
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 19 August 2024
Sec. Functional and Applied Plant Genomics
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1421221
 Hexiang Luan1*
Hexiang Luan1* Daiqiao Song1
Daiqiao Song1 Kai Huang1
Kai Huang1 Shuxin Li1
Shuxin Li1 Hao Xu2
Hao Xu2 Pradeep Kachroo3
Pradeep Kachroo3 Aardra Kachroo3*
Aardra Kachroo3* Longgang Zhao4*
Longgang Zhao4*Eukaryotic elongation factors (eEFs) are protein factors that mediate the extension of peptide chain, among which eukaryotic elongation factor 1 alpha (eEF1A) is one of the most abundant protein synthesis factors. Previously we showed that the P3 protein of Soybean mosaic virus (SMV), one of the most destructive and successful viral pathogens of soybean, targets a component of the soybean translation elongation complex to facilitate its pathogenesis. Here, we conducted a systematic analyses of the soybean eEF (GmeEF) gene family in soybean and examinedits role in virus resistance. In this study, GmeEF family members were identified and characterized based on sequence analysis. The 42 members, which were unevenly distributed across the 15 chromosomes, were renamed according to their chromosomal locations. The GmeEF members were further divided into 12 subgroups based on conserved motif, gene structure, and phylogenetic analyses. Analysis of the promoter regions showed conspicuous presence of myelocytomatosis (MYC) and ethylene-responsive (ERE) cis-acting elements, which are typically involved in drought and phytohormone response, respectively, and thereby in plant stress response signaling. Transcriptome data showed that the expression of 15 GmeEF gene family members changed significantly in response to SMV infection. To further examine EF1A function in pathogen response, three different Arabidopsis mutants carrying T-DNA insertions in orthologous genes were analyzed for their response to Turnip crinkle virus (TCV) and Cucumber mosaic virus (CMV). Results showed that there was no difference in viral response between the mutants and the wild type plants. This study provides a systematic analysis of the GmeEF gene family through analysis of expression patterns and predicted protein features. Our results lay a foundation for understanding the role of eEF gene in soybean anti-viral response.
Eukaryotic elongation factors (eEFs) is a GTP-binding protein and play a central role in the protein biosynthesis, which elongate the nascent polypeptide chain by one amino acid at a time. In prokaryotic cells, there are three types of elongation factors, named EF-Tu, EF-Ts, and EF-G respectively and in eukaryotic cells, called eukaryotic elongation factor 1 alpha (eEF1A), eEF1B, and eEF2. eEF1A is a multimeric protein that with molecular masses of around 50 kDa and has three distinct domains: domain I (binds GTP), domain II (binds the aminoacyl end of the aminoacylated tRNA and actin), and domain III (binds actin) (Kanibolotsky et al., 2008; Soares et al., 2009; Abbas et al., 2015; Juette et al., 2022). Domain I is made up of Rossmann-fold topology. Domain II and domain III are made up of entirely from beta-strands, each domain contains two beta sheets that form a beta barrel (Soares et al., 2009). eEF1A is an important and highly conserved protein with a typical role in binding and delivering aminoacylated tRNA to the A-site of the ribosomes in a GTP-dependent manner. After tRNA transfer, eEF1A is recycled to its active GTP-bound form by the guanine nucleotide exchange factor, eEF1B (Gromadski et al., 2007; Maruyama et al., 2019; Mills and Gago, 2021). eEF1A proteins are multifunctional because they have been implicated in non-canonical roles besides its function protein synthesis. It has been showed that eEF1A participate in the growth and proliferation of cells, including construction of cytoskeleton, nucleocytoplasmic trafficking, endoplasmic reticulum (ER) stress, signal transduction, apoptosis, and protein degradation (Hetherington et al., 2016; Luan et al., 2016; Migliaccio et al., 2016; Małecki et al., 2017; Liu et al., 2020; Liu et al., 2019; Akram et al., 2020; Hou et al., 2021; Romaus-Sanjurjo et al., 2022).
Chaperone activity of elongation factors may be important in protecting the plants from stress factors of the environment that lead to denaturation of proteins. eEF1A may be involved in the response process of plants to stress, which is closely related to the plant resistance, such as high temperature, high salinity, and drought (Guo et al., 2014; Momcilovic et al., 2016; Djukić et al., 2019; Sun et al., 2020; Markovic et al., 2021). Identically, a body of data demonstrates the potential role of components of the eEF1A in plant virus infection. Their functions mainly include promoting viral RNA translation, regulating viral RNA replication, and accelerating viral infection. The work reveals that silencing of a homolog of the eEF1A (NbS00023178g0001.1) suppressed both Tomato spotted wilt virus (TSWV) disease symptom development and systemic spread of the virus (Garcia-Ruiz, 2019; Helderman et al., 2021). eEF1A can facilitate Chinese wheat mosaic virus (CWMV) infection in plants via its binding to the 3′-untranslated region (UTR) of CWMV genomic RNAs (Chen et al., 2021); eEF1A binds to the 3′-end of the Tomato bushy stunt virus (TBSV) RNA as well as to TBSV p33 replication co-factor to enhanced TBSV replication (Li et al., 2009; Gamarnik et al., 2010). There is verified enhanced susceptibility to Cucumber mosaic virus (CMV) and Tobacco rattle virus (TRV) in LreEF1A4-overexpressing transgenic petunia plants (Sun et al., 2020). Research shows that eEF1A can not only promote virus replication but also inhibit virus replication. The study demonstrated the key role of the eEF1A gene in sustaining the resistance against Tomato yellow leaf curl virus (TYLCV), most probably by inhibiting virus replication and/or movement (Wazwaz, 2013).
When reviewing the previous literatures, the controversy concerning the effect of eEF1As on virus propagation still exists, which eEF1A can both promote and inhibit virus infection. The mechanisms underlying these apparently conflicting findings are not yet well understood. What’s more, few studies have identified and characterized the eEF1A genes to clarify the mechanism of SMV infection. Here, we attempt to determine the role of the Arabidopsis eEF1A homologous mutant in the pathogenesis of SMV. Given the completion of soybean genome sequencing, we used soybean genome and transcriptome data to analyze the eEF family genes from the aspects of systematic evolution, gene structure, protein motif, and chromosome location and further analyzed the gene function of GmEF1A homologous mutants in Arabidopsis under CMV infection. This study provides a theoretical basis for further exploring the function of GmeEF1A resistance to virus.
The T-DNA insertion mutants generated by the Salk Institute Genomic Analysis Laboratory were obtained from the Arabidopsis Biological Resource Center. The genotypes of three mutant lines: SALK_050704C (AT1G07930), SALK_079753C (AT1G07920), SALK_063369C (AT5G60390) were in the Col-0 ecotype background. Homozygous T-DNA insertion lines were generated by self-crossing heterozygous lines and verified by PCR amplification with primers specific for the T-DNA LP in combination with RP and RP in combination with LBB1 on inserted genes. Arabidopsis mutants that were grown in MTPS 144 Conviron (Winnipeg, MB, Canada) walk-in-chambers at 22°C, 65% relative humidity, and 14 h photoperiod. Soybean [Glycine max (L.)] cultivar NN1138-2 were grown in an aphid-free greenhouse at 25°C day and 20°C night, in 65% relative humidity and during a 14 h photoperiod. Transcripts synthesized in vitro from a cloned cDNA of TCV using T7 RNA polymerase were used for viral infections. For inoculations, 5 mg of the viral transcript was suspended in 80 μl inoculating buffer and DEPC water up to 200 μl. Healthy leaves of Arabidopsis were inoculated with 2μl inoculating mixture by rub-inoculation and then transferred to a Conviron MTR30 reach-in chamber.
First, we searched the genes in soybean transcriptome data to obtain gene ID and then searched in NCBI to obtain the eEF genes and protein sequences. Next, the physical and chemical properties of eEF proteins including the molecular weights (in kDa) and isoelectric points (pI), were analyzed using ExPASy (https://web.expasy.org/compute_pi/). Motif location was analyzed using MEME (https://meme-suite.org/meme/doc/meme.html?man_type=web). The conserved domains of eEF protein were evaluated using conservative structure domain tools in the NCBI database (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Two thousand base pairs upstream regions from the start codon site of GmeEF genes were sumbitted to Plant CARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to predict cis-element of GmeEF. The results of protein phylogeny, gene structure, and protein motif analysis were displayed using TBtools.
The eEF protein sequences of rice and Arabidopsis were extracted from Ensembl plant database (https://plants.ensembl.org/) for phylogenetic analysis. The phylogenetic trees of eEF proteins from 42 Glycine max, 29 Arabidopsis thaliana, and 30 Oryza sativa were constructed using the MEGA7.0.26 adjacency method with the following parameters: neighbor-joining (NJ) method, JTT+G model, and 1000 bootstrap replicates. The trees were visualized and modified using FigTree version v1.4.4 (https://github.com/rambaut/figtree/).
The RNA-seq data obtained from the JGI database (https://phytozome-next.jgi.doe.gov/report/gene/Gmax_Wm82_a2_v1/Glyma.02G182800) were used to explore the spatiotemporal expression patterns of GmeEF1A gene in pods, root-hairs, leaves, roots, nodules, seeds, shoot apical meristem(sam), stems, and flowers. The heat map of expression of GmeEF1A genes was plotted using TBtools.
Mechanical inoculation of SMV SC3 virus on soybean fully expanded unifoliate leaves. Samples were collected at 0 d, 7 d, and 14 d after treatment and stored at -80°C. Three biological replicates were analyzed for each treatment and control.
The three Arabidopsis mutants were ordered from Arabidopsis Biological Resource Center and the homozygous were confirmed by the primers in Supplementary Table 1. For TCV or CMV inoculations, transcripts synthesized in vitro from a cloned cDNA of TCV using T7 RNA polymerase were used for viral infections (Dempsey et al., 1993; Oh et al., 1995). Both the TCV and CMV transcript were suspended in an inoculation buffer then were used for rub-inoculation of leaves. Samples collected include leaves inoculated for 2 d and 3 d, and systemic leaves inoculated for 6 d and 9 d for western blot analysis. Respectively spot-treated with 5, 10, 15, and 20 mM paraquat (20 μl droplets each/leaf) for cell death assays. Phenotypic changes were observed after 1 day of inoculation. For paraquat treatments, paraquat was prepared in sterile water and leaves were spot inoculated with 10 mL of 5, 10, 15, 25, and 50 mM solutions. Lesion sizes were measured 48 h after paraquat application using vernier calipers. Results presented are representative of two or three separate treatments.
Detailed information of GmeEFs, including gene name, gene ID, predicted protein length, molecular weight, and theoretical isoelectric point distribution is listed in Table 1. The 42 GmeEFs genes ranged in size from 465 (GmeEF39) to 4068 bp (GmeEF42) and the corresponding predicted proteins ranged from 154 (GmeEF39) to 1355 (GmeEF42) amino acids. GmeEF39 showed the lowest molecular weight of 16929.51 Da, while GmeEF42 had the highest molecular weight of 148018.65 Da. Their isoelectric points (pI) varied from 5.02 (GmeEF34) to 9.22 (GmeEF39) with 27 members exhibiting pI values < 7, and 15 members with pI values > 7, suggesting that most of these genes encode acidic proteins. In addition, the basic protein was mainly concentrated in groups D1, D3, and E2.
We next conducted phylogenetic analysis to investigate the evolutionary relationships amongst eEF members from Glycine max, Oryza sativa, and Arabidopsis thaliana. For this, we constructed a neighbor connection phylogenetic tree using MEGA X and default parameters. 29 genes from Arabidopsis thaliana and 30 genes from Oryza sativa were analyzed along with the 42 GmeEF genes (Figure 1).The results indicate that eEF from three species can be divided into four groups named A to E. Group D was the largest and was further divided into 5 subgroups. Subgroup E2 was the largest and was annotated as eEF1A by CD-Search. Group A, group B, and group E were divided into 2 subgroups, respectively. Group C was the smallest with only 8 eEFs members. Each subgroup had eEFs from all three species. Notably, GmeEF30, GmeEF31, GmeEF16, GmeEF32, OseEF10, OseEF19, OseEF4, OseEF5, AteEF10, and AteEF19 did not belong to any subtype of the gene family.
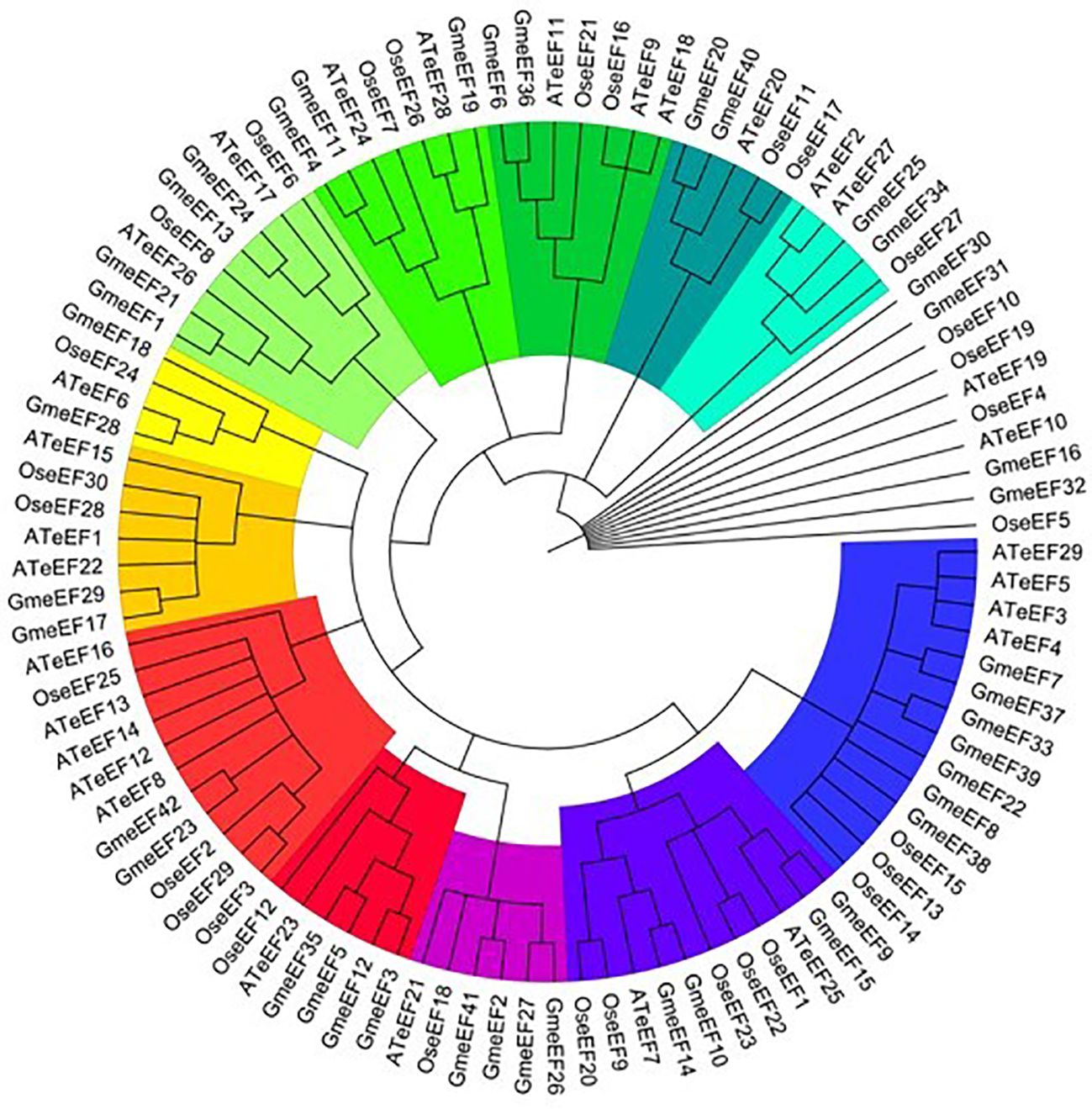
Figure 1. Phylogenetic tree of eEFs from Glycine max, Oryza sativa, and Arabidopsis thaliana was constructed by MEGA X according to NJ method. These proteins were divided into 12 subgroups and highlighted in different colors.
The phylogenetic relationship of GmeEFs neighbor-joining phylogenetic tree was constructed by MEGA7.0 software with the Poisson model and 1000 bootstrap replications (Figure 2A). Multiple Expectation Maximization for Motif Elicitation (MEME) software identified 10 conserved motifs in the GmeEF family protein sequences (Figure 2B). Motifs 2 and 4 were found in all GmeEF sequences except GmeEF39, which only contained motifs 3 and 9. In addition, except GmeEF27 and 39, all family members contain motif 1. Presumably, motifs 1, 2, and 4 were relatively conserved during the expansion of the GmeEF family and played an important role in the functions of GmeEF proteins. Motif 10 was only found in members of group E. In group D, only the D4 subgroup contains motif 8. In addition, motif 8 was contained in all groups except group D, suggesting functional differences within GmeEF family members. Exon–intron structural diversity often plays a key role in the evolution of gene families and can provide additional evidence to support phylogenetic grouping. Structures of GmeEF genes clustered in the same clade were very close, including number and position of exons and introns (Figure 2C). The GmeEF gene family contained 0-22 introns, of which GmeEF40, 12, and 5 were intronless. All genes in subgroup B2 contained the most introns, with the highest in GmeEF4 (22 introns). It was speculated that most eEF members contain different transcripts.
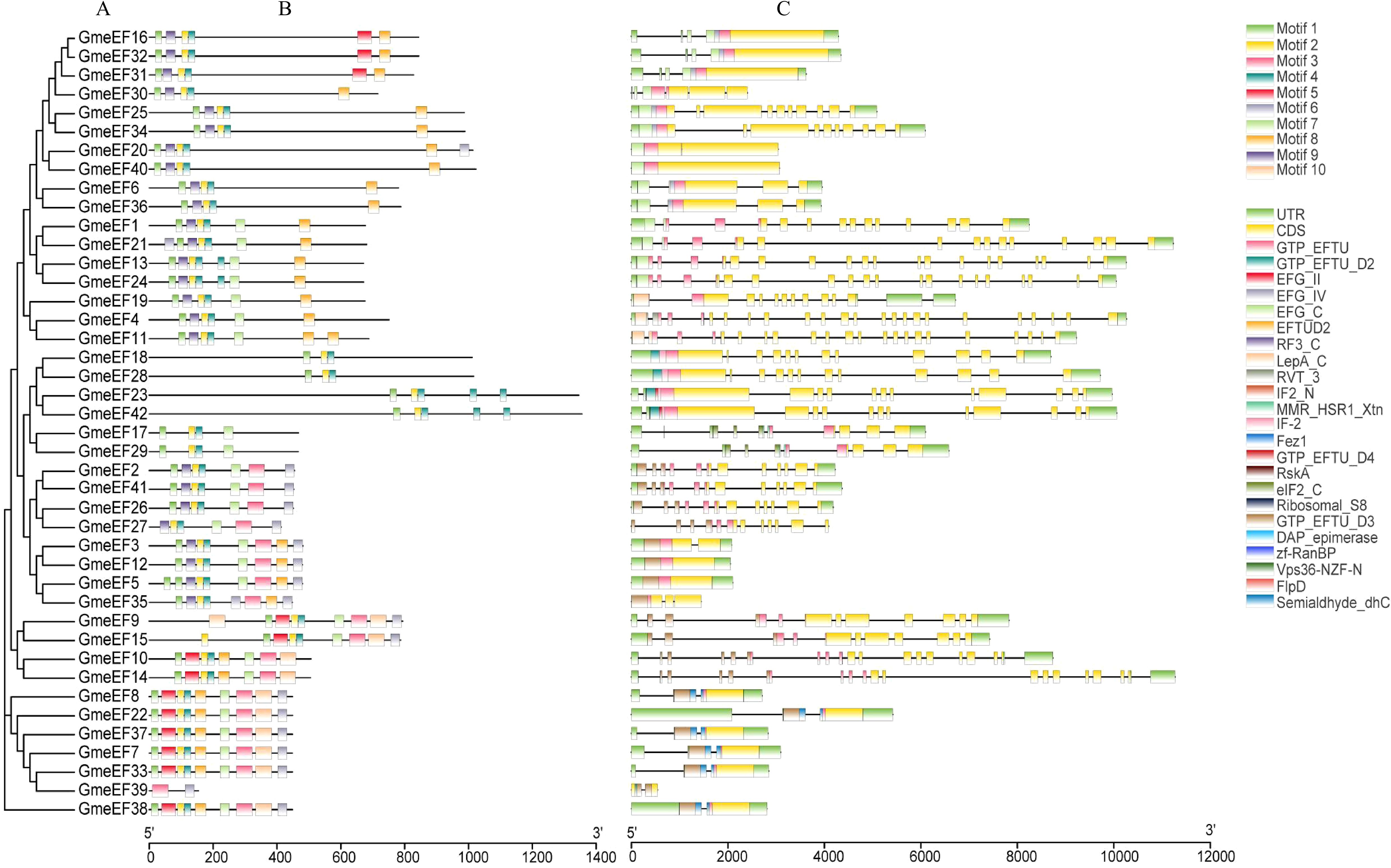
Figure 2. The phylogenetic tree, motif patten and gene structures of GmeEF. (A) The phylogenetic tree was constructed based on the full-length sequence of GmeEF proteins. (B) Conserved motifs of GmeEF proteins were identified by MEME suite. Each motif is represented by a specific color, with a total of 1–10 numbers. (C) Exon-intron organizations of GmeEF genes. Green boxes represent UTRs from 5’or 3’, yellow boxes represent CDS and black lines represent the introns.
To identify members of the GmeEF family, we used local protein BLAST on GmeEFs protein sequences from soybean and conducted Hidden Markov Model (HMM) analyses with conserved model (SLAC1.hmm) as the query. A total of 42 predicted GmeEF proteins were identified and designated GmeEF1-GmeEF42 based on their chromosomal locations (Figure 3). It was evident that the GmeEFs were unevenly distributed across the chromosomes. Specifically, chromosomes 5 and 15 contained the most GmeEF members with six; followed by chromosome 8 with five GmeEF members; chromosomes 17 and 19 with four GmeEF members each, chromosomes 6 and 10 each with three GmeEF members, chromosomes 4, 9, and 13 with two GmeEF members, chromosomes 2, 3, 12, 16, and 20 with one GmeEF gene each. No GmeEF genes were identified on chromosomes 1, 7, 11, 14, and 18. Moreover, one tandem duplication region comprising GmeEF30 and GmeEF31 was identified on chromosome 15.
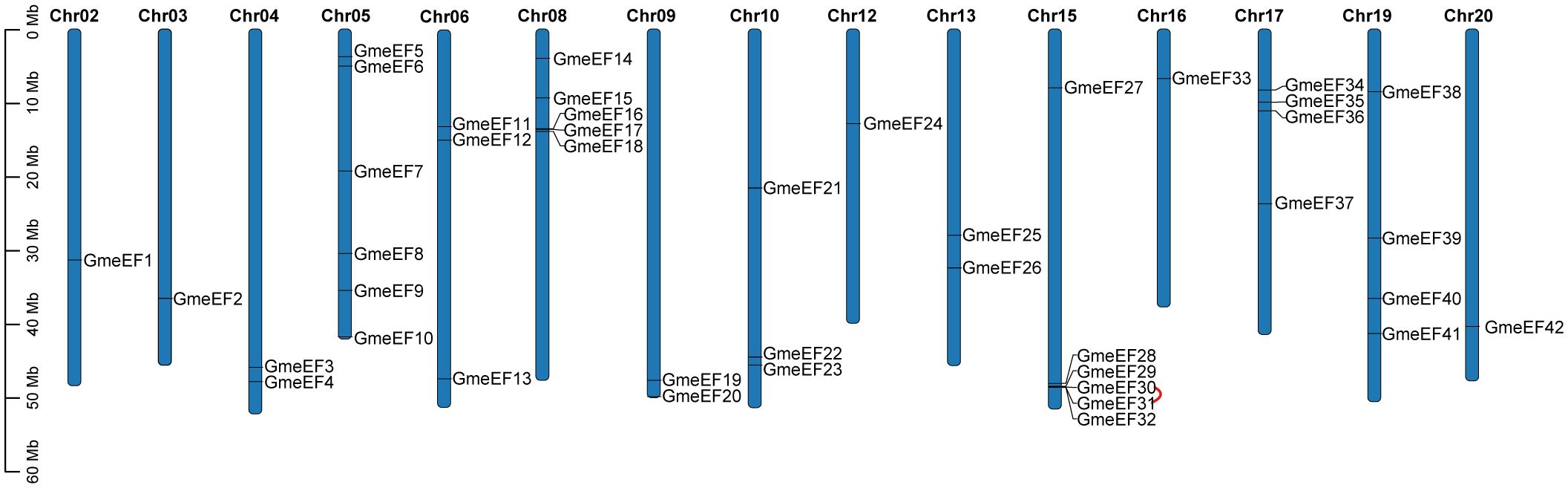
Figure 3. Mapping of GmeEF sequences on soybean chromosomes. The red sign represents a tandem duplication event region involving GmeEF30 and GmeEF31 on chromosome 15.
The control over gene transcription via upstream cis-acting regulatory elements (CAREs) is one of the most prominent mechanisms that regulates gene expression. To analyze the potential cis-elements, 2 kb regions upstream of GmeEF gene sequences were submitted to the Plant-CARE database. The type and position of cis-elements were marked as different icons. A total of 60 cis-acting elements were obtained, and 20 of them were selected for further analysis using TBtools (Figure 4). These cis-acting elements were related to phytohormone responsiveness, plant development, and stress responsive elements.
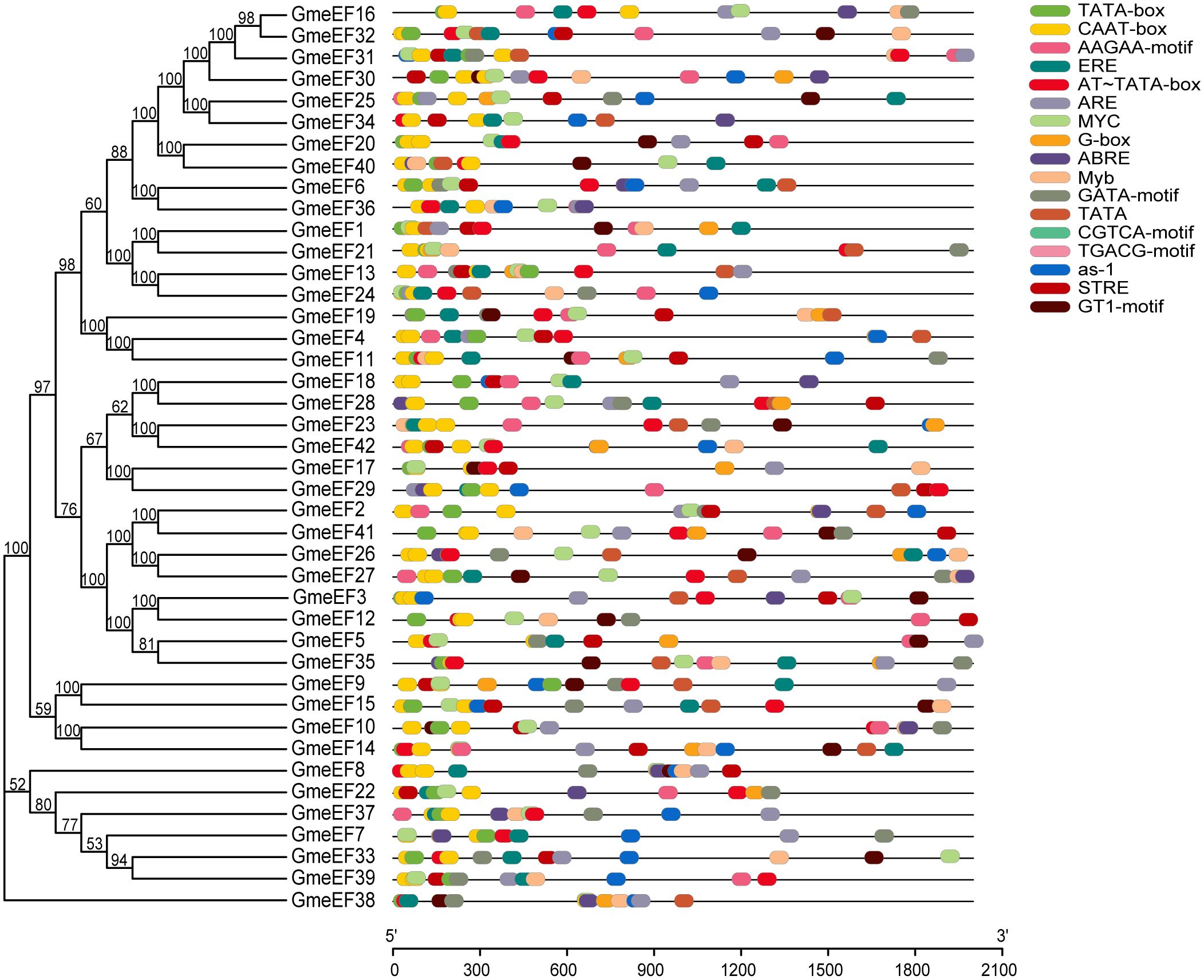
Figure 4. Distribution of cis-acting elements in soybean GmeEF promoter sequence. The figure shows 20 cis-acting elements that were selected and analyzed, and different colors indicate the elements related to growth and development, phytohormone responsive and stress responsive.
It is worth noting that the two promoters with the highest proportion in the GmeEF gene are CAAT box (45.32%) and TATA box (26.63%) (Table 2). Thus, CAAT and TATA box elements constituted the core promoter elements of GmeEF family members. Besides the cis-regulatory elements, AT~TATA-box, and TATA were also identified in majority of the GmeEF genes. Additionally, phytohormone responsive elements like methyl-jasmonate (MeJA)-responsive (CGTCA and TGACG), abscisic acid (ABA)-responsive element (ABRE), and ethylene-responsive elements (ERE) were identified. ERE participates in numerous development and stress responses (leaf development, senescence, fruit ripening, stimulation of germination, and oxidative stress). Other stress/development-associated motifs identified include, AAGAA-motif, GTI-motif, and activating sequence-1 (as-1); light response elements GATA-motif and G-box; drought-responsive myelocytomatosis (MYC) and myeloblastosis (MYB); antioxidant response element (ARE); and stress-responsive cis-elements (STRE). All in all, each GmeEF gene possessed different kinds and number of cis-elements, which are indicative of their unique functions or tissue-specific expression patterns.
To further examine GmeEF gene function, we investigated their expression profiles in various organs and at different growth stages of soybean. The transcriptome data of GmeEF genes obtained from the JGI database was used for comprehensive transcriptome analysis of different tissues including pods, root-hairs, leaves, roots, nodules, seeds, shoot apical meristem(sam), stems, and flowers (Figure 5). According to the results, GmeEF8, GmeEF37, GmeEF7, and GmeEF38 showed relatively high expression in most tissues than others, which shared similar expression patterns in specific tissue and showed relatively high expression level in root-hairs and seeds. Among them, GmeEF37 had the highest expression. Interestingly, all four genes belong to the E2 subgroup, which shared similar expression patterns in specific tissue. The transcripts of the GmeEF16, 17, and 32 genes were weakly expressed in all tissues. In addition to these, other family members also showed expression in leaf tissue, though their expression levels were negligible.
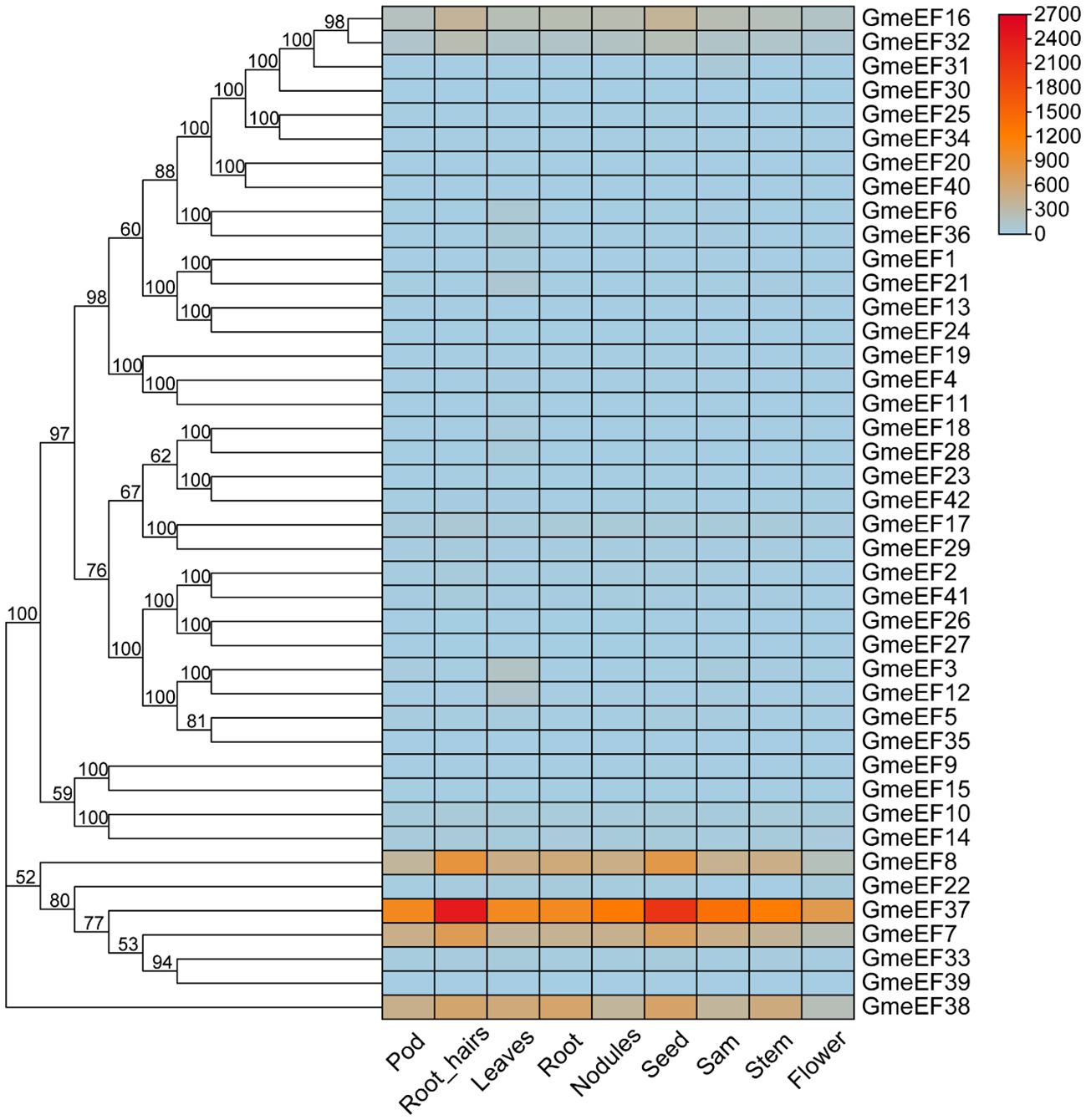
Figure 5. Expression analysis of GmeEFs during growth and development of different tissues of soybean. The RNA sequencing (RNA-Seq) dataset of 9 tissues of soybean collected from different development stages were obtained from JGI database to generate heatmap. The samples are listed at the bottom of each lane, and the color scale is shown at the right. The color box from blue to red indicate an increased expression level.
Synteny analysis was conducted among the soybean, rice, and Arabidopsis sequences (Figure 6). The results showed that majority of the Arabidopsis genes showed synteny with GmeEF syntenic genes. In contrast, synteny was detected for only two GmeEF genes in rice. There were more collinear genes and blocks between soybean and Arabidopsis than between soybean and rice. We found 12 GmeEFs (GmeEF1, GmeEF3, GmeEF4, GmeEF19, GmeEF20, GmeEF21, GmeEF22, GmeEF23, GmeEF24, and GmeEF42) did not have orthologs in either rice or Arabidopsis. As a dicotyledonous plant, soybean possessed superior synteny with Arabidopsis than rice, indicating that most eEF genes were formed after the differentiation of dicotyledonous and monocotyledonous plants.
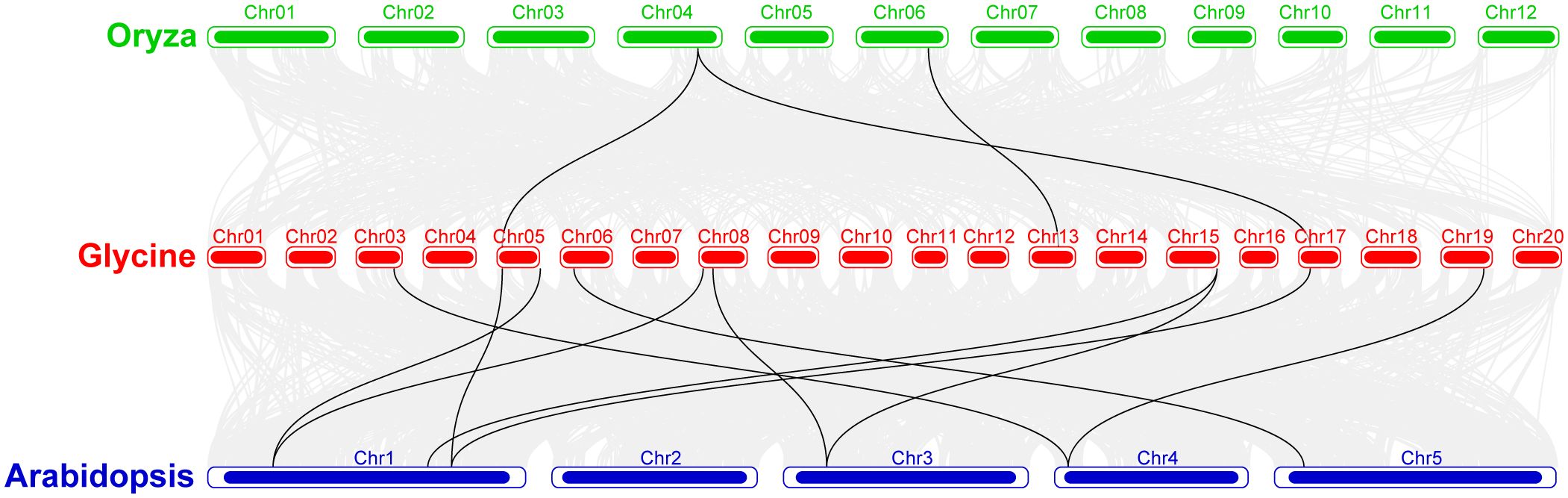
Figure 6. The collinearity analysis of the chromosomes from Glycine max, Oryza sativa, and Arabidopsis thaliana. The gray lines indicated all syntenic blocks within these three species genomes, and black lines indicated the syntenic GmeEF gene pairs.
To investigate the involvement of GmeEF genes in SMV pathogenesis, we tested their expression at various time points post SMV infection (0, 7, and 14 dpi) using RNA-seq analysis. The results showed that a large number of genes were suppressed in response to virus infection. GmeEF8 and GmeEF38 were significantly downregulated at 7 dpi; GmeEF22 presented unimodal change along with time, increasing on 7 dpi and then decreasing on 14 dpi; Eight genes (GmeEF1, 3, 5, 6, 12, 18, 21, 28, 32, and 36) had an opposite trend of decreasing on 7 dpi and then increasing on 14 dpi. These genes may play different roles in the process of SC3 infection (Figure 7). There was no significant change in transcript levels of other genes, indicating that these genes may not involve in SC3 infection. Taken together, the variational expression of GmeEF genes under SMV treatment implied that this gene family involved in virus infection in a complicated way.
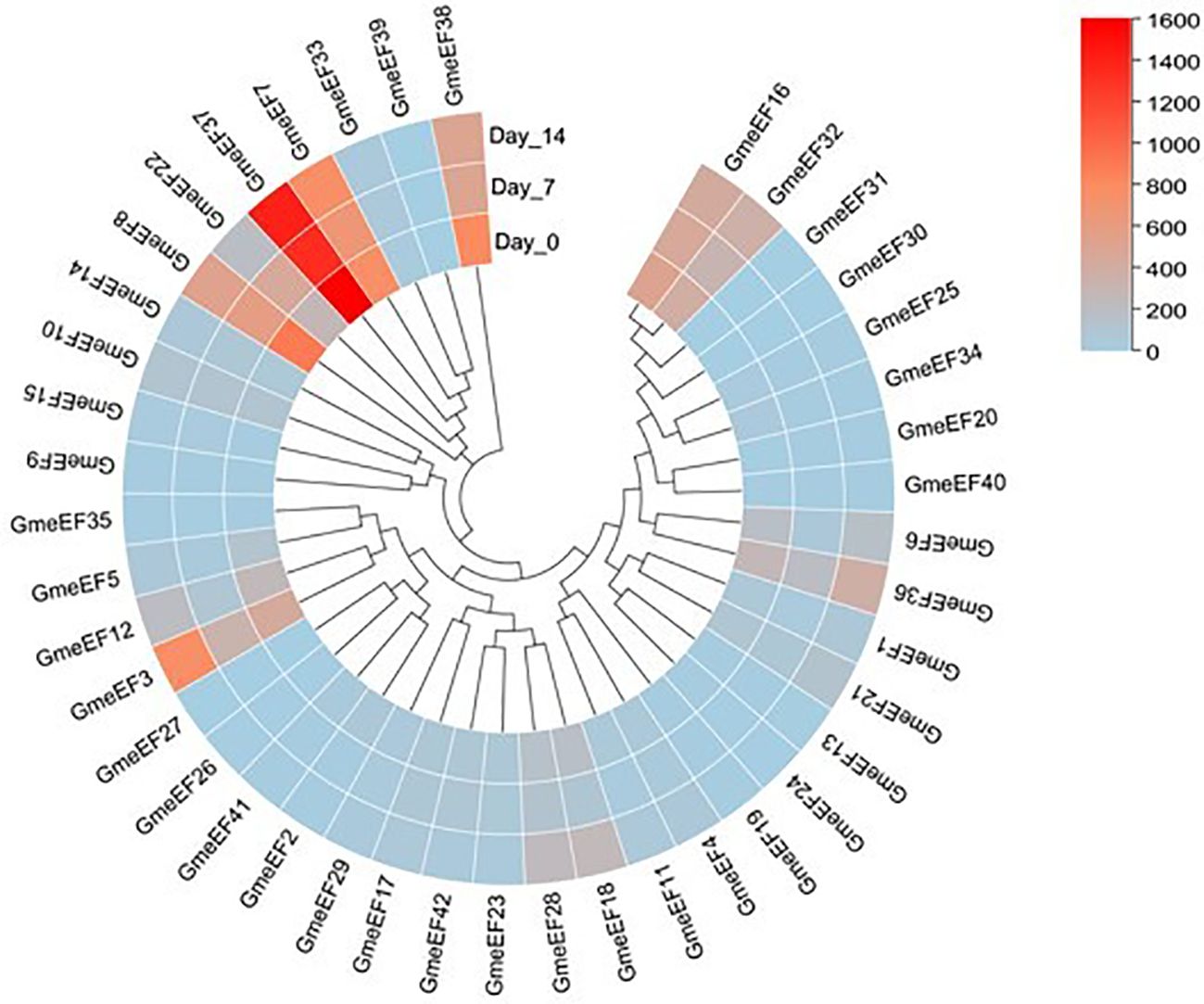
Figure 7. Expression analysis of GmeEF genes in response to the SMV strain SC3 infection. The heatmap of GmeEF genes was generated by the RNA-seq data which were obtained from the soybean plants inoculated with SMV SC3 at 0, 7, and 14 day post inoculation respectively. Three biological replicate experiments were performed for each treatment.
According to the summary of the systematic genome characterization of the eEF family, we could realize that the E2 subgroup members were mentioned many times, which harbored cis-acting elements related to stress resistance in most members. In addition, GmeEF8, GmeEF37, GmeEF7, and GmeEF38 had a relatively high expression in most tissues than others. In our previous research, the GmeEF37 transcript expression was induced after SMV infection and the knockout lines of GmeEF37 showed reduced amount of SMV, which indicated its negative regulation in SMV infection (Luan et al., 2016). Therefore, eEF1A was chosen for further studies in Arabidopsis.
Three homozygous SALK lines target to AteEF3 (AT1G07920), AteEF4 (AT1G07930), AteEF29 (AT5G60390) were genotyped by PCR with left border (LP) and right border (RP) primers as well as LBB1 and RP primers. Results showed that these three lines could not be amplified by LP and RP primers while the wide type plants showed band size over 1350 bp. The primer LBB1 in combination with RP could generate bands and wild type plants did not (Figures 8A, B). As we tested the expression of each specific genes in these three mutant lines, only the corresponding target gene was knocked out and the other two genes were not affected with the amplified size of 1350 bp length band, and tublin was control (Figure 8C). In order to explore the function of the Arabidopsis eEF1A in response to virus and chemical stress, the mutants were challenged with Turnip crinkle virus (TCV), Cucumber mosaic virus (CMV), and paraquat (1,1’-dimethyl-4,4’-bipyridinium dichloride) that induces cell death by promoting the formation of reactive oxygen species (Melicher et al., 2024). Western blot showed TCV and CMV accumulated the same amount in the local leaves of 2 and 4 dpi, as well as the distal leaves of 6 and 9 dpi of three mutant lines and did not show any difference with wild type plants (Figure 8D). The all mutants and wild type plants also exhibited impaired cell death when treated with paraquat (Figure 8E). All these data indicated that knocked out single copy of AteEF1A did not affect the replication of the TCV and CMV, as well as the response to paraquat. However complementation studies are required to make firm conclusions about functional similarities between soybean and Arabidopsis eEF1A proteins.
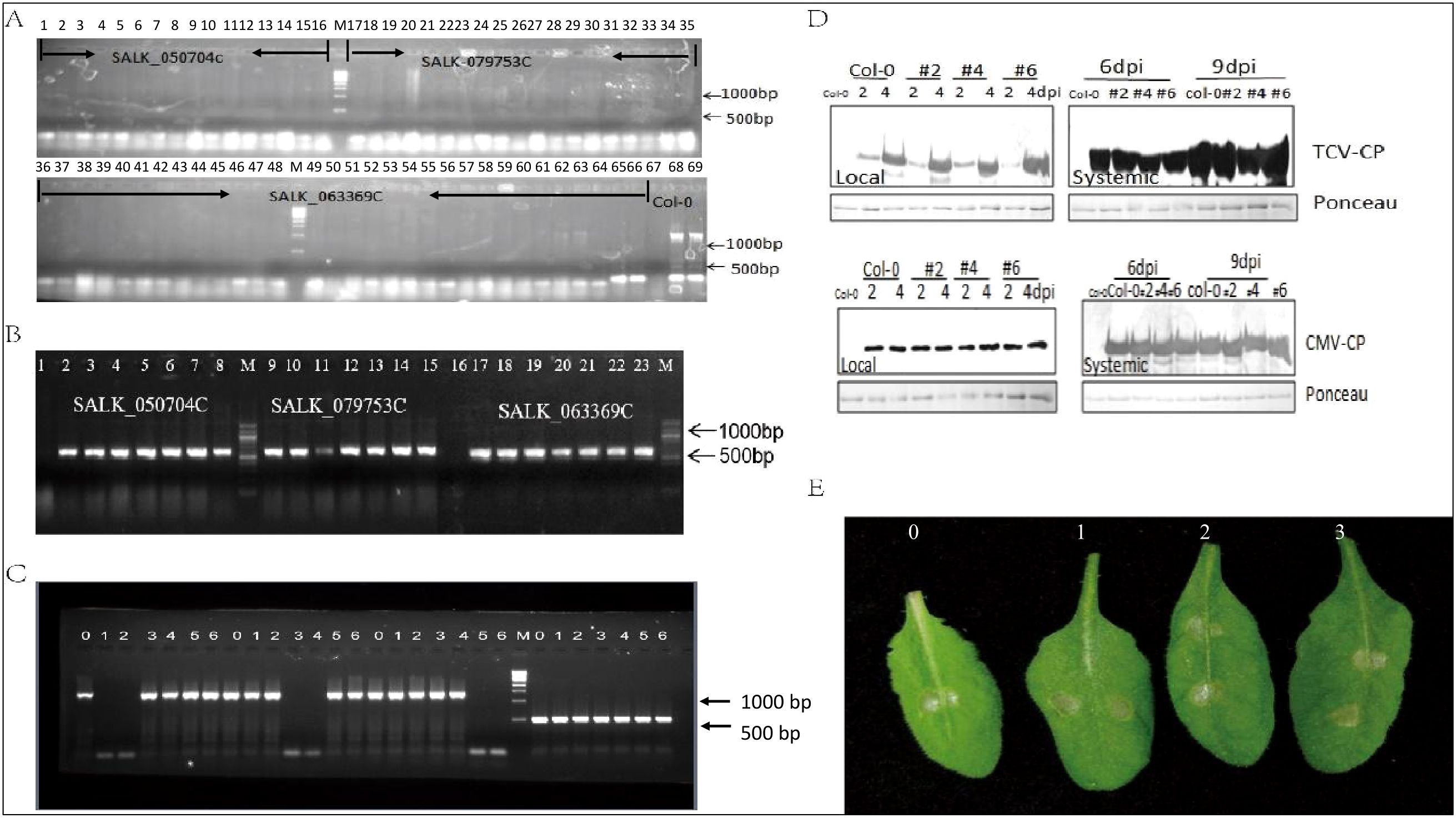
Figure 8. Elongation factor family function analysis. (A) Amplification of target genes by LP and RP primers: 1-16 SALK_050704C (AT1G07930); 16-35: SALK_079753C (AT1G07920); 36-66: SALK_063369C (AT5G60390). 67 empty; 68-69 Col-0. (B) Amplification of target genes expression by LBB1+RP. 1-8: SALK_050704C (AT1G07930); 9-15: SALK_079753C (AT1G07920); 17-23: SALK_063369C (AT5G60390). (C) Transcript level of 3 target genes. 0: Col-0; 1-2:SALK_050704C (AT1G07930); 3-4: SALK_079753C (AT1G07920); 5-6: SALK_063369C (AT5G60390). (D) Western blot analysis of TCV and CMV coat protein (CP) amount in three mutants. (E) Symptoms of three mutants after paraquat treatment, 0 is the Col-0, 1 is SALK_050704C, 2 is SALK_079753C, 3 is SALK_063369C.
It was well recognized that the eEF1A family comprises multiple copies in cotton, Arabidopsis, tomato, lily, and cassava (Pokalsky et al., 1989; Xu et al., 2007; Ransom-Hodgkins, 2009; Suhandono et al., 2014; Sun et al., 2020). eEF1A was well-known as an important component for protein biosynthesis (Xu et al., 2022) and a host factor in viral pathogenesis. In this study, 42 eEF family members were identified and systematically analyzed in soybean, their gene characteristics, chromosomal locations, phylogenetic relationships, domain composition, and cis-elements were predicted, tissue expression patterns and response to virus infection were carried out, which provide an opportunity to better understand of the eEF proteins. In the present study, 42, 30, and 29 eEF genes were identified in soybean, rice, and Arabidopsis respectively. To explore the evolutionary process of eEFs, phylogenetic analysis was conducted with comparing the eEF genes of soybean, rice, and Arabidopsis revealing 5 groups designated A to D. Subsequently, 5 groups were divided into 12 subgroups. It is worth noting that GmeEF30, GmeEF31, GmeEF16, GmeEF32, OseeEF10, OseeEF19, OseeEF4, OseeEF5, AteEF19, and OseeEF10 do not belong to any subtype of the gene family. Based on this, we speculate that the common ancestor of eEFs may evolve independently among different species from monocotyledon to dicotyledon.
The domains and motifs of transcription factors play an important role in protein interaction and DNA binding (Corbella et al., 2021). GmeEFs were identified in soybean with ten motifs. Motifs 2 and 4 were found highly conserved in almost all of the GmeEFs. Gene structure analysis of GmeEFs revealed the intron-exon structure was highly conserved in the same group and had similar intron region distribution. Almost all GmeEFs contained one or more introns, except for GmeEF40, 12, and 5 which were intronless, indicating GmeEFs might have functional diversity by multiple splicing during growth and development in plants. In addition, different kinds of cis-elements were analyzed including stress responsiveness, phytohormone response, light response, and growth regulation. We also collected transcriptome data from different growth stages and tissues of soybeans, i.e, pods, root hairs, leaves, roots, nodules, seeds, sam, stems, and flowers and conducted comprehensive transcriptome analysis. The results revealed that the expression patterns of GmeEF genes were diverse. GmeEF16, GmeEF23, GmeEF8, GmeEF37, GmeEF7, and GmeEF38 had a relatively high expression in most tissues than others, which shared similar expression patterns in specific tissue and showed relatively high expression levels in root-hairs and seeds, suggesting that these subgroup members might play important roles in root-hairs and seed development. A small number of GmeEF genes were expressed in leaves and the amount of expression was also very small. The majority of the GmeEF gene family members showed little expression. The tissue-specific/preferential expressions of eEF1A genes were found in other plants. In Arabidopsis thaliana, the highest level of eEF1A expression was found in specific tissue types including seeds, embryos, and roots (Ransom-Hodgkins, 2009). It may be because there are more polyribosomes in the meristem or the region of rapid division.
It has been previously demonstrated that eEF1A is involved in the regulation of responses to abiotic and biotic stresses. eEFs potential influence and connection with heat tolerance of plants has been the subject of many studies. The study have shown that a significant, positive, linear correlation was found between the expression of eEF1A and small grains productivity under heat-stress conditions (Djukić et al., 2019).eEF1A was upregulated in peanut worms from the M and L tidal flats, which response to sedimentary environments that have lower porosity and greater organic matter or even a high abundance of pathogens (Li et al., 2023). Virus invasion is a kind of biological stress. eEFs predominantly eEF1A, act in partially characterized complexes sometimes involving additional eEFs so as to enhance or inhibit virus replication (Chen et al., 2021; Helderman et al., 2021; Gan et al., 2024). Although plant viruses encode a number of essential proteins their genome is exceptionally small and they must depend on host resources and factors for their genome replication and movement (Wu et al., 2024). As an abundant, multifunctional cellular protein, the cellular factor eEF1A plays an important role in the regulation of F-actin stress fiber formation required for Respiratory syncytial virus assembly and release (Snape et al., 2018); eEF1A, an essential protein in the translation machinery, interacted with two proteins of a fish rhabdovirus, Siniperca chuatsi rhabdovirus, and inhibited virus infection via two different mechanisms (Meng et al., 2023);eEF1A and other subunits of the eEF1 complex have been suggested to be essential for RNA virus replication (Zeenko et al., 2002; Yamaji et al., 2006, Yamaji et al., 2009; Komoda et al., 2014). Both eEF1A and eEF1B play essential roles in the multiplication of Potato virus X (PVX) in pepper (Hwang et al., 2015); A2 of the four eEF1A was increased expression from infection with Tobacco mosaic virus (TMV) and Phytophthora infections in Arabidopsis thaliana (Ransom-Hodgkins, 2009); In soybean, the results of a previous study showed that SMV-P3 targets host elongation factors resulting in UPR, which in turn facilitates SMV replication (Luan et al., 2016). In this study, we investigated whether SMV SC3 would affect the expression patterns of GmeEF family genes. RNA-seq results showed that a large members of the GmeEF gene family were induced or repressed at 7 or 14 dpi. GmeEF3 induced significantly on 14 dpi, and GmeEF8 and 38 were significantly down-regulated on 7 dpi. The transcription levels of other genes remain unchanged under SC3 infection. The results indicated that the pathogenesis of the virus came from the complex interaction between SMV SC3 and eEFs.
In addition, several eEF genes were selected for functional analysis experiments. In order to explore the function of the Arabidopsis eEF1A in response to the virus and chemical stress, the mutants (At1G07920, At1G07930, and At5G60390) were challenged with TCV, CMV, and paraquat. Unfortunately, the knockout of a single copy of AteEF1A did not affect the replication of TCV and CMV as well as the response to paraquat. The completion of the virus life cycle depends on the intricate interplay between virus and host. However, our knowledge of the identities and functions of such host factors remains largely unknown. The dynamics of virus-host interaction networks will help us to better understand plant resistance responses and develop resistant cultivars to viruses.
In this study, 42 eEF genes were identified in soybean. Subsequently, we performed a systematic genome characterization of the eEF family genes. These GmeEF genes were located on 15 chromosomes and divided into 12 subgroups according to the phylogenetic tree. According to the analysis of conservative domains, gene structures, cis-acting elements, and the expression of GmeEFs in different parts of different periods, we found that GmeEFs belonging to the same subgroup had similar gene structures and motif compositions. We found that the GmeEF family was highly expressed in root-hairs and seeds. The function of elongation factor family was analyzed in Arabidopsis. The expression of each specific gene was tested in the three EF1a mutants and results showed that each mutant only knock out the corresponding gene and had no effect on other copies. Three mutant lines were used to test response to TCV, CMV, and paraquat respectively. We found that there was no difference between the wild type plants and the mutants in the viral expression or morphology. In a word, EF1a family does not involved in pathogens response in Arabidopsis, or it may involve in, but a single mutant is not enough. Other isoforms may confer it defect. This has laid the foundation for the pathogenesis of SMV and provided new ideas for the prevention and treatment of SMV.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
HL: Writing – review & editing, Writing – original draft, Supervision, Funding acquisition, Conceptualization. DS: Writing – original draft, Validation, Methodology, Investigation, Formal Analysis, Data curation. KH: Writing – original draft, Visualization, Validation, Investigation. SL: Writing – original draft, Validation, Investigation. HX: Writing – original draft, Validation, Investigation. PK: Writing – review & editing, Project administration, Conceptualization. AK: Funding acquisition, Writing – review & editing, Project administration. LZ: Supervision, Writing – review & editing, Funding acquisition.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The authors acknowledge support from the National Natural Science Foundation of China (32301895), Open project of National Key Laboratory for Crop Genetics and Germplasm Enhancement (ZW202005) and Natural Science Foundation of Shandong Province (ZR2020QC117) to HL, National key research and development program (2022YFD2300101-1), Seed-Industrialized Development Program in Shandong Province (2021LZGC003) and Science & Technology Specific Projects in Agricultural High-tech Industrial Demonstration Area of the Yellow River Delta (2022SZX18) to LZ, Seed-Industrialized Development Program in Shandong Province (2023LZGC008-001) and Shandong Soybean Industrial Technology System of China (SDAIT-28-02) to LZ and HL. This article is based upon work supported by (while serving at) the National Science Foundation for Aardra Kachroo.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1421221/full#supplementary-material
ABRE, ABA-responsive element; ARE, Antioxidant response element; as-1, Activating sequence-1; CAREs, Cis-acting regulatory elements; CDS, Coding sequences; CMV, Cucumber mosaic virus; CWMV, Chinese wheat mosaic virus; eEF, Eukaryotic elongation factor; eEF1A, Eukaryotic elongation factor 1 alpha; ER, Endoplasmic reticulum; ERE, Ethylene-responsive; HMM, Hidden Markov Model; LP, Left border; MeJA, Methyl-jasmonate; MEME, Multiple Expectation Maximization for Motif Elicitation; MYB, Myeloblastosis; MYC, Myelocytomatosis; NJ, Neighbor-joining; ORF, Open reading frame; PI, Isoelectric point; PVX, Potato virus X; RNA-Seq, RNA sequencing; RP, Right border; SMV, Soybean mosaic virus; STRE, Stress-responsive cis-elements; TAIR, The Arabidopsis Information Resource; TBSV, Tomato bushy stunt virus; TCV, Turnip crinkle virus; TRV, Tobacco rattle virus; TMV, Tobacco mosaic virus; TSWV, Tomato spotted wiltorthotospo virus; TYLCV, Tomato yellow leaf curl virus; UTR, Untranslated region.
Abbas, W., Kumar, A., Herbein, G. (2015). The eEF1A proteins: at the crossroads of oncogenesis, apoptosis, and viral infections. Front. Oncol. 5. doi: 10.3389/fonc.2015.00075
Akram, Z., Ahmed, I., Mack, H., Kaur, R., Silva, R. C., Castilho, B. A., et al. (2020). Yeast as a model to understand actin-mediated cellular functions in mammals-illustrated with four actin cytoskeleton proteins. Cells. 9, 627. doi: 10.3390/cells9030672
Chen, X., He, L., Xu, M., Yang, J., Li, J., Zhang, T., et al. (2021). Binding between elongation factor 1A and the 3′-UTR of Chinese wheat mosaic virus is crucial for virus infection. Mol. Plant Pathology. 22, 1383–1398. doi: 10.1111/mpp.13120
Corbella, M., Liao, Q., Moreira, C., Parracino, A., Kasson, P. M., Kamerlin, S. C. L. (2021). The N-terminal helix-turn-helix motif of transcription factors marA and rob drives DNA recognition. J. Phys. Chem. B 125, 6791–6806. doi: 10.1021/acs.jpcb.1c00771
Dempsey, D., Wobbe, K. K., Klessig, D. F. (1993). Resistance and susceptible responses of Arabidopsis thaliana to Turnip crinkle virus. Phytopathology. 83, 1021–1029. doi: 10.1105/tpc.12.5.677
Djukić, N., Knežević, D., Pantelić, D., Živančev, D., Torbica, A., Markovic, S. (2019). Expression of protein synthesis elongation factors in winter wheat and oat in response to heat stress. J. Plant Physiol. 240, 153015. doi: 10.1016/j.jplph.2019.153015
Gamarnik, A. V., Li, Z., Pogany, J., Tupman, S., Esposito, A. M., Kinzy, T. G., et al. (2010). Translation elongation factor 1A facilitates the assembly of the tombusvirus replicase and stimulates minus-strand synthesis. PloS Pathogens. 6, e1001175. doi: 10.1371/journal.ppat.1001175
Gan, H., Zhou, X., Lei, Q., Wu, L., Niu, J., Zheng, Q. (2024). RNA-dependent RNA polymerase of SARS-CoV-2 regulate host mRNA translation efficiency by hijacking eEF1A factors. Biochim. Biophys. Acta (BBA)-Molecular Basis Dis. 1870, 166871. doi: 10.1016/j.bbadis.2023.166871
Garcia-Ruiz, H. (2019). Host factors against plant viruses. Mol. Plant Pathology. 20, 1588–1601. doi: 10.1111/mpp.12851
Gromadski, K. B., Schümmer, T., Strømgaard, A., Knudsen, C. R., Kinzy, T. G., Rodnina, M. V. (2007). Kinetics of the interactions between yeast elongation factors 1A and 1Bα, guanine nucleotides, and aminoacyl-tRNA. J. Biol. Chem. 282, 35629–35637. doi: 10.1074/jbc.M707245200
Guo, J., Ling, H., Wu, Q., Xu, L., Que, Y. (2014). The choice of reference genes for assessing gene expression in sugarcane under salinity and drought stresses. Sci. Rep. 4, 7042. doi: 10.1038/srep07042
Helderman, T., Deurhof, L., Bertran, A., Boeren, S., Fokkens, L., Kormelink, R., et al. (2021). An isoform of the eukaryotic translation elongation factor 1A (eEF1a) acts as a pro-viral factor required for Tomato spotted wilt virus disease in Nicotiana benthamiana. Viruses. 13, 2190. doi: 10.3390/v13112190
Hetherington, A. M., Sawyez, C. G., Sutherland, B. G., Robson, D. L., Arya, R., Kelly, K., et al. (2016). Treatment with didemnin B, an elongation factor 1A inhibitor, improves hepatic lipotoxicity in obese mice. Physiol. Rep. 4, e12963. doi: 10.14814/phy2.12963
Hou, Y., Liu, R., Xia, M., Sun, C., Zhong, B., Yu, J., et al. (2021). Nannocystin ax, an eEF1A inhibitor, induces G1 cell cycle arrest and caspase-independent apoptosis through cyclin D1 downregulation in colon cancer in vivo. Pharmacol. Res. 173, 105870. doi: 10.1016/j.phrs.2021.105870
Hwang, J., Lee, S., Lee, J.-H., Kang, W.-H., Kang, J.-H. (2015). Plant translation elongation factor 1Bβ facilitates (PVX) infection and interacts with PVX triple gene block protein 1. PloS One 10, e0128014. doi: 10.1371/journal.pone.0128014
Kanibolotsky, D. S., Novosyl’na, O. V., Abbott, C. M., Negrutskii, B. S., El’skaya, A. V. (2008). Multiple molecular dynamics simulation of the isoforms of human translation elongation factor 1A reveals reversible fluctuations between “open” and “closed” conformations and suggests specific for eEF1A1 affinity for Ca2+-calmodulin. BMC Struct. Biol. 8, 1–16. doi: 10.1186/1472-6807-8-4
Komoda, K., Ishibashi, K., Kawamura-Nagaya, K., Ishikawa, M. (2014). Possible involvement of eEF1A in Tomato spotted wilt virus RNA synthesis. Virology. 468-470, 81–87. doi: 10.1016/j.virol.2014.07.053
Li, J., Wen, J., Hu, R., Pei, S., Li, T., Shan, B., et al. (2023). Transcriptome responses to different environments in intertidal zones in the peanut worm sipunculus nudus. Biology 12, 1182. doi: 10.3390/biology12091182
Li, Z., Pogany, J., Panavas, T., Xu, K., Esposito, A. M., Kinzy, T. G., et al. (2009). Translation elongation factor 1A is a component of the tombusvirus replicase complex and affects the stability of the p33 replication co-factor. Virology. 385, 245–260. doi: 10.1016/j.virol.2008.11.041
Liu, S., Hausmann, S., Carlson, S. M., Fuentes, M. E., Francis, J. W., Pillai, R., et al. (2019). METTL13 methylation of eEF1A increases translational output to promote tumorigenesis. Cell. 176, 491–504.e421. doi: 10.1016/j.cell.2018.11.038
Liu, C., Yang, Z., Cai, M., Shi, Y., Cui, H., Yuan, J. (2021). Generation of Plasmodium yoelii malaria parasite for conditional degradation of proteins. Mol. Biochem. Parasitology. 241, 111346. doi: 10.1016/j.molbiopara.2020.111346
Luan, H., Shine, M. B., Cui, X., Chen, X., Ma, N., Kachroo, P., et al. (2016). The Potyviral P3 protein targets eukaryotic elongation factor 1A to promote the unfolded protein response and viral pathogenesis. Plant Physiol. 172, 221–234. doi: 10.1104/pp.16.00505
Małecki, J., Aileni, V. K., Angela, Y. Y., Schwarz, J., Moen, A., Sørensen, V., et al. (2017). The novel lysine specific methyltransferase METTL21B affects mRNA translation through inducible and dynamic methylation of Lys-165 in human eukaryotic elongation factor 1 alpha (eEF1A). Nucleic Acids Res. 45, 4370–4389. doi: 10.1093/nar/gkx002
Markovic, S. M., Živančev, D., Horvat, D., Torbica, A., Jovankić, J., Djukić, N. H. (2021). Correlation of elongation factor 1A accumulation with photosynthetic pigment content and yield in winter wheat varieties under heat stress conditions. Plant Physiol. Biochem. 166, 572–581. doi: 10.1016/j.plaphy.2021.06.035
Maruyama, K., Imai, H., Kawamura, M., Ishino, S., Ishino, Y., Ito, K., et al. (2019). Switch of the interactions between the ribosomal stalk and EF1A in the GTP- and GDP-bound conformations. Sci. Rep. 9, 14761. doi: 10.1038/s41598-019-51266-x
Melicher, P., Dvořák, P., Řehák, J., Šamajová, O., Pechan, T., Šamaj, J., et al. (2024). Methyl viologen-induced changes in the Arabidopsis proteome implicate PATELLIN 4 in oxidative stress responses. J. Exp. Botany. 75, 405–421. doi: 10.1093/jxb/erad363
Meng, X.-Y., Jiang, Q.-Q., Yu, X.-D., Zhang, Q.-Y., Ke, F. (2023). Eukaryotic translation elongation factor 1 alpha (eEF1A) inhibits Siniperca chuatsi rhabdovirus (SCRV) infection through two distinct mechanisms. J. Virol. 97, e01226–e01223. doi: 10.1128/jvi.01226-23
Migliaccio, N., Martucci, N. M., Ruggiero, I., Sanges, C., Ohkubo, S., Lamberti, A., et al. (2016). Ser/Thr kinases and polyamines in the regulation of non-canonical functions of elongation factor 1A. Amino Acids 48, 2339–2352. doi: 10.1007/s00726-016-2311-3
Mills, A., Gago, F. (2021). On the need to tell apart fraternal twins eEF1A1 and eEF1A2, and their respective outfits. Int. J. Mol. Sci. 22, 6973. doi: 10.3390/ijms22136973
Momcilovic, I., Pantelić, D., Zdravković-Korać, S., Oljača, J., Rudić, J., Fu, J. (2016). Heat-induced accumulation of protein synthesis elongation factor 1A implies an important role in heat tolerance in potato. Planta. 244, 671–679. doi: 10.1007/s00425-016-2534-2
Oh, J.-W., Kong, Q., Song, C., Carpenter, C., Simon, A. E. (1995). Open reading frames of turnip crinkle virus involved in satellite symptom expression and incompatibility with Arabidopsis thaliana ecotype Dijon. MPMI-Molecular Plant Microbe Interactions. 8, 979–987. doi: 10.1094/mpmi-8-0979
Pokalsky, A., Hiatt, W., Ridge, N., Rasmussen, R., Houck, C., Shewmaker, C. (1989). Structure and expression of elongation factor 1α in tomato. Nucleic Acids Res. 17, 4661–4673. doi: 10.1093/nar/17.12.4661
Ransom-Hodgkins, W. D. (2009). The application of expression analysis in elucidating the eukaryotic elongation factor one alpha gene family in Arabidopsis thaliana. Mol. Genet. Genomics 281, 391–405. doi: 10.1007/s00438-008-0418-2
Romaus-Sanjurjo, D., Saikia, J. M., Kim, H. J., Tsai, K. M., Le, G. Q., Zheng, B. (2022). Overexpressing eukaryotic elongation factor 1 alpha (eEF1A) proteins to promote corticospinal axon repair after injury. Cell Death Discovery. 8, 390. doi: 10.1038/s41420-022-01186-z
Juette, M. F., Carelli, J. D., Rundlet, E. J., Brown, A., Shao, S., Ferguson, A., et al. (2022). Didemnin B and ternatin-4 differentially inhibit conformational changes in eEF1A required for aminoacyl-tRNA accommodation into mammalian ribosomes. eLife. 11, e81608. doi: 10.7554/eLife.81608
Snape, N., Li, D., Wei, T., Jin, H., Lor, M., Rawle, D. J., et al. (2018). The eukaryotic translation elongation factor 1A regulation of actin stress fibers is important for infectious RSV production. Virol. J. 15, 1–10. doi: 10.1186/s12985-018-1091-7
Soares, D. C., Barlow, P. N., Newbery, H. J., Porteous, D. J., Abbott, C. M. (2009). Structural models of human eEF1A1 and eEF1A2 reveal two distinct surface clusters of sequence variation and potential differences in phosphorylation. PloS one 4(7), e6315. doi: 10.1371/journal.pone.0006315
Suhandono, S., Apriyanto, A., Ihsani, N. (2014). Isolation and characterization of three cassava elongation factor 1 apha (MeEF1A) promoters. PloS One 9, e84692. doi: 10.1371/journal.pone.0084692
Sun, D., Ji, X., Jia, Y., Huo, D., Si, S., Zeng, L., et al. (2020). LreEF1A4, a translation elongation factor from lilium regale, is pivotal for Cucumber mosaic virus and Tobacco rattle virus infections and tolerance to salt and drought. Int. J. Mol. Sci. 21, 2083. doi: 10.3390/ijms21062083
Wazwaz, A. T. H. (2013). Collapse of resistance to Tomato yellow leaf curl virus in tomato upon silencing the elongation factor 1-alpha gene. Master of Science Thesis (Bethlehem, Palestine: Bethlehem University).
Wu, J., Zhang, Y., Li, F., Zhang, X., Ye, J., Wei, T., et al. (2024). Plant virology in the 21st century in China: Recent advances and future directions. J. Integr. Plant Biol. 66, 579–622. doi: 10.1111/jipb.13580
Xu, B., Liu, L., Song, G. (2022). Functions and regulation of translation elongation factors. Front. Mol. Biosciences. 8. doi: 10.3389/fmolb.2021.816398
Xu, W.-L., Wang, X.-L., Wang, H., Li, X.-B. (2007). Molecular characterization and expression analysis of nine cotton GhEF1A genes encoding translation elongation factor 1A. Gene. 389, 27–35. doi: 10.1016/j.gene.2006.09.014
Yamaji, Y., Kobayashi, T., Hamada, K., Sakurai, K., Yoshii, A., Suzuki, M., et al. (2006). In vivo interaction between Tobacco mosaic virus RNA-dependent RNA polymerase and host translation elongation factor 1A. Virology. 347, 100–108. doi: 10.1016/j.virol.2005.11.031
Yamaji, Y., Sakurai, K., Hamada, K., Komatsu, K., Ozeki, J., Yoshida, A., et al. (2009). Significance of eukaryotic translation elongation factor 1A in Tobacco mosaic virus infection. Arch. Virology. 155, 263–268. doi: 10.1007/s00705-009-0571-x
Zeenko, V. V., Ryabova, L. A., Spirin, A. S., Rothnie, H. M., Hess, D., Browning, K. S., et al. (2002). Eukaryotic elongation factor 1A interacts with the upstream pseudoknot domain in the 3′ untranslated region of Tobacco mosaic virus RNA. J. Virology. 76, 5678–5691. doi: 10.1128/jvi.76.11.5678-5691.2002
Keywords: GmeEF, eEF1A, bioinformatics, soybean, virus infection
Citation: Luan H, Song D, Huang K, Li S, Xu H, Kachroo P, Kachroo A and Zhao L (2024) Genome-wide analysis of the soybean eEF gene family and its involvement in virus resistance. Front. Plant Sci. 15:1421221. doi: 10.3389/fpls.2024.1421221
Received: 22 April 2024; Accepted: 31 July 2024;
Published: 19 August 2024.
Edited by:
Kai Wang, Nantong University, ChinaReviewed by:
Karthikeyan Adhimoolam, Jeju National University, Republic of KoreaCopyright © 2024 Luan, Song, Huang, Li, Xu, Kachroo, Kachroo and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hexiang Luan, aGV4aWFuZ2xAcWF1LmVkdS5jbg==; Aardra Kachroo, QWFyZHJhLkthY2hyb29AdWt5LmVkdQ==; Longgang Zhao, emhhb2xvbmdnYW5nQHFhdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.