
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 17 July 2024
Sec. Plant Breeding
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1417314
This article is part of the Research TopicManipulation of Plant Architecture for Crop ProductionView all 10 articles
 Zhuonan Dai1†
Zhuonan Dai1† Shaoyun Dong1†
Shaoyun Dong1† Hexu Cai1
Hexu Cai1 Diane M. Beckles2
Diane M. Beckles2 Jiantao Guan1
Jiantao Guan1 Xiaoping Liu1
Xiaoping Liu1 Xingfang Gu1
Xingfang Gu1 Han Miao1*
Han Miao1* Shengping Zhang1*
Shengping Zhang1*Background: The plant root system is critical for the absorption of water and nutrients, and have a direct influence on growth and yield. In cucumber, a globally consumed crop, the molecular mechanism of root development remains unclear, and this has implications for developing stress tolerant varieties. This study sought to determine the genetic patterns and related genes of cucumber root weight. A core cucumber germplasms population was used to do the GWAS analysis in three environments.
Results: Here, we investigated four root-weight related traits including root fresh weight (RFW), root dry weight (RDW), ratio of root dry weight to root fresh weight (RDFW) and the comprehensive evaluation index, D-value of root weight (DRW) deduced based on the above three traits for the core germplasm of the cucumber global repository. According to the D-value, we identified 21 and 16 accessions with light and heavy-root, respectively. We also found that the East Asian ecotype accessions had significantly heavier root than other three ecotypes. The genome-wide association study (GWAS) for these four traits reveals that 4 of 10 significant loci (gDRW3.1, gDRW3.2, gDRW4.1 and gDRW5.1) were repeatedly detected for at least two traits. Further haplotype and expression analysis for protein-coding genes positioned within these 4 loci between light and heavy-root accessions predicted five candidate genes (i.e., Csa3G132020 and Csa3G132520 both encoding F-box protein PP2-B1 for gDRW3.1, Csa3G629240 encoding a B-cell receptor-associated protein for gDRW3.2, Csa4G499330 encodes a GTP binding protein for gDRW4.1, and Csa5G286040 encodes a proteinase inhibitor for gDRW5.1).
Conclusions: We conducted a systematic analysis of the root genetic basis and characteristics of cucumber core germplasms population. We detected four novel loci, which regulate the root weight in cucumber. Our study provides valuable candidate genes and haplotypes for the improvement of root system in cucumber breeding.
Cucumber belongs to the Cucurbitaceae and is an important vegetable crop in the world. In 2021, the harvested area of cucumber reached 3,464,737 hectares, producing 169,126,456 tons worldwide (FAOSTAT, 2021). However, cucumber has a shallow root system that weakly absorbs water and fertilizer (Beyaert et al., 2007). It is usually necessary to increase cucumber yield through grafting (Huang et al., 2009; Li et al., 2014; Usanmaz and Abak, 2019). In recent years, breeders have made efforts to further improve crop yield through root improvement (Dorlodot et al., 2007; Uga et al., 2013). Some studies have examined the root architecture and branching patterns in cucumber (Zhang et al., 2012; Kiryushkin et al., 2024; Cai et al., 2021). It is known that cucumbers have longer taproots and shallower fibrous roots. The lateral roots branching out from the primary root (Mao et al., 2003). However, the specific genetic factors and regulatory mechanisms controlling root architecture in cucumber are not yet fully characterized. Compared to other crops such as Oryza sativa L (Kamoshita et al., 2002; Courtois et al., 2009), Zea mays L (Hochholdinger and Tuberosa, 2009; Burton et al., 2015), Glycine max (Liang et al., 2014, Liang et al., 2017) and the model plant Arabidopsis thaliana (Fukaki et al., 2002; Knight, 2007), there is less research on the genetic diversity and key regulatory genes of cucumber roots, which hinders future genetic breeding.
Root development is influenced by many factors, such as internal and external environments. As one of the endogenous factors, phytohormones [i.e., ethylene, auxin, gibberellin (GA), cytokinin (CK), jasmonic acid (JA), and brassinosteroid (BR)] and their crosstalk have been shown to play vital roles in the regulation of root growth (Li et al., 2022). Auxin, as one of the well-studied phytohormones, plays a critical role in regulating the elongation of primary and lateral roots (Ljung et al., 2005). Himanen et al. (2002) reported that auxin can promote lateral root formation by inhibiting the expression of cell division inhibitors KRP1 and KRP2. BR can also act synergically with auxin, and regulate the polar transport of auxin to promote lateral root development (Bao et al., 2004). Auxin can also disrupt the stability of the DELLA protein by regulating gibberellin (GA), thus affecting the elongation of cells in the root elongation zone (Fu and Harberd, 2003). Ethylene participates in the differentiation of root hair cells and can promote the elongation of root hair (Tanimoto et al., 1995; Pitts et al., 1998). JA plays a positive role in root development. F-box protein COI1 is a JA receptor, which positively regulates JA response. Some studies show that COI1/JAZ mediated JA signaling pathway is involved in the regulation of root hair elongation (Han et al., 2020). CK can regulate the development of root hairs by regulating the expression of C2H2 zinc finger protein ZFP5 (An et al., 2012). Applying exogenous CK can increase the length and density of hair (Zhang et al., 2016; Huang et al., 2020). The proper concentration of GA3 treatment had a strong promoting effect on the root dry weight and the root/shoot dry weight ratio of cucumber (Cai et al., 2021). In addition, studies have shown that exogenous ethylene can induce the development of cucumber adventitious roots (Deng et al., 2022).
Multiple regulatory genes involved in root formation have also been previously identified in other species. In Arabidopsis, SCR and SHR can regulate the expression of the auxin influx vector LAX3, and promote the development of primary and lateral roots (Aida et al., 2004). The PTL class of transcription factors are responsive to auxin accumulation signals, and co-regulate root stem cell differentiation with auxin response factors (ARFs) (Aida et al., 2004; Ding and Friml, 2010). Dig6 can regulate lateral root development by affecting the transport and distribution of auxin (Zhao et al., 2015). In rice, the OsCAND1 gene is important for taproot formation in rice, and controls taproot growth by participating in auxin signaling and maintaining the G2/M cell cycle transition in meristem (Wang et al., 2011). The SOLITARY-ROOT/IAA14 gene is associated with lateral root development (Fukaki et al., 2002). Members of the CRL family have been found to perform different biological functions in root formation. For example, CRL1 inhibits adventitious root formation; CRL2 is related to the formation of taproot and lateral root primordia; CRL3 is related to the formation of taproot primordia; CRL4 promotes adventitious root formation; and CRL5 can be induced by auxin to form taproots (Inukai et al., 2001, Inukai et al., 2005; Liu et al., 2009; Kitomi et al., 2011; Woo et al., 2018). In cucumber, Yan et al. (2018) found that CsGPA1 controlled root growth by promoting the cell size and meristem of cucumber tip cells through the study of G protein. CsCEP4 peptide can promote the growth of cucumber primary roots by regulating reactive oxygen species (Liu et al., 2021). The amino acid transporter CsAAP2 can mediate polar auxin transport in cucumber root tip to influence root development (Yao et al., 2023). However, the key regulators of root growth still remained unknown in cucumber, hindering the exploration of the regulatory network and its further genetic improvement.
In recent years, with the development of resequencing technology, GWAS has been widely used to identify loci that influence plant root traits. Beyer et al. (2019) evaluated five root traits of 211 hexaploid wheat materials at the seedling stage and identified 63 marker-trait associations (MTA). Wang et al. (2019) carried out a genome-wide association mapping of 13 root traits and three aboveground traits with 297 maize inbred lines, and three pleiotropic QTLs involving five root traits were detected. Pace et al. (2015) used 384 maize inbred lines to conduct a genome-wide association analysis on 22 root architecture of seedlings, and significant SNPs for multiple traits were located on gene model GRMZM2G153722. Li et al. (2021) measured 280 Brassica napus accessions with five consecutive nutrient stages and identified 16 persistent and 32 stage-specific quantitative trait loci (QTLs) through GWAS. Chen et al. (2021) re-sequenced 220 alfalfa core germplasm, and identified 26 loci for fresh root weight, 35 for dry root weight, 3 for root length, and 3 for root number through GWAS. In cucumber, Qi et al. (2013) has re-sequenced a core collection of 115 cucumber accessions that capture 77.2% of the total genetic diversity estimated for 3,342 accessions from a wide geographic distribution, which provided a rich germplasm panel representing the high diversity and suitable for the genetic dissection of root-related traits.
Here, we performed GWAS for four root-weight related traits of 96 core germplasm accessions and identified 10 significant loci. Furthermore, using haplotype and expression analysis between heavy and light-root lines, five candidate genes positioned within four repeatedly detected loci were determined. This study therefore establishes new loci that may be used for breeding new cucumber varieties with novel and robust roots trait, and provides new ideas for elucidating mechanisms related to root development in cucumber.
A total of 96 accessions, a representative collection selected from the global 3,342 accessions, were used for the GWAS (Supplementary Table S1), which includes all the four geographic groups including East Asian, Eurasian, Indian, and Xishuangbanna (Qi et al., 2013). These materials have been used for genome-wide association analysis of various cucumber traits (Liu et al., 2020, Liu et al., 2021a; Han et al., 2023; Li et al., 2023). These accessions were cultivated at the Institute of Vegetables and Flowers (IVF), Chinese Academy of Agricultural Sciences (CAAS) in three distinct seasons (spring and autumn at 2017 and spring at 2020). Seedlings were grown in a soilless culture with vermiculite as the growing medium. The seeds were sown in a 4×8 seedling tray and irrigated with water. Seven days after sowing, the seedlings were irrigated with 500 mL of the Hoagland working nutrient solution (Supplementary Table S2), per seedling plate per day. All the experiments adopted a random block design with three replicates, and five plants planted for each replicate, for each accession. The tested 96 accessions were provided by Cucumber Breeding Group of IVF, CAAS.
Four root-weight related traits were evaluated for the 96 accessions including root fresh weight (RFW), root dry weight (RDW), root dry weight/root fresh weight (RDFW), and D-value of root weight (DRW). When the seedlings grew with two fully expanded true leaves at 14 days after sowing, the roots were washed with water, and all moisture on the surface of the root was removed with absorbent paper. RFW was determined using an analytical balance. RDW was determined after drying at 60℃ until the root weight was constant (Nayyeripasand et al., 2021). When a trait is evaluated by multiple indicators, a comprehensive evaluation indicator D-value can be introduced through principal component analysis and membership function analysis (Xie et al., 2021). In this study, the D-value of root weight (DRW), as the comprehensive index to evaluate root weight, was computed using the membership function values from the principal component analysis of RFW, RDW and RDFW. Principal component weight Wj = Ij/∑Ij (j = 1, 2, …, n), where Ij represents the contribution rate of the Jth principal component. Membership function U (Xj) = (Xj − Xmin)/(Xmax −Xmin) (j = 1, 2, …, n), where Xj represents the Jth principal component value, Xmin and Xmax represent the minimum and maximum values of the Jth principal component in different strains, respectively. The D-value was calculated as D = ∑ (Uj × Wj) (j = 1, 2, …, n). Therefore, a total of four traits (RFW, RDW, RDFW, DRW) were involved in the evaluation and analysis of root weight (Supplementary Table S3).
Best Linear Unbiased Prediction (BLUP) has been reported to effectively integrate multiple environmental data, remove environmental effects, and obtain stable genetic phenotypes of individuals (Cumbie et al., 2011; Liu et al., 2022; Han et al., 2023). Therefore, we used the BLUP values across the three environments for each of the four root-weight related traits. The ‘lme40’ package of R V3.6.1 software (www.r-project.org) was used to calculate BLUP values (Liu et al., 2022). SAS V9.4 software was used for statistical analysis of phenotypic data. TBtools V1.0692 software was used to perform phylogenetic analysis and heat map plotting for the DRW (Chen et al., 2018).
Genotypic data of 96 cucumber accessions were obtained from publicly available sequence data NCBI Short Read Archive (SRA) under accession SRA056480 (http://www.ncbi.nlm.nih.gov/sra?term=SRA056480) (Qi et al., 2013). The Fast-LMM (factored spectrally transformed linear mixed model) software was used to perform the GWAS (Lippert et al., 2011) with an estimated relatedness matrix estimated using 1,547,181 SNPs. Compared to other methods, Fast-LMM can capture confounding factors and address confounding factors by using genetic similarity methods (Runcie and Crawford, 2019). In this study, GWAS were performed for both phenotypic data and BLUP data from three seasons. The genome-wide significance threshold was established through Bonferroni correction (i.e., corrected P = 1/n, where n represents the number of independent SNPs across the genome), a method frequently applied in various studies (Yang et al., 2014; Huang et al., 2017; Liu et al., 2021b; You et al., 2023). The value of n was determined by SNP dataset pruning with PLINK v1.9 using parameters ‘1000kb 10kb 0.2’, resulting in the retention of 55,152 independent SNPs. Thus, the significance threshold selected was -log10 P = 5.00. Manhattan plot was generated using R package qqman (Turner, 2014). Plink software was used to analyze the linkage disequilibrium (LD) block and calculate the linkage disequilibrium (LD) decay coefficients (r2) among high-density SNPs, and was used to evaluate LD decay (Purcell et al., 2007). An r2 ≥ 0.6 was chosen as the threshold to define the LD block (Li et al., 2023). The pairwise linkage disequilibrium heat map was plotted using the LDheatmap package (Shin et al., 2006).
The LD blocks containing the peak SNP was considered as the candidate region for further analysis. Annotations of the protein-coding genes within the candidate region for each locus were determined using the Chinese Long V2 genome on Cucumber genome website (http://cucurbitgenomics.org/). The analysis of candidate protein coding genes was conducted by using qPCR to analyze the relative expression levels of 4 light root germplasm (‘CG54’, ‘CG77’, ‘CG49’, ‘4795’) and 3 heavy root germplasm (‘CG108’, ‘CG28’, ‘3691’), and the relative expression levels of light-root ‘4795’ and heavy-root ‘3691’ in multiple tissues. The roots of ‘4795’ and ‘3691’ accessions showed differences in mass from the cotyledon stage (Supplementary Figure S1). Roots, stems, cotyledons, and the true leaves were collected at the two true leaves flattening stage. Samples were stored at -80°C until further analyzed. Total RNA of samples was extracted using Plant RNA Extraction Kit (TaKaRa MiniBEST), and first-strand cDNA was synthesized using HiScripRIII RT SuperMix for qPCR (Vazyme Biotech). ChamQ Universal SYBR qPCR Master Mix (Vazyme Biotech) was used for qRT-PCR. The expression level of CsActin1 (Csa3G806800) was used as the standardized control, and the 2−ΔCt method was used to calculate gene expression levels (Schmittgen and Zakrajsek, 2000). Three biological replicates and three technical replicates were set up to obtain the expression level of each gene. Primer3.0 was used to design gene-specific primers (https://primer3.ut.ee/). All primer information was listed in Supplementary Table S4.
The resequencing data of 16 heavy and 17 light-root accessions excavated by systematic cluster analysis were used for haplotype analysis of each candidate gene. The cis-acting elements at the putative promoter region (< 2.0kb from start codon ATG) were predicted using the PlantCARE tool (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Li et al., 2023).
We measured the fresh weight (RFW) and dry weight (RDW) data of roots for the 96 core accessions in three different environments (i.e., spring and autumn of 2017 as well as the spring of 2020) and also computed the RDFW value. In order to comprehensively evaluate the root weight related traits, we calculated the D-value of root weight (DRW) index based on principal component analysis (PCA) and membership function analysis. We also estimated the BLUP values for RFW, RDW, and RDFW to eliminate environmental influences for further analysis. Through PCA of the above three phenotypes (RFW, RDW and RDFW), three new indicators (Prin1, Prin2 and Prin3) were obtained. In this study, Prin1 mainly explained 59.99% RDW, 75.71% RFW, and -25.85% RDFW. Prin2 explained 54.58% RDW, 82.34% RDFW, and -15.21% RFW. Prin1 mainly represents root dry weight and root fresh weight, and Prin2 mainly represents root dry weight and root dry-fresh ratio. The cumulative contribution rate of Prin1 and Prin2 reached up to 95.86%. Thus, Prin1 and Prin2 were used to calculate DRW (Supplementary Table S4).
We next evaluated the phenotypic variation of these four traits. RDW ranged from 0.0176g to 0.0922g (with a mean of 0.0550g), RFW ranged from 0.4490g to 0.6571g (with a mean of 0.5274g), RDFW from 0.1151 to 0.1658 (with a mean of 0.1251), and DRW from 0.0702 to 0.6223 (with a mean of 0.3365). RDW and DRW had higher coefficient of variation (CV) values (31.4288% and 38.0553%, respectively), compared with the other two traits (6.4387% and 5.8995% for RFW and RDFW) (Table 1). Pearson correlation analysis between any two of these four traits showed significantly positive correlation (P < 0.001 using two-sided student’s t-tests), except for the negative correlation between RFW and RDFW. Also, RDW had the strongest correlation with DRW (r = 0.98) (Figure 1).
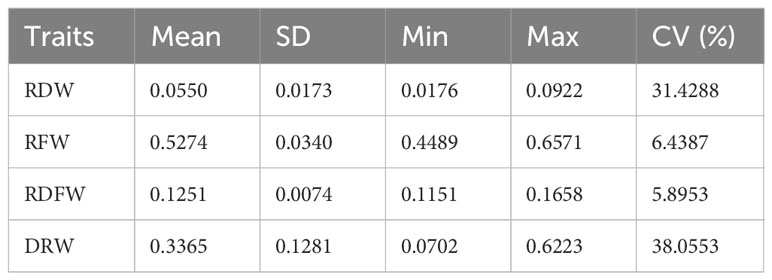
Table 1 Phenotypic variations (Standard deviation, SD; Minimum, Min; Maximum, Max; Coefficient of Variation, CV) of RDW, RFW, RDFW, and DRW.
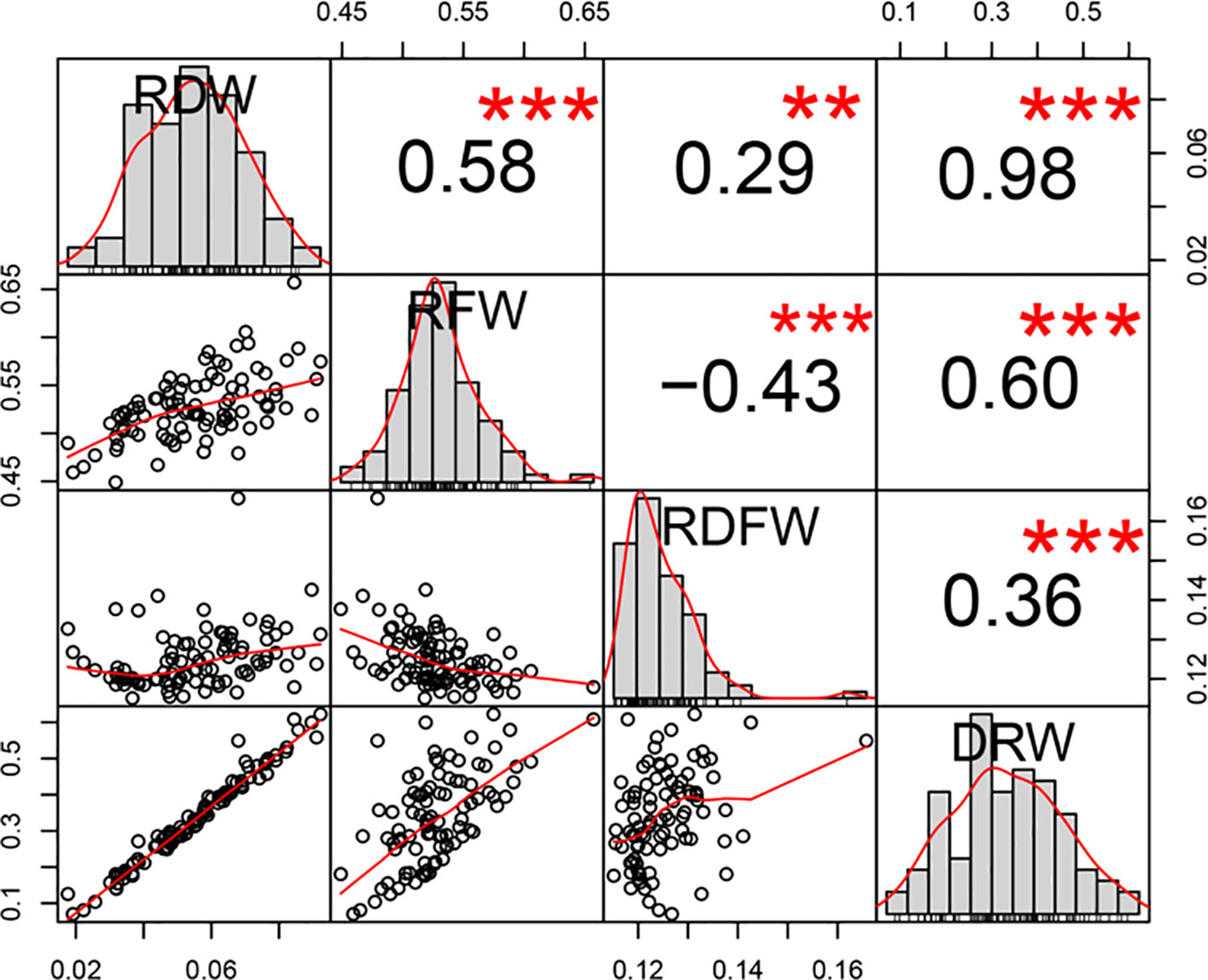
Figure 1 Distribution and Pearson correlation analysis among the four root-weight related traits in 96 cucumber core germplasm. **P < 0.01, ***P < 0.001 using two-sided student’s t-tests.
According to the geographical source, the 96 core accessions could be classified into four types including East Asian, Eurasian, Indian, and Xishuangbanna (Qi et al., 2013). Comparisons of RFW and RDW between different geographical types showed that the root of the East Asian group was the heaviest among the four geographical types (P < 0.05 using two-sided student’s t-tests) (Figures 2A–C). To further identify the germplasm exhibiting extremely heavy and light root, we perform clustering analysis using DRW data. The results showed that the core germplasm could be divided into three groups (Figure 2D): group I consisting of 21 light-root germplasm (LRG) with the lower mean values of RFW (0.5013 ± 0.0239 g), RDW (0.0321 ± 0.0061 g), RDFW (0.1219 ± 0.0051), and DRW (0.1645 ± 0.0417); group II comprising 16 heavy-root germplasm (HRG) with higher mean values of RFW (0.5593 ± 0.0417 g), RDW (0.0798 ± 0.0075 g), RDFW (0.1301 ± 0.0114), and DRW (0.5309 ± 0.0490). Other 57 medium-root germplasm (MRG) in group III showed intermediate mean values for RFW (0.5280 ± 0.0265 g), RDW (0.0570 ± 0.0091 g), RDFW (0.1249 ± 0.0059), and DRW (0.3453 ± 0.0632) (Figures 2G–J). Notably, 50% of LRG in group I were Indian types, whereas, 56.25% of HRG in group II were East Asian types (Figures 2E, F). This was consistent with the aforementioned results that East Asian type accessions showed heaviest root weight, but Indian type the lightest root weight. Both results demonstrated that the East Asian type accessions with heavy root may be preferred by local breeders or producers with heavier root and suggest that these materials have huge potential for utilization in improving the root weight traits in the further genetic breeding.
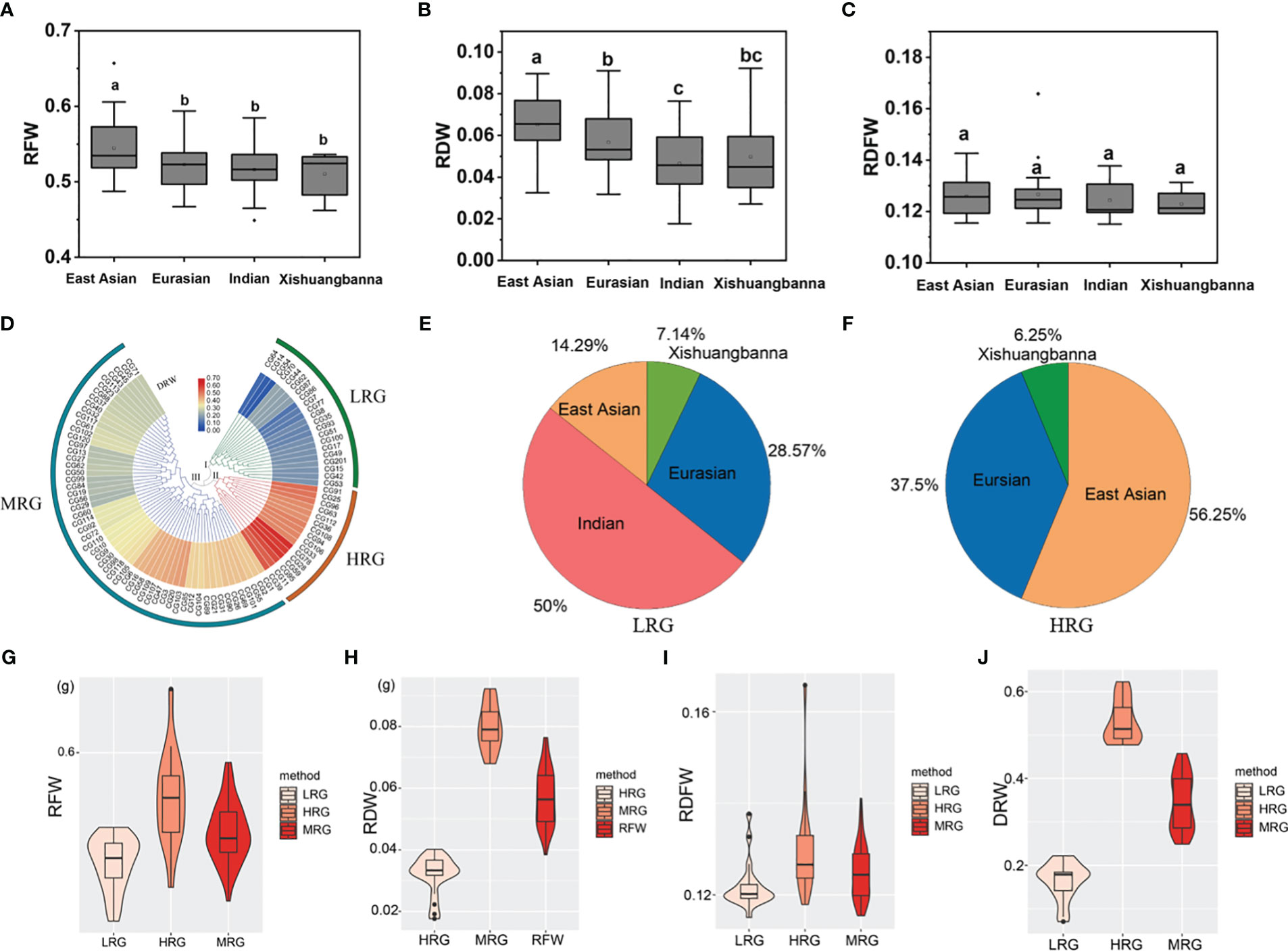
Figure 2 Cluster analysis of D-value of root weight (DRW) data derived from the cucumber core germplasm. (A–B) A histogram of RFW, RDW, and RDFW of four main geographic groups. a, b, and c represent the level of significant difference from high to low. (D) A heatmap of DRW of the cucumber core germplasm. (E) Distribution of four major geographic groups in light-root germplasm (LRG). (F) Distribution of four major geographic groups in heavy-root germplasm (HRG). (G–J) A histogram of RFW, RDW, RDFW, and DRW of LRG, HRG, and MRG.
The 96 core germplasm used in our study has been sequenced using Illumina platform (Qi et al., 2013) and 1,547,181 high-quality SNPs which were identified based on the ChineseLong V2.0 reference genome (http://cucurbitgenomics.org/). Based on the SNPs and phenotypic data, we used Fast-LMM (factored spectrally transformed linear mixed model) software to perform the GWAS analysis for RFW, RDW, RDFW and DRW traits to dissect the genetic basis of root-weight related traits. A total of 9 distinct loci were detected through three root traits: RFW, RDW, and RDFW (4 in RFW, 7 in RDW, and 2 in RDFW) in three experiments (2017S, 2017A, and 2020S) (Supplementary Figures S2–4; Supplementary Table S5). Then, GWAS analysis was performed based on the SNPs and BLUP data (Runcie and Crawford, 2019). A total of 10 distinct loci were detected for the four traits (4 in RDW, 3 in RFW, 3 in RDFW, and 7 in DRW). More loci information was shown in Table 2. Among them, 4 loci were repeatedly identified on chromosome 3, 4, and 5 for at least two traits (gRDW3.1, gRDW3.2, gRDW4.1, and gRDW5.1 for RDW, gRFW3.1, and gRFW5.1 for RFW, gRDFW3.2 for RDFW, gDRW3.1, gDRW3.2, gDRW4.1, and gDRW5.1 for DRW.) (Figure 3). Only these four stable and reliable loci were used for further analysis of candidate genes. Overall, the loci obtained from the BLUP data were highly similar as those obtained from the phenotypic data of the three seasons. (Table 2; Supplementary Table S5), indicating that these loci were repeatable and less influenced by the environment.
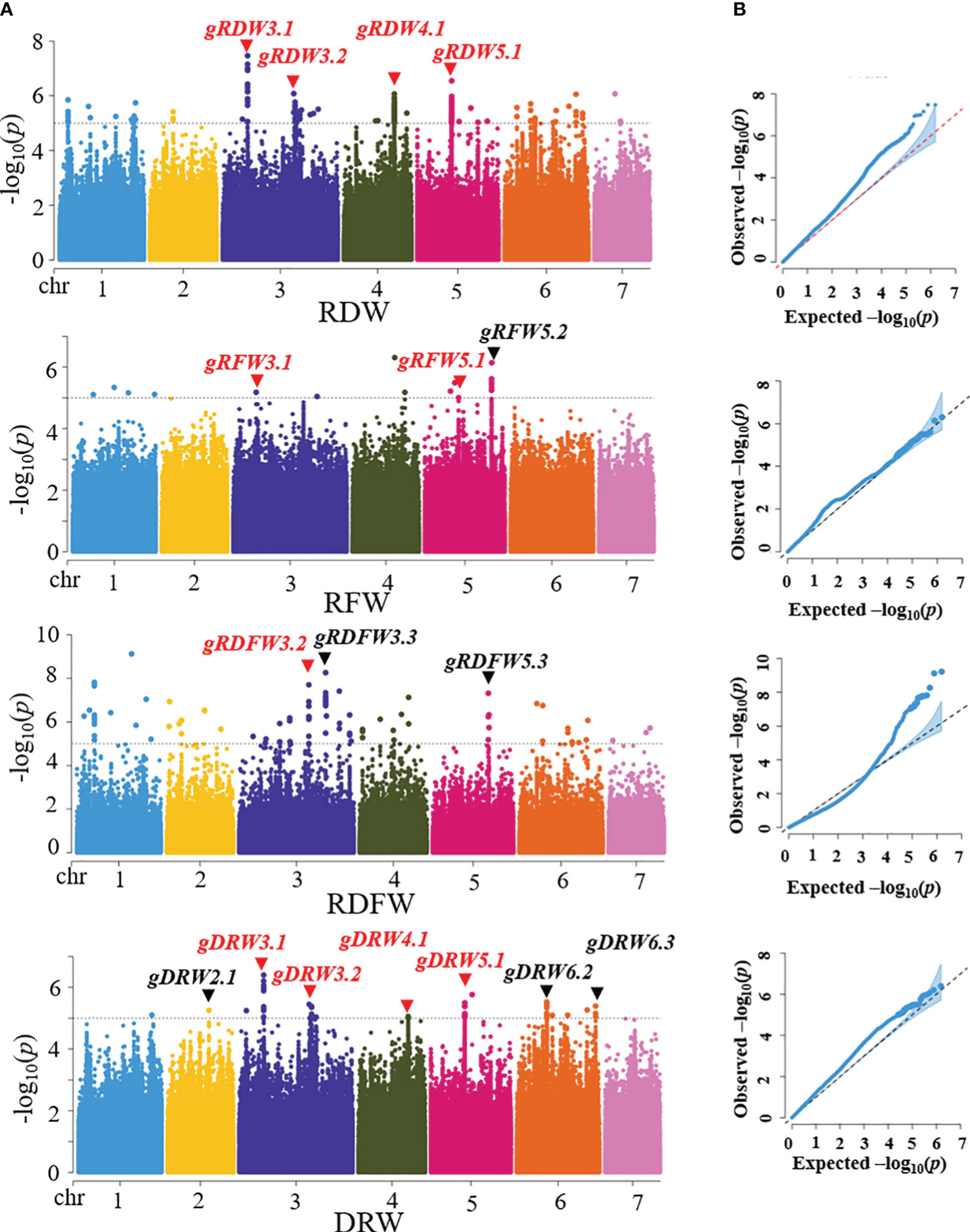
Figure 3 Genome-wide association analysis (GWAS) for 96 core germplasm. (A) Manhattan plots of RDW, RFW, RDFW, and DRW based on BLUP. Dashed line represents the significance threshold (−log10 P = 5.00). The loci marked in red were repeatedly detected for at least two root traits. (B) QQ plots of RDW, RFW, RDFW, and DRW based on BLUP.
In our study, we focused on stable and reliable loci that were identified through GWAS for additive effect genetic analysis. Interestingly, we observed that as the number of alters of heavy root increased, there was a corresponding increase in root weight. Based on these findings, we propose that the different loci identified in our study have additive effects on root size (Supplementary Figure S5).
Since all the repeated loci were identified by GWAS using DRW, we used it for candidate gene analysis. We first determined the LD blocks harboring the peak SNP using Plink software (Purcell et al., 2007), and then identified candidate genes positioned within the LD block. Next, we used qPCR to analyze the relative expression levels (RELs) of candidate genes in different haplotype accessions and multiple tissues (root, stem, true leaf, cotyledon) of light-root ‘4795’ and heavy-root ‘3691’ accessions. Finally, we analyzed all the reference allele and alternative allele of 21 LRGS and 16 HRGS identified by aforementioned clustering analysis, and obtained different gene haplotypes based on the mutations compared with reference gemoe. The phenotypes and gene expression levels of accessions carrying different haplotypes were then statistically analyzed. For gDRW3.1, we identified 5 genes (Csa3G131990, Csa3G132000, Csa3G132010, Csa3G132020, Csa3G132520) positioned within the LD block (8.575-8.610 Mb) (Figure 4A). Both Csa3G132020 and Csa3G132520 had sequence variation in the promoter region (Figure 4B). The qRT-PCR analysis showed that the RELs of Csa3G132020 and Csa3G132520 both encoding F-box protein PP2-B1differed between the HAP1 and HAP2 accessions (Figure 4C). Notably, these two genes expressed higher in roots than in cotyledons and leaves (P<0.01) (Figure 4D). Through the haplotype analysis, for each of the two genes, we could identify two haplotypes exhibiting significantly different DRW (Figure 4E; Supplementary Table S6). As the haplotypes of two genes located in the putative promoter region (< 2.0 Kb from the start codon), we then investigated the potential effect of haplotypes on the cis-binding element using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). For haplotypes of Csa3G132020, one of the SNPs at nucleotide position -1967 was located within the ERE element which is a typical ethylene-responsive element (Supplementary Figure S6A) (Shinshi, 1995; Adie et al., 2007; Liu et al., 2013). For gene Csa3G132520, SNP from the haplotype at nucleotide 682 was located in the CAAT-box (Supplementary Figure S6B). Previous studies have demonstrated that the 70-bp domain around the CAAT-box is necessary for gene expression in the dermatogen and meristematic cells of the root cortex Expression in different populations of cells of the root meristem is controlled by different domains of the rolB promoter (Capone et al., 1994).
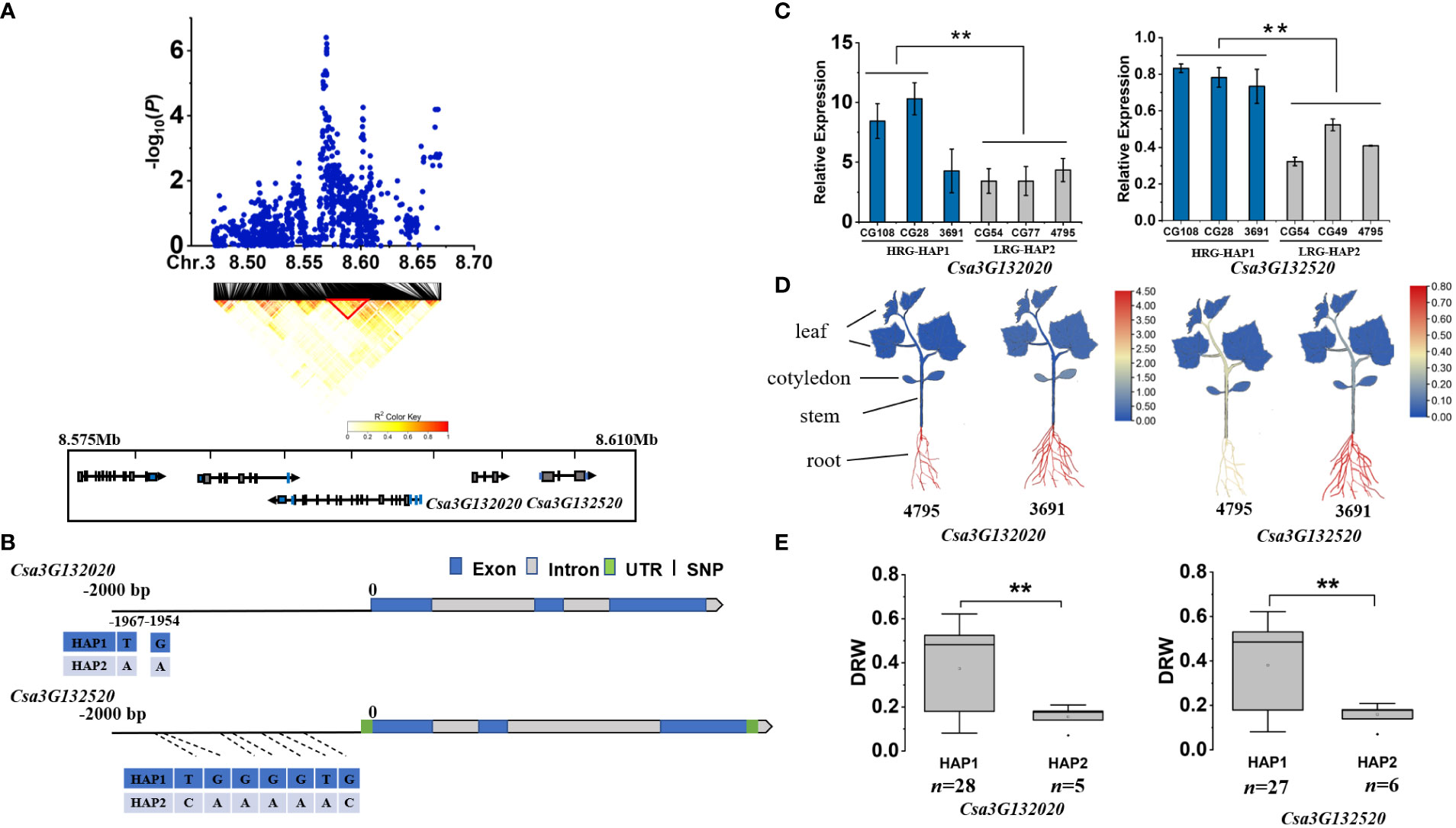
Figure 4 Identifying and assessing candidate genes at the gDRW3.1 locus. (A) Local Manhattan plot and LD heatmap surrounding the peak (top) and genes(bottom) in LD block represented by the red tri-angle (bottom). (B) Gene structure of candidate gene Csa3G132020 and Csa3G132520. (C) Relative expression levels in root of Csa3G132020 and Csa3G132520 in the HAP1 and HAP2 accessions through qRT-PCR. (D) Spatiotemporal expression of Csa3G132020 and Csa3G132520 in the light-root (‘4795’) and heavy-root (‘3691’) lines through qRT-PCR. (E) Box plots for LRG and HRG based on the haplotypes. n indicates the number of accessions with the same genotype. **P < 0.01 using two-sided student’s t-tests.
For gDRW3.2, we identified two genes (Csa3G629240 and Csa3G629740) within the LD block at 24.500-24.525 Mb on Chromosome 3 (Figure 5A). Csa3G629740 was almost unexpressed in all haplotype accessions. However, the Csa3G629240 encoding a B-cell receptor-associated protein. Three base variations exist in the promoter region of Csa3G629240 (Figure 5B). The expression of Csa3G629240 in HAP1 accessions were significantly higher than that in HAP2 accessions (Figure 5C), and its expression in roots and leaves was higher than that in cotyledons and stem (P<0.01) (Figure 5D). The DRW of accessions with HAP1 was significantly higher than that of accessions with HAP2 (Figure 5E; Supplementary Table S6). The SNPs of haplotype at nucleotide position -1654 and -1762 on the promoter of the gene Csa3G629240 were found to be located on the motif sequence of CAAT-box (Supplementary Figure S7). Thus, the above evidences all supported that Csa3G629240 was the candidate gene for gDRW3.2 locus.
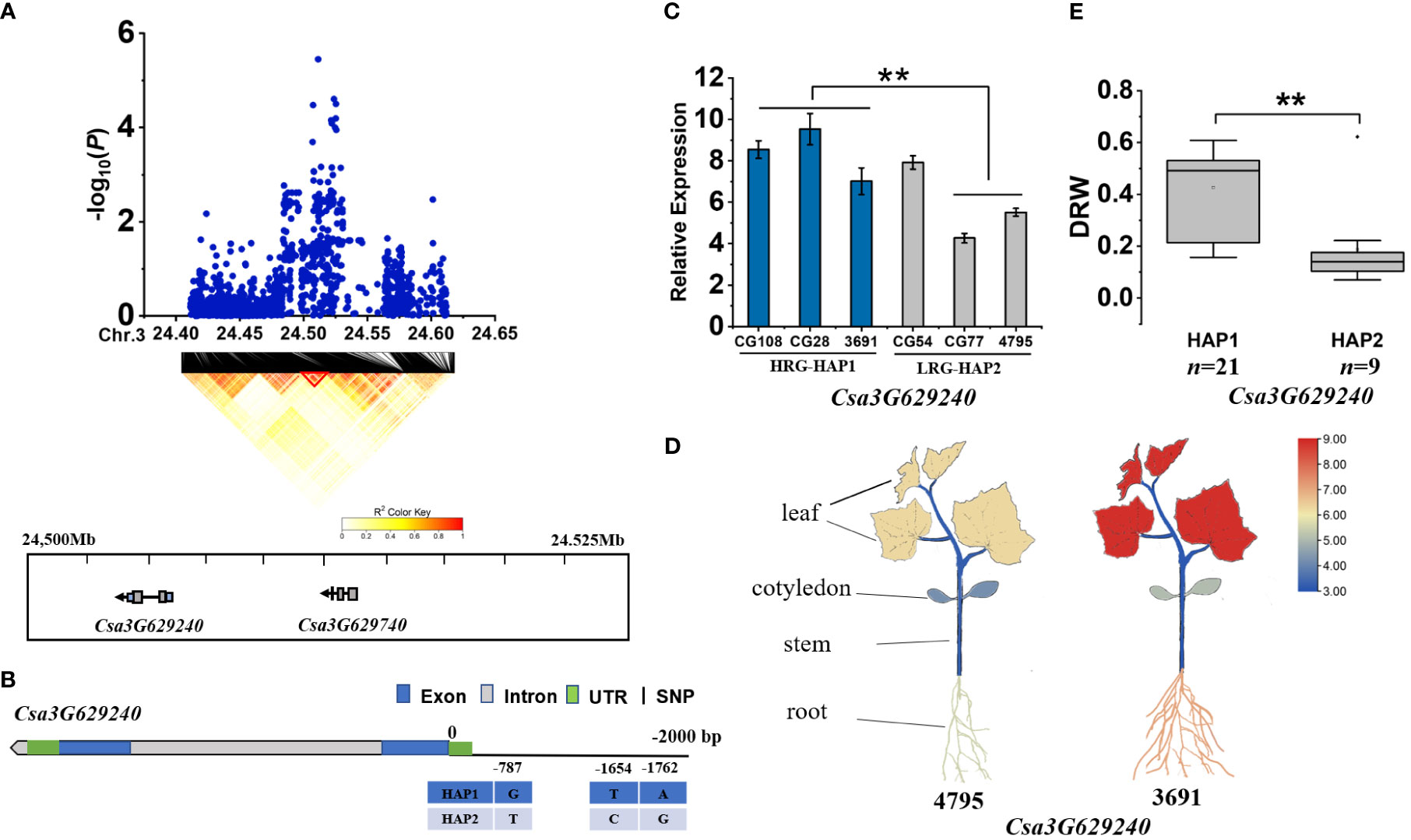
Figure 5 Identifying and assessing a candidate gene at the gDRW3.2 locus. (A) Local Manhattan plot and LD heatmap surrounding the peak (top) and genes in LD block represented by the red tri-angle (bottom). (B) Gene structure of Csa3G629240. (C) Expression levels in root of Csa3G629240 in the HAP1 and HAP2 accessions through qRT-PCR. (D) Spatiotemporal expression of Csa3G629240 and Csa3G132520 in the light-root (‘4795’) and heavy-root (‘3691’) lines through qRT-PCR. (E) Box plots for LRG and HRG based on the haplotypes. n indicates the number of accessions with the same genotype. **P < 0.01 using two-sided student’s t-tests.
For gDRW4.1 locus, two candidate genes (Csa4G499320 and Csa4G499330) were identified within the LD block (17.425-17.465 Mb) on Chromosome 4 (Figure 6A). Among them, the expression of Csa4G499320, encoding an unknown protein, was different at the cotyledon flattening stage. However, gene Csa4G499330 showed significantly different expression levels between the HAP1 and HAP2 accessions (Figure 6C), and its expression in roots was lower than that in the cotyledons and leaves (P<0.01), but higher than in the stem (P<0.01) (Figure 6D). Csa4G499330 encodes a GTP binding protein with its homolog AT1G08410 (Drought Inhibited Growth of Lateral roots 6, DIG6) in Arabidopsis thaliana regulating multiple auxin-mediated developmental processes and promote lateral root development (Zhao et al., 2015). Gene-based association analysis revealed that HAP1 was mainly found in accessions with a higher DRW, while HAP2 mainly occurs in accessions with a lower DRW (Figure 6E; Supplementary Table S6). The SNP at nucleotide position +4964 on the exon caused an amino acid change, from Lys to Glu (Figure 6B). Therefore, through expression analysis and gene annotation, Csa4G499330 may be promising candidate gene of gDRW4.1.
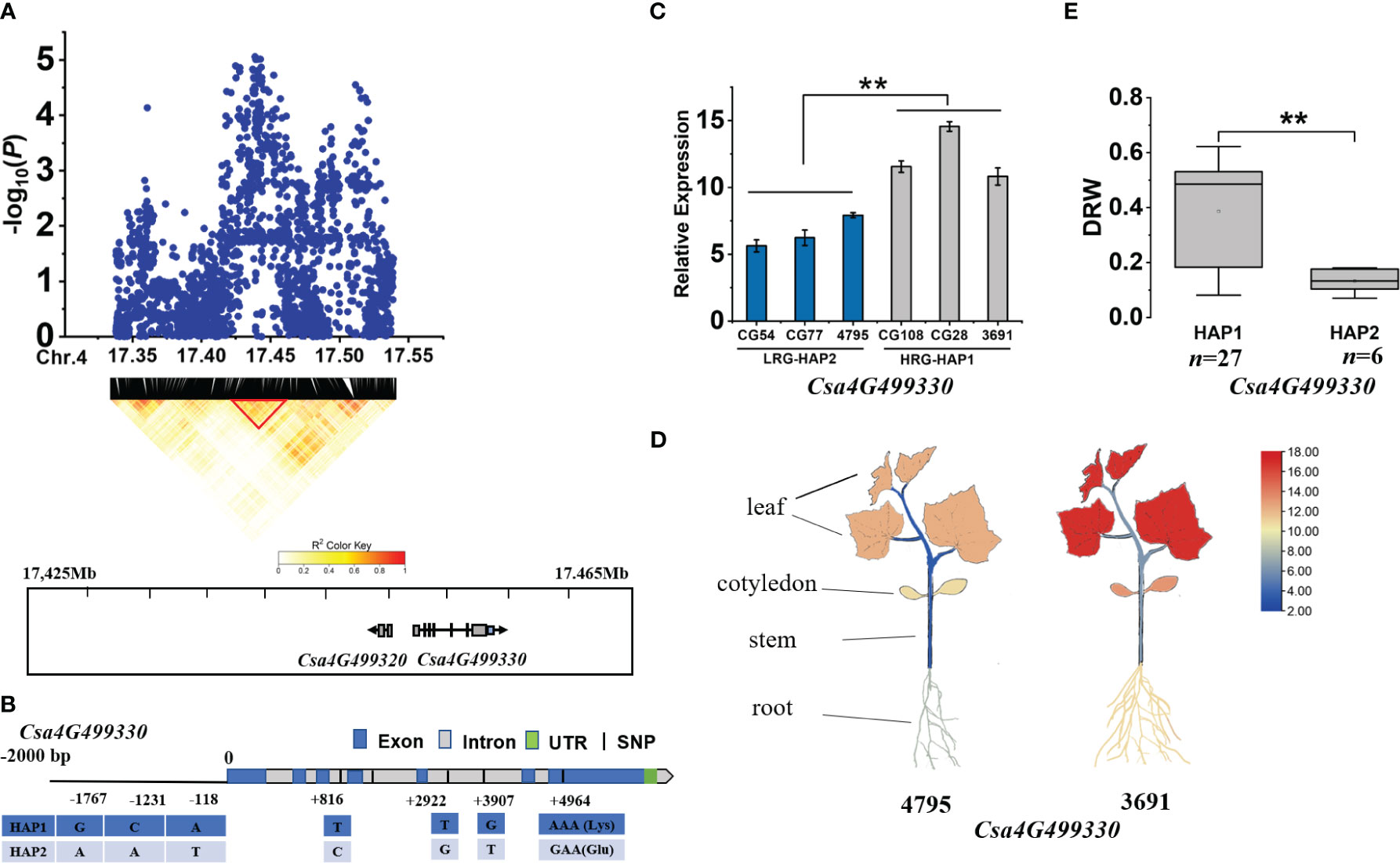
Figure 6 Identifying and assessing candidate genes at the gDRW4.1 locus. (A) Local Manhattan plot and LD heatmap surrounding the peak (top) and Genes in LD block represented by the red tri-angle (bottom). (B) Gene structure of Csa4G499330. (C) Relative expression levels in root of Csa4G499330 in the HAP1 and HAP2 accessions through qRT-PCR. (D) Spatiotemporal expression of Csa4G499330 and Csa4G499330 in the light-root (‘4795’) and heavy-root (‘3691’) lines through qRT-PCR. (E) Box plots for LRG and HRG based on the haplotypes. n indicates the number of accessions with the same genotype. **P < 0.01 using two-sided student’s t-tests.
Within the gDRW5.1, according to the LD block, we identified one gene (Csa5G286040) at 11.830-11.870 Mb on Chromosome 5 (Figure 7A). Multiple base variations exist in the promoter region of Csa5G286040 (Figure 7B). The expression of Csa5G286040 in HAP2 accessions were significantly higher than that in HAP1 accessions (Figure 7C). It is noteworthy that the expression level of Csa5G286040 was higher in roots than in other tissues (Figure 7D). Csa5G286040 encodes a proteinase inhibitor. The DRW of accessions with HAP1 was significantly higher than that of accessions with HAP2 (Figure 7E; Supplementary Table S6). The -1370 SNP of the promoter region is located on the G-box, and the -1389 SNP and -1391 SNP were both located within the CAAT-box (Supplementary Figure S8). In tomato, G-box elements are targeted by MYC2, a core TF of the jasmonic acid (JA) signaling pathway that regulates root growth (Wang et al., 2020).
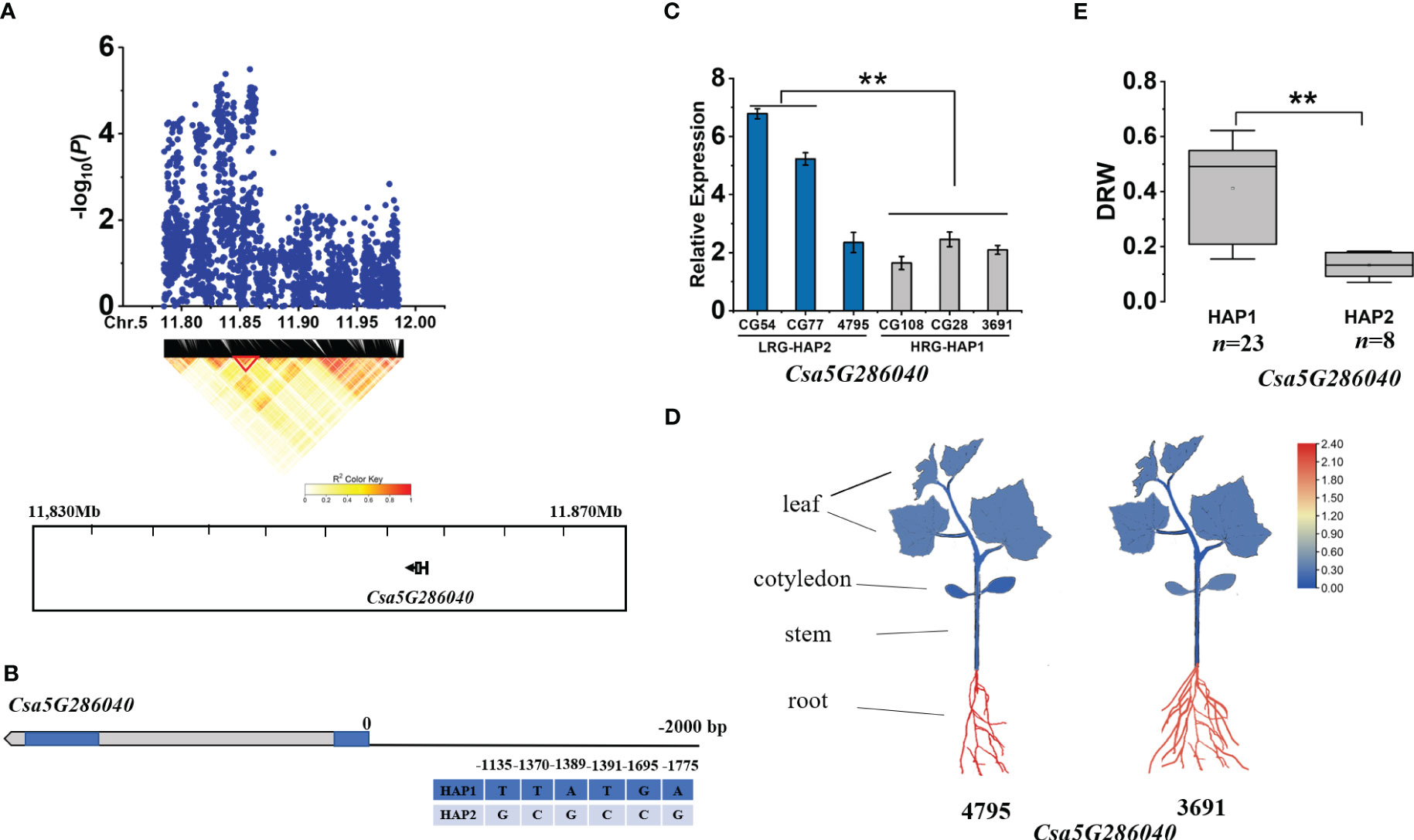
Figure 7 Identifying and assessing candidate genes within the gDRW5.1 locus. (A) Local Manhattan plot and LD heatmap surrounding the peak (top) and Genes in LD block represented by the red tri-angle (bottom). (B) Gene structure of Csa5G286040. (C) Relative expression levels in root of Csa5G286040 in HAP1 and HAP2 accessions through qRT-PCR. (D) Spatiotemporal expression of Csa5G286040 and Csa5G286040 in the light-root (‘4795’) and heavy-root (‘3691’) lines through qRT-PCR. (E) Box plots for LRG and HRG based on the haplotypes. n indicates the number of accessions with the same genotype. **P < 0.01 using two-sided student’s t-tests.
The study of genetic diversity is an important step in the exploration, conservation and utilization of cucumber germplasm resources. Cucumber is a shallow root vegetable crop and usually requires grafting rootstocks to improve its tolerance (Huang et al., 2010; Liu et al., 2016). At present, there are few reports about germplasm resources screening for cucumber root traits. Walters and Wehner (1994) evaluated the root length of 857 cultivated cucumber varieties. In our study, four traits related to root weight were investigated, among which RDW had the strongest correlation with DRW. So RDW is more accurate and reliable for the subjective evaluation of root weight. However, RDFW was not strongly correlated with RDW and DRW, which might be due to the difference in water content between different roots, which was also reported in other studies (Haq et al., 2008). Using the RDW index, we discovered heavy-root germplasm (16 accessions) and light-root germplasm (21 accessions), and found 56.25 percentage of heavy-root germplasm were collected from East Asian. The root weight of East Asian cucumber genotypes was also found to be significantly higher than that of other three cucumber ecotypes, thus the East Asian cucumber could be considered for use as elite resources in further breeding programs.
GWAS analysis of root traits have mainly focused on field crops, and there have been relatively few studies on vegetable crops like cucumber. Wang et al. (2018) analyzed SNPs from 1234 cucumber germplasm. The genetic diversity and population structure within the collection, the phylogenetic relationships, and linkage disequilibrium were characterized. Genomic regions significantly associated with 13 important horticultural traits were identified by GWAS, but no SNPs linked to root size were identified. This may be attributed to the low level of variation among root traits within the population, which would be unsuitable for a GWAS. The materials used in this study were selected from 3342 accessions collected worldwide and had a rich rate of genetic variation (Qi et al., 2013). Ten significant SNPs were detected by GWAS using four root weight-related traits.
Root development is regulated by many genes. In this study, candidate genes were predicted for four stable loci related to root weight obtained by GWAS analysis. The excavation of root related genes is of great significance for the analysis of root development mechanism and breeding of new strong root varieties.
Csa3G132020 and Csa3G132520 both encode the F-box protein PP2-B1. Yan et al. (2011) found in rice that overexpression of the F-box protein gene can promote root growth. Cheah et al. (2021) found six upregulated genes in an RNA-seq study of zinc stress in rice, and speculated that the F-box protein PP2-B1 (Os04g0280500) might be involved in the ubiquitination and proteasome degradation of target proteins in the auxin signaling pathway that regulates root development. Csa3G132020 and Csa3G132520 encode homologous proteins, and their expression in the heavy-root genotype was higher than that with light-roots. Moreover, Csa3G132020 and Csa3G132520 were highly expressed in roots compared to other tissues. Therefore, it is speculated that these two genes may play the same role in controlling root development.
Csa3G629240 encodes a B-cell receptor-associated protein. The function of B-cell receptors has been rarely reported in plants, however in animals, a B-cell receptor is a molecule located on the surface of B-cells responsible for the specific recognition and binding of antigens (Casola et al., 2004). In this study, the cucumber homologue Csa3G629240 was highly expressed in roots and leaves of the heavy-root material, and may positively regulate root development.
Csa4G499330 encodes a GTP binding protein, and its Arabidopsis thaliana homologue AT1G08410 (AtLSG1-2) encodes a large 60S subunit nuclear output GTPase 1, which is involved in ribosomal biogenesis, and which affects development processes regulated by various auxins. The expression level of the AtLSG1-2 was found to be highly expressed in dry seeds of Arabidopsis thaliana, and it exhibited a significantly higher expression in roots compared to other tissues in 1-day-old seedling (https://www.arabidopsis.org/). Zhao et al. (2015) isolated an Arabidopsis mutant dig6, defective in the ATLSG1-2 gene which showed a significant reduction in the number of lateral roots. Further studies showed that ATLSG1-2 was highly expressed in regions where the auxin accumulated, and that ribosomal biogenesis was impaired in the mutant. ATLSG1-2 deficiency resulted in altered auxin distribution, response, and transport in plants. Thus, ATLSG1-2 plays an indispensable role in ribosome biogenesis, with ensuing effects on auxin homeostasis and lateral root development. In this study, the expression of Csa4G499330 in the heavy-root genotypes was higher than that in the light-root genotypes. In addition, spatiotemporal expression of Csa4G499330 in the heavy-root genotypes was higher than that in the light-root genotypes. The expression of Csa4G499330 in leaves was significantly higher than that in roots, which may be because it has other function in the leaves. Therefore, it is speculated that Csa4G499330 may positively regulate plant development including root system through influencing auxin pathway.
Csa5G286040 encodes a proteinase inhibitor. Studies have shown that in tomato, root protease inhibitors can be induced by auxin, and that auxin induction only exists in roots and hypocotyls, but not in cotyledons and hypocotyls (Taylor et al., 1993). Thus, induction may be related to the initiation of lateral and adventitious roots. Therefore, Csa5G286040 may be induced by auxin to regulate root development. The Csa5G286040 homologue in Arabidopsis thaliana is a member of the PR-6 proteinase inhibitor family, encoding a PR (disease-related) peptide. Proteinase inhibitors are activated when plants are attacked by insects, fungi, or bacteria, along with reactive oxygen species (ROS) and hormones, e.g., ethylene (ET), jasmonic acid (JA) and salicylic acid (SA). These emergency responses are usually contradictory to the normal growth and development of plants (Sels et al., 2008). In this study, in the absence of biological stress, Csa5G286040 expression was limited to roots, and its expression in the light-root material was significantly higher than that of heavy-root material. Therefore, it can be speculated that the high expression of Csa5G286040 in the root system disturbed the hormone balance and thus hindered root development.
In summary, we evaluated the phenotypic variations for the root-weight related traits of a well-known core germplasm and identified a total of 16 accessions with heavy-roots and 21 with light-roots based on a comprehensive index for the assessment of root systems which we developed, i.e., the DRW. The East Asian ecotype accessions exhibited heavier root compared with other ecotypes. Ten genomic sites related to root weight were discovered by GWAS. Further analysis indicated that Csa3G132020, Csa3G132520, Csa3G629240, Csa4G499330, and Csa5G286040 were candidate genes that might be involved in root development. This work has identified novel cucumber cultivars that can serve as useful germplasm for breeding varieties with strong roots, furthermore, it points to candidate genes and the molecular mechanisms by which they may influence cucumber root development.
This study used the illumina data were obtained from publicly available sequence data NCBI Short Read Archive (SRA) under accession SRA056480 (http://www.ncbi.nlm.nih.gov/sra?term=SRA056480).
ZD: Data curation, Software, Writing – original draft, Writing – review & editing. SD: Formal Analysis, Writing – review & editing. HC: Investigation, Methodology, Writing – review & editing. DB: Supervision, Writing – review & editing. JG: Methodology, Writing – review & editing. XL: Investigation, Methodology, Writing – review & editing. XG: Software, Writing – review & editing. HM: Data curation, Writing – review & editing. SZ: Funding acquisition, Methodology, Resources, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Beijing Joint Research Program for Germplasm Innovation and New Variety Breeding (G20220628003), the Earmarked fund for Modern Agro-industry Technology Research System (CARS-23) and the Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture and Rural Affairs, People’s Republic of China.
The authors thank the Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture, China.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1417314/full#supplementary-material
Adie, B., Chico, J. M., Rubio-Somoza, I., Solano, R. (2007). Modulation of plant defenses by ethylene. J. Plant Growth Regulation. 26, 160–177. doi: 10.1007/s00344-007-0012-6
Aida, M., Beis, D., Heidstra, R., Willemsen, V., Blilou, I., Galinha, C., et al. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell. 119, 109–120. doi: 10.1016/j.cell.2004.09.018
An, L., Zhou, Z., Sun, L., Yan, A., Xi, W., Yu, N., et al. (2012). A zinc finger protein gene ZFP5 integrates phytohormone signaling to control root hair development in Arabidopsis. Plant J. 72, 474–490. doi: 10.1111/j.1365-313X.2012.05094.x
Bao, F., Shen, J., Brady, S. R., Muday, G. K., Asami, T., Yang, Z. (2004). Brassinosteroids interact with auxin to promote lateral root development in Arabidopsis. Plant Physiol. 134, 1624–1631. doi: 10.1104/pp.103.036897
Beyaert, R. P., Roy, R. C., Ball Coelho, B. R. (2007). Irrigation and fertilizer management effects on processing cucumber productivity and water use efficiency. Can. J. Plant Science. 87, 355–362. doi: 10.4141/P06-012
Beyer, S., Daba, S., Tyagi, P., Bockelman, H., Brown-Guedira, G., Mohammadi, M. (2019). Loci and candidate genes controlling root traits in wheat seedlings—a wheat root GWAS. Funct. Integr. Genomics 19, 91–107. doi: 10.1007/s10142-018-0630-z
Burton, A. L., Johnson, J., Foerster, J., Hanlon, M. T., Kaeppler, S. M., Lynch, J. P., et al. (2015). QTL mapping and phenotypic variation of root anatomical traits in maize (Zea mays L.). Theor. Appl. Genet. 128, 93–106. doi: 10.1007/s00122-014-2414-8
Cai, B., Wang, T., Sun, H., Liu, C., Chu, J., Ren, Z., et al. (2021). Gibberellins regulate lateral root development that is associated with auxin and cell wall metabolisms in cucumber. Plant Science. 317, 110995. doi: 10.1016/j.plantsci.2021.110995
Capone, I., Frugis, G., Costantino, P., Cardarelli, M. (1994). Expression in different populations of cells of the root meristem is controlled by different domains of the rolB promoter. Plant Mol. Biol. 25, 681–691. doi: 10.1007/BF00029606
Casola, S., Otipoby, K. L., Alimzhanov, M., Humme, S., Uyttersprot, N., Kutok, J. L., et al. (2004). B cell receptor signal strength determines B cell fate. Nat. Immunol. 5, 317–327. doi: 10.1038/ni1036
Cheah, B. H., Chen, Y. L., Lo, J. C., Tang, I. C., Yeh, K. C., Lin, Y. F. (2021). Divalent nutrient cations: Friend and foe during zinc stress in rice. Plant Cell Environment. 44, 3358–3375. doi: 10.1111/pce.14154
Chen, C., Chen, H., He, Y., Xia, R. (2018). TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv. 289660, 289660.
Chen, L., He, F., Long, R., Zhang, F., Li, M., Wang, Z., et al. (2021). A global alfalfa diversity panel reveals genomic selection signatures in Chinese varieties and genomic associations with root development. J. Integr. Plant Biol. 63, 1937–1951. doi: 10.1111/jipb.13172
Courtois, B., Ahmadi, N., Khowaja, F., Price, A. H., Rami, J.-F., Frouin, J., et al. (2009). Rice root genetic architecture: meta-analysis from a drought QTL database. Rice.2 2-3), 115–128. doi: 10.1007/s12284-009-9028-9
Cumbie, W., Eckert, A., Wegrzyn, J., Whetten, R., Neale, D., Goldfarb, B. (2011). Association genetics of carbon isotope discrimination, height and foliar nitrogen in a natural population of Pinus taeda L. Heredity. 107, 105–114. doi: 10.1038/hdy.2010.168
Deng, Y., Wang, C., Zhang, M., Wei, L., Liao, W. (2022). Identification of key genes during ethylene-induced adventitious root development in cucumber (Cucumis sativus L.). Int. J. Mol. Sci. 23, 12981. doi: 10.3390/ijms232112981
Ding, Z., Friml, J. (2010). Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc. Natl. Acad. Sciences.107 26), 12046–12051. doi: 10.1073/pnas.100067210
Dorlodot, S. D., Forster, B., Pagès, L., Price, A., Tuberosa, R., Draye, X. (2007). Root system architecture: opportunities and constraints for genetic improvement of crops. Trends Plant Science. 12, 474–481. doi: 10.1016/j.tplants.2007.08.012
FAOSTAT (2021)Available online at: http://www.fao.org/faostat/en/#data
Fu, X., Harberd, N. (2003). Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature. 421, 740–743. doi: 10.1038/nature01387
Fukaki, H., Tameda, S., Masuda, H., Tasaka, M. (2002). Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 29, 153–168. doi: 10.1046/j.0960-7412.2001.01201.x
Han, J., Dong, S., Shi, Y., Dai, Z., Miao, H., Li, B., et al. (2023). Genome-wide association study reveals candidate genes for gummy stem blight resistance in cucumber. Hortic. Plant J. 9, 261–272. doi: 10.1016/j.hpj.2022.06.004
Han, X., Zhang, M., Yang, M., Hu, Y. (2020). Arabidopsis JAZ proteins interact with and suppress RHD6 transcription factor to regulate jasmonatestimulated root hair development. Plant Cell. 32, 1049–1062. doi: 10.1105/tpc.19.00617
Haq, T. U., Akhtar, J., Gorham, J., Steele, K. A., Khalid, M. (2008). Genetic mapping of QTLs, controlling shoot fresh and dry weight under salt stress in rice (Oryza sativa L.) cross between Co39× moroberekan. Pakistan J. Botany. 40, 2369–2381.
Himanen, K., Boucheron, E., Vanneste, S., de Almeida Engler, J., Inzé, D., Beeckman, T. (2002). Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell. 14, 2339–2351. doi: 10.1105/tpc.004960
Hochholdinger, F., Tuberosa, R. (2009). Genetic and genomic dissection of maize root development and architecture. Curr. Opin. Plant Biol. 12, 172–177. doi: 10.1016/j.pbi.2008.12.002
Huang, Y., Bie, Z., He, S., Hua, B., Zhen, A., Liu, Z. (2010). Improving cucumber tolerance to major nutrients induced salinity by grafting onto Cucurbita ficifolia. Environ. Exp. Botany. 69, 32–38. doi: 10.1016/j.envexpbot.2010.02.002
Huang, L., Jiang, Q., Wu, J., An, L., Zhou, Z., Wong, C., et al. (2020). Zinc finger protein 5 (ZFP5) associates with ethylene signaling to regulate the phosphate and potassium deficiency-induced root hair development in Arabidopsis. Plant Mol. Biol. 102, 143–158. doi: 10.1007/s11103-019-00937-4
Huang, C., Nie, X., Shen, C., You, C., Li, W., Zhao, W., et al. (2017). Population structure and genetic basis of the agronomic traits of upland cotton in China revealed by a genome-wide association study using high-density SNPs. Plant Biotechnol. J. 15, 1374–1386. doi: 10.1111/pbi.12722
Huang, Y., Tang, R., Cao, Q., Bie, Z. (2009). Improving the fruit yield and quality of cucumber by grafting onto the salt tolerant rootstock under NaCl stress. Scientia Horticulturae. 122, 26–31. doi: 10.1016/j.scienta.2009.04.004
Inukai, Y., Miwa, M., Nagato, Y., Kitano, H., Yamauchi, A. (2001). RRL1, RRL2 and CRL2 loci regulating root elongation in rice. Breed. science. 51, 231–239. doi: 10.1270/jsbbs.51.231
Inukai, Y., Sakamoto, T., Ueguchi-Tanaka, M., Shibata, Y., Gomi, K., Umemura, I., et al. (2005). Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell. 17, 1387–1396. doi: 10.1105/tpc.105.030981
Kamoshita, A., Wade, L., Ali, M., Pathan, M., Zhang, J., Sarkarung, S., et al. (2002). Mapping QTLs for root morphology of a rice population adapted to rainfed lowland conditions. Theor. Appl. Genet. 104, 880–893. doi: 10.1007/s00122-001-0837-5
Kiryushkin, A. S., Ilina, E. L., Kiikova, T. Y., Pawlowski, K., Demchenkol, K. N. (2024). Do DEEPER ROOTING 1 Homologs Regulate the Lateral Root Slope Angle in Cucumber (Cucumis sativus)? Int. J. Mol. Sci. 25 (4), 1975. doi: 10.3390/ijms25041975
Kitomi, Y., Ito, H., Hobo, T., Aya, K., Kitano, H., Inukai, Y. (2011). The auxin responsive AP2/ERF transcription factor CROWN ROOTLESS5 is involved in crown root initiation in rice through the induction of OsRR1, a type-A response regulator of cytokinin signaling. Plant J. 67, 472–484. doi: 10.1111/j.1365-313X.2011.04610.x
Knight, M. R. (2007). New ideas on root hair growth appear from the flanks. Proc. Natl. Acad. Sci. 104, 20649–20650. doi: 10.1073/pnas.071063210
Li, C., Dong, S., Beckles, D. M., Liu, X., Guan, J., Gu, X., et al. (2023). GWAS reveals novel loci and identifies a pentatricopeptide repeat-containing protein (CsPPR) that improves low temperature germination in cucumber. Front. Plant Science. 14. doi: 10.3389/fpls.2023.1116214
Li, C., Li, Y., Bai, L., Zhang, T., He, C., Yan, Y., et al. (2014). Grafting-responsive miRNAs in cucumber and pumpkin seedlings identified by high-throughput sequencing at whole genome level. Physiologia plantarum. 151, 406–422. doi: 10.1111/ppl.12122
Li, K., Wang, J., Kuang, L., Tian, Z., Wang, X., Dun, X., et al. (2021). Genome-wide association study and transcriptome analysis reveal key genes affecting root growth dynamics in rapeseed. Biotechnol. biofuels. 14, 1–20. doi: 10.1186/s13068-021-02032-7
Li, M., Zhu, Y., Li, S., Zhang, W., Yin, C., Lin, Y. (2022). Regulation of phytohormones on the growth and development of plant root hair. Front. Plant Science. 13. doi: 10.3389/fpls.2022.865302
Liang, H., Dong, W., Xu, L., Yu, Y., Yang, H., Tan, Z., et al. (2017). QTL mapping for main root length and lateral root number in soybean at the seedling stage different N, P and in K environments. Scientia Agricultura Sinica. 50, 3450–3460. doi: 10.3864/j.issn.0578-1752.2017.18.002
Liang, H., Yu, Y., Yang, H., Xu, L., Dong, W., Du, H., et al. (2014). Inheritance and QTL mapping of related root traits in soybean at the seedling stage. Theor. Appl. Genet. 127, 2127–2137. doi: 10.1007/s00122-014-2366-z
Lippert, C., Listgarten, J., Liu, Y., Kadie, C. M., Davidson, R. I., Heckerman, D. (2011). FaST linear mixed models for genome-wide association studies. Nat. Methods 8, 833–835. doi: 10.1038/nmeth.1681
Liu, X., Gu, X., Lu, H., Liu, P., Miao, H., Bai, Y., et al. (2021a). Identification of novel loci and candidate genes for resistance to powdery mildew in a resequenced cucumber germplasm. Genes. 12, 584. doi: 10.3390/genes12040584
Liu, X., Lu, H., Liu, P., Miao, H., Bai, Y., Gu, X., et al. (2020). Identification of novel loci and candidate genes for cucumber downy mildew resistance using GWAS. Plants. 9, 1659. doi: 10.3390/plants9121659
Liu, S., Lv, X., Ahammed, G. J., Xia, X., Zhou, J., et al. (2016). Grafting cucumber onto luffa improves drought tolerance by increasing ABA biosynthesis and sensitivity. Sci. Rep. 6, 20212. doi: 10.1038/srep20212
Liu, Y., Shao, L., Zhou, J., Li, R., Pandey, M. K., Han, Y., et al. (2022). Genomic insights into the genetic signatures of selection and seed trait loci in cultivated peanut. J. Advanced Res. 42, 237–248. doi: 10.1016/j.jare.2022.01.016
Liu, Y., Wang, H., Jiang, Z., Wang, W., Xu, R., Wang, Q., et al. (2021b). Genomic basis of geographical adaptation to soil nitrogen in rice. Nature. 590, 600–605. doi: 10.1038/s41586-020-03091-w
Liu, S., Wang, J., Wang, L., Wang, X., Xue, Y., Wu, P., et al. (2009). Adventitious root formation in rice requires OsGNOM1 and is mediated by the OsPINs family. Cell Res. 19, 1110–1119. doi: 10.1038/cr.2009.70
Liu, Z., Wu, Y., Yang, F., Zhang, Y., Chen, S., Xie, Q., et al. (2013). BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc. Natl. Acad. Sci. 110, 6205–6210. doi: 10.1073/pnas.1215543110
Liu, Y., Zuo, T., Qiu, Z., Zhuang, K., Hu, S., Han, H. (2021). Genome-wide identification reveals the function of CEP peptide in cucumber root development. Plant Physiol. Biochem. 169, 119–126. doi: 10.1016/j.plaphy.2021.11.007
Ljung, K., Hull, A. K., Celenza, J., Yamada, M., Estelle, M., Normanly, J., et al. (2005). Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell. 17, 1090–1104. doi: 10.1105/tpc.104.029272
Mao, X., Liu, M., Wang, X., Liu, C., Hou, Z., Shi, J. (2003). Effects of deficit irrigation on yield and water use of greenhouse grown cucumber in the North China Plain. Agric. Water management. 61, 219–228. doi: 10.1016/S0378-3774(03)00022-2
Nayyeripasand, L., Garoosi, G. A., Ahmadikhah, A. (2021). Genome-wide association study (GWAS) to identify salt-tolerance QTLs carrying novel candidate genes in rice during early vegetative stage. Rice. 14, 1–21. doi: 10.1186/s12284-020-00433-0
Pace, J., Gardner, C., Romay, C., Ganapathysubramanian, B., Lübberstedt, T. (2015). Genome-wide association analysis of seedling root development in maize (Zea mays L.). BMC Genomics 16, 1–12. doi: 10.1186/s12864-015-1226-9
Pitts, R. J., Cernac, A., Estelle, M. (1998). Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J. 16, 553–560. doi: 10.1046/j.1365-313x.1998.00321.x
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A., Bender, D., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. doi: 10.1086/519795
Qi, J., Liu, X., Shen, D., Miao, H., Xie, B., Li, X., et al. (2013). A genomic variation map provides insights into the genetic basis of cucumber domestication and diversity. Nat. Genet. 45, 1510–1515. doi: 10.1038/ng.2801
Runcie, D. E., Crawford, L. (2019). Fast and flexible linear mixed models for genome-wide genetics. PloS Genet. 15, e1007978. doi: 10.1371/journal.pgen.1007978
Schmittgen, T. D., Zakrajsek, B. A. (2000). Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J. Biochem. Biophys. Methods 46, 69–81. doi: 10.1016/S0165-022X(00)00129-9
Sels, J., Mathys, J., De Coninck, B. M., Cammue, B. P., De Bolle, M. F. (2008). Plant pathogenesis-related (PR) proteins: a focus on PR peptides. Plant Physiol. Biochem. 46, 941–950. doi: 10.1016/j.plaphy.2008.06.011
Shin, J.-H., Blay, S., McNeney, B., Graham, J. (2006). LDheatmap: an R function for graphical display of pairwise linkage disequilibria between single nucleotide polymorphisms. J. Stat. Software 16, 1–9. doi: 10.18637/jss.v016.c03
Shinshi, O. T. (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell. 7, 173–182. doi: 10.1105/tpc.7.2.173
Tanimoto, M., Roberts, K., Dolan, L. (1995). Ethylene is a positive regulator of root hair development in Arabidopsis thaliana. Plant J. 8, 943–948. doi: 10.1046/j.1365-313X.1995.8060943.x
Taylor, B. H., Young, R. J., Scheuring, C. F. (1993). Induction of a proteinase inhibitor II-class gene by auxin in tomato roots. Plant Mol. Biol. 23, 1005–1014. doi: 10.1007/BF00021815
Turner, S. D. (2014). qqman: an R package for visualizing GWAS results using q-q and manhattan plots. Biorxiv. 3 (25), 1731. doi: 10.21105/joss.00731
Uga, Y., Sugimoto, K., Ogawa, S., Rane, J., Ishitani, M., Hara, N., et al. (2013). Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 45, 1097. doi: 10.1038/ng.2725
Usanmaz, S., Abak, K. (2019). Plant growth and yield of cucumber plants grafted on different commercial and local rootstocks grown under salinity stress. Saudi J. Biol. Sci. 26, 1134–1139. doi: 10.1016/j.sjbs.2018.07.010
Walters, S. A., Wehner, T. C. (1994). Evaluation of the US cucumber germplasm collection for root size using a subjective rating technique. Euphytica. 79, 39–43. doi: 10.1007/BF00023574
Wang, X., Bao, K., Reddy, U. K., Bai, Y., Hammar, S. A., Jiao, C., et al. (2018). The USDA cucumber (Cucumis sativus L.) collection: genetic diversity, population structure, genome-wide association studies, and core collection development. Horticulture Res. 5, 64. doi: 10.1038/s41438-018-0080-8
Wang, X., He, F., Ma, X., Mao, C., Hodgman, C., Lu, C., et al. (2011). OsCAND1 is required for crown root emergence in rice. Mol. Plant 4, 289–299. doi: 10.1093/mp/ssq068
Wang, Z., Liu, L., Su, H., Guo, L., Zhang, J., Li, Y., et al. (2020). Jasmonate and aluminum crosstalk in tomato: Identification and expression analysis of WRKYs and ALMTs during JA/Al-regulated root growth. Plant Physiol. Biochem. 154, 409–418. doi: 10.1016/j.plaphy.2020.06.026
Wang, H., Wei, J., Li, P., Wang, Y., Ge, Z., Qian, J., et al. (2019). Integrating GWAS and gene expression analysis identifies candidate genes for root morphology traits in maize at the seedling stage. Genes. 10, 773. doi: 10.3390/genes10100773
Woo, O., Kim, S., Cho, S. K., Kim, S., Lee, H. N., Chung, T., et al. (2018). BPH1, a novel substrate receptor of CRL3, plays a repressive role in ABA signal transduction. Plant Mol. Biol. 96, 593–606. doi: 10.1007/s11103-018-0717-x
Xie, H., Li, M., Chen, Y., Jia, Z. (2021). Important physiological changes due to drought stress on oat. Front. Ecol. Evolution. 9. doi: 10.3389/fevo.2021.644726
Yan, Y., Chen, X., Yang, K., Sun, Z., Fu, Y., Zhang, Y., et al. (2011). Overexpression of an F-box protein gene reduces abiotic stress tolerance and promotes root growth in rice. Mol. Plant 4, 190–197. doi: 10.1093/mp/ssq066
Yan, Y., Zhang, W., Li, Y., He, C., Gao, L., Yu, X. (2018). Functions of CsGPA1 on the hypocotyl elongation and root growth of cucumbers. Sci. Rep. 8, 15583. doi: 10.1038/s41598-018-33782-4
Yang, W., Guo, Z., Huang, C., Duan, L., Chen, G., Jiang, N., et al. (2014). Combining high-throughput phenotyping and genome-wide association studies to reveal natural genetic variation in rice. Nat. Commun. 5, 5087. doi: 10.1038/ncomms6087
Yao, X., Li, H., Nie, J., Liu, H., Guo, Y., Lv, L., et al. (2023). Disruption of the amino acid transporter CsAAP2 inhibits auxin-mediated root development in cucumber. New Phytologist. 239, 639–659. doi: 10.1111/nph.18947
You, J., Liu, Z., Qi, Z., Ma, Y., Sun, M., Su, L., et al. (2023). Regulatory controls of duplicated gene expression during fiber development in allotetraploid cotton. Nat. Genet. 55, 1987–1997. doi: 10.1038/s41588-023-01530-8
Zhang, B. G., Chen, Q. X., Luo, S. B., Zhang, C. Y., Yang, Q., Liu, K. D. (2012). Effects of NPK deficiencies on root architecture and growth of cucumber. Int. J. Agric. Biol. 14, 145–148. doi: 10.1016/j.compag.2011.09.003
Zhang, S., Huang, L., Yan, A., Liu, Y., Liu, B., Yu, C., et al. (2016). Multiple phytohormones promote root hair elongation by regulating a similar set of genes in the root epidermis in Arabidopsis. J. Exp. botany. 67, 6363–6372. doi: 10.1093/jxb/erw400
Zhao, H., Lü, S., Li, R., Chen, T., Zhang, H., Cui, P., et al. (2015). The Arabidopsis gene DIG6 encodes a large 60S subunit nuclear export GTPase 1 that is involved in ribosome biogenesis and affects multiple auxin-regulated development processes. J. Exp. botany. 66, 6863–6875. doi: 10.1093/jxb/erv391
Keywords: cucumber (Cucumis sativus L.), root, GWAS (genome-wide association study), candidate genes, haplotypes
Citation: Dai Z, Dong S, Cai H, Beckles DM, Guan J, Liu X, Gu X, Miao H and Zhang S (2024) Genome-wide association analysis reveal candidate genes and haplotypes related to root weight in cucumber (Cucumis sativus L.). Front. Plant Sci. 15:1417314. doi: 10.3389/fpls.2024.1417314
Received: 14 April 2024; Accepted: 25 June 2024;
Published: 17 July 2024.
Edited by:
Panagiotis Kalaitzis, Mediterranean Agronomic Institute of Chania, GreeceReviewed by:
Jingtao Nie, Zhejiang Agriculture and Forestry University, ChinaCopyright © 2024 Dai, Dong, Cai, Beckles, Guan, Liu, Gu, Miao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Han Miao, bWlhb2hhbkBjYWFzLmNu; Shengping Zhang, emhhbmdzaGVuZ3BpbmdAY2Fhcy5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.