
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 08 July 2024
Sec. Aquatic Photosynthetic Organisms
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1418049
This article is part of the Research Topic Photosynthetic Regulation and Carbon/Nitrogen Metabolism in Genetically Engineered Algae View all 5 articles
Many microbes accumulate energy storage molecules such as triglycerides (TAG) and starch during nutrient limitation. In eukaryotic green algae grown under nitrogen-limiting conditions, triglyceride accumulation is coupled with chlorosis and growth arrest. In this study, we show that reactive oxygen species (ROS) actively accumulate during nitrogen limitation in the microalga Chlorella sorokiniana. Accumulation of ROS is mediated by the downregulation of genes encoding ROS-quenching enzymes, such as superoxide dismutases, catalase, peroxiredoxin, and glutathione peroxidase-like, and by the upregulation of enzymes involved in generating ROS, such as NADPH oxidase, xanthine oxidase, and amine oxidases. The expression of genes involved in ascorbate and glutathione metabolism is also affected under this condition. ROS accumulation contributes to the degradation of monogalactosyl diacylglycerol (MGDG) and thylakoid membrane remodeling, leading to chlorosis. Quenching ROS under nitrogen limitation reduces the degradation of MGDG and the accumulation of TAG. This work shows that ROS accumulation, membrane remodeling, and TAG accumulation under nitrogen limitation are intricately linked in the microalga C. sorokiniana.
Microalgae are a potential source of renewable biofuels, as they are fast growing and do not compete with crops for arable land (Hu et al., 2008). Significant investments have been made in the past two decades toward developing algae as a reliable biofuel and bioproduct feedstock, by coupling algal biomass production to waste remediation (Allen et al., 2018). Microalgae, like many other microbes, accumulate storage molecules under unfavorable growth conditions (Zhu et al., 2016), and starch and oil are products of interest as sources of biofuel. Under nutrient limitation conditions, microalgae like Chlorella spp., Chlamydomonas (Chlamydomonas reinhardtii), and Nannochloropsis spp., accumulate oil (Miller et al., 2010; Stephenson et al., 2010; Wase et al., 2014; Ma X. N. et al., 2016; Jiang et al., 2019). Many efforts have been made to understand the mechanisms associated with metabolic remodeling and the regulation of oil accumulation during nitrogen (N) starvation (Miller et al., 2010; Wase et al., 2017; Wase et al., 2019). Many enzymes participating in the conversion of photosynthates into oil have been identified (Boyle et al., 2012; Shtaida et al., 2014). N limitation is a strong inducer of oil accumulation in different microalgae but often is associated with slower growth and severely compromised carbon fixation. This growth penalty constitutes an unfavorable factor when considering the industrial use of algae as a feedstock for waste remediation and biofuel production. Furthermore, growth arrest and accumulation of oil are accompanied by a decrease in chlorophyll content (Zhang et al., 2013) and chlorosis, also known as the “degreening” of cells.
Under N-limiting conditions, fatty acids for the biosynthesis of triacylglycerol (TAG) are derived from both a de novo synthesis route and by the remodeling of glycerolipids from preexisting membranes. Various enzymes involved in membrane remodeling have been reported in Chlamydomonas (Li et al., 2012a; Li et al., 2012b; Yoon et al., 2012) that act on different membrane glycerolipids to release fatty acid for TAG biosynthesis. One of the membrane lipids that experiences the most dramatic changes in abundance in response to N limitation is monogalactosyldiacylglycerol (MGDG), a plastid membrane lipid. Fatty acid from MGDG is channeled for triacylglycerol (TAG) synthesis (Li et al., 2012b).
All aerobic organisms use oxygen as an electron acceptor for respiration. Various pathways lead to the production of reactive oxygen species (ROS), such as mitochondrial electron transport, nitric oxide synthase, and peroxisomal β-oxidation (Møller, 2001). Different forms of ROS accumulate in cells, such as superoxide (O2–), hydroxyl radical (•OH–), singlet oxygen (1O2), and hydrogen peroxide (H2O2). Each of these reactive species have different half-lives and chemical properties that affect cellular biochemistry and physiology (Foyer and Noctor, 2009).
Cellular redox and ROS homeostasis are carefully balanced by an elaborate antioxidant defense system that includes the enzymes superoxide dismutase (SOD), catalase, glutathione peroxidase, and ascorbate peroxidase as well as low-molecular-weight scavengers such as uric acid, ascorbate, glutathione, carotenoids, and flavonoids (Czarnocka and Karpiński, 2018). Studies over the past two decades in plants and animals have revealed ROS as a fine-tuned signaling system that elicits both localized and global responses. Under different biotic and abiotic stress conditions, ROS signaling has been implicated in playing a key role in stress responses and acclimation (Ben Rejeb et al., 2015; Gupta et al., 2013; He et al., 2017) in both animals and plants (Foyer and Noctor, 2009; Schieber and Chandel, 2014).
In this study, we provide evidence that ROS signaling participates in the N starvation response in Chlorella sorokiniana, an industrial microalga of the Chlorophyceae family. We show that C. sorokiniana uses transcriptional mechanisms to control ROS accumulation and that ROS signaling is essential for chloroplast membrane remodeling, leading to chlorosis and oil accumulation.
Chlorella sorokiniana cells were grown in Tris acetate phosphate (TAP) medium (Gorman and Levine, 1965) at 25°C under continuous light of 40 μE. For nitrogen (N) limitation, parental cultures were grown for 2 days and then washed three times in the appropriate medium (TAP or TAP-N) before inoculation at a final concentration of 107 cells/mL. Cells were inoculated into either control (TAP) or N-limited medium of 10 mM or 250 µM ammonium chloride, respectively. Culture growth was recorded as optical density (OD) at 750 nm.
C. sorokiniana cells grown in appropriate conditions were first adjusted to an OD of 0.2 in respective media before 10 µL of a 100 µg/mL stock of Nile red (Sigma Aldrich) in DMSO was added to 100 µL of diluted cell suspension. After 10 min of incubation, the cells were visualized by fluorescence microscopy and fluorescence intensity was measured. Microscopy was conducted using an EVOS-fl epifluorescence microscope with a GFP light cube. Quantitative fluorescence measurements were performed on a Synergy H1 hybrid reader with excitation and emission wavelengths of 455 nm and 560 nm, respectively; fluorescence values were normalized to the optical density.
For transmission electron microscopy (TEM) analysis, cells were grown in the appropriate medium for 4 days and then fixed in 1% formaldehyde before processing the samples. Sample preparation and analysis were carried out as described by Gojkovic et al. (2014.) Imaging was carried out on a Hitachi H-7500 microscope.
Two days after inoculation, parental cultures were supplemented with 14C acetate (106 cpm/mL of culture-specific activity of 56 mCi/mmol, supplied by ARC Inc.) for 8 h. The cells were washed and inoculated in appropriate medium (TAP or TAP-N) at a concentration of 107 cells/mL. One milliliter of culture was collected at each time point, including at time zero. Cell pellets were collected by centrifugation at 4,000 rpm for 5 min at 25°C. Bligh and Dyer extraction was performed on the samples by partitioning lipids into the organic phase of a chloroform/methanol/0.2 M KCl solvent system (1:1:0.8, v/v/v). Glycerolipids were dried under N2 and sequentially separated by thin-layer chromatography (TLC) on a silica G60 plate (EMD-Millipore) in two solvent systems. The first ascent was performed until two-thirds the height of the plate in a solvent system of chloroform, methanol, acetic acid, and water (85:12.5:12.5:3, v/v/v/v) to separate polar lipids. The plate was then dried, and a second ascent to the full height of the plate was performed in petroleum ether, diethyl ether, and acetic acid (80:20:1) to separate neutral lipids such as TAG, steryl esters, and free sterols. After drying, plates were exposed to a storage phosphor screen for 24 h, and radioactivity was detected on a GE-Typhoon FLA 9500 scanner. Images were analyzed by ImageQuant TL v8.1, and individual bands were identified by reference to standards of known Rf and plotted as a percentage of initial counts in the specific lipid.
For RNA-seq analysis, two biological replicates grown at the same time but from different parent cultures in different flasks (independently grown cultures) were used. Samples were collected by pelleting the cells at 5,000 rpm for 3 min before freezing them in liquid nitrogen. Total RNA was extracted with the TRIzol reagent according to the manufacturer’s protocol. Briefly, frozen cells were gently resuspended in 500 µL of TRIzol reagent (Ambion TRIzol LS reagent), to which 600 µL of chloroform was added, gently mixed, and phase-separated at 12,000 rpm for 10 min at 4°C. Total RNA was precipitated by adding 40 µL of 5 M NaCl and 500 µL isopropanol to the supernatant and incubating at 4°C for 10 min. RNA was pelleted by centrifugation at 13.8 × g for 20 min at 4°C, followed by washing the pellet with ethanol and further centrifugation for 3 min at 13.8 × g in 4°C. The pellet was carefully dried and resuspended in RNase-free water. RNA sequencing library preparation and Illumina sequencing (75-bp, single-end reads) were carried out by GeneWiz (https://www.genewiz.com/). Two separate libraries were prepared and sequenced on different lanes per sample. Libraries were prepared using the Illumina TruSeq RNA sample preparation kit according to the manufacturer’s guidelines. Reads were aligned to our in-house genome sequence of C. sorokiniana UTEX 1230 (tentative id: SUB1977110) using the Rsubread package (Liao et al., 2013). Reads were mapped and counted using the featureCounts function with default setting. Differential expression analysis was performed using the edgeR-limma package. Weighted trimmed mean of M-values (TMM) were used to normalize the count data. Firstly, low-expressing transcripts were filtered out and 15,382 transcripts were retained. The linear model was fitted and differential expression analysis was performed using the eBayes function of limma package, and expression data for all genes are included as Supplementary Data File 1 (Robinson et al., 2009; Ritchie et al., 2015). Raw reads have been submitted to NCBI under the BioProject with accession number PRJNA1111962.
The quantification of ROS was carried out using 2′, 7′-dichlorodihydrofluorescein diacetate (DCFDA) cellular ROS detection assay kit (Abcam, product number AB113851) following the manufacturer’s protocol. Briefly, cells under different treatments were washed twice in the assay buffer and then adjusted to an OD of 0.2 before incubation with DCFDA at a final concentration of 10 µM for 1 h. The cells were then washed twice with the appropriate medium (TAP or TAP-N) to remove excess dye and incubated for one more hour. Fluorescence intensity was measured with excitation and emission wavelengths of 485 nm and 535 nm, respectively on a BioTek SynergyH1 reader. Values were normalized to the optical density.
A working solution of 250 µM of diethyldithiocarbamate (Sigma-Aldrich) solubilized in DMSO was used to inhibit superoxide dismutase and induce ROS accumulation. N-limited medium was supplemented with 500 µM ascorbate (Sigma Aldrich) to quench hydrogen peroxide. NADPH oxidase was inhibited by the addition of 50 µM 2-acetylphenothiazine (ML171, Selleck Chem) dissolved in DMSO.
Nitrogen (N) is an essential macronutrient for the growth of all organisms, as it provides the building blocks for the biosynthesis of nucleic acids, amino acids, and the amino acid–derived head groups of certain lipids like phosphatidylcholine and phosphatidylethanolamine. N limitation led to growth arrest of Chlorella sorokiniana (Figure 1A). We observed a decrease in growth as early as 9 h after inoculation into N-limited (250 µM ammonium chloride) acetate-containing medium, compared with cells grown in N-replete medium (10 mM ammonium chloride). C. sorokiniana, like many other microalgae, accumulated oil droplets when deprived of N, as illustrated by the green puncta observed specifically in N-limited C. sorokiniana cells stained with Nile red (Supplementary Figure 1A). N limitation was also associated with chlorosis in C. sorokiniana (Figure 1A, inset).
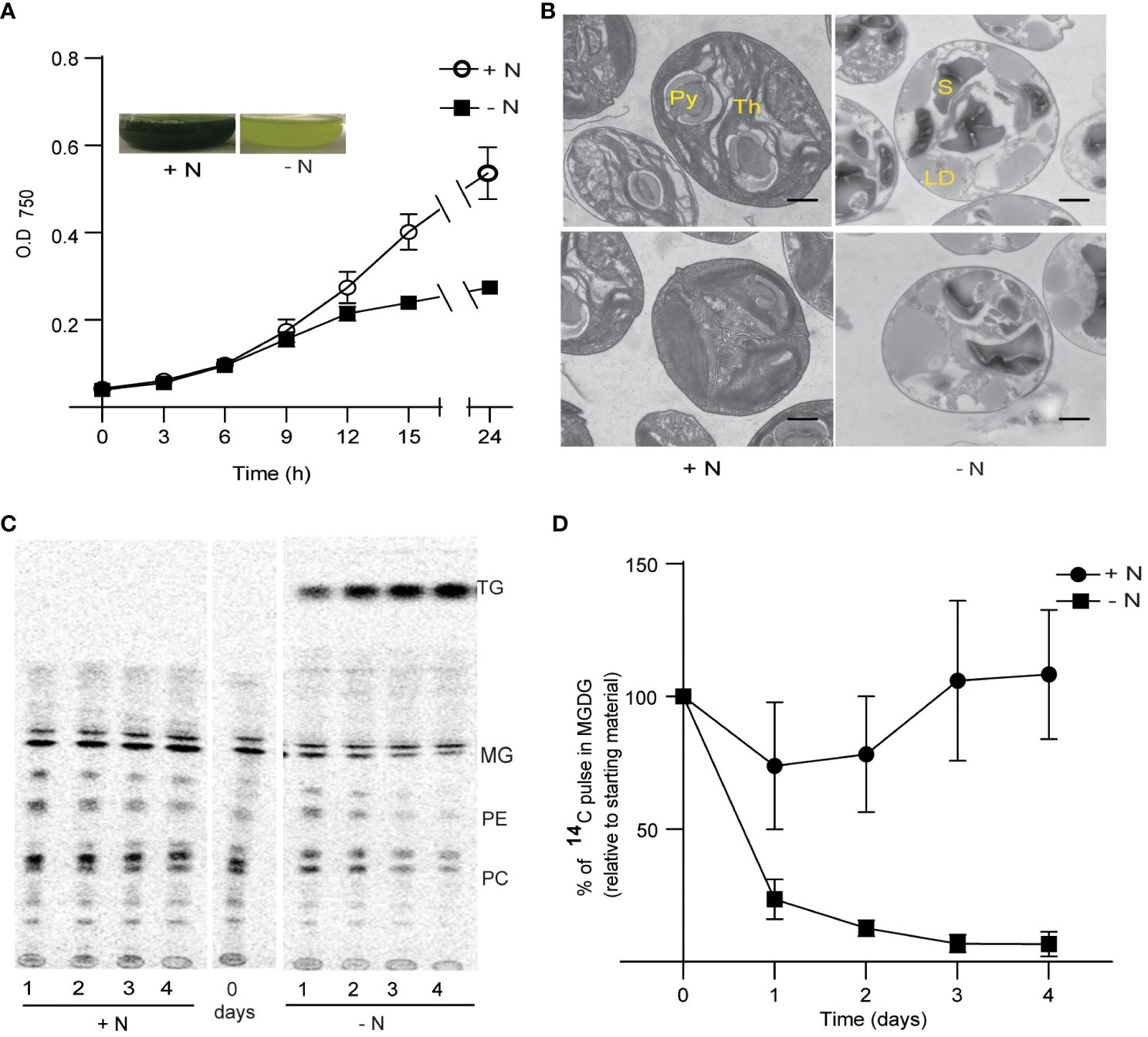
Figure 1 Growth and thylakoid membrane remodeling in Chlorella sorokiniana under nitrogen limitation. (A) Growth curves of Chlorella sorokiniana in N-replete (N+, 7.5 mM) and N-limited media (N−, 0.25 mM). Cultures were inoculated with 107 cells/ml, and the mean +/− SD of three biological replicates is presented. Inset: representative cultures under N-replete (N+) and N-deplete (N−) condition after 48 h of growth. (B) TEM image of Chlorella sorokiniana cells under N+ or N− conditions 4 days post inoculation. Scale bar indicates 800 nm. (C) Representative TLC radiograph of cells prelabeled with 14C acetate and then grown in N+ or N− media, indicating the degradation of preformed lipids under N− conditions. TAG, triacylglycerol; MG, monogalactosyldiacylglycerol; PE, phosphatidylethanolamine; PC, phosphatidylcholine. Lipid droplet accumulation under N limitation. (D) Quantitative measurement of 14C-containing MGDG degradation upon N limitation. Measurements are relative to the amount to 14C pulse present in MGDG at the initial time point (t0).
Chlorosis is generally caused by the degeneration of chloroplast membranes and is accompanied by chlorophyll degradation (Li et al., 2012b). We thus carried out transmission electron microscopy (TEM) observations of the ultrastructure of chloroplast membranes (Figure 1B). We observed a decrease in the number of dense thylakoid membranes in N-starved cells (after 4 days), relative to the control cells (N+). One of the characteristics of N limitation in Chlamydomonas is the substantial reduction in MGDG levels (Li et al., 2012b). To assess the extent of MGDG degradation under N limitation in C. sorokiniana, we labeled membrane lipids from N-replete cultures to saturation with 1,2-14C-acetate. We determined that MGDG is degraded when cells are transferred to N-limited conditions, as seen during a chase period with the replacement of labeled acetate with unlabeled acetate (Figures 1C, D). Other lipids, such as phosphatidylcholine and phosphatidylethanolamine, also appeared to be degraded upon transfer to N limitation, but not when cells were maintained in N-replete medium. Fatty acids released from these lipids were channeled into TAG, as evidenced by the increase in radiolabeled TAG during the N-limited chase period (Figure 1C).
To explore the mechanisms underlying chloroplast membrane degradation and oil accumulation under N limitation, we performed a transcriptome deep sequencing (RNA-seq) analysis. Accordingly, we collected cells grown in N-replete conditions or transferred to N-limited conditions for 9 h, when we first observed a decline in growth rate relative to N-replete cultures (Figure 1A); we also collected cells after 24 h in N-limited conditions as a late time point. We determined that 16% of all genes are differentially expressed 9 h into N limitation and reached 22% at the 24-h time point (Table 1). Differentially expressed gene sets of 9 h and 24 h significantly overlap (Supplementary Figure 2). The substantial rewiring of the transcriptome observed here indicated that the N starvation response in C. sorokiniana starts earlier than 9 h after the onset of N limitation, i.e., before a significant reduction in growth is noticeable. This is expected as nutrient sensing and initiation of starvation response would precede the decrease in growth rate.
From the RNA-seq data, we determined that the expression of many genes encoding enzymes involved in quenching ROS are decreased during N limitation. Indeed, the expression of superoxide dismutase genes (SOD1, SOD2, and SOD3) and catalase genes was decreased at 9 h and 24 h into N-limited conditions (Figure 2A). Enzymes such as SOD, catalase, and glutathione peroxidase quench and detoxify ROS generated during various cellular processes such as photosynthesis and mitochondrial electron transport (Møller, 2001; Asada, 2006). SOD converts superoxide radicals to H2O2, which is further degraded into less harmful substances by a variety of enzymes, such as peroxidases and catalases (Czarnocka and Karpiński, 2018).
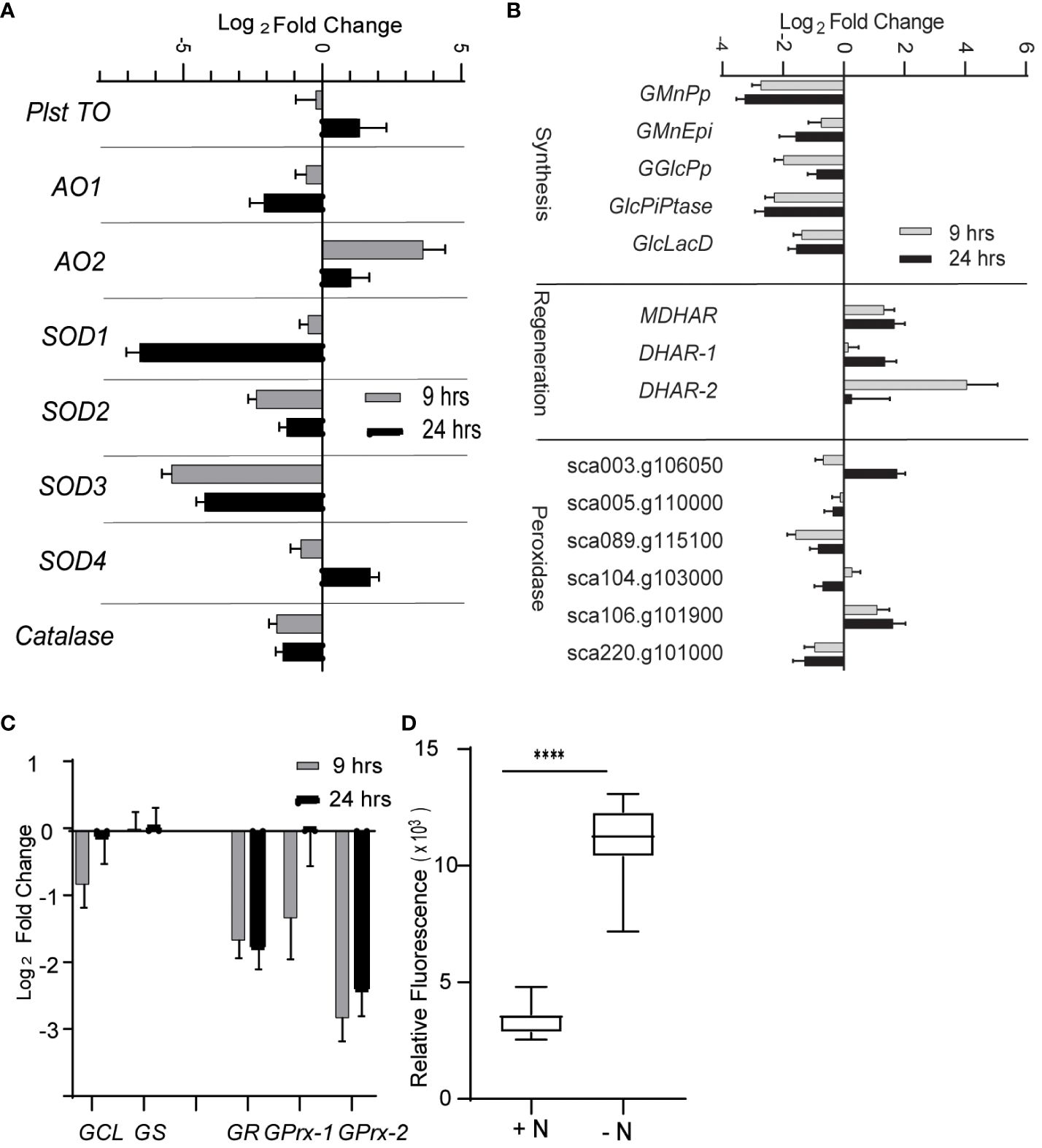
Figure 2 Reactive oxygen species under nitrogen limitation. (A) Expression of enzymes involved scavenging ROS. AO, alternative oxidase; SOD, superoxide dismutase; Plst TO, plastidial terminal oxidase. (B) Expression of genes involved in ascorbate (AsA) metabolism. Genes are classified as those involved in AsA synthesis, regeneration, and AsA-dependent peroxidases. GMnPp, GDP-d-mannose pyrophosphorylase; GMnEpi, GDP-mannose 3,5-pimerase 1; GGlcPp, GDP-L-galactose phosphorylase 1; GlcPiPtase: L-galactose-1-phosphate phosphatase; GlcLacD, L-galactono-1,4-lactone dehydrogenase; MDHAR, monodehydroascorbate reductase; DHAR, dehydroascorbate reductase. (C) Expression of genes involved in glutathione metabolism. GSH1, gamma-glutamylcysteine synthetase; GSH2, glutathione synthetase are involved in glutathione synthesis; GR, glutathione reductase regenerates reduced glutathione (GSH) from oxidized form of glutathione (GSSG); GPrx, glutathione peroxidases oxidizes GSH to quench ROS and form GSSG. (D) ROS level as measured by DCFDA fluorescence under N-limited condition after 24 h of inoculation in appropriate media. The values are normalized to optical density. Error bars indicate SD (n = 3). **** P value < 0.0001.
The expression of genes involved in the metabolism of ascorbate (AsA), a key antioxidant, was also significantly altered (Figure 2B). Specifically, genes involved in AsA biosynthesis from GDP-mannose were decreased in abundance under N limitation, including those encoding GDP-d-mannose pyrophosphorylase (GMnPp), GDP-mannose-3,5-epimerase 1 (GMnEpi), GDP-L-galactose phosphorylase 1 (GGlcPp), L-galactose-1-phosphate phosphatase (GlcPiPtase), and L-galactono-1,4-lactone dehydrogenase (GlcLacD) (Smirnoff, 2018). This observation suggested that AsA biosynthesis possibly diminishes upon N limitation. Conversely, the expression of genes encoding AsA-reducing enzymes such as monodehydroascorbate reductase (MDHAR) and dehydroascorbate reductase (DHAR) was upregulated in N-limited conditions (Figure 2B), indicating that the lower availability of AsA may be partially compensated for by increased regeneration of oxidized monodehydroascorbate or dehydroascorbate. Glutathione reductase (GR) expression, whose encoding enzyme regenerates glutathione (GSH) by reducing its oxidized form GSSG using NADH as a substrate (Foyer and Noctor, 2009), was also decreased in N-limited cells (Figure 2C). The expression of glutathione peroxidase-like genes (GPrxL-1 and GPrxL-2) was lower in N-limited cells compared with N-replete cells; the encoded enzymes possibly use GSH to reduce H2O2. This result indicated that GSH-mediated quenching of H2O2 may be lower in C. sorokiniana N-limited cells as well.
Decreased transcript abundance of genes encoding key enzymes involved in ROS quenching suggested that C. sorokiniana cells experiencing N limitation accumulate ROS in a deliberate, transcriptionally regulated manner. To test this hypothesis, we measured intracellular ROS levels in N-limited cells using the cell-permeable ROS-sensitive fluorescent dye 2′-7′-dichlorodihydrofluorescein diacetate (DCFDA). Indeed, we determined that ROS levels are significantly higher in N-limited C. sorokiniana cells than in the control cells (Figure 2D).
To gain a more complete picture of redox regulation under N limitation, we analyzed the expression profile of various genes encoding members of the ferredoxin, peroxiredoxin, thioredoxin, and glutaredoxin families. We observed significant changes in the expression of many Ferredoxin genes (Table 2). Their encoded proteins can function as a reservoir of redox potential, accepting or donating electrons to various biochemical processes, including photosynthesis (Schürmann and Buchanan, 2008). The expression of four Peroxiredoxin genes was significantly decreased upon N limitation (Table 2). Along with catalases and peroxidases, peroxiredoxins scavenge hydrogen peroxide (Rhee and Woo, 2011). The lower expression levels of Peroxiredoxin genes suggested that yet another mechanism of ROS scavenging might be perturbed in cells exposed to N limitation. The expression of many Thioredoxin genes also changed upon N limitation (Table 2). Thioredoxins contribute to cell signaling by modifying cysteine residues in proteins (Baumann and Juttner, 2002). We also noticed decreased abundance of genes encoding thioredoxin-reducing enzymes such as ferredoxin-thioredoxin reductases 1 and 2 (Frdxn TrxRed1 and 2) (Schürmann and Buchanan, 2008) (Table 2), indicating that thioredoxin-mediated redox signaling is altered under N-limitation conditions. Genes encoding glutaredoxins, which share functional similarity to thioredoxins (Meyer et al., 2008), were also differentially regulated (Table 2). However, we identified these genes based on sequence similarity to previously characterized members of this gene family; determining their putative function requires further characterization and validation.
In light of the observation that genes encoding SODs, catalase, peroxidases, ferredoxins, and peroxiredoxins are downregulated upon N limitation, we propose that ROS accumulation in N-limited C. sorokiniana cells is mediated, at least partially, by the downregulation of ROS scavenging enzymes leading to a more oxidized cellular environment.
We next asked if the observed increase in ROS levels contributed to the degradation of MGDG and other membrane remodeling events observed during N-limiting growth. To this end, we treated cells grown under N-replete conditions with diethyldithiocarbamate (DDC), a known inhibitor of SOD (Heikkila et al., 1976). Treatment with DDC induced ROS accumulation (Supplementary Figure 3). Treatment of cells with 250 µM DDC resulted in MGDG degradation (Figure 3A), although not to the same extent as in N-limited cells. The conversion of superoxide to hydrogen peroxide catalyzed by SOD can also occur spontaneously at a slower rate, and other ROS quenching mechanisms are active as well, which may decrease the accumulation of ROS relative to that of N-limited cells. Conversely, we wished to test if preventing the accumulation of ROS under N-limitation conditions might prevent membrane remodeling. To test this hypothesis, we supplemented the medium with 500 µM ascorbate to quench cellular hydrogen peroxide. We established that ROS levels are lower in ascorbate-treated cells 24 h after the onset of N-limitation (Figure 3B). These lower ROS levels under N limitation were coupled with a decrease in MGDG degradation (Figure 3C). A time-course analysis indicated that while N-limited cultures supplemented with ascorbate still degraded MGDG, their rate of degradation was slower than that of control N-limited cultures (Figure 3C). This is congruent with the observation that MGDG levels are higher in N-limited cultures when treated with ascorbate (Figure 3D). We also noticed that TAG accumulates to lower levels in N-limited cells treated with ascorbate after 24 h of N limitation (Figure 3E). Taken together, these results indicate that ROS accumulation under N limitation plays a vital role in chloroplast membrane remodeling degradation of MGDG and channeling fatty acids for TAG biosynthesis.
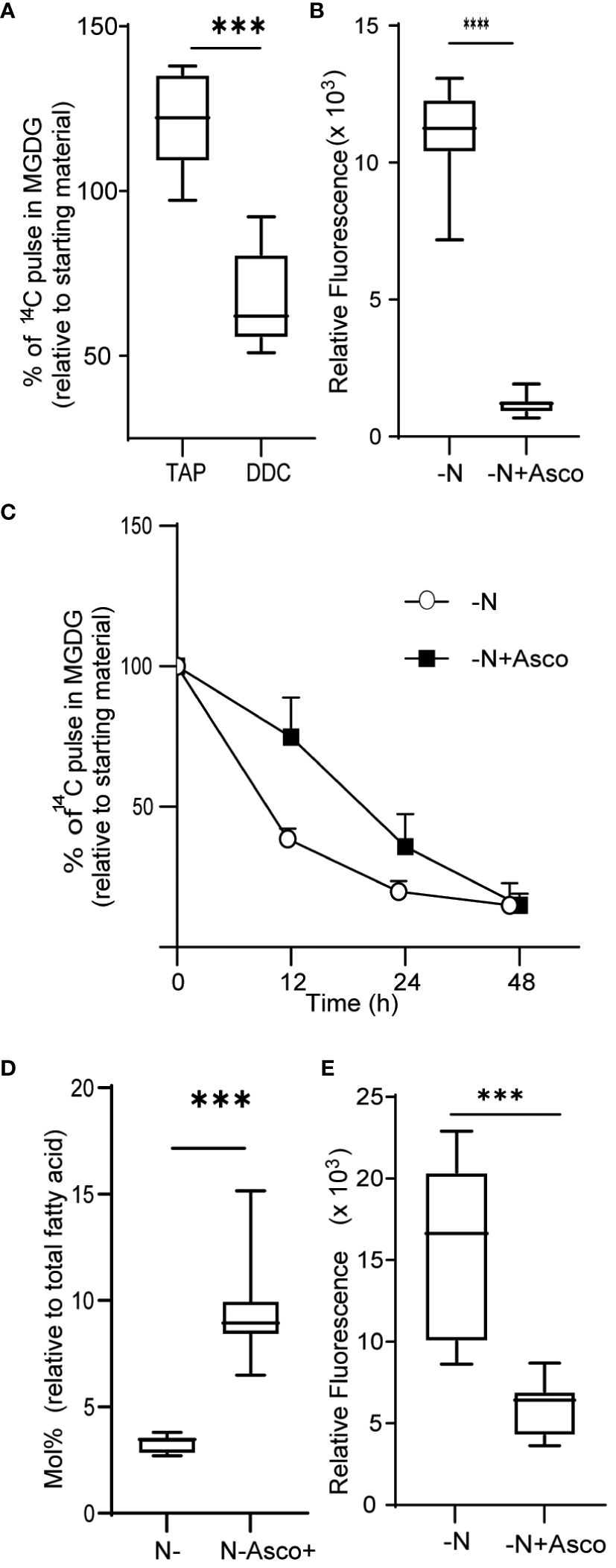
Figure 3 Quenching of ROS under nitrogen limitation and induction of ROS under nitrogen replete conditions. (A) Quantitative measurement of MGDG degradation in nitrogen replete media (TAP) when treated with 250 µM diethyldithiocarbamate (DDC) for 24 h. (B) ROS level under nitrogen-limited media fed with ascorbate, as measured by DCFDA fluorescence after 24 h of inoculation. Fluorescence values are normalized to O.D. (C) Quantitative measurement of MGDG degradation under N lim media when fed with 500 μM ascorbate. (D) Mole percentage of MGDG relative to total fatty acid from cultures under N limitation with or without ascorbate. (E) Level of TAG accumulation as measured by Nile red, under nitrogen limitation after 24 h of inoculation when fed with ascorbate. In all experiments n>7. Asterix indicates the statistical significance of the student t-test. *** indicates <0.001 and **** indicates <0.0001.
We sought to identify the possible source of ROS. Photosynthesis is a prominent source of superoxide in all photosynthetic organisms (Czarnocka and Karpiński, 2018). We thus tested the hypothesis that the increase in ROS levels observed in N-limited cells is derived from photosynthesis. To this end, we measured ROS levels in cells grown under continuous light or continuous darkness in N-limited conditions. We predicted that ROS levels would be lower in N-limited cells grown in the dark. However, we measured no appreciable difference in ROS levels between light- and dark-grown cells (Figure 4A). This result was also reflected in the very similar lipid profiles of the two types of cultures, as revealed by autoradiography after thin-layer chromatography separation of lipids (Figure 4B). We also quantified 1,2-14C-acetate-labeled MGDG levels from the autoradiograms and observed no differences between light- and dark-grown cultures (Figure 4C). These results collectively indicated that ROS involved in lipid remodeling in C. sorokiniana are from light-independent sources.
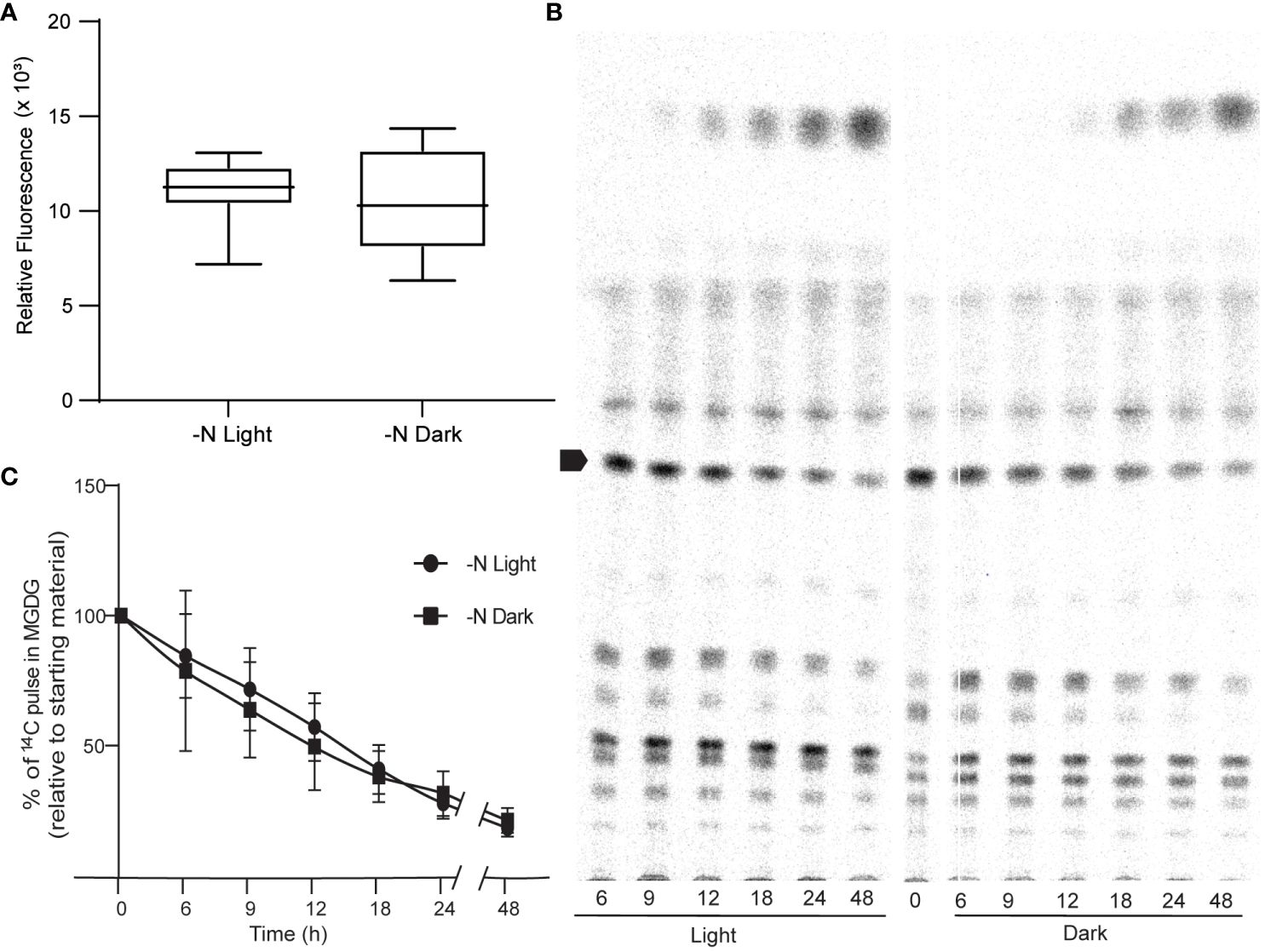
Figure 4 Photosynthesis as a source of ROS. (A) ROS levels of N lim culture grown under light (N-lim-L) and dark (N-lim-D) regimes as measured by DCFDA fluorescence after 24 h of inoculation. Fluorescence relative to O.D. (B) A representative TLC radiograph showing the remodeling of different lipids under nitrogen limitation in light or dark regime. Arrow indicates MGDG. (C) Quantitative measurement of degradation of preformed MGDG relative to the starting levels of 14C pulse in MGDG in N-lim cultures grown in light and dark regime. Error bars indicate SD (n = 3).
We next queried the transcriptome to identify genes encoding ROS-generating enzymes that are differentially expressed under N limitation. We determined that the expression of genes such as NADPH oxidase, Xanthine oxidase/dehydrogenase, Putrescine oxidase-like, and Copper amine oxidase (Tyramine oxidase-like) is upregulated under N limitation (Figure 5). The encoded enzymes all catalyze reactions that produce hydrogen peroxide or superoxide (Ma X. et al., 2016; Tavladoraki et al., 2016; Zandalinas and Mittler, 2018). Thus, the altered gene expression profiles we observed in genes with assigned roles in ROS generation suggested that ROS accumulation is enhanced by both the repression of ROS-quenching activities and the induction of ROS-generating activities.
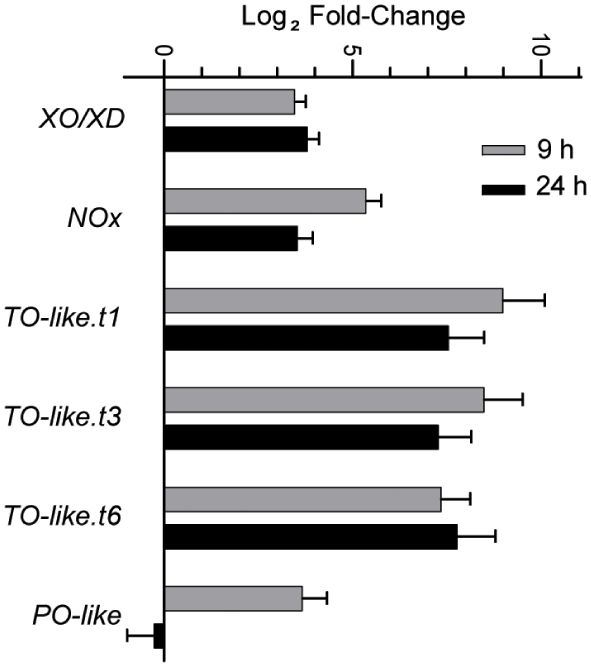
Figure 5 Expression of different ROS generating enzymes that increase during N-limited growth as detected in the RNA-seq studies. Error bars indicate SD (n = 2). XO/XD, xanthine oxidase/dehydrogenase; NOx, NADPH oxidase; TO-like, tyramine oxidase-like; PO-like, putrescine oxidase-like. TO-like.t1, t3 and t6 indicate the three different splice variants encoded by the same loci.
NADPH oxidases (NOXs) are well-studied enzymes due to their various physiological roles in human health. Their functions are well researched in mammalian systems and are important drug targets to counteract various disorders (Meitzler et al., 2014). Multiple NOX inhibitors are therefore available to manipulate NOX activity levels (Altenhöfer et al., 2015). Here, we used 2-acetyl phenothiazine, also known as ML171, to inhibit NOX activity and, thus, NOX-mediated ROS production (Gianni et al., 2010). When cultures were treated with 50 µM ML171 under N limitation, we observed a decrease in MGDG degradation (Figure 6A). Decrease in MGDG degradation leads to increased amount of MGDG as a percentage of total lipids (Figure 6B). This is coupled with lower TAG accumulation (Figure 6C). However, we did not see a significant decrease in ROS levels (Figure 6D), as measured by DCFDA fluorescence. The fact that MGDG degradation and TAG accumulation are decreased upon treatment with ML171 indicated that NADPH oxidase may play a role in the N starvation response.
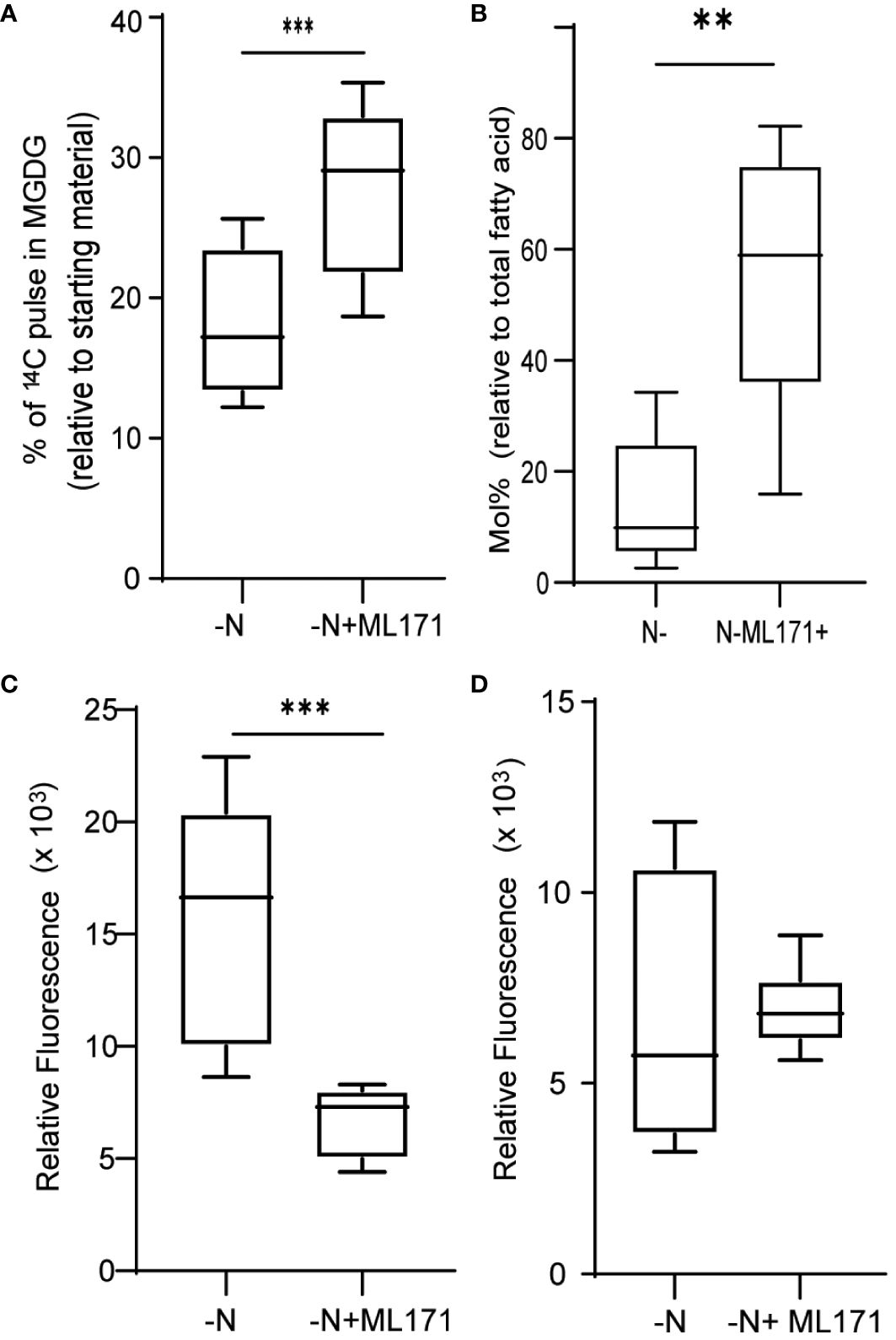
Figure 6 Role of NADPH oxidase in ROS generation and membrane remodeling. (A) MGDG degradation as measured by the remaining amount of preformed MGDG (24 h after inoculation), under nitrogen limitation when treated with 50 µM ML171-NADPH oxidase inhibitor. (B) Mol% of MGDG relative to total fatty acids in the cultures under N limitation treated with or without ML171. (C) TAG levels, measured as Nile red fluorescence, at 24 h post inoculation in cultures treated with ML171. (D) ROS levels, as measured by DCFDA fluorescence, under N-lim condition when treated with, ML171 (50 µM). ROS assay was carried out after 24 h of inoculation. Fluorescence is normalized to O.D. All error bars indicate S.D (n = 3). ** indicates a p-value < 0.001 and *** Indicates a p-value < 0.0001 in a paired student t-test.
Macronutrient limitation has been studied for decades as a potent inducer of storage compound (oil and starch) accumulation in oleaginous photosynthetic microorganisms. However, the industrial application of this mechanism of oil accumulation is invariably limited by lower photosynthetic capacity and biomass yield. Finding ways to stimulate oil accumulation without a growth penalty or diminished biomass accumulation has been a major goal of the research community. Indeed, a major driver of the US Department of Energy Aquatic Species Program (Sheehan, 1998) was the identification of a hypothetical “lipid trigger” that could be induced under optimal growth conditions, leading to the reproducible and high-yielding production of desired products. This hypothetical trigger has proven elusive, and the manipulation of single enzymes or transcriptional regulators has been only marginally effective in achieving the desired goals (Ajjawi et al., 2017).
Another recently proposed approach for the “lipid trigger” is the use of synthetic chemicals that induce storage lipid production with minimal retardation in growth, although the precise molecular mechanisms by which these compounds induce lipogenesis are, at present, poorly defined (Wase et al., 2014; Wase et al., 2015; Wase et al., 2018; Wase et al., 2019). Nevertheless, significant progress has been made in understanding the enzymology and certain aspects of the regulation of TAG biosynthesis and degradation in algal model systems such as Chlamydomonas (Li-Beisson et al., 2015; Li-Beisson et al., 2019) and Nannochloropsis (Ma X. N. et al., 2016). Clearly, the existence and nature of a hypothetical “lipid trigger” remain tenuous. This study presents several new insights related to abiotic stress signals that likely play an underappreciated role in stress-induced TAG accumulation in oleaginous algae.
First, we showed that, as in other algal species, N-limitation in C. sorokiniana UTEX-1230 is a potent inducer of thylakoid membrane degradation and channeling of fatty acids from chloroplast lipids into TAG (Figure 7). Our transcriptome analysis revealed that the expression of genes encoding many key enzymes involved in regulating ROS levels in cells, such as superoxide dismutase, catalase, and stromal ascorbate peroxidase, is downregulated, concomitantly with an increase in ROS levels under N limitation. This observation strongly suggests that cells actively accumulate ROS in a regulated fashion under these conditions. This is achieved by lowering the transcript abundance of enzymes involved in ROS quenching. The DCFDA assay provides a quantitative measurement of total ROS but does not indicate which specific ROS species (superoxides, hydrogen peroxide, or others) accumulate or which is linked to membrane remodeling and TAG accumulation. It is also possible that each ROS species plays an independent role in the various branches of the N starvation response.
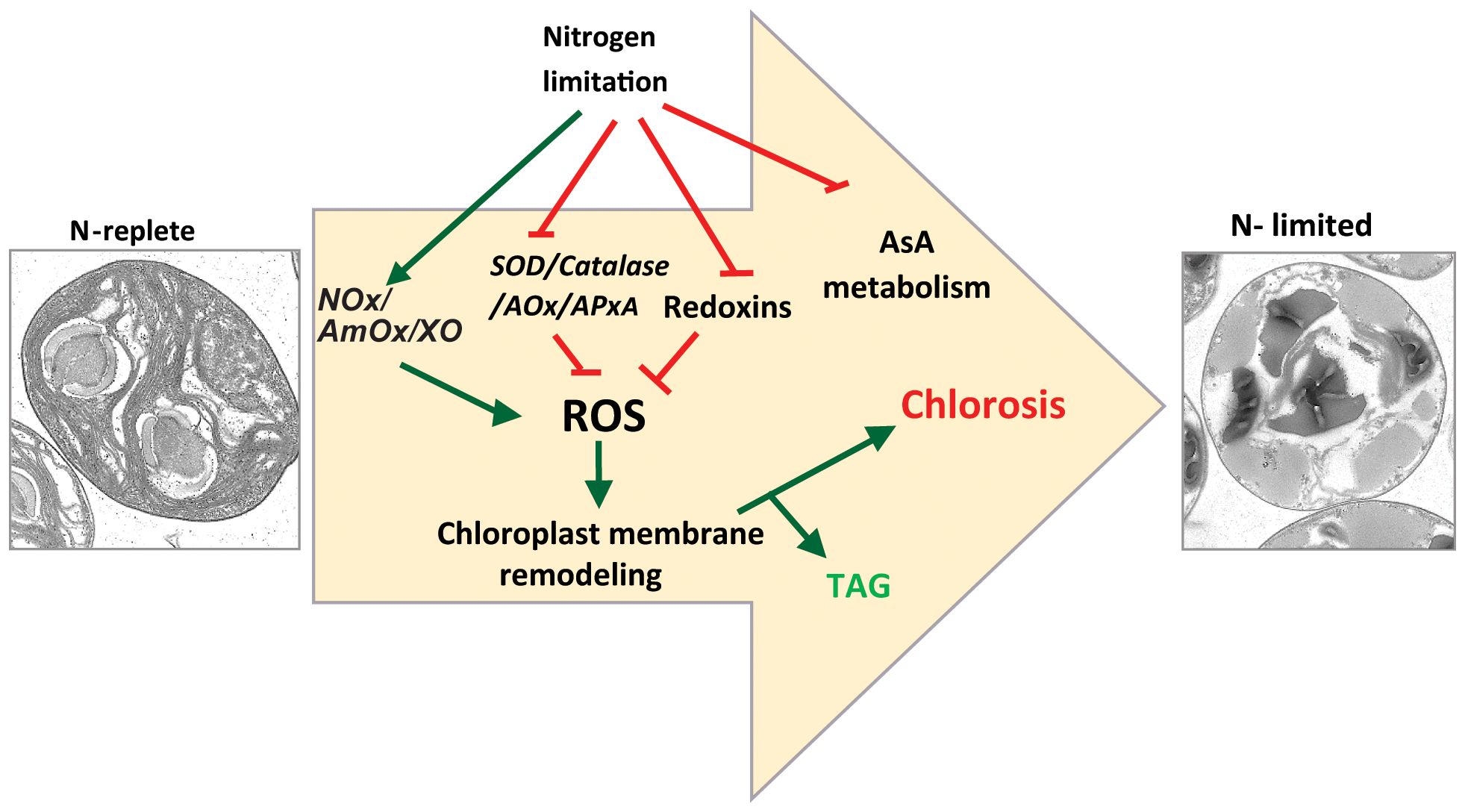
Figure 7 Proposed model for nitrogen starvation mediated ROS accumulation leading to chlorosis and TAG accumulation. NOx, NADPH oxidase; AmOx, amine oxidase; XO, xanthine oxidase; SOD, superoxide dismutase; AOx, alternate oxidase; APx, ascorbate oxidase.
We cannot ascertain the subcellular localization of ROS quenching enzymes, as C. sorokiniana is not yet amenable to genetic transformation. However, the separation of cells into various organelle preparation and biochemical assays showed that a Fe-SOD is present in the chloroplast, whereas a Mn-SOD is present in mitochondria in Chlamydomonas (Kitayama et al., 1999). In the case of C. sorokiniana, we determined that the expression of the genes encoding both Fe-SOD (SOD2 and SOD3) and Mn-SOD (SOD1) is downregulated upon N limitation. While we observed that the expression of SOD4, encoding a Mn-SOD, increased under our N-limited conditions, the relevance of this increase is unclear. Catalase is a mitochondrial protein in Chlamydomonas (Kato et al., 1997). The lower expression of genes encoding ROS-scavenging enzymes localizing to both chloroplast and mitochondria indicates the possibility that more than one organelle may be a source of ROS under N-limiting conditions.
The expression of genes encoding multiple ferredoxins and peroxiredoxins known to be involved in maintaining redox homeostasis changed upon N limitation. Additionally, we observed that the expression of genes related to ROS generation such as Amine oxidases, Xanthine oxidase, and NOXs increased. This result indicated that C. sorokiniana cells may undertake a two-pronged approach to ROS accumulation by downregulating scavenging systems and upregulating ROS-producing processes, further indicating that ROS accumulation is an active and regulated process.
The data presented in this study pertaining to ROS accumulation and redox signaling are based on gene expression profiles, which may not always correlate with the abundance or the activity of the gene product, which can depend on absolute protein levels and protein modifications. However, we saw a coordinated decrease in the expression levels of many key genes related to ROS accumulation and redox signaling. These observations, coupled with an increase in ROS levels in the cell, indicate that this response is likely a transcriptionally regulated process and that there is an underappreciated role for ROS and redox signaling under N limitation. Higher ROS levels under N limitation were also observed in the diatom Phaeodactylum tricornutum (Mizrachi et al., 2019). Accumulation of ROS under N limitation is thus not limited to C. sorokiniana and may confer a physiological advantage that has enabled this phenotype to be selected and maintained in these distantly related photosynthetic eukaryotes.
The ectopic induction of ROS accumulation by inhibiting SOD leads to MGDG degradation, whereas ROS quenching under N limitation limits MGDG degradation. The concomitant decrease in TAG levels in ascorbate-treated N-limited cells is thus likely due to a lower rate of degradation of membrane lipids such as MGDG and phospholipids. From these observations, it is clear that membrane remodeling–mediated TAG accumulation is closely linked to redox signaling. This result is in line with Zhang et al. (2013) who observed that exogenous application of hydrogen peroxide was sufficient to induce TAG accumulation in C. sorokiniana C3. In Chlamydomonas, a substantial flux of fatty acid for TAG accumulation is derived from preexisting glycerolipids (Young and Shachar-Hill, 2021). Indeed, mutations in genes encoding enzymes such as PGD1 or PDAT1 lead to lower TAG accumulation under N limitation. PGD1 and PDAT1 enzymes are involved in the turnover of fatty acid from MGDG and phosphatidylcholine to TAG, respectively (Li et al., 2012b; Yoon et al., 2012). In sum, our study strongly implicates N-limitation induced ROS accumulation as playing a crucial role in the metabolic reprogramming that leads to TAG accumulation (Figure 7). Future studies will be aimed at identifying the components that sense N limitation and alter ROS accumulation, as well as identifying the enzymes that act downstream of the ROS burst to regulate membrane turnover and TAG accumulation, thus providing additional details regarding the molecular trigger(s) for macronutrient-limited TAG accumulation in microalgae.
The data presented in the study are deposited in the NCBI repository, accession number PRJNA1111962.
JV: Conceptualization, Data curation, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. NW: Data curation, Methodology, Visualization, Writing – review & editing. KL: Methodology, Resources, Writing – review & editing. WM: Methodology, Writing – review & editing. CZ: Methodology, Resources, Writing – review & editing. WR: Conceptualization, Funding acquisition, Investigation, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Science Foundation award number EPS-1004094 (to WR), NASA award number 80NSSC17K0737 (to WR), and startup funding from the University of Nebraska-Lincoln College of Arts and Sciences and School of Biological Sciences (to WR).
We thank Dr. Rebecca Roston for her critical reading of the manuscript. This article has been submitted to bioRxiv as a preprint (Vijayan et al., 2021).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1418049/full#supplementary-material
Ajjawi, I., Verruto, J., Aqui, M., Soriaga, L. B., Coppersmith, J., Kwok, K., et al. (2017). Lipid production in Nannochloropsis gaditana is doubled by decreasing expression of a single transcriptional regulator. Nat. Biotechnol. 35, 647–652. doi: 10.1038/nbt.3865
Allen, J., Black, P. N., Riekhof, W. R., Allen, J., Unlu, S., Demirel, Y., et al. (2018). Integration of biology, ecology and engineering for sustainable algal-based biofuel and bioproduct biorefinery Integration of biology, ecology and engineering for sustainable algal-based biofuel and bioproduct biorefinery. Bioresour. Bioprocess. 5, 47 doi: 10.1186/s40643-018-0233-5
Altenhöfer, S., Radermacher, K. A., Kleikers, P. W. M., Wingler, K., Schmidt, H. H. H. W. (2015). Evolution of NADPH oxidase inhibitors: Selectivity and mechanisms for target engagement. Antioxidants Redox Signaling 23, 406–427. doi: 10.1089/ars.2013.5814
Asada, K. (2006). Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 141, 391–396. doi: 10.1104/pp.106.082040
Baumann, U., Juttner, J. (2002). Plant thioredoxins: the multiplicity conundrum. CMLS, Cell. Mol. Life Sci. 59, 1042–1057. doi: 10.1007/s00018-002-8485-8
Ben Rejeb, K., Benzarti, M., Debez, A., Bailly, C., Savouré, A., Abdelly, C. (2015). NADPH oxidase-dependent H2O2 production is required for salt-induced antioxidant defense in Arabidopsis thaliana. J. Plant Physiol. 174, 5–15. doi: 10.1016/j.jplph.2014.08.022
Boyle, N. R., Page, M. D., Liu, B., Blaby, I. K., Casero, D., Kropat, J., et al. (2012). Three acyltransferases and nitrogen-responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas. J. Biol. Chem. 287, 15811–15825. doi: 10.1074/jbc.M111.334052
Czarnocka, W., Karpiński, S. (2018). Friend or foe? Reactive oxygen species production, scavenging and signaling in plant response to environmental stresses. Free Radical Biol. Med. 122, 4–20. doi: 10.1016/j.freeradbiomed.2018.01.011
Foyer, C. H., Noctor, G. (2009). Redox regulation in photosynthetic organisms: Signaling, Acclimation, and Practical Implications. Antioxidants & Redox Signaling 11, 861–905. doi: 10.1089/ars.2008.2177
Gianni, D., Taulet, N., Zhang, H., Dermardirossian, C., Kister, J., Martinez, L. (2010). A novel and specific NADPH oxidase-1 (Nox1) small-molecule inhibitor blocks the formation of functional invadopodia in human colon cancer cells. ACS Chem. Biol. 5, 981–993. doi: 10.1021/cb100219n
Gojkovic, Ž., Vílchez, C., Torronteras, R., Vigara, J., Gómez-Jacinto, V., Janzer, N., et al. (2014). Effect of selenate on viability and selenomethionine accumulation of Chlorella sorokiniana grown in batch culture. Sci. World J. 401265, 13. doi: 10.1155/2014/401265
Gorman, D. S., Levine, R. P. (1965). Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardii. Proc. Natl. Acad. Sci. U.S.A 54, 1665–1669. doi: 10.1073/pnas.54.6.1665
Gupta, D. K., Palma, J. M., Corpas, F. J. (2013). Heavy Metal Stress In Plants Springer. doi: 10.1007/978-3-642-38469-1
He, H., Yan, J., Yu, X., Liang, Y., Fang, L., Scheller, H. V., et al. (2017). The NADPH-oxidase AtRbohI plays a positive role in drought-stress response in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 491, 834–839. doi: 10.1016/j.bbrc.2017.05.131
Heikkila, R. E., Cabbat, F. S., Cohen, G. (1976). In vivo inhibition of superoxide dismutase in mice by diethyldithiocarbamate. J. Biol. Chem. 251, 2182–2185. doi: 10.1016/S0021-9258(17)33675-X
Hu, Q., Sommerfeld, M., Jarvis, E., Ghirardi, M., Posewitz, M., Seibert, M., et al. (2008). Microalgal triacylglycerols as feedstocks for biofuel production: Perspectives and advances. Plant J. 54, 621–639. doi: 10.1111/j.1365-313X.2008.03492.x
Jiang, G., Li, F., Wang, S., Sun, J., Liu, Z. (2019). Positive correlation between lipid accumulation and gene expression of a copper-containing amine oxidase gene in Chlorella under nitrogen starvation. Algal Res. 40, 101504. doi: 10.1016/j.algal.2019.101504
Kato, J., Yamahara, T., Tanaka, K., Takio, S., Satoh, T. (1997). Characterization of catalase from green algae Chlamydomonas reinhardtii. J. Plant Physiol. 151, 262–268. doi: 10.1016/S0176-1617(97)80251-9
Kitayama, K., Kitayama, M., Togasaki, R. K. (1999). Subcellular localization of iron and manganese superoxide dismutase in Chlamydomonas reinhardtii (chlorophyceae). J. Phycology. 142, 136–142. doi: 10.1046/j.1529-8817.1999.3510136.x
Li, X., Benning, C., Kuo, M. H. (2012a). Rapid triacylglycerol turnover in Chlamydomonas reinhardtii requires a lipase with broad substrate specificity. Eukaryotic Cell. 11, 1451–1462. doi: 10.1128/EC.00268-12
Li, X., Moellering, E. R., Liu, B., Johnny, C., Fedewa, M., Sears, B. B., et al. (2012b). A galactoglycerolipid lipase is required for triacylglycerol accumulation and survival following nitrogen deprivation in Chlamydomonas reinhardtii. Plant Cell 24, 4670–4686. doi: 10.1105/tpc.17.00446
Liao, Y., Smyth, G. K., Shi, W. (2013). The Subread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41 (10), e108. doi: 10.1093/nar/gkt214
Li-Beisson, Y., Beisson, F., Riekhof, W. (2015). Metabolism of acyl-lipids in Chlamydomonas reinhardtii. Plant J. 82, 504–522. doi: 10.1111/tpj.12787
Li-Beisson, Y., Thelen, J. J., Fedosejevs, E., Harwood, J. L. (2019). The lipid biochemistry of eukaryotic algae. Prog. Lipid Res. 74, 31–68. doi: 10.1016/j.plipres.2019.01.003
Ma, X. N., Chen, T. P., Yang, B., Liu, J., Chen, F. (2016). Lipid production from nannochloropsis. Mar. Drugs 14. doi: 10.3390/md14040061
Ma, X., Wang, W., Bittner, F., Schmidt, N., Berkey, R., Zhang, L., et al. (2016). Dual and opposing roles of xanthine dehydrogenase in defense-associated reactive oxygen species metabolism in arabidopsis. Plant Cell. 28, 1108–1126. doi: 10.1105/tpc.15.00880
Meitzler, J. L., Antony, S., Wu, Y., Juhasz, A., Liu, H., Jiang, G., et al. (2014). NADPH oxidases: A perspective on reactive oxygen species production in tumor biology. Antioxidants Redox Signaling 20, 2873–2889. doi: 10.1089/ars.2013.5603
Meyer, Y., Siala, W., Bashandy, T., Riondet, C., Vignols, F., Reichheld, J. P. (2008). Glutaredoxins and thioredoxins in plants. Biochim. Biophys. Acta - Mol. Cell Res. 1783, 589–600. doi: 10.1016/j.bbamcr.2007.10.017
Miller, R., Wu, G., Deshpande, R. R., Vieler, A., Gartner, K., Li, X., et al. (2010). Changes in transcript abundance in Chlamydomonas reinhardtii following nitrogen deprivation predict diversion of metabolism. Plant Physiol. 154, 1737–1752. doi: 10.1104/pp.110.165159
Mizrachi, A., Graff van Creveld, S., Shapiro, O. H., Rosenwasser, S., Vardi, A. (2019). Light-dependent single-cell heterogeneity in the chloroplast redox state regulates cell fate in a marine diatom. eLife. 8, 1–27. doi: 10.7554/eLife.47732
Møller, I. M. (2001). P LANT M ITOCHONDRIA AND O XIDATIVE S TRESS: electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 561–591. doi: 10.1146/annurev.arplant.52.1.561
Rhee, S. G., Woo, H. A. (2011). Multiple functions of peroxiredoxins: Peroxidases, sensors and regulators of the intracellular messenger H2O2, and protein chaperones. Antioxidants Redox Signaling 15, 781–794. doi: 10.1089/ars.2010.3393
Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W., et al. (2015). Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. doi: 10.1093/nar/gkv007
Robinson, M. D., McCarthy, D. J., Smyth, G. K. (2009). edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26, 139–140. doi: 10.1093/bioinformatics/btp616
Schieber, M., Chandel, N. S. (2014). ROS function in redox signaling and oxidative stress. CURBIO. 24, R453–R462. doi: 10.1016/j.cub.2014.03.034
Schürmann, P., Buchanan, B. B. (2008). The ferredoxin/thioredoxin system of oxygenic photosynthesis. Antioxidants Redox Signaling 10, 1235–1273. doi: 10.1089/ars.2007.1931
Sheehan, J., Dunahay, T., Benemann, J., Roessler, P. (1998). A look back at the u. s. department of energy’s aquatic species program: biodiesel from algae. Natl. Renewable Energy Lab.
Shtaida, N., Khozin-Goldberg, I., Solovchenko, A., Chekanov, K., Didi-Cohen, S., Leu, S., et al. (2014). Downregulation of a putative plastid PDC E1α subunit impairs photosynthetic activity and triacylglycerol accumulation in nitrogen-starved photoautotrophic Chlamydomonas reinhardtii. J. Exp. Bot. 65, 6563–6576. doi: 10.1093/jxb/eru374
Smirnoff, N. (2018). Ascorbic acid metabolism and functions: A comparison of plants and mammals. Free Radical Biol. Med. 122, 116–129. doi: 10.1016/j.freeradbiomed.2018.03.033
Stephenson, A. L., Dennis, J. S., Howe, C. J., Scott, S. A., Smith, A. G. (2010). Influence of nitrogen-limitation regime on the production by Chlorella vulgaris of lipids for biodiesel feedstocks. Biofuels. 1, 47–58. doi: 10.4155/bfs.09.1
Tavladoraki, P., Cona, A., Angelini, R. (2016). Copper-containing amine oxidases and FAD-dependent polyamine oxidases are key players in plant tissue differentiation and organ development. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00824
Vijayan, J., Wase, N., Liu, K., Zhang, C., Riekhof, W. R. (2021). Reactive oxygen species mediate thylakoid membrane remodeling and triacylglycerol synthesis under nitrogen starvation in the alga Chlorella sorokiniana. bioRxiv Preprint. doi: 10.1101/2021.05.15.444036
Wase, N., Black, P., DiRusso, C. (2018). Innovations in improving lipid production: Algal chemical genetics. Prog. Lipid Res. 71, 101–123. doi: 10.1016/j.plipres.2018.07.001
Wase, N., Black, P. N., Stanley, B. A., Dirusso, C. C. (2014). Integrated quantitative analysis of nitrogen stress response in Chlamydomonas reinhardtii using metabolite and protein profiling. J. Proteome Res. 13, 1373–1396. doi: 10.1021/pr400952z
Wase, N., Tu, B., Black, P. N., DiRusso, C. C. (2015). Phenotypic screening identifies Brefeldin A/Ascotoxin as an inducer of lipid storage in the algae Chlamydomonas reinhardtii. Algal Res. 11, 74–84. doi: 10.1016/j.algal.2015.06.002
Wase, N., Tu, B., Allen, J. W., Black, P. N., DiRusso, C. C. (2017). Identification and metabolite profiling of chemical activators of lipid accumulation in green algae. Plant Physiol. 174, 2146–2165. doi: 10.1104/pp.17.00433
Wase, N., Tu, B., Rasineni, G. K., Cerny, R., Grove, R., Adamec, J., et al. (2019). Remodeling of Chlamydomonas metabolism using synthetic inducers results in lipid storage during growth. Plant Physiol. 181, 00758.2019. doi: 10.1104/pp.19.00758
Yoon, K., Han, D., Li, Y., Sommerfeld, M., Hu, Q. (2012). Phospholipid: Diacylglycerol acyltransferase is a multifunctional enzyme involved in membrane lipid turnover and degradation while synthesizing triacylglycerol in the unicellular green microalga Chlamydomonas reinhardtii. Plant Cell 24, 3708–3724. doi: 10.1105/tpc.112.100701
Young, D. Y., Shachar-Hill, Y. (2021). Large fluxes of fatty acids from membranes to triacylglycerol and back during N-deprivation and recovery in Chlamydomonas. Plant Physiol. 185, 796–814. doi: 10.1093/plphys/kiaa071
Zandalinas, S. I., Mittler, R. (2018). ROS-induced ROS release in plant and animal cells. Free Radical Biol. Med. 122, 21–27. doi: 10.1016/j.freeradbiomed.2017.11.028
Zhang, Y. M., Chen, H., He, C. L., Wang, Q. (2013). Nitrogen starvation induced oxidative stress in an oil-producing green alga chlorella sorokiniana C3. PloS One 8, 1–12. doi: 10.1371/journal.pone.0069225
Keywords: microalgae, oil accumulation, nitrogen limitation, ROS, lipid metabolism, membrane remodeling, biofuels
Citation: Vijayan J, Wase N, Liu K, Morse W, Zhang C and Riekhof WR (2024) ROS-mediated thylakoid membrane remodeling and triacylglycerol biosynthesis under nitrogen starvation in the alga Chlorella sorokiniana. Front. Plant Sci. 15:1418049. doi: 10.3389/fpls.2024.1418049
Received: 15 April 2024; Accepted: 11 June 2024;
Published: 08 July 2024.
Edited by:
Jianping Yu, National Renewable Energy Laboratory (DOE), United StatesReviewed by:
Seth Steichen, National Renewable Energy Laboratory (DOE), United StatesCopyright © 2024 Vijayan, Wase, Liu, Morse, Zhang and Riekhof. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jithesh Vijayan, aml0aGVzaGJ0QGh1c2tlcnMudW5sLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.