- 1State Key Laboratory of Crop Gene Exploration and Utilization in Southwest China, Rice Research Institute, Sichuan Agricultural University, Chengdu, China
- 2College of Agricultural Science, Panxi Crops Research and Utilization Key Laboratory of Sichuan Province, Xichang University, Liangshan, China
- 3Mianyang Academy of Agricultural Sciences, Crop Characteristic Resources Creation and Utilization Key Laboratory of Sichuan Province, Mianyang, China
In plants, carbohydrates are central products of photosynthesis. Rice is a staple that contributes to the daily calorie intake for over half of the world’s population. Hence, the primary objective of rice cultivation is to maximize carbohydrate production. The “source-sink” theory is proposed as a valuable principle for guiding crop breeding. However, the “flow” research lag, especially in sugar transport, has hindered high-yield rice breeding progress. This review concentrates on the genetic and molecular foundations of sugar transport and its regulation, enhancing the fundamental understanding of sugar transport processes in plants. We illustrate that the apoplastic pathway is predominant over the symplastic pathway during phloem loading in rice. Sugar transport proteins, such as SUTs and SWEETs, are essential carriers for sugar transportation in the apoplastic pathway. Additionally, we have summarized a regulatory pathway for sugar transport genes in rice, highlighting the roles of transcription factors (OsDOF11, OsNF-YB1, OsNF-YC12, OsbZIP72, Nhd1), OsRRM (RNA Recognition Motif containing protein), and GFD1 (Grain Filling Duration 1). Recognizing that the research shortfall in this area stems from a lack of advanced research methods, we discuss cutting-edge analytical techniques such as Mass Spectrometry Imaging and single-cell RNA sequencing, which could provide profound insights into the dynamics of sugar distribution and the associated regulatory mechanisms. In summary, this comprehensive review serves as a valuable guide, directing researchers toward a deep understanding and future study of the intricate mechanisms governing sugar transport.
1 Introduction
In plants, the primary photosynthetic products are carbohydrates, mainly monosaccharides (glucose and fructose), disaccharides (sucrose), and polysaccharides (starch), providing essential energy sources and matter for living organisms’ survival and growth. In cereal production activities, harvesting more edible carbohydrates is the ultimate goal (Alexandratos and Bruinsma, 2012; Julius et al., 2017). For high-yield crop breeding, the “source-sink” theory proposed by Mason M.L. and Maskell E.J. in 1928 has become an essential guiding theory (Mason and Maskell, 1928). The transportation and distribution of carbohydrates are the most critical components in this theory. The “source” refers to organs producing and exporting carbohydrates, such as leaves. In contrast, the “sink” represents organs that receive these sugars and store them as starch, such as seeds. The “flow” corresponds to the organs involved in sugar transportation between the “source” and “sink,” notably the phloem in the stem. According to this theory, high-yield breeding primarily focuses on optimizing the relationships of source-flow-sink, involving the enhancement of source organs to produce more photosynthetic products, increasing the storage capacity of sink organs, and improving the transport capacity of flow organs (Julius et al., 2017; Durand et al., 2018; Zhang and Turgeon, 2018).
Rice (Oryza sativa L.) plays a vital role as a primary provider of carbohydrates in the human diet and feeds more than half of the world’s population. Enhancing yield per unit area remains the foremost objective in rice breeding. Since the first Green Revolution and the introduction of hybrid rice technology, there has been no significant breakthrough in rice yield for years. In agricultural production, some rice varieties have large sinks and potent sources. However, their grains are not filled fully after maturity, with a low seed-setting rate, which seriously affects yield due to poor “flow” capacity (Mohammad et al., 1993; Mathan et al., 2021a; Li et al., 2022e). Furthermore, a recent study has shown that a more robust flow function may be a critical factor in the higher yield of cultivated rice than wild rice. While wild rice exhibits a more vital photosynthetic capacity and outputs more sucrose in the leaves, a weaker vascular system and reduced sucrose transporter activity limit the efficiency of transporting sucrose to the grains and yields (Mathan et al., 2021a).
Over the past decade, research on “source” and “sink” has been more in-depth, with a large number of genes related to photosynthetic efficiency and panicle traits being cloned and studied (Chen et al., 2022). However, our understanding of “flow” regulation has lagged despite the progress in the sugar transport of rice. Therefore, it is critical to explore essential genes related to sugar transport, elucidate their functions, and uncover the regulatory mechanisms (Ma et al., 2023). In this review, we reviewed the research progress and outlined a research prospect for sugar transport to facilitate further advancements in guiding high-yield rice breeding.
2 Core carbohydrates for study: sucrose and starch
Sucrose and starch, which are fundamental components of the plant’s sugar metabolism, are central indicators in flow study. By closely monitoring the levels and distribution of sucrose and starch, researchers could gain valuable insights into the dynamics of sugar transport and allocation in plants.
Sucrose, produced by photosynthesis, is the most prevalent disaccharide in plants. It is the primary transport form of sugar over long-distance in the phloem. Subsequently, a portion is allocated to support growth and development, with the remainder stored as starch in the sink (Patrick et al., 2013; Li et al., 2022c). Sucrose is a non-reducing sugar, rendering it more resistant to degradation, particularly during long-distance transport within plant vascular systems. Its low viscosity guarantees swift flow during transportation with higher speeds ranging from 0.5 to 3 m·h−1. Furthermore, sucrose is known for its remarkable solubility, with phloem concentrations typically falling within 200 to 1600 mmol·L−1, creating exceptionally high osmotic potential (van Bel, 1996; Kühn et al., 1999). These characteristics are crucial for enabling the efficient long-distance transport of carbon assimilates as sucrose within plants.
Starch, a polysaccharide composed of glucose molecules, is the primary storage form of sugar in plants. Its glucose molecule arrangement facilitates efficient hydrolysis, releasing energy when needed. This characteristic enables plants to store energy during periods of abundant photosynthesis and utilize it to support growth when necessary (Vandeputte and Delcour, 2004; Bao, 2019).
3 Process of sucrose transport
Sucrose transport in plants involves a highly coordinated process of three essential components: phloem loading, vascular transport, and unloading in the sink. These components ensure the successful delivery of sugars to where they are needed for growth, energy, and various metabolic processes (Figure 1). Furthermore, this extensive journey necessitates precise regulation and adaptability to meet plants’ growth and survival demands (Williams et al., 2000; Wen et al., 2022; Li et al., 2022c; Singh et al., 2023).
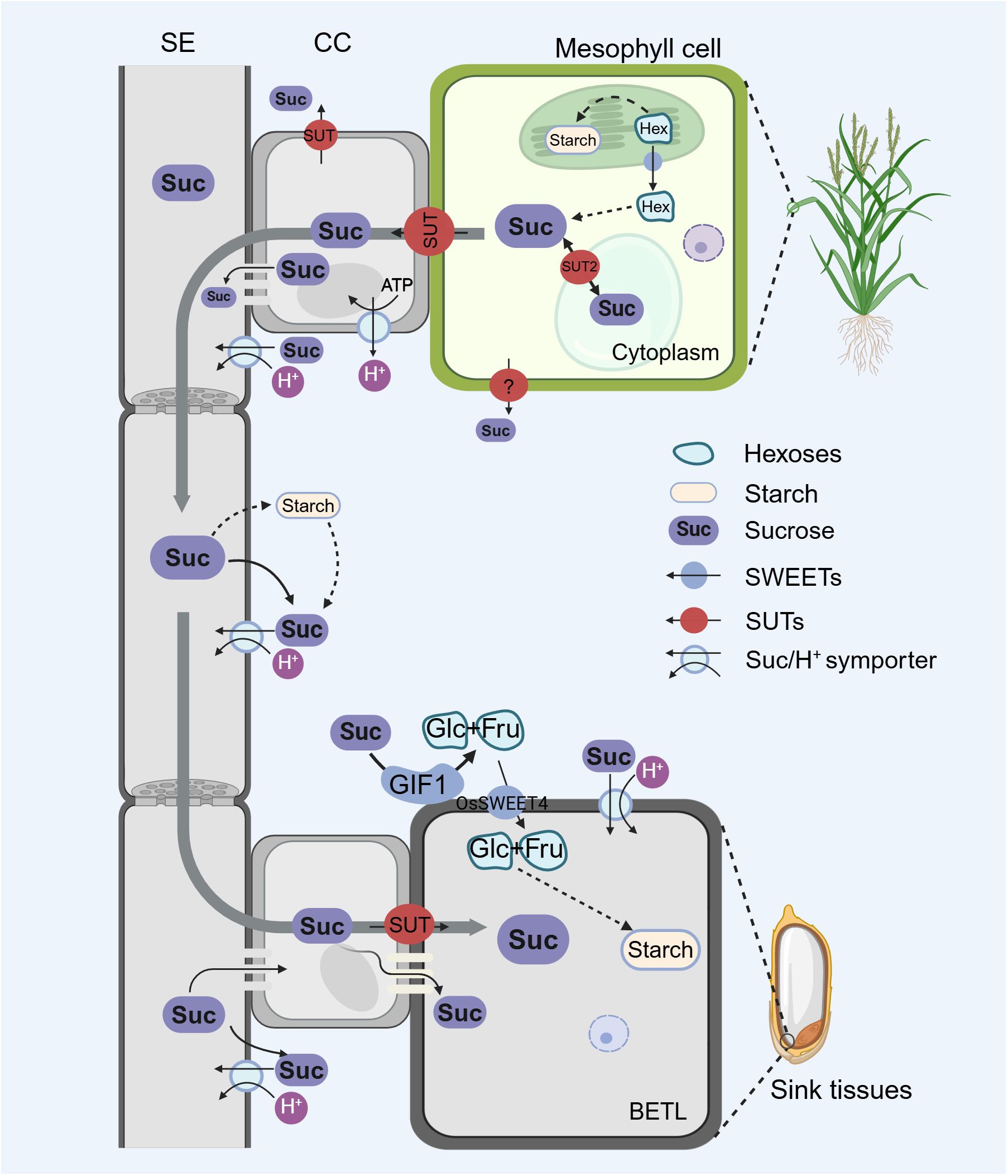
Figure 1. Long-distance transport of sucrose from source leaves to sink tissues, adapted from (Aoki et al., 2012). Sugars are produced in the mesophyll cells of source leaves, including hexoses, and are exported into the cytoplasm via sugar transport proteins. After undergoing a series of enzymatic catalysis reactions, hexoses are converted into sucrose. Sucrose enters the CC through symplastic and apoplastic pathways and is loaded into the SE of the phloem by these CC, then transported to sink tissues along the pressure potential gradient within the sieve elements. Sucrose is subsequently transported from the SE-CC complex to the surrounding parenchyma cells through plasmodesmata. In the apoplastic unloading pathway, sucrose is cleaved by cell wall invertase, generating hexoses such as Glc (Glucose) and Fru (Fructose). The produced Glc and Fru molecules are then transported into the BETL (Basal Endosperm Transfer Layer) for starch synthesis via the sugar transport protein OsSWEET4. Alternatively, sucrose could also be directly transported into storage cells. The figure was created with BioRender.com.
3.1 Phloem loading
The first crucial step in sucrose transport is loading it into the phloem, which occurs in source tissues such as leaves. Generally, plants have three major loading pathways: symplastic, apoplastic, and polymer trap pathways (Zhang and Turgeon, 2018).
3.1.1 Symplastic pathway
The symplastic pathway could be the primary route initially taken for sucrose transport in the leaves. This pathway utilizes plasmodesmata, which are intercellular connections that provide channels for cytoplasmic continuity. By these channels, sucrose enters the phloem SE-CC (Sieve Element-Companion Cell) complex through passive diffusion along a concentration gradient. It does not require sucrose transport carrier proteins to cross membranes and does not consume energy, but it may face additional biological limitations, such as the distribution and connectivity of plasmodesmata (Miras et al., 2022). However, since sucrose could leak out of the cell during its transport in the phloem, sucrose transporters are still necessary to recover sucrose into the partition and maintain a higher chemical concentration gradient during symplastic transport (Braun et al., 2014).
3.1.2 Apoplastic pathway
In some plants, the SE-CC complex lacks plasmodesmata connections with surrounding cells, so the apoplastic pathway is typically used for phloem loading. Unlike the symplastic pathway, which relies on plasmodesmata, the apoplastic pathway transports sugars through the cell apoplast. This pathway may offer greater flexibility for sugar transport, particularly during long-distance movement and in response to environmental changes (Aoki et al., 2012). In this pathway, the sucrose/H+ symporter is believed to mediate the transport of sucrose from the apoplast into SE-CC, and SWEET (Sugars Will Eventually be Exported Transporters) and SUT/SUC (Sucrose Transporters) are proved to be essential carriers. With the assistance of SWEET, sucrose is transported across membranes into the cell apoplast and then further transported to the SE-CC complex by SUT. This process entails active transport against a concentration gradient, which necessitates energy consumption, thus the efficiency of the apoplastic pathway may be influenced by factors such as pH (Hu et al., 2021).
3.1.3 Polymer trap pathway
In the polymer trap pathway, sucrose is not directly loaded into the sieve tubes; instead, it is converted into oligosaccharides, such as raffinose and stachyose, in intermediary cells. These oligosaccharides then enter the SE-CC complex through larger plasmodesmata connections that rely on proton motive force for transport. Significantly, oligosaccharides like raffinose and stachyose could not diffuse back into mesophyll cells due to their larger molecular weight, thereby forming a kind of “trap” in the sieve tubes, which facilitates the loading of sugars (Comtet et al., 2017). This pathway plays a key role in ensuring that carbohydrates are efficiently conveyed to regions requiring them the most, thus facilitating growth and development.
3.2 Vascular transport
After phloem loading, sucrose enters the transport process within the vascular bundles. In general, vascular transport primarily involves long-distance translocation, during which metabolites are transported from the leaf vasculature to satisfy the diverse requirements of various plant tissues. The vascular bundles comprising the xylem and phloem tissues are the structural framework for metabolite transport. The phloem, primarily composed of sieve tubes and companion cells, forms an uninterrupted network throughout the plant, playing a crucial role in sucrose transport (White and Ding, 2023).
3.3 Unloading in sink
The final component is sucrose release from the phloem into sink tissues. Sink tissues can include developing grains, roots, and other organs that require a supply of sugars for growth and development. The phloem unloading process is also tightly regulated and can occur through symplastic, apoplastic pathways, or a combination of both. In the symplastic pathway, sucrose is transported from the SE-CC complex to the surrounding parenchyma cells through plasmodesmata. In the apoplastic unloading pathway, sucrose is subject to enzymatic cleavage by cell wall invertase, producing glucose and fructose. These resulting glucose and fructose molecules are then transported into the storage cells. Alternatively, sucrose can be directly transported into the storage cells (Braun, 2022; Sun et al., 2022; Wen et al., 2022; Li et al., 2022c).
4 Rice sucrose loading pathway
In grass crops, wheat, barley, and maize have been proven to use the apoplasmic loading pathway in sucrose transport, while it is still not entirely clear for rice (Julius et al., 2017). It has been reported that rice phloem sap collected by severing stylets of brown planthoppers contains primarily only one sugar, sucrose (Fukumorita and Chino, 1982). Therefore, rice does not employ the polymer trap pathway for sucrose phloem loading. The symplastic pathway or the apoplastic pathway could be used in rice.
4.1 Symplastic loading pathway in rice
Some studies suggest that rice has the fundamentals of cell structure for the symplastic loading of sucrose. Unlike maize, plasmodesmata could connect the parenchyma cells with companion cells in the leaf vascular bundles of rice (Botha et al., 2008; Eom et al., 2012). Using the characteristic that CFDA (Carboxyfluorescein Diacetate) can only be transported within the plastid after entering the plant body, the dye-feeding experiments showed that there is a continuous connection, known as symplastic continuity, between the phloem and the surrounding parenchyma cells (Scofield et al., 2007b). However, a later study reported that the frequency of such plasmodesmata connecting the cytoplasms of vascular bundles and companion cells accounted for only 1.4% of the total plasmodesmata measured across cell types (Braun et al., 2014). Though sucrose loading via a symplastic pathway may exist in rice, such loading capacity of sucrose may be relatively low. Therefore, a symplastic pathway may not be the primary pathway for sucrose transport in rice (Julius et al., 2017).
4.2 Apoplastic loading pathways in rice
Several recent studies support that sucrose phloem loading in rice predominantly takes the apoplastic pathway (Braun, 2022; Li et al., 2022b; Scofield et al., 2007a; Wang et al., 2021). Based on studies in Arabidopsis, it was hypothesized that if the membrane specificity of proton pyrophosphatases is localized within the SE-CC complex of leaves, there would be the potential existence of an apoplastic transport pathway in the plant (Gaxiola et al., 2012; Pizzio et al., 2015). In rice, immunogold labeling revealed that proton pyrophosphatases primarily localized to the plasma membrane of SE-CC complexes in the minor vein of rice leaves, reinforcing the notion of apoplastic loading. This interpretation was further supported by sucrose synthase-specific immunogold labeling at the SE-CC complex (Regmi et al., 2016).
Moreover, transgenic rice plants with overexpressed yeast invertase displayed reduced sucrose loading in the phloem, resulting in sucrose and starch accumulation in the leaves, akin to the growth-inhibited phenotype observed in ossut1 mutants. The application of PCMBS (p-chloromercuribenzenesulfonic acid), an inhibitor of sugar transport protein function, notably hindered sucrose transport to rice phloem (Giaquinta, 1979; Wang et al., 2021). These results demonstrate the essential role of sucrose transport proteins in rice sucrose loading. Through electron microscopy, phloem tracer transportation, and 13C labeling, an additional study observed that the SE-CC complex did not exhibit a symplastic link with adjacent parenchyma cells in both leaves and stems. Moreover, the reduction in 13C isotope remobilization from leaves to stems and panicles upon PCMBS treatment indicated sucrose’s active transportation into the rice phloem (Li et al., 2022b). Therefore, rice is more inclined to use the apoplastic strategy for phloem loading.
5 Sucrose unloading pathway in rice
Till now, the sucrose unloading pathway in rice has been partially elucidated. Existing research indicates that, before unloading, the sucrose is initially enzymatically cleaved into glucose and fructose, with a crucial role played by a cell-wall invertase GIF1 (Grain Incomplete Filling 1) (Wang et al., 2008). Subsequently, glucose and fructose are transported into the endosperm via the sugar transporter protein OsSWEET4, which facilitates the transfer of glucose and fructose from the basal endosperm transfer layer to the endosperm for starch synthesis. Both gif1 and ossweet4 showed grain-filling defects, and ossweet4 even resulted in defective seed filling, suggesting that the primary unloading pathway in rice grain might be the apoplastic pathway (Sosso et al., 2015).
6 Sugar transporters and their regulators in rice
Based on the above analysis, the primary pathway for sucrose transport in rice is the apoplastic pathway, underscoring the pivotal role of sugar transporters in this physiological process. In plants, sugar transport is facilitated by various sugar transporters, primarily including the SWEET family and MFS (Major Facilitator Superfamily). The MFS can be further categorized based on the substrate specificity into the DST (Disaccharide Transporter) family and the MST (Monosaccharide Transporter) family (Williams et al., 2000; Büttner, 2007; Deng et al., 2019; Zhou M. et al., 2023). In addition, there are still some other transporters specific to various forms of sugar, which are beyond the current scope of this review.
In rice, SWEETs and the disaccharide transporter SUTs, have been recognized as crucial facilitators in sugar transport (Eom et al., 2011; Julius et al., 2017; Mathan et al., 2021a). Although some other types of sugar-related transport proteins have been reported, such as OsMEX1 (Maltose Excess 1), OsTMTs (Tonoplast Monosaccharide Transporters), OsNope1 (No perception 1), OsBT1 (Brittle 1) and so on, further introduction is not provided here because these studies are currently neither in-depth nor systematic enough (Cho et al., 2010; Ryoo et al., 2013; Cakir et al., 2016; Nadal et al., 2017; Li et al., 2017; Song et al., 2020).
6.1 SUT family
SUT proteins have 12 transmembrane domains, forming a pore to transport sucrose across the plasma membrane (Riesmeier et al., 1992). The SUT family can be divided into three clades (Aoki et al., 2003). Type I SUTs are exclusive to eudicots, while type II and III SUTs are found in all plants. The rice genome encodes five SUTs; four are type II, and one (OsSUT2) is type III (Aoki et al., 2003). The five SUTs possess diverse roles in both sink and source tissues (Aoki et al., 2003). OsSUT1, OsSUT3, OsSUT4, and OsSUT5 encode proteins in the plasma membrane, whereas OsSUT2 encodes a protein in the vacuolar membrane (Aoki et al., 2003; Eom et al., 2011; Siao et al., 2011).
In Arabidopsis, AtSUC2 mediates efflux and retrieval via the apoplast pathway (Srivastava et al., 2008, 2009). Interestingly, OsSUT1, the type II SUT like AtSUC2, was able to complement the atsuc2 mutant, indicating that OsSUT1 is a functional homolog of AtSUC2 for sucrose transporter (Matsukura et al., 2000; Furbank et al., 2001; Scofield et al., 2007a, 2007b; Hirose et al., 2010; Eom et al., 2016). OsSUT1 is highly expressed in the endosperm, leaf sheaths, nodes, vascular parenchyma, and nucellar projection (Matsukura et al., 2000). However, it is curious that no visible phenotype was observed in ossut1 knockdown plants at the beginning. Typical phenotypes of sugar transporter mutants, such as sugar accumulation within leaves and retarded growth, are not followed (Scofield et al., 2002; Hirose et al., 2010). Further study exhibited that it might be due to the low demand for sugar transport during the growth and development of rice in the greenhouse with low light. In the field-grown conditions, the ossut1 mutant showed reduced growth and grain yield due to sucrose and starch accumulation in the leaves (Feng et al., 2018). Therefore, OsSUT1 could function to load sucrose from the apoplast into the phloem in rice leaves.
OsSUT2 is highly expressed in lateral roots, inflorescence, stems, seed coats, leaf mesophyll, and bundle sheath cells but not in the veins (Aoki et al., 2003; Eom et al., 2011; Siao et al., 2011; Eom et al., 2012). OsSUT2 may be involved in vacuolar-to-cytosolic sucrose translocation, influencing sucrose movement into the phloem (Sun et al., 2008; Eom et al., 2011; Siao et al., 2011; Eom et al., 2012). The mutant ossut2 demonstrated distinctive growth-inhibited characteristics, shedding light on the crucial role of OsSUT2 in rice sugar transport mechanisms. Sugar export from the leaf was also reported to be significantly reduced in ossut2 mutants, associated with notable accumulations of sucrose, glucose, and fructose in mature leaves (Eom et al., 2011, 2012). OsSUT3 is preferentially expressed in pollen, indicating its involvement in pollen development and maturity rather than phloem loading in source leaves (Aoki et al., 2003; Scofield et al., 2007b; Li et al., 2020). OsSUT4 shows expression dominance in leaves and stems. The ossut4 mutant deficiency exhibited an accumulation of sucrose and starch in leaves and a significant reduction in net photosynthetic rate, which are the typical phenotypes of phloem loading obstruction in the vascular tissue (Mengzhu et al., 2020). OsSUT5 exhibits the highest expression level in storage leaves (Aoki et al., 2003). Interestingly, through biochemical activity studies in Xenopus laevis oocytes, OsSUT5 was found to have a higher substrate affinity for sucrose and less substrate specificity than OsSUT1. Thus, OsSUT5 could potentially replace the function of OsSUT1 in sucrose phloem loading (Sun et al., 2010).
6.2 SWEET family
SWEETs are identified as sugar transporters containing seven transmembrane domains, which are present in all organism kingdoms (Yuan and Wang, 2013; Anjali et al., 2020; Singh et al., 2023). SWEETs mediate the bidirectional transmembrane transport of sugar substances, playing crucial roles in various physiological processes throughout plant growth and development. Among the 21 paralogs in rice, OsSWEET4 and OsSWEET11 are believed to play a predominant role in grain filling and yield formation (Zhou et al., 2014; Morii et al., 2020; Li et al., 2022d). Similar to OsSWEET4, OsSWEET11 is crucial in facilitating sucrose transport from maternal tissue to the maternal-filial interface in the stages of caryopsis development. Knockout of OsSWEET11 led to decreased sucrose concentration in embryo sacs. Still, it increased the sucrose content and reduced starch levels in mature caryopses (Ma et al., 2017; Li et al., 2022d).
OsSWEET15 was found to share a similar function with OsSWEET11 in regulating the efflux of sucrose across the nucellar epidermis/aleurone interface. Notably, in the double mutant ossweet11 15, starch accumulation was observed in the pericarp, while a reduction in starch content was evident in the endosperm. This dynamic led to the development of shriveled caryopses (Ma et al., 2017; Yang et al., 2018). The single knockout mutants of ossweet14 did not exhibit discernible phenotypic changes. In contrast, the double mutant ossweet11 14 displayed considerably exacerbated phenotypes compared to the single-knockout mutants of ossweet11. These severe manifestations included a significant grain weight and yield reduction, a slower grain-filling rate, and an elevated accumulation of starch in the pericarp (Ma et al., 2017; Eom et al., 2019; Fei et al., 2021). Paradoxically, other studies have shown that ossweet14 had a shorter plant phenotype and produced smaller seeds (Antony et al., 2010; Li et al., 2022d).
OsSWEET5, which acts as a galactose transporter, is believed to regulate plant growth and senescence by interacting with auxin signaling pathways (Zhou et al., 2014). Overexpressed OsSWEET5 plants exhibited growth retardation and early senescence during the seedling stage. However, no discernible phenotype differences were observed in OsSWEET5-knockdown lines, suggesting the functional redundancy of galactose transporters existed in rice (Zhou et al., 2014). OsSWEET3a likely serves as a key player in facilitating the loading of glucose and GA (Gibberellin Acid) from the endosperm into the phloem for subsequent transport, thereby influencing early shoot development in rice. OsSWEET3a is primarily expressed in the vascular bundles of basal rice seedlings, while mutants lacking OsSWEET3a have significant deficiencies during germination and the initial stages of shoot development. Intriguingly, these impairments showed improvement when exogenous GA was introduced (Morii et al., 2020).
It is worth noting that overexpressing sugar transport proteins in a rough manner did not increase yield in rice. Some studies overexpressed OsSWEET11 or OsSWEET14. However, the transgenic lines showed unfavorable phenotypes (Yuan et al., 2009; Gao et al., 2018; Singh et al., 2021; Kim et al., 2021b). The overexpression of OsSWEET11 plants was markedly shorter and had fewer tillers than the wild type (Yuan et al., 2009), and OsSWEET14 overexpressed plants resulted in a dwarf phenotype, reduced 1,000‐grain weight and grain number per panicle, and tillers (Kim et al., 2021b). Another study even co-expressed three sugar transporters, OsSWEET11, OsSWEET14, and OsSUT1, which got similar results to the plants that overexpress either OsSWEET11 or OsSWEET14 (Singh et al., 2021). These results showed that simply overexpressing sugar transporters does not improve the transport capacity of the flow but may have the opposite effect.
6.3 Transcriptional regulation of sugar transporters
Sugar transport through the phloem can be affected by many environmental factors that alter source-sink relationships, including abiotic and biotic, such as drought, saline-alkali, heat, mineral deficiency, pathogenic microbes, viruses, and so on. However, most of the exact mechanisms are far from being completely known at the molecular level (Krasensky and Jonak, 2012; Lemoine et al., 2013; Mathan et al., 2021a; Salvi et al., 2022).
Recent reports indicate that sugar transporters are intricately connected with various regulatory pathways involving ABA (Abscisic Acid), GA, stress responses, nitrogen uptake, circadian rhythm, etc. Several transcription factors regulate the expression of rice sugar transporter genes, thereby controlling the sugar transport rate into various tissues (Yanagisawa and Sheen, 1998; Laloum et al., 2013; Li et al., 2013; Bai et al., 2016; Wu et al., 2018; Xiong et al., 2019; Liu et al., 2020; Sun et al., 2021; Zhang et al., 2021a; Kim et al., 2021b; Mathan et al., 2021b; Lee et al., 2022; Li et al., 2022a, 2023) (Figure 2).
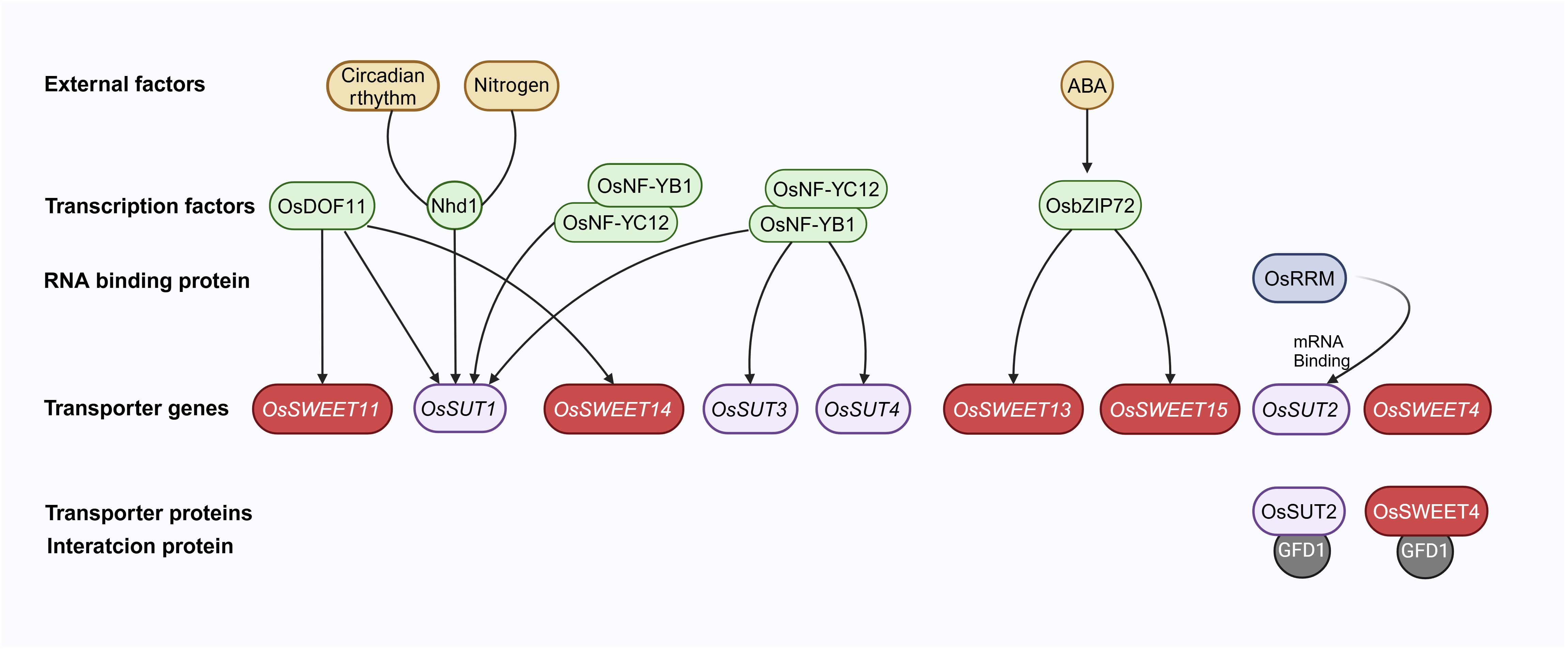
Figure 2. A simple regulation pathway of sugar transporters in rice. Transcription factors, such as OsDOF11, OsNF-YB1, OsNF-YC12, OsbZIP72, and Nhd1, directly regulate sugar transporter genes. These transcription factors are linked to the circadian clock, nitrogen absorption, and ABA stress signaling. The mRNA of OsSUT2 could be directly bound by an RNA Recognition Motif containing protein, OsRRM. The GFD1 protein directly interacts with both OsSUT2 and OsSWEET4. Arrows represent positive regulation of sugar transporter genes. The figure was created with BioRender.com.
DOF (DNA binding with one finger) transcription factors can recognize promoter elements containing (A/T)AAAG and subsequently regulate various aspects of plant development (Yanagisawa and Sheen, 1998). OsDOF11 is identified as a regulator of SUT and SWEET genes. It exhibits expression in vascular cells within photosynthetic organs and various sink tissues. The osdof11 mutants displayed a semi-dwarf phenotype with reduced tiller numbers and smaller panicles, ultimately leading to a decreased sucrose transport rate compared to the wild type. Furthermore, the expression profiles of transporter genes, including OsSUT1, OsSUT3, OsSUT4, OsSUT5, OsSWEET11, and OsSWEET14, are significantly altered in the osdof11 mutant. Chromatin immunoprecipitation assays have provided conclusive evidence for direct binding of OsDOF11 to the promoter regions of OsSUT1, OsSWEET11, and OsSWEET14. Interestingly, overexpression of OsDOF11 enhanced resistance to sheath blight, while reducing yield production. Alternatively, tissue-specific activation of OsDOF11 via VP16 significantly improves yield production and enhances sheath blight, resistance, underscoring the pivotal role of tissue-specific in optimizing sugar transport-related genes (Li et al., 2013; Kim et al., 2021b).
NF-Y (Nuclear factor-Y) is a class of conserved transcription factors that binds to the promoter’s CCAAT box (Laloum et al., 2013). In rice, the osnf-yb1 mutant exhibited chalky endosperm and defective grain-filling. RNA in situ hybridization assays displayed that OsNF-YB1 was localized to the aleurone layer. EMSA (Electrophoretic Mobility Shift Assays) demonstrated that OsNF-YB1 could directly bind to the CCAAT box in the promoters of OsSUT1, OsSUT3, and OsSUT4 (Bai et al., 2016). Another study showed that OsNF-YC12 was directly bound to the promoter of OsSUT1 using various ChIP (Chromatin Immunoprecipitation) and Y1H (Yeast One-Hybrid assays). Furthermore, OsNF-YC12 could also physically interact with OsNF-YB1 (Xiong et al., 2019). Therefore, OsNF-YB1 and OsNF-YC12 dimers could regulate the expression of OsSUTs in the aleurone layer during the grain-filling stage.
OsbZIP72 encodes a transcription factor responsive to ABA. Transactivation experiments have demonstrated that OsbZIP72 could directly bind to the promoters of OsSWEET13 and OsSWEET15 and promote their expressions. This finding suggests a plausible mechanism for regulating sucrose transport and allocation in response to abiotic stresses, and such a mechanism can potentially maintain sugar homeostasis in rice when faced with challenges such as drought and salinity (Mathan et al., 2021b).
An MYB transcription factor Nhd1 (N-mediated heading date-1), also known as a core clock component factor OsLHY (Late Elongated Hypocotyl), has been reported to regulate nitrogen uptake and flowering time (Sun et al., 2021; Zhang et al., 2021a; Lee et al., 2022; Li et al., 2022a). A subsequent study uncovered that the deletion of Nhd1 led to the inhibition of OsSUT1 expression. Notably, when compared to the wild type, both nhd1 and ossut1 mutants displayed similar characteristics, including reduced plant height, decreased shoot biomass, and lower sucrose concentration. Intriguingly, the overexpression of OsSUT1 could restore the impaired sucrose transport, partially alleviating the compromised growth observed in nhd1 mutants. EMSA, transient transactivation assays, and ChIP-qPCR in rice leaf protoplasts provided evidence that Nhd1 could directly activate the transcriptional expression of OsSUT1, suggesting a conventional regulatory module involving Nhd1 and OsSUT1 that maintains carbon and nitrogen balance in rice (Li et al., 2023).
A recent study identified a novel mechanism for the expression regulation of rice sugar transporter. An OsRRM (RNA Recognition Motif) containing protein could interact with mRNAs of sugar transporter genes and positively regulate their expression levels, such as OsSUT2 (Liu et al., 2020). To summarize, sugar transporter genes’ expression in rice is dynamically controlled by transcription factors and RNA-binding proteins to fine-tune sugar homeostasis (Dhungana and Braun, 2021).
6.4 Interacting proteins of sugar transporters
The transport activity of many transporters depends on the interactions of membrane proteins (Lalonde et al., 2010). For example, Arabidopsis nitrate transporter proteins NRT2.1 and NRT2.2 require interaction with the nitrate assimilation-related protein NAR2 to form a 150 kDa plasma membrane complex, which enables high-affinity nitrate transport (Quesada et al., 1994; Boone et al., 2007; Wirth et al., 2007; Lalonde et al., 2008; Jonikas et al., 2009; Yong et al., 2010; Jones et al., 2014; Sun et al., 2023). In Arabidopsis, the MIND (Membrane-based Interaction Network Database) revealed many interacting proteins of sugar transporter proteins (Cusick et al., 2005). However, their physiological and genetic functions are rarely reported in plants (Bitterlich et al., 2014; Jones et al., 2014; Abelenda et al., 2019; Xu et al., 2020; Wipf et al., 2021).
In rice, the interacting proteins of sugar transporters were unknown before a MATE (Multidrug and toxic compound extrusion) transporter protein GFD1 (Grain Filling Duration 1) was studied in our recent study (Sun et al., 2023). MATE transporter is one of the most prominent families of transporter proteins for cation transportation in most organisms (Omote et al., 2006; Takanashi et al., 2014; Upadhyay et al., 2019; Qin et al., 2021b; Zhang et al., 2021b; Zhou C. et al., 2023). Initially, due to the 12 transmembrane domains shared with SUT proteins, it was classified as a member of MFS (Marger and Saier, 1993; Omote et al., 2006). However, due to differences in protein sequence, it was later defined as an independent MATE transporter class (Brown et al., 1999).
In our recent study, GFD1 was identified as a MATE transporter protein that may be associated with sugar transport and starch accumulation in rice (Sun et al., 2023). The mutant gfd1 disrupted the balance of carbohydrate distribution in the stem and grain but not in the leaves. The mutant showed a prolonged grain filling period, increased grain size, and reduced number of spikelets. Through Y2H (Yeast Two-Hybrid), LCI (Firefly luciferase Complementation Imaging assay), and BiFC (Bimolecular Fluorescence Complementation), it was proven that GFD1 could interact with two sugar transporters, OsSWEET4 and OsSUT2. Genetic analysis using double mutants indicated that GFD1 might control the grain filling period through OsSWEET4 and regulate grain size through OsSUT2, while working with OsSUT2 and OsSWEET4 to handle the number of grains per spike. However, the interaction effects on the functions of OsSUT2 and OsSWEET4 remain unclear (Figure 2).
On the other hand, in addition to the photosynthetic products from the leaves, the formation of rice grain yield also relies significantly on the starch stored in the stem before flowering. Stem starch can contribute up to 30% of rice yield, and this contribution rate increases with increased storage capacity, particularly under adverse environmental conditions. When rice photosynthesis is impaired, the stem becomes a primary source of assimilates for grain filling, significantly mitigating the negative effects of stress on yield (CocK and Yoshida, 1972). Furthermore, stored starch in the stem could also be a crucial factor contributing to the increased yield of cultivated rice compared to wild rice (Mathan et al., 2021a). Therefore, increasing the accumulation of starch in rice stems and improving the transport efficiency are essential to enhance rice yield. However, the molecular mechanism of starch accumulation in the stem remains unclear. Our study showed that GFD1 is an important regulatory factor for starch accumulation in the stem, and the deficient mutant gfd1 significantly accumulates starch particles in the stem before grain filling (Sun et al., 2023). Therefore, GFD1 plays a significant role in the long-distance sugar transport within the stem.
7 Prospects for sugar transport study and application strategies in rice
As biotechnology advances, focusing solely on studying sugar transport in organs such as roots, stems, leaves, and panicles is no longer sufficient. The development of single-cell sequencing and MSI (Mass Spectrometry Imaging) opens up new possibilities for a more precise understanding of the sugar transport mechanism. In order to facilitate a better understanding of the molecular mechanisms underlying sugar transport, we propose a study prospect here (Figure 3).
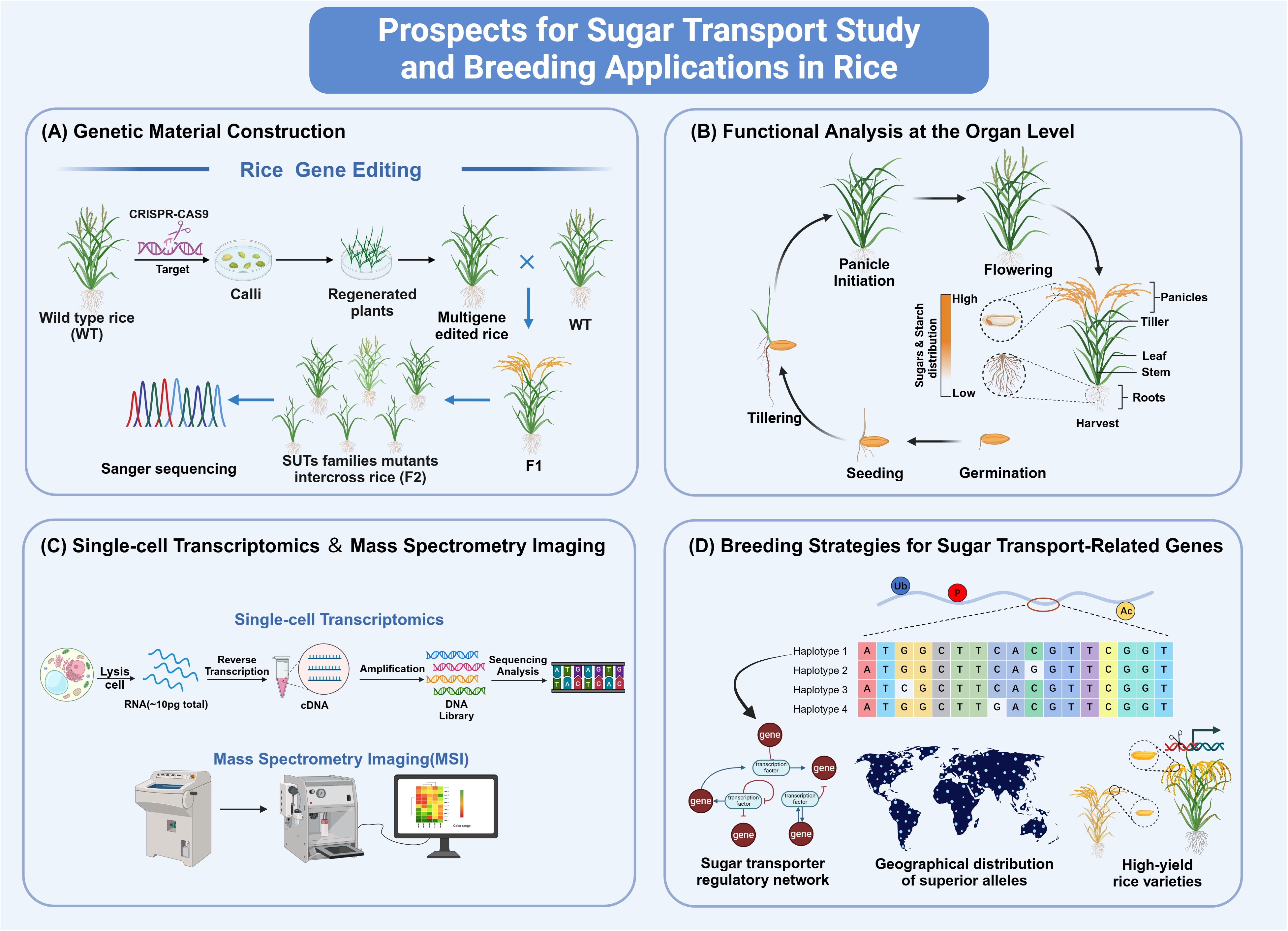
Figure 3. Prospects for sugar transport study and breeding applications in rice. (A) Gene editing technologies such as CRISPR/Cas9 have simplified the creation of mutants for SUT and SWEET families, facilitating the study of interactions between sugar transport proteins. (B) Measuring sugar and starch in tissues at various stages can reveal differences in their distribution. (C) Applying Single-cell transcriptomics and Spatial metabolomics allows for precise examination of multiple cells. (D) Using tissue-specific promoters, gene editing, and synthetic breeding with rice databases to select superior alleles can optimize sugar transport genes and boost rice yield, which is crucial for breeding. The figure was created with BioRender.com.
7.1 Genetic material construction
With the rapid advancement of gene editing technologies (Cong et al., 2013), obtaining a complete set of mutants for the genes of SUT and SWEET families has become more accessible. For instance, researchers have extensively employed CRISPR/Cas9 knockout techniques to generate mutants for most SWEETs and conducted comprehensive studies on these mutants (Li et al., 2022d). Furthermore, due to the redundancy of sugar transport proteins, it is now possible to create double, triple, or multiple mutants, thereby providing a rich genetic resource for investigating the precise interactions between these sugar transporters.
7.2 Functional analysis at the organ level
To elucidate the function of sugar transporters, measuring the sugar and starch content in different tissues at various developmental stages is essential. It could help uncover the differences in the distribution of sugars and starches across tissues (Sun et al., 2023). Such analyses will lay the foundation for further in-depth investigations into their microscopic functional differences using single-cell transcriptomics and spatial metabolomics techniques.
7.3 Single-cell transcriptomics
Our expanding knowledge highlights the remarkable heterogeneity in each cell, even within the same cell type. This intricate diversity and the various cell types in tissues amplify the need for spatially resolved single-cell omics techniques (Gilmore et al., 2019). In recent years, high-throughput single-cell transcriptomics has significantly progressed (Potter, 2018). Profiling multiple cells allows for a precise examination, unveiling unique insights into developmental processes, transcriptional pathways, and the molecular intricacies within tissues (Denyer et al., 2019). A notable investigation centered on vascular cells within Arabidopsis leaves, culminating in establishing a single-cell transcriptome of leaf vasculature. This comprehensive study successfully identified a minimum of 19 distinct cell clusters. Remarkably, among these clusters, PP cells exhibited a substantial enrichment of transporter genes, with prominent candidates including SWEET11 and SWEET12 (Kim et al., 2021a).
Leveraging this technology allows for the precise determination of the expression patterns of each sugar transporter gene at a single-cell resolution. Investigating these patterns is vital, encompassing the examination of single-cell expression patterns of sugar transporter genes, analysis of changes in downstream gene expression within sugar transporter mutants, and exploration of single-cell expression variations in upstream genes of the mutants. Furthermore, employing additional techniques such as in situ hybridization and immunofluorescence is pivotal and will substantially expedite the study of rice sugar transport (Qin et al., 2021b).
7.4 High-resolution spatial metabolomics technology
In recent decades, MSI has emerged as a powerful tool for non-targeted, label-free chemical imaging. It eliminates the need to pre-select specific molecular species for analysis, making detecting compounds without chemical modification or labeling agents possible. MSI encompasses various techniques such as MALDI-MSI (Matrix-assisted Laser Desorption Ionization Mass Spectrometry Imaging), DESI-MSI (Desorption Electrospray Ionization Mass Spectrometry Imaging), LAESI-MSI (Laser Ablation Electrospray Ionization Mass Spectrometry Imaging), SIMS (Imaging Secondary Ion Mass Spectrometry) and so on (Spengler, 2015; Buchberger et al., 2018).
By employing MSI, we can gain precise insights into the spatiotemporal distribution variations of sugars, including sucrose, fructose, and glucose (Hu et al., 2017). Utilizing MALDI-MSI, a recent study accurately depicted the spatial distribution of free soluble sugars (glucose, fructose, sucrose, and maltose) in rice seeds during the dough stage. These four free soluble sugars exhibited similar spatial distribution patterns, primarily concentrated in the seed cortex and embryo with high abundance (Zhao et al., 2023).
7.5 Breeding strategies for sugar transport-related genes
Merely elevating the expression of sugar transporters may not inevitably augment phloem transport capacity and has the potential to yield undesired outcomes. As demonstrated, the continuous overexpression of OsDOF11 leads to decreased yield, while the tissue-specific activation of OsDOF11 significantly enhances yield (Wu et al., 2018; Kim et al., 2021b). This underscores the crucial role of tissue-specific approaches when optimizing genes associated with sugar transport. Consequently, the strategic utilization of tissue-specific and site-specific promoters bears substantial significance.
In addition, the extensive publication of rice germplasm resources and genomic data has laid the foundation for identifying superior alleles of sugar transport-related genes (Wang et al., 2018; Qin et al., 2021a; Wang et al., 2023). Firstly, screening variant sites (including SNPs and InDels) within the promoters and coding regions of sugar transport-related genes and clustering analysis of their Haps (Haplotypes). Secondly, utilizing available databases of the yield-related traits, including 1000-grain weight, panicle grain number, and sugar distribution characteristics, such as 600 rice resources database (http://ricevarmap.ncpgr.cn/), analyzed their association with haplotypes of sugar transport-related genes for superior allelic variants identification. Thirdly, superior allelic variants enable functional validation and the creation of breeding materials in major rice cultivars. This could be achieved through precise gene editing and integrated breeding techniques, which facilitate the precise management of the balance of source-sink-flow.
In conclusion, the union of knowledge and methodological approaches concerning the cellular and tissue-level partitioning of sugar transporters, the elucidation of sugar transporters regulating networks, and their crosstalk with developmental mechanisms is imperative for a more sophisticated comprehension and precise manipulation of sugar transport mechanisms. This integrative perspective not only refines our grasp of the underlying regulatory models governing sugar transport but also prompts the optimization of rice germplasm. The strategic application of such knowledge is pivotal for enhancing rice yield potential and for stimulating its response to the environment.
Author contributions
JL: Writing – original draft, Writing – review & editing. CH: Writing – original draft, Writing – review & editing. SL: Writing – review & editing. YG: Writing – review & editing. YZ: Writing – review & editing. LZ: Writing – review & editing. XZ: Writing – review & editing. DX: Writing – review & editing. XL: Writing – review & editing. HL: Writing – review & editing. XY: Writing – review & editing. YW: Writing – review & editing. JS: Writing – review & editing. BY: Writing – review & editing. JW: Writing – review & editing. PW: Writing – review & editing. XD: Funding acquisition, Investigation, Supervision, Writing – review & editing. CS: Conceptualization, Funding acquisition, Investigation, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by grants from the Natural Science Foundation of Sichuan Province (2023NSFSC0213, 2024NSFTD0022, and 2022NSFSC1748), National Natural Science Foundation of China (32172022, 31371602, 31401358, and 32260502), Sichuan Science and Technology Program (2020YJ0408), Sichuan Rice Innovation Team of National Modern Agricultural Industry Technology System (CXTD-2024-01).
Acknowledgments
The figures were created with BioRender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abelenda, J. A., Bergonzi, S., Oortwijn, M., Sonnewald, S., Du, M., Visser, R. G. F., et al. (2019). Source-sink regulation is mediated by interaction of an FT homolog with a SWEET protein in potato. Curr. Biol. 29, 1178–1186 e1176. doi: 10.1016/j.cub.2019.02.018
Alexandratos, N., Bruinsma, J. (2012). World agriculture towards 2030/2050: the 2012 revision. Rome: ESA Working Paper. doi: 10.22004/ag.econ.288998
Anjali, A., Fatima, U., Manu, M. S., Ramasamy, S., Senthil-Kumar, M. (2020). Structure and regulation of SWEET transporters in plants: An update. Plant Physiol. Biochem. 156, 1–6. doi: 10.1016/j.plaphy.2020.08.043
Antony, G., Zhou, J., Huang, S., Li, T., Liu, B., White, F., et al. (2010). Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os-11N3. Plant Cell 22, 3864–3876. doi: 10.1105/tpc.110.078964
Aoki, N., Hirose, T., Furbank, R. T. (2012). “Sucrose transport in higher plants: from source to sink,” in Photosynthesis. Advances in Photosynthesis and Respiration, vol. 34 . Eds. Eaton-Rye, J., Tripathy, B., Sharkey, T. (Springer, Dordrecht). doi: 10.1007/978-94-007-1579-0_28
Aoki, N., Hirose, T., Scofield, G. N., Whitfeld, P. R., Furbank, R. T. (2003). The sucrose transporter gene family in rice. Plant Cell Physiol. 44, 223–232. doi: 10.1093/pcp/pcg030
Bai, A. N., Lu, X. D., Li, D. Q., Liu, J. X., Liu, C. M. (2016). NF-YB1-regulated expression of sucrose transporters in aleurone facilitates sugar loading to rice endosperm. Cell Res. 26, 384–388. doi: 10.1038/cr.2015.116
Bitterlich, M., Krugel, U., Boldt-Burisch, K., Franken, P., Kuhn, C. (2014). The sucrose transporter SlSUT2 from tomato interacts with brassinosteroid functioning and affects arbuscular mycorrhiza formation. Plant J. 78, 877–889. doi: 10.1111/tpj.12515
Boone, C., Bussey, H., Andrews, B. J. (2007). Exploring genetic interactions and networks with yeast. Nat. Rev. Genet. 8, 437–449. doi: 10.1038/nrg2085
Botha, C. E., Aoki, N., Scofield, G. N., Liu, L., Furbank, R. T., White, R. G. (2008). A xylem sap retrieval pathway in rice leaf blades: evidence of a role for endocytosis? J. Exp. Bot. 59, 2945–2954. doi: 10.1093/jxb/ern150
Braun, D. M. (2022). Phloem loading and unloading of sucrose: what a long, strange trip from source to sink. Annu. Rev. Plant Biol. 73, 553–584. doi: 10.1146/annurev-arplant-070721-083240
Braun, D. M., Wang, L., Ruan, Y. L. (2014). Understanding and manipulating sucrose phloem loading, unloading, metabolism, and signalling to enhance crop yield and food security. J. Exp. Bot. 65, 1713–1735. doi: 10.1093/jxb/ert416
Brown, M. H., Paulsen, I. T., Skurray, R. A. (1999). The multidrug efflux protein NorM is a prototype of a new family of transporters. Mol. Microbiol. 31, 394–395. doi: 10.1046/j.1365-2958.1999.01162.x
Buchberger, A. R., DeLaney, K., Johnson, J., Li, L. (2018). Mass spectrometry imaging: a review of emerging advancements and future insights. Anal. Chem. 90, 240. doi: 10.1021/acs.analchem.7b04733
Büttner, M. (2007). The monosaccharide transporter (-like) gene family in Arabidopsis. FEBS Lett. 581, 2318–2324. doi: 10.1016/j.febslet.2007.03.016
Cakir, B., Shiraishi, S., Tuncel, A., Matsusaka, H., Satoh, R., Singh, S., et al. (2016). Analysis of the rice ADP-glucose transporter (OsBT1) indicates the presence of regulatory processes in the amyloplast stroma that control ADP-glucose flux into starch. Plant Physiol. 170, 1271–1283. doi: 10.1104/pp.15.01911
Chen, R., Deng, Y., Ding, Y., Guo, J., Qiu, J., Wang, B., et al. (2022). Rice functional genomics: decades’ efforts and roads ahead. Sci. China Life Sci. 65, 33–92. doi: 10.1007/s11427-021-2024-0
Cho, J. I., Burla, B., Lee, D. W., Ryoo, N., Hong, S. K., Kim, H. B., et al. (2010). Expression analysis and functional characterization of the monosaccharide transporters, OsTMTs, involving vacuolar sugar transport in rice (Oryza sativa). New Phytol. 186, 657–668. doi: 10.1111/j.1469-8137.2010.03194.x
CocK, J. H., Yoshida, S. (1972). Accumulation of 14C-labelled carbohydrate before flowering and its subsequent redistribution and respiration in the rice plant. J. Crop Sci. 41, 226–234. doi: 10.1626/jcs.41.226
Comtet, J., Turgeon, R., Stroock, A. D. (2017). Phloem loading through plasmodesmata: A biophysical analysis. Plant Physiol. 175, 904–915. doi: 10.1104/pp.16.01041
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi: 10.1126/science.1231143
Cusick, M. E., Klitgord, N., Vidal, M., Hill, D. E. (2005). Interactome: gateway into systems biology. Hum. Mol. Genet. 14 Spec No. 2, R171–R181. doi: 10.1093/hmg/ddi335
Deng, X., An, B., Zhong, H., Yang, J., Kong, W., Li, Y. (2019). A novel insight into functional divergence of the MST gene family in rice based on comprehensive expression patterns. Genes 10, 239. doi: 10.3390/genes10030239
Denyer, T., Ma, X., Klesen, S., Scacchi, E., Nieselt, K., Timmermans, M. C. P. (2019). Spatiotemporal developmental trajectories in the arabidopsis root revealed using high-throughput single-cell RNA sequencing. Dev. Cell 48, 840–852 e845. doi: 10.1016/j.devcel.2019.02.022
Dhungana, S. R., Braun, D. M. (2021). Sugar transporters in grasses: Function and modulation in source and storage tissues. J. Plant Physiol. 266, 153541. doi: 10.1016/j.jplph.2021.153541
Durand, M., Mainson, D., Porcheron, B., Maurousset, L., Lemoine, R., Pourtau, N. (2018). Carbon source-sink relationship in Arabidopsis thaliana: the role of sucrose transporters. Planta 247, 587–611. doi: 10.1007/s00425-017-2807-4
Eom, J. S., Cho, J. I., Reinders, A., Lee, S. W., Yoo, Y., Tuan, P. Q., et al. (2011). Impaired function of the tonoplast-localized sucrose transporter in rice, OsSUT2, limits the transport of vacuolar reserve sucrose and affects plant growth. Plant Physiol. 157, 109–119. doi: 10.1104/pp.111.176982
Eom, J. S., Choi, S. B., Ward, J. M., Jeon, J. S. (2012). The mechanism of phloem loading in rice (Oryza sativa). Mol. Cells 33, 431–438. doi: 10.1007/s10059-012-0071-9
Eom, J. S., Luo, D., Atienza-Grande, G., Yang, J., Ji, C., Thi Luu, V., et al. (2019). Diagnostic kit for rice blight resistance. Nat. Biotechnol. 37, 1372–1379. doi: 10.1038/s41587-019-0268-y
Eom, J. S., Nguyen, C. D., Lee, D. W., Lee, S. K., Jeon, J. S. (2016). Genetic complementation analysis of rice sucrose transporter genes in Arabidopsis mutant. J. Plant Biol. 59, 231–237. doi: 10.1007/s12374-016-0015-6
Fei, H., Yang, Z., Lu, Q., Wen, X., Zhang, Y., Zhang, A., et al. (2021). OsSWEET14 cooperates with OsSWEET11 to contribute to grain filling in rice. Plant Sci. 306, 110851. doi: 10.1016/j.plantsci.2021.110851
Feng, B., Sun, Y., Ai, H., Liu, X., Yang, J., Liu, L., et al. (2018). Overexpression of sucrose transporter OsSUT1 affects rice morphology and physiology. CJRS 32, 549–556. doi: 10.16819/j.1001-7216.2018.8015
Fukumorita, T., Chino, M. (1982). Sugar, amino acid and inorganic contents in rice phloem sap. Plant Cell Physiol. 23, 273–283. doi: 10.1093/oxfordjournals.pcp.a076347
Furbank, R. T., Scofield, G. N., Hirose, T., Wang, X.-D., Patrick, J. W., Offler, C. E. (2001). Cellular localisation and function of a sucrose transporter OsSUT1 in developing rice grains. Funct. Plant Biol. 28, 1187–1196. doi: 10.1071/pp01111
Gao, Y., Zhang, C., Han, X., Wang, Z. Y., Ma, L., Yuan, P., et al. (2018). Inhibition of OsSWEET11 function in mesophyll cells improves resistance of rice to sheath blight disease. Mol. Plant Pathol. 19, 2149–2161. doi: 10.1111/mpp.12689
Gaxiola, R. A., Sanchez, C. A., Paez-Valencia, J., Ayre, B. G., Elser, J. J. (2012). Genetic manipulation of a “vacuolar” H(+)-PPase: from salt tolerance to yield enhancement under phosphorus-deficient soils. Plant Physiol. 159, 3–11. doi: 10.1104/pp.112.195701
Giaquinta, R. T. (1979). Phloem loading of sucrose: involvement of membrane ATPase and proton transport. Plant Physiol. 63, 744–748. doi: 10.1104/pp.63.4.744
Gilmore, I. S., Heiles, S., Pieterse, C. L. (2019). Metabolic imaging at the single-cell scale: recent advances in mass spectrometry imaging. Annu. Rev. Anal. Chem. (Palo Alto Calif) 12, 201–224. doi: 10.1146/annurev-anchem-061318-115516
Hirose, T., Zhang, Z., Miyao, A., Hirochika, H., Ohsugi, R., Terao, T. (2010). Disruption of a gene for rice sucrose transporter, OsSUT1, impairs pollen function but pollen maturation is unaffected. J. Exp. Bot. 61, 3639–3646. doi: 10.1093/jxb/erq175
Hu, X., Fang, C., Lu, L., Hu, Z., Shao, Y., Zhu, Z. (2017). Determination of soluble sugar profile in rice. J. Chromatogr. B 1058, 19–23. doi: 10.1016/j.jchromb.2017.05.001
Hu, Z., Tang, Z., Zhang, Y., Niu, L., Yang, F., Zhang, D., et al. (2021). Rice SUT and SWEET transporters. Int. J. Mol. Sci. 22, 11198. doi: 10.3390/ijms222011198
Jones, A. M., Xuan, Y., Xu, M., Wang, R. S., Ho, C. H., Lalonde, S., et al. (2014). Border control–a membrane-linked interactome of Arabidopsis. Science 344, 711–716. doi: 10.1126/science.1251358
Jonikas, M. C., Collins, S. R., Denic, V., Oh, E., Quan, E. M., Schmid, V., et al. (2009). Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science 323, 1693–1697. doi: 10.1126/science.1167983
Julius, B. T., Leach, K. A., Tran, T. M., Mertz, R. A., Braun, D. M. (2017). Sugar transporters in plants: new insights and discoveries. Plant Cell Physiol. 58, 1442–1460. doi: 10.1093/pcp/pcx090
Kim, J. Y., Symeonidi, E., Pang, T. Y., Denyer, T., Weidauer, D., Bezrutczyk, M., et al. (2021a). Distinct identities of leaf phloem cells revealed by single cell transcriptomics. Plant Cell 33, 511–530. doi: 10.1093/plcell/koaa060
Kim, P., Xue, C. Y., Song, H. D., Gao, Y., Feng, L., Li, Y., et al. (2021b). Tissue-specific activation of DOF11 promotes rice resistance to sheath blight disease and increases grain weight via activation of SWEET14. Plant Biotechnol. J. 19, 409–411. doi: 10.1111/pbi.13489
Krasensky, J., Jonak, C. (2012). Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 63, 1593–1608. doi: 10.1093/jxb/err460
Kühn, C., Barker, L., Bürkle, L., Frommer, W.-B. (1999). Update on sucrose transport in higher plants. J. Exp. Bot. 50, 935–953. doi: 10.1093/jexbot/50.suppl_1.935
Lalonde, S., Ehrhardt, D. W., Loque, D., Chen, J., Rhee, S. Y., Frommer, W. B. (2008). Molecular and cellular approaches for the detection of protein-protein interactions: latest techniques and current limitations. Plant J. 53, 610–635. doi: 10.1111/j.1365-313X.2007.03332.x
Lalonde, S., Sero, A., Pratelli, R., Pilot, G., Chen, J., Sardi, M. I., et al. (2010). A membrane protein/signaling protein interaction network for Arabidopsis version AMPv2. Front. Physiol. 1. doi: 10.3389/fphys.2010.00024
Laloum, T., De Mita, S., Gamas, P., Baudin, M., Niebel, A. (2013). CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci. 18, 157–166. doi: 10.1016/j.tplants.2012.07.004
Lee, S. J., Kang, K., Lim, J. H., Paek, N. C. (2022). Natural alleles of CIRCADIAN CLOCK ASSOCIATED1 contribute to rice cultivation by fine-tuning flowering time. Plant Physiol. 190, 640–656. doi: 10.1093/plphys/kiac296
Lemoine, R., Camera, S. L., Atanassova, R., Dédaldéchamp, F., Allario, T., Pourtau, N., et al. (2013). Source-to-sink transport of sugar and regulation by environmental factors. Front. Plant Sci. 4, 272. doi: 10.3389/fpls.2013.00272
Li, J., Blue, R., Zeitler, B., Strange, T. L., Pearl, J. R., Huizinga, D. H., et al. (2013). Activation domains for controlling plant gene expression using designed transcription factors. Plant Biotechnol. J. 11, 671–680. doi: 10.1111/pbi.12057
Li, J., Kim, Y. J., Zhang, D. (2022c). Source-to-sink transport of sugar and its role in male reproductive development. Genes (Basel) 13, 1323. doi: 10.3390/genes13081323
Li, C., Liu, X. J., Yan, Y., Alam, M. S., Liu, Z., Yang, Z. K., et al. (2022a). OsLHY is involved in regulating flowering through the Hd1- and Ehd1- mediated pathways in rice (Oryza sativa L.). Plant Sci. 315, 111145. doi: 10.1016/j.plantsci.2021.111145
Li, K., Tang, S., Zhang, S., Tian, Y., Qu, H., Gu, M., et al. (2023). Rice circadian clock regulator Nhd1 controls the expression of the sucrose transporter gene OsSUT1 and impacts carbon-nitrogen balance. J. Exp. Bot. 74, 1460–1474. doi: 10.1093/jxb/erac494
Li, P., Wang, L. H., Liu, H. B., Yuan, M. (2022d). Impaired-mediated sugar transportation impacts starch metabolism in developing rice seeds. Crop J. 10, 98–108. doi: 10.1016/j.cj.2021.04.012
Li, S., Wei, X., Ren, Y., Qiu, J., Jiao, G., Guo, X., et al. (2017). OsBT1 encodes an ADP-glucose transporter involved in starch synthesis and compound granule formation in rice endosperm. Sci. Rep. 7, 40124. doi: 10.1038/srep40124
Li, Z., Wei, X., Tong, X., Zhao, J., Liu, X., Wang, H., et al. (2022e). The OsNAC23-Tre6P-SnRK1a feed-forward loop regulates sugar homeostasis and grain yield in rice. Mol. Plant 15, 706–722. doi: 10.1016/j.molp.2022.01.016
Li, D., Xu, R., Lv, D., Zhang, C., Yang, H., Zhang, J., et al. (2020). Identification of the core pollen-specific regulation in the rice osSUT3 promoter. Int. J. Mol. Sci. 21, 1909. doi: 10.3390/ijms21061909
Li, G., Zhou, C., Yang, Z., Zhang, C., Dai, Q., Huo, Z., et al. (2022b). Low nitrogen enhances apoplastic phloem loading and improves the translocation of photoassimilates in rice leaves and stems. Plant Cell Physiol. 63, 991–1007. doi: 10.1093/pcp/pcac066
Liu, D., Xu, L., Wang, W., Jia, S., Jin, S., Gao, J. (2020). OsRRM, an RNA-binding protein, modulates sugar transport in rice (Oryza sativa L.). Front. Plant Sci. 11. doi: 10.3389/fpls.2020.605276
Ma, B., Zhang, L., He, Z. (2023). Understanding the regulation of cereal grain filling: The way forward. J. Integr. Plant Biol. 65, 526–547. doi: 10.1111/jipb.13456
Ma, L., Zhang, D., Miao, Q., Yang, J., Xuan, Y., Hu, Y. (2017). Essential role of sugar transporter osSWEET11 during the early stage of rice grain filling. Plant Cell Physiol. 58, 863–873. doi: 10.1093/pcp/pcx040
Marger, M. D., Saier, M. H., Jr. (1993). A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends Biochem. Sci. 18, 13–20. doi: 10.1016/0968-0004(93)90081-w
Mason, T., Maskell, E. (1928). Studies on the transport of carbohydrates in the cotton plant: II. The factors determining the rate and the direction of movement of sugars. Ann. Bot. 42, 571–636. doi: 10.1093/oxfordjournals.aob.a090131
Mathan, J., Singh, A., Ranjan, A. (2021a). Sucrose transport and metabolism control carbon partitioning between stem and grain in rice. J. Exp. Bot. 72, 4355–4372. doi: 10.1093/jxb/erab066
Mathan, J., Singh, A., Ranjan, A. (2021b). Sucrose transport in response to drought and salt stress involves ABA-mediated induction of OsSWEET13 and OsSWEET15 in rice. Physiol. Plant 171, 620–637. doi: 10.1111/ppl.13210
Matsukura, C., Saitoh, T., Hirose, T., Ohsugi, R., Perata, P., Yamaguchi, J. (2000). Sugar uptake and transport in rice embryo. Expression of companion cell-specific sucrose transporter (OsSUT1) induced by sugar and light. Plant Physiol. 124, 85–93. doi: 10.1104/pp.124.1.85
Mengzhu, L., Gaopeng, W., Yue, W., Yi, R., Ganghua, L., Zhenghui, L., et al. (2020). Function analysis of sucrose transporter OsSUT4 in sucrose transport in rice. Chin. J. Rice Sci. 34, 491. doi: 10.16819/j.1001-7216.2020.0316
Miras, M., Pottier, M., Schladt, T. M., Ejike, J. O., Redzich, L., Frommer, W. B., et al. (2022). Plasmodesmata and their role in assimilate translocation. J. Plant Physiol. 270, 153633. doi: 10.1016/j.jplph.2022.153633
Mohammad, A., Muhammad, A., Muhammad, S. (1993). “Genetic improvement in physiological traits of rice yield,” in Genetic improvement of field crops (Boca Raton, Florida: CRC Press), 413–455.
Morii, M., Sugihara, A., Takehara, S., Kanno, Y., Kawai, K., Hobo, T., et al. (2020). The dual function of osSWEET3a as a gibberellin and glucose transporter is important for young shoot development in rice. Plant Cell Physiol. 61, 1935–1945. doi: 10.1093/pcp/pcaa130
Nadal, M., Sawers, R., Naseem, S., Bassin, B., Kulicke, C., Sharman, A., et al. (2017). An N-acetylglucosamine transporter required for arbuscular mycorrhizal symbioses in rice and maize. Nat. Plants 3, 1–7. doi: 10.1038/nplants.2017.73
Omote, H., Hiasa, M., Matsumoto, T., Otsuka, M., Moriyama, Y. (2006). The MATE proteins as fundamental transporters of metabolic and xenobiotic organic cations. Trends Pharmacol. Sci. 27, 587–593. doi: 10.1016/j.tips.2006.09.001
Patrick, J. W., Botha, F. C., Birch, R. G. (2013). Metabolic engineering of sugars and simple sugar derivatives in plants. Plant Biotechnol. J. 11, 142–156. doi: 10.1111/pbi.12002
Pizzio, G. A., Paez-Valencia, J., Khadilkar, A. S., Regmi, K., Patron-Soberano, A., Zhang, S., et al. (2015). Arabidopsis type I proton-pumping pyrophosphatase expresses strongly in phloem, where it is required for pyrophosphate metabolism and photosynthate partitioning. Plant Physiol. 167, 1541–1553. doi: 10.1104/pp.114.254342
Potter, S. S. (2018). Single-cell RNA sequencing for the study of development, physiology and disease. Nat. Rev. Nephrol. 14, 479–492. doi: 10.1038/s41581-018-0021-7
Qin, P., Lu, H., Du, H., Wang, H., Chen, W., Chen, Z., et al. (2021a). Pan-genome analysis of 33 genetically diverse rice accessions reveals hidden genomic variations. Cell 184, 3542–3558 e3516. doi: 10.1016/j.cell.2021.04.046
Qin, P., Zhang, G., Hu, B., Wu, J., Chen, W., Ren, Z., et al. (2021b). Leaf-derived ABA regulates rice seed development via a transporter-mediated and temperature-sensitive mechanism. Sci. Adv. 7, eabc8873. doi: 10.1126/sciadv.abc8873
Quesada, A., Galvan, A., Fernandez, E. (1994). Identification of nitrate transporter genes in Chlamydomonas reinhardtii. Plant J. 5, 407–419. doi: 10.1111/j.1365-313x.1994.00407.x
Regmi, K. C., Zhang, S., Gaxiola, R. A. (2016). Apoplasmic loading in the rice phloem supported by the presence of sucrose synthase and plasma membrane-localized proton pyrophosphatase. Ann. Bot. 117, 257–268. doi: 10.1093/aob/mcv174
Riesmeier, J. W., Willmitzer, L., Frommer, W. B. (1992). Isolation and characterization of a sucrose carrier cDNA from spinach by functional expression in yeast. EMBO J. 11, 4705–4713. doi: 10.1002/j.1460-2075.1992.tb05575.x
Ryoo, N., Eom, J. S., Kim, H. B., Vo, B. T., Lee, S. W., Hahn, T. R., et al. (2013). Expression and functional analysis of rice plastidic maltose transporter, OsMEX1. J. Korean Soc. Appl. Bl. 56, 149–155. doi: 10.1007/s13765-012-3266-z
Salvi, P., Agarrwal, R., Gandass, N., Manna, M., Kaur, H., Deshmukh, R. (2022). Sugar transporters and their molecular tradeoffs during abiotic stress responses in plants. Physiol. Plant 174, e13652. doi: 10.1111/ppl.13652
Scofield, G. N., Aoki, N., Hirose, T., Takano, M., Jenkins, C. L., Furbank, R. T. (2007a). The role of the sucrose transporter, OsSUT1, in germination and early seedling growth and development of rice plants. J. Exp. Bot. 58, 483–495. doi: 10.1093/jxb/erl217
Scofield, G. N., Hirose, T., Aoki, N., Furbank, R. T. (2007b). Involvement of the sucrose transporter, OsSUT1, in the long-distance pathway for assimilate transport in rice. J. Exp. Bot. 58, 3155–3169. doi: 10.1093/jxb/erm153
Scofield, G. N., Hirose, T., Gaudron, J. A., Furbank, R. T., Upadhyaya, N. M., Ohsugi, R. (2002). Antisense suppression of the rice transporter gene, OsSUT1, leads to impaired grain filling and germination but does not affect photosynthesis. Funct. Plant Biol. 29, 815–826. doi: 10.1071/PP01204
Siao, W., Chen, J. Y., Hsiao, H. H., Chung, P., Wang, S. J. (2011). Characterization of osSUT2 expression and regulation in germinating embryos of rice seeds. Rice 4, 39–49. doi: 10.1007/s12284-011-9063-1
Singh, J., Das, S., Jagadis Gupta, K., Ranjan, A., Foyer, C. H., Thakur, J. K. (2023). Physiological implications of SWEETs in plants and their potential applications in improving source-sink relationships for enhanced yield. Plant Biotechnol. J. 21, 1528–1541. doi: 10.1111/pbi.13982
Singh, J., James, D., Murali Achary, V. M., Patel, M. K., Thakur, J. K., Reddy, M., et al. (2021). Coordinated overexpression of OsSUT1, OsSWEET11 and OsSWEET14 in rice impairs carbohydrate metabolism that has implications in plant growth, yield and susceptibility to Xanthomonas oryzae pv oryzae (Xoo). bioRxiv 2021. doi: 10.1101/2021.01.07.425507v1.abstract
Song, W., Hao, Q., Cai, M., Wang, Y., Zhu, X., Liu, X., et al. (2020). Rice OsBT1 regulates seed dormancy through the glycometabolism pathway. Plant Physiol. bioch. 151, 469–476. doi: 10.1016/j.plaphy.2020.03.055
Sosso, D., Luo, D., Li, Q. B., Sasse, J., Yang, J., Gendrot, G., et al. (2015). Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat. Genet. 47, 1489–1493. doi: 10.1038/ng.3422
Spengler, B. (2015). Mass spectrometry imaging of biomolecular information. Anal. Chem. 87, 64–82. doi: 10.1021/ac504543v
Srivastava, A. C., Dasgupta, K., Ajieren, E., Costilla, G., McGarry, R. C., Ayre, B. G. (2009). Arabidopsis plants harbouring a mutation in AtSUC2, encoding the predominant sucrose/proton symporter necessary for efficient phloem transport, are able to complete their life cycle and produce viable seed. Ann. Bot. 104, 1121–1128. doi: 10.1093/aob/mcp215
Srivastava, A. C., Ganesan, S., Ismail, I. O., Ayre, B. G. (2008). Functional characterization of the Arabidopsis AtSUC2 Sucrose/H+ symporter by tissue-specific complementation reveals an essential role in phloem loading but not in long-distance transport. Plant Physiol. 148, 200–211. doi: 10.1104/pp.108.124776
Sun, L., Deng, R., Liu, J., Lai, M., Wu, J., Liu, X., et al. (2022). An overview of sucrose transporter (SUT) genes family in rice. Mol. Biol. Rep. 49, 5685–5695. doi: 10.1007/s11033-022-07611-x
Sun, Y., Reinders, A., LaFleur, K. R., Mori, T., Ward, J. M. (2010). Transport activity of rice sucrose transporters OsSUT1 and OsSUT5. Plant Cell Physiol. 51, 114–122. doi: 10.1093/pcp/pcp172
Sun, C., Wang, Y., Yang, X., Tang, L., Wan, C., Liu, J., et al. (2023). MATE transporter GFD1 cooperates with sugar transporters, mediates carbohydrate partitioning and controls grain-filling duration, grain size and number in rice. Plant Biotechnol. J. 21, 621–634. doi: 10.1111/pbi.13976
Sun, A. J., Xu, H. L., Gong, W. K., Zhai, H. L., Meng, K., Wang, Y. Q., et al. (2008). Cloning and expression analysis of rice sucrose transporter genes OsSUT2M and OsSUT5Z. J. Integr. Plant Biol. 50, 62–75. doi: 10.1111/j.1744-7909.2007.00596.x
Sun, C., Zhang, K., Zhou, Y., Xiang, L., He, C., Zhong, C., et al. (2021). Dual function of clock component OsLHY sets critical day length for photoperiodic flowering in rice. Plant Biotechnol. J. 19, 1644–1657. doi: 10.1111/pbi.13580
Takanashi, K., Shitan, N., Yazaki, K. (2014). The multidrug and toxic compound extrusion (MATE) family in plants. Plant Biotechnol. 31, 417–430. doi: 10.5511/plantbiotechnology.14.0904a
Upadhyay, N., Kar, D., Deepak Mahajan, B., Nanda, S., Rahiman, R., Panchakshari, N., et al. (2019). The multitasking abilities of MATE transporters in plants. J. Exp. Bot. 70, 4643–4656. doi: 10.1093/jxb/erz246
van Bel, A. J. (1996). Interaction between sieve element and companion cell and the consequences for photoassimilate distribution. Two structural hardware frames with associated physiological software packages in dicotyledons? J. Exp. Bot. 47, 1129–1140. doi: 10.1093/jxb/47.Special_Issue.1129
Vandeputte, G., Delcour, J. A. (2004). From sucrose to starch granule to starch physical behaviour: a focus on rice starch. Carbohydr. Polym. 58, 245–266. doi: 10.1016/j.carbpol.2004.06.003
Wang, T., He, W., Li, X., Zhang, C., He, H., Yuan, Q., et al. (2023). A rice variation map derived from 10 548 rice accessions reveals the importance of rare variants. Nucleic Acids Res. 51, gkad840. doi: 10.1093/nar/gkad840
Wang, W., Mauleon, R., Hu, Z., Chebotarov, D., Tai, S., Wu, Z., et al. (2018). Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 557, 43–49. doi: 10.1038/s41586-018-0063-9
Wang, E., Wang, J., Zhu, X., Hao, W., Wang, L., Li, Q., et al. (2008). Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat. Genet. 40, 1370–1374. doi: 10.1038/ng.220
Wang, G., Wu, Y., Ma, L., Lin, Y., Hu, Y., Li, M., et al. (2021). Phloem loading in rice leaves depends strongly on the apoplastic pathway. J. Exp. Bot. 72, 3723–3738. doi: 10.1093/jxb/erab085
Wen, S., Neuhaus, H. E., Cheng, J., Bie, Z. (2022). Contributions of sugar transporters to crop yield and fruit quality. J. Exp. Bot. 73, 2275–2289. doi: 10.1093/jxb/erac043
White, P. J., Ding, G. (2023). “Long-distance transport in the xylem and phloem,” in Marschner’s Mineral Nutrition of Plants (Amsterdam: Elsevier), 73–104.
Williams, L. E., Lemoine, R., Sauer, N. (2000). Sugar transporters in higher plants–a diversity of roles and complex regulation. Trends Plant Sci. 5, 283–290. doi: 10.1016/s1360-1385(00)01681-2
Wipf, D., Pfister, C., Mounier, A., Leborgne-Castel, N., Frommer, W. B., Courty, P. E. (2021). Identification of putative interactors of arabidopsis sugar transporters. Trends Plant Sci. 26, 13–22. doi: 10.1016/j.tplants.2020.09.009
Wirth, J., Chopin, F., Santoni, V., Viennois, G., Tillard, P., Krapp, A., et al. (2007). Regulation of root nitrate uptake at the NRT2.1 protein level in Arabidopsis thaliana. J. Biol. Chem. 282, 23541–23552. doi: 10.1074/jbc.M700901200
Wu, Y., Lee, S. K., Yoo, Y., Wei, J., Kwon, S. Y., Lee, S. W., et al. (2018). Rice transcription factor osDOF11 modulates sugar transport by promoting expression of sucrose transporter and SWEET genes. Mol. Plant 11, 833–845. doi: 10.1016/j.molp.2018.04.002
Xiong, Y., Ren, Y., Li, W., Wu, F., Yang, W., Huang, X., et al. (2019). NF-YC12 is a key multi-functional regulator of accumulation of seed storage substances in rice. J. Exp. Bot. 70, 3765–3780. doi: 10.1093/jxb/erz168
Xu, Q., Yin, S., Ma, Y., Song, M., Song, Y., Mu, S., et al. (2020). Carbon export from leaves is controlled via ubiquitination and phosphorylation of sucrose transporter SUC2. Proc. Natl. Acad. Sci. U.S.A. 117, 6223–6230. doi: 10.1073/pnas.1912754117
Yanagisawa, S., Sheen, J. (1998). Involvement of maize Dof zinc finger proteins in tissue-specific and light-regulated gene expression. Plant Cell 10, 75–89. doi: 10.1105/tpc.10.1.75
Yang, J., Luo, D., Yang, B., Frommer, W. B., Eom, J. S. (2018). SWEET11 and 15 as key players in seed filling in rice. New Phytol. 218, 604–615. doi: 10.1111/nph.15004
Yong, Z., Kotur, Z., Glass, A. D. (2010). Characterization of an intact two-component high-affinity nitrate transporter from Arabidopsis roots. Plant J. 63, 739–748. doi: 10.1111/j.1365-313X.2010.04278.x
Yuan, M., Chu, Z., Li, X., Xu, C., Wang, S. (2009). Pathogen-induced expressional loss of function is the key factor in race-specific bacterial resistance conferred by a recessive R gene xa13 in rice. Plant Cell Physiol. 50, 947–955. doi: 10.1093/pcp/pcp046
Yuan, M., Wang, S. (2013). Rice MtN3/saliva/SWEET family genes and their homologs in cellular organisms. Mol. Plant 6, 665–674. doi: 10.1093/mp/sst035
Zhang, C., Turgeon, R. (2018). Mechanisms of phloem loading. Curr. Opin. Plant Biol. 43, 71–75. doi: 10.1016/j.pbi.2018.01.009
Zhang, Y.-M., Yu, H.-X., Ye, W.-W., Shan, J.-X., Dong, N.-Q., Guo, T., et al. (2021b). A rice QTL GS3. 1 regulates grain size through metabolic-flux distribution between flavonoid and lignin metabolons without affecting stress tolerance. Commun. Biol. 4, 1171. doi: 10.1038/s42003-021-02686-x
Zhang, S., Zhang, Y., Li, K., Yan, M., Zhang, J., Yu, M., et al. (2021a). Nitrogen mediates flowering time and nitrogen use efficiency via floral regulators in rice. Curr. Biol. 31, 671–683 e675. doi: 10.1016/j.cub.2020.10.095
Zhao, Y., Hu, J., Zhang, Y., Tao, H., Li, L., He, Y., et al. (2023). Unveiling targeted spatial metabolome of rice seed at the dough stage using Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry imaging. Food Res. Int. 174, 113578. doi: 10.1016/j.foodres.2023.113578
Zhou, C., Lin, Q., Ren, Y., Lan, J., Miao, R., Feng, M., et al. (2023). A CYP78As–SMG4–COPII pathway promotes grain size in rice. Plant Cell 35, 4325. doi: 10.1093/plcell/koad239
Zhou, M., Deng, X., Jiang, Y., Zhou, G., Chen, J. (2023). Genome-wide identification and an evolution analysis of tonoplast Monosaccharide Transporter (TMT) genes in seven gramineae crops and their expression profiling in rice. Genes 14, 1140. doi: 10.3390/genes14061140
Keywords: rice (Oryza sativa L.), carbohydrates, “source-sink” theory, sugar transporters, regulatory pathway, high-yield
Citation: Li J, He C, Liu S, Guo Y, Zhang Y, Zhang L, Zhou X, Xu D, Luo X, Liu H, Yang X, Wang Y, Shi J, Yang B, Wang J, Wang P, Deng X and Sun C (2024) Research progress and application strategies of sugar transport mechanisms in rice. Front. Plant Sci. 15:1454615. doi: 10.3389/fpls.2024.1454615
Received: 25 June 2024; Accepted: 05 August 2024;
Published: 21 August 2024.
Edited by:
Hao Zhou, Sichuan Agricultural University, ChinaReviewed by:
Yu Ling, Guangdong Ocean University, ChinaGuanfu Fu, China National Rice Research Institute (CAAS), China
Copyright © 2024 Li, He, Liu, Guo, Zhang, Zhang, Zhou, Xu, Luo, Liu, Yang, Wang, Shi, Yang, Wang, Wang, Deng and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaojian Deng, eGpkZW5nQHNpY2F1LmVkdS5jbg==; Changhui Sun, c3VuaHVpMDMwN0AxNjMuY29t
†These authors have contributed equally to this work
 Jun Li
Jun Li Changcai He
Changcai He Shihang Liu1
Shihang Liu1 Jun Shi
Jun Shi Pingrong Wang
Pingrong Wang Xiaojian Deng
Xiaojian Deng Changhui Sun
Changhui Sun