- 1Division of Plant Pathology, ICAR-Indian Agricultural Research Institute, New Delhi, India
- 2ICAR-National Institute for Plant Biotechnology, New Delhi, India
- 3ICAR-Directorate of Rapeseed-Mustard Research, Bharatpur, India
White rust disease caused by a biotrophic oomycete Albugo candida is one of the most serious impediments in realizing the production potential of Brassica juncea. Due to the obligate nature of the pathogen, R-gene-based resistance is unstable as the newer virulent races emerge quickly. For this, a deep understanding of the molecular basis of resistance is essential for developing durable resistant varieties. In this study, we selected one susceptible cultivar, ‘Pusa Jaikisan’ and its single R gene based resistant NIL, ‘Pusa Jaikisan WRR as the source of understanding the defense mechanism in B. juncea against A. candida. Comparative histochemical analysis at 12 dpi showed higher callose deposition in the resistant cultivar than in the susceptible which hints towards its possible role in defense mechanism. Based on the biochemical markers observation, total protein was found to have a negative correlation with the resistance. The antioxidant enzymes (POX, CAT, and SOD) and non-enzymatic ROS scavenging compounds such as polyphenols and proline showed a positive correlation with the white rust resistance. Polyphenol Oxidase (PPO) total chlorophyll and total carotenoids were also found to be more abundant in the ‘Pusa Jaikisan WRR’. Based on the heat map analysis, PAL was identified to be the comparatively most induced enzyme involved in the defense mechanism. The polyphenol oxidase, total chlorophyll and total carotenoids were also found to show higher activity in the ‘Pusa Jaikisan WRR’. Furthermore, to study the defense response of ‘Pusa Jaikisan WRR’ compared to ‘Pusa Jaikisan’ against A. candida infection, the gene expression analyses of salicylic acid (SA)-marker PR protein genes (PR1 and PR2) and jasmonic acid (JA)-marker PR protein genes (PR3 and PR12) were done by qRT-PCR. Based on the results, PR2 emerged as the best possible gene for defense against A. candida followed by PR1. PR3 and PR12 also showed positive correlation with the disease resistance which may be due to the JA pathway acting complementary to the SA pathway in case of B. juncea-A. candida interaction. This provides evidence for the JA-SA hormonal crosstalk to be synergistic in case of the white rust resistance.
1 Introduction
Regarding economic significance, oilseeds are second only to cereal crops. The global trends of area, production, and yield in the year 2021-22 were 41.95 mha, 88.35 mt and 2110 kg/ha, respectively (ICAR-DRMR, 2023). In India, rapeseed mustard has a share of one-fourth of total edible oil production and cultivated area. Out of this, Indian mustard [Brassica juncea (L.) Czern. And Coss.] has the lion’s share (about 36%) in India’s edible oil kitty (ICAR-DRMR, 2023). India imported 1.3 mt of edible oil in June 2023, which is alarming (The Solvent Extractors’ Association of India, 2023). On the other hand, many biotic and abiotic stresses are serious impediments to realizing the potential production of B. juncea. Among all these, white rust disease caused by Albugo candida is a globally important problem. It is known to cause losses ranging from 1% to 90% which depends on a multitude of factors such as plant population, host genotype, nutrition (Saharan and Verma, 1992). Chemicals such as Mancozeb and a combination of metalaxyl and mancozeb have proven to be quick and effective methods of disease management but they come with their own set of economically and environmentally negative spillovers (Gairola and Tewari, 2019). Breeding for disease resistance is the best strategy for managing the disease (Ren et al., 2016). A. candida being an obligate biotroph shares a strong co-evolutionary relationship with the host and this enables quick racial evolution and breaks the already existing R gene-based resistance. Given these factors, genetic engineering for the broad-spectrum non-host resistance (NHR) genes such as pathogenesis-related protein (PRs) genes is the most apt method for managing the dreaded white rust (Grover, 2012; Rai et al., 2023). Their expression pattern can be useful in identifying their role in resistance and future use. Further, a set of biochemical markers such as antioxidant enzymes, act as surrogate measures of resistance. These can be coupled with the histochemical parameters such as callose deposition in the inner walls which acts as determinants of resistance activation (Kaur et al., 1984). Together these three can be utilized for developing a thorough understanding of the defense mechanism against A. candida and developing durable transgenics.
As plants are sessile, they have evolved an array of mechanisms to thwart the growth of pathogens upon invasion. Among these, callose deposition, induction of pathogenesis related proteins (PR), catalase, superoxide dismutase and peroxidase, production of phenolics, and reactive oxygen species (ROS) are the foremost (Torres et al., 2006; Doughari, 2015). When a plant encounters the pathogen, it initiates a two-fold resistance response. The primary or basal response is mediated by recognizing the pathogen-associated molecular patterns (PAMPs) through the PAMP recognition receptors (PRRs). This is termed as the PAMP-triggered immunity (PTI) (Jones and Dangl, 2006). PTI induces the production of PR proteins, ROS, cell wall reinforcement and antimicrobial compounds produced through the mitogen-activated protein kinases (MAPKs) mediated signaling pathways (Bigeard et al., 2015). Cell death caused by ROS is usually favored by the pathogen in plant-necrotroph interaction but is an important component of disease resistance in the case of plant-biotroph interaction (Govrin and Levine, 2000; Pietrowska et al., 2015. Cell death is a necessary process, but too much can be detrimental to the plant itself. Hydrogen peroxide (H2O2) is one of the most important components of ROS and accumulates after the plant recognizes the pathogen. Hydrogen peroxide is converted into water and oxygen in peroxisome by catalase (CAT) (EC 1.11.1.6) (Afiyanti and Chen, 2014), whereas peroxidases (POX) (EC 1.11.1.7) reduce H2O2 along with many phenolics and non-phenolics substrates. Superoxide dismutase (SOD) (EC 1.15.1.1) negates the harmful effect of superoxide radicals by dismutation into oxygen and water (Bittner et al., 2017). The polyphenols play an important role by neutralizing the ROS, while polyphenol oxidase (PPO) (EC 1.10.3.2) catalyzes the oxygen-dependent oxidation of polyphenols to quinones. Together these two maintain ROS-homeostasis, which helps in plant defense (Arora et al., 2000; Kaur et al., 2017). Proline is an important non-enzymatic antioxidant that can also neutralize ROS and act as a balancing wheel (Verbruggen and Hermans, 2008). Phenylalanine ammonia-lyase (PAL) (EC 4.3.1.24) is one of the most crucial enzymes in the defense pathway. It catalyzes the deamination process of phenylalanine into secondary phenylpropanoid metabolism (Hahlbrock and Scheel, 1989). For producing these enzymes, the protein is broken down hence decreasing its concentration upon resistance activation (Bolwell and Wojtaszek, 1997). The relationship of chlorophyll and carotenoids with resistance have not been strongly ascertained and need future studies.
PR proteins along with hormonal responses induced by complex signaling pathways are the major determinants of plant resistance and are strongly induced upon pathogen inoculation (van Loon et al., 2006; Grover, 2012). These are small proteins which are accumulated and induced in response to an array of biotic and abiotic stresses (Ebrahim et al., 2011). At present, PR proteins are classified into 17 families and among them, the major ones are PR1 (antifungal), PR2 (β-1,3-glucanase), PR3 (chitinase), PR5 (thaumatin-like protein), PR9 (peroxidases), PR12 (plant defensin), PR13 (thionins) (van Loon et al., 2006). Regarding hormonal signaling-mediated resistance, salicylic acid (SA) and jasmonic acid/ethylene (JA/ET) are important components. They are thought to be mutually antagonistic. The JA/ET pathway is initiated upon attack by a necrotroph while in the case of a biotroph, the SA pathway is predominant. Multiple reports hint at a positive role of JA/ET in the case of biotroph attacks where they complement the SA pathway (Glazebrook, 2005). PR1, PR2 and PR5 are the SA-marker genes, while PR3, PR12 and PR13 are the JA/ET-marker genes (Thomma et al., 2001). As, B. juncea suffers from a heterogeneous group of pathogens which includes both biotrophs and necrotrophs, the signaling pathway genes may help in developing broad-spectrum durable resistance. Several transgenic plants over-expressing PR genes have been developed against B. juncea pathogens imparting disease resistance (Huang et al., 2016; Dong et al., 2017; Ali et al., 2018).
A comparative study of the disease resistance factors in resistant and susceptible cultivars of B. juncea is essential to develop durable transgenics. Here we have chosen B. juncea cv. Pusa Jaikisan (susceptible) and B. juncea cv. Pusa Jaikisan WRR (resistant) (Arora et al., 2019) source for white rust resistance and studied the disease progression, callose deposition after infection, defense-related enzymes and non-enzymatic compounds, and the role of PR genes as markers of the hormonal signaling pathway in resistance against A. candida.
2 Materials and methods
2.1 Plant materials and fungal materials
Two cultivars of Indian mustard (B. juncea), ‘Pusa Jaikisan’ and ‘Pusa Jaikisan WRR’ were used as plant materials and the seeds were obtained from the ICAR-Directorate of Rapeseed Mustard Research, Bharatpur (India). These experiments were carried out from November 2022 to March 2023 at the Division of Plant Pathology, ICAR- Indian Agricultural Research Institute, New Delhi (India). The research complied with the institutional, national, and international guidelines and legislation.
Albugo candida of New Delhi isolate Ac-Dli was originally isolated from ‘Pusa Jaikisan’ (B. juncea) from the field of Division of Genetics, ICAR- Indian Agricultural Research Institute. For maintaining the Ac-Dli isolate, seedlings of ‘Pusa Jaikisan’ were used. Seven-day-old plants were inoculated by spraying Ac-Dli with the concentration of 1 X 105 zoosporangia/ml, followed by incubating in a moist chamber for 24 h at 220C under the dark conditions and then in a growth chamber for 16 h light and 8 h dark at 210C, with regular irrigation. The process was repeated sufficiently for maintenance. The isolate Ac-Dli was maintained in the Division of Plant Pathology, ICAR- Indian Agricultural Research Institute.
2.2 Inoculation test
Seeds of ‘Pusa Jaikisan’ and ‘Pusa Jaikisan WRR’ were sown in trays in three replications each and were kept under 16 h light alternated with 8 h dark at 210C. Seven-day-old plants were inoculated by spraying Ac-Dli with a concentration of 1 X 105 zoosporangia/ml. To confirm the successful inoculation, plants were incubated in a dark growth chamber for 24 h at 220C under dark conditions and then in a growth chamber for 16 h light and 8 h dark at 210C, with regular irrigation. After 12 days, ‘Pusa Jaikisan’ showed characteristic white-colored zoosporangial pustules on both the adaxial and abaxial surface of the cotyledons, while no symptoms were seen on the ‘Pusa Jaikisan WRR’ on either side.
The susceptibility of ‘Pusa Jaikisan’ and resistance of ‘Pusa Jaikisan WRR’ were confirmed by maintaining the plants in a dark growth chamber for 24 h at 220C with 100% humidity. The plants were then moved into a growth chamber and provided controlled conditions for 16 h light and 8 h dark at 210C, with regular irrigation. For gene expression profiling and biochemical assays, 14-day-old seedlings were inoculated by spraying Ac-Dli with a concentration of 1 X 105 zoosporangia/ml. The infected leaf samples were harvested from ‘Pusa Jaikisan’ and ‘Pusa Jaikisan WRR’ at 12, 24, 48, 72 and 96 hpi, snap chilled in liquid nitrogen and stored at -800C for further analyses.
2.3 Disease parameters
Disease resistance or susceptibility were assayed on the leaves of the three plants at 12 dpi. A white rust disease scoring scale of 0 to 9 was used to characterize the plants in which 0, 1, 3, 5, 7 and 9 correspond to 0, <5, 5-10, 11-25, 26-50 and >50% diseased area, respectively (Bits et al., 2018). The diseased area comprised white-colored zoosporangial pustules. The experiment was carried out in three replications and for a single replication, five leaves were taken from each plant.
2.4 Confocal microscopy
Callose deposition near plasmodesmata was observed according to Zavaliev and Epel (2014). For tissue staining and sample preparation, the leaf sample was collected at 12 dpi from both ‘Pusa Jaikisan’ and ‘Pusa Jaikisan WRR’. The entire leaf was cut by holding it along the petiole and submerged in 96% ethanol solution. The container having the sample was covered and sealed with parafilm. Incubation was done at room temperature (RT) to bleach the sample. The bleached leaf was removed from the ethanol solution. The leaf was placed on a flat surface and cut strips 5 mm wide using a razor blade. Utmost care was taken to sample the same regions of both leaves as there may be differences in callose deposition in different regions. The sample was then rehydrated by sinking the strips with the help of DDW with 0.01% Tween-20 followed by incubation at RT for 1h. Strips were removed from DDW and placed in a 25 ml glass tube filled to 1/3 with aniline blue solution by using a wire loop. The vacuum desiccator was used for 10 min to get good dye penetration. Tubes were wrapped with aluminum foil and incubated at RT for 2 h. Specimens were then taken for microscopy and were mounted on microscopic slides and observed under a confocal microscope at ICAR- Indian Agricultural Research Institute. (Model: Leica TCS SP52). Configuration of confocal microscopy was built in a single-track mode. The 405 nm diode laser was used for excitation, while the 475-525 nm band-passed emission filter for the aniline blue fluorescence. A white pseudo color was used for aniline blue emission. The important area under the microscope showing disease progression was captured by a camera mounted on the system and photomicrographs were processed in Image J software (Collins, 2007).
2.5 Biochemical assays
The biochemical assay was carried out from the infected sample at 0, 12, 24, 48, 72 and 96 hpi. The enzyme extract was prepared by homogenizing 200 mg of leaf sample in liquid Nitrogen. Ice cold 1.5 ml of 50mM Sodium phosphate buffer (pH 7.0), 2mM EDTA, 5mM β-mercaptoethanol and 4% Polyvinylpyrrolidone (PVP) were added to the homogenized powder (except for TPC where no PVP was used). The homogenate was centrifuged at 3000 rpm for 30 min at 40C. The supernatant was stored in aliquots at -400C. This enzyme extract was used for total protein, polyphenols, PPO, PAL, CAT, SOD, and POX estimation.
2.5.1 Protein content
The total protein content was determined by using the Bradford's approach (1976). The bovine serum albumin served as the standard. The assay combination included 2 ml Bradford reagent, 990 µl water, and 10 µl enzyme extract. The absorbance was recorded at 595 nm by using a UV-visible spectrophotometer. Total protein content was expressed as mg/g dry weight.
2.5.2 POX activity
The POX activity was determined by following the Chance and Maehly (1955) methodology. In this, three solutions; Solution A; Sodium phosphate buffer (100mM, pH 7.0), Solution B: 10 mM H2O2 solution and Solution C: 20mM guaiacols were freshly prepared. 1.5 ml Solution A and 0.12 ml Solution B were taken in a cuvette. Absorbance increment was recorded at 470 nm wavelength for 3 min at intervals of 1 min each. The reaction mixture without enzyme served as blank. Tetra guaiacol (26.6 mM-1cm-1) was used as the extinction coefficient of the oxidation product.
2.5.3 CAT activity
CAT activity was determined by following the methodology described by Aebi (1984). 0.1 ml enzyme extract was mixed with 1.5 ml solution A and 0.4 ml distilled water. The decrease in absorbance was recorded for 3 min at 30 s intervals at 240 nm just after the addition of 1ml solution B. The extinction coefficient of H2O2 (39.4 mM-1 cm-1) was used for quantification.
2.5.4 Superoxide dismutase activity
Superoxide dismutase activity was measured following the Beauchamp and Fridovich (1971) method. The assay solution contained 27 ml of 50 mM sodium phosphate buffer (pH 7.8), 1 ml of 1.72 mM nitroblue tetrazolium (NBT), 1.5 ml of 201 mM methionine, and 0.75 ml of 1% Triton X-100. To prepare the enzyme extract, plant tissues were homogenized in the sodium phosphate buffer and centrifuged at 10,000 g for 15 minutes at 4°C. The supernatant was used for the assay. For the assay, 1 ml of the solution mix, 0.1 ml of enzyme extract, and 0.03 ml of 20 μM riboflavin were combined in a test tube and incubated in a foil-lined box with a 40W fluorescent bulb for 10 minutes. The reaction was terminated by switching off the light, and absorbance was measured at 560 nm. A reaction mixture without the enzyme extract served as the blank, representing 100% NBT reduction.
2.5.5 Total polyphenol content
The total polyphenol content was determined by Singleton and Rossi (1965) method. A diluted test sample (0.5 ml) was reacted with 0.2 ml Folin-Ciocalteau reagent, and the reaction was neutralized with sodium carbonate (0.5 ml). 10 ml distilled water was used to make up the volume. The absorbance of the blue color was measured at 760 nm. The gallic acid standard curve was used for quantification.
2.5.6 PPO activity
Polyphenol oxidase activity was measured following Meyer et al. (1998) method. Fresh leaves (0.2 g) were homogenized in 2 ml of pH 6.5 0.1 M sodium phosphate buffer and centrifuged at 16,000 rpm for 15 min at 40C. The enzyme was obtained from the supernatant. 200 µl of enzyme extract and 1.5 ml of 0.1 phosphate buffer (pH 6.5) were used for the process. The reaction was started by using 200 µl of 0.01 M catechol, and the enzyme activity was measured at 495 nm/min/mg protein.
2.5.7 PAL activity
PAL activity was measured following the methodology described by Edward and Kessmann (1992). 25 mM tris buffer (pH 8.8) was used as the homogenization buffer. After adding 0.1 ml of enzyme extract and 0.4 ml of 0.05 M tris HCL buffer (PH 8.8) having 0.2 mM phenylalanine, the reaction mixture was incubated in a water bath at 370C for 60 min. To stop the reaction, 0.1 ml of 0.5 N HCL was added. 2 ml of toluene was used to extract the trans-cinnamic acid. The absorbance was measured at 412 nm followed by calculating enzymatic activity.
2.5.8 Proline content
The total proline content was calculated following the methodology described by Koca et al. (2007). 0.2 g of fresh leaf sample was homogenized in 2 ml of 3% w/v distilled water sulfosalicylic acid and centrifuged at 10,000 rpm for 30 min at 4°C. 1 ml of supernatant was combined with 1 ml of glacial acetic acid and 1 ml of acid ninhydrin reagent and incubated for 1 hour at 100°C in a water bath before being immediately plunged into an ice bath to stop the reaction. 2 ml of toluene was after that transferred to each reaction mixture tube. The chromophore was finally extracted by vigorously swirling on a vortex mixer. A spectrophotometer was used to measure the absorbance of the chromophore-comprising toluene layer at 520 nm.
2.5.9 Chlorophyll content
The chlorophyll content ( mg/g fresh weight) was calculated by placing the 10 mg of fresh tissue in a 2 µl microcentrifuge and 700 ul of preheated DMSO was added to the tube containing the leaf tissue. For 30 min, the tube was placed in a 650C water bath. Following incubation, 300 µl of DMSO was added to achieve a volume of 1 ml. The absorbance was measured at three different wavelengths: 470 nm, 645 nm, and 663 nm. Arnon’s equations were used to calculate the chlorophyll content (Richardson et al., 2002). Lichtenthaler and Welburn’s (1983) methodology was used to determine the total carotenoid content (mg/g fresh weight) by using the same enzyme extract.
2.6 Gene expression profiling
For the gene expression profiling, RNA was isolated from the infected samples at 0, 12, 24, 48, 72 and 96 hpi by RNeasy® Plant Mini Kit QIAGEN (Germany) as described by the manufacturer’s protocol. For determining the purity and concentration of RNA, the Nanodrop spectrophotometer (NanoDrop2000 Thermo Scientific, Wilmington, DE) was used. First-strand cDNA was synthesized using total RNA isolated from the leaf samples by Promega ImProm-IITM Reverse Transcription system (USA) following the manufacturer’s protocol. The oligoanalyzer software was used for designing the primers of PR1, PR2, PR3 and PR12 genes. For this purpose, the conserved regions were used (Table 1). The genes PR1 and PR2 are SA-marker genes, while PR3 and PR12 are JA/ET-marker genes and are used for ascertaining their role in the defense pathway of B. juncea-A. candida interaction. The qRT-PCR reaction mixture contained 2 µl of cDNA, 5 µl of SYBR green qRT-PCR master mix (Takara kit, Japan) and 0.5 µl of each primer. Thermal conditions were kept at 940C for 3 min, followed by 40 cycles of 940C for 30 s, 600C for 30 s, and 720C for 30 s. Alpha-tubulin was used as the housekeeping gene which acted as internal control. The reactions were performed in triplicates and repeated using three biological replicates and the delta CT method was used to analyze the relative expression level of each gene (Livak and Schmittgen, 2001). Fold changes with p ≤ 0.05 were taken as the significant level of expression.
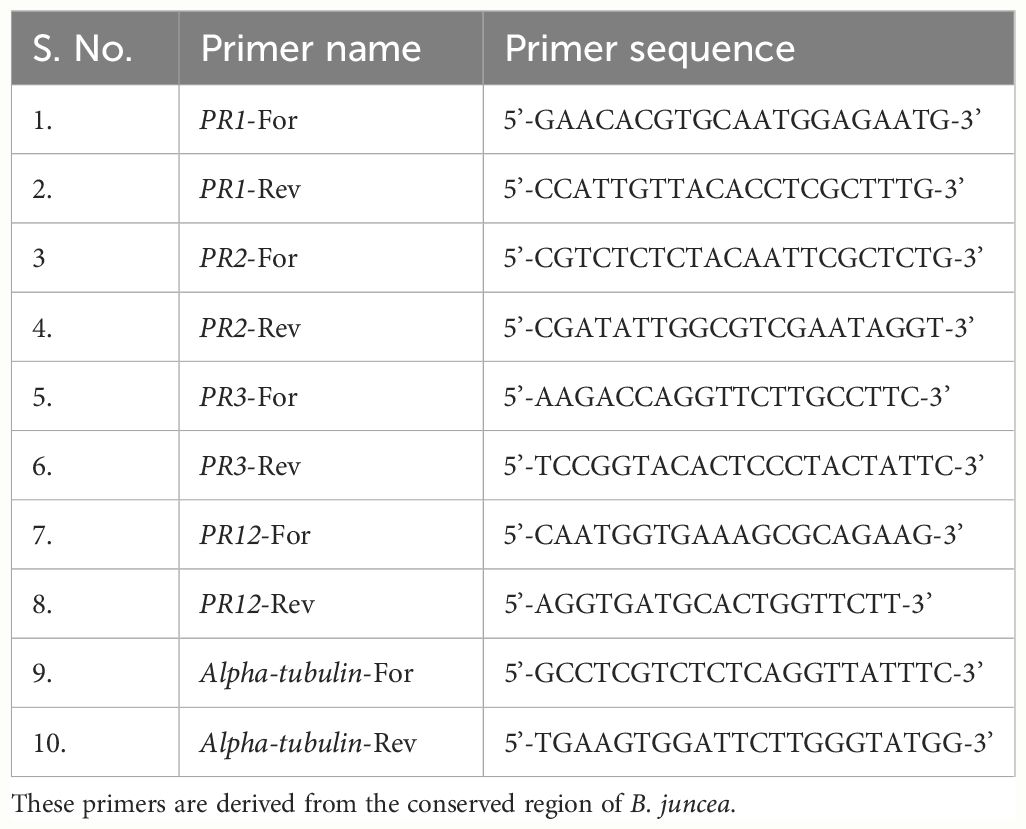
Table 1 List of primers used for the qRT-PCR analysis of PR1, PR2, PR3 and PR12 genes at 0, 12, 24, 48, 72 and 96 hpi.
2.7 Statistical analyses
A complete randomized design was set up for the experiment and the disease was scored by taking three replicates from each infected plant. The results were analyzed by using the one-way analysis of variance (ANOVA) p ≤ 0.05 level of significance through Microsoft Excel.
3 Results
3.1 Disease severity
Disease severity was recorded at 12 dpi. ‘Pusa Jaikisan’ was found to be highly susceptible with more than 50% area covered by pustules and a rating score of 9, while ‘Pusa Jaikisan WRR’ had no pustules on both adaxial and abaxial surface with 0% leaf area covered and rating of 0 on the 0-9 scale (Figure 1) (Bits et al., 2018). Disease reaction was found to be immune in the case of ‘Pusa Jaikisan WRR’. A susceptible disease reaction was found in ‘Pusa Jaikisan’.
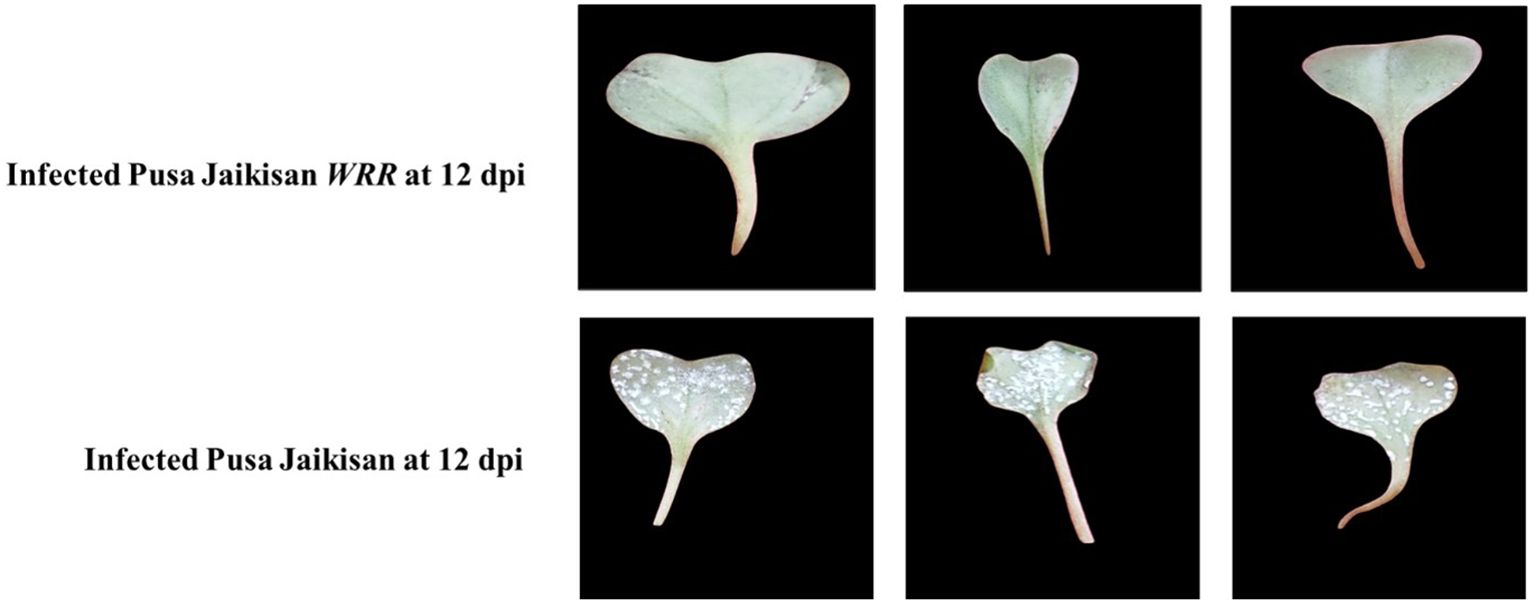
Figure 1 Comparative disease parameters in B. juncea cv. Pusa Jaikisan and B. juncea cv. Pusa Jaikisan WRR. The experiment was carried out in three replications Diseased area was more than 50% in the case of ‘Pusa Jaikisan WRR’, while it was 0% for ‘Pusa Jaikisan’.
3.2 Comparative callose deposition
Callose deposition is an important determinant of resistance and was histochemically analyzed through aniline blue staining. Based on the confocal microscopic examinations of both the specimens at 12 dpi, i.e., infected ‘Pusa Jaikisan’ and ‘Pusa Jaikisan WRR’, we found the callose to be deposited at plasmodesmata, between the plant cell wall and plasma membrane, and on other plant tissues. The callose deposition was observed at different scales i.e., 250 μm and 100 μm for ‘Pusa Jaikisan WRR’ and 250 μm and 75 μm in the case of ‘Pusa Jaikisan’. The quantum of callose deposition at the site of pathogen intrusion was found to be way higher in the case of ‘Pusa Jaikisan WRR’ as compared to ‘Pusa Jaikisan’ (Figure 2). This hinted towards its possible role in the defense against white rust.
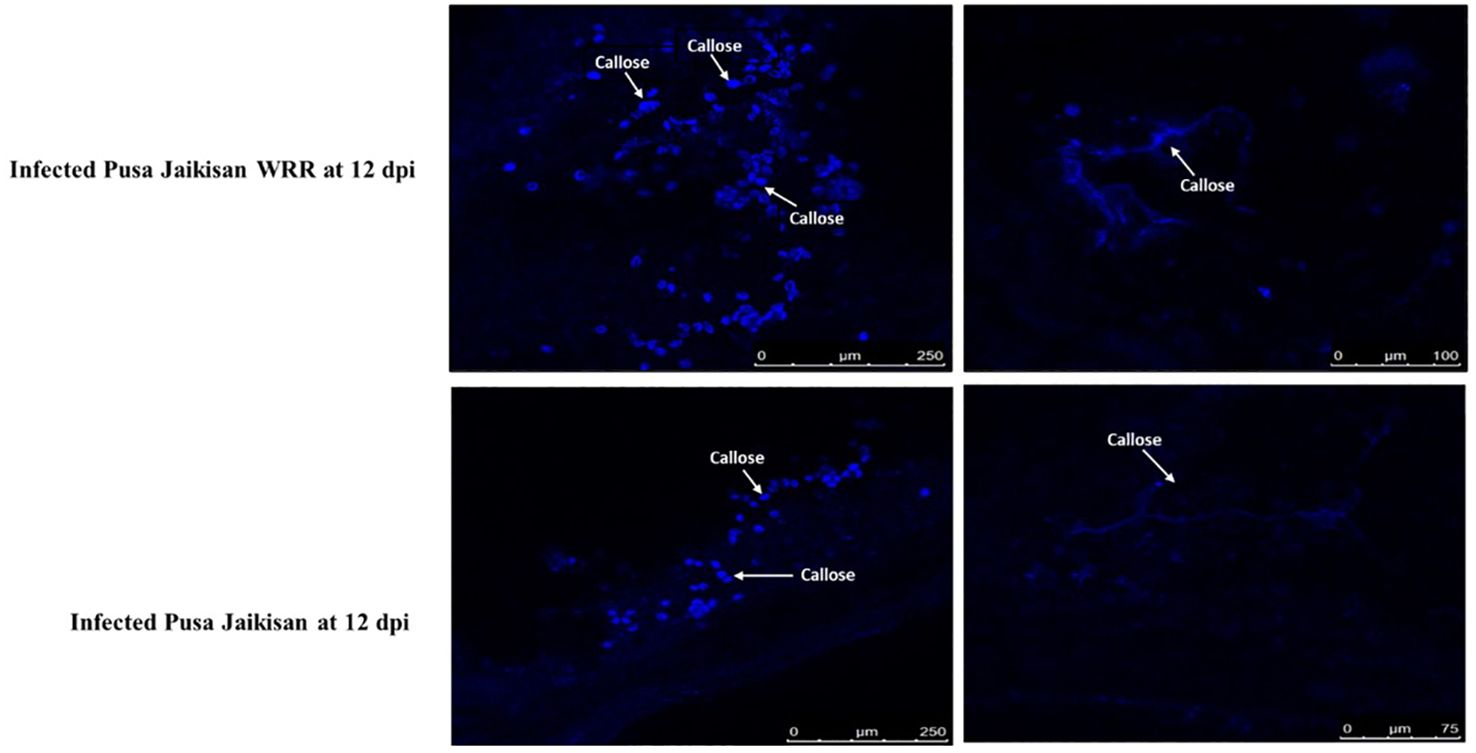
Figure 2 Comparative callose deposition in B. juncea cv. Pusa Jaikisan and B. juncea cv. Pusa Jaikisan WRR. The aniline blue-based histochemical analysis yielded that the callose deposition was higher in ‘Pusa Jaikisan WRR’ than in the ‘Pusa Jaikisan’.
3.3 Comparative estimation of defense-related biochemical parameters
Total protein content was higher in ‘Pusa Jaikisan’ than in the ‘Pusa Jaikisan WRR’ at all the time points. Across the time intervals, it reduced gradually in ‘Pusa Jaikisan’ but in the case of ‘Pusa Jaikisan WRR’, it decreased till 48 hpi and increased after that at both 72 hpi and 96 hpi. However, the protein content of the latter remained lower than the former (Figure 3A). Peroxidase was found to be higher in ‘Pusa Jaikisan WRR’ as compared to the ‘Pusa Jaikisan’ at all-time intervals. It gradually increased in Pusa Jaikisan from 12 to 96 hpi. But in the case of ‘Pusa Jaikisan WRR’, it attained its peak activity at 48 hpi and decreased thereafter (Figure 3B). The CAT was observed to be increased in the ‘Pusa Jaikisan WRR’ as compared to ‘Pusa Jaikisan’ at all-time intervals. In the case of ‘Pusa Jaikisan WRR’, CAT activity was recorded to be highest at 48 hpi and decreased thereafter. It saw a steady rise in the case of ‘Pusa Jaikisan’ (Figure 3C). SOD activity was recorded to be higher in ‘Pusa Jaikisan WRR’ as compared to ‘Pusa Jaikisan’ at all-time intervals. In the case of ‘Pusa Jaikisan WRR’, enzyme activity reached its maximum quantum at 48 hpi and then saw a gradual decline, whereas it kept on increasing at a steady pace in Pusa Jaikisan (Figure 3D).
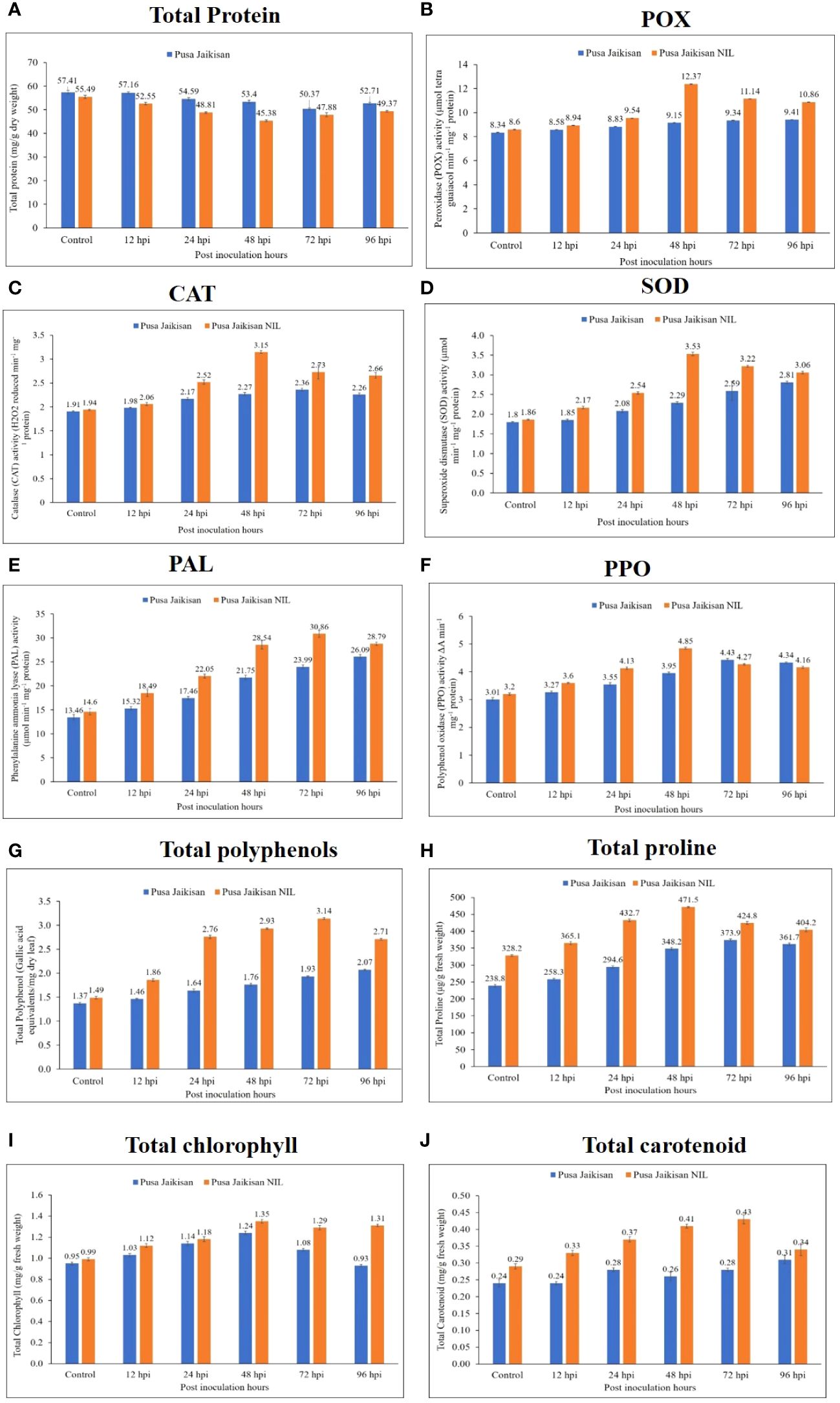
Figure 3 Changes in the biochemical markers in the leaves of ‘Pusa Jaikisan WRR’ and ‘Pusa Jaikisan’ after A. candida infection at 0, 12, 24, 48, 72 and 96 hpi. Values are mean of three replicates ± SE and indicating significant difference against 0 hpi (p<0.05). The biochemical parameters are (A) Total protein; (B) POX; (C) CAT; (D) SOD; (E) PAL; (F) PPO; (G) Total polyphenols; (H) Total proline; (I) Total chlorophyll; (J) Total carotenoids.
PAL activity was recorded to be higher in the ‘Pusa Jaikisan WRR’ as compared to ‘Pusa Jaikisan” at all the time intervals. The enzyme activity was found to be maximum at 72 hpi in the case of ‘Pusa Jaikisan WRR’ and decreased at 96 hpi. However, a steady rise in the activity of PAL was seen in ‘Pusa Jaikisan’ (Figure 3E). The PPO was recorded to be higher in ‘Pusa Jaikisan WRR’ at 12, 24 and 48 hpi, whereas it was slightly higher in ‘Pusa Jaikisan’ at 72 hpi and 96 hpi than the former. The maximum PPO activity in ‘Pusa Jaikisan WRR’ was shown at 48 hpi, and a steady decline was seen thereafter. It kept on increasing across all the time points in ‘Pusa Jaikisan’ (Figure 3F). In the case of total polyphenol, the activity was found higher in the ‘Pusa Jaikisan WRR’ as compared to the ‘Pusa Jaikisan’ at all the time intervals. The activity was found to be maximum at 72 hpi in the case of ‘Pusa Jaikisan WRR’ and it decreased at 96 hpi. There was a steady rise in ‘Pusa Jaikisan across all the time points (Figure 3G).
The proline content was found to be higher in the ‘Pusa Jaikisan WRR’ as compared to the ‘Pusa Jaikisan’. There was a huge difference in the enzyme activity of the 0 hpi sample of both cultivars. In the case of ‘Pusa Jaikisan WRR’, the maximum value was attained at 48 hpi followed by a steady decline. It kept on increasing till 72 hpi in ‘Pusa Jaikisan’ and there was a slight decline at 96 hpi (Figure 3H). Total chlorophyll content was found to be higher in ‘Pusa Jaikisan WRR’ than in ‘Pusa Jaikisan’ at all the time intervals. In the case of Pusa Jaikisan’, the activity increased up to 48 hpi and then decreased from 72 hpi, whereas there was an increment up to 48 hpi followed by a decline at 72 hpi and then again, an increase at 96 hpi in the case of ‘Pusa Jaikisan WRR’ (Figure 3I). The total carotenoid content was recorded to be higher in ‘Pusa Jaikisan WRR’ as compared to the ‘Pusa Jaikisan’ at all the time intervals. In the case of ‘Pusa Jaikisan’, it kept on increasing across the time intervals, whereas in ‘Pusa Jaikisan WRR’ it attained the peak value at 72 hpi and decreased thereafter (Figure 3J).
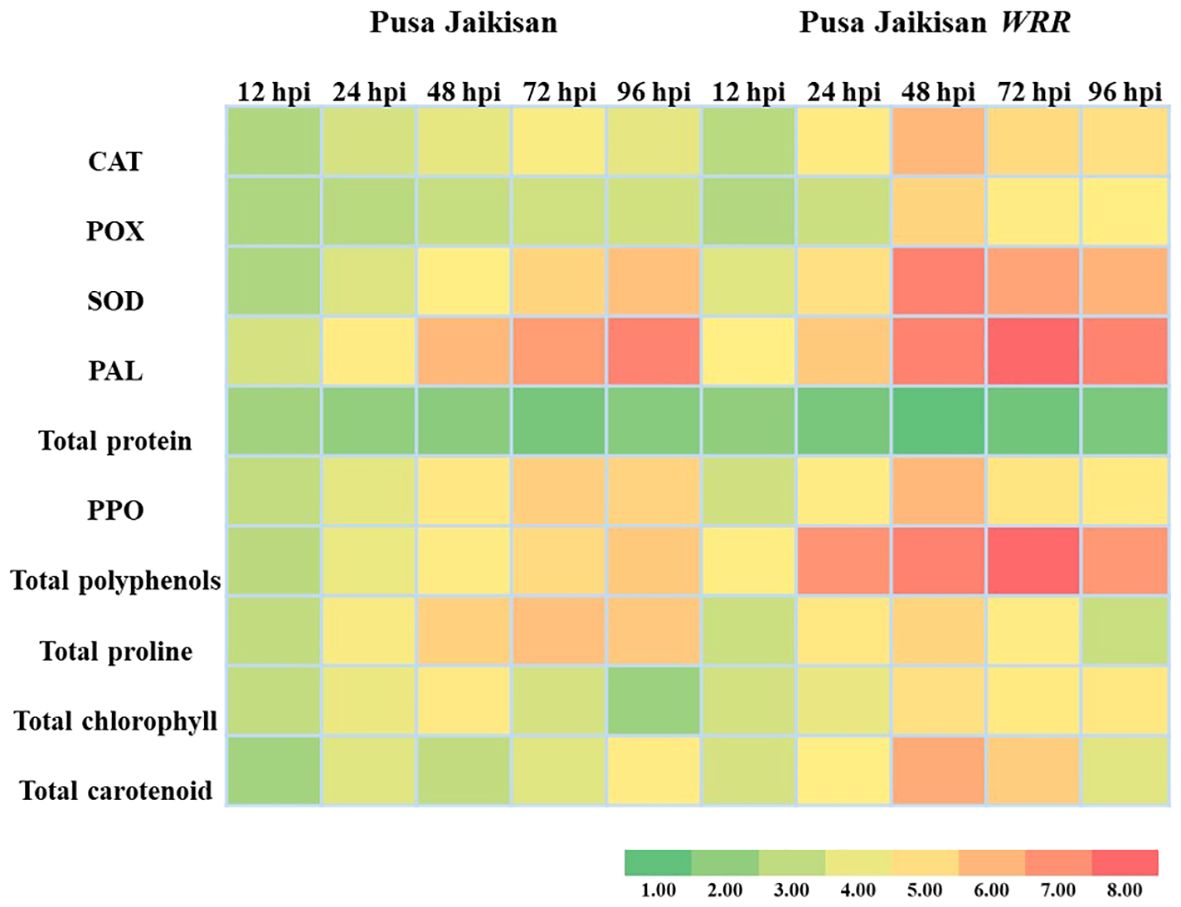
The above findings hint towards a positive correlation between resistance and that of antioxidant enzymes (POX, CAT, and SOD), PAL, PPO, polyphenols, proline, chlorophyll, and carotenoids. However, a negative correlation of total protein with disease resistance was observed, which may be due to the breaking down of proteins to produce defense-related enzymes, thereby decreasing protein concentration in plants. Heat map analysis was done for different time intervals against 0 hpi to determine the enzyme with the highest response and best time interval for resistance induction (Figure 4). Among all the biochemical markers, SOD, PAL, total polyphenols, and total carotenoids showed a higher quantum of jump as compared to the others. Most of the markers had their highest activity at 48 hpi or 72 hpi. So, these time intervals were the most promising ones for defense response.
3.4 Comparative PR gene expression
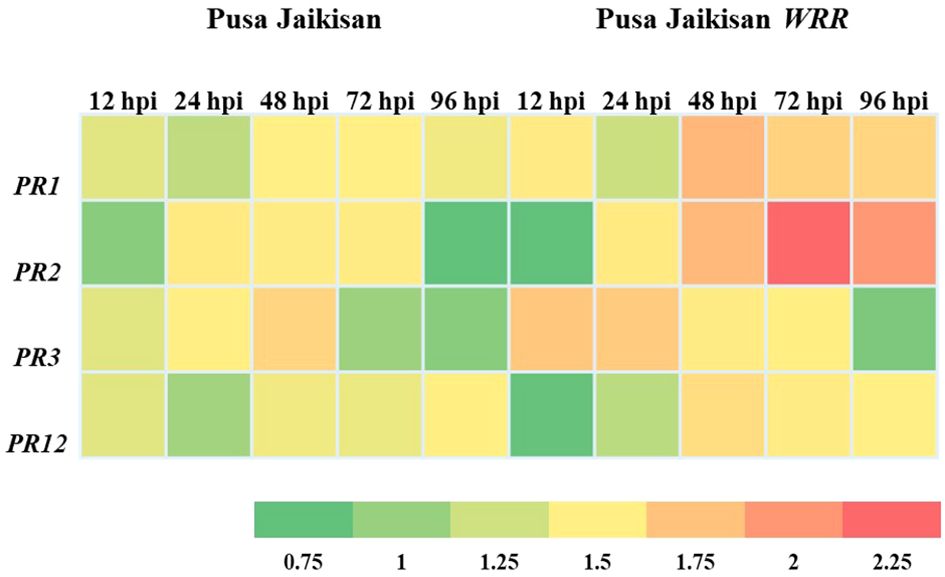
PR proteins are an important part of the plant’s stress response machinery. PR1 and PR2 are the SA pathway genes which play a vital role in defense against the Biotrophs. The PR3 and PR12 are JA/ET pathway genes, which have a positive role in defense against necrotrophic pathogens. A higher expression of the PR1 gene was found in the ‘Pusa Jaikisan WRR’ as compared to the ‘Pusa Jaikisan’ at all time intervals. In ‘Pusa Jaikisan WRR’, the maximum expression (4.24-fold) of PR1 occurred at 48 hpi. Thereafter, it gradually decreased (2.97-fold at 72 hpi and 2.84-fold at 96 hpi). In ‘Pusa Jaikisan’, the expression was found to be nearly the same at all the time intervals which hovered from a range of 1.41-fold to 1.63-fold (Figure 5A). In the case of PR2 gene expression, ‘Pusa Jaikisan WRR’ showed a higher expression at all the time intervals except at 12 hpi as compared to the ‘Pusa Jaikisan’. The highest expression in ‘Pusa Jaikisan WRR was seen at 72 hpi (7.82-fold) and decreased at 96 hpi to 5.66-fold. In Pusa Jaikisan, there was not a significant change and the maximum expression occurred at 24 hpi (Figure 5B). There was an interesting trend in the JA/ET-marker PR3 gene expression, where the expression was higher in ‘Pusa Jaikisan WRR’ at 12, 24 and 72 hpi, whereas it was reversed at 48 and 96 hpi. In the case of ‘Pusa Jaikisan WRR’, maximum expression was seen at 12 hpi (3.49-fold) and 24 hpi (2.84-fold), while for ‘Pusa Jaikisan’ it occurred at 48 hpi (2.84-fold). At 96 hpi, the expression was similar in both cultivars (Figure 5C). The expression pattern of PR12 didn’t show much fluctuation in ‘Pusa Jaikisan’ and hovered around 1.32-fold to 1.62-fold. In ‘Pusa Jaikisan WRR’, the highest expression (2.53-fold) occurred at 48 hpi. The resistant cultivar had higher expression at all the time intervals except at 12 hpi. Both the cultivars had at par expression at 96 hpi (Figure 5D). The findings of this study hint towards a positive role of PR1 and PR2 against A. candida. Also, the higher expression of PR3 in the initial stages and higher expression of PR12 in ‘Pusa Jaikisan WRR’ may be due to their complementary role in resistance to the SA-marker genes. This shows a possible synergistic role of both SA and JA/ET signaling pathways in white rust resistance. Heat map analysis was done for different time intervals against 0 hpi to determine the PR gene with the highest response and best time interval for resistance induction (Figure 6). Among all the four genes, PR2 showed the highest relative expression. As with the biochemical markers, gene expression analysis also showed that 48 hpi and 72 hpi are the most active time intervals for defense-response.
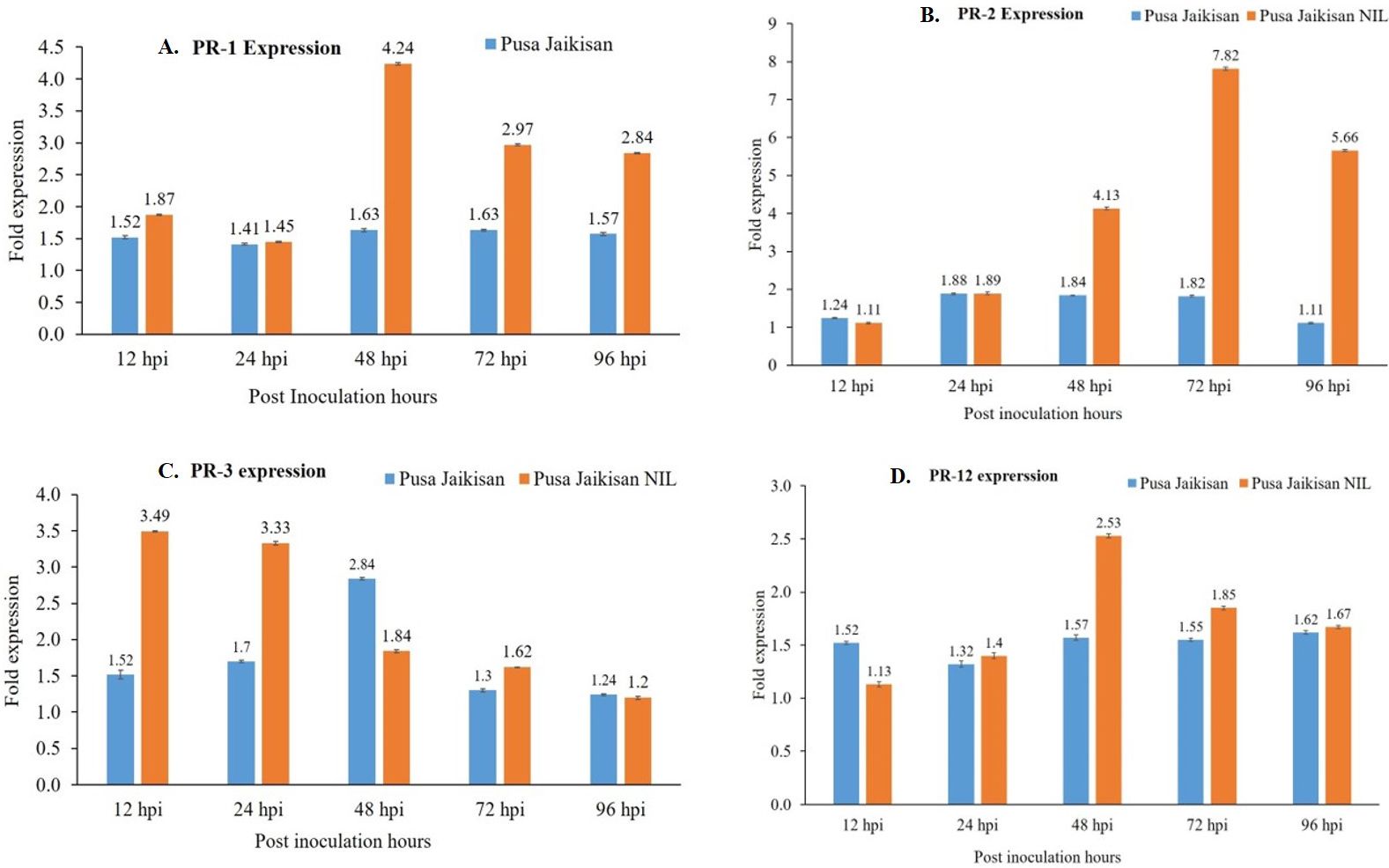
Figure 5 Differential gene expression of PR1 (A), PR2 (B), PR3 (C), and PR12 (D) genes in the leaves of ‘Pusa Jaikisan WRR’ and ‘Pusa Jaikisan’ after A. candida infection at 0, 12, 24, 48, 72 and 96 hpi. Values are mean of three biological replicates ± SE and indicating significant difference against 0 hpi at p < 0.05. The relative expression levels of each gene were analyzed by delta CT method.
4 Discussion
The host-pathogen interaction is the most important aspect of any study on plant disease as disease is termed as the manifestation of the interaction between the two entities in a conducive environment. The interaction may be compatible or incompatible based on certain factors out of which host resistance is the foremost. Histological studies are the primary method to establish this binary outcome. The current study emphasizes the role of callose, biochemical markers and PR protein genes in plant defense. In present study deposition of callose in both resistant and susceptible cultivars was observed, but the quantum of deposition was higher in the resistant one. Similar conclusions were made by Kaur et al., 1984, who reported activation of resistance due to the deposition of callose on inner walls. However, there is still a lack of evidence of callose being an important determinant of resistance, as the role may be ascertained due to its easily detectable nature.
The plant-pathogen interaction is characterized by a very well-organized and structured response by the plants to counter the pathogen. Reactive oxygen species (ROS) hold a very important place in B. juncea-A. candida interaction as the pathogen is an obligate biotroph and the resultant cell death due to the ROS favors the plant. The resistant cultivar ‘Pusa Jaikisan WRR’ varied only for a single R gene and whatever differential response it showed was due to the transferred gene. Different biochemical parameters were taken viz., total protein, POX, CAT, SOD, PAL, PPO, total polyphenols, total proline, total chlorophyll, and total carotenoids. The total protein and disease resistance in mustard is quite well-studied. Bolwell and coworkers suggested a relationship between the cell wall-invading pathogen and protein content in plants (Bolwell and Wojtaszek, 1997). In our study, we observed the total protein content to be lower in ‘Pusa Jaikisan WRR’ as compared to the ‘Pusa Jaikisan’. Similar reports have been made by many researchers. Mishra et al. (2009) evaluated the biochemical response of Brassica genotypes against A. candida and observed a negative correlation between disease and total proteins. The protein content was maximum in the cotyledonary leaves of susceptible cultivars. Chatterjee et al. (2022) also found fluctuation of significant biochemical parameters such as total proteins, sugars, and phenols, superoxide dismutase, and hydrogen peroxide during the A. candida infection in B. juncea.
POX catalyzes the oxidation of a variety of phenolic and non-phenolic electron donor substrates with H2O2 breakdown and is among the first enzymes to show differential response upon induction of biotic stress (Kozłowska et al., 2001). In this study, we observed the POX activity to be higher in the ‘Pusa Jaikisan WRR’ than in ‘Pusa Jaikisan’ at all the time intervals. In the resistant cultivar, the maximum activity was seen at 48 hpi followed by a gradual decline. Singh et al. (2011) reported an increment in POX concentration when inoculated with A. candida. Faizal canola, a resistant cultivar was found to harbor increased POX activity (Asif et al., 2018). Upadhyay et al. (2023) observed a significant increase in the POX concentration at 48 hpi in all the cultivars upon A. candida inoculation. Donskaja, a resistant cultivar showed maximum increase while the susceptible cultivars such as Pusa Bold and Varuna showed a minimal increment. CAT converts the excess H2O2 produced during developmental and environmental stresses in peroxisome into water and oxygen in all aerobic species. In the present study, CAT activity was found higher in ‘Pusa Jaikisan WRR’ than in ‘Pusa Jaikisan’ at all the time intervals. The peak in resistant cultivar was seen at 48 hpi. Similar observations have been made by other researchers. Sapna et al. (2009) found CAT to be higher in the resistant genotype, RH 781 as compared to the susceptible, Varuna upon A. candida inoculation. Asif et al. (2018) also noted higher CAT activity in the resistant cultivar, Faisal Canola when challenged with A. candida. SOD is an important antioxidant enzyme as it dismutates superoxide radicals into O2 and H2O2. In the present study, the SOD activity was seen to be higher in the resistant cultivar than in susceptible cultivar at all the time intervals. Like the previous antioxidant enzymes, maximum activity in resistant cultivar was seen at 48 hpi. Asif et al. (2018) observed similar results in the resistant cultivars, Faisal canola and Punjab Canola.
PAL catalyzes the deamination process of phenylalanine from the primary metabolism into the crucial secondary phenylpropanoid metabolism in plants. In present study a higher PAL activity was observed in ‘Pusa Jaikisan WRR’ than in ‘Pusa Jaikisan’ at all the time intervals. Maximum enzymatic activity was seen at 72 hpi in the ‘Pusa Jaikisan WRR’. Singh et al. (1999) had similar findings in which they documented that upon A. candida inoculation, B. juncea accumulates the PAL. Awasthi (2011) elucidated the positive role of PAL in resistance against white rust as the PAL activity was higher in incompatible reactions as compared to the compatible ones. A negative correlation was also established between PAL concentration and disease severity in the case of Alternaria leaf blight and white rust (Kaur et al., 2020). PPO catalyzes the oxygen-dependent oxidation of phenols to quinone and contributes to the plant defense against plant disease and insect pests. A higher PPO activity was observed in ‘Pusa Jaikisan WRR’ than in ‘Pusa Jaikisan’ at 12, 24 and 48 hpi whereas a reverse trend was seen at 72 and 96 hpi. The present findings are in line with Tirmali and Kolte (2013) who concluded that PPO has a positive correlation with the induction of host resistance in otherwise susceptible cultivar, EC-399301 against white rust. Banga et al. (2004) observed a similar trend where they introduced white rust resistance genes into B. juncea cv. RL 1359 from B. napus, B. carinata and B. tournefortii. They observed PPO to be positively associated with the white rust-resistant trait.
Phenols are thought to play diverse functions in stressed plants, including the neutralization of ROS, cell wall lignification, and anti-nutritional activity. In the present study total polyphenols content was found to be more in ‘Pusa Jaikisan WRR’ than in ‘Pusa Jaikisan’ at all the time intervals. Maximum activity was seen in ‘Pusa Jaikisan WRR’ at 72 hpi. Many researchers have reported similar trends. Yadav et al. (1996) found that total phenols and other phenolic compounds such as polyphenols have a positive role in the Indian mustard against white rust. Phenolic compounds and polyphenols lead to increased deposition of waxes on the leaf surface of B. juncea cultivars, and this acted as a structural barrier against the invading A. candida. Upadhyay et al. (2023) also showed that the total polyphenols content rises with the disease progression. Donskaja had the maximum polyphenol activity at 24 hpi. Proline is thought to be a potent non-enzymatic antioxidant that can neutralize the detrimental effects of several ROS members. Plants accumulate high amounts of proline in response to stress (Verbruggen and Hermans, 2008). We recorded a higher proline content in ‘Pusa Jaikisan WRR’ as compared to the ‘Pusa Jaikisan’ at all the time intervals. There was a huge difference between both cultivars in the proline content at 0 hpi. This hints towards a possible role of proline as an in-built resistance compound. The proline content was highest at 72 hpi and 48 hpi in ‘Pusa Jaikisan’ and ‘Pusa Jaikisan WRR’ respectively. Similar reports were also made by Tasleem et al. (2017), where they assessed tolerance to Alternaria blight disease by measuring the activity of oxidative enzymes in a transgenic line (BjV5) of B. juncea. Chaurasia et al. (2019) found that in the case of resistant cultivars, A. brassicae triggers proteolysis and generates cell-protecting antioxidants as seen in variety, PM-30. This results in higher proline accumulation in this genotype.
In the present study, total chlorophyll content was more in ‘Pusa Jaikisan WRR’ as compared to the ‘Pusa Jaikisan’ at all the time intervals. In ‘Pusa Jaikisan’, the activity saw an increment up to 48 hpi and then gradually declined. For ‘Pusa Jaikisan WRR’, the activity increased up to 48 hpi, then decreased at 72 hpi and again increased at 96 hpi. These findings are in line with former studies where Gupta et al. (1997) concluded a positive impact of chlorophyll on white rust resistance, and it should be a factor of consideration when screening for white rust resistance genotypes. We recorded and analyzed carotenoid content in both the resistant and susceptible cultivars and found it to be higher in the former at all time intervals. The maximum activity in the resistant cultivar, ‘Pusa Jaikisan WRR’ was seen at 72 hpi. Gupta et al. (2012) reported a positive relationship between total carotenoids and the disease intensity in Alternaria brassicae infected plants.
The above findings indicate a positive correlation between resistance and that of antioxidant enzymes (POX, CAT, and SOD), PAL, PPO, polyphenols, proline, chlorophyll, and carotenoids. The antioxidant enzymes such as POX, CAT, and SOD and the non-enzymatic ROS scavenging compounds such as polyphenols and proline increase in both resistant and susceptible cultivars, but the quantum of increase is much higher in the former. This may stem from the fact that ROS is an important determinant of cell death, and it occurs much more quickly in resistant cultivars, which ultimately necessitates faster scavenging of ROS to save the plants from excessive cell death. PAL is the first committed enzyme of the cinnamate-related secondary metabolism and is instrumental in resistance. Thus, it was found higher in the resistant cultivars. PPO was found to be increased in the resistant cultivars due to the role it plays in converting the phenols into toxic quinones which further stops the growth of the pathogen. Chlorophyll was reported to increase significantly in both the cultivars. This may be due to the induction of “green islands” by A. candida, which can fix 5 times more CO2 than uninfected plants (Harding et al., 1968). Carotenoids were found in higher concentrations in the resistant cultivar due to the reason that they are potent ROS scavengers. We also reported a negative correlation of total protein with disease resistance. A possible reason may be that for the synthesis of most of the defense-related enzymes, the proteins are broken down thus decreasing their concentration in plants.
PR proteins are an important component of disease resistance along with the hormonal responses that are induced by complex signaling and networking. The PR-protein induction has been reported in many plant-fungi interactions (Grover, 2012). Salicylic acid (SA) and Jasmonic acid (JA)/Ethylene (ET) are two important pillars of hormonal signaling-mediated resistance. Both these are generally thought to be antagonistic. In the case of biotrophs, the SA-mediated resistance is activated while in the case of necrotrophic attack, there is an induction of the JA/ET pathway. The gene-for-gene concept is positively applied to the biotrophs where the interaction between an Avr gene and an R gene results in resistance. This is further manifested by the activation of SAR and SA-dependent signaling. For this reason, both the concepts were integrated and PR1 and PR2 were taken as SA-marker genes and PR3 and PR12 were taken as JA/ET-marker genes to check whether both pathways are synergistic or antagonistic and which set of PR genes play a positive role in white rust resistance against B. juncea.
We found that the resistant cultivar ‘Pusa Jaikisan WRR’ had a higher expression of the PR1 gene at all the time intervals as compared to the susceptible one. In the resistant cultivar, the maximum expression occurred at 48 hpi. Thereafter, it gradually declined. In the case of ‘Pusa Jaikisan’, the expression was found to be similar at all the time intervals. Similar findings were observed by other workers such as Niderman et al. (1995) reported the PR1 protein isolated from tobacco and other solanaceous plants effectively reduced spore germination and pathogen growth in the plants. Overexpression of numerous PR1 genes in various plant species increased resistance to oomycetes (Broekaert et al., 2000), but the effect on other pathogens taxa is unknown. PR1 has recently been demonstrated to bind sterols, indicating a protective mode of action based on the limitation of this key nutrient for oomycetes (Gamir et al., 2017). In the case of PR2 gene expression, the ‘Pusa Jaikisan WRR’ showed a higher relative expression than in ‘Pusa Jaikisan’ at all the time intervals except at 12 hpi. Maximum expression in ‘Pusa Jaikisan WRR’ occurred at 72 hpi. There was not much significant rise in the ‘Pusa Jaikisan’ and the expression was relatively the same throughout. Several incidences of improved resistance to oomycetes in plants overexpressing PR2 have been documented since oomycetes have β-1,3-Glucanase in their cell walls (Broekaert et al., 2000).
Chitinase (PR3) catalyzes the hydrolytic cleavage of chitin (the planet’s second most prevalent biopolymer after cellulose) and is a significant antifungal enzyme We saw a very interesting trend in PR3 gene expression where it was more in ‘Pusa Jaikisan WRR’ at 12,24 and 72 hpi, while at 48 and 96 hpi it was more in the susceptible cultivar, ‘Pusa Jaikisan’. Maximum expression in ‘Pusa Jaikisan WRR’ occurred in the initial stages at 12 and 24 hpi. For the ‘Pusa Jaikisan WRR’, the peak was observed at 48 hpi. At 96 hpi both had a very similar expression. PR12 (also known as ‘plant defensin’) are the most important PR proteins for necrotrophic pathogen resistance. The expression pattern of PR12 in the current study didn’t show much fluctuation in the ‘Pusa Jaikisan’. In ‘Pusa Jaikisan WRR’, the peak expression was seen at 48 hpi. The ‘Pusa Jaikisan WRR’ had higher expression at all time points except at 12 hpi. At 96 hpi, just like the PR3, PR12 also had similar expression in both the cultivars. The role of PR3 and PR12 in disease resistance was elucidated by Yadav et al. (2020) as they conducted a gene expression analysis that revealed upregulation of PR3 and PR12 genes only in C. sativa and S. alba as compared to B. juncea, implying their role in Alternaria resistance. This may hint towards the involvement of PR3 in general resistance mechanism against both biotrophs and necrotrophs.
The above findings indicate a positive role of PR1 and PR2 genes in the white rust resistance of B. juncea. Also, the higher expression of PR3 in the initial stages and higher expression of PR12 in the resistant cultivar may be due to the positive role of JA/ET signaling that acts complementary to the SA pathway. Due to the tendency of SA and JA/ET signaling to be antagonistic to one another, JA/ET signaling is anticipated to negatively impact resistance to these pathogens. These findings are in line with the observation of Glazebrook (2005), who reported that the SA signaling is crucial for resistance against biotrophs such as Peronospora parasitica, Erysiphe species and Pseudomonas syringae. However, JA/ET signaling may also be instrumental in resistance if it is active, particularly in the case of P. parasitica and Erysiphe species. The biotrophs are generally known to be stopped by SA signaling and JA/ET acts against necrotrophs and insects. These two pathways are thought to be antagonistic but there is a lot of evidence that the JA/ET pathway is potentiating the SA pathway in resistance against the biotrophs as well. Incompatible reaction in Plasmopara viticola, a biotrophic oomycete was mediated through the JA pathway (Guerreiro et al., 2016). This may be the case with A. candida- B. juncea interaction, where both pathways possibly play synergistic roles.
5 Conclusion
B. juncea is one of the most important edible oilseed crops and A. candida is a serious impediment in realizing its production potential. Given the pathogen’s status of being an obligate biotroph, the R gene based resistance is easily overcome by more virulent races. So, to develop durable resistant varieties, we need to explore the factors behind resistance. In the present study, we studied the morphological, histochemical, biochemical, and molecular parameters and their role in white rust resistance in resistant (Pusa Jaikisan WRR) and susceptible (Pusa Jaikisan) cultivars of B. juncea. Morphological studies confirmed the resistant nature of ‘Pusa Jaikisan WRR’ as there was no spot on either the adaxial or abaxial surface. Histochemical studies hinted towards a positive role of callose deposition in resistance as it was deposited in a higher quantum in ‘Pusa Jaikisan WRR’. Biochemical studies established a positive correlation between resistance and POX, CAT, SOD, PAL, PPO, polyphenols, proline, chlorophyll, and carotenoids, while there was a negative correlation between total protein and white rust resistance. In addition to these, the gene expression analysis revealed that the SA-marker genes played a crucial role in resistance, while the JA-responsive genes may also have a positive role in the white rust defense mechanism. The comparative histochemical, biochemical, and molecular studies showed that the ‘Pusa Jaikisan WRR’ activated defense response by early detection of the pathogen. Results obtained in this study will pave the way for developing strategies to induce resistance against biotrophic pathogens in susceptible cultivars by developing transgenic plants over-expressing the PR1 and PR2 genes. The role of PR3 and PR12 should also be established in the case of other biotrophic pathogens of B. juncea such as Erysiphe polygoni and Hyaloperonospora parasitica. If the role of these JA-marker genes is positive in these cases as well, then these genes can be utilized by over-expression for a broad-spectrum resistance against both biotrophic and necrotrophic pathogen complexes of B. juncea.
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article. The data is part of thesis submitted in MS Swaminathan Library (T-11213). Raw data supporting the findings of this study are available from the corresponding author upon resonable request.
Author contributions
PR: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Resources, Software, Writing – original draft, Writing – review & editing. LP: Conceptualization, Data curation, Formal analysis, Supervision, Writing – review & editing. SM: Methodology, Supervision, Writing – review & editing. PY: Methodology, Supervision, Validation, Writing – review & editing. AS: Data curation, Formal analysis, Methodology, Resources, Supervision, Writing – review & editing. DM: Data curation, Methodology, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. PR acknowledges ICAR for providing financial support through the ICAR PG Fellowship to carryout the research work.
Conflict of interest
PR is Ph.D. student in Division of Plant Pathology, ICAR-IARI, New Delhi.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aebi, H. (1984). “[13] Catalase in vitro,” in Methods in enzymology, vol. 105. (USA: Academic press), 121–126). doi: 10.1016/s0076-6879(84)05016-3
Afiyanti, M., Chen, H. J. (2014). Catalase activity is modulated by calcium and calmodulin in detached mature leaves of sweet potato. J. Plant Physiol. 171, 35–47. doi: 10.1016/j.jplph.2013.10.003
Ali, S., Ganai, B. A., Kamili, A. N., Bhat, A. A., Mir, Z. A., Bhat, J. A., et al. (2018). Pathogenesis-related proteins, and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiological Res. 212–213, 29–37. doi: 10.1016/j.micres.2018.04.008
Arora, A., Byrem, T. M., Nair, M., Strasburg, G. M. (2000). Modulation of liposomal membrane fluidity by flavonoids and isoflavonoids. Arch. Biochem. Biophysics 373, 102–109. doi: 10.1006/abbi.1999.1525
Arora, H., Padmaja, K. L., Paritosh, K., Mukhi, N., Tiwari, A. K., Mukhopadhyay, A., et al. (2019). BjuWRR1, a CC-NB-LRR gene identified in Brassica juncea, confers resistance to white rust caused by Albugo candida. Theor. Appl. Genet. 132, 2223–2236. doi: 10.1007/s00122-019-03350-z
Asif, M., Atiq, M., Bashir, M. R., Yasin, O., Rajput, N. A., Ali, Y., et al. (2018). Biochemical alterations: markers for the identification of source of resistance in brassica germplasm against white rust. Int. J. Biosci. 13, 364-376. doi: 10.12692/ijb/13.1.364-376
Awasthi, R. P. (2011). Induced Systemic Resistance against White Rust of Mustard by Pre-or Coinoculation with an Incompatible Isolate (G.B. Pant University of Agriculture and Technology, Pantnagar, India-263145: Centre of Advanced Faculty Training in Plant Pathology).
Banga, S. S., Kaur, K., Ahuja, K. L., Banga, S. K. (2004). “Introgression and biochemical manifestation of the gene (s) for white rust resistance in Indian mustard (Brassica juncea (L.) Coss.),” in Proceedings 4th international crop science congress. (Brisbane, Australia)
Beauchamp, C., Fridovich, I. (1971). Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Analytical Biochem. 44, 276–287. doi: 10.1016/0003-2697(71)90370-8
Bigeard, J., Colcombet, J., Hirt, H. (2015). Signalling mechanisms in pattern-triggered immunity (PTI). Mol. Plant 8, 521–539. doi: 10.1016/j.molp.2014.12.022
Bits, K. S., Rana, M., Upadhyay, P., Awasthi, R. P. (2018). Disease assessment key for white rust disease caused by Albugo Candida, in rapeseed-mustard. Plant Pathol. J. 17, 11–18. doi: 10.3923/ppj.2018.11.18
Bittner, N., Trauer-Kizilelma, U., Hilker, M. (2017). Early plant defence against insect attack: involvement of reactive oxygen species in plant responses to insect egg deposition. Planta 245, 993–1007. doi: 10.1007/s00425-017-2654-3
Bolwell, G. P., Wojtaszek, P. (1997). Mechanisms for the generation of reactive oxygen species in plant defence–a broad perspective. Physiol. Mol. Plant Pathol. 51, 347–366. doi: 10.1006/pmpp.1997.0129
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochem. 72, 248–254. doi: 10.1006/abio.1976.9999
Broekaert, W. F., Terras, F. R. G., Cammue, B. P. A. (2000). Induced and preformed antimicrobial proteins. In: Mech. Resistance to Plant Siseases 371-477. doi: 10.1007/978-94-011-3937-3_11
Chance, B., Maehly, A. C. (1955). Assay of catalases and peroxidases. Methods Enzymol. 2, 764–775. doi: 10.1016/s0076-6879(55)02300-8
Chatterjee, A., Nirwan, S., Mohapatra, S., Sharma, P., Agnihotri, A., Shrivastava, N. (2022). Biochemical aspects of pathogenic variability in white rust infected Indian mustard. Mycologia 114, 757–768. doi: 10.1080/00275514.2022.2060007
Chaurasia, D., Srivashtav, V., Kumar, S., Pahuja, S. (2019). Screening of defense response against Alternaria brassicae (Berk.) Sacc. causing Leaf Spot in Indian mustard (Brassica juncea L.). Bull. Environ. Pharmacol. Life Sci. 9, 91–97.
Dong, X., Zhao, Y., Ran, X., Guo, L., Zhao, D. G. (2017). Overexpression of a new chitinase gene EuCHIT2 enhances resistance to Erysiphe cichoracearum DC. @ in tobacco plants. Int. J. Mol. Sci. 18, 2361. doi: 10.3390/ijms18112361
Doughari, J. (2015). An overview of plant immunity. J. Plant Pathol. Microbiol. 6, 10–4172. doi: 10.4172/2157-7471.1000322
DRMR (2023). Available online at: https://www.drmr.res.in.
Ebrahim, S., Usha, K. U., Singh, B. (2011). Pathogenesis-related (PR) proteins in plant defense mechanism. Sci. Against Microbial Pathog. 2, 1043–1054. doi: 10.1016/j.scienta.2011.09.014
Edward, E. A., Kessmann, H. (1992). Isoflavonoid phytoalexins and their biosynthetic enzymes. Molecular Plant Pathology. A Pract. Approach 2, 45–62. doi: 10.1093/oso/9780199633524.003.0006
Gairola, K., Tewari, A. K. (2019). Management of white rust (Albugo candida) in Indian mustard by fungicides and garlic extract. Pesticide Res. J. 31, 60–65. doi: 10.5958/2249-524X.2019.00006.2
Gamir, J., Darwiche, R., Van’t Hof, P., Choudhary, V., Stumpe, M., Schneiter, R., et al. (2017). The sterol-binding activity of Pathogenesis-Related Protein 1 reveals the mode of action of an antimicrobial protein. Plant J. 89, 502–509. doi: 10.1111/tpj.13398
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. doi: 10.1146/annurev.phyto.43.040204.135923
Govrin, E. M., Levine, A. (2000). The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr. Biol. 10, 751–757. doi: 10.1016/S0960-9822(00)00560-1
Grover, A. (2012). Plant chitinases: genetic diversity and physiological roles. Crit. Rev. Plant Sci. 31, 57–73. doi: 10.1080/07352689.2011.616043
Guerreiro, A., Figueiredo, J., Sousa Silva, M., Figueiredo, A. (2016). Linking jasmonic acid to grapevine resistance against the biotrophic oomycete Plasmopara viticola. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00565
Gupta, M. L., Singh, G., Raheja, R. K., Ahuja, K. L., Banga, S. K. (1997). Chlorophyll content in relation to white rust (Albugo candida) resistance in Indian mustard. Cruciferae Newslett. 19, 105–106.
Gupta, M., Summuna, B., Gupta, S., Mallick, S. A. (2012). Assessing the role of biochemical constituents in resistance to Alternaria blight in rapeseed-mustard. J. Mycol. Plant Pathol. 42, 463–468.
Hahlbrock, K., Scheel, D. (1989). Physiology and molecular biology of phenylpropanoid metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 40, 347–369. doi: 10.1146/annurev.pp.40.060189.002023
Harding, H., Williams, P. H., McNabola, S. S. (1968). Chlorophyll changes, photosynthesis, and ultrastructure of chloroplasts in Albugo candida induced “green islands” on detached Brassica juncea cotyledons. Can. J. Bot. 46, 1229–1234. doi: 10.1139/b68-165
Huang, L. F., Lin, K. H., He, S. L., Chen, J. L., Jiang, J. Z., Chen, B. H., et al. (2016). Multiple patterns of regulation and overexpression of a ribonucleaseLike pathogenesis-related protein gene, osPR10a, conferring disease resistance in rice and Arabidopsis. PloS One 11, e0156414. doi: 10.1371/journal.pone.0156414
Jones, J. G. D., Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kaur, D., Bhandari, N. N., Mukerji, K. G. (1984). A histochemical study of cytoplasmic changes during wall layer formation in the oospore of Albugo candida. J. Phytopathol. 109, 117–130. doi: 10.1111/j.1439-0434.1984.tb00699.x
Kaur, P., Gupta, S., Kaur Sanghag, M., Kaur, G. (2020). Effect of fungal infection and hugh temperature stress on antioxidant compounds in Brassica juncea recombinant inbred lines. Agric. Res. J. 57, 664–672. doi: 10.5958/2395-146x.2020.00098.8
Kaur, H., Salh, P. K., Singh, B. (2017). Role of defense enzymes and phenolics in resistance of wheat crop (Triticum aestivum L.) towards aphid complex. J. Plant Interact. 12, 304–311. doi: 10.1080/17429145.2017.1353653
Koca, H., Bor, M., Özdemir, F., Türkan, İ. (2007). The effect of salt stress on lipid peroxidation, antioxidative enzymes and proline content of sesame cultivars. Environ. Exp. Bot. 60, 344–351. doi: 10.1016/j.envexpbot.2006.12.005
Kozłowska, M., Fryder, K., Wolko, B. (2001). Peroxidase involvement in the defense response of red raspberry to Didymella applanata (Niessl/Sacc.). Acta Physiologiae Plantarum 23, 303–310. doi: 10.1007/s11738-001-0037-6
Lichtenthaler, H. K., Wellburn, A. R. (1983). Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc Trans. 11, 591–592. doi: 10.1042/bst0110591
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2 (–Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Meyer, G. D., Bigirimana, J., Elad, Y., Höfte, M. (1998). Induced systemic resistance in Trichoderma harzianum T39 biocontrol of Botrytis cinerea. Eur. J. Plant Pathol. 104, 279–286. doi: 10.1023/A:1008628806616
Mishra, K. K., Kolte, S. J., Nashaat, N. I., Awasthi, R. P. (2009). Pathological and biochemical changes in Brassica juncea (mustard) infected with Albugo candida (white rust). Plant Pathol. 58, 80–86. doi: 10.1111/j.1365-3059.2008.01939.x
Niderman, T., Genetet, I., Bruyere, T., Gees, R., Stintzi, A., Legrand, M., et al. (1995). Pathogenesis-Related PR-1 Proteins Are Antifungal (Isolation and Characterization of Three 14-Kilodalton Proteins of Tomato and of a Basic PR-1 of Tobacco with Inhibitory Activity against Phytophthora infestans). Plant Physiol. 108 (1), 17–27. doi: 10.1104/pp.108.1.17
Pietrowska, E., Różalska, S., Kaźmierczak, A., Nawrocka, J., Małolepsza, U. (2015). Reactive oxygen and nitrogen (ROS and RNS) species generation and cell death in tomato suspension cultures—Botrytis cinerea interaction. Protoplasma 252, 307–319. doi: 10.1007/s00709-014-0680-6
Rai, P., Prasad, L., Rai, P. K. (2023). Fungal effectors versus defense-related genes of B. juncea and the status of resistant transgenics against fungal pathogens. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1139009
Ren, L., Xu, L., Liu, F., Chen, K., Sun, C., Li, J., et al. (2016). Host range of Plasmodiophora brassicae on cruciferous crops and weeds in China. Plant Dis. 100, 933–939. doi: 10.1094/PDIS-09-15-1082-RE
Richardson, A. D., Duigan, S. P., Berlyn, G. P. (2002). An evaluation of noninvasive methods to estimate foliar chlorophyll content. New Phytol. 153, 185–194. doi: 10.1046/j.0028-646x.2001.00289.x
Saharan, G. S., Verma, P. R. (1992). White rusts: a review of economically important species (Ottawa, ON, CA: IDRC). doi: 10.1002/j.1537-2197.1992.tb14634.x
Sapna, Jain, S., Jain, V., Rathi, A. S. (2009). Changes in antioxidant pathway enzymes in Indian mustard leaves due to white rust infection. Indian Phytopathol. 62, 499–504.
Singh, U. S., Doughty, K. J., Nashaat, N. I., Bennett, R. N., Kolte, S. J. (1999). Induction of systemic resistance to Albugo candida in Brassica juncea by pre-or coinoculation with an incompatible isolate. Phytopathology 89, 1226–1232. doi: 10.1094/phyto.1999.89.12.1226
Singh, Y., Rao, D. V., Batra, A. (2011). Enzyme activity changes in Brassica juncea (L.) Czern. & Coss. in response to Albugo candida Kuntz. (Pers.). J. Chem. Pharm. Res. 3, 18–24.
Singleton, V. L., Rossi, J. A. (1965). Colorimetry of total phenolics with phosphomolybdic-phosphotungstic acid reagents. Am. J. Enol. Viticulture 16, 144–158. doi: 10.5344/ajev.1965.16.3.144
Tasleem, M., Baunthiyal, M., Kumar, A., Taj, G. (2017). Determination of antioxidant activity in overexpressed MPK3 transgenic Brassica juncea for induction of defense against Alternaria blight disease. J. Pharmacognosy Phytochem. 6, 2579–2582.
The Solvent Extractors’ Association of India (2023). Available online at: https://seaofIndia.com.
Thomma, B. P., Penninckx, I. A., Cammue, B. P., Broekaert, W. F. (2001). The complexity of disease signalling in Arabidopsis. Curr. Opin. Immunol. 13, 63–68. doi: 10.1016/s0952-7915(00)00183-7
Tirmali, A., Kolte, S. (2013). Effect of pre and post inoculation of amino acids on induction of resistance and polyphenol oxidase activity against Albugo candida in mustard. J. Plant Dis. Sci. 8, 63–69.
Torres, M. A., Jones, D. G. J., Dangl, J. L. (2006). Reactive oxygen species signalling in response to pathogens. Plant Physiol. 141 (2), 373–378. doi: 10.1104/pp.106.079467
Upadhyay, P., Tewari, A. K., Punetha, H. (2023). Biochemical estimation of polyphenol and peroxidases in resistant sources against white rust disease of rapeseed mustard incited by Albugo candida. Pharma Innovation 12, 4964–4974.
van Loon, L. C., Rep, M., Pieterse, C. M. J. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. doi: 10.1146/annurev.phyto.44.070505.143425
Verbruggen, N., Hermans, C. (2008). Proline accumulation in plants: a review. Amino Acids 35, 753–759. doi: 10.1007/s00726-008-0061-6
Yadav, P., Mir, Z. A., Ali, S., Papolu, P. K., Grover, A. (2020). A combined transcriptional, biochemical and histopathological study unravels the complexity of Alternaria resistance and susceptibility in Brassica coenospecies. Fungal Biol. 124, 44–53. doi: 10.1016/j.funbio.2019.11.002
Yadav, O. P., Yadava, T. P., Kumar, P., Gupta, S. K. (1996). Inheritance of phenols and protein in relation to white rust (Albugo candida) resistance in Indian mustard. Indian J. Genet. Plant Breed. 56, 256–261.
Keywords: Albugo candida, biotrophic, Brassica juncea, PR protein genes, biochemical, callose, Reactive Oxygen Species
Citation: Rai P, Prasad L, Mehta S, Yadav P, Sharma A and Mishra DN (2024) Transcriptional, biochemical, and histochemical response of resistant and susceptible cultivars of Brassica juncea against Albugo candida infection. Front. Plant Sci. 15:1426302. doi: 10.3389/fpls.2024.1426302
Received: 01 May 2024; Accepted: 08 July 2024;
Published: 01 August 2024.
Edited by:
Katarzyna Otulak-Kozieł, Warsaw University of Life Sciences, PolandReviewed by:
Francisco Roberto Quiroz-Figueroa, National Polytechnic Institute (IPN), MexicoDr. Rakesh Yadav, Guru Jambheshwar University of Science and Technology, India
Copyright © 2024 Rai, Prasad, Mehta, Yadav, Sharma and Mishra. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Prajjwal Rai, cHJhamp3YWxyYWlyYWk3QGdtYWlsLmNvbQ==; Lakshman Prasad, bGF4bWFucHJhc2FkMjVAeWFob28uY29t
 Prajjwal Rai
Prajjwal Rai Lakshman Prasad
Lakshman Prasad Samridhi Mehta
Samridhi Mehta Prashant Yadav
Prashant Yadav Anubhuti Sharma3
Anubhuti Sharma3 Deep Narayan Mishra
Deep Narayan Mishra