- 1School of Life and Environmental Sciences, The University of Sydney, Sydney, NSW, Australia
- 2The Sydney Institute of Agriculture, The University of Sydney, Sydney, NSW, Australia
- 3The Plant Breeding Institute, The University of Sydney, Sydney, NSW, Australia
Chickpeas are a vital source of protein and starch for a large portion of the world’s population and are known to be impacted by heat stress at every life stage. Previously known as an “Orphan Legume”, little is known of the genetic control of heat stress tolerance, and most previous research has focused on heat avoidance rather than tolerance. This study utilised a population of 148 chickpea genotypes, primarily Kabulis, in 12 field trials conducted at 2 locations, two sowing periods, and across 3 years. Physiology was examined, and data was paired with Diversity Arrays Technology (DArT) sequencing to perform a Genome Wide Association Study to connect phenotypic and genotypic regions. Fourteen QTL related to yield, seed size, time to flowering, time to maturity, and final canopy closure were found. Among these, are the first Quantitative Trait Loci (QTL) ever identified for canopy closure in chickpea, along with a QTL that is likely linked to early flowering under heat stress. Early flowering in this case refers to a cultivar flowering significantly earlier than the others in the genotype set. Additionally, several other QTL provide validation of previous research. These QTL hotspots that can be targeted for selective breeding of several traits concurrently. Overall, new targets for genome assisted breeding for heat tolerance in chickpea were identified and can be utilised by the breeder community to improve the status of selective breeding for heat tolerance in this crop.
1 Introduction
Chickpea (Cicer arietinum) is a crucial source of protein and starch in many countries (Abbo et al., 2003), making up roughly 20% of global pulse production (FAO, 2020). Chickpeas are a nutrient dense food, with high protein, iron, and starch contents, and are an agronomically strong crop, surviving on low nutrient soils through highly efficient nitrogen extraction via symbiosis with rhizobial bacteria (Sharma et al., 2013). Heat stress impacts every stage of plant growth and development (Jeffrey et al., 2021) and the latest Intergovernmental Panel on Climate Change (IPCC) report has found that between 2021-2040 it is very likely that the global surface temperature will increase by 1.5 °C relative to 1850-1900 (IPCC, 2021). It has been shown that high temperatures can decrease potential yield of this crop by 10-15% for every degree that temperature increases beyond the minimum stress level (Upadhyaya et al., 2011), with extreme heat events leading to a complete loss of yield (Kumar et al., 2013). With the increased urgency of mitigating the effect of climate change on global cropping systems, the last two decades have seen an increased focus on breeding for heat and drought tolerance in chickpea (Devasirvatham et al., 2015; Maphosa et al., 2020).
One of the main issues in chickpea genomic research is the relative paucity of sequence information compared to other crops such as wheat (Alonge et al., 2020), even though the chickpea genome is small at roughly 12% that of wheat (Varshney et al., 2017). In the last decade there has been a surge in research linking genetic markers to traits of interest driven by advancements in genotypic technologies (Thudi et al., 2014; Farahani et al., 2019). High density genotyping including over 10,000 SNP markers has been accessible since 2012 which has expediated genetic research in chickpea (Hiremath et al., 2012). This technology has been utilised in a number of studies to further understand genetic diversity, linkage disequilibrium and allelic diversity, essential for chickpea improvement (Thudi et al., 2014; Roorkiwal et al., 2016; Farahani et al., 2019). The most recent genome-wide marker map was produced using Single-Nucleotide Polymorphisms (SNPs) in 2013 (Varshney et al., 2013) with the previous iteration produced with microsatellite Simple Sequence Repeats (SSRs) in 2000 (Winter et al., 2000). Because of this, there is significant inconsistency in the markers used in research since 2000, making it difficult to understand the locations of previously reported marker trait associations (MTA) and quantitative trait loci (QTL).
All Cicer species are diploid, with 16 somatic chromosomes, but there are sufficient differences between chromosome length and chromosomal constrictions to distinguish C. arietinum from the wild progenitor C. reticulatum, while maintaining a large degree of intraspecific variation in karyotype (Varshney et al., 2017). Due to this speciation, early attempts to form heterotic hybrids have been mostly unsuccessful, due to post fertilisation abortion (Ahmad and Slinkard, 2004). As such, the primary direction for crop improvement via genomic technologies seems to be improvement of existing map resources, with the goal of developing enhanced genomic tools to assist breeding (GAB) programs (Pandey et al., 2016).
With the accelerating impacts of climate change, improved understanding of the chickpea genome and validated MTAs for GAB programs, the genetics underlying the physiological response of chickpea to heat stress needs to be further investigated. There have been markers found for traits that are known to be linked to performance under heat stress, such as flowering time, seed size, and plant height (Kumar and Abbo, 2001; Thudi et al., 2014). However, the extent of this research is limited, and many of these studies have been performed on only a small subset of genotypes, missing the full diversity of physiological responses possible in the crop.
This study used 148 chickpea genotypes in two commercial growing regions in Australia to assess physiological traits under typical and heat-stressed conditions. With these physiological trait data, a Genome-Wide Association Study (GWAS) was completed. The aim was to identify key QTL associated with traits related to or impacted by heat stress, to advance future genetic improvement and research into heat tolerance in this key crop. Additionally, the findings of previous research were combined to produce a chromosome map showing key genes and QTL associated with heat stress and similar physiological traits. The objective was to create a simplified resource that depicts known genomic regions of interest, regardless of the marker type that was used to find them.
2 Materials and methods
2.1 Source of germplasm
Germplasm from an international genebank, consisting of 148 genotypes were examined, comprising 10 desi and 137 kabuli types. These genotypes originated from Australia (9), ICARDA (129), and ICRISAT (9), and are a mix of released cultivars and elite breeding lines (Appendix 1). These genotypes were selected to represent a diverse range of responses to heat stress.
2.2 Field trials
In total, data was collected across 3 years, 2 locations, and 2 sowing times, resulting in 12 field trials. Field trials were conducted in Kununurra, Western Australia (15.7783° S, 128.7439° E), and Narrabri, New South Wales (30.2737° S, 149.7350° E), using agronomic management typical of each region. Thus, management practices were not the same at each location. These locations were selected to represent the historical mediterranean environment used for cultivating chickpeas (Narrabri) and a higher temperature environment (Kununurra). Genotypes were sown according to commercial standards (~169 seeds and ~381 seeds per plot in Narrabri and Kununurra respectively) in randomised complete block designs of 2 replicates for each time of sowing (TOS) in plots of 8 m2. Trials in Kununurra were flood-irrigated fortnightly, with regular Nitrogen side-dressing throughout the season, whereas Narrabri trials were irrigated to simulate a typical year, inoculated with rhizobium and a pre-emergent herbicide applied. Temperature was collected in hourly intervals at each location, and daily minima and maxima has been provided in Appendix 2. These data can be found in raw form through weather.agric.wa.gov.au (Kununurra site) and ozforcast.com.au (Narrabri site). Delayed sowing trials were carried out annually between 2018-2020 inclusive, with the first sowing (TOS1) corresponding to the regional commercial season, and the second (TOS2) delayed by 1 month in Kununurra, and 2 in Narrabri. This variation in sowing delay between the regions is due to the shorter season length in Kununurra, caused by consistently higher temperatures than those of Narrabri (Appendix 2).
2.3 Measured traits and data curation
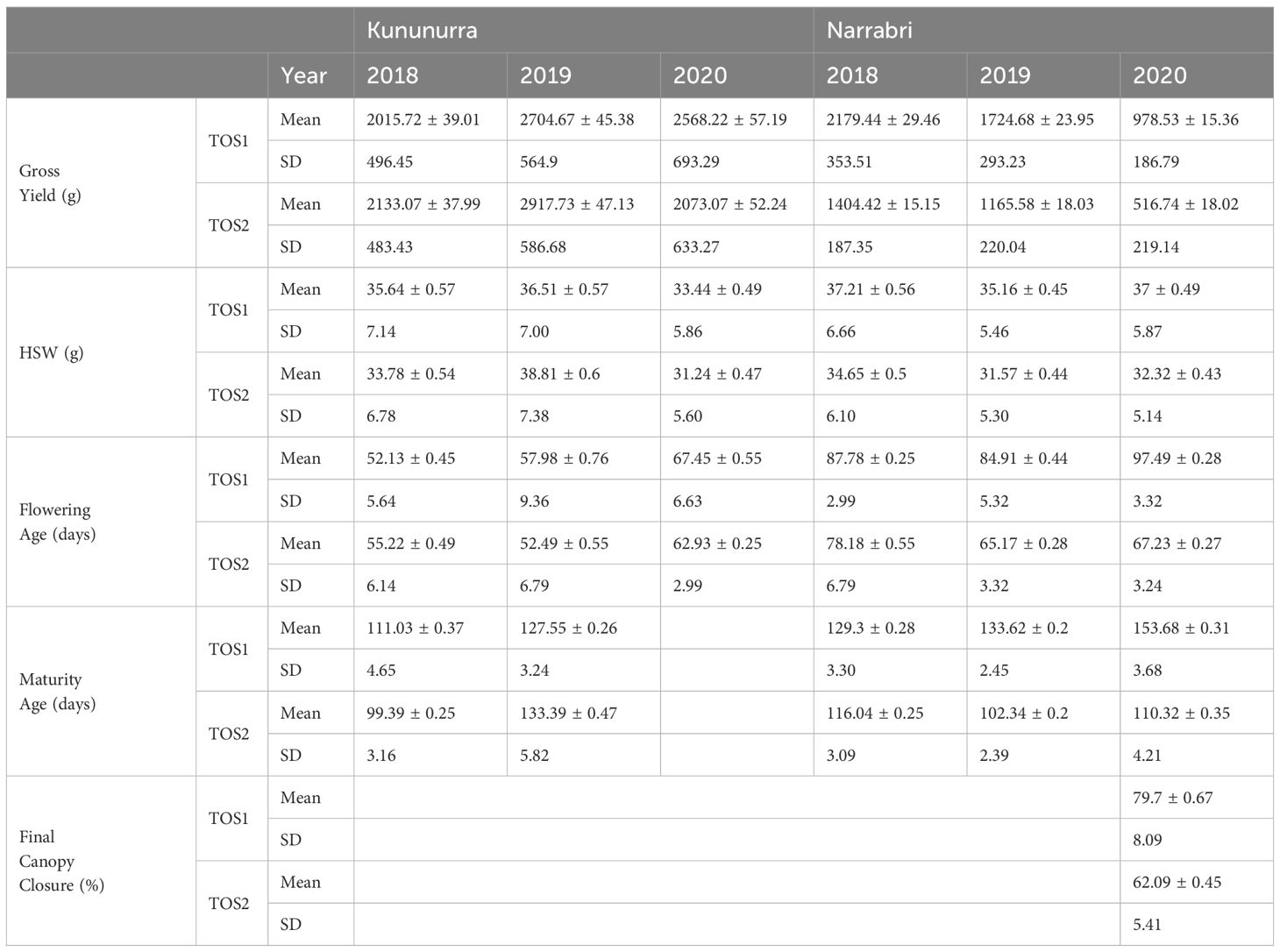
For all 3 years, sowing times and locations, the following traits were measured: Gross yield per plot (grams), hundred seed weight (HSW) (grams), flowering time, and maturity time. Flowering and maturity time were measured via a count of the number of days between sowing and the reaching of this set lifestage, as described in ICRISAT guidelines (IBPGR, ICRISAT, 1993). In addition, canopy closure was measured in the 2019 Narrabri trials using the “canopeo” app (Patrignani and Ochsner, 2015). Canopy closure measurements were taken towards the end of the podding phase, in order to ascertain the maximum canopy closure as a percentage of ground cover. To assess flowering and maturity time, plots were inspected as regularly as possible, typically every 2-3 days, to determine the date at which 50% of the plants in each plot had at least 1 flower and the date at which 50% of each plot was desiccated and a brown colour. These dates were then used to calculate the number of days from sowing to flowering and maturity (Pundir et al., 1988).
Phenotypic data is summarised in Table 1. The predicted means were calculated using linear mixed models in Genstat software (VSN-International, 2022), and subsequent data was organised into Quartile-Quartile (QQ) plots in R, grouped by location and year (Appendix 3). Year, genotypes, and sowing dates were arranged as fixed effects for each location, with range, and row within reps and sowing dates within years arranged as random effects. This was done to determine data conformity to a normal distribution, to assess quality for GWAS analysis.
2.4 DNA extraction
Seeds were sprouted in petri dishes until large enough to produce sufficient leaves for extraction. The first 3-4 young leaves per individual were collected and dried in an Eppendorf tube with silica gel for 2-3 days. Two ball bearings were added to each tube, and tissue was subsequently crushed with a lyser for 2min at 20 rpm. Ball bearings were then removed, and 800 µL of CTAB buffer added to each tube, with samples incubated at 65°C for 30-40 min, followed by 5 min at room temperature. 600 µL Phenol-chloroform (24:1 ratio) was added to each tube, and samples were mixed for 2 min and then centrifuged at 10,000 rpm for 10 min. 600 µL supernatant was removed and placed in a 1.5 mL microcentrifuge tube, followed by 500 µL cold isopropanol. Samples were then chilled at -20°C for 20 min, and then centrifuged at 10,000 rpm for 10 min. Supernatant was discarded, and remaining ethanol was removed through drying. The DNA pellet was washed via the addition of 500 µL washing to each tube, and the use of a centrifuge at 7,000 rpm for 10 min. Samples were then dried, and 100 µL TE (pH 8) and RNAse (1:100 ratio) was added to each tube, before incubation at 37°C for 1-2 hours. DNA samples were diluted with double-distilled autoclaved water to 50 ng/µL and stored at -20°C until use.
2.5 Marker filtering
DNA samples were sent for DArT-SeqTM genotyping through Diversity Arrays technologies Pty Ltd (Canberra, Australia), which returned 3,339 SNP markers. Data was curated by removing poor-quality markers. Markers with minor allele frequency < 5%, markers that failed to provide information for > 20% of lines, and markers without mapped positions on the Cicer arietinum CDC Frontier genome v1.0 map (BioProject PRJNA175619) (Varshney et al., 2013) were removed. 2,361 reported SNPs and 1,919 mapped positions were used for the GWAS analysis.
2.6 Genetic population structure
Population structure was investigated for the 148 genotypes and filtered marker set, principal component analysis (PCA) and phylogenetic cluster dendrogram were used to assess genetic relationships between individuals. Kinship matrix was calculated using the “synbreed” package in R (Wimmer et al., 2012), and visualised as a PCA using ggplot2 (Wickham, 2016) with genotypes colour coded by Desi and Kabuli type chickpeas (Appendix 1, Figure 1).
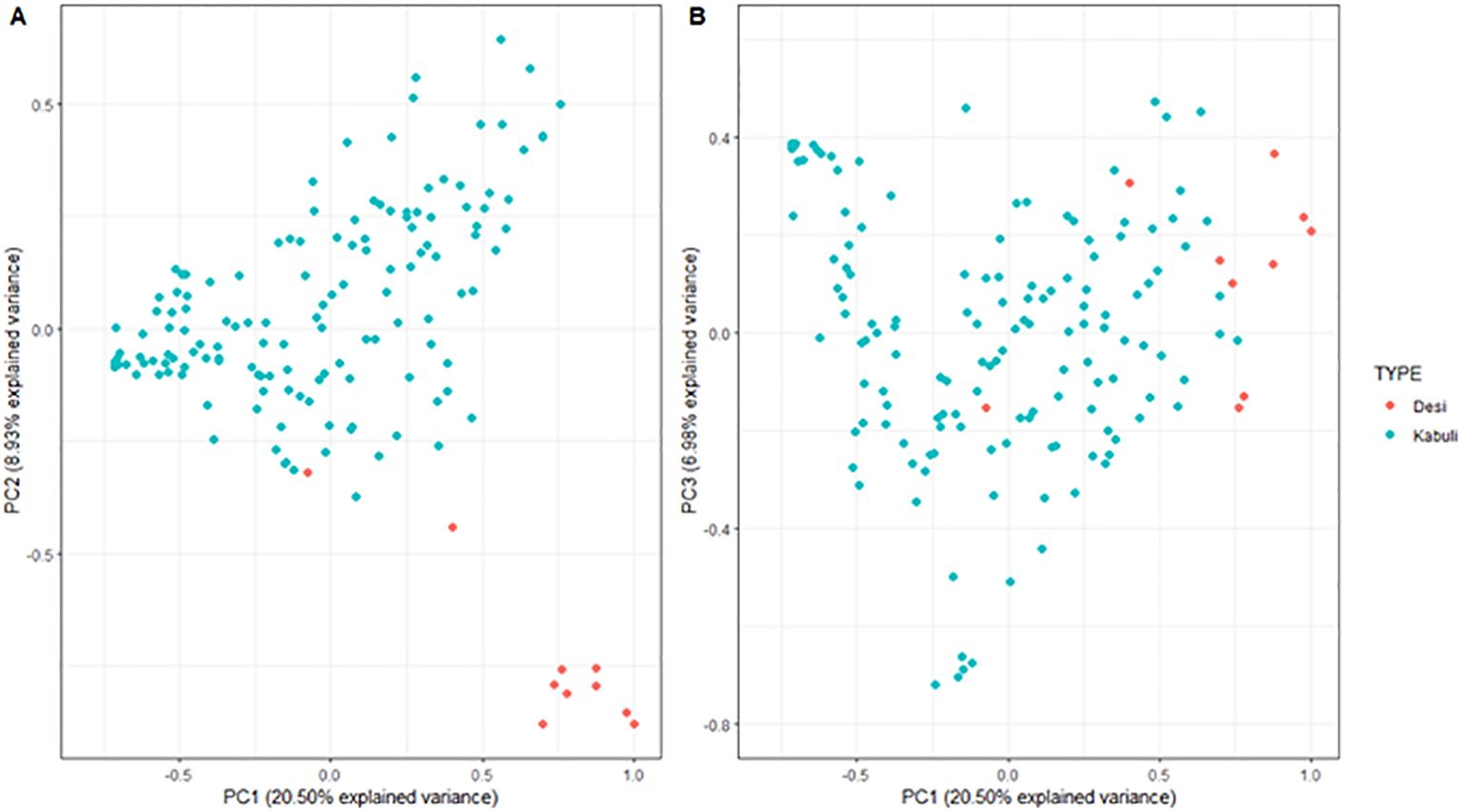
Figure 1. Principal component analysis (PCA) of the kinship matrix visualizing the genetic relationships between147 desi and kabuli chickpea genotypes. The figure on the left (A) represents the first principal component (PC1; x-axis) and the second principal compnent (PC2; y-axis), and the figure on the right (B) represents PC1 and the third principal component (PC3; y-axis). In both plots, genotypes are coloured according to chickpea type.
2.7 Genome wide association analysis and marker projection
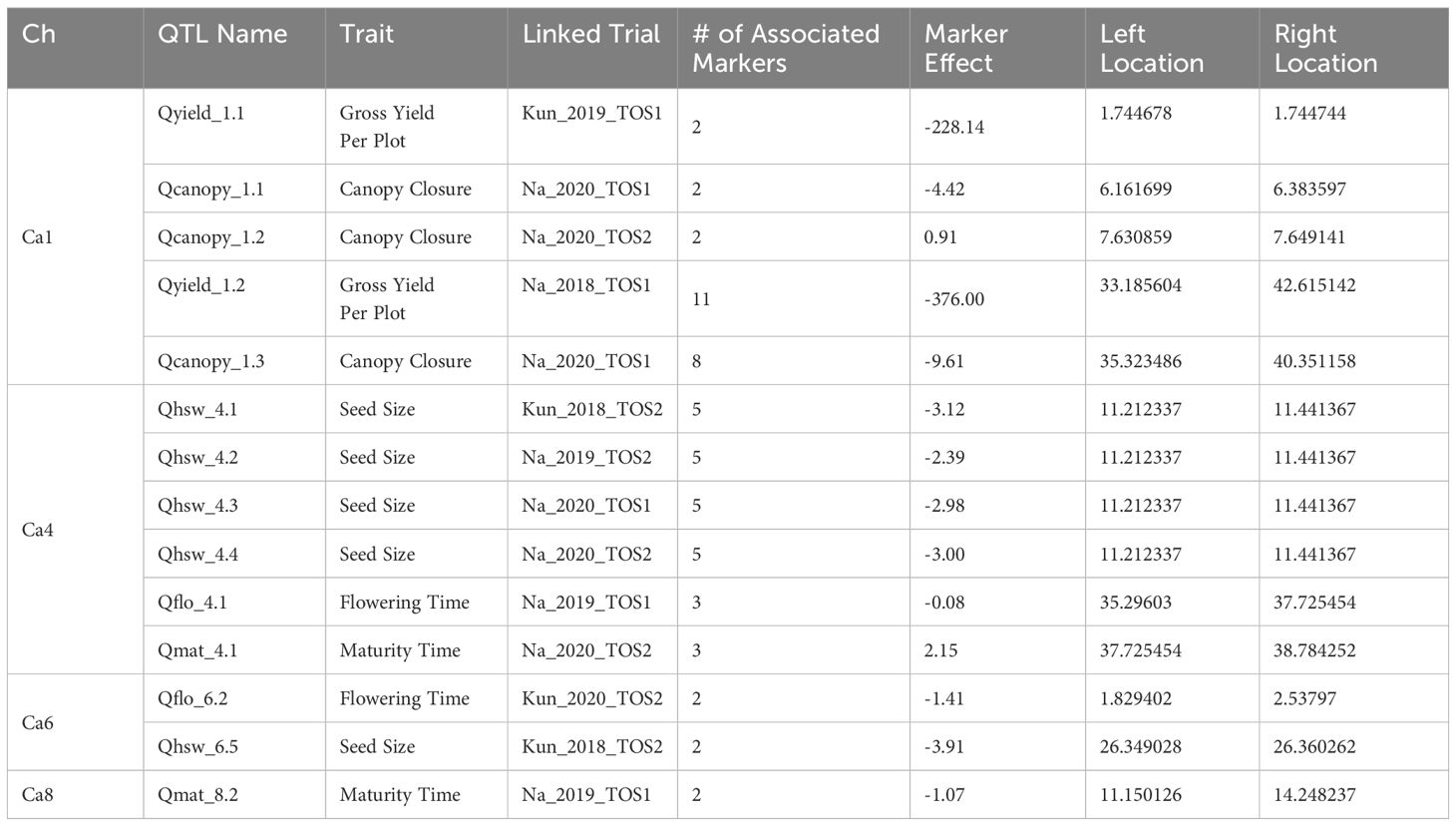
GWAS analysis was used to identify QTL, which refers to an identifiable genomic region that can be associated with the presentation of a particular phenotype (Abiola et al., 2003). GWAS analysis was conducted using the “rrBLUP” package (Endelman, 2011), this package implements the unified mixed-model approach of Yu et al. (2006). Phenotypic data was reasonably normally distributed therefore transformations did not result in any significant changes in the association results. This stability suggests that the rrBLUP model effectively managed deviations from normality and other potential biases in the raw data. Four approaches were trialled, and the most appropriate model selected and implemented. Models tested include: i) naïve model with no measures of relatedness, ii) K model, incorporating the kinship structure, iii) QK model, building on relatedness by chickpea type and origin as a measure of population structure, and iv) QK model using Principal Components (PCs) as a measure of population structure. Final model selected was a QK fixed for chickpea type and including 3 PCs. QQ plots were generated for the -log10 transformed p-values derived from the association tests performed on each phenotypic trait (Appendix 3). Deviations of the points from the red line indicate potential genetic associations with the trait. Upward deviations, particularly at the tail end, suggest the presence of true genetic signals, while flatter regions near the middle indicate a good fit to the null hypothesis. Markers associated with traits at significance level of −log10p ≥ 3 were referred to as MTAs (marker-trait associations), regions with >1 significant marker within 10mbp were considered putative QTL. Manhattan plots were created by plotting the -log10(p) values of the marker-trait associations against their chromosomal positions using the R package ‘qqman’ (Turner, 2018). Genome-wide significance thresholds were set at -log10(p) = 3 (Supplementary Figure 3). Marker effects were estimated utilising the GWASPoly package (Rosyara et al., 2016). Visualisation was performed using Mapchart 2.32 (Voorrips, 2002). Known genes and QTL were located from existing literature, and their sequences identified using the NCBI Basic Local Alignment Search Tool (BLAST) (Sayers et al., 2022). These genes were added to the same map to determine validity of data and existence of novel QTL. Novel QTL were named according to their identified chromosome, their order, and the physiological trait associated. In total, 48 GWAS (148 individuals, across 2 environments, 3 years, 2 TOS, and 5 traits, minus 12 missing environments in Table 1) were conducted. From this, 56 significant MTA were identified, contributing to 14 QTL identified across 4 chromosomes. QTL were defined within each analysis, leading to different trials producing different QTL.
3 Results
3.1 Genotypic and phenotypic diversity
Principal component analysis PC1 and PC2 explained the accumulated genotypic variation of 20.50% and 8.93%, respectively (Figure 1). Genotypic marker data, clustered by chickpea type (Desi and Kabuli), there was sufficient overlap to allow interbreeding among genotypes, and the population has suitable diversity to mine for QTL. Along with this genetic variation, there was a strong relationship between environment and trait performance, demonstrated by the variation among all treatment levels (Year, TOS, Environment) and across all traits (Table 1), while phenotypic data remained within the limits of normal distribution.
3.2 Novel and validated QTL identified by this study
This study identified a total of 14 QTL across 8 genomic regions, relating to gross yield, HSW, flowering age, maturity age, and final canopy closure. The QTL were located on chromosomes 1, 4, 6 and 8 (Table 2; Figure 2). Five QTL, one for each trait; were novel, located in regions that have not previously been linked to traits of interest, two of which were co-located (Figure 2). For each of the measured traits, there was at least 1 QTL aligning with genetic region associated with previously described QTL (Figure 2). QTL for flowering are the most extensively reported in literature (Table 3), whereas for this study, the trait with the highest representation of QTL was HSW (Figure 2, Table 2). Most QTL were related to the Narrabri field trials (10 of 14) particularly TOS1 (6), conversely there were more linked to the Kununurra TOS2 trials (3) than TOS1 (1) (Table 2). QTL associated with yield were detected in TOS1 trials, one at each location, whereas those for HSW span multiple trials (Table 2).
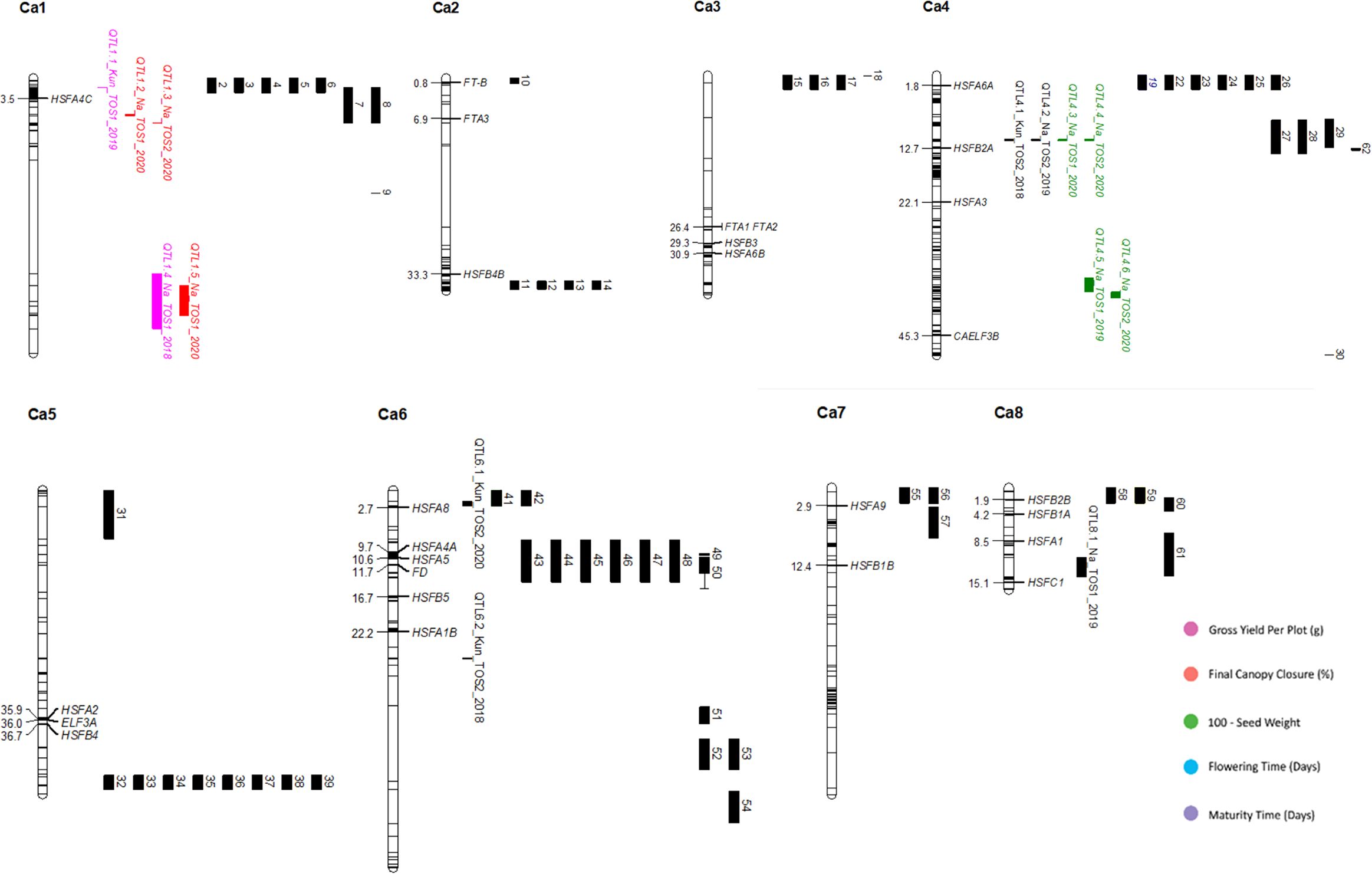
Figure 2. Physical map of QTLs identified in this study, along with the location of relevant and related gene and QTL findings from previous research. Chromosomes are represented as white vertical lines, with mapping markers denoted by black lines across the chromosomes. Genes are positioned according to Mb, and external QTLs are represented by black bands, numbered in order of chromosome and position, to the right of their relevant chromosome. QTLs identified by this study are represented by coloured bands, and labelled according to their linked trait. Positions are based on the CDC Frontier genome assembly (BioProject ID: PRJNA175619, NCBI Reference: NC_011163.1). A complete list of genes and QTLs, including their original position formats can be found in Supplementary Data Sheet 1. Colours denote putative roles in the following traits: Pink, total yield per plot (grams); Orange, final canopy closure (percentage); Green, 100-seed weight; Blue, days to 50% plot flowering; Purple, days to 50% maturation.
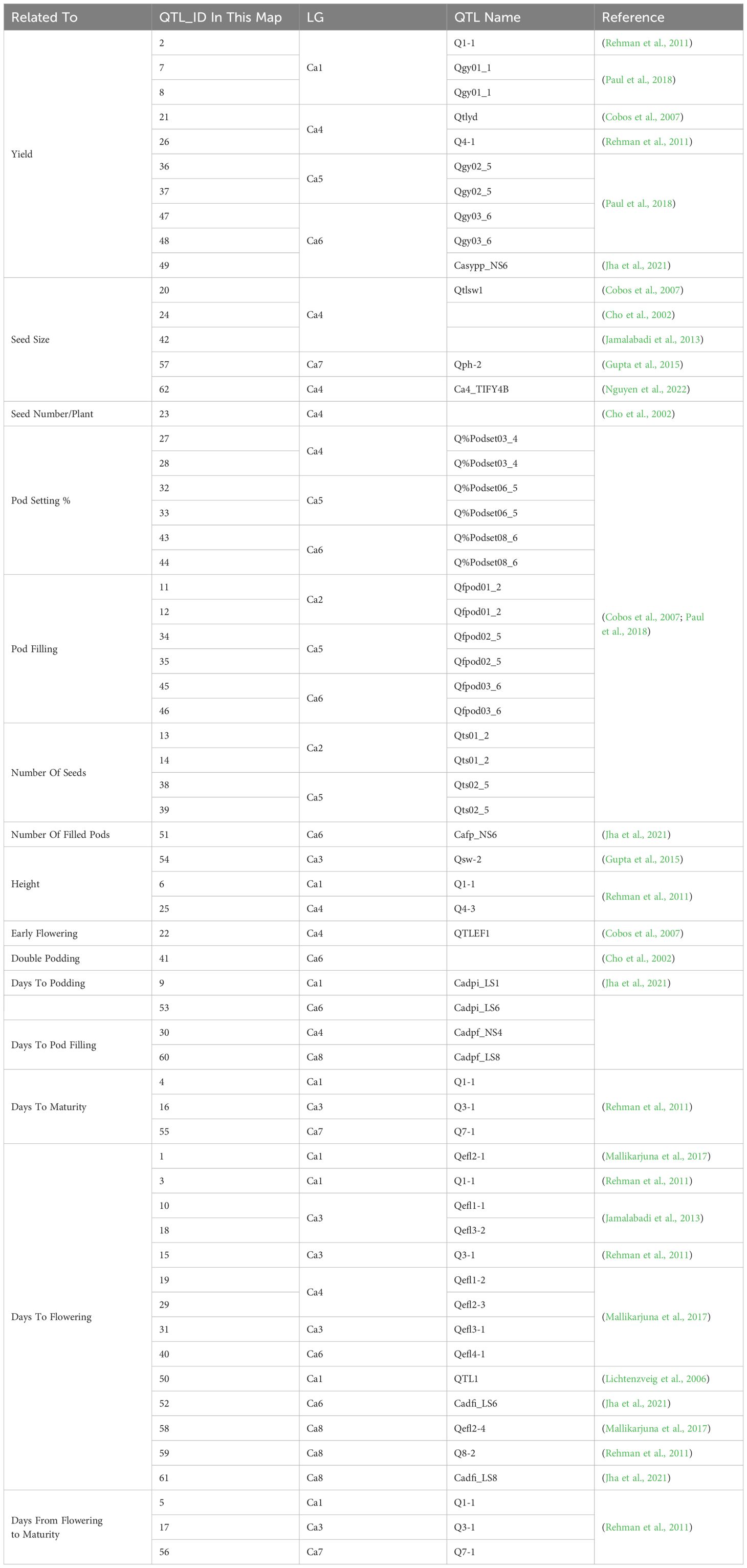
Table 3. QTLs identified by external literature for chickpea, related to traits measured in this study.
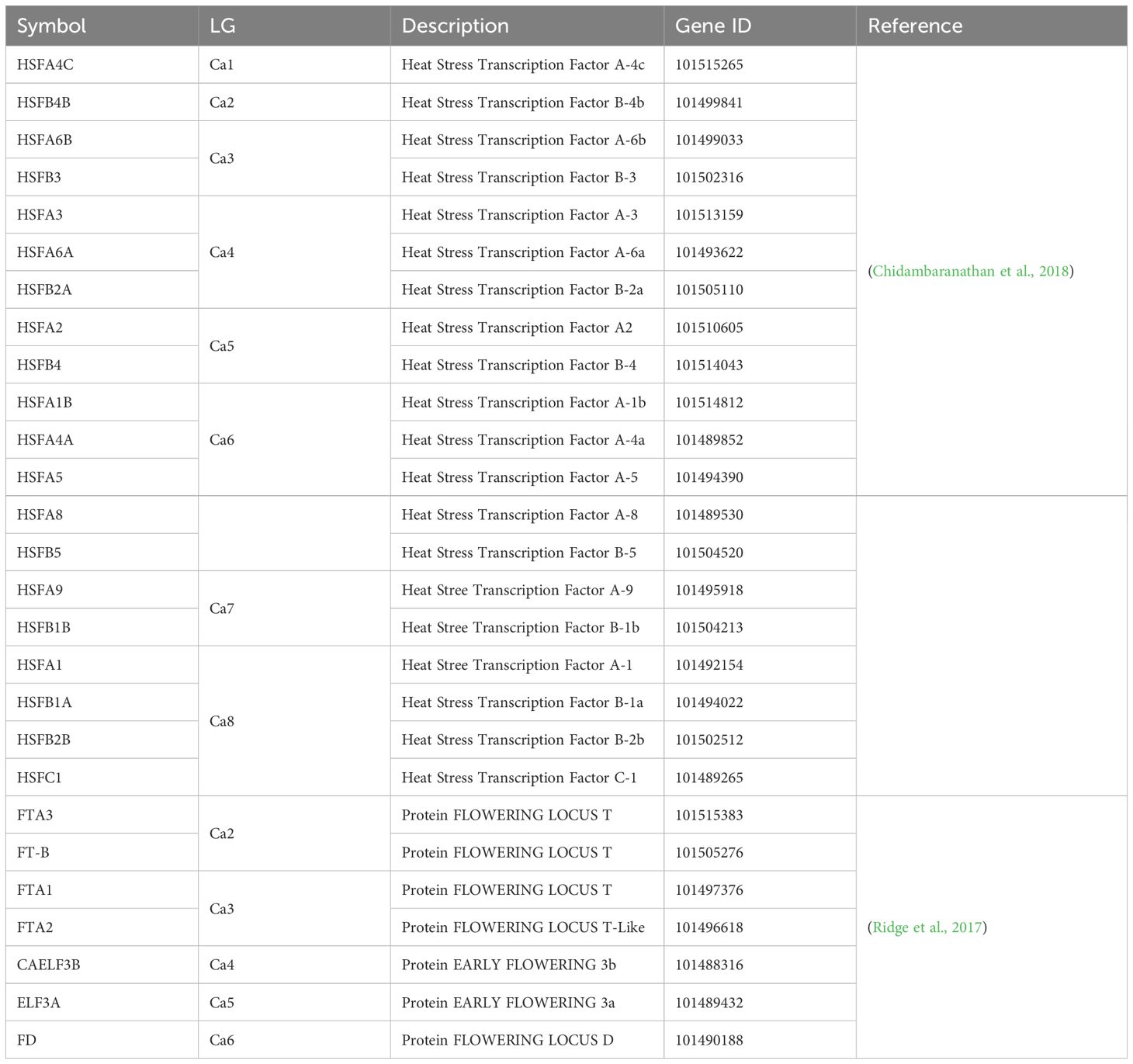
Several of QTL identified overlap with QTL and genes identified in previous research, such as the co-location of Qyield_1.1 and Q1-1 (Rehman et al., 2011). In total, there are 5 regions where QTL have overlapping positions with mapped QTL and genes found in previous literature. These are as follows: (1) Qyield_1.1, overlapping with QTL IDs 1-6, linked to days to flowering, days to maturity, days from flowering to maturity, and height ( (Rehman et al., 2011; Mallikarjuna et al., 2017), and HSFA4C (Chidambaranathan et al., 2018), (2) Qcanopy_1.1 and Qcanopy_1.2, overlapping with QTL IDs 7 and 8, linked to yield (Paul et al., 2018), (3) Qhsw_4.1-4.4, overlapping with QTL IDs 27-29, linked to pod setting and days to flowering (Cobos et al., 2007; Mallikarjuna et al., 2017), and HSFB2A (Chidambaranathan et al., 2018), (4) Qflo_6.2, overlapping with QTL IDs 41 and 42, linked to double podding and seed size (Cho et al., 2002; Jamalabadi et al., 2013), and HSFA8 (Chidambaranathan et al., 2018), (5) Qmat_8.2, overlapping with QTL ID 61 linked to days to flowering (Jha et al., 2021) (Figure 2, Tables 2–4).
4 Discussion
The QTL identified have the potential to advance current knowledge of the chickpea genome. The overlap of new and previously discovered QTL on chromosomes 1, 4, 6 and 8 suggest the presence of highly valuable QTL hotspots, which could be targeted for marker-assisted breeding for future improvement of the crop (Figure 2, Tables 2, 3). This is particularly exciting, considering the range of traits these QTL are associated with.
Due to the large number of trials on which these findings are based, it is also possible to infer and predict the expression of each QTL, based on the presence of a stressor. This provides a deeper understanding of the plasticity of these traits, and improved understanding of the expression of these traits under particular conditions. Most QTL found in this study are linked to Narrabri field trials, particularly those of TOS1, and these are linked both to physiological and yield traits (Table 2). This may be related to the domestication of chickpeas to a Mediterranean environment, which is similar in climate to Narrabri (Kumar and Abbo, 2001), reflecting selection pressure for this environment type. However, the location of Qflo_6.2, linked to flowering time in Kununurra TOS2 2020, indicates the expression of a genomic region linked to flowering under this particular heat stress, given that it does not co-locate with other mapped QTL for flowering time (Lichtenzveig et al., 2006) (Figure 2, Table 3). Qflo_6.2 overlaps with the mapped gene HSFA8, which is a HSF linked to shoot apical meristem and floral tissue development under heat stress (Chidambaranathan et al., 2018) (Figure 2, Table 3). This overlap suggests that Qflo_6.2 is linked to a developmental acceleration under heat stress, particularly as early flowering, or “escape”, is a known physiological strategy for coping with heat stress (Kumar and Abbo, 2001), and plant growth was accelerated under the higher temperatures in Kununurra. This is further suggested by the presence of the other QTL for flowering time, Qflo4.1, which was identified in Narrabri TOS1 2019, overlapping with Qmat_4.1 (Figure 2, Table 2). This could be caused by differential protein expression that is dependent on the environmental conditions. In contrast, the QTL identified for HSW predominately share an overlapping location on chromosome 4, though they are linked to different trials (Figure 2). This suggests that the expression of the genomic region responsible for seed size is not strongly impacted by environment, making it desirable for genomic selection (Pierce, 2020). This is supported by an investigation by Jeffrey et al. (under review)1, which found HSW to be highly heritable, particularly compared to yield. Additionally, the overlap of Qhsw_4.1-4.4 and the gene HSFB2A may indicate that under heat stress, plants divert energy away from new growth and into existing structures, such as seeds. HSFB2A is an HSF that is upregulated under heat stress, linked to the development of all tissue types except those of shoots, and linked most heavily with the development of young pods (Chidambaranathan et al., 2018). Similarly, the QTL associated with yield in this study were both linked to TOS1 trials, although different locations, suggesting that yield may be more heritable under stress-free conditions (Figure 2, Table 2). This suggestion validates the need for lab-based markers related to yield in this species, as it would assist breeders in marker assisted selection (MAS).
4.1 Canopy closure – A recommendation for future research
This study is the first to describe genomic regions associated with final canopy closure in chickpea. This is an exciting discovery both from the perspective of novel QTL discovery, and the potential to build on research that is focused on stress management. Much previous research has focused on heat escape, examining traits such as earliness in flowering and maturity (Berger et al., 2011). However, while breeding for heat escape can be beneficial, this is only a solution if seasons remain predictable, and temperature shifts minimal, which does not align with recent predictions (NASA, 2022). It has been known for some time that canopy climate is crucial for the maintenance of healthy plant tissues, particularly reproductive organs (Gates, 1968). The creation of shade via canopy closure has the potential to protect root systems, and their rhizobial symbionts, from overheating (Lewis et al., 2000). Additionally, as effective transpiration can cool leaves from 6-10°C, canopy closure can create a cooler zone in which pods and other delicate tissues can grow with reduced heat stress (Mathur et al., 2014). This study identified 3 novel QTL linked to final canopy closure, which are also linked to yield QTL found by both this and previous research (Paul et al., 2018; Jha et al., 2021). This doesn’t just give the potential to produce new chickpea genotypes that survive under high temperature, but also the potential to create genotypes that can manage their own stress response and thrive through physiological processes, without sacrificing yield. Finally, Qyield_1.1, Qcanopy_1.1, and Qcanopy_1.2 overlap with a QTL for flowering date (Mallikarjuna et al., 2017), a QTL for yield, flowering time, height, and maturity (Rehman et al., 2011), and a QTL for yield (Paul et al., 2018), along with gene HSFA4C, which is linked to a heat stress transcription factor (HSF), and expressed in the presence of a fusarium wilt infection (Chidambaranathan et al., 2018) (Figure 2, Tables 2, 3). This overlap suggests that these particular traits are linked or pleiotropic, suggesting the potential for GAB to vastly improve the performance of future chickpea genotypes (Pierce, 2020).
The discovery of QTL linked to this trait creates the potential for further understanding of its expression, control, and breeding potential, and this could lead to the development of genotypes that can respond to and manage stress at a significantly more efficient and effective rate. Additionally, as transpiration efficiency has been found to have a heritability rate of up to 70% (Thudi et al., 2014), and stomatal conductance is seen to be higher in heat tolerance genotypes (Kaushal et al., 2013), breeding plans can be created that target all of these traits, enhancing the tools we currently have for breeding.
Based on the review of existing literature (Table 3), this study identified the first QTL associated with final canopy closure in Chickpea, with Qcanopy_1.1, 1.2, and 1.3 identified on chromosome 1, and closely linked to yield-linked QTL Qyield_1.1 and 1.2, and those discovered in previous literature designated QTL 7 and 8, also linked to yield (Paul et al., 2018) (Figure 2, Table 2). These QTL were found across both trials in which the trait was measured (Table 2).
Data availability statement
Data is available within Supplementary Data Sheet 1 and at NCBI with accession number: 14822344.
Author contributions
CJ: Writing – review & editing, Writing – original draft. BK: Writing – review & editing, Supervision. RT: Writing – review & editing. LZ: Writing – review & editing, Writing – original draft, Methodology, Formal analysis, Data curation, Conceptualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Produced with funding assistance from the CRC-P58604, "Developing sustainable cropping systems for cotton, grains and fodder", in conjunction with the Northern Australia Crop Research Alliance (NACRA).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1458250/full#supplementary-material
Footnotes
- ^ Jeffrey, C., Ziems, L., Kaiser, B., and Trethowan, R. A Growing Degree Day (GDD) model determines the effect of temperature stress on diverse chickpea genotypes. Front. Plant Sci. (under review).
References
Abbo, S., Berger, J., Turner, N. C. (2003). Evolution of cultivated chickpea: four bottlenecks limit diversity and constrain adaptation. Funct. Plant Biol. 30, 1081–1087. doi: 10.1071/FP03084
Abiola, O., Angel, J. M., Avner, P., Bachmanov, A. A., Belknap, J. K., Bennett, B. (2003). The nature and identification of quantitative trait loci: a community's view. Nat. Rev. Genet. 4, 911–916. doi: 10.1038/nrg1206
Ahmad, F., Slinkard, A. (2004). The extent of embryo and endosperm growth following interspecific hybridization between Cicer arietinum L. and related annual wild species. Genet. Resour. - Crop Evol. 51, 765–772. doi: 10.1023/B:GRES.0000034580.67728.e4
Alonge, M., Shumate, A., Puiu, D., Zimin, A. V., Salzberg, S. L. (2020). Chromosome-scale assembly of the bread wheat genome reveals thousands of additional gene copies. Genetics 216, 599–608. doi: 10.1534/genetics.120.303501
Berger, J., Milroy, S., Turner, N., Siddique, K., Imtiaz, M., Malhotra, R. (2011). Chickpea evolution has selected for contrasting phenological mechanisms among different habitats. Euphytica 180, 1–15. doi: 10.1007/s10681-011-0391-4
Chidambaranathan, P., Jagannadham, P. T. K., Satheesh, V., Kohli, D. (2018). Genome-wide analysis identifies chickpea (Cicer arietinum) heat stress transcription factors (Hsfs) responsive to heat stress at the pod development stage. J. Plant Res. 131, 525–542. doi: 10.1007/s10265-017-0948-y
Cho, S., Kumar, J., Shultz, J. L., Anupama, K., Tefera, F., Muehlbauer, F. J. J. E. (2002). Mapping genes for double podding and other morphological traits in chickpea. Euphytica. 128, 285–292. doi: 10.1023/A:1020872009306
Cobos, M. J., Rubio, J., Fernández-Romero, M. D., Garza, R., Moreno, M. T., Millán, T., et al. (2007). Genetic analysis of seed size, yield and days to flowering in a chickpea recombinant inbred line population derived from a Kabuli × Desi cross. Annals of Applied Biology. 151, 33–42. doi: 10.1111/j.1744-7348.2007.00152.x
Devasirvatham, V., Tan, D. K., Gaur, P. M., Trethowan, R. M. (2015). Chickpea and temperature stress: An overview. Legumes under Environ. Stress: Yield Improve. Adaptations, 81–90. doi: 10.1002/9781118917091.ch5
Endelman, J. (2011). Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Genome. doi: 10.3835/plantgenome2011.08.0024
Farahani, S., Maleki, M., Mehrabi, R., Kanouni, H. (2019). Whole genome diversity, population structure, and linkage disequilibrium analysis of chickpea (Cicer arietinum L.) genotypes using genome-wide DArTseq-based SNP markers. Genes 10, 676.
Gates, D. M. (1968). Transpiration and leaf temperature. Annu. Rev. Plant Physiol. 19, 211–238. doi: 10.1146/annurev.pp.19.060168.001235
Gupta, S., Kumar, T., Verma, S., Bharadwaj, C., Bhatia, S. (2015). Development of gene-based markers for use in construction of the chickpea (Cicer arietinum L.) genetic linkage map and identification of QTLs associated with seed weight and plant height. Molecular biology reports 42, 1571–1580.
Hiremath, P. J., Kumar, A., Penmetsa, R. V., Farmer, A., Schlueter, J. A. (2012). Large-scale development of cost-effective SNP marker assays for diversity assessment and genetic mapping in chickpea and comparative mapping in legumes. Plant Biotechnol. J. 10, 716–732. doi: 10.1111/j.1467-7652.2012.00710.x
IPCC (2021). “Climate Change 2021: The Physical Science Basis,” in Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change. Eds. Arias, P. A., Bellouin, N., Coppola, E., Jones, R. G., Krinner, G., Marotzke, J., Naik, V., Palmer, M. D., Plattner, G. K., Rogelj, J., Rojas, M., Sillmann, J., Storelvmo, T., Thorne, P. W., Trewin, B., Rao, K.A., Adhikary, B., Allan, R. P., Armour, K., Bala, G., Barimalala, R., Berger, S., Canadell, J. G., Cassou, C., Cherchi, A., Collins, W., Collins, W. D., Connors, S. L., Corti, S., Cruz, F., Dentener, F. J., Dereczynski, C., Luca, A., Niang, A.D., Doblas-Reyes, F. J., Dosio, A., Douville, H., Engelbrecht, F., Eyring, V., Fischer, E., Forster, P., Fox-Kemper, B., Fuglestvedt, J. S., Fyfe, J. C., Gillett, N. P., Goldfarb, L., Gorodetskaya, I., Gutierrez, J. M., Hamdi, R., Hawkins, E., Hewitt, H. T., Hope, P., Islam, A. S., Jones, C., Kaufman, D. S., Kopp, R. E., Kosaka, Y., Kossin, J., Krakovska, S., Lee, J. Y., Li, J., Mauritsen, T., Maycock, T. K., Meinshausen, M., Min, S. K., Monteiro, P. M. S., Ngo-Duc, T., Otto, F., Pinto, I., Pirani, A., Raghavan, K., Ranasinghe, R., Ruane, A. C., Ruiz, L., Sallée, J. B., Samset, B. H., Sathyendranath, S., Seneviratne, S. I., Sörensson, A. A., Szopa, S., Takayabu, I., Tréguier, A.-M., Hurk, B.V. D., Vautard, R., Schuckmann, K.V., Zaehle, S.(Cambridge, United Kingdom, and New York, NY, USA: The IPCC Secretariat).
Jamalabadi, J. G., Saidi, A., Karami, E., Kharkesh, M., Talebi, R. (2013). Molecular mapping and characterization of genes governing time to flowering, seed weight, and plant height in an intraspecific genetic linkage map of chickpea (Cicer arietinum). Biochem. Genet. 51, 387–397. doi: 10.1007/s10528-013-9571-3
Jeffrey, C., Trethowan, R., Kaiser, B. (2021). Chickpea tolerance to temperature stress: Status and opportunity for improvement. J. Plant Physiol. 267, 153555. doi: 10.1016/j.jplph.2021.153555
Jha, U. C., Nayyar, H., Palakurthi, R., Jha, R. (2021). Major QTLs and potential candidate genes for heat stress tolerance identified in chickpea (Cicer arietinum L.). Front. Plant Sci. 12, 655103. doi: 10.3389/fpls.2021.655103
Kaushal, N., Awasthi, R., Gupta, K., Gaur, P. (2013). Heat-stress-induced reproductive failures in chickpea (Cicer arietinum) are associated with impaired sucrose metabolism in leaves and anthers. Funct. Plant Biol. 40, 1334–1349. doi: 10.1071/FP13082
Kumar, J., Abbo, S. (2001). “Genetics of flowering time in chickpea and its bearing on productivity in semiarid environments,” in Advances in Agronomy (Basel, Switzerland: International Crops Research Institute for the Semi Arid Tropics).
Kumar, S., Thakur, P., Kaushal, N., Malik, J. A., Gaur, P., Nayyar, H. (2013). Effect of varying high temperatures during reproductive growth on reproductive function, oxidative stress and seed yield in chickpea genotypes differing in heat sensitivity. Arch. Agron. Soil Sci. 59, 823–843. doi: 10.1080/03650340.2012.683424
Lewis, C. E., Noctor, G., Causton, D., Foyer, C. H. (2000). Regulation of assimilate partitioning in leaves. Funct. Plant Biol. 27, 507–519. doi: 10.1071/PP99177
Lichtenzveig, J., Bonfil, D. J., Zhang, H.-B., Shtienberg, D., Abbo, S. (2006). Mapping quantitative trait loci in chickpea associated with time to flowering and resistance to Didymella rabiei the causal agent of Ascochyta blight. Theor. Appl. Genet. 113, 1357–1369. doi: 10.1007/s00122-006-0390-3
Mallikarjuna, B. P., Samineni, S., Thudi, M., Sajja, S. B. (2017). Molecular mapping of flowering time major genes and QTLs in chickpea (Cicer arietinum L.). Front. Plant Sci. 8, 1140. doi: 10.3389/fpls.2017.01140
Maphosa, L., Richards, M. F., Norton, S. L., Nguyen, G. N. (2020). Breeding for abiotic stress adaptation in chickpea (Cicer arietinum L.): A comprehensive review. Crop Breed. Genet. Genomics 4, 1–39. doi: 10.20900/cbgg20200015
Mathur, S., Agrawal, D., Jajoo, A. (2014). Photosynthesis: Response to high temperature stress. J. Photochem. Photobiol. B: Biol. 137, 116–126. doi: 10.1016/j.jphotobiol.2014.01.010
NASA (2022). Global Climate Change Impact on Crops Expected Within 10 Years. Available online at: https://climate.nasa.gov/news/3124/global-climate-change-impact-on-crops-expected-within-10-years-nasa-study-finds/:~:text=en%20espa%C3%B1ol%20aqui.-,Climate%20change%20may%20affect%20the%20production%20of%20maize%20(corn)%20and,see%20growth%20of%20about%2017%25 (accessed October 1, 2022).
Nguyen, D. T., Hayes, J. E., Harris, J., Sutton, T. (2022). Fine mapping of a vigor QTL in chickpea (Cicer arietinum L.) reveals a potential role for Ca4_TIFY4B in regulating leaf and seed size 13, 829566. doi: 10.3389/fpls.2022.829566
Pandey, M. K., Roorkiwal, M., Singh, V. K., Ramalingam, A., Kudapa, H. (2016). Emerging genomic tools for legume breeding: current status and future prospects. Front. Plant Sci. 7, 1–18. doi: 10.3389/fpls.2016.00455
Patrignani, A., Ochsner, T. E. (2015). Canopeo: A powerful new tool for measuring fractional green canopy cover. Biometry Model. Stat 107, 2312–2320. doi: 10.2134/agronj15.0150
Paul, P. J., Samineni, S., Thudi, M., Sajja, S. B., Rathore, A. (2018). Molecular mapping of QTLs for heat tolerance in chickpea. Int. J. Mol. Sci. 19, 2166. doi: 10.3390/ijms19082166
Pierce, B. A. (2020). Genetics: a conceptual approach. 7th edn (New York, USA: WH Freeman New York).
Pundir, R., Reddy, K., Mengesha, M. H. (1988). ICRISAT chickpea germplasm catalog: evaluation and analysis (India: International Crops Research Institute for the Semi-Arid Tropics).
Rehman, A. U., Malhotra, R. S., Bett, K., Tar'an, B. (2011). Mapping QTL Associated with Traits Affecting Grain Yield in Chickpea (Cicer arietinum L.) under Terminal Drought Stress. Crop Science. 51, 450–463. doi: 10.2135/cropsci2010.03.0129
Ridge, S., Deokar, A., Lee, R., Daba, K., Macknight, R. C., Weller, J. L., et al. (2017). The chickpea Early Flowering 1 (Efl1) locus is an ortholog of Arabidopsis ELF3. Plant physiology 175 (2), 802–815.
Roorkiwal, M., Rathore, A., Das, R. R., Singh, M. K., Jain, A. (2016). Genome-enabled prediction models for yield related traits in chickpea. Front. Plant Sci. 7, 1666. doi: 10.3389/fpls.2016.01666
Rosyara, U. R., De Jong, W. S., Douches, D. S., Endelman, J. B. (2016). Software for genome-wide association studies in autopolyploids and its application to potato. PlantGenome 9, 2. doi: 10.3835/plantgenome2015.08.0073
Sayers, E. W., Bolton, E. E., Brister, J. R., Canese, K., Chan, J., Comeau, D. C., et al. (2022). Database resources of the national center for biotechnology information. Nucleic Acids Res. 50, D20–d26. doi: 10.1093/nar/gkab1112
Sharma, S., Upadhyaya, H. D., Roorkiwal, M., Varshney, R. K., Gowda, C. L. L. (2013). “4 - Chickpea,” in Genetic and Genomic Resources of Grain Legume Improvement. Eds. Singh, M., Upadhyaya, H. D., Bisht, I. S. (Elsevier, Oxford).
Thudi, M., Upadhyaya, H. D., Rathore, A., Gaur, P. M., Krishnamurthy, L., Roorkiwal, M., et al. (2014). Genetic dissection of drought and heat tolerance in chickpea through genome-wide and candidate gene-based association mapping approaches. PloS One 9, e96758. doi: 10.1371/journal.pone.0096758
Turner, S. (2018). qqman: an R package for visualizing GWAS results using QQ and Manhattan plots. J. Open Source Softw. 3, 731. doi: 10.1101/005165
Upadhyaya, H. D., Dronavalli, N., Gowda, C., Singh, S. (2011). Identification and evaluation of chickpea germplasm for tolerance to heat stress. Crop Sci. 51, 2079–2094. doi: 10.2135/cropsci2011.01.0018
Varshney, R. K., Song, C., Saxena, R. K., Azam, S. (2013). Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat. Biotechnol. 31, 240. doi: 10.1038/nbt.2491
Varshney, R. K., Thudi, M., Muehlbauer, F. J. (2017). “The Chickpea Genome: An Introduction,” in The Chickpea Genome. Eds. Varshney, R. K., Thudi, M., Muehlbauer, F. (Springer International Publishing, Cham).
Voorrips, R. E. (2002). MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
VSN-International (2022). Genstat for Windows 22nd Edition (Hemel Hempstead, UK: VSN International).
Wimmer, V., Albrecht, T., Auinger, Hj., Schoen, Cc. (2012). synbreed: a framework for the analysis of genomic prediction data using R. Bioinformatics. 28, 2086–2087. doi: 10.1093/bioinformatics/bts335
Winter, P., Benko-Iseppon, A.-M., Hüttel, B., Ratnaparkhe, M. (2000). A linkage map of the chickpea (Cicer arietinum L.) genome based on recombinant inbred lines from a C. arietinum× C. reticulatum cross: localization of resistance genes for fusarium wilt races 4 and 5. Theor. Appl. Genet. 101, 1155–1163. doi: 10.1007/s001220051592
Keywords: Cicer arietinum, heat stress, abiotic trait, GWAS, quantitative genetics
Citation: Jeffrey C, Kaiser B, Trethowan R and Ziems L (2024) Genome-wide association study reveals heat tolerance QTL for canopy-closure and early flowering in chickpea. Front. Plant Sci. 15:1458250. doi: 10.3389/fpls.2024.1458250
Received: 02 July 2024; Accepted: 05 November 2024;
Published: 17 December 2024.
Edited by:
Jiří Fajkus, Masaryk University, CzechiaReviewed by:
Jacob Olagbenro Popoola, Bowen University, NigeriaMuraleedhar S. Aski, Indian Agricultural Research Institute (ICAR), India
R. Varma Penmetsa, University of California, Davis, United States
Copyright © 2024 Jeffrey, Kaiser, Trethowan and Ziems. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cara Jeffrey, Y2FyYS5qZWZmcmV5QG91dGxvb2suY29tLmF1
 Cara Jeffrey
Cara Jeffrey Brent Kaiser
Brent Kaiser Richard Trethowan
Richard Trethowan Laura Ziems
Laura Ziems