- 1Genomics and Molecular Breeding Lab, Global Center of Excellence on Millets (Shree Anna), ICAR-Indian Institute of Millets Research, Hyderabad, India
- 2Department of Biotechnology and Bioinformatics, Jaypee University of Information Technology, Waknaghat, Solan, India
Pearl millet is a nutri-cereal that is mostly grown in harsh environments, making it an ideal crop to study heat tolerance mechanisms at the molecular level. Despite having a better-inbuilt tolerance to high temperatures than other crops, heat stress negatively affects the crop, posing a threat to productivity gain. Hence, to understand the heat-responsive genes, the leaf and root samples of two contrasting pearl millet inbreds, EGTB 1034 (heat tolerant) and EGTB 1091 (heat sensitive), were subjected to heat-treated conditions and generated genome-wide transcriptomes. We discovered 13,464 differentially expressed genes (DEGs), of which 6932 were down-regulated and 6532 up-regulated in leaf and root tissues. The pairwise analysis of the tissue-based transcriptome data of the two genotypes demonstrated distinctive genotype and tissue-specific expression of genes. The root exhibited a higher number of DEGs compared to the leaf, emphasizing different adaptive strategies of pearl millet. A large number of genes encoding ROS scavenging enzymes, WRKY, NAC, enzymes involved in nutrient uptake, protein kinases, photosynthetic enzymes, and heat shock proteins (HSPs) and several transcription factors (TFs) involved in cross-talking of temperature stress responsive mechanisms were activated in the stress conditions. Ribosomal proteins emerged as pivotal hub genes, highly interactive with key genes expressed and involved in heat stress response. The synthesis of secondary metabolites and metabolic pathways of pearl millet were significantly enriched under heat stress. Comparative synteny analysis of HSPs and TFs in the foxtail millet genome demonstrated greater collinearity with pearl millet compared to proso millet, rice, sorghum, and maize. In this study, 1906 unannotated DEGs were identified, providing insight into novel participants in the molecular response to heat stress. The identified genes hold promise for expediting varietal development for heat tolerance in pearl millet and similar crops, fostering resilience and enhancing grain yield in heat-prone environments.
Introduction
Pearl millet [Pennisetum glaucum (L.R. Br)] belongs to the Poaceae family. It is a crop widely grown in the arid and semi-arid regions of Sub-Saharan Africa and the Indian subcontinent, where other cereals fail to achieve an economic yield (Sun et al., 2020). India is the world’s largest pearl millet producer, with a total cultivated area of 7.41 million hectares and a production output of 10.3 million tons in 2020-2021 (Indiastat, 2020). It is a hardy crop that can withstand the unpredictable effects of climate change. Climate change endangers agricultural production, raising serious worries about global food security. Temperature fluctuations are a significant component that significantly impacts crop growth and development. Climate models predict that the production of pearl millet in Sub-Saharan Africa will reduce from 17% to 7% by 2050 (Schlenker and David, 2010).
Heat stress is a major environmental threat that reduces crop productivity and results in yield reduction (Jagadish et al., 2021). In comparison to other crops, pearl millet has a high tolerance level to abiotic stresses such as heat, drought, salinity, and nutrient deficiency (Vadez et al., 2012), which allows it to produce higher yields under the same conditions and is critical in ensuring food and nutritional security in fluctuating climatic conditions. It is a climate-resilient crop that can survive high temperatures of up to 42°C; however, when exposed to temperatures exceeding 42°C, crop carbohydrate reserves are depleted, and plant starvation occurs (Djanaguiraman et al., 2009). Furthermore, it results in decreased growth due to a loss of cellular water content and an overall reduction in cell size. This crop struggles to survive under prolonged heat stress and suffers from adverse effects such as compromised cell membrane integrity, a significant drop in chlorophyll content, and a decrease in antioxidant enzymes, resulting in the accumulation of free radicals that cause cell damage and apoptosis (Hasanuzzaman et al., 2013). Therefore, it is imperative to develop heat-tolerant varieties that can endure changes caused by high temperatures in the production ecologies in which they are cultivated.
Studies have revealed that pearl millet has outperformed maize regarding morphological and physiological indices such as relative growth rate and net assimilation rate (NAR) at high temperatures (Ashraf and Hafeez, 2004). The molecular mechanism entails the activation of transcription factors (TFs), heat shock proteins (HSPs), metabolite synthesis, and other heat stress-related genes (He et al., 2023). The repository of genes in pearl millet distinguishes it from other crops. Mwadzingeni et al., 2016 described heat stress as a complicated process mediated by an intricate interplay of many genes and their regulated expression (Mwadzingeni et al., 2016). Transcriptome analysis has emerged as a valuable methodology to investigate gene expression and complex regulatory networks. Its application has been beneficial in unravelling the molecular mechanisms operative in crops when exposed to heat stress (Frey et al., 2015). Recent studies have effectively used transcriptome-based approaches in several crops to elucidate the molecular function of abiotic stress tolerance namely, maize (Qian et al., 2019), rice (Wang et al., 2019), wheat (Rangan et al., 2020), eggplant (Zhang et al., 2019) and pearl millet (Goud et al., 2022).
Pearl millet genotypes show a wide level of variation in heat tolerance when compared to other cereal crops. The present study was designed to mine the heat-stress-responsive genes from such genetic variation. Here, two pearl millet inbreds contrasting to heat tolerance behavior were used to discover DEGs through a genome-wide RNA-Seq approach. We discovered DEGs that encode important transcription factors, ion transporters, and metabolic pathway regulators from the leaf and root tissues. Our findings establish the groundwork for mining essential genes linked with heat tolerance in pearl millet and elucidating the molecular mechanisms operating in pearl millet in response to heat stress. It will also provide valuable insights into improving pearl millet productivity under heat-stress ecologies in the changing climate scenarios.
Materials and methods
Plant materials and treatment condition
The study used two contrasting pearl millet genotypes, namely EGTB 1034 (heat-tolerant) and EGTB 1091 (heat-sensitive), developed from the pearl millet breeding program at ICAR-IIMR, Hyderabad. Several genotypes were systematically evaluated under heat stress in both pre-and post-flowering stages, and the above-contrasting genotypes were selected based on phenotypic performances for further characterization through transcriptomes. The seeds of these genotypes were grown in cups under a photoperiod of 16 hours of light and 8 hours of darkness at room temperature with 90% relative humidity. Heat stress was induced on seven-day-old seedlings in a controlled growth chamber with a constant temperature of 45°C for 24 hours, whereas, for control samples, the same procedure was performed at room temperature 30°C. Immediately after 24 hours, leaf and root samples were collected with three biological replicates from both control and heat-treated conditions of HTG and HSG and used for RNA sequencing, independently.
RNA extraction and library preparation
Total RNA from the leaf and root of the three biological replicates of both control and treated genotypes was extracted using TRIzol reagent (RNAiso-plus) (ThermoFisher Scientific, United States), following the protocol of the manufacturer and further purified using MN Nucleospin RNA clean up kit (Macherey-Nagel, Germany). RNA quality and integrity were checked in 1% agarose (Lonza, Belgium) and Nanodrop 2000 (Thermofisher Scientific, Massachusetts, USA). Furthermore, the samples with RNA integrity number ≥7 were processed for analysis (Hosseini et al., 2021). The libraries were constructed using the KAPA HyperPrep Kit for the cDNA Synthesis & Amplification module, and length assessment was carried out using a bioanalyzer (Agilent Technologies, Santa Clara, California, USA). Then, the RNA-Seq libraries were sequenced on the Illumina sequencing platform (NovaSeq 6000) using a paired-end approach.
Transcriptome sequencing analysis
Following RNA-Seq, a quality assessment of the reads was performed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and adapter sequences and Illumina-specific sequences were removed using Trimmomatic (Version 0.36) (Bolger et al., 2014). The remaining cleaned reads were mapped to the reference genome of pearl millet (Table 1), obtained from the International Pearl Millet Genome Sequence Consortium (https://cegsb.icrisat.org/ipmgsc/genome.html) using the alignment tool Hisat2 ver. 2.0.4. Finally, the number of reads for each gene was counted using the featureCounts tool (version 2.0.0) (Liao et al., 2014).
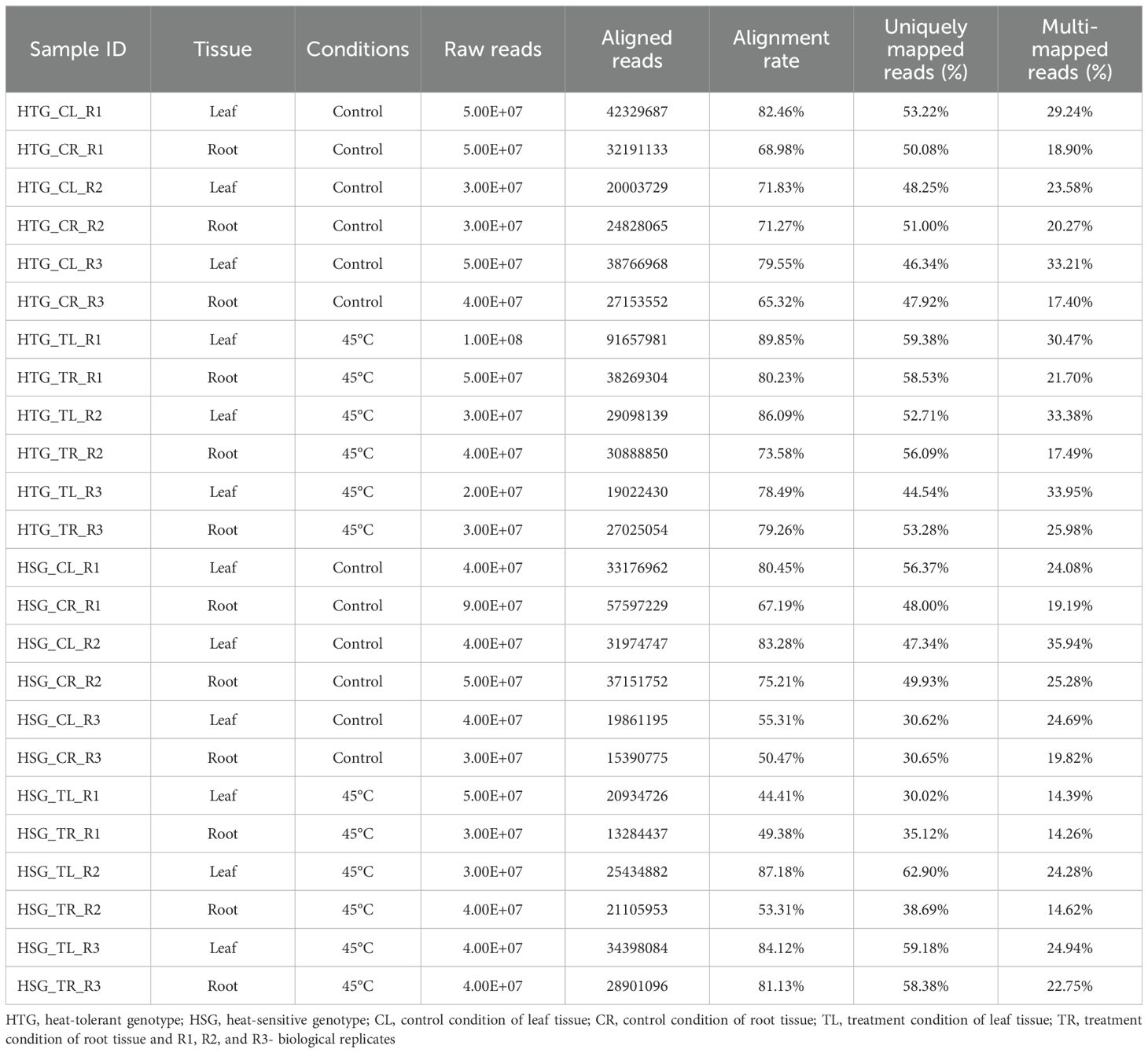
Table 1. Summary of RNA-Seq data sets acquired from 24 samples of heat-tolerant and sensitive genotypes under heat stress conditions mapped to pearl millet reference genome.
Identification of differentially expressed genes
DEGs were identified using the R bioconductor package edgeR version 3.42.4 (Robinson et al., 2010). TMM (Trimmed Mean of M-values) normalization method was applied to account for library size and composition differences across samples. The genes with the threshold of log2 fold change (FC) cutoff ≥2 and adjusted p-value ≤0.01 were selected as significant DEGs for further analyses and interpretations.
Functional annotation of DEGs
To identify putative biological functions and pathways for the DEGs, the Gene Ontology (GO) and Kyoto Encyclopedia Of Genes And Genomes (KEGG) databases were searched for annotation using the Database for Annotation, Visualization and Integrated Discovery (DAVID, version 6.8) (https://david.ncifcrf.gov/summary.jsp) and SRplot (https://www.bioinformatics.com.cn) (Huang et al., 2007). This analysis provided all GO terms significantly enriched in DEGs compared to the genome background and filtered the DEGs corresponding to the biological functions. Significant GO and KEGG pathways were identified with the criterion of FDR-corrected p-value <0.05 (Yoav and Hochberg, 2000). Hyper-geometric statistical tests and Bonferroni correction methods were also applied (Sidak, 1967).
Enrichment analysis of transcription factors
To identify enriched TF families functioning between genotypes under stressed and control conditions, a TF enrichment analysis was conducted. The identified DEGs were used as input and compared against the Setaria italica TF database from PlantRegMap (Tian et al., 2020). The DEGs were screened against 2,410 TFs, classified into 56 families, with a stringency p-value ≤0.01.
Identifying the hub genes involved in the protein interaction network
Protein-Protein Interactions (PPI) are crucial to most biological processes, and understanding them is imperative for unravelling the molecular mechanisms underlying DEGs in transcriptomics. The DEGs obtained under heat stress were utilized to construct a network of PPIs. The STRING (Search Tool for the Retrieval of Interacting Genes and Proteins) database was employed and the required interaction score for the physical sub-network was set to default parameters to identify both validated and predicted protein-protein interactions to investigate protein functional relationships (http://string-db.org) (Szklarczyk et al., 2023). The resulting interactions were utilized to construct the PPIs network, which was then analyzed and visualized using Cytoscape v3.10.0 (https://cytoscape.org/) (Shannon et al., 2003). The gene network was examined using average path length to determine key global centrality parameters such as proximity and betweenness, centrality, and average degree. The PPIs network was designed to identify significant players or hub genes (nodes with the highest degree) involved in heat stress tolerance. The hub genes were identified and ranked based on degree using the Cytoscape plugin cytoHubba (Chin et al., 2014). The degree algorithm calculates the number of direct interactions of each gene in the PPI network. Hub genes were recognized based on the higher number of connections or degrees over other genes.
Results
Genome-wide transcriptome data statistics
Genome-wide transcriptome profiling was conducted in leaf and root tissues of HTG and HSG under control and heat-treated conditions to examine the transcriptome regulation of tissue-specific genes in response to heat stress. We extracted a total of one billion raw reads using Illumina sequencing technology, of which 760 million reads, after rigorous quality testing and data cleaning, were mapped to the pearl millet reference genome from established 24 RNA-Seq cDNA libraries. Over 72.43% of the high-quality reads were effectively mapped to the pearl millet reference genome, of which 48% were uniquely mapped, whereas 23% illustrated multiple genomics locations (Table 1).
Gene expression profile analysis of HTG and HSG under heat stress
The expressed genes were generated and discovered using a stringent log2 FC ≥2 threshold and p-value <0.01 to comprehend their biological function better. Table 2 presents statistics of the DEGs between the twelve possible combinations. A thorough examination of tissue-specific comparisons between control and stress conditions in both genotypes produced 13,464 DEGs, of which 6,932 showed down-regulation and 6,532 showed up-regulation. In the leaf category, it was found that 1,665 genes were down-regulated, and 914 genes were up-regulated out of a total of 2,579 genes identified. Comparatively, 2,016 genes were induced in the root category, while 1,907 genes were suppressed among the 3,923 genes examined. The comparison between leaf and root tissues revealed that the most DEGs, with 3,360 showing lower expression and 3,602 displaying higher expression, out of 6,962 genes studied. In addition, tissue-specific evaluation of HTG and HSG revealed that 489 and 15 genes were commonly up-regulated and down-regulated, respectively (Figure 1).
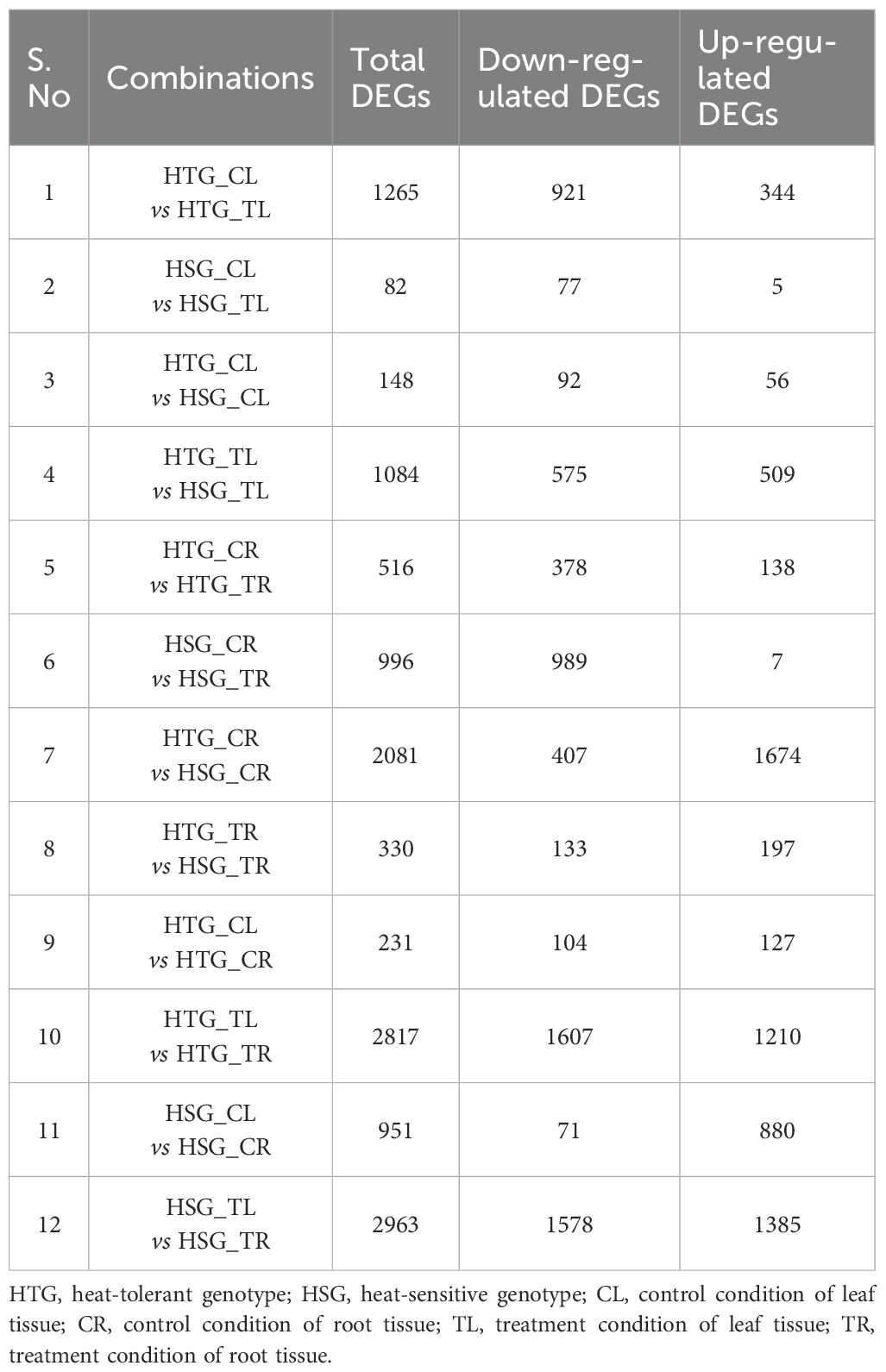
Table 2. Pairwise comparison of differentially expressed genes (DEGs) in heat-tolerant and heat-sensitive genotypes under different heat stress conditions.
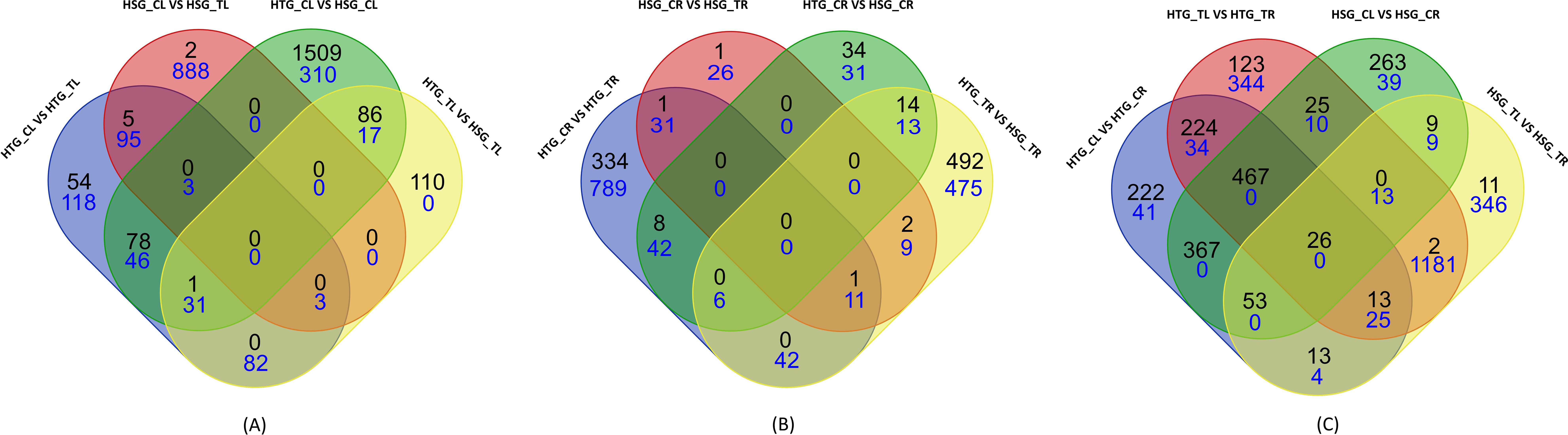
Figure 1. Up- (black) and down-regulated (blue) DEGs across comparisons in (A) leaf, (B) root, and (C) leaf vs root under heat stress treatments.
Identification of heat-responsive DEGs in leaf
The leaf transcriptome analysis identified 2,579 expressed transcripts, with 1,665 showing down-regulation and 914 showing up-regulation (Table 2). The Venn diagram (Figure 1) represents the total DEGs identified in the leaf tissues of both genotypes under control and treated conditions. Significant expression patterns were observed in various comparisons. For instance, in HTG_CL vs HSG_CL, 92 genes were down-regulated, and 56 genes were up-regulated. Similarly, in HTG_CL vs HTG_TL, we found 921 suppressed and 344 induced genes. Moreover, in HSG_CL vs HSG_TL, 77 genes showed lower expression while only five genes were over-expressed, and in HTG_TL vs HSG_TL, 575 genes were down-regulated while 509 were up-regulated.
The comparisons indicate a trend where genes responsible for various mechanisms and pathways in pearl millet leaves tend to experience suppression under stress conditions. The sensitive genotype showed a significant increase in down-regulated DEGs, suggesting its heightened vulnerability to heat stress. This implies that the HSG genotype is more sensitive to the negative impacts of high temperatures than the HTG genotype.
On comparing the transcript abundance profile among all four combinations, we identified that HATPase domain-containing protein, SHSP, protein kinases, lipoxygenase, chlorophyll a-b binding protein, and BHLH were significantly up-regulated. In contrast, stachyose synthase, BURP, lipase_3, lipase_GDSL domain-containing protein, cytochrome C and b559, BZIP, dirigent protein, and lipoxygenase showed significant down-regulation. Four of the top 10 highest up-regulated genes were expressed in HTG_TL vs HSG_TL, and of the top 10 down-regulated genes, HSG_CL vs HSG_TL included five highly-expressed genes. The significant up-regulation and down-regulation of expressed genes in the leaf are summarized in Table 2. Among all the genes expressed, lipase was expressed in all the combinations except HSG_CL vs HSG_TL, which was down-regulated (-11.5 FC) in the sensitive genotype. The comparison between tolerant and sensitive genotypes under control conditions revealed down-regulation of Receptor-like serine/threonine-protein kinase, AP2/ERF domain-containing protein, and ABC transporter. In contrast, there was a significant up-regulation, ranging from -11 to 14-fold, of Lipase_3 domain-containing protein, NAD(P)H dehydrogenase, and Laccase. Peroxidase, jmjC protein (chromatin remodeling and histone modification), WRKY, 24.1kDa HSP, LEA protein (prevents protein aggregation under stress), chitinase, elongation factor, NAC, and DNA helicase were induced several folds. On the other hand, NAD(P)H dehydrogenase, AAI domain-containing protein, bidirectional sugar transporter SWEET, FE2OG dioxygenase, phytocyanin, and ABC transporter genes were down-regulated across most of the combinations.
Chlorophyll a-b binding protein involved in photosynthetic activity was highly up-regulated in the tolerant genotype under treatment conditions. Peroxidase and thioredoxin enzymes belonging to the hydrogen peroxide catabolic process were more repressed in HSG. The lipoxygenase that plays a role in fatty acid biosynthesis and lipid oxidation was down-regulated across all combinations except HTG_TL vs HSG_TL, where it was five-fold induced. Genes involved in glutathione metabolic processes, such as glutathione synthetase and glutathione transferase, were mostly repressed in the treated sensitive genotype. Cytochrome b599 and photosystem I and II are cellular components of the chloroplast thylakoid membrane and were mostly down-regulated in the stressed HSG. In contrast, these genes displayed less suppression in the tolerant genotype than in the sensitive genotype. Extracellular region enzymes expansin, involved in cell wall organization, was up-regulated threefold in HTG_TL vs HSG_TL. In contrast, L-ascorbate oxidase was down-regulated threefold in HTG_CL vs HTG_TL. Bidirectional sugar transporter SWEET was down-regulated several folds in the HTG_TL vs HSG_TL and up-regulated in the HTG_CL vs HTG_TL. Amino acid permease involved in the transmembrane transporter activity of amino acid, alpha-amylase and PsbP domain-containing protein, which is part of PSII, was more down-regulated in the sensitive genotype than in the tolerant genotype under heat stress.
Identification of heat-responsive DEGs in root
Transcriptome analysis of all comparisons of root samples identified 3923 genes, of which 2016 and 1907 genes were induced and repressed, respectively (Figure 1). The Venn diagram represents commonly up-regulated and down-regulated genes under control and stress conditions in the roots of HTG and HSG. While analyzing the expression dynamics, on comparing HTG control with treatment, we found that 138 genes were up-regulated, and 378 genes were down-regulated. We observed more down-regulated genes (989) and fewer induced genes (Ashraf and Hafeez, 2004) in the control vs treatment of sensitive genotype. Comparing the control conditions of both genotypes, it was observed that 1674 genes were up-regulated while 407 genes were down-regulated. When comparing the treatment conditions of both genotypes, 197 genes showed over-expression, while 133 transcripts showed under-expression (Table 2).
In contrast to the leaf samples, the root tissues showed a higher number of up-regulated DEGs (2016). This up-regulation of genes in the roots under heat stress conditions suggests a different molecular response than the leaves. The HSG exhibited a notable number of down-regulated DEGs in the roots, indicating a suppression of gene expression, specifically in this genotype under heat stress.
From the present study, we identified DEGs encoding peroxidase, protein kinase and AAI domain-containing proteins that were significantly expressed across all combinations. Aldehyde oxygenase, involved in lipid biosynthesis, bidirectional sugar transporter SWEET, and Fe2OG dioxygenase domain-containing protein, was up-regulated in the HTG combinations, whereas in the sensitive genotype, these genes were down-regulated.
In the sensitive genotypes treatment condition, it was revealed that 17.9kDa HSP, chlorophyll a-b binding protein, PSII, RuBisCO, thioredoxin, malate dehydrogenase, potassium transporter, ABC transporter, copper transporter, glutamine synthetase, glutathione transferase, nitrate reductase, PsbP protein, Clp protease, temperature-induced lipocalin (TIL) that protects chloroplasts from ion toxicity, PSI, phosphate transporter, MAPK, MYB, RING-type E3 ubiquitin transferase and zinc finger proteins were down-regulated several folds. Enzymes lipase and lipoxygenase, laccase and stachyose synthase, and the fructose bisphosphate aldolase that participates in carbohydrate degradation in the glycolysis cycle were primarily down-regulated in the sensitive genotypes. Under control conditions, comparison between genotypes revealed suppressed expression of Glutathione S-transferase, Protein kinase, and Sucrose synthase. Conversely, there was up-regulation of terpene synthase, which is involved in the synthesis of secondary metabolites, as well as Phosphoenolpyruvate carboxykinase and Fructose-bisphosphate aldolase, both of which are involved in carbohydrate biosynthesis. In the treatment comparison of both the genotypes, stromal 70kDa HSP was down-regulated, in contrast to calcium-binding protein 60, which was induced two-fold. Xyloglucan endotransglucosylase/hydrolase was down-regulated across all the combinations, but the level of expression was found more suppressed (-6 folds) in the sensitive genotype in comparison to the tolerant. The results from the present study align with the comparative transcriptomic research conducted on Agrostis species, which revealed the activation of root antioxidant enzymes, genes involved in respiration, HSPs, and transcription factors aided tolerant genotype to adapt better to heat stress by maintaining growth and development (Huang, Bingru et al., 2012).
Comparison of heat-responsive DEGs in leaf vs root
Leaf and root stress treatments were compared to identify the tissue-specific expression of genes involved in transcriptional regulation of both pearl millet genotype’s responses to heat stress. The comparative transcriptomic analysis between leaf and root tissues revealed 6,962 expressed genes, with 3,602 showing up-regulation and 3,360 displaying down-regulation (Figure 1). Under control conditions, the comparison indicated 127 up-regulated genes and 104 down-regulated in the tolerant genotype, whereas 880 genes were induced and 71 genes were suppressed in the sensitive genotype. Under heat stress, HTG exhibited 1,210 up-regulated genes, while 1607 genes were down-regulated in the root. Meanwhile, 1385 genes were up-regulated, and 1,578 genes were down-regulated in the HSG (Table 2). The analysis of expression dynamics demonstrated distinct transcriptional responses to heat stress in the different tissues of both genotypes, highlighting their divergent molecular mechanisms in coping with environmental challenges.
AP2/ERF, auxin response factor, WRKY, NAC, lipoxygenase, lipase-GDSL, calmodulin-binding protein, PIP26, PIP11, respiratory burst oxidase, ring-type E3 ubiquitin transferase, sucrose synthase, terpene synthase, patatin, cytokinin dehydrogenase, bidirectional sugar transporter SWEET, and calcium uniporter showed induced expression across all the comparisons except in control condition of the tolerant genotype. In comparing leaf and root tissues under control conditions, the tolerant genotype showed enhanced expression of dirigent protein and Photosystem I P700. Conversely, in the sensitive genotype, protein kinase, peroxidase, and the bidirectional sugar transporter SWEET were significantly down-regulated. Some of the genes Burp, laccase, lipase, BZIP, potassium transporter, protein kinase, peroxidase, and glutathione transferase displayed distinct expression patterns across all combinations of HTG under control and stress conditions. Notably, FAD-binding protein, xyloglucan endotransglucosylase/hydrolase, and BHLH expression were positively regulated, indicating they were significantly more active in tolerant genotype’s roots.
Genes such as PEPC, chlorophyll a-b binding protein, cytochrome p450, fructose-bisphosphate aldolase, B-box zinc finger, ferredoxin–NADP reductase, PsbP, PSI, PSII, RNA helicase, Ring-CH-type domain-containing protein, MYB, sHsp17.0A, superoxide dismutase, stromal HSP70, thioredoxin, HSF protein, glutathione peroxidase, copper transporter, and CP12 domain-containing protein conversely demonstrated suppression of transcription and notable decrease in the expression levels. Furthermore, zeaxanthin epoxidase, zinc finger protein, RAP domain-containing protein, HATPase, GrpE protein, elongation factor, and catalase expression were down-regulated several folds in the root tissues.
Functional annotation and pathways enrichment of expressed genes
GO enrichment analysis was conducted to identify and describe putative DEGs between tolerant and sensitive genotypes under heat stress. A complete set of all DEGs was aligned against the DAVID GO database, resulting in the classification of annotated genes to three fundamental GO components: cellular component (CC) and molecular functions (MF) biological process (BP). Annotated genes included 452 for 19 CC, 331 for 22 MF, and 251 for 36 BP. The most substantially over-represented GO terms for the up-regulated genes were chloroplast thylakoid membrane, heme binding, and hydrogen peroxide catabolic process in the CC, MF, and BP categories, respectively. Similarly, for down-regulated genes, chloroplast, peroxidase activity, and hydrogen peroxide catabolic process were prevalent in each component.
The GO classification of expressed genes in different tissue-specific categories for both genotypes is represented in Supplementary Figure S1. In the leaf, out of 2552 genes analyzed, 1259 were sorted into three functional categories: 610 in BP, accounting for 48.5%, 644 in CC, making up 51.2%, and 870 in MF, representing 69.1%. For root tissues, the analysis revealed that out of 3885 genes studied, 1628 were classified as follows: 777 (47.7%) in BP, 858 (52.7%) in CC, and 1100 (67.6%) in MF. While comparing leaf versus root categories with a total of 6879 genes, 2569 were distributed across three classes: 1218 (47.4%) in BP, 1361 (53%) in CC, and 1777 (69.2%) in MF.
In our study, KEGG analysis was used to assign biological pathways to the identified expressed genes. The comparative study of the tolerant and sensitive genotypes under both control and stressed conditions suggested a significant enrichment of genes that regulate metabolic and biosynthesis of secondary metabolites pathways. Under control conditions, the protein modification pathway in leaves and the carotenoid biosynthesis pathway in roots were enriched in the tolerant compared to the sensitive genotype. Under stressed conditions, tissue-specific differences in pathway enrichment were observed. In leaf tissues of the tolerant genotype, genes associated with photosynthesis, cysteine and methionine metabolism, and phenylpropanoid biosynthesis pathways were significantly enriched, whereas, in roots, the isoquinoline alkaloid biosynthesis pathway was enriched. When comparing control and stressed conditions within the tolerant genotype, genes involved in photosynthesis, linoleic acid metabolism, and carotenoid biosynthesis pathways were expressed in leaf tissues, while in roots, genes involved in the biosynthesis of amino acids and starch and sucrose metabolism pathways were expressed (Figure 2). The pathway enrichment analysis revealed that 1,172 of the expressed genes in the tolerant genotype were associated with 17 significant pathways. Additionally, metabolic pathways, biosynthesis of secondary metabolites, and photosynthetic pathways were highly enriched in both overexpressed and underexpressed genes (Figure 2). These findings provide valuable insights into the specific functions, processes, and pathways associated with the heat stress response in pearl millet.
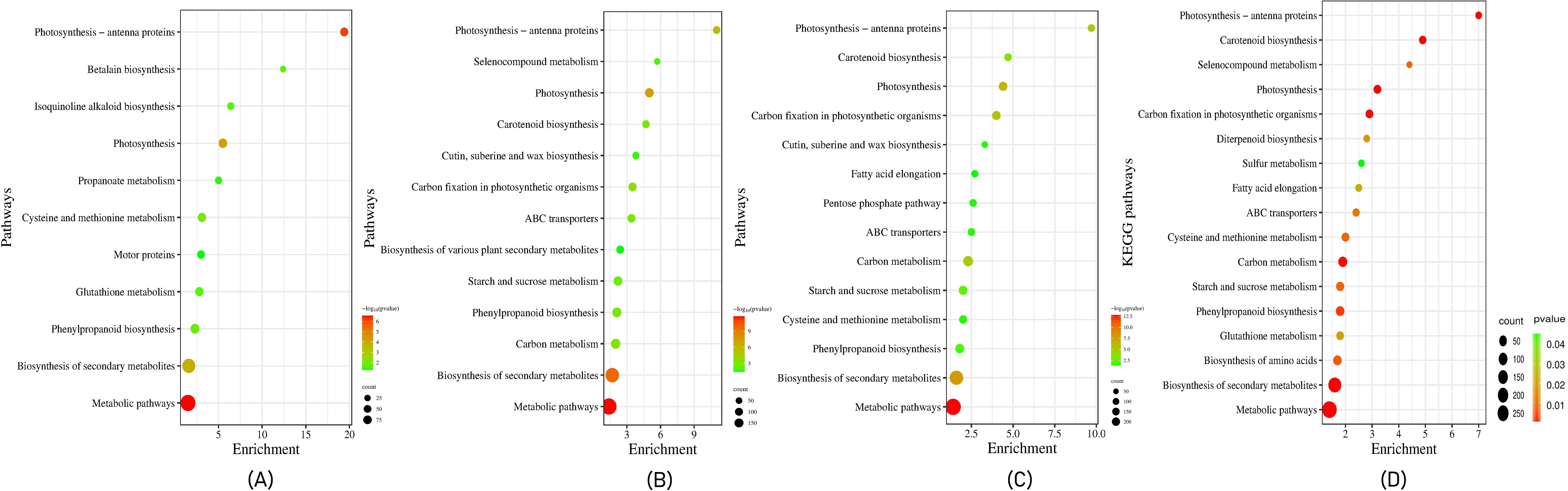
Figure 2. Biological pathways enrichment of the DEGs in (A) the leaves and roots of tolerant vs sensitive genotypes under heat stress conditions. (B) tolerant genotype leaf vs root (C) sensitive genotypes leaf vs root and (D) top 17 biological pathways enrichment of the DEGs in the leaves and roots of pearl millet genotypes under heat stress conditions. The size of the dot represents the number of the DEGs involved in each pathway. The color of the dot represents the p-value of each pathway; the pathways with p value ≤ 0.05 were significantly enriched pathways.
We successfully annotated a subset of the DEGs, revealing valuable information about their molecular functions, biological processes, and cellular components. The analysis also highlighted the presence of significant number of 1906 uncharacterized proteins, indicating potential novel genes associated with the heat stress response in pearl millet. A considerable proportion of the 60 transcripts remained unannotated, highlighting the need for future investigations to unravel their roles and functions. The identification of a substantial number of DEGs across various comparisons underscores the significant impact of heat stress on gene expression. It highlights the intricate molecular mechanisms underlying the plant’s response to heat stress.
Enrichment analysis of TFs under heat stress
To better understand the molecular mechanisms underlying the response to heat stress in pearl millet, TF enrichment analysis was performed on DEGs from HTG and HSG under both control and stressed conditions. This analysis revealed several TF families with significant enrichment, suggesting their potential regulatory roles under heat stress. The top five enriched TFs and their associated GO terms from each comparison category are summarized in Table 3. The comparisons included HTG vs HSG under both control and stressed conditions, as well as HTG and HSG stressed vs control conditions.

Table 3. Enriched transcription factors in tolerant and sensitive genotypes under control and stress conditions.
The significantly enriched TF families included bZIP, MYB, bHLH, G2-like, and NAC. The bZIP family was prominently enriched across all comparisons. In the tolerant vs sensitive genotype under stress, the bZIP family had 48 target genes, with GO terms indicating its involvement in responses to abiotic stresses. Additionally, the G2-like family, with 39 target genes, along with MYB and NAC families, with 15 and 25 target genes respectively, were significantly enriched and predominantly found in the tolerant genotype under heat stress. These TFs are involved in various biological processes and molecular functions related to transcription regulation, abiotic stress responses, and developmental processes. These findings suggest the crucial role of these TF families in gene regulation under heat stress in pearl millet, offering valuable targets for further functional studies to elucidate their roles in stress response and plant development.
Network analysis of core heat stress-responsive genes: hub genes, functional enrichment, and molecular insights
The PPI network of core heat stress-responsive genes comprised 260 nodes and 3336 edges, forming two distinct clusters− one large and one small. The genes in the larger clusters were most significantly enriched in functions related to ribosomes, metabolic pathways, carbon metabolism and biosynthesis of secondary metabolites. The genes in smaller clusters were predominantly enriched in starch and sucrose metabolism along with metabolic pathways.
Key hub genes exhibiting high connectivity (degree >70) included KOW domain-containing proteins, various ribosomal proteins (such as uS12, bL36, and bL17), S5 DRBM domain-containing proteins, elongation factor Tu (EF Tu), and several uncharacterized proteins. These hub genes interacted with other crucial proteins, such as BAG domain-containing protein, Superoxide dismutase, PsbP domain-containing protein, HATPase_c, WRKY, ERF, Fes1 domain-containing protein, Pyruvate kinase, Acyl carrier protein, chlorophyll a-b binding protein, Thioredoxin domain-containing protein, and EF Ts.
In this study, one notable hub gene, namely the KOW domain-containing protein, is a nuclear RNA binding protein essential for plants’ innate immunity against various biotic and abiotic stresses (Aksaas et al., 2011). We observed 39 genes related to ribosomal proteins, including S5 DRBM domain-containing protein, exhibited high connectivity ranging from 32 to 91 degrees (Figure 3). The results indicated that heat stress caused detrimental effects on the expression of ribosomal proteins of the large subunit genes due to their decreased stability. Ribosomal proteins are involved in the selective synthesis of important proteins in response to heat stress. These ribosomal proteins modulate protein accumulation under stress conditions and their regulation reduces energy consumption.
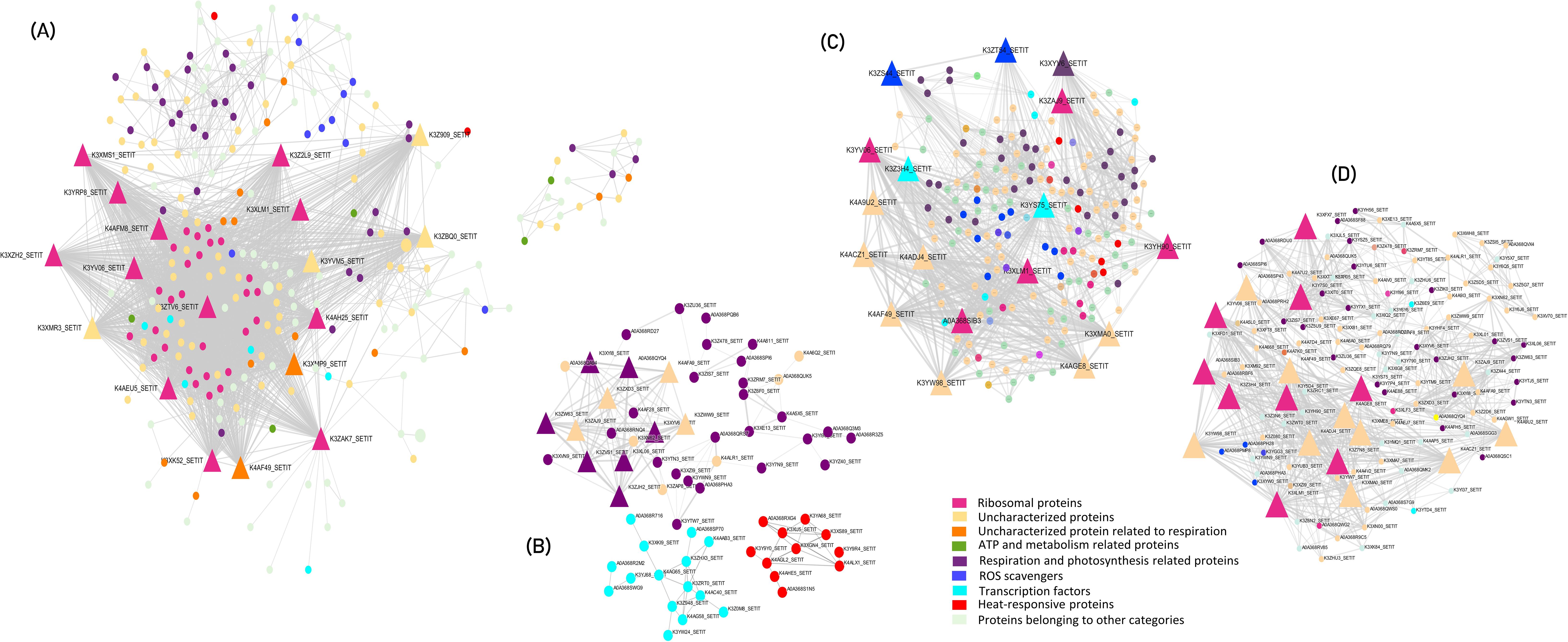
Figure 3. Protein-Protein Interaction network of (A) overall genes expressed in response to heat stress (B) DEGs in leaf and root of tolerant vs sensitive genotype (C) DEGs in the leaf vs root of tolerant genotype and (D) DEGs in the leaf vs root of sensitive genotype. Triangular nodes represent the hub genes identified and circular nodes represent the crucial genes involved in heat stress responsive mechanisms.
Another significant hub gene, EF Tu with a degree of 71, is a highly conserved GTP-binding protein essential for translation in many species, including prokaryotes and eukaryotes (Xifeng et al., 2018). Studies in spring wheat showed its accumulation in response to heat stress, with higher levels correlating with improved heat tolerance (Bukovnik et al., 2009). Notably, the recombinant maize pre-EF-Tu was stable at 45°C and acted as a molecular chaperone, reserving protein stability under heat stress by preventing thermal protein aggregation (Rao et al., 2004).
Genes associated with HATPase, photosynthesis, carbon metabolism and TFs including WRKY and ERF interacted with the hub genes. BAG domain-containg protein, recruited molecular chaperones using their domains under stress conditions to target proteins and changed their function by altering the protein conformation (Ding et al., 2020). BAG proteins regulate various physiological processes such as apoptosis, tumor induction, stress response and cell cycle. BAGs also regulate HSP chaperone proteins (positively or negatively) and form complexes with various transcription factors. At the transcriptional level, BAG family genes in plants have key roles in the PCD processes which range from growth, and tolerance to fungi to abiotic stress tolerance.
Comparative analysis of tolerant vs sensitive genotypes under heat stress conditions revealed a primary cluster and two sub-clusters, where hub genes in the primary cluster (degree >10) were predominantly associated with respiration and photosynthetic pathways. The two sub-clusters were enriched for transcription factors, such as NAC, and stress-responsive proteins, including small heat shock proteins (sHSPs). In the leaf vs root comparison of the tolerant genotype, hub genes identified were elongation factor Tu (EF Tu) with a degree of 53 and several uncharacterized ribosomal proteins. Conversely, in the sensitive genotype, hub genes comprised S5 DRBM domain-containing proteins, EF Tu, and other uncharacterized proteins, with degrees ranging from 20 to 42 (Figure 3). PPI network highlighted the intricate interplay among various proteins involved in the heat-stress response and explained potential mechanisms underlying heat tolerance in pearl millet.
Unveiling the evolutionary significance of heat shock proteins and transcription factors across related crop species under heat stress
Our transcriptome results highlighted the crucial involvement of HSPs and TFs in the heat stress response of pearl millet. Therefore, 15 HSP and 179 TF-related genes identified in pearl millet were searched against rice, maize, proso millet, sorghum and foxtail millet genomes using BLAST for the identification of orthologous genes. Foxtail millet showed the maximum gene homology, sharing 11 HSP-related genes with pearl millet, followed by proso millet, rice, maize, and sorghum (Chandel et al., 2013; Nagaraju et al., 2015; Singh et al., 2016; Li and Howell, 2021; Barthakur and Bharadwaj, 2022). TF-related genes also showed maximum homology, with foxtail millet sharing 137 genes, followed by proso millet, sorghum, rice and maize (Chandel et al., 2013; Nagaraju et al., 2015; Singh et al., 2016; Li and Howell, 2021; Barthakur and Bharadwaj, 2022). Annotations were assigned to the identified orthologues to understand their functionality in the respective species.
Our analysis revealed that several genes had more than one orthologous sequence across different crops. Specifically, in proso millet, out of the total of 132 identified genes associated with TF, 109 genes had more than one ortholog sequence. Similarly, out of the 10 identified genes related to HSP, 7 genes had more than one ortholog sequence. (Figure 4). All crops except sorghum had 2 genes associated with HSP, while sorghum had only one gene with more than one ortholog. Rice, sorghum, foxtail millet, and maize had 48, 43, 45, and 58 TFs, respectively, had more than one ortholog sequence.
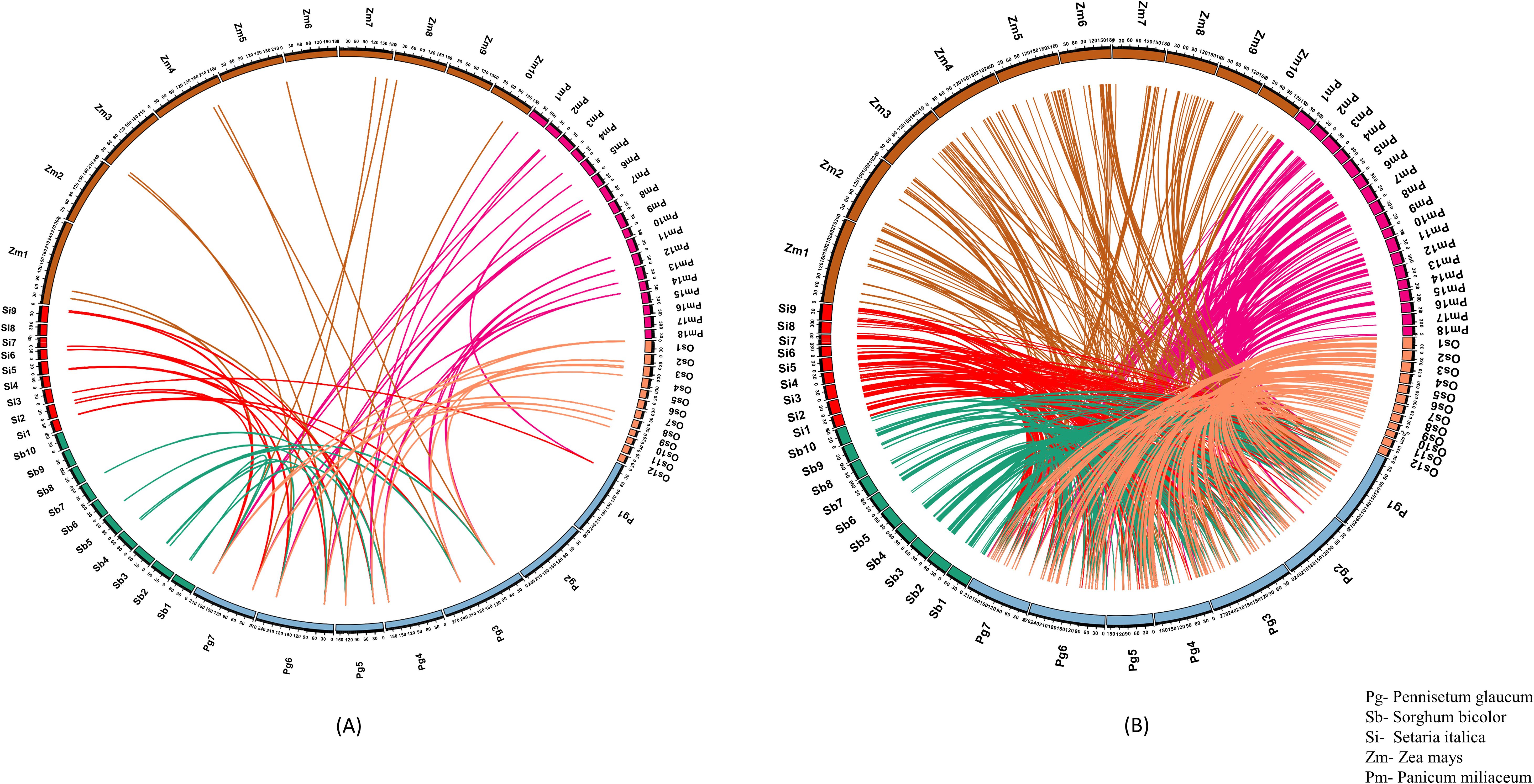
Figure 4. Comparative synteny plot demonstrating the orthologous genes related to (A) HSPs and (B) TFs in pearl millet to rice, maize, proso millet, sorghum and foxtail millet. The numeric values in the plot represent the chromosome number.
Foxtail millet showed the highest similarity to HSPs and TFs by capturing 13 and 184 ortholog sequences from 15 HSPs and 179 TFs, respectively (Figure 4). In proso millet, 18 ortholog sequences were identified for 10 HSPs. Out of these, 13 sequences showed more than 85% similarity and three sequences showed more than 70% similarity to pearl millet HSPs. Similarly, 285 ortholog sequences were identified for 132 TFs, of which 91 sequences showed more than 85% similarity and 81 sequences showed more than 70% similarity to pearl millet TFs.
In the study, it was found that Sorghum had nine ortholog sequences for eight HSPs. Out of these, six sequences showed more than 85% similarity and three sequences showed more than 70% similarity. On the other hand, 172 ortholog sequences were identified for TFs, of which 32 sequences showed more than 85% similarity and 44 sequences showed more than 70% similarity (Figure 4). The comparison between pearl millet and maize resulted in the identification of 11 ortholog sequences, which were mapped to nine genes related to HSP. Additionally, 205 ortholog sequences were detected for 125 genes related to TF. Our research explained the conservation and diversity of HSPs and TFs engaged in the response to heat stress among different crop species.
Discussion
Our results provide an understanding of the transcriptional responses of pearl millet to heat stress in leaf and root tissues. We discovered a total of 13,464 DEGs across all comparisons using comprehensive analysis, revealing the considerable influence of heat stress on gene expression patterns in different tissues of pearl millet. Figure 5 illustrates the expression pattern of DEGs across all the pairwise combinations of HTG and HSG.
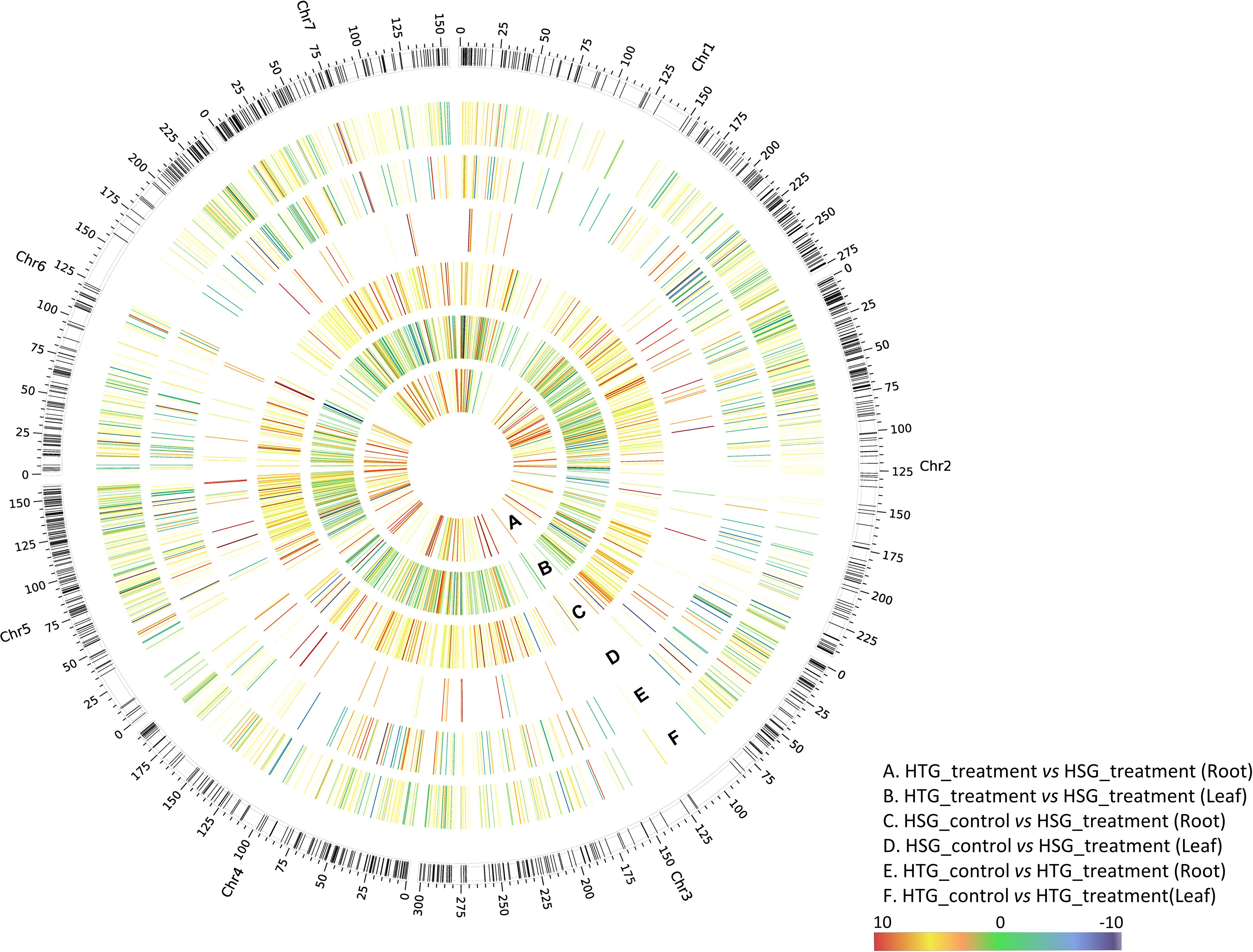
Figure 5. Circos plot represents differential expression pattern of heat-responsive genes in the HTG and HSG under control and treatment conditions. The outermost ring shows pearl millet chromosomes. The six rings namely, (A–F), explain the expression pattern of genes across different pairwise comparisons of HTG and HSG.
Uniquely expressed genes and their regulation associated with photosynthesis and CO2 assimilation
Photosynthesis, photochemical reactions, chlorophyll biosynthesis, NADPH and ATP synthesis, and respiration are all vital physiological processes in plants that help them adapt to heat stress. However, photosynthesis becomes susceptible at high temperatures, with its components, particularly PS II, being extremely sensitive (Ashraf and Harris, 2013). This susceptibility reduces photosynthetic efficiency, limiting plant development. Our study discovered 347 genes associated with respiration and photosynthesis pathways, including 90 uncharacterized proteins. Heat stress drastically reduced the expression of genes involved in CO2 assimilation and photosynthesis in the sensitive than tolerant genotype (Figure 5).
Several vital genes involved in electron transport, chlorophyll production, and carbohydrate metabolism, including chlorophyll-binding a/b proteins, PS I and PS II components, PsbP domain-containing protein, 2-hydroxy-acid oxidase, cytochrome P450, phosphoenolpyruvate carboxykinase (PEPC), phosphoglucomutase, stachyose synthase, phosphoribulokinase (PRK), malate dehydrogenase, phosphoglycerate kinase and ferredoxins, were significantly more down-regulated under heat stress in the sensitive over the tolerant genotype (Table 4). Xu and Huang also reported that in response to drought, heat and combined stress, chlorophyll-binding proteins were down-regulated in both the tolerant and sensitive Kentucky bluegrass genotypes, but the level of expression in the tolerant was less suppressed than the sensitive genotype (Xu and Bingru, 2012).
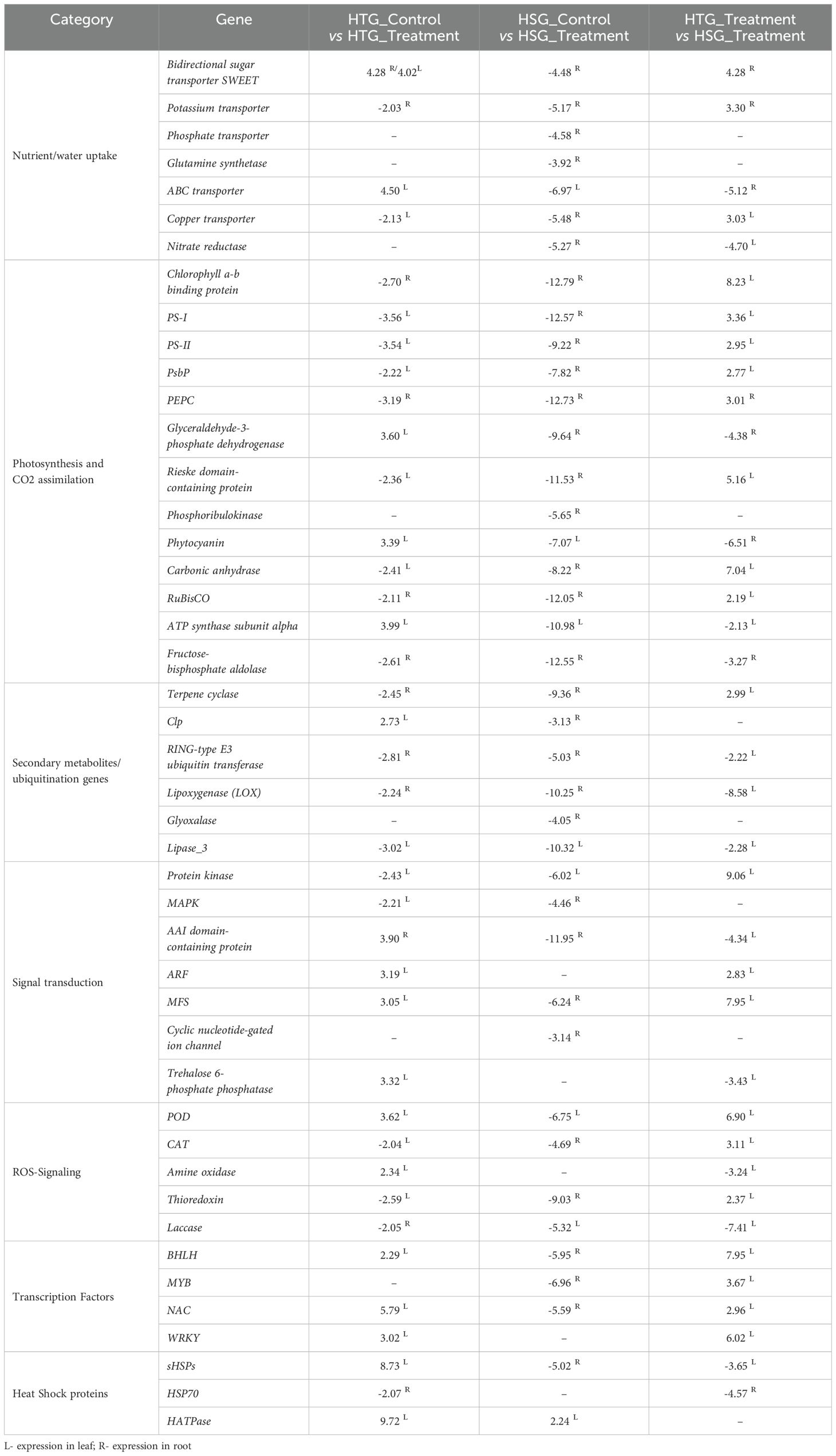
Table 4. Expression pattern of selected differentially expressed genes operating in important functional pathways under heat stress.
Genes encoding phytocyanins are pivotal in facilitating electron transfer exhibited induced expression in treated heat-tolerant genotypes. This upregulation suggests a potential role for phytocyanins in aiding stress adaptation mechanisms. The Rieske protein of cytochrome b6/f complex is a component of the photosynthetic electron transport chain in the chloroplast and was induced in the leaves of Pinellia ternate under heat stress (Zhu et al., 2013). The leaves of HTG recorded Rieske protein down-regulation to 2 folds. In contrast, the HSG was suppressed to -11 folds, indicating that the tolerant genotype under stress conditions adapts by conserving energy to cope with the adverse effects.
Under heat stress conditions, a significant decrease in gene expression related to photosynthesis and respiration pathways was evident. This decline was more pronounced in the sensitive genotype compared to the tolerant genotype (Table 4). This disparity suggests a higher susceptibility of sensitive genotypes to heat stress, resulting in more pronounced transcriptional suppression and a consequent reduction in respiratory processes. Heat affects the production of ATP and NADPH from the light reactions, and these, in turn, significantly affect the photosynthetic enzymes such as RuBisCO, carbonic anhydrase and PRK. ATP synthase α subunit expression was over-expressed in HTG and suppressed in HSG, which aligns with a study on T. aestivum, where ATP synthase α subunit activity was reduced in heat-sensitive and increased in the tolerant genotype (Wang et al., 2015). Carbonic anhydrase in G. max and Agrostis species was increased in the tolerant species than the sensitive ones under heat stress, suggesting it should play an important role in imparting tolerance (Xu and Huang, 2010; Das et al., 2016). Most of the genes involved in the photosynthesis process and CO2 assimilation under stress showed high connectivity with the significant hub genes (Figure 3).
The enzymes that play a key role in carbon flux in the Calvin cycle and in determining carbon assimilation, such as PRK, glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and fructose-bisphosphate aldolase (FBPA), were more suppressed in sensitive genotype than HTG (Figure 6). Previous studies have demonstrated the same results: PRK in rice, A. stolonifera and A. scabra, GAPDH in A. stolonifera, and A. scabra and FBPA in G. max and M. sinensis were suppressed under heat stress. Most of the genes involved in glycolysis and in the TCA cycle were down-regulated under heat stress in HSG, resulting in reduced respiratory electron transfer and oxidative phosphorylation in the sensitive than in the tolerant genotype. Similarly, genes involved in the Calvin cycle were also suppressed in the sensitive genotype. The tolerant genotype displayed less suppression of the above-mentioned genes in their leaves under heat stress conditions, indicating that respiratory carbon metabolism is significantly less inhibited under stress. This differential gene expression pattern favors the tolerant genotype, potentially aiding their adaptation to heat stress.
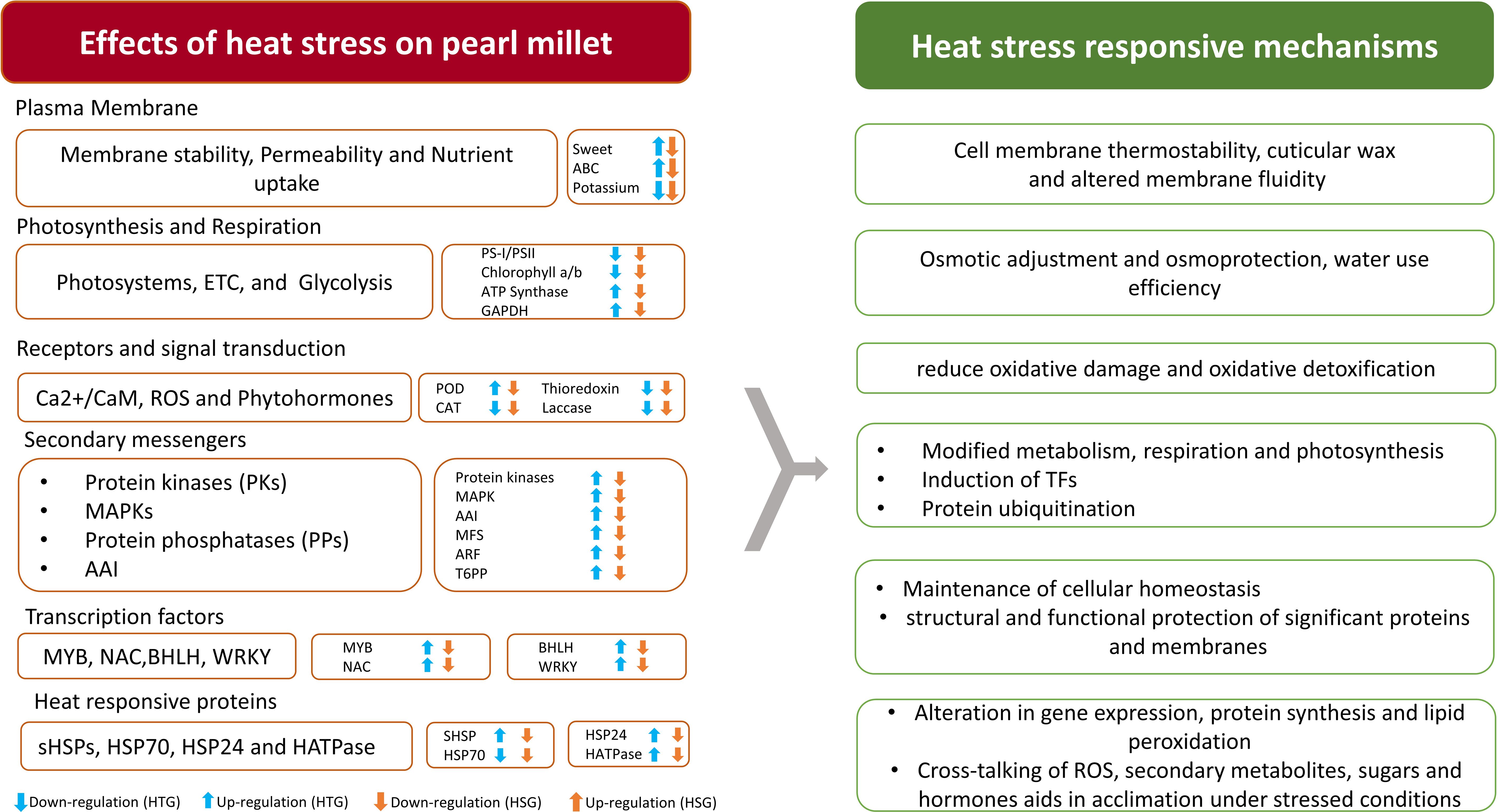
Figure 6. Elucidating the important genes operating in major pathways leading to heat stress tolerance in pearl millet.
This observation aligns with previous findings (Kurimoto et al., 2004) and suggests that the genes responsible for glycolysis, the Calvin cycle, and the TCA cycle exhibit a more resilient expression profile in the leaves of the tolerant genotype, contributing to their better adaptation to heat stress by conserving carbon resources. Reduction in respiratory and photosynthetic activity is correlated to a decrease in grain yield. At the cellular level, heat stress generates ROS that disrupt chloroplast membranes and the plasma membrane, leading to photosystem deactivation, reduced photosynthesis, and RuBisCO inactivation (Takahashi et al., 2007). This hampers the production and allocation of photo-assimilates, affecting grain’s anthesis, filling, size, number, and maturity, ultimately reducing crop productivity. Genes associated with chloroplast, photosynthesis, photosystem II, plant-type cell wall, and chloroplast thylakoid membrane were considerably enriched based on GO enrichment.
Investigating the impacts of heat stress on nutrient and water uptake genes
Plant growth and development depend on adequate nutrient and water availability, primarily controlled by roots via mineral cycling and phytohormone signaling (Freschet et al., 2018; Kong et al., 2012). Heat stress dramatically impairs the synthesis of proteins involved in the uptake and transportation of nutrients. Our research discovered critical gene expressions that regulate amino acids, carbohydrates, and vital micronutrients (e.g., zinc, potassium, magnesium, boron) and macronutrients (e.g., phosphate, copper) transport across different parts of a plant (Fahad et al., 2017; Zelazny and Vert, 2014).
The bidirectional sugar transporter SWEET promotes carbohydrate transport across membranes, influencing plant resistance to osmotic stress (Darko et al., 2019). Klemens (2013) demonstrated that over-expression of AtSWEET16 in Arabidopsis improved osmotic and cold tolerance (Klemens et al., 2013). The SWEET gene showed up-regulation in the leaf and root tissues of HTG. It is involved in the regulation of abiotic stresses such as drought and heat in different crops (Yuan and Wang, 2013). Sensitive genotype roots displayed diminished nutrient absorption under heat stress, associated with reduced expression of phosphate transporter, bidirectional sugar transporter SWEET, and copper transporters in contrast to their up-regulation under control conditions (Figure 6). Similarly, in the roots of HSG, potassium transporters and enzymes involved in nutritional assimilation, such as nitrate reductase and glutamine synthetase, exhibited suppressed expression under stress conditions (Table 4). Potassium fluxes are essentially required to regulate the transpiration process, so the down-regulation of potassium transporter may affect the plant’s response to heat and oxidative stress (Mulet et al., 2023). Furthermore, Rieske domain-containing proteins (-11.5 FC) involved in metal ion binding and nitrate assimilation were significantly down-regulated, particularly in the heat-sensitive genotype (Molik et al., 2001).
The ABC transporter domain-containing protein, an integral membrane component involved in ATPase-coupled transmembrane transporter activity and ATP binding (Dahuja et al., 2021), was repressed in the roots of the heat-treated sensitive genotype and induced in the leaf of HTG. ABC transporters are involved in the regulation of various metabolisms, growth, development and environmental responses. ABC transporters in model plants such as Arabidopsis and rice have been identified to resist biotic and abiotic stress (Moon and Jung, 2014). The down-regulation of genes associated with nutrient uptake, primarily observed in the HSG combination, led to decreased nutrient uptake, resulting in reduced root and shoot biomass of sensitive than in tolerant genotype.
Aquaporin PIP2, a membrane channel protein in the roots of the tolerant genotype, contributed to ionic balance maintenance, which is essential for water and solute transport, and showed enhanced expression, assisting heat tolerance by regulating water balance (Maurel et al., 2008; Giri et al., 2017). Previous studies suggested that PIPs have a role in modulating plant root’s water intake and function in plant heat tolerance (Vandeleur et al., 2005). Obaid et al. (2016) discovered that over-expression of three AQP genes increased water utilization and induced heat tolerance in Rhazya stricta (Obaid et al., 2016). Efforts have been made over the last decade to understand the function of PIPs, and a few studies have demonstrated that PIP gene over-expression is favorable in imparting tolerance under heat-stress conditions. The GO study discovered novel genes involved in water channel function and cellular response to water scarcity, revealing the active engagement of diverse cellular components in nutrition and water intakes, such as chloroplast structures and sugar transporters. Kegg pathway analysis revealed that 39 genes participated significantly in carbon metabolism, aiding in combating heat stress by providing the energy required for the maintenance of metabolic and cellular responses.
Genotype-specific responses of secondary metabolites biosynthesis and protein ubiquitination pathways
Our findings revealed unique gene expression patterns associated with 19 pathways corresponding to tolerant and sensitive genotypes, spanning critical biological activities. Pathways involved in amino acid, carbohydrate, photosynthesis, glycan, lipid, phenyl-propanoid metabolism, protein ubiquitination, and secondary metabolite production were significantly enriched. Notably, genes implicated in protein ubiquitination/modification pathways in the tolerant genotype, including caseinolytic protease (Clp) and ring type E3 ubiquitin transferase, displayed induced expression in the tolerant than sensitive genotype. Clp are chaperones involved in protein disaggregation; these proteases are required to protect against oxidative stress (Pulido et al., 2017). The PPI network revealed 39 ribosomal proteins that interacted with the key genes involved in the cellular response to heat stress (Figure 3). The intricate network of interactions between ribosomal proteins and stress-responsive genes explains the complex mechanisms that plants use to adapt and thrive under stress conditions (Figure 6). These findings are congruent with earlier research emphasizing varying crop responses to heat stress. High-temperature treatment affects the physiological functions of ER, hence affecting the protein’s synthesis, modification and proper folding.
Stress-modulated lipid metabolism, with lipoxygenase (LOX), lipase_3, lipase_GDSL domain-containing and patatin enzymes were significantly more down-regulated in sensitive genotypes, indicating repressed lipid metabolic pathways (Table 4). LOX is documented in several crops to catalyze the synthesis of plant’s defense-related 9(S)-hydroperoxy-octa-deca-trienoic acid (Göbel et al., 2001). In research on tomato seedlings, increased lipoxygenase activity was correlated with salt tolerance, and in pepper, the CaLOX1 gene was reported to modulate the abiotic stress responses via activation of defense-related marker genes and scavenging of ROS (Lim et al., 2015). Lipase_GDSL plays an important role in plants’ defensive mechanism. For example, AtLTL1 encoding lipase_GDSL in Arabidopsis was recorded to enhance salt tolerance (Naranjo et al., 2006).
Secondary metabolites have multifaceted roles in plant-environment interactions and provide pigmentation to various plant parts. However, terpene cyclase, terpene synthase, involved in the synthesis of metabolites that assist in the regulation of homeostasis and plant’s response to biotic and abiotic stress and glyoxalase, important in secondary metabolite biosynthesis, displayed less suppression in tolerant genotype while experiencing significant more down-regulation in sensitive genotype (Pérez-Llorca et al., 2023). Secondary metabolites in some plants function as osmolytes and growth precursors to help plants recover from heat stress.
Phenylpropanoids play a crucial role in lignin synthesis, particularly under high temperatures. This observation suggests a modulation in the production of compounds responsible for lignin synthesis, a key aspect of plant stress response, in the heat-tolerant genotype. The decrease in secondary metabolite biosynthesis in tolerant genotype at high temperatures might signify a strategic energy conservation response under high-temperature conditions. These changes in gene expression related to secondary metabolites and protein ubiquitination pathways point to a nuanced adaptive response to heat stress. These alterations emphasize the genotype-dependent regulation of significant stress adaptation pathways in pearl millet, reflecting the diverse strategies employed by different genotypes in coping with heat stress conditions. Kegg pathways represent the involvement of 180 genes in the biosynthesis of secondary metabolites which includes 3-ketoacyl-CoA synthase displaying up-regulation and down-regulation in the control vs treatment comparison of tolerant and sensitive genotype, respectively.
Increased temperature alters gene expression in signal transduction pathways
Elevated temperatures promote significant changes in gene expression within signal transduction pathways in plants, particularly when subjected to abiotic stress such as heat. These changes activate sophisticated regulatory networks, which trigger innate defense mechanisms. Protein kinases, particularly MAPKs, are important in orchestrating physiological adaptations by transducing environmental stimuli to the nucleus, thereby protecting plants from diverse biotic and abiotic stresses (Morrison, 2012). Furthermore, the Cysteine-rich receptor-like protein, a member of the RLK family, is involved in plant immunology, stress response, and growth and development (Tanaka et al., 2012). Importantly, elevated temperatures cause an increase in calcium influx, which is one of the first cellular alterations that occurs after a heat shock.
In this study, we found a significant down-regulation of genes encoding MAPK, protein kinase (-5.7 to -10.2 times), ABC1 domain-containing protein associated with protein kinase activity, calmodulin-binding protein 60, and cysteine-rich receptor-like protein in the sensitive genotype (Table 4). Heat stress triggers the MAPK signaling cascades in the plant system. MAPK is a master regulator that operates various physiological and cellular activities in response to heat stress. In wheat, MAPK triggers different genes that impart heat tolerance under terminal heat stress (Banerjee et al., 2020). This evidence indicates that heat stress has a more adverse impact on the expression of signaling genes in sensitive genotypes.
Integral membrane proteins, particularly ABC transporter proteins that use ATP as energy source, displayed reduced expression in most combinations except in tolerant genotype under stress conditions (Figure 6). However, membrane transport protein-encoding MFS (Major facilitator superfamily) genes, which facilitate compound transport across cell membranes using electrochemical gradients, were more down-regulated in various HSG combinations than in HTG, implying potential disruptions in membrane transport functions within leaves and subsequent effects on distinct cellular activities (Niño-González et al., 2019).
Further, cyclic nucleotide-gated ion channels (CNGCs), which are involved in calcium signal transduction, expression was repressed in the roots of the heat-sensitive plants under stress, similar to the findings in Arabidopsis (Alqurashi et al., 2016). This down-regulation is consistent with previous findings in Arabidopsis seedlings, implying a negative impact on thermo-tolerance by increasing ROS enzyme activity (Gao et al., 2012). Additionally, the observed suppression of gene expression for trehalose 6 phosphate phosphatase (T6PP) in sensitive genotype under stress conditions suggests a plausible reduction in trehalose levels induced by heat stress, potentially disrupting carbohydrate transport mechanisms critical for stress tolerance (Lunn et al., 2006; Ruan, 2014). The tolerant genotype displayed induced expression of T6PP in leaf compared to HSG. T6PP acts as a sugar signal and induces the expression of genes associated with stress injury (Lyu et al., 2018).
Plant growth regulators, also known as phytohormones, are essential in reacting to abiotic stress, particularly heat stress. Recent studies reveal that hormones such as auxin, cytokinin, ethylene, and abscisic acid (ABA) are actively implicated in heat response. ABA, a crucial stress-related hormone, and its association with the up-regulation of AAI (an abscisic acid-inducible protein) across the combinations of the tolerant genotype imply its potential involvement in conferring heat stress tolerance (Table 4). ABA boosts tolerance by regulating the transcript level of HSPs and engaging in spatial and temporal interactions with ROS (Suzuki et al., 2016). Auxin, responsible for cell wall synthesis and nucleic acid metabolism, activates multiple genes involved in auxin-mediated signaling pathways, including ARF, auxin efflux carriers, and short auxin-up RNA (SAUR) (Fenqi et al., 2023). The research discovered a two-fold increase in ARF gene expression in the stressed leaves of the tolerant genotype. In the heat-sensitive genotype, the SAUR 36 gene, which is related to leaf senescence and cell elongation suppression, was down-regulated (Jia et al., 2020).
Earlier findings revealed complex interactions between ethylene, ABA, and brassinosteroids in regulating heat stress responses. We identified the activation of CASP proteins and 19 undiscovered proteins associated with the brassinosteroids signaling pathway. Heat stress decreases cytokinin synthesis, influencing cell division and elongation, with down-regulation recorded in genes involved in cytokinin biosynthesis and zeaxanthin epoxidase, a key player in hormone synthesis (Hoang et al., 2020). The suppression was more pronounced in sensitive genotype under heat stress. These findings suggest that the tolerant genotype employs distinct adaptive mechanisms in response to heat stress compared to the sensitive genotype.
ROS scavenging: a key pathway for modulating heat stress response mechanisms
When exposed to high temperatures, plants overproduce ROS, a pivotal signaling component that causes oxidative stress by destroying the cell structure, particularly the membrane structure (Slimen et al., 2014). Plants have ROS scavenging strategies to mitigate the damage, essential for cellular recovery and redox equilibrium (Kotak et al., 2007). In this study, we noticed differences in the expression of ROS-scavenging enzymes in response to heat stress in both genotypes.
Several ROS scavenging genes namely, superoxide dismutase (SOD), ascorbate peroxidase (APX), catalase (CAT), peroxidase (PRX), glutathione peroxidase (GPX), amine oxidase, amino oxidase, respiratory burst oxidase, thioredoxin, and glutaredoxin activated under the stress condition (Figure 6). We discovered 52 peroxidase-related genes involved in suberin and lignin synthesis, stomatal closure control, and stress-induced heat shock protein (HSP) expression (Havaux, 1993). This enzyme is involved in scavenging ROS, which are produced in response to heat stress. Peroxidase gene expression was significantly suppressed in the HSG treated conditions, and in the leaf of tolerant genotype under stress, it displayed induced expression (Table 4). In the roots, SOD, which is important for dismutating superoxide radicals (O2-) into oxygen (O2) and hydrogen peroxide (H2O2) and thereby preserving photosynthetic organelles, was down-regulated in HSG under stress (Zang et al., 2020). Over-expression of SOD in Avicennia marina and Alfalfa confers tolerance to abiotic stress (Rubio et al., 2002; Prashanth et al., 2008).
Amine oxidase, involved in quinone binding and amine metabolism, influences plant responses to environmental stress and demonstrated induced expression patterns across tolerant genotype combinations (Gholizadeh and Mirzaghaderi, 2020). APX, which regulates hydrogen peroxide levels under heat and oxidative stress, displayed varying expression patterns across the tissues in both genotypes. The expression of APX in Arabidopsis resulted in chloroplast protection during heat stress (Panchuk et al., 2002). DEGs encoding APX were primarily more down-regulated in the sensitive than the tolerant genotype, indicating that in response to heat stress, HTG over-produced ROS, resulting in increased APX activity. The down-regulation of these genes suggests a possible disruption of metabolic adjustments in the leaf and roots of pearl millet under heat stress, primarily in the sensitive genotype.
Genes related to ROS detoxification, including thioredoxin involved in ROS signaling, respiratory burst oxidase, CAT, GPX, and laccase, which contributes to oxidoreductase activity in the apoplast, displayed down-regulation predominantly in the sensitive genotype under stress conditions (Mittler, 2017). This down-regulation indicates heightened oxidative stress in sensitive genotypes. In contrast, the tolerant genotype exhibited reduced oxidative stress despite the down-regulation of these genes. This observation aligns with recent studies that reported decreased expression of ROS-scavenging genes under heat stress. GO demonstrated a significant representation of 60 oxidative stress-related genes in the category of oxidoreductase activity, highlighting the importance of ROS buildup on plant responses.
Role of transcription factors in heat stress resilience
TFs are essential in regulating transcriptional responses to heat stress, and various TFs implicated in heat stress acclimation were found in multiple crops. Previous studies have reported the differences in the expression of different TF families, including WRKY, NAC, AP2/ERF, MYB, EF, GATA, bZIP, MADS-box, DEAD-box ATP-dependent RNA helicase, zinc finger protein, C2H2, and C3H domain-containing protein, highlighting their role in the heat stress response (Ward and Schroeder, 1994; Seki et al., 2003). Figure 7 represents the expression of different TFs across the combinations of HTG and HSG. Among the identified TFs, MYB, a significant player in chromosomal structure and stress interactions and is involved in the biosynthesis of secondary metabolites, exhibited distinct expression patterns (Samad et al., 2017). El-Kereamy et al., 2012 found that heat stress triggered the activation of OsMYB55 (El-Kereamy et al., 2012). Overexpressing OsMYB55 alleviated the adverse effects of high temperatures on grain yield by enhancing amino acid metabolism and improving rice’s heat stress tolerance. Seventeen differentially expressed MYB genes displayed reduced activity (Table 4) primarily in leaf and root tissues of the sensitive genotype, but upregulation in the tolerant genotype, indicating a vital function in regulating transcription under heat stress conditions (Chakraborty et al., 2022).
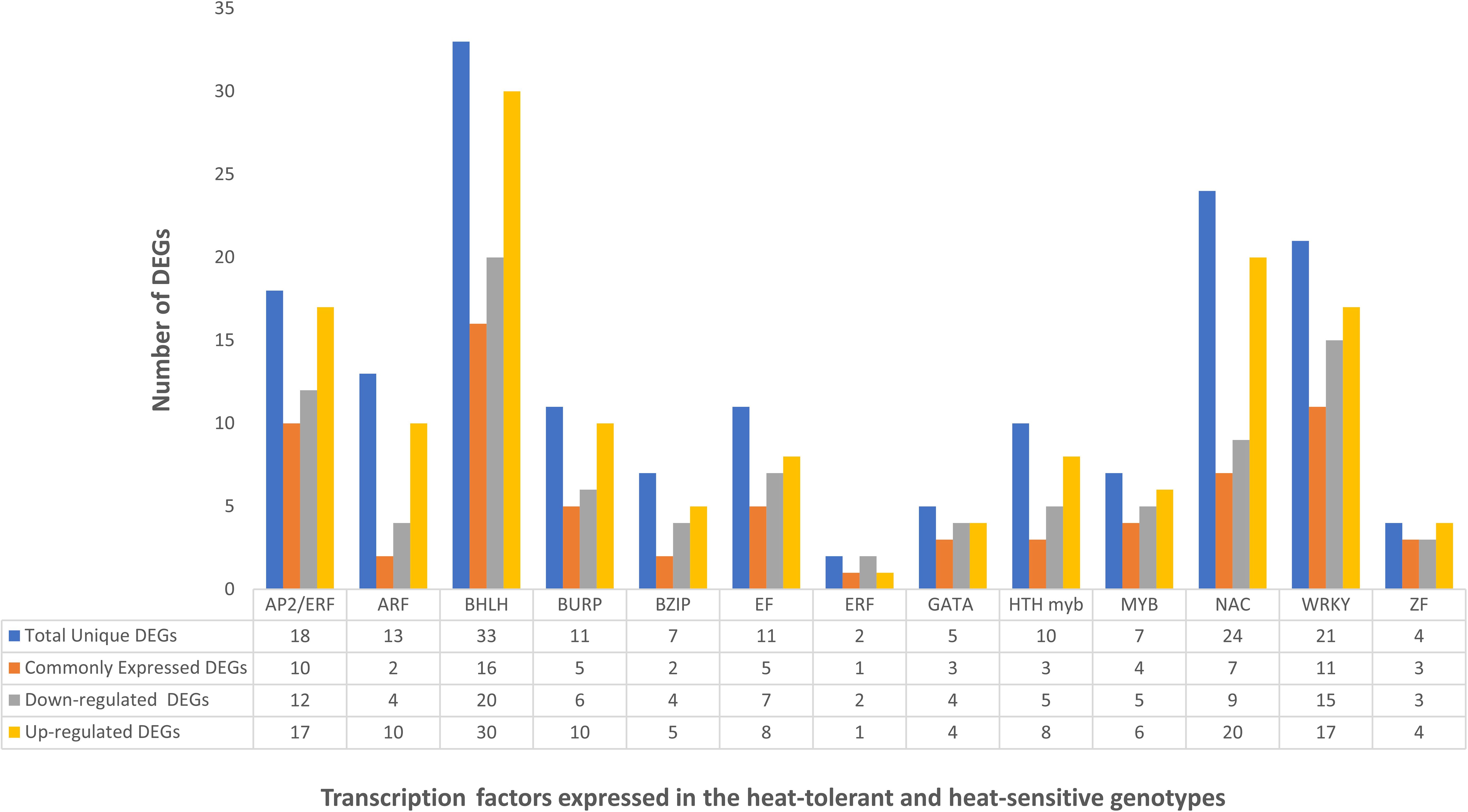
Figure 7. Expression of important transcription factors in the leaf and root tissues of heat-tolerant and heat-sensitive genotypes.
The C2H2 TF family and a single C3H gene, implicated in growth, development, and abiotic stress responses, showed different expression profiles (Thirunavukkarasu et al., 2017; Hu et al., 2019). WRKY, NAC, AP2/ERF, BHLH and BURP genes were significantly up-regulated in the tissues of the tolerant genotype (Table 3). In contrast, the expression of bZIP, MADS-box, BHLH, DEAD-box ATP-dependent RNA helicase, EF, and zinc finger proteins were drastically reduced in sensitive genotype leaf and root tissues. The role of WRKY in regulating gene expression during heat and drought stress is consistent with our findings, indicating their potential for improving heat stress tolerance in Arabidopsis and rice (Wu et al., 2009; He et al., 2016). Similarly, enhanced expression of NAC and AP2/ERF in the tolerant genotype supports the finding of the earlier research demonstrating their significance in imparting tolerance to multiple abiotic stresses across many crops (Sakuma et al., 2002; Nakano et al., 2006; Shiriga et al., 2014). The AP2/ERF family, comprising plant-specific TFs, possesses a conserved DNA-binding domain. This family contains DRE-binding proteins that activate stress-responsive genes by specifically binding to the dehydration-responsive element/C-repeat (DRE/CRT) in gene promoters (Sakuma et al., 2002; Nakano et al., 2006). The AP2-ERF super-family significantly influences plant growth, development, hormonal regulation, and responses to diverse environmental stresses, notably heat stress (Mizoi et al., 2012). SNAC3 in rice enhances the tolerance to heat and drought stress by modulating the ROS balance (Xi et al., 2022). A study on switchgrass showed that DEAD-box ATP-dependent RNA helicases may function as RNA chaperons (Li et al., 2013).
Moreover, the over-expression of BURP in the leaf of tolerant genotypes implies their participation in plant hormone signaling and adaptation to environmental stressors (Shu et al., 2018). In alignment with previous research, the down-regulation of bZIP and zinc finger protein expression in sensitive genotypes indicates that heat stress has a negative impact on growth and many signaling pathways. In the reproductive stage of Arabidopsis, bZIP regulates heat tolerance (Gao et al., 2022). BHLH play diverse roles in plant development and stress responses. The significant inhibition of BHLH gene expression in sensitive genotype’s leaf and root tissues suggested that it plays an essential role in regulatory networks responding to heat stress. Studies revealing the drought-responsive behavior of bHLH genes, such as MdbHLH130 in apples, and their participation in strengthening plant resistance highlight their potential significance in heat stress response pathways (Guo et al., 2021).
A study conducted on foxtail millet reveals the involvement of numerous bHLH genes in promoting drought tolerance, and Fe2OG dioxygenases are involved in various metabolic processes (Wang et al., 2018). Enrichment analysis of the TFs revealed that the TF-associated GO terms provide valuable insights into the underlying regulatory mechanisms and offer targets for further functional studies to elucidate their specific roles in plant stress response and development. Our research highlighted the conservation and diversity of TFs involved in heat stress response across various crops, and found that the foxtail millet is closely related to pearl millet TFs (Figure 4). The differential expression of TFs in both genotypes’ leaf and root tissues suggests that they play an important role in regulatory pathways and transcriptome reconfiguration during heat stress in pearl millet. It implies that these TFs have a complicated interplay in the plant’s response to heat stress, necessitating additional research into their precise regulatory roles.
The elevated response of heat shock proteins and heat shock factors in pearl millet under stressed conditions
Heat stress affects plant cell membrane integrity, alters protein structure, causes misfolding of native proteins, and promotes the accumulation of aberrant proteins. Plants have evolved different defense mechanisms in response to heat stress, including generating heat stress factors critical in regulating HSPs (Mittler et al., 2012). These HSPs function as molecular chaperones essential for maintaining cellular homeostasis. Several heat-related genes (Figure 6), including HSP70, HSP24, HSP10kDa, sHsp17.6, sHSP domain-containing protein, sHsp17.0, HSF domain-containing protein, and HATPase_c domain-containing protein were over-expressed. Figure 8 represents the expression pattern of HSPs and TFs across the pairwise comparisons of HTG and HSG. When exposed to heat, pearl millet synthesize stress proteins, specifically HSPs, which act as molecular chaperones, promoting protein folding and structural stability (Mukesh Sankar et al., 2021).
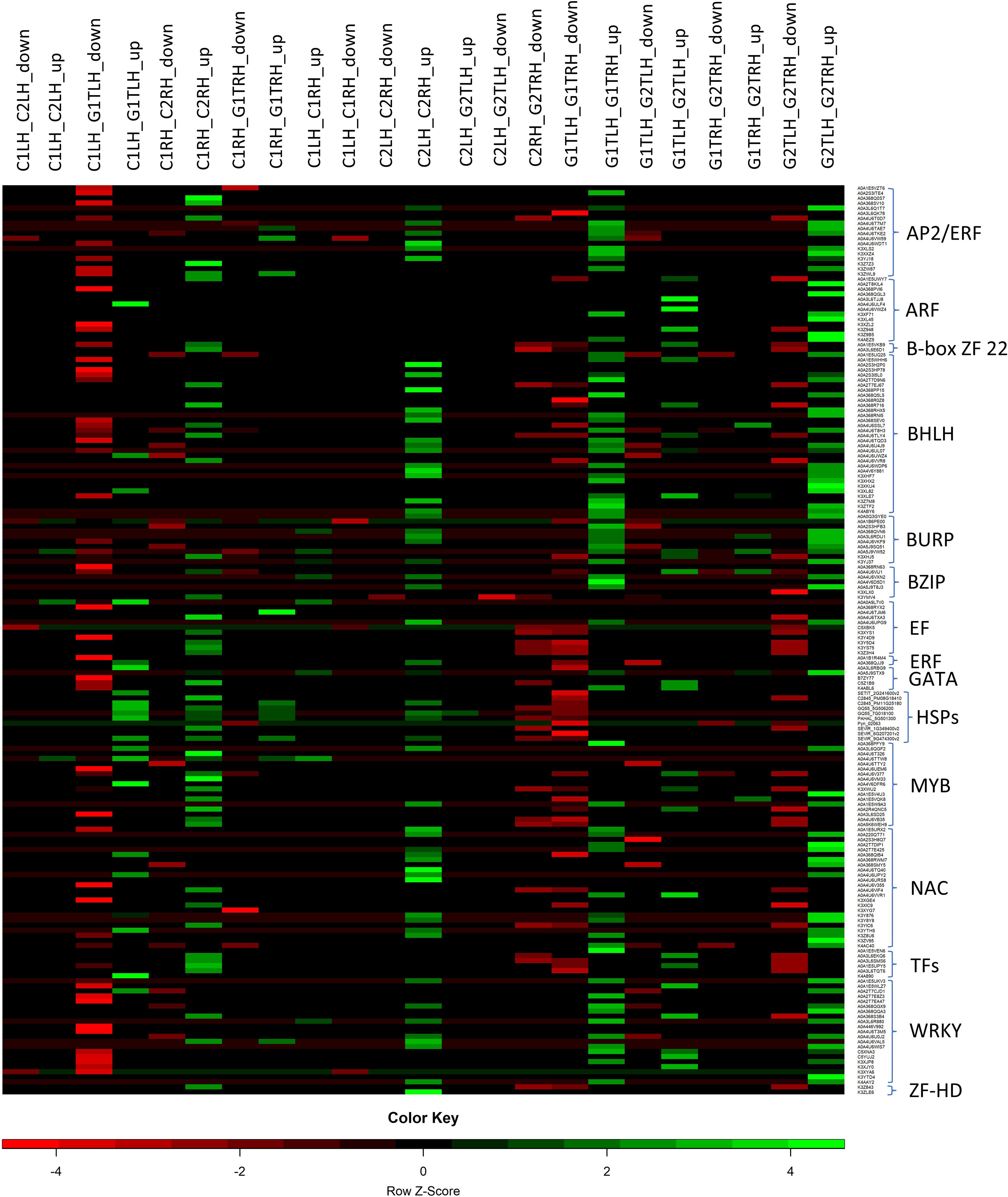
Figure 8. Heatmap of the selected TFs and HSPs operating under control and heat stress conditions in leaf, root and between leaf and root combinations of HTG and HSG. C1 and G1: HTG control and treatment conditions, C2 and G2: HSG control and treatment conditions, LH: leaf and RH: root, up: up-regulation and down: down-regulation.
Our findings revealed significant up-regulation of sHsp17.0, sHsp17.6, and sHSP domain-containing proteins in the leaf and roots of the tolerant genotype (Table 4). The expression of sHsp17.0 was prominently observed in root tissues of HTG under stressed conditions, and sHsp17.0 exhibited significant up-regulation in roots of treated conditions in the tolerant genotype (Figure 8). Several studies in Arabidopsis, P. pastoris, and woody plants have demonstrated that sHSPs confer heat and drought tolerance (Yang et al., 2017; Wang et al., 2017). Additionally, sHSP domain-containing proteins displayed up-regulation in the leaf and root tissues of HTG. The enhanced activity of HSP70, which is prevalent under stress, was significant, with 70kDa stromal HSPs expressing in both HSG and HTG roots under stress, indicating a more significant transcriptional response in roots compared to leaves.
Stromal 70kDa HSP participates in transport pathways between the endoplasmic reticulum, mitochondria, and chloroplastic pre-protein intake. According to previous research on various plants, including Hevea brasiliensis, rice, tobacco, and wheat, HSP70 plays an important role for cellular response to heat stress (Zhang et al., 2009; Wang et al., 2016; Wahab et al., 2020). HSPs in pearl millet, particularly HSP70 and HSP90, have been studied for their role in heat and drought adaptation processes (Donald et al., 2015). It has been found that HSP70 is involved in molecular chaperone activity under stress and plays a function in photo-protection and photosystem II repair (Schroda et al., 1999; Sung et al., 2001). HATPase, a member of the HSP90 family, showed over-expression in tolerant genotype combinations, signifying its function in ATP binding and ATPase activity (Figure 8). Five genes related to HATPase were identified in soybean and rice, with the majority exhibiting up-regulation in the tolerant genotype combination (Schroda et al., 1999; Sung et al., 2001; Li et al., 2020). A comparative analysis of genes related to HSPs in pearl millet revealed orthologue sequences in rice, foxtail millet, sorghum, proso millet, and maize (Figure 4). This suggests that these genes are expressed in response to stress in various crops. The prevalence of HSPs, especially across HTG genotype combinations, highlights their crucial role in maintaining protein structure, ATP binding, and hydrolysis in response to heat stress and raising heat stress tolerance.
HSPs and heat-related genes are also regulated by heat shock transcription factors (HSFs), which orchestrate the plant’s response to heat stress (Guo et al., 2016). Our findings revealed that HSF-related genes were up-regulated in tolerant genotypes and down-regulated in sensitive cultivars. Zhu et al., 2006 showed that over-expression of GmHSFA1 in soybeans confers thermo-tolerance, possibly due to the activation of sHSP and HSP70 (Zhu et al., 2006). This implies that they play an important role in tolerant genotype survival under heat-stress conditions. The PPI network reveals that the expression of HSP is related to different important TFs and HSF in response to heat stress (Figure 3). The prominent expression of HSPs and HSF-related genes in the tolerant genotype emphasizes their critical engagement in cellular and molecular processes during heat stress, which aligns with recent research across many crops. The pattern of HSP expression in pearl millet under stress conditions supports previous research on heat-induced HSPs, emphasizing their critical role in plants’ response to heat stress.
Our research revealed substantial differences in the expression of genes involved in various physiological and biochemical processes, specifically in the tolerant genotype. We found significant differences in the expression of genes crucial in water, nutrient, and ion transport (bidirectional sugar transporter SWEET, ABC transporter domain-containing protein, PIP, potassium transporters, nitrate reductase and glutamine synthetase), cell wall maintenance (expansin), photosynthesis (RuBisCO, carbonic anhydrase and ATP synthase α subunit), transcription factors (BZIP, BHLH, MYB, AP2/ERF, and BURP), signal transduction (protein kinases, MAPK, ABA, and AAI), ROS scavenging (SOD, peroxidases, GPX, and amine oxidase), HSPs (SHSPs, HATPase, and HSP 10kDa), secondary metabolite biosynthesis (LOX, Lipase_GDSL, terpene cyclase, terpene synthase, and 3-ketoacyl-CoA synthase) and protein modification pathways (Clp and ring type E3 ubiquitin transferase), implying that pearl millet regulates the synthesis and expression of these genes to survive under heat stress.
Conclusions
A genome-wide transcriptome study was performed in two pearl millet genotypes (HTG and HSG) to examine the molecular mechanisms in response to heat stress. We comprehensively analyzed the leaf and root samples in three categories: between genotypes, within genotypes, and between tissues. These DEGs from pairwise comparisons of the treated and control samples provide substantial insight into the effects of heat stress on pearl millet.
The analysis of heat-responsive genes and hub genes revealed that protein processing in the endoplasmic reticulum is one of the critical pathways involved in heat stress. The protective impact of HSPs could be related to the chaperone mechanism network, in which multiple chaperones work together. Under stress, many structural proteins undergo adverse structural and functional modifications. As a result, refolding of denatured proteins and maintaining their function is crucial for cell survival in stress conditions. These findings could help in our understanding of the role of the genes implicated in heat tolerance.
We discovered a significant number of DEGs belonging to uncharacterized proteins, revealing possible new genes involved in heat stress response in pearl millet. Our study provides fresh insights into the transcriptional modifications in different tissues of tolerant and sensitive genotypes, which aids in decoding the underlying mechanism to enhance crop resilience. This study will set the groundwork for identifying and utilizing key genes that are explicitly expressed in tissues to investigate the mechanisms of heat stress tolerance in pearl millet. These genes will also serve as appropriate molecular indicators for screening accessions for heat tolerance and speed up the variety of development programs in pearl millet and similar millet crops.
Data availability statement
The data presented in the study are deposited in the NCBI Sequence Read Archive database with accession number PRJNA1062049.
Author contributions
SS: Writing – review & editing, Writing – original draft, Visualization, Validation, Software, Resources, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. AV: Writing – review & editing, Methodology. AC: Writing – review & editing, Data curation. NN: Writing – review & editing, Data curation. RM: Writing – review & editing, Resources. JJ: Writing – review & editing, Methodology. SM: Writing – review & editing, Visualization. TC: Writing – review & editing. NT: Writing – review & editing, Supervision, Funding acquisition, Conceptualization.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The experiment was funded by the Bill and Melinda Gates Foundation project (INV-008187), the ICAR-Indian Institute of Millets Research project (CI/2018-23/120) and the Global Centre of Excellence on Millets (Shree Anna).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1443681/full#supplementary-material
References
Aksaas, K., Larsen, A. A. C. V., Rogne, M., Rosendal, K., Kvissel, A.-K., Skålhegg, BjørnS. (2011). G-patch domain and KOW motifs-containing protein, GPKOW; a nuclear RNA-binding protein regulated by protein kinase A. J. Mol. Signaling 6, 1–14. doi: 10.1186/1750-2187-6-10
Alqurashi, M., Gehring, C., Marondedze, C. (2016). Changes in the Arabidopsis thaliana proteome implicate cAMP in biotic and abiotic stress responses and changes in energy metabolism. Int. J. Mol. Sci. 17, 852. doi: 10.3390/ijms17060852
Ashraf, M., Hafeez, M. (2004). Thermotolerance of pearl millet and maize at early growth stages: growth and nutrient relations. Biol. Plantarum 48, 81–86. doi: 10.1023/B:BIOP.0000024279.44013.61
Ashraf, M. H. P. J. C., Harris, P. J. C. (2013). Photosynthesis under stressful environments: An overview. Photosynthetica 51, 163–190. doi: 10.1007/s11099-013-0021-6
Banerjee, G., Singh, D., Sinha, A. K. (2020). Plant cell cycle regulators: Mitogen-activated protein kinase, a new regulating switch? Plant Sci. 294, 110660. doi: 10.1016/j.plantsci.2020.110660
Barthakur, S., Bharadwaj, N. (2022). “Exploring genome-wide analysis of heat shock proteins (HSPs) in small millets as potential candidates for development of multistress tolerant crop plants,” in Omics of Climate Resilient Small Millets (Singapore: Springer Nature Singapore), 337–355. doi: 10.1007/978-981-19-3907-5_17
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bukovnik, Urška, Fu, J., Bennett, M., Prasad, P. V. V., Ristic., Z. (2009). Heat tolerance and expression of protein synthesis elongation factors, EF-Tu and EF-1α, in spring wheat. Funct. Plant Biol. 36, 234–241. doi: 10.1071/FP08266
Chakraborty, A., Viswanath, A., Malipatil, R., Semalaiyappan, J., Shah, P., Ronanki, S., et al. (2022). Identification of candidate genes regulating drought tolerance in Pearl Millet. Int. J. Mol. Sci. 23, 6907. doi: 10.3390/ijms23136907
Chandel, G., Dubey, M., Meena, X. X. X. R. (2013). Differential expression of heat shock proteins and heat stress transcription factor genes in rice exposed to different levels of heat stress. J. Plant Biochem. Biotechnol. 22, 277–285. doi: 10.1007/s13562-012-0156-8
Chin, C.-H., Chen, S.-H., Wu, H.-H., Ho, C.-W., Ko, M.-T., et al. (2014). cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 8, 1–7. doi: 10.1186/1752-0509-8-S4-S11
Dahuja, A., Kumar, R. R., Sakhare, A., Watts, A., Singh, B., Goswami, S., et al. (2021). Role of ATP-binding cassette transporters in maintaining plant homeostasis under abiotic and biotic stresses. Physiologia Plantarum 171, 785–801. doi: 10.1111/ppl.13302
Darko, E., Végh, Balázs, Khalil, R., Marček, T., Szalai, G., Pál, M., et al. (2019). Metabolic responses of wheat seedlings to osmotic stress induced by various osmolytes under iso-osmotic conditions. PloS One. 14, e0226151. doi: 10.137/journal.ponr.0226151
Das, A., Eldakak, M., Paudel, B., Kim, D.-W., Hemmati, H., Basu, C., et al. (2016). Leaf proteome analysis reveals prospective drought and heat stress response mechanisms in soybean. BioMed. Res. Int. 2016, 6021047. doi: 10.1155/2016/6021047
Ding, H., Mo, S., Qian, Y., Yuan, G., Wu, X., Ge, C. (2020). Integrated proteome and transcriptome analyses revealed key factors involved in tomato (Solanum lycopersicum) under high temperature stress. Food Energy Secur. 9, e239. doi: 10.1002/fes3.239
Djanaguiraman, M., Sheeba, J.A., Devi, D.D., Bangarusamy, U. (2009). Cotton leaf senescence can be delayed by nitrophenolate spray through enhanced antioxidant defense system. J. Agron. Crop Sci. 195, 213–224. doi: 10.1111/j.1439-037X.2009.00360.x
Donald, J., Tarafdar, A., Biswas, K., Sathyavathi, T. C., Padaria, J. C., Kumar, P.A. (2015). Development and characterization of a high temperature stress responsive subtractive cDNA library in Pearl Millet (Pennisetum glaucum (L.) R. Br). Indian J. Exp. Biol. 53, 543-550.
El-Kereamy, A., Bi, Y.-M., Perrin, H., Allen, G. (2012). Good, and Steven J. Rothstein. The rice R2R3-MYB transcription factor OsMYB55 is involved in the tolerance to high temperature and modulates amino acid metabolism. PloS One 7, e52030. doi: 10.1371/journal.pone.0052030
Fahad, S., Bajwa, A. A., Nazir, U., Anjum, S. A., Farooq, A., Zohaib, A., et al. (2017). Crop production under drought and heat stress: plant responses and management options. Front. Plant Sci. 8, 1147. doi: 10.3389/fpls.2017.01147
Fenqi, C., Zhang, J., Ha, X. (2023). and Huiling Ma. Genome-wide identification and expression analysis of the Auxin-Response factor (ARF) gene family in Medicago sativa under abiotic stress. BMC Genomics 24, 498. doi: 10.1186/s12864-023-09610-z
Freschet, G. T., Violle, C., Malo, Y., Bourget, M., Florian, F. (2018). Allocation, morphology, physiology, architecture: The multiple facets of plant above-and below-ground responses to resource stress. New Phytol. 219, 1338–1352. doi: 10.1111/nph.15225
Frey, F. P., Urbany, C., Hüttel, B., Reinhardt, R., Stich, B. (2015). Genome-wide expression profiling and phenotypic evaluation of European maize inbreds at seedling stage in response to heat stress. BMC Genomics 16, 1–15. doi: 10.1186/s12864-015-1282-1
Gao, F., Han, X., Wu, J., Zheng, S., Shang, Z., Sun, D., et al. (2012). A heat-activated calcium-permeable channel–Arabidopsis cyclic nucleotide-gated ion channel 6–is involved in heat shock responses. Plant J. 70, 1056–1069. doi: 10.1111/j.1365-313X.2012.04969.x
Gao, J., Wang, M.-J., Wang, J.-J., Lu, H.-P., Jian-Xiang, L. (2022). bZIP17 regulates heat stress tolerance at reproductive stage in Arabidopsis. Abiotech 3, 1–11. doi: 10.1007/s42994-021-00062-1
Gholizadeh, F., Mirzaghaderi, G. (2020). Genome-wide analysis of the polyamine oxidase gene family in wheat (Triticum aestivum L.) reveals involvement in temperature stress response. PloS One 15, e0236226. doi: 10.1371/journal.pone.0236226
Giri, A., Heckathorn, S., Mishra, S., Krause, C. (2017). Heat stress decreases levels of nutrient-uptake and-assimilation proteins in tomato roots. Plants 6, 6. doi: 10.3390/plants6010006
Göbel, C., Feussner, I., Schmidt, A., Scheel, D., Sanchez-Serrano, J., Hamberg, M., et al. (2001). Oxylipin profiling reveals the preferential stimulation of the 9-lipoxygenase pathway in elicitor-treated potato cells. J. Biol. Chem. 276, 6267–6273. doi: 10.1074/jbc.M008606200
Goud, Anjali, C., Satturu, V., Malipatil, R., Viswanath, A., Semalaiyappan, J., et al. (2022). Identification of iron and zinc responsive genes in pearl millet using genome-wide RNA-sequencing approach. Front. Nutr. 9, 884381. doi: 10.3389/fnut.2022.884381
Guo, M., Liu, J.-H., Ma, X., Luo, D.-X., Gong, Z.-H., Lu., M.-H. (2016). The plant heat stress transcription factors (HSFs: structure, regulation, and function in response to abiotic stresses. Front. Plant Sci. 7, 114. doi: 10.3389/fpls.2016.00114
Guo, J., Sun, B., He, H., Zhang, Y., Tian, H., Wang, B. (2021). Current understanding of bHLH transcription factors in plant abiotic stress tolerance. Int. J. Mol. Sci. 22, 4921. doi: 10.3390/ijms22094921
Hasanuzzaman, M., Nahar, K., Alam, Md M., Roychowdhury, R., Fujita, M. (2013). Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int. J. Mol. Sci. 14, 9643–9684. doi: 10.3390/ijms14059643
Havaux, M. (1993). Rapid photosynthetic adaptation to heat stress triggered in potato leaves by moderately elevated temperatures. Plant Cell Environ. 16, 461–467. doi: 10.1111/j.1365-3040.1993.tb00893.x
He, Y., Guan, H., Li, Bo, Zhang, S., Xu, Y., Yao, Y., et al. (2023). Transcriptome analysis reveals the dynamic and rapid transcriptional reprogramming involved in heat stress and identification of heat response genes in rice. Int. J. Mol. Sci. 24, 14802. doi: 10.3390/ijms241914802
He, G.-H., Xu, J.-Y., Wang, Y.-X., Liu, J.-M., Li, P.-S., Chen, M., et al. (2016). Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC Plant Biol. 16, 1–16. doi: 10.1186/s12870-016-0806-4
Hoang, M. H., Kim, H.-S., Zulfugarov, I. S., Lee, C.-H. (2020). Down-regulation of zeaxanthin epoxidation in vascular plant leaves under normal and photooxidative stress conditions. J. Plant Biol. 63, 331–336. doi: 10.1007/s12374-020-09260-8
Hosseini, Zahra, S., Ismaili, A., Nazarian-Firouzabadi, F., Fallahi, H., Nejad, A. R., et al. (2021). Dissecting the molecular responses of lentil to individual and combined drought and heat stresses by comparative transcriptomic analysis. Genomics 113, 693–705. doi: 10.1016/j.ygeno.2020.12.038
Hu, X., Zhu, L., Zhang, Y., Xu, L., Li, N., Zhang, X., et al. (2019). Genome-wide identification of C2H2 zinc-finger genes and their expression patterns under heat stress in tomato (Solanum lycopersicum L.). PeerJ 7, e7929. doi: 10.7717/peerj.7929
Huang, W., Sherman, B. T., Tan, Q., Collins, J. R., Alvord, W. G., Roayaei, J., et al. (2007). The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 8, 1–16. doi: 10.1186/gb-2007-8-9-r183
Huang, B., Rachmilevitch, S., Xu, J. (2012). Root carbon and protein metabolism associated with heat tolerance. J. Exp. Bot. 63, 3455–3465. doi: 10.1093/jxb/ers003
Indiastat. (2020). Agricultural production (New Delhi: Indiastat: Datanet India Pvt. Ltd) Available online at: http://www.Indiastat.com/searchresult.aspx (Accessed February 27, 2020).
Jagadish, S. V., Way, K. D. A., Sharkey, T. D. (2021). Plant heat stress: Concepts directing future research. Plant Cell Environ. 44, 1992–2005. doi: 10.1111/pce.14050
Jia, S., Li, H., Jiang, Y., Tang, Y., Zhao, G., Zhang, Y., et al. (2020). Transcriptomic analysis of female panicles reveals gene expression responses to drought stress in maize (Zea mays L.). Agronomy 10, 313. doi: 10.3390/agronomy10020313
Klemens, A. W., Patzke, K., Deitmer, J., Spinner, L., Hir, R. Le, Bellini, C., et al. (2013). Overexpression of the vacuolar sugar carrier AtSWEET16 modifies germination, growth, and stress tolerance in Arabidopsis. Plant Physiol. 163, 1338–1352. doi: 10.1104/pp.113.224972
Kong, X., Luo, Z., Dong, H., Eneji, A. E. (2012). and Weijiang Li. Effects of non-uniform root zone salinity on water use, Na+ recirculation, and Na+ and H+ flux in cotton. J. Exp. Bot. 63, 2105–2116. doi: 10.1093/jxb/err420
Kotak, S., Larkindale, J., Lee, U., Koskull-Döring, P. v., Vierling, E., Scharf, K.-D. (2007). Complexity of the heat stress response in plants. Curr. Opin. Plant Biol. 10, 310–316. doi: 10.1016/j.pbi.2007.04.011
Kurimoto, K., Day, D. A., Lambers, H., Noguchi., Ko (, 2004). Effect of respiratory homeostasis on plant growth in cultivars of wheat and rice. Plant Cell Environ. 27, 853–862. doi: 10.1111/j.1365-3040.2004.01191.x
Li, Z., Howell, S. H. (2021). Heat stress responses and thermotolerance in maize. Int. J. Mol. Sci. 22, 948. doi: 10.3390/ijms22020948
Li, J., Nadeem, M., Chen, L., Wang, M., Wan, M., Qiu, L., et al. (2020). Differential proteomic analysis of soybean anthers by iTRAQ under high-temperature stress. J. Proteomics 103968. doi: 10.1016/j.jprot.2020.103968
Li, Y.-F., Wang, Y., Tang, Y., Kakani, V. G., Ramamurthy, M. (2013). Transcriptome analysis of heat stress response in switchgrass (Panicum virgatum L.). BMC Plant Biol. 13, 1–12. doi: 10.1186/1471-2229-13-153
Liao, Y., Smyth, G. K., Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Lim, Woo, C., Han, S.-W., Hwang, In S., Kim, D. S., Hwang, B. K., et al. (2015). The pepper lipoxygenase CaLOX1 plays a role in osmotic, drought and high salinity stress response. Plant Cell Physiol. 56, 930–942. doi: 10.1093/pcp/pcv020
Lunn, J. E., Feil, R., Hendriks, J. H. M., Gibon, Y., Morcuende, R., Osuna, D., et al. (2006). Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. Biochem. J. 397, 139–148. doi: 10.1042/BJ20060083
Lyu, J. Il, Park, Ji H., Kim, J.-K., Bae, C.-H., Jeong, W.-J., Min, S. R., et al. (2018). Enhanced tolerance to heat stress in transgenic tomato seeds and seedlings overexpressing a trehalose-6-phosphate synthase/phosphatase fusion gene. Plant Biotechnol. Rep. 12, 399–408. doi: 10.1007/s11816-018-0505-8
Maurel, C., Verdoucq, L., Luu, D.-T., Santoni, Véronique (2008). Plant aquaporins: membrane channels with multiple integrated functions. Annu. Rev. Plant Biol. 59, 595–624. doi: 10.1146/annurev.arplant.59.032607.092734
Mittler, R., Finka, A., Goloubinoff, P. (2012). How do plants feel the heat. Trends Biochem. Sci. 37, 118–125. doi: 10.1016/j.tibs.2011.11.007
Mizoi, J., Shinozaki, K., Yamaguchi-Shinozaki, K. (2012). AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 1817, 86–96. doi: 10.1016/j.bbagrm.2011.08.004
Molik, S., Karnauchov, I., Weidlich, C., Herrmann, R. G., Kloüsgen, R. B. (2001). The Rieske Fe/S Protein of the Cytochromeb 6/f Complex in Chloroplasts: Missing link in the evolution of protein transport pathways in chloroplasts? J. Biol. Chem. 276, 42761–42766. doi: 10.1074/jbc.M106690200
Moon, S., Jung, K.-H. (2014). Genome-wide expression analysis of rice ABC transporter family across spatio-temporal samples and in response to abiotic stresses. J. Plant Physiol., 1276–1288. doi: 10.1016/j.jplph.2014.05.006
Morrison, D. K. (2012). MAP kinase pathways. Cold Spring Harbor Perspect. Biol. 4, a011254. doi: 10.1101/cshperspect.a011254
Mukesh Sankar, S., Satyavathi, C.T., Barthakur, S., Singh, S. P., Bharadwaj, C. (2021). Differential modulation of heat-inducible genes across diverse genotypes and molecular cloning of a sHSP from pearl millet (Pennisetum glaucum (L.) R. Br.). Front. Plant Sci. 12, 659893. doi: 10.3389/fpls.2021.659893
Mulet, Jose, M., Porcel, R., Yenush, L. (2023). Modulation of potassium transport to increase abiotic stress tolerance in plants. J. Exp. Bot. 74, 5989–6005. doi: 10.1093/jxb/erad333
Mwadzingeni, L., Shimelis, H., Dube, E., Laing, M. D., Tsilo., T. J. (2016). Breeding wheat for drought tolerance: Progress and technologies. J. Integr. Agric. 15, 935–943. doi: 10.1016/S2095-3119(15)61102-9
Nagaraju, M., Reddy, P. S., Kumar, S.A., Srivastava, R. K., Kishor, P.B.K., Manohar Rao, X. X. X. D. (2015). Genome-wide scanning and characterization of Sorghum bicolor L. heat shock transcription factors. Curr. Genomics 16, 279–291.
Nakano, T., Suzuki, K., Fujimura, T., Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140, 411–432. doi: 10.1104/pp.105.073783
Naranjo, M. A., Forment, J., Roldan, M., Serrano, R., Vicente, O. (2006). Overexpression of Arabidopsis thaliana LTL1, a salt-induced gene encoding a GDSL-motif lipase, increases salt tolerance in yeast and transgenic plants. Plant Cell Environ. 47, 1890–1900. doi: 10.1093/pcp/pcv020
Niño-González, María, Novo-Uzal, E., Richardson, D. N., Barros, P. M., Duque, P. (2019). More transporters, more substrates: the Arabidopsis major facilitator superfamily revisited. Mol. Plant 12, 1182–1202. doi: 10.1016/j.molp.2019.07.003
Obaid, A. Y., Sabir, J. S. M., Atef, A., Liu, X., Edris, S., El-Domyati, F. M., et al. (2016). Analysis of transcriptional response to heat stress in Rhazya stricta. BMC Plant Biol. 16, 1–18. doi: 10.1186/s12870-016-0938-6
Panchuk, I. I., Volkov, R. A., Schöffl, F. (2002). Heat stress-and heat shock transcription factor-dependent expression and activity of ascorbate peroxidase in Arabidopsis. Plant Physiol. 129, 838–853. doi: 10.1104/pp.001362
Pérez-Llorca, M., Pollmann, S., Müller, M. (2023). Ethylene and jasmonates signaling network mediating secondary metabolites under abiotic stress. Int. J. Mol. Sci. 24, 5990. doi: 10.3390/ijms24065990
Prashanth, S. R., Sadhasivam, V., Parida, A. (2008). and Ajay Parida. Over expression of cytosolic copper/zinc superoxide dismutase from a mangrove plant Avicennia marina in indica rice var Pusa Basmati-1 confers abiotic stress tolerance. Transgenic Res. 17, 281–291. doi: 10.1007/s11248-007-9099-6
Pulido, P., Llamas, E., Rodriguez-Concepcion, M. (2017). Both Hsp70 chaperone and Clp protease plastidial systems are required for protection against oxidative stress. Plant Signaling Behav. 12, e1290039. doi: 10.1080/15592324.2017.1290039
Qian, Y., Ren, Q., Zhang, J., Chen., L. (2019). Transcriptomic analysis of the maize (Zea mays L.). inbred line B73 response to heat stress at the seedling stage. Gene 692, 68–78. doi: 10.1016/j.gene.2018.12.062
Rangan, P., Furtado, A., Robert, H. (2020). Transcriptome profiling of wheat genotypes under heat stress during grain-filling. J. Cereal Sci. 91, 102895. doi: 10.1016/j.jcs.2019.102895
Rao, D., Momcilovic, I., Kobayashi, S., Callegari, E., Ristic, Z. (2004). Chaperone activity of recombinant maize chloroplast protein synthesis elongation factor, EF-Tu. Eur. J. Biochem. 271, 3684–3692. doi: 10.1111/j.1432-1033.2004.04309.x
Robinson, M. D., McCarthy, D. J., Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Ruan, Y.-L. (2014). Sucrose metabolism: gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 65, 33–67. doi: 10.1146/annurev-arplant-050213-040251
Rubio, M. C., González, E. M., Minchin, F. R., Webb, K.J., Arrese-Igor, C., Ramos, J., et al. (2002). Effects of water stress on antioxidant enzymes of leaves and nodules of transgenic alfalfa overexpressing superoxide dismutases. Physiologia Plantarum 115, 531–540. doi: 10.1034/j.1399-3054.2002.1150407.x
Sakuma, Y., Liu, Q., Dubouzet, J. G., Abe, H., Shinozaki, K., Kazuko Yamaguchi-, S. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration-and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290, 998–1009. doi: 10.1006/bbrc.2001.6299
Samad, A. F. A., Muhammad, S., Nazaruddin, N., Izzat, A. (2017). Fauzi, Abdul MA Murad, Zamri Zainal, and Ismanizan Ismail. MicroRNA and transcription factor: key players in plant regulatory network. Front. Plant Sci. 8, 565. doi: 10.3389/fpls.2017.00565
Schlenker, W., David, B. (2010). Lobell. Robust negative impacts of climate change on African agriculture. Environ. Res. Lett. 5, 014010. doi: 10.1088/1748-9326/5/1/014010
Schroda, M., Vallon, O., Wollman, Francis-André, Beck, C. F. (1999). A chloroplast-targeted heat shock protein 70 (HSP70) contributes to the photoprotection and repair of photosystem II during and after photoinhibition. Plant Cell 11, 1165–1178. doi: 10.1105/tpc.11.6.1165
Seki, M., Kamei, A., Yamaguchi-Shinozaki, K., Shinozaki, K. (2003). Molecular responses to drought, salinity and frost: common and different paths for plant protection. Curr. Opin. Biotechnol. 14, 194–199. doi: 10.1016/S0958-1669(03)00030-2
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shiriga, K., Sharma, R., Kumar, K., Yadav, S. K., Hossain, F., Thirunavukkarasu, N. (2014). Genome-wide identification and expression pattern of drought-responsive members of the NAC family in maize. Meta Gene 2, 407–417. doi: 10.1016/j.mgene.2014.05.001
Shu, Y., Li, W., Zhao, J., Liu, Y., Guo, C. (2018). Transcriptome sequencing and expression profiling of genes involved in the response to abiotic stress in Medicago ruthenica. Genet. Mol. Biol. 41, 638–648. doi: 10.1590/1678-4685-gmb-2017-0284
Sidak, Z. (1967). Rectangular confidence regions for the means of multivariate normal distributions. J. Am. Stat. Assoc. 62, 626–633. doi: 10.2307/2283989
Singh, R. K., Jaishankar, J., Muthamilarasan, M., Shweta, S., Dangi, A., Prasad, M. (2016). Genome-wide analysis of heat shock proteins in C4 model, foxtail millet identifies potential candidates for crop improvement under abiotic stress. Sci. Rep. 6, 32641. doi: 10.1038/srep32641
Slimen, B., Najar, M. T., Ghram, A., Dabbebi, H., Mrad, M. B., Manef, A. (2014). Reactive oxygen species, heat stress and oxidative-induced mitochondrial damage. A review of hyperthermia. Int. J. 30, 513–523. doi: 10.3109/02656736.2014.971446
Sun, M., Huang, D., Zhang, A., Khan, I., Yan, H., Wang, X., et al. (2020). Transcriptome analysis of heat stress and drought stress in pearl millet based on Pacbio full-length transcriptome sequencing. BMC Plant Biol. 20, 1–15. doi: 10.1186/s12870-020-02530-0
Sung, D. Y., Yul, D., Vierling, E., Guy, C. L. (2001). Comprehensive expression profile analysis of the Arabidopsis Hsp70 gene family. Plant Physiol. 126, 789–800. doi: 10.1104/pp.126.2.789
Suzuki, N., Bassil, E., Hamilton, J. S., Inupakutika, M. A., Zandalinas, S. I., Tripathy, D., et al. (2016). ABA is required for plant acclimation to a combination of salt and heat stress. PloS One. 11, e0147625. doi: 10.1371/journal.pone.0147625
Szklarczyk, D., Kirsch, R., Koutrouli, M., Nastou, K., Mehryary, F., Hachilif, R., et al. (2023). The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51, D638–D646. doi: 10.1093/nar/gkac1000
Takahashi, S., Bauwe, H., Badger, M. (2007). Impairment of the photorespiratory pathway accelerates photo-inhibition of photosystem II by suppression of repair but not acceleration of damage processes in Arabidopsis. Plant Physiol. 60, 487–494. doi: 10.1104/pp.107.097253
Tanaka, H., Osakabe, Y., Katsura, S., Mizuno, S., Maruyama, K., Kusakabe, K., et al. (2012). Abiotic stress-inducible receptor-like kinases negatively control ABA signaling in Arabidopsis. Plant J. 70, 599–613. doi: 10.1111/j.1365-313X.2012.04901.x
Thirunavukkarasu, N., Sharma, R., Singh, N., Shiriga, K., Mohan, S., Mittal, S., et al. (2017). Genomewide expression and functional interactions of genes under drought stress in maize. Int. J. Genomics. 2017, 2568706. doi: 10.1155/2017/2568706
Tian, F., Yang, D. C., Meng, Y. Q., Jin, J., Gao, G. (2020). PlantRegMap: charting functional regulatory maps in plants. Nucleic Acids Res. 48, D1104–D1113. doi: 10.1093/nar/gkz972
Vadez, V., Hash, T., Bidinger, F. R., Jana, Kholova (2012). II. 1.5 Phenotyping pearl millet for adaptation to drought. Front. Physiol. 3, 386. doi: 10.3389/fphys.2012.00386
Vandeleur, R., Niemietz, C., Tilbrook, J., Tyerman, S. D. (2005). Roles of aquaporins in root responses to irrigation. Plant Soil 274, 141–161. doi: 10.1007/s11104-004-8070-z
Wahab, M. M. S., Srividhya, A., Shanthi, P., Latha, X. X. X. P. (2020). Identification of differentially expressed genes under heat stress conditions in rice (Oryza sativa L.). Mol. Biol. Rep. 47, 1935–1948. doi: 10.1007/s11033-020-05291-z
Wang, X., Dinler, B. S., Vignjevic, M., Jacobsen, S., Wollenweber, B. (2015). Physiological and proteome studies of responses to heat stress during grain filling in contrasting wheat cultivars. Plant Sci. 230, 33–50. doi: 10.1016/j.plantsci.2014.10.009
Wang, P., Wang, H., Wang, Y., Ren, F., Liu, W. (2018). Analysis of bHLH genes from foxtail millet (Setaria italica) and their potential relevance to drought stress. PloS One 13, e0207344. doi: 10.1371/journal.pone.0207344
Wang, X., Yan, B., Shi, M., Zhou, W., Zekria, D., Wang, H., et al. (2016). Overexpression of a Brassica campestris HSP70 in tobacco confers enhanced tolerance to heat stress. Protoplasma 253, 637–645. doi: 10.1007/s00709-015-0867-5
Wang, Y., Zhang, Y., Zhang, Q., Cui, Y., Xiang, J., Chen, H., et al. (2019). Comparative transcriptome analysis of panicle development under heat stress in two rice (Oryza sativa L.). cultivars differing in heat tolerance. PeerJ 7, e7595. doi: 10.7717/peerj.7595
Wang, M., Zou, Z., Li, Q., Xin, H., Zhu, X., Chen, X., et al. (2017). Heterologous expression of three Camellia sinensis small heat shock protein genes confers temperature stress tolerance in yeast and. Arabidopsis thaliana. Plant Cell Rep. 36, 1125–1135. doi: 10.1007/s00299-017-2143-y
Ward, J. M., Schroeder, J. I. (1994). Calcium-activated K+ channels and calcium-induced calcium release by slow vacuolar ion channels in guard cell vacuoles implicated in the control of stomatal closure. Plant Cell 6, 669–683. doi: 10.1105/tpc.6.5.669
Wu, X., Shiroto, Y., Kishitani, S., Ito, Y., Toriyama, K. (2009). Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 28, 21–30. doi: 10.1007/s00299-008-0614-x
Xi, Y., Ling, Q., Zhou, Y., Liu, X., Qian, Y. (2022). ZmNAC074, a maize stress-responsive NAC transcription factor, confers heat stress tolerance in transgenic Arabidopsis. Front. Plant Sci. 13, 986628. doi: 10.1016/j.bbagrm.2011.08.004
Xifeng, L., Cai, C., Wang, Z., Fan, B., Zhu, C., Chen, Z. (2018). Plastid translation elongation factor Tu is prone to heat-induced aggregation despite its critical role in plant heat tolerance. Plant Physiol. 176, 3027–3045. doi: 10.1104/pp.17.01672
Xu, C., Bingru, H. (2012). Comparative analysis of proteomic responses to single and simultaneous drought and heat stress for two Kentucky bluegrass cultivars. Crop Sci. 152, 1246–1260. doi: 10.2135/cropsci2011.10.0551
Xu, C., Huang, B. (2010). Differential proteomic response to heat stress in thermal Agrostis scabra and heat-sensitive Agrostis stolonifera. Physiologia Plantarum 139, 192–204. doi: 10.1111/j.1399-3054.2010.01357.x
Yang, M., Zhang, Y., Zhang, H., Wang, H., Wei, T., Che, S., et al. (2017). Identification of MsHsp20 gene family in Malus sieversii and functional characterization of MsHsp16. 9 in heat tolerance. Front. Plant Sci. 8, 1761. doi: 10.3389/fpls.2017.01761
Yoav, B., Hochberg, Y. (2000). On the adaptive control of the false discovery rate in multiple testing with independent statistics. J. Educ. Behav. Stat 25, 60–83. doi: 10.3102/10769986025001060
Yuan, M., Wang, S. (2013). Rice MtN3/saliva/SWEET family genes and their homologs in cellular organisms. Mol. Plant 6, 665–674. doi: 10.1093/mp/sst035
Zang, Yu, Jun, C., Li, R. (2020). Shuai Shang, and Xuexi Tang. Genome-wide analysis of the superoxide dismutase (SOD) gene family in Zostera marina and expression profile analysis under temperature stress. PeerJ 8, e9063. doi: 10.7717/peerj.9063
Zelazny, E., Vert, Grégory (2014). Plant nutrition: root transporters on the move. Plant Physiol. 166, 500–508. doi: 10.1104/pp.114.244475
Zhang, S., Zhang, A., Wu, X., Zhu, Z., Yang, Z., Zhu, Y., et al. (2019). Transcriptome analysis revealed expression of genes related to anthocyanin biosynthesis in eggplant (Solanum melongena L.). under high-temperature stress. BMC Plant Biol. 19, 1–13. doi: 10.1186/s12870-019-1960-2
Zhang, Z.-L., Zhu, J.-H., Zhang, Q.-Q., Cai, Y.-B. (2009). Molecular characterization of an ethephon-induced Hsp70 involved in high and low-temperature responses in Hevea brasiliensis. Plant Physiol. Biochem. 47, 954–959. doi: 10.1016/j.plaphy.2009.06.003
Zhu, B., Ye, C., Lü, H., Chen, X., Chai, G., Chen, J., et al. (2006). Identification and characterization of a novel heat shock transcription factor gene, GmHsfA1, in soybeans (Glycine max). J. Plant Res. 119, 247–256. doi: 10.1007/s10265-006-0267-1
Keywords: abiotic stress, climate resilience, functional genes, heat stress, RNAseq, pearl millet, transcriptomes
Citation: Singh S, Viswanath A, Chakraborty A, Narayanan N, Malipatil R, Jacob J, Mittal S, Satyavathi TC and Thirunavukkarasu N (2024) Identification of key genes and molecular pathways regulating heat stress tolerance in pearl millet to sustain productivity in challenging ecologies. Front. Plant Sci. 15:1443681. doi: 10.3389/fpls.2024.1443681
Received: 04 June 2024; Accepted: 29 July 2024;
Published: 22 August 2024.
Edited by:
Zemin Wang, Gansu Agricultural University, ChinaReviewed by:
Rinku Sharma, Brigham and Women’s Hospital, United StatesAttila Fábián, Hungarian Research Network, Hungary
Copyright © 2024 Singh, Viswanath, Chakraborty, Narayanan, Malipatil, Jacob, Mittal, Satyavathi and Thirunavukkarasu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nepolean Thirunavukkarasu, bmVwb2xlYW5AbWlsbGV0cy5yZXMuaW4=
 Swati Singh
Swati Singh Aswini Viswanath
Aswini Viswanath Animikha Chakraborty
Animikha Chakraborty Neha Narayanan
Neha Narayanan Renuka Malipatil
Renuka Malipatil Jinu Jacob
Jinu Jacob Shikha Mittal
Shikha Mittal Tara C. Satyavathi
Tara C. Satyavathi Nepolean Thirunavukkarasu
Nepolean Thirunavukkarasu