
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 26 July 2024
Sec. Plant Metabolism and Chemodiversity
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1419796
This article is part of the Research Topic Biosynthesis and Metabolic Engineering of Living Stocks (Lignin) for Biofuels View all 4 articles
Forage maize is a versatile crop extensively utilized for animal nutrition in agriculture and holds promise as a valuable resource for the production of fermentable sugars in the biorefinery sector. Within this context, the carbohydrate fraction of the lignocellulosic biomass undergoes deconstruction during ruminal digestion and the saccharification process. However, the cell wall’s natural resistance towards enzymatic degradation poses a significant challenge during both processes. This so-called biomass recalcitrance is primarily attributed to the presence of lignin and ferulates in the cell walls. Consequently, maize varieties with a reduced lignin or ferulate content or an altered lignin composition can have important beneficial effects on cell wall digestibility. Considerable efforts in genetic improvement have been dedicated towards enhancing cell wall digestibility, benefiting agriculture, the biorefinery sector and the environment. In part I of this paper, we review conventional and advanced breeding methods used in the genetic improvement of maize germplasm. In part II, we zoom in on maize mutants with altered lignin for improved digestibility and biomass processing.
Maize, also known as corn, plays a multifaceted role in agriculture, serving as a crucial resource for food, feed and the production of basic chemicals such as fuels, plastics, etc. Maize silage represents the areal part of maize – including leaves, stems, cobs and seeds – stored under anaerobic conditions for conservation, to be used as ruminant feed (Barrière, 2017). The primary energy source in maize silage comes from starch in the kernels and from the cell wall carbohydrates. The cell wall, in the animal feed field known as neutral detergent fiber (NDF), is composed mainly of two types of carbohydrates, cellulose and hemicellulose, and the aromatic heteropolymer, lignin. NDF digestibility is influenced by the genetic background, the environmental conditions, field management and timing of harvest, and typically varies between 40 to 50% (Allen et al., 2003; Barrière, 2017). At the cell wall level, NDF digestibility is primary influenced by the lignin content, lignin composition and the ferulate cross-linkages between lignin and hemicelluloses (Wolf et al., 1993; Fontaine et al., 2003; Grabber et al., 2009; Courtial et al., 2013; Barrière, 2017). Consequently, breeding efforts to improve the NDF digestibility often target the lignin characteristics1.
Besides its use as ruminant feed, maize lignocellulosic biomass (i.e., maize stover consisting of the leaves, stems and cobs without seeds) has been identified as a promising resource for the biorefinery (Torney et al., 2007; Vermerris et al., 2007; Lorenzana et al., 2010; van der Weijde et al., 2013; Torres et al., 2015b; Saratale et al., 2019). During biorefining, the carbohydrate fraction of the lignocellulosic biomass is enzymatically deconstructed into primary sugars, a process called saccharification. These primary sugars can then be used in fermentation reactions to produce renewable materials and biofuels (Lips, 2022). Similar to ruminal digestibility, the intrinsic resistance of the maize stover to enzymatic degradation, also known as biomass recalcitrance, is mainly caused by the presence of lignin (Torres et al., 2015a). Therefore, breeding towards a reduced lignin amount, an altered lignin composition, or an altered interaction between lignin and hemicelluloses can be advantageous to improve both feed digestibility and the industrial saccharification process (Torres et al., 2016). In this review, we start by providing a brief history of conventional maize breeding methods, followed by an overview of more advanced breeding strategies. Next, we focus on maize mutants and transgenic lines with a modified lignin content and composition, and their effect on digestibility and biomass processing efficiency.
Maize (Zea mays L. spp. mays) is currently one of the most important staple crops, along with rice and wheat, worldwide (FAO, 2023). The origin of maize has been studied extensively; phylogenetic analyses and archaeological research show a direct ancestral link to two wild grass subspecies commonly known as teosinte (Z. mays ssp. parviglumis and Z. mays ssp. mexicana) (Yang et al., 2023). Maize domestication started 7.000 to 10.000 years ago in the tropical lowlands of present-day Mexico when indigenous Americans discovered its potential as food. From its origin, maize spread northwards and southwards throughout the American continent, giving rise to Northern Flint and Southern Dent lineages (see later) (Tenaillon and Charcosset, 2011). This spread caused maize to diversify under genetic drift and selection, resulting in varieties adapted to different climates and soil types, from sea level to the high altitudes in the Andean mountains (Vigouroux et al., 2008; Bouchet et al., 2013; Andorf et al., 2019). In the 15th and 16th century, maize was introduced into Europe, Africa and Asia via European explorers and traders (Barrière et al., 2006). The early maize breeders on each of the continents played a pivotal role in the domestication process and the development of maize cultivars as we know them today. Recent advances in biotechnology and genomics provide new tools for maize breeders to further improve their cultivars and speed up the breeding process (Singh et al., 2021b).
In the 19th century, farmers selected the best ears from the most productive and healthy plants. Their seeds were then sown in the next growth season and the selection process was repeated. This so-called mass selection is the oldest form of maize breeding. This method is simple and effective for fixing traits with high heritability, but not very effective for traits with low heritability, such as yield (Choo and Kannenberg, 1981; Hallauer et al., 2010). Hopkins introduced the ear-to-row selection method to speed up maize breeding (Hopkins, 1899). In this method, a number of maize plants with desirable phenotypes are identified and their seeds harvested separately. About 50 seeds from a single ear are then grown in a single progeny row, and allowed to open-pollinate. From these progeny rows, again the best plants are identified and allowed to open-pollinate. This process is repeated for three to six generations until the individual plants from this open-pollinated variety (OPV) start showing similarity for the desired trait (Awata et al., 2019). In the late 19th century, hybridization became a game-changing method in maize breeding. It was noticed that the offspring (i.e., hybrids) of two different open-pollinated maize cultivars had up to 53% higher yield as compared to either parent (Beal, 1878). This phenomenon, called “heterosis” or “hybrid vigor”, has been extensively exploited in breeding programs even though the molecular basis is still poorly understood (Labroo et al., 2021; Yu et al., 2021). Breeders have primarily relied on ‘heterotic groups’ to select parents to make hybrid combinations. A heterotic group is a collection of germplasm, that, when crossed with germplasm from another heterotic group, tends to exhibit a higher degree of heterosis (on the average) than when crossed with a member of its own group (Lee, 1995). The genetic diversity of the germplasm within one heterotic group is too small to give the desired hybrid vigor effect (Akinwale, 2021). Early 20th century, Shull and East independently discovered that both heterozygous and homozygous loci were present in OPVs and that fully homozygous lines can be obtained after five to seven generations of self-pollination (East, 1908; Shull, 1908, 1909). These inbred lines were often weak due to inbreeding depression, but vigor could be restored in the hybrid offspring of two different inbred lines. By continued improvement of the inbred lines, the production of ‘hybrid maize’ became a reality and hundreds of inbred lines were developed for the production of hybrids (Bennetzen and Hake, 2009; Hallauer et al., 2010). Therefore, the modern hybrid maize breeders have two main activities. First, genetically improving inbred lines by recombination and introgression of interesting alleles and, secondly, testing the combining ability between inbred lines to produce outstanding hybrids. Subsequently, these hybrids are extensively evaluated for their agronomic performance before commercialization (Lee and Tollenaar, 2007).
Inbred lines can be made in various ways, e.g., via pedigree breeding, backcross breeding or doubled haploid breeding. Pedigree breeding is a method to gradually improve a population by concentrating desirable alleles through a selection process of the best hybrids (Beckett et al., 2019; Singh et al., 2021a). In short, parents with desirable traits are crossed to generate hybrid seedstocks (in case the parental lines are heterozygous, their offspring will be highly heterogeneous). The resulting hybrids are then evaluated and the best-performing plants are self-pollinated for several generations to create inbreds. In each generation, plants with obvious defects are removed and the promising inbred lines are crossed with tester lines from different heterotic groups to evaluate their general combining ability. The newly generated inbreds are retained and the cycle is repeated until the inbreds are ready for use in cultivar development (Lee and Tollenaar, 2007).
Backcross breeding allows the breeder to transfer a desired trait obtained from a donor parent into a favored elite background (also called recurrent parent). Here, the objective is to genetically recover the recurrent parent except for the desired trait. Theoretically, in each backcross (BC) generation, about 50% of the recurrent parent genome is recovered (F1 – 50%, BC1 – 75%, BC2 – 87.5%, BC3 – 93.7%, BC4 – 96.9%, BC5 – 98.4%). Donor genes can reside for example in exotic germplasm, transgenic genotypes or mutants. Typically, breeders try to recover at least 98% of the recurrent parent genome (Vogel, 2009). This process is generally slow but molecular markers can be used to reduce the number of BC generations (Figure 1A). Marker technology can be carried out at the seedling stage and allows to characterize multiple loci at once and minimize linkage drag, thus speeding up the breeding process (Hasan et al., 2021).
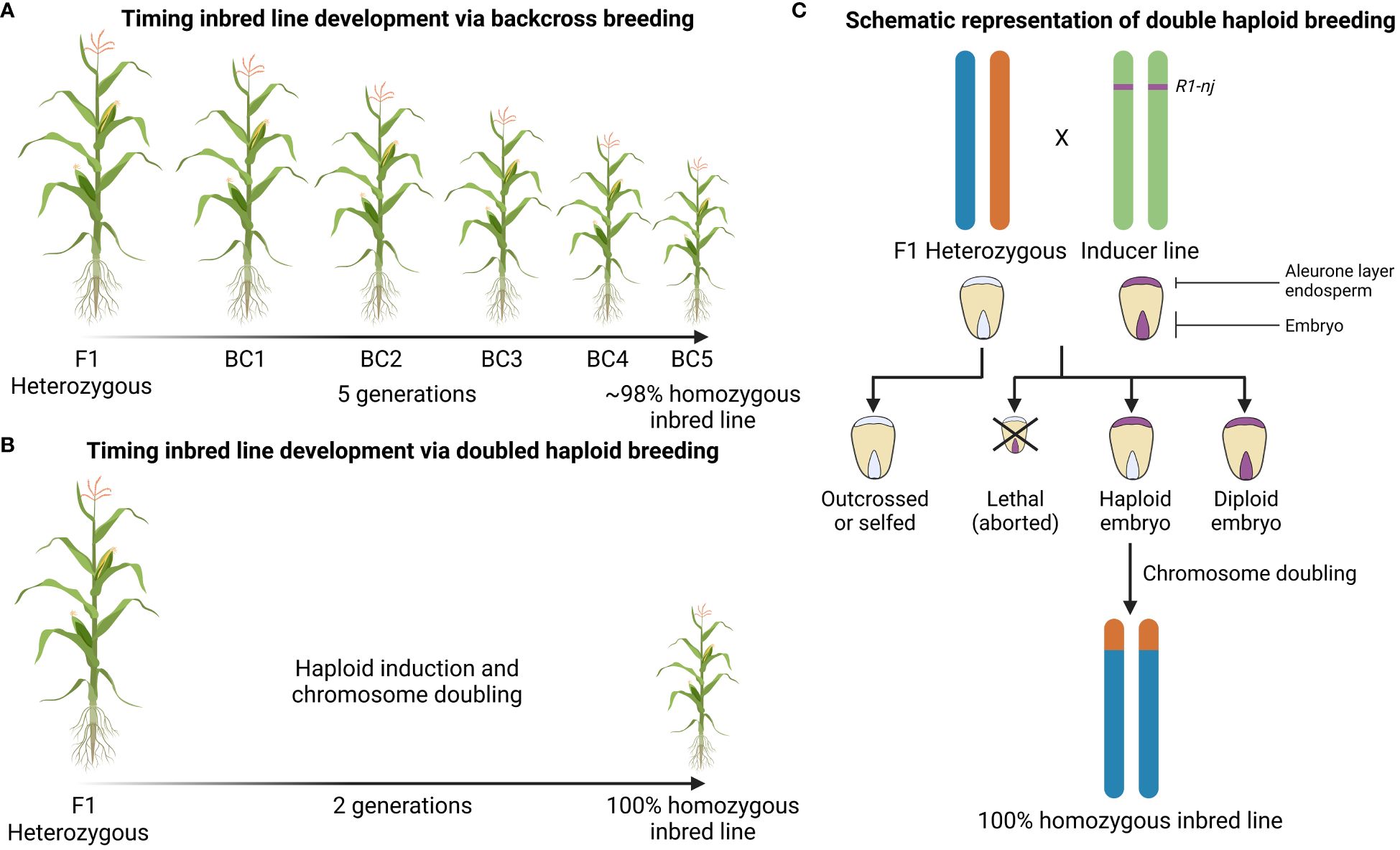
Figure 1 Graphical representation of inbred line development via conventional backcross breeding or doubled haploid technology. (A) Number of generations necessary to obtain a homozygous inbred line using conventional backcross breeding and (B) using doubled haploid breeding. (C) Identification of haploid seeds using the R1-nj color marker. The colored bars represent chromosomes. The inducer line carries the R1-nj dominant marker gene; the location on the green chromosomes does not reflect the actual physical location, but signifies its presence in the inducer line’s genome. Chromosome doubling, either artificial or spontaneous, after haploid induction is essential to obtain fertile homozygous inbred lines. The homozygous inbred line shown is just one of many possibilities, derived from a single recombinant gamete from the hybrid parent.
Doubled haploid breeding is a method to obtain pure inbred lines within a single generation (Ren et al., 2017) (Figure 1B). Typically, the production of stable and uniform hybrids relies on inbred line development and the latter takes multiple generations of self-crossing or back-crossing as described above. This labor intensive and time-consuming process can be overcome by the use of a genetic trick. Haploids can be generated in different ways (tissue culture from mega- and microsporocytes, natural generation by androgenesis, CENH3-mediated haploid induction and via haploid inducer lines) but making use of inducer lines is the most popular method (Ren et al., 2017; Maqbool et al., 2020; Meng et al., 2021). Coe discovered that one of his maize stocks, “stock 6”, produced 2 to 3% haploid plants when used as a male parent in crosses (Coe, 1959). As Coe’s “stock 6” was able to produce haploid progeny, it was named a “haploid inducer (HI)”. The process of haploid induction is also known as gynogenesis and is an asexual way of reproduction in which the male gamete triggers the development of an unfertilized egg into a haploid embryo (Gilles et al., 2017). In order to ease haploid identification, the HI is combined with the R1-navajo (R1-nj) gene, a dominant marker that results in a purple anthocyanin color in the kernel. This gene is expressed in the aleurone layer of the endosperm and in the embryo and allows to discriminate haploid seeds from diploid seeds (Rádi et al., 2020). The haploids will have no purple color in the embryo but will show a coloration in the aleurone layer (Figure 1C) (Bennetzen and Hake, 2009). Haploids are usually highly sterile and cannot undergo meiosis, but will produce pure and fertile diploid inbreds or doubled haploids after spontaneous or artificial chromosome doubling (Jackson, 2017).
The underlying major causal gene for the haploid induction in “stock 6” has been independently identified by three different research groups (Gilles et al., 2017; Kelliher et al., 2017; Liu et al., 2017). The mutation was mapped to a patatin-like phospholipase gene expressed in mature pollen and the pollen tube. This same gene was named by the three groups NOT LIKE DAD (NLD), MATRILINEAL (MTL), and ZmPHOSPHOLIPASE A1 (ZmPLA1). Recently, additional mutations have been identified that are able to further boost the haploid induction rate of NLD/MTL/ZmPLA1 mutants (Zhong et al., 2019; Li et al., 2021; Jiang et al., 2022). Modern inducer lines have 7 to 16% haploid progeny depending on the genetic background and can speed up the maize breeding process tremendously, especially when combined with genome-editing techniques such as CRISPR-Cas9 (Kalinowska et al., 2019; Jacquier et al., 2021; Impens et al., 2023).
Although maize has gone through some evolutionary bottlenecks during domestication and directional selection, the maize breeding germplasm still has standing variability with a wide phenotypic diversity, nutritional qualities and resistance against (a)biotic stresses. Two groups in particular are very popular in maize breeding in North America and Europe: the Northern Flints and Southern Dents (Troyer, 1999; Barrière et al., 2005). The Northern Flints are described as cold tolerant, early flowering and mature, having long slender ears with undented round kernels. The Southern Dents are more heat tolerant with later maturity, being taller, higher-yielding, and having wide ears with higher kernel rows and dented rectangular kernels. Crosses between the two groups provide better adapted and higher-yielding varieties in a range of environments (Troyer, 1999).
The success of highly adapted maize varieties is the result of ingenious farmers who contributed to population improvement and developed prominent OPVs, including, but not limited to, the Reid Yellow Dent, Lancaster Sure Crop, Minnesota 13, Leaming Corn, Northwestern Dent and Longfellow Flint (Sprague and Dudley, 1988). This heterogeneous group of OPVs was classified by breeders into heterotic groups. The major heterotic groups present in current maize breeding programs can be divided into the female heterotic groups, showing high kernel yield and smaller tassels, and the male heterotic groups, with more pollen and longer pollen shedding duration. Heterotic groups PA and Stiff Stalk (represented by inbreds such as B73 and B104) are mainly used as female heterotic groups, while PB, SPT, Non-Stiff Stalk (represented by inbreds such as Mo17, Oh43 and H99) and Iodent (represented by inbreds such as PH207) are predominantly used as male heterotic groups (Li et al., 2022). These heterotic groups continually expand by the creation of new inbred lines and the occasional introgression of exotic germplasm. Phenotypic and genetic evaluation of recent inbred lines from various heterotic groups revealed that advantageous alleles for traits accumulate within the heterotic groups during selection. Some traits evolved divergently between the female and male heterotic groups (e.g. tassel height, kernel weight) and other convergently (e.g. stress tolerance) (Li et al., 2022). Notably, some alleles showing fixation within specific heterotic groups remain genetically heterogenous between heterotic groups, potentially contributing to heterosis upon crossing inbred lines from different groups (Gerke et al., 2015; Li et al., 2022). Maintaining or even increasing allelic diversity among heterotic groups is key for maize improvement, with novel breeding techniques offering potential acceleration of this process (see later).
Allelic diversity occurs spontaneously in germline cells and is derived from natural processes such as UV radiation, errors in DNA replication during cell division or in DNA repair after breakage, or via transposable elements (see later) (Martínez-Fortún et al., 2022). Studies have estimated the spontaneous mutation rate in maize to be 2.2 to 3.9 x 10-8 per site per generation (Yang et al., 2017b). The exploitation of spontaneous mutations in breeding has led to variations in in vivo NDF digestibility in hybrids, ranging between 36 and 60% (Méchin et al., 2000; Barrière et al., 2004a; Barrière et al., 2009b). Lignin is a major contributor to this variation in cell wall degradability. Although the exact genetic cause for the variation in lignin content among different genotypes is often unclear, quantitative trait loci (QTL) studies have shown that various loci collectively contribute to this trait (Barrière et al., 2015). However, exceptions exist where the genetic cause of certain historical maize lignin mutants turned out to be monogenic. These lignin mutants typically show a brown midrib (bm) phenotype, facilitating their study due to its easy-to-see phenotype. However, it was not until the 90’s that the genetic cause for the bm phenotype and its associated increase in digestibility were elucidated (see later).
Conventional breeding happens in a relatively uncontrolled manner. The breeder chooses and crosses parental plants but the results are unpredictable at the phenotypic and genetic level (Wieczorek and Wright, 2012). Additionally, crossing and backcrossing of the hybrids to obtain elite lines is a laborious and time-consuming process. Marker-assisted selection accelerates the prediction and selection process in maize breeding, facilitating the identification of varieties with desired traits such as improved digestibility (Barrière et al., 2016; López-Malvar et al., 2019; Vinayan et al., 2021). Although conventional breeding relies on genetic diversity found in commercial varieties and landraces, this diversity can sometimes be limited. Consequently, modern biotechnological tools allowing to alter the plant’s DNA provide valuable alternatives (Hamdan et al., 2022). To mimic the process of random mutagenesis and increase genetic variation, the concept of mutation breeding was developed [reviewed by Ma et al. (2021)]. Mutation breeding is based on introducing mutations via radiation or chemical mutagens. Mutant populations can either be screened phenotypically, or by the use of techniques such as TILLING (Targeting Induced Local Lesions IN Genomes) that allow identifying specific mutations in genes of interest (Till et al., 2004). Another mutagenesis approach involves the use of Mutator (Mu) and Activator (Ac) lines. Mu transposons, often referred to as ‘jumping genes’, have the ability to randomly transpose within the genome. When activated by Ac genes, Mu transposons jump to new locations, thereby increasing the genetic variation (Bennetzen et al., 1993; May et al., 2003; Marcon et al., 2020).
In the 1990’s, another milestone in agriculture was reached with the development of transgenic breeding techniques, utilizing recombinant DNA technology and the ability of Agrobacterium tumefaciens to transfer DNA into host genomes to produce herbicide- and pest-resistant maize (Woo et al., 1997; Lundquist and Walters, 2001). RNA interference (RNAi) is a gene mechanism that allows to downregulate an endogenous gene by introducing part of that same gene in reverse orientation leading to double-stranded RNA that is degraded by the cellular machinery (Lindbo, 2012). Although currently no commercial RNAi lines for maize are available on the market, RNAi has been used to downregulate genes involved in lignification to study their effect on cell wall composition and degradability (see later). Although both mutation breeding approaches and transgenic breeding techniques modify the plant’s genome, there is a big difference between the two methods. Mutation breeding involves the introduction of mutations in the plant’s genome to achieve the desired traits, while recombinant DNA technology introduces foreign genes to impart new traits to the species.
Unlike the mutation techniques mentioned above, which may introduce unpredictable, random and unwanted changes, the use of sequence-specific nucleases enables the editing of a pre-defined DNA sequence in the host plant by introducing a double stranded break (DSB) at or near the target site. Subsequently, the DNA repair machinery induces errors, leading to a mutation (Hamdan et al., 2022). The first sequence-specific nucleases used for genome editing in maize were zinc finger nucleases and transcription activator-like effector nucleases (Shukla et al., 2009; Liang et al., 2014). These techniques were successful but relatively laborious, because protein engineering was necessary to adjust the enzyme for every new target gene (Bortesi and Fischer, 2015). More recently, CRISPR-Cas [clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR associated proteins (Cas)] has revolutionized the field by enabling precise gene editing without introducing exogenous DNA in the final product (Van Vu et al., 2022). Hence, the emphasis has shifted towards gene editing techniques, with legislation showing a more favorable stance towards gene editing as compared to its transgenic counterpart (Wang et al., 2023; Stokstad, 2024).
The CRISPR-Cas system uses a DNA nuclease and a guide RNA to introduce a DSB, which is repaired via the error-prone non-homologous end-joining (NHEJ) or the error-free homologous directed repair (HDR) pathways (Xue and Greene, 2021). NHEJ often introduces short insertions or deletions (indels), leading to a gene knock-out (loss-of-function) or truncated proteins (Piatek et al., 2018). However, the mutation outcome after NHEJ is not predictable. This urged for the development of more precise editing techniques such as base and prime editing, both of which have been successfully demonstrated in maize (Jiang et al., 2020; Li et al., 2020). HDR is an alternative repair pathway to precisely restore the DSB using a DNA template derived from a homologous chromosome or to introduce a genetic modification using an artificial DNA repair template (Piatek et al., 2018). Despite the potential of HDR for gene replacement or gene insertion, its inefficiency limits its exploitation for crop improvement (Hamdan et al., 2022). Various studies have used this technology for diverse purposes, e.g. reducing smut susceptibility, introducing herbicide resistance, enhancing grain yield and developing more drought-tolerant maize varieties (Svitashev et al., 2015; Shi et al., 2017; Pathi et al., 2020; Liu et al., 2021a). Currently, there are no published CRISPR mutants affecting the lignin biosynthesis in maize. However, a comprehensive list of CRISPR applications in maize is available on EU-SAGE (http://www.eu-sage.eu).
Beyond single-gene mutagenesis, CRISPR-Cas9 also allows for simultaneous editing of numerous genes, including members of the same gene family. A notable multiplex genome editing approach, BREEDIT, was developed to rapidly generate a collection of multiplex edited plants. This method employs a single construct that simultaneously targets up to twelve genes, followed by self-pollination or crossing to achieve even higher order mutants. This strategy enables identification of promising gene combinations that can later be used in breeding programs (Lorenzo et al., 2023).
The delivery of plasmid DNA encoding a CRISPR-Cas construct in maize primarily relies on Agrobacterium-mediated transformation or biolistic delivery. The major advantage of Agrobacterium-mediated T-DNA delivery is its ability to integrate a single or low copy number of relatively large DNA fragments (up to 150 kb) into the plant’s genome (Frame et al., 2006; Yadava et al., 2017). Biolistics or particle bombardment is a genotype-independent T-DNA delivery method that physically breaches the plant cell with gold or tungsten particles coated with an expression vector, DNA fragments or ribonucleoprotein complexes (Yadava et al., 2017; Liang et al., 2019). Biolistics also presents some challenges such as the introduction of multiple copies and complex integration events of the vector (Jackson et al., 2013). Therefore, Agrobacterium-mediated transformation is generally the method of choice for maize transformation (Peterson et al., 2021). However, it is important to note that genotype-associated recalcitrance is also related to the tissue culture procedure, explant material and the Agrobacterium strain used (Yassitepe et al., 2021). Significant innovations have been made to overcome this genotype-associated recalcitrance, with one advancement being the codelivery of the morphogenetic regulators BABY BOOM (BBM) and WUSCHEL (WUS). The co-expression of BBM/WUS induces somatic embryogenesis, resulting in improved transformation efficiency (Lowe et al., 2016). For example, the transformation frequency has increased from 0% up to 15% in the B73 inbred line, known to be highly recalcitrant towards transformation (Mookkan et al., 2017). Although the codelivery of these morphogenic regulators has also increased the transformation efficiency of the B104 inbred line, continuous expression of BBM/WUS leads to pleiotropic developmental effects and sterility (Aesaert et al., 2022), which can be mitigated using gene excision systems such as CRE/loxP and/or inducible promoters (Lowe et al., 2016; Mookkan et al., 2017; Aesaert et al., 2022).
Although CRISPR-based genome editing holds great potential in plant breeding, the genotype-associated recalcitrance mentioned earlier often limits its large-scale application for the development of new commercial maize lines. Maize lines such as Hi-II and B104 are amenable to the standard transformation protocol, however these lines are usually not suitable for commercial applications (Hernandes-Lopes et al., 2023). Moreover, commercial maize varieties are typically hybrids derived from a cross between distinct parental elite inbred lines. Consequently, introducing traits requires both parental elite inbred lines to be edited. To bypass the transformation procedure and thus the genotype-associated recalcitrance, transgenerational gene editing presents an alternative method to deliver the CRISPR-Cas machinery in elite inbred lines (Li et al., 2017; Wang et al., 2018). In short, a transgenic maize plant containing a CRISPR-Cas9 T-DNA is crossed with a recalcitrant genotype (Figure 2A). Within the resulting hybrid, CRISPR-Cas can then edit the target gene in trans. The advantage of this approach is that mutations are introduced in the recalcitrant background without the need for introgression of an allele derived from another variety, thus avoiding linkage drag (Impens et al., 2022). Nonetheless, multiple backcrosses are still required to restore the elite background, which is both a time-consuming and laborious process (Wolter et al., 2019). For example, maize plants transformed with a CRISPR-Cas construct targeting the LIGULELESS1 (LG1) gene were crossed with a recalcitrant elite inbred line, resulting in an in trans mutation frequency of 20% in the F1 generation (Li et al., 2017). Subsequent rounds of marker-assisted backcrossing were then conducted to recover the elite background. In another example, the GRANULE BOUND STARCH SYNTHASE I (GBSS I) or Wx locus was trans-edited in two parental lines to rapidly generate a single hybrid waxy maize (Qi et al., 2020). Variations of transgenerational gene editing, such as the HI-edit or HI-mediated genome editing (IMGE) methods (see Figure 2B), have been developed in maize (Wang et al., 2018; Kelliher et al., 2019). The HI-Edit/IMGE methods involve transient expression of the CRISPR construct from the paternal HI line to edit the maternal genome in trans, followed by the elimination of the paternal genome in the zygote phase (Yassitepe et al., 2021). Subsequently, the edited haploid progeny undergoes artificial chromosome doubling to produce transgene-free doubled haploids (Wang et al., 2018; Kelliher et al., 2019). However, efficiencies remain low because modern HIs typically produce between 7 to 16% haploids and only 2 to 4% of the haploids are edited in trans. Consequently, less than 1% of the progeny are edited haploids (Wang et al., 2018; Kalinowska et al., 2019; Kelliher et al., 2019; Impens et al., 2022).
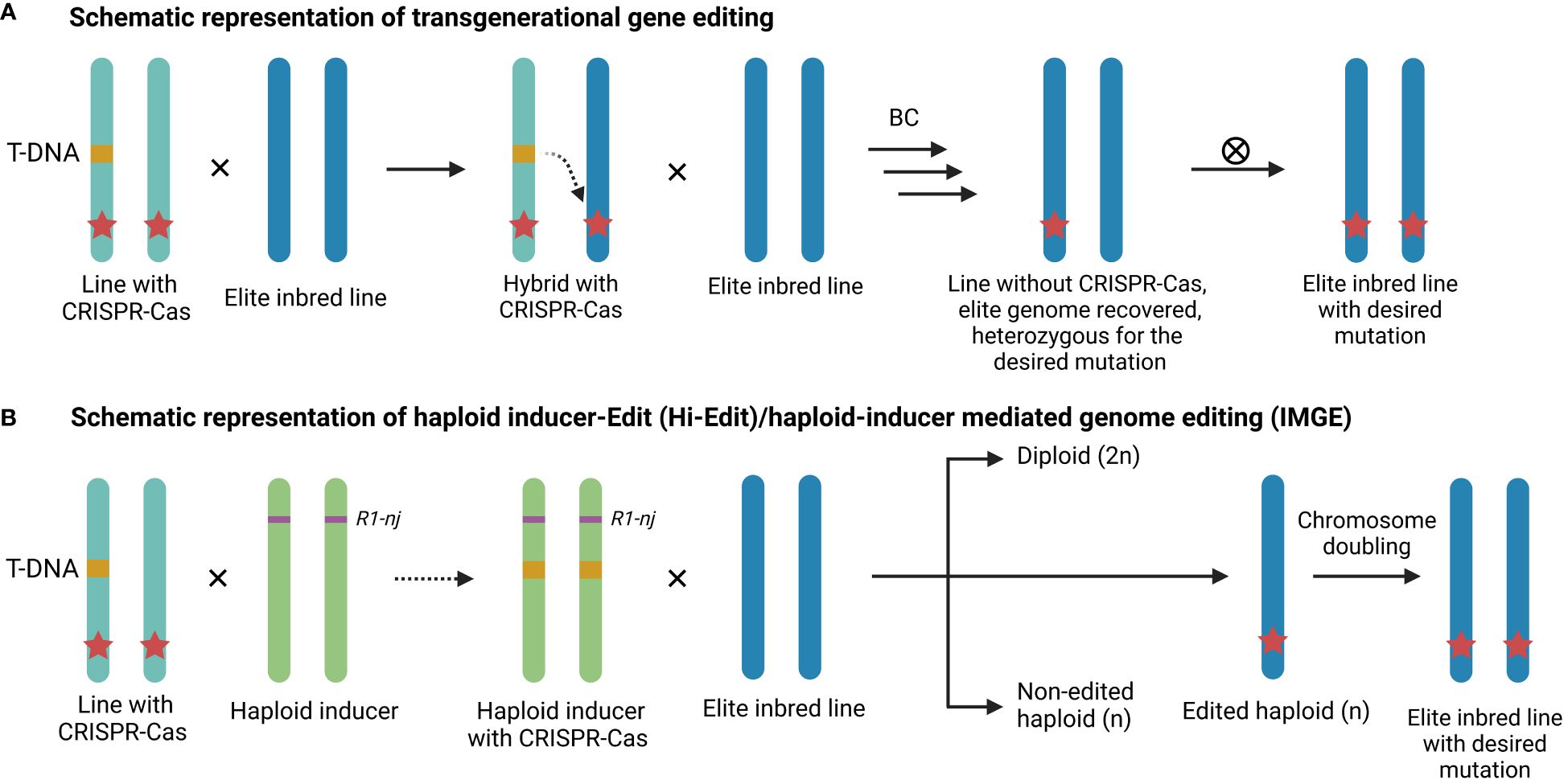
Figure 2 Schematic representation of transgenerational gene editing. (A) A maize plant containing a CRISPR-Cas T-DNA (orange) is crossed with an elite inbred line, resulting in an edited allele (red star) in the elite inbred line. The T-DNA free plants are retained and the elite background is restored via multiple rounds of backcrossing (BC). Finally, elite inbreds with homozygously edited alleles are screened for after selfing (⊗). (B) A CRISPR-Cas construct is introgressed or transformed into a haploid inducer, which is crossed with an elite inbred line and the edited haploids are identified. An elite inbred line homozygous for the edit is obtained after chromosome doubling.
Increasing the nutritional value of feed and replacing fossil fuels to produce bio-based products have been the driving forces behind the increased interest in cell wall biosynthesis and its enzymatic degradation. Primary targets in maize breeding for feed purposes have been related to grain yield and whole plant biomass yield. A comparison of ten different forage maize cultivars released between 1972 and 2020 revealed an average annual yield increase of 0.13 ton dry matter (DM)/ha (Taube et al., 2020). Comparable trends in yield increase were observed in forage hybrids released between 1950 and 1980 (0.07 ton DM/ha) and from 1991 to 2003 (0.18 ton DM/ha) (Barrière et al., 1987; Luciani, 2004). However, while substantial progress has been made in improving yield, improvements in forage quality remain modest. Barrière et al. (2006) reported even a decrease in cell wall digestibility during the breeding period of forage maize in Europe from 1958 to 2002, and subsequent research by Taube et al. (2020) found no significant increase in cell wall digestibility among cultivars released between 1972 and 2020. Although a range of naturally occurring genetic variants and induced mutants with increased biomass digestibility have been identified, including the aforementioned bm mutants having defects in lignin biosynthesis, it has been difficult to exploit them in modern cultivars due to their pleiotropic effects (prone to lodging and reduced yield) (Vignols et al., 1995; Halpin et al., 1998; Chen et al., 2012b; Tang et al., 2014; Li et al., 2015; Xiong et al., 2019). Here it is important to note that not all maize lines with altered lignin show a bm phenotype (Tamasloukht et al., 2011; Fornalé et al., 2012; Li et al., 2013; Marita et al., 2014), implying that numerous lignin-modified maize mutants may have been overlooked in breeding programs that primarily screened for this characteristic bm phenotype. Therefore, research has become increasingly focused on investigating mutants and transgenic plants perturbed in specific steps of the lignin biosynthesis pathway. Ideally, modifying these genes or their expression levels would reduce the biomass recalcitrance without compromising the biomass yield. Below, we describe the biosynthesis of lignin and its interaction with hemicellulose, followed by an overview of mutant and transgenic maize lines with altered lignin and the resulting improvement in digestibility.
Lignin is a structural component of the cell wall that accounts for up to 17.2% of the lignocellulosic biomass in maize (Zhu et al., 2005). Lignin offers support to the cell wall, facilitates water transport and protects the carbohydrates from being digested by pathogens and insects. It is a complex aromatic heteropolymer made from phenolic monomers that are biosynthesized in the cytosol and translocated to the apoplast prior to polymerization via oxidative combinatorial coupling (Boerjan et al., 2003; Dixon and Barros, 2019; Vanholme et al., 2019). The traditional monolignols are p-coumaryl alcohol, coniferyl alcohol and sinapyl alcohol, that give rise to the p-hydroxyphenyl (H), guaiacyl (G) and syringyl (S) units in the lignin polymer, respectively (Boerjan et al., 2003; Ralph et al., 2004; Vanholme et al., 2010; Dixon and Barros, 2019; Vanholme et al., 2019). In addition, maize lignin also incorporates the flavonoid tricin, coniferaldehyde, ferulic acid and acylated monolignols such as coniferyl acetate, coniferyl p-coumarate, sinapyl p-coumarate, coniferyl ferulate and sinapyl ferulate (Lu and Ralph, 1999; Grabber et al., 2008; Ralph et al., 2008; Hatfield et al., 2009; Marita et al., 2014; Lan et al., 2015; Vanholme et al., 2019). Lignin is linked to hemicelluloses through coupling with ferulate moieties that are esterified on the arabinoxylan (Hatfield et al., 2017).
The elucidation of the lignin biosynthetic pathway in maize is a work in progress. Figure 3 summarizes the latest insights into the general and the monolignol-specific pathway and compiles which enzymatic steps have been proven to occur in maize and which are based on findings in other grass species. Lignin is synthesized in a series of enzymatic steps starting from phenylalanine and tyrosine (Boerjan et al., 2003; Vanholme et al., 2010, 2019; Barros and Dixon, 2020). The first step of the general phenylpropanoid pathway is the conversion of phenylalanine by PHENYLALANINE AMMONIA LYASE (PAL) to cinnamic acid. Next, cinnamic acid is hydroxylated by CINNAMIC ACID 4-HYDROXYLASE (C4H) to form p-coumaric acid (Ôba and Conn, 1988). PALs in grasses, including maize, can be bifunctional and also have TYROSINE AMMONIA LYASE (TAL) activity that allows the conversion of tyrosine into p-coumaric acid (Rösler et al., 1997; Barros et al., 2016). Subsequently, p-coumaric acid is converted into p-coumaroyl-CoA through 4-COUMARATE:CoA LIGASE (4CL) (Yun et al., 2005; Xiong et al., 2019). Next, p-coumaroyl-CoA is esterified into its corresponding shikimic or quinic ester derivative catalyzed by p-HYDROXYCINNAMOYL-CoA: QUINATE/SHIKIMATE p-HYDROXYCINNAMOYLTRANSFERASE (HCT). This enzymatic conversion has not yet been proven to occur in maize, although it has been identified in other grass species like Brachypodium, switchgrass and sorghum (Shadle et al., 2007; Walker et al., 2013; Escamilla-Treviño et al., 2014; Serrani-Yarce et al., 2021). In turn, p-coumaroyl shikimate is hydroxylated by p-COUMAROYL-CoA 3’-HYDROXYLASE (C3’H) to produce caffeoyl shikimate. Currently, two C3’H-encoding genes, namely C3’H1 and C3’H2, have been found in maize (Barrière et al., 2007; Guillaumie et al., 2007); the role of C3’H1 in lignin biosynthesis has already been demonstrated through reverse genetics (Fornalé et al., 2015). Caffeoyl shikimate is then further esterified by HCT into caffeoyl-CoA (Serrani-Yarce et al., 2021). Alternatively to the biosynthetic route via 4CL, HCT, C3’H and again HCT, p-coumaric acid can also be converted into caffeoyl-CoA via a two-step pathway; hydroxylation by COUMARATE 3-HYDROXYLASE (C3H) (Barros et al., 2019) and ligation to CoA by 4CL. In the last step of the general phenylpropanoid pathway, caffeoyl-CoA is methylated by CAFFEOYL-CoA O-METHYLTRANSFERASE (CCoAOMT) into feruloyl-CoA (Wu et al., 2018).
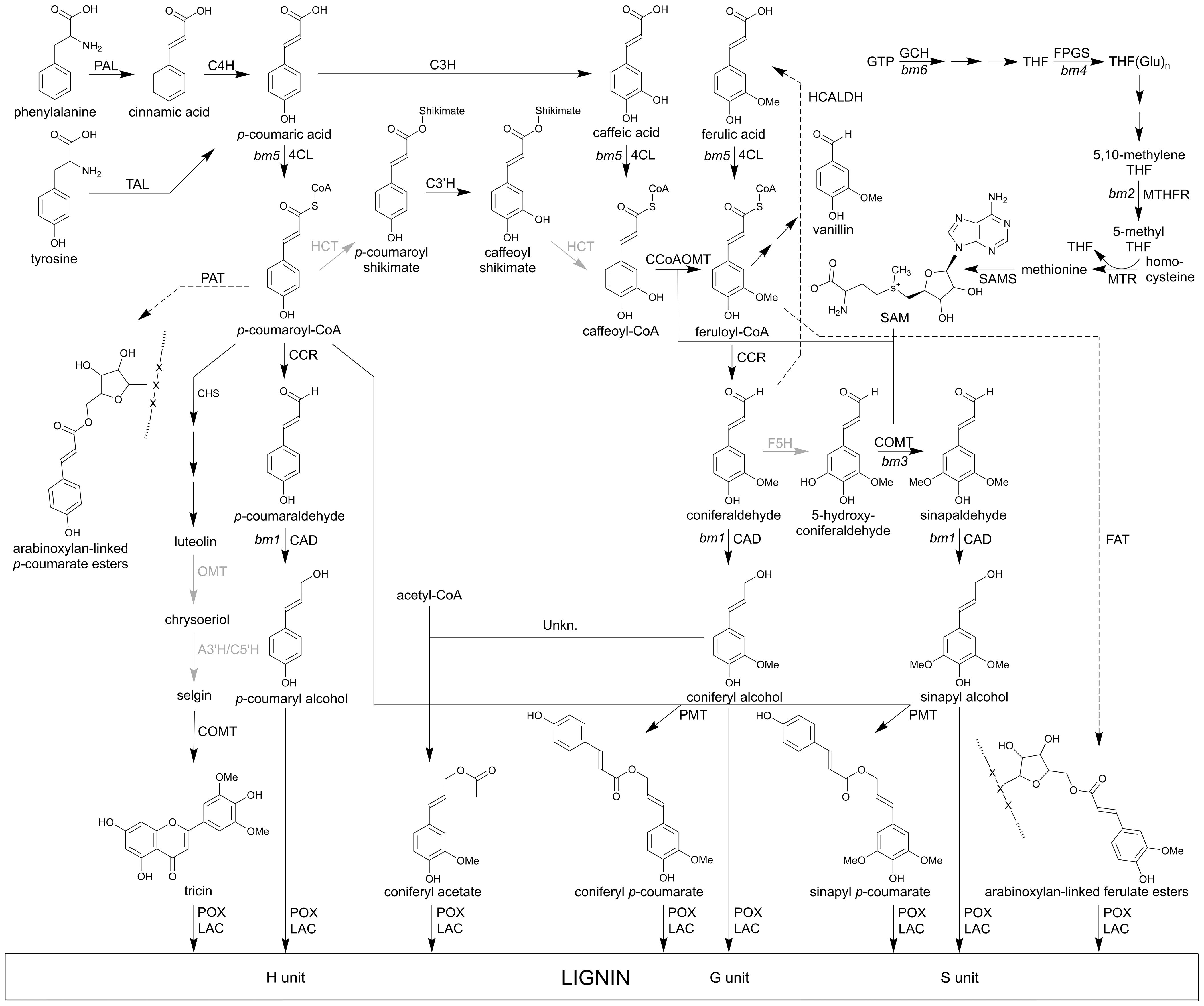
Figure 3 The main pathway of the lignin monomers in maize. Many metabolites are derived from the phenylpropanoid biosynthesis pathway. Here, only the routes to lignin monomers are shown. Solid arrows represent enzymatic steps evidenced by at least in vitro activity, dashed arrows indicate suggested conversions. Successive arrows indicate two or multiple metabolic conversions. The enzymatic conversions indicated in black are proven in maize, those that are shown in gray occur with certainty in grasses. PAL, PHENYLALANINE AMMONIA-LYASE; TAL, TYROSINE AMMONIA LYASE; C4H, CINNAMATE 4-HYDROXYLASE; C3H, p-COUMARATE 3-HYDROXYLASE; 4CL, 4-COUMARATE:CoA LIGASE; HCT, p-HYDROXYCINNAMOYL-CoA:QUINATE/SHIKIMATE p-HYDROXYCINNAMOYLTRANSFERASE; C3’H, p-COUMAROYL SHIKIMATE 3’-HYDROXYLASE; CCoAOMT, CAFFEOYL-CoA O-METHYLTRANSFERASE; CCR, CINNAMOYL-CoA REDUCTASE; F5H, FERULATE 5-HYDROXYLASE; COMT, CAFFEIC ACID O-METHYLTRANSFERASE; CAD, CINNAMYL ALCOHOL DEHYDROGENASE; HCALDH, HYDROXYCINNAMALDEHYDE DEHYDROGENASE; PMT, p-COUMAROYL-CoA MONOLIGNOL TRANSFERASE; CHS, CHALCONE SYNTHASE; A3’H/C5’H, APIGENIN 3’-HYDROXYLASE/CHRYSOERIOL 5’-HYDROXYLASE; POX, PEROXIDASE; LAC, LACCASE; PAT, p-COUMAROYL-CoA ARABINOFURANOSE TRANSFERASE; FAT, FERULOYL ARABINOFURANOSE TRANSFERASE; GTP, guanosine triphosphate; GCH1, GTP CYCLOHYDROLASE; THF, tetrahydrofolate; FPGS, FOLYLPOLYGLUTAMATE SYNTHASE; MTHFR, METHYLENETETRAHYDROFOLATE REDUCTASE; MTR, 5-METHYLTETRAHYDROFOLATE HOMOCYSTEINE METHYLTRANSFERASE; SAMS, S-ADENOSYLMETHIONINE SYNTHETASE; SAM, S-adenosyl-L-methionine. bm; brown midrib mutants.
The first committed enzyme of the monolignol-specific pathway is CINNAMOYL-CoA REDUCTASE (CCR) that converts hydroxycinnamoyl-CoA esters (p-coumaroyl-CoA and feruloyl-CoA) into their corresponding hydroxycinnamaldehydes (p-coumaraldehyde and coniferaldehyde) (Pichon et al., 1998). Coniferaldehyde can be converted into 5-hydroxyconiferaldehyde through FERULATE 5-HYDROXYLASE (F5H) (Kim et al., 2006; Wu et al., 2019). The maize genome encodes two F5H genes (Guillaumie et al., 2007), of which F5H1 was previously described as being associated with a QTL for digestibility by Puigdomenech et al. (2001), but no functional proof was given for either of the two genes through reverse genetics. In turn, 5-hydroxyconiferaldehyde is methylated by CAFFEIC ACID O-METHYLTRANSFERASE (COMT) into sinapaldehyde (Piquemal et al., 2002). Lastly, CINNAMYL ALCOHOL DEHYDROGENASE (CAD) catalyzes the conversion of the hydroxycinnamaldehydes into the traditional monolignols, p-coumaryl, coniferyl and sinapyl alcohol (Halpin et al., 1998). The respective hydroxycinnamyl alcohols can function either directly as lignin monomers or as substrates for acyltransferase reactions, resulting in γ-O-acylated monolignol conjugates that can also be incorporated into the lignin polymer. In maize, similar to other grasses, coniferyl alcohol is acylated with acetate via an enzyme yet to be identified, and primarily sinapyl alcohol is acylated with p-coumaric acid (pCA) by p-COUMAROYL-CoA: MONOLIGNOL TRANSFERASE (PMT) (Withers et al., 2012; Marita et al., 2014; Karlen et al., 2018). Monolignols (mainly sinapyl alcohol) can also be acylated with ferulic acid, via FERULOYL-CoA MONOLIGNOL TRANSFERASE (FMT) activity, although such conjugates are less abundant (Karlen et al., 2016; Smith et al., 2017).
The phenylpropanoid pathway is connected with other biosynthetic routes, some of which result in other lignin monomers, such as flavonoids, or molecules that connect lignin with hemicelluloses (Vanholme et al., 2019). For example, p-coumaroyl-CoA is the substrate of CHALCONE SYNTHASE (CHS), the key enzyme in regulating the flux toward the biosynthesis of flavonoids, including the lignin monomer tricin. Tricin is known to be incorporated in lignin, where it acts as an initiation site for lignin polymerization (Lan et al., 2015, 2016; Eloy et al., 2017). In grass cell walls, ferulate and to a lesser extent p-coumarate, can acylate the hemicellulose polymer via the C5-hydroxyl of the α-L-arabinosyl side chains of glucuronoarabinoxylan (GAX). These ferulate groups originate from feruloyl-CoA and are coupled to the arabinose residue by a FERULOYL ARABINOFURANOSE TRANSFERASE (FAT) belonging to the BAHD acyltransferases (Figure 3) (Schmitt et al., 1991; Guo et al., 2001; de Oliveira et al., 2015; Fanelli et al., 2021; Chandrakanth et al., 2023). Feruloyl-CoA is a phenylpropanoid pathway intermediate, and thus readily available in lignifying cells.
After their biosynthesis, the lignin monomers are thought to passively diffuse through the plasma membrane into the apoplast (Vermaas et al., 2019; Perkins et al., 2022). In the cell wall, the monomers are oxidized by peroxidases (POXs) and/or laccases (LACs) into their corresponding radicals (Figure 3) (Guillet-Claude et al., 2004; Zhao et al., 2013; Perkins et al., 2022). During lignin polymerization, these monomer radicals will undergo combinatorial coupling, resulting in a variety of chemical bonds, of which the β-aryl (8-O-4), resinol (8–8) and phenylcoumaran (8–5) bonds are the most frequent ones (Ralph et al., 2019). LAC4 has been described to be involved in lignification of the maize cob and to affect the ear length (Bi et al., 2024). Due to the broad substrate specificity of LACs and POXs, and the possibility of radical transfer, the ferulates present on GAX can also undergo oxidation and radical coupling, resulting in the formation of dehydrodiferulate dimers or oligomers, which crosslink the polysaccharide chains. Additionally, ferulates are also crosslinked to lignin polymers, forming covalently linked carbohydrate–lignin complexes (de Oliveira et al., 2015; Hatfield et al., 2017; Terrett and Dupree, 2019). Thereby, ferulates act as nucleation sites for lignin formation (Buanafina, 2009; Courtial et al., 2013; Hatfield et al., 2017; Terrett and Dupree, 2019). Furthermore, free ferulic acids can also be incorporated into the lignin polymer by 8-O-4 crosslinking, allowing new branching points with biphenyl structures to be formed (Ralph et al., 2008).
The transcriptional regulation of genes involved in lignin biosynthesis is tightly controlled by transcription factors of the MYB and NAC families (Barrière et al., 2015) (Zhao and Dixon, 2011; Zhong and Ye, 2015). But so far, few transcription factors have been evaluated by reverse genetics in grasses (Wang et al., 2016; Bhatia et al., 2017). Expression profiling of Arabidopsis overexpression lines has shown that maize ZmMYB31 and ZmMYB42 both act as repressors of COMT and potentially other lignin biosynthetic genes (Fornalé et al., 2006; Sonbol et al., 2009; Fornalé et al., 2010). It was later discovered that ZmMYB69 acts as an activator of ZmMYB31 and ZmMYB42 expression, and thus a repressor of lignin biosynthesis (Qiang et al., 2022). In contrast, maize ZmMYB46 is described as a master switch to activate cell wall biosynthesis, including cellulose, hemicellulose and lignin biosynthesis, based on overexpression experiments in Arabidopsis (Zhong et al., 2011). Furthermore, there is evidence that both ZmMYB5 and ZmMYB152 (which are also called ZmMYB148 and ZmMYB111, respectively) are activators of lignin biosynthesis, but their function has not yet been evaluated in planta (Zhang et al., 2016; Yang et al., 2017a). Based on overexpression in Brachypodium and maize, also ZmMYB167 has been identified as an activator of lignin biosynthesis (Bhatia et al., 2019). NAC transcription factors regulate the expression of downstream MYBs (Xiao et al., 2018). Specifically, NAC SECONDARY WALL THICKENING PROMOTING FACTOR 3 (NST3) and NST4 regulate the expression of, amongst others, MYB109, MYB128 and MYB149, and enhance cell wall thickening (Xiao et al., 2018; Ren et al., 2020). Even though NST3 and NST4 are shown to activate lignin biosynthesis in maize, the transcriptional cascades involved are not yet described (Xiao et al., 2018; Ren et al., 2020).
The availability of genomic information and insights into the lignin biosynthetic pathway have made it possible to make perturbations in this pathway to modify and steer the biosynthesis of the monomers or the structure of the polymer itself (Mottiar et al., 2016; Ralph et al., 2019). Table 1 provides an overview of maize lines with altered lignin amount and composition, and their cell wall degradability and biomass yield. It should be noted that variation in the genetic background in which the various mutations were studied hinders straightforward comparisons of the specific effects of a particular perturbation (Chabbert et al., 1994; Marita et al., 2003). Indeed, different inbred lines already differ significantly in their lignin content and forage quality (Lundvall et al., 1994). Hence, mutations in different genetic backgrounds may result in different phenotypes. In addition, often different mutant alleles, e.g. knock-out and weak alleles, have been described for a given gene, or transgenic plants may have different degrees of downregulation of the target gene (Park et al., 2012; Barrière et al., 2013). This genetic variation is reflected in the range of mutant phenotypes associated with a single gene perturbation.
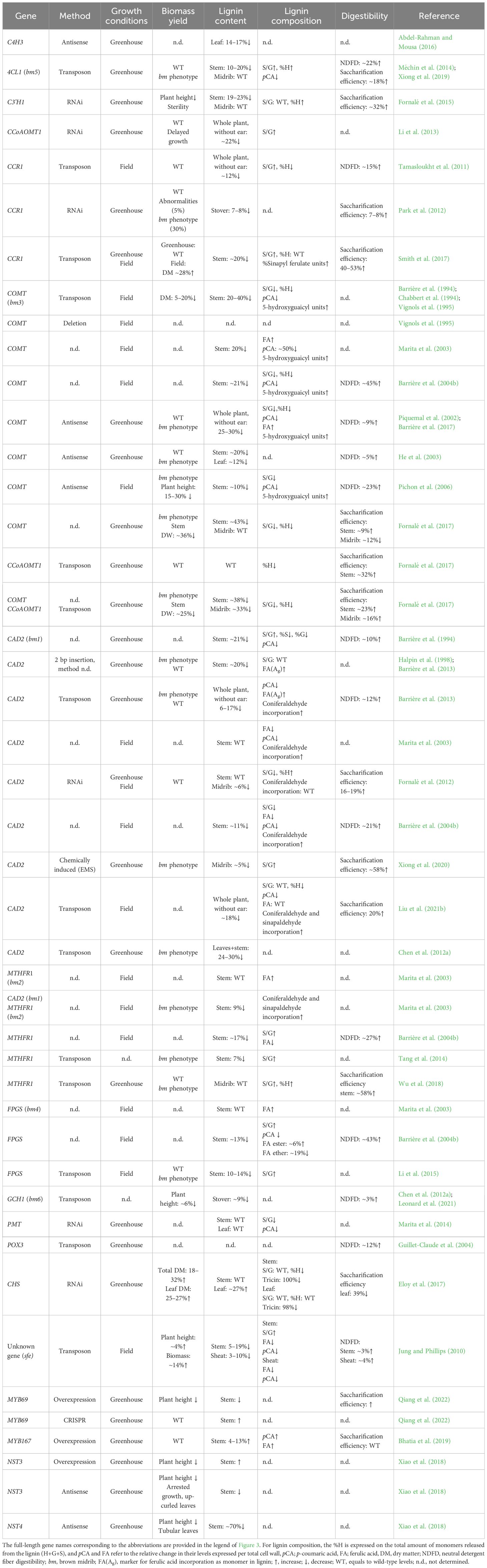
Table 1 List of lignin mutants in maize and the effects of the mutation on the lignin content, lignin composition, cell wall degradability and biomass yield.
Maize lines have been described that are mutated or downregulated in specific genes or gene family members of the general phenylpropanoid pathway, such as C4H3, 4CL1, C3’H1 and CCoAOMT (Table 1). Downregulation of C4H3 results in a decrease in lignin content of 14 to 17%, but no cell wall degradability tests have been reported for these lines (Abdel-Rahman and Mousa, 2016). The bm5 mutation that affects the 4CL1 gene results in a 10 to 20% decrease in lignin content without affecting the biomass yield (Méchin et al., 2014; Xiong et al., 2019). The neutral detergent fiber digestibility (NDFD) and saccharification efficiency of bm5 are increased on average by 18% and 22%, respectively (Xiong et al., 2019). Furthermore, the bm5 mutants show a 20 to 30% reduction in pCA ester levels and a 5- to 20-fold increase in the incorporation of ferulic acid into the lignin polymer (Méchin et al., 2014). Downregulation of C3’H1 by RNAi in maize results in a tendency towards a lower lignin content in stem tissue, while lignin content remains unchanged in the midrib (Fornalé et al., 2015). Additionally, the lignin composition in stem tissue shifts to an increased proportion of H units, a decreased frequency of S units and a tendency towards an increased proportion of G units. Moreover, although the in vitro digestibility is increased by 32%, the C3’H1-RNAi plants suffer from a growth reduction and male sterility (Fornalé et al., 2015). Downregulation of CCoAOMT1 in maize results in a decrease of 22% in lignin content and an increase in the S/G ratio. Besides a slight growth delay, the CCoAOMT1-RNAi lines are indistinguishable from wild-type (WT) plants (Li et al., 2013).
Maize lines with mutations in the monolignol-specific pathway have been reported for CCR, COMT and CAD (Table 1). Studies have shown that CCR-deficient plants have a lower lignin content, which translates into a significant increase in saccharification efficiency. For example, a Mu insertion in the first intron of the CCR1 gene results in a 31% reduction in CCR1 expression, leading to a 12% reduction in Klason lignin content, accompanied by an increase in the S/G ratio as well as a decreased frequency of H lignin units. These modifications are associated with a 15% improved in vitro digestibility. The ccr1 mutants do neither have a biomass yield penalty, nor a bm phenotype, when grown in the greenhouse (Tamasloukht et al., 2011). Similarly, when CCR1 is downregulated via an RNAi strategy, a reduction in Klason lignin content of 7 to 8% is accompanied by a 7 to 8% increased enzymatic conversion to fermentable sugars upon an ammonia fiber expansion pretreatment. However, six out of the twenty generated RNAi lines have a bm phenotype and a normal biomass yield, while 5% of the RNAi lines show stunted growth with curly leaves and aborted early flowering (Park et al., 2012). A third study examined the lignin content, composition and saccharification efficiency of a maize line with a Mu insertion in the fourth exon of CCR1. These maize lines have an approximately 20% lower lignin amount and show an up to 53% increased digestibility in a limited-extend digestibility test. These mutants have an increased S/G ratio and release more sinapyl ferulate units by DFRC relative to WT plants. These maize plants have neither a yield penalty nor a bm phenotype when grown in the greenhouse. Furthermore, the latter mutants have a 16% reduction in seed weight and an overall 28% increase in total biomass yield when grown in the field (Smith et al., 2017). Presumably, the differences in the phenotypes between the RNAi and stable ccr1 mutants are due to the possibility that in the RNAi lines, multiple CCR gene family members are simultaneously downregulated, resulting in further reductions in lignin content as compared to the lines where only CCR1 is mutated. These data also imply that one or more CCR gene family members other than CCR1 have a role in determining lignin content, and these gene family members still need to be pinpointed.
The bm3 mutants in maize have a defective COMT gene and exhibit an improved in vitro digestibility by up to 45% (Vignols et al., 1995). These mutants also have an average reduction of 20% in lignin content with a striking decrease in S units. Moreover, the esterified pCA content is consistently decreased, while in some instances, an increase in esterified ferulic acid levels is observed, but no differences in the level of crosslinking between arabinoxylans (Chabbert et al., 1994; Marita et al., 2003; Barrière et al., 2004b). Downregulation of CAD2 leads to a reduction in lignin content ranging from 4 to 20%, depending on the particular mutation or method used. Furthermore, a reduced content of pCA esters is observed, accompanied by an increase in coniferaldehyde, sinapaldehyde and ferulic acid incorporation into the lignin polymer (Ralph et al., 2008; Barrière et al., 2013; Liu et al., 2021b). Additionally, bm1 mutants, defective in CAD2, exhibit an improvement of up to 58% in saccharification efficiency (Xiong et al., 2020).
In addition, mutant maize lines have been described that have a reduced availability of S-adenosyl-methionine (SAM), the methyl donor for CCoAOMT and COMT (Figure 3). For example, bm2 mutants show a significant reduction in METHYLENE-TETRAHYDROFOLATE REDUCTASE 1 (MTHFR1) expression, resulting in a 7 to 17% decrease in lignin, an increase in the S/G ratio in stalks and a 21% increase in NDFD compared to WT plants (Barrière et al., 2004b; Tang et al., 2014). Furthermore, Wu et al. (2018) reported bm2 plants with lignin levels similar to WT but with an increase in the S/G ratio and fraction of H units. These changes lead to a notable 58% improvement in saccharification efficiency. On the other hand, the bm4 mutation impacts FOLYLPOLYGLUTAMATE SYNTHASE (FPGS) (Cossins and Chen, 1997; Mehrshahi et al., 2010). Similar to bm2, bm4 mutants accumulate 10 to 14% less lignin in the stalks, have an elevated S/G ratio and show an increase in NDFD by 43% (Barrière et al., 2004b; Li et al., 2015). The bm6 mutant phenotype is caused by a loss-of-function mutation in GTP-CYCLOHYDROLASE 1 (GCH1), encoding an enzyme functioning upstream of FPGS, where it mediates the first step in the tetrahydrofolate (THF) biosynthesis pathway. These mutants show a 6% reduction in lignin content and a 3% improvement in NDFD (Chen et al., 2012b; Leonard et al., 2021).
A transgenic line downregulated in PMT shows a substantial reduction in esterified pCA levels, accompanied by a decrease in S units (Marita et al., 2014). Furthermore, the naturally silenced colorless (c2) mutant, which is defective in the CHALCONE SYNTHASE gene, exhibits a 98% reduction in tricin incorporation in the leaves whereas tricin is below the detection limit in the mutant stems. Despite this, the c2 mutant exhibits WT lignin levels in the stem, although with a reduction in H units. Conversely, in leaf tissue, the c2 mutant shows a 27% increase in lignin content, which is accompanied by a decrease in saccharification efficiency of 39% (Eloy et al., 2017). Finally, maize lines with a defective allele of the peroxidase-encoding gene ZmPOX3, resulting from a MITE transposon insertion, exhibit higher forage digestibility compared to lines with similar genetic backgrounds but carrying a functional allele of ZmPOX3 (Guillet-Claude et al., 2004).
Only a limited number of maize lines has been described with altered levels of MYB or NAC transcription factors involved in lignin biosynthesis (Table 1). ZmMYB167 overexpression lines are not affected in their growth and development and exhibit an increase of 4 to 13% in lignin, 8 to 52% in pCA and 13 to 38% in ferulate esters in the internodes. Nevertheless, no changes in biomass recalcitrance are observed upon saccharification (Bhatia et al., 2019). Furthermore, maize lines in which ZmMYB69 is overexpressed display a decrease in plant height, as well as in vascular bundle cell wall thickness (Qiang et al. (2022). Significantly reduced levels of lignin are observed in the ZmMYB69 overexpression lines in comparison to the control, which is accompanied by increased saccharification efficiency. Conversely, thicker cell walls and a higher lignin content are observed in zmmyb69 loss-of-function lines, generated via CRISPR/Cas. These plants do not show any visible growth defects. In addition, both overexpression and downregulated lines were made for ZmNST3 and ZmNST4. The overexpression of ZmNST3 leads to a decrease in plant height and an increase in cell wall thickness, which is due to an increase in lignin and cellulose content in the internodes. Meanwhile, ZmNST4 overexpression appears to be lethal. The downregulation of either ZmNST3 or ZmNST4 results in a reduction in stem lignification and reduced plant height (Xiao et al., 2018).
Cell wall digestibility is a complex trait influenced primarily by factors such as lignin content, lignin composition and ferulate cross-linkages (Méchin et al., 2014; Torres et al., 2015b; Barrière, 2017). Plants with a lower lignin content generally have an improved digestibility, but it is not easy to discern the contribution of specific structural lignin alterations to the improvements in biomass digestibility (Halpin, 2019). For example, an increased proportion of H units may contribute to improved biomass digestibility in maize, by engendering smaller lignin polymers (Fornalé et al., 2012; Vanholme et al., 2012; Méchin et al., 2014; Fornalé et al., 2015; Wu et al., 2018). This has been achieved by suppressing C3’H1, but this engineering strategy additionally resulted in a reduction of total lignin (Fornalé et al., 2015). Furthermore, a positive correlation between S/G ratio and cell wall degradability in maize has been observed (Méchin et al., 2000, 2005; Zhang et al., 2011). This could likely be attributed to the different linkage type frequencies engendered by coupling of S and G units (Barrière et al., 2009a). Where lignin rich in S units is relatively linear and abundant in (8-O-4) ether bonds, lignin rich in G units has more carbon-carbon (8–5 and 5–5) linkages and branched structures (Ralph et al., 2008). Additionally, the observed positive correlation might also be caused by the confounding effect that S units, as compared to G units, are more prone to esterification into pCA esters, which have a positive effect on cell wall degradation (Méchin et al., 2000; Grabber et al., 2009; Zhang et al., 2011).
Maize lines mutant for enzymes acting upstream of coniferaldehyde (MTHFR, 4CL1, C3’H1, CCoAOMT, CCR1) show a reduction in both G and S lignin units, with a stronger decrease in G units, resulting in an increased S/G ratio, which could contribute to the increased cell wall digestibility (Barrière et al., 2004a; Tamasloukht et al., 2011; Smith et al., 2017; Xiong et al., 2020). In contrast, the decreased COMT activity in comt mutants strongly reduces the biosynthesis of sinapyl alcohol, resulting in a substantial decrease in the S/G ratio. The improved digestibility of comt mutants can primarily be attributed to their lower lignin content, while their reduced S/G lignin ratio would rather counteract the release of cell wall sugars (Fornalé et al., 2017). However, instead of S units, the lignin polymer contains 5-hydroxyguaiacyl (5-OH-guaiacyl) subunits, that give rise to benzodioxane structures, through the incorporation of 5-hydroxyconiferyl alcohol and 5-hydroxyconiferaldehyde (Marita et al., 2003). The increased presence of benzodioxane structures in the lignin polymer is thought to reduce cross-linking of lignin with cell wall carbohydrates, in this way contributing to the cell wall degradability (Weng et al., 2010; Vanholme et al., 2012).
In addition to 5-hydroxyconiferyl alcohol and 5-hydroxyconiferaldehyde, the incorporation of other intermediates from the lignin pathway can also facilitate cell wall processing. For example, a reduced CAD activity results in the integration of coniferaldehyde and sinapaldehyde into the lignin, thereby producing a lignin polymer with a higher proportion of free phenolic units (Lapierre et al., 1999; Ralph et al., 2001). Depending on the plant species and pretreatment used, incorporation of these hydroxycinnamaldehydes results in an improved cell wall processing (Fu et al., 2011; Fornalé et al., 2012; Van Acker et al., 2017; Liu et al., 2021b). In addition, reduced CAD activity also results in the incorporation of ferulic acid into the lignin polymer, presumably because its substrate coniferaldehyde is converted by HCALDH into ferulic acid (Nair et al., 2004; de Oliveira et al., 2015; Končitíková et al., 2015; Missihoun et al., 2016; Liu et al., 2021b). The incorporation of ferulic acid results – in contrast to GAX-bound ferulate moieties (see below) – in acetal bonds that are cleavable in acidic conditions (Ralph et al., 2008). However, in different lignin engineering strategies in a variety of plant species, the incorporation of ferulic acid appears to be marginal, presumably due to the low translocation efficiency of ferulic acid to the apoplast (Van Acker et al., 2013, 2014; Vermaas et al., 2019).
Considering that pCA is mainly esterified into sinapyl alcohol in maize, mutants with a reduced biosynthesis of sinapyl alcohol, such as cad2 and comt, show a reduced amount of pCA conjugates (Piquemal et al., 2002; Barrière et al., 2013; Méchin et al., 2014). However, the strongest reduction in cell wall bound pCA esters in maize was observed by downregulating PMT (Marita et al., 2014). The pCA moieties are pending groups with free phenolic ends that make the lignin polymer more soluble in alkaline pretreatment conditions (Hatfield et al., 2017; Lapierre et al., 2021). Therefore, the increase in pCA conjugates, e.g. by overexpression of PMT, is expected to improve the saccharification efficiency of the maize biomass.
GAX-bound ferulates allow cross-links between xylan chains and between xylans and lignin. Such cross-linkages have a negative effect on maize cell walls degradability (Grabber et al., 1998, 2009; de Oliveira et al., 2015). Downregulation of BAHD genes encoding acyltransferases with FAT activity in Brachypodium, rice and Setaria results in a drop in GAX-bound ferulates (Piston et al., 2010; Buanafina et al., 2016; de Souza et al., 2018). Expression analysis in maize hinted five genes encoding BAHD acyltransferases as potential FAT genes, however no functional analysis has been undertaken yet to validate their function (Chateigner-Boutin et al., 2016). The seedling ferulate ester (sfe) maize mutant was selected from a transposon mutant stock, by screening for seedlings with reduced ferulate ester content, but the mutated gene causing this phenotype is not yet identified (Table 1) (Jung and Phillips, 2010; Hatfield et al., 2018). The sfe mutant biomass does not only show reduced ferulate levels, but also has lower lignin levels, and improved rumen digestibility as evidenced by its performance in animal feeding trials (Jung et al., 2011).
Concerns often arise that plants with reduced lignin levels would have increased susceptibility to pests, diseases, reduced biomass yield and lodging. Indeed, lignin does play an important role in these agronomic traits and some maize mutants with altered lignin were reported to have such unfavorable characteristics (Table 1) (Oliveira et al., 2020; Ren et al., 2020). On the other hand, it seems that lignin content variations within the natural maize population have little discernible impact on its agronomic performances (Pedersen et al., 2005; Barrière, 2017; Manga-Robles et al., 2021). In addition, promising inbred lines have already been developed with enhanced fermentable sugar yields that rival or surpass the bm mutant. These lines exhibit superior tolerance to fall armyworm compared to their isogenic counterparts (Vermerris et al., 2007). Similarly, another study also observed no close correlation between insect susceptibility and the lignin content (Williams et al., 1998).
The bm3 mutants have demonstrated the most significant improvement in in vitro cell wall digestibility as compared to other bm mutants, making them the most promising among all bm mutants for breeding purposes. However, a bm3 mutation often results in a reduction of 15 to 20% in dry matter yield as compared to their non-mutant isogenic controls (Inoue and Kasuga, 1989). In contrast, two studies reported no biomass yield reductions associated with the bm3 mutation in certain genetic backgrounds as compared to their isogenic control lines (Weller et al., 1985; Gentinetta et al., 1990). The bm3 mutants are also often associated with reduced stalk strength, increased susceptibility towards pathogens and drought, and precocious senescence (Nicholson et al., 1976; Zuber et al., 1977; Vermerris et al., 2010; Akhter et al., 2018). However, several studies were not able to detect a correlation between increased lodging susceptibility and bm3 mutants (Weller et al., 1985; Inoue and Kasuga, 1989). Recently, some of these pleiotropic effects of a bm3 mutation, such as precocious senescence and low drought tolerance, were counteracted by the introgression of a BRACHYTIC2 (BR2) mutation, resulting in a bm3 br2 double mutant (Landoni et al., 2022). In conclusion, there are strong interactions between the gene of choice, the genetic background and the environment in which the plants are grown. Therefore, the agronomic performance will largely be determined by these factors (Pedersen et al., 2005).
Lignin plays a fundamental role in various agronomically relevant traits. Lignin offers tolerance against biotic stresses, provides structural support to stems to prevent lodging, yet it also acts as a biomass recalcitrance factor limiting cell wall degradability. Current evidence suggests that reducing lignin content in maize can be achieved without compromising plant growth and biomass yield by choosing the right gene or by introgression of the corresponding mutation into the most appropriate genetic background. There is still considerable potential for new lignin-engineering strategies in maize, as many genes involved in lignin biosynthesis and its regulation have yet to be investigated (Barrière et al., 2015; Penning et al., 2019). Genomic studies have shown that lignin biosynthesis genes are often part of multigene families, of which in most cases only a single member has been analyzed. Knocking-out multiple members of the same gene family, which has only now become possible by CRISPR-based gene editing, is a promising strategy to further improve the processability of lignocellulosic biomass. In addition, CRISPR-based gene editing allows the generation of a range of alleles, not only knock-out alleles, but also weak alleles that should allow the tuning of lignin content and composition without affecting yield, as illustrated by editing CCR2 in poplar (De Meester et al., 2020). Furthermore, by analyzing lignin mutants and transgenics, it has become clear that lignin is tolerant towards large compositional shifts. This opens perspectives to redirect the lignin pathway towards the overproduction of rare natural monomers, or even to let the plant synthesize alternative monomers by expressing exotic genes. For example, ectopic overexpression of the two penultimate genes of the scopoletin biosynthesis pathway in lignifying cells in Arabidopsis has resulted in the incorporation of scopoletin into the lignin polymer, making it more susceptible to alkaline pretreatments (Hoengenaert et al., 2022). Several other examples and strategies exist and have been reviewed (Mottiar et al., 2016; Vanholme et al., 2019; de Vries et al., 2021).
For future research and applications, a few critical factors need to be considered with regard to the genotype to work with, and the environment where the experiments are carried out. First, it needs to be recognized that most studies on the effects of lignin engineering on digestibility have been conducted in greenhouses. The main reason for this is that the effect of a genetic modification is easier to determine in a stable greenhouse environment. Experiments with transgenic plants under field conditions require a regulatory permit, which discourages most researchers from conducting field trials. Nevertheless, field trials are essential to validate the effect of the mutation. Field trials represent a realistic environment with fluctuating weather conditions (wind, UV radiation, rain, drought), exposure to abiotic stressors and variations in soil type and soil microorganisms. Furthermore, the planting density in the field differs from that of plants grown in pots under the controlled greenhouse settings. Thus, results obtained in the greenhouse may not always align with observations made under field conditions (Tuberosa, 2012; Nelissen et al., 2014). When improving a commercially relevant trait, it is therefore advisable to conduct the experiments directly in the field to select the most promising mutants, and only then study their more subtle effects in a greenhouse setting for purely scientific purposes. Second, it is important to recognize that genotypes typically used in experiments and transformation (laboratory strains) are not optimized for cell wall degradability. As a result, genomic modifications can lead to substantial improvements in the digestibility in these genotypes, whereas the improvement does not necessarily translate into elite inbreds or their hybrids. It will be essential to either edit elite inbred lines if transformation protocols exist for them, or to use other methods such as transgenerational editing. Importantly, when the mutations are recessive, both elite inbred lines will need to be edited in order to create a mutant hybrid. Only when the mutant hybrid outperforms, and preferentially in different environments, the strategy can be considered successful. Navigating the future of maize breeding requires embracing cutting-edge gene editing technology. By addressing the challenges associated with genotype selection and field validation, we can adopt a sharper approach to develop modern maize varieties that contribute to a sustainable agriculture and bio-based economy.
YV: Writing – original draft, Writing – review & editing. ADM: Writing – original draft, Writing – review & editing. HM: Writing – original draft, Writing – review & editing. RV: Writing – original draft, Writing – review & editing. WB: Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the Energy Transition Fund (ADV-BIO) to ADM. YV is indebted to VLAIO for a predoctoral fellowship (HBC.2019.2159).
We would like to thank Annick Bleys for proofreading the manuscript. Figures 1, 2 were created with BioRender.com.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdel-Rahman, M. M., Mousa, I. E. (2016). Effects of down regulation of lignin content in maize (Zea mays L.) plants expressing C4H3 gene in the antisense orientation. Biofuels 7, 289–294. doi: 10.1080/17597269.2015.1132374
Aesaert, S., Impens, L., Coussens, G., Van Lerberge, E., Vanderhaeghen, R., Desmet, L., et al. (2022). Optimized transformation and gene editing of the B104 public maize inbred by improved tissue culture and use of morphogenic regulators. Front. Plant Sci. 13, 883847. doi: 10.3389/fpls.2022.883847
Akhter, D., Qin, R., Nath, U. K., Alamin, M., Jin, X., Shi, C. (2018). The brown midrib leaf (bml) mutation in rice (Oryza sativa l.) causes premature leaf senescence and the induction of defense responses. Genes 9, 203. doi: 10.3390/genes9040203
Akinwale, R. O. (2021). Heterosis and heterotic grouping among tropical maize germplasm. Cereal Grains 2, 59–67. doi: 10.5772/intechopen.98742
Allen, M. S., Coors, J. G., Roth, G. W. (2003). Corn silage. Silage Sci. Technol. 42, 547–608. doi: 10.2134/agronmonogr42.c12
Andorf, C., Beavis, W. D., Hufford, M., Smith, S., Suza, W. P., Wang, K., et al. (2019). Technological advances in maize breeding: past, present and future. Theor. Appl. Genet. 132, 817–849. doi: 10.1007/s00122-019-03306-3
Awata, L. A. O., Tongoona, P., Danquah, E., Ifie, B. E., Mahabaleswara, S. L., Jumbo, M. B., et al. (2019). Understanding tropical maize (Zea mays L.): The major monocot in modernization and sustainability of agriculture in sub-Saharan Africa. Int. J. Adv. Agric. Res. 7, 32–77. doi: 10.33500/ijaar.2019.07.004
Barrière, Y., Riboulet, C., Méchin, V., Maltese, S., Pichon, M., Cardinal, A., et al. (2007). Genetics and genomics of lignification in grass cell walls based on maize as model species. Genes Genomes Genomics 1, 133–156.
Barrière, Y. (2017). Brown-midrib genes in maize and their efficiency in dairy cow feeding. Perspectives for breeding improved silage maize targeting gene modifications in the monolignol and p-hydroxycinnamate pathways. Maydica 62, 1–19.
Barrière, Y., Alber, D., Dolstra, O., Lapierre, C., Motto, M., Ordás Pérez, A., et al. (2005). Past and prospects of forage maize breeding in Europe. I. The grass cell wall as a basis of genetic variation and future improvements in feeding value. Maydica 50, 259–274.
Barrière, Y., Alber, D., Dolstra, O., Lapierre, C., Motto, M., Ordás Pérez, A., et al. (2006). Past and prospects of forage maize breeding in Europe. II. History, germplasm evolution and correlative agronomic changes. Maydica 51, 435–449.
Barrière, Y., Argillier, O., Chabbert, B., Tollier, M. T., Monties, B. (1994). Breeding silage maize with brown-midrib genes. Feeding value and biochemical characteristics. Agronomie 14, 15–25. doi: 10.1051/agro:19940102
Barrière, Y. Y., Chavigneau, H., Delaunay, S., Courtial, A. A., Bosio, M., Lassagne, H., et al. (2013). Different mutations in the ZmCAD2 gene underlie the maize brown-midrib1 (bm1) phenotype with similar effects on lignin characteristics and have potential interest for bioenergy production. Maydica 58, 6–20.
Barrière, Y., Courtial, A., Chateigner-Boutin, A.-L., Denoue, D., Grima-Pettenati, J. (2016). Breeding maize for silage and biofuel production, an illustration of a step forward with the genome sequence. Plant Sci. 242, 310–329. doi: 10.1016/j.plantsci.2015.08.007
Barrière, Y., Courtial, A., Soler, M., Grima-Pettenati, J. (2015). Toward the identification of genes underlying maize QTLs for lignin content, focusing on colocalizations with lignin biosynthetic genes and their regulatory MYB and NAC transcription factors. Mol. Breed. 35, 87. doi: 10.1007/s11032-015-0275-8
Barrière, Y., Emile, J. C., Traineau, R., Surault, F., Briand, M., Gallais, A. (2004a). Genetic variation for organic matter and cell wall digestibility in silage maize. Lessons from a 34-year long experiment with sheep in digestibility crates. Maydica 49, 115–126.
Barrière, Y., Gallais, A., Derieux, M., Panouillé, A. (1987). Etude de la valeur agronomique en plante entière au stade de récolte ensilage de différentes variétés de maïs grain sélectionnées entre 1950 et 1980. Agronomie 7, 73–79. doi: 10.1051/agro:19870201
Barrière, Y., Guillaumie, S., Denoue, D., Pichon, M., Goffner, D., Martinant, J.-P. (2017). Investigating the unusually high cell wall digestibility of the old INRA early flint F4 maize inbred line. Maydica 62, M31.
Barrière, Y., Guillaumie, S., Pichon, M., Emile, J. C. (2009a). “Breeding for silage quality traits,” in Handbook of Plant Breeding. Ed. Carena, M. (Springer, New York, NY), 367–394. doi: 10.1007/978-0-387-72297-9
Barrière, Y., Méchin, V., Lafarguette, F., Manicacci, D., Guillon, F., Wang, H., et al. (2009b). Toward the discovery of maize cell wall genes involved in silage quality and capacity to biofuel production. Maydica 54, 161–198.
Barrière, Y., Ralph, J., Méchin, V., Guillaumie, S., Grabber, J. H., Argillier, O., et al. (2004b). Genetic and molecular basis of grass cell wall biosynthesis and degradability. II. Lessons from brown-midrib mutants. C. R. Biol. 327, 847–860. doi: 10.1016/j.crvi.2004.05.010
Barros, J., Dixon, R. A. (2020). Plant phenylalanine/tyrosine ammonia-lyases. Trends Plant Sci. 25, 66–79. doi: 10.1016/j.tplants.2019.09.011
Barros, J., Escamilla-Trevino, L., Song, L., Rao, X., Serrani-Yarce, J. C., Palacios, M. D., et al. (2019). 4-Coumarate 3-hydroxylase in the lignin biosynthesis pathway is a cytosolic ascorbate peroxidase. Nat. Commun. 10, 1994. doi: 10.1038/s41467-019-10082-7
Barros, J., Serrani-Yarce, J. C., Chen, F., Baxter, D., Venables, B. J., Dixon, R. A. (2016). Role of bifunctional ammonia-lyase in grass cell wall biosynthesis. Nat. Plants 2, 16050. doi: 10.1038/nplants.2016.50
Beal, W. J. (1878). The improvement of grains, fruits, and vegetables. Seventeenth An. Rep. State Board Agr. Mich 450.
Beckett, T. J., Rocheford, T. R., Mohammadi, M. (2019). Reimagining maize inbred potential: identifying breeding crosses using genetic variance of simulated progeny. Crop Sci. 59, 1457–1468. doi: 10.2135/cropsci2018.08.0508
Bennetzen, J. L., Hake, S. (2009). Handbook of maize: genetics and genomics (New York: Springer). doi: 10.1007/978-0-387-77863-1
Bennetzen, J. L., Springer, P. S., Cresse, A. D., Hendrickx, M. (1993). Specificity and regulation of the mutator transposable element system in maize. Crit. Rev. Plant Sci. 12, 57–95. doi: 10.1080/07352689309382356
Bhatia, R., Dalton, S., Roberts, L. A., Moron-Garcia, O. M., Iacono, R., Kosik, O., et al. (2019). Modified expression of ZmMYB167 in Brachypodium distachyon and Zea mays leads to increased cell wall lignin and phenolic content. Sci. Rep. 9, 8800. doi: 10.1038/s41598-019-45225-9
Bhatia, R., Gallagher, J. A., Gomez, L. D., Bosch, M. (2017). Genetic engineering of grass cell wall polysaccharides for biorefining. Plant Biotechnol. J. 15, 1071–1092. doi: 10.1111/pbi.12764
Bi, Y., Jiang, F., Zhang, Y., Li, Z., Kuang, T., Shaw, R. K., et al. (2024). Identification of a novel marker and its associated laccase gene for regulating ear length in tropical and subtropical maize lines. Theor. Appl. Genet. 137, 94. doi: 10.1007/s00122-024-04587-z
Boerjan, W., Ralph, J., Baucher, M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54, 519–546. doi: 10.1146/annurev.arplant.54.031902.134938
Bortesi, L., Fischer, R. (2015). The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 33, 41–52. doi: 10.1016/j.biotechadv.2014.12.006
Bouchet, S., Servin, B., Bertin, P., Madur, D., Combes, V., Dumas, F., et al. (2013). Adaptation of maize to temperate climates: mid-density genome-wide association genetics and diversity patterns reveal key genomic regions, with a major contribution of the Vgt2 (ZCN8) locus. PloS One 8, e71377. doi: 10.1371/journal.pone.0071377
Buanafina, M.M.d. O. (2009). Feruloylation in grasses: current and future perspectives. Mol. Plant 2, 861–872. doi: 10.1093/mp/ssp067
Buanafina, M.M.d. O., Fescemyer, H. W., Sharma, M., Shearer, E. A. (2016). Functional testing of a PF02458 homologue of putative rice arabinoxylan feruloyl transferase genes in Brachypodium distachyon. Planta 243, 659–674. doi: 10.1007/s00425-015-2430-1
Chabbert, B., Tollier, M. T., Monties, B., Barrière, Y., Argillier, O. (1994). Biological variability in lignification of maize: expression of the brown midrib bm3 mutation in three maize cultivars. J. Sci. Food Agric. 64, 349–355. doi: 10.1002/jsfa.2740640317
Chandrakanth, N. N., Zhang, C., Freeman, J., de Souza, W. R., Bartley, L. E., Mitchell, R. A. C. (2023). Modification of plant cell walls with hydroxycinnamic acids by BAHD acyltransferases. Front. Plant Sci. 13, 1088879. doi: 10.3389/fpls.2022.1088879
Chateigner-Boutin, A.-L., Ordaz-Ortiz, J. J., Alvarado, C., Bouchet, B., Durand, S., Verhertbruggen, Y., et al. (2016). Developing pericarp of maize: a model to study arabinoxylan synthesis and feruloylation. Front. Plant Sci. 7, 1476. doi: 10.3389/fpls.2016.01476
Chen, Y., Liu, H., Ali, F., Scott, M. P., Ji, Q., Frei, U. K., et al. (2012b). Genetic and physical fine mapping of the novel brown midrib gene bm6 in maize (Zea mays L.) to a 180 kb region on chromosome 2. Theor. Appl. Genet. 125, 1223–1235. doi: 10.1007/s00122-012-1908-5
Chen, W., VanOpdorp, N., Fitzl, D., Tewari, J., Friedemann, P., Greene, T., et al. (2012a). Transposon insertion in a cinnamyl alcohol dehydrogenase gene is responsible for a brown midrib1 mutation in maize. Plant Mol. Biol. 80, 289–297. doi: 10.1007/s11103-012-9948-4
Choo, T., Kannenberg, L. (1981). Comparison of predicted and simulated responses to mass selection in a diploid, cross-fertilized species. Can. J. Plant Sci. 61, 1–8. doi: 10.4141/cjps81-001
Coe, E. H., Jr (1959). A line of maize with high haploid frequency. Am. Nat. 93, 381–382. doi: 10.1086/282098
Cossins, E. A., Chen, L. (1997). Folates and one-carbon metabolism in plants and fungi. Phytochemistry 45, 437–452. doi: 10.1016/S0031-9422(96)00833-3
Courtial, A., Soler, M., Chateigner-Boutin, A.-L., Rey-Mond, M., Méchin, V., Wang, H., et al. (2013). Breeding grasses for capacity to biofuel production or silage feeding value: an updated list of genes involved in maize secondary cell wall biosynthesis and assembly. Maydica 58, 67–102.
De Meester, B., Madariaga Calderón, B., de Vries, L., Pollier, J., Goeminne, G., Van Doorsselaere, J., et al. (2020). Tailoring poplar lignin without yield penalty by combining a null and haploinsufficient CINNAMOYL-CoA REDUCTASE2 allele. Nat. Commun. 11, 5020. doi: 10.1038/s41467-020-18822-w
de Oliveira, D. M., Finger-Teixeira, A., Rodrigues Mota, T., Salvador, V. H., Moreira-Vilar, F. C., Correa Molinari, H. B., et al. (2015). Ferulic acid: a key component in grass lignocellulose recalcitrance to hydrolysis. Plant Biotechnol. J. 13, 1224–1232. doi: 10.1111/pbi.12292
de Souza, W. R., Martins, P. K., Freeman, J., Pellny, T. K., Michaelson, L. V., Sampaio, B. L., et al. (2018). Suppression of a single BAHD gene in Setaria viridis causes large, stable decreases in cell wall feruloylation and increases biomass digestibility. New Phytol. 218, 81–93. doi: 10.1111/nph.14970
de Vries, L., Guevara-Rozo, S., Cho, M., Liu, L.-Y., Renneckar, S., Mansfield, S. D. (2021). Tailoring renewable materials via plant biotechnology. Biotechnol. Biofuels 14, 167. doi: 10.1186/s13068-021-02010-z
Dixon, R. A., Barros, J. (2019). Lignin biosynthesis: old roads revisited and new roads explored. Open Biol. 9, 190215. doi: 10.1098/rsob.190215
Dixon, K. A., Michelsen, M. K., Carpenter, C. L. (2023). Modern diets and the health of our planet: An investigation into the environmental impacts of food choices. Nutrients 15, 692. doi: 10.3390/nu15030692
Eloy, N. B., Voorend, W., Lan, W., Saleme, M. D. S., Cesarino, I., Vanholme, R., et al. (2017). Silencing CHALCONE SYNTHASE in maize impedes the incorporation of tricin into lignin and increases lignin content. Plant Physiol. 173, 998–1016. doi: 10.1104/pp.16.01108
Escamilla-Treviño, L., Shen, H., Hernandez, T., Yin, Y., Xu, Y., Dixon, R. A. (2014). Early lignin pathway enzymes and routes to chlorogenic acid in switchgrass (Panicum virgatum L.). Plant Mol. Biol. 84, 565–576. doi: 10.1007/s11103-013-0152-y
Fanelli, A., Rancour, D. M., Sullivan, M., Karlen, S. D., Ralph, J., Riaño-Pachón, D. M., et al. (2021). Overexpression of a sugarcane BAHD acyltransferase alters hydroxycinnamate content in maize cell wall. Front. Plant Sci. 12, 626168. doi: 10.3389/fpls.2021.626168
Fontaine, A.-S., Briand, M., Barrière, Y. (2003). Genetic variation and QTL mapping of para-coumaric and ferulic acid contents in maize stover at silage harvest. Maydica 48, 75–84.
Fornalé, S., Capellades, M., Encina, A., Wang, K., Irar, S., Lapierre, C., et al. (2012). Altered lignin biosynthesis improves cellulosic bioethanol production in transgenic maize plants down-regulated for cinnamyl alcohol dehydrogenase. Mol. Plant 5, 817–830. doi: 10.1093/mp/ssr097
Fornalé, S., Rencoret, J., Garcia-Calvo, L., Capellades, M., Encina, A., Santiago, R., et al. (2015). Cell wall modifications triggered by the down-regulation of Coumarate 3-hydroxylase-1 in maize. Plant Sci. 236, 272–282. doi: 10.1016/j.plantsci.2015.04.007
Fornalé, S., Rencoret, J., García-Calvo, L., Encina, A., Rigau, J., Gutiérrez, A., et al. (2017). Changes in cell wall polymers and degradability in maizemutants lacking 3'- and 5'-O-methyltransferases involvedin lignin biosynthesis. Plant Cell Physiol. 58, 240–255. doi: 10.1093/pcp/pcw198
Fornalé, S., Shi, X., Chai, C., Encina, A., Irar, S., Capellades, M., et al. (2010). ZmMYB31 directly represses maize lignin genes and redirects the phenylpropanoid metabolic flux. Plant J. 64, 633–644. doi: 10.1111/tpj.2010.64.issue-4
Fornalé, S., Sonbol, F. M., Maes, T., Capellades, M., Puigdomènech, P., Rigau, J., et al. (2006). Down-regulation of the maize and Arabidopsis thaliana caffeic acid O-methyl-transferase genes by two new maize R2R3-MYB transcription factors. Plant Mol. Biol. 62, 809–823. doi: 10.1007/s11103-006-9058-2
Frame, B. R., McMurray, J. M., Fonger, T. M., Main, M. L., Taylor, K. W., Torney, F. J., et al. (2006). Improved Agrobacterium-mediated transformation of three maize inbred lines using MS salts. Plant Cell Rep. 25, 1024–1034. doi: 10.1007/s00299-006-0145-2
Fu, C., Xiao, X., Xi, Y., Ge, Y., Chen, F., Bouton, J., et al. (2011). Downregulation of cinnamyl alcohol dehydrogenase (CAD) leads to improved saccharification efficiency in switchgrass. Bioenergy Res. 4, 153–164. doi: 10.1007/s12155-010-9109-z
Gentinetta, E., Bertolini, M., Rossi, I., Lorenzoni, C., Motto, M. (1990). Effect of brown midrib-3 mutant on forage and yield in maize. J. Genet. Breed. 44, 21–26.
Gerke, J. P., Edwards, J. W., Guill, K. E., Ross-Ibarra, J., McMullen, M. D. (2015). The genomic impacts of drift and selection for hybrid performance in maize. Genetics 201, 1201–1211. doi: 10.1534/genetics.115.182410
Gilles, L. M., Khaled, A., Laffaire, J. B., Chaignon, S., Gendrot, G., Laplaige, J., et al. (2017). Loss of pollen-specific phospholipase NOT LIKE DAD triggers gynogenesis in maize. EMBO J. 36, 707–717. doi: 10.15252/embj.201796603
Grabber, J. H., Hatfield, R. D., Lu, F., Ralph, J. (2008). Coniferyl ferulate incorporation into lignin enhances the alkaline delignification and enzymatic degradation of cell walls. Biomacromolecules 9, 2510–2516. doi: 10.1021/bm800528f
Grabber, J. H., Mertens, D. R., Kim, H., Funk, C., Lu, F., Ralph, J. (2009). Cell wall fermentation kinetics are impacted more by lignin content and ferulate cross-linking than by lignin composition. J. Sci. Food Agric. 89, 122–129. doi: 10.1002/jsfa.3418
Grabber, J. H., Ralph, J., Hatfield, R. D. (1998). Ferulate cross-links limit the enzymatic degradation of synthetically lignified primary walls of maize. J. Agric. Food Chem. 46, 2609–2614. doi: 10.1021/jf9800099
Guillaumie, S., San-Clemente, H., Deswarte, C., Martinez, Y., Lapierre, C., Murigneux, A., et al. (2007). MAIZEWALL. Database and developmental gene expression profiling of cell wall biosynthesis and assembly in maize. Plant Physiol. 143, 339–363. doi: 10.1104/pp.106.086405
Guillet-Claude, C., Birolleau-Touchard, C., Manicacci, D., Rogowsky, P. M., Rigau, J., Murigneux, A., et al. (2004). Nucleotide diversity of the ZmPox3 maize peroxidase gene: relationships between a MITE insertion in exon 2 and variation in forage maize digestibility. BMC Genet. 5, 19. doi: 10.1186/1471-2156-5-19
Guo, D., Chen, F., Inoue, K., Blount, J. W., Dixon, R. A. (2001). Downregulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic alfalfa: impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell 13, 73–88. doi: 10.1105/tpc.13.1.73
Hallauer, A. R., Carena, M. J., Miranda Filho, J. B. (2010). Quantitative genetics in maize breeding (New York: Springer). doi: 10.1007/978-1-4419-0766-0
Halpin, C. (2019). Lignin engineering to improve saccharification and digestibility in grasses. Curr. Opin. Biotechnol. 56, 223–229. doi: 10.1016/j.copbio.2019.02.013
Halpin, C., Holt, K., Chojecki, J., Oliver, D., Chabbert, B., Monties, B., et al. (1998). Brown-midrib maize (bm1)-a mutation affecting the cinnamyl alcohol dehydrogenase gene. Plant J. 14, 545–553. doi: 10.1046/j.1365-313X.1998.00153.x
Hamdan, M. F., Mohd Noor, S. N., Abd-Aziz, N., Pua, T.-L., Tan, B. C. (2022). Green revolution to gene revolution: technological advances in agriculture to feed the world. Plants 11, 1297. doi: 10.3390/plants11101297
Hasan, N., Choudhary, S., Naaz, N., Sharma, N., Laskar, R. A. (2021). Recent advancements in molecular marker-assisted selection and applications in plant breeding programs. J. Genet. Eng. Biotechnol. 19, 128. doi: 10.1186/s43141-021-00231-1
Hatfield, R. D., Jung, H., Marita, J. M., Kim, H. (2018). Cell wall characteristics of a maize mutant selected for decreased ferulates. Am. J. Plant Sci. 9, 446–466. doi: 10.4236/ajps.2018.93034
Hatfield, R. D., Marita, J. M., Frost, K., Grabber, J., Ralph, J., Lu, F., et al. (2009). Grass lignin acylation: p-coumaroyl transferase activity and cell wall characteristics of C3 and C4 grasses. Planta 229, 1253–1267. doi: 10.1007/s00425-009-0900-z
Hatfield, R. D., Rancour, D. M., Marita, J. M. (2017). Grass cell walls: A story of cross-linking. Front. Plant Sci. 7, 2056. doi: 10.3389/fpls.2016.02056
He, X., Hall, M. B., Gallo-Meagher, M., Smith, R. L. (2003). Improvement of forage quality by downregulation of maize O-methyltransferase. Crop Sci. 43, 2240–2251. doi: 10.2135/cropsci2003.2240
Henchion, M., Zimmermann, J. (2021). Animal food products: Policy, market and social issues and their influence on demand and supply of meat. Proc. Nutr. Soc. 80, 252–263. doi: 10.1017/S0029665120007971
Hernandes-Lopes, J., de Carvalho Yassitepe, J. E., Koltun, A., Pauwels, L., Heinzen da Silva, V. C., Dante, R. A., et al. (2023). Genome editing in maize: Toward improving complex traits in a global crop. Genet. Mol. Biol. 46, e20220217. doi: 10.1590/1678-4685-gmb-2022-0217
Hoengenaert, L., Wouters, M., Kim, H., De Meester, B., Morreel, K., Vandersyppe, S., et al. (2022). Overexpression of the scopoletin biosynthetic pathway enhances lignocellulosic biomass processing. Sci. Adv. 8, abo5738. doi: 10.1126/sciadv.abo5738
Hopkins, C. G. (1899). Improvement in the chemical composition of the corn kernel. J. Am. Chem. Soc 21, 1039–1057. doi: 10.1021/ja02061a012
Impens, L., Jacobs, T. B., Nelissen, H., Inze, D., Pauwels, L. (2022). Mini-review: Transgenerational CRISPR/Cas9 gene editing in plants. Front. Genome Ed. 4, 825042. doi: 10.3389/fgeed.2022.825042
Impens, L., Lorenzo, C. D., Vandeputte, W., Wytynck, P., Debray, K., Haeghebaert, J., et al. (2023). Combining multiplex gene editing and doubled haploid technology in maize. New Phytol. 239, 1521–1532. doi: 10.1111/nph.19021
Inoue, N., Kasuga, S. (1989). Agronomic traits and nutritive value of stover in brown midrib-3 maize hybrids. J. Japanese Soc. Grassland Sci. 35, 220–227.
Jackson, D. (2017). No sex please, we're (in)breeding. EMBO J. 36, 703–704. doi: 10.15252/embj.201796735
Jackson, M. A., Anderson, D. J., Birch, R. G. (2013). Comparison of Agrobacterium and particle bombardment using whole plasmid or minimal cassette for production of high-expressing, low-copy transgenic plants. Transgenic Res. 22, 143–151. doi: 10.1007/s11248-012-9639-6
Jacquier, N. M. A., Gilles, L. M., Martinant, J.-P., Rogowsky, P. M., Widiez, T. (2021). Maize in planta haploid inducer lines: A cornerstone for doubled haploid technology. Methods Mol. Biol. 2288, 25–48. doi: 10.1007/978-1-0716-1335-1_2
Jiang, Y.-Y., Chai, Y.-P., Lu, M.-H., Han, X.-L., Lin, Q., Zhang, Y., et al. (2020). Prime editing efficiently generates W542L and S621I double mutations in two ALS genes in maize. Genome Biol. 21, 257. doi: 10.1186/s13059-020-02170-5
Jiang, C., Sun, J., Li, R., Yan, S., Chen, W., Guo, L., et al. (2022). A reactive oxygen species burst causes haploid induction in maize. Mol. Plant 15, 943–955. doi: 10.1016/j.molp.2022.04.001
Jung, H. G., Mertens, D. R., Phillips, R. L. (2011). Effect of reduced ferulate-mediated lignin/arabinoxylan cross-linking in corn silage on feed intake, digestibility, and milk production. J. Dairy Sci. 94, 5124–5137. doi: 10.3168/jds.2011-4495
Jung, H. G., Phillips, R. L. (2010). Putative seedling ferulate ester (sfe) maize mutant: morphology, biomass yield, and stover cell wall composition and rumen degradability. Crop Sci. 50, 403–418. doi: 10.2135/cropsci2009.04.0191
Kalinowska, K., Chamas, S., Unkel, K., Demidov, D., Lermontova, I., Dresselhaus, T., et al. (2019). State-of-the-art and novel developments of in vivo haploid technologies. Theor. Appl. Genet. 132, 593–605. doi: 10.1007/s00122-018-3261-9
Karlen, S. D., Free, H. C. A., Padmakshan, D., Smith, B. G., Ralph, J., Harris, P. J. (2018). Commelinid monocotyledon lignins are acylated by p-coumarate. Plant Physiol. 177, 513–521. doi: 10.1104/pp.18.00298
Karlen, S. D., Zhang, C., Peck, M. L., Smith, R. A., Padmakshan, D., Helmich, K. E., et al. (2016). Monolignol ferulate conjugates are naturally incorporated into plant lignins. Sci. Adv. 2, e1600393. doi: 10.1126/sciadv.1600393
Kelliher, T., Starr, D., Richbourg, L., Chintamanani, S., Delzer, B., Nuccio, M. L., et al. (2017). MATRILINEAL, a sperm-specific phospholipase, triggers maize haploid induction. Nature 542, 105–109. doi: 10.1038/nature20827
Kelliher, T., Starr, D., Su, X., Tang, G., Chen, Z., Carter, J., et al. (2019). One-step genome editing of elite crop germplasm during haploid induction. Nat. Biotechnol. 37, 287–292. doi: 10.1038/s41587-019-0038-x
Kim, J.-H., Hwa Yang, D., Kim, J.-S., Baek, M.-H., Mi Pal, Y., Gon Wi, S., et al. (2006). Cloning, characterization, and expression of two cDNA clones for a rice ferulate-5-hydroxylase gene, a cytochrome P450-dependent monooxygenase. J. Plant Biol. 49, 200–204. doi: 10.1007/BF03030533
Končitíková, R., Vigouroux, A., Kopečná, M., Andree, T., Bartoš, J., Šebela, M., et al. (2015). Role and structural characterization of plant aldehyde dehydrogenases from family 2 and family 7. Biochem. J. 468, 109–123. doi: 10.1042/BJ20150009
Labroo, M. R., Studer, A. J., Rutkoski, J. E. (2021). Heterosis and hybrid crop breeding: A multidisciplinary review. Front. Genet. 12, 643761. doi: 10.3389/fgene.2021.643761
Lan, W., Lu, F., Regner, M., Zhu, Y., Rencoret, J., Ralph, S. A., et al. (2015). Tricin, a flavonoid monomer in monocot lignification. Plant Physiol. 167, 1284–1295. doi: 10.1104/pp.114.253757
Lan, W., Morreel, K., Lu, F., Rencoret, J., del Río, J. C., Voorend, W., et al. (2016). Maize tricin-oligolignol metabolites and their implications for monocot lignification. Plant Physiol. 171, 810–820. doi: 10.1104/pp.16.02012
Landoni, M., Cassani, E., Ghidoli, M., Colombo, F., Sangiorgio, S., Papa, G., et al. (2022). Brachytic2 mutation is able to counteract the main pleiotropic effects of brown midrib3 mutant in maize. Sci. Rep. 12, 2446. doi: 10.1038/s41598-022-06428-9
Lapierre, C., Pollet, B., Petit-Conil, M., Toval, G., Romero, J., Pilate, G., et al. (1999). Structural alterations of lignins in transgenic poplars with depressed cinnamyl alcohol dehydrogenase or caffeic acid O-methyltransferase activity have an opposite impact on the efficiency of industrial kraft pulping. Plant Physiol. 119, 153–164. doi: 10.1104/pp.119.1.153
Lapierre, C., Sibout, R., Laurans, F., Lesage-Descauses, M.-C., Déjardin, A., Pilate, G. (2021). p-Coumaroylation of poplar lignins impacts lignin structure and improves wood saccharification. Plant Physiol. 187, 1374–1386. doi: 10.1093/plphys/kiab359
Lee, M. (1995). DNA markers and plant breeding programs. Adv. Agron. 55, 265–344. doi: 10.1016/S0065-2113(08)60542-8
Lee, E. A., Tollenaar, M. (2007). Physiological basis of successful breeding strategies for maize grain yield. Crop Sci. 47, S–202-S-215. doi: 10.2135/cropsci2007.04.0010IPBS
Leonard, A., Jiao, S., Heetland, L., Jaqueth, J., Hudson, T., Simcox, K. D., et al. (2021). The maize brown midrib6 (bm6) mutation encodes a functional GTP Cyclohydrolase1. Maydica 66, M26.
Li, X., Chen, W., Zhao, Y., Xiang, Y., Jiang, H., Zhu, S., et al. (2013). Downregulation of caffeoyl-CoA O-methyltransferase (CCoAOMT) by RNA interference leads to reduced lignin production in maize straw. Genet. Mol. Biol. 36, 540–546. doi: 10.1590/S1415-47572013005000039
Li, C., Guan, H., Jing, X., Li, Y., Wang, B., Li, Y., et al. (2022). Genomic insights into historical improvement of heterotic groups during modern hybrid maize breeding. Nat. Plants 8, 750–763. doi: 10.1038/s41477-022-01190-2
Li, L., Hill-Skinner, S., Liu, S., Beuchle, D., Tang, H. M., Yeh, C. T., et al. (2015). The maize brown midrib4 (bm4) gene encodes a functional folylpolyglutamate synthase. Plant J. 81, 493–504. doi: 10.1111/tpj.12745
Li, Y., Lin, Z., Yue, Y., Zhao, H., Fei, X., E, L., et al. (2021). Loss-of-function alleles of ZmPLD3 cause haploid induction in maize. Nat. Plants 7, 1579–1588. doi: 10.1038/s41477-021-01037-2
Li, C., Liu, C., Qi, X., Wu, Y., Fei, X., Mao, L., et al. (2017). RNA-guided Cas9 as an in vivo desired-target mutator in maize. Plant Biotechnol. J. 15, 1566–1576. doi: 10.1111/pbi.12739
Li, Y., Zhu, J., Wu, H., Liu, C., Huang, C., Lan, J., et al. (2020). Precise base editing of non-allelic acetolactate synthase genes confers sulfonylurea herbicide resistance in maize. Crop J. 8, 449–456. doi: 10.1016/j.cj.2019.10.001
Liang, Z., Chen, K., Gao, C. (2019). Biolistic delivery of CRISPR/Cas9 with ribonucleoprotein complex in wheat. Methods Mol. Biol. 1917, 327–335. doi: 10.1007/978-1-4939-8991-1_24
Liang, Z., Zhang, K., Chen, K., Gao, C. (2014). Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J. Genet. Genomics 41, 63–68. doi: 10.1016/j.jgg.2013.12.001
Lindbo, J. A. (2012). A historical overview of RNAi in plants. Methods Mol. Biol. 894, 1–16. doi: 10.1007/978-1-61779-882-5_1
Lips, D. (2022). Fueling the future of sustainable sugar fermentation across generations. Eng. Biol. 6, 3–16. doi: 10.1049/enb2.12017
Liu, L., Gallagher, J., Arevalo, E. D., Chen, R., Skopelitis, T., Wu, Q., et al. (2021a). Enhancing grain-yield-related traits by CRISPR–Cas9 promoter editing of maize CLE genes. Nat. Plants 7, 287–294. doi: 10.1038/s41477-021-00858-5
Liu, C., Li, X., Meng, D., Zhong, Y., Chen, C., Dong, X., et al. (2017). A 4-bp insertion at ZmPLA1 encoding a putative phospholipase A generates haploid induction in maize. Mol. Plant 10, 520–522. doi: 10.1016/j.molp.2017.01.011
Liu, X., Van Acker, R., Voorend, W., Pallidis, A., Goeminne, G., Pollier, J., et al. (2021b). Rewired phenolic metabolism and improved saccharification efficiency of a Zea mays cinnamyl alcohol dehydrogenase 2 (zmcad2) mutant. Plant J. 105, 1240–1257. doi: 10.1111/tpj.15108
López-Malvar, A., Butrón, A., Samayoa, L. F., Figueroa-Garrido, D. J., Malvar, R. A., Santiago, R. (2019). Genome-wide association analysis for maize stem cell wall-bound hydroxycinnamates. BMC Plant Biol. 19, 519. doi: 10.1186/s12870-019-2135-x
Lorenzana, R. E., Lewis, M. F., Jung, H. J. G., Bernardo, R. (2010). Quantitative trait loci and trait correlations for maize stover cell wall composition and glucose release for cellulosic ethanol. Crop Sci. 50, 541–555. doi: 10.2135/cropsci2009.04.0182
Lorenzo, C. D., Debray, K., Herwegh, D., Develtere, W., Impens, L., Schaumont, D., et al. (2023). BREEDIT: a multiplex genome editing strategy to improve complex quantitative traits in maize. Plant Cell 23, 218–238. doi: 10.1093/plcell/koac243
Lowe, K., Wu, E., Wang, N., Hoerster, G., Hastings, C., Cho, M. J., et al. (2016). Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell 28, 1998–2015. doi: 10.1105/tpc.16.00124
Lu, F., Ralph, J. (1999). Detection and determination of p-coumaroylated units in lignins. J. Agric. Food Chem. 47, 1988–1992. doi: 10.1021/jf981140j
Luciani, A. (2004). Etude du progrès génétique chez différentes espèces de grandes cultures. GEVES: Ed La Minière.
Lundvall, J. P., Buxton, D. R., Hallauer, A. R., George, J. R. (1994). Forage quality variation among maize inbreds: In vitro digestibility and cell-wall components. Crop Sci. 34, 1672–1678. doi: 10.2135/cropsci1994.0011183X003400060046x
Ma, L., Kong, F., Sun, K., Wang, T., Guo, T. (2021). From classical radiation to modern radiation: Past, present, and future of radiation mutation breeding. Front. Public Health 9, 768071. doi: 10.3389/fpubh.2021.768071
Manga-Robles, A., Santiago, R., Malvar, R. A., Moreno-González, V., Fornalé, S., López, I., et al. (2021). Elucidating compositional factors of maize cell walls contributing to stalk strength and lodging resistance. Plant Sci. 307, 110882. doi: 10.1016/j.plantsci.2021.110882
Maqbool, M. A., Beshir, A., Khokhar, E. S. (2020). Doubled haploids in maize: Development, deployment, and challenges. Crop Sci. 60, 2815–2840. doi: 10.1002/csc2.20261
Marcon, C., Altrogge, L., Win, Y. N., Stöcker, T., Gardiner, J. M., Portwood, J. L., II, et al. (2020). BonnMu: a sequence-indexed resource of transposon-induced maize mutations for functional genomics studies. Plant Physiol. 184, 620–631. doi: 10.1104/pp.20.00478
Marita, J. M., Hatfield, R. D., Rancour, D. M., Frost, K. E. (2014). Identification and suppression of the p-coumaroyl CoA:hydroxycinnamyl alcohol transferase in Zea mays L. Plant J. 78, 850–864. doi: 10.1111/tpj.12510
Marita, J. M., Vermerris, W., Ralph, J., Hatfield, R. D. (2003). Variations in the cell wall composition of maize brown midrib mutants. J. Agric. Food Chem. 51, 1313–1321. doi: 10.1021/jf0260592
Martínez-Fortún, J., Phillips, D. W., Jones, H. D. (2022). Natural and artificial sources of genetic variation used in crop breeding: A baseline comparator for genome editing. Front. Genome Ed. 4, 937853. doi: 10.3389/fgeed.2022.937853
May, B. P., Liu, H., Vollbrecht, E., Senior, L., Rabinowicz, P. D., Roh, D., et al. (2003). Maize-targeted mutagenesis: A knockout resource for maize. Proc. Natl. Acad. Sci. U.S.A. 100, 11541–11546. doi: 10.1073/pnas.1831119100
Méchin, V., Argillier, O., Menanteau, V., Barriere, Y., Mila, I., Pollet, B., et al. (2000). Relationship of cell wall composition to in vitro cell wall digestibility of maize inbred line stems. J. Sci. Food Agric. 80, 574–580. doi: 10.1002/(SICI)1097-0010(200004)80:5<574::AID-JSFA575>3.0.CO;2-R
Méchin, V., Argillier, O., Rocher, F., Hébert, Y., Mila, I., Pollet, B., et al. (2005). In search of a maize ideotype for cell wall enzymatic degradability using histological and biochemical lignin characterization. J. Agric. Food Chem. 53, 5872–5881. doi: 10.1021/jf050722f
Méchin, V., Laluc, A., Legée, F., Cézard, L., Denoue, D., Barrière, Y., et al. (2014). Impact of the brown-midrib bm5 mutation on maize lignins. J. Agric. Food Chem. 62, 5102–5107. doi: 10.1021/jf5019998
Mehrshahi, P., Gonzalez-Jorge, S., Akhtar, T. A., Ward, J. L., Santoyo-Castelazo, A., Marcus, S. E., et al. (2010). Functional analysis of folate polyglutamylation and its essential role in plant metabolism and development. Plant J. 64, 267–279. doi: 10.1111/tpj.2010.64.issue-2
Meng, D., Liu, C., Chen, S., Jin, W. (2021). Haploid induction and its application in maize breeding. Mol. Breed. 41, 20. doi: 10.1007/s11032-021-01204-5
Missihoun, T. D., Kotchoni, S. O., Bartels, D. (2016). Active sites of Reduced Epidermal Fluorescence1 (REF1) isoforms contain amino acid substitutions that are different between monocots and dicots. PloS One 11, e0165867. doi: 10.1371/journal.pone.0165867
Mookkan, M., Nelson-Vasilchik, K., Hague, J., Zhang, Z. J., Kausch, A. P. (2017). Selectable marker independent transformation of recalcitrant maize inbred B73 and sorghum P898012 mediated by morphogenic regulators BABY BOOM and WUSCHEL2. Plant Cell Rep. 36, 1477–1491. doi: 10.1007/s00299-017-2169-1
Mottiar, Y., Vanholme, R., Boerjan, W., Ralph, J., Mansfield, S. D. (2016). Designer lignins: harnessing the plasticity of lignification. Curr. Opin. Biotechnol. 37, 190–200. doi: 10.1016/j.copbio.2015.10.009
Nair, R. B., Bastress, K. L., Ruegger, M. O., Denault, J. W., Chapple, C. (2004). The Arabidopsis thaliana REDUCED EPIDERMAL FLUORESCENCE1 gene encodes an aldehyde dehydrogenase involved in ferulic acid and sinapic acid biosynthesis. Plant Cell 16, 544–554. doi: 10.1105/tpc.017509
Nelissen, H., Moloney, M., Inzé, D. (2014). Translational research: from pot to plot. Plant Biotechnol. J. 12, 277–285. doi: 10.1111/pbi.12176
Nicholson, R., Bauman, L., Warren, H. (1976). Association of Fusarium moniliforme with brown midrib maize. Plant Dis. Rep. 60, 908–910.
Ôba, K., Conn, E. E. (1988). Induction of cinnamic acid 4-hydroxylase in developing maize seedlings. Phytochemistry 27, 2447–2450. doi: 10.1016/0031-9422(88)87010-9
Oliveira, D. M., Mota, T. R., Salatta, F. V., Sinzker, R. C., Končitíková, R., Kopečný, D., et al. (2020). Cell wall remodeling under salt stress: Insights into changes in polysaccharides, feruloylation, lignification, and phenolic metabolism in maize. Plant Cell Environ. 43, 2172–2191. doi: 10.1111/pce.13805
Park, S. H., Mei, C., Pauly, M., Ong, R. G., Dale, B. E., Sabzikar, R., et al. (2012). Downregulation of maize cinnamoyl-coenzyme a reductase via RNA interference technology causes brown midrib and improves ammonia fiber expansion-pretreated conversion into fermentable sugars for biofuels. Crop Sci. 52, 2687–2701. doi: 10.2135/cropsci2012.04.0253
Pathi, K. M., Rink, P., Budhagatapalli, N., Betz, R., Saado, I., Hiekel, S., et al. (2020). Engineering smut resistance in maize by site-directed mutagenesis of LIPOXYGENASE 3. Front. Plant Sci. 11, 543895. doi: 10.3389/fpls.2020.543895
Pedersen, J. F., Vogel, K. P., Funnell, D. L. (2005). Impact of reduced lignin on plant fitness. Crop Sci. 45, 812–819. doi: 10.2135/cropsci2004.0155
Penning, B. W., McCann, M. C., Carpita, N. C. (2019). Evolution of the cell wall gene families of grasses. Front. Plant Sci. 10, 1205. doi: 10.3389/fpls.2019.01205
Perkins, M. L., Schuetz, M., Unda, F., Chen, K. T., Bally, M. B., Kulkarni, J. A., et al. (2022). Monolignol export by diffusion down a polymerization-induced concentration gradient. Plant Cell 34, p2080–2095. doi: 10.1093/plcell/koac051
Peterson, D., Barone, P., Lenderts, B., Schwartz, C., Feigenbutz, L., St Clair, G., et al. (2021). Advances in Agrobacterium transformation and vector design result in high-frequency targeted gene insertion in maize. Plant Biotechnol. J. 19, 2000–2010. doi: 10.1111/pbi.13613
Piatek, A. A., Lenaghan, S. C., Neal Stewart, C., Jr (2018). Advanced editing of the nuclear and plastid genomes in plants. Plant Sci. 273, 42–49. doi: 10.1016/j.plantsci.2018.02.025
Pichon, M., Courbou, I., Beckert, M., Boudet, A.-M., Grima-Pettenati, J. (1998). Cloning and characterization of two maize cDNAs encoding Cinnamoyl-CoA Reductase (CCR) and differential expression of the corresponding genes. Plant Mol. Biol. 38, 671–676. doi: 10.1023/A:1006060101866
Pichon, M., Deswartes, C., Gerentes, D., Guillaumie, S., Lapierre, C., Toppan, A., et al. (2006). Variation in lignin and cell wall digestibility in caffeic acid O-methyltransferase down-regulated maize half-sib progenies in field experiments. Mol. Breed. 18, 253–261. doi: 10.1007/s11032-006-9033-2
Piquemal, J., Chamayou, S., Nadaud, I., Beckert, M., Barrière, Y., Mila, I., et al. (2002). Down-regulation of caffeic acid O-methyltransferase in maize revisited using a transgenic approach. Plant Physiol. 130, 1675–1685. doi: 10.1104/pp.012237
Piston, F., Uauy, C., Fu, L., Langston, J., Labavitch, J., Dubcovsky, J. (2010). Down-regulation of four putative arabinoxylan feruloyl transferase genes from family PF02458 reduces ester-linked ferulate content in rice cell walls. Planta 231, 677–691. doi: 10.1007/s00425-009-1077-1
Puigdomenech, P., Perez, P., Murigneux, A., Martinant, J.-P., Tixier, M.-H., Rigau, J., et al. (2001). WIPO. US10/182,113.
Qi, X., Wu, H., Jiang, H., Zhu, J., Huang, C., Zhang, X., et al. (2020). Conversion of a normal maize hybrid into a waxy version using in vivo CRISPR/Cas9 targeted mutation activity. Crop J. 8, 440–448. doi: 10.1016/j.cj.2020.01.006
Qiang, Z., Sun, H., Ge, F., Li, W., Li, C., Wang, S., et al. (2022). The transcription factor ZmMYB69 represses lignin biosynthesis by activating ZmMYB31/42 expression in maize. Plant Physiol. 189, 1916–1919. doi: 10.1093/plphys/kiac233
Rádi, F., Török, K., Nagymihály, M., Kereszt, A., Dudits, D. (2020). Improved reliability in production of maize inbred lines by the combination of the R1-navajo marker with flow cytometry or microsatellite genotyping. Cereal Res. Commun. 48, 423–430. doi: 10.1007/s42976-020-00054-9
Ralph, J., Kim, H., Lu, F., Grabber, J. H., Leplé, J.-C., Berrio-Sierra, J., et al. (2008). Identification of the structure and origin of a thioacidolysis marker compound for ferulic acid incorporation into angiosperm lignins (and an indicator for cinnamoyl CoA reductase deficiency). Plant J. 53, 368–379. doi: 10.1111/j.1365-313X.2007.03345.x
Ralph, J., Lapierre, C., Boerjan, W. (2019). Lignin structure and its engineering. Curr. Opin. Biotechnol. 56, 240–249. doi: 10.1016/j.copbio.2019.02.019
Ralph, J., Lapierre, C., Marita, J. M., Kim, H., Lu, F., Hatfield, R. D., et al. (2001). Elucidation of new structures in lignins of CAD- and COMT-deficient plants by NMR. Phytochemistry 57, 993–1003. doi: 10.1016/S0031-9422(01)00109-1
Ralph, J., Lundquist, K., Brunow, G., Lu, F., Kim, H., Schatz, P. F., et al. (2004). Lignins: natural polymers from oxidative coupling of 4-hydroxyphenyl-propanoids. Phytochem. Rev. 3, 29–60. doi: 10.1023/B:PHYT.0000047809.65444.a4
Ren, J., Wu, P., Trampe, B., Tian, X., Lubberstedt, T., Chen, S. (2017). Novel technologies in doubled haploid line development. Plant Biotechnol. J. 15, 1361–1370. doi: 10.1111/pbi.12805
Ren, Z., Zhang, D., Cao, L., Zhang, W., Zheng, H., Liu, Z., et al. (2020). Functions and regulatory framework of ZmNST3 in maize under lodging and drought stress. Plant Cell Environ. 43, 2272–2286. doi: 10.1111/pce.13829
Rösler, J., Krekel, F., Amrhein, N., Schmid, J. (1997). Maize phenylalanine ammonia-lyase has tyrosine ammonia-lyase activity. Plant Physiol. 113, 175–179. doi: 10.1104/pp.113.1.175
Saratale, G. D., Saratale, R. G., Banu, J. R., Chang, J.-S. (2019). “Biohydrogen production from renewable biomass resources,” in Biohydrogen, 2nd ed. Eds. Pandey, A., Mohan, S. V., Chang, J.-S., Hallenbeck, P. C., Larroche, C. (Elsevier, Amsterdam, Netherlands), 247–277.
Schmitt, D., Pakusch, A.-E., Matern, U. (1991). Molecular cloning, induction, and taxonomic distribution of caffeoyl-CoA 3-O-methyltransferase, an enzyme involved in disease resistance. J. Biol. Chem. 266, 17416–17423. doi: 10.1016/S0021-9258(19)47389-4
Serrani-Yarce, J. C., Escamilla-Trevino, L., Barros, J., Gallego-Giraldo, L., Pu, Y., Ragauskas, A., et al. (2021). Targeting hydroxycinnamoyl CoA: shikimate hydroxycinnamoyl transferase for lignin modification in Brachypodium distachyon. Biotechnol. Biofuels 50. doi: 10.1186/s13068-021-01905-1
Shadle, G., Chen, F., Reddy, M. S. S., Jackson, L., Nakashima, J., Dixon, R. A. (2007). Down-regulation of hydroxycinnamoyl CoA: shikimate hydroxycinnamoyl transferase in transgenic alfalfa affects lignification, development and forage quality. Phytochemistry 68, 1521–1529. doi: 10.1016/j.phytochem.2007.03.022
Shi, J., Gao, H., Wang, H., Lafitte, H. R., Archibald, R. L., Yang, M., et al. (2017). ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 15, 207–216. doi: 10.1111/pbi.12603
Shukla, V. K., Doyon, Y., Miller, J. C., DeKelver, R. C., Moehle, E. A., Worden, S. E., et al. (2009). Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 459, 437–441. doi: 10.1038/nature07992
Shull, G. H. (1908). The composition of a field of maize. J. Hered. 4, 296–301. doi: 10.1093/jhered/os-4.1.296
Shull, G. H. (1909). A pure-line method in corn breeding. Am. Breeders Assoc. Rep. 5, 51–59. doi: 10.1093/jhered/os-5.1.51
Singh, D. P., Singh, A. K., Singh, A. (2021a). “Mass and pure line selection,” in Plant Breeding and Cultivar Development. Eds. Singh, D. P., Singh, A. K., Singh, A. (Academic Press, London, UK), 211–222. doi: 10.1016/B978-0-12-817563-7.00018-0
Singh, D. P., Singh, A. K., Singh, A. (2021b). “Plant breeding: past, present, and future perspectives,” in Plant Breeding and Cultivar Development. Eds. Singh, D. P., Singh, A. K., Singh, A. (Academic Press, London, UK), 1–24. doi: 10.1016/B978-0-12-817563-7.00004-0
Smith, R. A., Cass, C. L., Mazaheri, M., Sekhon, R. S., Heckwolf, M., Kaeppler, H., et al. (2017). Suppression of CINNAMOYL-CoA REDUCTASE increases the level of monolignol ferulates incorporated into maize lignins. Biotechnol. Biofuels 10, 109. doi: 10.1186/s13068-017-0793-1
Sonbol, F.-M., Fornalé, S., Capellades, M., Encina, A., Touriño, S., Torres, J.-L., et al. (2009). The maize ZmMYB42 represses the phenylpropanoid pathway and affects the cell wall structure, composition and degradability in Arabidopsis thaliana. Plant Mol. Biol. 70, 283–296. doi: 10.1007/s11103-009-9473-2
Sprague, G. F., Dudley, J. W. (1988). Corn and corn improvement (Madison, WI: American Society of Agronomy, Inc). doi: 10.2134/agronmonogr18.3ed
Stokstad, E. (2024). European Parliament votes to ease regulation of gene-edited crops. ScienceInsider.
Svitashev, S., Young, J. K., Schwartz, C., Gao, H., Falco, S. C., Cigan, A. M. (2015). Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 169, 931–945. doi: 10.1104/pp.15.00793
Tamasloukht, B., Wong Quai Lam, M. S., Martinez, Y., Tozo, K., Barbier, O., Jourda, C., et al. (2011). Characterization of a cinnamoyl-CoA reductase 1 (CCR1) mutant in maize: effects on lignification, fiber development, and global gene expression. J. Exp. Bot. 62, 3837–3848. doi: 10.1093/jxb/err077
Tang, H. M., Liu, S., Hill-Skinner, S., Wu, W., Reed, D., Yeh, C.-T., et al. (2014). The maize brown midrib2 (bm2) gene encodes a methylenetetrahydrofolate reductase that contributes to lignin accumulation. Plant J. 77, 380–392. doi: 10.1111/tpj.12394
Taube, F., Vogeler, I., Kluß, C., Herrmann, A., Hasler, M., Rath, J., et al. (2020). Yield progress in forage maize in NW Europe—breeding progress or climate change effects? Front. Plant Sci. 11, 1214. doi: 10.3389/fpls.2020.01214
Tenaillon, M. I., Charcosset, A. (2011). A European perspective on maize history. C. R. Biol. 334, 221–218. doi: 10.1016/j.crvi.2010.12.015
Terrett, O. M., Dupree, P. (2019). Covalent interactions between lignin and hemicelluloses in plant secondary cell walls. Curr. Opin. Biotechnol. 56, 97–104. doi: 10.1016/j.copbio.2018.10.010
Till, B. J., Reynolds, S. H., Weil, C., Springer, N., Burtner, C., Young, K., et al. (2004). Discovery of induced point mutations in maize genes by TILLING. BMC Plant Biol. 4, 12. doi: 10.1186/1471-2229-4-12
Torney, F., Moeller, L., Scarpa, A., Wang, K. (2007). Genetic engineering approaches to improve bioethanol production from maize. Curr. Opin. Biotechnol. 18, 193–199. doi: 10.1016/j.copbio.2007.03.006
Torres, A. F., Noordam-Boot, C. M., Dolstra, O., van der Weijde, T., Combes, E., Dufour, P., et al. (2015a). Cell wall diversity in forage maize: genetic complexity and bioenergy potential. Bioenergy Res. 8, 187–202. doi: 10.1007/s12155-014-9507-8
Torres, A. F., Slegers, P. M., Noordam-Boot, C. M. M., Dolstra, O., Vlaswinkel, L., van Boxtel, A. J. B., et al. (2016). Maize feedstocks with improved digestibility reduce the costs and environmental impacts of biomass pretreatment and saccharification. Biotechnol. Biofuels 9, 63. doi: 10.1186/s13068-016-0479-0
Torres, A. F., Visser, R. G. F., Trindade, L. M. (2015b). Bioethanol from maize cell walls: genes, molecular tools, and breeding prospects. GCB Bioenergy 7, 591–607. doi: 10.1111/gcbb.12164
Troyer, A. F. (1999). Background of U.S. hybrid corn. Crop Sci. 39, 601–626. doi: 10.2135/cropsci1999.0011183X003900020001xa
Tuberosa, R. (2012). Phenotyping for drought tolerance of crops in the genomics era. Front. Physiol. 3, 347. doi: 10.3389/fphys.2012.00347
Van Acker, R., Déjardin, A., Desmet, S., Hoengenaert, L., Vanholme, R., Morreel, K., et al. (2017). Different routes for conifer- and sinapaldehyde and higher saccharification upon deficiency in the dehydrogenase CAD1. Plant Physiol. 175, 1018–1039. doi: 10.1104/pp.17.00834
Van Acker, R., Leplé, J.-C., Aerts, D., Storme, V., Goeminne, G., Ivens, B., et al. (2014). Improved saccharification and ethanol yield from field-grown transgenic poplar deficient in cinnamoyl-CoA reductase. Proc. Natl. Acad. Sci. U.S.A. 111, 845–850. doi: 10.1073/pnas.1321673111
Van Acker, R., Vanholme, R., Storme, V., Mortimer, J. C., Dupree, P., Boerjan, W. (2013). Lignin biosynthesis perturbations affect secondary cell wall composition and saccharification yield in Arabidopsis thaliana. Biotechnol. Biofuels 6, 46. doi: 10.1186/1754-6834-6-46
van der Weijde, T., Alvim Kamei, C. L., Torres, A. F., Vermerris, W., Dolstra, O., Visser, R. G. F., et al. (2013). The potential of C4 grasses for cellulosic biofuel production. Front. Plant Sci. 4, 107. doi: 10.3389/fpls.2013.00107
Vanholme, R., Demedts, B., Morreel, K., Ralph, J., Boerjan, W. (2010). Lignin biosynthesis and structure. Plant Physiol. 153, 895–905. doi: 10.1104/pp.110.155119
Vanholme, R., De Meester, B., Ralph, J., Boerjan, W. (2019). Lignin biosynthesis and its integration into metabolism. Curr. Opin. Biotechnol. 56, 230–239. doi: 10.1016/j.copbio.2019.02.018
Vanholme, R., Morreel, K., Darrah, C., Oyarce, P., Grabber, J. H., Ralph, J., et al. (2012). Metabolic engineering of novel lignin in biomass crops. New Phytol. 196, 978–1000. doi: 10.1111/j.1469-8137.2012.04337.x
Van Vu, T., Das, S., Hensel, G., Kim, J.-Y. (2022). Genome editing and beyond: what does it mean for the future of plant breeding? Planta 255, 130. doi: 10.1007/s00425-022-03906-2
Vermaas, J. V., Dixon, R. A., Chen, F., Mansfield, S. D., Boerjan, W., Ralph, J., et al. (2019). Passive membrane transport of lignin-related compounds. Proc. Natl. Acad. Sci. U.S.A. 116, 23117–23123. doi: 10.1073/pnas.1904643116
Vermerris, W., Saballos, A., Ejeta, G., Mosier, N. S., Ladisch, M. R., Carpita, N. C. (2007). Molecular breeding to enhance ethanol production from corn and sorghum stover. Crop Sci. 47, S142–S153. doi: 10.2135/cropsci2007.04.0013IPBS
Vermerris, W., Sherman, D. M., McIntyre, L. M. (2010). Phenotypic plasticity in cell walls of maize brown midrib mutants is limited by lignin composition. J. Exp. Bot. 61, 2479–2490. doi: 10.1093/jxb/erq093
Vignols, F., Rigau, J., Torres, M. A., Capellades, M., Puigdomènech, P. (1995). The brown midrib3 (bm3) mutation in maize occurs in the gene encoding caffeic acid O-methyltransferase. Plant Cell 7, 407–416. doi: 10.1105/tpc.7.4.407
Vigouroux, Y., Glaubitz, J. C., Matsuoka, Y., Goodman, M. M., Sánchez G., J., Doebley, J. (2008). Population structure and genetic diversity of New World maize races assessed by DNA microsatellites. Am. J. Bot. 95, 1240–1253. doi: 10.3732/ajb.0800097
Vinayan, M., Seetharam, K., Babu, R., Zaidi, P. H., Blummel, M., Nair, S. K. (2021). Genome wide association study and genomic prediction for stover quality traits in tropical maize (Zea mays L.). Sci. Rep. 11, 686. doi: 10.1038/s41598-020-80118-2
Vogel, K. E. (2009). Backcross breeding. Methods Mol. Biol. 526, 161–169. doi: 10.1007/978-1-59745-494-0_14
Walker, A. M., Hayes, R. P., Youn, B., Vermerris, W., Sattler, S. E., Kang, C. (2013). Elucidation of the structure and reaction mechanism of sorghum hydroxycinnamoyltransferase and its structural relationship to other coenzyme A-dependent transferases and synthases. Plant Physiol. 162, 640–651. doi: 10.1104/pp.113.217836
Wang, H., Li, K., Hu, X., Liu, Z., Wu, Y., Huang, C. (2016). Genome-wide association analysis of forage quality in maize mature stalk. BMC Plant Biol. 16, 227. doi: 10.1186/s12870-016-0919-9
Wang, Y., Mu, Y., Yan, L., Tang, B., Jiang, F. (2023). Regulatory policies of genome editing products around the world. J. Biomed. Res. Environ. Sci. 4, 1447–1454. doi: 10.37871/jbres
Wang, W., Pan, Q., He, F., Akhunova, A., Chao, S., Trick, H., et al. (2018). Transgenerational CRISPR-Cas9 activity facilitates multiplex gene editing in allopolyploid wheat. CRISPR J. 1, 65–74. doi: 10.1089/crispr.2017.0010
Weller, R. F., Phipps, R. H., Cooper, A. (1985). The effect of the brown midrib-3 gene on the maturity and yield of forage maize. Grass Forage Sci. 40, 335–339. doi: 10.1111/j.1365-2494.1985.tb01761.x
Weng, J.-K., Mo, H., Chapple, C. (2010). Over-expression of F5H in COMT-deficient Arabidopsis leads to enrichment of an unusual lignin and disruption of pollen wall formation. Plant J. 64, 898–911. doi: 10.1111/tpj.2010.64.issue-6
Wieczorek, A., Wright, M. (2012). History of agricultural biotechnology: How crop development has evolved. Nat. Educ. Knowledge 3, 9.
Williams, P. W., Davis, F. M., Buckley, P. M., Hedin, P. A., Baker, G. T., Luthe, D. S. (1998). Factors associated with resistance to fall armyworm (Lepidoptera: Noctuidae) and southwestern corn borer (Lepidoptera: Crambidae) in corn at different vegetative stages. J. Econ. Entomol. 91, 1471–1480. doi: 10.1093/jee/91.6.1471
Withers, S., Lu, F., Kim, H., Zhu, Y., Ralph, J., Wilkerson, C. G. (2012). Identification of grass-specific enzyme that acylates monolignols with p-coumarate. J. Biol. Chem. 287, 8347–8355. doi: 10.1074/jbc.M111.284497
Wolf, D. P., Coors, J. G., Albrecht, K. A., Undersander, D. J., Carter, P. R. (1993). Forage quality of maize genotypes selected for extreme fiber concentrations. Crop Sci. 33, 1353–1359. doi: 10.2135/cropsci1993.0011183X003300060046x
Wolter, F., Schindele, P., Puchta, H. (2019). Plant breeding at the speed of light: the power of CRISPR/Cas to generate directed genetic diversity at multiple sites. BMC Plant Biol. 19, 176. doi: 10.1186/s12870-019-1775-1
Woo, E. P., Inbasekaran, M., Shiang, W., Roof, G. R. (1997). International Patent Application WO 97/05 184.
Wu, Z., Ren, H., Xiong, W., Roje, S., Liu, Y., Su, K., et al. (2018). Methylenetetrahydrofolate reductase modulates methyl metabolism and lignin monomer methylation in maize. J. Exp. Bot. 69, 3963–3973. doi: 10.1093/jxb/ery208
Wu, Z., Wang, N., Hisano, H., Cao, Y., Wu, F., Liu, W., et al. (2019). Simultaneous regulation of F5H in COMT-RNAi transgenic switchgrass alters effects of COMT suppression on syringyl lignin biosynthesis. Plant Biotechnol. J. 17, 836–845. doi: 10.1111/pbi.13019
Xiao, W., Yang, Y., Yu, J. (2018). ZmNST3 and ZmNST4 are master switches for secondary wall deposition in maize (Zea mays L.). Plant Sci. 266, 83–94. doi: 10.1016/j.plantsci.2017.03.012
Xiong, W., Li, Y., Wu, Z., Ma, L., Liu, Y., Qin, L., et al. (2020). Characterization of two new brown midrib1 mutations from an EMS-mutagenic maize population for lignocellulosic biomass utilization. Front. Plant Sci. 11, 594798. doi: 10.3389/fpls.2020.594798
Xiong, W., Wu, Z., Liu, Y., Li, Y., Su, K., Bai, Z., et al. (2019). Mutation of 4-coumarate: coenzyme A ligase 1 gene affects lignin biosynthesis and increases the cell wall digestibility in maize brown midrib5 mutants. Biotechnol. Biofuels 12, 82. doi: 10.1186/s13068-019-1421-z
Xue, C., Greene, E. C. (2021). DNA repair pathway choices in CRISPR-Cas9-mediated genome editing. Trends Genet. 37, 639–656. doi: 10.1016/j.tig.2021.02.008
Yadava, P., Abhishek, A., Singh, R., Singh, I., Kaul, T., Pattanayak, A., et al. (2017). Advances in maize transformation technologies and development of transgenic maize. Front. Plant Sci. 7, 1949. doi: 10.3389/fpls.2016.01949
Yang, F., Li, W., Jiang, N., Yu, H., Morohashi, K., Ouma, W. Z., et al. (2017a). A maize gene regulatory network for phenolic metabolism. Mol. Plant 10, 498–515. doi: 10.1016/j.molp.2016.10.020
Yang, N., Wang, Y., Liu, X., Jin, M., Vallebueno-Estrada, M., Calfee, E., et al. (2023). Two teosintes made modern maize. Science 382, eadg8940. doi: 10.1126/science.adg8940
Yang, N., Xu, X.-W., Wang, R.-R., Peng, W.-L., Cai, L., Song, J.-M., et al. (2017b). Contributions of Zea mays subspecies mexicana haplotypes to modern maize. Nat. Commun. 8, 1874. doi: 10.1038/s41467-017-02063-5
Yassitepe, J. E., d., C. T., Heinzen da Silva, V. C., Hernandes-Lopes, J., Dante, R. A., Gerhardt, I. R., et al. (2021). Maize transformation: From plant material to the release of genetically modified and edited varieties. Front. Plant Sci. 12, 766702. doi: 10.3389/fpls.2021.766702
Yu, D., Gu, X., Zhang, S., Dong, S., Miao, H., Gebretsadik, K., et al. (2021). Molecular basis of heterosis and related breeding strategies reveal its importance in vegetable breeding. Hortic. Res. 8, 120. doi: 10.1038/s41438-021-00552-9
Yun, M.-S., Chen, W., Deng, F., Yogo, Y. (2005). Differential properties of 4-coumarate: CoA ligase related to growth suppression by chalcone in maize and rice. Plant Growth Regul. 46, 169–176. doi: 10.1007/s10725-005-6832-0
Zhang, Y., Culhaoglu, T., Pollet, B., Melin, C., Denoue, D., Barrière, Y., et al. (2011). Impact of lignin structure and cell wall reticulation on maize cell wall degradability. J. Agric. Food Chem. 59, 10129–10135. doi: 10.1021/jf2028279
Zhang, J., Zhang, S., Li, H., Du, H., Huang, H., Li, Y., et al. (2016). Identification of transcription factors ZmMYB111 and ZmMYB148 involved in phenylpropanoid metabolism. Front. Plant Sci. 7, 148. doi: 10.3389/fpls.2016.00148
Zhao, Q., Dixon, R. A. (2011). Transcriptional networks for lignin biosynthesis: more complex than we thought? Trends Plant Sci. 16, 227–233. doi: 10.1016/j.tplants.2010.12.005
Zhao, Q., Nakashima, J., Chen, F., Yin, Y., Fu, C., Yun, J., et al. (2013). LACCASE is necessary and nonredundant with PEROXIDASE for lignin polymerization during vascular development in Arabidopsis. Plant Cell 25, 3976–3987. doi: 10.1105/tpc.113.117770
Zhong, R., Lee, C., McCarthy, R. L., Reeves, C. K., Jones, E. G., Ye, Z.-H. (2011). Transcriptional activation of secondary wall biosynthesis by rice and maize NAC and MYB transcription factors. Plant Cell Physiol. 52, 1856–1871. doi: 10.1093/pcp/pcr123
Zhong, Y., Liu, C., Qi, X., Jiao, Y. Y., Wang, D., Wang, Y., et al. (2019). Mutation of ZmDMP enhances haploid induction in maize. Nat. Plants 5, 575–580. doi: 10.1038/s41477-019-0443-7
Zhong, R., Ye, Z.-H. (2015). Secondary cell walls: Biosynthesis, patterned deposition and transcriptional regulation. Plant Cell Physiol. 56, 195–214. doi: 10.1093/pcp/pcu140
Zhu, Y., Lee, Y. Y., Elander, R. T. (2005). “Optimization of dilute-acid pretreatment of corn stover using high-solids percolation reactor,” in Twenty-Sixth Symposium on Biotechnology for Fuels and Chemicals (Applied Biochemistry and Biotechnology 121–124). Eds. Davison, B. H., Evans, B. R., Finkelstein, M., Mcmillan, J. D. (Humana Press, Totowa, NJ), 1045–1054.
Keywords: maize, lignin, breeding, lignin engineering, digestibility, saccharification
Citation: Vanhevel Y, De Moor A, Muylle H, Vanholme R and Boerjan W (2024) Breeding for improved digestibility and processing of lignocellulosic biomass in Zea mays. Front. Plant Sci. 15:1419796. doi: 10.3389/fpls.2024.1419796
Received: 18 April 2024; Accepted: 10 June 2024;
Published: 26 July 2024.
Edited by:
Deyu Xie, North Carolina State University, United StatesReviewed by:
Xianhai Zhao, Brookhaven National Laboratory (DOE), United StatesCopyright © 2024 Vanhevel, De Moor, Muylle, Vanholme and Boerjan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wout Boerjan, d291dC5ib2VyamFuQHBzYi52aWItdWdlbnQuYmU=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.