- 1Shenzhen Research Institute, Northwest A&F University, Shenzhen, China
- 2State Key Laboratory for Crop Stress Resistance and High-Efficiency Production, College of Agronomy, Northwest A&F University, Yangling, China
Growth-regulating factors (GRFs) are transcription factors that play a pivotal role in plant growth and development. This study identifies 12 Solanum tuberosum GRF transcription factors (StGRFs) and analyzes their physicochemical properties, phylogenetic relationships, gene structures and gene expression patterns using bioinformatics. The StGRFs exhibit a length range of 266 to 599 amino acids, with a molecular weight of 26.02 to 64.52 kDa. The majority of StGRFs possess three introns. The promoter regions contain a plethora of cis-acting elements related to plant growth and development, as well as environmental stress and hormone response. All the members of the StGRF family contain conserved WRC and QLQ domains, with the sequences of these two conserved domain modules exhibiting high levels of conservation. Transcriptomic data indicates that StGRFs play a significant role in the growth and development of stamens, roots, young tubers, and other tissues or organs in potatoes. Furthermore, a few StGRFs exhibit differential expression patterns in response to Phytophthora infestans, chemical elicitors, heat, salt, and drought stresses, as well as multiple hormone treatments. The results of the expression analysis indicate that StGRF1, StGRF2, StGRF5, StGRF7, StGRF10 and StGRF12 are involved in the process of tuber sprouting, while StGRF4 and StGRF9 may play a role in tuber dormancy. These findings offer valuable insights that can be used to investigate the roles of StGRFs during potato tuber dormancy and sprouting.
Introduction
Transcription factors (TFs), also referred to as trans-acting elements, can bind to specific sequences (cis-acting elements) in the gene promoter region to regulate the expression of a target gene. TFs exhibit a wide range of functions and play crucial roles in numerous biological processes and regulatory pathways in plants. To date, 320,370 transcription factors have been identified and classified into 58 families across 165 species (Jin et al., 2017). Among these families, growth-regulating factors (GRFs) are a plant-specific type of TF that were originally identified for their roles in stem and leaf development in rice (van der Knaap et al., 2000). To date, members of the GRF family have been identified in a number of plant species, including thale cress (Arabidopsis thaliana), maize (Zea mays), rice (Oryza sativa), oilseed rape (Brassica napus), soybean (Glycine max), tomato (Solanum lycopersicum), and moss (Physcomitrella patens) (Kim et al., 2003; Choi et al., 2004; Zhang et al., 2008; Khatun et al., 2017; Ma et al., 2017; Chen et al., 2019). The GRF proteins share common features, including the QLQ (Gln, Leu, Gln) and WRC (Trp, Arg, Cys) domains at the N-terminus, and the relatively variable regions at the C-terminus. They form complexes with their co-activators, known as GRF-interacting factors (GIFs), which can bind to the cis-acting region of downstream target genes and regulate their expression, inferring a role in transcriptional regulation.
GRF family genes are involved in the growth, development, and regeneration of plants. In Arabidopsis, AtGRF1, AtGRF2, and AtGRF3 are predominantly expressed in shoots, roots, and stems (Kim et al., 2003). AtGRF4 and AtGRF6 are expressed in the midvein of leaves, while AtGRF5 is expressed in leaf primordia (Horiguchi et al., 2005). AtGRF7 is expressed in the blades and petioles of true leaves, while AtGRF7 and AtGRF8 are predominantly expressed in the shoot tips (Kim et al., 2003). Additionally, AtGRF1, AtGRF2, AtGRF4, AtGRF5, AtGRF6, AtGRF7, and AtGRF9 are expressed in the flower (http://bar.utoronto.ca/eplant/). AtGRF1, AtGRF2, AtGRF3, AtGRF4, AtGRF5, and AtGRF7 are strongly expressed in the meristematic zone (Kim et al., 2003; Rodriguez et al., 2010; Kim et al., 2012; Bao et al., 2014; Pajoro et al., 2014). GRFs have been reported to alter leaf cell numbers, thereby affecting the leaf size and longevity (Debernardi et al., 2014; Wu et al., 2014; Nelissen et al., 2015; Vercruyssen et al., 2015), stem elongation (van der Knaap et al., 2000; Kuijt et al., 2014), root development (Hewezi et al., 2012; Bao et al., 2014), floral organ development and regulation of flowering time (Pajoro et al., 2014), and seed oil content (Liu et al., 2012). GIFs act as transcriptional co-regulators of GRFs. GIF1 interacts with AtGRF1, AtGRF2, AtGRF4, AtGRF5 and AtGRF9 through its conserved QLQ domain (Kim and Kende, 2004; Horiguchi et al., 2005). The overexpression of GIF1/AN3 leads to an increase in leaf area due to an increase in cell number (Horiguchi et al., 2005). Conversely, a moderate reduction in GIF1 expression results in smaller leaves due to a reduction in cell number. Moreover, a number of GRF genes contain the miRNA396 target site, and it is known that miRNA396 plays a role in regulating GRF genes during plant development (Rodriguez et al., 2010; Debernardi et al., 2012). In Arabidopsis, miR396a and miR396b regulate leaf growth and development by post-transcriptional repression of GRF genes (Rodriguez et al., 2010; Wang et al., 2011). OsmiR396d targets OsGRF6 and OsGRF10, and overexpression of OsmiR396 results in an abnormal number of stigmas and stamens (Liu et al., 2014). Furthermore, evidences indicate that GRFs play a role in the plant adaptation to stress (Kim et al., 2012; Casadevall et al., 2013; Liu et al., 2014), and are closely associated with plant hormones (Choi et al., 2004; Bazin et al., 2013).
Potato (Solanum tuberosum L.) is the third most important food crop worldwide, after rice and wheat. It plays a vital role in ensuring global food security, particularly in light of the growing population and the concomitant increase in hunger. It is estimated that over one billion people worldwide consume potatoes, with global production exceeding 300 million metric tons. The release of the potato’s complete genome sequence has enabled a comprehensive analysis of the GRF genes (Xu et al., 2011). GRFs are involved in regulating the growth of different plant tissues and organs. The potato tuber is a swollen underground stem formed by shortened internodes and nodes that develop into tuber eyes. Meristematic activity in the tuber eyes is completely suppressed during the development of the tuber. Even when placed in optimal conditions for sprouting, such as warm temperature, darkness, and high humidity, the tuber buds are generally dormant and will not sprout or grow. Subsequently, the tuber enters a period of dormancy, during which the eye exhibits visible growth of a bud. As tuber dormancy is an important agronomic trait, a short dormancy period renders potato tubers challenging to store for an extended period, whereas a long dormancy period makes them difficult to plant in a timely manner. Tuber sprouting typically originates from the tuber apical meristem (TAM), although GRFs are among the most crucial regulators of meristem development and cell differentiation restriction in the shoot apical meristem (SAM). Nevertheless, the role of GRF genes in potato tuber dormancy and sprouting remains elusive. In order to gain a better understanding of the role of GRF transcription factors in potato, a genome-wide identification and analysis of the potato (S. tuberosum) GRF family members (StGRFs) was conducted. In the present study, a total of 12 GRF genes in the potato genome were identified. The expression patterns of StGRFs in different tissues were analyzed. Furthermore, the expression profiles of StGRFs under multiple external stimuli and hormones, as well as during tuber dormancy were examined. The results demonstrated that StGRF genes exhibited distinct expression patterns in different tissues, and that their transcription was induced by diverse biotic and abiotic stresses. Further analysis of the expression patterns of these StGRFs revealed that they may play a significant role in regulating the release of tuber dormancy.
Materials and methods
Plant materials
A diploid potato line EB063 derived from a cross between parent E (ED25) and B (CW2–1) (Xiao et al., 2018) was utilized in the present study. The sprouted seed tubers were planted at the Yulin potato breeding station, located in the Shaanxi Province of China. Local cultural practices were employed to ensure optimal plant growth. The tubers were harvested at the appropriate stage of maturity and stored in the dark at room temperature for a period of two weeks, during which time they underwent wound healing. Subsequently, tubers of a similar size were then placed in light-proof boxes at a temperature of 22 ± 2°C in order to facilitate the release of dormancy. The tubers were designated as dormant tubers (DT) at the point at which they were fully mature (0 day at dormancy release array), while any sprouts that were ≥2 mm in length were considered to be sprouting tubers (ST). For the purpose of sample collection, the apical bud meristems (also known as the dormancy eye) were excised at day 0 of the dormancy release array. The sprouting sprouts and sprouting sprout bases were collected at five weeks of the dormancy release array, respectively, from three to five tubers using a 6 mm size cork borer. The samples were immediately frozen in liquid nitrogen and stored at −80°C, after which they were subjected to RNA isolation.
Identification and physicochemical properties analysis of the GRF family members in potato
The potato genome files (version number DM v6.1) were downloaded from the Spud DB database (http://spuddb.uga.edu/). Amino acid sequences of Arabidopsis (TAIR, http://www.arabidopsis.org/) and rice (RGAP, http://rice.uga.edu/) GRF proteins were employed as queries in a BlastP homology search to identify candidate GRF proteins against the potato genome DM v6.1. All retrieved amino acid sequences of potato were subjected to verification for the presence of both the QLQ and WRC (PF08880, PF08879, respectively; http://pfam.xfam.org/) domains through the CDD (Conserved Domains Database; https://www.ncbi.nlm.nih.gov/cdd/) and SMART (http://smart.embl-heidelberg.de/) programs. The physiochemical properties of StGRF proteins including the amino acid number, molecular weight (MW), isoelectric point (pI), and grand average of hydropathicity (GRAVY), were analyzed by the ExPASy ProtParam tool (http://www.expasy.org/protparam/). The subcellular localization of StGRF proteins was determined by web-server predictors in the Cell-PLoc package (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/).
Phylogenetic analysis, gene structure, and conserved motifs of StGRFs
To visualize the evolutionary relationship of GRF family members, full-length amino acid sequences of StGRFs, AtGRFs, OsGRFs and PtGRFs were then aligned using ClustalW. A phylogenetic tree was constructed using the neighbor-joining method in MEGA11 software (https://www.megasoftware.net/) and the bootstrap test was carried out with 1000 replicates. A homology analysis of potato GRF proteins was conducted by aligning of the amino acid sequences using ClustalW (https://www.genome.jp/tools-bin/clustalw), and the resulting alignment was visualized using the SnapGene tool. The map of exon-intron structures of the StGRF genes were visualized using GSDS2.0 (Gene Structure Display Server 2.0, http://gsds.cbi.pku.edu.cn/index.php). Furthermore, the MEME tool (https://meme-suite.org/meme/tools/meme) was employed to predict conserved motifs of the potato GRF family member. The number of motifs was set to 8 and zero or one occurrence per sequence.
Cis-acting element analysis of the StGRF promoters
The 2000 bp upstream sequences of StGRF genes were extracted using TBtools software (Chen et al., 2020) based on the full-length DNA sequence of the potato genome. The cis-acting elements in the potential promoter regions were identified using the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). The potential interactions of transcription factors in the 2000 bp upstream regions of StGRF genes were predicted using the Plant Transcriptional Regulatory Map (http://plantregmap.gao-lab.org/) with the following parameters: the P-value was set to 1e-6, and Arabidopsis thaliana as set to as the reference species. The results of the prediction were visualized using Cytoscape software (v3.9.1). The occurrence frequencies of transcription factors were employed to generate a wordcloud using the wordcloud package in the R project.
Expression profiles of StGRF genes
The expression profiles of StGRF genes were determined using RNA-Seq data available at the SpudDB (http://spuddb.uga.edu/). The tissue specificity of different tissues (tuber cortex, tuber pith, tuber peel and shoot apex) and organs (leaf, stem, flower, root, stamen, petiole, stolon, mature tuber and young tuber) and potato plants subjected to diverse stresses (microbial pathogen infection, salt, heat, BAP, IAA, ABA and GA3) was analyzed. Gene expression levels of StGRFs were represented by reads per kb per million reads (RFKM). The heatmap of the expression patterns was constructed using TBtools (https://github.com/CJ-Chen/TBtools/).
RNA isolation and RT-qPCR
Total RNA was extracted using the RNA simple Total RNA Kit (Cat. No. DP419, TIANGEN) in accordance with the manufacturer’s instructions. RNA quality was analyzed using a NanoDrop One spectrophotometer and agarose gel electrophoresis. One microgram of total RNA was employed to synthesize the first strand of cDNA using the HiFiScript gDNA Removal cDNA Synthesis Kit (Cat. No. CW2020M, CWBIO) according to the manufacturer’s instructions. The polymerase chain reaction (PCR) was conducted in a total volume of 10 µL, comprising 5 µL of 2×qPCR Smart Mix (SYBR Green) (Cat. No. DY20302, DEEYEE), 0.5 µL (10 µM) of each gene-specific primer, 3.5 µL of ddH2O, and 1 µL of cDNA. The target gene was detected using the QuantStudioTM7 Flex System (Applied Biosystems, Thermo Fisher Scientific, USA). StUBI3 was employed as an internal control, and the primers used for the qPCR are listed in Supplementary Table S1. The 2-ΔΔCT method was used to calculate the relative gene expression, and all RT-qPCR experiments were performed in triplicate.
Results
Identification of StGRF genes in the potato genome
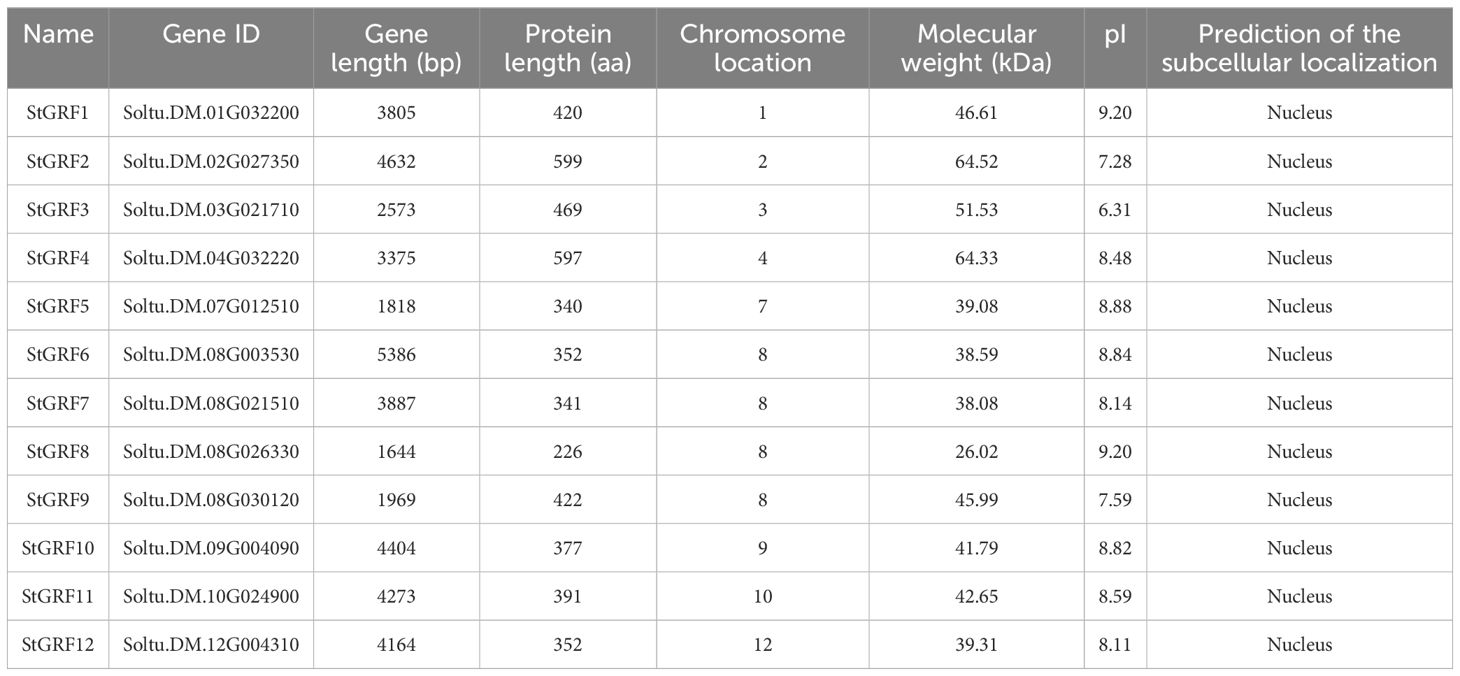
A total of 12 GRF family members in potato were identified and named StGRF1 to StGRF12 according to their physical locations on the chromosomes (Table 1) through the use of bi-directional BLAST and conserved domain analysis. Amino acid length analysis revealed a considerable range in the amino acid lengths of the StGRF proteins, with StGRF8 having the shortest length of 226 amino acids and StGRF2 having the longest length of 599 amino acids (Table 1). The molecular weight of the StGRFs exhibited a similarly wide range, from 26.02 kDa (StGRF8) to 64.52 kDa (StGRF2), while the isoelectric point of StGRFs displayed a similar range, from 6.31 (StGRF3) to 9.20 (StGRF1 and StGRF8) (Table 1). The 12 StGRF genes were unevenly distributed across nine chromosomes (Table 1). Among the 12 StGRF genes, the chromosome 8 contains four StGRF genes, namely StGRF6, StGRF7, StGRF8 and StGRF9. In contrast, the remaining chromosomes each contain only one StGRF gene (Table 1). Subcellular localization prediction indicated that all the StGRF proteins were localized in the nucleus (Table 1).
Phylogenetic, conserved motif and gene structure analysis
To explore the evolutionary relationships and sequence homology among GRF proteins from potato, Arabidopsis (Kim et al., 2003), rice (Choi et al., 2004) and poplar (Wang et al., 2020), a neighbor-joining phylogenetic tree was constructed using MEGA11 software (Figure 1). The evolutionary relationships of the GRFs were analyzed and a total of 52 GRFs from four plant species were clustered into five subgroups with supported bootstrap values. These were designated as Group I, Group II, Group III, Group IV and Group V (Figure 1). A total of three, one, two, two, and four StGRF members were assigned to subgroups I, II, III, IV and V, respectively (Figure 1). The phylogenetic tree indicates that the StGRFs are more closely related to PtGRFs and AtGRFs than with to OsGRFs (Figure 1). This could be attributed to the fact that potato, poplar and Arabidopsis are dicotyledonous plants. In order to ascertain the structural diversity and functional prediction of StGRFs, multiple alignments of the amino acid sequences of StGRF family members were performed (Figure 2A). A total of eight conserved motifs were identified, with the length of these motifs ranging from 9 to 41 amino acids (Supplementary Table S2). Among these, motifs 1 and 2 were respectively annotated as the WRC and QLQ domain, which were included in all StGRF family members (Figures 2A, B), indicating that two domains are highly conserved. All family members, with the exception of StGRF1, StGRF8 and StGRF9, contained a TQL (Thr, Gln, Leu) domain at the C-terminus (Figures 2A, B). In contrast, the C-termini of all StGRF members, with the exception of StGRF1, StGRF3, StGRF8 and StGRF9, were found to contain an FFD (Phe, Phe, Asp) domain (Figures 2A, B). StGRF2, StGRF3, StGRF4, StGRF9, StGRF10, and StGRF11 possess a GGPL (Gly, Gly, Pro, Leu) domain at the C-terminus (Figures 2A, B). A multiple sequence alignment of the core QLQ and WRC domain of StGRFs is presented in Supplementary Figure S1. It is notable that the characteristics of these motifs were consistent within the same cluster (Figures 2A, B). For example, StGRF6, StGRF7 and StGRF12 exhibited seven common conserved motifs (motifs 1, 2, 3, 4, 5, 6 and 7) (Figures 2A, B). The gene structure of the StGRF gene family was further analyzed using the GSDS online tool. The StGRFs exhibited a length range of 1000 bp to 6000 bp, with the number of introns was found to be 2 to 3 (Figure 2C). StGRFs from the same clusters exhibited similar exon/intron structure patterns (Figure 2), suggesting that phylogenetic relationships among gene family members are highly correlated with gene structure. Furthermore, StGRF7 has only a 3’ untranslated region (UTR), whereas the other 11 StGRF genes possess 5’- and 3’-UTRs at both ends.
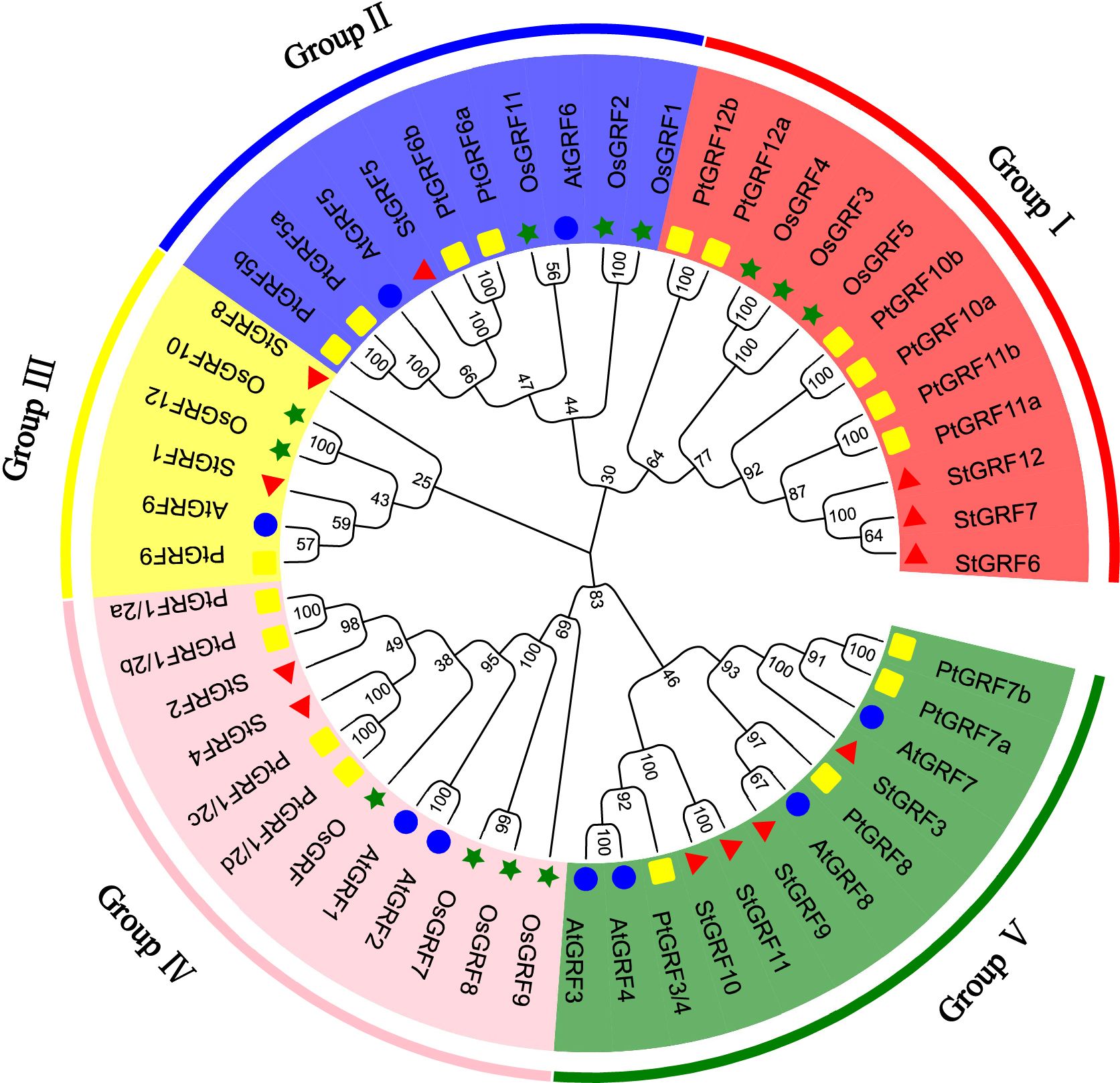
Figure 1 Phylogenetic tree of GRF proteins from Arabidopsis thaliana (At), Oryza sativa (Os), Solanum tuberosum (St), and Populus trichocarpa (Pt). Blue circle: Arabidopsis thaliana (At), green pentagram: Oryza sativa (Os), red triangle: Solanum tuberosum (St), yellow square: Populus trichocarpa (Pt).
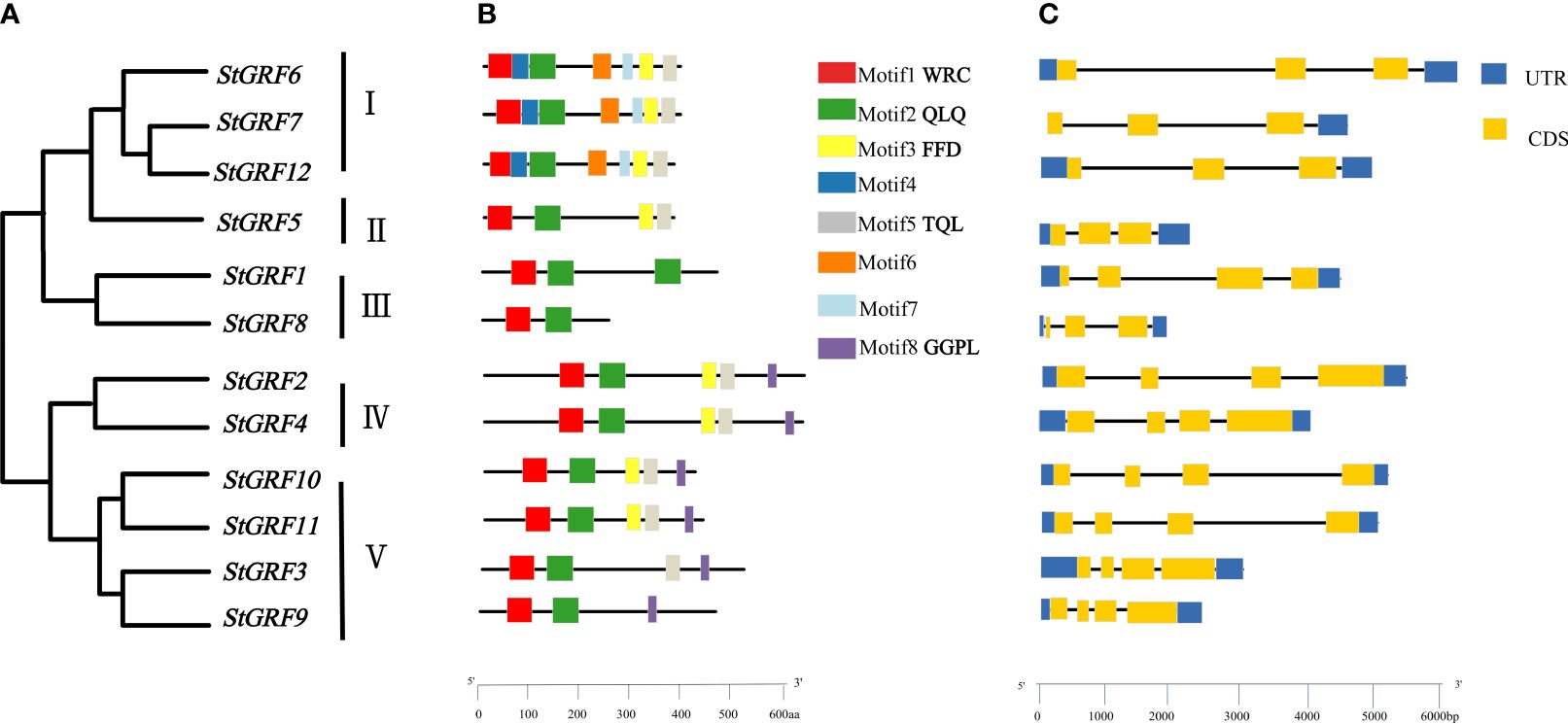
Figure 2 Phylogenetic tree, gene structure and motif of the GRF gene family in potato (Solanum tuberosum). (A) Phylogenetic relationship of StGRF proteins. (B)The distribution of eight conserved motifs in StGRF proteins, identified by the MEME program, as indicated by different colored blocks. (C) Exon/intron structures of StGRFs. The exons and introns were represented by yellow boxes and black lines, respectively. The dark blue boxes indicate the upstream and downstream untranslated regions, respectively.
Cis-acting elements in the promoter regions of StGRF genes
Cis-acting elements within the gene promoters are specific binding sites for proteins involved in the initiation and regulation of gene transcription. To gain further insights into the functions of cis-acting elements within the promoter region of StGRFs, the promoter sequences for StGRFs were submitted to PlantCARE for prediction (Figure 3). The cis-acting elements within the StGRFs promoters are primarily responsible for plant growth and development, as well as responses to hormones and abiotic or biotic stresses (Figure 3). StGRF2, StGRF9 and StGRF12 were found to contain several cis-acting elements closely associated with hormone responses, including ABRE (related to the abscisic acid responsiveness), AuxRR-core (auxin responsiveness), TCA-element (involved in salicylic acid responsiveness), CGTCA- and TGACG-motif (involved in MeJA-responsiveness), TATC-box and P-box (gibberellin-responsive element) (Figure 3). The majority of StGRF genes contained MeJA-responsive elements, CGTCA and TGACG motifs, which were observed 22 times, representing 42% of the hormone-related cis-acting elements. ABRE, the abscisic acid responsiveness elements, were observed 13 times, which represented 25% of the hormone-responsive elements of StGRFs (Figure 3). The promoter regions of nine StGRFs contain cis-acting elements related to environmental stresses (Figure 3). For example, LTR, a low-temperature responsiveness cis-acting element identified in StGRF5 and StGRF12. MBS, a drought-inducibility cis-acting element identified in StGRF2, StGRF3, StGRF11 and StGRF12. The TC-rich repeats and wound-responsive element 3 (WRE 3), which are known to be involved in wounding and pathogen response, have been identified in StGRF1, StGRF2, StGRF4, StGRF5, StGRF11 and StGRF12. The ARE motif, which is essential for the anaerobic induction, has been identified on nine occasions, representing for 31% of the stress-related cis-acting elements (Figure 3). Furthermore, the promoters of 11 StGRFs carry cis-acting elements related to plant growth and development (Figure 3). The promoters of StGRF5, StGRF7, StGRF8, and StGRF9 contain the CAT-box, which is associated with meristem development (Figure 3). Similarly, the promoters of StGRF1, StGRF3, StGRF10, and StGRF11 contain the GCN4 motif, which is involved in plant endosperm development (Figure 3). The promoters of StGRF6, StGRF8, StGRF9, StGRF10, StGRF11, and StGRF12 contain the O2-site, which is involved in the regulation of zein metabolism (Figure 3). The O2-site and GCN4 motifs were identified on 8 and 7 occasions, respectively, and accounted for 50% of the plant growth and development-related motifs (Figure 3). Additionally, cis-acting elements related to circadian control (circadian) and light response elements (G-box and Sp1) were also identified in the promoters of the StGRFs (Figure 3).
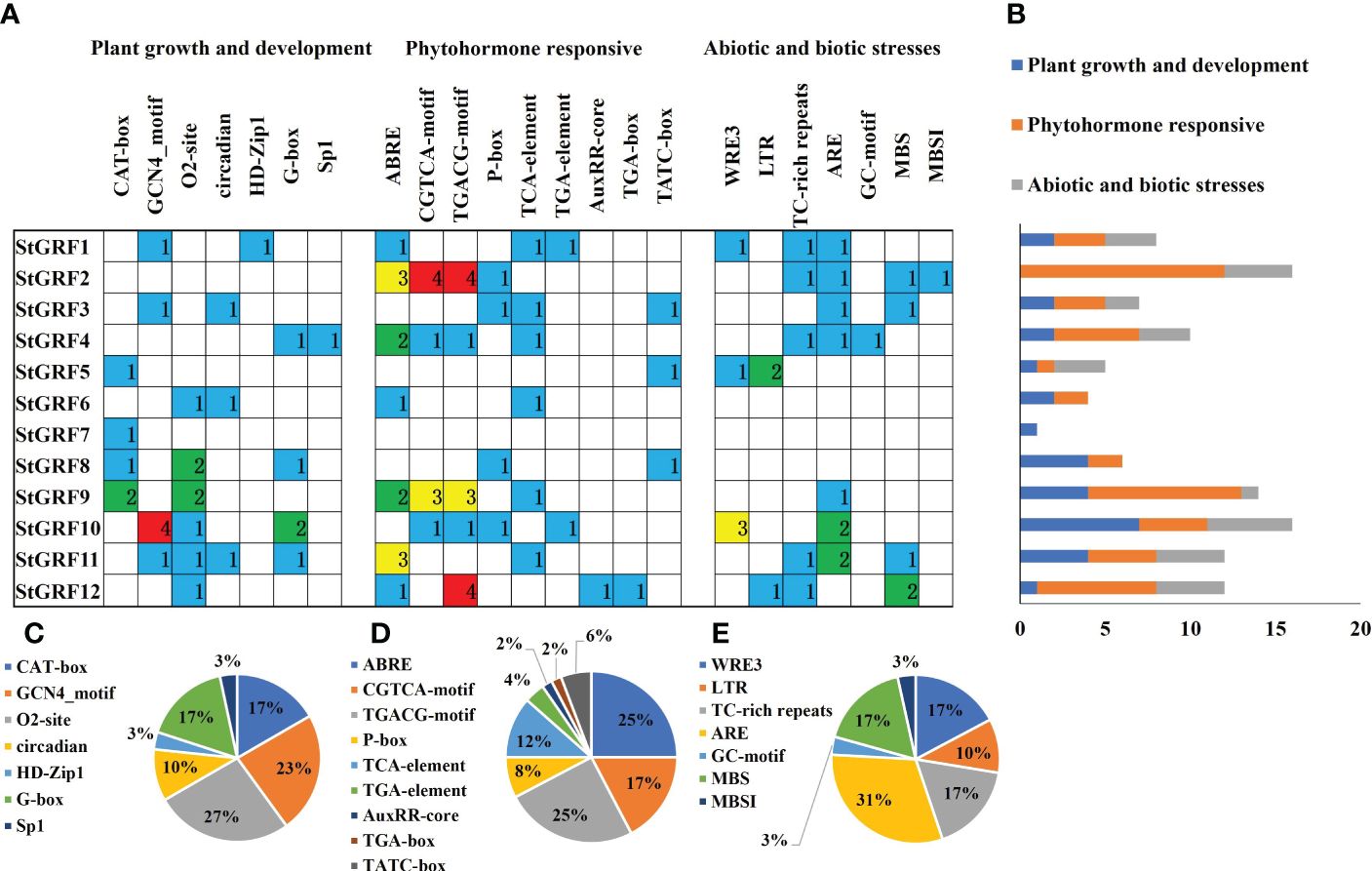
Figure 3 Cis-acting element analysis of the potato GRF family genes. The different colors represent different cis-acting elements. (A) Investigation of the number of cis-acting elements in the StGRF promoter regions. (B) Statistics for the number of the promoter elements in three major subfamilies. (C–E) The proportion of each promoter in the category.
Bioinformatic analysis of StGRFs-mediated regulatory network
To predict the potential roles of StGRFs in potato, a StGRFs-mediated regulatory network was further constructed. Data indicated that a total of 52 transcription factors belonging to 15 different TF families, including Dof, MYB, C2H2, BBR-BPC, MIKC_MADS, AP2, etc., were identified as the potential regulators of StGRFs (Figure 4A, Supplementary Table S3). The predicted TFs were found to be most abundant in the Dof family (54), followed by the MIKC_MADS family (33), the BBR-BPC family (32) and the C2H2 family (20) (Figure 4B, Supplementary Table S3). In contrast, the least abundant TF families contain only a few members, including the Trihelix family (1), the TCP family (1), the C3H family (1) and the GATA family (2), the CPP family (2) (Supplementary Table S3). The predictions indicate that StGRF6 has the largest class of TFs among all StGRFs (10 TFs), followed by StGRF7 (7 TFs), StGRF1 (6 TFs) and StGRF4 (5 TFs) (Supplementary Table S3). These enriched transcription factor families may play an essential role in regulating the expression of StGRFs in potato. Collectively, the predicted regulatory network of StGRFs indicates that they may be involved in a number of biological processes, including plant growth and development, biotic and abiotic stress responses, and network associations.
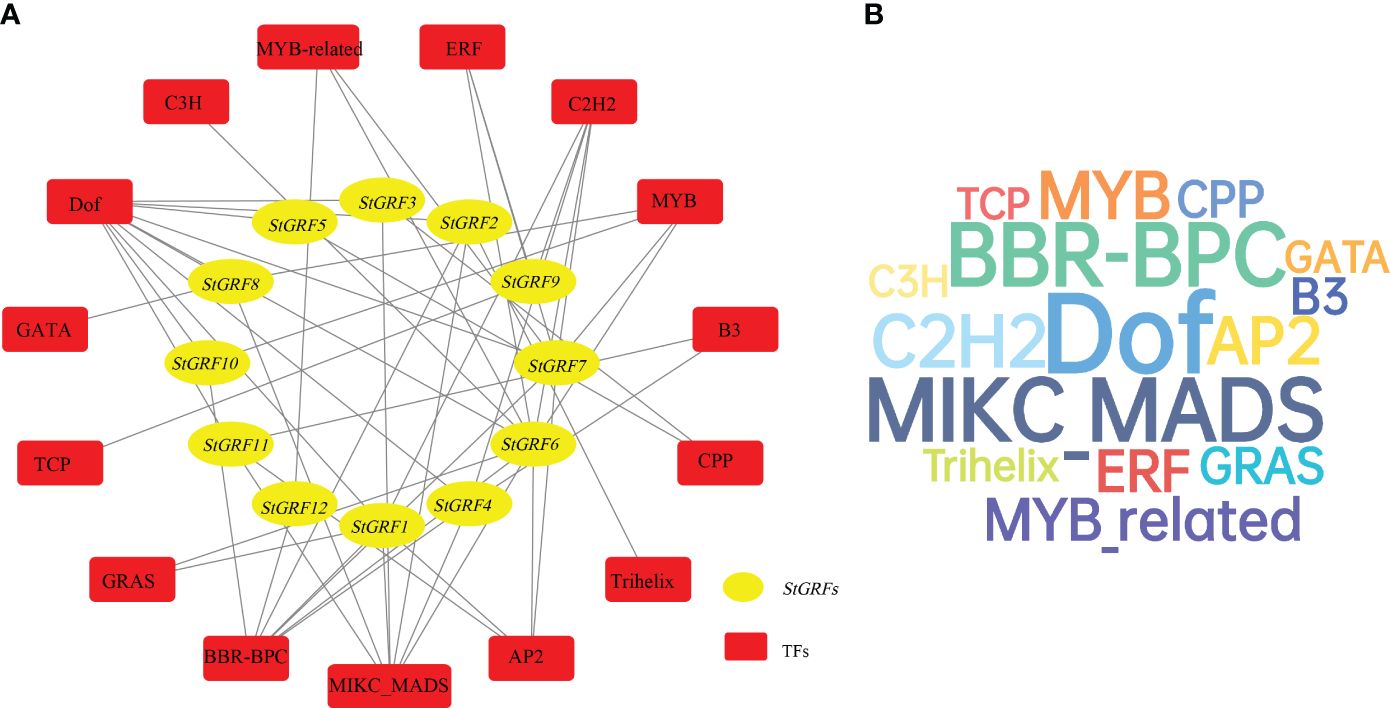
Figure 4 (A) The putative transcription factors (TFs) regulatory network analysis of StGRFs. Red rounded rectangle nodes represent TFs; yellow ellipse nodes represent StGRFs. (B) Wordcloud for TFs. The font size is positively correlated with the number of corresponding TFs.
Tissue-specific expression of StGRFs in potato
A comparative analysis of the tissue-specific expression of the 12 StGRFs revealed differential expression in different potato tissues and developmental stages (Figure 5). The majority of StGRFs exhibited low expression in stamens, mature flowers, and mature fruits, whereas high expression was observed in immature tubers, including StGRF2, StGRF4, StGRF6, StGRF10 and StGRF12 (Figure 5). In heterozygous diploids, all StGRFs except for StGRF1, StGRF5, StGRF8 and StGRF11 were expressed in different organs, including roots, stolons, petioles, flowers and tubers, with varying levels of expression. The expression of different StGRF members exhibited tissue-specific characteristics. StGRF4 exhibited high expression in stolons and young tubers, while lower expression was observed in stems and stamens (Figure 5). StGRF12 was predominantly expressed in roots, stolons, tuber sprouts and young tubers (Figure 5). StGRF9 exhibited high expression in flowers, stolons, and leaves, suggesting an essential role in flower growth and development (Figure 5). Notably, StGRF6 exhibited high expression in four tissues, yet displayed a lower transcript level in stamens, leaves and stems (Figure 5). StGRF3 was highly specifically expressed in stolons, tuber piths and mature tubers, whereas StGRF7 was highly expressed in stolons and tuber sprouts (Figure 5). StGRF5 showed high expression in stolons, tuber sprouts and young tubers, with very weak expression in other tissues. StGRF2 and StGRF10 were highly expressed in young tubers, tuber sprouts, and stolons, with minimal expression in other tissues. StGRF1, StGRF8, and StGRF11 showed weak expression in the majority of the examined tissues, with the exception of stolons (Figure 5). Collectively, all the 12 StGRFs exhibited distinct expression patterns according to the SpudDB database, indicating that they may have diverse biological functions in various tissues.
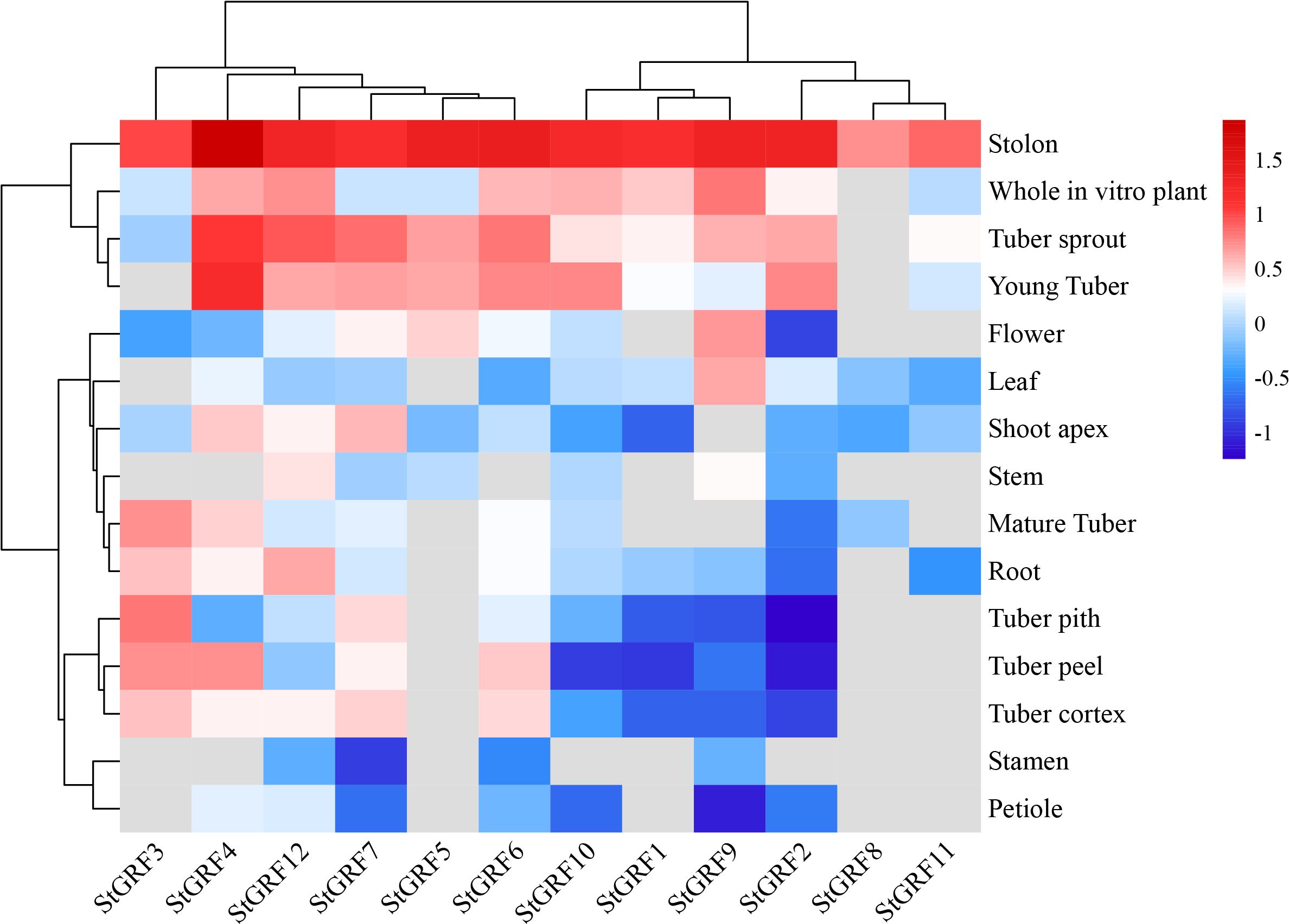
Figure 5 Expression profile analysis of StGRF genes in different potato tissues and organs. The color spectrum, ranging from red to blue, represents the expression levels of the genes, with red indicating high expression and blue indicating low expression. Grey indicates the value of zero in the original RNA-Seq data, which the software automatically recognizes this part of the data as ‘missing’.
Expression analysis of StGRF genes under multiple external stimuli and hormones
The expression patterns of the StGRF genes in response to abiotic stresses (salt, heat, osmotic shock) were investigated. Salt stress induced up-regulation of StGRF1, StGRF3, StGRF4, StGRF5, StGRF6, StGRF7, StGRF8 and StGRF10, StGRF11 and StGRF12, and down-regulation of StGRF9, whereas StGRF2 showed negligible changes under the NaCl treatment (Figure 6A). A rapid increase in the transcript levels of StGRF1, StGRF3, StGRF4, StGRF5, StGRF6, StGRF7, StGRF9, StGRF10 and StGRF11 was observed during osmotic shock induced by the mannitol treatment (Figure 6A). Upon exposure to heat stress, the mRNA abundance of StGRF1, StGRF2, StGRF3, StGRF4, StGRF5, StGRF6, StGRF8, StGRF9, StGRF10 and StGRF12 was found to be down-regulated, whereas StGRF7 and StGRF11 were up-regulated (Figure 6B). To ascertain whether StGRFs are involved in the hormone response, the expression patterns of StGRF genes in potato upon treatment with ABA (abscisic acid), GA3 (gibberellic acid), IAA (indole-3-acetic acid) and BAP (6-benzylaminopurine) were investigated (Figure 6C). The expression levels of StGRF6, StGRF7, and StGRF9 were significantly elevated in response to ABA stimulation, whereas the expression levels of StGRF2, StGRF4, StGRF8, StGRF10, StGRF11 and StGRF12 were significantly reduced, and StGRF1 showed no significant changes (Figure 6C). Upon IAA treatment, the expression levels of StGRF1, StGRF2, StGRF3, StGRF4, StGRF5, StGRF6, StGRF8 and StGRF12 were down-regulated (Figure 6C). The application of GA3 led to the up-regulation of StGRF5 and StGRF9, while the expression levels of StGRF1, StGRF2, StGRF4, StGRF6, StGRF7, StGRF8, StGRF10, StGRF11 and StGRF12 were downregulated (Figure 6C). Treatment with BAP induced an increase in StGRF5, StGRF8 and StGRF9, while the steady-state mRNA levels of other StGRF members exhibited a decrease (Figure 6C). Notably, the assays also revealed that the up-regulation of StGRF1, StGRF3, StGRF4 and StGRF9 was observed in leaves infected with the biotic factor Phytophthora infestans. Furthermore, except for StGRF1, the down-regulation of StGRF2, StGRF4, StGRF6, StGRF7, StGRF9 was observed. The expression of StGRF10 and StGRF11 was observed in leaves treated with the chemical elicitor BABA (β-aminobutyric acid), whereas the expression of StGRF1, StGRF6, StGRF9 and StGRF12 was repressed in leaves treated with the chemical elicitor BTH (benzothiadiazole) (Figure 6D). This indicates that StGRF1 and StGRF9 play an essential role in potato defense against invading microbes. Taken together, the detailed expression analyses suggest that StGRF genes may be involved in the regulation of biotic and abiotic stresses in potato.
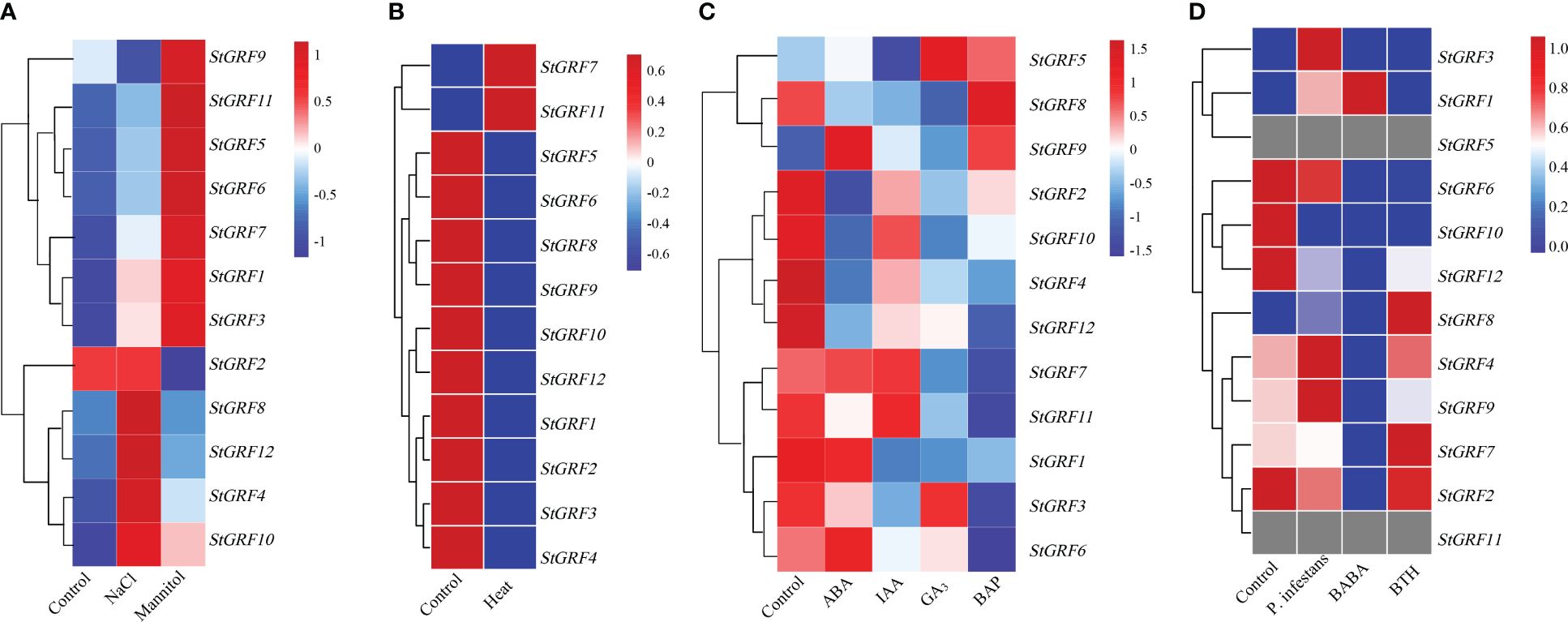
Figure 6 Heatmaps of the expression profiles of ten StGRF genes under ten different biotic or abiotic stresses. The transcripts were identified through the use of RNA-Seq technology. (A) Salt stresses include 150 mM NaCl for 24 h, and 260 μM mannitol for 24 h. (B) The heat stress treatment was 35°C for 24 h. (C) The phytohormone treatments included 50 μM ABA (abscisic acid) for 24 h, 10 μM IAA (indole-3-acetic acid) for 24 h, 50 μM GA3 (gibberellic acid) for 24 h and 10 μM BAP (6-benzylaminopurine) for 24 h. (D) Biotic stresses include Phytophthora infestans, the stress elicitors BTH (benzothiadiazole), and BABA (β-aminobutyric acid), which are applied to leaves for 48 hours each. The color patterns, ranging from red (up-regulated expression level) to green (down-regulated expression level), provide an indication of the expression levels of the detected genes under the given conditions. The grey color indicates the value of zero in the original RNA-Seq data, and the software automatically recognizes this part of the data as ‘missing’. The conditions (vertical) and genes (horizontal) with similar profiles were hierarchically clustered (Pearson correlation, average linkage).
Expression profiles of StGRFs during the process of potato tuber dormancy release
The expression of StGRF genes was evaluated based on TPM (transcripts per kilobase of exon model per million mapped reads) values during the process of tuber dormancy release in publicly available RNA-Seq datasets from potato cultivars Longshu3 and Russet Burbank (Spud DB). In the case of the potato cultivar Longshu3, the transcript levels of the StGRF1, StGRF2, StGRF5, StGRF6, StGRF7, StGRF10, StGRF11 and StGRF12 increased in the dormancy-release tuber and sprouting tuber compared to the dormant tuber. In contrast, the transcript levels of StGRF3 and StGRF4 showed a decrease after tuber dormancy release, while the transcription of StGRF8 was practically undetectable (Figure 7A). In the potato cultivar Russet Burbank, the expression profiles of StGRF2, StGRF5, StGRF7, StGRF8, StGRF10 and StGRF12 are up-regulated after tuber dormancy release or in nondormant tuber, whereas the expression profiles of StGRF3, StGRF4, StGRF6, StGRF9 and StGRF11 decrease after tuber dormancy release or in nondormant tuber (Figure 7B). A reduction in the expression of StGRF3 and StGRF4 was observed in potato cultivars Longshu3 and Russet Burbank following the release of tuber dormancy. This indicates that these genes may be involved in maintaining tuber dormancy. An up-regulation of StGRF2, StGRF5, StGRF7, StGRF10 and StGRF12 was observed in cultivars Longshu3 and Russet Burbank after tuber dormancy release, suggesting an association of these genes with the breaking of tuber dormancy and sprouting. To investigate whether StGRF genes were implicated in tuber dormancy, an RT-qPCR assay was conducted on all members of the StGRF family to ascertain their respective RNA accumulation profiles during the process of potato tuber dormancy release (Figure 8). The mRNA levels of StGRF5, StGRF6, StGRF8, and StGRF12 were observed to increase after the tuber dormancy release in sprouts, and remained high in apical parts lacking sprouts compared to dormant apical parts. The mRNA levels of StGRF1, StGRF7, and StGRF11 were observed to increase following the release of tuber dormancy in sprouts but decreased in apical parts without sprouts. This indicates that these StGRFs may be involved in promoting bud outgrowth. The expression of StGRF4 and StGRF9 decreased dramatically during tuber dormancy release, indicating that they play a role in of the regulation of tuber sprouting. No transcription was observed for StGRF3 in sprouts and apical sections. The relative abundance levels of these StGRFs as determined by qPCR were found to be generally consistent with those generated from the RNA-Seq datasets (Figures 7, 8), suggesting that StGRF1, StGRF2, StGRF5, StGRF7, StGRF10 and StGRF12 may be sprout-related genes, while StGRF4 and StGRF9 may be dormancy-related genes.
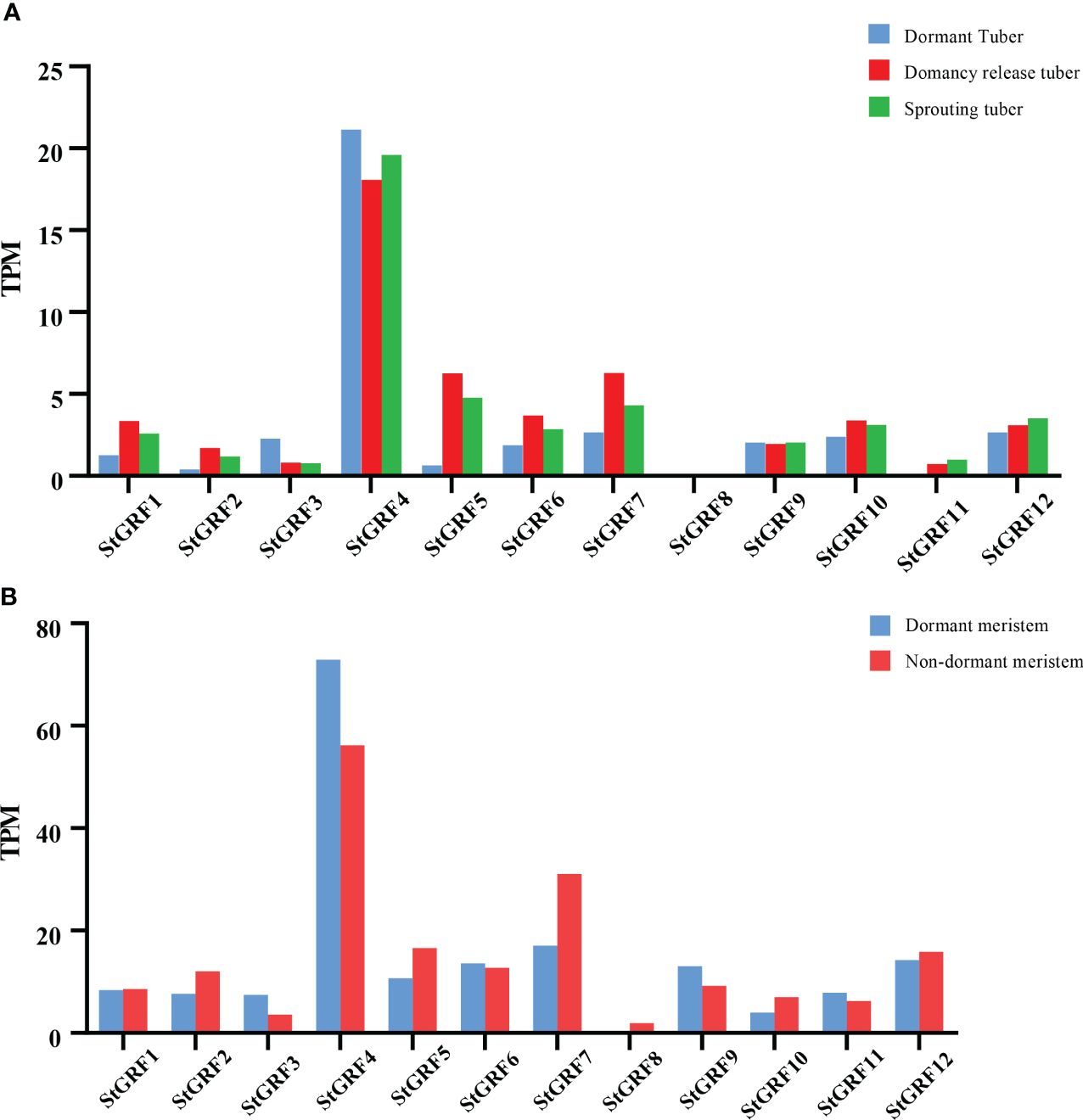
Figure 7 Expression profiles of StGRFs during dormancy release. Transcripts from the potato cultivars Longshu3 (A), and Russet Burbank (B) were detected using RNA-Seq technology, as described by Liu et al. (Liu et al., 2015) and Campbell et al. (Campbell et al., 2008).
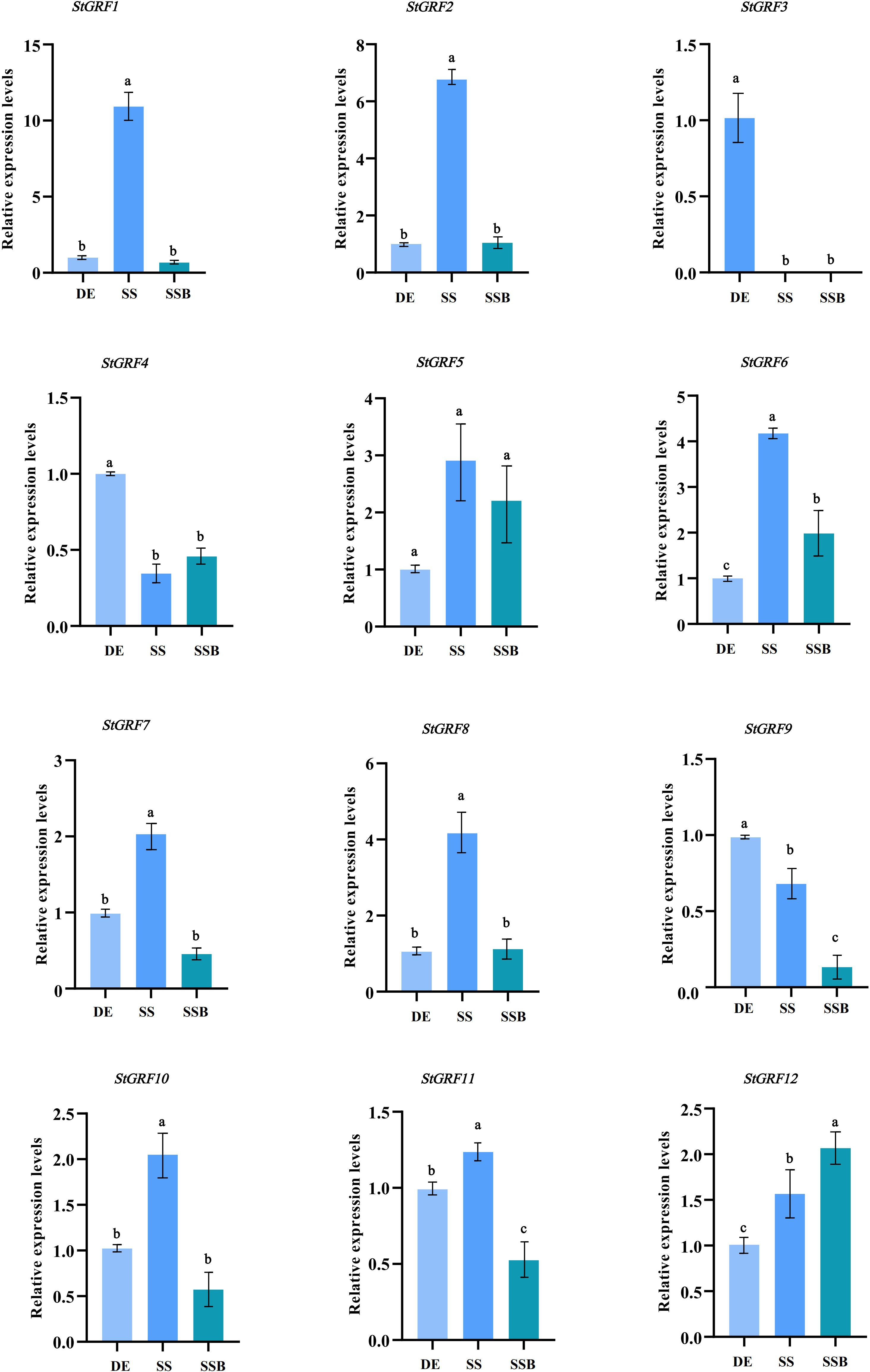
Figure 8 The expression levels of StGRF genes from the diploid potato line EB063 during dormancy release were determined by quantitative reverse transcription-polymerase chain reaction (RT-qPCR). Using the dormant eye as a control (DE), data represent the mean ± SEM (n=3). Analysis of variance and multiple comparisons (Tukey) were performed using the software GraphPad Prism9.0. The different letters indicate significant differences between samples (p < 0.05), and the same letter indicates that there is no significant difference between samples (p ≥ 0.05).
Discussion
In this study, 12 potato GRF genes (StGRFs) were identified through database analysis (Figure 1), followed by an in-depth investigation of gene structure, protein motifs, phylogenetic relationships, and gene expression patterns. StGRF proteins contain conserved WRC (Motif1) and QLQ (Motif2) domains (Figure 2; Supplementary Figure S1). The QLQ domain is responsible for protein-protein interactions, whereas the WRC domain is involved in DNA binding and nuclear targeting of the transcription factor (van der Knaap et al., 2000). Notably, StGRF1 contains a second WRC domain in its C-terminal region, similar to BrGRF12 from the Chinese cabbage Brassica rapa (Wang et al., 2014). In addition to the N-terminal conserved WRC and QLQ domains, the StGRF proteins carry additional C-terminal motifs, namely FFD, TQL, and GGPL, located in Motif 3, Motif 5, and Motif 8, respectively, in addition to the N-terminal conserved WRC and QLQ domains. The TQL motif, which has been described in AtGRF1 to AtGRF4, as well as in OsGRF1 to OsGRF5 (Omidbakhshfard et al., 2015), is present in StGRF2 to StGRF7, StGRF10 to StGRF12 (Figure 2). The FFD motif reported in the sweet orange (Citrus sinensis) GRF family members (Fu et al., 2024) is present in StGRF family members StGRF2, StGRF4, StGRF5, StGRF6, StGRF7, StGRF10, StGRF11 and StGRF12 (Figure 2). Similarly, the GGPL motif, which was reported in AtGRF1 to AtGRF4, AtGRF7, and AtGRF8 (van der Knaap et al., 2000; Kim et al., 2003; Choi et al., 2004), has been found to be located in StGRF2 to StGRF4, StGRF9 to StGRF11 (Figure 2). These TQL, FFD and GGPL motifs are also present in several GRFs from other plant species and are crucial for GRF function in plant tissues and organs (Omidbakhshfard et al., 2015). It is noteworthy that motifs 4, 6 and 7 are exclusive to the StGRF family members StGRF6, StGRF7 and StGRF12, which were categorized as subgroup I (Figures 1, 2). This suggests that these three motifs may be specific to subgroup I. The divergent C-terminal motifs of GRFs act as binding sites for transcriptional co-regulators and also have transcriptional transactivation activities (van der Knaap et al., 2000). Consequently, the conservation of both the N-terminal and C-terminal motifs is of significant importance for the evolutionary expansion and functional conservation of GRF family members in potato.
GRF genes are typically highly expressed in relatively active tissues such as germinating seeds and buds (Kim et al., 2003; Horiguchi et al., 2005; Kim et al., 2012; Zhang et al., 2018). RNA-Seq data show that StGRF genes are expressed at higher levels in the roots, shoot apices, flowers, and young tubers, where cell proliferation is strong in Arabidopsis (Kim et al., 2003; Rodriguez et al., 2010). The increased expression of StGRF9 in the leaf, flower and stolon, in comparison to the root, suggests a role for this gene in these organs, analogous to its presumed orthologue AtGRF8 (Kim et al., 2003). StGRF5 is expressed in the flower (Figure 5), in a manner analogous to its putative orthologue AtGRF5, which is predominantly expressed in the floral meristem (Pajoro et al., 2014). The overexpression of AtGRF5 in Arabidopsis results in an increase in cell proliferation in leaf primordia, leading to the formation of larger leaves than those observed in wild-type plants (Horiguchi et al., 2005). StGRF3 is highly expressed in the root and clusters with AtGRF3, AtGRF4 and AtGRF7 (Figures 1, 5), suggesting that it may perform similar functions in the root. AtGRF3 is expressed in the meristematic zone and elongation zone of primary and emerging lateral roots (Gorlova et al., 2014; Hepworth and Lenhard, 2014; Lee et al., 2018). AtGRF4 has been identified in carpels and roots, with the highest levels of expression observed in the root tip and differential zone (Bao et al., 2014). AtGRF4 is strongly expressed in the meristematic zone but weakly expressed in the elongation zone (Bao et al., 2014). AtGRF7 is mainly expressed in flowers and shoot tips, with minimal expression in roots (Kim et al., 2003). Furthermore, AtGRF7 is also expressed in developing tissues, including the vascular tissues, the inflorescence meristem, the bud pistil, the silique replum, the veins of leaf blades, and the petioles of true leaves (Kim et al., 2012). The same subfamily, StGRF1 and StGRF8, demonstrated weak expression in all tissues examined (Figures 1, 5). However, StGRF6, StGRF7 and StGRF12, which belong to the same subfamily as their homologs OsGRF3, OsGRF4, and OsGRF5, exhibited high expression in the young tubers, tuber sprouts and stolons (Figures 1, 5). This high expression may be involved in biological pathways that contribute to plant growth and development. The tissue-specific expression profiles of genes are typically associated with cis-regulatory elements (Saeed et al., 2006). It was observed that most of the StGRFs contain one or more than one cis-element related to plant growth and development in their promoter regions (Figure 3). The 12 StGRF genes containing these cis-elements may be involved in a variety of functions (Figure 3). The diversity of their functions and the distribution of cis-elements in the promoter regions of these genes suggest that StGRFs may differentially regulate the expression of genes involved in potato tuber development. Further functional characterization of the StGRF genes is essential to gain new insights into the molecular mechanism of tuber development in potato. Tuber initiation and development are of significant importance in potato production. The initiation and development of tubers are regulated by members of the FLOWERING LOCUS T (FT) clade and multiple environmental factors (Navarro et al., 2011). The high expression of the majority of StGRF genes in young tubers suggests their involvement in tuber formation and development (Figure 5), potentially through the association with other tuber-specific expression genes. For instance, StGRF4 expression was significantly higher in young tubers than in mature tubers (Figure 5), which is consistent with previous findings indicating that GRFs are involved in the initial stages of growth and development in various tissues.
A number of studies have investigated the impact of abiotic stresses, including heat, drought, and salinity, on potato yield, tuber quality, and market value (Liu et al., 2009; Liang et al., 2014; Dahal et al., 2019; Tiwari et al., 2022). Moreover, it has been demonstrated that GRFs may play a role in response to drought and salt stress (Kim et al., 2012). In comparison to the wild-type and AtGRF7-overexpressor lines, the atgrf7 mutant line exhibits increased tolerance to drought and salt stress. Conversely, AtGRF7 suppresses the expression of osmotic stress-responsive genes, including DREB2A, even in the absence of stress treatment (Liu et al., 1998; Kim et al., 2012). This indicates that abiotic stress causes the repression of AtGRF7 expression, which in turn induces the activation of osmotic stress-responsive genes. It was shown that salt stress resulted in the down-regulation of five GhGRF genes (GhGRF3, GhGRF4, GhGRF5, GhGRF7, and GhGRF16) in Gossypium hirsutum (Liu et al., 2022). However, the majority of StGRF genes were found to be up-regulated under NaCl stress, with the exception of StGRF2 and StGRF9 (Figure 6A). Furthermore, the expression levels of StGRF1, StGRF3, StGRF5, StGRF6, StGRF7, StGRF9 and StGRF11 genes showed a significant increase following mannitol treatment (Figure 6A). This indicates that osmotic stress is responsible for the activation of StGRF gene expression. It was demonstrated that poplar GRF15 was induced by heat stress. Compared to wild-type poplar plants, the transgenic lines overexpressing PtGRF15 and lacking the miR396a target sites exhibited enhanced photosynthetic efficiency and heat tolerance (Zhao et al., 2021). The expression of most StGRF genes was suppressed under heat stress, while StGRF7 and StGRF11 genes exhibited a slight induction (Figure 6B). Nevertheless, further research is required to elucidate the underlying mechanisms of these phenomena.
Phytohormones exert a profound influence on the growth, differentiation, and development of plants. Previous reports have shown that GA3 treatment increases the expression of several GRF genes in rice and cabbage (van der Knaap et al., 2000; Choi et al., 2004; Wang et al., 2014), whereas AtGRFs do not appear to be induced by GA3 (Kim et al., 2003). KNOTTED1-like homeobox (KNOX) transcription factors are known to restrict cell differentiation and are important regulators of meristem development. GRFs have been demonstrated to act as upstream repressors of KNOX genes, which inhibit GA biosynthesis (Kuijt et al., 2014). The application of GA3 induces the expression of KNOX, which subsequently leads to the up-regulation of GRFs at low levels of KNOX expression (Kuijt et al., 2014). The expression of several NtabGRF genes in Nicotiana tabacum is triggered by a number of different treatments, including GA3, IAA, BR, ABA, and BAP (Zhang et al., 2018). Further analysis revealed that the NtabGRF promoters contain several hormone-related cis-elements, including the GARE-motif, TATC-box, and P-box (gibberellin responsiveness) elements in GA3-inducible NtabGRF genes and ABRE (abscisic acid responsiveness) elements in ABA-inducible NtabGRF genes (Zhang et al., 2018). In Camellia sinensis, only one GRF gene was induced by GA3 treatment, whereas most of GRFs were induced by SA or IAA (Wu et al., 2017). This study revealed that the expression levels of StGRF2, StGRF4, StGRF8 and StGRF12 were simultaneously downregulated, while StGRF9 was induced by GA3, IAA, and ABA (Figure 6C). The StGRF12 gene was also differentially downregulated by BAP treatment (Figure 6C), suggesting that StGRF12 may be a key negative regulator of potato in response to GA3, IAA, ABA, and BAP treatments. It can be concluded that these StGRFs play a crucial role in regulating hormone feedback mechanisms in potato. A promoter analysis revealed that the potential functions of StGRFs are induced by different hormones (Figure 3). The promoter regions of StGRF2, StGRF3, StGRF8 and StGRF10 contain GA-responsive elements, specifically P-box motifs (Figure 3). Similarly, StGRF12 contains auxin-responsive TGA-box motifs (Figure 3). Furthermore, the ABA-responsive ABRE motifs were identified in StGRF1, StGRF2, StGRF4, StGRF6, StGRF9, StGRF11, and StGRF12 (Figure 3). It is therefore postulated that StGRFs may have a role in regulating physiological processes related to hormonal feedback mechanisms in potato.
Potato tubers are formed from shortened internodes, which subsequently undergo swelling and the formation of tuber eyes. Meristematic activity in the tuber eyes is completely ceased, resulting in the tuber entering a period of dormancy. Consequently, potato tuber dormancy is confined to the tuber eyes, where the meristem is located, while the remainder of the tuber continues to undergo physiological metabolic activity (Aksenova et al., 2013). It is postulated that changes in meristematic activity represent a pivotal factor in the process of dormancy release. It is hypothesized that the reactivation of meristematic function coincides with the breaking of dormancy. Meristematic activity typically resumes prior to tuber bud emergence, which is referred to as the breaking of dormancy or tuber dormancy release. Subsequently, an increase in cell division results in the visible growth of the bud, which is known as tuber sprouting. This often occurs with buds that are larger than 2 mm. GRFs are frequently highly expressed in actively growing tissues. The data indicate that AtGRFs are highly expressed in developing and growing tissues where cell proliferation occurs, in different parts of Arabidopsis roots and shoots (Kim et al., 2003). Gene expression profiles provide valuable insights into the potential functions of genes. In the present study, publicly available RNA-Seq data and RT-qPCR were used to elucidate the expression of StGRFs during potato tuber dormancy and sprouting (Figures 7, 8). The gene expression patterns of StGRFs in dormant and sprouting tubers were analyzed in order to identify the complex expression patterns exhibited by these genes (Figure 8). The StGRF genes displayed spatiotemporally specific patterns (Figures 7, 8). StGRF2, StGRF5, StGRF7, StGRF10, and StGRF12 showed high expression levels following dormancy release, indicating their expression in growing tissues (Figures 7, 8). AtGRF1, AtGRF2, and AtGRF3 have been shown to regulate cell proliferation, leaf, and cotyledon development (Kim et al., 2003). AtGRF5 overexpression results in larger leaf areas due to increased cell number (Horiguchi et al., 2005). Conversely, ZmGRF10 overexpression in maize leads to a decrease in plant height and leaf size, particularly leaf length, accompanied by impaired cell proliferation (Wu et al., 2014). This indicates that the reduction in leaf size observed in transgenic maize is likely due to a limitation in cell proliferation. Transcriptome data indicated a reduction in the expression level of the StGRF3 gene following germination, while RT-qPCR results demonstrated that the expression level of the StGRF3 gene was reduced to zero after germination (Figures 7, 8). This suggests that the StGRF3 gene may act as a negative regulator in the process of tuber dormancy. The expression of the StGRF8 gene was not detected during the germination of the potato cultivar Longshu 3. In contrast, in the diploid potato line EB063, StGRF8 gene expression increased with the release of dormancy, which was consistent with the expression observed in the potato cultivar Russert Burbank (Figures 7, 8). These findings indicate that StGRF8 exhibits distinct expression patterns during the germination process in different potato genotypes. Therefore, it is therefore necessary to further verify its specific role. Furthermore, StGRF4 and StGRF9 exhibited comparable reduced expression levels after the release of dormancy (Figure 8), indicating that these two StGRF genes may be involved in bud dormancy.
Conclusions
In this study, 12 members of the potato GRF family were identified and their conserved N-terminal WRC and QLQ domains, as well as C-terminal FFD, TQL, and GGPL motifs were characterized. The intron–exon organization was analyzed, and the evolutionary relationships between the StGRFs and their homologs from two representative plant species were investigated. A total of 30 cis-acting elements related to plant growth and development, 30 cis-acting elements related to abiotic stress, and 40 cis-acting elements related to hormone response were identified in the promoter regions of StGRFs. A StGRFs-mediated regulatory network was constructed, comprising 52 transcription factors belonging to 15 different TF families, which were identified as the potential regulators of StGRFs. Furthermore, we examined tissue-specific gene expression patterns, as well as gene expression patterns induced by the heat, salt, drought stress, several phytohormones, Phytophthora infestans, and chemical elicitors. The involvement of seven StGRF genes, StGRF1, StGRF2, StGRF5, StGRF6, StGRF7, StGRF10 and StGRF12, in tuber sprouting was confirmed. Furthermore, it was demonstrated that two StGRF genes, StGRF4 and StGRF9, may be associated with tuber dormancy. The results of our analysis of gene structure, phylogenetic relationships, and transcript expression profiles suggest that StGRF genes may have specific roles in potato developmental processes and environmental stresses, particularly during potato tuber dormancy and sprouting.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
DC: Writing – original draft. YS: Writing – review & editing. WJ: Writing – original draft. HY: Writing – original draft, Writing – review & editing. SW: Writing – original draft. LY: Supervision, Writing – review & editing. BL: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Key-Area R&D Program of Guangdong Province in China (grant number 2022B0202060001), Key R&D Program of Shaanxi Province in China (grant number 2018ZDCXL-NY-03-03, 2022NY-174).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1417204/full#supplementary-material
Supplementary Figure 1 | Multiple sequence alignment (A) and composition (B-C) of conserved domains (QLQ and WRC domains) in StGRF proteins.
References
Aksenova, N. P., Sergeeva, L. I., Konstantinova, T. N., Golyanovskaya, S. A., Kolachevskaya, O. O., Romanov, G. A. (2013). Regulation of potato tuber dormancy and sprouting. Russ. J. Plant Physiol. 60, 301–312. doi: 10.1134/s1021443713030023
Bao, M. L., Bian, H. W., Zha, Y. L., Li, F. Y., Sun, Y. Z., Bai, B., et al. (2014). miR396a-mediated basic helix-loop-helix transcription factor bHLH74 repression acts as a regulator for root growth in Arabidopsis seedlings. Plant Cell Physiol. 55, 1343–1353. doi: 10.1093/pcp/pcu058
Bazin, J., Khan, G. A., Combier, J. P., Bustos-Sanmamed, P., Debernardi, J. M., Rodriguez, R., et al. (2013). miR396 affects mycorrhization and root meristem activity in the legume Medicago truncatula. Plant J. 74, 920–934. doi: 10.1111/tpj.12178
Campbell, M., Segear, E., Beers, L., Knauber, D., Suttle, J. (2008). Dormancy in potato tuber meristems: chemically induced cessation in dormancy matches the natural process based on transcript profiles. Funct. Integr. Genomics 8 (4), 317–328. doi: 10.1007/s10142-008-0079-6
Casadevall, R., Rodriguez, R. E., Debernardi, J. M., Palatnik, J. F., Casati, P. (2013). Repression of growth regulating factors by the MicroRNA396 inhibits cell proliferation by uv-b radiation in Arabidopsis leaves. Plant Cell 25, 3570–3583. doi: 10.1105/tpc.113.117473
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, F., Yang, Y. Z., Luo, X. F., Zhou, W. G., Dai, Y. J., Zheng, C., et al. (2019). Genome-wide identification of GRF transcription factors in soybean and expression analysis of GmGRF family under shade stress. BMC Plant Biol. 19 (1), 269. doi: 10.1186/s12870–019-1861–4
Choi, D., Kim, J. H., Kende, H. (2004). Whole genome analysis of the OsGRF gene family encoding plant-specific putative transcription activators in rice (Oryza sativa L.). Plant Cell Physiol. 45, 897–904. doi: 10.1093/pcp/pch098
Dahal, K., Li, X. Q., Tai, H., Creelman, A., Bizimungu, B. (2019). Improving potato stress tolerance and tuber yield under a climate change scenario - A current overview. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00563
Debernardi, J. M., Mecchia, M. A., Vercruyssen, L., Smaczniak, C., Kaufmann, K., Inze, D., et al. (2014). Post-transcriptional control of GRF transcription factors by microRNA miR396 and GIF co-activator affects leaf size and longevity. Plant J. 79, 413–426. doi: 10.1111/tpj.12567
Debernardi, J. M., Rodriguez, R. E., Mecchia, M. A., Palatnik, J. F. (2012). Functional specialization of the plant mir396 regulatory network through distinct microrna-target interactions. PloS Genet. 8 (1), e1002419. doi: 10.1371/journal.pgen.1002419
Fu, M. K., He, Y. N., Yang, X. Y., Tang, X., Wang, M., Dai, W. S. (2024). Genome-wide identification of the GRF family in sweet orange (Citrus sinensis) and functional analysis of the CsGRF04 in response to multiple abiotic stresses. BMC Genom. 25 (1), 37. doi: 10.1186/s12864–023-09952–8
Gorlova, O., Fedorov, A., Logothetis, C., Amos, C., Gorlov, I. (2014). Genes with a large intronic burden show greater evolutionary conservation on the protein level. BMC Evol. Biol. 14 (1), 50. doi: 10.1186/1471–2148-14–50
Hepworth, J., Lenhard, M. (2014). Regulation of plant lateral-organ growth by modulating cell number and size. Curr. Opin. Plant Biol. 17, 36–42. doi: 10.1016/j.pbi.2013.11.005
Hewezi, T., Maier, T. R., Nettleton, D., Baum, T. J. (2012). The Arabidopsis MicroRNA396-GRF1/GRF3 regulatory module acts as a developmental regulator in the reprogramming of root cells during cyst nematode infection. Plant Physiol. 159, 321–335. doi: 10.1104/pp.112.193649
Horiguchi, G., Kim, G. T., Tsukaya, H. (2005). The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 43, 68–78. doi: 10.1111/j.1365–313X.2005.02429.x
Jin, J. P., Tian, F., Yang, D. C., Meng, Y. Q., Kong, L., Luo, J. C., et al. (2017). PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 45, D1040–D1045. doi: 10.1093/nar/gkw982
Khatun, K., Robin, A. H. K., Park, J. I., Nath, U. K., Kim, C. K., Lim, K. B., et al. (2017). Molecular characterization and expression profiling of tomato GRF transcription factor family genes in response to abiotic stresses and phytohormones. Int. J. Mol. Sci. 18 (5), 1056. doi: 10.3390/ijms18051056
Kim, J. H., Choi, D. S., Kende, H. (2003). The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 36, 94–104. doi: 10.1046/j.1365–313X.2003.01862.x
Kim, J. H., Kende, H. (2004). A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc.Natl. Acad. Sci. U.S.A. 101, 13374–13379. doi: 10.1073/pnas.0405450101
Kim, J. S., Mizoi, J., Kidokoro, S., Maruyama, K., Nakajima, J., Nakashima, K., et al. (2012). Arabidopsis GROWTH-REGULATING FACTOR7 functions as a transcriptional repressor of abscisic acid- and osmotic stress-responsive genes, including DREB2A. Plant Cell 24, 3393–3405. doi: 10.1105/tpc.112.100933
Kuijt, S. J. H., Greco, R., Agalou, A., Shao, J. X., t Hoen, C. C. J., Övernäs, E., et al. (2014). Interaction between the GROWTH-REGULATING FACTOR and KNOTTED1-LIKE HOMEOBOX families of transcription factors. Plant Physiol. 164, 1952–1966. doi: 10.1104/pp.113.222836
Lee, S. J., Lee, B. H., Jung, J. H., Park, S. K., Song, J. T., Kim, J. H. (2018). GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR specify meristematic cells of gynoecia and anthers. Plant Physiol. 176, 717–729. doi: 10.1104/pp.17.00960
Liang, G., He, H., Li, Y., Wang, F., Yu, D. Q. (2014). Molecular mechanism of microRNA396 mediating pistil development in Arabidopsis. Plant Physiol. 164, 249–258. doi: 10.1104/pp.113.225144
Liu, B., Zhang, N., Wen, Y., Jin, X., Yang, J., Si, H., et al. (2015). Transcriptomic changes during tuber dormancy release process revealed by RNA sequencing in potato. J. Biotechnol. 198, 17–30. doi: 10.1016/j.jbiotec.2015.01.019
Liu, D. M., Song, Y., Chen, Z. X., Yu, D. Q. (2009). Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiol. Plantarum 136, 223–236. doi: 10.1111/j.1399–3054.2009.01229.x
Liu, H. H., Guo, S. Y., Xu, Y. Y., Li, C. H., Zhang, Z. Y., Zhang, D. J., et al. (2014). OsmiR396d-regulated OsGRFs function in floral organogenesis in rice through binding to their targets OsJMJ706 and OsCR4. Plant Physiol. 165, 160–174. doi: 10.1104/pp.114.235564
Liu, J., Hua, W., Yang, H. L., Zhan, G. M., Li, R. J., Deng, L. B., et al. (2012). The BnGRF2 gene (GRF2-like gene from Brassica napus) enhances seed oil production through regulating cell number and plant photosynthesis. J. Exp. Bot. 63, 3727–3740. doi: 10.1093/jxb/ers066
Liu, K., Kabir, N., Wei, Z. Z., Sun, Z. J., Wang, J., Qi, J., et al. (2022). Genome-wide identification and expression profile of GhGRF gene family in Gossypium hirsutum L. Peerj 10, e13372. doi: 10.7717/peerj.13372
Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., et al. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10, 1391–1406. doi: 10.2307/3870648
Ma, J. Q., Jian, H. J., Yang, B., Lu, K., Zhang, A. X., Liu, P., et al. (2017). Genome-wide analysis and expression profiling of the GRF gene family in oilseed rape (Brassica napus L.). Gene 620, 36–45. doi: 10.1016/j.gene.2017.03.030
Navarro, C., Abelenda, J. A., Cruz-Oró, E., Cuéllar, C. A., Tamaki, S., Silva, J., et al. (2011). Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature 478, 119–U132. doi: 10.1038/nature10431
Nelissen, H., Eeckhout, D., Demuynck, K., Persiau, G., Walton, A., van Bel, M., et al. (2015). Dynamic changes in ANGUSTIFOLIA3 complex composition reveal a growth regulatory mechanism in the maize leaf. Plant Cell 27, 1605–1619. doi: 10.1105/tpc.15.00269
Omidbakhshfard, M. A., Proost, S., Fujikura, U., Mueller-Roeber, B. (2015). Growth-regulating factors (GRFs): A small transcription factor family with important functions in plant biology. Mol. Plant 8 (7), 998–1010. doi: 10.1016/j.molp.2015.01.013
Pajoro, A., Madrigal, P., Muiño, J. M., Matus, J. T., Jin, J., Mecchia, M. A., et al. (2014). Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biol. 15 (3), R41. doi: 10.1186/gb-2014–15-3-r41
Rodriguez, R. E., Mecchia, M. A., Debernardi, J. M., Schommer, C., Weigel, D., Palatnik, J. F. (2010). Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137, 103–112. doi: 10.1242/dev.043067
Saeed, A. I., Hagabati, N. K., Braisted, J. C., Liang, W., Sharov, V., Howe, E. A., et al. (2006). TM4 microarray software suite. Methods Enzymol. 411, 134–193. doi: 10.1016/S0076-6879(06)11009-5
Tiwari, J. K., Buckseth, T., Zinta, R., Bhatia, N., Dalamu, D., Naik, S., et al. (2022). Germplasm, breeding, and genomics in potato improvement of biotic and abiotic stresses tolerance. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.805671
van der Knaap, E., Kim, J. H., Kende, H. (2000). A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiol. 122, 695–704. doi: 10.1104/pp.122.3.695
Vercruyssen, L., Tognetti, V. B., Gonzalez, N., Van Dingenen, J., De Milde, L., Bielach, A., et al. (2015). GROWTH REGULATING FACTOR5 stimulates Arabidopsis chloroplast division, photosynthesis, and leaf longevity. Plant Physiol. 167, 817. doi: 10.1104/pp.114.256180
Wang, L., Gu, X. L., Xu, D. Y., Wang, W., Wang, H., Zeng, M. H., et al. (2011). miR396-targeted AtGRF transcription factors are required for coordination of cell division and differentiation during leaf development in Arabidopsis. J. Exp.Bot. 62, 761–773. doi: 10.1093/jxb/erq307
Wang, F. D., Qiu, N. W., Ding, Q., Li, J. J., Zhang, Y. H., Li, H. Y., et al. (2014). Genome-wide identification and analysis of the growth-regulating factor family in Chinese cabbage (Brassica rapa L. ssp. pekinensis). BMC Genom. 15 (1), 807. doi: 10.1186/1471–2164-15–807
Wang, J., Zhou, H., Zhao, Y., Sun, P., Tang, F., Song, X., et al. (2020). Characterization of poplar growth-regulating factors and analysis of their function in leaf size control. BMC Plant Biol. 20 (1), 509. doi: 10.1186/s12870–020-02699–4
Wu, Z. J., Wang, W. L., Zhuang, J. (2017). Developmental processes and responses to hormonal stimuli in tea plant (Camellia sinensis) leaves are controlled by GRF and GIF gene families. Funct. Integr. Genomic 17, 503–512. doi: 10.1007/s10142–017-0553–0
Wu, L., Zhang, D. F., Xue, M., Qian, J. J., He, Y., Wang, S. C. (2014). Overexpression of the maize GRF10, an endogenous truncated growth-regulating factor protein, leads to reduction in leaf size and plant height. J. Integr. Plant Biol. 56, 1053–1063. doi: 10.1111/jipb.12220
Xiao, G., Huang, W., Cao, H., Tu, W., Wang, H., Zheng, X., et al. (2018). Genetic loci conferring reducing sugar accumulation and conversion of cold-stored potato tubers revealed by QTL analysis in a diploid population. Front. Plant Sci. 9, 315. doi: 10.3389/fpls.2018.00315
Xu, X., Pan, S. K., Cheng, S. F., Zhang, B., Mu, D. S., Ni, P. X., et al. (2011). Genome sequence and analysis of the tuber crop potato. Nature 475, 189–U194. doi: 10.1038/nature10158
Zhang, D. F., Li, B., Jia, G. Q., Zhang, T. F., Dai, J. R., Li, J. S., et al. (2008). Isolation and characterization of genes encoding GRF transcription factors and GIF transcriptional coactivators in maize (Zea mays L.). Plant Sci. 175, 809–817. doi: 10.1016/j.plantsci.2008.08.002
Zhang, J. F., Li, Z. F., Jin, J. J., Xie, X. D., Zhang, H., Chen, Q. S., et al. (2018). Genome-wide identification and analysis of the growth-regulating factor family in tobacco (Nicotiana tabacum). Gene 639, 117–127. doi: 10.1016/j.gene.2017.09.070
Keywords: GRF transcription factors, gene expression, regulatory network, tuber dormancy, sprouting, potato
Citation: Cui D, Song Y, Jiang W, Ye H, Wang S, Yuan L and Liu B (2024) Genome-wide characterization of the GRF transcription factors in potato (Solanum tuberosum L.) and expression analysis of StGRF genes during potato tuber dormancy and sprouting. Front. Plant Sci. 15:1417204. doi: 10.3389/fpls.2024.1417204
Received: 14 April 2024; Accepted: 03 June 2024;
Published: 24 June 2024.
Edited by:
Yi Wang, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Tengfei Liu, Huazhong Agricultural University, ChinaWen Huang, The Sainsbury Laboratory, United Kingdom
Copyright © 2024 Cui, Song, Jiang, Ye, Wang, Yuan and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bailin Liu, bGl1YmxAbndhZnUuZWR1LmNu
†These authors have contributed equally to this work
 Danni Cui1,2†
Danni Cui1,2† Yin Song
Yin Song Bailin Liu
Bailin Liu