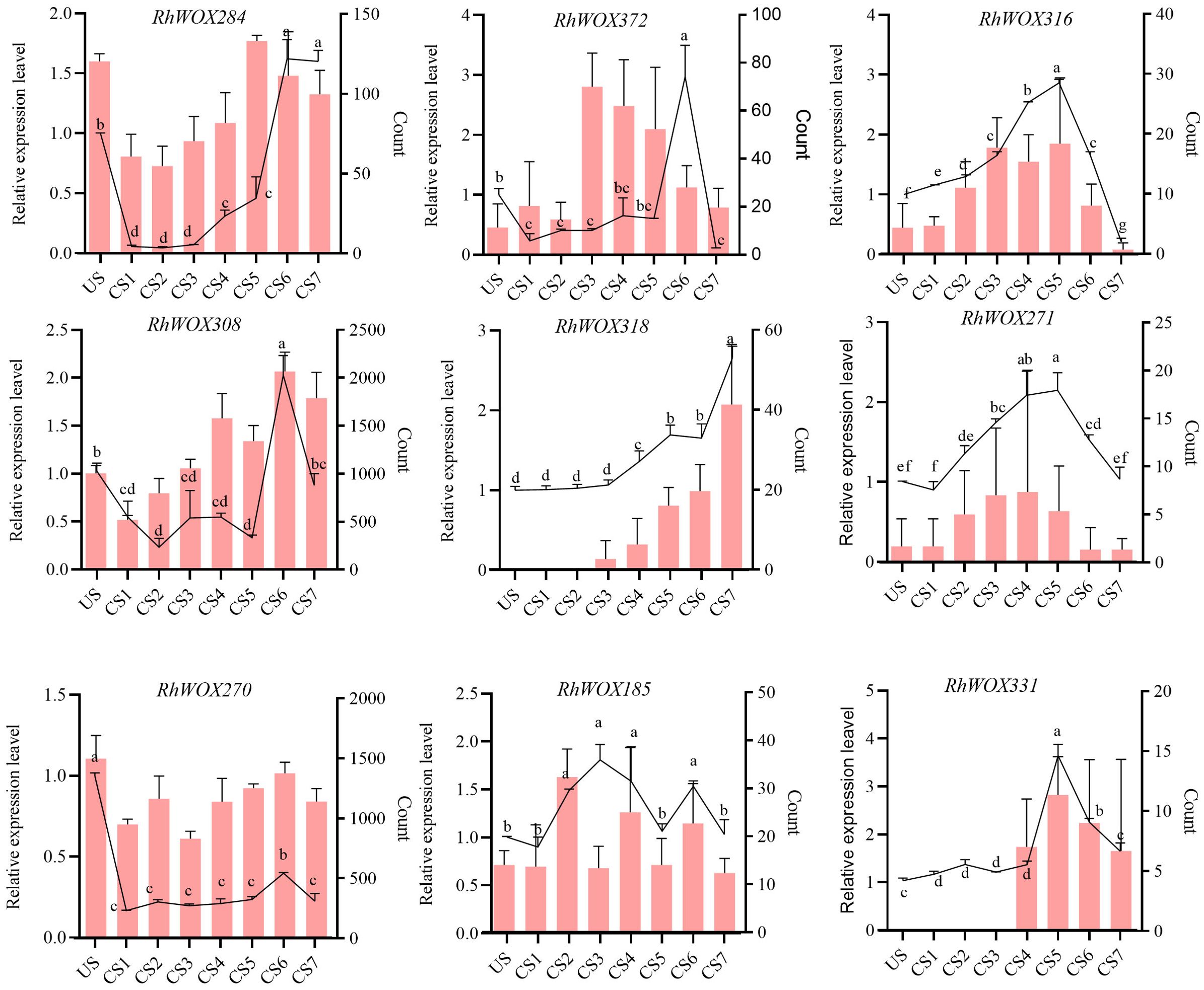
Figure 4. Relative expression of the RhWOX gene family during rooting at the stem nodes of R. ‘ The Fairy ‘ scions. The line graph shows the relative expression by RT-qPCR and the bar graph shows the gene count values determined by RNA-seq. The horizontal coordinates indicate the time of cuttings of R. ‘The Fairy’. US: 0 d; CS1: 15 min; CS2: 1 d; CS3: 3 d; CS4: 5 d; CS5: 10 d; CS6: 15 d; CS7: 20 d. Different lowercase letters indicate significant differences among different treatments.
The expression of RhWOX331 showed tissue-specificity, with the highest expression in roots, followed by stems, and the lowest expression in flowers (Figure 5A). Exogenous application of IBA promotes adventitious root formation in roses, whereas NPA application suppresses it. By the 10th day of cutting, exogenous IBA significantly increased the expression of RhWOX331 to 1.3 times that of the hormone-free control, while exogenous NPA significantly reduced RhWOX331 expression to 0.8 times that of the hormone-free control (Figure 5B). IBA promoted the expression of RhWOX331 continuously. In the absence of hormones, expression of RhWOX331 in cuttings remained almost unchanged after 5 d of cultivation. The gene was up-regulated from 5 to 10 days and then down-regulated from 15 to 20 days. After the application of exogenous IBA, RhWOX331 showed upregulated expression as early as 5 d after culture initiation. At each time point thereafter, the expression level of this gene was significantly higher compared to the control without any hormone addition (Figure 5C).
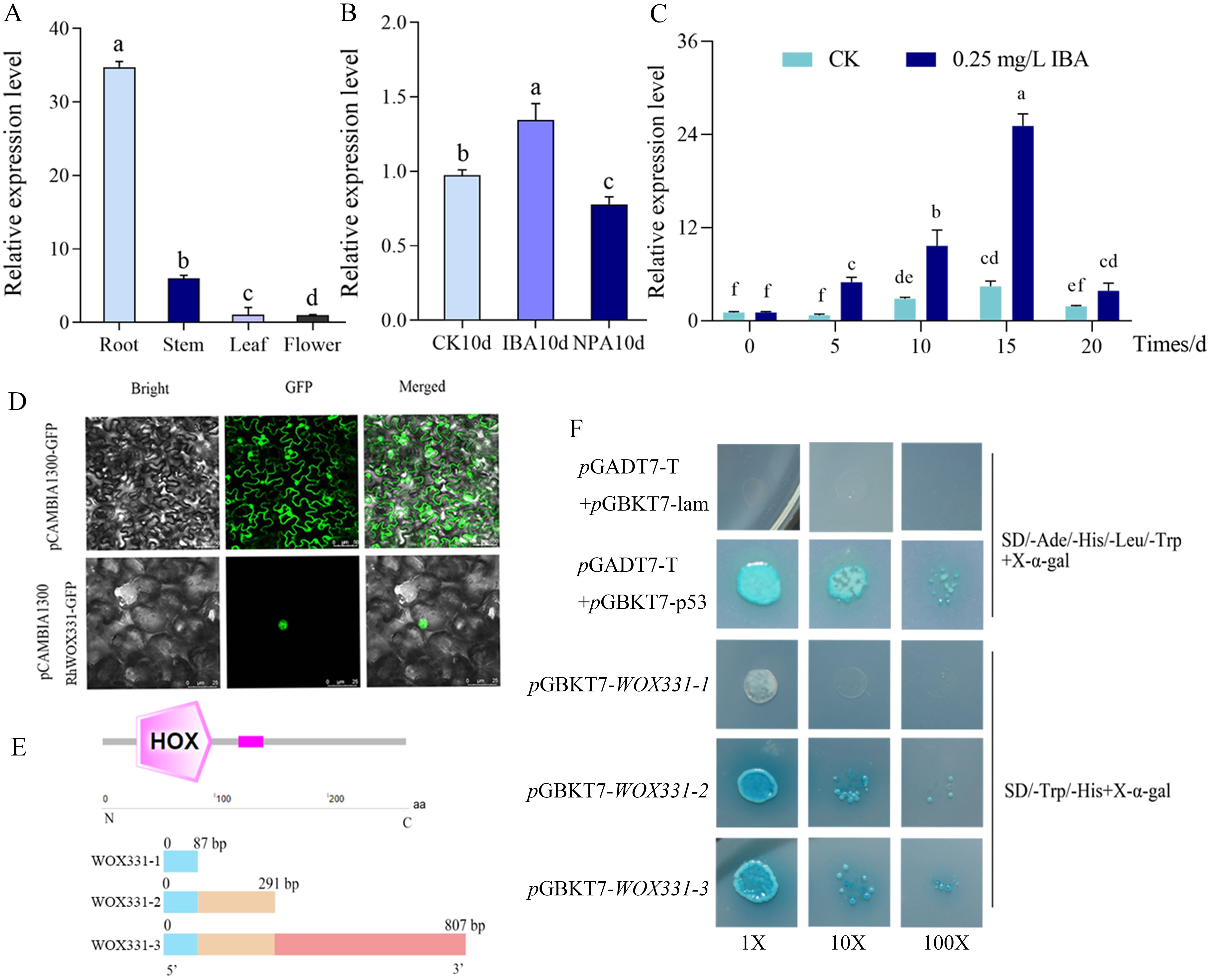
Figure 5. Characterization of RhWOX331. (A) Expression of RhWOX331 in different tissues of R. ‘The Fairy’ (B) Relative expression of RhWOX331 gene in 0.25 mg/L IBA, 0.2 mg/L NPA-treated and untreated rose cuttings. (C) Trend of RhWOX331 gene expression during adventitious root primordia formation in R. ‘The Fairy’ cuttings. (D) Subcellular localization of RhWOX331. pCAMBIA1300-GFP empty vector as a control. (E) Schematic diagram of yeast self-activation vector construction for RhWOX331. (F) Verification of yeast self-activation of RhWOX331. pGADT7-T+pGBKT7-p53 served as the positive control, while pGADT7-T+pGBKT7-lam was utilized as the negative control. Different lowercase letters indicate significant differences among different treatments.
Subcellular localization analysis revealed that in the control tobacco leaf cells, green fluorescence can be observed simultaneously in both the cell membrane and nucleus. In the RhWOX331 group, only green fluorescence was observed within the nucleus, confirming the nuclear localization of RhWOX331 (Figure 5D). To verify the transcriptional activation activity of WOX331, three segments of the WOX331 gene were constructed into the pGBKT7 vector (Figure 5E). On SD/-Ade/-His/-Leu/-Trp medium, the negative control yeast did not grow, while the positive control yeast grew and turned blue after adding X-α-gal. The yeast that transformed pGBKT7-WOX331-1 did not grow, while the yeast that transformed pGBKT7-WOX331-2 and pGBKT7-WOX331-3, which both containing the HOX domain grew normally and turned blue after adding X-α-gal (Figure 5F). This indicates that the transcription factor RhWOX331 possesses self-activation activity, which may be attributed to the HOX domain spanning amino acids 87 to 807.
RhWOX331-overexpressing A. thaliana lines were obtained and identified to investigate the influence of RhWOX331 on root development (Supplementary Figure S3). There was no significant difference in root length between wild-type (WT) and RhWOX331-overexpressing A. thaliana plants on hormone-free medium or medium containing 0.5 mg/L 6-BA or 1 mg/L GA3 (Figure 6B). The average root length of 14-day-old plants was approximately 6.78 cm in the CK, 0.74 cm in the 6-BA group and 4.6cm in the GA3 group. Interestingly, on medium containing 0.25 mg/L IBA and 10 μM NPA, A. thaliana growth was inhibited, showing differences in root length between WT and transgenic plants (Figures 6A, C, F). The root lengths of WT plants were 1.34 cm and 1.91 cm, respectively. However, overexpression of RhWOX331 alleviated the inhibitory effects of these high concentrations of exogenous hormones, resulting in primary root lengths of 3.08 cm and 2.86 cm, respectively. The number of lateral roots on the primary root had also significantly increased. Moreover, it was found that both the plant height and the height between the capsules and the rosette were increased after overexpressing RhWOX331 (Figures 6G, H, I). It was different that the number of capsules did not increase (Figure 6J). These results indicate that overexpression of RhWOX331 did not promote elongation of primary roots in A. thaliana, but enhanced lateral root formation. It also alleviated the inhibitory effects of high concentrations of auxin and auxin inhibitors on primary root elongation. Moreover, RhWOX331 increased plant height by raising the height between the capsules and the rosettes rather than increasing the number of flowers.
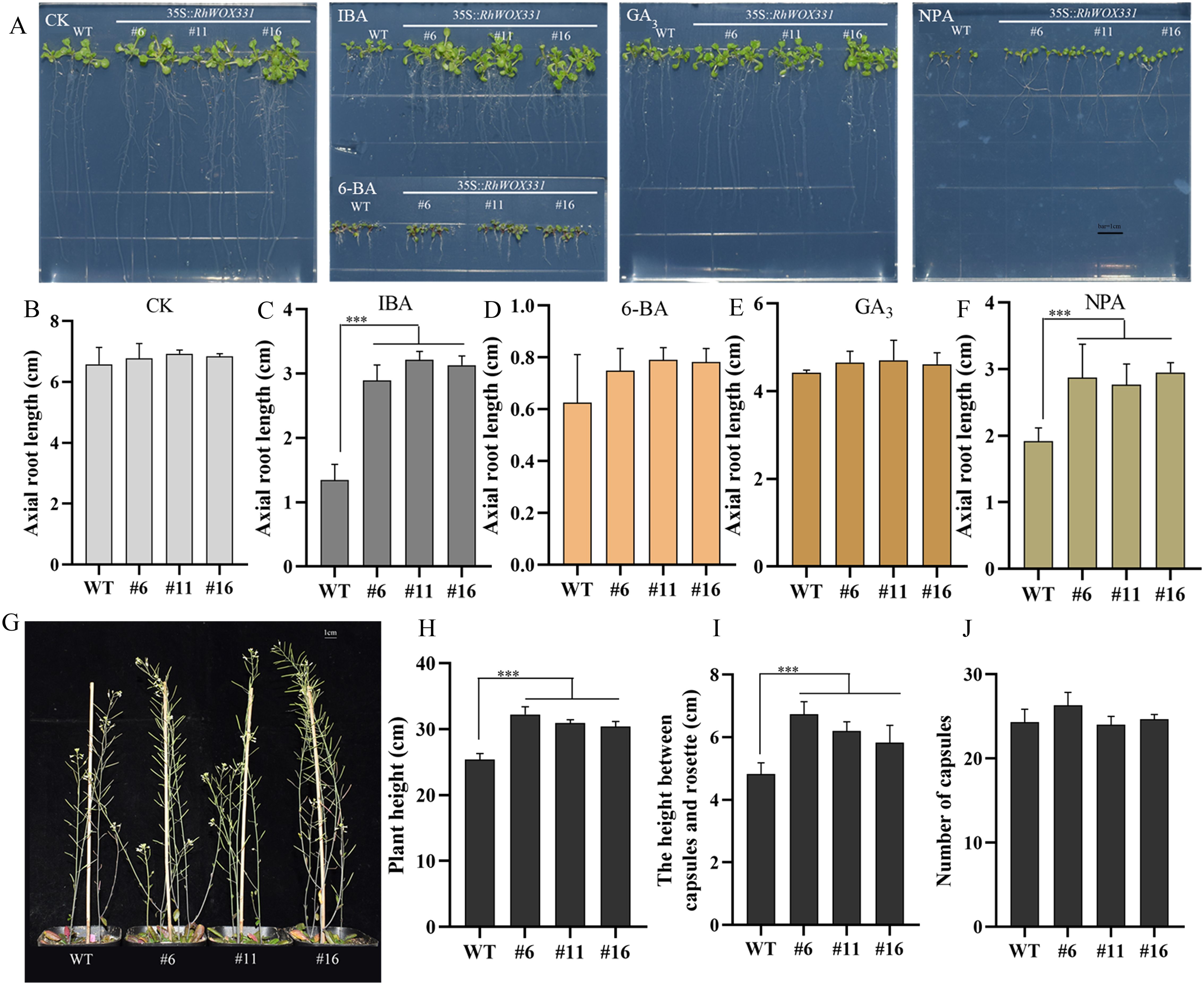
Figure 6. Growth of A thaliana seeds overexpressing RhWOX331. (A) Primary root length of WT and RhWOX331 overexpressing A. thaliana seeds after 14 days of cultivation in different mediums. CK) hormone-free medium; IBA) medium containing 0.25 mg/L IBA; 6-BA) medium containing 0.5 mg/L 6-BA; GA3) medium containing 1 mg/L GA3; NPA) medium containing 10 μM NPA. (B-F) Primary root length of WT and RhWOX331 overexpressing A. thaliana seeds in different medium. Bar = 1 cm. (B) hormone-free medium; (C) medium containing 0.25 mg/L IBA; (D) medium containing 0.5 mg/L 6-BA; (E) medium containing 1 mg/L GA3; (F) medium containing 10 μM NPA. (G) The phenotypes of mature WT and RhWOX331 overexpressing A. thaliana plants. Bar = 1 cm. (H) The plant height of mature WT and RhWOX331 overexpressing A. thaliana plants. (I) The height between the capsules and the rosette of mature WT and RhWOX331 overexpressing A. thaliana plants. (J) the number of capsules of mature WT and RhWOX331 overexpressing A. thaliana plants. The *** mark indicates significant difference between WT and transgenic lines.
The primary roots of 14-day-old A. thaliana were removed and cultivated on B5 medium containing different hormones. Adventitious root formation in A. thaliana was enhanced on hormone-free medium and medium containing 0.25 mg/L IBA. Overexpression lines exhibited earlier adventitious root emergence, with a greater number and longer lengths of adventitious roots compared to the WT (Figures 7A–C, G, H). The difference in the number of adventitious roots was particularly pronounced. On the medium containing 0.5 mg/L 6-BA, 1 mg/L GA3, and 10 μM NPA, WT plants almost did not form roots after 10 days of culture, whereas RhWOX331 overexpressing plants developed some adventitious roots (Figures 7A, D–F, I–K). In terms of both the number and length of adventitious roots, RhWOX331 overexpressing A. thaliana demonstrated a stronger rooting ability. These results suggest that overexpression of RhWOX331 promotes adventitious root formation in A. thaliana and alleviates the inhibitory effects of some hormones on adventitious root development.
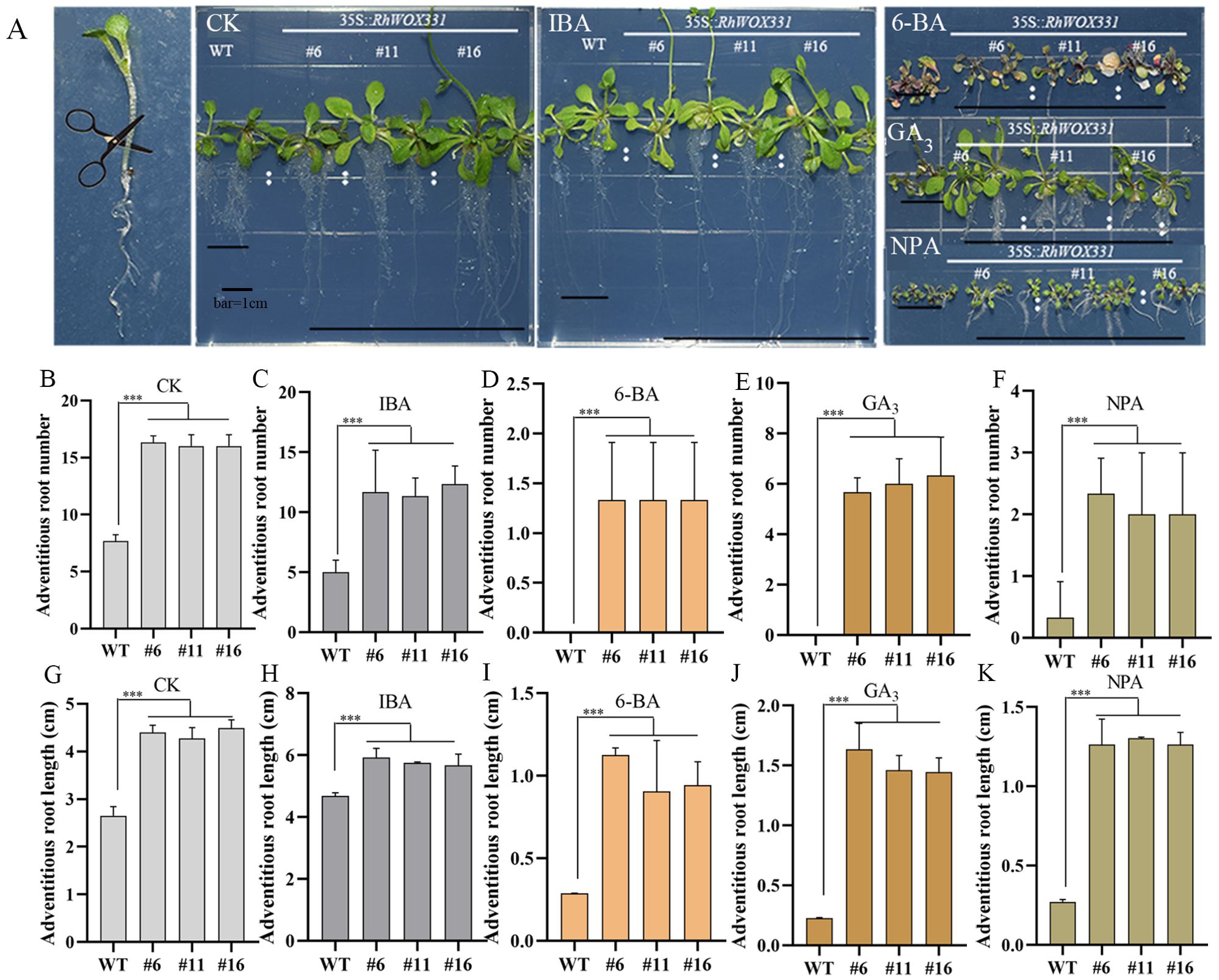
Figure 7. Growth of adventitious roots of WT and RhWOX331 overexpressing A. thaliana in medium containing different hormones. (A) After removal of the primary root, WT and RhWOX331 overexpressing A. thaliana developed adventitious roots on medium containing different hormones: CK) hormone-free medium; IBA) medium containing 0.25 mg/L IBA; 6-BA) medium containing 0.5 mg/L 6-BA; GA3) medium containing 1 mg/L GA3; NPA) medium containing 10 μM NPA. (B-F) The number of adventitious roots occurring in WT and overexpressed RhWOX331 A thaliana in different medium. Bar = 1 cm. (B) hormone-free medium; (C) medium containing 0.25 mg/L IBA; (D) medium containing 0.5 mg/L 6-BA; (E) medium containing 1 mg/L GA3; (F) medium containing 10 μM NPA. (G-K) Length of adventitious roots occurring in WT and overexpressed RhWOX331 A. thaliana in different mediums. (G) hormone-free medium; (H) medium containing 0.25 mg/L IBA; (I) medium containing 0.5 mg/L 6-BA; (J) medium containing 1 mg/L GA3; (K) medium containing 10 μM NPA. The *** mark indicates significant difference between WT and transgenic lines.
By predicting the approximately 2000bp sequence upstream of the RhWOX331 gene start codon, it was found that this sequence contained abundant cis-regulatory elements (Supplementary Figure S2), which may be one of the reasons that RhWOX331 was regulated by many hormones, such as IBA. According to the position of the TATA-box, the 2113bp sequence was divided into three segments (Figure 8A), pWOX331-1::GUS, pWOX331-2::GUS, and pWOX331-3::GUS vectors were constructed and transformed into A. thaliana (Figure 8B). P1, P2, and P3 represent A. thaliana transformed with pWOX331-1:: GUS, pWOX331-2:: GUS, and pWOX331-3:: GUS, respectively. Observation of GUS staining in 7-day-old A. thaliana seedlings revealed no blue spots in the WT plants, while GUS signals were detected at the shoot apical meristem and cotyledonary node in overexpressing plants carrying pWOX331-1::GUS and pWOX331-2::GUS vectors (Figure 8C). GUS activity of RhWOX331 promoter showed the same results (Figure 8D). Considering that no GUS signal was detected in transgenic A. thaliana after adventitious root formation, it was speculated that WOX331 played a role before visible adventitious root formation. These results indicate that the promoter of WOX331 is located between 731bp and 2113bp. In addition to regulating adventitious root formation, RhWOX331 also plays a role in the growth point of A. thaliana cotyledons.
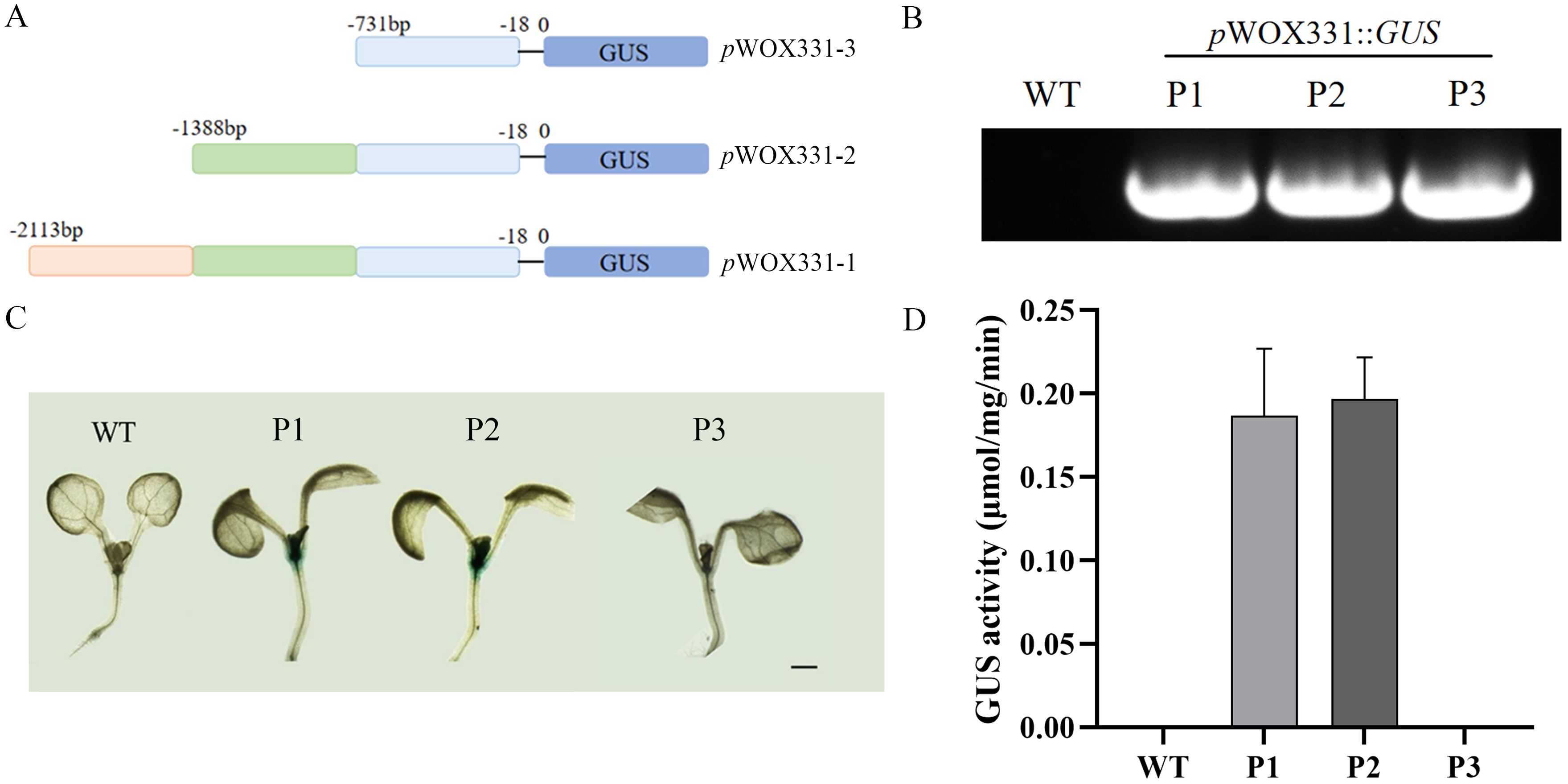
Figure 8. Analysis of GUS activity of RhWOX331. (A) Schematic diagram of RhWOX331 GUS vector construction. (B) Identification of A. thaliana transformed with promoter of RhWOX331. P1, P2, and P3 represent A. thaliana transformed with pWOX331-1::GUS, pWOX331-2::GUS and pWOX331-3::GUS, respectively. (C) GUS staining of WT and A. thaliana overexpressing pWOX331. Bar = 1 mm. (D) GUS activity of WT and A. thaliana overexpressing pWOX331.
A total of 381 WOX genes were identified in rose, a number significantly higher than that found in other species, including 18 in A. thaliana, 28 in N. tabacum, 26 in P. trichocarpa (Figure 1), 18 in Eriobotrya japonica (Yu et al., 2022) and 33 in Glycine max (Hao et al., 2019). The occurrence of more than 100 members in the WOX gene family was not unique to roses. Other species within the Rosa genus which had published genomes also had a relatively large number of WOX genes. Rosa multiflora contained 170 WOX genes, and 105 WOX genes were identified in Rosa rugosa. The number of WOX genes in rose was substantially higher than that in other species, but in other Rosaceae species, the number of WOX genes was not particularly high. There were 9-14 WOX gene family members in Pyrus bretschneideri and other Rosaceae species (Cao et al., 2017). Lv identified WOX gene family members in nine Prunus species, ranging from 6 to 40 (Lv et al., 2023). The number of WOX genes in R. chinensis, R. multiflora and R. rugosa was also above average, suggesting that the large-scale expansion of the WOX gene family was a phenomenon specific to the genus Rosa. The genetic background of R. chinesis was relatively complex, and Rosa multiflora and Rosa rugosa might be involved in the breeding process of this species (Cui et al., 2022). The WOX gene may have replicated during this process. At present, there is no analysis on the WOX function of roses, and more genetic functional evidence is needed to determine the specific significance of this replication process. Whole genome duplications (WGD) are the primary driver of WOX family evolution (Cao et al., 2017). In Bromeliaceae plants, the CAM-related gene families had experienced accelerated expansion, supporting gene family evolution as a driver of CAM evolution (Groot Crego et al., 2024). Abubakar identified four segmental duplications and one tandem duplication of WOX gene family in Boehmeria nivea (Abubakar et al., 2023), which suggested that whole-genome duplication (WGD) had contributed to the expansion of the WOX gene family in B. nivea. During the Paleocene-Eocene boundary, Rosaceae underwent a WGD event, leading to extensive gene duplication (Xiang et al., 2017). The entire Malus genus experienced a WGD event, resulting in the duplication of several MADS-box genes potentially linked to pome formation during that period (Zhang et al., 2023). We hypothesize that roses might have undergone WGD during long-term evolution, leading to the expansion of the RcWOX gene family, enabling them to adapt to various complex growth environments. After analyzing the collinearity within the rose genome, it is found that the number of collinear genes in rose is 0. Therefore, further data evidence is needed to explain the significant expansion of the WOX gene family in rose.
The rooting process of roses is jointly regulated by many genes, but not all members of the WOX gene family are involved in this process. Apart from classical clades, most of the WOX genes in clades I-V showed no expression during the rose rooting process. Among the 364 WOX members from clades I to V, 359 members showed almost no expression during the rooting process of rose, with only RhWOX276, RhWOX51, RhWOX33, RhWOX284, and RhWOX372 genes exhibiting transcriptional expression counts higher than 10 during three or more periods. The classical clades in RcWOX gene family members in rose demonstrated similar structures and the presence of UTR in most cases. Conversely, the majority of RcWOX family members in clades I-V exhibited UTR loss (Supplementary Figure S1). Similar to classical clades in rose, 14 out of 16 pairs of homologous genes in the soybean GmWOX gene family exhibited relatively conserved exon/intron structures (Hao et al., 2019). Many genes in WOX gene family of rose did not function during the formation of adventitious roots, while the genes in classical clades exhibited relatively high expression levels, suggesting that these genes might play a role in the rose cutting rooting process. Multiple WOX gene family members in different stages of rose rooting responded to cutting signals, such as RcaWOX1 in R. canina callus tissue formation at an early stage (Gao et al., 2014), similar to the expression pattern of RhWOX185 in R. ‘The Fairy’. The homologous gene MdWOX11 of RhWOX331 in apple cuttings reached its highest expression level at 3 days, and its expression was inhibited by 6-BA (Mao et al., 2023), corresponding with the expression trends of genes RhWOX372, RhWOX316, and RhWOX271 in rose. In conclusion, the WOX gene of clades I-V regulating the functions of other aspects of roses require further investigation.
Further research on the expression pattern and function of RhWOX331 in plants revealed that it not only played a role in adventitious root development, but may also be related to plant meristem activity and regulated the development of aboveground and underground parts of plants. Compared to other tissues, the expression level of RhWOX331 gene in rose roots was significantly increased (Figure 5A), similarly, WOX genes in poplar were primarily expressed in roots and leaves (Liu et al., 2014). In Triticum aestivum, the homologous gene TaWOX11 of RhWOX331 was also highly expressed in roots compared to other tissues. In addition, both TaWUS and TaWOX9 were transcriptional activators and the transcription activation regions were located at the C-terminus (Li et al., 2020).
Following IBA signaling, the expression of RhWOX331 was upregulated and its functional role was advanced during the rooting process (Figure 5C). Overexpression of RhWOX331 in A. thaliana demonstrated enhanced primary root and adventitious root formation, indicating the role of RhWOX331 in promoting primary root elongation and adventitious root development in plants (Figures 6, 7). Similarly, in A. thaliana, AtWOX11 and AtWOX12 responded to auxin signals, inducing fate transition of stem cells from the pericycle cells to root founder cells, thereby inducing adventitious root formation (Liu and Xu, 2018). AtWOX11 was involved in the transition of vascular cambium cells to new lateral root primordia primordial cells (Baesso et al., 2018).
The RhWOX331 promoter, pWOX331-1 and pWOX331-2, triggers GUS protein expression in the meristematic region, indicating the gene’s regulation of plant meristematic activity (Figure 8). Additionally, auxin signaling can be detected in this area during A. thaliana embryogenesis (Baesso et al., 2018), suggesting that pWOX331-2 may overlap with auxin signaling to regulate embryonic development. Indeed, during adventitious root formation in A. thaliana, the distribution of auxin response coincides with the expression region of WOX11, directly responding to the maximum auxin level in the wound-induced pericycle. In rice crown root development, WOX11 might integrate auxin and cytokinin signaling to regulate the expression of RR2 (Type-A cytokinin-responsive regulator) genes in the crown root primordium, thereby regulating cell proliferation (Zhao et al., 2009). WOX gene family played an important role in embryogenesis and shoot apical meristem establishment in conifers (Bueno et al., 2021). Therefore, we propose that RhWOX331 can respond to auxin signals, regulate plant meristematic activity, and positively correlate with the development of both aboveground and underground parts of plants.
The study identified 381 WOX genes in Rosa chinensis through whole-genome bioinformatics analysis. Phylogenetic analysis and evolutionary tree construction classified the RcWOX gene family into eight clades. Gene structure and promoter cis-element analysis revealed that genes within the same clade exhibit similar structures and functions. Chromosomal localization of RcWOX genes in roses indicated significant expansion on chromosome 2. Relative expression analysis of nine WOX gene family members during rose rooting identified several genes with significant expression changes in this process. The RhWOX331 gene, potentially associated with rooting, was identified through tissue-specific expression analysis, showing high expression in roots and inducibility by IBA while being suppressed by NPA. RhWOX331 located to the nucleus and exhibited yeast self-activation activity. Overexpression of the RhWOX331 gene significantly increased the number of lateral roots on the primary root and enhanced the height of A. thaliana. Additionally, it accelerated adventitious root formation and alleviated the inhibition of adventitious root initiation by certain hormones. This gene functioned at the growth point of A. thaliana cotyledons. Our study provides initial insights into the role of RhWOX331 in the process of adventitious root formation in R. ‘The Fairy’, offering direction and inspiration for future research on the WOX gene family of rose.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
LD: Formal analysis, Investigation, Validation, Writing – original draft, Visualization, Writing – review & editing. ZH: Investigation, Validation, Writing – review & editing. WZ: Formal analysis, Writing – review & editing. SL: Investigation, Writing – review & editing. MH: Formal analysis, Writing – review & editing. JZ: Resources, Writing – review & editing. TY: Resources, Writing – review & editing. JD: Methodology, Supervision, Writing – review & editing. DC: Conceptualization, Funding acquisition, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the China Postdoctoral Science Foundation (2023M730535) and National Natural Science Foundation of China (No. 31971700).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1461322/full#supplementary-material
Supplementary Figure 1 | Conserved motifs and conserved domains of RcWOXs in Rosa chinensis. (A) Motif composition of RcWOX proteins, with different colors representing twenty distinct motifs. (B) Conserved domains of RcWOXs, with various colors indicating different structural domains. (C) Green rectangles denote untranslated regions (UTRs); yellow rectangles represent coding sequences (CDS) or exons; black lines indicate introns.
Supplementary Figure 2 | Cis-acting element analysis of RcWOXs. Each cis-acting element is indicated by a different color.
Supplementary Figure 3 | Characterization of A. thaliana overexpressing RhWOX331.
Abubakar, A. S., Wu, Y., Chen, F., Zhu, A., Chen, P., Chen, K., et al. (2023). Comprehensive analysis of WUSCEL-related homeobox gene family in Ramie (Boehmeria nivea) indicates its potential role in adventitious root development. Biology 12, 1475. doi: 10.3390/biology12121475
Baesso, B., Chiatante, D., Terzaghi, M., Zenga, D., Nieminen, K., Mahonen, A. P., et al. (2018). Transcription factors PRE3 and WOX11 are involved in the formation of new lateral roots from secondary growth taproot in A. thaliana. Plant Biol. Stuttg. Ger. 20, 426–432. doi: 10.1111/plb.12711
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Bannoud, F., Bellini, C. (2021). Adventitious rooting in populus species: update and perspectives. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.668837
Bellini, C., Pacurar, D. I., Perrone, I. (2014). Adventitious roots and lateral roots: similarities and differences. Annu. Rev. Plant Biol. 65, 639–666. doi: 10.1146/annurev-arplant-050213-035645
Bent, A. (2006). Arabidopsis thaliana floral dip transformation method. Methods Mol. Biol. Clifton NJ 343, 87–103. doi: 10.1385/1-59745-130-4:87
Bueno, N., Cuesta, C., Centeno, M. L., Ordás, R. J., Alvarez, J. M. (2021). In vitro plant regeneration in conifers: the role of WOX and KNOX gene families. Genes 12, 438. doi: 10.3390/genes12030438
Cao, Y., Han, Y., Meng, D., Li, G., Li, D., Abdullah, M., et al. (2017). Genome-Wide Analysis Suggests the Relaxed Purifying Selection Affect the Evolution of WOX Genes in Pyrus bretschneideri, Prunus persica, Prunus mume, and Fragaria vesca. Front. Genet. 8. doi: 10.3389/fgene.2017.00078
Che, G., Gu, R., Zhao, J., Liu, X., Song, X., Zi, H., et al. (2020). Gene regulatory network controlling carpel number variation in cucumber. Dev. Camb. Engl. 147, dev184788. doi: 10.1242/dev.184788
Chen, C., Wu, Y., Li, J., Wang, X., Zeng, Z., Xu, J., et al. (2023). TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 16, 1733–1742. doi: 10.1016/j.molp.2023.09.010
Cheng, S., Huang, Y., Zhu, N., Zhao, Y. (2014). The rice WUSCHEL-related homeobox genes are involved in reproductive organ development, hormone signaling and abiotic stress response. Gene 549, 266–274. doi: 10.1016/j.gene.2014.08.003
Cui, W.-H., Du, X.-Y., Zhong, M.-C., Fang, W., Suo, Z.-Q., Wang, D., et al. (2022). Complex and reticulate origin of edible roses (Rosa, Rosaceae) in China. Hortic. Res. 9, uhab051. doi: 10.1093/hr/uhab051
De Klerk, G.-J., van der Krieken, W., De Jong, J. C. (1999). Review the formation of adventitious roots: New concepts, new possibilities. Vitro Cell. Dev. Biol. - Plant 35, 189–199. doi: 10.1007/s11627-999-0076-z
Deyhle, F., Sarkar, A. K., Tucker, E. J., Laux, T. (2007). WUSCHEL regulates cell differentiation during anther development. Dev. Biol. 302, 154–159. doi: 10.1016/j.ydbio.2006.09.013
Dong, J., Cao, L., Zhang, X., Zhang, W., Yang, T., Zhang, J., et al. (2021). An R2R3-MYB transcription factor rmMYB108 responds to chilling stress of rosa multiflora and conferred cold tolerance of arabidopsis. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.696919
Fan, Y., Gao, P., Zhou, T., Pang, S., Zhang, J., Yang, T., et al. (2023). Genome-Wide Identification and Expression Analysis of the Trehalose-6-phosphate Synthase and Trehalose-6-phosphate Phosphatase Gene Families in Rose (Rosa hybrida cv ‘Carola’) under Different Light Conditions. Plants Basel Switz. 13, 114. doi: 10.3390/plants13010114
Gao, B., Wen, C., Fan, L., Kou, Y., Ma, N., Zhao, L. (2014). A Rosa canina WUSCHEL-related homeobox gene, RcWOX1, is involved in auxin-induced rhizoid formation. Plant Mol. Biol. 86, 671–679. doi: 10.1007/s11103-014-0255-0
Ge, Y., Liu, J., Zeng, M., He, J., Qin, P., Huang, H., et al. (2016). Identification of WOX family genes in selaginella kraussiana for studies on stem cells and regeneration in lycophytes. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00093
Groot Crego, C., Hess, J., Yardeni, G., de la Harpe, M., Priemer, C., Beclin, F., et al. (2024). CAM evolution is associated with gene family expansion in an explosive bromeliad radiation. Plant Cell koae130. doi: 10.1093/plcell/koae130
Hao, Q., Zhang, L., Yang, Y., Shan, Z., Zhou, X. (2019). Genome-wide analysis of the WOX gene family and function exploration of gmWOX18 in soybean. Plants 8, 215. doi: 10.3390/plants8070215
Hu, B., Jin, J., Guo, A.-Y., Zhang, H., Luo, J., Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Jiang, F. X., Liu, F.-L., Zhao, L. J. (2012). Overexpression of RaWUS gene of Rosa canina regeneration from root tip of transgenic inducing shoot tobacco. Sci. Silvae Sin. 47, 43–52. doi: 10.1097/RLU.0b013e3181f49ac7
Jung, S., Lee, T., Cheng, C.-H., Buble, K., Zheng, P., Yu, J., et al. (2019). 15 years of GDR: New data and functionality in the Genome Database for Rosaceae. Nucleic Acids Res. 47, D1137–D1145. doi: 10.1093/nar/gky1000
Koo, J., Kim, Y., Kim, J., Yeom, M., Lee, I. C., Nam, H. G. (2007). A GUS/luciferase fusion reporter for plant gene trapping and for assay of promoter activity with luciferin-dependent control of the reporter protein stability. Plant Cell Physiol. 48, 1121–1131. doi: 10.1093/pcp/pcm081
Lamesch, P., Berardini, T. Z., Li, D., Swarbreck, D., Wilks, C., Sasidharan, R., et al. (2012). The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40, D1202–D1210. doi: 10.1093/nar/gkr1090
Laux, T., Mayer, K. F., Berger, J., Jürgens, G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Dev. Camb. Engl. 122, 87–96. doi: 10.1242/dev.122.1.87
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, R., Gao, P., Yang, T., Dong, J., Chen, Y., Xie, Y., et al. (2023). Genome-wide analysis of the SWEET transporters and their potential role in response to cold stress in Rosa rugosa. Horticulturae 9, 1212. doi: 10.3390/horticulturae9111212
Li, J., Jia, H., Zhang, J., Liu, B., Hu, J., Wang, L., et al. (2018). Effect of Overexpression of Populus tomentosa WUSCHEL-related homeobox 4 (PtoWOX4a) on the Secondary Growth of Poplar. Linye KexueScientia Silvae Sin. 54, 52–59. doi: 10.11707/j.1001-7488.20180206
Li, Z., Liu, D., Xia, Y., Li, Z., Jing, D., Du, J., et al. (2020). Identification of the WUSCHEL-related homeobox (WOX) gene family, and interaction and functional analysis of taWOX9 and taWUS in wheat. Int. J. Mol. Sci. 21, 1581. doi: 10.3390/ijms21051581
Liu, B., Wang, L., Zhang, J., Li, J., Zheng, H., Chen, J., et al. (2014). WUSCHEL-related Homeobox genes in Populus tomentosa: diversified expression patterns and a functional similarity in adventitious root formation. BMC Genomics 15, 296. doi: 10.1186/1471-2164-15-296
Liu, W., Xu, L. (2018). Recruitment of IC-WOX genes in root evolution. Trends Plant Sci. 23, 490–496. doi: 10.1016/j.tplants.2018.03.011
Lv, J., Feng, Y., Jiang, L., Zhang, G., Wu, T., Zhang, X., et al. (2023). Genome-wide identification of WOX family members in nine Rosaceae species and a functional analysis of MdWOX13-1 in drought resistance. Plant Sci. 328, 111564. doi: 10.1016/j.plantsci.2022.111564
Mao, J., Niu, C., Li, K., Fan, L., Liu, Z., Li, S., et al. (2023). Cytokinin-responsive MdTCP17 interacts with MdWOX11 to repress adventitious root primordium formation in apple rootstocks. Plant Cell 35, 1202–1221. doi: 10.1093/plcell/koac369
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
Ohmori, Y., Tanaka, W., Kojima, M., Sakakibara, H., Hirano, H.-Y. (2013). WUSCHEL-RELATED HOMEOBOX4 is involved in meristem maintenance and is negatively regulated by the CLE gene FCP1 in rice. Plant Cell 25, 229–241. doi: 10.1105/tpc.112.103432
Palovaara, J., Hallberg, H., Stasolla, C., Hakman, I. (2010). Comparative expression pattern analysis of WUSCHEL-related homeobox 2 (WOX2) and WOX8/9 in developing seeds and somatic embryos of the gymnosperm Picea abies. New Phytol. 188, 122–135. doi: 10.1111/j.1469-8137.2010.03336.x
Raymond, O., Gouzy, J., Just, J., Badouin, H., Verdenaud, M., Lemainque, A., et al. (2018). The Rosa genome provides new insights into the domestication of modern roses. Nat. Genet. 50, 772. doi: 10.1038/s41588-018-0110-3
Reiser, L., Modrusan, Z., Margossian, L., Samach, A., Ohad, N., Haughn, G. W., et al. (1995). The BELL1 gene encodes a homeodomain protein involved in pattern formation in the Arabidopsis ovule primordium. Cell 83, 735–742. doi: 10.1016/0092-8674(95)90186-8
Schmittgen, T. D., Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Shuang, W., Yang, Z., Meng-Xuan, R., Ying-Ying, L., Zhi-Gang, W. (2019). Genome-wide analysis of the WOX family reveals their involvement in stem growth of populus trichocarpa. Bull. Bot. Res. 39, 568. doi: 10.7525/J.ISSN.1673-5102.2019.04.011
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tanaka, W., Ohmori, Y., Ushijima, T., Matsusaka, H., Matsushita, T., Kumamaru, T., et al. (2015). Axillary meristem formation in rice requires the WUSCHEL ortholog TILLERS ABSENT1. Plant Cell 27, 1173–1184. doi: 10.1105/tpc.15.00074
van der Graaff, E., Laux, T., Rensing, S. A. (2009). The WUS homeobox-containing (WOX) protein family. Genome Biol. 10, 248. doi: 10.1186/gb-2009-10-12-248
Walker, J. M. (Ed.) (2005). The Proteomics Protocols Handbook (Totowa, NJ: Humana Press). doi: 10.1385/1592598900
Wang, J., Chitsaz, F., Derbyshire, M. K., Gonzales, N. R., Gwadz, M., Lu, S., et al. (2023). The conserved domain database in 2023. Nucleic Acids Res. 51, D384–D388. doi: 10.1093/nar/gkac1096
Xiang, Y., Huang, C.-H., Hu, Y., Wen, J., Li, S., Yi, T., et al. (2017). Evolution of rosaceae fruit types based on nuclear phylogeny in the context of geological times and genome duplication. Mol. Biol. Evol. 34, 262–281. doi: 10.1093/molbev/msw242
Yu, Y., Yang, M., Liu, X., Xia, Y., Hu, R., Xia, Q., et al. (2022). Genome-wide analysis of the WOX gene family and the role of EjWUSa in regulating flowering in loquat (Eriobotrya japonica). Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1024515
Zhang, J., Eswaran, G., Alonso-Serra, J., Kucukoglu, M., Xiang, J., Yang, W., et al. (2019). Transcriptional regulatory framework for vascular cambium development in Arabidopsis roots. Nat. Plants 5, 1033–1042. doi: 10.1038/s41477-019-0522-9
Zhang, Y., Jiao, Y., Jiao, H., Zhao, H., Zhu, Y.-X. (2017). Two-step functional innovation of the stem-cell factors WUS/WOX5 during plant evolution. Mol. Biol. Evol. 34, 640–653. doi: 10.1093/molbev/msw263
Zhang, L., Morales-Briones, D. F., Li, Y., Zhang, G., Zhang, T., Huang, C.-H., et al. (2023). Phylogenomics insights into gene evolution, rapid species diversification, and morphological innovation of the apple tribe (Maleae, Rosaceae). New Phytol. 240, 2102–2120. doi: 10.1111/nph.19175
Keywords: Rosa hybrida, WOX gene family, RhWOX331, adventitious roots, gene function analysis
Citation: Duan L, Hou Z, Zhang W, Liang S, Huangfu M, Zhang J, Yang T, Dong J and Che D (2024) Genome-wide analysis of the WOX gene family and function exploration of RhWOX331 in rose (R. ‘The Fairy’). Front. Plant Sci. 15:1461322. doi: 10.3389/fpls.2024.1461322
Received: 08 July 2024; Accepted: 13 August 2024;
Published: 03 September 2024.
Edited by:
Huihui Li, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Tangren Cheng, Beijing Forestry University, ChinaCopyright © 2024 Duan, Hou, Zhang, Liang, Huangfu, Zhang, Yang, Dong and Che. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jie Dong, amllZG9uZ0BudWF1LmVkdS5jbg==; Daidi Che, ZGFpZGljaGVAbmVhdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.