- 1College of Agricultural Science and Engineering, Liaocheng University, Liaocheng, China
- 2National Nanfan Research Institute, Chinese Academy of Agricultural Sciences, Sanya, China
- 3State Key Laboratory of Rice Biology and Breeding, China National Rice Research Institute, Hangzhou, China
Pentatricopeptide repeat (PPR) proteins constitute one of the largest protein families in land plants, with over 300 members in various species. Nearly all PPR proteins are nuclear-encoded and targeted to the chloroplast and mitochondria, modulating organellar gene expression by participating in RNA metabolism, including mRNA stability, RNA editing, RNA splicing, and translation initiation. Organelle RNA metabolism significantly influences chloroplast and mitochondria functions, impacting plant photosynthesis, respiration, and environmental responses. Over the past decades, PPR proteins have emerged as a research focus in molecular biology due to their diverse roles throughout plant life. This review summarizes recent progress in understanding the roles and molecular mechanisms of PPR proteins, emphasizing their functions in fertility, abiotic and biotic stress, grain quality, and chloroplast development in rice. Furthermore, we discuss prospects for PPR family research in rice, aiming to provide a theoretical foundation for future investigations and applications.
1 Introduction
PPR proteins is one of the largest protein families in higher plants, which are characterized by 2–30 tandemly arranged repeats. Each repeat consists of degenerated 30–40 amino acid PPR motifs (Qin et al., 2021). Each PPR motif forms a hairpin structure with a pair of anti-paralleled α-helixes, recognizing nucleotides in RNA through interactions with residues at position 6′ and 1′ of the motif (Barkan and Small, 2014). These proteins have been identified in various plant species, such as Arabidopsis (Lurin et al., 2004), foxtail millet (Xing et al., 2018), poplar (Liu et al., 2016), and maize (Chen et al., 2018a, b), with 441, 486, 626, and 491 members, respectively. PPR proteins are classified into two main subfamilies, P and PPR-like (PLS), based on PPR motif architectures (Cheng et al., 2016). The ancestral P subfamily proteins contain canonical 35 amino acid P (PPR) motifs. PLS subfamily proteins have P-, L- (long PPR, 35 or 36 amino acids), and S- (short PPR, 31 or 32 amino acids) tandem repeats. The PLS subfamily is further divided into PLS, E+, E, and DYW subclasses according to the characteristic C-terminal domain (Lurin et al., 2004; Li et al., 2021).
Eukaryotic nuclear genome encodes PPR proteins, which are primarily post-translationally transported to mitochondria or chloroplast (Barkan et al., 2012; Cheng et al., 2016). PPR proteins play critical roles in the plant organelle RNA processes. PPRs could not only act as site recognition factors but also bind to cis-elements specifically. The resultant PPR-RNA complex participates in RNA editing, RNA splicing, RNA stability, RNA cleavage, and RNA translation (Figure 1). P-type PPR proteins predominantly participate in organelle RNA transcript splicing, stability, and translation. PLS-type PPR proteins mainly function in mitochondrial and chloroplastic RNA editing (Gruttner et al., 2022). These two PPR protein types may exhibit partially overlapping functions in regulating RNA splicing and editing.
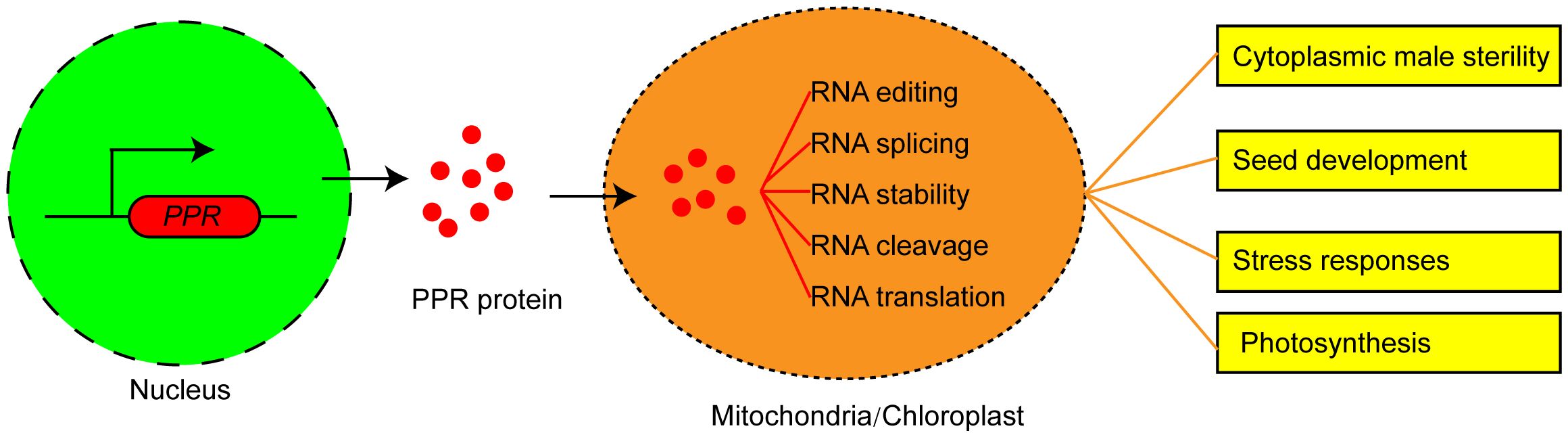
Figure 1 The functions and molecular mechanism of PPR proteins in plants. PPR proteins are encoded by nuclear genes, translated in the cytoplasm, and then imported into mitochondrion or chloroplast to mediate multiple steps of RNA processing and determine the plant phenotype.
Extensive research has demonstrated the diverse functions of PPR proteins in plant growth and development, including cytoplasmic male sterility (CMS), seed development, photosynthesis, and responses to biotic and abiotic stresses (Li et al., 2021). Most chloroplast-localized PPRs are associated with photosynthesis, mutations of which result in photosynthetic defects, aberrant leaf development and decreased leaf pigmentation (Hao et al., 2019). Some mitochondrial-localized PPRs are involved in seed development. These PPRs mutation generally causes floury/defective endosperm and retarded growth (Yang et al., 2017; Wang et al., 2019). CMS is considered to be jointly regulated by mitochondrial genes and their corresponding restorer of fertility (Rf) genes. Most Rf genes belong to the PPR genes family, which can restore fertility by suppressing the production of mitochondrial CMS associated proteins (Li et al., 2021). RNA editing contributes to the adaptation of land plants to extreme temperature, UV, and oxidative stress. Increasing evidences have showed that biotic and abiotic stresses change the expression patterns of PPR genes. To date, many PPR genes have been reported to be associated with salt stress, drought stress, cold stress, and defense response (Jiang et al., 2015; Qiu et al., 2021).
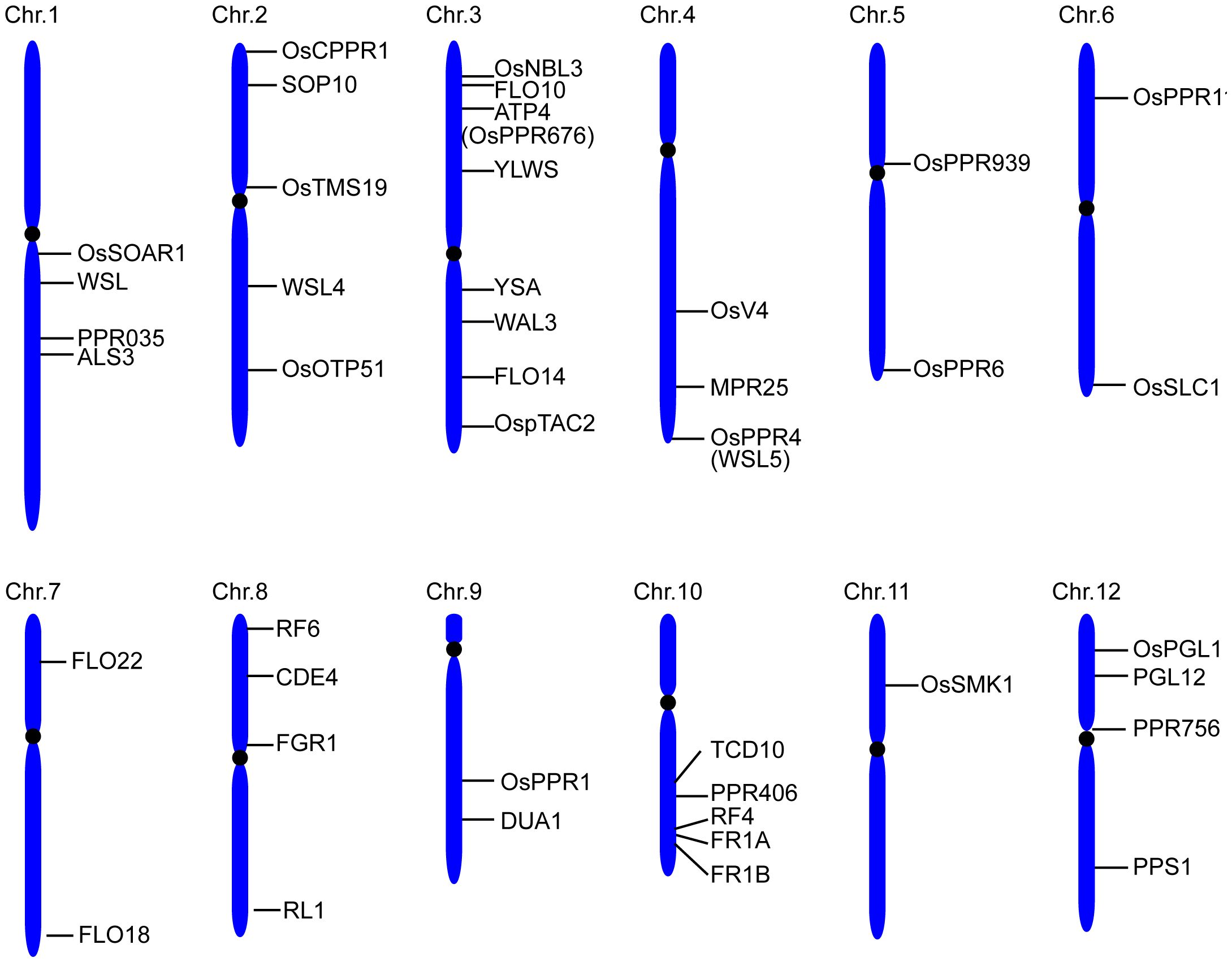
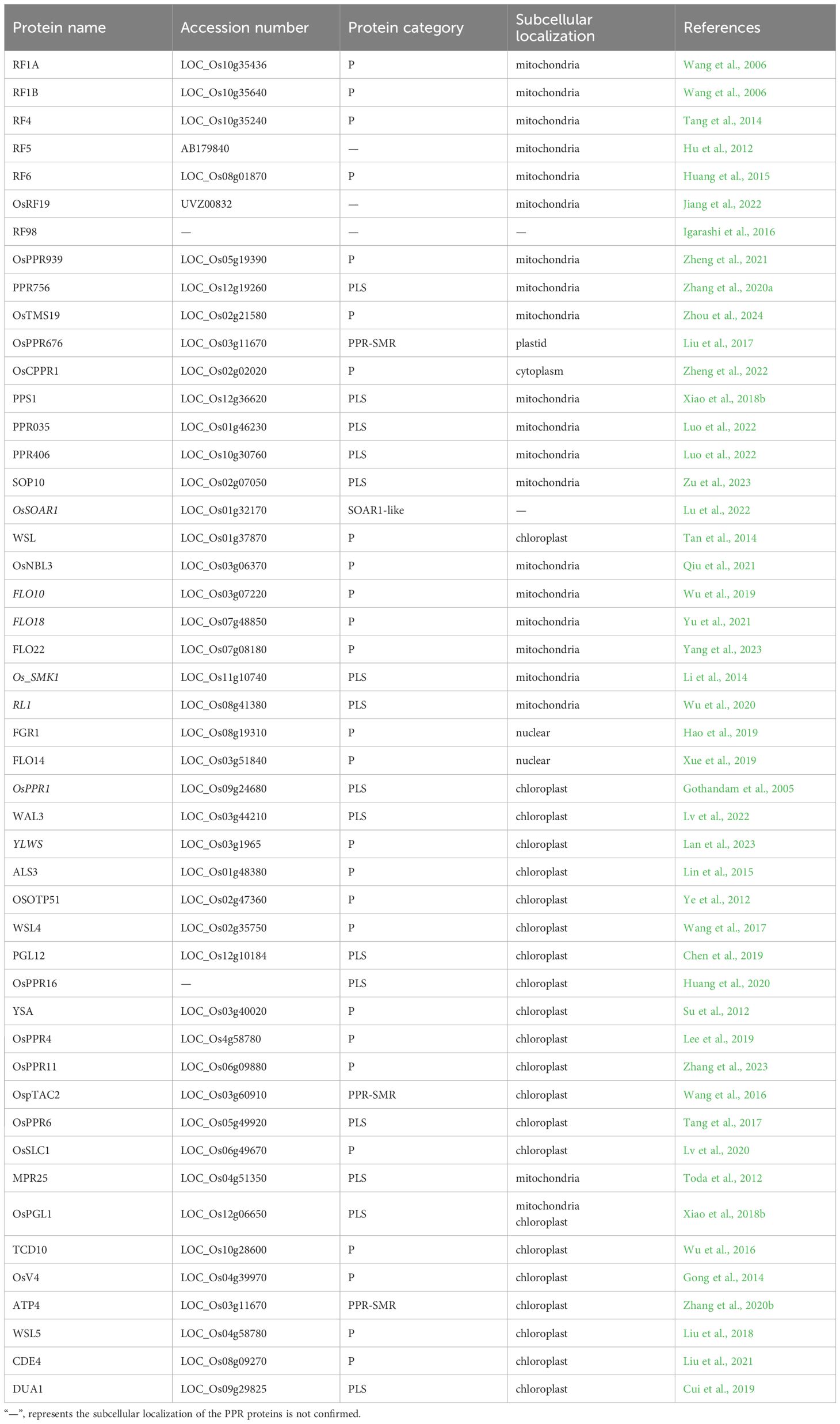
A total of 491 PPR genes were identified in the rice genome, distributed across all 12 rice chromosomes. Predictions revealed that most PPR proteins were targeted to either mitochondria (54%) or plastid (28%). Among them, 245 and 246 PPR proteins belong to the PLS and P subfamilies, respectively. Structural analysis indicated that in rice, 319 PPR genes lack introns, 79 genes contain one intron, and 93 genes harbor more than one intron (Chen et al., 2018a). To date, at least 48 PPR genes distributed in all 12 rice chromosomes, have been cloned and identified in rice (Figure 2, Table 1). Among the 48 PPR genes, most of them are confirmed to regulate rice development by directly or indirectly influencing the RNA splicing, RNA editing, RNA cleavage, RNA degradation, and RNA translation. Most cloned PPR proteins are targeted to mitochondria and chloroplast, with only a few being targeted to the nucleus and cytoplasm (Figure 3). This review summarizes the recent progress on PPR proteins in rice and discusses their roles and molecular mechanisms in fertility, stress response, grain quality, and chloroplast development.
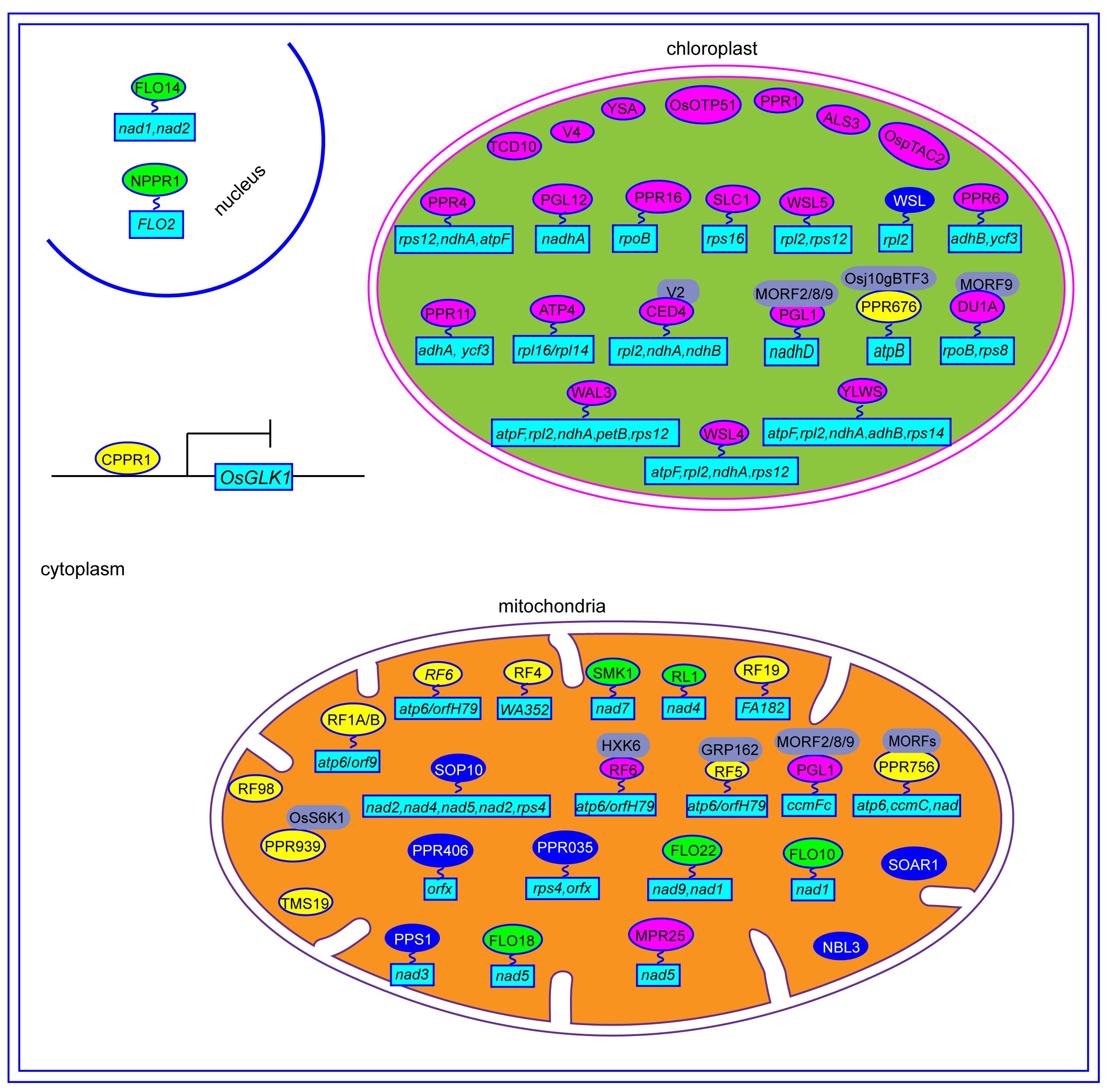
Figure 3 The regulation mechanisms of PPR proteins in rice. The yellow ovals represent PPR protein involved in CMS. The green ovals represent PPR protein involved in grain quality. The blue ovals represent PPR protein involved in stress responses. The red ovals represent PPR protein involved in chloroplast development. The gray ovals represent the interaction protein of PPR. The light blue rectangles represent the genes that are regulated by PPR proteins.
2 Functions of PPR proteins in rice fertility
CMS is a prevalent maternally inherited trait in flowering plants, characterized by female fertility and abortive pollen (Liu et al., 2017). Mitochondrial genes and their corresponding nuclear-encoded restorer of fertility (Rf) genes reportedly control CMS by regulating the expression of CMS-related genes in mitochondria (Wang et al., 2006). The CMS/Rf system is not only essential for hybrid seed production but also serves as an ideal genetic tool to investigate heterosis and mitochondrial-nuclear genetic interactions (Chen and Liu, 2014). To date, numerous rice Rf genes have been cloned and characterized, mostly belonging to PPR proteins (Figure 4). Rf1A and Rf1B, containing 19 and 11 PPR repeats, respectively, are mitochondrial and restore male sterility by inhibiting the cytotoxic ORF79 peptide production, which causes gametophytic male sterility. Rf1a and Rf1b restore male fertility via the cleavage and degradation of mitochondrial chimeric gene B-atp6/orf79 mRNA, respectively (Wang et al., 2006). Rf4, encodes a mitochondrial P-subfamily protein with 19 PPR repeats, rescuing Wild-Abortive CMS (WA-CMS) by suppressing CMS-associated gene WA352 transcription (Tang et al., 2014). Rf5, the rice fertility restoration gene of Hongliian CMS (HL-CMS), physically interacts with GRP162, which binds atp6-orfH79 via an RNA recognition motif, thereby promoting the atp6-orfH79 transcript processing (Hu et al., 2012). The interaction between the PPR proteins OsRF6 and hexokinase OsHXK6 separates the mitochondrial chimeric gene atp6-orfH79 into two fragments, atp6 and orfH79, to restore the fertility of HL-CMS lines (Huang et al., 2015). OsRF19 rescues male sterility of Fujian Abortive CMS (FA-CMS) by mediating the RNA cleavage of the chimeric gene FA182. OsRf19 originates from a recent duplication in wild rice relatives and has a common ancestor with OsRf1a/OsRf5 (Jiang et al., 2022). Rf98 (PPR762) is an essential fertility restorer gene for RT98-type CMS, which only restores partial fertility, thereby implying the presence of additional genes near the Rf98 locus (Igarashi et al., 2016).
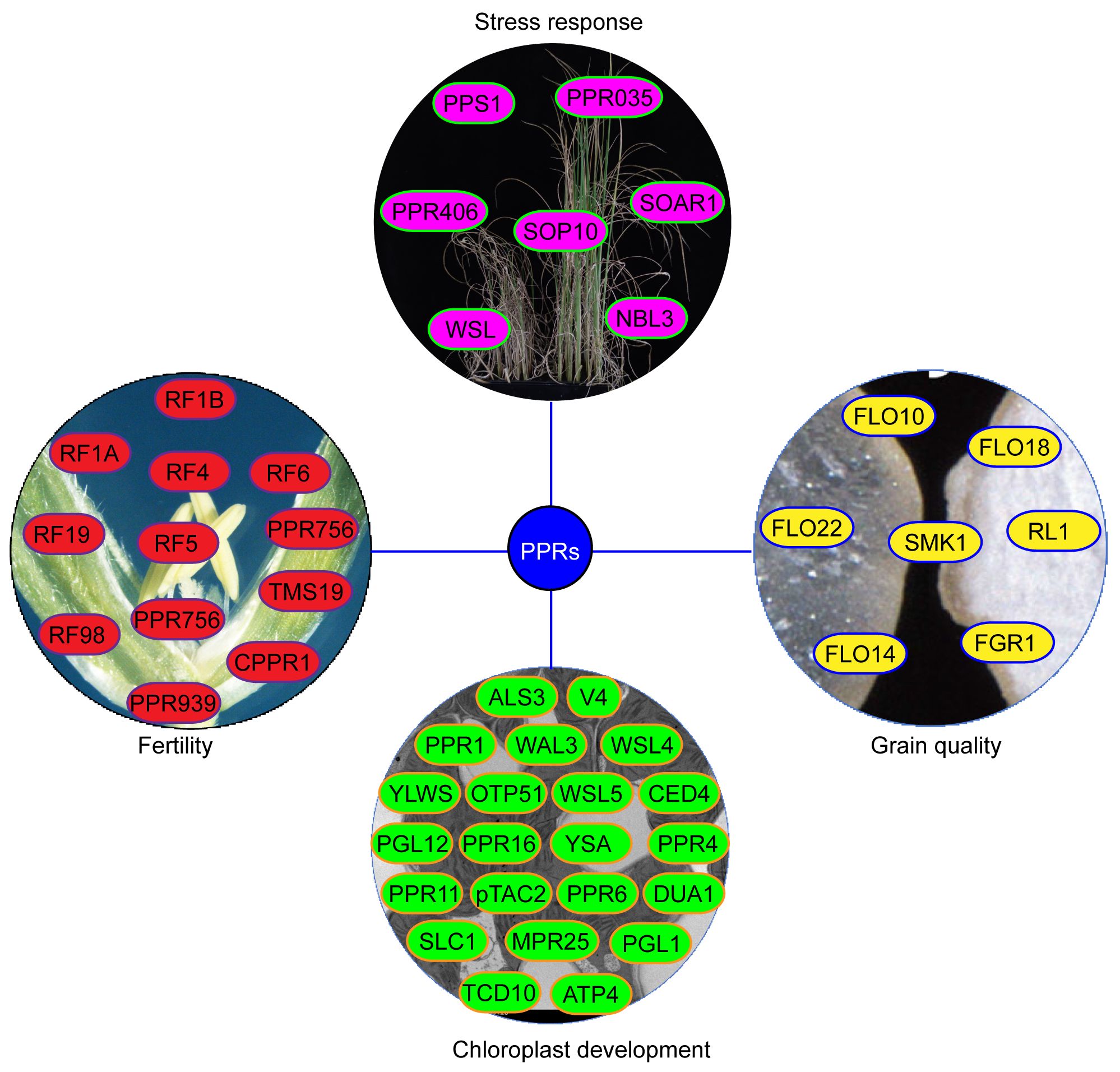
Figure 4 The functions of PPR proteins in rice. The yellow proteins represent PPR protein involved in grain quality. The green proteins represent PPR protein involved in chloroplast development. The red proteins represent PPR protein involved in CMS. The light red proteins represent PPR protein involved in stress responses.
Besides the Rf genes, some genes play a role in regulating pollen development. For instance, OsPPR939 is crucial for splicing of mitochondrial nad5 introns 1, 2, and 3. The OsPPR939 protein can be phosphorylated by OsS6K1, which facilitates its mitochondrial import. Its disruption results in growth retardation and pollen sterility (Zheng et al., 2021). PPR756 participates in editing mitochondrial genes atp6, ccmC, and nad and interacts with organellar RNA editing factors OsMORF1, OsMORF8–1, and OsMORF8–2 to form editosome complexes. Loss of PPR756 function abolishes RNA editing of atp6, ccmC, and nad, thereby causing retarded growth and pollen sterility (Zhang et al., 2020a). A point mutation (T-A) in the OsTMS19 exon is responsible for photo/thermo-sensitive genic male sterility. Excessive ROS accumulation in ostms19 anthers leads to male sterility, whereas effective ROS scavenging restores fertility (Zhou et al., 2024).
The PPR proteins regulating rice fertility, as previously mentioned, are primarily targeted to the mitochondria. However, two other PPR-SMR proteins, OsPPR676 and OsCPPR1, are localized to the plastid and cytoplasm, respectively, while also being associated with pollen development. OsPPR676 interacts with Osj10gBTF3, a NAC protein involved in pollen development regulation. The osppr676 mutant exhibits disrupted atpB mRNA translation, growth retardation, and partial pollen sterility (Liu et al., 2017). OsCPPR1 directly binds to single-stranded regions of OsGLK1, thereby regulating its transcription. In the oscppr1 mutant, OsGLK1 expression is significantly upregulated, which causes abnormal plastid development, prolonged tapetal programmed cell death, and tapetum degradation. Consequently, pollen fertility is significantly decreased (Zheng et al., 2022).
3 Functions of PPR proteins in biotic and abiotic stress responses
Multiple studies have demonstrated the involvement of numerous PPR proteins in rice’s response to biotic and abiotic stresses. Chen et al. (2018a) reported up-regulation of 75 and 73 PPR genes under salt and drought stresses, respectively, compared to control. In a separate study, Luo et al. (2022) analyzed PPR gene expression across different stress treatments and identified 16/81, 15/127, and 27/35 PPR genes that were upregulated/downregulated in response to osmotic, salt, and oxidative stress, respectively (Luo et al., 2022).
Several PPR proteins have been identified to play roles in the biotic and abiotic stresses. Except WSL, most PPR proteins are primarily localized in the mitochondria (Figure 3). PPS1, a PLS-type PPR protein, participates in C-to-U RNA editing of nad3 transcripts by binding to cis-elements near its five conserved RNA-editing sites. PPS1-RNAi plants exhibit decreased editing efficiency, increased ROS accumulation, and heightened sensitivity to abiotic stresses such as salinity and ABA, when compared to WT (Xiao et al., 2018a, 2021). Both PPR035 and PPR406 are localized in mitochondria, with PPR035 affecting the editing efficiency of rps4-C926 and orfx-C406, while PPR406 affects the editing efficiency of orfx-C355. Although both prp035 and ppr406 mutants improve drought and salt stress resistance, their underlying molecular mechanisms remain unclear (Luo et al., 2022). Recent study showed that SOP10 affects splicing efficiency of nad4 and nad5 introns and RNA editing efficiency of nad2, nad6, and rps4 transcripts in the mitochondria. Mutation of SOP10 leads to mitochondrial complex I deficiency, which inhibits ROS production and enhances cold tolerance in indica rice varieties (Zu et al., 2023). The transgenic rice lines overexpressing OsSOAR1 demonstrate enhanced salt tolerance during seedling growth, along with increased chlorophyll content and reduced ion leakage compared to WT (Lu et al., 2022).
Belonging to the P-subfamily of PPR proteins, WSL is localized in the chloroplast and is involved in chloroplast transcript rpl2 splicing. In wsl mutants, PEP-dependent plastid gene expression is significantly down-regulated, and plastid rRNAs and translation products accumulate to very low levels compared to WT. Mutant wsl exhibits defective rpl2 splicing, which generates a white striped phenotype in seedlings, and increases sensitivity to ABA and salinity stress (Tan et al., 2014).
Additionally, PPR proteins are implicated in biotic stress. OsNBL3 encodes a mitochondrion-localized P-type PPR protein, with nbl3 mutants displaying a lesion mimic phenotype, spontaneous cell death, enhanced resistance to Magnaporthe oryzae and Xanthomonas oryzae pv. Oryzae, and improved salt tolerance. OsNBL3 is involved in splicing mitochondrial gene nad5 intron 4. Disruption of OsNBL3 reportedly reduced complex I activity, increased alternative respiratory pathways, and damaged mitochondrial morphology (Qiu et al., 2021). These findings contribute to a better understanding of PPR genes and their functions in biotic and abiotic stresses. Therefore, we can use gene engineering or traditional hybridization concentrated on the PPR gene to improving rice adaptability.
4 Effects of PPR proteins on rice grain quality
Endosperm, the primary storage organ in cereal grains, significantly influences grain yield and quality. Several PPR proteins reportedly regulate endosperm development in rice (Figure 4). FLO10 and FLO18 encode mitochondrion-targeted P-type PPR proteins with 26 and 15 PPR motifs, respectively. Loss of FLO10 (WBG1) disrupts splicing of mitochondrial nad1 intron 1 and increases accumulation of nad1 exon 1 and exons 2–5 precursors (Wu et al., 2019). Disruption of FLO18 function impairs 5′-end processing of mitochondrial nad5 mRNA (Yu et al., 2021). Mutants of both FLO10 and FLO18 exhibit reduced assembly and activity of mitochondrial complex I in the electron transport pathway, altering mitochondrial morphology and resulting in abnormal endosperm development characterized by smaller starch grains, reduced starch content, and abnormal aleurone cells (Wu et al., 2019; Yu et al., 2021; Wu et al., 2023). The mitochondrion-tagged P-type PPR protein FLO22 directly binds to the”GAAGUGGAAG”sequence of nad1, thereby influencing the splicing and editing efficiency of nad1 and nad9 mRNA, respectively. FLO22 interacts with DYW3, a DYW-type PPR protein, forming a complex that likely synergistically functions in mitochondrial RNA editing. Mutation of FLO22 leads to alterations in complex I activity, respiration rate, mitochondrial morphology, and function, resulting in opaque, floury mature grains (Yang et al., 2023). Both Os_SMK1 and RL1 encode mitochondrion-targeted PPR-E subclass proteins, which are involved in C-U editing of nad7–836 and splicing of mitochondrial nad4 intron 1, respectively. Mutants of both Os_SMK1 and RL1 display chalky endosperm (Li et al., 2014; Wu et al., 2020).
Contrary to earlier findings suggesting chloroplast or/and mitochondrial localization for most PPR proteins, the FGR1 (OsNPPR1) and FLO14 (OsNPPR3) proteins, from the P-subfamily PPR proteins, are instead nuclear-localized (Hao et al., 2019; Xue et al., 2019). OsNPPR1 directly binds to the”CUCAC” motif, whose mutation alters splicing of certain nuclear genes linked to mitochondrial functions, thereby causing intron retention. The fgr1 mutant exhibits opaque endosperm with numerous smaller, single starch grains (SGs) and reduced amylose content (Hao et al., 2019). Furthermore, FLO14 mutation reduces splicing efficiency of mitochondrial genome-encoded transcripts, nad1–2 and nad2. The flo14 mutant displays a chalky endosperm phenotype with immature, smaller, and more scattered starch granules and lower starch content compared to WT (Xue et al., 2019).
5 Roles of PPR proteins in rice chloroplast development
Chloroplasts, essential for plant growth and development, generate energy for respiration and other physiological processes by fixing carbon and releasing oxygen (Lan et al., 2023). As a semi-autonomous organelle with its own genome, chloroplasts development rely on coordinated expressions of both chloroplast and nuclear genes (Woodson and Chory, 2008). Proteins encoded by nuclear genes are transported into organelles to regulate chloroplast gene expression at various levels, including transcriptional and post-transcriptional mechanisms such as RNA splicing, editing, and translation (Xin et al., 2021). Notably, PPR proteins serve as vital cofactors in chloroplast development and gene regulation (Figure 4). The OsPPR1 gene was the first rice PPR gene involving in chloroplast development in 2005. Transgenic plants expressing antisense OsPPR1 exhibit abnormal chloroplast shape and reduced chlorophyll contents, leading to an albinism and lethal phenotypes (Gothandam et al., 2005). Subsequent research, employing forward or reverse genetics approaches, has identified numerous other PPR genes contributing to rice chloroplast development (Table 1; Figure 3).
WAL3, a PLS-type PPR protein, is crucial for rice chloroplast development. Its mutation causes an albino lethal phenotype by disrupting splicing of multiple group II introns, including atpF, ndhA, petB, rps12, and rpl2. This mutation affects chlorophyll synthesis and photosynthetic metabolic pathways (Lv et al., 2022). YLWS, a P-type PPR protein with 11 PPR motifs, directly binds to specific sites on atpF, rpl2, and ndhA pre-mRNAs, regulating their intron splicing efficiency. YLWS also impacts ndhA, ndhB, and rps14 transcript editing. Disruption of YLWS leads to defective chloroplast development, characterized by vacuolated plastids and disorganized thylakoid membranes, resulting in the white-striped leaf phenotype (Lan et al., 2023). ALS3 regulates transcriptional levels of plastid translation machinery-associated genes and photosynthesis. Its mutation causes albino seedling lethality (Lin et al., 2015). OSOTP51, functionally conserved among higher plants, affects chloroplast ycf3 mRNA intron splicing. OSOTP51 mutation induces widespread changes in PSI structure and function and leads to severe photoinhibition, and albino phenotype (Ye et al., 2012).
WSL4, a P-family PPR protein, localizes to chloroplasts. Mutation of WSL4 disrupts splicing of four group II introns (ndhA, atpF, rpl2, and rps12), resulting in white-striped leaves in rice seedlings (Wang et al., 2017). PGL12 is involved in ndhA splicing and 16S rRNA processing. The pgl12 mutant exhibit yellow-green leaves, gradually becoming pale green as plants grow (Chen et al., 2019). OsPPR16, a chloroplast-targeted PLS-DYW subfamily protein with 14 PPR motifs, edits the chloroplast rpoB mRNA. Knockout of OsPPR16 reduces rpoB accumulation and PEP-dependent gene expression, thereby leading to a pale phenotype (Huang et al., 2020). YSA, a P-type PPR protein with 16 tandem PPR motifs, is localized in chloroplasts. The ysa mutants shows albino leaves before the three-leaf stage, which gradually return to normal green by the six-leaf stage. Although chloroplast development is affected in ysa mutant, the main agronomic traits such as plant height, grain weight, and seed setting rate remain unaltered compared to WT. Therefore, in hybrid rice production, ysa mutants serve as a selective marker for effectively identifying and eliminating false hybrids (Su et al., 2012).
OsPPR4 regulates photosynthesis, chlorophyll, and chloroplast biosynthesis by directly binding to a specific sequence of chloroplast rps12 intron 1, thereby affecting its splicing, and indirectly influencing the splicing of rps12, ndhA, atpF, and petB introns. The loss-of-function osppr4 mutant exhibits an albino phenotype and fails to survive past the young seedling stage (Lee et al., 2019). OsPPR11, a P-type PPR protein, is responsible for splicing ndhA and ycf3–1 introns. The osppr11 mutant displays yellowing leaves and defective chloroplast development (Zhang et al., 2023). OspTAC2 plays a critical role in chloroplast development, with its mutation causes reduced chlorophyll content, electron transport, and photochemical reactions of photosynthesis. Its mutant exhibits albino seedlings after germination, with death occurring about two weeks later (Wang et al., 2016). OsSLC1, a member of the P subgroup of PPR proteins, is involved in splicing rps16 introns. The slc1 mutant displayed chlorosis (Lv et al., 2020). OsPPR6 encodes a plastid-localized PLS subfamily protein, which is involved in ndhB transcript editing and ycf3 transcript splicing. The osppr6 mutant exhibits early chloroplast developmental defects, albino leaves, and seedling death (Tang et al., 2017).
The chloroplast-targeted PPR proteins discussed earlier are involved in chloroplast development. Additionally, two mitochondria-targeted PPR proteins, MPR25 and OsPGL1, also play roles in this process. MPR25, a member of the PPR family’s E subgroup, participates in C-U RNA editing of nad5 transcripts by directly interacting with the editing site. Mutation in MPR25 prevents C-U RNA editing, resulting in pale-green leaves with reduced chlorophyll (Toda et al., 2012). OsPGL1, which targets both chloroplasts and mitochondria, edits ndhD-878 in chloroplasts and ccmFc-543 in mitochondria. It interacts with three OsMORFs (OsMORF2/8/9), which suggests involvement in RNA editing via an editosome. The loss-of-function in ospgl1 leads to reduced chlorophyll content and defective chloroplast development, resulting in pale green leaves (Xiao et al., 2018b).
Under cold stress in rice, several PPR proteins, including TCD10, OsV4, ATP4, WSL5, CDE4, and DUA1, are essential for chloroplast development. TCD10 and OsV4 encode chloroplast-localized PPR proteins with 27 and 4 PPR motifs, respectively. Mutants of these genes exhibit reduced expression of chloroplast-associated genes, defective chloroplast development, and albino phenotypes under cold stress (Gong et al., 2014; Wu et al., 2016). ATP4, a PPR-SMR protein, is crucial for accumulating dicistronic rpl16-rpl14 transcripts and the C-U editing of rps8 transcripts. Loss of ATP4 function results in a chlorotic phenotype (Zhang et al., 2020b). Both wsl5 and ced4 mutants showed leaf albino phenotypes at low temperatures. WSL5 primarily regulates chloroplast development by affecting the splicing of group II introns, rpl2, and rps12 (Liu et al., 2018). CDE4, a P-type PPR protein, binds directly to pre-mRNA of rpl2, ndhA, and ndhB in chloroplasts, thereby regulating their intron splicing. It interacts physically with guanylate kinase V2. Overexpressing V2 in cde4 mutants restores group II intron splicing efficiency and mutant phenotype. Under cold stress, V2 likely stabilizes CDE4 protein, ensuring normal intron splicing that is necessary for chloroplast development (Liu et al., 2021). DUA1, localized in chloroplasts, interacts with RNA editing cofactor WSP1 and chloroplast sigma factor OsSIG1, thereby regulating editing efficiency of rpoB-C545, rpoB-C560, and rps8-C182 sites. Among which, DUA1 directly binds rps8 transcripts. WSP1 enhances DUA1 protein stability under cold stress (Cui et al., 2019). Additionally, DUA1 interacts with a multiple organellar RNA editing factor OsMORF9, a determinant of chloroplast development (Zhang et al., 2021). The dua1 mutants exhibited defective chloroplast development, chlorophyll biosynthesis, and albino phenotype at low temperatures (Cui et al., 2019; Du et al., 2021).
6 Conclusions and perspectives
In conclusion, PPR proteins play critical roles in fertility, biotic and abiotic stress responses, grain quality, and chloroplast development in rice (Figure 4). Research focusing on PPR protein function provides valuable gene resources for rice breeding. Among the 491 PPR proteins identified in rice, only ~9.7% (about 48) have been functionally characterized (Figure 4; Table 1), leaving the roles of the remaining PPR proteins unclear. It is imperative to comprehensively understand PPR protein functions. An effective strategy for elucidating PPR protein function is employing emerging genome-editing technologies like CRISPR/Cas9, which allow researchers to knockout candidate genes and determine their functions. Additionally, nearly half of the identified PPR proteins are implicated in chloroplast development. Mutations in these proteins lead to chlorophyll-deficient phenotypes and impair photosynthesis (Figure 4). Thus, investigating whether overexpression of specific PPR genes can enhance photosynthesis and rice yield is warranted. In rice, function losses of 5 PPR genes including PPR405, PPR406, SOP10, OsSOAR1, and OsNBL3 improve the abilities to tolerant biotic and abiotic stress. Thus we can use the CRISPR/Cas9 technology to seek the PPR genes that can improve rice stress tolerance without yield loss.
PPR proteins, encoded by the nuclear genome, are targeted post-translationally to chloroplasts and mitochondria, where they participate in organelle RNA processing. Numerous studies indicate their role in regulating rice growth and development by editing, stabilizing, and splicing RNA transcripts in these organelles. Thus, identifying additional mitochondria and chloroplast RNA transcripts regulated by PPR proteins could serve as an ideal model for understanding interactions between organelle RNA and nuclear genes, thereby shedding light on signal transduction between the nucleus and cytoplasm. Furthermore, studying the expression of organelle genes regulated by PPR proteins can reveal key target genes influencing organelle development and agronomic traits in rice. For example, knocking out the rice mitochondrial gene orf79 using mitoTALENs technology was found to successfully generate a new CMS line (Kazama et al., 2016).
Some PPR proteins have been observed to interact with OsMORFs, forming editosome complexes and participating in organelle RNA editing processes. However, to date, most identified PPR proteins function independently, without interactions with other proteins. Consequently, it is necessary to investigate whether PPR proteins interact with or form spliceosome complexes with other proteins to regulate organelle RNA processing in rice. In rice, most PPR proteins are targeted in mitochondria or chloroplast, whereas several PPR proteins including OsPPR676, OsCPPR1, WSL, FGR1, and FLO14 are targeted in nucleus and cytoplasm. Are there any differences in structure and regulation mechanism between these two types of proteins that warrant further study. Furthermore, it is intriguing that different types of PPR proteins are sometimes required for the expression of the same organellar genes. Therefore, this raises the question that whether these PPR proteins interact with or regulate each other. Thus, focusing future research on these aspects will be instrumental in unraveling the regulatory mechanisms of PPR proteins.
Author contributions
LM: Writing – original draft. MD: Writing – original draft. TZ: Writing – original draft. GL: Writing – original draft. YD: Writing – original draft. QZ: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was financially supported by the Natural Science Foundation of Shandong Province (K22LB56), Doctoral Research Startup Funds, Liaocheng University (318052165), the Open Project of Liaocheng University Landscape Architecture Discipline (319462212) and the Innovation and Entrepreneurship Training Program for College Students of Liaocheng University (S202310447034).
Acknowledgments
The authors would like to express their gratitude to EditSprings (https://www.editsprings.cn) for the expert linguistic services provided.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Barkan, A., Rojas, M., Fujii, S., Yap, A., Chong, Y. S., Bond, C. S., et al. (2012). A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PloS Genet. 8, e1002910. doi: 10.1371/journal.pgen.1002910
Barkan, A., Small, I. (2014). Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 65, 415–442. doi: 10.1146/annurev-arplant-050213-040159
Chen, L., Huang, L., Dai, L., Gao, Y., Zou, W., Lu, X., et al. (2019). PALE-GREEN LEAF12 encodes a novel pentatricopeptide repeat protein required for chloroplast development and 16S rRNA processing in rice. Plant Cell Physiol. 60, 587–598. doi: 10.1093/pcp/pcy229
Chen, L., Li, Y. X., Li, C., Shi, Y., Song, Y., Zhang, D., et al. (2018b). Genome-wide analysis of the pentatricopeptide repeat gene family in different maize genomes and its important role in kernel development. BMC Plant Biol. 18, 366. doi: 10.1186/s12870-018-1572-2
Chen, L., Liu, Y. G. (2014). Male sterility and fertility restoration in crops. Annu. Rev. Plant Biol. 65, 579–606. doi: 10.1146/annurev-arplant-050213-040119
Chen, G., Zou, Y., Hu, J., Ding, Y. (2018a). Genome-wide analysis of the rice PPR gene family and their expression profiles under different stress treatments. BMC Genomics 19, 720. doi: 10.1186/s12864-018-5088-9
Cheng, S., Gutmann, B., Zhong, X., Ye, Y., Fisher, M. F., Bai, F., et al. (2016). Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. Plant J. 85, 532–547. doi: 10.1111/tpj.13121
Cui, X., Wang, Y., Wu, J., Han, X., Gu, X., Lu, T., et al. (2019). The RNA editing factor DUA1 is crucial to chloroplast development at low temperature in rice. New Phytol. 221, 834–849. doi: 10.1111/nph.15448
Du, Y., Mo, W., Ma, T., Tang, W., Tian, L., Lin, R. (2021). A pentatricopeptide repeat protein DUA1 interacts with sigma factor 1 to regulate chloroplast gene expression in Rice. Photosynth. Res. 147, 131–143. doi: 10.1007/s11120-020-00793-0
Gong, X., Su, Q., Lin, D., Jiang, Q., Xu, J., Zhang, J., et al. (2014). The rice OsV4 encoding a novel pentatricopeptide repeat protein is required for chloroplast development during the early leaf stage under cold stress. J. Integr. Plant Biol. 56, 400–410. doi: 10.1111/jipb.12138
Gothandam, K. M., Kim, E. S., Cho, H., Chung, Y. Y. (2005). OsPPR1, a pentatricopeptide repeat protein of rice is essential for the chloroplast biogenesis. Plant Mol. Biol. 58, 421–433. doi: 10.1007/s11103-005-5702-5
Gruttner, S., Nguyen, T. T., Bruhs, A., Mireau, H., Kempken, F. (2022). The P-type pentatricopeptide repeat protein DWEORG1 is a non-previously reported rPPR protein of Arabidopsis mitochondria. Sci. Rep. 12, 12492. doi: 10.1038/s41598-022-16812-0
Hao, Y., Wang, Y., Wu, M., Zhu, X., Teng, X., Sun, Y., et al. (2019). The nuclear-localized PPR protein OsNPPR1 is important for mitochondrial function and endosperm development in rice. J. Exp. Bot. 70, 4705–4720. doi: 10.1093/jxb/erz226
Hu, J., Wang, K., Huang, W., Liu, G., Gao, Y., Wang, J., et al. (2012). The rice pentatricopeptide repeat protein RF5 restores fertility in Hong-Lian cytoplasmic male-sterile lines via a complex with the glycine-rich protein GRP162. Plant Cell 24, 109–122. doi: 10.1105/tpc.111.093211
Huang, W., Yu, C., Hu, J., Wang, L., Dan, Z., Zhou, W., et al. (2015). Pentatricopeptide-repeat family protein RF6 functions with hexokinase 6 to rescue rice cytoplasmic male sterility. Proc. Natl. Acad. Sci. U. S. A. 112, 14984–14989. doi: 10.1073/pnas.1511748112
Huang, W., Zhang, Y., Shen, L., Fang, Q., Liu, Q., Gong, C., et al. (2020). Accumulation of the RNA polymerase subunit RpoB depends on RNA editing by OsPPR16 and affects chloroplast development during early leaf development in rice. New Phytol. 228, 1401–1416. doi: 10.1111/nph.16769
Igarashi, K., Kazama, T., Toriyama, K. (2016). A gene encoding pentatricopeptide repeat protein partially restores fertility in RT98-Type cytoplasmic male-sterile rice. Plant Cell Physiol. 57, 2187–2193. doi: 10.1093/pcp/pcw135
Jiang, H., Lu, Q., Qiu, S., Yu, H., Wang, Z., Yu, Z., et al. (2022). Fujian cytoplasmic male sterility and the fertility restorer gene OsRf19 provide a promising breeding system for hybrid rice. Proc. Natl. Acad. Sci. U. S. A. 119, e2208759119. doi: 10.1073/pnas.2208759119
Jiang, S. C., Mei, C., Liang, S., Yu, Y. T., Lu, K., Wu, Z., et al. (2015). Crucial roles of the pentatricopeptide repeat protein SOAR1 in Arabidopsis response to drought, salt and cold stresses. Plant Mol. Biol. 88, 369–385. doi: 10.1007/s11103-015-0327-9
Kazama, T., Itabashi, E., Fujii, S., Nakamura, T., Toriyama, K. (2016). Mitochondrial ORF79 levels determine pollen abortion in cytoplasmic male sterile rice. Plant J. 85, 707–716. doi: 10.1111/tpj.13135
Lan, J., Lin, Q., Zhou, C., Liu, X., Miao, R., Ma, T., et al. (2023). Young Leaf White Stripe encodes a P-type PPR protein required for chloroplast development. J. Integr. Plant Biol. 65, 1687–1702. doi: 10.1111/jipb.13477
Lee, K., Park, S. J., Colas des Francs-Small, C., Whitby, M., Small, I., Kang, H. (2019). The coordinated action of PPR4 and EMB2654 on each intron half mediates trans-splicing of rps12 transcripts in plant chloroplasts. Plant J. 100, 1193–1207. doi: 10.1111/tpj.14509
Li, X., Sun, M., Liu, S., Teng, Q., Li, S., Jiang, Y. (2021). Functions of PPR proteins in plant growth and development. Int. J. Mol. Sci. 22, 11274. doi: 10.3390/ijms222011274
Li, X. J., Zhang, Y. F., Hou, M., Sun, F., Shen, Y., Xiu, Z. H., et al. (2014). Small kernel 1 encodes a pentatricopeptide repeat protein required for mitochondrial nad7 transcript editing and seed development in maize (Zea mays) and rice (Oryza sativa). Plant J. 79, 797–809. doi: 10.1111/tpj.12584
Lin, D., Gong, X., Jiang, Q., Zheng, K., Zhou, H., Xu, J., et al. (2015). The rice ALS3 encoding a novel pentatricopeptide repeat protein is required for chloroplast development and seedling growth. Rice (N Y) 8, 17. doi: 10.1186/s12284-015-0050-9
Liu, X., Lan, J., Huang, Y., Cao, P., Zhou, C., Ren, Y., et al. (2018). WSL5, a pentatricopeptide repeat protein, is essential for chloroplast biogenesis in rice under cold stress. J. Exp. Bot. 69, 3949–3961. doi: 10.1093/jxb/ery214
Liu, Y. J., Liu, X., Chen, H., Zheng, P., Wang, W., Wang, L., et al. (2017). A plastid-localized pentatricopeptide repeat protein is required for both pollen development and plant growth in rice. Sci. Rep. 7, 11484. doi: 10.1038/s41598-017-10727-x
Liu, J. M., Xu, Z. S., Lu, P. P., Li, W. W., Chen, M., Guo, C. H., et al. (2016). Genome-wide investigation and expression analyses of the pentatricopeptide repeat protein gene family in foxtail millet. BMC Genomics 17, 840. doi: 10.1186/s12864-016-3184-2
Liu, X., Zhang, X., Cao, R., Jiao, G., Hu, S., Shao, G., et al. (2021). CDE4 encodes a pentatricopeptide repeat protein involved in chloroplast RNA splicing and affects chloroplast development under low-temperature conditions in rice. J. Integr. Plant Biol. 63, 1724–1739. doi: 10.1111/jipb.13147
Lu, K., Li, C., Guan, J., Liang, W. H., Chen, T., Zhao, Q. Y., et al. (2022). The PPR-domain protein SOAR1 regulates salt tolerance in rice. Rice (N Y) 15, 62. doi: 10.1186/s12284-022-00608-x
Luo, Z., Xiong, J., Xia, H., Wang, L., Hou, G., Li, Z., et al. (2022). Pentatricopeptide repeat gene-mediated mitochondrial RNA editing impacts on rice drought tolerance. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.926285
Lurin, C., Andres, C., Aubourg, S., Bellaoui, M., Bitton, F., Bruyere, C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16, 2089–2103. doi: 10.1105/tpc.104.022236
Lv, J., Shang, L., Chen, Y., Han, Y., Yang, X., Xie, S., et al. (2020). OsSLC1 encodes a pentatricopeptide repeat protein essential for early chloroplast development and seedling survival. Rice (N Y) 13, 25. doi: 10.1186/s12284-020-00385-5
Lv, Y., Wang, Y., Zhang, Q., Chen, C., Qian, Q., Guo, L. (2022). WAL3 encoding a PLS-type PPR protein regulates chloroplast development in rice. Plant Sci. 323, 111382. doi: 10.1016/j.plantsci.2022.111382
Qin, T., Zhao, P., Sun, J., Zhao, Y., Zhang, Y., Yang, Q., et al. (2021). Research progress of PPR proteins in RNA editing, stress response, plant growth and development. Front. Genet. 12. doi: 10.3389/fgene.2021.765580
Qiu, T., Zhao, X., Feng, H., Qi, L., Yang, J., Peng, Y. L., et al. (2021). OsNBL3, a mitochondrion-localized pentatricopeptide repeat protein, is involved in splicing nad5 intron 4 and its disruption causes lesion mimic phenotype with enhanced resistance to biotic and abiotic stresses. Plant Biotechnol. J. 19, 2277–2290. doi: 10.1111/pbi.13659
Su, N., Hu, M. L., Wu, D. X., Wu, F. Q., Fei, G. L., Lan, Y., et al. (2012). Disruption of a rice pentatricopeptide repeat protein causes a seedling-specific albino phenotype and its utilization to enhance seed purity in hybrid rice production. Plant Physiol. 159, 227–238. doi: 10.1104/pp.112.195081
Tan, J., Tan, Z., Wu, F., Sheng, P., Heng, Y., Wang, X., et al. (2014). A novel chloroplast-localized pentatricopeptide repeat protein involved in splicing affects chloroplast development and abiotic stress response in rice. Mol. Plant 7, 1329–1349. doi: 10.1093/mp/ssu054
Tang, H., Luo, D., Zhou, D., Zhang, Q., Tian, D., Zheng, X., et al. (2014). The rice restorer Rf4 for wild-abortive cytoplasmic male sterility encodes a mitochondrial-localized PPR protein that functions in reduction of WA352 transcripts. Mol. Plant 7, 1497–1500. doi: 10.1093/mp/ssu047
Tang, J., Zhang, W., Wen, K., Chen, G., Sun, J., Tian, Y., et al. (2017). OsPPR6, a pentatricopeptide repeat protein involved in editing and splicing chloroplast RNA, is required for chloroplast biogenesis in rice. Plant Mol. Biol. 95, 345–357. doi: 10.1007/s11103-017-0654-0
Toda, T., Fujii, S., Noguchi, K., Kazama, T., Toriyama, K. (2012). Rice MPR25 encodes a pentatricopeptide repeat protein and is essential for RNA editing of nad5 transcripts in mitochondria. Plant J. 72, 450–460. doi: 10.1111/j.1365-313X.2012.05091.x
Wang, Y., Liu, X. Y., Yang, Y. Z., Huang, J., Sun, F., Lin, J., et al. (2019). Empty Pericarp21 encodes a novel PPR-DYW protein that is required for mitochondrial RNA editing at multiple sites, complexes I and V biogenesis, and seed development in maize. PloS Genet. 15, e1008305. doi: 10.1371/journal.pgen.1008305
Wang, D., Liu, H., Zhai, G., Wang, L., Shao, J., Tao, Y. (2016). OspTAC2 encodes a pentatricopeptide repeat protein and regulates rice chloroplast development. J. Genet. Genomics 43, 601–608. doi: 10.1016/j.jgg.2016.09.002
Wang, Y., Ren, Y., Zhou, K., Liu, L., Wang, J., Xu, Y., et al. (2017). WHITE STRIPE LEAF4 encodes a novel P-type PPR protein required for chloroplast biogenesis during early leaf development. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01116
Wang, Z., Zou, Y., Li, X., Zhang, Q., Chen, L., Wu, H., et al. (2006). Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell 18, 676–687. doi: 10.1105/tpc.105.038240
Woodson, J. D., Chory, J. (2008). Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 9, 383–395. doi: 10.1038/nrg2348
Wu, M., Cai, M., Zhai, R., Ye, J., Zhu, G., Yu, F., et al. (2023). mitochondrion-associated PPR protein, WBG1, regulates grain chalkiness in rice. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1136849
Wu, M., Ren, Y., Cai, M., Wang, Y., Zhu, S., Zhu, J., et al. (2019). Rice FLOURY ENDOSPERM10 encodes a pentatricopeptide repeat protein that is essential for the trans-splicing of mitochondrial nad1 intron 1 and endosperm development. New Phytol. 223, 736–750. doi: 10.1111/nph.15814
Wu, L., Wu, J., Liu, Y., Gong, X., Xu, J., Lin, D., et al. (2016). The rice pentatricopeptide repeat gene TCD10 is needed for chloroplast development under cold stress. Rice (N Y) 9, 67. doi: 10.1186/s12284-016-0134-1
Wu, M. W., Zhao, H., Zhang, J. D., Guo, L., Liu, C. M. (2020). RADICLELESS 1 (RL1)-mediated nad4 intron 1 splicing is crucial for embryo and endosperm development in rice (Oryza sativa L.). Biochem. Biophys. Res. Commun. 523, 220–225. doi: 10.1016/j.bbrc.2019.11.084
Xiao, H., Liu, Z., Zou, X., Xu, Y., Peng, L., Hu, J., et al. (2021). Silencing of rice PPR gene PPS1 exhibited enhanced sensibility to abiotic stress and remarkable accumulation of ROS. J. Plant Physiol. 258-259, 153361. doi: 10.1016/j.jplph.2020.153361
Xiao, H., Xu, Y., Ni, C., Zhang, Q., Zhong, F., Huang, J., et al. (2018b). A rice dual-localized pentatricopeptide repeat protein is involved in organellar RNA editing together with OsMORFs. J. Exp. Bot. 69, 2923–2936. doi: 10.1093/jxb/ery108
Xiao, H., Zhang, Q., Qin, X., Xu, Y., Ni, C., Huang, J., et al. (2018a). Rice PPS1 encodes a DYW motif-containing pentatricopeptide repeat protein required for five consecutive RNA-editing sites of nad3 in mitochondria. New Phytol. 220, 878–892. doi: 10.1111/nph.15347
Xin, K., Pan, T., Gao, S., Yan, S. (2021). A transcription factor regulates gene expression in chloroplasts. Int. J. Mol. Sci. 22, 6769. doi: 10.3390/ijms22136769
Xing, H., Fu, X., Yang, C., Tang, X., Guo, L., Li, C., et al. (2018). Genome-wide investigation of pentatricopeptide repeat gene family in poplar and their expression analysis in response to biotic and abiotic stresses. Sci. Rep. 8, 2817. doi: 10.1038/s41598-018-21269-1
Xue, M., Liu, L., Yu, Y., Zhu, J., Gao, H., Wang, Y., et al. (2019). Lose-of-function of a rice nucleolus-localized pentatricopeptide repeat protein is responsible for the floury endosperm14 mutant phenotypes. Rice (N Y) 12, 100. doi: 10.1186/s12284-019-0359-x
Yang, Y. Z., Ding, S., Wang, H. C., Sun, F., Huang, W. L., Song, S., et al. (2017). The pentatricopeptide repeat protein EMP9 is required for mitochondrial ccmB and rps4 transcript editing, mitochondrial complex biogenesis and seed development in maize. New Phytol. 214, 782–795. doi: 10.1111/nph.14424
Yang, H., Wang, Y., Tian, Y., Teng, X., Lv, Z., Lei, J., et al. (2023). Rice FLOURY ENDOSPERM22, encoding a pentatricopeptide repeat protein, is involved in both mitochondrial RNA splicing and editing and is crucial for endosperm development. J. Integr. Plant Biol. 65, 755–771. doi: 10.1111/jipb.13402
Ye, J. W., Gong, Z. Y., Chen, C. G., Mi, H. L., Chen, G. Y. (2012). A mutation of OSOTP51 leads to impairment of photosystem I complex assembly and serious photo-damage in rice. J. Integr. Plant Biol. 54, 87–98. doi: 10.1111/j.1744-7909.2012.01094.x
Yu, M., Wu, M., Ren, Y., Wang, Y., Li, J., Lei, C., et al. (2021). Rice FLOURY ENDOSPERM 18 encodes a pentatricopeptide repeat protein required for 5’ processing of mitochondrial nad5 messenger RNA and endosperm development. J. Integr. Plant Biol. 63, 834–847. doi: 10.1111/jipb.13049
Zhang, Q., Chen, C., Wang, Y., He, M., Li, Z., Shen, L., et al. (2023). OsPPR11 encoding P-type PPR protein that affects group II intron splicing and chloroplast development. Plant Cell. Rep. 42, 421–431. doi: 10.1007/s00299-022-02968-6
Zhang, J., Guo, Y., Fang, Q., Zhu, Y., Zhang, Y., Liu, X., et al. (2020b). The PPR-SMR protein ATP4 is required for editing the chloroplast rps8 mRNA in rice and maize. Plant Physiol. 184, 2011–2021. doi: 10.1104/pp.20.00849
Zhang, Q., Wang, Y., Xie, W., Chen, C., Ren, D., Hu, J., et al. (2021). OsMORF9 is necessary for chloroplast development and seedling survival in rice. Plant Sci. 307, 110907. doi: 10.1016/j.plantsci.2021.110907
Zhang, Q., Xu, Y., Huang, J., Zhang, K., Xiao, H., Qin, X., et al. (2020a). The rice pentatricopeptide repeat protein PPR756 is involved in pollen development by affecting multiple RNA editing in mitochondria. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00749
Zheng, S., Dong, J., Lu, J., Li, J., Jiang, D., Yu, H., et al. (2022). A cytosolic pentatricopeptide repeat protein is essential for tapetal plastid development by regulating OsGLK1 transcript levels in rice. New Phytol. 234, 1678–1695. doi: 10.1111/nph.18105
Zheng, P., Liu, Y., Liu, X., Huang, Y., Sun, F., Wang, W., et al. (2021). OsPPR939, a nad5 splicing factor, is essential for plant growth and pollen development in rice. Theor. Appl. Genet. 134, 923–940. doi: 10.1007/s00122-020-03742-6
Zhou, L., Mao, Y. C., Yang, Y. M., Wang, J. J., Zhong, X., Han, Y., et al. (2024). Temperature and light reverse the fertility of rice P/TGMS line ostms19 via reactive oxygen species homeostasis. Plant Biotechnol. J. 22, 2020–2032. doi: 10.1111/pbi.14322
Keywords: PPR proteins, RNA metabolism, chloroplast, mitochondria, stress response
Citation: Meng L, Du M, Zhu T, Li G, Ding Y and Zhang Q (2024) PPR proteins in plants: roles, mechanisms, and prospects for rice research. Front. Plant Sci. 15:1416742. doi: 10.3389/fpls.2024.1416742
Received: 26 April 2024; Accepted: 13 June 2024;
Published: 27 June 2024.
Edited by:
SangJin Kim, Michigan State University, United StatesReviewed by:
Ghazala Mustafa, Quaid-i-Azam University, PakistanNaveen Sharma, Michigan State University, United States
Copyright © 2024 Meng, Du, Zhu, Li, Ding and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiang Zhang, emhhbmdxaWFuZzkwMjRAMTI2LmNvbQ==
†These authors have contributed equally to this work
 Lingzhi Meng
Lingzhi Meng Mengxue Du1†
Mengxue Du1† Qiang Zhang
Qiang Zhang