- 1School of Agriculture and Biotechnology, Hunan University of Humanities, Science and Technology, Loudi, China
- 2Shuangfeng Agriculture and Rural Bureau, Loudi, Hunan, China
Heavy metal pollution has become a serious concern across the globe due to their persistent nature, higher toxicity, and recalcitrance. These toxic metals threaten the stability of the environment and the health of all living beings. Heavy metals also enter the human food chain by eating contaminated foods and cause toxic effects on human health. Thus, remediation of HMs polluted soils is mandatory and it needs to be addressed at higher priority. The use of microbes is considered as a promising approach to combat the adverse impacts of HMs. Microbes aided in the restoration of deteriorated environments to their natural condition, with long-term environmental effects. Microbial remediation prevents the leaching and mobilization of HMs and they also make the extraction of HMs simple. Therefore, in this context recent technological advancement allowed to use of bioremediation as an imperative approach to remediate polluted soils. Microbes use different mechanisms including bio-sorption, bioaccumulation, bioleaching, bio-transformation, bio-volatilization and bio-mineralization to mitigate toxic the effects of HMs. Thus, keeping in the view toxic HMs here in this review explores the role of bacteria, fungi and algae in bioremediation of polluted soils. This review also discusses the various approaches that can be used to improve the efficiency of microbes to remediate HMs polluted soils. It also highlights different research gaps that must be solved in future study programs to improve bioremediation efficency.
Introduction
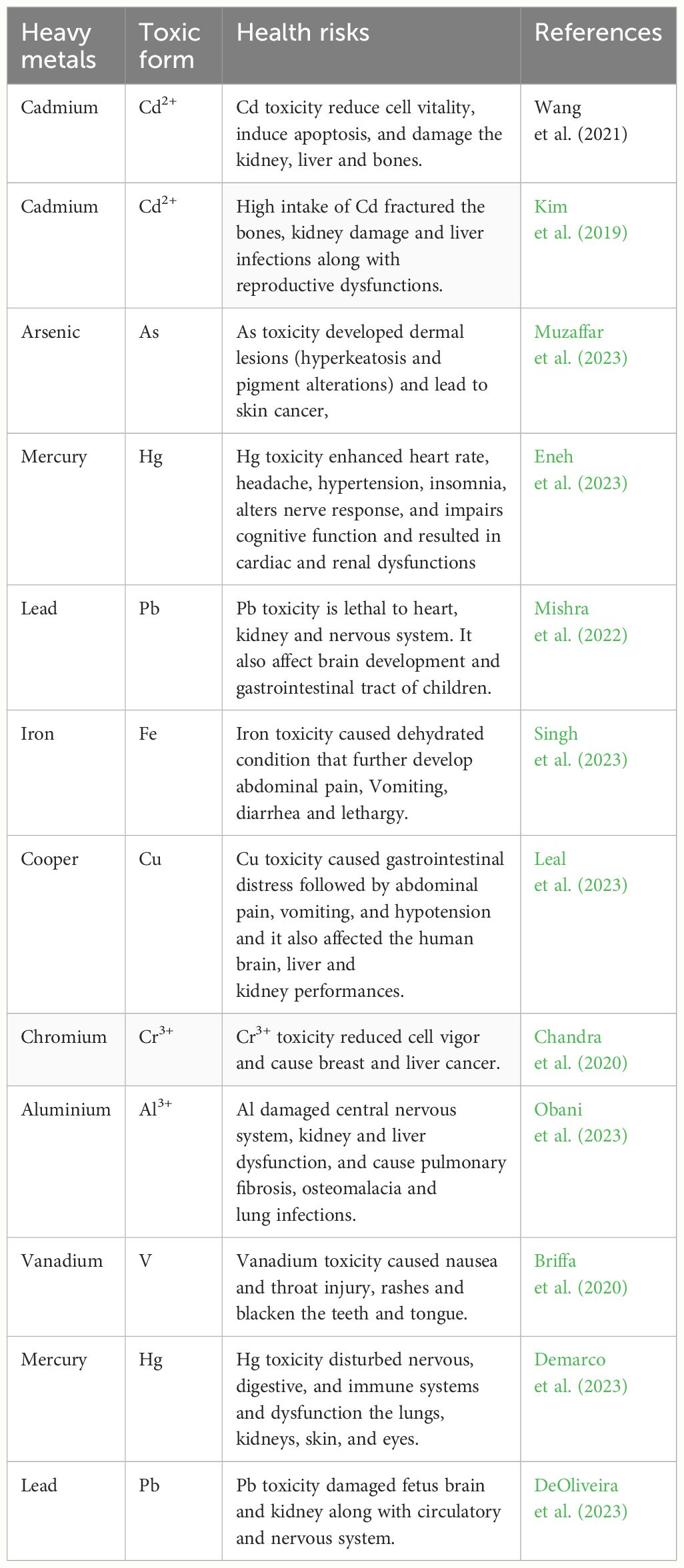
The world’s population is continuously growing up with a corresponding increase in food demands (Yaashikaa and Kumar, 2022). The recent increase in industrialization and anthropogenic activities are a serious threat to crop production owing to the fact they negatively soil fertility and productivity (Yaashikaa and Kumar, 2022). Various industries excrete toxic heavy metals (HMs) that enter into the soil and negatively affect soil fertility, microbial activities, and crop productivity and these HMs also induce serious effects on humans (Table 1) by eating the contaminated foods (Asyakina et al., 2021; Debonne et al., 2021; Nizamutdinov et al., 2022). Global agricultural communities have serious concerns about contamination of agricultural soils with HMs. These HMs are very toxic and they can persist in the soils over a long time period. Different HMs including cadmium (Cd), lead (Pb), zinc (Zn) and copper (Cu) enter into agricultural soils with organic and inorganic fertilizers, while arsenic (As) and mercury (Hg) enter into agricultural soils through nearby located industrial enterprises (Uchimiya et al., 2020; Guan et al., 2022).
Heavy metals are known to accumulate in plants and they negatively affect the plant’s physiological and biochemical processes and consequently cause serious yield losses (Yan et al., 2020). HMs reduce seed germination by negatively affecting the germination related processes which in turn reduce the overall stand establishment (Hassan et al., 2019). HMs also disturb the plant water status, membrane stability and increase the losses of important osmolytes through excessive production of malondialdehyde (MDA) and hydrogen peroxide (H2O2). Further, HMs also induce excessive reactive oxygen species (ROS) production which damages the proteins, lipids and DNA (Hassan et al., 2013).
Globally, different chemical, physical and biological methods are being used to remove the HMs from soils. Physical methods like thermal treatments, soil washing, vitri-fication, and chemical methods like the application of lime, organic amendments and phosphate compounds are being used to treat the HMs polluted soils (Gong et al., 2018). The physical and chemical methods are quick and efficient; however, they have major limitations. For instance, they are expensive and laborious and they can cause drastic changes in soil quality therefore, these methods offer no optimal solution to treat HMs polluted soils (Gong et al., 2018). Thus, in this context, biological methods offer an alternative solution owing to environmental friendly nature and they are less expensive. The biological methods involve the use of plants (phytoremediation) and microorganisms (bioremediation) to treat the HMs polluted soils (Khalid et al., 2017). The biological methods are economical and environment friendly, and they have appreciable applicability and efficiency as compared to physical and chemical methods (Yan et al., 2020). However, these methods also have some limitations like lengthy periods, environmental sensitivity, and contaminant toxicity (Liu et al., 2023). The use of microbes (bioremediation) got a great scientific attraction across the globe in recent times. The microbes remove the HMs from soil through different mechanisms including bio-sorption, bio-accumulation, bio-volatilization, bio-mineralization, oxidation and reduction, bio-leaching and production of bio-surfactants (Rahman and Singh, 2020). Micro-organism can protect from the negative effects of HMs; however, many HMs destroy membranes of microbial cells. Thus, the ability of microbes to survive under the effect of HMs is an area of decisive importance (Ayangbenro and Babalola, 2017). It has been reported that HMs toxicity and mobility is depend on the degree of oxidation of HMs (Haque et al., 2022). Microbe use HMs pollutants as a food source and change their redox potential (Faskhutdinova et al., 2021). Under HMs stress, some microbes also secrete different substances including polysaccharides, proteins, and lipids that can bind HMs ions and therefore reduce their availability (Martis et al., 2021).
Microbes also reduce the concentration of HMs in soil; for instance, Aspergillus niger showed an appreciable ability to for bioaccumulation of Cd and Cr (Khan et al., 2019), similarly, Stenotrophomonas rhizophila also significantly removed Pb and Cu by 76.9% and 83.4% (Sun et al., 2021). Due to small size microbes also provide a large surface area to adsorb the HMs which reduces the overall availability of HMs (Rajput et al., 2022). Further, microbes also accelerate the bio-adsorption of toxic HMs which makes them an excellent amendment to remediate HMs contaminated soils (Srivastav et al., 2018). Microbes can also multiply quickly; thus, the use of microbes could be an important amendment to treat the HMs polluted soils (Singh et al., 2020). The recent advancement in microbial bioremediation techniques has shown promising results to remediate polluted soils. For instance, different bioinformatics are being used to develop more effective remediation technologies. These tools are using different databases to explore the underlying mechanisms of degradation (Zheng et al., 2018). Recently, bio-remediation also used genomics, transcriptomics, metabolomics, and proteomics which is added to the evaluation processes of in-situ bio-remediation (Villegas-Plazas et al., 2019). Moreover, genomic studies have also allowed us to analyze the genetic information of microbes within the cell which ensures to development of better microbes for remediation (Hakeem et al., 2020). Additionally, recent advancements in synthetic biology also showed promising results and genetically modified organisms (GMOs) have shown appreciable results in removing pesticides, and xenobiotics from the environment (Bala et al., 2022). There are many reviews available regarding the role of microbes in remediating metals polluted soils. Nonetheless, there is no comprehensive review available describing the role of microbes in remediating the antimony, arsenic, cadmium, chromium, mercury, lead, and nickel-contaminated soils. The aforementioned metals/metalloids are highly toxic and their concentration is rapidly increasing in the environment. Recently, bio-remediation got appreciable attention across the globe, therefore, we have discussed the role and mechanisms of microbes to remediate soils polluted by these toxic metals. The current review also discusses the different research gaps that must be filled besides the appreciable progress in the field of bio-remediation. This review provides insights to boost microbial functioning for the remediation of polluted soils.
Sources of heavy metals entry into soils
Recent industrialization is meeting population food demands but also posing a severe hazard to the environment by excreting poisonous compounds such as HMs (Aluko et al., 2021). These toxic HMs enter into the human food chain by eating the contaminated foods (Sayyed et al., 2019). Among HMs, As, Cr, Cd, Pb and Hg got a serious attention across the globe because their concentrations in many terrestrial, marine, and aerial systems exceed the safety threshold (WHO 1990; Rahman and Singh, 2019). HMs have both natural and anthropogenic origins and they can be found in the atmosphere, water, soil and biological organisms (Yin et al., 2021). HMs generation from human sources is permanent and constant while the generation of HMs from natural sources is also affected by natural sources (Armah et al., 2014). The major human sources of HMs are agriculture, industries and urbanization (Li et al., 2021). Textiles, tanneries, fertilizers, galvanizing factories, metallurgic factories, varnishes, pharmaceuticals and pesticide companies are major sources of HMs pollution (Verasoundarapandian et al., 2022).
In the mining process, a significant amount of waste rocks is produced which contains a low quantity of HMs. These HMs are carried into ground and water areas by biological and chemical leaching and are then enters into the human food chain (Li and Yu, 2015). Agriculture activities also add a significant amount of HMs into soil owing to the continuous use of inorganic chemicals. Natural phosphate contains impurities in the form of HMs, and different HMs such as As, Cd, Ni, Cr, and Zn have been identified in higher concentrations in over 200 phosphate fertilizers used worldwide (Nziguheba and Smolders, 2008). Likewise, pesticides also contained impurities in the form of HMs and it has been found that different pesticides contained Hg, As, Cu and Pb as an active elements. Different pesticides containing Hg (II) and Pb(II) has been banned owing to their higher toxicity (Kothe et al., 2010). The application of industrial and municipal wastewater is also common practice and the constant application of these waste waters also leads to the accumulation of HMs in soil (Ren et al., 2015; Li et al., 2017b). Electronic waste also has a significant contribution in HMs pollution. For instance, in China in electronic waste recycling site has a significant amount of Cd and Cu greater than the threshold levels (Wu et al., 2015). Heavy metals from natural sources include mineral deposition, eruption of volcanic pathogenic processes, and oceanic evaporation (Zhang et al., 2012). Mining is an important source of HMs release (Acosta et al., 2011), and in China mining is produces around 12 lakh ha of wasteland per year with an annual increase of approximately 47,000 ha (Zhuang et al., 2009).
Effects of HMs on agro-ecosystem
Soil biology is crucial for maintaining soil quality, which is critical for agricultural sustainability. Human activities are a major source of HMs and they disturb soil microbes, soil fertility, and productivity (Sharma et al., 2017). The survival of microbes is negatively corrected with prolonged exposure of HMs like Pb (Yuan et al., 2015). Similarly, coal mining activities also cause a decrease in microbial abundance, biomass, and variability (Nayak et al., 2015). Heavy metals also slow down the breakdown of litter resulting in uneven deposition of litter on the soil (Marschner and Kalbitz, 2003). Furthermore, HMs have a deleterious impact on the breakdown of stream litter (Hogsden and Harding, 2012; Ferreira et al., 2016). Moreover, HMs also induce a negative effect on soil microbes and a negative correlation has been reported between the concentration of HMs and microbial respiration (Nwuche and Ugoji, 2008). Depending on the soil parameters, substrate concentration, and HM exposure, heavy metals can either accelerate or inhibit N mineralization. The toxicity of HMs also disrupts the N transformation pathways which consequently affect the mineralization of HMs (Hamsa et al., 2017). Further, HM pollution also induces a negative effect on N mineralization and nitrification and both these processes decrease with increasing the amount of HMs pollutants. Further, nitrification is considered to be more susceptible to HMs as compared to mineralization (Bewley and Stotzky, 1983). Moreover, HMs also affect the soil enzymatic activities and microbial abundance (Xian et al., 2015) and it has been found that HMs reduce the soil enzymatic and microbial activities and soil microbial abundance (Pan and Yu, 2011; Xian et al., 2015).
For instance, Li et al. (2020) documented that HMs reduced bioactivity, richness, and microbial diversity. They found that heavy metals (Cu, Cr, Ni, Pb, Zn, and Mn) showed total variations of 87.7%, 56.6%, 83.0%, and 55.1% α-diversity, and community composition, predicted by PICRUSt. In another study, it was documented that Pb stress altered the bacterial community structure. These authors found that Pb 2.5% and 5% increased Actinobacteria abundance by 118.56 and 147.25% while 5% Pb stress Bacteroidota and Myxococcota increased abundance by 280.76 and 138.54%, respectively (Meng et al., 2023). In another study, a significant change in microbial abundance and diversity was observed in Cd-polluted soil. Cadmium toxicity (50 mg kg-1) increased Bacteroidota and Proteobacteria by 2 and 0.3 folds while Cd toxicity decreased the abundance of Acidobacteriota, Firmicutes, Chloroflexi, Myxococcota, and Gemmatimonadota by 0.3, 0.5, 1.7, 2.2 and 2.4 folds (Bandara et al., 2022). The studies have documented that long-term exposure to heavy metals negatively affects soil health. For instance, Cheng et al. (2022) long-term Cd toxicity decreased the soil organic matter, nitrogen, phosphorus, and potassium availability. The other group of authors found that long-term As toxicity showed a negative showed a negative impact on soil enzymatic activities and soil properties. They found that As toxicity reduced the urease and dehydrogenase activities and soil nitrogen, SOM and clay were the main factors affecting the soil enzyme activity (Nurzhan et al., 2022). Some studies also reported that microbial species show resilience in response to HMs. For instance, Philippot et al. (2008) found higher resilience of nitrate reduction rates to Hg stress (100 mg kg-1). Brandt et al. (2010) noted that soil bacterial communities showed structural and functional resilience to Cd exposure (0, 40, 150, and 500 mg·kg−1). They found that the observed increase in Cu tolerance against higher concentrations of Cu was involved in the phenotypic adaption and selection at the micro-diversity level. HMs-mediated disruption in soil microbial activities also negatively soil properties and microbial activities. For instance, soils contaminated with HMs are associated with insufficient nutrients, organic matter, and water retention capacity (Singh and Kalamdhad, 2011). The increase in toxicity of heavy decreases the microbial abundance and diversity and indirectly affects soil enzyme activities by changing microbial community synthesizing enzymes (Singh and Kalamdhad, 2016). Moreover, heavy metals also inhibit soil enzymatic activities and reduce the mineralization of SOM and nutrient nutrient cycle (Bakshi et al., 2018). Globally, different including physical, synthetic, and natural remediation techniques (in situ and ex-situ) are used to remediate polluted soils. The use of genetically modified microbes has received appreciable attention to cleanup metal-contaminated soils and improve stress tolerance (Narayanan and Ma, 2023).
Plant responses to heavy metals
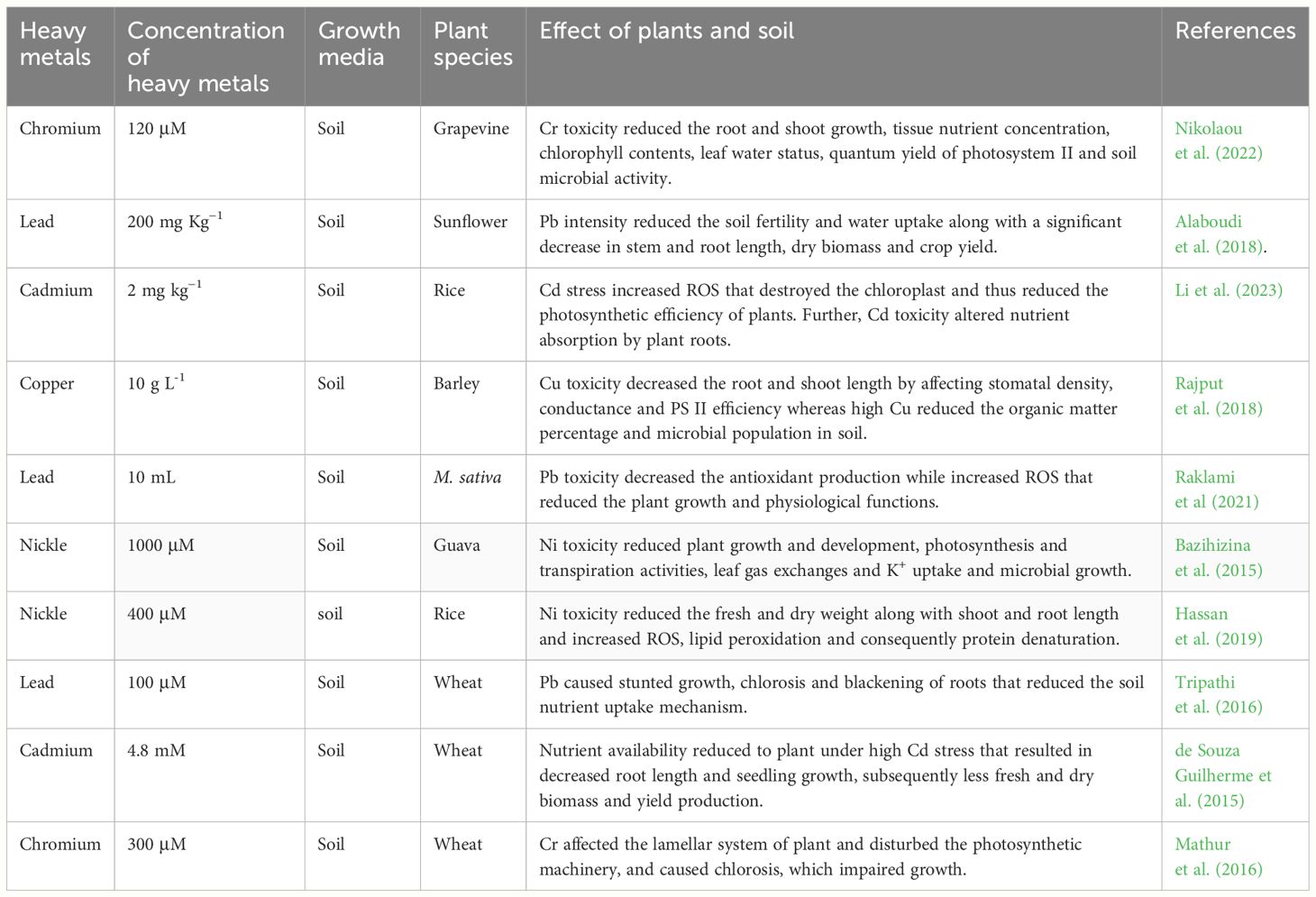
Heavy metals seriously affect plants and the effects of HMs on plants can be seen from germination to senescence (Table 2). Seed germination is one of the most critical stages of plant life and a mediated decrease in seed germination declines seedling growth and subsequent stand establishment (Adrees et al., 2015). For instance, in a study, it was found that combined Cu and Cd stress reduce seed germination, growth of seedlings, and lateral growth rate (Neelima and Reddy, 2003). The exact mechanism through which HMs change seed physiology is not well understood, and different authors reported that HMs inhibit the activities of various enzymes that cause a reduction in seed germination (Figure 1). For instance, Hg induced a decrease in seed germination owing to the direct interaction of Hg with HS group proteins that leads to the formation of an S-Hg-S bridge thus causing a loss in enzymatic activities (Cui et al., 2014).
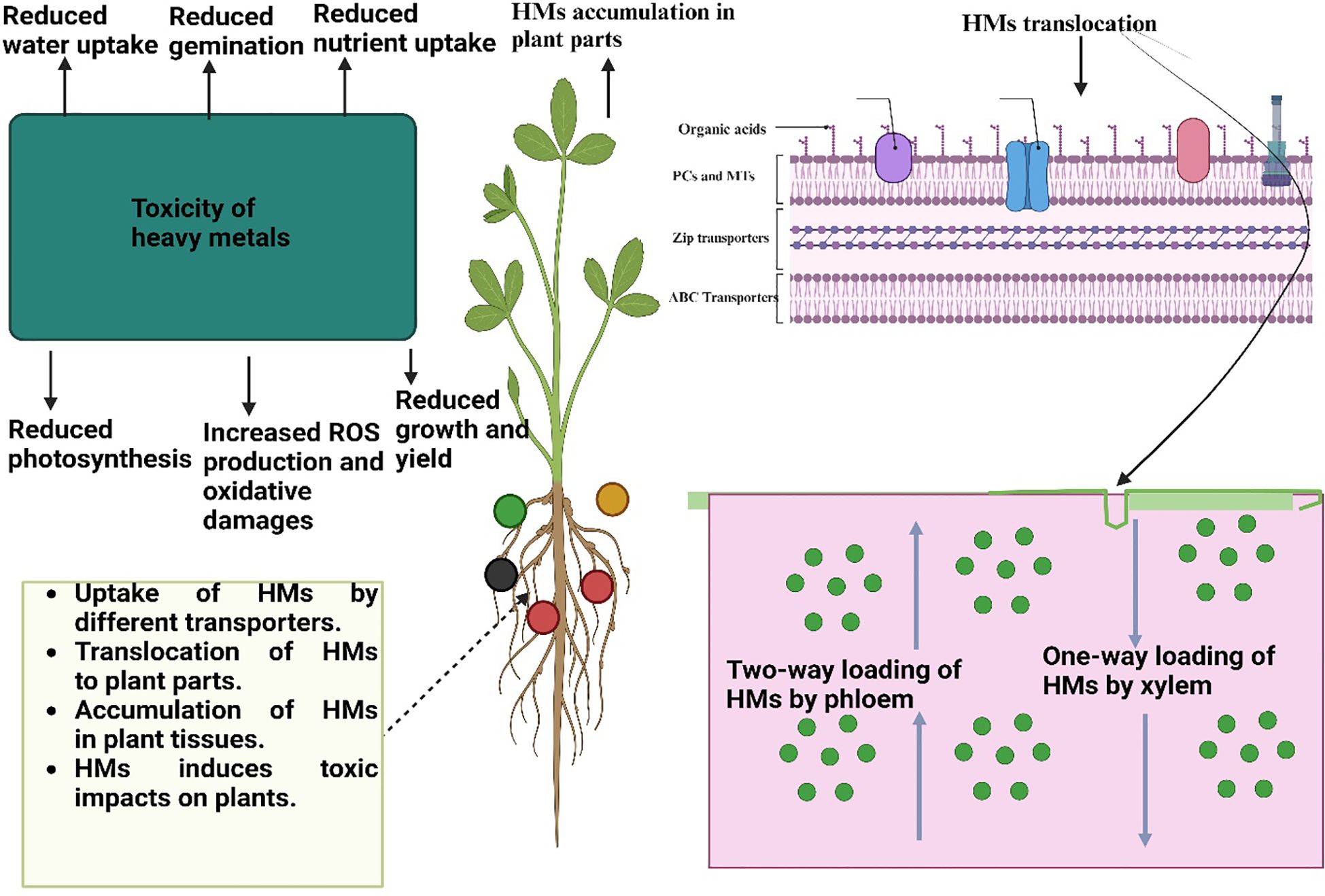
Figure 1 Toxic effects of heavy metals on plants. Heavy metals are absorbed by plants roots and then they are moved to above ground plant by different transporters and then accumulated in above ground parts. The accumulation of HMs in plant parts induce ROS production, necrosis, and decrease nutrient and water uptake and caused protein degradation, cell detoxification thus reduce the plant growth and development.
Apart from seed germinations, HMs also change the root architecture and this effect has been reported in plants. In particular, HMs decreased the root elongation (3-4 folds) and enhanced the formation of lateral roots (2-3 folds) in the presence of different HMs like Cu, Pb, Cr, Zn, and Cd (Sofo et al., 2017). The formation of lateral roots is the initial symptom of HMs toxicity which consequently impairs the uptake of nutrients and water thereby reducing subsequent plant growth (Rucińska-Sobkowiak, 2016). Along with root inhibition, HMS also causes a reduction in plant growth. HMs transport from roots to aerial parts and accumulates in plant cells which interfere with cellular metabolism and thus cause a reduction in plant growth (Shanker et al., 2005; Wang et al., 2020). As a result of their interactions with the central atom (Mg) of the porphyrin ring, heavy metals also break down the chlorophyll molecules, severely reducing photosynthesis and ultimately impairing plant growth (Yadav et al., 2014). Moreover, HMs like Cu also cause lignification of both roots and shoots which reduces biomass production owing to impaired cell development (Martins et al., 2020). Additionally, HMs hurt the water relationships which in turn affects a variety of physiological activities like photosynthesis and transpiration (Alsokari and Aldesuquy, 2011). A recent study showed that Cd stress (100 uM) decreased the plant height by 69% and 73% in sorghum cultivars JS-2002 and Chakwal sorghum (Hassan et al., 2019). Further, Cd toxicity also increased MDA concentration by 39% and 43% respectively in both cultivars (Hassan et al., 2019). In another study, it was witnessed that Pb stress decreased the photosynthetic rate, carbon dioxide concentration, transpiration rate, and WUE by 50.5, 73.2, 48.6, and 148.8% respectively (Qin et al., 2023). Heavy metal toxicity also negatively effect nutrient uptake by plants. For instance, Fava bean plants’ Cd toxicity (150 mg/L) decreased the Ca and Mg concentration by 1.82 and 1.27 times while Cd toxicity (300 mg/L) decreased the Cd and Mg concentration by 2.278 and 2.25 folds (Piršelová and Ondrušková, 2021).In plants like Helianthus annuus and Vigna radiata, HMs (As) increased the number of stomata followed by the development of abnormal, arrested, and fused stomata (Gomes et al., 2011; Gupta and Bhatnagar, 2015). Heavy metals also affect the xylem vessels’ parenchymatous and mesophyll cells and resultantly change the plant water relations and are considered to be responsible for the decrease in leaf growth. Heavy metals also negatively affect the photosynthetic machinery however, it depends on the concentration of HMs. Moreover, HMs also negatively affect the light-harvesting, transport of electrons, and RuBisCo activity which in turn reduce the overall plant photosynthetic efficiency (Paunov et al., 2018; Latif et al., 2020). Besides this HMs (Cd) also reduced the photochemical efficiency (Fv/Fm), the effective quantum yield of photosystem II (φPSII), and chlorophyll florescence thereby leading to the inhibition of photosynthesis (Gao et al., 2020; Yotsova et al., 2020).
Generally, HMs (Hg, Cu, Pb, Ni, Cd, and Zn) target the plant chlorophyll in three different ways by increasing the activity of chlorophyllase enzyme, causing oxidation of chlorophyll through increased ROS production, and inhibiting biosynthesis of chlorophyll biosynthesis (Gill et al., 2012; Shahzad et al., 2018; Sharma et al., 2020). HMs not only affect the chlorophyll molecules but also the membranes of the chloroplast and thylakoid cells. For example, swelled thylakoids, degraded chloroplast membranes, and loss of chloroplast membrane were noted in barley plants under Pb stress (Wang et al., 2017). Moreover, HMs also inhibit the light reactions by decreasing the efficiencies of PS-I and PS-II and they also decrease the dark reactions owing to decreased activities of enzymes linked with the Calvin cycle (Souri et al., 2019). Heavy metals also induce overproduction of ROS that damage proteins, DNA, and lipids and lead to the induction of oxidative stress (Foyer and Noctor, 2016). However, plants also activate excellent deference and they also accumulate various osmolytes to counter the toxic effects of HMs. For example, Chowardhara et al. (2020) found that activities of CAT, GST, GR, APX, and POD and accumulation of proline and ascorbic acid were increased in response to Cd toxicity in B. juncea. It has been reported that HMs also decrease the uptake of water and nutrient which in turn cause significant growth losses (Rucińska-Sobkowiak, 2016; Wang et al., 2017). For instance, Cd toxicity competes with calcium (Ca), iron (Fe), and magnesium (mg) which caused a significant reduction in growth and biomass production (Raza, 2022).
Under the harmful effects of HMs, nitrogen metabolism is essential for plant growth and development. According to reports, HMs decrease the nitrate and ammonia assimilation enzymes by increasing protease activity. MicroRNAs play an imperative role in HMs toxicity by regulating the plant antioxidant responses, chelations, and auxin and cytokinin signaling (Ding et al., 2020). For instance, Casarrubia et al. (2020) found that mycorrhizal and microRNA played a significant role in Cd tolerance in Vaccinium myrtillus. In another study, it was found that MicroRNA expression significantly improved the Cd and Al tolerance in tobacco (Cedillo-Jimenez et al., 2020). Heavy metals also negatively affect the quality of crops and it has been found that Cd toxicity in rice reduced the rice protein contents, and milling degree and increased the kernel chalkiness (Imran et al., 2021).
Microorganisms responsible for bioremediation
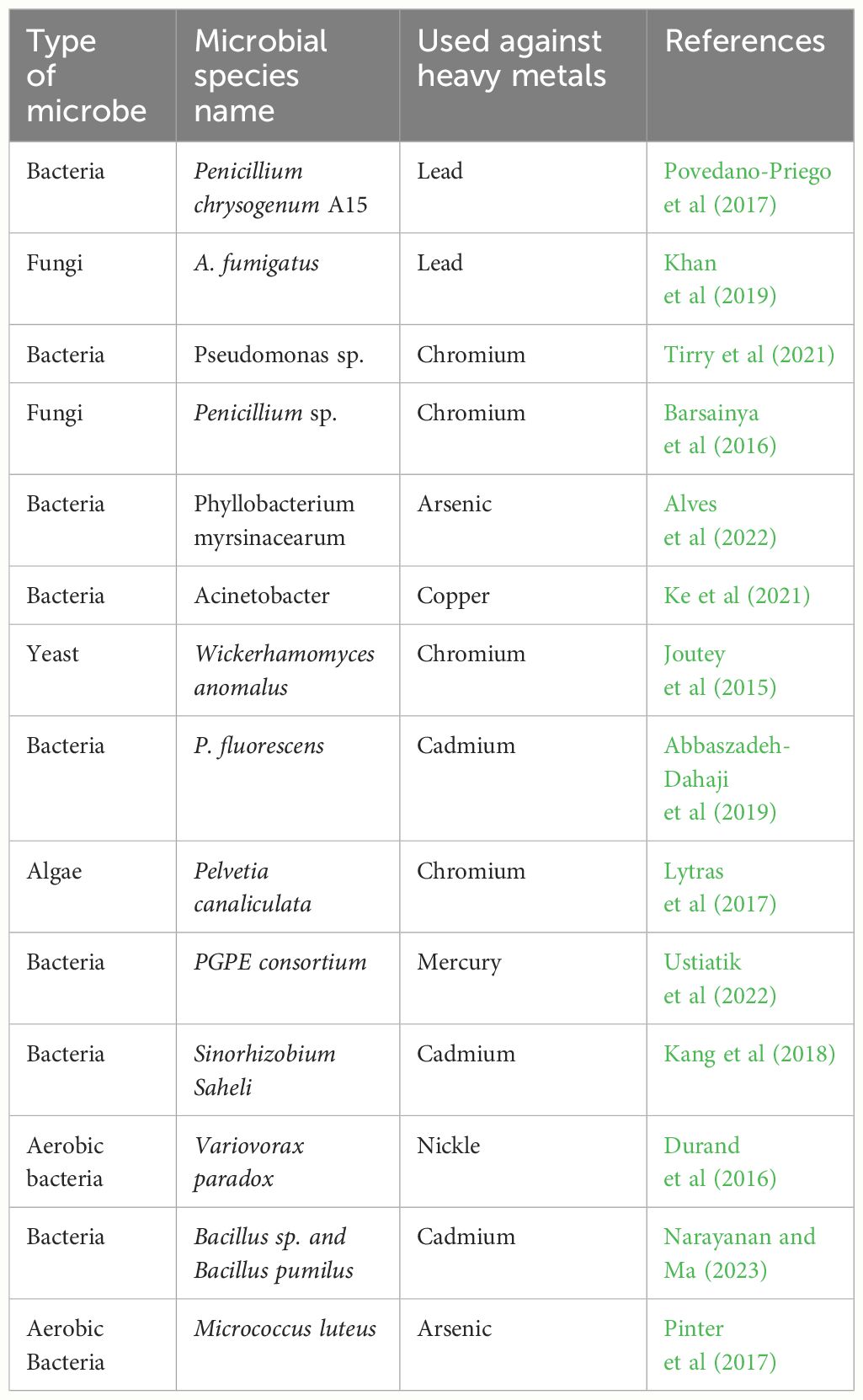
Heavy metal pollution poses a severe threat to public health by contaminating food supplies and drinking water on a global scale (Huang et al., 2020). Microbial remediation is an imperative approach and it has appreciable potential to improve crop productivity, and human health and restore the ecosystem (Narayanan and Ma, 2023). The microbial-mediated bio-accumulation and bio-magnification are very successful in removing the pollutant to ensure safe and sustainable crop production (Manorma et al., 2023). Different microbes including, algae, bacteria, and fungi are being used to clean up the HMs contaminated soils (Table 3).
Bacteria
The interaction of microbes with HMs occurs through different mechanism which depends on metal and microbe type and surrounding environment. Different factors including temperature, pH, nutrient source, and metal ions play an important role in the mobility and bioavailability of HMs for microbial transformation. Bacteria’s small size, rapid growth, and ease of cultivation allow them to thrive in a variety of environmental situations. HMs often connect to functional groups including amino, carboxyl, sulfate, and phosphate groups that are present on the layers of bacterial cell walls (Yue et al., 2015). The potential of bacteria for HMs uptake can vary from 1-500 mg/g. For instance, Hg resistant pseudomonas aeruginosa strain absorbed the Hg uptake 180 mg/g (Yin et al., 2016). Likewise, different microbes like Bacillus sp. PZ-1 and Pseudomonas also absorb the Pb from wastewater (Li et al., 2017a). On the other hand Arthrobacter viscosus can absorb the Cr and it also has an excellent capacity to transfer the Cr (VI) into Cr (III) (Hlihor et al., 2017).
Rhodobacter capsulatus also showed a maximum capacity of 164 mg/g to absorb the Zn (II) (Magnin et al., 2014) while Bacillus ceres showed a maximum bio-sorption capacity of 31.95 mg/g and 24.01 mg for Cd (II) in dead and living cells (Huang et al., 2013). Extracellular polymeric substances (EPS) protect the microorganism from the toxic effects of HMs by restricting entry of HMs into the cell. It has been discovered that EPS has both anion and cationic functional groups, which help to accumulate HM ions like Cd, Hg, Cu, and cobalt (Fang et al., 2017). After adsorption HMs are converted to diverse ionic states in bacterial cells that reduce their toxicity. Pseudomonas putidais an important microbe and it can absorb 100% Hg from the marine environment it also reduces the Hg(II) into Hg(0) (Sheng et al., 2018). The findings of Zhang et al. (2012) showed that a new microbial strain Acinetobacter sp. showed an excellent ability to detoxify the Cr. In another; authors screened 72 acidothermophilicautotrophic microbes for their ability to tolerate and bio-absorb the HMs and these authors found that the ATh-14 strain showed an appreciable potential and it showed absorption capacity of 85.82% for solubilization of copper (Umrania, 2006). Bacteria are better bio-sorbents as compared to other microbes due to their size, ubiquity resilience, and ability to grow under a wide range of conditions (Hlihor et al., 2017).
Fungi
Fungi also have an excellent ability to remediate the HMs polluted soils. The presence of chitin, polysaccharides, phosphate, and glucuronic acid in fungal cells is essential for the adsorption of HMs (Purchase et al., 2009). Different functional groups and fungal strains had a significant impact on the adsorption rate of HMs (Iram et al., 2015). In a study, it was found that Termitomyces clypeatus detoxified the Cr(VI) by adsorbing Cr on its surface through carboxyl, imidazole, hydroxyl, phosphate, and sulfhydryl groups (Ramrakhiani et al., 2011). Further, Amirnia et al. (2015) found that Saccharomyces cerevisiae eliminated the Cu(II) from water sources, while Talukdar et al. (2020) found that Aspergillus flavus fungal species removed the Cr by more than 70%. Moreover, Aspergillus fumigates also showed an appreciable potential to remove the Cd, Cr, Cu, Ni, and Zn from the contaminated soils (Shazia et al., 2013). In another investigation, three different fungal species including Penicillium citrinum, Trichoderma viride, and Penicillium showed a significant potential (250 mg/L) to adsorb the Cr(VI) (Zapana-Huarache et al., 2020).
Algae
Algae have also shown a good potential to remediate HM-polluted sites owing to the fact algae produce various peptides that help the accumulation of HMs and defend against the HMs (Bilal et al., 2018). For instance, Fucus vesiculosus showed a tremendous potential to adsorb the Pb(II) (Demey et al., 2018), likewise, Cladophora fascicularis also showed a significant potential to remediate the Pb(II) from wastewater. Similarly, Sargassum marine algae also showed a significant potential to detoxify the Cu (II) from the aqueous solution (Barquilha et al., 2017). In another study Christoforidis et al. (2015) tested the absorption capacity of Cystoseira crinitophylla for copper and found that this algae showed a maximum capacity of 160 mg/g to adsorb Cu (Christoforidis et al., 2015). On the other hand, authors noted that Saccharina fusiforme and Saccharina japonica substantially detoxify the Zn(II), Cd(II), and Cu(II) (Poo et al., 2018) while Desmodesmus also showed an appreciable potential to remove the Cu(II) and Ni(II) from the wastewaters (Rugnini et al., 2018). The study findings of Aslam et al. (2019) showed that microalgae showed promising results in the accumulation Mn, Cu, and Zn (Freitas et al., 2011). Moreover, the findings of Freitas et al. (2011) showed that algal biomass showed an appreciable potential for HMs like Fe.
Factors affecting the bioremediation process
Different factors including metal concentration, valance state, metals bioavailability, redox potential, soil temperature, and pH affect the bioremediation process (Bandowe et al., 2014). The pH of the soil has an impact on bacterial enzymatic activity as well as microbial bio-sorption (Morton-Bermea et al., 2002). Soil pH also changes the surface charge of microbes by affecting the ability of microbes to absorb the HM ions (Galiulin and Galiulina, 2008). Soil pH substantially affects both the transportation and hydration of HM ions in soil (Dermont et al., 2008) and it has been documented that the rate of HMs removal is increased with increasing pH over a certain rate and after this, the rate of removal starts declining (Wierzba, 2015). The ideal pH range for most bacteria is 5.5-6.5 (Wang et al., 2001), however, some bacteria like Bacillus jeotgali can thrive at a pH of 7 (Rodríguez-Tirado et al., 2012). Another significant component that influences the absorption of HMs is temperature; which influences the development and proliferation of microorganisms (Fang et al., 2011). Different bacteria require different temperatures to carry out their functions (Acar and Malkoc, 2004). However, HM ions, soil additives, and soil type all have an impact on microbial activity. It is challenging to achieve microbial adsorption due to the low mobility of HM ions caused by soil adsorption and retention of HM ions (Hu et al., 2010).
Soil pH is an important factor that affects microbial growth. For instance, unfavorable pH affects enzyme activity which lowers the rate of microbial metabolism and it also affects the binding capacity between HMs and adsorbants (Bandowe et al., 2014). The changes in pH also affect the mobility and hydration of metals (Bandowe et al., 2014). For instance, the adsorption capacity of Zn and Pb was increased with increasing pH, and an increase in soil pH above 5.5 decreased the removal of Pb and Zn (Wierzba, 2015). Other authors also documented that soil acidification increased the mobility of metals in the following order Cd>Zn>Pb. These authors also document that soil pH affects mobility, causes metal ions to become more or less active, and increases or decreases their environmental risk (Kicińska et al., 2022). Temperature is also a factor that affects microbial growth (Fang et al., 2011). The increase in temperature affects the diffusion of metals and increases the bioavailability of metals. However, optimum degradation temperature can vary according to metal types, for instance, Cd bio-degradation by Bacillus jeotgali was maximum at 35°C while bio-degradation by the same bacteria was higher at 30°C (Chanmugathas and Bollag, 1988). The adsorption efficiency is also affected by soil organic matter, for instance, organic matter tends to fix the metals in soil which reduces the availability to metals (Wang et al., 2022). A short-term study investigates the response of different temperatures (5, 15, and 25oC) Cd, Cu, Pb, and Zn removal by Carex pseudocyperus, C. riparia, and Phalaris arundinacea. Low temperatures reduce the removal capacity of all the metals and an increase in temperature increases the removal capacity of all the metals (Schück and Greger, 2023). Climate change also induces a significant impact on soil microbial activities. For instance, climate-induced variation in soil temperature, and humidity affect the decomposition of SOM and nutrient cycling (Burns et al., 2013), and it partially or fully depends on microbial activity. The change in soil temperature and moisture can change the growth, structure, function, composition, and interaction among microbes for the degradation of pollutants in soils (Alkorta et al., 2017).
Bioremediation is generally limited to bio-degradable compounds, and it is also susceptible to rapid degradation which more toxic compounds. Besides it, bio-remediation also needs extensive monitoring and it has major drawbacks in terms of environmental growth conditions, nutrient requirement, temperature, and pH conditions. Therefore, it is essential to find ways to identify the microbes having a wider adaptability under a wide range of temperature and pH conditions for an efficient remediation process. On a long-term basis, microbial mediation remediation is a simple, cheap, and environmental method and it can improve the overall soil fertility, ecosystem health, and safer and sustainable food production. Nonetheless, implantation of bioremediation needs a comprehensive understanding of soil microbial communities, properties of contaminants, and environmental conditions as these factors play a critical role in getting effective results.
Microbial mediated remediation of heavy metals polluted soils
The use of microbes is considered as an effective way to treat the HMs in polluted soils, as these microbes absorb HMs and also convert them into less toxic forms (Gupta et al., 2016). Microorganisms play a critical role in remediating HMs polluted soils owing to the fact they can with stand metal toxicity. Numerous HMs have been reported to be precipitated, undergo oxidation state changes, and be sequestered by microbes (Figure 2; Kang et al., 2016).
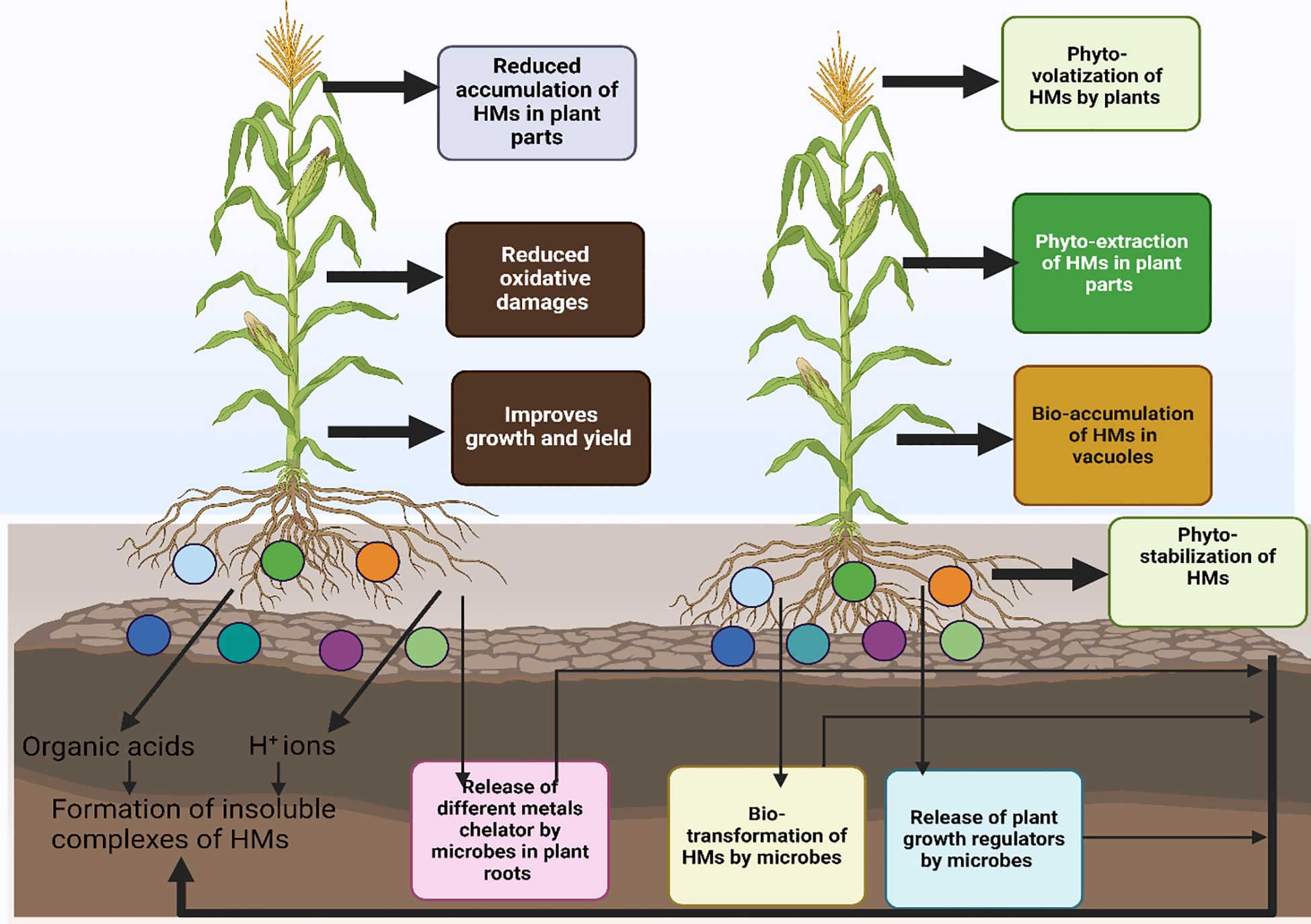
Figure 2 Different mechanism used by microbes to induce heavy metals toxicity in plants. Microbes use different mechanisms bio-sorption, bio-mineralization, bio-accumulation, bio-leaching and bio-transformation to remediate polluted soils. They also increase the availability of nutrients by increasing production of IAA and ACC deaminase, and siderophores thus resulting in better growth under polluted soils.
Microbial mediated remediation of antimony contaminated soils
Microorganisms are crucial for remediating Sb polluted soils and they reduce the toxicity of Sb through different ways including, bio-reduction and bio-oxidation (Jeyasundar et al., 2021). Many bacteria have been identified that can be used to remediate the Sb-polluted soils (He et al., 2019). For instance, two bacteria Shinella and Ensifer discovered from Sb-contaminated soils showed a tremendous potential to oxidase Sb (Choi et al., 2017) while the bacterial Bacillales strain also showed marked results to change the Sb-V into Sb-III (Lai et al., 2018). Similarly, fungi have been also used to remediate the Sb polluted soils, and study findings of Xi et al. (2022) showed that AMF increased plant antioxidant activities by reducing the retention of Sb in plant parts. The findings of Liu et al. (2013) showed that the bacterial strain Pseudomonas substantially increased the plant growth, and microbial activity and decreased Sb availability (Liu et al., 2013). In another study Zhang et al. (2012) found that microbes isolated from the rice field contributed significantly towards the oxidation of Sb-III likewise, Li and Yu (2015) also found that Agrobacterium tumefaciens contributed towards the oxidation of Sb-III.
Some environmental microorganisms, particularly those that thrive in anaerobic environments, are capable of converting Sb(V) to Sb(III). For instance, Hockmann et al. (2014) noted that microbes converted the SB-V to Sb-III with the help of lactate as an electron donor. Similarly, Kulp et al. (2014) found that microbes in Sb-polluted mines reduced Sb-V to Sb-III. In the case of flooded mine pit soils group of researchers from China found that autotrophic bacteria reduced the Sb-V and generated Sb2O3 by using hydrogen gas (H2) as an electron donor (Lai et al., 2016). Additionally, Huang et al. (2022) found that after 60 days of injection of the B. cereus solution into plant roots; the concentration of As and Sb in soil was significantly reduced as compared to soil without bacteria solution this indicates that this strain promoted the absorption of As and Sb from soil (Huang et al., 2022).
Microbial mediated remediation of arsenic contaminated soils
Arsenic occurs in the environment in different inorganic forms including As-0, As-III, and As-V, and organic forms like dimethylarsinic acid (DMA), monomethylarsonic acid (MMA), trimethylarsine oxide (TMAO) and arsenobetaine. It has been found that bacteria, algae, and fungi can methylate As-III into methylated species (Yang and Rosen, 2016; De Francisco et al., 2021). Different fungal species like Aspergillus, Candida, Scopulariopsis, and Penicillium can also cause a change in the methylate inorganic As to the organic As species (Bentley and Chasteen, 2002). It is important to keep in mind is that the ability of certain microorganisms to methylate and volatilize depends on soil organic matter (SOM), soil chemistry, and As concentration (Mestrot et al., 2011). Spagnoletti et al. (2016) tested the impact of AMF (R. intraradices) on soybean plants under As stress and found a marked improvement in plant biomass and a reduction in As accumulation. Likewise, Chan et al. (2013) also found that AFM (Geosporum) enhanced the phosphorus uptake and reduced the As concentration in rice grains.
Transgenic microbes are also an effective way to treat As toxicity. For instance, transgenic microbes with expressed arsM showed an ability of 2.2-4.5% to remove As from soil while the same microbe showed an ability of 10-fold in nutrient solution (Liu et al., 2011). In another study, Huang et al. (2015) found that thermophilic strain Bacillus subtilis 168 was unable to do methylation and volatilization of As. They genetically modified this bacteria with CmarsM gene and found that genetically modified bacteria caused methylation and volatilization of As it occurred within 48 hours in As-contaminated organic compost. Moreover, Villadangos et al. (2014) also modified the As-resistant bacteria named Corynebacterium glutamicum by ArsC1 and ArsC2. These authors found that As(V) was significantly increased after the introduction of genetically modified bacteria. Additionally, Preetha et al. (2023) also prepared the mutant C. glutamicum strain and found that this strain showed an ability of 15 folds and 30 folds more to accumulate As-III and As-V as compared to the control treatment.
Microbial mediated remediation of cadmium contaminated soils
Cadmium is a very toxic HM posing a serious threat to human health and the environment. The application of microbes is an effective and promising technique to treat Cd-polluted soils. In a study, Ma et al. (2020) discovered the Cd immobilization PGPR (TZ5) and found that this bacteria significantly increased ryegrass weight by 77.78% and decreased the concentration of Cd in ryegrass by 48.49%. Further, the application of this bacteria also increased the soil enzymatic activities and microbial growth which indicates that this bacterial strain (TZ5) can provide a practical approach to remediate Cd-polluted soils (Ma et al., 2020). Limited studies are conducted to determine the impacts of single and co-inoculation of Bacillus mycoides and Micrococcus roseus on growth and nutrient uptake of maize grown under Cd stress (100 and 200 mg kg-1). These authors found that all bacterial treatments appreciably improved the plant growth and biomass and the combination of both bacteria reduced the root and shoot Cd uptake and transfer and translocation as compared to control (Malekzadeh et al., 2012).
In Cd-contaminated soils, Cd-tolerant bacteria play an important role (Bravo, 2022). The microbes use various mechanisms including biosorption and intra-cellular accumulation to mitigate the adverse impacts of Cd stress (Ghosh et al., 2022). Recently, genetically modified organisms also played an important role in remediating Cd-polluted soils (Abbas et al., 2018). Different genetically modified organisms (CdtB Enterobacter and Klebsiella variicola) showed an appreciable potential to remediate Cd polluted soils (Feria-Cáceres et al., 2022; Quiroga-Mateus et al., 2022). Similarly, Arce-Inga et al. (2022) found that the application of Theobroma cacao (CCN51) significantly decreased the uptake of Cd, and its translocation to plant parts. Feng et al. (2023) studied the impact of mixotrophic acidophiles under Cd-contaminated soils. These authors also found that soil solution pH and reduction level of glucose affected the abundance of Acidithiobacillus which contributes significantly towards removal of Cd (Feng et al., 2023). On the other hand, the fungal strain belonging to Purpureocillium lilacinum tolerated the Cd stress up to 12000 mg/L. The SEM analysis indicated Cd can be accumulated on the mycelial surface generating plenty of metal precipitation particles. Further, these authors also found that in pot experiments this fungal strain also reduced the soil Cd concentration in soil by 12.56% and promoted plant growth this indicates that this fungal could be an important candidate to remediate Cd polluted soils (Deng et al., 2021).
Microbial mediated remediation of chromium contaminated soils
Chromium is released into the environment as a result of human and anthropogenic activities which pose a serious threat to living organisms. Microbial remediation is an effective approach to treatment the Cr polluted soils. For instance, in a study authors tested the impact of Nostoc linckia to remediate Cr polluted soils. They found that this microbe showed an appreciable potential to accumulate Cr and suggested that this bacteria could be an effective candidate to remediate Cr-polluted soils (Cepoi et al., 2021). In another study, edaphic cyanobacteria were tested for Cr remediation and it was found that these bacteria produce polysaccharides, glycoproteins, lipopolysaccharides, and ionic functional groups that can coordinate with Cr and reduce its availability (Cheung and Gu, 2007). Moreover, different Cr-tolerant bacteria including Bacillus, Enterobacter, Pseudomonas, and Streptomyceshave been identified and they can remove the Cr by 50-90% (Ramesh and Winkler, 2010; Bansal et al., 2019; Elahi and Rehman, 2019; Murthy et al., 2022). The study findings of Wen et al. (2023) showed that the addition of SR-2, PA-1, and LB-5 improved the plant fresh weight by 10.3%, 13.5%, and 14.2% and increased the soil enzymatic (catalase and sucrose) activities and significantly decreased the shoot Cr concentration by 19.2-83.6%.
Chen et al. (2021) studied the impact of B. cereus WHX-1 on mitigating Cr toxicity. They found that this microbial species improved the soil physicochemical properties, soil bulk density and decreased the redox potential. They also found that this microbial species transferred the Cr-IV by 94.225 into Cr-III increasing the residual fraction of Cr by 63.38%. Further, these authors also found that the application of B. cereus improved the growth and biomass production of ryegrass. In another study, Ahmed (2018) studied the impact of chromium-tolerant auxin-producing rhizobacteria on growth characteristics of Lens culinaris growing under different Cr concentrations (0, 50, 100, 200, 400, and 500 µgml−1). The results of their study findings showed that Bacillus species mitigated the deleterious impacts of Cr reduced the Cr accumulation in soil and reduced Cr availability to plants.
Microbial mediated remediation of lead contaminated soils
Bioremediation with microbes is considered an effective approach is a promising technique to remediate the Pb-contaminated soils. For instance, a pot study conducted on wheat showed that R. sphaeroidesreduce the Pb concentration in root and lead by 14.78% and 24.01% (Li et al., 2016). On the other hand, Rhee et al. (2012) found that two fungal species Paecilomyces javanicus and Metarhizium anisopliae isolated from mining produced organic acids that resulted in precipitation of Pb. In another research study Sun et al. (2017) found that soil inoculation with M. circinelloides significantly increased the Pb removal by S. nigrum L. These authors also found that soil fertility was also increased after inoculating the soil with S. nigrum (Sun et al., 2017). Likewise, Zhou et al. (2016) added WH16-1 strain in Pb2+ contaminated paddy soil and found that this bacterial strain decreased the exchangeable and carbonate-bound Pb in the paddy soil 14.04 and 10.69% (Zhou et al., 2016).
The study findings of Puyen et al. (2012) showed that Micrococcus luteus marked decreased Pb concentration in soil, likewise, findings of Kalita and Joshi (2017) showed that Pseudomonas aeruginosa application to Pb-polluted soil appreciably reduction the concentration of Pb with 40 mg g-1 sorption capacity (Kalita and Joshi, 2017). Shanab et al. (2012) tested the potential of different algae to remediate Pb-polluted soils and they found that different algae isolates like Phormidium ambiguum, Pseudochlorococcum typicum, and Scenedesmus significantly reduced the Pb toxicity. Fungi is also an effective candidate for reducing Pb toxicity (Fawzy et al., 2017) application of AMF under Pb stress effectively increased the sunflower biomass and mitigated the toxic effects of Pb (Hassan et al., 2013). In addition to producing various organic acids, polyphosphates, peptides, and sulfur compounds, fungi also do cell wall binding, and make chelate, and precipitate that decreases Pb toxicity (Bellion et al., 2006).
Microbial mediated remediation of mercury contaminated soils
Mercury microbial remediation needs the microbial species to withstand and remove the Hg over extended periods. Various authors noted that microbes effectively remediate the Hg-contaminated soils. For instance, Vigna unguiculata inoculated with Photobacterium and grown on Hg-contaminated soil (27 mg/kg) showed increased root growth (11%), seed production (33%), leaf numbers (50%), Hg uptake in roots (25%) and decreased Hg concentration in aerial plant organs (55%) as compared to un-inoculated control (Mathew et al., 2015). Similarly, two bacterial strains like Brevundimonas diminuta and Alcaligenes faecalis applied to Hg and Pb-contaminated soil increased the phyto-accumulation of Pb and Hg by roots and shoots (Hamzah et al., 2015). In another study Hg resistant microbials including Enterobacter ludwigii and Klebsiella pneumoniae, promoted plant growth and decreased proline concentration, MDA concentration and electrolyte leakage in wheat seedlings growing under Hg stress 75 μM; (Gontia-Mishra et al., 2016).
In another study, bacteria inoculation significantly improved maize growth and reduced the Hg uptake by maize plants growing under Hg (Mariano et al., 2020). Fungi have also shown an appreciable potential to remediate Hg-contaminated soils and it has been found that AMF inoculation increased the plant growth, P uptake, and reduced Hg uptake as well as translocation in Lactuca sativa growing under Hg stress under (10 mg/kg) (Cozzolino et al., 2016). Moreover, commercial AMF like Glomus, Entrophospora and Scutellospora genera, appreciably improved the seedling growth and root elongation of rice plants growing under Hg toxicity (Vargas Aguirre et al., 2018). Another group of authors also found that commercial AMF also promoted plant growth and stimulated the uptake of Hg in Lolium perenne and rice plants growing under Hg toxicity (Leudo et al., 2020). Likewise, Pietro-Souza et al. (2020) found that compared with Chrysopogon zizanioides plants growing with AMF under Hg stress showed a marked improvement in plant growth, root and shoot biomass, chlorophyll concentration and showed a reduction in Hg accumulation. Moreover, Aspergillus and Curvularia geniculata also appreciably increased the maize root growth, root dry weight, shoot dry weight, chlorophyll, and Hg accumulation by 40% and 34% respectively (Pietro-Souza et al., 2020).
Microbial mediated remediation of nickel contaminated soils
Microorganisms are extremely important for the bioremediation of Ni-polluted soils owing to the fact this method is very economically effective against Ni toxicity (Hassan et al., 2019). Various bacterial strains including Bacillus thuringiensis and Bacillus cereus have shown promising results in treating the Ni contaminated soils (Zhu et al., 2016). Cabello-Conejo et al (2014) recorded that Arthrobacter nicotinovorans appreciably improved plant growth and increased the phyto-extraction of Ni from polluted soil. Zaidi et al. (2006) documented that Bacillus subtilis decreased the toxicity of nickel while noticeably boosting mustard growth and nickel phyto-extraction. Other authors also found that inoculation with Trichoderma atrovirideand Glomus intraradices improved the Ni phyto-extraction and reduced the Ni toxicity in linseed and mustard (Cao et al., 2008).
In another study, Alboghobeish et al. (2014) tested the potential of bacterial strain (Klebsiella oxytoca) and found that this strain showed a Ni tolerance of 24 mM. Likewise, Enterobacter asburiae from industrial water depicted the Ni tolerance to a 15 mM concentration and it removed the 75% Ni by bio-accumulation (Paul and Mukherjee, 2016). Heidari and his colleagues found that a Microbacterium oxydans strain showed a Ni removal efficiency of 83-91% (Heidari et al., 2020) while Das et al. (2014) reported that Bacillus thuringiensis found that removed the Ni by 82% through bio-sorption process. According to Costa and Tavares (2017), Alternaria and Penicillium species have respective Ni biosorption potentials of 11.3 and 13.1 mg g-1. Trichoderma and Aspergillus inoculation also considerably increased the effectiveness of Ni’s phytoextraction (Jiang et al., 2008) and Stenotrophomonas from industrial waste also showed an appreciable potential to remove the Ni (Aslam et al., 2020). However, the biosorption of Ni by microbes significantly affects microbial strain, pH, temperature, and initial Ni concentration (Heidari et al., 2020).
Microbial resistance to heavy metals and their mechanisms
During HMs stress microbes either die owing to toxicity developed by HMs or they thrive in this condition through different resistance mechanisms against HMs (Table 4). Microbes develop different mechanisms including, extra and inter-cellular sequestration, and extracellular barriers, and they actively transport the metal ions to tolerate HMs toxicity. On the surface of bacteria, there are several barriers such as cell walls, plasma membranes, and other structures like EPS that prevent HMs from entering bacterial cells (Bhati et al., 2019). The research findings of Kumar et al. (2014) indicated that bacteria and fungi cause the bio-sorption of metals like Cu, Pb, and Cr. Microbial biofilms contain polymers that accumulate HM ions and protect the inside bacterial cells and the presence of biofilm on Pseudomonas aeruginosa showed tolerance against, Cu, Pb, and Zn (Teitzel and Parsek, 2003). Further, the presence of biofilms also increased the elimination efficiency of HMs (Grujic et al., 2017). Additionally, cell walls and EPS also work as an excellent barrier and they substantially adsorb the metal ions like Pb and Cr (Kushwaha et al., 2017).
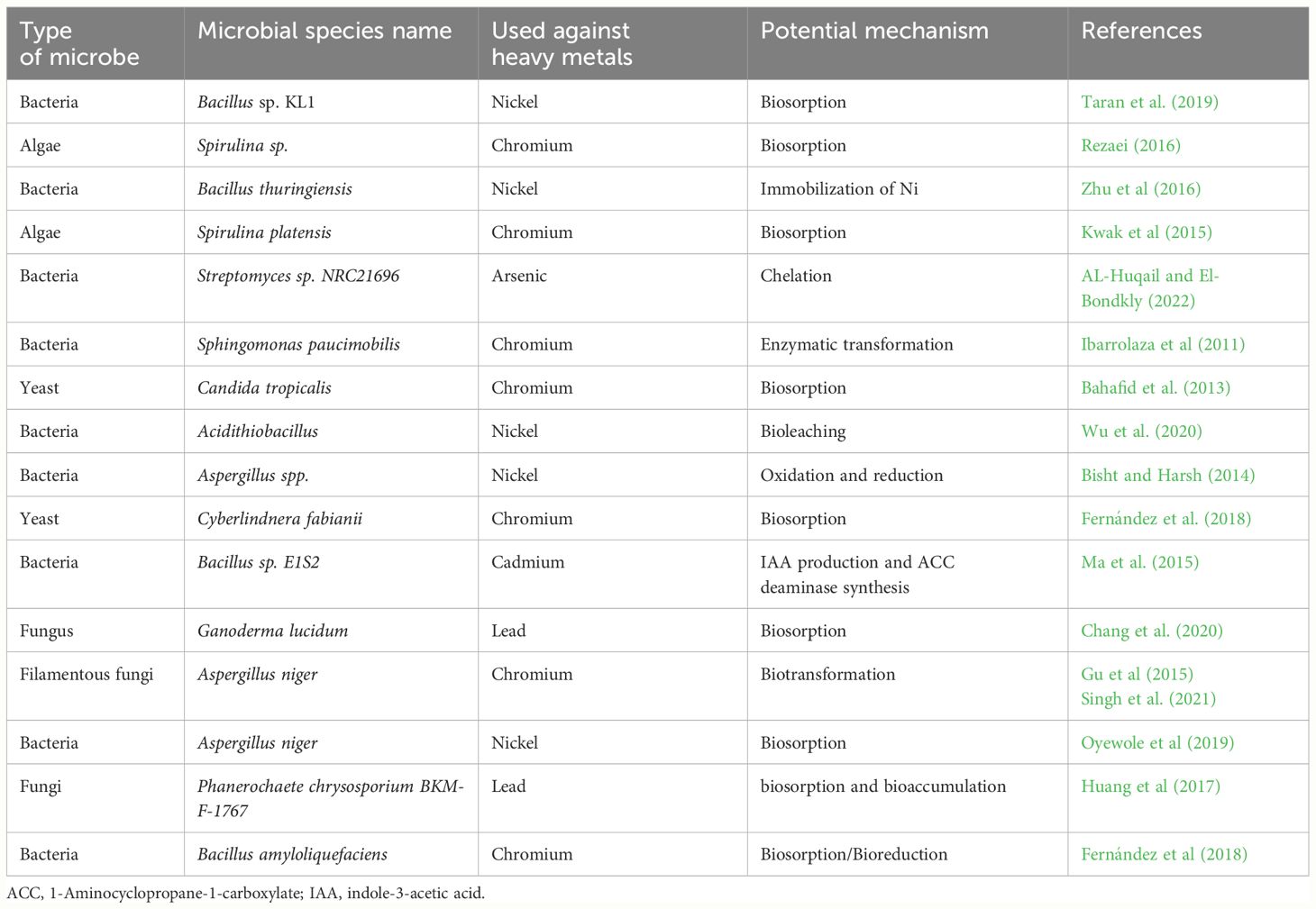
Table 4 Microbial remediation of heavy metals contaminated soils and different mechanism used by microbes to remediate heavy metals contaminated soils.
The cellular membranes of microorganisms contain additional proteins and metabolic products that interact with HMs to decrease their availability. Microbes also develop extracellular sequestration which involves the complexation of metal ions as insoluble compounds and this mechanism is an important way to reduce the HMs toxicity (Thelwell et al., 1998). Microbes also develop intra-cellular sequestration in the metal ions form complexes with distinct compounds in the cell cytoplasm and this is a very common mechanism used by microbes to withstand the toxicity of HMs. Microbes with the aid of low molecular proteins like cysteine accumulation HMs like Cu, Cd and Zn intra-cellularly (Higham et al., 1986) and other microbes like Rhizobium leguminosarum use glutathione to accumulate HMs (Cd) intra-cellularly (Lima et al., 2006). The cell wall of fungi is made of lipids, chitin, polysaccharide, polyphosphates, and proteins which help them to accumulate HMs both intracellularly and extra-cellularly (Remenar et al., 2018).
Numerous metal exporting proteins, including ABC transporters, P-type efflux ATPase, cation diffusion facilitator, and proton-cation anti-porters, are found in microorganisms and assist in the efflux of harmful metals (Soto et al., 2019). ABC transporters also help microorganisms tolerate the stress brought on by HM by facilitating the ions’ transfer across membranes (Lerebours et al., 2016; Zammit et al., 2016). The microbial resistance to HM is also contributed by enzymes that transfer the HMs ions from hazardous to less toxic forms (Giovanella et al., 2016; Liu et al., 2017). Microbes use different mechanisms including, biotransformation, extrusion, EPS production, and proteins to survive the toxicity of metals (Wu et al., 2009). They also produce different proteins like metallothioneins that bind heavy metals thereby reducing HMs toxicity (Wu et al., 2010). Further, EPS produced by microbes is a mixture of proteins, nucleic acid, and polysaccharides that find metals and reduce their concentration in the surrounding environment. Different mechanisms including electrostatic interaction, ion exchange, precipitation, redox process, and surface complexation are involved in processes (Yang et al., 2015). The enzymes transfer metals into less toxic forms in cells through oxidation, reduction, complexation, sequestration, methylation, and de-methylation. Different enzymes like arsenite oxidase, mercuric reductase, chromate reductase, and nickel-coenzyme m reductase have been identified to convert the metals Lerebours et al., 2016; Zammit et al., 2016).
Microbial mechanism used clean up HMs polluted soils
Different mechanisms were used by microbes to clean up the HM-polluted soils. Microbes play a critical role in the oxidation of metals, for instance, Thiobacillus ferrooxidans) can promote the oxidation of metal sulfides to enhance the release of HMs. Microbes mediate the transformation of metal sulfides by sulfur oxidation. In this process, microbes oxidize sulfide ores into metal ions by the process of biological leaching (Kaksonen et al., 2020). Microbes also cause the reduction of metals to reduce their toxicity. The removal capacity of HM-nFeS against Cr-VI was 12-20% lowest as compared to DM-nFeS which was linked with the capacity of both HM-nFeS and DM-nFeS to reduce the Cr (Du et al., 2016). The details of various mechanisms used by microbes to remediate the HMs polluted soils are discussed below.
Bioaccumulation and biosorption
Bioaccumulation and biosorption are the most common mechanisms used by microbes to remediate polluted soils and in both mechanisms, microbes bound the HMs from the surrounding environment (Joutey et al., 2015). In bio-sorption microbes use cellular structure to capture the HM ions and then absorb these HMs on the binding sites of cell walls (Malik, 2004). Microbes also used adsorption mechanisms as bioremediation of HM. Different microbes including Magnetospirillum gryphiswaldense, Bacillus subtilis, microalgae, Chaetomorphalinum, Rhizopus arrhizus, and Saccharomyces cerevisiae produce biosorbents for remediation of HM (Zhou et al., 2012). In comparison to other microbes, bacteria are thought to be superior bio-sorbents because of their larger surface-to-volume ratio and variety of chemosorption sites in their cell walls, including teichoic acid (Beveridge, 1989). Dead bacterial strains also have good biosorbent properties and it has been found that dead Bacillus sphaericus showed 13-20% more bio-sorption capacity for Cr as compared to living cell of the same strain (Velásquez and Dussan, 2009). On the other hand, bio-accumulation depends on an import storage mechanism. This process is known as active bio-accumulation and it involves the movement of HM ions across the lipid bilayer of the cell membrane and into the cytoplasm or intracellular regions. The bioaccumulation of HM in bacterial membranes is mediated via a variety of ionic channels, carrier-mediated transports, permeation, and lipid permeation (Shahpiri and Mohammadzadeh, 2018). In literature, it has been well documented that microbes cause bioaccumulation of Pb, Ni, Hg, Cd, and Cr (Rani and Goel, 2009; Sher and Rehman, 2019; Naskar et al., 2020). Different researchers also identified the micro-bacterium that shows resistance to HMs. For instance, Henson et al. (2015) reported that Microbacterium sp. (Cr-K29) reduced the Cr-IV uptake by 88% while Pattanapipitpaisal et al. (2001) found that Microbacterium liquefaciens eliminated the Cr by 90-95%. These microbes use heavy metal ions in order to facilitate their metabolic activities or they also use enzymes produced by bacterial cells to detoxify ions of HMs (Kubrak et al., 2010).
Bioleaching
Bioleaching is another important mechanism used by a wide range of microbes to remediate polluted soils. For instance, in a research study, authors found that Acidophiles and chemolithotrophs oxidized the Fe-II to Fe-III and reduced sulfur to sulfuric acid. The production of sulfuric acid leads to the synthesis of ferric ions as well as protons which helps to extract metals through solubilizing oxides and sulfides of metal (Srichandan et al., 2014). Microbes are utilized in bioleaching as reduction agents, but they can also be used to extract and recover HMs (Wang and Zhao, 2009). Bio-remediation has been offered as an excellent tool to recover raw materials from effluents (Gadd, 2010). Using an Annona squamosa-based absorbent with 0.1 M HCl, Isaac and Sivakumar (2013) achieved Cd recovery efficiency of 98.7%. Contrarily, matrix-immobilized P. putida cells demonstrated 100% recovery for Cu while Pseudomonas aeruginosa biomass demonstrated 82% recovery efficiency for Cd (Hammaini et al., 2007). In another study, co-application Pseudomonas aeruginosa biomass, and hydrochloric acid (0.1 M HCl) achieved the Cd recovery rate by 82% (Dickerhof et al., 2019), whileP. putida achieved a Cu recovery rate of 100%. Further, autochthonous variant Enterobacter brought an exceeding recovery of >90% for Cu and Pb (Bayramoglu and Arica, 2011). Acidphiles produce different acids through their metabolic process which aids in the dissolution of metal ores, thereby reducing the availability of metals. On the other hand, chemolithotrophs cause oxidation and reduction of sulfur compounds which provide energy to them and also increase the production of acids subsequently increasing solubilization of metals. The bio-leaching carried by both acidophiles and chemolithotrophs is eco-friendly and it can be carried at lower temperatures along with additional benefits of energy saving (Adetunji et al., 2023).
Biotransformation
In biotransformation, microbes converted the toxic metal ions to less hazardous forms (Pervaiz et al., 2013). To adapt to environmental changes, bacteria have developed bio-transformation mechanisms. Production of carbon bonds, isomerization, functional groups, oxidation, reduction, condensation, hydrolysis, methylation, and demethylation help the microbes to transform the HMs. These are all processes that can be used to alter HM in microbes. Microbes cause the transformation of HM and Nagvenkar and Ramaiah (2010) noted that Micrococcusand Acinetobacter caused the oxidation of As-III into less soluble and non-toxic form. Moreover, Thatoi et al. (2014) documented that Cr (VI) tolerant Bacillusspecies cause the biotransformation of Cr (VI) and changed it into a less hazardous form of Cr (III). Both Micrococcus and Acinetobacter reduce the toxicity of metals by causing oxidation, reduction, biological chelation, and inducing the metabolic transformation and bio-film formations (Adetunji et al., 2023).
Bio-volatilization
Bio-volatilization is a process where microbes convert the HMs into volatile compounds enzymatically. This process significantly reduced the availability and toxicity of metals in soil and water. Bio-volatilization uses enzymatic reduction and methylation to convert toxic metals into less toxic forms. Different enzymes like Arsenic methyltransferase, Mercury reductase, and Antimony methyltransferase are involved in the bio-volatilization of As, Hg, and Sb. This method is considered to be suitable for HMs like Hg, As, and Sb, and in this process, these HMs are converted into non-toxic compounds by bio-volatilization (Boriová et al., 2014). Bacterial enzymes like methyltransferases transfer the As(V) into the mono, di, and tri-methylated As species which is then transferred into the atmosphere owing to its volatile nature. In another study, enzymes like reductase (MerA) and mercurial lyase present in archaea and eubacteria caused bio-volatilization (Freedman et al., 2012). Similarly, Scopulariopsis brevicaulis, also showed promising results to convert the As(V) and Hg(II) to their nontoxic states (Urík et al., 2007; Boriová et al., 2014).
Bio-mineralization
In the bio-mineralization process, microbes activate the synthesis of minerals and microbes to tackle with HMs. Different bacteria cause immobilization of Pb and Cr by carbon mineralization (He et al., 2019). Similarly, another bacterial strain Sporosarcina ginsengisoli caused immobilization of different HM calcite, aragonite, and vaterite biomineralization (Achal et al., 2012; Cheng and Holman, 2012). Fungal species also showed promising results for bio-mineralization, for example, Penicillium chrysogenum causes mineralization of Pb and Cr (Qian et al., 2017). Likewise, Penicillium chrysogenum effectively causes bio-mineralization of Pb (Povedano-Priego et al., 2017) additionally, due to the synthesis of PO43- that is released during the breakdown of Pb, Bacillus subtilis triggered bio-mineralization of Pb (Lin et al., 2016). Moreover, other authors reported that Pseudomonas putida forms the carbonate and phosphate minerals which speed up Cd precipitation (Li et al., 2016). Microbes play a critical role in the bio-mineralization process as this process involves the production of mineral deposits to immobilize HMs. The microbes produce EPS, specific metabolites, and organic acids which promote the formation of mineral deposits thereby leading to the immobilization of HMs (Qian et al., 2017). The siderophores and polysaccharides produced by microbes bind the HMs by forming complexes with metals thereby reducing uptake and accumulation of metals by plants. Besides this, they also facilitate the sequestration of metals in soil thereby reducing toxicity of metals on plants.
Modern approaches used to remediate HMs contaminated soils
Different techniques are being applied globally to clean up HM-polluted soils. The role of modern approaches to remediate HM-contaminated soils is discussed below.
Phyto-microbial system for remediation of polluted soils
The application of plants and microbes has emerged as an excellent tool to remediate HM-polluted soils. The use of PGPR has been tested as an effective, and environmentally friendly way to eliminate HMs (Sati et al., 2023). Different microbes like bacteria and fungi can help the plants absorb the HMs (Bojórquez et al., 2016). For instance, Joner and Leyval (1997) noted that fungal inoculated plants uptake more Cd by 90, 127, and 131% growing under different Cd levels (1, 10, and 100 mg/kg) as compared to un-inoculated plants. Similarly, fungal inoculation improves the plant’s ability to absorb Cu, Cd, and Zn (Sati et al., 2023). Different PGPB also produce polysaccharides which increase the transformation, immobilization, and chelation of HM thus reducing their availability. PGPB decreases soil pH by increasing the production of organic acids which helps to remove the HM ions, further, these PGPB also provide nutrients to plants thus reducing the negative effects of HM on plants. Siderophore is also an important microbe and it has shown an appreciable ability to form complexes with different metals like Al, Cd, Cu, Zn, and Pb (Rajkumar et al., 2010). When bio-augmentation and phytoremediation are used together, they produce noticeable results and can also get around some of the challenges that arise with using them alone. The plant also showed significant results to remediate polluted soils and according to Wang et al. (2021), planting Salix in soils with Cd contamination increased the diversity of helpful fungi and microorganisms and contributed to impressive bioremediation outcomes. Plant growth-promoting rhizobacteria (PGPR) interact with plants to increase their ability to absorb HMs through a different mechanism like the production of chelators, increased nutrient uptake, volatilization, transformation, and phytostabilization. This technique is considered sustainable and eco-friendly which can help to mitigate the HMs pollution in agricultural settings.
Genetically engineered microbes: key player to remediated HM polluted soils
The recent advancements in genetic engineering and the production of genetically modified microbes have shown promising results for the remediation of polluted soils. Molecular biology involves understanding and changing the genes to improve the bio-remediation process. It has been documented that different microbes possess resistance mechanisms against HMs (Jaiswal et al., 2019). This includes genes that encode different metal proteins, transporters, and enzymes involved the detoxification (Malla et al., 2018). Thus, engineering these genes can allow for an increase in the microbial ability to effectively carry the microbial remediation process. The recent advance in CRISPR-Cas9 also ensured the editing of microbial genes and the introduction of new genes resulting in improved performance of microbes against HMs (Lee and Lee, 2021). The microbial metabolic pathways can also be modified which can enhance the microbe’s ability, while fine-tuning genes is also leading to better remediation capabilities. Moreover, omics and microbial consortia engineering also provided insights into the response of microbes to HMs (Peña-Castro et al., 2023). This can help to identify different genes, regulatory pathways, and elements to improve the remediation process (Peña-Castro et al., 2023).
The literature shows that genetically modified microbes have a better capacity to remove the HMs (Bhatt et al., 2022). The editing of a single gene and changing the sequence of the gene are important practices used to produce genetically modified microbes (Diep et al., 2018). Different HMs like Cd, Cu, Hg, Ni, and Fe are eliminated by engineered bacteria (Azad et al., 2014) however, the degradation rate largely depends on enzymes present in bacterial cells (Kang, 2014). Moreover, the use of recombinant DNA technology and the introduction of foreign genes has also allowed to develop the genetically modified microbes. For instance, the use of genetically modified Pseudomonas putida and Escherichia colie effectively removed the Hg from polluted soils (Deckwer et al., 2004), similarly, the addition of mer operon from Escherichia coli to bacterium Deinococcus geothemalis also reduce the Hg pollution even at higher temperature (Dixit et al., 2015).
Cupriavidus metallidurans modified genetically with pTP6 plasmid also significantly reduced the Hg from polluted soils (Dixit et al., 2015). The use pMR68 plasmid to introduce novel genes into Pseudomonas also led to the development of Hg resistance (Sone et al., 2013). To enhance the bioremediation of HM, microbial membrane transporters can also be genetically engineered and in this context, transporters and binding mechanisms play a critical role to remediate polluted soils (Manoj et al., 2020). When HMs enter the cell, several phytochelatins, metallothioneins, and polyphosphates collaborate to sequester the HM and alter the HM key storage system, enhancing their ability to take HMs from soil and water (Diep et al., 2018).
The use of genetically modified microbes (GEMs) has speeded up the remediation process. For the successful implementation of implementation of GEMs bacteria must be capable of tolerating the antagonism induced by other native bacterial species (Dixit et al., 2015). Therefore, more novel approaches to screening as well as isolation of microbes for remediation of polluted soils must be used. Recently, different approaches like genomics, metagenomics, metabolomics, proteomics transcriptomics, and computational biology have been used to develop the GEMs for the remediation of HMs (Raza et al., 2024). The recent advancement in high throughput techniques has allowed us to identify the genes involved in the bio-remediation of diverse metals. Further, recent techniques like CRISPR-Cas also made it possible to create GEMs containing genes that can break down the HMs. Besides this, it also made it easy to transfer the desired set of information into microbial genomes to develop the microbes with better ability (Miglani, 2017).
CRISPR-Cas9 techniques have also allowed to development of microbes with appreciable precision, high efficiency, and targeting multiple metals. Genetically modified microbes can provide better results to remediate polluted soils. For instance, genetically modified microbes enhance metal uptake capacity, and they have better metal tolerance and resistance with minimal environmental impacts. They also have appreciable sequestration, transformation, detoxification, and uptake abilities which make them effective tools to mitigate metals toxicity. However, many potential ethical and environmental implications must be considered when using genetically modified microbes for the remediation of polluted soils. For instance, it includes proper regulation and monitoring of genetically modified organisms to balance the benefits of reduction in contamination along with potential risks. Other concerns could be human health, environmental quality, and the negative effects of farming practices. Many environmental considerations must be used while using genetically modified microbes. These microbes should not disrupt biodiversity, food webs, and ecosystem health.
Use of nano-technology for microbial remediation of HM polluted soils
Nano-materials have documented appreciable results in remediating polluted soils owing to their higher surface area, reactivity, and surface chemistry (Khati et al., 2017; Baragaño et al., 2020. Different types of nano-materials including zero-valent metals, metal oxide nanoparticles, carbon-based nano-materials, nano-composites, and nano-biosensors are used around the globe to remediate polluted soils (Aliyari et al., 2023). The nano-materials serve as electron donors in the microbial reduction process and they promote the reduction of toxic metals. On the other hand, nano-particles serve as absorbents and they also favor HMs degradation (Dhanapal et al., 2024). Further, carbon-based produces also enhance the transfer of electrons among metal ions and microbes which in turn increases the efficiency of bio-remediation. Recently, nano-biosensors have also shown appreciable results in detecting HMS which has allowed the monitoring of the remediation process (Dhanapal et al., 2024). Nano-biosorbents can be employed as a substitute for conventional bio-sorbents (Alviz-Gazitua et al., 2019). There are various functional groups found in NPs, including NH2, -COOH, and -OH, and customizing the right functional groups by activating them physically or chemically or by altering their surfaces has produced promising results for the elimination of HMs. Additionally, bacterial strains produce the NPs that can aid in the bio-remediation of the HMs (Arshad et al., 2019). It has been shown that using NPs in conjunction with microorganisms boosted the reduction of HMs, producing more beneficial benefits than using them alone. Nano-particles have a higher surface area, ion exchange, reduction and stabilization capacity, mobility, and delivery which enhance the remediation efficacy. The interaction between NPs and microorganisms is, however, influenced by a variety of factors, including NPs’ chemical properties, size and shape, coating qualities, crystalline phase, level of contamination, and resistance to hazardous elements (Tan et al., 2018). Their tailored properties and enhanced adsorption capacities make them promising candidates for sustainable and efficient remediation strategies, provided that environmental and safety considerations are carefully addressed in their application. However, nano-sorbents must be tested for their environmental impacts in terms of stability and NPs release into the environment. Microbes trapped with nano-materials produce the nano-composite, a combination of Halomonasand iron oxide NPs substantially eliminated the Cd-II and Pb-II (Cao et al., 2020). Since, separation and recovery of HMs from nano-materials is laborious and time-consuming thus magnetic NPs gained significant attention in recent times, wherein surface amendment, coating of diverse materials, and encapsulation focused on simple separation of HMs.
Conclusion and future research directions
Heavy metals pollution is a serious issue across the globe and it is considered the biggest challenge of this century. Heavy metal pollution has drastic effects on soil quality, soil fertility, microbial activities, and diversity, and it also impost deleterious impacts on human health by entering the food chain. Globally, different physiochemical strategies are used to remediate the HMs polluted soils. However, these strategies are very expensive, difficult to application, inefficient in certain conditions and they can also alter the soil quality. Therefore, new biological methods have been developed to remediate polluted soils. Among biological methods, the use of microbes is considered as an effective, economical, and eco-feasible measure to remediate polluted soils. The microbes use different mechanisms to remediate polluted soils and recently engineered microbes provided excellent results for bioremediation which makes them an effective measure to be used on polluted soils.
The use of a single strategy could be both noneffective and inefficient in reclaiming polluted soils. Therefore, a combination of microbes and plants, nano-particles, and additives could also be an important approach to remediate polluted soil. Moreover, a combination of microbes with other strategies including organic and carbon-based materials must also be tested. Additionally, to create the HM tolerance in microbes more focus must be done to understand the physiochemical, biological, and molecular characteristics of microorganisms in soil and water habitats where HMs are prevalent. In the literature, no studies are available about the long-term effects of altering soil pH, temperature, and redox conditions for bioremediation efforts on soil health, microbial diversity, and the persistence of heavy metals over a long period. Therefore, efforts must be made to study the long-term impacts of soil pH, temperature, and redox conditions on on soil health, microbial diversity, and the persistence of heavy metals. Besides this, there is also a lack of information about the interactions between metal concentration, pH, redox potential, and temperature affecting the efficiency and effectiveness of microbial bioremediation processes in contaminated soils. Thus, it is interesting to study the interactions between metal concentration, pH, redox potential, and temperature affecting the effectiveness of the remediation process.
To identify prospective metal resistance and detoxification genes that can be regulated in other species to improve their particular performance, meta-genomic techniques, and microbial metabolic studies are required. Additionally, genetic study is required to comprehend the routes and mechanisms that plants and microorganisms use to tolerate and detoxify heavy metals. The recent advance in omics-based approach can also help to develop the strains tolerant against the prevalent environmental conditions. Recently, yeast has been modified and it showed promising hyper-accumulation capacity, therefore, other bacteria can also be developed in the same way to clean the polluted soils. Future research should pay more attention to the usage of algae since it may be a promising strategy for the sorption of heavy metals. The application of nanotechnology in combination with microbes can also promote microbial use and their efficiency on polluted soil.
Author contributions
HT: Writing – original draft. GX: Writing – original draft, Writing – review & editing. WX: Writing – review & editing. ZY: Writing – review & editing. BZ: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported and funded by National Natural Science Foundation of China (32060318); Hunan Provincial Natural Science Foundation of China (2023JJ50474, 2023JJ50475); 2023 Industry-University Collaborative Education Project of the Higher Education Department of the Ministry of Education (230801720230445). 2023 Hunan University Students Innovation and Entrepreneurship Training Program (S202310553008). 2024 Ministry of Education Supply and Demand docking Employment Education Project: Research on Employment-oriented Practical Teaching Model Reform of Plant Production Courses (2024011153277).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abbas, S., Rafatullah, M., Hossain, K., Ismail, N., Tajarudin, H., Abdul Khalil, H. (2018). A review on mechanism and future perspectives of cadmium-resistant bacteria. Int. J. Environ. Sci. Technol. 15, 243–262. doi: 10.1007/s13762-017-1400-5
Abbaszadeh-Dahaji, P., Baniasad-Asgari, A., Hamidpour, M. (2019). The effect of Cu-resistant plant growth-promoting rhizobacteria and EDTA on phytoremediation efficiency of plants in a Cu-contaminated soil. Environ. Sci. pollut. Res. 26, 31822–31833. doi: 10.1007/s11356-019-06334-0
Acar, F., Malkoc, E. (2004). The removal of chromium (VI) from aqueous solutions by Fagus orientalis L. Bioresour Technol. 94, 13–15. doi: 10.1016/j.biortech.2003.10.032
Achal, V., Pan, X., Fu, Q., Zhang, D. (2012). Biomineralization based remediation of As (III) contaminated soil by Sporosarcina ginsengisoli. J. Hazard Mater 201, 178–184. doi: 10.1016/j.jhazmat.2011.11.067
Acosta, J., Faz, A., Martínez-Martínez, S., Zornoza, R., Carmona, D., Kabas, S. (2011). Multivariate statistical and GIS-based approach to evaluate heavy metals behavior in mine sites for future reclamation. J. Geochem Explor. 109, 8–17. doi: 10.1016/j.gexplo.2011.01.004
Adetunji, A. I., Oberholster, P. J., Erasmus, M. (2023). Bioleaching of metals from e-waste using microorganisms: A review. Minerals 13, 828. doi: 10.3390/min13060828
Adrees, M., Ali, S., Rizwan, M., Ibrahim, M., Abbas, F., Farid, M., et al. (2015). The effect of excess copper on growth and physiology of important food crops: a review. Environ. Sci. pollut. Res. 22, 8148–8162. doi: 10.1007/s11356-015-4496-5
Aguirre, C. F. V., Páez, F. A. R., Vargas, S. E. (2018). Effect of arbuscular mycorrhizae and mercury on Lactuca sativa (Asteraceae) seedling morpho—histology. Environ. Exp. Bot. 156, 197–202.
Alaboudi, K. A., Ahmed, B., Brodie, G. (2018). Phytoremediation of Pb and Cd contaminated soils by using sunflower (Helianthus annuus) plant. Ann. Agric. Sci. 63, 123–127. doi: 10.1016/j.aoas.2018.05.007
Alboghobeish, H., Tahmourespour, A., Doudi, M. (2014). The study of Nickel Resistant Bacteria (NiRB) isolated from wastewaters polluted with different industrial sources. J. Environ. Heal Sci. Eng. 12, 1–7. doi: 10.1186/2052-336X-12-44
AL-Huqail, A. A., El-Bondkly, A. (2022). Improvement of Zea mays L. growth parameters under chromium and arsenic stress by the heavy metal-resistant Streptomyces sp. NRC21696. Int. J. Environ. Sci. 19, 5301–5322. doi: 10.1007/s13762-021-03532-7
Aliyari, R. S., Nobaharan, K., Pashapoor, N., Pandey, J., Dehghanian, Z., Senapathi, V., et al. (2023). Nano-microbial remediation of polluted soil: a brief insight. Sustain. 15, 876. doi: 10.3390/su15010876
Alkorta, I., Epelde, L., Garbisu, C. (2017). Environmental parameters altered by climate change affect the activity of soil microorganisms involved in bioremediation. FEMS Microbiol. Lett. 364, 200. doi: 10.1093/femsle/fnx200
Alsokari, S., Aldesuquy, H. (2011). Synergistic effect of polyamines and waste water on leaf turgidity, heavy metals accumulation in relation to grain yield. J. Appl. Sci. Res. 7, 376–384.
Aluko, O. A., Opoku, E. E. O., Ibrahim, M. (2021). Investigating the environmental effect of globalization: Insights from selected industrialized countries. J. Environ. Manag 281, 111892. doi: 10.1016/j.jenvman.2020.111892
Alves, A. R., Yin, Q., Oliveira, R. S., Silva, E. F., Novo, L. A. (2022). Plant growth-promoting bacteria in phytoremediation of metal-polluted soils: Current knowledge and future directions. Sci. Total Environ. 838, 156435. doi: 10.1016/j.scitotenv.2022.156435
Alviz-Gazitua, P., Fuentes-Alburquenque, S., Rojas, L. A., Turner, R. J., Guiliani, N., Seeger, M. (2019). The response of Cupriavidus metallidurans CH34 to cadmium involves inhibition of the initiation of biofilm formation, decrease in intracellular c-di-GMP levels, and a novel metal regulated phosphodiesterase. Front. Microbiol. 10, 1499. doi: 10.3389/fmicb.2019.01499
Amirnia, S., Ray, M. B., Margaritis, A. (2015). Heavy metals removal from aqueous solutions using Saccharomyces cerevisiae in a novel continuous bioreactor–biosorption system. Chem. Eng. J. 264, 863–872. doi: 10.1016/j.cej.2014.12.016
Arce-Inga, M., González-Pérez, A. R., Hernandez-Diaz, E., Chuquibala-Checan, B., Chavez-Jalk, A., Llanos-Gomez, K. J., et al. (2022). Bioremediation Potential of Native Bacillus sp. Strains as a Sustainable Strategy for Cadmium Accumulation of Theobroma cacao in Amazonas Region. Microorganisms 10, 2108.
Armah, F. A., Quansah, R., Luginaah, I. (2014). A systematic review of heavy metals of anthropogenic origin in environmental media and biota in the context of gold mining in Ghana. Int. Sch Res. Not 2014 (1), 252148. doi: 10.1155/2014/252148
Arshad, F., Selvaraj, M., Zain, J., Banat, F., Haija, M. A. (2019). Polyethylenimine modified graphene oxide hydrogel composite as an efficient adsorbent for heavy metal ions. Sep Purif Technol. 209, 870–880. doi: 10.1016/j.seppur.2018.06.035
Aslam, F., Yasmin, A., Sohail, S. (2020). Bioaccumulation of lead, chromium, and nickel by bacteria from three different genera isolated from industrial effluent. Int. Microbiol. 23, 253–261. doi: 10.1007/s10123-019-00098-w
Asyakina, L. K., Dyshlyuk, L. S., Prosekov, A. Y. (2021). Reclamation of post-technological landscapes: International experience. doi: 10.21603/2074-9414-2021-4
Ayangbenro, A. S., Babalola, O. O. (2017). A new strategy for heavy metal polluted environments: a review of microbial biosorbents. Int. J. Env. Res. Public Health 14, 94. doi: 10.3390/ijerph14010094
Azad, M. A. K., Amin, L., Sidik, N. M. (2014). Genetically engineered organisms for bioremediation of pollutants in contaminated sites. Chin. Sci. Bull. 59, 703–714. doi: 10.1007/s11434-013-0058-8
Bahafid, W., Tahri Joutey, N., Sayel, H., Boularab, I., El Ghachtouli, N. (2013). Bioaugmentation of chromium-polluted soil microcosms with Candida tropicalis diminishes phytoavailable chromium. J. Appl. Microbiol. 115, 727–734. doi: 10.1111/jam.12282
Bakshi, S., Banik, C., He, Z. (2018). “The impact of heavy metal contamination on soil health,” in Managing soil health for sustainable agriculture, vol. 2 . Ed. Reicosky, 1–36.
Bala, S., Garg, D., Thirumalesh, B. V., Sharma, M., Sridhar, K., Inbaraj, B. S., et al. (2022). Recent strategies for bioremediation of emerging pollutants: a review for a green and sustainable environment. Toxics 10 (8), 484. doi: 10.3390/toxics10080484
Bandara, T., Krohn, C., Jin, J., Chathurika, J. B. A. J., Franks, A., Xu, J., et al. (2022). The effects of biochar aging on rhizosphere microbial communities in cadmium-contaminated acid soil. Chemosphere 303, 135153. doi: 10.1016/j.chemosphere.2022.135153
Bandowe, B. A. M., Bigalke, M., Boamah, L., Nyarko, E., Saalia, F. K., Wilcke, W. (2014). Polycyclic aromatic compounds (PAHs and oxygenated PAHs) and trace metals in fish species from Ghana (West Africa): bioaccumulation and health risk assessment. Environ. Int. 65, 135–146. doi: 10.1016/j.envint.2013.12.018
Bansal, N., Coetzee, J. J., Chirwa, E. M. (2019). In situ bioremediation of hexavalent chromium in presence of iron by dried sludge bacteria exposed to high chromium concentration. Ecotoxicol Environ. Saf. 172, 281–289. doi: 10.1016/j.ecoenv.2019.01.094
Baragaño, D., Forján, R., Welte, L., Gallego, J. L. R. (2020). Nanoremediation of As and metals polluted soils by means of graphene oxide nanoparticles. Sci. Rep. 10, 1896. doi: 10.1038/s41598-020-58852-4
Barquilha, C., Cossich, E., Tavares, C., Silva, E. (2017). Biosorption of nickel (II) and copper (II) ions in batch and fixed-bed columns by free and immobilized marine algae Sargassum sp. J. Clean Prod 150, 58–64. doi: 10.1016/j.jclepro.2017.02.199
Barsainya, M., Chandra, P., Singh, D. P. (2016). Investigation of Cr (VI) uptake in saline condition using psychrophilic and mesophilic Penicillium sp. Int. J. Curr. Microbiol. Appl. Sci. 5, 274–288. doi: 10.20546/ijcmas
Bayramoglu, G., Arica, M. Y. (2011). Preparation of a composite biosorbent using scenedesmus quadricauda biomass and alginate/polyvinyl alcohol for removal of Cu(II) and Cd(II) Ions: isotherms, kinetics, and thermodynamic studies. Water Air Soil pollut. 221, 391–403. doi: 10.1007/s11270-011-0798-5
Bazihizina, N., Redwan, M., Taiti, C., Giordano, C., Monetti, E., Masi, E., et al. (2015). Root based responses account for Psidium guajava survival at high nickel concentration. J. Plant Physiol. 174, 137–146. doi: 10.1016/j.jplph.2014.10.011
Bellion, M., Courbot, M., Jacob, C., Blaudez, D., Chalot, M. (2006). Extracellular and cellular mechanisms sustaining metal tolerance in ectomycorrhizal fungi. FEMS Microbiol. Lett. 254, 173–181. doi: 10.1111/fml.2006.254.issue-2
Bentley, R., Chasteen, T. G. (2002). Microbial methylation of metalloids: arsenic, antimony, and bismuth. Microbiol. Mol. Biol. Rev. 66, 250–271. doi: 10.1128/MMBR.66.2.250-271.2002
Beveridge, T. J. (1989). Role of cellular design in bacterial metal accumulation and mineralization. Annu. Rev. Microbiol. 43, 147–171. doi: 10.1146/annurev.mi.43.100189.001051
Bewley, R., Stotzky, G. (1983). Effects of cadmium and zinc on microbial activity in soil; influence of clay minerals. Part I: metals added individually. Sci. Total Environ. 31, 41–55. doi: 10.1016/0048-9697(83)90055-4
Bhati, T., Gupta, R., Yadav, N., Singh, R., Fuloria, A., Waziri, A., et al. (2019). Assessment of bioremediation potential of Cellulosimicrobium sp. for treatment of multiple heavy metals. Microbiol. Biotechnol. Lett. 47, 269–277. doi: 10.4014/mbl.1808.08006
Bhatt, P., Sethi, K., GAngola, S., Bhandari, G., Verma, A., Adnan, M., et al (2022). Modeling and simulation of atrazine biodegradation in bacteria and its effect in other living systems. J. Biomol Struct. Dyn 40, 3285–3295. doi: 10.1080/07391102.2020.1846623
Bilal, M., Rasheed, T., Sosa-Hernández, J. E., Raza, A., Nabeel, F., Iqbal, H. M. (2018). Biosorption: an interplay between marine algae and potentially toxic elements—a review. Mar. Drugs 16, 65. doi: 10.3390/md16020065
Bisht, J., Harsh, N. (2014). Utilizing Aspergillus Niger for bioremediation of tannery effluent. Environ. Res. J. 2 (1), 77–81.
Bojórquez, C., Frías Espericueta, M. G., Voltolina, D. (2016). Removal of cadmium and lead by adapted strains of Pseudomonas aeruginosa and Enterobacter cloacae. Rev. Int. Contaminación Ambiental 32, 407–412. doi: 10.20937/RICA
Boriová, K., Čerňanský, S., Matúš, P., Bujdoš, M., Šimonovičová, A. (2014). Bioaccumulation and biovolatilization of various elements using filamentous fungus Scopulariopsis brevicaulis. Lett. Appl. Microbiol. 59, 217–223. doi: 10.1111/lam.12266
Brandt, K. K., Frandsen, R. J. N., Holm, P. E., Nybroe, O. (2010). Development of pollution-induced community tolerance is linked to structural and functional resilience of a soil bacterial community following a five-year field exposure to copper. Soil Biol. Biochem. 49, 748–757. doi: 10.1016/j.soilbio.2010.01.008
Bravo, D. (2022). Bacterial cadmium-immobilization activity measured by isothermal microcalorimetry in cacao-growing soils from Colombia. Front. Environ. Sci. 10, 910234.
Briffa, J., Sinagra, E., Blundell, R. (2020). Heavy metal pollution in the environment and their toxicological effects on humans. Heliyon 6 (9), E04691. doi: 10.1016/j.heliyon.2020.e04691
Burns, R. G., DeForest, J. L., Marxsen, J., Sinsabaugh, R. L., Stromberger, M. E., Wallenstein, M. D., et al. (2013). Soil enzymes in a changing environment: current knowledge and future directions. Soil Biol. Biochem. 58, 216–234. doi: 10.1016/j.soilbio.2012.11.009
Cabello-Conejo, M., Becerra-Castro, C., Prieto-Fernández, A., Monterroso, C., Saavedra-Ferro, A., Mench, M., et al. (2014). Rhizobacterial inoculants can improve nickel phytoextraction by the hyperaccumulator Alyssum pintodasilvae. Plant Soil 379, 35–50. doi: 10.1007/s11104-014-2043-7
Cao, X., Alabresm, A., Chen, Y. P., Decho, A. W., Lead, J. (2020). Improved metal remediation using a combined bacterial and nanoscience approach. Sci. Total Environ. 704, 135378. doi: 10.1016/j.scitotenv.2019.135378
Cao, L., Jiang, M., Zeng, Z., Du, A., Tan, H., Liu, Y. (2008). Trichoderma atroviride F6 improves phytoextraction efficiency of mustard (Brassica juncea L.) Coss. Var. foliosa Bailey) Cd Ni contaminated soils. Chemosphere 71, 1769–1773. doi: 10.1016/j.chemosphere.2008.01.066
Casarrubia, S., Martino, E., Daghino, S., Kohler, A., Morin, E., Khouja, H.-R., et al. (2020). Modulation of plant and fungal gene expression upon cd exposure and symbiosis in ericoid mycorrhizal Vaccinium myrtillus. Front. Microbiol. 11, 341. doi: 10.3389/fmicb.2020.00341
Cepoi, L., Zinicovscaia, I., Valuta, A., Codreanu, L., Rudi, L., Chiriac, T., et al. (2021). Bioremediation capacity of edaphic cyanobacteria Nostoc linckia for chromium in association with other heavy-metals-contaminated soils. Environ 9, 1. doi: 10.3390/environments9010001
Chan, W., Li, H., Wu, F., Wu, S., Wong, M. H. (2013). Arsenic uptake in upland rice inoculated with a combination or single arbuscular mycorrhizal fungi. J. Hazard Mater 262, 1116–1122. doi: 10.1016/j.jhazmat.2012.08.020
Chandra, R., Manna, A. K., Sahu, M., Rout, K., Patra, G. K. (2020). Simple salicylaldimine-functionalized dipodal bis Schiff base chromogenic and fluorogenic chemosensors for selective and sensitive detection of Al3+ and Cr3+. Inorg Chim. Acta 499, 119192. doi: 10.1016/j.ica.2019.119192
Chang, J., Zhang, H., Cheng, H., Yan, Y., Chang, M., Cao, Y., et al. (2020). Spent Ganoderma lucidum substrate derived biochar as a new bio-adsorbent for Pb2+/Cd2+ removal in water. Chemosphere 241, 125121. doi: 10.1016/j.chemosphere.2019.125121
Chanmugathas, P., Bollag, J. M. (1988). A column study of the biological mobilization and speciation of cadmium in soil. Arch. Environ. Contam Toxicol. 17, 229–237. doi: 10.1007/BF01056029
Chen, Y., Wu, H., Sun, P., Liu, J., Qiao, S., Zhang, D., et al. (2021). Remediation of chromium-contaminated soil based on Bacillus cereus WHX-1 immobilized on biochar: Cr (VI) transformation and functional microbial enrichment. Front. Microbiol. 12, 641913. doi: 10.3389/fmicb.2021.641913
Cheng, Z., Shi, J., He, Y., Wu, L., Xu, J. (2022). Assembly of root-associated bacterial community in cadmium contaminated soil following five-year consecutive application of soil amendments: Evidences for improved soil health. J. Hazard Mat 426, 128095. doi: 10.1016/j.jhazmat.2021.128095
Cheng, Y., Holman, H.-Y., Lin, Z. (2012). Remediation of chromium and uranium contamination by microbial activity. Elements. 8, 107–112.
Cheung, K., Gu, J.-D. (2007). Mechanism of hexavalent chromium detoxification by microorganisms and bioremediation application potential: a review. Int. Biodeterior Biodegrad 59, 8–15. doi: 10.1016/j.ibiod.2006.05.002
Choi, W., Yu, J., Lee, T. (2017). Microbial oxidation of antimonite and arsenite by bacteria isolated from antimony-contaminated soils. Int. J. Hydrogen Energy 42, 27832–27842. doi: 10.1016/j.ijhydene.2017.08.056
Chowardhara, B., Borgohain, P., Saha, B., Awasthi, J. P., Panda, S. K. (2020). Differential oxidative stress responses in Brassica juncea L. Czern and Coss cultivars induced by cadmium at germination and early seedling stage. Acta Physiol. Plant 42, 1–12. doi: 10.1007/s11738-020-03094-0
Christoforidis, A., Orfanidis, S., Papageorgiou, S., Lazaridou, A., Favvas, E., Mitropoulos, A. C. (2015). Study of Cu (II) removal by Cystoseira crinitophylla biomass in batch and continuous flow biosorption. Chem. Eng. J. 277, 334–340. doi: 10.1016/j.cej.2015.04.138
Costa, F., Tavares, T. (2017). Bioremoval of Ni and Cd in the presence of diethylketone by fungi and by bacteria–A comparative study. Int. Biodeterior Biodegrad 120, 115–123. doi: 10.1016/j.ibiod.2017.02.018
Cozzolino, V., De Martino, A., Nebbioso, A., Di Meo, V., Salluzzo, A., Piccolo, A. (2016). Plant tolerance to mercury in a contaminated soil is enhanced by the combined effects of humic matter addition and inoculation with arbuscular mycorrhizal fungi. Environ. Sci. pollut. Res. 23, 11312–11322. doi: 10.1007/s11356-016-6337-6
Cui, L., Feng, X., Lin, C. J., Wang, X., Meng, B., Wang, X., et al. (2014). Accumulation and translocation of 198Hg in four crop species. Environ. Toxicol. Chem. 33, 334–340. doi: 10.1002/etc.2443
Das, P., Sinha, S., Mukherjee, S. K. (2014). Nickel bioremediation potential of Bacillus thuringiensis KUNi1 and some environmental factors in nickel removal. Bioremediation J. 18, 169–177. doi: 10.1080/10889868.2014.889071
Debonne, N., Van Vliet, J., Metternicht, G., Verburg, P. (2021). Agency shifts in agricultural land governance and their implications for land degradation neutrality. Global Environ. Change 66, 102221. doi: 10.1016/j.gloenvcha.2020.102221
Deckwer, W.-D., Becker, F., Ledakowicz, S., Wagner-Döbler, I. (2004). Microbial removal of ionic mercury in a three-phase fluidized bed reactor. Environ. Sci. Technol. 38, 1858–1865. doi: 10.1021/es0300517
De Francisco, P., Martín-González, A., Rodriguez-Martín, D., Díaz, S. (2021). Interactions with arsenic: mechanisms of toxicity and cellular resistance in eukaryotic microorganisms. Int. J. Env. Res. Public Health 18, 12226. doi: 10.3390/ijerph182212226
de Souza Guilherme, M. d. F., de Oliveira, H. M., da Silva, E. (2015). Cadmium toxicity on seed germination and seedling growth of wheat triticum aestivum. Acta Sci. Biol. Sci. 37, 499–504.
Demarco, C. F., Quadro, M. S., Selau Carlos, F., Pieniz, S., Morselli, L. B. G. A., Andreazza, R. (2023). Bioremediation of aquatic environments contaminated with heavy metals: a review of mechanisms, solutions and perspectives. Sustainability 15, 1411. doi: 10.3390/su15021411
Demey, H., Vincent, T., Guibal, E. (2018). A novel algal-based sorbent for heavy metal removal. Chem. Eng. J. 332, 582–595. doi: 10.1016/j.cej.2017.09.083
Deng, Y., Huang, H., Fu, S., Jiang, L., Liang, Y., Liu, X., et al. (2021). Cadmium uptake and growth responses of potted vegetables to the cd-contaminated soil inoculated with cd-tolerant purpureocillium lilacinum N1. Minerals 11, 622. doi: 10.3390/min11060622
DeOliveira, E. C. M., Pires, L. P., Santos, V. S. V., Caixeta, E. S., Bravo, J. V. M., Pereira, B. B. (2023). Phytoremediation, bioaccessibility and ecotoxicological risk assessment of arsenic in a gold mining area. Chemosphere 319, 138030.
Dermont, G., Bergeron, M., Mercier, G., Richer-Laflèche, M. (2008). Soil washing for metal removal: a review of physical/chemical technologies and field applications. J. Hazard Mater 152, 1–31. doi: 10.1016/j.jhazmat.2007.10.043
Dhanapal, A. R., Thiruvengadam, M., Vairavanathan, J., Venkidasamy, B., Easwaran, M., Ghorbanpour, M. (2024). Nanotechnology approaches for the remediation of agricultural polluted soils. ACS Omega 9, 13522–13533. doi: 10.1021/acsomega.3c09776
Dickerhof, N., Isles, V., Pattemore, P., Hampton, M. B., Kettle, A. J. (2019). Exposure of Pseudomonas aeruginosa to bactericidal hypochlorous acid during neutrophil phagocytosis is compromised in cystic fibrosis. J. Biol. Chem. 294, 13502–13514. doi: 10.1074/jbc.RA119.009934
Diep, P., Mahadevan, R., Yakunin, A. F. (2018). Heavy metal removal by bioaccumulation using genetically engineered microorganisms. Front. Bioeng Biotechnol. 6, 157. doi: 10.3389/fbioe.2018.00157
Ding, Y., Ding, L., Xia, Y., Wang, F., Zhu, C. (2020). Emerging roles of microRNAs in plant heavy metal tolerance and homeostasis. J. Agric. Food Chem. 68, 1958–1965. doi: 10.1021/acs.jafc.9b07468
Dixit, R., Wasiullah, X., Malaviya, D., Pandiyan, K., Singh, U. B., Sahu, A., et al. (2015). Bioremediation of heavy metals from soil and aquatic environment: an overview of principles and criteria of fundamental processes. Sustainability 7, 2189–2212. doi: 10.3390/su7022189
Du, J., Bao, J., Lu, C., Werner, D. (2016). Reductive sequestration of chromate by hierarchical FeS@ Fe0 particles. Water Res. 102, 73–81. doi: 10.1016/j.watres.2016.06.009
Durand, A., Piutti, S., Rue, M., Morel, J., Echevarria, G., Benizri, E. (2016). Improving nickel phytoextraction by co-cropping hyperaccumulator plants inoculated by plant growth promoting rhizobacteria. Plant Soil 399, 179–192. doi: 10.1007/s11104-015-2691-2
Elahi, A., Rehman, A. (2019). Comparative behavior of two gram positive Cr6+ resistant bacterial strains Bacillus aerius S1 and Brevibacterium iodinum S2 under hexavalent chromium stress. Biotechnol. Rep. 21, e00307. doi: 10.1016/j.btre.2019.e00307
Eneh, O. C., Eneh, C. A., Eneonwo, C. I., Okosun, A., Emenuga, V., Obi, N. I., et al. (2023). Mitigating potential public health risks and challenges from hazardous materials contained in electronic waste items in a developing country setting. Environ. Anal. Health Toxicol. 38 (1), e2023001. doi: 10.5620/eaht.2023001
Fang, L., Zhou, C., Cai, P., Chen, W., Rong, X., Dai, K., et al. (2011). Binding charac teristics of copper and cadmium by cyanobacterium Spirulina platensis. . J. Hazard Mater 190, 810–815. doi: 10.1016/j.jhazmat.2011.03.122
Faskhutdinova, E. R., Osintseva, M. A., Neverova, O. A. (2021). Prospects of using soil microbiome of mine tips for remediationof anthropogenically disturbed ecosystems. Food Processing: Techniques and Technology 4, 883–904. doi: 10.21603/2074-9414-2021-4-883-904
Fawzy, E. M., Abdel-Motaal, F. F., El-zayat, S. A. (2017). Biosorption of heavy metals onto different eco-friendly substrates. J. Toxicol. Environ. Health 9, 35–44. doi: 10.4172/2155-6199
Feng, G., Chen, Z., Zhu, P., Yan, L., Hao, X., Xiao, Y. (2023). The potential roles of free and attached microbial community in decreasing cadmium level from cadmium-contaminated soils by mixotrophic acidophiles of different scale-up cultivation stages. Minerals 13, 546. doi: 10.3390/min13040546
Feria-Cáceres, P. F., Penagos-Velez, L., Moreno-Herrera, C. X. (2022). Tolerance and cadmium (Cd) immobilization by native bacteria isolated in cocoa soils with increased metal content. Microbiol. Res. 13, 556–573. doi: 10.3390/microbiolres13030039
Fernández, P. M., Viñarta, S. C., Bernal, A. R., Cruz, E. L., Figueroa, L. I. (2018). Bioremediation strategies for chromium removal: current research, scale-up approach and future perspectives. Chemosphere 208, 139–148. doi: 10.1016/j.chemosphere.2018.05.166
Ferreira, V., Koricheva, J., Duarte, S., Niyogi, D. K., Guérold, F. (2016). Effects of anthropogenic heavy metal contamination on litter decomposition in streams–a meta-analysis. Environ. pollut. 210, 261–270. doi: 10.1016/j.envpol.2015.12.060
Foyer, C. H., Noctor, G. (2016). Stress-triggered redox signalling: what’s in pROSpect? Plant Cell Environ. 39, 951–964.
Freedman, Z., Zhu, C., Barkay, T. (2012). Mercury resistance and mercuric reductase activities and expression among chemotrophic thermophilic Aquificae. Appl. Environ. Microbiol. 78, 6568–6575. doi: 10.1128/AEM.01060-12
Freitas, A. P. P., Schneider, I. A. H., Schwartzbold, A. (2011). Biosorption of heavy metals by algal communities in water streams affected by the acid mine drainage in the coal-mining region of Santa Catarina state, Brazil. Miner Eng. 24, 1215–1218. doi: 10.1016/j.mineng.2011.04.013
Gadd, G. M. (2010). Metals, minerals and microbes: geomicrobiology and bioremediation. Microbiology 156, 609–643. doi: 10.1099/mic.0.037143-0
Galiulin, R., Galiulina, R. (2008). Removing heavy metals from soil with plants. Herald Russ Acad. Sci. 78, 141–143. doi: 10.1134/S1019331608020044
Gao, M., Chang, X., Yang, Y., Song, Z. (2020). Foliar graphene oxide treatment increases photosynthetic capacity and reduces oxidative stress in cadmium-stressed lettuce. Plant Physiol. Biochem. 154, 287–294. doi: 10.1016/j.plaphy.2020.06.021
Ghosh, S., Bhattacharya, J., Nitnavare, R., Webster, T. J. (2022). “Heavy metal removal by Bacillus for sustainable agriculture,” in Bacilli in Agrobiotechnology: Plant stress tolerance, bioremediation, and bioprospecting (Springer), 1–30.
Gill, S. S., Anjum, N. A., Ahmad, I., Thangavel, P., Sridevi, G., Pacheco, M., et al. (2012). “Metal hyperaccumulation and tolerance in Alyssum, Arabidopsis and Thlaspi: An overview,” in The plant family Brassicaceae: contribution towards phytoremediation, 99–137.
Giovanella, P., Cabral, L., Bento, F. M., Gianello, C., Camargo, F. A. O. (2016). Mercury (II) removal by resistant bacterial isolates and mercuric (II) reductase activity in a new strain of Pseudomonas sp. B50A. New Biotechnol. 33, 216–223. doi: 10.1016/j.nbt.2015.05.006
Gomes, M. P., MdOG, N., EMd, C., Soares, ÂM (2011). Ecophysiological and anatomical changes due to uptake and accumulation of heavy metal in Brachiaria decumbens. Sci. Agric. 68, 566–573. doi: 10.1590/S0103-90162011000500009
Gong, Y., Zhao, D., Wang, Q. (2018). An overview of field-scale studies on remediation of soil contaminated with heavy metals and metalloids: Technical progress over the last decade. Water Res. 147, 440–460. doi: 10.1016/j.watres.2018.10.024
Gontia-Mishra, I., Sapre, S., Sharma, A., Tiwari, S. (2016). Alleviation of mercury toxicity in wheat by the interaction of mercury-tolerant plant growth-promoting rhizobacteria. J. Plant Growth Regul. 35, 1000–1012. doi: 10.1007/s00344-016-9598-x
Grujic, S., Vasić, S., Radojević, I., Čomić, L., Ostojić, A. (2017). Comparison of the Rhodotorula mucilaginosa biofilm and planktonic culture on heavy metal susceptibility and removal potential. Water Air Soil pollut. 228, 1–8. doi: 10.1007/s11270-017-3259-y
Gu, Y., Xu, W., Liu, Y., Zeng, G., Huang, J., Tan, X., et al. (2015). Mechanism of Cr (VI) reduction by Aspergillus Niger: enzymatic characteristic, oxidative stress response, and reduction product. Environ. Sci. pollut. Res. 22, 6271–6279. doi: 10.1007/s11356-014-3856-x
Guan, Q., Liu, Z., Shao, W., Tian, J., Luo, H., Ni, F., et al. (2022). Probabilistic risk assessment of heavy metals in urban farmland soils of a typical oasis city in northwest China. Sci. Total Environ. 833, 155096. doi: 10.1016/j.scitotenv.2022.155096
Gupta, P., Bhatnagar, A. (2015). Spatial distribution of arsenic in different leaf tissues and its effect on structure and development of stomata and trichomes in mung bean, Vigna radiata (L.) Wilczek. Environ. Exp. Bot. 109, 12–22. doi: 10.1016/j.envexpbot.2014.08.001
Gupta, A., Joia, J., Sood, A., Sood, R., Sidhu, C., Kaur, G. (2016). Microbes as potential tool for remediation of heavy metals: a review. J. Microb. Biochem. Technol. 8, 364–372. doi: 10.4172/1948-5948
Hakeem, K. R., Bhat, R. A., Qadri, H. (2020). Bioremediation and biotechnology (Springer: Cham)Switzerland doi: 10.1007/978-3-030-35691-0
Hammaini, A., González, F., Ballester, A., Blázquez, M., Munoz, J. (2007). Biosorption of heavy metals by activated sludge and their desorption characteristics. J. Environ. Manag 84, 419–426. doi: 10.1016/j.jenvman.2006.06.015
Hamsa, N., Yogesh, G., Koushik, U., Patil, L. (2017). Nitrogen transformation in soil: effect of heavy metals. Int. J. Curr. Microbiol. Appl. Sci. 6, 816–832. doi: 10.20546/ijcmas
Hamzah, A., Sarmani, S., Yatim, N. (2015). Phytoremediation of Pb and Hg by using Scirpus mucronatus with addition of bacterial inoculums. J. Radioanal Nucl. Chem. 304, 151–155. doi: 10.1007/s10967-014-3775-9
Haque, S., Srivastava, N., Pal, D. B., Alkhanani, M. F., Almalki, A. H., Areeshi, M. Y., et al. (2022). Functional microbiome strategies for the bioremediation of petroleum-hydrocarbon and heavy metal contaminated soils: A review. Sci. Total Environ. 833, 155222. doi: 10.1016/j.scitotenv.2022.155222
Hassan, M. U., Chattha, M. U., Khan, I., Chattha, M. B., Aamer, M., Nawaz, M., et al. (2019). Nickel toxicity in plants: reasons, toxic effects, tolerance mechanisms, and remediation possibilities—a review. Environ. Sci. pollut. Res. 26, 12673–12688. doi: 10.1007/s11356-019-04892-x
Hassan, S. E., Hijri, M., St-Arnaud, M. (2013). Effect of arbuscular mycorrhizal fungi on trace metal uptake by sunflower plants grown on cadmium contaminated soil. New Biotechnol. 30, 780–787. doi: 10.1016/j.nbt.2013.07.002
He, J., Chen, X., Zhang, Q., Achal, V. (2019). More effective immobilization of divalent lead than hexavalent chromium through carbonate mineralization by Staphylococcus epidermidis HJ2. Int. Biodeterior Biodegrad 140, 67–71. doi: 10.1016/j.ibiod.2019.03.012
Heidari, J., Amooaghaie, R., Kiani, S. (2020). Impact of chitosan on nickel bioavailability in soil, the accumulation and tolerance of nickel in Calendula tripterocarpa. Int. J. Phytoremediation 22, 1175–1184. doi: 10.1080/15226514.2020.1748564
Henson, M. W., Santo, D. J. W., Kourtev, P. S., Jensen, R. V., Dunn, J. A., Learman, D. R. (2015). Metabolic and genomic analysis elucidates strain-level variation in Microbacterium spp. isolated chromate contaminated sediment. PeerJ 3, 1395–1412. doi: 10.7717/peerj.1395
Higham, D. P., Sadler, P. J., Scawen, M. D. (1986). Cadmium-binding proteins in Pseudomonas putida: pseudothioneins. Environ. Health Perspect. 65, 5–11.
Hlihor, R. M., Figueiredo, H., Tavares, T., Gavrilescu, M. (2017). Biosorption potential of dead and living Arthrobacter viscosus biomass in the removal of Cr (VI): Batch and column studies. Process Saf. Environ. Prot 108, 44–56. doi: 10.1016/j.psep.2016.06.016
Hockmann, K., Lenz, M., Tandy, S., Nachtegaal, M., Janousch, M., Schulin, R. (2014). Release of antimony from contaminated soil induced by redox changes. J. hazardous materials 275, 215–221.
Hogsden, K. L., Harding, J. S. (2012). Consequences of acid mine drainage for the structure and function of benthic stream communities: a review. Freshw. Sci. 31, 108–120. doi: 10.1899/11-091.1
Hu, N., Luo, Y., Song, J., Wu, L., Zhang, H. (2010). Influences of soil organic matter, pH and temperature on Pb sorption by four soils in Yangtze River Delta. Acta Pedol Sin. 47, 246–252.
Huang, K., Chen, C., Shen, Q., Rosen, B. P., Zhao, F.-J. (2015). Genetically engineering Bacillus subtilis with a heat-resistant arsenite methyltransferase for bioremediation of arsenic-contaminated organic waste. Appl. Environ. Microbiol. 81, 6718–6724. doi: 10.1128/AEM.01535-15
Huang, F., Dang, Z., Guo, C.-L., Lu, G.-N., Gu, R. R., Liu, H.-J., et al. (2013). Biosorption of Cd (II) by live and dead cells of Bacillus cereus RC-1 isolated from cadmium-contaminated soil. Colloids Surf B Biointerfaces 107, 11–18. doi: 10.1016/j.colsurfb.2013.01.062
Huang, H., Fan, L., Zhao, Y., Jin, Q., Yang, G., Zhao, D., et al. (2022). Integrating broussonetia papyrifera and two bacillus species to repair soil antimony pollutions. Front. Microbiol. 13, 871581. doi: 10.3389/fmicb.2022.871581
Huang, H., Jia, Q., Jing, W., Dahms, H.-U., Wang, L. (2020). Screening strains for microbial biosorption technology of cadmium. Chemosphere 251, 126428. doi: 10.1016/j.chemosphere.2020.126428
Huang, C., Lai, C., Xu, P., Zeng, G., Huang, D., Zhang, J., et al. (2017). Lead-induced oxidative stress and antioxidant response provide insight into the tolerance of Phanerochaete chrysosporium to lead exposure. Chemosphere 187, 70–77. doi: 10.1016/j.chemosphere.2017.08.104
Ibarrolaza, A., Coppotelli, B. M., Del Panno, M. T., Donati, E. R., Morelli, I. S. (2011). Application of the knowledge-based approach to strain selection for a bioaugmentation process of phenanthrene-and Cr (VI)-contaminated soil. J. Appl. Microbiol. 111, 26–35. doi: 10.1111/jam.2011.111.issue-1
Imran, K., Seleiman, M. F., Chattha, M. U., Jalal, R. S., Mahmood, F., Hassan, F. A., et al. (2021). Enhancing antioxidant defense system of mung bean with a salicylic acid exogenous application to mitigate cadmium toxicity. Not Bot. Horti Agrobot Cluj Napoca 49, 12303–12303. doi: 10.15835/nbha49212303
Iram, S., Shabbir, R., Zafar, H., Javaid, M. (2015). Biosorption and bioaccumulation of copper and lead by heavy metal-resistant fungal isolates. Arab J. Sci. Eng. 40, 1867–1873. doi: 10.1007/s13369-015-1702-1
Isaac, C. P. J., Sivakumar, A. (2013). Removal of lead and cadmium ions from water using Annona squamosa shell: kinetic and equilibrium studies. Desalin Water Treat 51, 7700–7709. doi: 10.1080/19443994.2013.778218
Jaiswal, S., Singh, D. K., Shukla, P. (2019). Gene editing and systems biology tools for pesticide bioremediation: a review. Front. Microb. 10, 421095. doi: 10.3389/fmicb.2019.00087
Jeyasundar, P. G. S. A., Ali, A., Azeem, M., Li, Y., Guo, D., Sikdar, A., et al. (2021). Green remediation of toxic metals contaminated mining soil using bacterial consortium and Brassica juncea. Environ. pollut. 277, 116789. doi: 10.1016/j.envpol.2021.116789
Jiang, M., Cao, L., Zhang, R. (2008). Effects of Acacia (Acacia auriculaeformis A. Cunn)-associated fungi on mustard (Brassica juncea (L.) Coss. var. foliosa Bailey) growth in Cd-and Ni-contaminated soils. Lett. Appl. Microbiol. 47, 561–565. doi: 10.1111/lam.2008.47.issue-6
Joutey, N. T., Sayel, H., Bahafid, W., El Ghachtouli, N. (2015). Mechanisms of hexavalent chromium resistance and removal by microorganisms. Rev. Environ. Contam Toxicol. 233, 45–69.
Joner, E. J., Leyval, C. (1997). Uptake of 109Cd by roots and hyphae of a glomus mosseae/trifolium subterraneum mycorrhiza from soil amended with high and low concentrations of cadmium. New Phytol. 135, 353–360. doi: 10.1046/j.1469-8137.1997.00633.x
Kaksonen, A. H., Aino-Maija, L., Olli, H. T. (2020). Acid and ferric sulfate bioleaching of uranium ores: A review. J. Cleaner Prod 264, 121586. doi: 10.1016/j.jclepro.2020.121586
Kalita, D., Joshi, S. (2017). Study on bioremediation of Lead by exopolysaccharide producing metallophilic bacterium isolated from extreme habitat. Biotechnol. Rep. 16, 48–57. doi: 10.1016/j.btre.2017.11.003
Kang, J. W. (2014). Removing environmental organic pollutants with bioremediation and phytoremediation. Biotechnol. Lett. 36, 1129–1139. doi: 10.1007/s10529-014-1466-9
Kang, C.-H., Kwon, Y.-J., So, J.-S. (2016). Bioremediation of heavy metals by using bacterial mixtures. Ecol. Eng. 89, 64–69. doi: 10.1016/j.ecoleng.2016.01.023
Kang, X., Yu, X., Zhang, Y., Cui, Y., Tu, W., Wang, Q., et al. (2018). Inoculation of Sinorhizobium saheli YH1 leads to reduced metal uptake for Leucaena leucocephala grown in mine tailings and metal-polluted soils. Front. Microbiol. 9, 1853. doi: 10.3389/fmicb.2018.01853
Ke, T., Guo, G., Liu, J., Zhang, C., Tao, Y., Wang, P., et al. (2021). Improvement of the Cu and Cd phytostabilization efficiency of perennial ryegrass through the inoculation of three metal-resistant PGPR strains. Environ. pollut. 271, 116314. doi: 10.1016/j.envpol.2020.116314
Khalid, S., Shahid, M., Niazi, N. K., Murtaza, B., Bibi, I., Dumat, C. (2017). A comparison of technologies for remediation of heavy metal contaminated soils. J. Geochem Explor. 182, 247–268. doi: 10.1016/j.gexplo.2016.11.021
Khan, I., Aftab, M., Shakir, S., Ali, M., Qayyum, S., Rehman, M. U., et al. (2019). Mycoremediation of heavy metal (Cd and Cr)–polluted soil through indigenous metallotolerant fungal isolates. Environ. Monit Assess. 191, 1–11. doi: 10.1007/s10661-019-7769-5
Khati, P., Sharma, A., GAngola, S., Kumar, R., Bhatt, P., Kumar, G. (2017). Impact of agri-usable nanocompounds on soil microbial activity: an indicator of soil health. CLEAN–Soil Air Water 45, 1600458. doi: 10.1002/clen.201600458
Kicińska, A., Pomykała, R., Izquierdo-Diaz, M. (2022). Changes in soil pH and mobility of heavy metals in contaminated soils. Eur. J. Soil Sci. 73, e13203.
Kim, J.-J., Kim, Y.-S., Kumar, V. (2019). Heavy metal toxicity: An update of chelating therapeutic strategies. J. Trace Elem Med. Biol. 54, 226–231. doi: 10.1016/j.jtemb.2019.05.003
Kothe, E., Dimkpa, C., Haferburg, G., Schmidt, A., Schmidt, A., Schütze, E. (2010). Streptomycete heavy metal resistance: extracellular and intracellular mechanisms. Soil heavy metals 225-235. doi: 10.1007/978-3-642-02436-8
Kubrak, O. I., Lushchak, O. V., Lushchak, J. V., Torous, I. M., Storey, J. M., Storey, K. B., et al. (2010). Chromium effects on free radical processes in goldfish tissues: Comparison of Cr (III) and Cr (VI) exposures on oxidative stress markers, glutathione status and antioxidant enzymes. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 152, 360–370. doi: 10.1016/j.cbpc.2010.06.003
Kulp, T. R., Miller, L. G., Braiotta, F., Webb, S. M., Kocar, B. D., Blum, J. S., et al. (2014). Microbiological reduction of Sb(V) in anoxic freshwater sediments. Environ. Sci. Technol. 48, 218–226. doi: 10.1021/es403312j
Kumar, R., Singh, P., Dhir, B., Sharma, A. K., Mehta, D. (2014). Potential of some fungal and bacterial species in bioremediation of heavy metals. J. Nucl. Physics Material Sciences Radiat. Appl. 1 (2), 213–223.
Kushwaha, A., Rani, R., Kumar, S., Thomas, T., David, A. A., Ahmed, M. (2017). A new insight to adsorption and accumulation of high lead concentration by exopolymer and whole cells of lead-resistant bacterium Acinetobacter junii L. Pb1 isolated coal mine dump. Environ. Sci. pollut. Res. 24, 10652–10661. doi: 10.1007/s11356-017-8752-8
Kwak, H. W., Kim, M. K., Lee, J. Y., Yun, H., Kim, M. H., Park, Y. H., et al. (2015). Preparation of bead-type biosorbent from water-soluble Spirulina platensis extracts for chromium (VI) removal. Algal Res. 7, 92–99. doi: 10.1016/j.algal.2014.12.006
Lai, C.-Y., Dong, Q.-Y., Rittmann, B. E., Zhao, H.-P. (2018). Bioreduction of antimonate by anaerobic methane oxidation in a membrane biofilm batch reactor. Environ. Sci. Technol. 52, 8693–8700. doi: 10.1021/acs.est.8b02035
Lai, C.-Y., Wen, L.-L., Zhang, Y., Luo, S.-S., Wang, Q.-Y., Luo, Y.-H., et al. (2016). Autotrophic antimonate bio-reduction using hydrogen as the electron donor. Water Res. 88, 467–474. doi: 10.1016/j.watres.2015.10.042
Latif, U., Farid, M., Rizwan, M., Ishaq, H. K., Farid, S., Ali, S., et al. (2020). Physiological and biochemical response of Alternanthera bettzickiana (Regel) G. Nicholson under acetic acid assisted phytoextraction of lead. Plants 9, 1084.
Leal, M. F. C., Catarino, R. I., Pimenta, A. M., Souto, M. R. S. (2023). The influence of the biometals Cu, Fe, and Zn and the toxic metals Cd and Pb on human health and disease. Trace Elem Electrolytes 40, 1. doi: 10.5414/TE500038
Lee, H. J., Lee, S. J. (2021). Advances in accurate microbial genome-editing CRISPR technologies. J. Microbiol. Biotech. 31, 903. doi: 10.4014/jmb.2106.06056
Lerebours, A., To, V. V., Bourdineaud, J. P. (2016). Danio rerio ABC transporter genes abcb3 and abcb7 play a protecting role against metal contamination. J. Appl. Toxicol. 36, 1551–1557. doi: 10.1002/jat.3313
Leudo, A. M., Cruz, Y., Montoya-Ruiz, C., MdP, D., Saldarriaga, J. F. (2020). Mercury phytoremediation with Lolium perenne-Mycorrhizae in contaminated soils. Sustainability 12, 3795. doi: 10.3390/su12093795
Li, X., Peng, W., Jia, Y., Lu, L., Fan, W. (2016). Bioremediation of lead contaminated soil with Rhodobacter sphaeroides. Chemosphere 156, 228–235. doi: 10.1016/j.chemosphere.2016.04.098
Li, C., Quan, Q., Gan, Y., Dong, J., Fang, J., Wang, L., et al. (2020). Effects of heavy metals on microbial communities in sediments and establishment of bioindicators based on microbial taxa and function for environmental monitoring and management. Sci. Total Environ. 749, 141555. doi: 10.1016/j.scitotenv.2020.141555
Li, Y., Rahman, S. U., Qiu, Z., Shahzad, S. M., Nawaz, M. F., Huang, J., et al. (2023). Toxic effects of cadmium on the physiological and biochemical attributes of plants, and phytoremediation strategies: A review. Environ. pollut. 325, 121433. doi: 10.1016/j.envpol.2023.121433
Li, J., Shi, Z., Liu, M., Wang, G., Liu, F., Wang, Y. (2021). Identifying anthropogenic sources of groundwater contamination by natural background levels and stable isotope application in Pinggu basin, China. J. Hydrol 596, 126092. doi: 10.1016/j.jhydrol.2021.126092
Li, D., Xu, X., Yu, H., Han, X. (2017a). Characterization of Pb2+ biosorption by psychrotrophic strain Pseudomonas sp. I3 isolated from permafrost soil of Mohe wetland in Northeast China. J. Environ. Manag 196, 8–15. doi: 10.1016/j.jenvman.2017.02.076
Li, X., Yin, X., Lian, B. (2017b). The degradation of dimethoate and the mineral immobilizing function for Cd2+ by Pseudomonas putida. Geomicrobiol J. 34, 346–354. doi: 10.1080/01490451.2016.1193571
Li, W.-W., Yu, H.-Q. (2015). Stimulating sediment bioremediation with benthic microbial fuel cells. Biotechnol. Adv. 33, 1–12. doi: 10.1016/j.biotechadv.2014.12.011
Lima, A. I. G., Corticeiro, S. C., Figueira, E. (2006). Glutathione-mediated cadmium sequestration in Rhizobium leguminosarum. Enzyme Microb. Technol. 39, 763–769. doi: 10.1016/j.enzmictec.2005.12.009
Lin, W., Huang, Z., Li, X., Liu, M., Cheng, Y. (2016). Bio-remediation of acephate–Pb (II) compound contaminants by Bacillus subtilis FZUL-33. J. Environ. Sci. 45, 94–99. doi: 10.1016/j.jes.2015.12.010
Liu, C. J., Deng, S. G., Hu, C. Y., Gao, P., Khan, E., Yu, C. P., et al. (2023). Applications of bioremediation and phytoremediation in contaminated soils and waters: CREST publications during 2018–2022. Crit. Rev. Environ. Sci. Technol. 53, 723–732. doi: 10.1080/10643389.2023.2168365
Liu, W., Sun, J., Ding, L., Luo, Y., Chen, M., Tang, C. (2013). Rhizobacteria (Pseudomonas sp. SB) assist phytoremediation of oily-sludge-contaminated soil by tall fescue (Testuca arundinacea L.). Plant Soil 371, 533–542. doi: 10.1007/s11104-013-1717-x
Liu, S.-H., Zeng, G.-M., Niu, Q.-Y., Liu, Y., Zhou, L., Jiang, L.-H., et al. (2017). Bioremediation mechanisms of combined pollution of PAHs and heavy metals by bacteria and fungi: A mini review. Bioresour Technol. 224, 25–33. doi: 10.1016/j.biortech.2016.11.095
Liu, S., Zhang, F., Chen, J., Sun, G. (2011). Arsenic removal from contaminated soil via biovolatilization by genetically engineered bacteria under laboratory conditions. J. Environ. Sci. 23, 1544–1550. doi: 10.1016/S1001-0742(10)60570-0
Lytras, G., Lytras, C., Argyropoulou, D., Dimopoulos, N., Malavetas, G., Lyberatos, G. (2017). A novel two-phase bioreactor for microbial hexavalent chromium removal from wastewater. J. Hazard Mater 336, 41–51. doi: 10.1016/j.jhazmat.2017.04.049
Ma, Y., Oliveira, R. S., Nai, F., Rajkumar, M., Luo, Y., Rocha, I., et al. (2015). The hyperaccumulator Sedum plumbizincicola harbors metal-resistant endophytic bacteria that improve its phytoextraction capacity in multi-metal contaminated soil. J. Environ. Managet 156, 62–69. doi: 10.1016/j.jenvman.2015.03.024
Ma, H., Wei, M., Wang, Z., Hou, S., Li, X., Xu, H. (2020). Bioremediation of cadmium polluted soil using a novel cadmium immobilizing plant growth promotion strain Bacillus sp. TZ5 loaded on biochar. J. Hazard Mater 388, 122065. doi: 10.1016/j.jhazmat.2020.122065
Magnin, J.-P., Gondrexon, N., Willison, J. C. (2014). Zinc biosorption by the purple non-sulfur bacterium Rhodobacter capsulatus. Canad J. Microbiol. 60, 829–837. doi: 10.1139/cjm-2014-0231
Malekzadeh, E., Alikhani, H., Savaghebi-Firoozabadi, G., Zarei, M. (2012). Bioremediation of cadmium-contaminated soil through cultivation of maize inoculated with plant growth–promoting rhizobacteria. Bioremediation J. 16, 204–211. doi: 10.1080/10889868.2012.703258
Malik, A. (2004). Metal bioremediation through growing cells. Environ. Int. 30, 261–278. doi: 10.1016/j.envint.2003.08.001
Malla, M. A., Dubey, A., Yadav, S., Kumar, A., Hashem, A. (2018). Understanding and designing the strategies for the microbe-mediated remediation of environmental contaminants using omics approaches. Front. Microbiol. 9, 1132. doi: 10.3389/fmicb.2018.01132
Manoj, S. R., Karthik, C., Kadirvelu, K., Arulselvi, P. I., Shanmugasundaram, T., Bruno, B., et al. (2020). Understanding the molecular mechanisms for the enhanced phytoremediation of heavy metals through plant growth promoting rhizobacteria: A review. J. Environ. Manag 254, 109779. doi: 10.1016/j.jenvman.2019.109779
Manorma, K., Sharma, S., Sharma, A., Chauhan, P. K. (2023). Potential of microbes for the remediation of heavy metal–contaminated soil. In Integr. Strategies Bioremediation Environ. Contaminants 2, 113–138.
Mariano, C., Mello, I. S., Barros, B. M., da Silva, G. F., Terezo, A. J., Soares, M. A. (2020). Mercury alters the rhizobacterial community in Brazilian wetlands and it can be bioremediated by the plant-bacteria association. Environ. Sci. pollut. Res. 27, 13550–13564. doi: 10.1007/s11356-020-07913-2
Marschner, B., Kalbitz, K. (2003). Controls of bioavailability and biodegradability of dissolved organic matter in soils. Geoderma 113, 211–235. doi: 10.1016/S0016-7061(02)00362-2
Martins, J. P. R., LLd, V., PdCdS, B., FP, R., LT, C., LCdA, R., et al. (2020). Morphophysiological responses, bioaccumulation and tolerance of Alternanthera tenella Colla (Amaranthaceae) to excess copper under in vitro conditions. Plant Cell Tissue Organ Cult 143, 303–318. doi: 10.1007/s11240-020-01917-z
Martis, S., Mohan, A. K., Chiplunkar, S., Kamath, S., Goveas, L. C., Rao, C. V. (2021). Bacterium isolated from coffee waste pulp biosorps lead: Investigation of EPS mediated mechanism. Curr. Res. Microb. Sci. 2, 100029. doi: 10.1016/j.crmicr.2021.100029
Mathew, D. C., Ho, Y.-N., Gicana, R. G., Mathew, G. M., Chien, M.-C., Huang, C.-C. (2015). A rhizosphere-associated symbiont, Photobacterium spp. strain MELD1, and its targeted synergistic activity for phytoprotection against mercury. PloS One 10, e0121178.
Mathur, S., Kalaji, H., Jajoo, A. (2016). Investigation of deleterious effects of chromium phytotoxicity and photosynthesis in wheat plant. Photosynthetica 54, 185–192. doi: 10.1007/s11099-016-0198-6
Meng, L., Wu, Y., Mu, M., Wang, Z., Chen, Z., Wang, L., et al. (2023). Effects of different concentrations of biochar amendments and Pb toxicity on rhizosphere soil characteristics and bacterial community of red clover (Trifolium pretense L.). Front. Plant Sci. 14, 1112002. doi: 10.3389/fpls.2023.1112002
Mestrot, A., Feldmann, J., Krupp, E. M., Hossain, M. S., Roman-Ross, G., Meharg, A. A. (2011). Field fluxes and speciation of arsines emanating from soils. Environ. Sci. Technol. 45, 1798–1804. doi: 10.1021/es103463d
Miglani, G. S. (2017). Genome editing in crop improvement: Present scenario and future prospects. J. Crop Improv 31, 453–559. doi: 10.1080/15427528.2017.1333192
Mishra, D. K., Awasthi, H., Srivastava, D., Fatima, Z. (2022). Phytochemical: A treatment option for heavy metal induced neurotoxicity. J. Complementary Integr. Med. 19, 513–530. doi: 10.1515/jcim-2020-0325
Morton-Bermea, O., Hernandez Alvarez, E., Gaso, I., Segovia, N. (2002). Heavy metal concentrations in surface soils from Mexico City. Bull. Environ. Contam. Toxicol. 68, 383–388. doi: 10.1007/s00128-001-0265-x
Murthy, M. K., Khandayataray, P., Padhiary, S., Samal, D. (2022). A review on chromium health hazards and molecular mechanism of chromium bioremediation. Rev Environ Health. 38 (3), 461–478. doi: 10.1515/reveh-2021-0139
Muzaffar, S., Khan, J., Srivastava, R., Gorbatyuk, M. S., Athar, M. (2023). Mechanistic understanding of the toxic effects of arsenic and warfare arsenicals on human health and environment. Cell Biol. Toxicol. 39, 85–110. doi: 10.1007/s10565-022-09710-8
Nagvenkar, G. S., Ramaiah, N. (2010). Arsenite tolerance and biotransformation potential in estuarine bacteria. Ecotoxicol 19, 604–613. doi: 10.1007/s10646-009-0429-8
Narayanan, M., Ma, Y. (2023). Mitigation of heavy metal stress in the soil through optimized interaction between plants and microbes. J. Environ. Managet 345, 118732. doi: 10.1016/j.jenvman.2023.118732
Naskar, A., Majumder, R., Goswami, M. (2020). Bioaccumulation of Ni (II) on growing cells of Bacillus sp.: response surface modeling and mechanistic insight. Environ. Technol. Innov. 20, 101057.
Nayak, A., Raja, R., Rao, K., Shukla, A., Mohanty, S., Shahid, M., et al. (2015). Effect of fly ash application on soil microbial response and heavy metal accumulation in soil and rice plant. Ecotoxicol Environ. Saf. 114, 257–262. doi: 10.1016/j.ecoenv.2014.03.033
Neelima, P., Reddy, K. J. (2003). Differential effect of cadmium and mercury on growth and metabolism of Solanum melongena L. seedlings. J. Environ. Biol. 24, 453–460.
Nikolaou, K.-E., Chatzistathis, T., Theocharis, S., Argiriou, A., Koundouras, S., Zioziou, E. (2022). Effects of chromium toxicity on physiological performance and nutrient uptake in two grapevine cultivars (Vitis vinifera L.) growing on own roots or grafted onto different rootstocks. Hortic 8, 493.
Nizamutdinov, S. A. R., EN, M., NV, D., Abakumov, E. V. (2022). Ecotoxicological analysis of fallow soils at the yamal experimental agricultural station. Food Process Tech Technol. 52 (2), 2022. doi: 10.21603/2074-9414-2022-2
Nurzhan, A., Tian, H., Nuralykyzy, B., He, W. (2022). Soil enzyme activities and enzyme activity indices in long-term arsenic-contaminated soils. Eurasian Soil Sci. 55, 1425–1435. doi: 10.1134/S106422932210012X
Nwuche, C., Ugoji, E. O. (2008). Effects of heavy metal pollution on the soil microbial activity. Int. J. Environ. Sci. Technol. 5, 409–414. doi: 10.1007/BF03326036
Nziguheba, G., Smolders, E. (2008). Inputs of trace elements in agricultural soils via phosphate fertilizers in European countries. Sci. Total Environ. 390, 53–57. doi: 10.1016/j.scitotenv.2007.09.031
Obani, I. S., Anyachor, C. P., Okereke, I. (2023). Preclinical and epidemiological mucoskeletal evidence of aluminium toxicity: A systematic review. IPS J. Public Health 3, 10–24.
Oyewole, O. A., Zobeashia, SSL-T, Oladoja, E. O., Raji, R. O., Odiniya, E. E., Musa, A. M. (2019). Biosorption of heavy metal polluted soil using bacteria and fungi isolated from soil. SN Appl. Sci. 1, 1–8.
Pan, J., Yu, L. (2011). Effects of Cd or/and Pb on soil enzyme activities and microbial community structure. Ecol. Eng. 37, 1889–1894. doi: 10.1016/j.ecoleng.2011.07.002
Pattanapipitpaisal, P., Brown, N. L., Macaskie, L. E. (2001). Chromate reduction and 16S rRNA identification of bacteria isolated from a Cr (VI)-contaminated site. Appl. Microbiol. Biotechnol. 57, 257–261.
Paul, A., Mukherjee, S. K. (2016). KUNi5, a nickel resistant bacterium for possible bioremediation of nickel contaminated sites. Polish J. Microbiol. 65, 115–118. doi: 10.5604/17331331.1197284
Paunov, M., Koleva, L., Vassilev, A., Vangronsveld, J., Goltsev, V. (2018). Effects of different metals on photosynthesis: cadmium and zinc affect chlorophyll fluorescence in durum wheat. Int. J. Mol. Sci. 19, 787. doi: 10.3390/ijms19030787
Peña-Castro, J. M., Muñoz-Páez, K. M., Robledo-Narvaez, P. N., Vázquez-Núñez, E. (2023). Engineering the metabolic landscape of microorganisms for lignocellulosic conversion. Microorganisms 11, 2197. doi: 10.3390/microorganisms11092197
Pervaiz, I., Ahmad, S., Madni, M., Ahmad, H., Khaliq, F. (2013). Microbial biotransformation: a tool for drug designing. Appl. Biochem. Microbiol. 49, 437–450. doi: 10.1134/S0003683813050098
Philippot, L., Cregut, M., Chèneby, D., Bressan, M., Dequiet, S., Martin-Laurent, F., et al. (2008). Effect of primary mild stresses on resilience and resistance of the nitrate reducer community to a subsequent severe stress. FEMS Microbiol. Lett. 285, 51–57. doi: 10.1111/fml.2008.285.issue-1
Pietro-Souza, W., de Campos Pereira, F., Mello, I. S., Stachack, F. F. F., Terezo, A. J., da Cunha, C. N., et al. (2020). Mercury resistance and bioremediation mediated by endophytic fungi. Chemosphere 240, 124874. doi: 10.1016/j.chemosphere.2019.124874
Pinter, I. F., Salomon, M. V., Berli, F., Bottini, R., Piccoli, P. (2017). Characterization of the As (III) tolerance conferred by plant growth promoting rhizobacteria to in vitro-grown grapevine. Appl. Soil Ecol. 109, 60–68.
Piršelová, B., Ondrušková, E. (2021). Effect of cadmium chloride and cadmium nitrate on growth and mineral nutrient content in the root of fava bean (Vicia faba L.). Plants 10, 1007. doi: 10.3390/plants10051007
Poo, K.-M., Son, E.-B., Chang, J.-S., Ren, X., Choi, Y.-J., Chae, K.-J. (2018). Biochars derived from wasted marine macro-algae (Saccharina japonica and Sargassum fusiforme) and their potential for heavy metal removal in aqueous solution. J. Environ. Manag 206, 364–372. doi: 10.1016/j.jenvman.2017.10.056
Povedano-Priego, C., Martín-Sánchez, I., Jroundi, F., Sánchez-Castro, I., Merroun, M. L. (2017). Fungal biomineralization of lead phosphates on the surface of lead metal. Miner Eng. 106, 46–54. doi: 10.1016/j.mineng.2016.11.007
Preetha, J. S. Y., Arun, M., Vidya, N., Kowsalya, K., Halka, J., Ondrasek, G. (2023). Biotechnology advances in bioremediation of arsenic: A review. Molecules 28, 1474. doi: 10.3390/molecules28031474
Purchase, D., Scholes, L. N., Revitt, D. M., Shutes, R. B. E. (2009). Effects of temperature on metal tolerance and the accumulation of Zn and Pb by metal-tolerant fungi isolated from urban runoff treatment wetlands. J. Appl. Microbiol. 106, 1163–1174. doi: 10.1111/jam.2009.106.issue-4
Puyen, Z. M., Villagrasa, E., Maldonado, J., Diestra, E., Esteve, I., Solé, A. (2012). Biosorption of lead and copper by heavy-metal tolerant Micrococcus luteus DE2008. Bioresour Technol. 126, 233–237. doi: 10.1016/j.biortech.2012.09.036
Qian, X., Fang, C., Huang, M., Achal, V. (2017). Characterization of fungal-mediated carbonate precipitation in the biomineralization of chromate and lead from an aqueous solution and soil. J. Clean Prod 164, 198–208. doi: 10.1016/j.jclepro.2017.06.195
Qin, J., Jiang, X., Qin, J., Zhao, H., Dai, M., Liu, H., et al. (2023). Effects of lead pollution on photosynthetic characteristics and chlorophyll fluorescence parameters of different populations of Miscanthus floridulus. Processes 11, 1562. doi: 10.3390/pr11051562
Quiroga-Mateus, R., López-Zuleta, S., Chávez, E., Bravo, D. (2022). Cadmium-tolerant bacteria in cacao farms from antioquia, Colombia: isolation, characterization and potential use to mitigate cadmium contamination. Processes 10, 1457. doi: 10.3390/pr10081457
Rahman, Z., Singh, V. P. (2019). The relative impact of toxic heavy metals (THMs)(arsenic (As), cadmium (Cd), chromium (Cr)(VI), mercury (Hg), and lead (Pb)) on the total environment: an overview. Environ. Monit Assess. 191, 1–21. doi: 10.1007/s10661-019-7528-7
Rahman, Z., Singh, V. P. (2020). Bioremediation of toxic heavy metals (THMs) contaminated sites: concepts, applications and challenges. Environ. Sci. pollut. Res. 27, 27563–27581. doi: 10.1007/s11356-020-08903-0
Rajkumar, M., Ae, N., Prasad, M. N. V., Freitas, H. (2010). Potential of siderophore-producing bacteria for improving heavy metal phytoextraction. Trends Biotechnol. 28, 142–149. doi: 10.1016/j.tibtech.2009.12.002
Rajput, V., Minkina, T., Fedorenko, A., Sushkova, S., Mandzhieva, S., Lysenko, V., et al. (2018). Toxicity of copper oxide nanoparticles on spring barley (Hordeum sativum distichum). Sci. Total Environ. 645, 1103–1113. doi: 10.1016/j.scitotenv.2018.07.211
Rajput, V. D., Minkina, T., Upadhyay, S. K., Kumari, A., Ranjan, A., Mandzhieva, S., et al. (2022). Nanotechnology in the restoration of polluted soil. Nanomaterials 12, 769. doi: 10.3390/nano12050769
Raklami, A., A-i, T., Bechtaoui, N., Pajuelo, E., Baslam, M., Meddich, A., et al. (2021). Restoring the plant productivity of heavy metal-contaminated soil using phosphate sludge, marble waste, and beneficial microorganisms. J. Environ. Sci. 99, 210–221. doi: 10.1016/j.jes.2020.06.032
Ramesh, A., Winkler, W. C. (2010). Magnesium-sensing riboswitches in bacteria. RNA Biol. 7, 77–83. doi: 10.4161/rna.7.1.10490
Ramrakhiani, L., Majumder, R., Khowala, S. (2011). Removal of hexavalent chromium by heat inactivated fungal biomass of Termitomyces clypeatus: Surface characterization and mechanism of biosorption. Chem. Eng. J. 171, 1060–1068. doi: 10.1016/j.cej.2011.05.002
Rani, A., Goel, R. (2009). Strategies for crop improvement in contaminated soils using metal-tolerant bioinoculants. Microbial strategies Crop improvement, 85–104.
Raza, A. (2022). Metabolomics: a systems biology approach for enhancing heat stress tolerance in plants. Plant Cell Rep. 41, 741–763. doi: 10.1007/s00299-020-02635-8
Raza, A., Salehi, H., Bashir, S., Tabassum, J., Jamla, M., Charagh, S., et al. (2024). Transcriptomics, proteomics, and metabolomics interventions prompt crop improvement against metal (loid) toxicity. Plant Cell Rep. 43 (3), 80.
Remenar, M., Kamlarova, A., Harichova, J., Zámocký, M., Ferianc, P. (2018). The heavy-metal resistance determinant of newly isolated bacterium from a nickel-contaminated soil in Southwest Slovakia. Pol. J. Microbiol. 67, 191–201. doi: 10.21307/pjm-2018-022
Ren, G., Jin, Y., Zhang, C., Gu, H., Qu, J. (2015). Characteristics of bacillus sp. PZ-1 its biosorption to Pb (II). Ecotoxicol Environ. Saf. 117, 141–148. doi: 10.1016/j.ecoenv.2015.03.033
Rezaei, H. (2016). Biosorption of chromium by using Spirulina sp. Arab J. Chem. 9, 846–853. doi: 10.1016/j.arabjc.2013.11.008
Rhee, Y. J., Hillier, S., Gadd, G. M. (2012). Lead transformation to pyromorphite by fungi. Curr. Biol. 22, 237–241. doi: 10.1016/j.cub.2011.12.017
Rodríguez-Tirado, V., Green-Ruiz, C., Gómez-Gil, B. (2012). Cu and Pb biosorption on Bacillus thioparans strain U3 in aqueous solution: Kinetic and equilibrium studies. Chem. Eng. J. 181, 352–359. doi: 10.1016/j.cej.2011.11.091
Rucińska-Sobkowiak, R. (2016). Water relations in plants subjected to heavy metal stresses. Acta Physiol. Plant 38, 1–13. doi: 10.1007/s11738-016-2277-5
Rugnini, L., Costa, G., Congestri, R., Antonaroli, S., Di Toppi, L. S., Bruno, L. (2018). Phosphorus and metal removal combined with lipid production by the green microalga Desmodesmus sp.: An integrated approach. Plant Physiol. Biochem. 125, 45–51. doi: 10.1016/j.plaphy.2018.01.032
Sati, D., Pande, V., Pandey, S. C., Samant, M. (2023). Recent advances in PGPR and molecular mechanisms involved in drought stress resistance. J. Soil Sci. Plant Nutr. 23, 106–124. doi: 10.1007/s42729-021-00724-5
Sayyed, R., Seifi, S., Patel, P., Shaikh, S., Jadhav, H., Enshasy, H. E. (2019). Siderophore production in groundnut rhizosphere isolate, Achromobacter sp RZS2 influenced by physicochemical factors and metal ions. Environ. Sustain 2, 117–124. doi: 10.1007/s42398-019-00070-4
Schück, M., Greger, M. (2023). Salinity and temperature influence removal levels of heavy metals and chloride from water by wetland plants. Environ. Sci. Poll Res. 30, 58030–58040. doi: 10.1007/s11356-023-26490-8
Shahpiri, A., Mohammadzadeh, A. (2018). Mercury removal by engineered Escherichia coli cells expressing different rice metallothionein isoforms. Ann. Microbiol. 68, 145–152. doi: 10.1007/s13213-018-1326-2
Shahzad, B., Tanveer, M., Rehman, A., Cheema, S. A., Fahad, S., Rehman, S., et al. (2018). Nickel; whether toxic or essential for plants and environment-A review. Plant Physiol. Biochem. 132, 641–651. doi: 10.1016/j.plaphy.2018.10.014
Shanab, S., Essa, A., Shalaby, E. (2012). Bioremoval capacity of three heavy metals by some microalgae species (Egyptian Isolates). Plant Signal Behav. 7, 392–399. doi: 10.4161/psb.19173
Shanker, A. K., Cervantes, C., Loza-Tavera, H., Avudainayagam, S. (2005). Chromium toxicity in plants. Environ. Int. 31, 739–753. doi: 10.1016/j.envint.2005.02.003
Sharma, A., Kumar, V., Shahzad, B., Ramakrishnan, M., Singh Sidhu, G. P., Bali, A. S., et al. (2020). Photosynthetic response of plants under different abiotic stresses: a review. J. Plant Growth Regul. 39, 509–531. doi: 10.1007/s00344-019-10018-x
Sharma, B., Sarkar, A., Singh, P., Singh, R. P. (2017). Agricultural utilization of biosolids: A review on potential effects on soil and plant grown. Waste Manag 64, 117–132. doi: 10.1016/j.wasman.2017.03.002
Shazia, I., Uzma, S. G., Talat, A. (2013). Bioremediation of heavy metals using isolates of filamentous fungus Aspergillus fumigatus collected from polluted soil of Kasur, Pakistan. Int. Res. J. Biol. Sci. 2, 66–73.
Sher, S., Rehman, A. (2019). Use of heavy metals resistant bacteria—a strategy for arsenic bioremediation. Appl. Microbiol. Biotechnol. 103, 6007–6021. doi: 10.1007/s00253-019-09933-6
Singh, R., Behera, M., Kumar, S. (2020). “Nano-bioremediation: An innovative remediation technology for treatment and management of contaminated sites,” in Bioremediation of industrial waste for environmental safety: volume II: biological agents and methods for industrial waste manag, 165–182.
Singh, S., Chaurasia, S., Pandey, R. K. (2023). Suicidal acute iron poisoning in adolescent females–a case series. J. Popul Ther. Clin. Pharmacol. 30, 504–510.
Singh, P., Itankar, N., Patil, Y. (2021). Biomanagement of hexavalent chromium: Current trends and promising perspectives. J. Environ. Managet 279, 111547. doi: 10.1016/j.jenvman.2020.111547
Singh, J., Kalamdhad, A. S. (2011). Effects of heavy metals on soil, plants, human health and aquatic life. Intern. J. Res. Chem. Environ. 2, 15–21.
Sofo, A., Bochicchio, R., Amato, M., Rendina, N., Vitti, A., Nuzzaci, M., et al. (2017). Plant architecture, auxin homeostasis and phenol content in Arabidopsis thaliana grown in cadmium-and zinc-enriched media. J. Plant Physiol. 216, 174–180. doi: 10.1016/j.jplph.2017.06.008
Sone, Y., Mochizuki, Y., Koizawa, K., Nakamura, R., Pan-Hou, H., Itoh, T., et al. (2013). Mercurial-resistance determinants in Pseudomonas strain K-62 plasmid pMR68. AMB Express 3, 1–7. doi: 10.1186/2191-0855-3-41
Soto, D. F., Recalde, A., Orell, A., Albers, S.-V., Paradela, A., Navarro, C. A., et al. (2019). Global effect of the lack of inorganic polyphosphate in the extremophilic archaeon Sulfolobus solfataricus: a proteomic approach. J. Proteomics 191, 143–152. doi: 10.1016/j.jprot.2018.02.024
Souri, Z., Cardoso, A. A., da-Silva, C. J., de Oliveira, L. M., Dari, B., Sihi, D., et al. (2019). “Heavy metals and photosynthesis: Recent developments,” in Photosynthesis, productivity and environmental stress, 107–134.
Spagnoletti, F. N., Balestrasse, K., Lavado, R. S., Giacometti, R. (2016). Arbuscular mycorrhiza detoxifying response against arsenic and pathogenic fungus in soybean. Ecotoxicol Environ. Saf. 133, 47–56. doi: 10.1016/j.ecoenv.2016.06.012
Srichandan, H., Pathak, A., Singh, S., Blight, K., Kim, D.-J., Lee, S. W. (2014). Sequential leaching of metals from spent refinery catalyst in bioleaching–bioleaching and bioleaching–chemical leaching reactor: comparative study. Hydrometallurgy 150, 130–143. doi: 10.1016/j.hydromet.2014.09.019
Srivastav, A., Yadav, K. K., Yadav, S., Gupta, N., Singh, J. K., Katiyar, R., et al. (2018). Nano-phytoremediation of pollutants from contaminated soil environment: current scenario and future prospects. Phytoremediation: Manage. Environ. Contaminants Volume 6, 383–401. doi: 10.1007/978-3-319-99651-6
Sun, L., Cao, X., Li, M., Zhang, X., Li, X., Cui, Z. (2017). Enhanced bioremediation of lead-contaminated soil by Solanum nigrum L. with Mucor circinelloides. Environ. Sci. pollut. Res. 24, 9681–9689. doi: 10.1007/s11356-017-8637-x
Sun, S.-C., Chen, J.-X., Wang, Y.-G., Leng, F.-F., Zhao, J., Chen, K., et al. (2021). Molecular mechanisms of heavy metals resistance of Stenotrophomonas rhizophila JC1 by whole genome sequencing. Arch. Microbiol. 203, 2699–2709. doi: 10.1007/s00203-021-02271-0
Talukdar, D., Jasrotia, T., Sharma, R., Jaglan, S., Kumar, R., Vats, R., et al. (2020). Evaluation of novel indigenous fungal consortium for enhanced bioremediation of heavy metals from contaminated sites. Environ. Technol. Innov. 20, 101050. doi: 10.1016/j.eti.2020.101050
Tan, W., Peralta-Videa, J. R., Gardea-Torresdey, J. L. (2018). Interaction of titanium dioxide nanoparticles with soil components and plants: current knowledge and future research needs–a critical review. Environ. Sci. Nano 5, 257–278. doi: 10.1039/C7EN00985B
Taran, M., Fateh, R., Rezaei, S., Gholi, M. K. (2019). Isolation of arsenic accumulating bacteria from garbage leachates for possible application in bioremediation. Iran J. Microbiol. 11, 60. doi: 10.18502/ijm.v11i1.707
Teitzel, G. M., Parsek, M. R. (2003). Heavy metal resistance of biofilm and planktonic Pseudomonas aeruginosa. Appl. Environ. Microbiol. 69, 2313–2320. doi: 10.1128/AEM.69.4.2313-2320.2003
Thatoi, H., Das, S., Mishra, J., Rath, B. P., Das, N. (2014). Bacterial chromate reductase, a potential enzyme for bioremediation of hexavalent chromium: a review. J. Environ. Manag 146, 383–399. doi: 10.1016/j.jenvman.2014.07.014
Thelwell, C., Robinson, N. J., Turner-Cavet, J. S. (1998). An SmtB-like repressor from Synechocystis PCC 6803 regulates a zinc exporter. Proc. Natl. Acad. Sci. 95, 10728–10733. doi: 10.1073/pnas.95.18.10728
Tirry, N., Kouchou, A., El Omari, B., Ferioun, M., El Ghachtouli, N. (2021). Improved chromium tolerance of Medicago sativa by plant growth-promoting rhizobacteria (PGPR). J. Genet. Eng. Biotechnol. 19, 1–14. doi: 10.1186/s43141-021-00254-8
Tripathi, D. K., Singh, V. P., Prasad, S. M., Dubey, N. K., Chauhan, D. K., Rai, A. K. (2016). LIB spectroscopic and biochemical analysis to characterize lead toxicity alleviative nature of silicon in wheat (Triticum aestivum L.) seedlings. J. Photochem. Photobiol. B: Biol. 154, 89–98. doi: 10.1016/j.jphotobiol.2015.11.008
Uchimiya, M., Bannon, D., Nakanishi, H., McBride, M. B., Williams, M. A., Yoshihara, T. (2020). Chemical speciation, plant uptake, and toxicity of heavy metals in agricultural soils. J. Agric. Food Chem. 68, 12856–12869. doi: 10.1021/acs.jafc.0c00183
Umrania, V. V. (2006). Bioremediation of toxic heavy metals using acidothermophilic autotrophes. Bioresour Technol. 97, 1237–1242. doi: 10.1016/j.biortech.2005.04.048
Urík, M., Čerňanský, S., Ševc, J., Šimonovičová, A., Littera, P. (2007). Biovolatilization of arsenic by different fungal strains. Water Air Soil Poll 186, 337–342. doi: 10.1007/s11270-007-9489-7
Ustiatik, R., Nuraini, Y., Suharjono, S., Jeyakumar, P., Anderson, C. W., Handayanto, E. (2022). Mercury resistance and plant growth promoting traits of endophytic bacteria isolated from mercury-contaminated soil. Bioremediation J. 26, 208–227. doi: 10.1080/10889868.2021.1973950
Velásquez, L., Dussan, J. (2009). Biosorption and bioaccumulation of heavy metals on dead and living biomass of Bacillus sphaericus. J. Hazard Mater 167, 713–716. doi: 10.1016/j.jhazmat.2009.01.044
Verasoundarapandian, G., Lim, Z. S., Radziff, S. B. M., Taufik, S. H., Puasa, N. A., Shaharuddin, N. A., et al. (2022). Remediation of pesticides by microalgae as feasible approach in agriculture: Bibliometric strategies. Agron 12, 117. doi: 10.3390/agronomy12010117
Villadangos, A. F., Ordóñez, E., Pedre, B., Messens, J., Gil, J. A., Mateos, L. M. (2014). Engineered coryneform bacteria as a bio-tool for arsenic remediation. Appl. Microbiol. Biotechnol. 98, 10143–10152. doi: 10.1007/s00253-014-6055-2
Villegas-Plazas, M., Sanabria, J., Junca, H. A. (2019). Composite taxonomical and functional framework of microbiomes under acid mine drainage bioremediation systems. J. Env. Manag 251, 109581. doi: 10.1016/j.jenvman.2019.109581
Wang, Y., Guo, J., Liu, R. (2001). Biosorption of heavy metals by bacteria isolated from activated sludge. Huan Jing Ke Xue 22, 72–75.
Wang, L., Liu, S., Li, J., Li, S. (2022). Effects of several organic fertilizers on heavy metal passivation in Cd-contaminated gray-purple soil. Front. Environ. Sci. 10, 895646. doi: 10.3389/fenvs.2022.895646
Wang, X., Ma, R., Cui, D., Cao, Q., Shan, Z., Jiao, Z. (2017). Physio-biochemical and molecular mechanism underlying the enhanced heavy metal tolerance in highland barley seedlings pre-treated with low-dose gamma irradiation. Sci. Rep. 7, 14233. doi: 10.1038/s41598-017-14601-8
Wang, Y., Zhang, L., Wang, J., Lv, J. (2020). Identifying quantitative sources and spatial distributions of potentially toxic elements in soils by using three receptor models and sequential indicator simulation. Chemosphere 242, 125266. doi: 10.1016/j.chemosphere.2019.125266
Wang, S., Zhao, X. (2009). On the potential of biological treatment for arsenic contaminated soils and groundwater. J. Environ. Manag 90, 2367–2376. doi: 10.1016/j.jenvman.2009.02.001
Wen, Z., Liu, Q., Yu, C., Huang, L., Liu, Y., Sa, Xu, et al. (2023). The difference between rhizosphere and endophytic bacteria on the safe cultivation of lettuce in cr-contaminated farmland. Toxics 11, 371. doi: 10.3390/toxics11040371
Wierzba, S. (2015). Biosorption of lead (II), zinc (II) and nickel (II) from industrial wastewater by Stenotrophomonas maltophilia and Bacillus subtilis. Pol. J. Chem. Technol. 17, 79–87. doi: 10.1515/pjct-2015-0012
Wu, C., Jiang, M., Hsieh, L., Cai, Y., Shen, Y., Wang, H., et al. (2020). Feasibility of bioleaching of heavy metals from sediment with indigenous bacteria using agricultural sulfur soil conditioners. Sci. Total Environ. 703, 134812. doi: 10.1016/j.scitotenv.2019.134812
Wu, G., Kang, H., Zhang, X., Shao, H., Chu, L., Ruan, C. (2010). A critical review on the bio-removal of hazardous heavy metals from contaminated soils: Issues, progress, eco-environmental concerns and opportunities. J. Hazard Mat 174, 1–8. doi: 10.1016/j.jhazmat.2009.09.113
Wu, Q., Leung, J. Y., Geng, X., Chen, S., Huang, X., Li, H., et al. (2015). Heavy metal contamination of soil and water in the vicinity of an abandoned e-waste recycling site: implications for dissemination of heavy metals. Sci. Total Environ. 506, 217–225. doi: 10.1016/j.scitotenv.2014.10.121
Xi, L., Shen, Y., Zhao, X., Zhou, M., Mi, Y., Li, X., et al (2022). Effects of arbuscular mycorrhizal fungi on frond antimony enrichment, morphology, and proteomics in Pteris cretica var nervosa during antimony phytoremediation. Sci. Total Environ. 804, 149904. doi: 10.1016/j.scitotenv.2021.149904
Xian, Y., Wang, M., Chen, W. (2015). Quantitative assessment on soil enzyme activities of heavy metal contaminated soils with various soil properties. Chemosphere 139, 604–608. doi: 10.1016/j.chemosphere.2014.12.060
Yaashikaa, P., Kumar, P. S. (2022). Bioremediation of hazardous pollutants from agricultural soils: A sustainable approach for Waste Manag towards urban sustainability. Environ. pollut., 120031. doi: 10.1016/j.envpol.2022.120031
Yadav, G., Srivastava, P. K., Singh, V. P., Prasad, S. M. (2014). Light intensity alters the extent of arsenic toxicity in Helianthus annuus L. seedlings. Biol. Trace Elem Res. 158, 410–421. doi: 10.1007/s12011-014-9950-6
Yan, A., Wang, Y., Tan, S. N., Mohd, Y. M. L., Ghosh, S., Chen, Z. (2020). Phytoremediation: a promising approach for revegetation of heavy metal-polluted land. Front. Plant Sci. 11, 359. doi: 10.3389/fpls.2020.00359
Yang, T., Chen, M., Wang, J. (2015). Genetic and chemical modification of cells for selective separation and analysis of heavy metals of biological or environmental significance. TrAC Trends Anal. Chemi 66, 90–102. doi: 10.1016/j.trac.2014.11.016
Yang, H.-C., Rosen, B. P. (2016). New mechanisms of bacterial arsenic resistance. BioMed. J. 39, 5–13. doi: 10.1016/j.bj.2015.08.003
Yin, K., Lv, M., Wang, Q., Wu, Y., Liao, C., Zhang, W., et al. (2016). Simultaneous bioremediation and biodetection of mercury ion through surface display of carboxylesterase E2 from Pseudomonas aeruginosa PA1. Water Res. 103, 383–390. doi: 10.1016/j.watres.2016.07.053
Yin, X., Wei, R., Chen, H., Zhu, C., Liu, Y., Wen, H., et al. (2021). Cadmium isotope constraints on heavy metal sources in a riverine system impacted by multiple anthropogenic activities. Sci. Total Environ. 750, 141233. doi: 10.1016/j.scitotenv.2020.141233
Yotsova, E., Dobrikova, A., Stefanov, M., Misheva, S., Bardáčová, M., Matušíková, I., et al. (2020). Effects of cadmium on two wheat cultivars depending on different nitrogen supply. Plant Physiol. Biochem. 155, 789–799. doi: 10.1016/j.plaphy.2020.06.042
Yuan, L., Zhi, W., Liu, Y., Karyala, S., Vikesland, P. J., Chen, X., et al. (2015). Lead toxicity to the performance, viability, and community composition of activated sludge microorganisms. Environ. Sci. Technol. 49, 824–830. doi: 10.1021/es504207c
Yue, Z.-B., Li, Q., C-c, Li, Wang, J. (2015). Component analysis and heavy metal adsorption ability of extracellular polymeric substances (EPS) from sulfate reducing bacteria. Bioresour Technol. 194, 399–402. doi: 10.1016/j.biortech.2015.07.042
Zaidi, S., Usmani, S., Singh, B. R., Musarrat, J. (2006). Significance of Bacillus subtilis strain SJ-101 as a bioinoculant for concurrent plant growth promotion and nickel accumulation in Brassica juncea. Chemosphere 64, 991–997. doi: 10.1016/j.chemosphere.2005.12.057
Zammit, C. M., Weiland, F., Brugger, J., Wade, B., Winderbaum, L. J., Nies, D. H., et al. (2016). Proteomic responses to gold (iii)-toxicity in the bacterium Cupriavidus metallidurans CH34. Metallomics 8, 1204–1216. doi: 10.1039/C6MT00142D
Zapana-Huarache, S., Romero-Sánchez, C., Gonza, A. D., Torres-Huaco, F. D., Rivera, A. L. (2020). Chromium (VI) bioremediation potential of filamentous fungi isolated from Peruvian tannery industry effluents. Braz. J. Microbiol. 51, 271–278. doi: 10.1007/s42770-019-00209-9
Zhang, W., Chen, L., Liu, D. (2012). Characterization of a marine-isolated mercury-resistant Pseudomonas putida strain SP1 and its potential application in marine mercury reduction. Appl. Microbiol. Biotechnol. 93, 1305–1314. doi: 10.1007/s00253-011-3454-5
Zheng, Y., Li, Y., Long, H., Zhao, X., Jia, K., Li, J., et al. (2018). bifA regulates biofilm development of Pseudomonas putida MnB1 as a primary response to H2O2 and Mn2+. Front. Microbiol. 9, 1490. doi: 10.3389/fmicb.2018.01490
Zhou, G., Xia, X., Wang, H., Li, L., Wang, G., Zheng, S., et al. (2016). Immobilization of lead by Alishewanella sp. WH16-1 in pot experiments of Pb-contaminated paddy soil. Water Air Soil pollut. 227, 1–11. doi: 10.1007/s11270-016-3040-7
Zhou, W., Zhang, Y., Ding, X., Liu, Y., Shen, F., Zhang, X., et al. (2012). Magnetotactic bacteria: promising biosorbents for heavy metals. Appl. Microbiol. Biotechnol. 95, 1097–1104. doi: 10.1007/s00253-012-4245-3
Zhu, X., Li, W., Zhan, L., Huang, M., Zhang, Q., Achal, V. (2016). The large-scale process of microbial carbonate precipitation for nickel remediation from an industrial soil. Environ. pollut. 219, 149–155. doi: 10.1016/j.envpol.2016.10.047
Keywords: bio-sorption, genetic engineering, heavy metals, bioremediation, nano-particles
Citation: Tang H, Xiang G, Xiao W, Yang Z and Zhao B (2024) Microbial mediated remediation of heavy metals toxicity: mechanisms and future prospects. Front. Plant Sci. 15:1420408. doi: 10.3389/fpls.2024.1420408
Received: 20 April 2024; Accepted: 28 June 2024;
Published: 19 July 2024.
Edited by:
Dinesh Yadav, Deen Dayal Upadhyay Gorakhpur University, IndiaReviewed by:
Peiman Zandi, Yibin University, ChinaFernanda Maria Policarpo Tonelli, Universidade Federal de São João del-Rei, Brazil
Copyright © 2024 Tang, Xiang, Xiao, Yang and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zeliang Yang, YmlvaW5mb0BxcS5jb20=; Baoyi Zhao, emJ5MTMzOTczODg4MDdAMTYzLmNvbQ==
 Haiying Tang
Haiying Tang Guohong Xiang1
Guohong Xiang1