- 1Citrus Research and Education Center, Institute of Food and Agricultural Sciences, University of Florida, Lake Alfred, FL, United States
- 2Department of Microbiology and Cell Science, Institute of Food and Agricultural Sciences, University of Florida, Lake Alfred, FL, United States
- 3Department of Horticultural Sciences, Institute of Food and Agricultural Sciences, University of Florida, Lake Alfred, FL, United States
Introduction: Citrus canker, caused by Xanthomonas citri subsp. citri (Xcc), is a devastating disease worldwide. Previously, we successfully generated canker-resistant Citrus sinensis cv. Hamlin lines in the T0 generation. This was achieved through the transformation of embryogenic protoplasts using the ribonucleoprotein (RNP) containing Cas12a and one crRNA to edit the canker susceptibility gene, CsLOB1, which led to small indels.
Methods: Here, we transformed embryogenic protoplasts of Hamlin with RNP containing Cas12a and three crRNAs.
Results: Among the 10 transgene-free genome-edited lines, long deletions were obtained in five lines. Additionally, inversions were observed in three of the five edited lines with long deletions, but not in any edited lines with short indel mutations, suggesting long deletions maybe required for inversions. Biallelic mutations were observed for each of the three target sites in four of the 10 edited lines when three crRNAs were used, demonstrating that transformation of embryogenic citrus protoplasts with Cas12a and three crRNAs RNP can be very efficient for multiplex editing. Our analysis revealed the absence of off-target mutations in the edited lines. These cslob1 mutant lines were canker- resistant and no canker symptoms were observed after inoculation with Xcc and Xcc growth was significantly reduced in the cslob1 mutant lines compared to the wild type plants.
Discussion: Taken together, RNP (Cas12a and three crRNAs) transformation of embryogenic protoplasts of citrus provides a promising solution for transgene-free multiplex genome editing with high efficiency and for deletion of long fragments.
Introduction
CRISPR/Cas-mediated genome editing has shown enormous potential in genetic improvements of crops. It has been used to improve disease resistance, yield, quality, and tolerance against abiotic stress among others (Zhu et al., 2020). For the adoption of the genome-edited crops, the plants are usually required to be transgene-free to address the public concerns and regulatory issues (Turnbull et al., 2021). In addition, other putative impacts have also been reported to result from transgenic expression of CRISPR/Cas including unintended genome editing owing to the consistent expression of CRISPR/Cas and disruption of gene functions at the insertion site (O'Malley and Ecker, 2010; He et al., 2018; Sturme et al., 2022). Most genome-edited plants were generated via Agrobacterium-mediated transformation resulting in transgenic plants. For the transgenic genome-edited plants, foreign genes can be removed relatively easily for annual crops by segregation through selfing or backcrossing (Bhattacharjee et al., 2023) and using a transgene-killer strategy (He et al., 2018). In addition, multiple technologies have been reported to generate transgene-free genome-edited plants in the T0 generation. For instance, transient expression of ribonucleoproteins of Cas9/gRNA or Cas12a/crRNA, as well as CRISPR/Cas DNA and RNA have been successfully used to generate transgene-free genome edited Arabidopsis thaliana, tobacco, lettuce, rice, wheat grape, apple, soybean, and cabbage (Woo et al., 2015; Malnoy et al., 2016; Svitashev et al., 2016; Zhang et al., 2016; Kim et al., 2017; Liang et al., 2017; Murovec et al., 2018). Viral vectors have also been used to generate transgene-free plants (Ma et al., 2020).
Citrus is one of the top three tree crops worldwide. Traditional citrus breeding usually takes 20-30 years owing to the long juvenility (Gmitter et al., 2012). Cas9/sgRNA was firstly used for citrus genome editing in 2013 (Jia and Wang, 2014a). Significant progress in citrus genome editing has since been made (Jia and Wang, 2014b; Jia et al., 2016, Jia et al, 2017; Leblanc et al., 2017; Peng et al., 2017; Zhang et al., 2017b; Jia et al., 2019; Zhu et al., 2019; Zou et al., 2019; Dutt et al., 2020; Huang et al., 2020;Huang et al., 2021; Jia et al., 2022; Mahmoud et al., 2022). It was reported that the biallelic/homozygous mutation rates was 89% for Carrizo citrange and 79% for Hamlin sweet orange via Agrobacterium-mediated transformation of epicotyls (Huang et al., 2021). Interestingly, genome editing via hairy root genetic transformation using Agrobacterium rhizogenes has also shown promises (Irigoyen et al., 2020; Ma et al., 2022; Wang et al., 2023).
CRISPR/Cas genome editing has successfully improved disease resistance against citrus canker by mutating LOB1, the canker susceptibility gene (Hu et al., 2014; Jia et al., 2016, Jia et al, 2017; Peng et al., 2017; Huang et al., 2021). Citrus canker caused by Xanthomonas citri pv. citri (Xcc) is arguably one of the top two citrus diseases in the world (Gottwald et al., 2002; Ference et al., 2018). The LOB1-edited pummelo, grapefruit, and sweet orange all showed no canker symptoms when inoculated with Xcc (Jia et al., 2016, Jia et al, 2017; Peng et al., 2017; Huang et al., 2021;Huang et al, 2022;Jia et al., 2022). Importantly, we have generated transgene-free canker resistant C. sinensis cv. Hamlin through transformation of embryogenic protoplasts with Cas12a/crRNA RNP (Su et al., 2023). In addition, a co-editing strategy based on Agrobacterium transformation has also been successfully used to generate transgene-free genome edited pummelo (Huang et al., 2023) and sweet orange (Jia et al., 2024).
In a previous study, one crRNA was used to guide the genome editing of the canker susceptibility gene LOB1 to generate transgene-free canker-resistant C. sinensis cv. Hamlin (Su et al., 2023). The mutations were mostly short deletions. Multiple gRNAs have been used to improve genome editing efficiency and generate long deletions (Xie et al., 2015). Here, we have conducted genome editing of the LOB1 gene in C. sinensis cv. Hamlin using three crRNAs through transformation of embryogenic protoplasts with Cas12a/3 crRNAs RNP. Three crRNAs indeed achieved high genome editing efficiency, led to multiplex genome editing, long deletions as well as inversions.
Materials and methods
Citrus plants and cell culture conditions
C. sinensis seedlings were grown in a greenhouse at the Citrus Research and Education Center in Lake Alfred, Florida. Embryogenic callus lines originating from immature ovules of C. sinensis cv. Hamlin were established and maintained using the Murashige and Tucker (1969, MT) medium (PhytoTech Labs, Lenexa, KS, USA). This medium was supplemented with 5.0 mg/L of kinetin (KIN) and 500 mg/L of malt extract. Meanwhile, the suspension cell culture of C. sinensis cv. Hamlin was maintained in darkness at 22°C and sub-cultured biweekly. The cultivation medium was the H+H medium (Omar et al., 2016a). After 7-10 days following subculturing, the suspension cells were used for protoplast isolation.
Protoplast isolation
Protoplast isolation was conducted as described previously (Su et al., 2023). Briefly, embryogenic C. sinensis cv. Hamlin protoplasts were isolated from the suspension cells with digestion solution (2.5 × volume BH3 (Omar et al., 2016a) and 1.5× volume enzyme solution (0.7 M mannitol, 24 mM CaCl2, 6.15 mM MES buffer, 2.4% (w/v) Cellulase Onozuka RS (MX7353, Yakult Honsha, Minato-ku, Tokyo, Japan), 2.4% (w/v) Macerozyme R-10 (MX7351, Yakult Honsha), pH 5.6) for 16-20 hours at 28°C. The digested protoplast mixture was then filtered with a 40 μm cell strainer (431750, Corning, Durham, NC, USA) into a 50 mL Falcon tube, which were centrifuged at 60 g for 7 min. The pellets were resuspended with BH3 medium to rinse the protoplast. After repeating the washing step, the protoplasts were resuspended in 2 mL BH3 medium and diluted to 1 ×106 cell/mL and kept in dark at room temperature for 1 hour.
Transformation of embryogenic citrus protoplasts and plant regeneration
Protoplast transformation with RNP and plant regeneration were conducted as previously described (Omar et al., 2016a; Liu et al., 2023). Briefly, for RNP assembly, 0.81 nmol LbCas12U protein and 0.45 nmol of each of three crRNAs were assembled in 1 x Nuclease Reaction Buffer (NEB). Protein and crRNAs were mixed and incubated for 10 min at room temperature. For transfection, 1 mL protoplast cells, preassembled RNP, and 1 mL PEG-CaCl2 (0.4 M mannitol, 100 mM CaCl2, and 40% PEG-4000) were mixed and kept at room temperature for 15 min in dark followed by washing with BH3 medium twice. Transfected citrus protoplast cells were kept in liquid medium (1:1:1 (v:v:v) mixture of BH3 and EME sucrose 0.6 M and EME sucrose 0.15 M) for 3 – 4 weeks at 28°C in dark without shaking. Then citrus cells were transferred to EME sucrose medium supplemented with 1:2 mixture of BH3 and EME maltose 0.15 M and kept at 28°C for 3–4 weeks in dark. Calli were regenerated from protoplasts and transferred to EME maltose solid medium supplemented with 1:2 mixture of BH3 and EME maltose 0.15 M and kept at 28°C in dark for 3 – 4 weeks to generate embryos. Embryos were transferred to EME maltose solid medium and kept at room temperature under light for 3 – 4 weeks, and then to solid EME1500 medium and kept at room temperature under light for 3 – 4 months to generate shoots. Small plantlets were transferred to MS medium and kept at room temperature for 3 – 4 weeks. The regenerated shoots were micro-grafted onto Carrizo citrange rootstock in liquid media and kept in tissue culture room at 25°C under light for 3 – 4 weeks, grown in stonewool cubes in a growth chamber at 25°C under light for 1 month, then planted in soil.
Mutation identification and off-target test
Genomic DNA was isolated from leaves of both wild-type and cslob1-edited lines of C. sinensis cv. Hamlin. The Supplementary Table S1 lists the primers employed in the PCR. For PCR amplification, the CloneAmp HiFi PCR Premix (639298, TakaraBio USA, San Jose, CA, USA) was utilized, in accordance with the manufacturer’s guidelines. The amplification procedure consisted of an initial step at 98°C for 30 seconds, followed by 40 cycles at 98°C for 10 seconds, 54°C for 10 seconds, and 72°C for 45 seconds. A final extension was performed at 72°C for 5 minutes. The resulting PCR amplicons were cloned and subjected to sequencing employing the amplifying primers. The cloning was conducted using the Zero Blunt TOPO PCR Cloning Kit (450245, Thermo Fisher, San Jose, CA, USA), followed by transformation into Stellar Competent Cells from Takara. For the amplification and Sanger sequencing of single colonies, M13-F (GTAAAACGACGGCCAGTG) and M13-R (CAGGAAACAGCTATGACC) primers were used.
The off-target candidate sites sequences were listed in Supplementary Table S1. The off-target sites were predicted using CRISPR-P 2.0 program (Liu et al., 2017). Then the predicted off-target sites with TTTV PAM site were further confirmed as off-target candidate sites and aligning target sequence with whole genome using BLAST program. Based on the whole genome sequencing mapping results, mutations of off-target sites were detected using the SAMtools package version 1.2 and deepvariant program version 1.4.0. There were no any off-target mutations detected in mutated plants.
DNA library construction, sequencing, and data analysis
Following the manufacturer’s protocol of short read DNA sequencing from Illumina, the library was prepared. 150-bp paired-end reads were generated using the Illumina NovaSeq 6000 platform according to the manufacturer’s instructions at Novogene, China. The raw paired-end reads were filtered to remove low-quality reads using fastp program version 0.22.0 (Chen et al., 2018b). On average, more than 17.21 Gb of high-quality data was generated for each edited sweet orange lines (Supplementary Table S2). To identify the mutations (single nucleotide polymorphisms, deletions and insertions) in the edited plant genomes, high quality paired-end short genomic reads were mapped to sweet orange (Wang et al., 2021) reference genome using Bowtie2 software version 2.2.6 (Langmead and Salzberg, 2012). Based on the mapping results, mutations were identified using the SAMtools package version 1.2 (Li et al., 2009) and deepvariant program version 1.4.0 (Poplin et al., 2018) The identified mutations were filtered by quality and sequence depth (mapping quality > 10 and mapping depth >10). The mutations of target sites were visualized using IGV software version 2.15.4 (Robinson et al., 2011).
Quantitative reverse-transcription PCR
Xcc strain 306 was infiltrated into wild type C. sinensis cv. Hamlin and transgene-free cslob1 mutants at the concentration of 1 × 107 cfu/mL. The infiltration areas of the leaf samples were collected at 9 days post-inoculation (dpi) for RNA isolation. Four biological replicates were used, each leaf is one biological replicate. Total RNA was extracted by TRIzol Reagent (15596026, Thermo-Fisher) following the manufacturer’s instructions. cDNA was synthesized by qScript cDNA SuperMix (101414, Quantabio, Beverly, MA, USA). Primers used for qRT-PCR were listed in Supplementary Table S1. Briefly, qPCR was performed with QuantiStudio3 (Thermo-Fisher) using SYBR Green Real-Time PCR Master Mix (4309155, Thermo-Fisher) in a 10 μL reaction. The standard amplification protocol was 95°C for 3 min followed by 40 cycles of 95°C 15 s, 60°C for 60 s. The CsGAPDH gene was used as an endogenous control. All reactions were performed in triplicate. Relative gene expression and statistical analysis were calculated using the 2-ΔΔCT method (Livak and Schmittgen, 2001). qRT-PCR was repeated twice with similar results.
Microscopy assay
The infiltration areas of Xcc-infiltrated leaves and non-Xcc-infiltrated leaves from both the wild type C. sinensis cv. Hamlin and the cslob1 mutant were carefully excised using sterilized blades. These excised portions were fixed using 4% paraformaldehyde for a minimum of 2 hours. Subsequently, the specimens underwent a dehydration process before embedded into paraffin chips. The paraffin-embedded chips were cut using a Leica 2155 microtome. Each section was crafted to a thickness of 8 μm. These thin ribbons were positioned onto glass slides and incubated at 37°C overnight to ensure proper heat fixation. Following this, a process of dewaxing and rehydration was performed. The slides were subsequently stained using a 0.05% toluidine blue solution for 30 seconds. After staining, they were rinsed in ddH2O, subjected to dehydration, and then a drop of mounting medium was added before covering with a coverslip. Once the mounting medium had solidified, photographs of the slides were captured using the Leica LasX software (Leica Biosystems Inc., Lincolnshire, IL, USA). These images were taken under a bright-field microscope (Olympus BX61; Olympus Corporation, Shinjuku City, Tokyo, Japan).
Xcc growth assay
For Xcc infiltration assay, the pictures were captured at 9 days after infiltration. For Xcc spraying assay, Xcc strain 306 was spraying onto wild type C. sinensis cv. Hamlin and transgene-free cslob1 mutants at the concentration of 5 × 108 cfu/mL. and the leaves were punctured with syringes, creating 8 wounds/leave before spray. After spraying, the plants were covered with plastic bag to maintain the humidity for 24 hours. Images were taken at 18 days after spray. Leaf discs with a diameter of 0.5 cm were collected from the leaves of the plants and leaf discs were ground in 0.2 mL of sterilized H2O. To facilitate the assessment of bacterial concentration, serial dilutions of the grinding suspensions, each amounting to 100 μL, were meticulously spread across NA plates (dilutions ranging from 10-1 to 10-6). Bacterial colonies were counted after 48 h and the number of CFU (colony-forming units) per cm2 of leaf disc was calculated and presented with Prism GraphPad software.
Quantification of H2O2 concentration
H2O2 concentration measurement was conducted (Sels et al., 2008; Liu et al., 2023). Briefly, four 0.5-cm-diameter leaf disks from the same leaf that had been injected with water or Xcc (1 × 107 cfu/mL) were pooled and stored in a 1.5 mL tube with 0.5 mL of double-distilled (DD) water. The samples were rotated on a platform at 20 rpm for 30 min, and ddH2O was replenished with fresh ddH2O. Samples were incubated for an additional 6 h on a rotating platform at 20 rpm. H2O2 concentration was quantified in the supernatants using the Pierce Quantification Peroxide Assay Kit (23280, Thermo Fisher Scientific, Waltham, MA, USA).
Data availability
The raw reads of genome resequencing for sweet orange plants generated in this study were deposited in the NCBI Bioproject database under the accession number PRJNA1077621. The reference genome of sweet orange was downloaded from Citrus Pan-genome to Breeding Database [http://citrus.hzau.edu.cn/index.php].
Results
Genome editing efficacy via Cas12a/crRNAs RNP using 3 crRNAs
For evaluating the gene-editing efficacy via Cas12a/crRNAs RNP using 3 crRNAs, we targeted the CsLOB1 gene (canker susceptibility gene) by designing three crRNAs. The three crRNAs were located at the promoter region (crRNA1), exon1 region (crRNA2), and exon2 region (crRNA3) (Figure 1A). LbCas12aU/3 crRNAs RNP mixture was transfected into C. sinensis cv. Hamlin protoplast cells via PEG-mediated protoplast transformation (Omar et al., 2016b; Su et al., 2023). The DNA samples of transfected protoplast cells at 3 days post transformation (DPT) were isolated and PCR-amplified. The corresponding PCR products were shorter than wild type Hamlin (Figure 1B), which indicated long deletion in the CsLOB1 gene caused by the Cas12a/3 crRNAs RNP transformation. The short DNA band (Figure 1B) was cut and subjected to colony PCR and Sanger sequencing. All eight colonies were confirmed to harbor different long deletions (Figure 1C).
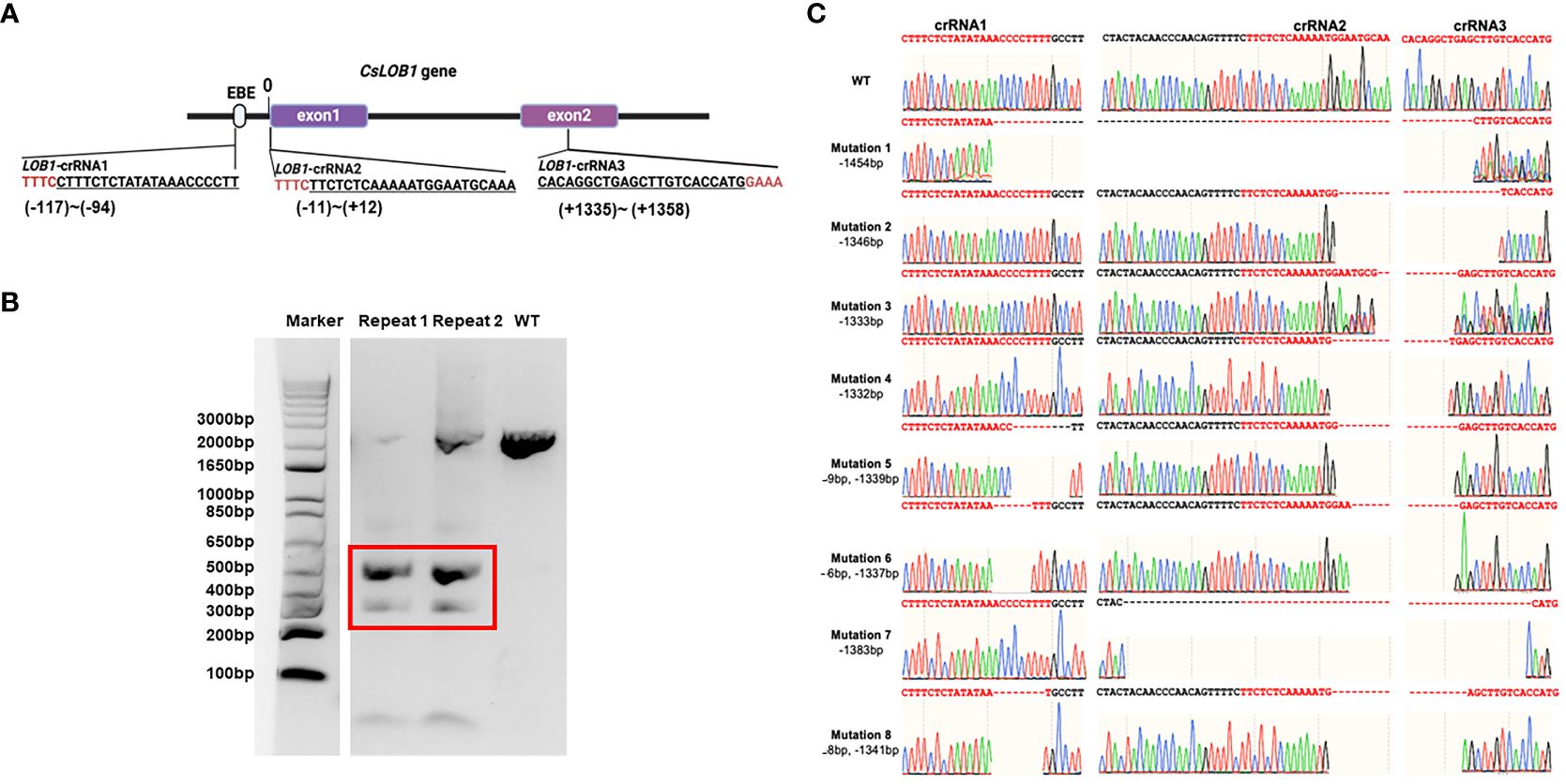
Figure 1 Schematic representation of 3 crRNAs targeting CsLOB1 gene and evaluation activity of Cas12a RNPs containing three crRNAs in C. sinensis cv. Hamlin protoplast at 3 days post transformation (dpt). (A) The schematic representation of the CsLOB1 gene and crRNAs. EBE: effector-binding element. Line fragments indicate introns. The PAM (protospacer adjacent motif), either TTTC or GAAA, is highlighted in red. The corresponding crRNAs’ location are labeled below the crRNA sequence. The space between promoter region and the start codon is ‘0’. (B) Gel picture of CsLOB1 PCR product amplified from protoplast at 3 days post transformation (dpt). Electrophoresis was performed using a 2% agarose gel. The bands enclosed in the red box correspond to long deletion PCR products, which were subsequently excised, purified, and employed for ligation and colony PCR. (C) Sequencing results of colonies from PCR products that were excised from the red box region in panel (B). Among the 8 sequencing reactions, all revealed distinct long deletions associated with crRNAs. “-” indicates deletion at target sites.
Transgene-free genome editing of the CsLOB1 gene in C. sinensis cv. Hamlin
Next, we employed Cas12a/crRNAs RNP to generate transgene-free canker-resistant C. sinensis cv. Hamlin by editing both promoter region and coding region with 3 crRNAs (Figure 1A). Approximately 9 to 15 months post transformation, we obtained a total of 10 individual regenerated lines from transformed embryogenic citrus protoplasts (Table 1). The 10 regenerated lines exhibited normal growth characteristics akin to the wild type (Figure 2A). Sanger sequencing analysis and whole genome sequencing demonstrated that the 10 lines were mutated in the CsLOB1 gene (Figure 2B, Supplementary Figure S1–S20). Among them, 7 lines (#1, 2, 3, 6, 7, 11 and 12) contained biallelic mutations (Table 1). Three lines (#4, 8, and 13) were chimeric (Table 1). Nevertheless, all the edited lines showed 100% mutation rate regardless of being biallelic or chimeric.
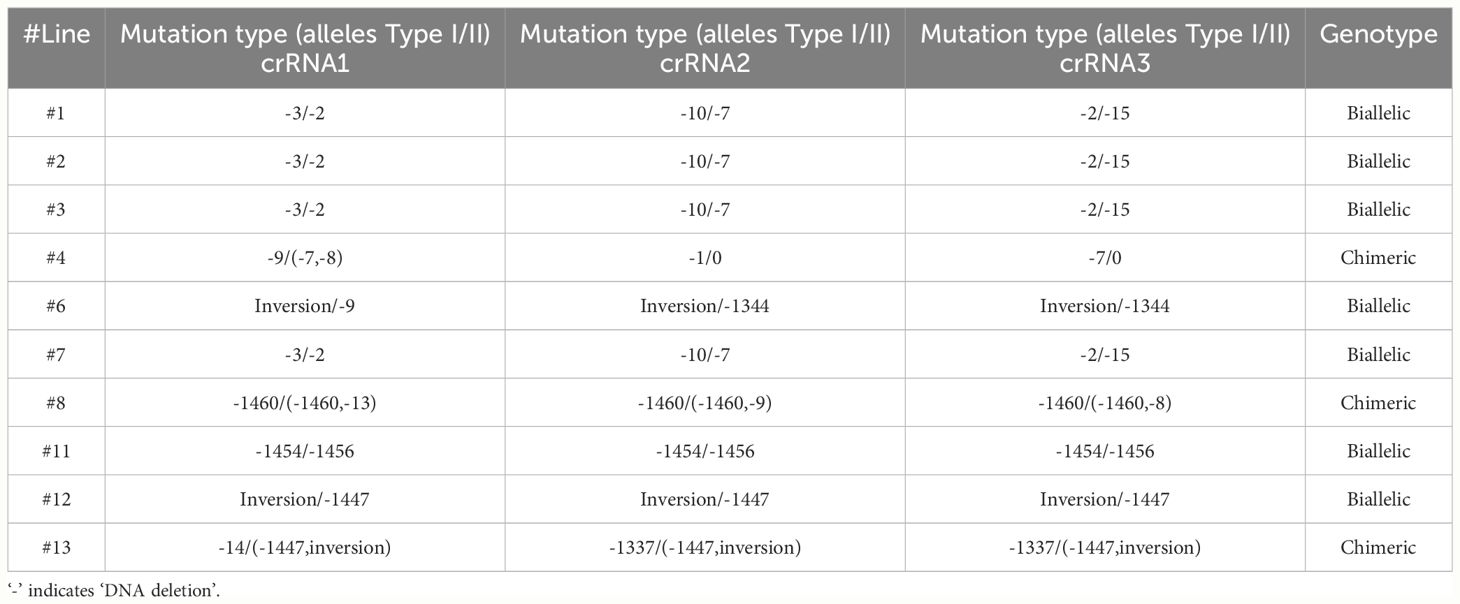
Table 1 Summary of transgene-free CsLOB1-edited C. sinensis cv. Hamlin plants generated by LbCas12a-CsLOB1(3crRNAs) transformation of embryogenic protoplasts.
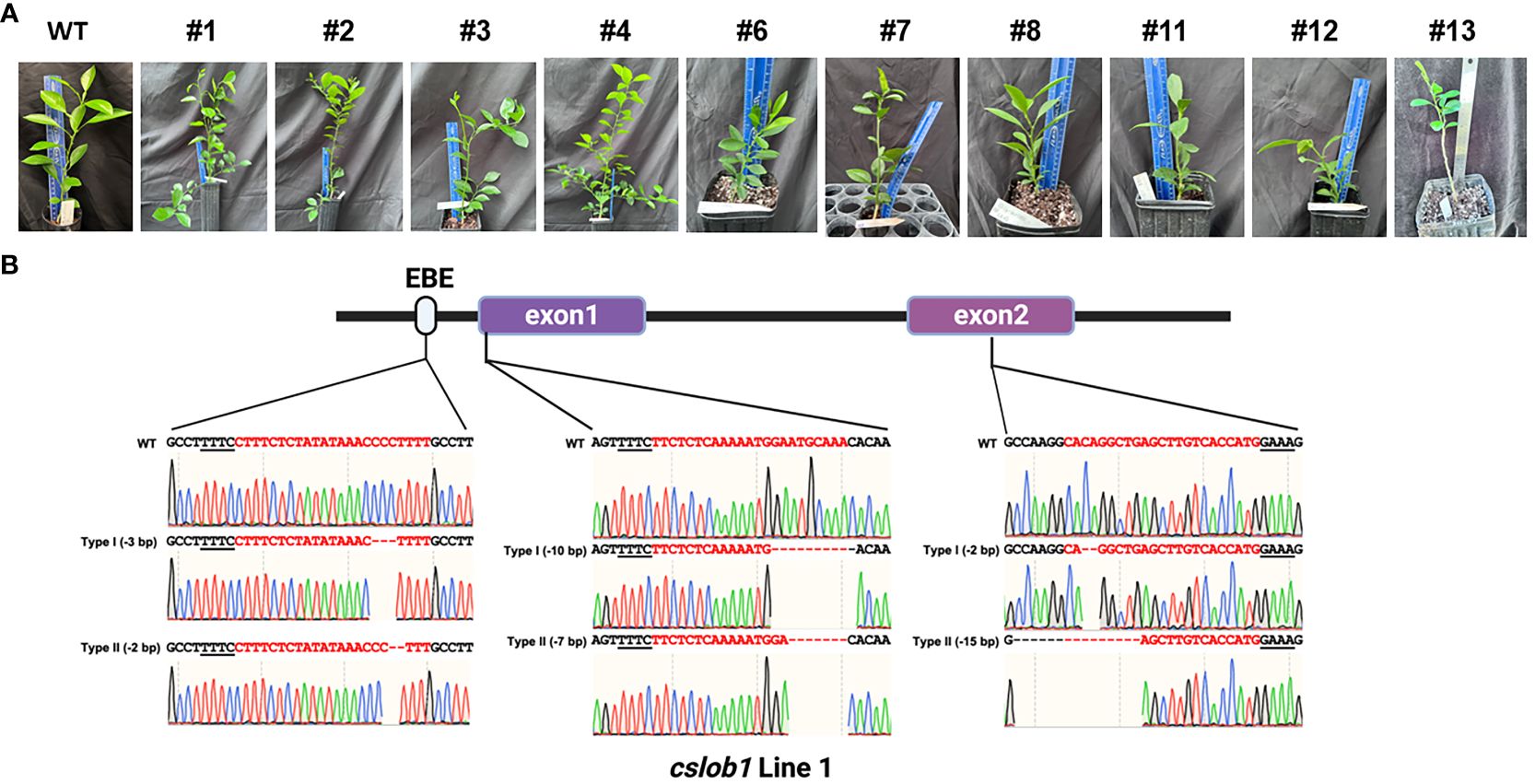
Figure 2 Identification of transgene-free cslob1 mutants of C. sinensis cv. Hamlin via Sanger sequencing. (A) Transgene-free cslob1 mutants of C. sinensis cv. Hamlin were grafted on Carrizo citrange (Poncirus trifoliate × Citrus sinensis) rootstock and kept in greenhouse. The genotypes of the mutants were shown. Wild type Hamlin generated from seed was grafted on the same rootstock as a control. The pictures were taken 20 months after potting. (B) Representative sequencing chromatogram results of transgene-free cslob1 mutant line 1, which has -3 bp, -10 bp, and -2 bp deletions in type I allele and -2 bp, -7 bp, and -15 bp deletions in type II allele. “-” indicates deletion at target sites.
Notably, 5 of the 10 lines contained long deletions, among which 3 lines displayed large inversions (Table 1). Two distinct types of large sequence inversions were identified (Figure 3, Supplementary Figures S5, S10, S12). One type (e.g., line 6) involved a large deletion (over 100 bp) between two target sites, followed by inversion (Figure 3A, Supplementary Figure S5), while the other type (e.g., line 12) displayed a short deletion occurring at three target sites, followed by inversion (Figure 3B, Supplementary Figure S10).
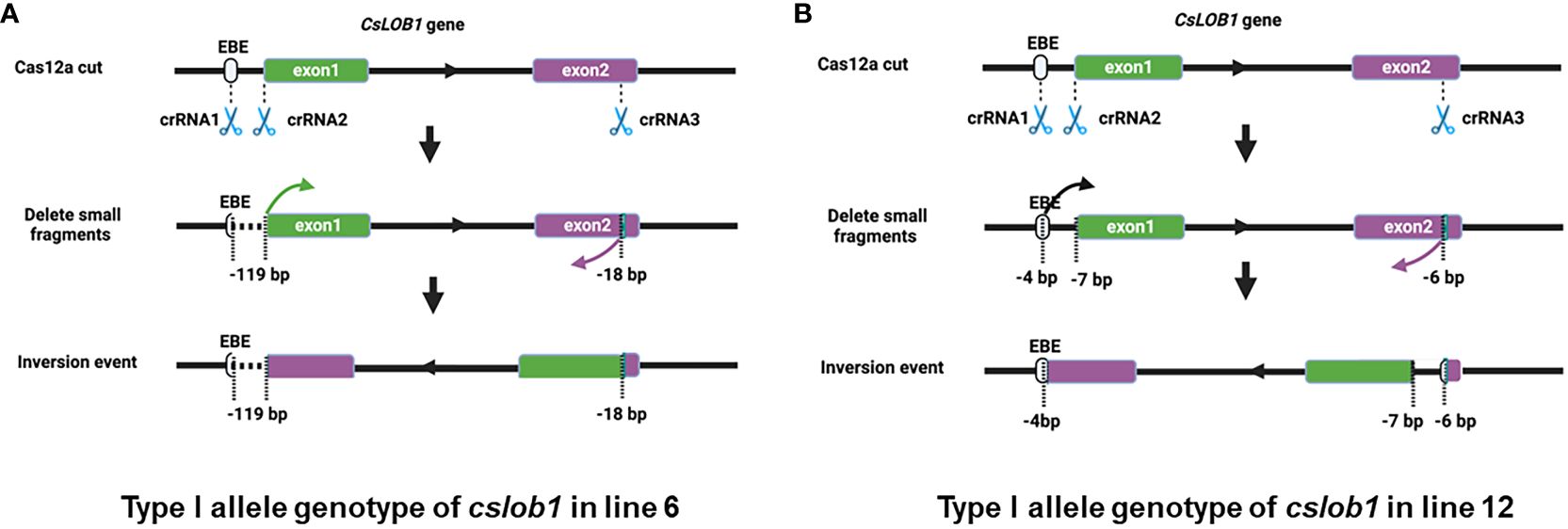
Figure 3 The representative genotypes of inversion events in the cslob1 mutants. (A) Type I allele contains inversion in line 6. The three crRNAs resulted in a deletion of 119 bp between the crRNA1 and crRNA2 sites and a deletion of 18 bp at the crRNA3 site. A large fragment as indicated by two arrows was inverted and ligated between the crRNA1 and crRNA3 sites. (B) Type I allele contains inversion in line 12. The three crRNAs resulted in deletions of 4 bp at the crRNA1 site, 7 bp at the crRNA2 site and 6 bp at the crRNA3 site. A large fragment as indicated by two arrows was inverted and ligated between crRNA1 and crRNA3 sites. “-” indicates deletion at target sites.
To assess potential off-target mutations, we employed the CRISPR-P v2.0 program (Liu et al., 2017) to identify putative off-target sites with up to 4 nucleotides mismatches with the TTTV PAM site within the citrus genome for the three crRNAs. The analysis revealed zero off-target candidate sites for crRNA1, two candidate sites for crRNA2, and one candidate site for crRNA3 (Supplementary Table S1). However, off-target mutations were not identified via whole-genome sequencing in all 10 lines.
Evaluation of resistance to citrus canker in cslob1 mutants
Subsequently, we assessed whether the cslob1 mutants were resistant to Xcc. Following Xcc inoculation for 9 days, the wild type displayed typical canker symptoms, while the cslob1 mutants exhibited no canker symptoms similar as the non-inoculated leaves (Figures 4A, C, 5). Quantification of Xcc titers further corroborated the findings, showing that at 4 dpi and 9 dpi, Xcc titers in the wild type were significantly higher than those in the cslob1 mutants (Figures 4B, 5). To simulate Xcc infection in the natural setting, we conducted Xcc inoculation via foliar spray. Typical canker symptoms were observed on wild-type leaves 18 days after spray, whereas no canker symptoms were observed on the cslob1mutants (Figure 4D). Moreover, the Xcc titer was significantly reduced in the cslob1 mutants compared to the wild-type at 18 days after spray (Figure 4E).
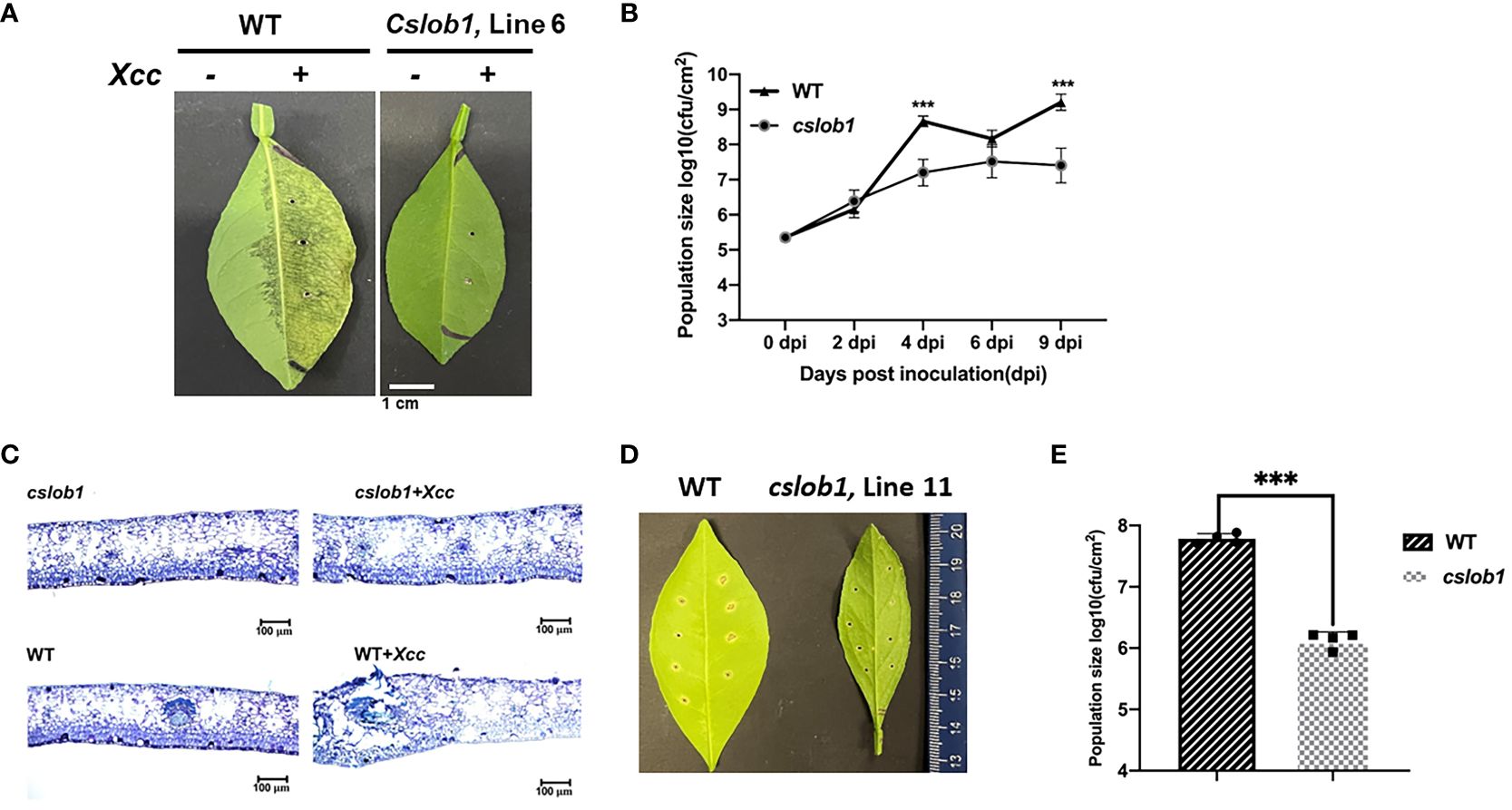
Figure 4 Assessment of disease resistance of the cslob1 mutant line 6 of C. sinensis cv. Hamlin against Xcc via injection and foliar spray. (A) Symptoms of citrus canker on both wild-type and the cslob1 mutant line 6 of C. sinensis cv. Hamlin. The plants were inoculated with Xcc through injection using needleless syringes. Inoculation was performed by injecting Xcc at a concentration of 1 × 107 CFU/ml into fully expanded young leaves using needleless syringes. The photograph was captured 9 days post inoculation, and a representative image was displayed. The experiment was conducted two times with 3 biological replicates for each experiment with similar results. (B) Xcc growth curve in citrus leaves. Three biological replicates were employed for each experiment and the experiment was repeated two times with similar results. Statistical analysis was conducted using Student’s t-test. Significant differences were indicated by asterisks (P-value<0.05). ‘***’ indicates P-value<0.001. (P-values at 0 dpi, 2 dpi, 4 dpi, 6 dpi and 9 dpi were 0.8636, 0.2787, 0.0003, 0.0664, and 0.0006, respectively). (C) Thin cross-section images of (A). A typical hypertrophy and hyperplasia phenotype in leaf tissues caused by Xcc can be observed in wild-type plants treated with Xcc. (D, E). Canker symptom and Xcc growth of wild-type C. sinensis cv. Hamlin and the cslob1 mutant line 11 after Xcc inoculation via spray. Foliar spray was conducted at a concentration of 5×108 cfu/mL and the leaves were punctured with syringes to make 8 wounds/leave before spray. The plants were covered with plastic bag to maintain the humidity for 24 hours after spray. The image was captured at 18 days after spray (D) and the Xcc titer of (D) was presented €. Student’ s t-test was used for statistical analysis (E). ***indicates P-value <0.001.
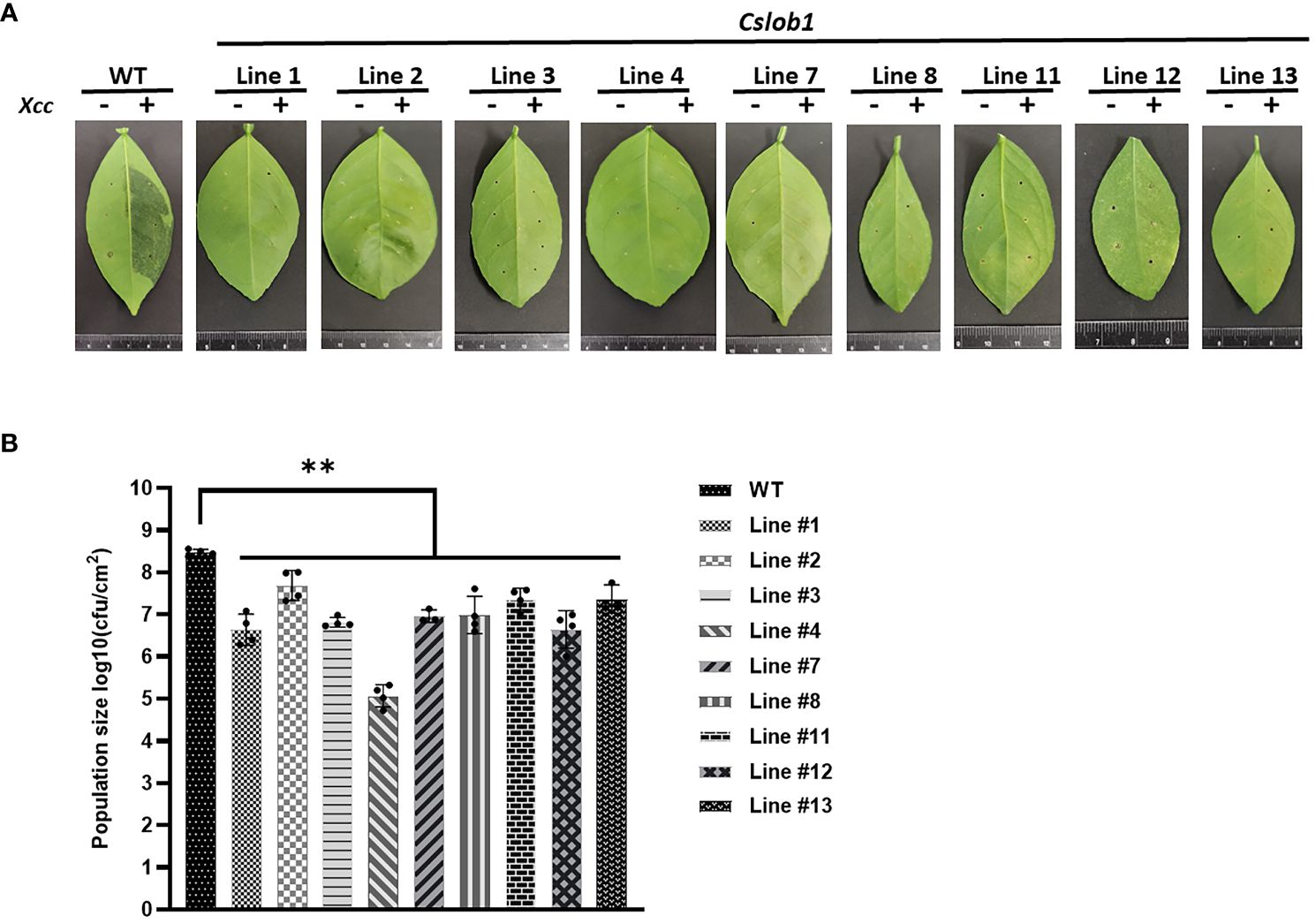
Figure 5 Evaluation of canker resistance of multiple cslob1 mutants of C. sinensis cv. Hamlin via injection. Inoculation was performed by injecting Xcc at a concentration of 1 × 107 CFU/ml into fully expanded young leaves using needleless syringes. The photograph was captured 9 days after inoculation (dpi), and a representative image was displayed for each line (A). The experiment included three biological replicates, with similar results. (B) Xcc growth in citrus leaves at 9 dpi. Four biological replicates were used, and the mean values ± SD (n = 4) are displayed. The experiments were conducted two times with similar results. Statistical analysis was conducted using Student’s t-test, revealing significant differences indicated by asterisks(P-value<0.05). ‘**’ indicates P-value < 0.01.
Concomitantly, we analyzed the gene expression levels of Cs7g32410 (expansin), Cs6g17190 (RSI-1), and Cs9g17380 (PAR1), known to be up-regulated by CsLOB1 during Xcc infection, 9 days post-Xcc inoculation (Zhang et al., 2017c; Duan et al., 2018; Zou et al., 2021). The expression levels of these three genes were significantly reduced in the cslob1 mutants compared to the wild type plants (Figure 6A). Concurrently, the H2O2 levels showed no obvious differences in the cslob1 mutants compared to the wild type (Figure 6B).
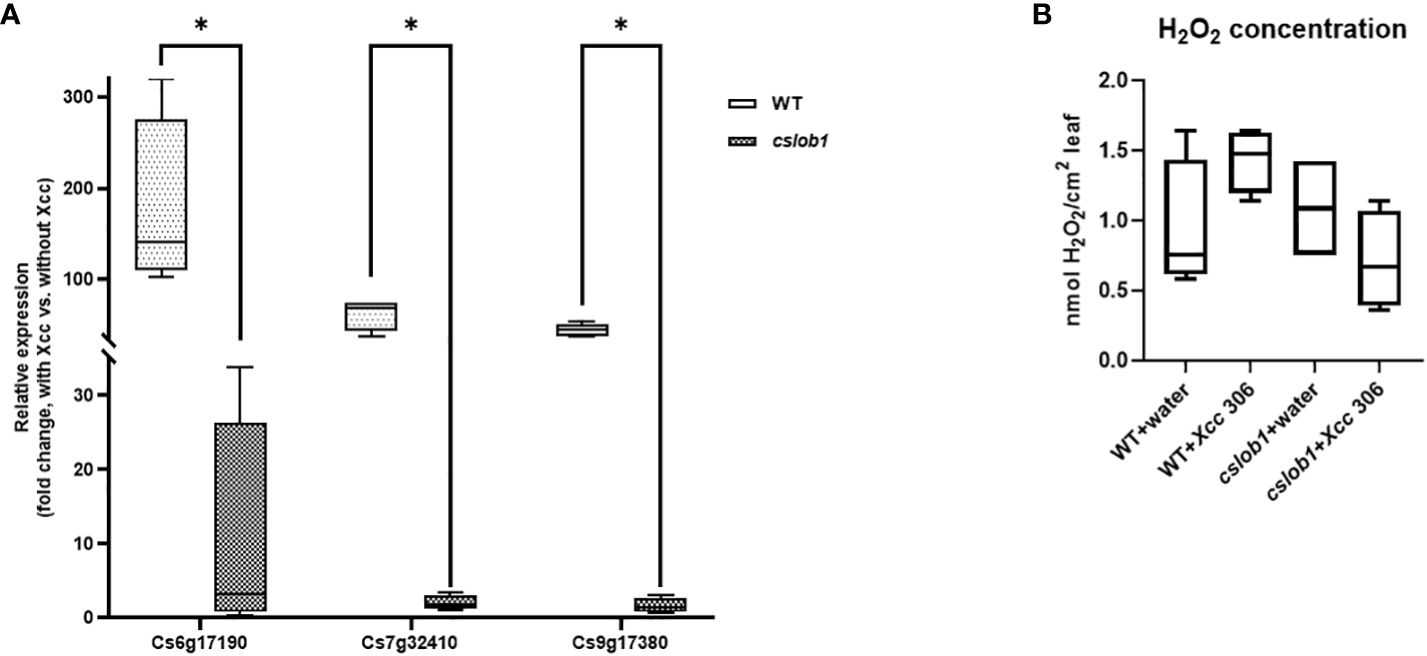
Figure 6 Gene expression and H2O2 concentration comparison between wild-type and the cslob1 mutant of C. sinensis cv. Hamlin plants in response to Xcc inoculation. (A) The expression levels of Cs6g17190, Cs7g32410, and Cs9g17380 genes known to be up-regulated by CsLOB1 during Xcc infection, were examined in both the cslob1 mutant and wild-type C. sinensis cv. Hamlin. This assessment was performed under conditions of Xcc inoculation at a concentration of 1 × 107 cfu/mL using syringes. CsGAPDH, a housekeeping gene encoding glyceraldehyde-3-phosphate dehydrogenase in citrus, served as the endogenous control. Mean values ± SD (n=4) from four biological replicates were presented. Statistical analysis was conducted using a two-sided Student’s t-test, with single asterisks (*) denoting significant differences (P values of Cs6g17190, Cs7g32410, and Cs9g17380 were 0.0412, 0.000447, and 0.0000179, respectively). The experiments were replicated two times with similar results. (B) Quantification of H2O2 concentration was performed one day post-inoculation. Each experiment was conducted with four biological replicates. Both the wild type and cslob1 mutant of C. sinensis leaves were subjected to Xcc inoculation (1 × 107 cfu/mL) or water, utilizing needleless syringes. The values presented depict means ± SD (n=4). This experiment was repeated twice with similar results. All box plots encompass a median line, where the box represents the interquartile range (IQR), and the whiskers delineate the remainder of the data distribution, excluding outliers. The lower and upper hinges of the boxes correspond to the 25th and 75th percentiles.
Discussion
In this study, we have demonstrated that transformation of embryogenic protoplasts of citrus with Cas12a/3 crRNAs RNP successfully generates transgene-free biallelic mutants. This is another example of transgene-free genome editing of citrus using this method in addition to a previous report (Su et al., 2023). In the previous report, 38 of 39 of the edited lines were biallelic/homozygous mutants, representing 97.5% mutation rate. In this study, all the edited lines achieved 100% mutation rate with 7 being biallelic mutants whereas 3 being chimeric mutants. It is probable that multiple crRNAs reduce the editing efficacy of individual crRNAs owing to competing for Cas12a but increase the overall editing efficacy. This is consistent with a previous report that higher number of gRNAs leads to reduction of editing efficiency which is likely due to the competition for Cas9 among gRNAs (Xie et al., 2015). Similarly, multiple sgRNAs cause dCas9 bottlenecks in CRISPRi targeted repression by dropping off roughly 1/N, where N is the number of sgRNAs expressed (Chen et al., 2018). On the other hand, it was reported that more gRNAs are more efficient than one gRNA in genome editing (Huang et al., 2023). The high genome editing efficacy of transformation of embryogenic protoplasts of citrus with Cas12a/crRNA was also reported by Zhang et al (Zhang et al., 2022). Despite few reports on generating transgene-free genome-edited citrus through transformation of embryogenic protoplasts with Cas12a/crRNA RNP (Zhang et al., 2022; Su et al., 2023), it has been successfully used in other plants including Arabidopsis thaliana, tobacco, lettuce, rice (Woo et al., 2015), grapevine and apple (Malnoy et al., 2016), maize (Svitashev et al., 2016), soybean and tobacco (Kim et al., 2017), wheat (Liang et al., 2017), and cabbage (Murovec et al., 2018), demonstrating enormous potential for its application in genetic improvements of plants.
As expected, transgene-free LOB1-edited lines with long deletions were obtained using 3 crRNAs in the RNP complex. Multiple crRNAs induce two or more DSBs in a single chromosome, which is known to mainly lead to deletions (Siebert and Puchta, 2002) and sometimes inversions (Qi et al., 2013; Zhang et al., 2017a). Long deletions were obtained in 5 of the 10 edited lines when 3 crRNAs were used for citrus genome editing, but none of the mutations were long deletions when only one crRNA was used (Su et al., 2023). Additionally, 3 edited lines contained inversions. Importantly, these 3 lines all contained long deletions whereas only two edited lines with long deletions did not contain inversions. This suggests that long deletions maybe required for occurrence of inversions. The occurrence of inversions suggests that knockin is highly possible when donors are provided in combination with multiple gRNAs, e.g., 3 crRNAs.
Biallelic/homozygous/chimeric mutations were observed for each of the three sites in 4 of the 10 edited lines (#1, #2, #3 and #7) when 3 crRNAs were used. Our data demonstrate that transformation of embryogenic citrus protoplasts with Cas12a/crRNA RNP can be very efficient for multiplex editing. Importantly, this approach avoids the complicated and lengthy process for construction of multiplex vectors or multiple individual vectors (Xie et al., 2015; Čermák et al., 2017; Huang et al., 2020).
CRISPR genome editing has been widely used for genetic improvements of crops (Zhu et al., 2020). However, most of the genome edited plants have not been commercialized despite improved traits such as yield, quality, and disease resistance. This is because most of the genome-edited plants were transgenic, which require lengthy and expensive deregulation process and are also a concern of consumers. Transgene-free genome editing, on the other hand, can address those issues (Gong et al., 2021; Turnbull et al., 2021; Bhattacharjee et al., 2023). We have previously generated transgene-free canker resistant C. sinensis cv. Hamlin plants which were exempted from the regulation of APHIS and is in the process of registration and commercialization (Su et al., 2023). The newly generated transgene-free canker resistant C. sinensis cv. Hamlin lines contain long deletions, indels and inversions and need to go through rigorous field testing for canker resistance and other horticultural traits. Despite different mutation types, the long deletions, indels, and inversions in the lob1 gene are all loss-of-function mutations of the single gene locus (LOB1) without introducing foreign DNAs, thus meeting the requirements of exempt of USDA Animal and Plant Health Inspection Service (APHIS). As expected, the CsLOB1 mutations abolish the induction of genes known to be induced by Xcc (Hu et al., 2014; Zhang et al., 2017c; Duan et al., 2018; Zou et al., 2021). The cslob1 mutants were resistant to citrus canker by reducing growth of Xcc. This reduction probably results from limiting hypertrophy and hyperplasia which are known to provide nutrients to Xcc growth, without affecting ROS production as indicted by H2O2 (Brunings and Gabriel, 2003). Additionally, the lob1 mutants generated using the three crRNAs RNP approach in this study exhibit resistance to canker similar as the lob1 mutants produced using one crRNA RNP method in our previous work (Su et al., 2023).
Overall, Cas12a/3 crRNAs RNP transformation of embryogenic protoplasts of citrus can be used for transgene-free multiplex genome editing with high efficiency. It can be used for deletion of long fragment and has potential for knockin of useful genes.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA1077621.
Author contributions
HS: Data curation, Investigation, Methodology, Supervision, Writing – original draft, Writing – review & editing. YW: Data curation, Investigation, Methodology, Writing – original draft, Writing – review & editing. JX: Data curation, Investigation, Methodology, Software, Writing – original draft, Writing – review & editing. AO: Investigation, Methodology, Resources, Writing – original draft, Writing – review & editing. JG: Investigation, Methodology, Resources, Writing – original draft, Writing – review & editing. NW: Conceptualization, Funding acquisition, Investigation, Methodology, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This project was supported by funding from Florida Citrus Initiative Program, Citrus Research and Development Foundation 18-025, U.S. Department of Agriculture National Institute of Food and Agriculture grants 2023-70029-41280, 2022-70029-38471, 2021-67013-34588, and 2018-70016-27412, FDACS Specialty Crop Block Grant Program AM22SCBPFL1125, and Hatch project (FLA-CRC-005979) to NW.
Acknowledgments
We thank Wang lab members for constructive suggestions and insightful discussions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1448807/full#supplementary-material
References
Bhattacharjee, S., Bhowmick, R., Kant, L., Paul, K. (2023). Strategic transgene-free approaches of CRISPR-based genome editing in plants. Mol. Genet. Genomics 298, 507–520. doi: 10.1007/s00438-023-01998-3
Brunings, A. M., Gabriel, D. W. (2003). Xanthomonas citri: breaking the surface. Mol. Plant Pathol. 4, 141–157. doi: 10.1046/j.1364-3703.2003.00163.x
Čermák, T., Curtin, S. J., Gil-Humanes, J., Čegan, R., Kono, T. J. Y., Konečná, E., et al. (2017). A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell 29, 1196–1217. doi: 10.1105/tpc.16.00922
Chen, P. Y., Qian, Y., Vecchio, D. D. (2018). “A model for resource competition in CRISPR-mediated gene repression,” in 2018 IEEE Conference on Decision and Control (CDC), Miami, Florida. 4333–4338. doi: 10.1109/CDC.2018.8619016
Chen, S., Zhou, Y., Chen, Y., Gu, J. (2018b). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Duan, S., Jia, H., Pang, Z., Teper, D., White, F., Jones, J., et al. (2018). Functional characterization of the citrus canker susceptibility gene CsLOB1. Mol. Plant Pathol. 19, 1908–1916. doi: 10.1111/mpp.12667
Dutt, M., Mou, Z., Zhang, X., Tanwir, S. E., Grosser, J. W. (2020). Efficient CRISPR/Cas9 genome editing with Citrus embryogenic cell cultures. BMC Biotechnol. 20, 58. doi: 10.1186/s12896-020-00652-9
Ference, C. M., Gochez, A. M., Behlau, F., Wang, N., Graham, J. H., Jones, J. B. (2018). Recent advances in the understanding of Xanthomonas citri ssp. citri pathogenesis and citrus canker disease management. Mol. Plant Pathol. 19, 1302–1318. doi: 10.1111/mpp.12638
Gmitter, F. G., Chen, C., MaChado, M. A., De Souza, A. A., Ollitrault, P., Froehlicher, Y., et al. (2012). Citrus genomics. Tree Genet. Genomes 8, 611–626. doi: 10.1007/s11295-012-0499-2
Gong, Z., Cheng, M., Botella, J. R. (2021). Non-GM genome editing approaches in crops. Front. Genome Ed 3, 817279.doi: 10.3389/fgeed.2021.817279
Gottwald, T. R., Graham, J. H., Schubert, T. S. (2002). Citrus canker: the pathogen and its impact. Plant Health Prog. 3, 15. doi: 10.1094/PHP-2002-0812-01-RV
He, Y., Zhu, M., Wang, L., Wu, J., Wang, Q., Wang, R., et al. (2018). Programmed self-elimination of the CRISPR/cas9 construct greatly accelerates the isolation of edited and transgene-free rice plants. Mol. Plant 11, 1210–1213. doi: 10.1016/j.molp.2018.05.005
Hu, Y., Zhang, J., Jia, H., Sosso, D., Li, T., Frommer, W. B., et al. (2014). Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc. Natl. Acad. Sci. U.S.A. 111, E521–E529. doi: 10.1073/pnas.1313271111
Huang, X., Jia, H., Xu, J., Wang, Y., Wen, J., Wang, N. (2023). Efficient transgene-free genome editing in plants in the T0 generation based on a co-editing strategy. bioRxiv. 9, 1591–1597. doi: 10.1101/2023.03.02.530790
Huang, X., Wang, Y., Wang, N. (2021). Highly efficient generation of canker-resistant sweet orange enabled by an improved CRISPR/cas9 system. Front. Plant Sci. 12, 769907. doi: 10.3389/fpls.2021.769907
Huang, X., Wang, Y., Wang, N. (2022). Base editors for citrus gene editing. Front. Genome Ed 4, 852867.doi: 10.3389/fgeed.2022.852867
Huang, X., Wang, Y., Xu, J., Wang, N. (2020). Development of multiplex genome editing toolkits for citrus with high efficacy in biallelic and homozygous mutations. Plant Mol. Biol. 104, 297–307. doi: 10.1007/s11103-020-01043-6
Irigoyen, S., Ramasamy, M., Pant, S., Niraula, P., Bedre, R., Gurung, M., et al. (2020). Plant hairy roots enable high throughput identification of antimicrobials against Candidatus Liberibacter spp. Nat. Commun. 11, 5802. doi: 10.1038/s41467-020-19631-x
Jia, H., Omar, A. A., Xu, J., Dalmendray, J., Wang, Y., Feng, Y., et al. (2024). Generation of transgene-free canker-resistant Citrus sinensis cv. Hamlin in the T0 generation through Cas12a/CBE co-editing. Front. Plant Sci. 15, 1385768. doi: 10.3389/fpls.2024.1385768
Jia, H., Orbovic, V., Jones, J. B., Wang, N. (2016). Modification of the PthA4 effector binding elements in Type I CsLOB1 promoter using Cas9/sgRNA to produce transgenic Duncan grapefruit alleviating XccΔpthA4:dCsLOB1.3 infection. Plant Biotechnol. J. 14, 1291–1301. doi: 10.1111/pbi.12495
Jia, H., Wang, N. (2014a). Targeted genome editing of sweet orange using cas9/sgRNA. PloS One 9, e93806. doi: 10.1371/journal.pone.0093806
Jia, H., Wang, N. (2014b). Xcc-facilitated agroinfiltration of citrus leaves: a tool for rapid functional analysis of transgenes in citrus leaves. Plant Cell Rep. 33, 1993–2001. doi: 10.1007/s00299-014-1673-9
Jia, H., Wang, Y., Su, H., Huang, X., Wang, N. (2022). LbCas12a-D156R efficiently edits LOB1 effector binding elements to generate canker-resistant citrus plants. Cells 11, 315. doi: 10.3390/cells11030315
Jia, H., Zhang, Y., Orbović, V., Xu, J., White, F. F., Jones, J. B., et al. (2017). Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol. J. 15, 817–823. doi: 10.1111/pbi.12677
Jia, H., Zou, X., Orbovic, V., Wang, N. (2019). Genome editing in citrus tree with CRISPR/cas9. Methods Mol. Biol. 1917, 235–241. doi: 10.1007/978-1-4939-8991-1_17
Kim, H., Kim, S. T., Ryu, J., Kang, B. C., Kim, J. S., Kim, S. G. (2017). CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat. Commun. 8, 14406. doi: 10.1038/ncomms14406
Langmead, B., Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Leblanc, C., Zhang, F., Mendez, J., Lozano, Y., Chatpar, K., Irish, V., et al. (2017). Increased efficiency of targeted mutagenesis by CRISPR/Cas9 in plants using heat stress. Plant J. 93, 377–386. doi: 10.1111/tpj.13782
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Liang, Z., Chen, K., Li, T., Zhang, Y., Wang, Y., Zhao, Q., et al. (2017). Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat. Commun. 8, 14261. doi: 10.1038/ncomms14261
Liu, H., Ding, Y., Zhou, Y., Jin, W., Xie, K., Chen, L. L. (2017). CRISPR-P 2.0: an improved CRISPR-cas9 tool for genome editing in plants. Mol. Plant 10, 530–532. doi: 10.1016/j.molp.2017.01.003
Liu, Q., Zhao, C., Sun, K., Deng, Y., Li, Z. (2023). Engineered biocontainable RNA virus vectors for non-transgenic genome editing across crop species and genotypes. Mol. Plant. 16, 616–631. doi: 10.1016/j.molp.2023.02.003
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, H., Meng, X., Xu, K., Li, M., Gmitter, F. G., Jr., Liu, N., et al. (2022). Highly efficient hairy root genetic transformation and applications in citrus. Front. Plant Sci. 13, 1039094.doi: 10.3389/fpls.2022.1039094
Ma, X., Zhang, X., Liu, H., Li, Z. (2020). Highly efficient DNA-free plant genome editing using virally delivered CRISPR–Cas9. Nat. Plants 6, 773–779. doi: 10.1038/s41477-020-0704-5
Mahmoud, L. M., Kaur, P., Stanton, D., Grosser, J. W., Dutt, M. (2022). A cationic lipid mediated CRISPR/Cas9 technique for the production of stable genome edited citrus plants. Plant Methods 18, 33. doi: 10.1186/s13007-022-00870-6
Malnoy, M., Viola, R., Jung, M. H., Koo, O. J., Kim, S., Kim, J. S., et al. (2016). DNA-free genetically edited grapevine and apple protoplast using CRISPR/cas9 ribonucleoproteins. Front. Plant Sci. 7, 1904.doi: 10.3389/fpls.2016.01904
Murovec, J., Guček, K., Bohanec, B., Avbelj, M., Jerala, R. (2018). DNA-Free Genome Editing of Brassica oleracea and B. rapa Protoplasts Using CRISPR-Cas9 Ribonucleoprotein Complexes. Front. Plant Sci. 9, 1594. doi: 10.3389/fpls.2018.01594
O'Malley, R. C., Ecker, J. R. (2010). Linking genotype to phenotype using the Arabidopsis unimutant collection. Plant J. 61, 928–940. doi: 10.1111/j.1365-313X.2010.04119.x
Omar, A. A., Dutt, M., Gmitter, F. G., Grosser, J. W. (2016). “Somatic Embryogenesis: Still a Relevant Technique in Citrus Improvement,” in In Vitro Embryogenesis in Higher Plants, eds. Germana, M. A., Lambardi, M., (New York, NY: Springer New York), 289–327. doi: 10.1007/978-1-4939-3061-6
Peng, A., Chen, S., Lei, T., Xu, L., He, Y., Wu, L., et al. (2017). Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 15, 1509–1519. doi: 10.1111/pbi.12733
Poplin, R., Chang, P. C., Alexander, D., Schwartz, S., Colthurst, T., Ku, A., et al. (2018). A universal SNP and small-indel variant caller using deep neural networks. Nat. Biotechnol. 36, 983–987. doi: 10.1038/nbt.4235
Qi, Y., Li, X., Zhang, Y., Starker, C. G., Baltes, N. J., Zhang, F., et al. (2013)Targeted deletion and inversion of tandemly arrayed genes in arabidopsis thaliana using zinc finger nucleases. G3 (Bethesda). 3, 1707–1715 doi: 10.1534/g3.113.006270
Robinson, J. T., Thorvaldsdottir, H., Winckler, W., Guttman, M., Lander, E. S., Getz, G., et al. (2011). Integrative genomics viewer. Nat. Biotechnol. 29, 24–26. doi: 10.1038/nbt.1754
Sels, J., Mathys, J., De Coninck, B. M., Cammue, B. P., De Bolle, M. F. (2008). Plant pathogenesis-related (PR) proteins: a focus on PR peptides. Plant Physiol. Biochem. 46, 941–950. doi: 10.1016/j.plaphy.2008.06.011
Siebert, R., Puchta, H. (2002). Efficient repair of genomic double-strand breaks by homologous recombination between directly repeated sequences in the plant genome. Plant Cell 14, 1121–1131. doi: 10.1105/tpc.001727
Sturme, M. H. J., van der Berg, J. P., Bouwman, L. M. S., De Schrijver, A., De Maagd, R. A., Kleter, G. A., et al. (2022). Occurrence and nature of off-target modifications by CRISPR-cas genome editing in plants. ACS Agric. Sci. Technol. 2, 192–201. doi: 10.1021/acsagscitech.1c00270
Su, H., Wang, Y., Xu, J., Omar, A. A., Grosser, J. W., Calovic, M., et al. (2023). Generation of the transgene-free canker-resistant Citrus sinensis using Cas12a/crRNA ribonucleoprotein in the T0 generation. Nat. Commun. 14, 3957. doi: 10.1038/s41467-023-39714-9
Svitashev, S., Schwartz, C., Lenderts, B., Young, J. K., Mark Cigan, A. (2016). Genome editing in maize directed by CRISPR-Cas9 ribonucleoprotein complexes. Nat. Commun. 7, 13274. doi: 10.1038/ncomms13274
Turnbull, C., Lillemo, M., Hvoslef-Eide, T. (2021). Global regulation of genetically modified crops amid the gene edited crop boom - A review. Front. Plant Sci. 12, 630396.doi: 10.3389/fpls.2021.630396
Wang, L., Huang, Y., Liu, Z., He, J., Jiang, X., He, F., et al. (2021). Somatic variations led to the selection of acidic and acidless orange cultivars. Nat. Plants 7, 954–965. doi: 10.1038/s41477-021-00941-x
Wang, M., Qin, Y. Y., Wei, N. N., Xue, H. Y., Dai, W. S. (2023). Highly efficient Agrobacterium rhizogenes-mediated hairy root transformation in citrus seeds and its application in gene functional analysis. Front. Plant Sci. 14, 1293374.doi: 10.3389/fpls.2023.1293374
Woo, J. W., Kim, J., Kwon, S. I., Corvalán, C., Cho, S. W., Kim, H., et al. (2015). DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 33, 1162–1164. doi: 10.1038/nbt.3389
Xie, K., Minkenberg, B., Yang, Y. (2015). Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. U.S.A. 112, 3570–3575. doi: 10.1073/pnas.1420294112
Zhang, Y., Cheng, Y., Fang, H., Roberts, N., Zhang, L., Vakulskas, C. A., et al. (2022). Highly efficient genome editing in plant protoplasts by ribonucleoprotein delivery of CRISPR-cas12a nucleases. Front. Genome Ed 4, 780238.doi: 10.3389/fgeed.2022.780238
Zhang, J., Huguet-Tapia, J. C., Hu, Y., Jones, J., Wang, N., Liu, S., et al. (2017c). Homologues of CsLOB1 in citrus function as disease susceptibility genes in citrus canker. Mol. Plant Pathol. 18, 798–810. doi: 10.1111/mpp.12441
Zhang, F., Leblanc, C., Irish, V. F., Jacob, Y. (2017b). Rapid and efficient CRISPR/Cas9 gene editing in Citrus using the YAO promoter. Plant Cell Rep. 36, 1883–1887. doi: 10.1007/s00299-017-2202-4
Zhang, Y., Liang, Z., Zong, Y., Wang, Y., Liu, J., Chen, K., et al. (2016). Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 7, 12617. doi: 10.1038/ncomms12617
Zhang, C., Liu, C., Weng, J., Cheng, B., Liu, F., Li, X., et al. (2017a). Creation of targeted inversion mutations in plants using an RNA-guided endonuclease. Crop J. 5, 83–88. doi: 10.1016/j.cj.2016.08.001
Zhu, H., Li, C., Gao, C. (2020). Applications of CRISPR–Cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol. 21, 661–677. doi: 10.1038/s41580-020-00288-9
Zhu, C., Zheng, X., Huang, Y., Ye, J., Chen, P., Zhang, C., et al. (2019). Genome sequencing and CRISPR/Cas9 gene editing of an early flowering Mini-Citrus (Fortunella hindsii). Plant Biotechnol. J. 17, 2199–2210. doi: 10.1111/pbi.13132
Zou, X., Du, M., Liu, Y., Wu, L., Xu, L., Long, Q., et al. (2021). CsLOB1 regulates susceptibility to citrus canker through promoting cell proliferation in citrus. Plant J. 106, 1039–1057. doi: 10.1111/tpj.15217
Keywords: CRISPR/Cas12a, genome editing, non-transgenic, inversion, Xanthomonas, CsLOB1, disease resistance
Citation: Su H, Wang Y, Xu J, Omar AA, Grosser JW and Wang N (2024) Cas12a RNP-mediated co-transformation enables transgene-free multiplex genome editing, long deletions, and inversions in citrus chromosome. Front. Plant Sci. 15:1448807. doi: 10.3389/fpls.2024.1448807
Received: 13 June 2024; Accepted: 19 July 2024;
Published: 01 August 2024.
Edited by:
Aytug Tuncel, University of Maryland, United StatesReviewed by:
Xiuping Zou, Southwest University, ChinaJaindra Nath Tripathi, International Institute of Tropical Agriculture (IITA), Kenya
Copyright © 2024 Su, Wang, Xu, Omar, Grosser and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nian Wang, bmlhbndhbmdAdWZsLmVkdQ==
†These authors have contributed equally to this work
 Hang Su
Hang Su Yuanchun Wang
Yuanchun Wang Jin Xu
Jin Xu Ahmad A. Omar
Ahmad A. Omar Jude W. Grosser
Jude W. Grosser Nian Wang
Nian Wang