- 1College of Agronomy and Biotechnology, Yunnan Agricultural University, Kunming, China
- 2The Key Laboratory for Crop Production and Smart Agriculture of Yunnan Province, Yunnan Agricultural University, Kunming, China
Low-temperature stress significantly impacts plant growth, development, yield, and geographical distribution. However, during the long-term process of evolution, plants have evolved complicated mechanisms to resist low-temperature stress. The cold tolerance trait is regulated by multiple pathways, such as the Ca2+ signaling cascade, mitogen-activated protein kinase (MAPK) cascade, inducer of CBF expression 1 (ICE1)-C-repeat binding factor (CBF)-cold-reulated gene (COR) transcriptional cascade, reactive oxygen species (ROS) homeostasis regulation, and plant hormone signaling. However, the specific responses of these pathways to cold stress and their interactions are not fully understood. This review summarizes the response mechanisms of plants to cold stress from four aspects, including cold signal perception and transduction, ICE1-CBF-COR transcription cascade regulation, ROS homeostasis regulation and plant hormone signal regulation. It also elucidates the mechanism of cold stress perception and Ca2+ signal transduction in plants, and proposes the important roles of transcription factors (TFs), post-translational modifications (PTMs), light signals, circadian clock factors, and interaction proteins in the ICE1-CBF-COR transcription cascade. Additionally, we analyze the importance of ROS homeostasis and plant hormone signaling pathways in plant cold stress response, and explore the cross interconnections among the ICE1-CBF-COR cascade, ROS homeostasis, and plant hormone signaling. This comprehensive review enhances our understanding of the mechanism of plant cold tolerance and provides a molecular basis for genetic strategies to improve plant cold tolerance.
Introduction
Low-temperature stress significantly affects the growth, development, yield and geographical distribution of plants (Adhikari et al., 2022; Guan et al., 2023). Accumulating evidence indicates that low temperature can increase membrane permeability, impair photosynthesis, cause oxidative damage, and even lead to abundant cell death (Orvar et al., 2000; Lukatkin, 2003; Theocharis et al., 2012; Uritani et al., 2014; Liu et al., 2018). In addition, continuous low-temperature stress can cause severe damage to plants and even lead to death. In the process of crop production, low-temperature stress inhibits seed vigor and the germination rate, slows crop growth, and decreases crop yield and quality (Rutayisire et al., 2020; Xu et al., 2022). However, during the long-term process of evolution, plants have evolved complicated mechanisms to tolerate low-temperature stress.
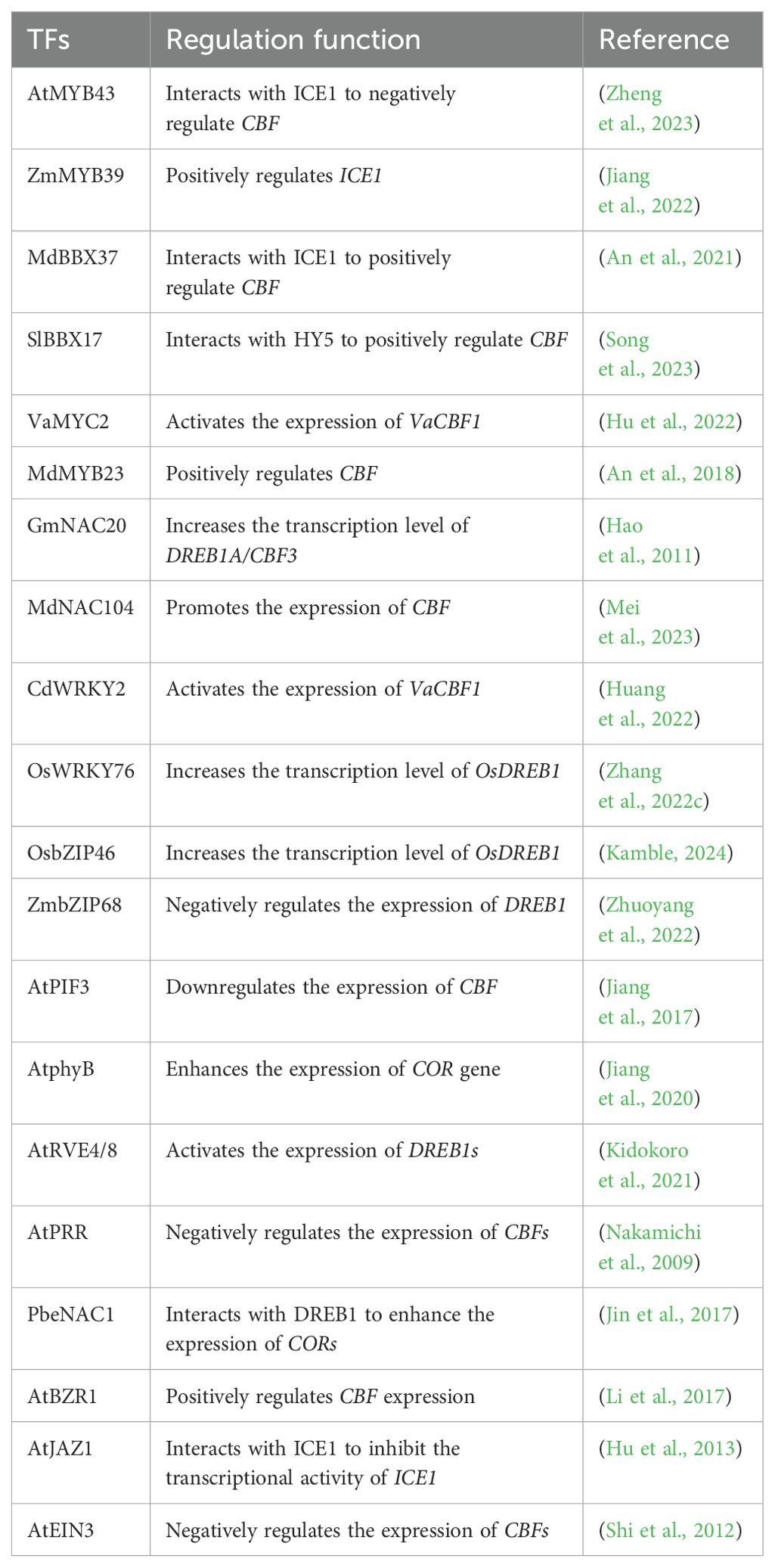
The freezing tolerance of many plants can be increased by cold acclimation, a process involving preexposure to low, nonfreezing temperatures (Zhao et al., 2016; Qi et al., 2022). Cold acclimation is a multifaceted process that encompasses the perception and transduction of environmental cold stress signals, the expression of cold tolerance genes, and specific metabolic and physiological changes (Xiong et al., 2002; Zhu, 2016; Chang et al., 2020). Usually, cold receptors on the plant cell membrane sense low-temperature signals and trigger calcium influx to activate the calcium (Ca2+) signaling cascade network (Ma et al., 2015) and the phosphorylation cascade regulated by protein kinases (Zhao et al., 2017), ultimately activating the response of the downstream ICE1-CBF-COR signaling cascade pathway (Gusain et al., 2023). The ICE1-CBF-COR pathway is considered the most important and archetypal cold stress response pathway in plants. However, the transcriptional activation of the ICE1-CBF-COR cascade is a complex network involving direct or indirect regulation of transcription factors (TFs) (Zhang et al., 2022c; Kamble, 2024), post-translational modifications (PTMs) (Zhao et al., 2017; Ding et al., 2019), light signals (Jiang et al., 2020; Zhang et al., 2020a), circadian clock factors (Kidokoro et al., 2021, Kidokoro et al., 2023) and interacting proteins (Jin et al., 2017; Lee et al., 2021).
In general, plant cells improve cold tolerance by accumulating osmotic regulators such as proline (Pro), soluble sugar (Ss), and soluble protein (Sp), as well as increasing the activities of antioxidant enzymes including catalase (CAT), superoxide dismutase (SOD), ascorbate peroxidase (APX), and peroxidase (POD) (Yan et al., 2019; Ritonga and Chen, 2020; Zhang et al., 2020b; Ramazan et al., 2021). These changes in physiological activities help maintain cell behavior and activity, protect against damage from reactive oxygen species (ROS), and maintain the stability of biologically active cell membranes and protein structures (Ritonga and Chen, 2020). TFs such as basic leucine zipper (bZIP) (Bai et al., 2022), myeloblastosis (MYB) (Yang et al., 2022a) and teosinte branched 1/cycloidea/proliferating cell factor (TCP) (Li et al., 2022b) bind to the promoters of genes encoding antioxidant enzymes, thereby increasing their expression and ultimately enhancing plant cold tolerance. Interestingly, maintaining appropriate ROS levels is essential for plant cold tolerance. Because ROS also act as signaling molecule that activate CBF transcription (Liu et al., 2022), this suggests an interaction between ROS homeostasis and the CBF signaling pathway.
Plant hormones, including abscisic acid (ABA) (Hauser et al., 2017), salicylic acid (SA) (Wang et al., 2020), strigolactones (SL) (Cooper et al., 2018), brassinosteroid (BR) (An et al., 2023), ethylene (ETH) (Wang et al., 2021a) and jasmonic acid (JA) (Hu et al., 2017), also play important roles in plant growth and development and stress response. Cold stress can promote the biosynthetic pathway of these hormones, thereby activating their signaling pathways to respond to cold stress. In addition, there are also interactions between these hormone signaling pathways, such as brassinazole resistant 1 (BZR1, a positive regulator of the BR signaling pathway) enhances 9-cis epoxycarotenoid dioxygenase (NCED1, a key enzyme for the process of ABA synthesis) gene expression and increases the level of ABA (An et al., 2023), thereby increasing cold tolerance in tomato plants. Interestingly, in these plant hormone signaling pathways, ABA regulation of cold tolerance is CBF-independent (Li et al., 2021), whereas BR, SA, SL, ETH, and JA depend on CBF (Ye et al., 2019; Wang et al., 2020, Wang et al., 2021a, Wang et al., 2023b; Yang et al., 2023). In addition, ABA, SA and SL also promote ROS scavenging and regulate cold tolerance (Pan et al., 2020; Li et al., 2021; Wang et al., 2023b). These studies revealed interactions among the plant hormone signaling pathways, CBF transcription and ROS homeostasis.
Although the functions and regulatory mechanisms of many cold tolerance-related genes have been revealed, there is still a lack of systematic reviews on how to integrate these new ideas into the complex mechanism network of the plant response to low temperature. In this review, we summarize four aspects of the response mechanism of plants to low-temperature stress (I. Plant perception and transduction of cold stress signals; II. ICE1-CBF-COR transcription cascade regulation; III. ROS homeostasis regulation; and IV. plant hormone signal regulation), and presents the cross interconnections between these pathways (Figure 1). This comprehensive overview enhances our understanding of plant cold tolerance mechanisms and offers insights for genetic improvements in enhancing plant cold tolerance.
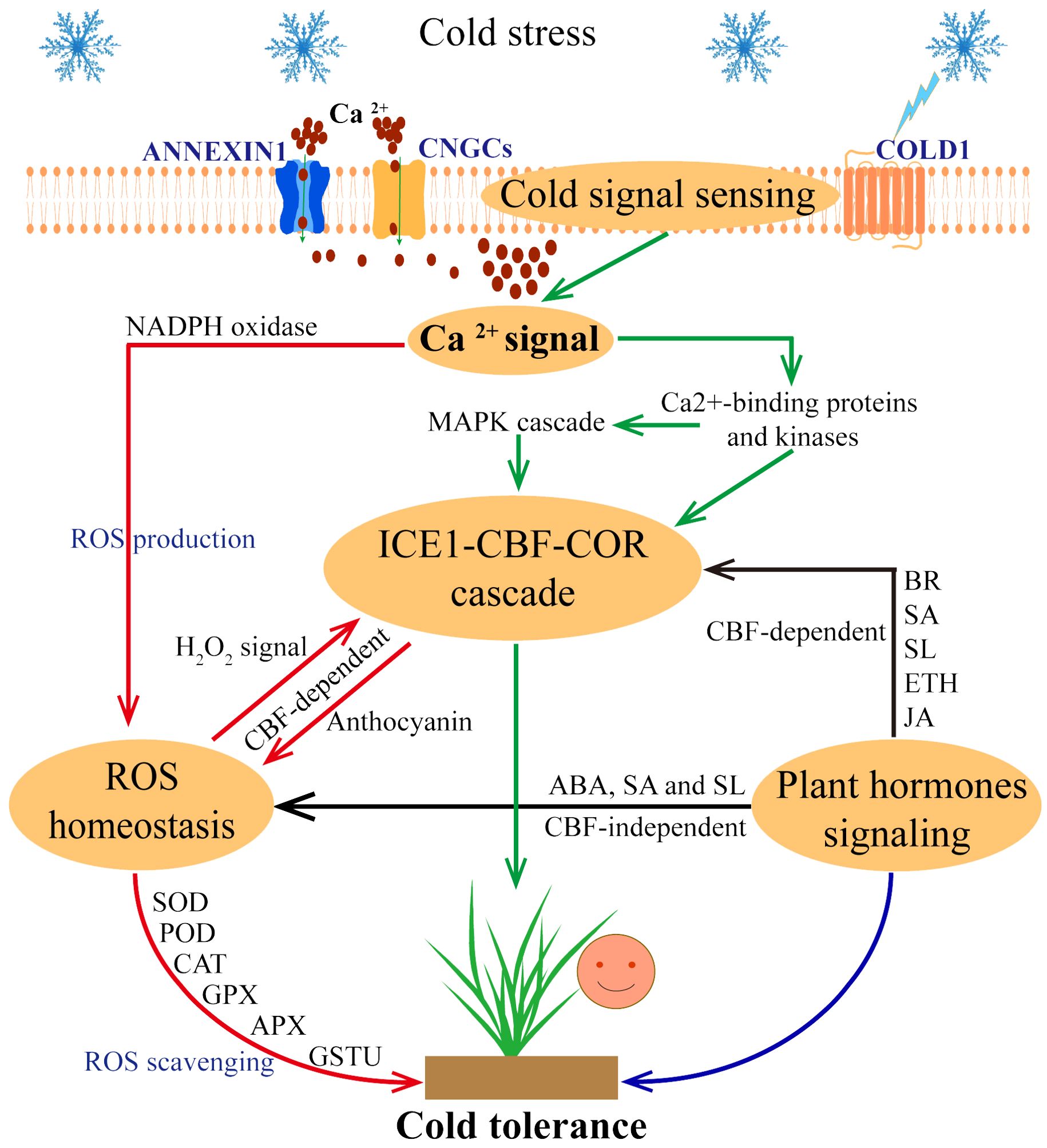
Figure 1. Response strategies of plants to cold stress. When cold stress occurs, plant cells can sense and transduce cold signals, COLD1 senses low temperature and triggers Ca2+ channels (ANNEXIN1 and CNGCs) to transport Ca2+ into the cytoplasm. Further, Ca2+ signaling pathway activates the ICE1-CBF-COR cascade (through the Ca2+-binding proteins and kinases, MAPK cascade pathway) and ROS homeostasis (through the NADPH oxidase pathway). In this process, ICE1-CBF-COR cascade and ROS homeostasis are involved in CBF-dependent ways to respond to cold stress. In addition, plant hormone signaling pathways also regulate plant cold tolerance through CBF-dependence and ROS homeostasis, indicating a cross interconnections between these pathways.
Cold signal perception and transduction
Plants sense low-temperature stress signals, which in turn regulate downstream Ca2+ signaling networks and activate the expression of cold-responsive genes (Figure 2). When plants are exposed to low temperature, chilling-tolerance divergence 1 (COLD1) perceives cold signals, subsequently interacting with G-protein α subunit 1 (RGA1) to activate Ca2+ channels and trigger Ca2+ influx (Ma et al., 2015; Shi and Yang, 2015). Current studies have reported that some Ca2+ channel proteins are activated by cold stress, including cyclic nucleotide-gated channels (CNGCs), Ca2+-permeable transporter (ANNEXIN1), mid1 complementing activity (MCA1) and mid2 complementing activity (MCA2), which mediate Ca2+ transport and positively regulate plant cold tolerance (Wang et al., 2021b; Peng et al., 2024; Mori et al., 2018; Liu et al., 2021). It is well known that at normal temperatures, the activity of these Ca2+ channel proteins is inhibited to maintain lower concentrations of Ca2+ in plant cells, and only when cold stress occurs are they activated to increase the Ca2+ influx. Therefore, it is necessary to further understand the mechanism by which cold stress activates Ca2+ channel protein activity. Interestingly, recent studies have suggested that the activity of CNGCs is activated by the phosphorylation of plant peptide containing sulfated tyrosine1 receptor (PSY1R) and osmotic stress/aba-activated protein kinase 8 (OsSAPK8) under cold stress (Wang et al., 2021b; Peng et al., 2024). In addition, Ca2+ transport activity of ANNEXIN1 was activated by OST phosphorylation under cold stress (Liu et al., 2021). However, CNGC20 is degraded by cold-responsive protein kinase 1 (CRPK1)-mediated phosphorylation under long-term cold stress. Researchers believe that Ca2+ transport mediated by Ca2+ channel proteins is an early event in the plant response to cold stress (Peng et al., 2024). Therefore, it is worth considering how plants in long-term snow-covered areas maintain the transmission of cold signals, which clearly requires further research to reveal.
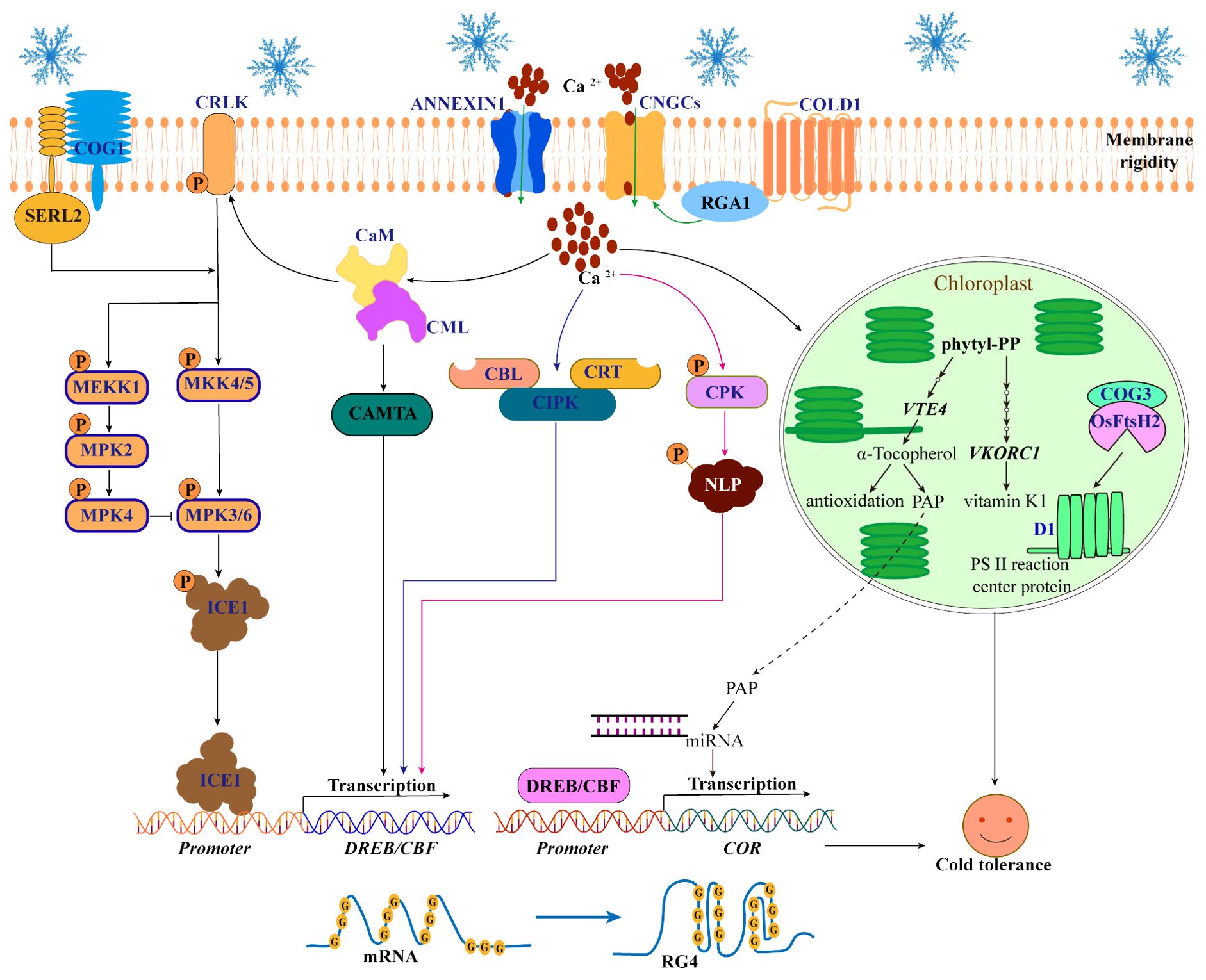
Figure 2. Perception and transduction of cold signals in plants. When cold stress occurs, the COLD1 senses cold signals and then interacts with RGA1 to activate Ca2+ channels (ANNEXIN1 and CNGCs) to release Ca2+ into the cytoplasm. An increase in Ca2+ in the cytoplasm can further activate the regulatory pathways of three Ca2+-binding proteins (CaM/CML, CBLs and CDPK/CPK) to enhance the cold tolerance of plants. First, CaM/CML activates CRLK1 and CAMTA to regulate the MAPK-ICE1-CBF cascade pathway. Second, CBLs and CRT interact with CIPKs to increase the expression levels of DREB/CBF, respectively. Third, CPK phosphorylates NLP7 and promotes its ability to regulate the expression of CBFs. In addition, Ca2+ signaling triggered by COLD1 promotes the biosynthesis pathway of vitamins E and K (By increasing the expression of VTE4 and VKORC1), leading to the production of PAP and its transport into the nucleus to regulate the miRNAs in chloroplasts. Moreover, cold-induced COG3 interaction with OsFtsH2 maintains the balance of photosystem II protein D1 in chloroplasts. Interestingly, the RG4 structure of mRNA in the nucleus and the COG1-OsSERL2 complex in the plasma membrane also function as cold receptors.
As a secondary messenger, Ca2+ plays an important role in the transduction of cold signals in plants (Liu et al., 2021). The increase of Ca2+ levels in cytoplasmic can further activate the regulatory pathways of three Ca2+-binding proteins (calmodulin/calmodulin-like protein (CaM/CML) (Yang et al., 2010a), Ca2+ binding to calcineurin B-like protein (CBL) (Huang et al., 2011) and calcium-dependent protein kinase (CDPK/CPK) (Ding et al., 2022)) to increase the cold tolerance (Figure 2). First, Ca2+ binds to CaM and CML to activate Ca2+/CaM-regulated receptor-like kinase 1 (CRLK1) (Yuan et al., 2018) and calmodulin-binding transcription activator (CAMTA) (Doherty et al., 2009). In Arabidopsis, CRLK1 regulates the mitogen-activated protein kinases (MAPK) cascade pathway: CRLK1 phosphorylation activates the MKK4/5-MPK3/6 cascade to promote the degradation of ICE1 and negatively regulates cold tolerance. However, CRLK1 phosphorylation activates the MEKK1-MKK2-MPK4 cascade to inhibit MPK3/6 phosphorylation to degrade ICE1 and positively regulate cold tolerance (Yang et al., 2010b; Zhao et al., 2017; Duan et al., 2018. In addition, CaM/CML promotes CAMTA3 binding to the DNA motifs (CM2) cis-element on the CBF promoter to positively regulate the expression of CBF and increase cold tolerance (Doherty et al., 2009; Kim et al., 2013). Second, CBL promotes interactions with CBL protein kinases (CIPKs) to increase the expression levels of DREB/CBF and enhance cold tolerance in Arabidopsis (Huang et al., 2011). Moreover, calreticulin 3 (OsCRT3) conformational changes to promote interaction with OsCIPK7 play an important role in increasing the expression levels of CBF in rice (Zhang et al., 2019a; Guo et al., 2023). Third, Ca2+ binds to CDPK/CPK to increase the CPK phosphorylation of nin-like protein 7 (NLP7) and promotes the transfer of NLP7 from the cytoplasm to the nucleus to regulate the expression of the CBF and COR genes, increasing cold tolerance in Arabidopsis (Ding et al., 2022).
In addition to activating pathways regulated by Ca2+-binding proteins, Ca2+ also regulates the cold response pathway in chloroplasts. For example, Ca2+ signaling triggered by COLD1 increases the expression of VTE4 (the gene responsible for α-tocopherol (vitamin E)) and VKORC1 (the key gene in the vitamin K1 pathway) and promotes the biosynthesis pathway of vitamins E and K in chloroplasts, leading to the production of 3′-phosphoadenosine 5′-phosphate (PAP) and its transport into the nucleus to regulate the production of miRNAs, ultimately promoting the expression of cold-responsive genes and improving cold tolerance in rice (Luo et al., 2021). However, the mechanism by which Ca2+ regulates the vitamin network remains unclear. Additionally, chilling-tolerance in Geng/japonica rice 3 (COG3), a putative calmodulin-binding protein, interacts with the proteolytic enzyme OsFtsH2 to remove the cold-damaged photosystem II protein D1 and balance the turnover of the D1 protein in chloroplasts, thereby maintaining photosynthetic efficiency and regulating cold tolerance in rice (Liu et al., 2024). However, it is not clear whether the function of the COG3 is regulated by Ca2+ signaling pathway. Therefore, further elucidation of the mechanism by which Ca2+ signaling activates the cold response pathway in chloroplasts is warranted.
COLD1 is a low-temperature sensor that activates the early Ca2+ signaling network. However, a single low-temperature sensor is not enough to realize the sensing and transduction of cold signals in complex networks. Other cold sensors have also been identified. In Arabidopsis, cold stress triggers the folding of guanine (G-rich) sequences of mRNAs into RNA G-quadruplex (RG4) structures, which inhibits mRNA degradation and increases cold tolerance (Yang et al., 2022b). The author proposed that the RG4 structure, as a low-temperature sensor, can maintain the stability of mRNAs under cold stress. This study demonstrated the importance of the RG4 structure in plant cold tolerance, which is helpful for the excavation and utilization of cold-tolerant germplasm resources. However, we hypothesized whether the RG4 structure is related to the physiological activity and structure of protoplasts under cold stress. In addition, the chilling tolerance in Gengdao 1 (COG1) protein, which is induced by cold stress, interacts with somatic embryogenesis receptor-like kinase 2 (OsSERL2) to form the COG1-OsSERL2 complex in the cell membrane. This complex subsequently activates the MAPK signaling cascade, contributing to enhanced cold tolerance in rice (Changxuan et al., 2023). Both the COG1-OsSERL2 complex and COLD1 are membrane-localized low-temperature sensors that together promote the MAPK cascade, but the COG1-OsSERL2 complex is independent of the Ca2+ signaling pathway (Figure 2).
The ICE1-CBF-COR transcriptional cascade enhances plant cold tolerance
The ICE1-CBF-COR transcriptional cascade, a key component of the downstream COLD1-Ca2+-binding protein-MAPK signaling cascade, is widely recognized as the prototypical and evolutionarily conserved response pathway to cold stress in plants. In general, the transcriptional activity of the ICE1-CBF-COR cascade in cold-tolerant germplasms is greater than that in cold-sensitive germplasms (Todorovska et al., 2014). ICE1, a MYC-like basic helix-loop-helix (bHLH) transcription factor, serves as the central regulator of cold-responsive gene expression (Wang et al., 2023c). In Arabidopsis, ICE1/2 binds to the MYC binding site (CANNTG) in the CBF1/2/3 gene promoter, leading to the upregulation of the CBF1/2/3 gene, which in turn controls the transcription of COR genes and positively regulates cold tolerance (Kim et al., 2015). This transcriptional cascade also plays a role in enhancing cold tolerance in transgenic plants of rice (Verma et al., 2020), Zea mays (Lu et al., 2017) and Zoysia japonica (Zuo et al., 2019). Furthermore, ICE1 not only regulates CBF but can also directly interacts with the COR gene promoter to facilitate COR gene transcription (Tang et al., 2020).
The dehydration-responsive element binding protein/C-repeat binding factors DREB/CBFs belong to the APETALA2/ethylene- responsive element binding factor (AP2/ERF) superfamily of TFs and are pivotal in the ICE1-CBF-COR cascade pathway. In Arabidopsis and rice, DREB1A/CBF3, DREB1B/CBF1, and DREB1C/CBF2 are known as typical cold-inducible genes that are crucial for plant cold stress resistance (Liu et al., 1998; Ito et al., 2006; Zhao et al., 2016). Approximately 10-20% of COR genes in Arabidopsis are estimated to be directly regulated by CBFs (Kosova et al., 2021), which include low temperature-induced (LTI), cold-inducible (KIN), response-to-dehydration (RD) and early dehydration inducible (ERD) gene families (Thomashow, 1998). These families include COR78, COR15, COR6.6, and COR47, with COR47 encoding late embryogenesis-abundant (LEA) family homologous proteins dehydrin (DHN) (Thomashow, 1998). The expression of these COR genes results in the production of hydrophilic peptides that protect cells from freezing damage. In addition, many studies have indicated that the overexpression of these COR genes in transgenic plants increases cold tolerance by scavenging ROS and increasing membrane stability (Vladan et al., 2013; Liu et al., 2015; Guo et al., 2019; Chen et al., 2022). These studies demonstrated the importance of the ICE1-CBF-COR transcriptional cascade in the plant cold stress response. Therefore, identification of the transcriptional activity of the ICE1-CBF-COR transcriptional cascade can be used as molecular evidence for the evaluation of cold-tolerant germplasms.
Regulation of the ICE1-CBF-COR transcriptional cascade
Recent studies revealed that the ICE1-CBF-COR cascade pathway is regulated by various TFs (Table 1). These TFs directly target the ICE1-CBF-COR transcriptional cascade and regulate cold tolerance in plants. For example, MYBs not only bind to the promoter of ICE1 but also interact with the promoter of CBFs to increase the transcription level of ICE1 and CBFs (An et al., 2018; Jiang et al., 2022). In addition, the B-box (BBX), NAC (no apical meristem (NAM), Arabidopsis transcription activation factor (ATAF) and cup-shaped cotyledon (CUC)), WRKY and basic leucine zipper (bZIP) TFs are also involved in regulating CBF transcription (Hao et al., 2011; An et al., 2021; Zhang et al., 2022c; Kamble, 2024). Moreover, some TFs related to light signaling (phytochrome interacting factors (PIF) and phytochrome B (phyB)), circadian clock (REVEILLE4 (RVE) and pseudoresponse regulators (PRRs)), and plant hormone signaling (brassinazole resistant 1 (BZR1), jasmonate zim-domain 1 (JAZ1) and ethylene-insensitive 3 (EIN3)) also play a role in regulating the ICE1-CBF-COR transcriptional cascade (Table 1). Therefore, the transcription of the ICE1-CBF-COR cascade involves a complex network involving many TFs and regulatory pathways. Further analysis of the promoter functional elements will help to elucidate the transcriptional regulatory network upstream of the ICE1-CBF-COR cascade.
Post-translational modifications (PTMs), including phosphorylation, myristoylation and ubiquitination, can change the structure, stability, activity and function of proteins, which is an important reason for protein diversity. These modifications can generally be induced by cold stress and directly or indirectly regulate the ICE1-CBF-COR transcriptional cascade (Ding et al., 2019; Ye et al., 2019; Wang et al., 2022) (Figure 3A). For example, the MPK cascade phosphorylates ICE1 to regulate the stability of ICE1, which is very important for the response of Arabidopsis and rice to cold stress (Zhao et al., 2017). Independent of the MPK cascade, open stomata 1 (OST1) phosphorylates ICE1 to increase its stability, whereas brassinosteroid insensitive 2 (BIN2) phosphorylates ICE1 to promote its degradation (Zhan et al., 2015; Ye et al., 2019). Notably, the phosphorylation of ICE1 by OST1 is regulated by myristoylation and ubiquitination, which may be important strategies for balanced plant growth and cold tolerance. Under normal temperature, clade-E growth-regulating 2 (EGR2) is N‐myristoylated by N-myristoyltransferase 1 (NMT1) to form m-EGR2. The interaction between m-EGR2 and OST1 inhibits the activity of OST1 kinase, which is beneficial for the normal growth of plants. However, under cold stress, EGR2 myristoylation is suppressed, leading to the dominance of unmyristoylated EGR2 (u-EGR2). u-EGR2 inhibits interaction between m-EGR2 and OST1 increasing OST1 activity, facilitating ICE1 phosphorylation by OST1, and maintaining the stability of ICE1 to enhance cold tolerance in Arabidopsis (Ding et al., 2019). In addition, the E3 ubiquitin ligase HOS1 (high expression of osmotically responsive gene 1) can promote ICE1 degradation in Arabidopsis, but this process is inhibited by OST1 under cold stress (Ding et al., 2015). Therefore, the activity of OST1 is regulated by myristoylation and ubiquitination, which can be considered important mechanisms by which ICE1 accurately regulates plant cold tolerance. Moreover, in apple, the U-box-type E3 ubiquitin ligase MdPUB23 promotes the degradation of MdICE1 through the 26S proteasome pathway and negatively regulates cold tolerance (Wang et al., 2022). These studies have shown that phosphorylation, myristoylation and ubiquitination are involved in the transduction of cold signals and directly or indirectly regulate the ICE1-CBF-COR transcriptional cascade. Therefore, investigating the mechanism by which PTMs regulate cold signals will help elucidate the cold response mechanism of plants. However, the types of PTMs are abundant in plant cells (Zhang et al., 2022a), and it is not clear whether other PTMs regulate the ICE1-CBF-COR transcriptional cascade.
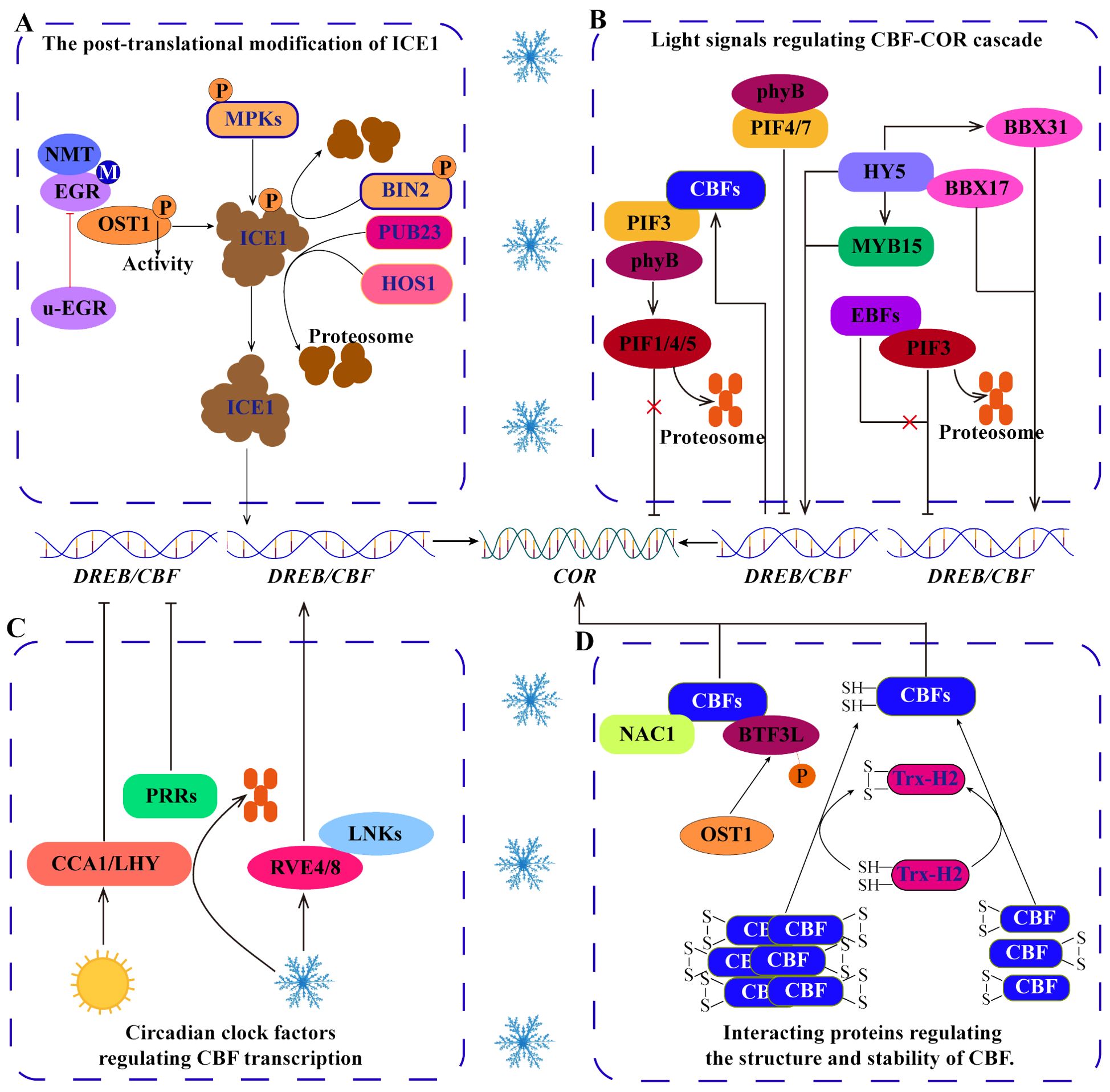
Figure 3. Regulation of the ICE1-CBF-COR signaling cascade. (A) Posttranslational modifications, including phosphorylation, myristoylation and ubiquitination, regulate the stability of the ICE1 protein under cold stress. (B) Light signal pathway TFs regulate the CBF-COR cascade. (C) Circadian clock factors regulate the expression of CBFs. (D) Some proteins interact with CBFs to regulate their structure and stability.
In addition to participating in growth and development, light signaling pathways are also involved in regulating CBF expression and responding to cold stress (Jiang et al., 2017, Jiang et al., 2020) (Figure 3B). Phytochrome interacting factors (PIFs) act as negative regulators that target CBF, with PIF3 binding to CBF promoters to downregulate their expression and negatively regulate cold tolerance. However, the E3 ubiquitin ligase ein3-binding f-box 1/2 (EBF1/2) can interact with PIF3 to promote its degradation through the 26S proteasome pathway (Jiang et al., 2017). In addition, phytochrome B (phyB) interacts with PIFs to precisely regulate CBF expression under long-day and short-day conditions (Lee and Thomashow, 2012). However, how phyB and PIFs perceive low-temperature signals is not clear. Interestingly, the feedback regulation of CBF, where CBF interacts with PIF3 to inhibit the codegradation of the PIF3 and phyB proteins, increases the stability of the phyB protein to promote the degradation of PIF1/4/5 via the 26S proteasome pathway and enhances the expression of the COR gene (Jiang et al., 2020). These studies have shown that the interactions among CBF, PIF and phyB integrate low temperature and light signals and play important roles in balancing plant photomorphogenesis and cold tolerance. Furthermore, elongated hypocotyl 5 (SIHY5), a bZIP-type light-temperature signal integration factor, not only directly regulates the expression of CBFs (Zhang et al., 2020a), but also indirectly increases the level of CBF transcripts by activating MYB and BBX in tomato (Zhang et al., 2020a; Song et al., 2023; Zhu et al., 2023). This information indicates that the integration of light and temperature signaling pathways is critical for CBF transcription.
The expression of cold tolerance genes is usually induced by cold stress, with the expression being inhibited at normal temperatures. Increasing evidence shows that the expression of CBF is also regulated by circadian clock factors (Kidokoro et al., 2023; Kim et al., 2024) (Figure 3C). In Arabidopsis, clock-related MYB TFs (RVE4/LCL1 and RVE8/LCL5) are rapidly transferred from the cytoplasm to the nucleus in response to cold stress, where they activate the expression of DREB1s. In contrast, the central oscillator components of the circadian clock, circadian clock associated 1 (CCA1) and late elongated hypocotyl (LHY), inhibit the expression of DREB1s under normal temperature (Kidokoro et al., 2021). In that process, cold stress enhances the phosphorylation of night light-inducible and clock-regulated (LNK) proteins and promotes the interaction between LNKs and RVEs to positively regulate CBF expression (Kidokoro et al., 2023). In addition, pseudoresponse regulators (PRRs), components of the circadian clock, negatively regulate the expression of CBFs via the circadian rhythm (Kidokoro et al., 2023). Interestingly, HOS15-mediated PRR7 degradation increases the expression of CBF1 and COR15A under dark and cold stress (Kim et al., 2024). These evidences indicate that the circadian clock factors, as important switch for the expression of cold-responsive genes, accurately regulate the cold response of plants. However, it is not clear how circadian clock factors precisely regulate CBF and whether this regulation depends on other modification pathways. PTMs may represent a gap in understanding this pathway, as current evidence suggests that phosphorylation and ubiquitination mediated by LNK and HOS, respectively, are involved in this pathway.
CBF/DREB can bind to CRT/DRE (CCGAC) in the COR promoter to regulate the expression of COR genes and improve cold tolerance (Hwarari et al., 2022). During this process, DREB/CBFs need to interact with other proteins to enhance this regulation (Lee et al., 2021) (Figure 3D). For example, PbeNAC1 from Pyrus betulifolia interacts with PbeDREB1 to upregulate the transcription of CORs and increase cold tolerance in transgenic plants (Jin et al., 2017). In banana, the interaction between MaNAC1 and MaCBF1 enhances cold tolerance (Shan et al., 2014). In addition, thioredoxin h2 (Trx-H2) interacts with CBFs to modify the structure of CBF into monomers under cold stress, enhancing the regulation of COR expression by CBFs (Lee et al., 2021). Moreover, OST1 phosphorylates BTF3‐like protein (BTF3L, a β‐subunit of NAC) to facilitate the interaction of BTF3L with CBFs, increasing the stability of CBF under cold stress and thereby increasing the expression of CORs and cold tolerance in Arabidopsis (Ding et al., 2018). These studies have shown that interacting proteins are very important for the conformational changes and stability of CBF. However, the current understanding of CBF-interacting proteins is still insufficient. Generally, protein structure and stability are closely related to PTMs. Therefore, it is necessary to further study whether PTMs directly act on CBFs.
The ICE1-CBF-COR cascade is clearly a central pathway for the synergistic regulation of different signaling pathways to increase resistance to cold stress. In this complex network, various TFs contribute to the transcriptional level of the ICE1-CBF-COR cascade, and modifications at the protein level regulate its stability. The enhancement of the ICE1-CBF-COR cascade pathway may be one of the strategies for future genetic improvements in plant cold tolerance.
ROS homeostasis regulates plant cold tolerance
Low temperatures can lead to the accumulation of reactive oxygen species (ROS), causing oxidative damage and cell death. To combat cold-induced damage, plants must effectively remove excess ROS. Typically, plants achieve this by increasing the activity of antioxidant enzymes (including SOD, POD, CAT, glutathione peroxidase (GPX), APX, and glutathione S-Transferase (GST)) (He et al., 2019; Ritonga and Chen, 2020; Yang et al., 2022a; Ye et al., 2024) and increasing the accumulation of antioxidants (such as L-ascorbic acid (AsA), Glutathione (GSH) and anthocyanin) (Zhou et al., 2020; Ma et al., 2021; Liu et al., 2023a). The physiological activity and level of antioxidant enzymes are usually induced by cold stress, which is important for ROS scavenging. In this process, TFs such as bZIP, MYB, TCP, and NAC bind to the promoters of genes that encode antioxidant enzymes, thereby promoting their expression to remove excessive ROS (Figure 4). For example, cold-induced AcePosF21 (a bZIP TF) interacts with AceMYB102, directly binds to the promoter of GDP-L-galactose phosphorylase 3 (AceGGP3), a key regulatory gene in AsA biosynthesis, and promotes AsA production, thereby eliminating excessive ROS produced by chilling injury and improving cold tolerance in kiwifruit (Liu et al., 2023a). In chrysanthemum, DgMYBs directly target the promoters of GPX and POD to increase the activity of GPX and POD and reduce the accumulation of ROS (Yang et al., 2022a; Luo et al., 2024b). Similarly, DgbZIP3 and DgTCP1 are capable of binding to the promoter of DgPOD to promote the expression of DgPOD and reduce the accumulation of ROS, thus improving cold tolerance in chrysanthemum (Bai et al., 2022; Li et al., 2022b). In addition, the translocation of de-S-acylated MtNAC80 to the nucleus is mediated by acyl protein thioesterases (MtAPT1), and MtNAC80 subsequently binds to the promoter of MtGSTU1 and promotes its expression, thereby reducing the accumulation of H2O2 and improving the cold resistance of Medicago truncatula (Ye et al., 2024). These studies have shown that TFs are critical for the regulation of antioxidase activity. Therefore, further exploration of cold-inducible TFs that target antioxidant enzyme activities will help to elucidate the antioxidant protection mechanism of plants under cold stress.
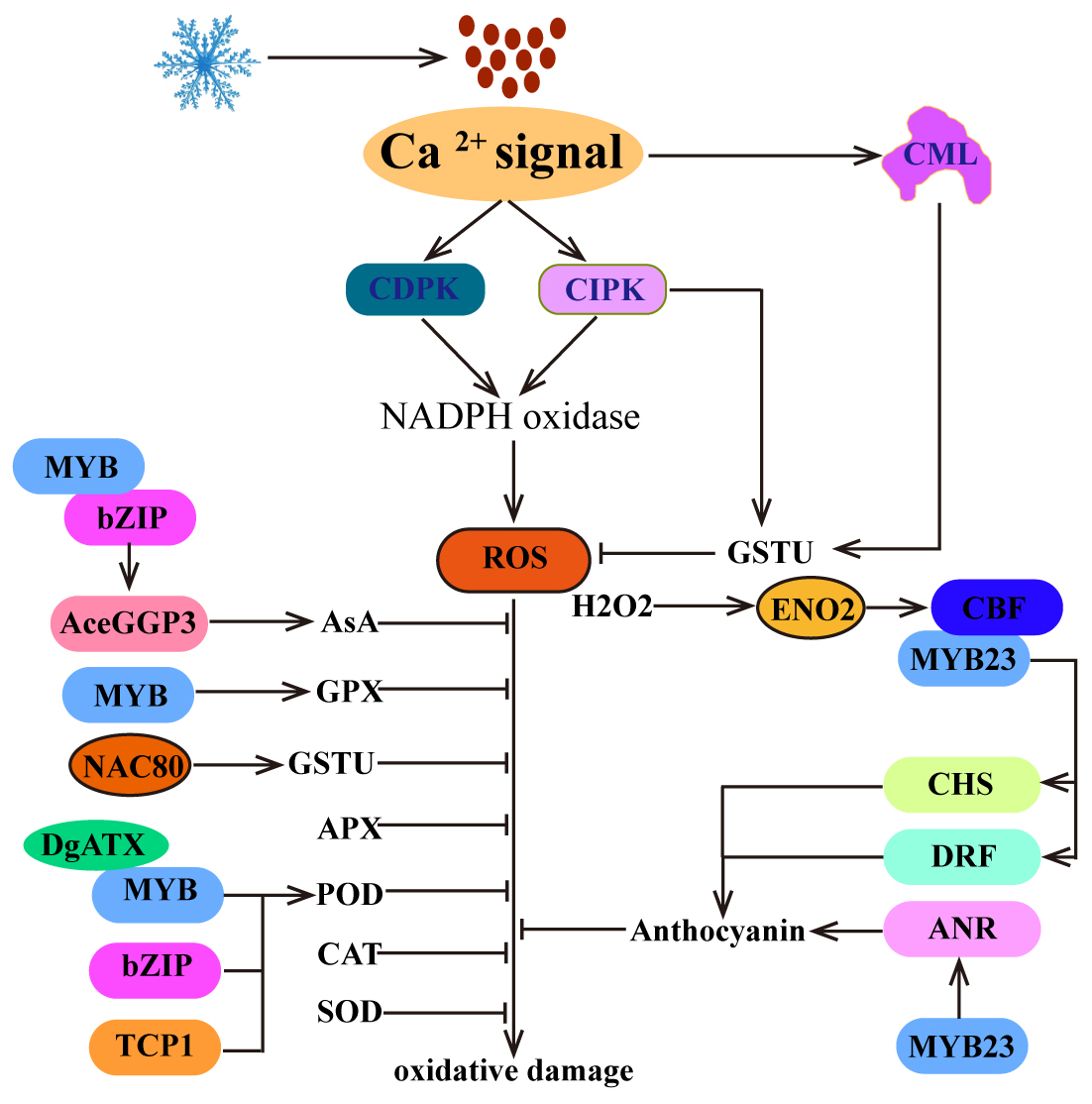
Figure 4. ROS homeostasis regulates plant cold tolerance. The Ca2+ signal is not only associated with the generation of early ROS signals but also promotes the clearance of excessive ROS. In addition, plants can scavenge excess ROS by increasing the activity of antioxidant enzymes, such as SOD, POD, CAT, GPX, APX, GSTU, and AsA. Moreover, H2O2, as a signaling molecule, positively regulates the expression of CBF1. Furthermore, CBF is involved in the biosynthetic pathway of anthocyanins and promotes ROS clearance.
Moreover, cold stress can also induce the biosynthesis of anthocyanins to eliminate the accumulation of ROS, thereby increasing cold tolerance in plants (Figure 4). In this process, MYBs bind to the promoters of anthocyanin biosynthesis-related genes (chalcone synthase (CHS), dihydroflavonol 4-reductase (DRF) and anthocyanidin reductase (ANR)) to promote their expression and anthocyanin biosynthesis. For example, MdMYB23 binds to the promoter of MdANR and activates its expression to promote proanthocyanidin accumulation and ROS scavenging (An et al., 2018). In addition, SmCBFs interact with SmMYB113 to regulate the expression of SmCHS and SmDRF, promoting anthocyanin biosynthesis and ROS scavenging and thereby increasing cold tolerance in transgenic plants (Zhou et al., 2020). Interestingly, in strawberry, FveDREB1B promotes anthocyanin accumulation by directly activating FveCHS (Luo et al., 2024a). These studies have shown that CBF directly or indirectly regulates the cold-induced anthocyanin biosynthesis pathway and promotes ROS scavenging.
Previous studies have shown that cold stress-induced ROS production is derived from nicotinamide adenine dinucleotide phosphate-oxidase (NADPH oxidase) in plant cells (Kawarazaki et al., 2013). The activity of NADPH oxidase is regulated by CDPK and CIPK (Kobayashi et al., 2007; Kimura et al., 2013) (Figure 4). These findings indicate that Ca2+ is closely related to the production of early ROS signals. Interestingly, in Medicago sativa, MsCML10 is activated by cold-induced Ca2+ signaling and interacts with MsGSTU8 to maintain ROS homeostasis and enhance cold tolerance (Yu et al., 2022). In Camellia sinensis, CsCIPK11 phosphorylates CsGSTU23, enhances its stability and positively regulates cold tolerance (Di et al., 2024). These findings suggest that Ca2+ signaling plays a crucial role in the regulation of ROS homeostasis.
While the accumulation of ROS in plants can potentially result in oxidative damage, various studies have shown that ROS also play a crucial role in transmitting signals related to abiotic stress (You and Chan, 2015). In Arabidopsis, increasing H2O2 levels can improve cold tolerance, whereas decreasing H2O2 contents can diminish cold tolerance (Figure 4). This phenomenon is attributed to the fact that cold-induced H2O2 increases the nuclear import of cytosolic enolase 2 (ENO2) by sulfenylating cysteine 408, allowing it to directly bind to the promoter of the CBF1 gene and activate its expression, enhancing cold stress tolerance (Liu et al., 2022). Although there is currently no more evidence to explain the relationship between CBF and ROS, these findings emphasize the importance of H2O2 as a signaling molecule in regulating the expression of CBF, indicating an indirect regulatory relationship between CBF and ROS homeostasis.
Response of hormone signaling pathways to cold stress
Abscisic acid (ABA) is a plant hormone that plays an important role in plant growth, development and stress response. It is derived from zeaxanthin, a process catalyzed by the ABA1 (zeaxanthin epoxidase), ABA4 (neoxanthin synthase), NCED (9-cis-epoxycarotenoid dioxygenase), ABA2 (xanthoxin dehydrogenase) and ABA3 (molybdenum cofactor sulfurase) enzymes in five steps (Hauser et al., 2017). Many studies have shown that appropriate regulation of exogenous or endogenous ABA levels plays an important role in plant cold stress responses (Miura and Nozawa, 2014; Mega et al., 2015; Zhang et al., 2019b; Lim and Lee, 2020; Cai et al., 2021; Guo et al., 2021; Shen et al., 2022). Exogenous ABA treatment can reduce the damage caused by low temperature to cell membranes, increase the endogenous ABA content and improve the cold tolerance in sugarcane (Huang et al., 2015). Similarly, exogenous ABA treatment can increase the activities of POD, CAT, and APX, reduce membrane lipid peroxidation damage and induce endogenous ABA accumulation, and increase cold tolerance in melon (Li et al., 2021). These studies indicate that ABA positively regulates cold tolerance in plants. Cold stress induces the expression of the ABA biosynthesis-related gene NCED and downregulates the ABA degradation gene CYP707A (encoding ABA 8’-hydroxylase), thereby increasing the content of endogenous ABA and improving cold tolerance in Fragaria species (Shen et al., 2022). The overexpression of NCED3 in melon promotes ABA accumulation and triggers the expression of ABRE-binding factor (ABFs), which enhances cold tolerance (Li et al., 2022a). The ABA-responsive element binding protein/ABRE-binding factor (AREB/ABF) is a key TF downstream of ABA signaling. Exogenous ABA treatment and the accumulation of endogenous ABA can increase ScAREB4 transcription, leading to increased cold tolerance in potato plants (Liu et al., 2023b). ABA is oxidized by the ABA8ox enzyme (mainly encoded by the CYP707A1, CYP707A2, CYP707A3 and CYP707A4 genes) into phaseic acid (PA), which is further reduced to inactive dihydrophaseic acid (DPA) (Cutler and Krochko, 2000; Dong et al., 2015). The overexpression of the OsABA8ox1 can reduce the endogenous ABA content and increase the sensitivity of rice to low-temperature stress (Mega et al., 2015). These studies revealed that increasing ABA biosynthesis and preventing ABA catabolism can increase plant cold tolerance. Furthermore, ABA can bind to the Pyrabactin resistance (PYR)/pyrabactin resistance-like (PYL)/regulatory component of ABA receptors (RCAR) receptors, inhibit the activity of protein phosphatase 2C (PP2C), maintain the phosphorylation of SnRK2/OST1 protein kinases, and activate the expression of downstream cold response genes (CORs) (Ma et al., 2009; Boneh et al., 2012; Lim and Lee, 2020). These studies revealed that the ABA biosynthesis pathway gene NCED is upregulated, whereas the ABA degradation gene CYP707A is downregulated in response to cold stress, resulting in endogenous ABA accumulation. Furthermore, ABA binds to PYR/PYL/RCAR receptors, inhibiting PP2C protein phosphatase activity and activating SnRK2/OST1 protein kinases (sucrose-non-fermenting-1-related protein kinase 2) to regulate the expression of downstream cold response genes, ultimately increasing plant cold tolerance (Figure 5). These studies fully demonstrate that ABA positively regulates plant cold tolerance independent of CBF.
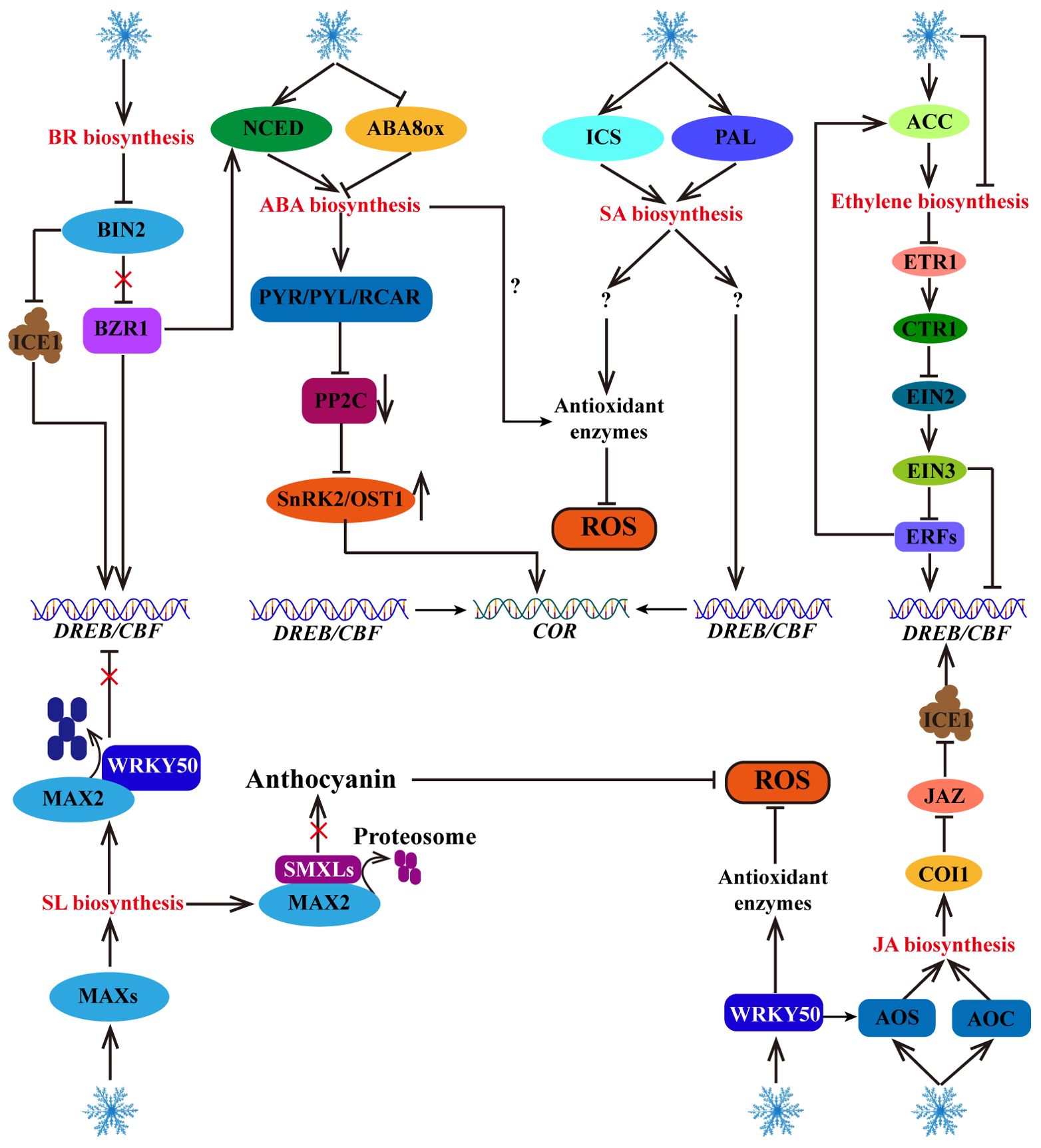
Figure 5. Response of hormone signaling pathways to cold stress. Cold stress increases the biosynthesis of plant hormones, including ABA, BR, JA, SA, ethylene and SL. These plant hormones further activate their signaling pathways to directly or indirectly regulate the CBF-COR cascade. In addition, some plant hormones (such as ABA, SA, SL and JA) contribute to ROS scavenging.
Brassinosteroid (BR) is an important plant hormone that play important roles in the cold stress response. Cold-induced DWARF (DWF, a BR synthesis gene) and brassinazole resistant 1 (BZR1, a positive regulator of the BR signaling pathway), can increase NCED1 gene expression to increase the level of ABA, thereby enhancing cold tolerance in tomato plants (An et al., 2023). In Arabidopsis, BZR1 positively regulates freezing tolerance through CBF-dependent and CBF-independent pathways: low temperature promotes the dephosphorylation of BZR1 to regulate CBF1/CBF2 expression. In addition, dephosphorylated BZR1 regulates the expression of cold response genes (such as WKRY6, PYL6, suppressor of overexpression of constans1 (SOC1), jasmonic acid carboxyl methyltransferase (JMT) and Senescence Associated Gene 21 (SAG21)) through a CBF-independent pathway (Li et al., 2017). Brassinosteroid-insensitive 2 (BIN2) is a negative regulator of the BR signaling pathway that can inhibit BZR1 and CBF, thereby negatively regulating cold tolerance. Cold stress and BR can inhibit the transcription of BIN2 protein kinase, thereby increasing the transcription level of BZR1 and promoting the expression of the NCED1 gene to increase ABA accumulation in tomato plants (An et al., 2023). Additionally, BIN2 can phosphorylate ICE1, facilitating the interaction of ICE1 with the E3 ubiquitin ligase HOS1, thereby promoting ICE1 degradation and negatively regulating CBF gene expression in Arabidopsis (Ye et al., 2019). These studies have shown that BR positively regulates plant cold tolerance through the ABA and CBF pathways (Figure 5).
As an important plant hormone, salicylic acid (SA) not only regulates plant defense immunity but also promotes plant cold tolerance. The synthesis of endogenous SA is derived from the isochorismate synthase (ICS) and phenylalanine ammonia-lyase (PAL) pathways. Cold stress can induce these two pathways and promote SA accumulation (Kim et al., 2013; Dong et al., 2014). Many studies have shown that under low-temperature stress, exogenous SA treatment and endogenous SA accumulation can improve plant antioxidant enzyme activity and ROS scavenging ability, promote the expression of cold response genes, such as WRKY, CBFs, and CORs, and increase plant cold tolerance (Ding et al., 2016; Aazami and Mahna, 2017; Pan et al., 2020; Wang et al., 2020; Li and Wang, 2021). Although SA positively regulates plant cold tolerance through the CBF-dependent pathway and ROS homeostasis regulation, how does SA activate CBF? How can the activity of antioxidant enzymes be promoted? These regulatory mechanisms are not yet clear (Figure 5).
Jasmonic acid (JA) is an important plant hormone involved in the plant response to cold stress. Exogenous treatment with exogenous JA has been shown to increase plant cold tolerance, and the biosynthesis of endogenous JA is activated under cold stress (Hu et al., 2017). In tomato, cold stress-induced SlWRKY50 can bind to the promoter of the allene oxide synthase (SlAOS) gene, a vital enzyme in the JA synthesis pathway, to positively regulate its transcription and facilitate JA biosynthesis (Wang et al., 2023a). In addition, cold stress induces the expression of allene oxide cyclase (MfAOC2), another key enzyme in JA biosynthesis, to promote the accumulation of JA, which increases the expression levels of CBF genes and enhances cold tolerance in Medicago truncatula (Yang et al., 2023). Moreover, under normal temperatures, jasmonate zim-domain (JAZ), an inhibitor of the JA signaling pathway, can interact with ICE1/2 to inhibit the transcriptional activity of ICE1. However, under cold stress conditions, the JA receptor coronatine insensitive 1 (COI1) can degrade JAZs, thereby activating the transcriptional activity of ICE1, promoting CBF gene expression, and improving cold tolerance in Arabidopsis (Hu et al., 2013). These findings demonstrate that cold stress induces the expression of genes involved in JA biosynthesis, leading to JA accumulation and the positive regulation of plant cold tolerance through the ICE1-CBF-COR pathway (Figure 5).
Strigolactones (SLs) are carotenoid phytohormones that not only participate in plant morphogenesis but also play important roles in the cold stress response (Cooper et al., 2018). Cold stress triggers the expression of more axillary growth (MAX) genes to promote the accumulation of SLs (Chi et al., 2021). Furthermore, SLs positively regulate frost resistance through the CBF-dependent pathway (SLs promote the interaction between MAX2 and WRKY41, mediate the degradation of WRKY41 through the 26S proteasome pathway, and relieve the inhibitory effect of WRKY41 on the expression of CBFs) and the CBF-independent pathway (SLs facilitate the interaction between MAX2 and suppressor of max2‐like (SMXLs), increase the degradation of SMXLs, and promote the accumulation of anthocyanin) in Arabidopsis (Wang et al., 2023b) (Figure 5).
Ethylene plays dual roles in the cold stress response of various plants (Figure 5). In tomato, cold-induced SlNAM3 activates the transcription of ACC (1-aminocyclo-propane-1-carboxylic acid) synthase genes (1-Aminocyclopropane-1-carboxylic acid synthase (SlACS1) and 1-aminocyclopropane-1-carboxylic acid oxidase (SlACO)) by directly binding to their promoters to promote ethylene production and increase cold tolerance, indicating that ethylene positively regulates cold tolerance (Dong et al., 2022). In contrast, in Arabidopsis, endogenous ethylene overproduction and exogenous ACC treatment can reduce cold tolerance under plate culture conditions, but cold tolerance is increased by the application of an ethylene biosynthesis inhibitor, indicating that high concentrations of ethylene are not conducive to cold tolerance in Arabidopsis (Shi et al., 2012). In addition, cold stress inhibits ethylene biosynthesis and negatively regulates the ethylene signaling pathway (ACS-ethylene-ethylene receptor (ETR)-constitutive triple response 1 (CTR1)-ethylene insensitive 2 (EIN2)-EIN3-CBF) to increase the transcription level of CBF, indicating that the ethylene signaling pathway negatively regulates CBF in Arabidopsis (Shi et al., 2012). However, it has been found that maintaining adequate levels of endogenous ethylene also contributes to resistance to cold stress in Arabidopsis. For example, under potting soil conditions, cold-induced rare cold inducible 1a (RCI1A) interacts with ACS to decrease ACS stability, leading to a decrease in ethylene biosynthesis and cold tolerance in Arabidopsis (Catala et al., 2014). Since ethylene is a volatile gas, high-humidity plate culture conditions may inhibit the release of ethylene. Unlike in Arabidopsis, the cold tolerance of apple increases after treatment with exogenous ACC but decreases after the application of an ethylene biosynthesis inhibitor (Wang et al., 2021a). Under cold stress, MdERF1B, an ethylene-responsive factor, binds to the promoter of MdACO1 to promote its expression and increase ethylene production, and its cold tolerance further improves via ERF-mediated activation of CBF expression (Wang et al., 2021a). Moreover, cold-induced PtrERF9 enhances ethylene biosynthesis by activating PtrACS1, thereby positively regulating cold tolerance in Poncirus trifoliata (Zhang et al., 2022b). These studies highlight the role of ERF positive feedback in regulating the cold-induced ethylene signaling pathway, and also indicate that ERF-mediated ethylene signaling regulates CBF transcription.
In summary, plant hormones, including ABA, BR, JA, SA, ethylene, and SL, play crucial roles in the response of plants to cold stress. These hormones are involved in CBF-dependent pathways and also contribute to ROS scavenging. The response of the ethylene signaling pathway to cold stress appears to vary among different plant species. However, how cold stress signals activate plant hormone signaling pathways remains unclear.
Conclusion and future perspectives
The response of plants to cold stress is a typical quantitative genetic trait involving the regulation of multiple genes. Cold stress signals are detected by plants through cold sensors, and the transduction and amplification of these signals are mediated by Ca2+ signaling and protein kinase pathways. The ICE1-CBF-COR transcriptional cascade is a key pathway activated to combat low temperature stress. This process involves various factors such as TFs, PTMs, light signals, circadian clock factors, and interacting proteins. Additionally, ROS homeostasis and plant hormone signaling pathways play important roles in the response to cold stress in both CBF-dependent and CBF-independent manners, highlighting the interconnected nature of the ICE1-CBF-COR cascade, ROS homeostasis, and plant hormone signaling.
It is evident from the existing studies that the plant response to cold stress involves a complex regulatory network with interconnected signaling pathways. To gain a better understanding of this network, it is crucial to elucidate the molecular mechanisms underlying the interactions involved in the cold tolerance pathways. Moreover, PTMs, such as phosphorylation, ubiquitination and myristoylation, play a role in regulating TFs and cold-tolerant proteins under cold stress. Further integration of multiomics data, including genome, transcriptome, proteome, and PTMs data, will be essential for a comprehensive investigation of the cold stress regulatory network in plants.
Many studies have shown that manipulating the expression of a few genes can affect the cold tolerance of plants, particularly during the seedling stage under ideal conditions. However, further research is needed to investigate whether these genes can increase cold tolerance throughout the entire growth period in natural environments. Research has indicated that the overexpression of genes related to cold tolerance can lead to a reduction in the biological yield of transgenic plants due to increased energy consumption. For example, the overexpression of DREB/CBF genes in Arabidopsis and rice resulted in dwarf phenotypes (Liu et al., 1998; Ito et al., 2006). This phenomenon is attributed to the constitutive expression of cold tolerance genes, which necessitates energy expenditure. Therefore, the identification of cold-inducible promoters that regulate the expression of cold-tolerant genes, allowing plants to activate these genes selectively in response to cold stress, represents a crucial approach for the future breeding of cold-tolerant crop varieties.
Author contributions
ZQ: Conceptualization, Writing – original draft. LH: Writing – review & editing. FL: Conceptualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The author(s) declare that this work was supported by the Special Project of the Key Laboratory for Crop Production and Smart Agriculture of Yunnan Province (202105AG070007), the Major Science and Technology Project of Yunnan Province (202202AE090021), the National Natural Science Foundation Project of China (31960451) and the Scientific Research Fund Project of Yunnan Provincial Department of Education (2024Y258).
Acknowledgments
We thank Professor Wu Qibin and Professor Que Youxiong, of Fujian Agriculture and Forestry University for guiding and reviewing this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
ABA: abscisic acid
ACO: 1-aminocyclopropane-1-carboxylic acid oxidase
ACS: 1-Aminocyclopropane-1-carboxylic acid synthase
ANNEXIN1: Ca2+-permeable transporter
ANR: anthocyanidin reductase
AOC: allene oxide cyclase
AOS: allene oxide synthase
AP2/ERF: APETALA2/ethylene- responsive element binding factor
APT: acyl protein thioesterases
AREB/ABF: ABA-responsive element binding protein/ABRE-binding factor
AsA: L-ascorbic acid
BBX: B-box
bHLH: basic helix-loop-helix
BIN2: brassinosteroid insensitive 2
BR: brassinosteroid
bZIP: basic leucine zipper
BZR1: brassinazole resistant 1
CaM/CML: calmodulin/calmodulin-like protein
CAMTA: calmodulin-binding transcription activator
CBF: C-repeat binding factor
CBL: calcineurin B-like protein
CCA1: circadian clock associated 1
CDPK/CPK: calcium-dependent protein kinase
CHS: chalcone synthase
CIPKs: CBL protein kinases
CNGCs: cyclic nucleotide-gated channels
COG3: Geng/japonica rice 3
COI1: coronatine insensitive 1
COLD1: chilling-tolerance divergence 1
COR: cold-reulated gene
CRLK1: Ca2+/CaM-regulated receptor-like kinase 1
CRPK1: cold-responsive protein kinase 1
CRT3: calreticulin 3
CTR: constitutive triple response
CYP707A: ABA 8’-hydroxylase
DRF: dihydroflavonol 4-reductase
EBF1/2: ein3-binding f-box 1/2
EGR2: clade-E growth-regulating 2
EIN: ethylene insensitive
ENO2: cytosolic enolase 2
ETH: ethylene
ETR: ethylene receptor
GGP3: GDP-L-galactose phosphorylase 3
GPX: glutathione peroxidase
GSH: Glutathione
GST: glutathione S-Transferase
HOS1: high expression of osmotically responsive gene 1
ICE1: inducer of CBF expression 1
ICS: isochorismate synthase
JA: jasmonic acid
JAZ: jasmonate zim-domain
JMT: jasmonic acid carboxyl methyltransferase
LHY: late elongated hypocotyl
LNK: night light-inducible and clock-regulated
MAPK: mitogen-activated protein kinase
MAX: more axillary growth
MCA1: mid1 complementing activity
MYB: myeloblastosis
NAC: (no apical meristem (NAM), Arabidopsis transcription activation factor (ATAF) and cup-shaped cotyledon (CUC))
NADPH: nicotinamide adenine dinucleotide phosphate
NCED1: 9-cis epoxycarotenoid dioxygenase
NLP7: nin-like protein 7
NMT1: N-myristoyltransferase 1
SERL2: somatic embryogenesis receptor-like kinase 2
OST1: open stomata 1
PAL: phenylalanine ammonia-lyase
phyB: phytochrome B
PIF: phytochrome interacting factors
PP2C: protein phosphatase 2C
PRRs: pseudoresponse regulators
PSY1R: sulfated tyrosine1 receptor
PTMs: post-translational modifications
RCI1A: rare cold inducible 1a
RG4: RNA G-quadruplex
RGA1: G-protein α
ROS: reactive oxygen species
RVE: REVEILLE
SA: salicylic acid
HY5: elongated hypocotyl 5
SL: strigolactones
SMXLs: suppressor of max2‐like
SnRK2: sucrose-non-fermenting-1-related protein kinase 2
SOC1: constans1
TCP: teosinte branched 1/cycloidea/proliferating cell factor
TFs: transcription factors
Trx-H2: thioredoxin h2
References
Aazami, M. A., Mahna, N. (2017). Salicylic acid affects the expression of vvcbf4 gene in grapes subjected to low temperature. J. Genet. Eng. Biotechnol. 15, 257–261. doi: 10.1016/j.jgeb.2017.01.005
Adhikari, L., Baral, R., Paudel, D., Min, D., Makaju, S. O., Poudel, H. P., et al. (2022). Cold stress in plants: Strategies to improve cold tolerance in forage species. Plant Stress. 4, 100081. doi: 10.1016/j.stress.2022.100081
An, J. P., Li, R., Qu, F. J., You, C. X., Wang, X. F., Hao, Y. J. (2018). R2r3-myb transcription factor mdmyb23 is involved in the cold tolerance and proanthocyanidin accumulation in apple. Plant J. 96, 562–577. doi: 10.1111/tpj.14050
An, J. P., Wang, X. F., Zhang, X. W., You, C. X., Hao, Y. J. (2021). Apple b-box protein bbx37 regulates jasmonic acid mediated cold tolerance through the jaz-bbx37-ice1-cbf pathway and undergoes miel1-mediated ubiquitination and degradation. New Phytol. 229, 2707–2729. doi: 10.1111/nph.17050
An, S., Liu, Y., Sang, K., Wang, T., Yu, J., Zhou, Y., et al. (2023). Brassinosteroid signaling positively regulates abscisic acid biosynthesis in response to chilling stress in tomato. J. Integr. Plant Biol. 65, 10–24. doi: 10.1111/jipb.13356
Bai, H., Liao, X., Li, X., Wang, B., Luo, Y., Yang, X., et al. (2022). Dgbzip3 interacts with dgbzip2 to increase the expression of dgpod for cold stress tolerance in chrysanthemum. Hortic. Res. 9, uhac105. doi: 10.1093/hr/uhac105
Boneh, U., Biton, I., Schwartz, A., Ben-Ari, G. (2012). Characterization of the aba signal transduction pathway in vitis vinifera. Plant Sci. 187, 89–96. doi: 10.1016/j.plantsci.2012.01.015
Cai, S., Wang, G., Xu, H., Liu, J., Luo, J., Shen, Y. (2021). Exogenous spermidine improves chilling tolerance in sweet corn seedlings by regulation on abscisic acid, ros and ca 2+ pathways. J. Plant Biol. 64, 1–13. doi: 10.1007/S12374-021-09319-0
Catala, R., Lopez-Cobollo, R., Mar, C. M., Angosto, T., Alonso, J. M., Ecker, J. R., et al. (2014). The arabidopsis 14-3-3 protein rare cold inducible 1a links low-temperature response and ethylene biosynthesis to regulate freezing tolerance and cold acclimation. Plant Cell. 26, 3326–3342. doi: 10.1105/tpc.114.127605
Chang, Y., Zhang, J., Bao, G., Yan, B., Qu, Y., Zhang, M., et al. (2020). Physiological responses of highland barley seedlings to nacl, drought, and freeze-thaw stress. J. Plant Growth Regul. 40, 1–8. doi: 10.1007/s00344-020-10085-5
Changxuan, X., Guohua, L., Kang, C., Yunyuan, X. (2023). The cog1-osserl2 complex senses cold to trigger signaling network for chilling tolerance in japonica rice. Nat. Commun. 14, 3104. doi: 10.1038/S41467-023-38860-4
Chen, N., Fan, X., Wang, C., Jiao, P., Jiang, Z., Ma, Y., et al. (2022). Overexpression of zmdhn15 enhances cold tolerance in yeast and arabidopsis. Int. J. Mol. Sci. 24, 480. doi: 10.3390/ijms24010480
Chi, C., Xu, X., Wang, M., Zhang, H., Fang, P., Zhou, J., et al. (2021). Strigolactones positively regulate abscisic acid-dependent heat and cold tolerance in tomato. Hortic. Res. 8, 237. doi: 10.1038/s41438-021-00668-y
Cooper, J. W., Hu, Y., Beyyoudh, L., Yildiz, D. H., Kunert, K., Beveridge, C. A., et al. (2018). Strigolactones positively regulate chilling tolerance in pea and in arabidopsis. Plant Cell Environ. 41, 1298–1310. doi: 10.1111/pce.13147
Cutler, A. J., Krochko, J. E. (2000). Formation and breakdown of aba. Trends Plant Sci. 4, 472–478. doi: 10.1016/s1360-1385(99)01497-1
Di, T., Wu, Y., Feng, X., He, M., Lei, L., Wang, J., et al. (2024). Cipk11 phosphorylates gstu23 to promote cold tolerance in camellia sinensis. Plant Cell Environ. doi: 10.1111/pce.15070
Ding, Y., Jia, Y., Shi, Y., Zhang, X., Song, C., Gong, Z., et al. (2018). Ost1-mediated btf3l phosphorylation positively regulates cbfs during plant cold responses. Embo. J. 37, e98228. doi: 10.15252/embj.201798228
Ding, Y., Li, H., Zhang, X., Xie, Q., Gong, Z., Yang, S. (2015). Ost1 kinase modulates freezing tolerance by enhancing ice1 stability in arabidopsis. Dev. Cell 32, 278–289. doi: 10.1016/j.devcel.2014.12.023
Ding, Y., Lv, J., Shi, Y., Gao, J., Hua, J., Song, C., et al. (2019). Egr2 phosphatase regulates ost1 kinase activity and freezing tolerance in arabidopsis. Embo. J. 38, e99819. doi: 10.15252/embj.201899819
Ding, Y., Yang, H., Wu, S., Fu, D., Li, M., Gong, Z., et al. (2022). Cpk28-nlp7 module integrates cold-induced ca(2+) signal and transcriptional reprogramming in arabidopsis. Sci. Adv. 8, eabn7901. doi: 10.1126/sciadv.abn7901
Ding, Y., Zhao, J., Nie, Y., Fan, B., Wu, S., Zhang, Y., et al. (2016). Salicylic-acid-induced chilling- and oxidative-stress tolerance in relation to gibberellin homeostasis, c-repeat/dehydration-responsive element binding factor pathway, and antioxidant enzyme systems in cold-stored tomato fruit. J. Agric. Food. Chem. 64, 8200–8206. doi: 10.1021/acs.jafc.6b02902
Doherty, C. J., Van Buskirk, H. A., Myers, S. J., Thomashow, M. F. (2009). Roles for arabidopsis camta transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell. 21, 972–984. doi: 10.1105/tpc.108.063958
Dong, C. J., Li, L., Shang, Q. M., Liu, X. Y., Zhang, Z. G. (2014). Endogenous salicylic acid accumulation is required for chilling tolerance in cucumber (cucumis sativus l.) Seedlings. Planta 240, 687–700. doi: 10.1007/s00425-014-2115-1
Dong, T., Park, Y., Hwang, I. (2015). Abscisic acid: biosynthesis, inactivation, homoeostasis and signalling. Essays. Biochem. 58, 29–48. doi: 10.1042/bse0580029
Dong, Y., Tang, M., Huang, Z., Song, J., Xu, J., Ahammed, G. J., et al. (2022). The mir164a-nam3 module confers cold tolerance by inducing ethylene production in tomato. Plant J. 111, 440–456. doi: 10.1111/tpj.15807
Duan, Z., Qin, X., Zhu, X., Song, C. (2018). Making sense of cold signaling: ice is cold or not cold? Chin. Bull. Bot. 53, 149–153. doi: 10.11983/CBB18039
Guan, Y. L., Hwarari, D., Mateko, K. H., Baseer, A., Cao, Y. W., Movahedi, A., et al. (2023). Low temperature stress-induced perception and molecular signaling pathways in plants. Environ. Exp. Bot. 207, 105190. doi: 10.1016/J.ENVEXPBOT.2022.105190
Guo, Y., Yan, J., Su, Z., Chang, J., Yang, J., Wei, C., et al. (2021). Abscisic acid mediates grafting-induced cold tolerance of watermelon via interaction with melatonin and methyl jasmonate. Front. Plant Sci. 12, 785317. doi: 10.3389/fpls.2021.785317
Guo, X., Zhang, D., Wang, Z., Xu, S., Batistic, O., Steinhorst, L., et al. (2023). Cold-induced calreticulin oscrt3 conformational changes promote oscipk7 binding and temperature sensing in rice. Embo. J. 42, e110518. doi: 10.15252/embj.2021110518
Guo, X., Zhang, L., Wang, X., Zhang, M., Xi, Y., Wang, A., et al. (2019). Overexpression of saussurea involucrata dehydrin gene sidhn promotes cold and drought tolerance in transgenic tomato plants. PloS One 14, e0225090. doi: 10.1371/journal.pone.0225090
Gusain, S., Joshi, S., Joshi, R. (2023). Sensing, signalling, and regulatory mechanism of cold-stress tolerance in plants. Plant Physiol. Biochem. 197, 107646. doi: 10.1016/j.plaphy.2023.107646
Hao, Y. J., Wei, W., Song, Q. X., Chen, H. W., Zhang, Y. Q., Wang, F, et al. (2011). Soybean nac transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 68, 302–313. doi: 10.1111/j.1365-313X.2011.04687.x
Hauser, F., Li, Z., Waadt, R., Schroeder, J. I. (2017). Snapshot: abscisic acid signaling. Cell 171, 1708. doi: 10.1016/j.cell.2017.11.045
He, F., Li, H. G., Wang, J. J., Su, Y., Wang, H. L., Feng, C. H., et al. (2019). Pestz1, a c2h2-type zinc finger transcription factor from populus euphratica, enhances freezing tolerance through modulation of ros scavenging by directly regulating peapx2. Plant Biotechnol. J. 17, 2169–2183. doi: 10.1111/pbi.13130
Hu, Y., Jiang, Y., Han, X., Wang, H., Pan, J., Yu, D. (2017). Jasmonate regulates leaf senescence and tolerance to cold stress: crosstalk with other phytohormones. J. Exp. Bot. 68, 1361–1369. doi: 10.1093/jxb/erx004
Hu, Y., Jiang, L., Wang, F., Yu, D. (2013). Jasmonate regulates the inducer of cbf expression-c-repeat binding factor/dre binding factor1 cascade and freezing tolerance in arabidopsis. Plant Cell. 25, 2907–2924. doi: 10.1105/tpc.113.112631
Hu, Y., Zhang, H., Gu, B., Zhang, J. (2022). The transcription factor vamyc2 from chinese wild vitis amurensis enhances cold tolerance of grape (v. Vinifera) by up-regulating vacbf1 and vap5cs. Plant Physiol. Biochem. 192, 218–229. doi: 10.1016/j.plaphy.2022.10.011
Huang, X., Cao, L., Fan, J., Ma, G., Chen, L. (2022). Cdwrky2-mediated sucrose biosynthesis and cbf-signalling pathways coordinately contribute to cold tolerance in Bermudagrass. Plant Biotechnol. J. 20, 660–675. doi: 10.1111/pbi.13745
Huang, X., Chen, M., Yang, L., Li, Y., Wu, J. (2015). Effects of exogenous abscisic acid on cell membrane and endogenous hormone contents in leaves of sugarcane seedlings under cold stress. Sugar Tech. 17, 59–64. doi: 10.1007/s12355-014-0343-0
Huang, C., Ding, S., Zhang, H., Du, H., An, L. (2011). Cipk7 is involved in cold response by interacting with cbl1 in arabidopsis thaliana. Plant Sci. 181, 57–64. doi: 10.1016/j.plantsci.2011.03.011
Hwarari, D., Guan, Y., Ahmad, B., Movahedi, A., Min, T., Hao, Z., et al. (2022). Ice-cbf-cor signaling cascade and its regulation in plants responding to cold stress. Int. J. Mol. Sci. 23, 1549. doi: 10.3390/ijms23031549
Ito, Y., Katsura, K., Maruyama, K., Taji, T., Kobayashi, M., Seki, M., et al. (2006). Functional analysis of rice dreb1/cbf-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 47, 141–153. doi: 10.1093/pcp/pci230
Jiang, H., Shi, Y., Liu, J., Li, Z., Fu, D., Wu, S., et al. (2022). Natural polymorphism of zmice1 contributes to amino acid metabolism that impacts cold tolerance in maize. Nat. Plants 8, 1176–1190. doi: 10.1038/s41477-022-01254-3
Jiang, B., Shi, Y., Peng, Y., Jia, Y., Yan, Y., Dong, X., et al. (2020). Cold-induced cbf-pif3 interaction enhances freezing tolerance by stabilizing the phyb thermosensor in arabidopsis. Mol. Plant 13, 894–906. doi: 10.1016/j.molp.2020.04.006
Jiang, B., Shi, Y., Zhang, X., Xin, X., Qi, L., Guo, H., et al. (2017). Pif3 is a negative regulator of the cbf pathway and freezing tolerance in arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 114, E6695–E6702. doi: 10.1073/pnas.1706226114
Jin, C., Li, K. Q., Xu, X. Y., Zhang, H. P., Chen, H. X., Chen, Y. H., et al. (2017). A novel nac transcription factor, pbenac1, of pyrus betulifolia confers cold and drought tolerance via interacting with pbedrebs and activating the expression of stress-responsive genes. Front. Plant Sci. 8, 1049. doi: 10.3389/fpls.2017.01049
Kamble, N. U. (2024). Histone deacetylase oshda716 chilling out with osbzip46: antagonistically regulating cold stress tolerance in rice. Plant Cell 36, 1592–1593. doi: 10.1093/plcell/koae025
Kawarazaki, T., Kimura, S., Iizuka, A., Hanamata, S., Nibori, H., Michikawa, M., et al. (2013). A low temperature-inducible protein atsrc2 enhances the ros-producing activity of nadph oxidase atrbohf. Biochim. Biophys. Acta 1833, 2775–2780. doi: 10.1016/j.bbamcr.2013.06.024
Kidokoro, S., Hayashi, K., Haraguchi, H., Ishikawa, T., Soma, F., Konoura, L., et al. (2021). Posttranslational regulation of multiple clock-related transcription factors triggers cold-inducible gene expression in arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 118, e2021048118. doi: 10.1073/pnas.2021048118
Kidokoro, S., Konoura, I., Soma, F., Suzuki, T., Miyakawa, T., Tanokura, M., et al. (2023). Clock-regulated coactivators selectively control gene expression in response to different temperature stress conditions in arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 120, e2078784176. doi: 10.1073/pnas.2216183120
Kim, Y. J., Kim, W. Y., Somers, D. E. (2024). Hos15-mediated turnover of prr7 enhances freezing tolerance. New Phytol. 244, 798–810. doi: 10.1111/nph.20062
Kim, Y. S., Lee, M., Lee, J. H., Lee, H. J., Park, C. M. (2015). The unified ice-cbf pathway provides a transcriptional feedback control of freezing tolerance during cold acclimation in arabidopsis. Plant Mol. Biol. 89, 187–201. doi: 10.1007/s11103-015-0365-3
Kim, Y., Park, S., Gilmour, S. J., Thomashow, M. F. (2013). Roles of camta transcription factors and salicylic acid in configuring the low-temperature transcriptome and freezing tolerance of arabidopsis. Plant J. 75, 364–376. doi: 10.1111/tpj.12205
Kimura, S., Kawarazaki, T., Nibori, H., Michikawa, M., Imai, A., Kaya, H., et al. (2013). The cbl-interacting protein kinase cipk26 is a novel interactor of arabidopsis nadph oxidase atrbohf that negatively modulates its ros-producing activity in a heterologous expression system. J. Biochem. 153, 191–195. doi: 10.1093/jb/mvs132
Kobayashi, M., Ohura, I., Kawakita, K., Yokota, N., Fujiwara, M., Shimamoto, K., et al. (2007). Calcium-dependent protein kinases regulate the production of reactive oxygen species by potato nadph oxidase. Plant Cell. 19, 1065–1080. doi: 10.1105/tpc.106.048884
Kosova, K., Klima, M., Prasil, I. T., Vitamvas, P. (2021). Cor/lea proteins as indicators of frost tolerance in triticeae: a comparison of controlled versus field conditions. Plants 10, 789. doi: 10.3390/plants10040789
Lee, E. S., Park, J. H., Wi, S. D., Kang, C. H., Chi, Y. H., Chae, H. B., et al. (2021). Redox-dependent structural switch and cbf activation confer freezing tolerance in plants. Nat. Plants 7, 914–922. doi: 10.1038/s41477-021-00944-8
Lee, C. M., Thomashow, M. F. (2012). Photoperiodic regulation of the c-repeat binding factor (cbf) cold acclimation pathway and freezing tolerance in arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 109, 15054–15059. doi: 10.1073/pnas.1211295109
Li, M., Duan, X. D., Gao, G., Liu, T., Qi, H. (2022a). Cmabf1 and cmcbf4 cooperatively regulate putrescine synthesis to improve cold tolerance of melon seedlings. Hortic. Res. 9, uhac002. doi: 10.1093/hr/uhac002
Li, B., Wang, W. (2021). Salicylic acid induces tolerance of vitisripariaxv.labrusca to chilling stress by altered photosynthetic, antioxidant mechanisms and expression of cold stress responsive genes. Plant Signal. Behav. 16, 1973711. doi: 10.1080/15592324.2021.1973711
Li, M., Wang, C., Shi, J., Zhang, Y., Liu, T., Qi, H. (2021). Abscisic acid and putrescine synergistically regulate the cold tolerance of melon seedlings. Plant Physiol. Biochem. 166, 1054–1064. doi: 10.1016/j.plaphy.2021.07.011
Li, X., Yang, Q., Liao, X., Tian, Y., Zhang, F., Zhang, L., et al. (2022b). A natural antisense rna improves chrysanthemum cold tolerance by regulating the transcription factor dgtcp1. Plant Physiol. 190, 605–620. doi: 10.1093/plphys/kiac267
Li, H., Ye, K., Shi, Y., Cheng, J., Zhang, X., Yang, S. H. (2017). Bzr1 positively regulates freezing tolerance via cbf-dependent and cbf-independent pathways in arabidopsis. Mol. Plant 10, 545–559. doi: 10.1016/j.molp.2017.01.004
Lim, C. W., Lee, S. C. (2020). Aba-dependent and aba-independent functions of rcar5/pyl11 in response to cold stress. Front. Plant Sci. 11, 587620. doi: 10.3389/fpls.2020.587620
Liu, X., Bulley, S. M., Varkonyi-Gasic, E., Zhong, C., Li, D. (2023a). Kiwifruit bzip transcription factor aceposf21 elicits ascorbic acid biosynthesis during cold stress. Plant Physiol. 192, 982–999. doi: 10.1093/plphys/kiad121
Liu, Q., Ding, Y., Shi, Y., Ma, L., Wang, Y., Song, C., et al. (2021). The calcium transporter annexin1 mediates cold-induced calcium signaling and freezing tolerance in plants. Embo. J. 40, e104559. doi: 10.15252/embj.2020104559
Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Shinozaki, K. Y., et al. (1998). Two transcription factors, dreb1 and dreb2, with an erebp/ap2 dna binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in arabidopsis. Plant Cell. 10, 1391–1406. doi: 10.1105/tpc.10.8.1391
Liu, D., Luo, S., Li, Z., Liang, G., Guo, Y., Xu, Y., et al. (2024). Cog3 confers the chilling tolerance to mediate osftsh2-d1 module in rice. New Phytol. 241, 2143–2157. doi: 10.1111/nph.19514
Liu, W. C., Song, R. F., Qiu, Y. M., Zheng, S. Q., Li, T. T., Wu, Y., et al. (2022). Sulfenylation of enolase2 facilitates h(2)o(2)-conferred freezing tolerance in arabidopsis. Dev. Cell 57, 1883–1898. doi: 10.1016/j.devcel.2022.06.012
Liu, T., Wang, J., Chen, L., Liu, S., Liu, T., Yu, L., et al. (2023b). Scareb4 promotes potato constitutive and acclimated freezing tolerance associated with enhancing trehalose synthesis and oxidative stress tolerance. Plant Cell Environ. 46, 3839–3857. doi: 10.1111/pce.14707
Liu, H., Yu, C., Li, H., Ouyang, B., Wang, T., Zhang, J., et al. (2015). Overexpression of shdhn, a dehydrin gene from solanum habrochaites enhances tolerance to multiple abiotic stresses in tomato. Plant Sci. 231, 198–211. doi: 10.1016/j.plantsci.2014.12.006
Liu, X., Zhou, Y., Xiao, J., Bao, F. (2018). Effects of chilling on the structure, function and development of chloroplasts. Front. Plant Sci. 9, 1715. doi: 10.3389/fpls.2018.01715
Lu, X., Yang, L., Yu, M., Lai, J., Wang, C., McNeil, D., et al. (2017). A novel zea mays ssp. Mexicana l. Myc-type ice-like transcription factor gene zmmice1, enhances freezing tolerance in transgenic arabidopsis thaliana. Plant Physiol. Biochem. 113, 78–88. doi: 10.1016/j.plaphy.2017.02.002
Lukatkin, A. S. (2003). Contribution of oxidative stress to the development of cold-induced damage to leaves of chilling-sensitive plants: 3. Injury of cell membranes by chilling temperatures. Russ. J. Plant Physiol. 50, 243–246. doi: 10.1023/A:1022985500733
Luo, H., Guan, Y., Zhang, Z., Zhang, Z., Zhang, Z., Li, H. (2024a). Fvedreb1b improves cold tolerance of woodland strawberry by positively regulating fvescl23 and fvechs. Plant Cell Environ. doi: 10.1111/pce.15052
Luo, W., Huan, Q., Xu, Y., Qian, W., Chong, K., Zhang, J. (2021). Integrated global analysis reveals a vitamin e-vitamin k1 sub-network, downstream of cold1, underlying rice chilling tolerance divergence. Cell Rep. 36, 109397. doi: 10.1016/j.celrep.2021.109397
Luo, Y., Wang, Y., Li, X., Yang, X., Bai, H., Liao, X., et al. (2024b). Transcription factor dgmyb recruits h3k4me3 methylase to dgperoxidase to enhance chrysanthemum cold tolerance. Plant Physiol. 194, 1104–1119. doi: 10.1093/plphys/kiad479
Ma, Y., Dai, X., Xu, Y., Luo, W., Zheng, X., Zeng, D., et al. (2015). Cold1 confers chilling tolerance in rice. Cell 160, 1209–1221. doi: 10.1016/j.cell.2015.01.046
Ma, B., Suo, Y., Zhang, J., Xing, N., Gao, Z., Lin, X., et al. (2021). Glutaredoxin like protein (rtgrl1) regulates h(2)o(2) and na(+) accumulation by maintaining the glutathione pool during abiotic stress. Plant Physiol. Biochem. 159, 135–147. doi: 10.1016/j.plaphy.2020.11.040
Ma, Y., Szostkiewicz, I., Korte, A., Moes, D., Yang, Y., Christmann, A., et al. (2009). Regulators of pp2c phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. doi: 10.1126/science.1172408
Mega, R., Meguro-Maoka, A., Endo, A., Shimosaka, E., Murayama, S., Nambara, E., et al. (2015). Sustained low abscisic acid levels increase seedling vigor under cold stress in rice (oryza sativa l.). Sci. Rep. 5, 13819. doi: 10.1038/srep13819
Mei, C., Yang, J., Mei, Q., Jia, D., Yan, P., Feng, B., et al. (2023). Mdnac104 positively regulates apple cold tolerance via cbf-dependent and cbf-independent pathways. Plant Biotechnol. J. 21, 2057–2073. doi: 10.1111/pbi.14112
Miura, K., Nozawa, R. (2014). Overexpression of siz1 enhances tolerance to cold and salt stresses and attenuates response to abscisic acid in arabidopsis thaliana. Plant Biotechnol. 31, 167–172. doi: 10.5511/plantbiotechnology.14.0109a
Mori, K., Renhu, N., Naito, M., Nakamura, A., Miura, K. (2018). Ca2+-permeable mechanosensitive channels mca1 and mca2 mediate cold-induced cytosolic ca2+ increase and cold tolerance in arabidopsis. Sci. Rep. 8, 550. doi: 10.1038/s41598-017-17483-y
Nakamichi, N., Kusano, M., Fukushima, A., Kita, M., Ito, S., Yamashino, T., et al. (2009). Transcript profiling of an arabidopsis pseudo response regulator arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 50, 447–462. doi: 10.1093/pcp/pcp004
Orvar, B. L., Sangwan, V., Omann, F., Dhindsa, R. S. (2000). Early steps in cold sensing by plant cells: the role of actin cytoskeleton and membrane fluidity. Plant J. 23, 785–794. doi: 10.1046/j.1365-313x.2000.00845.x
Pan, D. Y., Fu, X., Zhang, X. W., Liu, F. J., Bi, H. G., Ai, X. Z. (2020). Hydrogen sulfide is required for salicylic acid-induced chilling tolerance of cucumber seedlings. Protoplasma 257, 1543–1557. doi: 10.1007/s00709-020-01531-y
Peng, Y., Ming, Y., Jiang, B., Zhang, X., Fu, D., Lin, Q., et al. (2024). Differential phosphorylation of ca2+-permeable channel cngc20 modulates calcium-mediated freezing tolerance in arabidopsis. Plant Cell. 36, 4356–4371. doi: 10.1093/plcell/koae177
Qi, C., Dong, D., Li, Y., Wang, X., Guo, L., Liu, L., et al. (2022). Heat shock-induced cold acclimation in cucumber through cshsfa1d-activated ja biosynthesis and signaling. Plant J. 111, 85–102. doi: 10.1111/tpj.15780
Ramazan, S., Qazi, H. A., Dar, Z. A., John, R. (2021). Low temperature elicits differential biochemical and antioxidant responses in maize (zea mays) genotypes with different susceptibility to low temperature stress. Physiol. Mol. Biol. Plants 27, 1395–1412. doi: 10.1007/s12298-021-01020-3
Ritonga, F. N., Chen, S. (2020). Physiological and molecular mechanism involved in cold stress tolerance in plants. Plants 9, 560. doi: 10.3390/plants9050560
Rutayisire, A., Mukayiranga, A., Habineza, J. C., Avosa, M., Edema, R., Lubadde, G. (2020). Effect of low temperature stress on field performance of highland sorghum (sorghum bicolor (l.) Moench) at flowering stages. J. Plant Breed. Crop Sci. 12, 25–33. doi: 10.5897/jpbcs2019.0844
Shan, W., Kuang, J. F., Lu, W. J., Chen, J. Y. (2014). Banana fruit nac transcription factor manac1 is a direct target of maice1 and involved in cold stress through interacting with macbf1. Plant Cell Environ. 37, 2116–2127. doi: 10.1111/pce.12303
Shen, J., Liu, J., Yuan, Y., Chen, L., Ma, J., Li, X., et al. (2022). The mechanism of abscisic acid regulation of wild fragaria species in response to cold stress. BMC Genomics 23, 670. doi: 10.1186/s12864-022-08889-8
Shi, Y., Tian, S., Hou, L., Huang, X., Zhang, X., Guo, H., et al. (2012). Ethylene signaling negatively regulates freezing tolerance by repressing expression of cbf and type-a arr genes in arabidopsis. Plant Cell. 24, 2578–2595. doi: 10.1105/tpc.112.098640
Shi, Y., Yang, S. (2015). Cold1: a cold sensor in rice. Sci. China Life Sci. 58, 409–410. doi: 10.1007/s11427-015-4831-6
Song, J., Lin, R., Tang, M., Wang, L., Fan, P., Xia, X., et al. (2023). Slmpk1- and slmpk2-mediated slbbx17 phosphorylation positively regulates cbf-dependent cold tolerance in tomato. New Phytol. 239, 1887–1902. doi: 10.1111/nph.19072
Tang, K., Zhao, L., Ren, Y. Y., Yang, S. H., Zhu, J. K., Zhao, C. (2020). The transcription factor ice1 functions in cold stress response by binding to the promoters of cbf and cor genes. J. Integr. Plant Biol. 62, 258–263. doi: 10.1111/jipb.12918
Theocharis, A., Clement, C., Barka, E. A. (2012). Physiological and molecular changes in plants grown at low temperatures. Planta 235, 1091–1105. doi: 10.1007/s00425-012-1641-y
Thomashow, M. F. (1998). Role of cold-responsive genes in plant freezing tolerance. Plant Physiol. 118, 1–8. doi: 10.1104/pp.118.1.1
Todorovska, E. G., Kolev, S., Christov, N. K., Balint, A., Kocsy, G., Vágújfalvi, A., et al. (2014). The expression of CBF genes at Fr-2 locus is associated with the level of frost tolerance in Bulgarian winter wheat cultivars. Biotechnol. Biotechnol. Equip. 28, 392–401. doi: 10.1080/13102818.2014.944401
Uritani, I., Hyodo, H., Kuwano, M. (2014). Mechanism of cold injury in sweet potatoes. Agric. Biol. Chem. 35, 1248–1253. doi: 10.1080/00021369.1971.10860060
Verma, R. K., Kumar, V., Yadav, S. K., Kumar, T. S., Rao, M. V., Chinnusamy, V. (2020). Overexpression of arabidopsis ice1 enhances yield and multiple abiotic stress tolerance in indica rice. Plant Signal. Behav. 15, 1814547. doi: 10.1080/15592324.2020.1814547
Vladan, B., Jan, S., Jürgen, S., Carsten, K. (2013). Dehydrins (lti29, lti30, cor47) from arabidopsis thaliana expressed in escherichia coli protect thylakoid membrane during freezing. J. Serb. Chem. Soc 78, 1149–1160. doi: 10.2298/JSC121127017B
Wang, L., Chen, H., Chen, G., Luo, G., Shen, X., Ouyang, B., et al. (2023a). Transcription factor slwrky50 enhances cold tolerance in tomato by activating the jasmonic acid signaling. Plant Physiol. 194, 1075–1090. doi: 10.1093/plphys/kiad578
Wang, Y., Jiang, H., Mao, Z., Liu, W., Jiang, S., Xu, H., et al. (2021a). Ethylene increases the cold tolerance of apple via the mderf1b-mdcibhlh1 regulatory module. Plant J. 106, 379–393. doi: 10.1111/tpj.15170
Wang, X., Li, Z., Shi, Y., Liu, Z., Zhang, X., Gong, Z., et al. (2023b). Strigolactones promote plant freezing tolerance by releasing the wrky41-mediated inhibition of cbf/dreb1 expression. Embo. J. 42, e112999. doi: 10.15252/embj.2022112999
Wang, K. N., Liu, H. Y., Mei, Q. L., Yang, J., Ma, F. W., Mao, K (2023c). Characteristics of bhlh transcription factors and their roles in the abiotic stress responses of horticultural crops. Sci. Hortic. 310, 111710. doi: 10.1016/J.SCIENTA.2022.111710
Wang, J., Ren, Y., Liu, X., Luo, S., Zhang, X., Liu, X., et al. (2021b). Transcriptional activation and phosphorylation of oscngc9 confer enhanced chilling tolerance in rice. Mol. Plant 14, 315–329. doi: 10.1016/j.molp.2020.11.022
Wang, W., Wang, X., Zhang, J., Huang, M., Cai, J., Dai, T., et al. (2020). Salicylic acid and cold priming induce late-spring freezing tolerance by maintaining cellular redox homeostasis and protecting photosynthetic apparatus in wheat. Plant Growth Regulation: Int. J. On Plant Growth Dev. 90, 109–121. doi: 10.1007/s10725-019-00553-8
Wang, D. R., Zhang, X. W., Xu, R. R., Wang, G. L., You, C. X., An, J. P (2022). Apple u-box-type e3 ubiquitin ligase mdpub23 reduces cold-stress tolerance by degrading the cold-stress regulatory protein mdice1. Hortic. Res. 9, uhac171. doi: 10.1093/hr/uhac171
Xiong, L., Schumaker, K. S., Zhu, J. K. (2002). Cell signaling during cold, drought, and salt stress. Plant Cell. 14 Suppl, S165–S183. doi: 10.1105/tpc.000596
Xu, H., Hassan, M. A., Sun, D., Wu, Z., Jiang, G., Liu, B. (2022). Effects of low temperature stress on source-sink organs in wheat and phosphorus mitigation strategies. Front. Plant Sci. 13, 807844. doi: 10.3389/fpls.2022.807844
Yan, L., Tariq, S., Cheng, Y., Lü, Y., Zhang, X., Zuo, X. (2019). Physiological and molecular responses to cold stress in rapeseed(brassica napus l.). J. Integr. Agric. 18, 2742–2752. doi: 10.1016/S2095-3119(18)62147-1
Yang, T., Chaudhuri, S., Yang, L., Du, L., Poovaiah, B. W. (2010a). A calcium/calmodulin-regulated member of the receptor-like kinase family confers cold tolerance in plants. J. Biol. Chem. 285, 7119–7126. doi: 10.1074/jbc.M109.035659
Yang, X., Luo, Y., Bai, H., Li, X., Tang, S., Liao, X., et al. (2022a). Dgmyb2 improves cold resistance in chrysanthemum by directly targeting dggpx1. Hortic. Res. 9, uhab028. doi: 10.1093/hr/uhab028
Yang, T., Shad, A. G., Yang, L., Du, L., Reddy, A. S., Poovaiah, B. W. (2010b). Calcium/calmodulin-regulated receptor-like kinase crlk1 interacts with mekk1 in plants. Plant Signal. Behav. 5, 991–994. doi: 10.4161/psb.5.8.12225
Yang, L., Sun, Q., Geng, B., Shi, J., Zhu, H., Sun, Y., et al. (2023). Jasmonate biosynthesis enzyme allene oxide cyclase 2 mediates cold tolerance and pathogen resistance. Plant Physiol. 193, 1621–1634. doi: 10.1093/plphys/kiad362
Yang, X., Yu, H., Duncan, S., Zhang, Y., Cheema, J., Liu, H., et al. (2022b). Rna g-quadruplex structure contributes to cold adaptation in plants. Nat. Commun. 13, 6224. doi: 10.1038/s41467-022-34040-y
Ye, K., Li, H., Ding, Y., Shi, Y., Song, C., Gong, Z., et al. (2019). Brassinosteroid-insensitive2 negatively regulates the stability of transcription factor ice1 in response to cold stress in arabidopsis. Plant Cell. 31, 2682–2696. doi: 10.1105/tpc.19.00058
Ye, Q., Zheng, L., Liu, P., Liu, Q., Ji, T., Liu, J., et al. (2024). The s-acylation cycle of transcription factor mtnac80 influences cold stress responses in medicago truncatula. Plant Cell. 36, 2629–2651. doi: 10.1093/plcell/koae103
You, J., Chan, Z. (2015). Ros regulation during abiotic stress responses in crop plants. Front. Plant Sci. 6, 1092. doi: 10.3389/fpls.2015.01092
Yu, S., Wu, J., Sun, Y., Zhu, H., Sun, Q., Zhao, P., et al. (2022). A calmodulin-like protein (cml10) interacts with cytosolic enzymes gstu8 and fba6 to regulate cold tolerance. Plant Physiol. 190, 1321–1333. doi: 10.1093/plphys/kiac311
Yuan, P., Yang, T., Poovaiah, B. W. (2018). Calcium signaling-mediated plant response to cold stress. Int. J. Mol. Sci. 19, 3896. doi: 10.3390/ijms19123896
Zhan, X., Zhu, J. K., Lang, Z. (2015). Increasing freezing tolerance: kinase regulation of ice1. Dev. Cell 32, 257–258. doi: 10.1016/j.devcel.2015.01.004
Zhang, D., Guo, X., Xu, Y., Li, H., Ma, L., Weng, Y., et al. (2019a). Oscipk7 point-mutation leads to conformation and kinase-activity change for sensing cold response. J. Integr. Plant Biol. 61, 1194–1200. doi: 10.1111/jipb.12800
Zhang, L., Jiang, X., Liu, Q., Ahammed, G. J., Lin, R., Zhang, L., et al. (2020a). The hy5 and myb15 transcription factors positively regulate cold tolerance in tomato via the cbf pathway. Plant Cell Environ. 43, 2712–2726. doi: 10.1111/pce.13868
Zhang, B., Li, S., Shui, W. (2022a). Post-translational modifications of g protein-coupled receptors revealed by proteomics and structural biology. Front. Chem. 10, 843502. doi: 10.3389/fchem.2022.843502
Zhang, Y., Ming, R., Khan, M., Wang, Y., Dahro, B., Xiao, W., et al. (2022b). Erf9 of poncirus trifoliata (l.) Raf. Undergoes feedback regulation by ethylene and modulates cold tolerance via regulating a glutathione s-transferase u17 gene. Plant Biotechnol. J. 20, 183–200. doi: 10.1111/pbi.13705
Zhang, W., Wang, J., Huang, Z., Mi, L., Xu, K., Wu, J., et al. (2019b). Effects of low temperature at booting stage on sucrose metabolism and endogenous hormone contents in winter wheat spikelet. Front. Plant Sci. 10, 498. doi: 10.3389/fpls.2019.00498
Zhang, X., Zhang, L., Sun, Y., Zheng, S., Wang, J., Zhang, T., et al. (2020b). Hydrogen peroxide is involved in strigolactone induced low temperature stress tolerance in rape seedlings (brassica rapa l.). Plant Physiol. Biochem. 157, 402–415. doi: 10.1016/j.plaphy.2020.11.006
Zhang, M., Zhao, R., Huang, K., Huang, S., Wang, H., Wei, Z., et al. (2022c). The oswrky63-oswrky76-osdreb1b module regulates chilling tolerance in rice. Plant J. 112, 383–398. doi: 10.1111/tpj.15950
Zhao, C., Wang, P., Si, T., Hsu, C. C., Wang, L., Zayed, O., et al. (2017). Map kinase cascades regulate the cold response by modulating ice1 protein stability. Dev. Cell 43, 618–629. doi: 10.1016/j.devcel.2017.09.024
Zhao, C., Zhang, Z., Xie, S., Si, T., Li, Y., Zhu, J. K. (2016). Mutational evidence for the critical role of cbf transcription factors in cold acclimation in arabidopsis. Plant Physiol. 171, 2744–2759. doi: 10.1104/pp.16.00533
Zheng, P., Cao, L., Zhang, C., Fang, X., Wang, L., Miao, M., et al. (2023). The transcription factor myb43 antagonizes with ice1 to regulate freezing tolerance in arabidopsis. New Phytol. 238, 2440–2459. doi: 10.1111/nph.18882
Zhou, L., He, Y., Li, J., Liu, Y., Chen, H. (2020). Cbfs function in anthocyanin biosynthesis by interacting with myb113 in eggplant (solanum melongena l.). Plant Cell Physiol. 61, 416–426. doi: 10.1093/pcp/pcz209
Zhu, J. K. (2016). Abiotic stress signaling and responses in plants. Cell 167, 313–324. doi: 10.1016/j.cell.2016.08.029
Zhu, Y., Zhu, G., Xu, R., Jiao, Z., Yang, J., Lin, T., et al. (2023). A natural promoter variation of slbbx31 confers enhanced cold tolerance during tomato domestication. Plant Biotechnol. J. 21, 1033–1043. doi: 10.1111/pbi.14016
Zhuoyang, L., Diyi, F., Xi, W., Rong, Z., Xuan, Z., Tian, J., et al. (2022). Natural variation in the bzip68 promoter modulates cold tolerance and was targeted during maize domestication. Plant Cell 34, 2833–2851. doi: 10.1093/plcell/koac137
Keywords: cold stress, cold signal perception and transduction, ICE1-CBF-COR transcription cascade, ROS homeostasis, plant hormone signal
Citation: Qian Z, He L and Li F (2024) Understanding cold stress response mechanisms in plants: an overview. Front. Plant Sci. 15:1443317. doi: 10.3389/fpls.2024.1443317
Received: 03 June 2024; Accepted: 09 October 2024;
Published: 06 November 2024.
Edited by:
Balpreet Kaur Dhatt, Bayer Crop Science, United StatesReviewed by:
Muhammad Ahmad Hassan, Anhui Academy of Agricultural Sciences, ChinaLaxman Adhikari, King Abdullah University of Science and Technology, Saudi Arabia
Copyright © 2024 Qian, He and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fusheng Li, bGZzODEwQHNpbmEuY29t
 Zhenfeng Qian1
Zhenfeng Qian1 Fusheng Li
Fusheng Li