- 1Guangdong Key Laboratory for New Technology Research of Vegetables, Vegetable Research Institute, Guangdong Academy of Agricultural Sciences, Guangzhou, China
- 2College of Horticulture, South China Agricultural University, Guangzhou, China
Fruit color is a crucial attribute of fruit quality in peppers (Capsicum spp.). However, few studies have focused on the mechanism of color formation in immature pepper fruits. In this study, the light-yellow color observed in immature CSJ009 fruits compared to CSJ010 could be attributed to decreased chlorophyll and carotenoid pigments. Through integrated analysis of the transcriptome and metabolome of CSJ009 and CSJ010, we identified 23,930 differentially expressed genes (DEGs) and 345 differentially accumulated metabolites (DAMs). Furthermore, integrated analysis revealed a strong correlation between the HCT-like gene and metabolite MWS0178 (chlorogenic acid). Paraffin section assay revealed that the epidermal cells of immature CSJ010 fruits exhibited a more compact arrangement with significantly greater length than those of CSJ009. Quantitative determination of carotenoids showed that lutein emerged as the predominant carotenoid in immature pepper fruits. Additionally, missense mutation of LCYB2 is likely to lead to a decrease in β-carotene content in immature CSJ009 fruits, whereas CCS may directly catalyze the conversion of lycopene to β-carotene in mature fruits. The null mutation in CCS promoted the biosynthesis of β,ϵ-branch carotenoids leading to lutein being the most abundant carotenoid found in orange CSJ010 fruits. These findings provide important insights into the mechanism underlying color formation in pepper fruits and establish a foundation for the further exploration of color-related genes.
1 Introduction
Fruit color is a crucial trait for assessing fruit quality in peppers (Capsicum spp.). The colors of immature pepper fruit primarily include green, purple, and black, which can be attributed to the accumulation of various pigments such as chlorophylls, flavonoids, and carotenoids (Lightbourn et al., 2008). Chlorophylls are localized within chloroplasts, whereas flavonoids are found in cell vacuoles (Lightbourn et al., 2008). Carotenoids are contained within chromoplasts that develop from pre-existing chloroplasts during fruit ripening (Egea et al., 2011). The green color typically arises from the accumulation of chlorophylls, along with carotenoids, such as lutein, β-carotene, and violaxanthin (Deli et al., 1996; Paran and van der Knaap, 2007). Purple and black fruits mainly contain anthocyanins, chlorophyll, β-carotene, lutein, and violaxanthin (Lightbourn et al., 2008).
During the ripening process, the color of pepper fruit gradually transitions to red, orange, or yellow (Brand et al., 2012), which is determined by the amount and composition of carotenoids accumulated within chromoplasts (Ha et al., 2007). In red fruits, there is a significant increase in the total carotenoid content (Matus et al., 1991), and capsanthin is the predominant carotenoid (Bouvier et al., 1994), followed by β-cryptoxanthin, β-carotene, capsorubin, zeaxanthin, and antheraxanthin (Wahyuni et al., 2011); whereas chlorophyll levels decrease to zero (Deli et al., 1996). Yellow fruits do not contain detectable levels of capsanthin and capsorubin (Wahyuni et al., 2011), mainly due to the accumulation of α- and β-carotene, zeaxanthin, β-cryptoxanthin (Gómez-García and Ochoa-Alejo, 2013), violaxanthin, and lutein (Wahyuni et al., 2011). The orange-colored fruits primarily arise from the accumulation of β-carotene (Guzman et al., 2010), while also contain low but significant levels of red carotenoids, such as capsanthin, β-cryptoxanthin, and antheraxanthin (Wahyuni et al., 2011).
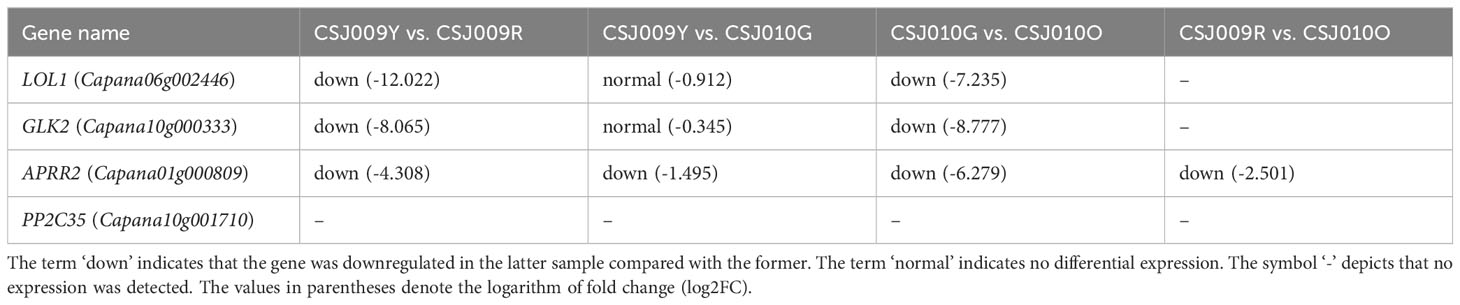
Chlorophyll is the most abundant tetrapyrrole molecule in photosynthetic organisms (Masuda and Fujita, 2008). Glutamyl-tRNA is activated by glutamyl-tRNA reductase (HEMA) to synthesize 5-aminolevulinic acid (ALA), the initial precursor of chlorophyll biosynthesis, which forms protoporphyrin IX (Masuda and Fujita, 2008). Mg-chelatase insert Mg2+ into protoporphyrin IX to produce Mg-protoporphyrin IX, which catalyzes the formation of chlorophyll a by a series of enzymes, such as magnesium chelatase subunit H (CHLH), magnesium protoporphyrin IX methyltransferase (CHLM), Mg-protoIX methyl ester oxidative cyclase (CRD), Genomes uncoupled 4 (GUN4), and protochlorophyllide oxidoreductase (POR). A portion of chlorophyll a undergoes conversion to chlorophyll b through the activity of chlorophyllide a oxygenase (CAO) (Masuda and Fujita, 2008). In Capsicum, Capana10g001710 (CaPP2C35) has been identified as controlling the light-green immature fruit color (Wu et al., 2022). In addition, two quantitative trait loci (QTLs) pc8.1 and pc10.1 (Brand et al., 2012), along with three transcription factors (TFs), Lsd one like1 zinc finger (LOL1) (Borovsky et al., 2019), Golden2-like (GLK2) (Brand et al., 2014) and Arabidopsis pseudo-response regulator2 (APRR2) (Jeong et al., 2020; Lee et al., 2020; Pan et al., 2013) have been reported to regulate immature fruit color via modulating chlorophyll content and chloroplast development. In our recent study, we successfully mapped the locus ly responsible for the light-yellow immature fruit color into a 6.51Mbp region on chromosome 9 (Song et al., 2022).
Flavonoids are polyphenolic compounds widespread in the plant kingdom (Gil and Couto, 2013). To date, approximately 9000 different flavonoids have been found in all higher plants (Ferrer et al., 2008), which can be classified into six subclasses: flavonols, flavones, isoflavones, flavanones, anthocyanidins, and flavanols (Manach et al., 2004). Flavonoids are synthesized via from phenylalanine phenylpropanoid and flavonoid pathways. Phenylalanine is catalyzed by enzymes such as phenylalanine lyase (PAL), cinnamic acid 4-hydroxylase (C4H), and 4-coumadin-CoA ligase (4CL) to produce p-coumaroyl-CoA as the initial substrate for flavonoid synthesis (Liu et al., 2020). On one hand, p-coumaroyl-CoA undergoes enzymatic reactions involving chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavonoid-3’-hydroxylase (F3’H), and flavonoid 3’,5’-hydroxylase (F3’5’H) to generate dihydroflavonols (Liu et al., 2019). On the other hand, p-coumaroyl-CoA can also be converted to caffeoyl-CoA by shikimate O-hydroxycinnamoyltransferase (HCT) and coumaroylquinate 3-monooxygenase (C3H), which is subsequently activated by caffeoyl-CoA O-methyltransferase (CCOAOMT) to form feruloyl-CoA as an intermediate for dihydroflavonol biosynthesis (Pascual et al., 2016; Zhang et al., 2017). Dihydroflavonols are transformed into various metabolites via dihydroflavonol 4-reductase (DFR) and flavonol synthase (FLS) (Zhou et al., 2020). DFR catalyzes the stereospecific reduction of dihydroflavonols to leucoanthocyanidins (Huang et al., 2012; Martens et al., 2003). FLS converts dihydroflavonols into flavonols, which compete with DFR for common substrates at a key branch in the flavonoid pathway (Luo et al., 2016). Regulation of this complex flavonoid biosynthesis process relies mainly on MYB proteins (Nabavi et al., 2020). In Capsicum, anthocyanin accumulation in immature fruit is controlled by a MYB TF homologous to Anthocyanin2 (An2) found in Petunia (Borovsky et al., 2004).
Carotenoids are polyisoprenoid compounds that can be classified as carotenes and xanthophylls (Rodríguez-Bernaldo de Quirós and Costa, 2006). To date, over 700 distinct carotenoids have been characterized (Nishino et al., 2009). Geranylgeranyl diphosphate (GGPP) is a common substrate used for the biosynthesis of carotenoids and chlorophylls (Wang et al., 2019). Phytoene synthase (PSY) catalyzes the initial committed condensation step from GGPP to generate colorless C40 phytoene, thereby directing metabolic flux towards carotenoid biosynthesis (Wang et al., 2019). Subsequently, phytoene is converted into acyclic lycopene by a series of key enzymes including phytoene desaturase (PDS), ζ-carotene isomerase (Z-ISO), ζ-carotene desaturase (ZDS), and carotenoid isomerase (CRTISO) (Wang et al., 2019). All-trans-lycopene undergoes cyclization by lycopene β-cyclases (LCYBs) to produce all-trans-β-carotene (β,β-branch), which is further catalyzed by enzymes such as β-carotene hydroxylase (CHYB), zeaxanthin epoxidase (ZEP), violaxanthin de-epoxidase (VDE), and capsanthin-capsorubin synthase (CCS), to form capsanthin and capsorubin (Yuan et al., 2015). All-trans-β-carotene can also be converted into 9-cis-β-carotene via β-carotene isomerase D27 (D27), thus directing metabolic flux towards strigolactones biosynthesis (Alder et al., 2012). On the other hand, the open end of lycopene can also be cyclized by lycopene ϵ-cyclase (LCYE) and LCYB to form α-carotene (β,ϵ-branch). This α-carotene then undergoes cyclization by β-hydroxylase (HYDB) and carotene ϵ-monooxygenase (CYP97C, namely LUT1) to produce lutein (Yuan et al., 2015). The degradation of carotenoids involves carotenoid cleavage dioxygenase (CCD) and 9-cis epoxy carotenoid cleavage dioxygenase (NCED) enzymes (Zhang et al., 2020). In pepper fruit, β-carotene and zeaxanthin are synthesized throughout the ripening process, whereas capsanthin in the β,β-branch is synthesized only in mature fruits, and lutein in the β,ϵ-branch is usually found in immature fruits (Wang et al., 2019).
However, to date, few studies have focused on identifying the transcriptional regulators and metabolites that influence immature fruit color in pepper. In this study, we conducted an integrative analysis of the transcriptome and metabolome to elucidate the mechanism underlying fruit color formation in Capsicum. Our integrative analysis revealed a strong correlation between the HCT-like gene and metabolite MWS0178. The qRT-PCR assay demonstrated negligible expression of HCT-like in immature light-yellow CSJ009 fruits, likely due to a null mutation identified through whole-genome re-sequencing (WGRS) analysis. Furthermore, WGRS analysis identified several mutations in carotenoid-biosynthesis genes, including LCYB2, APRR2 and CCS. Quantitative determination provided robust evidence to validate the function of these color-related genes and clearly illustrated the differences in carotenoid concentrations between CSJ009 and CSJ010 fruits at different developmental stages. Our findings not only provide novel insights into the mechanism underlying color formation in pepper fruits but also lay a foundation for further exploration of color-related genes.
2 Materials and methods
2.1 Plant growth and sampling
For this study, two independent highly inbred lines of C. annuum, namely ‘CSJ009’ and ‘CSJ010’ were used after undergoing multiple generations of self-crossing. The immature fruits (40 d post-anthesis) of CSJ009 and CSJ010 are light-yellow (Figure 1A; hereafter referred to as CSJ009Y) and green (Figure 1B; hereafter referred to as CSJ010G), respectively. Upon maturation (80 d post-anthesis), the fruit color changed to red for CSJ009 (Figure 1C; hereafter referred to as CSJ009R) and orange for CSJ010 (Figure 1D; hereafter referred to as CSJ010O). Both inbred lines were cultivated under identical environmental conditions with consistent management practices in an open field located at the DaFeng Research Base, Vegetable Research Institute, Guangdong Academy of Agricultural Sciences, Guangzhou, China.
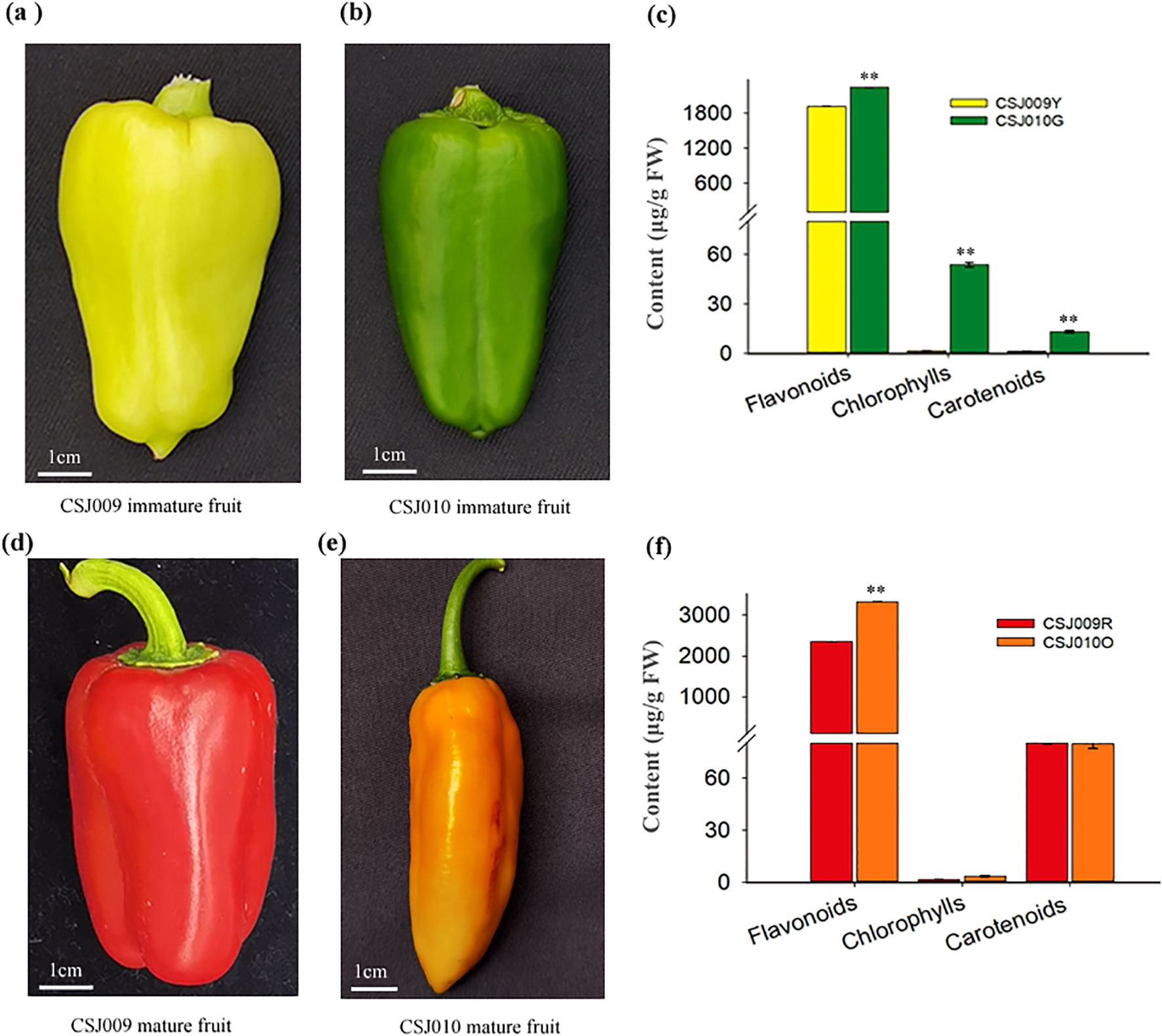
Figure 1. Fruit color and pigment content in CSJ009 and CSJ010. Subfigures (A) and (B) present the immature fruit colors of CSJ009 and CSJ010, respectively. Subfigures (D) and (E) show the mature fruit colors of CSJ009 and CSJ010, respectively. Subfigures (C) and (F) present the variation of chlorophylls, flavonoids, and carotenoids in immature and mature fruits, respectively. The data are presented as mean ± standard deviation (n = 12). Different capital letters indicate significant difference (**P < 0.01) in pigment content between the two samples in Student’s t-test.
The young leaves of the two inbred lines were collected for WGRS. As a biological replicate of immature-stage samples, fruit peels (1 cm wide in the middle) from three fruits of different individual plants per inbred line were collected 40 d post-anthesis on November 4, 2020. The mature-stage samples were prepared 80 d post-anthesis using the aforementioned method on December 14, 2020. Three biological replicates per inbred line were sampled in both the immature and mature stages, resulting in a total of 12 samples that were submitted to biotech companies for transcriptome, metabolome, and pigment analyses.
2.2 Determination of chlorophylls, carotenoids and flavonoids
The chlorophyll and carotenoid contents were quantified following the methodology described by Wang et al. (2020) and Song et al. (2022). The total flavonoid content was determined using a NaNO2-Al(NO3)3-NaOH colorimetric assay (Jang et al., 2024).
2.3 Scanning and transmission electron microscopy
The pericarps of fresh fruit from each inbred line were dissected into small fragments and fixed using a stationary liquid 40 d post-anthesis. Samples were cut into paraffin sections, stained with toluidine blue, and observed using a PANNORAMIC microscope (3DHISTECH, Hungary). Another set of samples was visualized using a HITACHI HF7700 system (Hitachi, Japan) for the TEM assay.
2.4 Metabolome analysis
Metabolite profiling was conducted by the Wuhan Metware Biotechnology Co. Ltd. (Wuhan, China). Carotenoid extracts were quantified using a UPLC-APCI-MS/MS system (ExionLC™ AD; MS, Applied Biosystems 6500 Triple Quadrupole). The sample was freeze-dried, ground into a powder (30 Hz, 1.5 min), and stored at -80°C. The powder (50 mg) was weighed and extracted with 0.5 mL mixed solution of n-hexane: acetone: ethanol (1:1:1, v/v/v). The extract was vortexed for 20 min at room temperature. The supernatants were collected after centrifuged at 12000 r/min for 5 min at 4°C. The residue was reextracted by repeating the aforementioned steps under the same conditions. The mixture was evaporated to dryness, and reconstituted in a mixed solution of MeOH/MTBE (1:1, v/v). The solution was filtered through a 0.22 μm membrane filter for further LC-MS/MS analysis (Inbaraj et al., 2008).
Widely targeted metabolomic analysis of metabolites (mainly flavonoids) was carried out using a UPLC-ESI-MS/MS system (UPLC, Shim-pack UFLC SHIMADZU CBM30A system; MS, Applied Biosystems 4500 QTRAP). In brief, the freeze-dried fruit peel was crushed into powder using a mixer mill (MM 400, Retsch) with zirconia bead for 1.5 min at 30 Hz. Subsequently, 100 mg of the powder was extracted overnight at 4°C with 0.6 mL 70% aqueous methanol. After centrifugation at 10,000 × g for 10 min, the extracts were absorbed and filtered before LC-MS/MS analysis (Chen et al., 2013). Metabolite quantification was performed in the multiple reaction monitoring (MRM) mode. A combination of principal component analysis (PCA), fold change (FC) and orthogonal projections to latent structures discriminant (OPLSDA) analyses was used to identify differentially accumulated metabolites (DAMs) based on three criteria: VIP (variable importance in projection) ≥ 1, FC ≥ 2 or ≤ 0.5, and P value ≤ 0.05 (Yun et al., 2015).
2.5 Transcriptome analysis and WGRS analysis
Transcriptome profiling was performed using Biomarker Technologies Co. Ltd. (Beijing, China). Total RNA was extracted from 12 frozen peel samples using TRIZOL reagent (Invitrogen). Approximately 1 μg of RNA per sample was used as the input material for RNA sample preparations. Sequencing libraries were generated and index codes were added to attribute sequences to each sample. Purified mRNAs were cleaved into small fragments using divalent cations under elevated temperatures and reverse-transcribed into first-strand cDNAs using random hexamer primers and M-MuLV Reverse Transcriptase. Subsequently, double-stranded cDNAs were synthesized, purified, end-repaired, and ligated to the adaptors. Finally, cDNA libraries were constructed by PCR for paired-end sequencing. After clustering the index-coded samples, library preparations were sequenced on an Illumina XtenPE150 platform.
Hisat2 (Daehwan et al., 2015) was used to map the clean reads onto C. annuum Zunla-1 genome sequence (Qin et al., 2014). Picard and samtools (Li et al., 2009) were used to sort and remove duplicated reads and merge the alignment results of each sample. Gene function was annotated using BLAST (Altschul et al., 1997) by aligning their sequences to GO, COG, Pfam, KEGG, NR, and Swiss-Prot databases. The gene expression level was calculated and normalized using the FPKM (Fragments Per Kilobase of exon model per Million mapped fragments) method (Li and Dewey, 2011). To control the false discovery rate (FDR), Benjamini and Hochberg’s approach (Benjamini and Hochberg, 1995) was applied to adjust the resulting P values, while DESeq2 (Love et al., 2014) was utilized to identify DEGs based on two criteria: FC ≥ 2 or ≤ 0.5 and FDR (false discovery rate) ≤ 0.01). The identified DEGs were further subjected to the GO, COG, and KEGG databases for enrichment analysis. To explore the correlation between these DEGs and DAMs, canonical correlation analysis (CCA) and a network graph of correlation were performed.
Total genomic DNA was extracted from the young leaves of the two inbred lines using an improved CTAB method (Allen et al., 2006). DNA was randomly fragmented and fragments ranging from 300 to 400 bp were selected for amplification. These amplified fragments were used to construct paired-end libraries, which were subjected to paired-end sequencing on an Illumina HiSeq ™ 2500 platform. Finally, the obtained DNA sequences were optimized, aligned, and annotated using the same procedure as that used for the RNA sequencing.
2.6 Quantitative real-time PCR validation
Total RNA was extracted from the fruit peels of CSJ009 and CSJ010 at 40 d post-anthesis using TRIzol reagent (Invitrogen) and then purified with a DNA-free RNA kit. First-strand cDNA was synthesized using the Fermentas RevertAid First Strand cDNA Synthesis Kit. Ca-Actin (GQ339766) served as a reference gene for the normalization of gene expression (Wan et al., 2011). All primers were designed using Primer Premier 6.0 (PREMIER Biosoft, USA), and their details are provided in Supplementary Table S1. qRT-PCR was conducted using a Roche Light Cycler 2.10 with a 2×SYBR Green I PCR Master Mix according to the following procedure: 95°C for 3 min; 39 cycles of 95°C for 10 s, 58°C for 30 s; and 72°C for 4 s, and quantitative data of gene expression were analyzed using the 2-ΔΔCT method (Livak and Schmittgen, 2001).
2.7 Statistical analysis
All experiments were performed in triplicate, and the results are expressed as mean ± SE. Analysis of variance was performed using SPSS (version 22.0; SPSS Institute Inc., United States), and treatment means were compared using Student’s t-test. Two subfigures (C and F) in Figure 1 were prepared using SigmaPlot 14.0 (Systat Software, Inc., Germany). Other statistical charts were generated using Microsoft Excel 2013 (Microsoft Corporation, United States).
3 Results
3.1 Variation of pigment content
CSJ010 bears green immature and orange mature fruits, whereas CSJ009 bears light-yellow immature and red mature fruits (Figure 1). Chlorophyll was barely detected in CSJ009R and CSJ010O. In contrast, the levels of chlorophylls in CSJ010G (53.87 μg g-1 FW) were 34 times higher compared to those in CSJ009Y (1.61μg g-1 FW). Similarly, the total carotenoid content of CSJ010G (13.21 μg g-1 FW) was significantly higher (P < 0.01) than that of CSJ009Y (1.43 μg g-1 FW). However, the total carotenoid level in CSJ009R (80.05 μg g-1 FW) was comparable to that in CSJ010O (79.74 μg g-1 FW). The total flavonoid content of CSJ010G (2242.79 μg g-1 FW) was 17% higher than that of CSJ009Y (1919.37 μg g-1 FW).
3.2 Phenotype analysis of immature fruit skin in pepper
Paraffin section assay revealed that the epidermal cells of CSJ010G exhibit a more compact arrangement (Figures 2A, B) and significantly greater length (Figure 2C) than those of CSJ009Y. The difference between CSJ009Y and CSJ010G in the length and width of subepidermal cells was smaller than that of epidermal cells (Figure 2C).
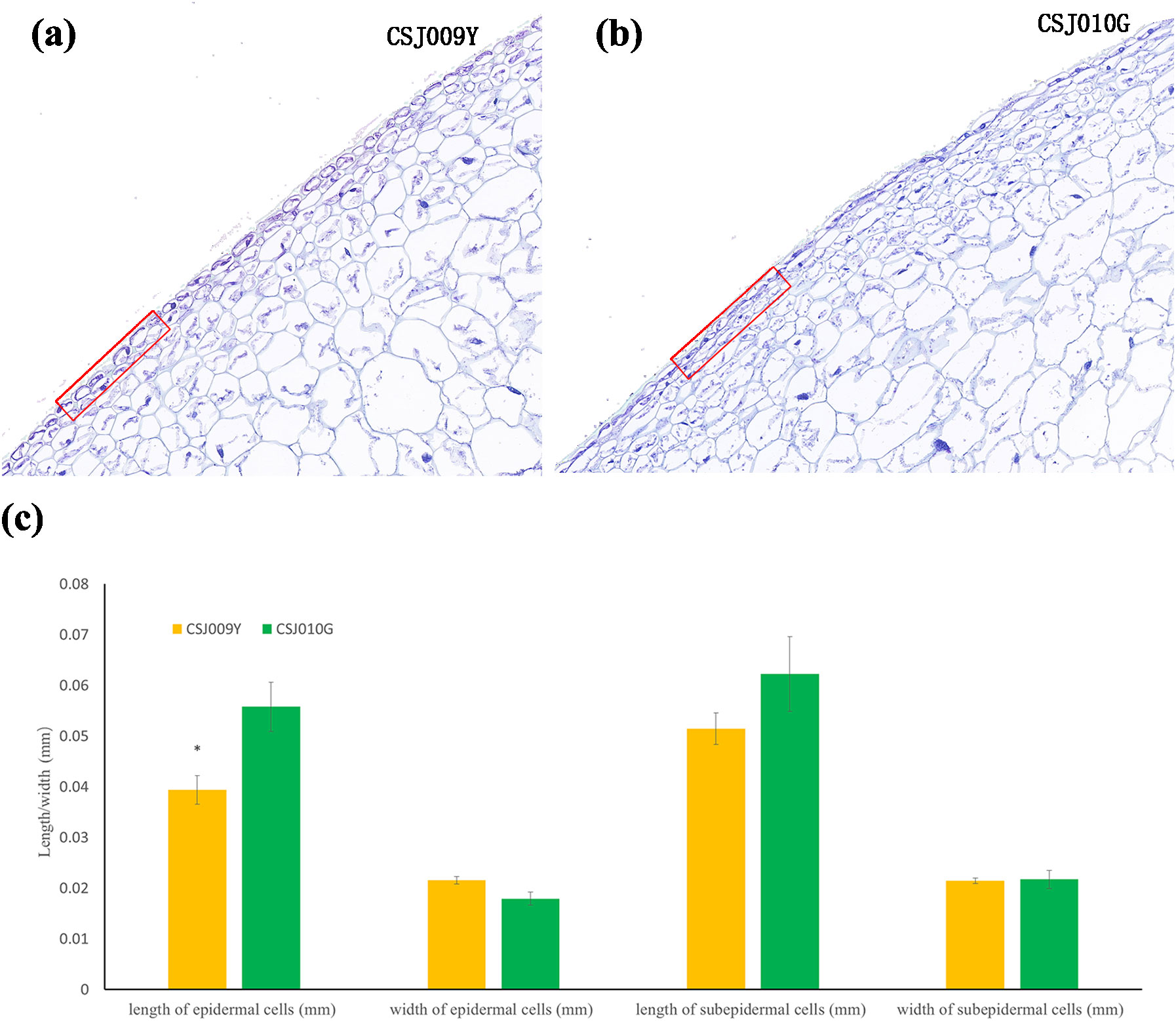
Figure 2. Paraffin section assay of the fruit skin in CSJ009Y and CSJ010G. Subfigures (A, B), the arrangement of skin cells. Subfigure (C) shows the difference in the length and width of the skin cells between CSJ009Y and CSJ010G. *P < 0.05 in Student’s t-test.
We also examined the inner structures of plastids in immature fruits using TEM. Chloroplasts exhibiting the characteristic thylakoid membrane structure were clearly observed in CJS010G (Figures 3D–F), whereas they were barely detectable even in the 20× magnified images in CSJ009Y (Figures 3A–C). The presence of a greater number of chloroplasts and thylakoids within the pericarp cells of CSJ010G, compared to CSJ009Y, suggests a higher accumulation of chlorophyll in the immature fruit skins of CSJ010 than those of CSJ009, which is consistent with the pigment substance determination results.
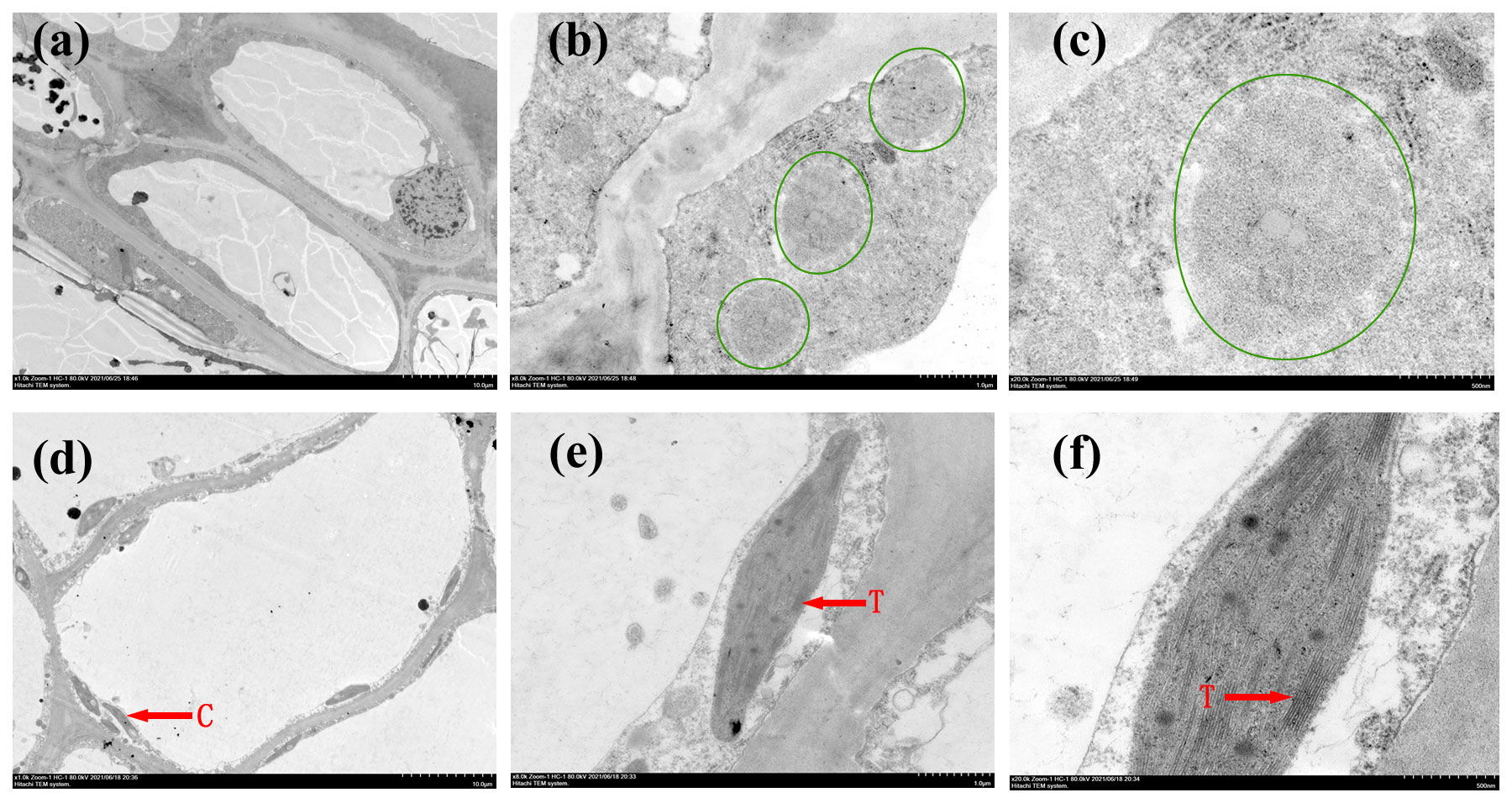
Figure 3. Transmission electron microscopy observations of the fruit skin in CSJ009Y and CSJ010G. Subfigures (A–C), TEM images of CSJ009 pericarp cells. Subfigures (D–F), TEM images of CSJ010 pericarp cells. The tissue in the green circle represents a proplastid. ‘C’ represents chloroplast. ‘T’ represents thylakoid. Scale bar in (A) and (D) 10.0 μm, (B) and (E) 1.0 μm, (C) and (F) 500.0 nm.
3.3 Identification of differently expressed genes
After conducting mRNA sequencing on 12 samples, a total of 91.79 Gbp clean data were generated, with a minimum of 6.77 Gbp per sample. A total of 8,152 new genes were discovered, of which 79.8% were successfully annotated. Through differential expression analysis, we identified 23,930 DEGs across the four groups (Supplementary Table S2), with DEG numbers ranging from 1,212 to 5,744 between any two groups. From these DEGs, we classified 726 TFs into 59 families, among which AP2/ERF-ERF, bHLH, C2H2, MYB, and B3 were the top five TF families (Supplementary Figure S1).
In the immature-fruit group (CSJ009Y vs CSJ010G), we found a total of 2,520 DEGs, comprising 1,332 upregulated genes and 1,188 downregulated genes. The volcano plot clearly illustrates the differences in the gene expression levels between the two groups (Supplementary Figure S2). KEGG analysis revealed that 441 DEGs were enriched in 108 pathways, with 23 DEGs specifically identified within the three color-related pathways (Supplementary Table S3). Additionally, in the mature-fruit group (CSJ009R vs. CSJ010O), we identified a set of 2,280 DEGs, comprising 1,212 upregulated and 1,068 downregulated genes.
As shown in Figure 4, the majority of genes associated with chlorophyll and carotenoid synthesis (e.g., HEMA1, CHLM, CRD1, POR1, LCYB2, LCYE1, CHYB1 and LUT1) were upregulated in CSJ010G compared to CSJ009Y. Interestingly, most genes involved in flavonoid-biosynthesis (e.g., HCT, FLS, CCOAOMT, and At4g26220) were downregulated in CSJ010G, except for HCT-like. Furthermore, both PSY2 and CCS involved in carotenoid synthesis were downregulated in mature orange CSJ010O fruit relative to red CSJ009R fruit. Three genes (CHLP, POR1 and DVR) implicated in chlorophyll synthesis were upregulated.
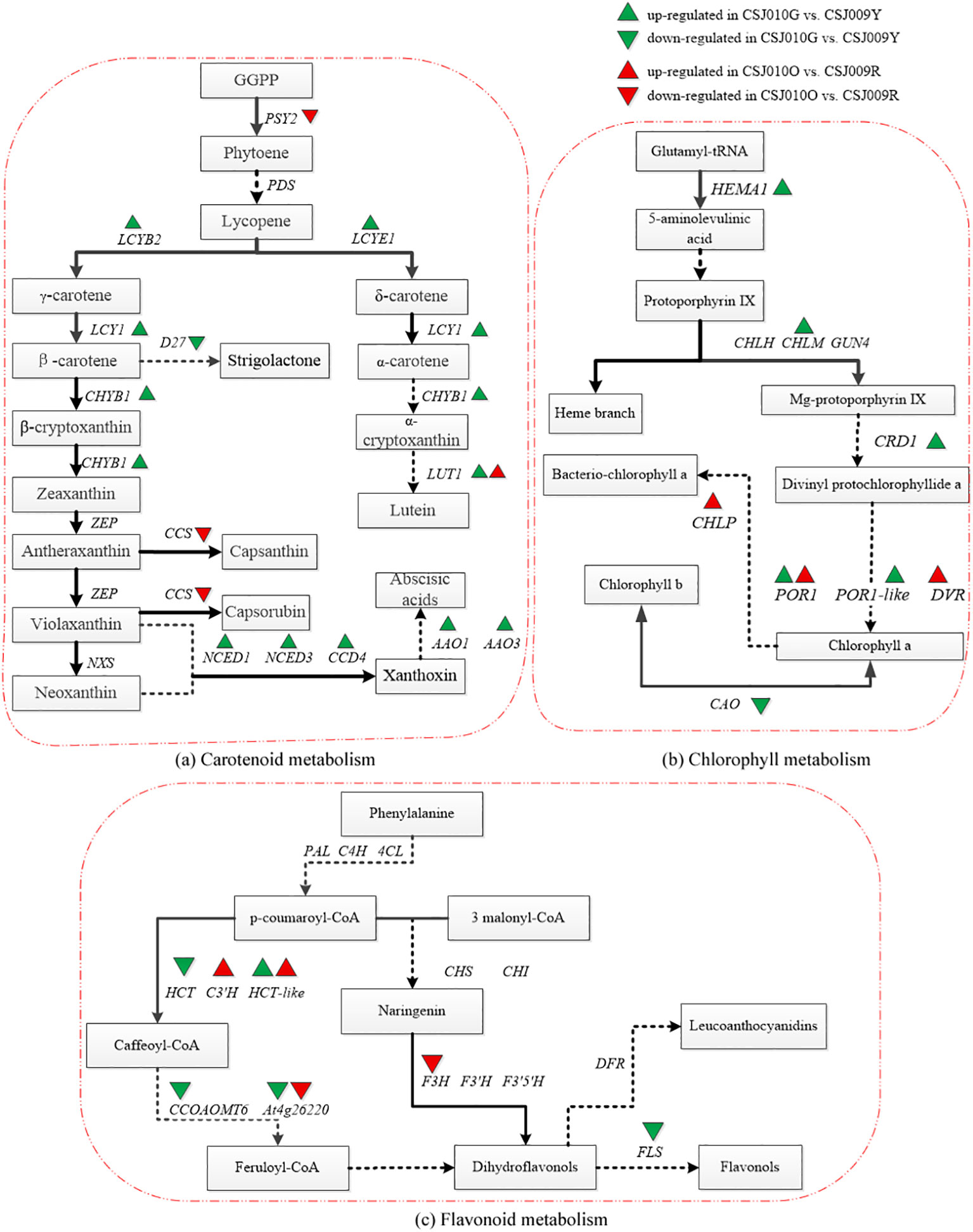
Figure 4. Pathway map of DEGs related to chlorophyll, carotenoid and flavonoid metabolism in KEGG annotation analysis. Subfigure (A), the pathway map of carotenoid metabolism was plotted according to the differential expression analysis and KEGG annotation analysis with reference to Yuan’s study (Yuan et al., 2015). PSY2 (Phytoene desaturase 2, Capana02g002284), PDS (Phytoene desaturase), LCYB2 (Lycopene β-cyclase, Capana10g002320), LCYE1 (LCYE: Lycopene ϵ-cyclase, Capana09g000177), LUT1 (Carotene epsilon-monooxygenase, Capana10g001912), ZEP (Zeaxanthin epoxidase), CCS (Capsanthin-capsorubin synthase, Capana06g000615), NXS (Neoxanthin synthase), CCD4 (Carotenoid cleavage dioxygenase 4, Capana01g000948), NCED1 (9-cis-epoxy carotenoid dioxygenase 1, Capana00g003114), NCED3 (9-cis-epoxy carotenoid dioxygenase 3, Capana01g003704), AAO1 (Indole-3-acetaldehyde oxidase 1, Capana11g000423), AAO3 (Abscisic-aldehyde oxidase 3, Capana11g000422), CHYB1 (β-carotene hydroxylase1, Capana03g002170). Subfigure (B), the pathway map of chlorophyll metabolism was plotted according to the differential expression analysis and KEGG annotation analysis with reference to Masuda’s study (Masuda and Fujita, 2008). CAO (Chlorophyllide a oxygenase, Capana06g001725), CHLM (Magnesium protoporphyrin IX methyltransferase, Capana03g000633), CRD1 (Magnesium-protoporphyrin IX monomethyl ester, Capana10g002504), POR1 (Protochlorophyllide oxidoreductase1, Capana00g004560), POR1-like (Protochlorophyllide oxidoreductase 1-like, Capana10g000065), HEMA1 (Glutamyl-tRNA reductase1, Capana04g000804), CHLP (Geranylgeranyl diphosphate reductase, Capana03g000791), DVR (Divinyl chlorophyllide a 8-vinyl-reductase, Capana01g003169), CHLH (Magnesium chelatase subunit H), GUN4 (Genomes uncoupled4). Subfigure (C), the pathway map of flavonoid metabolism was plotted according to the differential expression analysis and KEGG annotation analysis with reference to Yuan’s study (Yuan et al., 2015). HCT (Shikimate O-hydroxycinnamoyltransferase, Capana03g000549), HCT-like (Shikimate O-hydroxycinnamoyltransferase -like, Capana09g000120), PAL (Phenylalanine lyase), C4H (Cinnamic acid 4-hydroxylase), 4CL (4-coumadin-CoA ligase), CHS (Chalcone synthase), CHI (Chalcone isomerase), F3H (Flavanone 3-hydroxylase, Capana02g002586), F3’H (Flavonoid-3’-hydroxylase), F3’5’H (Flavonoid 3’,5’-hydroxylase), C3’H (Coumaroylquinate 3’-monooxygenase, Capana10g002482), CCOAOMT (Caffeoyl-CoA O-methyltransferase, Capana08g002351), At4g26220 (Caffeoyl-CoA O-methyltransferase At4g26220 isoform X1, Capana00g004448), DFR (Dihydroflavonol 4-reductase), FLS (Flavonol synthase, Capana09g002174). The green upward or downward triangle indicates that the gene was upregulated or downregulated in CSJ010G compared to CSJ009Y. The red upward or downward triangle indicates that the gene was upregulated or downregulated in CSJ010O compared to CSJ009R.
Color-related genes in immature fruits were the focus of our study. PP2C35 was not detected in any of the four groups (Table 1). The expression levels of the three TFs were drastically downregulated (P < 0.01) in mature fruits compared to immature fruits, indicating their predominant expression in immature fruits. There were no significant differences in the expression levels of GLK2 and LOL1 between immature- and mature-fruit groups. APRR2 expression was significantly higher (P < 0.01) in CSJ009 than in CSJ010.
3.4 Identification of differently accumulated metabolites
A total of 568 metabolites were identified using widely targeted metabolomic analysis, which can be classified into 11 known classes of metabolites (Supplementary Figure S4). The top five metabolites were flavonoids (18.8%), lipids (16.5%), amino acids (12.7%), phenolic acids (12.1%), and alkaloids (9.7%). Through a conjoint analysis of PCA, FC and OPLSDA, 345 metabolites were identified as DAMs among the four groups (Supplementary Table S4). Among the 345 DAMs, 86 and 121 were detected in the mature- and immature-fruit groups, respectively.
Eighteen differentially accumulated flavonoids (DAFs) associated with pepper fruit color were identified as representative DAMs (Supplementary Table S5). In the immature-fruit group, most flavones and flavonols were upregulated in CSJ009Y compared with CSJ010G, except for epicatechin glucoside, kaempferol-3-O-(6’’-malonyl) galactoside, and kaempferol-3-O-galactoside (trifolin). In the mature-fruit group, half of the DAFs were upregulated in CSJ009R vs. CSJ010O.
As depicted in the volcano plot (Supplementary Figure S3A), chlorogenic acid was the only metabolite downregulated in CSJ009Y vs. CSJ010G. Within the mature-fruit group, all top five DAMs were upregulated in CSJ009R vs. CSJ010O (Supplementary Figure S3B). Among the three color-related metabolic pathways, only DAMs and DEGs involved in the flavonoid biosynthesis pathway fulfilled the criteria for correlation analysis. As illustrated in Supplementary Figure S3, Capana09g000120 was the exclusive gene that positively correlated with MWS0178 (chlorogenic acid).
3.5 Several mutations of color-related genes were identified via WGRS
The differential expression of GLK2, LOL1, and APRR2 in the transcriptome analysis promoted us to investigate their coding sequences. WGRS data revealed a G-to-A substitution in the coding DNA sequence (CDS) 8 region of APRR2, resulting in a premature stop codon in CSJ009 (Figure 5A), whereas no non-synonymous single nucleotide variants (SNV) were found in PP2C35, GLK2, and LOL1. The nonsense mutation observed in APRR2 was identical to that reported previously in C. annuum IT158782 (Lee et al., 2020) and white-fruited pepper lines (Pan et al., 2013).
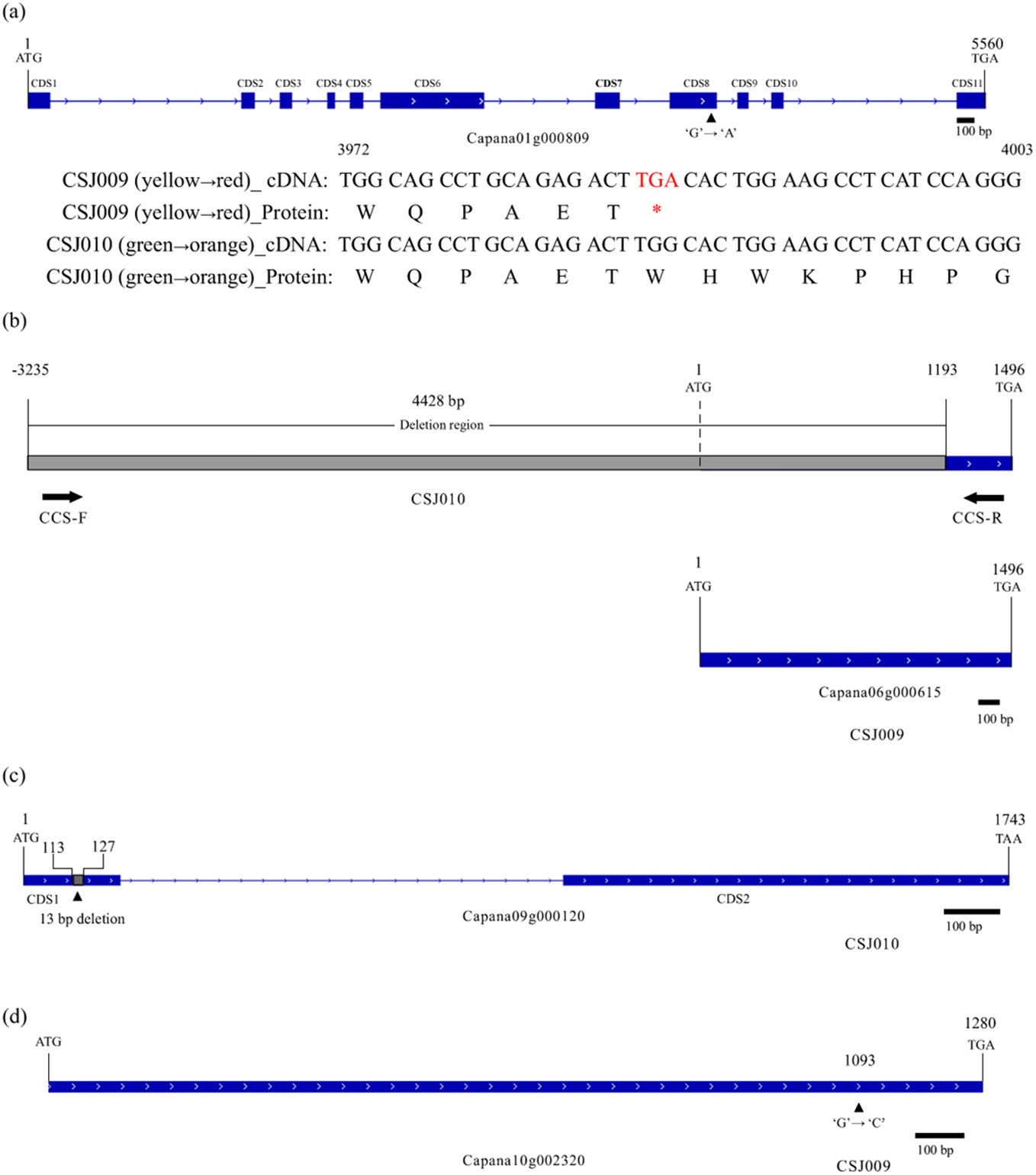
Figure 5. Mutations in APRR2, CCS, HCT-like and LCYB2. Subfigure (A), Nonsense mutation of APRR2 (Capana01g000809) in CSJ009. The symbol '*' means 'TGA' is a terminator. Subfigure (B), Null mutation of CCS (Capana06g000615) in CSJ010. Subfigure (C), Null mutation of HCT-like (Capana09g000120) in CSJ009. Subfigure (D), Missense mutation of LCYB2 (Capana10g002320) in CSJ009. Blue and gray rectangles indicate the coding and deletion region, respectively.
Several mutations in carotenoid-biosynthesis genes have been subsequently identified in re-sequencing data. A 4,428 bp deletion encompassing the promoter and coding regions of CCS (Capana06g000615) was detected in CSJ010 (Figure 5B). In the CDS1 region of HCT-like (Capana09g000120), a 13 bp coding sequence was deleted (Figure 5C). In addition, one non-synonymous SNV was observed in the coding regions of LCYB2 (Capana10g002320, Figure 5D) and PIF1 (Capana09g001161) in CSJ009. To verify these mutations, PCR amplification of related genes was performed for CSJ009 and CSJ010, and sequencing results showed that the amplified fragment sequences were consistent with the WGRS results, except for the CCS gene. We designed several primers for CCS gene but failed to amplify them successfully in CSJ010.
3.6 Identification of differently accumulated carotenoids
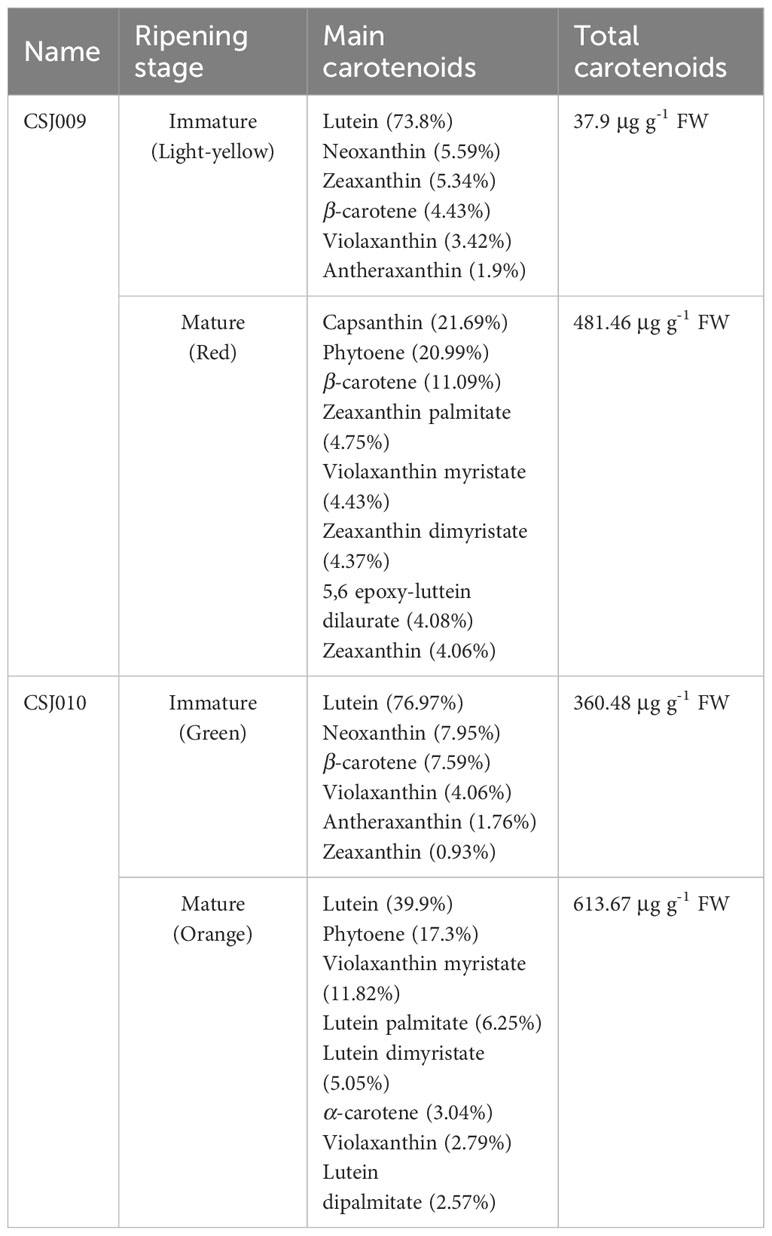
A total of 68 carotenoids were identified in the four samples (Supplementary Table S6). In immature fruits, 20 and 17 carotenoids were assayed in the CSJ009 and CSJ010, respectively. Lutein has emerged as the predominant carotenoid, constituting over 73% of total carotenoid content. Notably, lutein, neoxanthin, zeaxanthin, β-carotene, violaxanthin, and antheraxanthin ranked among the top six carotenoids in both accessions. The immature fruits of CSJ009 contained a significantly lower amount of total carotenoids (37.9 μg g-1 FW) than those of CSJ010 (360.48 μg g-1 FW), representing only 10.5% of its counterpart.
In mature fruits, 44 carotenoids were measured in both the inbred lines. However, the predominant carotenoid components exhibited significant differences (Table 2). The red CSJ009 fruits displayed the highest concentration of capsanthin (21.69%), followed by phytoene (20.99%), β-carotene (11.09%), zeaxanthin palmitate (4.75%), violaxanthin myristate (4.43%), and zeaxanthin dimyristate (4.37%). Lutein (39.9%) and phytoene (17.3%) were the major constituents of orange CSJ010 fruit, whereas violaxanthin myristate (11.82%), lutein palmitate (6.25%), lutein dimyristate (5.05%), and α-carotene (3.04%) followed. Capsanthin (only 0.087 μg g-1 FW) was barely detected in CSJ010O mature fruits, while the total carotenoid content of CSJ010O reached 613.67 μg g-1 FW, surpassing that of CSJ009R (481.46 μg g-1 FW). During the fruit maturation stages, there was an increase of 1170.47% and 70.24% in the total carotenoid content for CSJ009 and CSJ010 fruits, respectively.
As the fruit ripens, the amount and composition of carotenoids undergo drastic changes. A total of 28 differentially accumulated carotenoids (DACs) were detected in the four groups (Supplementary Table S7). Zeaxanthin dimyristate, violaxanthin myristate, and β-carotene exhibited remarkable increases of 617.83-fold, 72.93-fold and 31.83-fold, respectively, in CSJ009R vs. CSJ009Y. Conversely, lutein level decreased dramatically by 97.68% in CSJ009R compared to CSJ009Y; however, no significant change (FC = 0.88) was observed in CSJ010 fruit during this period. The levels of lutein dilaurate, lutein dimyristate, and violaxanthin myristate in CSJ010O were significantly increased by 677.41-fold, 528.23-fold, and 459.95-fold, respectively, compared to those in CSJ010G. In contrast to the upregulated expression of β-carotene in CSJ009R vs. CSJ009Y (FC = 31.8), the level of β-carotene in CSJ010O was only 41% of that in CSJ010G (FC = 0.41).
The composition and content of carotenoids in the fruits at the same growth-time fruits exhibited significant variations. In mature fruits, 27 DACs have been identified in CSJ009R and CSJ010O. Lutein, violaxanthin, and their fatty-acid esters (dilaurate, dimyristate, dipalmitate, myristate, myristate, and laurate) were upregulated. In contrast, zeaxanthin and its esters (palmitate, laurate, myristate, and dimyristate) were downregulated in this group. In particular, lutein increased by a remarkable 377-fold, whereas capsanthin decreased by an astonishing 1199-fold in CSJ010O compared to CSJ009R. In immature fruits, only two lutein esters (dilaurate and dimyristate) displayed down-regulation in CSJ010G vs. CSJ009Y.
3.7 qRT−PCR analysis of color-related genes
qRT-PCR analyses of representative genes, including 22 DEGs enriched in three color-related metabolic pathways (Supplementary Table S3) and four color-related genes (Table 1), were performed for CSJ009Y and CSJ010G. The majority of chlorophyll and carotenoid biosynthesis genes exhibited upregulated expression levels in the immature fruits of CSJ010G compared to CSJ009Y (Figure 6), except for CAO and D27. Additionally, the expression of three carotenoid degradation genes (CCD4, NCED1, and NCED3) was also upregulated in CSJ010G vs. CSJ009Y. However, APRR2 was significantly upregulated (P < 0.01) in CSJ009Y vs. CSJ010G, whereas almost no PP2C35 expression was detected in CSJ009Y and CSJ010G. To confirm this observation, three independent qRT−PCR assays were repeated, which confirmed that HCT-like were highly expressed in CSJ010G but not in CSJ009Y. These results indicate that the downregulated expressions of genes involved in chlorophyll and carotenoid biosynthesis led to reduced levels of chlorophyll and carotenoids in CSJ009Y compared to CSJ010G, which is in agreement with the transcriptome data.
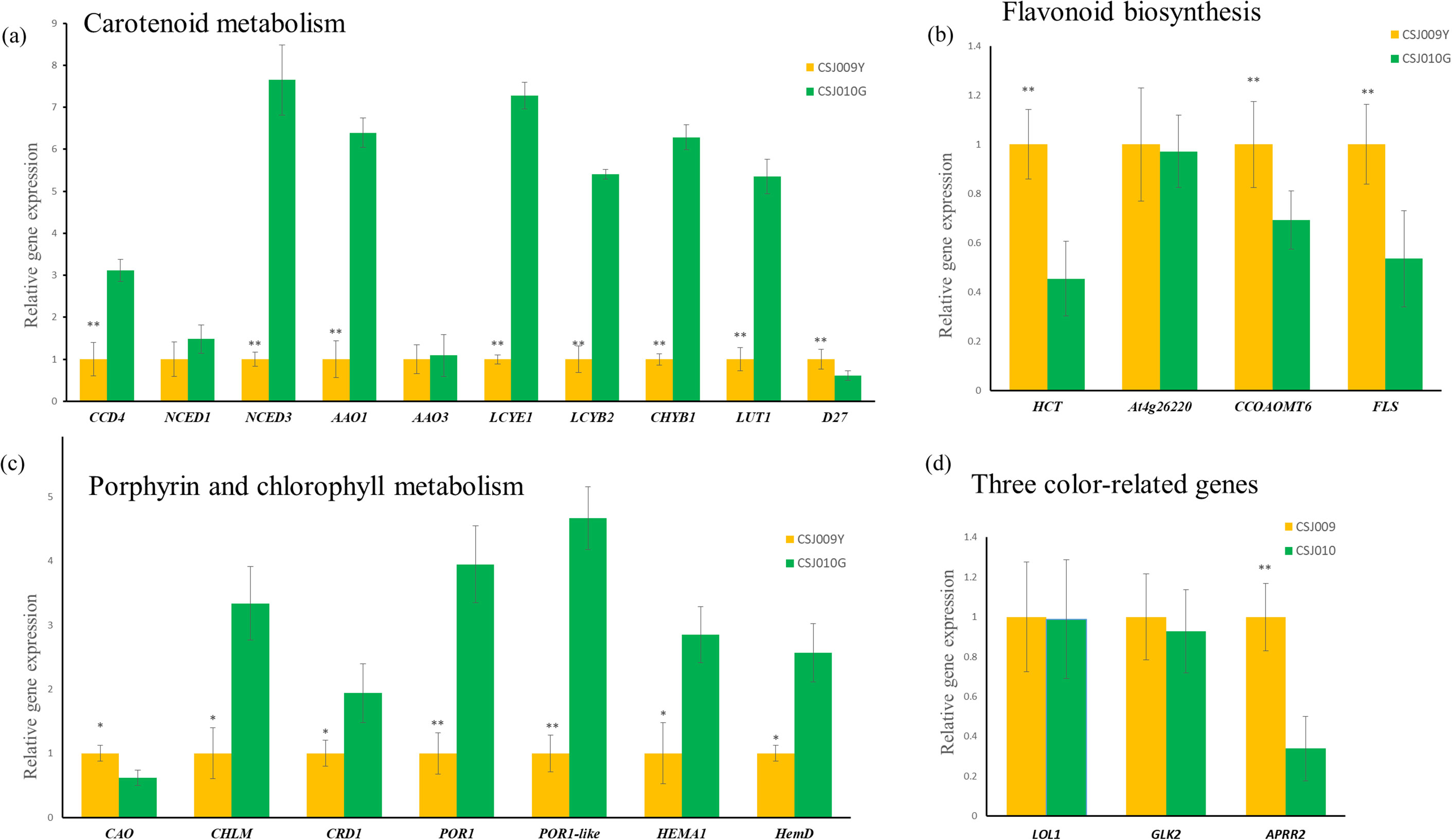
Figure 6. qRT-PCR expression analysis of color-related genes between CSJ009Y and CSJ010G. The X-axis genes depicted in subfigures (A–C) are interpreted as illustrated in Figure 4. Subfigure (D), The X-axis represents genes: Lsd one like1 (LOL1, Capana06g002446), Golden2-like (GLK2, Capana10g000333), Arabidopsis pseudo response regulator2 (APRR2, Capana01g000809). The Y-axis in subfigures (A–D) represents the relative gene expression. Error bar is standard error. *P < 0.05 and **P < 0.01 in Student’s t-test.
4 Discussion
Pepper (Capsicum spp.) is an appealing crop for fruit color research owing to its rich color. Fruit of the two inbred lines CSJ009 and CSJ010, exhibited distinct colors at both immature and ripe stages. According to the prevailing theory, the colors of immature and ripe pepper fruits are controlled by a different series of genes. Mature fruit color is governed by three loci: y, c1, and c2 (Kormos and Kormos, 1960; Wang and Bosland, 2006). CCS, APRR2, and PSY have been identified as candidate genes for y, c1, and c2, respectively (Huh et al., 2001; Lee et al., 2020; Lefebvre et al., 1998). In contrast, green to ivory immature fruit colors are attributed to three loci: sw1, sw2, and sw3 (Lightbourn et al., 2008). However, no candidate genes were identified for these three loci. Only two QTLs pc8.1 (renamed pc1), pc10.1 (Borovsky et al., 2019; Brand et al., 2012), along with four genes, including GLK2 (Brand et al., 2014), LOL1 (Borovsky et al., 2019), APRR2 (Jeong et al., 2020; Lee et al., 2020), and PP2C35 (Wu et al., 2022) are thought to regulate immature fruit color. In this study, we selected two pepper inbred lines that displayed varying colors for both immature and mature fruits as our research objects. An integrative strategy of transcriptome and metabolome analysis combined with re-sequencing and quantitative determination was adopted to investigate the regulation of color-related genes in pepper fruits.
4.1 Decreased chlorophylls and carotenoids should be responsible for light-yellow immature pepper fruit
The formation of various fruit colors in pepper is primarily attributed to the accumulation of chlorophylls, flavonoids and carotenoids in varying proportions (Lightbourn et al., 2008). The pigment determination assay revealed that flavonoids were the predominant pigments in CSJ009Y, whereas the levels of chlorophylls and carotenoids were significantly lower (P < 0.01) than those in CSJ010G. This observation prompted us to investigate the differences in the epidermal cells and chloroplasts within the fruit skin between CSJ009Y and CSJ010G.
Chlorophylls and carotenoids are chloroplast-specific pigments, whereas flavonoids are located within the cell vacuoles (Lightbourn et al., 2008). Carotenoid biosynthetic enzymes are predominant in thylakoid membranes (Sun et al., 2018), which are specialized organelles responsible for carotenoid synthesis and sequestration (Nisar et al., 2015). Therefore, maintaining an intact chloroplast structure is crucial for carotenoid and chlorophyll accumulation. The spherical organelles observed in CSJ009Y (marked with green circles in Figure 3) highly resembled the progenitors of chloroplasts (proplastids) as described by Mullet (1988). In Arabidopsis, the loss of PIF1 leads to the delayed development of chloroplasts (Pogson and Albrecht, 2011). Similarly, the missense mutation of PIF1 (Capana09g001161) in CSJ009 may impede chloroplast development and subsequently decrease carotenoid and chlorophyll levels in immature pepper fruits. Additionally, the loosely arranged epidermal cells observed in CSJ009Y resembled those observed in the white cucumber fruit skin (Wang et al., 2020). Consequently, a combination of reduced chlorophyll and carotenoids, along with abundant flavonoids contributes to the light-yellow appearance of immature fruits in CSJ009 through the regulation of cell structure and chloroplast development.
Most chlorophyll and carotenoid biosynthesis genes were upregulated in green-fruited CSJ010. Interestingly, most of the identified chlorophyll biosynthesis genes in this study, including CHLM, POR1-like, HEMA1, CRD1, and POR1 (except CAO) were downregulated in CSJ009Y compared with CSJ010G. This result further supports the upregulation of most chlorophyll biosynthesis genes in dark green fruits compared to pale green fruits in pepper (Lee et al., 2020). Additionally, LUT1, LCYB, and LCYE have been identified as crucial genes involved in lutein and zeaxanthin formation (Zhang et al., 2020). CHYB hydroxylates β-carotene to produce zeaxanthin (Diretto et al., 2007). As shown in Supplementary Table S3, several carotenoids biosynthesis-related DEGs, including LUT1, CHYB1, LCYB2, and LCYE1 (except D27) were significant upregulated in CSJ010G compared to CSJ009Y. The observed upregulated expression levels of these key genes associated with both chlorophyll and carotenoid biosynthesis may contribute to the higher content of both chlorophyll and carotenoids in CSJ010G relative to CSJ009Y.
Degradation processes also influenced the accumulation of carotenoids. CCD plays a pivotal role in the degradation of various xanthophylls, including zeaxanthin, lutein, and other carotenoids in strawberry (García-Limones et al., 2008), marigold (Zhang et al., 2020) and chrysanthemum (Ohmiya et al., 2006). In contrast, NCED specifically catalyzes the degradation of zeaxanthin (Zhang et al., 2020). In this study, three carotenoid degradation genes CCD, NCED1 and NCED3, were upregulated in CSJ010G compared to CSJ009Y. Notably, the upregulation of carotenoid biosynthetic genes observed in green-fruited CSJ010G was accompanied by a simultaneous increase in the expression levels of degradation genes compared to light-yellow-fruited CSJ009Y.
4.2 HCT may regulate the chlorogenic acid biosynthesis in green pepper fruit
The HCT gene is involved in phenylpropanoid biosynthesis (Hoffmann et al., 2004), which regulates chlorogenic acid biosynthesis in tomato (Aseel et al., 2019). In the present study, the total flavonoid content of immature and mature fruits of CSJ010 was significantly higher (P < 0.01) than that of CSJ009 (Figure 1C). Among the five DEGs identified in the flavonoid biosynthesis pathway in the immature-fruit group (Supplementary Table S3), HCT, CCOAOMT6, FLS, and At4g26220 were all downregulated in CSJ010G vs. CSJ009Y. However, HCT-like (Capana09g000120) was the only upregulated gene (FPKM = 4.8) in CSJ010G; and no expression (FPKM = 0) was detected in CSJ009Y.
Furthermore, integrative analysis of the metabolome and transcriptome revealed a significant correlation between HCT-like gene and chlorogenic acid (MWS0178) levels (Supplementary Figure S3). These findings unraveled that HCT-like may regulate chlorogenic acid biosynthesis specifically in green-fruited CSJ010 but not yellow-fruited CSJ009 due to the null mutation of HCT-like observed in CSJ009. Although chlorogenic acid and chlorophyll content are significantly positively correlated and chlorophyll synthesis interacts with chlorogenic acid synthesis in tobacco leaves (Sheen, 1973), few studies on chlorogenic acid content changing plant fruit color have been reported so far.
4.3 APRR2 may regulate the immature fruit color via reducing carotenoid content
GLK2, LOL1, APRR2, and PP2C35 (Borovsky et al., 2019; Brand et al., 2014; Lee et al., 2020; Wu et al., 2022) were thought to be related to chlorophyll biosynthesis in immature fruits. WGRS analysis revealed no mutations of GLK2, LOL1 or PP2C35 in CSJ009. LOL1 and GLK were exclusively detected in the immature-fruit group during the transcriptome analysis (Table 1). Furthermore, no significant difference was observed in the expression levels of GLK2, LOL1, and PP2C35 between CSJ009 and CSJ010, according to qRT-PCR analysis (Figure 6). In contrast, APRR2 exhibited differential expression within the immature- and mature-fruit groups. Among the four color-related genes analyzed, APRR2 was the only DEG identified in immature-fruit group (Table 1). This finding is consistent with previous results showing that APRR2 regulates both immature and mature fruit color in pepper (Lee et al., 2020).
APRR2 has been demonstrated to be closely associated with immature fruit color in a double haploid population (Pan et al., 2013) and an F2 population (Lee et al., 2020). APRR2 regulates the immature fruit color via reducing chloroplast size and thylakoid numbers to decrease the levels of chlorophyll and carotenoids (Lee et al., 2020). However, the role of APRR2 in governing pepper fruit color appears to be inconsistent. In C. frutescens, APRR2 specifically expressed in immature fruits while being almost undetectable in mature fruits (Jeong et al., 2020). The chlorophyll level of immature fruit in the light-green-fruited line, which harbors a nonsense mutation in APRR2, was determined by CcLOL1 rather than by APRR2 (Borovsky et al., 2019). Another dark-green-fruited line 165-2 also carried the same nonsense mutation in APRR2 (Borovsky et al., 2019). These results suggest that reduced chlorophyll content may not be an inescapable consequence of a nonsense mutation in APRR2.
WGRS suggests that the nonsense mutation of APRR2 in CSJ009 is consistent with those found in C. annuum ‘IT158782’ (Lee et al., 2020) and APRR2-Like gene (Pan et al., 2013), but differs from those observed in C. frutescens ‘AC08-201’ (Jeong et al., 2020). The immature fruit color of CSJ009 resembles that of blocky pepper harboring APRR2-like, albeit being much paler yellow compared to C. frutescens ‘AC08-201’ and C. annuum ‘IT158782’. Our quantification demonstrated that the total carotenoid content in the immature fruits of CSJ009 accounted for only 10% of the CSJ010 content. qRT-PCR analysis revealed a significant upregulation of APRR2 in the immature fruits of CSJ009 compared to CSJ010 (Figure 6). Collectively, these findings suggest that APRR2 may regulate immature fruit color by reducing carotenoid content.
In peppers, the peach fruit has been suggested to be determined by the c1 locus (Thorup et al., 2000). The regulatory factor APRR2 regulates mature fruit color via reducing the amount of carotenoids (Lee et al., 2020) and has been recognized as a candidate for the c1 locus in C. chinense, C. annuum (Lee et al., 2020), and C. frutescens (Jeong et al., 2020). The c2 (PSY1) mutation has been found to be responsible for the ripe orange fruit color in C. chinense ‘Habanero Orange’ (Kim et al., 2010). The c1 (APRR2) mutation coupled with the c2 (PSY1) mutation formed peach fruit color in C. chinense ‘Habanero Peach’ and no y (CCS) mutation was found in the line (Lee et al., 2020). The null mutation of y (CCS), coupled with the nonsense mutation of c1 (APRR2) and the null mutation of c2 (PSY1), produced white mature fruits in C. annuum ‘IT158782’ (Lee et al., 2020) and C. frutescens ‘AC08-201’ (Jeong et al., 2020). In this study, the mature fruit color of CSJ009 was red, implying the absence of any mutations in c2 (PSY1) and y (CCS), which was further supported by the WGRS analysis (Figure 5B). A premature stop codon was identified in the 8th CDS region of APRR2 (Figure 5A), and the total carotenoid content of red CSJ009 fruit was lower than that of orange CSJ010 fruit (Supplementary Table S6), suggesting that the c1 (APRR2) mutation presents in CSJ009. These results indicate that red-fruited CSJ009 contains the c1C2Y genotype, orange-fruited CSJ010 harbors the C1C2y genotype, peach-fruited pepper has the c1c2Y genotype, and white-fruited pepper has the c1c2y genotype. In summary, the nonsense mutation of APRR2 is not an inevitable result of the formation of a mature peach fruit, but a prerequisite.
4.4 The null mutation of CCS may enhance the biosynthesis of β,ϵ-branch carotenoids
The types and levels of carotenoids vary widely among pepper fruits. Red mature CSJ009 fruits predominantly accumulated capsanthin, accompanied by phytoene, β-carotene, zeaxanthin palmitate, and violaxanthin myristate (Supplementary Table S6). Similarly, mature orange CSJ010 fruits also primarily accumulated lutein, along with phytoene, violaxanthin myristate, lutein palmitate, lutein dimyristate, and α-carotene. Phytoene is the main carotenoid in red and orange pepper fruits, next to capsanthin and lutein, thereby challenging previous conclusions that β-carotene is the most abundant carotenoid in orange fruits (Guzman et al., 2010), and lutein is usually only found in immature fruits (Gómez-García and Ochoa-Alejo, 2013).
Transcriptional regulation is a major determinant for carotenoid production in the classical model systems of tomato and pepper during fruit ripening in response to developmental signals (Yuan et al., 2015). The accumulation of capsanthin during pepper fruit ripening from green to red is linked with the transcriptional upregulation of CCS, along with PSY, PDS, and BCH (Hugueney et al., 1996). Upon comparing the DEGs involved in carotenoid biosynthesis between the mature- and immature-fruit groups, only CCS (Capana06g000615) and PSY2 (Capana02g002284) were detected in the mature-fruit group. The PSY2 gene is typically expressed in leaves but also at low levels in ripening fruit in tomato (Paran and van der Knaap, 2007). However, an ortholog of PSY2 has not yet been identified in pepper, and it has been postulated to exist in pepper and contribute to carotenoid synthesis in mature pepper fruits (Paran and van der Knaap, 2007). In line with this hypothesis, our results demonstrated that the expression of PSY2 was downregulated in CSJ010O vs. CSJ009R, further supporting its potential role as a key regulator of carotenoid accumulation, specifically in red pepper fruits.
Interestingly, while CSJ009 showed a high expression level of CCS (FPKM = 9343), almost no expression was detected in CSJ010 (FPKM = 0.72). The null mutation of CCS gene in CSJ010 was identical to the mutations found in CCS-P1 (Jeong et al., 2019) and IT158782 (Lee et al., 2020). The CCS-P1 mutation produces mature orange or yellow fruits (Jeong et al., 2019). Considering the quantification results for capsanthin, it can be inferred that this null mutation in CCS leads to the development of orange CSJ010 fruits. Notably, all accessions exhibiting this null mutation belonged to C. annuum lines. These results showed that CCS was specifically expressed in mature pepper fruits and regulates their red and orange coloration.
α-Carotene and β-carotene are downstream products of lycopene cyclized by LCY genes in β,ϵ- and β,β-branches, respectively (Zhang et al., 2020). The expression of LCYB and LCYE has been demonstrated to regulate the bifurcation of the metabolic flux from lycopene to its downstream β,β- and β,ϵ-branches (Cunningham and Gantt, 1998). The silencing of LCYE increased the contents of β-carotene in potato tubers (Diretto et al., 2006). While contents of both β-carotene and lutein were found to be downregulated when LCYB was silenced in tomato (Ma et al., 2011). LCYB1 (Capana05g000023), LCYB2 (Capana10g002320) and LCYE1 (Capana09g000177) have been identified and characterized in pepper, and both LCYB1 and LCYB2 had higher expression levels than LCYE1 in any of the ripening stages (Camara and Dogbo, 1986; Wang et al., 2019). In immature fruits, the α-carotene and β-carotene contents of CSJ009Y were found to be 12.09% and 6.13% of CSJ010G, respectively. In mature fruits, the α-carotene content in CSJ009R (2.61 μg g-1 FW) remained lower than that in CSJ010O (18.65 μg g-1 FW), while the β-carotene content was 4.78 times higher in CSJ009R vs. CSJ010O. The reduced level of β-carotene in CSJ009Y is likely attributed to a missense mutation affects LCYB2 function. Furthermore, the relative expression of LCYB2 was significantly higher (P < 0.01) in CSJ010G than in CSJ009Y (Figure 6). These findings strongly suggested that LCYB2 is impaired in CSJ009 and plays a partial role in carotenoid synthesis in immature pepper fruits. In addition, no CCS gene expression was detected in orange CSJ010O; however, a small amount of capsaicin (0.087 μg g-1 FW) was still present. This may be attributed to the potential CCS activity of LCYB2 gene, similar to that observed in tomato (Ronen et al., 2000).
The induction of both LCYB and CCS in competing against LCYE involved for synthesizing carotenoids of the β,β-branch (Wang et al., 2019). Although the expression of the key carotenoid-biosynthesis genes CCS and LCYB2 was significantly impaired in CSJ010O and CSJ009R, respectively, there was no significant difference in the total carotenoid content between CSJ010O and CSJ009R (Figure 1F). Quantification analysis revealed that the major constituents of CSJ009R were carotenoids of the β,β-branch, whereas the products of the β,ϵ-branch were dominant in CSJ010O. Lutein was the most abundant carotenoid in immature fruits of both CSJ009 and CSJ010, which is consistent with previous studies indicating its prevalence in these stages (Gómez-García and Ochoa-Alejo, 2013). Our quantification demonstrates that the strong expression of CCS in CSJ009R resulted in the highest production of capsanthin (104.42 μg g-1 FW), while lutein levels significantly decreased to a negligible amount (0.65 μg g-1 FW). Conversely, CSJ010O contained lutein as the predominant carotenoid (244.88 μg g-1 FW). This is likely attributed to the direct catalytic effect of CCS on lycopene and its inhibitory effect on LCYE expression in mature pepper fruits (Wang et al., 2019). CCS has high sequence identity with LCYB and belongs to the lycopene cyclase family (Jeknić et al., 2012). In pepper, the enzyme CCS was found to possess the ability to catalyze lycopene cyclization to produce β-carotene (Hugueney et al., 1995). This process was proposed to compete against LCYE and repress the synthesis of carotenoids in the β,ϵ-branch (Wang et al., 2019). The null mutation of CCS leads to a reduced ability for repression of LCYE and the biosynthesis of capsanthin in the β,β-branch (Lefebvre et al., 1998; Li et al., 2013), thereby promoting the biosynthesis of α-carotene and lutein as products of thr β,ϵ-branch in CSJ010O.
Data availability statement
The datasets presented in this study can be found in online repositories. Transcriptome data of 12 samples (BioProject accession: PRJNA1095014) have been uploaded to the NCBI (https://www.ncbi.nlm.nih.gov/).
Author contributions
ZS: Writing – original draft. XX: Writing – review & editing. XC: Writing – review & editing. JJC: Writing – review & editing. JL: Writing – review & editing. JWC: Writing – review & editing. BZ: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was funded by the Natural Science Foundation of Guangdong Province (2023A1515012694), National Key Research and Development Program of China (Grant No. 2023YFD1201502), Guangzhou Basic and Applied Basic Research Project (202201010706), and the National Natural Science Foundation of China (32172564, U22A20497), Guangdong Provincial Department of Agriculture and Rural Affairs (2024-440000-87020100-8753) and Key-Area Research and Development Program of Guangdong Province (2022B0202080001).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1448060/full#supplementary-material
Abbreviations
CCA, canonical correlation analysis; COG, cluster of orthologous groups of proteins database (http://www.ncbi.nlm.nih.gov/COG); DACs, differentially accumulated carotenoids; DAFs, differentially accumulated flavonoids; DEGs, differential expression genes; DAMs, differentially accumulated metabolites; FC, fold change; FDR, false discovery rate; GO, gene ontology database (http://www.geneontology.org); KEGG, the database of kyoto encyclopedia of genes and genomes (http://www.genome.jp/kegg); OPLSDA, orthogonal projections to latent structures discriminant analysis; PCA, principal component analysis; Pfam, the database of homologous protein family (http://pfam.xfam.org); PIF1, phytochrome-interacting factors 1; TFs, transcription factors; MRM, multiple reaction monitoring; NR, NCBI protein database (ftp://ftp.ncbi.nih.gov/blast/db); Swiss-Prot, manually annotated database of non-redundant protein (http://www.uniprot.org); WGRS, whole-genome re-sequencing; VIP, variable importance in projection; qRT-PCR, quantitative Real-Time PCR.
References
Alder, A., Jamil, M., Marzorati, M., Bruno, M., Vermathen, M., Bigler, P., et al. (2012). The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335, 1348–1351. doi: 10.1126/science.1218094
Allen, G., Flores-Vergara, M., Krasynanski, S., Kumar, S., Thompson, W. (2006). A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 1, 2320–2325. doi: 10.1038/nprot.2006.384
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Aseel, D. G., Rashad, Y. M., Hammad, S. M. (2019). Arbuscular mycorrhizal fungi trigger transcriptional expression of flavonoid and chlorogenic acid biosynthetic pathways genes in tomato against tomato mosaic virus. Sci. Rep. 9, 9692. doi: 10.1038/s41598-019-46281-x
Benjamini, Y., Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Society: Ser. B (Methodological). 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Borovsky, Y., Monsonego, N., Mohan, V., Shabtai, S., Kamara, I., Faigenboim, A., et al. (2019). The zinc-finger transcription factor CcLOL1 controls chloroplast development and immature pepper fruit color in Capsicum chinense and its function is conserved in tomato. Plant J. 99, 41–55. doi: 10.1111/tpj.2019.99.issue-1
Borovsky, Y., Oren-Shamir, M., Ovadia, R., De Jong, W., Paran, I. (2004). The A locus that controls anthocyanin accumulation in pepper encodes a MYB transcription factor homologous to Anthocyanin2 of Petunia. Theor. Appl. Genet. 109, 23–29. doi: 10.1007/s00122-004-1625-9
Bouvier, F., Hugueney, P., d’Harlingue, A., Kuntz, M., Camara, B. (1994). Xanthophyll biosynthesis in chromoplasts: isolation and molecular cloning of an enzyme catalyzing the conversion of 5, 6-epoxycarotenoid into ketocarotenoid. Plant J. 6, 45–54. doi: 10.1046/j.1365-313X.1994.6010045.x
Brand, A., Borovsky, Y., Hill, T., Rahman, K. A., Bellalou, A., Van Deynze, A., et al. (2014). CaGLK2 regulates natural variation of chlorophyll content and fruit color in pepper fruit. Theor. Appl. Genet. 127, 2139–2148. doi: 10.1007/s00122-014-2367-y
Brand, A., Borovsky, Y., Meir, S., Rogachev, I., Aharoni, A., Paran, I. (2012). pc8.1, a major QTL for pigment content in pepper fruit, is associated with variation in plastid compartment size. Planta 235, 579–588. doi: 10.1007/s00425-011-1530-9
Camara, B., Dogbo, O. (1986). Demonstration and solubilization of lycopene cyclase from capsicum chromoplast membranes. Plant Physiol. 80, 172–174. doi: 10.1104/pp.80.1.172
Chen, W., Gong, L., Guo, Z., Wang, W., Zhang, H., Liu, X., et al. (2013). A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Mol. Plant 6, 1769–1780. doi: 10.1093/mp/sst080
Cunningham, F., Jr., Gantt, E. (1998). Genes and enzymes of carotenoid biosynthesis in plants. Annu. Rev. Plant Biol. 49, 557–583. doi: 10.1146/annurev.arplant.49.1.557
Daehwan, K., Ben, L., Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Deli, J., Matus, Z., Tóth, G. (1996). Carotenoid composition in the fruits of Capsicum annuum Cv. Szentesi Kosszarvú during ripening. J. Agric. Food Chem. 44, 711–716. doi: 10.1021/jf950354n
Diretto, G., Tavazza, R., Welsch, R., Pizzichini, D., Mourgues, F., Papacchioli, V., et al. (2006). Metabolic engineering of potato tuber carotenoids through tuber-specific silencing of lycopene epsilon cyclase. BMC Plant Biol. 6, 13. doi: 10.1186/1471-2229-6-13
Diretto, G., Welsch, R., Tavazza, R., Mourgues, F., Pizzichini, D., Beyer, P., et al. (2007). Silencing of beta-carotene hydroxylase increases total carotenoid and beta-carotene levels in potato tubers. BMC Plant Biol. 7, 11. doi: 10.1186/1471-2229-7-11
Egea, I., Bian, W., Barsan, C., Jauneau, A., Pech, J.-C., Latché, A., et al. (2011). Chloroplast to chromoplast transition in tomato fruit: spectral confocal microscopy analyses of carotenoids and chlorophylls in isolated plastids and time-lapse recording on intact live tissue. Ann. Botany. 108, 291–297. doi: 10.1093/aob/mcr140
Ferrer, J. L., Austin, M. B., Stewart, C., Noel, J. P. (2008). Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol. Biochem. 46, 356–370. doi: 10.1016/j.plaphy.2007.12.009
García-Limones, C., Schnäbele, K., Blanco-Portales, R., Luz Bellido, M., Caballero, J. L., Schwab, W., et al. (2008). Functional characterization of FaCCD1: a carotenoid cleavage dioxygenase from strawberry involved in lutein degradation during fruit ripening. J. Agric. Food Chem. 56, 9277–9285. doi: 10.1021/jf801096t
Gil, E. S., Couto, R. O. (2013). Flavonoid electrochemistry: a review on the electroanalytical applications. Rev. Bras. Farmacognosia. 23, 542–558. doi: 10.1590/S0102-695X2013005000031
Gómez-García, M., Ochoa-Alejo, N. (2013). Biochemistry and molecular biology of carotenoid biosynthesis in chili peppers (Capsicum spp.). Int. J. Mol. Sci. 14, 19025–19053. doi: 10.3390/ijms140919025
Guzman, I., Hamby, S., Romero, J., Bosland, P. W., O’Connell, M. A. (2010). Variability of carotenoid biosynthesis in orange colored Capsicum spp. Plant Sci. (Amsterdam Netherlands). 179, 49–59. doi: 10.1016/j.plantsci.2010.04.014
Ha, S. H., Kim, J. B., Park, J. S., Lee, S. W., Cho, K. J. (2007). A comparison of the carotenoid accumulation in Capsicum varieties that show different ripening colours: deletion of the capsanthin-capsorubin synthase gene is not a prerequisite for the formation of a yellow pepper. J. Exp. Botany. 58, 3135–3144. doi: 10.1093/jxb/erm132
Hoffmann, L., Besseau, S., Geoffroy, P., Ritzenthaler, C., Meyer, D., Lapierre, C., et al. (2004). Silencing of hydroxycinnamoyl-coenzyme A shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell. 16, 1446–1465. doi: 10.1105/tpc.020297
Huang, Y., Gou, J., Jia, Z., Yang, L., Sun, Y., Xiao, X., et al. (2012). Molecular cloning and characterization of two genes encoding dihydroflavonol-4-reductase from Populus trichocarpa. PloS One 7, e30364. doi: 10.1371/journal.pone.0030364
Hugueney, P., Badillo, A., Chen, H. C., Klein, A., Hirschberg, J., Camara, B., et al. (1995). Metabolism of cyclic carotenoids: a model for the alteration of this biosynthetic pathway in Capsicum annuum chromoplasts. Plant J. 8, 417–424. doi: 10.1046/j.1365-313X.1995.08030417.x
Hugueney, P., Bouvier, F., Badillo, A., Quennemet, J., d’Harlingue, A., Camara, B. (1996). Developmental and stress regulation of gene expression for plastid and cytosolic isoprenoid pathways in pepper fruits. Plant Physiol. 111, 619–626. doi: 10.1104/pp.111.2.619
Huh, H. J., Kang, C. B., Nahm, H. S., Kim, S., Ha, S. K., Lee, H. M., et al. (2001). A candidate gene approach identified phytoene synthase as the locus for mature fruit color in red pepper (Capsicum spp.). Theor. Appl. Genet. 102, 524–530. doi: 10.1007/s001220051677
Inbaraj, B. S., Lu, H., Hung, C. F., Wu, W. B., Lin, C. L., Chen, B. H. (2008). Determination of carotenoids and their esters in fruits of Lycium barbarum Linnaeus by HPLC–DAD–APCI–MS. J. Pharm. Biomed. Analysis. 47, 812–818. doi: 10.1016/j.jpba.2008.04.001
Jang, H., Choi, M., Jang, K.-S. (2024). Comprehensive phytochemical profiles and antioxidant activity of Korean local cultivars of red chili pepper (Capsicum annuum L.). Front. Plant Science. 15. doi: 10.3389/fpls.2024.1333035
Jeknić, Z., Morré, J. T., Jeknić, S., Jevremović, S., Subotić, A., Chen, T. H. (2012). Cloning and functional characterization of a gene for capsanthin-capsorubin synthase from tiger lily (Lilium lancifolium Thunb.’Splendens’). Plant Cell Physiol. 53, 1899–1912. doi: 10.1093/pcp/pcs128
Jeong, H.-B., Jang, S.-J., Kang, M.-Y., Kim, S., Kwon, J.-K., Kang, B.-C. (2020). Candidate gene analysis reveals that the fruit color locus C1 Corresponds to PRR2 in pepper (Capsicum frutescens). Front. Plant Science. 11, 1–14. doi: 10.3389/fpls.2020.00399
Jeong, H.-B., Kang, M.-Y., Jung, A., Han, K., Lee, J.-H., Jo, J., et al. (2019). Single-molecule real-time sequencing reveals diverse allelic variations in carotenoid biosynthetic genes in pepper (Capsicum spp.). Plant Biotechnol. J. 17, 1081–1093. doi: 10.1111/pbi.2019.17.issue-6
Kim, O. R., Cho, M.-C., Kim, B.-D., Huh, J. H. (2010). A splicing mutation in the gene encoding phytoene synthase causes orange coloration in Habanero pepper fruits. Molecules Cells 30, 569–574. doi: 10.1007/s10059-010-0154-4
Kormos, J., Kormos, K. (1960). The genetic types of the carotenoid system of the red-pepper fruit. Acta Botanica Academiae Scientiarum Hungaricae. 6, 305–319.
Lee, S. B., Kim, J. E., Kim, H. T., Lee, G. M., Kim, B. S., Lee, J. M. (2020). Genetic mapping of the c1 locus by GBS-based BSA-seq revealed Pseudo-Response Regulator 2 as a candidate gene controlling pepper fruit color. Theor. Appl. Genet. 133, 1897–1910. doi: 10.1007/s00122-020-03565-5
Lefebvre, V., Kuntz, M., Camara, B., Palloix, A. (1998). The capsanthin-capsorubin synthase gene: a candidate gene for the y locus controlling the red fruit colour in pepper. Plant Mol. Biol. 36, 785–789. doi: 10.1023/A:1005966313415
Li, B., Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 12, 323. doi: 10.1186/1471-2105-12-323
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Li, Z., Wang, S., Gui, X. L., Chang, X. B., Gong, Z. H. (2013). A further analysis of the relationship between yellow ripe-fruit color and the capsanthin-capsorubin synthase gene in pepper (Capsicum sp.) indicated a new mutant variant in C. annuum and a tandem repeat structure in promoter region. PloS One 8, e61996. doi: 10.1371/journal.pone.0061996
Lightbourn, G. J., Griesbach, R. J., Novotny, J. A., Clevidence, B. A., Rao, D. D., Stommel, J. R. (2008). Effects of anthocyanin and carotenoid combinations on foliage and immature fruit color of Capsicum annuum L. J. Heredity. 99, 105–111. doi: 10.1093/jhered/esm108
Liu, H., Lou, Q., Ma, J., Su, B., Gao, Z., Liu, Y. (2019). Cloning and functional characterization of dihydroflavonol 4-reductase gene involved in anthocyanidin biosynthesis of Grape Hyacinth. Int. J. Mol. Sci. 20, 4743. doi: 10.3390/ijms20194743
Liu, Y., Lv, J., Liu, Z., Wang, J., Yang, B., Chen, W., et al. (2020). Integrative analysis of metabolome and transcriptome reveals the mechanism of color formation in pepper fruit (Capsicum annuum L.). Food Chem. 306, 125629. doi: 10.1016/j.foodchem.2019.125629
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using Real-Time Quantitative PCR and the 2–ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Luo, P., Ning, G., Wang, Z., Shen, Y., Jin, H., Li, P., et al. (2016). Disequilibrium of flavonol synthase and dihydroflavonol-4-reductase expression associated tightly to white vs. red color flower formation in plants. Front. Plant Science. 6, 1–12. doi: 10.3389/fpls.2015.01257
Ma, C., Ma, B., He, J., Hao, Q., Lu, X., Wang, L. (2011). Regulation of carotenoid content in tomato by silencing of lycopene β/ϵ-cyclase genes. Plant Mol. Biol. Reporter. 29, 117–124. doi: 10.1007/s11105-010-0211-3
Manach, C., Scalbert, A., Morand, C., Rémésy, C., Jiménez, L. (2004). Polyphenols: food sources and bioavailability. Am. J. Clin. Nutr. 79, 727–747. doi: 10.1093/ajcn/79.5.727
Martens, S., Knott, J., Seitz, C. A., Janvari, L., Yu, S.-N., Forkmann, G. (2003). Impact of biochemical pre-studies on specific metabolic engineering strategies of flavonoid biosynthesis in plant tissues. Biochem. Eng. J. 14, 227–235. doi: 10.1016/S1369-703X(02)00224-3
Masuda, T., Fujita, Y. (2008). Regulation and evolution of chlorophyll metabolism. Photochemical Photobiological Sci. 7, 1131–1149. doi: 10.1039/b807210h
Matus, Z., Deli, J., Szabolcs, J. (1991). Carotenoid composition of yellow pepper during ripening: isolation of.beta.-cryptoxanthin 5,6-epoxide. J. Agric. Food Chem. 39, 1907–1914. doi: 10.1021/jf00011a003
Mullet, J. E. (1988). Chloroplast development and gene expression. Annu. Rev. Plant Physiol. Plant Mol. Biol. 39, 475–502. doi: 10.1146/annurev.pp.39.060188.002355
Nabavi, S. M., Šamec, D., Tomczyk, M., Milella, L., Russo, D., Habtemariam, S., et al. (2020). Flavonoid biosynthetic pathways in plants: Versatile targets for metabolic engineering. Biotechnol. Advances. 38, 107316. doi: 10.1016/j.biotechadv.2018.11.005
Nisar, N., Li, L., Lu, S., Nay, C., Barry, J. (2015). Carotenoid metabolism in plants. Mol. Plant 8, 68–82. doi: 10.1016/j.molp.2014.12.007
Nishino, H., Murakoshi, M., Tokuda, H., Satomi, Y. (2009). Cancer prevention by carotenoids. Arch. Biochem. Biophysics. 483, 165–168. doi: 10.1016/j.abb.2008.09.011
Ohmiya, A., Kishimoto, S., Aida, R., Yoshioka, S., Sumitomo, K. (2006). Carotenoid cleavage dioxygenase (CmCCD4a) contributes to white color formation in chrysanthemum petals. Plant Physiol. 142, 1193–1201. doi: 10.1104/pp.106.087130
Pan, Y., Bradley, G., Pyke, K., Ball, G., Lu, C., Fray, R., et al. (2013). Network inference analysis identifies an APRR2-like gene linked to pigment accumulation in tomato and pepper fruits. Plant Physiol. 161, 1476–1485. doi: 10.1104/pp.112.212654
Paran, I., van der Knaap, E. (2007). Genetic and molecular regulation of fruit and plant domestication traits in tomato and pepper. J. Exp. Botany. 58, 3841–3852. doi: 10.1093/jxb/erm257
Pascual, M. B., El-Azaz, J., de la Torre, F. N., Cañas, R. A., Avila, C., Cánovas, F. M. (2016). Biosynthesis and metabolic fate of phenylalanine in conifers. Front. Plant Science. 7, 1–13. doi: 10.3389/fpls.2016.01030
Pogson, B. J., Albrecht, V. (2011). Genetic dissection of chloroplast biogenesis and development: An overview. Plant Physiol. 155, 1545–1551. doi: 10.1104/pp.110.170365
Qin, C., Yu, C., Shen, Y., Fang, X., Chen, L., Min, J., et al. (2014). Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. 111, 5135–5140. doi: 10.1073/pnas.1400975111
Rodríguez-Bernaldo de Quirós, A., Costa, H. S. (2006). Analysis of carotenoids in vegetable and plasma samples: A review. J. Food Composition Analysis. 19, 97–111. doi: 10.1016/j.jfca.2005.04.004
Ronen, G., Carmel-Goren, L., Zamir, D., Hirschberg, J. (2000). An alternative pathway to β-carotene formation in plant chromoplasts discovered by map-based cloning of Beta and old-gold color mutations in tomato. Proc. Natl. Acad. Sci. 97, 11102–11107. doi: 10.1073/pnas.190177497
Sheen, S. J. (1973). Correlation between chlorophyll and chlorogenic acid content in tobacco leaves. Plant Physiol. 52, 422–426. doi: 10.1104/pp.52.5.422
Song, Z., Zhong, J., Dong, J., Hu, F., Zhang, B., Cheng, J., et al. (2022). Mapping immature fruit colour-related genes via bulked segregant analysis combined with whole-genome re-sequencing in pepper (Capsicum annuum). Plant Breeding. 141, 277–285. doi: 10.1111/pbr.v141.2
Sun, T., Yuan, H., Cao, H., Yazdani, M., Tadmor, Y., Li, L. (2018). Carotenoid metabolism in plants: the role of plastids. Mol. Plant 11, 58–74. doi: 10.1016/j.molp.2017.09.010
Thorup, T. A., Tanyolac, B., Livingstone, K. D., Popovsky, S., Paran, I., Jahn, M. (2000). Candidate gene analysis of organ pigmentation loci in the Solanaceae. Proc. Natl. Acad. Sci. 97, 11192–11197. doi: 10.1073/pnas.97.21.11192
Wahyuni, Y., Ballester, A.-R., Sudarmonowati, E., Bino, R. J., Bovy, A. G. (2011). Metabolite biodiversity in pepper (Capsicum) fruits of thirty-two diverse accessions: variation in health-related compounds and implications for breeding. Phytochemistry 72, 1358–1370. doi: 10.1016/j.phytochem.2011.03.016
Wan, H., Yuan, W., Ruan, M., Ye, Q., Wang, R., Li, Z., et al. (2011). Identification of reference genes for reverse transcription quantitative real-time PCR normalization in pepper (Capsicum annuum L.). Biochem. Biophys. Res. Commun. 416, 24–30. doi: 10.1016/j.bbrc.2011.10.105
Wang, D., Bosland, P. W. (2006). The genes of capsicum. HortScience 41, 1169–1187. doi: 10.21273/HORTSCI.41.5.1169
Wang, Q., Cao, T.-J., Zheng, H., Zhou, C.-F., Wang, Z., Wang, R., et al. (2019). Manipulation of carotenoid metabolic flux by lycopene cyclization in ripening red pepper (Capsicum annuum var. conoides) fruits. J. Agric. Food Chem. 67, 4300–4310. doi: 10.1021/acs.jafc.9b00756
Wang, M., Chen, L., Liang, Z., He, X., Liu, W., Jiang, B., et al. (2020). Metabolome and transcriptome analyses reveal chlorophyll and anthocyanin metabolism pathway associated with cucumber fruit skin color. BMC Plant Biol. 20, 1–13. doi: 10.1186/s12870-020-02597-9
Wu, L., Wang, H., Liu, S., Liu, M., Liu, J., Wang, Y., et al. (2022). Mapping of CaPP2C35 involved in the formation of light-green immature pepper (Capsicum annuum L.) fruits via GWAS and BSA. Theor. Appl. Genet. 135, 591–604. doi: 10.1007/s00122-021-03987-9
Yuan, H., Zhang, J., Nageswaran, D., Li, L. (2015). Carotenoid metabolism and regulation in horticultural crops. Horticulture Res. 2, 15036. doi: 10.1038/hortres.2015.36
Yun, Y.-H., Liang, F., Deng, B.-C., Lai, G.-B., Vicente Gonçalves, C. M., Lu, H.-M., et al. (2015). Informative metabolites identification by variable importance analysis based on random variable combination. Metabolomics 11, 1539–1551. doi: 10.1007/s11306-015-0803-x
Zhang, J., Cheng, X., Jin, Q., Su, X., Li, M., Yan, C., et al. (2017). Comparison of the transcriptomic analysis between two Chinese white pear (Pyrus bretschneideri Rehd.) genotypes of different stone cells contents. PloS One 12, e0187114. doi: 10.1371/journal.pone.0187114
Zhang, H., Zhang, S., Zhang, H., Chen, X., Liang, F., Qin, H., et al. (2020). Carotenoid metabolite and transcriptome dynamics underlying flower color in marigold (Tagetes erecta L.). Sci. Rep. 10, 16835. doi: 10.1038/s41598-020-73859-7
Keywords: Capsicum, fruit color, transcriptome, widely targeted metabolome, quantitative determination, whole-genome re-sequence
Citation: Song Z, Xu X, Chen X, Chang J, Li J, Cheng J and Zhang B (2024) Multi-omics analysis provides insights into the mechanism underlying fruit color formation in Capsicum. Front. Plant Sci. 15:1448060. doi: 10.3389/fpls.2024.1448060
Received: 12 June 2024; Accepted: 15 October 2024;
Published: 06 November 2024.
Edited by:
Rebecca Grumet, Michigan State University, United StatesReviewed by:
Renata Rivera-Madrid, Scientific Research Center of Yucatán (CICY), MexicoNeftali Ochoa-Alejo, Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional, Mexico
Copyright © 2024 Song, Xu, Chen, Chang, Li, Cheng and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhao Song, c29uZ3poYW9AZ2RhYXMuY24=; Jiaowen Cheng, amlhb3dlbmNoZW5nQHNjYXUuZWR1LmNu; Baige Zhang, emhhbmdiYWlnZUBnZGFhcy5jbg==
 Zhao Song
Zhao Song Xiaowan Xu1
Xiaowan Xu1